- 1Department of Biological Chemistry, University of Michigan, Ann Arbor, MI, United States
- 2Department of Molecular, Cellular, and Developmental Biology, University of Michigan, Ann Arbor, MI, United States
Cytoplasmic dynein-1 (dynein) is the primary microtubule minus-end directed molecular motor in most eukaryotes. As such, dynein has a broad array of functions that range from driving retrograde-directed cargo trafficking to forming and focusing the mitotic spindle. Dynein does not function in isolation. Instead, a network of regulatory proteins mediate dynein’s interaction with cargo and modulate dynein’s ability to engage with and move on the microtubule track. A flurry of research over the past decade has revealed the function and mechanism of many of dynein’s regulators, including Lis1, dynactin, and a family of proteins called activating adaptors. However, the mechanistic details of two of dynein’s important binding partners, the paralogs Nde1 and Ndel1, have remained elusive. While genetic studies have firmly established Nde1/Ndel1 as players in the dynein transport pathway, the nature of how they regulate dynein activity is unknown. In this review, we will compare Ndel1 and Nde1 with a focus on discerning if the proteins are functionally redundant, outline the data that places Nde1/Ndel1 in the dynein transport pathway, and explore the literature supporting and opposing the predominant hypothesis about Nde1/Ndel1’s molecular effect on dynein activity.
Introduction
The microtubule-associated motor proteins, cytoplasmic dynein-1 (dynein) and kinesins, promote many types of cellular movements and facilitate intracellular organization in eukaryotes. Dynein and kinesin motors engage with cellular cargos and move along microtubules to power cargo movements that ultimately underlie the spatial and temporal organization of the eukaryotic cytoplasm. Each motor moves in opposite directions on the polar microtubule track: dynein moves in a retrograde fashion toward the microtubule minus-end and nearly all kinesins move in an anterograde manner towards the microtubule plus-end (one exception is the kinesin-14 family, which move towards the minus-end of microtubules (She and Yang, 2017)). Cellular cargos trafficked by dynein and kinesins include, but are not limited to, membrane-bound vesicles, organelles, mRNAs, proteins, and viruses that hijack the motor machinery (Kardon and Vale, 2009; Reck-Peterson et al., 2018). Dynein and kinesins also provide the power to build and separate the mitotic spindle. Dynein localized to the spindle pole body and cortex promotes spindle focusing and alignment (Heald et al., 1996; Gaglio et al., 1997). Dynein at the kinetochore promotes the transition from metaphase to anaphase by trafficking mitotic-spindle checkpoint proteins away from the kinetochore to facilitate progression through metaphase (Vaughan, 2012).
Although both dynein and kinesins use ATP hydrolysis to move along the microtubule track, their evolutionary origins are distinct. Gene duplication and divergence gave rise to a superfamily of kinesins, with each motor specialized for a specific function (Hirokawa and Noda, 2008; Hirokawa et al., 2009). In stark contrast, only a single dynein motor traffics cargo in the cytoplasm and builds the mitotic spindle (Pfister et al., 2006). How dynein can recognize and transport such a diverse array of cargo is only beginning to become clear: a host of dynein regulatory proteins facilitate access to cargo and promote diverse dynein activities.
Over the past 20 years, it has become increasingly apparent that regulation of dynein motility is integral to dynein function. Dynein’s affinity for the microtubule track, ability to move processively, or ability to interact with cargoes are regulated by dynein binding proteins (Kardon and Vale, 2009; Cianfrocco et al., 2015). Unlike many kinesins, which are inherently processive, mammalian dynein must bind to at least two additional regulatory partners to achieve processive movement on microtubules. The multi-subunit complex, dynactin, and one of a class of proteins called activating adaptors convert dynein into a processive motor and together form what we will refer to as the activated dynein complex (McKenney et al., 2014; Schlager et al., 2014) (Figure 1). In addition to promoting dynein activation, each activating adaptor links dynein to specific subsets of cargo (reviewed recently (Reck-Peterson et al., 2018)). Lis1, a protein best known for association with the developmental disease lissencephaly, is an additional dynein regulator that seems to have two functions: Lis1 modulates dynein’s affinity for microtubules in vitro and promotes dynein’s association with dynactin and an activating adaptor (McKenney et al., 2010; Huang et al., 2012; Reiner and Sapir, 2013; Baumbach et al., 2017; DeSantis et al., 2017; Elshenawy et al., 2020; Htet et al., 2020). Together, dynactin, activating adaptors, and Lis1 are three out of four of dynein’s proposed “ubiquitous” regulators, termed such as they are likely involved in all of dynein’s cellular functions (Kardon and Vale, 2009). The fourth ubiquitous regulator designation is shared by the paralogs Nde1 and Ndel1. While there is strong evidence that Nde1 and Ndel1 operate within the dynein transport pathway, the molecular functions of Nde1/Ndel1 remain poorly understood. In this review we will compare Nde1 and Ndel1’s proposed functions, highlight data that places both proteins in the dynein transport pathway, and discuss hypotheses about Nde1/Ndel1’s molecular mechanism.
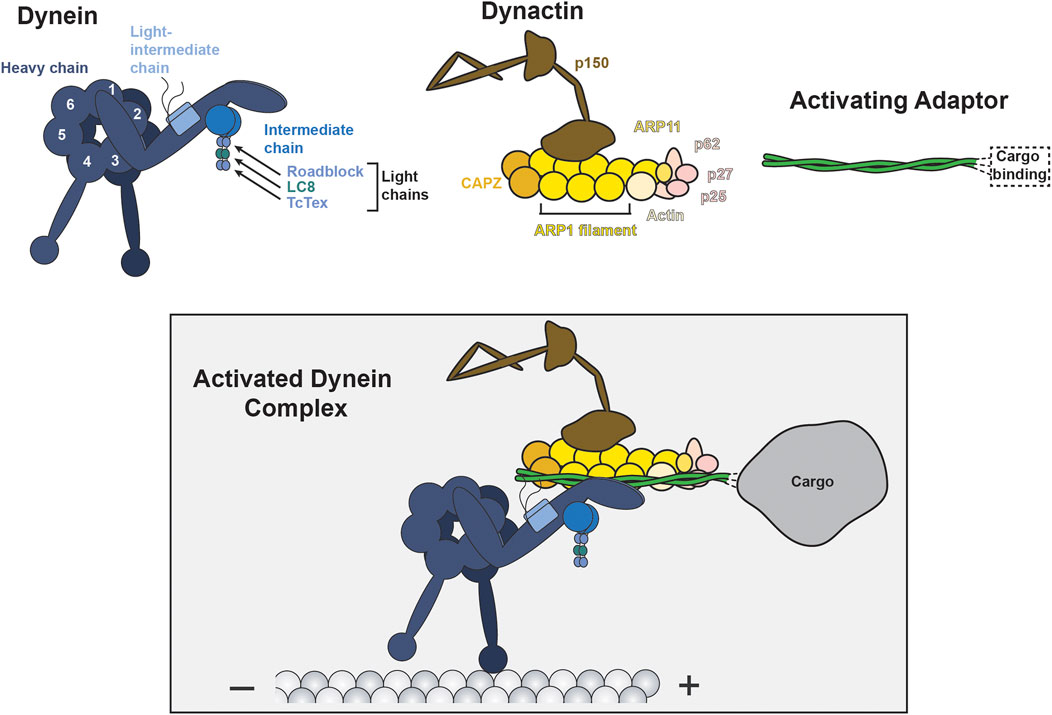
FIGURE 1. Components of the activated dynein complex. The activated dynein complex (boxed) is the active and processive motile complex driven by dynein motor activity. The activated dynein complex is comprised of dynein (blue), dynactin (yellow/brown/pink), and an activating adaptor (green). Dynein is comprised of six dimeric subunits: heavy chain, intermediate chain, light-intermediate chain, and three distinct light chains. The ATPase motor is contained in the heavy chain and is comprised of six separate ATPase Associated with various cellular Activities (AAA) domains (numbered 1–6), three of which (AAA1, AAA3, and AAA4) are capable of actively hydrolyzing ATP. Dynein’s C-terminal tail extends from AAA6, behind the motor ring, and is not shown in this schematic. Dynactin is a 23-polypeptide complex, with some key subunits indicated. All known activating adaptors are coiled coils, bind dynein and dynactin via domains in the N-terminus, and engage with cargos via domains in the C-terminus (reviewed recently (Reck-Peterson et al., 2018)).
Nde1 and Ndel1 are Critical to Development
Nde1/Ndel1 (previously referred to as NudE/Nudel) are conserved from yeast to human. Budding yeast and filamentous fungi contain just one Nde1/Ndel1-type gene, while higher eukaryotes, including zebrafish and Xenopus, have two distinct genes (Drerup et al., 2007; Ma et al., 2009). Although very similar in sequence (human Nde1 and Ndel1 are 56% identical and 69% similar) (Figure 2) (Altschul et al., 2005; Sievers et al., 2011), Nde1 and Ndel1 are expressed differently. While both human proteins are expressed in all tissues, Nde1 expression is highest for a few months immediately after fertilization and then tapers down to a relatively low level in most tissues (Cardoso-Moreira et al., 2019). Ndel1 expression is high and relatively constant throughout life (Bradshaw et al., 2013; Cardoso-Moreira et al., 2019). Consistent with an important role throughout the entire developmental timeline, Ndel1 knockout mice are not viable (Sasaki et al., 2005), while Nde1 knockout mice survive (Feng and Walsh, 2004). Despite differences in viability, both proteins appear to regulate the same process in neurodevelopment: Ndel1 conditional knockout mice and Nde1 knockout mice both display cortical thinning resulting from neuron progenitor migration defects (Feng and Walsh, 2004; Youn et al., 2009; Soto-Perez et al., 2020).
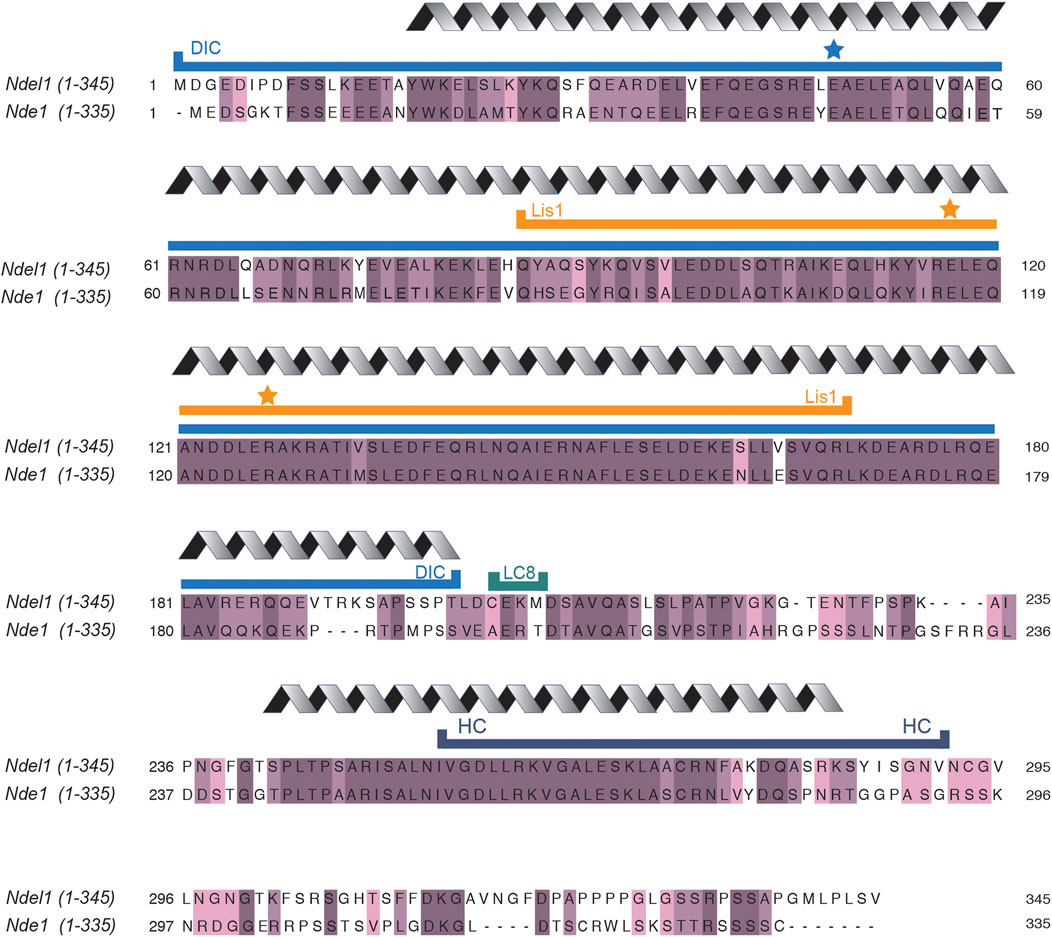
FIGURE 2. Ndel1 and Nde1 sequence alignments. Sequence alignments of human Ndel1 and Nde1 were performed with Clustal Omega (Sievers et al., 2011). Dark purple indicates residues that are fully conserved, medium purple indicates amino acids that are well-conserved with respect to side chain properties, light purple indicates residues that are weakly conserved. Grey coil above sequence indicates regions that are predicted or observed to be coiled-coils (Derewenda et al., 2007; Ye et al., 2020). All other regions not marked with grey coil are predicted to be random coil or disordered. Minimal regions required to bind dynein intermediate chain (blue line labeled with DIC), Lis1 (orange line labeled with Lis1), dynein light chain 8 (teal line labeled with LC8), and dynein heavy chain (dark blue line labeled with HC) are indicated. Blue star indicates residues that, when mutated, ablate binding to dynein intermediate chain (Derewenda et al., 2007; Wang and Zheng, 2011). Orange stars indicate residues that, when mutated, ablate binding to Lis1 (Wang and Zheng, 2011; Żyłkiewicz et al., 2011; Wang et al., 2013).
