- 1Center for Reproductive Health, School of Pharmaceutical Sciences, Hangzhou Medical College (Zhejiang Academy of Medical Sciences), Hangzhou, China
- 2Department of Urology and Andrology, Sir Run-Run Shaw Hospital, Zhejiang University School of Medicine, Hangzhou, China
- 3Zhejiang Provincial Laboratory of Experimental Animal’s & Nonclinical Laboratory Studies, Hangzhou Medical College, Hangzhou, China
In mammalian testes, the apical cytoplasm of each Sertoli cell holds up to several dozens of germ cells, especially spermatids that are transported up and down the seminiferous epithelium. The blood-testis barrier (BTB) established by neighboring Sertoli cells in the basal compartment restructures on a regular basis to allow preleptotene/leptotene spermatocytes to pass through. The timely transfer of germ cells and other cellular organelles such as residual bodies, phagosomes, and lysosomes across the epithelium to facilitate spermatogenesis is important and requires the microtubule-based cytoskeleton in Sertoli cells. Kinesins, a superfamily of the microtubule-dependent motor proteins, are abundantly and preferentially expressed in the testis, but their functions are poorly understood. This review summarizes recent findings on kinesins in mammalian spermatogenesis, highlighting their potential role in germ cell traversing through the BTB and the remodeling of Sertoli cell-spermatid junctions to advance spermatid transport. The possibility of kinesins acting as a mediator and/or synchronizer for cell cycle progression, germ cell transit, and junctional rearrangement and turnover is also discussed. We mostly cover findings in rodents, but we also make special remarks regarding humans. We anticipate that this information will provide a framework for future research in the field.
Introduction
Kinesins, dyneins, and myosins are three large superfamilies of motor proteins that are involved in intracellular transport. Kinesins and dyneins, which are powered by the hydrolysis of adenosine triphosphate (ATP), generally move in opposite directions along microtubule tracks, toward the microtubule plus and minus ends (i.e., away from or towards the center of the cell), respectively. Myosins, on the other hand, hydrolyze ATP to enable its movement on filamentous actin (F-actin), towards the fast-growing “barbed” end (plus end) of the polar microfilaments [for reviews (Kato et al., 2018; Sweeney and Holzbaur, 2018),]. Here, we focus on kinesin superfamily proteins (KIFs) and their evolving functions in mammalian spermatogenesis.
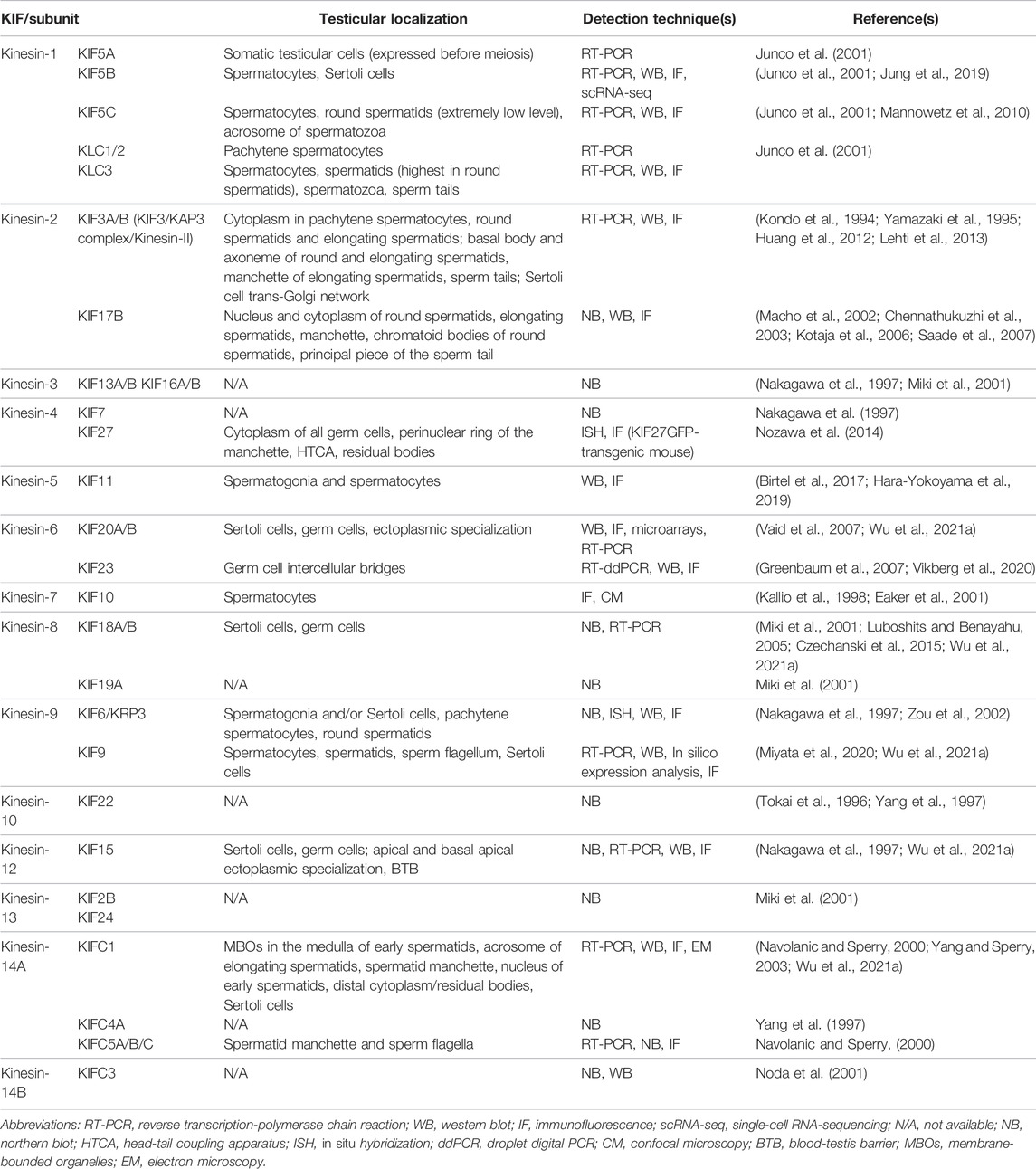
The first kinesin (also known as conventional kinesin or kinesin-1) was discovered in 1985 (Vale et al., 1985; Brady, 1985). Since then, dozens more have been identified, and before a unified nomenclature was proposed in 2004, there were numerous designations and redundant names for a single KIF (Lawrence et al., 2004; Endow et al., 2010). For example, KIF23, a member of the kinesin-6 family, was also known as CHO1/MKLP1 (human), ZEN-4/CeMKLP1 (C. elegans), and Pavarotti/MKLP1 (Pav-KLP, Drosophila) (Kuriyama et al., 2002). KIFs appear to be present in all eukaryotes, from fungi to plants and animals, as well as in different cell types. A typical KIF molecule has a motor domain—the head—that contains the ATP and microtubule binding sites and is referred to as the catalytic core; a tail domain opposite the motor that is involved in adaptor or cargo binding; and a long filamentous coiled-coil stalk in between the head and the tail that allows oligomerization and self-interaction, as well as regulation of microtubule binding and subcellular localization of the motor and cargo (Figure 1) (Miki et al., 2001; Marx et al., 2009; Hirokawa and Tanaka, 2015; Hirokawa et al., 2020). The motor domains of KIFs share considerable homology, which defines the superfamily. Outside of this conserved region, KIF members have minimal commonalities, with each carrying class-specific domains that recognize different payloads. KIFs come in a variety of shapes, including monomers, heterologous or homologous dimers, trimers, and tetramers (Figure 2). For instance, the founding member of the superfamily, kinesin-1, is a heterotetrameric motor made up of two kinesin heavy chain (KHC) and two kinesin light chain (KLC) subunits. The motor domain is housed in the KHCs, which interacts with the KLCs that connects to the cargo (Figure 1). KIFs have been shown to transport a wide variety of cargoes through the cell, including proteins, lipid droplets, mRNAs, chromosomes, organelles, and vesicles such as mitochondria, endoplasmic reticulum, early endosomes, late endosomes, and lysosomes. They have a role in pathogenesis of metabolic diseases, brain development, neurotransmitter transport, cilia and flagella assembly, and chromosome and spindle function during mitosis and meiosis. To date, over 45 mouse and human KIFs have been identified, which are further divided into 14 classes of three major types based on the position of the motor domain, namely N-kinesins with the motor domain in the N-terminal region (kinesin-1–12), M-kinesins with the motor domain located internally in the middle of the polypeptide chain (kinesin-13), and C-kinesins with the motor domain located C-terminally (kinesin-14) (Figure 2). Interestingly, the direction of cargo transport mirrors this functional classification, with N- and C-kinesins driving microtubule plus and minus end-directed transport, respectively, whereas M-kinesins displaying non-motor activities of depolymerizing microtubules (Miki et al., 2001; Marx et al., 2009; Hirokawa and Tanaka, 2015; Hirokawa et al., 2020). Some N- and C-kinesins also perform non-motor functions. Kinesin-8, for example, is a depolymerizing kinesin that regulates the length and disassembly of microtubules at the plus-ends (Ohi et al., 2013). Kinesin-5 has crosslinking and sliding properties for anti-parallel microtubules (Kapitein et al., 2005; van den Wildenberg et al., 2008).
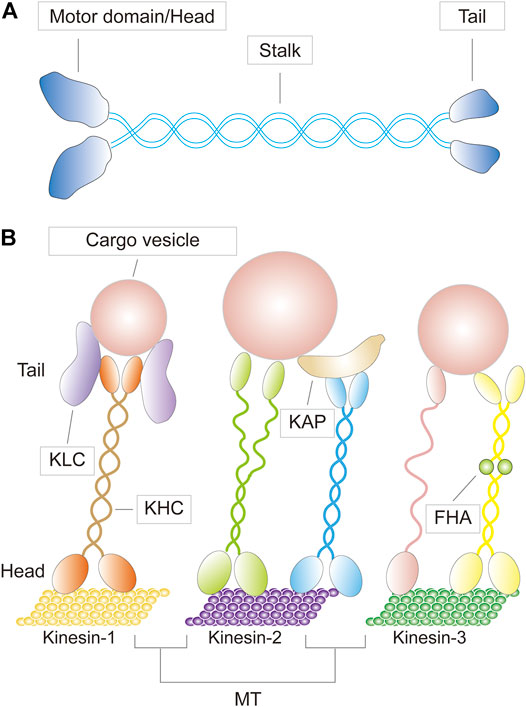
FIGURE 1. (A) A schematic diagram depicting the basic structure of a kinesin motor (B) Three representative kinesins (kinesin-1,-2, and -3) bearing cargoes traveling on microtubule (MT) tracks, with their specific molecular structures shown. KLC, kinesin light chain; KHC, kinesin heavy chain; KAP, kinesin associated protein; FHA, forkhead associated domain.

FIGURE 2. The domain structure and classifications of kinesins discovered in each of the human, mouse, and rat genomes (except that KIF16A and KIF19B are predictive in the rat). KIF11 is also known as KIF8 in the mouse, and KIF6 is also known as kinesin-related protein 3 (KRP3) in the rat. KIFC1 has isoforms KIFC4 and KIFC5 in the mouse. Kinesins share a common structure but also have domains that are unique to each KIF member. KLC, kinesin light chain; KHC, kinesin heavy chain; TPR, tetratricopeptide repeat; WD40 repeats, protein interaction motifs of approximately 40 amino acids that usually terminate in tryptophan-aspartic acid (WD); FHA, forkhead associated; PH, Pleckstrin homology; CAP-Gly, cytoskeleton-associated protein glycine-rich; PX, PhoX homologous; HhH1, Helix-hairpin-Helix DNA-binding motif class 1.
Using Northern blotting, PCR analysis with cDNA, and/or immunoblotting, most subfamilies of KIFs have been shown to be preferentially expressed in rodent testes (Table 1). Many KIFs, including KIF6, KIF7, KIF9, KIF17B, KIF18B, KIFC1, and KIFC3, are predominantly, if not exclusively, expressed in the testis (Nakagawa et al., 1997; Miki et al., 2001; Noda et al., 2001; Yang and Sperry, 2003; Wong-Riley and Besharse, 2012). The presence of such large amounts of KIFs in the testis clearly indicates that the testis has a very robust intracellular transport system that relies on KIF participation. Mammalian spermatogenesis, which occurs in the testis, is one of the most important developmental processes in nature. During this process, KIFs are involved in spindle formation and chromosome segregation during meiosis, acrosome biogenesis, nuclear shaping of the sperm head, flagellar movements, spermatid transcription, and spermatid transport within Sertoli cell crypts (Vogl et al., 2008; Tang et al., 2016a; Ma et al., 2017). Mutations in KIFs have been shown to impact male fertility in mouse models (Table 2). Their function in the mammalian testis, however, is unclear. Although the testis contains a large number of mitotically and meiotically active germ cells, as well as highly polarized Sertoli cells as one type of specialized epithelial cell, KIF has received less attention in testicular biology than in mitosis or other polarized cell settings such as neurons and epithelial/endothelial cells. The goal of this review is to summarize recent KIF findings related to mammalian spermatogenesis. We discuss herein the significance of this superfamily of microtubule motor proteins in the testis, with a focus on their probable function in the remodeling of the blood-testis barrier (BTB) and the Sertoli cell-spermatid junction dynamics to facilitate germ cell transport. We anticipate that this information will serve as a basis for future studies in the field.
