- 1Department of Medicine, Section of Dermatology, University of Chicago, Chicago, IL, United States
- 2Committee on Cancer Biology, University of Chicago, Chicago, IL, United States
Chemical modifications of RNA molecules regulate both RNA metabolism and fate. The deposition and function of these modifications are mediated by the actions of writer, reader, and eraser proteins. At the cellular level, RNA modifications regulate several cellular processes including cell death, proliferation, senescence, differentiation, migration, metabolism, autophagy, the DNA damage response, and liquid-liquid phase separation. Emerging evidence demonstrates that RNA modifications play active roles in the physiology and etiology of multiple diseases due to their pervasive roles in cellular functions. Here, we will summarize recent advances in the regulatory and functional role of RNA modifications in these cellular functions, emphasizing the context-specific roles of RNA modifications in mammalian systems. As m6A is the best studied RNA modification in biological processes, this review will summarize the emerging advances on the diverse roles of m6A in cellular functions. In addition, we will also provide an overview for the cellular functions of other RNA modifications, including m5C and m1A. Furthermore, we will also discuss the roles of RNA modifications within the context of disease etiologies and highlight recent advances in the development of therapeutics that target RNA modifications. Elucidating these context-specific functions will increase our understanding of how these modifications become dysregulated during disease pathogenesis and may provide new opportunities for improving disease prevention and therapy by targeting these pathways.
Introduction
Many RNA modifications are reversible modifications that are deposited onto RNA molecules, including mRNAs, tRNAs, rRNAs, and non-coding RNAs. To date, over 100 RNA modifications have been identified, including m6A, m5C, and m1A on mRNA (Roundtree et al., 2017). Of these modifications, N6-methyladenosine (m6A) is the most abundant internal mRNA modification (Roundtree et al., 2017). Since m6A is the best-studied mRNA modification, we will focus on the cellular functions of m6A in mammalian systems in this review. Other RNA modifications, such as m5C and m1A, have been studied in the context of cellular processes as well, and will be summarized here. The role of m6A and other RNA modifications in non-mammalian systems, including plants and yeast, is beyond the scope of this review and is detailed elsewhere (Schwartz et al., 2013; Shen et al., 2019; Yue et al., 2019).
RNA modifications are deposited onto, and erased from, RNA molecules through the actions of writer and eraser enzymes. m6A writers and cofactors include METTL3, METTL14, WTAP, VIRMA/KIAA1429, RBM15/15B, ZC3H13, and METTL16 (Yang et al., 2018). Together, METTL3, METTL14, WTAP, VIRMA/KIAA1429, RBM15/15B, and ZC3H13 make up the methyltransferase complex (MTC) (Yang et al., 2018). Within the MTC, METTL3 serves as the catalytic subunit, while METTL14 serves as the RNA-binding subunit (Yang et al., 2018). m6A erasers include FTO and ALKBH5 (Yang et al., 2018). RNA modifications influence various mechanisms of RNA metabolism, including nuclear processing, mRNA decay, and translation, through the action of reader enzymes (Roundtree et al., 2017). m6A readers include YTHDF1-3, YTHDC1-2, IGF2BP1-3, HNRNPA2B1, and eIF3 (Meyer et al., 2015; Yang et al., 2018). m6A mRNA writer, eraser and reader proteins are highlighted in Figure 1. While the writers and erasers can install or remove modifications in RNAs, it is the regulatory effect of the readers that ultimately controls the RNA fate and gene expression.
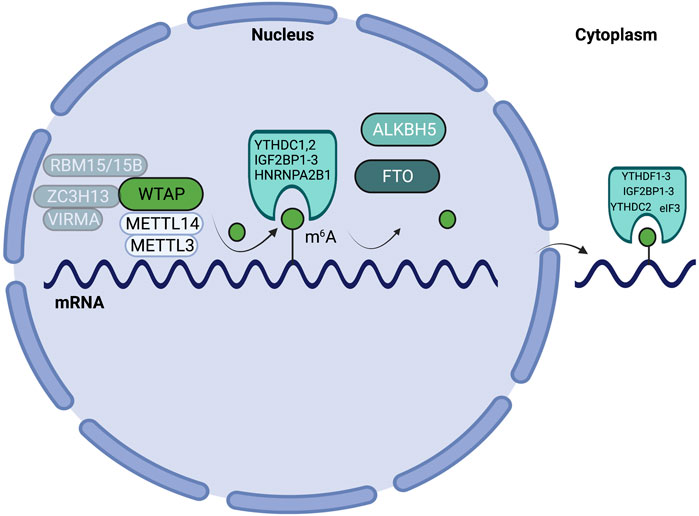
FIGURE 1. Schematic highlighting m6A writers, erasers, and readers on mRNA. m6A patterning on mRNA is mediated by the actions of writers (METTL3, METTL14, WTAP, VIRMA/KIAA1429, RBM15/15B, ZC3H13), erasers (ALKBH5, FTO), and reader enzymes (YTHDF1-3, YTHDC1/2, IGF2BP1-3, HNRNPA2B1, and eIF3).
