- 1School of Life Science and Technology, China Pharmaceutical University, Nanjing, China
- 2Key Laboratory of Pathogen Biology of Jiangsu Province, Nanjing, China
Long noncoding RNAs (lncRNAs) have emerged as vital regulators of gene expression during embryonic stem cell (ESC) self-renewal and differentiation. Here, we systemically analyzed the differentially regulated lncRNAs during ESC-derived cardiomyocyte (CM) differentiation. We established a perspicuous profile of lncRNA expression at four critical developmental stages and found that the differentially expressed lncRNAs were grouped into six distinct clusters. The cluster with specific expression in ESC enriches the largest number of lncRNAs. Investigation of lncRNA-protein interaction network revealed that they are not only controlled by classic key transcription factors, but also modulated by epigenetic and epitranscriptomic factors including N6-methyladenosine (m6A) effector machineries. A detailed inspection revealed that 28 out of 385 lncRNAs were modified by methylation as well as directly recruited by the nuclear m6A reader protein Ythdc1. Unlike other 27 non-coding transcripts, the ESC-specific lncRNA Gm2379, located in both nucleus and cytoplasm, becomes dramatically upregulated in response to the depletion of m6A or Ythdc1. Consistent with the role of m6A in cell fate regulation, depletion of Gm2379 results in dysregulated expressions of pluripotent genes and crucial genes required for the formation of three germ layers. Collectively, our study provides a foundation for understanding the dynamic regulation of lncRNA transcriptomes during ESC differentiation and identifies the interplay between epitranscriptomic modification and key lncRNAs in the regulation of cell fate decision.
Introduction
Embryonic stem cells (ESCs) are pluripotent stem cells derived from the inner cell mass of the blastocyst. They possess dual ability to self-renew unlimitedly and differentiate into multiple tissue types found in the adult animal. Both ESC proliferation and differentiation are complex processes requiring the synergetic regulation by transcription factors (TFs), epigenetic factors, histone modification and chromatin remodelers (Martello and Smith, 2014; Atlasi and Stunnenberg, 2017; Chen et al., 2017; Zhao and Jin, 2017). Apart from protein-mediated transcriptional and epigenetic networks, long non-coding RNAs (lncRNAs) emerge as new players which guide the fate regulation of pluripotency and stem cell differentiation (Fico et al., 2019; Chen et al., 2020; Constanty and Shkumatava, 2021). They are a major group of non-coding RNAs with over 200 nucleotides in length but generally lack of translated open reading frames and obvious protein coding capacity (Statello et al., 2021). Many lncRNAs are dynamically regulated during ESC differentiation and expressed highly at specific developmental stage (Luo et al., 2016; Li et al., 2017). Therefore, establishment of lncRNA expression profiles at different developmental stages will facilitate our understanding of gene regulatory events during normal development at additional layers.
Distinct from coding mRNAs, lncRNAs themselves serve as the functional unit to modulate the expression of target genes depending on their subcellular localization and interacting molecules (Carlevaro-Fita and Johnson, 2019; Yao et al., 2019). In the cytoplasm, lncRNAs generally function to regulate mRNA stability, mediate mRNA translation and act as competing endogenous RNAs (Statello et al., 2021). In comparison, the functional roles of nucleus-localized lncRNAs have been more extensively studied. The specific interacting proteins present in the nucleus endow lncRNAs with modulation of chromatin architecture and transcriptional activity (Bergmann and Spector, 2014; Rutenberg-Schoenberg et al., 2016; Statello et al., 2021). For example, XIST lncRNA is able to recruit the polycomb repressive complex (PRC) to trigger the PRC1-guided mono-ubiquitylation of histone H2AK119 and the PRC2-dependent H3K27me3, resulting in silencing of gene transcription (Plath et al., 2003; Fang et al., 2004). In addition, nuclear lncRNA also participate in establishment of the nuclear compartment organization via scaffolding. For example, MALAT1 lncRNA functions as a scaffold molecule in nuclear speckles to regulate splicing via recruiting serine/arginine (SR) splicing factors (Tripathi et al., 2010). Thus, investigation of the interactions between lncRNAs with RNA-binding proteins (RBPs) are crucial for deciphering their functional roles in diverse biological processes.
In addition to the structural features, the modifications embedded in non-coding RNAs are also able to regulate target gene expression through recruiting a subgroup of specific RBPs (Liu et al., 2020, 2021; Chen et al., 2021; Lee et al., 2021; Wei and He, 2021). N6-methyladenosine (m6A), the most abundant modification identified in mRNA, has been recently found to exist on lncRNAs and associate with specific “reader” proteins to modulate their functionality. m6A residues on the XIST are able to recruit nuclear RBP YTH domain containing 1 (YTHDC1) and required for XIST-mediated transcriptional gene silencing (Patil et al., 2016). Besides, the interaction of YTHDC1 with methylated adenosines on MALAT1 has been identified to regulate homeostasis of nuclear speckles and cancer metastasis (Wang et al., 2021). During heat shock stress response, a novel lncRNA Heat silences stress gene expression via the m6A-YTHDC1 association (Ji et al., 2021). Despite the emergence of certain studies for m6A-guided lncRNA functionality, how the interplay between m6A and lncRNA regulate stem cell pluripotency and differentiation remains largely unknown.
In this study, we systematically analyzed the lncRNA expression at different stages during ESC-derived cardiomyocyte (CM) differentiation. We found that the largest group of lncRNAs enriched was expressed specifically in ESC. They were not merely modulated by cell fate-associated TFs, but also able to recruit RNA m6A epigenetic factors. Indeed, a subgroup of these lncRNAs were obviously modified by m6A and bound with the reader protein YTHDC1. Intriguingly, only one ESC-specific lncRNA Gm2379 was sensitive to the dysfunction of m6A-regulatory proteins. Depletion of m6A-modified Gm2379 resulted in dysregulated expressions of crucial genes required for ESC pluripotency maintenance and differentiation.
Materials and Methods
Cell Lines and Reagents
J-1 male mouse embryonic stem cells (ESCs) were purchased from the Cell Bank of Chinese Academy of Sciences. Wild-type and Mettl3-KO J-1 mouse ESCs were provided by Howard Y. Chang in Stanford University (Batista et al., 2014). Ythdc1flox/flox (Control) and Ythdc1-cKO (Ythdc1-KO) mouse ESCs were provided by Yawei Gao in Tongji University (Chen et al., 2021). Mycoplasma testing was performed routinely and cells tested negative were used in further experiments. Tissue culture plates were initially treated with 0.2% gelatin for at least 30 min at 37°C. ESCs were subsequently grown in gelatinized plates containing 15% KnockOut Serum Replacement (Gibco), 1% non-essential amino acids (Gibco), 1% Glutamax (Gibco), 1% Penn-Strep (Life technologies), 0.1 mM β-mercaptoethanol (Invitrogen) and 1000 U/ml ESGRO Recombinant Mouse leukemia inhibitory factor (LIF) Protein (Millipore).
Cardiomyocyte Differentiation
To generate embryoid bodies (EBs), suspension of each cell line at a concentration of 10 × 104cells per milliliter in differentiating medium (Knock-out DMEM supplemented with 15% FBS (Gibco), 1% non-essential amino acids, 1% Glutamax, 1% Penn-Strep, 0.1 mm β-mercaptoethanol) was deposited in 20 μL hanging drops in Petri dishes. After 2 days, the suspension-cultured cells were plated onto 0.2% gelatin-coated dishes in cardiomyocyte differentiation media (CMD) containing DMEM (BI), 15% FBS (Gibco), 1% Penn-Strep, 1% non-essential amino acids (Gibco), 1% Glutamine (Gibco), 0.1 mm β-mercaptoethanol and 1 mm Ascorbic Acid (Sigma) for continued differentiation. Cell culture media was changed every other day and contracting patches of cells were collected at indicated time points.
Targeted Knockdown of Gm2379 Long Noncoding RNAs in Mouse Embryonic Stem Cells
The shRNA targeting sequences were cloned into pRSI9-U6-(sh)-UbiC-TagRFP-2A-Blasticidin. Lentiviral particles were packaged using Lenti-X 293T cells. Virus-containing supernatants were collected at 48 h after transfection and filtered to eliminate cell debris. Wild-type and Mettl3-KO mESCs were infected by the lentivirus for 48 h and stable mESC knockdown lines were generated using blasticidin (MCE) selection (10 µg/ml). shRNA targeting sequences are listed below.
Gm2379 (target sequence 1): 5′- GCACGGCATTACAGAATTTCA-3′; Gm2379 (target sequence 3): 5′- GAAAGCCTTCATAGGAAACAG-3′; Scramble Target Sequence: 5′- AATTCTCCGAACGTGTCACGT-3′.
