
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Cell Dev. Biol. , 03 March 2022
Sec. Cell Growth and Division
Volume 10 - 2022 | https://doi.org/10.3389/fcell.2022.867870
This article is part of the Research Topic Centrosomes and other Microtubule-Organizing Centers View all 7 articles
Microtubules are dynamic, filamentous polymers composed of α- and β-tubulin. Arrays of microtubules that have a specific polarity and distribution mediate essential processes such as intracellular transport and mitotic chromosome segregation. Microtubule arrays are generated with the help of microtubule organizing centers (MTOC). MTOCs typically combine two principal activities, the de novo formation of microtubules, termed nucleation, and the immobilization of one of the two ends of microtubules, termed anchoring. Nucleation is mediated by the γ-tubulin ring complex (γTuRC), which, in cooperation with its recruitment and activation factors, provides a template for α- and β-tubulin assembly, facilitating formation of microtubule polymer. In contrast, the molecules and mechanisms that anchor newly formed microtubules at MTOCs are less well characterized. Here we discuss the mechanistic challenges underlying microtubule anchoring, how this is linked with the molecular activities of known and proposed anchoring factors, and what consequences defective microtubule anchoring has at the cellular and organismal level.
Microtubules, elongated, cylindrical polymers assembled from heterodimers of α- and β-tubulin, are major elements of the cytoskeleton that mediate a wide range of functions in cycling as well as post-mitotic, differentiated cells. The orientation of tubulin dimers within the microtubule lattice provides microtubules with an intrinsic polarity, exposing β-tubulin at the “plus-end” and α-tubulin at the “minus-end” (Alushin et al., 2014). Microtubule polarity is recognized by motor proteins to allow directed transport. Whereas most kinesins are plus-end-directed, the dynein motor and a few kinesins move towards the minus-end. Other proteins interact specifically with either of the two ends to modulate its dynamic behaviour through stabilization or destabilization (Akhmanova and Steinmetz, 2015). To function efficiently and to fulfil the specific needs of different cell types and cell cycle stages, microtubules are arranged into various types of arrays. These arrays differ in shape and distribution and may contain microtubules of uniform or mixed polarity and of variable dynamicity (Sallee and Feldman, 2021). To generate different types of microtubule arrays, cells employ microtubule organizing centers (MTOCs) (Sanchez and Feldman, 2017; Wu and Akhmanova, 2017; Paz and Lüders, 2018). MTOCs can be assembled at the cytoplasmic surfaces of various organelles. The best-known example is the centrosomal MTOC, which is assembled around centrioles, but other, typically membrane-bound organelles such as the Golgi or the nuclear envelope, can also acquire MTOC activity.
A key component of most MTOCs is the microtubule nucleator γTuRC, which allows generation of new microtubules. Mimicking the structure of a microtubule in cross-section, the circular, helical arrangement of γ-tubulins in the γTuRC provides docking sites for tubulin heterodimers, promoting their lateral interactions and facilitating polymer formation (Moritz et al., 2000; Consolati et al., 2020; Liu et al., 2020; Thawani et al., 2020; Wieczorek et al., 2020; Zimmermann et al., 2020). Since soluble native and recombinant γTuRCs are intrinsically asymmetric and do not perfectly match the microtubule symmetry, they likely require activation, for example through a conformational change (Kollman et al., 2015). Alternatively, nucleation may be stimulated by cofactors that interact with and stabilize tubulin assembly intermediates on γTuRC such as members of the XMAP215 family of tubulin polymerases (Wieczorek et al., 2015; Flor-Parra et al., 2018; Gunzelmann et al., 2018; Thawani et al., 2018).
Apart from nucleation, the second, possibly most important activity of MTOCs is their ability to anchor microtubules. While most MTOCs also nucleate microtubules, MTOC activity could, in principle, also be carried out without nucleation, by capturing and anchoring of microtubules that were nucleated elsewhere. Indeed, it is the anchoring of microtubules at MTOCs that ultimately confers specific polarity and shape to microtubule arrays (Sanchez and Feldman, 2017; Wu and Akhmanova, 2017; Paz and Lüders, 2018). Several anchoring factors have been identified (Table 1), but in most cases their molecular functions are unclear and a mechanistic picture is still missing.
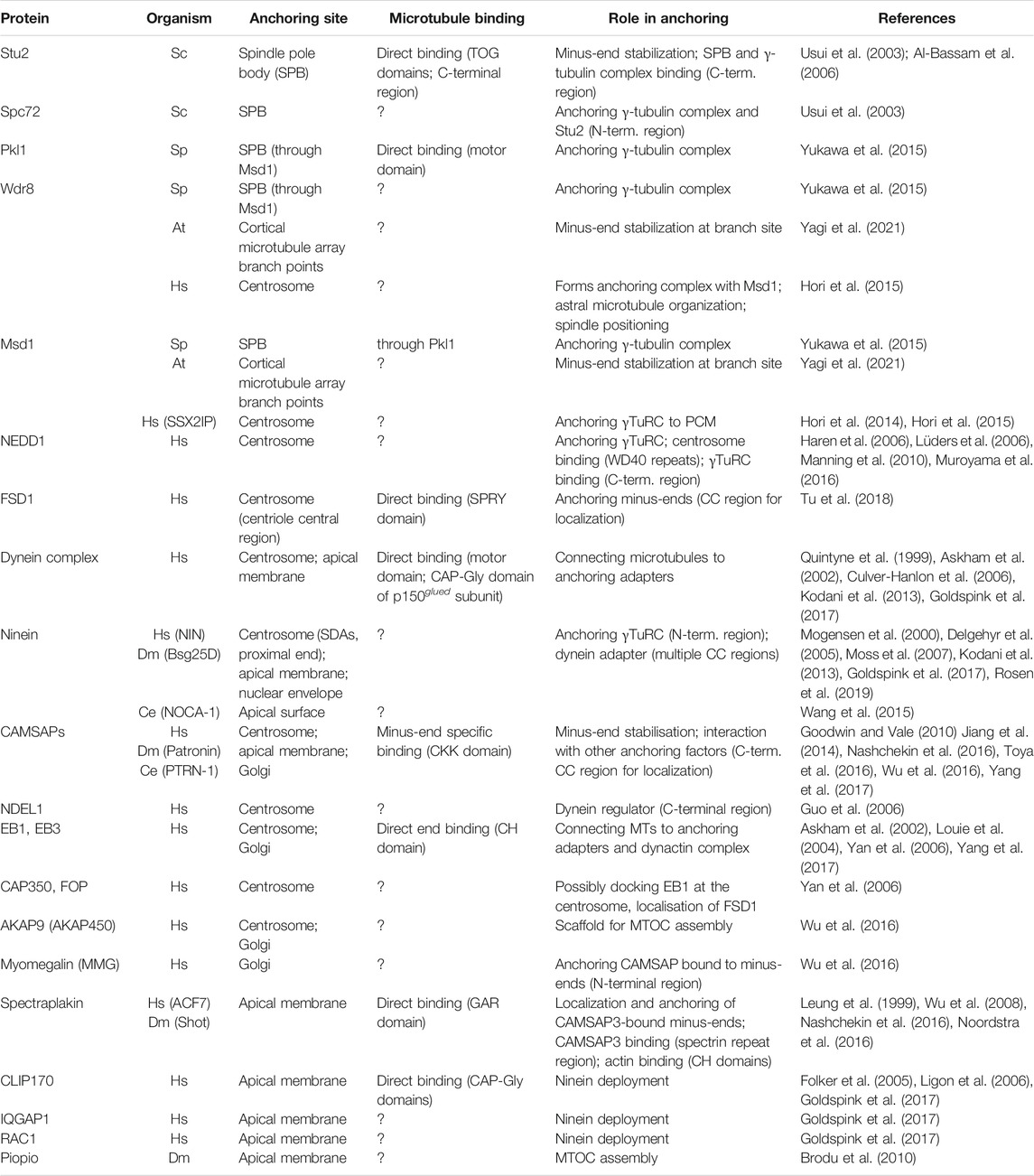
TABLE 1. Major anchoring factors. Factors reported to be involved in anchoring microtubules at different MTCOs. Factors that likely affect anchoring more indirectly were not included. For each anchoring factor, identified by both the common name and organism-specific name, we indicate the ability to directly bind microtubules, the proposed role in anchoring, and the domains/regions involved in these functions. Question marks indicate cases where experimental data is not available. Abbreviations used: Sc–S. cerevisiae, Sp–S. pompe, At–A. thaliana, Hs–H. sapiens, Dm–D. melanogaster, Ce–C. elegans, SDA–Subdistal appendages, CC–Coiled coil.
Here, we focus on the anchoring of microtubules at MTOCs and discuss our current knowledge regarding the molecules and mechanisms involved in this process. For an in-depth discussion of MTOCs and associated microtubule nucleation, we refer the reader to several recent reviews (Lee and Liu, 2019; Valenzuela et al., 2020; Wilkes and Moore, 2020; Lüders, 2021; Sallee and Feldman, 2021). We begin, by outlining conceptually how anchoring of microtubules may be achieved, and then, using various types of MTOCs and anchoring factors in different organisms as examples, discuss how available evidence supports these concepts. Finally, we highlight how defective microtubule anchoring impairs the microtubule cytoskeleton and may impair organismal development.
