- School of Biomedical Sciences, Faculty of Medicine, The University of Hong Kong, Pokfulam, Hong Kong SAR, China
Microtubules are cytoskeletal filaments underlying the morphology and functions of all eukaryotic cells. In higher eukaryotes, the basic building blocks of these non-covalent polymers, ɑ- and β-tubulins, are encoded by expanded tubulin family genes (i.e., isotypes) at distinct loci in the genome. While ɑ/β-tubulin heterodimers have been isolated and examined for more than 50 years, how tubulin isotypes contribute to the microtubule organization and functions that support diverse cellular architectures remains a fundamental question. To address this knowledge gap, in vitro reconstitution of microtubules with purified ɑ/β-tubulin proteins has been employed for biochemical and biophysical characterization. These in vitro assays have provided mechanistic insights into the regulation of microtubule dynamics, stability, and interactions with other associated proteins. Here we survey the evolving strategies of generating purified ɑ/β-tubulin heterodimers and highlight the advances in tubulin protein biochemistry that shed light on the roles of tubulin isotypes in determining microtubule structures and properties.
Introduction
α/β-tubulin heterodimers polymerize into microtubules that are fundamental to various cellular processes, including cell division, migration, and organelle transport [reviewed in (Nogales, 2000)]. However, not all cells form microtubules with the same composition. Cells can express multiple tubulin isotypes that are different from each other in amino acid sequences (Ludueña and Banerjee, 2008). Humans have at least nine α- and ten β-tubulin isotypes (Findeisen et al., 2014), and most of them can acquire a variety of post-translational modifications, including acetylation, polyglutamylation, and de-tyrosination (Janke and Bulinski, 2011). This diversity in tubulin is important; specialized cells, such as neurons, often express specific tubulin isotypes (Ludueña and Banerjee, 2008), and accumulation of α-tubulin acetylation is a marker for long-lived, stable microtubules (Janke and Bulinski, 2011). Exactly how heterogeneous microtubule composition is established and used by cells to facilitate functional outputs is still an open question.
In vivo genetics and cell biology studies have revealed the critical roles of tubulin isotypes and tubulin post-translational modifications in forming functional cellular microtubule architectures (i.e., the tubulin code) (Sullivan, 1988; Wilson and Borisy, 1997; Verhey and Gaertig, 2007; Janke and Bulinski, 2011). In particular, among tubulin variants that cause phenotypes in a wide variety of eukaryotes, mutations in specific tubulin isotypes have been associated with human diseases such as neurological disorders, impaired oocyte maturation, and defective platelet formation (Gadadhar et al., 2017; Pham and Morrissette, 2019). However, the challenge of generating biochemically pure tubulin has limited our ability to reconstitute microtubules with a defined tubulin composition for quantitative in vitro biochemical and biophysical characterization. How tubulin isotypes determine microtubule properties (e.g., dynamics and post-translational modifications) remains unclear. This review focuses on the milestones in protein biochemistry that have advanced our understanding of microtubule biology (Figure 1). From the initial isolation of α/β-tubulin protein heterodimers as the building block of microtubules, the characterization of purified tubulin variants, to the recent breakthrough in generating recombinant tubulin, the decades-long effort is now ready to decipher the molecular mechanisms underlying the biological functions of tubulin isotypes.
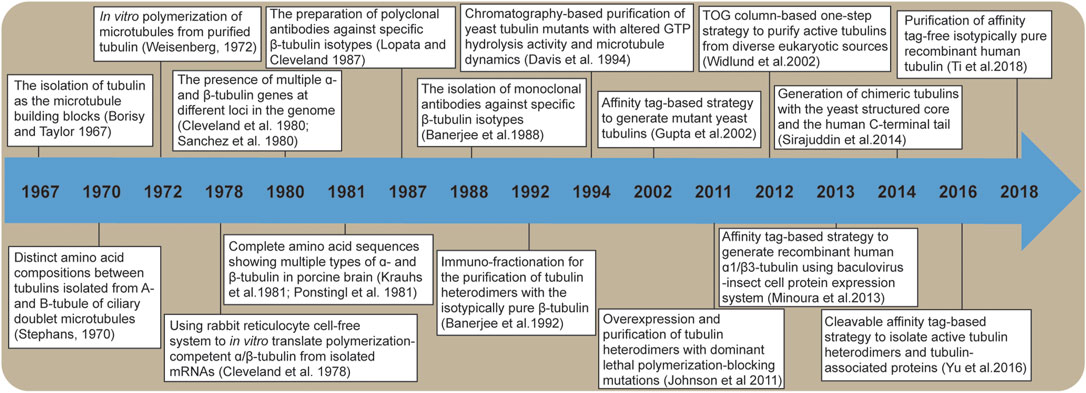
FIGURE 1. A timeline of major breakthroughs that have advanced our understanding of tubulin isotypes.
The Isolation of “Tubulin” as the Building Blocks of Microtubules
In the early 1960s, negative stain electron microscopy images observed the ubiquitous tubular filaments (i.e., microtubules) in diverse cell types across species (Ledbetter and Porter, 1963; Slautterback, 1963). Further examinations, with improved fixing reagents and negative staining strategies, described microtubules as thirteen beaded profibrils surrounding the long axis of the filaments (Pease, 1963; Ledbetter and Porter, 1964; Gall, 1966; Phillips, 1966) (Figure 2). While these fine features are coherent to the modern structural model of microtubules, identifying the building blocks was challenging, mainly due to the lack of effective assays for tracing a microtubule-associated property during biochemical fractionation of the cell lysate.
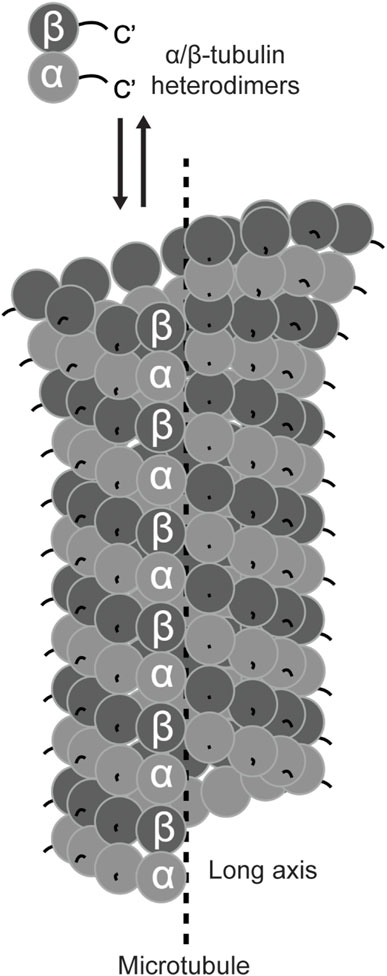
FIGURE 2. Schematic of α/β-tubulin heterodimers and a microtubule. The structured cores of α- (light grey) and β- (dark grey) tubulin are shown in circles. The unstructured C-terminal tails are exposed on the surface of the microtubule.
Colchicine disrupts diverse cellular functions without inhibiting DNA, RNA, and protein synthesis (Taylor, 1965). To reveal the mechanism of action, Borisy and Taylor tracked the radioactivity of tritium-labeled colchicine in fractionated cell homogenates. They found that colchicine targeted a 6S protein enriched in cells or tissues with abundant microtubules, suggesting the colchicine-binding protein to be a subunit of the filaments (Borisy and Taylor, 1967). The following early characterization of the microtubule subunit proteins depended on studies using cilia and flagella, which have the unique 9 + 2 arrangement of microtubules that can be solubilized and fractionated in mild conditions (Gibbons and Grimstone, 1960; Gibbons, 1963). Guided by colchicine-binding activity as well as the electron micrographs of cilia and flagella, the microtubule building blocks were isolated as a dimeric protein of two 55 kDa components, and each protein dimer bound two molecules of guanine nucleotide and one molecule of colchicine (Shelanski and Taylor, 1967, 1968; Stephens et al., 1967; Renaud et al., 1968). Further analyses identified the microtubule subunits as heterodimers of two different kinds of proteins, α- and β-tubulin (Bryan and Wilson, 1971; Feit et al., 1971; Olmsted et al., 1971). With the name ‘tubulin’ (Mohri, 1968), the purified microtubule subunit proteins have led to in vitro biochemical and biophysical studies providing mechanistic insights into diverse cellular processes, such as cell division, cell migration, and intracellular cargo transport.
The Formulation of the “Multi-Tubulin Hypothesis”
Compared to the colchicine-binding protein isolated from the mammalian brain, the purified cilia and flagella tubulin showed the similarity of microtubule subunits in molecular weight, amino acid composition, and guanine nucleotide-binding activity (Weisenberg et al., 1968). This study not only revealed the conservation of microtubule building blocks but also established the critical knowledge (e.g., time sensitivity, the necessity of Mg-GTP, and limited exposure to high salt) for employing brain tissues as the source to purify a large quantity of active tubulin. In particular, the rapid exchange of one tubulin-bound guanine nucleotide with free nucleotides confers the necessity of including GTP during the purification process to maintain the native conformation of tubulin (Weisenberg et al., 1968).
The initial characterization indicated the biochemical similarity in tubulin isolated from different sources. However, the variations in the stability of cellular architectures when subjected to chemical (e.g., colchicine) and physical (e.g., exposure to non-physiological temperature) treatment suggested microtubules with different tubulin compositions. The evidence supporting this hypothesis originated from ciliary and flagellar cold-resistant doublet microtubules, where A-tubules had higher thermostability than B-tubules and remained intact when treated with the elevated temperature (Behnke and Forer, 1967). Temperature-dependent fractionation (i.e., thermal fractionation) of flagellar doublet microtubules revealed the distinct amino acid compositions between tubulins isolated from A- and B-tubules (Stephens, 1970). Further study demonstrated the presence of different types of tubulin in cilia and neuronal cells (Olmsted et al., 1971). These early biochemical studies formulated the ‘multi-tubulin hypothesis’ that the diversity of tubulin proteins underlies the formation of microtubule networks for various biological functions in cells (Fulton and Simpson, 1976).
