- 1Orthopedic Department, Taizhou Hospital Affiliated to Wenzhou Medical University, Linhai, China
- 2Enze Medical Research Center, Taizhou Hospital, Wenzhou Medical University, Linhai, China
- 3Department of Medical Oncology, The Second Affiliated Hospital, Zhejiang University School of Medicine, Hangzhou, China
Mammalian cell membranes are decorated by the glycocalyx, which offer versatile means of generating biochemical signals. By manipulating the set of glycans displayed on cell surface, it is vital for gaining insight into the cellular behavior modulation and medical and biotechnological adhibition. Although genetic engineering is proven to be an effective approach for cell surface modification, the technique is only suitable for natural and genetically encoded molecules. To circumvent these limitations, non-genetic approaches are developed for modifying cell surfaces with unnatural but functional groups. Here, we review latest development of metabolic glycoengineering (MGE), which enriches the chemical functions of the cell surface and is becoming an intriguing new tool for regenerative medicine and tissue engineering. Particular emphasis of this review is placed on discussing current applications and perspectives of MGE.
Introduction
The cell surfaces are dominated by multiple functional molecules, which play essential roles in regulating intercellular communications, molecules selective transportation and intracellular associated signaling pathways (Abbina et al., 2018). Understanding and manipulating the cell surface have aroused crucial topics in fundamental researches on cell behaviors, novel biotechnical applications and therapeutic exploitation (Nischan and Kohler, 2016). Compared with other molecules on cell surface, glycans signify the function of a cell and specify how it interacts with its surroundings (Du and Yarema, 2010). Although there are abundant chemical groups on the cell surface, only a handful of functional groups can be used for direct covalent bond formation reactions under suitable conditions. Since cell membrane modification can be exploited to rapidly provide additional cell functionality, therefore, it is critical for endowing or improving cellular biological behaviors, including immuno-evasion, adhesion and homing.
Redecorating cell surfaces with genes encoding peptides or proteins using conventional genetic engineering is a well-established strategy, which has been extensively applied in basic research and translational medicine (Rao et al., 2020). For example, previous reports have shown that using surface-displayed genetically engineered chimeric antigen receptor (CAR) T cells to recognize cancer antigens for the treatment of acute lymphoblastic leukemia (Kuehn, 2017). Although quite practical, such traditional genetic manipulation approaches are limited to displaying natural genetic encoded molecules on cellular surface, but cannot display non-genetically encoded molecules, such as cytotoxic drugs, fluorophores, spectroscopic probes, affinity tags, and vaccine adjuvants (Plumet et al., 2021). In addition, viral vectors are associated with high risks of genetic integration which may cause tumorigenesis and stimulate immunogenic responses (Lee et al., 2018).
Metabolic glycoengineering (MGE) - a technique where monosaccharide analogs are introduced into the metabolic pathways of a cell and are biosynthetically incorporated into the glycocalyx on cell membrane - has been shown to be precious for engineering cellular surface properties to allow manipulation of cellular behaviour and function using natural or unnatural functional groups (Du et al., 2009; Nagasundaram et al., 2020). Over the last 2 decades, more than 20 unnatural mannosamines that are suitable for MGE, have been introduced. These N-acetyl d-mannosamine (ManNAc) analogs can be divided into two groups, namely aliphatic and bioorthogonal analogues (Wratil et al., 2016). MGE can equip cell surfaces with various unnatural functional groups, such as ketone-, azide-, thiol-, or alkyne-modified glycans, which can then be combined with abundant ligands via bioorthogonal chemoselective ligation reactions, thereby extremely boosting versatility of this strategy. Specifically, MGE has various advantages, they are: 1) stable under physiologic conditions; 2) applicable to all cell types; 3) innocuous to the modified cell; and 4) reversible upon administration of a controlled precursor (Csizmar et al., 2018). Currently, cell membrane glycoengineering is a powerful tool that has been widely applied across the following fields: 1) endowment of cells with new abilities (Wang et al., 2020); 2) glycoengineered membrane self-assembly servers as vehicles to coat nanoparticles (Rao et al., 2020); 3) in vivo cell tracking (Lim et al., 2019); 4) metabolic glycan labelling (Hu et al., 2018b; Lim et al., 2019; Costa et al., 2020; Wang and Mooney, 2020); 5) cell and drug delivery (Lee et al., 2018; Xia et al., 2020); and 6) single cell encapsulation (Kim et al., 2018; Oh et al., 2020).
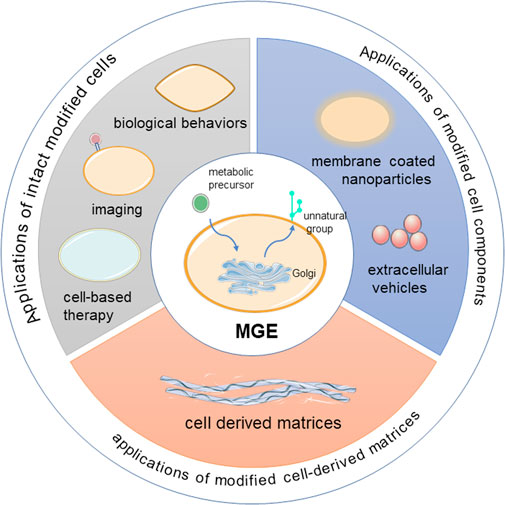
Currently, metabolic glycoengineering has numerous applications. If divided according to the cell structure, the applications can be divided into the following three broad types, namely the applications of intact cells, cell components and cell-derived matrices (Scheme 1). This review systematically summarizes current approaches and applications for cell surface glycoengineering, both in vitro and in vivo. We highlight key examples of MGE in each section, and provide perspectives and future trends of this rapidly growing field.