There are two basic requirements to be fulfilled by microtubule anchoring factors (Figure 1A). The anchoring proteins or protein complexes have to bind microtubules and, at the same time, interact with an MTOC. This linkage needs to be not only robust but also flexible, to resist the variable mechanical forces that act on microtubules as they extend away from the MTOC and serve as tracks for motor proteins. Importantly, to allow arrangement of microtubules with a specific orientation, binding of the anchoring factor to the microtubule needs to occur specifically at only one of the two ends. An additional challenge is the dynamic nature of microtubules. In microtubules assembled from pure tubulin in vitro, both minus- and plus-ends are dynamic and can undergo phases of growth and shrinkage. Thus, anchoring of microtubules likely involves stabilization and inhibition of microtubule end dynamics (Hendershott and Vale, 2014; Jiang et al., 2014; Akhmanova and Steinmetz, 2015). In the case of microtubules that are nucleated by γTuRC, the nucleator may form a stabilizing cap at their minus-end (Wiese and Zheng, 2000), leaving only the plus-end free to grow or shrink. This is consistent with the observation that anchoring of microtubules to MTOCs typically occurs via their minus-ends, whereas the plus-ends extend away from it and are more dynamic. Assuming that γTuRC remains bound to the minus-ends of newly nucleated microtubules, the nucleator itself could also provide anchoring function, as observed in reconstitution assays in vitro (Consolati et al., 2020). However, it seems that in cells anchoring usually involves additional factors (Figures 1B–E).
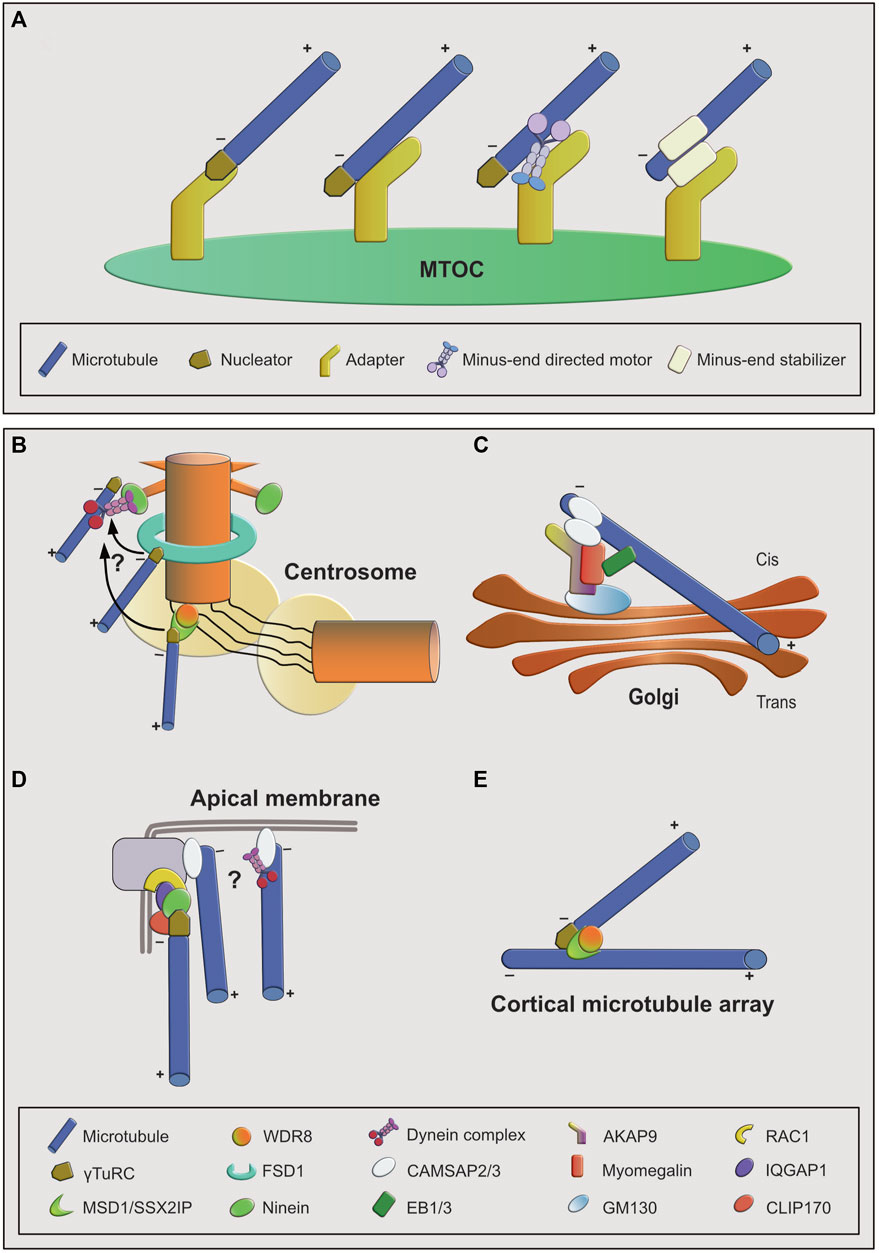
FIGURE 1. Overview of microtubule anchoring sites and mechanisms. (A): Conceptual overview of mechanisms by which microtubules can be anchored at MTOCs. From left to right: first, the nucleator may be part of the anchoring complex as a stabilizing minus-end cap. Anchoring to the MTOC may be achieved through an MTOC-bound adapter that interacts with the minus-end cap or with the microtubule lattice. Lattice interaction could be direct or indirect via a minus-end directed motor. Second, the nucleator may not be part of the anchoring complex. In this case anchoring is facilitated by an adapter protein interacting with a minus-end-bound, stabilizing protein. (B–E): Examples of MTOCs and associated anchoring factors. (B): At the interphase centrosome anchoring to the mother centriole is achieved through multiple mechanisms, involving ninein-dynein at the subdistal appendages, FSD1 in the central region, and MSD1-WDR8 in the proximal/PCM region. FSD1-and MSD1-mediated anchoring may be transient and minus-ends may be transferred to subdistal appendages. (C): Anchoring at cis-Golgi membranes involves the AKAP9-myomegalin complex as adapter and CAMSAP2 as stabilizer at the microtubule minus-end that connects it to the adapter complex. EB proteins provide an additional way of connecting microtubules to the adapter complex through myomegalin. (D): At apical junction complexes and membranes in epithelial cells both ninein- and CAMSAP-mediated anchoring mechanisms may act in parallel. (E): At branch points on plant cortical microtubules, MSD1-WDR8 complexes stabilize and anchor the minus-ends of newly nucleated microtubule branches to the lattice of the pre-existing microtubules.
A relatively simple microtubule network is found in budding yeast, where microtubules are organized by the spindle pole body (SPB), a single MTOC that is equivalent to the centrosome. Here anchoring of cytoplasmic microtubules was shown to involve ternary complexes composed of the SPB-bound adapter Spc72, the γ-tubulin-containing nucleation complex, and Stu2, a member of the XMAP215 family that also functions as nucleation stimulator (Usui et al., 2003; Gunzelmann et al., 2018). Following nucleation at the SPB, minus-ends are capped by γ-tubulin complexes and Stu2 may simultaneously interact with the nucleation complex and with the proximal wall of the newly nucleated microtubule (Usui et al., 2003). Similarly, in fission yeast γ-tubulin complexes cooperate with the XMAP215 homolog Alp14 to nucleate microtubules from the SPB and from the nuclear envelope, a second interphase MTOC, but a role of Alp14 in anchoring has not been described (Flor-Parra et al., 2018; Liu et al., 2019). Specifically during mitosis minus-ends of mitotic spindle microtubules are anchored through the coiled-coil protein Msd1 (Toya et al., 2007). Msd1 functions as part of a ternary complex with two other proteins, Wdr8 and the minus end-directed kinesin-14 motor Pkl1 (Yukawa et al., 2015). Minus-end-directed motor activity of Pkl1 is used to transport Msd1 and Wdr8 towards the SPB, where the ternary complex interacts with γ-tubulin complexes and promotes minus-end anchoring. Interestingly, Pkl1, artificially tethered to the SPB, provided partial anchoring function, even in the absence of Msd1-Wdr8 and using a motor-defective rigor mutant of Pkl1 (Yukawa et al., 2015). One could speculate that in this scenario mutant Pkl1 may still be able to provide the two basic functions of anchoring factors outline above: localization to the SPB and interaction with microtubules. The role of Msd1 in anchoring is conserved. Human MSD1, also known as SSX2IP, was shown to interact with the nucleator γTuRC and promote microtubule anchoring at the centrosome, both in interphase and mitosis (Bärenz et al., 2013; Hori et al., 2014, 2015). Interestingly, at the centrosome MSD1 partially colocalizes with γTuRC, but does not colocalize with ninein/NIN, an established anchoring factor and component of subdistal appendages, structures specific to the mother centriole that have been implicated in microtubule anchoring. Moreover, a C-terminal MSD1 fragment that was sufficient to interact with γTuRC provides anchoring activity when artificially tethered to the pericentriolar material (PCM) in the proximal part of centrioles (Hori et al., 2014). This suggests that microtubules may be anchored not only at subdistal appendages and that anchoring could be coupled with nucleation. However, while human MSD1 also interacts with WDR8 (Hori et al., 2015), the molecular basis of its anchoring activity has not been revealed.
