
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Cell Dev. Biol. , 24 March 2022
Sec. Cellular Biochemistry
Volume 10 - 2022 | https://doi.org/10.3389/fcell.2022.857237
This article is part of the Research Topic Bioinorganic Chemistry of Metals in Cell Function and Disease View all 13 articles
Iron deficiency anemia can be treated with oral or intravenous Fe supplementation. Such supplementation has considerable effects on the human microbiome, and on opportunistic pathogenic micro-organisms. Molecular understanding of the control and regulation of Fe availability at the host-microbe interface is crucial to interpreting the side effects of Fe supplementation. Here, we provide a concise overview of the regulation of Fe by the opportunistic pathogen Staphylococcus aureus. Ferric uptake regulator (Fur) plays a central role in controlling Fe uptake, utilization and storage in order to maintain a required value. The micro-organism has a strong preference for heme iron as an Fe source, which is enabled by the Iron-regulated surface determinant (Isd) system. The strategies it employs to overcome Fe restriction imposed by the host include: hijacking host proteins, replacing metal cofactors, and replacing functions by non-metal dependent enzymes. We propose that integrated omics approaches, which include metalloproteomics, are necessary to provide a comprehensive understanding of the metal tug of war at the host-microbe interface down to the molecular level.
Iron deficiency is a major health concern worldwide, resulting in over one billion cases of iron-deficiency anemia (Gardner and Kassebaum 2020; Pasricha et al., 2021). Currently, the first-line treatment for iron deficiency anemia is the use of oral iron supplements. However, many side effects have been reported following their use: e.g., 30–70% of the patients report gastrointestinal problems (DeLoughery 2019). The supplemented iron is only partially absorbed by the human body, resulting in a significantly increased amount of iron available to the human gut microbiome (DeLoughery 2019; Finlayson-Trick et al., 2020). The microbiome of an individual plays an important role in human health, and metal compounds are known to affect the survival and reproduction of bacteria (Sheldon and Skaar 2019). Recognition of these side effects and the discovery that intravenously administered iron causes only minor adverse effects, after having been (incorrectly) considered more dangerous for decades, has sparked the use of intravenous Fe supplementation (Auerbach, Gafter-Gvili, and Macdougall 2020; Schaefer et al., 2020). However, following intravenous iron administration, blood borne pathogens will be exposed to excess iron. One of these pathogens, responsible for a wide variety of clinical diseases, is Staphylococcus aureus of which the methicillin resistant (MRSA) strain represents a global human health challenge (David and Daum 2010; DeLeo et al., 2010; Ganz et al., 2020).
The effect of supplemented iron on S. aureus proliferation has been investigated in a few patient studies only, where bacterial growth assays were performed on serum samples taken from the subjects following iron supplementation. In this way, Cross et al. (2015) found supplemented oral iron to significantly increase transferrin saturation (TSAT) in the serum samples. However, while gram-negative bacteria, including E. coli, and the gram-positive Staphylococcus epidermis, showed elevated growth rates, S. aureus appeared unaffected (Cross et al., 2015). The authors suggested this to be caused by a preference of S. aureus for heme iron over transferrin-bound iron (Cross et al., 2015), which is consistent with other studies (Barton Pai et al., 2006; Suffredini et al., 2017; Skaar et al., 2004). In fact, in hemodialysis patients, which have significantly lower transferrin levels, intravenous iron sucrose administration was found to correlate with increased non-transferrin-bound iron (NTBI) levels in the patients’ serum. Significantly increased S. aureus growth was observed on the serum samples of these patients, compared to the NTBI-negative subjects (Barton Pai et al., 2006). This indicates that the molecular form of iron in the blood influences its uptake by S. aureus, which seems to prefer NTBI and heme iron, but is less responsive to transferrin-bound iron.
While multiple reviews have recently been published on the interaction of supplemented iron and enteric pathogens on a molecular level (Yilmaz and Li 2018; Finlayson-Trick et al., 2020; Qi et al., 2020), investigations focusing on the impact of supplemented intravenous iron on blood borne pathogens such as S. aureus are lacking. Therefore, in this mini-review, we aim to give an overview of recent insights into iron and S. aureus in the context of excess iron and iron-limiting conditions imposed by the host (nutritional immunity) during S. aureus infections. Here, we will first look in detail at the regulation systems S. aureus uses to control uptake of both free and heme iron, and to regulate the intracellular Fe levels, and then describe how the pathogen is able to survive under iron starvation conditions.
In engineering, control systems regulate the operation of devices and their processes using control loops. For a functioning control loop, you need to measure a process value which can be either below or above a target set point. The device or process then needs to be adjusted to attain the desired process value. In a similar manner, bacteria have evolved to control the intracellular concentrations of nutrients and metabolites, including iron, to pre-set conditions required for growth and/or maintenance. For this process, called homeostasis, bacteria produce sensors that measure the amount of intracellular iron, and a control system (or systems) that can change the expression of proteins and the functionality of enzymes in order to reach the target value. This control occurs at: transcriptional (DNA→ mRNA), post-transcriptional (stabilizing or degradation of mRNA), translational (mRNA→ protein) or post-translational level (protein degradation, modification, and allosteric interaction). These four levels allow very precise tuning and distribution of iron, depending on necessity and environmental conditions. Precise tuning is important because Fe is essential for life, while at the same time Fe2+ can generate toxic reactive oxygen species (ROS) with O2, and Fe3+ is insoluble under neutral aqueous conditions.
