- 1The Cancer Institute, The Affiliated Hospital of Qingdao University, Qingdao, China
- 2School of Public Health, Qingdao University, Qingdao, China
When eukaryotic cells enter mitosis, dispersed chromosomes move to the cell center along microtubules to form a metaphase plate which facilitates the accurate chromosome segregation. Meanwhile, kinetochores not stably attached by microtubules activate the spindle assembly checkpoint and generate a wait signal to delay the initiation of anaphase. These events are highly coordinated. Disruption of the coordination will cause severe problems like chromosome gain or loss. Bub1, a conserved serine/threonine kinase, plays important roles in mitosis. After extensive studies in the last three decades, the role of Bub1 on checkpoint has achieved a comprehensive understanding; its role on chromosome alignment also starts to emerge. In this review, we summarize the latest development of Bub1 on supporting the two mitotic events. The essentiality of Bub1 in higher eukaryotic cells is also discussed. At the end, some undissolved questions are raised for future study.
Introduction
Genetic materials need to be accurately passed to the next generation which ensures the continuation of life. In eukaryotes, the chromosomes carrying the genetic information are replicated first and then equally segregated into daughter cells during mitosis. To secure the equal segregation, each pair of replicated chromatids need to be correctly attached by the spindle microtubules on the kinetochores, which is a proteinaceous structure built on centromeric chromosome. Afterwards, the physical connection between the sister chromatids is cleaved and each chromatid move towards the spindle pole driven by the shortening of microtubules attached to kinetochore. Finally, an actomyosin-based contractile ring forms at the cortex between the spindle poles and constricts to divide the cell into two. Due to the existence of multiple chromosomes in eukaryotic cells, the generation of stable attachment between kinetochores of all the chromosomes and spindle microtubules needs a certain time. Only when all the kinetochores have been properly attached by microtubules, chromosome segregation starts. Otherwise, a premature segregation in the presence of unattached chromosomes will cause gain or loss of chromosomes in daughter cells which could result in severe problems like cell death, miscarriage, developmental defects or tumorigenesis. To fulfill the temporal requirement, eukaryotic cells have evolved a monitoring mechanism called the spindle assembly checkpoint (SAC). Unattached kinetochores activate SAC which catalyzes the formation a protein complex termed mitotic checkpoint complex (MCC). MCC is composed by three checkpoint proteins BubR1, Bub3, Mad2 and cell division cycle 20 (Cdc20), which is a coactivator of E3 ubiquitin ligase Anaphase-Promoting Complex or Cyclosome (APC/C). Cdc20 activated APC/C promotes the transition from metaphase to anaphase by degrading a few key mitotic regulators. MCC binds and inhibits APC/C, thus delays the onset of anaphase until the proper kinetochore-microtubule attachments are established. Once the stable attachment is generated, kinetochores stop producing MCC and existing MCC gets disassembled. Only now, the cells could enter anaphase (reviewed in Musacchio, 2015; Kapanidou et al., 2017; Maiato et al., 2017; Lara-Gonzalez et al., 2021a).
Budding Uninhibited by Benzimidazoles 1 (BUB1) was first identified from the pioneer screening for mitotic checkpoint genes (Hoyt et al., 1991; Li et al., 1991). Bub1 protein was soon characterized as a serine/threonine kinase by the ability to phosphorylate itself and associated Bub3 protein (Roberts et al., 1994). Besides its role on SAC, early study also discovered another crucial function of Bub1 in maintaining genome stability (Bernard et al., 1998). Since then, numerous studies have greatly advanced our knowledge of the functions Bub1 plays in mitosis. Among the multiple functions, the role of Bub1 in activating the spindle assembly checkpoint is the most studied. Now it is clear that Bub1 is required for the full activation of the checkpoint through multiple mechanisms though the extent of requirement is context-dependent. How does Bub1 promote chromosome alignment is not fully understood. Recent studies have shed some lights on this functionality. Besides these two roles, Bub1 is also involved in chromatid cohesion protection through recruiting Sgo1/PP2A and this role seems more essential in meiosis than in mitosis (Kitajima et al., 2004; Tang et al., 2004; Kitajima et al., 2005; Tang et al., 2006; Perera et al., 2007; Schliekelman et al., 2009; Miyazaki et al., 2017; Yi et al., 2019; Carvalhal et al., 2022). Bub1 may be required for telomere replication (Li et al., 2018) and activation of DNA damage response (Hein et al., 2009). Due to the size limit of this review, we only discuss the latest development of Bub1 on activating SAC and promoting chromosome alignment. We try to dissect the functions through examining individual interactions along with the kinase activity. At the end we discuss the essentiality of Bub1 in human cells and the questions remain to be answered.
The Role of Bub1 in the Spindle Assembly Checkpoint
The molecular mechanism of the SAC has been thoroughly reviewed and we refer the reader to excellent reviews on the topic (Musacchio, 2015; Lara-Gonzalez et al., 2021a). Here we focus on the checkpoint protein Bub1 with an emphasis on recent discoveries.
Unoccupied kinetochores activate SAC. In line with this, all the SAC proteins associate dynamically with the outer kinetochores in early mitosis. Understanding the molecular mechanisms of the kinetochore localization is key to comprehend the SAC activation and silencing. Bub1 was speculated to be one of the first checkpoint components on kinetochores (Jablonski et al., 1998). The kinetochore localization of Bub1 was found to depend on its association with Bub3. Bub1 makes a stable complex with Bub3 through a domain in the N-terminal region of Bub1 (Taylor et al., 1998). Subsequent studies found that KNL1 was required for Bub1 kinetochore localization and this was suggested to be driven by an interaction between a KI motif in KNL1 and the N terminal tetratricho-peptide repeat (TPR) domain of Bub1 (Kiyomitsu et al., 2007; Kiyomitsu et al., 2011). However, this model did not readily explain the requirement for Bub3 and subsequent work questioned the relevance of the KI-TPR domain interaction for Bub1 localization (Krenn et al., 2012). Further studies showed that a phosphorylation-dependent interaction between KNL1 and Bub1-Bub3 was required for Bub1 kinetochore localization. The checkpoint kinase Mps1 phosphorylates a repetitive motif composed of the consensus sequence [M/I] [E/D/N] [I/L/M] [S/T] (named MELT motif) on KNL1. The phosphorylated MELT motifs provide docking sites for Bub1/Bub3 via a direct interaction with Bub1-Bub3 (London et al., 2012; Shepperd et al., 2012; Yamagishi et al., 2012; Primorac et al., 2013; Zhang et al., 2014; Vleugel et al., 2015b; Roy et al., 2020). This interaction is tightly regulated by protein phosphatases PP2A/B56 and PP1 to ensure ontime SAC silencing (London et al., 2012; Espert et al., 2014; Nijenhuis et al., 2014; Zhang et al., 2014; Roy et al., 2019; Braga et al., 2020). Since the establishment of how Bub1 is recruited to kinetochores, the hierarchy of the SAC has become clear. Now it is known that Bub1 is a major hub for assembly of the SAC machinery through direct interaction with BubR1, Mad1-Mad2, Cdc20, as well as interaction with the RZZ complex. These interactions and their function in the SAC are discussed in an order from the N-terminus to the C-terminus of Bub1 in the subsequent sections (Figure 1).
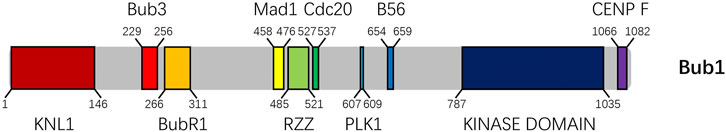
FIGURE 1. Schematic of the domains on human Bub1 protein. From left to right, the domains interacting with KNL1, Bub3, BubR1, Mad1, RZZ, Cdc20, Plk1, B56 (putative) and CENP-F are marked with reported boundaries. The kinase domain is also shown. The numbers below or above the schematic indicate the start and end of the boundaries.
Bub1-BubR1 Interaction Contributes to Spindle Assembly Checkpoint Silencing
BubR1 is the key component of MCC which inhibits APC/C directly by blocking the access of the substrates. Interestingly, BUBR1 and BUB1 are paralog genes and the gene products share highly structural similarity. Sub-functionalization during evolution confers the scaffold functionalities to Bub1 and MCC functionalities to BubR1 (Kops et al., 2020). Early studies suggested an interaction between Bub1 and BubR1 (Millband and Hardwick, 2002; Johnson et al., 2004; Rischitor et al., 2007; Kiyomitsu et al., 2011). The interaction was later revealed to be through direct binding of the regions adjacent to the Bub3 binding domain on each protein (Overlack et al., 2015; Zhang et al., 2015). This interaction is required for efficient kinetochore localization of BubR1. The reason BubR1-Bub3 does not efficiently localize to kinetochores is because there are direct contacts between Bub1 and the phosphorylated MELT motifs. This direct contact is mediated by a unique loop in Bub1 that increases the binding affinity with phosphorylated MELT motifs (Overlack et al., 2015). Disrupting the direct interaction between Bub1-BubR1 did not reduce but increased the checkpoint strength which likely reflects the role of checkpoint silencing by the protein phosphatase PP2A-B56 associated with BubR1 (Espert et al., 2014; Zhang et al., 2015; Zhang et al., 2016; Qian et al., 2017).
One interesting question raised here is whether the kinetochore localized BubR1 is important for the checkpoint activity. Based on the fact that low amounts of BubR1 remain on kinetochores following complete removal of Bub1, we proposed a model that two populations of BubR1 exist on kinetochores. The Bub1-dependent BubR1 contributes to the chromosome alignment and the Bub1-independent BubR1 is important for the checkpoint activation (Zhang et al., 2016). The existence of a Bub1-independent pool of BubR1 has been observed on kinetochores in many studies (Overlack et al., 2015; Vleugel et al., 2015a; Zhang et al., 2016; Raaijmakers and Medema, 2019; Chen et al., 2021). FRAP analysis also identified a slow population and a fast population of BubR1 on unattached kinetochores. The t1/2 of the slow BubR1 is very close to the t1/2 of Bub1 (Howell et al., 2004). Besides, Bub1 alone does not saturate the phosphorylated MELT motifs which leaves spaces for the binding of BubR1-Bub3 to the vacant MELT motifs (Overlack et al., 2015). In fission yeast BubR1 (Mad3) expresses three to four times to Bub1 which is also likely the case in human cells (Heinrich et al., 2013). All these aspects indicate a direct binding of BubR1-Bub3 with phosphorylated MELT motif could co-exist with the heterodimerization of BubR1-Bub3 with Bub1-Bub3. As discussed below, this more dynamic pool of BubR1-Bub3 could interact with newly formed Mad2-Cdc20 complex in close proximity and form MCC which leaves kinetochores to inhibit the APC/C.
Bub1-Mad1 Interaction is Critical for Spindle Assembly Checkpoint Activation
How unattached kinetochores catalyze the generation of MCC is the key question in understanding SAC signaling. The prevailing model is centered on the Mad1:Mad2 complex as a key catalyst for loading Mad2 onto Cdc20. Mad1 contains a long predicted coiled-coil in the N-terminal region and a RWD (RING finger-, WD-repeat-, and DEAD-like proteins) domain at the C-terminus. Dimerized Mad1 forms a stable tetramer with two molecules of Mad2. There are two conformations of the Mad2 protein, open Mad2 (O-Mad2) and closed Mad2 (C-Mad2), and Mad2 exists in the closed form when bound to its ligands Mad1 and Cdc20. The kinetochore localized Mad1:C-Mad2 recruits O-Mad2 from the cytoplasm through heterodimerization and converts it into an empty C-Mad2 intermediate which rapidly binds Cdc20 to form the Mad2-Cdc20 complex (Luo et al., 2002; Sironi et al., 2002; Shah et al., 2004; De Antoni et al., 2005; Mapelli et al., 2006; Mapelli et al., 2007; Luo and Yu, 2008; Yang et al., 2008; Kim et al., 2012; Hara et al., 2015). Afterwards, BubR1-Bub3 binds Mad2-Cdc20 to form the MCC complex which can diffuse and inhibit the APC/C (Kulukian et al., 2009). Obviously, discovering the mechanism of Mad1 kinetochore localization is important to test the above model and infer further mechanistic insight into SAC signaling. In mammalian cells, multiple proteins have been proposed as the kinetochore receptors for Mad1 but work is converging on Bub1 and the RZZ complex as the relevant receptors (reviewed in Luo et al., 2018). Recent studies confirmed these two distinct but integrated pathways responsible for Mad1 kinetochore localization and functioning (Silió et al., 2015; Rodriguez-Rodriguez et al., 2018; Zhang et al., 2019). The two pathways will be discussed separately in the following sections.
