- 1Dipartimento di Medicina Sperimentale, Università degli Studi della Campania L. Vanvitelli, Naples, Italy
- 2CNRS, IFCE, INRAE, Université de Tours, PRC, Nouzilly, France
- 3Dipartimento di Scienze Anatomiche, Istologiche, Medico Legali e dell’Apparato Locomotore, “Sapienza” Università di Roma, Roma, Italy
- 4Dipartimento di Scienze Motorie e del Benessere, Università di Napoli Parthenope, Napoli, Italy
Kisspeptins are involved in the regulation of hypothalamic-pituitary-gonadal axis, Leydig cell functions, and testosterone secretion, acting as endogenous ligands of the KISS1 receptor. ANKRD31 protein participates in male fertility, regulating meiotic progression, and epididymal sperm maturation. Here, we show that in Leydig cells, KISS1 receptor and ANKRD31 proteins physically interact; the formation of this protein complex is enhanced by Kisspeptin-10 that also modulates F-actin synthesis, favoring histone acetylation in chromatin and gene expression via the cytoskeletal–nucleoskeletal pathway. Kp/KISS1R system deregulation, expression impairment of cytoskeletal–nucleoskeletal mediators, Leydig gene targets, and the decreased testosterone secretion in Ankrd31−/− testis strongly supported our hypothesis. Furthermore, cytochalasin D treatment subverted the gene expression induction dependent on Kisspeptin-10 action. In conclusion, the current work highlights a novel role for the Kisspeptin-10 in the induction of the cytoskeletal–nucleoskeletal route, downstream a physical interaction between KISS1 receptor and ANKRD31, with gene expression activation as final effect, in Leydig cells.
Introduction
Kisspeptins (Kps), encoded by Kiss-1 gene, are a group of neuropeptides involved in the neuroendocrine control of reproduction because they are able to stimulate the secretion of gonadotrophin-releasing hormone (GnRH), luteinizing hormone (LH), and follicle-stimulating hormone (FSH), through the activation of a G protein-coupled receptor, KISS1R (Oakley et al., 2009; Pinilla et al., 2012).
The action of Kps on GnRH neurons is tightly linked to the expression of KISS1R receptor on themselves (Colledge, 2009). KISS1R activation, dependent on Kisspeptin binding, is able to induce a potent GnRH neuron depolarization, associated with an intrinsic Kp-dependent gonadotrophin-releasing regulation, electing so, the Kp/KISS1R system act as a key mediator in the regulation of hypothalamic-pituitary-gonadal (HPG) axis at puberty and during adulthood (Thompson et al., 2004; Han et al., 2005; Navarro and Tena-Sempere, 2011). Indeed, Kps stimulate GnRH secretion, associated with a prominent LH-releasing downstream effect, in several species, including rodents and primates (Shahab et al., 2005; Chianese et al., 2016). Accordingly, the lack of Kp signaling results in hypogonadotropic hypogonadism and delayed sexual maturation (de Roux et al., 2003; Seminara et al., 2003; León et al., 2016). Despite the primary effect of Kps in the regulation of the HPG axis at the central level, a peripheral expression of the Kp/KISS1R system in mammalian and non-mammalian gonads has been reported (Gaytan et al., 2007; Chianese et al., 2013, 2015, 2017; Calder et al., 2014; Dorfman et al., 2014; Salehi et al., 2015; Meccariello et al., 2020; Gloria et al., 2021), although its testicular biological role needs to be further explored (Sharma et al., 2020). In this regard, recent findings support the involvement of Kps in Leydig cell functions, including testosterone secretion (Salehi et al., 2015). In particular, progressive Kp expression during mouse pubertal development, especially in Leydig cells, confirms a testicular local effect of Kps. In addition, isolated Leydig cells, in vitro treated with Kps, show enhanced KISS1/KISS1R gene expression and testosterone secretion, suggesting an autocrine role of the Kp/KISS1R system in these cells (Han et al., 2020).
Ankyrin repeat domain-containing proteins (ANKRDs) are sub-membranous proteins that favor the direct interaction between membrane- and cytoskeletal proteins, acting as a link-protein scaffold (Batrukova et al., 2000; Hryniewicz-Jankowska et al., 2002; Manfrevola et al., 2021a). This intricate physical protein interaction regulates several biological functions, such as cellular adhesion and cytoarchitecture (Cunhaa and Mohler, 2009). The favorite ANKRD cytoskeletal protein interactors are the SPECTRINS and, in turn, the actins, through which mechanobiological pathways are drawn (Xu et al., 2013; Saito et al., 2015; Dou et al., 2018).
In particular, mechanotransduction pathways, through the formation of cytoskeletal–nucleoskeletal connections, transduce extracellular mechanical forces into biochemical signals propagating them along the cytoskeleton to the nuclear envelope, in order to regulate chromatin organization and gene expression, as the final biological effect (Uhler and Shivashankar, 2017; Manfrevola et al., 2021a). New insights regarding the involvement of ANKRDs in mechanotransduction have been reported as ANKRDs, interacting with mechanosensitive channels, are required for the cytoplasmic transmission of mechanosensory activity in neurons (Argudo et al., 2019; Tang et al., 2020). In addition, ANKRD responsiveness to Kp/KISS1R signaling has been unveiled in GnRH neurons (Soga et al., 2016).
Interestingly, an emerging role of ANKRDs in male reproduction has also been reported. Recent studies have highlighted the involvement of ANKRD31 in male sterility, focusing the attention on the main role of ANKRD31 in homologous recombination, meiosis progression, blood–epididymal barrier integrity, and epididymal sperm maturation (Boekhout et al., 2019; Acquaviva et al., 2020; Manfrevola et al., 2021b).
A putative involvement of ANKRD31, in mediating Kp signaling in the regulation of Leydig cell functions and gene expression, through the cytoskeletal–nucleoskeletal pathway, is a novel aspect investigated here.
In the current work, we report the testicular responsiveness to Kp/KISS1R system stimulation, through the analysis of cytoskeletal–nucleoskeletal modulators and Leydig gene targets. In detail, we confirm the expression of the Kp/KISS1R system in murine Leydig cells, and we show that Kisspeptin-10 (Kp-10), via KISS1R activation, induces the expression of KISS1R and ANKRD31, with an upregulation of cytoskeletal–nucleoskeletal actors. Testicular KISS1R activation promotes the expression of Leydig cell genes, a biological effect that we found correlated with an increasing histone acetylation.
Based on these observations, we hypothesized that the Kp/KISS1R system could regulate Leydig cell functions and gene expression via ANKRD31 and the cytoskeletal–nucleoskeletal pathway. In support to this hypothesis, we demonstrate a complete Kp/KISS1R system deregulation in Ankrd31−/− testis associated with the expression impairment of cytoskeletal–nucleoskeletal mediators, as well as of Leydig gene targets and testosterone secretion. In addition, we show, for the first time, the interaction among KISS1R, ANKRD31, and F-actin proteins in murine primary Leydig cell cultures. Furthermore, the stimulation of Leydig cells with Kp-10 induces F-actin synthesis and, sequentially, enhances nuclear histone acetylation with a subsequent increase in gene expression, leading us to suppose a direct involvement of F-actin and the cytoskeletal–nucleoskeletal pathway in this biological effect. Cytochalasin D treatment carried out in primary Leydig cell cultures subverted the Kp-dependent induction of both histone acetylation and Leydig gene expression, confirming that Kp-10 enhances KISS1R/ANKRD31 interaction and, in turn, cytoskeletal–nucleoskeletal actors, in order to remodel chromatin and regulate Leydig gene expression.
Materials and Methods
Experimental Animals
C57BL/6 male mice (Charles River Laboratory, Lecco, Italy) were used in this study. All animals were housed as three per cage under controlled illumination (12 h light/dark cycle; light on 6:00 a.m.) and standard environmental conditions (ambient temperature 20–22°C, humidity 55%–60%) and were maintained on a standard pellet diet with free access to water, before the beginning of experimental procedures. The number of the enrolled animals was determined by the parameters adopted for the G*Power analysis required to get the permission for in vivo experiments, which is suggested by the legal entity giving the permission. For experimental procedures, adult males (3–5 months) were sacrificed under anesthesia, by cardiac perfusion with PBS (pH 7.6), to clean peripheral tissues from blood contaminants, or by cervical dislocation, depending on the experimental designs. Animals were placed in a plexiglas chamber with 4% isoflurane (Iso-Vet, Piramal Healthcare, United Kingdom) for 5 min and were sacrificed when fully sedated, as measured by a lack of heartbeat and active paw reflex. Testes were rapidly removed and collected depending on the experimental procedure, as described later.
In addition, wild-type (WT) male mice and males carrying Ankrd31 null mutation (Ankrd31−/−) (Manfrevola et al., 2021b) were used in this study. Heterozygous mice were bred on a C57BL/6 background before generating WT and Ankrd31−/− male mice. For experimental procedures, WT and Ankrd31−/− adult males (3–5 months) were sacrificed, under anesthesia, by cervical dislocation. Testes and blood samples were rapidly removed and collected depending on the experimental procedures.
Chemicals
Kp-10 (Metastin 45–54, H-YNWNSFGLRF-NH2) of human origin and Kp-234, a specific KISS1R antagonist (Roseweir et al., 2009), were purchased from DBA Italia (Milan, Italy). Kp-10 of human origin was chosen on the basis of its high homology with the murine peptide sequence. Cytochalasin D (C8273) was obtained from Sigma-Aldrich (Milan, Italy). The drugs were dissolved in dimethylsulfoxide (DMSO), according to the manufacturer’s instructions. NB4 collagenase was obtained from Serva (Heidbergh, Germany).
Histology and Immunocytochemistry Analysis
Testes collected from WT and Ankrd31−/− mice (n = 5 for each group) were fixed overnight in Bouin’s solution, dehydrated in ethanol, cleared in xylene, and embedded in paraffin using standard procedures. Microtome serial sections (7 μm thick) were cut and processed for hematoxylin/eosin (H&E) staining and immunocytochemistry staining analyses. For H&E staining, the sections were deparaffinized and processed using standard procedures. For immunocytochemistry staining, testes sections were deparaffinized, rehydrated, and permeabilized with PBS pH 7.4 containing 0.1% Triton X-100. A citrate buffer of 0.01 M (pH 6.0) was used for antigen retrieval. After blocking with PBS containing 5% BSA and normal goat serum (diluted 1:5), sections were incubated with anti-KISS1 antibody (PA5-50513, Invitrogen; diluted 1:100), anti-KISS1R antibody (PA5-96221, Invitrogen; diluted 1:100), anti-F-actin antibody (MA1-80729, Invitrogen; diluted 1:100), anti-H3K14ac antibody (703894, Invitrogen; diluted 1:100), and anti-StAR antibody (sc-166821, Santa Cruz, diluted 1:100) overnight at 4°C. Immunoreactivity was revealed using the avidin/biotin complex system and H2O2/DAB as the substrate/chromogen. The specificity of immunoreactions is routinely checked by omitting primary antibodies. The histological observations and analysis were carried out under a light microscope (Leica CTR500, Leica Microsystems Inc., Milan, Italy), and images were captured using a high-resolution digital camera (Leica DC300F).