Real-Time Quantitative PCR
Total RNA from different cells was isolated from PBS-washed cells using Trizol reagent (Invitrogen). Isolated RNAs were used as templates for reverse transcription based on the manual for HiScript III 1st Strand cDNA Synthesis Kit plus gDNA wiper (Vazyme Biotech). Real-time PCR analysis was conducted with ChamQ SYBR qPCR Master Mix (Vazyme Biotech) and carried on a QuantStudio three Real-Time PCR System (Applied Biosystems). Primers for amplifying each target were listed in Supplementary Table S1.
Cell Fractionation Assay
ESCs were pelleted by centrifugation at 500 g for 3 min at 4°C. After washing with phosphate-buffered saline (PBS) buffer twice, cells were lysed in ice-cold cytoplasmic lysis buffer (10 mM HEPES pH 7.5, 0.05%TritonX-100, 5 mM MgCl2, 10 U SUPERase-In and 1x protease inhibitor cocktail) on ice for 5 min. Samples were centrifuged at 2,000 g at 4°C for 10 min. The supernatant was collected as cytoplasmic fraction. The remaining nuclear pellets were washed with PBS twice and centrifuged at 2,000 g at 4°C for 2 min. After removing the supernatant, the nuclear pellet was resuspended in RIPA buffer (50 mM Tris-HCl, pH7.5, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 10 U SUPERase-In and 1 × protease inhibitor cocktail). This sample was collected as nuclear fraction. RNA from different fractions was isolated using TRIzol reagent (Invirtrogen), and the protein sample was prepared by adding SDS loading buffer.
RNA-Seq
Total RNA was isolated using TRIzol reagent (Invitrogen). Ribosomal RNAs depletion, strand-specific library preparation and sequencing were performed at the Annoroad Gene Technology (Beijing, China). Two biological replicates were prepared for each differentiation phases. The libraries were sequenced on an Illumina Novaseq 6,000 system using the 2 × 150-bp paired-end read mode.
Long Noncoding RNA-Seq Data Processing
The lncRNA-seq data has been deposited in NCBI’s Gene Expression Omnibus and is accessible through GEO Series accession number GSE197205. Trim Galore (v.0.6.6) was used to trim the low quality sequences (Martin, 2011). The paired-end reads were aligned to mouse genome (GRCm38) with HISAT2 aligner (v2.2.1) with --rna-strandness RF parameters and Ensemble genes (GRCm38.102) transcriptome annotation (Pertea et al., 2016). Then gene expression were quantified using featureCounts (v2.0.1) with -s 2 parameter (Liao et al., 2014), and differentially expressed genes were identifified using DESeq2 (v1.34.0) R package (Love et al., 2014). The FoldChange > 2 or < 0.5 and ajusted p-value < 0.05 genes were considered as differentially expressed genes. The TPM (Transcripts Per Kilobase of exonmodel per Million mapped reads) was used to produce heatmap.
Fuzzy C-Means Clustering
LncRNAs with average TPM expressions in replicates from four differentiation stages were grouped into six different clusters using Mfuzz (v2.54.0) package in R with fuzzy c-means algorithm (Kumar and Futschik, 2007).
Regulatory Motif and Functional Analysis of Embryonic Stem Cells-specific Long Noncoding RNAs
HOMER’s findMotifs.pl program was used to identify motifs in the promoters of lncRNA genes (encompassing regions 2000 bp upstream and 500 bp downstream of the transcription start site) from each cluster. If one lncRNA has multiple transcript isoforms, the longest transcript isoform was used for analysis and bedtools getfasta (v2.29.2) was used to extract sequnences. Enrichment analysis was performed on Gene Ontology website (Ashburner et al., 2000). Network plot was produced by Cytoscape (v3.8.2) (Paul Shannon et al., 1971).
MeRIP-Seq and YTH Domain Containing 1 RIP-Seq Data Analysis
MeRIP-seq and RIP-seq datasets were obtained from the Gene Expression Omnibus (GEO) public database under accession number GSE145315 and GSE157265, respectively (Chen et al., 2021; Liu et al., 2021). Quality control of MeRIP-seq and RIP-seq data were performed using FastQC (v.0.11.9). Low-quality bases and adaptor contamination were removed by Trim Galore. Remaining reads were aligned to mouse genome version GRCm38 with HISAT2 aligner with default parameters. For RIP-seq data SAMtools (v1.13) was used to remove the reads with mapping quality below 30 (Li et al., 2009). m6A peaks and differential m6A peaks were identified using exomePeak2 (v1.6.1) R package with p_cutoff = 0.00001 and log2FC_cutoff = 1 parameters (Meng et al., 2014). Log2FoldChange < −1 and ajusted p value < 0.05 was considered as significantly downregulated peaks. For biological replicates, bedtools (v2.29.2) was used to intersect peaks with overlaped proportion > 0.5 (Meng et al., 2014). Peaks were annotated by HOMER’s annotatePeaks.pl program. The distribution of m6A peaks was plotted by R package Guitar (v.1.5.0) (Cui et al., 2016). The final bam files were transformed to bigwig format by using Deeptools bamCoverage (v3.5.1) with --normalizeUsing RPKM parameters (Ramírez et al., 2014). IGV (Integrative genomics viewer, v2.11.1) was used to visualize the aligned data (Thorvaldsdóttir et al., 2013). DESeq2 was used to identify Ythdc1 binding genes. The FoldChange > 2 and ajusted p-value < 0.05 genes were considered as Ythdc1 binding RNAs.
RNA-Seq Data Analysis
Ythdc1‐KO and Mettl3‐KO RNA-seq datasets were obtained from the Gene Expression Omnibus (GEO) public database under accession number GSE157262. The expression analysis was performed based on the available processed fpkm-matrix files in the database.
Results
Dynamic Long Noncoding RNA Transcriptome During Embryonic Stem Cells-Derived Cardiac Differentiation
To investigate the global expression profiles of lncRNA transcriptome during ESC-derived cardiac differentiation, we collected the samples at four different phases including undifferentiated ESC, mesoderm (MES), cardiac progenitor (CP), and CM. Purified RNA after removal of ribosomal RNA was utilized for sequencing. The reproducible data revealed that 18,749 mRNAs and 6,061 lncRNAs were expressed during cardiac differentiation (Supplementary Figure S1A). As stage-specific marker genes, Nanog and Mesp1 were highly expressed at ESC and MES stages, while Mef2c and Tnnt2 were expressed at higher levels in the CP and CM phases (Figure 1A), indicating the high effectiveness of lncRNA-seq datasets. Comparative analysis of the expression patterns of mRNAs and lncRNAs showed that the expression of lncRNAs was lower than that of mRNAs. Over 50% of mRNAs had TPM values > 20 at four distinct stages, whereas only less than 10% of lncRNAs had similar TPM values (Supplementary Figure S1B). Mapping of these lncRNAs to genomic regions revealed that about half of the lncRNAs were derived from intergenic regions (Figure 1B).
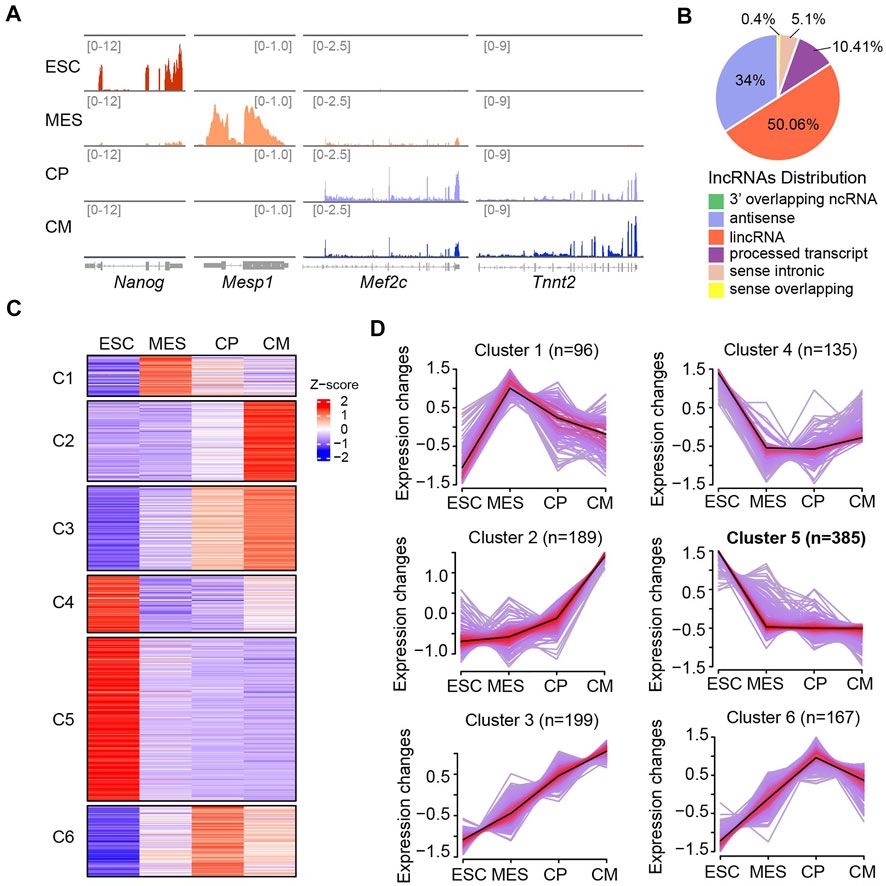
FIGURE 1. Dynamic lncRNA expression pattern during ESC-derived cardiac differentiation. (A) Integrative Genomics Viewer (IGV) of read coverage for specific marker genes at distinct differentiation phases including ESC (Day 0), MES (Day 4), CP (Day 6) and CM (Day 10). The number of reads indicate the peak heights in corresponding genomic regions. (B) Genomic annotation of lncRNAs (n = 6,061) showing that the majority of lncRNAs were mapped to lincRNA (50.06%) and antisense (34%). (C) Heat map showing the expression patterns of lncRNAs were grouped into six different groups (C1-C6) across four differentiation stages. (D) Clustering analysis showing the expression patterns of lncRNAs were divided into six different groups (C1-C6) as shown in (C). The average trend is indicated by black lines. The number of genes is indicated for each cluster. The ESC-specific cluster (C5) contains the largest population of lncRNAs (n = 385).