The Bub1-Mad1 interaction was originally reported to be crucial for checkpoint activation in budding yeast (Brady and Hardwick, 2000). A conserved Arg-Leu-Lys sequence (RLK motif) on Mad1 was found to be required for the interaction while the corresponding motif on Bub1 was only revealed recently. In fission yeast, the middle region of Bub1 bound directly to Mad1 C-terminal region and the interaction required that Bub1 was phosphorylated by Mps1 on a conserved site. The binding was critical for Mad1 kinetochore localization and SAC activation (London and Biggins, 2014; Mora-Santos et al., 2016). In mammalian cells, the region on Bub1 involved in the interaction was narrowed down to a conserved domain 1 (CD1) (Klebig et al., 2009; Ji et al., 2017; Zhang et al., 2017; Qian et al., 2017). Within the CD1 domain, two phosphorylation happens sequentially at S459 and T461. The phosphorylation of S459 by CDK1 primes the phosphorylation of T461 by Mps1 which then mediates the interaction with Mad1 RLK motif. However, even with double phosphorylation, the affinity between Bub1 CD1 peptide and recombinant Mad1 C-terminal protein is relatively low which explains the difficulty to detect the Bub1-Mad1 interaction in human cells (Ji et al., 2017; Zhang et al., 2017). Subsequent dephosphorylation of the two sites by PP2A-B56 makes the detection even more difficult (Qian et al., 2017). An interesting feature of CD1 domain is the presence of a theoretical alpha helix starting at T464 following the two phosphorylation sites. Mutagenesis of residues within the helix region abolishes the Bub1-Mad1 interaction and the checkpoint indicating more complicated interactions besides the electrostatic interaction (Zhang et al., 2017). In a recent structure study, Fischer et al. (2021) found pT461, but not pS459 formed a direct contact with R617 on Mad1 (Figures 2A,B). Interestingly, the contact between pT461 and R617 caused conformational changes on Bub1 CD1 domain into an alpha-helix starting from pT461 instead of the predicted T464. A simultaneous conformation rearrangement also happened on Mad1 after bound to phosphorylated Bub1 which further enhanced the interaction with the formation of a hydrophobic pocket and multiple contacts between the CD1 alpha-helix and Mad1 homodimer (Fischer et al., 2021; Figures 2C,D). The structural study is fully consistent to the previous functional analysis (Ji et al., 2017; Zhang et al., 2017; Fischer et al., 2021). Besides CDK1 and Mps1, Aurora B was recently reported to promote Bub1-Mad1 interaction when it was induced to dimerize with Bub1 (Roy et al., 2022). It will be interesting to test the model under physiological conditions in future work.
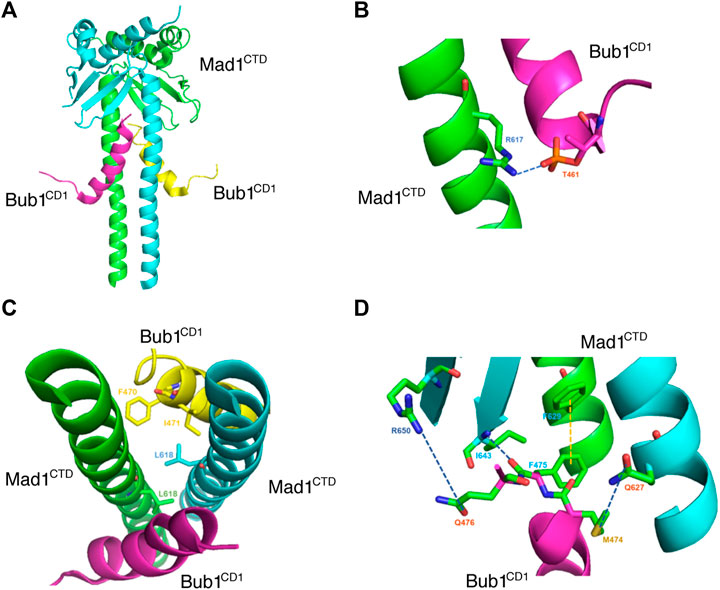
FIGURE 2. Crystal structure of Mad1CTD-Bub1CD1 shows multiple direct interactions. (A) Crystal structure of Mad1CTD dimer (green/light blue) bound to two Bub1CD1 molecules (purple/yellow) from PDB: 7B1F (Fischer et al., 2021). (B) Close-up view of the interaction between Mad1 R617 with Bub1 pT461. The dark blue dashes indicate the hydrogen bonding. (C) Top view of the hydrophobic pocket formed by Mad1 L618 and Bub1 F470 and I471. (D) Close-up view of the interactions between Mad1 R650 and Bub1 Q476, Mad1 I643 and Bub1 F475, Mad1 F629 and Bub1 F475, Mad1 Q627 and Bub1 M474. The dark blue dashes indicate hydrogen bonding and the yellow dashes indicate π–π stacking. Pymol was used to visualize the structure and interactions between residues.
Adjacent to the CD1 domain is a Cdc20 binding motif (ABBA motif or Phe box) which directly interacts with a binding pocket on Cdc20 WD 40 repeats and recruits Cdc20 onto kinetochores (Di Fiore et al., 2015; Diaz-Martinez et al., 2015). Interestingly, a basic patch on the N-terminal region of Cdc20 is able to bind the C-terminus of Mad1 after Mad1 is phosphorylated by Mps1. The binding relieves the intramolecular interaction within Cdc20, thus exposes the Mad2 interacting motif (MIM) to the ligand-free intermediate C-Mad2. Therefore, the simultaneously binding of Cdc20 and Mad1 with Bub1 facilitates the initial assembly of Mad2-Cdc20 complex (Ji et al., 2017; Faesen et al., 2017; Piano et al., 2021; Lara-Gonzalez et al., 2021b; Figure 3). The remaining question is how intermediate C-Mad2 meet Cdc20. The newly resolved Bub1-Mad1 structure positions Bub1-bound Cdc20 in between the Mad2 binding domain and the C-terminus of Mad1. So intermediate C-Mad2 may leave Mad1-Mad2 molecules and capture the close-by exposed MIM on Cdc20. There are evidences showing Cdc20 is one of the kinetochore receptors of C-Mad2 but not vice versa (Lischetti, et al., 2014; Zhang et al., 2018; Figure 3). After the initial formation, Mad2-Cdc20 complex will rapidly bind BubR1/Bub3 to form the final MCC to regulate the mitotic progression. Intriguing results from Lara-Gonzalez et al. (2021b) showed depletion of BubR1 increased Mad2-GFP signals on kinetochores indicating BubR1 is able to release Mad2-Cdc20 from kinetochores by replacing Bub1 on binding to the β-propeller of Cdc20. As discussed in the Bub1-BubR1 section, the second pool of BubR1 is more dynamic and suitable to interact with newly formed Mad2-Cdc20 complex in close proximity than the Bub1-dependent pool.
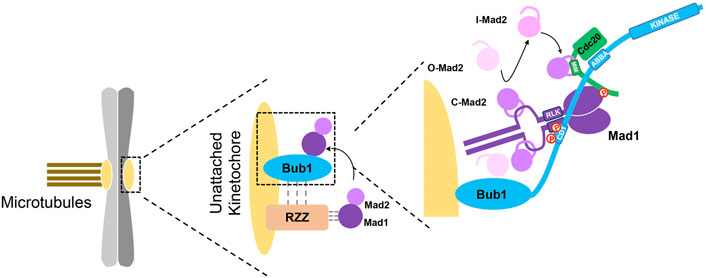
FIGURE 3. Bub1 activates SAC via interactions with multiple checkpoint proteins. The cartoon describes the interactions of Bub1 with Mad1, RZZ and Cdc20 in activating SAC signaling on unattached kinetochores. The left part shows a pair of chromatids with one kinetochore attached by microtubules and the other one unattached. The middle part shows recruiting Mad1/Mad2 onto kinetochores through two distinct and integrated pathways, Bub1-Mad1 and RZZ-Mad1. The dashed lines indicates dependency of RZZ on Bub1 and Mad1 on RZZ whereas the physical interactions have not been confirmed. RZZ binding with Mad1 promotes the binding between Bub1 and Mad1, which further catalyzes the formation of Mad2-Cdc20 complex as shown in the right part. Simultaneous interaction of Bub1 with Mad1-Mad2 and Cdc20 brings Mad2 close to Cdc20. The phosphorylation-mediated binding of the N-terminal Cdc20 with the C-terminal Mad1 exposes the Mad2 interacting motif on Cdc20 and facilitates the formation of Mad2-Cdc20 complex. The minor role of the kinase activity of Bub1 in SAC activation is not described here. Adapted from Lara-Gonzalez et al. (2021b).
Bub1-RZZ Interaction may Contribute to Spindle Assembly Checkpoint Activation
Composed by Rod, Zwilch and ZW10, RZZ complex localizes on unattached kinetochores and provides another layer of SAC regulation in higher eukaryotic cells. So far, no RZZ homologs have been identified in yeast. Together with Spindly, an adaptor protein for dynein-dynactin, RZZ assembles and expands the outmost region of kinetochores to form a fibrous corona, which maximizes the chances of binding with microtubules. Meanwhile RZZ recruits Mad1-Mad2 to the outer kinetochores to monitor the interaction with microtubules (Basto et al., 2000; Chan et al., 2000; Buffin et al., 2005; Kops et al., 2005; Gama et al., 2017; Mosalaganti et al., 2017; Pereira et al., 2018; Rodriguez-Rodriguez et al., 2018; Sacristan et al., 2018; Zhang et al., 2019; Raisch et al., 2021; Barbosa et al., 2022). Once kinetochores are attached to microtubules, RZZ and Mad1-Mad2 are transported away from kinetochores via the Spindly/dynein/dynactin to silence SAC (Barisic et al., 2010; Gama et al., 2017; Gassmann et al., 2010; Pereira et al., 2018; Sacristan et al., 2018; reviewed in McHugh and Welburn, 2017).
How RZZ localizes onto kinetochores is not fully understood and currently direct protein interactions responsible for this is missing. Zwint, the chaperon protein of KNL1 was originally proposed as the kinetochore receptor for RZZ (Wang et al., 2004; Kops et al., 2005; Famulski et al., 2008; Vos et al., 2011). Later it was realized that the dependency is indirect due to the instability of KNL1 in the absence of Zwint (Zhang et al., 2015; van Hooff et al., 2017). Quantification of RZZ kinetochore signals in cells complemented with different KNL1 mutants revealed a requirement for Bub1-MELT interactions for efficient RZZ kinetochore localization. Further truncation analysis of Bub1 identified a region encompassing CD1 domain required for the robust RZZ kinetochore localization. Since efficient Bub1 depletion by RNAi could only remove 65%–85% of ZW10 from kinetochores, there are obviously other receptors for RZZ recruitment (Caldas et al., 2015; Zhang et al., 2015). In Caenorhabditis elegans, the unstructured N-terminal tail of Ndc80 interacts with Rod (Cheerambathur et al., 2013). Whether this interaction is conserved in mammalian cells and anchors RZZ on kinetochores needs to be characterized.
As discussed in Bub1-Mad1 section, the Bub1 CD1 domain interacts with the Mad1 C-terminal domain through multiple contacts. Is it Mad1 recruited by Bub1 that is responsible for RZZ kinetochore localization? Single amino acid mutation disrupting the CD1-Mad1 interaction did not reduce RZZ kinetochore signals which argues against Mad1 playing a direct role in RZZ recruitment (Zhang et al., 2019). The molecular mechanism of Bub1 promoting RZZ kinetochore localization needs further investigation.
Similar as Bub1, depletion of Rod removed around 50% of Mad1/Mad2 from kinetochores in cells treated with nocodazole. In either case, the SAC was significantly impaired but not completely abolished (Caldas et al., 2015; Silió et al., 2015; Rodriguez-Rodriguez et al., 2018; Zhang et al., 2019). When both Rod and Bub1 were depleted, Mad1 could not localize onto kinetochores anymore and the checkpoint was completely inactivated. Interestingly, tethering Mad1 onto kinetochores by fusing it to kinetochore protein Mis12 or KNL1 could fully restore the checkpoint defect in the absence of RZZ, but not in the absence of Bub1. This clearly shows distinct roles of RZZ and Bub1 in the SAC signaling (Rodriguez-Rodriguez et al., 2018; Zhang et al., 2019). Based on these observations, it is proposed that RZZ recruitment of Mad1 facilitates the interaction between Bub1 and Mad1 which catalyzes the production of MCC (Zhang et al., 2019; Figure 3). How Mad1 interacts with RZZ is not known yet. A recent study identified a region on Mad1 to be involved in interacting with RZZ has shed light on this question (Herman et al., 2021). The mechanism of RZZ-Mad1 facilitating Bub1-Mad1 interaction is also missing. The finding that the CD1 C-terminal end and the downstream region of Bub1 promote RZZ kinetochore localization may be part of the mechanism.
