
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell Dev. Biol., 09 August 2022
Sec. Molecular and Cellular Pathology
Volume 10 - 2022 | https://doi.org/10.3389/fcell.2022.837000
This article is part of the Research TopicDecipher Cellular and Molecular Mechanism of the Development and Degeneration of Functional Spinal Unit with Potential Therapeutic ApproachesView all 18 articles
 Lijun Shu1‡
Lijun Shu1‡ Cong Wang1‡
Cong Wang1‡ Zhengzheng Ding1‡
Zhengzheng Ding1‡ Jianjiao Tang1‡
Jianjiao Tang1‡ Yuanyuan Zhu1
Yuanyuan Zhu1 Liuxin Wu1
Liuxin Wu1 Zheyue Wang2
Zheyue Wang2 Tingting Zhang1
Tingting Zhang1 Tianjun Wang1
Tianjun Wang1 Yetao Xu1*†
Yetao Xu1*† Lizhou Sun1*†
Lizhou Sun1*†Preeclampsia (PE) is the predominant medical condition leading to maternal and fetal mortality, and the lack of effective treatment increases its risk to the public health. Among the numerous predisposing factors, the ineffectual remodeling of the uterine spiral arteries, which can induce abnormal placental angiogenesis, has been focused to solve the pathogenesis of PE. According to the preceding research results, abnormal expression of long non-coding RNAs (lncRNA)s could be associated with the pathological changes inducing PE. To be more specific, lncRNA HIF1A-AS2 was proposed for its potential to participate in the molecular mechanisms underlying PE. In vitro, in trophoblast cell lines HTR-8/SVneo and human umbilical vein endothelial cells HUVECs, HIF1A-AS2 knockdown inhibited cell proliferation, migration and tube formation. Mechanistically, transcription factor FOXP1 could regulate the expression of HIF1A-AS2. Moreover, a series of assays, including RNA pull down and mass spectrometry, RNA immunoprecipitation and chromatin immunoprecipitation assay, revealed that HIF1A-AS2 interacted with Lamin A/C (LMNA) to inhibit ANGPTL4 expression in trophoblast cells, thus further participating in the progression of PE. Taken together, these findings suggested that further analysis on HIF1A-AS2 could contribute to the development of prospective therapeutic strategy for PE.
Preeclampsia (PE) is a hypertensive disorder specifically occurred in pregnancy, which is characterized by the new onset of increased blood pressure and proteinuria after 20 weeks of pregnancy in a previously normotensive woman. According to the recent epidemiological estimations, PE impacts 3%–8% of pregnancy women worldwide (Bouter and Duvekot, 2020) and causes serious hazard to the health of mothers and their fetuses. Moreover, it has been generally admitted that PE has remarkable causal relationship with maternal and perinatal mortality, especially in low-income and middle-income countries (Ives et al., 2020; Aukes et al., 2021). Globally, PE could result in more than 50,000 maternal deaths annually (Ananth et al., 2021). Concerning the limitations of the existing medical treatment, the conventional choice of drug for the alleviation of PE is aspirin, but it is unable to prevent the development of the disease. Although novel therapies for PE are being developed, placenta delivery is still the main and only effective treatment for PE (Mol et al., 2016).
Because PE originate in the placenta, placental dysfunction underlies a spectrum of PE-related symptoms (Phipps et al., 2019; Stepan et al., 2020). To induce PE, the aberrant development and dysfunction of placenta can be associated with abnormal remodeling of maternal uterine arteries, the main cause of which is impaired extravillous trophoblast (EVT) functional phenotype. Many papers proposed that inadequate EVT invasion could prevent the proper engrafting and remodeling of uterine spiral arteries, consequently inducing the occurrence of PE (Redline and Patterson, 1995; Staff, 2019; Staff et al., 2020). Overall, it is suggested to further explore the mechanisms underlying the abnormal EVT functional phenotypes and spiral artery remodeling, thereby expanding the theoretical basis for improving the clinical treatment of PE.
Long non-coding RNA (lncRNA) is defined by its length (more than 200 nucleotides) and the lack of protein-coding capability. The family of lncRNAs has been increasingly focused by various studies in recent years (Delas and Hannon, 2017; Palazzo and Koonin, 2020). To date, numerous lncRNAs have been recognized as critical factors to modulate biological functions, such as chromatin modifiers recruitment (Davidovich and Cech, 2015), immune response (Jiang et al., 2018), cell differentiation (Jiang et al., 2017), and disease-related dysfunctions (Wu et al., 2017; Hu et al., 2020). Aberrant expressions of lncRNAs are known to be closely related to various human diseases, as well as to the different stages of the diseases, such as the occurrence, the development, and the prevention, especially in PE (Xu et al., 2018a; Wu et al., 2018; Xu et al., 2020). For instance, the lncRNA TUG1 modulates RND3 expression by binding to EZH2, in order to modulate cell proliferation, migration and network formation in vitro (Xu et al., 2017). Therefore, to improve the diagnosis and the treatment for PE, the key lncRNAs in the relevant molecular pathways should be identified, and their functions should be thoroughly investigated, which would contribute to the understanding of PE pathogenesis.
Hypoxia-inducible factor 1 alpha-antisense RNA 2 (HIF1A-AS2), a 2051-bp lncRNA gene, locates on chromosome14q23.2. Our previous study has reported that HIF1A-AS2 expression in PE placental tissues were dramatically downregulated. HIF1A-AS2 can participate in the invasive apoptotic biological process of trophoblast cells, in which it inhibits PHLDA1 expression by binding to LSD1 (Wu et al., 2019). Although lncRNAs can modulate the biological functions of cells through multiple mechanistic pathways, our particular interest is to explore how HIF1A-AS2 participates in the pathways relating to angiogenesis. To reveal furthermore details of the relevant molecular pathways, this study was also designed to identify the upstream and downstream regulatory factors for HIF1A-AS2 inducing the abnormal cellular activities in PE trophoblast cells, in order to recognize novel instructive clues for the improvement of clinical application in the future. Based on those findings, the association between PE and abnormal HIF1A-AS2 expression was evaluated, in order to recognize novel instructive clues for the improvement of clinical application in the future.
Previously, our team demonstrated that HIF1A-AS2 was dramatically down-expressed in PE placental tissue compared to the expression level of normal pregnant women. Meanwhile, the enhancement of HIF1A-AS2 expression promoted the proliferation of trophoblast cells and thereby contribute to the uterine spiral artery recasting. The further analyses focused on the correlation between HIF1A-AS2 expression and placental vasculature. The first step was to select trophoblast cell line (HTR-8/SVneo) and Human umbilical vein endothelial cells (HUVECs), which should be associated with placental angiogenesis and thus enable the demonstration of the relevant regulatory functions of HIF1A-AS2. To evaluate the influences of the changes in HIF1A-AS2 expression on the placental blood vessels, HIF1A-AS2 was silenced with specific siRNAs or overexpressed with plasmid (pcDNA3.1-HIF1A-AS2) in the two selected cell lines, respectively. To exclude off-target effects, three different siRNAs were specifically designed. After the transfection with siRNAs into the HTR-8/SVneo and HUVEC, qPCR assays confirmed that HIF1A-AS2 expression was silenced in the cells treated with si-HIF1A-AS2 1#, siHIF1A-AS2 2# and siHIF1A-AS2 3#, respectively (Figures 1A,B). Therefore, si-HIF1A-AS2-1# and/or si-HIF1A-AS2-3# were used for subsequent experiments. Depending on the application of pcDNA3.1-HIF1A-AS2 plasmid, the upregulation of HIF1A-AS2 expression was also achieved (Figures 1C,D).
