- 1Department of Oncology, Affiliated Sixth People’s Hospital, Shanghai Jiaotong University, Shanghai, China
- 2Department of Endocrinology and Metabolism, Zhongshan Hospital, Key Laboratory of Metabolism and Molecular Medicine, the Ministry of Education, Fudan University, Shanghai, China
- 3Institute of Clinical Science, Zhongshan Hospital, Fudan University, Shanghai, China
- 4Jinshan Hospital Center for Tumor Diagnosis and Therapy, Jinshan Hospital, Fudan University, Shanghai, China
Background: Melanoma is a highly malignant and aggressive tumor. The search for new and effective biomarkers facilitates early diagnosis and treatment, ultimately improving the prognosis of melanoma patients. Although the transmembrane protein TMEM176B has been linked to a number of cancers, its role in cancer immunity remains unknown.
Methods: Expression levels of TMEM176B in normal tissues and several cancers, including Skin Cutaneous Melanoma (SKCM), were collected from TCGA and GTEx. We used Receiver operating characteristic and Kaplan–Meier survival curves and performed regression analysis to elucidate the link between TMEM176B and clinicopathological features of SKCM in order to determine the prognostic significance of TMEM176B in SKCM. We then used the GEPIA and STRING websites to search for proteins and associated top genes that may interact with TMEM176B and enriched them for analysis. The link between TMEM176B and immune cells infiltration was then investigated using TIMER, CIBERSORT algorithm and GSVA package of R (v3.6.3). Finally, animal tests were conducted to confirm the expression of Tmem176b and its influence on T-cell immune infiltration.
Results: TMEM176B expression was considerably elevated in SKCM compared to normal tissues. Particularly, TMEM176B expression was also linked to pathological stage, tumor ulceration and radiation therapy. Patients with elevated TMEM176B expression had a better prognosis, according to the survival analysis. The majority of tumor infiltrating lymphocytes (TILs) especially T cells in SKCM was positively linked with TMEM176B expression. Our animal experiments also verified that the T-cell infiltration was significantly inhibited in local melanoma tissue of Tmem176b knockout mice. At the same time deleting Tmem176b accelerated tumor progress and impaired T cells effector function.
Conclusion: Upregulated expression of TMEM176B in SKCM is associated with a better prognosis and it has the potential to serve as a diagnostic and prognostic marker for the disease. It may serve as a target for SKCM immunotherapy by regulating CD8+ T cells although it requires more evidence.
Introduction
Melanoma, which is aggressive, is a malignant tumor that develops from melanocytes (Schadendorf et al., 2015; Shain and Bastian, 2016; Lin and Fisher, 2017). Cutaneous melanoma is most common in Western countries, but the global incidence has gradually increased in recent years (Hollestein et al., 2012). Ultraviolet light (UV) radiation is an important pathogenic factor in the development of melanoma (Schadendorf et al., 2015; Michielin et al., 2019). Early diagnosis is made easier by general clinical and pathological aspects due to the specific color characteristics of melanoma and the fact that it occurs on the skin surface, although diagnostic hurdles still exist (Schadendorf et al., 2015). Patients with advanced metastases have a poor prognosis, despite the fact that early-stage tumors can be surgically excised with a high cure rate (Lin and Fisher, 2017). As a result, new diagnostic and prognostic biomarkers are needed to increase the survival rate of melanoma patients.
To achieve complete control of tumors, the antitumor effector cells in the immune system must coordinate with each other to overcome tumor immune evasion (Huntington et al., 2020). T cells are one of the most important anti-tumor effector cells because they may directly attack cancer cells. Immune checkpoint therapy targeting T cells, for instance, cytotoxic T lymphocyte-associated antigen 4 (CTLA4) and programmed cell death protein 1 (PD1) has shown promising results in patients with melanoma in recent years, but only some tumor types have benefited, the degree of tumor infiltration and immune cell activation, particularly CD8+ T cells, are both factors that influence immune checkpoint therapy efficacy (Kim et al., 2020). Exploring the immune phenotype and interactions in the tumor microenvironment is therefore critical for discovering new immunotherapeutic targets in melanoma.
Some transmembrane proteins have been found to be overexpressed in tumors and to be linked to prognosis, suggesting that they could be useful targets for immunotherapy (Choi et al., 2007; Tomimaru et al., 2015). TMEM176B (Transmembrane protein 176B), which was formerly known as TORID (Tolerance-related and Inducible Transcript) (Louvet et al., 2005), is a gene that is, highly overexpressed in tolerance allografts (Viklicky et al., 2013). Its human homolog was originally found to be expressed in a subset of lung fibroblasts, the mouse homolog Clast1 was upregulated after B cells were activated by CD40 ligand (Lurton et al., 1999). TMEM176B, like its paralog TMEM176A (Transmembrane protein 176A), has four transmembrane domains and belongs to the MS4A family (Eon Kuek et al., 2016). It is mostly expressed in the lungs, kidneys, lymph nodes, and spleen, among different immune cell subtypes, it has been observed that the high expression of Tmem176b and Tmem176a in BMDCs (Bone Marrow-Derived Dendritic Cells) and cDCs (Classic Dendritic Cells), and is associated with the immature state of DCs (Dendritic Cells) (Condamine et al., 2010).
The expression level of TMEM176B was higher in gastric cancer tissues than in normal tissues, and according to survival analysis, higher expression levels were related to poorer prognosis (Sun et al., 2018). The expression of TMEM176B in lymphoma and its control tissues also differed significantly (Cuajungco et al., 2012; Ma and Li, 2021). The expression of TMEM176B is up-regulated in the tumor blood vessels of human renal cell carcinoma specimens, suggesting that it is implicated in tumor angiogenesis and could be a target of anti-angiogenesis therapy for cancer patients (Otsubo et al., 2014). However, the role of TMEM176B in cancers has yet to be determined, or we do not yet have a complete grasp of its function in malignancies.
In addition, the correlation between TMEM176B and tumor immunity and its role in melanoma is less reported. Tmem176b and its homolog Tmem176a has been found to be highly expressed in CD4+ Th17 cells (Drujont et al., 2016), this study also found that these two molecules play the same ion channel function and are co-localized near the Golgi apparatus (Drujont et al., 2016). The expression of some costimulatory molecules in T cells can be inhibited by transfection of Tmem176b into immature dendritic cells (Louvet et al., 2005). It revealed that suppressing inflammasomes by knocking down Tmem176b in mice or utilizing TMEM176B inhibitors can improve the anti-tumor effect of CD8+ T and the efficacy of anti-CTLA-4 and anti-PD1 therapy (Segovia et al., 2019). However, cancer is a very heterogeneous disease and melanoma has its unique immune microenvironment. It is unknown the role of TMEM176B in regulating CD8+ T cells biology and the course of melanoma.
We explored the expression of TMEM176B in SKCM and its relationship with tumor patients’ prognosis and immune infiltration. Furthermore, we found the regulatory effect of TMEM176B on tumor-infiltrating CD8+ T cells. Therefore, TMEM176B is a potential diagnostic or prognostic biomarker for melanoma.
Methods
Gene Expression Analysis
We utilized R package “ggplot2” to analysis the differential expression of TMEM176B in various malignancies or cancer subtypes. Extraction of SKCM in TCGA and corresponding normal tissue data in GTEx has been analyzed for differential expression of TMEM176B in cutaneous melanoma. Click “repository” in TCGA database and make the following settings: under File directory (select “Gene Expression Quantification” under Data Category module, select “HTseq-FPKM” under Workflow Type module); under Cases (under Primary Site module, select “skin,” and under Project, select “TCGA-SKCM”); under Cases (select “skin” under the Primary Site module and “TCGA-SKCM” under Project) to get the clinical information of SKCM.
Survival Prognosis Analysis
R package “survminer” and “survival” were utilized to analyze the prognostic value of the expression level of TMEM176B in different tumors. We collected the data from the TCGA database (Liu et al., 2018) and compared the overall survival (OS) of cancer patients separated by the median expression level of TMEM176B. The “pROC” package and the “ggplot2” package were used to extract and analyze the SKCM data of TCGA and the corresponding normal tissue data in GTEx to obtain the receiver operating characteristic (ROC) curves.