To detect expression changes of lncRNAs during ESC-derived cardiac differentiation, we performed Fuzzy C-means Clustering based on the TPM values. This analysis led to the identification of six different gene clusters with differential lncRNA expression patterns across the differentiation phases (Figures 1C,D). Cluster one and six contained a batch of lncRNAs that were specifically expressed at MES and CP stages, respectively. Both cluster two and three comprised genes that were specifically expressed at the CM stage. However, lncRNAs in cluster two showed mildly increased expression from the ESC to CP stage but dramatically upregulated expression from the CP to CM stage. In contrast, the expressions of lncRNAs in cluster three were increased gradually with the progression of cardiac differentiation (Figures 1C,D). LncRNAs in cluster four showed decreased expressions from the ESC to CP stage and slightly recovered expression at the CM stage, while cluster five contained largest population of lncRNAs (n = 385) which expressed at highest levels in ESC and considerably downregulated afterwards. Altogether, our results suggested lncRNA transcriptome underwent dynamic regulation during ESC-derived cardiac differentiation and identified a batch of potential non-coding regulators involved in cardiogenesis.
Regulatory Properties and Cellular Functions of Embryonic Stem Cells-specific Long Noncoding RNAs
It has been well documented that pluripotent genes such as Nanog and Sox2, are restricted to pluripotent cells and downregulated upon differentiation (Zhao and Jin, 2017). Similar as the regulatory pattern of pluripotent genes, we identified that 385 lncRNAs were expressed specifically in ESC and downregulated upon differentiation. Close examination of these lncRNAs revealed that the majority of them were less than 4,000 nucleotides in length (Figure 2A). Furthermore, 85.2% of ESC-specific lncRNAs harbored poly(A) tails, which might be associated with their stabilization in the cellular compartment. As canonical non-coding regulators of cell fate in the nucleus, Xist, Lncenc1, and Gas5, were included in the subgroup of polyadenylated lncRNAs specifically expressed in ESC (Figure 2B), further suggesting the accuracy and validity of our lncRNA-seq data. Intriguingly, this cluster also contained a large number of members from two lncRNA families, Platr and Snhg, some of which have been reported to regulate the stem cell fate transition (Bergmann et al., 2015; Lu et al., 2019; Li et al., 2020; Du et al., 2021; Yan et al., 2021).
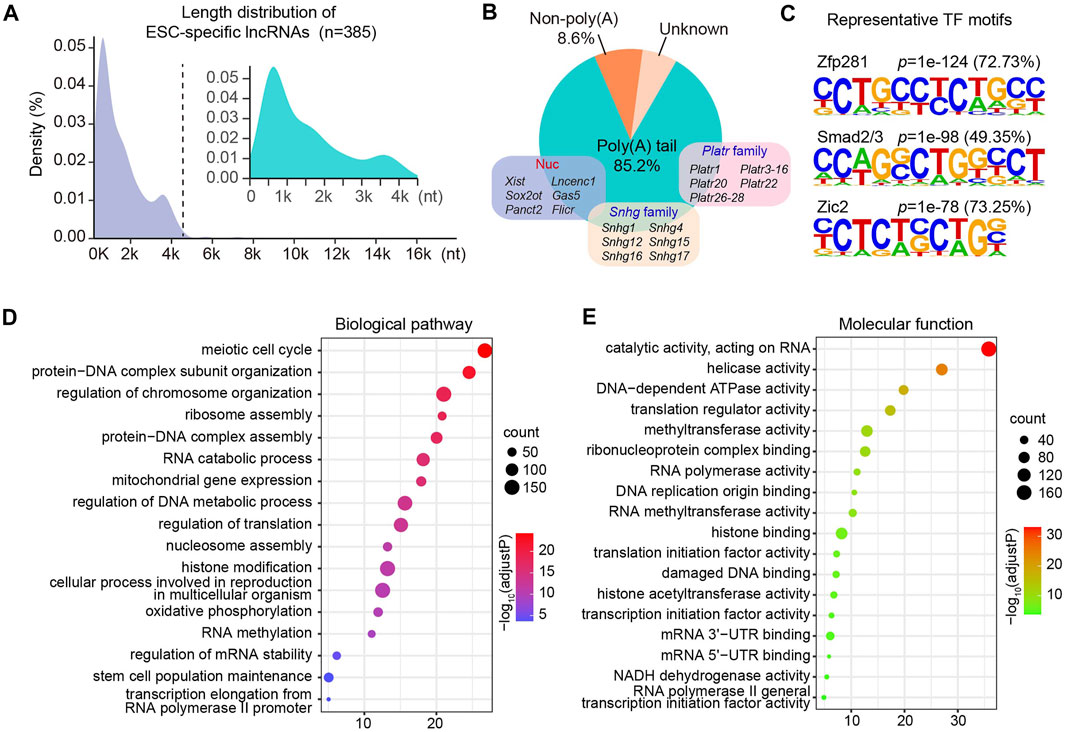
FIGURE 2. Regulatory features and cellular functions of ESC-specific lncRNAs. (A) Length distribution of ESC-specific lncRNA transcripts. (B) Polyadenylation features of ESC-specific lncRNAs. (C) Motif enrichment analysis showing representative TF motifs identified in the promoter region of genes coding ESC-specific lncRNAs. The selected motifs are linked to their corresponding TF. (D,E) Bubble plots show the biological pathways (D) and molecular functions (E) enriched by ESC-specific mRNAs co-expressed with lncRNAs. The size of a bubble is proportional to the number of genes. The intensity of the circle color is corresponding to the significance level which is indicated as -log10 (adjust p-value).
To understand the key regulators that control the gene expressions of these ESC-specific lncRNAs, we next assessed the TF-binding cis-elements imbedded in the promoter region of their coding genes. We observed that about 72.73% of the lncRNAs could be bounded by Zfp281 (Figure 2C), which has been reported to be not required for maintenance of ESCs but indispensable for proper differentiation by targeting core pluripotent genes (Fidalgo et al., 2011). In addition, Smad2/3 and Zic2 have been implicated in the self-renewal and ESC specification (Sakaki-Yumoto et al., 2013; Luo et al., 2015), and our analysis suggested that approximately 49.35 and 73.25% of ESC-specific lncRNAs were enriched for the Smad2/3 and Zic2 motif, respectively (Figure 2C).
The clustering analysis also led to identification of ten distinct expression patterns of protein-coding genes across four differentiation phases (Supplementary Figure S2). We then performed gene ontology (GO) enrichment analysis of ESC-specific protein-coding genes in cluster two that were co-expressed with specifically expressed lncRNAs in ESC. Our analysis revealed that they were significantly enriched in biological processes including meiotic cell cycle, protein-DNA complex assembly, ribosome assembly and protein translation. Interestingly, these co-expressed lncRNAs were also involved in RNA methylation and histone modification (Figure 2D). Consistent with their potential biological processes, these ESC-specific lncRNAs were co-expressed with genes encoding proteins with translation regulator activity, methyltransferase activity and histone acetyltransferase activity (Figure 2E). These results suggest that the ESC-retained lncRNAs are forming regulatory networks with stage-specific TFs and protein-coding genes to modulate ESC fate determination. Supporting this notion, a human ESC-specific lncRNA, lncPRESS1, has been reported to be not only directly regulated by transcription factor P53, but also interact with SIRT6 and modulate histone H3K56 and H3K9 acetylation to effect ESC state (Jain et al., 2016).
Identification of Embryonic Stem Cell-specific Long Noncoding RNA-Binding Proteins
LncRNA generally modulates target gene expression through recruiting specific functional proteins (Statello et al., 2021). We therefore searched for ESC-specific lncRNA-binding proteins by integrative analysis with two RNA-binding protein databases from both NPinter and RNAinter (Teng et al., 2020; Kang et al., 2022). Eventually we identified 1719 pairs of lncRNA-protein interactions. GO analysis of these binding proteins revealed that they harbored molecular activities of single- and double-stranded RNA binding, RNA helicase and chromatin binding. Surprisingly, m6A-containing RNA binding activity was also found in these lncRNA-interacting proteins (Figure 3A).
