- Malaria Parasite Molecular Laboratory, Department of Biochemistry, Genetics and Microbiology, Institute for Sustainable Malaria Control, University of Pretoria, Pretoria, South Africa
Plasmodium falciparum remains the deadliest parasite species in the world, responsible for 229 million cases of human malaria in 2019. The ability of the P. falciparum parasite to progress through multiple life cycle stages and thrive in diverse host and vector species hinges on sophisticated mechanisms of epigenetic regulation of gene expression. Emerging evidence indicates such epigenetic control exists in concentric layers, revolving around core histone post-translational modification (PTM) landscapes. Here, we provide a necessary update of recent epigenome research in malaria parasites, focusing specifically on the ability of dynamic histone PTM landscapes to orchestrate the divergent development and differentiation pathways in P. falciparum parasites. In addition to individual histone PTMs, we discuss recent findings that imply functional importance for combinatorial PTMs in P. falciparum parasites, representing an operational histone code. Finally, this review highlights the remaining gaps and provides strategies to address these to obtain a more thorough understanding of the histone modification landscapes that are at the center of epigenetic regulation in human malaria parasites.
Introduction
Malaria persists as a global burden to public health and in 2019, was responsible for 409000 deaths. Of the five Plasmodium species that infect humans, P. falciparum is the most likely to cause severe disease and accounts for the vast majority of deaths from malaria (WHO, 2020). The complexities of the P. falciparum life cycle are evident from the array of developmental stages and cycles that are compartmentalized into diverse host cell types within both the human and the mosquito vector (Sherman, 1979). The unique biology of each stage is underpinned by the expression of stage-specific gene sets, regulated at the epigenetic, transcriptional, post-transcriptional, and post-translational levels (Bozdech et al., 2003; Young et al., 2005; Lopez-Barragan et al., 2011; van Biljon et al., 2019). Recent studies involving a diverse set of chromatin-based technologies, including a combination of chromatin immunoprecipitation coupled with next-generation sequencing (ChIP-seq), quantitative mass spectrometry (MS), fluorescence in situ hybridization (FISH), and histone mutagenesis and transcriptomics (microarrays and RNA-sequencing), have revealed that histone post-translational modifications (PTMs) are foundational in generating these stage-specific gene expression fingerprints and are thus proposed to be central to P. falciparum parasite life cycle regulation (Comeaux and Duraisingh, 2007; Issar et al., 2009; Trelle et al., 2009; Crowley et al., 2011; Kaur et al., 2016; Saraf et al., 2016; Coetzee et al., 2017; Gomez-Diaz et al., 2017; Gupta and Bozdech, 2017; Gupta et al., 2017; Ngwa et al., 2017; Herrera-Solorio et al., 2019; Witmer et al., 2020; Connacher et al., 2021; Kumar et al., 2021; Ngwa et al., 2021). Here, we review the current knowledge regarding epigenetic regulation in P. falciparum parasites with a specific focus on the recent developments in our understanding of how histone PTM landscapes orchestrate life cycle progression.
P. Falciparum Parasite Life Cycle
Malaria infections are initiated after sporozoite infection of liver cells. After exoerythrocytic development (EDC), merozoites are released into the peripheral circulation to invade erythrocytes, initiating a new asexual intraerythrocytic developmental cycle (IDC) (Figure 1) (Kappe et al., 2004). Here, ring, trophozoite and schizont development leads to population expansion and the release of new daughter merozoites to repeat the cycle (Dvorak et al., 1975). Parasite survival and transmission are ensured by the divergence of a small proportion (≤10%) of the parasites that commit to sexual differentiation. In P. falciparum, this process is uniquely associated with stage differentiation with stage I gametocytes sequestering in the bone marrow to maturate into stages II-IV (Figure 1) (Sinden et al., 1978; Carter and Graves, 1988; Aguilar et al., 2014; Joice et al., 2014). This process of gametocytogenesis yields mature stage V male and female gametocytes that re-enter the host’s circulatory system for transmission to female Anopheles mosquitoes during feeding (Smalley et al., 1981; Aguilar et al., 2014). Since stage V gametocytes are the only transmissible forms of the malaria parasite, they have become the focus of the discovery and development of transmission-blocking compounds to aid in malaria elimination strategies (Delves, 2012; Birkholtz et al., 2016). Following gametogenesis of micro- (male) and macro- (female) gametocytes in the mosquito midgut, haploid gametes are fertilized to produce a diploid zygote that transforms into a tetraploid motile ookinete (Figure 1). The maturation of ookinetes into oocysts results in the formation of sporozoites, which can be transmitted to a new host during feeding (Sinden et al., 1978; Billker et al., 1998).
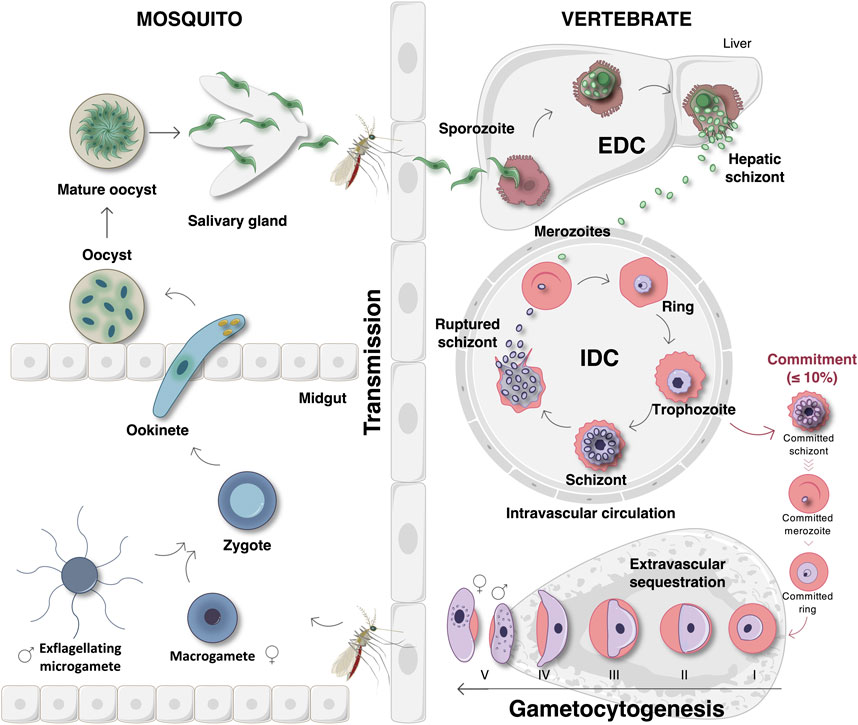
FIGURE 1. The life cycle of P. falciparum parasites. The life cycle begins when an Anopheles mosquito vector injects sporozoites into the circulatory system of a human host. The sporozoites then travel to and invade liver cells to initiate the exoerythrocytic developmental cycle (EDC) that results in the formation hepatic schizonts. Once mature, these hepatic schizonts release merozoites into the peripheral circulation. Merozoites invade erythrocytes and initiate the 48-hour intraerythrocytic developmental cycle (IDC) that involves asexual proliferation through ring, trophozoite, and schizont stages. Schizonts then rupture and release new daughter merozoites that repeat this cycle once again. Within each IDC, a small proportion (≤10%) of the parasites will deviate from this fate and instead commit to gametocytogenesis. Sexually committed merozoites invade erythrocytes, forming stage I gametocytes that sequester in the bone marrow where subsequent maturation into gametocyte stages II–IV occurs. Gametocytogenesis yields mature stage V gametocytes that re-enter into the host’s circulatory system where they are ideally situated for transmission to the mosquito during feeding. Once taken up during a blood meal, gametogenesis ensues in the mosquito midgut in which female macrogametes are formed and male gametocytes undergo exflagellation to form microgametes. Thereafter, the microgametes fertilize macrogametes, forming diploid zygotes. Zygote development involves maturation into an ookinete and then an oocyst that contains new, maturing sporozoites. Finally, the mature oocysts rupture, releasing sporozoites that travel to the mosquito’s salivary glands where they will be transmitted to a new host during feeding. Image was created with BioRender.com.
Nucleosome and Chromatin Structure in P. Falciparum
The P. falciparum nucleosome consists of 147 bp of DNA associated with an octamer of four histone proteins, typically the H2A, H2B, H3, and H4 core histones, each of which display particular functional and stage-specific characteristics (Figure 2) (Miao et al., 2006; Trelle et al., 2009). Within certain nucleosomes, these core histones may be exchanged for their variants, namely, H2A.Z, H2B.Z, H3.3, and H3Cen, which may also be modified by an array of chemical groups. These variant histones are typically incorporated to demarcate or influence the properties of specific chromatin domains. For example, distinct intergenic regions of the P. falciparum genome are demarcated by double-variant nucleosomes containing H2A.Z and H2B.Z while H3.3 is incorporated to index euchromatic coding regions and subtelomeric repeat regions (Bartfai et al., 2010; Hoeijmakers et al., 2013; Petter et al., 2013; Fraschka et al., 2016). Despite sequence divergence, P. falciparum histones have not adapted increased binding affinity for the extremely A + T-rich (∼80%) genome (Gardner et al., 2002; Silberhorn et al., 2016). Combined with the apicomplexan-specific absence of the H1 linker histone (Gardner et al., 2002; Sullivan et al., 2006) and short nucleosome repeat length (155 bp) (Silberhorn et al., 2016), this evolution results in a general lack of stable nucleosome patterns in asexual P. falciparum parasites (Hoeijmakers et al., 2012). Except for a few internal gene clusters that are brought into proximity with subtelomeric heterochromatin via chromatin looping, typical eukaryotic topologically associating domains are therefore largely absent in P. falciparum parasites (Lemieux et al., 2013; Bunnik et al., 2014; Bunnik et al., 2018).
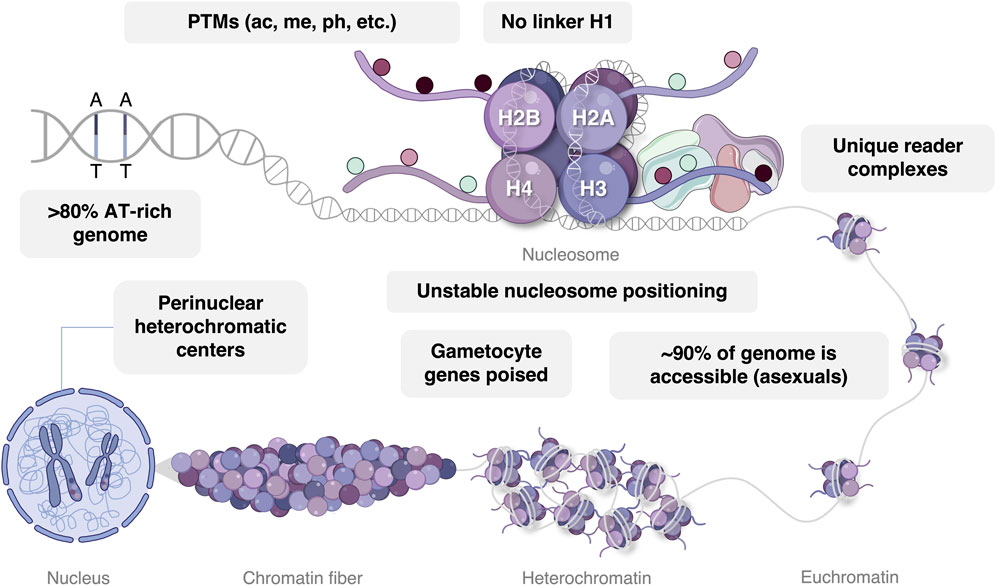
FIGURE 2. Epigenetic gene regulation in P. falciparum parasites. DNA with an AT bias is wrapped around a nucleosome with conventional eukaryotic histone proteins, including H2A, H2B, H3, and H4. The majority of histone post-translational modifications (PTMs) are read by unique epigenetic reader complexes consisting of somewhat divergent writer and eraser enzymes. In asexual parasites, the majority of chromatin exists in an euchromatic conformational state, while gametocyte chromatin exhibits an expansion of H3K9me3-mediated heterochromatin and is stage-specifically poised for transcriptional activation. Perinuclear heterochromatic centers reside in the nucleus associated with var gene regulation. Image was created with BioRender.com.
