
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell Dev. Biol. , 23 June 2022
Sec. Signaling
Volume 10 - 2022 | https://doi.org/10.3389/fcell.2022.873278
 Jakob Mitgau
1
†
Jakob Mitgau
1
† Julius Franke1
†
Julius Franke1
† Camilla Schinner2
Camilla Schinner2 Gabriele Stephan3
Gabriele Stephan3
 Sandra Berndt
1
Sandra Berndt
1 Dimitris G. Placantonakis3
Dimitris G. Placantonakis3
 Hermann Kalwa
4
Hermann Kalwa
4
 Volker Spindler
2
Volker Spindler
2
 Caroline Wilde
1
Caroline Wilde
1
 Ines Liebscher
1*
Ines Liebscher
1*The adhesion G protein–coupled receptor (aGPCR) GPR126/ADGRG6 plays an important role in several physiological functions, such as myelination or peripheral nerve repair. This renders the receptor an attractive pharmacological target. GPR126 is a mechano-sensor that translates the binding of extracellular matrix (ECM) molecules to its N terminus into a metabotropic intracellular signal. To date, the structural requirements and the character of the forces needed for this ECM-mediated receptor activation are largely unknown. In this study, we provide this information by combining classic second-messenger detection with single-cell atomic force microscopy. We established a monoclonal antibody targeting the N terminus to stimulate GPR126 and compared it to the activation through its known ECM ligands, collagen IV and laminin 211. As each ligand uses a distinct mode of action, the N terminus can be regarded as an allosteric module that can fine-tune receptor activation in a context-specific manner.
The adhesion G protein–coupled receptor (aGPCR) GPR126/ADGRG6 plays an essential role in several important physiologic and pathogenic processes, including myelination (Monk et al., 2009; Monk et al., 2011; Mogha et al., 2013; Ravenscroft et al., 2015), peripheral nerve injury and repair (Mogha et al., 2016), the development of the peripheral nervous system (PNS) (Ravenscroft et al., 2015), and the differentiation of osteoblasts (Sun et al., 2020) and adipocytes (Suchý et al., 2020). Furthermore, an association of GPR126 variants with the development of scoliosis was found in humans and mice (Kou et al., 2013; Karner et al., 2015; Xu et al., 2015; Qin et al., 2017; Kou et al., 2018; Liu et al., 2018; Xu et al., 2019a; Xu et al., 2019b; Xu et al., 2019c; Man et al., 2019; Takeda et al., 2019). Thus, pharmacological targeting of this receptor is of high interest. The physiological implications are mainly attributed to the modulation of cAMP levels by the receptor, which is achieved through the receptor’s coupling to Gs protein (Mogha et al., 2013).
Like other aGPCRs, GPR126 harbors an endogenous tethered agonistic sequence located distal of the GPS cleavage motif, termed the Stachel sequence (Liebscher et al., 2014). Synthetic peptides derived from this integral agonist can be used as agonists on the receptor (Liebscher et al., 2014; Demberg et al., 2017). More recently, small-molecule agonists have been identified (Bradley et al., 2019; Diamantopoulou et al., 2019), but similar to agonistic peptides, they lack specificity for the receptor (Demberg et al., 2017; Bradley et al., 2019). Additional activation can be achieved through the receptor’s N-terminal ligands collagen IV (Paavola et al., 2014), prion protein PrPC (Küffer et al., 2016), and laminin 211 (Petersen et al., 2015). Yet, none of these agonists is specific for GPR126, and ECM proteins lack characteristics of a classic receptor agonist as they are long-lived, stable molecules with essentially no diffusivity (Bassilana et al., 2019; Baxendale et al., 2021). Thus, it is unclear how the interaction between aGPCR and the ECM can be interpreted as a specific signal to modulate receptor activity levels. Mechanical forces are suggested to facilitate this interaction (Liebscher et al., 2014; Stoveken et al., 2015; Scholz et al., 2017; Dannhäuser et al., 2020). However, the force input as well as the structural components needed for activation have not been defined.
Targeting the large N terminus of an aGPCR with an antibody provides a specific way for receptor activation, which has been successfully shown for two other representatives of this receptor class (Yona et al., 2008; Bhudia et al., 2020; Chatterjee et al., 2021). The mechanism behind this N-terminal mediated signal is currently as unclear as the signals mediated by the ECM ligands. Understanding these fundamental activation processes will not only increase our understanding of the physiological circumstances that govern GPR126-mediated functions, but it will also set the stage for allosteric pharmaceutical targeting of this (Lu and Zhang, 2019) and potentially other aGPCRs. Allosteric modulation of a GPCR is of high pharmaceutical interest as it induces intermediate activation states, which potentially form the basis for activity-specific and biased modulation of signaling pathways (Ni et al., 2020; Lu et al., 2021a; Lu et al., 2021b; Wang et al., 2022).
In the absence of a specific antibody recognizing the N terminus of GPR126, we used an antibody recognizing an N-terminal HA epitope in GPR126, which was sufficient to activate the receptor. Our study characterized the structure–function prerequisites for this antibody-mediated activation and describes in real time the type and strength of the mechanical input needed to activate GPR126 through either an antibody or endogenous ligands, collagen IV and laminin 211. We conclude that the activation through the antibody is most likely mediated through cross-linking of the receptor, while collagen IV and laminin 211 need specific pushing and pulling forces. As the occurrence of ECM molecules is timely and locally regulated in tissues, a temporal–spatial and force-specific activation of GPR126 can be achieved through the N terminus as an allosteric force integrator.
If not stated otherwise, all standard substances were purchased from Sigma Aldrich (Taufkirchen, Germany), Merck (Darmstadt, Germany), and C. Roth GmbH (Karlsruhe, Germany). The cell culture material was obtained from Thermo Fisher Scientific (Schwerte, Germany), and primers were obtained from Microsynth Seqlab GmbH (Göttingen, Germany).
The magnetic stimulator was created by embedding N45 NdFeB magnets (https://www.supermagnete.de/data_sheet_S-08-05-N.pdf) into a custom 3D-printed holder matching the outer diameter of a corresponding cell culture plate.
The constructs of human GPR126 and ∆CUB, ∆PTX, and ∆CUB∆PTX mutants have been described previously (Mogha et al., 2013; Liebscher et al., 2014; Petersen et al., 2015). Point mutations for R468A and H839R constructs were inserted by quick change mutagenesis. In brief, plasmid DNA was amplified with PCR and then digested with DpnI restriction enzyme for 4 h at 37°C prior to heat shock transformation in E. coli. The mRuby epitope was inserted into the human GPR126 construct after the hemagglutinin (HA) epitope by a PCR-based site-directed mutagenesis and fragment replacement strategy. The sequences of all generated mutants of human GPR126 were verified by Sanger sequencing (Microsynth Seqlab, Göttingen, Germany). The GPR126/pULTRA construct was generated using one-step isothermal DNA assembly (Gibson et al., 2009). The reaction buffer was prepared according to the protocol. All enzymes were acquired from NEB: T5 exonuclease (M0363), Taq DNA Ligase (M0208), and Phusion DNA Polymerase (M0530). The WT receptor DNA for the assembly was obtained via PCR using the previously described construct of the full-length human GPR126 in the pcDps vector (Liebscher et al., 2014). The pULTRA [pULTRA was a gift from Malcolm Moore, Addgene plasmid #24129 (Lou et al., 2012)] vector was restriction-digested using Xba1 (NEB, R0145).
Sequences encoding Anti-HA Fab (clone 12CA5) heavy and light chains were codon-optimized for human cells and synthesized and cloned into pcDNA3.4 (Thermo Fisher Scientific) by Genscript. The DNA of heavy and light chains was mixed in the ratio 1:1, transfected into Expi293 cells (Thermo Fisher Scientific) using PEI Max (Polysciences), and expressed at 37°C for 7 days. Fab was purified from clarified culture supernatants using a CaptureSelect CH1-XL column (Thermo Fisher Scientific) and buffer-exchanged into Tris-buffered saline (100 mM NaCl, 20 mM Tris, pH7.5) using a HiPrep 26/10 desalting column (Cytiva, Freiburg, Germany). Binding of the Fab fragment to the present HA-tag was proven to be concentration-dependent on P2Y12-transfected cells, which served as receptor expression positive control (Supplementary Figure S1A).