The Discovery of Tubulin Variants With Distinct Amino Acid Sequences
For several years after the success in tubulin isolation, it had been challenging to reconstitute and characterize the mechanisms of microtubule assembly and disassembly, which play a fundamental role in the cellular functions of the cytoskeletal filaments. To address this limitation, a critical study with detailed biochemical characterization recognized the key factors [magnesium ions (Mg2+), EGTA, and warm temperature (35°C)] stimulating in vitro microtubule assembly from purified tubulin proteins (Weisenberg, 1972). In contrast, elements like calcium ions (Ca2+), EDTA, and cold temperature (0°C) inhibited the formation of the filaments (Weisenberg, 1972). Further characterization identified a high concentration of glycerol [4 M, or about 37% (w/v)] as a reagent that conferred rapid nucleation and superior stability of the tubulin polymers, allowing the development of a strategy to purify tubulin proteins by reversible microtubule polymerization (Shelanski et al., 1973). With phosphocellulose chromatography removing the tubulin-associated proteins, the temperature-dependent microtubule polymerization and depolymerization provided a robust methodology for isolating a large quantity of brain tubulin (Borisy et al., 1975; Weingarten et al., 1975). Since then, purified mammalian brain tubulin (usually from porcine or bovine) has served as a popular material for in vitro assays to dissect the molecular basis of microtubule structure, polymerization dynamics, and interaction with microtubule-associated proteins. By improving the efficiency of in vitro microtubule polymerization, a recent protocol can generate tubulin with controlled post-translational modifications from cell lines or brain tissues of genetically engineered mice (Souphron et al., 2019).
Cycles of microtubule assembly and disassembly have been the core of protocols for obtaining tubulin from non-neuronal cell lines (Doenges et al., 1977; Nagle et al., 1977; Weber et al., 1977; Weatherbee et al., 1978; Doenges et al., 1979; Newton et al., 2002) as well as from different tissues of a variety of species, such as sea urchin (Binder and Rosenbaum, 1978; Farrell and Wilson, 1978; Keller et al., 1982; Detrich and Wilson, 1983), fungi (Kilmartin, 1981; Yoon and Oakley, 1995; Braun et al., 2009; Drummond et al., 2011), nematodes (Dawson et al., 1983), surf clam (Suprenant and Rebhun, 1984), and cold-water fish (Langford, 1978; Williams et al., 1985; Detrich and Overton, 1986; Detrich et al., 1989; Detrich et al., 2000). These pioneering studies revealed the source-dependent variation in the intrinsic properties of the filaments (e.g., stability, critical concentration, and protofilament numbers), the optimal temperature for polymer assembly, and the responses to microtubule-destabilizing small molecules. In vitro polymerization of microtubules started showing the various properties of tubulin purified from different biological contexts.
Advances in the purification strategy provided high quality and enough material for analyzing tubulin polymorphism. Electrofocusing analysis of brain tubulin showed a heterogeneous mixture of a dozen polypeptides with distinct isoelectric points, suggesting the presence of subspecies of tubulin (Feit et al., 1971; George et al., 1981). Tryptic peptide mapping revealed the significant diversity of the primary sequences among tubulin subspecies (George et al., 1981). Peptide sequencing by Edman degradation further supported the heterogeneity in the primary sequences of brain tubulin, which contained at least four types of α- and two types of β-tubulin polypeptides with aspartic and glutamic residues enriched at the carboxy-terminus (i.e., the C-terminal tail) (Krauhs et al., 1981; Ponstingl et al., 1981). These studies proposed that the acidic C-terminal tails could interact with positively charged domains of microtubule-associated proteins (Krauhs et al., 1981; Ponstingl et al., 1981). Meanwhile, the cloning of mRNAs and genomic DNA analysis disclosed multiple α- and β-tubulin genes (i.e., isotypes) at different loci in the genome (Cleveland et al., 1978; Cleveland et al., 1980; Sanchez et al., 1980; Cleveland et al., 1981a). Together, the polymorphism in the tubulin primary sequences likely regulates the properties of microtubule-based cellular architectures.
Immunofractionation of Tubulin Heterodimers With Specific β-Tubulin Isotypes
According to the molecular genetic analyses of the tubulin genes, the most divergent region between tubulin isotypes is the ∼15-residue polypeptide chain at the C-terminal tail (Sullivan and Cleveland, 1986; Villasante et al., 1986; Wang et al., 1986; Pratt et al., 1987). In particular, the amino acid sequence of the C-terminal tail is not only evolutionarily conserved across different vertebrate species but also characteristic to each β-tubulin isotype (Sullivan and Cleveland, 1986; Wang et al., 1986). While genetics and cell biology studies suggested the distinct biological functions of tubulin isotypes, the regulatory roles of tubulin isotype compositions on intrinsic microtubule properties (e.g., polymerization dynamics) was unknown due to the significant challenge of generating isotypically pure tubulin for in vitro biochemical and biophysical assays (Cleveland, 1987).
As the C-terminal tail has the characteristic amino acid sequence of each tubulin isotype, synthetic peptides corresponding to the tail domain can be the antigen for acquiring isotype-specific antibodies (Lopata and Cleveland, 1987). By using peptide-derived polyclonal antibodies against each of the six vertebrate β-tubulin isotypes, immunofluorescence mapped the spatial distribution of β-tubulin isotypes in cultured cells (Lopata and Cleveland, 1987). The success of this antibody-mediated approach further motivated the isolation of monoclonal antibodies for in vitro protein biochemistry studies of tubulin isotypes, establishing that bovine brain β-tubulin is a mixture of four subspecies: type I (βI, 3%), type II (βII, 58%), type III (βIII, 25%) and type IV (βIV, 13%) (Banerjee et al., 1988). The immunodepletion of βIII-tubulin conferred the fractionated brain tubulin an increased rate and a greater extent of microtubule assembly, suggesting the regulatory roles of tubulin isotype compositions in the microtubule polymerization properties (Banerjee et al., 1990). By employing monoclonal antibodies against βII-, βIII- and βIV-tubulin, carefully designed immunoaffinity chromatography of bovine brain tubulin allowed the purification of tubulin heterodimers with isotypically pure β-tubulin, αβII-, αβIII- and αβIV-tubulin (Banerjee et al., 1992).
The immunofractionation of bovine brain tubulin led the way for tubulin isotypes’ functional studies. Thermodynamic characterization revealed the effects of β-tubulin isotypes on the binding affinity of antimitotic alkaloid colchicine for tubulin heterodimers (Banerjee and Luduena, 1992). Compared to the affinity for αβIII-tubulin, colchicine bound to αβII- and αβIV-tubulin about 2-fold and 30-fold tighter, respectively, (Banerjee and Luduena, 1992). In addition, the characterization of microtubule assembly showed that β-tubulin isotype compositions determined the critical concentration for polymer nucleation and the elongation behavior of the filaments (Banerjee et al., 1992; Lu and Luduena, 1994). By using differential interference contrast (DIC) video microscopy to observe single dynamic microtubules, the detailed quantification established that, in comparison to αβII- and αβIV-tubulin, αβIII-tubulin assembled into filaments with higher dynamicity (Panda et al., 1994). The compositions of β-tubulin isotypes can modulate the dynamic instability parameters (e.g., growth rate, shortening rate, and catastrophe frequency) of microtubules (Panda et al., 1994).
In vitro reconstitution using immunofractionated brain tubulin allowed the generation of microtubules with a defined tubulin isotype composition and opened a new avenue toward dissecting the multi-tubulin hypothesis. However, three limitations prevented the general adoption of this immunoaffinity approach for studying tubulin isotypes. First, the low variance in the C-terminal tails limits the availability of antibodies targeting specific α-tubulin isotypes. This restriction makes it challenging to understand how the crosstalk between isotypes of α- and β-tubulin determines the microtubule properties. Second, the purification of brain tubulin usually requires microtubule polymerization cycles, which are selective for tubulin isotypes that favor this process. Third, tubulin is not abundant in most non-neuronal cells. It has been challenging to achieve the critical concentration for microtubule polymerization in tubulin purification procedures. The low tubulin recovery efficiency of polymerization cycles further hinders the generation of enough tubulin from other cell or tissue types for immunofractionation. Tubulin purification strategies with higher efficiency and flexibility would be essential for dissecting the underlying molecular mechanisms by which tubulin isotypes regulate microtubule functions and structures.
Tubulin-Affinity Chromatography For Efficient Isolation of α/β-Tubulin Heterodimers
One approach exploits tubulin-binding ligands that can be immobilized on the stationary phase as an affinity chromatography column to isolate α/β-tubulin heterodimers from complex cell extracts. In cells, conserved XMAP215/Dis1 family proteins are processive microtubule polymerases that employ the tumor overexpressed gene (TOG) domains to specifically recruit α/β-tubulin from the cytoplasm onto the growing filament ends (Al-Bassam et al., 2006; Al-Bassam et al., 2007; Brouhard et al., 2008; Widlund et al., 2011). Due to the selective and reversible binding to tubulin, the immobilized TOG domains serve as an optimal purification matrix (i.e., TOG-column) to sequester native tubulin from cell lines or tissues of various species (Widlund et al., 2012). This one-step affinity chromatography strategy allows the rapid and efficient isolation of α/β-tubulin heterodimers from extracts of cells with low tubulin expression levels, for example, S. cerevisiae (about 0.05% of the total protein) (Kilmartin, 1981; Widlund et al., 2012).
The success of tubulin purification using a TOG-column overcomes the following two significant drawbacks of in vitro biochemical reconstitution assays using mammalian brain tubulin. First, while tubulin is conserved in eukaryotes (S. cerevisiae and human α-tubulin protein primary sequences are about 75% identical), microtubule-associated proteins behave differently in assays with mammalian brain tubulin or with tubulin purified from homologous species (Alonso et al., 2007; Kollman et al., 2015). The TOG-column-mediated affinity chromatography strategy is revolutionary as the purified native tubulin from corresponding biological contexts is handy for homologous in vitro reconstitution assays. In particular, current studies have reconstituted flagellar sliding using axonemal tubulin and dynein (Alper J. et al., 2013; Alper J. D. et al., 2013; Alper et al., 2014), dissected the length regulation mechanisms of S. cerevisiae, D. melanogaster, or A. thaliana microtubules (Fujita et al., 2013; Podolski et al., 2014; Hibbel et al., 2015; Hotta et al., 2016; Moriwaki and Goshima, 2016; Otani et al., 2018; Edzuka and Goshima, 2019), elucidated the structural insight into the binding of motors and microtubule-associated proteins to S. pombe or human filaments (Atherton et al., 2019; von Loeffelholz et al., 2019; von Loeffelholz et al., 2017), revealed the molecular basis of the unique polymerization dynamics of C. elegans microtubules (Chaaban et al., 2018), identified parasite-specific small molecules targeting microtubule polymerization (Hirst et al., 2022), as well as established the roles of microtubule dynamics in the control of spindle morphology of Xenopus species (Hirst et al., 2020; Biswas et al., 2021).
Second, tubulin purified from mammalian brain tissues has various post-translational modifications (Janke and Bulinski, 2011). It has been difficult to use brain tubulin for studying the molecular mechanisms by which individual tubulin modifications regulate microtubule properties and functions. TOG-affinity chromatography allows the purification of tubulin from sources that have a clean profile of tubulin post-translational modifications, such as a human embryonic kidney cell line (tsA201) (Vemu et al., 2014; Vemu et al., 2017). The mass spectrometric analysis showed that the purified tsA201 tubulin contained α1B-, βI-, βII- and βIVB-tubulin with no detectable post-translational modification (i.e., naïve tubulin) (Vemu et al., 2014). This ‘naïve’ tsA201 tubulin can then be selectively modified by recombinant tubulin-modifying enzymes (Vemu et al., 2014). This enzymatic approach to generating filaments with a defined composition of post-translational modifications has led to mechanistic insights into the roles of tubulin’s chemical adducts in the regulation of microtubule structures and functions (Garnham et al., 2015; Valenstein and Roll-Mecak, 2016; Garnham et al., 2017; Mahalingan et al., 2020; Zheng et al., 2022).
