- 1Department of Medicine, University of Illinois at Chicago, Chicago, IL, United States
- 2Department of Preventive Medicine, Northwestern University, Chicago, IL, United States
- 3Research Informatics Core, Research Resources Center, University of Illinois at Chicago, Chicago, IL, United States
- 4Department of Neurology, Mayo Clinic, Rochester, MN, United States
The voltage-gated proton channel HVCN1 is a member of the voltage-gated ion channel family. HVCN1 channel controls acid extrusion and regulates pH homeostasis in various cell types. Recent evidence indicated that the HVCN1 channel was associated with cardiac function. To investigate the role of HVCN1 in cardiac myocytes, we performed an RNA sequencing analysis of murine hearts and showed that HVCN1 null hearts exhibited a differential transcriptome profile compared with wild-type hearts. The RNA-seq data indicating impaired pH homeostasis in HVCN1 null hearts were the downregulated NADPH oxidoreductases (NOXs) and decreased expression of Cl−/HCO3− exchanger, indicating HVCN1 is a regulator of gene transcriptional networks controlling NOX signaling and CO2 homeostasis in the heart. Additionally, HVCN1 null hearts exhibited differential expression of cardiac ion channels, suggesting a potential role of HVCN1 in cardiac electrophysiological remodeling. The study highlights the importance of HVCN1 in cardiac function and may present a novel target associated with heart diseases.
Introduction
The voltage-gated proton channel HVCN1 is a member of the voltage-gated ion channel family (Ramsey et al., 2006; Sasaki et al., 2006). It is composed of two subunits. Each subunit contains a proton-permeable voltage-sensing domain and lacks the pore domain typical of other voltage-gated ion channels. The HVCN1 proton channel is highly selective for H+ (Berger and Isacoff, 2011; Musset et al., 2011), and plays a crucial role in regulating pH homeostasis in various cell types (DeCoursey, 2013).
The HVCN1 channel extrudes protons during the respiratory burst of the NADPH oxidoreductases (NOXs) in phagocytes. It provides charge and pH compensation and controls the production of reactive oxygen species (ROS) and H2O2 by NOX (Seredenina et al., 2015). In the nervous system, HVCN1 is required for NOX-dependent ROS/H2O2 generation in brain microglia in the central nervous system (Wu et al., 2012; Peng et al., 2021). HVCN1 is a sperm flagellar regulator of intracellular pH and plays a crucial role in sperm capacitation (Lishko et al., 2010). It mediates H+ efflux at the pulmonary alveolar cell membrane and acidifies excessively alkaline airway surface liquid in the airway cells (Iovannisci et al., 2010). In addition, HVCN1 activity is required for acid extrusion to shape action potentials in snail neurons (Thomas and Meech, 1982), and contributes to efficient proton efflux in algal cells to sustain intracellular calcification in coccolithophores (Taylor et al., 2011). We previously developed guanidine derivatives as HVCN1 inhibitors (Hong et al., 2013; Hong et al., 2014; Zhao et al., 2021a; Zhao et al., 2021b). Pharmacologic inhibition of HVCN1 activity by these blockers has been reported to alter sperm motility (Matamoros-Volante and Trevino, 2020; Yeste et al., 2020), promote leukemic Jurkat T cell apoptosis (Asuaje et al., 2017), and inhibit breast cancer progression (Ventura et al., 2020).
Recently, accumulating evidence indicated that the HVCN1 channel was involved in cardiac function. A previous study reported that a voltage-activated proton current in human cardiac fibroblasts regulated intracellular pH and membrane potential (El Chemaly et al., 2006). Another study showed that the change of cardiac pHi was voltage-sensitive (Yamamoto et al., 2005). Recent transcriptome analyses indicated that the voltage-gated proton channel HVCN1 mRNAs were expressed in the heart at high levels comparable to Cl−/HCO3− exchangers (Brawand et al., 2011; Yu et al., 2014; Vairamani et al., 2017). These studies suggest a potential role of voltage-gated proton channel HVCN1 in cardiac pH homeostasis.
To explore the effects of HVCN1 gene deletion on cardiac function, we performed an RNA-profiling analysis of HVCN1 null and wild-type mouse hearts. The HVCN1−/− hearts showed a significantly differential transcriptome profile. The deletion of HVCN1 altered the expression of cardiac NADPH oxidoreductases, bicarbonate transporter, and ion channels associated with the cardiac electrophysiological profile.
Materials and Methods
Animals
The experimental procedures in mice and the protocol used in this study were approved by the University of Illinois at Chicago (UIC) Animal Care Committee (ACC No. 19-178). All animal studies were performed according to approved guidelines for the use and care of live animals, and conformed with animal care policies and procedures of UIC. The HVCN1−/− and wild-type (C57BL/6 background) mice were used in the study. Mice were housed in temperature- and humidity-controlled rooms with 12-h light-dark cycles in the animal care facility at the UIC. We studied male mice, similarly aged HVCN1−/− mice (n = 4, 3 months old) and wild-type control mice (n = 4, 3 months old) were euthanized and heart samples were isolated for analysis.
RNA-Seq Analysis
The RNA-seq analysis was performed as previously described (Hong et al., 2021). We first extracted RNA with Maxwell® RSC simplyRNA Cells Kit (Promega AS1390) based on manufacturer’s instructions, using Maxwell® RSC Instrument (Promega AS4500), and then RNA was quantified using Qubit 4.0 Fluorometer (Invitrogen) with the Qubit RNA HS Assay Kit (REF Q32855) analyzed for integrity using the Agilent 4200 TapeStation RNA ScreenTape assay (Agilent 5067–5576, 5067–5577). RNA samples were normalized to 250ng, and library prep was carried out using the universal Plus mRNA-Seq kit (NuGen 0520-A01) as written in the product manual (NuGen M01485 v5). In brief, RNA underwent poly-A selection, enzymatic fragmentation, and generation of double-stranded cDNA using a mixture of Oligo (dT) and random priming. The cDNA underwent end repair, ligation of dual-index adaptors, strand selection, and 15 cycles of PCR amplification. We determined the number of cycles by qPCR of a small aliquot of un-amplified libraries. All intermediate purification steps, and final library purification were carried out using Agencourt AMPure XP Beads (Beckman Coulter A63881). We measured purified library concentrations with the Qubit 1X dsDNA HS Assay Kit (Invitrogen Q33231), and fragment size distribution was confirmed using the D5000 ScreenTape assay (Agilent 5067–5588, 5067–5589), and libraries were pooled in equimolar amounts based on the Qubit concentration and TapeStation average size and run on MiniSeq for index balancing. The libraries were re-pooled with corrected inputs based on the % Reads Identified (PF) results from the MiniSeq run, and the new pool was purified with the Agencourt AMPure XP Beads (Beckman Coulter A63881). The final, purified pool was quantified by qPCR using the KAPA Library Quantification Kit and run on a NovaSeq6000 SP flow cell, 2 × 50 nt, one lane, at the University of Illinois Roy J. Carver Biotechnology Center High-Throughput Sequencing and Genotyping Unit. Raw reads were aligned to the reference genome hg38 using STAR (Dobin et al., 2013). ENSEMBL gene expression was quantified using FeatureCounts (Liao et al., 2014). Normalized and differential expression statistics were computed using edgeR (Robinson et al., 2010; McCarthy et al., 2012), and p-values were adjusted for multiple testing using the false discovery rate (FDR) correction of Benjamini and Hochberg. We performed unsupervised hierarchical clustering of all differentially expressed genes using the Euclidean distance and complete linkage method, and generated volcano plots using R. Up- and down-regulated genes were analyzed separately using the DAVID functional annotation tool and the Gene Ontology Biological Process (GO BP) database. Bar charts of the top 20 most significant upregulated and downregulated pathways, based on FDR-corrected p-value, were constructed with the ggplot2 R package. We constructed heatmaps for the cardiac conduction and calcium ion transport into cytosol pathways with the ComplexHeatmap R package.
