- 1Department of Neurology, Shanghai Children’s Hospital, Shanghai JiaoTong University, Shanghai, China
- 2Department of Clinical Laboratory, Shanghai Children’s Hospital, Shanghai Jiao Tong University, Shanghai, China
- 3NHC Key Laboratory of Medical Embryogenesis and Developmental Molecular Biology and Shanghai Key Laboratory of Embryo and Reproduction Engineering, Shanghai, China
Background: The chromodomain helicase DNA-binding protein 2 (CHD2) gene, is an ATPase and part of the CHD family of chromatin remodelers. Mutations in the CHD2 gene are inherited in an autosomal-dominant manner and can lead to intellectual disability, epilepsy, and autism. We investigated the clinical characteristics of CHD2-related conditions and their possible pathogenesis.
Methods: We collected and analysed the clinical data of patients that were identified as having CHD2 mutations. Genetic testing was performed using targeted sequencing or whole-exome sequencing. We analysed the expression of CHD2 and repressor element 1-silencing transcription factor (REST) in blood samples using quantitative PCR and the conservation of the mutations. The CHD2 mutations we identified were compared with the known mutations reported in the CHD2-related literature.
Results: Eight patients with CHD2 gene mutations were analysed. Six mutations were identified; four were unreported previously (c.670C>T; c.4012A>C; c.2416dup; c.1727–1728insAT), and two were known mutations: c.5035C>T (two cases) and c.4173dup (two cases). Among these mutations, seven were de novo mutations, and one could not be determined because the parents refused genetic testing. The clinical manifestations included mild or severe intellectual disability, epilepsy, and behavioural abnormalities. Quantitative PCR showed that the CHD2 gene expression levels among the patients, parents, and the controls were not significantly different. The levels of REST gene expression in the patients were significantly higher than those of the controls; thus, mutation of the CHD2 gene led to an increase in the expression level of the REST gene. The mutations reported were all located in conserved positions in different species. Among the various medications administered for treatment, valproate showed the best results for the treatment of epilepsy caused by CHD2 gene mutation.
Conclusion: Mutation in CHD2 did not lead to a significant decrease in its expression level, indicating that the clinical phenotype was unrelated to its expression level, and the mutant protein may retain some function. Most of the mutations relatively stable. In addition, the clinical manifestations from the same mutation in the CHD2 gene were different among the known cases; this may be related to the regulation of REST or other regulatory factors.
Introduction
The chromodomain helicase DNA-binding protein 2 (CHD2) is a member of the ATP-dependent chromatin remodelling family of proteins that are critical for the assembly and regulation of chromatin (Lamar and Carvill, 2018). The CHD2 gene is located on the long arm of chromosome 15 and encodes a member of the CHD family of proteins (Wilson et al., 2021). The CHD2 gene contains 42 exons that encode 1828 amino acids. The CHD2 protein is composed of several functional domains, including two chromodomains at the N-terminus, an ATPase/helicase domain, and a DNA-binding domain. The N-terminal region of CHD2, which contains tandem chromodomains, serves an auto inhibitory role in both the DNA-binding and ATPase activities of CHD2 (Liu et al., 2015).
In 2009, a microdeletion of 15q26.1 was identified in a 30-month-old female who had refractory myoclonic epilepsy with intellectual disability, growth retardation, and a distinctive facial appearance (Veredice et al., 2009). In 2013, the gene primarily responsible for the epileptic component, and the disruption of neural developmental directly caused by seizures that resulted from the 15q26 defect, was CHD2 (Suls et al., 2013; Capelli et al., 2012; Epi4K Consortium et al., 2013; Poot et al., 2013). Since then, several mutations in the CHD2 gene have been identified and associated with developmental delay, microcephaly, autism, and epilepsy (Epi4K Consortium et al., 2013; Poot et al., 2013; Carvill et al., 2013; O'Roak et al., 2014; Deciphering Developmental Disorders Study, 2017). Seizures typically develop as early as 6 months and before 4 years of age; the multiple seizure types include myoclonus, myoclonic-absence seizures, and drop attacks (Capelli et al., 2012; Lamar and Carvill, 2018; Nieto-Estevez and Hsieh, 2018; Chen et al., 2020).
This study reports the cases of eight patients with CHD2 gene mutations and describes their clinical characteristics, laboratory examinations, electroencephalogram (EEG) results, mutational genotypes, and mRNA expression levels of CHD2 and repressor element 1-silencing transcription factor (REST). To our knowledge, all CHD2 mutations reported so far are heterozygous and the vast majority are new mutations. At present, the mechanism of CHD2 mutation to autosomal dominance is not clear. In this paper, we will preliminarily study and speculate the cause. Besides we found that the expression of REST gene was significantly increased in patients with CHD2 mutation, but the specific mechanism is still unclear and needs further study.
Materials and Methods
Patients
Venous blood samples from eight patients suspected of having CHD2 mutations were collected in the Shanghai Children’s Hospital from September 2019 to June 2021 after informed consent was obtained from the patients and/or their families. Data about age at seizure onset, seizure types, birth history, family history, EEG and brain magnetic resonance imaging (MRI) results, and treatment were also collected. This study was approved by the Ethics Committee of Shanghai Children’s Hospital, and all protocols complied with Chinese bioethics laws and the Declaration of Helsinki.