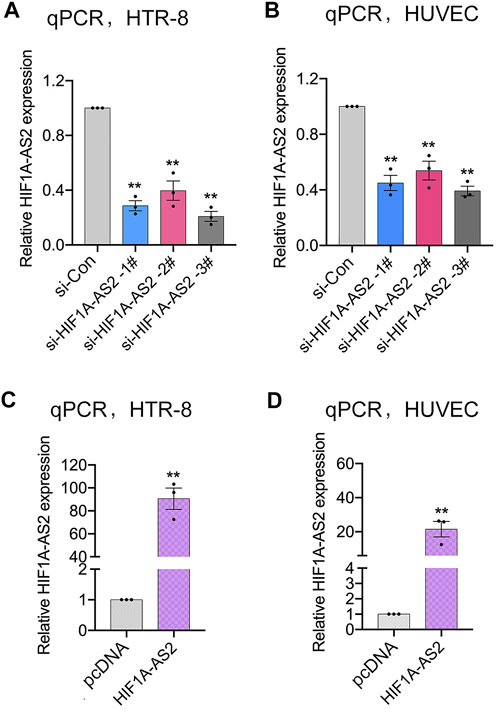
FIGURE 1. Modulation of HIF1A-AS2 expression in trophoblasts. qPCR analysis of HIF1A-AS2 expression in HTR-8/SVneo (A) and HUVECs (B) after transfected with HIF1A-AS2–specific siRNAs, respectively. HIF1A-AS2 expression levels were tested by qPCR after treating with empty vector and pcDNA3.1-HIF1A-AS2 plasmid in HTR-8/SVneo (C) and HUVECs (D). Error bars are mean with SEM of technical replicates (n = 3). *p < 0.05, **p < 0.01; significance by Student’s t test.
In response to abnormal HIF1A-AS2 expression levels, the cellular activities of trophoblasts were expected to exhibit corresponding changes. To verify the association, transwell experiments were conducted to evaluate the invasion and migration abilities of HTR-8/SVneo and HUVEC cells. For establishing blood flow between maternal and fetal systems, appropriate cellular motility and adhesion of trophoblasts is essential, whereas the impaired placental vascular development of PE patient is induced by suppressed trophoblast activities. The silencing of HIF1A-AS2 could inhibit the invasion and migration of HTR-8/SVneo and HUVECs (Figures 2A,C), while the upregulated HIF1A-AS2 expression presented causal relationship with boosted trophoblast invasion and migration (Figures 2B,D).
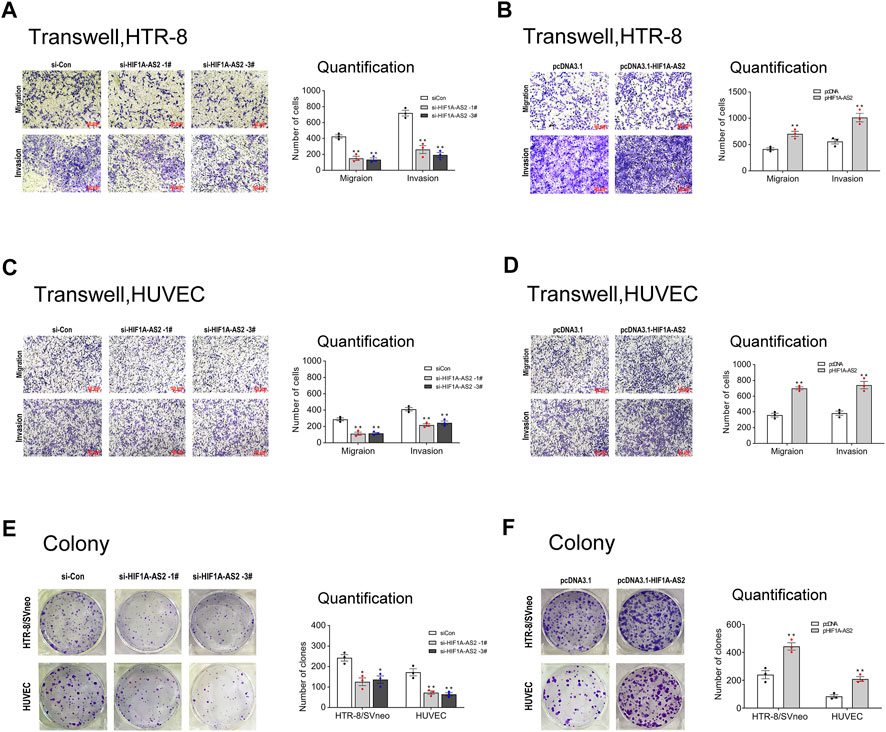
FIGURE 2. HIF1A-AS2 promotes trophoblast proliferation, migration and invasion in vitro. Transwell assays showed the migratory and invasive abilities of siHIF1A-AS2-treated in HTR-8/SVneo (A) and HUVECs (C). The quantity of cells was markedly decreased compared to siCon control. With the treatment of overexpressed HIF1A-AS2, the 2 cell lines showed significant increase than that in pcDNA control (B,D). Colony formation assay to assess the ability of proliferation in HTR-8/SVneo and HUVEC cells, transfected with siRNAs or plasmid. Silenced HIF1A-AS2 expression in HTR-8/SVneo and HUVEC cells (E) and overexpressed-HIF1A-AS2 (F). The statistics of the relevant data is on the right side of the corresponding graph. Error bars are mean with SEM of technical replicates (n = 3), *p < 0.05, **p < 0.01; significance by Student’s t test.
Next, in the colony formation assay, when HIF1A-AS2 was silenced, both tested cell lines showed decreased clone numbers (Figure 2E). Conversely, the elevated HIF1A-AS2 expression facilitated cell number increases in vitro (Figure 2F), confirming the positive relationship between HIF1A-AS2 expression and the proliferation of the tested cell lines. Thus, verified by these noticeable findings, HIF1A-AS2 could effectively modulate the proliferation, invasion and migration of trophoblast cells and umbilical vein endothelial cells.
Abnormal development of uterine spiral artery remodeling in placenta is one of the main predisposing conditions of PE, which can induce ischemia, hypoxia, oxidative stress, and excessive release of various cytokines into the maternal blood circulation, leading to systemic Inflammatory immune overactivation and vascular endothelial damage. In order to recognize the association between HIF1A-AS2 and vascular remodeling, for the two essential cells in the placental blood vessels recasting, tube formation assay was applied. HIF1A-AS2 silencing inhibited the tube formation ability of HTR-8/SVneo cells and HUVECs (Figure 3A). Consistently, the numbers of segments and total segments lengths were both decreased compared to the controls. HIF1A-AS2 overexpression resulted in the opposite trend (Figure 3B). The presence of red plugs in the tested cell lines was significantly different from that of the vector control cell sample, confirming that enhanced HIF1A-AS2 expression promoted the angiogenic activities of HTR-8/SVneo and HUVECs cells (Figure 3D). Knock-down of HIF1A-AS2 led to the opposite outcome (Figure 3C). To summarize these observations, HIF1A-AS2 played a pivotal role in proangiogenic activities of HTR-8/SVneo cells and HUVECs.