Anchoring of microtubules at the vertebrate centrosome involves subdistal appendages and the activity of the subdistal appendage protein ninein (Bouckson-Castaing et al., 1996; Mogensen et al., 2000; Dammermann and Merdes, 2002; Ou et al., 2002; Delgehyr et al., 2005; Lin et al., 2006). Several proteins described to affect anchoring may do so through altering subdistal appendage structure and/or ninein recruitment (Ibi et al., 2011; Kodani et al., 2013; Huang et al., 2017). Anchoring at subdistal appendages would imply that microtubules nucleated by γTuRC in the more proximally located PCM, which is considered to be the main nucleation site, would be transferred to the subdistal appendages for stable anchoring, possibly with γTuRC as a stabilizing minus-end cap (Delgehyr et al., 2005; Hori et al., 2014). However, γTuRC localizes not only to the PCM and there is some evidence that nucleation may also occur directly at subdistal appendages (Schweizer and Lüders, 2021). The finding that γTuRC interacts with ninein would be consistent with both models (Delgehyr et al., 2005). It should also be noted that centrosomal ninein is not restricted to subdistal appendages, but is also present at the proximal ends of both mother and daughter centrioles. The significance of this localization is not entirely clear but it may be related to ninein’s role in centrosome cohesion (Mazo et al., 2016). How does ninein mediate microtubule anchoring? Ninein’s N- and C-terminal regions were shown to mediate γTuRC-binding and centrosome targeting, respectively, but whether ninein can bind microtubules was not investigated (Delgehyr et al., 2005; Lin et al., 2006). However, ninein was shown to bind dynein (Casenghi et al., 2005) and, more recently, to function as dynein activator (Redwine et al., 2017). Dynein has been implicated in the centrosome targeting of several proteins, in some cases in the form of particles known as centriolar satellites (Kubo et al., 1999; Dammermann and Merdes, 2002; Prosser and Pelletier, 2020). In this case, however, dynein’s ability to bind microtubules and move towards their minus-ends may be invoked by centrosome-bound ninein to anchor microtubules. Consistent with this possibility, several studies have linked dynein complexes with centrosomal microtubule anchoring (Quintyne et al., 1999; Guo et al., 2006; Kodani et al., 2013). Such a mechanism would have to ensure that dynein does not run off the microtubule once it has reached its minus-end. Indeed, at least in vitro, certain dynein complexes were observed to remain bound and accumulate at minus-ends (McKenney et al., 2014; Soundararajan and Bullock, 2014). Clearly, further work is needed to elucidate the potential cooperation between ninein and dynein in centrosomal minus-end anchoring.
Apart from MSD1 and ninein discussed above, a recent study has revealed another centrosomal protein, FSD1 (also known as MIR1 and GLFND), as microtubule anchoring factor (Tu et al., 2018). A comprehensive analysis showed that a coiled-coil domain at its N-terminus is sufficient for centrosome localization and that the B30.2/SPRY domain in the C-terminal part directly binds to and is required for anchoring of microtubules at the centrosome. Interestingly, FSD1 localizes in a circular fashion around centrioles, similar to subdistal appendage proteins, but positioned more proximally (Tu et al., 2018). Even though FSD1 localizes also around the daughter centriole, it promotes microtubule anchoring only at the mother centriole, pointing at the involvement of additional factors specific to the mother centriole. Notably, FSD1 and ninein are not dependent on each other for their specific localisations (Tu et al., 2018). The data suggest that FSD1, similar to MSD1, either extends the mother centriole-specific microtubule anchoring activity to the central portion of the cylinder or that it may be involved in the transfer of minus ends from proximally located nucleation sites to the subdistal appendage region for stable anchoring. Additional work is needed to clarify this issue.
Some anchoring factors share the ability to interact with γ-tubulin-containing nucleation complexes. This observation may indicate a mechanistic link between nucleation and anchoring and/or that γTuRC has two separate functions. Apart from providing a nucleation template, it may form a cap structure at minus-ends (Wiese and Zheng, 2000) that is used for microtubule anchoring. If so, distinct subpopulations of γTuRC may exist at centrosomes to mediate nucleation and anchoring, respectively. This was recently suggested to be the case in keratinocytes. In contrast to other cell types, where NEDD1 depletion robustly impairs centrosomal nucleation (Haren et al., 2006; Lüders et al., 2006), in keratinocytes nucleation activity is largely dependent on γTuRC in complex with the PCM protein CDK5RAP2, whereas γTuRC associated with NEDD1 is mainly used for anchoring (Muroyama et al., 2016).
Even in the presence of an active centrosome, MTOC activity associated with the Golgi may significantly contribute to microtubule network organization. This activity may be further enhanced when centrosome activity is compromised (Efimov et al., 2007; Miller et al., 2009; Rivero et al., 2009; Wu et al., 2016; Gavilan et al., 2018). CLASPs were initially proposed to provide anchoring function to microtubules associated with the trans-Golgi network (Efimov et al., 2007), but more recent work suggested that they merely function in stabilization (Wu et al., 2016). AKAP9/AKAP450 is a central organizer of the MTOC at the cis-Golgi that recruits both nucleation and anchoring factors. γTuRC is used to nucleate Golgi-associated microtubules but does not seem to remain bound to their minus-ends. Instead, these are bound by the minus-end-stabilizing protein CAMSAP2, and tethered to Golgi membranes via myomegalin (Wu et al., 2016). Curiously, end-binding proteins EB1 and EB3, known as plus-end regulators, were shown to participate in tethering microtubules to Golgi membranes (Yang et al., 2017). Importantly, apart from Golgi-nucleated microtubules, CAMSAP2-decorated microtubules from other sites (e.g., nucleated and released from the centrosome) (Keating et al., 1997; Mogensen, 1999; Dong et al., 2017) can be captured and attached to the Golgi MTOC (Jiang et al., 2014; Wu et al., 2016). While the CAMSAP2-mediated minus-end binding mechanism is quite well understood (Atherton et al., 2017), the interplay with myomegalin and EBs for anchoring at the Golgi much less so.
In metazoans, during cell differentiation the centrosome frequently loses its role as central microtubule organizer. As a result, in many specialized cell types microtubules are nucleated and anchored at more broadly distributed, non-centrosomal MTOCs (Paz and Lüders, 2018; Sallee and Feldman, 2021). An extreme case are plants, which lack centrioles altogether. In the plant interphase cortical microtubule array, for example, new microtubules are nucleated as branches from the lattice of pre-existing microtubules. Here the conserved Msd1-Wdr8 module was recently shown to anchor and stabilize microtubule minus-ends at the branch sites (Yagi et al., 2021). In addition, the Msd1-Wdr8 complex recruits katanin to the branch site, to allow severing and release of the newly nucleated microtubule branch. These activities are important for proper cortical microtubule array organization (Yagi et al., 2021).
Early work showed that during the differentiation of vertebrate polarized epithelia ninein expression is essential for cell polarization and formation of the apicobasal array of microtubules in these cells. As cells convert their centrosomal microtubule array to an apico-basal array, ninein is released from the centrosome to relocate anchoring function to an apical, non-centrosomal MTOC (Lechler and Fuchs, 2007; Moss et al., 2007; Bellett et al., 2009; Goldspink et al., 2017). In the epidermis this MTOC is formed in association with desmosomes at cell-cell junctions and is mediated by desmoplakin (Lechler and Fuchs, 2007). In columnar epithelial cells, ninein colocalizes with the adherens junction protein β-catenin. During differentiation, ninein associates with microtubules to be deployed at the apical MTOC in a process that depends on the plus-end interactor CLIP170 and cortical IQGAP1 and active Rac1 (Goldspink et al., 2017). Interestingly, once established, maintenance of the apico-basal microtubule array no longer required ninein. Experimental loss of ninein may be compensated for by apically localized CAMSAP2, and the dynactin subunit p150Glued (Goldspink et al., 2017), which has been implicated previously in anchoring at the centrosome (Quintyne et al., 1999; Kodani et al., 2013). Thus, different anchoring factors and mechanisms contribute and provide redundancy to apical anchoring of microtubules.
Apart from the apical membrane in polarized epithelial cells, ninein has also been identified at other non-centrosomal MTOCs, suggesting a broader role in microtubule anchoring. In mammalian multi-nucleated myotubes and in cardiomyocytes, ninein was identified as part of a non-centrosomal MTOC that forms during differentation at the nuclear envelope (Tassin et al., 1985; Bugnard et al., 2005; Srsen et al., 2009; Vergarajauregui et al., 2020; Becker et al., 2021). In muscle cells from Drosophila larvae ninein was also found in association with the perinuclear MTOC (Zheng et al., 2016). Later it was shown that in fly embryonic myotubes ninein cooperates with ensconsin/MAP7 in positioning nuclei along the myotube, which is important for muscle function (Rosen et al., 2019). More recently, a nuclear envelope-associated MTOC containing ninein was also described in Drosophila fat body cells, a cell type equivalent to liver adipocytes (Zheng et al., 2020). However, a formal demonstration that ninein mediates anchoring of microtubule minus-ends at these sites, is still lacking.