Applications of Intact Modified Cells
Modulation of Cell Biological Behaviors
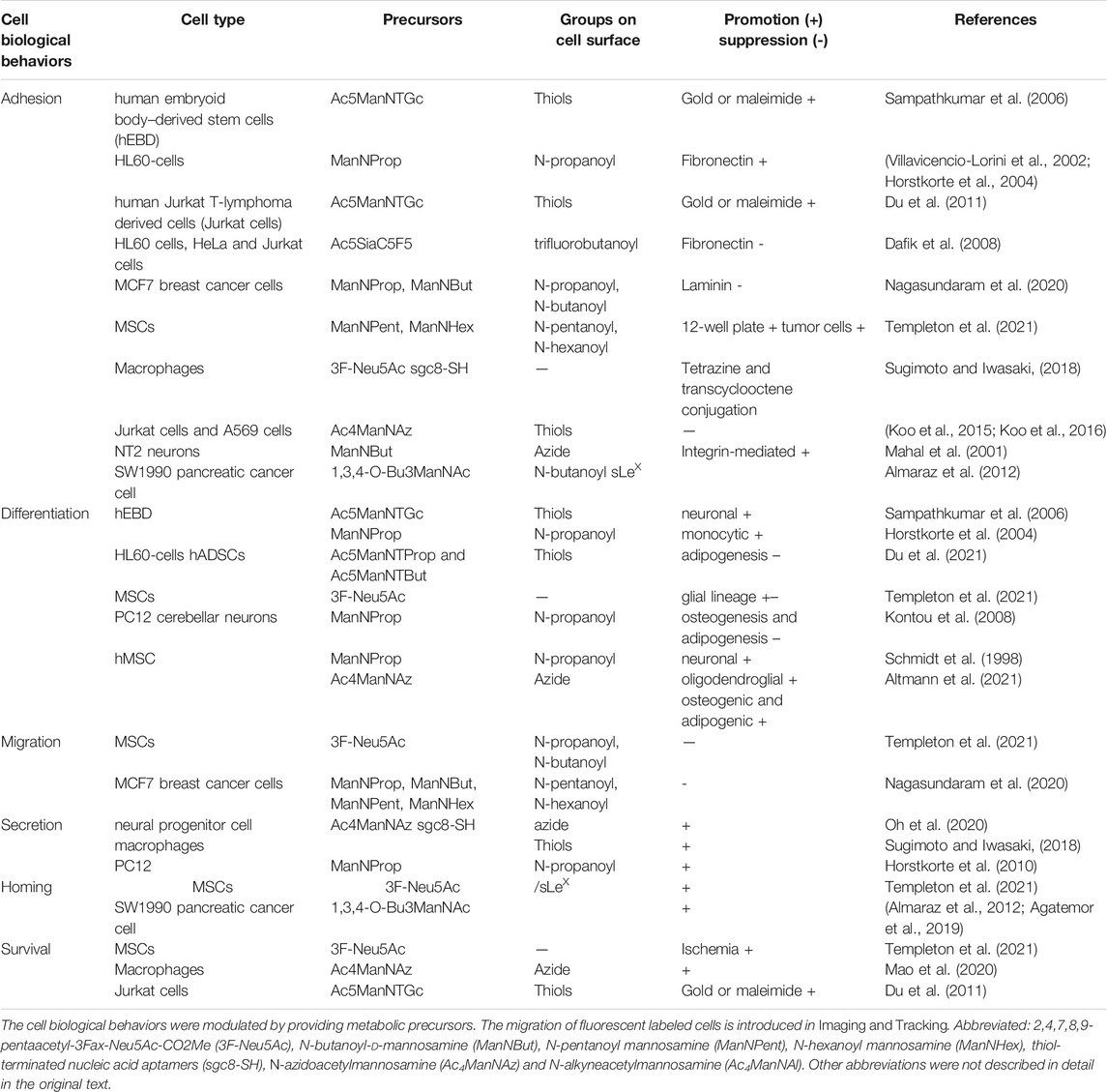
Previous evidences have shown that most cellular glycans on the outer surface of membrane mediate or modulate cell-cell, cell-molecule, and cell-matrix interactions, which are essential for the development and functioning of cells (Du and Yarema, 2010). Pretreatment of peracetylated N-thiolglycolyl-d-mannosamine (Ac5ManNTGc) resulted in expression of thiol on cell surface, which stimulated cell adhesion to complementary gold or maleimide-derivatized substrates (Sampathkumar et al., 2006). In addition, treatment of HL60-cells with N-propanoylmannosamine (ManNProp) increased cell adhesion to fibronectin by activating β1-integrin (Villavicencio-Lorini et al., 2002). Findings from other research have revealed significant improvement in adhesion ability of leukocytes, Jurkat cells, neural cells, mesenchymal stem cells (MSCs) and A549 cells after treatment with ManNAc analogs (Du et al., 2009; Du et al., 2011; Koo et al., 2015; Koo et al., 2016; Lo et al., 2016). However, Nagasundaram et al.(Nagasundaram et al., 2020) found a marked reduction in adhesion of MCF7 breast cancer cells to laminin upon treatment with different ManNAc analogs. In addition, they found that application of all non-natural sialic acid precursors downregulated N-acetylneuraminic acid (Neu5Ac) and polysialic acids (polySia), and further suppressed adhesion and migration.
MGE can also change the differentiation ability of cells. For example, supplement of human stem cells with Ac5ManNTGc was found to improve their differentiation towards a neural lineage (Sampathkumar et al., 2006). ManNProp reportedly promoted monocytic differentiation of HL60-cells (Horstkorte et al., 2004). Both Ac5ManNTProp and Ac5ManNTBut, which display thiol groups on the cell surface, were found to suppress adipogenic differentiation in hADSCs, but they did not interfere with differentiation to a glial lineage (Du et al., 2021). Moreover, both osteogenic and adipogenic differentiation were inhibited, when MSCs were pre- or continuous treated with 3F-Neu5Ac (Templeton et al., 2021).
MSCs incubated with ManNProp significantly upregulates sialyl Lewis X (sLeX) (Natunen et al., 2013), an epitope that promotes osteotropism (Dykstra et al., 2016; Sánchez-Martínez et al., 2021), augments neurotropism (Merzaban et al., 2015) and improves ischemia-reperfusion by increased homing efficacy towards porcine heart (Lo et al., 2016). In another study, Byeongtaek et al. (Oh et al., 2020) coated each neural progenitor cell (NPC) with a layer of polymer, via click-chemistry and glycoengineering. The cells enhanced trophic factor release by optimizing stiffness of the polymer coating, which reduced the required number of cells through augmenting the efficacy of each NPC. On the other hand, supplementation with 3F-Neu5Ac was found to improve adhesion and elevate the rate of migration of MSCs, thereby promoting their survival in an in vitro ischemia model (Templeton et al., 2021). In summary, MGE can effectively modulate cell biological behaviors (Table 1). Also, we conclude that the effects of MGE on cellular activities are dependent on cell types and different kinds of ManNAc analogs.
Imaging and Tracking
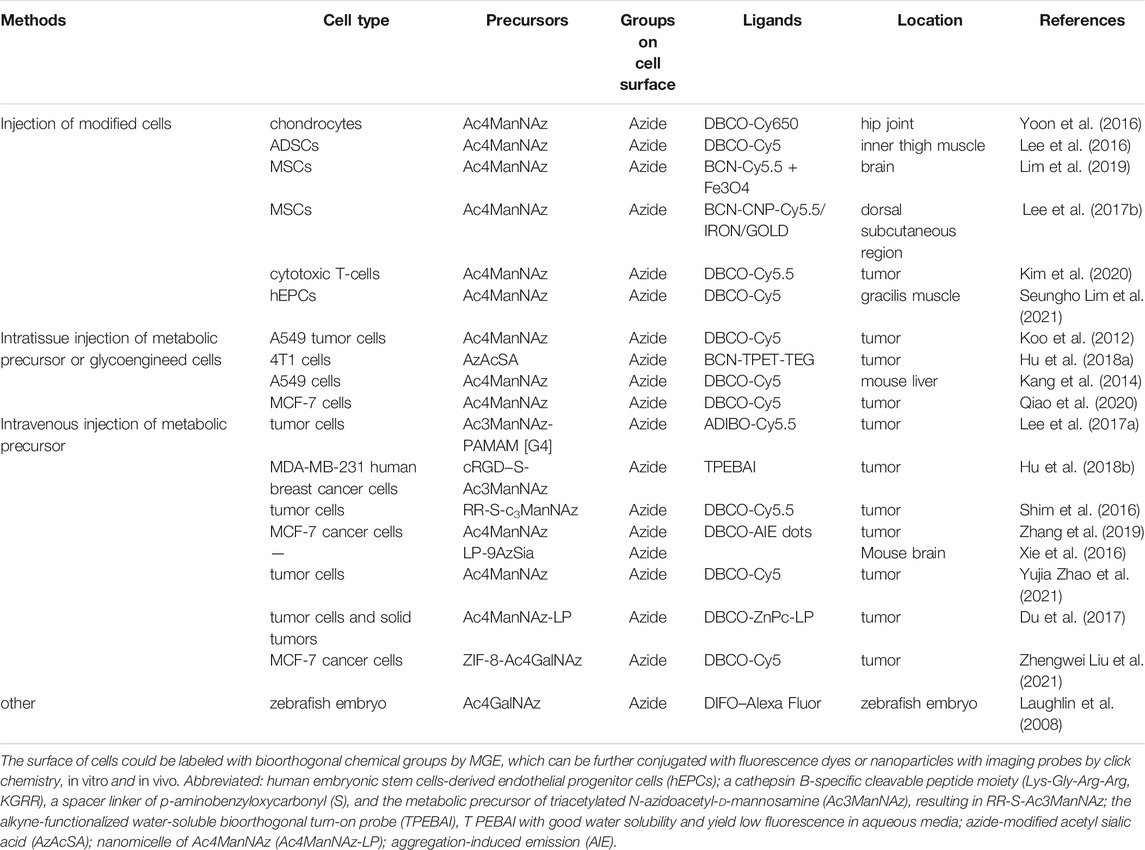
Various targetable chemical groups, such as azides (N3-), alkynes, thiols, and ketones can be successfully generated into cell glycans via MGE (Du et al., 2009). Imaging molecules, like fluorescence dyes or imaging agents, are then labeled with the generated chemical groups by click chemistry. Notably, N3-containing artificial chemical precursors are the most widely used, owing to the fact that these metabolites (N3-groups) can be specifically conjugated with various bioorthogonal agents, such as dibenzylcyclooctyne (DBCO) and bicyclo [6.1.0], nonyne (BCN) via bioorthogonal copper-free click chemistry under both in vitro and in vivo conditions. In addition, their inherent properties, including nontoxicity, high stability, and rapid reaction time under physiological conditions, make them highly suitable for labeling of live cells (Lim et al., 2019). To date, two ways of cell imaging or tracking in vivo have been described. The first involves direct injection of glycoengineered cells labeled with fluorescent dye (Seungho Lim et al., 2021), while the second entails intravenous (Lim et al., 2019), peritoneal (Prescher et al., 2004), or intratumoral (Koo et al., 2012) injection of precursors. After injection, the generated azide groups are able to bind DBCO/BCN group-modified nanoparticles in vivo. These nanoparticles can comprise fluorescence dyes, superparamagnetic iron oxide and so on. Apart from fluorescence, magnetic resonance imaging and computed tomography can also be used for cell imaging. Besides imaging tumors in living animals, cancer diagnosis is also the promising outlook for MGE, where it can be used for identification of biomarkers based on the capture and analysis of bioorthogonally modified glycoconjugates (Badr et al., 2017). Interestingly, Laughlin et al. (2008) achieved three-dimensional spatiotemporal imaging by treating zebrafish embryos with N-azidoacetylgalactosamine (Ac4GalNAz). The azide groups could only be generated on embryo surface that was newly grown and then conjugated different color of Alexa Fluor, which could be used to study embryo development in zebrafish. Overall, this indicates that unnatural groups generated exogenously can be used as target molecules, and can then be applied for imaging of live cells in vivo (Table 2).