Another modification on mRNA, tRNA, and other non-coding RNAs, is 5-methylcytosine (m5C). m5C writers include DNMT2 and the NSUN (NSUN1-7) family proteins (Xue et al., 2020). The NSUN proteins contain an RNA recognition motif and a catalytic core that houses S-adenosylmethionine (SAM) (Bohnsack et al., 2019). Similarly, DNMT2 contains a catalytic site and a SAM binding site (Xue et al., 2020). The detailed biochemical mechanisms of how m5C methyltransferases mediate methyl group transfers are discussed elsewhere (Bohnsack et al., 2019). m5C readers, or m5C binding-proteins, include ALYFREF and YBX1 (Xue et al., 2020). The TET family of enzymes have been hypothesized to serve as m5C erasers (Xue et al., 2020). The function of m5C in regulating RNA metabolism and expression is summarized elsewhere (Xue et al., 2020).
In addition to m6A and m5C, N1-methyladenosine (m1A) is another modification found on mRNA, tRNA, rRNA, and non-coding RNA (Xiong et al., 2018). m1A writers include TRMT10C, TRMT6, TRMT61A, and TRMT61B (Xiong et al., 2018). TRMT6/TRMT61A form a heterotetrameric complex wherein TRMT61A functions as the catalytic subunit and TRMT6 is required for its methyltransferase function (Shi et al., 2020; Graille, 2022). m1A erasers include ALKBH1 and ALKBH3 (Xiong et al., 2018).
Other RNA modification writer, eraser and reader enzymes are discussed elsewhere (Esteve-Puig et al., 2020). Elucidating the role of RNA modifications in mediating the RNA metabolism of diverse RNA species remains an active area of research.
RNA modifications, and their respective writer, eraser, and reader proteins, also play a role in a number of cellular functions. Here, we summarize the role of RNA modifications in such cellular processes as cell death, proliferation, differentiation, migration, metabolism, autophagy, and liquid-liquid phase separation in mammalian systems. Additionally, we also discuss the cell-type specific targets of these enzymes within these cellular processes.
Due to the pervasive roles of RNA modifications in numerous cellular functions, dysregulated RNA modifications have contributed to the pathogenesis of many diseases and can serve as attractive therapeutic targets due to the reversible nature of these modifications. The role of RNA modifications in diseases is covered in detail elsewhere (Wilkinson et al., 2021). Increasing our knowledge of RNA modifications in cellular processes will increase our understanding of the roles that RNA modifications play in disease etiology and will aid in identifying new therapeutic targets. Clinical success of therapeutics targeting RNA modifications has not been reached and may reflect an incomplete understanding of the role that these modifications play in cellular functions.
The Role of m6A in Diverse Cellular Functions
The writer, eraser, and reader proteins that regulate m6A have been well studied in several cellular processes. Of the writer proteins, we will focus on the role of METTL3 and METTL14 in cellular functions, as they are best studied in cellular functions. The role of WTAP and VIRMA is summarized as well.
Cell Death
Apoptosis is a mechanism of programmed cell death (Elmore, 2007). This process involves coordination and communication across intracellular signaling pathways that ultimately result in the cellular decision to undergo cell death (Elmore, 2007). Apoptosis is initiated in response to pathogens or cellular stressors, immune stimulation, and within embryonic development (Elmore, 2007; Yan et al., 2020). While apoptosis is not the sole mechanism of cell death, it is the best-studied mechanism in the context of m6A. The role of m6A in specialized forms of cell death remains an active area of research.
Writers
The m6A writer METTL3 can inhibit apoptosis, as several studies have shown that decreased Mettl3 expression and methyltransferase activity resulted in increased apoptosis. As previously mentioned, coordinated apoptosis is required for embryonic development (Elmore, 2007; Yan et al., 2020). Accordingly, knockdown of Mettl3 resulted in decreased m6A levels, which increased the mRNA half-lives of neuronal apoptosis-associated genes, including Dapk1, Fadd, and Ngfr, in mouse cerebral granular cells (CGCs) (Wang C.-X. et al., 2018). Increased mRNA half-lives of these genes led to increased apoptosis in CGCs and contributed to severe developmental defects in mouse cerebella (Wang C.-X. et al., 2018).
Furthermore, several recent studies have established that METTL3 may play an oncogenic role in cell death by negatively regulating and reducing the translation of apoptosis-associated proteins, thereby promoting cell survival (Vu et al., 2017; Choe et al., 2018; Huang et al., 2020). Accordingly, knockdown of Mettl3 resulted in increased expression of pro-apoptotic proteins in several cancer cell lines, emphasizing that METTL3 can function as a negative regulator of apoptosis (Vu et al., 2017; Zhou et al., 2020). In the MOLM-13 leukemia cell line, knockdown of Mettl3 increased protein expression of pro-apoptotic proteins CASP3, CASP7, and BIM (Vu et al., 2017). Expression of CASP3 and BAX, other pro-apoptotic proteins, were also increased in Mettl3 knockdown-osteosarcoma cell lines (Zhou et al., 2020). While it is unclear whether METTL3 regulates apoptosis in an m6A-dependent manner, both studies provide evidence that METTL3 may inhibit apoptosis by regulating BCL-2 family proteins and caspase expression (Vu et al., 2017; Zhou et al., 2020). Knockdown of Mettl3 in prostate cancer cells also increased protein expression of pro-apoptotic proteins BAK and BAX, CASP3 and CASP7 activity, and PARP cleavage (Cai et al., 2019). Additionally, knockdown of Mettl3 in prostate cancer cells decreased protein expression of anti-apoptotic genes BCL-2 and BCL-XL (Cai et al., 2019). Furthermore, knockdown of Mettl3 decreased expression of GLI1, a component of the Sonic hedgehog (SHH) signaling pathway, which prostate cancer cells are dependent on for survival (Regl et al., 2002; Chen et al., 2011; Cai et al., 2019). Due to prostate cancer cells’ dependence on SHH signaling, decreased expression of GLI1 deprived prostate cancer cells of SHH signaling and forced apoptosis (Cai et al., 2019). METTL3 regulated the expression of GLI1 in an m6A-dependent manner, as expression of GLI1 was rescued by the re-expression of wild-type, but not catalytically-inactive mutant, METTL3 (Cai et al., 2019). In gastric cancer, METTL3 promoted the mRNA stability of Sec62, which functions as a negative regulator of apoptosis, in an m6A/IGF2BP1 -dependent manner (He et al., 2019). Increased Sec62 mRNA stability and expression subsequently lead to decreased apoptosis and increased gastric cancer cell survival (He et al., 2019). The role of METTL3 in apoptosis is summarized in Figure 2.