Cox Regression
We performed Cox regression analysis to identify the association between SKCM disease characteristics and the expression level of TMEM176B.
Survival Analysis in PrognoScan
In PrognoScan, the association between TMEM176B expression and prognosis of cancer patients was analyzed, including distant metastasis-free survival (DMFS), disease-free survival (DFS), relapse-free survival (RFS), OS, and distant recurrence-free survival (DRFS).
Relationship Between TMEM176B Expression and Immune Cell Infiltration
TIMER is a fully functional public platform for analyzing of immune infiltration in tumors. Correlations between TMEM176B expression and immune infiltration levels in more than 30 different cancer types in TCGA were obtained in the gene module of TIMER. We also investigated the connection between tumor purity and TMEM176B expression. And we utilized R package “GSVA” to obtain the link among TMEM176B and other immunocytes (Bindea et al., 2013). Next “CIBERSORT algorithm” was used to explore the immune infiltration between high- and low-expression with TMEM176B to obtain enriched immunocytes between the two groups.
TMEM176B-Related Gene Enrichment Analysis and Single Gene Co-Expression Analysis
We acquired the top 100 genes associated with TMEM176B in SKCM cases using the “Similar Gene Detection” module of GEPIA2 and performed correlation analysis on the top 5 genes using “Correlation Analysis” in GEPIA2. We then obtained protein networks that may interact with TMEM176B in the STRING database. In addition, we subjected the 100 genes obtained in GEPIA2 to GO and KEGG pathway analysis using the “clusterProfiler” and “ggplot2” R packages.
R package “ggplot2” was used to determine the link between the expression level of TMEM176B in SKCM and other genes, including CD274, CTLA4, GZMA, GAMB, IFNG, PRF1, and so on.
Animals
Aged 6–8 weeks Tmem176b−/− mice and C57BL/6J mice were purchased from GemPharmatech Co. Ltd. and Shanghai Jie Si Jie Laboratory Animal Co. Ltd., respectively.
Cell Culture
B16-OVA melanoma cells were cultured in DMEM medium, which contains 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin.
Tumor Model
We injected 1 × 106 B16-OVA cells into the right flank of WT or Tmem176b−/− mice subcutaneously and used vernier calipers to measure tumor volume every 1–2 days. On the 14th day, the mice were euthanized and their tumor tissues, spleen, lymph nodes, and peripheral blood were taken for further research.
Flow Cytometry
Cells were stained with CD3 (BioLegend, clone:17A2), CD4 (BioLegend, clone: GK1.5), CD8 (BioLegend, clone:53-6.7), GZMB (Biolegend, clone: QA16A02), IFN-γ (Biolegend, clone: XMG1.2), Ki-67 (BioLegend, clone: 16A8), PD-1(Biolegend, clone:29F.1A12), PD-L1 (BioLegend, clone:10F.9G2), CD44 (Biolegend, clone: IM7), CD62L (BioLegend, clone: MEL-14), CD25(BioLegend, clone: 3C7), CD69(BioLegend, H1.2F3), BCL2 (Cell Signaling Technology, clone: 124), PRF(BioLegend, clone: S16009A). For intracellular staining, cells were given PMA/ionomycin (BioLegend, Cat. No. 423303) re-stimulation for 4–5 h, fixed, permeabilized, and then stained. FACS analysis was performed on a BD FACS Aria III flow cytometer and analyzed by FlowJo V.10 software. BD FACS Aria III flow cytometer was used to perform FACS analysis and data analysis was performed in FlowJo V.10 software.
Statistical Analysis
Graphpad Prism (v8) and R language version (3.6.3) were utilized for designing figures and statistical analysis. Wilcoxon rank sum test and Student’s t tests were used to compare the expression of TMEM176B in different groups. When p values were less than 0.05, differences were considered significant.
Results
Clinical Characteristics of Patients
We extracted clinical information from the TCGA, including TNM stage, pathologic stage, age, race, weight, gender and whether or not they had received radiation. Supplementary Table S1 summarizes all the information. The table manifested that the expression of TMEM176B is closely related to the TNM stage of SKCM, the pathological stage and radiation therapy.
The Expression of TMEM176B in SKCM Was Increased
We demonstrated the expression status of TMEM176B across various types of cancer. TMEM176B expression was considerably enhanced in some malignancies, as demonstrated in Figure 1A, which further demonstrates that heterogeneity is an important attribute of cancer and a major contributor to tumor progression. Compared to normal tissue, TMEM176B expression was higher in SKCM (p <0.001) (Figure 1B). The correlation analysis revealed that there was a statistical difference between TMEM176B and T stage, pathologic stage (p <0.05), and melanoma ulceration, radiation therapy, breslow depth (p <0.001) (Figures 1C–G).
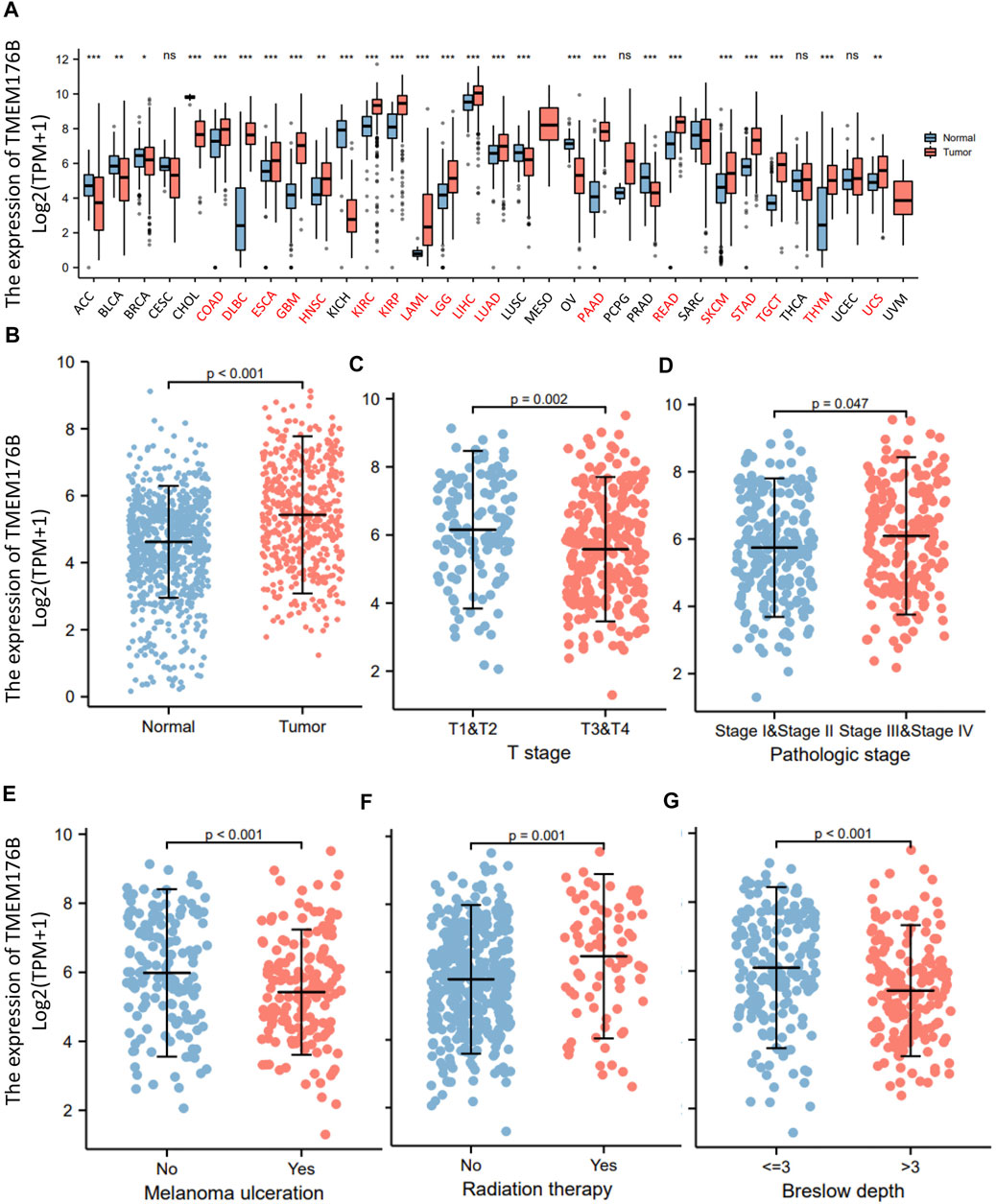
FIGURE 1. The expression of TMEM176B in malignancies. (A) The expression status of the TMEM176B across various cancer types. *p < 0.05; **p < 0.01; ***p < 0.001. (B) The expression level of TMEM176B in SKCM was significantly increased compared to the normal tissue (p < 0.001). (C–G) Association between TMEM176B and several clinical features in SKCM.