Human Chorionic Gonadotropin (hCG) Administration in vivo
WT (n = 5) and Ankrd31−/− (n = 5) male mice (3 months old) were injected with hCG (15 IU/Kg; Chorulon Grovet) intraperitoneally. After 2 h from hCG injection, blood samples were collected and plasma was obtained by centrifugation at 1,000× g for 10 min. Plasma samples were stored at −20°C for further testosterone dosage.
Testosterone EIA Assay
Plasma TT levels of WT (n = 5) and Ankrd31−/− (n = 5) male mice, at basal conditions and following hCG injection, were measured by a competitive enzyme immunoassay (EIA) designed by the Phenotyping Endocrinology Laboratory (Research unit: Physiology of Reproduction and Behavours, Nouzilly, France). The sample size used for the assay was 8 µL of undiluted sample. The minimal detectable TT concentration was 0.15 ng/ml, and all samples were analyzed in the same assay. The intra- and inter-assay coefficients of variation were 4.7% and 7.1%, respectively. All determinations were made in triplicate for each plasma sample analyzed.
Kp-10 In Vitro Treatment of Mouse Testis
C57BL/6 testes (n = 5 for each experimental group) were incubated in PBS for 90 min at room temperature (RT), with vehicle (0.005% DMSO; control group, CTRL) or with Kp-10 at 0.01 μM, 0.1 μM, and 1 μM. After treatment, the testes were placed at −80°C for following molecular investigations.
Isolation and Culture of Mouse Leydig Cells
Leydig cells were isolated as previously described by Sun et al. (2011). In brief, testes from adult mice (n = 5) were harvested, and tunica albuginea was removed. Then, testes were digested at 37°C for 15 min in 0.03% collagenase NB4 in a thermomixer imposing 150 rpm vibration. After this first digestion, the supernatant was discarded. Then, testes were incubated again in 0.03% collagenase NB4 in the thermomixer for 15 min, lowering the vibration speed to 130 rpm. Isolated cells were counted and approximately 1 × 105 cells per testis were obtained. Isolated cells were cultured in low glucose DMEM with 10% of fetal bovine serum (FBS) plating them in culture plates (60 mm or 35 mm culture dish Falcon) for molecular analysis and in eight well chamber slides (Ibidi) for immunofluorescence experiments. STAR staining was carried out to identify the cells and to assess their purity (data not shown). When the Leydig cells were grown to 90% confluence were treated, when indicated, with Kp-10 (0.1 µM), Kp-234 (1 µM), and cytochalasin D (10 µM) alone or in combination with Kp-10. Cytochalasin D and Kp-234 were added 30 min before Kp-10 treatment.
Immunofluorescence and Confocal Microscopy Analysis
For in situ analyses, Leydig cells, cultured in eight well chamber slides, were fixed in 4% paraformaldehyde in PBS (pH 7.4) at 4°C overnight. To detect F-actin and H3K14ac, immunofluorescence experiments were performed. Fixed cells were permeabilized in PBS supplemented with 1% BSA and 0.1% Triton for 2 h and then quenched with 5% donkey serum in PBS/BSA/Triton. Samples were then incubated overnight with anti-H3K14ac antibody diluted 1:100 (703894, Invitrogen Life Technologies, Paisley, United Kingdom). Then, samples were washed three times in PBS/BSA/Triton for 30 min and incubated with the appropriate secondary antibody: Cy5 conjugated anti-rabbit IgG was used to detect H3K14ac immunocomplexes (Jackson Immuno Research, Cambridge, United Kingdom). TO-PRO3 iodide fluorescent dye 642/661 (1:5000 in PBS, Invitrogen Life Technologies, Paisley, United Kingdom) was used for nuclei staining. As a negative control, the primary antibody was omitted. For the detection of F-actin, a FITC-conjugated phalloidin (Invitrogen, Life Technologies, Paisley, United Kingdom) was used.
Immunofluorescence experiments were analyzed using a Leica confocal microscope (Laser Scanning TCS SP2 equipped with Kr/Ar and He/Ne lasers, Mannheim, Germany). Laser lines were 488, 543, and 633 nm for FITC, Cy5, and TO-PRO3 excitation, respectively. The images were scanned under a ×20 or ×40 oil immersion objectives. To perform quantitative analysis of fluorescence, optical spatial series with a step size of 1 µm were recovered with fixed laser intensities, and the sum of fluorescence intensity (SUM(I)) was determined in maximum projection image of each series using Leica confocal software. Three independent experiments in triplicate were analyzed.
Determination of TT Levels in Leydig Cell Culture Media
Culture media of Leydig cells in vitro treated with Kp-10 (0.1 µM) alone or in combination with Kp-234 (1 µM) were collected to perform TT determination. TT was extracted from cell culture media using methanol as an organic solvent (liquid/liquid extraction) and purified by solid phase extraction (SPE) using specific cartridges for steroids (AFFINIMIP, Affinisep, France). The procedure described by Errico et al. (2019) was modified specifically for cell culture media including the quality control system used to monitor method performance and to prevent assay contamination. The LC-MS/MS analysis was performed using a Dionex UltiMate3000 HPLC system (Thermo Fisher Scientific Inc, Monza, Italy), coupled to an ESI-triple quadrupole mass spectrometer (API 2000, Sciex, Germany). A Kinetex F5 (×100 4.6 mm, 2.6 µm) column (Phenomenex, Italy) was used for reversed-phase separations. Chromatographic separation and instrumental parameters were reported in Errico et al., 2019. The analyte identification was based on multiple reactions monitoring in a positive mode. The TT was specifically identified not only by the retention time but also by monitoring the following ion transitions: m/z 291.3 → m/z 97.2 (quantifier) and m/z 291.3 → m/z 108.9 (qualifier). The linearity of the detector response was verified over the concentration range 0.100–50 ng/ml. All samples were analyzed in triplicate, for each experimental group, with relative standard deviations (RSDs) less than 13%. HPLC grade reagents, including ultrapure water, acetonitrile (ACN), and methanol (MeOH), were purchased from Romil (ROMIL Ltd., United Kingdom). TT was purchased from Merch (Germany). AFFINIMIP cartridges were purchased from Affinisep (France).
Total RNA Preparation
The TRIzol® reagent (Invitrogen Life Technologies, Paisley, United Kingdom) was used to extract total RNA from C57BL/6 testes in vitro treated with Kp-10 (0.01 μM, 0.1 μM, and 10 μM) and from primary murine Leydig cells in vitro treated with Kp-10 (0.1 μM) and cytochalasin D (10 μM) alone or in combination with Kp-10 (0.1 μM). In brief, the testes were homogenized in the TRIzol reagent and incubated for 5 min at 20°C. Then, 0.2 ml chloroform/ml Trizol reagent were added, and the samples were centrifuged at 12,000× g for 15 min at 4°C. The aqueous phase was transferred to a fresh tube and total RNA was precipitated by mixing with isopropyl alcohol (0.5 ml/ml Trizol reagent) and 1 μl of glycogen (20 mg/ml). After centrifugation at 12,000× g for 10 min at 4°C, the RNA pellet was washed with 75% ethanol, centrifuged at 7,500× g for 10 min at 4°C, and dissolved in DEPC-H2O. Total RNAs were assessed with a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, United States) to quantify concentration (ng/μl) and purity (260/280 and 260/230 ratios). Then, RNA aliquots (10 μg) were treated with 2U DNase I (RNase-free DNase I, Ambion, Thermo Fisher Scientific, Massachusetts, United States) to remove potential contamination of genomic DNA and finally preserved at −80°C until the next step.
RNA Expression Analysis by One-Step Evagreen qRT-PCR
According to the manufacturer’s instructions, a kit containing quantitative real-time polymerase chain reaction (qRT-PCR) enzyme mix and an Evagreen qPCR Mastermix (Applied Biological Materials Inc., Richmond, Canada) was used for gene expression analysis in i) testes in vitro treated with Kp-10 (0.01 μM, 0.1 μM, and 10 μM) (n = 5 animals for each experimental group) and in ii) primary murine Leydig cells in vitro treated with Kp-10 (0.1 μM) and cytochalasin D (10 μM) alone or in combination with Kp-10 (0.1 μM) (n = 5 samples for each experimental group). A concentration of 50 ng of total RNA was used for all reactions on a CFX-96 Real-Time Polymerase Chain Reaction (PCR) System (Biorad, Milan, Italy). A negative control, without RNA, was included. The qRT-PCR in triplicate from each experimental group was analyzed. A gene expression analysis, corrected for PCR efficiency, and normalized toward the reference gene (Rp18S), was performed by CFX Manager software (Biorad, Milan, Italy). Normalized fold expression (n.f.e) of mRNAs was calculated by applying the 2–ΔΔCT method.
PCR Primer Design
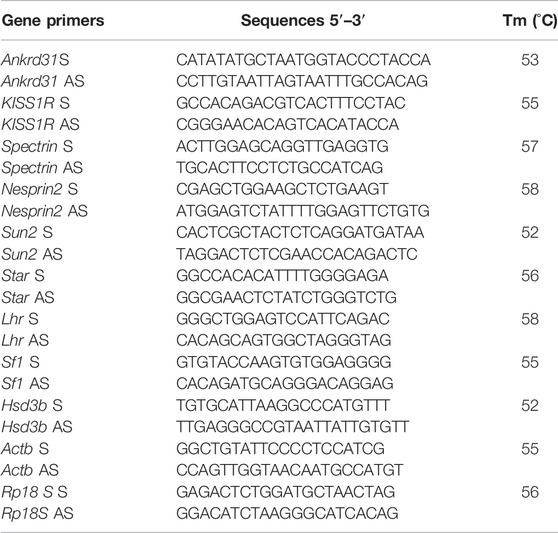
Primers to amplify selected RNAs were designed through the online tool Primer-BLAST1. Primers for mouse genes are shown in Table 1.
Protein Extraction and Western Blot Analysis
Total protein extraction was performed for i) testes in vitro treated with Kp-10 (0.01 μM, 0.1 μM, and 10 μM) (n = 5 animals for each experimental group), ii) primary murine Leydig cells in vitro treated with Kp-10 (0.1 μM) alone or in combination with the specific antagonist Kp-234 (1 μM) (n = 5 samples for each experimental group), and iii) primary murine Leydig cells in vitro treated with Kp-10 (0.1 μM) and cytochalasin D (10 μM) alone or in combination with Kp-10 (0.1 μM) (n = 5 samples for each experimental group). The samples were separately homogenized in RIPA buffer [PBS, pH 7.4, 10 mM of dithiothreitol, 0.02% sodium azide, 0.1% SDS, 1% NP-40, and 0.5% sodium deoxycholate, in the presence of protease inhibitors (10 μg/ml of leupeptin, aprotinin, pepstatin A, chymostatin, and 5 μg/ml of TPCK)] and sonicated three times for 30 s bursts, each at 60 mW. Proteins were separated by SDS-PAGE and transferred to a polyvinylidene difluoride membrane (GE Healthcare, Milan, Italy) at 280 mA for 2.5 h, at 4°C. The filters were treated for 2.5 h with blocking solution [5% non-fat milk, 0.25% Tween 20 in Tris-buffered saline (TBS, pH 7.6)] and incubated with different primary antibodies in TBS-milk buffer (TBS pH 7.6, 3% non-fat milk) overnight, at 4°C. The filters were washed in 0.25% Tween 20–TBS and incubated with secondary antibodies diluted 1:1,000 in TBS-milk buffer and then washed again. An enhanced chemiluminescence–Western blotting detection system (Amersham ECL Western Blotting Detection Reagent, cod: RPN2106, GE Healthcare, Milan, Italy) was used to detect the immune complexes.