Nde1 and, to a lesser extent, Ndel1 are associated with neurodevelopmental diseases. Biallelic Nde1 mutation or deletion is associated with severe brain malformations, including microcephaly, microlissencephaly, and microhydrancephaly (Alkuraya et al., 2011; Bakircioglu et al., 2011; Guven et al., 2012; Lipka et al., 2013; Paciorkowski et al., 2013; Abdel‐Hamid et al., 2019). Copy number variation of the locus containing Nde1 is associated with epilepsy, autism, and intellectual disability (Ullmann et al., 2007; Hannes et al., 2009; Heinzen et al., 2010; Mefford et al., 2010; Tropeano et al., 2013). Schizophrenia is associated with duplication or deletion of the gene locus containing Nde1 and Ndel1 (Kirov et al., 2009; Ingason et al., 2011; Malhotra and Sebat, 2012; Rees et al., 2014; Johnstone et al., 2015). It is unclear if Nde1’s stronger link to disease phenotypes is due to differences in the two proteins’ functions or simply due to differences in expression during development. Given that Ndel1 knockout is embryonic lethal, substantial copy number variation or mutation of Ndel1 may not be permissive to development (Sasaki et al., 2005).
The Molecular Function of Nde1/Ndel1 Requires Further Study
The molecular mechanism of Nde1/Ndel1 remains ill-defined. There are two primary reasons for the uncertainty surrounding Nde1/Ndel1’s molecular function. First, experiments probing Nde1/Ndel1 function have been conducted in many different model organisms, cell culture systems, and in vitro, and have yielded contradictory findings. For example, mammalian Nde1 appears to negatively regulate dynein’s microtubule binding affinity, while the yeast Nde1/Ndel1 homolog does not (Yamada et al., 2008; McKenney et al., 2010; McKenney et al., 2011; Torisawa et al., 2011; Huang et al., 2012). It is not clear if contradictory results arise from bona fide functional differences across species, differences in experimental systems, or reflect functional plasticity inherent in Nde1/Ndel1. Secondly, Nde1/Ndel1, in conjunction with dynein, regulate multiple and diverse cellular processes ranging from promoting spindle assembly during division to driving nuclear oscillations in dividing neural progenitor cells. It is not clear if Nde1/Ndel1 fulfill the same role during each process they support.
Despite relatively high homology and a shared importance in neurodevelopment, in some contexts, Nde1 and Ndel1 seem to regulate different cellular events (Vergnolle and Taylor, 2007; Doobin et al., 2016). The extent to which Nde1 and Ndel1 have divergent or redundant functionality has not been firmly established. Both proteins are subject to extensive and often unique post-translational modifications (reviewed in (Bradshaw et al., 2013)). In cases where the proteins receive the same post-translational modification on the same amino acid, the modification can have different effects on the association of Nde1 and Ndel1 with dynein. For example, while both Nde1 and Ndel1 can be palmitoylated, this modification negatively regulates Ndel1 binding to dynein, but does not affect Nde1’s interaction with dynein (Shmueli et al., 2010). Finally, while both proteins bind directly to dynein and some of its regulatory proteins (discussed in detail below), Nde1 and Ndel1 each interact with a unique subset of other proteins (reviewed in (Bradshaw et al., 2013)), supporting the idea that Nde1 and Ndel1 may enable dynein to access different cellular pathways. Below we will discuss Nde1 and Ndel1’s cellular function, with specific emphasis on work that probes Nde1/Ndel1’s function with respect to dynein. We will also highlight, whenever possible, evidence that Nde1 and Ndel1 serve similar or disparate cellular roles.
Cellular Evidence for Nde1/Ndel1’s Role as a Dynein Regulator
Dynein is a remarkably multifunctional motor protein. As the primary retrograde microtubule motor in most eukaryotes, dynein must engage with hundreds of different types of cargo and provide the power to reshape and focus the mitotic spindle. There is evidence that Nde1 and Ndel1 are involved in most dynein-dependent processes (Simões et al., 2018).
Nuclear Positioning
One of the first observable functions of dynein was to promote proper nuclear positioning in fungi (Xiang et al., 1994; Plamann et al., 1994; Eshel et al., 1993; Li et al., 1993). A Nde1/Ndel1-type protein was first identified in mutagenesis screens seeking to identify proteins that, like dynein, promote nuclear positioning in the filamentous fungi N. crassa and A. nidulans (Minke et al., 1999; Efimov and Morris, 2000; Bruno et al., 1996). Mutants of the Nde1/Ndel1-type proteins identified (called ro-11 in N. crassa and NudE in A. nidulans) display defective hyphal growth and nuclear distribution defects, with nuclei failing to migrate into the hyphae and accumulating closer to the spore body (Figure 3A). This phenotype placed ro-11 and NudE in the dynein regulatory pathway and was the first evidence that a Nde1/Ndel1-type protein may work in conjunction with dynein to promote nuclear distribution (Li et al., 1993; Plamann et al., 1994; Xiang et al., 1994; Xiang et al., 1995).
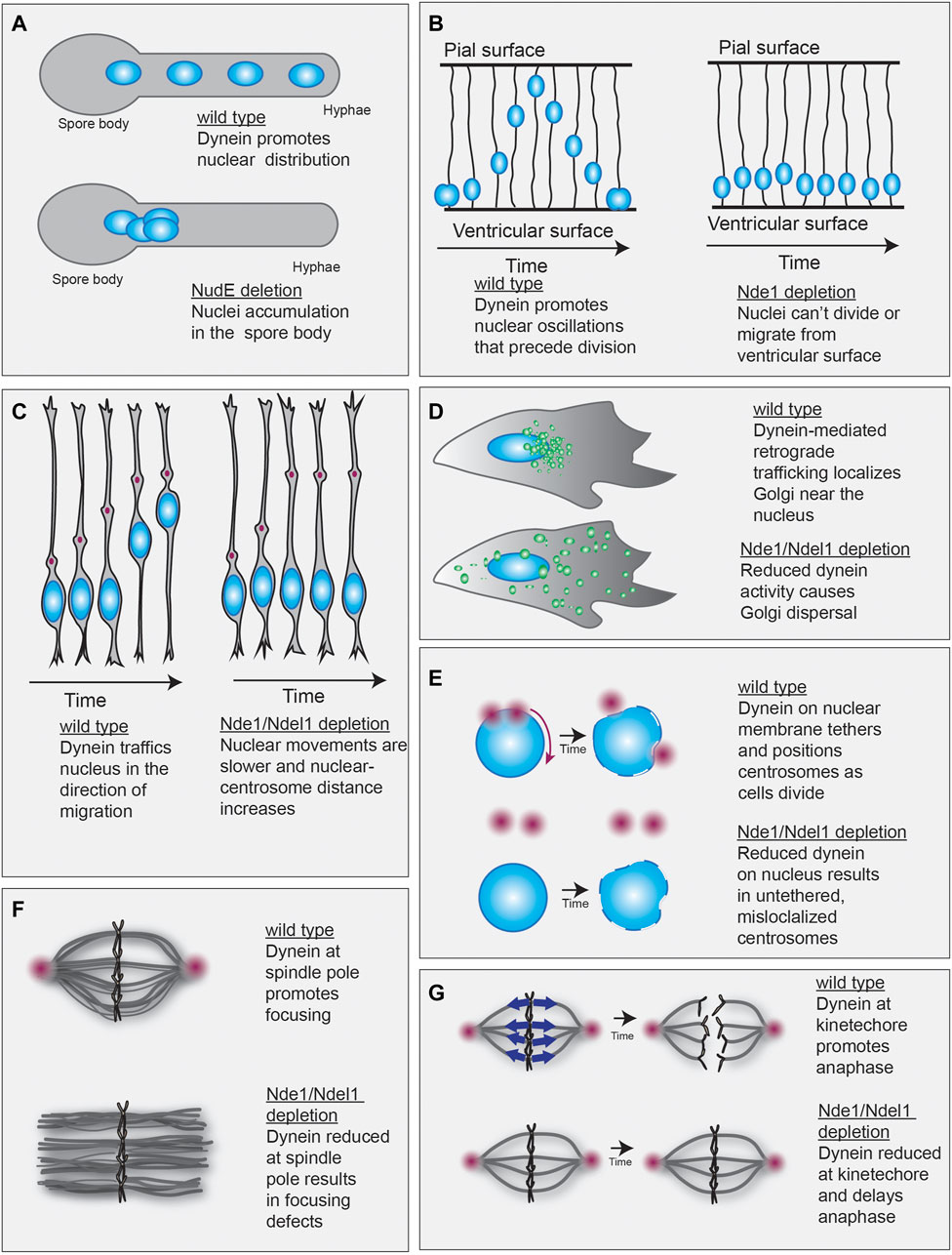
FIGURE 3. Evidence of a role for Nde1/Ndel1 in dynein-mediated transport. (A) Dynein supports localization of nucleus (blue spheres) along the hyphae in filamentous fungi (top). NudE or ro-11 depletion causes nuclei to accumulate in the spore body (bottom) (Bruno et al., 1996; Minke et al., 1999; Efimov and Morris, 2000). (B) Dynein drives the ventricular-directed movement of nuclei in neural progenitors. These oscillations are required for cell division (left). Nde1 depletion prevents nuclear oscillatory motions, likely by inhibiting mitosis (right) (Doobin et al., 2016). (C) After division, neural progenitor cells migrate away from the ventricular zone. During this type of migration, the centrosome (red) is oriented in the direction of migration and its forward movement precedes nucleokinesis (left). Nde1/Ndel1 depletion prevents dynein-driven nucleokinesis, resulting in slow migration velocities and increased distance between the centrosome and nucleus (right) (Shu et al., 2004; Youn et al., 2009; Doobin et al., 2016). (D) Retrograde trafficking mediated by dynein promotes the juxta-nuclear localization of the Golgi (green) (top). Nde1/Ndel1 depletion results in Golgi dispersal, suggesting dynein transport defects (bottom) (Liang et al., 2004; Lam et al., 2010). (E) Dynein anchored at the nuclear envelope helps position and anchor centrosomes during cell division (top). Nde1/Ndel1 depletion reduces dynein at the nuclear envelope, which leads to impaired centrosome localization (bottom). (F) Dynein promotes spindle focusing (top). Depletion of Nde1 and Ndel1 in xenopus egg extracts generates spindle focusing defects (bottom) (Wang and Zheng, 2011; Wang et al., 2013). (G) Dynein localized to the kinetochore primarily functions to inactivate the mitotic spindle checkpoint to promote transition to anaphase. Dynein (blue arrows) accomplishes the checkpoint inactivation by trafficking check-point proteins away from the kinetochore and towards the spindle pole, where microtubule minus-ends are clustered (red sphere) (left). Nde1 depletion reduces dynein localization at the kinetochore resulting in prolonged mitosis (right) (Vergnolle and Taylor, 2007).
Nde1 and Ndel1 are implicated in nuclear positioning in other organisms, as well. Nde1 promotes the nuclear oscillations that precede radial glial progenitor cell division. This process, called interkinetic nuclear migration (INM), occurs in the ventricular zone of the developing cortex (reviewed in (Bertipaglia et al., 2018)). Although the developmental function of INM is unknown, it is a conserved process (Kishimoto et al., 2013; Leung et al., 2011; Meyer et al., 2011; Tsai et al., 2010; Azizi et al., 2020). Dynein, dynactin, and Lis1 promote the apical-directed nuclear movements during INM (Tsai et al., 2010; Tsai et al., 2005; Tsai et al., 2007; Del Bene et al., 2008). Nde1 depletion also impairs apical-directed nuclear migration, suggesting a shared function with dynein transport machinery (Doobin et al., 2016) (Figure 3B). Interestingly, Ndel1 depletion does not effect INM, supporting distinct functionality of these proteins during this process (Doobin et al., 2016).
After INM, neural progenitor cells migrate out of the ventricular zone to the outer strata of the brain. During this process, dynein-dependent nucleokinesis occurs, where dynein powers the transport of the nucleus in the direction of migration. Lis1 depletion impairs nucleokinesis during neuron migration, supporting its role in dynein activation (Youn et al., 2009; Tsai et al., 2007). Consistent with this, Ndel1 depletion increases the centrosome to nucleus distance in migrating mouse neurons, which is a hallmark of impaired nucleokinesis (Youn et al., 2009; Shu et al., 2004). Similarly, depletion of Nde1 impairs migration velocity, which is consistent with what is seen upon Ndel1 depletion (Doobin et al., 2016; Shu et al., 2004) (Figure 3C). Furthermore, exogenous Nde1 expression can rescue Ndel1 depletion defects during migration, supporting an overlapping role during migratory nucleokinesis (Doobin et al., 2016).