In Staphylococcus aureus, Fur (ferric uptake regulator) is the major control system for iron (Figure 1). The Fur protein is homodimeric, with each monomer consisting of an N-terminal DNA binding domain and a C-terminal dimerization domain (Price and Boyd 2020). Between the two domains is a metal ion binding site, which was recently shown to bind a [2Fe-2S] cluster in E. coli (Fontenot et al., 2020). S. aureus Fur has been described as binding two separate Fe2+ ions in the hinge regions between the N- and C-terminal domains. E. coli and S. aureus Fur share 30% sequence identity and 49% sequence similarity, which includes three conserved Cysteines (Supplementary Material). The precise nature of the Fe-bound form of S. aureus Fur remains to be established. Upon dimerization, the DNA binding part of Fe-bound Fur is a transcriptional repressor of a range of genes related to iron homeostasis. It functions by binding to a so-called Fur-box upstream of the coding genes. These genes involve Fe transporters and many other genes, as discussed below. Some are established virulence factors, which means they are involved in disease processes. Related proteins, called Fur family proteins, with affinity to other metal ions or compounds have been discovered, such as Zur (zinc uptake regulator) for Zn2+ and PerR (peroxide operon regulator) which is a metal-dependent regulator for hydrogen peroxide. The Fe2+ and Mn2+ dependence of PerR highlights crosslinks between the different control systems (Horsburgh, Ingham, and Foster 2001).
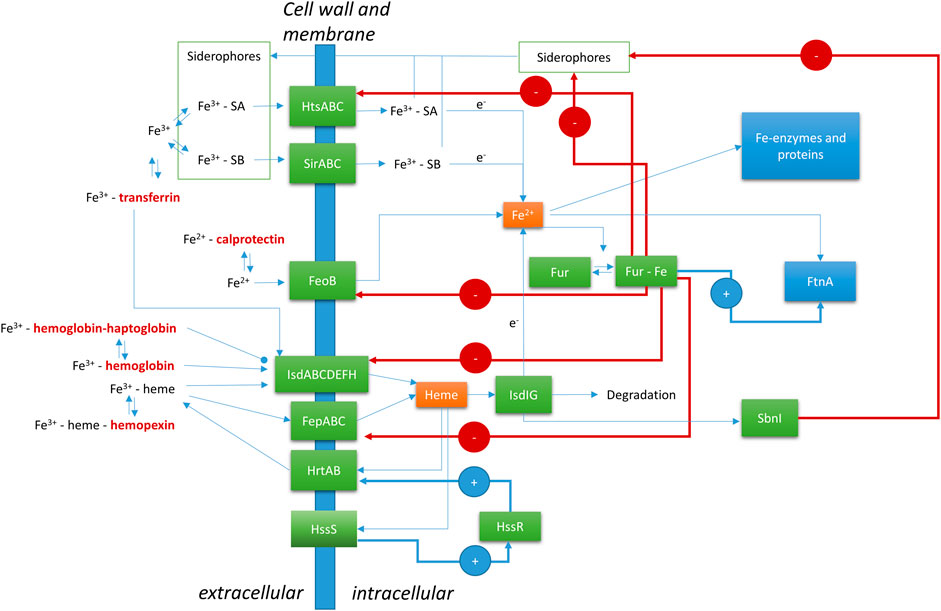
FIGURE 1. Regulation of Fe by Staphylococcus aureus at the host-microbe interface. FeoB, ferrous Fe transporter; FepABC, Fe dependent peroxidase transporter; FtnA, ferritin; Fur, ferric uptake regulator; HrtAB, heme regulator transporter efflux pump; HssR, heme sensing two-component regulator regulatory protein; HssS, heme sensing two-component regulator sensor protein; HtsABC, heme transport system involved in Fe-SA uptake; IsdABCDEFGHI, iron-regulated surface determinant system; SA, Staphyloferrin A; SB, Staphyloferrin B; SbnI, L-serine kinase and heme responsive regulator of SB biosynthesis. Human (host) proteins are in bold and dark red font.
Ferrous iron uptake by transporter FeoB is still poorly characterized, although recently inhibitors have been identified that may have important medical implications as novel antibiotics against MRSA and other multidrug resistant Gram-positive bacteria (Shin et al., 2021). Fur and PerR differentially regulate the S. aureus ferritin gene (FtnA), which encodes the Fe-storage protein ferritin (Morrissey et al., 2004). Ferritin can take up circa 4,000 Fe atoms in the form of a ferrihydrite mineral nanoparticle inside a 24-meric protein sphere (Honarmand Ebrahimi, Hagedoorn, and Hagen 2015). Iron storage by ferritin involves oxidation of Fe2+ to Fe3+ and concerted incorporation into a growing ferrihydrite mineral core. Upon reduction, by a mechanism that has not yet been established, Fe can also be released from ferritin as Fe2+.
Another Fur controlled Fe uptake system is the FepABC (Fe dependent peroxidase) transporter. The transporter has not been well characterized, but it has been implicated in Fe and possibly heme uptake. The transporter consists of FepA, a predicted membrane anchored lipoprotein that may act as an Fe (compound) binding protein, peroxidase FepB, and integral membrane protein FepC (Biswas et al., 2009). FepB can bind heme and protoporphyrin IX (heme without Fe) and has a low peroxidase activity (Turlin et al., 2013). FepB is a substrate of the Twin-Arginine Translocation pathway, which allows membrane translocation of fully folded cofactor bound proteins (Biswas et al., 2009). Heterologous expression of the S. aureus FepAB in E. coli allowed heme utilization in this organism (Turlin et al., 2013).
Siderophores are small, extracellular, peptide-derived compounds with a high affinity for Fe3+. S. aureus produces two siderophores: Staphyloferrin A (SA) and Staphyloferrin B (SB). SA is produced using the gene cluster sfa, and SB is produced using the sbn gene cluster (Marchetti et al., 2020). The gene clusters are both transcriptionally repressed by Fur. Recently, a heme sensitive regulator of siderophore production was identified: SbnI (Laakso et al., 2016; Verstraete et al., 2019). The gene product of SbnI is an enzyme producing a precursor to the siderophore. Furthermore, the protein can bind DNA and upon dimerization contains a heme binding domain. Heme transfer from IsdI to SnbI has been suggested to help control the production of siderophores, thereby shifting focus to heme utilization rather than free iron uptake. Fe3+ bound SA is taken up using the ABC transporter HtsABC (heme transport system). Despite the name, it is unclear if the Hts transporter is involved in heme uptake, and if so by which mechanism (Price and Boyd 2020). Hts transcription is regulated by Fur. SB is taken up by the ABC transporter SirABC (Grigg et al., 2010a).