Antibodies and relative dilutions are reported in Table 2. The specificity of the immunoreactions was routinely checked by omitting all primary antibodies used in this study (data not shown). Western blot experimental triplicates from each experimental group were quantified by densitometry analysis, adjusted relatively to Ponceau S staining, and reported as O.D. fold change (mean ± SEM).
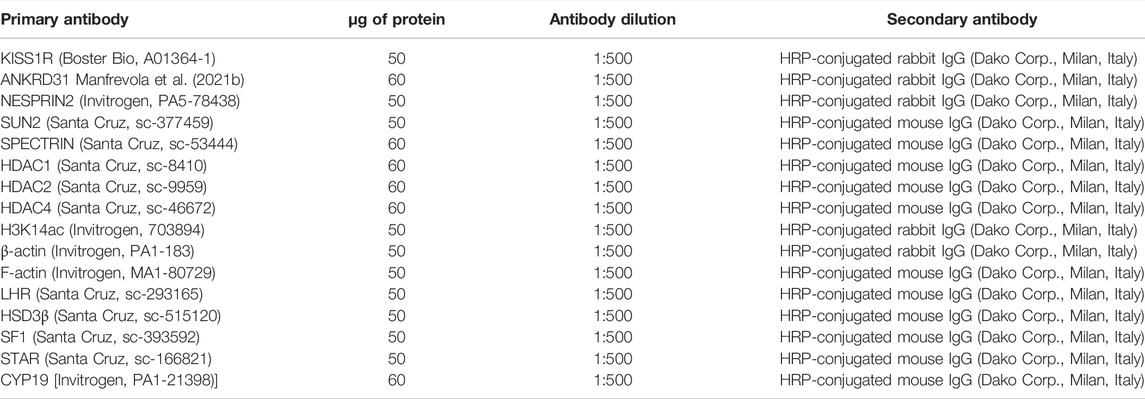
TABLE 2. Primary antibodies, protein amounts, antibody dilution, and secondary antibodies used for Western blot analysis.
Protein Immunoprecipitation Assay
For IP, primary murine Leydig cells in vitro treated with Kp-10 (0.1 μM) alone or in combination with the specific antagonist Kp-234 (1 μM) were lyzed in RIPA buffer, in the presence of protease inhibitors (10 μg/ml of leupeptin, aprotinin, pepstatin A, chymostatin, and 5 μg/ml of TPCK), sonicated three times for 30 s bursts, each at 60 mW, and then incubated on ice for 30 min. After centrifugation at 20,000× g for 30 min at 4°C, the protein supernatant was collected. A concentration of 500 μg of supernatant proteins from each sample was incubated with 2 μg of KISS1R antibody (IP-KISS1R), ANKRD31 antibody (IP-ANKRD31), or IgG as negative control (12370, Sigma-Aldrich, Milan, Italy), under rotary agitation, overnight at 4°C. Afterward, Protein A/G PLUS Agarose Beads (sc-2003, Santa Cruz Biotechnology, Cambridge, United Kingdom) were added to each sample and incubated overnight under rotary agitation at 4°C. After bead incubation, samples were washed three times (3,000× g for 3 min at 4°C) in TBS pH 7.6 and boiled in Laemmli sample buffer for 10 min to be later analyzed by SDS-PAGE.
Statistical Analysis
ANOVA followed by Student’s t-test (for two independent group comparisons) and Tukey test (for multi group comparison) was used to identify groups having different mean. Differences with p < 0.05 were considered statistically significant. Data were expressed as the mean ± SEM from at least five independent animals for each genotype or experimental group. For qRT-PCR, Western blot, LC-MS/MS, and EIA assay, triplicates from five animals/genotypes or experimental groups each were considered.
Results
Kp-10 Positively Regulates Testicular KISS1R Expression and the Cytoskeletal–Nucleoskeletal Pathway
In order to study the effects of Kps in the regulation of the Kp/KISS1R system and the cytoskeletal–nucleoskeletal pathway in Leydig cells, we evaluated i) the expression of the Kp/KISS1R system in WT mouse testis, by an immunocytochemistry analysis, and ii) the expression of several cytoskeletal–nucleoskeletal mediators, in testes in vitro treated with different doses of Kp-10 (0.01 µM, 0.1 µM, and 1 µM), by qRT-PCR. Results showed a striking KISS1 and KISS1R protein localization in Leydig cells (Figure 1A). Interestingly, higher expression levels of Kiss1R, at all the chosen doses of Kp-10, were observed in comparison to the CTRL group (Figure 1B) (p < 0.01). In addition, a significant increase in Ankrd31, Spectrin, and β-Actin expression levels was also observed at all the chosen doses of Kp-10, in comparison to the CTRL group (Figures 1C–E) (p < 0.05; p < 0.01), whereas lower expression levels of Nesprin2 and Sun2 were observed following Kp-10 in vitro treatment (Figures 1F,G) (p < 0.01).
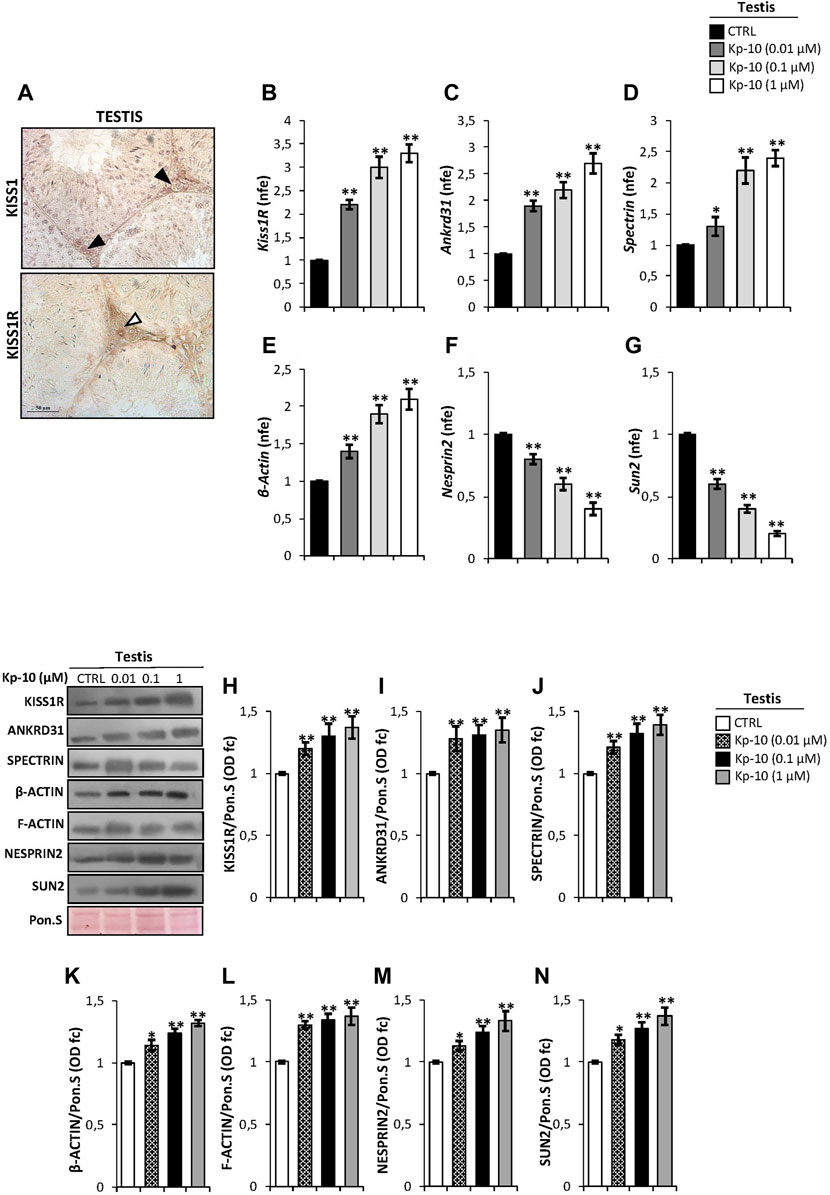
FIGURE 1. (A) Immunocytochemistry of KISS1 and KISS1R in Bouin’s fixed C57BL/6 testis sections (7 μm). The KISS1 and KISS1R protein localization in Leydig cells was indicated by black and white arrowheads, respectively. Scale bar: 50 μm. (B–G) Differential expression analysis of KissR and cytoskeletal–nucleoskeletal pathway mediator mRNAs in mice testes in vitro treated with different doses of Kp-10 (0.01 µM, 0.1 µM, and 1 µM), by qRT-PCR. (B) Kiss1R, (C) Ankrd31, (D) Spectrin, (E) β-Actin, (F) Nesprin2, and (G) Sun2 expression levels were normalized using Rp18S as a housekeeping gene and expressed as normalized fold expression (n.f.e.), relatively to the CTRL group. All data are reported as mean value ± S.E.M; *p < 0.05; **p < 0.01. Western blot analysis of (H) KISS1R, (I) ANKRD31, (J) SPECTRIN-α-II, (K) β-actin, (L) F-actin, (M) NESPRIN2, and (N) SUN2 proteins levels in mice testes in vitro treated with different doses of Kp-10 (0.01 µM, 0.1 µM, and 1 µM). Signals were quantified by the densitometry analysis and normalized to Ponceau Red (Pon.S). Data are expressed in O.D. values as fold change (O.D. fc), relatively to the CTRL group, and reported as mean ± SEM; *p < 0.05; **p < 0.01.
Western blot analysis was carried out to investigate the testicular protein levels of KISS1R receptor and cytoskeletal–nucleoskeletal modulators, following Kp-10 in vitro treatment. As reported, data showed that all the analyzed proteins were significantly increased following Kp-10 treatment. In details, a significant increase of testicular KISS1R, ANKRD31, and SPECTRIN protein amount was observed at all the chosen doses of Kp-10, relatively to the CTRL group (Figures 1H–J) (p < 0.01). In agreement, the levels of cytoskeletal modulators, as β-actin and F-actin, were higher in testes in vitro treated with Kp-10 than the CTRL group (Figures 1K,L) (p < 0.05; p < 0.01) and, finally, a significant increase of NESPRIN2 and SUN2 protein levels was also observed at all the doses, relatively to the CTRL group (Figures 1M–N) (p < 0.05; p < 0.01), suggesting that Kp-10 in vitro treatment positively modulates the expression of both KISS1R receptor and cytoskeletal–nucleoskeletal pathway modulators, having Leydig cells as the main target.
In Testis Kp-10 Enhances the Expression of Leydig Cell Genes and Epigenetic Markers
Considering that in mouse testis KISS1R is expressed in Leydig cells (Anjum et al., 2012; Salehi et al., 2015), as confirmed by our immunocytochemistry analysis, we evaluated a possible effect of Kps on the expression of key modulators of steroidogenesis. Results showed a significant increase of Lhr and Hsd3b expression levels at the Kp-10 doses of 0.1 and 1 µM (p < 0.01), (Figures 2A,B), in comparison with the CTRL group, whereas no effect was observed at the Kp-10 dose of 0.01 µM (Figures 2A,B). Similar results were observed for Star and Sf1 (Figures 2C,D).