The human N- and C-terminally tagged GPR126 was heterologously expressed in COS-7 cells grown in Dulbecco’s minimum essential medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 100 units/ml penicillin, and 100 μg/ml streptomycin or the GripTite 293 MSR Cell Line (Thermo Fisher Scientific) (HEK-GT) grown in DMEM supplemented with 10% fetal bovine serum, 1% G418 (Thermo Fisher Scientific, 10131035) and 1% Non-Essential Amino Acids (Thermo Fisher Scientific, 11140050) at 37°C and 5% CO2 in a humidified atmosphere. The used variant of the mouse GPR114 (ADGRG5) is tagged just like GPR126 and has been published before (Wilde et al., 2016). All steps of the assays were performed the same way for both receptors: the cells were split into 48-well plates (3×104 cells/well for COS-7 or 1.3 × 105 cells/well for HEK-GT) for antibody stimulation assays or into 96-well plates (1.5 × 104 cells/well for COS-7 or 4.5 × 104/well for HEK-GT) for peptide stimulation. Transfection was carried out with Lipofectamine 2000 (Thermo Fisher Scientific) according to the manufacturer’s protocol using 50 ng (96-well plates) or 500 ng (48-well plates) of receptor plasmid DNA/well. 48 h after transfection, GPR126 and empty vector–transfected cells were stimulated with the indicated concentrations of anti–HA antibody (H3663, stock concentration 1 mg/ml, Sigma-Aldrich), anti-HA Fab fragment (stock concentration 1 mg/ml, kind gift from T. Schiffner), anti–HA conjugated super paramagnetic Dynabeads® (14311D, stock concentration 10 mg/ml, Thermo Fisher Scientific) or anti–FLAG antibody (F1804, Sigma-Aldrich) in DMEM for 1 h, followed by incubation with 3-isobutylmethyl-xanthine (1 mM)-containing medium or including the secondary antibody anti-mouse IgG (Fc specific, M2650, Sigma-Aldrich) for 30 min. For peptide stimulation, a Stachel-sequence derived peptide was diluted in IBMX-containing medium. A peptide solution from purified powder was achieved by preparing a 100 mM in 100% DMSO solution, which was further diluted into 10 mM stocks using a 50 mM pH 8 Tris buffer and finally pH-controlled. Peptide concentrations used in assays are 1 mM. The 1 mM peptide solution contains 1% DMSO and 10% Tris buffer for dilution. After stimulation, the cells were lysed in LI buffer (PerkinElmer, Rodgau, Germany) and frozen at −80°C until measurement. To measure cAMP concentration, the Alpha Screen cAMP assay kit (PerkinElmer) was used according to the manufacturer’s protocol. The accumulated cAMP was measured in 384-well white OptiPlate microplates (PerkinElmer) with the EnVision Multilabel Reader (PerkinElmer). Super paramagnetic Dynabeads® were conjugated with anti–HA antibody according to the manufacturer’s protocol. Briefly, Dynabeads were weighed, washed with C1 solution, mixed with an appropriate amount of anti–HA antibody in C1 solution, C2 solution was added, and the mixture was incubated overnight at 37°C on a roller, followed by washing afterward.
The cells were split into 48-well plates (3 × 104 cells/well for COS-7 or 1.3 × 105 cells/well for HEK-GT). To estimate cell surface expression of receptors carrying an N-terminal HA tag, an indirect cellular enzyme-linked immunosorbent assay (ELISA) was used (Schöneberg et al., 1998). Briefly, the cells were transfected with the indicated constructs. 48 h after transfection, the cells were fixed with 4% formaldehyde, washed with PBS, blocked with 10% FBS medium, and incubated with anti–HA POD-conjugated antibody followed by o-phenylendiamine treatment. Optical densities were measured at a wavelength of 492 nm with the EnVision Multilabel Reader (PerkinElmer).
For Western blot analysis, the cells were split into 24-well plates (6 × 104 cells/well for COS-7) and transfected with either 250 ng of empty pcDps vector, GPR126, GPR126 R468A, or GPR126 R468A/H839R using the standard protocols described earlier. Every other transfected construct was incubated with the primary anti–HA antibody for an hour as explained before. Supernatants were harvested and cells were lysed with the addition of 300 μL of 2x SDS loading dye (#S3401, Sigma-Aldrich). After freeze–thaw cycling, the lysates were run on a 10% SDS-PAGE gel, followed by Western blotting. The PVDF membranes were activated using 100% methanol, and transfer was performed for 1 h at 80 V. The membranes were blocked for 1 h with 5% non-fat dry milk in TBST buffer, washed three times with TBST buffer, and incubated overnight with either primary antibody (rabbit ant-HA, #3724 and anti-GAPDH, #97166 antibody, Cell Signaling). The following day, the membranes were again washed with TBST buffer and incubated with secondary anti-rabbit antibody, which has an HRP conjugation (#7074, Cell Signaling) for 1 h at room temperature. Following three washing steps with TBST buffer, SuperSignal West Pico PLUS Chemiluminescent Substrate (Thermo Fisher Scientific) was added to membranes to visualize protein bands using the Bio-Rad Gel Doc Imager.
For all atomic force microscopy (AFM) experiments, HEK-GT cells were cultured at the same conditions as for the in vitro functional assays. Cells were seeded on 24-mm glass coverslips (coated with Poly-L-Lysin (Sigma-Aldrich, P4707), (1% PLL solution incubated at 37°C for 5 min, then washed with PBS and dried under a sterile hood) in 6-well plates (1.5 × 106 cells/well) and co-transfected with the cAMP sensor Pink Flamindo (Addgene plasmid #102356) and either an empty vector or the given GPR126 construct in the pULTRA vector on the next day using Lipofectamine 2000 according to the manufacturer’s protocol. The media were changed to the described culture media ∼24 h after transfection and AFM measurements took place ∼48 h after transfection: the coverslips were transferred into a 35-mm cell culture dish and washed with DMEM without phenol red three times and placed into AFM coverslip holders. 750 µL of culture media without phenol red was added, and the cells were stored at 37°C with 5% CO2 until right before the measurements took place.
Tipless silicon nitride AFM cantilevers (NanoWorld, PNP-TR-TL) were coated with monoclonal anti–HA antibodies produced in mouse (H3663, Sigma-Aldrich) or Fc-control for the antibody-based mechano-activation experiments using flexible PEG spacers as described before (Ebner et al., 2007). Recombinant human Fc was expressed by transfected Chinese hamster ovary (CHO) cells. The supernatant was collected and purification was carried out via His-tag using HisLink Protein Purification Resin (V8823, Promega) according to the manufacturer’s instructions. Protein purity was confirmed by Western blot analysis. For the ligand-based mechano-activation experiments, tipless silicon nitride AFM cantilevers were washed twice in chloroform and dried. The cantilevers were then placed into a collagen IV (C6745, Sigma Aldrich) or laminin-211 (LN221, BioLamina, Sundbyberg, Sweden) solution (0.15 mg/ml) and incubated at 4°C overnight. The next day, the cantilevers were washed in HBSS twice and stored in HBSS at 4°C until use.
AFM measurements were performed using a Nanowizard IV AFM (Bruker, Billerica, MA) mounted on an IX 83 inverted optical microscope equipped with a 63x PL APO NA 1.42 oil objective (both Olympus Life Sciences, Wallisellen, Switzerland) and coupled to an X-Cite Exacte Light source (Excelitas Technologies, Waltham, MA).
Cantilevers were calibrated using the thermal noise method according to (Slattery et al., 2013).
Successfully double-transfected cells were identified by the GFP from the pULTRA vector and the Pink Flamindo fluorescence signal. The cell was then imaged three times in the following order: Brightfield, GFP, and Pink Flamindo using a Zyla sCMOS camera (Andor Technology, Belfast, Northern Ireland). The AFM cantilever was then placed centrally on the cell and approached until it contacted the cell surface. Proper positioning was verified by a brightfield image, and the baseline Pink Flamindo signal was recorded. During stimulation, all light sources except the AFM laser were turned off. Immediately after the stimulation was finished, another image of the Pink Flamindo signal was obtained using the same exposure time as before.
For force clamp measurements, the cantilevers were initially pressed onto the cell with a force of 1 nN for 5 s in order to allow antibody/ligands and the receptor to bind. The cantilever was then retracted (1 µm/s) until the desired clamp force was reached. This value was kept constant for the indicated times before the cantilever was fully retracted. The extend and retract length was 15 µm, and extend and retract speed was 5 µm/s for all experiments.
For analysis, the Pink Flamindo images taken before and after the stimulation were compared by measuring the mean intensity of a rectangular area on the stimulated cell using Fiji ImageJ (Schindelin et al., 2012). To account for variations affecting all cells independently from the stimulation, such as bleaching, a rectangular area was measured on five other cells that were not touched by the cantilever but expressed Pink Flamindo. The average of the changes in these five reference cells was subtracted from the measured change in the stimulated cell to isolate the effect the AFM cantilever stimulation has on the receptor-transfected cell (Supplementary Figure S3A).
To evaluate the mechano-independent changes of the Pink Flamindo fluorescence signal, HEK-GT cells were split into 96-well plates (4.5 × 104/well) and transfected the following day, analogous to the cAMP accumulation assays described earlier. Two days after transfection, the media were removed from the wells and 40 µL of DMEM without phenol-red was added to each well. Then, the GFP signal (transfection control) as well as the Pink Flamindo fluorescence signal was imaged using a Celigo Image Cytometer (Nexcelom Bioscience). Forskolin, pGPR126, and anti–HA antibody diluted in DMEM without phenol-red were added to a final volume of 50 µL/well and the following concentrations: 10 µM forskolin, 1 µM anti-HA antibody and 1 mM pGPR126. The cells were imaged again 60 s after the addition of the respective stimulus, and the intensity of the Pink Flamindo signal before and after was compared.
Receptor expression and activation were analyzed using one- and two-way ANOVA as well as t-test as indicated at each figure legend. p values < 0.05 were considered statistically significant (*p < 0.05; **p < 0.01; ***p < 0.001). All statistical analyses were performed by GraphPad Prism version 6.00 for Windows (GraphPad Software, Inc., La Jolla, United States) or Microsoft Excel 2016 (Microsoft Corporation, Redmond, United States).
Since GPR126 can be activated through interaction with its extracellular ligands and mechanical stimuli, it can be assumed that the N terminus plays a decisive role in mediating these signals. However, our understanding of these dynamic processes is limited. In order to establish a specific N-terminal interacting partner of GPR126, we established an antibody-based approach. Due to the lack of antibodies targeting the endogenous GPR126 sequence, we used the hemagglutinin (HA) epitope for our experimental setup and inserted it right after the predicted signal peptide of the receptor (Figure 1A). Surprisingly, increasing concentrations of the commercially available anti–HA antibody significantly elevated cAMP levels in COS-7 cells transfected with the HA-tagged wild type (WT) GPR126 (Figure 1B) but not in the empty vector transfected control cells. In a control experiment with an anti–FLAG antibody targeting the C-terminal epitope of GPR126, no change in cAMP levels was observed (Supplementary Figure S1B).