Cargo Trafficking
One of dynein’s primary functions in somatic cells is cargo trafficking. In rat dorsal root ganglia, Ndel1 depletion has little effect on retrograde cargo trafficking flux, but when combined with Lis1 depletion, both retrograde and anterograde trafficking is severely impaired (Pandey and Smith, 2011). This result is similar to when the dynein heavy chain is depleted (Pandey and Smith, 2011). A common hallmark of dynein malfunction is Golgi dispersal (Burkhardt et al., 1997; Corthésy-Theulaz et al., 1992). In HeLa or HEK293T cells, Nde1 or Ndel1 depletion results in mild Golgi dispersal (Liang et al., 2004; Lam et al., 2010). Significant dispersal occurs when both Nde1 and Ndel1 are depleted simultaneously (Lam et al., 2010) (Figure 3D). Exogenous expression of either Nde1 or Ndel1 can rescue the dispersal phenotype, suggesting an overlapping function of the paralogs (Lam et al., 2010). Similarly, microinjection of a function-blocking anti-Nde1/Ndel1 antibody causes rapid dispersal of acidic organelles, suggesting acute inhibition of retrograde, dynein-mediated trafficking (Yi et al., 2011). In COS1 cells, Nde1 and Ndel1 depletion reduced microtubule-motor-dependent movement of lipid droplets. Specifically, overall movement and run lengths of lipid droplets were reduced upon simultaneous Nde1 and Ndel1 depletion (Reddy et al., 2016). Together, this body of evidence supports a role for both Nde1 and Ndel1 in cargo trafficking.
Mitotic Functions
Some of the most substantial evidence that Nde1/Ndel1 promote dynein activity comes from studies probing dynein activity during cell division. Dynein promotes multiple steps of cell division (Dwivedi and Sharma, 2018). First, dynein activity at the nuclear pore complex promotes nuclear envelope breakdown that precedes division (Salina et al., 2002; Li et al., 2010). Second, dynein facilitates duplicated centrosome positioning and helps assemble and focus the mitotic spindle (Robinson et al., 1999; Sitaram et al., 2012; Titus and Wadsworth, 2012). Finally, dynein promotes the transition from metaphase to anaphase by transporting checkpoint proteins away from kinetochores (Gassmann et al., 2010; Howell et al., 2001). There is evidence that Nde1/Ndel1 support nearly all of dynein’s mitotic dynein functions. Both Nde1 and Ndel1 drive dynein localization to the nuclear envelope to promote nuclear envelope breakdown and positioning of centrosomes, (Figure 3E), promote spindle formation and focusing (Figure 3F), and facilitate dynein localization at the kinetochore in prometaphase (Figure 3G) (Liang et al., 2007; Mori et al., 2007; Stehman et al., 2007; Vergnolle and Taylor, 2007; Hebbar et al., 2008; Wainman et al., 2009; Bolhy et al., 2011; Wang and Zheng, 2011; Żyłkiewicz et al., 2011; Monda and Cheeseman, 2018; Wynne and Vallee, 2018). Although Nde1 and Ndel1 seem capable of fulfilling similar roles during cell division, Nde1 drives dynein localization at kinetochores to a greater extent than Ndel1 (Vergnolle and Taylor, 2007). Additionally, Nde1 depletion impairs cell division more than Ndel1 depletion in neural progenitors (Doobin et al., 2016). Together, these results suggest that Nde1 and Ndel1 may have subtly distinct roles during cell division.
Additional Functions of Nde1/Ndel1
Though this review is focused on Nde1/Ndel1’s role in dynein activity, it is important to emphasize that Nde1 and Ndel1 have additional cellular functions, some of which may be dynein-independent. One of the best characterized interaction partners of Nde1 and Ndel1 is the protein Disrupted-In-Schizophrenia-1 (DISC1) (Porteous et al., 2006). DISC1 is involved in the pathology of psychiatric disorders including schizophrenia, bipolar disorder, and depression (Korth, 2009; Soares et al., 2011). DISC1 is part of a large interaction network with proteins that are involved in several key signaling pathways (Millar et al., 2005; Soares et al., 2011; Bradshaw and Porteous, 2012). The protein binding interaction between Nde1, Ndel1, and DISC1 is well-established and the genetic interaction between Nde1/Ndel1 and DISC1 is associated with an increased risk for schizophrenia (Morris, 2003; Ozeki et al., 2003; Brandon et al., 2004; Burdick et al., 2008; Ye et al., 2020). However, the molecular outcome of Nde1/Ndel1 binding to DISC1 is not yet fully elucidated. There is evidence that DISC1 may promote Nde1’s localization to kinetochores, which may in turn promote dynein localization (Ye et al., 2020). However, more work is required to determine if Nde1 or Ndel1 function to modulate DISC1 activity, to promote DISC1 access to the dynein transport pathway, or if the role that Nde1 and Ndel1 play in DISC1 biology represent a largely dynein-independent process.
Nde1 plays a role in safeguarding the genome of neural progenitor cells against DNA damage (Houlihan and Feng, 2014). During DNA replication in S-phase, Nde1 homozygous deletion knockout mice (Nde1_/_) accrue double stranded DNA breaks that result in elevated levels of apoptosis (Houlihan and Feng, 2014). It is hypothesized that nuclear-localized Nde1 interacts with the chromatin associated proteins, like cohesin and the remodeler SNF2h, to aid in DNA remodeling during replication (Houlihan and Feng, 2014). Given the absence of dynein from the nucleus during S-phase, it is unlikely that dynein is involved in this Nde1 functionality.
Nde1 is negative regulator of primary cilia length, with excess expression of Nde1 promoting short cilia (Kim et al., 2011; Maskey et al., 2015). To modulate ciliary length, Nde1 protein levels are tightly controlled during G1/G0 and CDK5 phosphorylation of Nde1 ultimately promotes its proteasomal degradation (Maskey et al., 2015). The role of dynein in this Nde1 activity is unknown.
Ndel1 is a serine oligopeptidase and can cleave unstructured peptides (Hayashi et al., 2000; Hayashi et al., 2005). The peptidase activity is inhibited by DISC1 binding, suggesting that Ndel1 peptidase activity is under tight regulation (Hayashi et al., 2005). How or if Ndel1’s oligopeptidase activity effects Ndel1-mediated dynein regulation remains to be determined.
The Molecular Determinants of Nde1/Ndel1’s Regulation of Dynein
Despite evidence that Nde1 and Ndel1 function in the dynein transport pathway, their molecular function with respect to dynein activity remains elusive. Several in vitro studies have used purified proteins to investigate the functional outcome of Ndel1 and Nde1’s interaction with dynein or its regulatory partners. However, it is still not clear how or if Nde1/Ndel1 directly affect dynein motility. It is worth noting that the majority of in vitro work designed to understand how Nde1/Ndel1 influence dynein motility was conducted before the discovery that dynein requires both dynactin and an activating adaptor to achieve true processive motility (McKenney et al., 2014; Schlager et al., 2014). Below, we will outline the data that describe how Nde1 or Ndel1 interact with dynein and its regulatory proteins, then discuss the evidence for and against the current model for how Nde1/Ndel1 modulate dynein activity.
Nde1/Ndel1 Structure
Nde1 and Ndel1 have similar secondary structures, with both proteins containing long stretches of coiled-coil and random coil (Figure 2). Both proteins are dimers and may assemble into tetramers under some conditions (Derewenda et al., 2007; Soares et al., 2012). There is evidence that Nde1 and Ndel1 can form heterodimers with each other, as well, although it is not clear what proportion of the Nde1 and Ndel1 population are homodimers vs heterodimers in vivo (Bradshaw et al., 2009).
Nde1/Ndel1 Bind to Lis1
Nde1 and Ndel1 and Lis1 binding is well-described and well-characterized (McKenney et al., 2010; Efimov and Morris, 2000; Wang and Zheng, 2011; Derewenda et al., 2007; Tarricone et al., 2004; Feng et al., 2000). The binding interface for Lis1 is contained within amino acids 85–169 of Ndel1 (a region that is highly conserved with Nde1) (Derewenda et al., 2007). The interaction of Ndel1 with Lis1 is relatively strong, with a high nanomolar dissociation constant (Tarricone et al., 2004). On Ndel1, the interaction has been mapped with high precision: both Ndel1E116A and Ndel1R127A completely abolish binding to Lis1, as demonstrated with human and Xenopus proteins (indicated by orange stars in Figure 2) (Derewenda et al., 2007; Wang and Zheng, 2011). The interaction with Nde1/Ndel1 has been mapped to one side of the Lis1 beta-propeller, and yeast two-hybrid experiments have implicated the Lis1 residues Lis1S169 and Lis1H149 as key mediators of the interaction with Nde1 and Ndel1 (Feng et al., 2000; Tarricone et al., 2004).
Nde1/Ndel1 Bind to Dynein
There is evidence that Nde1 and Ndel1 bind directly to multiple sites on the dynein motor (Figure 4A). One of the binding sites for Nde1/Ndel1 is on the dynein intermediate chain (McKenney et al., 2011; Wang and Zheng, 2011; Żyłkiewicz et al., 2011; Wang et al., 2013; Nyarko et al., 2012). Nde1 and Ndel1 constructs containing the first ∼200 amino acids are sufficient to bind dynein intermediate chain robustly, and the point mutant, Ndel1E48A, inhibits binding (marked by a blue star in Figure 2) (Wang and Zheng, 2011; Żyłkiewicz et al., 2011; Wang et al., 2013). The binding site for Nde1/Ndel1 is within the first 40 amino acids of dynein intermediate chain (McKenney et al., 2011; Nyarko et al., 2012). This interaction is well-supported as much of the work validating it was performed with pure proteins (McKenney et al., 2011; Wang and Zheng, 2011; Nyarko et al., 2012; Wang et al., 2013), allowing for detailed affinity measurement showing a dissociation constant in the low micromolar range (Nyarko et al., 2012; Wang et al., 2013).
