- 1Department of Anatomy, Physiology, and Pharmacology, University of Saskatchewan, Saskatoon, SK, Canada
- 2Department of Computer Science, University of Calgary, Calgary, AB, Canada
To explain how cartilage appeared in different parts of the vertebrate body at discrete times during evolution, we hypothesize that different embryonic populations co-opted expression of a core gene regulatory network (GRN) driving chondrocyte differentiation. To test this hypothesis, laser-capture microdissection coupled with RNA-seq was used to reveal chondrocyte transcriptomes in the developing chick humerus and ceratobranchial, which are mesoderm- and neural crest-derived, respectively. During endochondral ossification, two general types of chondrocytes differentiate. Immature chondrocytes (IMM) represent the early stages of cartilage differentiation, while mature chondrocytes (MAT) undergo additional stages of differentiation, including hypertrophy and stimulating matrix mineralization and degradation. Venn diagram analyses generally revealed a high degree of conservation between chondrocyte transcriptomes of the limb and head, including SOX9, COL2A1, and ACAN expression. Typical maturation genes, such as COL10A1, IBSP, and SPP1, were upregulated in MAT compared to IMM in both limb and head chondrocytes. Gene co-expression network (GCN) analyses of limb and head chondrocyte transcriptomes estimated the core GRN governing cartilage differentiation. Two discrete portions of the GCN contained genes that were differentially expressed in limb or head chondrocytes, but these genes were enriched for biological processes related to limb/forelimb morphogenesis or neural crest-dependent processes, respectively, perhaps simply reflecting the embryonic origin of the cells. A core GRN driving cartilage differentiation in limb and head was revealed that included typical chondrocyte differentiation and maturation markers, as well as putative novel “chondrocyte” genes. Conservation of a core transcriptional program during chondrocyte differentiation in both the limb and head suggest that the same core GRN was co-opted when cartilage appeared in different regions of the skeleton during vertebrate evolution.
Introduction
The formation of cartilage is a trait with an interesting evolutionary history. While initially considered a vertebrate novelty, the presence of cartilage in vertebrate outgroups, such as hemichordates and cephalochordates, indicates that cartilage was very likely present in the ancestor to vertebrates (Rychel et al., 2006; Rychel and Swalla 2007). During vertebrate evolution, however, cartilage appeared in different parts of the body at different times. For example, the cranial skeleton appeared before the appendicular skeleton (Janvier 1996). A tantalizing hypothesis to explain this phenomenon is that a core gene regulatory network (GRN) driving chondrocyte differentiation in the head was later co-opted in the paired appendages. If true, then both limb and head chondrocytes might express a core transcriptional program underlying chondrocyte differentiation. As proof of principle, shared chondrocyte gene expression in amphioxus and vertebrates suggested that neural crest-derived cartilage evolved by co-opting a chondrocyte GRN from mesoderm or endoderm (Meulemans and Bronner-Fraser 2007; Hall and Gillis 2013; Jandzik et al., 2015). An understanding of chondrocyte differentiation, including the chondrocyte GRN, and embryonic origins of limb and head mesenchyme is required to test this hypothesis.
Endochondral ossification generally involves the differentiation of two types of chondrocyte: immature (IMM) and mature (MAT; Eames et al., 2003; Eames et al., 2004; Tamamura et al., 2005; Gentili and Cancedda 2009). Examples of IMM are proliferative and resting chondrocytes that deposit Col2 fibers and proteoglycans in the extracellular matrix, whereas examples of MAT include pre-hypertrophic and hypertrophic chondrocytes that modify immature cartilage extracellular matrix, such as by depositing Col10 fibers and mineralizing the matrix (Leboy et al., 1988; Farquharson et al., 1994; Takeda et al., 2001).
The GRN driving chondrocyte differentiation has been refined over the years (Cole and Hall 2009; Kerkhofs et al., 2012; Oh et al., 2014; Liu and Lefebvre 2015; Ohba et al., 2015; He et al., 2016; Tan et al., 2018; Hojo and Ohba 2019). Initially, a chondrocyte GRN inferred from published mammalian literature reflected the regulatory importance of SOX9 and RUNX2 on their downstream targets (Cole 2011; Kerkhofs et al., 2012). During IMM differentiation, for example, SOX9 binds to its cofactors SOX5 and SOX6 to activate expression of important cartilage differentiation markers, such as Col2a1, Col9a1, and Acan (Lefebvre et al., 2001; Akiyama et al., 2002; Liu and Lefebvre 2015). During MAT differentiation, SOX9 levels decrease, and RUNX2 levels increase to activate the expression of such genes as Col10a1, Mef2c, Mmp13, Spp1, and Ibsp (Ducy et al., 1997; Komori et al., 1997; Inada et al., 1999; Lee et al., 2000; Arnold et al., 2007; Li et al., 2011; Lu et al., 2014; Niu et al., 2017; Komori 2018). Since cartilage maturation-like changes, such as hypertrophy and matrix degradation, play a role at different stages during osteoarthritis (OA), several of these MAT genes have also been linked to this skeletal pathology (Lamas et al., 2010; van der Kraan and van den Berg 2012; Wang et al., 2013; Lv et al., 2015; Chen et al., 2020). Later microarray analyses coupled to ChIP-seq from developing cartilage refined the GRN further. For example, analyses of newborn mouse tibia revealed interactions of SOX9 with GLI1, GLI3, and FOXA2 (Tan et al., 2018). Many studies provide valuable insight into the chondrocyte GRN, but they mostly focus on limb cartilage, so whether expression of this GRN is conserved throughout the body remains unclear.
Cartilages in the limb and head of vertebrates can have two distinct embryonic origins: mesoderm and neural crest. The appendicular skeleton within fins or limbs derives from lateral plate mesoderm, whereas cranial neural crest-derived ectomesenchyme gives rise to a large portion of the cranial skeleton, including the jaws, anterior calvarium, palate, and hyoid bone (Couly et al., 1993; Knight and Schilling 2006; Fonseca et al., 2017). IMM and MAT are present in both the limb and head, so mesenchyme derived from both mesoderm and neural crest can produce both types of chondrocyte.
Previous studies of a targetted subset of molecular markers suggested that the same GRN driving chondrocyte differentiation is expressed regardless of its embryonic origin or location in the body. For example, IMM from both limb and head express Sox9, Sox5, Sox6, Acan, and Col2a1 (Lefebvre and de Crombrugghe 1998; Smits et al., 2001; Akiyama et al., 2002; Eames et al., 2004; Smits et al., 2004; Dale and Topczewski 2011; Lefebvre and Dvir-Ginzberg 2017; Xiong et al., 2018), while MAT from both limb and head express Runx2, Col10a1, and Ihh (Eames et al., 2004; Yoshida et al., 2004; Young et al., 2006). Moreover, if the function of any of these genes is perturbed, then chondrocytes are affected throughout the body, suggesting that the same core GRN driving chondrocyte differentiation might be expressed in both limb and head (Komori et al., 1997; Bi et al., 1999; Smits et al., 2001; Smits et al., 2004; Yoshida et al., 2004). Unbiased studies comparing gene expression globally between limb and head are needed to verify if the GRN underlying chondrocyte differentiation is the same throughout the body.
Comparative transcriptomics has revealed differences between mesenchymal precursors of limb mesoderm and cranial neural crest, but a differentiated cell type can express a core set of genes in both limb and head. Gene expression profiles from mesenchyme derived from neural crest and mesoderm that were isolated from the first pharyngeal arch using laser-capture microdissection (LCM) revealed 140 differentially expressed genes (Bhattacherjee et al., 2007). Very few studies have used transcriptomics to reveal how gene expression in skeletal cells from head and limb might vary. In perhaps the most relevant study, osteoblasts were obtained from mouse calvaria and hindlimb cortical bones, and scRNA-seq revealed that the transcriptomes of head and limb osteoblasts were highly similar (Ayturk et al., 2020). Typical osteoblast differentiation markers, including Col1a1, Col1a2, Bglap, Ifitm5, and Dmp1, were conserved in head and limb, suggesting that regardless of embryonic origin and location, osteoblasts employ a core GRN during differentiation (Ayturk et al., 2020).
To test the hypothesis that a core GRN is expressed during chondrocyte differentiation in the limb and head, LCM coupled with RNA-seq was used to generate transcriptomes of IMM and MAT from a limb cartilage (i.e., humerus) and a head cartilage (i.e., ceratobranchial) in the chick embryo (Figure 1). Analyses of the resulting data highlight a core transcriptional program (e.g., SOX9, SOX5, SOX6, COL2A1, ACAN, and COL10A1) that drives chondrocyte differentiation throughout the body. GRN estimates from gene co-expression network (GCN) analyses showed that enriched IMM and MAT genes were negatively correlated in both the limb and head. We discuss that many genes commonly described as cartilage genes that were enriched in limb or head chondrocytes (e.g., ID1, PAX7, ZIC1, HOXA, HOXD, and SHOX2) might actually only serve that purpose in specific regions of the body, and should not be considered part of the core chondrocyte GRN. Together these data support the hypothesis that cartilage appeared in different regions of the vertebrate body during evolution by co-opting the same core GRN driving chondrocyte differentiation.
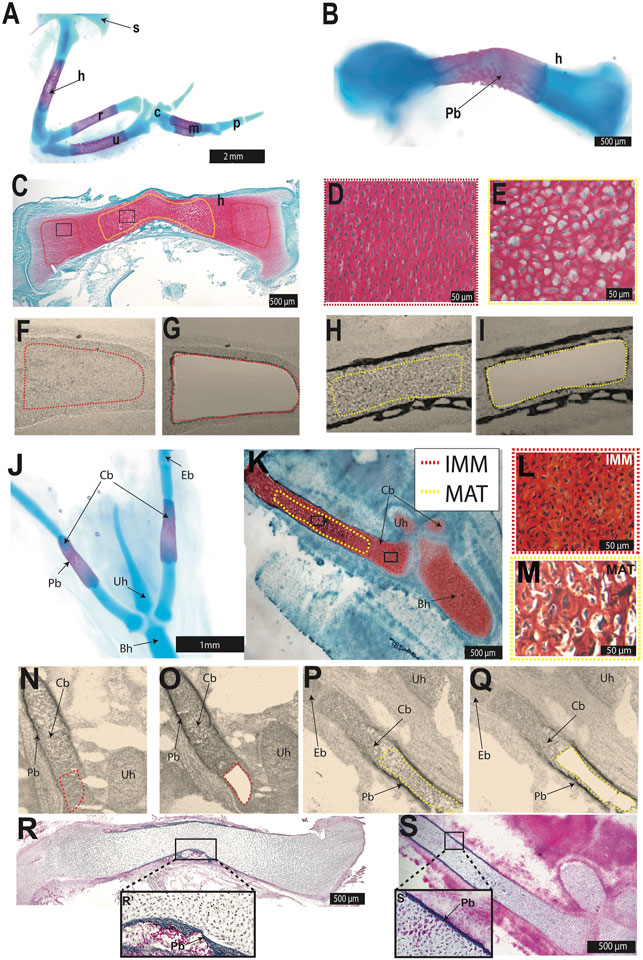
FIGURE 1. Laser capture microdissection was used to isolate chondrocytes from the chick HH36 humerus and ceratobranchial. (A,B,J) Whole-mount Alcian blue and Alizarin red staining identified cartilage and perichondral bone in chondral bones of the chick forelimb (A,B) or hyoid (J). (C,K) Safranin O-stained section of HH36 humerus or ceratobranchial highlighted immature (IMM, red dotted outline) and mature cartilage (MAT, yellow dotted outline). (D,E,L,M) High-magnification images of IMM (D,L) and MAT (E,M) from black boxes in (C) or (K) (F–I,N–Q) Unstained sections of HH36 chick humerus or ceratobranchial before (F,N) and after (G,O) laser capture of IMM, and before (H,P) and after (I,Q) laser capture of MAT. (R,S) Trichrome-stained section of HH36 humerus or ceratobranchial showed Aniline blue staining of bone matrix in perichondral bone. Abbreviations: Bh = basihyal; c = carpal; Cb = ceratobranchial; Eb = epibranchial; h = humerus; IMM = immature chondrocytes; m = metacarpals; MAT = mature chondrocytes; p = phalanges; Pb = perichondral bone; r = radius; s = scapula; u = ulna; Uh = urohyal.
Results
Identification and Isolation of Limb and Head Immature and Mature Chondrocyte Transcriptomes
IMM and MAT were obtained from two chondral bones, the humerus and ceratobranchial (Figure 1), which are derived from lateral plate mesoderm and cranial neural crest, respectively (Noden 1982; Couly et al., 1993; Le Douarin et al., 1999; Burke and Nowicki 2001; Tani et al., 2020). In chick, the epiphyseal growth plate of such long bones as the humerus and ceratobranchial should contain IMM and MAT at HH36 (E10; Milz et al., 2002; Conen et al., 2009; Lui et al., 2014). Alcian blue identified cartilage, whereas Alizarin Red identified perichondral bone in whole-mount stains of both skeletal elements at HH36 (Figures 1A,B,J). Perichondral bone is often associated with underlying MAT, and Safranin O staining of histological sections demonstrated that MAT, underneath perichondral bone, have undergone hypertrophy in the mid-diaphyseal region (i.e., shaft of a long bone) of the humerus and ceratobranchial (Figures 1C–E,K–M). LCM was used to isolate IMM and MAT from the humerus and ceratobranchial (Figures 1F–I,N–Q). At HH36, vascular invasion had not yet occurred, and bone matrix was only detected in perichondral bone in both the humerus and ceratobranchial, as shown by Trichrome staining (Figures 1R,S), suggesting that no transdifferentiation of chondrocytes to osteoblasts had yet occurred (Conen et al., 2009; Zhou et al., 2014; Qin et al., 2020). RNA-seq was then carried out on RNA isolated from IMM and MAT.
Comparative Transcriptomics Revealed Expression of a Core Set of Genes Underlying Chondrocyte Differentiation in Limb and Head
To test the hypothesis that a core GRN is expressed during chondrocyte differentiation in the limb and head, chondrocyte transcriptomes from the HH36 chick humerus and ceratobranchial were compared (Figure 2; Supplementary Figure S1). To identify similarities and differences among limb and head chondrocyte transcriptomes, a principal component analysis (PCA) was performed (Supplementary Figure S1). The variation in the samples was captured with two components (39% variance explained by PC1 and PC2; Supplementary Figure S1). The limb IMM and MAT transcriptomes were separated from other samples in PC1/PC2 with 95% confidence, while head IMM and MAT transcriptomes overlapped in PC1/PC2 (Supplementary Figure S1).