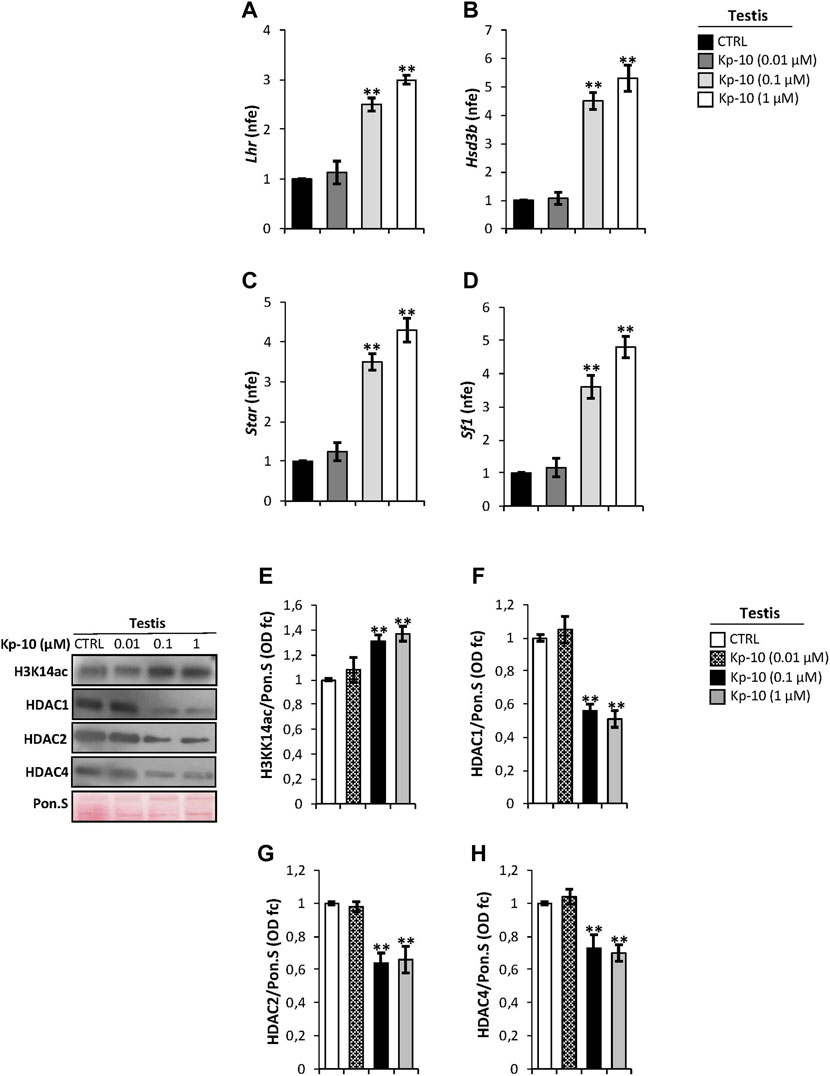
FIGURE 2. (A–D) Differential expression analysis of Leydig cell genes in mice testes in vitro treated with different doses of Kp-10 (0.01 µM, 0.1 µM, and 1 µM), by qRT-PCR. (A) Lhr, (B) Hsd3b, (C) Star, and (D) Sf1 expression levels were normalized using Rp18S as a housekeeping gene and expressed as normalized fold expression (n.f.e.), relatively to the CTRL group. All data are reported as mean value ± S.E.M; **p < 0.01. Western blot analysis of (E) H3K14ac, (F) HDAC1, (G) HDAC2, and (H) HDAC4 protein levels in mice testes in vitro treated with different doses of Kp-10 (0.01 µM, 0.1 µM, and 1 µM). Signals were quantified by the densitometry analysis and normalized to Ponceau Red (Pon.S). Data are expressed in O.D. values as fold change (O.D. fc), relatively to the CTRL group, and reported as mean ± SEM; **p < 0.01.
Western blot analysis was carried out to investigate a possible responsiveness of epigenetic markers following Kp in vitro treatment. As showed, relatively to the CTRL group, a significant increase of H3K14ac protein levels, chosen as a marker of active gene transcription, was observed at Kp-10 doses of 0.1 µM and 1 µM (p < 0.01), whereas no effect was observed at the dose of 0.01 µM (Figure 2E). Analogously, testicular in vitro treatment with the doses 0.1 and 1 µM of Kp-10 significantly decreased protein levels of HDAC1, HADAC2, and HDAC4 (p < 0.01) than the CTRL group (Figures 2F,G), whereas no effect was observed at the dose of 0.01 µM for all HDAC types investigated, suggesting that Kps might positively regulate Leydig cell gene expression modulating epigenetic markers.
Ankrd31 Gene Deletion Negatively Affects Kp/KISS1R System and Leydig Functions
In order to demonstrate that the Kp/KISS1R system could regulate Leydig cell functions and gene expression via ANKRD31, and in turn by the cytoskeletal–nucleoskeletal pathway, we carried out immunocytochemistry and molecular analyses in Ankrd31−/− mouse testes, chosen as putative model of cytoskeletal disruption. A histological analysis performed by H&E staining on testicular sections of both WT and Ankrd31−/− showed an impairment of interstitial environment in Ankrd31−/− compared to WT (Figure 3A), suggesting that the loss of ANKRD31 protein could affect Leydig cell functions. Interestingly, the immunocytochemistry analysis showed a lower signal of both KISS1 and KISS1R in Ankrd31−/− Leydig cells than WT, confirming a deregulation of the Kp/KISS1R system (Figures 3B,C). In addition, the immunocytochemistry analysis of F-actin, H3K14ac as well as STAR evidenced a strong signal reduction of all markers analyzed in Ankrd31−/− compared to WT Leydig cells (Figures 3D–F). To confirm Kp/KISS1R system deregulation, the Western blot analysis of KISS1R was carried out in WT and Ankrd31−/− testis. As reported, a significant reduction of KISS1R protein content was observed in Ankrd31−/− in comparison to WT testis (p < 0.01) (Figure 3G). In order to assess if the deregulation of the Kp/KISS1R system observed in Ankrd31−/− Leydig cells occurred due to their compromised functions, we evaluated the expression of key modulators of steroidogenesis by the Western blot analysis. The results showed a significant decrease of LHR, HSD3β, STAR, and SF1 protein contents in Ankrd31−/− in comparison to WT (p < 0.01), (Figures 3H–K). In agreement, plasma TT levels were significantly lower in Ankrd31−/− than WT (p < 0.01) in both basal conditions as well as following hCG stimulation (Figure 3L), confirming that i) Ankrd31−/− Leydig cells possessed an impaired ability to respond to LH stimulation and ii) the deregulation of Kp/KISS1R system and of steroidogenesis modulators negatively affected TT production in Ankrd31−/− mice.
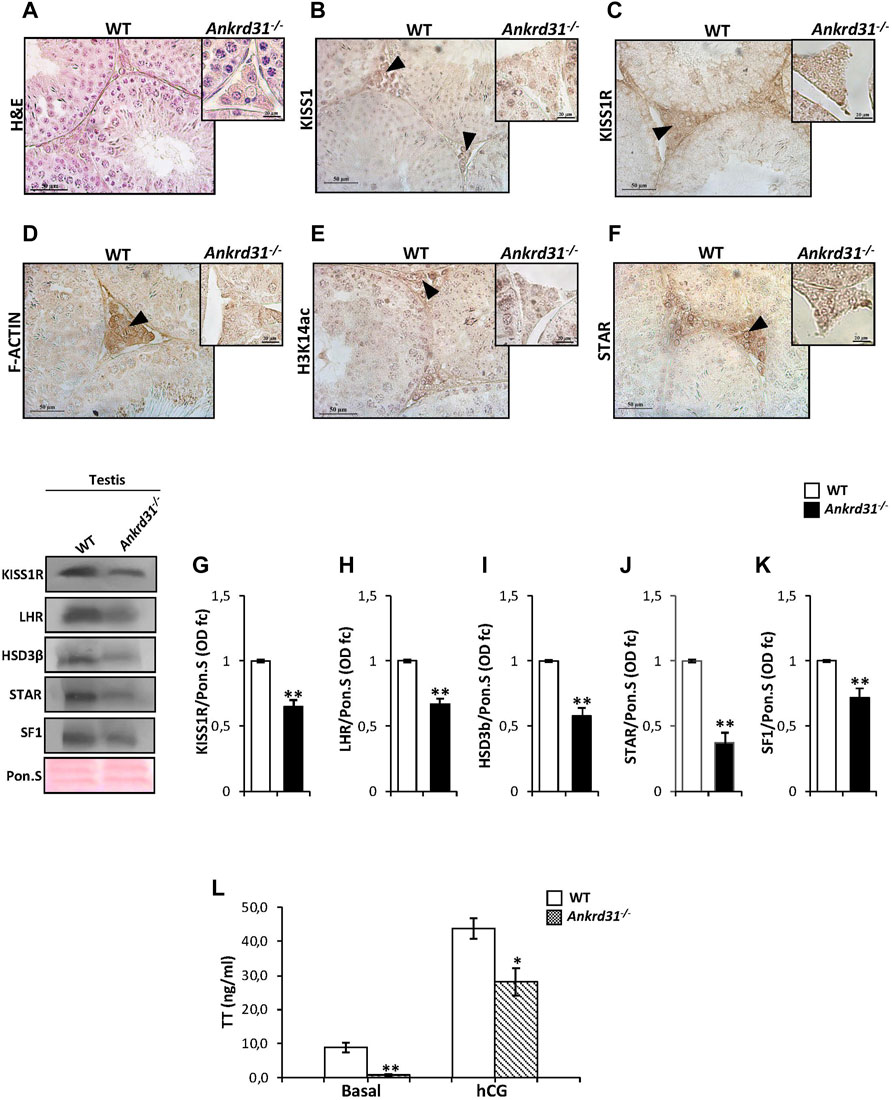
FIGURE 3. (A) H&E staining of Bouin’s fixed WT and Ankrd31−/− testes sections (7 μm). Leydig cells were indicated by black arrowheads. Scale bar: 50 μm. (B–F) Immunocytochemistry of (B) KISS1, (C) KISS1R, (D) F-actin, (E) H3K14ac, (F) STAR in Bouin’s fixed WT, and Ankrd31−/− testes sections (7 μm). The protein localization in Leydig cells was indicated by black arrowheads. Scale bar: 50 μm; scale bar inset: 50 μm. (G–K). Western blot analysis of (G) KISS1R, (H) LHR, (I) HSD3β, (J) STAR, and (K) SF1 in WT and Ankrd31−/− testes. Signals were quantified by the densitometry analysis and normalized to Ponceau Red (Pon.S). Data were expressed in O.D. values as fold change and reported as mean ± SEM; **p < 0.01. (L) Plasma testosterone (TT) levels, at basal condition and following hCG stimulation, in WT and Ankrd31−/− mice by EIA assay; data were reported as mean ± SEM; **p < 0.01.
Kp-10 Enhances KISS1R–ANKRD31 Protein Interaction and Actin Polymerization in Leydig Cells
In order to investigate if a stimulation of KISS1R may be able to regulate Leydig cell gene expression, via the cytoskeletal–nucleoskeletal pathway, IP experiments were carried out in murine primary Leydig cell cultures to show a physical interaction among KISS1R, ANKRD31, and F-actin.