Bub1-Cdc20 Plays Important Role on Spindle Assembly Checkpoint Activation
Cdc20 is a coactivator of APC/C as well as a component of MCC. It contains a disordered N-terminal region and a C-terminal β-propeller composed by seven WD40 repeats. Using bioinformatic and biochemical tools, a Cdc20 binding motif was identified in several proteins including cyclin A, BubR1, Bub1 and Acm1. The consensus of the motif is Fx [ILV] [FHY] × [DE] and was named as ABBA motif or Phe box (Di Fiore et al., 2015; Diaz-Martinez et al., 2015). The ABBA motif on Bub1 locates exactly after the region required for Mad1 and RZZ kinetochore localization (Figure 1). Functional analysis found the motif is important for checkpoint activation, though less critical as CD1 domain (Di Fiore et al., 2015; Zhang et al., 2017; Roy et al., 2022). As discussed in Bub1-Mad1 section, Bub1 positions Cdc20 close to Mad1-Mad2 and favors the formation of Mad2-Cdc20 (Figure 3). Interestingly, the kinetochore recruitment of Cdc20 by Bub1 ABBA motif may delay anaphase onset by other mechanism beyond MCC formation. There are evidences that Bub1, together with the bound kinase Plk1 is able to phosphorylate Cdc20 associated with Bub1 ABBA motif. The phosphorylated Cdc20 then binds and inhibits APC/C by preventing its interaction with the E2 ubiquitin-conjugating enzyme Ube2S (Tang et al., 2004; Craney et al., 2016; Jia et al., 2016). It is worth noting that Bub1-Cdc20 interaction may also accelerate APC/C activation under different circumstances. For example, in C. elegans embryos, once kinetochore-microtubule attachment is generated, protein phosphatase one binds to KNL1 and eliminates the phosphorylation on Cdc20 bound with Bub1. Unphosphorylated Cdc20 then works as an activator of APC/C to promote the metaphase-anaphase transition (Kim et al., 2017). Interestingly, in mammalian cells, both PP1 and PP2A-B56 were reported to dephosphorylate Cdc20 (Bancroft et al., 2020; Hein et al., 2021).
Bub1-Plk1 Interaction Plays Minor Roles on Spindle Assembly Checkpoint Activation
Plk1 is an important mitotic kinase playing multiple roles on mitotic entry, centrosome maturation, bipolar spindle assembly, chromatid cohesioin, kinetochore-microtubule attachment and mitotic exit (reviewed in Combes et al., 2017). Plk1 binds to Bub1 through the C-terminal Polo-box domain (PBD) and the PBD binding motif on Bub1 (Qi et al., 2006; Ikeda and Tanaka, 2017). Emerging evidences indicate that Plk1 is involved in activating and maintaining the SAC signaling. When Plk1 alone gets inhibited, there is very limited effect on the checkpoint. The effect on checkpoint only becomes pronounced when Mps1 or Aurora B is partially inhibited simultaneously with Plk1 inhibition (Espeut et al., 2015; O’Connor et al., 2015; von Schubert et al., 2015; Ikeda and Tanaka, 2017). Two mechanisms have been proposed based on these evidences. The first one bypasses the MCC formation. Plk1 is able to phosphorylate Cdc20 on multiple sites, and the phosphorylated Cdc20 inhibits APC/C directly as discussed in Bub1-Cdc20 section (Craney et al., 2016; Jia et al., 2016). The second mechanism is through promoting the MCC formation. Plk1 phosphorylates Mps1 for its full activation as well as KNL1 on the MELT motifs to promote the kinetochore localization of checkpoint proteins and subsequent MCC production (Espeut et al., 2015; O’Connor et al., 2015; von Schubert et al., 2015; Ikeda and Tanaka, 2017; Cordeiro et al., 2020).
Bub Kinase Plays Minor Roles on Spindle Assembly Checkpoint Activation
Bub1 is a bona fide kinase with a few substrates identified (Roberts et al., 1994; Tang et al., 2004; Kawashima et al., 2010; Asghar et al., 2015). The most characterized substrate is histone H2A. Phosphorylation of H2A on T120 provides centromere docking site for TOP2A and Sgo1, the latter further recruits Aurora B and PP2A to promote an accurate chromosome segregation (Kitajima et al., 2005; Tang et al., 2006; Boyarchuk et al., 2007; Fernius et al., 2007; Kawashima et al., 2010; Tsukahara et al., 2010; Broad et al., 2020; Hadders et al., 2020; Zhang et al., 2020). Whether the kinase activity is required for SAC is highly debated (Roberts et al., 1994; Sharp-Baker and Chen, 2001; Warren et al., 2002; Yamaguchi et al., 2003; Chen et al., 2004; Tang et al., 2004; Vanoosthuyse et al., 2004; Fernius et al., 2007; Klebig et al., 2009; Kawashima et al., 2010; Perera et al., 2010; Ricke et al., 2012; Baron et al., 2016; Faesen et al., 2017; Siemeister et al., 2018; Roy et al., 2019; Hadders et al., 2020). Careful examination of the above studies indeed revealed a limited role of the kinase on activating the checkpoint which might be through multiple mechanisms. By phosphorylating the N-terminal region of Cdc20, Bub1 could inhibit APC/C beyond the formation of MCC (Tang et al., 2004). By phosphorylating H2A T120, Bub1 promotes the centromere enrichment of Aurora B which further activates other players like Mps1 and Plk1 to fully activate the checkpoint (Saurin et al., 2011; Carmena et al., 2012; Shao et al., 2015; Singh et al., 2021). The third possibility is the auto-phosphorylation which may regulate interactions with other checkpoint proteins and kinetochore kinetics of Bub1 (Asghar et al., 2015).
Bub1 in Chromosome Alignment
After nuclear envelope breaks down, the dispersed chromosomes need to attach to the microtubules and migrate to the cell center to form the metaphase plate. Aligning chromosomes at the center favors the accurate chromatids segregation. Disturbance of the congression or the failure to maintain the alignment will cause chromosome aligning defects and chromatids segregation errors. The mechanisms behind chromosome congression has been comprehensively reviewed (Maiato et al., 2017).
The role of Bub1 in supporting chromosome alignment was first observed almost two decades ago (Johnson et al., 2004; Meraldi and Sorger 2005; Perera et al., 2007), but the mechanism only starts to emerge recently. In the following, we discuss the known and the possible roles of the Bub1 interactors as well as the kinase activity on promoting chromosome alignment.
Bub1-BubR1 Interaction Is Important for Chromosome Alignment
The mechanism of BubR1 in supporting chromosome alignment has been extensively studied. In summary, through direct interaction of a conserved LxxIxE motif on BubR1 with a surface-exposed pocket on B56, BubR1 recruits protein phosphatase PP2A-B56 onto kinetochores to antagonize the phosphorylation of outer kinetochore proteins like Ndc80, Dsn1, KNL1 and Ska3 (Suijkerbuijk et al., 2012; Kruse et a., 2013; Xu et al., 2013; Espert et al., 2014; Hertz et al., 2016; Wang et al., 2016; Maciejowski et al., 2017). Reducing the phosphorylation level of outer kinetochore proteins promotes stable attachment of kinetochores with microtubules. There are a few studies showing Bub1-dependent BubR1 is required for efficient chromosome alignment during mitotic progression (Zhang et al., 2016; Carvalhal et al., 2022; Figure 4A). Whether Bub1-BubR1 interaction plays the major role of Bub1 to promote chromosome alignment needs further investigation.
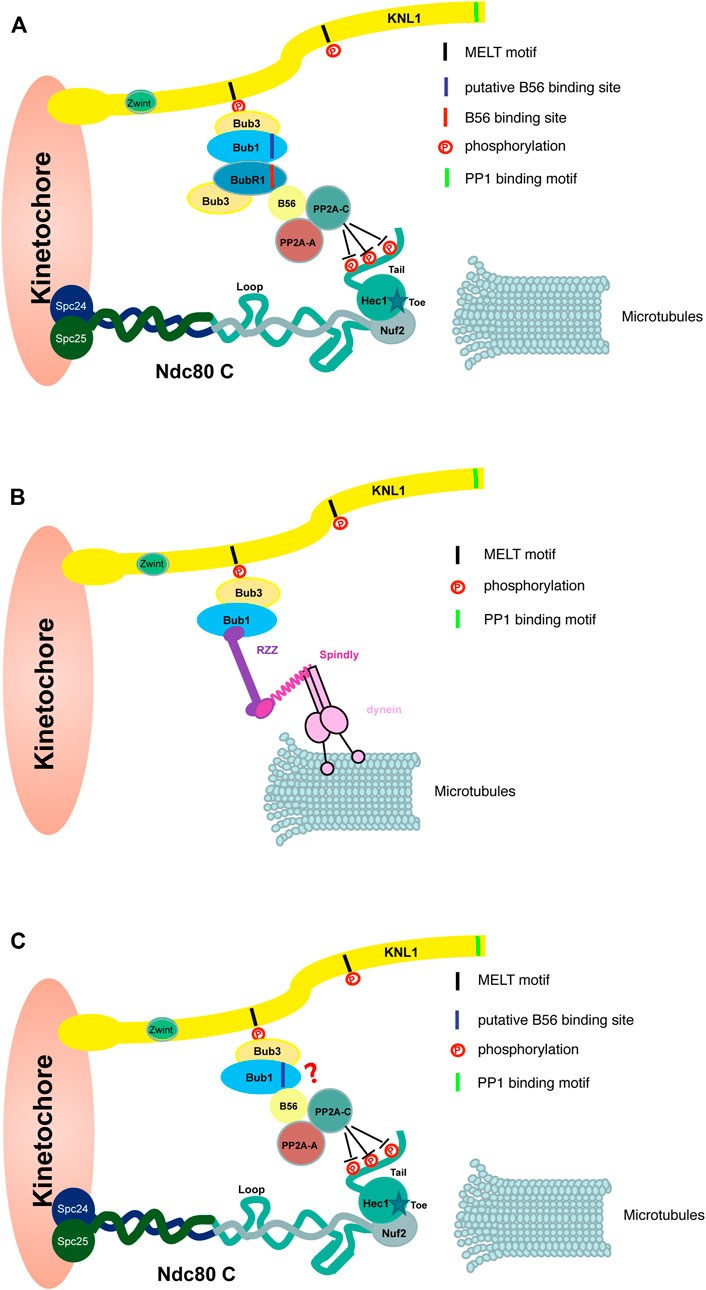
FIGURE 4. Bub1 promotes kinetochore-microtubule attachment through multiple possible pathways. The cartoon shows Bub1 promotes kinetochore-microtubule attachment through a few possible pathways. (A) Bub1 interacts BubR1 directly and the latter brings protein phosphatase PP2A-B56 onto outer kinetochores. The PP2A-B56 complex dephosphorylates multiple outer kinetochore proteins like Ndc80 shown here, which stabilizes the attachment between kinetochores and microtubules. (B) Bub1 promotes the kinetochore localization of RZZ complex. Through an interaction with dynein complex mediated by Spindly protein, Bub1-RZZ may facilitate the initial lateral interaction between kinetochores and microtubules. RZZ-Spindly is able to oligomerize into a fibrous corona which is not shown here. (C) Bub1 may interact PP2A/B56 directly through the putative B56 binding motif. PP2A/B56 promotes a stable kinetochore-microtubule attachment as described in (A) The contribution of the kinase activity is not shown here.
Bub1-RZZ may Contribute to Chromosome Alignment
As mentioned in the above sections, Bub1 promotes RZZ complex localization onto unattached kinetochores (Caldas et al., 2015; Zhang et al., 2015). RZZ complex plays multiple roles to facilitate the kinetochore-microtubule attachment. For example, RZZ recruits Spindly and dynein-dynactin complex onto kinetochores (Gassmann et al., 2008; Barisic et al., 2010; Gama et al., 2017; Mosalaganti et al., 2017; Sacristan et al., 2018). Dynein-dynactin, the minus-end directed motor, facilitates the initial lateral kinetochore-microtubule attachment and a fast poleward movement of chromosomes. This is important for chromosomes, especially the ones close to the spindle poles to complete the congression (Li et al., 2007; Yang et al., 2007; Vorozhko et al., 2008). Together with Spindly, RZZ oligomerizes into a fibrous meshwork which drives the formation of the corona on outmost region of kinetochores (Pereira et al., 2018; Sacristan et al., 2018). The fibrous corona is able to facilitate the kinetochores-microtubules attachment, reduce merotelic attachment and promote spindle assembly (reviewed in Kops and Gassmann, 2020; Barbosa et al., 2022). How much does Bub1-dependent RZZ contribute to the process is less known. One recent study in C. elegans embryos demonstrated that Bub1 indeed regulates chromosome congression via recruiting RZZ/Spindly/dynein/dynactin onto kinetochores (Edwards et al., 2018; Figure 4B). Whether this is also the case in mammalian cells awaits further studies.
Bub1-B56 Interaction Awaits Further Examination
Similar to its paralog BubR1, Bub1 also harbors a putative B56 binding motif (FSPIQE) at the similar position. An in vitro peptide binding assay suggests Bub1 may not bind B56 as efficiently as BubR1 (Braga et al., 2020). In contrast, the study in C. elegans oocytes found Bub1 was able to recruit PP2A/B56 via the B56 binding motif and the Bub1-B56 interaction was important for chromosome congression in meiosis I (Borja et al., 2020; Figure 4C). Again, a functional analysis in mammalian cells is needed to examine whether this motif is involved in promoting the chromosome alignment in mitosis.
Bub1-Plk1 Interaction may be Dispensable for Chromosome Alignment
Plk1 kinase plays essential roles during mitosis including promoting chromosome alignment (reviewed in Combes et al., 2017). Whether the Bub1-Plk1 interaction is needed for chromosome alignment is not clear till recently. Three studies have identified Bub1 and CENP-U as the major kinetochore receptors for Plk1. Simultaneously blocking the two receptors caused strong chromosome aligning defects while disrupting either one alone did not (Chen et al., 2021; Nguyen et al., 2021; Singh et al., 2021). Interestingly, BubR1 also binds Plk1 which further phosphorylates BubR1 to enhance the binding with B56 (Elowe et al., 2007; Suijkerbuijk et al., 2012; Wang et al., 2016). Probably due to the negative feedback from PP2A/B56, BubR1 contributes less on Plk1 kinetochore localization than Bub1 or CENP-U (Singh et al., 2021). Different to Bub1 and CENP-U, disrupting the Plk1 docking on BubR1 severely impairs chromosome alignment (Elowe et al., 2007). Very likely, the Plk1 bound to Bub1 and CENP-U has different targets other than BubR1 to regulate the interaction between microtubules-kinetochores. It will be interesting to identify the substrates in future study.