Cell-Based Therapy
Cell-based therapy plays a tremendous role in cancer immunotherapy, drug delivery, and tissue regeneration. Modification of cells, coupled with manipulation of their functions based on purposive therapeutic designs have generated numerous scientific interests in biomedical research. Cell-based therapy that alters cell biological activity via MGE has been discussed above (Modulation of Cell Biological Behaviors). This section discusses application of metabolic glycoengineered cells in cancer therapy.
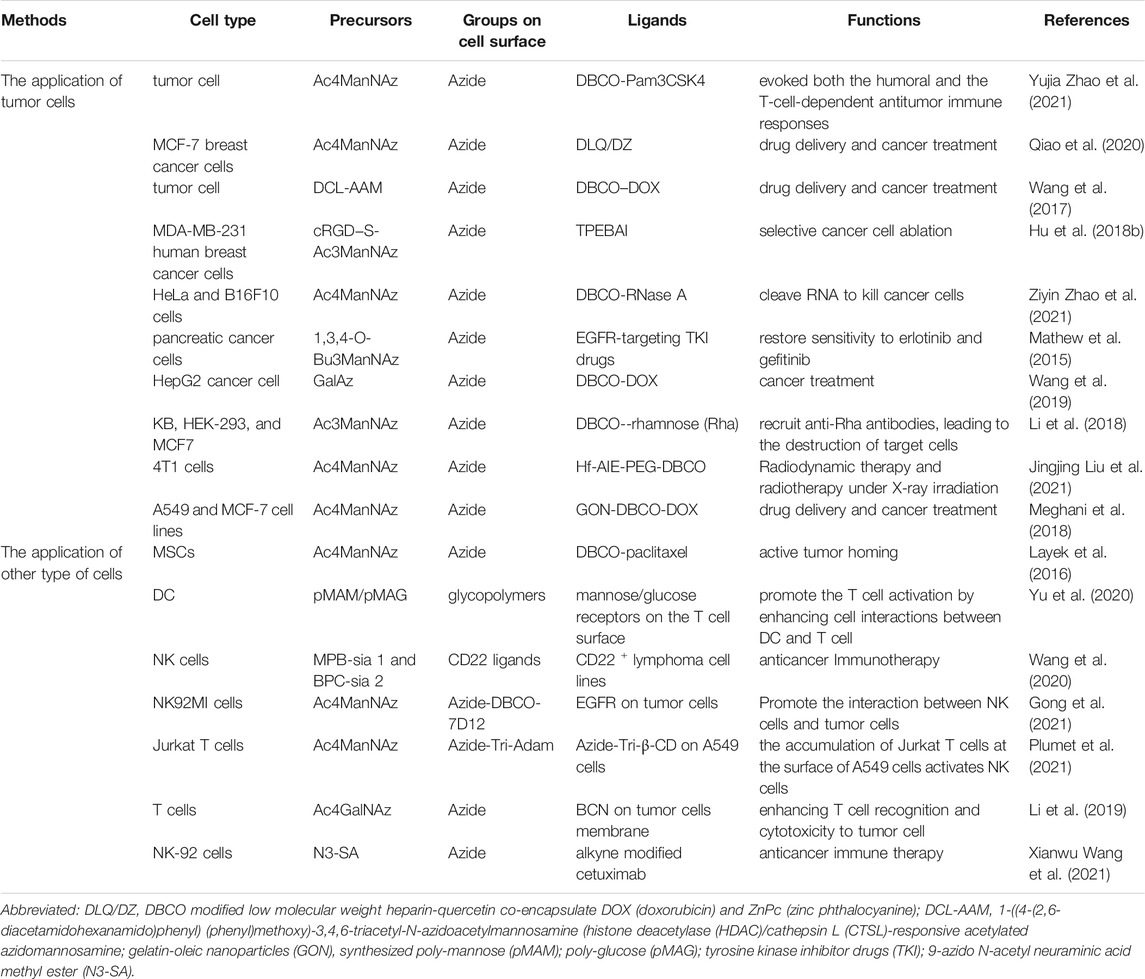
Immunotherapy for cancer treatment is mainly constrained by lack of tumor specific antigens and immune tolerance. Manual introduction of chemical receptors onto the cell surface can well solve these drawbacks. Use of metabolic glycoengineered cells in the field of tumor therapy has been documented. Generally, target non-natural molecules on tumor cell membrane are generated using two approaches, namely intravenous administration (Lee et al., 2017a) and intratumoral injection (Koo et al., 2012) of metabolic precursors. In addition, several methods, such as generating a neoantigen (Yujia Zhao et al., 2021), chemo-photothermal therapy (Du et al., 2017), drug therapy (Layek et al., 2016) and synergistic therapy (Qiao et al., 2020), among others, are applied for tumor treatment.
Metabolic precursors accumulate in tumors usually through tumor-specific enhanced permeability and retention (EPR) effect after i. v. injection (Qiao et al., 2020). Although this method can easily introduce functional groups on the cell surface, it is still a great challenge to selectively label interested cell types in vitro and in vivo. Wang et al. (2017) took advantages of cancer-overexpressed enzymes to selectively cleave caged ether bonds converted by anomeric acetyl groups in tetraacetyl-N-azidoacetylmannosamine (Ac4ManAz) and found azide groups were introduced on the surface of cancer cells. On the other hand, Hu et al. (2018b) utilized over-expressed αvβ3 integrin on tumor cell membranes and developed a dual-responsive metabolic precursor, termed cRGD-S-Ac3ManNAz. Functionally, cRGD can be specifically recognized by αvβ3 integrin, while the disulfide group is responsive to intracellular glutathione. Both of the above methods can selectively generate unnatural glycans on cell membranes.