The Prognostic Value of TMEM176B in SKCM
Since the significant differential expression within tumor and normal tissues, we investigated the effect of TMEM176B expression on the prognosis of tumor patients. Cancer cases were separated into high- and low-expression groups based on TMEM176B median expression levels to evaluate how TMEM176B expression influences prognosis of patients in SKCM. The area under the curve can be derived from the ROC curve as 0.668 (Figure 2A). High TMEM176B level is linked with a better prognosis, according to the Kaplan-Meier survival analysis (Figure 2B). We also summarized in Supplementary Figures 1, 2 that the expression level of TMEM176B is closely associated with the prognosis of patients with various malignancies. We next combined clinical and pathological data, and analyzed the association between TMEM176B expression and a number of clinical parameters in SKCM patients. For OS, univariate Cox regression showed that TMEM176B played a detrimental role in patients with SKCM with the following characteristics: age >60 (n = 210, HR = 1.656, 95% CI from 1.251 to 2.192, p < 0.001), T stage 3 and 4 (n = 243, HR = 2.085, 95% CI from 1.501 to 2.895, p < 0.001), N stage 2 and 3 (n = 105, HR = 1.818, 95% CI from 1.310 to 2.523, p < 0.001), M stage 1 (n = 24, HR = 1.897, 95% CI from 1.029 to 3.496, p < 0.05), and Pathologic stage III and IV (n = 193, HR = 1.617, 95% CI from 1.207 to 2.165, p = 0.001) (Figure 2C). It is clear from this that TMEM176B plays an important role and serves as a reliable prognostic marker in SKCM.
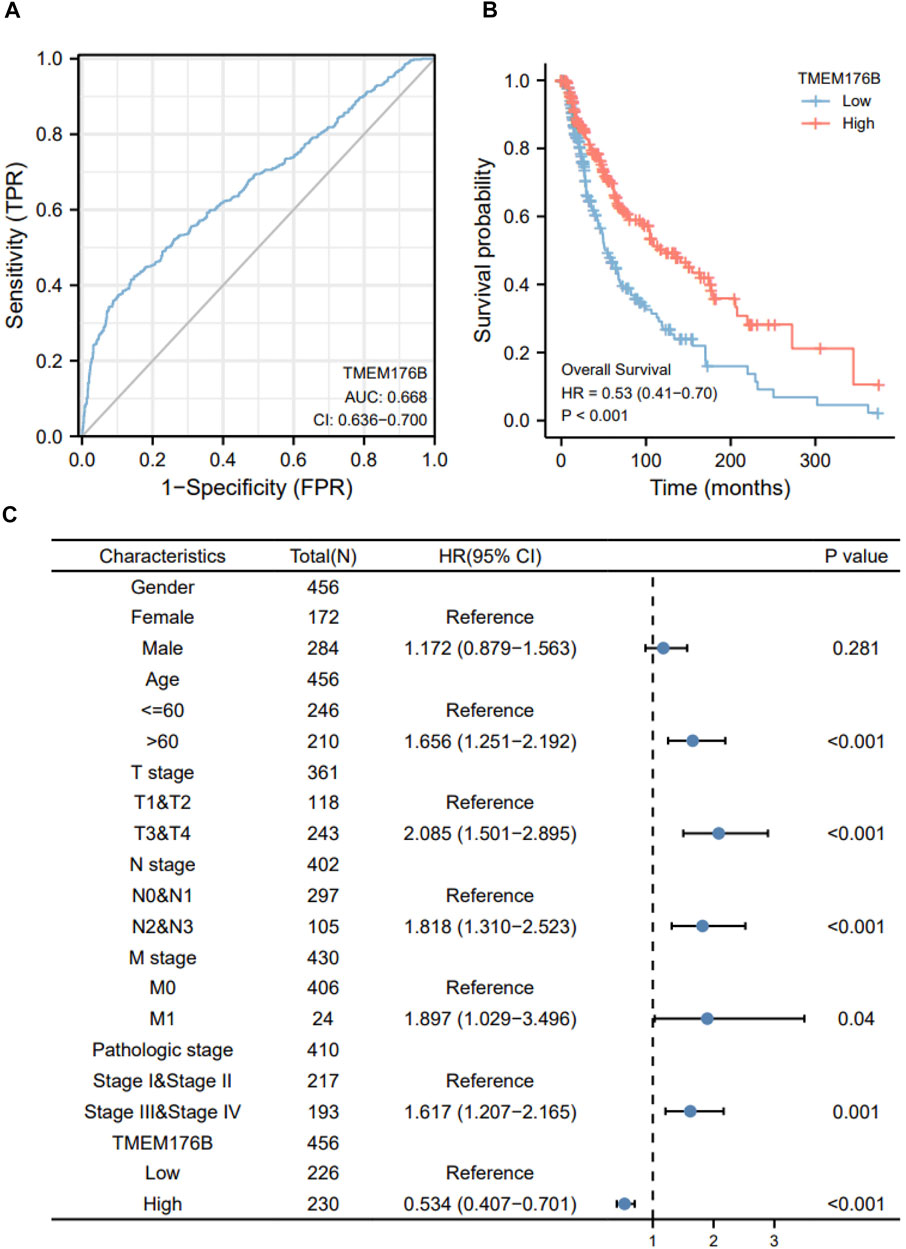
FIGURE 2. The prognostic value of TMEM176B. (A) ROC analysis showed that TMEM176B was able to identify tumor and normal tissue, the AUC was 0.668. (B) Survival evaluation indicators OS, via the expression level of the TMEM176B gene in SKCM. (C) Univariate Cox regression analysis of TMEM176B and clinicopathological variables with OS in SKCM.
TMEM176B-Related Gene Enrichment Analysis
We screened for proteins or related genes that might bind to TMEM176B in order to learn more about how it plays a role in carcinogenesis. GEPIA2 was used to identify the top 100 genes with the strongest relationship to TMEM176B in SKCM. The expression level of TMEM176B is positively linked with TMEM176A (Transmembrane Protein 176A) (R = 0.94), LAIR1 (Leukocyte Associated Immunoglobulin Like Receptor 1) (R = 0.79), SLC7A7 (Solute Carrier Family 7 Member 7) (R = 0.78), ITGB2 (Integrin Subunit Beta 2) (R = 0.77), HCK (HCK Proto-Oncogene, Src Family Tyrosine Kinase) (R = 0.77), as illustrated in Figures 3A–E. The STRING program was also used to generate a network of proteins that interact with TMEM176B (Figure 3F). We subjected the top 100 genes obtained from GEPIA2 to GO/KEGG analysis, Figures 3G,H suggest these gene enriched in “regulation of lymphocyte activation” and “phagocytosis” pathway.