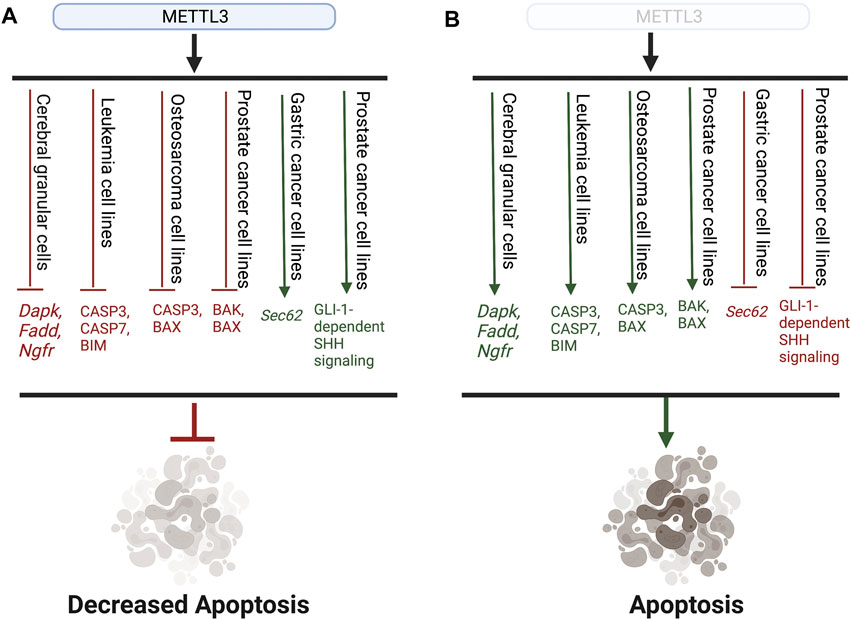
FIGURE 2. Representative schematic highlighting the role of METTL3 in apoptosis. (A) METTL3 prohibits the expression of both pro-apoptotic genes and proteins (red) as well as promotes the expression of anti-apoptotic genes and pathways (green), leading to overall decreased apoptosis. (B) Without METTL3 expression, pro-apoptotic genes and proteins are expressed (green), while anti-apoptotic pathways are inhibited, leading to the induction of apoptosis.
In contrast, the role of METTL14 in mediating apoptosis is not as widely explored and presents an area of research that requires future study. One study found that knockdown of Mettl14 promoted apoptosis in human AML cell lines (Weng et al., 2018). Mechanistically, METTL14 was found to promote cell survival by promoting the mRNA stability and translation of pro-survival proteins MYB and MYC in an m6A-dependent manner (Weng et al., 2018). Therefore, in the absence of METTL14, MYB and MYC expression was decreased, leading to the induction of apoptosis (Weng et al., 2018).
Few studies have explored the role of m6A writer-associated protein WTAP in cell death. One study found that WTAP expression was increased upon exposure to hypoxia/reoxygenation (H/R), resulting in the induction of ER stress and apoptosis in cardiomyocyte cells (Wang J. et al., 2021). Accordingly, H/R exposure in Wtap knockdown cells abrogated the induction of ER stress and apoptosis, suggesting that WTAP may regulate these processes upon H/R exposure (Wang J. et al., 2021). Mechanistically, WTAP was found to promote the mRNA stability of ER stress-response gene, Atf4, in an m6A-dependent manner (Wang J. et al., 2021).
VIRMA, another m6A writer-associated protein, has not been well-studied in the context of cell death. VIRMA has been found to serve an oncogenic role in several cancers, and was found to promote resistance to apoptosis in HCC (Lan et al., 2019). Mechanistically, VIRMA mediated the deposition of m6A onto the 3′-untranslated region (UTR) of Gata3, a tumor suppressor, resulting in decreased GATA3 expression and promoted resistance to apoptosis (Lan et al., 2019). The oncogenic role of VIRMA in several cancers is covered elsewhere in detail (Zhu W. et al., 2021).