Three-dimensional chromatin structure influences the accessibility of DNA to chromatin-binding proteins and transcription factors, thereby enhancing or repressing gene expression (Zheng and Xie, 2019). As such, the rearrangement of genome spatial organization occurs throughout the parasite’s life cycle and corresponds with changes in transcriptional activity (Ay et al., 2014; Bunnik et al., 2014). A relatively simple and dichotomous nuclear organization characterizes the asexual parasite with the majority of the genome (∼90%) in a euchromatic state and with the remainder of the DNA present in perinuclear heterochromatic centers (Hoeijmakers et al., 2012; Brancucci et al., 2014; Coleman et al., 2014). Trophozoites exhibit the most euchromatic and accessible genome organization, in line with the increased level of transcriptional activity required in preparation of schizogony (Ay et al., 2014). Contrasting with this restricted distribution of heterochromatin in the asexual parasite stages, repressive factors become more prolific in the gametocyte stages (Brancucci et al., 2014; Coetzee et al., 2017; Bunnik et al., 2018; Fraschka et al., 2018), reflecting the specific transcriptional environment that underlies sexual differentiation and development (Poran et al., 2017; Walzer et al., 2018; van Biljon et al., 2019).
Chromatin status is largely influenced by post-translational modification of the N-terminal tails of histone proteins that make up the core of the nucleosome (Goll and Bestor, 2002). Quantitative mass spectrometry and antibody-based techniques such as Western blotting has been used to identify histone PTMs in P. falciparum parasites with acetylated, methylated, phosphorylated, ubiquitinated, and SUMOylated histones quantitatively detected throughout the life cycle (Miao et al., 2006; Salcedo-Amaya et al., 2009; Trelle et al., 2009; Treeck et al., 2011; Dastidar et al., 2013; Gupta et al., 2013; Ukaegbu et al., 2014; Cobbold et al., 2016; Kaur et al., 2016; Saraf et al., 2016; Coetzee et al., 2017; Gupta et al., 2017; Bui et al., 2019; Green et al., 2020). Acetylation and methylation marks indeed form the major component of histone PTMs in P. falciparum and have been accurately detected and quantified over multiple life cycle stages. This provides confidence as to their biological relevance, and indeed, a large number of these marks have been extensively functionally validated. The same is not true for the less prevalent marks, particularly for crotonylation and formylation. Quantitative detection of less abundant (or perhaps time-point specific) histone PTMs, including crotonylation, require antibody-based enrichment of histone PTMs prior to mass spectrometric analysis where, for example, H2AK3crK5cr, H2BK18cr, and H4K78cr was identified in P. falciparum asexual parasites (Wang and Zhang, 2020). However, besides detection, no information on the importance of crotonylation to parasite development is currently available and it remains to be seen if this histone PTM is functionally distinct from acetylation in the parasite. Remarkably, lysine crotonylation catalyzed by p300 increased transcriptional activation more potently than lysine acetylation in a cell-free system (Sabari et al., 2015) but such functional validation of a biological role for this histone PTM in Plasmodium is lacking.
In addition to individual PTMs, histones of mammalian cells for example are also readily modified with distinct patterns of co-existing PTMs at multiple sites, e.g., methylation of lysine 4 with lysine 9 acetylation of histone H3 (Muller and Muir, 2015). While the functional relevance of a relatively small number of single histone PTMs is well documented, evidence indicating that histone PTMs act in concert with one another to regulate transcriptional programs suggests that combinatorial histone PTMs landscapes contribute an additional layer of regulation in eukaryotes (Berger, 2002; Zhao and Garcia, 2015). The compendium of co-existing histone PTMs on specifically histone H3 and H3.3 in P. falciparum parasites was recently updated and revealed that PTM combinations are prevalent in asexual parasites and gametocytes and are considerably diverse and stage-stratified, as determined by quantitative mass spectrometric analysis of whole histone N-terminal tails (von Grüning et al., 2022). Histone PTM crosstalk influences the addition, removal, and binding of effector proteins between histone PTMs in combination (Hunter, 2007). While the enzymes responsible for depositing and removing these individual and combinatorial PTMs are somewhat divergent from other organisms based on homology (Miao et al., 2006; Cui et al., 2008a; Miao et al., 2010a), P. falciparum parasites employ a typical eukaryotic histone PTMs repertoire (Miao et al., 2006; Chung et al., 2009; Saraf et al., 2016; Coetzee et al., 2017), including functional acetylation, methylation and phosphorylation (Figure 3) to expand the regulatory capacity of nucleosomes using a limited set of effector proteins (summarized in Table 1).
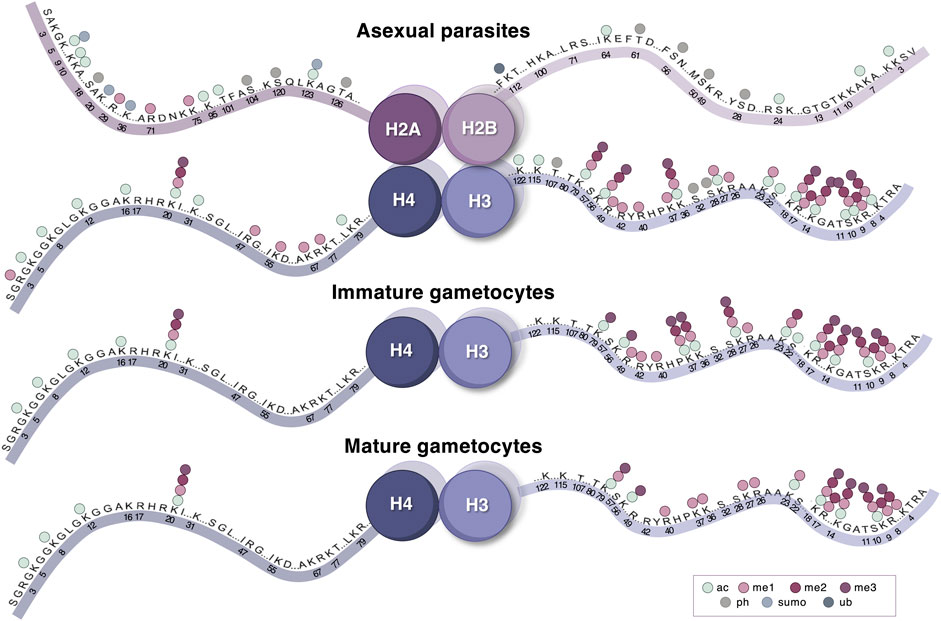
FIGURE 3. The histone PTM landscape of P. falciparum parasites. Histone PTMs (acetylation, different levels of methylation, phosphorylation, SUMOylation, and ubiquitination) are indicated on all positions for the four histones where quantitative detection has been described. This is presented across three life cycle stages—asexual parasites, immature and mature gametocytes. Image was created with BioRender.com.
Histone Modifying Enzymes in P. Falciparum
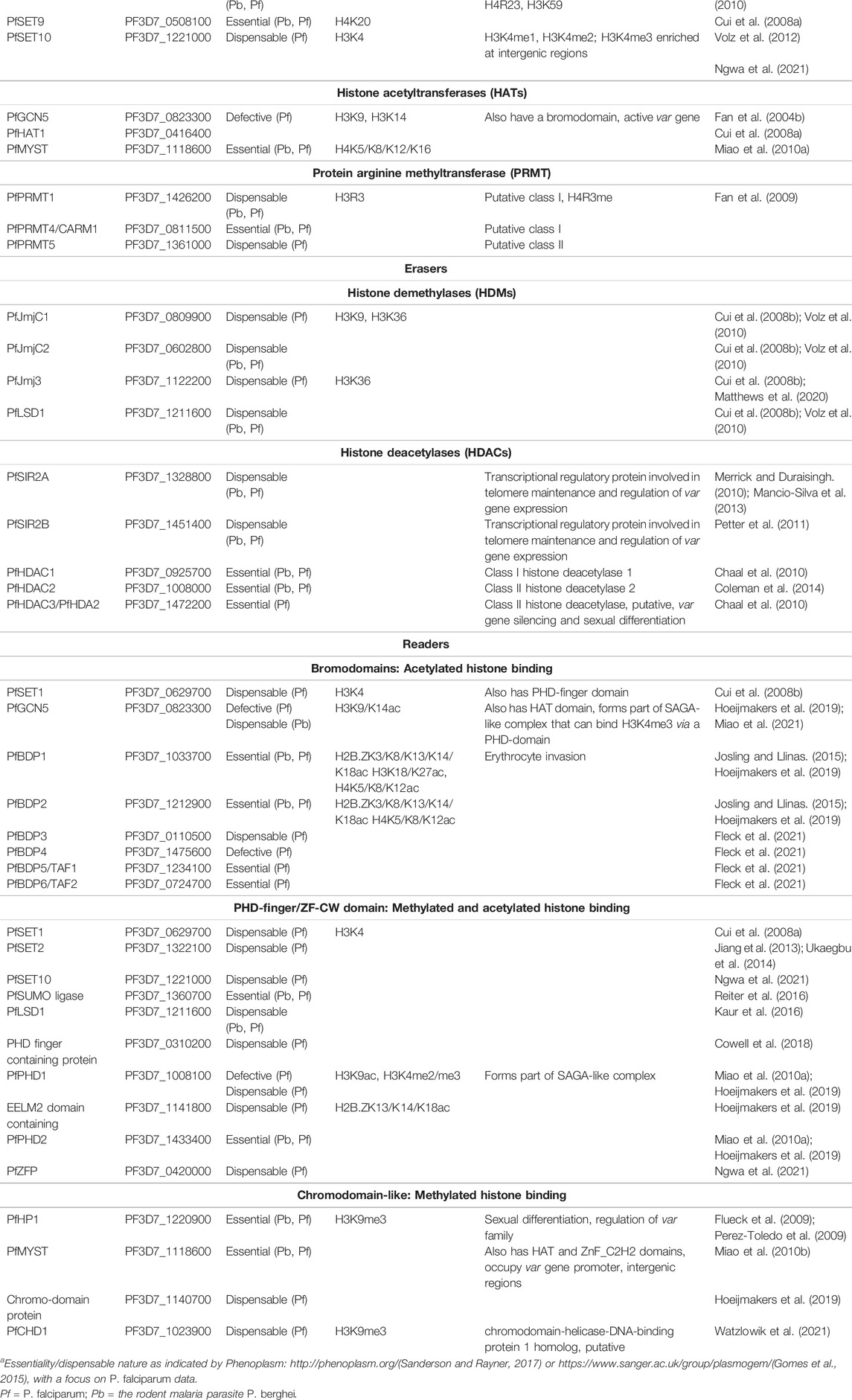
The methylation of histones in P. falciparum is mediated by ten histone methyltransferases (HMTs), all of which belong to the evolutionarily conserved SET [Su (var)3-9, Enhancer of Zeste and Trithorax] domain-containing protein family (Cui et al., 2008a). The site specificities of all, except two (SET5 and SET9) of these HMTs have been determined through recombinant protein expression, pull-down proteomics, mutagenesis, and histone lysine methyltransferase (Cui et al., 2008a; Volz et al., 2012; Jiang et al., 2013; Chen et al., 2016; Ngwa et al., 2021) (Table 1). Less is known regarding the three Jumonji-C (JmjC) domain-containing histone demethylases (HDMs) and the one lysine-specific demethylase 1 (LSD1) that demethylate histones in P. falciparum, however, JMJC1 and JMJ3 have predicted and known H3K36-specificity (Cui et al., 2008a; Matthews et al., 2020). Although the precise mechanisms by which histone methyltransferases and demethylases modify histones remain relatively unclear, several studies have demonstrated their importance for asexual and sexual stage development through genetic disruption and chemical interrogation (Jiang et al., 2013; Ukaegbu et al., 2014; Zhang et al., 2018; Coetzee et al., 2020; Matthews et al., 2020; Connacher et al., 2021; Reader et al., 2021) (Table 1).