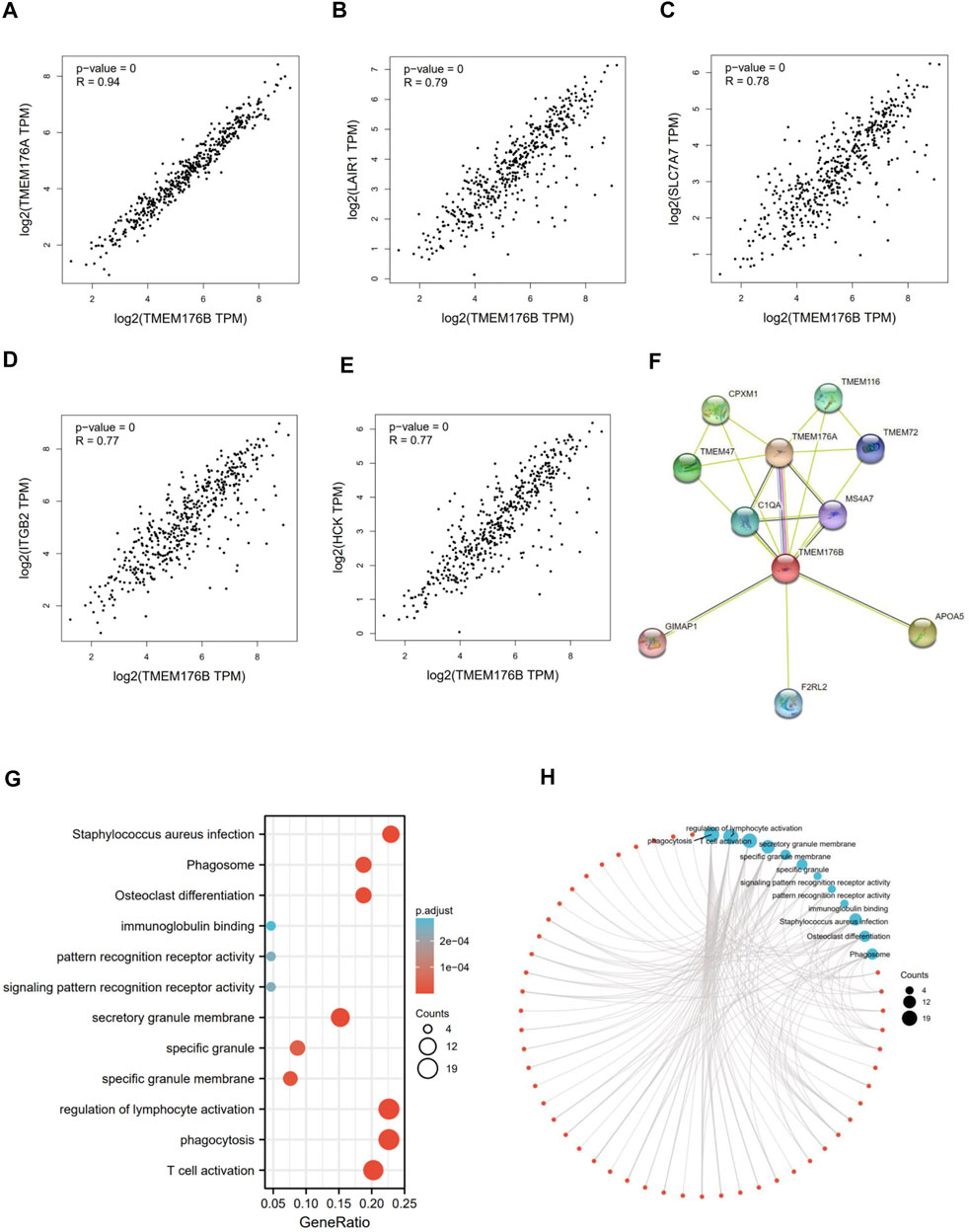
FIGURE 3. TMEM176B-related gene enrichment analysis. (A–E) Using GEPIA2, we identified the top 100 genes related with TMEM176B in SKCM and examined the expression connection between TMEM176B and five genes, includingTMEM176A, LAIR1, SLC7A7, ITGB2, and HCK. (F) Protein networks that may interact with TMEM176B, data from STRING. (G,H) Enrichment analysis of TMEM176B-related genes.
Correlation Between TMEM176B and Tumor Immune Infiltration
As can be seen from the above, TMEM176B was highly expressed in tumor tissues of SKCM and that higher expression correlated with better prognosis of SKCM patients. A variety of immune cells with different functions are present in the tumor microenvironment, including immunosuppressive cells and cells that promote tumor immunity (Quail and Joyce, 2013). As a result, it is necessary to continually explore the relationship between TMEM176B and the infiltration of immune cells. Data from TIMER suggests statistical significance between TMEM176B expression and immune cells, with B cells, CD4+ T cells, CD8+ T cells, macrophages, neutrophils, and dendritic cells were significantly positively correlated with the expression level of TMEM176B in SKCM (Figure 4A). TMEM176B expression levels, on the other hand, were significantly adversely associated with tumor purity (Figure 4A). We can also derive the association among TMEM176B expression and other immune cells from Figure 4B. Supplementary Figures S3A–C demonstrated correlation of TMEM176B expression and immune infiltration in different cancers of TCGA except SKCM. Among them, high expression of TMEM176B is associated with a higher degree of immune infiltration in most tumors. However, in LIHC, COAD, MESO and DLBC, the correlations were negative.
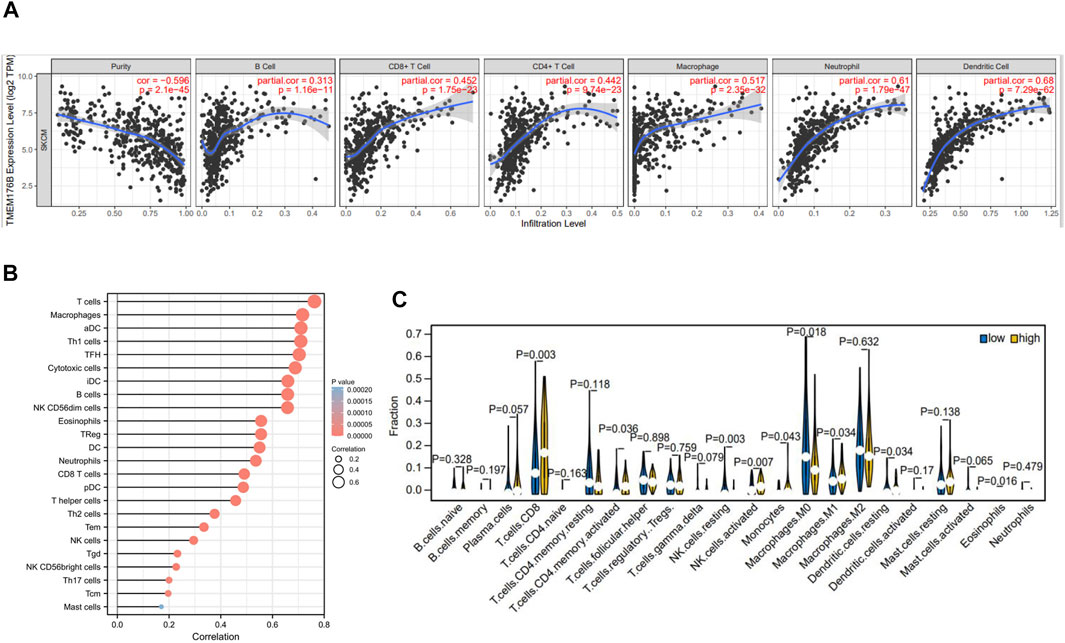
FIGURE 4. Correlation of TMEM176B expression and the level of immune infiltration in SKCM. (A,B) In SKCM, the infiltration levels of B cells, CD8+ T cells, CD4+ T cells, macrophages, neutrophils, dendritic cells and other immune cells with TMEM176B expression. p < 0.05 is considered significant. (C) The varied proportions of 22 subtypes of immune cells in high and low TMEM176B expression groups in tumor samples.
Using the CIBERSORT method, we investigated TMEM176B expression profiles to assess the level of 22 different immunocytes (Figure 4C). Our findings revealed that 9 kinds of immunocytes, including CD8+ T cells, activated CD4 memory T cells, resting and activated NK cells, Monocytes, Macrophages, resting dendritic cells and eosinophils were influenced by TMEM176B expression, among these cells CD8+ T cells and macrophages were significantly different between the TMEM176B high and low groups. The results of the high expression group compared to the low expression group showed that CD8+ T cells increased, whereas M0 Macrophages decreased. These findings imply that TMEM176B modulates patient survival via immunological infiltrative interactions in the tumor microenvironment. CD8+ cytotoxic T lymphocytes (CTLs) are essential in antitumor immunity. Therefore, the impact of TMEM176B on CD8+ T cell infiltration level and the prognosis of tumor patients deserves close attention.
