- Department of Cell Biology and Human Anatomy, University of California, Davis, Davis, CA, United States
A broad repertoire of transcription factors and other genes display oscillatory patterns of expression, typically ranging from 30 min to 24 h. These oscillations are associated with a variety of biological processes, including the circadian cycle, somite segmentation, cell cycle, and metabolism. These rhythmic behaviors are often prompted by transcriptional feedback loops in which transcriptional activities are inhibited by their corresponding gene target products. Oscillatory transcriptional patterns have been proposed as a mechanism to drive biological clocks, the molecular machinery that transforms temporal information into accurate spatial patterning during development. Notably, several microRNAs (miRNAs) -small non-coding RNA molecules-have been recently shown to both exhibit rhythmic expression patterns and regulate oscillatory activities. Here, we discuss some of these new findings in the context of the developing retina. We propose that miRNA oscillations are a powerful mechanism to coordinate signaling pathways and gene expression, and that addressing the dynamic interplay between miRNA expression and their target genes could be key for a more complete understanding of many developmental processes.
Introduction
The surge of new techniques to survey the transcriptome over the last few decades has led to the identification of numerous types of non-coding RNAs. While protein-coding sequences constitute less than 1.5% of the human genome, large-scale screenings have revealed that virtually the entire genome is transcribed to generate myriads of non-coding RNAs (Kapranov et al., 2002; Carninci et al., 2005; Mattick and Makunin, 2006; Birney et al., 2007). These RNA molecules are differentially expressed in distinct cell types and dynamically regulated during development (Hangauer et al., 2013; Mercer and Mattick, 2013).
Among non-protein coding RNAs, microRNAs (miRNAs) have emerged as key post-transcriptional regulators of gene expression (Bushati and Cohen, 2007; Bartel, 2009; Chekulaeva and Filipowicz, 2009). MiRNAs are small (∼22-nucleotide (nt) long), evolutionarily conserved molecules. First described in Caenorhabditis elegans (Lee et al., 1993), miRNAs are also present in a wide diversity of organisms in the bacteria, archaea, and eukaryote domains (Dexheimer and Cochella, 2020).
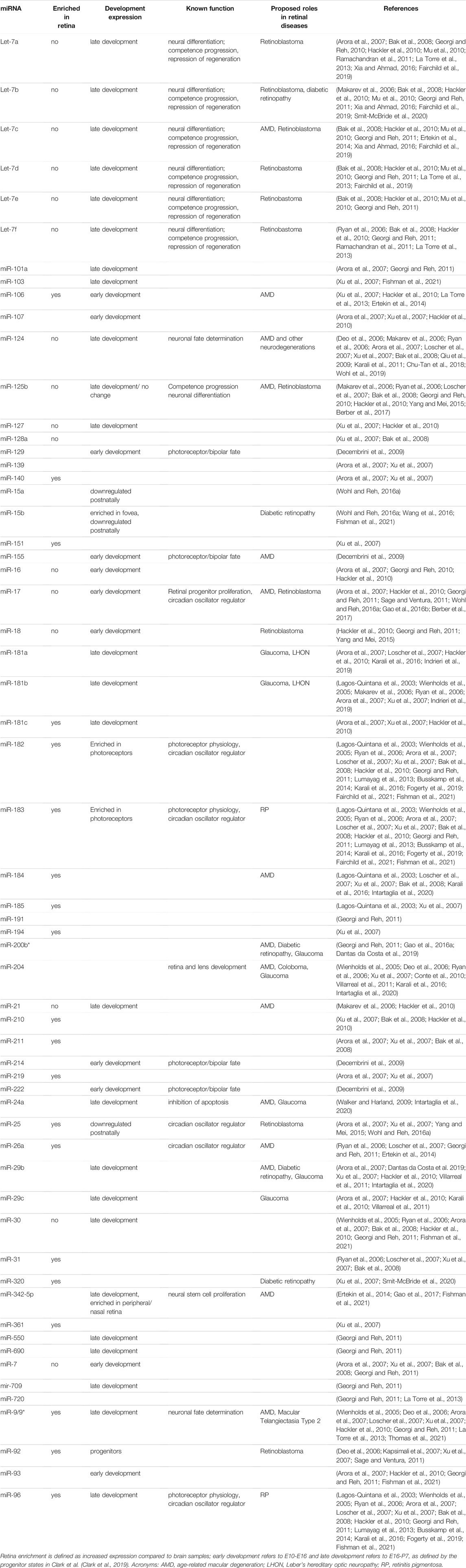
MiRNAs are transcribed from DNA sequences as long transcripts called primary miRNAs (pri-miRNAs) that contain double-stranded hairpin-like structures in which at least one of the two strands includes a mature miRNA (Figure 1). About half of all currently identified miRNAs are intergenic, mostly localized in introns, and controlled by the regulatory elements of the host gene; the other half are intragenic and are regulated independently by their own promoters (Ha and Kim, 2014). About 25% of all miRNAs are arranged in clusters and transcribed as longer transcripts that contain more than one mature miRNA sequence. Intergenic miRNAs are processed by the splicing machinery, while intragenic pri-miRNAs are cleaved by the microprocessor complex that includes Drosha ribonuclease and DiGeorge critical region 8 (DGCR8). In both cases, this first cleavage step produces a precursor miRNA (pre-miRNA) of about 70-nt that is exported out of the nucleus. Pre-miRNAs are further processed by the enzyme Dicer, which removes the loop of the hairpin, yielding a mature miRNA duplex that can be loaded onto the RNA-Induced Silencing Complex (RISC, Figure 1). Mature miRNAs bind to their target mRNAs, usually to the 3′ untranslated region (3′UTR), through imperfect base-pairing, hindering the stability and translation of their target mRNAs (Eulalio et al., 2008). Hence, miRNAs are part of complex networks where one individual miRNA can regulate a large number of genes, frequently from a similar biochemical pathway, and where a single target mRNA can be regulated concomitantly by multiple miRNAs. Thus far, about 2,500 mature miRNAs have been identified in the human genome (miRBase.org) (Kozomara and Griffiths-Jones, 2011), and bioinformatics studies have estimated that over 60% of the human transcriptome is regulated by miRNAs (Friedman et al., 2009).
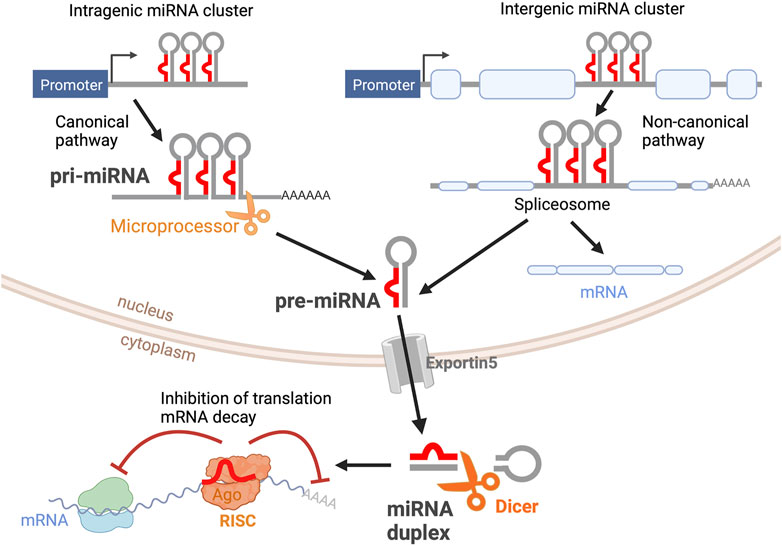
FIGURE 1. miRNA biogenesis. Primary miRNAs (pri-miRNAs) are transcribed as double-stranded hairpin-like structures. Intragenic pri-miRNAs are processed via the canonical pathway, where the clusters of hairpin-like structures are cleaved into individual precursor miRNAs (pre-miRNAs) by the microprocessor complex. Intergenic pri-miRNAs are processed by the splicing machinery. The resultant pre-miRNA from both pathways is an individual hairpin-like structure of 70-nt. After being exported from the nucleus via Exportin5, pre-miRNAs are further processed by Dicer into mature miRNA duplexes. One miRNA strand is loaded into the RNA-Induced Silencing Complex (RISC). Mature miRNA binds to its target mRNA, inhibiting mRNA translation and inducing mRNA decay. This figure was created with BioRender.com.
A large body of research suggests that this previously unknown miRNA-based regulation is crucial for many physiological and pathophysiological events and that the complex interactions between transcription factors and miRNAs could be instrumental in delineating developmental programs.
miRNAs in the Developing Retina
To gain further understanding of the roles of miRNAs in ocular tissues, several groups have attempted to characterize the retina miRNome by in situ hybridization, computational predictions, and profiling techniques. Hundreds of different miRNAs have been identified in the retina of different species (Loscher et al., 2007; Xu et al., 2007; Bak et al., 2008; Hackler et al., 2010; Karali et al., 2010, 2016; Fishman et al., 2021) and several miRNAs show a significantly enriched expression in the retina compared to other tissues (Table 1). Two early reports by Hackler et al. (Hackler et al., 2010) and Xu et al. (Xu et al., 2007) compared miRNA expression patterns at different developmental ages in the mouse retina and brain. Consistent with other studies (Lewis et al., 2003), the authors found that miRNAs with identical seed sequences exhibited highly similar expression profiles. Additionally, these studies and others have defined the repertoire of miRNAs expressed at different time points during retinal development (Table 1). Two main miRNA categories have been consistently identified: miRNAs expressed primarily at early developmental stages (embryonic day (E)10-E16 in the mouse) and miRNAs present at later stages of retinal development (E16-postnatal day (P)7) and maturation (>P7). Specifically, miR-17, miR-18, miR-19, miR-20, miR-93, miR-106, and miR-130 are down-regulated throughout development, while the let-7 family, miR-7, miR-9, miR-9*, miR-96, miR-101, miR-124, miR-181, miR-182, and miR-183 are some of the miRNAs that increase during retinal development from E10 to adulthood in mice. Additional studies have also identified cell-specific expression of subsets of these miRNAs (Georgi and Reh, 2010; Ohana et al., 2015; Wohl and Reh, 2016a) (Table 1).
Dicer and DGCR8 transgenic models (Damiani et al., 2008; Decembrini et al., 2008; Georgi and Reh, 2010, 2011; Pinter and Hindges, 2010; Davis et al., 2011; Iida et al., 2011; La Torre et al., 2013; Busskamp et al., 2014; Sundermeier et al., 2017), miRNA mutants (Lumayag et al., 2013; Barbato et al., 2017; Fogerty et al., 2019), sponge strategies (Zhu et al., 2011), and miRNA inhibitors (Decembrini et al., 2009; La Torre et al., 2013; Taylor et al., 2019; Wohl et al., 2019) have been extensively used to shed some light on the specific roles of miRNAs during retinal development. While many miRNA functions have been elucidated using these strategies, far less is known about miRNA target genes and the specific circuits that regulate development and pathophysiological processes in the retina. Furthermore, these global analyses do not capture the dynamic nature of miRNA expression and activity. Importantly, several miRNAs are involved in complex feedback and feed-forward regulations with their target genes, allowing for increased robustness of protein expression towards gene background noise (Borenstein and Ruppin, 2006). MiRNAs also participate in negative feedback loops, where target mRNAs regulate miRNA expression, leading to the occurrence of biological rhythms. Correspondingly, miRNAs have been shown to display rhythmic behaviors in the retina and other organs, and to regulate the circadian clock, the cell cycle, and the Hes1 ultradian oscillator (Figure 2). Here, we summarize some of the recent findings on miRNA oscillatory behaviors, their regulatory mechanisms, and some of their possible functions during retinal development.