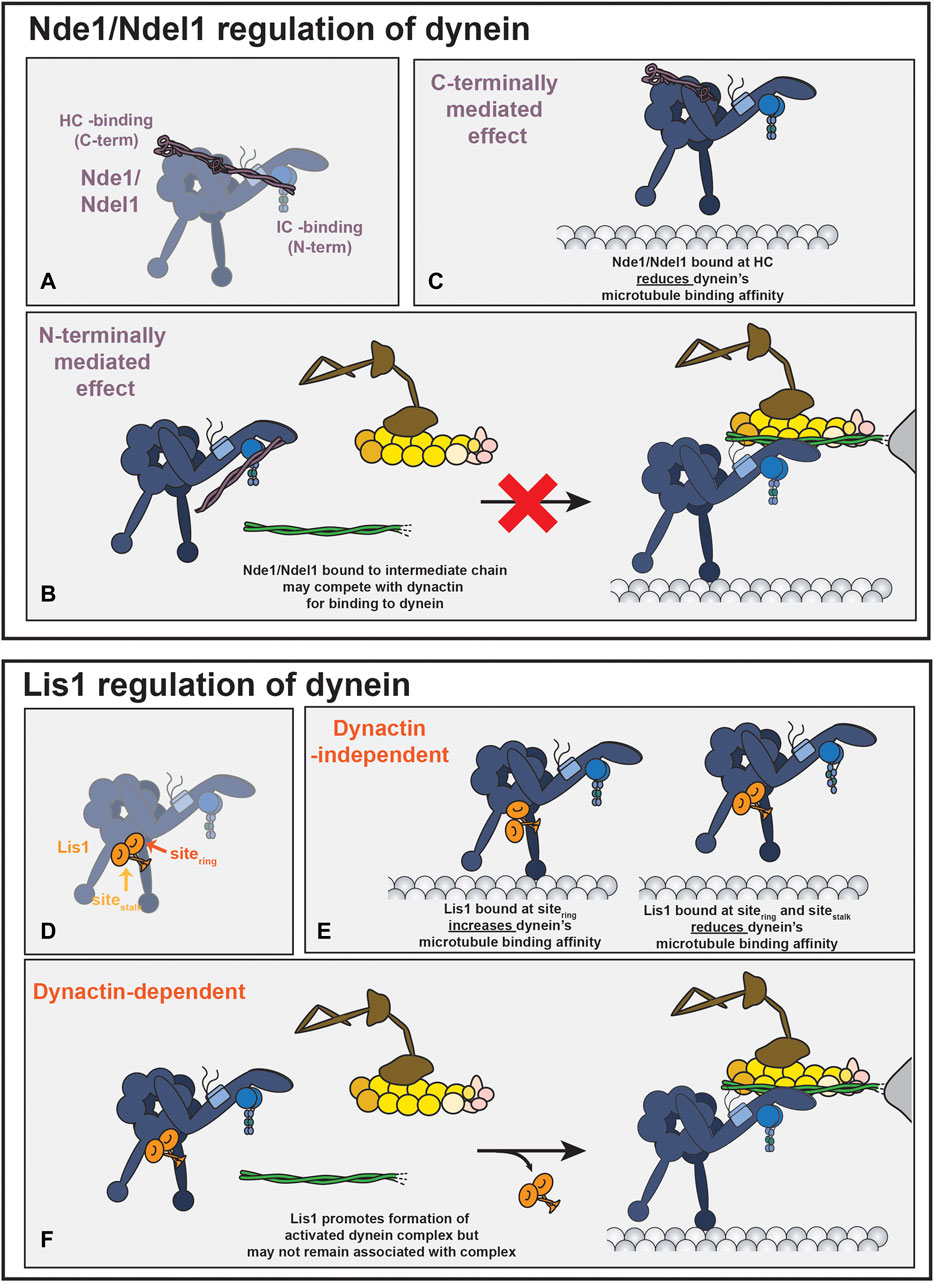
FIGURE 4. Nde1/Ndel1 and Lis1 regulation of dynein. (A) Nde1/Ndel1, shown in purple, binds to up to three places on the dynein motor. The binding with the intermediate chain (IC) is mediated by Nde1/Ndel1’s N-terminus and binding with the heavy chain (HC) is mediated by Ndel1’s C-terminus (Liang et al., 2004; Sasaki et al., 2005; McKenney et al., 2011; Wang and Zheng, 2011; Żyłkiewicz et al., 2011; Nyarko et al., 2012; Wang et al., 2013). Binding between Nde1/Ndel1 and dynein’s LC8 chain is not shown (Stehman et al., 2007; McKenney et al., 2011). Although this schematic shows Nde1/Ndel1 binding to both the intermediate chain and the heavy chain simultaneously, this has not been observed directly. (B) By binding competitively with dynactin for dynein’s intermediate chain, Nde1/Ndel1’s N-terminus could potentially inhibit activated dynein complex formation (McKenney et al., 2011; Nyarko et al., 2012; Jie et al., 2017). (C) The interaction between Nde1/Ndel1’s C-terminal fragment and dynein reduces dynein’s interaction with the microtubule track (Torisawa et al., 2011). (D) Lis1 binds to two places on the dynein motor domain. Sitering (indicated with dark orange arrow) is located on the motor domain between AAA3 and AAA4. Sitestalk, indicated with a light orange arrow, is located on the stalk insertion in AAA4. Two binding sites have been observed with yeast and human proteins (DeSantis et al., 2017; Htet et al., 2020). (E) Lis1 binding at just sitering increases dynein’s affinity for microtubules. Lis1 binding at both sites reduces dynein’s microtubule binding affinity. The low-affinity regulator state, however, has only been observed with yeast proteins. (F) Lis1, likely by binding to both sites, promotes dynein’s association with dynactin and an activating adaptor (Gutierrez et al., 2017; Elshenawy et al., 2020; Htet et al., 2020). Lis1 likely promotes association with dynactin and activating adaptor by disfavoring a conformation of dynein that has a reduced affinity for dynactin (not shown) (Zhang et al., 2017; Qiu et al., 2019; Elshenawy et al., 2020; Htet et al., 2020; Marzo et al., 2020). Once the activated dynein complex is formed, it is not clear if Lis1 remains bound to the activated dynein complex, as some studies have observed that Lis1 dissociates after complex formation and others have observed that Lis1 remains bound (Baumbach et al., 2017; Gutierrez et al., 2017; Jha et al., 2017; Elshenawy et al., 2020; Htet et al., 2020). Lis1 also promotes formation of activated dynein complexes with two dynein dimers (not shown) (Elshenawy et al., 2020; Htet et al., 2020). While not illustrated in this figure, the tether model suggests that Nde1/Ndel1 associated with dynein IC can promote Lis1 binding on the dynein motor domain.
In addition to dynein intermediate chain, Ndel1 is also reported to bind to dynein heavy chain. Yeast two-hybrid assays and immunoprecipitations from HEK293 cell lysate support a binding interface between Ndel1 amino acids 256–291 and dynein’s heavy chain, potentially near one of dynein’s primary ATPase domains (AAA1) or the C-terminus (Liang et al., 2004; Sasaki et al., 2000). Despite relatively high conservation with Ndel1 in the proposed binding site, purified Nde1 and dynein motor domain do not bind in vitro (Figure 2) (McKenney et al., 2010).
Finally, Nde1 (amino acids 200–203) binds directly to the dynein light chain, LC8 (Stehman et al., 2007; McKenney et al., 2011). Intriguingly, these residues are not completely conserved between Nde1 and Ndel1, highlighting another potential difference in dynein association between the two paralogs.
Nde1 and Ndel1 May Bind Directly to Some Dynein Cargo
Nde1 and Ndel1 bind directly to GTP-Rab9A with low micromolar affinity (Zhang et al., 2022). The interaction between Rab9a and Nde1/Ndel1 likely promotes dynein’s association to Rab9-positive late endosomes (Zhang et al., 2022). This finding suggest that Nde1/Ndel1 may function as cargo adaptors in some contexts and directly link the dynein transport machinery to certain cargos.
Nde1/Ndel1 Compete with Dynactin for Binding to Dynein Intermediate Chain
In addition to binding to Nde1 and Ndel1, dynein intermediate chain interacts with the p150 (also called p150Glued) subunit in dynactin (Nyarko et al., 2012; Jie et al., 2017; Karki and Holzbaur, 1995; Siglin et al., 2013; Vaughan and Vallee, 1995; Paschal et al., 1993). p150 and Nde1/Ndel1 associate with dynein intermediate chain via overlapping, yet not identical binding sites and Nde1 appears to bind competitively with p150 (McKenney et al., 2011; Nyarko et al., 2012; Jie et al., 2017). The competitive nature of these interactions suggest that Nde1/Ndel1 may affect or regulate the formation of the activated dynein complex (Figure 4B). However, the regions of p150 and dynein intermediate chain that interact are not resolved in the structures of the activated dynein complex, making it difficult to conclude how Nde1/Ndel1 and p150 competition for dynein intermediate chain would affect dynein activation (Urnavicius et al., 2015; Grotjahn et al., 2018; Urnavicius et al., 2018).
Nde1/Ndel1 May Modulate Dynein’s Microtubule-Binding Affinity
Several studies show that mammalian Nde1 and Ndel1 negatively regulate dynein’s microtubule-binding affinity (McKenney et al., 2010; McKenney et al., 2011; Yamada et al., 2008; Torisawa et al., 2011). C-terminal truncations of Ndel1 are sufficient to recapitulate the reduction in dynein’s microtubule binding affinity (Torisawa et al., 2011) (Figure 4C). The mechanistic basis or physiological relevance for the change in microtubule-binding affinity is unknown. Nde1/Ndel1 from budding yeast (called Ndl1) lacks the C-terminal domain and does not modulate yeast dynein’s microtubule-binding affinity (Huang et al., 2012), suggesting that the ability for Nde1 and Ndel1 to regulate dynein’s microtubule-binding affinity is not shared by all organisms.
Support For and Against the Model That Nde1/Ndel1 Tethers Lis1 to Dynein
Lis1 is a catalytically inactive subunit of the platelet-activating factor acetylhydrolase (PAF-AH) and a well-studied dynein regulatory protein (reviewed recently (Markus et al., 2020)). Several studies over the last decade have revealed how Lis1 regulates dynein (Huang et al., 2012; DeSantis et al., 2017; Htet et al., 2020; Elshenawy et al., 2020; Baumbach et al., 2017; Splinter et al., 2012; Marzo et al., 2020; Qiu et al., 2019; Gutierrez et al., 2017; Jha et al., 2017) (Figures 4D–F). Lis1 interaction with dynein has two general outcomes, which are not necessarily mutually exclusive: first, in the absence of dynactin and an activating adaptor, Lis1 modulates dynein’s apparent affinity for microtubules (Huang et al., 2012; DeSantis et al., 2017; Htet et al., 2020; McKenney et al., 2010; Toropova et al., 2014) (Figure 4E). Secondly, in the presence of dynactin and an activating adaptor, Lis1 increases the formation of the activated dynein complex by disfavoring dynein’s autoinhibited conformation (Htet et al., 2020; Elshenawy et al., 2020; Marzo et al., 2020; Qiu et al., 2019; Zhang et al., 2017) (Figure 4F). Lis1 also promotes the formation of activated dynein complexes that contain two dynein dimers, which results in an activated dynein complex that moves with an elevated velocity (Htet et al., 2020; Elshenawy et al., 2020). Lis1 binds to two different sites on the dynein motor, although it is currently unclear what contribution each binding site has to each Lis1 function (Figure 4D) (DeSantis et al., 2017; Htet et al., 2020; Gillies et al., 2022). One of the cellular outcomes of Lis1’s interaction with dynein is to promote dynein’s association with dynactin and cargo-bound activating adaptors (Coquelle et al., 2002; Lee et al., 2003; Sheeman et al., 2003; Markus et al., 2009; Splinter et al., 2012). It is currently unclear how Nde1/Ndel1 fit into this schema.
The prevailing model for Nde1/Ndel1’s function in the dynein transport pathway is to increase Lis1’s association with dynein. Nde1/Ndel1 are hypothesized to tether dynein and Lis1 together (Tarricone et al., 2004; McKenney et al., 2010; Wang and Zheng, 2011; Żyłkiewicz et al., 2011). In this model, Nde1/Ndel1 act as an amplifier of the Lis1 effect by increasing the effective affinity between Lis1 and dynein. There is significant evidence that supports the tether model. In budding yeast, filamentous fungi, Xenopus egg extract, several cell culture lines, and in migrating neurons, the deleterious effects of Nde1/Ndel1 depletion can be rescued by Lis1 overexpression, which is consistent with a model where Nde1/Ndel1’s purpose is to tether Lis1 to dynein (Efimov, 2003; Shu et al., 2004; Li et al., 2005; Lam et al., 2010; Wang and Zheng, 2011; Moon et al., 2014). Overexpression of the PAF-AH catalytic subunits, α1 and α2, inactivates dynein, likely by competing for Lis1 binding (Ding et al., 2009). Ndel1 overexpression rescues the α1 and α2-induced dynein inactivation, suggesting that Ndel1 can promote Lis1-dynein association (Ding et al., 2009). In A. nidulans, mutations in dynein that promote its ability to form the activated dynein complex partially rescue the effect of NudE depletion, bolstering the idea that NudE works in conjunction with Lis1 to activate dynein (Qiu et al., 2019). In further support of a tethering function, experiments conducted with purified mammalian proteins have shown that Nde1 increases the association of Lis1 and dynein (McKenney et al., 2011). Consistent with a tethering role, with purified S. cerevisiae proteins, Ndl1 decreases the amount of Lis1 (called Pac1 in yeast) required to slow the velocity with which dynein moves on microtubules, suggesting that Ndl1 acts to increase the effective concentration of Pac1 (Huang et al., 2012). (It is important to note that yeast dynein is processive in the absence of dynactin and an activating adaptor and that Pac1 slows the velocity of walking dynein (Reck-Peterson et al., 2006; Huang et al., 2012).) Altogether, there is compelling data that support a role for Nde1/Ndel1 in stabilizing or promoting the interaction with dynein and Lis1 in a way that amplifies Lis1’s effect on dynein.
However, not all studies support a simple tether model of Nde1/Ndel1 function. Lis1 overexpression does not rescue the axon swelling caused by Ndel1 depletion in neurons (Kuijpers et al., 2016). Furthermore, the centrosome localization defect caused by Ndel1 depletion can be rescued by exogenous expression of a Ndel1 mutant that cannot bind Lis1 (Monda and Cheeseman, 2018). Similarly, anaphase onset delays induced by Nde1 depletion are largely rescued by expression of a Nde1 mutant that doesn’t bind Lis1 (Wynne and Vallee, 2018). Importantly, a dynein binding-deficient Nde1 construct does not rescue the anaphase onset delays, supporting a dynein-dependent role for Nde1 during cell cycle progression (Wynne and Vallee, 2018). Additionally, Lis1 mutation and Nde1 mutation are associated with distinct neurodevelopmental diseases. Nde1 mutation is largely associated with microcephaly, while Lis1 mutation is associated with lissencephaly (Lipka et al., 2013; Reiner and Sapir, 2013), suggesting that Lis1 and Nde1/Ndel1 have unique functions during development.
The strongest evidence that opposes the tether model comes from in vitro studies with mammalian proteins. The Lis1-mediated increase in dynein’s microtubule-binding affinity is not amplified by the presence of Nde1 or Ndel1 as the tether model predicts. Instead, the decrease in microtubule-binding affinity caused by Nde1 and Ndel1 and the increase caused by Lis1 is additive (Yamada et al., 2008; McKenney et al., 2010; Torisawa et al., 2011). Additionally, Lis1 promotes the association of dynein with dynactin, while Nde1/Ndel1 may compete with dynactin for dynein binding (McKenney et al., 2011; Nyarko et al., 2012; Splinter et al., 2012; Elshenawy et al., 2020; Htet et al., 2020).