Genetic Analysis and cDNA Synthesis
The screening for mutations in CHD2 (RefSeq NM_001271.3) was performed using epilepsy-targeted next-generation sequencing (P6) or whole-exome sequencing (WES, P1-P5 and P7-P8). This study was approved by the Ethics Committee of Shanghai Children’s Hospital (approval No. 2019R071-F03). Variants calling and interpretation have been described elsewhere (Xiaozhen et al., 2021).
Total RNA was extracted from 0.25 ml of fresh blood using an RNA extraction kit (YEASEN Biotech Co., Ltd., Shanghai, China) according to the manufacturer’s instructions (Qiagen). The concentration and purity of the total RNA were detected using a nucleic acid–protein analyser. Next, cDNA was synthesized using the total RNA as the template for reverse transcription according to the manufacturer’s instructions of the reverse transcription kit. The reaction contents were 5 μl template RNA (≤500 ng), 2 μl Primer Script RT Enzyme Mix, and 3 μl RNase-free water. The samples were incubated at 37°C for 15 min and then 85°C for 5 s.
Quantitative PCR Analysis of CHD2 and REST Gene Expression
Because two families refused a second collection of blood samples, cDNA made from the blood of the core members of the remaining six families and control peers was used for fluorescence quantitative PCR (qPCR), which was carried out using a real-time fluorescence qPCR kit (Takara Biomedical Technology) according to the manufacturer’s instructions. Due to an insufficient blood sample in Case 5, the REST gene expression level could not be determined. The primers used to measure the expression of CHD2 were (forward) 5′-CGAAAACAGGCACTGGACCACT-3′ and (reverse) 5′-GATGACGACTGTGTCCGCTGAA-3′. The REST primers were (forward) 5′-CCCCTTCACATGGAGCCAAT-3′ and (reverse) 5′-CTTCCGTGCCCTTTCACTCT-3′. The reaction (20 μl) contained cDNA (2 μl), forward primer (10 μM; 0.4 μl), reverse primer (10 μM; 0.4 μl), 2× SYBR Premix Ex Taq (10 μl), and nanopure water (7.2 μl). Housekeeping gene GAPDH was used as a relative quantity. Statistical analysis. Data are presented as means ± s.e.m. from three independent experiments. Each experiment was performed in duplicate. Data were analyzed using the Student’s t-test, and p < 0.05 was considered to be statistically significant.
Conserved Sequence Analysis
We analyzed the mutations in the CHD2 protein sequence for H.Sapiens, M.Mulatta, C.Lupus, M.Muschulus, R.Norvegicus, G.Gallus, D.Rerio and D.Melanogaster to predict sequence conservation at these loci.
Results
Clinical Data
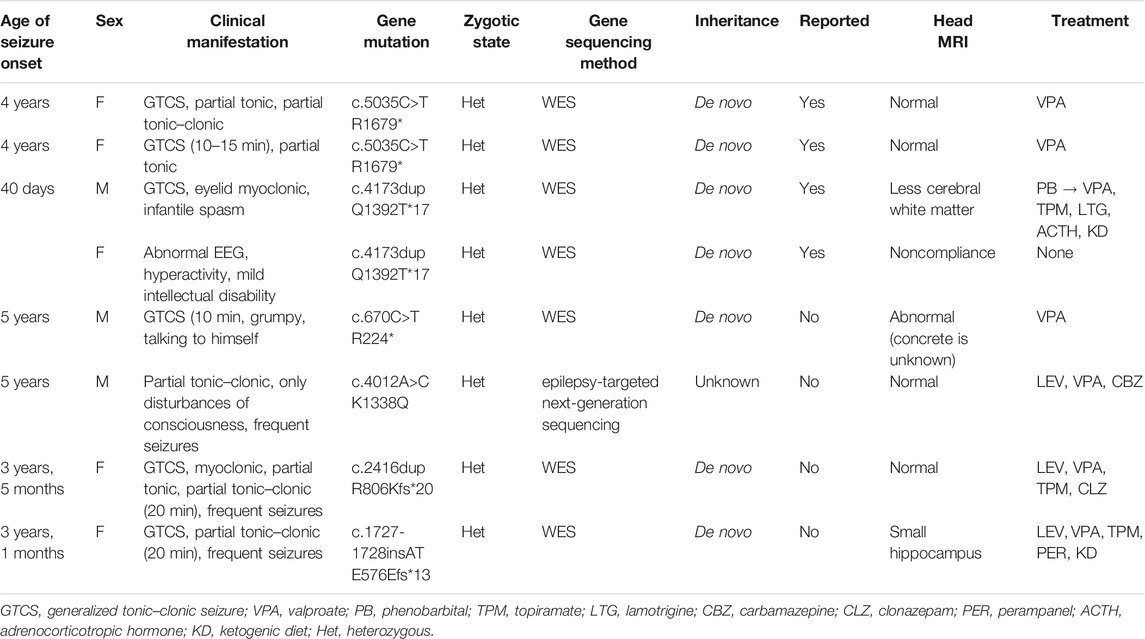
The clinical characteristics of the patients, the CHD2 gene mutations identified in the patients, and their treatments are presented in Table 1. In all, eight children with CHD2 gene mutations in seven families were analysed. These included three males and five females, with six mutations, including two mutations that had been reported previously (c.5035C>T, p. R 1679*; and c.4173dup, p. Q 1392 T*17), and four mutations had not been reported (c.670C> T, p. R 224*; c4012A>C, p. K 1338Q; c.2416dup, p. R806Kfs*20; and c.1727–1728insAT, p. E576Efs*13) (Figure 1). All of these mutations were de novo in the children; in Case 6, the parents refused genetic examination.