While the TOG affinity-based approach can effectively isolate soluble tubulin from any cell lysate with no preference to specific tubulin isotypes (Widlund et al., 2012), limitations remained on characterizing how the variance in tubulin primary sequences (i.e., isotypes or mutations) could impact microtubule properties (e.g., structure, polymerization dynamics, as well as interactions with motors and microtubule-associated proteins). A recombinant protein strategy of expressing and purifying tubulin variants is essential. However, the generation of recombinant tubulin has been somewhat challenging.
Significant Barriers to Achieving Recombinant Tubulin Stem From Its Complex Biosynthesis
In vitro transcription and translation system using a rabbit reticulocyte lysate system allows the purification of a small quantity of recombinant tubulin (Cleveland et al., 1978). However, achieving a yield at the milligram scale of recombinant tubulin is not trivial, likely due to the following two cellular mechanisms regulating tubulin biosynthesis and homeostasis.
First, the formation of native α/β-tubulin heterodimers requires the newly synthesized tubulin polypeptides to go through the tubulin-folding pathway that employs a complex chaperone system including prefoldin, cytosolic chaperonin, and tubulin-specific folding cofactors (Gao et al., 1992; Yaffe et al., 1992; Tian et al., 1996; Lewis et al., 1997; Tian et al., 1997; Vainberg et al., 1998; Bhamidipati et al., 2000). As common prokaryotic organisms for producing recombinant proteins (e.g., E. coli) lack these chaperone components, the overexpressed tubulin polypeptides of interest form non-functional aggregates in these organisms. Second, the tubulin biosynthesis is under a tight regulation that employs a negative feedback loop to self-regulate the stability of tubulin mRNAs in response to the concentration of soluble α/β-tubulin heterodimers (Ben-Ze’ev et al., 1979; Cleveland, 1989; Cleveland et al., 1981b; Gasic et al., 2019). This negative correlation, also known as tubulin autoregulation, involves a ribosome-associating factor, TTC5, that binds to the N-terminus of nascent tubulin polypeptides and stimulates co-translational degradation of tubulin mRNA following increased soluble tubulin concentration (Yen et al., 1988; Theodorakis and Cleveland, 1992; Lin et al., 2020).
Together, due to the complex eukaryotic machinery that controls the folding and the concentration of soluble tubulin in the cytoplasm, the yield of recombinant tubulin is irrelevant to the ectopic overexpression level of the tubulin genes of interest. It has been challenging to generate recombinant tubulin in the native state for dissecting how variation in the primary protein sequences affects the structures and functions of microtubules.
Exploiting Yeast Cells to Express and Purify Recombinant Tubulin
The genomes of budding yeast S. cerevisiae and fission yeast S. pombe encode two α- and one β-tubulin isotypes, which the yeast genetic tools can engineer to construct strains harboring modified tubulin genes. With the established methodology to purify milligram quantities of wild-type yeast tubulin proteins (Barnes et al., 1992; Davis et al., 1993), the relatively simple tubulin isotype composition has made these unicellular fungi a powerful platform to access mutant tubulin proteins. In particular, the stable haploid and diploid states of yeast cells provide an opportunity to characterize tubulin harboring lethal mutations (Davis et al., 1994). Furthermore, the genetically modified strains with only one α-tubulin isotype have been the source for generating isotypically pure yeast α/β-tubulin heterodimers, which allows the assembly of microtubules with a defined tubulin isotype composition (Bode et al., 2003; Braun et al., 2009; des Georges et al., 2008; Uchimura et al., 2010; Uchimura et al., 2006).
To isolate the mutant tubulin from yeast cells for in vitro assays, researchers engineered the yeast β-tubulin C-terminus to include a hexahistidine tag for affinity purification or to alter the quantities of negatively charged glutamic acid residues for ion-exchange chromatography (Davis et al., 1994; Gupta et al., 2002). These yeast strains expressed α- and β-tubulin proteins from the endogenous gene loci or extra copies of the α- and β-tubulin genes controlled by a galactose-induced overexpression promoter. The chromatography-based strategy and affinity purification allowed the isolation of mutant tubulin without cycles of polymerization and depolymerization (Davis et al., 1994; Gupta et al., 2002). The mutagenesis analyses of yeast tubulin have provided mechanistic insights into the roles of GTP hydrolysis activity in microtubule dynamics (Davis et al., 1994; Dougherty et al., 2001), investigated the structure-activity relationship of tubulin-targeting small molecules (Gupta et al., 2002; Gupta et al., 2003), and examined the microtubule-binding site regulating the kinesin motor activity (Uchimura et al., 2006; Uchimura et al., 2010). However, the excess amounts of β-tubulin (alone or together with α-tubulin) causes cell cycle arrest, chromosome losses, and depolymerization of cellular microtubules (Burke et al., 1989). The lethality limited the yield of recombinant yeast tubulin and restricted access to dominant loss-of-function mutant tubulin for in vitro biochemical and biophysical characterization. The full capacity of the yeast protein expression system remained unexplored.
A robust strategy has unleashed the power of using S. cerevisiae to express and purify recombinant mutant tubulin (Johnson et al., 2011). The success of this chromatography-based approach depends on a transient (three to five hours) but strong protein overexpression from galactose-inducible promoters in high-copy-number plasmids. By significantly improving the final yield while bypassing the lethality due to excess α- and β-tubulin, this strategy opens a new avenue to characterizing tubulin constructs harboring dominant polymerization-blocking mutations, which have explicated the principles of protein machinery that regulates microtubule growth and stability (Ayaz et al., 2012; Ayaz et al., 2014; Geyer et al., 2018; Majumdar et al., 2018). Further mutagenesis analyses of recombinant yeast tubulin revealed the long-overdue molecular bases of microtubule dynamics, such as the allosteric effects of nucleotide states and the regulatory roles of nucleotide exchange on growing filament ends (Geyer et al., 2015; Piedra et al., 2016), demonstrated the effects of disease-related tubulin mutations on microtubule properties (Denarier et al., 2021; Park et al., 2021), and elucidated the mechanism of kinesin-8 family depolymerase activity (Arellano-Santoyo et al., 2017).
This established recombinant yeast tubulin system also has stimulated the development of new methodologies to label tubulin for revealing molecular features of microtubules. For example, site- and topology-specifically labeled yeast tubulin with probes or tags can facilitate studies of how proteins, small molecules, and post-translation modifications interact and regulate microtubules (Kleiner et al., 2013). Furthermore, strategies tagging the C-terminus of yeast β-tubulin with a microbead or a gold nanoparticle have significantly improved the resolution of light microscopy for characterizing in vitro reconstituted microtubules (Driver et al., 2017; Mickolajczyk et al., 2019). Direct measurements by laser tweezers determined the strain energy stored in microtubule protofilaments (Driver et al., 2017), while direct observation of tubulin subunits association and dissociation at growing filament ends provided quantitative insights into microtubule dynamics (Mickolajczyk et al., 2019).
Using this yeast-based strategy to generate recombinant human tubulin was unsuccessful (Sirajuddin et al., 2014). Instead, an alternative approach employed chimeric proteins consisting of the folded yeast tubulin core and the human tubulin unstructured C-terminal tail that contains sites for several post-translational modifications and interacts with most microtubule-associated proteins (Sirajuddin et al., 2014; Janke and Magiera, 2020). These chimeric tubulin proteins were purified by affinity chromatography using a hexahistidine-tag in the acetylation loop of the α-tubulin subunit. These tubulin chimeras have been a powerful tool to characterize the regulatory roles of tubulin isotypes and post-translational modifications in protein activities such as the processivity and velocity of microtubule motors (Sirajuddin et al., 2014) and the permeability of voltage-dependent anion channels (Rostovtseva et al., 2018).
The Recent Breakthrough in Making Recombinant Higher Eukaryotic Tubulin
The genome of higher eukaryotes encodes a substantially expanded number of α- and β-tubulin isotypes. For example, the human genome encodes at least nine α- and ten β-tubulin isotypes that show cell-type-specific expression profiles (Ludueña and Banerjee, 2008; Findeisen et al., 2014). While genetics and cell biology studies suggested that each tubulin gene could have unique cellular functions (Janke and Magiera, 2020), it remains unclear how tubulin isotypes modulate microtubule structures and functions. Due to the complex cellular machinery that regulates tubulin biosynthesis and homeostasis, it has been challenging to purify active human tubulin in the recombinant form.
In 2013, a pioneering strategy used the baculovirus-insect cell protein expression system to generate polymerization competent recombinant human tubulin (Minoura et al., 2013). By employing a hexahistidine-tag fused to the C-terminus of α-tubulin and a FLAG-tag at the C-terminus of β-tubulin, this affinity chromatography-based workflow allowed co-expressing both tubulin and generating high purity of active human α1/β3-tubulin, mouse α1/β2-tubulin and Drosophila α1/β1-tubulin (Minoura et al., 2013; Ayukawa et al., 2021; Diao et al., 2021). Later, another approach used an internal hexahistidine-tag at α-tubulin (Sirajuddin et al., 2014) together with a protease-cleavable FLAG-tag at the C-terminus of β-tubulin also successfully produced isotypically pure human α1/β3-tubulin (Vemu et al., 2016). The success in generating active α/β-tubulin isotypes of higher eukaryotes has offered the opportunities to dissect the molecular mechanisms by which high eukaryotic tubulins regulate microtubule properties. In particular, recombinant human α1/β3-tubulin has become a popular material for characterizing the impacts of disease-related tubulin mutations on the behaviors of kinesin motors (Minoura et al., 2016), the modulation of microtubule dynamics by tubulin isotype composition (Vemu et al., 2017), the incorporation of soluble GTP-tubulin into damaged sites along the microtubule shaft (Vemu et al., 2018), and the effects of GTP hydrolysis on microtubule structures and dynamics (Roostalu et al., 2020; LaFrance et al., 2022). However, these recombinant proteins contain uncleavable charged affinity tags fused to the tubulin domains that interact with microtubule-associated proteins (e.g., the C-terminal tail) or inter-tubulin contacts within the microtubule lattice (e.g., the acetylation loop). It will be favorable to access recombinant tubulin with cleavable affinity tags.