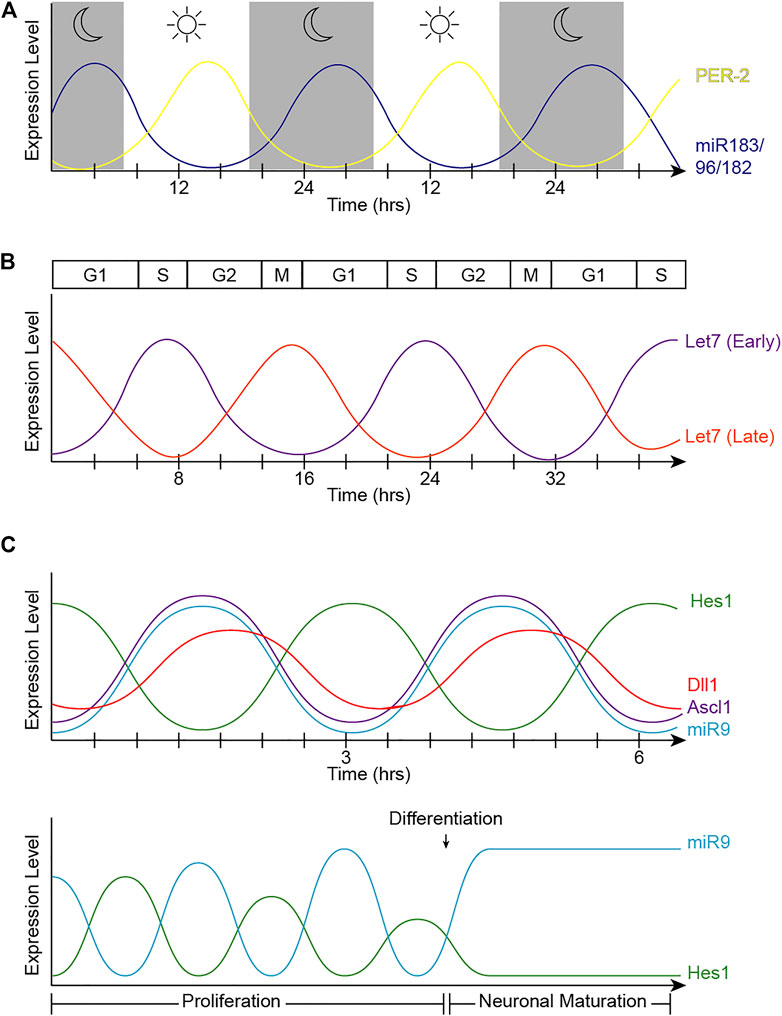
FIGURE 2. Oscillatory patterns of miRNAs in biological processes. (A) Oscillatory behavior of the miR-183 complex. miR-96 directly targets PER2, causing an out-of-phase oscillation pattern with PER-2 peaking during light and miR-183/96/182 peaking in dark hours. (B) Let-7 oscillatory behavior. The fluctuation of let-7 expression in accordance with the cell cycle changes at different stages of development. Early in development (let-7 early), let-7 expression is at its lowest at the start of the cell cycle in G1, and peaks in S-phase. The phase of oscillation shifts later in development (let-7 late), when let-7 expression is at its highest in mitosis and lowest in S-phase. (C) Hes1/miR-9 ultradian oscillator. (Top) Hes1 oscillation is self-driven with a rhythmicity of 2–3 h. The Hes1 oscillator represses Ascl1 and Notch ligands, consequently driving their oscillation patterns. MiR-9 and Hes1 participate in a negative feedback loop. (Bottom) Hes1 and miR-9 have out-of-phase expression patterns and are dependent on one another. As miR-9 continues to accumulate during proliferation, Hes1 is consequently dampened. RPC differentiation is induced when miR-9 levels reach a threshold to maintain high, steady levels while dulling Hes1 oscillations, resulting in neuronal maturation.
miR-183, -96, -182 and the Circadian Clock
The textbook view of the circadian clock consists of a light-dark pattern of approximately 24 h (Figure 2A) that governs rhythmicity within the organism and is regulated by two interwoven feedback loops with positive and negative components (Figure 3). One of these regulatory mechanisms involves the heterodimeric transcriptional activators CLOCK and BMAL1, which trigger the expression of repressors such as Period (PER1, PER2, and PER3) and Cryptochrome (CRY1 and CRY2) that, in turn, will repress the transcriptional activity of their activators (Sangoram et al., 1998; Zylka et al., 1998; Lowrey and Takahashi, 2004). The second loop involves the expression of Rev-Erbα and Rorα genes also regulated by CLOCK and BMAL1. Subsequently, REV-ERBα and RORα proteins compete for binding to the Bmal1 promoter (Lowrey and Takahashi, 2004). These self-sustaining feedback clocks are reset by fluctuating inputs, including light, temperature, or feeding patterns, to synchronize the molecular clock with the environment and the Earth’s rotation. This timing mechanism is controlled by a master pacemaker in the suprachiasmatic nuclei (SCN) of the hypothalamus, but independent circadian oscillators are present throughout the organism. Studies in the early 80’s already demonstrated that the circadian clock was present in the Xenopus retina (Besharse and Iuvone, 1983), and further analyses have added that the retinal circadian rhythm controls many aspects of the vertebrate ocular physiology, including melatonin and dopamine synthesis, photoreceptor disk shedding, visual sensitivity, and intraocular pressure (LaVail and Ward, 1978; Doyle et al., 2002; Maeda et al., 2006; Storch et al., 2007; Tosini et al., 2008). Dysregulation of these retinal circadian clocks can lead to ocular diseases and have impacts on the circadian rhythm within the whole body (Ko, 2020).
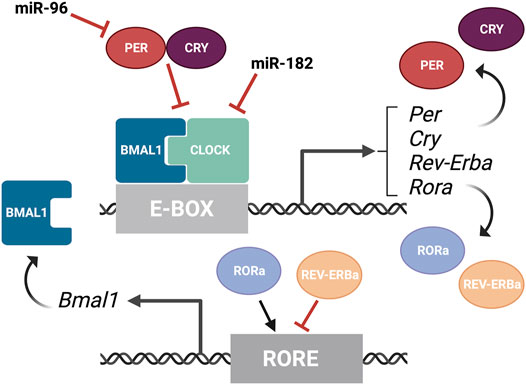
FIGURE 3. Overview of the molecular components of the circadian rhythm. Circadian rhythm is regulated by two interwoven feedback loops. The first loop involves CLOCK and BMAL1 activating regulatory elements containing E-boxes to induce expression of repressors. PER and CRY proteins bind to CLOCK/BMAL1 to repress the transcriptional activity of their activators. MiR-182 targets CLOCK, among other circadian rhythm regulators, and miR-9 targets PER-2. The second loop involves REV-ERBα and RORα competing for binding on RORE binding elements, which promotes Bmal1 transcription. This figure was created with BioRender.com.
Mathematical modeling predicted decades ago that the regulation of mRNA stability is essential for rhythmic protein output (Wuarin et al., 1992). More recently, high-throughput analyses have shown that 25–50% of all rhythmically expressed proteins do not exhibit transcriptional rhythmicity (Mauvoisin et al., 2014). Accordingly, instead of the simplified transcription-translation view, the circadian rhythm undergoes very complex and dynamic regulatory processes that include polyadenylation, RNA splicing, and miRNA regulation.
Numerous miRNAs exhibit circadian rhythmicity, although the mechanisms that regulate these oscillations often remain unclear. In some cases, miRNA coding regions contain E-Box or RORE upstream elements that could be regulated by the core components of the circadian clock (Cheng et al., 2007). Dicer expression has also been reported to display diurnal rhythmicity (Yan et al., 2013), which could lead to a rhythmic pattern of miRNA maturation.
By means of microarray technologies and other tools, early screenings identified the miR-183 cluster (miR-183, miR-96, and miR-182) as miRNAs robustly regulated by the circadian clock (Xu et al., 2007; Yang et al., 2008). For instance, circadian fluctuations in dme-miR-263a and dme-miR-263b expression, the Drosophila orthologues of the miR-183 cluster, were detected in wild type flies and the levels of these miRNAs were significantly reduced in the arrhythmic clock mutant cyc01 (Yang et al., 2008). Likewise, in the adult mouse retina, the expression of these miRNAs obeys a circadian rhythm, with the miRNA levels being significantly higher during zeitgeber time (ZT) 17 (midnight) compared to ZT 5 (noon) (Xu et al., 2007). The expression of these miRNAs is also regulated by light in the mammalian retina and the total levels of miR-183, miR-96, and miR-182 shift quickly (within 30 min) after light or dark adaptation (Krol et al., 2010).
MiR-183, -96, and -182 are part of a highly-conserved polycystronic miRNA cluster that plays multiple roles in sensory tissues including the retina (Lagos-Quintana et al., 2002; Xu et al., 2007), the inner ear (Weston et al., 2006), and the olfactory epithelium (Xu et al., 2007). In the vertebrate retina, this cluster has been shown to elicit neuroprotective functions in photoreceptors, modulate outer segment maintenance, and enhance light responses in stem cell-derived retinal organoids (Loscher et al., 2007; Krol et al., 2010; Lumayag et al., 2013; Busskamp et al., 2014). Many reports indicate that the miR-183 cluster is a key regulator of apoptosis and programmed cell death and validated target genes include CASP2, FOXO1, SLC1A1, and PDCD4 (Zuzic et al., 2019). Recent studies have indicated that the miR-183 cluster is also an important morphogenetic factor regulating multiple signaling pathways involved in photoreceptor differentiation and maintenance. In this direction, the miR-183 cluster targets PAX6 (Peskova et al., 2020), a highly conserved paired-box transcription factor that is critical for eye morphogenesis in a wide range of species (Glaser et al., 1992, 1994; Lauderdale et al., 2000; Davis et al., 2021).
The exact mechanisms that regulate the oscillatory expression of this miRNA cluster are not well understood. The putative promoter region of the miR-183 cluster contains several binding sites for transcription factors known to regulate the circadian rhythm in the eye, including RORα (Xu et al., 2007), but there is currently no experimental data to confirm this transcriptional regulation. Genetic variants with abnormal processing of pre-miR-182 have been described (Saus et al., 2010) and neuronal miRNAs have been shown to have very quick turn-over ratios (Krol et al., 2010) compared to nonneuronal cells (Bhattacharyya et al., 2006; Hwang et al., 2007; Krol et al., 2010). Thus, the regulation of miRNA processing and/or degradation could also play important roles in its oscillatory behavior.
A recent phenotype-driven genome-wide miRNA screen using reporter human cell lines identified several miRNAs with the potential to modulate circadian rhythms (Zhou et al., 2021). Among 989 miRNAs tested, this study identified 120 miRNAs that significantly changed the period length in a dose-dependent manner, including let-7, miR-17, and the miR-183 cluster. Importantly, these changes were tissue-specific and the inactivation of the miR-183 cluster shortened the circadian period in the retina but did not change the period length of the SCN in mice. All three members of the miR-183 cluster can modulate circadian rhythms and luciferase-based assays have shown that miR-182 potentially targets CLOCK (Saus et al., 2010) as well as the circadian rhythm regulators ADCY6 and MITF (Xu et al., 2007), while miR-96 directly targets PER-2 (Zhou et al., 2021) (Figure 3). Similarly, experimental evidence in zebrafish indicates that miR-183 targets other circadian regulators such as E4BP4-6 and AANAT2 (Ben-Moshe et al., 2014). However, these results do not exclude possible additional regulation through non-cell autonomous mechanisms.
Finally, while it is not known whether the oscillatory behavior of miR-183 has any effects on retinal development, the removal of circadian clock genes led to defective dorso-ventral patterning of cones, thinner inner retinal nuclear and plexiform layers, and reduced photoreceptor viability (Ait-Hmyed et al., 2013; Baba et al., 2018). Future studies might shed light on the role of the miR-183 cluster in these phenotypes and the interplay between the circadian rhythmicity and miRNA roles in photoreceptor differentiation and function.