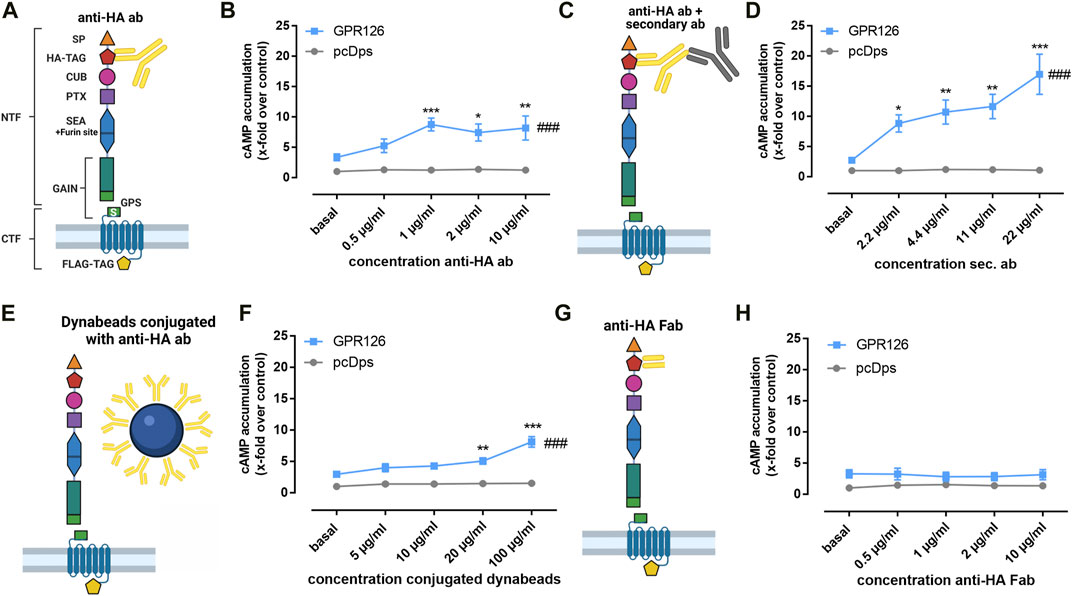
FIGURE 1. Antibodies against the N terminus activate GPR126. (A) Schematic setup for the anti–HA antibody (anti-HA ab) stimulation of full-length WT GPR126 in cAMP accumulation assays. The N terminus contains the signal peptide (SP, orange triangle), a complement C1r/C1s-Uegf-BMP1 domain (CUB, magenta oval), a pentraxin domain (PTX, purple square), and the sperm protein, enterokinase and agrin (SEA) domain (blue hexagon), including the furin site. The highly conserved GAIN domain (green rectangle) contains the GPS, which is followed by the Stachel sequence (S). In our receptor constructs, we inserted an N-terminal hemagglutinin tag (HA-TAG, red pentagon) immediately distal to the signal peptide and a C-terminal Flag tag (FLAG-TAG, yellow pentagon) right before the stop codon. (B). Different concentrations of anti–HA antibody (0.5 µg/ml ≙ 3.3 µM; 1 µg/ml ≙ 6.67 µM; 2 µg/ml ≙ 13.34 µM; 10 µg/ml ≙ 66.7 µM) were used to treat vector control (pcDps) and GPR126-transfected COS-7 cells (effect of construct p = 0.0410, effect of concentration p < 0.0001, interaction construct × concentration p = 0.0506; two-way ANOVA). Basal cAMP level in pcDps transfected cells: 8.9 ± 0.8 nM/well. (C,D) Amplification of the cAMP signal of GPR126 with 1 µg/ml of anti–HA antibody and subsequent incubation with different concentrations of secondary antibody (2.2 µg/ml ≙ 14.7 µM; 4.4 µg/ml ≙ 29.3 µM; 11 µg/ml ≙ 73.3 µM; 22 µg/ml ≙ 146.7 µM) on vector control and GPR126-transfected COS-7 cells (effect of construct p = 0.0104, effect of concentration p < 0.0001, interaction construct × concentration p = 0.0072; two-way ANOVA). Basal cAMP level in pcDps transfected cells: 4.2 ± 1.0 nM/well. (E,F) Paramagnetic Dynabeads® were conjugated with anti–HA ab and used in different concentrations for stimulation of vector control and GPR126-transfected COS-7 cells (effect of construct p = 0.0001, effect of concentration p < 0.0001, interaction construct × concentration p < 0.0001; two-way ANOVA). An empty vector (pcDps) served as a negative control. Basal cAMP level in pcDps transfected cells: 5.8 ± 0.4 nM/well. (G,H) cAMP accumulation upon incubation with indicated concentrations of Fab fragment (0.5 µg/ml ≙ 10 μM; 1 µg/ml ≙ 20 μM; 2 µg/ml ≙ 40 μM; 10 µg/ml ≙ 200 µM) on vector control and GPR126-transfected COS-7 cells (effect of construct p < 0.0001, effect of concentration p = 0.9920, interaction construct x concentration p = 0.9032; two-way ANOVA). Basal cAMP level in pcDps transfected cells: 7.7 ± 1.2 nM/well. All data are given as means ± SEM of three–five independent experiments each performed in triplicates. Statistics were performed by applying a two-way ANOVA followed by Dunnett’s post hoc analysis; *p < 0.05; **p < 0.01; ***p < 0.001. All significances given as stars (*) above individual points in the graphs show the result of the post hoc analysis, while # indicates significant concentration-dependent effects (### p < 0.001). Corresponding raw data can be found in the repository. Schematic images were created with BioRender.com.
Adding increasing concentrations of a secondary antibody to a fixed concentration of 1 µg/ml of the anti–HA antibody yielded an even stronger activation of the receptor (Figures 1C, D). We also observed the activation of GPR126 via the anti–HA antibody and the further modulation by the secondary antibody to be receptor-specific as we tested the experimental setup on a different mechano-sensitive aGPCR, GPR114 (ADGRG5) (Wilde et al., 2016). Neither the anti–HA antibody alone nor the addition of the secondary antibody led to any activation of the receptor (Supplementary Figure S1C). In order to elucidate whether the observed activation is a consequence of receptor cross-linking through the antibodies or due to the additional weight being attached to the receptor’s N terminus, we added anti–HA antibody–conjugated paramagnetic Dynabeads®, which are decisively larger in size than antibodies (Figure 1E). We observed a significant increase in cAMP levels (Figure 1F), which was comparable with anti–HA antibody–mediated activation alone (Figure 1B) but lower than the combination of primary and secondary antibody (Figure 1D), indicating that simply adding weight is not the sole key to GPR126 activation. In line with this observation, exposure to a 700-Gs magnetic field placed below the cell layer cannot further enhance conjugated Dynabead-mediated activation (Supplementary Figure S1D). Crosslinking, on the other hand, appears to be a key element to anti–HA-antibody–mediated activation as incubation with the respective monomeric Fab fragment cannot induce cAMP accumulation (Figures 1G, H).
The N terminus of GPR126 includes five structurally different domains (Leon et al., 2020), which can be subject to tissue-specific splicing (Knierim et al., 2019) and serve to interact with the known ligands collagen IV (Paavola et al., 2014), laminin 211 (Petersen et al., 2015), and prion protein (Küffer et al., 2016). To investigate their role in mechano-sensing, we used different N-terminal deletion mutants of GPR126, whose basal activity and expression levels were previously reported (Petersen et al., 2015). The constructs generated were ΔCUB (lacking 109 aa compared to the WT), ΔPTX (lacking 206 aa), and ΔCUBΔPTX (lacking 315 aa). In addition, we generated an N-terminal prolonged variant of GPR126 containing an mRuby-tag (236 aa prolongation compared to the WT) (Figure 2A). Reliable cell surface expression levels of the mutants were demonstrated with ELISA (Supplementary Figure S2A). The ability of receptor activation for each mutant was measured in cAMP accumulation assays using the synthetic GPR126 Stachel peptide as a stimulus, thereby ensuring undisturbed signaling (Supplementary Figure S2B). We stimulated these mutants with the same antibody-based setup as described in Figure 1 and found that deletion of the CUB and PTX domains yielded results highly similar to those of WT GPR126 (Figure 2B). The N-terminally prolonged mRuby construct, despite showing WT-like expression and peptide activation (Supplementary Figures S2A, B), could not be activated through antibodies (Figure 2B).