Quantitative Real-Time PCR (RT-PCR) Analysis
RT-PCR analysis was performed as previously described (Kim et al., 2014). Total RNA was isolated from hearts of adult wild-type or HVCN1−/− mice using Trizol RNA extraction agent (Invitrogen) according to the manufacturer’s instructions. For the reverse transcription, total RNA was quantified using the Nanodrop spectrophotometer (Thermofisher Scientific). Synthesis of cDNA was carried out with SuperScript IV RNase Reverse transcriptase (Invitrogen) and primers with 2 μg of total RNA as template. Real-time PCR was performed on an Applied Biosystems (Thermofisher Scientific) to determine mRNA levels of differentially expressed genes according to the manufacturer’s recommended protocol using SYBR green assays. For each group, data were collected from three independent samples; three replicas were performed for each sample. The primer sequences used for SYBR green-based fluorescence were GAPDH: 5′-AGGTCGGTGTGAACGGATTTG-3′ and 5′-TGTAGACCATGTAGTTGAGGTCA-3′; NOX1: 5′-TTCCTCACTGGCTGGGATAG-3′ and 5′-AGTCCGAGGGCCACATAAGA-3′; NOX2: 5′-TGGCGATCTCAGCAAAAGGTGG-3′ and 5′-GTACTGTCCCACCTCCATCTTG-3′; NOX4: 5′-TCTGGAAAACCTTCCTGCTG-3′ and 5′-CCGGCACATAGGTAAAAGGA-3′; SLC4A1: 5′-CCGTGAACTCTTCATTGCTGCC-3′ and 5′-ACCAGGAACAGCAAGCTCATGC-3′; SLC4A2: 5′-GCACCTCCATTCTGTTTGCGGT-3′ and 5′-GCTCACGAACTTCCAACACAGC-3′; SLC4A3: 5′-GGTGCTGATTGCCTTCTCCAGT-3′ and 5′-GACAACGAAGCCAGAGGAGAAG-3′; SLC26A6: 5′-TACCGTGTGGACAGTAACCAGG-3′ and 5′-CCTGTACCAAGCTCCGAGACAT-3′.
Data and Statistical Analysis
All data were presented as the mean ± SEM. The normality in each group was determined by the Shapiro-Wilk test, and p > 0.05 was considered to indicate normally distributed. Homoscedasticity was determined by the two-sample F test. For equal variances between two groups, significance between means was determined by Student’s t-test. For unequal variances between groups, significance between means was determined by Welch’s t-test. p < 0.05 was considered to indicate a statistically significant difference.
Results
RNA Sequencing Analysis Revealed a Differential Transcriptome Profile in HVCN1−/− Mouse Hearts
To explore the potential function of HVCN1 in the heart, we performed a transcriptomic analysis of wild-type (WT) and HVCN1−/− mouse hearts. The HVCN1 null hearts exhibited a significantly different transcriptome profile compared with WT hearts (Figure 1 and Supplementary Table S1). Hierarchical clustering of differentially expressed genes (DEGs) in WT and HVCN1−/− hearts showed two main clusters with samples of the same group clustered together (Figures 1A,B). A total of 206 DEGs were identified with 67 downregulated and 139 upregulated (Figure 1C). The major Gene Ontology (GO) categories identified by RNA sequencing and the most significantly modulated in the HVCN1−/− mouse hearts included immunological activity, ion and receptor binding, metabolic process, and ion channel activity (Figures 1D,E).
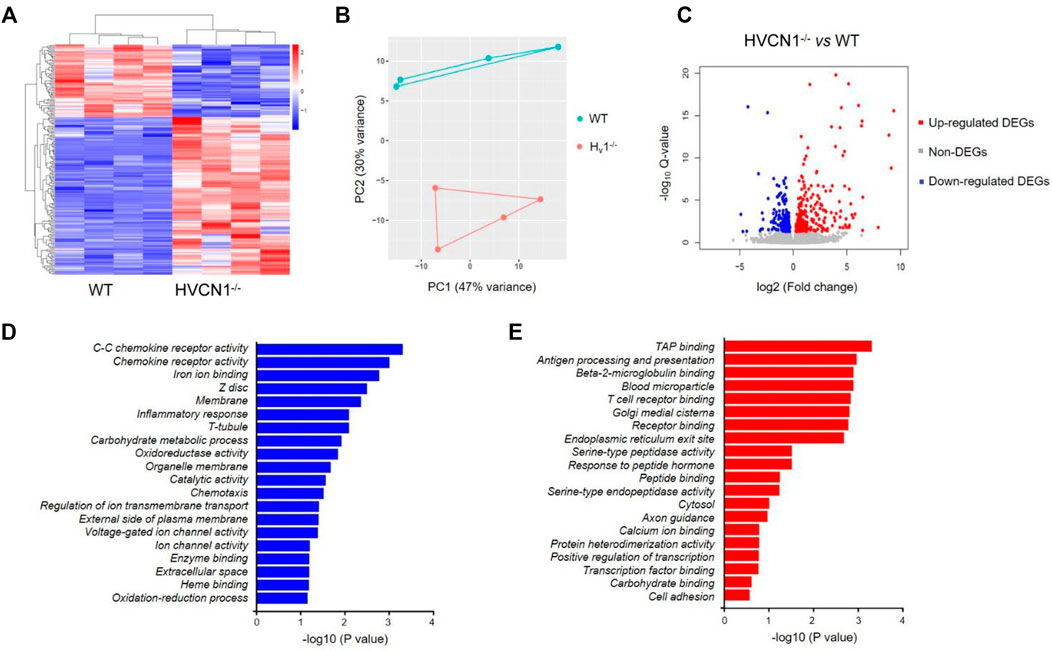
FIGURE 1. RNA sequencing analysis revealed a differential transcriptome profile in HVCN1−/− mouse heart. (A) Heatmap and hierarchical clustering of DEGs in the wild-type and HVCN1−/− hearts, n = 4 hearts for each group. (B) Principal component analysis of wild-type and HVCN1−/− hearts. (C) The volcano plot has red points representing upregulated DEGs, blue representing downregulated DEGs, and gray points representing non-DEGs. (D-E) Gene Ontology enrichment analysis of top significant gene groups either downregulated (D) or upregulated (E).