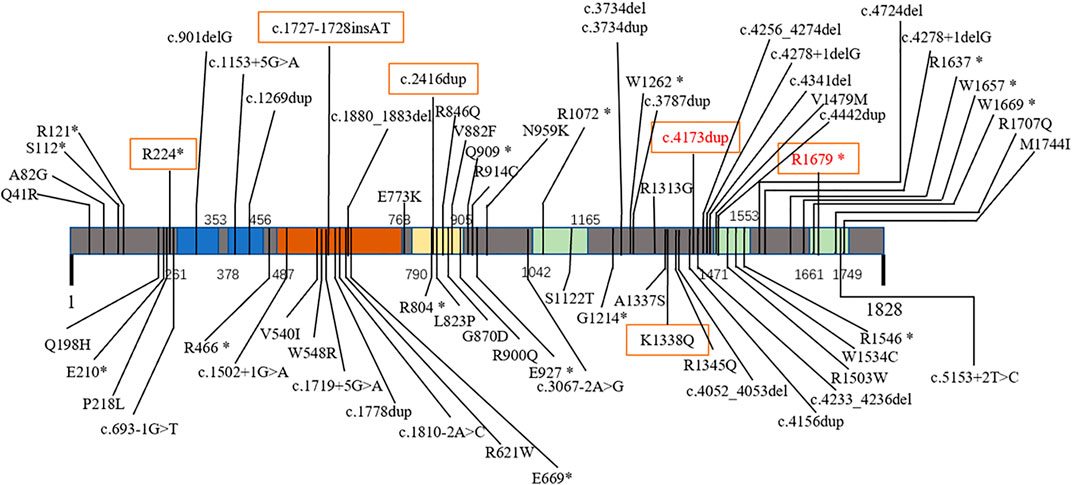
FIGURE 1. Mutations of CHD2-related epilepsy in different domains. Primary structure and mutations in the CHD2 protein: chromodomain 1 (261–353) and 2 (378–456) (blue); SNF2-related helicase domain (487–768, brown) with DEAD-like helicase superfamily (503–648) and helicase ATP-binding (496–666) domains; ATP helicase domain (790–905, yellow); and domains of unknown function (1042–1165, 1471–1553, 1661–1749, green). Red text: previously reported mutations. Boxed text, mutations in this report.
Each of the eight patients had neurological abnormalities; seven had seizures, and the other one had growth retardation with multiple abnormal EEG discharges. The initial seizure age ranged from 40 days to 5 years. The types of epilepsy were generalized tonic–clonic seizure, eyelid myoclonic, myoclonic, tonic, partial tonic, partial tonic–clonic, consciousness-only disturbances, and infantile spasm. Five patients had status epilepticus, and four patients had frequent onset. Case 1 and Case 2 were identical twin sisters; the other children were unrelated. In Case 3, a caesarean section was performed before birth due to insufficient foetal movement. The child had a history of hypoglycaemia 3 days after birth, and spasticity occurred 40 days after birth. Case 6 had polydactyl deformity. The other children had no abnormal birth history. The EEG results were consistent with the clinical patterns.
Case 1 and Case 2 were twin sisters who were delivered by caesarean section at 36 weeks of gestation. Oxygen was administered through a nasal catheter after birth; the rest of the birth history was normal. They carried a de novo c.5035C>T (p.R 1679*) CHD2 mutation, which has been reported previously (Wang et al., 2017).Originally they had status epilepticus, and EEG suggests epileptic discharge, and photosensitivity. The two patients had mild developmental delays before onset and an IQ of 70; after controlling the epilepsy using valproate (VPA), an EEG examination did not find abnormalities.
Case 3 was a male with no abnormal birth history, an age of seizure onset of 40 days, who carried a de novo c.4173dup (p. Q 1392T*17) CHD2 mutation, which has been reported previously (Chen et al., 2020). He was admitted to the neonatal intensive care unit 3 days after birth due to hypoglycaemia and developed seizures 40 days after birth with clinical manifestations of eyelid myoclonus, tonic–clonus, tonics, and infantile spasms. An EEG showed hypsarrhythmia. For treatment, phenobarbital → VPA + topiramate (TPM) + adrenocorticotropic hormone (ACTH) + lamotrigine (LTG) → pulse methylprednisolone + ACTH + ketogenic diet (KD) were successively administered. After the seizure was controlled, the EEG was improved, but there was still developmental delay; she could not sit up without assistance at 11 months of age, and a head MRI exam suggested less white matter.