Siderophores play a very important role in Staphylococcus biofilm formation to ensure Fe availability (Johnson et al., 2005; Oliveira et al., 2021). Fe chelators that compete with siderophores can disturb biofilm formation and may therefore be of medical importance (Richter et al., 2017; Coraça-Huber et al., 2018).
Heme obtained from red blood cells is a major source of iron for Staphylococcus aureus during infections (Skaar et al., 2004). There is evidence that S. aureus has evolved a specificity towards human hemoglobin versus other mammalian orthologs that is unique among pathogenic bacteria (Pishchany et al., 2010). However, a high level of intracellular heme is dangerous due to its potential to form reactive oxygen species (ROS). S. aureus uses the two-component regulator HssRS (Figure 1) (Heme sensing two-component system) to control the intracellular level of free heme (Price and Boyd 2020; Stauff, Torres, and Skaar 2007; Stauff and Skaar 2009). HssS is a transmembrane protein, which responds to the heme level by an unknown mechanism. Upon activation, HssS acts as a histine kinase to phosphorylate the histidine of HssR, thereby activating the protein as a transcriptional activator of the heme efflux transporter HrtAB (Heme regulator transporter efflux pump). Whether the precise compound that is expelled by HrtAB is heme or a heme metabolite is unknown (Price and Boyd 2020). Additional targets of HssR have not been identified to date.
The cell wall of S. aureus contains a unique system to acquire heme, which is called Isd (Iron-regulated surface determinant system) (Skaar and Schneewind 2004; Grigg et al., 2010b; Mazmanian et al., 2003). Similar systems are present in other Gram-positive pathogenic bacteria, such as Listeria monocystogenes and Clostridium tetani. This system takes up heme from human (host) hemoproteins. The Isd system involves nine different proteins, of which four are bound to the cell wall: IsdA, IsdB, IsdC, and IsdH. Two proteins, IsdE and IsdF, constitute an ABC-transporter for the heme cofactor. IsdD is a transmembrane protein of unknown function. And the final two proteins, IsdI and IsdG, are soluble intracellular heme degrading enzymes. The outer cell wall proteins IsdB and IsdH bind free heme, methemoglobin and hemoglobin-haptoglobin complexes from the host. The cell wall proteins IsdC and IsdA are involved in heme transport through the 15–30 nm thick cell wall to the ligand binding component of the ABC-transporter IsdE. After translocation of the heme to the cytoplasm, the cofactor is degraded by the heme degrading enzymes IsdI and IsdG. These enzymes are distantly related to well-characterized heme oxygenases and have been found to release Fe from the cofactor, yet the precise reaction mechanism remains to be solved (Grigg et al., 2010b). Fur regulates the expression of the genes for IsdA, IsdB, IsdC and IsdH. The gene for the enzyme sortase B (SrtB) is part of the same transcriptional unit as IsdC, and therefore also regulated by Fur. Sortase B is involved in the cell wall anchoring of the Isd components.
Upon infection, the host starts the immune response. Macrophages are activated by interaction with S. aureus via Toll-like receptors (TLRs) (Pidwill et al., 2021). This starts signal transduction cascades which include mechanisms to limit the availability of iron in blood (Pandur et al., 2021). Central in the regulation of these processes is the hormone hepcidin. Hepcidin interacts with the host Fe efflux protein ferroportin, thereby limiting Fe export from macrophages (Theurl et al., 2008). Interestingly the same exposed S. aureus lipoproteins that support Fe acquisition, for example, via the Isd system, are also recognized by the TLRs, thereby evoking inflammation responses in the host (Schmaler et al., 2009; Sheldon and Heinrichs 2012). One of the cellular host responses involved is the endocytosis and degradation of erythrocytes by the macrophages in a process called erythrophagocytosis (Knutson et al., 2005). The Fe retained by the host cells is also put to good use, as Fe2+ enabled production of ROS is used to kill bacteria taken up by these cells (Rosen et al., 1995; Haschka, Hoffmann, and Weiss 2021). Interestingly. ROS also induce antibiotic resistance in S. aureus, indicating a negative side-effect of our innate immune response (Rowe et al., 2020).
In the context of sepsis, S. aureus is capable of lysing erythrocytes by secreting hemolytic toxins to free hemoglobin and obtain it through the Isd system (Torres et al., 2010). The S. aureus heme-oxygenases IsdG and IsdI have been shown to be important for full virulence with heme as the primary iron source (Reniere and Skaar 2008). Host heme oxygenase 1 (HO1) catalyzes the rate-limiting step in heme degradation, producing biliverdin, Fe2+ and CO (carbon monoxide) (Singh et al., 2018). CO can act as a messenger in various protective cascades. Links between HO1 and protective effects on S. aureus infection have been shown (MacGarvey et al., 2012; Gahlot et al., 2017).