Tubulin with a cleavable affinity tag can be a powerful tool for identifying tubulin-associated proteins (Yu et al., 2016; Yu and Galjart, 2018). A SUMO protease cleavable biotinylation tag at the N-terminus of human α1-or β3-tubulin mediated the isolation of tubulin and the associated tubulin-binding proteins from the cell lysate (Yu et al., 2016; Yu and Galjart, 2018). The expression of the tubulin constructs together with bacterial biotin ligase (BirA) in HEK293T cells led to the generation of biotinylated human tubulin. After streptavidin-coupled matrix-based enrichment, SUMO protease treatment facilitated the release of biotin-tagged tubulin and the tubulin-associated proteins for further mass spectrometry analyses. While this approach provides an opportunity to reveal the interaction proteome of human tubulin isotypes or disease-related mutant tubulins (Yu and Galjart, 2018), the relatively low yield has limited the application of this strategy to generate recombinant human tubulin for in vitro reconstitution of microtubules.
To obtain affinity tag-free recombinant tubulin, we reasoned that the cleavable affinity tag must be at the N- or C-terminus of tubulin for enzymatic removal of the peptide tag after affinity chromatography. While it is promising to fuse the affinity peptide ligand to the C-terminus of β-tubulin, the initial attempts to tag either end of α-tubulin significantly reduced the amount of recombinant human tubulin obtained. By employing the baculovirus-insect cell system, we expressed untagged human α1B-tubulin together with human β2-or β3-tubulin fused at the C-terminus with a tobacco etch virus (TEV) protease cleavable hexahistidine-tag (Ti et al., 2016). Our three-step tubulin purification strategy involved nickel-affinity chromatography followed by tag removal and the final TOG-column affinity chromatography. This approach yielded isotypically pure human β-tubulin dimerized with either recombinant human α1B-tubulin or endogenous insect α-tubulin, indicating that the C-terminal hexahistidine tag is sufficient to isolate specifically human β-tubulin from the complex cell lysate. The characterization of the isotypically pure recombinant human β-tubulin revealed how disease-related β-tubulin mutations, human β-tubulin isotypes, and tubulin allosteric conformational changes affect microtubule dynamics (Pamula et al., 2016; Ti et al., 2016; Ye et al., 2020).
Our systematic evaluation indicated that both the composition and position (N- or C-terminus) of the fused polypeptide are critical for the yield of functional human tubulin. To achieve optimal cleavage efficiency of the affinity tags, we incorporated a TEV-cleavable decahistidine tag with an Ala-Pro dipeptide linker to the N-terminus of human α1B-tubulin and a TEV-cleavable strep tag with a Gly-Gly-Ser-Gly-Gly pentapeptide linker to the C-terminus of human β2-and β3-tubulin (Ti et al., 2018; Ti et al., 2020). We note that the enzymatic digestion gets rid of the affinity tags but leaves residual ‘scars’ at the N-terminus of the α-tubulin (Gly-Ala-Pro) and the C-terminus of the β-tubulin (Glu-Asn-Leu-Tyr-Phe-Gln). We speculate that combining our approach with other protein engineering tools (e.g., protein ligation) will generate recombinant human tubulin with native sequence.
With these constructs, we recently developed an affinity chromatography-based purification strategy that allows the routine preparation of affinity tag-free recombinant human tubulin. As the sequential isolation of human α- and β-tubulin depends solely on the affinity tags, this approach applies to studies of tubulin variants (e.g., isotypes and mutants) that could impact the binding to TOG domains or the microtubule polymerization properties. By employing this strategy, current studies have revealed the effects of human β-tubulin isotypes on the microtubule stability and protofilament numbers (Ti et al., 2018) as well as dissected the molecular mechanisms by which methyltransferases modify human α-tubulin (Kearns et al., 2021). Together, the ability to obtain biochemically pure higher eukaryotic tubulin has paved the way to deciphering the functions of tubulin diversity and a clearer understanding of microtubule biology.
Conclusion and Perspectives
Tubulin protein biochemistry has been evolving since more than 50 years ago, when the colchicine-binding activity led to the isolation of the building blocks of endogenous microtubules (Borisy and Taylor, 1967; Shelanski and Taylor, 1967). With recently established affinity tag-based strategies of generating recombinant α/β-tubulin with defined primary sequences (Figure 3), it becomes feasible to correlate in vivo tubulin mutagenesis analyses with in vitro biochemical and biophysical characterization of mutant tubulin. This integrative approach is potentially applicable to the mutagenesis analysis of any tubulin isotype of interest for a mechanistic understanding of how tubulin diversity regulates cellular microtubule structures and functions.
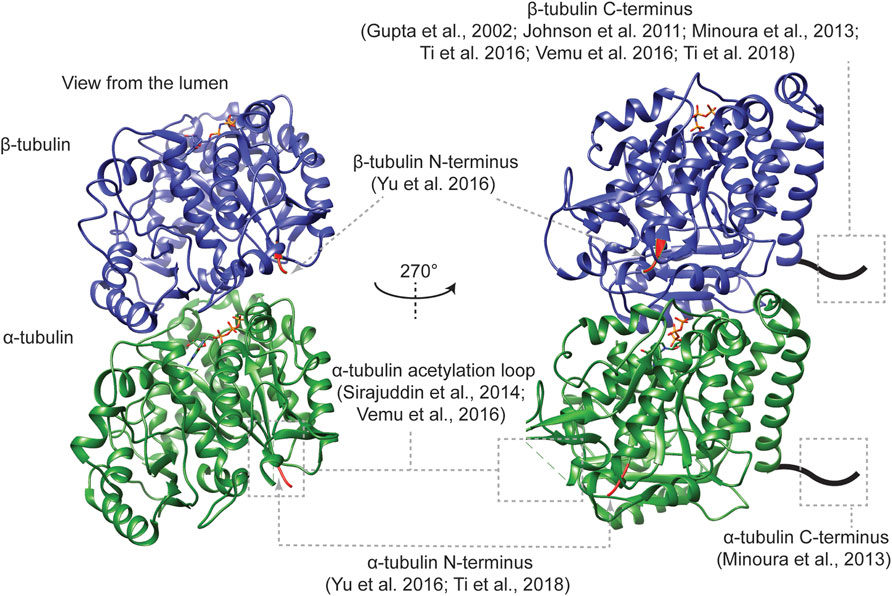
FIGURE 3. The location of affinity tags for the purification of recombinant tubulin. The ribbon diagrams show the structure of an α/β-tubulin heterodimer (PDB ID: 6e7b). The first three residues at the N-termini of each tubulin are in red, while the rest of α- and β-tubulin are in green and blue, respectively. The stick model represents the tubulin-bound nucleotides.
The advances in tubulin protein biochemistry also provide opportunities to address some fundamental questions in microtubule biology by developing the needed tools such as 1) isotype-specific antibodies/nanobodies to characterize the spatial distribution of tubulin isotypes in cellular microtubules, 2) recombinant tubulin incorporated with a probe at a specific site for the identification of small molecules or protein binders targeting explicit tubulin isotypes, 3) engineered tubulin harboring defined modifications to investigate the crosstalk between tubulin isotypes and post-translation modifications (i.e., the tubulin code), and 4) small molecules targeting tubulin isotypes of interest not only for dissecting the biological functions but also for novel chemotherapeutic agents. By combining recombinant tubulin with a chemical biology toolbox for protein engineering (e.g., amber suppression and protein ligation), these technology breakthroughs will expand our ability to tackle the challenges in the field. We speculate that decades of research have set the stage to unveil the molecular basis of how cells establish and use the heterogeneous microtubule composition to facilitate the functional outputs.
Author Contributions
S-CT prepared the manuscript and secured the funding that supported this research.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We are grateful for the support from the Research Grants Council of Hong Kong (ECS 27124120 and GRF 17118421) and the University of Hong Kong (Seed Fund 201909185025, 202010160017, and 202011159046).