The Effects of Tmem176b on CD8+ T Cells in the TME
We would like to further study the regulatory relationship between TMEM176B and CD8+ T cells in the tumor microenvironment (TME). We investigated the role of Tmem176b in a mouse melanoma model and sought to continue to explore the role of Tmem176b in the regulation of CD8+ T cells in the tumor microenvironment. We constructed a mouse tumor model (Figure 5A) to figure out how Tmem176b regulates the differentiation, function, and immune infiltration levels of CD8+ T cells in the tumor microenvironment. After implanting the tumor subcutaneously for 14 days, we euthanized the mice for flow cytometry analysis. The tumor volume of mice following Tmem176b knockout was similarly greater and grew quicker than that of WT mice (p < 0.05) (Figure 5B).
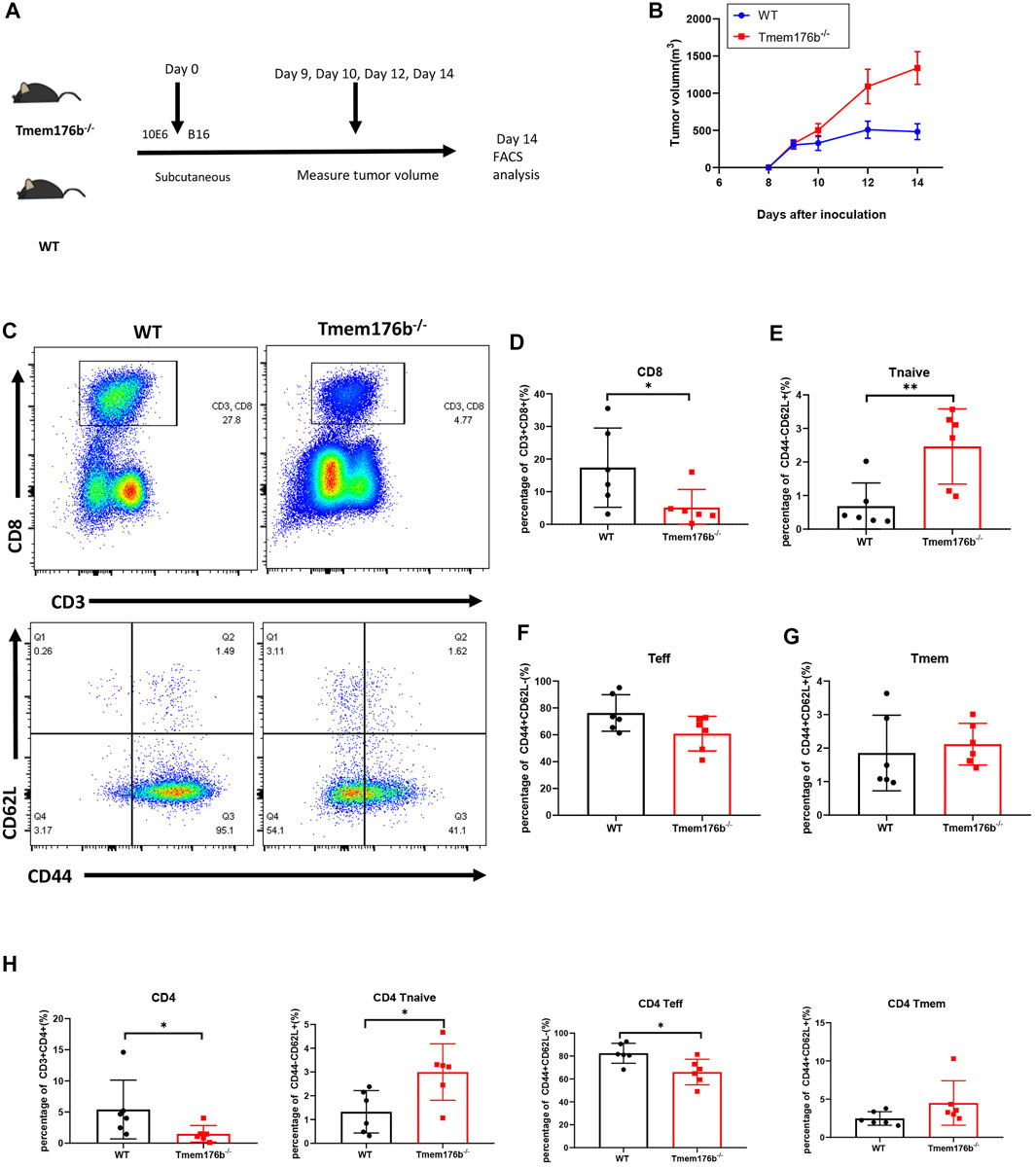
FIGURE 5. The regulatory effect of Tmem176b on T cells after tumor implantation. (A) Schematic diagram of mouse tumor model experiment. Six female mice per group. (B) Tumor growth curves of WT and Tmem176b−/− mice injected subcutaneously with 1 × 106 B16-OVA melanoma cells (per group n = 6). (C) Flow cytometry of tumor-infiltrating lymphocytes TIL CD8+ T cells (top rows) and its subgroup (bottom rows) from WT and Tmem176b−/− mice at day 14 after subcutaneous injection with 1 × 106 B16-OVA melanoma cells (n = 6). (D–G) Percentage of tumor-infiltrating CD3+ CD8+ T cells and their subpopulations Tnaive, Teff, Tmem from mice as in (C) (n = 6). (H) Percentage of tumor-infiltrating CD3+ CD4+ T cells and their subpopulations Tnaive, Teff, Tmem from mice as in (C) (n = 6). Each symbol represents a single mouse; data are mean SD from six independent samples. ns, not significant (two-tailed unpaired Student’s t test). *p < 0.05, **p < 0.01, and ***p < 0.001.
Our findings revealed that the proportion of CD3+ CD8+ T cells in Tumor-infiltrating T lymphocytes (TILS), is dramatically reduced after Tmem176b knockout (Figures 5C,D), the same trend can be seen in CD3+ CD4+ T cells (Figure 5H). This downward trend was also found in peripheral blood, spleen, and lymph nodes, although we did not obtain statistical differences in all tissues (Supplementary Figure S4A), and this difference is more pronounced in CD8+ T cells, consistent with our previous results that higher expression of TMEM176B in SKCM was associated with higher levels of infiltration of immune cells (Figure 4). CD8+ T cells are subdivided into naive T cells (Tnaive), effector T cells (Teff) and memory T cells (Tmem) subsets. In our study, compared to wild type mice, more CD8+ Tnaive and less CD8+ Teff presented in tumor of Tmem176b−/− mice (Figures 5E,F), and the same change had also been observed in tumor-infiltrated CD4+ T cells (Figure 5H). However, Tmem remained unchanged between groups (Figures 5G,H). The influence of Tmem176b on the differentiation of T cells was not significant in other non-tumor organs (Supplementary Figures S4B,C). It suggested that T cell populations were normal in non-tumor organs and T cell homeostasis was altered in response to immunogenic stress exhibited by the facts that increased Tnaive cell number and decreased Teff cell number were presented in the local tumor of Tmem176b−/− mice.
We also looked at the expression of some immunosuppressive molecules following tumor transplantation. Comparing to WT mice, our findings revealed that the expression of multiple activation or effector function-related molecules were inhibited in tumor-infiltrated CD8+ T cells of Tmem176b−/− mice (Figure 6) although the trend was not consistent in blood, spleen or lymph node (Supplementary Figures S4D, S5). Our findings showed that after tumor implantation, knocking down Tmem176b has effect on T cell activation rather than anti-apoptosis, or proliferation (Supplementary Figure S5).