The host restricts the availability of iron in its different forms further by producing the hemoglobin binding protein haptoglobin, the heme binding protein hemopexin, the free Fe3+ binding proteins transferrin and lactoferrin, and the free Fe2+ binding protein calprotectin (Nakashige et al., 2015; Marchetti et al., 2020). Haptoglobin binding to hemoglobin inhibits uptake of heme by the Isd system of S. aureus, although the protein still binds to IsdH (Mikkelsen, Runager, and Andersen 2020). Calprotectin (CP) was originally identified to be involved in Mn2+ limitation by the host, but was more recently found to bind Fe2+ efficiently in the presence of Ca2+ (Nakashige et al., 2015). It has been demonstrated that CP induces Fe starvation in S. aureus cultures (Obisesan, Zygiel, and Nolan 2021; Zygiel et al., 2021). The ability of S. aureus to efficiently incorporate heme affords protection against CP induced Fe starvation (Zygiel et al., 2021). In the preceding sections we have described the mechanisms through which S. aureus controls intracellular Fe levels in response to iron sources in the human host. However, these control systems may be insufficient when Fe availability is strongly reduced. Pathogenic bacteria such as S. aureus have evolved several strategies to tackle the metal restrictions imposed through nutritional immunity: 1) hijacking host proteins, 2) replacing metal cofactors, and 3) replacing functions by non-metal dependent enzymes. We will discuss examples of each strategy below.
S. aureus cannot use hemopexin as a heme source using the Isd system. However, it has been reported to take up iron from host transferrin using a transferrin receptor. The nature of the transferrin receptor of S. aureus is convoluted in literature. This cell-wall associated protein was first identified as a functional glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (Modun, Morrissey, and Williams 2000). However, this was shown to be incorrect, and the protein was identified as staphylococcal transferrin-binding protein StbA (Taylor and Heinrichs 2002). Later, it was shown that StbA is the same protein as IsdA, part of the Isd system for heme uptake described above (Clarke, Wiltshire, and Foster 2004; Maresso and Schneewind 2006). The ongoing tug of war for Fe between host transferrin and bacterial transferrin receptors has caused rapid evolutionary development of the involved proteins (Barber and Elde 2014). The presence of a transferrin receptor indicates that S. aureus can take up Fe from transferrin, at least to some extent, although the major Fe source from the host is heme.
A well-established example of replacing metal cofactors by S. aureus are the Mn-dependent superoxide dismutases (SODs). Neutrophils, and other host immune cells, can induce oxidative bursts as a defensive strategy against S. aureus (Rigby and DeLeo 2012). This process generates high levels of damaging ROS, including superoxide (Forrester et al., 2018; Jakubczyk et al., 2020). The expression of SODs is one way in which S. aureus can combat ROS. S. aureus has two superoxide dismutases, SodA and SodM. SodA can incorporate only Mn as metal cofactor while SodM can use either Fe or Mn depending on the conditions (Garcia et al., 2017). As part of the host immune response, the neutrophil protein CP sequesters trace metals, including Mn. The action of CP disturbs the correct metalation of SodA (Kehl-Fie et al., 2013). However, S. aureus encodes an additional Mn-dependent SOD, SodM, which can substitute its metal cofactor for Fe under Mn-limiting conditions (Garcia et al., 2017; Treffon et al., 2020). In this way, S. aureus can retain sufficient SOD activity despite CP activity and maintain virulence. Small non-coding regulatory RNA molecule RsaC (co-transcribed with Mn transporter MntABC) represses the translation of the SodA coding mRNA under Mn limiting conditions. So, if there is a shortage of Mn (Mn uptake by MntABC needed), the Mn SOD is suppressed in favor of the Fe containing SOD. A clinically relevant example of a highly oxidative stressful environment with strong CP presence are the airways of cystic fibrosis patients, where S. aureus can cause persistent infections for years. Investigation of gene expression demonstrated significantly elevated SodM expression levels in clinical isolates compared to laboratory strains (Treffon et al., 2020).
An alternative strategy of S. aureus to respond to nutritional immunity is to use a protein variant that lacks a metal cofactor altogether. The consumption of glucose through glycolysis is a process fundamental to many life forms. For bacteria, some of the enzymes involved are Mn-dependent. Yet even under Mn-limited conditions, S. aureus was shown to prefer glucose as main carbon source despite its burden on cellular Mn demand (Radin et al., 2019b). It has recently been demonstrated that S. aureus can express a Mn-independent variant of phosphoglycerate mutase to maintain glucose consumption under Mn-stress (Radin et al., 2019a). The discovery of metal-independent variants is not unique to S. aureus and may indicate a broader pattern among bacteria.
Despite the wealth of knowledge on metal homeostasis in S. aureus at the host-microbe interface, a comprehensive overview of the complex interactions between different metal control systems is currently lacking. Many individual proteins and pathways have been identified. However, in most cases, the molecular mechanisms of action of all these different proteins are not fully established. We envision that an integrated omics approach to determine the changes in the proteome, metabolome and metalloproteome will offer a comprehensive view of the pathogen response to nutritional immunity as well as Fe overload. Recently, integrated omics approaches have been successfully used to explore the molecular action of metallodrugs such as platinum and ruthenium anticancer drugs (Wang, Li, and Sun 2019; Steel and Hartinger 2020). Of key importance will be metalloproteomics, which will provide information on the changing intracellular metal distribution among S. aureus’ proteins. Such experiments have been performed for Fe in E. coli using the metalloproteomic approach Metal Isotope Radio Autography in Gel Electrophoresis (MIRAGE), which showed substantial changes in Fe distribution among proteins under normal and high Fe conditions (Sevcenco et al., 2011; 2009). It will be especially interesting to focus on the complex interplay between Fe and Mn homeostasis in this respect, as described above for SOD. A comprehensive understanding down to the molecular level will provide a new basis for the development of treatments against pathogenic bacteria, including MRSA. At the same time, it will allow clinicians to take pathogen response into account when treating iron-deficient patients, paving the way towards personalized treatment. This new understanding is crucial, especially considering the continuous increase of multidrug-resistant bacteria and their far-reaching impact on human health globally.
