- 1Key Lab of Freshwater Animal Breeding, College of Fisheries, Ministry of Agriculture and Rural Affairs/Key Lab of Agricultural Animal Genetics, Breeding and Reproduction of Ministry of Education/Engineering Technology Research Center for Fish Breeding and Culture in Hubei Province/Engineering Research Center of Green Development for Conventional Aquatic Biological Industry in the Yangtze River Economic Belt of Ministry of Education, Huazhong Agricultural University, Wuhan, China
- 2Hubei Hongshan Laboratory, Wuhan, China
- 3CAS Key Laboratory of Tropical Marine Bio-Resources and Ecology, South China Sea Institute of Oceanology, Innovation Academy of South China Sea Ecology and Environmental Engineering, Chinese Academy of Sciences, Guangzhou, China
- 4Guangdong Laboratory for Lingnan Modern Agriculture, Guangzhou, China
Bone morphogenetic protein 7 (BMP7) belongs to the transforming growth factor β (TGF-β) family, which not only induces cartilage and bone formation, but also regulates eye development and melanoma tumorigenesis in mammals. In teleosts, BMP7 differentiates into two subtypes, bmp7a and bmp7b, which have clearly differentiated structures. To fully understand the functional differentiation of bmp7a and bmp7b in fish species, we successfully constructed bmp7a and bmp7b gene deletion mutants in zebrafish using CRISPR/Cas9-mediated gene editing technology. Our results showed that bmp7a mutation caused abnormal development of the embryo’s dorsal-ventral pattern that led to death; bmp7b mutation induced growth inhibition and increased melanin production in the skin and eye of mutants. Histological analysis revealed that melanin in the retina of the eyes in bmp7b mutants increased, and behavioral observation showed that the vision and sensitivity to food of the mutants were reduced. Transcriptome analysis of the skin and eye tissues showed that the expression changes of wnt7ba and gna14 in bmp7b mutants might promote the increase of melanin. Additionally, the eye transcriptome analysis indicated that changes in the structure of the eyes in bmp7b mutants led to defects in phototransduction, and seven DEGs (rgs9a, rgs9b, rcvrn2, guca1d, grk1b, opn1mw4, and gc2) were identified as key candidate genes that affected the photonic response of the eyes. The study revealed the functional differentiation of bmp7a and bmp7b in teleosts and the first report about the inhibitory effect of bmp7b on melanogenesis may provide useful information for the future research on human melanoma-related diseases.
Introduction
Bone morphogenetic protein 7 [BMP7 (OP-1)], a member of the transforming growth factor-β (TGF-β) superfamily, is a 35 kDa homodimeric protein that is involved in the differentiation of mesenchymal cells into osteoblasts and promotes ectopic bone formation (Celeste et al., 1990; Ozkaynak et al., 1990; Ducy and Karsenty, 2000). BMP7 mainly binds to type I and type II BMP receptors expressed on the cell surface to activate receptor-regulated SMADs (R-SMADs, including SMAD1, 2, 3, 5, and 8). The activated R-SMADs interact with SMAD4 (Co-SMAD) to form a complex, which then translocates to the nucleus to regulate the transcription of the target gene (Bragdon et al., 2011). In addition to bone formation, previous studies have proved that BMP7 also has an indispensable role in the growth, differentiation, and apoptosis of various tissues (Ducy and Karsenty, 2000). In humans and mammals, BMP7 is involved in the regulation of hair follicle development and hair growth (Lv et al., 2016), and the size and spatial distribution of hair (Michon et al., 2008). In the early development of birds, BMP7 signaling mediates the ability of the epidermis to respond to inductive signals from the dermis to form feathers and scales (Harris et al., 2004). In recent years, BMP7 was also shown to be involved in regulating the production of melanin and was strongly expressed in metastatic and primary melanoma (Rothhammer et al., 2005; Vitic et al., 2021).
Furthermore, BMP7 is crucial for numerous biological processes in vertebrates. For example, the bmp7 gene plays an osteoinductive role in the development and reconstruction of human and mouse teeth and alveolar bone (Zouvelou et al., 2009; Kim et al., 2019); 25% of bmp7 homozygous mutant mice showed small size, eye deformities, kidney hypoplasia, and anterior axis polydactyly on the hind limbs and forelimbs of certain embryos, and died 48 h after birth (Luo et al., 1995). A previous study had reported that during evolution, two BMP7 orthologs, bmp7a and bmp7b, have emerged in some bony fish such as zebrafish (Danio rerio) (Shawi and Serluca, 2008). The bmp7a and bmp7b may have functional differentiation in fish species, loss of BMP7 gene function or acquisition of new functions. Until now, researches on the BMP7 gene in fish is limited, and the dynamic changes during fish development are unclear. In zebrafish, bmp7a could form heterodimers with other proteins in the same family and played an important role in the development of dorsal-ventral axis of the embryos (Schmid et al., 2000), while bmp7b was expressed in several structures, including the digestive system, fin, optic cup and sensory system (Shawi and Serluca, 2008). Although the gene expression data indicated that BMP7 may play a role in the development of many organs and tissues of vertebrates, mostly previous researches had been focused on the eyes and bones development of mammals. The role of BMP7 in teleosts is not clear and needs to be investigated to improve our understanding of BMP7 functions in vertebrates.
Here, we describe the differential expression of bmp7a and bmp7b in embryonic, juvenile, and adult zebrafish. In order to explore the roles of bmp7a and bmp7b in fish development and growth, we produced bmp7a and bmp7b mutant zebrafish through using CRISPR/Cas9 gene editing technology. The bmp7a mutants died during the gastrulation stage due to defective development of dorsal-ventral axis. The bmp7b mutants were found to be viable but showed slow growth, and the color of their body and iris became darker throughout the growth and development process. In addition, defects in the retinal structure of the eyes led to slower feeding responses of the bmp7b mutant zebrafish. Comparative transcriptome analysis was conducted to reveal the possible molecular regulation about the impact of bmp7b on skin melanin production and eye structural defects. The study helps us understand the functional differentiation of bmp7a and bmp7b in teleosts and may provide a new perspective for studying the molecular mechanism of fish pigmentation.
Materials and Methods
All animals experiments were approved by the Institutional Animal Care and Use Ethics Committee of Huazhong Agricultural University (HZAUDO-2016-005, 2016-10-26) and conducted in accordance with the “Guidelines for Experimental Animals” of the Ministry of Science and Technology (Beijing, China).
Phylogenetic Analysis and Gene Structure Analysis
The sequences of all species (Homo sapiens, Mus musculus, Gallus, Xenopus tropicalis, Danio rerio, Latimeria chalumnae, Oryzias latipes, Oreochromis niloticus, Takifugu rubripes, and Gasterosteus aculeatus) for the phylogenetic analysis were obtained from NCBI (https://www.ncbi.nlm.nih.gov). All the sequence IDs are available in Supplementary Table S1. GSDS2.0 (http://gsds.gao-lab.org/index.php) and MEME (https://meme-suite.org/meme/tools/meme) were used to reveal the homology of gene structure among species. The SWISS_MODEL (https://swissmodel.expasy.org/interactive) was used to predict the protein tertiary structures of bmp7a and bmp7b.
Generation of Mutant Zebrafish Lines and Maintenance
The transgenic zebrafish parent labeled with green fluorescent protein for osteoblast-specific transcription factor (Osterix GFP) used in this experiment was provided by Professor Xiao Chongde from Chung Yuan Christian University, Taiwan. Fish were kept at 26–28°C under a controlled light cycle (14 h light, 10 h dark) to induce spawning. CRISPR-Cas9 genome editing technology was used to target zebrafish bmp7a and bmp7b. The sequencing of gRNAs and PCR primers are listed in Supplementary Table S2. The mutations of bmp7a and bmp7b were predicted to cause a translational frame shift and induced premature transcription termination.
In Situ Hybridization
The zebrafish embryos were fixed with 4% paraformaldehyde for 24 h. Fragments (length between 300 and 600 bp) of bmp7a and bmp7b were cloned using the primers shown in Supplementary Table S2. The PCR products were purified, and then the whole-mount in situ hybridization probes were synthesized in accordance with the manufacturer’s instructions using the DIG RNA Labeling Kit (SP6/T7) (Roche Diagnostics GmbH, Mannheim, Germany, SKU: 11175025910). The steps of whole-mount in situ hybridization were performed according to the method described by Thisse and Thisse (2008). Anti-DIG-AP,Fab fragments (Roche, SKU: 11093274910) from Sigma-Aldrich were used as antibodies.
Histology Analysis
The eye tissues at 30 days post-fertilization (dpf) and skin tissues of wild types and bmp7b mutants at 60 dpf (3 samples each) were fixed in 4% paraformaldehyde for 24 h. After that, the samples were dehydrated through a series of graded ethanol solutions (70–100%), embedded in paraffin, and sectioned at 5 μm thickness. These sections were stained with hematoxylin and eosin stain; then analyzed and photographed using an Olympus BX53 Upright microscope (Olympus, Beijing, China).
Bone Staining
Bone staining with alizarin red was performed as previously described (Nie et al., 2019) for 90 dpf wild types and bmp7b mutants (10 samples each). The stained specimens were analyzed and photographed under an Olympus SZX2 Stereo microscope (Olympus, Beijing, China).
Determination of Skin Melanin Content
To study the change in melanin content, we stripped the skin (0.2 g) of wild type and mutant fish (20 samples each) after anesthetizing, and extracted the melanin referring to a modified method (Yuan et al., 2008). A UV spectrophotometer UV-3100 (Mapada, Shanghai, China) was used to measure the photometric value of the solution containing melanin. Melanin content was expressed as A290 nm/g (wet weight).