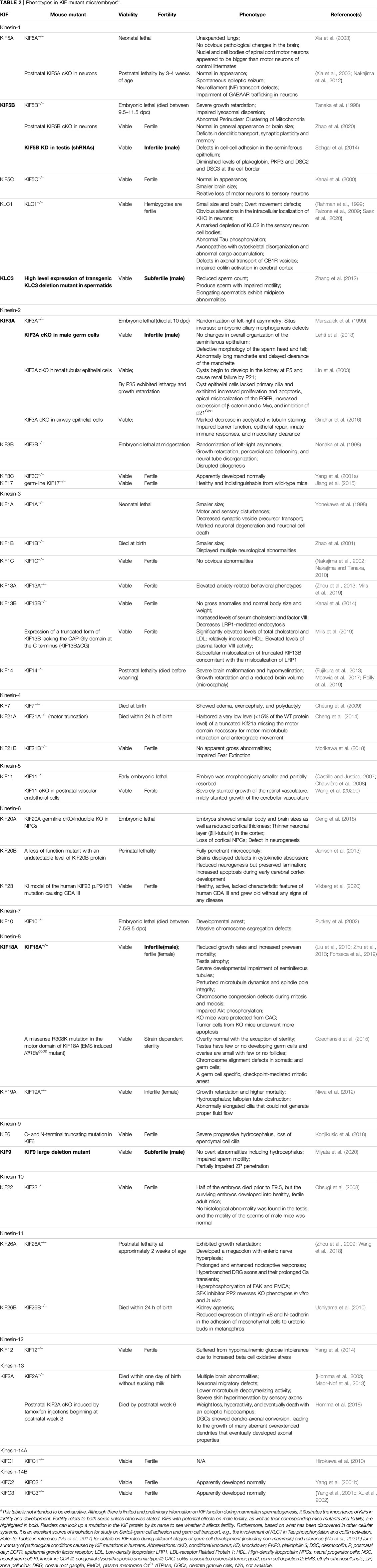
TABLE 2. Phenotypes in KIF mutant mice/embryosa.
Kinesins in Germ Cell Development in Mammalian Testes
Cell Division Regulation
Mammalian testes produce sperm over a male’s lifetime. Diploid spermatogonia evolve into mature haploid spermatozoa in a cyclic process called spermatogenesis. Each spermatogenic wave is characterized by three distinct functional phases: 1) spermatogonia (2N, 2C) proliferation by mitosis, 2) a single primary spermatocyte (2N, 4C) to yield two secondary spermatocytes (1N, 2C) by meiosis I, followed by four haploid round spermatids (1N, 1C) by meiosis II, and 3) transformation of spermatids into streamlined, flagellated, and functionally competent spermatozoa by spermiogenesis (Figure 3) (Griswold, 2016; de Kretser et al., 1998). Mitosis and meiosis, the two successive cell divisions in spermatogenesis, are tightly regulated processes, with the latter being a fundamental element of gametogenesis (Griswold, 2016; de Kretser et al., 1998). Kinesins are involved in spindle morphogenesis, chromosomal alignment and segregation during mitosis and meiosis in mammalian testes. For example, KIF11 (Eg5/kinesin-5) is a known mitotic kinesin that regulates microtubule sliding in mammalian somatic cells to facilitate bipolar spindle assembly (Hara-Yokoyama et al., 2019). During mouse spermatogenesis, KIF11 is closely associated with mitosis and meiosis and can serve as a marker of spermatogenic progression, even in mutant mice with arrested spermatogenesis, by giving a timeline of spindle formation (Hara-Yokoyama et al., 2019). When a specific/selective KIF11 inhibitor such as Monastrol, S-trityl-l-cysteine, or Dimethylenastron was injected into mouse testis, dividing spermatocytes exhibited impaired spindle bipolarity following KIF11 suppression. KIF11 inhibition also causes a reduction in spermatid count and defective sperm in mice (She et al., 2020a). By immunofluorescent staining, KIF10 from kinesin-7 family was revealed to be expressed in germ cells in the mouse testis. Following either intraperitoneal or intratesticular injection of a selective KIF10 inhibitor GSK923295, spermatogenesis in the mouse testis was disrupted, possibly due to a mechanism in which KIF10 malfunction causes problems in chromosome alignment and spindle assembly during meiosis (She et al., 2020b). KIF20A, a member of the kinesin-6 family, was also discovered to be required for central spindle organization and cytokinesis in meiosis, as well as acrosome formation during mouse spermatogenesis, according to the same research group (She et al., 2020c). Despite this, more research into the cellular expression and localization of KIFs in the testis is needed, as many of the KIFs that are known to be expressed in the testis have an ambiguous cellular localization, according to the summary of findings in Table 1. As in the case of KIF11, it remains to be verified whether it is restricted to germ cells (esp. spermatogonia and spermatocytes) rather than somatic Sertoli cells of the mouse testis (Hara-Yokoyama et al., 2019; She et al., 2020a), e.g., by immunoblotting and/or mRNA analysis utilizing purified germ or Sertoli cells isolated from rodent testes.

FIGURE 3. (Left) A diagram depicting the cell cycle progression and different germ cell types during spermatogenesis. (Right) A schematic drawing that illustrates the cross section of the seminiferous tubule in the rat testis. Four types of Sertoli cell-cell junctions at the blood-testis barrier (BTB), namely tight junction (TJ), basal ectoplasmic specialization, gap junction (GJ), and desmosome, as well as cell-cell junctions at the Sertoli-germ cell interface are illustrated. Spermatogonia, spermatocytes, and steps 1–7 round spermatids are connected to Sertoli cells via GJ/desmosome. More advanced spermatids (steps 8 and later) are connected to Sertoli cells via apical ectoplasmic specialization. The BTB, which is created by two adjacent Sertoli cells, divides the seminiferous epithelium into two compartments: adluminal and basal. At stage VIII of the epithelial cycle, the “new” BTB forms before the “old” BTB disassembles to allow the transport of preleptotene/leptotene spermatocytes. Also, near the seminiferous tubule lumen, apical ectoplasmic specialization disassembles to release the elongated spermatids during spermiation. The apical ectoplasmic specialization is hypothesized to be associated with microtubule (MT)-based motors (kinesins and dyneins) that allow “down and up” movement in the apical seminiferous epithelium of maturing spermatids.
The relevance of kinesins in cell cycle progression is further demonstrated in studies employing knockout mice (Table 2). KIF18A (Kinesin-8 family) activity is required for correct chromosome congression during cell division, and its absence in mice causes significant abnormalities in microtubule dynamics, spindle integrity, and checkpoint activation, resulting in germinal cell aplasia (Liu et al., 2010). Understanding the temporal and spatial expression patterns of KIFs in the testis (e.g., before or after meiosis, as well as expression and intensity in different germ cell types) may also help to elucidate their role in the cell cycle. Mammalian kinesin-1 subtypes [KLC (KLC1, 2 and 3) and KHC (KIF5A, B, and C)], for example, exhibit highly distinct spatiotemporal testicular patterns. KLC3 and KIF5C are expressed in spermatids after meiosis, whereas KLC1, KLC2, KIF5A, and KIF5B are expressed prior to meiosis, with KIF5A identified in somatic testicular cells and the others in spermatocytes (Junco et al., 2001). This suggests that different combinations of kinesin-1 subunits may function at different stages of germ cell differentiation. For a more comprehensive review regarding the role of kinesins in mitosis and meiosis (including non-mammalian), please see (Ma et al., 2017).
Spermiogenesis
The morphogenesis of the sperm head and flagellum are two important aspects of spermiogenesis, and the shaping of the spermatid head begins with the formation of the acrosome. The acrosome is a Golgi-derived secretory organelle that covers the anterior half of the sperm head. It is a cap-shaped structure that contains digestive enzymes needed for fertilization. The F-actin-based acroplaxome in the subacrosomal region, as well as the transitory microtubule- and F-actin-containing manchette that envelops the spermatid nucleus, are all part of the spermatid head shaping machinery and work with the acrosome as an endogenous regulating force to remodel the spermatid head (Kierszenbaum and Tres, 2004; Dunleavy et al., 2019; Khawar et al., 2019). KIFC1, for example, which associates with the manchette and the nuclear membrane, is known to play a role in vesicular trafficking during acrosome biogenesis and nuclear shaping in the rat, possibly through a nucleocytoplasmic transport mechanism (Yang and Sperry, 2003; Yang et al., 2006). In humans, reduced testicular KIFC1 expression has been associated to globozoospermia, a male infertility syndrome typified by the presence of round-headed spermatozoa lacking the acrosome (Zhi et al., 2016). KIF3A, KIF5C, KIF17B, KIFC5A, Kinesin-related protein (KRP)3A, and 3B (KIF6) in acrosome biogenesis and nuclear shaping; as well as KLC3, KIF3A, and KIF17B in sperm tail formation, have been reviewed recently and are not reiterated here (Ma et al., 2017; Pleuger et al., 2020). Multiple KIFs are believed to be related to the manchette, including KLC3, KIF3A, KIF3B, KIF5C, KIF10, KIF17B, KIF27, and KIFC5A (Ma et al., 2017; She et al., 2020b; Pleuger et al., 2020).
It is conceivable that large amounts of spermatid/sperm proteins must be synthesized due to the massive morphological and structural changes that occur during spermiogenesis. A highly specialized approach to postmeiotic gene expression has evolved in the testis. During mid-spermiogenesis, for example, the nucleus enters a transcriptional pause, and mRNAs required for spermatid differentiation are saved for later use until the nucleus resumes translation in elongate spermatids. Premature mRNA translation, which interrupts spermiogenesis, causes fertility problems (Hecht, 1998; Braun, 1998; O'Donnell, 2014; Geisinger et al., 2021). KIF17B, a testis-specific and enriched kinesin, has been demonstrated to be crucial for CREM (cAMP-responsive element modulator, a transcriptional activator)-dependent transcription and translation in spermatids. In the mouse testis, KIF17B functionally interacts with the transcriptional coactivator ACT (activator of CREM in testis) (Macho et al., 2002; Kotaja et al., 2005; Hogeveen and Sassone-Corsi, 2006) and mRNAs of ACT/CREM target genes (RNA-binding proteins such as TB-RBP and Miwi, which bind directly to KIF17B, form complexes with those mRNAs and act as adaptor, repressor, and/or stabilizer proteins) (Deng and Lin, 2002; Chennathukuzhi et al., 2003; Hogeveen and Sassone-Corsi, 2006; Kotaja et al., 2006), and KIF17B regulates their nucleocytoplasmic trafficking in post-meiotic spermatids. Thus, by attaching or detaching such gene expression elements and transferring them between the nucleus and the cytoplasm, KIF17B modulates the time of transcription termination and/or translation initiation. In the case of the ACT, KIF17B shuttle is driven by protein kinase A (PKA)-dependent phosphorylation rather than a motor- or microtubule-dependent mechanism, as deletion of the motor domain or disruption of the cellular microtubule network have no effect on KIF17B mobility (Kotaja et al., 2005; Hogeveen and Sassone-Corsi, 2006).
Germ Cell Transport Across the Seminiferous Epithelium
Sertoli-Germ and Sertoli-Sertoli Cell Junctions in the Seminiferous Tubules
Unlike motile cells, male germ cells do not have the motile appendages formed by dynamic actin structures, such as lamellipodia and filopodia in fibroblasts; therefore, they are non-motile and metabolically inactive before exiting the seminiferous tubule to reach the epididymis. Sertoli cells are the “mother” or “nurse” cells that are entirely responsible for the nourishment and transfer of germ cells. Characterized by a teardrop-shaped nucleus and an irregular cell outline, the Sertoli cell stretches from the basement membrane to the lumen of the seminiferous tubules and contains 30–50 germ cells at various stages of differentiation (Figure 3). In rodent testes, spermatogonia, spermatocytes, and steps 1–7 round spermatids are connected to Sertoli cells via intermediate filament-based desmosomes and/or actin-based gap junctions (Pointis and Segretain, 2005; Kopera et al., 2010; Mruk and Cheng, 2011; Xiao et al., 2014a; Rode et al., 2021). When the nucleus polarizes to one side of the cytoplasm and the acrosomal region makes contact with the cell surface (typically the basal side facing the basement membrane), step 8 round spermatids in stage VIII tubules begin to elongate (O'Donnell, 2014), and the desmosome/gap junction is replaced with a type of actin-based adherens junction (AJ) unique to the testis known as apical ectoplasmic specialization (Grove and Vogl, 1989; Vogl et al., 2000; Lee and Cheng, 2004; Cheng and Mruk, 2010a). The apical ectoplasmic specialization disintegrates at the end of spermiogenesis, allowing mature sperm to be released from the seminiferous epithelium at spermiation (O'Donnell, 2014). Throughout their development, the germ cells differentiate while remaining attached to the Sertoli cell, and substantial crosstalk occurs at the Sertoli-germ cell junctions to aid in germ cell trafficking across the seminiferous epithelium (Mruk and Cheng, 2004; Wu et al., 2020).