Erasers
The role of the m6A eraser ALKBH5 in apoptosis is cell-type dependent. Alkbh5-deficient mice exhibited increased apoptosis and developmental defects, potentially through a p53-mediated mechanism that is not entirely understood (Zheng et al., 2013). Conversely, Alkbh5 knockdown in human ovarian granulosa (hGCs) cells had no effect on the apoptosis rate; rather, knockdown of Fto resulted in increased apoptosis in hGCs (Ding et al., 2018).
The role of FTO in apoptosis may also be cell-type-dependent. In leukemia, FTO is preferentially inhibited by R-2-hydroxyglutarate (R-2HG), an oncometabolite produced by mutant IDH1/2 enzymes (Ye et al., 2013; Su et al., 2018). Fto depletion decreased the mRNA stability and mRNA expression of downstream targets Myc and Cebpa through increased m6A accumulation (Su et al., 2018). Decreased expression of Myc and Cebpa resulted in decreased expression of downstream MYC targets, including the E2F transcription factors, which are major regulators of cell cycle, thereby preventing cells from entering the cell cycle and forcing apoptosis or cell cycle arrest (Su et al., 2018). The trend wherein knockdown of Fto resulted in increased apoptosis was also seen in breast cancer and melanoma, suggesting that FTO may also serve an oncogenic function in these cancers by inhibiting apoptosis (Yang S. et al., 2019; Niu et al., 2019). In breast cancer, FTO mediated the demethylation of m6A at the 3′-UTR of Bnip3, a pro-apoptotic gene, resulting in YTHDF2-mediated degradation of the Bnip3 transcript (Niu et al., 2019). In melanoma, FTO promoted resistance to IFN γ-mediated cell death through m6A demethylation of pro-tumorigenic genes Pdcd-1, Cxcr4, and Sox10 (Yang S. et al., 2019). Demethylation of Pdcd-1, Cxcr4, and Sox10 prevented downstream YTHDF2-mediated mRNA decay, resulting in increased expression of these melanoma-promoting genes (Yang S. et al., 2019).
Readers
Few studies have examined the role of only m6A readers in mediating cell death. One study found that YTHDF2 negatively regulates apoptosis in TNBC (Einstein et al., 2021). Mechanistically, YTHDF2 was found to mediate the degradation of the Prss23 mRNA transcript, a gene involved in translation, in an m6A-dependent manner (Einstein et al., 2021). More generally, YTHDF2-mediated mRNA decay provides a mechanism by which to control the number of translated mRNAs by degrading mRNA transcripts, therefore resulting in translational control (Einstein et al., 2021). Knockdown of Ythdf2 subsequently lead to an increase in PRSS23 expression and protein translation, triggering ER stress and inducing proteotoxic cell death (Einstein et al., 2021). Furthermore, in chondrocytes, YTHDF1 was found to negatively regulate apoptosis by promoting the mRNA stability of anti-apoptotic Bcl-2 in a METTL3/m6A-dependent manner (He et al., 2022).
While significant research efforts have been made to establish the role of RNA modifications in cell death, there remain significant gaps in knowledge surrounding this topic. Compelling evidence suggests a critical role of m6A RNA methylation in other forms of cell death. For example, Guo et al. have recently established that m6A may regulate macrophage pyroptosis, an inflammation-induced form of cell death, in circular RNAs in patients with acute coronary syndrome (Bergsbaken et al., 2009; Guo et al., 2020). However, the contribution of the m6A machinery in this cellular process remains to be investigated.
Proliferation
The role of m6A in regulating cellular proliferation has been best-studied in the context of cancer. Accordingly, m6A writers, erasers, and readers may represent viable therapeutic targets for their role in promoting cell proliferation.
Writers
While the m6A writer METTL3 has been well-studied within the context of apoptosis, METTL3 has been found to either promote or inhibit cellular proliferation, depending on the cellular context.
METTL3 was shown to inhibit cellular proliferation in endometrial cancer, as METTL3-mediated m6A promoted the translation of PHLPP2, a negative regulator of pro-proliferative AKT signaling (Liu et al., 2018). Accordingly, knockdown of Mettl3 resulted in increased cellular proliferation through decreased m6A-dependent translation of PHLPP2, thereby promoting AKT signaling (Liu et al., 2018). Furthermore, in renal cell carcinoma (RCC), decreased Mettl3 expression resulted in increased proliferation through induction of the PI3K/AKT/mTOR pathway (Li X. et al., 2017). Whether the activation of the PI3K/AKT/mTOR pathway upon Mettl3 knockdown is m6A-dependent was not explored (Li X. et al., 2017).