Bub1 Kinase Plays Minor Roles on Chromosome Alignment
Similar as in SAC signaling, the role of Bub1 kinase on the chromosome alignment is also highly inconsistent (Warren et al., 2002; Fernius et al., 2007; Klebig et al., 2009; Kawashima et al., 2010; Perera et al., 2010; Ricke et al., 2012; Baron et al., 2016; Siemeister et al., 2018; Broad et al., 2020; Hadders et al., 2020; Liang et al., 2020; Zhang et al., 2020; Chen et al., 2021; Carvalhal et al., 2022). For example, the kinase dead Bub1 could not rescue the chromosome alignment defects caused by depleting endogenous Bub1 in HeLa and RPE cells (Klebig et al., 2009). This indicates the kinase activity is a key regulator for chromosome alignment. In contrast, HeLa cells with Bub1 kinase inactivated by CRISPR/Cas9 only displays mild mitotic defects (Chen et al., 2021). The mitosis took 30 min longer than control cells due to the delayed chromosome alignment. Eventually all chromosomes aligned on the metaphase plate and segregated correctly. It has to be noted that in these cells, the Bub1 protein was expressed at low levels (3%–10% to parental cells). The delayed chromosome alignment may be caused by both low Bub1 protein level and kinase inactivation. In this case, the mild mitotic defects does not support an important role for the kinase activity on chromosome alignment.
The discrepancy also exists in the studies with mouse embryonic fibroblasts cells. Perera et al. (2010) found kinase dead Bub1 could fully restore the chromosome alignment and segregation defects in mouse embryo fibroblasts (MEFs) when BUB1 was conditionally inactivated. Another study of Bub1KD/KD (KD means kinase dead) MEFs found a significant increase of rates with misaligned chromosomes at the onset of anaphase compared with wild type cells. However, there were still around 80% of Bub1KD/KD MEFs without chromosome alignment defects or mitotic delay (Ricke et al., 2012). Compared with the Bub1 knockout MEFs, the alignment defect is much milder in Bub1KD/KD MEFs indicating the kinase activity of Bub1 may contribute to the chromosome alignment, but not as the main driving force. Consistently, Bub1KD/KD mice were viable while Bub1Δ/Δ died after day E3.5 (Perera et al., 2007).
Recently, the first two patients with biallelic germline BUB1 mutations were reported (Carvalhal et al., 2022). One of the patients harbors a short deletion in the kinase domain. In fibroblast cells derived from the patient, the protein level of the kinase-inactive Bub1 was significantly reduced and the mitotic process was 24 min delayed with a high frequency of segregation errors in anaphase. Again, the phenotype could be a combined result of both low protein level and kinase inactivation. Besides the genetic inactivation, a series of studies using chemical inhibition of the Bub1 kinase activity revealed either none or marginal mitotic defects (Baron et al., 2016; Siemeister et al., 2018; Broad et al., 2020; Hadders et al., 2020; Liang et al., 2020; Zhang et al., 2020).
In general, majority evidences do not favor an important role of Bub1 kinase activity in promoting chromosome alignment. The minor role of Bub1 kinase may play in this function could be through recruiting Aurora B onto centromeres since artificial centromeric tethering of INCENP, an interactor of Aurora B, could fully rescue the chromosome misalignment in Bub1KD/KD MEFs (Ricke et al., 2012). The mechanism of the rescue is not clear yet. The prevailing model proposes the centromere localized Aurora B phosphorylates outer kinetochore proteins to destabilize and correct erroneous attachment (Liu et al., 2009; Lampson and Cheeseman, 2011). Therefore, inactivation of Bub1 kinase results in Aurora B delocalization and impairs its ability to correct attachment errors. In contrast to this model, the phosphorylation levels of outer kinetochore proteins like Hec1 pS44 were not affected or only moderately reduced (25%–40%) in the cells without centromere Aurora B after simultaneous inhibition of both Bub1 and Haspin (Hadders et al., 2020; Broad et al., 2020; Liang et al., 2020). Since most of the phosphorylation remains on kinetochores, a kinetochore pool of Aurora B has been suggested responsible for outer kinetochore protein phosphorylation. Interestingly, Bub1 itself has been proposed as one kinetochore receptor for Aurora B which needs to be verified in the future (Broad et al., 2020).
Another possibility is the full activation of Plk1 requires centromeric Aurora B (Carmena et al., 2012; Shao et al., 2015; Singh et al., 2021). Plk1 plays important roles on the kinetochore-microtubule attachment as discussed in Bub1-Plk1 section. Thus, delocalized Aurora B reduces the kinase activity of Plk1 when Bub1 kinase is under inhibition. Less active Plk1 may not efficiently phosphorylate BubR1 or other substrates to promote the stable kinetochore-microtubule attachment.
One interesting observation from the patient cells without Bub1 kinase activity is the chromatid bridges with centromeres separated which indicates errors on chromatid arms instead of on kinetochores. Inactivation of Bub1 kinase activity causes delocalization of Sgo1/PP2A from centromeres to the chromosome arms which may protect cohesin complex on chromosome arms from being removed by Plk1 and Aurora B (Kitajima et al., 2005; Tang et al., 2006; Fernius et al., 2007; Kawashima et al., 2010; Carvalhal et al., 2022). Bub1 kinase is also required for TOP2A centromere enrichment. Delocalized TOP2A can not efficiently resolve sister chromatid entanglements which results in ultra-fine bridges (UFBs) during anaphase (Zhang et al., 2020).
Bub1-CENP-F Interaction may be Dispensible for Chromosome Alignment
CENP-F is a long coiled-coil protein containing two microtubule binding domains. In mitosis, CENP-F localizes on kinetochores through direct interaction with the C-terminus of Bub1 (Rattner et al., 1993; Liao et al., 1995; Johnson et al., 2004; Feng et al., 2006; Musinipally et al., 2013; Volkov et al., 2015; Ciossani et al., 2018; Raaijmakers et al., 2018). Furthermore, CENP-F recruits dynein through Nde1/Ndel1/Lis1 complex (Stehman, et al., 2007; Vergnolle et al., 2007; Wynne et al., 2018). Early studies discovered important roles of CENP-F on chromosome alignment by RNA interference (Bomont et al., 2005; Holt et al., 2005; Yang et al., 2005; Feng et al., 2006). The work in C. elegans embryos also found Bub1 contributes to the kinetochore-microtubule attachment via CENP-F/CLASP axis (Edwards et al., 2018). However, recent studies by knocking out CENP-F showed different results. First, CENP-F knockout mice were viable which questions the essentiality of the gene (Haley et al., 2019). Second, the CENP-F knockout cell lines displayed none mitotic phenotype or very mild effect on mitotic progression (Raaijmakers et al., 2018; Auckland et al., 2020). Though lack of enough evidences, an important role of Bub1-CENP-F interaction on chromosome alignment is disfavored in mammalian cells.
Bub1 Essentiality
Considering the critical roles Bub1 plays in mitosis as discussed above, it is hardly to question its essentiality in higher eukaryotes. Indeed, in mouse studies, knockout of BUB1 resulted in embryonic lethality after day E3.5. Late inactivation on day E10.5 quickly shut down further development. In MEFs cells, knocking out of Bub1 severely impaired SAC and chromosome alignment and the cell proliferation stopped due to the strong chromosome segregation errors (Perera et al., 2007; Tilston et al., 2009). BUB1 hypomorphic mice expressing minimal level of Bub1 truncate (less than 5% to wild type mice) were viable with increased tumorigenesis (Schliekelman et al., 2009). In humans, low Bub1 expression correlates with spontaneous miscarriage (Shi et al., 2011). Recently, two patients with biallelic germline BUB1 mutations were reported. Patient one has homozygous point mutation in the start codon of BUB1 resulting in residual levels of Bub1 protein beyong detection by regular methods. The existence of H2A pT120 signals on kinetochores indicates the presence of a functional Bub1 protein in this patient. The other patient expresses a low level of truncated Bub1 protein without kinase activity. Both patients suffered microcephaly, delayed development and other abnormalities (Carvalhal et al., 2022).
The above studies clearly show BUB1 is essential in higher eukaryotes. However, the successful generation of BUB1 knockout human cells seriously questioned the essentiality (Currie et al., 2018; Raaijmakers et al., 2018; Rodriguez- Rodriguez et al., 2018; Zhang et al., 2019; Chen et al., 2021). Further studies found all these BUB1 knockout cell lines still expressed low level of Bub1 truncates by nonsense-associated alternative splicing (Rodriguez- Rodriguez et al., 2018; Zhang et al., 2019; Chen et al., 2021). The residual Bub1 (3%–30% of parental cells) was able to fully support the SAC and largely promote chromosome alignment in the cells. Thus, the mitotic progression was only moderately elongated with one or two chromosomes delayed on joining the metaphase plate. The functional checkpoint ensured the delayed chromosomes could align properly before cells entered anaphase without severe chromosome segregation errors. One interesting feature of these cells is the super sensitivity to RNA interference. Treatment of the cells with siRNA oligos against Bub1 resulted in severe impairment on both SAC signaling and chromosome alignment and cells entered anaphase with massive unaligned chromosomes which was hardly achieved in normal cells (Zhang et al., 2019; Chen et al., 2021). Importantly, supplemented with siRNA resistant wild type Bub1 fully rescued these defects indicating Bub1 is indeed required for faithful mitosis (Zhang et al., 2019). So far, by introducing indels in either exons for N-terminal region or exons for C-terminal region failed to completely inactivate Bub1. The effort to generate a full deletion of BUB1 in diploid or aneuploid human cells also failed. The failure could be from the technology difficulty or from the possibility that diploid or aneuploid cells can not survive the full BUB1 deletion. Only in near-haploid HAP1 cells, a full BUB1 deletion by simultaneously targeting exon 1 and exon 25 was achieved showing BUB1 is not essential in HAP1 cells. In these cells, the SAC strength was reduced at least in the presence of low concentration of microtubule toxin (Raaijmakers and Medema, 2019). It needs to be noted that having only one set of genome may confer different response to the loss of essential genes in HAP1 cells than in diploid or aneuploid cells.
An alternative strategy to confirm the essentiality of BUB1 in human cells is introducing indels in the exons encoding domains with important functions (McKinley and Cheeseman, 2017). Alternative splicing normally escapes part or the whole exon Cas9 targeted. Thus, the truncated proteins if possibly produced, are not able to execute these functions. Examining the viability of the cells following the Cas9 cutting will provide direct answer on the essentiality of BUB1. Exons 8, 9, and 12 encoding Bub3, BubR1, Mad1 binding domains are particularly interesting for this purpose. Disrupting exon 8 will prevent Bub1 truncate from kinetochores which is very likely lethal as Perera et al. (2010) showed in MEFs cells. Singly cutting exon 9 or 12 will impair the chromosome congression or SAC signaling which may not be enough to cause lethality due to the protection by full or partial functional checkpoint. Simultaneously targeting exon 9 and 12 is more likely to be intolerable for cells (Figure 5).
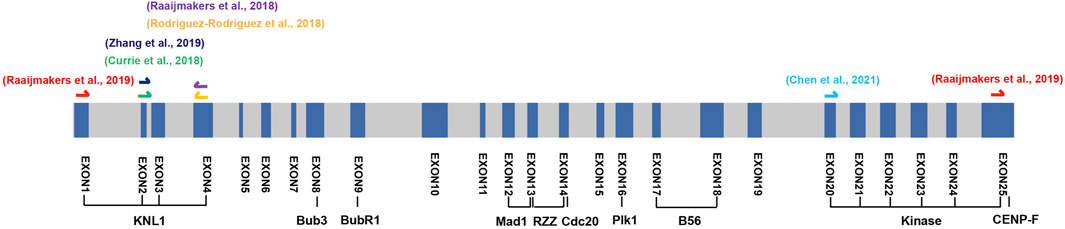
FIGURE 5. Schematic of the targeted exons of the Bub1 knockout cell lines. The schematic shows all the exons of BUB1 in human genome. The exons targeted by CRISPR/Cas9 from a number of studies are marked on top of the schematic. We propose targeting exon 8 (GLEBS domain) or co-targeting exon 9 (BubR1 binding domain) and exon 12 (CD1 domain) may cause lethality. The lengths of exons, but not introns are proportional to the actual lengths on genome.
In general, after 30 years’ extensive investigation, the importance of Bub1 in mitosis has been well documented and largely appreciated with some questions undissolved yet. On the checkpoint: how does Bub1 promote RZZ kinetochore localization? what is the molecular mechanism that RZZ-Mad1 facilitates Bub1-Mad1 interaction? On chromosome alignment: are there more domains of Bub1 required for chromosome alignment? what is the role of the putative B56 binding motif on Bub1? are the two B56 binding motifs on Bub1 and BubR1 redundant? On the essentiality: can human cells tolerate the complete loss of Bub1? Answer these questions will help us further understand the roles of Bub1 in mitosis.