What could be the function of Nde1/Ndel1 if they promote Lis1-dynein binding while simultaneously inhibiting Lis1’s ability to increase dynein’s microtubule-binding affinity or recruit dynactin? If Nde1/Ndel1 are not functioning as a simple tether, what is their function? One possibility is that Nde1/Ndel1 promote Lis1 association with dynein in a step that is temporally separated from Lis1-mediated dynein activation. Indeed, during INM, two separate pools of dynein are recruited to the nuclear envelope. The activating adaptor BicD2 recruits one pool of dynein to the nuclear envelope (Hu et al., 2013). Dynein recruited in this stage is likely bound to both BicD2 and dynactin in the activated dynein complex (Splinter et al., 2010). A second pool of dynein is sequentially recruited to the nuclear envelope in a Nde1/Ndel1 dependent manner (Hu et al., 2013). Given that Lis1 depletion effects INM at multiple stages, it is likely that Lis1 is involved in recruiting both pools of dynein (Tsai et al., 2005). This may suggest that Nde1/Ndel1’s function in the dynein transport pathway isn’t to promote binding of dynein to dynactin and an activating adaptor, as Lis1 does, but to fulfill some other, unknown role that is temporally separated from Lis1-mediated activation. Since dynein and Lis1 localization are often dependent on Nde1/Ndel1’s proper localization, we hypothesize that Nde1/Ndel1 may function as scaffolds to colocalize dynein and Lis1 while preventing Lis1 from acting on dynein. Because Nde1/Ndel1 may bind competitively with dynactin and reduce dynein’s microtubule binding affinity, we postulate that Nde1/Ndel1 may promote dynein and Lis1 association yet keep dynein and Lis1 in an inactive conformation until some unknown event triggers Lis1-induced dynein activation, dynactin and activating adaptor association, and Nde1/Ndel1 release.
Finally, both Nde1 and Ndel1 are highly phosphorylated by numerous kinases (reviewed in (Bradshaw et al., 2013)). Most exploration of the effect of phosphorylation on Nde1/Ndel1 activity comes from cell-based studies. These studies have shown that phosphorylation of Nde1 and Ndel1 is critically important to their localization, association with dynein and Lis1, and affects the ability of Nde1 and Ndel1 to promote the dynein-Lis1 interaction (Yan et al., 2003; Hebbar et al., 2008; Bradshaw et al., 2011; Bradshaw et al., 2013; Klinman and Holzbaur, 2015; Klinman et al., 2017; Wynne and Vallee, 2018). For example, phosphorylation by Cdk1 promotes Nde1 localization to the nuclear envelope and kinetochores, while Aurora-A kinase phosphorylation increases Ndel1 localization to centrosomes (Mori et al., 2007; Wynne and Vallee, 2018). Cdk5 phosphorylation of Ndel1 regulates Ndel1’s ability to bind dynein and promotes dynein’s association with microtubules (Hebbar et al., 2008; Klinman and Holzbaur, 2015; Klinman et al., 2017). Cdc2, Erk1, and Erk2 activity increase association between Ndel1 and Lis1 (Yan et al., 2003). PKA phosphorylation of Nde1 increases binding to Lis1 and promotes heterodimerization or tetramerization between Nde1 and Ndel1 (Bradshaw et al., 2011). Altogether, there is significant evidence that, in cells, post-translational modification of Nde1 and Ndel1 alter the mechanistic outcome of Nde1 and Ndel1 binding to dynein and its regulatory partners. However, most in vitro studies to date have ignored Nde1/Ndel1 phosphorylation as a variable in determining their molecular activity. This may account for some of the discrepancies observed between Nde1 and Ndel1’s activities in cells and in vitro.
There are many outstanding questions about Nde1 and Ndel1 function and interaction with the dynein transport machinery. Some of these questions include: 1) How do Nde1/Ndel1 affect dynein motility in the presence of dynactin, an activating adaptor, and Lis1? 2) How do post-translational modifications alter Nde1 and Ndel1’s ability to interact with dynein and its regulators? 3) Do Nde1 and Ndel1 modulate dynein the same way or do the differences that underlie the proteins’ cellular functions reveal differences in their regulation of dynein? 4) Do Lis1 and Nde1/Ndel1 always function in tandem or are there Lis1-independent functions of Nde1/Ndel1? Recent advances in our understanding of dynein regulation by dynactin and Lis1 required the integration of biochemistry, structural biology, and cell biology studies. A similar approach will be required to develop a unified theory of Nde1 and Ndel1 regulation and to fully unpack Nde1 and Ndel1’s molecular mechanism.
Author Contributions
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Drs. Samara Reck-Peterson, Michael Cianfrocco, and Richard Baker for helpful feedback. We also thank the editor and reviewers for their insightful feedback. This work was supported by NIH Grant R00-GM127757 (MD).
References
Abdel‐Hamid, M. S., El‐Dessouky, S. H., Ateya, M. I., Gaafar, H. M., and Abdel‐Salam, G. M. H. (2019). Phenotypic Spectrum of NDE1 ‐related Disorders: from Microlissencephaly to Microhydranencephaly. Am. J. Med. Genet. 179 (3), 494–497. doi:10.1002/ajmg.a.61035
Alkuraya, F. S., Cai, X., Emery, C., Mochida, G. H., Al-Dosari, M. S., Felie, J. M., et al. (2011). Human Mutations in NDE1 Cause Extreme Microcephaly with Lissencephaly. Am. J. Hum. Genet. 88 (5), 536–547. doi:10.1016/j.ajhg.2011.04.003
Altschul, S. F., Wootton, J. C., Gertz, E. M., Agarwala, R., Morgulis, A., Schaffer, A. A., et al. (2005). Protein Database Searches Using Compositionally Adjusted Substitution Matrices. FEBS J. 272 (20), 5101–5109. doi:10.1111/j.1742-4658.2005.04945.x
Azizi, A., Herrmann, A., Wan, Y., Buse, S. J., Keller, P. J., Goldstein, R. E., et al. (2020). Nuclear Crowding and Nonlinear Diffusion during Interkinetic Nuclear Migration in the Zebrafish Retina. eLife 9, e58635. doi:10.7554/eLife.58635
Bakircioglu, M., Carvalho, O. P., Khurshid, M., Cox, J. J., Tuysuz, B., Barak, T., et al. (2011). The Essential Role of Centrosomal NDE1 in Human Cerebral Cortex Neurogenesis. Am. J. Hum. Genet. 88 (5), 523–535. doi:10.1016/j.ajhg.2011.03.019
Baumbach, J., Murthy, A., McClintock, M. A., Dix, C. I., Zalyte, R., Hoang, H. T., et al. (2017). Lissencephaly-1 Is a Context-dependent Regulator of the Human Dynein Complex. eLife 6, e21768. doi:10.7554/eLife.21768
Bertipaglia, C., Gonçalves, J. C., and Vallee, R. B. (2018). Nuclear Migration in Mammalian Brain Development. Semin. Cel Dev. Biol. 82, 57–66. doi:10.1016/j.semcdb.2017.11.033
Bolhy, S., Bouhlel, I., Dultz, E., Nayak, T., Zuccolo, M., Gatti, X., et al. (2011). A Nup133-dependent NPC-Anchored Network Tethers Centrosomes to the Nuclear Envelope in Prophase. J. Cel Biol. 192 (5), 855–871. doi:10.1083/jcb.201007118
Bradshaw, N. J., Christie, S., Soares, D. C., Carlyle, B. C., Porteous, D. J., and Millar, J. K. (2009). NDE1 and NDEL1: Multimerisation, Alternate Splicing and DISC1 Interaction. Neurosci. Lett. 449 (3), 228–233. doi:10.1016/j.neulet.2008.10.095
Bradshaw, N. J., Hennah, W., and Soares, D. C. (2013). NDE1 and NDEL1: Twin Neurodevelopmental Proteins with Similar 'nature' but Different 'nurture'. BioMolecular Concepts 4 (5), 447–464. doi:10.1515/bmc-2013-0023
Bradshaw, N. J., and Porteous, D. J. (2012). DISC1-binding Proteins in Neural Development, Signalling and Schizophrenia. Neuropharmacology 62 (3), 1230–1241. doi:10.1016/j.neuropharm.2010.12.027
Bradshaw, N. J., Soares, D. C., Carlyle, B. C., Ogawa, F., Davidson-Smith, H., Christie, S., et al. (2011). PKA Phosphorylation of NDE1 Is DISC1/PDE4 Dependent and Modulates its Interaction with LIS1 and NDEL1. J. Neurosci. 31 (24), 9043–9054. doi:10.1523/JNEUROSCI.5410-10.2011
Brandon, N. J., Handford, E. J., Schurov, I., Rain, J.-C., Pelling, M., Duran-Jimeniz, B., et al. (2004). Disrupted in Schizophrenia 1 and Nudel Form a Neurodevelopmentally Regulated Protein Complex: Implications for Schizophrenia and Other Major Neurological Disorders. Mol. Cell Neurosci. 25 (1), 42–55. doi:10.1016/j.mcn.2003.09.009
Bruno, K. S., Tinsley, J. H., Minke, P. F., and Plamann, M. (1996). Genetic Interactions Among Cytoplasmic Dynein, Dynactin, and Nuclear Distribution Mutants of Neurospora Crassa. Proc. Natl. Acad. Sci. U.S.A. 93 (10), 4775–4780. doi:10.1073/pnas.93.10.4775
Burdick, K. E., Kamiya, A., Hodgkinson, C. A., Lencz, T., DeRosse, P., Ishizuka, K., et al. (2008). Elucidating the Relationship between DISC1, NDEL1 and NDE1 and the Risk for Schizophrenia: Evidence of Epistasis and Competitive Binding. Hum. Mol. Genet. 17 (16), 2462–2473. doi:10.1093/hmg/ddn146
Burkhardt, J. K., Echeverri, C. J., Nilsson, T., and Vallee, R. B. (1997). Overexpression of the Dynamitin (P50) Subunit of the Dynactin Complex Disrupts Dynein-dependent Maintenance of Membrane Organelle Distribution. J. Cel Biol. 139 (2), 469–484. doi:10.1083/jcb.139.2.469
Cardoso-Moreira, M., Halbert, J., Valloton, D., Velten, B., Chen, C., Shao, Y., et al. (2019). Gene Expression across Mammalian Organ Development. Nature 571 (7766), 505–509. doi:10.1038/s41586-019-1338-5
Cianfrocco, M. A., DeSantis, M. E., Leschziner, A. E., and Reck-Peterson, S. L. (2015). Mechanism and Regulation of Cytoplasmic Dynein. Annu. Rev. Cel Dev. Biol. 31 (1), 83–108. doi:10.1146/annurev-cellbio-100814-125438
Coquelle, F. M., Caspi, M., Cordelières, F. P., Dompierre, J. P., Dujardin, D. L., Koifman, C., et al. (2002). LIS1, CLIP-170's Key to the Dynein/Dynactin Pathway. Mol. Cel Biol 22 (9), 3089–3102. doi:10.1128/MCB.22.9.3089-3102.2002
Corthésy-Theulaz, I., Pauloin, A., and Pfeffer, S. R. (1992). Cytoplasmic Dynein Participates in the Centrosomal Localization of the Golgi Complex. J. Cel Biol. 118 (6), 1333–1345. doi:10.1083/jcb.118.6.1333
Del Bene, F., Wehman, A. M., Link, B. A., and Baier, H. (2008). Regulation of Neurogenesis by Interkinetic Nuclear Migration through an Apical-Basal Notch Gradient. Cell 134 (6), 1055–1065. doi:10.1016/j.cell.2008.07.017
Derewenda, U., Tarricone, C., Choi, W. C., Cooper, D. R., Lukasik, S., Perrina, F., et al. (2007). The Structure of the Coiled-Coil Domain of Ndel1 and the Basis of its Interaction with Lis1, the Causal Protein of Miller-Dieker Lissencephaly. Structure 15 (11), 1467–1481. doi:10.1016/j.str.2007.09.015