Behavioral Observation
Observations of feeding behavior was performed according to previously study (Miller et al., 1993). Fish were starved for 12 h before the experiments. Wild types and bmp7b mutants at 90 dpf were separately transferred to an experimental tank (length 30 cm × height 24 cm × width 12 cm filled with 2 L of clean water) and allowed to acclimatize to the environment for at least 1 h. On one side of the tank, 5 ml of brine shrimp (Artemia salina) was added at a speed of 15 ml/min to stimulate the fish. A digital video format (Sony, Shanghai, China) was used to record videos of 5 min before and after adding brine shrimp, and EthoVision XT 14 software (Noldus Information Technology, Beijing, China) was used to analyze the behavioral changes (speed, swimming route and reaction time) between the wild-type and mutant fish. The wild type and mutants each had six biological replicates. According to report on zebrafish behavior from Fero et al. (2011), we tested the photosensitivity of the wild-type and mutant fish. Wild types and mutants (all 5 dpf) were separately transferred to an experimental tank (diameter 35 mm, height 10 mm) and allowed to acclimatize to the environment for at least 15 min in the light. Then the light was turned off and the behavior of the fish was recorded. All experiments were performed in EthoVision XT 14 software and there were six samples for wild-type and mutant fish, respectively. During the experiments, the room and water temperatures were maintained at 25 ± 1°C.
Comparative Transcriptome Analysis
Total RNA was isolated from the eyes and skin of 30 dpf wild types and bmp7b mutants with three biological replicates using RNAiso Plus Reagent (TaKaRa, Beijing, China) according to the manufacturer’s protocol. Each sample consisted of tissues from six fish. Subsequently, total RNA was quantified using a 2100 Bioanalyzer system (Agilent Technology, CA, United States) and the samples that met the quality requirements were used for RNA-seq library preparation. After RNA quality and quantity control, 12 transcriptome sequencing libraries were generated using NEBNext® Ultra™ RNA Library Prep Kit for Illumina® (NEB, Beijing, China) following the manufacturer’s recommendations, and index codes were added to attribute sequences to each sample. After cluster generation, the library preparations were sequenced on an Illumina Hiseq platform. After quality control of raw data, paired-end clean reads were aligned to the zebrafish reference genome (ftp://ftp.ensembl.org/pub/release-92/fasta/danio_rerio/) using Hisat2 v2.0.4 (Kim et al., 2015). HTSeq v0.9.1 was subsequently used to count the read numbers mapped to each gene, and then the FPKM of each gene was calculated (Trapnell et al., 2010). Differential expression analysis of two conditions/groups (three biological replicates per condition) was performed using the DESeq R package (1.18.0) (Wang et al., 2009). Genes with a p-value < 0.05, adjusted using the Benjamini and Hochberg’s approach for controlling the false discovery rate, were defined as differentially expressed genes (DEGs). KEGG enrichment analysis of DEGs was performed and KOBAS software was used to test the statistical enrichment of DEGs in the KEGG pathways (http://www.genome.jp/kegg/) (Mao et al., 2005).
Quantitative Real-Time PCR
The NCBI primer tool (https://www.ncbi.nlm.nih.gov/tools/primer-blast/) was used to design primers for the tested genes (rgs9a, rgs9b, rcvrn2, guca1d, grk1b, opn1mw4, gc2, wnt7ba, gna14, erbb3b, bambia, bmp7b, β-actin, gh1, ghra/b, igf-1/2a/2b, igf1ra/b, and igf2r). We used the PrimeScript RT Reagent Kit with gDNA Eraser (TaKaRa, code No. RR047A) to synthesize the cDNA. The qRT-PCR experiments were performed using TB Green® Premix Ex Taq™ II (Tli RNaseH Plus) (TaKaRa, code No. RR820A) and the QuantStudio™ 6 Flex qRT-PCR system (ABI, Foster City, CA, United States). Thermal cycling conditions were as follows: 5 min at 95°C, followed by 40 cycles at 95°C for 20 s, and at 60°C for 25 s. Using the β-actin gene of zebrafish as a reference gene, we used the comparative 2−△△Ct method to determine the relative gene expression between the wild types and mutants (Livak and Schmittgen, 2001). IBM SPSS Statistics software (version 22.0) was used to perform a Student’s t-test to compare the differences between the two groups. Statistical significance was set at p < 0.05, and statistical high significance at p < 0.01. The fold-change values are the average of three biological replicates in each group. The primer sequences are listed in Supplementary Table S2.
Results
Phylogenetic Analysis of BMP7 in Vertebrates
The evolutionary origin and phylogenetic distribution of BMP7 showed that BMP7 orthologs, which evolved from the same ancestral gene family of vertebrates, had duplications in teleosts after the evolution of L. chalumnae, named bmp7a and bmp7b. The bmp7b clade was clustered into one group of humans and mammals (Figure 1A). The gene sequence and structure analysis of BMP7 genes showed that the zebrafish bmp7b gene is highly homologous to the BMP7 gene in humans and mammals. Although the exon numbers of these genes were the same, zebrafish bmp7b (100.69 kb) was closer in length to human (97.87 kb) and mouse (72.31 kb) BMP7 genes, and was significantly larger than zebrafish bmp7a (31.76 kb) (Figure 1B). In addition, these species have a common sequence encoded by exon 6 and exon 7, called the TGF-β domain (103aa). The TGF-β domain-encoding sequences were highly conserved across different species (Figure 1C).
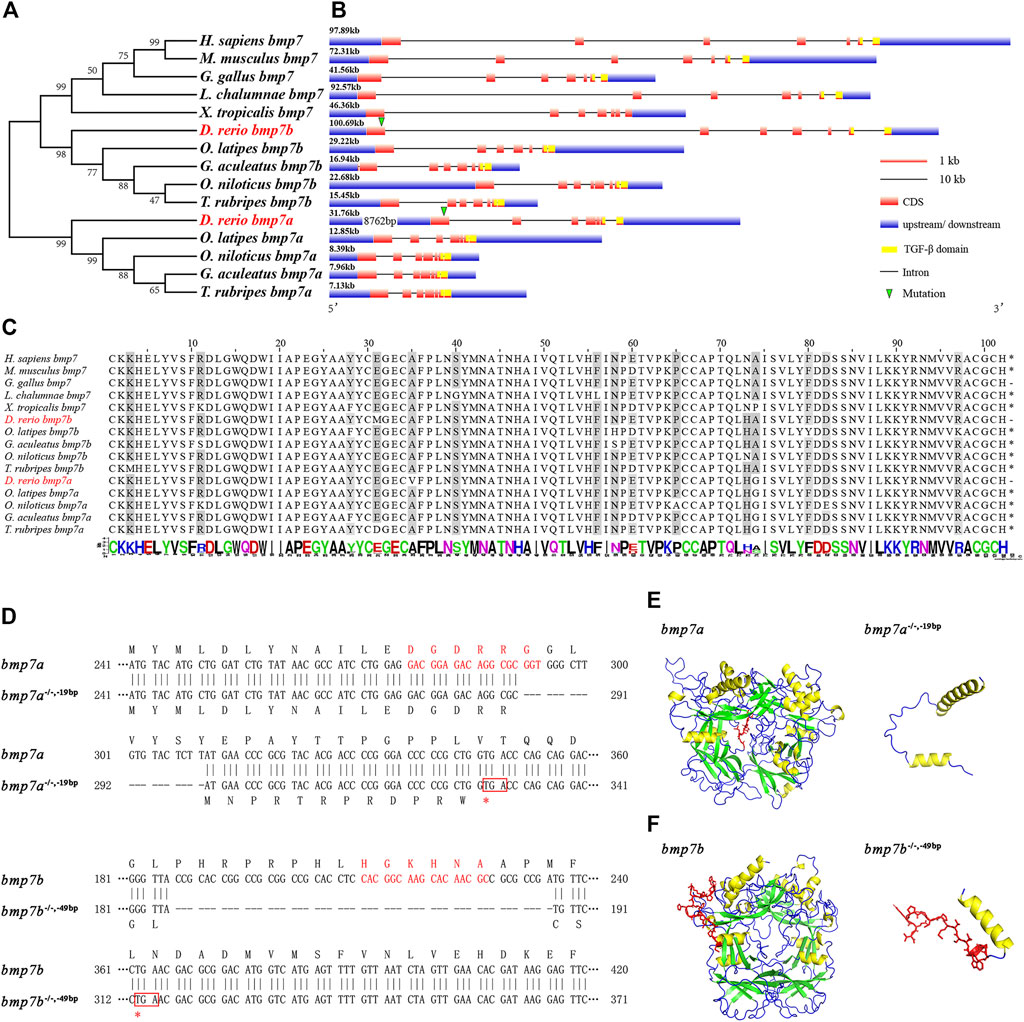
FIGURE 1. Phylogenetic analysis, conservative sequence, and protein structure prediction of BMP7 genes. (A) Phylogenetic analysis of BMP7 genes among different species. (B) The structural analysis of BMP7 genes among different species. The green triangle represents the knockout site of bmp7a and bmp7b. (C) Conservation analysis of amino acids in BMP7 genes’ domains among different species. (D) Schematic representation of bmp7a and bmp7b DNA and protein sequences, and the mutant alleles for bmp7a and bmp7b. CRISPR-Cas9 target sequence is marked with red letters in the wild and mutant DNA sequences. The missing nucleotides are represented by (-) in the mutant, and the red box represents the stop codon. (E) The bmp7a protein tertiary structure prediction of wild type and mutation with -19 bp deletion. The yellow area represents the α-helix; the green area represents the β-sheet; the red area represents the missing part of the mutation. (F) The bmp7b protein tertiary structure prediction of wild type and mutation with -49 bp deletion. The yellow area represents the α-helix; the green area represents the β-sheet; the red area represents the missing part of the mutation.