Let-7 Levels Oscillate With the Cell Cycle in the Embryonic Retina
The cell cycle is a precisely regulated oscillatory process essential for growth and maintenance of tissues as well as for coordinating the timing of major cellular events during development. The cell cycle is classically divided into four different phases: Gap1 (G1), DNA Synthesis (S), Gap2 (G2), and Mitosis (M) (Norbury and Nurse, 1992). The ability of the cells to progress though these phases to ultimately produce two daughter cells is generally attributed to two classes of molecules: Cyclin-dependent kinases (CDKs), a large family of serine/threonine kinases, and their binding partners named Cyclins because their concentration varies in a cyclical manner (Malumbres and Barbacid, 2001). The abundance of individual Cyclins, and the consequent activation of the appropriate CDKs at specific phases, orchestrates the orderly completion of DNA replication and cell division and constitutes the core cell cycle oscillator (Figure 4). Thus, CyclinD/CDK4,6 activity ensures G1 progression, CyclinE/CDK2 promotes the G1/S transition, while CyclinA/CDK2 regulates the transition between S and G2. Finally, CyclinB/CDK1 warrants the G2/M transition and entry of cell into mitosis (Malumbres and Barbacid, 2001). However, cell cycle progression is not only regulated by the rise and fall of Cyclin molecules’ concentrations, but is tightly regulated at several levels and through many different mechanisms (Figure 4).
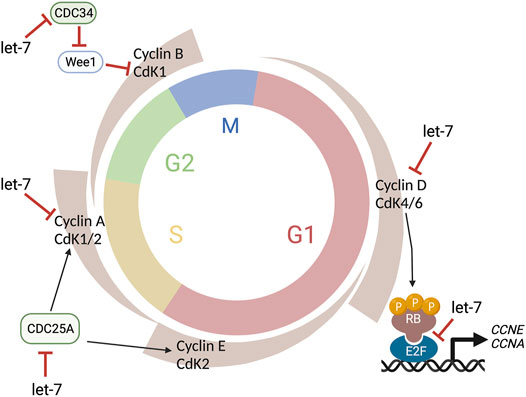
FIGURE 4. Let-7 regulation of the cell cycle. Let-7 regulates the cell cycle kinetics by both promoting cell cycle exit and lengthening distinct phases. Let-7 targets Cyclin D, CDK4, and CDK6, CDC25A, Cyclin A, and CDC34, affecting the G1/S, S/G2, and G2/M transitions, respectively. This figure was created with BioRender.com.
The first studies on miRNAs published three decades ago already suggested a role for the miRNA let-7 in the cell cycle of C. elegans (Ambros, 2001; Lee and Ambros, 2001). Let-7 is part of the heterochronic pathway required in the nematode seam cells to determine the timing of stage-specific developmental events (Ambros and Horvitz, 1984; Moss et al., 1997). Since then, numerous studies have revealed that let-7 is a master regulator of cell proliferation. Accordingly, let-7 alters cell cycle progression, controls the timing of cell cycle exit, and inhibits self-renewal, and disruptions in let-7 coding genes can enhance oncogenic transformation (Johnson et al., 2005; Sampson et al., 2007; Zhao et al., 2010).
Elegant genetic studies from the Ruvkun laboratory (Pasquinelli et al., 2000; Reinhart et al., 2000) revealed that let-7 is a genetic switch that controls major developmental transitions in bilaterally symmetrical animals, from flies and worms to vertebrates. In the developing mammalian retina, let-7 regulates the developmental transition that allows the retinal progenitors to generate the late cell types (amacrine cells, rod photoreceptors, bipolar cells, and Müller glia) (La Torre et al., 2013; Xia and Ahmad, 2016), and also plays a central role in Müller glia-dependent regeneration (Ramachandran et al., 2010; Wohl et al., 2019). Similar roles have been described in other parts of the developing CNS, where let-7 is required for the generation of the later cell populations in different species (Wu et al., 2012; Patterson et al., 2014; Shu et al., 2019).
In the developing retina, let-7 regulates cell cycle kinetics by both promoting cell cycle exit and lengthening S/G2 phases (Fairchild et al., 2019). Notably, no differences were detected in G1 length in time-lapse experiments using the fluorescent reporter FUCCI (Fluorescence Ubiquitination-based Cell Cycle Indicator) in combination with gain-of-function or loss-of-function of let-7 (Fairchild et al., 2019). Given that let-7 levels normally increase throughout developmental time in the retina (Arora et al., 2007; La Torre et al., 2013; Xia and Ahmad, 2016) (Table 1), these data correlate with classic experiments using 3H-thymidine cumulative labeling that indicated that the cell cycle lengthens during retinal development mainly due to an increase in S-phase length (Alexiades and Cepko, 1996).
The cell cycle proteins CDC25A, CDC34, CDK4, CDK6, Cyclin A, Cyclin D1, D2, and D3 are known let-7 targets (Bueno and Malumbres, 2011) (Figure 4) as well as TLX (Zhao et al., 2010), another cell cycle regulator, and oncogenic chromatin proteins such as HMGA1 and HMGA2 (Lee and Dutta, 2007; Xia and Ahmad, 2016). However, since the specific effects of let-7 overexpression or inhibition are different in different experimental paradigms (e.g., induction of cell cycle arrest vs cell cycle lengthening), let-7’s ability to target these genes may be concentration and/or context dependent. Similarly, the human genome contains 10 different mature miRNAs in the let-7 family (let-7a, let-7b, let-7c, let-7d, let-7e, let-7f, let-7g, let-7i, mir-98, miR-202), produced from 13 precursor sequences. As each of these miRNAs have identical seed sequences and highly conserved regions for target recognition and thus, the individual roles and targets for each let-7 are not well characterized.
Strikingly, not only does let-7 regulate developmental transitions and cell cycle, but its expression and activities also oscillate concurrently with the cell cycle across the developing CNS, including the retina (Fairchild et al., 2019) (Figure 2B). Neural progenitors undergo interkinetic nuclear migration between the apical and basal surfaces in concert with the cell cycle (Sauer, 1935; Miyata, 2008; Norden et al., 2009). Thus, mitotic cell bodies are only found in the apical surface, and cell somas move basally in G1. Cells in S-phase are found at the most basal positions, which move again apically in G2. Intriguingly, let-7 levels also fluctuate within these regions, suggesting that let-7 oscillates in coordination with cell cycle (Fairchild et al., 2019). Mathematical modeling also supports that oscillatory levels of let-7 are required for the complex balance between let-7 and Cyclin/CDK complexes (Gerard et al., 2019) and more recently, these fluctuations have been validated by flow cytometry analyses and time-lapse imaging (Fairchild et al., 2019).
The cell cycle-dependent fluctuation of let-7 suggests that some cell cycle genes may be regulating its expression; however, given that the let-7 family is located in 13 different loci in the genome, the transcriptional regulation of these miRNAs is still poorly understood. E2F transcription factors have been shown to directly regulate let7a-d and let-7i expression and c-MYC represses the expression of several let-7 clusters (Bueno and Malumbres, 2011). Additionally, CyclinD1 can regulate the expression of Dicer (Yu et al., 2013) and thus, cell cycle-dependent miRNA processing may have an impact on let-7 fluctuations. Consistent with this idea of negative feedback loops, the miRNA machinery can be directly regulated by miRNAs, for example, a loop involving let-7 and Ago2 is critical to maintain pluripotency (Liu et al., 2021). Importantly, miRNA stability and turn-over rates could also be regulated in a cell cycle-dependent manner.
The specific role(s) of the periodicity of let-7 expression and activity have not been previously explored but it can be speculated that cell cycle-coupled miRNA oscillatory circuits may be an important strategy to coordinate division rates with complex cellular activities as well as the timing of cell cycle exit and fate decisions.
MiR-9 Is Part of the Notch Ultradian Oscillator
Proper retina development relies on the tight balance between retinal progenitor cell (RPC) proliferation and differentiation. It is well-documented that Notch activation perpetuates RPC maintenance, whereas Notch pathway disruption leads to neuronal differentiation (Dorsky et al., 1995; Tomita et al., 1996; Jadhav et al., 2006; Nelson et al., 2007; Kaufman et al., 2019). Notch also regulates neural patterning (Baek et al., 2006; Bosze et al., 2020), cell fate specification (Yaron et al., 2006; Riesenberg et al., 2009; Chen and Emerson, 2021), is essential for Müller glia development (Furukawa et al., 2000; Bernardos et al., 2005; Nelson et al., 2011), and a key mediator of regeneration (Conner et al., 2014; Sahu et al., 2021). Together, a growing body of literature supports the notion that the Notch pathway is dynamic and remarkably pleiotropic, and that the timing and levels of Notch signaling must be precisely regulated to maintain the temporal control driving normal retinal development.
Since the Notch receptor was first identified in Drosophila over 100 years ago (Dexter, 1914), genetic and molecular interaction studies have helped map the Notch signaling pathway that is recognized today (Figure 5) (reviewed in Louvi and Artavanis-Tsakonas, 2006; Pierfelice et al., 2011; Bray, 2016). Briefly, the intracellular signaling pathway is initiated by cell-cell contacts, where the transmembrane Notch receptor (Notch1–4) on one cell is activated by a ligand (Delta-like (Dll1, Dll3, and Dll4), or Jagged (Jag1 and Jag2)) on a neighboring cell. Ligand binding prompts a series of proteolytic cleavage events that culminates in the release of the Notch receptor’s intracellular domain (NICD). NICD translocates into the nucleus, where it forms a transcriptional complex with Rbpj (recombination signal-binding protein for immunoglobulin kappa J region) and Maml1 (Mastermind-like transcriptional co-activator 1) to activate gene expression. The best characterized Notch targets are the Hes (Hes1, Hes3, and Hes5) and related Hey genes (Ohtsuka et al., 1999), which encode inhibitory basic helix-loop-helix (bHLH) proteins that suppress pro-neural bHLH genes Ngn1, Ngn2 (Neurogenins 1–2), NeuroD1, NeuroD2, NeuroD4, NeuroD6 (Neuronal Differentiation 1-2,4,6), and Ascl1 (Acheate-Scute) (Taylor et al., 2015; Dennis et al., 2019). Importantly, Hes proteins also repress the expression of Notch ligands, affecting the Notch activity of their neighbors (Jarriault et al., 1995).
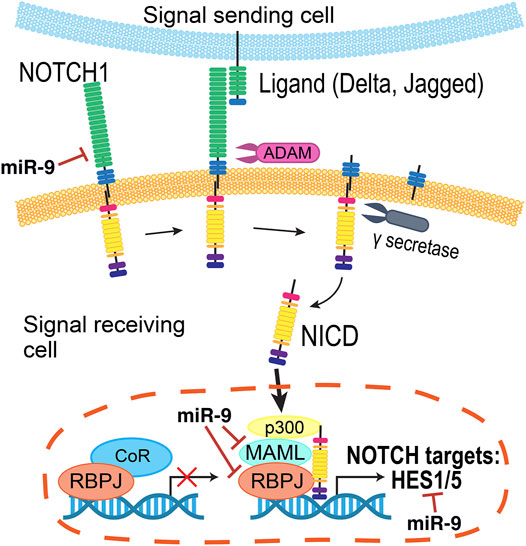
FIGURE 5. Summary of the Notch pathway. Notch signaling is initiated when a transmembrane Notch receptor (Notch1-4) on one cell is activated by a neighboring cell’s ligand (Delta, Jagged/Serrate), prompting proteolytic cleavage events by ADAM and γ-secretase to release the Notch receptor’s intracellular domain (NICD). Inside the cell, NICD translocates into the nucleus to form a transcriptional complex with a number of co-activators to in turn activate the expression of genes, including the Hes and Hey families. The miRNA miR-9 regulates several members of this pathway.