References
Al-Bassam, J., Larsen, N. A., Hyman, A. A., and Harrison, S. C. (2007). Crystal Structure of a TOG Domain: Conserved Features of XMAP215/Dis1-Family TOG Domains and Implications for Tubulin Binding. Structure 15, 355–362. doi:10.1016/j.str.2007.01.012
Al-Bassam, J., van Breugel, M., Harrison, S. C., and Hyman, A. (2006). Stu2p Binds Tubulin and Undergoes an Open-To-Closed Conformational Change. J. Cel Biol 172, 1009–1022. doi:10.1083/jcb.200511010
Alonso, M. C., Drummond, D. R., Kain, S., Hoeng, J., Amos, L., and Cross, R. A. (2007). An ATP Gate Controls Tubulin Binding by the Tethered Head of Kinesin-1. Science 316, 120–123. doi:10.1126/science.1136985
Alper, J. D., Decker, F., Agana, B., and Howard, J. (2014). The Motility of Axonemal Dynein Is Regulated by the Tubulin Code. Biophysical J. 107, 2872–2880. doi:10.1016/j.bpj.2014.10.061
Alper, J. D., Tovar, M., and Howard, J. (2013b). Displacement-weighted Velocity Analysis of Gliding Assays Reveals that Chlamydomonas Axonemal Dynein Preferentially Moves Conspecific Microtubules. Biophysical J. 104, 1989–1998. doi:10.1016/j.bpj.2013.03.041
Alper, J., Geyer, V., Mukundan, V., and Howard, J. (2013a). Reconstitution of Flagellar Sliding. Methods Enzymol. 524, 343–369. doi:10.1016/b978-0-12-397945-2.00019-6
Arellano-Santoyo, H., Geyer, E. A., Stokasimov, E., Chen, G.-Y., Su, X., Hancock, W., et al. (2017). A Tubulin Binding Switch Underlies Kip3/Kinesin-8 Depolymerase Activity. Develop. Cel 42, 37–51. doi:10.1016/j.devcel.2017.06.011
Atherton, J., Luo, Y., Xiang, S., Yang, C., Rai, A., Jiang, K., et al. (2019). Structural Determinants of Microtubule Minus End Preference in CAMSAP CKK Domains. Nat. Commun. 10, 5236. doi:10.1038/s41467-019-13247-6
Ayaz, P., Munyoki, S., Geyer, E. A., Piedra, F. A., Vu, E. S., Bromberg, R., et al. (2014). A Tethered Delivery Mechanism Explains the Catalytic Action of a Microtubule Polymerase. Elife 3, e03069. doi:10.7554/eLife.03069
Ayaz, P., Ye, X., Huddleston, P., Brautigam, C. A., and Rice, L. M. (2012). A TOG:αβ-tubulin Complex Structure Reveals Conformation-Based Mechanisms for a Microtubule Polymerase. Science 337, 857–860. doi:10.1126/science.1221698
Ayukawa, R., Iwata, S., Imai, H., Kamimura, S., Hayashi, M., Ngo, K. X., et al. (2021). GTP-dependent Formation of Straight Tubulin Oligomers Leads to Microtubule Nucleation. J. Cel Biol 220. doi:10.1083/jcb.202007033
Banerjee, A., and Luduena, R. F. (1992). Kinetics of Colchicine Binding to Purified Beta-Tubulin Isotypes from Bovine Brain. J. Biol. Chem. 267, 13335–13339. doi:10.1016/s0021-9258(18)42215-6
Banerjee, A., Roach, M. C., Trcka, P., and Ludueña, R. F. (1990). Increased Microtubule Assembly in Bovine Brain Tubulin Lacking the Type III Isotype of Beta-Tubulin. J. Biol. Chem. 265, 1794–1799. doi:10.1016/s0021-9258(19)40087-2
Banerjee, A., Roach, M. C., Trcka, P., and Luduena, R. F. (1992). Preparation of a Monoclonal Antibody Specific for the Class IV Isotype of Beta-Tubulin. Purification and Assembly of Alpha Beta II, Alpha Beta III, and Alpha Beta IV Tubulin Dimers from Bovine Brain. J. Biol. Chem. 267, 5625–5630. doi:10.1016/s0021-9258(18)42811-6
Banerjee, A., Roach, M. C., Wall, K. A., Lopata, M. A., Cleveland, D. W., and Ludueña, R. F. (1988). A Monoclonal Antibody against the Type II Isotype of Beta-Tubulin. Preparation of Isotypically Altered Tubulin. J. Biol. Chem. 263, 3029–3034. doi:10.1016/s0021-9258(18)69171-9
Barnes, G., Louie, K. A., and Botstein, D. (1992). Yeast Proteins Associated with Microtubules In Vitro and In Vivo. MBoC 3, 29–47. doi:10.1091/mbc.3.1.29
Behnke, O., and Forer, A. (1967). Evidence for Four Classes of Microtubules in Individual Cells. J. Cel Sci 2, 169–192. doi:10.1242/jcs.2.2.169
Ben-Ze'ev, A., Farmer, S. R., and Penman, S. (1979). Mechanisms of Regulating Tubulin Synthesis in Cultured Mammalian Cells. Cell 17, 319–325. doi:10.1016/0092-8674(79)90157-0
Bhamidipati, A., Lewis, S. A., and Cowan, N. J. (2000). ADP Ribosylation Factor-like Protein 2 (Arl2) Regulates the Interaction of Tubulin-Folding Cofactor D with Native Tubulin. J. Cel Biol 149, 1087–1096. doi:10.1083/jcb.149.5.1087
Binder, L. I., and Rosenbaum, J. L. (1978). The In Vitro Assembly of Flagellar Outer Doublet Tubulin. J. Cel Biol 79, 500–515. doi:10.1083/jcb.79.2.500
Biswas, A., Kim, K., Cojoc, G., Guck, J., and Reber, S. (2021). The Xenopus Spindle Is as Dense as the Surrounding Cytoplasm. Develop. Cel 56, 967–975. doi:10.1016/j.devcel.2021.03.013
Bode, C. J., Gupta, M. L., Suprenant, K. A., and Himes, R. H. (2003). The Two α‐tubulin Isotypes in Budding Yeast Have Opposing Effects on Microtubule Dynamics In Vitro. EMBO Rep. 4, 94–99. doi:10.1038/sj.embor.embor716
Borisy, G. G., Marcum, J. M., Olmsted, J. B., Murphy, D. B., and Johnson, K. A. (1975). Purification of Tubulin and Associated High Molecular Weight Proteins from Porcine Brain and Characterization of Microtubule Assembly In Vitro. Ann. NY Acad. Sci. 253, 107–132. doi:10.1111/j.1749-6632.1975.tb19196.x
Borisy, G. G., and Taylor, E. W. (1967). The Mechanism of Action of Colchicine. J. Cel Biol 34, 525–533. doi:10.1083/jcb.34.2.525
Braun, M., Drummond, D. R., Cross, R. A., and McAinsh, A. D. (2009). The Kinesin-14 Klp2 Organizes Microtubules into Parallel Bundles by an ATP-dependent Sorting Mechanism. Nat. Cel Biol 11, 724–730. doi:10.1038/ncb1878
Brouhard, G. J., Stear, J. H., Noetzel, T. L., Al-Bassam, J., Kinoshita, K., Harrison, S. C., et al. (2008). XMAP215 Is a Processive Microtubule Polymerase. Cell 132, 79–88. doi:10.1016/j.cell.2007.11.043
Bryan, J., and Wilson, L. (1971). Are Cytoplasmic Microtubules Heteropolymers? Proc. Natl. Acad. Sci. U.S.A. 68, 1762–1766. doi:10.1073/pnas.68.8.1762
Burke, D., Gasdaska, P., and Hartwell, L. (1989). Dominant Effects of Tubulin Overexpression in Saccharomyces cerevisiae. Mol. Cel Biol 9, 1049–1059. doi:10.1128/mcb.9.3.1049-1059.1989
Chaaban, S., Jariwala, S., Hsu, C.-T., Redemann, S., Kollman, J. M., Müller-Reichert, T., et al. (2018). The Structure and Dynamics of C. elegans Tubulin Reveals the Mechanistic Basis of Microtubule Growth. Develop. Cel 47, 191–204. doi:10.1016/j.devcel.2018.08.023
Cleveland, D. W. (1989). Autoregulated Control of Tubilin Synthesis in Animal Cells. Curr. Opin. Cel Biol. 1, 10–14. doi:10.1016/s0955-0674(89)80030-4
Cleveland, D. W., Hughes, S. H., Stubblefield, E., Kirschner, M. W., and Varmus, H. E. (1981a). Multiple Alpha and Beta Tubulin Genes Represent Unlinked and Dispersed Gene Families. J. Biol. Chem. 256, 3130–3134. doi:10.1016/s0021-9258(19)69734-6
Cleveland, D. W., Kirschner, M. W., and Cowan, N. J. (1978). Isolation of Separate mRNAs for α- and β-Tubulin and Characterization of the Corresponding In Vitro Translation Products. Cell 15, 1021–1031. doi:10.1016/0092-8674(78)90286-6
Cleveland, D. W., Lopata, M. A., MacDonald, R. J., Cowan, N. J., Rutter, W. J., and Kirschner, M. W. (1980). Number and Evolutionary Conservation of α- and β-tubulin and Cytoplasmic β- and γ-actin Genes Using Specific Cloned cDNA Probes. Cell 20, 95–105. doi:10.1016/0092-8674(80)90238-x
Cleveland, D. W., Lopata, M. A., Sherline, P., and Kirschner, M. W. (1981b). Unpolymerized Tubulin Modulates the Level of Tubulin mRNAs. Cell 25, 537–546. doi:10.1016/0092-8674(81)90072-6
Cleveland, D. W. (1987). The Multitubulin Hypothesis Revisited: what Have We Learned? J. Cel Biol 104, 381–383. doi:10.1083/jcb.104.3.381
Davis, A., Sage, C. R., Dougherty, C. A., and Farrell, K. W. (1994). Microtubule Dynamics Modulated by Guanosine Triphosphate Hydrolysis Activity of β-Tubulin. Science 264, 839–842. doi:10.1126/science.8171338
Davis, A., Sage, C. R., Wilson, L., and Farrell, K. W. (1993). Purification and Biochemical Characterization of Tubulin from the Budding Yeast Saccharomyces cerevisiae. Biochemistry 32, 8823–8835. doi:10.1021/bi00085a013
Dawson, P. J., Gutteridge, W. E., and Gull, K. (1983). Purification and Characterisation of Tubulin from the Parasitic Nematode, Ascaridia Galli. Mol. Biochem. Parasitol. 7, 267–277. doi:10.1016/0166-6851(83)90026-9
Denarier, E., Ecklund, K. H., Berthier, G., Favier, A., O’Toole, E. T., Gory-Fauré, S., et al. (2021). Modeling a Disease-Correlated Tubulin Mutation in Budding Yeast Reveals Insight into MAP-Mediated Dynein Function. MBoC 32, ar10. doi:10.1091/mbc.e21-05-0237
des Georges, A., Katsuki, M., Drummond, D. R., Osei, M., Cross, R. A., and Amos, L. A. (2008). Mal3, the Schizosaccharomyces pombe Homolog of EB1, Changes the Microtubule Lattice. Nat. Struct. Mol. Biol. 15, 1102–1108. doi:10.1038/nsmb.1482
Detrich, H. W., Johnson, K. A., and Marchese-Ragona, S. P. (1989). Polymerization of Antarctic Fish Tubulins at Low Temperatures: Energetic Aspects. Biochemistry 28, 10085–10093. doi:10.1021/bi00452a031
Detrich, H. W., and Overton, S. A. (1986). Heterogeneity and Structure of Brain Tubulins from Cold-Adapted Antarctic Fishes. Comparison to Brain Tubulins from a Temperate Fish and a Mammal. J. Biol. Chem. 261, 10922–10930. doi:10.1016/s0021-9258(18)67475-7
Detrich, H. W., Parker, S. K., Williams, R. C., Nogales, E., and Downing, K. H. (2000). Cold Adaptation of Microtubule Assembly and Dynamics. J. Biol. Chem. 275, 37038–37047. doi:10.1074/jbc.m005699200
Detrich, H. W., and Wilson, L. (1983). Purification, Characterization, and Assembly Properties of Tubulin from Unfertilized Eggs of the Sea Urchin Strongylocentrotus purpuratus. Biochemistry 22, 2453–2462. doi:10.1021/bi00279a023
Diao, L., Liu, M. Y., Song, Y. L., Zhang, X., Liang, X., and Bao, L. (2021). alpha1A and alpha1C Form Microtubules to Display Distinct Properties Mainly Mediated by Their C-Terminal Tails. J. Mol. Cel Biol 13, 864. doi:10.1093/jmcb/mjab062
Doenges, K. H., Nagle, B. W., Uhlmann, A., and Bryan, J. (1977). In Vitro assembly of Tubulin from Nonneural Cells (Ehrlich Ascites Tumor Cells). Biochemistry 16, 3455–3459. doi:10.1021/bi00634a025
Doenges, K. H., Weissinger, M., Fritzsche, R., and Schroeter, D. (1979). Assembly of Nonneural Microtubules in the Absence of Glycerol and Microtubule-Associated Proteins. Biochemistry 18, 1698–1702. doi:10.1021/bi00576a010
Dougherty, C. A., Sage, C. R., Davis, A., and Farrell, K. W. (2001). Mutation in the β-Tubulin Signature Motif Suppresses Microtubule GTPase Activity and Dynamics, and Slows Mitosis. Biochemistry 40, 15725–15732. doi:10.1021/bi010070y
Driver, J. W., Geyer, E. A., Bailey, M. E., Rice, L. M., and Asbury, C. L. (2017). Direct Measurement of Conformational Strain Energy in Protofilaments Curling Outward from Disassembling Microtubule Tips. Elife 6. doi:10.7554/eLife.28433
Drummond, D. R., Kain, S., Newcombe, A., Hoey, C., Katsuki, M., and Cross, R. A. (2011). Purification of Tubulin from the Fission Yeast Schizosaccharomyces pombe. Methods Mol. Biol. 777, 29–55. doi:10.1007/978-1-61779-252-6_3
Edzuka, T., and Goshima, G. (2019). Drosophila Kinesin-8 Stabilizes the Kinetochore-Microtubule Interaction. J. Cel Biol 218, 474–488. doi:10.1083/jcb.201807077
Farrell, K. W., and Wilson, L. (1978). Microtubule Reassembly In Vitro of Strongylocentrotus, Purpuratus Sperm Tail Outer Doublet Tubulin. J. Mol. Biol. 121, 393–410. doi:10.1016/0022-2836(78)90371-6
Feit, H., Slusarek, L., and Shelanski, M. L. (1971). Heterogeneity of Tubulin Subunits. Proc. Natl. Acad. Sci. U.S.A. 68, 2028–2031. doi:10.1073/pnas.68.9.2028
Findeisen, P., Mühlhausen, S., Dempewolf, S., Hertzog, J., Zietlow, A., Carlomagno, T., et al. (2014). Six Subgroups and Extensive Recent Duplications Characterize the Evolution of the Eukaryotic Tubulin Protein Family. Genome Biol. Evol. 6, 2274–2288. doi:10.1093/gbe/evu187
Fujita, S., Pytela, J., Hotta, T., Kato, T., Hamada, T., Akamatsu, R., et al. (2013). An Atypical Tubulin Kinase Mediates Stress-Induced Microtubule Depolymerization in Arabidopsis. Curr. Biol. 23, 1969–1978. doi:10.1016/j.cub.2013.08.006
Fulton, C., and Simpson, P. A. (1976). Selective Synthesis and Utilization of Flagellar Tubulin. The Multi-Tubulin Hypothesis. Cel Motil. 3, 987–1005.