Apart from modifying tumor cells via MGE, glycoengineered immunocytes can also be used in tumor treatment. Particularly, T cells treated with Ac4GalNAz can introduce N3 on cell surface, thereby specific tumor targeting was initiated through a bio-orthogonal click reaction between N3 and BCN. This artificial targeting strategy has been shown to remarkably promote recognition and migration of T cells to tumor cells, thereby elevating cytotoxicity of T cells against different types of tumor cells by 2–4 times (Li et al., 2019). CAR T-cells neither express sLeX nor bind E-selectin. Results from a previous study showed that fucosylated human CAR-T cells upregulated sLeX expression upon exposure to type 2 sialylLacNAc, a precursor of sLeX. In fact, these fucosylated cells infiltrate the marrow at an efficiency that is 10-fold than that in unfucosylated cells (Mondal et al., 2019). Dendritic cell (DC) modification for enhancement of antigen presentation has emerged as a highly successful strategy in tumor immune therapy. Previous studies demonstrated that synthetic glycopolymer modified on the DC surface through facilitating DC interaction with T cells markedly elevated T cell activation and results in higher tumor toxicity (Yu et al., 2020).
Natural killer (NK) cells are cytotoxic cells with important functions in antitumor immunity, proposed as a promising alternative approaches to extend T cell-based therapy (Mehta and Rezvani, 2018). However, NK cells do not possess inherent targeting abilities towards cancer cells, and are also known to adversely affect endogenous gene uptake, a phenomenon that causes low transgene expression (Matosevic, 2018). CD22, a sialic acid binding protein, is lowly expressed on B cells. Previous studies showed that although antibody-based agents targeting CD22 on B lymphoma and leukemia cells were clinically efficacious against these malignancies, they also attacked normal B cells resulting in immune deficiency (Müller and Nitschke, 2014; Hong et al., 2020). Whereas, the abilities of glycoengineered of NK cells to bind and kill CD22 + lymphoma cells were significantly improved when cells were modified with 9-O modified sialic acid-based CD22 ligands (Haso et al., 2013; Ereño-Orbea et al., 2017). For example, NK cells were found to uptake metabolic precursors MPB-sia one and BPC-sia two and transform them into CD22 ligands through the cellular glycosylation machinery (Wang et al., 2020). Overall, these evidences (Table 3) indicate that cancer immunotherapy was potentiated by providing a simple and general metabolic glycoengineering-based cell therapy.
Applications of Modified Cell Components
Cell component modification is recently applied; thus, this approach is not as common as that of intact cells. In this section, we highlight the modification of cell components in two parts for the first time: extracellular vehicles (EVs) and cell membrane. EVs, endogenously secreted by cells, have bioactive components that correspond to unique biological functions (van den Boorn et al., 2013). Unfortunately, most of bare EVs are excreted by the reticuloendothelial system via the liver and spleen, when they are systemically administered (Smyth et al., 2015). Therefore, understanding biodistribution of EVs in vivo, coupled with their therapeutic efficacy and potential toxicity are imperative to their therapeutic application. Previous studies have shown that glycosylation of EVs can effectively solve the biodistribution of EVs (Williams et al., 2018). For example, Lim et al. decorated EV-secreting donor cells with an azide group via MGE using Ac4ManNAz and prepared DBCO-terminated PEGylated hyaluronic acid (DBCO-PHA) to specifically label the N3 group generated exogenously on the cells. PHA not only prolongs blood circulation, but also exhibits specific binding affinity to CD44 (Choi et al., 2011). Consequently, PHA-decorated EVs were found to accumulate in CD44-abundant tissues, such as rheumatoid arthritis and tumor (Gyeong Taek Lim et al., 2021). In addition, You et al. (2021) reported the fine surface editing of EVs by the MGE of ADSCs to target activated macrophages and promote M1-M2 polarization.
Cell membrane-coating has emerged as a promising nano-delivery system for drugs, owing to various ideal characteristics that include small size, safety, biocompatibility, biorecognition, high stability and target specificity (Zhu et al., 2016; Zhen et al., 2019). Consequently, previous reports have documented the wide application of genetically engineered cell-membrane-coated nanoparticles in cancer immunotherapy and drug delivery (Rao et al., 2020; Xia et al., 2020). Notably, a specific artificial targeting strategy in vitro and in vivo based on MGE-click chemistry was developed with the aim of addressing limitations associated with endogenous protein receptors dependent. For example, Hao Wang et al. (2021) encapsulated photosensitizer IR-780 into the N3-labeled cell membrane, by incubating cells with Ac4ManNAz, and endowed cells in psoriatic lesions with DBCO groups via subcutaneous injection with Ac4ManN-DBCO. The bioorthogonal click chemistry between the N3 and DBCO groups allowed efficient accumulation of IR-780 in lesion skin, thereby promoting photodynamic and photothermal therapy. On the other hand, Xiao et al. (2021) coated an N3-labeled DC membrane on imiquimod-loaded polymeric nanoparticles, and sequentially modified anti-CD3ε antibody via click chemistry. The nanoscale artificial antigen-presenting cells exhibited improved distribution in lymph nodes, and also stimulated T cells and resident antigen-presenting cells.
Applications of Modified Cell-Derived Matrices
In recent years, the application of cell-derived matrices (CDMs) as biomaterials evolves rapidly. CDMs contains complex biomolecules, resulting in its highly bioactivity and biocompatibility. However, the scarcity of specific addressable functional groups greatly hinders its biological application (Ruff et al., 2017). In 2017, Ruff et al. developed a novel method, for the first time, for overcoming this limitation, by specifically using MGE to incorporate azide groups into the cellular glycoconjugates of CDMs (Ruff et al., 2017). Similarly, Gutmann et al.(Gutmann et al., 2018; Gutmann et al., 2019) successfully modified the ECM of NIH3T3 fibroblasts with azide groups using the glucosamine derivate 2-azidoacetylamino-2-deoxy-(1,3,4,6)-tetra-O-acetyl-d-glucopyranoside (Ac4GlcNAz). Recently, Keller et al. (2020) used an azide-modified ECM to create homogeneous, dense, stable and highly bioactive cell substrates which can be used for bioconjugation. At present, azide groups are widely used for CDMs metabolic modification, mainly because of their small size, ease of incorporation, absence in nature, and ability to selectively react with alkynes in bioorthogonal 1,3-dipolar Huisgen cycloadditions.
Prospects
In the near future, we envisage the trend of applications of MGE are as follows: 1) MGE will be used in other new disease models, not just in tumor therapy, such as type 1 diabetes (Au et al., 2021); 2) There will be rapid discovery of new precursors for MGE. Lu et al. (2015) labeled cell surface GPIs and GPI-anchored proteins with artificial inositol derivatives, while Wang et al. (2020) labeled NK cells with CD22 ligands using MPB-sia one and BPC-sia 2; 3) As mentioned above, azide groups are wildly used for MGE, the application of other reporter groups (alkynes, alkenes … ) will be explored in the future; 4) The stability and half-life issue of the functional molecules on the cell could be ameliorated for certain applications, due to cell division and protease degradation; 5) Most of the research in this field has been carried out in cell culture and also in animals. Translating MGE from the laboratory into clinical practice is on the way.
Conclusion
This safe and reversible MGE strategy endows natural cell membrane with additional chemical functionalities, thereby offering unprecedented opportunities for cellular biological functions modulation and novel therapeutics development. This systematic review of recent applications of MGE affirms this approach’s potential in tissue engineering and regenerative medicine. In the future, new chemical reporter groups and bioorthogonal ligation reactions will further expand the development and application of MGE.
Author Contributions
LY, JX: data collecting, writing original draft, revision DH: data collecting QZ, ZH: conceptualization, supervision.