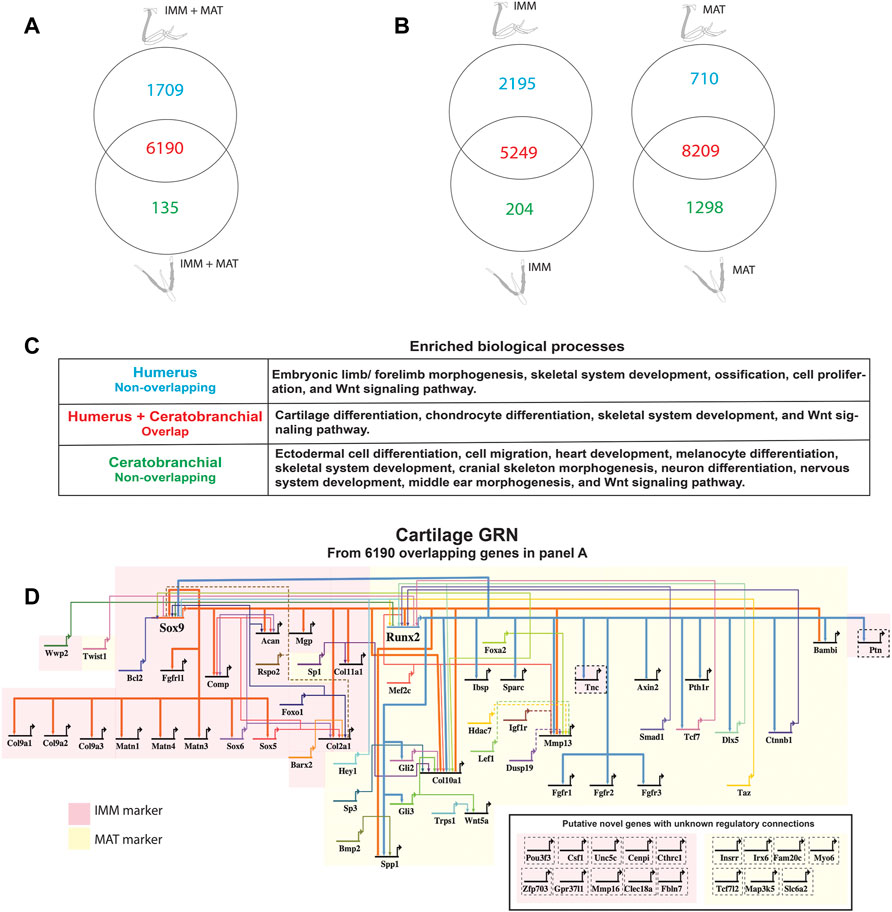
FIGURE 2. A core GRN drives chondrocyte differentiation in the limb and head. (A) Venn diagram demonstrated that limb and head chondrocytes shared almost 80% of the genes expressed above threshold (limb and head data combined before normalization and cutoff calculations). (B) Venn diagrams of IMM or MAT (separately normalized) showed that limb and head chondrocytes again shared expression of most genes. (C) Enriched biological processes in the overlap of panel A included cartilage differentiation processes, whereas limb morphogenesis and neural crest-related processes were enriched in the non-overlapping humerus and ceratobranchial portions, respectively. (D) A core cartilage gene regulatory network (GRN), compiled from the overlapping portion of panel A and published literature, included typical IMM and MAT markers highlighted in pink and yellow, respectively. Arrowheads represent positive interaction, whereas negative interactions are depicted as –|. Interactions included in this network could be direct (solid lines) or indirect (dashed lines). Novel putative cartilage genes with unknown regulatory interactions were included as single-nodes surrounded by dashed lines to the bottom right of the network, except for PTN and TNC whose regulatory interactions have been confirmed.
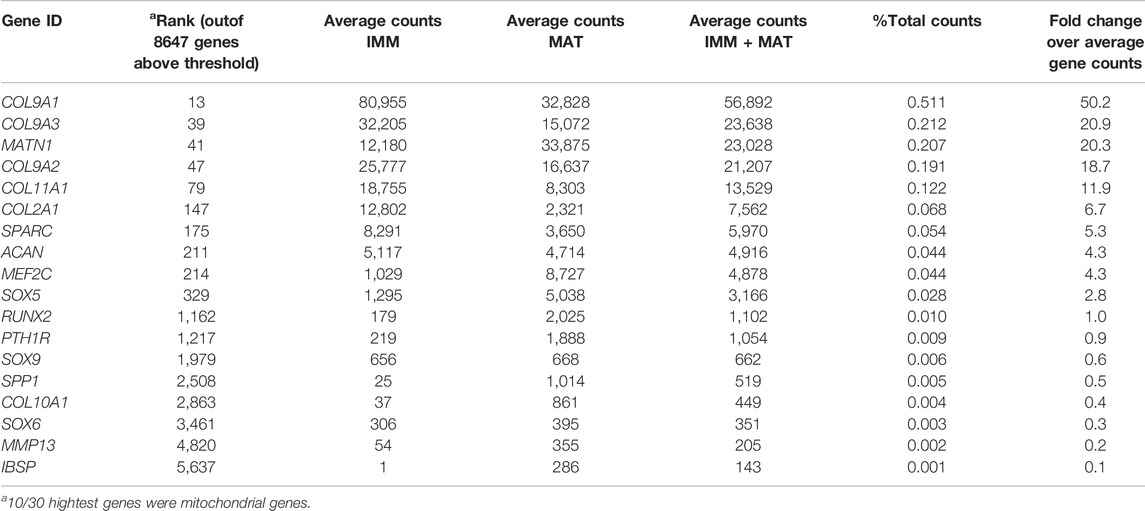
When IMM and MAT datasets from limb and head were combined before normalization (i.e., IMM and MAT were considered as the same cell type) to reflect generally the chick “chondrocyte”, limb and head chondrocytes shared 77% of genes expressed above threshold (6190/8034 genes; Figure 2A; see normalization techniques in Methods). Limb and head chondrocytes expressed typical IMM genes, such as SOX9, SOX5, SOX6, ACAN, and COL2A1, and typical MAT genes, including RUNX2, COL10A1, MEF2C, MMP13, IBSP, and SPP1 at high levels (Tables 1, 2; Vortkamp et al., 1996; Zhao et al., 1997; Bridgewater et al., 1998; Watanabe et al., 1998; Bi et al., 1999; Smits et al., 2001; Smits et al., 2004; Arnold et al., 2007; Nicolae et al., 2007; Dy et al., 2012; Lu et al., 2014; Nakatani and Partridge 2017).
When IMM and MAT were each compared separately between the limb and head before normalization (i.e., IMM and MAT were considered as different cell types), each cell type still shared the vast majority of genes expressed above threshold in both the limb and head (for IMM: 5249/7648 = 69%; for MAT: 8209/10,217 = 80%; Figure 2B). Overlapping genes in the limb and head again included many typical cartilage genes, such as SOX9, COL2A1, ACAN, and COL9A1 for IMM, and RUNX2, COL10A1, SPP1, MMP13, and IBSP for MAT (Tables 1, 2). Gene ontogeny analyses on IMM and MAT from both limb and head demonstrated that cartilage-specific processes were enriched and conserved between the limb and head, even though genes associated exclusively with these processes only comprise approximately 2% of the GO term-associated genes (Figure 2C). The most enriched biological processes (>60% genes expressed above threshold) were related to basic cellular processes, such as cell proliferation, cell differentiation, transcription, and translation. Collectively, these data suggest that a core GRN driving chondrocyte differentiation is expressed in different regions of the skeleton.
Using published work on mouse, chick, frog, and fish, regulatory interactions among important genes from chick chondrocyte transcriptomes were summarized into a GRN (Figure 2D; Longabaugh et al., 2005). As master regulators of IMM and MAT, respectively, SOX9 and RUNX2 were placed at the top of the GRN hierarchy (Figure 2D; Komori et al., 1997; Bi et al., 1999; Lian and Stein 2003; Eames et al., 2004). In the IMM portion of the GRN (highlighted in pink in Figure 2D), SOX9 binds to SOX5 and SOX6 during early stages of chondrocyte differentiation and activates the expression of typical IMM markers, such as COL2A1, COL9A1, and ACAN (Lefebvre et al., 2001; Akiyama et al., 2002; Liu and Lefebvre 2015). In the MAT portion of the GRN (highlighted in yellow in Figure 2D), RUNX2 activates the expression of typical MAT markers, including COL10A1, MMP13, SPP1, and IBSP (Ducy et al., 1997; Komori et al., 1997; Inada et al., 1999; Inada et al., 1999; Lee et al., 2000; Wang et al., 2009; Li et al., 2011; Peacock et al., 2011; Lu et al., 2014). Since SOX9 and RUNX2 generally exhibit an antagonistic relationship, SOX9 inhibits the expression of MAT markers, such as RUNX2, SPP1, and IBSP, while RUNX2 inhibits the expression of SOX9 and thus likely other IMM markers (Figure 2D; Zhou et al., 2006; Cheng and Genever 2010; Peacock et al., 2011; Lui et al., 2019).
Genes expressed above threshold only in limb or head might reflect the embryonic origin of the cells. For genes located in the humerus portion of the Venn diagrams, such as TBX5, DLX6, SALL4, HOXA10, and HOXD10, embryonic limb/forelimb morphogenesis was an enriched process, and these genes are all known regulators of limb development (Figure 2B; Wahba et al., 2001; Robledo et al., 2002; Zakany and Duboule 2007; Vieux-Rochas et al., 2013; Neufeld et al., 2014; Akiyama et al., 2015). More general skeletal processes, such as skeletal system development and ossification, were also enriched in limb chondrocytes. Many neural crest-dependent biological processes were enriched specifically in ceratobranchial IMM and MAT transcriptomes, such as cranial skeleton morphogenesis, cell migration, neuron differentiation, middle ear morphogenesis, heart development, and melanocyte differentiation (Figure 2B). Many orthologs of a proposed GRN driving neural crest-derived cartilage, such as PAX7, SIX1, and ID1, were also identified (Figure 2B; Meulemans and Bronner-Fraser 2005; Meulemans and Bronner-Fraser 2007; Betancur et al., 2010; Murdoch et al., 2012; Wu et al., 2019).
Differentially Expressed Genes in Immature Chondrocytes Were Negatively Correlated With Those of Mature Chondrocytes During Differentiation in Both the Limb and Head
GRNs rely upon functional data to verify regulatory interactions, but such studies are limited (Figure 2D; Su et al., 2009; Peter and Davidson 2011). To infer regulatory interactions underlying chondrocyte differentiation, and to compare GRN organization in the limb and head, GRNs were estimated using Cytoscape to graph gene co-expression networks (GCNs) of chondrocyte transcriptomes of the HH36 humerus or ceratobranchial (McCall 2013; Khosravi et al., 2015). All genes expressed above threshold were used to construct these GCNs (>8,000 genes, Figures 2A,B). GRNs of both limb and head data were organized into two large groups of genes expressed during chondrocyte differentiation (Figures 3A,B). Expression within each group was positively correlated (red lines in Figures 3A,B), but expression between the two groups was negatively correlated (blue lines in Figures 3A,B). One group was enriched for genes that were differentially expressed in IMM, and the other group was enriched for genes that were differentially expressed in MAT (for ease of view, selected IMM and MAT differentially expressed genes of limb and head chondrocytes are depicted in Figures 3C,D, respectively).
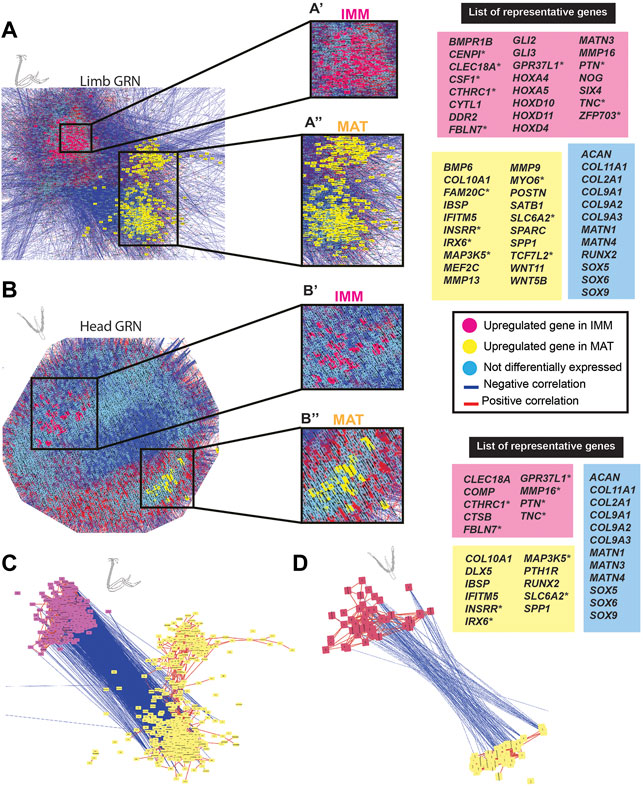
FIGURE 3. GCN of either limb or head chondrocyte data was organized into groups of enriched IMM or MAT genes that were negatively correlated with each other. (A,B) A GRN underlying chondrocyte differentiation was estimated from a gene co-expression network (GCN) from limb (A) or head (B) chondrocytes (separately normalized). In both limb and head GRNs, one portion contained genes upregulated in IMM (pink nodes), while another portion contained genes upregulated in MAT (yellow nodes). Typical chondrocyte differentiation genes included in the network (light blue nodes) were not differentially expressed between IMM and MAT. Representative lists of genes in each of these three categories are shown to the side of the GRNs. These candidate genes were selected because they are known to have a role during cartilage differentiation in different vertebrates, and they are listed in alphabetical order. Putative novel genes are indicated by an asterisk next to the gene name. (C,D) Isolated portions of IMM- and MAT-enriched genes from the limb (C) or head (D) GRNs. Most upregulated genes in IMM or MAT were connected by positive interactions (red lines), whereas IMM and MAT genes were mostly connected by negative interactions (blue lines).
In the limb, these two portions of the GRN included 859 genes [absolute log2 fold change greater than 2 (p < 0.01)] that were differentially expressed between IMM and MAT of the HH36 chick humerus (see Supplementary Tables S1, S2 for a full list of genes). A total of 263 genes were upregulated in IMM, whereas 596 genes were upregulated in MAT (Figure 3A; Supplementary Tables S1, S2). Upregulated genes in IMM included MATN3, GLI2, GLI3, DDR2, and NOG, and also some HOX genes that have a role during chondrocyte differentiation in the limb (Figure 3A, labelled pink in GRN; Koziel et al., 2005; Kruger and Kappen 2010; Tan et al., 2018; Yamamoto et al., 2019). Typical maturation genes were upregulated in MAT, including COL10A1, MMP13, SPP1, MEF2C, IBSP, and SPARC (Figure 3A, labelled yellow in GRN; Bianco et al., 1991; D'Angelo et al., 2000; Arnold et al., 2007; Peacock et al., 2011; Lu et al., 2014; Rosset and Bradshaw 2016). These differences in gene expression patterns were also demonstrated by unsupervised model-based clustering analysis (Supplementary Table S2). Some clusters showed enriched expression of genes in IMM including hallmark cartilage genes, while others showed enhanced expression in MAT including several important maturation markers (Supplementary Table S2). Typical chondrocyte differentiation genes, such as SOX9, SOX5, SOX6, COL2A1, and ACAN showed high expression levels in both IMM and MAT of the HH36 chick humerus (Figure 3A, labelled blue in GRN; Tables 1, 2).
In the head, the two portions of the GRN included 118 genes that were differentially expressed between IMM and MAT of the HH36 chick ceratobranchial (see Supplementary Tables S3, S4 for a full list of genes). A total of 70 genes were upregulated in IMM, whereas 48 genes were upregulated in MAT (Figure 3B; Supplementary Tables S3, S4). Upregulated genes in IMM included COMP and MMP16 (Figure 3B, labelled pink in GRN; Hecht et al., 2005). Typical maturation genes were upregulated in MAT, including RUNX2, COL10A1, IBSP, and SPP1 (Figure 3B, labelled yellow in GRN; Bianco et al., 1991; Komori and Kishimoto 1998; Arnold et al., 2007; Peacock et al., 2011; Lu et al., 2014).
To directly visualize an estimate of the general chick chondrocyte GRN, limb and head data were combined before normalization and graphed as a GCN. The overall organization of this GRN was similar to that for just limb or head chondrocyte data. Two portions of positively correlated genes that were differentially expressed in each type of chondrocyte (i.e., IMM or MAT) were negatively correlated with each other (Figures 4A,B). A total of 458 genes were differentially expressed between IMM and MAT (see Supplementary Tables S5, S6 for a full list of genes). Of these, 195 genes were upregulated in IMM, and 263 genes were upregulated in MAT (Supplementary Tables S5, S6). Upregulated genes in IMM and MAT included many of the same genes that were upregulated in either limb or head data alone (compare genes labelled pink in Figures 4A,B to Figures 3A,B). Other upregulated IMM genes were not previously implicated in chondrocyte differentiation, such as those encoding the transcription factors POU3F3 and ZFP703, growth factors CSF1 and PTN, and the Netrin receptor UNC5C (Figures 4A,B, labelled pink; Cecchini et al., 1997; Tare et al., 2002; Srivatsa et al., 2014; Kumar A. et al., 2016; Kumar S. et al., 2016). Upregulated genes in MAT included IFITM5, whose role had only been studied previously in osteoblast differentiation, and many other signalling pathway genes previously implicated in cartilage maturation, including BMP4, FGF9, HES5, TEK, TGFBR2, TGFBR3, WNT5B, and WNT11 (Figures 4A,B, labelled yellow; Hoffmann and Gross 2001; Yang et al., 2003; Karlsson et al., 2010; Shen et al., 2013; Usami et al., 2016; Zhang et al., 2021). Additional genes upregulated in MAT had never been associated previously with chondrocyte maturation, including those encoding the kinase FAM20C and transcription factor TCF7L2 (Hirose et al., 2020; Li et al., 2021).