Case 4 was a female with no abnormal birth history who was admitted to our department due to hyperactivity and intellectual disability. Her IQ was 79. Her EEG was abnormal: epileptic discharge in the Rolandic area during sleep; previously, the head predominated with slightly more epileptic discharge, and the slow complex wave index in non-rapid eye movement sleep was about 60–65%. The CHD2 gene had a de novo c.4173dup (p. Q 1392T*17) mutation, which is the same as in Case 3. However, unlike Case 3, the child presented only developmental delay and abnormal behaviour but no seizures.
Case 5 was a male with no abnormal birth history who developed a generalized tonic–clonic seizure at the age of 5 years. An EEG showed that in the two occipital regions, there were several bursts of diffuse irregular spikes/spines/multi-spines that were emitted in short segments. After treatment with VPA, the seizures were controlled. After 4 years of drug withdrawal, one seizure occurred, and there was abnormal mental behaviour. The patient carried a de novo c.670C>T (p.R 224*) CHD2 mutation, and his IQ was 63.
Case 6 was a 5-year-old male who underwent caesarean section due to insufficient foetal movement at birth. He had multiple malformations of the little finger/toe of his right hand and right foot, and his growth and development showed regression. The epilepsy showed disturbances of consciousness and partial ankylosis, with frequent seizures at the beginning of the disease; the seizures continued after LEV+VPA treatment but were controlled with the addition of carbamazepine. The patient carried a c.4012A>C (p. K 1338 Q) CHD2 mutation; the parental genes could not be analysed due to refused consent.
Case 7 was a 3.5-year-old female with no abnormal birth history. She had frequent seizures at 3.5 years, with general tonic–clonic as the clinical manifestation. Her growth and development before onset were normal for her age, and seizures were frequent and difficult to control at the beginning of the disease. After LEV+VPA+TPM+pirampanet+KD treatment, the seizures were controlled. She carried a de novo c.2416dup (p.R806Kfs*20) CHD2 mutation.
Case 8 was a 3-year-old female with no abnormal birth history. She developed tonic–clonic seizures at the age of 3 years, with frequent seizures and initial difficulty in control. After treatment with LEV+VPA+TPM+CLZ, she had seizure control. After the onset of the seizures, there was developmental regression and mild mental behaviour abnormalities. She carried a de novo c.2416dup (p.R806Kfs*20) mutation in CHD2.
qPCR Analysis of CHD2 and REST Gene Expression
The expression levels of CHD2 in the six families and the controls were not significantly different (Figure 2A). The REST gene expression levels in the five patients were significantly higher than those of the controls (Figure 2B).
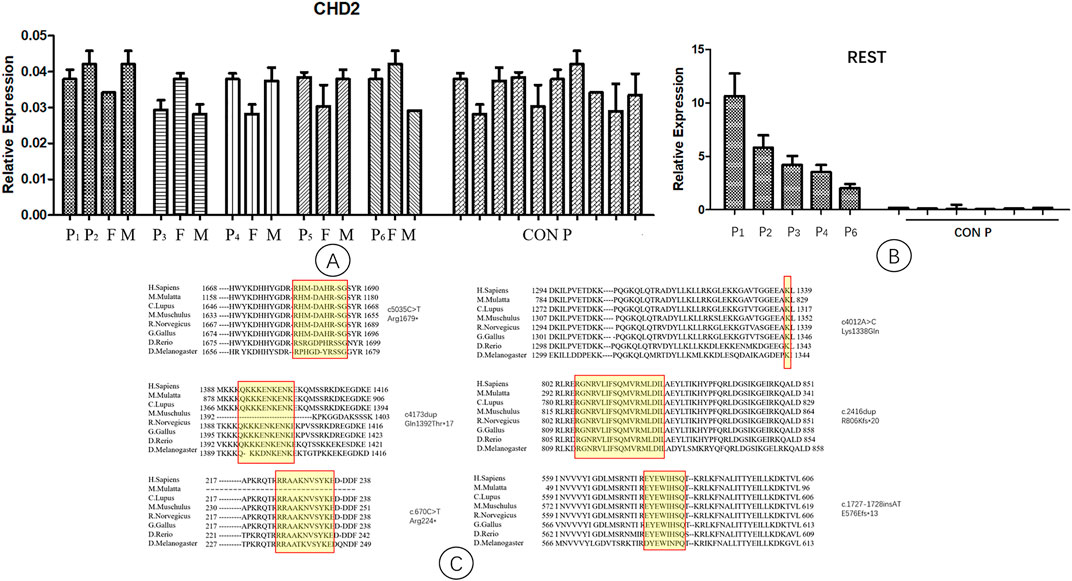
FIGURE 2. Quantitative PCR analysis of CHD2 and REST gene expression. P1-P6, patient 1–6; F, father; M, mother; CON P, control group. Arabic numerals represent the age of the control group. (A) The expression levels of the CHD2 gene in patients, parents, and the controls. They were not significantly different. (B) The expression levels of the REST gene in patients and the controls. The levels of REST gene expression in the patients were significantly higher than those of the controls. (C) Conservation studies were conducted for CHD2 mutations reported in this manuscript in H.Sapiens, M.Mulatta, C.Lupus, M.Muschulus, R.Norvegicus, G.Gallus, D.Rerio, D.Melanogaster. All the mutations in this study were relatively stable.