Funding
This work was supported by grants from National Natural Science Foundation of China, Grant/Award Number: 81272059; Medical and Health Technology Program of Zhejiang Province, Grant/Award Number:2012KYA188.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abbina, S., Siren, E. M. J., Moon, H., and Kizhakkedathu, J. N. (2018). Surface Engineering for Cell-Based Therapies: Techniques for Manipulating Mammalian Cell Surfaces. ACS Biomater. Sci. Eng. 4 (11), 3658–3677. doi:10.1021/acsbiomaterials.7b00514
Agatemor, C., Buettner, M. J., Ariss, R., Muthiah, K., Saeui, C. T., and Yarema, K. J. (2019). Exploiting Metabolic Glycoengineering to advance Healthcare. Nat. Rev. Chem. 3 (10), 605–620. doi:10.1038/s41570-019-0126-y
Almaraz, R. T., Tian, Y., Bhattarcharya, R., Tan, E., Chen, S.-H., Dallas, M. R., et al. (2012). Metabolic Flux Increases Glycoprotein Sialylation: Implications for Cell Adhesion and Cancer Metastasis. Mol. Cell Proteomics 11 (7), 017558. doi:10.1074/mcp.M112.017558
Altmann, S., Mut, J., Wolf, N., Meißner-Weigl, J., Rudert, M., Jakob, F., et al. (2021). Metabolic Glycoengineering in hMSC-TERT as a Model for Skeletal Precursors by Using Modified Azide/Alkyne Monosaccharides. Int. J. Mol. Sci. 22 (6), 2820. doi:10.3390/ijms22062820
Au, K. M., Medik, Y., Ke, Q., Tisch, R., and Wang, A. Z. (2021). Immune Checkpoint‐Bioengineered Beta Cell Vaccine Reverses Early‐Onset Type 1 Diabetes. Adv. Mater. 33 (25), 2101253. doi:10.1002/adma.202101253
Badr, H. A., AlSadek, D. M. M., El-Houseini, M. E., Saeui, C. T., Mathew, M. P., Yarema, K. J., et al. (2017). Harnessing Cancer Cell Metabolism for Theranostic Applications Using Metabolic Glycoengineering of Sialic Acid in Breast Cancer as a Pioneering Example. Biomaterials 116, 158–173. doi:10.1016/j.biomaterials.2016.11.044
Choi, K. Y., Yoon, H. Y., Kim, J.-H., Bae, S. M., Park, R.-W., Kang, Y. M., et al. (2011). Smart Nanocarrier Based on PEGylated Hyaluronic Acid for Cancer Therapy. ACS Nano 5 (11), 8591–8599. doi:10.1021/nn202070n
Costa, A. F., Campos, D., Reis, C. A., and Gomes, C. (2020). Targeting Glycosylation: A New Road for Cancer Drug Discovery. Trends Cancer 6 (9), 757–766. doi:10.1016/j.trecan.2020.04.002
Csizmar, C. M., Petersburg, J. R., and Wagner, C. R. (2018). Programming Cell-Cell Interactions through Non-genetic Membrane Engineering. Cel Chem. Biol. 25 (8), 931–940. doi:10.1016/j.chembiol.2018.05.009
Dafik, L., d’Alarcao, M., and Kumar, K. (2008). Fluorination of Mammalian Cell Surfaces via the Sialic Acid Biosynthetic Pathway. Bioorg. Med. Chem. Lett. 18 (22), 5945–5947. doi:10.1016/j.bmcl.2008.09.010
Du, J., Meledeo, M. A., Wang, Z., Khanna, H. S., Paruchuri, V. D. P., and Yarema, K. J. (2009). Metabolic Glycoengineering: Sialic Acid and beyond. Glycobiology 19 (12), 1382–1401. doi:10.1093/glycob/cwp115
Du, J., Che, P.-L., Wang, Z.-Y., Aich, U., and Yarema, K. J. (2011). Designing a Binding Interface for Control of Cancer Cell Adhesion via 3D Topography and Metabolic Oligosaccharide Engineering. Biomaterials 32 (23), 5427–5437. doi:10.1016/j.biomaterials.2011.04.005
Du, J., Agatemor, C., Saeui, C. T., Bhattacharya, R., Jia, X., and Yarema, K. J. (2021). Glycoengineering Human Neural and Adipose Stem Cells with Novel Thiol-Modified N-Acetylmannosamine (ManNAc) Analogs. Cells 10 (2), 377. doi:10.3390/cells10020377
Du, J., and Yarema, K. J. (2010). Carbohydrate Engineered Cells for Regenerative Medicine☆. Adv. Drug Deliv. Rev. 62 (7-8), 671–682. doi:10.1016/j.addr.2010.01.003
Du, L., Qin, H., Ma, T., Zhang, T., and Xing, D. (2017). In Vivo Imaging-Guided Photothermal/Photoacoustic Synergistic Therapy with Bioorthogonal Metabolic Glycoengineering-Activated Tumor Targeting Nanoparticles. ACS Nano 11 (9), 8930–8943. doi:10.1021/acsnano.7b03226
Dykstra, B., Lee, J., Mortensen, L. J., Yu, H., Wu, Z. L., Lin, C. P., et al. (2016). Glycoengineering of E-Selectin Ligands by Intracellular versus Extracellular Fucosylation Differentially Affects Osteotropism of Human Mesenchymal Stem Cells. Stem Cells 34 (10), 2501–2511. doi:10.1002/stem.2435
Ereño-Orbea, J., Sicard, T., Cui, H., Mazhab-Jafari, M. T., Benlekbir, S., Guarné, A., et al. (2017). Molecular Basis of Human CD22 Function and Therapeutic Targeting. Nat. Commun. 8 (1), 764. doi:10.1038/s41467-017-00836-6
Gong, L., Li, Y., Cui, K., Chen, Y., Hong, H., Li, J., et al. (2021). Nanobody‐Engineered Natural Killer Cell Conjugates for Solid Tumor Adoptive Immunotherapy. Small 17 (45), 2103463. doi:10.1002/smll.202103463
Gutmann, M., Bechold, J., Seibel, J., Meinel, L., and Lühmann, T. (2019). Metabolic Glycoengineering of Cell-Derived Matrices and Cell Surfaces: A Combination of Key Principles and Step-by-step Procedures. ACS Biomater. Sci. Eng. 5 (1), 215–233. doi:10.1021/acsbiomaterials.8b00865
Gutmann, M., Braun, A., Seibel, J., and Lühmann, T. (2018). Bioorthogonal Modification of Cell Derived Matrices by Metabolic Glycoengineering. ACS Biomater. Sci. Eng. 4 (4), 1300–1306. doi:10.1021/acsbiomaterials.8b00264
Gyeong Taek Lim, G. T., You, D. G., Han, H. S., Lee, H., Shin, S., Oh, B. H., et al. (2021). Bioorthogonally Surface‐edited Extracellular Vesicles Based on Metabolic Glycoengineering for CD44‐mediated Targeting of Inflammatory Diseases. J. Extracellular Vesicles 10 (5), e12077. doi:10.1002/jev2.12077
Hao Wang, H., Su, D., Huang, R., Shu, F., Cheng, F., and Zheng, G. (2021). Cellular Nanovesicles with Bioorthogonal Targeting Enhance Photodynamic/photothermal Therapy in Psoriasis. Acta Biomater. 134, 674–685. doi:10.1016/j.actbio.2021.07.068
Haso, W., Lee, D. W., Shah, N. N., Stetler-Stevenson, M., Yuan, C. M., Pastan, I. H., et al. (2013). Anti-CD22-chimeric Antigen Receptors Targeting B-Cell Precursor Acute Lymphoblastic Leukemia. Blood 121 (7), 1165–1174. doi:10.1182/blood-2012-06-438002
Hong, S., Yu, C., Wang, P., Shi, Y., Cao, W., Cheng, B., et al. (2020). Glycoengineering of NK Cells with Glycan Ligands of CD22 and Selectins for B‐Cell Lymphoma Therapy. Angew. Chem. Int. Ed. 60, 3603–3610. doi:10.1002/anie.202005934
