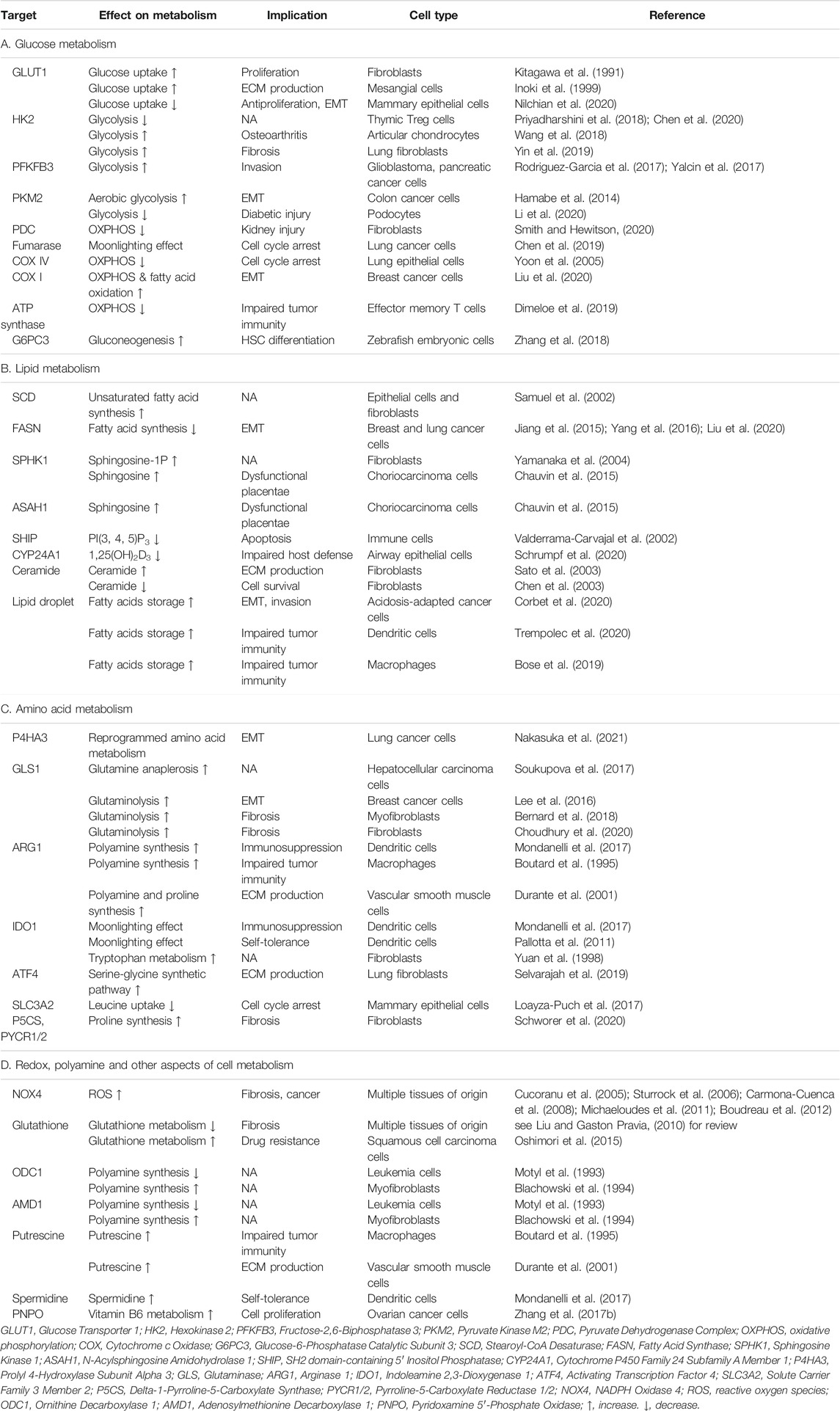
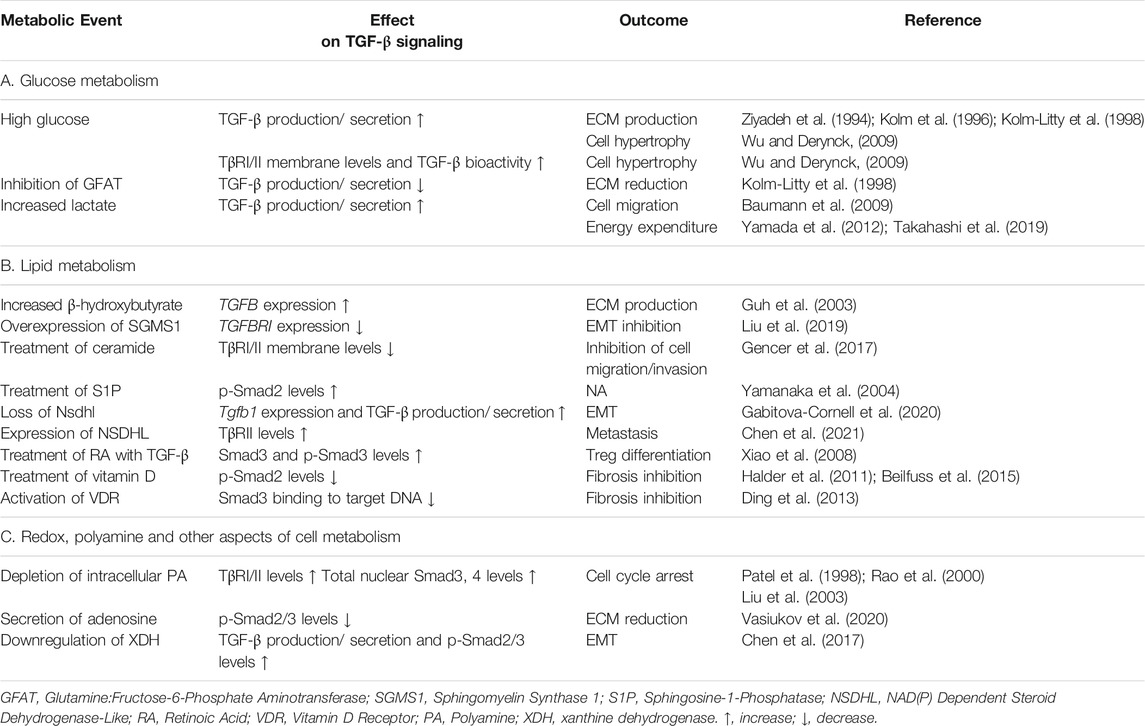
- The State Key Laboratory of Membrane Biology, Tsinghua-Peking Center for Life Sciences, School of Life Sciences, Tsinghua University, Beijing, 100084, China
The transforming growth factor-β (TGF-β) signaling plays a critical role in the development and tissue homeostasis in metazoans, and deregulation of TGF-β signaling leads to many pathological conditions. Mounting evidence suggests that TGF-β signaling can actively alter metabolism in diverse cell types. Furthermore, metabolic pathways, beyond simply regarded as biochemical reactions, are closely intertwined with signal transduction. Here, we discuss the role of TGF-β in glucose, lipid, amino acid, redox and polyamine metabolism with an emphasis on how TGF-β can act as a metabolic modulator and how metabolic changes can influence TGF-β signaling. We also describe how interplay between TGF-β signaling and cell metabolism regulates cellular homeostasis as well as the progression of multiple diseases, including cancer.
Introduction
Comprising 33 members in mammalian cells, the transforming growth factor-β (TGF-β) superfamily is distinct from other cytokines owning to its more widespread and pleiotropic effects (Morikawa et al., 2016). The TGF-β signaling pathway contributes to a broad range of physiological and pathological processes, and its key roles in development, immunity, wound healing, cancer, fibrosis, skeletal and cardiac diseases have been extensively studied (Massague, 2008; Wu and Hill, 2009; Dobaczewski et al., 2011; Travis and Sheppard, 2014; Meng et al., 2016; Morikawa et al., 2016; Salazar et al., 2016; Kiritsi and Nystrom, 2018; Derynck et al., 2020). A plethora of cellular activities, including cell proliferation, differentiation, apoptosis, adhesion and migration, are controlled by TGF-β superfamily members in a context-dependent manner (Feng and Derynck, 2005; Moustakas and Heldin, 2009; Massague, 2012; David and Massague, 2018). Although cellular responses to TGF-β signaling are mainly induced via its transcriptional regulation of genes (Massague and Chen, 2000; Massague and Wotton, 2000), other means have been recognized for TGF-β signaling to shape cell behavior, such as epigenetic modification, mRNA splicing and miRNA expression (Derynck and Budi, 2019). In addition, accumulating evidence indicates that TGF-β signal can also remodel cell metabolism.
As a network of chemical reactions essential for sustaining life, metabolism has long been centered in energy provision, building of blocks for biomacromolecules and elimination of compounds that are otherwise toxic to the organism. Studies in the past decades, especially with the aid of metabolomics, have further unraveled the profound interactions between metabolism and the regulation of protein activity and genes expression (Rinschen et al., 2019). Metabolic substrates, beyond serving as “ingredients” or biomarkers, are able to modify the chromatin structure and regulate gene expression (Li et al., 2018). On the other hand, metabolic enzymes, in response to signaling cues, can fulfill many moonlighting functions other than catalyzing (Xu et al., 2021). Therefore, these non-metabolic roles of metabolites and metabolic enzymes have been shown to play a critical role in signal transduction.
In this review, we discuss the current knowledge of how TGF-β signaling functions by altering various facets of cell metabolism and how metabolic changes can result in modulation of TGF-β signaling, thereby affecting an array of cellular processes. Such interplay between TGF-β signaling and cell metabolism is thought to be instrumental in maintaining homeostasis, and its aberration contributes to disease development. Due to the large number of TGF-β superfamily members, the scope of this review is restricted to the TGF-β ligands (TGF-β1, 2 and 3), which have been most extensively studied.
Basics of the TGF-β Signaling Pathway
Based on the similarities in protein sequence and structure, the mammalian TGF-β members, with a few exceptions, can be classified into three major groups: the TGF-β family, the inhibin/activin family and the BMP (bone morphogenic protein)/GDF (growth and differentiation factor) family (Morikawa et al., 2016). The TGF-β family consists of TGF-β1, 2 and 3 that have largely redundant functions. Each isoform contains nine highly conserved cysteine residues, mediating the formation of inter- or intramolecular disulfide bonds that interlock two TGF-β polypeptides as a dimer (Hinck et al., 2016). The dimeric TGF-β ligand associates with the pro-region-derived latency-associated peptide (LAP) and a latent TGF-β binding protein (LTBP) and forms a large latent complex (LLC), which is trapped in the extracellular matrix (ECM) (Robertson and Rifkin, 2016). Activation of TGF-β ligands is mediated by different proteins in various tissues, serving as a way to ensure the precision of signal presentation (Rifkin, 2005).
Once activated, the dimeric TGF-β initiates signaling by promoting the assembly of two type I (TβRI) and two type II (TβRI) transmembrane receptors (Hata and Chen, 2016) (Figure 1). In the absence of TGF-β ligands, both TβRI and TβRII exist as monomers (Zhang et al., 2009; Zhang et al., 2010), although early studies reported that they exist as homodimers (Chen and Derynck, 1994; Henis et al., 1994; Gilboa et al., 1998), most likely due to their overexpression. Both of TβRI and TβRII possess Ser/Thr kinase activity in the cytoplasmic domain. Ligand binding results in the tetramer receptor complex formation with two TβRI and two TβRII, in which TβRI is activated via phosphorylation of Thr and Ser residues in its GS domain (TTSGSGSG) by the constitutively active TβRII (Wrana et al., 1994). The phosphorylation-induced conformational change activates the TβRI kinase that relays the signal to the effector Smad proteins (Huse et al., 1999; Huse et al., 2001; Chaikuad and Bullock, 2016; Hata and Chen, 2016) (Figure 1A).