Histone acetylation is regulated by four histone acetyltransferases (HATs) including proteins from the MYST and GNAT families (Cui et al., 2008a). The acetyltransferase activities of GCN5 and MYST are specific to the lysine residues of H3 and H4, respectively (Cui et al., 2007b; Miao et al., 2010a), with the functional importance of these enzymes evident from the transcriptional deregulation arising from their genetic and chemical disruption (Cui et al., 2007b; Cui et al., 2008b; Chaal et al., 2010; Bhowmick et al., 2020; Miao et al., 2021) (Table 1). Acetyltransferase activities are antagonized by a repertoire of three histone deacetylases (HDACs) and two sirtuin histone deacetylase proteins in P. falciparum (Kanyal et al., 2018). Interestingly, the P. falciparum genome encodes for an essential YEATS-domain protein (PF3D7_0807000). Its orthologues in mammalians and yeast are readers of crotonylation. It would therefore be interesting to see if similar activity is present in Plasmodia and if so, crotonylation could be important to biological processes in the parasite despite it being a rarely identified PTM (Table 1).
The chromatin reorganization induced by these enzymes results either from a direct physical change in nucleosome structure or the recruitment of epigenetic “reader complexes” that mediate subsequent chromatin remodeling (Latchman, 2010; Abel and Le Roch, 2019) (Table 1). To date, five putative reader complexes have been identified in P. falciparum parasites, however, the vast majority remain to be functionally validated (Hoeijmakers et al., 2019). These P. falciparum readers largely exist in complexes with other reader proteins and/or transcription factors that would presumably require sequential binding occurrences for recruitment (Hoeijmakers et al., 2019). Such cooperation between epigenetic regulatory proteins is exemplified in the recruitment of the GCN5/ADA2 reader complex by H3K4me3 via interaction with PHD1 (Hoeijmakers et al., 2019). Additionally, evidence suggests that as in other organisms, epigenetic complexes in P falciparum may be recruited to or “flavored” to specific histone PTMs or combinations of PTMs, as determined by peptide pull-down coupled with mass spectrometry. For example, GCN5/ADA2 core reader complex has at least two distinct “flavors” with a unique composition of reader proteins, such as the presence or absence of PHD1 or PHD2 (Hoeijmakers et al., 2019; Miao et al., 2021). Additionally, the intricate nature of multiprotein reader complexes supports the existence of unique sets of functional histone PTMs combinations in P. falciparum parasites whereby different reader proteins bind to PTMs using reader domains. Whether these modifications directly influence chromatin structure via the recruitment of such readers or by virtue of their biochemical properties, HMs are undoubtedly at the core of epigenetic regulation.
Histone PTM Landscapes in P. Falciparum
Although up to 230 histone PTMs have been identified in various qualitative and quantitative proteomic based approaches P. falciparum parasites (Salcedo-Amaya et al., 2009; Trelle et al., 2009; Saraf et al., 2016), only 46 of these have been quantitatively validated (Coetzee et al., 2017) (Figure 3). The P. falciparum histone PTM repertoire expands beyond the typical eukaryotic histone modifications (e.g., H3K4me3, H3K9me3, and H3K36me3) and contains several less common modifications, including H4K20ac, H3.3K79me3, and H3K23me1 (Coetzee et al., 2017). Similar to the hierarchical cascades of gene expression that are associated with each asexual and sexual stage (Bozdech et al., 2003; van Biljon et al., 2019), strikingly unique and stage-specific histone PTM landscapes characterize each stage of the P. falciparum parasite life cycle (Figure 3) (Coetzee et al., 2017).
Asexual Blood Stage PTM Landscape
The greatest proportion of histone PTMs in the asexual parasite stages consists of acetylation and mono-methylation, both of which are associated with transcriptionally permissive chromatin (Saraf et al., 2016; Coetzee et al., 2017). Throughout the IDC, the mostly euchromatic nature of the genome is marked by nucleosomes containing acetylated histones H3 (K9, K14, and K56) and H4 (K5, K8, K12, and K16) in addition to H3K4me3 and H4K20me1, most of which are positively correlated with gene expression in P. falciparum (Gupta et al., 2013). The large transcriptional disruption (∼60% of the genome) and effects on growth that arise from inhibiting histone acetylation and methylation in P. falciparum parasites, highlight the crucial nature of these histone PTMs for parasite regulation (Cui et al., 2007a; Cui et al., 2008b; Matthews et al., 2020; Connacher et al., 2021). Regarding combinatorial histone PTMs in the asexual parasite stages, co-existing H3K4me3, H3K9ac, and H3K14ac on histone H3 are achieved through the individual actions of PfGCN5 and PfSET7 (Fan et al., 2004a; Fan et al., 2004b; Salcedo-Amaya et al., 2009; Cui and Miao, 2010; Chen et al., 2016).
Throughout the asexual life cycle, dynamic patterns of H3K18ac and H3K27ac mark the transcriptional start sites of active genes with the AP2-I transcription factor and bromodomain protein 1 (BDP1) reader protein exhibiting similar temporal enrichment at these sites as determined by ChIP-seq and RNA-seq (Tang et al., 2020). Ring and schizonts stages have increased abundance of H4K20me1 and H4K8ac, respectively while acetylation of H3K9 and H3K14 become highly abundant in trophozoites (Gupta et al., 2013; Coetzee et al., 2017). H3K37me1, a novel histone PTMs in P. falciparum which has been associated with licensing of replication origins in yeast (Santos-Rosa et al., 2021; von Grüning et al., 2022), is prominent in trophozoite stages. Similarly, on histone H3.3, K37me3 steadily increases in abundance from asexual parasites and immature gametocytes to mature gametocytes (von Grüning et al., 2022). Interestingly, the methylation of H3R17 and H3R40 is more prevalent compared to H3K4me3, suggesting that these histone PTMs may be additional to the canonical acetylation modifications (e.g., H3K9a and H3K14ac) that are involved in transcriptional activation in trophozoites (von Grüning et al., 2022). In addition to this, the connectivity between histone PTMs is exemplified by H3K9acK14ac that is present, and together with the binding of effector proteins, likely contributes to the euchromatic nature of the trophozoite genome (von Grüning et al., 2022). H3K9ac also co-exists with H3K36me3 and exhibits the most notable crosstalk in the trophozoite stages, highlighting the exclusive nature of these particular combinations for asexual parasites, evidenced by quantitative middle-down mass spectrometry (von Grüning et al., 2022). Overall, PTMs on variant histones have also been identified, yet not reproducibly quantified (Saraf et al., 2016; Coetzee et al., 2017). Nonetheless, the C-terminal SQ motif of histone H2A has been shown to be phosphorylated in response to DNA damage (Goyal et al., 2021) in addition to the qualitative detection of serine phosphorylation and lysine acetylation (Coetzee et al., 2017). Similarly, lysine acetylation and methylation, and serine phosphorylation, have also been detected on H3.3, and variant histones H2A.Z, H2B.Z, and centromeric H3 (Saraf et al., 2016; Coetzee et al., 2017; von Grüning et al., 2022). SUMOylation, a PTM that is linked to antagonizing transcription and chromatin remodeling, has been predicted on a combined ten lysine sites on the N-termini of histone H2A and H2A.Z, with histone H4 being a putative target for SUMOylation as well (Issar et al., 2008; Ryu and Hochstrasser, 2021). This suggests that SUMOylation might well be an integral part of the histone PTM language of the parasite. Together with the abundance of individual activating modifications, these combinations provide a critical roadmap that underpins the “just-in-time” transcription of the asexual stages. Interestingly, another ApiAP2 transcription factor family member that is thought to act downstream of AP2-G, AP2-G2, occupancy is highly correlated with repressive H3K36me3, H4K20me3 and H3K27me3 in asexual parasites as determined by pairwise Pearson correlation comparing read counts (Singh et al., 2020). Although AP2-G2 is not essential for asexual proliferation, it is, however, essential for gametocyte maturation where AP2-G2 functions by repressing genes necessary for asexual replication (Singh et al., 2020). Together, this indicates that although present at lower abundance, repressive PTMs are an equally important facet of transcriptional regulation during asexual proliferation. Interestingly, negative crosstalk has been observed between the novel arginine PTMs, H3R40me1, and H3R17me1 in the trophozoite stages (von Grüning et al., 2022). It remains to be seen if H3R40me1 is important to microgametogenesis, similar to its involvement in sporulation in yeast, a process similar to male gametogenesis (Govin et al., 2010) and if H3R17me1 serves independently from canonical euchromatic PTMs in trophozoites to de-represses silenced chromatin as in mammalian cells (Miller and Grant, 2013). In model organisms, H3K56me1&2 have been implicated as docking sites for DNA replication and repair enzymes (Lee et al., 2012; Yu et al., 2012; Janssen et al., 2019), inviting the question as to whether the peak abundance of these histone PTMs in the ring-stage parasites may be indicative of the parasite’s preparation for genome replication in the proceeding stages.
The most notable increase in stage-specific histone PTM abundance is present during the schizont stages (Coetzee et al., 2017), specifically for H3K4ac and H4K16ac, both of which have roles in transcriptional activation and is therefore unsurprising given the increased metabolic and transcriptional activity in schizonts (Bozdech et al., 2003; van Biljon et al., 2019). H3K4ac is abundant in active promoter regions with the levels of this PTM regulated by H3K4 methylation in higher eukaryotes (Guillemette et al., 2011). In line with this, H3K4me2&3 are also present in high abundance in schizonts (Coetzee et al., 2017), with functional data indicating H3K4 methylation also positively influences transcription via the recruitment of the GCN5/ADA2 and the associated BDP1/2 reader complexes (Fan et al., 2004a; Hoeijmakers et al., 2019). Although the roles of H4K16ac are less clear, studies in other organisms have documented this PTM as a high-affinity docking site for chromatin-remodeling complexes that promote transcription (Ruthenburg et al., 2011), with the abundance of H4K16ac in asexual parasites, therefore, presenting a possible core mechanism that promotes euchromatin formation. H4K20me3 also exhibits a schizont stage-specific peak in abundance (Coetzee et al., 2017) and was recently identified by machine learning algorithms to be highly indicative of active gene expression in P. falciparum parasites (Read et al., 2019).
Histone PTMs That Regulate Clonally Variant Gene Expression and Phenotypic Plasticity
The involvement of epigenetic regulatory mechanisms in variant expression of several gene families have been extensively studied in P. falciparum. Epigenetic regulation of processes such as immune evasion (e.g., var, rifin, and stevor), erythrocyte remodeling (e.g., PfMC-2TM) and lipid metabolism (e.g., acyl-CoA binding proteins) (Rovira-Graells et al., 2012; Gomez-Diaz et al., 2017) ensures heritability and rapid adaptation in response to environmental changes. For example, single var gene expression is ensured by H2A.Z/H2B.Z double-variant histones, H3.3 and H3K9ac occupancy (Petter et al., 2013), and heritable transmission is ensured by a poised state associated with H3K4me2 (Volz et al., 2012). Additionally, H3.3 is also present in the promoter region and has thus been proposed to be involved in regulating the single active or poised var gene (Fraschka et al., 2016). The remainder of the var gene family in perinuclear repressive centers is actively silenced with H3K9me3/HP1 and H3K36me3 (Chookajorn et al., 2007; Flueck et al., 2009; Lopez-Rubio et al., 2009; Salcedo-Amaya et al., 2009; Trelle et al., 2009; Jiang et al., 2013; Ay et al., 2014; Ukaegbu et al., 2014; Bunnik et al., 2019). H3K9me3/HP1 have similar strong mechanistic links with the regulation of the rif, stevor, pfmc-2tm, and lysophospholipase families as well as genes that direct the transition from asexual proliferation to sexual differentiation in P. falciparum parasites (Rovira-Graells et al., 2012; Brancucci et al., 2014; Gomez-Diaz et al., 2017).