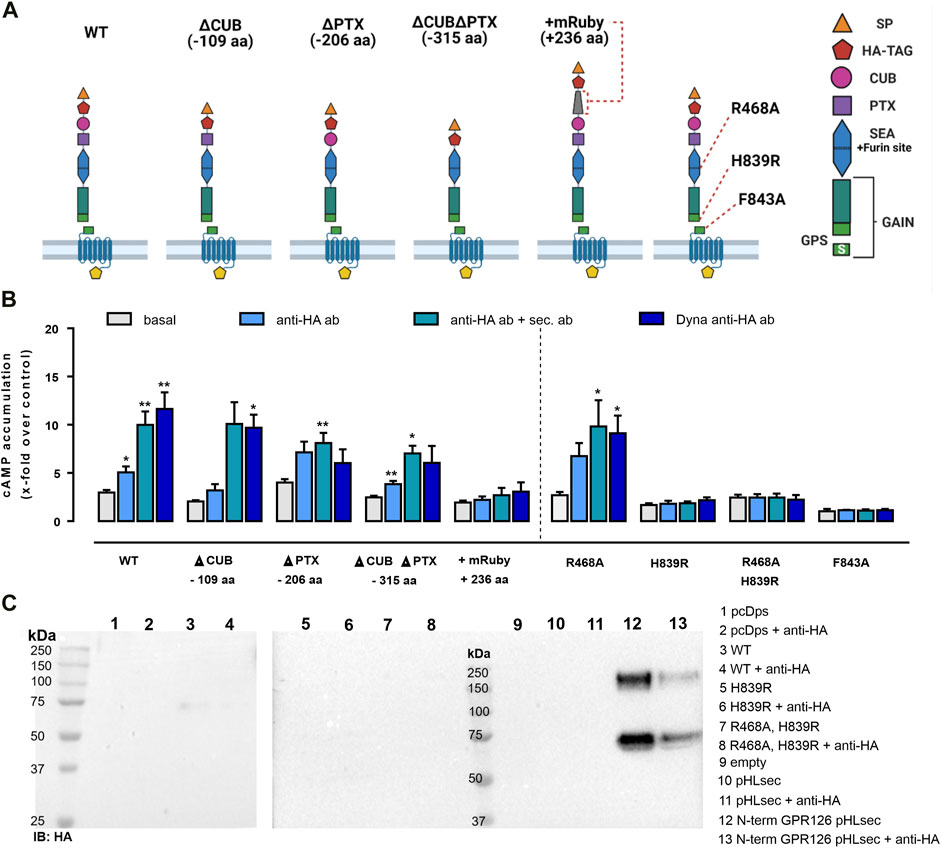
FIGURE 2. Antibody-mediated activation of GPR126 depends on autoproteolytic cleavage and is obliterated in a construct with an elongated N terminus. (A) Domain architecture of the human GPR126 WT and respective mutants is depicted. The positions of the furin-deficient receptor mutant R468A, the GPS cleavage mutant H839R, and the tethered agonist mutant F843A within the N terminus are displayed. Images were created with BioRender.com. (B) Receptor variants were transfected into COS-7 cells and analyzed with the same antibody-based approach as used in Figure 1 and tested in cAMP assays. An empty vector (pcDps) served as a negative control (cAMP level: 4.3 ± 0.7 nM/well). Data are given as mean ± SEM of three–eight different experiments each performed in triplicates. Statistics were performed by applying one-way ANOVA followed by Dunnett’s post hoc analysis; *p < 0.05; **p < 0.01; ***p < 0.001. All shown statistical significance compare the condition to the basal cAMP accumulation of the same construct. Corresponding raw data can be found in the repository. (C) COS-7 cells were transfected with the indicated constructs and treated with 1 µg/ml mouse anti–HA ab for 1 h, 48 h after transfection. Then, supernatants were harvested and analyzed by Western blot. Membranes were incubated with rabbit anti-HA ab and secondary HRP-conjugated anti-rabbit ab. A corresponding Western blot of cell lysates can be found in Supplementary Figure S2C. The secreted N-terminal fragment of GPR126 was only detected when the secretion vector pHLsec containing just the N-terminal fragment of GPR126 as a positive control was transfected.
The complex architecture of the N terminus of GPR126 includes two cleavage sites; the GPCR proteolysis site (GPS) within the GAIN domain (Araç et al., 2012), at which the receptor is cleaved into an N-terminal fragment (NTF) and a C-terminal fragment (CTF) at the conserved HLT motif (Lin et al., 2004; Moriguchi et al., 2004), and a furin site, located in the SEA domain (Figure 2A). To analyze whether cleavage at either position may be required for antibody-mediated activation, we generated the furin-cleavage-deficient receptor mutant R468A, the GPS-cleavage-deficient mutant H839R, and the double-deficient mutant R468A H838R to test in antibody-mediated activation. Proper protein expression, activation capacity through Stachel peptide, as well as cleavage-deficiency of the mutants were confirmed (Supplementary Figure S2A–C). While mutation of the furin site (R468A) has no effect on the antibody-mediated stimulation approach, deletion of the GPS cleavage (H839R) abolishes signaling capacity similar to the previously described tethered agonist mutant F843A (Liebscher et al., 2014).
This observation could support the notion that antibody-mediated activation of this aGPCR requires the dissociation of the NTF from the CTF, as has been suggested for the activation of GPR133 (Frenster et al., 2021). To test this assumption, we harvested the supernatants from the empty vector and GPR126 transfected cells with or without anti–HA antibody stimulation and subjected them to Western blot analysis (Figure 2C). As a positive control, we used the N terminus of GPR126 encoded on the secretion vector pHLsec. No soluble NTF was found in the supernatants except for the positive control (Figure 2C). However, only a faint band was visible for the WT GPR126 of approximately 75 kDa, suggesting residual removal of the N terminus due to furin cleavage as seen for the cell lysate of the WT construct (Supplementary Figure S2C). We observed no increase in band intensities after prior stimulation with the mouse anti–HA antibody. Thus, even though the autoproteolytic procession of GPR126 at the GPS site is a prerequisite for antibody-mediated activation, it does not lead to NTF removal. It is conceivable that proteolytic processing at the GPS results in a favorable orientation of the Stachel sequence, which is indispensable for GPR126 activity.
To evaluate whether cross-linking alone is sufficient or if additional mechanical forces are needed to activate GPR126 through the anti–HA antibody, we used an atomic force microscopy (AFM) approach. We theorized that the antibody could exert either pulling (through cross-linking) or pushing forces (through residing on the cell layer) on the receptor and that both could be quantified with AFM. To do so, tipless AFM cantilevers were coated with anti–HA antibody using PEG linkers according to a well-established protocol (Ebner et al., 2007). In contrast to cantilevers with a tip, this approach allows for multiple antibodies to bind and thus interact with the cell surface. Therefore, the mechanical force induced by the cantilever and mediated by the antibodies is applied to multiple receptors expressed on the cell surface simultaneously. The coated cantilevers were then used to apply pressure (pushing) or a force-clamp (pulling) to individual cells that were successfully co-transfected with the given receptor construct and the Pink Flamindo cAMP sensor (Harada et al., 2017) (Figure 3A). This setup allows for the simultaneous application of force and the detection of changes in intracellular cAMP levels. Single cells within a confluent monolayer were chosen for the measurements based on the detection of the GFP signal from the pULTRA vector, which allows for bicistronic expression of EGFP and GPR126 (Lou et al., 2012) and the fluorescent signal from the Pink Flamindo cAMP sensor (Harada et al., 2017) (Supplementary Figure S3A). Since COS-7 cells are not suited for AFM experiments due to their weak adherence to glass coverslips, we used GripTite™ 293 MSR (HEK-GT) cells instead. Receptor cell surface expression and activation in HEK-GT cells was confirmed prior to AFM experiments by ELISA and cAMP accumulation assays, respectively (Supplementary Figures S3B, C). Relative possible Pink Flamindo fluorescence changes upon stimulation of GPR126 with either forskolin, anti–HA antibody, or Stachel peptide pGPR126 are displayed in Supplementary Figure S3D.
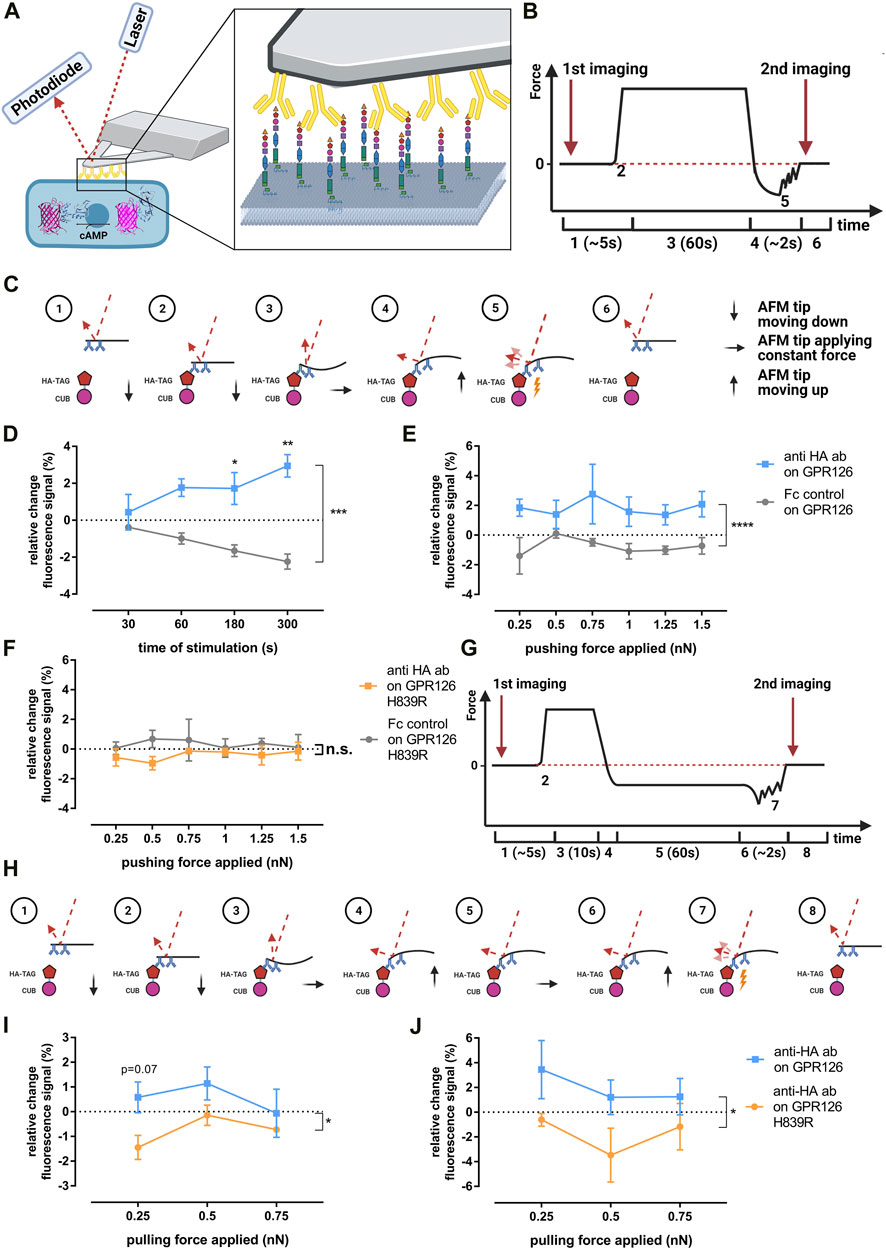
FIGURE 3. Evaluating potential mechano-activation of GPR126 via anti-HA antibodies. (A) Schematic AFM experiment setup with a coated tipless cantilever pressing on a cell co-transfected with GPR126 and the Pink Flamindo cAMP sensor. (B,C) Course of the AFM cantilever deflection during the pushing experiments and the forces applied to the receptor and corresponding laser deflection: (Monk et al., 2009) cantilever approaches cell; (Monk et al., 2011) point of contact between the cantilever and cell; (Mogha et al., 2013) constant pressure being applied to the cell; (Ravenscroft et al., 2015) cantilever is retracted from the cell; (Mogha et al., 2016) rupture of cantilever bindings; (Sun et al., 2020) all bindings are ruptured and the cantilever is back in its starting position. (D) Changes in Pink Flamindo fluorescence intensity after applying a pushing force of 1 nN on the WT receptor at different time points (effect of time p = 0.775, effect of coating p = 0.0002, interaction time × coating p = 0.065; two-way ANOVA). (E,F) Changes in Pink Flamindo fluorescence intensity after applying indicated pushing forces for 60s on the WT receptor (E) (effect of force applied p = 0.878, effect of coating p < 0.001, interaction force × coating p = 0.882; two-way ANOVA) and the H839R mutant (F) (effect