However, in several other cancer cell types, METTL3 was shown to promote cell proliferation. In colorectal cancer, METTL3 promoted GLUT1 translation in an m6A-dependent manner, which resulted in downstream activation of mTORC1 and increased cell survival and proliferation (Chen et al., 2021). In hepatoblastoma cells, increased METTL3 activity resulted in increased m6A deposition on Ctnnb1, leading to aberrant activation of the WNT/CTNNB1 pathway, which promoted hepatoblastoma cell growth (Liu et al., 2019). Another study in hepatocellular carcinoma (HCC) cells found that METTL3 promoted proliferation by inhibiting the expression of SOCS2, a transcription factor that can negatively regulate cell proliferation, through m6A-dependent/YTHDF2-mediated mRNA degradation (Chen M. et al., 2018). Furthermore, in breast cancer cells, METTL3 was found to participate in a feedback loop with HBXIP, a co-factor of anti-apoptotic protein SURVIVIN, wherein HBXIP up-regulated METTL3 expression by suppressing METTL3 inhibitor let-7g, an miRNA (Garcia-Saez et al., 2011; Cai et al., 2018). Increased METTL3 expression then further promoted HBXIP expression in an m6A-dependent manner and drove cell proliferation (Cai et al., 2018). However, whether METTL3-mediated m6A on Hbxip promotes mRNA stability or translation was not explored (Cai et al., 2018). In bladder cancer, METTL3 promoted the m6A-mediated maturation of pri-miR221/222, a PTEN antagonist, resulting in loss of cell cycle control and increased proliferation (Han et al., 2019). Similarly, METTL3 also drove proliferation in ovarian cancer through regulation of the receptor tyrosine kinase AXL; however, the regulatory mechanism by which METTL3 up-regulates AXL was not established (Hua et al., 2018). While the role of METTL3 in promoting proliferation in many cancer types has been well-established, the role of METTL3 in pancreatic cancer proliferation remains controversial as two independent studies found contrasting roles for METTL3 in promoting pancreatic cancer cell proliferation (Taketo et al., 2018; Xia et al., 2019). Authors of these studies reconcile differences in results based on differences in proliferation assays and cell lines used (Taketo et al., 2018; Xia et al., 2019).
Other m6A writer co-factors have also been found to regulate cellular proliferation in a context-dependent manner. METTL14 expression was found to be decreased in colorectal cancer patients, and Mettl14 knockdown in vitro resulted in decreased m6A deposition on downstream target Xist, a long non-coding RNA (lncRNA) that has been found to promote proliferation (Yang et al., 2020). Decreased m6A on Xist prevented YTHDF2-mediated mRNA degradation, resulting in increased Xist expression and increased cell proliferation (Yang et al., 2020). Similarly, in gastric cancer, METTL14 decreased cell proliferation by negatively regulating the pro-proliferative PI3K/AKT/mTOR pathway, emphasizing the cell-type-specific role of METTL14 in this cellular process (Liu X. et al., 2021). Whether METTL14 regulates the PI3K/AKT/mTOR pathway in an m6A-dependent manner remains unclear (Liu X. et al., 2021). METTL14 can also promote cell proliferation. In breast cancer, METTL14 was recruited by oncogenic lncRNA LINC00942 to increase the m6A-mediated mRNA stability and protein expression of two downstream targets, CXCR4 and CYP1B1, which resulted in increased cell proliferation and tumorigenesis (Sun et al., 2020). In AML, METTL14 promoted cell survival and proliferation by regulating the mRNA stability and translation of two pro-proliferative downstream targets, MYB and MYC, in an m6A-dependent manner (Weng et al., 2018). In skin cancer, METTL14 was also found to promote cell proliferation, as knockdown of Mettl14 in human keratinocytes resulted in decreased cell proliferation; however, the mechanism by which METTL14 promotes proliferation in this context remains unclear (Yang Z. et al., 2021).
WTAP, another m6A writer co-factor, has been found to promote proliferation. However, studies examining the role of WTAP in regulating proliferation do not detail whether WTAP promotes proliferation in an m6A-dependent manner. In renal cell carcinoma (RCC), WTAP promoted the mRNA stability of Cdk2, a regulator of cell cycle control over the G1/S and S/G2 transition, by directly binding to the Cdk2 transcript at the 3′-UTR (Tang et al., 2018). Furthermore, in primary AML patient samples and AML cell lines, reverse phase protein array (RPPA) analysis revealed that WTAP is positively associated with pro-proliferative cyclins and HSP90, as well as anti-apoptotic proteins, such as BCL-2 (Bansal et al., 2014). The mechanism by which WTAP regulates the expression of these pro-proliferative proteins, and whether this regulation is m6A-dependent, was not explored in this study (Bansal et al., 2014).
Additionally, VIRMA was found to promote non-small cell lung cancer (NSCLC) and increased NSCLC proliferation in vitro and in vivo (Xu et al., 2021). Mechanistically, VIRMA promoted the mRNA decay of tumor suppressor Dapk3 through an m6A-dependent YTHDF2/YTHDF3-mediated mechanism (Xu et al., 2021). Furthermore, VIRMA was found to promote breast cancer progression by promoting the mRNA stability of Cdk1 in an m6A-independent manner (Qian et al., 2019).
Erasers
The pro-proliferative role of the m6A eraser FTO is well-studied within the context of cancer. In leukemia, FTO promoted the proliferation of AML cells by reducing m6A levels at the 3′-UTR of Asb2 and 3′ and 5′-UTR of Rara, two mediators of hematopoiesis and differentiation, resulting in decreased ASB2 and RARA protein expression (Li Z. et al., 2017). In melanoma, FTO promoted cell proliferation and overall tumorigenicity by demethylating m6A on melanoma-promoting genes Pdcd1, Cxcr4, and Sox10 (Yang S. et al., 2019). Furthermore, exposure to arsenic, a known human carcinogen, resulted in increased FTO stability and abundance in human keratinocytes, ultimately leading to increased proliferation and tumorigenesis (Cui et al., 2021). Furthermore, FTO and MYC have also been found to cooperate to drive cell proliferation in both pancreatic and cervical cancer (Tang et al., 2019; Zou et al., 2019). In pancreatic cancer, FTO mediated the m6A demethylation of the c-Myc transcript, resulting in increased c-MYC expression (Tang et al., 2019). In cervical cancer, FTO was found to promote MYC translation; however, whether this mechanism was m6A-dependent was not established (Zou et al., 2019). FTO also promoted the proliferation of NSCLC cells by demethylating and increasing the mRNA stability of the ubiquitinase Usp7, resulting in increased USP7 protein expression (Li et al., 2019). Future studies are needed to define the role of USP7 in mediating cell proliferation (Li et al., 2019).