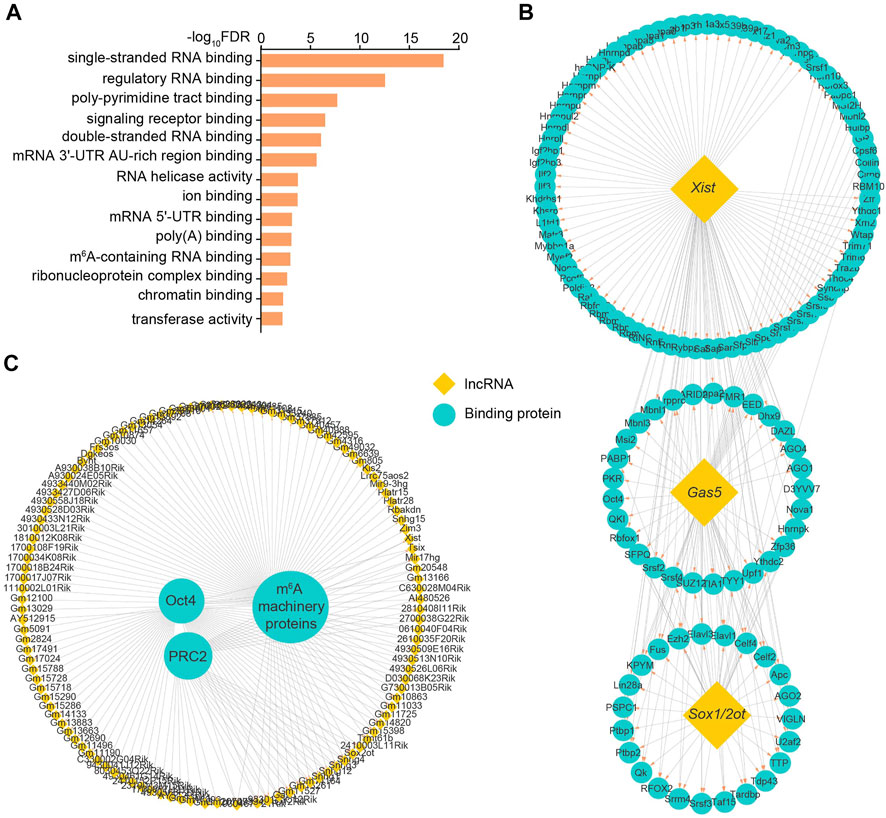
FIGURE 3. Identification of ESC-specific lncRNA-binding proteins. (A) GO analysis of molecular functions enriched by identified binding proteins for ESC-specific lncRNAs. (B) The lncRNA-protein interaction network identifies the candidate proteins binding with ESC-specific lncRNAs Xist, Gas5 or Sox1/2ot. (C) The lncRNA-protein interaction network identifies the candidate lncRNAs binding with pluripotency regulators Oct4, PRC2 and m6A machinery proteins. Yellow and green respectively represent lncRNAs and the interacting proteins of lncRNAs. See also Supplementary Table S2.
For individual lncRNAs, Gas5 and Sox1/2ot, which are linked to the regulation of pluripotency and specific linage differentiation (Knauss et al., 2018; Tu et al., 2018; Xi et al., 2022), were able to recruit many functional factors, such as Ago and Srsf family proteins (Figure 3B). Consistent with multi-faceted roles reported previously (Plath et al., 2003; Fang et al., 2004; Patil et al., 2016), Xist lncRNA could guide the regulation of gene expression through coupling with a variety of proteins including the key components of PRC and m6A machineries (Figure 3B). Among the identified proteins, the most representative ones are the core pluripotency component Oct4, histone modification activator PRC2 and m6A machinery proteins such as Igf2bp1/2/3 and Ythdc1/2, which modulate lncRNA functions potentially through direct interaction (Figure 3C). These results suggest that these ESC-specific lncRNAs mediate multi-layered regulation of gene expression through transcription, epigenetic and epitranscriptomic modifications.
Modification of Embryonic Stem Cell-specific Long Noncoding RNAs by m6A
Given that the ESC-specific lncRNAs are directly recognized by m6A machinery proteins, we therefore hypothesized that at least some of them could be modified by RNA methylation. Indeed, 97 out of 385 lncRNAs contained typical m6A peaks. Distinct from m6A distribution in the vicinity of the stop codons of mRNAs (Dominissini et al., 2012; Meyer et al., 2012), m6As in lncRNAs were highly enriched in close proximity to the middle of the transcript body (Figures 4A,C). In addition, over 85% of mRNA m6As were subject to the regulation of core methyltransferase Mettl3. While, only less than 13% of m6As in ESC-associated lncRNAs were regulated by Mettl3 (Figure 4B), suggesting the existence of additional methyltransferase catalyzing the m6A formation of these lncRNAs.
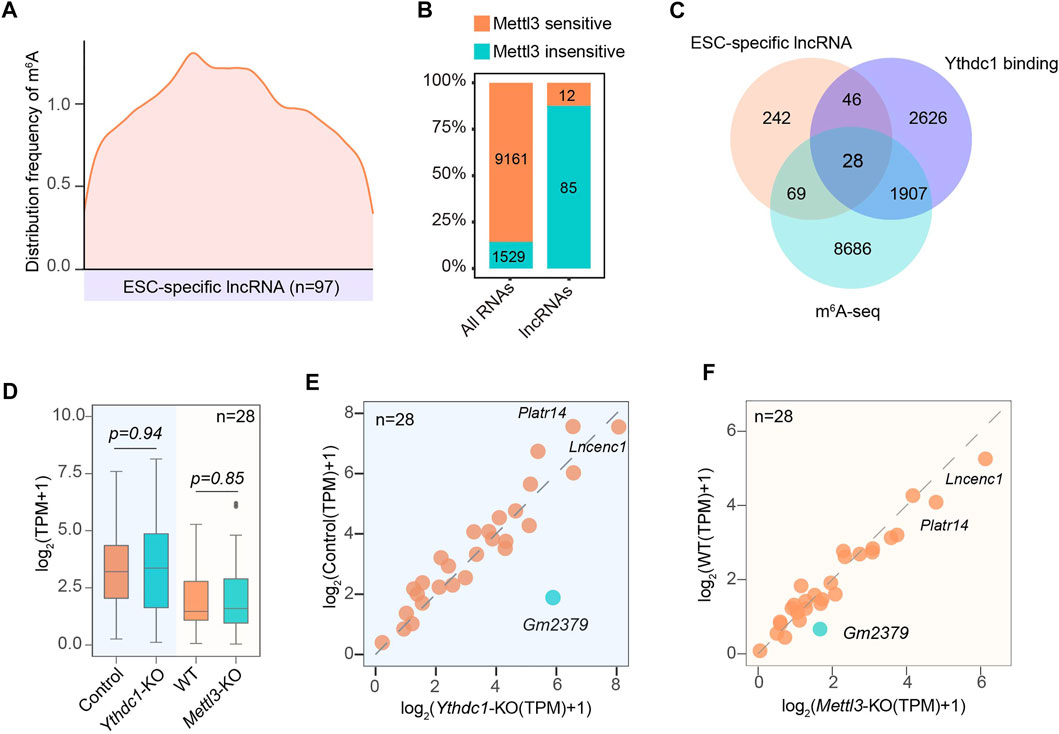
FIGURE 4. Characterization of the modification status on ESC-specific lncRNAs by m6A. (A) Transcriptome-wide distribution pattern of m6A for ESC-specific m6A-containing lncRNAs. (B) The percentage and number of ESC-specific lncRNAs containing m6A peaks that are regulated by Mettl3. (C) A Venn diagram shows the overlapping of lncRNA transcripts containing m6A sites and binding with nuclear m6A “reader” Ythdc1. (D) A box plot shows the expression of 28 overlapped lncRNAs (C) containing m6A sites and binding with Ythdc1 in response to the depletion of Mettl3 or Ythdc1. (E) A scatter plot showing the expression changes of 28 overlapped lncRNAs (C) in the presence or absence of Ythdc1. (F) A scatter plot showing the expression changes of 28 overlapped lncRNAs (C) in the presence or absence of Mettl3. Gm2379 is highlighted in green.