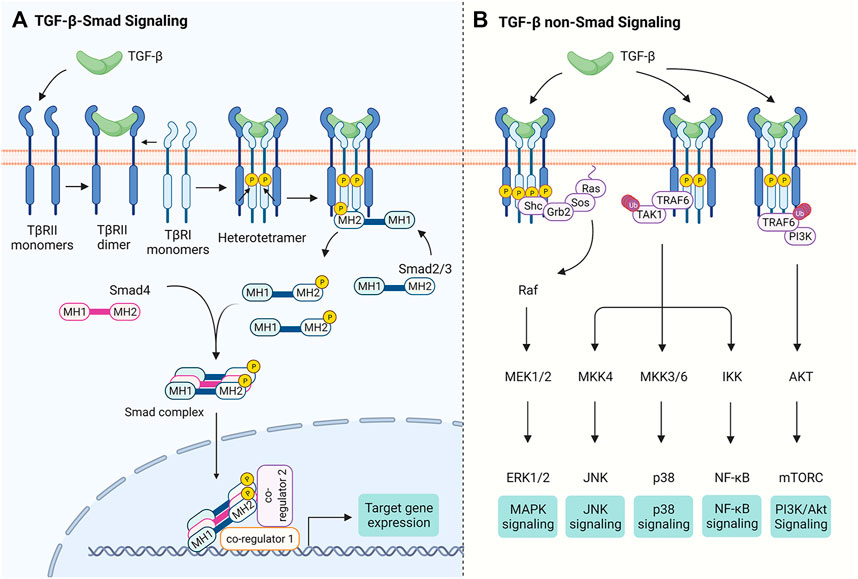
FIGURE 1. The TGF-β Signaling Pathways. Binding of TGF-β to TβRII leads to the tetramer assembly of monomeric TβRII and TβRI receptors. (A) In Smad-dependent TGF-β signal transduction, TβRII trans-phosphorylates TβRI and activates its kinase activity, which in turn phosphorylates Smad2/3 at the C-terminal tail. Phosphorylated Smad2/3 form a trimeric complex with Smad4 and is translocated into the nucleus. The Smad complex binds DNA via their MH1 domains and controls gene expression through interacting with other transcription co-regulators. (B) TGF-β receptors trigger non-Smad signaling pathways. For instance, TGF-β receptors have been reported to recruit Shc, Grb2 and Sos to activate Ras, thereby initiating MAPK signaling. TGF-β receptors also activate TAK1 through TRAF6, which is required for TGF-β-induced JNK, p38 and NF-κB activation. It has been proposed that interaction of TRAF6 with TβRI also leads to PI3K/Akt activation. Figure is created with Biorender.com.
There are three subgroups of Smad proteins: the receptor-activated Smads (R-Smads, Smad2/3 for TGF-β/activin/inhibin receptors and Smad1/5/8 for BMP/GDF receptors), the common-mediator Smad (Co-Smad, i.e., Smad4) that interacts with R-Smads, and the inhibitory Smads (I-Smads, Smad6 and Smad7). Both R- and Co-Smads propagate signals, while I-Smads dampen the signal transduction (Hata and Chen, 2016). All R-Smad proteins contain a highly conserved C-terminal MH2 domain that, via an inner L3 loop, engages in Smad-receptor and Smad-Smad interactions (Lo et al., 1998). The conserved N-terminal MH1 domain in R-Smads and Co-Smad has a nuclear localization signal and a DNA-binding β-hairpin (Hata and Chen, 2016; Chaikuad and Bullock, 2016; Shi et al., 1998; Hill, 2016). Upon activation of TβRI kinase activity, Smad2/3 is phosphorylated at two serine residues in the SSXS motif and subsequently is dissociated from the TβRI kinase domain, forming a trimeric Smad complex composed of two Smad2/3 and one Smad4 (Hata and Chen, 2016; Chaikuad and Bullock, 2016; Kawabata et al., 1998; Chacko et al., 2004; Xu et al., 2016). This Smad complex is then accumulated in the nucleus and acts as a transcription factor to regulate contextual expression of target genes through collaboration with diverse co-factors (Moustakas and Heldin, 2009; Massague et al., 2005) (Figure 1A). TGF-β ligands can also signal independently of Smad proteins through crosstalk with other signaling pathways (see Zhang, 2017; Derynck and Budi, 2019) (Figure 1B).
While it is clear that TGF-β signaling targets genes related to cell cycle progression, ECM production and epithelial-mesenchymal transition (EMT), a panoramic view of metabolic genes whose transcription directly controlled by TGF-β signaling are not attained. It remains even more obscure precisely how metabolic changes regulate the TGF-β signal transduction. In the following sections, we will illustrate the interplay between TGF-β signaling and multiple aspects of cell metabolism with a discussion on their important physiological or pathological roles in mammalian cells.
TGF-β Signaling and Glucose Metabolism
The first evidence that TGF-β regulates glucose metabolism perhaps comes from work on Swiss mouse 3T3 cells demonstrating TGF-β treatment upregulates Glut1 (glucose transporter type 1) mRNA level and increases glucose uptake (Kitagawa et al., 1991). This observation is later reproduced in rat glomerular mesangial cells and is associated with excessive glucose uptake-induced overproduction of ECM proteins (Inoki et al., 1999), which is a hallmark of diabetic nephropathy. In a different model using mouse normal mammary gland (NMuMG) cells to study TGF-β-induced EMT, however, Glut1 expression is reduced at both the protein and mRNA levels during short-term TGF-β exposure but is later restored, which may be explained by differential effects of TGF-β on proliferation of epithelial and mesenchymal cells through regulation of glucose uptake (Nilchian et al., 2020). In mesangial cells, high glucose can potently increase autocrine secretion of TGF-β (Ziyadeh et al., 1994; Kolm et al., 1996; Kolm-Litty et al., 1998) (Figure 2). It seems that a positive feedback loop, in which elevated glucose levels stimulate TGF-β production and TGF-β, in turn, enhances glucose uptake, may pathologically contribute to the progression of diabetic nephropathy. Interestingly, production of TGF-β induced by high glucose is impaired by inhibition of Gfat (Glutamine:fructose-6-phosphate aminotransferase, the rate-limiting enzyme that converts fructose-6-phosphate into glucosamine-6-phosphate) (Kolm-Litty et al., 1998), suggesting a potential role of glucosamine-6-phosphotse in regulating TGF-β expression (Figure 2). In addition to promote TGF-β ligand production, high glucose is shown to increase cell membrane levels of both TβRI and TβRII and to induce latent-TGF-β activation by matrix metalloproteinases, leading to activation of the Akt-mTOR pathway and consequently causing cell hypertrophy in fibroblasts and epithelial cells (Wu and Derynck, 2009).
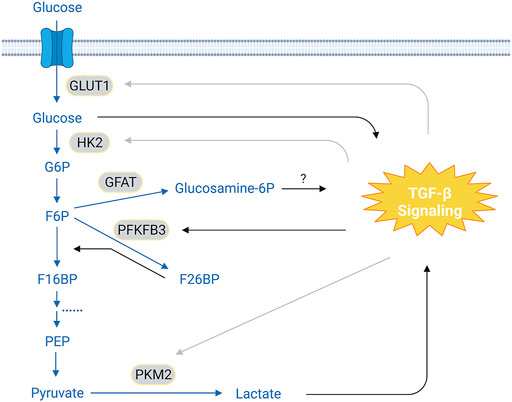
FIGURE 2. Crosstalk Between TGF-β Signaling and Glycolysis. In the glycolytic pathway, glucose is converted to pyruvates via a cascade of enzymatic reactions. It has been reported that TGF-β signaling can either increase or decrease the expression of GLUT1 and HK2, depending on the cell types. This cell type-context effect is also seen on PKM2, an enzyme that catalyzes pyruvate into lactate. TGF-β signaling upregulates PFKFB3, leading to increased F26BP levels, which, in turn, accelerate conversion of F6P to F16BP. It has been suggested that glucose and lactate can promote TGF-β signaling; and inhibition of GFAT prevents glucose-induced expression of TGF-β ligands, implying a potential role of glucosamine-6-phosphotase in mediating this process. The conversion of F16BP to pyruvate has been omitted for clarity. Blue texts and arrows, glycolysis and its branches; gray arrows, cell-type dependent effect. Abbreviations: G6P, glucose-6-phosphate; F6P, fructose-6-phosphate; F16BP, fructose-1,6-biphosphate; F26BP, fructose-2,6-biphosphate; PEP, phosphoenolpyruvate; GLUT1, glucose transport 1; HK2, hexokinase 2; GFAT, glutamine:fructose-6-phosphate aminotransferase; PFKFB3, 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3; PKM2, pyruvate kinase M2. Figure is created with BioRender.com.
TGF-β signaling also regulates other components in the glycolytic pathway (Figure 2; Table 1A). For example, TGF-β treatment significantly decreases hexokinase 2 (HK2) expression in murine thymic-derived Tregs (Priyadharshini et al., 2018; Chen et al., 2020). However, HK2 levels are slightly increased in TGF-β-treated articular chondrocytes from patients with osteoarthritis (Wang et al., 2018). TGF-β stimulation also specifically increases HK2 abundance in murine and human lung fibroblasts, which is required for profibrotic actions of TGF-β possibly through upregulating YAP/TAZ protein levels by an unknown mechanism (Yin et al., 2019). These results together suggest a cell-type dependent effect of TGF-β signaling on HK2 regulation. Phosphofructokinase 2 (PFK2), an enzyme that generates fructose-2,6-biphosphate that allosterically activates phosphofructokinases, is overexpressed in many cancer cells. TGF-β induces PFK2 expression in glioblastoma and pancreatic cancer cells (Rodriguez-Garcia et al., 2017; Yalcin et al., 2017), which is required for activation of SNAI1 transcription and promotes cell invasion (Yalcin et al., 2017). In SW480 colon cancer cells, increased pyruvate kinase M2 (PKM2) expression by TGF-β and EGF has been reported to be indispensable for EMT (Hamabe et al., 2014). In podocytes, the interaction of Smad4 with PKM2 interrupts the active PKM2 tetramer and reduces glycolysis activity (Li et al., 2020).
Lactate, the product of anaerobic glycolysis generated from pyruvate, appears to positively modulate TGF-β signaling (Figure 2; Table 2A). For instance, lactate induces TGF-β2 expression in glioma cells and knockdown of lactate dehydrogenase A (LDHA), an enzyme that catalyzes lactate production, downregulates TGF-β2 levels (Baumann et al., 2009). Lactate generated during exercising is associated with increased bioactive TGF-β concentration in rat cerebrospinal fluid (Yamada et al., 2012). Consistently, injection of lactate into mice results in elevated serum TGF-β2 levels, and incubation of adipocytes with lactate causes increased TGF-β2 concentrations in the media (Takahashi et al., 2019), though the underlying mechanism remains to be determined.
When oxygen is plentiful, pyruvate generally enters the TCA cycle, and most ATP is produced via oxidative phosphorylation (OXPHOS). TGF-β signaling has been shown to attenuate pyruvate dehydrogenase complex (PDC) activity in fibroblasts from injured kidneys and reduces free acetyl-CoA levels (Smith and Hewitson, 2020). TGF-β also causes phosphorylation of fumarase at T90 via the p38 pathway (Chen et al., 2019). Although the phosphorylated fumarase seems to retain normal catalytic activity, it gains non-metabolic functions and can shuttle into the nucleus to activate p21 expression through interaction with the CSL/RBPJ-p53 complex, thereby facilitating cell cycle arrest (Chen et al., 2019). TGF-β signaling also targets OXPHOS (Table 1A). In murine and human natural killer cells, TGF-β signaling dampens cell metabolism and represses OXPHOS (Viel et al., 2016; Zaiatz-Bittencourt et al., 2018; Slattery et al., 2021), in a mTOR signaling-dependent (Viel et al., 2016) or -independent manner (Zaiatz-Bittencourt et al., 2018). In addition, TGF-β suppresses the activity of ATP synthase in effector memory CD4+ T cells and therefore reduces mitochondria respiratory capacity (Dimeloe et al., 2019). Since mitochondria are critical to many key immune functions (Mills et al., 2017), these inhibitory effects on OXPHOS in immune cells may underlie some negative effects of TGF-β in immunity. In mink lung epithelial Mv1Lu cells, TGF-β inhibits mitochondria complex IV activity and increases intracellular ROS accumulation, leading to senescence (Yoon et al., 2005). However, TGF-β has also been reported to enhance OXPHOS. For instance, in MCF-7 breast cancer cells, TGF-β increases the expression of OXPHOS-associated proteins, including NADH:ubiquinone oxidoreductase subunit B8 (NDUFB8), cytochrome c oxidase subunit I (COX I) and mitochondrial transcription factor A (TFAM) during EMT, a cellular process that is thought to promote metastasis (Liu et al., 2020). In addition, TGF-β signaling in precursors of exhausted effector T cells promotes OXPHOS by repressing mTOR, enabling the preservation of mitochondrial metabolism that supports long-term T cell responses during chronic infection (Gabriel et al., 2021).