Generation of bmp7a and bmp7b Mutant Zebrafish
To explore the role of BMP7 gene in fish species and the differences between orthologous genes, we constructed bmp7a and bmp7b mutant zebrafish using CRISPR-Cas9 technology. One bmp7a nonsense allele with a 19 bp deletion in the first exon and two bmp7b nonsense alleles with 49 and 25 bp deletions in the first exon were generated. The bmp7a and bmp7b nonsense alleles all led to premature translation termination (Figure 1D). Additionally, protein structure prediction showed that the TGF-β domains of bmp7a and bmp7b were destroyed because of the missing sequence and could not form a complete protein structure (Figures 1E,F). Since the phenotypic consistency of different mutant lines of the same gene was confirmed in subsequent experiments, the homozygous mutant lines of bmp7a and bmp7b were referred to as bmp7a-/- and bmp7b-/-, respectively, unless otherwise stated.
Bmp7a Deletion Causes Early Embryonic Lethality
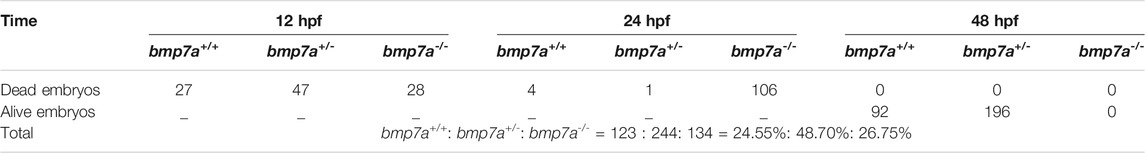
We did not find any mutants when we tested the 48 hpf embryos from the bmp7a heterozygote cross. Therefore, we suspected that bmp7a deletion is lethal and would cause early embryonic death in the bmp7a-/- mutants. To confirm this, we detected embryos at different stages of development, and all genotypes containing a homozygous knockout of bmp7a were found to be lethal at 12–24 hpf. After all the surviving embryos were tested, no homozygous embryos were found to survive at 48 hpf (Table 1). To elucidate how a homozygous bmp7a deletion causes early embryonic death, we examined the morphology of the wild-type and bmp7a-/- embryos. After a large-scale screening of the 501 embryos from three pairs of heterozygous parents, we counted the number of three genotypes. The proportions (bmp7b+/+: bmp7b+/-: bmp7b-/- = 24.55%: 48.70%: 26.75%) of genotypes met Mendel’s laws of inheritance. Moreover, we simultaneously observed the morphology of these embryos and found that bmp7a-/- embryos had defects in the dorsal-ventral pattern. The bmp7a-/- embryos were morphologically distinguishable as distinctly ovoid-shaped embryos at 11 hpf. In the initial stage of somite formation (13–15 hpf), all somites of the mutants extended to the ventral side, whereas the tail did not develop to the ventral side. At the 10-somite stage (16 hpf), the ventral fusion of somites caused the yolk cells to rupture and eventually led to death (Figure 2A). In addition, the tails of bmp7a-/- mutants showed severe deformities at 16 hpf. Approximately, 34% of the bmp7a-/- mutants developed their tails to the ventral side, 66% to the dorsal side (Figure 2B). Moreover, we investigated the spatiotemporal expression pattern of bmp7a using whole-mount in situ hybridization. In 12 hpf wild-type embryos, bmp7a was expressed on the entire blastoderm, especially on the ventral side. In 14 hpf wild-type embryos, the head and tail ends were significantly expressed, but no expression was observed in the dorsal area. In 24 hpf wild-type embryos, bmp7a was significantly expressed in the head and abdomen, but not in the tail. However, in the mutant embryos, the expression of bmp7a disappeared at alive time points (Figure 2C).
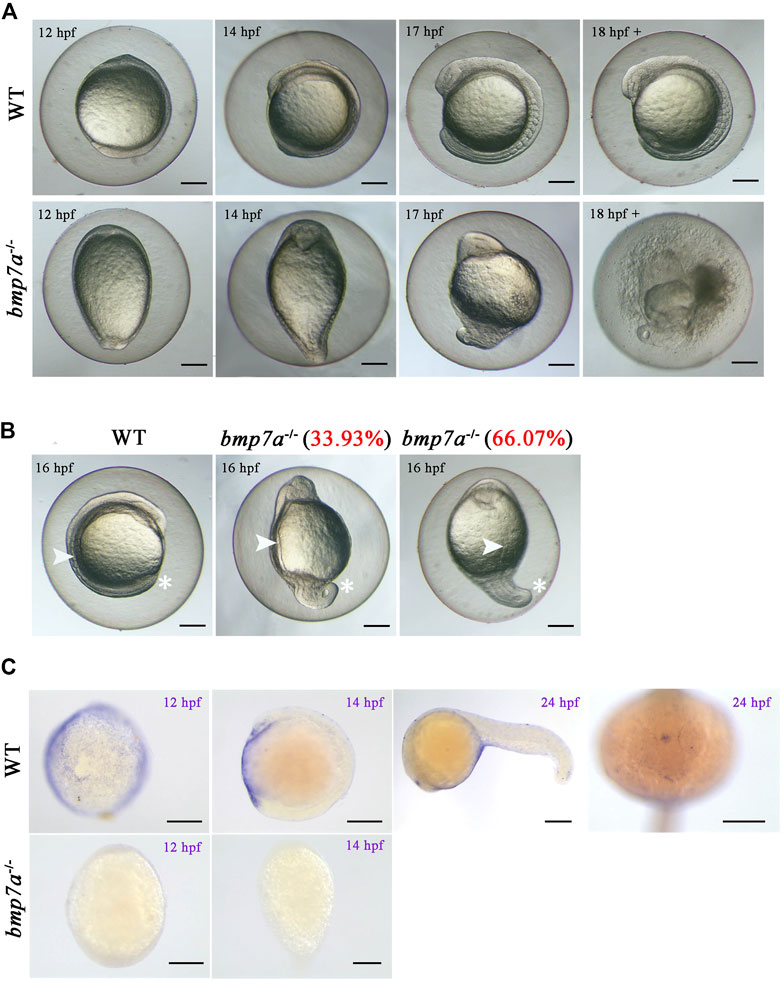
FIGURE 2. Observation of the homozygous lethal phenotype and gene expression in bmp7a embryos. (A) Morphological features of the wild-type and bmp7a-/- embryos at 12 hpf, 14 hpf, 17 hpf and after 18 hpf. (B) Morphological features of the wild-type and bmp7a-/- embryos at 16 hpf. The proportion of mutant embryos with different morphology in mutant embryos at 16 hpf was shown in red font. (C) Whole-mount in situ hybridization of the bmp7a gene at 12 hpf, 16 hpf wild-type and bmp7a-/- embryos, and whole-mount in situ hybridization of the bmp7a at 24 hpf wild-type embryos. Scale bars, 200 μm.
Bmp7b-/- Mutants Show Bending of the Haemal Arches and Growth Retardation
We studied the effect of the bmp7b gene on bone development by staining adult fish with whole-mount alizarin red. The results showed that approximately 80% of adult bmp7b-/- (n = 20) had bend haemal arches and nodules in the intermuscular bones (Figure 3A). Compared to the wild type fish, the external morphology of bmp7b-/- mutants was significantly different. That is, the growth was significantly slower, and these mutants had a darker body surface color (Figure 3B). To compare the effects of bmp7b deletion on growth and development, we measured the growth data (body length and weight) of the bmp7b-/- and wild-type zebrafish at three time points (30, 60, and 90 dpf). The results showed that the body length and weight of the bmp7b-/- zebrafish were significantly lower than those of wild types (p < 0.05) (Figure 3C).
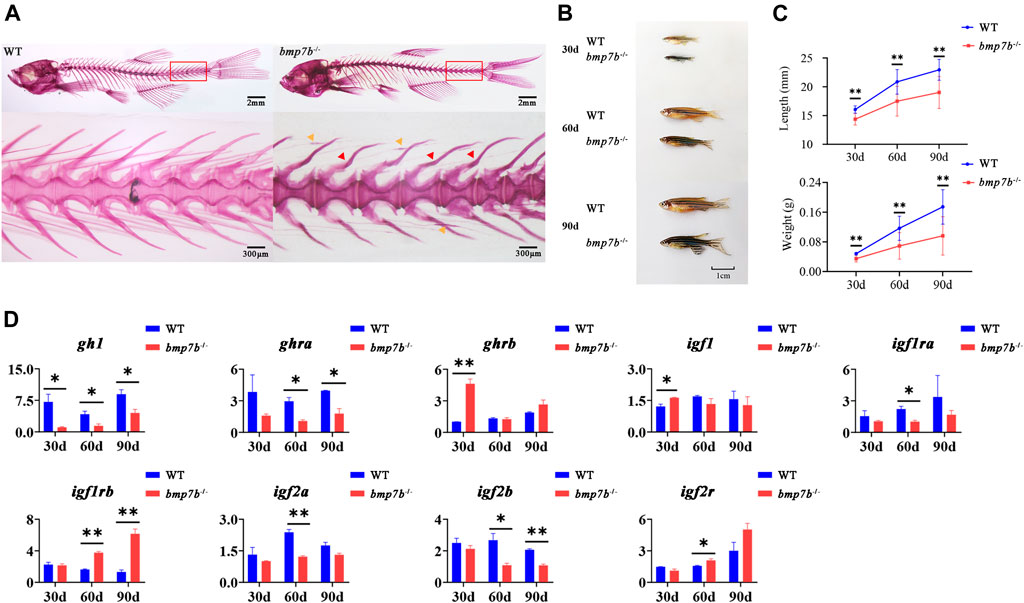
FIGURE 3. The bone and growth phenotype, and gene expression differences between the wild-type and bmp7b-/- zebrafish. (A) Alizarin red staining for mineralized bones at 3 months old fish. The red arrow indicates the severely curved part of the haemal arches, and the yellow arrow indicates the nodular part of the intermuscular bone. (B) The external physical characteristics. (C) The differences of body length and weight. (D) Quantitative expression analysis of growth related genes. Statistically significant differences are marked as *p < 0.05 and **p < 0.01.