Since Ythdc1 has recently been identified to regulate the functionality of a number of non-coding RNAs (Wei and He, 2021), we next analyzed whether these m6A-containing lncRNAs could recruit Ythdc1 as the “reader” protein. As shown in Figure 4C, 28 out of 97 lncRNAs harbored m6A peaks that were targeted for regulation by Ythdc1 (Figure 4C). The remaining 69 lncRNAs may be regulated by other “reader” proteins. Surprisingly, although methylated adenosines of the 28 lncRNAs were directly bound by Ythdc1, almost all of them did not significantly change their expressions upon loss of Mettl3 or Ythdc1 (Figures 4D–F). A recent study has indicated that the RNA transcribed from long interspersed nuclear element-1 (LINE1) acts as a binding scaffold for Ythdc1 to regulate transcription activities in early embryo and mouse ESCs (Chen et al., 2021; Liu et al., 2021). It is possible that Ythdc1 may facilitate the formation of lncRNA-scaffold complex rather than regulate its abundance to modulate the nuclear gene expression in ESC. Intriguingly, one out of the 28 lncRNAs, Gm2379, was dually regulated by Mettl3 and Ythdc1, as loss of either m6A machinery protein resulted in obviously increased expression of Gm2379 (Figures 4E,F), suggesting that m6A may form different regulatory mode with lncRNAs to modulate their functionality.
Characterization of the Long Noncoding RNA Gm2379
We next focused on the m6A-containing transcript Gm2379, as it is the only lncRNA that responds to the loss of the m6A machinery proteins. We initially validated the expression pattern of Gm2379 during ESC-derived cardiac differentiation. As shown in Figure 5A, its expression was dramatically decreased at day 2 when embryonic bodies (EBs) were formed. Comparatively, the abundance of core pluripotency gene Nanog still maintained at high levels at this stage and declined after EBs generation. Nkx2-5 as one of the earliest markers of the cardiac differentiation, was found to be obviously upregulated at day 6. To decipher the function of Gm2379, we next probed its expression in separated nuclear and cytoplasmic fractions from cultured ESCs. Different from the nuclear 45S rRNA, Gm2379 was localized in both compartments (Figure 5B). Consistent with the global pattern of 97 ESC-specific lncRNAs, the two main m6A peaks of Gm2379 were distributed in proximity to the middle of the transcript body as well as subject to the regulation of Mettl3 and Ythdc1, as m6A signals of Gm2379 were overlapped with Ythdc1 RIP-seq signals and almost disappeared upon knockout of Mettl3 (Figure 5C). In addition, the transcript levels of Gm2379 were significantly increased in ESCs depleting Mettl3 or Ythdc1 (Figures 5D,E), which is in good agreement with previous data analysis. Altogether, our findings indicate that Gm2379 is specifically expressed in ESCs and dynamically regulated by nuclear “reader” protein Ythdc1 in a m6A-dependent manner.
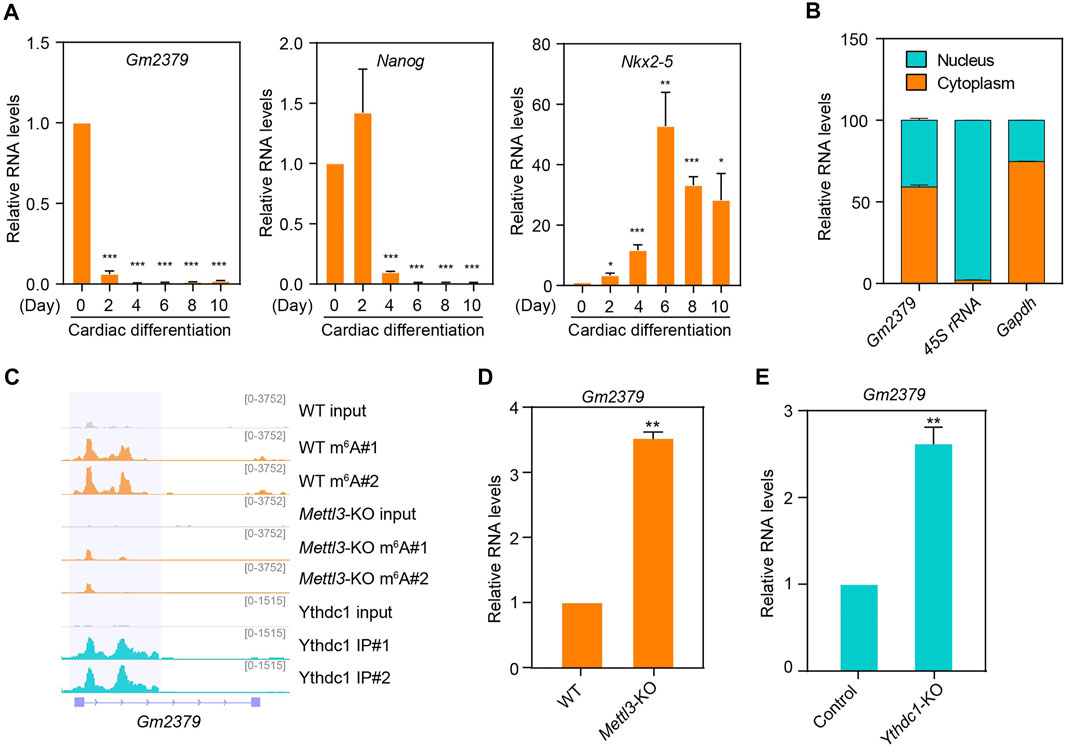
FIGURE 5. The m6A-harboring lncRNA Gm2379 is regulated by both Mettl3 and Ythdc1. (A) Detection of Gm2379 transcript levels during ESC-derived cardiac differentiation by RT-qPCR. Nanog and Nkx2-5 serve as pluripotent and cardiac maker genes. Error bars, mean ± SEM; n = 4 independent replicates; *p < 0.05, **p < 0.01, ***p < 0.001. (B) Detection of the distribution of Gm2379 transcript in ESCs by cell fractionation assay. The RT-qPCR data represented a percentage of the total amount of detected transcripts. Error bars, mean + SEM; n = 2 independent replicates. (C) Genome browser view of a representative genomic region showing m6A read density and Ythdc1-binding feature of the Gm2379 transcript. (D) Detection of Gm2379 transcript levels in WT and Mettl3-KO ESCs by RT-qPCR. Error bars, mean + SEM; n = 3 independent replicates; **p < 0.01. (E) Detection of Gm2379 transcript levels in control and Ythdc1-KO ESCs by RT-qPCR. Error bars, mean + SEM; n = 4 independent replicates; **p < 0.01.
Gm2379 Modulates the Expressions of Pluripotent and Germ-Layer Genes
m6A has been implicated in the regulation of ESC self-renewal and differentiation. Depletion of Mettl3 results in m6A erasure on target pluripotent mRNAs, facilitating ESC self-renewal and impairing directed differentiation (Batista et al., 2014; Geula et al., 2015). The nuclear “reader” Ythdc1 has been found to be required for the maintenance of ESC pluripotency in a m6A-dependent manner (Chen et al., 2021; Liu et al., 2021). We have revealed that the expression of Gm2379 lncRNA is regulated by both Mettl3 and Ythdc1, serving as a potential target of m6A in ESC. To determine whether Gm2379 is directly involved in the regulation of pluripotency, we generated stable ESC lines with shRNA-mediated gene knockdown of Gm2379 (Figure 6A). LIF is indispensable to maintain the pluripotent state of ESC (Martello and Smith, 2014). In the presence of LIF, depletion of Gm2379 selectively affected the expression of pluripotency genes. The transcript levels of Nanog and Dppa3 were significantly decreased in Gm2379-knock down (KD) cells, while Klf2 was upregulated in Gm2379-KD cells (Figure 6B). However, in the absence of LIF, the expression of Nanog and Klf2 rather than Dppa3 was comparable in control and Gm2379-KD ESCs, suggesting the selective regulation of pluripotency genes by Gm2379 lncRNA in a LIF-dependent manner.
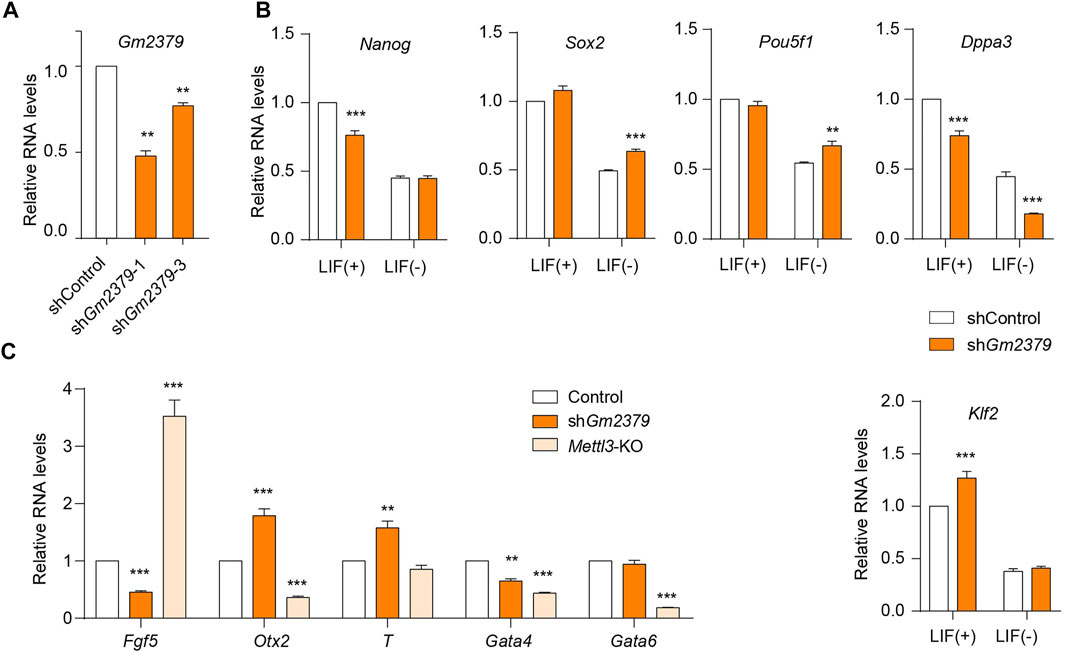
FIGURE 6. Gm2379 modulates the expressions of pluripotent and germ-layer genes. (A) Generation of Gm2379-KD ESCs. (B) Detection of transcript levels of pluripotent genes in control and Gm2379-KD ESCs in the presence or absence of LIF by RT-qPCR. (C) Detection of transcript levels of three germ-layer genes in control, Gm2379-KD and Mettl3-KO ESCs by RT-qPCR. shGm2379-1 ESCs were used in (B) and (C). Error bars, mean + SEM; n = 4 independent replicates; *p < 0.05, **p < 0.01, ***p < 0.001.