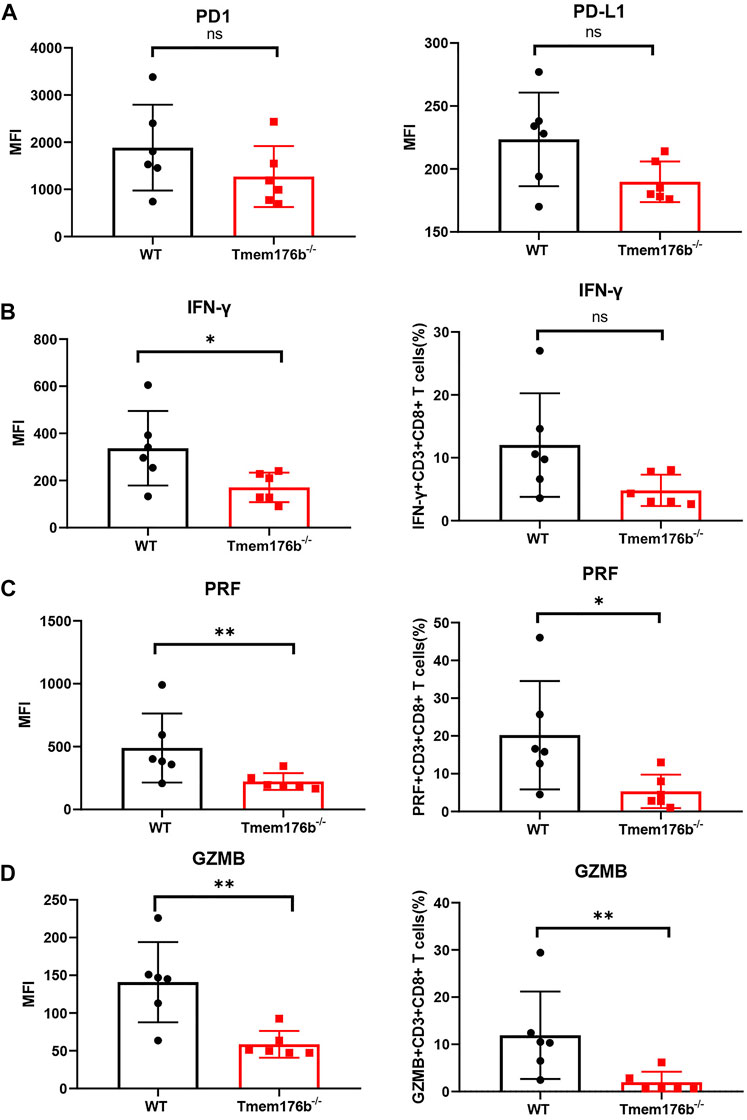
FIGURE 6. The regulatory effect of Tmem176b on T cells after tumor implantation. (A) The expression level of PD1 and PD-L1 of tumor-infiltrating CD8+ T lymphocytes (TIL CD8+ T cells) from WT and Tmem176b−/− mice (n = 6). (B–D) The MFI (left row) level of IFN-γ, PRF and GZMB of TIL CD8+ T cells, and frequency (right row) of IFN-γ+, PRF+ or GZMB+ (among TIL CD8+ T cells) from mice as in (A) (n = 6). Each symbol represents a single mouse; data are mean SD from six independent samples. ns, not significant (two-tailed unpaired Student’s t test). *p < 0.05, **p < 0.01, and ***p < 0.001.
We could conclude from Figure 6 that the expression levels of cytotoxic molecules such as IFN-γ, PRF1, and GZMB in tumor-infiltrating CD8+ T cells was reduced following Tmem176b deletion, indicating that the function of tumor-infiltrating T cells was significantly compromised. It is the possible reason for the larger tumor size in Tmem176b knockout mice. To verify our results, we researched the correlation between the expression of TMEM176B and some molecules in SKCM. As shown in Supplementary Figure S6, the expression of TMEM176B was significantly positive related to the expression levels of PD-L1 (CD274), CTLA4, LAG3, GZMK, GZMB, GZMA, GZMM, PRF1, TNF and IFNG in SKCM. PD1 is a hallmark of T-cell exhaustion related to co-expression of other checkpoint molecules such as LAG-3 and TIM-3, as well as an activation marker for T-cells including antigen specific responses. The results of this part indicate that Tmem176b positively regulates the effector function and activation of tumor-infiltrating CD8+ T cells.
Discussion
Immune cells play an essential role in tumor immunity, however, to date, there are few reports on the association of TMEM176B with immune cells. Tmem176b is predominantly expressed in BMDCs and cDC, and is involved in maintaining the immature state of DC (Condamine et al., 2010). In addition, strong expression of Tmem176b in a family of retinoic acid-related orphan receptor RORγt+ lymphocytes have been reported (Drujont et al., 2016; Lancien et al., 2021). Our research discovered a link between the expression of TMEM176B and the infiltration degree of immunocytes in cancers. TMEM176B was found to be substantially connected with the purity of most tumors in TIMER, as well as the infiltration of B cells, CD4+ T cells, CD8+ T cells, macrophages, neutrophils, and dendritic cells. In SKCM, our results indicated that TMEM176B was significantly and positively correlated with the infiltration of most immune cells. Robust infiltration of lymphocytes in melanoma correlates with a reduced risk of tumor growth (Galon et al., 2006).
Next, we used an animal model to identify the connection between Tmem176b and immune cell infiltration. After tumor implantation, we tested the effect of deleting Tmem176b on T lymphocyte development and function in diverse tissues and immunological organs. Lancien, M. et al. reported that Tmem176a and Tmem176b (Tmem176a/b) deficiency has no effect on tumor growth in MCA101-sOVA fibrosarcoma and B16-OVA melanoma (Lancien et al., 2021). However, our results showed that in comparison to wild-type mice, Tmem176b knockout mice had a greater volume of tumors. The study has (Segovia et al., 2019) reported that targeting TMEM176B controls tumor growth. The observed differences and inconsistency may due to the fact of tumor heterogeneity in which diverse roles of certain gene in different tumor models.
It was found that deletion of Tmem176a and Tmem176b does not affect CD8+ T cell responses but plays an irreplaceable role in the optimal initiation of initial CD4+ T cells by DCs (Lancien et al., 2021). Our results showed that the overall proportion of CD3+ CD4+ T and CD3+ CD8+ T cells in tissues was reduced, and tumor-infiltrating T lymphocytes (TIL CD4+ T, TIL CD8+ T) were significantly reduced in Tmem176b −/−mice, which is consistent with our findings from public databases, where the higher the expression of TMEM176B, the higher the degree of T cell infiltration in SKCM. The reduced proportion of CD8+ T cells in Tmem176b knockout mouse could explain our finding of the accelerating progress and a poor prognosis in same mice model. Because high levels of CD8+ T cell infiltration have been shown to facilitate tumor treatment in tumors such as breast cancer and uroepithelial carcinoma (Sharma et al., 2007; Mahmoud et al., 2011).
In addition, we observed that the expression of some immunosuppressive molecules such as PD1, PD-L1 in Tmem176b knockout mice was lower than WT mice, and this downward trend was more significant at the level of PD-L1 expression. Although sometimes high PD-L1 levels predict suppression of anti-cancer immune responses upon binding to PD-1, PD-L1 expression is not necessarily equivalent to immune escape (Taube et al., 2014). A clinical study has shown that high PD-L1 expression on immune cells in uremic patients is associated with an increased probability of response to atezolizumab (An immune checkpoint blocker targeting PD-L1) (Rosenberg et al., 2016). Therefore, it is necessary to clarify that although high expression of immunosuppressive molecules or their ligands in tumors usually indicates the failure of the immune response that accompanies disease progression, they may also be a marker for the effectiveness of blockade therapies (Galluzzi et al., 2018). It has been explained that transcriptional regulation of PD-L1 may promote immune inflammation in the local tumor microenvironment, thus making tumors more sensitive to immune checkpoint blockade therapies (Gao et al., 2020). In addition, the increased expression of PD-1 in CD8+ T cells does not exactly indicate that the cells are in an exhaustion state, but also symbolize the activation of the cells (Wherry and Kurachi, 2015).