Downregulation of the NOX Family of ROS-Generating NADPH Oxidases in HVCN1−/− Hearts
It is noted that both oxidoreductase activity and oxidation-reduction process were downregulated in HVCN1 null hearts from the GO enrichment analysis of top significant gene groups (Figure 1D). The oxidoreductase utilizes NADP+ or NAD+ as cofactors to catalyze the transfer of electron between molecules, and HVCN1 provides charge and pH compensation during the NADPH oxidoreductases (NOXs) activation (Seredenina et al., 2015). Since HVCN1 was coupled with NOX activity, we determined the effects of HVCN1 deletion on the expression of NOX family. We detected three isoforms of the NOX family in the hearts, including NOX1, NOX2, and NOX4 (Figure 2). The RNA-seq results showed that three isoforms were downregulated in HVCN1 null hearts (Figures 2A–C). RT-PCR confirmed changes in RNA-seq, and the mRNA expression levels of all three NOX isoforms were markedly reduced in the HVCN1−/− hearts compared with the ones in the WT hearts (Figures 2D–F), indicating that deletion of HVCN1 was associated with gene expression of the NOX family.
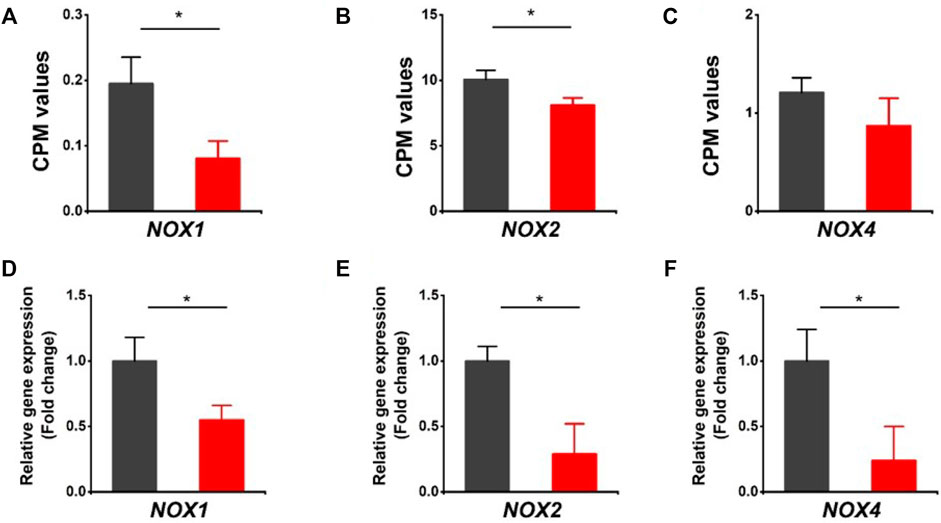
FIGURE 2. Downregulation of the NOX family of ROS-generating NADPH oxidases in HVCN1−/− hearts. Comparing relative gene expression levels of NOX1, NOX2, and NOX4 between WT (black) and HVCN1−/− (red) detected by RNA-seq (A-C) and qPCR (D-F). n = 4 independent experiments for RNA-seq; n = 3 independent experiments for qPCR; *p < 0.05.
Differentially Expressed Genes Related to Cl−/HCO3− Exchangers
The HVCN1 has been proposed to combine with HCO3− exchangers in regulating transport-mediated CO2 disposal in the heart (Vairamani et al., 2017). We assessed the expression of bicarbonate transporters in the HVCN1 null hearts. The SLC4A1 (encoding Anion Exchanger 1, AE1), SLC4A2 (encoding Anion Exchanger 2, AE2), SLC4A3 (encoding Anion Exchanger 3, AE3), and SLC26A6 are the most abundant Cl−/HCO3− exchangers expressed in hearts (Figures 3A–D). The RNA-profiling and RT-PCR results showed HVCN1−/− significantly downregulated SLC26A6 (Figures 3D–H), but did not significantly affect the AE1 and AE3 (Figures 3A,C,E, G). Although RNA-seq data indicated increased AE2 in HVCN1 null hearts (Figure 3B), the RT-PCR analysis showed no significant alterations of AE2 expression between groups (Figure 3F). These data suggested that HVCN1 might affect the expression of SLC26A6 in the regulation of cardiac CO2 homeostasis.
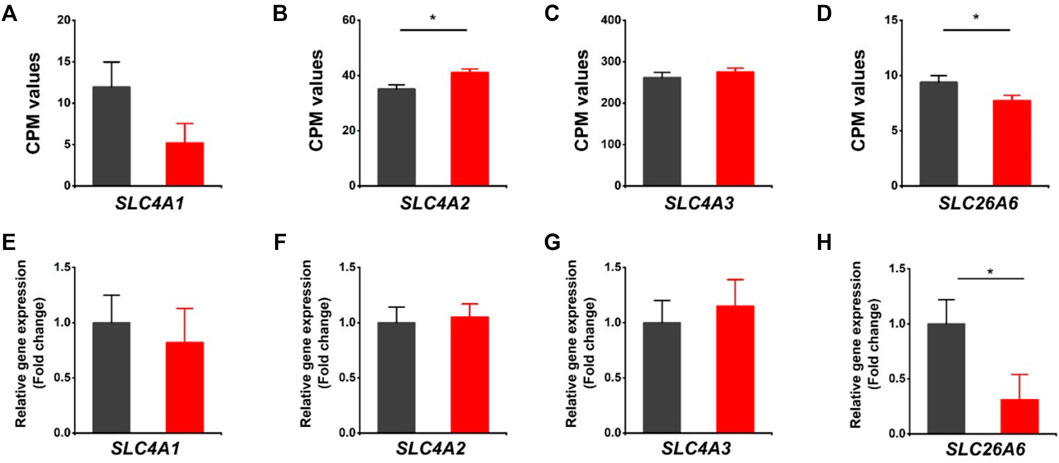
FIGURE 3. Differentially expressed genes related to Cl−/HCO3- exchangers in HVCN1−/− hearts. Comparing relative gene expression levels of SLC4A1 (AE1), SLC4A2 (AE2), SLC4A3 (AE3), SLC26A6 between WT (black) and HVCN1−/− (red) detected by RNA-seq (A-D) and qPCR (E-H). n = 4 independent experiments for RNA-seq; n = 3 independent experiments for qPCR; *p < 0.05.