Fish growth is mainly regulated by the hypothalamus-pituitary-liver growth axis (GH/IGF axis). To explore the causes for the differences in growth between the wild-type and bmp7b-/- zebrafish, we compared the expression levels of genes related to growth (growth hormone 1, gh1; growth hormone receptor a/b, ghra/b; insulin-like growth factor-1/2a/2b, igf-1/2a/2b; insulin-like growth factor 1 receptor a/b; igf1ra/b; and insulin like growth factor 2 receptor; igf2r) at different points (30, 60, and 90 dpf). The results showed that in the bmp7b-/- mutants, the expression of gh1 decreased significantly at all three points, and ghra and igf2b decreased significantly at both 60 and 90 dpf (p < 0.05); however, igf-1 and ghrb were increased significantly at 30 dpf, and igf1rb was increased at both 60 and 90 dpf (Figure 3D).
Bmp7b-/- Mutants Exhibit Color Darkening
We were intrigued by the darker body color of bmp7b-/- mutants; therefore, we stained the transverse section of the skin at 30 dpf with HE. We compared the structure of the stripes in bmp7b-/- mutants and wild types in four parts (Figure 4A), and found that the skin of the bmp7b-/- mutants had a higher number of melanocytes with a wider distribution (Figure 4B). In addition, the melanin content in the skin of adult bmp7b-/- mutants and wild types were measured. The melanin content in the skin of wild types was 32.987 ± 0.477 A290/g, and that in the bmp7b-/- mutants was 42.800 ± 0.904 A290/g, indicating a significant difference between the both (p < 0.05) (Figure 4C). These results confirmed that the increase in melanin content in the skin of bmp7b-/- mutants led to the darkening of the body color.
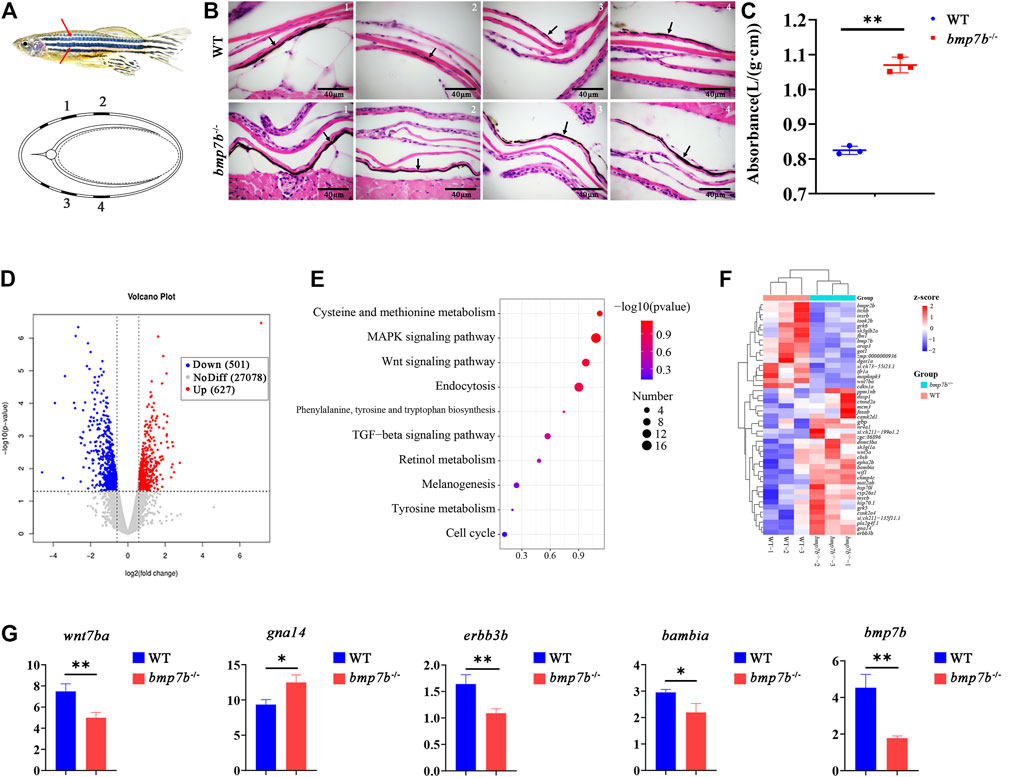
FIGURE 4. The skin structure, melanin content and gene expression differences between the wild-type and bmp7b-/- zebrafish. (A) Schematic diagram. Top: The part of skin melanin tissue section. The red arrow represents the position on the fish body where the tissue section was observed and analyzed. Bottom: the cross-sectional schematic diagram, 1, 2, 3, and 4 correspond to the observation position of the tissue section picture in turn. (B) Skin tissue section. The arrow points to the melanin area. (C) Determination of the melanin content of the skin. The absorbance/wet weight (A290/g) represents the melanin content. (D) The volcano map of DEGs. The abscissa represents multiple changes in gene expression in wild-type and bmp7b-/- fish. The ordinate represents the statistical significance of differences in gene expression. (E) KEGG enrichment analysis of skin transcriptome. (F) Heat map of differential genes enriched in skin transcriptome KEGG. (G) qPCR of key genes (wnt7ba, gna14, erbb3b, bambia and bmp7b). Statistically significant differences are marked as *p < 0.05 and **p < 0.01.
To further explore the causes for the difference in body color, we compared the transcriptomes of the skin of wild types and bmp7b-/- mutants at 30 dpf using RNA-seq. After quality control of the original data, reference genome mapping, and gene expression level quantification, a total of 1128 DEGs between the two groups, including 627 upregulated genes and 501 downregulated genes in mutants, were identified based on an adjusted p-value < 0.05 (Figure 4D, Supplementary Table S3). Subsequently, KEGG pathway analysis revealed important signalling pathways that were significantly enriched in DEGs, such as the mitogen-activated protein kinase (MAPK) (KO: dre 04010), Wnt (KO: dre 04310), melanogenesis (KO: dre 04916), and tyrosine metabolism (KO: dre 00350) signaling pathways, associated with melanin production (Figure 4E, Supplementary Table S4). The upregulation of gna14 and downregulation of wnt7ba, erbb3b, and bambia might promote an increase in melanin production in the skin (Figure 4F). The expression of candidate genes was verified by qPCR (Figure 4G), and the results were consistent with the transcriptome data.
Bmp7b-/- Mutants Have Abnormal Eye Development
The results of whole-mount in situ hybridization of wild-type embryos showed that bmp7b was significantly expressed in the eyes in 24 hpf embryos, confirming that bmp7b may have a role in eye development in fish. Additionally, we also observed that the expression of bmp7b in the mutants was significantly reduced (Figure 5A). The eyes of bmp7b-/- mutants showed a larger iris (Figure 5B), and the ratio of the iris diameter to the eye diameter also increased significantly (p < 0.05) and gradually increased with age (Figure 5C).
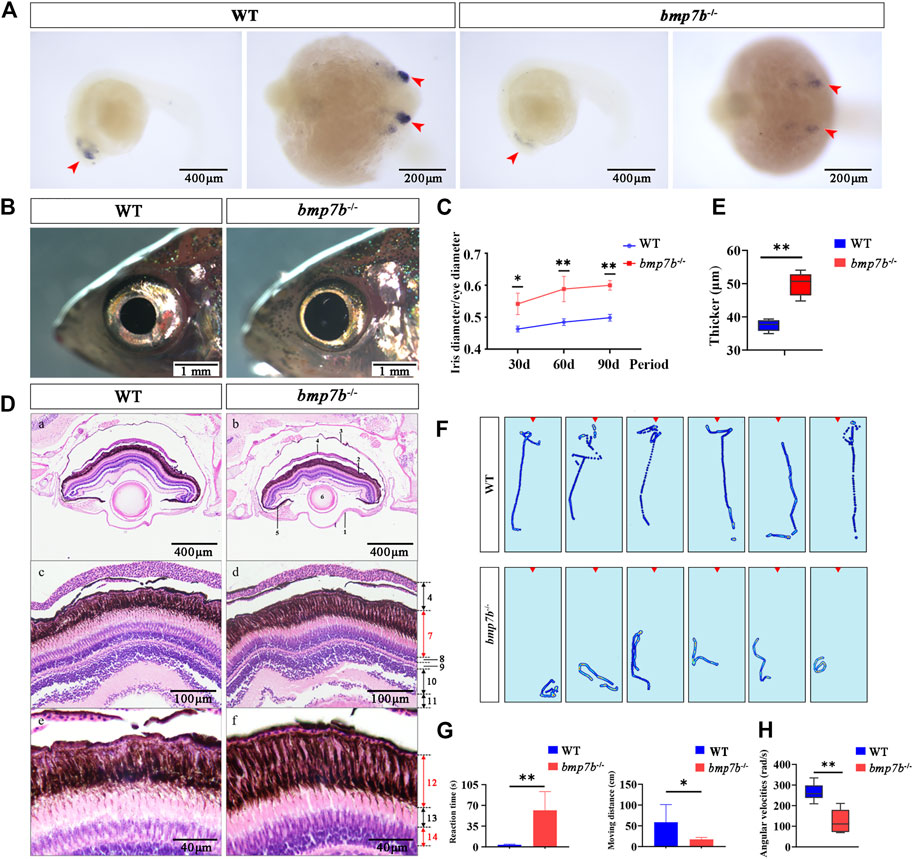
FIGURE 5. The eye structure, feeding behavior and gene expression differences between the wild-type and bmp7b-/- zebrafish. (A) Whole-mount in situ hybridization of the bmp7b at 24 hpf. The blue area represents the zebrafish eyes. (B) The external physical characteristics of eyes. (C) Comparative analysis of the proportion of iris diameter in eye diameter in different points (30, 60, and 90 dpf). (D) HE staining results of eye tissue sections. 1, cornea; 2, retina; 3, sclera; 4, choroid; 5, iris; 6, lens; 7, pigment cell epithelium and photosensitive layer; 8, outer granular layer; 9, outer reticulum layer; 10, inner granule Layer; 11, inner reticulum layer; 12, pigment cell layer; 13, rod cell layer; 14, cone cell layer. (E) Thickness measurement of melanin layer in different regions of eye tissue slices (n = 6). (F) Heat map showing trajectories of adults within 5 s after stimulation (adding brine shrimp) (six repetitions). The red triangle area represents the location of adding brine shrimp. (G) Reaction time and movement distance of adult zebrafish within 5 s after adding brine shrimp. (H) Angular velocities of juvenile zebrafish within 5 s after the light intensity was reduced (from light to dark). Statistically significant differences are marked as *p < 0.05 and **p < 0.01.