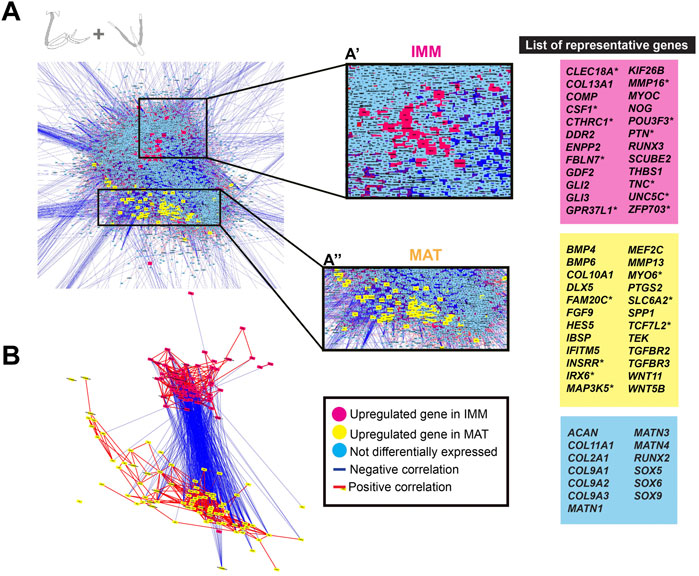
FIGURE 4. GCN of combined limb and head chondrocyte data was organized into groups of enriched IMM or MAT genes that were negatively correlated with each other. (A) A GRN underlying chondrocyte differentiation was estimated from a GCN from limb and head chondrocytes (normalized together). One portion of the GRN contained genes upregulated in IMM (pink nodes), while another portion contained genes upregulated in MAT (yellow nodes). Typical chondrocyte differentiation genes included in the network (light blue nodes) were not differentially expressed between IMM and MAT. Representative lists of genes in each of these three categories are shown to the side of the GRNs. These candidate genes were selected because they are known to have a role during cartilage differentiation in different vertebrates, and they are listed in alphabetical order. Putative novel genes are indicated by an asterisk next to the gene name. (B) Isolated portions of IMM- and MAT-enriched genes in the GRN. Most upregulated genes in IMM or MAT were connected by positive interactions (red lines), whereas IMM and MAT genes were mostly connected by negative interactions (blue lines).
Although typical chondrocyte differentiation genes, such as SOX9 and COL2A1, were not differentially expressed between limb and head (Figures 3, 4), differences in gene expression were obvious when the normalized counts were compared (Tables 1, 2). In the limb, for instance, the SOX trio (SOX9, SOX5, and SOX6) showed higher expression levels in IMM, and expression levels decreased in MAT, as reported by others (Ikegami et al., 2011; Lui et al., 2019). In the head, however, expression levels of SOX9 and SOX6 remained at comparable levels in IMM and MAT, and MAT even showed higher levels of SOX5 expression compared to IMM. A similar situation is observed when RUNX2 levels are compared between limb and head. In the head, RUNX2 expression strikingly increases during IMM to MAT transition (Tables 1, 2; Figure 3B), while in the limb this increase in RUNX2 expression is not as as dramatic (Tables 1, 2). These differences in the expression might be the result of variation in the timing of cartilage maturation among skeletal elements, as well as differences related to growth and shape of endochondral bones in distinct locations of the body (Chiba et al., 1995; Patton and Kaufman 1995; Mitgutsch et al., 2011). Together, these data demonstrated that the GRN driving chondrocyte differentiation is organized similarly throughout the body and provided novel genes that might regulate IMM and MAT differentiation.
Patterning Genes Expressed During Chondrocyte Differentiation Were Enriched in the Limb or Head
To identify genes that might influence which type of chondrocyte differentiates in specific embryonic regions, IMM or MAT data from the limb and head was used to estimate a GRN underlying IMM or MAT differentiation by graphing a GCN. Two portions of each GRN were identified, this time enriched for genes that were differentially expressed in chondrocytes of the limb or the head (Figures 5A,B). Many of these region-specific genes were positively correlated to each other, while negatively correlated to genes enriched in chondrocytes from the other embryonic region (Figures 5A–D). Many of these genes were patterning genes of the limb or head.
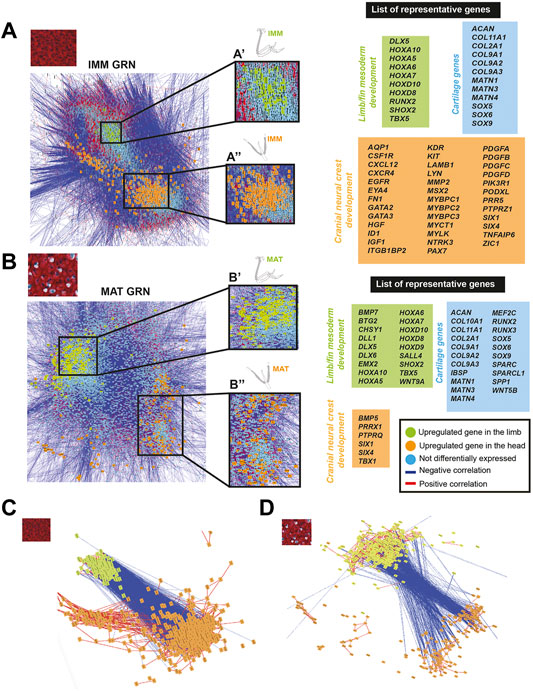
FIGURE 5. GCN of either IMM or MAT chondrocyte data was organized into groups of enriched limb or head genes that were negatively correlated with each other. (A,B) A GRN underlying IMM (A) or MAT (B) differentiation was estimated from a GCN from limb and head chondrocytes (IMM and MAT data separately normalized). In both IMM and MAT GRNs, one portion contained genes upregulated in the limb (green nodes), while another portion contained genes upregulated in the head (orange nodes). Typical chondrocyte differentiation genes included in the network (light blue nodes) were not differentially expressed between limb and head. Representative lists of genes in each of these three categories are shown to the side of the GRNs. (C,D) Isolated portions of limb- and head-enriched genes in the IMM (C) and MAT (D) GRNs. Most upregulated genes in limb or head were connected by positive interactions (red lines), whereas limb and head genes were mostly connected by negative interactions (blue lines).
Genes upregulated in limb chondrocytes were very similar for the IMM and MAT data, while genes upregulated in head chondrocytes varied among IMM and MAT data. The two portions of the IMM GRN included 1223 genes that were differentially expressed between limb and head chondrocytes (Figure 5A; see Supplementary Tables S7, S8 for a full list of genes). A total of 216 genes were upregulated in limb IMM, while 1,007 genes were upregulated in head IMM (Supplementary Tables S7, S8). The two portions of the MAT GRN included 1,215 genes that were differentially expressed between limb and head chondrocytes (Figure 5B; Supplementary Tables S9, S10). A total of 664 genes were upregulated in limb MAT, whereas 551 genes were upregulated in head MAT (Supplementary Tables S9, S10). Upregulated genes in limb IMM and MAT included several HOX genes, DLX5, TBX5, and SHOX2, all of which are known to have a role during limb morphogenesis (Figures 5A,B, labelled green in GRN; Agarwal et al., 2003; Koziel et al., 2005; Yu et al., 2007; Kruger and Kappen 2010; Vieux-Rochas et al., 2013; Tan et al., 2018; Yamamoto et al., 2019). The other portion of the IMM GRN included genes upregulated in the head that are involved in neural crest related processes, such as ZIC1, SIX1, SIX4, PAX7, ID1, GATA2, GATA3, and MSX2, as identified by gene ontology analyses and previous studies (Figure 5A, labelled orange in the GRN; Han et al., 2007; Murdoch et al., 2012; Simões-Costa and Bronner 2013; Garcez et al., 2014; Plouhinec et al., 2014; Simões-Costa and Bronner 2015; Wu et al., 2019; Seal and Monsoro-Burq 2020). Similar to the IMM GRN, SIX1, and SIX4 were also upregulated in the head in one portion of the MAT GRN. Also upregulated in head MAT were TBX1 and PRRX1, which actually can regulate both limb and cranial neural crest (Figure 5B, labelled orange in GRN; Moraes et al., 2005; Balic et al., 2009; Simões-Costa and Bronner 2015). Similar results were obtained when IMM and MAT data from the limb and head were considered as four separate datasets before normalization, and included in the same GRN (Supplementary Figure S3). Unsupervised model-based clustering analyses also identified specific categories of gene expression when comparing limb and head chondrocyte data (Supplementary Figure S2). Some clusters showed enriched expression of genes in the limb, including several classic limb patterning genes, while other clusters showed increased expression in the head, including many genes involved in cranial neural crest differentiation (Supplementary Figure S2). Again, comparisons between limb and head transcriptomes revealed that typical IMM and MAT genes, including SOX9, COL2A1, RUNX2, COL10A1, and SPP1, were not differentially expressed between head and limb, supporting the hypothesis that a core transcriptional program driving chondrocyte differentiation is expressed wherever cartilage forms in the body.
Discussion
Perhaps the most interesting chapter in the story of cartilage evolution is that cartilage appeared in different parts of the body at different times during vertebrate evolution. How could this have happened? Once the ability to differentiate cartilage was encoded in an ancestor’s genome as a core GRN, then adding cartilage to another location in the body might only require that a different population of cells co-opt expression of this GRN (Eames et al., 2020). This hypothesis is realistic, because SOX9 sits at the top of the chondrocyte GRN hierarchy, so perhaps during evolution only a few regulatory elements were added to SOX9 for cartilage to form in another embryonic region.
As proof of principle, data argues that cartilage appeared in a different embryonic population at the origin of vertebrates by GRN co-option. The GRN driving chondrocyte differentiation of invertebrates in cranial mesoderm or endoderm contains Sox9-like (SoxE), Sox5/6-like (SoxD), and Col2a1-like (ColA) genes (Rychel et al., 2006; Cattell et al., 2011; Jandzik et al., 2015; Tarazona et al., 2016). Regulatory connections in the chondrocyte GRN seem conserved among invertebrates and vertebrates (Figure 6). For example, SoxE from a hard-shell invertebrate activated expression of the human COL2A1 gene (Tarazona et al., 2016). In the ancestral vertebrate, SoxE regulatory elements, and thus the core cartilage GRN, might have been co-opted from cranial mesoderm or endoderm by cranial neural crest to expand cartilage in the ancestral vertebrate head (Meulemans and Bronner-Fraser 2007; Hall and Gillis 2013; Jandzik et al., 2015).
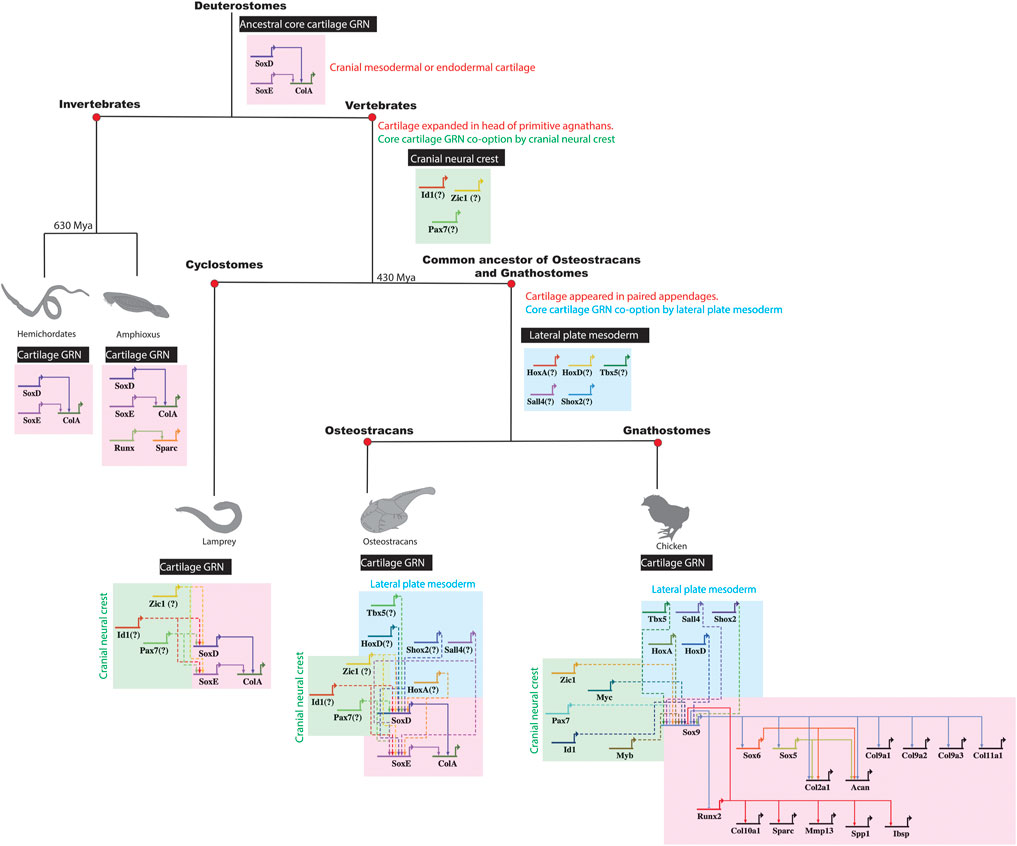
FIGURE 6. Co-option of a core cartilage GRN by different embryonic populations during vertebrate evolution. The ancestral core cartilage GRN, shared in the common ancestor of vertebrates and their sister invertebrates, likely included such genes as SoxD, SoxE, and ColA expressed in cranial mesoderm or endoderm. In primitive agnathan vertebrates, cranial cartilage expanded when cranial neural crest cells co-opted a core cartilage GRN, likely building new regulatory connections between cranial neural crest genes and SoxE orthologs. Later, when paired appendages with cartilage evolved in the common ancestor of osteostracans and gnathostomes, a core cartilage GRN was additionally co-opted by lateral plate mesoderm, likely by establishing new regulatory connections between lateral plate mesoderm genes and SoxE orthologs.
After the origin of vertebrates, did lateral plate mesoderm similarly co-opt a core chondrocyte GRN? Cartilage was in the head of primitive agnathans, such as ancestors of cyclostomes, before the evolution of paired appendages, such as pelvic or pectoral fins (Figure 6; Janvier 1996; Berendsen and Olsen 2015). While extant cyclostomes lack paired appendages, the fossil record reveals that some extinct agnathans (e.g., osteostracans) had evolved paired appendages with cartilage (Janvier et al., 2004; Adachi et al., 2016). If cartilage in paired appendages of osteostracans was derived from lateral plate mesenchyme, and if the common ancestor of osteostracans and gnathostomes shared this feature, then all subsequent lineages of vertebrates might retain features of lateral plate mesoderm co-opting expression of a core GRN underlying chondrocyte differentiation (Figure 6). In this case, chondrocytes in the limb and head of all living gnathostomes might express the same core transcriptional program making cartilage.
Before discussing our data testing similarities between limb and head chondrocyte transcriptomes, what is meant by “core GRN”? Ultimately, GRNs derive from an organism’s genome, which is the same in most cells of the body. Here, we propose that the core GRN of a chondrocyte is the set of genes and their regulatory connections that are required for the organism to produce this cell type (Figure 6). In creating cartilage, the chondrocyte largely functions to secrete ECM, so genes encoding ECM molecules and the transcription factors that regulate them are likely key components of the chondrocyte core GRN (Gray and Williams, 1989; Gentili and Cancedda 2009; Hojo and Ohba 2019; Neefjes et al., 2020). In support of this definition, ECM genes were among the most highly expressed genes in all chondrocyte transcriptome data presented here. Regarding the idea that genes in the core GRN are required for an organism to produce a chondrocyte, we also discuss below examples of genes that are expressed in a region-specific manner. While debatable, we have argued that such genes are not part of the core chondrocyte GRN, because their loss-of-function produces only a region-specific loss of cartilage (Eames et al., 2020). Such genes, which often include transcription factors and growth factors that might act as region-specific cartilage competency or morphogenesis factors, would not be required for an organism to produce a chondrocyte, since chondrocytes are still produced in other parts of the organism’s body. Admittedly, according to another definition of core GRN, conclusions from our data might differ, but we hope this discussion helps to focus efforts on understanding the evolution of cell types (Arendt 2008; Achim and Arendt 2014; Sachkova and Burkhardt 2019; Callier 2020).