During neuronal differentiation ninein was observed to relocate from centrosomes to the cytoplasm in different neuronal compartments in the form of small granules, but no specific MTOC was identified (Baird et al., 2004; Ohama and Hayashi, 2009). Subsequently, ninein was revealed as a major transcriptional target of Sip1, a regulator of nervous system development. Loss of ninein phenocopied Sip1 deletion, and exogenous ninein expression was shown to rescue Sip1 deletion phenotypes, promoting axonal growth and branching by enhancing microtubule growth and stability (Srivatsa et al., 2015). It remains unclear though, whether these effects are related to a function of ninein in minus-end anchoring.
CAMSAP family members, which are not present in yeast and plants, can specifically recognize and stabilize minus-ends of non-centrosomal microtubules (Meng et al., 2008; Baines et al., 2009; Goodwin and Vale, 2010; Jiang et al., 2014; Atherton et al., 2017). Consistently, CAMSAPs are also associated with non-centrosomal MTOCs. In polarized epithelial cells in flies, worms and mammals, CAMSAP homologs were shown to contribute to the organization of apico-basal microtubule arrays that have their minus-ends anchored at non-centrosomal, apical MTOCs (Meng et al., 2008; Tanaka et al., 2012; Wang et al., 2015; Nashchekin et al., 2016; Ning et al., 2016; Noordstra et al., 2016; Toya et al., 2016). The contribution of CAMSAPs to apical minus-end anchoring may involve their ability to decorate microtubule minus-ends and to interact with spectraplakins that tether microtubules to the cortical actin network (Khanal et al., 2016; Nashchekin et al., 2016; Sanchez et al., 2021). In the larval epidermis in C. elegans, the CAMSAP homolog PTRN-1 functions redundantly with NOCA-1, a worm ninein homolog. Whereas NOCA-1 seems to work together with γ-tubulin, PTRN-1 likely stabilizes minus-ends in the absence of γ-tubulin (Wang et al., 2015). Similarly, in Drosophila fat body cells, ninein and patronin, the fly CAMSAP, function in parallel in organizing microtubule minus-ends at the nuclear envelope-associated MTOC. This function did not require γ-tubulin, even though it was also present at the nuclear envelope (Zheng et al., 2020). Recent testing by induced degradation of a panel of candidate factors in C. elegans embryonic intestinal epithelial cells has confirmed significant redundancy in apical MTOC assembly and anchoring mechanisms (Sallee et al., 2018). A novel type of MTOC that lacked detectable γ-tubulin was recently described within varicosities of the basal process of highly polarized neural progenitors/radial glial cells in the brain (Coquand et al., 2021). CAMSAPs accumulated in the varicosities and knockdown of CAMSAP1/2 reduced microtubule growth from these sites and destabilized the entire basal process. Since the varicosities were positive for trans-Golgi and trans-Golgi-network markers, the microtubule-anchoring structures may be similar to those of the Golgi-associated MTOC (Wu et al., 2016; Coquand et al., 2021).
In cycling cells, centrosomal anchoring defects are expected to reduce the fidelity of mitotic spindle assembly, and impair the positioning of spindles, which relies on astral microtubule anchoring around centrosomes at the spindle poles. Anchoring defects at non-centrosomal MTOCs during differentiation, will likely interfere with proper microtubule network remodelling, which is required for the morphological and functional adaptations that cells undergo to carry out specific functions. Indeed, ninein depletion in cultured human cells prevents the organization of a radial, centrosome-centered interphase microtubule array, and causes multipolar spindles in mitosis (Dammermann and Merdes, 2002; Logarinho et al., 2012). In the early fly embryo, maternally provided ninein is required for proper mitotic spindle assembly, but it is not essential at later developmental stages (Kowanda et al., 2016; Zheng et al., 2016). Ninein in neural progenitors of the developing mammalian brain has a role in progenitor interkinetic nuclear migration, asymmetric centrosome inheritance, and progenitor maintenance (Wang et al., 2009; Shinohara et al., 2013). Depletion of the neural progenitor pool by mitotic defects has been shown to cause microcephaly in mouse models of Seckel syndrome, a developmental disorder that is caused by mutations in genes encoding centrosome proteins including ninein (Dauber et al., 2012; Marjanović et al., 2015). Additional work in ninein KO mice has revealed defects in the skin. Ninein loss was found to disrupt correctly oriented progenitor cell divisions and, during epidermal cell differentiation, the formation of non-centrosomal cortical microtubule arrays, impeding desmosome assembly and skin barrier formation. These defects are reminiscent of epidermis defects observed in C. elegans NOCA-1 (ninein) and PTRN-1 (CAMSAP) double loss-of-function mutants (Wang et al., 2015).
Several of the factors that contribute to anchoring microtubules at the interphase centrosome have also been implicated in the assembly of primary cilia, surface-exposed signalling organelles that form as an extension of the distal end of the mother centriole. Ciliary defects cause a group of developmental disorders known as ciliopathies. FSD1, ninein and KIF3A promote assembly of the ciliary transition zone, a critical step in ciliogenesis. At least in part this involves the formation and trafficking of centriolar satellites along mother centriole-anchored microtubules (Kubo et al., 1999; Kodani et al., 2013; Tu et al., 2018; Odabasi et al., 2019). Similar observations were made for MSD1, which is required for ciliogenesis in cultured cells and in zebrafish embryos (Hori et al., 2014). Subdistal appendage anchoring of microtubules is also important for proper positioning of cilia, which allows surface exposure of primary cilia (Mazo et al., 2016) and, in the case of motile cilia, coordination of ciliary beating (Kunimoto et al., 2012).
Loss of CAMSAP family members does not seem to affect centrosomes but rather non-centrosomal MTOCs. In flies, cortical patronin helps to define the anterior-posterior axis in the oocyte and, during abdominal epidermis formation, it is required for epithelial remodelling and proper abdomen development (Nashchekin et al., 2016; Panzade and Matis, 2021). Homozygous deletion of CAMSAP3’s microtubule-binding domain in mice resulted in growth defects and, at the cellular level, in mispositioning of organelles. The architecture of polarized intestinal epithelial cells was only mildly affected, consistent with redundancy in apico-basal polarity organization (Toya et al., 2016). Analysis of CAMSAP loss-of-function in invertebrate and vertebrate models has revealed a wide range of phenotypes such as axon and dendrite growth and branching defects, reduced cell survival and organ size, or loss of ciliary motility (Chuang et al., 2014; Marcette et al., 2014; Richardson et al., 2014; Robinson et al., 2020; Liu et al., 2021; Yang and Choi, 2021). However, since CAMSAPs are likely general minus-end stabilizers rather than dedicated anchoring factors, some of these phenotypes may not necessarily result from anchoring defects, but, for example, from an overall reduction in microtubule density in CAMSAP-deficient cells (Jiang et al., 2014).
The increasing interest in microtubule anchoring mechanisms has led to several important discoveries during recent years. This has also been facilitated by the use of invertebrate models such as Drosophila melanogaster and C. elegans, which are particularly useful for studying non-centrosomal MTOCs in the context of differentiated cells and tissues. The emerging picture is that MTOCs use multiple anchoring factors and mechanisms, often resulting in redundancy. While some mechanisms depend on the nucleator γTuRC, presumably as a stabilizing minus-end cap, others rely on γTuRC-independent anchoring, employing alternative minus-end stabilizers such as CAMSAP family members.
Important open questions are how microtubule minus-end binding is achieved, in particular for anchoring factors that do not directly bind to microtubules, and whether the presence of multiple anchoring mechanisms at a single MTOC simply provides redundancy or, alternatively, may indicate the presence of distinct anchoring sites that are specific for subsets of microtubules (Sallee et al., 2018). For example, dynamic microtubules may be anchored differently than more stable microtubules. This distinction may depend on the nucleation mechanism and site used to generate these microtubules, and may also involve specific post-translation modifications on their lattice (Janke and Magiera, 2020).
One major obstacle in studying minus-end organization at MTOCs is the crowded nature of these areas. Thus, when addressing the above questions, the consequent use of super resolution techniques including expansion microscopy should enable researchers to probe anchoring sites with improved spatial resolution, to dissect single microtubule minus-ends, their post-translational modifications, and their associated molecules.
CV drafted the manuscript and prepared figures. JL conceived the review and prepared the final manuscript text.