The blood-testis barrier (BTB), which is created by the basolateral borders of two neighboring Sertoli cells, is another location with complex cell junction dynamics (Figure 3). BTB is an ultrastructure composed of different types of junctions that coexist and interact with one another, namely tight junctions, basal ectoplasmic specialization [although ultrastructurally identical to the apical ectoplasmic specialization, it exhibits differences; for a review, see (Lee and Cheng, 2004)], desmosomes, and gap junctions (Cheng and Mruk, 2012). The BTB physically splits the seminiferous epithelium into two different compartments: adluminal and basal. A-single (As) spermatogonia present as spermatogonial stem cells (SSCs) in the basal compartment. After several rounds of mitosis, type B spermatogonia are produced, followed by preleptotene/leptotene spermatocytes, the only types of germ cells transported apically through the BTB at stages VIII-IX of the seminiferous epithelial cycle. Therefore, the BTB must undergo cyclic remodeling at the appropriate time to ensure that primary spermatocytes enter the adluminal compartment from the basal compartment without compromising the integrity of the BTB (Figure 3) (Dym and Fawcett, 1970; Clermont, 1972; Russell, 1977; de Rooij, 2001; Cheng and Mruk, 2012).
Cell cycle progression and sperm morphogenesis are proposed to be synchronized with germ cell movement and junction restructuring during the seminiferous epithelial cycle of spermatogenesis (Cheng and Mruk, 2010b; Lie et al., 2009). Very little is known about the role of kinesins in these orchestrated processes. In the aforementioned nucleocytoplasmic transport of ACT, KIF17B migrates into the cytoplasm with ACT at the commencement of spermatid elongation around stages VIII-IX of the seminiferous epithelial cycle in the mouse testis, and its cytoplasmic localization is determined by PKA-dependent phosphorylation (Kotaja et al., 2005). At the same stages as KIF17B relocation, a series of coordinated cytoskeletal and junctional events occur simultaneously in the epithelium: 1) disassembly of the apical ectoplasmic specialization for spermiation; 2) assembly of the new apical ectoplasmic specialization at the step 8 spermatid-Sertoli cell interface; and 3) BTB reorganization with “new” BTB assembly and “old” BTB disassembly for spermatocyte transport across the BTB (Figure 3). The cell junctions and cytoskeletons undergo significant turnover, as does communication between Sertoli and germ cells, necessitating extensive and timely transport of proteins, mRNAs, organelles, and vesicles (Cheng and Mruk, 2010a; Cheng and Mruk, 2012). PKA activation has been shown to perturb the Sertoli cell permeability barrier in vitro, suggesting that it may contribute to the breakdown of the “old” BTB (Li et al., 2001). Although KIF17B has proven to be a versatile kinesin, its involvement in these signaling events has yet to be characterized, such as whether its translocation and phosphorylation by PKA trigger any cytoskeletal and junctional events. Another such situation is KIF5B. According to a study (Sehgal et al., 2014), KIF5B couples with KLC1 in the mouse testis. KIF5B deficiency induces male sterility, while its knockdown substantially affects testicular cell adhesion. KIF5B regulates plakoglobin transport to the site of desmosome formation in the mouse testis by forming a complex with 14-3-3γ. The desmosome is a form of intermediate filament-based anchoring junction in the testis that is less well understood. As previously noted, desmosomes are located between Sertoli cells at the BTB, as well as between Sertoli cells and germ cells at early stages, such as spermatogonia and spermatocytes (Mruk and Cheng, 2011). Correspondingly, KIF5B and KLC1 were discovered to be expressed in spermatocytes prior to meiosis in mice and rats (Junco et al., 2001). A recent study, however, demonstrated KIF5B to be present in mouse Sertoli cells, where it co-localized with vimentin intermediate filaments (Jung et al., 2019). Therefore, in addition to identifying whether KIF5B/KLC1 is an integrated component of the BTB and/or confers cell adhesion of the spermatocytes to Sertoli cells, more research is needed to examine whether KIF5B plays any role in synchronization of the spermatocyte differentiation and its transport across the seminiferous epithelium (e.g., regulating the cell cycle of spermatocytes vs initiating “new” desmosome formation at the BTB and/or between Sertoli cells and spermatocytes).
Germ Cell Transport Supported by Actin- and Microtubule-Based Cytoskeletons
The cytoskeleton, which includes actin, microtubule, intermediate filament, and septin networks, is essential for the formation and disassembly of Sertoli-germ and Sertoli-Sertoli cell junctions, as well as the regulation of germ cell development and transport across the seminiferous epithelium (Dunleavy et al., 2019; Wu et al., 2021a). Although the role of intermediate and septin filaments in the testis has not been thoroughly studied, there is a better understanding of the actin- and microtubule-based cytoskeletons, which we will discuss below.
During mammalian spermatogenesis, the timely transport of a wide range of organelles and vesicles, including developing germ cells, across the seminiferous epithelium is vital, and it is largely accomplished by microtubule and actin tracks and their associated motors (kinesin, dynein, and myosin superfamilies). Long-distance longitudinal transport of cellular cargoes is assumed to be propelled along microtubule paths, whereas short-distance transverse transport is facilitated by F-actin. Sertoli cells feature massive networks of microtubule and F-actin at different stages of the epithelial cycle to ensure proper cargo delivery (Dunleavy et al., 2019; Hirokawa et al., 2010; Wu et al., 2021b; Barlan and Gelfand, 2017; Wen et al., 2016). F-actin was used for attachment by cell junction proteins at the apical ectoplasmic specialization, as well as tight junction, basal ectoplasmic specialization, and gap junctions at the BTB. At stage VIII of the seminiferous epithelial cycle, transport of preleptotene/leptotene spermatocyte from the basal to the adluminal compartment is dependent on the restructuring of Sertoli cell junctions at the BTB to guarantee meiosis at stage XIV in the rat or XII in the mouse (Figure 3). F-actin tracks across the seminiferous epithelium are only most visible in late stage VIII tubules, implying that actin-based traffic is likely transient (Wang et al., 2020a; Chenouard et al., 2020). Microtubule bundles, on the other hand, run perpendicular to the basement membrane (parallel to the direction of spermatid transport) and stretch over the entire seminiferous epithelium during various stages of the epithelial cycle, with the plus ends at the base and the minus ends at the apex of the Sertoli cell. Microtubule tracks are used for spermatid transport and are intimately linked to the apical ectoplasmic specialization. Developing spermatids travel “down and up” the seminiferous epithelium in the adluminal compartment before returning to the apical regions for spermiation. This is considered to mirror the movement of apical ectoplasmic specialization along the nearby microtubule by engaging either plus or minus end motor proteins (Figure 3) (Dunleavy et al., 2019; Wen et al., 2016; Tang et al., 2016b; Tang et al., 2015; Vaid et al., 2007; Guttman et al., 2000; Beach and Vogl, 1999). Indeed, several kinesins have been implicated in spermatid translocation in the rat/mouse testis, including KIF6/KRP3, KIF20, and KIF15, since they appear to be associated with apical ectoplasmic specialization, elongating spermatids, and/or Sertoli cells (Table 1) (Zou et al., 2002; Vaid et al., 2007). However, functional interactions between these kinesins and components of the apical ectoplasmic specialization can be tested using assays like co-immunoprecipitation to refine the findings. The microtubule track is critical for the delivery of elongated spermatids to the tubule lumen and for the timely completion of spermiation. It is involved in disruption of the apical ectoplasmic specialization upon sperm release (Li et al., 2017; Dunleavy et al., 2019). Furthermore, a major microtubule minus end-directed motor protein, Dynein 1, has been shown to maintain proper F-actin structure in cultured primary Sertoli cells and in adult rat testes. This demonstrates that microtubules also contribute to the integrity of the F-actin network in the testis (Wen et al., 2018).
Kinesins are emerging as regulators or components of cytoskeletons other than microtubules, indicating the coordination of multiple cytoskeletal systems. According to a study combining an in vitro model system and single-molecule techniques, kinesin may collaborate with myosin to deliver a specific cargo on both microtubule and actin cytoskeletal tracks (Ali et al., 2008). In Xenopus oocytes, a nuclear and meiotic actin-bundling kinesin (NabKin) directly links microtubules with F-actin and coordinates the interactions of these two cytoskeletons. Its somatic paralogue is the actin-binding domain-containing kinesin KIF14 (Samwer et al., 2013). Septin 9, a microtubule-associated septin, has been uncovered to affect interactions between KIF17 and its membrane cargo, as well as the dendritic entry of KIF5 and KIF1 in neurons (Bai et al., 2016; Karasmanis et al., 2018). It has long been known that conventional kinesin is essential for the interplay of vimentin intermediate filaments with microtubules in various cells (Gyoeva and Gelfand, 1991; Liao and Gundersen, 1998; Prahlad et al., 1998). Research into mammalian testes, on the other hand, is still in the early stages. A recent study has shown that, KIF15 knockdown not only impairs Sertoli cell barrier function in vitro, but also disturbs the homeostasis of the four cytoskeletons in Sertoli cells: microtubule, actin, vimentin, and septin (Wu et al., 2021a), demonstrating that kinesins have a more profound and extensive role in mammalian testis with its regulatory effects that extend beyond microtubules. Whether kinesins are required for germ cell transport facilitated by multiple cytoskeletons, as well as how kinesins select which track to use and promote cytoskeleton coordination in the testis, merit further investigation.
Kinesins in Regulation of Epithelial Junctions
Sertoli cell junctions at the BTB and spermatid adhesion onto Sertoli cells, as stated previously, underlie mammalian spermatogenesis and confer Sertoli cell function in the basal and adluminal compartments, respectively. Although the evidence for KIF participation in BTB and apical ectoplasmic specialization is sparse, other epithelial cells, cancer cells, and different migratory cells can provide insight.
Focal adhesion (FA) or focal contact links the internal actin cytoskeleton of an interacting cell with the extracellular matrix (ECM), which is important to cell adhesion and migration. FAs are large, dynamic protein complexes comprising transmembrane integrins and their ECM ligands (such as fibronectin, collagen and laminin), as well as a number of adaptor, scaffold, docking, and signaling proteins between integrins and F-actin [such as talin, vinculin, paxillin, p130Cas, non-receptor tyrosine kinases FAK (focal adhesion kinase) and SFKs (SRC family kinases)], all of which are involved in the dynamic association with the actin cytoskeleton (Wozniak et al., 2004; Mitra et al., 2005; Martino et al., 2018). KIF5A enhances the transport and secretion of type I collagen for FA assembly, according to recent research in lung adenocarcinoma cells (Tan et al., 2022). To increase endoplasmic reticulum (ER) transport to the cell surface and boost FA growth and maturation during cell migration, KIF5B forms a complex with the ER-resident protein kinectin-1 and the small GTPase Rab18 in human osteosarcoma epithelial cells (Guadagno et al., 2020). KIF13A has an impact on FA dynamics in lung adenocarcinoma cells (Banerjee et al., 2021). Various miRNAs can control KIFs to support FA function in gastric cancer cells (Ma et al., 2021). The testis, on the other hand, lacks focal contact. But the same FA component proteins, such as FAK, SFKs, integrins, and laminins, have been identified at the Sertoli-germ and Sertoli-Sertoli cell junctions in the rat testis. The presence of laminin and collagen chains in the basement membrane of the seminiferous tubule has also been reported in new research (Li et al., 2021). FAK and SRC, for instance, are found in the tight junctions and basal ectoplasmic specialization at the BTB, as well as the apical ectoplasmic specialization at the spermatid-Sertoli cell interface. Both FAK and SRC are structurally coupled with the occludin/ZO-1 adhesion complex located at the BTB. The α6β1-integrin/laminin-α3β3γ3 complex is the major component of the apical ectoplasmic specialization, and FAK physically interacts with β1-integrin, which is important during spermiation for sperm release from the seminiferous epithelium (Cheng and Mruk, 2010a; Cheng and Mruk, 2012). This is analogous to how FAK and SRC collaborate to promote FA disassembly in other epithelia or cancer cells (Mitra et al., 2005).