of force applied p = 0.981, effect of coating p = 0.0867, interaction force × coating p = 0.924; two-way ANOVA). (G,H) Course of the AFM cantilever deflection during the force clamp experiments and the forces applied to the receptor and corresponding laser deflection: (Monk et al., 2009) cantilever approaches cell; (Monk et al., 2011) point of contact between cantilever and cell; (Mogha et al., 2013) initial pressure being applied to allow binding between cantilever and cell; (Ravenscroft et al., 2015) cantilever is retracted from the cell until the desired pulling force is applied; (Mogha et al., 2016) constant pulling force is applied; (Sun et al., 2020) cantilever is retracted from the cell completely; (Suchý et al., 2020) rupture of cantilever bindings; (Karner et al., 2015), all bindings are ruptured and the cantilever is back in its starting position. Changes in Pink Flamindo fluorescence intensity after applying indicated pulling forces over (I) 60s (effect of force applied p = 0.297, effect of receptor variant p = 0.0135, interaction force × receptor variant p = 0.497; two-way ANOVA) and (J) 300s (effect of force applied p = 0.405, effect of receptor variant p = 0.0386, interaction force × receptor variant p = 0.804; two-way ANOVA). Data are given as means ± SEM of three-five different experiments each measuring three individual cells for each force and time. Statistics were performed as two-way ANOVA followed by Tukey’s post hoc analysis; *p < 0.05; **p < 0.01. All significances given as stars (*) above individual points in the graphs show the result of the post hoc analysis, while # indicates significant coating-dependent (D,E) or receptor-variant-dependent (I,J) effects (# p < 0.05; ### p < 0.001). Corresponding raw data can be found in the repository. Schematic images (A–C,G–H) were created with BioRender.com. The Pink Flamindo depiction in A was taken from (Harada et al., 2017).
The course of the cantilever deflection and therefore the force applied to the cell for the pushing setup is depicted in Figures 3B,C. The cAMP-evoked changes in the Pink Flamindo fluorescence were monitored by imaging right before the AFM cantilever applied pressure or a pulling force and immediately after its retraction (Figure 3B). A cantilever coated with human Fc-protein instead of anti–HA antibodies was used as a negative control.
When a pushing force of 1 nN was applied over varying times, a significant increase in the Pink Flamindo fluorescence signal could be observed in GPR126-transfected cells compared to the negative control. This effect got stronger and more significant over time, suggesting continuous stimulation of the receptor (Figure 3D). The control condition responded with a reduction of cAMP sensor intensity over time, presumably due to bleaching effects induced by the AFM detection laser. This bleaching effect was confirmed in a control experiment in which the cells were transfected with only the empty vector and Fc-protein–coated cantilevers were used to approach the cells without applying any force (Supplementary Figure S3E). Having established that a significant increase in intracellular cAMP can be achieved by pressure application via anti-HA antibody–coated tips, we applied varying forces over a constant time in order to quantify the strength of the pushing force needed to activate GPR126 (Figure 3E). Applying varying pushing forces (0.25–1.5 nN) with anti-HA antibody–coated tips over 60 s led to a significant increase in cAMP levels consistently over the whole range of applied forces. This indicates that either the binding or respective cross-linking of the antibody to the receptor or the pushing forces that occur during the encounter between the coated cantilever and receptor are already sufficient to activate GPR126. When we performed the same experiment with the cleavage-deficient mutant H839R (Figure 3F), which showed no activation in cAMP accumulation assays using antibodies and dynabeads (Figure 2B), we again observed no change in the cAMP-mediated Pink Flamindo signal regardless of the pressure applied.
To investigate how pulling forces affect GPR126 activation, we used the force-clamp setup as shown in Figures 3G,H. An Fc-control antibody-coated cantilever could not be used as a negative control in this case as it does not bind to the receptor or the HA-epitope and therefore no pulling forces would be applied. Instead, we compared the WT receptor to the insensitive H839R mutant, which was still able to bind the anti–HA antibody. We applied varying pulling forces (0.25–0.75 nN) over 60 s (Figure 3I) and 300 s as fold changes were very low after the shorter pulling time period (Figure 3J). There was a significant difference in the responses observed in WT and cleavage-deficient receptors. However, highest fluorescence signals were detected for the lowest pulling force (0.25 nN), while an increase in this stimulus tended to reduce cAMP production. The magnitude of the observed Pink Flamindo signal corresponds to that seen in the pushing approach. Thus, the detected increase under low pulling conditions might be due to the same reasons as the observed pushing signal (cross-linking or initial interaction push), while applying stronger pulling forces might actually inactivate the receptor.
We set to investigate the forces needed to stimulate GPR126 using its natural ligands. Several ligands have been shown to modulate GPR126 activity (Figure 4A): collagen IV has been described as a directly activating ligand (Paavola et al., 2014), which could be interpreted as a pushing force on the receptor as it ‘sits’ on it. Laminin 211, on the other hand, was reported to require mechanical stimuli such as shaking or vibration to induce cAMP signaling (Petersen et al., 2015), which could be a proxy for pulling forces. To test these assumptions, tipless AFM cantilevers were coated with each ligand (Figures 4B, E).
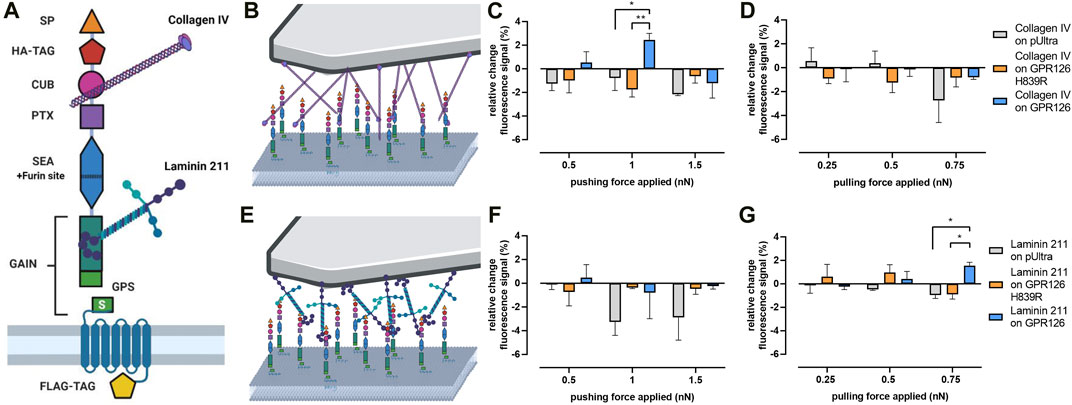
FIGURE 4. Mechano-activation of GPR126 via its ligands. (A) Domain architecture of human GPR126 with the binding sites of its ligands, collagen IV and laminin 211, is depicted. (B,E) Schematic demonstrating the AFM setup with (B) collagen IV- and (E) laminin 211-coated cantilevers. (C,D) Changes in Pink Flamindo fluorescence intensity after applying a varying pushing (C) or pulling (D) force over 60s with a collagen IV–coated cantilever. (F,G) Changes in Pink Flamindo fluorescence intensity after applying a pushing (F) or pulling (G) force over 60s with a laminin 211-coated cantilever. Data are given as the means ± SEM of three different experiments, each measuring three individual cells for each force. Statistics were performed as two-way ANOVA (C): effect of construct p = 0.1452, effect of force applied p = 0.0635, interaction construct × force applied p = 0.0298; (D): effect of construct p = 0.8096, effect of force applied p = 0.1748, interaction construct × force applied p = 0.2744; (F): effect of construct p = 0.0537, effect of force applied p = 0.4536, interaction construct × force applied p = 0,7286; (G): effect of construct p = 0.2225, effect of force applied p = 0.5884, interaction construct × force applied p = 0.0246) in combination with Tukey’s post hoc analysis; *p < 0.05; **p < 0.01. Significances in the graphs show the results of the post hoc analysis. Corresponding raw data can be found in the repository. Schematic images were created with BioRender.com.