DeSantis, M. E., Cianfrocco, M. A., Htet, Z. M., Tran, P. T., Reck-Peterson, S. L., and Leschziner, A. E. (2017). Lis1 Has Two Opposing Modes of Regulating Cytoplasmic Dynein. Cell 170 (6), 1197–1208. e12. doi:10.1016/j.cell.2017.08.037
Ding, C., Liang, X., Ma, L., Yuan, X., and Zhu, X. (2009). Opposing Effects of Ndel1 and α1 or α2 on Cytoplasmic Dynein through Competitive Binding to Lis1. J. Cel Sci. 122 (16), 2820–2827. doi:10.1242/jcs.048777
Doobin, D. J., Kemal, S., Dantas, T. J., and Vallee, R. B. (2016). Severe NDE1-Mediated Microcephaly Results from Neural Progenitor Cell Cycle Arrests at Multiple Specific Stages. Nat. Commun. 7, 12551. doi:10.1038/ncomms12551
Drerup, C. M., Ahlgren, S. C., and Morris, J. A. (2007). Expression Profiles of Ndel1a and Ndel1b, Two Orthologs of the NudE-like Gene, in the Zebrafishthe Zebrafish. Gene Expr. Patterns 7 (6), 672–679. doi:10.1016/j.modgep.2007.03.003
Dwivedi, D., and Sharma, M. (2018). Multiple Roles, Multiple Adaptors: Dynein during Cell Cycle. Adv. Exp. Med. Biol. 1112, 13–30. doi:10.1007/978-981-13-3065-0_2
Efimov, V. P., and Morris, N. R. (2000). The Lis1-Related Nudf Protein of Aspergillus nidulans Interacts with the Coiled-Coil Domain of the Nude/Ro11 Protein. J. Cel Biol. 150 (3), 681–688. doi:10.1083/jcb.150.3.681
Efimov, V. P. (2003). Roles of NUDE and NUDF Proteins ofAspergillus Nidulans: Insights from Intracellular Localization and Overexpression Effects. MBoC 14 (3), 871–888. doi:10.1091/mbc.e02-06-0359
Elshenawy, M. M., Kusakci, E., Volz, S., Baumbach, J., Bullock, S. L., and Yildiz, A. (2020). Lis1 Activates Dynein Motility by Modulating its Pairing with Dynactin. Nat. Cel Biol 22 (5), 570–578. doi:10.1038/s41556-020-0501-4
Eshel, D., Urrestarazu, L. A., Vissers, S., Jauniaux, J. C., van Vliet-Reedijk, J. C., Planta, R. J., et al. (1993). Cytoplasmic Dynein Is Required for normal Nuclear Segregation in Yeast. Proc. Natl. Acad. Sci. U.S.A. 90 (23), 11172–11176. doi:10.1073/pnas.90.23.11172
Feng, Y., Olson, E. C., Stukenberg, P. T., Flanagan, L. A., Kirschner, M. W., and Walsh, C. A. (2000). LIS1 Regulates CNS Lamination by Interacting with mNudE, a Central Component of the Centrosome. Neuron 28 (3), 665–679. doi:10.1016/S0896-6273(00)00145-8
Feng, Y., and Walsh, C. A. (2004). Mitotic Spindle Regulation by Nde1 Controls Cerebral Cortical Size. Neuron 44 (2), 279–293. doi:10.1016/j.neuron.2004.09.023
Gaglio, T., Dionne, M. A., and Compton, D. A. (1997). Mitotic Spindle Poles Are Organized by Structural and Motor Proteins in Addition to Centrosomes. J. Cel Biol. 138 (5), 1055–1066. doi:10.1083/jcb.138.5.1055
Gassmann, R., Holland, A. J., Varma, D., Wan, X., Çivril, F., Cleveland, D. W., et al. (2010). Removal of Spindly from Microtubule-Attached Kinetochores Controls Spindle Checkpoint Silencing in Human Cells. Genes Dev. 24 (9), 957–971. doi:10.1101/gad.1886810
Gillies, J. P., Reimer, J. M., Karasmanis, E. P., Lahiri, I., Htet, Z. M., Leschziner, A. E., et al. (2022). Structural Basis for Cytoplasmic Dynein-1 Regulation by Lis1. eLife 11, e71229. doi:10.7554/eLife.71229
Grotjahn, D. A., Chowdhury, S., Xu, Y., McKenney, R. J., Schroer, T. A., and Lander, G. C. (2018). Cryo-electron Tomography Reveals that Dynactin Recruits a Team of Dyneins for Processive Motility. Nat. Struct. Mol. Biol. 25 (3), 203–207. doi:10.1038/s41594-018-0027-7
Gutierrez, P. A., Ackermann, B. E., Vershinin, M., and McKenney, R. J. (2017). Differential Effects of the Dynein-Regulatory Factor Lissencephaly-1 on Processive Dynein-Dynactin Motility. J. Biol. Chem. 292 (29), 12245–12255. doi:10.1074/jbc.M117.790048
Guven, A., Gunduz, A., Bozoglu, T. M., Yalcinkaya, C., and Tolun, A. (2012). Novel NDE1 Homozygous Mutation Resulting in Microhydranencephaly and Not Microlyssencephaly. neurogenetics 13 (3), 189–194. doi:10.1007/s10048-012-0326-9
Hannes, F. D., Sharp, A. J., Mefford, H. C., de Ravel, T., Ruivenkamp, C. A., Breuning, M. H., et al. (2009). Recurrent Reciprocal Deletions and Duplications of 16p13.11: the Deletion Is a Risk Factor for MR/MCA while the Duplication May Be a Rare Benign Variant. J. Med. Genet. 46 (4), 223–232. doi:10.1136/jmg.2007.055202
Hayashi, M. A. F., Portaro, F. C. V., Bastos, M. F., Guerreiro, J. R., Oliveira, V., Gorrão, S. S., et al. (2005). Inhibition of NUDEL (Nuclear Distribution Element-Like)-Oligopeptidase Activity by Disrupted-In-Schizophrenia 1. Proc. Natl. Acad. Sci. U.S.A. 102 (10), 3828–3833. doi:10.1073/pnas.0500330102
Hayashi, M. A. F., Portaro, F. C. V., Tambourgi, D. V., Sucupira, M., Yamane, T., Fernandes, B. L., et al. (2000). Molecular and Immunochemical Evidences Demonstrate that Endooligopeptidase A Is the Predominant Cytosolic Oligopeptidase of Rabbit Brain. Biochem. Biophysical Res. Commun. 269 (1), 7–13. doi:10.1006/bbrc.2000.2243
Heald, R., Tournebize, R., Blank, T., Sandaltzopoulos, R., Becker, P., Hyman, A., et al. (1996). Self-organization of Microtubules into Bipolar Spindles Around Artificial Chromosomes in Xenopus Egg Extracts. Nature 382 (6590), 420–425. doi:10.1038/382420a0
Hebbar, S., Mesngon, M. T., Guillotte, A. M., Desai, B., Ayala, R., and Smith, D. S. (2008). Lis1 and Ndel1 Influence the Timing of Nuclear Envelope Breakdown in Neural Stem Cells. J. Cel Biol. 182 (6), 1063–1071. doi:10.1083/jcb.200803071
Heinzen, E. L., Radtke, R. A., Urban, T. J., Cavalleri, G. L., Depondt, C., Need, A. C., et al. (2010). Rare Deletions at 16p13.11 Predispose to a Diverse Spectrum of Sporadic Epilepsy Syndromes. Am. J. Hum. Genet. 86 (5), 707–718. doi:10.1016/j.ajhg.2010.03.018
Hirokawa, N., and Noda, Y. (2008). Intracellular Transport and Kinesin Superfamily Proteins, KIFs: Structure, Function, and Dynamics. Physiol. Rev. 88 (3), 1089–1118. doi:10.1152/physrev.00023.2007
Hirokawa, N., Noda, Y., Tanaka, Y., and Niwa, S. (2009). Kinesin Superfamily Motor Proteins and Intracellular Transport. Nat. Rev. Mol. Cel Biol 10 (10), 682–696. doi:10.1038/nrm2774
Houlihan, S. L., and Feng, Y. (2014). The Scaffold Protein Nde1 Safeguards the Brain Genome during S Phase of Early Neural Progenitor Differentiation. eLife 3, e03297. doi:10.7554/eLife.03297
Howell, B. J., McEwen, B. F., Canman, J. C., Hoffman, D. B., Farrar, E. M., Rieder, C. L., et al. (2001). Cytoplasmic Dynein/dynactin Drives Kinetochore Protein Transport to the Spindle Poles and Has a Role in Mitotic Spindle Checkpoint Inactivation. J. Cel Biol. 155 (7), 1159–1172. doi:10.1083/jcb.200105093
Htet, Z. M., Gillies, J. P., Baker, R. W., Leschziner, A. E., DeSantis, M. E., and Reck-Peterson, S. L. (2020). LIS1 Promotes the Formation of Activated Cytoplasmic Dynein-1 Complexes. Nat. Cel Biol 22 (5), 518–525. doi:10.1038/s41556-020-0506-z
Hu, D. J.-K., Baffet, A. D., Nayak, T., Akhmanova, A., Doye, V., and Vallee, R. B. (2013). Dynein Recruitment to Nuclear Pores Activates Apical Nuclear Migration and Mitotic Entry in Brain Progenitor Cells. Cell 154 (6), 1300–1313. doi:10.1016/j.cell.2013.08.02410.1016/j.cell.2013.08.024
Huang, J., Roberts, A. J., Leschziner, A. E., and Reck-Peterson, S. L. (2012). Lis1 Acts as a "Clutch" between the ATPase and Microtubule-Binding Domains of the Dynein Motor. Cell 150 (5), 975–986. doi:10.1016/j.cell.2012.07.022
Ingason, A., Rujescu, D., Rujescu, D., Cichon, S., Sigurdsson, E., Sigmundsson, T., et al. (2011). Copy Number Variations of Chromosome 16p13.1 Region Associated with Schizophrenia. Mol. Psychiatry 16 (1), 17–25. doi:10.1038/mp.2009.101
Jha, R., Roostalu, J., Cade, N. I., Trokter, M., and Surrey, T. (2017). Combinatorial Regulation of the Balance between Dynein Microtubule End Accumulation and Initiation of Directed Motility. Embo J. 36 (22), 3387–3404. doi:10.15252/embj.201797077
Jie, J., Löhr, F., and Barbar, E. (2017). Dynein Binding of Competitive Regulators Dynactin and NudE Involves Novel Interplay between Phosphorylation Site and Disordered Spliced Linkers. Structure 25 (3), 421–433. doi:10.1016/j.str.2017.01.003
Johnstone, M., Maclean, A., Heyrman, L., Lenaerts, A.-S., Nordin, A., Nilsson, L.-G., et al. (2015). Copy Number Variations in DISC1 and DISC1-Interacting Partners in Major Mental Illness. Mol. Neuropsychiatry 1 (3), 175–190. doi:10.1159/000438788
Kardon, J. R., and Vale, R. D. (2009). Regulators of the Cytoplasmic Dynein Motor. Nat. Rev. Mol. Cel Biol 10 (12), 854–865. doi:10.1038/nrm2804
Karki, S., and Holzbaur, E. L. F. (1995). Affinity Chromatography Demonstrates a Direct Binding between Cytoplasmic Dynein and the Dynactin Complex. J. Biol. Chem. 270 (48), 28806–28811. doi:10.1074/jbc.270.48.28806
Kim, S., Zaghloul, N. A., Bubenshchikova, E., Oh, E. C., Rankin, S., Katsanis, N., et al. (2011). Nde1-mediated Inhibition of Ciliogenesis Affects Cell Cycle Re-entry. Nat. Cel Biol 13 (4), 351–360. doi:10.1038/ncb2183
Kirov, G., Grozeva, D., Norton, N., Ivanov, D., Mantripragada, K. K., Holmans, P., et al. (2009). Ivanov D, Mantripragada KK, Holmans P, International Schizophrenia Consortium, Wellcome Trust Case Control Consortium, Craddock N, Owen MJ, O’Donovan MCSupport for the Involvement of Large Copy Number Variants in the Pathogenesis of Schizophrenia. Hum. Mol. Genet. 18 (8), 1497–1503. doi:10.1093/hmg/ddp043
Kishimoto, T., Fugo, K., and Kiyokawa, T. (2013). Intracellular Position of G2/M-phase Nuclei in Neoplastic and Non-neoplastic Pseudostratified Glands Suggests the Occurrence of Interkinetic Nuclear Migration. Med. Mol. Morphol. 46 (4), 210–216. doi:10.1007/s00795-013-0026-z
Klinman, E., and Holzbaur, E. L. F. (2015). Stress-Induced CDK5 Activation Disrupts Axonal Transport via Lis1/Ndel1/Dynein. Cel Rep. 12 (3), 462–473. doi:10.1016/j.celrep.2015.06.032
Klinman, E., Tokito, M., and Holzbaur, E. L. F. (2017). CDK5-dependent Activation of Dynein in the Axon Initial Segment Regulates Polarized Cargo Transport in Neurons. Traffic 18 (12), 808–824. doi:10.1111/tra.12529
Korth, C. (2009). DISCopathies: Brain Disorders Related to DISC1 Dysfunction. Rev. Neurosciences 20 (5–6), 321–330. doi:10.1515/revneuro.2009.20.5-6.321
Kuijpers, M., van de Willige, D., Freal, A., Chazeau, A., Franker, M. A., Hofenk, J., et al. (2016). Dynein Regulator NDEL1 Controls Polarized Cargo Transport at the Axon Initial Segment. Neuron 89 (3), 461–471. doi:10.1016/j.neuron.2016.01.022