Here, comparative transcriptomics supported the hypothesis that a core GRN driving chondrocyte differentiation is expressed in cartilage of the limb and head. Venn diagram analyses revealed a large overlap (∼75%) in gene expression between the transcriptomes of chick chondrocytes isolated from the humerus and the ceratobranchial. Gene ontology analyses revealed that biological processes related to cartilage differentiation were enriched in genes shared between limb and head chondrocytes. The master chondrocyte differentiation gene, SOX9, as well as many other genes that are regulated by this transcription factor, such as SOX5, SOX6, COL2A1, COL9A1, COL10A1, and ACAN, were not differentially expressed between limb and head chondrocytes, as previously suggested by others, however differences in gene expression were identified when comparing normalized gene counts in limb and head (Tables 1, 2; Lefebvre et al., 1997; Zhao et al., 1997; Bi et al., 1999; Sekiya et al., 2000; Smits et al., 2001; Akiyama et al., 2002; Zhang et al., 2003; Eames and Helms 2004; Dy et al., 2012; Gu et al., 2012). Since the timing of cartilage maturation and ossification can vary among skeletal elements, future studies should assess these differences in gene expression by providing a detailed timeline of maturation in limb versus head, given the limitations of the current data. Importantly, the core GRN presented here also shows deep conservation when comparing the present data with mouse limb RNA-seq and scRNA-seq datasets. Many of the typical cartilage genes included in the core GRN, such as Sox9, Sox5, Sox6, Acan, Col2a1, Col10a1, and Runx2, were conserved between mouse and chick (Ayturk et al., 2020; Duan et al., 2020; Sebastian et al., 2021). These data clearly support the hypothesis that a core GRN driving chondrocyte differentiation is expressed throughout the body, and perhaps it has not been modified dramatically during evolution.
Interestingly, some other genes that have little or no known role in chondrocyte differentiation showed high expression levels in both the limb and head. For example, novel putative cartilage genes in IMM include those encoding the transcription factors POU3F3 and ZFP703 and the growth factor signalling members CSF1, PTN, and UNC5C, and other genes, such as CENPI, CLEC18A, CTHRC1, FBLN7, GPR37L1, MMP16, and TNC. Putative mature cartilage genes upregulated in MAT included INSRR, IRX6, MAP3K5, MYO6, SLC6A2, FAM20C, and the transcription factor TCF7L2 (Figure 2D). Importantly, several of these putative novel genes, including Ptn, Tnc, Unc5c, and Fbln7, were also found to be expressed above threshold in mouse limb RNA-seq and scRNA-seq datasets (Ayturk et al., 2020; Duan et al., 2020; Sebastian et al., 2021). In addition, orthologs for most of these novel candidate genes are present in the genome of other vertebrates, including bony fishes, reptiles, amphibians, and cartilaginous fishes, and even some invertebrates, but only a few have been shown to be expressed in chondral bones of non-mammalian vertebrates, such as frog and fish (Nalbant et al., 2005; Geurtzen et al., 2014; Russell et al., 2014; Square et al., 2015; Barske et al., 2020; Kraus et al., 2022). Also, many of these novel putative core cartilage GRN genes have been found to be expressed in OA cartilage, and even to have a role during the progression of this skeletal disease, further supporting the importance of these genes during cartilage development (Mentlein 2007; Johnson et al., 2015; Sun et al., 2018; Zhang et al., 2018; Bhattaram and Jones 2019; Rice et al., 2019; Chakraborty et al., 2020; Hasegawa et al., 2020; Shi et al., 2022). Future loss of function experiments in these and other vertebrates will be required to assess whether these genes indeed qualify as cartilage core genes.
The GRN driving chondrocyte differentiation, derived from both traditional methods and from GCN analyses, also had very similar organization using transcriptomic data from limb or head chondrocytes. Traditionally, GRNs are derived from functional experiments that identify positive or negative regulatory relationships between genes involved in a given biological process, such as mesoderm formation (Davidson and Erwin 2006; Peter and Davidson 2011; Erkenbrack 2016). Using published data on regulatory interactions among genes that were shared from our limb and head chondrocyte transcriptomes, we expanded upon the first published traditional chondrocyte GRN (Figure 2C; Cole 2011). Seven transcription factors (ATF3, DLX6, FOXA3, FOXK2, FOXN2, FOXO1, RUNX3, and SOX8) with no known regulatory connections and many other genes were incorporated into the chondrocyte GRN. This GRN also featured inhibition of MAT genes by Sox9, an IMM gene (Zhou et al., 2006; Peacock et al., 2011; Lui et al., 2019).
GCNs also can reveal regulatory relationships among genes (Stuart et al., 2003; McCall 2013; Khosravi et al., 2015), and independent estimation of the chondrocyte GRN from GCNs of our transcriptomic data confirmed and expanded the traditional approach. For both limb and head data, positive correlations between cell-type enriched genes were observed within the same cell type (IMM or MAT), likely driven by Sox9 and Runx2 activity for IMM and MAT, respectively (Eames et al., 2004; Cole 2011; Oh et al., 2014; Wu et al., 2014; Gomez-Picos and Eames 2015; Ohba et al., 2015; Tan et al., 2018). On the other hand, negative correlations were predominant between different cell types (IMM vs. MAT). While Sox9 inhibition of MAT genes can explain some of the cross-inhibition between IMM and MAT genes, additional molecular mechanisms, such as epigenetic switches, should be investigated.
In the chondrogenic program presented here, SOX9 and RUNX2 were placed at the top of the hierarchy of the GRN (Figure 2D). Molecular genetic experiments have shown that both SOX9 and RUNX2 drive expression of mature chondrocyte genes in limb and head, while only RUNX2 is known to drive osteoblast genes (Eames et al., 2004; Ding et al., 2012; Dy et al., 2012). Several MAT genes included here as part of the core cartilage GRN are also likely part of an osteogenic core GRN. However, a few of these genes (i.e., COL10A1) are only expressed in mature chondrocytes, not osteoblasts, of chick and other tetrapods (Bendall et al., 2003; Conen et al., 2009; Leung et al., 2011; Gu et al., 2014). Also, at this early timepoint of collection, only chondrocytes were identified in both the humerus and ceratobranchial, while osteoblasts were restricted to perichondral bone, not within cartilage itself, suggesting no transdifferentiation from chondrocytes to osteoblasts had yet occurred (Figure 1; Zhou et al., 2014; Qin et al., 2020).
Many genes currently associated with cartilage differentiation, and thus the core cartilage GRN, might only be region-specific cartilage competency or morphogenesis factors. Enriched GO terms from humerus or ceratobranchial chondrocytes reflected the embryonic origin of the cells, including limb/forelimb morphogenesis or neural crest-derived processes, respectively. Given region-specific expression, some commonly described cartilage genes, such as ID genes, HOX genes, Tbx5, Sall4, and Shox2, might only serve that purpose in specific regions of the body (Thornemo et al., 1996; Asp et al., 1998; Jung and Tsonis 1998; Yu et al., 2007; Gross et al., 2012; Yamamoto et al., 2019). In principle, establishing regulatory interactions between these transcription factors and the core cartilage GRN might have been crucial to stabilizing cartilage formation in new areas of the vertebrate body during evolution (Figure 6). Interestingly, genes enriched in different embryonic regions were negatively correlated to each other, suggesting cross-inhibition of region-specific transcriptional programs.
In conjunction with region-specific transcription factors, region-specific signalling pathways might activate or stabilize expression of a core cartilage GRN in different regions of the body (Eames and Helms 2004). Both limb and head chondrocytes showed enriched expression of genes involved in BMP, FGF, interleukin (IL), and TGF-B signalling, but the specific upregulated genes were different in each embryonic region. Signalling genes upregulated in the limb included BMP7, FGF2, FGFRL1, IL13RA2, and TGFBR2, while those upregulated in the head included BMP5, FGF13, FGF18, FGF23, IL18RAP, IL1RAPL1, and TGFBI. Insulin growth factors (i.e., IGF2BP2) were only upregulated in limb chondrocytes, whereas genes involved in EGF signalling (EGF, EGFL6, and EGFR), PDGF signalling (i.e., PDGFA, PDGFB, PDGFC, and PDGFD), and VEGF (VEGFC) signalling pathways were only upregulated in head chondrocytes.
With respect to clinical applications, the data presented here supports the idea that the origin of cells does not influence the type of cartilage formed. Therefore, if chondrocytes from one location were to be transplanted into a new location in the body, then genes required for these chondrocytes to properly differentiate in that new environment will ultimately be expressed, and cartilage will grow and differentiate in this new location in the body. Indeed, previous work has shown that when nasal chondrocytes are transplanted into an osteoarthritic knee, they can efficiently adapt to this new environment, and successfully contribute to cartilage repair (Pelttari et al., 2020; Acevedo Rua et al., 2021).
In summary, these comparative transcriptomic results demonstrate that a core transcriptional program is expressed during chondrocyte differentiation of the limb and head. Therefore, when cartilage was added to different regions of the vertebrate skeleton, a core GRN might have been co-opted to drive chondrocyte differentiation (Figure 6). While identifying conserved chondrocyte genes is crucial for developing new therapies for cartilage injuries and disorders, expanding transcriptomic comparisons across more clades will provide valuable insights into the evolutionary development of cartilage.
Materials and Methods
Embryo Collection and Tissue Processing
All animal procedures were performed according to guidelines approved by the University of Saskatchewan Animal Care and Use Committee. White leghorn chicken eggs were incubated in a humified incubator at a constant temperature of 37°C. Embryos were harvested at Hamburger-Hamilton stage 36 (∼E10; Hamburger and Hamilton 1951). Each embryo was decapitated, and the forelimbs and lower jaws were dissected and either fixed in 4% paraformaldehyde overnight or immediately placed in 1X PBS/DEPC, followed by embedding in OCT (Tissue Tek, Torrance, CA, United States), and immediately flash-frozen using liquid N2 and 2-Methylbutane (isopentane).
Histology
Chick HH36 embryos were stained with Alcian blue and Alizarin red using an acid-free solution that included MgCl2 to differentiate staining, and then cleared in glycerol/KOH as described elsewhere (Eames et al., 2011). Importantly, in our hands, various sources of Alcian blue do not work with the acid-free protocol, but one that does is from Acros Organics (Alcian Blue 8GX). Safranin O/Fast Green staining was performed on 10 μm thick frozen sections of the HH36 chick humeri and ceratobranchial, as described previously (Ferguson et al., 1998). Trichrome staining was performed on 10 μm thick frozen sections of the HH36 chick mandible, as described elsewhere (Ashique et al., 2022).
Laser Capture Microdissection
LCM was performed on a Laser Microdissection—Molecular Machines & Industries (MMI) CellCut apparatus. Immature and mature chondrocytes were captured from developing chick HH36 humeri (IMM, n = 4; MAT, n = 5) and ceratobranchial (IMM, n = 3; MAT, n = 3). At this early stage of development, perichondral bone was evident in both the humerus and ceratobranchial, but no osteoblasts or other cell types were present in the cartilage template, and invasion by the vasculature had not yet occurred. Tissue slices were processed and sequenced individually (not pooled at any stage), and the captured cells were collected onto the inner lid of 0.5 ml MMI IsolationCaps (either Diffuser caps (Prod#50202) or Transparent caps (Prod#50204; MMI Molecular Machines & Industries).
RNA Isolation and Amplification
RNA was isolated from laser-captured tissue using the ARCTURUS PicoPure RNA Isolation Kit (ThermoFisher Scientific; Cat# KIT0204), according to the manufacturer’s instructions, and DNase treatment was done using RNase-Free DNase (Qiagen; Cat#79254). RNA was then amplified with one round using MessageAmp II aRNA Kit (ThermoFisher Scientific; Cat# AM1751). RNA integrity was evaluated on the observation of a signature eletropherogram pattern (Bioanalyzer).
Library Preparation and Deep RNA Sequencing
RNA-seq libraries were prepared by the National Research Council (NRC, Saskatoon) using the Illumina TruSeq RNA Sample Prep Kit v2 with the following modification: the protocol was started at the Elute, Prime, Fragment step using 5 µl amplified mRNA [minimum amount was 5–10 ng mRNA as determined using Quant-iT RiboGreen RNA Assay Kit (Invitrogen)]. The quality of each cDNA library was checked on a DNA 1,000 chip using the 2,100 Bioanalyzer (Agilent Technologies Inc.). In average, the sequencing depth was 21-million reads per sample (min. 14-million reads per sample; max. 31- million reads per sample).
Reads Preprocessing, Mapping, Quantitation, and Primary Analysis of RNA-Seq Data
The paired-end Illumina reads were trimmed using a Java -based tool, Trimmomatic v0.30 (Bolger et al., 2014), and the reads were then mapped to the chicken genome on Ensembl using STAR v 2.5.2 (Dobin et al., 2012). The location of each read was matched to genome annotation using HTSeq-count (Anders et al., 2015). The distribution of average log2 expression across three replicates of each tissue produced three bimodal distributions, which were used to set the count thresholds to 142 and 23 for IMM and MAT isolated from the ceratobranchial, and 37 and 58 for IMM and MAT isolated from the humerus. When head and limb data were combined before normalization, thresholds were set to 64 and 35 for IMM and MAT, respectively. Including different sets of samples before normalization affects the number of genes expressed above threshold, because the gene counts will change depending on the exact samples they are normalized against. Differential expression analysis was performed using EdgeR after excluding genes with zero or very low counts (less than three counts for all cell types) across the cell type. Pairwise comparisons between tissues were made with Fisher’s exact test, and a gene was considered differentially expressed if it had an absolute log2 fold change greater than 2 (p < 0.01). Venn diagrams were constructed using gplots v3.0.1 for isoforms and RNA-seq expression data.
Principal Component Analysis
To evaluate similarities and differences among the IMM and MAT transcriptomes obtained from limb and head chondrocytes, a principal component analysis (PCA) was performed. PCA was performed on the data using prcomp from the stats library in R to determine if the biological replicates of each cell type separated into distinct groups based on gene expression variance. The 95% confidence ellipses were included using R package factoextra version 1.0.7. The variation in the samples was captured with two components (39% variance explained by PC1 and PC2; Supplementary Figure S1).
Cluster Analysis
The algorithms from MBCluster.Seq 1.0 package in R were used to cluster the genes from our RNA-seq data (Si et al., 2014). Genes were assigned to 10 clusters based on expression profiles across all IMM and MAT isolated from limb and head (Supplementary Figure S2).
Validated Chondrocyte GRN
The skeletal cell GRN was constructed using BioTapestry version 7.1.2 (www.BioTapestry.org/) following developer’s protocol (Longabaugh et al., 2005). Regulatory interactions were validated based on published studies including genetic molecular experiments and cis-regulatory analyses performed in bones of mouse, chick, frog, and zebrafish (Ducy et al., 1997; Komori et al., 1997; Lecanda et al., 1997; Zhao et al., 1997; Bridgewater et al., 1998; Drissi et al., 2000; Sekiya et al., 2000; Akiyama et al., 2002; Zhang et al., 2003; Zheng et al., 2003; Akiyama et al., 2004; Bastepe et al., 2004; Stock et al., 2004; Yang et al., 2004; Hong et al., 2005; Magee et al., 2005; Meech et al., 2005; Yagi et al., 2005; Arnold et al., 2007; Holleville et al., 2007; Liu et al., 2007; Yun and Im 2007; Grogan et al., 2008; Higashihori et al., 2008; Vincourt et al., 2008; Teplyuk et al., 2009; Wang et al., 2009; Dao et al., 2010; Fazenda et al., 2010; Higashiyama et al., 2010; Leung et al., 2011; Nagy et al., 2011; Peacock et al., 2011; Gu et al., 2012; Ionescu et al., 2012; McGee-Lawrence et al., 2013; Oh et al., 2014; Liu and Lefebvre 2015; Ohba et al., 2015; Heilig et al., 2016; Takegami et al., 2016; Watanabe et al., 2016; Komori 2017; Yao et al., 2017; Kawane et al., 2018; Komori 2018; Liu et al., 2018; Qin et al., 2018; Tan et al., 2018; Xu et al., 2018; Komori 2019; Kurakazu et al., 2019; Mokuda et al., 2019; Yamashita et al., 2019; Wuelling et al., 2020).