Histone PTMs That Orchestrate the Asexual to Sexual Stage Transition
Several extrinsic drivers of the switch from proliferation to sexual differentiation in P. falciparum have been investigated including increased parasitaemia, antimalarial drugs, and lysophosphatidylcholine (lysoPC) depletion (Trager and Gill, 1992; Buckling et al., 1999; Williams, 1999; Dyer and Day, 2000; Brancucci et al., 2017). Currently, the specific mechanism linking these factors with the switch to sexual differentiation is unclear. Nevertheless, several essential genetic factors have reproducibly been shown to be upregulated upon this cue and drive sexual differentiation in Plasmodium parasites (Kafsack et al., 2014; Sinha et al., 2014). The commitment process is largely driven by AP2-G, a member of the apicomplexan-specific Apetala2 (ApiAP2) DNA binding domain-containing protein family (Balaji et al., 2005; Kafsack et al., 2014). During the IDC, the ap2-g locus remains silenced within H3K9me3/HP1-mediated heterochromatin (Brancucci et al., 2014; Filarsky et al., 2018) and the expression of gametocyte development protein 1 gene (gdv1) is inhibited by its own antisense RNA. The gametocyte essential factor, AP2-G5, further prevents sexual commitment by binding ap2-g upstream and exonic regions that are responsible for maintaining local heterochromatin which suppresses ap2-g gene expression (Shang et al., 2021). Upon the cue for commitment, GDV1 is upregulated and evicts H3K9me3/HP1, resulting in the de-repression of the ap2-g locus (Poran et al., 2017; Filarsky et al., 2018). Sequence-specific binding of AP2-G to promoters results the transcriptional activation of hundreds of genes that are predominantly involved in commitment and early-stage gametocyte development, including those encoding invasion proteins that are co-regulated by a second AP2 TF, AP2-I (Kafsack et al., 2014; Poran et al., 2017; Josling et al., 2020). Following sexual commitment, AP2-G5 downregulates AP2-G as well as the early gametocyte genes activated by AP2-G, resulting in a cascade of gene on/off switching that is required for gametocyte maturation (Shang et al., 2021). Aside from the abovementioned H3K9 modifications, the histone PTM landscape dynamics of commitment remain unknown. It is possible that the interaction between H3K9me3 and PfHP1 is influenced by PTMs that co-exist with H3K9me3, most notably, H3S10ph. Although not proven yet, there may be a negative crosstalk relationship between H3K9me3 and H3S10ph in Plasmodium, similar to how H3S10ph evicts HP1 from H3K9me3 and prevents the spread of H3K9me2/me3 across the genome in mammalian cells (Fischle et al., 2005; Hirota et al., 2005; Chen et al., 2018). Interestingly, H3.3 carries less PTMs in combination than H3 in trophozoite stages but increase to up to seven PTMs co-existing on the H3.3 histone tail in immature gametocytes and could be linked to the drastic shift in transcriptional program in immature gametocytes (von Grüning et al., 2022).
In model organisms, differentiation consists of multiple stages, each with characteristic epigenome signatures (Bhanu et al., 2016). Elucidating whether the histone PTMs that characterize differentiation initiation in other eukaryotes, such as H3K4 H3K9, H3K20 methylation (Bhanu et al., 2016) and global histone acetylation (Meshorer et al., 2006; Tan et al., 2013), is also associated with sexual commitment in P. falciparum remains an interesting question. The impact of the metabolic shift between asexual and gametocytes on the chromatin milieu also warrants clarification, with the indication that loss of function of acetyl-CoA synthetase leads to chromatin hypoacetylation and that the restriction of S-adenosyl-L-methionine availability can cause heterochromatin formation (Prata et al., 2021; Summers et al., 2021).
Histone PTM Landscapes That Direct Gametocytogenesis
The histone PTM landscapes associated with gametocyte development stand in strong contrast with those involved in asexual stage-specific transcriptional regulation (Coetzee et al., 2017) (Figure 3). Gametocytogenesis is characterized by fewer typical euchromatic PTMs and an increased abundance of repressive di- and tri-methylation on histone H3 (Coetzee et al., 2017). The presence of these PTMs is particularly notable in the early (I-III) stages of gametocyte development and include H3K9me3, H3K27me2&3, and H3K36me2 (Coetzee et al., 2017), the latter of which has been linked to global repression in asexual stages (Karmodiya et al., 2015). Other less well-characterized PTMs including H3K37me1, H3R17me1 and H3R17me2 are also prominent in the early-stage gametocytes (von Grüning et al., 2022).
During gametocytogenesis, H3K9me3/HP1 patterns undergo substantial changes, leading to large heterochromatin reorganization events (Bunnik et al., 2018; Fraschka et al., 2018). Together with a subtelomeric stretch of chromosome 10, genes encoding merozoite invasion proteins become strongly associated with the distinct perinuclear H3K9me3/HP1-enriched heterochromatin clusters (Brancucci et al., 2014; Bunnik et al., 2018). Additionally, heterochromatin expansion across the subtelomeric region of chromosome 14 leads to the formation of a topologically associated superdomain (>800 kb) with a strong boundary that is postulated to mediate the expression of cell growth and division inhibitors during gametocyte development (Bunnik et al., 2018), similar to what has been observed in other organisms (Dixon et al., 2012). During gametocytogenesis, certain genomic regions show reduced H3K9me3/HP1 occupancy, including those containing sexual development genes (e.g., ap2-g, pfg14-744, and pfg14-748) (Bunnik et al., 2018; Fraschka et al., 2018).
Transcriptional repression of asexual-stage associated genes that become obsolete in the immature gametocyte stages has been associated with several PTMs including H3K36me2&3 (Connacher et al., 2021). Despite the strong link between H3K36me3 and AP2-G2 occupancy in the asexual stages, fewer H3K36me2&3 enriched genes are associated with AP2-G2 in the early-stage gametocyte, indicating that, similar to the divergent transcriptional programs, regulation by histone PTM landscapes is highly specific to gametocytogenesis. The histone PTM landscape of immature gametocytes is also characterized by an increased abundance of repressive H3K27me2&3, H4K20me3 and H3K27ac, the latter two of which co-occur with AP2-G2 in asexual parasites (Singh et al., 2020). While it is tempting to speculate that these histone PTMs are similarly involved in recruiting transcription factors and chromatin modifying enzymes in P. falciparum as in other eukaryotes (Carrozza et al., 2005; Keogh et al., 2005; Maltby et al., 2012; Ren et al., 2021), the functional and mechanistic links between these epigenetic factors remain interesting points for further study.
Arginine methylation combinations may represent a key feature of epigenetic regulation for gametocyte development, exemplified by the exclusive combinations of H3R42me1 with H3K37me1 and H3R40me1 (von Grüning et al., 2022). The abundance of combinatorial H3K14me2 with H3K9me1 or H3K18 methylation offers the first suggestion that co-existing PTMs may also modulate heterochromatin establishment in early-stage gametocytes (von Grüning et al., 2022). PTM crosstalk in the late-stage gametocyte reflects a unique relationship between PTMs compared to other stages of the parasite’s life cycle. Late-stage (stages IV/V) gametocytes are transcriptionally poised for subsequent gamete formation, as reflected by equal distribution of hetero- and euchromatin marks and the use of translational repression mechanisms in female gametocytes (Lasonder et al., 2016; Coetzee et al., 2017). The combination of H3K27me1K36me1 (Coetzee et al., 2017; von Grüning et al., 2022), may further contribute towards generating poised mRNAs, congruent with the dependence of H3K27me1 on H3K36me1 to induce transcription in embryonic stem cells (Jung et al., 2013). Ultimately, the histone PTM landscape of the late-stage gametocytes presents a suitable environment in which certain genes remain transcriptionally poised in preparation for rapid fertilization in the mosquito.
Conclusion and Future Directions
Given the central nature of histone PTMs for P. falciparum reviewed here, further understanding the mechanistic relevance of individual and combinatorial PTMs will help identification of the most suited epigenetic targets for drug-based intervention. Although the stage-specific functional relevance of certain individual histone PTMs has been elucidated, the remaining PTMs and combinations with stage-specific patterns should be investigated. Specifically, the influence of these PTMs on asexual and sexual development and the writers, erasers and readers of the histone PTM landscapes and thus insights into the direct or indirect consequences on transcription. In the future, we foresee the use of the combinatorial histone code as a “barcode” that could define parasite life cycle stages or ongoing biological processes. This barcode is dynamic and associated with remarkable changes to allow shifts in chromatin status in different developmental stages of the parasite. In-depth critical analyses of the available data clearly implicate this changing histone PTM landscape as higher-order mechanisms of regulation of gene expression, providing a clear blueprint associated with required phenotypic changes in the parasite to adapt to host responses (antigenic variation), allow massive population expansion in the host (asexual proliferation) and exploiting host environments to allow differentiation and species continuation (sexual development). However, as histone PTMs are not isolated but co-exist to fine-tune binding of effector proteins, the PTM landscape and combinatorial code may indeed guide a highly specific transcriptional program. Lastly, the cause and/or effect relationship between nutrient metabolism and available substrates to modify the histones requires deconvolution. Taken together, the findings discussed in this necessary update demonstrate that although many factors, including nucleosome positioning, chromatin structure and transcription factors participate in transcriptional regulation in P. falciparum parasites, the histone PTM landscapes represent the road maps that are central to the specific biology of each stage. The demonstration of histone PTMs as key determinants of both asexual proliferation and sexual differentiation of P. falciparum parasites highlights the individual factors as novel targets for disrupting malaria transmission and underscore the importance of a more thorough understanding of these critical regulators.
Author Contributions
LB conceived the work and JC, HvG, and LB wrote and edited the paper.
Funding
This work was supported by the South African Research Chairs Initiative of the Department of Science and Innovation, administered through the South African National Research Foundation (UID 84627) to LB. The UP ISMC acknowledges the South African Medical Research Council (SA MRC) as Collaborating Centre for Malaria Research.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
Ac, acetylation; EDC, exoerythrocytic developmental cycle; gdv1, gametocyte development protein 1; HAT, histone acetyltransferase; HDAC, histone deacetylase; HDM: histone demethylase; histone PTMs, histone post-translational modifications; HMT, histone methyltransferase; HP1, heterochromatin protein 1; IDC, intraerythrocytic developmental cycle; JMJC, jumonji-C containing demethylase; LSD, lysine-specific demethylase; me, methylation.