Horstkorte, R., Rau, K., Laabs, S., Danker, K., and Reutter, W. (2004). Biochemical Engineering of theN-Acyl Side Chain of Sialic Acid Leads to Increased Calcium Influx from Intracellular Compartments and Promotes Differentiation of HL60 Cells. FEBS Lett. 571 (1-3), 99–102. doi:10.1016/j.febslet.2004.06.067
Horstkorte, R., Reinke, S., Bauer, C., Reutter, W., and Kontou, M. (2010). N-Propionylmannosamine-induced Over-expression and Secretion of Thioredoxin Leads to Neurite Outgrowth of PC12 Cells. Biochem. Biophysical Res. Commun. 395 (3), 296–300. doi:10.1016/j.bbrc.2010.03.113
Hu, F., Mao, D., KenryCai, X., Cai, X., Wu, W., Kong, D., et al. (2018a). A Light-Up Probe with Aggregation-Induced Emission for Real-Time Bio-Orthogonal Tumor Labeling and Image-Guided Photodynamic Therapy. Angew. Chem. Int. Ed. 57 (32), 10182–10186. doi:10.1002/anie.201805446
Hu, F., Yuan, Y., Wu, W., Mao, D., and Liu, B. (2018b). Dual-Responsive Metabolic Precursor and Light-Up AIEgen for Cancer Cell Bio-Orthogonal Labeling and Precise Ablation. Anal. Chem. 90 (11), 6718–6724. doi:10.1021/acs.analchem.8b00547
Jingjing Liu, J., Hu, F., Wu, M., Tian, L., Gong, F., Zhong, X., et al. (2021). Bioorthogonal Coordination Polymer Nanoparticles with Aggregation‐Induced Emission for Deep Tumor‐Penetrating Radio‐ and Radiodynamic Therapy. Adv. Mater. 33 (9), 2007888. doi:10.1002/adma.202007888
Kang, S.-W., Lee, S., Na, J. H., Yoon, H. I., Lee, D.-E., Koo, H., et al. (2014). Cell Labeling and Tracking Method without Distorted Signals by Phagocytosis of Macrophages. Theranostics 4 (4), 420–431. doi:10.7150/thno.7265
Keller, S., Wörgötter, K., Liedek, A., Kluger, P. J., Bach, M., Tovar, G. E. M., et al. (2020). Azide-Functional Extracellular Matrix Coatings as a Bioactive Platform for Bioconjugation. ACS Appl. Mater. Inter. 12 (24), 26868–26879. doi:10.1021/acsami.0c04579
Kim, H., Shin, K., Park, O. K., Choi, D., Kim, H. D., Baik, S., et al. (2018). General and Facile Coating of Single Cells via Mild Reduction. J. Am. Chem. Soc. 140 (4), 1199–1202. doi:10.1021/jacs.7b08440
Kim, W., Yoon, H. Y., Lim, S., Stayton, P. S., Kim, I.-S., Kim, K., et al. (2021). In Vivo tracking of Bioorthogonally Labeled T-Cells for Predicting Therapeutic Efficacy of Adoptive T-Cell Therapy. J. Controlled Release 329, 223–236. doi:10.1016/j.jconrel.2020.12.002
Kontou, M., Bauer, C., Reutter, W., and Horstkorte, R. (2008). Sialic Acid Metabolism Is Involved in the Regulation of Gene Expression during Neuronal Differentiation of PC12 Cells. Glycoconj J. 25 (3), 237–244. doi:10.1007/s10719-008-9104-1
Koo, H., Lee, S., Na, J. H., Kim, S. H., Hahn, S. K., Choi, K., et al. (2012). Bioorthogonal Copper-free Click Chemistry In Vivo for Tumor-Targeted Delivery of Nanoparticles. Angew. Chem. Int. Ed. 51 (47), 11836–11840. doi:10.1002/anie.201206703
Koo, H., Choi, M., Kim, E., Hahn, S. K., Weissleder, R., and Yun, S. H. (2015). Bioorthogonal Click Chemistry-Based Synthetic Cell Glue. Small 11 (48), 6458–6466. doi:10.1002/smll.201502972
Koo, H., Hahn, S. K., and Yun, S. H. (2016). Controlled Detachment of Chemically Glued Cells. Bioconjug. Chem. 27 (11), 2601–2604. doi:10.1021/acs.bioconjchem.6b00546
Kuehn, B. M. (2017). The Promise and Challenges of CAR-T Gene Therapy. JAMA 318 (22), 2167–2169. doi:10.1001/jama.2017.15605
Laughlin, S. T., Baskin, J. M., Amacher, S. L., and Bertozzi, C. R. (2008). In Vivo imaging of Membrane-Associated Glycans in Developing Zebrafish. Science 320 (5876), 664–667. doi:10.1126/science.1155106
Layek, B., Sadhukha, T., and Prabha, S. (2016). Glycoengineered Mesenchymal Stem Cells as an Enabling Platform for Two-step Targeting of Solid Tumors. Biomaterials 88, 97–109. doi:10.1016/j.biomaterials.2016.02.024
Lee, S. Y., Lee, S., Lee, J., Yhee, J. Y., Yoon, H. I., Park, S.-J., et al. (2016). Non-invasive Stem Cell Tracking in Hindlimb Ischemia Animal Model Using Bio-Orthogonal Copper-free Click Chemistry. Biochem. Biophysical Res. Commun. 479 (4), 779–786. doi:10.1016/j.bbrc.2016.09.132
Lee, S., Jung, S., Koo, H., Na, J. H., Yoon, H. Y., Shim, M. K., et al. (2017a). Nano-sized Metabolic Precursors for Heterogeneous Tumor-Targeting Strategy Using Bioorthogonal Click Chemistry In Vivo. Biomaterials 148, 1–15. doi:10.1016/j.biomaterials.2017.09.025
Lee, S., Yoon, H. I., Na, J. H., Jeon, S., Lim, S., Koo, H., et al. (2017b). In Vivo stem Cell Tracking with Imageable Nanoparticles that Bind Bioorthogonal Chemical Receptors on the Stem Cell Surface. Biomaterials 139, 12–29. doi:10.1016/j.biomaterials.2017.05.050
Lee, D. Y., Cha, B.-H., Jung, M., Kim, A. S., Bull, D. A., and Won, Y.-W. (2018). Cell Surface Engineering and Application in Cell Delivery to Heart Diseases. J. Biol. Eng. 12, 28. doi:10.1186/s13036-018-0123-6
Li, S., Yu, B., Wang, J., Zheng, Y., Zhang, H., Walker, M. J., et al. (2018). Biomarker-Based Metabolic Labeling for Redirected and Enhanced Immune Response. ACS Chem. Biol. 13 (6), 1686–1694. doi:10.1021/acschembio.8b00350
Li, W., Pan, H., He, H., Meng, X., Ren, Q., Gong, P., et al. (2019). Bio-Orthogonal T Cell Targeting Strategy for Robustly Enhancing Cytotoxicity against Tumor Cells. Small 15 (4), 1804383. doi:10.1002/smll.201804383
Lim, S., Yoon, H. Y., Jang, H. J., Song, S., Kim, W., Park, J., et al. (2019). Dual-Modal Imaging-Guided Precise Tracking of Bioorthogonally Labeled Mesenchymal Stem Cells in Mouse Brain Stroke. ACS Nano 13 (10), 10991–11007. doi:10.1021/acsnano.9b02173
Lo, C. Y., Weil, B. R., Palka, B. A., Momeni, A., Canty, J. M., and Neelamegham, S. (2016). Cell Surface Glycoengineering Improves Selectin-Mediated Adhesion of Mesenchymal Stem Cells (MSCs) and Cardiosphere-Derived Cells (CDCs): Pilot Validation in Porcine Ischemia-Reperfusion Model. Biomaterials 74, 19–30. doi:10.1016/j.biomaterials.2015.09.026
Lu, L., Gao, J., and Guo, Z. (2015). Labeling Cell Surface GPIs and GPI-Anchored Proteins through Metabolic Engineering with Artificial Inositol Derivatives. Angew. Chem. Int. Ed. 54 (33), 9679–9682. doi:10.1002/anie.201503814