Gadadhar, S., Bodakuntla, S., Natarajan, K., and Janke, C. (2017). The Tubulin Code at a Glance. J. Cel Sci 130, 1347–1353. doi:10.1242/jcs.199471
Gao, Y., Thomas, J. O., Chow, R. L., Lee, G.-H., and Cowan, N. J. (1992). A Cytoplasmic Chaperonin that Catalyzes β-actin Folding. Cell 69, 1043–1050. doi:10.1016/0092-8674(92)90622-j
Garnham, C. P., Vemu, A., Wilson-Kubalek, E. M., Yu, I., Szyk, A., Lander, G. C., et al. (2015). Multivalent Microtubule Recognition by Tubulin Tyrosine Ligase-like Family Glutamylases. Cell 161, 1112–1123. doi:10.1016/j.cell.2015.04.003
Garnham, C. P., Yu, I., Li, Y., and Roll-Mecak, A. (2017). Crystal Structure of Tubulin Tyrosine Ligase-like 3 Reveals Essential Architectural Elements Unique to Tubulin Monoglycylases. Proc. Natl. Acad. Sci. U.S.A. 114, 6545–6550. doi:10.1073/pnas.1617286114
Gasic, I., Boswell, S. A., and Mitchison, T. J. (2019). Tubulin mRNA Stability Is Sensitive to Change in Microtubule Dynamics Caused by Multiple Physiological and Toxic Cues. Plos Biol. 17, e3000225. doi:10.1371/journal.pbio.3000225
George, H. J., Misra, L., Field, D. J., and Lee, J. C. (1981). Polymorphism of Brain Tubulin. Biochemistry 20, 2402–2409. doi:10.1021/bi00512a006
Geyer, E. A., Burns, A., Lalonde, B. A., Ye, X., Piedra, F. A., Huffaker, T. C., et al. (2015). A Mutation Uncouples the Tubulin Conformational and GTPase Cycles, Revealing Allosteric Control of Microtubule Dynamics. Elife 4, e10113. doi:10.7554/eLife.10113
Geyer, E. A., Miller, M. P., Brautigam, C. A., Biggins, S., and Rice, L. M. (2018). Design Principles of a Microtubule Polymerase. Elife 7, e34574. doi:10.7554/eLife.34574
Gibbons, I. R., and Grimstone, A. V. (1960). On Flagellar Structure in Certain Flagellates. J. Biophys. Biochem. Cytol. 7, 697–716. doi:10.1083/jcb.7.4.697
Gibbons, I. R. (1963). Studies on the Protein Components of Cilia from Tetrahymena Pyriformis. Proc. Natl. Acad. Sci. U.S.A. 50, 1002–1010. doi:10.1073/pnas.50.5.1002
Gupta, M. L., Bode, C. J., Georg, G. I., and Himes, R. H. (2003). Understanding Tubulin-Taxol Interactions: Mutations that Impart Taxol Binding to Yeast Tubulin. Proc. Natl. Acad. Sci. U.S.A. 100, 6394–6397. doi:10.1073/pnas.1131967100
Gupta, M. L., Bode, C. J., Thrower, D. A., Pearson, C. G., Suprenant, K. A., Bloom, K. S., et al. (2002). β-Tubulin C354 Mutations that Severely Decrease Microtubule Dynamics Do Not Prevent Nuclear Migration in Yeast. MBoC 13, 2919–2932. doi:10.1091/mbc.e02-01-0003
Hibbel, A., Bogdanova, A., Mahamdeh, M., Jannasch, A., Storch, M., Schäffer, E., et al. (2015). Kinesin Kip2 Enhances Microtubule Growth In Vitro through Length-dependent Feedback on Polymerization and Catastrophe. Elife 4, e10542. doi:10.7554/eLife.10542
Hirst, W. G., Biswas, A., Mahalingan, K. K., and Reber, S. (2020). Differences in Intrinsic Tubulin Dynamic Properties Contribute to Spindle Length Control in Xenopus Species. Curr. Biol. 30, 2184–2190. e2185. doi:10.1016/j.cub.2020.03.067
Hirst, W. G., Fachet, D., Kuropka, B., Weise, C., Saliba, K. J., and Reber, S. (2022). Purification of Functional Plasmodium Falciparum Tubulin Allows for the Identification of Parasite-specific Microtubule Inhibitors. Curr. Biol. 32, 919. doi:10.1016/j.cub.2021.12.049
Hotta, T., Fujita, S., Uchimura, S., Noguchi, M., Demura, T., Muto, E., et al. (2016). Affinity Purification and Characterization of Functional Tubulin from Cell Suspension Cultures of Arabidopsis and Tobacco. Plant Physiol. 170, 1189–1205. doi:10.1104/pp.15.01173
Janke, C., and Chloë Bulinski, J. (2011). Post-translational Regulation of the Microtubule Cytoskeleton: Mechanisms and Functions. Nat. Rev. Mol. Cel Biol 12, 773–786. doi:10.1038/nrm3227
Janke, C., and Magiera, M. M. (2020). The Tubulin Code and its Role in Controlling Microtubule Properties and Functions. Nat. Rev. Mol. Cel Biol 21, 307–326. doi:10.1038/s41580-020-0214-3
Johnson, V., Ayaz, P., Huddleston, P., and Rice, L. M. (2011). Design, Overexpression, and Purification of Polymerization-Blocked Yeast αβ-Tubulin Mutants. Biochemistry 50, 8636–8644. doi:10.1021/bi2005174
Kearns, S., Mason, F. M., Rathmell, W. K., Park, I. Y., Walker, C., Verhey, K. J., et al. (2021). Molecular Determinants for α-tubulin Methylation by SETD2. J. Biol. Chem. 297, 100898. doi:10.1016/j.jbc.2021.100898
Keller, T. C., Jemiolo, D. K., Burgess, W. H., and Rebhun, L. I. (1982). Strongylocentrotus purpuratus Spindle Tubulin. II. Characteristics of its Sensitivity to Ca++ and the Effects of Calmodulin Isolated from Bovine Brain and S. purpuratus Eggs. J. Cel Biol 93, 797–803. doi:10.1083/jcb.93.3.797
Kilmartin, J. V. (1981). Purification of Yeast Tubulin by Self-Assembly In Vitro. Biochemistry 20, 3629–3633. doi:10.1021/bi00515a050
Kleiner, R. E., Ti, S.-C., and Kapoor, T. M. (2013). Site-specific Chemistry on the Microtubule Polymer. J. Am. Chem. Soc. 135, 12520–12523. doi:10.1021/ja405199h
Kollman, J. M., Greenberg, C. H., Li, S., Moritz, M., Zelter, A., Fong, K. K., et al. (2015). Ring Closure Activates Yeast γTuRC for Species-specific Microtubule Nucleation. Nat. Struct. Mol. Biol. 22, 132–137. doi:10.1038/nsmb.2953
Krauhs, E., Little, M., Kempf, T., Hofer-Warbinek, R., Ade, W., and Ponstingl, H. (1981). Complete Amino Acid Sequence of Beta-Tubulin from Porcine Brain. Proc. Natl. Acad. Sci. U.S.A. 78, 4156–4160. doi:10.1073/pnas.78.7.4156
LaFrance, B. J., Roostalu, J., Henkin, G., Greber, B. J., Zhang, R., Normanno, D., et al. (2022). Structural Transitions in the GTP Cap Visualized by Cryo-Electron Microscopy of Catalytically Inactive Microtubules. Proc. Natl. Acad. Sci. U S A. 119, e2114994119. doi:10.1073/pnas.2114994119
Langford, G. M. (1978). In Vitro assembly of Dogfish Brain Tubulin and the Induction of Coiled Ribbon Polymers by Calcium. Exp. Cel Res. 111, 139–151. doi:10.1016/0014-4827(78)90244-6
Ledbetter, M. C., and Porter, K. R. (1963). A "Microtubule" in Plant Cell Fine Structure. J. Cel Biol 19, 239–250. doi:10.1083/jcb.19.1.239
Ledbetter, M. C., and Porter, K. R. (1964). Morphology of Microtubules of Plant Cell. Science 144, 872–874. doi:10.1126/science.144.3620.872
Lewis, S. A., Tian, G., and Cowan, N. J. (1997). The α- and β-tubulin Folding Pathways. Trends Cell Biology 7, 479–484. doi:10.1016/s0962-8924(97)01168-9
Lin, Z., Gasic, I., Chandrasekaran, V., Peters, N., Shao, S., Mitchison, T. J., et al. (2020). TTC5 Mediates Autoregulation of Tubulin via mRNA Degradation. Science 367, 100–104. doi:10.1126/science.aaz4352
Lopata, M. A., and Cleveland, D. W. (1987). In Vivo microtubules Are Copolymers of Available Beta-Tubulin Isotypes: Localization of Each of Six Vertebrate Beta-Tubulin Isotypes Using Polyclonal Antibodies Elicited by Synthetic Peptide Antigens. J. Cel Biol 105, 1707–1720. doi:10.1083/jcb.105.4.1707
Lu, Q., and Luduena, R. F. (1994). In Vitro analysis of Microtubule Assembly of Isotypically Pure Tubulin Dimers. Intrinsic Differences in the Assembly Properties of Alpha Beta II, Alpha Beta III, and Alpha Beta IV Tubulin Dimers in the Absence of Microtubule-Associated Proteins. J. Biol. Chem. 269, 2041–2047. doi:10.1016/s0021-9258(17)42132-6
Ludueña, R. F., and Banerjee, A. (2008). “The Isotypes of Tubulin,” in The Role of Microtubules in Cell Biology, Neurobiology, and Oncology. Editor T. Fojo (Totowa, NJ, USA: Humana Press), 123–175.