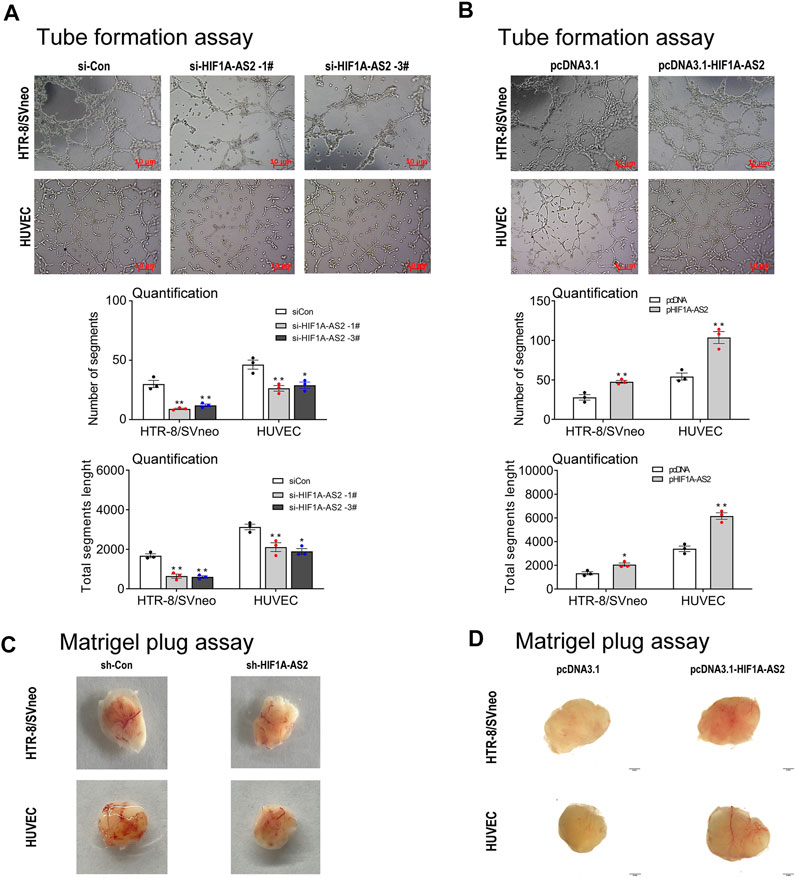
FIGURE 3. HIF1A-AS2 affects angiogenesis of HTR-8/SVneo trophoblast and HUVEC cells in vitro and in vivo. Consequent to network formation, the number of segments and total segments length of the network were decreased in HIF1A-AS2 with siRNAs (A) and increased in overexpressed with pcDNA3.1-HIF1A-AS2 (B) in HTR-8/SVneo and HUVECs, respectively. The quantity of cells is shown below the corresponding graph. The Matrigel plug assay showed that HIF1A-AS2-related siRNA inhibited angiogenesis in nude mice (C), and overexpression of HIF1A-AS2 promoted angiogenesis (D). Error bars are mean with SEM of technical replicates (n = 3). *p < 0.05, **p < 0.01; significance by Student’s t test.
The expression patterns of various lncRNAs had been proved to be mediated by transcription factors in previous research (Sato et al., 2010; Xu et al., 2020). Relying on the bioinformatic approach, the JASPAR database (http://jaspar2014.genereg.net) could be utilized to identify the candidate factors activating or inactivating HIF1A-AS2 expression. As listed in Table 1, in HIF1A-AS2 promoter region, there were 15 Forkhead box P1 (FOXP1)-binding putative sites, implying that HIF1A-AS2 may be regulated by transcription factor FOXP1 in trophoblast cells. In our previous study, we analyzed the expression levels of FOXP1 in 64 pairs of PE samples and adjacent normal samples with normalization to GAPDH by quantitative real-time PCR assay. The expression of FOXP1 was significantly downregulated in PE placenta compared with that in control. The same trend was also obtained in the animal model of preeclampsia (Xu et al., 2022). Subsequently, FOXP1 expression at both RNA and protein levels in the HTR-8/SVneo were silenced with siRNAs (Figures 4A,B), and in response to the manipulation, HIF1A-AS2 expression showed obvious decreases (Figure 4C). The putative binding sites of FOXP1 on HIF1A-AS2 promoter region, and four potentially relevant binding sequences were acquired by using the JASPAR website (Figure 4D).
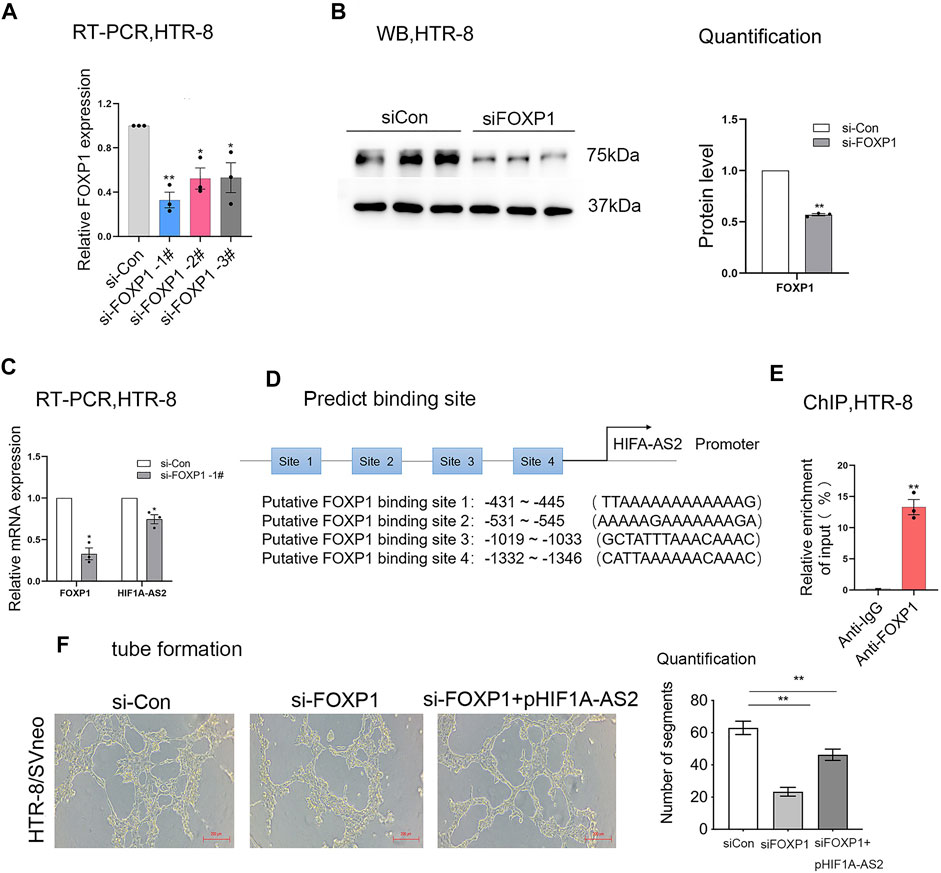
FIGURE 4. HIF1A-AS2 interacts with the transcription factor FOXP1. FOXP1 expression level was detected at RNA and protein level by qPCR (A,C) and Western blotting (B), respectively, after FOXP1-specific siRNAs were transfected into HTR-8/SVneo. After the treatment with siRNAs against HIF1A-AS2, the lncRNA expression was tested by qPCR (C). The putative binding sites of FOXP1 on HIF1A-AS2 promoter region, and four potentially relevant binding sequences were acquired by using the JASPAR website (D). ChIP assay confirmed the binding of FOXP1to the promoter of HIF1A-AS2 in HTR-8/SVneo. qPCR data were compared with IgG negative control (E). Error bars are mean with SEM of technical replicates (n = 3). *p < 0.05, **p < 0.01; significance by Student’s t test. Performing network formation, cells transfected with FOXP1 siRNAs and HIF1A-AS2 plasmid showed an increase in node numbers as compared with silencing of FOXP1 in HTR-8/SVneo (F). Error bars are mean with SEM of technical replicates (n = 3). *p < 0.05 and **p < 0.01; significance by one-way ANOVA with Tukey post-test. All data are representative of at least two independent experiments.