Aerobic glycolysis, or the Warburg effect, is widely adopted in many cancer cells (Hanahan and Weinberg, 2011), which is characterized by the preference of glycolysis over oxidative phosphorylation as a major source of energy production even when oxygen is abundant. Aerobic glycolysis can be induced in normal mammary fibroblasts by overexpression of constitutively active TβRI, powering the metabolically reprogrammed fibroblasts to fuel growth of cancer cells via energy transfer (Guido et al., 2012; Martinez-Outschoorn et al., 2012). In prostate cancer cells, overexpression of Smad2/3 enhances aerobic glycolysis independently of TGF-β stimulation but requires PKCε-mediated phosphorylation of the Smad3 linker region, which assists binding of Smad3 to the promoter of glycolytic genes (Xu et al., 2018). However, most of the studies were carried out in cell lines, and whether endogenous activation of TGF-β signaling promotes aerobic glycolysis in tumor cells awaits further investigation.
Compared to glucose catabolism, TGF-β1 has been documented to increase gluconeogenesis via the c-Jun/G6PC3 (glucose-6-phosphatase catalytic subunit 3) axis in zebrafish embryos, which fosters the nascent hematopoietic stem cells (Zhang et al., 2018). It would be worth exploring whether this mechanism can be applied to mammals or humans. Furthermore, there are many other metabolic pathways other than glycolysis that require glucose, including the pentose phosphate pathway, the hexosamine pathway, glycogenesis, the serine biosynthesis pathway and its many branches (Hay, 2016). Whether TGF-β signaling interacts with these pathways is unclear.
TGF-β Signaling and Lipid Metabolism
Lipids are a large group of water-insoluble molecules that, according to their diverse cellular functions, can be roughly divided into three categories represented by triglycerides that store energy; phosphoglycerides, sphingolipids and sterols that build the main structure of biological membrane; and many derivatives that actively engage in signal transduction and enzymatic reaction (Ridgway and McLeod, 2008).
Fatty acids can be released from triglycerides and provide the energetic needs through fatty acid oxidation (β-oxidation) in mitochondria. Blocking TGF-β signaling in mice via Smad3 ablation promotes brown adipogenesis within white adipose tissue and boosts mitochondria biogenesis in adipocytes, causing a significant elevation in fatty acid oxidation (Yadav et al., 2011). Conditional knockout of Tgfbr2 in hepatocytes ameliorates CDAA (choline-deficient l-amino acid-defined) diet-induced steatohepatitis in mice, prevents CDAA-induced expression of genes related to lipogenesis, and enhances gene expression involved in β-oxidation (Yang et al., 2014). As inhibition of TGF-β signaling promotes fatty acid oxidation, it is plausible to postulate that TGF-β signaling activates the synthesis of fatty acids. Indeed, all three types of the TGF-β ligands, but not other members of the TGF-β superfamily, are shown to increase stearoyl-CoA desaturase expression in a Smad-dependent way in many human cell lines (Samuel et al., 2002). However, other studies demonstrate that the effect of TGF-β on fatty acid oxidation or synthesis is context-dependent (see Table 1B for details). Many studies report that TGF-β suppresses the expression of fatty acid synthase during the induction of EMT in cancer cells (Jiang et al., 2015; Yang et al., 2016; Liu et al., 2020). In Hep3B cells, TGF-β causes a significant reduction in carnitine-conjugated fatty acids, which coincides with upregulation of fatty acid transporter genes, implying increased carnitine-mediated entry of fatty acids into mitochondria that are destined for β-oxidation (Soukupova et al., 2021). In addition, TGF-β2 or TGF-β3, but not TGF-β1, is shown to reinforce fatty acid oxidation in myotubules and adipocytes (Takahashi et al., 2019). Ketone bodies are formed in the liver from acetyl-CoA produced by oxidation of fatty acids. As a major form of ketone body, β-hydroxybutyrate has been reported to increase TGF-β expression in HK-2 renal cells (Guh et al., 2003). However, the effect of TGF-β on the ketone bodies remains unknown.
TGF-β signaling also regulates the metabolism of some structural lipids that define the membrane architecture (Table 1B). Sphingolipids are a large class of membrane lipids, among which ceramide is the only one that can be de novo synthesized and serves as the structural precursor of higher sphingolipid members (Figure 3). Ceramide can be hydrolyzed by N-acylsphingosine amidohydrolase 1 (ASAH1) into sphingosine, which can be phosphorylated by sphingosine kinase 1/2 (SPHK1/2) into sphingosine-1-phosphate (S1P) to regulate a variety of physiological and pathological processes (Maceyka et al., 2012). In NMuMG cells and human normal bladder HCV29 cells, TGF-β can rewire glycosphingolipid composition to promote EMT by reducing intracellular levels of gangliotetraosylceramide or GM2 (Guan et al., 2009). TGF-β enhances the activity and expression of SPHK1 in human fibroblasts that are important for the expression of TIMP-1 (Yamanaka et al., 2004). In contrast, TGF-β1 and TGF-β3 downregulates SPHK1 expression but upregulates ASAH1 expression in the human choriocarcinoma JEG-3 cell line, leading to aberrant sphingosine accumulation characteristic of dysfunctional placentae in intrauterine growth restriction (Chauvin et al., 2015). TGF-β can also diminish ceramide production to inhibit apoptosis in NIH3T3 cells during serum starvation (Chen et al., 2003), while increasing ceramide levels in human dermal fibroblasts and Mv1Lu cells (Sato et al., 2003). The increased ceramide is shown to act as a positive regulator of TGF-β signaling by facilitating TGF-β-induced COL1A2 expression in foreskin fibroblasts (Sato et al., 2003). Ceramide has also been reported to inhibit TβRI/II trafficking to primary cilia by stabilizing the TβRI-Smad7 interaction, thereby attenuating cell migration and metastasis (Gencer et al., 2017). Consistent with the observation that TGF-β induces SPHK1 expression, exogenous sphingosine 1-phosphate can elevate phosphorylated Smad2 levels and increase TIMP-1 expression in rat renal mesangial cells (Yamanaka et al., 2004). Moreover, overexpression of sphingomyelin synthase 1, a key enzyme that converts ceramides into sphingomyelins, downregulates TβRI expression and thus impairs TGF-β-induced EMT in breast cancer cell lines (Liu et al., 2019). Aside from sphingolipid metabolism, the metabolic pathway of cholesterol has been shown to regulate TGF-β signaling (Table 2B). Cholesterol is enriched in lipid rafts, a membrane microdomain which modulates TGF-β signaling. TGF-β receptors can be internalized via lipid raft-dependent endocytosis and transported to lysosome for degradation (Chen, 2009), while the location at lipid rafts of TGF-β receptors is required for TGF-β activation of MAP kinases (Zuo and Chen, 2009). Cholesterol depletion specifically inhibits TGF-β-induced activation of extracellular signal-regulated kinase (ERK) and p38 and therefore impairs EMT and cell migration (Zuo and Chen, 2009). In addition, loss of the rate limiting enzyme Nsdhl (NAD(P)-dependent steroid dehydrogenase-like) involved in cholesterol synthesis in mouse pancreatic ductal adenocarcinoma cells activates Srebp1 (sterol regulatory element-binding protein 1), which enhances TGF-β1 expression and secretion and consequently facilitates EMT (Gabitova-Cornell et al., 2020). However, another study reported an opposite observation in human breast cancer cells: NSDHL expression augments TGF-β signaling by inhibiting TβRII degradation and therefore promotes cell migration (Chen et al., 2021). Hence, like in many other cases, this regulation is cell-specific.
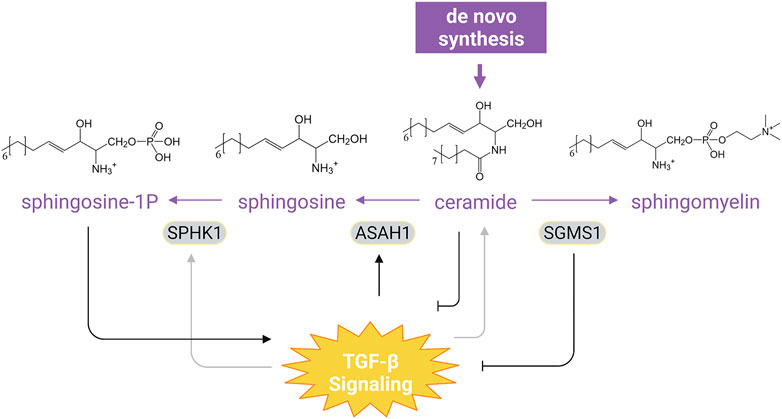
FIGURE 3. Interaction of TGF-β Signaling with Sphingolipid Metabolism. TGF-β differentially regulates SPHK1 expression in a context-dependent manner and can upregulate ASAH1 to promote aberrant accumulation of sphingosine. Ceramide, the only sphingolipid that can be de novo synthesized, has been shown to constrain TGF-β signaling. Overexpression of SGMS1, which catalyzes synthesis of sphingomyelin from ceramide, also inhibits TGF-β signal transduction. Moreover, sphingosine-1P can evoke TGF-β-like responses in cells (see text). For simplicity, synthesis of other sphingolipid and the downstream catabolism of sphingosine-1P have been omitted. Purple texts and arrows, the sphingolipid metabolic pathway; gray arrows, cell-type dependent effect. Abbreviations: SPHK1, sphingosine kinase 1; ASAH1, N-acylsphingosine amidohydrolase 1; SGMS1, sphingomyelin synthase 1. Figure is created with BioRender.com.
The phosphorylated derivatives of phosphatidylinositol (PI) play a critical role in intracellular signal transduction. Phosphorylation of PI(4, 5)P2 to PI(3, 4, 5)P3 by PI-3-kinase triggers activation of Akt, inhibiting apoptosis and promoting cell survival (Manning and Toker, 2017). In hematopoietic cells, TGF-β signals are shown to counteract Akt signaling and promote apoptosis by upregulating the expression of the SH2-containing inositol phosphatase SHIP (Valderrama-Carvajal et al., 2002), which breaks down PI(3, 4, 5)P3 to PI(3, 4)P2. The interplay between TGF-β signaling and lipophilic hormones such as retinoic acid and vitamin D is well studied (Tables 1B, 2B). Retinoic acid has been shown to synergistically increase the expression and phosphorylation of Smad3 in the presence of TGF-β during differentiation of CD4+ T cells toward Treg (Xiao et al., 2008). The biologically active form of vitamin D, 1α,25-dihydroxyvitamin D3 (1,25(OH)2D3), has been reported to revert TGF-β-increased OXPHOS and reactive oxygen species (ROS) in human bronchial epithelial cells (Ricca et al., 2019). 1,25(OH)2D3 has also been shown to antagonize TGF-β-mediated fibrogenesis. In the presence of the ligands, the vitamin D receptor (VDR) occupies Smad3-binding sites at profibrotic genes and reduces TGF-β-mediated hepatic fibrosis (Ding et al., 2013). Similarly, VDR ablation abolishes the antagonistic effect of 1,25(OH)2D3 on TGF-β-promoted hepatic fibrosis (Beilfuss et al., 2015). In human leiomyoma cells, 1,25(OH)2D3 can reduce Smad2 expression or activation by TGF-β and thus expression of profibrotic genes (Halder et al., 2011). In hepatic stellate cells, vitamin D supplementation also showed similar effects (Beilfuss et al., 2015). Reversely, TGF-β can cause vitamin D catabolism through upregulation of the vitamin D-24A-hydroxylase CYP24A1, resulting in undermined host defense in airway epithelium (Schrumpf et al., 2020). Interestingly, Smad3 can form a complex with VDR in a ligand-dependent manner and enhances its transactivation activity (Yanagisawa et al., 1999).