References
Abel, S., and Le Roch, K. G. (2019). The Role of Epigenetics and Chromatin Structure in Transcriptional Regulation in Malaria Parasites. Brief. Funct. Genomics 18, 302–313. doi:10.1093/bfgp/elz005
Aguilar, R., Magallon-Tejada, A., Achtman, A. H., Moraleda, C., Joice, R., Cisteró, P., et al. (2014). Molecular Evidence for the Localization of Plasmodium Falciparum Immature Gametocytes in Bone Marrow. Blood 123, 959–966. doi:10.1182/blood-2013-08-520767
Ay, F., Bunnik, E. M., Varoquaux, N., Bol, S. M., Prudhomme, J., Vert, J.-P., et al. (2014). Three-dimensional Modeling of the P. Falciparum Genome during the Erythrocytic Cycle Reveals a strong Connection between Genome Architecture and Gene Expression. Genome Res. 24, 974–988. doi:10.1101/gr.169417.113
Balaji, S., Babu, M. M., Iyer, L. M., and Aravind, L. (2005). Discovery of the Principal Specific Transcription Factors of Apicomplexa and Their Implication for the Evolution of the AP2-Integrase DNA Binding Domains. Nucleic Acids Res. 33, 3994–4006. doi:10.1093/nar/gki709
Bártfai, R., Hoeijmakers, W. A. M., Salcedo-Amaya, A. M., Smits, A. H., Janssen-Megens, E., Kaan, A., et al. (2010). H2A.Z Demarcates Intergenic Regions of the Plasmodium Falciparum Epigenome that Are Dynamically Marked by H3K9ac and H3K4me3. Plos Pathog. 6, e1001223. doi:10.1371/journal.ppat.1001223
Berger, S. L. (2002). Histone Modifications in Transcriptional Regulation. Curr. Opin. Genet. Dev. 12, 142–148. doi:10.1016/s0959-437x(02)00279-4
Bhanu, N. V., Sidoli, S., and Garcia, B. A. (2016). Histone Modification Profiling Reveals Differential Signatures Associated with Human Embryonic Stem Cell Self-Renewal and Differentiation. Proteomics 16, 448–458. doi:10.1002/pmic.201500231
Bhowmick, K., Tehlan, A., SunitaSudhakar, R., Sudhakar, R., Kaur, I., Sijwali, P. S., et al. (2020). Plasmodium Falciparum GCN5 Acetyltransferase Follows a Novel Proteolytic Processing Pathway that Is Essential for its Function. J. Cel Sci 133. doi:10.1242/jcs.236489
Billker, O., Lindo, V., Panico, M., Etienne, A. E., Paxton, T., Dell, A., et al. (1998). Identification of Xanthurenic Acid as the Putative Inducer of Malaria Development in the Mosquito. Nature 392, 289–292. doi:10.1038/32667
Birkholtz, L.-M., Coetzer, T. L., Mancama, D., Leroy, D., and Alano, P. (2016). Discovering New Transmission-Blocking Antimalarial Compounds: Challenges and Opportunities. Trends Parasitol. 32, 669–681. doi:10.1016/j.pt.2016.04.017
Bozdech, Z., Llinás, M., Pulliam, B. L., Wong, E. D., Zhu, J., and Derisi, J. L. (2003). The Transcriptome of the Intraerythrocytic Developmental Cycle of Plasmodium Falciparum. Plos Biol. 1, E5. doi:10.1371/journal.pbio.0000005
Brancucci, N. M. B., Bertschi, N. L., Zhu, L., Niederwieser, I., Chin, W. H., Wampfler, R., et al. (2014). Heterochromatin Protein 1 Secures Survival and Transmission of Malaria Parasites. Cell Host & Microbe 16, 165–176. doi:10.1016/j.chom.2014.07.004
Brancucci, N. M. B., Gerdt, J. P., Wang, C., De Niz, M., Philip, N., Adapa, S. R., et al. (2017). Lysophosphatidylcholine Regulates Sexual Stage Differentiation in the Human Malaria Parasite Plasmodium Falciparum. Cell 171, 1532–1544. doi:10.1016/j.cell.2017.10.020
Buckling, A., Ranford-Cartwright, L. C., Miles, A., and Read, A. F. (1999). Chloroquine Increases Plasmodium Falciparum Gametocytogenesis In Vitro. Parasitology 118 (Pt 4), 339–346. doi:10.1017/s0031182099003960
Bui, H. T. N., Niederwieser, I., Bird, M. J., Dai, W., Brancucci, N. M. B., Moes, S., et al. (2019). Mapping and Functional Analysis of Heterochromatin Protein 1 Phosphorylation in the Malaria Parasite Plasmodium Falciparum. Sci. Rep. 9, 16720. doi:10.1038/s41598-019-53325-9
Bunnik, E. M., Cook, K. B., Varoquaux, N., Batugedara, G., Prudhomme, J., Cort, A., et al. (2018). Changes in Genome Organization of Parasite-specific Gene Families during the Plasmodium Transmission Stages. Nat. Commun. 9, 1910. doi:10.1038/s41467-018-04295-5
Bunnik, E. M., Polishko, A., Prudhomme, J., Ponts, N., Gill, S. S., Lonardi, S., et al. (2014). DNA-encoded Nucleosome Occupancy Is Associated with Transcription Levels in the Human Malaria Parasite Plasmodium Falciparum. BMC Genomics 15, 347. doi:10.1186/1471-2164-15-347
Bunnik, E. M., Venkat, A., Shao, J., Mcgovern, K. E., Batugedara, G., Worth, D., et al. (2019). Comparative 3D Genome Organization in Apicomplexan Parasites. Proc. Natl. Acad. Sci. USA 116, 3183–3192. doi:10.1073/pnas.1810815116
Carrozza, M. J., Li, B., Florens, L., Suganuma, T., Swanson, S. K., Lee, K. K., et al. (2005). Histone H3 Methylation by Set2 Directs Deacetylation of Coding Regions by Rpd3S to Suppress Spurious Intragenic Transcription. Cell 123, 581–592. doi:10.1016/j.cell.2005.10.023
Carter, R., and Graves, P. M. (1988). “Gametocytes,” in Malaria: Principles and Practice of Malariology. Editor I. Mcgregor (London: Churchill Livingstone), 253–305.
Chaal, B. K., Gupta, A. P., Wastuwidyaningtyas, B. D., Luah, Y.-H., and Bozdech, Z. (2010). Histone Deacetylases Play a Major Role in the Transcriptional Regulation of the Plasmodium Falciparum Life Cycle. Plos Pathog. 6, e1000737. doi:10.1371/journal.ppat.1000737
Chen, C. C. L., Goyal, P., Karimi, M. M., Abildgaard, M. H., Kimura, H., and Lorincz, M. C. (2018). H3S10ph Broadly marks Early-Replicating Domains in Interphase ESCs and Shows Reciprocal Antagonism with H3K9me2. Genome Res. 28, 37–51. doi:10.1101/gr.224717.117
Chen, P. B., Ding, S., Zanghì, G., Soulard, V., Dimaggio, P. A., Fuchter, M. J., et al. (2016). Plasmodium Falciparum PfSET7: Enzymatic Characterization and Cellular Localization of a Novel Protein Methyltransferase in Sporozoite, Liver and Erythrocytic Stage Parasites. Sci. Rep. 6, 21802. doi:10.1038/srep21802
Chookajorn, T., Dzikowski, R., Frank, M., Li, F., Jiwani, A. Z., Hartl, D. L., et al. (2007). Epigenetic Memory at Malaria Virulence Genes. Proc. Natl. Acad. Sci. U.S.A. 104, 899–902. doi:10.1073/pnas.0609084103
Cobbold, S. A., Santos, J. M., Ochoa, A., Perlman, D. H., and Llinás, M. (2016). Proteome-wide Analysis Reveals Widespread Lysine Acetylation of Major Protein Complexes in the Malaria Parasite. Sci. Rep. 6, 19722. doi:10.1038/srep19722
Coetzee, N., Sidoli, S., Van Biljon, R., Painter, H., Llinás, M., Garcia, B. A., et al. (2017). Quantitative Chromatin Proteomics Reveals a Dynamic Histone post-translational Modification Landscape that Defines Asexual and Sexual Plasmodium Falciparum Parasites. Sci. Rep. 7, 607. doi:10.1038/s41598-017-00687-7
Coetzee, N., von Grüning, H., Opperman, D., Van Der Watt, M., Reader, J., and Birkholtz, L.-M. (2020). Epigenetic Inhibitors Target Multiple Stages of Plasmodium Falciparum Parasites. Sci. Rep. 10, 2355. doi:10.1038/s41598-020-59298-4
Coleman, B. I., Skillman, K. M., Jiang, R. H. Y., Childs, L. M., Altenhofen, L. M., Ganter, M., et al. (2014). A Plasmodium Falciparum Histone Deacetylase Regulates Antigenic Variation and Gametocyte Conversion. Cell Host & Microbe 16, 177–186. doi:10.1016/j.chom.2014.06.014
Comeaux, C. A., and Duraisingh, M. T. (2007). Unravelling a Histone Code for Malaria Virulence. Mol. Microbiol. 66, 1291–1295. doi:10.1111/j.1365-2958.2007.06038.x
Connacher, J., Josling, G. A., Orchard, L. M., Reader, J., Llinás, M., and Birkholtz, L.-M. (2021). H3K36 Methylation Reprograms Gene Expression to Drive Early Gametocyte Development in Plasmodium Falciparum. Epigenetics & Chromatin 14, 19. doi:10.1186/s13072-021-00393-9
Cowell, A. N., Istvan, E. S., Lukens, A. K., Gomez-Lorenzo, M. G., Vanaerschot, M., Sakata-Kato, T., et al. (2018). Mapping the Malaria Parasite Druggable Genome by Using In Vitro Evolution and Chemogenomics. Science 359, 191–199. doi:10.1126/science.aan4472
Crowley, V. M., Rovira-Graells, N., de Pouplana, L. R., and Cortés, A. (2011). Heterochromatin Formation in Bistable Chromatin Domains Controls the Epigenetic Repression of Clonally Variant Plasmodium Falciparum Genes Linked to Erythrocyte Invasion. Mol. Microbiol. 80, 391–406. doi:10.1111/j.1365-2958.2011.07574.x
Cui, L., Fan, Q., Cui, L., and Miao, J. (2008a). Histone Lysine Methyltransferases and Demethylases in Plasmodium Falciparum. Int. J. Parasitol. 38, 1083–1097. doi:10.1016/j.ijpara.2008.01.002
Cui, L., and Miao, J. (2010). Chromatin-mediated Epigenetic Regulation in the Malaria Parasite Plasmodium Falciparum. Eukaryot. Cel 9, 1138–1149. doi:10.1128/ec.00036-10
Cui, L., Miao, J., and Cui, L. (2007a). Cytotoxic Effect of Curcumin on Malaria Parasite Plasmodium Falciparum : Inhibition of Histone Acetylation and Generation of Reactive Oxygen Species. Antimicrob. Agents Chemother. 51, 488–494. doi:10.1128/aac.01238-06
Cui, L., Miao, J., Furuya, T., Fan, Q., Li, X., Rathod, P. K., et al. (2008b). Histone Acetyltransferase Inhibitor Anacardic Acid Causes Changes in Global Gene Expression during In Vitro Plasmodium Falciparum Development. Eukaryot. Cel 7, 1200–1210. doi:10.1128/ec.00063-08
Cui, L., Miao, J., Furuya, T., Li, X., Su, X.-z., and Cui, L. (2007b). PfGCN5-mediated Histone H3 Acetylation Plays a Key Role in Gene Expression in Plasmodium Falciparum. Eukaryot. Cel 6, 1219–1227. doi:10.1128/ec.00062-07
Dastidar, E. G., Dzeyk, K., Krijgsveld, J., Malmquist, N. A., Doerig, C., Scherf, A., et al. (2013). Comprehensive Histone Phosphorylation Analysis and Identification of Pf14-3-3 Protein as a Histone H3 Phosphorylation Reader in Malaria Parasites. PLoS One 8, e53179. doi:10.1371/journal.pone.0053179
Delves, M. J. (2012). Plasmodium Cell Biology Should Inform Strategies Used in the Development of Antimalarial Transmission-Blocking Drugs. Future Med. Chem. 4, 2251–2263. doi:10.4155/fmc.12.182
Dixon, J. R., Selvaraj, S., Yue, F., Kim, A., Li, Y., Shen, Y., et al. (2012). Topological Domains in Mammalian Genomes Identified by Analysis of Chromatin Interactions. Nature 485, 376–380. doi:10.1038/nature11082
Doug Chung, D.-W., Ponts, N., Cervantes, S., and Le Roch, K. G. (2009). Post-translational Modifications in Plasmodium: More Than You Think! Mol. Biochem. Parasitol. 168, 123–134. doi:10.1016/j.molbiopara.2009.08.001
Dvorak, J. A., Miller, L. H., Whitehouse, W. C., and Shiroishi, T. (1975). Invasion of Erythrocytes by Malaria Merozoites. Science 187, 748–750. doi:10.1126/science.803712