MCD, RMK, and P-LH conceptualized, wrote and edited the manuscript and approved the submitted version of the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2022.857237/full#supplementary-material
Auerbach, M., Gafter-Gvili, A., and Macdougall, I. C. (2020). Intravenous Iron: A Framework for Changing the Management of Iron Deficiency. Lancet Haematol. 7, e342. doi:10.1016/S2352-3026(19)30264-9
Barber, M. F., and Elde, N. C. (2014). Escape from Bacterial Iron Piracy through Rapid Evolution of Transferrin. Science 346 (6215), 1362–1366. doi:10.1126/science.1259329
Barton Pai, A., Pai, M. P., Depczynski, J., McQuade, C. R., and Mercier, R.-C. (2006). Non-Transferrin-Bound Iron Is Associated with Enhanced Staphylococcus aureus Growth in Hemodialysis Patients Receiving Intravenous Iron Sucrose. Am. J. Nephrol. 26 (3), 304–309. doi:10.1159/000094343
Biswas, L., Biswas, R., Nerz, C., Ohlsen, K., Schlag, M., Schafer, T., et al. (2009). Role of the Twin-Arginine Translocation Pathway in Staphylococcus. J. Bacteriol. 191 (19), 5921–5929. doi:10.1128/JB.00642-09
Clarke, S. R., Wiltshire, M. D., and Foster, S. J. (2004). IsdA of Staphylococcus aureus Is a Broad Spectrum, Iron-Regulated Adhesin. Mol. Microbiol. 51 (5), 1509–1519. doi:10.1111/j.1365-2958.2003.03938.x
Coraça-Huber, D. C., Dichtl, S., Steixner, S., Nogler, M., and Weiss, G. (2018). Iron Chelation Destabilizes Bacterial Biofilms and Potentiates the Antimicrobial Activity of Antibiotics against Coagulase-Negative Staphylococci. Pathog. Dis. 76 (5), 52. doi:10.1093/femspd/fty052
Cross, J. H., Bradbury, R. S., Fulford, A. J., Jallow, A. T., Wegmüller, R., Prentice, A. M., et al. (2015). Oral Iron Acutely Elevates Bacterial Growth in Human Serum. Sci. Rep. 5 (1), 1–7. doi:10.1038/srep16670
David, M. Z., and Daum, R. S. (2010). Community-Associated Methicillin-Resistant Staphylococcus Aureus : Epidemiology and Clinical Consequences of an Emerging Epidemic. Clin. Microbiol. Rev. 23 (3), 616–687. doi:10.1128/CMR.00081-09
DeLeo, F. R., Otto, M., Kreiswirth, B. N., and Chambers, H. F. (2010). Community-Associated Meticillin-Resistant Staphylococcus aureus. The Lancet 375 (9725), 1557–1568. doi:10.1016/S0140-6736(09)61999-1
DeLoughery, T. G. (2019). Safety of Oral and Intravenous Iron. Acta Haematol. 142 (1), 8–12. doi:10.1159/000496966
Finlayson-Trick, E. C. L., Fischer, J. A., Goldfarb, D. M., and Karakochuk, C. D. (2020). The Effects of Iron Supplementation and Fortification on the Gut Microbiota: A Review. Gastrointest. Disord. 2 (4), 327–340. doi:10.3390/gidisord2040030
Fontenot, C. R., Tasnim, H., Valdes, K. A., Popescu, C. V., and Ding, H. (2020). Ferric Uptake Regulator (Fur) Reversibly Binds a [2Fe-2S] Cluster to Sense Intracellular Iron Homeostasis in Escherichia Coli. J. Biol. Chem. 295 (46), 15454–15463. doi:10.1074/jbc.RA120.014814
Forrester, S. J., Kikuchi, D. S., Hernandes, M. S., Xu, Q., and Griendling, K. K. (2018). Reactive Oxygen Species in Metabolic and Inflammatory Signaling. Circ. Res. 122 (6), 877–902. doi:10.1161/CIRCRESAHA.117.311401
Gahlot, S., Nasreen, N., Johnson, J. A., Sahn, S. A., and Mohammed, K. A. (2017). Heme Oxygenase-1 Deficiency Diminishes Methicillin-Resistant Staphylococcus aureus Clearance Due to Reduced TLR9 Expression in Pleural Mesothelial Cells. PLOS ONE 12 (1), e0169245. doi:10.1371/journal.pone.0169245
Ganz, T., Aronoff, G. R., Goodnough, L. T., Macdougall, I. C., Mayer, G., Porto, G., et al. (2020). Iron Administration, Infection, and Anemia Management in CKD: Untangling the Effects of Intravenous Iron Therapy on Immunity and Infection Risk. Kidney Med. 2, 341–353. doi:10.1016/j.xkme.2020.01.006
Garcia, Y. M., Barwinska-Sendra, A., Tarrant, E., Skaar, E. P., Waldron, K. J., and Kehl-Fie, T. E. (2017). A Superoxide Dismutase Capable of Functioning with Iron or Manganese Promotes the Resistance of Staphylococcus aureus to Calprotectin and Nutritional Immunity. Plos Pathog. 13 (1), e1006125–19. doi:10.1371/journal.ppat.1006125
Gardner, W., and Kassebaum, N. (2020). Global, Regional, and National Prevalence of Anemia and its Causes in 204 Countries and Territories, 1990-2019. Curr. Dev. Nutr. 4 (Supplement_2), 830. doi:10.1093/cdn/nzaa053_035
Grigg, J. C., Cheung, J., Heinrichs, D. E., and Murphy, M. E. P. (2010a). Specificity of Staphyloferrin B Recognition by the SirA Receptor from Staphylococcus aureus. J. Biol. Chem. 285 (45), 34579–34588. doi:10.1074/jbc.M110.172924
Grigg, J. C., Ukpabi, G., Gaudin, C. F. M., and Murphy, M. E. P. 2010b. “Structural Biology of Heme Binding in the Staphylococcus aureus Isd System.” J. Inorg. Biochem. 104 (3): 341–348. doi:10.1016/j.jinorgbio.2009.09.012
Haschka, D., Hoffmann, A., and Weiss, G. (2021). Iron in Immune Cell Function and Host Defense. Semin. Cel Develop. Biol. 115 (July), 27–36. doi:10.1016/j.semcdb.2020.12.005
Honarmand Ebrahimi, K., Hagedoorn, P.-L., and Hagen, W. R. (2015). Unity in the Biochemistry of the Iron-Storage Proteins Ferritin and Bacterioferritin. Chem. Rev. 115 (1), 295–326. doi:10.1021/cr5004908
Horsburgh, M. J., Ingham, E., and Foster, S. J. 2001, In Staphylococcus aureus , Fur Is an Interactive Regulator with PerR, Contributes to Virulence, and Is Necessary for Oxidative Stress Resistance through Positive Regulation of Catalase and Iron Homeostasis, J. Bacteriol., 183 (2): 468–475. doi:10.1128/JB.183.2.468-475.2001
Jakubczyk, K., Dec, K., Kałduńska, J., Kawczuga, D., Kochman, J., and Janda, K. (2020). Reactive Oxygen Species - Sources, Functions, Oxidative Damage. Pol. Merkur Lekarski 48 (284), 124–127.