In addition to the pluripotent genes, the crucial genes involved in the formation of three germ layers (ectoderm, mesoderm and endoderm) were dramatically dysregulated in Gm2379-KD ESCs (Figure 6C). Specifically, the ectoderm gene Fgf5 and endodermal gene Gata4 were significantly downregulated while the transcripts of the mesoderm gene T and ectoderm gene Otx2 were obviously increased in response to Gm2379 depletion. Mettl3, as the core m6A methyltransferase, has been demonstrated to regulate ESC pluripotency and differentiation (Batista et al., 2014; Geula et al., 2015). In contrast to Gm2379 KD, loss of Mettl3 resulted in increased expression of Fgf5 and decreased expression of Otx2 (Figure 6C). Considering the negative regulation of Gm2379 by Mettl3 shown above, it is very possible that Mettl3 directly targets Gm2379 lncRNA to modulate the expression of ectodermal genes.
Discussion
LncRNA as genomic “dark matter” has been implicated to play crucial roles in multiple cellular processes by forming signaling regulatory networks with other functional molecules. In the process of cell fate regulation, only a small fraction of lncRNAs has been characterized (Fico et al., 2019; Chen et al., 2020). Therefore, it could be expected that many more non-coding transcripts will participate in the regulation of pluripotency and differentiation processes. In this study, we performed a global investigation of lncRNA expression profiles across four different phases of ESC-derived cardiac differentiation. Our findings showed that the lncRNA transcriptome changed dramatically during cardiac differentiation (Figures 1C,D), revealing the dynamic and complex regulation of cell differentiation at additional layers.
Among the six lncRNAs clusters with distinct expression patterns we identified, the cluster five contained the largest number of non-coding transcripts which expressed at most abundant levels in ESC and dramatically downregulated at MES stage and subsequent phases during cardiac differentiation (Figures 1C,D), suggesting the crucial roles of lncRNA in the regulation of ESC gene expression. Careful inspection revealed that many reported lncRNAs in the regulation of cell fate were included in this subgroup, such as Gas5 and Lncenc1. Depletion of Gas5 has been found to dysregulate a number of critical genes involved in self-renewal and endodermal differentiation through forming interaction networks with pluripotent transcriptional factors and epigenetic factors (Tu et al., 2018). Lncenc1 maintains the self-renewal of naive ESCs through cooperation with PTBP1 and HNRNPK to regulate the expression of glycolytic genes (Sun et al., 2018). Therefore, it is reasonable to speculate that the highly specific expression of lncRNAs in ESC is tightly associated with their molecular functions in the regulation of pluripotency and differentiation. In addition, our analysis also revealed a number of previously unappreciated transcripts from same lncRNA families (Figure 2B), which will facilitate our understanding of dynamics and complexity of lncRNA regulatory networks governing cell fate determination.
The function of lncRNAs is accurately exerted dependent on the formation of both cis-regulatory modules with TFs and trans-regulatory modules with RBPs (Statello et al., 2021). An increasing number of studies have been absorbed in deciphering how lncRNA regulates target gene expression, while few researches investigate how lncRNA itself is transcriptionally regulated (Tompkins et al., 2016). Consistent with the stage-specific transcriptional features, our analytic data showed that a large portion of ESC-specific lncRNAs were directly bounded and regulated by the pluripotency TF regulators Zfp281, Smad2/3 and Zic2 (Figure 2C). In addition, a number of these non-coding transcripts were found to cooperate with stage-specific trans RBPs in the regulation of ESC gene expression, such as Oct4 and PRC2 (Figure 3C). Surprisingly, m6A machinery proteins were also identified as direct interaction partners of ESC-specific lncRNAs. These observations collectively suggest that the ESC-retained lncRNAs exhibit subtle signaling networks with crucial TF regulator and trans RBPs to modulate cell pluripotency and differentiation progression.
Considering the m6A machineries identified as representative lncRNA-binding proteins, we thus evaluated the methylation status of these ESC-specific lncRNAs. Approximately 25% of these non-coding transcripts were modified by m6A. Whereas, only a small portion of m6A-containing lncRNAs were subject to the regulation of m6A-catalyzer Mettl3 (Figure 4B). Consistent with this observation, a recent study also found a subgroup of chromatin-associated LINE1 regulatory RNAs harbored m6A peaks that are insensitive to Mettl3 (Chen et al., 2021). It is possible that other methyltransferases such as Mettl16, mediate m6A formation for those Mettl3-insensitive ESC-specific lncRNAs. An increasing number of studies have been identified Ythdc1 as a primary m6A “reader” to regulate molecular functions of non-coding transcripts (Patil et al., 2016; Liu et al., 2020, 2021; Chen et al., 2021; Ji et al., 2021; Lee et al., 2021; Ninomiya et al., 2021; Wang et al., 2021). However, our survey revealed that only about 28% of m6A-containing lncRNAs were directly targeted by Ythdc1. More interestingly, only one out of the 28 Ythdc1-binding m6A-containing lncRNAs, Gm2379, dramatically altered its expression in response to the deletion of Mettl3 or Ythdc1 (Figures 4E,F), suggesting that majority of these non-coding transcripts may act as dynamic structural scaffolds for recruiting Ythdc1, which in turn does not regulate their expressions.
We next focused on the characterization of the novel lncRNA Gm2379. Consistent with the lncRNA-seq data, the m6A-containing lncRNA Gm2379 was dramatically downregulated during ESC-derived cardiac differentiation (Figures 5A,C). Furthermore, depletion of Mettl3 or Ythdc1 leads to significantly increased expression of Gm2379 (Figures 5D,E). In addition to the nuclear “reader” Ythdc1, it is very likely subject to the regulation of additional m6A-binding proteins in the cytoplasm, as Gm2379 was found in both nuclear and cytoplasmic compartments (Figure 5B). Functional identification showed that depletion of Gm2379 resulted in limited but significant alteration of pluripotent gene expressions. Instead, the key genes required for the formation of three germ layers, were more dramatically dysregulated upon knockdown of Gm2379 (Figure 6). The opposite regulation of ectodermal genes by Mettl3 and Gm2379 suggest that Gm2379 lncRNA might function as a direct target of Mettl3 to govern the formation of ectoderm. Further investigation should focus on the examination of additional cooperating factors that are required for Gm2379-mediated ESC differentiation.
In summary, our findings provide global identification and characterization of lncRNA expression profiles during ESC differentiation. Investigation of cis and trans regulatory network suggest that ESC-specific lncRNAs are governed by key TFs as well as m6A effector machineries. Subsequent identification of a specific m6A-containing lncRNA on the molecular level further demonstrates the crucial interplay between epitranscriptomic modification and key lncRNAs in the regulation of cell state maintenance and transition.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: GEO, accession number GSE197205 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE197205).
Author Contributions
Conceptualization, X-ML; Methodology, SW and JZha; Investigation, SW, YD, HZ, XW, LH, and JH; Data Analysis, JZha; Writing, X-ML; Review and Editing, X-ML, SW, and JZha; Funding Acquisition, X-ML and JZho; Resources, X-ML and JZho; Supervision, X-ML and JZho. All authors read and approved the final version.