In addition to FTO, m6A eraser ALKBH5 drove proliferation in glioblastoma stem cells by demethylating nascent mRNA transcripts of Foxm1, a transcription factor involved in cell-cycle control and proliferation, resulting in increased FOXM1 expression and activity in an m6A-dependent manner (Zona et al., 2014; Zhang S. et al., 2017).
Readers
The m6A reader YTHDF2 promotes cell proliferation across different cell types and through distinct mechanisms. In pancreatic cancer, YTHDF2 promoted cell growth through activation of the AKT/GSK3β/CCND1 pathway (Chen et al., 2017). However, it is unclear whether YTHDF2 mediates pancreatic cancer growth in an m6A-dependent manner (Chen et al., 2017). In leukemia, YTHDF2 increased cell proliferation by promoting the m6A-dependent mRNA decay of Wee1, which regulates mitotic entry and serves as a negative cell-cycle regulator (Fei et al., 2020).
Additionally, PRRC2A, an m6A-binding protein, promoted the proliferation of oligodendrocytes, a class of glial cells found in the brain and central nervous system, by binding and stabilizing the Olig2 mRNA transcript in an m6A-dependent manner (Wu et al., 2019).
While the role of m6A in proliferation is widely studied in the context of cancer, cell proliferation is critical for other biological processes, such as wound repair and development, and is dysregulated in many diseases. Future studies are needed to address the role of m6A in proliferation in these contexts.
Senescence
Senescence is a cellular mechanism wherein cells permanently undergo cell cycle arrest in response to cellular stress or other stimuli (Kumari and Jat, 2021). Intracellularly, senescent cells undergo metabolic and genomic changes that promote cell survival, yet in a growth-arrested state (Kumari and Jat, 2021). Extracellularly, senescent cells communicate with neighboring cells through a variety of secreted factors, including cytokines and chemokines, and assume a senescence-associated secretory phenotype (SASP) (Kumari and Jat, 2021). The role of m6A in senescence has been studied in a variety of contexts, including tumorigenesis and aging, and is reviewed in detail elsewhere (Casella et al., 2019). In this section, we will summarize recent advances on the role of m6A in senescence.
Writers
Liu et al. established that the m6A writers METTL3 and METTL14 promoted SASP in lung embryonic fibroblasts in an m6A-independent manner (Liu P. et al., 2021). During cellular senescence, METTL14 was found to localize to enhancer subunits, while METTL3 localized to promoters of SASP genes (Liu P. et al., 2021). Interestingly, WTAP was found to be required for the nuclear localization of METTL3 and METTL14 during senescence (Liu P. et al., 2021). However, METTL3 may function to inhibit senescence in human mesenchymal stem cells (hMSCs), as knockdown of Mettl3 in hMSCs resulted in accelerated senescence (Wu et al., 2020). Overexpression of Mettl3 in hMSCs reversed the phenotype seen in Mettl3-deficient hMSCs and delayed senescence induction through m6A/IGF2BP2-mediated stabilization of the pro-proliferative gene Mis12 (Wu et al., 2020). Furthermore, in human nucleus pulposus cells, METTL14 positively regulated TNFα-induced cellular senescence by promoting the maturation of miR-34a-5p, which inhibits SIRT1, a negative regulator of senescence (Zhu H. et al., 2021). However, the role of miR-34a-5p in senescence is not completely understood.
Erasers
The m6A eraser FTO has been found to serve as a negative regulator of senescence in various contexts. Accordingly, FTO negatively regulated cellular senescence in granulosa-cell-induced ovarian aging in an m6A-dependent manner (Jiang et al., 2021). In this context, expression of catalytically inactive mutant FTO, which lacks demethylase activity, increased m6A on the 3′-UTR of Fos, a transcription factor that promotes aging, preventing the m6A-mediated degradation of Fos mRNA and increasing FOS translation (Jiang et al., 2021). Similar deactivating mutations in FTO resulted in increased senescence in skin fibroblasts (Boissel et al., 2009). While the mechanism by which FTO inhibits senescence was not delineated, these studies suggest that the demethylase activity of FTO is required to inhibit senescence (Boissel et al., 2009; Jiang et al., 2021).
Readers
The role of readers in cellular senescence is not well-studied. In human ovarian epithelial cells, RAS activation resulted in increases in Reactive Oxygen Species (ROS), which led to decreased expression of YTHDF2 (Zhu et al., 2020). Decreased expression of YTHDF2, which functions to mediate mRNA decay, resulted in downstream activation of the MAPK pathway and prevented the mRNA decay of Map2k4 and Map4k4 (Zhu et al., 2020). Activation of the MAPK pathway then led to downstream activation of NF-κB signaling pathways, resulting in the induction of SASP and senescence (Zhu et al., 2020).