Johnson, M., Cockayne, A., Williams, P. H., and Morrissey, J. A. 2005. “Iron-Responsive Regulation of Biofilm Formation in Staphylococcus Aureus Involves Fur-Dependent and Fur-Independent Mechanisms.” J. Bacteriol. 187 (23): 8211–8215. doi:10.1128/JB.187.23.8211-8215.2005
Kehl-Fie, T. E., Chitayat, S., Hood, M. I., Damo, S., Restrepo, N., Garcia, C., et al. (2011). Nutrient Metal Sequestration by Calprotectin Inhibits Bacterial Superoxide Defense, Enhancing Neutrophil Killing of Staphylococcus aureus. Cell Host & Microbe 10 (2), 158–164. doi:10.1016/j.chom.2011.07.004
Knutson, M. D., Oukka, M., Koss, L. M., Aydemir, F., and Wessling-Resnick, M. (2005). Iron Release from Macrophages after Erythrophagocytosis Is Up-Regulated by Ferroportin 1 Overexpression and Down-Regulated by Hepcidin. Proc. Natl. Acad. Sci. 102 (5), 1324–1328. doi:10.1073/pnas.0409409102
Laakso, H. A., Marolda, C. L., Pinter, T. B., Stillman, M. J., and Heinrichs, D. E. (2016). A Heme-Responsive Regulator Controls Synthesis of Staphyloferrin B in Staphylococcus aureus. J. Biol. Chem. 291 (1), 29–40. doi:10.1074/jbc.M115.696625
MacGarvey, N. C., Suliman, H. B., Bartz, R. R., Fu, P., Withers, C. M., Welty-Wolf, K. E., et al. (2012). Activation of Mitochondrial Biogenesis by Heme Oxygenase-1-Mediated NF-E2-Related Factor-2 Induction Rescues Mice from Lethal Staphylococcus aureus Sepsis. Am. J. Respir. Crit. Care Med. 185 (8), 851–861. doi:10.1164/rccm.201106-1152OC
Marchetti, M., de Bei, O., Bettati, S., Campanini, B., Kovachka, S., Gianquinto, E., et al. (2020). Iron Metabolism at the Interface between Host and Pathogen: From Nutritional Immunity to Antibacterial Development. Int. J. Mol. Sci. 21 (6), 2145. doi:10.3390/ijms21062145
Maresso, A. W., and Schneewind., O. (2006). Iron Acquisition and Transport in Staphylococcus aureus. BioMetals 19 (2), 193–203. doi:10.1007/s10534-005-4863-7
Mazmanian, S. K., Skaar, E. P., Gaspar, A. H., Humayun, M., Gornicki, P., Jelenska, J., et al. (2003). Passage of Heme-Iron across the Envelope of Staphylococcus Aureus. Science 299 (5608), 906–909. doi:10.1126/science.1081147
Mikkelsen, J. H., Runager, K., and Andersen, C. B. F. (2020). The Human Protein Haptoglobin Inhibits IsdH-Mediated Heme-Sequestering by Staphylococcus aureus. J. Biol. Chem. 295 (7), 1781–1791. doi:10.1074/jbc.RA119.011612
Modun, B., Morrissey, J., and Williams, P. (2000). The Staphylococcal Transferrin Receptor: A Glycolytic Enzyme with Novel Functions. Trends Microbiol. 8 (5), 231–237. doi:10.1016/S0966-842X(00)01728-5
Morrissey, J. A., Cockayne, A., Brummell, K., and Williams, P. 2004. “The Staphylococcal Ferritins Are Differentially Regulated in Response to Iron and Manganese and via PerR and Fur.” Infect. Immun. 72 (2): 972–979. doi:10.1128/IAI.72.2.972-979.2004
Nakashige, T. G., Zhang, B., Krebs, C., and Nolan, E. M. (2015). Human Calprotectin Is an Iron-Sequestering Host-Defense Protein. Nat. Chem. Biol. 11 (10), 765–771. doi:10.1038/nchembio.1891
Obisesan, A. O., Zygiel, E. M., and Nolan, E. M. (2021). Bacterial Responses to Iron Withholding by Calprotectin. Biochemistry 60 (45), 3337–3346. doi:10.1021/acs.biochem.1c00572
Oliveira, F., Rohde, H., Vilanova, M., and Cerca, N. (2021). Fighting Staphylococcus epidermidis Biofilm-Associated Infections: Can Iron Be the Key to Success? Front. Cel. Infect. Microbiol. 11 563. doi:10.3389/fcimb.2021.798563
Pandur, E., Tamási, K., Pap, R., Jánosa, G., and Sipos, K. (2021). Distinct Effects of Escherichia coli, Pseudomonas aeruginosa and Staphylococcus aureus Cell Wall Component-Induced Inflammation on the Iron Metabolism of THP-1 Cells. Int. J. Mol. Sci. 22 (3), 1497. doi:10.3390/ijms22031497
Pasricha, Sant. Rayn., Tye-Din, Jason., Muckenthaler, Martina. U., and Swinkels, Dorine. W. (2021). Iron Deficiency. The Lancet 397, 233. doi:10.1016/S0140-6736(20)32594-0
Pidwill, G. R., Gibson, J. F., Cole, J., Renshaw, S. A., and Foster, S. J. (2021). The Role of Macrophages in Staphylococcus aureus Infection. Front. Immunol. 11 (January). doi:10.3389/fimmu.2020.620339