In the study, we found that, in comparison to wild-type animals, Tmem176b knockout lowered the expression of IFN-γ, PRF1 and GZMB in TIL CD8+ T cells, indicating that the function of tumor-infiltrating T cells was suppressed. As a result of that enhanced expression of IFN-γ and GZMB in the tumor microenvironment can achieve optimal tumor clearance (Lin et al., 2014). We analyzed the expression of TMEM176B in SKCM with the expression of some T cell-related genes to confirm our findings in the mouse tumor model, and we discovered that the expression of TMEM176B was significantly and positively linked with the expression of immunosuppressive molecules CD274 (PD-L1), CTLA4, LAG3, and cytotoxic molecules associated with T cell function TNF, PRF1, GZMA, GZMK, and GZMM, and most cytotoxic molecules, have been validated to act mainly as anti-tumoral and anti-infectious factors (Arias et al., 2017).
Conclusion
Taken together, as one of the aggressive malignancies, the survival rate of melanoma varies depending on the stage of diagnosis. We discovered that TMEM176B expression was higher in SKCM than in normal tissue, and enhanced expression was linked to better OS. Also, TMEM176B was positively linked to immune cell infiltration in SKCM, which may explain that TMEM176B affects the prognosis of tumor patients. These findings lead us to hypothesize that TMEM176B can be employed as a diagnostic and prognostic marker. To validate our hypothesis, we developed melanoma model in Tmem176b knockout mice and revealed that T cell infiltration and effector function were significantly inhibited. It suggests that TMEM176B plays an essential role in anti-tumor function of CD8+ T cells, but the underlying mechanisms need to be further investigated.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics Statement
The animal study was reviewed and approved by the Zhongshan Hospital, Fudan University Ethical Committee.
Author Contributions
DW and ZS gained funding, designed the study and revised the manuscript. LJ and YY performed experiments and wrote the manuscript; FL and MM analyzed the data; JG, LS, and YC performed experiments and drew figures.
Funding
This study was supported by the National Natural Science Foundation of China (No. 81771672), “Cross key project of mathematics and medical health” of the National Natural Science Foundation of China (No. 12026608), Open fund project of Shenzhen BGI Institute of Life Science (No. BGIRSZ20200004) and Special Fund for Clinical Research of Zhongshan Hospital, Fudan University (No. 2020ZSLC07), Key Subject Construction Program of Shanghai Health Administrative Authority (ZK 2019B30), and Shanghai Engineering Research Center of Tumor Multi-Target Gene Diagnosis (20DZ2254300).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2022.859958/full#supplementary-material
Supplementary Table 1 | Clinical characteristics of the SKCM patients based on TCGA.
Supplementary Table 2 | The number of cases used in each cancer types.
References
Arias, M., Martínez-Lostao, L., Santiago, L., Ferrandez, A., Granville, D. J., and Pardo, J. (2017). The Untold Story of Granzymes in Oncoimmunology: Novel Opportunities with Old Acquaintances. Trends Cancer 3 (6), 407–422. doi:10.1016/j.trecan.2017.04.001
Bindea, G., Mlecnik, B., Tosolini, M., Kirilovsky, A., Waldner, M., Obenauf, A. C., et al. (2013). Spatiotemporal Dynamics of Intratumoral Immune Cells Reveal the Immune Landscape in Human Cancer. Immunity 39 (4), 782–795. doi:10.1016/j.immuni.2013.10.003
Choi, Y., Jeon, Y.-H., Kang, J.-H., Chung, J.-K., Schmidt, M., and Kim, a. C.-W. (2007). MIDGE/hNIS Vaccination Generates Antigen-Associated CD8+IFN-γ+ T Cells and Enhances Protective Antitumor Immunity. Int. J. Cancer 120 (9), 1942–1950. doi:10.1002/ijc.22567
Condamine, T., Le Texier, L., Howie, D., Lavault, A., Hill, M., Halary, F., et al. (2010). Tmem176B and Tmem176A Are Associated with the Immature State of Dendritic Cells. J. Leukoc. Biol. 88 (3), 507–515. doi:10.1189/jlb.1109738
Cuajungco, M. P., Podevin, W., Valluri, V. K., Bui, Q., Nguyen, V. H., and Taylor, K. (2012). Abnormal Accumulation of Human Transmembrane (TMEM)-176A and 176B Proteins Is Associated with Cancer Pathology. Acta Histochem. 114 (7), 705–712. doi:10.1016/j.acthis.2011.12.006
Drujont, L., Lemoine, A., Moreau, A., Bienvenu, G., Lancien, M., Cens, T., et al. (2016). RORγt+ Cells Selectively Express Redundant Cation Channels Linked to the Golgi Apparatus. Sci. Rep. 6, 23682. doi:10.1038/srep23682
Eon Kuek, L., Leffler, M., Mackay, G. A., and Hulett, M. D. (2016). The MS4A Family: Counting Past 1, 2 and 3. Immunol. Cel Biol 94 (1), 11–23. doi:10.1038/icb.2015.48
Galluzzi, L., Chan, T. A., Kroemer, G., Wolchok, J. D., and López-Soto, A. (2018). The Hallmarks of Successful Anticancer Immunotherapy. Sci. Transl Med. 10 (459), eaat7807. doi:10.1126/scitranslmed.aat7807
Galon, J., Costes, A., Sanchez-Cabo, F., Kirilovsky, A., Mlecnik, B., Lagorce-Pagès, C., et al. (2006). Type, Density, and Location of Immune Cells within Human Colorectal Tumors Predict Clinical Outcome. Science 313 (5795), 1960–1964. doi:10.1126/science.1129139
Gao, Y., Nihira, N. T., Bu, X., Chu, C., Zhang, J., Kolodziejczyk, A., et al. (2020). Acetylation-Dependent Regulation of PD-L1 Nuclear Translocation Dictates the Efficacy of Anti-PD-1 Immunotherapy. Nat. Cel Biol 22 (9), 1064–1075. doi:10.1038/s41556-020-0562-4
Hollestein, L. M., van den Akker, S. A. W., Nijsten, T., Karim-Kos, H. E., Coebergh, J. W., and de Vries, E. (2012). Trends of Cutaneous Melanoma in The Netherlands: Increasing Incidence Rates Among All Breslow Thickness Categories and Rising Mortality Rates since 1989. Ann. Oncol. 23 (2), 524–530. doi:10.1093/annonc/mdr128
Huntington, N. D., Cursons, J., and Rautela, J. (2020). The Cancer-Natural Killer Cell Immunity Cycle. Nat. Rev. Cancer 20 (8), 437–454. doi:10.1038/s41568-020-0272-z
Kim, H., Kim, H., Feng, Y., Li, Y., Tamiya, H., Tocci, S., et al. (2020). PRMT5 Control of cGAS/STING and NLRC5 Pathways Defines Melanoma Response to Antitumor Immunity. Sci. Transl Med. 12 (551), eaaz5683. doi:10.1126/scitranslmed.aaz5683
Lancien, M., Bienvenu, G., Salle, S., Gueno, L., Feyeux, M., Merieau, E., et al. (2021). Dendritic Cells Require TMEM176A/B Ion Channels for Optimal MHC Class II Antigen Presentation to Naive CD4+ T Cells. J. Immunol. 207 (2), 421–435. doi:10.4049/jimmunol.2000498
Lin, R., Chen, L., Chen, G., Hu, C., Jiang, S., Sevilla, J., et al. (2014). Targeting miR-23a in CD8+ Cytotoxic T Lymphocytes Prevents Tumor-dependent Immunosuppression. J. Clin. Invest. 124 (12), 5352–5367. doi:10.1172/jci76561