GO Analysis
DAVID v6.8 (http://david.abcc.ncifcrf.gov/home.jsp) functional annotation analysis was performed on genes expressed above threshold in head and limb. The GO term biological process (BP) in DAVID was used to perform the gene-annotation enrichment analysis.
Gene Co-Expression Network Analyses
For constructing GCNs, the Pearson correlation between genes was calculated using the TMM normalized gene expression data. Thresholding the edge weights (+/−0.85) was then performed to remove potentially irrelevant edge weights before visualization. All processing and normalization of the RNA-seq counts were performed using the edgeR package using R version 4.0.0. GCNs were visualized in Cytoscape version 3.8.2 and all color coding of edges and nodes was performed using Cytoscape.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI’s Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/) and are accessible through GEO Series accession number GSE186980.
Ethics Statement
The animal study was reviewed and approved by University of Saskatchewan Animal Care and Use Committee.
Author Contributions
PG-P performed all biological experiments required to generate IMM and MAT transcriptomes, wrote the paper, interpreted the data, and generated the figures. KO generated gene co-expression network analysis and bioinformatically analyzed the data. BE conceived the project, supervised the work, and assisted with revision of the manuscript. All authors contributed to the article and approved the submitted version.
Funding
We acknowledge the support of the Natural Sciences and Engineering Research Council of Canada (NSERC) (funding reference Nos. 435655-201 and 2019-05977). No funding body played any roles in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2022.876825/full#supplementary-material
References
Acevedo Rua, L., Mumme, M., Manferdini, C., Darwiche, S., Khalil, A., Hilpert, M., et al. (2021). Engineered Nasal Cartilage for the Repair of Osteoarthritic Knee Cartilage Defects. Sci. Transl. Med. 13, eaaz4499. doi:10.1126/scitranslmed.aaz4499
Achim, K., and Arendt, D. (2014). Structural Evolution of Cell Types by Step-wise Assembly of Cellular Modules. Curr. Opin. Genet. Dev. 27, 102–108. doi:10.1016/j.gde.2014.05.001
Adachi, N., Robinson, M., Goolsbee, A., and Shubin, N. H. (2016). Regulatory Evolution of Tbx5 and the Origin of Paired Appendages. Proc. Natl. Acad. Sci. U.S.A. 113, 10115–10120. doi:10.1073/pnas.1609997113
Agarwal, P., Wylie, J. N., Galceran, J., Arkhitko, O., Li, C., Deng, C., et al. (2003). Tbx5 Is Essential for Forelimb Bud Initiation Following Patterning of the Limb Field in the Mouse Embryo. Development 130, 623–633. doi:10.1242/dev.00191
Akiyama, H., Chaboissier, M.-C., Martin, J. F., Schedl, A., and de Crombrugghe, B. (2002). The Transcription Factor Sox9 Has Essential Roles in Successive Steps of the Chondrocyte Differentiation Pathway and Is Required for Expression of Sox5 and Sox6. Genes Dev. 16, 2813–2828. doi:10.1101/gad.1017802
Akiyama, H., Lyons, J. P., Mori-Akiyama, Y., Yang, X., Zhang, R., Zhang, Z., et al. (2004). Interactions between Sox9 and β-catenin Control Chondrocyte Differentiation. Genes Dev. 18, 1072–1087. doi:10.1101/gad.1171104
Akiyama, R., Kawakami, H., Wong, J., Oishi, I., Nishinakamura, R., and Kawakami, Y. (2015). Sall4-Gli3 System in Early Limb Progenitors Is Essential for the Development of Limb Skeletal Elements. Proc. Natl. Acad. Sci. U.S.A. 112, 5075–5080. doi:10.1073/pnas.1421949112
Anders, S., Pyl, P. T., and Huber, W. (2015). HTSeq--a Python Framework to Work with High-Throughput Sequencing Data. Bioinformatics 31, 166–169. doi:10.1093/bioinformatics/btu638
Arendt, D. (2008). The Evolution of Cell Types in Animals: Emerging Principles from Molecular Studies. Nat. Rev. Genet. 9, 868–882. doi:10.1038/nrg2416
Arnold, M. A., Kim, Y., Czubryt, M. P., Phan, D., McAnally, J., Qi, X., et al. (2007). MEF2C Transcription Factor Controls Chondrocyte Hypertrophy and Bone Development. Dev. Cell 12, 377–389. doi:10.1016/j.devcel.2007.02.004
Ashique, A. M., Atake, O. J., Ovens, K., Guo, R., Pratt, I. V., Detrich, H. W., et al. (2022). Bone Microstructure and Bone Mineral Density Are Not Systemically Different in Antarctic Icefishes and Related Antarctic Notothenioids. J. Anat. 240, 34–49. doi:10.1111/joa.13537
Asp, J., Thornemo, M., Inerot, S., and Lindahl, A. (1998). The Helix-Loop-Helix Transcription Factors Id1 and Id3 Have a Functional Role in Control of Cell Division in Human Normal and Neoplastic Chondrocytes. FEBS Lett. 438, 85–90. doi:10.1016/s0014-5793(98)01268-x
Ayturk, U. M., Scollan, J. P., Goz Ayturk, D., Suh, E. S., Vesprey, A., Jacobsen, C. M., et al. (2020). Single˗Cell RNA Sequencing of Calvarial and Long˗Bone Endocortical Cells. J. Bone Min. Res. 35, 1981–1991. doi:10.1002/jbmr.4052
Balic, A., Adams, D., and Mina, M. (2009). Prx1andPrx2cooperatively Regulate the Morphogenesis of the Medial Region of the Mandibular Process. Dev. Dyn. official Publ. Am. Assoc. Anatomists 238, 2599–2613. doi:10.1002/dvdy.22092
Barske, L., Fabian, P., Hirschberger, C., Jandzik, D., Square, T., Xu, P., et al. (2020). Evolution of Vertebrate Gill Covers via Shifts in an Ancient Pou3f3 Enhancer. Proc. Natl. Acad. Sci. U.S.A. 117, 24876–24884. doi:10.1073/pnas.2011531117
Bastepe, M., Weinstein, L. S., Ogata, N., Kawaguchi, H., Jüppner, H., Kronenberg, H. M., et al. (2004). Stimulatory G Protein Directly Regulates Hypertrophic Differentiation of Growth Plate Cartilage In Vivo. Proc. Natl. Acad. Sci. U.S.A. 101, 14794–14799. doi:10.1073/pnas.0405091101
Bendall, A. J., Hu, G., Levi, G., and Abate-Shen, C. (2003). Dlx5 Regulates Chondrocyte Differentiation at Multiple Stages. Int. J. Dev. Biol. 47, 335–344.
Berendsen, A. D., and Olsen, B. R. (2015). Bone Development. Bone 80, 14–18. doi:10.1016/j.bone.2015.04.035
Betancur, P., Bronner-Fraser, M., and Sauka-Spengler, T. (2010). Assembling Neural Crest Regulatory Circuits into a Gene Regulatory Network. Annu. Rev. Cell Dev. Biol. 26, 581–603. doi:10.1146/annurev.cellbio.042308.113245
Bhattacherjee, V., Mukhopadhyay, P., Singh, S., Johnson, C., Philipose, J. T., Warner, C. P., et al. (2007). Neural Crest and Mesoderm Lineage-dependent Gene Expression in Orofacial Development. Differentiation 75, 463–477. doi:10.1111/j.1432-0436.2006.00145.x
Bhattaram, P., and Jones, K. (2019). Regulation of Fibroblast-like Synoviocyte Transformation by Transcription Factors in Arthritic Diseases. Biochem. Pharmacol. 165, 145–151. doi:10.1016/j.bcp.2019.03.018
Bi, W., Deng, J. M., Zhang, Z., Behringer, R. R., and de Crombrugghe, B. (1999). Sox9 Is Required for Cartilage Formation. Nat. Genet. 22, 85–89. doi:10.1038/8792
Bianco, P., Fisher, L. W., Young, M. F., Termine, J. D., and Robey, P. G. (1991). Expression of Bone Sialoprotein (BSP) in Developing Human Tissues. Calcif. Tissue Int. 49, 421–426. doi:10.1007/bf02555854
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a Flexible Trimmer for Illumina Sequence Data. Bioinformatics 30, 2114–2120. doi:10.1093/bioinformatics/btu170
Bridgewater, L. C., Lefebvre, V., and de Crombrugghe, B. (1998). Chondrocyte-specific Enhancer Elements in the Col11a2 Gene Resemble the Col2a1 Tissue-specific Enhancer. J. Biol. Chem. 273, 14998–15006. doi:10.1074/jbc.273.24.14998
Burke, A. C., and Nowicki, J. L. (2001). HoxGenes and Axial Specification in Vertebrates. Am. Zool. 41, 687611–687697. doi:10.1093/icb/41.3.687
Callier, V. (2020). Inner Workings: Understanding the Evolution of Cell Types to Explain the Roots of Animal Diversity. Proc. Natl. Acad. Sci. U.S.A. 117, 5547–5549. doi:10.1073/pnas.2002403117
Cattell, M., Lai, S., Cerny, R., and Medeiros, D. M. (2011). A New Mechanistic Scenario for the Origin and Evolution of Vertebrate Cartilage. PLoS One 6, e22474. doi:10.1371/journal.pone.0022474
Cecchini, M. G., Hofstetter, W., Halasy, J., Wetterwald, A., and Felix, R. (1997). Role of CSF-1 in Bone and Bone Marrow Development. Mol. Reprod. Dev. 46, 75–84. ; discussion 83-74. doi:10.1002/(sici)1098-2795(199701)46:1<75::aid-mrd12>3.0.co;2-2
Chakraborty, P., Dash, S. P., and Sarangi, P. P. (2020). The Role of Adhesion Protein Fibulin7 in Development and Diseases. Mol. Med. 26, 47. doi:10.1186/s10020-020-00169-z
Chen, D., Kim, D. J., Shen, J., Zou, Z., and O'Keefe, R. J. (2020). Runx2 Plays a Central Role in Osteoarthritis Development. J. Orthop. Transl. 23, 132–139. doi:10.1016/j.jot.2019.11.008
Cheng, A., and Genever, P. G. (2010). SOX9 Determines RUNX2 Transactivity by Directing Intracellular Degradation. J. Bone Min. Res. 25, 2680–2689. doi:10.1002/jbmr.174
Chiba, K., Rahman, M. E., Ishikawa, H., and Endo, A. (1995). The Timing of Appearance of Ossification Centers of Carpal and Tarsal Bones in Mouse Newborns. Congenit. Anomalies 35, 189–197. doi:10.1111/j.1741-4520.1995.tb00610.x
Cole, A. G., and Hall, B. K. (2009). Cartilage Differentiation in Cephalopod Molluscs. Zoology 112, 2–15. doi:10.1016/j.zool.2008.01.003
Cole, A. (2011). A Review of Diversity in the Evolution and Development of Cartilage: the Search for the Origin of the Chondrocyte. eCM 21, 122–129. doi:10.22203/ecm.v021a10
Conen, K. L., Nishimori, S., Provot, S., and Kronenberg, H. M. (2009). The Transcriptional Cofactor Lbh Regulates Angiogenesis and Endochondral Bone Formation during Fetal Bone Development. Dev. Biol. 333, 348–358. doi:10.1016/j.ydbio.2009.07.003
Couly, G. F., Coltey, P. M., and Le Douarin, N. M. (1993). The Triple Origin of Skull in Higher Vertebrates: a Study in Quail-Chick Chimeras. Development 117, 409–429. doi:10.1242/dev.117.2.409
Dale, R. M., and Topczewski, J. (2011). Identification of an Evolutionarily Conserved Regulatory Element of the Zebrafish Col2a1a Gene. Dev. Biol. 357, 518–531. doi:10.1016/j.ydbio.2011.06.020
D'Angelo, M., Yan, Z., Nooreyazdan, M., Pacifici, M., Sarment, D. S., Billings, P. C., et al. (2000). MMP-13 Is Induced during Chondrocyte Hypertrophy. J. Cell. Biochem. 77, 678–693. doi:10.1002/(sici)1097-4644(20000615)77:4<678::aid-jcb15>3.0.co;2-p
Dao, D. Y., Yang, X., Flick, L. M., Chen, D., Hilton, M. J., and O'Keefe, R. J. (2010). Axin2 Regulates Chondrocyte Maturation and Axial Skeletal Development. J. Orthop. Res. 28, 89–95. doi:10.1002/jor.20954
Davidson, E. H., and Erwin, D. H. (2006). Gene Regulatory Networks and the Evolution of Animal Body Plans. Science 311, 796–800. doi:10.1126/science.1113832
Ding, M., Lu, Y., Abbassi, S., Li, F., Li, X., Song, Y., et al. (2012). Targeting Runx2 Expression in Hypertrophic Chondrocytes Impairs Endochondral Ossification during Early Skeletal Development. J. Cell. Physiol. 227, 3446–3456. doi:10.1002/jcp.24045
Dobin, A., Davis, C. A., Schlesinger, F., Drenkow, J., Zaleski, C., Jha, S., et al. (2012). STAR: Ultrafast Universal RNA-Seq Aligner. Bioinformatics 29, 15–21. doi:10.1093/bioinformatics/bts635
Drissi, H., Luc, Q., Shakoori, R., Chuva De Sousa Lopes, S., Choi, J.-Y., Terry, A., et al. (2000). Transcriptional Autoregulation of the Bone Related CBFA1/RUNX2 Gene. J. Cell. Physiol. 184, 341–350. doi:10.1002/1097-4652(200009)184:3<341::aid-jcp8>3.0.co;2-z
Duan, X., Cai, L., Schmidt, E. J., Shen, J., Tycksen, E. D., O'Keefe, R. J., et al. (2020). RNA-seq Analysis of Chondrocyte Transcriptome Reveals Genetic Heterogeneity in LG/J and SM/J Murine Strains. Osteoarthr. Cartil. 28, 516–527. doi:10.1016/j.joca.2020.01.001
Ducy, P., Zhang, R., Geoffroy, V., Ridall, A. L., and Karsenty, G. (1997). Osf2/Cbfa1: a Transcriptional Activator of Osteoblast Differentiation. Cell 89, 747–754. doi:10.1016/s0092-8674(00)80257-3
Dy, P., Wang, W., Bhattaram, P., Wang, Q., Wang, L., Ballock, R. T., et al. (2012). Sox9 Directs Hypertrophic Maturation and Blocks Osteoblast Differentiation of Growth Plate Chondrocytes. Dev. Cell 22, 597–609. doi:10.1016/j.devcel.2011.12.024
Eames, B. F., and Helms, J. A. (2004). Conserved Molecular Program Regulating Cranial and Appendicular Skeletogenesis. Dev. Dyn. 231, 4–13. doi:10.1002/dvdy.20134
Eames, B. F., de la Fuente, L., and Helms, J. A. (2003). Molecular Ontogeny of the Skeleton. Birth Defect Res. C 69, 93–101. doi:10.1002/bdrc.10016
Eames, B. F., Sharpe, P. T., and Helms, J. A. (2004). Hierarchy Revealed in the Specification of Three Skeletal Fates by Sox9 and Runx2. Dev. Biol. 274, 188–200. doi:10.1016/j.ydbio.2004.07.006
Eames, B. F., Yan, Y.-L., Swartz, M. E., Levic, D. S., Knapik, E. W., Postlethwait, J. H., et al. (2011). Mutations in Fam20b and Xylt1 Reveal that Cartilage Matrix Controls Timing of Endochondral Ossification by Inhibiting Chondrocyte Maturation. PLoS Genet. 7, e1002246. doi:10.1371/journal.pgen.1002246
Eames, B. F., Medeiros, D. M., and Adameyko, I. (2020). Evolving Neural Crest Cells. Boca Raton, Florida, United States: CRC Press.