Next, the interpretation of ChIP assays supported that the binding of FOXP1 to the HIF1A-AS2 promoter region could upregulate the lncRNA at RNA level (Figure 4E). Then we performed tube formation assay in HTR-8/SVneo, the results showed that the inhibition of FOXP1 and upregulation of HIF1A-AS2 can promote antiogenesis compare with silencing of FOXP1 (Figure 4F). Therefore, FOXP1 was suggested to be the most potent candidate factor for the upstream regulation on HIF1A-AS2 activation.
Our research group had carried out RNA transcriptome sequencing for the siRNA (siCon) and siHIF1A-AS2-transfected HTR-8/SVneo cells with the presence of negative control, in order to identify the downstream influences of altered HIF1A-AS2 expression (Wu et al., 2019). Relying on the Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases, the biological pathways relevant to HIF1A-AS2 activation could be identified. In response to HIF1A-AS2 silencing, a bunch of genes responsible for vascular development were significantly affected in their expression levels (Wu et al., 2019). Through qPCR, Angiopoietin Like 4 (ANGPTL4), a transcript related to vascular development was selected, the expression of which showed significant downregulation based upon HIF1A-AS2 overexpression (Figure 5A) and upregulation based upon HIF1A-AS2 knockdown in HTR-8/SVneo (Figure 5B).
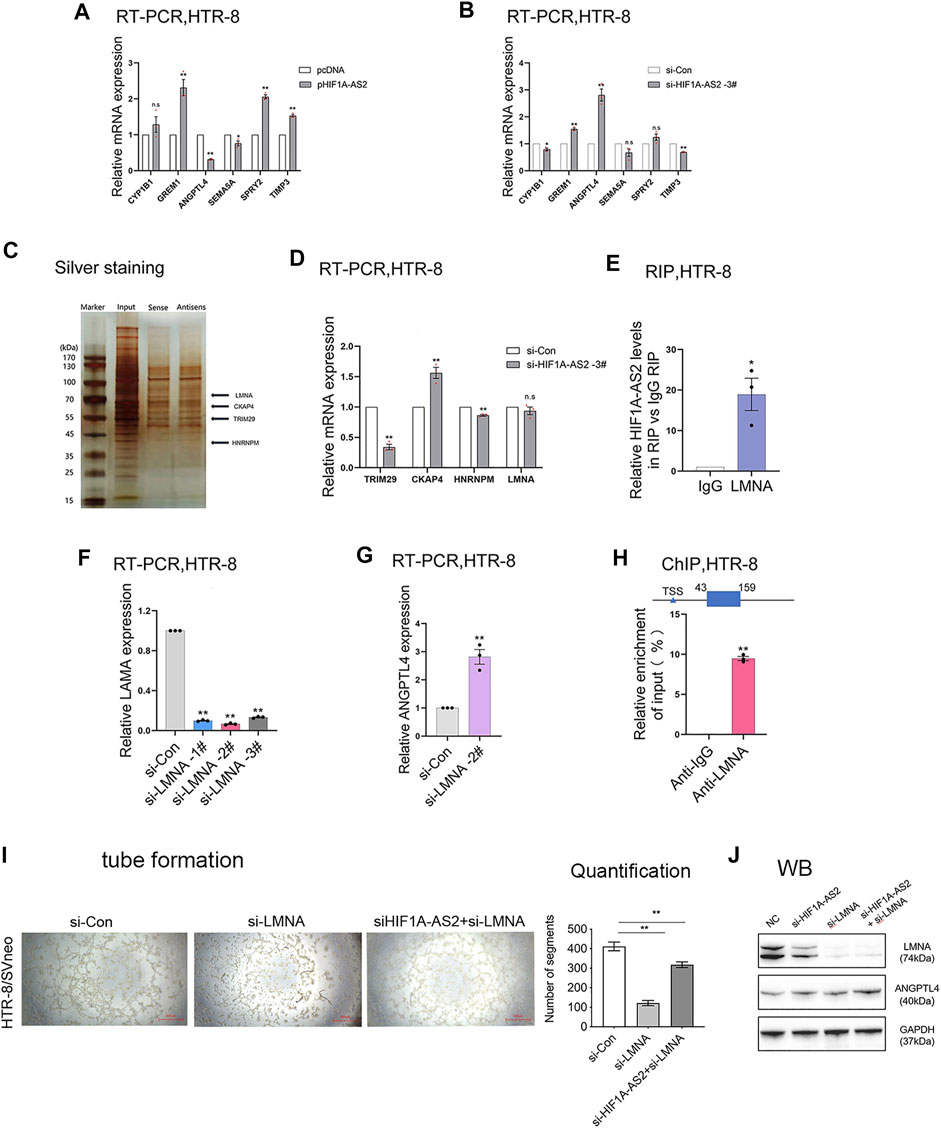
FIGURE 5. HIF1A-AS2 binds to LMNA to inhibit ANGPTL4 expression. qPCR analysis of HTR-8/SVneo cells uncovered the relative mRNA expression of vascularized genes, with (A) HIF1A-AS2 overexpression or (B) knockdown by siHIF1A-AS2. RNA pull down, silver staining and mass spectrometry analysis were combined to analyze several potential binding proteins (C). After transfected with siHIF1A-AS2, the changes in the mRNA expression of several possible binding proteins were analyzed by qPCR (D). RIP results showed that HIF1A-AS2 could bind LMNA in HTR-8/SVneo cells (E). After LMNA knockdown, the expression of ANGPTL4 was increased at RNA levels (F,G). Error bars are mean with SEM of technical replicates (n = 3). *p < 0.05, **p < 0.01; significance by Student’s t test. ChIP assays revealed the enrichment of LMNA in ANGPTL4 promoter region (H). Consequent to tube formation, the number of segments and total segments length of the tube were decreased in the inhibition of HIF1A-AS2 and LMNA (I); ANGPTL4 and LMNA expression levels were detected at protein level by WB in HTR-8/SVneo (J). Error bars are mean with SEM of technical replicates (n = 3). *p < 0.05 and **p < 0.01; significance by one-way ANOVA with Tukey post-test. All data are representative of at least two independent experiments.
Our previous studies had clarified the distribution of HIF1A-AS2 RNA, mainly in the nucleus of HTR-8/SVneo cells (Wu et al., 2019). Accordingly, HIF1A-AS2 might have functions in transcriptional regulation via recruiting RNA-binding proteins. To examine the prediction, RNA pull down assay and mass spectrometry analysis were conducted, and the results showed that HIF1A-AS2 might interact with Lamin A/C (LMNA) (Figure 5C). However, qPCR assays found that there was almost no change in LMNA expression at the RNA level (Figure 5D) after knocked down HIF1A-AS2. Subsequently, relying on the results of RNA immunoprecipitation (RIP) analysis, it was also proved that HIF1A-AS2 could recruit LMNA in HTR-8/SVneo (Figure 5E).