Differentially Expressed Genes Related to Other Proton Channels and Proton-Coupled Transporters/Antiporters
The effects of HVCN1 proton channel deletion on other proton channels/exchangers and proton-coupled transporters/antiporters were investigated. The sodium-hydrogen exchanger 1 (NHE1) has been reported to regulate intracellular pH in cardiomyocytes (Sundset et al., 2003). Our RNA-seq revealed that NHE1 was upregulated in HVCN1 null hearts. The increased expression of NHE1 might be a functional compensation for the H+ transport burden due to the deletion of the HVCN1 proton channel (Figure 4A). Additionally, proton pump (V-type proton ATPase, ATP6V0A1) and proton channel otopetrin 1 (OTOP1) were not altered in the HVCN1 null hearts (Figures 4B,C).
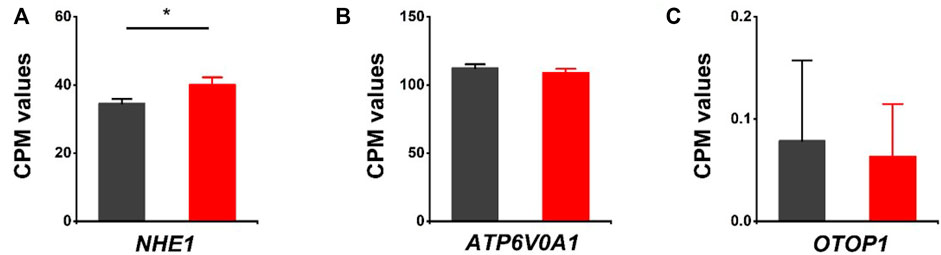
FIGURE 4. Differentially expressed genes related to other proton transporters/channels in HVCN1−/− hearts. Comparing relative gene expression levels of proton transporters between WT (black) and HVCN1−/− (red) including sodium-hydrogen exchanger 1 (NHE1) (A), a1-subunit of the V0 domain of V-type proton ATPase (ATP6V0A1) (B), and otopetrin proton channel 1 (OTOP1) (C), n = 4 independent experiments for RNA-seq; *p < 0.05.
We further assessed the expression of acid-sensing ion channels (ASIC1, ASIC2, ASIC3, ASIC4), proton activated chloride channel (TMEM206), proton-coupled metal-ion transporters (SLC11A1, SLC11A2), proton-coupled amino acid transporters (SLC36A1, SLC36A2, SLC36A4), and proton/amine antiporters (SLC18A1, SLC18A2). Except for ASIC3, there were no notable changes in those genes associated with the HVCN1 deletion (Figures 5A–L). Moreover, the proton-dependent oligopeptide transporters showed differential expression linked with the HVCN1−/−. Although the expression of SLC15A2 was not changed, the expression of SLC15A3 was decreased, and SLC15A4 was increased (Figures 5M–O), indicating a role of HVCN1 in differential regulation of proton-dependent oligopeptide transport.
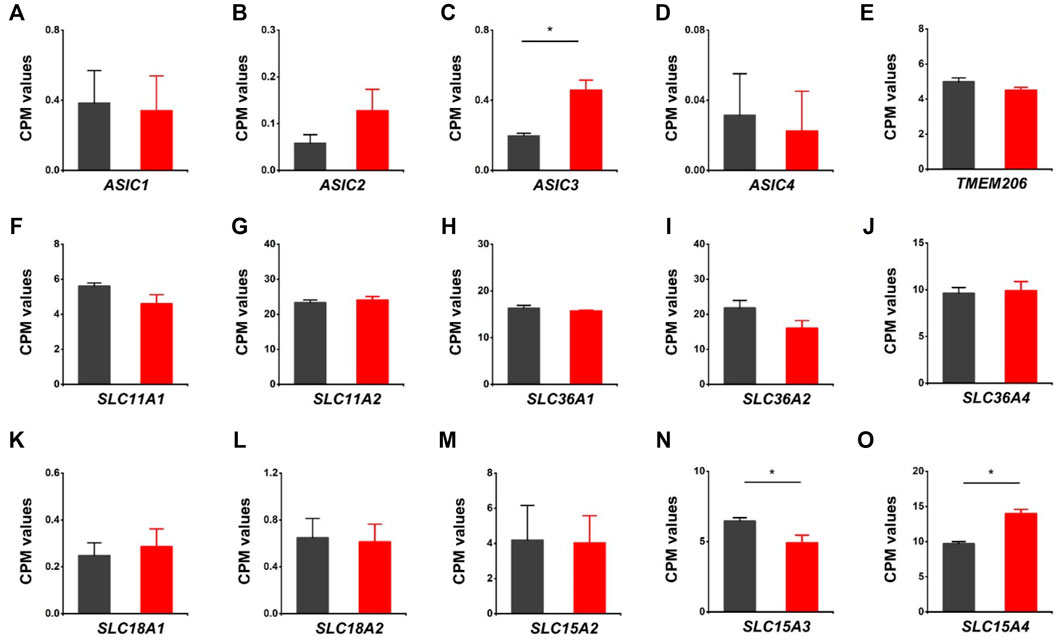
FIGURE 5. Differentially expressed genes related to proton-coupled channels/transporters/antiporters in HVCN1−/− hearts. Comparing relative gene expression levels of proton-coupled transporters between WT (black) and HVCN1−/− (red) including acid-sensing ion channels ASIC1, ASIC2, ASIC3, ASIC4 (A-D), proton activated chloride channel TMEM206 (E), proton-coupled metal-ion transporters SLC11A1, SLC11A2 (F-G), proton-coupled amino acid transporters SLC36A1, SLC36A2, SLC36A4 (H-J), proton/amine antiporters SLC18A1, SLC18A2 (K-L), and proton-dependent oligopeptide transporters SLC15A2, SLC15A3, SLC15A4 (M-O). n = 4 independent experiments for RNA-seq; *p < 0.05.
Differentially Expressed Genes Related to Cardiac Ion Channels in HVCN1−/− hearts
We evaluated the effects of HVCN1 deletion on the expression profiles of cardiac ion channels, including cardiac sodium channel alpha subunit (SCN5A), sodium channel beta subunits SCN1B and SCN2B, L-type and T-type calcium channels (CACNA1C, CNCNA1G), IKs, IKr, Ito, IK1, IKur potassium channels (KCNQ1, KCNH2, KCNE1, KCNE3, KCNE4, KCND2, KCND3, KCNA4, KCNJ2, KCNJ12, KCNA5), If channels (HCN2, HCN4), and sarcoplasmic reticulum calcium handling proteins (RYR2, SERCA pump) (Figure 6). The knockout hearts exhibited upregulation of L-type calcium channel CACNA1C, potassium channel beta subunit KCNE3, IKur potassium channel KCNA5, If channel HCN2 (Figures 6D,I, P, Q), and downregulation of SERCA pump ATPA2 (Figure 6T). The data supported that HVCN1 was involved in the regulation of cardiac electrophysiology, and HVCN1 deletion potentially remodeled action potential profiles in cardiomyocytes.