Lipid droplets (LD) are a type of organelle instrumental in lipid and energy homeostasis and also involved in diverse cellular activities other than lipid metabolism (Olzmann and Carvalho, 2019; Walther and Farese, 2012). TGF-β has been demonstrated to induce its formation in many cell types (Table 1B). TGF-β2 induces fatty acids storage and LD formation in acidosis-adapted cancer cells, which meets cellular energetic needs for EMT and cell invasion (Corbet et al., 2020). It also increases LD content in dendritic cells under acidic circumstances (Trempolec et al., 2020). In addition, treatment of murine macrophages with TGF-β causes LD accumulation, accompanied by a shift of macrophages from M1 phenotype to the pathological M2 phenotype (Bose et al., 2019). However, the mechanisms underlying TGF-β-induced LD formation are currently unclear.
TGF-β Signaling and Amino Acid Metabolism
It has come to appreciate that amino acids, besides their fundamental role as substrates for protein synthesis, also perform multifarious cellular functions including energy homeostasis, cell growth and immune response (Wu, 2009). Taking advantage of metabolomics, Nakasuka and others have nicely demonstrated that TGF-β can change intracellular amino acid levels in non-small cell lung cancer cells (Nakasuka et al., 2021). Depletion of a particular amino acid (e.g., Phe, Thr, Leu, Ile, or Tyr), whose intracellular concentrations are significantly decreased by TGF-β, in culture media, induces EMT-like elongated morphology. They further showed that TGF-β induces the expression of prolyl 4-hydroxylase subunit alpha 3, an enzyme catalyzing proline to 4-hydroxylproline, whose knockdown abrogates TGF-β-induced amino acid changes and EMT (Nakasuka et al., 2021). It would be intriguing to know how altered expression of one gene involved in proline metabolism can cause global changes of amino acid levels.
In addition to its comprehensive effects on amino acid metabolism, TGF-β signaling also specifically mediates certain amino acid metabolic pathways (Table 1C). For instance, TGF-β modulates glutamine metabolism, which takes a key part in tumor development (Zhang et al., 2017a). In hepatocellular carcinoma cells, TGF-β augments glutamine metabolism by inducing the expression of glutamine transporter and glutaminase 1 (GLS1) and reduces oxidative metabolism, concomitant with enhanced EMT and cell migration (Soukupova et al., 2017). Interestingly, the way TGF-β induces GLS1 expression seems to cell type-specific. In MCF-7 cells, TGF-β-induced GLS1 expression is mediated by the transcription factor Dlx-2, leading to enhanced glutamine metabolism that contributes to EMT (Lee et al., 2016). In myofibroblasts, however, TGF-β upregulates GLS1 expression via Smad3 and p38 and promotes myofibroblast differentiation (Bernard et al., 2018). Furthermore, TGF-β elevates GLS1 levels in AKR-2B mouse fibroblasts by repressing the transcription factor SIRT7 and FOXO4, and the process requires Smad2/3 as well as mTOR (Choudhury et al., 2020).
Tryptophan metabolism, especially in immune cells, exemplifies another aspect of amino acid metabolism modulated by TGF-β signaling (Table 1C). At the core of tryptophan metabolism lies the kynurenine pathway, in which kynurenine is generated from tryptophan, serving as the common precursor for the synthesis of various downstream metabolites including NAD+ (Kolodziej et al., 2011). Two serial enzymatic reactions convert tryptophan to kynurenine, and the first and rate-limiting step is catalyzed by three different enzymes: IDO-1 (indoleamine 2,3-dioxygenase-1), IDO-2 (indoleamine 2,3-dioxygenase-2) or TDO (tryptophan 2,3-dioxygenase). TGF-β prominently abolishes IFN-γ-induced IDO expression in human fibroblasts (Yuan et al., 1998). In contrast, IDO expression is upregulated by TGF-β in dendritic cells, which relies on the TGF-β-induced expression of arginase 1 and increased abundance of spermidine (Mondanelli et al., 2017). Importantly, TGF-β also confers IDO immunoregulatory function independently of its metabolic activity. By inducing phosphorylation of IDO at the putative immunoreceptor tyrosine-based inhibitory motifs (ITIMs) via the kinase Fyn, TGF-β promotes the complex formation of IDO with two tyrosine phosphatase SHP-1 and SHP-2, thereby activating a circuit of downstream signaling events required to maintain self-tolerance (Pallotta et al., 2011).
TGF-β also regulates other amino acid metabolic pathways (Table 1C). In human lung fibroblasts, TGF-β activates expression of ATF4, a master transcription factor of amino acid metabolism (Ameri and Harris, 2008; Kilberg et al., 2009), and leads to upregulation of PHGDH, PSAT1, PSPH and SHMT2, which are key players involved in glycine-serine synthesis (Nigdelioglu et al., 2016; Selvarajah et al., 2019). TGF-β inhibits leucine transporter SLC3A2 expression and therefore impairs leucine uptake, contributing to TGF-β-induced cell cycle arrest of mammary epithelial cells (Loayza-Puch et al., 2017). In NIH3T3 fibroblasts, the TGF-β/Smad signaling stimulates proline synthesis from glutamate by elevating protein levels of pyrroline-5-carboxylate synthase and pyrroline-5-carboxylate reductase 1/2 in the synthetic pathway to buffer mitochondrial redox stress (Schworer et al., 2020).
TGF-β Signaling and Other Aspects of Cell Metabolism
Homeostasis of redox metabolism is crucial to an extensive range of cellular and physiological conditions. The redox imbalance, often arises from aberrant accumulation of ROS and is marked by oxidative stress, can promote progression of multiple diseases (Sies et al., 2017). The crosstalk between redox metabolism and TGF-β signaling during cancer and fibrosis is comprehensively reviewed elsewhere (Richter et al., 2015; Ramundo et al., 2021). Noteworthy, the enzyme NADPH oxidase 4 (NOX4) appears to play a main role in mediating TGF-β-induced ROS generation under many circumstances (Cucoranu et al., 2005; Sturrock et al., 2006; Carmona-Cuenca et al., 2008; Michaeloudes et al., 2011; Boudreau et al., 2012). TGF-β can also inhibit the key antioxidant systems by downregulating glutathione (GSH) metabolism (Liu and Gaston Pravia, 2010). However, increased GSH metabolism and alleviated ROS levels are also observed in TGF-β-mediated drug resistance of squamous cell carcinoma cells (Oshimori et al., 2015).
The polyamine metabolic pathway attracts great interests in the past decade due to their roles in cell biology beyond early described importance for cell proliferation (Miller-Fleming et al., 2015). Depending on the cell types, TGF-β signaling can differentially regulate the activity of the two rate-limiting enzymes ornithine decarboxylase (ODC1) and adenosylmethionine decarboxylase 1 (AMD1) in polyamine synthesis (Figure 4; Table 1D). TGF-β suppresses the enzymatic activity of ODC1 and AMD1 in leukemic cells (Motyl et al., 1993), while stimulating their activities in myofibroblasts (Blachowski et al., 1994). TGF-β can also indirectly regulates the polyamine synthesis in immune cells and vascular smooth muscle cells. By inducing the expression of arginase 1, an enzyme that converts arginine to ornithine that serves as the common precursor for polyamine synthesis, TGF-β is able to increase the arginine-dependent production of specific polyamines (Mondanelli et al., 2017; Boutard et al., 1995; Durante et al., 2001). However, a recent study reported that the TGF-β blunt the increased influx of arginine to putrescine and spermidine during polarization of CD4+ naïve T cells (Puleston et al., 2021). Polyamine metabolism can also modulate the TGF-β signal transduction (Table 2C). In mouse intestinal epithelial cells, depletion of intracellular polyamines by an ODC1 inhibitor DFMO leads to increased expression of TGF-β, TβRI, Smad3 and Smad4 as well as nuclear accumulation of these Smads, sensitizing cells to TGF-β-induced cytostasis (Patel et al., 1998; Rao et al., 2000; Liu et al., 2003).
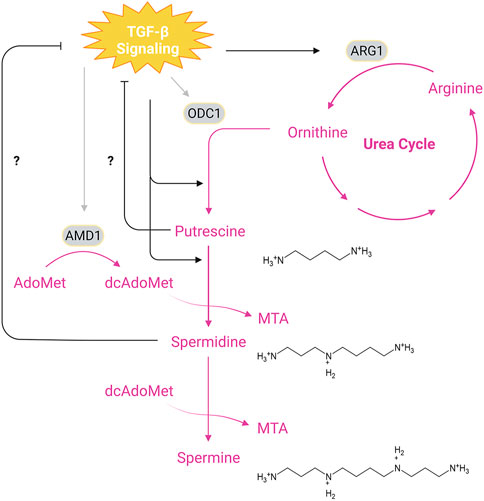
FIGURE 4. Interplay Between TGF-β Signaling and Polyamine Synthesis. The main biosynthetic pathway of polyamines begins with ODC1-catalyzed formation of putrescine from ornithine, a product of the urea cycle, which is generated from arginine through hydrolysis by ARG1. Synthesis of spermidine and spermine requires transfer of aminopropyl groups from dcAdoMet, a decarboxylated product of AdoMet (S-adenosylmethionine) catalyzed by AMD1. Putrescine and spermidine may inhibit TGF-β signaling since depletion of cellular putrescine and spermidine has been shown to enhance TβRI/II levels and Smad nuclear translocation, while TGF-β signaling promotes synthesis of putrescine and spermidine through upregulation of ARG1. In addition, TGF-β signals can stimulate or dampen the activity of ODC1 and AMD1 depending on the cell types. Pink texts and arrows, the urea cycle and polyamine synthesis. Abbreviations: MTA, methylthioadenosine; ARG1, arginase 1; ODC1, ornithine decarboxylase 1; AMD1, adenosylmethionine decarboxylase 1. Figure is created with BioRender.com.
In addition, TGF-β is able to upregulate the expression of PNPO (pyridoxamine 5′-phosphate oxidase), which encodes the rate-limiting enzyme in vitamin B6 metabolism, to produce active forms of vitamin B6 that may promote ovarian cancer progression (Zhang et al., 2017b). Adenosine secreted from myeloid cells is shown to modulate TGF-β signaling in proximal fibroblasts by reducing phosphorylation of Smad2/3 and to affect ECM deposition and therefore influence the tumor microenvironment of mammary carcinoma (Vasiukov et al., 2020). Furthermore, downregulation of the purine catabolism enzyme xanthine dehydrogenase increases TGF-β2/3 and phosphorylated Smad2/3 levels and contributes to EMT and cell migration in hepatocellular carcinoma cell lines (Chen et al., 2017). How these metabolic alterations convey their regulatory instructions to TGF-β signaling awaits further investigation.
Concluding Remarks
As summarized above, TGF-β signaling can exert its cellular and physiological effects through reprograming of cell metabolism. It controls the activity of many metabolic pathways as wells as the production of functional metabolites by regulating the expression of key metabolic proteins or enzymatic activities (Motyl et al., 1993; Blachowski et al., 1994; Yoon et al., 2005; Dimeloe et al., 2019; Hua et al., 2020; Li et al., 2020; Smith and Hewitson, 2020). In addition, TGF-β signaling is able to reprogram cell metabolism by conferring enzymes non-metabolic functions through post-translational modification (Pallotta et al., 2011; Chen et al., 2019). Of note, the metabolic outputs of TGF-β signaling in cells are context-dependent and highly specific to the cell type, which probably result from the different epigenetic landscapes of distinct cell types, or the different Smad-interacting transcriptional cofactors (Feng and Derynck, 2005; Massague, 2012; David and Massague, 2018). Importantly, rather than being passively regulated by TGF-β signaling, cell metabolism can also modulate TGF-β signaling. Intracellular metabolites and metabolic proteins affect the production or bioactivity of the TGF-β ligands, influence the expression and membrane levels of TGF-β receptors (Rao et al., 2000; Wu and Derynck, 2009; Gencer et al., 2017; Liu et al., 2019; Gabitova-Cornell et al., 2020; Chen et al., 2021), regulate phosphorylation or the abundance of Smad proteins (Inoki et al., 1999; Xiao et al., 2008; Halder et al., 2011; Beilfuss et al., 2015; Chen et al., 2017; Vasiukov et al., 2020), and impact translocation of Smad complex or their binding to TGF-β-target genes (Liu et al., 2003; Ding et al., 2013).