Furthermore, we observed the histological characteristics of the eyes in wild-type and bmp7b-/- zebrafish at 60 dpf by longitudinal tissue sections and found an increase in the thickness of the pigment cell layer of the bmp7b-/- mutants (Figures 5D,E). Therefore, we speculated that the changes in the retinal structure caused by bmp7b deletion may impact the vision of bmp7b-/- mutants. To verify this hypothesis, we analyzed the feeding behavior of bmp7b-/- mutants by observing their reaction after the addition of food (brine shrimp). The response of the wild types to food was instant while that of the bmp7b-/- mutants was more sluggish (Figure 5F), consistent with their trajectory behavior. After adding brine shrimp, the wild types quickly found it and swam towards it, while the bmp7b-/- mutants took a longer time to discover it. Additionally, we also analyzed the behavior 5s after adding brine shrimp and the sensitivity to food and the moving distance of bmp7b-/- mutants was significantly lower than that of the wild types (p < 0.05) (Figure 5G). In order to further verify whether the eye development of the mutant was affected, we examined the angular velocities of wild type and bmp7b-/- mutants within 5 s after the light intensity was reduced (from light to dark). The results showed that the angular velocity of the mutant was significantly reduced (p < 0.05) (Figure 5H).
The transcriptomes of the eye tissues between the wild-type and bmp7b-/- zebrafish at 30 dpf were compared using RNA-seq. Transcriptomic analysis indicated significant differences between the wild-type and bmp7b-/- zebrafish (Figure 6A). Over 3,552 DEGs were detected in the eyes of bmp7b-/- fish compared to wild types, with 1,090 DEGs were upregulated and 2,462 DEGs were downregulated (Figure 6B, Supplementary Table S5). KEGG analysis showed that 20 enriched pathways for the DEGs between wild-type and bmp7b-/- mutants were clustered. These included six pathways related to diseases or immune responses (Supplementary Table S6), including apoptosis (KO: dre00010), necroptosis (KO: dre04217), phagosome (KO: dre04145), cell cycle (KO: dre04110), cellular senescence (KO: dre04218), and herpes simplex infection (KO: dre05168), suggesting that inflammatory and eye or retina-related diseases appeared during the development of the eye. In addition, changes in the structure of the eye and disease pathways may lead to the changes in phototransduction (KO: dre04744), which has been associated with dramatic morphological, structural, and functional changes in eye or retinal development. This resulted in a decrease in the vision of the mutants, which was also consistent with the behavioral results. Wnt (KO: dre04310), TGF-β (KO: dre04350), tyrosine metabolism (KO: dre00350), and MAPK signaling pathways (KO: dre04010) were all involved in the production and development of melanin (Figure 6C). Rgs9a, rgs9b, rcvrn2, guca1d, grk1b, opn1mw4, and gc2 genes might be associated with the light-sensing ability of the eye, while wnt7ba, gna14, erbb3b, and bambia genes might promote an increase in melanin in the iris of the eyes. The expression of candidate genes was verified by qPCR (Figure 6D), and the results were consistent with the transcriptome data.
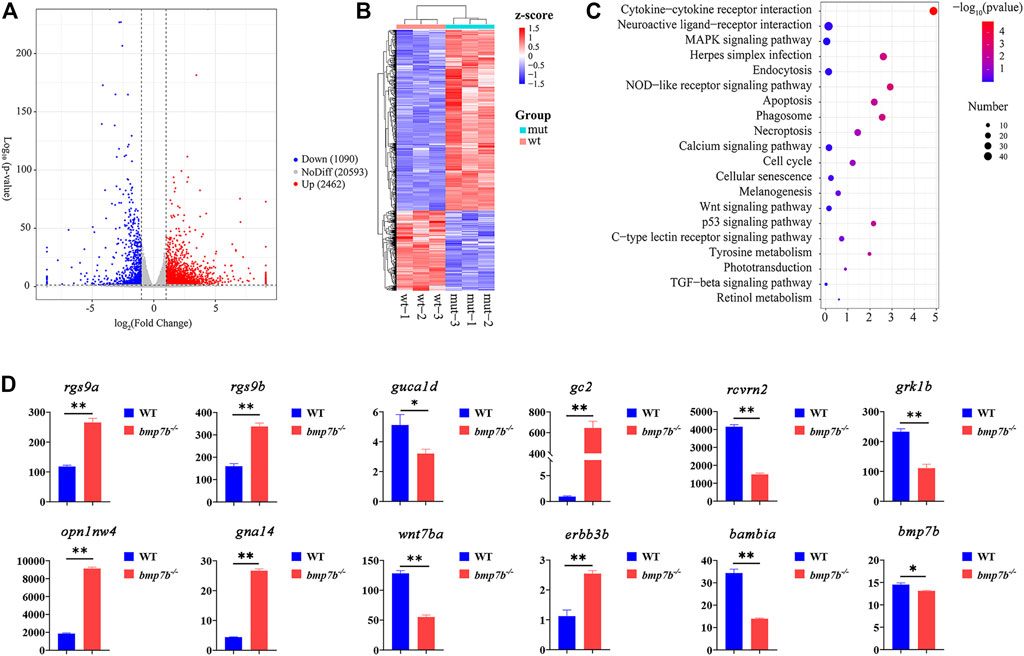
FIGURE 6. The transcriptome and gene qRT-PCR analysis of eye tissues between the wild-type and bmp7b-/- zebrafish at 30 days. (A) DEG’s volcano map. (B) DEG’s Heat map. (C) KEGG enrichment analysis of the eye transcriptome. (D) QPCR of key genes (rgs9a, rgs9b, rcvrn2, guca1d, grk1b, opn1mw4, gc2, and bmp7b). Statistically significant differences from the wildtypes and bmp7b-/- mutants are marked as *p < 0.05 and **p < 0.01.
Discussion
Compared to mammals, birds and amphibians, which have only one BMP7 gene, fish-specific genome duplication (FSGD) led to the formation of BMP7 homologs, bmp7a and bmp7b, in zebrafish and other Actinopterygii. L. chalumnae, which belongs to Sarcopterygii, only has one BMP7 gene. Therefore, it was speculated that BMP7 homologs might have appeared after the differentiation of the Actinopterygii and the Sarcopterygii. Previous genome replication studies have shown that gene homologs might lose, silence, or gain new functions (Lynch and Force, 2000; Glasauer and Neuhauss, 2014). For example, although the dmrt2a and dmrt2b domains are highly conserved in zebrafish, the functions of these two genes are not consistent. Dmrt2a is required for symmetric somite formation, whereas dmrt2b regulates somite differentiation, impacting slow muscle development in zebrafish (Johnsen and Andersen, 2012). Similar to dmrt2, the sequence homology analysis of bmp7a and bmp7b proteins revealed that the protein similarity reached 70%, and the structural domain was highly conserved. However, the significantly reduced gene scale and changed protein coding of the bmp7a gene (bmp7a, 3384 bp; bmp7b, 1694 bp) indicate that the gene is likely to evolve into new functions. Our results showed that bmp7a was involved in the development of the dorsal-ventral pattern; while bmp7b was involved in the development of eyes and bones, which also mediates the production of skin melanin, indicating that bmp7b has a new function in fish species.
The zebrafish snailhouse mutants displayed the strongest dorsalization in the dorso-ventral patterning during gastrulation, which was verified by morpholino knockdown study that bmp7a mutation caused the dorsalized phenotype (Lele et al., 2001). The precisely coordinated induction and morphogenesis processes of gastrulation establish the basis of vertebrate bodies (Keller et al., 2000). BMP2, BMP4, and BMP7 have crucial roles in the formation of the dorsal-ventral pattern of the early embryonic mesoderm. Previous studies on Xenopus laevis have shown that overexpression of BMP7 led to ventralization of early embryos and loss of dorsal tissues (Graff et al., 1996). The bmp7a-/- mutants that we constructed died during the segmentation period due to severe dorsal-ventral axis defects in the embryos. The mutants continued to develop in an ovoid shape after gastrulation, then the dorsal somites and tails of bmp7a-/- mutants could not develop normally. At the same time, bmp7a was abundantly expressed in the abdomen of early embryos, indicating that bmp7a plays a key role in the development of the dorsal-ventral pattern of early embryos. Interestingly, BMP signaling is involved in the growth and pattern formation of the brain (Miyake et al., 2017). We found that bmp7a was not only expressed in the abdomen but also in the brain area of wild-type embryos at 24 hpf. Our results suggest that bmp7a is involved in the formation of the dorsal-ventral pattern of the early embryonic mesoderm, as well as the development of the brain during the embryonic period.
BMP7 is also a growth factor and a secretory signaling molecule. Previous studies have shown that BMP7 is a possible candidate gene for bone and body growth characteristics of chicken (Gallus gallus) and large yellow croaker (Larimichthys crocea) (Wang et al., 2018; Zhou et al., 2019). Compared with the wild types, bmp7b-/- mutants showed a significant decrease in body length and weight during growth (p < 0.05). In contrast to other vertebrates, most fish continue to grow during their life cycle (Johnston et al., 2011). In fish, growth hormones (gh) are synthesized by the pituitary and released into the blood, then act on certain organs by binding to the growth hormone receptor (ghr) on the target cell membrane to regulate growth and development (Kittilson et al., 2011; McMenamin et al., 2013; Hu et al., 2019). Our data showed that bmp7b knockout in zebrafish resulted in a significant downregulation of gh1, and its receptor ghra was also significantly downregulated. In addition, the main target tissue of insulin-like growth factors, igf1 and igf2, is the skeletal muscle in mammals (Duan et al., 2010), and they exert their biological effects by binding to igf1r (Glass, 2005; Wu and Zhu, 2011). The significantly reduced expression of igf2a/2b at 60 and 90 dpf (p < 0.05) might lead to a slower growth of muscle cells in mutants. Correspondingly, the increased expression of igf1rb was due to a negative feedback regulation in response to the downregulation of igf2a/2b expression, but it still could not prevent the negative effects of igf2a/2b expression decline on muscle growth. In summary, bmp7b deletion affected the expression of GH/IGF axis genes, thereby inhibiting the growth of bmp7b-/- mutants.