The protein products of the LMNA gene, Lamin A/C proteins, are essential for the normal functions of the nuclear lamina due to their multiple cellular functions, including the modulation of DNA transcription, the maintenance of chromatin organization and nuclear stability, the regulation of cell cycle, and the initiation of apoptosis (Nishikawa et al., 2021). Then, our results revealed that the expression of ANGPTL4 was increased after down-regulating LMNA with specific-siRNAs at RNA level (Figures 5F,G). Moreover, we performed ChIP assays and revealed that LMNA could directly interact with ANGPTL4 promoter regions (Figure 5H). If considering the downstream interactions as an integrated mechanism, it suggested that HIF1A-AS2 might regulate the expression of ANGPTL4 in trophoblasts through interacting with LMNA. Then we performed tube formation assay, and the results indicated that the inhibition of HIF1A-AS2 and LNMA can promote antiogenesis compare with silencing of LNMA (Figure 5I). Also, the western blotting assays were conducted, and the expression of ANGPTL4 was significantly elevated in the inhibition of HIF1A-AS2 and LMNA compare with that in the silencing of LNMA (Figure 5J).
To evaluate the relevance between ANGPTL4 and PE, mRNA expression levels of ANGPTL4 were compared between placental tissue samples from severe PE patients and healthy pregnant participants. qPCR assays were carried out, and the results suggested increased ANGPTL4 mRNA expression in severe PE tissue samples compared with the control (Figure 6A). Furthermore, we performed immunohistochemistry assay and consequently confirmed that ANGPTL4 expression levels of the PE-positive placental samples were elevated at protein level (Figures 6B,C). In conclusion, for the patients of severe PE, ANGPTL4 expression level had significant negative correlation with that of HIF1A-AS2 in their placental tissue samples.
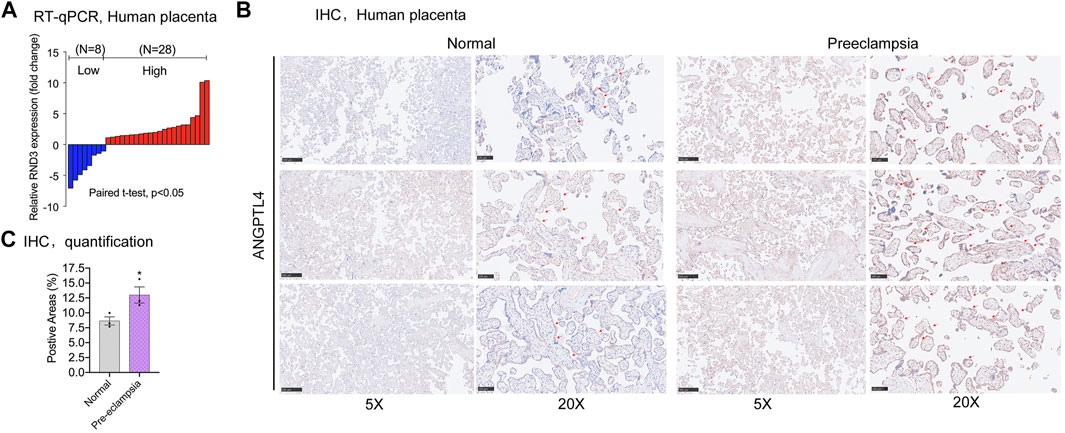
FIGURE 6. Expression of ANGPTL4 in preeclampsia placental tissues. qPCR results showed the fold change of placental tissue in PE patients compared to healthy pregnant women (n = 34) (A). The abundance of ANGPTL4 protein was detected by immunohistochemistry (B), quantification was performed by positive areas (C). All data are representative of at least three independent experiments. *p < 0.05, **p < 0.01; significance by Student’s t test.
Among the large variety of non-coding RNAs existing in human genome, the two major types, microRNAs (miRNAs) and lncRNAs (>200 nucleotides), are currently sorted by the lengths, and their total number is approximately 16.000. Due to the necessity of their functions to maintain the normal running of relevant biological processes, the correlations between the abnormal expression of specific lncRNAs and the occurrences of diseases had been increasingly noticed (Sun et al., 2018; Statello et al., 2021). To be specific, the interest of our group was the association between the candidate lncRNAs and the pathogenesis of PE. Our published papers had identified some PE-related lncRNAs, including TUG1, PVT1, and HOXA11-AS1 (Xu et al., 2017; Xu et al., 2018a; Xu et al., 2018b). Based on the precedent studies, this study aimed to verify the relationship between HIF1A-AS2 and PE, thereby revealing more details of the pathogenetic mechanisms at molecular level and proposing the new candidates for further analyses to evaluate their potential in clinical practice against PE.
Multiple preceding studies had revealed that HIF1A-AS2 could effectively participate in the pathogenetic mechanisms of various malignancies (Wang et al., 2019a; Wang et al., 2019b; Si et al., 2021). Combined with our existing research work, the PE-related functions of HIF1A-AS2 could be preliminarily identified. To obtain more details at molecular level, this study was designed to further evaluate the influences of abnormal HIF1A-AS2 expression on HTR-8/SVneo and HUVEC. In vitro, the silencing of HIF1A-AS2 expression resulted in remarkably suppressed proliferation, invasion, migration and vascularization of trophoblast cells, and consistently, overexpressed HIF1A-AS2 also induced significant changes in these cellular activities at the opposite direction. In vivo, we observed that upregulation of HIF1A-AS2 could promote angiogenesis in trophoblast cells and vascular endothelial cells. The observations supported the effective participance of HIF1A-AS2 in the development of PE, which depended on the regulatory functions of the lncRNA, and might indicate its clinical significance in PE. Overall, our further analyses on the underlying molecular mechanisms should be promising.
Next, we performed high-throughput sequencing (RNA-seq) to explore the downstream objects of HIF1A-AS2. According to the in-vitro tests, after suppression of HIF1A-AS2, noticeable increases of the cellular inactivator ANGPTL4 could be detected at both the mRNA and proteins level. ANGPTL4, located on chromosome 19p13.2, encodes a peptide hormone belonging to the family of adipokines. Its protein product has a variety of biological functions, including the modulatory effects on cell cycle progression, metastasis and angiogenesis (Guo et al., 2014; Yang et al., 2020). However, the exact mechanisms establishing the association between ANGPTL4, and preeclampsia remains a mystery. For the accessibility between ANGPTL4 and HIF1A-AS2, our previous study had reported the predominant nuclear localization of HIF1A-AS2 in trophoblast cells (Wu et al., 2019). The subsequent mechanism experiments included RIP assay, RNA pull down and mass spectrometry analysis. The results provided more details to confirm the interaction between HIF1A-AS2 and LMNA in trophoblasts. According to the findings, in the pathogenetic mechanism of PE, a part of the downstream pathways for HIF1A-AS2 might rely on its binding to LMNA, thereby suppressing ANGPTL4 expression in trophoblasts.
Mounting studies stated that transcript factor could activate/inactivate lncRNA transcription further regulating target expression which accelerates the progression of PE (Ma et al., 2014; Ghosh et al., 2018; Huh et al., 2019). For example, Xu et al. (2019) reported that KLF5 could act as transcriptional factor and modulate lncRNA 00346-prompted gastric cancer progression. Reported by our published work, the transcript factor FOXP1 could contribute to the activation of AGAP2-AS1 expression in trophoblasts (Xu et al., 2020). Accordingly, for this study, the bioinformatic analysis also suggested that FOXP1 might promote HIF1A-AS2 expression at transcriptional level (Xu et al., 2020). To verify this conjecture, ChIP-PCR results demonstrated that FOXP1 could directly bind to the promoter region of HIF1A-AS2 in HTR-8/SVneo. Furthermore, FOXP1 upregulation could enhance HIF1A-AS2 transcription in trophoblasts. Together, these findings established that HIF1A-AS2 could be activated by FOXP1, emphasizing the complicity of the upstream regulation on the expression patterns of lncRNAs.