Conservation of Mutations
Conservation studies were conducted for all the mutations of this study in H.Sapiens, M.Mulatta, C.Lupus, M.Muschulus, R.Norvegicus, G.Gallus, D.Rerio and D.Melanogaster (Figure 2C). These analyses suggested that the mutations relatively stable.
Discussion
Mutations in the CHD2 gene can lead to neurodevelopmental delay, intellectual disability, epilepsy, and behavioural problems. Hanly et al. (Hanly et al., 2021) analysed 56 CHD2 gene mutations through a literature search and found that 80.3% (45 cases) of the patients had epilepsy; the incidence of epilepsy in our study was 87.5%. While most CHD2 mutations are de novo, a few are inherited from asymptomatic parents. To our knowledge, only Petersen et al. (Petersen et al., 2018) has reported a case of an inherited symptomatic CHD2 mutation, which affected both mother and daughter.
A search of the Human Gene Mutation Database (2020.6) shows that 128 cases of CHD2 mutation have been reported with 103 mutation sites. Missense mutations are the most common, followed by frameshift and nonsense mutations; splice mutations are uncommon, and other mutations are rare (Table 2). CHD2 is a relatively conserved gene, and many loci are relatively conserved in multiple species. These missense mutations that have been reported are mostly at these conserved loci. Among the 128 cases reported, the associated CHD2 mutations included c.693-1G>T (two cases), c.1809+1delG (three cases), c.1942C>T (two cases), c.2567A>G (three cases), c.2895_2898delAGAA (three cases), c.3521G>A (two cases), c.3734delA (two cases), c.3787dupG (two cases), c.4173dupA (four cases), c.4233_4236delAGAA (two cases), c.4799_4814delACCTTCACCCTCAGAA (three cases), c.4909C>T (four cases), c.4949dupG (three cases), c.5035C>T (three cases), and other mutations (one case). There seems to be no difference in the number of mutation sites of CHD2-related epilepsy in different domains (Figure 1).
Photosensitivity is often present, and self-induced seizures are seen in some patients (Suls et al., 2013; Galizia et al., 2015; Thomas et al., 2015). In this study, Case 1 and Case 2 showed photosensitivity, which is consistent with previous reports. Zebrafish larvae with a CHD2 knockout exhibit epileptic behaviour (Suls et al., 2013); CHD2 morpholino-injected larvae display body curvature, excessive body pigmentation, and a developmental delay (Suls et al., 2013). CHD2+/− mice showed an increase in pyramidal neuron action potential discharge, a decrease in spinous process adaptation, significantly increased amplitude of micro-excitatory postsynaptic currents, and a significantly decreased frequency of micro-inhibitory postsynaptic currents. EEG recordings from the somatosensory neocortex of CHD2+/− mice show increased frequency ranges of α, δ, θ, and γ waves, suggesting that CHD2 affects synaptic transmission of glutamate and γ-aminobutyric acid (GABA) and cortical rhythmgenesis in the adult hippocampus (Kim et al., 2018; Nieto-Estevez and Hsieh, 2018). This suggests that CHD2 mutation leads to a prominent reduction of GABA and an increase in neuronal excitability that results in seizures; this could be verified by further studies.
Two cases of the previously reported c5035C>T mutation were found in identical twin sisters (Cases 1 and 2). Their age of epilepsy onset was 4 years, and their clinical manifestations included complex partial seizures that initially presented as a persistent epileptic state and an EEG that suggested photosensitivity; these are consistent with previous reports (Wang et al., 2017).
Two cases of the p. Q 1392T*17 mutation were reported in this study, and five cases have been previously reported at this site (Lund et al., 2014; Galizia et al., 2015; Pinto et al., 2016; Chen et al., 2020), all of which were de novo and presented mainly as Lennox–Gastaut syndrome and autistic disorder. The reported manifestations included eyelid myoclonus, myoclonus, atonia, and atypical absence; all of the patients carrying this mutation had intellectual disability. However, the two cases we report in the present study had different clinical manifestations. One presented infantile spasm, while the other presented only mild intellectual disability and behavioural abnormalities, an abnormal EEG, and no seizures; these indicate that there are different clinical manifestations of the same gene mutation at the same site. Children with epilepsy inherited from asymptomatic fathers have been reported in the past (Chen et al., 2020). A CHD2 mutation that affected both mother and daughter has been reported with varying clinical manifestations (Petersen et al., 2018). This further supports the lack of a genotype–phenotype correlation among individuals with the same genetic variant, and suggests that CHD2 gene effects might be mediated by other mechanisms. It has been demonstrated that CHD2 mRNA is regulated in some tissues by the ubiquitous splicing regulator Rbfox2 (Cho et al., 2019), and CHD2 probably binds directly to REST genomic DNA to regulate the expression of REST (Shen et al., 2015). This suggests that different clinical manifestations may be related to these regulatory factors.