These cross-regulatory activities raise a hypothetical problem: in the absence of Hes1, cells prematurely differentiate into neurons, but then how is the progenitor pool maintained if Hes1 reduces Notch signaling in neighboring cells? Several pioneering studies from the Kageyama laboratory solved the conundrum and showed that the expression of Hes1, Ascl1, and Dll1 display oscillatory behaviors (Figure 2C top) using luciferase-based reporters in several contexts (Masamizu et al., 2006), including neural progenitors (Shimojo et al., 2008; Imayoshi et al., 2013). These oscillatory expression patterns are driven by the Hes1 oscillator (Hirata et al., 2002). Hes1 protein represses its own expression by binding to N-box regulatory elements in the Hes1 promoter, and both Hes1 protein and mRNA have very short half-lives. Thus, upon repression, the levels of Hes1 decline rapidly, leading to the reactivation of Hes1 transcription with a rhythmicity of 2–3 h (Hirata et al., 2002). These oscillations are key in maintaining pools of progenitor cells from precociously differentiating; when Hes1 oscillations are quenched, even if the Notch signaling pathway can still be activated, neural progenitors undergo premature cell cycle exit (Shimojo et al., 2016). Importantly, these rhythmic patterns can in part explain the heterogeneity of gene expression observed in individual RPCs with “snapshot” techniques such as immunostaining and sequencing (Cepko, 2014; Dixit et al., 2014; Clark et al., 2019; Sridhar et al., 2020).
Many signaling pathways are common beneficiaries of miRNA-mediated regulation, and the Notch pathway is no exception (Inui et al., 2010; Roese-Koerner et al., 2016). In fact, functional relationships between Notch and miRNA pathways have been described in the developing retina as Dicer conditional knockout mice showed downregulation of Notch pathway components and at the same time, overexpression of NICD in Dicer-null retinas did not lead to classic “Notchy” phenotypes such as induction of glial fates (Georgi and Reh, 2011).
MiR-9, a miRNA highly expressed in the developing CNS, interacts with Notch components in several organisms (Tan et al., 2012). Target prediction analyses have shown that miR-9 may directly target components of the Notch pathway, including Notch ligands, Rbpj, and Maml1 (Roese-Koerner et al., 2017). Additionally, bioinformatics analyses have identified miR-9-binding sites in mouse, rat, and human Hes1 (Baek et al., 2006), zebrafish her5 and her9 (Leucht et al., 2008), and Xenopus hairy1 (Bonev et al., 2011). Manipulation of miR-9 activity by antisense inhibitors resulted in increased levels of Hes1, and overexpression of miR-9 conversely reduced the half-life of Hes1 (Bonev et al., 2012).
In humans, miR-9 is transcribed from three independent genomic loci (pri-miR-9-1, pri-miR-9-2, and pri-miR-9-3) that give rise to two functional miRNAs, miR-9-5p and miR-9-3p. Hes1 reduces miR-9 expression, as observed by in situ hybridization of pri-miR-9-2 in the mouse cortex and binds to several N-boxes in the putative miR-9 promoters of all three miR-9 genes (Figure 6). However, Hes1 only regulates the expression of pri-miR-9-1 and pri-miR-9-2, but not pri-miR-9-3 (Bonev et al., 2012). These promoters are embedded within CpG islands and thus, they could also be regulated by epigenetic mechanisms or other indirect means.
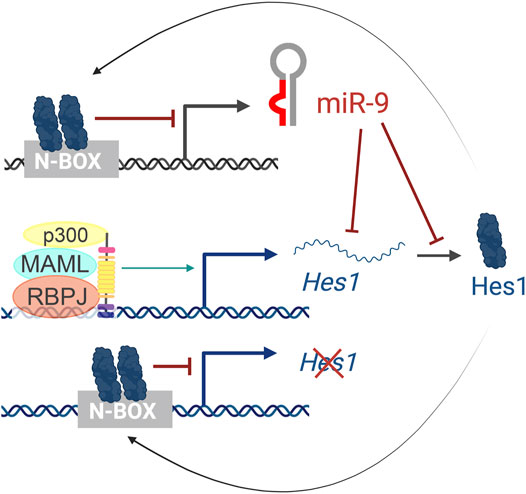
FIGURE 6. The Hes1/miR-9 oscillator. Activation of the Notch pathway leads to the activation of Hes1 transcription. Hes1 protein then dimerizes and binds to N-box domains to repress its own expression as well as miR-9 transcription. In turn, miR-9 reduces Hes1 levels by controlling the stability of Hes1 mRNA and inhibiting its translation, resulting in oscillatory behaviors.
Importantly, the cross-regulations between miR-9 and Hes1 (Figure 6) also contribute to the Hes1 oscillator, and overexpressing or inhibiting miR-9 has been shown to reduce Hes1 oscillations (Bonev et al., 2012). The negative feedback loops between miR-9 and Hes1 creates an out-of-phase oscillatory pattern of expression (Figure 2C top), which is important for limiting Hes1 oscillations (Roese-Koerner et al., 2016). As development continues, miR-9 accumulates (due to its longer half-life than the less-stable Hes1 mRNA and protein) until it reaches a threshold for differentiation (Shimojo et al., 2016). At that point, Hes1 oscillations are dampened, and miR-9 maintains high, steady levels allowing for neural differentiation to proceed (Figure 2C bottom) (Bonev et al., 2012). Although oscillations of miR-9 have not been observed directly, mathematical modelling analyses incorporating miR-9 into the Hes1 oscillator recapitulate the behaviors observed experimentally (Goodfellow et al., 2014).
During retinal development, miR-9 expression increases in RPCs over developmental time and regulates cell fate acquisition (La Torre et al., 2013). Mir-9 is also important in the mature mouse retina to maintain homeostasis of the Müller glia (Wohl et al., 2017) and can potentiate Müller glia conversion into progenitor-like cells in culture in combination with miR-124 (Wohl and Reh, 2016b). While the molecular mechanisms downstream of these functional roles remain widely unexplored, it can be speculated that the oscillatory interplay between Notch and miR-9 in combination with the increasing levels of mature miR-9 over time may be one of the mechanisms that enables cells with an ability to keep track of time while maintaining the ability to adapt to external stimuli. This model accommodates the existing evidence indicating that fate decisions during retinal development are both cell autonomous and strongly influenced by external factors.
Recently, a novel CIS-regulatory element of pri-miR-9-2 has been described for its association with retinal diseases such as Macular Telangiectasia Type 2 and Macular Degeneration (Thomas et al., 2021). Interestingly, the deletion of this enhancer leads to reduced miR-9 levels, a decrease in the number of rod photoreceptors, and perturbation of Müller glia homeostasis in human retinal organoids. Transcriptional data indicates that the Notch pathway is affected in these cells, but the exact nature of this regulation and whether the oscillatory behaviors and feedback loops between Notch and miR-9 play any roles have not yet been investigated.
Conclusions and Perspectives
During normal development, different cell fates are specified with exquisite spatial and temporal accuracy. Oscillatory feedback circuits that integrate temporal cues are part of the machinery that establishes the robustness of developmental transitions and progenitor outcomes. It is now obvious that miRNAs are functionally integrated in many of these oscillatory pathways. Beyond the examples offered in this review, a reciprocal relationship between miRNAs and transcription factors that in turn regulate miRNA expression may be a common theme in a variety of developmental contexts.
Despite all the evidence accumulated in the last few years, we are only starting to understand the relevance of these rhythmic behaviors, largely because most of the miRNA expression data to date comes from studies that used sequencing technologies that do not capture dynamic changes of expression within a cell. Thus, efforts to develop tools to show miRNA levels longitudinally with cellular resolution need to be advanced.
In addition, our understanding of miRNA transcriptional regulation is still quite limited. The complex regulation of miRNA processing and turn-over may similarly open new avenues to further understand the regulatory networks that govern neural development.
Author Contributions
EF and AL wrote the manuscript, JH and AL made the figures, and EF, JH, and AL edited the manuscript.
Funding
This work was supported by a grant from NIH (R0EY026942; AL) and support from a training grant (T32 5T32EY015387-20 to EF).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank all the members of the La Torre and Simo laboratories for their support.