KIF22 binds to CAR (coxsackievirus and adenovirus receptor) and its regulation of microtubule dynamics influences CAR localization at the lung cancer cell-cell junctions (Pike et al., 2018). KIF26A modulates FA and cell adhesion possibly via its effect on protein levels of FAK/p-FAK, E-cadherin/N-cadherin, and p-β-catenin in gastric cancer and mesenchymal cells. KIF26A has also been demonstrated to interact directly with FAK and to drive a pathway that inhibits integrin-SFK-FAK pathway signaling in primary cultured neurons and mouse brains (Combes et al., 2015; Wang et al., 2018; Ma et al., 2021). KIF17 not only stabilizes microtubule, but also enhances cell-cell adhesions by boosting apical actin network assembly and junctional E-cadherin accumulation, and this activity is independent of microtubule binding in epithelial cells (Acharya et al., 2016; Kreitzer and Myat, 2018). While not being engaged in E-cadherin transport, KIFC3 reduces E-cadherin degradation by recruiting the ubiquitin-specific protease 47 (USP47, which deubiquitinates E-cadherin) to AJ, which prevents E-cadherin degradation and maintains the epithelial integrity (Meng et al., 2008; Sako-Kubota et al., 2014). In cultured epithelial cells, kinesin-1 is abundant in the apical junctional complex (AJC), and it similarly accumulates at the epithelial cell-cell contacts in normal human colonic mucosa, where it is coupled with the E-cadherin-catenin complex (Ivanov et al., 2006). Desmoglein-2 (DSG2) and desmocollin-2 (DSC2) are transported by kinesin-1 and -2, respectively, for desmosome assembly in epithelial cells (Nekrasova et al., 2011). KLC and KIF3A/B have both been implicated in endothelial/epithelial barrier function, with the latter involved in tight-junction protein transport and interaction with β-catenin (Wu et al., 2007; Aguado-Fraile et al., 2012; Giridhar et al., 2016; Kreitzer and Myat, 2018; Ichinose et al., 2019; Johansson et al., 2019; Stevens et al., 2020). These integral membrane proteins (CAR, E-cadherin/N-cadherin, DSG2/DSC2), as well as the signaling and adaptor proteins, are all established integrated components of the BTB and apical ectoplasmic specialization (Wu et al., 1993; Wang and Cheng, 2007; Cheng and Mruk, 2010a; Lie et al., 2010; Cheng and Mruk, 2012). As a result, it is worth investigating whether similar interactions occur in the testis. These findings illustrate that a single KIF is likely to favor a specific mode of cargo delivery and/or junction formation, as well as highlight the regulatory effects of KIF on cytoskeletons based on microtubule, actin, and intermediate filaments.
Kinesins in Human Male Reproduction
The involvement of kinesin in reproduction is an underappreciated topic that is only getting started. There is even less information available about humans. However, growing evidence indicates that kinesins can have an important role in human male reproduction. By analyzing Gene Expression Omnibus (GEO) datasets, KIF2C, and related miRNAs have been connected to spermatogenetic failure and non-obstructive azoospermia in patients (Cao et al., 2021). KIFC1 is linked to globozoospermia in human testes as previously mentioned (Zhi et al., 2016). KIFC1 and KIF3B are enriched in tissues of human seminoma, one of the most frequent testis cancers affecting young men, with a role in cell division regulation in seminoma (Xiao et al., 2017; Shen et al., 2017). Several kinesins, including KIF4, KIF5A, and KIF21B, have been proposed as candidates for human immunopathogenesis (Bernasconi et al., 2008; Alcina et al., 2010; International Multiple Sclerosis Genetics Consortium (IMSGC), 2010), but their contribution to male autoimmune infertility has not been established. On the other hand, KIF1C has been reported to confer dominant resistance to experimental autoimmune orchitis (EAO) in mice, which remains to be studied in human males (del Rio et al., 2012). KIF1A is predominantly expressed in the brain and nerves, and knockout mice died within 1 day of birth (Table 2). KIF1A-related disorders are relatively rare in the general population, but males with pathogenic KIF1A variations have been observed to have tiny testes or penises, as well as cryptorchidism (Kaur et al., 2020; Boyle et al., 2021; Pennings et al., 2020). Similarly, knockout mice for KIF7 died at birth (Table 2), and a boy with acrocallosal syndrome (ACLS) who carries a novel homozygous KIF7 nonsense mutation had unilateral maldescensus testis (Ibisler et al., 2015). These findings suggest that kinesins play a crucial role in human testis development, while it may be difficult to discern their functions from the phenotypes of experimental animal models. Cell lines or cultured primary cells obtained from humans could be a viable alternative to animal models, providing mechanistic insights without demanding fresh primary cultures from human testes. TCam-2, a human tumor cell line derived from a primary testicular seminoma of a 35-year-old men and assumed to reflect human male germ cells, was utilized to investigate KIF18A activity in proliferation, cell cycle and apoptosis in the human cell line (Smialek et al., 2020). The use of a well-established human Sertoli cell in vitro system sourced from cadaveric testes of males aged 12–36 years could be a useful tool for studying the role of kinesins in human Sertoli cell function and BTB dynamics (Chui et al., 2011; Xiao et al., 2014b; Chen et al., 2017).
Concluding Remarks
Herein, we review current findings on kinesins in mammalian spermatogenesis, including their role in cell cycle and sperm morphogenesis, as well as their putative function in germ cell transport through the BTB and the remodeling of cell junctions and underlying cytoskeletons to expedite germ cell transport across the seminiferous epithelium. We also propose a role of kinesins as mediators and/or synchronizers of cell cycle progression, germ cell transport, and junctional turnover in mammalian testes. Yet many aspects of kinesin function in the mammalian testis remain unexplored, such as the mechanism(s) by which they affect spermatogenesis globally and promote Sertoli-germ cell interactions during spermatogenesis. The loss of function of many kinesins resulted in lethality as well as infertility or subfertility in mice (Table 2), but the underlying pathway(s) or molecule(s) is unknown. Nonetheless, it might be able to help us better grasp the role of kinesins in human disorders or as disease markers, providing insight into male factor infertility and male autoimmune diseases. In vitro culture of germ cells (or germ cell-like cells) and Sertoli cells derived from human males is also a good supplement to animal model studies.
Author Contributions
XX conceived the ideas and laid the concepts for this review; MY and XX conducted the literature search and critically evaluated published findings in the field; MY, HQ, YH, CYC and XX critically discussed current concepts relevant to this review; MY and XX prepared the figures and tables; XX drafted, edited, and revised the manuscript; and MY, HQ, YH, CYC and XX approved the final version.
Funding
This work was supported by grants from the National Natural Science Foundation of China (U20A20133 and 31371176), Zhejiang Provincial Natural Science Foundation of China (LY21H040005), Zhejiang Provincial Department of Education (Y202045395), Health Commission of Zhejiang Province (2020RC052).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Acharya, B. R., Espenel, C., Libanje, F., Raingeaud, J., Morgan, J., Jaulin, F., et al. (2016). KIF17 Regulates RhoA-dependent Actin Remodeling at Epithelial Cell-Cell Adhesions. J. Cell Sci 129 (5), 957–970. doi:10.1242/jcs.173674
Aguado-Fraile, E., Ramos, E., Sáenz-Morales, D., Conde, E., Blanco-Sánchez, I., Stamatakis, K., et al. (2012). miR-127 Protects Proximal Tubule Cells against Ischemia/reperfusion: Identification of Kinesin Family Member 3B as miR-127 Target. PLoS One 7 (9), e44305. doi:10.1371/journal.pone.0044305
Alcina, A., Vandenbroeck, K., Otaegui, D., Saiz, A., Gonzalez, J. R., Fernandez, O., et al. (2010). The Autoimmune Disease-Associated KIF5A, CD226 and SH2B3 Gene Variants Confer Susceptibility for Multiple Sclerosis. Genes Immun. 11 (5), 439–445. doi:10.1038/gene.2010.30
Ali, M. Y., Lu, H., Bookwalter, C. S., Warshaw, D. M., and Trybus, K. M. (2008). Myosin V and Kinesin Act as Tethers to Enhance Each Others' Processivity. Proc. Natl. Acad. Sci. U.S.A. 105 (12), 4691–4696. doi:10.1073/pnas.0711531105
Bai, X., Karasmanis, E. P., and Spiliotis, E. T. (2016). Septin 9 Interacts with Kinesin KIF17 and Interferes with the Mechanism of NMDA Receptor Cargo Binding and Transport. MBoC 27 (6), 897–906. doi:10.1091/mbc.e15-07-0493
Banerjee, P., Xiao, G.-Y., Tan, X., Zheng, V. J., Shi, L., Rabassedas, M. N. B., et al. (2021). The EMT Activator ZEB1 Accelerates Endosomal Trafficking to Establish a Polarity axis in Lung Adenocarcinoma Cells. Nat. Commun. 12 (1), 6354. doi:10.1038/s41467-021-26677-y
Barlan, K., and Gelfand, V. I. (2017). Microtubule-Based Transport and the Distribution, Tethering, and Organization of Organelles. Cold Spring Harb Perspect. Biol. 9 (5), a025817. doi:10.1101/cshperspect.a025817
Beach, S. F., and Vogl, A. W. (1999). Spermatid Translocation in the Rat Seminiferous Epithelium: Coupling Membrane Trafficking Machinery to a Junction Plaque1. Biol. Reprod. 60 (4), 1036–1046. doi:10.1095/biolreprod60.4.1036
Bernasconi, P., Cappelletti, C., Navone, F., Nessi, V., Baggi, F., Vernos, I., et al. (2008). The Kinesin Superfamily Motor Protein KIF4 Is Associated with Immune Cell Activation in Idiopathic Inflammatory Myopathies. J. Neuropathol. Exp. Neurol. 67 (6), 624–632. doi:10.1097/nen.0b013e318177e5fd
Birtel, J., Gliem, M., Mangold, E., Tebbe, L., Spier, I., Müller, P. L., et al. (2017). Novel Insights into the Phenotypical Spectrum of KIF11-Associated Retinopathy, Including a New Form of Retinal Ciliopathy. Invest. Ophthalmol. Vis. Sci. 58 (10), 3950–3959. doi:10.1167/iovs.17-21679
Boyle, L., Rao, L., Kaur, S., Fan, X., Mebane, C., Hamm, L., et al. (2021). Genotype and Defects in Microtubule-Based Motility Correlate with Clinical Severity in KIF1A-Associated Neurological Disorder. HGG Adv. 2 (2). doi:10.1016/j.xhgg.2021.100026
Brady, S. T. (1985). A Novel Brain ATPase with Properties Expected for the Fast Axonal Transport Motor. Nature 317 (6032), 73–75. doi:10.1038/317073a0