Funding
This work was supported by the grants from the National Natural Science Foundation of China (32070868 and 81974527), the Natural Science Foundation of Jiangsu Province, China (BK20211219 and BK20190553), and Jiangsu Province Laboratory of Pathogen Biology (KLPBJP-K202008).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2022.880674/full#supplementary-material
References
Ashburner, M., Ball, C. A., Blake, J. A., Botstein, D., Butler, H., Cherry, J. M., et al. (2000). Gene Ontology: Tool for the Unification of Biology. Nat. Genet. 25, 25–29. doi:10.1038/75556
Atlasi, Y., and Stunnenberg, H. G. (2017). The Interplay of Epigenetic Marks during Stem Cell Differentiation and Development. Nat. Rev. Genet. 18, 643–658. doi:10.1038/nrg.2017.57
Batista, P. J., Molinie, B., Wang, J., Qu, K., Zhang, J., Li, L., et al. (2014). m6A RNA Modification Controls Cell Fate Transition in Mammalian Embryonic Stem Cells. Cell. Stem Cell. 15, 707–719. doi:10.1016/j.stem.2014.09.019
Bergmann, J. H., Li, J., Eckersley-Maslin, M. A., Rigo, F., Freier, S. M., and Spector, D. L. (2015). Regulation of the ESC Transcriptome by Nuclear Long Noncoding RNAs. Genome Res. 25, 1336–1346. doi:10.1101/gr.189027.114
Bergmann, J. H., and Spector, D. L. (2014). Long Non-coding RNAs: Modulators of Nuclear Structure and Function. Curr. Opin. Cell. Biol. 26, 10–18. doi:10.1016/j.ceb.2013.08.005
Carlevaro-Fita, J., and Johnson, R. (2019). Global Positioning System: Understanding Long Noncoding RNAs through Subcellular Localization. Mol. Cell. 73, 869–883. doi:10.1016/j.molcel.2019.02.008
Chen, C.-Y., Cheng, Y.-Y., Yen, C. Y. T., and Hsieh, P. C. H. (2017). Mechanisms of Pluripotency Maintenance in Mouse Embryonic Stem Cells. Cell. Mol. Life Sci. 74, 1805–1817. doi:10.1007/s00018-016-2438-0
Chen, C., Liu, W., Guo, J., Liu, Y., Liu, X., Liu, J., et al. (2021). Nuclear m6A Reader YTHDC1 Regulates the Scaffold Function of LINE1 RNA in Mouse ESCs and Early Embryos. Protein Cell. 12, 455–474. doi:10.1007/s13238-021-00837-8
Chen, J., Wang, Y., Wang, C., Hu, J.-F., and Li, W. (2020). LncRNA Functions as a New Emerging Epigenetic Factor in Determining the Fate of Stem Cells. Front. Genet. 11, 1–12. doi:10.3389/fgene.2020.00277
Constanty, F., and Shkumatava, A. (2021). LncRNAs in Development and Differentiation: From Sequence Motifs to Functional Characterization. Development 148, dev182741. doi:10.1242/dev.182741
Cui, X., Wei, Z., Zhang, L., Liu, H., Sun, L., Zhang, S.-W., et al. (2016). Guitar: An R/Bioconductor Package for Gene Annotation Guided Transcriptomic Analysis of RNA-Related Genomic Features. BioMed Res. Int. 2016, 1–8. doi:10.1155/2016/8367534
Dominissini, D., Moshitch-Moshkovitz, S., Schwartz, S., Salmon-Divon, M., Ungar, L., Osenberg, S., et al. (2012). Topology of the Human and Mouse m6A RNA Methylomes Revealed by m6A-Seq. Nature 485, 201–206. doi:10.1038/nature11112
Du, Z., Wen, X., Wang, Y., Jia, L., Zhang, S., Liu, Y., et al. (2021). Chromatin lncRNA Platr10 Controls Stem Cell Pluripotency by Coordinating an Intrachromosomal Regulatory Network. Genome Biol. 22, 1–29. doi:10.1186/s13059-021-02444-6
Fang, J., Chen, T., Chadwick, B., Li, E., and Zhang, Y. (2004). Ring1b-mediated H2A Ubiquitination Associates with Inactive X Chromosomes and Is Involved in Initiation of X Inactivation. J. Biol. Chem. 279, 52812–52815. doi:10.1074/jbc.C400493200
Fico, A., Fiorenzano, A., Pascale, E., Patriarca, E. J., and Minchiotti, G. (2019). Long Non-coding RNA in Stem Cell Pluripotency and Lineage Commitment: Functions and Evolutionary Conservation. Cell. Mol. Life Sci. 76, 1459–1471. doi:10.1007/s00018-018-3000-z
Fidalgo, M., Shekar, P. C., Ang, Y.-S., Fujiwara, Y., Orkin, S. H., and Wang, J. (2011). Zfp281 Functions as a Transcriptional Repressor for Pluripotency of Mouse Embryonic Stem Cells. Stem Cells 29, 1705–1716. doi:10.1002/stem.736
Geula, S., Moshitch-Moshkovitz, S., Dominissini, D., Mansour, A. A., Kol, N., Salmon-Divon, M., et al. (2015). m 6 A mRNA Methylation Facilitates Resolution of Naïve Pluripotency toward Differentiation. Science 347, 1002–1006. doi:10.1126/science.1261417
Jain, A. K., Xi, Y., McCarthy, R., Allton, K., Akdemir, K. C., Patel, L. R., et al. (2016). LncPRESS1 Is a P53-Regulated LncRNA that Safeguards Pluripotency by Disrupting SIRT6-Mediated De-acetylation of Histone H3K56. Mol. Cell. 64, 967–981. doi:10.1016/j.molcel.2016.10.039
Ji, Q., Zong, X., Mao, Y., and Qian, S.-B. (2021). A Heat Shock-Responsive lncRNA Heat Acts as a HSF1-Directed Transcriptional Brake via M 6 A Modification. Proc. Natl. Acad. Sci. U.S.A. 118, 1–11. doi:10.1073/pnas.2102175118
Kang, J., Tang, Q., He, J., Li, L., Yang, N., Yu, S., et al. (2022). RNAInter v4.0: RNA Interactome Repository with Redefined Confidence Scoring System and Improved Accessibility. Nucleic Acids Res. 50, D326–D332. doi:10.1093/nar/gkab997
Knauss, J. L., Miao, N., Kim, S.-N., Nie, Y., Shi, Y., Wu, T., et al. (2018). Long Noncoding RNA Sox2ot and Transcription Factor YY1 Co-regulate the Differentiation of Cortical Neural Progenitors by Repressing Sox2. Cell. Death Dis. 9, 799. doi:10.1038/s41419-018-0840-2
Kumar, L., and Futschik, M. E. (2007). Mfuzz: A Software Package for Soft Clustering of Microarray Data. Bioinformation 2, 5–7. doi:10.6026/97320630002005
Lee, J.-H., Wang, R., Xiong, F., Krakowiak, J., Liao, Z., Nguyen, P. T., et al. (2021). Enhancer RNA m6A Methylation Facilitates Transcriptional Condensate Formation and Gene Activation. Mol. Cell. 81, 3368–3385. e9. doi:10.1016/j.molcel.2021.07.024
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., et al. (2009). The Sequence Alignment/Map Format and SAMtools. Bioinformatics 25, 2078–2079. doi:10.1093/bioinformatics/btp352
Li, Y., Zhang, J., Huo, C., Ding, N., Li, J., Xiao, J., et al. (2017). Dynamic Organization of lncRNA and Circular RNA Regulators Collectively Controlled Cardiac Differentiation in Humans. EBioMedicine 24, 137–146. doi:10.1016/j.ebiom.2017.09.015
Li, Z., Guo, X., and Wu, S. (2020). Epigenetic Silencing of KLF2 by Long Non-coding RNA SNHG1 Inhibits Periodontal Ligament Stem Cell Osteogenesis Differentiation. Stem Cell. Res. Ther. 11, 1–11. doi:10.1186/s13287-020-01953-8
Liao, Y., Smyth, G. K., and Shi, W. (2014). FeatureCounts: An Efficient General Purpose Program for Assigning Sequence Reads to Genomic Features. Bioinformatics 30, 923–930. doi:10.1093/bioinformatics/btt656
Liu, J., Dou, X., Chen, C., Chen, C., Liu, C., Xu, M. M., et al. (2020). N 6 -methyladenosine of Chromosome-Associated Regulatory RNA Regulates Chromatin State and Transcription. Science 367, 580–586. doi:10.1126/science.aay6018
Liu, J., Gao, M., He, J., Wu, K., Lin, S., Jin, L., et al. (2021). The RNA m6A Reader YTHDC1 Silences Retrotransposons and Guards ES Cell Identity. Nature 591, 322–326. doi:10.1038/s41586-021-03313-9
Love, M. I., Huber, W., and Anders, S. (2014). Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 15, 1–21. doi:10.1186/s13059-014-0550-8