To summarize the discoveries in this study, among the pathogenetic mechanisms of PE, the abnormal HIF1A-AS2 downregulation can induce the enhancement of ANGPTL4 expression via the LMNA-mediated regulatory pathway, resulting in aggravated dysfunction of angiogenesis (Figure 7). Thus, HIF1A-AS2 has the potential to be investigated as a candidate molecular target which may facilitate the improvement of clinical strategies against PE. However, for the limitation of this study, the potential biological phenotypes of HIF1A-AS2 have not been completely explored, while the identification of other achievable downstream and regulatory mechanisms could still be expected. Further studies are required to illuminate other relevant upstream and downstream pathways, through which HIF1A-AS2 can exert its regulatory functions on the cellular activities of trophoblasts in PE.
Human trophoblast cell (HTR-8/SVneo) and human umbilical vein endothelial cells HUVECs were offered by the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). RPMI 1640 medium, added by 10% fetal bovine serum (FBS, GIBCO, United States) and antibiotics (100 U/ml of penicillin and 100 mg/ml of streptomycin), was the choice to culture HTR-8/SVneo cell. HUVEC cells were cultured in ECM medium. The temperature set for cell growth was 37.0°C, with high humidity and 5% CO2 in the incubating condition.
DNA Midiprep kits (QIAGEN, Hilden, Germany) were chosen, which enable the plasmid vectors (pcDNA3.1-HIF1A-AS2) to arrayed. The sequences of siRNAs, including HIF1A-AS2-specific siRNAs, FOXP1 siRNA, LAMA siRNA and scrambled siCon, were designed by Gene Pharma; their details were listed in Supplementary Table S1. After HUVECs or HTR-8/SVneo cells were inoculated on 12-well plates, transfection could be carried out when the degree of cell fusion reached 70%–80%. Reagent Lipofectamine 2000 (Invitrogen, United States) was applied for siRNA transfection. FuGENE HD Transfection reagent (Roche, Pennsburg, Germany) was applied to treat the plasmid. The processing steps complied with manufacturer’s instructions. After 24 h, the treated cells were collected for further analyses.
FastPure Cell/Tissue Total RNA Isolation Kit V2 (Vazyme, Nanjing, China) was chosen to conduct total RNA extraction. The following measurements of RNA concentration and purity was achieved with Onedrop. RNA was reversely transcribed into cDNA using HiScript®III RT SuperMix for qPCR (+gDNA Wiper (Vazyme, Nanjing, China). ChamQ SYBR qPCR Master Mix (High ROX Premixed) (Vazyme, Nanjing, China) was applied to detect the expression of HIF1A-AS2 and LAMA, and the mixture was configured according to the instructions of StepOnePlus™ (Thermo Fisher Scientific, Waltham, MA, United States) instrument. The qPCR data were analyzed by StepOne™ software v2.3. The PCR primers were listed in Supplementary Table S1.
For colony formation capability, each cell sample had been treated with a specific siRNA or the plasmid before the seeding. Each well of the 6-well plate was filled with 600, 800 or 1,000 cells and had the addition of 2 ml 10% FBS complete medium. About 10–14 days after the formation of colony, with the fixation used methanol and the staining used 0.1% crystal violet. The capability of colony formation was quantified as the number of stained colonies.
To assess cell migration and invasion, the transwell chamber (pore size 8 μm) was placed in a 24 well plate. Briefly, the 4–6 × 104 cells suspended in 300 μl empty medium were seeded into the chamber inside, and 700 μl complete medium containing 10% FBS was infused into the chamber outside. The diluted Matrigel Matrix (Corning, United States) was coated or uncoated on top of the membrane for invasion or migration assays, respectively. Culturing for 24–48 h, the cells below the membrane were fixed with methanol and stained with 0.5% crystal violet. Under the light microscopy, from each chamber, 3-5 areas were picked up by random selection for photographing and data recording.
The precooled Matrigel Matrix (Corning, United States) was added to the wells in the 96-well plates, in order to achieve polymerization by incubation at 37°C for half an hour. HUVECs or HTR-8/SVneo cells were inoculated onto 12-well plates, followed by being transfected with 3 nM siRNAs or 2 μg plasmid for 24 h. The treated HUVECs or HTR-8/SVneo were digested. Subsequently, each sample was suspended in 150 μl conditioned medium and then transferred to a coated 96-well plate (4 × 104 cells/well or 6 × 104 cells/well). The incubation conditions were set at 37°C in 5% CO2 for 8–16 h. Under light microscopy, photographs were taken. The analysis of the formed tube was conducted by an ImageJ Angiogenesis Analyzer.
Total proteins in cells or tissues were extracted with cold lysates and separated by 10% TGX FastCast acrylamide gel (Bio-Rad, United States), followed by being transferred to 0.45 μm PVDF membranes (Millipore, United States). Subsequently, the PVDF membrane was sealed with a blocking solution for 15 min at room temperature. The appropriate dilution antibody (FOXP1, CST, Catalog No. D35D10, United States) and anti-GAPDH antibody (Proteintech, Catalog No. 10494-1-AP, United States) were applied for overnight incubation on a shaker in a 4°C fridge. After washing, the secondary antibody was applied for 1 h incubation. The chemiluminescence (Thermo) fluid was applied for exposure. Quantity One software (Bio-RAD) read the protein bands and interpreted the patterns into numeric data.
After the linearization of the plasmid templates, the mMESSAGE mMACHINE T7 Transcription Kit (Life Technologies, United States) was applied for in vitro transcription. Subsequently, complying with the manufacturer’s instructions, the major steps of RNA pull down assay was conducted as below. A single biotinylated nucleotide was attached to label RNA 3′terminus with T4 RNA ligase. Labeled HIF1A-AS2 capture was achieved with streptavidin magnetic beads, which was then incubated with cell protein lysates from HTR-8/SVneo. After silver staining, the protein eluted solution was ready for mass spectrometry analysis on Q Exactive mass spectrometer (Proxeon Biosystems, now Thermo Fisher Scientific), Analysis of the mass spectrometry data are listed in Supplementary Table S2.
EZ-Magna ChIP A kit (Catalog No. 17–408, Merck & Millipore) was applied to for the ChIP assay. The steps complied with the manufacturer’s instructions. After growth, HTR-8/SVneo cells were treated with formaldehyde, which crosslinks proteins to DNA to ensure co-precipitation. Then, the cells were broken open, which allowed the sonication to shear the chromatins into 200-1,000 bp DNA pieces. Immunoprecipitation was performed using primary antibodies or control IgG. Next, after the chromatin proteins were removed by DNA purification, qPCR was applied. The detailed information about specific ChIP primers were exhibited as Supplementary Table S1.
Following the procedures introduced by Xu et al. (2018a), RIP assays were set up. The producer of the LMNA antibodies was Millipore (Billerica, MA, Catalog No. 17–701 Merck & Millipore United States). For statistical analyses, qPCR was conducted to interpret the presence of HIF1A-AS2 and IgG into numeric data.