Despite reasonable knowledge have been acquired to date, many questions about the interconnection between TGF-β signaling and cell metabolism still remain. First, we lack a characterization of TGF-β-responsive metabolic gene signature across different cell types, and we do not know how many metabolites or metabolic enzymes can also function as signaling effectors in response to TGF-β. A combination of transcriptomics, untargeted metabolomics and phosphoproteomics will considerably aid in handling this problem. Second, the underlying mechanisms by which metabolites regulates TGF-β signaling remain poorly understood. Since control of gene expression appears to be a mainstay of metabolite-mediated regulation of TGF-β signaling, it would be worthy to investigate if epigenetic regulation by metabolites could account for their modulatory effects (Li et al., 2018). Last, the majority of experiments were conducted in vitro using cell lines and whether these findings could be reproduced at a physiological level are currently unknown. Hence, it is of great importance to develop mouse models to examine if the interactions between TGF-β and cell metabolism are indeed physiologically and pathologically relevant. These emerging problems at the interface between TGF-β signaling and cellular metabolism might offer new avenues for future research and bring therapeutic benefits to treat diseases.
Author Contributions
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
Funding
The research in YGC’s lab has been supported by grants from the National Natural Science Foundation of China (31988101 and 31730056) and the National Key Research and Development Program of China (2017YFA0103601).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Ameri, K., and Harris, A. L. (2008). Activating Transcription Factor 4. Int. J. Biochem. Cel Biol 40 (1), 14–21. doi:10.1016/j.biocel.2007.01.020
Baumann, F., Leukel, P., Doerfelt, A., Beier, C. P., Dettmer, K., Oefner, P. J., et al. (2009). Lactate Promotes Glioma Migration by TGF-Beta 2-dependent Regulation of Matrix Metalloproteinase-2. Neuro-Oncology 11 (4), 368–380. doi:10.1215/15228517-2008-106
Beilfuss, A., Sowa, J.-P., Sydor, S., Beste, M., Bechmann, L. P., Schlattjan, M., et al. (2015). Vitamin D Counteracts Fibrogenic TGF-Beta Signalling in Human Hepatic Stellate Cells Both Receptor-Dependently and Independently. Gut 64 (5), 791–799. doi:10.1136/gutjnl-2014-307024
Bernard, K., Logsdon, N. J., Benavides, G. A., Sanders, Y., Zhang, J. H., Darley-Usmar, V. M., et al. (2018). Glutaminolysis Is Required for Transforming Growth Factor-Beta 1-induced Myofibroblast Differentiation and Activation. J. Biol. Chem. 293 (4), 1218–1228. doi:10.1074/jbc.RA117.000444
Blachowski, S., Motyl, T., Grzelkowska, K., Kasterka, M., Orzechowski, A., and Interewicz, B. (1994). Involvement of Polyamines in Epidermal Growth-Factor (Egf), Transforming Growth-Factor (Tgf)-Alpha and (Tgf)-Beta-1 Action on Culture of L6 and Fetal Bovine Myoblasts. Int. J. Biochem. 26 (7), 891–897. doi:10.1016/0020-711x(94)90082-5
Bose, D., Banerjee, S., Chatterjee, N., Das, S., Saha, M., and Das Saha, K. (2019). Inhibition of TGF-Beta Induced Lipid Droplets Switches M2 Macrophages to M1 Phenotype. Toxicol. Vitro 58, 207–214. doi:10.1016/j.tiv.2019.03.037
Boudreau, H. E., Casterline, B. W., Rada, B., Korzeniowska, A., and Leto, T. L. (2012). Nox4 Involvement in TGF-Beta and SMAD3-Driven Induction of the Epithelial-To-Mesenchymal Transition and Migration of Breast Epithelial Cells. Free Radic. Biol. Med. 53 (7), 1489–1499. doi:10.1016/j.freeradbiomed.2012.06.016
Boutard, V., Havouis, R., Fouqueray, B., Philippe, C., Moulinoux, J. P., and Baud, L. (1995). Transforming Growth-Factor-Beta Stimulates Arginase Activity in Macrophages - Implications for the Regulation of Macrophage Cytotoxicity. J. Immunol. 155 (4), 2077–2084.
Carmona-Cuenca, I., Roncero, C., Sancho, P., Caja, L., Fausto, N., Fernandez, M., et al. (2008). Upregulation of the NADPH Oxidase NOX4 by TGF-Beta in Hepatocytes Is Required for its Pro-apoptotic Activity. J. Hepatol. 49 (6), 965–976. doi:10.1016/j.jhep.2008.07.021
Chacko, B. M., Qin, B. Y., Tiwari, A., Shi, G., Lam, S., Hayward, L. J., et al. (2004). Structural Basis of Heteromeric Smad Protein Assembly in TGF-Beta Signaling. Mol. Cel 15 (5), 813–823. doi:10.1016/j.molcel.2004.07.016
Chaikuad, A., and Bullock, A. N. (2016). Structural Basis of Intracellular TGF-Beta Signaling: Receptors and Smads. Cold Spring Harb Perspect. Biol. 8 (11). doi:10.1101/cshperspect.a022111
Chauvin, S., Yinon, Y., Xu, J., Ermini, L., Sallais, J., Tagliaferro, A., et al. (2015). Aberrant TGF Beta Signalling Contributes to Dysregulation of Sphingolipid Metabolism in Intrauterine Growth Restriction. J. Clin. Endocrinol. Metab. 100 (7), E986–E996. doi:10.1210/jc.2015-1288
Chen, G. L., Ye, T., Chen, H. L., Zhao, Z. Y., Tang, W. Q., Wang, L. S., et al. (2017). Xanthine Dehydrogenase Downregulation Promotes TGF Beta Signaling and Cancer Stem Cell-Related Gene Expression in Hepatocellular Carcinoma. Oncogenesis 6. doi:10.1038/oncsis.2017.81
Chen, H. H., Zhao, S., and Song, J. G. (2003). TGF-beta1 Suppresses Apoptosis via Differential Regulation of MAP Kinases and Ceramide Production. Cell Death Differ 10 (5), 516–527. doi:10.1038/sj.cdd.4401171
Chen, M., Zhao, Y., Yang, X., Zhao, Y., Liu, Q., Liu, Y., et al. (2021). NSDHL Promotes Triple-Negative Breast Cancer Metastasis through the TGF Beta Signaling Pathway and Cholesterol Biosynthesis. Breast Cancer Res. Treat. 187 (2), 349–362. doi:10.1007/s10549-021-06213-8
Chen, R. H., and Derynck, R. (1994). Homomeric Interactions between Type II Transforming Growth Factor-Beta Receptors. J. Biol. Chem. 269 (36), 22868–22874. doi:10.1016/s0021-9258(17)31725-8
Chen, T., Wang, T., Liang, W., Zhao, Q., Yu, Q., Ma, C.-M., et al. (2019). PAK4 Phosphorylates Fumarase and Blocks TGF Beta-Induced Cell Growth Arrest in Lung Cancer Cells. Cancer Res. 79 (7), 1383–1397. doi:10.1158/0008-5472.Can-18-2575
Chen, X., Feng, L., Li, S., Long, D., Shan, J., and Li, Y. (2020). TGF-beta 1 Maintains Foxp3 Expression and Inhibits Glycolysis in Natural Regulatory T Cells via PP2A-Mediated Suppression of mTOR Signaling. Immunol. Lett. 226, 31–37. doi:10.1016/j.imlet.2020.06.016
Chen, Y. G. (2009). Endocytic Regulation of TGF-Beta Signaling. Cell Res 19 (1), 58–70. doi:10.1038/cr.2008.315
Choudhury, M., Yin, X., Schaefbauer, K. J., Kang, J.-H., Roy, B., Kottom, T. J., et al. (2020). SIRT7-mediated Modulation of Glutaminase 1 Regulates TGF-Beta-Induced Pulmonary Fibrosis. Faseb J. 34 (7), 8920–8940. doi:10.1096/fj.202000564R
Corbet, C., Bastien, E., de Jesus, J. P. S., Dierge, E., Martherus, R., Vander Linden, C., et al. (2020). TGF Beta 2-induced Formation of Lipid Droplets Supports Acidosis-Driven EMT and the Metastatic Spreading of Cancer Cells. Nat. Commun. 11 (1). doi:10.1038/s41467-019-14262-3
Cucoranu, I., Clempus, R., Dikalova, A., Phelan, P. J., Ariyan, S., Dikalov, S., et al. (2005). NAD(P)H Oxidase 4 Mediates Transforming Growth Factor-Beta1-Induced Differentiation of Cardiac Fibroblasts into Myofibroblasts. Circ. Res. 97 (9), 900–907. doi:10.1161/01.RES.0000187457.24338.3D
David, C. J., and Massague, J. (2018). Contextual Determinants of TGF Beta Action in Development, Immunity and Cancer. Nat. Rev. Mol. Cel Biol. 19 (7), 419–435. doi:10.1038/s41580-018-0007-0
Derynck, R., and Budi, E. H. (2019). Specificity, Versatility, and Control of TGF-Beta Family Signaling. Sci. Signaling 12 (570). doi:10.1126/scisignal.aav5183
Derynck, R., Turley, S. J., and Akhurst, R. J. (2020). TGFbeta Biology in Cancer Progression and Immunotherapy. Nat. Rev. Clin. Oncol. doi:10.1038/s41571-020-0403-1
Dimeloe, S., Gubser, P., Loeliger, J., Frick, C., Develioglu, L., Fischer, M., et al. (2019). Tumor-derived TGF-Beta Inhibits Mitochondrial Respiration to Suppress IFN-Gamma Production by Human CD4(+) T Cells. Sci. Signaling 12 (599). doi:10.1126/scisignal.aav3334
Ding, N., Yu, R. T., Subramaniam, N., Sherman, M. H., Wilson, C., Rao, R., et al. (2013). A Vitamin D Receptor/SMAD Genomic Circuit gates Hepatic Fibrotic Response. Cell 153 (3), 601–613. doi:10.1016/j.cell.2013.03.028
Dobaczewski, M., Chen, W., and Frangogiannis, N. G. (2011). Transforming Growth Factor (TGF)-beta Signaling in Cardiac Remodeling. J. Mol. Cel Cardiol 51 (4), 600–606. doi:10.1016/j.yjmcc.2010.10.033
Durante, W., Liao, L., Reyna, S. V., Peyton, K. J., and Schafer, A. I. (2001). Transforming Growth Factor-Beta(1) Stimulates L-Arginine Transport and Metabolism in Vascular Smooth Muscle Cells - Role in Polyamine and Collagen Synthesis. Circulation 103 (8), 1121–1127. doi:10.1161/01.Cir.103.8.1121
Feng, X. H., and Derynck, R. (2005). Specificity and Versatility in TGF-Beta Signaling through Smads. Annu. Rev. Cel Dev Biol 21, 659–693. doi:10.1146/annurev.cellbio.21.022404.142018