A current gap in knowledge in this field revolves around our understanding of the m6A machinery in cell fate decisions. Future studies should be centered on understanding the dynamic nature of m6A in initiating cellular senescence and quiescence, as well as the changes in m6A that are needed for the cell to re-enter the cell cycle. Elucidating the roles of m6A machinery in cell fate decisions has broad-standing implications in understanding stem cell biology, cancer stem cell formation and maintenance, and cell cycle control.
Differentiation
Cell differentiation is the process of transformation into specialized cell types and is essential for development. The hematopoietic system is a well-established model which emphasizes the cell-type and stage-specific role of m6A in differentiation. Outside of hematopoiesis, m6A has been found to be a critical regulator in stem cell fate, neuronal development, and skin development. The role of m6A in development and stem cell biology is reviewed extensively elsewhere (Frye et al., 2018; Malla et al., 2019; Rosselló-Tortella et al., 2020; Vasic et al., 2020; Song et al., 2021). Here we will summarize the role of m6A in differentiation in several contexts.
Writers
The role of RNA modifications within hematopoietic differentiation is stage-specific. At early stages, m6A is necessary for differentiation during the endothelial to hematopoietic transition (EHT), which mediates early-stage hematopoietic stem and progenitor cell (HSPC) differentiation (Zhang C. et al., 2017). The necessity of m6A within EHT is demonstrated by mettl3−/− zebrafish, which display disrupted HSPC development (Zhang C. et al., 2017). Mechanistically, mettl3−/− zebrafish show continuous Notch activation, as depletion of m6A on the notch1a transcript prevents Ythdf2-mediated notch1a mRNA decay (Zhang C. et al., 2017). Continuous Notch activation in mettl3−/− zebrafish promotes an endothelial cell lineage, thereby inhibiting EHT and preventing the HPSC generation (Zhang C. et al., 2017). In mice, conditional Mettl3 knockout promoted hematopoietic stem cell (HSC) accumulation in the bone marrow, suggesting that HSC differentiation was unable to progress without METTL3 or m6A (Lee et al., 2019). Mechanistically, METTL3-mediated m6A is believed to promote the mRNA translation of downstream target Myc, which regulates differentiation; Mettl3−/− mice therefore display a differentiation block due to decreased MYC translation (Lee et al., 2019). Other independent studies have also noted blocks in HSC differentiation in Mettl3−/− mice, establishing a pervasive role for METTL3-mediated m6A within differentiation (Cheng et al., 2019). However, knockdown of Mettl3 in HSPCs resulted in increased cellular differentiation, emphasizing the stage-specific function of m6A within differentiation (Vu et al., 2017). Furthermore, METTL3-mediated m6A was found to inhibit differentiation in AML cells, which suggests that m6A may have distinct functions upon oncogenic transformation in AML cell lines (Lee et al., 2019).
In embryonic stem cells, m6A was found to be critical for mediating the mRNA decay and turnover of transcripts within differentiation (Batista et al., 2014). Similarly, in the context of neuronal development, Yoon et al. identified m6A to be a critical factor in mediating neurogenesis, as m6A was found to promote the mRNA decay of transcription factors involved in this process (Yoon et al., 2017). m6A was also found to regulate embryonic neural stem cell renewal and differentiation through regulation of histone modifications, which may further influence the transcription or expression of transcription factors involved in neuronal development (Wang Y. et al., 2018). PRRC2A, an m6A-binding protein, also promoted the fate determination of oligodendrocytes through stabilization of the Olig2 mRNA transcript in an m6A-dependent manner (Wu et al., 2019). Together, these studies establish the critical role of m6A in mediating the coordination in gene expression events in stem cell differentiation.
Differentiation is a key process in skin development, homeostasis, and wound repair (Lopez-Pajares et al., 2013). Accordingly, Lee et al. determined that METTL14-dependent m6A methylation on lncRNA Pvt1 regulates stemness in epidermal progenitor cells, promoting both Pvt1-MYC interactions and MYC protein stabilization (Lee J. et al., 2021).
Furthermore, WTAP was found to be an essential factor for mediating the differentiation of endoderm and mesoderm as mouse embryos lacking Wtap failed to differentiate into endoderm and mesoderm and were embryonic lethal during the gastrulation phase of development (Fukusumi et al., 2008). Horiuchi et al. also found that loss of Wtap resulted in embryonic lethality at day 6.5 (Horiuchi et al., 2006). Mechanistically, this study found that WTAP promoted the stabilization of Cyclin A2 mRNA, which regulates the G2/M transition, and that loss of Wtap resulted in G2 accumulation and subsequent lethality (Horiuchi et al., 2006).
Erasers
The m6A eraser FTO may regulate differentiation across different cell types. However, the role of FTO in promoting, or inhibiting, differentiation is cell-type dependent. Knockout of Fto in adult neural stem cells (aNSCs) resulted in increased aNSC proliferation and differentiation through aberrant activation of the STAT3 pathway, resulting in inhibited neurogenesis and dysregulated neuronal development (Cao et al., 2019). Mechanistically, the STAT3 pathway was activated through increased m6A enrichment on Pdgfrα and Socs5 mRNA transcripts, due to decreased FTO expression and activity (Cao et al., 2019). Interestingly, decreased FTO expression and activity resulted in increased PDGFRα protein expression and decreased SOCS5 protein expression, which, together, promote the phosphorylation and activation of STAT3 (Cao et al., 2019). The differences between m6A-dependent regulation of PDGFRα and SOCS5 protein expression were not explored in this study (Cao et al., 2019).