JL acknowledges support by grants BFU2015-69275-P (MINECO/FEDER), PGC2018-099562-B-I00 (MICINN), network grants 2017 SGR 1089 (AGAUR) and RED2018-102723-T (MICINN), and by intramural funds of IRB Barcelona, recipient of a Severo Ochoa Centre of Excellence Award from the Spanish Ministry of Science and Innovation and supported by CERCA (Generalitat de Catalunya). CV was supported by a doctoral fellowship (PRE2018-083390; MICINN).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Akhmanova, A., and Steinmetz, M. O. (2015). Control of Microtubule Organization and Dynamics: Two Ends in the Limelight. Nat. Rev. Mol. Cell Biol 16, 711–726. doi:10.1038/nrm4084
Al-Bassam, J., van Breugel, M., Harrison, S. C., and Hyman, A. (2006). Stu2p Binds Tubulin and Undergoes an Open-To-Closed Conformational Change. J. Cell Biol. 172, 1009–1022. doi:10.1083/jcb.200511010
Alushin, G. M., Lander, G. C., Kellogg, E. H., Zhang, R., Baker, D., and Nogales, E. (2014). High-Resolution Microtubule Structures Reveal the Structural Transitions in αβ-Tubulin upon GTP Hydrolysis. Cell 157, 1117–1129. doi:10.1016/j.cell.2014.03.053
Askham, J. M., Vaughan, K. T., Goodson, H. V., and Morrison, E. E. (2002). Evidence that an Interaction between EB1 and p150GluedIs Required for the Formation and Maintenance of a Radial Microtubule Array Anchored at the Centrosome. MBoC 13, 3627–3645. doi:10.1091/mbc.e02-01-0061
Atherton, J., Jiang, K., Stangier, M. M., Luo, Y., Hua, S., Houben, K., et al. (2017). A Structural Model for Microtubule Minus-End Recognition and protection by CAMSAP Proteins. Nat. Struct. Mol. Biol. 24, 931–943. doi:10.1038/nsmb.3483
Baines, A. J., Bignone, P. A., King, M. D. A., Maggs, A. M., Bennett, P. M., Pinder, J. C., et al. (2009). The CKK Domain (DUF1781) Binds Microtubules and Defines the CAMSAP/ssp4 Family of Animal Proteins. Mol. Biol. Evol. 26, 2005–2014. doi:10.1093/molbev/msp115
Baird, D. H., Myers, K. A., Mogensen, M., Moss, D., and Baas, P. W. (2004). Distribution of the Microtubule-Related Protein Ninein in Developing Neurons. Neuropharmacology 47, 677–683. doi:10.1016/j.neuropharm.2004.07.016
Bärenz, F., Inoue, D., Yokoyama, H., Tegha-Dunghu, J., Freiss, S., Draeger, S., et al. (2013). The Centriolar Satellite Protein SSX2IP Promotes Centrosome Maturation. J. Cell Biol. 202, 81–95. doi:10.1083/jcb.201302122
Becker, R., Vergarajauregui, S., Billing, F., Sharkova, M., Lippolis, E., Mamchaoui, K., et al. (2021). Myogenin Controls via AKAP6 Non-centrosomal Microtubule-Organizing center Formation at the Nuclear Envelope. Elife 10, e65672. doi:10.7554/eLife.65672
Bellett, G., Carter, J. M., Keynton, J., Goldspink, D., James, C., Moss, D. K., et al. (2009). Microtubule Plus-End and Minus-End Capture at Adherens Junctions Is Involved in the Assembly of Apico-Basal Arrays in Polarised Epithelial Cells. Cell Motil. Cytoskeleton 66, 893–908. doi:10.1002/cm.20393
Bouckson-Castaing, V., Moudjou, M., Ferguson, D. J., Mucklow, S., Belkaid, Y., Milon, G., et al. (1996). Molecular Characterisation of Ninein, a New Coiled-Coil Protein of the Centrosome. J. Cell Sci. 109 (Pt 1), 179–190. doi:10.1242/jcs.109.1.179
Brodu, V., Baffet, A. D., Le Droguen, P.-M., Casanova, J., and Guichet, A. (2010). A Developmentally Regulated Two-step Process Generates a Noncentrosomal Microtubule Network in Drosophila Tracheal Cells. Developmental Cell 18, 790–801. doi:10.1016/j.devcel.2010.03.015
Bugnard, E., Zaal, K. J. M., and Ralston, E. (2005). Reorganization of Microtubule Nucleation during Muscle Differentiation. Cell Motil. Cytoskeleton 60, 1–13. doi:10.1002/cm.20042
Casenghi, M., Barr, F. A., Nigg, E. A., Barr, F. A., Nigg, E. A., and Nigg, E. A. (2005). Phosphorylation of Nlp by Plk1 Negatively Regulates its Dynein-dynactin-dependent Targeting to the Centrosome. J. Cell Sci. 118, 5101–5108. doi:10.1242/jcs.02622
Chuang, M., Goncharov, A., Wang, S., Oegema, K., Jin, Y., and Chisholm, A. D. (2014). The Microtubule Minus-End-Binding Protein patronin/PTRN-1 Is Required for Axon Regeneration in C. elegans. Cell Rep. 9, 874–883. doi:10.1016/j.celrep.2014.09.054
Consolati, T., Locke, J., Roostalu, J., Chen, Z. A., Gannon, J., Asthana, J., et al. (2020). Microtubule Nucleation Properties of Single Human γTuRCs Explained by Their Cryo-EM Structure. Developmental Cell 53, 603–617. e8. doi:10.1016/j.devcel.2020.04.019
Coquand, L., Victoria, G. S., Tata, A., Carpentieri, J. A., Brault, J.-B., Guimiot, F., et al. (2021). CAMSAPs Organize an Acentrosomal Microtubule Network from Basal Varicosities in Radial Glial Cells. J. Cell Biol. 220, e202003151. doi:10.1083/jcb.202003151
Culver–Hanlon, T. L., Lex, S. A., Stephens, A. D., Quintyne, N. J., and King, S. J. (2006). A Microtubule-Binding Domain in Dynactin Increases Dynein Processivity by Skating along Microtubules. Nat. Cell Biol. 8, 264–270. doi:10.1038/ncb1370
Dammermann, A., and Merdes, A. (2002). Assembly of Centrosomal Proteins and Microtubule Organization Depends on PCM-1. J. Cell Biol. 159, 255–266. doi:10.1083/jcb.200204023
Dauber, A., Lafranchi, S. H., Maliga, Z., Lui, J. C., Moon, J. E., McDeed, C., et al. (2012). Novel Microcephalic Primordial Dwarfism Disorder Associated with Variants in the Centrosomal Protein Ninein. J. Clin. Endocrinol. Metab. 97, E2140–E2151. doi:10.1210/jc.2012-2150
Delgehyr, N., Sillibourne, J., and Bornens, M. (2005). Microtubule Nucleation and Anchoring at the Centrosome Are Independent Processes Linked by Ninein Function. J. Cell Sci. 118, 1565–1575. doi:10.1242/jcs.02302
Dong, C., Xu, H., Zhang, R., Tanaka, N., Takeichi, M., and Meng, W. (2017). CAMSAP3 Accumulates in the Pericentrosomal Area and Accompanies Microtubules Release from the Centrosome via Katanin. J. Cell Sci 130, 1709–1715. doi:10.1242/jcs.198010
Efimov, A., Kharitonov, A., Efimova, N., Loncarek, J., Miller, P. M., Andreyeva, N., et al. (2007). Asymmetric CLASP-dependent Nucleation of Noncentrosomal Microtubules at the Trans-golgi Network. Developmental Cell 12, 917–930. doi:10.1016/j.devcel.2007.04.002
Flor-Parra, I., Iglesias-Romero, A. B., and Chang, F. (2018). The XMAP215 Ortholog Alp14 Promotes Microtubule Nucleation in Fission Yeast. Curr. Biol. 28, 1681–1691. e4. doi:10.1016/j.cub.2018.04.008
Folker, E. S., Baker, B. M., and Goodson, H. V. (2005). Interactions between CLIP-170, Tubulin, and Microtubules: Implications for the Mechanism of Clip-170 Plus-End Tracking Behavior. MBoC 16, 5373–5384. doi:10.1091/mbc.e04-12-1106
Gavilan, M. P., Gandolfo, P., Balestra, F. R., Arias, F., Bornens, M., and Rios, R. M. (2018). The Dual Role of the Centrosome in Organizing the Microtubule Network in Interphase. EMBO Rep. 19, e45942. doi:10.15252/embr.201845942
Goldspink, D. A., Rookyard, C., Tyrrell, B. J., Gadsby, J., Perkins, J., Lund, E. K., et al. (2017). Ninein Is Essential for Apico-Basal Microtubule Formation and CLIP-170 Facilitates its Redeployment to Non-centrosomal Microtubule Organizing Centres. Open Biol. 7, 160274. doi:10.1098/rsob.160274