All experiments on mice complied with the guidance of Laboratory Animal Management Regulations, the national law of China and the instruction of the local authority. The approval of Zoopery was issued by the Ethics Committee of Nanjing Medical University Animal Welfare (IACUC-2106050). The plug assay was carried out as the conventional method reported by previous studies (Aktas et al., 2017; Ding et al., 2019). Briefly, female BALB/c nude mice (4–6 weeks after birth) were qualified for the experiments. The cells were resuspended with 0.2 ml serum-free medium and evenly mixed with equal volume of High Concentration Matrigel (BD Biosciences). Immediately, the mixture was injected subcutaneously. After the injection, 2 weeks later, Matrigel plugs were removed from the euthanized mice.
The data came from three independent experiments. GraphPad Prism version 8.0 (GraphPad Software) conducted the statistical calculations. Adobe Photoshop 2020 contributed to the graphic presentation. The evaluation of results passed two-tailed Student’s t test and expressed as mean ± S.E.M. **p < 0.01, *p < 0.05 were considered significant.
The original contributions presented in the study are included in the article/Supplementary Materials, further inquiries can be directed to the corresponding authors.
The animal study was reviewed and approved by the Ethics Committee of the First Affiliated Hospital of Nanjing Medical University, China.
LSh and CW carried out all experiments, analyzed the data, and prepared the pictures. ZD, YZ, TW, and JT collected the clinical data and placenta tissues. LSu and YX conceived this research. LSu and YX contributed to study design and data interpretation and the manuscript preparation. Lsh and YX wrote this paper with the help of all the authors. LSh and LSu provided intellectual insights and critical discussion of this project. All listed authors contributed to the manuscript and approved the submitted version.
This project was supported by grants (BK20201078 to YX) from the Natural Science Foundation of Jiangsu Province, the National Natural Science Foundation of China (No. 82001578 to YX), and Postdoctoral Fund Project in Jiangsu Province (No. 2020Z087 to YX), and China Postdoctoral Fund Project (No. 2020M671394 to YX), and Postdoctoral Fund Project in Nanjing (No. 2021BSH209) to YX, Maternal and child health “young talents” project in Jiangsu (No. FRC202151) to YX.
Trophoblast Cells HTR-8/SVneo, which derived from primary extra villous trophoblast cell, was friendly offered by Dr. Charies Graham, Queen’s University, Canada. We would like to thank 51 runse for the English language editing service.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2022.837000/full#supplementary-material
Supplementary Table S1 | The list of PCR/ChIP-PCR primers and siRNAs sequences.
Supplementary Table S2 | Analysis of the mass spectrometry data.
lncRNA, long non-coding RNA; PE, Preeclampsia; ANGPTL4, Angiopoietin-like 4; LMNA, Lamin A/C; RIP, RNA immunoprecipitation; ChIP, Chromatin immunoprecipitation.
Aktas, T., Avsar Ilik, I., Maticzka, D., Bhardwaj, V., Pessoa Rodrigues, C., Mittler, G., et al. (2017). DHX9 suppresses RNA processing defects originating from the Alu invasion of the human genome. Nature 544, 115–119. doi:10.1038/nature21715
Ananth, C. V., Brandt, J. S., Hill, J., Graham, H. L., Grover, S., Schuster, M., et al. (2021). Historical and recent changes in maternal mortality due to hypertensive disorders in the United States, 1979 to 2018. Hypertension 78, 1414–1422. doi:10.1161/HYPERTENSIONAHA.121.17661
Aukes, A. M., Arion, K., Bone, J. N., Li, J., Vidler, M., Bellad, M. B., et al. (2021). Causes and circumstances of maternal death: A secondary analysis of the community-level interventions for pre-eclampsia (CLIP) trials cohort. Lancet. Glob. Health 9, e1242–e1251. doi:10.1016/S2214-109X(21)00263-1
Bouter, A. R., and Duvekot, J. J. (2020). Evaluation of the clinical impact of the revised ISSHP and ACOG definitions on preeclampsia. Pregnancy Hypertens. 19, 206–211. doi:10.1016/j.preghy.2019.11.011
Davidovich, C., and Cech, T. R. (2015). The recruitment of chromatin modifiers by long noncoding RNAs: Lessons from PRC2. RNA 21, 2007–2022. doi:10.1261/rna.053918.115
Delas, M. J., and Hannon, G. J. (2017). lncRNAs in development and disease: from functions to mechanisms. Open Biol. 7, 170121. doi:10.1098/rsob.170121
Ding, X., Jia, X., Wang, C., Xu, J., Gao, S. J., Lu, C., et al. (2019). A DHX9-lncRNA-MDM2 interaction regulates cell invasion and angiogenesis of cervical cancer. Cell. Death Differ. 26, 1750–1765. doi:10.1038/s41418-018-0242-0
Ghosh, S., Roy-Chowdhuri, S., Kang, K., Im, S. H., and Rudra, D. (2018). The transcription factor Foxp1 preserves integrity of an active Foxp3 locus in extrathymic Treg cells. Nat. Commun. 9, 4473. doi:10.1038/s41467-018-07018-y
Guo, L., Li, S. Y., Ji, F. Y., Zhao, Y. F., Zhong, Y., Lv, X. J., et al. (2014). Role of Angptl4 in vascular permeability and inflammation. Inflamm. Res. 63, 13–22. doi:10.1007/s00011-013-0678-0
Hu, M., Wu, Y., Yang, C., Wang, X., Wang, W., Zhou, L., et al. (2020). Novel long noncoding RNA lnc-URIDS delays diabetic wound healing by targeting Plod1. Diabetes 69, 2144–2156. doi:10.2337/db20-0147
Huh, H. D., Kim, D. H., Jeong, H. S., and Park, H. W. (2019). Regulation of TEAD transcription factors in cancer Biology. Cells 8, E600. doi:10.3390/cells8060600
Ives, C. W., Sinkey, R., Rajapreyar, I., Tita, A. T. N., and Oparil, S. (2020). Preeclampsia-pathophysiology and clinical presentations: JACC state-of-the-art review. J. Am. Coll. Cardiol. 76, 1690–1702. doi:10.1016/j.jacc.2020.08.014
Jiang, M., Zhang, S., Yang, Z., Lin, H., Zhu, J., Liu, L., et al. (2018). Self-recognition of an inducible host lncRNA by RIG-I feedback restricts innate immune response. Cell. 173, 906–919. doi:10.1016/j.cell.2018.03.064
Jiang, R., Tang, J., Chen, Y., Deng, L., Ji, J., Xie, Y., et al. (2017). The long noncoding RNA lnc-EGFR stimulates T-regulatory cells differentiation thus promoting hepatocellular carcinoma immune evasion. Nat. Commun. 8, 15129. doi:10.1038/ncomms15129
Ma, M. Z., Li, C. X., Zhang, Y., Weng, M. Z., Zhang, M. D., Qin, Y. Y., et al. (2014). Long non-coding RNA HOTAIR, a c-Myc activated driver of malignancy, negatively regulates miRNA-130a in gallbladder cancer. Mol. Cancer 13, 156. doi:10.1186/1476-4598-13-156