The importance of the BMP7 signaling pathway in vertebrate retinal development has been repeatedly demonstrated through gene inactivation and functional gain experiments in mice and birds (Hogan, 1996; Zhang et al., 2013). During zebrafish embryonic development, bmp7b is first expressed in the eye (Shawi and Serluca, 2008), and it can significantly increase the number of outer nodal protrusions formed by rod-shaped cells and cone cells (Sehgal et al., 2006). We found that pigment layers in the eye tissue sections of bmp7b-/- mutants at 60 dpf was thicker than that of the wild types. Therefore, we hypothesized that the vision of bmp7b-/- mutants was affected. Both adult and juvenile zebrafish depend on their vision for feeding (McElligott and O’Malley, 2005; Howe et al., 2018). Therefore, we observed the feeding behavior of zebrafish and found that the reaction time of bmp7b-/- mutants to food stimulation was much longer than that of the wild type, thereby confirming that bmp7b deletion reduced the ability of mutant zebrafish to observe food and the visual sensitivity. To further explore whether the eye development process in mutants was affected, we examined the angular velocities of wild types and bmp7b-/- mutants within 5 s after the light intensity was reduced (from light to dark) (Fero et al., 2011). The angular velocity of the bmp7b-/- mutants was significantly reduced (p < 0.05), indicating that the response of the mutants to light intensity was less sensitive than that of the wild types. In addition, reduced swimming speed of mutants also might be due to skeletal defects. Taking together, bmp7b-/- mutants had skeletal defects and visual dysfunction, which led to a reduction in feeding ability.
Moreover, we also found that bmp7b deletion resulted in an increase in melanin content in the zebrafish skin and eyes. In human hair and skin, BMP7 protein expression was found to change in a hair cycle-dependent manner, which suggests that BMP7 may be involved in the regulation of skin development and function (Adly et al., 2006). Although BMP7 is not a traditional pigment gene, it directly or indirectly affects some pigmentation pathways. It can additionally inhibit normal melanocyte growth, tumor growth, and metastasis of human uveal melanoma (Notting et al., 2007; Hsu et al., 2008; Na et al., 2009), but its dual role in cancer indicates that high BMP7 expression promotes the development of melanoma (Hsu et al., 2005; Rothhammer et al., 2005). BMP7 can affect the production of pheomelanin by interacting with melanocortin during the differentiation and thermogenesis of brown adipocytes (Baxter et al., 2009; Richard and Picard, 2011). In addition, the upregulation of BMP7 is related to hyperpigmentation of the visceral peritoneum and combs (Luo et al., 2013; Dong et al., 2019). Our results indicated that bmp7b inhibited the production of zebrafish melanin, while bmp7b knockout suppressed this inhibitory effect and caused an increase in melanin production and darkening of zebrafish body color.
The signaling pathways associated with eye defects of bmp7b-/- mutants were revealed through a comprehensive transcriptomic analysis, and the changes in the expression of many key genes involved in phototransduction. Phototransduction is the process by which light is converted into electrical signals via photoreceptors (rods and cones) in the retina, which mediates vision in vertebrates (Fu and Yau, 2007). In this process, the reduction in rcvrn2 expression accelerates cone photoresponse recovery (Zang et al., 2015). The concentration of rgs9 is a potentially important determinant of faster response kinetics and lower sensitivity of cones, as compared with rods (Cowan et al., 1998). In bmp7b-/- mutants, the expression changes of rcvrn2, rgs9a, and rgs9b genes affected the sensitivity of light response recovery. The decreased expression of guca1d (gcap4), which is uniquely present in cone photoreceptors, indicated that its reception of light was hindered in bmp7b-/- mutants (Imanishi et al., 2014), whereas the increased expression of opn1mw4 in the green cones leads to an inaccurate detection of red-shifted down-welling light in bmp7b-/- mutants (Ogawa and Corbo, 2021). In addition, it is possible that the decreased expression of grk1b had an impact on the formation of functional cone mosaics. Therefore, the spatial resolution of zebrafish is affected (Chrispell et al., 2018). The changes in genes related to retinol metabolism (pathway ID: dre00830) also play a pivotal role in phototransduction (Parker and Crouch, 2010; Li et al., 2018). In short, the function of cones and rods was based on the phototransduction, and the deletion of bmp7b led to the expression changes of genes related to phototransduction. Therefore, it was suggested that the sensitivity of bmp7b-/- mutants to light was decreased, which in turn affected feeding behavior.
Similar to the eye transcriptome, their differential genes in skin tissues were jointly enriched in signaling pathways, such as the MAPK, Wnt, melanogenesis, and TGF-β signaling pathways. In melanogenesis, gna14 and wnt7ba genes showed the same trend of changes in the skin and eyes, and these changes might be key factors leading to an increase in melanin. The Wnt signaling pathway plays a vital role via the microphthalmia-associated transcription factor (Mitf) in the development of melanophores (Jin et al., 2001). Wnt7b is a direct target of canonical BMP signaling in hair follicle stem cells, and wnt7b expression reaches a peak during the development of black and white striped feather hair follicles in the neck of eight-week-old chicken (Kandyba and Kobielak, 2014; Wang et al., 2019). In Ctenophorus decresii, wnt7b showed stronger expression in skin with a high melanin concentration (Mclean et al., 2017). In addition, wnt7b is highly expressed in all benign nevi and some large cell melanomas (Pham et al., 2003). Gna14 is another important gene involved in melanin production; the cell pellets of newborn human melanocytes with gna14 mutations were lighter in color than those of wild types (Lim et al., 2016). Moreover, gna14 is one of the candidate genes that caused differences in coat color between white rex rabbits and purple-blue rex rabbits (Qin, 2014). Gna14 can also be coupled to melanin and participates in melanopsin phototransduction in vivo (Hughes et al., 2015). Melanin production was also affected by changes in gene expression in other pathways; signals mediated by the tyrosine kinase erbb3b establish the latent precursors metamorphic melanophores, contributing to melanophore development (Budi et al., 2008). Bambi is regulated by TGF-β signaling and is overexpressed in melanoma skin compared to normal skin (Hagedorn et al., 2016). Melanin production involves several signalling pathways and the regulation of transcription factors. In this study, we confirmed that bmp7b is a key inhibitor of melanogenesis that exhibits its effects by regulating the expression of the above mentioned genes.
In summary, we constructed two stable mutant strains, bmp7a and bmp7b, and revealed their functional differentiation. Bmp7a plays a vital role in the development of the dorsal-ventral pattern of the embryo, while bmp7b impacts eye development and melanin production. The first report about the inhibitory effect of bmp7b on melanogenesis may provide useful information for the future research on human melanoma-related diseases.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI PRJNA788440.
Ethics Statement
The animal study was reviewed and approved by Research Ethics Committee, Huazhong Agricultural University, Wuhan, China (HZAUDO-2016-005, 2016-10-26). Written informed consent was obtained from the owners for the participation of their animals in this study.