Mahalingan, K. K., Keith Keenan, E., Strickland, M., Li, Y., Liu, Y., Ball, H. L., et al. (2020). Structural Basis for Polyglutamate Chain Initiation and Elongation by TTLL Family Enzymes. Nat. Struct. Mol. Biol. 27, 802–813. doi:10.1038/s41594-020-0462-0
Majumdar, S., Kim, T., Chen, Z., Munyoki, S., Tso, S.-C., Brautigam, C. A., et al. (2018). An Isolated CLASP TOG Domain Suppresses Microtubule Catastrophe and Promotes rescue. MBoC 29, 1359–1375. doi:10.1091/mbc.e17-12-0748
Mickolajczyk, K. J., Geyer, E. A., Kim, T., Rice, L. M., and Hancock, W. O. (2019). Direct Observation of Individual Tubulin Dimers Binding to Growing Microtubules. Proc. Natl. Acad. Sci. U.S.A. 116, 7314–7322. doi:10.1073/pnas.1815823116
Minoura, I., Hachikubo, Y., Yamakita, Y., Takazaki, H., Ayukawa, R., Uchimura, S., et al. (2013). Overexpression, Purification, and Functional Analysis of Recombinant Human Tubulin Dimer. FEBS Lett. 587, 3450–3455. doi:10.1016/j.febslet.2013.08.032
Minoura, I., Takazaki, H., Ayukawa, R., Saruta, C., Hachikubo, Y., Uchimura, S., et al. (2016). Reversal of Axonal Growth Defects in an Extraocular Fibrosis Model by Engineering the Kinesin-Microtubule Interface. Nat. Commun. 7, 10058. doi:10.1038/ncomms10058
Mohri, H. (1968). Amino-acid Composition of "Tubulin" Constituting Microtubules of Sperm Flagella. Nature 217, 1053–1054. doi:10.1038/2171053a0
Moriwaki, T., and Goshima, G. (2016). Five Factors Can Reconstitute All Three Phases of Microtubule Polymerization Dynamics. J. Cel Biol 215, 357–368. doi:10.1083/jcb.201604118
Nagle, B. W., Doenges, K. H., and Bryan, J. (1977). Assembly of Tubulin from Cultured Cells and Comparison with the Neurotubulin Model. Cell 12, 573–586. doi:10.1016/0092-8674(77)90258-6
Newton, C. N., DeLuca, J. G., Himes, R. H., Miller, H. P., Jordan, M. A., and Wilson, L. (2002). Intrinsically Slow Dynamic Instability of HeLa Cell Microtubules In Vitro. J. Biol. Chem. 277, 42456–42462. doi:10.1074/jbc.m207134200
Nogales, E. (2000). Structural Insights into Microtubule Function. Annu. Rev. Biochem. 69, 277–302. doi:10.1146/annurev.biochem.69.1.277
Olmsted, J. B., Witman, G. B., Carlson, K., and Rosenbaum, J. L. (1971). Comparison of the Microtubule Proteins of Neuroblastoma Cells, Brain, and Chlamydomonas Flagella. Proc. Natl. Acad. Sci. U.S.A. 68, 2273–2277. doi:10.1073/pnas.68.9.2273
Otani, K., Ishizaki, K., Nishihama, R., Takatani, S., Kohchi, T., Takahashi, T., et al. (2018). An Evolutionarily Conserved NIMA-Related Kinase Directs Rhizoid Tip Growth in the Basal Land Plant Marchantia Polymorpha. Development 145, 154617. doi:10.1242/dev.154617
Pamula, M. C., Ti, S.-C., and Kapoor, T. M. (2016). The Structured Core of Human β Tubulin Confers Isotype-specific Polymerization Properties. J. Cel Biol 213, 425–433. doi:10.1083/jcb.201603050
Panda, D., Miller, H. P., Banerjee, A., Ludueña, R. F., and Wilson, L. (1994). Microtubule Dynamics In Vitro Are Regulated by the Tubulin Isotype Composition. Proc. Natl. Acad. Sci. U.S.A. 91, 11358–11362. doi:10.1073/pnas.91.24.11358
Park, K., Hoff, K. J., Wethekam, L., Stence, N., Saenz, M., and Moore, J. K. (2021). Kinetically Stabilizing Mutations in Beta Tubulins Create Isotype-specific Brain Malformations. Front. Cel Dev. Biol. 9, 765992. doi:10.3389/fcell.2021.765992
Pease, D. C. (1963). The Ultrastructure of Flagellar Fibrils. J. Cel Biol 18, 313–326. doi:10.1083/jcb.18.2.313
Pham, C. L., and Morrissette, N. S. (2019). The Tubulin Mutation Database: A Resource for the Cytoskeleton Community. Cytoskeleton 76, 186–191. doi:10.1002/cm.21514
Phillips, D. M. (1966). Substructure of Flagellar Tubules. J. Cel Biol 31, 635–638. doi:10.1083/jcb.31.3.635
Piedra, F.-A., Kim, T., Garza, E. S., Geyer, E. A., Burns, A., Ye, X., et al. (2016). GDP-to-GTP Exchange on the Microtubule End Can Contribute to the Frequency of Catastrophe. MBoC 27, 3515–3525. doi:10.1091/mbc.e16-03-0199
Podolski, M., Mahamdeh, M., and Howard, J. (2014). Stu2, the Budding Yeast XMAP215/Dis1 Homolog, Promotes Assembly of Yeast Microtubules by Increasing Growth Rate and Decreasing Catastrophe Frequency. J. Biol. Chem. 289, 28087–28093. doi:10.1074/jbc.m114.584300
Ponstingl, H., Krauhs, E., Little, M., and Kempf, T. (1981). Complete Amino Acid Sequence of Alpha-Tubulin from Porcine Brain. Proc. Natl. Acad. Sci. U.S.A. 78, 2757–2761. doi:10.1073/pnas.78.5.2757
Pratt, L. F., Okamura, S., and Cleveland, D. W. (1987). A Divergent Testis-specific Alpha-Tubulin Isotype that Does Not Contain a Coded C-Terminal Tyrosine. Mol. Cel. Biol. 7, 552–555. doi:10.1128/mcb.7.1.552
Renaud, F. L., Rowe, A. J., and Gibbons, I. R. (1968). Some Properties of the Protein Forming the Outer Fibers of Cilia. J. Cel Biol 36, 79–90. doi:10.1083/jcb.36.1.79
Roostalu, J., Thomas, C., Cade, N. I., Kunzelmann, S., Taylor, I. A., and Surrey, T. (2020). The Speed of GTP Hydrolysis Determines GTP Cap Size and Controls Microtubule Stability. Elife 9, e51992. doi:10.7554/eLife.51992
Rostovtseva, T. K., Gurnev, P. A., Hoogerheide, D. P., Rovini, A., Sirajuddin, M., and Bezrukov, S. M. (2018). Sequence Diversity of Tubulin Isotypes in Regulation of the Mitochondrial Voltage-dependent Anion Channel. J. Biol. Chem. 293, 10949–10962. doi:10.1074/jbc.ra117.001569
Sánchez, F., Natzle, J. E., Cleveland, D. W., Kirschner, M. W., and McCarthy, B. J. (1980). A Dispersed Multigene Family Encoding Tubulin in Drosophila melanogaster. Cell 22, 845–854. doi:10.1016/0092-8674(80)90561-9
Shelanski, M. L., Gaskin, F., and Cantor, C. R. (1973). Microtubule Assembly in the Absence of Added Nucleotides. Proc. Natl. Acad. Sci. U.S.A. 70, 765–768. doi:10.1073/pnas.70.3.765
Shelanski, M. L., and Taylor, E. W. (1967). Isolation of a Protein Subunit from Microtubules. J. Cel Biol 34, 549–554. doi:10.1083/jcb.34.2.549
Shelanski, M. L., and Taylor, E. W. (1968). Properties of the Protein Subunit of central-pair and Outer-Doublet Microtubules of Sea Urchin Flagella. J. Cel Biol 38, 304–315. doi:10.1083/jcb.38.2.304
Sirajuddin, M., Rice, L. M., and Vale, R. D. (2014). Regulation of Microtubule Motors by Tubulin Isotypes and post-translational Modifications. Nat. Cel Biol 16, 335–344. doi:10.1038/ncb2920
Slautterback, D. B. (1963). Cytoplasmic Microtubules. J. Cel Biol 18, 367–388. doi:10.1083/jcb.18.2.367
Souphron, J., Bodakuntla, S., Jijumon, A. S., Lakisic, G., Gautreau, A. M., Janke, C., et al. (2019). Purification of Tubulin with Controlled post-translational Modifications by Polymerization-Depolymerization Cycles. Nat. Protoc. 14, 1634–1660. doi:10.1038/s41596-019-0153-7
Stephens, R. E., Renaud, F. L., Gibbons, I. R., and Stevens, R. E. (1967). Guanine Nucleotide Associated with the Protein of the Outer Fibers of Flagella and Cilia. Science 156, 1606–1608. doi:10.1126/science.156.3782.1606
Stephens, R. E. (1970). Thermal Fractionation of Outer Fiber Doublet Microtubules into A- and B-Subfiber Components: A- and B-Tubulin. J. Mol. Biol. 47, 353–363. doi:10.1016/0022-2836(70)90307-4
Sullivan, K. F., and Cleveland, D. W. (1986). Identification of Conserved Isotype-Defining Variable Region Sequences for Four Vertebrate Beta Tubulin Polypeptide Classes. Proc. Natl. Acad. Sci. U.S.A. 83, 4327–4331. doi:10.1073/pnas.83.12.4327
Sullivan, K. F. (1988). Structure and Utilization of Tubulin Isotypes. Annu. Rev. Cel. Biol. 4, 687–716. doi:10.1146/annurev.cb.04.110188.003351
Suprenant, K. A., and Rebhun, L. I. (1984). Purification and Characterization of Oocyte Cytoplasmic Tubulin and Meiotic Spindle Tubulin of the Surf Clam Spisula solidissima. J. Cel Biol 98, 253–266. doi:10.1083/jcb.98.1.253
Taylor, E. W. (1965). The Mechanism of Colchicine Inhibition of Mitosis. I. Kinetics of Inhibition and the Binding of H3-Colchicine. J. Cel Biol 25 (Suppl. l), 145–160. doi:10.1083/jcb.25.1.145
Theodorakis, N. G., and Cleveland, D. W. (1992). Physical Evidence for Cotranslational Regulation of Beta-Tubulin mRNA Degradation. Mol. Cel. Biol. 12, 791–799. doi:10.1128/mcb.12.2.791