Mahal, L. K., Charter, N. W., Angata, K., Fukuda, M., Koshland, D. E., and Bertozzi, C. R. (2001). A Small-Molecule Modulator of Poly-Α2,8-Sialic Acid Expression on Cultured Neurons and Tumor Cells. Science 294 (5541), 380–381. doi:10.1126/science.1062192
Mao, D., Zhang, C., KenryLiu, J., Liu, J., Wang, X., Li, B., et al. (2020). Bio-orthogonal Click Reaction-Enabled Highly Specific In Situ Cellularization of Tissue Engineering Scaffolds. Biomaterials 230, 119615. doi:10.1016/j.biomaterials.2019.119615
Mathew, M. P., Tan, E., Saeui, C. T., Bovonratwet, P., Liu, L., Bhattacharya, R., et al. (2015). Metabolic Glycoengineering Sensitizes Drug-Resistant Pancreatic Cancer Cells to Tyrosine Kinase Inhibitors Erlotinib and Gefitinib. Bioorg. Med. Chem. Lett. 25 (6), 1223–1227. doi:10.1016/j.bmcl.2015.01.060
Matosevic, S. (2018). Viral and Nonviral Engineering of Natural Killer Cells as Emerging Adoptive Cancer Immunotherapies. J. Immunol. Res. 2018, 1–20. doi:10.1155/2018/4054815
Meghani, N. M., Amin, H. H., Park, C., Park, J.-B., Cui, J.-H., Cao, Q.-R., et al. (2018). Design and Evaluation of Clickable Gelatin-Oleic Nanoparticles Using Fattigation-Platform for Cancer Therapy. Int. J. Pharmaceutics 545 (1-2), 101–112. doi:10.1016/j.ijpharm.2018.04.047
Mehta, R. S., and Rezvani, K. (2018). Chimeric Antigen Receptor Expressing Natural Killer Cells for the Immunotherapy of Cancer. Front. Immunol. 9, 283. doi:10.3389/fimmu.2018.00283
Merzaban, J. S., Imitola, J., Starossom, S. C., Zhu, B., Wang, Y., Lee, J., et al. (2015). Cell Surface Glycan Engineering of Neural Stem Cells Augments Neurotropism and Improves Recovery in a Murine Model of Multiple Sclerosis. Glycobiology 25 (12), 1392–1409. doi:10.1093/glycob/cwv046
Mondal, N., Silva, M., Castano, A. P., Maus, M. V., and Sackstein, R. (2019). Glycoengineering of Chimeric Antigen Receptor (CAR) T-Cells to Enforce E-Selectin Binding. J. Biol. Chem. 294 (48), 18465–18474. doi:10.1074/jbc.RA119.011134
Müller, J., and Nitschke, L. (2014). The Role of CD22 and Siglec-G in B-Cell Tolerance and Autoimmune Disease. Nat. Rev. Rheumatol. 10 (7), 422–428. doi:10.1038/nrrheum.2014.54
Nagasundaram, M., Horstkorte, R., and Gnanapragassam, V. S. (2020). Sialic Acid Metabolic Engineering of Breast Cancer Cells Interferes with Adhesion and Migration. Molecules 25 (11), 2632. doi:10.3390/molecules25112632
Natunen, S., Lampinen, M., Suila, H., Ritamo, I., Pitkänen, V., Nairn, A. V., et al. (2013). Metabolic Glycoengineering of Mesenchymal Stromal Cells with N-Propanoylmannosamine. Glycobiology 23 (8), 1004–1012. doi:10.1093/glycob/cwt039
Nischan, N., and Kohler, J. J. (2016). Advances in Cell Surface Glycoengineering Reveal Biological Function. Glycobiology 26 (8), 789–796. doi:10.1093/glycob/cww045
Oh, B., Swaminathan, V., Malkovskiy, A., Santhanam, S., McConnell, K., and George, P. M. (2020). Single‐Cell Encapsulation via Click‐Chemistry Alters Production of Paracrine Factors from Neural Progenitor Cells. Adv. Sci. 7 (8), 1902573. doi:10.1002/advs.201902573
Plumet, C., Mohamed, A. S., Vendeuvre, T., Renoux, B., Clarhaut, J., and Papot, S. (2021). Cell-cell Interactions via Non-covalent Click Chemistry. Chem. Sci. 12 (26), 9017–9021. doi:10.1039/d1sc01637g
Prescher, J. A., Dube, D. H., and Bertozzi, C. R. (2004). Chemical Remodelling of Cell Surfaces in Living Animals. Nature 430 (7002), 873–877. doi:10.1038/nature02791
Qiao, J., Tian, F., Deng, Y., Shang, Y., Chen, S., Chang, E., et al. (2020). Bio-orthogonal Click-Targeting Nanocomposites for Chemo-Photothermal Synergistic Therapy in Breast Cancer. Theranostics 10 (12), 5305–5321. doi:10.7150/thno.42445
Rao, L., Zhao, S. K., Wen, C., Tian, R., Lin, L., Cai, B., et al. (2020). Activating Macrophage‐Mediated Cancer Immunotherapy by Genetically Edited Nanoparticles. Adv. Mater. 32, 2004853. doi:10.1002/adma.202004853
Ruff, S. M., Keller, S., Wieland, D. E., Wittmann, V., Tovar, G. E. M., Bach, M., et al. (2017). clickECM: Development of a Cell-Derived Extracellular Matrix with Azide Functionalities. Acta Biomater. 52, 159–170. doi:10.1016/j.actbio.2016.12.022
Sampathkumar, S.-G., Li, A. V., Jones, M. B., Sun, Z., and Yarema, K. J. (2006). Metabolic Installation of Thiols into Sialic Acid Modulates Adhesion and Stem Cell Biology. Nat. Chem. Biol. 2 (3), 149–152. doi:10.1038/nchembio770
Sánchez‐Martínez, D., Gutiérrez‐Agüera, F., Romecin, P., Vinyoles, M., Palomo, M., Tirado, N., et al. (2021). Enforced sialyl‐Lewis‐X (sLeX) Display in E‐selectin Ligands by Exofucosylation Is Dispensable for CD19‐CAR T‐cell Activity and Bone Marrow Homing. Clin. Translational Med. 11 (2), e280. doi:10.1002/ctm2.280
Schmidt, C., Stehling, P., Schnitzer, J., Reutter, W., and Horstkorte, R. (1998). Biochemical Engineering of Neural Cell Surfaces by the SyntheticN-Propanoyl-Substituted Neuraminic Acid Precursor. J. Biol. Chem. 273 (30), 19146–19152. doi:10.1074/jbc.273.30.19146
Seungho Lim, S., Yoon, H. Y., Park, S.-J., Song, S., Shim, M. K., Yang, S., et al. (2021). Predicting In Vivo Therapeutic Efficacy of Bioorthogonally Labeled Endothelial Progenitor Cells in Hind Limb Ischemia Models via Non-invasive Fluorescence Molecular Tomography. Biomaterials 266, 120472. doi:10.1016/j.biomaterials.2020.120472
Shim, M. K., Yoon, H. Y., Ryu, J. H., Koo, H., Lee, S., Park, J. H., et al. (2016). Cathepsin B‐Specific Metabolic Precursor for In Vivo Tumor‐Specific Fluorescence Imaging. Angew. Chem. Int. Ed. 55 (47), 14698–14703. doi:10.1002/anie.20160850410.1002/anie.201608504
Smyth, T., Kullberg, M., Malik, N., Smith-Jones, P., Graner, M. W., and Anchordoquy, T. J. (2015). Biodistribution and Delivery Efficiency of Unmodified Tumor-Derived Exosomes. J. Controlled Release 199, 145–155. doi:10.1016/j.jconrel.2014.12.013