Author Contributions
Z-XG conceived the project. X-RD, S-MW, J-JZ, and J-HD collected the samples. X-RD conducted the biological experiments, analyzed the data and wrote the draft. C-HN, Y-LC, S-MW, and Z-XG modified the manuscript and discussed the data. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by Hubei Hongshan Laboratory (2021hszd001), NKRDP 2018YFD0900102, the China Agriculture Research System of MOF and MARA (No. CARS-45-08), National Natural Science Foundation of China (31872559), Wuhan Applied Foundational Frontier Project 2020020601012253.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
Thanks to all members of the Zexia Gao’s Laboratories for their support and discussion.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2022.838721/full#supplementary-material
References
Adly, M. A., Assaf, H. A., and Hussein, M. R. (2006). Expression of Bone Morphogenetic Protein-7 in Human Scalp Skin and Hair Follicles. Br. J. Dermatol. 154, 551–554. doi:10.1111/j.1365-2133.2005.07064.x
Baxter, L. L., Loftus, S. K., and Pavan, W. J. (2009). Networks and Pathways in Pigmentation, Health, and Disease. Wires Syst. Biol. Med. 1, 359–371. doi:10.1002/wsbm.20
Bragdon, B., Moseychuk, O., Saldanha, S., King, D., Julian, J., and Nohe, A. (2011). Bone Morphogenetic Proteins: A Critical Review. Cell Signal. 23, 609–620. doi:10.1016/j.cellsig.2010.10.003
Budi, E. H., Patterson, L. B., and Parichy, D. M. (2008). Embryonic Requirements for ErbB Signaling in Neural Crest Development and Adult Pigment Pattern Formation. Development 135, 2603–2614. doi:10.1242/dev.019299
Celeste, A. J., Iannazzi, J. A., Taylor, R. C., Hewick, R. M., Rosen, V., Wang, E. A., et al. (1990). Identification of Transforming Growth Factor Beta Family Members Present in Bone-Inductive Protein Purified from Bovine Bone. Proc. Natl. Acad. Sci. 87, 9843–9847. doi:10.1073/pnas.87.24.9843
Chrispell, J. D., Dong, E., Osawa, S., Liu, J., Cameron, D. J., and Weiss, E. R. (2018). Grk1b and Grk7a Both Contribute to the Recovery of the Isolated Cone Photoresponse in Larval Zebrafish. Invest. Ophthalmol. Vis. Sci. 59, 5116–5124. doi:10.1167/iovs.18-24455
Cowan, C. W., Fariss, R. N., Sokal, I., Palczewski, K., and Wensel, T. G. (1998). High Expression Levels in Cones of RGS9, the Predominant GTPase Accelerating Protein of Rods. Proc. Natl. Acad. Sci. 95, 5351–5356. doi:10.1073/pnas.95.9.5351
Dong, X., Li, J., Zhang, Y., Han, D., Hua, G., Wang, J., et al. (2019). Genomic Analysis Reveals Pleiotropic Alleles at EDN3 and BMP7 Involved in Chicken Comb Color and Egg Production. Front. Genet. 10, 612. doi:10.3389/fgene.2019.00612
Duan, C., Ren, H., and Gao, S. (2010). Insulin-like Growth Factors (IGFs), IGF Receptors, and IGF-Binding Proteins: Roles in Skeletal Muscle Growth and Differentiation. Gen. Comp. Endocrinol. 167, 344–351. doi:10.1016/j.ygcen.2010.04.009
Ducy, P., and Karsenty, G. (2000). The Family of Bone Morphogenetic Proteins. Kidney Int. 57, 2207–2214. doi:10.1046/j.1523-1755.2000.00081.x
Fero, K., Yokogawa, T., and Burgess, H. A. (2011). “The Behavioral Repertoire of Larval Zebrafish,” in Zebrafish Models in Neurobehavioral Research. Editors A. V. Kalueff, and J. M. Cachat (Totowa, NJ: Humana Press), 249–291. doi:10.1007/978-1-60761-922-2_12
Fu, Y., and Yau, K. W. (2007). Phototransduction in Mouse Rods and Cones. Pflugers Arch. - Eur. J. Physiol. 454, 805–819. doi:10.1007/s00424-006-0194-y
Glasauer, S. M. K., and Neuhauss, S. C. F. (2014). Whole-Genome Duplication in Teleost Fishes and its Evolutionary Consequences. Mol. Genet. Genomics 289, 1045–1060. doi:10.1007/s00438-014-0889-2
Glass, D. J. (2005). Skeletal Muscle Hypertrophy and Atrophy Signaling Pathways. Int. J. Biochem. Cel Biol. 37, 1974–1984. doi:10.1016/j.biocel.2005.04.018
Graff, J. M., Bansal, A., and Melton, D. A. (1996). Xenopus Mad Proteins Transduce Distinct Subsets of Signals for the TGFβ Superfamily. Cell 85, 479–487. doi:10.1016/s0092-8674(00)81249-0
Hagedorn, M., Siegfried, G., Hooks, K. B., and Khatib, A.-M. (2016). Integration of Zebrafish Fin Regeneration Genes with Expression Data of Human Tumors In Silico Uncovers Potential Novel Melanoma Markers. Oncotarget 7, 71567–71579. doi:10.18632/oncotarget.12257
Harris, M. P., Linkhart, B. L., and Fallon, J. F. (2004). Bmp7 Mediates Early Signaling Events during Induction of Chick Epidermal Organs. Dev. Dyn. 231, 22–32. doi:10.1002/dvdy.20096
Hogan, B. L. (1996). Bone Morphogenetic Proteins in Development. Curr. Opin. Genet. Develop. 6, 432–438. doi:10.1016/s0959-437x(96)80064-5
Howe, H. B., McIntyre, P. B., and Wolman, M. A. (2018). Adult Zebrafish Primarily Use Vision to Guide Piscivorous Foraging Behavior. Behav. Process. 157, 230–237. doi:10.1016/j.beproc.2018.10.005
Hsu, M. Y., Rovinsky, S. A., Lai, C. Y., Qasem, S., Liu, X., How, J., et al. (2008). Aggressive Melanoma Cells Escape from BMP7-Mediated Autocrine Growth Inhibition through Coordinated Noggin Upregulation. Lab. Invest. 88, 842–855. doi:10.1038/labinvest.2008.55
Hsu, M.-Y., Rovinsky, S., Penmatcha, S., Herlyn, M., and Muirhead, D. (2005). Bone Morphogenetic Proteins in Melanoma: Angel or Devil. Cancer Metastasis Rev. 24, 251–263. doi:10.1007/s10555-005-1575-y
Hu, Z., Ai, N., Chen, W., Wong, Q. W. L., and Ge, W. (2019). Loss of Growth Hormone Gene (Gh1) in Zebrafish Arrests Folliculogenesis in Females and Delays Spermatogenesis in Males. Endocrinology 160, 568–586. doi:10.1210/en.2018-00878
Hughes, S., Jagannath, A., Hickey, D., Gatti, S., Wood, M., Peirson, S. N., et al. (2015). Using siRNA to Define Functional Interactions between Melanopsin and Multiple G Protein Partners. Cell. Mol. Life Sci. 72, 165–179. doi:10.1007/s00018-014-1664-6
Imanishi, Y., Yang, L., Sokal, I., Filipek, S. a., Palczewski, K., and Baehr, W. (2004). Diversity of Guanylate Cyclase-Activating Proteins (GCAPs) in Teleost Fish: Characterization of Three Novel GCAPs (GCAP4, GCAP5, GCAP7) from Zebrafish (Danio rerio) and Prediction of Eight GCAPs (GCAP1-8) in Pufferfish (Fugu Rubripes). J. Mol. Evol. 59, 204–217. doi:10.1007/s00239-004-2614-y
Jin, E. J., Erickson, C. A., Takada, S., and Burrus, L. W. (2001). Wnt and BMP Signaling Govern Lineage Segregation of Melanocytes in the Avian Embryo. Develop. Biol. 233, 22–37. doi:10.1006/dbio.2001.0222
Johnsen, H., and Andersen, Ø. (2012). Sex Dimorphic Expression of Five Dmrt Genes Identified in the Atlantic Cod Genome. The Fish-specific Dmrt2b Diverged from Dmrt2a before the Fish Whole-Genome Duplication. Gene 505, 221–232. doi:10.1016/j.gene.2012.06.021
Johnston, I. A., Bower, N. I., and Macqueen, D. J. (2011). Growth and the Regulation of Myotomal Muscle Mass in Teleost Fish. J. Exp. Biol. 214, 1617–1628. doi:10.1242/jeb.038620
Kandyba, E., and Kobielak, K. (2014). Wnt7b Is an Important Intrinsic Regulator of Hair Follicle Stem Cell Homeostasis and Hair Follicle Cycling. Stem Cells 32, 886–901. doi:10.1002/stem.1599
Keller, R., Davidson, L., Edlund, A., Elul, T., Ezin, M., Shook, D., et al. (2000). Mechanisms of Convergence and Extension by Cell Intercalation. Phil. Trans. R. Soc. Lond. B. 355, 897–922. doi:10.1098/rstb.2000.0626
Kim, D., Langmead, B., and Salzberg, S. L. (2015). HISAT: a Fast Spliced Aligner with Low Memory Requirements. Nat. Methods 12, 357–360. doi:10.1038/nmeth.3317
Kim, H.-S., Neugebauer, J., McKnite, A., Tilak, A., and Christian, J. L. (2019). BMP7 Functions Predominantly as a Heterodimer with BMP2 or BMP4 during Mammalian Embryogenesis. Elife 8, e48872. doi:10.7554/eLife.48872
Kittilson, J. D., Jones, E., and Sheridan, M. A. (2011). ERK, Akt, and STAT5 Are Differentially Activated by the Two Growth Hormone Receptor Subtypes of a Teleost Fish (Oncorhynchus Mykiss). Front. Endocrin. 2, 30. doi:10.3389/fendo.2011.00030
Lele, Z., Bakkers, J., and Hammerschmidt, M. (2001). Morpholino Phenocopies Ofthe Swirl,snailhouse,somitabun,minifin,silberblick, Andpipetail Mutations. Genesis 30, 190–194. doi:10.1002/gene.1063
Li, B., Rognon, G. T., Mattinson, T., Vachali, P. P., Gorusupudi, A., Chang, F.-Y., et al. (2018). Supplementation with Macular Carotenoids Improves Visual Performance of Transgenic Mice. Arch. Biochem. Biophys. 649, 22–28. doi:10.1016/j.abb.2018.05.003
Lim, Y. H., Bacchiocchi, A., Qiu, J., Straub, R., Bruckner, A., Bercovitch, L., et al. (2016). GNA14 Somatic Mutation Causes Congenital and Sporadic Vascular Tumors by Mapk Activation. Am. J. Hum. Genet. 99, 443–450. doi:10.1016/j.ajhg.2016.06.010