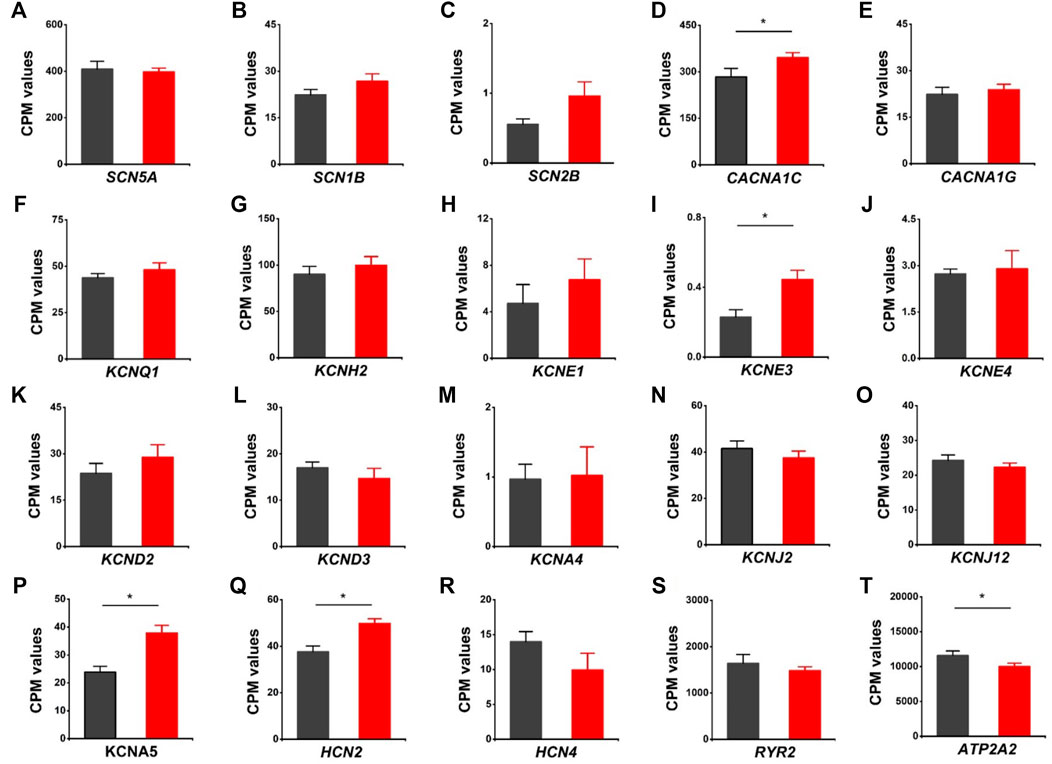
FIGURE 6. Differentially expressed genes related to cardiac ion channels in HVCN1−/− hearts. Comparing relative gene expression levels of cardiac ion channels between WT (black) and HVCN1−/− (red) including cardiac sodium channel SCN5A (A), sodium channel beta subunits SCN1B and SCN2B (B-C), cardiac L-type calcium channel CACNA1C (D), T-type calcium channel CACNA1G (E), cardiac potassium channels KCNQ1 (F), KCNH2 (G), potassium channel beta subunits KCNE1, KCNE3, KCNE4 (H-J), Ito channels KNCD2, KCND3, KCNA4 (K-M), IK1 channels KCNJ2, KCNJ12 (N-O), Ikur channel KCNA5 (P), If channels HCN2, HCN4 (Q-R), and cardiac ryanodine receptor RYR2 (S) and SERCA pump ATPA2 (T). n = 4 independent experiments for RNA-seq; *p < 0.05.
Discussion
The present study suggested that HVCN1 regulated gene transcriptional networks controlling NOX signaling and CO2 homeostasis in the heart. The RNA-profiling data indicating impaired pH homeostasis in the HVCN1−/− hearts were the downregulated NADPH oxidoreductases (NOXs), decreased expression of Cl−/HCO3− exchanger SLC26A6, and differential expression of cardiac ion transporters/channels.
HVCN1 channel regulates NOX-mediated respiratory burst to control the production of ROS/H2O2 in the cells. The NOX family has seven members, including NOX1, NOX2, NOX3, NOX4, NOX5, Duox1, and Duox2 (Bedard et al., 2015). During the respiratory burst, NOX uses NADPH as an electron donor and molecular oxygen as an electron acceptor to produce O2−. This activity leaves excess proton (H+) in the cytoplasm. Proton extrusion by the HVCN1 channel compensates for the electrogenic activity of NOX and limits the inhibition of the enzyme induced by cytoplasmic acidification, thus favoring the efficient generation of ROS/H2O2 from O2 (Seredenina et al., 2015).
Three isoforms, including NOX1, NOX2, and NOX4, were detected in mouse hearts. We showed that HVCN1 deletion significantly influenced the expression of the NOX family, and mRNA expression levels of all NOX isoforms were reduced in the HVCN1−/− hearts. The downregulated NOXs likely generate disturbance in the production of H2O2. The NOX-medicated ROS signaling pathways regulate heart diseases’ physiological or pathophysiological processes (Pena et al., 2020; Zhang et al., 2020). It was reported that NOX2 activity was associated with angiotensin II-induced myocyte hypertrophy (Hingtgen et al., 2006), and NOX1 mediated myocyte death under stress situations (Matsuno et al., 2012). Moreover, high-level expression of NOX4 was shown to play essential roles in hypoxia and heart failure (Kuroda et al., 2010; Zhang et al., 2010). Our results suggested a potential role of the HVCN1 channel in heart diseases, such as hypertrophy and heart failure associated with NOX-medicated ROS signaling pathways. In pathophysiological conditions, inflammation and excessive stress cause cardiac dysfunction. As compensation, myocyte cells generate more ROS to activate cellular signaling pathways to counteract the abnormal stress and stimulation. In the long run, the activation of these pathways leads to myocardial hypertrophy via H2O2 (Zafari et al., 1998), which is the primary ROS regulated by HVCN1.