Lam, C., Vergnolle, M. A. S., Thorpe, L., Woodman, P. G., and Allan, V. J. (2010). Functional Interplay between LIS1, NDE1 and NDEL1 in Dynein-dependent Organelle Positioning. J. Cel Sci. 123 (2), 202–212. doi:10.1242/jcs.059337
Lee, W.-L., Oberle, J. R., and Cooper, J. A. (2003). The Role of the Lissencephaly Protein Pac1 during Nuclear Migration in Budding Yeast. J. Cel Biol. 160 (3), 355–364. doi:10.1083/jcb.200209022
Leung, L., Klopper, A. V., Grill, S. W., Harris, W. A., and Norden, C. (2011). Apical Migration of Nuclei during G2 Is a Prerequisite for All Nuclear Motion in Zebrafish Neuroepithelia. Development (Cambridge, England) 138 (22), 5003–5013. doi:10.1242/dev.071522
Li, H., Liu, X. S., Yang, X., Song, B., Wang, Y., and Liu, X. (2010). Polo-like Kinase 1 Phosphorylation of P150 Glued Facilitates Nuclear Envelope Breakdown during Prophase. Proc. Natl. Acad. Sci. U.S.A. 107 (33), 14633–14638. doi:10.1073/pnas.1006615107
Li, J., Lee, W.-L., and Cooper, J. A. (2005). NudEL Targets Dynein to Microtubule Ends through LIS1. Nat. Cel Biol 7 (7), 686–690. doi:10.1038/ncb1273
Li, Y. Y., Yeh, E., Hays, T., and Bloom, K. (1993). Disruption of Mitotic Spindle Orientation in a Yeast Dynein Mutant. Proc. Natl. Acad. Sci. U.S.A. 90 (21), 10096–10100. doi:10.1073/pnas.90.21.10096
Liang, Y., Yu, W., Li, Y., Yang, Z., Yan, X., Huang, Q., et al. (2004). Nudel Functions in Membrane Traffic Mainly through Association with Lis1 and Cytoplasmic Dynein. J. Cel Biol. 164 (4), 557–566. doi:10.1083/jcb.200308058
Liang, Y., Yu, W., Li, Y., Yu, L., Zhang, Q., Wang, F., et al. (2007). Nudel Modulates Kinetochore Association and Function of Cytoplasmic Dynein in M Phase. MBoC 18 (7), 2656–2666. doi:10.1091/mbc.E06-04-0345
Lipka, J., Kuijpers, M., Jaworski, J., and Hoogenraad, C. C. (2013). Mutations in Cytoplasmic Dynein and its Regulators Cause Malformations of Cortical Development and Neurodegenerative Diseases. Biochem. Soc. Trans. 41 (6), 1605–1612. doi:10.1042/BST20130188
Ma, L., Tsai, M.-Y., Wang, S., Lu, B., Chen, R., Iii, J. R. Y., et al. (2009). Requirement for Nudel and Dynein for Assembly of the Lamin B Spindle Matrix. Nat. Cel Biol 11 (3), 247–256. doi:10.1038/ncb1832
Malhotra, D., and Sebat, J. (2012). CNVs: Harbingers of a Rare Variant Revolution in Psychiatric Genetics. Cell 148 (6), 1223–1241. doi:10.1016/j.cell.2012.02.039
Markus, S. M., Marzo, M. G., and McKenney, R. J. (2020). New Insights into the Mechanism of Dynein Motor Regulation by Lissencephaly-1. eLife 9, e59737. doi:10.7554/eLife.59737
Markus, S. M., Punch, J. J., and Lee, W.-L. (2009). Motor- and Tail-dependent Targeting of Dynein to Microtubule Plus Ends and the Cell Cortex. Curr. Biol. 19 (3), 196–205. doi:10.1016/j.cub.2008.12.047
Marzo, M. G., Griswold, J. M., and Markus, S. M. (2020). Pac1/LIS1 Stabilizes an Uninhibited Conformation of Dynein to Coordinate its Localization and Activity. Nat. Cel Biol 22 (5), 559–569. doi:10.1038/s41556-020-0492-1
Maskey, D., Marlin, M. C., Kim, S., Kim, S., Ong, E. C., Li, G., et al. (2015). Cell Cycle-dependent Ubiquitylation and Destruction of NDE1 by CDK5-FBW7 Regulates Ciliary Length. EMBO J. 34 (19), 2424–2440. doi:10.15252/embj.201490831
McKenney, R. J., Huynh, W., Tanenbaum, M. E., Bhabha, G., and Vale, R. D. (2014). Activation of Cytoplasmic Dynein Motility by Dynactin-Cargo Adapter Complexes. Science 345 (6194), 337–341. doi:10.1126/science.1254198
McKenney, R. J., Vershinin, M., Kunwar, A., Vallee, R. B., and Gross, S. P. (2010). LIS1 and NudE Induce a Persistent Dynein Force-Producing State. Cell 141 (2), 304–314. doi:10.1016/j.cell.2010.02.035
McKenney, R. J., Weil, S. J., Scherer, J., and Vallee, R. B. (2011). Mutually Exclusive Cytoplasmic Dynein Regulation by NudE-Lis1 and Dynactin. J. Biol. Chem. 286 (45), 39615–39622. doi:10.1074/jbc.M111.289017
Mefford, H. C., Muhle, H., Ostertag, P., von Spiczak, S., Buysse, K., Baker, C., et al. (2010). Genome-Wide Copy Number Variation in Epilepsy: Novel Susceptibility Loci in Idiopathic Generalized and Focal Epilepsies. Plos Genet. 6 (5), e1000962. doi:10.1371/journal.pgen.1000962
Meyer, E. J., Ikmi, A., and Gibson, M. C. (2011). Interkinetic Nuclear Migration Is a Broadly Conserved Feature of Cell Division in Pseudostratified Epithelia. Curr. Biol. 21 (6), 485–491. doi:10.1016/j.cub.2011.02.002
Millar, J. K., Pickard, B. S., Mackie, S., James, R., Christie, S., Buchanan, S. R., et al. (2005). DISC1 and PDE4B Are Interacting Genetic Factors in Schizophrenia that Regulate cAMP Signaling. Science 310 (5751), 1187–1191. doi:10.1126/science.1112915
Minke, P. F., Lee, I. H., Tinsley, J. H., Bruno, K. S., and Plamann, M. (1999). Neurospora Crassa Ro-10 and Ro-11 Genes Encode Novel Proteins Required for Nuclear Distribution. Mol. Microbiol. 32 (5), 1065–1076. doi:10.1046/j.1365-2958.1999.01421.x
Monda, J. K., and Cheeseman, I. M. (2018). Nde1 Promotes Diverse Dynein Functions through Differential Interactions and Exhibits an Isoform-specific Proteasome Association. MBoC 29 (19), 2336–2345. doi:10.1091/mbc.E18-07-0418
Moon, H. M., Youn, Y. H., Pemble, H., Yingling, J., Wittmann, T., and Wynshaw-Boris, A. (2014). LIS1 Controls Mitosis and Mitotic Spindle Organization via the LIS1-NDEL1-Dynein Complex. Hum. Mol. Genet. 23 (2), 449–466. doi:10.1093/hmg/ddt436
Mori, D., Yano, Y., Toyo-oka, K., Yoshida, N., Yamada, M., Muramatsu, M., et al. (2007). NDEL1 Phosphorylation by Aurora-A Kinase Is Essential for Centrosomal Maturation, Separation, and TACC3 Recruitment. Mol. Cel Biol 27, 352–367. doi:10.1128/MCB.00878-06
Morris, J. A. (2003). DISC1 (Disrupted-In-Schizophrenia 1) Is a Centrosome-Associated Protein that Interacts with MAP1A, MIPT3, ATF4/5 and NUDEL: Regulation and Loss of Interaction with Mutation. Hum. Mol. Genet. 12 (13), 1591–1608. doi:10.1093/hmg/ddg162
Nyarko, A., Song, Y., and Barbar, E. (2012). Intrinsic Disorder in Dynein Intermediate Chain Modulates its Interactions with NudE and Dynactin *. J. Biol. Chem. 287 (30), 24884–24893. doi:10.1074/jbc.M112.376038
Ozeki, Y., Tomoda, T., Kleiderlein, J., Kamiya, A., Bord, L., Fujii, K., et al. (2003). Disrupted-in-Schizophrenia-1 (DISC-1): Mutant Truncation Prevents Binding to NudE-like (NUDEL) and Inhibits Neurite Outgrowth. Proc. Natl. Acad. Sci. U.S.A. 100 (1), 289–294. doi:10.1073/pnas.0136913100
Paciorkowski, A. R., Keppler‐Noreuil, K., Robinson, L., Sullivan, C., Sajan, S., Christian, S. L., et al. (2013). Deletion 16p13.11 Uncovers NDE1 Mutations on the Non‐deleted Homolog and Extends the Spectrum of Severe Microcephaly to Include Fetal Brain Disruption. Am. J. Med. Genet. 161 (7), 1523–1530. doi:10.1002/ajmg.a.35969
Pandey, J. P., and Smith, D. S. (2011). A Cdk5-dependent Switch Regulates Lis1/Ndel1/Dynein-Driven Organelle Transport in Adult Axons. J. Neurosci. 31 (47), 17207–17219. doi:10.1523/JNEUROSCI.4108-11.2011
Paschal, B. M., Holzbaur, E. L., Pfister, K. K., Clark, S., Meyer, D. I., and Vallee, R. B. (1993). Characterization of a 50-kDa Polypeptide in Cytoplasmic Dynein Preparations Reveals a Complex with p150GLUED and a Novel Actin. J. Biol. Chem. 268 (20), 15318–15323. doi:10.1016/s0021-9258(18)82472-3
Pfister, K. K., Shah, P. R., Hummerich, H., Russ, A., Cotton, J., Annuar, A. A., et al. (2006). Genetic Analysis of the Cytoplasmic Dynein Subunit Families. Plos Genet. 2 (1), e1. doi:10.1371/journal.pgen.0020001
Plamann, M., Minke, P. F., Tinsley, J. H., and Bruno, K. S. (1994). Cytoplasmic Dynein and Actin-Related Protein Arp1 Are Required for normal Nuclear Distribution in Filamentous Fungi. J. Cel Biol. 127 (1), 139–149. doi:10.1083/jcb.127.1.139
Porteous, D. J., Thomson, P., Brandon, N. J., and Millar, J. K. (2006). The Genetics and Biology of Disc1-An Emerging Role in Psychosis and Cognition. Biol. Psychiatry 60 (2), 123–131. doi:10.1016/j.biopsych.2006.04.008
Qiu, R., Zhang, J., and Xiang, X. (2019). LIS1 Regulates Cargo-Adapter-Mediated Activation of Dynein by Overcoming its Autoinhibition In Vivo. J. Cel Biol. 218 (11), 3630–3646. doi:10.1083/jcb.201905178
Reck-Peterson, S. L., Redwine, W. B., Vale, R. D., and Carter, A. P. (2018). The Cytoplasmic Dynein Transport Machinery and its many Cargoes. Nat. Rev. Mol. Cel Biol 19 (6), 382–398. doi:10.1038/s41580-018-0004-3
Reck-Peterson, S. L., Yildiz, A., Carter, A. P., Gennerich, A., Zhang, N., and Vale, R. D. (2006). Single-Molecule Analysis of Dynein Processivity and Stepping Behavior. Cell 126 (2), 335–348. doi:10.1016/j.cell.2006.05.046
Reddy, B. J. N., Mattson, M., Wynne, C. L., Vadpey, O., Durra, A., Chapman, D., et al. (2016). Load-induced Enhancement of Dynein Force Production by LIS1-NudE In Vivo and In Vitro. Nat. Commun. 7 (1), 12259. doi:10.1038/ncomms12259
Rees, E., Walters, J. T. R., Georgieva, L., Isles, A. R., Chambert, K. D., Richards, A. L., et al. (2014). Analysis of Copy Number Variations at 15 Schizophrenia-Associated Loci. Br. J. Psychiatry 204 (2), 108–114. doi:10.1192/bjp.bp.113.131052
Reiner, O., and Sapir, T. (2013). LIS1 Functions in normal Development and Disease. Curr. Opin. Neurobiol. 23 (6), 951–956. doi:10.1016/j.conb.2013.08.001
Robinson, J. T., Wojcik, E. J., Sanders, M. A., McGrail, M., and Hays, T. S. (1999). Cytoplasmic Dynein Is Required for the Nuclear Attachment and Migration of Centrosomes during Mitosis in Drosophila. J. Cel Biol. 146 (3), 597–608. doi:10.1083/jcb.146.3.597
Salina, D., Bodoor, K., Eckley, D. M., Schroer, T. A., Rattner, J. B., and Burke, B. (2002). Cytoplasmic Dynein as a Facilitator of Nuclear Envelope Breakdown. Cell 108 (1), 97–107. doi:10.1016/S0092-8674(01)00628-6
Sasaki, S., Mori, D., Toyo-oka, K., Chen, A., Garrett-Beal, L., Muramatsu, M., et al. (2005). Complete Loss of Ndel1 Results in Neuronal Migration Defects and Early Embryonic Lethality. Mol. Cel Biol 25 (17), 7812–7827. doi:10.1128/MCB.25.17.7812-7827.2005
Sasaki, S., Shionoya, A., Ishida, M., Gambello, M. J., Yingling, J., Wynshaw-Boris, A., et al. (2000). A LIS1/NUDEL/Cytoplasmic Dynein Heavy Chain Complex in the Developing and Adult Nervous System. Neuron 28 (3), 681–696. doi:10.1016/S0896-6273(00)00146-X
Schlager, M. A., Hoang, H. T., Urnavicius, L., Bullock, S. L., and Carter, A. P. (2014). In Vitro reconstitution of a Highly Processive Recombinant Human Dynein Complex. Embo J. 33 (17), 1855–1868. doi:10.15252/embj.201488792