When pushing onto WT GPR126 transfected cells with a collagen IV-coated cantilever (Figure 4B), it took specifically 1 nN to induce a significant increase in cAMP levels (Figure 4C). Lower or higher pressure did not activate the receptor. Again, GPR126 H839R could not be activated through this mechanical stimulus. Applying pulling forces with collagen IV did not produce any significant changes in the Pink Flamindo fluorescence signal for neither the WT receptor nor the cleavage-deficient mutant (Figure 4D). The laminin 211-coated cantilever (Figure 4E) did not activate GPR126 through pushing (Figure 4F), but a significant increase in the Pink Flamindo fluorescence signal was seen in the force-clamp setup when applying a pulling force of 0.75 nN (Figure 4G). Lower pulling forces were not sufficient to activate the receptor.
The aGPCR GPR126 can be activated through different mechanisms, including agonistic Stachel sequence-derived peptides (Liebscher et al., 2014); its ligands collagen IV (Paavola et al., 2014), laminin 211 (Petersen et al., 2015), and prion protein (Küffer et al., 2016); the small-molecule compound apomorphine (Bradley et al., 2019); and mechanical stimuli such as vibration or shaking (Petersen et al., 2015). Yet, all of these activators lack specificity for GPR126, which hampers their practicality as tools for in vivo experiments or potential therapeutic approaches. The agonistic peptide pGPR126 can, for example, cross-activate the aGPCR GPR64/ADGRG2 (Demberg et al., 2017), while the ECM molecules collagen and laminin can also bind and activate integrins (Keely et al., 1995) and apomorphin is an agonist on dopamine receptors (Millan et al., 2002). In this study, we show that a commonly used monoclonal antibody targeting an N-terminal HA epitope can serve as an activator of GPR126.
Antibody-mediated GPCR activation is a known phenomenon. For example, in several disease contexts, autoantibodies target GPCRs, such as the thyroid-stimulating hormone receptor, calcium-sensing receptor, and muscarinic M1 and M2 receptors (Unal et al., 2012). Previously, the aGPCRs EMR2 and GPR56 were shown to be activated through antibodies targeting the receptor N-termini (Bhudia et al., 2020; Chatterjee et al., 2021; Yona et al., 2008). In the absence of known antibodies against GPR126, we probed a commercial anti–HA antibody targeting an artificially introduced HA epitope at the N terminus of GPR126 and found that this universal antibody was, indeed, capable of activating the receptor (Figure 1B). We, thus, wondered how the anti–HA antibody mediates this activation. For example, agonistic properties of GPCR-targeting antibodies have been previously assigned to their interaction with the cognate ligand’s binding pocket or stabilization of ligand-induced active receptor conformations (Gupta et al., 2008), for example, through cross-linking/dimerization of the receptor as has been described for β1-adrenergic (β1AR) receptor (Hutchings et al., 2014). With respect to the known and anticipated activation scenarios for aGPCRs, it would also be conceivable that the antibody leads to a dissociation of the NTF, resulting in exposure of the tethered agonist (Petersen et al., 2015; Mathiasen et al., 2020; Frenster et al., 2021) or that it mediates mechanical stimuli such as pushing or pulling.
Our results support the notion that the anti–HA antibody–mediated activation is most likely due to cross-linking of the receptor as a monomeric Fab fragment of the anti-HA antibody alone cannot activate GPR126 (Figure 1H). The observation that the addition of a secondary antibody further enhanced cAMP production (Figure 1D) indicated that the receptor might also respond to the weight of molecules pushing on it. However, neither the paramagnetic Dynabeads® coated with the anti–HA antibody alone (Figure 1F) nor in combination with a magnet below the cell monolayer (Supplementary Figure S1B) enhanced cAMP levels compared to anti–HA antibody incubation alone. These results, as well as a lack of activation through direct pushing or pulling with an anti–HA antibody-coated cantilever in the AFM setup (Figure 3), demonstrate that cross-linking through the antibody is sufficient to activate GPR126, while no additional forces are required. Our mutagenesis data show that the anti–HA antibody–mediated stimulation of GPR126 depends on an intact tethered agonist sequence; thus, we can rule out the option that the antibody can directly interact with the endogenous agonist binding pocket (Figure 2B). Similarly, cleavage at the GPS is essential for this activation (Figure 2B), but we found no indication that this would lead to a dissociation of the NTF (Figure 2C). It can be speculated that this cleavage event would be required instead to induce a conformation that is necessary for the tethered agonist to reach its binding pocket. This is in contrast to other aGPCRs such as GPR56 (Chatterjee et al., 2021) and latrophilin (Scholz et al., 2015), whose antibody- and mechano-mediated activations, respectively, are not affected by mutations of their GPS cleavage motifs. Thus, it seems that autoproteolysis at the GPS serves distinct purposes among different receptors.
When studying aGPCR activation by ligands or antibodies, splice variants normally have to be considered since the complex exon–intron composition results in a large subset of functionally divergent receptors. As an example, activation of GPR56 through an antibody targeting the GAIN domain is dependent on a serine–threonine–proline-rich (STP) region that otherwise does not influence basal signaling levels of the receptor (Chatterjee et al., 2021). In the case of GPR126, alternative splicing influences the domain composition of the N terminus (Knierim et al., 2019). Within the N terminus, the CUB/PTX domain is of functional relevance as it serves as a point of interaction for collagen IV (Paavola et al., 2014); yet, for anti–HA antibody–mediated activation, it appears to be neglectable. In contrast, elongating the receptor’s N terminus through the addition of a fluorescent protein abolishes this activation. Similar observations have been made for the adhesion GPCR dCIRL, where elongation of the N terminus reduces the response to mechanical stimuli (Scholz et al., 2017). Thus, large artificial N-terminal domains influence signaling properties, even when they do not act as binding sites for the activating antibodies. It is currently unknown whether this is due to the simple change in length or an altered three-dimensional structure of the N terminus.
The endogenous interaction partners collagen IV and laminin 211 also bind to the N terminus of GPR126 but show different activation mechanisms. While incubation with collagen IV directly activates the receptor (Paavola et al., 2014), laminin 211 only induces cAMP production in combination with mechanical forces (Petersen et al., 2015). As the quality and the quantity of the required forces have not been defined at a single-cell level, it was hard to judge whether the in vitro findings could possibly be relevant in an in vivo context. Using the AFM approach, we found that laminin 211 requires increasing pulling forces of at least 0.75 nN (Figure 4G), while collagen IV only raises cAMP levels upon a pushing force of 1 nN (Figure 4C). Both mechano-stimulations require a cleavable GPR126. The activation patterns fit the physiological setting for these ligands in the process of myelination (Bunge et al., 1990; Paavola et al., 2014; Petersen et al., 2015), and the detected forces needed to induce a ligand-specific response are within the physiologic force range. They are below the traction forces that are normally transmitted by cell adhesions to the surrounding ECM (1–10 nN) (Balaban et al., 2001), indicating that already small changes can be detected by an aGPCR.
In summary, we were able to define the different ways in which a subset of structurally divergent molecules bound to the N terminus of GPR126 can modify the activity of this receptor. This establishes the N terminus as an allosteric integrator of signals coming from the immediate extracellular environment that is able to induce a spatio-temporal-force-dependent signal. It should, therefore, be considered a prime target for future pharmaceutical interventions as it provides the basis for receptor and potentially signaling-specific modulation of GPR126 activity.
The datasets generated for this study (x-folds as well as raw values of cAMP accumulation and fluorescence changes) can be found online in “zenodo,” https://zenodo.org/record/6489015 (DOI: 10.5281/zenodo.6489015).
JM, CW, CS, VS, DP, and IL designed the research plan. JF, GS, JM, HK, SB, and CW performed the experiments. JF, GS, JM, SB, CW, and IL analyzed the results. JM, CW, and IL wrote the manuscript with input from all authors. All authors edited and approved the manuscript.