The HVCN1 channel was implicated in HCO3− transport in lung epithelium and regulated airway surface liquid pH (Fischer, 2012). Studies reported that HCO3− and HCO3−-handling proteins played essential roles in regulating cardiac function (Wang et al., 2014). Recent transcriptome analyses indicated that the voltage-gated proton channel HVCN1 mRNAs were expressed in the heart at high levels comparable to those of Cl−/HCO3− exchangers (Brawand et al., 2011; Yu et al., 2014; Vairamani et al., 2017), and Cl−/HCO3− exchanger AE3 was proposed to combine with HVCN1-mediated H+ currents to generate transport-mediated CO2 disposal in the heart (Vairamani et al., 2017). The present study revealed that HVCN1−/− significantly downregulated SLC26A6 but did not influence the expression of AE3. We speculate that HVCN1 could alter the expression of SLC26A6 regulating cardiac CO2 homeostasis (Figure 7).
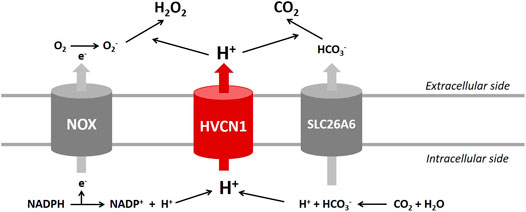
FIGURE 7. Proposed roles of HVCN1 in regulating pH homeostasis in the heart. Proton (H+) extrusion by the HVCN1 channel compensates for the electrogenic activity of NOX and regulates the efficient generation of H2O2. In addition, HVCN1-mediated H+ current is associated with transport-mediated CO2 disposal. Except for HVCN1 functional regulation of pH homeostasis, the present study revealed that HVCN1 regulated the genetic expression of NOXs and SLC26A6 in the heart.
Additionally, the RNA-seq results provided limited support that HVCN1 was involved in modulating cardiac electrophysiology. The HVCN1 knockout hearts exhibited differential expression of cardiac ion channels, including upregulated L-type calcium channel, IKur, HCN2, and downregulated SERCA pump. These results indicated that HVCN1 was associated with the electrophysiological remodeling in cardiomyocytes, and HVCN1 might regulate cardiac ion channels function like other channel partners (Hong et al., 2020; Wu and Hong, 2021). Previous studies reported that deficits of the HVCN1 channel by the gene deletion or pharmacological block produced intracellular acidification (low pHi) in a variety of cell types (Cherny and DeCoursey, 1999; Wu et al., 2012; Asuaje et al., 2017). The deletion of HVCN1 likely also alters the intracellular pH in myocytes. It was known that cellular pH was an essential modulator of cardiac function, and low pHi regulated many cardiac ion channel functions (Vaughan-Jones et al., 2009). The low pHi reduced transient outward K+ currents (Ito) current amplitude (Saegusa et al., 2013), stimulated L-type calcium channel (ICa,L) gating process (Saegusa et al., 2011), and weakened rectification of the cardiac KATP channel (Baukrowitz et al., 1999). Through effects on the Ca2+ and K+ conductance pathways, low pHi modified action potential (AP) profiles and induced cardiac arrhythmias. Moreover, the low pHi-induced Ca2+-overload could result in mitochondrial dysfunction and defects in contractility, accounting for cardiac hypertrophy and heart failure (Duchen et al., 2008). However, the effects of HVCN1-mediated pH homeostasis on the cardiac ion channel function remain unclear. Studies in the field will help explore the role of HVCN1 in cardiac electrophysiology.
In summary, the present study highlights the importance of HVCN1 in cardiac function and may present a novel target associated with heart diseases. The RNA-seq data indicated that HVCN1 was involved in cardiac pH homeostasis (Figure 7) and showed that HVCN1 regulated gene transcriptional networks controlling NOX signaling and CO2 homeostasis, suggesting a potential role of the HVCN1 channel in heart failure and cardiomyopathy, which are associated with abnormal expression of NOX and bicarbonate transporters. Additionally, HVCN1 dysfunction caused intracellular acidification in various cells. As the cellular pH has a crucial influence on cardiac contractility and rhythm, the pH disturbances causing reversible contractile dysfunction are linked with cardiac arrhythmias. Future functional studies on the effects of HVCN1 on cardiac electrophysiology are required to address these uncertainties.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: GEO, GSE195945.
Ethics Statement
The animal study was reviewed and approved by the University of Illinois at Chicago (UIC) Animal Care Committee.
Author Contributions
LH conceived the project and designed the experiments. XW performed experiments. XW and LH wrote the manuscript. LW provided HVCN1−/− mice, XW, YL, MM-C, LF, and LH analyzed RNA sequencing data. All authors revised the manuscript.
Funding
This work was supported in part by the National Institute of Health Grant R01GM139991 (LH), American Heart Association Grant 19CDA34630041 (LH), and NCATS UL1TR002003 (MM-C).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to thank Genome Research Core, a part of the UIC Research Resources Center, for assisting with the preparation of RNA samples and RNA sequencing.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2022.860502/full#supplementary-material
References
Asuaje, A., Smaldini, P., Martín, P., Enrique, N., Orlowski, A., Aiello, E. A., et al. (2017). The Inhibition of Voltage-Gated H+ Channel (HVCN1) Induces Acidification of Leukemic Jurkat T Cells Promoting Cell Death by Apoptosis. Pflugers Arch. - Eur. J. Physiol. 469, 251–261. doi:10.1007/s00424-016-1928-0
Baukrowitz, T., Tucker, S. J., Schulte, U., Benndorf, K., Ruppersberg, J. P., and Fakler, B. (1999). Inward Rectification in KATP Channels: a pH Switch in the Pore. EMBO J. 18, 847–853. doi:10.1093/emboj/18.4.847