Lu, W., Yu, J., Shi, F., Zhang, J., Huang, R., Yin, S., et al. (2019). The Long Non-coding RNA Snhg3 Is Essential for Mouse Embryonic Stem Cell Self-Renewal and Pluripotency. Stem Cell. Res. Ther. 10, 1–12. doi:10.1186/s13287-019-1270-5
Luo, S., Lu, J. Y., Liu, L., Yin, Y., Chen, C., Han, X., et al. (2016). Divergent lncRNAs Regulate Gene Expression and Lineage Differentiation in Pluripotent Cells. Cell. Stem Cell. 18, 637–652. doi:10.1016/j.stem.2016.01.024
Luo, Z., Gao, X., Lin, C., Smith, E. R., Marshall, S. A., Swanson, S. K., et al. (2015). Zic2 Is an Enhancer-Binding Factor Required for Embryonic Stem Cell Specification. Mol. Cell. 57, 685–694. doi:10.1016/j.molcel.2015.01.007
Martello, G., and Smith, A. (2014). The Nature of Embryonic Stem Cells. Annu. Rev. Cell. Dev. Biol. 30, 647–675. doi:10.1146/annurev-cellbio-100913-013116
Martin, M. (2011). Cutadapt Removes Adapter Sequences from High-Throughput Sequencing Reads. EMBnet J. 17, 10–12. doi:10.14806/ej.17.1.200
Meng, J., Lu, Z., Liu, H., Zhang, L., Zhang, S., Chen, Y., et al. (2014). A Protocol for RNA Methylation Differential Analysis with MeRIP-Seq Data and exomePeak R/Bioconductor Package. Methods 69, 274–281. doi:10.1016/j.ymeth.2014.06.008
Meyer, K. D., Saletore, Y., Zumbo, P., Elemento, O., Mason, C. E., and Jaffrey, S. R. (2012). Comprehensive Analysis of mRNA Methylation Reveals Enrichment in 3′ UTRs and Near Stop Codons. Cell. 149, 1635–1646. doi:10.1016/j.cell.2012.05.003
Ninomiya, K., Iwakiri, J., Aly, M. K., Sakaguchi, Y., Adachi, S., Natsume, T., et al. (2021). m 6 A Modification of HSATIII lncRNAs Regulates Temperature‐dependent Splicing. EMBO J. 40, e107976. doi:10.15252/embj.2021107976
Patil, D. P., Chen, C.-K., Pickering, B. F., Chow, A., Jackson, C., Guttman, M., et al. (2016). m6A RNA Methylation Promotes XIST-Mediated Transcriptional Repression. Nature 537, 369–373. doi:10.1038/nature19342
Paul, S., Andrew, M., Owen, O., Nitin, S., Jonathan, T. W., Daniel, R., et al. (1971). Cytoscape: A Software Environment for Integrated Models. Genome Res. 13, 426. doi:10.1101/gr.1239303.metabolite
Pertea, M., Kim, D., Pertea, G. M., Leek, J. T., and Salzberg, S. L. (2016). Transcript-level Expression Analysis of RNA-Seq Experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 11, 1650–1667. doi:10.1038/nprot.2016.095
Plath, K., Fang, J., Mlynarczyk-Evans, S. K., Cao, R., Worringer, K. A., Wang, H., et al. (2003). Role of Histone H3 Lysine 27 Methylation in X Inactivation. Science 300, 131–135. doi:10.1126/science.1084274
Ramírez, F., Dündar, F., Diehl, S., Grüning, B. A., and Manke, T. (2014). DeepTools: A Flexible Platform for Exploring Deep-Sequencing Data. Nucleic Acids Res. 42, W187–W191. doi:10.1093/nar/gku365
Rutenberg-Schoenberg, M., Sexton, A. N., and Simon, M. D. (2016). The Properties of Long Noncoding RNAs that Regulate Chromatin. Annu. Rev. Genom. Hum. Genet. 17, 69–94. doi:10.1146/annurev-genom-090314-024939
Sakaki-Yumoto, M., Liu, J., Ramalho-Santos, M., Yoshida, N., and Derynck, R. (2013). Smad2 Is Essential for Maintenance of the Human and Mouse Primed Pluripotent Stem Cell State. J. Biol. Chem. 288, 18546–18560. doi:10.1074/jbc.M112.446591
Statello, L., Guo, C.-J., Chen, L.-L., and Huarte, M. (2021). Gene Regulation by Long Non-coding RNAs and its Biological Functions. Nat. Rev. Mol. Cell. Biol. 22, 96–118. doi:10.1038/s41580-020-00315-9
Sun, Z., Zhu, M., Lv, P., Cheng, L., Wang, Q., Tian, P., et al. (2018). The Long Noncoding RNA Lncenc1 Maintains Naive States of Mouse ESCs by Promoting the Glycolysis Pathway. Stem Cell. Rep. 11, 741–755. doi:10.1016/j.stemcr.2018.08.001
Teng, X., Chen, X., Xue, H., Tang, Y., Zhang, P., Kang, Q., et al. (2020). NPInter v4.0: An Integrated Database of ncRNA Interactions. Nucleic Acids Res. 48, D160–D165. doi:10.1093/nar/gkz969
Thorvaldsdóttir, H., Robinson, J. T., and Mesirov, J. P. (2013). Integrative Genomics Viewer (IGV): High-Performance Genomics Data Visualization and Exploration. Briefings Bioinforma. 14, 178–192. doi:10.1093/bib/bbs017
Tompkins, J. D., Jung, M., Chen, C.-y., Lin, Z., Ye, J., Godatha, S., et al. (2016). Mapping Human Pluripotent-To-Cardiomyocyte Differentiation: Methylomes, Transcriptomes, and Exon DNA Methylation "Memories". EBioMedicine 4, 74–85. doi:10.1016/j.ebiom.2016.01.021
Tripathi, V., Ellis, J. D., Shen, Z., Song, D. Y., Pan, Q., Watt, A. T., et al. (2010). The Nuclear-Retained Noncoding RNA MALAT1 Regulates Alternative Splicing by Modulating SR Splicing Factor Phosphorylation. Mol. Cell. 39, 925–938. doi:10.1016/j.molcel.2010.08.011
Tu, J., Tian, G., Cheung, H.-H., Wei, W., and Lee, T.-l. (2018). Gas5 Is an Essential lncRNA Regulator for Self-Renewal and Pluripotency of Mouse Embryonic Stem Cells and Induced Pluripotent Stem Cells. Stem Cell. Res. Ther. 9, 1–13. doi:10.1186/s13287-018-0813-5
Wang, X., Liu, C., Zhang, S., Yan, H., Zhang, L., Jiang, A., et al. (2021). N6-methyladenosine Modification of MALAT1 Promotes Metastasis via Reshaping Nuclear Speckles. Dev. Cell. 56, 702–715. e8. doi:10.1016/j.devcel.2021.01.015
Wei, J., and He, C. (2021). Chromatin and Transcriptional Regulation by Reversible RNA Methylation. Curr. Opin. Cell. Biol. 70, 109–115. doi:10.1016/j.ceb.2020.11.005
Xi, J., Xu, Y., Guo, Z., Li, J., Wu, Y., Sun, Q., et al. (2022). LncRNA SOX1‐OT V1 Acts as a Decoy of HDAC10 to Promote SOX1‐dependent hESC Neuronal Differentiation. EMBO Rep. 23, e53015. doi:10.15252/embr.202153015
Yan, P., Lu, J. Y., Niu, J., Gao, J., Zhang, M. Q., Yin, Y., et al. (2021). LncRNA Platr22 Promotes Super-enhancer Activity and Stem Cell Pluripotency. J. Mol. Cell. Biol. 13, 295–313. doi:10.1093/jmcb/mjaa056
Yao, R.-W., Wang, Y., and Chen, L.-L. (2019). Cellular Functions of Long Noncoding RNAs. Nat. Cell. Biol. 21, 542–551. doi:10.1038/s41556-019-0311-8
Keywords: lncRNA, m6A modificaiton, embryonic stem cell, Gm2379, cell fate
Citation: Wang S, Zhang J, Ding Y, Zhang H, Wu X, Huang L, He J, Zhou J and Liu X-M (2022) Dynamic Transcriptome Profiling Reveals LncRNA-Centred Regulatory Networks in the Modulation of Pluripotency. Front. Cell Dev. Biol. 10:880674. doi: 10.3389/fcell.2022.880674
Received: 21 February 2022; Accepted: 20 April 2022;
Published: 11 May 2022.
Edited by:
Jun Liu, Peking University, ChinaReviewed by:
Ezgi Hacisuleyman, The Rockefeller University, United StatesSha Sun, University of California, Irvine, United States
Lin Zhang, University of California, San Diego, United States
Copyright © 2022 Wang, Zhang, Ding, Zhang, Wu, Huang, He, Zhou and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jun Zhou, ano1NzJAY3B1LmVkdS5jbg==; Xiao-Min Liu, bGl1eG02NDJAY3B1LmVkdS5jbg==
†These authors have contributed equally to this work
 Shen Wang
Shen Wang Jun Zhang
Jun Zhang Yu’an Ding
Yu’an Ding Haotian Zhang
Haotian Zhang Xiang Wu
Xiang Wu Lingci Huang
Lingci Huang Junjie He
Junjie He Jun Zhou
Jun Zhou Xiao-Min Liu
Xiao-Min Liu