Our research is funded by the German Research Foundation (CRC1052 project number 209933838 to IL, CRC1423 project number 421152132 B05 to IL, FOR2149 project number 246212759, P5 to IL), and (STE 2843/1-1 project number) to GS. Further funding comes from the European Union (European Social Fund) to IL and the Swiss National Science Foundation (SNF 310030_197764/1) to VS. Further support was provided by the junior research grant by the Medical Faculty, Leipzig University to CW and by the COST association providing an STSM grant within the COST action CA18240 to JM. DP was supported by NIH/NINDS R01 NS102665, NYSTEM (NY State Stem Cell Science) IIRP C32595GG, NIH/NIBIB R01 EB028774, NYU Grossman School of Medicine, and DFG FOR2149. This paper was supported by Open Access Publishing Fund of the Leipzig University.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank Torben Schiffner and Clara T. Schoeder for producing the anti-HA Fab fragment.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2022.873278/full#supplementary-material
Araç, D., Boucard, A. A., Bolliger, M. F., Nguyen, J., Soltis, S. M., Südhof, T. C., et al. (2012). A Novel Evolutionarily Conserved Domain of Cell-Adhesion GPCRs Mediates Autoproteolysis. EMBO J. 31, 1364–1378. doi:10.1038/emboj.2012.26
Balaban, N. Q., Schwarz, U. S., Riveline, D., Goichberg, P., Tzur, G., Sabanay, I., et al. (2001). Force and Focal Adhesion Assembly: a Close Relationship Studied Using Elastic Micropatterned Substrates. Nat. Cell Biol. 3, 466–472. doi:10.1038/35074532
Bassilana, F., Nash, M., and Ludwig, M.-G. (2019). Adhesion G Protein-Coupled Receptors: Opportunities for Drug Discovery. Nat. Rev. Drug Discov. 18, 869–884. doi:10.1038/s41573-019-0039-y
Baxendale, S., Asad, A., Shahidan, N. O., Wiggin, G. R., and Whitfield, T. T. (2021). The Adhesion GPCR Adgrg6 (Gpr126): Insights from the Zebrafish Model. Genesis 59, e23417. doi:10.1002/dvg.23417
Bhudia, N., Desai, S., King, N., Ancellin, N., Grillot, D., Barnes, A. A., et al. (2020). G Protein-Coupling of Adhesion GPCRs ADGRE2/EMR2 and ADGRE5/CD97, and Activation of G Protein Signalling by an Anti-EMR2 Antibody. Sci. Rep. 10, 1004. doi:10.1038/s41598-020-57989-6
Bradley, E. C., Cunningham, R. L., Wilde, C., Morgan, R. K., Klug, E. A., Letcher, S. M., et al. (2019). In Vivo identification of Small Molecules Mediating Gpr126/Adgrg6 Signaling during Schwann Cell Development. Ann. N.Y. Acad. Sci. 1456, 44–63. doi:10.1111/nyas.14233
Bunge, M. B., Clark, M. B., Dean, A. C., Eldridge, C. F., and Bunge, R. P. (1990). Schwann Cell Function Depends upon Axonal Signals and Basal Lamina Components. Ann. N. Y. Acad. Sci. 580, 281–287. doi:10.1111/j.1749-6632.1990.tb17937.x
Chatterjee, T., Zhang, S., Posey, T. A., Jacob, J., Wu, L., Yu, W., et al. (2021). Anti-GPR56 Monoclonal Antibody Potentiates GPR56-Mediated Src-Fak Signaling to Modulate Cell Adhesion. J. Biol. Chem. 296, 100261. doi:10.1016/j.jbc.2021.100261
Dannhäuser, S., Lux, T. J., Hu, C., Selcho, M., Chen, J. T.-C., Ehmann, N., et al. (2020). Antinociceptive Modulation by the Adhesion GPCR CIRL Promotes Mechanosensory Signal Discrimination. Elife 9, 2020. doi:10.7554/eLife.56738
Demberg, L. M., Winkler, J., Wilde, C., Simon, K.-U., Schön, J., Rothemund, S., et al. (2017). Activation of Adhesion G Protein-Coupled Receptors. J. Biol. Chem. 292, 4383–4394. doi:10.1074/jbc.M116.763656
Diamantopoulou, E., Baxendale, S., de la Vega de León, A., Asad, A., Holdsworth, C. J., Abbas, L., et al. (2019). Identification of Compounds that Rescue Otic and Myelination Defects in the Zebrafish Adgrg6 (Gpr126) Mutant. Elife 8. doi:10.7554/eLife.44889
Ebner, A., Wildling, L., Kamruzzahan, A. S. M., Rankl, C., Wruss, J., Hahn, C. D., et al. (2007). A New, Simple Method for Linking of Antibodies to Atomic Force Microscopy Tips. Bioconjugate Chem. 18, 1176–1184. doi:10.1021/bc070030s
Frenster, J. D., Stephan, G., Ravn-Boess, N., Bready, D., Wilcox, J., Kieslich, B., et al. (2021). Functional Impact of Intramolecular Cleavage and Dissociation of Adhesion G Protein-Coupled Receptor GPR133 (ADGRD1) on Canonical Signaling. J. Biol. Chem. 296, 100798. doi:10.1016/j.jbc.2021.100798
Gibson, D. G., Young, L., Chuang, R.-Y., Venter, J. C., Hutchison, C. A., and Smith, H. O. (2009). Enzymatic Assembly of DNA Molecules up to Several Hundred Kilobases. Nat. Methods 6, 343–345. doi:10.1038/nmeth.1318
Gupta, A., Heimann, A., Gomes, I., and Devi, L. (2008). Antibodies against G-Protein Coupled Receptors: Novel Uses in Screening and Drug Development. Cchts 11, 463–467. doi:10.2174/138620708784911465
Harada, K., Ito, M., Wang, X., Tanaka, M., Wongso, D., Konno, A., et al. (2017). Red Fluorescent Protein-Based cAMP Indicator Applicable to Optogenetics and In Vivo Imaging. Sci. Rep. 7, 7351. doi:10.1038/s41598-017-07820-6
Hutchings, C. J., Cseke, G., Osborne, G., Woolard, J., Zhukov, A., Koglin, M., et al. (2014). Monoclonal Anti-β1-adrenergic Receptor Antibodies Activate G Protein Signaling in the Absence of β-arrestin Recruitment. MAbs 6, 246–261. doi:10.4161/mabs.27226
Karner, C. M., Long, F., Solnica-Krezel, L., Monk, K. R., and Gray, R. S. (2015). Gpr126/Adgrg6deletion in Cartilage Models Idiopathic Scoliosis and Pectus Excavatum in Mice. Hum. Mol. Genet. 24, 4365–4373. doi:10.1093/hmg/ddv170
Keely, P. J., Wu, J. E., and Santoro, S. A. (1995). The Spatial and Temporal Expression of the α2β1 Integrin and its Ligands, Collagen I, Collagen IV, and Laminin, Suggest Important Roles in Mouse Mammary Morphogenesis. Differentiation 59, 1–13. doi:10.1046/j.1432-0436.1995.5910001.x
Knierim, A. B., Röthe, J., Çakir, M. V., Lede, V., Wilde, C., Liebscher, I., et al. (2019). Genetic Basis of Functional Variability in Adhesion G Protein-Coupled Receptors. Sci. Rep. 9, 11036. doi:10.1038/s41598-019-46265-x
Kou, I., Takahashi, Y., Johnson, T. A., Takahashi, A., Guo, L., Dai, J., et al. (2013). Genetic Variants in GPR126 Are Associated with Adolescent Idiopathic Scoliosis. Nat. Genet. 45, 676–679. doi:10.1038/ng.2639
Kou, I., Watanabe, K., Watanabe, K., Takahashi, Y., Momozawa, Y., Khanshour, A., et al. (2018). A Multi-Ethnic Meta-Analysis Confirms the Association of Rs6570507 with Adolescent Idiopathic Scoliosis. Sci. Rep. 8, 11575. doi:10.1038/s41598-018-29011-7
Küffer, A., Lakkaraju, A. K. K., Mogha, A., Petersen, S. C., Airich, K., Doucerain, C., et al. (2016). The Prion Protein Is an Agonistic Ligand of the G Protein-Coupled Receptor Adgrg6. Nature 536, 464–468. doi:10.1038/nature19312
Leon, K., Cunningham, R. L., Riback, J. A., Feldman, E., Li, J., Sosnick, T. R., et al. (2020). Structural Basis for Adhesion G Protein-Coupled Receptor Gpr126 Function. Nat. Commun. 11, 194. doi:10.1038/s41467-019-14040-1
Liebscher, I., Schön, J., Petersen, S. C., Fischer, L., Auerbach, N., Demberg, L. M., et al. (2014). A Tethered Agonist within the Ectodomain Activates the Adhesion G Protein-Coupled Receptors GPR126 and GPR133. Cell Rep. 9, 2018–2026. doi:10.1016/j.celrep.2014.11.036
Lin, H.-H., Chang, G.-W., Davies, J. Q., Stacey, M., Harris, J., and Gordon, S. (2004). Autocatalytic Cleavage of the EMR2 Receptor Occurs at a Conserved G Protein-Coupled Receptor Proteolytic Site Motif. J. Biol. Chem. 279, 31823–31832. doi:10.1074/jbc.M402974200
Liu, G., Liu, S., Lin, M., Li, X., Chen, W., Zuo, Y., et al. (2018). Genetic Polymorphisms of GPR126 Are Functionally Associated with PUMC Classifications of Adolescent Idiopathic Scoliosis in a Northern Han Population. J. Cell. Mol. Med. 22, 1964–1971. doi:10.1111/jcmm.13486
Lou, E., Fujisawa, S., Morozov, A., Barlas, A., Romin, Y., Dogan, Y., et al. (2012). Tunneling Nanotubes Provide a Unique Conduit for Intercellular Transfer of Cellular Contents in Human Malignant Pleural Mesothelioma. PLoS ONE 7, e33093. doi:10.1371/journal.pone.0033093
Lu, S., Chen, Y., Wei, J., Zhao, M., Ni, D., He, X., et al. (2021). Mechanism of Allosteric Activation of SIRT6 Revealed by the Action of Rationally Designed Activators. Acta Pharm. Sin. B 11, 1355–1361. doi:10.1016/j.apsb.2020.09.010
Lu, S., He, X., Yang, Z., Chai, Z., Zhou, S., Wang, J., et al. (2021). Activation Pathway of a G Protein-Coupled Receptor Uncovers Conformational Intermediates as Targets for Allosteric Drug Design. Nat. Commun. 12, 4721. doi:10.1038/s41467-021-25020-9