References
Ait-Hmyed, O., Felder-Schmittbuhl, M.-P., Garcia-Garrido, M., Beck, S., Seide, C., Sothilingam, V., et al. (2013). Mice Lacking Period 1 and Period 2 Circadian Clock Genes Exhibit Blue Cone Photoreceptor Defects. Eur. J. Neurosci. 37 (7), 1048–1060. doi:10.1111/ejn.12103
Alexiades, M. R., and Cepko, C. (1996). Quantitative Analysis of Proliferation and Cell Cycle Length During Development of the Rat Retina. Dev. Dyn. 205 (3), 293–307. doi:10.1002/(sici)1097-0177(199603)205::3<293aid-aja9>3.0.co;2-d
Ambros, V., and Horvitz, H. R. (1984). Heterochronic Mutants of the Nematode Caenorhabditis elegans. Science 226 (4673), 409–416. doi:10.1126/science.6494891
Arora, A., McKay, G. J., and Simpson, D. A. C. (2007). Prediction and Verification of miRNA Expression in Human and Rat Retinas. Invest. Ophthalmol. Vis. Sci. 48 (9), 3962–3967. doi:10.1167/iovs.06-1221
Baba, K., Piano, I., Lyuboslavsky, P., Chrenek, M. A., Sellers, J. T., Zhang, S., et al. (2018). Removal of Clock Gene Bmal1 from the Retina Affects Retinal Development and Accelerates Cone Photoreceptor Degeneration During Aging. Proc. Natl. Acad. Sci. USA 115 (51), 13099–13104. doi:10.1073/pnas.1808137115
Baek, J. H., Hatakeyama, J., Sakamoto, S., Ohtsuka, T., and Kageyama, R. (2006). Persistent and High Levels of Hes1 Expression Regulate Boundary Formation in the Developing Central Nervous System. Development 133 (13), 2467–2476. doi:10.1242/dev.02403
Bak, M., Silahtaroglu, A., Møller, M., Christensen, M., Rath, M. F., Skryabin, B., et al. (2008). MicroRNA Expression in the Adult Mouse Central Nervous System. RNA 14 (3), 432–444. doi:10.1261/rna.783108
Barbato, S., Marrocco, E., Intartaglia, D., Pizzo, M., Asteriti, S., Naso, F., et al. (2017). MiR-211 Is Essential for Adult Cone Photoreceptor Maintenance and Visual Function. Sci. Rep. 7 (1), 17004. doi:10.1038/s41598-017-17331-z
Bartel, D. P. (2009). MicroRNAs: Target Recognition and Regulatory Functions. Cell 136 (2), 215–233. doi:10.1016/j.cell.2009.01.002
Ben-Moshe, Z., Alon, S., Mracek, P., Faigenbloom, L., Tovin, A., Vatine, G. D., et al. (2014). The Light-Induced Transcriptome of the Zebrafish Pineal Gland Reveals Complex Regulation of the Circadian Clockwork by Light. Nucleic Acids Res. 42 (6), 3750–3767. doi:10.1093/nar/gkt1359
Berber, P., Grassmann, F., Kiel, C., and Weber, B. H. F. (2017). An Eye on Age-Related Macular Degeneration: The Role of MicroRNAs in Disease Pathology. Mol. Diagn. Ther. 21 (1), 31–43. doi:10.1007/s40291-016-0234-z
Bernardos, R. L., Lentz, S. I., Wolfe, M. S., and Raymond, P. A. (2005). Notch-Delta Signaling Is Required for Spatial Patterning and Müller Glia Differentiation in the Zebrafish Retina. Developmental Biol. 278 (2), 381–395. doi:10.1016/j.ydbio.2004.11.018
Besharse, J. C., and Iuvone, P. M. (1983). Circadian Clock in Xenopus Eye Controlling Retinal Serotonin N-Acetyltransferase. Nature 305 (5930), 133–135. doi:10.1038/305133a0
Bhattacharyya, S. N., Habermacher, R., Martine, U., Closs, E. I., and Filipowicz, W. (2006). Stress-induced Reversal of microRNA Repression and mRNA P-Body Localization in Human Cells. Cold Spring Harbor Symposia Quantitative Biol. 71, 513–521. doi:10.1101/sqb.2006.71.038
Birney, E., Birney, E., Stamatoyannopoulos, J. A., Dutta, A., Guigó, R., Gingeras, T. R., et al. (2007). Identification and Analysis of Functional Elements in 1% of the Human Genome by the ENCODE Pilot Project. Nature 447 (7146), 799–816. doi:10.1038/nature05874
Bonev, B., Pisco, A., and Papalopulu, N. (2011). MicroRNA-9 Reveals Regional Diversity of Neural Progenitors Along the Anterior-Posterior Axis. Developmental Cel. 20 (1), 19–32. doi:10.1016/j.devcel.2010.11.018
Bonev, B., Stanley, P., and Papalopulu, N. (2012). MicroRNA-9 Modulates Hes1 Ultradian Oscillations by Forming a Double-Negative Feedback Loop. Cel. Rep. 2 (1), 10–18. doi:10.1016/j.celrep.2012.05.017
Borenstein, E., and Ruppin, E. (2006). Direct Evolution of Genetic Robustness in microRNA. Proc. Natl. Acad. Sci. 103 (17), 6593–6598. doi:10.1073/pnas.0510600103
Bosze, B., Moon, M.-S., Kageyama, R., and Brown, N. L. (2020). Simultaneous Requirements for Hes1 in Retinal Neurogenesis and Optic Cup-Stalk Boundary Maintenance. J. Neurosci. 40 (7), 1501–1513. doi:10.1523/JNEUROSCI.2327-19.2020
Bray, S. J. (2016). Notch Signalling in Context. Nat. Rev. Mol. Cel. Biol. 17 (11), 722–735. doi:10.1038/nrm.2016.94
Bueno, M. J., and Malumbres, M. (2011). MicroRNAs and the Cell Cycle. Biochim. Biophys. Acta (Bba) - Mol. Basis Dis. 1812 (5), 592–601. doi:10.1016/j.bbadis.2011.02.002
Bushati, N., and Cohen, S. M. (2007). microRNA Functions. Annu. Rev. Cel. Dev. Biol. 23, 175–205. doi:10.1146/annurev.cellbio.23.090506.123406
Busskamp, V., Krol, J., Nelidova, D., Daum, J., Szikra, T., Tsuda, B., et al. (2014). miRNAs 182 and 183 Are Necessary to Maintain Adult Cone Photoreceptor Outer Segments and Visual Function. Neuron 83 (3), 586–600. doi:10.1016/j.neuron.2014.06.020
Carninci, P., Kasukawa, T., Katayama, S., Gough, J., Frith, M. C., Maeda, N., et al. (2005). The Transcriptional Landscape of the Mammalian Genome. Science 309 (5740), 1559–1563. doi:10.1126/science.1112014
Cepko, C. (2014). Intrinsically Different Retinal Progenitor Cells Produce Specific Types of Progeny. Nat. Rev. Neurosci. 15 (9), 615–627. doi:10.1038/nrn3767
Chekulaeva, M., and Filipowicz, W. (2009). Mechanisms of miRNA-Mediated post-transcriptional Regulation in Animal Cells. Curr. Opin. Cel. Biol. 21 (3), 452–460. doi:10.1016/j.ceb.2009.04.009
Chen, X., and Emerson, M. M. (2021). Notch Signaling Represses Cone Photoreceptor Formation Through the Regulation of Retinal Progenitor Cell States. Sci. Rep. 11 (1), 14525. doi:10.1038/s41598-021-93692-w
Cheng, H.-Y. M., Papp, J. W., Varlamova, O., Dziema, H., Russell, B., Curfman, J. P., et al. (2007). microRNA Modulation of Circadian-Clock Period and Entrainment. Neuron 54 (5), 813–829. doi:10.1016/j.neuron.2007.05.017
Chu-Tan, J. A., Rutar, M., Saxena, K., Aggio-Bruce, R., Essex, R. W., Valter, K., et al. (2018). MicroRNA-124 Dysregulation Is Associated with Retinal Inflammation and Photoreceptor Death in the Degenerating Retina. Invest. Ophthalmol. Vis. Sci. 59 (10), 4094–4105. doi:10.1167/iovs.18-24623
Clark, B. S., Stein-O’Brien, G. L., Shiau, F., Cannon, G. H., Davis-Marcisak, E., Sherman, T., et al. (2019). Single-Cell RNA-Seq Analysis of Retinal Development Identifies NFI Factors as Regulating Mitotic Exit and Late-Born Cell Specification. Neuron 102 (6), 1111–1126. doi:10.1016/j.neuron.2019.04.010
Conner, C., Ackerman, K. M., Lahne, M., Hobgood, J. S., and Hyde, D. R. (2014). Repressing Notch Signaling and Expressing TNF Are Sufficient to Mimic Retinal Regeneration by Inducing Muller Glial Proliferation to Generate Committed Progenitor Cells. J. Neurosci. 34 (43), 14403–14419. doi:10.1523/JNEUROSCI.0498-14.2014
Conte, I., Carrella, S., Avellino, R., Karali, M., Marco-Ferreres, R., Bovolenta, P., et al. (2010). miR-204 Is Required for Lens and Retinal Development via Meis2 Targeting. Proc. Natl. Acad. Sci. 107 (35), 15491–15496. doi:10.1073/pnas.0914785107
Damiani, D., Alexander, J. J., O'Rourke, J. R., McManus, M., Jadhav, A. P., Cepko, C. L., et al. (2008). Dicer Inactivation Leads to Progressive Functional and Structural Degeneration of the Mouse Retina. J. Neurosci. 28 (19), 4878–4887. doi:10.1523/JNEUROSCI.0828-08.2008
Dantas da Costa e Silva, M. E., Polina, E. R., Crispim, D., Sbruzzi, R. C., Lavinsky, D., Mallmann, F., et al. (2019). Plasma Levels of miR-29b and miR-200b in Type 2 Diabetic Retinopathy. J. Cel. Mol. Med. 23 (2), 1280–1287. doi:10.1111/jcmm.14030
Davis, E. S., Voss, G., Miesfeld, J. B., Zarate‐Sanchez, J., Voss, S. R., and Glaser, T. (2021). The Rax Homeobox Gene Is Mutated in the Eyeless Axolotl, Ambystoma mexicanum. Developmental Dyn. 250 (6), 807–821. doi:10.1002/dvdy.246
Davis, N., Mor, E., and Ashery-Padan, R. (2011). Roles for Dicer1 in the Patterning and Differentiation of the Optic Cup Neuroepithelium. Development 138 (1), 127–138. doi:10.1242/dev.053637
Decembrini, S., Andreazzoli, M., Barsacchi, G., and Cremisi, F. (2008). Dicer Inactivation Causes Heterochronic Retinogenesis in Xenopus laevis. Int. J. Dev. Biol. 52 (8), 1099–1103. doi:10.1387/ijdb.082646sd
Decembrini, S., Bressan, D., Vignali, R., Pitto, L., Mariotti, S., Rainaldi, G., et al. (2009). MicroRNAs Couple Cell Fate and Developmental Timing in Retina. Pnas 106 (50), 21179–21184. doi:10.1073/pnas.0909167106
Dennis, D. J., Han, S., and Schuurmans, C. (2019). bHLH Transcription Factors in Neural Development, Disease, and Reprogramming. Brain Res. 1705, 48–65. doi:10.1016/j.brainres.2018.03.013
Deo, M., Yu, J.-Y., Chung, K.-H., Tippens, M., and Turner, D. L. (2006). Detection of Mammalian microRNA Expression by In Situ Hybridization with RNA Oligonucleotides. Dev. Dyn. 235 (9), 2538–2548. doi:10.1002/dvdy.20847
Dexheimer, P. J., and Cochella, L. (2020). MicroRNAs: From Mechanism to Organism. Front. Cel. Dev. Biol. 8, 409. doi:10.3389/fcell.2020.00409
Dexter, J. S. (1914). The Analysis of a Case of Continuous Variation in Drosophila by a Study of its Linkage Relations. The Am. Naturalist 48 (576), 712–758. doi:10.1086/279446
Dixit, R., Tachibana, N., Touahri, Y., Zinyk, D., Logan, C., and Schuurmans, C. (2014). Gene Expression Is Dynamically Regulated in Retinal Progenitor Cells Prior to and During Overt Cellular Differentiation. Gene Expr. Patterns 14 (1), 42–54. doi:10.1016/j.gep.2013.10.003
Dorsky, R. I., Rapaport, D. H., and Harris, W. A. (1995). Xotch Inhibits Cell Differentiation in the Xenopus Retina. Neuron 14 (3), 487–496. doi:10.1016/0896-6273(95)90305-4
Doyle, S. E., Grace, M. S., McIvor, W., and Menaker, M. (2002). Circadian Rhythms of Dopamine in Mouse Retina: The Role of Melatonin. Vis. Neurosci. 19 (5), 593–601. doi:10.1017/s0952523802195058
Ertekin, S., Yıldırım, O., Dinç, E., Ayaz, L., Fidancı, S. B., and Tamer, L. (2014). Evaluation of Circulating miRNAs in Wet Age-Related Macular Degeneration. Mol. Vis. 20, 1057–1066. http://www.molvis.org/molvis/v20/1057/.