We first immunoprecipitated KISS1R from total proteins extracted from Leydig cells (IP-KISS1R), followed by immunoblotting with KISS1R, ANKRD31, and F-actin antibodies (Figure 4A). The results showed stronger KISS1R, ANKRD31, and F-actin signals in IP-KISS1R as compared with a significantly weaker control signals, suggesting a protein complex formation among KISS1R, ANKRD31, and F-actin in Leydig cells (Figure 4A). Accordingly, the immunoprecipitation of ANKRD31 (IP-ANKRD31) from total proteins extracted from Leydig cells and the immunoblotting with KISS1R, ANKRD31, and F-actin antibodies confirmed the existence of the protein complex (Figure 4B).
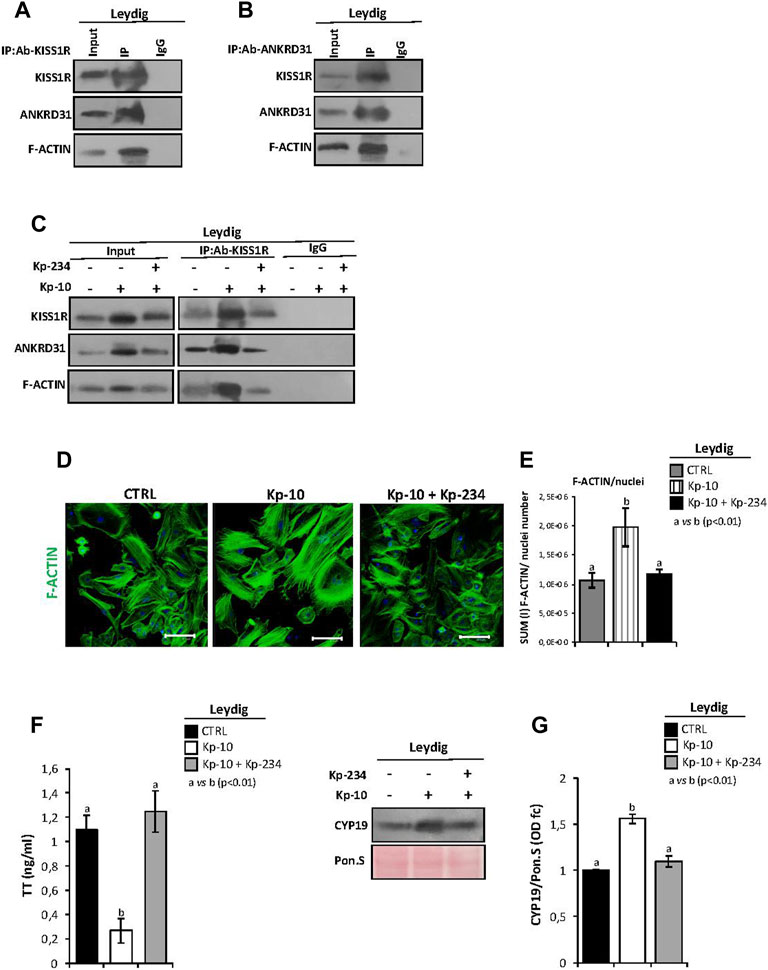
FIGURE 4. (A,B) IP in murine primary Leydig cells. Total proteins collected from murine primary Leydig cell cultures were immunoprecipitated using (A) KISS1R and (B) ANKRD31 antibodies, respectively. Protein interaction among KISS1R, ANKRD31, and F-actin was detected by Western blot analysis. (C) IP in murine primary Leydig cells in vitro treated with Kp-10 alone (0.1 µM) or in combination with the specific antagonist Kp-234 (1 µM) using KISS1R antibody. Protein interaction among KISS1R, ANKRD31, and F-actin was detected by Western blot analysis. (D) Immunofluorescence analysis of F-actin, using phalloidin staining (green) in murine primary Leydig cells in vitro treated with Kp-10 alone (0.1 µM) or in combination with the specific antagonist Kp-234 (1 µM). Nuclei were labeled with TO-PRO3 iodide (blue). Scale bar: 37.5 µm. (E) Quantitative immunofluorescence analysis: F-actin signals were normalized against nuclei number, expressed in SUM(I) values and reported as mean ± SEM; experimental groups with statistically significant differences (p < 0.01) were indicated with different letters; the experimental groups without statistically significant differences were indicated with the same letter. (F) Analysis of TT content (as ng/ml) in culture media of Leydig cells in vitro treated with Kp-10 alone (0.1 µM) or in combination with the specific antagonist Kp-234 (1 µM). All the data were reported as mean ± SEM; **p < 0.01. Experimental groups with statistically significant differences (p < 0.01) were indicated with different letters. (G) Western blot analysis of CYP19 in murine primary Leydig cells in vitro treated with Kp-10 alone (0.1 µM) or in combination with the specific antagonist Kp-234 (1 µM). Signals were quantified by the densitometry analysis and normalized to Ponceau Red (Pon.S). Data are expressed in O.D. values as fold change (O.D. fc), relatively to the CTRL group, and reported as mean ± SEM; experimental groups with statistically significant differences (p < 0.01) were indicated with different letters.
In addition, murine primary Leydig cell cultures were in vitro treated with Kp-10 alone (0.1 µM) or in combination with the specific antagonist Kp-234 (1 µM), with the aim to investigate the cytoskeletal–nucleoskeletal responsiveness to the Kp/KISS1R system activation in Leydig cells. Following the treatment, total Leydig cell protein extracts, derived from the three experimental groups (CTRL, Kp-10, and Kp-10+Kp-234), were used for KISS1R protein IP experiments. Immunoblotting with KISS1R, ANKRD31, and F-actin antibodies showed KISS1R, ANKRD31, and F-actin signals in IP-KISS1R carried out in all experimental groups (Figure 4C). Interestingly, results evidenced a strong increase of protein interaction among KISS1R, ANKRD31, and F-actin following Kp-10 treatment (Figure 4C). This increase was significantly counteracted by Kp-234 treatment and was dependent on the variations of total protein content, as confirmed by the analysis of input samples (total lysates isolated before the IP) (Figure 4C). Data suggested that Kp effectively increases the expression of KISS1R, ANKRD31, and F-actin and, consequentially, the formation of protein complex in Leydig cells.
Phalloidin staining was carried out in Leydig cells in vitro treated with Kp-10 alone or in combination with the specific antagonist Kp-234, in order to investigate the effect of Kp/KISS1R system activation on actin polymerization (Figure 4D). As showed, relatively to the CTRL group, a strong increase in phalloidin staining was observed in Leydig cells treated with Kp-10, whereas the signal intensity in the combined treatment Kp-10 + Kp-234 was similar to the control (Figure 4D). Consistently, phalloidin immunofluorescence quantification, relative to the CTRL group, confirmed a significant increase of the signal in Leydig cells treated with Kp-10 (p < 0.01); this effect was efficiently counteracted by Kp-234 (Figure 4E), suggesting that the Kp/KISS1R system activation in Leydig cells promoted an active F-actin synthesis, in addition to the enhancing of the protein complex among KISS1R, ANKRD31, and F-actin.
In order to show if KISS1R, ANKRD31, and F-actin protein complex may affect Leydig cell biological functions, TT levels were quantified in culture media of Leydig cells in vitro treated with Kp-10 alone (0.1 µM) or in combination with Kp-234 (1 µM) by the LC-MS/MS analysis. Interestingly, TT levels significantly decreased following Kp-10 treatment (p < 0.01), relatively to the CTRL group, and such an effect was significantly counteracted by Kp-234 treatment (Figure 4F). In addition, a significant increase of CYP19 protein content was observed after Kp-10 treatment (p < 0.01), in comparison to the CTRL group, by Western blot (Figure 4G). This effect was counteracted by Kp-234 (Figure 4G), suggesting that Kp/KISS1R system activation in Leydig cells may enhance testosterone conversion to estrogens.
Kp-10 Enhances Leydig Cell Gene Expression via the Cytoskeletal-Nucleoskeletal Pathway
With the purpose to demonstrate that Kps could regulate gene expression in Leydig cells, via the activation of the cytoskeletal–nucleoskeletal pathway, murine primary Leydig cell cultures were in vitro treated with Kp-10 (0.1 µM), cytochalasin D (10 µM), and, finally, with Kp-10 in combination with cytochalasin D. Following pharmacological treatments, co-immunofluorescence analysis was carried out to investigate the responsiveness of F-actin and H3K14ac to the different experimental conditions (Figure 5A).
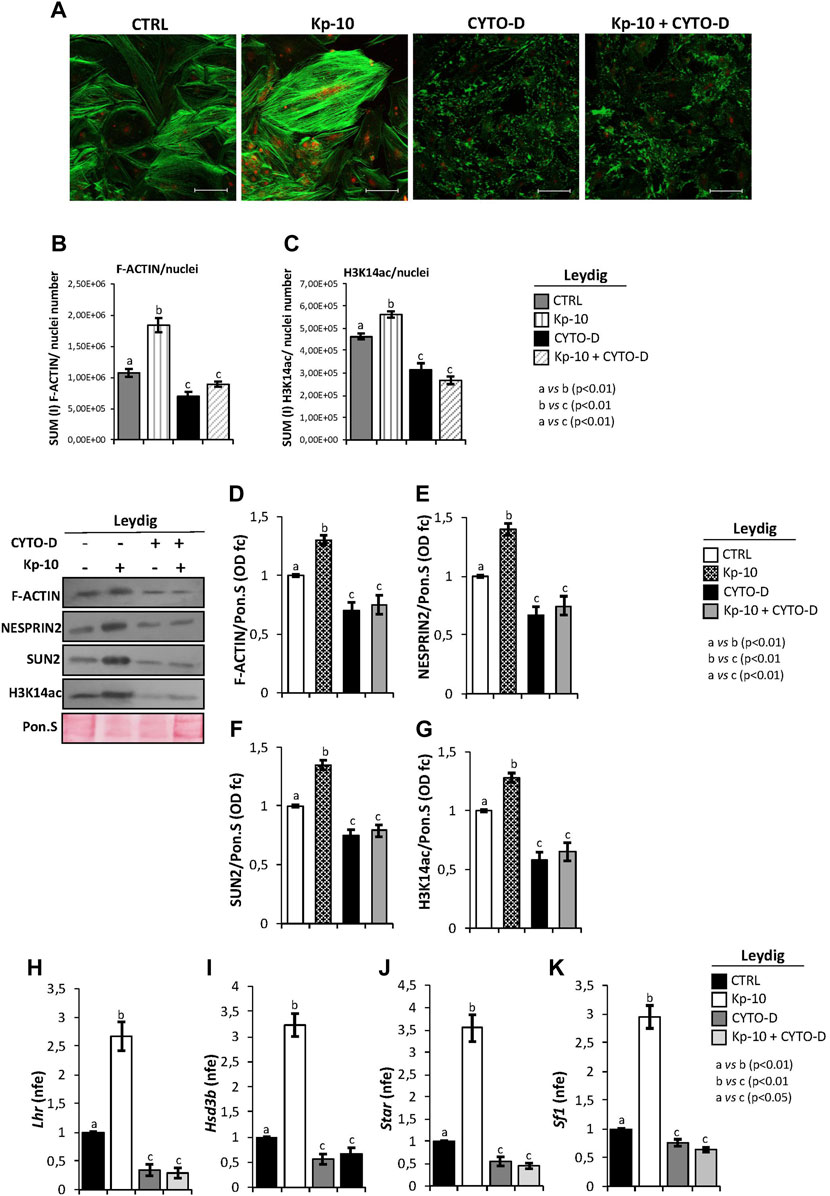
FIGURE 5. (A) Immunofluorescence analysis of F-actin (green) and H3K14ac (red) in murine primary Leydig cells in vitro treated with Kp-10 (0.1 µM) and cytochalasin D (CYTO-D) (10 µM) alone or in combination with Kp-10 (Kp-10+CYTO-D). Nuclei were labeled with TO-PRO3 iodide (blue). Scale bar: 37.5 µm. (B,C) Quantitative immunofluorescence analysis of F-actin and H3K14ac signals. Data were normalized against nuclei number, expressed in SUM(I) values and reported as mean ± SEM; experimental groups with statistically significant differences (p < 0.01) were indicated with different letters; the experimental groups without statistically significant differences were indicated with the same letter. Western blot analysis of (D) F-actin, (E) NESPRIN2, (F) SUN2, and (G) H3K14ac proteins levels in murine primary Leydig cells in vitro treated with Kp-10 (0.1 µM) and cytochalasin D (CYTO-D) (10 µM) alone or in combination with Kp-10 (Kp-10 + CYTO-D). Signals were quantified by the densitometry analysis and normalized to Ponceau Red (Pon.S). Data are expressed in O.D. values as fold change (O.D. fc), relatively to the CTRL group, and reported as mean ± SEM; experimental groups with statistically significant differences (p < 0.01) were indicated with different letters; the experimental groups without statistically significant differences were indicated with the same letter. The differential expression analysis of Leydig cell genes in primary Leydig cells in vitro treated with Kp-10 (0.1 µM) and cytochalasin D (CYTO-D) (10 µM) alone or in combination with Kp-10 (Kp-10+CYTO-D), by qRT-PCR. (H) Lhr, (I) Hsd3b, (J) Star, and (K) Sf1 expression levels were normalized using Rp18S as a housekeeping gene and expressed as normalized fold expression (n.f.e.), relatively to the CTRL group. All data are reported as mean value ± S.E.M; experimental groups with statistically significant differences (p < 0.01) were indicated with different letters; the experimental groups without statistically significant differences were indicated with the same letter.