Braun, R. E. (1998). Post-transcriptional Control of Gene Expression during Spermatogenesis. Semin. Cell Developmental Biol. 9 (4), 483–489. doi:10.1006/scdb.1998.0226
Cao, H., Wan, Z., Wang, F., Liu, Z., Li, X., and Hou, J. (2021). Downregulation of KIF2C and TEKT2 Is Associated with Male Infertility and Testicular Carcinoma. Aging 13 (19), 22898–22911. doi:10.18632/aging.203583
Castillo, A., and Justice, M. J. (2007). The Kinesin Related Motor Protein, Eg5, Is Essential for Maintenance of Pre-implantation Embryogenesis. Biochem. Biophysical Res. Commun. 357 (3), 694–699. doi:10.1016/j.bbrc.2007.04.021
Chauvière, M., Kress, C., and Kress, M. (2008). Disruption of the Mitotic Kinesin Eg5 Gene (Knsl1) Results in Early Embryonic Lethality. Biochem. Biophys. Res. Commun. 372 (4), 513–519. doi:10.1016/j.bbrc.2008.04.177
Chen, H., Gao, Y., Mruk, D. D., Xiao, X., John, C. M., Turek, P. J., et al. (2017). Rescue of PFOS-Induced Human Sertoli Cell Injury by Overexpressing a P-FAK-Y407e Phosphomimetic Mutant. Sci. Rep. 7 (1), 15810. doi:10.1038/s41598-017-15671-4
Cheng, C. Y., and Mruk, D. D. (2010). A Local Autocrine axis in the Testes that Regulates Spermatogenesis. Nat. Rev. Endocrinol. 6 (7), 380–395. doi:10.1038/nrendo.2010.71
Cheng, C. Y., and Mruk, D. D. (2010). The Biology of Spermatogenesis: the Past, Present and Future. Phil. Trans. R. Soc. B 365 (1546), 1459–1463. doi:10.1098/rstb.2010.0024
Cheng, C. Y., and Mruk, D. D. (2012). The Blood-Testis Barrier and its Implications for Male Contraception. Pharmacol. Rev. 64 (1), 16–64. doi:10.1124/pr.110.002790
Cheng, L., Desai, J., Miranda, C. J., Duncan, J. S., Qiu, W., Nugent, A. A., et al. (2014). Human CFEOM1 Mutations Attenuate KIF21A Autoinhibition and Cause Oculomotor Axon Stalling. Neuron 82 (2), 334–349. doi:10.1016/j.neuron.2014.02.038
Chennathukuzhi, V., Morales, C. R., El-Alfy, M., and Hecht, N. B. (2003). The Kinesin KIF17b and RNA-Binding Protein TB-RBP Transport Specific cAMP-Responsive Element Modulator-Regulated mRNAs in Male Germ Cells. Proc. Natl. Acad. Sci. U.S.A. 100 (26), 15566–15571. doi:10.1073/pnas.2536695100
Chenouard, N., Xuan, F., and Tsien, R. W. (2020). Synaptic Vesicle Traffic Is Supported by Transient Actin Filaments and Regulated by PKA and NO. Nat. Commun. 11 (1), 5318. doi:10.1038/s41467-020-19120-1
Cheung, H. O., Zhang, X., Ribeiro, A., Mo, R., Makino, S., Puviindran, V., et al. (2009). The Kinesin Protein Kif7 Is a Critical Regulator of Gli Transcription Factors in Mammalian Hedgehog Signaling. Sci. Signal. 2 (76), ra29. doi:10.1126/scisignal.2000405
Chui, K., Trivedi, A., Cheng, C. Y., Cherbavaz, D. B., Dazin, P. F., Huynh, A. L. T., et al. (2011). Characterization and Functionality of Proliferative Human Sertoli Cells. Cell Transpl. 20 (5), 619–635. doi:10.3727/096368910x536563
Clermont, Y. (1972). Kinetics of Spermatogenesis in Mammals: Seminiferous Epithelium Cycle and Spermatogonial Renewal. Physiol. Rev. 52 (1), 198–236. doi:10.1152/physrev.1972.52.1.198
Combes, A. N., Davies, J. A., and Little, M. H. (2015). Cell-cell Interactions Driving Kidney Morphogenesis. Curr. Top. Dev. Biol. 112, 467–508. doi:10.1016/bs.ctdb.2014.12.002
Czechanski, A., Kim, H., Byers, C., Greenstein, I., Stumpff, J., and Reinholdt, L. G. (2015). Kif18a Is Specifically Required for Mitotic Progression during Germ Line Development. Developmental Biol. 402 (2), 253–262. doi:10.1016/j.ydbio.2015.03.011
de Kretser, D. M., Loveland, K. L., Meinhardt, A., Simorangkir, D., and Wreford, N. (1998). Spermatogenesis. Hum. Reprod. 13 (Suppl. 1), 1–8. doi:10.1093/humrep/13.suppl_1.1
de Rooij, D. (2001). Proliferation and Differentiation of Spermatogonial Stem Cells. Reproduction 121 (3), 347–354. doi:10.1530/rep.0.1210347
del Rio, R., McAllister, R. D., Meeker, N. D., Wall, E. H., Bond, J. P., Kyttaris, V. C., et al. (2012). Identification of Orch3, a Locus Controlling Dominant Resistance to Autoimmune Orchitis, as Kinesin Family Member 1C. Plos Genet. 8 (12), e1003140. doi:10.1371/journal.pgen.1003140
Deng, W., and Lin, H. (2002). Miwi, a Murine Homolog of Piwi, Encodes a Cytoplasmic Protein Essential for Spermatogenesis. Developmental Cell 2 (6), 819–830. doi:10.1016/s1534-5807(02)00165-x
Dunleavy, J. E. M., O’Bryan, M. K., Stanton, P. G., and O’Donnell, L. (2019). The Cytoskeleton in Spermatogenesis. Reproduction 157 (2), R53–R72. doi:10.1530/rep-18-0457
Dym, M., and Fawcett, D. W. (1970). The Blood-Testis Barrier in the Rat and the Physiological Compartmentation of the Seminiferous Epithelium1. Biol. Reprod. 3 (3), 308–326. doi:10.1093/biolreprod/3.3.308
Eaker, S., Pyle, A., Cobb, J., and Handel, M. A. (2001). Evidence for Meiotic Spindle Checkpoint from Analysis of Spermatocytes from Robertsonian-Chromosome Heterozygous Mice. J. Cell Sci 114 (Pt 16), 2953–2965. doi:10.1242/jcs.114.16.2953
Endow, S. A., Kull, F. J., and Liu, H. (2010). Kinesins at a Glance. J. Cell Sci 123 (Pt 20), 3420–3424. doi:10.1242/jcs.064113
Falzone, T. L., Stokin, G. B., Lillo, C., Rodrigues, E. M., Westerman, E. L., Williams, D. S., et al. (2009). Axonal Stress Kinase Activation and Tau Misbehavior Induced by Kinesin-1 Transport Defects. J. Neurosci. 29 (18), 5758–5767. doi:10.1523/jneurosci.0780-09.2009
Fonseca, C. L., Malaby, H. L. H., Sepaniac, L. A., Martin, W., Byers, C., Czechanski, A., et al. (2019). Mitotic Chromosome Alignment Ensures Mitotic Fidelity by Promoting Interchromosomal Compaction during Anaphase. J. Cell Biol 218 (4), 1148–1163. doi:10.1083/jcb.201807228
Fujikura, K., Setsu, T., Tanigaki, K., Abe, T., Kiyonari, H., Terashima, T., et al. (2013). Kif14 Mutation Causes Severe Brain Malformation and Hypomyelination. PLoS One 8 (1), e53490. doi:10.1371/journal.pone.0053490
Geisinger, A., Rodríguez-Casuriaga, R., and Benavente, R. (2021). Transcriptomics of Meiosis in the Male Mouse. Front. Cell Dev. Biol. 9, 626020. doi:10.3389/fcell.2021.626020
Geng, A., Qiu, R., Murai, K., Liu, J., Wu, X., Zhang, H., et al. (2018). KIF20A/MKLP2 Regulates the Division Modes of Neural Progenitor Cells during Cortical Development. Nat. Commun. 9 (1), 2707. doi:10.1038/s41467-018-05152-1
Giridhar, P. V., Bell, S. M., Sridharan, A., Rajavelu, P., Kitzmiller, J. A., Na, C.-L., et al. (2016). Airway Epithelial KIF3A Regulates Th2 Responses to Aeroallergens. J.I. 197 (11), 4228–4239. doi:10.4049/jimmunol.1600926
Greenbaum, M. P., Ma, L., and Matzuk, M. M. (2007). Conversion of Midbodies into Germ Cell Intercellular Bridges. Developmental Biol. 305 (2), 389–396. doi:10.1016/j.ydbio.2007.02.025
Griswold, M. D. (2016). Spermatogenesis: The Commitment to Meiosis. Physiol. Rev. 96 (1), 1–17. doi:10.1152/physrev.00013.2015
Grove, B. D., and Vogl, A. W. (1989). Sertoli Cell Ectoplasmic Specializations: a Type of Actin-Associated Adhesion junction? J. Cell Sci 93 (Pt 2), 309–323. doi:10.1242/jcs.93.2.309
Guadagno, N. A., Margiotta, A., Bjørnestad, S. A., Haugen, L. H., Kjos, I., Xu, X., et al. (2020). Rab18 Regulates Focal Adhesion Dynamics by Interacting with Kinectin-1 at the Endoplasmic Reticulum. J. Cell Biol 219 (7), e201809020. doi:10.1083/jcb.201809020
Guttman, J. A., Kimel, G. H., and Vogl, A. W. (2000). Dynein and Plus-End Microtubule-dependent Motors Are Associated with Specialized Sertoli Cell junction Plaques (Ectoplasmic Specializations). J. Cell Sci 113 (Pt 12), 2167–2176. doi:10.1242/jcs.113.12.2167
Gyoeva, F. K., and Gelfand, V. I. (1991). Coalignment of Vimentin Intermediate Filaments with Microtubules Depends on Kinesin. Nature 353 (6343), 445–448. doi:10.1038/353445a0
Hara-Yokoyama, M., Kurihara, H., Ichinose, S., Matsuda, H., Ichinose, S., Kurosawa, M., et al. (2019). KIF11 as a Potential Marker of Spermatogenesis within Mouse Seminiferous Tubule Cross-Sections. J. Histochem. Cytochem. 67 (11), 813–824. doi:10.1369/0022155419871027
Hecht, N. B. (1998). Molecular Mechanisms of Male Germ Cell Differentiation. Bioessays 20 (7), 555–561. doi:10.1002/(sici)1521-1878(199807)20:7<555:aid-bies6>3.0.co;2-j
Hirokawa, N., and Miki, H. (2020). “Kinesin Superfamily Classification,” in Encyclopedia of Biophysics. Editors G. Roberts, and A. Watts (Berlin, Heidelberg: Springer Berlin Heidelberg), 1–10. doi:10.1007/978-3-642-35943-9_762-1
Hirokawa, N., Niwa, S., and Tanaka, Y. (2010). Molecular Motors in Neurons: Transport Mechanisms and Roles in Brain Function, Development, and Disease. Neuron 68 (4), 610–638. doi:10.1016/j.neuron.2010.09.039
Hirokawa, N., and Tanaka, Y. (2015). Kinesin Superfamily Proteins (KIFs): Various Functions and Their Relevance for Important Phenomena in Life and Diseases. Exp. Cell Res. 334 (1), 16–25. doi:10.1016/j.yexcr.2015.02.016
Hogeveen, K. N., and Sassone-Corsi, P. (2006). Regulation of Gene Expression in post-meiotic Male Germ Cells: CREM-Signalling Pathways and Male Fertility. Hum. Fertil. 9 (2), 73–79. doi:10.1080/14647270500463400
Homma, N., Ruyun, Z., Muhammad, I. N., Adeel, G. C., Mohammed, H. Al-Q., and Nobutaka, H. (2018). KIF2A Regulates the Development of Dentate Granule Cells and Postnatal Hippocampal Wiring. Elife 7, e30935. doi:10.7554/elife.30935
Homma, N., Takei, Y., Tanaka, Y., Nakata, T., Terada, S., Kikkawa, M., et al. (2003). Kinesin Superfamily Protein 2A (KIF2A) Functions in Suppression of Collateral branch Extension. Cell 114 (2), 229–239. doi:10.1016/s0092-8674(03)00522-1
Huang, C. J., Huang, C. C., and Chang, C. C. (2012). Association of the Testis-specific TRIM/RBCC Protein RNF33/TRIM60 with the Cytoplasmic Motor Proteins KIF3A and KIF3B. Mol. Cell Biochem 360 (1-2), 121–131. doi:10.1007/s11010-011-1050-8
Ibisler, A. l., Hehr, U., Barth, A., Koch, M., Epplen, J. r. T., and Hoffjan, S. (2015). Novel KIF7 Mutation in a Tunisian Boy with Acrocallosal Syndrome: Case Report and Review of the Literature. Mol. Syndromol 6 (4), 173–180. doi:10.1159/000439414
Ichinose, S., Ogawa, T., Jiang, X., and Hirokawa, N. (2019). The Spatiotemporal Construction of the Axon Initial Segment via KIF3/KAP3/TRIM46 Transport under MARK2 Signaling. Cell Rep. 28 (9), 2413–2426. doi:10.1016/j.celrep.2019.07.093
International Multiple Sclerosis Genetics Consortium (IMSGC) (2010). Comprehensive Follow-Up of the First Genome-wide Association Study of Multiple Sclerosis Identifies KIF21B and TMEM39A as Susceptibility Loci. Hum. Mol. Genet. 19 (5), 953–962. doi:10.1093/hmg/ddp542
Ivanov, A. I., McCall, I. C., Babbin, B., Samarin, S. N., Nusrat, A., and Parkos, C. A. (2006). Microtubules Regulate Disassembly of Epithelial Apical Junctions. BMC Cell Biol 7, 12. doi:10.1186/1471-2121-7-12