Author Contributions
GZ did the literature research and wrote the manuscript with JF; YQZ, CS, LW, HJ, YJZ, and YW assembled the figures and proofread the manuscript.
Funding
GZ lab is supported by Taishan Scholar Project (tsqn201812054) from Shandong, China and the National Natural Science Foundation of China (grant No. 31970666).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors thank Jakob Nilsson for constructive comments of the manuscript. We apologize for the studies on Bub1 not cited in this review due to the size limit of the manuscript.
References
Asghar, A., Lajeunesse, A., Dulla, K., Combes, G., Thebault, P., Nigg, E. A., et al. (2015). Bub1 Autophosphorylation Feeds Back to Regulate Kinetochore Docking and Promote Localized Substrate Phosphorylation. Nat. Commun. 6, 8364. doi:10.1038/ncomms9364
Auckland, P., Roscioli, E., Coker, H. L. E., and McAinsh, A. D. (2020). CENP-F Stabilizes Kinetochore-Microtubule Attachments and Limits Dynein Stripping of corona Cargoes. J. Cel Biol. 219, e201905018. doi:10.1083/jcb.201905018
Bancroft, J., Holder, J., Geraghty, Z., Alfonso-Pérez, T., Murphy, D., Barr, F. A., et al. (2020). PP1 Promotes Cyclin B Destruction and the Metaphase-Anaphase Transition by Dephosphorylating CDC20. MBoC 31, 2315–2330. doi:10.1091/mbc.E20-04-0252
Barbosa, J., Sunkel, C. E., and Conde, C. (2022). The Role of Mitotic Kinases and the RZZ Complex in Kinetochore-Microtubule Attachments: Doing the Right Link. Front. Cell Dev. Biol. 10, 787294. doi:10.3389/fcell.2022.787294
Barisic, M., Sohm, B., Mikolcevic, P., Wandke, C., Rauch, V., Ringer, T., et al. (2010). Spindly/CCDC99 Is Required for Efficient Chromosome Congression and Mitotic Checkpoint Regulation. MBoC 21, 1968–1981. doi:10.1091/mbc.e09-04-0356
Baron, A. P., von Schubert, C., Cubizolles, F., Siemeister, G., Hitchcock, M., Mengel, A., et al. (2016). Probing the Catalytic Functions of Bub1 Kinase Using the Small Molecule Inhibitors BAY-320 and BAY-524. Elife 5, e12187. doi:10.7554/eLife.12187
Basto, R., Gomes, R., and Karess, R. E. (2000). Rough deal and Zw10 Are Required for the Metaphase Checkpoint in Drosophila. Nat. Cel Biol. 2, 939–943. doi:10.1038/35046592
Bel Borja, L., Soubigou, F., Taylor, S. J. P., Fraguas Bringas, C., Budrewicz, J., Lara-Gonzalez, P., et al. (2020). BUB-1 Targets PP2A:B56 to Regulate Chromosome Congression during Meiosis I in C. elegans Oocytes. Elife 9, e65307. doi:10.7554/eLife.65307
Bernard, P., Hardwick, K., and Javerzat, J.-P. (1998). Fission Yeast Bub1 Is a Mitotic Centromere Protein Essential for the Spindle Checkpoint and the Preservation of Correct Ploidy through Mitosis. J. Cel Biol. 143, 1775–1787. doi:10.1083/jcb.143.7.1775
Bomont, P., Maddox, P., Shah, J. V., Desai, A. B., and Cleveland, D. W. (2005). Unstable Microtubule Capture at Kinetochores Depleted of the Centromere-Associated Protein CENP-F. EMBO J. 24, 3927–3939. doi:10.1038/sj.emboj.7600848
Boyarchuk, Y., Salic, A., Dasso, M., and Arnaoutov, A. (2007). Bub1 Is Essential for Assembly of the Functional Inner Centromere. J. Cel Biol. 176, 919–928. doi:10.1083/jcb.200609044
Brady, D. M., and Hardwick, K. G. (2000). Complex Formation between Mad1p, Bub1p and Bub3p Is Crucial for Spindle Checkpoint Function. Curr. Biol. 10, 675–678. doi:10.1016/s0960-9822(00)00515-7
Broad, A. J., DeLuca, K. F., and DeLuca, J. G. (2020). Aurora B Kinase Is Recruited to Multiple Discrete Kinetochore and Centromere Regions in Human Cells. J. Cel Biol. 219, e201905144. doi:10.1083/jcb.201905144
Buffin, E., Lefebvre, C., Huang, J., Gagou, M. E., and Karess, R. E. (2005). Recruitment of Mad2 to the Kinetochore Requires the Rod/Zw10 Complex. Curr. Biol. 15, 856–861. doi:10.1016/j.cub.2005.03.052
Caldas, G. V., Lynch, T. R., Anderson, R., Afreen, S., Varma, D., and DeLuca, J. G. (2015). The RZZ Complex Requires the N-Terminus of KNL1 to Mediate Optimal Mad1 Kinetochore Localization in Human Cells. Open Biol. 5, 150160. doi:10.1098/rsob.150160
Carmena, M., Pinson, X., Platani, M., Salloum, Z., Xu, Z., Clark, A., et al. (2012). The Chromosomal Passenger Complex Activates Polo Kinase at Centromeres. Plos Biol. 10, e1001250. doi:10.1371/journal.pbio.1001250
Carvalhal, S., Bader, I., Rooimans, M. A., Oostra, A. B., Balk, J. A., Feichtinger, R. G., et al. (2022). Biallelic BUB1 Mutations Cause Microcephaly, Developmental Delay, and Variable Effects on Cohesion and Chromosome Segregation. Sci. Adv. 8, eabk0114. doi:10.1126/sciadv.abk0114
Chan, G. K. T., Jablonski, S. A., Starr, D. A., Goldberg, M. L., and Yen, T. J. (2000). Human Zw10 and ROD Are Mitotic Checkpoint Proteins that Bind to Kinetochores. Nat. Cel Biol. 2, 944–947. doi:10.1038/35046598
Cheerambathur, D. K., Gassmann, R., Cook, B., Oegema, K., and Desai, A. (2013). Crosstalk between Microtubule Attachment Complexes Ensures Accurate Chromosome Segregation. Science 342, 1239–1242. doi:10.1126/science.1246232
Chen, Q., Zhang, M., Pan, X., Yuan, X., Zhou, L., Yan, L., et al. (2021). Bub1 and CENP-U Redundantly Recruit Plk1 to Stabilize Kinetochore-Microtubule Attachments and Ensure Accurate Chromosome Segregation. Cel Rep. 36, 109740. doi:10.1016/j.celrep.2021.109740
Chen, R.-H. (2004). Phosphorylation and Activation of Bub1 on Unattached Chromosomes Facilitate the Spindle Checkpoint. EMBO J. 23, 3113–3121. doi:10.1038/sj.emboj.7600308
Ciossani, G., Overlack, K., Petrovic, A., Huis in 't Veld, P. J., Koerner, C., Wohlgemuth, S., et al. (2018). The Kinetochore Proteins CENP-E and CENP-F Directly and Specifically Interact with Distinct BUB Mitotic Checkpoint Ser/Thr Kinases. J. Biol. Chem. 293, 10084–10101. doi:10.1074/jbc.RA118.003154
Combes, G., Alharbi, I., Braga, L. G., and Elowe, S. (2017). Playing polo during Mitosis: PLK1 Takes the lead. Oncogene 36, 4819–4827. doi:10.1038/onc.2017.113
Cordeiro, M. H., Smith, R. J., and Saurin, A. T. (2020). Kinetochore Phosphatases Suppress Autonomous Polo-like Kinase 1 Activity to Control the Mitotic Checkpoint. J. Cel Biol. 219, e202002020. doi:10.1083/jcb.202002020
Craney, A., Kelly, A., Jia, L., Fedrigo, I., Yu, H., and Rape, M. (2016). Control of APC/C-dependent Ubiquitin Chain Elongation by Reversible Phosphorylation. Proc. Natl. Acad. Sci. U S A. 113, 1540–1545. doi:10.1073/pnas.1522423113
Currie, C. E., Mora-Santos, M., Smith, C. A., McAinsh, A. D., and Millar, J. B. A. (2018). Bub1 Is Not Essential for the Checkpoint Response to Unattached Kinetochores in Diploid Human Cells. Curr. Biol. 28, R929–R930. doi:10.1016/j.cub.2018.07.040
De Antoni, A., Pearson, C. G., Cimini, D., Canman, J. C., Sala, V., Nezi, L., et al. (2005). The Mad1/Mad2 Complex as a Template for Mad2 Activation in the Spindle Assembly Checkpoint. Curr. Biol. 15, 214–225. doi:10.1016/j.cub.2005.01.038
Diaz-Martinez, L. A., Tian, W., Li, B., Warrington, R., Jia, L., Brautigam, C. A., et al. (2015). The Cdc20-Binding Phe Box of the Spindle Checkpoint Protein BubR1 Maintains the Mitotic Checkpoint Complex during Mitosis. J. Biol. Chem. 290, 2431–2443. doi:10.1074/jbc.M114.616490
Di Fiore, B., Davey, N. E., Hagting, A., Izawa, D., Mansfeld, J., Gibson, T. J., et al. (2015). The ABBA Motif Binds APC/C Activators and Is Shared by APC/C Substrates and Regulators. Dev. Cel 32, 358–372. doi:10.1016/j.devcel.2015.01.003
Edwards, F., Maton, G., Gareil, N., Canman, J. C., and Dumont, J. (2018). BUB-1 Promotes Amphitelic Chromosome Biorientation via Multiple Activities at the Kinetochore. Elife 7, e40690. doi:10.7554/eLife.40690
Elowe, S., Hümmer, S., Uldschmid, A., Li, X., and Nigg, E. A. (2007). Tension-sensitive Plk1 Phosphorylation on BubR1 Regulates the Stability of Kinetochore-Microtubule Interactions. Genes Dev. 21, 2205–2219. doi:10.1101/gad.436007
Espert, A., Uluocak, P., Bastos, R. N., Mangat, D., Graab, P., and Gruneberg, U. (2014). PP2A-B56 Opposes Mps1 Phosphorylation of Knl1 and Thereby Promotes Spindle Assembly Checkpoint Silencing. J. Cel Biol. 206, 833–842. doi:10.1083/jcb.201406109
Espeut, J., Lara-Gonzalez, P., Sassine, M., Shiau, A. K., Desai, A., and Abrieu, A. (2015). Natural Loss of Mps1 Kinase in Nematodes Uncovers a Role for Polo-like Kinase 1 in Spindle Checkpoint Initiation. Cel Rep. 12, 58–65. doi:10.1016/j.celrep.2015.05.039
Faesen, A. C., Thanasoula, M., Maffini, S., Breit, C., Müller, F., van Gerwen, S., et al. (2017). Basis of Catalytic Assembly of the Mitotic Checkpoint Complex. Nature 542, 498–502. doi:10.1038/nature21384
Famulski, J. K., Vos, L., Sun, X., and Chan, G. (2008). Stable hZW10 Kinetochore Residency, Mediated by hZwint-1 Interaction, Is Essential for the Mitotic Checkpoint. J. Cel Biol. 180, 507–520. doi:10.1083/jcb.200708021
Feng, J., Huang, H., and Yen, T. J. (2006). CENP-F Is a Novel Microtubule-Binding Protein that Is Essential for Kinetochore Attachments and Affects the Duration of the Mitotic Checkpoint Delay. Chromosoma 115, 320–329. doi:10.1007/s00412-006-0049-5
Fernius, J., and Hardwick, K. G. (2007). Bub1 Kinase Targets Sgo1 to Ensure Efficient Chromosome Biorientation in Budding Yeast Mitosis. Plos Genet. 3, e213. doi:10.1371/journal.pgen.0030213
Fischer, E. S., Yu, C. W. H., Bellini, D., McLaughlin, S. H., Orr, C. M., Wagner, A., et al. (2021). Molecular Mechanism of Mad1 Kinetochore Targeting by Phosphorylated Bub1. EMBO Rep. 22, e52242. doi:10.15252/embr.202052242
Gama Braga, L., Cisneros, A. F., Mathieu, M. M., Clerc, M., Garcia, P., Lottin, B., et al. (2020). BUBR1 Pseudokinase Domain Promotes Kinetochore PP2A-B56 Recruitment, Spindle Checkpoint Silencing, and Chromosome Alignment. Cel Rep. 33, 108397. doi:10.1016/j.celrep.2020.108397
Gama, J. B., Pereira, C., Simões, P. A., Celestino, R., Reis, R. M., Barbosa, D. J., et al. (2017). Molecular Mechanism of Dynein Recruitment to Kinetochores by the Rod-Zw10-Zwilch Complex and Spindly. J. Cel Biol. 216, 943–960. doi:10.1083/jcb.201610108
Gassmann, R., Essex, A., Hu, J.-S., Maddox, P. S., Motegi, F., Sugimoto, A., et al. (2008). A New Mechanism Controlling Kinetochore-Microtubule Interactions Revealed by Comparison of Two Dynein-Targeting Components: SPDL-1 and the Rod/Zwilch/Zw10 Complex. Genes Dev. 22, 2385–2399. doi:10.1101/gad.1687508