Mice with homozygous deletions of the C-terminus of CHD2 exhibit perinatal lethality, and heterozygous mice exhibit decreased survival rates, suggesting that CHD2 is an essential gene for normal development (Marfella et al., 2006). All the patients in this study had heterozygous mutations, and there were no homozygous or complex heterozygous mutations reported in the past, suggesting that the CHD2 gene is essential for human growth and development. Suls et al. (Suls et al., 2013) performed qPCR on two cases of nonsense mutations and found that the expression level was not decreased, which is consistent with the present study and suggests that the aberrant allele is not degraded by nonsense-mediated mRNA decay. Analysis using qPCR performed on a patient with a splicing mutation showed that both alleles were expressed (Suls et al., 2013). It is suggested that the CHD2 mutation does not affect the RNA expression level, and that the protein translated from the mutated CHD2 gene has some functionality; this mutant protein might affect the excitability of GABAergic neurons or other electrophysiological channels, resulting in increased neuronal excitability and leading to seizures.
Recent studies have shown that CHD2 is expressed throughout brain tissue, but varies in different cell subtypes at different developmental stages (Shen et al., 2015; Wilson et al., 2021). In mature mice (30 days after birth), CHD2 is significantly expressed in the olfactory bulb, neocortex, hippocampus, and cerebellum (Kim et al., 2018). In CHD2+/− mice, there are fewer GABAergic interneurons in the somatosensory cortex, and the density of GABA neurons in the cortex is decreased; the loss of GABA intermediate neurons affects hippocampal memory (Kim et al., 2018). CHD2 is critical for embryonic and adult neuronal differentiation, synaptic properties, and memory; this partially explains the symptoms of CHD2 patients (Kim et al., 2018). Brain MRI of Case 8 in this study suggested that a reduction of the hippocampal volume might be related to the CHD2 mutation. During embryonic development, there is a decrease in proliferative cells in the ventricular zone and subventricular zone as well as in NKX2.1+ progenitor cells in the medial and caudal ganglionic eminence (Kim et al., 2018). CHD2 is highly expressed in the Pax6+ radial glia but rarely expressed in Tbr2+ intermediate progenitors, suggesting that CHD2 deficiency suppresses the self-renewal capacity of the radial glia and instead promotes premature neuronal differentiation (Shen et al., 2015; Lamar and Carvill, 2018). This early differentiation probably leads to rapid depletion of the progenitor pool, resulting in a smaller cortex and defects in later-born neurons (Homem et al., 2015; Lamar and Carvill, 2018). In human cortical interneurons derived from human embryonic stem cell cultures, Meganathan and others (Shen et al., 2015) found that CHD2 expression increases during cortical interneuron differentiation.
Knockout of the CHD2 gene in human embryonic stem cell–derived interneurons results in a decrease in the number of neurons and a reduction in the neurite length (Meganathan et al., 2017). Among the eight patients in this report, head MRI indicated widening of the cerebral sulcus in four cases; in one case head MRI imaging data could not be obtained; cranial MRI did not show abnormalities in the other three cases. Except for two patients that had normal development before the onset of epilepsy, the rest of the patients had different degrees of slowed mental development; the two patients experienced mental regression after the seizure onset.
CHD2 probably binds directly to the REST genomic DNA to regulate the expression of REST; the in vitro overexpression of REST rescues the defect in neurogenesis caused by CHD2 knockdown (Shen et al., 2015). To evaluate the relationship between CHD2 and REST in vivo, we measured the expression levels of REST mRNA in patients with CHD2 mutations. We found that the expression level of REST mRNA in the peripheral blood of patients with CHD2 mutations was significantly higher than that of the control (wild type) group. This suggests that the mutation of the CHD2 gene could result in high expression of REST, which might alleviate the clinical symptoms caused by the CHD2 mutation and cause the lack of a genotype–phenotype correlation mentioned above.
A total of six CHD2 mutation sites were reported among eight cases; of these, four mutation sites were previously unreported; this identifies new sites for pathogenic CHD2 mutations. The c.4173dupA mutation was reported in five previous cases, and two more cases are reported in this paper, suggesting that this site may be a hotspot for mutation. The pathogenesis of epilepsy caused by CHD2 is still unclear and may be related to decreased GABA activity. In this study, seven patients with epilepsy were treated with VPA to control their seizures, and it has been reported that VPA has good effect in the treatment of CHD2 mutation (Chen et al., 2020), but for patients with refractory epilepsy, a combination of medications or other methods are needed to control epilepsy. At present, the pathogenesis caused by CHD2 gene mutations is not clear, and further research is needed to elucidate the mechanism. Two possible causes of the phenotypic effects of a CHD2 gene mutation are as follows: 1) the mutant protein might still have some function and can lead to the dysfunction of another normal protein, and 2) the gene mutation results in reduced GABAergic channel activity, which can lead to seizures. These speculations need to be tested by further study.
Data Availability Statement
Publicly available datasets were analyzed in this study. This data can be found here: http://archive.uwcm.ac.uk/uwcm/mg/hgmd0.html.
Ethics Statement
The studies involving human participants were reviewed and approved by Approval Letter of Ethics Review Committee, Children’s Hospital of Shanghai/Shanghai Children’s Hospital, Shanghai Jiao Tong University. Written informed consent to participate in this study was provided by the participants’ legal guardian/next of kin. Written informed consent was obtained from the individual(s), and minor(s)’ legal guardian/next of kin, for the publication of any potentially identifiable images or data included in this article.