Eulalio, A., Huntzinger, E., and Izaurralde, E. (2008). Getting to the Root of miRNA-Mediated Gene Silencing. Cell 132 (1), 9–14. doi:10.1016/j.cell.2007.12.024
Fairchild, C. L. A., Cheema, S. K., Wong, J., Hino, K., Simó, S., and La Torre, A. (2021). Author Correction: Let-7 Regulates Cell Cycle Dynamics in the Developing Cerebral Cortex and Retina. Sci. Rep. 11 (1), 2872. doi:10.1038/s41598-021-82224-1
Fairchild, C. L. A., Cheema, S. K., Wong, J., Hino, K., Simó, S., and La Torre, A. (2019). Let-7 Regulates Cell Cycle Dynamics in the Developing Cerebral Cortex and Retina. Sci. Rep. 9 (1), 15336. doi:10.1038/s41598-019-51703-x
Fishman, E. S., Louie, M., Miltner, A. M., Cheema, S. K., Wong, J., Schlaeger, N. M., et al. (2021). MicroRNA Signatures of the Developing Primate Fovea. Front. Cel. Dev. Biol. 9, 654385. doi:10.3389/fcell.2021.654385
Fogerty, J., Stepanyan, R., Cianciolo, L. T., Tooke, B. P., and Perkins, B. D. (2019). Genomic Non-redundancy of the Mir-183/96/182 Cluster and its Requirement for Hair Cell Maintenance. Sci. Rep. 9 (1), 10302. doi:10.1038/s41598-019-46593-y
Friedman, R. C., Farh, K. K.-H., Burge, C. B., and Bartel, D. P. (2009). Most Mammalian mRNAs Are Conserved Targets of microRNAs. Genome Res. 19 (1), 92–105. doi:10.1101/gr.082701.108
Furukawa, T., Mukherjee, S., Bao, Z.-Z., Morrow, E. M., and Cepko, C. L. (2000). Rax, Hes1, and Notch1 Promote the Formation of Müller Glia by Postnatal Retinal Progenitor Cells. Neuron 26 (2), 383–394. doi:10.1016/s0896-6273(00)81171-x
Gao, F., Zhang, Y.-F., Zhang, Z.-P., Fu, L.-A., Cao, X.-L., Zhang, Y.-Z., et al. (2017). miR-342-5p Regulates Neural Stem Cell Proliferation and Differentiation Downstream to Notch Signaling in Mice. Stem Cel. Rep. 8 (4), 1032–1045. doi:10.1016/j.stemcr.2017.02.017
Gao, L., Jiang, B., Lei, D., Zhou, X., and Yuan, H. (2016a). Expression Profiling of microRNAs in Optineurin (E50K) Mutant Transgenic Mice. Biomed. Rep. 4 (2), 193–196. doi:10.3892/br.2015.565
Gao, Q., Zhou, L., Yang, S.-Y., and Cao, J.-M. (2016b). A Novel Role of microRNA 17-5p in the Modulation of Circadian Rhythm. Sci. Rep. 6, 30070. doi:10.1038/srep30070
Georgi, S. A., and Reh, T. A. (2011). Dicer Is Required for the Maintenance of Notch Signaling and Gliogenic Competence During Mouse Retinal Development. Devel. Neurobio. 71 (12), 1153–1169. doi:10.1002/dneu.20899
Georgi, S. A., and Reh, T. A. (2010). Dicer Is Required for the Transition from Early to Late Progenitor State in the Developing Mouse Retina. J. Neurosci. 30 (11), 4048–4061. doi:10.1523/JNEUROSCI.4982-09.2010
Gérard, C., Lemaigre, F., and Gonze, D. (2019). Modeling the Dynamics of Let-7-Coupled Gene Regulatory Networks Linking Cell Proliferation to Malignant Transformation. Front. Physiol. 10, 848. doi:10.3389/fphys.2019.00848
Glaser, T., Jepeal, L., Edwards, J. G., Young, S. R., Favor, J., and Maas, R. L. (1994). PAX6 Gene Dosage Effect in a Family with Congenital Cataracts, Aniridia, Anophthalmia and central Nervous System Defects. Nat. Genet. 7 (4), 463–471. doi:10.1038/ng0894-463
Glaser, T., Walton, D. S., and Maas, R. L. (1992). Genomic Structure, Evolutionary Conservation and Aniridia Mutations in the Human PAX6 Gene. Nat. Genet. 2 (3), 232–239. doi:10.1038/ng1192-232
Goodfellow, M., Phillips, N. E., Manning, C., Galla, T., and Papalopulu, N. (2014). microRNA Input into a Neural Ultradian Oscillator Controls Emergence and Timing of Alternative Cell States. Nat. Commun. 5, 3399. doi:10.1038/ncomms4399
Ha, M., and Kim, V. N. (2014). Regulation of microRNA Biogenesis. Nat. Rev. Mol. Cel. Biol. 15 (8), 509–524. doi:10.1038/nrm3838
Hackler, L., Wan, J., Swaroop, A., Qian, J., and Zack, D. J. (2010). MicroRNA Profile of the Developing Mouse Retina. Invest. Ophthalmol. Vis. Sci. 51 (4), 1823–1831. doi:10.1167/iovs.09-4657
Hangauer, M. J., Vaughn, I. W., and McManus, M. T. (2013). Pervasive Transcription of the Human Genome Produces Thousands of Previously Unidentified Long Intergenic Noncoding RNAs. Plos Genet. 9 (6), e1003569. doi:10.1371/journal.pgen.1003569
Hirata, H., Yoshiura, S., Ohtsuka, T., Bessho, Y., Harada, T., Yoshikawa, K., et al. (2002). Oscillatory Expression of the bHLH Factor Hes1 Regulated by a Negative Feedback Loop. Science 298 (5594), 840–843. doi:10.1126/science.1074560
Hwang, H.-W., Wentzel, E. A., and Mendell, J. T. (2007). A Hexanucleotide Element Directs microRNA Nuclear Import. Science 315 (5808), 97–100. doi:10.1126/science.1136235
Iida, A., Shinoe, T., Baba, Y., Mano, H., and Watanabe, S. (2011). Dicer Plays Essential Roles for Retinal Development by Regulation of Survival and Differentiation. Invest. Ophthalmol. Vis. Sci. 52 (6), 3008–3017. doi:10.1167/iovs.10-6428
Imayoshi, I., Isomura, A., Harima, Y., Kawaguchi, K., Kori, H., Miyachi, H., et al. (2013). Oscillatory Control of Factors Determining Multipotency and Fate in Mouse Neural Progenitors. Science 342 (6163), 1203–1208. doi:10.1126/science.1242366
Indrieri, A., Carrella, S., Romano, A., Spaziano, A., Marrocco, E., Fernandez‐Vizarra, E., et al. (2019). miR‐181a/b Downregulation Exerts a Protective Action on Mitochondrial Disease Models. EMBO Mol. Med. 11 (5), e8734. doi:10.15252/emmm.201708734
Intartaglia, D., Giamundo, G., and Conte, I. (2020). The Impact of miRNAs in Health and Disease of Retinal Pigment Epithelium. Front. Cel Dev. Biol. 8, 589985. doi:10.3389/fcell.2020.589985
Inui, M., Martello, G., and Piccolo, S. (2010). MicroRNA Control of Signal Transduction. Nat. Rev. Mol. Cel Biol 11 (4), 252–263. doi:10.1038/nrm2868
Jadhav, A. P., Cho, S.-H., and Cepko, C. L. (2006). Notch Activity Permits Retinal Cells to Progress through Multiple Progenitor States and Acquire a Stem Cell Property. Proc. Natl. Acad. Sci. 103 (50), 18998–19003. doi:10.1073/pnas.0608155103
Jarriault, S., Brou, C., Logeat, F., Schroeter, E. H., Kopan, R., and Israel, A. (1995). Signalling Downstream of Activated Mammalian Notch. Nature 377 (6547), 355–358. doi:10.1038/377355a0
Johnson, S. M., Grosshans, H., Shingara, J., Byrom, M., Jarvis, R., Cheng, A., et al. (2005). RAS Is Regulated by the Let-7 microRNA Family. Cell 120 (5), 635–647. doi:10.1016/j.cell.2005.01.014
Kapranov, P., Cawley, S. E., Drenkow, J., Bekiranov, S., Strausberg, R. L., Fodor, S. P. A., et al. (2002). Large-scale Transcriptional Activity in Chromosomes 21 and 22. Science 296 (5569), 916–919. doi:10.1126/science.1068597
Kapsimali, M., Kloosterman, W. P., de Bruijn, E., Rosa, F., Plasterk, R. H., and Wilson, S. W. (2007). MicroRNAs Show a Wide Diversity of Expression Profiles in the Developing and Mature central Nervous System. Genome Biol. 8 (8), R173. doi:10.1186/gb-2007-8-8-r173
Karali, M., Manfredi, A., Puppo, A., Marrocco, E., Gargiulo, A., Allocca, M., et al. (2011). MicroRNA-restricted Transgene Expression in the Retina. PLoS One 6 (7), e22166. doi:10.1371/journal.pone.0022166
Karali, M., Peluso, I., Gennarino, V. A., Bilio, M., Verde, R., Lago, G., et al. (2010). miRNeye: A microRNA Expression Atlas of the Mouse Eye. BMC Genomics 11, 715. doi:10.1186/1471-2164-11-715
Karali, M., Persico, M., Mutarelli, M., Carissimo, A., Pizzo, M., Singh Marwah, V., et al. (2016). High-resolution Analysis of the Human Retina miRNome Reveals isomiR Variations and Novel microRNAs. Nucleic Acids Res. 44 (4), 1525–1540. doi:10.1093/nar/gkw039
Kaufman, M. L., Park, K. U., Goodson, N. B., Chew, S., Bersie, S., Jones, K. L., et al. (2019). Transcriptional Profiling of Murine Retinas Undergoing Semi-synchronous Cone Photoreceptor Differentiation. Developmental Biol. 453 (2), 155–167. doi:10.1016/j.ydbio.2019.05.016
Ko, G. Y. P. (2020). Circadian Regulation in the Retina: From Molecules to Network. Eur. J. Neurosci. 51 (1), 194–216. doi:10.1111/ejn.14185
Kozomara, A., and Griffiths-Jones, S. (2011). miRBase: Integrating microRNA Annotation and Deep-Sequencing Data. Nucleic Acids Res. 39, D152–D157. doi:10.1093/nar/gkq1027
Krol, J., Busskamp, V., Markiewicz, I., Stadler, M. B., Ribi, S., Richter, J., et al. (2010). Characterizing Light-Regulated Retinal microRNAs Reveals Rapid Turnover as a Common Property of Neuronal microRNAs. Cell 141 (4), 618–631. doi:10.1016/j.cell.2010.03.039
La Torre, A., Georgi, S., and Reh, T. A. (2013). Conserved microRNA Pathway Regulates Developmental Timing of Retinal Neurogenesis. Proc. Natl. Acad. Sci. 110 (26), E2362–E2370. doi:10.1073/pnas.1301837110
Lagos-Quintana, M., Rauhut, R., Meyer, J., Borkhardt, A., and Tuschl, T. (2003). New microRNAs from Mouse and Human. RNA 9 (2), 175–179. doi:10.1261/rna.2146903
Lagos-Quintana, M., Rauhut, R., Yalcin, A., Meyer, J., Lendeckel, W., and Tuschl, T. (2002). Identification of Tissue-specific microRNAs from Mouse. Curr. Biol. 12 (9), 735–739. doi:10.1016/s0960-9822(02)00809-6
Lauderdale, J. D., Wilensky, J. S., Oliver, E. R., Walton, D. S., and Glaser, T. (2000). 3' Deletions Cause Aniridia by Preventing PAX6 Gene Expression. Proc. Natl. Acad. Sci. 97 (25), 13755–13759. doi:10.1073/pnas.240398797
LaVail, M. M., and Ward, P. A. (1978). Studies on the Hormonal Control of Circadian Outer Segment Disc Shedding in the Rat Retina. Invest. Ophthalmol. Vis. Sci. 17 (12), 1189–1193. https://iovs.arvojournals.org/article.aspx?articleid=2158737.