Mol, B. W. J., Roberts, C. T., Thangaratinam, S., Magee, L. A., de Groot, C. J. M., Hofmeyr, G. J., et al. (2016). Pre-eclampsia. Lancet 387, 999–1011. doi:10.1016/S0140-6736(15)00070-7
Nishikawa, T., Kuwano, Y., Nakata, M., Rokutan, K., and Nishida, K. (2021). Multiple G-quadruplexes in the LMNA promoter regulate LMNA variant 6 transcription and promote colon cancer cell growth. Biochim. Biophys. Acta. Gene Regul. Mech. 1864, 194746. doi:10.1016/j.bbagrm.2021.194746
Palazzo, A. F., and Koonin, E. V. (2020). Functional long non-coding RNAs evolve from junk transcripts. Cell. 183, 1151–1161. doi:10.1016/j.cell.2020.09.047
Phipps, E. A., Thadhani, R., Benzing, T., and Karumanchi, S. A. (2019). Pre-eclampsia: Pathogenesis, novel diagnostics and therapies. Nat. Rev. Nephrol. 15, 275–289. doi:10.1038/s41581-019-0119-6
Redline, R. W., and Patterson, P. (1995). Pre-eclampsia is associated with an excess of proliferative immature intermediate trophoblast. Hum. Pathol. 26, 594–600. doi:10.1016/0046-8177(95)90162-0
Sato, K., Nakagawa, H., Tajima, A., Yoshida, K., and Inoue, I. (2010). ANRIL is implicated in the regulation of nucleus and potential transcriptional target of E2F1. Oncol. Rep. 24, 701–707. doi:10.3892/or_00000910
Si, J., Ma, Y., Lv, C., Hong, Y., Tan, H., Yang, Y., et al. (2021). HIF1A-AS2 induces osimertinib resistance in lung adenocarcinoma patients by regulating the miR-146b-5p/IL-6/STAT3 axis. Mol. Ther. Nucleic Acids 26, 613–624. doi:10.1016/j.omtn.2021.09.003
Staff, A. C., Fjeldstad, H. E., Fosheim, I. K., Moe, K., Turowski, G., Johnsen, G. M., et al. (2020). Failure of physiological transformation and spiral artery atherosis: Their roles in preeclampsia. Am. J. Obstet. Gynecol. 226, S895–S906. doi:10.1016/j.ajog.2020.09.026
Staff, A. C. (2019). The two-stage placental model of preeclampsia: An update. J. Reprod. Immunol. 134-135, 1–10. doi:10.1016/j.jri.2019.07.004
Statello, L., Guo, C. J., Chen, L. L., and Huarte, M. (2021). Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell. Biol. 22, 96–118. doi:10.1038/s41580-020-00315-9
Stepan, H., Hund, M., and Andraczek, T. (2020). Combining biomarkers to predict pregnancy complications and redefine preeclampsia: The angiogenic-placental syndrome. Hypertension 75, 918–926. doi:10.1161/HYPERTENSIONAHA.119.13763
Sun, Q., Hao, Q., and Prasanth, K. V. (2018). Nuclear long noncoding RNAs: Key regulators of gene expression. Trends Genet. 34, 142–157. doi:10.1016/j.tig.2017.11.005
Wang, X., Peng, L., Gong, X., Zhang, X., and Sun, R. (2019). LncRNA HIF1A-AS2 promotes osteosarcoma progression by acting as a sponge of miR-129-5p. Aging (Albany NY) 11, 11803–11813. doi:10.18632/aging.102448
Wang, Y., Zhang, G., and Han, J. (2019). HIF1A-AS2 predicts poor prognosis and regulates cell migration and invasion in triple-negative breast cancer. J. Cell. Biochem. 120, 10513–10518. doi:10.1002/jcb.28337
Wu, D., Xu, Y., Zou, Y., Zuo, Q., Huang, S., Wang, S., et al. (2018). Long noncoding RNA 00473 is involved in preeclampsia by LSD1 binding-regulated TFPI2 transcription in trophoblast cells. Mol. Ther. Nucleic Acids 12, 381–392. doi:10.1016/j.omtn.2018.05.020
Wu, D., Yang, N., Xu, Y., Wang, S., Zhang, Y., Sagnelli, M., et al. (2019). lncRNA HIF1A antisense RNA 2 modulates trophoblast cell invasion and proliferation through upregulating PHLDA1 expression. Mol. Ther. Nucleic Acids 16, 605–615. doi:10.1016/j.omtn.2019.04.009
Wu, K., Zhao, Z., Liu, K., Zhang, J., Li, G., Wang, L., et al. (2017). Long noncoding RNA lnc-sox5 modulates CRC tumorigenesis by unbalancing tumor microenvironment. Cell. Cycle 16, 1295–1301. doi:10.1080/15384101.2017.1317416
Xu, T. P., Ma, P., Wang, W. Y., Shuai, Y., Wang, Y. F., Yu, T., et al. (2019). KLF5 and MYC modulated LINC00346 contributes to gastric cancer progression through acting as a competing endogeous RNA and indicates poor outcome. Cell. Death Differ. 26, 2179–2193. doi:10.1038/s41418-018-0236-y
Xu, Y., Ge, Z., Zhang, E., Zuo, Q., Huang, S., Yang, N., et al. (2017). The lncRNA TUG1 modulates proliferation in trophoblast cells via epigenetic suppression of RND3. Cell. Death Dis. 8, e3104. doi:10.1038/cddis.2017.503
Xu, Y., Lian, Y., Zhang, Y., Huang, S., Zuo, Q., Yang, N., et al. (2018). The long non-coding RNA PVT1 represses ANGPTL4 transcription through binding with EZH2 in trophoblast cell. J. Cell. Mol. Med. 22, 1272–1282. doi:10.1111/jcmm.13405
Xu, Y., Wu, D., Hui, B., Shu, L., Tang, X., Wang, C., et al. (2022). A novel regulatory mechanism network mediated by lncRNA TUG1 that induces the impairment of spiral artery remodeling in preeclampsia. Mol. Ther. 30, 1692–1705. doi:10.1016/j.ymthe.2022.01.043
Xu, Y., Wu, D., Liu, J., Huang, S., Zuo, Q., Xia, X., et al. (2018). Downregulated lncRNA HOXA11-AS affects trophoblast cell proliferation and migration by regulating RND3 and HOXA7 expression in PE. Mol. Ther. Nucleic Acids 12, 195–206. doi:10.1016/j.omtn.2018.05.007
Xu, Y., Xia, X., Jiang, Y., Wu, D., Wang, S., Fu, S., et al. (2020). Down-regulated lncRNA AGAP2-AS1 contributes to pre-eclampsia as a competing endogenous RNA for JDP2 by impairing trophoblastic phenotype. J. Cell. Mol. Med. 24, 4557–4568. doi:10.1111/jcmm.15113
Keywords: preeclampsia, lncRNA, HIF1A-AS2, placental angiogenesis, ANGPTL4, LNMA
Citation: Shu L, Wang C, Ding Z, Tang J, Zhu Y, Wu L, Wang Z, Zhang T, Wang T, Xu Y and Sun L (2022) A novel regulated network mediated by downregulation HIF1A-AS2 lncRNA impairs placental angiogenesis by promoting ANGPTL4 expression in preeclampsia. Front. Cell Dev. Biol. 10:837000. doi: 10.3389/fcell.2022.837000
Received: 16 December 2021; Accepted: 30 June 2022;
Published: 09 August 2022.
Edited by:
Ramani Ramchandran, Medical College of Wisconsin, United StatesReviewed by:
Xing-Hong Ma, Northeast Agricultural University, ChinaCopyright © 2022 Shu, Wang, Ding, Tang, Zhu, Wu, Wang, Zhang, Wang, Xu and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lizhou Sun, c3VubGl6aG91MTAxQDE2My5jb20=; Yetao Xu, eHl0MTEwNzM5QDE2My5jb20=
†ORCID: Lizhou Sun, orcid.org/0000-0002-0465-8825; Yetao Xu, orcid.org/0000-0002-8653-3929
‡These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.