Goodwin, S. S., and Vale, R. D. (2010). Patronin Regulates the Microtubule Network by Protecting Microtubule Minus Ends. Cell 143, 263–274. doi:10.1016/j.cell.2010.09.022
Gunzelmann, J., Rüthnick, D., Lin, T.-C., Zhang, W., Neuner, A., Jäkle, U., et al. (2018). The Microtubule Polymerase Stu2 Promotes Oligomerization of the γ-TuSC for Cytoplasmic Microtubule Nucleation. Elife 7, e39932. doi:10.7554/eLife.39932
Guo, J., Yang, Z., Song, W., Chen, Q., Wang, F., Zhang, Q., et al. (2006). Nudel Contributes to Microtubule Anchoring at the Mother Centriole and Is Involved in Both Dynein-dependent and -independent Centrosomal Protein Assembly. MBoC 17, 680–689. doi:10.1091/mbc.e05-04-0360
Haren, L., Remy, M.-H., Bazin, I., Callebaut, I., Wright, M., and Merdes, A. (2006). NEDD1-dependent Recruitment of the γ-tubulin Ring Complex to the Centrosome Is Necessary for Centriole Duplication and Spindle Assembly. J. Cell Biol. 172, 505–515. doi:10.1083/jcb.200510028
Hendershott, M. C., and Vale, R. D. (2014). Regulation of Microtubule Minus-End Dynamics by CAMSAPs and Patronin. Proc. Natl. Acad. Sci. 111, 5860–5865. doi:10.1073/pnas.1404133111
Hori, A., Ikebe, C., Tada, M., and Toda, T. (2014). Msd1/SSX 2 IP ‐dependent Microtubule anchorage Ensures Spindle Orientation and Primary Cilia Formation. EMBO Rep. 15, 175–184. doi:10.1002/embr.201337929
Hori, A., Morand, A., Ikebe, C., Frith, D., Snijders, A. P., and Toda, T. (2015). The Conserved Wdr8-hMsd1/SSX2IP Complex Localises to the Centrosome and Ensures Proper Spindle Length and Orientation. Biochem. Biophysical Res. Commun. 468, 39–45. doi:10.1016/j.bbrc.2015.10.169
Huang, N., Xia, Y., Zhang, D., Wang, S., Bao, Y., He, R., et al. (2017). Hierarchical Assembly of Centriole Subdistal Appendages via Centrosome Binding Proteins CCDC120 and CCDC68. Nat. Commun. 8, 15057. doi:10.1038/ncomms15057
Ibi, M., Zou, P., Inoko, A., Shiromizu, T., Matsuyama, M., Hayashi, Y., et al. (2011). Trichoplein Controls Microtubule Anchoring at the Centrosome by Binding to Odf2 and Ninein. J. Cell Sci. 124, 857–864. doi:10.1242/jcs.075705
Janke, C., and Magiera, M. M. (2020). The Tubulin Code and its Role in Controlling Microtubule Properties and Functions. Nat. Rev. Mol. Cell Biol. 21, 307–326. doi:10.1038/s41580-020-0214-3
Jiang, K., Hua, S., Mohan, R., Grigoriev, I., Yau, K. W., Liu, Q., et al. (2014). Microtubule Minus-End Stabilization by Polymerization-Driven CAMSAP Deposition. Developmental Cell 28, 295–309. doi:10.1016/j.devcel.2014.01.001
Keating, T. J., Peloquin, J. G., Rodionov, V. I., Momcilovic, D., and Borisy, G. G. (1997). Microtubule Release from the Centrosome. Proc. Natl. Acad. Sci. 94, 5078–5083. doi:10.1073/pnas.94.10.5078
Khanal, I., Elbediwy, A., Diaz de la Loza, M. D. C., Fletcher, G. C., and Thompson, B. J. (2016). Shot and Patronin Polarise Microtubules to Direct Membrane Traffic and Biogenesis of Microvilli in Epithelia. J. Cell Sci. 129, 2651–2659. doi:10.1242/jcs.189076
Kodani, A., Salomé Sirerol-Piquer, M., Seol, A., Manuel Garcia-Verdugo, J., and Reiter, J. F. (2013). Kif3a Interacts with Dynactin Subunit p150Glued to Organize Centriole Subdistal Appendages. EMBO J. 32, 597–607. doi:10.1038/emboj.2013.3
Kollman, J. M., Greenberg, C. H., Li, S., Moritz, M., Zelter, A., Fong, K. K., et al. (2015). Ring Closure Activates Yeast γTuRC for Species-specific Microtubule Nucleation. Nat. Struct. Mol. Biol. 22, 132–137. doi:10.1038/nsmb.2953
Kowanda, M., Bergalet, J., Wieczorek, M., Brouhard, G., Lécuyer, É., and Lasko, P. (2016). Loss of Function of the Drosophila Ninein-Related Centrosomal Protein Bsg25D Causes Mitotic Defects and Impairs Embryonic Development. Biol. Open 5, 1040–1051. doi:10.1242/bio.019638
Kubo, A., Sasaki, H., Yuba-Kubo, A., Tsukita, S., and Shiina, N. (1999). Centriolar Satellites. J. Cell Biol. 147, 969–980. doi:10.1083/jcb.147.5.969
Kunimoto, K., Yamazaki, Y., Nishida, T., Shinohara, K., Ishikawa, H., Hasegawa, T., et al. (2012). Coordinated Ciliary Beating Requires Odf2-Mediated Polarization of Basal Bodies via Basal Feet. Cell 148, 189–200. doi:10.1016/j.cell.2011.10.052
Lechler, T., and Fuchs, E. (2007). Desmoplakin: an Unexpected Regulator of Microtubule Organization in the Epidermis. J. Cell Biol. 176, 147–154. doi:10.1083/jcb.200609109
Lee, Y. R. J., and Liu, B. (2019). Microtubule Nucleation for the Assembly of Acentrosomal Microtubule Arrays in Plant Cells. New Phytol. 222, 1705–1718. doi:10.1111/nph.15705
Leung, C. L., Sun, D., Zheng, M., Knowles, D. R., and Liem, R. K. H. (1999). Microtubule Actin Cross-Linking Factor (Macf). J. Cell Biol. 147, 1275–1286. doi:10.1083/jcb.147.6.1275
Ligon, L. A., Shelly, S. S., Tokito, M. K., and Holzbaur, E. L. F. (2006). Microtubule Binding Proteins CLIP-170, EB1, and p150Gluedform Distinct Plus-End Complexes. FEBS Lett. 580, 1327–1332. doi:10.1016/j.febslet.2006.01.050
Lin, C.-C., Cheng, T.-S., Hsu, C.-M., Wu, C.-H., Chang, L.-S., Shen, Z.-S., et al. (2006). Characterization and Functional Aspects of Human Ninein Isoforms that Regulated by Centrosomal Targeting Signals and Evidence for Docking Sites to Direct Gamma-Tubulin. Cell Cycle 5, 2517–2527. doi:10.4161/cc.5.21.3404
Liu, H., Zheng, J., Zhu, L., Xie, L., Chen, Y., Zhang, Y., et al. (2021). Wdr47, Camsaps, and Katanin Cooperate to Generate Ciliary central Microtubules. Nat. Commun. 12, 5796. doi:10.1038/s41467-021-26058-5
Liu, P., Zupa, E., Neuner, A., Böhler, A., Loerke, J., Flemming, D., et al. (2020). Insights into the Assembly and Activation of the Microtubule Nucleator γ-TuRC. Nature 578, 467–471. doi:10.1038/s41586-019-1896-6
Liu, W., Zheng, F., Wang, Y., and Fu, C. (2019). Alp7-Mto1 and Alp14 Synergize to Promote Interphase Microtubule Regrowth from the Nuclear Envelope. J. Mol. Cell Biol. 11, 944–955. doi:10.1093/jmcb/mjz038
Logarinho, E., Maffini, S., Barisic, M., Marques, A., Toso, A., Meraldi, P., et al. (2012). CLASPs Prevent Irreversible Multipolarity by Ensuring Spindle-Pole Resistance to Traction Forces during Chromosome Alignment. Nat. Cell Biol. 14, 295–303. doi:10.1038/ncb2423
Louie, R. K., Bahmanyar, S., Siemers, K. A., Votin, V., Chang, P., Stearns, T., et al. (2004). Adenomatous Polyposis Coli and EB1 Localize in Close Proximity of the Mother Centriole and EB1 Is a Functional Component of Centrosomes. J. Cell Sci. 117, 1117–1128. doi:10.1242/jcs.00939
Lüders, J. (2021). Nucleating Microtubules in Neurons: Challenges and Solutions. Develop. Neurobiol. 81, 273–283. doi:10.1002/dneu.22751
Lüders, J., Patel, U. K., and Stearns, T. (2006). GCP-WD Is a γ-tubulin Targeting Factor Required for Centrosomal and Chromatin-Mediated Microtubule Nucleation. Nat. Cell Biol. 8, 137–147. doi:10.1038/ncb1349
Manning, J. A., Shalini, S., Risk, J. M., Day, C. L., and Kumar, S. (2010). A Direct Interaction with NEDD1 Regulates γ-Tubulin Recruitment to the Centrosome. PLoS One 5, e9618. doi:10.1371/journal.pone.0009618
Marcette, J. D., Chen, J. J., and Nonet, M. L. (2014). The Caenorhabditis elegans Microtubule Minus-End Binding Homolog PTRN-1 Stabilizes Synapses and Neurites. Elife 3, e01637. doi:10.7554/eLife.01637