Dyer, M., and Day, K. (2000). Expression of Plasmodium Falciparum Trimeric G Proteins and Their Involvement in Switching to Sexual Development. Mol. Biochem. Parasitol. 110, 437–448. doi:10.1016/s0166-6851(00)00288-7
Fan, Q., An, L., and Cui, L. (2004a). PfADA2, a Plasmodium Falciparum Homologue of the Transcriptional Coactivator ADA2 and its In Vivo Association with the Histone Acetyltransferase PfGCN5. Gene 336, 251–261. doi:10.1016/j.gene.2004.04.005
Fan, Q., An, L., and Cui, L. (2004b). Plasmodium Falciparum Histone Acetyltransferase, a Yeast GCN5 Homologue Involved in Chromatin Remodeling. Eukaryot. Cel 3, 264–276. doi:10.1128/ec.3.2.264-276.2004
Fan, Q., Miao, J., Cui, L., and Cui, L. (2009). Characterization of PRMT1 from Plasmodium Falciparum. Biochem. J. 421, 107–118. doi:10.1042/bj20090185
Filarsky, M., Fraschka, S. A., Niederwieser, I., Brancucci, N. M. B., Carrington, E., Carrió, E., et al. (2018). GDV1 Induces Sexual Commitment of Malaria Parasites by Antagonizing HP1-dependent Gene Silencing. Science 359, 1259–1263. doi:10.1126/science.aan6042
Fischle, W., Tseng, B. S., Dormann, H. L., Ueberheide, B. M., Garcia, B. A., Shabanowitz, J., et al. (2005). Regulation of HP1-Chromatin Binding by Histone H3 Methylation and Phosphorylation. Nature 438, 1116–1122. doi:10.1038/nature04219
Fleck, K., Nitz, M., and Jeffers, V. (2021). "Reading" a New Chapter in Protozoan Parasite Transcriptional Regulation. Plos Pathog. 17, e1010056. doi:10.1371/journal.ppat.1010056
Flueck, C., Bartfai, R., Volz, J., Niederwieser, I., Salcedo-Amaya, A. M., Alako, B. T. F., et al. (2009). Plasmodium Falciparum Heterochromatin Protein 1 marks Genomic Loci Linked to Phenotypic Variation of Exported Virulence Factors. Plos Pathog. 5, e1000569. doi:10.1371/journal.ppat.1000569
Fraschka, S. A.-K., Henderson, R. W. M., and Bártfai, R. (2016). H3.3 Demarcates GC-Rich Coding and Subtelomeric Regions and Serves as Potential Memory Mark for Virulence Gene Expression in Plasmodium Falciparum. Sci. Rep. 6, 31965. doi:10.1038/srep31965
Fraschka, S. A., Filarsky, M., Hoo, R., Niederwieser, I., Yam, X. Y., Brancucci, N. M. B., et al. (2018). Comparative Heterochromatin Profiling Reveals Conserved and Unique Epigenome Signatures Linked to Adaptation and Development of Malaria Parasites. Cell Host & Microbe 23, 407–420. doi:10.1016/j.chom.2018.01.008
Gardner, M. J., Hall, N., Fung, E., White, O., Berriman, M., Hyman, R. W., et al. (2002). Genome Sequence of the Human Malaria Parasite Plasmodium Falciparum. Nature 419, 498–511. doi:10.1038/nature01097
Goll, M. G., and Bestor, T. H. (2002). Histone Modification and Replacement in Chromatin Activation: Figure 1. Genes Dev. 16, 1739–1742. doi:10.1101/gad.1013902
Gomes, A. R., Bushell, E., Schwach, F., Girling, G., Anar, B., Quail, M. A., et al. (2015). A Genome-Scale Vector Resource Enables High-Throughput Reverse Genetic Screening in a Malaria Parasite. Cell Host & Microbe 17, 404–413. doi:10.1016/j.chom.2015.01.014
Gómez-Díaz, E., Yerbanga, R. S., Lefèvre, T., Cohuet, A., Rowley, M. J., Ouedraogo, J. B., et al. (2017). Epigenetic Regulation of Plasmodium Falciparum Clonally Variant Gene Expression during Development in Anopheles gambiae. Sci. Rep. 7, 40655. doi:10.1038/srep40655
Govin, J., Dorsey, J., Gaucher, J., Rousseaux, S., Khochbin, S., and Berger, S. L. (2010). Systematic Screen Reveals New Functional Dynamics of Histones H3 and H4 during Gametogenesis. Genes Dev. 24, 1772–1786. doi:10.1101/gad.1954910
Goyal, M., Heinberg, A., Mitesser, V., Kandelis-Shalev, S., Singh, B. K., and Dzikowski, R. (2021). Phosphorylation of the Canonical Histone H2A Marks Foci of Damaged DNA in Malaria Parasites. mSphere 6, e01131. doi:10.1128/mSphere.01131-20
Green, J. L., Wu, Y., Encheva, V., Lasonder, E., Prommaban, A., Kunzelmann, S., et al. (2020). Ubiquitin Activation Is Essential for Schizont Maturation in Plasmodium Falciparum Blood-Stage Development. Plos Pathog. 16, e1008640. doi:10.1371/journal.ppat.1008640
Guillemette, B., Drogaris, P., Lin, H.-H. S., Armstrong, H., Hiragami-Hamada, K., Imhof, A., et al. (2011). H3 Lysine 4 Is Acetylated at Active Gene Promoters and Is Regulated by H3 Lysine 4 Methylation. Plos Genet. 7, e1001354. doi:10.1371/journal.pgen.1001354
Gupta, A. P., and Bozdech, Z. (2017). Epigenetic Landscapes Underlining Global Patterns of Gene Expression in the Human Malaria Parasite, Plasmodium Falciparum. Int. J. Parasitol. 47, 399–407. doi:10.1016/j.ijpara.2016.10.008
Gupta, A. P., Chin, W. H., Zhu, L., Mok, S., Luah, Y.-H., Lim, E.-H., et al. (2013). Dynamic Epigenetic Regulation of Gene Expression during the Life Cycle of Malaria Parasite Plasmodium Falciparum. Plos Pathog. 9, e1003170. doi:10.1371/journal.ppat.1003170
Gupta, A. P., Zhu, L., Tripathi, J., Kucharski, M., Patra, A., and Bozdech, Z. (2017). Histone 4 Lysine 8 Acetylation Regulates Proliferation and Host-Pathogen Interaction in Plasmodium Falciparum. Epigenetics & Chromatin 10, 40. doi:10.1186/s13072-017-0147-z
Herrera-Solorio, A. M., Vembar, S. S., Macpherson, C. R., Lozano-Amado, D., Meza, G. R., Xoconostle-Cazares, B., et al. (2019). Clipped Histone H3 Is Integrated into Nucleosomes of DNA Replication Genes in the Human Malaria Parasite Plasmodium Falciparum. EMBO Rep. 20, e46331. doi:10.15252/embr.201846331
Hirota, T., Lipp, J. J., Toh, B.-H., and Peters, J.-M. (2005). Histone H3 Serine 10 Phosphorylation by Aurora B Causes HP1 Dissociation from Heterochromatin. Nature 438, 1176–1180. doi:10.1038/nature04254
Hoeijmakers, W. A. M., Miao, J., Schmidt, S., Toenhake, C. G., Shrestha, S., Venhuizen, J., et al. (2019). Epigenetic Reader Complexes of the Human Malaria Parasite, Plasmodium Falciparum. Nucleic Acids Res. 47, 11574–11588. doi:10.1093/nar/gkz1044
Hoeijmakers, W. A. M., Salcedo‐Amaya, A. M., Smits, A. H., Françoijs, K. J., Treeck, M., Gilberger, T. W., et al. (2013). H 2 A . Z/H 2 B . Z Double‐variant Nucleosomes Inhabit the at ‐rich Promoter Regions of the P Lasmodium Falciparum Genome. Mol. Microbiol. 87, 1061–1073. doi:10.1111/mmi.12151
Hoeijmakers, W. A. M., Stunnenberg, H. G., and Bártfai, R. (2012). Placing the Plasmodium Falciparum Epigenome on the Map. Trends Parasitol. 28, 486–495. doi:10.1016/j.pt.2012.08.006
Hunter, T. (2007). The Age of Crosstalk: Phosphorylation, Ubiquitination, and beyond. Mol. Cel 28, 730–738. doi:10.1016/j.molcel.2007.11.019
Issar, N., Ralph, S. A., Mancio-Silva, L., Keeling, C., and Scherf, A. (2009). Differential Sub-nuclear Localisation of Repressive and Activating Histone Methyl Modifications in P. Falciparum. Microbes Infect. 11, 403–407. doi:10.1016/j.micinf.2008.12.010
Issar, N., Roux, E., Mattei, D., and Scherf, A. (2008). Identification of a Novel post-translational Modification inPlasmodium Falciparum: Protein Sumoylation in Different Cellular Compartments. Cell Microbiol 10, 1999–2011. doi:10.1111/j.1462-5822.2008.01183.x
Janssen, A., Colmenares, S. U., Lee, T., and Karpen, G. H. (2019). Timely Double-Strand Break Repair and Pathway Choice in Pericentromeric Heterochromatin Depend on the Histone Demethylase dKDM4A. Genes Dev. 33, 103–115. doi:10.1101/gad.317537.118
Jiang, L., Mu, J., Zhang, Q., Ni, T., Srinivasan, P., Rayavara, K., et al. (2013). PfSETvs Methylation of Histone H3K36 Represses Virulence Genes in Plasmodium Falciparum. Nature 499, 223–227. doi:10.1038/nature12361
Joice, R., Nilsson, S. K., Montgomery, J., Dankwa, S., Egan, E., Morahan, B., et al. (2014). Plasmodium Falciparum Transmission Stages Accumulate in the Human Bone Marrow. Sci. Transl Med. 6, 244re5. doi:10.1126/scitranslmed.3008882
Josling, G. A., and Llinás, M. (2015). Sexual Development in Plasmodium Parasites: Knowing when It's Time to Commit. Nat. Rev. Microbiol. 13, 573–587. doi:10.1038/nrmicro3519
Josling, G. A., Russell, T. J., Venezia, J., Orchard, L., Van Biljon, R., Painter, H. J., et al. (2020). Dissecting the Role of PfAP2-G in Malaria Gametocytogenesis. Nat. Commun. 11, 1503. doi:10.1038/s41467-020-15026-0
Jung, H. R., Sidoli, S., Haldbo, S., Sprenger, R. R., Schwämmle, V., Pasini, D., et al. (2013). Precision Mapping of Coexisting Modifications in Histone H3 Tails from Embryonic Stem Cells by ETD-MS/MS. Anal. Chem. 85, 8232–8239. doi:10.1021/ac401299w
Kafsack, B. F. C., Rovira-Graells, N., Clark, T. G., Bancells, C., Crowley, V. M., Campino, S. G., et al. (2014). A Transcriptional Switch Underlies Commitment to Sexual Development in Malaria Parasites. Nature 507, 248–252. doi:10.1038/nature12920
Kanyal, A., Rawat, M., Gurung, P., Choubey, D., Anamika, K., and Karmodiya, K. (2018). Genome‐wide Survey and Phylogenetic Analysis of Histone Acetyltransferases and Histone Deacetylases of Plasmodium Falciparum. FEBS J. 285, 1767–1782. doi:10.1111/febs.14376
Kappe, S. H. I., Buscaglia, C. A., and Nussenzweig, V. (2004). Plasmodium Sporozoite Molecular Cell Biology. Annu. Rev. Cel Dev. Biol. 20, 29–59. doi:10.1146/annurev.cellbio.20.011603.150935
Karmodiya, K., Pradhan, S. J., Joshi, B., Jangid, R., Reddy, P. C., and Galande, S. (2015). A Comprehensive Epigenome Map of Plasmodium Falciparum Reveals Unique Mechanisms of Transcriptional Regulation and Identifies H3K36me2 as a Global Mark of Gene Suppression. Epigenetics & Chromatin 8, 32. doi:10.1186/s13072-015-0029-1
Kaur, I., Zeeshan, M., Saini, E., Kaushik, A., Mohmmed, A., Gupta, D., et al. (2016). Widespread Occurrence of Lysine Methylation in Plasmodium Falciparum Proteins at Asexual Blood Stages. Sci. Rep. 6, 35432. doi:10.1038/srep35432
Keogh, M.-C., Kurdistani, S. K., Morris, S. A., Ahn, S. H., Podolny, V., Collins, S. R., et al. (2005). Cotranscriptional Set2 Methylation of Histone H3 Lysine 36 Recruits a Repressive Rpd3 Complex. Cell 123, 593–605. doi:10.1016/j.cell.2005.10.025
Kumar, M., Skillman, K., and Duraisingh, M. T. (2021). Linking Nutrient Sensing and Gene Expression in Plasmodium Falciparum Blood‐stage Parasites. Mol. Microbiol. 115, 891–900. doi:10.1111/mmi.14652
Lasonder, E., Rijpma, S. R., van Schaijk, B. C. L., Hoeijmakers, W. A. M., Kensche, P. R., Gresnigt, M. S., et al. (2016). Integrated Transcriptomic and Proteomic Analyses ofP. Falciparumgametocytes: Molecular Insight into Sex-specific Processes and Translational Repression. Nucleic Acids Res. 44, 6087–6101. doi:10.1093/nar/gkw536
Latchman, D. S. (2010). “Role of Chromatin Structure in Gene Control,,” in Gene Control. Editor E. Owen (New York: Garland Science), 66–73.