Lee, R. C., and Ambros, V. (2001). An Extensive Class of Small RNAs in Caenorhabditis elegans. Science 294 (5543), 862–864. doi:10.1126/science.1065329
Lee, R. C., Feinbaum, R. L., and Ambros, V. (1993). The C. elegans Heterochronic Gene Lin-4 Encodes Small RNAs with Antisense Complementarity to Lin-14. Cell 75 (5), 843–854. doi:10.1016/0092-8674(93)90529-y
Lee, Y. S., and Dutta, A. (2007). The Tumor Suppressor microRNA Let-7 Represses the HMGA2 Oncogene. Genes Dev. 21 (9), 1025–1030. doi:10.1101/gad.1540407
Leucht, C., Stigloher, C., Wizenmann, A., Klafke, R., Folchert, A., and Bally-Cuif, L. (2008). MicroRNA-9 Directs Late Organizer Activity of the Midbrain-Hindbrain Boundary. Nat. Neurosci. 11 (6), 641–648. doi:10.1038/nn.2115
Lewis, B. P., Shih, I.-h., Jones-Rhoades, M. W., Bartel, D. P., and Burge, C. B. (2003). Prediction of Mammalian microRNA Targets. Cell 115 (7), 787–798. doi:10.1016/s0092-8674(03)01018-3
Liu, Q., Novak, M. K., Pepin, R. M., Eich, T., and Hu, W. (2021). microRNA-Mediated Regulation of microRNA Machinery Controls Cell Fate Decisions. Elife 10. doi:10.7554/eLife.72289
Loscher, C. J., Hokamp, K., Kenna, P. F., Ivens, A. C., Humphries, P., Palfi, A., et al. (2007). Altered Retinal microRNA Expression Profile in a Mouse Model of Retinitis Pigmentosa. Genome Biol. 8 (11), R248. doi:10.1186/gb-2007-8-11-r248
Louvi, A., and Artavanis-Tsakonas, S. (2006). Notch Signalling in Vertebrate Neural Development. Nat. Rev. Neurosci. 7 (2), 93–102. doi:10.1038/nrn1847
Lowrey, P. L., and Takahashi, J. S. (2004). Mammalian Circadian Biology: Elucidating Genome-wide Levels of Temporal Organization. Annu. Rev. Genom. Hum. Genet. 5, 407–441. doi:10.1146/annurev.genom.5.061903.175925
Lumayag, S., Haldin, C. E., Corbett, N. J., Wahlin, K. J., Cowan, C., Turturro, S., et al. (2013). Inactivation of the microRNA-183/96/182 Cluster Results in Syndromic Retinal Degeneration. Proc. Natl. Acad. Sci. 110 (6), E507–E516. doi:10.1073/pnas.1212655110
Maeda, A., Tsujiya, S., Higashide, T., Toida, K., Todo, T., Ueyama, T., et al. (2006). Circadian Intraocular Pressure Rhythm Is Generated by Clock Genes. Invest. Ophthalmol. Vis. Sci. 47 (9), 4050–4052. doi:10.1167/iovs.06-0183
Makarev, E., Spence, J. R., Del Rio-Tsonis, K., and Tsonis, P. A. (2006). Identification of microRNAs and Other Small RNAs from the Adult Newt Eye. Mol. Vis. 12, 1386–1391. http://www.molvis.org/molvis/v12/a156/.
Malumbres, M., and Barbacid, M. (2001). To Cycle or Not to Cycle: A Critical Decision in Cancer. Nat. Rev. Cancer 1 (3), 222–231. doi:10.1038/35106065
Masamizu, Y., Ohtsuka, T., Takashima, Y., Nagahara, H., Takenaka, Y., Yoshikawa, K., et al. (2006). Real-time Imaging of the Somite Segmentation Clock: Revelation of Unstable Oscillators in the Individual Presomitic Mesoderm Cells. Proc. Natl. Acad. Sci. 103 (5), 1313–1318. doi:10.1073/pnas.0508658103
Mattick, J. S., and Makunin, I. V. (2006). Non-coding RNA. Hum. Mol. Genet. 15, R17–R29. doi:10.1093/hmg/ddl046
Mauvoisin, D., Wang, J., Jouffe, C., Martin, E., Atger, F., Waridel, P., et al. (2014). Circadian Clock-dependent and -independent Rhythmic Proteomes Implement Distinct Diurnal Functions in Mouse Liver. Proc. Natl. Acad. Sci. USA 111 (1), 167–172. doi:10.1073/pnas.1314066111
Mercer, T. R., and Mattick, J. S. (2013). Understanding the Regulatory and Transcriptional Complexity of the Genome Through Structure. Genome Res. 23 (7), 1081–1088. doi:10.1101/gr.156612.113
Miyata, T. (2008). Development of Three-Dimensional Architecture of the Neuroepithelium: Role of Pseudostratification and Cellular ‘community'. Dev. Growth Differ. 50 (Suppl. 1), S105–S112. doi:10.1111/j.1440-169X.2007.00980.x
Moss, E. G., Lee, R. C., and Ambros, V. (1997). The Cold Shock Domain Protein LIN-28 Controls Developmental Timing in C. elegans and Is Regulated by the Lin-4 RNA. Cell 88 (5), 637–646. doi:10.1016/s0092-8674(00)81906-6
Mu, G., Liu, H., Zhou, F., Xu, X., Jiang, H., Wang, Y., et al. (2010). Correlation of Overexpression of HMGA1 and HMGA2 with Poor Tumor Differentiation, Invasion, and Proliferation Associated with Let-7 Down-Regulation in Retinoblastomas. Hum. Pathol. 41 (4), 493–502. doi:10.1016/j.humpath.2009.08.022
Nelson, B. R., Hartman, B. H., Georgi, S. A., Lan, M. S., and Reh, T. A. (2007). Transient Inactivation of Notch Signaling Synchronizes Differentiation of Neural Progenitor Cells. Developmental Biol. 304 (2), 479–498. doi:10.1016/j.ydbio.2007.01.001
Nelson, B. R., Ueki, Y., Reardon, S., Karl, M. O., Georgi, S., Hartman, B. H., et al. (2011). Genome-Wide Analysis of Müller Glial Differentiation Reveals a Requirement for Notch Signaling in Postmitotic Cells to Maintain the Glial Fate. PLoS One 6 (8), e22817. doi:10.1371/journal.pone.0022817
Norbury, C., and Nurse, P. (1992). Animal Cell Cycles and Their Control. Annu. Rev. Biochem. 61, 441–468. doi:10.1146/annurev.bi.61.070192.002301
Norden, C., Young, S., Link, B. A., and Harris, W. A. (2009). Actomyosin Is the Main Driver of Interkinetic Nuclear Migration in the Retina. Cell 138 (6), 1195–1208. doi:10.1016/j.cell.2009.06.032
Ohana, R., Weiman-Kelman, B., Raviv, S., Tamm, E., Pasmanik-Chor, M., Rinon, A., et al. (2015). MicroRNAs of the RPE Are Essential for RPE Differentiation and Photoreceptor Maturation. Development 142 (14), 2487–2498. doi:10.1242/dev.121533
Ohtsuka, T., Ishibashi, M., Gradwohl, G., Nakanishi, S., Guillemot, F., and Kageyama, R. (1999). Hes1 and Hes5 as Notch Effectors in Mammalian Neuronal Differentiation. EMBO J. 18 (8), 2196–2207. doi:10.1093/emboj/18.8.2196
Pasquinelli, A. E., Reinhart, B. J., Slack, F., Martindale, M. Q., Kuroda, M. I., Maller, B., et al. (2000). Conservation of the Sequence and Temporal Expression of Let-7 Heterochronic Regulatory RNA. Nature 408 (6808), 86–89. doi:10.1038/35040556
Patterson, M., Gaeta, X., Loo, K., Edwards, M., Smale, S., Cinkornpumin, J., et al. (2014). let-7 miRNAs Can Act Through Notch to Regulate Human Gliogenesis. Stem Cel. Rep. 3 (5), 758–773. doi:10.1016/j.stemcr.2014.08.015
Peskova, L., Jurcikova, D., Vanova, T., Krivanek, J., Capandova, M., Sramkova, Z., et al. (2020). miR-183/96/182 Cluster Is an Important Morphogenetic Factor Targeting PAX6 Expression in Differentiating Human Retinal Organoids. Stem Cells 38, 1557–1567. doi:10.1002/stem.3272
Pierfelice, T., Alberi, L., and Gaiano, N. (2011). Notch in the Vertebrate Nervous System: An Old Dog with New Tricks. Neuron 69 (5), 840–855. doi:10.1016/j.neuron.2011.02.031
Pinter, R., and Hindges, R. (2010). Perturbations of microRNA Function in Mouse Dicer Mutants Produce Retinal Defects and Lead to Aberrant Axon Pathfinding at the Optic Chiasm. PLoS One 5 (4), e10021. doi:10.1371/journal.pone.0010021
Qiu, R., Liu, K., Liu, Y., Mo, W., Flynt, A. S., Patton, J. G., et al. (2009). The Role of miR-124a in Early Development of the Xenopus Eye. Mech. Development 126 (10), 804–816. doi:10.1016/j.mod.2009.08.002
Ramachandran, R., Fausett, B. V., and Goldman, D. (2010). Ascl1a Regulates Müller Glia Dedifferentiation and Retinal Regeneration Through a Lin-28-dependent, Let-7 microRNA Signalling Pathway. Nat. Cel. Biol. 12 (11), 1101–1107. doi:10.1038/ncb2115
Ramachandran, R., Zhao, X.-F., and Goldman, D. (2011). Ascl1a/Dkk/ -catenin Signaling Pathway Is Necessary and Glycogen Synthase Kinase-3 Inhibition Is Sufficient for Zebrafish Retina Regeneration. Proc. Natl. Acad. Sci. 108 (38), 15858–15863. doi:10.1073/pnas.1107220108
Reinhart, B. J., Slack, F. J., Basson, M., Pasquinelli, A. E., Bettinger, J. C., Rougvie, A. E., et al. (2000). The 21-nucleotide Let-7 RNA Regulates Developmental Timing in Caenorhabditis elegans. Nature 403 (6772), 901–906. doi:10.1038/35002607
Riesenberg, A. N., Liu, Z., Kopan, R., and Brown, N. L. (2009). Rbpj Cell Autonomous Regulation of Retinal Ganglion Cell and Cone Photoreceptor Fates in the Mouse Retina. J. Neurosci. 29 (41), 12865–12877. doi:10.1523/JNEUROSCI.3382-09.2009
Roese-Koerner, B., Stappert, L., Berger, T., Braun, N. C., Veltel, M., Jungverdorben, J., et al. (2016). Reciprocal Regulation between Bifunctional miR-9/9∗ and its Transcriptional Modulator Notch in Human Neural Stem Cell Self-Renewal and Differentiation. Stem Cel. Rep. 7 (2), 207–219. doi:10.1016/j.stemcr.2016.06.008
Roese-Koerner, B., Stappert, L., and Brüstle, O. (2017). Notch/Hes Signaling and miR-9 Engage in Complex Feedback Interactions Controlling Neural Progenitor Cell Proliferation and Differentiation. Neurogenesis 4 (1), e1313647. doi:10.1080/23262133.2017.1313647
Ryan, D. G., Oliveira-Fernandes, M., and Lavker, R. M. (2006). MicroRNAs of the Mammalian Eye Display Distinct and Overlapping Tissue Specificity. Mol. Vis. 12, 1175–1184. http://www.molvis.org/molvis/v12/a134/.