Gabitova-Cornell, L., Surumbayeva, A., Peri, S., Franco-Barraza, J., Restifo, D., Weitz, N., et al. (2020). Cholesterol Pathway Inhibition Induces TGF-Beta Signaling to Promote Basal Differentiation in Pancreatic Cancer. Cancer Cell 38 (4). doi:10.1016/j.ccell.2020.08.01510.1016/j.ccell.2020.08.015
Gabriel, S. S., Tsui, C., Chisanga, D., Weber, F., Llano-Leon, M., Gubser, P. M., et al. (2021). Transforming Growth Factor-Beta-Regulated mTOR Activity Preserves Cellular Metabolism to Maintain Long-Term T Cell Responses in Chronic Infection. Immunity 54 (8), 1698–1714. doi:10.1016/j.immuni.2021.06.007
Gencer, S., Oleinik, N., Kim, J., Selvam, S. P., De Palma, R., Dany, M., et al. (2017). TGF-beta Receptor I/II Trafficking and Signaling at Primary Cilia Are Inhibited by Ceramide to Attenuate Cell Migration and Tumor Metastasis. Sci. Signaling 10 (502). doi:10.1126/scisignal.aam7464
Gilboa, L., Wells, R. G., Lodish, H. F., and Henis, Y. I. (1998). Oligomeric Structure of Type I and Type II Transforming Growth Factor Beta Receptors: Homodimers Form in the ER and Persist at the Plasma Membrane. J. Cel Biol 140 (4), 767–777. doi:10.1083/jcb.140.4.767
Guan, F., Handa, K., and Hakomori, S. I. (2009). Specific Glycosphingolipids Mediate Epithelial-To-Mesenchymal Transition of Human and Mouse Epithelial Cell Lines. Proc. Natl. Acad. Sci. United States America 106 (18), 7461–7466. doi:10.1073/pnas.0902368106
Guh, J. Y., Chuang, T. D., Chen, H. C., Hung, W. C., Lai, Y. H., Shin, S. J., et al. (2003). Beta-Hydroxybutyrate-induced Growth Inhibition and Collagen Production in HK-2 Cells Are Dependent on TGF-Beta and Smad3. Kidney Int. 64 (6), 2041–2051. doi:10.1046/j.1523-1755.2003.00330.x
Guido, C., Whitaker-Menezes, D., Capparelli, C., Balliet, R., Lin, Z., Pestell, R. G., et al. (2012). Metabolic Reprogramming of Cancer-Associated Fibroblasts by TGF-Beta Drives Tumor Growth Connecting TGF-Beta Signaling with "Warburg-like" Cancer Metabolism and L-Lactate Production. Cell Cycle 11 (16), 3019–3035. doi:10.4161/cc.21384
Halder, S. K., Goodwin, J. S., and Al-Hendy, A. (2011). 1,25-Dihydroxyvitamin D3 Reduces TGF-Beta3-Induced Fibrosis-Related Gene Expression in Human Uterine Leiomyoma Cells. J. Clin. Endocrinol. Metab. 96 (4), E754–E762. doi:10.1210/jc.2010-2131
Hamabe, A., Konno, M., Tanuma, N., Shima, H., Tsunekuni, K., Kawamoto, K., et al. (2014). Role of Pyruvate Kinase M2 in Transcriptional Regulation Leading to Epithelial-Mesenchymal Transition. Proc. Natl. Acad. Sci. United States America 111 (43), 15526–15531. doi:10.1073/pnas.1407717111
Hanahan, D., and Weinberg, R. A. (2011). Hallmarks of Cancer: the Next Generation. Cell 144 (5), 646–674. doi:10.1016/j.cell.2011.02.013
Hata, A., and Chen, Y. G. (2016). TGF-beta Signaling from Receptors to Smads. Cold Spring Harb Perspect. Biol. 8 (9). doi:10.1101/cshperspect.a022061
Hay, N. (2016). Reprogramming Glucose Metabolism in Cancer: Can it Be Exploited for Cancer Therapy? Nat. Rev. Cancer 16 (10), 635–649. doi:10.1038/nrc.2016.77
Henis, Y. I., Moustakas, A., Lin, H. Y., and Lodish, H. F. (1994). The Types II and III Transforming Growth Factor-Beta Receptors Form Homo-Oligomers. J. Cel Biol 126 (1), 139–154. doi:10.1083/jcb.126.1.139
Hill, C. S. (2016). Transcriptional Control by the SMADs. Cold Spring Harb Perspect. Biol. 8 (10). doi:10.1101/cshperspect.a022079
Hinck, A. P., Mueller, T. D., and Springer, T. A. (2016). Structural Biology and Evolution of the TGF-Beta Family. Cold Spring Harb Perspect. Biol. 8 (12). doi:10.1101/cshperspect.a022103
Hua, W., Ten Dijke, P., Kostidis, S., Giera, M., and Hornsveld, M. (2020). TGFbeta-induced Metabolic Reprogramming during Epithelial-To-Mesenchymal Transition in Cancer. Cell Mol Life Sci 77 (11), 2103–2123. doi:10.1007/s00018-019-03398-6
Huse, M., Chen, Y.-G., Massagué, J., and Kuriyan, J. (1999). Crystal Structure of the Cytoplasmic Domain of the Type I TGF β Receptor in Complex with FKBP12. Cell 96 (3), 425–436. doi:10.1016/s0092-8674(00)80555-3
Huse, M., Muir, T. W., Xu, L., Chen, Y.-G., Kuriyan, J., and Massagué, J. (2001). The TGFβ Receptor Activation Process. Mol. Cel 8 (3), 671–682. doi:10.1016/s1097-2765(01)00332-x
Inoki, K., Haneda, M., Maeda, S., Koya, D., and Kikkawa, R. (1999). TGF-beta 1 Stimulates Glucose Uptake by Enhancing GLUT1 Expression in Mesangial Cells. Kidney Int. 55 (5), 1704–1712. doi:10.1046/j.1523-1755.1999.00438.x
Jiang, L., Xiao, L., Sugiura, H., Huang, X., Ali, A., Kuro-O, M., et al. (2015). Metabolic Reprogramming during TGF Beta 1-induced Epithelial-To-Mesenchymal Transition. Oncogene 34 (30), 3908–3916. doi:10.1038/onc.2014.321
Kawabata, M., Inoue, H., Hanyu, A., Imamura, T., and Miyazono, K. (1998). Smad Proteins Exist as Monomers In Vivo and Undergo Homo- and Hetero-Oligomerization upon Activation by Serine/threonine Kinase Receptors. EMBO J. 17 (14), 4056–4065. doi:10.1093/emboj/17.14.4056
Kilberg, M. S., Shan, J., and Su, N. (2009). ATF4-dependent Transcription Mediates Signaling of Amino Acid Limitation. Trends Endocrinol. Metab. 20 (9), 436–443. doi:10.1016/j.tem.2009.05.008
Kiritsi, D., and Nystrom, A. (2018). The Role of TGFbeta in Wound Healing Pathologies. Mech. Ageing Dev. 172, 51–58. doi:10.1016/j.mad.2017.11.004
Kitagawa, T., Masumi, A., and Akamatsu, Y. (1991). Transforming Growth Factor-Beta 1 Stimulates Glucose Uptake and the Expression of Glucose Transporter mRNA in Quiescent Swiss Mouse 3T3 Cells. J. Biol. Chem. 266 (27), 18066–18071. doi:10.1016/s0021-9258(18)55237-6
Kolm, V., Sauer, U., Olgemoller, B., and Schleicher, E. D. (1996). High Glucose-Induced TGF-Beta 1 Regulates Mesangial Production of Heparan Sulfate Proteoglycan. Am. J. Physiol. 270 (5 Pt 2), F812–F821. doi:10.1152/ajprenal.1996.270.5.F812
Kolm-Litty, V., Sauer, U., Nerlich, A., Lehmann, R., and Schleicher, E. D. (1998). High Glucose-Induced Transforming Growth Factor Beta1 Production Is Mediated by the Hexosamine Pathway in Porcine Glomerular Mesangial Cells. J. Clin. Invest. 101 (1), 160–169. doi:10.1172/JCI119875
Kolodziej, L. R., Paleolog, E. M., and Williams, R. O. (2011). Kynurenine Metabolism in Health and Disease. Amino Acids 41 (5), 1173–1183. doi:10.1007/s00726-010-0787-9
Lee, S. Y., Jeon, H. M., Ju, M. K., Jeong, E. K., Kim, C. H., Park, H. G., et al. (2016). Dlx-2 and Glutaminase Upregulate Epithelial-Mesenchymal Transition and Glycolytic Switch. Oncotarget 7 (7), 7925–7939. doi:10.18632/oncotarget.6879
Li, J. H., Sun, Y. B. Y., Chen, W. Y., Fan, J. J., Li, S. H., Qu, X. L., et al. (2020). Smad4 Promotes Diabetic Nephropathy by Modulating Glycolysis and OXPHOS. Embo Rep. 21 (2). doi:10.15252/embr.201948781
Li, X., Egervari, G., Wang, Y., Berger, S. L., and Lu, Z. (2018). Regulation of Chromatin and Gene Expression by Metabolic Enzymes and Metabolites. Nat. Rev. Mol. Cel Biol 19 (9), 563–578. doi:10.1038/s41580-018-0029-7
Liu, L., Santora, R., Rao, J. N., Guo, X., Zou, T. T., Zhang, H. F. M., et al. (2003). Activation of TGF-Beta-Smad Signaling Pathway Following Polyamine Depletion in Intestinal Epithelial Cells. Am. J. Physiology-Gastrointestinal Liver Physiol. 285 (5), G1056–G1067. doi:10.1152/ajpgi.00151.2003
Liu, Q.-Q., Huo, H.-Y., Ao, S., Liu, T., Yang, L., Fei, Z.-Y., et al. (2020). TGF-beta 1-induced Epithelial-Mesenchymal Transition Increases Fatty Acid Oxidation and OXPHOS Activity via the P-AMPK Pathway in Breast Cancer Cells. Oncol. Rep. 44 (3), 1206–1215. doi:10.3892/or.2020.7661
Liu, R. M., and Gaston Pravia, K. A. (2010). Oxidative Stress and Glutathione in TGF-Beta-Mediated Fibrogenesis. Free Radic. Biol. Med. 48 (1), 1–15. doi:10.1016/j.freeradbiomed.2009.09.026
Liu, S., Hou, H., Zhang, P. P., Wu, Y. F., He, X. H., Li, H., et al. (2019). Sphingomyelin Synthase 1 Regulates the Epithelial-To-Mesenchymal Transition Mediated by the TGF-/Smad Pathway in MDA-MB-231 Cells. Mol. Med. Rep. 19 (2), 1159–1167. doi:10.3892/mmr.2018.9722
Lo, R. S., Chen, Y. G., Shi, Y., Pavletich, N. P., and Massague, J. (1998). The L3 Loop: a Structural Motif Determining Specific Interactions between SMAD Proteins and TGF-Beta Receptors. EMBO J. 17 (4), 996–1005. doi:10.1093/emboj/17.4.996
Loayza-Puch, F., Rooijers, K., Zijlstra, J., Moumbeini, B., Zaal, E. A., Vrielink, J. F. O., et al. (2017). TGF Beta 1-induced Leucine Limitation Uncovered by Differential Ribosome Codon reading. Embo Rep. 18 (4), 549–557. doi:10.15252/embr.201744000
Maceyka, M., Harikumar, K. B., Milstien, S., and Spiegel, S. (2012). Sphingosine-1-phosphate Signaling and its Role in Disease. Trends Cel Biol 22 (1), 50–60. doi:10.1016/j.tcb.2011.09.003
Manning, B. D., and Toker, A. (2017). AKT/PKB Signaling: Navigating the Network. Cell 169 (3), 381–405. doi:10.1016/j.cell.2017.04.001
Martinez-Outschoorn, U. E., Sotgia, F., and Lisanti, M. P. (2012). Power Surge: Supporting Cells "fuel" Cancer Cell Mitochondria. Cell Metab 15 (1), 4–5. doi:10.1016/j.cmet.2011.12.011
Massague, J., and Chen, Y. G. (2000). Controlling TGF-Beta Signaling. Genes Dev. 14 (6), 627–644. doi:10.1101/gad.14.6.627
Massague, J., Seoane, J., and Wotton, D. (2005). Smad Transcription Factors. Genes Dev. 19 (23), 2783–2810. doi:10.1101/gad.1350705