Ti, S.-C., Alushin, G. M., and Kapoor, T. M. (2018). Human β-Tubulin Isotypes Can Regulate Microtubule Protofilament Number and Stability. Develop. Cel 47, 175–190. e175. doi:10.1016/j.devcel.2018.08.014
Ti, S.-C., Pamula, M. C., Howes, S. C., Duellberg, C., Cade, N. I., Kleiner, R. E., et al. (2016). Mutations in Human Tubulin Proximal to the Kinesin-Binding Site Alter Dynamic Instability at Microtubule Plus- and Minus-Ends. Develop. Cel 37, 72–84. doi:10.1016/j.devcel.2016.03.003
Ti, S. C., Wieczorek, M., and Kapoor, T. M. (2020). Purification of Affinity Tag-free Recombinant Tubulin from Insect Cells. STAR Protoc. 1, 100011. doi:10.1016/j.xpro.2019.100011
Tian, G., Huang, Y., Rommelaere, H., Vandekerckhove, J., Ampe, C., and Cowan, N. J. (1996). Pathway Leading to Correctly Folded β-Tubulin. Cell 86, 287–296. doi:10.1016/s0092-8674(00)80100-2
Tian, G., Lewis, S. A., Feierbach, B., Stearns, T., Rommelaere, H., Ampe, C., et al. (1997). Tubulin Subunits Exist in an Activated Conformational State Generated and Maintained by Protein Cofactors. J. Cel Biol 138, 821–832. doi:10.1083/jcb.138.4.821
Uchimura, S., Oguchi, Y., Hachikubo, Y., Ishiwata, S. i., and Muto, E. (2010). Key Residues on Microtubule Responsible for Activation of Kinesin ATPase. EMBO J. 29, 1167–1175. doi:10.1038/emboj.2010.25
Uchimura, S., Oguchi, Y., Katsuki, M., Usui, T., Osada, H., Nikawa, J.-i., et al. (2006). Identification of a strong Binding Site for Kinesin on the Microtubule Using Mutant Analysis of Tubulin. EMBO J. 25, 5932–5941. doi:10.1038/sj.emboj.7601442
Vainberg, I. E., Lewis, S. A., Rommelaere, H., Ampe, C., Vandekerckhove, J., Klein, H. L., et al. (1998). Prefoldin, a Chaperone that Delivers Unfolded Proteins to Cytosolic Chaperonin. Cell 93, 863–873. doi:10.1016/s0092-8674(00)81446-4
Valenstein, M. L., and Roll-Mecak, A. (2016). Graded Control of Microtubule Severing by Tubulin Glutamylation. Cell 164, 911–921. doi:10.1016/j.cell.2016.01.019
Vemu, A., Szczesna, E., Zehr, E. A., Spector, J. O., Grigorieff, N., Deaconescu, A. M., et al. (2018). Severing Enzymes Amplify Microtubule Arrays through Lattice GTP-Tubulin Incorporation. Science 361, 1504. doi:10.1126/science.aau1504
Vemu, A., Atherton, J., Spector, J. O., Moores, C. A., and Roll-Mecak, A. (2017). Tubulin Isoform Composition Tunes Microtubule Dynamics. MBoC 28, 3564–3572. doi:10.1091/mbc.e17-02-0124
Vemu, A., Atherton, J., Spector, J. O., Szyk, A., Moores, C. A., and Roll-Mecak, A. (2016). Structure and Dynamics of Single-Isoform Recombinant Neuronal Human Tubulin. J. Biol. Chem. 291, 12907–12915. doi:10.1074/jbc.c116.731133
Vemu, A., Garnham, C. P., Lee, D.-Y., and Roll-Mecak, A. (2014). Generation of Differentially Modified Microtubules Using In Vitro Enzymatic Approaches. Methods Enzymol. 540, 149–166. doi:10.1016/b978-0-12-397924-7.00009-1
Verhey, K. J., and Gaertig, J. (2007). The Tubulin Code. Cell Cycle 6, 2152–2160. doi:10.4161/cc.6.17.4633
Villasante, A., Wang, D., Dobner, P., Dolph, P., Lewis, S. A., and Cowan, N. J. (1986). Six Mouse Alpha-Tubulin mRNAs Encode Five Distinct Isotypes: Testis-specific Expression of Two Sister Genes. Mol. Cel. Biol. 6, 2409–2419. doi:10.1128/mcb.6.7.2409
von Loeffelholz, O., Peña, A., Drummond, D. R., Cross, R., and Moores, C. A. (2019). Cryo-EM Structure (4.5-Å) of Yeast Kinesin-5-Microtubule Complex Reveals a Distinct Binding Footprint and Mechanism of Drug Resistance. J. Mol. Biol. 431, 864–872. doi:10.1016/j.jmb.2019.01.011
von Loeffelholz, O., Venables, N. A., Drummond, D. R., Katsuki, M., Cross, R., and Moores, C. A. (2017). Nucleotide- and Mal3-dependent Changes in Fission Yeast Microtubules Suggest a Structural Plasticity View of Dynamics. Nat. Commun. 8, 2110. doi:10.1038/s41467-017-02241-5
Wang, D., Villasante, A., Lewis, S. A., and Cowan, N. J. (1986). The Mammalian Beta-Tubulin Repertoire: Hematopoietic Expression of a Novel, Heterologous Beta-Tubulin Isotype. J. Cel Biol 103, 1903–1910. doi:10.1083/jcb.103.5.1903
Weatherbee, J. A., Luftig, R. B., and Weihing, R. R. (1978). In Vitro polymerization of Microtubules from HeLa Cells. J. Cel Biol 78, 47–57. doi:10.1083/jcb.78.1.47
Weber, K., Koch, R., Herzog, W., and Vandekerckhove, J. (1977). The Isolation of Tubulin and Actin from Mouse 3T3 Cells Transformed by Simian Virus 40 (SV3T3 Cells), an Established Cell Line Growing in Culture. Eur. J. Biochem. 78, 27–32. doi:10.1111/j.1432-1033.1977.tb11710.x
Weingarten, M. D., Lockwood, A. H., Hwo, S. Y., and Kirschner, M. W. (1975). A Protein Factor Essential for Microtubule Assembly. Proc. Natl. Acad. Sci. U.S.A. 72, 1858–1862. doi:10.1073/pnas.72.5.1858
Weisenberg, R. C., Broisy, G. G., and Taylor, E. W. (1968). Colchicine-binding Protein of Mammalian Brain and its Relation to Microtubules. Biochemistry 7, 4466–4479. doi:10.1021/bi00852a043
Weisenberg, R. C. (1972). Microtubule Formation In Vitro in Solutions Containing Low Calcium Concentrations. Science 177, 1104–1105. doi:10.1126/science.177.4054.1104
Widlund, P. O., Podolski, M., Reber, S., Alper, J., Storch, M., Hyman, A. A., et al. (2012). One-step Purification of Assembly-Competent Tubulin from Diverse Eukaryotic Sources. MBoC 23, 4393–4401. doi:10.1091/mbc.e12-06-0444
Widlund, P. O., Stear, J. H., Pozniakovsky, A., Zanic, M., Reber, S., Brouhard, G. J., et al. (2011). XMAP215 Polymerase Activity Is Built by Combining Multiple Tubulin-Binding TOG Domains and a Basic Lattice-Binding Region. Proc. Natl. Acad. Sci. U.S.A. 108, 2741–2746. doi:10.1073/pnas.1016498108
Williams, R. C., Correia, J. J., and De Vries, A. L. (1985). Formation of Microtubules at Low Temperature by Tubulin from Antarctic Fish. Biochemistry 24, 2790–2798. doi:10.1021/bi00332a029
Wilson, P. G., and Borisy, G. G. (1997). Evolution of the Multi-Tubulin Hypothesis. Bioessays 19, 451–454. doi:10.1002/bies.950190603
Yaffe, M. B., Farr, G. W., Miklos, D., Horwich, A. L., Sternlicht, M. L., and Sternlicht, H. (1992). TCP1 Complex Is a Molecular Chaperone in Tubulin Biogenesis. Nature 358, 245–248. doi:10.1038/358245a0
Ye, X., Kim, T., Geyer, E. A., and Rice, L. M. (2020). Insights into Allosteric Control of Microtubule Dynamics from a Buried β‐tubulin Mutation that Causes Faster Growth and Slower Shrinkage. Protein Sci. 29, 1429–1439. doi:10.1002/pro.3842
Yen, T. J., Machlin, P. S., and Cleveland, D. W. (1988). Autoregulated Instability of β-tubulin mRNAs by Recognition of the Nascent Amino Terminus of βtubulin. Nature 334, 580–585. doi:10.1038/334580a0
Yoon, Y., and Oakley, B. R. (1995). Purification and Characterization of Assembly-Competent Tubulin from Aspergillus nidulans. Biochemistry 34, 6373–6381. doi:10.1021/bi00019a016
Yu, N., and Galjart, N. (2018). TAPping into the Treasures of Tubulin Using Novel Protein Production Methods. Essays Biochem. 62, 781–792. doi:10.1042/ebc20180033
Yu, N., Signorile, L., Basu, S., Ottema, S., Lebbink, J. H. G., Leslie, K., et al. (2016). Isolation of Functional Tubulin Dimers and of Tubulin-Associated Proteins from Mammalian Cells. Curr. Biol. 26, 1728–1736. doi:10.1016/j.cub.2016.04.069
Keywords: tubulin, microtubules, tubulin isotypes, tubulin protein biochemistry, recombinant tubulin
Citation: Ti S-C (2022) Reconstituting Microtubules: A Decades-Long Effort From Building Block Identification to the Generation of Recombinant α/β-Tubulin. Front. Cell Dev. Biol. 10:861648. doi: 10.3389/fcell.2022.861648
Received: 24 January 2022; Accepted: 18 April 2022;
Published: 28 April 2022.
Edited by:
Jeffrey Moore, University of Colorado Anschutz Medical Campus, United StatesReviewed by:
Luke Rice, University of Texas Southwestern Medical Center, United StatesJawdat Al-Bassam, University of California, Davis, United States
Mohan Gupta, Iowa State University, United States
Copyright © 2022 Ti. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shih-Chieh Ti, amVmZnRpQGhrdS5oaw==
 Shih-Chieh Ti
Shih-Chieh Ti