Lee, S.-B., Jasencakova, Z., and Groth, A. (2012). H3K56me1 marks a Spot for PCNA. Mol. Cel 46, 1–2. doi:10.1016/j.molcel.2012.03.022
Lemieux, J. E., Kyes, S. A., Otto, T. D., Feller, A. I., Eastman, R. T., Pinches, R. A., et al. (2013). Genome‐wide Profiling of Chromosome Interactions in P Lasmodium Falciparum Characterizes Nuclear Architecture and Reconfigurations Associated with Antigenic Variation. Mol. Microbiol. 90, 519–537. doi:10.1111/mmi.12381
López-Barragán, M. J., Lemieux, J., Quiñones, M., Williamson, K. C., Molina-Cruz, A., Cui, K., et al. (2011). Directional Gene Expression and Antisense Transcripts in Sexual and Asexual Stages of Plasmodium Falciparum. BMC Genomics 12, 587. doi:10.1186/1471-2164-12-587
Lopez-Rubio, J.-J., Mancio-Silva, L., and Scherf, A. (2009). Genome-wide Analysis of Heterochromatin Associates Clonally Variant Gene Regulation with Perinuclear Repressive Centers in Malaria Parasites. Cell Host & Microbe 5, 179–190. doi:10.1016/j.chom.2008.12.012
Maltby, V. E., Martin, B. J. E., Schulze, J. M., Johnson, I., Hentrich, T., Sharma, A., et al. (2012). Histone H3 Lysine 36 Methylation Targets the Isw1b Remodeling Complex to Chromatin. Mol. Cell Biol. 32, 3479–3485. doi:10.1128/mcb.00389-12
Mancio-Silva, L., Lopez-Rubio, J. J., Claes, A., and Scherf, A. (2013). Sir2a Regulates rDNA Transcription and Multiplication Rate in the Human Malaria Parasite Plasmodium Falciparum. Nat. Commun. 4, 1530. doi:10.1038/ncomms2539
Matthews, K. A., Senagbe, K. M., Nötzel, C., Gonzales, C. A., Tong, X., Rijo-Ferreira, F., et al. (2020). Disruption of the Plasmodium Falciparum Life Cycle through Transcriptional Reprogramming by Inhibitors of Jumonji Demethylases. ACS Infect. Dis. 6, 1058–1075. doi:10.1021/acsinfecdis.9b00455
Merrick, C. J., and Duraisingh, M. T. (2010). Epigenetics in Plasmodium: what Do We Really Know? Eukaryot. Cel 9, 1150–1158. doi:10.1128/ec.00093-10
Meshorer, E., Yellajoshula, D., George, E., Scambler, P. J., Brown, D. T., and Misteli, T. (2006). Hyperdynamic Plasticity of Chromatin Proteins in Pluripotent Embryonic Stem Cells. Develop. Cel 10, 105–116. doi:10.1016/j.devcel.2005.10.017
Miao, J., Fan, Q., Cui, L., Li, J., Li, J., and Cui, L. (2006). The Malaria Parasite Plasmodium Falciparum Histones: Organization, Expression, and Acetylation. Gene 369, 53–65. doi:10.1016/j.gene.2005.10.022
Miao, J., Fan, Q., Cui, L., Li, X., Wang, H., Ning, G., et al. (2010a). The MYST Family Histone Acetyltransferase Regulates Gene Expression and Cell Cycle in Malaria Parasite Plasmodium Falciparum. Mol. Microbiol. 78, 883–902. doi:10.1111/j.1365-2958.2010.07371.x
Miao, J., Li, J., Fan, Q., Li, X., Li, X., and Cui, L. (2010b). The Puf-Family RNA-Binding Protein PfPuf2 Regulates Sexual Development and Sex Differentiation in the Malaria parasitePlasmodium Falciparum. J. Cel Sci 123, 1039–1049. doi:10.1242/jcs.059824
Miao, J., Wang, C., Lucky, A. B., Liang, X., Min, H., Adapa, S. R., et al. (2021). A Unique GCN5 Histone Acetyltransferase Complex Controls Erythrocyte Invasion and Virulence in the Malaria Parasite Plasmodium Falciparum. Plos Pathog. 17, e1009351. doi:10.1371/journal.ppat.1009351
Miller, J. L., and Grant, P. A. (2013). The Role of DNA Methylation and Histone Modifications in Transcriptional Regulation in Humans. Subcell Biochem. 61, 289–317. doi:10.1007/978-94-007-4525-4_13
Müller, M. M., and Muir, T. W. (2015). Histones: at the Crossroads of Peptide and Protein Chemistry. Chem. Rev. 115, 2296–2349. doi:10.1021/cr5003529
Ngwa, C. J., Gross, M. R., Musabyimana, J. P., Pradel, G., and Deitsch, K. W. (2021). The Role of the Histone Methyltransferase PfSET10 in Antigenic Variation by Malaria Parasites: a Cautionary Tale. mSphere 6, e01217. doi:10.1128/mSphere.01217-20
Ngwa, C. J., Kiesow, M. J., Papst, O., Orchard, L. M., Filarsky, M., Rosinski, A. N., et al. (2017). Transcriptional Profiling Defines Histone Acetylation as a Regulator of Gene Expression during Human-To-Mosquito Transmission of the Malaria Parasite Plasmodium Falciparum. Front. Cel. Infect. Microbiol. 7, 320. doi:10.3389/fcimb.2017.00320
Perez-Toledo, K., Rojas-Meza, A. P., Mancio-Silva, L., Hernandez-Cuevas, N. A., Delgadillo, D. M., Vargas, M., et al. (2009). Plasmodium Falciparum Heterochromatin Protein 1 Binds to Tri-methylated Histone 3 Lysine 9 and Is Linked to Mutually Exclusive Expression of Var Genes. Nucleic Acids Res. 37, 2596–2606. doi:10.1093/nar/gkp115
Petter, M., Lee, C. C., Byrne, T. J., Boysen, K. E., Volz, J., Ralph, S. A., et al. (2011). Expression of P. Falciparum Var Genes Involves Exchange of the Histone Variant H2A.Z at the Promoter. Plos Pathog. 7, e1001292. doi:10.1371/journal.ppat.1001292
Petter, M., Selvarajah, S. A., Lee, C. C., Chin, W. H., Gupta, A. P., Bozdech, Z., et al. (2013). H2A.Z and H2B.Z Double-Variant Nucleosomes Define Intergenic Regions and Dynamically Occupyvargene Promoters in the Malaria parasitePlasmodium Falciparum. Mol. Microbiol. 87, 1167–1182. doi:10.1111/mmi.12154
Poran, A., Nötzel, C., Aly, O., Mencia-Trinchant, N., Harris, C. T., Guzman, M. L., et al. (2017). Single-cell RNA Sequencing Reveals a Signature of Sexual Commitment in Malaria Parasites. Nature 551, 95–99. doi:10.1038/nature24280
Prata, I. O., Cubillos, E. F. G., Krüger, A., Barbosa, D., Martins, J., Setubal, J. C., et al. (2021). Plasmodium Falciparum Acetyl-CoA Synthetase Is Essential for Parasite Intraerythrocytic Development and Chromatin Modification. ACS Infect. Dis. 7, 3224–3240. doi:10.1021/acsinfecdis.1c00414
Read, D. F., Cook, K., Lu, Y. Y., Le Roch, K. G., and Noble, W. S. (2019). Predicting Gene Expression in the Human Malaria Parasite Plasmodium Falciparum Using Histone Modification, Nucleosome Positioning, and 3D Localization Features. Plos Comput. Biol. 15, e1007329. doi:10.1371/journal.pcbi.1007329
Reader, J., Van Der Watt, M. E., Taylor, D., Le Manach, C., Mittal, N., Ottilie, S., et al. (2021). Multistage and Transmission-Blocking Targeted Antimalarials Discovered from the Open-Source MMV Pandemic Response Box. Nat. Commun. 12, 269. doi:10.1038/s41467-020-20629-8
Reiter, K. H., Ramachandran, A., Xia, X., Boucher, L. E., Bosch, J., and Matunis, M. J. (2016). Characterization and Structural Insights into Selective E1-E2 Interactions in the Human and Plasmodium Falciparum SUMO Conjugation Systems. J. Biol. Chem. 291, 3860–3870. doi:10.1074/jbc.m115.680801
Ren, W., Fan, H., Grimm, S. A., Kim, J. J., Li, L., Guo, Y., et al. (2021). DNMT1 Reads Heterochromatic H4K20me3 to Reinforce LINE-1 DNA Methylation. Nat. Commun. 12, 2490. doi:10.1038/s41467-021-22665-4
Rovira-Graells, N., Gupta, A. P., Planet, E., Crowley, V. M., Mok, S., Ribas De Pouplana, L., et al. (2012). Transcriptional Variation in the Malaria Parasite Plasmodium Falciparum. Genome Res. 22, 925–938. doi:10.1101/gr.129692.111
Ruthenburg, A. J., Li, H., Milne, T. A., Dewell, S., Mcginty, R. K., Yuen, M., et al. (2011). Recognition of a Mononucleosomal Histone Modification Pattern by BPTF via Multivalent Interactions. Cell 145, 692–706. doi:10.1016/j.cell.2011.03.053
Ryu, H.-Y., and Hochstrasser, M. (2021). Histone Sumoylation and Chromatin Dynamics. Nucleic Acids Res. 49, 6043–6052. doi:10.1093/nar/gkab280
Sabari, B. R., Tang, Z., Huang, H., Yong-Gonzalez, V., Molina, H., Kong, H. E., et al. (2015). Intracellular Crotonyl-CoA Stimulates Transcription through P300-Catalyzed Histone Crotonylation. Mol. Cel 58, 203–215. doi:10.1016/j.molcel.2015.02.029
Salcedo-Amaya, A. M., Van Driel, M. A., Alako, B. T., Trelle, M. B., Van Den Elzen, A. M. G., Cohen, A. M., et al. (2009). Dynamic Histone H3 Epigenome Marking during the Intraerythrocytic Cycle of Plasmodium Falciparum. Proc. Natl. Acad. Sci. U.S.A. 106, 9655–9660. doi:10.1073/pnas.0902515106
Sanderson, T., and Rayner, J. C. (2017). PhenoPlasm: a Database of Disruption Phenotypes for Malaria Parasite Genes. Wellcome Open Res. 2, 45. doi:10.12688/wellcomeopenres.11896.1
Santos-Rosa, H., Millán-Zambrano, G., Han, N., Leonardi, T., Klimontova, M., Nasiscionyte, S., et al. (2021). Methylation of Histone H3 at Lysine 37 by Set1 and Set2 Prevents Spurious DNA Replication. Mol. Cel 81, 2793–2807. doi:10.1016/j.molcel.2021.04.021