Accordingly, Kp-10 induced a strong increase of F-actin signal, as previously reported, whereas a complete disaggregation of F-actin was observed following cytochalasin D treatment. In addition, Kp-10 was not able to effectively counteract the depolymerization effect of cytochalasin D on F-actin filaments (Figure 5A). Interestingly, H3K14ac immunofluorescence signal was increased in Leydig cells treated with Kp-10, whereas a weak signal was detected in Leydig cells treated with cytochalasin D alone or in combination with Kp-10 (Figure 5A).
Immunofluorescence quantification data, relatively to the CTRL group, confirmed the significant increase of F-actin staining in Leydig cells treated with Kp-10 (p < 0.01) and its reduction after cytochalasin D alone or in combination with Kp-10 (p < 0.01) (Figure 5B). In addition, H3K14ac immunofluorescence quantification showed the same trend observed for F-actin. In details, a significant increase of H3K14ac amount was observed in Leydig cells treated with Kp-10 (p < 0.01) and, consistently, a strong reduction occurred when Leydig cells were treated with cytochalasin D alone or in combination with Kp-10 (p < 0.01) (Figure 5C), suggesting that Kp/KISS1R system activation modulates the epigenetic landscape of Leydig cells via F-actin synthesis.
To strongly demonstrate this hypothesis, Western blot analysis of F-actin, cytoskeletal–nucleoskeletal mediators, as NESPRIN2 and SUN2, and H3K14ac was carried out in murine primary Leydig cells in vitro treated with Kp-10 and cytochalasin D alone or in combination with Kp. For F-actin, results showed a significant increase following Kp-10 treatment (p < 0.01) and a significant reduction following cytochalasin D treatment alone or in combination with Kp-10 (p < 0.01), in comparison with the CTRL group (Figure 5D). In addition, both NESPRIN2 and SUN2 showed higher protein levels following Kp-10 treatment and lower protein levels following cytochalasin D treatment alone or in combination with Kp-10, than the CTRL group, respectively (p < 0.01) (Figures 5E,F). Finally, relatively to the CTRL group, a significant increase of H3K14ac protein levels was observed in Leydig cells treated with Kp-10 (p < 0.01), and a significant reduction was observed when Leydig cells were treated with cytochalasin D alone or in combination with Kp-10 (p < 0.01) (Figure 5G).
With the aim to highlight that Kps, via the cytoskeletal–nucleoskeletal pathway activation, could enhance H3K14ac and, in turn, Leydig gene expression, the expression analysis of several markers was carried out by qRT-PCR. The results showed a significant increase of Lhr, Hsd3b, Star, and Sf1 expression levels following Kp-10 treatment (p < 0.01), in comparison with the CTRL group (Figures 5H–K), whereas a significant reduction was observed following cytochalasin D treatment, alone or in combination with Kp-10 (p < 0.01) (Figures 5H–K), suggesting that Kps enhance Leydig cell gene expression increasing key modulators of the cytoskeletal–nucleoskeletal pathway and, then, epigenetic markers.
Discussion
The role of Kp/KISS1R system in the testis, especially in Leydig cells, has been deeply suggested (Anjum et al., 2012; Chianese et al., 2013, 2015, 2017; Salehi et al., 2015; Han et al., 2020; Meccariello et al., 2020; Gloria et al., 2021).
Important biological processes including cytoarchitecture organization, chromatin remodeling, and the optimal maintenance of male fertility are under the control of ANKRD proteins (Cunhaa and Mohler, 2009; Argudo et al., 2019; Tang et al., 2020; Manfrevola et al., 2021a). In this scenario, particular attention has been given to ANKRD31, a key regulator of male meiosis and epididymal sperm maturation (Boekhout et al., 2019; Acquaviva et al., 2020; Manfrevola et al., 2021b). Despite a clear effect of Kp/KISS1R signaling on ANKRD expression has been demonstrated in GnRH neurons, a possible crosstalk or a functional interaction among the Kp/KISS1R system and ANKRDs, outside the HPG axis, as well as in Leydig cells remain unknown (Soga et al., 2016). Hence, in this work, we have characterized the testicular responsiveness to Kp/KISS1R system activation, defining its role in Leydig cell physiology, via ANKRD31 and the cytoskeletal–nucleoskeletal pathway.
First, we confirmed the expression of the Kp/KISS1R system in murine Leydig cells. Then, we showed that Kp-10 in vitro treatment exerted a testicular effect on KISS1R, promoting both mRNA and protein expression. Analogously, Kp-10 treatment significantly stimulated the expression of ANKRD31, SPECTRIN, β-actin, and F-actin. A similar effect was observed in GnRH neurons where Kp signaling increased ANKRD26 gene expression (Soga et al., 2016). ANKRDs, acting as a scaffold useful for protein–protein interactions, may regulate complex cellular mechanisms via the cytoskeletal–nucleoskeletal pathway. Indeed, Kp inhibits GnRH neuronal movement through an intracellular mechanotransduction pathway, in turn dependent on ANKRD26 expression (Soga et al., 2016). These data lead us to hypothesize that a similar signaling can also occur in the testis. Actually, Kp-10 increased NESPRIN2 and SUN2 protein levels, the two fundamental actors of the linker of nucleoskeleton and cytoskeleton (LINC) complex (Crisp et al., 2006; Rajgor and Shanahan, 2013; Guilluy et al., 2014; Manfrevola et al., 2021a). However, in Kp-10 treatment, the mRNA and protein levels of NESPRIN2 and SUN2 were not perfectly correlated with each other probably due to other levels of regulation between transcript and protein products (Maier et al., 2009; Vogel and Marcotte, 2012).
Considering that our results confirmed the expression of the Kp/KISS1R system in Leydig cells, we wanted to assess i) the Kp-dependent cytoskeletal–nucleoskeletal pathway switch-on in Leydig cells and ii) the potential effect of Kp-10 on the expression levels of Leydig cell markers. With this in mind, we evaluated an increase in gene expression of Lhr, Hsd3b, Star, and finally Sf1, induced by Kp-10. As well known, mechanical forces could regulate gene expression through the modulation of nuclear chromatin and epigenetic landscapes, via the cytoskeletal–nucleoskeletal pathway (Li et al., 2020; Manfrevola et al., 2021a). Interestingly, testicular Kp-10 treatment was associated with a strong increase of H3K14ac, in correlation with a decrease of several HDACs, including HDAC1, HDAC2, and HDAC4. The epigenetic histone modification H3K14ac is defined as a canonical marker of active gene expression able to modulate chromatin folding in favor of transcriptional activation; furthermore, it participates to several stages of spermatogenesis and male germ cell maturation (Chioccarelli et al., 2020). It is of fundamental interest to specify that a dynamic interplay between histone H3 acetylation and HDAC activity in Leydig cells has been well reported. Indeed, studies on the LC540 Leydig cell in vitro system have demonstrated that HDAC inhibition increased histone H3 acetylation promoting the gene expression of steroidogenic markers Star, Hsd3b, and Hsd17b (Sadasivam et al., 2015). In agreement, H3K14ac acetylation in Star promoter regulates its gene expression in rat testes (Liang et al., 2012). Based on these intriguing observations, we suppose that Kp-10 enhances the expression of Leydig cell markers regulating histone acetylation, through the cytoskeletal–nucleoskeletal pathway activated by the KISS1R–ANKRD31 interaction.
To assess our hypothesis, we carried out several histological and molecular analyses in Ankrd31−/− testis, an experimental model useful to verify the responsiveness of the Kp/KISS1R system, and in turn of the Leydig cell functions, dependent on ANKRD31 loss. Interestingly, in Ankrd31−/− testis, a complete morphological deregulation of Leydig cells, associated with a severe Kp/KISS1R system disruption, was observed. Consistently, the observed reduction of F-actin and H3K14ac, in association with the impaired LHR, HSD3β, STAR, and SF1 expression levels, as well as the affected testosterone secretion, strongly confirmed that the lack of ANKRD31 negatively influenced the Kp/KISS1R system and, in turn, the cytoskeletal–nucleoskeletal actors needful for Leydig cell functions.
In order to deeply demonstrate the fundamental role of the KISS1R–ANKRD31 interaction in the enhancing of Leydig cell functions via the cytoskeletal–nucleoskeletal pathway, we isolated murine primary Leydig cells and showed, for the first time, a protein interaction among KISS1R, ANKRD31, and F-actin proteins. Then, we treated primary Leydig cell culture with Kp-10 alone or in combination with the specific antagonist of KISS1R, Kp-234, in order to evaluate their biological effects on such an interaction. Interestingly, Kp-10 induced the expression of KISS1R and ANKRD31, promoting not only the protein interaction between them but also, surprisingly, a strong F-actin synthesis. Kp-234 reverted the Kp-dependent increase. In support to IP experiments, phalloidin staining clearly showed a strong increase of the signal after Kp-10 stimulation in Leydig cells; this effect was counteracted when Leydig cells were treated with Kp-10 in combination with Kp-234. The responsiveness of Leydig cells to Kp-10 stimulation is in accordance with previous studies that showed i) the expression of Kp system, including KISS1R, in Leydig cells and ii) the increase of Kiss1/KISS1R expression in Leydig cells following Kp-10 treatment (Han et al., 2020). In addition, the reduction of TT levels following Kp-10 stimulation, associated with CYP19 increase, strongly suggested an enhanced testosterone-to-estrogen conversion, thus confirming the primary role of the Kp/KISS1R system in the regulation of Leydig cell biological functions. These data are consistent with our previous study carried out in anuran amphibian, where low doses of Kp-10 affected intratesticular TT levels in favor of its estrogen conversion, due to CYP19 increase. Interestingly, there as here, Kp-10 modulation of steroidogenesis may be suggested to be dose-dependent (Chianese et al., 2017).