Janisch, K. M., Vock, V. M., Fleming, M. S., Shrestha, A., Grimsley-Myers, C. M., Rasoul, B. A., et al. (2013). The Vertebrate-specific Kinesin-6, Kif20b, Is Required for normal Cytokinesis of Polarized Cortical Stem Cells and Cerebral Cortex Size. Development 140 (23), 4672–4682. doi:10.1242/dev.093286
Jiang, L., Tam, B. M., Ying, G., Wu, S., Hauswirth, W. W., Frederick, J. M., et al. (2015). Kinesin Family 17 (Osmotic Avoidance Abnormal‐3) Is Dispensable for Photoreceptor Morphology and Function. FASEB j. 29 (12), 4866–4880. doi:10.1096/fj.15-275677
Johansson, E., Biagini Myers, J. M., Martin, L. J., He, H., Ryan, P., LeMasters, G. K., et al. (2019). Identification of Two Early Life Eczema and Non‐eczema Phenotypes with High Risk for Asthma Development. Clin. Exp. Allergy 49 (6), 829–837. doi:10.1111/cea.13379
Junco, A., Bhullar, B., Tarnasky, H. A., and van der Hoorn, F. A. (2001). Kinesin Light-Chain KLC3 Expression in Testis Is Restricted to Spermatids1. Biol. Reprod. 64 (5), 1320–1330. doi:10.1095/biolreprod64.5.1320
Jung, M., Wells, D., Rusch, J., Ahmad, S., Marchini, J., Myers, S. R., et al. (2019). Unified Single-Cell Analysis of Testis Gene Regulation and Pathology in Five Mouse Strains. Elife 8, e43966. doi:10.7554/eLife.43966
Kallio, M., Mustalahti, T., Yen, T. J., and Lähdetie, J. (1998). Immunolocalization of α-Tubulin, γ-Tubulin, and CENP-E in Male Rat and Male Mouse Meiotic Divisions: Pathway of Meiosis I Spindle Formation in Mammalian Spermatocytes. Developmental Biol. 195 (1), 29–37. doi:10.1006/dbio.1997.8822
Kanai, Y., Okada, Y., Tanaka, Y., Harada, A., Terada, S., and Hirokawa, N. (2000). KIF5C, a Novel Neuronal Kinesin Enriched in Motor Neurons. J. Neurosci. 20 (17), 6374–6384. doi:10.1523/jneurosci.20-17-06374.2000
Kanai, Y., Wang, D., and Hirokawa, N. (2014). KIF13B Enhances the Endocytosis of LRP1 by Recruiting LRP1 to Caveolae. J. Cell Biol 204 (3), 395–408. doi:10.1083/jcb.201309066
Kapitein, L. C., Peterman, E. J. G., Kwok, B. H., Kim, J. H., Kapoor, T. M., and Schmidt, C. F. (2005). The Bipolar Mitotic Kinesin Eg5 Moves on Both Microtubules that it Crosslinks. Nature 435 (7038), 114–118. doi:10.1038/nature03503
Karasmanis, E. P., Phan, C.-T., Angelis, D., Kesisova, I. A., Hoogenraad, C. C., McKenney, R. J., et al. (2018). Polarity of Neuronal Membrane Traffic Requires Sorting of Kinesin Motor Cargo during Entry into Dendrites by a Microtubule-Associated Septin. Developmental Cell 46 (2), 204–218. doi:10.1016/j.devcel.2018.06.013
Kato, Y., Miyakawa, T., and Tanokura, M. (2018). Overview of the Mechanism of Cytoskeletal Motors Based on Structure. Biophys. Rev. 10 (2), 571–581. doi:10.1007/s12551-017-0368-1
Kaur, S., Van Bergen, N. J., Verhey, K. J., Nowell, C. J., Budaitis, B., Yue, Y., et al. (2020). Expansion of the Phenotypic Spectrum of De Novo Missense Variants in Kinesin Family Member 1A ( KIF1A ). Hum. Mutat. 41 (10), 1761–1774. doi:10.1002/humu.24079
Khawar, M. B., Gao, H., and Li, W. (2019). Mechanism of Acrosome Biogenesis in Mammals. Front. Cell Dev. Biol. 7, 195. doi:10.3389/fcell.2019.00195
Kierszenbaum, A. L., and Tres, L. L. (2004). The Acrosome-Acroplaxome-Manchette Complex and the Shaping of the Spermatid Head. Arch. Histology Cytol. 67 (4), 271–284. doi:10.1679/aohc.67.271
Kondo, S., Sato-Yoshitake, R., Noda, Y., Aizawa, H., Nakata, T., Matsuura, Y., et al. (1994). KIF3A Is a New Microtubule-Based Anterograde Motor in the Nerve Axon. J. Cell Biol 125 (5), 1095–1107. doi:10.1083/jcb.125.5.1095
Konjikusic, M. J., Yeetong, P., Boswell, C. W., Lee, C., Roberson, E. C., Ittiwut, R., et al. (2018). Mutations in Kinesin Family Member 6 Reveal Specific Role in Ependymal Cell Ciliogenesis and Human Neurological Development. Plos Genet. 14 (11), e1007817. doi:10.1371/journal.pgen.1007817
Kopera, I. A., Bilinska, B., Cheng, C. Y., and Mruk, D. D. (2010). Sertoli-germ Cell Junctions in the Testis: a Review of Recent Data. Phil. Trans. R. Soc. B 365 (1546), 1593–1605. doi:10.1098/rstb.2009.0251
Kotaja, N., Lin, H., Parvinen, M., and Sassone-Corsi, P. (2006). Interplay of PIWI/Argonaute Protein MIWI and Kinesin KIF17b in Chromatoid Bodies of Male Germ Cells. J. Cell Sci 119 (Pt 13), 2819–2825. doi:10.1242/jcs.03022
Kotaja, N., Macho, B., and Sassone-Corsi, P. (2005). Microtubule-independent and Protein Kinase A-Mediated Function of Kinesin KIF17b Controls the Intracellular Transport of Activator of CREM in Testis (ACT). J. Biol. Chem. 280 (36), 31739–31745. doi:10.1074/jbc.m505971200
Kreitzer, G., and Myat, M. M. (2018). Microtubule Motors in Establishment of Epithelial Cell Polarity. Cold Spring Harb Perspect. Biol. 10 (2). doi:10.1101/cshperspect.a027896
Kuriyama, R., Gustus, C., Terada, Y., Uetake, Y., and Matuliene, J. (2002). CHO1, a Mammalian Kinesin-like Protein, Interacts with F-Actin and Is Involved in the Terminal Phase of Cytokinesis. J. Cell Biol 156 (5), 783–790. doi:10.1083/jcb.200109090
Lawrence, C. J., Dawe, R. K., Christie, K. R., Cleveland, D. W., Dawson, S. C., Endow, S. A., et al. (2004). A Standardized Kinesin Nomenclature. J. Cell Biol 167 (1), 19–22. doi:10.1083/jcb.200408113
Lee, N. P. Y., and Cheng, C. Y. (2004). Ectoplasmic Specialization, a Testis-specific Cell-Cell Actin-Based Adherens junction Type: Is This a Potential Target for Male Contraceptive Development? Hum. Reprod. Update 10 (4), 349–369. doi:10.1093/humupd/dmh026
Lehti, M. S., Kotaja, N., and Sironen, A. (2013). KIF3A Is Essential for Sperm Tail Formation and Manchette Function. Mol. Cell Endocrinol 377 (1-2), 44–55. doi:10.1016/j.mce.2013.06.030
Li, J. C., Mruk, D., and Cheng, C. Y. (2001). The Inter-sertoli Tight junction Permeability Barrier Is Regulated by the Interplay of Protein Phosphatases and Kinases: an In Vitro Study. J. Androl. 22 (5), 847–856. doi:10.1002/j.1939-4640.2001.tb02590.x
Li, L., Huitao, L., Lingling, W., Tiao, B., Shiwen, L., Baiping, M., et al. (2021). A Local Regulatory Network in the Testis Mediated by Laminin and Collagen Fragments that Supports Spermatogenesis. Crit. Rev. Biochem. Mol. Biol. 56 (3), 236–254. doi:10.1080/10409238.2021.1901255
Li, L., Tang, E. I., Chen, H., Lian, Q., Ge, R., Silvestrini, B., et al. (2017). Sperm Release at Spermiation Is Regulated by Changes in the Organization of Actin- and Microtubule-Based Cytoskeletons at the Apical Ectoplasmic Specialization-A Study Using the Adjudin Model. Endocrinology 158 (12), 4300–4316. doi:10.1210/en.2017-00660
Liao, G., and Gundersen, G. G. (1998). Kinesin Is a Candidate for Cross-Bridging Microtubules and Intermediate Filaments. J. Biol. Chem. 273 (16), 9797–9803. doi:10.1074/jbc.273.16.9797
Lie, P. P., Cheng, C. Y., and Mruk, D. D. (2009). Coordinating Cellular Events during Spermatogenesis: a Biochemical Model. Trends Biochem. Sci. 34 (7), 366–373. doi:10.1016/j.tibs.2009.03.005
Lie, P. P. Y., Cheng, C. Y., and Mruk, D. D. (2010). Crosstalk between desmoglein-2/desmocollin-2/Src Kinase and coxsackie and Adenovirus receptor/ZO-1 Protein Complexes, Regulates Blood-Testis Barrier Dynamics. Int. J. Biochem. Cell Biol. 42 (6), 975–986. doi:10.1016/j.biocel.2010.02.010
Lin, F., Hiesberger, T., Cordes, K., Sinclair, A. M., Goldstein, L. S. B., Somlo, S., et al. (2003). Kidney-specific Inactivation of the KIF3A Subunit of Kinesin-II Inhibits Renal Ciliogenesis and Produces Polycystic Kidney Disease. Proc. Natl. Acad. Sci. U.S.A. 100 (9), 5286–5291. doi:10.1073/pnas.0836980100
Liu, X.-s., Zhao, X.-d., Wang, X., Yao, Y.-x., Zhang, L.-l., Shu, R.-z., et al. (2010). Germinal Cell Aplasia in Kif18a Mutant Male Mice Due to Impaired Chromosome Congression and Dysregulated BubR1 and CENP-E. Genes & Cancer 1 (1), 26–39. doi:10.1177/1947601909358184
Luboshits, G., and Benayahu, D. (2005). MS-KIF18A, New Kinesin; Structure and Cellular Expression. Gene 351, 19–28. doi:10.1016/j.gene.2005.02.009
Ma, D.-D., Wang, D.-H., and Yang, W.-X. (2017). Kinesins in Spermatogenesis†. Biol. Reprod. 96 (2), 267–276. doi:10.1095/biolreprod.116.144113
Ma, R.-R., Zhang, H., Chen, H.-F., Zhang, G.-H., Tian, Y.-R., and Gao, P. (2021). MiR-19a/miR-96-mediated Low Expression of KIF26A Suppresses Metastasis by Regulating FAK Pathway in Gastric Cancer. Oncogene 40 (14), 2524–2538. doi:10.1038/s41388-020-01610-7
Macho, B., Brancorsini, S., Fimia, G. M., Setou, M., Hirokawa, N., and Sassone-Corsi, P. (2002). CREM-dependent Transcription in Male Germ Cells Controlled by a Kinesin. Science 298 (5602), 2388–2390. doi:10.1126/science.1077265
Mannowetz, N., Kartarius, S., Wennemuth, G., and Montenarh, M. (2010). Protein Kinase CK2 and New Binding Partners during Spermatogenesis. Cell. Mol. Life Sci. 67 (22), 3905–3913. doi:10.1007/s00018-010-0412-9
Maor-Nof, M., Homma, N., Raanan, C., Nof, A., Hirokawa, N., and Yaron, A. (2013). Axonal Pruning Is Actively Regulated by the Microtubule-Destabilizing Protein Kinesin Superfamily Protein 2A. Cell Rep. 3 (4), 971–977. doi:10.1016/j.celrep.2013.03.005
Marszalek, J. R., Ruiz-Lozano, P., Roberts, E., Chien, K. R., and Goldstein, L. S. B. (1999). Situs Inversus and Embryonic Ciliary Morphogenesis Defects in Mouse Mutants Lacking the KIF3A Subunit of Kinesin-II. Proc. Natl. Acad. Sci. U.S.A. 96 (9), 5043–5048. doi:10.1073/pnas.96.9.5043
Martino, F., Perestrelo, A. R., Vinarský, V., Pagliari, S., and Forte, G. (2018). Cellular Mechanotransduction: From Tension to Function. Front. Physiol. 9, 824. doi:10.3389/fphys.2018.00824
Marx, A., Hoenger, A., and Mandelkow, E. (2009). Structures of Kinesin Motor Proteins. Cell Motil. Cytoskeleton 66 (11), 958–966. doi:10.1002/cm.20392
Meng, W., Mushika, Y., Ichii, T., and Takeichi, M. (2008). Anchorage of Microtubule Minus Ends to Adherens Junctions Regulates Epithelial Cell-Cell Contacts. Cell 135 (5), 948–959. doi:10.1016/j.cell.2008.09.040
Miki, H., Setou, M., Kaneshiro, K., and Hirokawa, N. (2001). All Kinesin Superfamily Protein, KIF, Genes in Mouse and Human. Proc. Natl. Acad. Sci. U.S.A. 98 (13), 7004–7011. doi:10.1073/pnas.111145398
Mills, J., Hanada, T., Hase, Y., Liscum, L., and Chishti, A. H. (2019). LDL Receptor Related Protein 1 Requires the I3 Domain of Discs-Large Homolog 1/DLG1 for Interaction with the Kinesin Motor Protein KIF13B. Biochim. Biophys. Acta (Bba) - Mol. Cell Res. 1866 (12), 118552. doi:10.1016/j.bbamcr.2019.118552
Mitra, S. K., Hanson, D. A., and Schlaepfer, D. D. (2005). Focal Adhesion Kinase: in Command and Control of Cell Motility. Nat. Rev. Mol. Cell Biol 6 (1), 56–68. doi:10.1038/nrm1549
Miyata, H., Shimada, K., Morohoshi, A., Oura, S., Matsumura, T., Xu, Z., et al. (2020). Testis‐enriched Kinesin KIF9 Is Important for Progressive Motility in Mouse Spermatozoa. FASEB j. 34 (4), 5389–5400. doi:10.1096/fj.201902755r