Sugimoto, S., and Iwasaki, Y. (2018). Surface Modification of Macrophages with Nucleic Acid Aptamers for Enhancing the Immune Response against Tumor Cells. Bioconjug. Chem. 29 (12), 4160–4167. doi:10.1021/acs.bioconjchem.8b00793
Templeton, K., Ramos, M., Rose, J., Le, B., Zhou, Q., Cressman, A., et al. (2021). Mesenchymal Stromal Cells Regulate Sialylations of N-Glycans, Affecting Cell Migration and Survival. Int. J. Mol. Sci. 22 (13), 6868. doi:10.3390/ijms22136868
van den Boorn, J. G., Dassler, J., Coch, C., Schlee, M., and Hartmann, G. (2013). Exosomes as Nucleic Acid Nanocarriers. Adv. Drug Deliv. Rev. 65 (3), 331–335. doi:10.1016/j.addr.2012.06.011
Villavicencio-Lorini, P., Laabs, S., Danker, K., Reutter, W., and Horstkorte, R. x. d. (2002). Biochemical Engineering of the Acyl Side Chain of Sialic Acids Stimulates Integrin-dependent Adhesion of HL60 Cells to Fibronectin. J. Mol. Med. 80 (10), 671–677. doi:10.1007/s00109-002-0382-y
Wang, H., and Mooney, D. J. (2020). Metabolic Glycan Labelling for Cancer-Targeted Therapy. Nat. Chem. 12 (12), 1102–1114. doi:10.1038/s41557-020-00587-w
Wang, H., Wang, R., Cai, K., He, H., Liu, Y., Yen, J., et al. (2017). Selective In Vivo Metabolic Cell-Labeling-Mediated Cancer Targeting. Nat. Chem. Biol. 13 (4), 415–424. doi:10.1038/nchembio.2297
Wang, H., Liu, Y., Xu, M., and Cheng, J. (2019). Azido-galactose Outperforms Azido-Mannose for Metabolic Labeling and Targeting of Hepatocellular Carcinoma. Biomater. Sci. 7 (10), 4166–4173. doi:10.1039/c9bm00898e
Wang, X., Lang, S., Tian, Y., Zhang, J., Yan, X., Fang, Z., et al. (2020). Glycoengineering of Natural Killer Cells with CD22 Ligands for Enhanced Anticancer Immunotherapy. ACS Cent. Sci. 6 (3), 382–389. doi:10.1021/acscentsci.9b00956
Williams, C., Royo, F., Aizpurua-Olaizola, O., Pazos, R., Boons, G.-J., Reichardt, N.-C., et al. (2018). Glycosylation of Extracellular Vesicles: Current Knowledge, Tools and Clinical Perspectives. J. Extracellular Vesicles 7 (1), 1442985. doi:10.1080/20013078.2018.1442985
Wratil, P. R., Horstkorte, R., and Reutter, W. (2016). Metabolic Glycoengineering withN-Acyl Side Chain Modified Mannosamines. Angew. Chem. Int. Ed. 55 (33), 9482–9512. doi:10.1002/anie.201601123
Xia, Y., Rao, L., Yao, H., Wang, Z., Ning, P., and Chen, X. (2020). Engineering Macrophages for Cancer Immunotherapy and Drug Delivery. Adv. Mater. 32 (40), 2002054. doi:10.1002/adma.202002054
Xianwu Wang, X., Luo, X., Tian, Y., Wu, T., Weng, J., Li, Z., et al. (2021). Equipping Natural Killer Cells with Cetuximab through Metabolic Glycoengineering and Bioorthogonal Reaction for Targeted Treatment of KRAS Mutant Colorectal Cancer. ACS Chem. Biol. 16 (4), 724–730. doi:10.1021/acschembio.1c00022
Xiao, P., Wang, J., Zhao, Z., Liu, X., Sun, X., Wang, D., et al. (2021). Engineering Nanoscale Artificial Antigen-Presenting Cells by Metabolic Dendritic Cell Labeling to Potentiate Cancer Immunotherapy. Nano Lett. 21 (5), 2094–2103. doi:10.1021/acs.nanolett.0c04783
Xie, R., Dong, L., Du, Y., Zhu, Y., Hua, R., Zhang, C., et al. (2016). In Vivo metabolic Labeling of Sialoglycans in the Mouse Brain by Using a Liposome-Assisted Bioorthogonal Reporter Strategy. Proc. Natl. Acad. Sci. USA 113 (19), 5173–5178. doi:10.1073/pnas.1516524113
Yoon, H. I., Yhee, J. Y., Na, J. H., Lee, S., Lee, H., Kang, S.-W., et al. (2016). Bioorthogonal Copper Free Click Chemistry for Labeling and Tracking of Chondrocytes In Vivo. Bioconjug. Chem. 27 (4), 927–936. doi:10.1021/acs.bioconjchem.6b00010
You, D. G., Lim, G. T., Kwon, S., Um, W., Oh, B. H., Song, S. H., et al. (2021). Metabolically Engineered Stem Cell-Derived Exosomes to Regulate Macrophage Heterogeneity in Rheumatoid Arthritis. Sci. Adv. 7 (23), eabe0083. doi:10.1126/sciadv.abe0083
Yu, L., Feng, R., Zhu, L., Hao, Q., Chu, J., Gu, Y., et al. (2020). Promoting the Activation of T Cells with Glycopolymer-Modified Dendritic Cells by Enhancing Cell Interactions. Sci. Adv. 6, eabb6595. doi:10.1126/sciadv.abb6595
Yujia Zhao, Y., Li, S., Lv, J., Liu, Y., Chen, Y., Liu, Y., et al. (2021). Generation of Triacyl Lipopeptide-Modified Glycoproteins by Metabolic Glycoengineering as the Neoantigen to Boost Anti-tumor Immune Response. Theranostics 11 (15), 7425–7438. doi:10.7150/thno.60211
Zhang, P., Jiang, T., Li, Y., Zhao, Z., Gong, P., Cai, L., et al. (2019). Bio‐orthogonal AIE Dots Based on Polyyne‐Bridged Red‐emissive AIEgen for Tumor Metabolic Labeling and Targeted Imaging. Chem. Asian J. 14 (6), 770–774. doi:10.1002/asia.201801609
Zhen, X., Cheng, P., and Pu, K. (2019). Recent Advances in Cell Membrane-Camouflaged Nanoparticles for Cancer Phototherapy. Small 15 (1), 1804105. doi:10.1002/smll.201804105
Zhengwei Liu, Z., Zhang, L., Cui, T., Ma, M., Ren, J., and Qu, X. (2021). A Nature‐Inspired Metal-Organic Framework Discriminator for Differential Diagnosis of Cancer Cell Subtypes. Angew. Chem. Int. Ed. 60 (28), 15436–15444. doi:10.1002/anie.202102286
Zhu, J.-Y., Zheng, D.-W., Zhang, M.-K., Yu, W.-Y., Qiu, W.-X., Hu, J.-J., et al. (2016). Preferential Cancer Cell Self-Recognition and Tumor Self-Targeting by Coating Nanoparticles with Homotypic Cancer Cell Membranes. Nano Lett. 16 (9), 5895–5901. doi:10.1021/acs.nanolett.6b02786
Keywords: metabolic glycoengineering., cell-based therapy, cell membrane modification, ManNAc analogs, imaging
Citation: Ying L, Xu J, Han D, Zhang Q and Hong Z (2022) The Applications of Metabolic Glycoengineering. Front. Cell Dev. Biol. 10:840831. doi: 10.3389/fcell.2022.840831
Received: 21 December 2021; Accepted: 28 January 2022;
Published: 17 February 2022.
Edited by:
Ayano Satoh, Okayama University, JapanReviewed by:
Hideaki Mabashi-Asazuma, Ochanomizu University, JapanRyo Okamoto, Osaka University, Japan
Copyright © 2022 Ying, Xu, Han, Zhang and Hong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qingguo Zhang, emhhbmdxZ0BlbnplbWVkLmNvbQ==; Zhenghua Hong, MDAwMWh6aEAxNjMuY29t
†These authors have contributed equally to this work
 Liwei Ying
Liwei Ying Junxi Xu
Junxi Xu Dawei Han
Dawei Han Qingguo Zhang
Qingguo Zhang Zhenghua Hong1,2*
Zhenghua Hong1,2*