At this point, we wondered if the induction of the cytoskeletal–nucleoskeletal pathway, Kp-dependent, could be responsible for an epigenetic modulation able to favor the gene expression of Leydig cell markers. To answer this question, we treated murine primary Leydig cell cultures with Kp-10, cytochalasin D, a potent F-actin depolymerization factor, and with a combination of both. Consistent with our previous data, Kp-10 induced a massive F-actin synthesis in association with increased NESPRIN2, SUN2, and H3K14ac levels. As expected, a significant reduction of F-actin synthesis occurred following cytochalasin D treatment. More interestingly, a reduction of NESPRIN2, SUN2, and H3K14ac levels was induced by cytochalasin D, an effect also displayed in the combined treatment with Kp-10.
The observed increase in the gene expression of Leydig cell markers Lhr, Hsd3b, Star, and Sf1, following Kp-10 treatment, and their respective reduction following the treatment with cytochalasin D alone or in combination with Kp-10, could be strongly dependent on H3K14ac levels, thus to conclude that the ignition of the mechanostraduction pathway, Kp-dependent, regulates the epigenetic landscape of Leydig cells, thus favoring active gene expression.
NESPRIN and SUN proteins are implicated in the organization of nuclear envelope, in the interaction of nucleus with cytoskeletal filaments and, finally, in the chromatin remodeling (Manfrevola et al., 2021a). In addition, the participation of NESPRIN2 and SUN2 in the mechanotransduction pathways useful for the regulation of histone posttranslational modifications has also been reported (Kandert et al., 2007; Camozzi et al., 2012; Alam et al., 2016). In this regard, fibroblast cells harboring Lmna gene mutations showed alteration in the NESPRIN2 localization and in the distribution of both phosphorylated RNA polymerase II and acetylated histones, with affected chromatin topology and transcriptional activity (Kandert et al., 2007). In the same scenario, skin fibroblasts harboring Lmna gene mutations showed a direct correlation between SUN2 mislocalization and the loss of histone methylation (Camozzi et al., 2012). A similar association among the NESPRIN2–SUN2 complex and histone H3 acetylation is not to be excluded in Leydig cells, as our data suggest.
Taken together, our results demonstrate how Kp/KISS1R system activation in Leydig cells is able to induce KISS1R/ANKRD31 interaction, downstream triggering the activation of the cytoskeletal–nucleoskeletal pathway. Meanwhile, we have showed the responsiveness of the epigenetic signature to the cytoskeletal–nucleoskeletal pathway, resulting in the switch-on of Leydig cell gene expression.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics Statement
The animal study was reviewed and approved by experiments involving C57BL/6 animals were approved by the Italian Ministry of Education and the Italian Ministry of Health, with authorization n°405/2021-PR. Procedure involving animal care were carried out in accordance with the National Research Council’s Publication Guide for Care and Use of Laboratory Animals (National of Institutes of Health Guide). Experiments involving WT and Ankrd31−/− animals were performed following French and European guidelines on the protection of animals used for scientific purposes and approved by an ethical committee for animal experimentation (CEEA Val de Loire Projet 12884).
Author Contributions
Conceptualization: GR, RC, FM, and RM (for the setup of Kp system and for the role of Kp system on the ANKRD expression); Methodology: GR, VGM, FM, RM, MM, and ND; Formal analysis and investigation: FG, GR, FM, and ND; Writing-Original Draft Preparation: GR, FM, and ND; Figure Preparation: GR and FM; Writing-Review & Editing: RC and RP; Visualization: FG, AC, TC, SF, and GC; Supervision: RP and RC; Funding Acquisition: RC, GC, RM, and RP.
Funding
This work was supported by the Italian Ministry of University and Research (Grant PRIN to RP 2017); Università degli Studi della Campania (Grant VALERE, Vanvitelli per la Ricerca 2019 to GC); Università degli Studi della Campania Luigi Vanvitelli (Grant Dip. Medicina Sperimentale-2020 to RC); and the Ministry of University and Research (Grant PRIN to RM 2017).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We are grateful to A. L. Lainé and C. Laclie (Phenotyping-Endocrinology Laboratory, Reproductive Physiology and Behavior Unit, INRAE CNRS, Nouzilly, France) for the hormonal assays.
References
Acquaviva, L., Boekhout, M., Karasu, M. E., Brick, K., Pratto, F., Li, T., et al. (2020). Ensuring Meiotic DNA Break Formation in the Mouse Pseudoautosomal Region. Nature 582 (7812), 426–431. doi:10.1038/s41586-020-2327-4
Alam, S. G., Zhang, Q., Prasad, N., Li, Y., Chamala, S., Kuchibhotla, R., et al. (2016). The Mammalian LINC Complex Regulates Genome Transcriptional Responses to Substrate Rigidity. Sci. Rep. 6, 38063. doi:10.1038/srep38063
Anjum, S., Krishna, A., Sridaran, R., and Tsutsui, K. (2012). Localization of Gonadotropin-Releasing Hormone (GnRH), Gonadotropin-Inhibitory Hormone (GnIH), Kisspeptin and GnRH Receptor and Their Possible Roles in Testicular Activities from Birth to Senescence in Mice. J. Exp. Zool. A Ecol. Genet. Physiol. 317 (10), 630–644. doi:10.1002/jez.1765
Argudo, D., Capponi, S., Bethel, N. P., and Grabe, M. (2019). A Multiscale Model of Mechanotransduction by the Ankyrin Chains of the NOMPC Channel. J. Gen. Physiol. 151 (3), 316–327. doi:10.1085/jgp.201812266
Batrukova, M. A., Betin, V. L., Rubtsov, A. M., and Lopina, O. D. (2000). Ankyrin: Structure, Properties, and Functions. Biochem. (Mosc) 65 (4), 395–408. doi:10.1002/jez.1765
Boekhout, M., Karasu, M. E., Wang, J., Acquaviva, L., Pratto, F., Brick, K., et al. (2019). REC114 Partner ANKRD31 Controls Number, Timing, and Location of Meiotic DNA Breaks. Mol. Cell 74 (5), 1053–1068. doi:10.1016/j.molcel.2019.03.023
Calder, M., Chan, Y.-M., Raj, R., Pampillo, M., Elbert, A., Noonan, M., et al. (2014). Implantation Failure in Female Kiss1−/− Mice Is Independent of Their Hypogonadic State and Can Be Partially Rescued by Leukemia Inhibitory Factor. Endocrinology 155 (8), 3065–3078. doi:10.1210/en.2013-1916
Camozzi, D., D’Apice, M. R., Schena, E., Cenni, V., Columbaro, M., Capanni, C., et al. (2012). Altered Chromatin Organization and SUN2 Localization in Mandibuloacral Dysplasia Are Rescued by Drug Treatment. Histochem Cell Biol. 138 (4), 643–651. doi:10.1007/s00418-012-0977-5
Chianese, R., Ciaramella, V., Fasano, S., Pierantoni, R., and Meccariello, R. (2013). Kisspeptin Receptor, GPR54, as a Candidate for the Regulation of Testicular Activity in the Frog Rana esculenta. Biol. Reprod. 88 (3), 73. doi:10.1095/biolreprod.112.103515
Chianese, R., Ciaramella, V., Fasano, S., Pierantoni, R., and Meccariello, R. (2014). Hypothalamus-pituitary axis: an Obligatory Target for Endocannabinoids to Inhibit Steroidogenesis in Frog Testis. General Comp. Endocrinol. 205, 88–93. doi:10.1016/j.ygcen.2014.02.010
Chianese, R., Ciaramella, V., Fasano, S., Pierantoni, R., and Meccariello, R. (2015). Kisspeptin Drives Germ Cell Progression in the Anuran Amphibian Pelophylax Esculentus: a Study Carried Out in Ex Vivo Testes. General Comp. Endocrinol. 211, 81–91. doi:10.1016/j.ygcen.2014.11.008
Chianese, R., Ciaramella, V., Fasano, S., Pierantoni, R., and Meccariello, R. (2017). Kisspeptin Regulates Steroidogenesis and Spermiation in Anuran Amphibian. Reproduction 154 (4), 403–414. doi:10.1530/rep-17-0030
Chianese, R., Cobellis, G., Chioccarelli, T., Ciaramella, V., Migliaccio, M., Fasano, S., et al. (2016). Kisspeptins, Estrogens and Male Fertility. Cmc 23 (36), 4070–4091. doi:10.2174/0929867323666160902155434
Chioccarelli, T., Pierantoni, R., Manfrevola, F., Porreca, V., Fasano, S., Chianese, R., et al. (2020). Histone Post-Translational Modifications and CircRNAs in Mouse and Human Spermatozoa: Potential Epigenetic Marks to Assess Human Sperm Quality. Jcm 9 (3), 640. doi:10.3390/jcm9030640
Colledge, W. H. (2009). Kisspeptins and GnRH Neuronal Signalling. Trends Endocrinol. Metabolism 20 (3), 115–121. doi:10.1016/j.tem.2008.10.005
Crisp, M., Liu, Q., Roux, K., Rattner, J. B., Shanahan, C., Burke, B., et al. (2006). Coupling of the Nucleus and Cytoplasm: Role of the LINC Complex. J. Cell Biol. 172, 41–53. doi:10.1083/jcb.200509124
Cunha, S. R., and Mohler, P. J. (2009). Ankyrin Protein Networks in Membrane Formation and Stabilization. J. Cell Mol. Med. 13 (11-12), 4364–4376. doi:10.1111/j.1582-4934.2009.00943.x
de Roux, N., Genin, E., Carel, J.-C., Matsuda, F., Chaussain, J.-L., and Milgrom, E. (2003). Hypogonadotropic Hypogonadism Due to Loss of Function of the KiSS1-Derived Peptide Receptor GPR54. Proc. Natl. Acad. Sci. U.S.A. 100 (19), 10972–10976. doi:10.1073/pnas.1834399100
Dorfman, M. D., Garcia-Rudaz, C., Alderman, Z., Kerr, B., Lomniczi, A., Dissen, G. A., et al. (2014). Loss of NTRK2/KISS1R Signaling in Oocytes Causes Premature Ovarian Failure. Endocrinology 155 (8), 3098–3111. doi:10.1210/en.2014-1111
Dou, X., Menkari, C., Mitsuyama, R., Foroud, T., Wetherill, L., Hammond, P., et al. (2018). L1 Coupling to Ankyrin and the Spectrin‐actin Cytoskeleton Modulates Ethanol Inhibition of L1 Adhesion and Ethanol teratogenesisL1 Coupling to Ankyrin and the Spectrin-Actin Cytoskeleton Modulates Ethanol Inhibition of L1 Adhesion and Ethanol Teratogenesis. FASEB J. 32 (3), 1364–1374. doi:10.1096/fj.201700970
Errico, S., Chioccarelli, T., Moggio, M., Diano, N., and Cobellis, G. (2019). A New LC-MS/MS Method for Simultaneous and Quantitative Detection of Bisphenol-A and Steroids in Target Tissues: A Power Tool to Characterize the Interference of Bisphenol-A Exposure on Steroid Levels. Molecules 25 (1), 48. doi:10.3390/molecules25010048
Gaytán, M., Castellano, J. M., Roa, J., Sánchez-Criado, J. E., Tena-Sempere, M., and Gaytán, F. (2007). Expression of KiSS-1 in Rat Oviduct: Possible Involvement in Prevention of Ectopic Implantation? Cell Tissue Res. 329 (3), 571–579. doi:10.1007/s00441-007-0426-2
Gloria, A., Contri, A., Mele, E., Fasano, S., Pierantoni, R., and Meccariello, R. (2021). Kisspeptin Receptor on the Sperm Surface Reflects Epididymal Maturation in the Dog. Ijms 22 (18), 10120. doi:10.3390/ijms221810120
Guilluy, C., Osborne, L. D., Van Landeghem, L., Sharek, L., Superfine, R., Garcia-Mata, R., et al. (2014). Isolated Nuclei Adapt to Force and Reveal a Mechanotransduction Pathway in the Nucleus. Nat. Cell Biol. 16, 376–381. doi:10.1038/ncb2927
Han, S.-K., Gottsch, M. L., Lee, K. J., Popa, S. M., Smith, J. T., Jakawich, S. K., et al. (2005). Activation of Gonadotropin-Releasing Hormone Neurons by Kisspeptin as a Neuroendocrine Switch for the Onset of Puberty. J. Neurosci. 25 (49), 11349–11356. doi:10.1523/jneurosci.3328-05.2005
Han, Y., Peng, X., Si, W., Liu, G., Han, Y., Jiang, X., et al. (2020). Local Expressions and Function of Kiss1/GPR54 in Goats' Testes. Gene 738, 144488. doi:10.1016/j.gene.2020.144488
Hryniewicz-Jankowska, A., Czogalla, A., Bok, E., and Sikorsk, A. F. (2002). Ankyrins, Multifunctional Proteins Involved in Many Cellular Pathways. Folia Histochem Cytobiol. 40 (3), 239–249.