Gassmann, R., Holland, A. J., Varma, D., Wan, X., Çivril, F., Cleveland, D. W., et al. (2010). Removal of Spindly from Microtubule-Attached Kinetochores Controls Spindle Checkpoint Silencing in Human Cells. Genes Dev. 24, 957–971. doi:10.1101/gad.1886810
Hadders, M. A., Hindriksen, S., Truong, M. A., Mhaskar, A. N., Wopken, J. P., Vromans, M. J. M., et al. (2020). Untangling the Contribution of Haspin and Bub1 to Aurora B Function during Mitosis. J. Cel Biol. 219, e201907087. doi:10.1083/jcb.201907087
Haley, C. O., Waters, A. M., and Bader, D. M. (2019). Malformations in the Murine Kidney Caused by Loss of CENP‐F Function. Anat. Rec. 302, 163–170. doi:10.1002/ar.24018
Hara, M., Özkan, E., Sun, H., Yu, H., and Luo, X. (2015). Structure of an Intermediate Conformer of the Spindle Checkpoint Protein Mad2. Proc. Natl. Acad. Sci. U S A. 112, 11252–11257. doi:10.1073/pnas.1512197112
Hein, J. B., Garvanska, D. H., Nasa, I., Kettenbach, A. N., and Nilsson, J. (2021). Coupling of Cdc20 Inhibition and Activation by BubR1. J. Cel Biol 220, e202012081. doi:10.1083/jcb.202012081
Hein, J., Boichuk, S., Wu, J., Cheng, Y., Freire, R., Jat, P. S., et al. (2009). Simian Virus 40 Large T Antigen Disrupts Genome Integrity and Activates a DNA Damage Response via Bub1 Binding. J. Virol. 83, 117–127. doi:10.1128/JVI.01515-08
Heinrich, S., Geissen, E.-M., Kamenz, J., Trautmann, S., Widmer, C., Drewe, P., et al. (2013). Determinants of Robustness in Spindle Assembly Checkpoint Signalling. Nat. Cel Biol. 15, 1328–1339. doi:10.1038/ncb2864
Herman, J. A., Carter, L., Arora, S., Zhu, J., Biggins, S., and Paddison, P. J. (2021). Functional Dissection of Human Mitotic Genes Using CRISPR-Cas9 Tiling Screens. bioRxiv. doi:10.1101/2021.05.20.445000
Hertz, E. P. T., Kruse, T., Davey, N. E., López-Méndez, B., Sigurðsson, J. O., Montoya, G., et al. (2016). A Conserved Motif Provides Binding Specificity to the PP2A-B56 Phosphatase. Mol. Cel 63, 686–695. doi:10.1016/j.molcel.2016.06.024
Holt, S. V., Vergnolle, M. A. S., Hussein, D., Wozniak, M. J., Allan, V. J., and Taylor, S. S. (2005). Silencing Cenp-F Weakens Centromeric Cohesion, Prevents Chromosome Alignment and Activates the Spindle Checkpoint. J. Cel Sci. 118, 4889–4900. doi:10.1242/jcs.02614
Hooff, J. J., Tromer, E., Wijk, L. M., Snel, B., and Kops, G. J. (2017). Evolutionary Dynamics of the Kinetochore Network in Eukaryotes as Revealed by Comparative Genomics. EMBO Rep. 18, 1559–1571. doi:10.15252/embr.201744102
Howell, B. J., Moree, B., Farrar, E. M., Stewart, S., Fang, G., and Salmon, E. D. (2004). Spindle Checkpoint Protein Dynamics at Kinetochores in Living Cells. Curr. Biol. 14, 953–964. doi:10.1016/j.cut.2004.05.05310.1016/j.cub.2004.05.053
Hoyt, M. A., Totis, L., and Roberts, B. T. (1991). S. cerevisiae Genes Required for Cell Cycle Arrest in Response to Loss of Microtubule Function. Cell 66, 507–517. doi:10.1016/0092-8674(81)90014-3
Ikeda, M., and Tanaka, K. (2017). Plk1 Bound to Bub1 Contributes to Spindle Assembly Checkpoint Activity during Mitosis. Sci. Rep. 7, 8794. doi:10.1038/s41598-017-09114-3
Jablonski, S. A., Chan, G. K. T., Cooke, C. A., Yen, T. J., and Earnshaw, W. C. (1998). The hBUB1 and hBUBR1 Kinases Sequentially Assemble onto Kinetochores during Prophase with hBUBR1 Concentrating at the Kinetochore Plates in Mitosis. Chromosoma 107, 386–396. doi:10.1007/s004120050322
Ji, Z., Gao, H., Jia, L., Li, B., and Yu, H. (2017). A Sequential Multi-Target Mps1 Phosphorylation cascade Promotes Spindle Checkpoint Signaling. Elife 6, e22513. doi:10.7554/eLife.22513
Jia, L., Li, B., and Yu, H. (2016). The Bub1-Plk1 Kinase Complex Promotes Spindle Checkpoint Signalling through Cdc20 Phosphorylation. Nat. Commun. 7, 10818. doi:10.1038/ncomms10818
Johnson, V. L., Scott, M. I. F., Holt, S. V., Hussein, D., and Taylor, S. S. (2004). Bub1 Is Required for Kinetochore Localization of BubR1, Cenp-E, Cenp-F and Mad2, and Chromosome Congression. J. Cel Sci. 117, 1577–1589. doi:10.1242/jcs.01006
Kapanidou, M., Curtis, N. L., and Bolanos-Garcia, V. M. (2017). Cdc20: at the Crossroads between Chromosome Segregation and Mitotic Exit. Trends Biochem. Sci. 42, 193–205. doi:10.1016/j.tibs.2016.12.001
Kawashima, S. A., Yamagishi, Y., Honda, T., Ishiguro, K.-i., and Watanabe, Y. (2010). Phosphorylation of H2A by Bub1 Prevents Chromosomal Instability through Localizing Shugoshin. Science 327, 172–177. doi:10.1126/science.1180189
Kim, S., Sun, H., Tomchick, D. R., Yu, H., and Luo, X. (2012). Structure of Human Mad1 C-Terminal Domain Reveals its Involvement in Kinetochore Targeting. Proc. Natl. Acad. Sci. U S A. 109, 6549–6554. doi:10.1073/pnas.1118210109
Kim, T., Lara-Gonzalez, P., Prevo, B., Meitinger, F., Cheerambathur, D. K., Oegema, K., et al. (2017). Kinetochores Accelerate or Delay APC/C Activation by Directing Cdc20 to Opposing Fates. Genes Dev. 31, 1089–1094. doi:10.1101/gad.302067.117
Kitajima, T. S., Hauf, S., Ohsugi, M., Yamamoto, T., and Watanabe, Y. (2005). Human Bub1 Defines the Persistent Cohesion Site along the Mitotic Chromosome by Affecting Shugoshin Localization. Curr. Biol. 15, 353–359. doi:10.1016/j.cub.2004.12.044
Kitajima, T. S., Kawashima, S. A., and Watanabe, Y. (2004). The Conserved Kinetochore Protein Shugoshin Protects Centromeric Cohesion during Meiosis. Nature 427, 510–517. doi:10.1038/nature02312
Kiyomitsu, T., Murakami, H., and Yanagida, M. (2011). Protein Interaction Domain Mapping of Human Kinetochore Protein Blinkin Reveals a Consensus Motif for Binding of Spindle Assembly Checkpoint Proteins Bub1 and BubR1. Mol. Cel Biol. 31, 998–1011. doi:10.1128/MCB.00815-10
Kiyomitsu, T., Obuse, C., and Yanagida, M. (2007). Human Blinkin/AF15q14 Is Required for Chromosome Alignment and the Mitotic Checkpoint through Direct Interaction with Bub1 and BubR1. Dev. Cel 13, 663–676. doi:10.1016/j.devcel.2007.09.005
Klebig, C., Korinth, D., and Meraldi, P. (2009). Bub1 Regulates Chromosome Segregation in a Kinetochore-independent Manner. J. Cel Biol. 185, 841–858. doi:10.1083/jcb.200902128
Kops, G. J. P. L., and Gassmann, R. (2020). Crowning the Kinetochore: the Fibrous corona in Chromosome Segregation. Trends Cel Biol. 30, 653–667. doi:10.1016/j.tcb.2020.04.006
Kops, G. J. P. L., Kim, Y., Weaver, B. A. A., Mao, Y., McLeod, I., Yates, J. R., et al. (2005). ZW10 Links Mitotic Checkpoint Signaling to the Structural Kinetochore. J. Cel Biol. 169, 49–60. doi:10.1083/jcb.200411118
Kops, G. J. P. L., Snel, B., and Tromer, E. C. (2020). Evolutionary Dynamics of the Spindle Assembly Checkpoint in Eukaryotes. Curr. Biol. 30, R589–R602. doi:10.1016/j.cub.2020.02.021
Krenn, V., Wehenkel, A., Li, X., Santaguida, S., and Musacchio, A. (2012). Structural Analysis Reveals Features of the Spindle Checkpoint Kinase Bub1-Kinetochore Subunit Knl1 Interaction. J. Cel Biol. 196, 451–467. doi:10.1083/jcb.201110013
Kruse, T., Zhang, G., Larsen, M. S. Y., Lischetti, T., Streicher, W., Kragh Nielsen, T., et al. (2013). Direct Binding between BubR1 and B56-Pp2a Phosphatase Complexes Regulate Mitotic Progression. J. Cel Sci. 126, 1086–1092. doi:10.1242/jcs.122481
Kulukian, A., Han, J. S., and Cleveland, D. W. (2009). Unattached Kinetochores Catalyze Production of an Anaphase Inhibitor that Requires a Mad2 Template to Prime Cdc20 for BubR1 Binding. Dev. Cel 16, 105–117. doi:10.1016/j.devcel.2008.11.005
Lampson, M. A., and Cheeseman, I. M. (2011). Sensing Centromere Tension: Aurora B and the Regulation of Kinetochore Function. Trends Cel Biol. 21, 133–140. doi:10.1016/j.tcb.2010.10.007
Lara-Gonzalez, P., Kim, T., Oegema, K., Corbett, K., and Desai, A. (2021b). A Tripartite Mechanism Catalyzes Mad2-Cdc20 Assembly at Unattached Kinetochores. Science 371, 64–67. doi:10.1126/science.abc1424
Lara-Gonzalez, P., Pines, J., and Desai, A. (2021a). Spindle Assembly Checkpoint Activation and Silencing at Kinetochores. Semin. Cel Dev. Biol. 117 (21), 86–98. doi:10.1016/j.semcdb.2021.06.009
Li, F., Kim, H., Ji, Z., Zhang, T., Chen, B., Ge, Y., et al. (2018). The BUB3-BUB1 Complex Promotes Telomere DNA Replication. Mol. Cel 70, 395–407. doi:10.1016/j.molcel.2018.03.032
Li, R., and Murray, A. W. (1991). Feedback Control of Mitosis in Budding Yeast. Cell 66, 519–531. doi:10.1016/0092-8674(81)90015-5
Li, Y., Yu, W., Liang, Y., and Zhu, X. (2007). Kinetochore Dynein Generates a Poleward Pulling Force to Facilitate Congression and Full Chromosome Alignment. Cell Res 17, 701–712. doi:10.1038/cr.2007.65
Liang, C., Zhang, Z., Chen, Q., Yan, H., Zhang, M., Zhou, L., et al. (2020). Centromere-localized Aurora B Kinase Is Required for the Fidelity of Chromosome Segregation. J. Cel Biol. 219, e201907092. doi:10.1083/jcb.201907092
Liao, H., Winkfein, R. J., Mack, G., Rattner, J. B., and Yen, T. J. (1995). CENP-F Is a Protein of the Nuclear Matrix that Assembles onto Kinetochores at Late G2 and Is Rapidly Degraded after Mitosis. J. Cel Biol. 130, 507–518. doi:10.1083/jcb.130.3.507
Lischetti, T., Zhang, G., Sedgwick, G. G., Bolanos-Garcia, V. M., and Nilsson, J. (2014). The Internal Cdc20 Binding Site in BubR1 Facilitates Both Spindle Assembly Checkpoint Signalling and Silencing. Nat. Commun. 5, 5563. doi:10.1038/ncomms6563
Liu, D., Vader, G., Vromans, M. J. M., Lampson, M. A., and Lens, S. M. A. (2009). Sensing Chromosome Bi-orientation by Spatial Separation of aurora B Kinase from Kinetochore Substrates. Science 323, 1350–1353. doi:10.1126/science.1167000
London, N., and Biggins, S. (2014). Mad1 Kinetochore Recruitment by Mps1-Mediated Phosphorylation of Bub1 Signals the Spindle Checkpoint. Genes Dev. 28, 140–152. doi:10.1101/gad.233700.113
London, N., Ceto, S., Ranish, J. A., and Biggins, S. (2012). Phosphoregulation of Spc105 by Mps1 and PP1 Regulates Bub1 Localization to Kinetochores. Curr. Biol. 22, 900–906. doi:10.1016/j.cub.2012.03.052