Lin, W. M., and Fisher, D. E. (2017). Signaling and Immune Regulation in Melanoma Development and Responses to Therapy. Annu. Rev. Pathol. Mech. Dis. 12, 75–102. doi:10.1146/annurev-pathol-052016-100208
Liu, J., Lichtenberg, T., Hoadley, K. A., Poisson, L. M., Lazar, A. J., Cherniack, A. D., et al. (2018). An Integrated TCGA Pan-Cancer Clinical Data Resource to Drive High-Quality Survival Outcome Analytics. Cell 173 (2), 400–416. doi:10.1016/j.cell.2018.02.052
Louvet, C., Chiffoleau, E., Heslan, M., Tesson, L., Heslan, J.-M., Brion, R., et al. (2005). Identification of a New Member of the CD20/FcepsilonRIbeta Family Overexpressed in Tolerated Allografts. Am. J. Transpl. 5 (9), 2143–2153. doi:10.1111/j.1600-6143.2005.01007.x
Lurton, J., Rose, T. M., Raghu, G., and Narayanan, A. S. (1999). Isolation of a Gene Product Expressed by a Subpopulation of Human Lung Fibroblasts by Differential Display. Am. J. Respir. Cel Mol Biol 20 (2), 327–331. doi:10.1165/ajrcmb.20.2.3368
Ma, C., and Li, H. (2021). Hub Gene and its Key Effects on Diffuse Large B-Cell Lymphoma by Weighted Gene Coexpression Network Analysis. Biomed. Res. Int. 2021, 8127145. doi:10.1155/2021/8127145
Mahmoud, S. M. A., Paish, E. C., Powe, D. G., Macmillan, R. D., Grainge, M. J., Lee, A. H. S., et al. (2011). Tumor-Infiltrating CD8+ Lymphocytes Predict Clinical Outcome in Breast Cancer. J. Clin. Oncol. 29 (15), 1949–1955. doi:10.1200/jco.2010.30.5037
Michielin, O., van Akkooi, A. C. J., Ascierto, P. A., Dummer, R., and Keilholz, U. (2019). Cutaneous Melanoma: ESMO Clinical Practice Guidelines for Diagnosis, Treatment and Follow-Up. Ann. Oncol. 30 (12), 1884–1901. doi:10.1093/annonc/mdz411
Otsubo, T., Hida, Y., Ohga, N., Sato, H., Kai, T., Matsuki, Y., et al. (2014). Identification of Novel Targets for Antiangiogenic Therapy by Comparing the Gene Expressions of Tumor and Normal Endothelial Cells. Cancer Sci. 105 (5), 560–567. doi:10.1111/cas.12394
Quail, D. F., and Joyce, J. A. (2013). Microenvironmental Regulation of Tumor Progression and Metastasis. Nat. Med. 19 (11), 1423–1437. doi:10.1038/nm.3394
Rosenberg, J. E., Hoffman-Censits, J., Powles, T., van der Heijden, M. S., Balar, A. V., Necchi, A., et al. (2016). Atezolizumab in Patients with Locally Advanced and Metastatic Urothelial Carcinoma Who Have Progressed Following Treatment with Platinum-Based Chemotherapy: A Single-Arm, Multicentre, Phase 2 Trial. Lancet 387 (10031), 1909–1920. doi:10.1016/S0140-6736(16)00561-4
Schadendorf, D., Fisher, D. E., Garbe, C., Gershenwald, J. E., Grob, J.-J., Halpern, A., et al. (2015). Melanoma. Nat. Rev. Dis. Primers 1, 15003. doi:10.1038/nrdp.2015.3
Segovia, M., Russo, S., Jeldres, M., Mahmoud, Y. D., Perez, V., Duhalde, M., et al. (2019). Targeting TMEM176B Enhances Antitumor Immunity and Augments the Efficacy of Immune Checkpoint Blockers by Unleashing Inflammasome Activation. Cancer Cell 35 (5), 767–781. doi:10.1016/j.ccell.2019.04.003
Shain, A. H., and Bastian, B. C. (2016). From Melanocytes to Melanomas. Nat. Rev. Cancer 16 (6), 345–358. doi:10.1038/nrc.2016.37
Sharma, P., Shen, Y., Wen, S., Yamada, S., Jungbluth, A. A., Gnjatic, S., et al. (2007). CD8 Tumor-Infiltrating Lymphocytes Are Predictive of Survival in Muscle-Invasive Urothelial Carcinoma. Proc. Natl. Acad. Sci. U.S.A. 104 (10), 3967–3972. doi:10.1073/pnas.0611618104
Sun, L., Zhang, Y., and Zhang, C. (2018). Distinct Expression and Prognostic Value of MS4A in Gastric Cancer. Open Med. (Wars) 13, 178–188. doi:10.1515/med-2018-0028
Taube, J. M., Klein, A., Brahmer, J. R., Xu, H., Pan, X., Kim, J. H., et al. (2014). Association of PD-1, PD-1 Ligands, and Other Features of the Tumor Immune Microenvironment with Response to Anti-PD-1 Therapy. Clin. Cancer Res. 20 (19), 5064–5074. doi:10.1158/1078-0432.ccr-13-3271
Tomimaru, Y., Mishra, S., Safran, H., Charpentier, K. P., Martin, W., De Groot, A. S., et al. (2015). Aspartate-β-Hydroxylase Induces Epitope-Specific T Cell Responses in Hepatocellular Carcinoma. Vaccine 33 (10), 1256–1266. doi:10.1016/j.vaccine.2015.01.037
Viklicky, O., Krystufkova, E., Brabcova, I., Sekerkova, A., Wohlfahrt, P., Hribova, P., et al. (2013). B-Cell-Related Biomarkers of Tolerance Are Up-Regulated in Rejection-Free Kidney Transplant Recipients. Transplantation 95 (1), 148–154. doi:10.1097/tp.0b013e3182789a24
Wherry, E. J., and Kurachi, M. (2015). Molecular and Cellular Insights into T Cell Exhaustion. Nat. Rev. Immunol. 15 (8), 486–499. doi:10.1038/nri3862
Glossary
ACC Adrenocortical carcinoma
BLCA Bladder urothelial carcinoma
BRCA Breast invasive carcinoma
CESC Cervical squamous cell carcinoma and endocervical adenocarcinoma
CHOL Cholangiocarcinoma
COAD Colon adenocarcinoma
DLBC Lymphoid neoplasm diffuse large B-cell lymphoma
ESCA Esophageal carcinoma
GBM Glioblastoma multiforme
GBMLGG Glioma
HNSC Head and neck squamous cell carcinoma
KICH Kidney chromophobe
KIRC Kidney renal clear cell carcinoma
KIRP Kidney renal papillary cell carcinoma
LAML Acute myeloid leukemia
LGG Brain lower grade glioma
LIHC Liver hepatocellular carcinoma
LUAD Lung adenocarcinoma
LUSC Lung squamous cell carcinoma
MESO Mesothelioma
OV Ovarian serous cystadenocarcinoma
PAAD Pancreatic adenocarcinoma
PCPG Pheochromocytoma and paraganglioma
PRAD Prostate adenocarcinoma
READ Rectum adenocarcinoma
SARC Sarcoma
SKCM Skin cutaneous melanoma
STAD Stomach adenocarcinoma
TGCT Testicular germ cell tumors
THCA Thyroid carcinoma
THYM Thymoma
UCEC Uterine corpus endometrial carcinoma
UCS Uterine carcinosarcoma
UVM Uveal melanoma
Keywords: TMEM176B, cancer, CD8+ T cell, tumor immunology, SKCM
Citation: Jiang L, Yang Y, Liu F, Ma M, Gao J, Sun L, Chen Y, Shen Z and Wu D (2022) A Potential Diagnostic and Prognostic Biomarker TMEM176B and Its Relationship With Immune Infiltration in Skin Cutaneous Melanoma. Front. Cell Dev. Biol. 10:859958. doi: 10.3389/fcell.2022.859958
Received: 22 January 2022; Accepted: 04 March 2022;
Published: 23 March 2022.
Edited by:
Geng Chen, GeneCast Biotechnology Co., Ltd., ChinaReviewed by:
Ting Li, National Center for Toxicological Research (FDA), United StatesLiqun He, Uppsala University, Sweden
Copyright © 2022 Jiang, Yang, Liu, Ma, Gao, Sun, Chen, Shen and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Duojiao Wu, d3UuZHVvamlhb0B6cy1ob3NwaXRhbC5zaC5jbg==; Zan Shen, c3NoZW56emFuQHZpcC5zaW5hLmNvbQ==
†These authors have contributed equally to this work and share first authorship
 Linlan Jiang
Linlan Jiang Yanyin Yang
Yanyin Yang Fangming Liu
Fangming Liu Mingyue Ma
Mingyue Ma Jie Gao
Jie Gao Lu Sun
Lu Sun Yuwen Chen
Yuwen Chen Zan Shen
Zan Shen Duojiao Wu
Duojiao Wu