Massague, J. (2012). TGF Beta Signalling in Context. Nat. Rev. Mol. Cel Biol. 13 (10), 616–630. doi:10.1038/nrm3434
Massague, J., and Wotton, D. (2000). Transcriptional Control by the TGF-beta/Smad Signaling System. Embo J. 19 (8), 1745–1754. doi:10.1093/emboj/19.8.1745
Meng, X.-m., Nikolic-Paterson, D. J., and Lan, H. Y. (2016). TGF-beta: the Master Regulator of Fibrosis. Nat. Rev. Nephrol. 12 (6), 325–338. doi:10.1038/nrneph.2016.48
Michaeloudes, C., Sukkar, M. B., Khorasani, N. M., Bhavsar, P. K., and Chung, K. F. (2011). TGF-beta Regulates Nox4, MnSOD and Catalase Expression, and IL-6 Release in Airway Smooth Muscle Cells. Am. J. Physiology-Lung Cell Mol. Physiol. 300 (2), L295–L304. doi:10.1152/ajplung.00134.2010
Miller-Fleming, L., Olin-Sandoval, V., Campbell, K., and Ralser, M. (2015). Remaining Mysteries of Molecular Biology: The Role of Polyamines in the Cell. J. Mol. Biol. 427 (21), 3389–3406. doi:10.1016/j.jmb.2015.06.020
Mills, E. L., Kelly, B., and O'Neill, L. A. J. (2017). Mitochondria Are the Powerhouses of Immunity. Nat. Immunol. 18 (5), 488–498. doi:10.1038/ni.3704
Mondanelli, G., Bianchi, R., Pallotta, M. T., Orabona, C., Albini, E., Iacono, A., et al. (2017). A Relay Pathway between Arginine and Tryptophan Metabolism Confers Immunosuppressive Properties on Dendritic Cells. Immunity 46 (2), 233–244. doi:10.1016/j.immuni.2017.01.005
Morikawa, M., Derynck, R., and Miyazono, K. (2016). TGF-beta and the TGF-Beta Family: Context-dependent Roles in Cell and Tissue Physiology. Cold Spring Harbor Perspect. Biol. 8 (5). doi:10.1101/cshperspect.a021873
Motyl, T., Kasterka, M., Grzelkowska, K., Blachowski, S., and Sysa, P. (1993). Tgf-Beta(1) Inhibits Polyamine Biosynthesis In K-562 Leukemic-Cells. Ann. Hematol. 67 (6), 285–288. doi:10.1007/bf01696348
Moustakas, A., and Heldin, C.-H. (2009). The Regulation of TGF Beta Signal Transduction. Development 136 (22), 3699–3714. doi:10.1242/dev.030338
Nakasuka, F., Tabata, S., Sakamoto, T., Hirayama, A., Ebi, H., Yamada, T., et al. (2021). TGF-beta-dependent Reprogramming of Amino Acid Metabolism Induces Epithelial-Mesenchymal Transition in Non-small Cell Lung Cancers. Commun. Biol. 4 (1). doi:10.1038/s42003-021-02323-7
Nigdelioglu, R., Hamanaka, R. B., Meliton, A. Y., O'Leary, E., Witt, L. J., Cho, T., et al. (2016). Transforming Growth Factor (TGF)-beta Promotes De Novo Serine Synthesis for Collagen Production. J. Biol. Chem. 291 (53), 27239–27251. doi:10.1074/jbc.M116.756247
Nilchian, A., Giotopoulou, N., Sun, W., and Fuxe, J. (2020). Different Regulation of Glut1 Expression and Glucose Uptake during the Induction and Chronic Stages of TGF Beta 1-Induced EMT in Breast Cancer Cells. Biomolecules 10 (12). doi:10.3390/biom10121621
Olzmann, J. A., and Carvalho, P. (2019). Dynamics and Functions of Lipid Droplets. Nat. Rev. Mol. Cel Biol 20 (3), 137–155. doi:10.1038/s41580-018-0085-z
Oshimori, N., Oristian, D., and Fuchs, E. (2015). TGF-beta Promotes Heterogeneity and Drug Resistance in Squamous Cell Carcinoma. Cell 160 (5), 963–976. doi:10.1016/j.cell.2015.01.043
Pallotta, M. T., Orabona, C., Volpi, C., Vacca, C., Belladonna, M. L., Bianchi, R., et al. (2011). Indoleamine 2,3-dioxygenase Is a Signaling Protein in Long-Term Tolerance by Dendritic Cells. Nat. Immunol. 12 (9), 870–878. doi:10.1038/ni.2077
Patel, A. R., Li, J., Bass, B. L., and Wang, J. Y. (1998). Expression of the Transforming Growth Factor-Beta Gene during Growth Inhibition Following Polyamine Depletion. Am. J. Physiol. 275 (2), C590–C598. doi:10.1152/ajpcell.1998.275.2.C590
Priyadharshini, B., Loschi, M., Newton, R. H., Zhang, J. W., Finn, K. K., Gerriets, V. A., et al. (2018). Cutting Edge: TGF-Beta and Phosphatidylinositol 3-Kinase Signals Modulate Distinct Metabolism of Regulatory T Cell Subsets. J. Immunol. 201 (8), 2215–2219. doi:10.4049/jimmunol.1800311
Puleston, D. J., Baixauli, F., Sanin, D. E., Edwards-Hicks, J., Villa, M., Kabat, A. M., et al. (2021). Polyamine Metabolism Is a central Determinant of Helper T Cell Lineage Fidelity. Cell. doi:10.1016/j.cell.2021.06.007
Ramundo, V., Giribaldi, G., and Aldieri, E. (2021). Transforming Growth Factor-Beta and Oxidative Stress in Cancer: A Crosstalk in Driving Tumor Transformation. Cancers 13 (12). doi:10.3390/cancers13123093
Rao, J. N., Li, L., Bass, B. L., and Wang, J. Y. (2000). Expression of the TGF-Beta Receptor Gene and Sensitivity to Growth Inhibition Following Polyamine Depletion. Am. J. Physiology-Cell Physiol. 279 (4), C1034–C1044. doi:10.1152/ajpcell.2000.279.4.c1034
Ricca, C., Aillon, A., Viano, M., Bergandi, L., Aldieri, E., and Silvagno, F. (2019). Vitamin D Inhibits the Epithelial-Mesenchymal Transition by a Negative Feedback Regulation of TGF-Beta Activity. J. Steroid Biochem. Mol. Biol. 187, 97–105. doi:10.1016/j.jsbmb.2018.11.006
Richter, K., Konzack, A., Pihlajaniemi, T., Heljasvaara, R., and Kietzmann, T. (2015). Redox-fibrosis: Impact of TGF Beta 1 on ROS Generators, Mediators and Functional Consequences. Redox Biol. 6, 344–352. doi:10.1016/j.redox.2015.08.015
Rifkin, D. B. (2005). Latent Transforming Growth Factor-Beta (TGF-Beta) Binding Proteins: Orchestrators of TGF-Beta Availability. J. Biol. Chem. 280 (9), 7409–7412. doi:10.1074/jbc.R400029200
Rinschen, M. M., Ivanisevic, J., Giera, M., and Siuzdak, G. (2019). Identification of Bioactive Metabolites Using Activity Metabolomics. Nat. Rev. Mol. Cel Biol 20 (6), 353–367. doi:10.1038/s41580-019-0108-4
Robertson, I. B., and Rifkin, D. B. (2016). Regulation of the Bioavailability of TGF-Beta and TGF-Beta-Related Proteins. Cold Spring Harb Perspect. Biol. 8 (6). doi:10.1101/cshperspect.a021907
Rodriguez-Garcia, A., Samso, P., Fontova, P., Simon-Molas, H., Manzano, A., Castano, E., et al. (2017). TGF-beta 1 Targets Smad, P38 MAPK, and PI3K/Akt Signaling Pathways to Induce PFKFB3 Gene Expression and Glycolysis in Glioblastoma Cells. Febs J. 284 (20), 3437–3454. doi:10.1111/febs.14201
Salazar, V. S., Gamer, L. W., and Rosen, V. (2016). BMP Signalling in Skeletal Development, Disease and Repair. Nat. Rev. Endocrinol. 12 (4), 203–221. doi:10.1038/nrendo.2016.12
Samuel, W., Nagineni, C. N., Kutty, R. K., Parks, W. T., Gordon, J. S., Prouty, S. M., et al. (2002). Transforming Growth Factor-Beta Regulates Stearoyl Coenzyme A Desaturase Expression through a Smad Signaling Pathway. J. Biol. Chem. 277 (1), 59–66. doi:10.1074/jbc.M108730200
Sato, M., Markiewicz, M., Yamanaka, M., Bielawska, A., Mao, C. G., Obeid, L. M., et al. (2003). Modulation of Transforming Growth Factor-Beta (TGF-Beta) Signaling by Endogenous Sphingolipid Mediators. J. Biol. Chem. 278 (11), 9276–9282. doi:10.1074/jbc.M211529200
Schrumpf, J. A., Ninaber, D. K., van der Does, A. M., and Hiemstra, P. S. (2020). TGF-beta 1 Impairs Vitamin D-Induced and Constitutive Airway Epithelial Host Defense Mechanisms. J. Innate Immun. 12 (1), 74–89. doi:10.1159/000497415
Schworer, S., Berisa, M., Violante, S., Qin, W. G., Zhu, J. J., Hendrickson, R. C., et al. (2020). Proline Biosynthesis Is a Vent for TGF Beta-Induced Mitochondrial Redox Stress. Embo J. 39 (8). doi:10.15252/embj.2019103334
Selvarajah, B., Azuelos, I., Plate, M., Guillotin, D., Forty, E. J., Contento, G., et al. (2019). mTORC1 Amplifies the ATF4-dependent De Novo Serine-glycine Pathway to Supply glycine during TGF-Beta 1-induced Collagen Biosynthesis. Sci. Signaling 12 (582). doi:10.1126/scisignal.aav3048
Shi, Y., Wang, Y.-F., Jayaraman, L., Yang, H., Massagué, J., and Pavletich, N. P. (1998). Crystal Structure of a Smad MH1 Domain Bound to DNA. Cell 94 (5), 585–594. doi:10.1016/s0092-8674(00)81600-1
Sies, H., Berndt, C., and Jones, D. P. (2017). Oxidative Stress. Annu. Rev. Biochem. 86, 715–748. doi:10.1146/annurev-biochem-061516-045037
Slattery, K., Woods, E., Zaiatz-Bittencourt, V., Marks, S., Chew, S., Conroy, M., et al. (2021). TGF Beta Drives NK Cell Metabolic Dysfunction in Human Metastatic Breast Cancer. J. Immunother. Cancer 9 (2), 2020–002044. doi:10.1136/jitc-2020-002044
Smith, E. R., and Hewitson, T. D. (2020). TGF-beta 1 Is a Regulator of the Pyruvate Dehydrogenase Complex in Fibroblasts. Scientific Rep. 10 (1). doi:10.1038/s41598-020-74919-8
Soukupova, J., Malfettone, A., Bertran, E., Hernandez-Alvarez, M. I., Penuelas-Haro, I., Dituri, F., et al. (2021). Epithelial-Mesenchymal Transition (EMT) Induced by TGF-Beta in Hepatocellular Carcinoma Cells Reprograms Lipid Metabolism. Int. J. Mol. Sci. 22 (11). doi:10.3390/ijms22115543
Soukupova, J., Malfettone, A., Hyrossova, P., Hernandez-Alvarez, M. I., Penuelas-Haro, I., Bertran, E., et al. (2017). Role of the Transforming Growth Factor-Beta in Regulating Hepatocellular Carcinoma Oxidative Metabolism. Sci. Rep. 7 (1), 12486. doi:10.1038/s41598-017-12837-y
Sturrock, A., Cahill, B., Norman, K., Huecksteadt, T. P., Hill, K., Sanders, K., et al. (2006). Transforming Growth Factor-Beta1 Induces Nox4 NAD(P)H Oxidase and Reactive Oxygen Species-dependent Proliferation in Human Pulmonary Artery Smooth Muscle Cells. Am. J. Physiol. Lung Cel Mol Physiol 290 (4), L661–L673. doi:10.1152/ajplung.00269.2005