Luo, X., Tang, Z., Rizo, J., and Yu, H. (2002). The Mad2 Spindle Checkpoint Protein Undergoes Similar Major Conformational Changes upon Binding to Either Mad1 or Cdc20. Mol. Cel 9, 59–71. doi:10.1016/s1097-2765(01)00435-x
Luo, X., and Yu, H. (2008). Protein Metamorphosis: the Two-State Behavior of Mad2. Structure 16, 1616–1625. doi:10.1016/j.str.2008.10.002
Luo, Y., Ahmad, E., and Liu, S.-T. (2018). MAD1: Kinetochore Receptors and Catalytic Mechanisms. Front. Cel Dev. Biol. 6, 51. doi:10.3389/fcell.2018.00051
Maciejowski, J., Drechsler, H., Grundner-Culemann, K., Ballister, E. R., Rodriguez-Rodriguez, J.-A., Rodriguez-Bravo, V., et al. (2017). Mps1 Regulates Kinetochore-Microtubule Attachment Stability via the Ska Complex to Ensure Error-free Chromosome Segregation. Dev. Cel 41, 143–156. e6. doi:10.1016/j.devcel.2017.03.025
Maiato, H., Gomes, A., Sousa, F., and Barisic, M. (2017). Mechanisms of Chromosome Congression during Mitosis. Biology 6, 13. doi:10.3390/biology6010013
Mapelli, M., Filipp, F. V., Rancati, G., Massimiliano, L., Nezi, L., Stier, G., et al. (2006). Determinants of Conformational Dimerization of Mad2 and its Inhibition by P31comet. EMBO J. 25, 1273–1284. doi:10.1038/sj.emboj.7601033
Mapelli, M., Massimiliano, L., Santaguida, S., and Musacchio, A. (2007). The Mad2 Conformational Dimer: Structure and Implications for the Spindle Assembly Checkpoint. Cell 131, 730–743. doi:10.1016/j.cell.2007.08.049
McHugh, T., and Welburn, J. P. I. (2017). Dynein at Kinetochores: Making the Connection. J. Cel Biol. 216, 855–857. doi:10.1083/jcb.201703054
McKinley, K. L., and Cheeseman, I. M. (2017). Large-Scale Analysis of CRISPR/Cas9 Cell-Cycle Knockouts Reveals the Diversity of P53-dependent Responses to Cell-Cycle Defects. Dev. Cel 40, 405–420. doi:10.1016/j.devcel.2017.01.012
Meraldi, P., and Sorger, P. K. (2005). A Dual Role for Bub1 in the Spindle Checkpoint and Chromosome Congression. EMBO J. 24, 1621–1633. doi:10.1038/sj.emboj.7600641
Millband, D. N., and Hardwick, K. G. (2002). Fission Yeast Mad3p Is Required for Mad2p to Inhibit the Anaphase-Promoting Complex and Localizes to Kinetochores in a Bub1p-, Bub3p-, and Mph1p-dependent Manner. Mol. Cel. Biol. 22, 2728–2742. doi:10.1128/MCB.22.8.2728-2742.2002
Miyazaki, S., Kim, J., Yamagishi, Y., Ishiguro, T., Okada, Y., Tanno, Y., et al. (2017). Meikin-associated polo-like Kinase Specifies Bub1 Distribution in Meiosis I. Genes Cells 22, 552–567. doi:10.1111/gtc.12496
Mora-Santos, M. d. M., Hervas-Aguilar, A., Sewart, K., Lancaster, T. C., Meadows, J. C., and Millar, J. B. A. (2016). Bub3-Bub1 Binding to Spc7/KNL1 Toggles the Spindle Checkpoint Switch by Licensing the Interaction of Bub1 with Mad1-Mad2. Curr. Biol. 26, 2642–2650. doi:10.1016/j.cub.2016.07.040
Mosalaganti, S., Keller, J., Altenfeld, A., Winzker, M., Rombaut, P., Saur, M., et al. (2017). Structure of the RZZ Complex and Molecular Basis of its Interaction with Spindly. J. Cel Biol. 216, 961–981. doi:10.1083/jcb.201611060
Musacchio, A. (2015). The Molecular Biology of Spindle Assembly Checkpoint Signaling Dynamics. Curr. Biol. 25, R1002–R1018. doi:10.1016/j.cub.2015.08.051
Musinipally, V., HowesAlushin, S. G. M., Alushin, G. M., and Nogales, E. (2013). The Microtubule Binding Properties of CENP-E's C-Terminus and CENP-F. J. Mol. Biol. 425, 4427–4441. doi:10.1016/j.jmb.2013.07.027
Nguyen, A. L., Fadel, M. D., and Cheeseman, I. M. (2021). Differential Requirements for the CENP-O Complex Reveal Parallel PLK1 Kinetochore Recruitment Pathways. MBoC 32, 712–721. doi:10.1091/mbc.E20-11-0751
Nijenhuis, W., Vallardi, G., Teixeira, A., Kops, G. J. P. L., and Saurin, A. T. (2014). Negative Feedback at Kinetochores Underlies a Responsive Spindle Checkpoint Signal. Nat. Cel Biol. 16, 1257–1264. doi:10.1038/ncb3065
O'Connor, A., Maffini, S., Rainey, M. D., Kaczmarczyk, A., Gaboriau, D., Musacchio, A., et al. (2015). Requirement for PLK1 Kinase Activity in the Maintenance of a Robust Spindle Assembly Checkpoint. Biol. Open. 5, 11–19. doi:10.1242/bio.014969
Overlack, K., Primorac, I., Vleugel, M., Krenn, V., Maffini, S., Hoffmann, I., et al. (2015). A Molecular Basis for the Differential Roles of Bub1 and BubR1 in the Spindle Assembly Checkpoint. Elife 4, e05269. doi:10.7554/eLife.05269
Pereira, C., Reis, R. M., Gama, J. B., Celestino, R., Cheerambathur, D. K., Carvalho, A. X., et al. (2018). Self-assembly of the RZZ Complex into Filaments Drives Kinetochore Expansion in the Absence of Microtubule Attachment. Curr. Biol. 28, 3408–3421. e8. doi:10.1016/j.cub.2018.08.056
Perera, D., and Taylor, S. S. (2010). Sgo1 Establishes the Centromeric Cohesion protection Mechanism in G2 before Subsequent Bub1-dependent Recruitment in Mitosis. J. Cel Sci. 123, 653–659. doi:10.1242/jcs.059501
Perera, D., Tilston, V., Hopwood, J. A., Barchi, M., Boot-Handford, R. P., and Taylor, S. S. (2007). Bub1 Maintains Centromeric Cohesion by Activation of the Spindle Checkpoint. Dev. Cel 13, 566–579. doi:10.1016/j.devcel.2007.08.008
Piano, V., Alex, A., Stege, P., Maffini, S., Stoppiello, G. A., Huis in ’t Veld, P. J., et al. (2021). CDC20 Assists its Catalytic Incorporation in the Mitotic Checkpoint Complex. Science 371, 67–71. doi:10.1126/science.abc1152
Primorac, I., Weir, J. R., Chiroli, E., Gross, F., Hoffmann, I., van Gerwen, S., et al. (2013). Bub3 Reads Phosphorylated MELT Repeats to Promote Spindle Assembly Checkpoint Signaling. Elife 2, e01030. doi:10.7554/eLife.01030
Qi, W., Tang, Z., and Yu, H. (2006). Phosphorylation- and polo-box-dependent Binding of Plk1 to Bub1 Is Required for the Kinetochore Localization of Plk1. MBoC 17, 3705–3716. doi:10.1091/mbc.e06-03-0240
Qian, J., García-Gimeno, M. A., Beullens, M., Manzione, M. G., Van der Hoeven, G., Igual, J. C., et al. (2017). An Attachment-independent Biochemical Timer of the Spindle Assembly Checkpoint. Mol. Cel 68, 715–730. doi:10.1016/j.molcel.2017.10.011
Raaijmakers, J. A., and Medema, R. H. (2019). Killing a Zombie: a Full Deletion of the BUB 1 Gene in HAP 1 Cells. EMBO J. 38, e102423. doi:10.15252/embj.2019102423
Raaijmakers, J. A., van Heesbeen, R. G. H. P., Blomen, V. A., Janssen, L. M. E., van Diemen, F., Brummelkamp, T. R., et al. (2018). BUB1 Is Essential for the Viability of Human Cells in Which the Spindle Assembly Checkpoint Is Compromised. Cel Rep. 22, 1424–1438. doi:10.1016/j.celrep.2018.01.034
Raisch, T., Ciossani, G., d’Amico, E., Cmentowski, V., Carmignani, S., Maffini, S., et al. (2021). Structure of the RZZ Complex and Molecular Basis of Spindly-Driven corona Assembly at Human Kinetochores. bioRxiv. doi:10.1101/2021.12.03.471119
Rattner, J. B., Rao, A., Fritzler, M. J., Valencia, D. W., and Yen, T. J. (1993). CENP-F Is a Ca 400 kDa Kinetochore Protein that Exhibits a Cell-Cycle Dependent Localization. Cell Motil. Cytoskeleton 26, 214–226. doi:10.1002/cm.970260305
Ricke, R. M., Jeganathan, K. B., Malureanu, L., Harrison, A. M., and van Deursen, J. M. (2012). Bub1 Kinase Activity Drives Error Correction and Mitotic Checkpoint Control but Not Tumor Suppression. J. Cel Biol. 199, 931–949. doi:10.1083/jcb.201205115
Rischitor, P. E., May, K. M., and Hardwick, K. G. (2007). Bub1 Is a Fission Yeast Kinetochore Scaffold Protein, and Is Sufficient to Recruit Other Spindle Checkpoint Proteins to Ectopic Sites on Chromosomes. PLoS One 2, e1342. doi:10.1371/journal.pone.0001342
Roberts, B. T., Farr, K. A., and Hoyt, M. A. (1994). The Saccharomyces cerevisiae Checkpoint Gene BUB1 Encodes a Novel Protein Kinase. Mol. Cel. Biol. 14, 8282–8291. doi:10.1128/mcb.14.12.8282-8291.1994
Rodriguez-Rodriguez, J.-A., Lewis, C., McKinley, K. L., Sikirzhytski, V., Corona, J., Maciejowski, J., et al. (2018). Distinct Roles of RZZ and Bub1-KNL1 in Mitotic Checkpoint Signaling and Kinetochore Expansion. Curr. Biol. 28, 3422–3429. doi:10.1016/j.cub.2018.10.006
Roy, B., Han, S. J., Fontan, A. N., and Joglekar, A. P. (2020). The Copy-Number and Varied Strengths of MELT Motifs in Spc105 Balance the Strength and Responsiveness of the Spindle Assembly Checkpoint. Elife 9, e55096. doi:10.7554/eLife.55096
Roy, B., Han, S. J. Y., Fontan, A. N., Jema, S., and Joglekar, A. P. (2022). Aurora B Phosphorylates Bub1 to Promote Spindle Assembly Checkpoint Signaling. Curr. Biol. 32, 237–247. e6. doi:10.1016/j.cub.2021.10.049
Roy, B., Verma, V., Sim, J., Fontan, A., and Joglekar, A. P. (2019). Delineating the Contribution of Spc105-Bound PP1 to Spindle Checkpoint Silencing and Kinetochore Microtubule Attachment Regulation. J. Cel Biol. 218, 3926–3942. doi:10.1083/jcb.201810172
Sacristan, C., Ahmad, M. U. D., Keller, J., Fermie, J., Groenewold, V., Tromer, E., et al. (2018). Dynamic Kinetochore Size Regulation Promotes Microtubule Capture and Chromosome Biorientation in Mitosis. Nat. Cel Biol. 20, 800–810. doi:10.1038/s41556-018-0130-3
Saurin, A. T., van der Waal, M. S., Medema, R. H., Lens, S. M. A., and Kops, G. J. P. L. (2011). Aurora B Potentiates Mps1 Activation to Ensure Rapid Checkpoint Establishment at the Onset of Mitosis. Nat. Commun. 2, 316. doi:10.1038/ncomms1319
Schliekelman, M., Cowley, D. O., O'Quinn, R., Oliver, T. G., Lu, L., Salmon, E. D., et al. (2009). Impaired Bub1 FunctionIn vivoCompromises Tension-dependent Checkpoint Function Leading to Aneuploidy and Tumorigenesis. Cancer Res. 69, 45–54. doi:10.1158/0008-5472.CAN-07-6330
Shah, J., Botvinick, E., Bonday, Z., Furnari, F., Berns, M., and Cleveland, D. (2004). Dynamics of Centromere and Kinetochore ProteinsImplications for Checkpoint Signaling and Silencing. Curr. Biol. 14, 942–952. doi:10.1016/j.cub.2004.05.04610.1016/s0960-9822(04)00381-1
Shao, H., Huang, Y., Zhang, L., Yuan, K., Chu, Y., Dou, Z., et al. (2015). Spatiotemporal Dynamics of Aurora B-PLK1-MCAK Signaling axis Orchestrates Kinetochore Bi-orientation and Faithful Chromosome Segregation. Sci. Rep. 5, 12204. doi:10.1038/srep12204
Sharp-Baker, H., and Chen, R.-H. (2001). Spindle Checkpoint Protein Bub1 Is Required for Kinetochore Localization of Mad1, Mad2, Bub3, and CENP-E, Independently of its Kinase Activity. J. Cel Biol. 153, 1239–1250. doi:10.1083/jcb.153.6.1239