Saraf, A., Cervantes, S., Bunnik, E. M., Ponts, N., Sardiu, M. E., Chung, D.-W. D., et al. (2016). Dynamic and Combinatorial Landscape of Histone Modifications during the Intraerythrocytic Developmental Cycle of the Malaria Parasite. J. Proteome Res. 15, 2787–2801. doi:10.1021/acs.jproteome.6b00366
Shang, X., Shen, S., Tang, J., He, X., Zhao, Y., Wang, C., et al. (2021). A cascade of Transcriptional Repression Determines Sexual Commitment and Development in Plasmodium Falciparum. Nucleic Acids Res. 49, 9264–9279. doi:10.1093/nar/gkab683
Sherman, I. W. (1979). Biochemistry of Plasmodium (Malarial Parasites). Microbiol. Rev. 43, 453–495. doi:10.1128/mr.43.4.453-495.1979
Silberhorn, E., Schwartz, U., Löffler, P., Schmitz, S., Symelka, A., De Koning-Ward, T., et al. (2016). Plasmodium Falciparum Nucleosomes Exhibit Reduced Stability and Lost Sequence Dependent Nucleosome Positioning. Plos Pathog. 12, e1006080. doi:10.1371/journal.ppat.1006080
Sinden, R. E., Canning, E. U., Bray, R. S., and Smalley, M. E. (1978). Gametocyte and Gamete Development in Plasmodium Falciparum. Proc. R. Soc. Lond. B Biol. Sci. 201, 375–399. doi:10.1098/rspb.1978.0051
Singh, S., Santos, J. M., Orchard, L. M., Yamada, N., Van Biljon, R., Painter, H. J., et al. (2020). The PfAP2-G2 Transcription Factor Is a Critical Regulator of Gametocyte Maturation. Mol. Microbiol. 115, 1005–1024. doi:10.1111/mmi.14676
Sinha, A., Hughes, K. R., Modrzynska, K. K., Otto, T. D., Pfander, C., Dickens, N. J., et al. (2014). A cascade of DNA-Binding Proteins for Sexual Commitment and Development in Plasmodium. Nature 507, 253–257. doi:10.1038/nature12970
Smalley, M. E., Abdalla, S., and Brown, J. (1981). The Distribution of Plasmodium Falciparum in the Peripheral Blood and Bone Marrow of Gambian Children. Trans. R. Soc. Trop. Med. Hyg. 75, 103–105. doi:10.1016/0035-9203(81)90019-5
Sullivan, W. J., Naguleswaran, A., and Angel, S. O. (2006). Histones and Histone Modifications in Protozoan Parasites. Cel Microbiol 8, 1850–1861. doi:10.1111/j.1462-5822.2006.00818.x
Summers, R. L., Pasaje, C. F. A., Pisco, J. P., Striepen, J., Luth, M. R., Kumpornsin, K., et al. (2021). Chemogenomics Identifies Acetyl-Coenzyme A Synthetase as a Target for Malaria Treatment and Prevention. Cell Chem Biol 29, 191. doi:10.1016/j.chembiol.2021.07.010
Tan, Y., Xue, Y., Song, C., and Grunstein, M. (2013). Acetylated Histone H3K56 Interacts with Oct4 to Promote Mouse Embryonic Stem Cell Pluripotency. Proc. Natl. Acad. Sci. U.S.A. 110, 11493–11498. doi:10.1073/pnas.1309914110
Tang, J., Chisholm, S. A., Yeoh, L. M., Gilson, P. R., Papenfuss, A. T., Day, K. P., et al. (2020). Histone Modifications Associated with Gene Expression and Genome Accessibility Are Dynamically Enriched at Plasmodium Falciparum Regulatory Sequences. Epigenetics & Chromatin 13, 50. doi:10.1186/s13072-020-00365-5
Trager, W., and Gill, G. S. (1992). Enhanced Gametocyte Formation in Young Erythrocytes byPlasmodium falciparumIn Vitro. J. Protozool 39, 429–432. doi:10.1111/j.1550-7408.1992.tb01476.x
Treeck, M., Sanders, J. L., Elias, J. E., and Boothroyd, J. C. (2011). The Phosphoproteomes of Plasmodium Falciparum and Toxoplasma Gondii Reveal Unusual Adaptations within and beyond the Parasites' Boundaries. Cell Host & Microbe 10, 410–419. doi:10.1016/j.chom.2011.09.004
Trelle, M. B., Salcedo-Amaya, A. M., Cohen, A. M., Stunnenberg, H. G., and Jensen, O. N. (2009). Global Histone Analysis by Mass Spectrometry Reveals a High Content of Acetylated Lysine Residues in the Malaria Parasite Plasmodium Falciparum. J. Proteome Res. 8, 3439–3450. doi:10.1021/pr9000898
Ukaegbu, U. E., Kishore, S. P., Kwiatkowski, D. L., Pandarinath, C., Dahan-Pasternak, N., Dzikowski, R., et al. (2014). Recruitment of PfSET2 by RNA Polymerase II to Variant Antigen Encoding Loci Contributes to Antigenic Variation in P. Falciparum. Plos Pathog. 10, e1003854. doi:10.1371/journal.ppat.1003854
Van Biljon, R., Van Wyk, R., Painter, H. J., Orchard, L., Reader, J., Niemand, J., et al. (2019). Hierarchical Transcriptional Control Regulates Plasmodium Falciparum Sexual Differentiation. BMC Genomics 20, 920. doi:10.1186/s12864-019-6322-9
Volz, J., Carvalho, T. G., Ralph, S. A., Gilson, P., Thompson, J., Tonkin, C. J., et al. (2010). Potential Epigenetic Regulatory Proteins Localise to Distinct Nuclear Sub-compartments in Plasmodium Falciparum. Int. J. Parasitol. 40, 109–121. doi:10.1016/j.ijpara.2009.09.002
Volz, J. C., Bártfai, R., Petter, M., Langer, C., Josling, G. A., Tsuboi, T., et al. (2012). PfSET10, a Plasmodium Falciparum Methyltransferase, Maintains the Active Var Gene in a Poised State during Parasite Division. Cell Host & Microbe 11, 7–18. doi:10.1016/j.chom.2011.11.011
Von Grüning, H., Coradin, M., Mendoza, M. R., Reader, J., Sidoli, S., Garcia, B. A., et al. (2022). A Dynamic and Combinatorial Histone Code Drives Malaria Parasite Asexual and Sexual Development. Mol. Cell Proteomics, 100199. doi:10.1016/j.mcpro.2022.100199
Walzer, K. A., Kubicki, D. M., Tang, X., and Chi, J. T. (2018). Single-Cell Analysis Reveals Distinct Gene Expression and Heterogeneity in Male and Female Plasmodium Falciparum Gametocytes. mSphere 3, e00130. doi:10.1128/mSphere.00130-18
Wang, W.-F., and Zhang, Y.-L. (2020). PfSWIB, a Potential Chromatin Regulator for Var Gene Regulation and Parasite Development in Plasmodium Falciparum. Parasites Vectors 13, 48. doi:10.1186/s13071-020-3918-5
Watzlowik, M. T., Das, S., Meissner, M., and Längst, G. (2021). Peculiarities of Plasmodium Falciparum Gene Regulation and Chromatin Structure. Int. J. Mol. Sci. 22, 5168. doi:10.3390/ijms22105168
WHO (2020). World Malaria Report 2020: 20 Years of Global Progress and Challenges. Geneva: World Health Organization.
Williams, J. L. (1999). Stimulation of Plasmodium Falciparum Gametocytogenesis by Conditioned Medium from Parasite Cultures. Am. J. Trop. Med. Hyg. 60, 7–13. doi:10.4269/ajtmh.1999.60.7
Witmer, K., Fraschka, S. A., Vlachou, D., Bártfai, R., and Christophides, G. K. (2020). An Epigenetic Map of Malaria Parasite Development from Host to Vector. Sci. Rep. 10, 6354. doi:10.1038/s41598-020-63121-5
Young, J. A., Fivelman, Q. L., Blair, P. L., De La Vega, P., Le Roch, K. G., Zhou, Y., et al. (2005). The Plasmodium Falciparum Sexual Development Transcriptome: a Microarray Analysis Using Ontology-Based Pattern Identification. Mol. Biochem. Parasitol. 143, 67–79. doi:10.1016/j.molbiopara.2005.05.007
Yu, Y., Song, C., Zhang, Q., Dimaggio, P. A., Garcia, B. A., York, A., et al. (2012). Histone H3 Lysine 56 Methylation Regulates DNA Replication through its Interaction with PCNA. Mol. Cel 46, 7–17. doi:10.1016/j.molcel.2012.01.019
Zhang, M., Wang, C., Otto, T. D., Oberstaller, J., Liao, X., Adapa, S. R., et al. (2018). Uncovering the Essential Genes of the Human Malaria Parasite Plasmodium Falciparum by Saturation Mutagenesis. Science 360, eaap7847. doi:10.1126/science.aap7847
Zhao, Y., and Garcia, B. A. (2015). Comprehensive Catalog of Currently Documented Histone Modifications. Cold Spring Harb Perspect. Biol. 7, a025064. doi:10.1101/cshperspect.a025064
Keywords: histone post-translational modifications, histone combinations, epigenetic regulation, malaria, Plasmodium, gametocyte
Citation: Connacher J, von Grüning H and Birkholtz L (2022) Histone Modification Landscapes as a Roadmap for Malaria Parasite Development. Front. Cell Dev. Biol. 10:848797. doi: 10.3389/fcell.2022.848797
Received: 05 January 2022; Accepted: 04 March 2022;
Published: 01 April 2022.
Edited by:
Till Voss, Swiss Tropical and Public Health Institute (Swiss TPH), SwitzerlandReviewed by:
Richard Bartfai, Radboud University Nijmegen, NetherlandsMichaela Petter, University Hospital Erlangen, Germany
Jun Miao, University of South Florida, United States
Copyright © 2022 Connacher, von Grüning and Birkholtz. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: L. Birkholtz, bGJpcmtob2x0ekB1cC5hYy56YQ==
†These authors have contributed equally to this work
 J. Connacher†
J. Connacher† L. Birkholtz
L. Birkholtz