Erkenbrack, E. M. (2016). Divergence of Ectodermal and Mesodermal Gene Regulatory Network Linkages in Early Development of Sea Urchins. Proc. Natl. Acad. Sci. U. S. A. 113, E7202–E7211. doi:10.1073/pnas.1612820113
Farquharson, C., Whitehead, C. C., and Loveridge, N. (1994). Alterations in Glycosaminoglycan Concentration and Sulfation during Chondrocyte Maturation. Calcif. Tissue Int. 54, 296–303. doi:10.1007/bf00295954
Fazenda, C., Simões, B., Kelsh, R. N., Cancela, M. L., and Conceição, N. (2010). Dual Transcriptional Regulation by Runx2 of Matrix Gla Protein in Xenopus laevis. Gene 450, 94–102. doi:10.1016/j.gene.2009.10.007
Ferguson, C. M., Miclau, T., Hu, D., Alpern, E., and Helms, J. A. (1998). Common Molecular Pathways in Skeletal Morphogenesis and Repair. Ann. N. Y. Acad. Sci. 857, 33–42. doi:10.1111/j.1749-6632.1998.tb10105.x
Fonseca, B. F., Couly, G., and Dupin, E. (2017). Respective Contribution of the Cephalic Neural Crest and Mesoderm to SIX1-Expressing Head Territories in the Avian Embryo. BMC Dev. Biol. 17, 13. doi:10.1186/s12861-017-0155-z
Garcez, R. C., Le Douarin, N. M., and Creuzet, S. E. (2014). Combinatorial Activity of Six1-2-4 Genes in Cephalic Neural Crest Cells Controls Craniofacial and Brain Development. Cell Mol. Life Sci. 71, 2149–2164. doi:10.1007/s00018-013-1477-z
Gentili/snm, C., >, R., and Cancedda, R. (2009). Cartilage and Bone Extracellular Matrix. Cpd 15, 1334–1348. doi:10.2174/138161209787846739
Geurtzen, K., Knopf, F., Wehner, D., Huitema, L. F. A., Schulte-Merker, S., and Weidinger, G. (2014). Mature Osteoblasts Dedifferentiate in Response to Traumatic Bone Injury in the Zebrafish Fin and Skull. Development 141, 2225–2234. doi:10.1242/dev.105817
Gómez-Picos, P., and Eames, B. F. (2015). On the Evolutionary Relationship between Chondrocytes and Osteoblasts. Front. Genet. 6, 297. doi:10.3389/fgene.2015.00297
Grogan, S. P., Olee, T., Hiraoka, K., and Lotz, M. K. (2008). Repression of Chondrogenesis through Binding of Notch Signaling Proteins HES-1 and HEY-1 to N-Box Domains in the COL2A1 Enhancer Site. Arthritis Rheum. 58, 2754–2763. doi:10.1002/art.23730
Gross, S., Krause, Y., Wuelling, M., and Vortkamp, A. (2012). Hoxa11 and Hoxd11 Regulate Chondrocyte Differentiation Upstream of Runx2 and Shox2 in Mice. PloS one 7, e43553. doi:10.1371/journal.pone.0043553
Gu, S., Boyer, T. G., and Naski, M. C. (2012). Basic Helix-Loop-Helix Transcription Factor Twist1 Inhibits Transactivator Function of Master Chondrogenic Regulator Sox9. J. Biol. Chem. 287, 21082–21092. doi:10.1074/jbc.m111.328567
Gu, J., Lu, Y., Li, F., Qiao, L., Wang, Q., Li, N., et al. (2014). Identification and Characterization of the Novel Col10a1 Regulatory Mechanism during Chondrocyte Hypertrophic Differentiation. Cell Death Dis. 5, e1469. doi:10.1038/cddis.2014.444
Hall, B. K., and Gillis, J. A. (2013). Incremental Evolution of the Neural Crest, Neural Crest Cells and Neural Crest-Derived Skeletal Tissues. J. Anat. 222, 19–31. doi:10.1111/j.1469-7580.2012.01495.x
Hamburger, V., and Hamilton, H. L. (1951). A Series of Normal Stages in the Development of the Chick Embryo. J. Morphol. 88, 49–92. doi:10.1002/jmor.1050880104
Han, J., Ishii, M., Bringas, P., Maas, R. L., Maxson, R. E., and Chai, Y. (2007). Concerted Action of Msx1 and Msx2 in Regulating Cranial Neural Crest Cell Differentiation during Frontal Bone Development. Mech. Dev. 124, 729–745. doi:10.1016/j.mod.2007.06.006
Hasegawa, M., Yoshida, T., and Sudo, A. (2020). Tenascin-C in Osteoarthritis and Rheumatoid Arthritis. Front. Immunol. 11, 577015. doi:10.3389/fimmu.2020.577015
He, X., Ohba, S., Hojo, H., and McMahon, A. P. (2016). AP-1 Family Members Act with Sox9 to Promote Chondrocyte Hypertrophy. Development 143, 3012–3023. doi:10.1242/dev.134502
Hecht, J., Hayes, E., Haynes, R., and Cole, W. (2005). COMP Mutations, Chondrocyte Function and Cartilage Matrix. Matrix Biol. 23, 525–533. doi:10.1016/j.matbio.2004.09.006
Heilig, J., Paulsson, M., and Zaucke, F. (2016). Insulin-like Growth Factor 1 Receptor (IGF1R) Signaling Regulates Osterix Expression and Cartilage Matrix Mineralization during Endochondral Ossification. Bone 83, 48–57. doi:10.1016/j.bone.2015.10.007
Higashihori, N., Song, Y., and Richman, J. M. (2008). Expression and Regulation of the Decoy Bone Morphogenetic Protein receptorBAMBIin the Developing Avian Face. Dev. Dyn. 237, 1500–1508. doi:10.1002/dvdy.21529
Higashiyama, R., Miyaki, S., Yamashita, S., Yoshitaka, T., Lindman, G., Ito, Y., et al. (2010). Correlation between MMP-13 and HDAC7 Expression in Human Knee Osteoarthritis. Mod. Rheumatol. 20, 11–17. doi:10.1007/s10165-009-0224-7
Hirose, K., Ishimoto, T., Usami, Y., Sato, S., Oya, K., Nakano, T., et al. (2020). Overexpression of Fam20C in Osteoblast In Vivo Leads to Increased Cortical Bone Formation and Osteoclastic Bone Resorption. Bone 138, 115414. doi:10.1016/j.bone.2020.115414
Hoffmann, A., and Gross, G. (2001). BMP Signaling Pathways in Cartilage and Bone Formation. Crit. Rev. Eukaryot. Gene Expr. 11, 23–45. doi:10.1615/critreveukargeneexpr.v11.i1-3.20
Hojo, H., and Ohba, S. (2019). Insights into Gene Regulatory Networks in Chondrocytes. Int. J. Mol. Sci. 20. 6324. doi:10.3390/ijms20246324
Holleville, N., Matéos, S., Bontoux, M., Bollerot, K., and Monsoro-Burq, A. H. (2007). Dlx5 Drives Runx2 Expression and Osteogenic Differentiation in Developing Cranial Suture Mesenchyme. Dev. Biol. 304, 860–874. doi:10.1016/j.ydbio.2007.01.003
Hong, J.-H., Hwang, E. S., McManus, M. T., Amsterdam, A., Tian, Y., Kalmukova, R., et al. (2005). TAZ, a Transcriptional Modulator of Mesenchymal Stem Cell Differentiation. Science 309, 1074–1078. doi:10.1126/science.1110955
Ikegami, D., Akiyama, H., Suzuki, A., Nakamura, T., Nakano, T., Yoshikawa, H., et al. (2011). Sox9 Sustains Chondrocyte Survival and Hypertrophy in Part through Pik3ca-Akt Pathways. Development 138, 1507–1519. doi:10.1242/dev.057802
Inada, M., Yasui, T., Nomura, S., Miyake, S., Deguchi, K., Himeno, M., et al. (1999). Maturational Disturbance of Chondrocytes inCbfa1-Deficient Mice. Dev. Dyn. 214, 279–290. doi:10.1002/(sici)1097-0177(199904)214:4<279::aid-aja1>3.0.co;2-w
Ionescu, A., Kozhemyakina, E., Nicolae, C., Kaestner, K. H., Olsen, B. R., and Lassar, A. B. (2012). FoxA Family Members Are Crucial Regulators of the Hypertrophic Chondrocyte Differentiation Program. Dev. Cell 22, 927–939. doi:10.1016/j.devcel.2012.03.011
Jandzik, D., Garnett, A. T., Square, T. A., Cattell, M. V., Yu, J.-K., and Medeiros, D. M. (2015). Evolution of the New Vertebrate Head by Co-option of an Ancient Chordate Skeletal Tissue. Nature 518, 534–537. doi:10.1038/nature14000
Janvier, P., Lund, R., and Grogan, E. D. (2004). Further Consideration of the Earliest Known lamprey,Hardistiella montanensisJanvier and Lund, 1983, from the Carboniferous of Bear Gulch, Montana, U.S.A. J. Vertebrate Paleontology 24, 742–743. doi:10.1671/0272-4634(2004)024[0742:fcotek]2.0.co;2
Johnson, K., Reynard, L. N., and Loughlin, J. (2015). Functional Characterisation of the Osteoarthritis Susceptibility Locus at Chromosome 6q14.1 Marked by the Polymorphism Rs9350591. BMC Med. Genet. 16, 81. doi:10.1186/s12881-015-0215-9
Jung, J. C., and Tsonis, P. A. (1998). Role of 5' HoxD Genes in Chondrogenesis In Vitro. Int. J. Dev. Biol. 42, 609–615.
Karlsson, C., Brantsing, C., Kageyama, R., and Lindahl, A. (2010). HES1 and HES5 Are Dispensable for Cartilage and Endochondral Bone Formation. Cells Tissues Organs 192, 17–27. doi:10.1159/000280416
Kawane, T., Qin, X., Jiang, Q., Miyazaki, T., Komori, H., Yoshida, C. A., et al. (2018). Runx2 Is Required for the Proliferation of Osteoblast Progenitors and Induces Proliferation by Regulating Fgfr2 and Fgfr3. Sci. Rep. 8, 13551. doi:10.1038/s41598-018-31853-0
Kerkhofs, J., Roberts, S. J., Luyten, F. P., Van Oosterwyck, H., and Geris, L. (2012). Relating the Chondrocyte Gene Network to Growth Plate Morphology: from Genes to Phenotype. PLoS One 7, e34729. doi:10.1371/journal.pone.0034729
Khosravi, P., Gazestani, V. H., Pirhaji, L., Law, B., Sadeghi, M., Goliaei, B., et al. (2015). Inferring Interaction Type in Gene Regulatory Networks Using Co-expression Data. Algorithms Mol. Biol. 10, 23. doi:10.1186/s13015-015-0054-4
Knight, R. D., and Schilling, T. F. (2006). Cranial Neural Crest and Development of the Head Skeleton. Adv. Exp. Med. Biol. 589, 120–133. doi:10.1007/978-0-387-46954-6_7
Komori, T., and Kishimoto, T. (1998). Cbfa1 in Bone Development. Curr. Opin. Genet. Dev. 8, 494–499. doi:10.1016/s0959-437x(98)80123-8
Komori, T., Yagi, H., Nomura, S., Yamaguchi, A., Sasaki, K., Deguchi, K., et al. (1997). Targeted Disruption of Cbfa1 Results in a Complete Lack of Bone Formation Owing to Maturational Arrest of Osteoblasts. Cell 89, 755–764. doi:10.1016/s0092-8674(00)80258-5
Komori, T. (2017). Roles of Runx2 in Skeletal Development. Adv. Exp. Med. Biol. 962, 83–93. doi:10.1007/978-981-10-3233-2_6
Komori, T. (2018). Runx2, an Inducer of Osteoblast and Chondrocyte Differentiation. Histochem Cell Biol. 149, 313–323. doi:10.1007/s00418-018-1640-6
Komori, T. (2019). Regulation of Proliferation, Differentiation and Functions of Osteoblasts by Runx2. Int. J. Mol. Sci. 20. 1694. doi:10.3390/ijms20071694
Koziel, L., Wuelling, M., Schneider, S., and Vortkamp, A. (2005). Gli3 Acts as a Repressor Downstream of Ihh in Regulating Two Distinct Steps of Chondrocyte Differentiation. Development 132, 5249–5260. doi:10.1242/dev.02097
Kraus, J. M., Giovannone, D., Rydzik, R., Balsbaugh, J. L., Moss, I. L., Schwedler, J. L., et al. (2022). Notch Signaling Enhances Bone Regeneration in the Zebrafish Mandible. Development 149. dev199995. doi:10.1242/dev.199995
Kruger, C., and Kappen, C. (2010). Expression of Cartilage Developmental Genes in Hoxc8- and Hoxd4-Transgenic Mice. PLoS One 5, e8978. doi:10.1371/journal.pone.0008978
Kumar, A., Chalamalasetty, R. B., Kennedy, M. W., Thomas, S., Inala, S. N., Garriock, R. J., et al. (2016a). Zfp703 Is a Wnt/β-Catenin Feedback Suppressor Targeting the β-Catenin/Tcf1 Complex. Mol. Cell Biol. 36, 1793–1802. doi:10.1128/mcb.01010-15
Kumar, S., Rathkolb, B., Kemter, E., Sabrautzki, S., Michel, D., Adler, T., et al. (2016b). Generation and Standardized, Systemic Phenotypic Analysis of Pou3f3L423P Mutant Mice. PLoS One 11, e0150472. doi:10.1371/journal.pone.0150472
Kurakazu, I., Akasaki, Y., Hayashida, M., Tsushima, H., Goto, N., Sueishi, T., et al. (2019). FOXO1 Transcription Factor Regulates Chondrogenic Differentiation through Transforming Growth Factor β1 Signaling. J. Biol. Chem. 294, 17555–17569. doi:10.1074/jbc.ra119.009409
Lamas, J. R., Rodríguez-Rodríguez, L., Vigo, A. G., Álvarez-Lafuente, R., López-Romero, P., Marco, F., et al. (2010). Large-scale Gene Expression in Bone Marrow Mesenchymal Stem Cells: a Putative Role for COL10A1 in Osteoarthritis. Ann. Rheumatic Dis. 69, 1880–1885. doi:10.1136/ard.2009.122564
Le Douarin, N., LeDouarin, N. M., and Kalcheim, C. (1999). The Neural Crest. Cambridge, United Kingdom: Cambridge University Press.