Kandert, S., Lüke, Y., Kleinhenz, T., Neumann, S., Lu, W., Jaeger, V. M., et al. (2007). Nesprin-2 Giant Safeguards Nuclear Envelope Architecture in LMNA S143F Progeria Cells. Hum. Mol. Genet. 16 (23), 2944–2959. doi:10.1093/hmg/ddm255
León, S., Barroso, A., Vázquez, M. J., García-Galiano, D., Manfredi-Lozano, M., Ruiz-Pino, F., et al. (201619206). Direct Actions of Kisspeptins on GnRH Neurons Permit Attainment of Fertility but Are Insufficient to Fully Preserve Gonadotropic Axis Activity. Sci. Rep. 6, 19206. doi:10.1038/srep19206
Li, S., Yang, D., Gao, L., Wang, Y., and Peng, Q. (2020). Epigenetic Regulation and Mechanobiology. Biophys. Rep. 6, 33–48. doi:10.1007/s41048-020-00106-x
Liang, J., Zhu, H., Li, C., Ding, Y., Zhou, Z., and Wu, Q. (2012). Neonatal Exposure to Benzo[a]pyrene Decreases the Levels of Serum Testosterone and Histone H3K14 Acetylation of the StAR Promoter in the Testes of SD Rats. Toxicology 302 (2-3), 285–291. doi:10.1016/j.tox.2012.08.010
Maier, T., Güell, M., and Serrano, L. (2009). Correlation of mRNA and Protein in Complex Biological Samples. FEBS Lett. 583 (24), 3966–3973. doi:10.1016/j.febslet.2009.10.036
Manfrevola, F., Guillou, F., Fasano, S., Pierantoni, R., and Chianese, R. (2021a). LINCking the Nuclear Envelope to Sperm Architecture. Genes 12 (5), 658. doi:10.3390/genes12050658
Manfrevola, F., Martinez, G., Coutton, C., Rocco, D., Reynaud, K., Le Vern, Y., et al. (2021b). Ankrd31 in Sperm and Epididymal Integrity. Front. Cell Dev. Biol. 9, 741975. doi:10.3389/fcell.2021.741975
Meccariello, R., Fasano, S., and Pierantoni, R. (2020). Kisspeptins, New Local Modulators of Male Reproduction: A Comparative Overview. General Comp. Endocrinol. 299, 113618. doi:10.1016/j.ygcen.2020.113618
Navarro, V. M., and Tena-Sempere, M. (2011). Kisspeptins and the Neuroendocrine Control of Reproduction. Front. Biosci. S3, 267–275. doi:10.2741/s150
Oakley, A. E., Clifton, D. K., and Steiner, R. A. (2009). Kisspeptin Signaling in the Brain. Endocr. Rev. 30 (6), 713–743. doi:10.1210/er.2009-0005
Pinilla, L., Aguilar, E., Dieguez, C., Millar, R. P., and Tena-Sempere, M. (2012). Kisspeptins and Reproduction: Physiological Roles and Regulatory Mechanisms. Physiol. Rev. 92 (3), 1235–1316. doi:10.1152/physrev.00037.2010
Rajgor, D., and Shanahan, C. M. (2013). Nesprins: from the Nuclear Envelope and beyond. Expert Rev. Mol. Med. 15, e5. doi:10.1017/erm.2013.6
Roseweir, A. K., Kauffman, A. S., Smith, J. T., Guerriero, K. A., Morgan, K., Pielecka-Fortuna, J., et al. (2009). Discovery of Potent Kisspeptin Antagonists Delineate Physiological Mechanisms of Gonadotropin Regulation. J. Neurosci. 29 (12), 3920–3929. doi:10.1523/jneurosci.5740-08.2009
Sadasivam, M., Ramatchandirin, B., Balakrishnan, S., and Prahalathan, C. (2015). HDAC7 Modulates TNF-α-Mediated Suppression of Leydig Cell Steroidogenesis. Mol. Cell Biochem. 406 (1-2), 83–90. doi:10.1007/s11010-015-2426-y
Saito, M., Watanabe-Nakayama, T., Machida, S., Osada, T., Afrin, R., and Ikai, A. (2015). Spectrin-ankyrin Interaction Mechanics: A Key Force Balance Factor in the Red Blood Cell Membrane Skeleton. Biophys. Chem. 200-201, 1–8. doi:10.1016/j.bpc.2015.03.007
Salehi, S., Adeshina, I., Chen, H., Zirkin, B. R., Hussain, M. A., Wondisford, F., et al. (2015). Developmental and Endocrine Regulation of Kisspeptin Expression in Mouse Leydig Cells. Endocrinology 156 (4), 1514–1522. doi:10.1210/en.2014-1606
Seminara, S. B., Messager, S., Chatzidaki, E. E., Thresher, R. R., Acierno, J. S., Shagoury, J. K., et al. (2003). TheGPR54Gene as a Regulator of Puberty. N. Engl. J. Med. 349 (17), 1614–1627. doi:10.1056/nejmoa035322
Shahab, M., Mastronardi, C., Seminara, S. B., Crowley, W. F., Ojeda, S. R., and Plant, T. M. (2005). Increased Hypothalamic GPR54 Signaling: A Potential Mechanism for Initiation of Puberty in Primates. Proc. Natl. Acad. Sci. U.S.A. 102 (6), 2129–2134. doi:10.1073/pnas.0409822102
Sharma, A., Thaventhiran, T., Minhas, S., Dhillo, W. S., and Jayasena, C. N. (2020). Kisspeptin and Testicular Function-Is it Necessary? Ijms 21 (8), 2958. doi:10.3390/ijms21082958
Soga, T., Lim, W. L., Khoo, A. S.-B., and Parhar, I. S. (2016). Kisspeptin Activates Ankrd 26 Gene Expression in Migrating Embryonic GnRH Neurons. Front. Endocrinol. 7, 15. doi:10.3389/fendo.2016.00015
Sun, J., Zhong, L., Zhu, Y., and Liu, G. (2011). Research on the Isolation of Mouse Leydig Cells Using Differential Digestion with a Low Concentration of Collagenase. J. Reproduction Dev. 57 (3), 433–436. doi:10.1262/jrd.10-123n
Tang, Y.-Q., Lee, S. A., Rahman, M., Vanapalli, S. A., Lu, H., and Schafer, W. R. (2020). Ankyrin Is an Intracellular Tether for TMC Mechanotransduction Channels. Neuron 107 (1), 112–125. doi:10.1016/j.neuron.2020.03.026
Thompson, E. L., Patterson, M., Murphy, K. G., Smith, K. L., Dhillo, W. S., Todd, J. F., et al. (2004). Central and Peripheral Administration of Kisspeptin-10 Stimulates the Hypothalamic-Pituitary-Gonadal axis. J. Neuroendocrinol. 16 (10), 850–858. doi:10.1111/j.1365-2826.2004.01240.x
Uhler, C., and Shivashankar, G. V. (2017). Regulation of Genome Organization and Gene Expression by Nuclear Mechanotransduction. Nat. Rev. Mol. Cell Biol. 18, 717–727. doi:10.1038/nrm.2017.101
Vogel, C., and Marcotte, E. M. (2012). Insights into the Regulation of Protein Abundance from Proteomic and Transcriptomic Analyses. Nat. Rev. Genet. 13 (4), 227–232. doi:10.1038/nrg3185
Keywords: kisspeptin, KISS1R, ankyrins, male fertility, Leydig cells, cytoskeletal–nucleoskeletal pathway, actin
Citation: Ricci G, Guillou F, Catizone A, Mele VG, Moggio M, Chioccarelli T, Diano N, Meccariello R, Pierantoni R, Fasano S, Cobellis G, Chianese R and Manfrevola F (2022) KISS1R and ANKRD31 Cooperate to Enhance Leydig Cell Gene Expression via the Cytoskeletal-Nucleoskeletal Pathway. Front. Cell Dev. Biol. 10:877270. doi: 10.3389/fcell.2022.877270
Received: 16 February 2022; Accepted: 05 May 2022;
Published: 23 June 2022.
Edited by:
Honoo Satake, Suntory Foundation for Life Sciences, JapanReviewed by:
Hitoshi Ozawa, Nippon Medical School, JapanSatoshi Ogawa, Monash University Malaysia, Malaysia
Copyright © 2022 Ricci, Guillou, Catizone, Mele, Moggio, Chioccarelli, Diano, Meccariello, Pierantoni, Fasano, Cobellis, Chianese and Manfrevola. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rosanna Chianese, cm9zYW5uYS5jaGlhbmVzZUB1bmljYW1wYW5pYS5pdA==
 Giulia Ricci
Giulia Ricci Florian Guillou
Florian Guillou Angela Catizone
Angela Catizone Vincenza Grazia Mele1
Vincenza Grazia Mele1 Teresa Chioccarelli
Teresa Chioccarelli Rosaria Meccariello
Rosaria Meccariello Riccardo Pierantoni
Riccardo Pierantoni Silvia Fasano
Silvia Fasano Gilda Cobellis
Gilda Cobellis Rosanna Chianese
Rosanna Chianese Francesco Manfrevola
Francesco Manfrevola