Takahashi, H., Alves, C. R. R., Stanford, K. I., Middelbeek, R. J. W., Nigro, P., Ryan, R. E., et al. (2019). TGF-beta 2 Is an Exercise-Induced Adipokine that Regulates Glucose and Fatty Acid Metabolism. Nat. Metab. 1 (2), 291–303. doi:10.1038/s42255-018-0030-7
Travis, M. A., and Sheppard, D. (2014). TGF-beta Activation and Function in Immunity. Annu. Rev. Immunol. Vol 32 (32), 51–82. doi:10.1146/annurev-immunol-032713-120257
Trempolec, N., Degavre, C., Doix, B., Brusa, D., Corbet, C., and Feron, O. (2020). Acidosis-Induced TGF-Beta 2 Production Promotes Lipid Droplet Formation in Dendritic Cells and Alters Their Potential to Support Anti-mesothelioma T Cell Response. Cancers 12 (5). doi:10.3390/cancers12051284
Valderrama-Carvajal, H., Cocolakis, E., Lacerte, A., Lee, E. H., Krystal, G., Ali, S., et al. (2002). Activin/TGF-beta Induce Apoptosis through Smad-dependent Expression of the Lipid Phosphatase SHIP. Nat. Cel Biol. 4 (12), 963–969. doi:10.1038/ncb885
Vasiukov, G., Novitskaya, T., Zijlstra, A., Owens, P., Ye, F., Zhao, Z., et al. (2020). Myeloid Cell-Derived TGF Beta Signaling Regulates ECM Deposition in Mammary Carcinoma via Adenosine-dependent Mechanisms. Cancer Res. 80 (12), 2628–2638. doi:10.1158/0008-5472.Can-19-3954
Viel, S., Marcais, A., Guimaraes, F. S., Loftus, R., Rabilloud, J., Grau, M., et al. (2016). TGF-beta Inhibits the Activation and Functions of NK Cells by Repressing the mTOR Pathway. Sci. Signal. 9(415), ra19. doi:10.1126/scisignal.aad1884
Walther, T. C., and Farese, R. V. (2012). Lipid Droplets and Cellular Lipid Metabolism. Annu. Rev. Biochem. 81, 687–714. doi:10.1146/annurev-biochem-061009-102430
Wang, C. C., Silverman, R. M., Shen, J., and O'Keefe, R. J. (2018). Distinct Metabolic Programs Induced by TGF-Beta 1 and BMP2 in Human Articular Chondrocytes with Osteoarthritis. J. Orthopaedic Translation 12, 66–73. doi:10.1016/j.jot.2017.12.004
Wrana, J. L., Attisano, L., Wieser, R., Ventura, F., and Massague, J. (1994). Mechanism of Activation of the TGF-Beta Receptor. Nature 370 (6488), 341–347. doi:10.1038/370341a0
Wu, G. (2009). Amino Acids: Metabolism, Functions, and Nutrition. Amino Acids 37 (1), 1–17. doi:10.1007/s00726-009-0269-0
Wu, L., and Derynck, R. (2009). Essential Role of TGF-Beta Signaling in Glucose-Induced Cell Hypertrophy. Dev. Cel 17 (1), 35–48. doi:10.1016/j.devcel.2009.05.010
Wu, M. Y., and Hill, C. S. (2009). TGF-beta Superfamily Signaling in Embryonic Development and Homeostasis. Dev. Cel 16 (3), 329–343. doi:10.1016/j.devcel.2009.02.012
Xiao, S., Jin, H., Korn, T., Liu, S. M., Oukka, M., Lim, B., et al. (2008). Retinoic Acid Increases Foxp3(+) Regulatory T Cells and Inhibits Development of Th17 Cells by Enhancing TGF-Beta-Driven Smad3 Signaling and Inhibiting IL-6 and IL-23 Receptor Expression. J. Immunol. 181 (4), 2277–2284. doi:10.4049/jimmunol.181.4.2277
Xu, D., Shao, F., Bian, X., Meng, Y., Liang, T., and Lu, Z. (2021). The Evolving Landscape of Noncanonical Functions of Metabolic Enzymes in Cancer and Other Pathologies. Cel Metab 33 (1), 33–50. doi:10.1016/j.cmet.2020.12.015
Xu, P., Lin, X., and Feng, X. H. (2016). Posttranslational Regulation of Smads. Cold Spring Harb Perspect. Biol. 8 (12). doi:10.1101/cshperspect.a022087
Xu, W. F., Zeng, F. Y., Li, S. Y., Li, G. H., Lai, X. J., Wang, Q. J., et al. (2018). Crosstalk of Protein Kinase C Epsilon with Smad2/3 Promotes Tumor Cell Proliferation in Prostate Cancer Cells by Enhancing Aerobic Glycolysis. Cell Mol. Life Sci. 75 (24), 4583–4598. doi:10.1007/s00018-018-2914-9
Yadav, H., Quijano, C., Kamaraju, A. K., Gavrilova, O., Malek, R., Chen, W., et al. (2011). Protection from Obesity and Diabetes by Blockade of TGF-beta/Smad3 Signaling. Cel Metab 14 (1), 67–79. doi:10.1016/j.cmet.2011.04.013
Yalcin, A., Solakoglu, T. H., Ozcan, S. C., Guzel, S., Peker, S., Celikler, S., et al. (2017). 6-phosphofructo-2-kinase/fructose 2,6-bisphosphatase-3 Is Required for Transforming Growth Factor Beta 1-enhanced Invasion of Panci Cells In Vitro. Biochem. Biophysical Res. Commun. 484 (3), 687–693. doi:10.1016/j.bbrc.2017.01.178
Yamada, H., Iwaki, Y., Kitaoka, R., Fujitani, M., Shibakusa, T., Fujikawa, T., et al. (2012). Blood Lactate Functions as a Signal for Enhancing Fatty Acid Metabolism during Exercise via TGF-Beta in the Brain. J. Nutr. Sci. Vitaminology 58 (2), 88–95. doi:10.3177/jnsv.58.88
Yamanaka, M., Shegogue, D., Pei, H., Bu, S., Bielawska, A., Bielawski, J., et al. (2004). Sphingosine Kinase 1 (SPHK1) Is Induced by Transforming Growth Factor-Beta and Mediates TIMP-1 Up-Regulation. J. Biol. Chem. 279 (52), 53994–54001. doi:10.1074/jbc.M410144200
Yanagisawa, J., Yanagi, Y., Masuhiro, Y., Suzawa, M., Watanabe, M., Kashiwagi, K., et al. (1999). Convergence of Transforming Growth Factor-Beta and Vitamin D Signaling Pathways on SMAD Transcriptional Coactivators. Science 283 (5406), 1317–1321. doi:10.1126/science.283.5406.1317
Yang, L., Roh, Y. S., Song, J., Zhang, B., Liu, C., Loomba, R., et al. (2014). Transforming Growth Factor Beta Signaling in Hepatocytes Participates in Steatohepatitis through Regulation of Cell Death and Lipid Metabolism in Mice. Hepatology 59 (2), 483–495. doi:10.1002/hep.26698
Yang, L., Zhang, F. Q., Wang, X., Tsai, Y., Chuang, K. H., Keng, P. C., et al. (2016). A FASN-TGF-Beta 1-FASN Regulatory Loop Contributes to High EMT/metastatic Potential of Cisplatin-Resistant Non-small Cell Lung Cancer. Oncotarget 7 (34), 55543–55554. doi:10.18632/oncotarget.10837
Yin, X. Q., Choudhury, M., Kang, J. H., Schaefbauer, K. J., Jung, M. Y., Andrianifahanana, M., et al. (2019). Hexokinase 2 Couples Glycolysis with the Profibrotic Actions of TGF-Beta. Sci. Signaling 12 (612). doi:10.1126/scisignal.aax4067
Yoon, Y. S., Lee, J. H., Hwang, S. C., Choi, K. S., and Yoon, G. (2005). TGF Beta1 Induces Prolonged Mitochondrial ROS Generation through Decreased Complex IV Activity with Senescent Arrest in Mv1Lu Cells. Oncogene 24 (11), 1895–1903. doi:10.1038/sj.onc.1208262
Yuan, W., Collado-Hidalgo, A., Yufit, T., Taylor, M., and Varga, J. (1998). Modulation of Cellular Tryptophan Metabolism in Human Fibroblasts by Transforming Growth Factor-β: Selective Inhibition of Indoleamine 2,3-dioxygenase and Tryptophanyl-tRNA Synthetase Gene Expression. J. Cell Physiol. 177 (1), 174–186. doi:10.1002/(sici)1097-4652(199810)177:1<174::Aid-jcp18>3.0.Co;2-d
Zaiatz-Bittencourt, V., Finlay, D. K., and Gardiner, C. M. (2018). Canonical TGF-Beta Signaling Pathway Represses Human NK Cell Metabolism. J. Immunol. 200 (12), 3934–3941. doi:10.4049/jimmunol.1701461
Zhang, C. Y., Yin, H. M., Wang, H., Su, D., Xia, Y., Yan, L. F., et al. (2018). Transforming Growth Factor-Beta 1 Regulates the Nascent Hematopoietic Stem Cell Niche by Promoting Gluconeogenesis. Leukemia 32 (2), 479–491. doi:10.1038/leu.2017.198
Zhang, J., Pavlova, N. N., and Thompson, C. B. (2017). Cancer Cell Metabolism: the Essential Role of the Nonessential Amino Acid, Glutamine. EMBO J. 36 (10), 1302–1315. doi:10.15252/embj.201696151
Zhang, L. Y., Zhou, D. B., Guan, W. C., Ren, W. M., Sun, W. W., Shi, J. M., et al. (2017). Pyridoxine 5 '-phosphate Oxidase Is a Novel Therapeutic Target and Regulated by the TGF-Beta Signalling Pathway in Epithelial Ovarian Cancer. Cel Death Dis. 8. doi:10.1038/s41419-017-0050-3
Zhang, W., Jiang, Y., Wang, Q., Ma, X., Xiao, Z., Zuo, W., et al. (2009). Single-molecule Imaging Reveals Transforming Growth Factor-Beta-Induced Type II Receptor Dimerization. Proc. Natl. Acad. Sci. U S A. 106 (37), 15679–15683. doi:10.1073/pnas.0908279106
Zhang, W., Yuan, J., Yang, Y., Xu, L., Wang, Q., Zuo, W., et al. (2010). Monomeric Type I and Type III Transforming Growth Factor-Beta Receptors and Their Dimerization Revealed by Single-Molecule Imaging. Cel Res 20 (11), 1216–1223. doi:10.1038/cr.2010.105
Zhang, Y. E. (2017). Non-Smad Signaling Pathways of the TGF-Beta Signaling. Cold Spring Harb Perspect. Biol. 9(2), a022129. doi:10.1038/cr.2008.328
Ziyadeh, F. N., Sharma, K., Ericksen, M., and Wolf, G. (1994). Stimulation of Collagen Gene Expression and Protein Synthesis in Murine Mesangial Cells by High Glucose Is Mediated by Autocrine Activation of Transforming Growth Factor-Beta. J. Clin. Invest. 93 (2), 536–542. doi:10.1172/JCI117004
Keywords: TGF-β signaling, Smad, glucose metabolism, lipid metabolism, amino acid metabolism
Citation: Liu H and Chen Y-G (2022) The Interplay Between TGF-β Signaling and Cell Metabolism. Front. Cell Dev. Biol. 10:846723. doi: 10.3389/fcell.2022.846723
Received: 31 December 2021; Accepted: 14 February 2022;
Published: 09 March 2022.
Edited by:
Long Zhang, Zhejiang University, ChinaReviewed by:
Terytty Yang Li, Swiss Federal Institute of Technology Lausanne, SwitzerlandVinay Bulusu, Indian Institute of Science Education and Research Berhampur (IISER), India
Copyright © 2022 Liu and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ye-Guang Chen, eWdjaGVuQHRzaW5naHVhLmVkdS5jbg==
 Huidong Liu
Huidong Liu Ye-Guang Chen
Ye-Guang Chen