Moawia, A., Shaheen, R., Rasool, S., Waseem, S. S., Ewida, N., Budde, B., et al. (2017). Mutations of KIF14 Cause Primary Microcephaly by Impairing Cytokinesis. Ann. Neurol. 82 (4), 562–577. doi:10.1002/ana.25044
Morikawa, M., Tanaka, Y., Cho, H.-S., Yoshihara, M., and Hirokawa, N. (2018). The Molecular Motor KIF21B Mediates Synaptic Plasticity and Fear Extinction by Terminating Rac1 Activation. Cell Rep. 23 (13), 3864–3877. doi:10.1016/j.celrep.2018.05.089
Mruk, D. D., and Cheng, C. Y. (2011). Desmosomes in the Testis. Spermatogenesis 1 (1), 47–51. doi:10.4161/spmg.1.1.15443
Mruk, D. D., and Cheng, C. Y. (2004). Sertoli-Sertoli and Sertoli-Germ Cell Interactions and Their Significance in Germ Cell Movement in the Seminiferous Epithelium during Spermatogenesis. Endocr. Rev. 25 (5), 747–806. doi:10.1210/er.2003-0022
Nakagawa, T., Tanaka, Y., Matsuoka, E., Kondo, S., Okada, Y., Noda, Y., et al. (1997). Identification and Classification of 16 New Kinesin Superfamily (KIF) Proteins in Mouse Genome. Proc. Natl. Acad. Sci. U.S.A. 94 (18), 9654–9659. doi:10.1073/pnas.94.18.9654
Nakajima, K., Takei, Y., Tanaka, Y., Nakagawa, T., Nakata, T., Noda, Y., et al. (2002). Molecular Motor KIF1C Is Not Essential for Mouse Survival and Motor-dependent Retrograde Golgi Apparatus-To-Endoplasmic Reticulum Transport. Mol. Cell Biol 22 (3), 866–873. doi:10.1128/mcb.22.3.866-873.2002
Nakajima, K., and Tanaka, Y. (2010). Exclusion of Kif1c as a Candidate Gene for Anthrax Toxin Susceptibility. Microb. Pathogenesis 48 (5), 188–190. doi:10.1016/j.micpath.2010.02.001
Nakajima, K., Yin, X., Takei, Y., Seog, D.-H., Homma, N., and Hirokawa, N. (2012). Molecular Motor KIF5A Is Essential for GABAA Receptor Transport, and KIF5A Deletion Causes Epilepsy. Neuron 76 (5), 945–961. doi:10.1016/j.neuron.2012.10.012
Navolanic, P. M., and Sperry, A. O. (2000). Identification of Isoforms of a Mitotic Motor in Mammalian Spermatogenesis1. Biol. Reprod. 62 (5), 1360–1369. doi:10.1095/biolreprod62.5.1360
Nekrasova, O. E., Amargo, E. V., Smith, W. O., Chen, J., Kreitzer, G. E., and Green, K. J. (2011). Desmosomal Cadherins Utilize Distinct Kinesins for Assembly into Desmosomes. J. Cell Biol 195 (7), 1185–1203. doi:10.1083/jcb.201106057
Niwa, S., Nakajima, K., Miki, H., Minato, Y., Wang, D., and Hirokawa, N. (2012). KIF19A Is a Microtubule-Depolymerizing Kinesin for Ciliary Length Control. Developmental Cell 23 (6), 1167–1175. doi:10.1016/j.devcel.2012.10.016
Noda, Y., Okada, Y., Saito, N., Setou, M., Xu, Y., Zhang, Z., et al. (2001). KIFC3, a Microtubule Minus End-Directed Motor for the Apical Transport of Annexin XIIIb-Associated Triton-Insoluble Membranes. J. Cell Biol 155 (1), 77–88. (0021-9525 (Print)). doi:10.1083/jcb.200108042
Nonaka, S., Tanaka, Y., Okada, Y., Takeda, S., Harada, A., Kanai, Y., et al. (1998). Randomization of Left-Right Asymmetry Due to Loss of Nodal Cilia Generating Leftward Flow of Extraembryonic Fluid in Mice Lacking KIF3B Motor Protein. Cell 95 (6), 829–837. doi:10.1016/s0092-8674(00)81705-5
Nozawa, Y. I., Yao, E., Gacayan, R., Xu, S.-M., and Chuang, P.-T. (2014). Mammalian Fused Is Essential for Sperm Head Shaping and Periaxonemal Structure Formation during Spermatogenesis. Developmental Biol. 388 (2), 170–180. doi:10.1016/j.ydbio.2014.02.002
O'Donnell, L. (2014). Mechanisms of Spermiogenesis and Spermiation and How They Are Disturbed. Spermatogenesis 4 (2), e979623. doi:10.4161/21565562.2014.979623
Ohi, R., and Wordeman, L. (2013). “Kinesins as Microtubule Disassembly Enzymes,” in Encyclopedia of Biological Chemistry. Editors W. J. Lennarz, and M. D. Lane. Second Edition (Waltham: Academic Press), 672–678. doi:10.1016/b978-0-12-378630-2.00505-3
Ohsugi, M., Adachi, K., Horai, R., Kakuta, S., Sudo, K., Kotaki, H., et al. (2008). Kid-mediated Chromosome Compaction Ensures Proper Nuclear Envelope Formation. Cell 132 (5), 771–782. doi:10.1016/j.cell.2008.01.029
Pennings, M., Schouten, M. I., van Gaalen, J., Meijer, R. P. P., de Bot, S. T., Kriek, M., et al. (2020). KIF1A Variants Are a Frequent Cause of Autosomal Dominant Hereditary Spastic Paraplegia. Eur. J. Hum. Genet. 28 (1), 40–49. doi:10.1038/s41431-019-0497-z
Pike, R., Ortiz-Zapater, E., Lumicisi, B., Santis, G., and Parsons, M. (2018). KIF22 Coordinates CAR and EGFR Dynamics to Promote Cancer Cell Proliferation. Sci. Signal. 11 (515). doi:10.1126/scisignal.aaq1060
Pleuger, C., Lehti, M. S., Dunleavy, J. E., Fietz, D., and O’Bryan, M. K. (2020). Haploid Male Germ Cells-The Grand Central Station of Protein Transport. Hum. Reprod. Update 26 (4), 474–500. doi:10.1093/humupd/dmaa004
Pointis, G., and Segretain, D. (2005). Role of Connexin-Based gap junction Channels in Testis. Trends Endocrinol. Metab. 16 (7), 300–306. doi:10.1016/j.tem.2005.07.001
Prahlad, V., Yoon, M., Moir, R. D., Vale, R. D., and Goldman, R. D. (1998). Rapid Movements of Vimentin on Microtubule Tracks: Kinesin-dependent Assembly of Intermediate Filament Networks. J. Cell Biol 143 (1), 159–170. doi:10.1083/jcb.143.1.159
Putkey, F. R., Cramer, T., Morphew, M. K., Silk, A. D., Johnson, R. S., McIntosh, J. R., et al. (2002). Unstable Kinetochore-Microtubule Capture and Chromosomal Instability Following Deletion of CENP-E. Developmental Cell 3 (3), 351–365. doi:10.1016/s1534-5807(02)00255-1
Rahman, A., Kamal, A., Roberts, E. A., and Goldstein, L. S. B. (1999). Defective Kinesin Heavy Chain Behavior in Mouse Kinesin Light Chain Mutants. J. Cell Biol 146 (6), 1277–1288. doi:10.1083/jcb.146.6.1277
Reilly, M. L., Stokman, M. F., Magry, V., Jeanpierre, C., Alves, M., Paydar, M., et al. (2019). Loss-of-function Mutations inKIF14cause Severe Microcephaly and Kidney Development Defects in Humans and Zebrafish. Hum. Mol. Genet. 28 (5), 778–795. doi:10.1093/hmg/ddy381
Rode, K., Langeheine, M., Seeger, B., and Brehm, R. (2021). Connexin43 in Germ Cells Seems to Be Dispensable for Murine Spermatogenesis. Int. J. Mol. Sci. 22 (15), 7924. doi:10.3390/ijms22157924
Russell, L. (1977). Movement of Spermatocytes from the Basal to the Adluminal Compartment of the Rat Testis. Am. J. Anat. 148 (3), 313–328. doi:10.1002/aja.1001480303
Saade, M., Irla, M., Govin, J., Victorero, G., Samson, M., and Nguyen, C. (2007). Dynamic Distribution of Spatial during Mouse Spermatogenesis and its Interaction with the Kinesin KIF17b. Exp. Cell Res. 313 (3), 614–626. doi:10.1016/j.yexcr.2006.11.011
Saez, T. M. M., Fernandez Bessone, I., Rodriguez, M. S., Alloatti, M., Otero, M. G., Cromberg, L. E., et al. (2020). Kinesin-1-mediated Axonal Transport of CB1 Receptors Is Required for Cannabinoid-dependent Axonal Growth and Guidance. Development 147 (8). doi:10.1242/dev.184069
Sako-Kubota, K., Tanaka, N., Nagae, S., Meng, W., and Takeichi, M. (2014). Minus End-Directed Motor KIFC3 Suppresses E-Cadherin Degradation by Recruiting USP47 to Adherens Junctions. MBoC 25 (24), 3851–3860. doi:10.1091/mbc.e14-07-1245
Samwer, M., Dehne, H.-J., Spira, F., Kollmar, M., Gerlich, D. W., Urlaub, H., et al. (2013). The Nuclear F-Actin Interactome of Xenopus Oocytes Reveals an Actin-Bundling Kinesin that Is Essential for Meiotic Cytokinesis. EMBO J. 32 (13), 1886–1902. doi:10.1038/emboj.2013.108
Sehgal, L., Mukhopadhyay, A., Rajan, A., Khapare, N., Sawant, M., Vishal, S. S., et al. (2014). 14-3-3γ-Mediated Transport of Plakoglobin to the Cell Border Is Required for the Initiation of Desmosome Assembly In Vitro and In Vivo. J. Cell Sci 127 (Pt 10), 2174–2188. doi:10.1242/jcs.125807
She, Z.-Y., Li, Y.-L., Lin, Y., Lu, M.-H., Wei, Y.-L., Yu, K.-W., et al. (2020). Kinesin-6 Family Motor KIF20A Regulates central Spindle Assembly and Acrosome Biogenesis in Mouse Spermatogenesis. Biochim. Biophys. Acta (Bba) - Mol. Cell Res. 1867 (4), 118636. doi:10.1016/j.bbamcr.2019.118636
She, Z.-Y., Yu, K.-W., Zhong, N., Xiao, Y., Wei, Y.-L., Lin, Y., et al. (2020). Kinesin-7 CENP-E Regulates Chromosome Alignment and Genome Stability of Spermatogenic Cells. Cell Death Discov. 6, 25. doi:10.1038/s41420-020-0261-8
She, Z.-Y., Zhong, N., Yu, K.-W., Xiao, Y., Wei, Y.-L., Lin, Y., et al. (2020). Kinesin-5 Eg5 Is Essential for Spindle Assembly and Chromosome Alignment of Mouse Spermatocytes. Cell Div 15, 6. doi:10.1186/s13008-020-00063-4
Shen, H.-Q., Xiao, Y.-X., She, Z.-Y., Tan, F.-Q., and Yang, W.-X. (2017). A Novel Role of KIF3b in the Seminoma Cell Cycle. Exp. Cell Res. 352 (1), 95–103. doi:10.1016/j.yexcr.2017.01.023
Smialek, M. J., Kuczynska, B., Ilaslan, E., Janecki, D. M., Sajek, M. P., Kusz-Zamelczyk, K., et al. (2020). Kinesin KIF18A Is a Novel PUM-Regulated Target Promoting Mitotic Progression and Survival of a Human Male Germ Cell Line. J. Cell Sci 133 (7). doi:10.1242/jcs.240986
Stevens, M. L., Zhang, Z., Johansson, E., Ray, S., Jagpal, A., Ruff, B. P., et al. (2020). Disease-associated KIF3A Variants Alter Gene Methylation and Expression Impacting Skin Barrier and Atopic Dermatitis Risk. Nat. Commun. 11 (1), 4092. doi:10.1038/s41467-020-17895-x
Sweeney, H. L., and Holzbaur, E. L. F. (2018). Motor Proteins. Cold Spring Harb Perspect. Biol. 10 (5). doi:10.1101/cshperspect.a021931
Tan, X., Priyam, B., Xin, L., Jiang, Y., Sieun, L., Young-Ho, A., et al. (2022). Transcriptional Control of a Collagen Deposition and Adhesion Process that Promotes Lung Adenocarcinoma Growth and Metastasis. JCI Insight 7 (1), e153948. doi:10.1172/jci.insight.153948
Tanaka, Y., Kanai, Y., Okada, Y., Nonaka, S., Takeda, S., Harada, A., et al. (1998). Targeted Disruption of Mouse Conventional Kinesin Heavy Chain kif5B, Results in Abnormal Perinuclear Clustering of Mitochondria. Cell 93 (7), 1147–1158. doi:10.1016/s0092-8674(00)81459-2
Tang, E. I., Lee, W. M., and Cheng, C. Y. (2016). Coordination of Actin- and Microtubule-Based Cytoskeletons Supports Transport of Spermatids and Residual Bodies/Phagosomes during Spermatogenesis in the Rat Testis. Endocrinology 157 (4), 1644–1659. doi:10.1210/en.2015-1962
Tang, E. I., Mok, K.-W., Lee, W. M., and Cheng, C. Y. (2015). EB1 Regulates Tubulin and Actin Cytoskeletal Networks at the Sertoli Cell Blood-Testis Barrier in Male Rats: an In Vitro Study. Endocrinology 156 (2), 680–693. doi:10.1210/en.2014-1720
Tang, E. I., Mruk, D. D., and Cheng, C. Y. (2016). Regulation of Microtubule (MT)-based Cytoskeleton in the Seminiferous Epithelium during Spermatogenesis. Semin. Cell Dev Biol 59, 35–45. doi:10.1016/j.semcdb.2016.01.004