Pishchany, G., McCoy, A. L., Torres, V. J., Krause, J. C., Crowe, J. E., Fabry, M. E., et al. (2010). Specificity for Human Hemoglobin Enhances Staphylococcus aureus Infection. Cell Host & Microbe 8 (6), 544–550. doi:10.1016/j.chom.2010.11.002
Price, E. E., and Boyd, J. M. (2020). Genetic Regulation of Metal Ion Homeostasis in Staphylococcus aureus. Trends Microbiol. 28 (10), 821–831. doi:10.1016/j.tim.2020.04.004
P. Skaar, E., and Schneewind, O. 2004. “Iron-Regulated Surface Determinants (Isd) of Staphylococcus aureus: Stealing Iron from Heme.” Microbes Infect. 6 (4): 390–397. doi:10.1016/j.micinf.2003.12.008
Qi, X., Zhang, Y., Guo, H., Hai, Y., Luo, Y., and Yue, T. (2020). Mechanism and Intervention Measures of Iron Side Effects on the Intestine. Crit. Rev. Food Sci. Nutr. 60 (12), 2113–2125. doi:10.1080/10408398.2019.1630599
Radin, J. N., Kelliher, J. L., Párraga Solórzano, P. K., Grim, K. P., Ramezanifard, R., Slauch, J. M., et al. (2019a). Metal-Independent Variants of Phosphoglycerate Mutase Promote Resistance to Nutritional Immunity and Retention of Glycolysis during Infection. Plos Pathog. 15 (7), e1007971–21. doi:10.1371/journal.ppat.1007971
Radin, J. N., Zhu, J., Brazel, E. B., McDevitt, C. A., and Kehl-Fie, T. E. (2019b). Synergy between Nutritional Immunity and Independent Host Defenses Contributes to the Importance of the MntABC Manganese Transporter during Staphylococcus aureus Infection. Infect. Immun. 87 (1). doi:10.1128/IAI.00642-18
Reniere, M. L., and Skaar, E. P. (2008). Staphylococcus aureus Haem Oxygenases Are Differentially Regulated by Iron and Haem. Mol. Microbiol. 69 (5), 1304–1315. doi:10.1111/j.1365-2958.2008.06363.x
Richter, K., Thomas, N., Claeys, J., McGuane, J., Prestidge, C. A., Coenye, T., et al. (2017). A Topical Hydrogel with Deferiprone and Gallium-Protoporphyrin Targets Bacterial Iron Metabolism and Has Antibiofilm Activity. Antimicrob. Agents Chemother. 61 (6). doi:10.1128/AAC.00481-17
Rigby, K. M., and DeLeo, F. R. (2012). Neutrophils in Innate Host Defense against Staphylococcus aureus Infections. Semin. Immunopathol 34 (2), 237–259. doi:10.1007/s00281-011-0295-3
Rosen, G. M., Pou, S., RamosCohen, C. L. Myron. S., Cohen, M. S., and Britigan, B. E. (1995). Free Radicals and Phagocytic Cells. FASEB J. 9 (2), 200–209. doi:10.1096/fasebj.9.2.7540156
Rowe, S. E., Wagner, N. J., Li, L., Beam, J. E., Wilkinson, A. D., Radlinski, L. C., et al. (2020). Reactive Oxygen Species Induce Antibiotic Tolerance during Systemic Staphylococcus aureus Infection. Nat. Microbiol. 5 (2), 282–290. doi:10.1038/s41564-019-0627-y
Schaefer, B., Meindl, E., Wagner, S., Tilg, H., and Zoller, H. (2020). Intravenous Iron Supplementation Therapy. Mol. Aspects Med. 75, 100862. doi:10.1016/j.mam.2020.100862
Schmaler, M., Jann, N. J., Ferracin, F., Landolt, L. Z., Biswas, L., Götz, F., et al. (2009). Lipoproteins in Staphylococcus aureus Mediate Inflammation by TLR2 and Iron-dependent Growth In Vivo. J. Immunol. 182 (11), 7110–7118. doi:10.4049/jimmunol.0804292
Sevcenco, A.-M., Krijger, G. C., Pinkse, M. W. H., Verhaert, P. D. E. M., Hagen, W. R., and Hagedoorn, P.-L. (2009). Development of a Generic Approach to Native Metalloproteomics: Application to the Quantitative Identification of Soluble Copper Proteins in Escherichia coli. J. Biol. Inorg. Chem. 14, 631–640. doi:10.1007/s00775-009-0477-9
Sevcenco, A.-M., Pinkse, M. W. H., Wolterbeek, H. T., Verhaert, P. D. E. M., Hagen, W. R., and Hagedoorn, P.-L. 2011. “Exploring the Microbial Metalloproteome Using MIRAGE.” Metallomics 3: 1324–1330. doi:10.1039/C1MT00154J
Sheldon, J. R., and Heinrichs, D. E. (2012). The Iron-Regulated Staphylococcal Lipoproteins. Front. Cel. Inf. Microbio. 2. doi:10.3389/fcimb.2012.00041
Sheldon, J. R., and Skaar, E. P. (2019). Metals as Phagocyte Antimicrobial Effectors. Curr. Opin. Immunol. 60 (October), 1–9. doi:10.1016/j.coi.2019.04.002
Shin, M., Jin, Y., Park, J., Mun, D., Kim, S. R., Payne, S. M., et al. (2021). Characterization of an Antibacterial Agent Targeting Ferrous Iron Transport Protein FeoB against Staphylococcus aureus and Gram-Positive Bacteria. ACS Chem. Biol. 16 (1), 136–149. doi:10.1021/acschembio.0c00842