Bedard, K., Whitehouse, S., and Jaquet, V. (2015). Challenges, Progresses, and Promises for Developing Future NADPH Oxidase Therapeutics. Antioxid. Redox Signaling 23, 355–357. doi:10.1089/ars.2015.6450
Berger, T. K., and Isacoff, E. Y. (2011). The Pore of the Voltage-Gated Proton Channel. Neuron 72, 991–1000. doi:10.1016/j.neuron.2011.11.014
Brawand, D., Soumillon, M., Necsulea, A., Julien, P., Csárdi, G., Harrigan, P., et al. (2011). The Evolution of Gene Expression Levels in Mammalian Organs. Nature 478, 343–348. doi:10.1038/nature10532
Cherny, V. V., and Decoursey, T. E. (1999). Ph-Dependent Inhibition of Voltage-Gated H+ Currents in Rat Alveolar Epithelial Cells by Zn2+ and Other Divalent Cations. J. Gen. Physiol. 114, 819–838. doi:10.1085/jgp.114.6.819
Decoursey, T. E. (2013). Voltage-Gated Proton Channels: Molecular Biology, Physiology, and Pathophysiology of the HVFamily. Physiol. Rev. 93, 599–652. doi:10.1152/physrev.00011.2012
Dobin, A., Davis, C. A., Schlesinger, F., Drenkow, J., Zaleski, C., Jha, S., et al. (2013). STAR: Ultrafast Universal RNA-Seq Aligner. Bioinformatics 29, 15–21. doi:10.1093/bioinformatics/bts635
Duchen, M., Verkhratsky, A., and Muallem, S. (2008). Mitochondria and Calcium in Health and Disease. Cell Calcium 44, 1–5. doi:10.1016/j.ceca.2008.02.001
El Chemaly, A., Guinamard, R., Demion, M., Fares, N., Jebara, V., Faivre, J.-F., et al. (2006). A Voltage-Activated Proton Current in Human Cardiac Fibroblasts. Biochem. Biophysical Res. Commun. 340, 512–516. doi:10.1016/j.bbrc.2005.12.038
Fischer, H. (2012). Function of Proton Channels in Lung Epithelia. Wires Membr. Transp Signal. 1, 247–258. doi:10.1002/wmts.17
Hingtgen, S. D., Tian, X., Yang, J., Dunlay, S. M., Peek, A. S., Wu, Y., et al. (2006). Nox2-containing NADPH Oxidase and Akt Activation Play a Key Role in Angiotensin II-Induced Cardiomyocyte Hypertrophy. Physiol. Genomics 26, 180–191. doi:10.1152/physiolgenomics.00029.2005
Hong, L., Kim, I. H., and Tombola, F. (2014). Molecular Determinants of Hv1 Proton Channel Inhibition by Guanidine Derivatives. Proc. Natl. Acad. Sci. USA 111, 9971–9976. doi:10.1073/pnas.1324012111
Hong, L., Pathak, M. M., Kim, I. H., Ta, D., and Tombola, F. (2013). Voltage-sensing Domain of Voltage-Gated Proton Channel Hv1 Shares Mechanism of Block with Pore Domains. Neuron 77, 274–287. doi:10.1016/j.neuron.2012.11.013
Hong, L., Zhang, M., Ly, O. T., Chen, H., Sridhar, A., Lambers, E., et al. (2021). Human Induced Pluripotent Stem Cell-Derived Atrial Cardiomyocytes Carrying an SCN5A Mutation Identify Nitric Oxide Signaling as a Mediator of Atrial Fibrillation. Stem Cel Rep. 16, 1542–1554. doi:10.1016/j.stemcr.2021.04.019
Hong, L., Zhang, M., Sridhar, A., and Darbar, D. (2020). Pathogenic Mutations Perturb Calmodulin Regulation of Nav1.8 Channel. Biochem. Biophys. Res. Commun. doi:10.1016/j.bbrc.2020.08.010
Iovannisci, D., Illek, B., and Fischer, H. (2010). Function of the HVCN1 Proton Channel in Airway Epithelia and a Naturally Occurring Mutation, M91T. J. Gen. Physiol. 136, 35–46. doi:10.1085/jgp.200910379
Kim, I. H., Hevezi, P., Varga, C., Pathak, M. M., Hong, L., Ta, D., et al. (2014). Evidence for Functional Diversity between the Voltage-Gated Proton Channel Hv1 and its Closest Related Protein HVRP1. PLoS One 9, e105926. doi:10.1371/journal.pone.0105926
Kuroda, J., Ago, T., Matsushima, S., Zhai, P., Schneider, M. D., and Sadoshima, J. (2010). NADPH Oxidase 4 (Nox4) Is a Major Source of Oxidative Stress in the Failing Heart. Proc. Natl. Acad. Sci. 107, 15565–15570. doi:10.1073/pnas.1002178107
Liao, Y., Smyth, G. K., and Shi, W. (2014). featureCounts: an Efficient General Purpose Program for Assigning Sequence Reads to Genomic Features. Bioinformatics 30, 923–930. doi:10.1093/bioinformatics/btt656
Lishko, P. V., Botchkina, I. L., Fedorenko, A., and Kirichok, Y. (2010). Acid Extrusion from Human Spermatozoa Is Mediated by Flagellar Voltage-Gated Proton Channel. Cell 140, 327–337. doi:10.1016/j.cell.2009.12.053
Matamoros-Volante, A., and Trevino, C. L. (2020). Capacitation-associated Alkalization in Human Sperm Is Differentially Controlled at the Subcellular Level. J. Cel Sci. doi:10.1242/jcs.238816
Matsuno, K., Iwata, K., Matsumoto, M., Katsuyama, M., Cui, W., Murata, A., et al. (2012). NOX1/NADPH Oxidase Is Involved in Endotoxin-Induced Cardiomyocyte Apoptosis. Free Radic. Biol. Med. 53, 1718–1728. doi:10.1016/j.freeradbiomed.2012.08.590
Mccarthy, D. J., Chen, Y., and Smyth, G. K. (2012). Differential Expression Analysis of Multifactor RNA-Seq Experiments with Respect to Biological Variation. Nucleic Acids Res. 40, 4288–4297. doi:10.1093/nar/gks042
Musset, B., Smith, S. M. E., Rajan, S., Morgan, D., Cherny, V. V., and Decoursey, T. E. (2011). Aspartate 112 Is the Selectivity Filter of the Human Voltage-Gated Proton Channel. Nature 480, 273–277. doi:10.1038/nature10557
Pena, E., Brito, J., El Alam, S., and Siques, P. (2020). Oxidative Stress, Kinase Activity and Inflammatory Implications in Right Ventricular Hypertrophy and Heart Failure under Hypobaric Hypoxia. Int. J. Mol. Sci. 21. doi:10.3390/ijms21176421
Peng, J., Yi, M.-H., Jeong, H., Mcewan, P. P., Zheng, J., Wu, G., et al. (2021). The Voltage-Gated Proton Channel Hv1 Promotes Microglia-Astrocyte Communication and Neuropathic Pain after Peripheral Nerve Injury. Mol. Brain 14, 99. doi:10.1186/s13041-021-00812-8
Ramsey, I. S., Moran, M. M., Chong, J. A., and Clapham, D. E. (2006). A Voltage-Gated Proton-Selective Channel Lacking the Pore Domain. Nature 440, 1213–1216. doi:10.1038/nature04700