Tokai, N., Fujimoto-Nishiyama, A., Toyoshima, Y., Yonemura, S., Tsukita, S., Inoue, J., et al. (1996). Kid, a Novel Kinesin-like DNA Binding Protein, Is Localized to Chromosomes and the Mitotic Spindle. EMBO J. 15 (3), 457–467. doi:10.1002/j.1460-2075.1996.tb00378.x
Uchiyama, Y., Sakaguchi, M., Terabayashi, T., Inenaga, T., Inoue, S., Kobayashi, C., et al. (2010). Kif26b , a Kinesin Family Gene, Regulates Adhesion of the Embryonic Kidney Mesenchyme. Proc. Natl. Acad. Sci. U.S.A. 107 (20), 9240–9245. doi:10.1073/pnas.0913748107
Vaid, K. S., Guttman, J. A., Singaraja, R. R., and Vogl, A. W. (2007). A Kinesin Is Present at Unique Sertoli/Spermatid Adherens Junctions in Rat and Mouse Testes1. Biol. Reprod. 77 (6), 1037–1048. doi:10.1095/biolreprod.107.063735
Vale, R., Reese, T., and Sheetz, M. (1985). Identification of a Novel Force-Generating Protein, Kinesin, Involved in Microtubule-Based Motility. Cell 42 (1), 39–50. doi:10.1016/s0092-8674(85)80099-4
van den Wildenberg, S. M. J. L., Tao, L., Kapitein, L. C., Schmidt, C. F., Scholey, J. M., and Peterman, E. J. G. (2008). The Homotetrameric Kinesin-5 KLP61F Preferentially Crosslinks Microtubules into Antiparallel Orientations. Curr. Biol. 18 (23), 1860–1864. doi:10.1016/j.cub.2008.10.026
Vikberg, A.-L., Malla, S., and Golovleva, I. (2020). Differential Tissue Specific Expression of Kif23 Alternative Transcripts in Mice with the Human Mutation Causing Congenital Dyserythropoietic Anemia Type III. Blood Cell Mol. Dis. 85, 102483. doi:10.1016/j.bcmd.2020.102483
Vogl, A. W., Vaid, K. S., and Guttman, J. A. (2008). The Sertoli Cell Cytoskeleton. Adv. Exp. Med. Biol. 636, 186–211. doi:10.1007/978-0-387-09597-4_11
Vogl, A. W., Pfeiffer, D. C., Mulholland, D., Kimel, G., and Guttman, J. (2000). Unique and Multifunctional Adhesion Junctions in the Testis. Ectoplasmic Specializations. Arch. Histology Cytol. 63 (1), 1–15. doi:10.1679/aohc.63.1
Wang, C. Q. F., and Cheng, C. Y. (2007). A Seamless Trespass: Germ Cell Migration across the Seminiferous Epithelium during Spermatogenesis. J. Cell Biol 178 (4), 549–556. doi:10.1083/jcb.200704061
Wang, L., Tanaka, Y., Wang, D., Morikawa, M., Zhou, R., Homma, N., et al. (2018). The Atypical Kinesin KIF26A Facilitates Termination of Nociceptive Responses by Sequestering Focal Adhesion Kinase. Cell Rep. 24 (11), 2894–2907. doi:10.1016/j.celrep.2018.05.075
Wang, L., Yan, M., Wu, S., Wu, X., Bu, T., Wong, C. K. C., et al. (2020). Actin Binding Proteins, Actin Cytoskeleton and Spermatogenesis - Lesson from Toxicant Models. Reprod. Toxicol. 96, 76–89. doi:10.1016/j.reprotox.2020.05.017
Wang, Y., Smallwood, P. M., Williams, J., and Nathans, J. (2020). A Mouse Model for Kinesin Family Member 11 (Kif11)-Associated Familial Exudative Vitreoretinopathy. Hum. Mol. Genet. 29 (7), 1121–1131. doi:10.1093/hmg/ddaa018
Wen, Q., Tang, E. I., Lui, W.-y., Lee, W. M., Wong, C. K. C., Silvestrini, B., et al. (2018). Dynein 1 Supports Spermatid Transport and Spermiation during Spermatogenesis in the Rat Testis. Am. J. Physiology-Endocrinology Metab. 315 (5), E924–E948. doi:10.1152/ajpendo.00114.2018
Wen, Q., Tang, E. I., Xiao, X., Gao, Y., Chu, D. S., Mruk, D. D., et al. (2016). Transport of Germ Cells across the Seminiferous Epithelium during Spermatogenesis-The Involvement of Both Actin- and Microtubule-Based Cytoskeletons. Tissue Barriers 4 (4), e1265042. doi:10.1080/21688370.2016.1265042
Wong-Riley, M. T. T., and Besharse, J. C. (2012). The Kinesin Superfamily Protein KIF17: One Protein with many Functions. Biomol. Concepts 3 (3), 267–282. doi:10.1515/bmc-2011-0064
Wozniak, M. A., Modzelewska, K., Kwong, L., and Keely, P. J. (2004). Focal Adhesion Regulation of Cell Behavior. Biochim. Biophys. Acta 1692 (2-3), 103–119. doi:10.1016/j.bbamcr.2004.04.007
Wu, J.-C., Gregory, C. W., and DePhilip, R. M. (1993). Expression of E-Cadherin in Immature Rat and Mouse Testis and in Rat Sertoli Cell Cultures1. Biol. Reprod. 49 (6), 1353–1361. doi:10.1095/biolreprod49.6.1353
Wu, S., Lv, L., Li, L., Wang, L., Mao, B., Li, J., et al. (2021). KIF15 Supports Spermatogenesis via its Effects on Sertoli Cell Microtubule, Actin, Vimentin, and Septin Cytoskeletons. Endocrinology 162 (4), bqab010. doi:10.1210/endocr/bqab010
Wu, S., Chen, H., Alexeyev, M. F., King, J. A. C., Moore, T. M., Stevens, T., et al. (2007). Microtubule Motors Regulate ISOC Activation Necessary to Increase Endothelial Cell Permeability. J. Biol. Chem. 282 (48), 34801–34808. doi:10.1074/jbc.m704522200
Wu, S., Li, H., Wang, L., Mak, N., Wu, X., Ge, R., et al. (2021). Motor Proteins and Spermatogenesis. Adv. Exp. Med. Biol. 1288, 131–159. doi:10.1007/978-3-030-77779-1_7
Wu, S., Yan, M., Ge, R., and Cheng, C. Y. (2020). Crosstalk between Sertoli and Germ Cells in Male Fertility. Trends Mol. Med. 26 (2), 215–231. doi:10.1016/j.molmed.2019.09.006
Xia, C.-H., Roberts, E. A., Her, L.-S., Liu, X., Williams, D. S., Cleveland, D. W., et al. (2003). Abnormal Neurofilament Transport Caused by Targeted Disruption of Neuronal Kinesin Heavy Chain KIF5A. J. Cell Biol 161 (1), 55–66. doi:10.1083/jcb.200301026
Xiao, X., Mruk, D. D., Tang, E. I., Wong, C. K. C., Lee, W. M., John, C. M., et al. (2014). Environmental Toxicants Perturb Human Sertoli Cell Adhesive Function via Changes in F-Actin Organization Mediated by Actin Regulatory Proteins. Hum. Reprod. 29 (6), 1279–1291. doi:10.1093/humrep/deu011
Xiao, X., Mruk, D. D., Wong, C. K. C., and Yan Cheng, C. (2014). Germ Cell Transport across the Seminiferous Epithelium during Spermatogenesis. Physiology 29 (4), 286–298. doi:10.1152/physiol.00001.2014
Xiao, Y.-X., Shen, H.-Q., She, Z.-Y., Sheng, L., Chen, Q.-Q., Chu, Y.-L., et al. (2017). C-terminal Kinesin Motor KIFC1 Participates in Facilitating Proper Cell Division of Human Seminoma. Oncotarget 8 (37), 61373–61384. doi:10.18632/oncotarget.18139
Xu, Y., Takeda, S., Nakata, T., Noda, Y., Tanaka, Y., and Hirokawa, N. (2002). Role of KIFC3 Motor Protein in Golgi Positioning and Integration. J. Cell Biol 158 (2), 293–303. doi:10.1083/jcb.200202058
Yamazaki, H., Nakata, T., Okada, Y., and Hirokawa, N. (1995). KIF3A/B: a Heterodimeric Kinesin Superfamily Protein that Works as a Microtubule Plus End-Directed Motor for Membrane Organelle Transport. J. Cell Biol 130 (6), 1387–1399. doi:10.1083/jcb.130.6.1387
Yang, W.-X., Jefferson, H., and Sperry, A. O. (2006). The Molecular Motor KIFC1 Associates with a Complex Containing Nucleoporin NUP62 that Is Regulated during Development and by the Small GTPase RAN1. Biol. Reprod. 74 (4), 684–690. doi:10.1095/biolreprod.105.049312
Yang, W.-X., and Sperry, A. O. (2003). C-terminal Kinesin Motor KIFC1 Participates in Acrosome Biogenesis and Vesicle Transport1. Biol. Reprod. 69 (5), 1719–1729. doi:10.1095/biolreprod.102.014878
Yang, W., Tanaka, Y., Bundo, M., and Hirokawa, N. (2014). Antioxidant Signaling Involving the Microtubule Motor KIF12 Is an Intracellular Target of Nutrition Excess in Beta Cells. Developmental Cell 31 (2), 202–214. doi:10.1016/j.devcel.2014.08.028
Yang, Z., Hanlon, D. W., Marszalek, J. R., and Goldstein, L. S. B. (1997). Identification, Partial Characterization, and Genetic Mapping of Kinesin-like Protein Genes in Mouse. Genomics 45 (1), 123–131. doi:10.1006/geno.1997.4901
Yang, Z., Roberts, E. A., and Goldstein, L. S. B. (2001). Functional Analysis of Mouse C-Terminal Kinesin Motor KifC2. Mol. Cell Biol 21 (7), 2463–2466. doi:10.1128/mcb.21.7.2463-2466.2001
Yang, Z., Roberts, E. A., and Goldstein, L. S. B. (2001). Functional Analysis of Mouse Kinesin Motor Kif3C. Mol. Cell Biol 21 (16), 5306–5311. doi:10.1128/mcb.21.16.5306-5311.2001
Yang, Z., Xia, C.-h., Roberts, E. A., Bush, K., Nigam, S. K., and Goldstein, L. S. B. (2001). Molecular Cloning and Functional Analysis of Mouse C-Terminal Kinesin Motor KifC3. Mol. Cell Biol 21 (3), 765–770. doi:10.1128/mcb.21.3.765-770.2001
Yonekawa, Y., Harada, A., Okada, Y., Funakoshi, T., Kanai, Y., Takei, Y., et al. (1998). Defect in Synaptic Vesicle Precursor Transport and Neuronal Cell Death in KIF1A Motor Protein-Deficient Mice. J. Cell Biol 141 (2), 431–441. doi:10.1083/jcb.141.2.431
Zhang, Y., Ou, Y., Cheng, M., Shojaei Saadi, H., Thundathil, J. C., and van der Hoorn, F. A. (2012). KLC3 Is Involved in Sperm Tail Midpiece Formation and Sperm Function. Developmental Biol. 366 (2), 101–110. doi:10.1016/j.ydbio.2012.04.026
Zhao, C., Takita, J., Tanaka, Y., Setou, M., Nakagawa, T., Takeda, S., et al. (2001). Charcot-Marie-Tooth Disease Type 2A Caused by Mutation in a Microtubule Motor KIF1Bβ. Cell 105 (5), 587–597. doi:10.1016/s0092-8674(01)00363-4
Zhao, J., Fok, A. H. K., Fan, R., Kwan, P. Y., Chan, H. L., Lo, L. H., et al. (2020). Specific Depletion of the Motor Protein KIF5B Leads to Deficits in Dendritic Transport, Synaptic Plasticity and Memory. Elife 9. doi:10.7554/eLife.53456
Zhi, E., Li, P., Chen, H., Xu, P., Zhu, X., Zhu, Z., et al. (2016). Decreased Expression of KIFC1 in Human Testes with Globozoospermic Defects. Genes (Basel) 7 (10), 75. doi:10.3390/genes7100075
Zhou, R., Niwa, S., Guillaud, L., Tong, Y., and Hirokawa, N. (2013). A Molecular Motor, KIF13A, Controls Anxiety by Transporting the Serotonin Type 1A Receptor. Cell Rep. 3 (2), 509–519. doi:10.1016/j.celrep.2013.01.014
Zhou, R., Niwa, S., Homma, N., Takei, Y., and Hirokawa, N. (2009). KIF26A Is an Unconventional Kinesin and Regulates GDNF-Ret Signaling in Enteric Neuronal Development. Cell 139 (4), 802–813. doi:10.1016/j.cell.2009.10.023
Zhu, H., Xu, W., Zhang, H., Liu, J., Xu, H., Lu, S., et al. (2013). Targeted Deletion of Kif18a Protects from Colitis-Associated Colorectal (CAC) Tumors in Mice through Impairing Akt Phosphorylation. Biochem. Biophysical Res. Commun. 438 (1), 97–102. doi:10.1016/j.bbrc.2013.07.032
Keywords: kinesin, testis, sertoli cells, spermatogenesis, cell junctions, blood-testis barrier, apical ectoplasmic specialization, cytoskeleton
Citation: Yao M, Qu H, Han Y, Cheng CY and Xiao X (2022) Kinesins in Mammalian Spermatogenesis and Germ Cell Transport. Front. Cell Dev. Biol. 10:837542. doi: 10.3389/fcell.2022.837542
Received: 16 December 2021; Accepted: 25 March 2022;
Published: 25 April 2022.
Edited by:
Debarun Roy, University of Illinois at Urbana-Champaign, United StatesReviewed by:
Ciler Celik-Ozenci, Koç University, TurkeyJessica Dunleavy, The University of Melbourne, Australia
Luís Crisóstomo, Universidade do Porto, Portugal
Copyright © 2022 Yao, Qu, Han, Cheng and Xiao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiang Xiao, eHhpYW9AemphbXMuY24=
 Mingxia Yao1
Mingxia Yao1 C. Yan Cheng
C. Yan Cheng Xiang Xiao
Xiang Xiao