Sage, J., and Ventura, A. (2011). miR Than Meets the Eye: Figure 1. Genes Dev. 25 (16), 1663–1667. doi:10.1101/gad.17454011
Sahu, A., Devi, S., Jui, J., and Goldman, D. (2021). Notch Signaling via Hey1 and Id2b Regulates Müller Glia's Regenerative Response to Retinal Injury. Glia 69 (12), 2882–2898. doi:10.1002/glia.24075
Sampson, V. B., Rong, N. H., Han, J., Yang, Q., Aris, V., Soteropoulos, P., et al. (2007). MicroRNA Let-7a Down-Regulates MYC and Reverts MYC-Induced Growth in Burkitt Lymphoma Cells. Cancer Res. 67 (20), 9762–9770. doi:10.1158/0008-5472.CAN-07-2462
Sangoram, A. M., Saez, L., Antoch, M. P., Gekakis, N., Staknis, D., Whiteley, A., et al. (1998). Mammalian Circadian Autoregulatory Loop. Neuron 21 (5), 1101–1113. doi:10.1016/s0896-6273(00)80627-3
Sauer, F. C. (1935). Mitosis in the Neural Tube. J. Comp. Neurol. 62 (2), 377–405. doi:10.1002/cne.900620207
Saus, E., Soria, V., Escaramís, G., Vivarelli, F., Crespo, J. M., Kagerbauer, B., et al. (2010). Genetic Variants and Abnormal Processing of Pre-miR-182, a Circadian Clock Modulator, in Major Depression Patients with Late Insomnia. Hum. Mol. Genet. 19 (20), 4017–4025. doi:10.1093/hmg/ddq316
Shimojo, H., Isomura, A., Ohtsuka, T., Kori, H., Miyachi, H., and Kageyama, R. (2016). Oscillatory Control of Delta-like1 in Cell Interactions Regulates Dynamic Gene Expression and Tissue Morphogenesis. Genes Dev. 30 (1), 102–116. doi:10.1101/gad.270785.115
Shimojo, H., Ohtsuka, T., and Kageyama, R. (2008). Oscillations in Notch Signaling Regulate Maintenance of Neural Progenitors. Neuron 58 (1), 52–64. doi:10.1016/j.neuron.2008.02.014
Shu, P., Wu, C., Ruan, X., Liu, W., Hou, L., Fu, H., et al. (2019). Opposing Gradients of MicroRNA Expression Temporally Pattern Layer Formation in the Developing Neocortex. Developmental Cel. 49 (5), 764–785. doi:10.1016/j.devcel.2019.04.017
Smit-McBride, Z., Nguyen, A. T., Yu, A. K., Modjtahedi, S. P., Hunter, A. A., Rashid, S., et al. (2020). Unique Molecular Signatures of microRNAs in Ocular Fluids and Plasma in Diabetic Retinopathy. PLoS One 15 (7), e0235541. doi:10.1371/journal.pone.0235541
Sridhar, A., Hoshino, A., Finkbeiner, C. R., Chitsazan, A., Dai, L., Haugan, A. K., et al. (2020). Single-Cell Transcriptomic Comparison of Human Fetal Retina, hPSC-Derived Retinal Organoids, and Long-Term Retinal Cultures. Cel. Rep. 30 (5), 1644–1659. doi:10.1016/j.celrep.2020.01.007
Storch, K.-F., Paz, C., Signorovitch, J., Raviola, E., Pawlyk, B., Li, T., et al. (2007). Intrinsic Circadian Clock of the Mammalian Retina: Importance for Retinal Processing of Visual Information. Cell 130 (4), 730–741. doi:10.1016/j.cell.2007.06.045
Sundermeier, T. R., Sakami, S., Sahu, B., Howell, S. J., Gao, S., Dong, Z., et al. (2017). MicroRNA-processing Enzymes Are Essential for Survival and Function of Mature Retinal Pigmented Epithelial Cells in Mice. J. Biol. Chem. 292 (8), 3366–3378. doi:10.1074/jbc.M116.770024
Tan, S.-L., Ohtsuka, T., González, A., and Kageyama, R. (2012). MicroRNA9 Regulates Neural Stem Cell Differentiation by Controlling Hes1 Expression Dynamics in the Developing Brain. Genes Cells 17 (12), 952–961. doi:10.1111/gtc.12009
Taylor, S. M., Alvarez-Delfin, K., Saade, C. J., Thomas, J. L., Thummel, R., Fadool, J. M., et al. (2015). The bHLH Transcription Factor NeuroD Governs Photoreceptor Genesis and Regeneration through Delta-Notch Signaling. Invest. Ophthalmol. Vis. Sci. 56 (12), 7496–7515. doi:10.1167/iovs.15-17616
Taylor, S. M., Giuffre, E., Moseley, P., and Hitchcock, P. F. (2019). The MicroRNA, miR-18a , Regulates NeuroD and Photoreceptor Differentiation in the Retina of Zebrafish. Develop Neurobiol. 79 (2), 202–219. doi:10.1002/dneu.22666
Thomas, E. D., Timms, A. E., Giles, S., Harkins-Perry, S., Lyu, P., Hoang, T., et al. (2021). Multi-omic Analysis of Developing Human Retina and Organoids Reveals Cell-specific Cis-Regulatory Elements and Mechanisms of Non-coding Genetic Disease Risk. bioRxiv 2007, 2031–454254. doi:10.1101/2021.07.31.454254
Tomita, K., Ishibashi, M., Nakahara, K., Ang, S.-L., Nakanishi, S., Guillemot, F., et al. (1996). Mammalian Hairy and Enhancer of Split Homolog 1 Regulates Differentiation of Retinal Neurons and Is Essential for Eye Morphogenesis. Neuron 16 (4), 723–734. doi:10.1016/s0896-6273(00)80093-8
Tosini, G., Pozdeyev, N., Sakamoto, K., and Iuvone, P. M. (2008). The Circadian Clock System in the Mammalian Retina. Bioessays 30 (7), 624–633. doi:10.1002/bies.20777
Villarreal, G., Oh, D.-J., Kang, M. H., and Rhee, D. J. (2011). Coordinated Regulation of Extracellular Matrix Synthesis by the microRNA-29 Family in the Trabecular Meshwork. Invest. Ophthalmol. Vis. Sci. 52 (6), 3391–3397. doi:10.1167/iovs.10-6165
Walker, J. C., and Harland, R. M. (2009). microRNA-24a Is Required to Repress Apoptosis in the Developing Neural Retina. Genes Dev. 23 (9), 1046–1051. doi:10.1101/gad.1777709
Wang, Q., Navitskaya, S., Chakravarthy, H., Huang, C., Kady, N., Lydic, T. A., et al. (2016). Dual Anti-inflammatory and Anti-angiogenic Action of miR-15a in Diabetic Retinopathy. EBioMedicine 11, 138–150. doi:10.1016/j.ebiom.2016.08.013
Weston, M. D., Pierce, M. L., Rocha-Sanchez, S., Beisel, K. W., and Soukup, G. A. (2006). MicroRNA Gene Expression in the Mouse Inner Ear. Brain Res. 1111 (1), 95–104. doi:10.1016/j.brainres.2006.07.006
Wienholds, E., Kloosterman, W. P., Miska, E., Alvarez-Saavedra, E., Berezikov, E., de Bruijn, E., et al. (2005). MicroRNA Expression in Zebrafish Embryonic Development. Science 309 (5732), 310–311. doi:10.1126/science.1114519
Wohl, S. G., Hooper, M. J., and Reh, T. A. (2019). MicroRNAs miR-25, Let-7 and miR-124 Regulate the Neurogenic Potential of Müller Glia in Mice. Development 146 (17). doi:10.1242/dev.179556
Wohl, S. G., Jorstad, N. L., Levine, E. M., and Reh, T. A. (2017). Müller Glial microRNAs Are Required for the Maintenance of Glial Homeostasis and Retinal Architecture. Nat. Commun. 8 (1), 1603. doi:10.1038/s41467-017-01624-y
Wohl, S. G., and Reh, T. A. (2016b). miR-124-9-9* Potentiates Ascl1-Induced Reprogramming of Cultured Müller Glia. Glia 64 (5), 743–762. doi:10.1002/glia.22958
Wohl, S. G., and Reh, T. A. (2016a). The microRNA Expression Profile of Mouse Müller Glia In Vivo and In Vitro. Sci. Rep. 6, 35423. doi:10.1038/srep35423
Wu, Y.-C., Chen, C.-H., Mercer, A., and Sokol, N. S. (2012). Let-7-complex microRNAs Regulate the Temporal Identity of Drosophila Mushroom Body Neurons via Chinmo. Developmental Cel. 23 (1), 202–209. doi:10.1016/j.devcel.2012.05.013
Wuarin, J., Falvey, E., Lavery, D., Talbot, D., Schmidt, E., Ossipow, V., et al. (1992). The Role of the Transcriptional Activator Protein DBP in Circadian Liver Gene Expression. J. Cel. Sci. Suppl. 1992, 123–127. doi:10.1242/jcs.1992.supplement_16.15
Xia, X., and Ahmad, I. (2016). let-7 microRNA Regulates Neurogliogenesis in the Mammalian Retina Through Hmga2. Developmental Biol. 410 (1), 70–85. doi:10.1016/j.ydbio.2015.12.010
Xu, S., Witmer, P. D., Lumayag, S., Kovacs, B., and Valle, D. (2007). MicroRNA (miRNA) Transcriptome of Mouse Retina and Identification of a Sensory Organ-specific miRNA Cluster. J. Biol. Chem. 282 (34), 25053–25066. doi:10.1074/jbc.M700501200
Yan, Y., Salazar, T. E., Dominguez, J. M., Nguyen, D. V., Li Calzi, S., Bhatwadekar, A. D., et al. (2013). Dicer Expression Exhibits a Tissue-specific Diurnal Pattern that Is Lost During Aging and in Diabetes. PLoS One 8 (11), e80029. doi:10.1371/journal.pone.0080029
Yang, M., Lee, J.-E., Padgett, R. W., and Edery, I. (2008). Circadian Regulation of a Limited Set of Conserved microRNAs in Drosophila. BMC Genomics 9, 83. doi:10.1186/1471-2164-9-83
Yang, Y., and Mei, Q. (2015). miRNA Signature Identification of Retinoblastoma and the Correlations Between Differentially Expressed miRNAs During Retinoblastoma Progression. Mol. Vis. 21, 1307–1317. http://www.molvis.org/molvis/v21/1307/.
Yaron, O., Farhy, C., Marquardt, T., Applebury, M., and Ashery-Padan, R. (2006). Notch1 Functions to Suppress Cone-Photoreceptor Fate Specification in the Developing Mouse Retina. Development 133 (7), 1367–1378. doi:10.1242/dev.02311
Yu, Z., Wang, L., Wang, C., Ju, X., Wang, M., Chen, K., et al. (2013). Cyclin D1 Induction of Dicer Governs microRNA Processing and Expression in Breast Cancer. Nat. Commun. 4, 2812. doi:10.1038/ncomms3812
Zhao, C., Sun, G., Li, S., Lang, M.-F., Yang, S., Li, W., et al. (2010). MicroRNA Let-7b Regulates Neural Stem Cell Proliferation and Differentiation by Targeting Nuclear Receptor TLX Signaling. Proc. Natl. Acad. Sci. USA 107 (5), 1876–1881. doi:10.1073/pnas.0908750107
Zhou, L., Miller, C., Miraglia, L. J., Romero, A., Mure, L. S., Panda, S., et al. (2021). A Genome-wide microRNA Screen Identifies the microRNA-183/96/182 Cluster as a Modulator of Circadian Rhythms. Proc. Natl. Acad. Sci. USA 118 (1), e2020454118. doi:10.1073/pnas.2020454118
Zhu, Q., Sun, W., Okano, K., Chen, Y., Zhang, N., Maeda, T., et al. (2011). Sponge Transgenic Mouse Model Reveals Important Roles for the microRNA-183 (miR-183)/96/182 Cluster in Postmitotic Photoreceptors of the Retina. J. Biol. Chem. 286 (36), 31749–31760. doi:10.1074/jbc.M111.259028
Zuzic, M., Rojo Arias, J. E., Wohl, S. G., and Busskamp, V. (2019). Retinal miRNA Functions in Health and Disease. Genes 10 (5), 377. doi:10.3390/genes10050377
Keywords: MiR-183 cluster, Let-7, miR-9, circadian rhythm, cell cycle, Notch
Citation: Fishman ES, Han JS and La Torre A (2022) Oscillatory Behaviors of microRNA Networks: Emerging Roles in Retinal Development. Front. Cell Dev. Biol. 10:831750. doi: 10.3389/fcell.2022.831750
Received: 08 December 2021; Accepted: 07 January 2022;
Published: 02 February 2022.
Edited by:
Taran Gujral, Fred Hutchinson Cancer Research Center, United StatesReviewed by:
Lucia Poggi, University of Trento, ItalySeth Blackshaw, Johns Hopkins University, United States
Copyright © 2022 Fishman, Han and La Torre. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Anna La Torre, YWxhdG9ycmVAdWNkYXZpcy5lZHU=
 Elizabeth S. Fishman
Elizabeth S. Fishman Jisoo S. Han
Jisoo S. Han Anna La Torre
Anna La Torre