Robinson, M. D., Mccarthy, D. J., and Smyth, G. K. (2010). edgeR: a Bioconductor Package for Differential Expression Analysis of Digital Gene Expression Data. Bioinformatics 26, 139–140. doi:10.1093/bioinformatics/btp616
Saegusa, N., Garg, V., and Spitzer, K. W. (2013). Modulation of Ventricular Transient Outward K+ Current by Acidosis and its Effects on Excitation-Contraction Coupling. Am. J. Physiology-Heart Circulatory Physiol. 304, H1680–H1696. doi:10.1152/ajpheart.00070.2013
Saegusa, N., Moorhouse, E., Vaughan-Jones, R. D., and Spitzer, K. W. (2011). Influence of pH on Ca2+ Current and its Control of Electrical and Ca2+ Signaling in Ventricular Myocytes. J. Gen. Physiol. 138, 537–559. doi:10.1085/jgp.201110658
Sasaki, M., Takagi, M., and Okamura, Y. (2006). A Voltage Sensor-Domain Protein Is a Voltage-Gated Proton Channel. Science 312, 589–592. doi:10.1126/science.1122352
Seredenina, T., Demaurex, N., and Krause, K.-H. (2015). Voltage-Gated Proton Channels as Novel Drug Targets: From NADPH Oxidase Regulation to Sperm Biology. Antioxid. Redox Signaling 23, 490–513. doi:10.1089/ars.2013.5806
Sundset, R., Bertelsen, G., and Ytrehus, K. (2003). Role of the Na+–H+exchanger (NHE1) in Heart Muscle Function during Transient Acidosis. A Study in Papillary Muscles from Rat and guinea Pig Hearts. Can. J. Physiol. Pharmacol. 81, 937–943. doi:10.1139/y03-091
Taylor, A. R., Chrachri, A., Wheeler, G., Goddard, H., and Brownlee, C. (2011). A Voltage-Gated H+ Channel Underlying pH Homeostasis in Calcifying Coccolithophores. Plos Biol. 9, e1001085. doi:10.1371/journal.pbio.1001085
Thomas, R. C., and Meech, R. W. (1982). Hydrogen Ion Currents and Intracellular pH in Depolarized Voltage-Clamped Snail Neurones. Nature 299, 826–828. doi:10.1038/299826a0
Vairamani, K., Wang, H.-S., Medvedovic, M., Lorenz, J. N., and Shull, G. E. (2017). RNA SEQ Analysis Indicates that the AE3 Cl−/HCO3 − Exchanger Contributes to Active Transport-Mediated CO2 Disposal in Heart. Sci. Rep. 7, 7264. doi:10.1038/s41598-017-07585-y
Vaughan-Jones, R. D., Spitzer, K. W., and Swietach, P. (2009). Intracellular pH Regulation in Heart. J. Mol. Cell Cardiol. 46, 318–331. doi:10.1016/j.yjmcc.2008.10.024
Ventura, C., Leon, I. E., Asuaje, A., Martin, P., Enrique, N., Nunez, M., et al. (2020). Differential Expression of the Long and Truncated Hv1 Isoforms in Breast-Cancer Cells. J. Cel Physiol.
Wang, H.-S., Chen, Y., Vairamani, K., and Shull, G. E. (2014). Critical Role of Bicarbonate and Bicarbonate Transporters in Cardiac Function. Wjbc 5, 334–345. doi:10.4331/wjbc.v5.i3.334
Wu, L.-J., Wu, G., Sharif, M. R. A., Baker, A., Jia, Y., Fahey, F. H., et al. (2012). The Voltage-Gated Proton Channel Hv1 Enhances Brain Damage from Ischemic Stroke. Nat. Neurosci. 15, 565–573. doi:10.1038/nn.3059
Wu, X., and Hong, L. (2021). Calmodulin Interactions with Voltage-Gated Sodium Channels. Int. J. Mol. Sci. 22. doi:10.3390/ijms22189798
Yamamoto, T., Swietach, P., Rossini, A., Loh, S.-H., Vaughan-Jones, R. D., and Spitzer, K. W. (2005). Functional Diversity of Electrogenic Na+-HCO3−cotransport in Ventricular Myocytes from Rat, Rabbit and guinea Pig. J. Physiol. 562, 455–475. doi:10.1113/jphysiol.2004.071068
Yeste, M., Llavanera, M., Mateo-Otero, Y., Catalan, J., Bonet, S., and Pinart, E. (2020). HVCN1 Channels Are Relevant for the Maintenance of Sperm Motility during In Vitro Capacitation of Pig Spermatozoa. Int. J. Mol. Sci. 21. doi:10.3390/ijms21093255
Yu, Y., Fuscoe, J. C., Zhao, C., Guo, C., Jia, M., Qing, T., et al. (2014). A Rat RNA-Seq Transcriptomic BodyMap across 11 Organs and 4 Developmental Stages. Nat. Commun. 5, 3230. doi:10.1038/ncomms4230
Zafari, A. M., Ushio-Fukai, M., Akers, M., Yin, Q., Shah, A., Harrison, D. G., et al. (1998). Role of NADH/NADPH Oxidase-Derived H 2 O 2 in Angiotensin II-Induced Vascular Hypertrophy. Hypertension 32, 488–495. doi:10.1161/01.hyp.32.3.488
Zhang, M., Brewer, A. C., Schroder, K., Santos, C. X. C., Grieve, D. J., Wang, M., et al. (2010). NADPH Oxidase-4 Mediates protection against Chronic Load-Induced Stress in Mouse Hearts by Enhancing Angiogenesis. Proc. Natl. Acad. Sci. 107, 18121–18126. doi:10.1073/pnas.1009700107
Zhang, Y., Murugesan, P., Huang, K., and Cai, H. (2020). NADPH Oxidases and Oxidase Crosstalk in Cardiovascular Diseases: Novel Therapeutic Targets. Nat. Rev. Cardiol. 17, 170–194. doi:10.1038/s41569-019-0260-8
Zhao, C., Hong, L., Galpin, J. D., Riahi, S., Lim, V. T., Webster, P. D., et al. (2021a). New Arginine Mimic Inhibitors of the Hv1 Channel with Improved VSD-Ligand Interactions. J. Gen. Physiol. 153.HIFs
Keywords: HVCN1, RNA-seq, NOX, pH homeostasis, heart
Citation: Wu X, Li Y, Maienschein-Cline M, Feferman L, Wu L and Hong L (2022) RNA-Seq Analyses Reveal Roles of the HVCN1 Proton Channel in Cardiac pH Homeostasis. Front. Cell Dev. Biol. 10:860502. doi: 10.3389/fcell.2022.860502
Received: 23 January 2022; Accepted: 16 February 2022;
Published: 16 March 2022.
Edited by:
Wanling Xuan, University of South Florida, United StatesReviewed by:
Yunxian Chen, Sun Yat-sen University, ChinaBin Liu, University of Arizona, United States
Elisabeth Pinart, University of Girona, Spain
Copyright © 2022 Wu, Li, Maienschein-Cline, Feferman, Wu and Hong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Liang Hong, aG9uZzIwMDRAdWljLmVkdQ==
 Xin Wu
Xin Wu Yawei Li
Yawei Li Mark Maienschein-Cline3
Mark Maienschein-Cline3 Longjun Wu
Longjun Wu Liang Hong
Liang Hong