Shepperd, L. A., Meadows, J. C., Sochaj, A. M., Lancaster, T. C., Zou, J., Buttrick, G. J., et al. (2012). Phosphodependent Recruitment of Bub1 and Bub3 to Spc7/KNL1 by Mph1 Kinase Maintains the Spindle Checkpoint. Curr. Biol. 22, 891–899. doi:10.1016/j.cub.2012.03.051
Shi, Q., Hu, M., Luo, M., Liu, Q., Jiang, F., Zhang, Y., et al. (2011). Reduced Expression of Mad2 and Bub1 Proteins Is Associated with Spontaneous Miscarriages. Mol. Hum. Repod. 17, 14–21. doi:10.1093/molehr/gaq065
Siemeister, G., Mengel, A., Fernández-Montalván, A. E., Bone, W., Schröder, J., Zitzmann-Kolbe, S., et al. (2018). Inhibition of Bub1 Kinase by BAY 1816032 Sensitizes Tumor Cells toward Taxanes, ATR, and PARP Inhibitors In Vitro and In Vivo. Clin. Cancer Res. 25, 1404–1414. doi:10.1158/1078-0432.CCR-18-0628
Silió, V., McAinsh, A. D., and Millar, J. B. (2015). KNL1-Bubs and RZZ Provide Two Separable Pathways for Checkpoint Activation at Human Kinetochores. Dev. Cel 35, 600–613. doi:10.1016/j.devcel.2015.11.012
Singh, P., Pesenti, M. E., Maffini, S., Carmignani, S., Hedtfeld, M., Petrovic, A., et al. (2021). BUB1 and CENP-U, Primed by CDK1, Are the Main PLK1 Kinetochore Receptors in Mitosis. Mol. Cel 81, 67–87. doi:10.1016/j.molcel.2020.10.040
Sironi, L., Mapelli, M., Knapp, S., De Antoni, A., Jeang, K., and Musacchio, A. (2002). Crystal Structure of the Tetrameric Mad1-Mad2 Core Complex: Implications of a 'safety belt' Binding Mechanism for the Spindle Checkpoint. EMBO J. 21, 2496–2506. doi:10.1093/emboj/21.10.2496
Stehman, S. A., Chen, Y., McKenney, R. J., and Vallee, R. B. (2007). NudE and NudEL Are Required for Mitotic Progression and Are Involved in Dynein Recruitment to Kinetochores. J. Cel Biol. 178, 583–594. doi:10.1083/jcb.200610112
Suijkerbuijk, S. J. E., Vleugel, M., Teixeira, A., and Kops, G. J. P. L. (2012). Integration of Kinase and Phosphatase Activities by BubR1 Ensures Formation of Stable Kinetochore-Microtubule Attachments. Dev. Cel 23, 745–755. doi:10.1016/j.devcel.2012.09.005
Tang, Z., Shu, H., Oncel, D., Chen, S., and Yu, H. (2004). Phosphorylation of Cdc20 by Bub1 Provides a Catalytic Mechanism for APC/C Inhibition by the Spindle Checkpoint. Mol. Cel 16, 387–397. doi:10.1016/j.molcel.2004.09.031
Tang, Z., Shu, H., Qi, W., Mahmood, N. A., Mumby, M. C., and Yu, H. (2006). PP2A Is Required for Centromeric Localization of Sgo1 and Proper Chromosome Segregation. Dev. Cel 10, 575–585. doi:10.1016/j.devcel.2006.03.010
Tang, Z., Sun, Y., Harley, S. E., Zou, H., and Yu, H. (2004). Human Bub1 Protects Centromeric Sister-Chromatid Cohesion through Shugoshin during Mitosis. Proc. Natl. Acad. Sci. U S A. 101, 18012–18017. doi:10.1073/pnas.0408600102
Taylor, S. S., Ha, E., and McKeon, F. (1998). The Human Homologue of Bub3 Is Required for Kinetochore Localization of Bub1 and a Mad3/Bub1-Related Protein Kinase. J. Cel Biol. 142, 1–11. doi:10.1083/jcb.142.1.1
Tilston, V., Taylor, S. S., and Perera, D. (2009). Inactivating the Spindle Checkpoint Kinase Bub1 during Embryonic Development Results in a Global Shutdown of Proliferation. BMC Res. Notes 2, 190. doi:10.1186/1756-0500-2-190
Tsukahara, T., Tanno, Y., and Watanabe, Y. (2010). Phosphorylation of the CPC by Cdk1 Promotes Chromosome Bi-orientation. Nature 467, 719–723. doi:10.1038/nature09390
Vanoosthuyse, V., Valsdottir, R., Javerzat, J.-P., and Hardwick, K. G. (2004). Kinetochore Targeting of Fission Yeast Mad and Bub Proteins Is Essential for Spindle Checkpoint Function but Not for All Chromosome Segregation Roles of Bub1p. Mol. Cel Biol. 24, 9786–9801. doi:10.1128/MCB.24.22.9786-9801.2004
Vergnolle, M. A. S., and Taylor, S. S. (2007). Cenp-F Links Kinetochores to Ndel1/Nde1/Lis1/dynein Microtubule Motor Complexes. Curr. Biol. 17, 1173–1179. doi:10.1016/j.cub.2007.05.077
Vleugel, M., Hoek, T., Tromer, E., Sliedrecht, T., Groenewold, V., Omerzu, M., et al. (2015a). Dissecting the Roles of Human BUB1 in the Spindle Assembly Checkpoint. J. Cel Sci. 128, 2975–2982. doi:10.1242/jcs.169821
Vleugel, M., Omerzu, M., Groenewold, V., Hadders, M. A., Lens, S. M. A., and Kops, G. J. P. L. (2015b). Sequential Multisite Phospho-Regulation of KNL1-BUB3 Interfaces at Mitotic Kinetochores. Mol. Cel 57, 824–835. doi:10.1016/j.molcel.2014.12.036
Volkov, V. A., Grissom, P. M., Arzhanik, V. K., Zaytsev, A. V., Renganathan, K., McClure-Begley, T., et al. (2015). Centromere Protein F Includes Two Sites that Couple Efficiently to Depolymerizing Microtubules. J. Cel Biol. 209, 813–828. doi:10.1083/jcb.201408083
von Schubert, C., Cubizolles, F., Bracher, J. M., Sliedrecht, T., Kops, G. J. P. L., and Nigg, E. A. (2015). Plk1 and Mps1 Cooperatively Regulate the Spindle Assembly Checkpoint in Human Cells. Cel Rep. 12, 66–78. doi:10.1016/j.celrep.2015.06.007
Vorozhko, V. V., Emanuele, M. J., Kallio, M. J., Stukenberg, P. T., and Gorbsky, G. J. (2008). Multiple Mechanisms of Chromosome Movement in Vertebrate Cells Mediated through the Ndc80 Complex and Dynein/dynactin. Chromosoma 117, 169–179. doi:10.1007/s00412-007-0135-3
Vos, L. J., Famulski, J. K., and Chan, G. K. T. (2011). hZwint-1 Bridges the Inner and Outer Kinetochore: Identification of the Kinetochore Localization Domain and the hZw10-Interaction Domain. Biochem. J. 436, 157–168. doi:10.1042/BJ20110137
Wang, H., Hu, X., Ding, X., Dou, Z., Yang, Z., Shaw, A. W., et al. (2004). Human Zwint-1 Specifies Localization of Zeste White 10 to Kinetochores and Is Essential for Mitotic Checkpoint Signaling. J. Biol. Chem. 279, 54590–54598. doi:10.1074/jbc.M407588200
Wang, X., Bajaj, R., Bollen, M., Peti, W., and Page, R. (2016). Expanding the PP2A Interactome by Defining a B56-specific SLiM. Structure 24, 2174–2181. doi:10.1016/j.str.2016.09.010
Warren, C. D., Brady, D. M., Johnston, R. C., Hanna, J. S., Hardwick, K. G., and Spencer, F. A. (2002). Distinct Chromosome Segregation Roles for Spindle Checkpoint Proteins. MBoC 13, 3029–3041. doi:10.1091/mbc.e02-04-0203
Wynne, C. L., and Vallee, R. B. (2018). Cdk1 Phosphorylation of the Dynein Adapter Nde1 Controls Cargo Binding from G2 to Anaphase. J. Cel Biol. 217, 3019–3029. doi:10.1083/jcb.201707081
Xu, P., Raetz, E. A., Kitagawa, M., Virshup, D. M., and Lee, S. H. (2013). BubR1 Recruits PP2A via the B56 Family of Targeting Subunits to Promote Chromosome Congression. Biol. Open. 2, 479–486. doi:10.1242/bio.20134051
Yamagishi, Y., Yang, C.-H., Tanno, Y., and Watanabe, Y. (2012). MPS1/Mph1 Phosphorylates the Kinetochore Protein KNL1/Spc7 to Recruit SAC Components. Nat. Cel Biol. 14, 746–752. doi:10.1038/ncb2515
Yamaguchi, S., Decottignies, A., and Nurse, P. (2003). Function of Cdc2p-dependent Bub1p Phosphorylation and Bub1p Kinase Activity in the Mitotic and Meiotic Spindle Checkpoint. EMBO J. 22, 1075–1087. doi:10.1093/emboj/cdg100
Yang, M., Li, B., Liu, C.-J., Tomchick, D. R., Machius, M., Rizo, J., et al. (2008). Insights into Mad2 Regulation in the Spindle Checkpoint Revealed by the crystal Structure of the Symmetric Mad2 Dimer. Plos Biol. 6, e50. doi:10.1371/journal.pbio.0060050
Yang, Z., Guo, J., Chen, Q., Ding, C., Du, J., and Zhu, X. (2005). Silencing Mitosin Induces Misaligned Chromosomes, Premature Chromosome Decondensation before Anaphase Onset, and Mitotic Cell Death. Mol. Cel Biol. 25, 4062–4074. doi:10.1128/MCB.25.10.4062-4074.2005
Yang, Z., Tulu, U. S., Wadsworth, P., and Rieder, C. L. (2007). Kinetochore Dynein Is Required for Chromosome Motion and Congression Independent of the Spindle Checkpoint. Curr. Biol. 17, 973–980. doi:10.1016/j.cub.2007.04.056
Yi, Q., Chen, Q., Yan, H., Zhang, M., Liang, C., Xiang, X., et al. (2019). Aurora B Kinase Activity-dependent and -independent Functions of the Chromosomal Passenger Complex in Regulating Sister Chromatid Cohesion. J. Biol. Chem. 294, 2021–2035. doi:10.1074/jbc.RA118.005978
Zhang, G., Kruse, T., Guasch Boldú, C., Garvanska, D. H., Coscia, F., Mann, M., et al. (2019). Efficient Mitotic Checkpoint Signaling Depends on Integrated Activities of Bub1 and the RZZ Complex. EMBO J. 38, e100977. doi:10.15252/embj.2018100977
Zhang, G., Kruse, T., López-Méndez, B., Sylvestersen, K. B., Garvanska, D. H., Schopper, S., et al. (2017). Bub1 Positions Mad1 Close to KNL1 MELT Repeats to Promote Checkpoint Signalling. Nat. Commun. 8, 15822. doi:10.1038/ncomms15822
Zhang, G., Lischetti, T., Hayward, D. G., and Nilsson, J. (2015). Distinct Domains in Bub1 Localize RZZ and BubR1 to Kinetochores to Regulate the Checkpoint. Nat. Commun. 6, 7162. doi:10.1038/ncomms8162
Zhang, G., Lischetti, T., and Nilsson, J. (2014). A Minimal Number of MELT Repeats Supports All Functions of KNL1 in Chromosome Segregation. J. Cel Sci. 127, 871–884. doi:10.1242/jcs.139725
Zhang, G., Mendez, B. L., Sedgwick, G. G., and Nilsson, J. (2016). Two Functionally Distinct Kinetochore Pools of BubR1 Ensure Accurate Chromosome Segregation. Nat. Commun. 7, 12256. doi:10.1038/ncomms12256
Zhang, G., and Nilsson, J. (2018). The Closed Form of Mad2 Is Bound to Mad1 and Cdc20 at Unattached Kinetochores. Cell Cycle 17, 1087–1091. doi:10.1080/15384101.2018.1480209
Keywords: mitosis, spindle assembly checkpoint, kinetochore, Bub1, chromosome alignment
Citation: Zhang Y, Song C, Wang L, Jiang H, Zhai Y, Wang Y, Fang J and Zhang G (2022) Zombies Never Die: The Double Life Bub1 Lives in Mitosis. Front. Cell Dev. Biol. 10:870745. doi: 10.3389/fcell.2022.870745
Received: 07 February 2022; Accepted: 06 April 2022;
Published: 13 May 2022.
Edited by:
Patrick Meraldi, Université de Genève, SwitzerlandReviewed by:
Raquel Oliveira, Gulbenkian Institute of Science (IGC), PortugalBabhrubahan Roy, Michigan Medicine, University of Michigan, United States
Copyright © 2022 Zhang, Song, Wang, Jiang, Zhai, Wang, Fang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jing Fang, amZhbmdAcWR1LmVkdS5jbg==; Gang Zhang, emhhbmdnYW5nX3NtYUBxZHUuZWR1LmNu
†These authors have contributed equally to this work
 Yuqing Zhang
Yuqing Zhang Chunlin Song
Chunlin Song Lei Wang1†
Lei Wang1† Hongfei Jiang
Hongfei Jiang Gang Zhang
Gang Zhang