Leboy, P. S., Shapiro, I. M., Uschmann, B. D., Oshima, O., and Lin, D. (1988). Gene Expression in Mineralizing Chick Epiphyseal Cartilage. J. Biol. Chem. 263, 8515–8520. doi:10.1016/s0021-9258(18)68508-4
Lecanda, F., Avioli, L. V., and Cheng, S.-L. (1997). Regulation of Bone Matrix Protein Expression and Induction of Differentiation of Human Osteoblasts and Human Bone Marrow Stromal Cells by Bone Morphogenetic Protein-2. J. Cell. Biochem. 67, 386–398. doi:10.1002/(sici)1097-4644(19971201)67:3<386::aid-jcb10>3.0.co;2-b
Lee, K.-S., Kim, H.-J., Li, Q.-L., Chi, X.-Z., Ueta, C., Komori, T., et al. (2000). Runx2 Is a Common Target of Transforming Growth Factor β1 and Bone Morphogenetic Protein 2, and Cooperation between Runx2 and Smad5 Induces Osteoblast-specific Gene Expression in the Pluripotent Mesenchymal Precursor Cell Line C2C12. Mol. Cell Biol. 20, 8783–8792. doi:10.1128/mcb.20.23.8783-8792.2000
Lefebvre, V., and de Crombrugghe, B. (1998). Toward Understanding SOX9 Function in Chondrocyte Differentiation. Matrix Biol. 16, 529–540. doi:10.1016/s0945-053x(98)90065-8
Lefebvre, V., and Dvir-Ginzberg, M. (2017). SOX9 and the Many Facets of its Regulation in the Chondrocyte Lineage. Connect. tissue Res. 58, 2–14. doi:10.1080/03008207.2016.1183667
Lefebvre, V., Huang, W., Harley, V. R., Goodfellow, P. N., and de Crombrugghe, B. (1997). SOX9 Is a Potent Activator of the Chondrocyte-specific Enhancer of the Pro alpha1(II) Collagen Gene. Mol. Cell Biol. 17, 2336–2346. doi:10.1128/mcb.17.4.2336
Lefebvre, V., Behringer, R. R., and de Crombrugghe, B. (2001). L-Sox5, Sox6 and Sox9 Control Essential Steps of the Chondrocyte Differentiation Pathway. Osteoarthr. Cartil. 9 (A), S69–S75. doi:10.1053/joca.2001.0447
Leung, V. Y. L., Gao, B., Leung, K. K. H., Melhado, I. G., Wynn, S. L., Au, T. Y. K., et al. (2011). SOX9 Governs Differentiation Stage-specific Gene Expression in Growth Plate Chondrocytes via Direct Concomitant Transactivation and Repression. PLoS Genet. 7, e1002356. doi:10.1371/journal.pgen.1002356
Li, S., Kong, H., Yao, N., Yu, Q., Wang, P., Lin, Y., et al. (2011). The Role of Runt-Related Transcription Factor 2 (Runx2) in the Late Stage of Odontoblast Differentiation and Dentin Formation. Biochem. Biophysical Res. Commun. 410, 698–704. doi:10.1016/j.bbrc.2011.06.065
Li, J., Zhou, L., Ouyang, X., and He, P. (2021). Transcription Factor-7-Like-2 (TCF7L2) in Atherosclerosis: A Potential Biomarker and Therapeutic Target. Front. Cardiovasc. Med. 8, 701279. doi:10.3389/fcvm.2021.701279
Lian, J., and Stein, G. (2003). Runx2/Cbfa1: a Multifunctional Regulator of Bone Formation. Cpd 9, 2677–2685. doi:10.2174/1381612033453659
Liu, C.-F., and Lefebvre, V. (2015). The Transcription Factors SOX9 and SOX5/SOX6 Cooperate Genome-wide through Super-enhancers to Drive Chondrogenesis. Nucleic Acids Res. 43, 8183–8203. doi:10.1093/nar/gkv688
Liu, C.-j., Zhang, Y., Xu, K., Parsons, D., Alfonso, D., and Di Cesare, P. E. (2007). Transcriptional Activation of Cartilage Oligomeric Matrix Protein by Sox9, Sox5, and Sox6 Transcription Factors and CBP/p300 Coactivators. Front. Biosci. a J. virtual Libr. 12, 3899–3910. doi:10.2741/2359
Liu, C. F., Angelozzi, M., Haseeb, A., and Lefebvre, V. (2018). SOX9 Is Dispensable for the Initiation of Epigenetic Remodeling and the Activation of Marker Genes at the Onset of Chondrogenesis. Development 145, dev164459. doi:10.1242/dev.164459
Longabaugh, W. J. R., Davidson, E. H., and Bolouri, H. (2005). Computational Representation of Developmental Genetic Regulatory Networks. Dev. Biol. 283, 1–16. doi:10.1016/j.ydbio.2005.04.023
Lu, Y., Qiao, L., Lei, G., Mira, R. R., Gu, J., and Zheng, Q. (2014). Col10a1 Gene Expression and Chondrocyte Hypertrophy during Skeletal Development and Disease. Front. Biol. 9, 195–204. doi:10.1007/s11515-014-1310-6
Lui, J. C., Nilsson, O., and Baron, J. (2014). Recent Research on the Growth Plate: Recent Insights into the Regulation of the Growth Plate. J. Mol. Endocrinol. 53, T1–T9. doi:10.1530/jme-14-0022
Lui, J. C., Yue, S., Lee, A., Kikani, B., Temnycky, A., Barnes, K. M., et al. (2019). Persistent Sox9 Expression in Hypertrophic Chondrocytes Suppresses Transdifferentiation into Osteoblasts. Bone 125, 169–177. doi:10.1016/j.bone.2019.05.027
Lv, C., Li, Y., Xu, J., Cao, H., Li, X., Ma, B., et al. (2015). Association of SPP1 Promoter Variants with Hip Osteoarthritis Susceptibility in Chinese Population. Gene 564, 9–13. doi:10.1016/j.gene.2015.03.036
Magee, C., Nurminskaya, M., Faverman, L., Galera, P., and Linsenmayer, T. F. (2005). SP3/SP1 Transcription Activity Regulates Specific Expression of CollagenType X in HypertrophicChondrocytes. J. Biol. Chem. 280, 25331–25338. doi:10.1074/jbc.m412549200
McCall, M. N. (2013). Estimation of Gene Regulatory Networks. Postdoc J. 1, 60–69. doi:10.14304/surya.jpr.v1n1.7
McGee-Lawrence, M. E., Li, X., Bledsoe, K. L., Wu, H., Hawse, J. R., Subramaniam, M., et al. (2013). Runx2 Protein Represses Axin2 Expression in Osteoblasts and Is Required for Craniosynostosis in Axin2-Deficient Mice*. J. Biol. Chem. 288, 5291–5302. doi:10.1074/jbc.m112.414995
Meech, R., Edelman, D. B., Jones, F. S., and Makarenkova, H. P. (2005). The Homeobox Transcription Factor Barx2 Regulates Chondrogenesis during Limb Development. Development 132, 2135–2146. doi:10.1242/dev.01811
Mentlein, R. (2007). Targeting Pleiotropin to Treat Osteoarthritis. Expert Opin. Ther. Targets 11, 861–867. doi:10.1517/14728222.11.7.861
Meulemans, D., and Bronner-Fraser, M. (2005). Central Role of Gene Cooption in Neural Crest Evolution. J. Exp. Zool. 304B, 298–303. doi:10.1002/jez.b.21047
Meulemans, D., and Bronner-Fraser, M. (2007). Insights from Amphioxus into the Evolution of Vertebrate Cartilage. PLoS One 2, e787. doi:10.1371/journal.pone.0000787
Milz, S., Boszczyk, A., and Putz, R. (2002). Development and Functional Structure of the Epiphyseal Plate. Orthopäde 31, 835–840. doi:10.1007/s00132-002-0359-0
Mitgutsch, C., Wimmer, C., Sánchez-Villagra, M. R., Hahnloser, R., and Schneider, R. A. (2011). Timing of Ossification in Duck, Quail, and Zebra Finch: Lntraspecific Variation, Heterochronies, and Life History Evolution. Zoological Sci. 28, 491410–491500. doi:10.2108/zsj.28.491
Mokuda, S., Nakamichi, R., Matsuzaki, T., Ito, Y., Sato, T., Miyata, K., et al. (2019). Wwp2 Maintains Cartilage Homeostasis through Regulation of Adamts5. Nat. Commun. 10, 2429. doi:10.1038/s41467-019-10177-1
Moraes, F., Nóvoa, A., Jerome-Majewska, L. A., Papaioannou, V. E., and Mallo, M. (2005). Tbx1 Is Required for Proper Neural Crest Migration and to Stabilize Spatial Patterns during Middle and Inner Ear Development. Mech. Dev. 122, 199–212. doi:10.1016/j.mod.2004.10.004
Murdoch, B., DelConte, C., and García-Castro, M. I. (2012). Pax7 Lineage Contributions to the Mammalian Neural Crest. PLoS One 7, e41089. doi:10.1371/journal.pone.0041089
Nagy, A., Kénesi, E., Rentsendorj, O., Molnár, A., Szénási, T., Sinkó, I., et al. (2011). Evolutionarily Conserved, Growth Plate Zone-specific Regulation of the Matrilin-1 Promoter: L-Sox5/Sox6 and Nfi Factors Bound Near TATA Finely Tune Activation by Sox9. Mol. Cell Biol. 31, 686–699. doi:10.1128/mcb.00019-10
Nakatani, T., and Partridge, N. C. (2017). MEF2C Interacts with C-FOS in PTH-Stimulated Mmp13 Gene Expression in Osteoblastic Cells. Endocrinology 158, 3778–3791. doi:10.1210/en.2017-00159
Nalbant, D., Youn, H., Nalbant, S. I., Sharma, S., Cobos, E., Beale, E. G., et al. (2005). FAM20: an Evolutionarily Conserved Family of Secreted Proteins Expressed in Hematopoietic Cells. BMC Genomics 6, 11. doi:10.1186/1471-2164-6-11
Neefjes, M., van Caam, A. P. M., and van der Kraan, P. M. (2020). Transcription Factors in Cartilage Homeostasis and Osteoarthritis. Biology 9, 290. doi:10.3390/biology9090290
Neufeld, S. J., Wang, F., and Cobb, J. (2014). Genetic Interactions between Shox2 and Hox Genes during the Regional Growth and Development of the Mouse Limb. Genetics 198, 1117–1126. doi:10.1534/genetics.114.167460
Nicolae, C., Ko, Y.-P., Miosge, N., Niehoff, A., Studer, D., Enggist, L., et al. (2007). Abnormal Collagen Fibrils in Cartilage of Matrilin-1/matrilin-3-Deficient Mice. J. Biol. Chem. 282, 22163–22175. doi:10.1074/jbc.m610994200
Niu, P., Zhong, Z., Wang, M., Huang, G., Xu, S., Hou, Y., et al. (2017). Zinc Finger Transcription Factor Sp7/Osterix Acts on Bone Formation and Regulates Col10a1a Expression in Zebrafish. Sci. Bull. 62, 174–184. doi:10.1016/j.scib.2017.01.009
Noden, D. M. (1982). Patterns and Organization of Craniofacial Skeletogenic and Myogenic Mesenchyme: a Perspective. Prog. Clin. Biol. Res. 101, 167–203.
Oh, C.-d., Lu, Y., Liang, S., Mori-Akiyama, Y., Chen, D., de Crombrugghe, B., et al. (2014). SOX9 Regulates Multiple Genes in Chondrocytes, Including Genes Encoding ECM Proteins, ECM Modification Enzymes, Receptors, and Transporters. PLoS One 9, e107577. doi:10.1371/journal.pone.0107577
Ohba, S., He, X., Hojo, H., and McMahon, A. P. (2015). Distinct Transcriptional Programs Underlie Sox9 Regulation of the Mammalian Chondrocyte. Cell Rep. 12, 229–243. doi:10.1016/j.celrep.2015.06.013
Patton, J. T., and Kaufman, M. H. (1995). The Timing of Ossification of the Limb Bones, and Growth Rates of Various Long Bones of the Fore and Hind Limbs of the Prenatal and Early Postnatal Laboratory Mouse. J. Anat. 186 (Pt 1), 175–185.
Peacock, J. D., Huk, D. J., Ediriweera, H. N., and Lincoln, J. (2011). Sox9 Transcriptionally Represses Spp1 to Prevent Matrix Mineralization in Maturing Heart Valves and Chondrocytes. PLoS One 6, e26769. doi:10.1371/journal.pone.0026769
Pelttari, K., Rua, L. A., Mumme, M., Manferdini, C., Darwiche, S., Khalil, A., et al. (2020). Engineered Nasal Cartilage for the Repair of Osteoarthritic Knee Cartilage Defects. Cytotherapy 22, S14. doi:10.1016/j.jcyt.2020.03.477
Peter, I. S., and Davidson, E. H. (2011). A Gene Regulatory Network Controlling the Embryonic Specification of Endoderm. Nature 474, 635–639. doi:10.1038/nature10100
Plouhinec, J.-L., Roche, D. D., Pegoraro, C., Figueiredo, A. L., Maczkowiak, F., Brunet, L. J., et al. (2014). Pax3 and Zic1 Trigger the Early Neural Crest Gene Regulatory Network by the Direct Activation of Multiple Key Neural Crest Specifiers. Dev. Biol. 386, 461–472. doi:10.1016/j.ydbio.2013.12.010
Qin, X., Jiang, Q., Miyazaki, T., and Komori, T. (2018). Runx2 Regulates Cranial Suture Closure by Inducing Hedgehog, Fgf, Wnt and Pthlh Signaling Pathway Gene Expressions in Suture Mesenchymal Cells. Hum. Mol. Genet. 28, 896–911. doi:10.1093/hmg/ddy386
Qin, X., Jiang, Q., Nagano, K., Moriishi, T., Miyazaki, T., Komori, H., et al. (2020). Runx2 Is Essential for the Transdifferentiation of Chondrocytes into Osteoblasts. PLoS Genet. 16, e1009169. doi:10.1371/journal.pgen.1009169
Rice, S. J., Cheung, K., Reynard, L. N., and Loughlin, J. (2019). Discovery and Analysis of Methylation Quantitative Trait Loci (mQTLs) Mapping to Novel Osteoarthritis Genetic Risk Signals. Osteoarthr. Cartil. 27, 1545–1556. doi:10.1016/j.joca.2019.05.017
Robledo, R. F., Rajan, L., Li, X., and Lufkin, T. (2002). The Dlx5 and Dlx6 Homeobox Genes Are Essential for Craniofacial, Axial, and Appendicular Skeletal Development. Genes Dev. 16, 1089–1101. doi:10.1101/gad.988402
Rosset, E. M., and Bradshaw, A. D. (2016). SPARC/osteonectin in Mineralized Tissue. Matrix Biol. 52-54, 78–87. doi:10.1016/j.matbio.2016.02.001
Russell, M. W., Raeker, M. O., Geisler, S. B., Thomas, P. E., Simmons, T. A., Bernat, J. A., et al. (2014). Functional Analysis of Candidate Genes in 2q13 Deletion Syndrome Implicates FBLN7 and TMEM87B Deficiency in Congenital Heart Defects and FBLN7 in Craniofacial Malformations. Hum. Mol. Genet. 23, 4272–4284. doi:10.1093/hmg/ddu144
Rychel, A. L., and Swalla, B. J. (2007). Development and Evolution of Chordate Cartilage. J. Exp. Zoology Part B Mol. Dev. Evol. 308B, 325–335. doi:10.1002/jez.b.21157
Rychel, A. L., Smith, S. E., Shimamoto, H. T., and Swalla, B. J. (2006). Evolution and Development of the Chordates: Collagen and Pharyngeal Cartilage. Mol. Biol. Evol. 23, 541–549. doi:10.1093/molbev/msj055
Sachkova, M., and Burkhardt, P. (2019). Exciting Times to Study the Identity and Evolution of Cell Types. Development 146. dev178996. doi:10.1242/dev.178996
Seal, S., and Monsoro-Burq, A. H. (2020). Insights into the Early Gene Regulatory Network Controlling Neural Crest and Placode Fate Choices at the Neural Border. Front. Physiol. 11, 608812. doi:10.3389/fphys.2020.608812
Sebastian, A., McCool, J. L., Hum, N. R., Murugesh, D. K., Wilson, S. P., Christiansen, B. A., et al. (2021). Single-Cell RNA-Seq Reveals Transcriptomic Heterogeneity and Post-Traumatic Osteoarthritis-Associated Early Molecular Changes in Mouse Articular Chondrocytes. Cells 10, 1462. doi:10.3390/cells10061462
Sekiya, I., Tsuji, K., Koopman, P., Watanabe, H., Yamada, Y., Shinomiya, K., et al. (2000). SOX9 Enhances Aggrecan Gene Promoter/enhancer Activity and Is Up-Regulated by Retinoic Acid in a Cartilage-Derived Cell Line, TC6. J. Biol. Chem. 275, 10738–10744. doi:10.1074/jbc.275.15.10738
Shen, J., Li, J., Wang, B., Jin, H., Wang, M., Zhang, Y., et al. (2013). Deletion of the Transforming Growth Factor β Receptor Type II Gene in Articular Chondrocytes Leads to a Progressive Osteoarthritis-like Phenotype in Mice. Arthritis & Rheumatism 65, 3107–3119. doi:10.1002/art.38122