Author Contributions
XL, LL, and XS performed laboratory experiments and drafted the manuscript. FY, SW, WZ, and XJ collected samples and analysed clinical data. SW, XL, YW, ML, JY, and FZ analysed the genomic data. XL and XS performed the formal analysis. LL, XL, and YW assisted with the software and methodology. JZ, XS, and XL performed the validation experiments. YC, SW, and XL edited the manuscript. YW has made a lot of contributions in the later revision work. Funds for the work were obtained by FY and XL. All authors have approved the final manuscript.
Funding
This work was supported by grants from the Shanghai Key Clinical Specialty Project (grant no. shslczdzk05705). Scientific Research Fund of China Association Against Epilepsy (CJ-A-2021-07) and National Fund Cultivation Special Project of Shanghai Children’s Hospital (2021YGZQ05).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to express our gratitude to the patients and their parents for their cooperation. Additionally, we would like to thank Wuhen Xu for her help in this study.
References
Capelli, L. P., Krepischi, A. C. V., Gurgel-Giannetti, J., Mendes, M. F. S., Rodrigues, T., Varela, M. C., et al. (2012). Deletion of the RMGA and CHD2 Genes in a Child with Epilepsy and Mental Deficiency. Eur. J. Med. Genet. 55, 132–134. doi:10.1016/j.ejmg.2011.10.004
Carvill, G. L., Heavin, S. B., Yendle, S. C., McMahon, J. M., O'Roak, B. J., Cook, J., et al. (2013). Targeted Resequencing in Epileptic Encephalopathies Identifies De Novo Mutations in CHD2 and SYNGAP1. Nat. Genet. 45, 825–830. doi:10.1038/ng.2646
Chen, J., Zhang, J., Liu, A., Zhang, L., Li, H., Zeng, Q., et al. (2020). CHD2 ‐related Epilepsy: Novel Mutations and New Phenotypes. Dev. Med. Child. Neurol. 62, 647–653. doi:10.1111/dmcn.14367
Cho, N., Kim, J. O., Lee, S., Choi, S., Kim, J., Ko, M.-S., et al. (2019). Alternative Splicing Induces Cytoplasmic Localization of RBFOX2 Protein in Calcific Tendinopathy. Exp. Mol. Pathol. 109, 36–41. doi:10.1016/j.yexmp.2019.104264
Deciphering Developmental Disorders Study (2017). Prevalence and Architecture of De Novo Mutations in Developmental Disorders. Nature 542, 433–438. doi:10.1038/nature21062
Epi4K ConsortiumEpilepsy Phenome/Genome Project Allen, A. S., Berkovic, S. F., Cossette, P., Delanty, N., et al. (2013). De Novo mutations in Epileptic Encephalopathies. Nature 501, 217–221. doi:10.1038/nature12439
Galizia, E. C., Myers, C. T., Leu, C., de Kovel, C. G. F., Afrikanova, T., Cordero-Maldonado, M. L., et al. (2015). CHD2variants Are a Risk Factor for Photosensitivity in Epilepsy. Brain 138, 1198–1208. doi:10.1093/brain/awv052
Hanly, C., Shah, H., Au, P. Y. B., and Murias, K. (2021). Description of Neurodevelopmental Phenotypes Associated with 10 Genetic Neurodevelopmental Disorders: a Scoping Review. Clin. Genet. 99, 335–346. doi:10.1111/cge.13882
Homem, C. C. F., Repic, M., and Knoblich, J. A. (2015). Proliferation Control in Neural Stem and Progenitor Cells. Nat. Rev. Neurosci. 16, 647–659. doi:10.1038/nrn4021
Kim, Y. J., Khoshkhoo, S., Frankowski, J. C., Zhu, B., Abbasi, S., Lee, S., et al. (2018). CHD2 Is Necessary for Neural Circuit Development and Long-Term Memory. Neuron 100, 1180–1193. doi:10.1016/j.neuron.2018.09.049
Lamar, K.-M. J., and Carvill, G. L. (2018). Chromatin Remodeling Proteins in Epilepsy: Lessons from CHD2-Associated Epilepsy. Front. Mol. Neurosci. 11, 208. doi:10.3389/fnmol.2018.00208
Liu, J. C., Ferreira, C. G., and Yusufzai, T. (2015). Human CHD2 Is a Chromatin Assembly ATPase Regulated by its Chromo- and DNA-Binding Domains. J. Biol. Chem. 290, 25–34. doi:10.1074/jbc.m114.609156
Lund, C., Brodtkorb, E., Øye, A.-M., Røsby, O., and Selmer, K. K. (2014). CHD2 Mutations in Lennox-Gastaut Syndrome. Epilepsy Behav. 33, 18–21. doi:10.1016/j.yebeh.2014.02.005
Marfella, C. G. A., Ohkawa, Y., Coles, A. H., Garlick, D. S., Jones, S. N., and Imbalzano, A. N. (2006). Mutation of the SNF2 Family Member CHD2 Affects Mouse Development and Survival. J. Cel. Physiol. 209, 162–171. doi:10.1002/jcp.20718