She, Z.-Y., and Yang, W.-X. (2017). Molecular Mechanisms of Kinesin-14 Motors in Spindle Assembly and Chromosome Segregation. J. Cel Sci. 130 (13), 2097–2110. doi:10.1242/jcs.200261
Sheeman, B., Carvalho, P., Sagot, I., Geiser, J., Kho, D., Hoyt, M. A., et al. (2003). Determinants of S. cerevisiae Dynein Localization and Activation. Curr. Biol. 13 (5), 364–372. doi:10.1016/S0960-9822(03)00013-7
Shmueli, A., Segal, M., Sapir, T., Tsutsumi, R., Noritake, J., Bar, A., et al. (2010). Ndel1 Palmitoylation: a New Mean to Regulate Cytoplasmic Dynein Activity. Embo J. 29 (1), 107–119. doi:10.1038/emboj.2009.325
Shu, T., Ayala, R., Nguyen, M.-D., Xie, Z., Gleeson, J. G., and Tsai, L.-H. (2004). Ndel1 Operates in a Common Pathway with LIS1 and Cytoplasmic Dynein to Regulate Cortical Neuronal Positioning. Neuron 44 (2), 263–277. doi:10.1016/j.neuron.2004.09.030
Sievers, F., Wilm, A., Dineen, D., Gibson, T. J., Karplus, K., Li, W., et al. (2011). Fast, Scalable Generation of High‐quality Protein Multiple Sequence Alignments Using Clustal Omega. Mol. Syst. Biol. 7, 539. doi:10.1038/msb.2011.75
Siglin, A. E., Sun, S., Moore, J. K., Tan, S., Poenie, M., Lear, J. D., et al. (2013). Dynein and Dynactin Leverage Their Bivalent Character to Form a High-Affinity Interaction. PLOS ONE 8 (4), e59453. doi:10.1371/journal.pone.0059453
Simões, P. A., Celestino, R., Carvalho, A. X., and Gassmann, R. (2018). NudE/L Regulates Dynein at Kinetochores but Is Dispensable for Other Dynein Functions in the C. elegans Early Embryo. J. Cel Sci. 131 (1), jcs212159. doi:10.1242/jcs.212159
Sitaram, P., Anderson, M. A., Jodoin, J. N., Lee, E., and Lee, L. A. (2012). Regulation of Dynein Localization and Centrosome Positioning by Lis-1 and Asunder during Drosophila Spermatogenesis. Development 139 (16), 2945–2954. doi:10.1242/dev.077511
Soares, D. C., Bradshaw, N. J., Zou, J., Kennaway, C. K., Hamilton, R. S., Chen, Z. A., et al. (2012). The Mitosis and Neurodevelopment Proteins NDE1 and NDEL1 Form Dimers, Tetramers, and Polymers with a Folded Back Structure in Solution. J. Biol. Chem. 287 (39), 32381–32393. doi:10.1074/jbc.M112.393439
Soares, D. C., Carlyle, B. C., Bradshaw, N. J., and Porteous, D. J. (2011). DISC1: Structure, Function, and Therapeutic Potential for Major Mental Illness. ACS Chem. Neurosci. 2 (11), 609–632. doi:10.1021/cn200062k
Soto-Perez, J., Baumgartner, M., and Kanadia, R. N. (2020). Role of NDE1 in the Development and Evolution of the Gyrified Cortex. Front. Neurosci. 14, 1267. doi:10.3389/fnins.2020.617513
Splinter, D., Razafsky, D. S., Schlager, M. A., Serra-Marques, A., Grigoriev, I., Demmers, J., et al. (2012). BICD2, Dynactin, and LIS1 Cooperate in Regulating Dynein Recruitment to Cellular Structures. MBoC 23 (21), 4226–4241. doi:10.1091/mbc.e12-03-0210
Splinter, D., Tanenbaum, M. E., Lindqvist, A., Jaarsma, D., Flotho, A., Yu, K. L., et al. (2010). Bicaudal D2, Dynein, and Kinesin-1 Associate with Nuclear Pore Complexes and Regulate Centrosome and Nuclear Positioning during Mitotic Entry. Plos Biol. 8 (4), e1000350. doi:10.1371/journal.pbio.1000350
Stehman, S. A., Chen, Y., McKenney, R. J., and Vallee, R. B. (2007). NudE and NudEL Are Required for Mitotic Progression and Are Involved in Dynein Recruitment to Kinetochores. J. Cel Biol. 178 (4), 583–594. doi:10.1083/jcb.200610112
Tarricone, C., Perrina, F., Monzani, S., Massimiliano, L., Kim, M., Derewenda, Z., et al. (2004). Coupling PAF Signaling to Dynein RegulationStructure of LIS1 in Complex with PAF-Acetylhydrolase. Neuron 44 (5), 809–821. doi:10.1016/S0896-6273(04)00751-2
Titus, J., and Wadsworth, P. (2012). “Mitotic Spindle Assembly: The Role of Motor Protein,” in eLS. Chichester: John Wiley and Sons, Ltd. doi:10.1002/9780470015902.a0022519
Torisawa, T., Nakayama, A., Furuta, K. y., Yamada, M., Hirotsune, S., and Toyoshima, Y. Y. (2011). Functional Dissection of LIS1 and NDEL1 towards Understanding the Molecular Mechanisms of Cytoplasmic Dynein Regulation. J. Biol. Chem. 286 (3), 1959–1965. doi:10.1074/jbc.M110.169847
Toropova, K., Zou, S., Roberts, A. J., Redwine, W. B., Goodman, B. S., Reck-Peterson, S. L., et al. (2014). Lis1 Regulates Dynein by Sterically Blocking its Mechanochemical Cycle. eLife 3, e03372. doi:10.7554/eLife.03372
Tropeano, M., Ahn, J. W., Dobson, R. J. B., Breen, G., Rucker, J., Dixit, A., et al. (2013). Male-biased Autosomal Effect of 16p13.11 Copy Number Variation in Neurodevelopmental Disorders. PLoS One 8 (4), e61365. doi:10.1371/journal.pone.0061365
Tsai, J.-W., Bremner, K. H., and Vallee, R. B. (2007). Dual Subcellular Roles for LIS1 and Dynein in Radial Neuronal Migration in Live Brain Tissue. Nat. Neurosci. 10 (8), 970–979. doi:10.1038/nn1934
Tsai, J.-W., Chen, Y., Kriegstein, A. R., and Vallee, R. B. (2005). LIS1 RNA Interference Blocks Neural Stem Cell Division, Morphogenesis, and Motility at Multiple Stages. J. Cel Biol. 170 (6), 935–945. doi:10.1083/jcb.200505166
Tsai, J.-W., Lian, W.-N., Kemal, S., Kriegstein, A. R., and Vallee, R. B. (2010). Kinesin 3 and Cytoplasmic Dynein Mediate Interkinetic Nuclear Migration in Neural Stem Cells. Nat. Neurosci. 13 (12), 1463–1471. doi:10.1038/nn.2665
Ullmann, R., Turner, G., Kirchhoff, M., Chen, W., Tonge, B., Rosenberg, C., et al. (2007). Array CGH Identifies Reciprocal 16p13.1 Duplications and Deletions that Predispose to Autism And/or Mental Retardation. Hum. Mutat. 28 (7), 674–682. doi:10.1002/humu.20546
Urnavicius, L., Lau, C. K., Elshenawy, M. M., Morales-Rios, E., Motz, C., Yildiz, A., et al. (2018). Cryo-EM Shows How Dynactin Recruits Two Dyneins for Faster Movement. Nature 554 (7691), 202–206. doi:10.1038/nature25462
Urnavicius, L., Zhang, K., Diamant, A. G., Motz, C., Schlager, M. A., Yu, M., et al. (2015). The Structure of the Dynactin Complex and its Interaction with Dynein. Science 347 (6229), 1441–1446. doi:10.1126/science.aaa4080
Vaughan, K. T. (2012). Roles of Cytoplasmic Dynein during Mitosis. Dyneins 2012, 522–535. doi:10.1016/B978-0-12-382004-4.10020-2
Vaughan, K. T., and Vallee, R. B. (1995). Cytoplasmic Dynein Binds Dynactin through a Direct Interaction between the Intermediate Chains and p150Glued. J. Cel Biol. 131 (6 Pt 1), 1507–1516. doi:10.1083/jcb.131.6.1507
Vergnolle, M. A. S., and Taylor, S. S. (2007). Cenp-F Links Kinetochores to Ndel1/Nde1/Lis1/Dynein Microtubule Motor Complexes. Curr. Biol. 17 (13), 1173–1179. doi:10.1016/j.cub.2007.05.077
Wainman, A., Creque, J., Williams, B., Williams, E. V., Bonaccorsi, S., Gatti, M., et al. (2009). Roles of theDrosophilaNudE Protein in Kinetochore Function and Centrosome Migration. J. Cel Sci. 122 (11), 1747–1758. doi:10.1242/jcs.041798
Wang, S., Ketcham, S. A., Schön, A., Goodman, B., Wang, Y., Yates, J., et al. (2013). Nudel/NudE and Lis1 Promote Dynein and Dynactin Interaction in the Context of Spindle Morphogenesis. MBoC 24 (22), 3522–3533. doi:10.1091/mbc.e13-05-0283
Wang, S., and Zheng, Y. (2011). Identification of a Novel Dynein Binding Domain in Nudel Essential for Spindle Pole Organization in Xenopus Egg Extract. J. Biol. Chem. 286 (1), 587–593. doi:10.1074/jbc.M110.181578
Wynne, C. L., and Vallee, R. B. (2018). Cdk1 Phosphorylation of the Dynein Adapter Nde1 Controls Cargo Binding from G2 to Anaphase. J. Cel Biol. 217 (9), 3019–3029. doi:10.1083/jcb.201707081
Xiang, X., Beckwith, S. M., and Morris, N. R. (1994). Cytoplasmic Dynein Is Involved in Nuclear Migration in Aspergillus nidulans. Proc. Natl. Acad. Sci. U.S.A. 91 (6), 2100–2104. doi:10.1073/pnas.91.6.2100
Xiang, X., Osmani, A. H., Osmani, S. A., Xin, M., and Morris, N. R. (1995). NudF, a Nuclear Migration Gene in Aspergillus nidulans, Is Similar to the Human LIS-1 Gene Required for Neuronal Migration. MBoC 6 (3), 297–310. doi:10.1091/mbc.6.3.297
Yamada, M., Toba, S., Yoshida, Y., Haratani, K., Mori, D., Yano, Y., et al. (2008). LIS1 and NDEL1 Coordinate the Plus-End-Directed Transport of Cytoplasmic Dynein. Embo J. 27 (19), 2471–2483. doi:10.1038/emboj.2008.182
Yan, X., Li, F., Liang, Y., Shen, Y., Zhao, X., Huang, Q., et al. (2003). Human Nudel and NudE as Regulators of Cytoplasmic Dynein in Poleward Protein Transport along the Mitotic Spindle. Mol. Cel Biol 23 (4), 1239–1250. doi:10.1128/MCB.23.4.1239-1250.2003
Ye, J., Li, J., Ye, F., Zhang, Y., Zhang, M., and Wang, C. (2020). Mechanistic Insights into the Interactions of Dynein Regulator Ndel1 with Neuronal Ankyrins and Implications in Polarity Maintenance. Proc. Natl. Acad. Sci. U.S.A. 117 (2), 1207–1215. doi:10.1073/pnas.1916987117
Yi, J. Y., Ori-McKenney, K. M., McKenney, R. J., Vershinin, M., Gross, S. P., and Vallee, R. B. (2011). High-resolution Imaging Reveals Indirect Coordination of Opposite Motors and a Role for LIS1 in High-Load Axonal Transport. J. Cel Biol. 195 (2), 193–201. doi:10.1083/jcb.201104076
Youn, Y. H., Pramparo, T., Hirotsune, S., and Wynshaw-Boris, A. (2009). Distinct Dose-dependent Cortical Neuronal Migration and Neurite Extension Defects in Lis1 and Ndel1 Mutant Mice. J. Neurosci. 29 (49), 15520–15530. doi:10.1523/JNEUROSCI.4630-09.2009
Zhang, K., Foster, H. E., Rondelet, A., Lacey, S. E., Bahi-Buisson, N., Bird, A. W., et al. (2017). Cryo-EM Reveals How Human Cytoplasmic Dynein Is Auto-Inhibited and Activated. Cell 169 (7), 1303–1314. e18. doi:10.1016/j.cell.2017.05.025
Zhang, Y., Chen, Z., Wang, F., Sun, H., Zhu, X., Ding, J., et al. (2022). Nde1 Is a Rab9 Effector for Loading Late Endosomes to Cytoplasmic Dynein Motor Complex. Structure 30, 386–395. doi:10.1016/j.str.2021.10.013
Keywords: NDE1, NDEL1, Lis1, cytoplasmic dynein 1, microtubule, motor protein
Citation: Garrott SR, Gillies JP and DeSantis ME (2022) Nde1 and Ndel1: Outstanding Mysteries in Dynein-Mediated Transport. Front. Cell Dev. Biol. 10:871935. doi: 10.3389/fcell.2022.871935
Received: 08 February 2022; Accepted: 17 March 2022;
Published: 12 April 2022.
Edited by:
Xin Xiang, Uniformed Services University of the Health Sciences, United StatesReviewed by:
Nicholas J. Bradshaw, University of Rijeka, CroatiaRichard McKenney, University of California, Davis, United States
Copyright © 2022 Garrott, Gillies and DeSantis. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Morgan E. DeSantis, bWRlc2FudEB1bWljaC5lZHU=
 Sharon R. Garrott
Sharon R. Garrott John P. Gillies2
John P. Gillies2 Morgan E. DeSantis
Morgan E. DeSantis