Marjanović, M., Sánchez-Huertas, C., Terré, B., Gómez, R., Scheel, J. F., Pacheco, S., et al. (2015). CEP63 Deficiency Promotes P53-dependent Microcephaly and Reveals a Role for the Centrosome in Meiotic Recombination. Nat. Commun. 6, 7676. doi:10.1038/ncomms8676
Mazo, G., Soplop, N., Wang, W.-J., Uryu, K., and Tsou, M.-F. B. (2016). Spatial Control of Primary Ciliogenesis by Subdistal Appendages Alters Sensation-Associated Properties of Cilia. Developmental Cell 39, 424–437. doi:10.1016/j.devcel.2016.10.006
McKenney, R. J., Huynh, W., Tanenbaum, M. E., Bhabha, G., and Vale, R. D. (2014). Activation of Cytoplasmic Dynein Motility by Dynactin-Cargo Adapter Complexes. Science 345, 337–341. doi:10.1126/science.1254198
Meng, W., Mushika, Y., Ichii, T., and Takeichi, M. (2008). Anchorage of Microtubule Minus Ends to Adherens Junctions Regulates Epithelial Cell-Cell Contacts. Cell 135, 948–959. doi:10.1016/j.cell.2008.09.040
Miller, P. M., Folkmann, A. W., Maia, A. R. R., Efimova, N., Efimov, A., and Kaverina, I. (2009). Golgi-derived CLASP-dependent Microtubules Control Golgi Organization and Polarized Trafficking in Motile Cells. Nat. Cell Biol. 11, 1069–1080. doi:10.1038/ncb1920
Mogensen, M. M., Malik, A., Piel, M., Bouckson-Castaing, V., and Bornens, M. (2000). Microtubule Minus-End anchorage at Centrosomal and Non-centrosomal Sites: the Role of Ninein. J. Cell Sci. 113, 3013–3023. doi:10.1242/jcs.113.17.3013
Mogensen, M. M. (1999). Microtubule Release and Capture in Epithelial Cells. Biol. Cell 91, 331–341. doi:10.1111/j.1768-322x.1999.tb01091.x
Moritz, M., Braunfeld, M. B., Guénebaut, V., Heuser, J., and Agard, D. A. (2000). Structure of the γ-tubulin Ring Complex: a Template for Microtubule Nucleation. Nat. Cell Biol. 2, 365–370. doi:10.1038/35014058
Moss, D. K., Bellett, G., Carter, J. M., Liovic, M., Keynton, J., Prescott, A. R., et al. (2007). Ninein Is Released from the Centrosome and Moves Bi-directionally along Microtubules. J. Cell Sci. 120, 3064–3074. doi:10.1242/jcs.010322
Muroyama, A., Seldin, L., and Lechler, T. (2016). Divergent Regulation of Functionally Distinct γ-tubulin Complexes during Differentiation. J. Cell Biol. 213, 679–692. doi:10.1083/jcb.201601099
Nashchekin, D., Fernandes, A. R., and St Johnston, D. (2016). Patronin/Shot Cortical Foci Assemble the Noncentrosomal Microtubule Array that Specifies the Drosophila Anterior-Posterior Axis. Developmental Cell 38, 61–72. doi:10.1016/j.devcel.2016.06.010
Ning, W., Yu, Y., Xu, H., Liu, X., Wang, D., Wang, J., et al. (2016). The CAMSAP3-ACF7 Complex Couples Noncentrosomal Microtubules with Actin Filaments to Coordinate Their Dynamics. Developmental Cell 39, 61–74. doi:10.1016/j.devcel.2016.09.003
Noordstra, I., Liu, Q., Nijenhuis, W., Hua, S., Jiang, K., Baars, M., et al. (2016). Control of Apico-Basal Epithelial Polarity by the Microtubule Minus-End Binding Protein CAMSAP3 and Spectraplakin ACF7. J. Cell Sci. 129, 4278–4288. doi:10.1242/jcs.194878
Odabasi, E., Gul, S., Kavakli, I. H., and Firat‐Karalar, E. N. (2019). Centriolar Satellites Are Required for Efficient Ciliogenesis and Ciliary Content Regulation. EMBO Rep. 20, e47723. doi:10.15252/embr.201947723
Ohama, Y., and Hayashi, K. (2009). Relocalization of a Microtubule-Anchoring Protein, Ninein, from the Centrosome to Dendrites during Differentiation of Mouse Neurons. Histochem. Cell Biol. 132, 515–524. doi:10.1007/s00418-009-0631-z
Ou, Y. Y., Mack, G. J., Zhang, M., and Rattner, J. B. (2002). CEP110 and Ninein Are Located in a Specific Domain of the Centrosome Associated with Centrosome Maturation. J. Cell Sci. 115, 1825–1835. doi:10.1242/jcs.115.9.1825
Panzade, S., and Matis, M. (2021). The Microtubule Minus-End Binding Protein Patronin Is Required for the Epithelial Remodeling in the Drosophila Abdomen. Front. Cell Dev. Biol. 9, 682083. doi:10.3389/fcell.2021.682083
Paz, J., and Lüders, J. (2018). Microtubule-Organizing Centers: Towards a Minimal Parts List. Trends Cell Biol. 28, 176–187. doi:10.1016/j.tcb.2017.10.005
Prosser, S. L., and Pelletier, L. (2020). Centriolar Satellite Biogenesis and Function in Vertebrate Cells. J. Cell Sci. 133, jcs239566. doi:10.1242/jcs.239566
Quintyne, N. J., Gill, S. R., Eckley, D. M., Crego, C. L., Compton, D. A., and Schroer, T. A. (1999). Dynactin Is Required for Microtubule Anchoring at Centrosomes. J. Cell Biol. 147, 321–334. doi:10.1083/jcb.147.2.321
Redwine, W. B., DeSantis, M. E., Hollyer, I., Htet, Z. M., Tran, P. T., Swanson, S. K., et al. (2017). The Human Cytoplasmic Dynein Interactome Reveals Novel Activators of Motility. Elife 6, e28257. doi:10.7554/eLife.28257
Richardson, C. E., Spilker, K. A., Cueva, J. G., Perrino, J., Goodman, M. B., and Shen, K. (2014). PTRN-1, a Microtubule Minus End-Binding CAMSAP Homolog, Promotes Microtubule Function in Caenorhabditis elegans Neurons. Elife 3, e01498. doi:10.7554/eLife.01498
Rivero, S., Cardenas, J., Bornens, M., and Rios, R. M. (2009). Microtubule Nucleation at the Cis-Side of the Golgi Apparatus Requires AKAP450 and GM130. EMBO J. 28, 1016–1028. doi:10.1038/emboj.2009.47
Robinson, A. M., Takahashi, S., Brotslaw, E. J., Ahmad, A., Ferrer, E., Procissi, D., et al. (2020). CAMSAP3 Facilitates Basal Body Polarity and the Formation of the central Pair of Microtubules in Motile Cilia. Proc. Natl. Acad. Sci. USA 117, 13571–13579. doi:10.1073/pnas.1907335117
Rosen, J. N., Azevedo, M., Soffar, D. B., Boyko, V. P., Brendel, M. B., Schulman, V. K., et al. (2019). The Drosophila Ninein Homologue Bsg25D Cooperates with Ensconsin in Myonuclear Positioning. J. Cell Biol. 218, 524–540. doi:10.1083/jcb.201808176
Sallee, M. D., and Feldman, J. L. (2021). Microtubule Organization across Cell Types and States. Curr. Biol. 31, R506–R511. doi:10.1016/j.cub.2021.01.042
Sallee, M. D., Zonka, J. C., Skokan, T. D., Raftrey, B. C., and Feldman, J. L. (2018). Tissue-specific Degradation of Essential Centrosome Components Reveals Distinct Microtubule Populations at Microtubule Organizing Centers. PLoS Biol. 16, e2005189. doi:10.1371/journal.pbio.2005189
Sanchez, A. D., Branon, T. C., Cote, L. E., Papagiannakis, A., Liang, X., Pickett, M. A., et al. (2021). Proximity Labeling Reveals Non-centrosomal Microtubule-Organizing center Components Required for Microtubule Growth and Localization. Curr. Biol. 31, 3586–3600. e11. doi:10.1016/j.cub.2021.06.021
Sanchez, A. D., and Feldman, J. L. (2017). Microtubule-organizing Centers: from the Centrosome to Non-centrosomal Sites. Curr. Opin. Cell Biol. 44, 93–101. doi:10.1016/j.ceb.2016.09.003
Schweizer, N., and Lüders, J. (2021). From Tip to Toe - Dressing Centrioles in γTuRC. J. Cell Sci. 134, jcs258397. doi:10.1242/jcs.258397
Shinohara, H., Sakayori, N., Takahashi, M., and Osumi, N. (2013). Ninein is Essential for the Maintenance of the Cortical Progenitor Character by Anchoring the Centrosome to Microtubules. Biol. Open 2, 739–749. doi:10.1242/bio.20135231