Shi, M., Sun, M., Wang, C., Shen, Y., Wang, Y., and Yan, S. 2022. Therapeutic Potential of POU3F3, a Novel Long Non-coding RNA Alleviates the Pathogenesis of Osteoarthritis by Regulating miR-29a-3p/FOXO3 axis. Curr. Gene Ther. 1. 1. doi:10.2174/1566523222666220309150722
Si, Y., Liu, P., Li, P., and Brutnell, T. P. (2014). Model-based Clustering for RNA-Seq Data. Bioinformatics 30, 197–205. doi:10.1093/bioinformatics/btt632
Simões-Costa, M., and Bronner, M. E. (2013). Insights into Neural Crest Development and Evolution from Genomic Analysis. Genome Res. 23, 1069–1080. doi:10.1101/gr.157586.113
Simões-Costa, M., and Bronner, M. E. (2015). Establishing Neural Crest Identity: a Gene Regulatory Recipe. Development 142, 242–257. doi:10.1242/dev.105445
Smits, P., Li, P., Mandel, J., Zhang, Z., Deng, J. M., Behringer, R. R., et al. (2001). The Transcription Factors L-Sox5 and Sox6 Are Essential for Cartilage Formation. Dev. Cell 1, 277–290. doi:10.1016/s1534-5807(01)00003-x
Smits, P., Dy, P., Mitra, S., and Lefebvre, V. (2004). Sox5 and Sox6 Are Needed to Develop and Maintain Source, Columnar, and Hypertrophic Chondrocytes in the Cartilage Growth Plate. J. Cell Biol. 164, 747–758. doi:10.1083/jcb.200312045
Square, T., Jandzik, D., Cattell, M., Coe, A., Doherty, J., and Medeiros, D. M. (2015). A Gene Expression Map of the Larval Xenopus laevis Head Reveals Developmental Changes Underlying the Evolution of New Skeletal Elements. Dev. Biol. 397, 293–304. doi:10.1016/j.ydbio.2014.10.016
Srivatsa, S., Parthasarathy, S., Britanova, O., Bormuth, I., Donahoo, A.-L., Ackerman, S. L., et al. (2014). Unc5C and DCC Act Downstream of Ctip2 and Satb2 and Contribute to Corpus Callosum Formation. Nat. Commun. 5, 3708. doi:10.1038/ncomms4708
Stock, M., Schäfer, H., Fliegauf, M., and Otto, F. (2004). Identification of Novel Target Genes of the Bone-specific Transcription Factor Runx2. J. Bone Min. Res. 19, 959–972. doi:10.1359/jbmr.2004.19.6.959
Stuart, J. M., Segal, E., Koller, D., and Kim, S. K. (2003). A Gene-Coexpression Network for Global Discovery of Conserved Genetic Modules. Science 302, 249–255. doi:10.1126/science.1087447
Su, Y.-H., Li, E., Geiss, G. K., Longabaugh, W. J. R., Krämer, A., and Davidson, E. H. (2009). A Perturbation Model of the Gene Regulatory Network for Oral and Aboral Ectoderm Specification in the Sea Urchin Embryo. Dev. Biol. 329, 410–421. doi:10.1016/j.ydbio.2009.02.029
Sun, J., Hong, J., Sun, S., Wang, X., Peng, Y., Zhou, J., et al. (2018). Transcription Factor 7-like 2 Controls Matrix Degradation through Nuclear Factor κB Signaling and Is Repressed by microRNA-155 in Nucleus Pulposus Cells. Biomed. Pharmacother. 108, 646–655. doi:10.1016/j.biopha.2018.09.076
Takeda, S., Bonnamy, J.-P., Owen, M. J., Ducy, P., and Karsenty, G. (2001). Continuous Expression of Cbfa1 in Nonhypertrophic Chondrocytes Uncovers its Ability to Induce Hypertrophic Chondrocyte Differentiation and Partially Rescues Cbfa1-Deficient Mice. Genes Dev. 15, 467–481. doi:10.1101/gad.845101
Takegami, Y., Ohkawara, B., Ito, M., Masuda, A., Nakashima, H., Ishiguro, N., et al. (2016). R-spondin 2 Facilitates Differentiation of Proliferating Chondrocytes into Hypertrophic Chondrocytes by Enhancing Wnt/β-Catenin Signaling in Endochondral Ossification. Biochem. Biophysical Res. Commun. 473, 255–264. doi:10.1016/j.bbrc.2016.03.089
Tamamura, Y., Otani, T., Kanatani, N., Koyama, E., Kitagaki, J., Komori, T., et al. (2005). Developmental Regulation of Wnt/β-Catenin Signals Is Required for Growth Plate Assembly, Cartilage Integrity, and Endochondral Ossification. J. Biol. Chem. 280, 19185–19195. doi:10.1074/jbc.m414275200
Tan, Z., Niu, B., Tsang, K. Y., Melhado, I. G., Ohba, S., He, X., et al. (2018). Synergistic Co-regulation and Competition by a SOX9-GLI-FOXA Phasic Transcriptional Network Coordinate Chondrocyte Differentiation Transitions. PLoS Genet. 14, e1007346. doi:10.1371/journal.pgen.1007346
Tani, S., Chung, U.-i., Ohba, S., and Hojo, H. (2020). Understanding Paraxial Mesoderm Development and Sclerotome Specification for Skeletal Repair. Exp. Mol. Med. 52, 1166–1177. doi:10.1038/s12276-020-0482-1
Tarazona, O. A., Slota, L. A., Lopez, D. H., Zhang, G., and Cohn, M. J. (2016). The Genetic Program for Cartilage Development Has Deep Homology within Bilateria. Nature 533, 86–89. doi:10.1038/nature17398
Tare, R. S., Oreffo, R. O. C., Clarke, N. M. P., and Roach, H. I. (2002). Pleiotrophin/Osteoblast-stimulating Factor 1: Dissecting its Diverse Functions in Bone Formation. J. Bone Min. Res. 17, 2009–2020. doi:10.1359/jbmr.2002.17.11.2009
Teplyuk, N. M., Haupt, L. M., Ling, L., Dombrowski, C., Mun, F. K., Nathan, S. S., et al. (2009). The Osteogenic Transcription Factor Runx2 Regulates Components of the Fibroblast Growth Factor/proteoglycan Signaling axis in Osteoblasts. J. Cell. Biochem. 107, 144–154. doi:10.1002/jcb.22108
Thornemo, M., Jansson, E. S., and Lindahl, A. (1996). Expression of the ID1 and ID3 Genes during Chondrocyte Differentiation. Ann. N. Y. Acad. Sci. 785, 337–339. doi:10.1111/j.1749-6632.1996.tb56302.x
Usami, Y., Gunawardena, A. T., Iwamoto, M., and Enomoto-Iwamoto, M. (2016). Wnt Signaling in Cartilage Development and Diseases: Lessons from Animal Studies. Lab. Invest. 96, 186–196. doi:10.1038/labinvest.2015.142
van der Kraan, P. M., and van den Berg, W. B. (2012). Chondrocyte Hypertrophy and Osteoarthritis: Role in Initiation and Progression of Cartilage Degeneration? Osteoarthr. Cartil. 20, 223–232. doi:10.1016/j.joca.2011.12.003
Vieux-Rochas, M., Bouhali, K., Mantero, S., Garaffo, G., Provero, P., Astigiano, S., et al. (2013). BMP-mediated Functional Cooperation between Dlx5;Dlx6 and Msx1;Msx2 during Mammalian Limb Development. PLoS One 8, e51700. doi:10.1371/journal.pone.0051700
Vincourt, J.-B., Vignaud, J.-M., Lionneton, F., Sirveaux, F., Kawaki, H., Marchal, S., et al. (2008). Increased Expression of Matrilin-3 Not Only in Osteoarthritic Articular Cartilage but Also in Cartilage-Forming Tumors, and Down-Regulation of SOX9 via Epidermal Growth Factor Domain 1-dependent Signaling. Arthritis Rheum. 58, 2798–2808. doi:10.1002/art.23761
Vortkamp, A., Lee, K., Lanske, B., Segre, G. V., Kronenberg, H. M., and Tabin, C. J. (1996). Regulation of Rate of Cartilage Differentiation by Indian Hedgehog and PTH-Related Protein. Science 273, 613–622. doi:10.1126/science.273.5275.613
Wahba, G. M., Hostikka, S. L., and Carpenter, E. M. (2001). The Paralogous Hox Genes Hoxa10 and Hoxd10 Interact to Pattern the Mouse Hindlimb Peripheral Nervous System and Skeleton. Dev. Biol. 231, 87–102. doi:10.1006/dbio.2000.0130
Wang, W., Lian, N., Li, L., Moss, H. E., Wang, W., Perrien, D. S., et al. (2009). Atf4 Regulates Chondrocyte Proliferation and Differentiation during Endochondral Ossification by Activating Ihh Transcription. Development 136, 4143–4153. doi:10.1242/dev.043281
Wang, M., Sampson, E. R., Jin, H., Li, J., Ke, Q. H., Im, H.-J., et al. (2013). MMP13 Is a Critical Target Gene during the Progression of Osteoarthritis. Arthritis Res. Ther. 15, R5. doi:10.1186/ar4133
Watanabe, H., Yamada, Y., and Kimata, K. (1998). Roles of Aggrecan, a Large Chondroitin Sulfate Proteoglycan, in Cartilage Structure and Function. J. Biochem. 124, 687–693. doi:10.1093/oxfordjournals.jbchem.a022166
Watanabe, K., Hida, M., Sasaki, T., Yano, H., Kawano, K., Yoshioka, H., et al. (2016). Sp1 Upregulates the Proximal Promoter Activity of the Mouse Collagen α1(XI) Gene (Col11a1) in Chondrocytes. Vitro Cell.Dev.Biol.-Animal 52, 235–242. doi:10.1007/s11626-015-9959-y
Wu, H., Whitfield, T. W., Gordon, J. A. R., Dobson, J. R., Tai, P. W. L., van Wijnen, A. J., et al. (2014). Genomic Occupancy of Runx2 with Global Expression Profiling Identifies a Novel Dimension to Control of Osteoblastogenesis. Genome Biol. 15, R52. doi:10.1186/gb-2014-15-3-r52
Wu, Z., Rao, Y., Zhang, S., Kim, E. J., Oki, S., Harada, H., et al. (2019). Cis˗ Control of Six1 Expression in Neural Crest Cells during Craniofacial Development. Dev. Dyn. 248, 1264–1272. doi:10.1002/dvdy.109
Wuelling, M., Schneider, S., Schröther, V. A., Waterkamp, C., Hoffmann, D., and Vortkamp, A. (2020). Wnt5a Is a Transcriptional Target of Gli3 and Trps1 at the Onset of Chondrocyte Hypertrophy. Dev. Biol. 457, 104–118. doi:10.1016/j.ydbio.2019.09.012
Xiong, Q., Liu, Y., Xue, Y., Liu, S., Wang, J., Li, P., et al. (2018). A Novel De Novo Mutation in COL2A1 Leading to Spondyloepiphyseal Dysplasia Congenita in a Chinese Family. Hum. Genome Var. 5, 17059. doi:10.1038/hgv.2017.59
Xu, P., Balczerski, B., Ciozda, A., Louie, K., Oralova, V., Huysseune, A., et al. (2018). Fox Proteins Are Modular Competency Factors for Facial Cartilage and Tooth Specification. Development 145. dev165498. doi:10.1242/dev.165498
Yagi, R., McBurney, D., and Horton, W. E. (2005). Bcl-2 Positively Regulates Sox9-dependent Chondrocyte Gene Expression by Suppressing the MEK-Erk1/2 Signaling Pathway. J. Biol. Chem. 280, 30517–30525. doi:10.1074/jbc.m502751200
Yamamoto, S., Uchida, Y., Ohtani, T., Nozaki, E., Yin, C., Gotoh, Y., et al. (2019). Hoxa13 Regulates Expression of Common Hox Target Genes Involved in Cartilage Development to Coordinate the Expansion of the Autopodal Anlage. Dev. Growth Differ. 61, 228–251. doi:10.1111/dgd.12601
Yamashita, S., Kataoka, K., Yamamoto, H., Kato, T., Hara, S., Yamaguchi, K., et al. (2019). Comparative Analysis Demonstrates Cell Type-specific Conservation of SOX9 Targets between Mouse and Chicken. Sci. Rep. 9, 12560. doi:10.1038/s41598-019-48979-4
Yang, Y., Topol, L., Lee, H., and Wu, J. (2003). Wnt5a and Wnt5b Exhibit Distinct Activities in Coordinating Chondrocyte Proliferation and Differentiation. Development 130, 1003–1015. doi:10.1242/dev.00324
Yang, X., Matsuda, K., Bialek, P., Jacquot, S., Masuoka, H. C., Schinke, T., et al. (2004). ATF4 Is a Substrate of RSK2 and an Essential Regulator of Osteoblast Biology. Cell 117, 387–398. doi:10.1016/s0092-8674(04)00344-7
Yao, Z.-Z., Hu, A.-X., and Liu, X.-S. (2017). DUSP19 Regulates IL-1β-induced Apoptosis and MMPs Expression in Rat Chondrocytes through JAK2/STAT3 Signaling Pathway. Biomed. Pharmacother. 96, 1209–1215. doi:10.1016/j.biopha.2017.11.097
Yoshida, C. A., Yamamoto, H., Fujita, T., Furuichi, T., Ito, K., Inoue, K.-i., et al. (2004). Runx2 and Runx3 Are Essential for Chondrocyte Maturation, and Runx2 Regulates Limb Growth through Induction of Indian Hedgehog. Genes Dev. 18, 952–963. doi:10.1101/gad.1174704
Young, B., Minugh-Purvis, N., Shimo, T., St-Jacques, B., Iwamoto, M., Enomoto-Iwamoto, M., et al. (2006). Indian and Sonic Hedgehogs Regulate Synchondrosis Growth Plate and Cranial Base Development and Function. Dev. Biol. 299, 272–282. doi:10.1016/j.ydbio.2006.07.028
Yu, L., Liu, H., Yan, M., Yang, J., Long, F., Muneoka, K., et al. (2007). Shox2 Is Required for Chondrocyte Proliferation and Maturation in Proximal Limb Skeleton. Dev. Biol. 306, 549–559. doi:10.1016/j.ydbio.2007.03.518
Yun, K., and Im, S.-H. (2007). Transcriptional Regulation of MMP13 by Lef1 in Chondrocytes. Biochem. Biophysical Res. Commun. 364, 1009–1014. doi:10.1016/j.bbrc.2007.10.121
Zakany, J., and Duboule, D. (2007). The Role of HOX Genes during Vertebrate Limb Development. Curr. Opin. Genet. Dev. 17, 359–366. doi:10.1016/j.gde.2007.05.011
Zhang, P., Jimenez, S. A., and Stokes, D. G. (2003). Regulation of Human COL9A1 Gene Expression. J. Biol. Chem. 278, 117–123. doi:10.1074/jbc.m208049200
Zhang, Q., Yin, Z. S., Zhang, F. W., Cao, K., and Sun, H. Y. (2018). CTHRC1 Mediates IL-1β-induced A-poptosis in C-hondrocytes via JNK1/2 S-ignaling. Int. J. Mol. Med. 41, 2270–2278. doi:10.3892/ijmm.2018.3403
Zhang, X., Weng, M., and Chen, Z. (2021). Fibroblast Growth Factor 9 (FGF9) Negatively Regulates the Early Stage of Chondrogenic Differentiation. PLoS One 16, e0241281. doi:10.1371/journal.pone.0241281
Zhao, Q., Eberspaecher, H., Lefebvre, V., and De Crombrugghe, B. (1997). Parallel Expression ofSox9 andCol2a1 in Cells Undergoing Chondrogenesis. Dev. Dyn. 209, 377–386. doi:10.1002/(sici)1097-0177(199708)209:4<377::aid-aja5>3.0.co;2-f
Zheng, Q., Zhou, G., Morello, R., Chen, Y., Garcia-Rojas, X., and Lee, B. (2003). Type X Collagen Gene Regulation by Runx2 Contributes Directly to its Hypertrophic Chondrocyte-specific Expression In Vivo. J. Cell Biol. 162, 833–842. doi:10.1083/jcb.200211089
Zhou, G., Zheng, Q., Engin, F., Munivez, E., Chen, Y., Sebald, E., et al. (2006). Dominance of SOX9 Function over RUNX2 during Skeletogenesis. Proc. Natl. Acad. Sci. U.S.A. 103, 19004–19009. doi:10.1073/pnas.0605170103
Keywords: chondrocytes, GRN evolution, limb cartilage, head cartilage, GRN co-option
Citation: Gomez-Picos P, Ovens K and Eames BF (2022) Limb Mesoderm and Head Ectomesenchyme Both Express a Core Transcriptional Program During Chondrocyte Differentiation. Front. Cell Dev. Biol. 10:876825. doi: 10.3389/fcell.2022.876825
Received: 15 February 2022; Accepted: 26 May 2022;
Published: 17 June 2022.
Edited by:
Jaime J. Carvajal, Andalusian Center for Development Biology (CSIC), SpainReviewed by:
Daniel Graf, University of Alberta, CanadaGretel Nusspaumer, Andalusian Center for Development Biology (CSIC), Spain
Copyright © 2022 Gomez-Picos, Ovens and Eames. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: B. Frank Eames, Yi5mcmFua0B1c2Fzay5jYQ==
 Patsy Gomez-Picos
Patsy Gomez-Picos Katie Ovens
Katie Ovens B. Frank Eames
B. Frank Eames