Singh, N., Ahmad, Z., Baid, N., and Kumar, A. (2018). Host Heme Oxygenase-1: Friend or Foe in Tackling Pathogens? IUBMB Life 70 (9), 869–880. doi:10.1002/iub.1868
Skaar, E. P., Humayun, M., Bae, T., DeBord, K. L., and Schneewind., O. (2004). Iron-Source Preference of Staphylococcus aureus Infections. Science 305 (5690), 1626–1628. doi:10.1126/science.1099930
Stauff, D. L., and Skaar, E. P. (2009). The Heme Sensor System of Staphylococcus aureus. Contrib. Microbiol. 16, 120–135. doi:10.1159/000219376
Stauff, D. L., Torres, V. J., and Skaar, E. P. (2007). Signaling and DNA-Binding Activities of the Staphylococcus aureus HssR-HssS Two-Component System Required for Heme Sensing. J. Biol. Chem. 282 (36), 26111–26121. doi:10.1074/jbc.M703797200
Steel, T. R., and Hartinger, C. G. (2020). Metalloproteomics for Molecular Target Identification of Protein-Binding Anticancer Metallodrugs. Metallomics 12 (11), 1627–1636. doi:10.1039/d0mt00196a
Suffredini, D. A., Xu, W., Sun, J., Barea‐Mendoza, J., Solomon, S. B., Brashears, S. L., et al. (2017). Parenteral Irons versus Transfused Red Blood Cells for Treatment of Anemia during Canine Experimental Bacterial Pneumonia. Transfusion 57 (10), 2338–2347. doi:10.1111/trf.14214
Taylor, J. M., and Heinrichs, D. E. (2002). Transferrin Binding in Staphylococcus aureus: Involvement of a Cell Wall-Anchored Protein. Mol. Microbiol. 43 (6), 1603–1614. doi:10.1046/j.1365-2958.2002.02850.x
Theurl, I., Theurl, M., Seifert, M., Mair, S., Nairz, M., Rumpold, H., et al. (2008). Autocrine Formation of Hepcidin Induces Iron Retention in Human Monocytes. Blood 111 (4), 2392–2399. doi:10.1182/blood-2007-05-090019
Torres, V. J., Attia, A. S., Mason, W. J., Hood, M. I., Corbin, B. D., Beasley, F. C., et al. (2010). Staphylococcus aureus Fur Regulates the Expression of Virulence Factors that Contribute to the Pathogenesis of Pneumonia. Infect. Immun. 78 (4), 1618–1628. doi:10.1128/IAI.01423-09
Treffon, J., Chaves‐Moreno, D., Niemann, S., Pieper, D. H., Vogl, T., Roth, J., et al. (2020). Importance of Superoxide Dismutases A and M for Protection of Staphylococcus aureus in the Oxidative Stressful Environment of Cystic Fibrosis Airways. Cell Microbiol. 22, 1–14. doi:10.1111/cmi.13158
Turlin, E., Débarbouillé, M., Augustyniak, K., Gilles, A.-M., and Wandersman, C. (2013). Staphylococcus aureus FepA and FepB Proteins Drive Heme Iron Utilization in Escherichia coli. PLoS ONE 8 (2), e56529. doi:10.1371/journal.pone.0056529
Verstraete, M. M., Morales, L. D., Kobylarz, M. J., Loutet, S. A., Laakso, H. A., Pinter, T. B., et al. (2019). The Heme-Sensitive Regulator SbnI Has a Bifunctional Role in Staphyloferrin B Production by Staphylococcus aureus. J. Biol. Chem. 294 (30), 11622–11636. doi:10.1074/jbc.RA119.007757
Wang, Y., Li, H., and Sun, H. (2019). Metalloproteomics for Unveiling the Mechanism of Action of Metallodrugs. Inorg. Chem. 58 (20), 13673–13685. doi:10.1021/acs.inorgchem.9b01199
Yilmaz, B., and Li, H. (2018). Gut Microbiota and Iron: The Crucial Actors in Health and Disease. Pharmaceuticals 11 (4), 98. doi:10.3390/PH11040098
Keywords: Staphylococcus aureus, MRSA, iron homeostasis, nutritional immunity, iron deficiency anemia, ferric uptake regulator, iron-regulated surface determinant system, heme
Citation: van Dijk MC, de Kruijff RM and Hagedoorn P- (2022) The Role of Iron in Staphylococcus aureus Infection and Human Disease: A Metal Tug of War at the Host—Microbe Interface. Front. Cell Dev. Biol. 10:857237. doi: 10.3389/fcell.2022.857237
Received: 18 January 2022; Accepted: 24 February 2022;
Published: 24 March 2022.
Edited by:
Fernando Antunes, University of Lisbon, PortugalReviewed by:
Guenter Weiss, Innsbruck Medical University, AustriaCopyright © 2022 van Dijk, de Kruijff and Hagedoorn. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Robin M. de Kruijff, ci5tLmRla3J1aWpmZkB0dWRlbGZ0Lm5s; Peter-Leon Hagedoorn, cC5sLmhhZ2Vkb29ybkB0dWRlbGZ0Lm5s
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.