Lu, S., and Zhang, J. (2019). Small Molecule Allosteric Modulators of G-Protein-Coupled Receptors: Drug-Target Interactions. J. Med. Chem. 62, 24–45. doi:10.1021/acs.jmedchem.7b01844
Man, G. C.-W., Tang, N. L.-S., Chan, T. F., Lam, T. P., Li, J. W., Ng, B. K.-W., et al. (2019). Replication Study for the Association of GWAS-Associated Loci with Adolescent Idiopathic Scoliosis Susceptibility and Curve Progression in a Chinese Population. Spine 44, 464–471. doi:10.1097/BRS.0000000000002866
Mathiasen, S., Palmisano, T., Perry, N. A., Stoveken, H. M., Vizurraga, A., McEwen, D. P., et al. (2020). G12/13 Is Activated by Acute Tethered Agonist Exposure in the Adhesion GPCR ADGRL3. Nat. Chem. Biol. 16, 1343–1350. doi:10.1038/s41589-020-0617-7
Millan, M. J., Maiofiss, L., Cussac, D., Audinot, V., Boutin, J.-A., and Newman-Tancredi, A. (2002). Differential Actions of Antiparkinson Agents at Multiple Classes of Monoaminergic Receptor. I. A Multivariate Analysis of the Binding Profiles of 14 Drugs at 21 Native and Cloned Human Receptor Subtypes. J. Pharmacol. Exp. Ther. 303, 791–804. doi:10.1124/jpet.102.039867
Mogha, A., Benesh, A. E., Patra, C., Engel, F. B., Schöneberg, T., Liebscher, I., et al. (2013). Gpr126 Functions in Schwann Cells to Control Differentiation and Myelination via G-Protein Activation. J. Neurosci. 33, 17976–17985. doi:10.1523/JNEUROSCI.1809-13.2013
Mogha, A., Harty, B. L., Carlin, D., Joseph, J., Sanchez, N. E., Suter, U., et al. (2016). Gpr126/Adgrg6 Has Schwann Cell Autonomous and Nonautonomous Functions in Peripheral Nerve Injury and Repair. J. Neurosci. 36, 12351–12367. doi:10.1523/JNEUROSCI.3854-15.2016
Monk, K. R., Naylor, S. G., Glenn, T. D., Mercurio, S., Perlin, J. R., Dominguez, C., et al. (2009). A G Protein-Coupled Receptor Is Essential for Schwann Cells to Initiate Myelination. Science 325, 1402–1405. doi:10.1126/science.1173474
Monk, K. R., Oshima, K., Jörs, S., Heller, S., and Talbot, W. S. (2011). Gpr126 Is Essential for Peripheral Nerve Development and Myelination in Mammals. Development 138, 2673–2680. doi:10.1242/dev.062224
Moriguchi, T., Haraguchi, K., Ueda, N., Okada, M., Furuya, T., and Akiyama, T. (2004). DREG, a Developmentally Regulated G Protein-Coupled Receptor Containing Two Conserved Proteolytic Cleavage Sites. Genes cells. 9, 549–560. doi:10.1111/j.1356-9597.2004.00743.x
Ni, D., Wei, J., He, X., Rehman, A. U., Li, X., Qiu, Y., et al. (2020). Discovery of Cryptic Allosteric Sites Using Reversed Allosteric Communication by a Combined Computational and Experimental Strategy. Chem. Sci. 12, 464–476. doi:10.1039/d0sc05131d
Paavola, K. J., Sidik, H., Zuchero, J. B., Eckart, M., and Talbot, W. S. (2014). Type IV Collagen Is an Activating Ligand for the Adhesion G Protein-Coupled Receptor GPR126. Sci. Signal. 7, ra76. doi:10.1126/scisignal.2005347
Petersen, S. C., Luo, R., Liebscher, I., Giera, S., Jeong, S.-J., Mogha, A., et al. (2015). The Adhesion GPCR GPR126 Has Distinct, Domain-dependent Functions in Schwann Cell Development Mediated by Interaction with Laminin-211. Neuron 85, 755–769. doi:10.1016/j.neuron.2014.12.057
Qin, X., Xu, L., Xia, C., Zhu, W., Sun, W., Liu, Z., et al. (2017). Genetic Variant of GPR126 Gene Is Functionally Associated with Adolescent Idiopathic Scoliosis in Chinese Population. Spine 42, E1098–E1103. doi:10.1097/BRS.0000000000002123
Ravenscroft, G., Nolent, F., Rajagopalan, S., Meireles, A. M., Paavola, K. J., Gaillard, D., et al. (2015). Mutations of GPR126 Are Responsible for Severe Arthrogryposis Multiplex Congenita. Am. J. Hum. Genet. 96, 955–961. doi:10.1016/j.ajhg.2015.04.014
Schindelin, J., Arganda-Carreras, I., Frise, E., Kaynig, V., Longair, M., Pietzsch, T., et al. (2012). Fiji: an Open-Source Platform for Biological-Image Analysis. Nat. Methods 9, 676–682. doi:10.1038/nmeth.2019
Scholz, N., Gehring, J., Guan, C., Ljaschenko, D., Fischer, R., Lakshmanan, V., et al. (2015). The Adhesion GPCR Latrophilin/CIRL Shapes Mechanosensation. Cell Rep. 11, 866–874. doi:10.1016/j.celrep.2015.04.008
Scholz, N., Guan, C., Nieberler, M., Grotemeyer, A., Maiellaro, I., Gao, S., et al. (2017). Mechano-dependent Signaling by Latrophilin/CIRL Quenches cAMP in Proprioceptive Neurons. Elife 6. doi:10.7554/eLife.28360
Schöneberg, T., Schulz, A., Biebermann, H., Grüters, A., Grimm, T., Hübschmann, K., et al. (1998). V2 Vasopressin Receptor Dysfunction in Nephrogenic Diabetes Insipidus Caused by Different Molecular Mechanisms. Hum. Mutat. 12, 196–205. doi:10.1002/(sici)1098-1004(1998)12:3<196::aid-humu7>3.0.co;2-f
Slattery, A. D., Blanch, A. J., Quinton, J. S., and Gibson, C. T. (2013). Accurate Measurement of Atomic Force Microscope Cantilever Deflection Excluding Tip-Surface Contact with Application to Force Calibration. Ultramicroscopy 131, 46–55. doi:10.1016/j.ultramic.2013.03.009
Stoveken, H. M., Hajduczok, A. G., Xu, L., and Tall, G. G. (2015). Adhesion G Protein-Coupled Receptors Are Activated by Exposure of a Cryptic Tethered Agonist. Proc. Natl. Acad. Sci. U.S.A. 112, 6194–6199. doi:10.1073/pnas.1421785112
Suchý, T., Zieschang, C., Popkova, Y., Kaczmarek, I., Weiner, J., Liebing, A.-D., et al. (2020). The Repertoire of Adhesion G Protein-Coupled Receptors in Adipocytes and Their Functional Relevance. Int. J. Obes. 44, 2124–2136. doi:10.1038/s41366-020-0570-2
Sun, P., He, L., Jia, K., Yue, Z., Li, S., Jin, Y., et al. (2020). Regulation of Body Length and Bone Mass by Gpr126/Adgrg6. Sci. Adv. 6, eaaz0368. doi:10.1126/sciadv.aaz0368
Takeda, K., Kou, I., Hosogane, N., Otomo, N., Yagi, M., Kaneko, S., et al. (2019). Association of Susceptibility Genes for Adolescent Idiopathic Scoliosis and Intervertebral Disc Degeneration with Adult Spinal Deformity. Spine 44, 1623–1629. doi:10.1097/BRS.0000000000003179
Unal, H., Jagannathan, R., and Karnik, S. S. (2012). Mechanism of GPCR-Directed Autoantibodies in Diseases. Adv. Exp. Med. Biol. 749, 187–199. doi:10.1007/978-1-4614-3381-1_13
Wang, Y., Li, M., Liang, W., Shi, X., Fan, J., Kong, R., et al. (2022). Delineating the Activation Mechanism and Conformational Landscape of a Class B G Protein-Coupled Receptor Glucagon Receptor. Comput. Struct. Biotechnol. J. 20, 628–639. doi:10.1016/j.csbj.2022.01.015
Wilde, C., Fischer, L., Lede, V., Kirchberger, J., Rothemund, S., Schöneberg, T., et al. (2016). The Constitutive Activity of the Adhesion GPCR GPR114/ADGRG5 Is Mediated by its Tethered Agonist. FASEB J. 30, 666–673. doi:10.1096/fj.15-276220
Xu, E., Lin, T., Jiang, H., Ji, Z., Shao, W., Meng, Y., et al. (2019). Asymmetric Expression of GPR126 in the Convex/concave Side of the Spine Is Associated with Spinal Skeletal Malformation in Adolescent Idiopathic Scoliosis Population. Eur. Spine J. 28, 1977–1986. doi:10.1007/s00586-019-06001-5
Xu, E., Shao, W., Jiang, H., Lin, T., Gao, R., and Zhou, X. (2019). A Genetic Variant in GPR126 Causing a Decreased Inclusion of Exon 6 Is Associated with Cartilage Development in Adolescent Idiopathic Scoliosis Population. BioMed Res. Int. 2019, 1–8. doi:10.1155/2019/4678969
Xu, J.-F., Yang, G.-h., Pan, X.-H., Zhang, S.-J., Zhao, C., Qiu, B.-S., et al. (2015). Association of GPR126 Gene Polymorphism with Adolescent Idiopathic Scoliosis in Chinese Populations. Genomics 105, 101–107. doi:10.1016/j.ygeno.2014.11.009
Xu, L., Wu, Z., Xia, C., Tang, N., Cheng, J. C. Y., Qiu, Y., et al. (2019). A Genetic Predictive Model Estimating the Risk of Developing Adolescent Idiopathic Scoliosis. Cg 20, 246–251. doi:10.2174/1389202920666190730132411
Keywords: adhesion GPCR, mechano-activation, signal transduction, allosteric modulator, activating antibody, extracellular matrix ligand
Citation: Mitgau J, Franke J, Schinner C, Stephan G, Berndt S, Placantonakis DG, Kalwa H, Spindler V, Wilde C and Liebscher I (2022) The N Terminus of Adhesion G Protein–Coupled Receptor GPR126/ADGRG6 as Allosteric Force Integrator. Front. Cell Dev. Biol. 10:873278. doi: 10.3389/fcell.2022.873278
Received: 10 February 2022; Accepted: 12 May 2022;
Published: 23 June 2022.
Edited by:
Nan-Shan Chang, National Cheng Kung University, TaiwanReviewed by:
Paul J. Kammermeier, University of Rochester, United StatesCopyright © 2022 Mitgau, Franke, Schinner, Stephan, Berndt, Placantonakis, Kalwa, Spindler, Wilde and Liebscher. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ines Liebscher, aW5lcy5saWVic2NoZXJAbWVkaXppbi51bmktbGVpcHppZy5kZQ==
† These authors share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.