Soundararajan, H. C., and Bullock, S. L. (2014). The Influence of Dynein Processivity Control, MAPs, and Microtubule Ends on Directional Movement of a Localising mRNA. Elife 3, e01596. doi:10.7554/eLife.01596
Srivatsa, S., Parthasarathy, S., Molnár, Z., and Tarabykin, V. (2015). Sip1 Downstream Effector Ninein Controls Neocortical Axonal Growth, Ipsilateral Branching, and Microtubule Growth and Stability. Neuron 85, 998–1012. doi:10.1016/j.neuron.2015.01.018
Srsen, V., Fant, X., Heald, R., Rabouille, C., and Merdes, A. (2009). Centrosome Proteins Form an Insoluble Perinuclear Matrix during Muscle Cell Differentiation. BMC Cell Biol. 10, 28. doi:10.1186/1471-2121-10-28
Tanaka, N., Meng, W., Nagae, S., and Takeichi, M. (2012). Nezha/CAMSAP3 and CAMSAP2 Cooperate in Epithelial-specific Organization of Noncentrosomal Microtubules. Proc. Natl. Acad. Sci. 109, 20029–20034. doi:10.1073/pnas.1218017109
Tassin, A. M., Maro, B., and Bornens, M. (1985). Fate of Microtubule-Organizing Centers during Myogenesis In Vitro. J. Cell Biol. 100, 35–46. doi:10.1083/jcb.100.1.35
Thawani, A., Kadzik, R. S., and Petry, S. (2018). XMAP215 Is a Microtubule Nucleation Factor that Functions Synergistically with the γ-tubulin Ring Complex. Nat. Cell Biol. 20, 575–585. doi:10.1038/s41556-018-0091-6
Thawani, A., Rale, M. J., Coudray, N., Bhabha, G., Stone, H. A., Shaevitz, J. W., et al. (2020). The Transition State and Regulation of γ-TuRC-mediated Microtubule Nucleation Revealed by Single Molecule Microscopy. Elife 9, e54253. doi:10.7554/eLife.54253
Toya, M., Kobayashi, S., Kawasaki, M., Shioi, G., Kaneko, M., Ishiuchi, T., et al. (2016). CAMSAP3 Orients the Apical-To-Basal Polarity of Microtubule Arrays in Epithelial Cells. Proc. Natl. Acad. Sci. USA 113, 332–337. doi:10.1073/pnas.1520638113
Toya, M., Sato, M., Haselmann, U., Asakawa, K., Brunner, D., Antony, C., et al. (2007). γ-Tubulin Complex-Mediated Anchoring of Spindle Microtubules to Spindle-Pole Bodies Requires Msd1 in Fission Yeast. Nat. Cell Biol. 9, 646–653. doi:10.1038/ncb1593
Tu, H.-Q., Qin, X.-H., Liu, Z.-B., Song, Z.-Q., Hu, H.-B., Zhang, Y.-C., et al. (2018). Microtubule Asters Anchored by FSD1 Control Axoneme Assembly and Ciliogenesis. Nat. Commun. 9, 5277. doi:10.1038/s41467-018-07664-2
Usui, T., Maekawa, H., Pereira, G., and Schiebel, E. (2003). The XMAP215 Homologue Stu2 at Yeast Spindle Pole Bodies Regulates Microtubule Dynamics and anchorage. EMBO J. 22, 4779–4793. doi:10.1093/emboj/cdg459
Valenzuela, A., Meservey, L., Nguyen, H., and Fu, M.-M. (2020). Golgi Outposts Nucleate Microtubules in Cells with Specialized Shapes. Trends Cell Biol. 30, 792–804. doi:10.1016/j.tcb.2020.07.004
Vergarajauregui, S., Becker, R., Steffen, U., Sharkova, M., Esser, T., Petzold, J., et al. (2020). AKAP6 Orchestrates the Nuclear Envelope Microtubule-Organizing center by Linking Golgi and Nucleus via AKAP9. Elife 9, e61669. doi:10.7554/eLife.61669
Wang, X., Tsai, J-W., Imai, J. H., Lian, W-N., Vallee, R. B., and Shi, S-H. (2009). Asymmetric Centrosome Inheritance Maintains Neural Progenitors in the Neocortex. Nature 461, 947–955. doi:10.1038/nature08435
Wang, S., Wu, D., Quintin, S., Green, R. A., Cheerambathur, D. K., Ochoa, S. D., et al. (2015). NOCA-1 Functions with γ-tubulin and in Parallel to Patronin to Assemble Non-centrosomal Microtubule Arrays in C. elegans. Elife 4, e08649. doi:10.7554/eLife.08649
Wieczorek, M., Bechstedt, S., Chaaban, S., and Brouhard, G. J. (2015). Microtubule-associated Proteins Control the Kinetics of Microtubule Nucleation. Nat. Cell Biol. 17, 907–916. doi:10.1038/ncb3188
Wieczorek, M., Urnavicius, L., Ti, S.-C., Molloy, K. R., Chait, B. T., and Kapoor, T. M. (2020). Asymmetric Molecular Architecture of the Human γ-Tubulin Ring Complex. Cell 180, 165–175. e16. doi:10.1016/j.cell.2019.12.007
Wiese, C., and Zheng, Y. (2000). A New Function for the γ -tubulin Ring Complex as a Microtubule Minus-End Cap. Nat. Cell Biol. 2, 358–364. doi:10.1038/35014051
Wilkes, O. R., and Moore, A. W. (2020). Distinct Microtubule Organizing Center Mechanisms Combine to Generate Neuron Polarity and Arbor Complexity. Front. Cell. Neurosci. 14, 594199. doi:10.3389/fncel.2020.594199
Wu, J., and Akhmanova, A. (2017). Microtubule-Organizing Centers. Annu. Rev. Cell Dev. Biol. 33, 51–75. doi:10.1146/annurev-cellbio-100616-060615
Wu, J., de Heus, C., Liu, Q., Bouchet, B. P., Noordstra, I., Jiang, K., et al. (2016). Molecular Pathway of Microtubule Organization at the Golgi Apparatus. Developmental Cell 39, 44–60. doi:10.1016/j.devcel.2016.08.009
Wu, X., Kodama, A., and Fuchs, E. (2008). ACF7 Regulates Cytoskeletal-Focal Adhesion Dynamics and Migration and Has ATPase Activity. Cell 135, 137–148. doi:10.1016/j.cell.2008.07.045
Yagi, N., Kato, T., Matsunaga, S., Ehrhardt, D. W., Nakamura, M., and Hashimoto, T. (2021). An Anchoring Complex Recruits Katanin for Microtubule Severing at the Plant Cortical Nucleation Sites. Nat. Commun. 12, 3687. doi:10.1038/s41467-021-24067-y
Yan, X., Habedanck, R., and Nigg, E. A. (2006). A Complex of Two Centrosomal Proteins, CAP350 and FOP, Cooperates with EB1 in Microtubule Anchoring. MBoC 17, 634–644. doi:10.1091/mbc.e05-08-0810
Yang, C., Wu, J., de Heus, C., Grigoriev, I., Liv, N., Yao, Y., et al. (2017). EB1 and EB3 Regulate Microtubule Minus End Organization and Golgi Morphology. J. Cell Biol. 216, 3179–3198. doi:10.1083/jcb.201701024
Yang, D.-W., and Choi, K.-W. (2021). Suppression of Patronin Deficiency by Altered Hippo Signaling in Drosophila Organ Development. Cell Death Differ 28, 233–250. doi:10.1038/s41418-020-0597-x
Yukawa, M., Ikebe, C., and Toda, T. (2015). The Msd1-Wdr8-Pkl1 Complex Anchors Microtubule Minus Ends to Fission Yeast Spindle Pole Bodies. J. Cell Biol. 209, 549–562. doi:10.1083/jcb.201412111
Zheng, Y., Buchwalter, R. A., Zheng, C., Wight, E. M., Chen, J. V., and Megraw, T. L. (2020). A Perinuclear Microtubule-Organizing centre Controls Nuclear Positioning and Basement Membrane Secretion. Nat. Cell Biol. 22, 297–309. doi:10.1038/s41556-020-0470-7
Zheng, Y., Mennella, V., Marks, S., Wildonger, J., Elnagdi, E., Agard, D. A., et al. (2016). The Seckel Syndrome and Centrosomal Protein Ninein Localizes Asymmetrically to Stem Cell Centrosomes but Is Not Required for normal Development, Behavior, or DNA Damage Response in Drosophila. MBoC 27, 1740–1752. doi:10.1091/mbc.E15-09-0655
Keywords: MTOC, anchoring, microtubule, centrosome, nucleation
Citation: Vineethakumari C and Lüders J (2022) Microtubule Anchoring: Attaching Dynamic Polymers to Cellular Structures. Front. Cell Dev. Biol. 10:867870. doi: 10.3389/fcell.2022.867870
Received: 01 February 2022; Accepted: 11 February 2022;
Published: 03 March 2022.
Edited by:
Chad G. Pearson, University of Colorado Anschutz Medical Campus, United StatesCopyright © 2022 Vineethakumari and Lüders. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jens Lüders, jens.luders@irbbarcelona.org
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.