Meganathan, K., Lewis, E. M. A., Gontarz, P., Liu, S., Stanley, E. G., Elefanty, A. G., et al. (2017). Regulatory Networks Specifying Cortical Interneurons from Human Embryonic Stem Cells Reveal Roles for CHD2 in Interneuron Development. Proc. Natl. Acad. Sci. U S A. 114, e11180–89. doi:10.1073/pnas.1712365115
Nieto-Estevez, V., and Hsieh, J. (2018). CHD2: One Gene, many Roles. Neuron 100, 1014–1016. doi:10.1016/j.neuron.2018.11.036
O'Roak, B. J., Stessman, H. A., Boyle, E. A., Witherspoon, K. T., Martin, B., Lee, C., et al. (2014). Recurrent De Novo Mutations Implicate Novel Genes Underlying Simplex Autism Risk. Nat. Commun. 5, 5595. doi:10.1038/ncomms6595
Petersen, A. K., Streff, H., Tokita, M., and Bostwick, B. L. (2018). The First Reported Case of an Inherited Pathogenic CHD2 Variant in a Clinically Affected Mother and Daughter. Am. J. Med. Genet. 176, 1667–1669. doi:10.1002/ajmg.a.38835
Pinto, A. M., Bianciardi, L., Mencarelli, M. A., Imperatore, V., Di Marco, C., Furini, S., et al. (2016). Exome Sequencing Analysis in a Pair of Monozygotic Twins Re-evaluates the Genetics behind Their Intellectual Disability and Reveals a CHD2 Mutation. Brain Develop. 38, 590–596. doi:10.1016/j.braindev.2015.12.006
Poot, M., Verrijn Stuart, A. A., van Daalen, E., van Iperen, A., van Binsbergen, E., and Hochstenbach, R. (2013). Variable Behavioural Phenotypes of Patients with Monosomies of 15q26 and a Review of 16 Cases. Eur. J. Med. Genet. 56, 346–350. doi:10.1016/j.ejmg.2013.04.001
Shen, T., Ji, F., Yuan, Z., and Jiao, J. (2015). CHD2 Is Required for Embryonic Neurogenesis in the Developing Cerebral Cortex. Stem Cells 33, 1794–1806. doi:10.1002/stem.2001
Suls, A., Jaehn, J. A., Kecskés, A., Weber, Y., Weckhuysen, S., Craiu, D. C., et al. (2013). De Novo loss-of-function Mutations in CHD2 Cause a Fever-Sensitive Myoclonic Epileptic Encephalopathy Sharing Features with Dravet Syndrome. Am. J. Hum. Genet. 93, 967–975. doi:10.1016/j.ajhg.2013.09.017
Thomas, R. H., Zhang, L. M., Carvill, G. L., Archer, J. S., Heavin, S. B., Mandelstam, S. A., et al. (2015). CHD2 Myoclonic Encephalopathy Is Frequently Associated with Self-Induced Seizures. Neurology 84, 951–958. doi:10.1212/wnl.0000000000001305
Veredice, C., Bianco, F., Contaldo, I., Orteschi, D., Stefanini, M. C., Battaglia, D., et al. (2009). Early Onset Myoclonic Epilepsy and 15q26 Microdeletion: Observation of the First Case. Epilepsia 50, 1810–1815. doi:10.1111/j.1528-1167.2009.02078.x
Wang, Y., Du, X., Bin, R., Yu, S., Xia, Z., Zheng, G., et al. (2017). Erratum: Corrigendum: Genetic Variants Identified from Epilepsy of Unknown Etiology in Chinese Children by Targeted Exome Sequencing. Sci. Rep. 7, 46520. doi:10.1038/srep46520
Wilson, M.-M., Henshall, D. C., Byrne, S. M., and Brennan, G. P. (2021). CHD2-related CNS Pathologies. Int. J. Mol. Sci. 22, 588. doi:10.3390/ijms22020588
Keywords: chromodomain helicase DNA-binding protein 2 (CHD2) gene, neurological diseases, epilepsy, repressor element 1-silencing transcription factor, expression
Citation: Luo X, Sun X, Wang Y, Lin L, Yuan F, Wang S, Zhang W, Ji X, Liu M, Wu S, Lan X, Zhang J, Yan J, Zeng F and Chen Y (2022) Clinical Study of 8 Cases of CHD2 Gene Mutation–Related Neurological Diseases and Their Mechanisms. Front. Cell Dev. Biol. 10:853127. doi: 10.3389/fcell.2022.853127
Received: 12 January 2022; Accepted: 01 March 2022;
Published: 21 March 2022.
Edited by:
Zi-Bing Jin, Capital Medical University, ChinaReviewed by:
Fei Yin, Xiangyang Central Hospital, ChinaKimberly Aldinger, Seattle Children’s Research Institute, United States
Copyright © 2022 Luo, Sun, Wang, Lin, Yuan, Wang, Zhang, Ji, Liu, Wu, Lan, Zhang, Yan, Zeng and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yucai Chen, Y2hlbnljQHNoY2hpbGRyZW4uY29tLmNu
†These authors have contributed equally to this work and share first authorship
 Xiaona Luo
Xiaona Luo Xiaoang Sun1†
Xiaoang Sun1† Yilin Wang
Yilin Wang Fang Yuan
Fang Yuan Simei Wang
Simei Wang Shengnan Wu
Shengnan Wu Yucai Chen
Yucai Chen