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 25, 402–408. doi:10.1006/meth.2001.1262
Luo, C., Qu, H., Wang, J., Wang, Y., Ma, J., Li, C., et al. (2013). Genetic Parameters and Genome-wide Association Study of Hyperpigmentation of the Visceral Peritoneum in Chickens. BMC Genomics 14, 334. doi:10.1186/1471-2164-14-334
Luo, G., Hofmann, C., Bronckers, A. L., Sohocki, M., Bradley, A., and Karsenty, G. (1995). BMP-7 Is an Inducer of Nephrogenesis, and Is Also Required for Eye Development and Skeletal Patterning. Genes Dev. 9, 2808–2820. doi:10.1101/gad.9.22.2808
Lv, X. Y., Ni, R., Sun, W., Su, R., Musa, H. H., Yin, J. F., et al. (2016). Candidate Genes for the Development of Hair Follicles in Hu Sheep. Genet. Mol. Res. 15, 15036877. doi:10.4238/gmr.15036877
Lynch, M., and Force, A. (2000). The Probability of Duplicate Gene Preservation by Subfunctionalization. Genetics 154, 459–473. doi:10.1093/genetics/154.1.459
Mao, X., Cai, T., Olyarchuk, J. G., and Wei, L. (2005). Automated Genome Annotation and Pathway Identification Using the KEGG Orthology (KO) as a Controlled Vocabulary. Bioinformatics 21, 3787–3793. doi:10.1093/bioinformatics/bti430
McElligott, M. B., and O’Malley, D. M. (2005). Prey Tracking by Larval Zebrafish: Axial Kinematics and Visual Control. Brain Behav. Evol. 66, 177–196. doi:10.1159/000087158
Mclean, C. A., Lutz, A., Rankin, K. J., Stuart-fox, D., and Moussalli, A. (2017). Revealing the Biochemical and Genetic Basis of Color Variation in a Polymorphic Lizard. Mol. Biol. Evol. 34, 1924–1935. doi:10.1093/molbev/msx136
McMenamin, S. K., Minchin, J. E. N., Gordon, T. N., Rawls, J. F., and Parichy, D. M. (2013). Dwarfism and Increased Adiposity in the Gh1 Mutant Zebrafish Vizzini. Endocrinology 154, 1476–1487. doi:10.1210/en.2012-1734
Michon, F., Forest, L., Collomb, E., Demongeot, J., and Dhouailly, D. (2008). BMP2 and BMP7 Play Antagonistic Roles in Feather Induction. Development 135, 2797–2805. doi:10.1242/dev.018341
Miller, T. J., Crowder, L. B., and Rice, J. A. (1993). Ontogenetic Changes in Behavioural and Histological Measures of Visual Acuity in Three Species of Fish. Environ. Biol. Fish. 37, 1–8. doi:10.1007/BF00000707
Miyake, A., Mekata, Y., Fujibayashi, H., Nakanishi, K., Konishi, M., and Itoh, N. (2017). Brorin Is Required for Neurogenesis, Gliogenesis, and Commissural Axon Guidance in the Zebrafish Forebrain. PLoS One 12, e0176036. doi:10.1371/journal.pone.0176036
Na, Y. R., Seok, S. H., Kim, D. J., Han, J. H., Kim, T. H., Jung, H., et al. (2009). Bone Morphogenetic Protein 7 Induces Mesenchymal-To-Epithelial Transition in Melanoma Cells, Leading to Inhibition of Metastasis. Cancer Sci. 100, 2218–2225. doi:10.1111/j.1349-7006.2009.01301.x
Nie, C. H., Hilsdorf, A. W. S., Wan, S. M., and Gao, Z. X. (2019). Understanding the Development of Intermuscular Bones in Teleost: Status and Future Directions for Aquaculture. Rev. Aquacult. 12, 759–772. doi:10.1111/raq.12348
Notting, I., Buijs, J., Mintardjo, R., van der Horst, G., Vukicevic, S., Lowik, C., et al. (2007). Bone Morphogenetic Protein 7 Inhibits Tumor Growth of Human Uveal Melanoma In Vivo. Invest. Ophthalmol. Vis. Sci. 48, 4882–4889. doi:10.1167/iovs.07-0505
Ogawa, Y., and Corbo, J. C. (2021). Partitioning of Gene Expression Among Zebrafish Photoreceptor Subtypes. Sci. Rep. 11, 17340. doi:10.1038/s41598-021-96837-z
Ozkaynak, E., Rueger, D. C., Drier, E. A., Corbett, C., Ridge, R. J., Sampath, T. K., et al. (1990). OP-1 cDNA Encodes an Osteogenic Protein in the TGF-Beta Family. EMBO J. 9, 2085–2093. doi:10.1002/j.1460-2075.1990.tb07376.x
Parker, R. O., and Crouch, R. K. (2010). Retinol Dehydrogenases (RDHs) in the Visual Cycle. Exp. Eye Res. 91, 788–792. doi:10.1016/j.exer.2010.08.013
Pham, K., Milovanovic, T., Barr, R. J., Truong, T., and Holcombe, R. F. (2003). Wnt Ligand Expression in Malignant Melanoma: Pilot Study Indicating Correlation with Histopathological Features. Mol. Pathol. 56, 280–285. doi:10.1136/mp.56.5.280
Qin, L. Z. (2014). Identification of Genes Related to Fur Color and Cloning and Diversity Analysis of Creb1 Gene in Rex Rabbit (Yangzhou, China: Yangzhou University). M.D. diss
Richard, D., and Picard, F. (2011). Brown Fat Biology and Thermogenesis. Front. Biosci. 16, 1233–1260. doi:10.2741/3786
Rothhammer, T., Poser, I., Soncin, F., Bataille, F., Moser, M., and Bosserhoff, A. K. (2005). Bone Morphogenic Proteins Are Overexpressed in Malignant Melanoma and Promote Cell Invasion and Migration. Cancer Res. 65, 448–456. doi:10.1158/0008-5472.448.65.2
Schmid, B., Furthauer, M., Connors, S. A., Trout, J., Thisse, B., Thisse, C., et al. (2000). Equivalent Genetic Roles for Bmp7/snailhouse and Bmp2b/swirl in Dorsoventral Pattern Formation. Development 127, 957–967. doi:10.1242/dev.127.5.957
Sehgal, R., Andres, D. J., Adler, R., and Belecky-Adams, T. L. (2006). Bone Morphogenetic Protein 7 Increases Chick Photoreceptor Outer Segment Initiation. Invest. Ophthalmol. Vis. Sci. 47, 3625–3634. doi:10.1167/iovs.06-0079
Shawi, M., and Serluca, F. C. (2008). Identification of a BMP7 Homolog in Zebrafish Expressed in Developing Organ Systems. Gene Expr. Patterns 8, 369–375. doi:10.1016/j.gep.2008.05.004
Thisse, C., and Thisse, B. (2008). High-resolution In Situ Hybridization to Whole-Mount Zebrafish Embryos. Nat. Protoc. 3, 59–69. doi:10.1038/nprot.2007.514
Trapnell, C., Williams, B. A., Pertea, G., Mortazavi, A., Kwan, G., van Baren, M. J., et al. (2010). Transcript Assembly and Quantification by RNA-Seq Reveals Unannotated Transcripts and Isoform Switching during Cell Differentiation. Nat. Biotechnol. 28, 511–515. doi:10.1038/nbt.1621
Vitic, Z., Safory, H., Jovanovic, V. M., Sarusi, Y., Stavsky, A., Kahn, J., et al. (2021). BMP5/7 Protect Dopaminergic Neurons in an α-synuclein Mouse Model of Parkinson's Disease. Brain 144, e15. doi:10.1093/brain/awaa368
Wang, L., Feng, Z., Wang, X., Wang, X., and Zhang, X. (2009). DEGseq: An R Package for Identifying Differentially Expressed Genes from RNA-Seq Data. Bioinformatics 26, 136–138. doi:10.1093/bioinformatics/btp612
Wang, X., Li, D., Song, S., Zhang, Y., Li, Y., Wang, X., et al. (2019). Combined Transcriptomics and Proteomics Forecast Analysis for Potential Genes Regulating the Columbian Plumage Color in Chickens. PLoS One 14, e0210850. doi:10.1371/journal.pone.0210850
Wang, Y., Guo, F., Qu, H., Luo, C., Wang, J., and Shu, D. (2018). Associations between Variants of Bone Morphogenetic Protein 7 Gene and Growth Traits in Chickens. Br. Poult. Sci. 59, 264–269. doi:10.1080/00071668.2018.1454586
Wu, J., and Zhu, A. X. (2011). Targeting Insulin-like Growth Factor axis in Hepatocellular Carcinoma. J. Hematol. Oncol. 4, 30. doi:10.1186/1756-8722-4-30
Yuan, L. Q., Ma, X. Z., Wang, W., and Ye, L. (2008). Effects of Dietary Lipid Levels Oil Growth and Body Pigmentation of Darkbarbel Catfish (Pelteobagrus Vachelli Richardson). J. Shanghai Ocean Univ. 17, 577–584. doi:10.3724/SP.J.1035.2008.00038
Zang, J., Keim, J., Kastenhuber, E., Gesemann, M., and Neuhauss, S. C. F. (2015). Recoverin Depletion Accelerates Cone Photoresponse Recovery. Open Biol. 5, 150086. doi:10.1098/rsob.150086
Zhang, Y., Liu, Y., Ho, C., and Wildsoet, C. F. (2013). Effects of Imposed Defocus of Opposite Sign on Temporal Gene Expression Patterns of BMP4 and BMP7 in Chick RPE. Exp. Eye Res. 109, 98–106. doi:10.1016/j.exer.2013.02.010
Zhou, Z., Han, K., Wu, Y., Bai, H., Ke, Q., Pu, F., et al. (2019). Genome-Wide Association Study of Growth and Body-Shape-Related Traits in Large Yellow Croaker (Larimichthys Crocea) Using ddRAD Sequencing. Mar. Biotechnol. 21, 655–670. doi:10.1007/s10126-019-09910-0
Keywords: bmp7a, bmp7b, growth difference, eye defect, melanin pigmentation, transcriptome, zebrafish
Citation: Dong X-, Wan S-, Zhou J-, Nie C-, Chen Y-, Diao J- and Gao Z- (2022) Functional Differentiation of BMP7 Genes in Zebrafish: bmp7a for Dorsal-Ventral Pattern and bmp7b for Melanin Synthesis and Eye Development. Front. Cell Dev. Biol. 10:838721. doi: 10.3389/fcell.2022.838721
Received: 18 December 2021; Accepted: 23 February 2022;
Published: 15 March 2022.
Edited by:
Yoshihiro Komatsu, University of Texas Medical School at Houston, United StatesCopyright © 2022 Dong, Wan, Zhou, Nie, Chen, Diao and Gao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ze-Xia Gao, Z2FvenhAbWFpbC5oemF1LmVkdS5jbg==
 Xiao-Ru Dong
Xiao-Ru Dong Shi-Ming Wan
Shi-Ming Wan Jia-Jia Zhou1,2
Jia-Jia Zhou1,2 Yu-Long Chen
Yu-Long Chen Ze-Xia Gao
Ze-Xia Gao