FTO is also involved in adipogenic differentiation. Accordingly, decreased FTO demethylase activity resulted in decreased preadipocyte differentiation in an m6A-dependent manner, and FTO-over-expressing mouse embryonic fibroblasts (MEFs) showed increased adipogenic differentiation (Merkestein et al., 2015; Zhang et al., 2015).
Readers
The m6A reader YTHDF2 was identified to function as the main regulator of mRNA decay of transcriptional regulators involved in hematopoiesis and self-renewal (Li et al., 2018). Ythdf2−/− HSPCs resulted in increased expansion of HSCs and increased mRNA expression of transcription factors involved in self-renewal, such as Gata2, Runx1, Tal1, and Stat5 (Li et al., 2018). Mechanistically, YTHDF2 is believed to negatively regulate HSC expansion by facilitating the mRNA decay of Gata2, Runx1, Tal1, and Stat5 in an m6A-dependent manner (Li et al., 2018). Furthermore, in mettl3−/− zebrafish, decreased m6A resulted in decreased Ythdf2-mediated mRNA decay of notch1a, a transcription factor that represses HSPC formation (Zhang C. et al., 2017).
YTHDC1 also serves a role in differentiation as Ythdc1 expression was increased in M0 undifferentiated acute myeloblastic leukemia cells, suggesting that YTHDC1 may be required to maintain an undifferentiated state (Cheng et al., 2021). Furthermore, knockdown of Ythdc1 in the OCIAML3 cell line resulted in increased differentiation (Cheng et al., 2021). Mechanistically, YTHDC1 is believed to inhibit differentiation through downstream m6A-dependent regulation of MYC (Cheng et al., 2021).
m6A plays crucial roles in differentiation in the hematopoietic system, as well as in stem cell fate, neuronal development, and skin development. Expanded studies should be employed to specifically address the dynamic changes in m6A machinery across totipotent, multipotent, and pluripotent stem cells.
Migration
Cell migration involves the coordination of biophysical and mechanical mechanisms that allow cells to migrate. Cell migration is also the major cellular process that drives wound healing, cancer progression, and metastasis.
Writers
METTL14 is the best-studied m6A writer in the context of migration. Many studies have found that METTL14 may serve as either a positive or negative regulator of migration and metastasis, depending on cellular context. In gastric cancer and endometrial cancer, knockdown of Mettl14 and decreased m6A levels increased cell migration and invasiveness, establishing METTL14 as a negative regulator of migration and metastasis in these contexts (Liu et al., 2018; Zhang C. et al., 2019). Furthermore, in colorectal cancer, METTL14 inhibited migration and metastasis through m6A/YTHDF2-mediated mRNA degradation of the epithelial-to-mesenchymal transition (EMT)-promoting transcription factor Sox4 (Chen X. et al., 2020). Furthermore, in papillary thyroid cancer, METTL14 inhibited migration by binding, and decreasing the expression of, lncRNA OIP5-As1, which promotes proliferation and migration through downstream regulation of the MEK/ERK, EGFR, and PI3K pathways (Zhang et al., 2021). However, it is unclear whether this mechanism is m6A-dependent (Zhang et al., 2021). Conversely, in keratinocytes and skin cancer cells, METTL14 promoted migration in an m6A dependent manner, as knockdown of Mettl14 decreased migration, while overexpression of wild-type, but not catalytically inactive mutant, Mettl14 resulted in increased migration (Yang Z. et al., 2021).
Similarly, METTL3 may promote or inhibit migration depending on the cellular context. Several studies in melanoma have found that METTL3 induced migration by increasing expression of pro-migratory proteins, c-MET and MMP2, in an m6A-dependent manner (Spina et al., 2015; Dahal et al., 2019; Luo et al., 2020). In lung cancer, METTL3 was increased during TGF-β-induced EMT (Wanna-Udom et al., 2020). Furthermore, in liver cancer, METTL3 was found to mediate increases in m6A levels during EMT, including specific m6A increases on the coding sequence (CDS) of EMT-associated transcription factor Snail, resulting in YTHDF1-mediated increases in SNAIL translation and EMT progression (Lin X. et al., 2019). Similarly, in bladder cancer, METTL3 deposited m6A on the 3′-UTR of Cdcp1, which has been found to promote migration across several cancer types, resulting in YTHDF1-mediated increases in CDCP1 translation and increased cellular migration (Yang F. et al., 2019). In NSCLC and gastric cancer, METTL3 promoted migration through downstream activation of PI3K/AKT; however, whether this mechanism is m6A-dependent is unclear (Lin S. et al., 2019; Wei et al., 2019). Interestingly, METTL3 expression in ovarian cancer also increased migration and induction of EMT through increased protein expression of the receptor tyrosine kinase AXL; however, while the Axl mRNA transcript contains fourteen m6A sites, METTL3 regulation of AXL translation is believed to be m6A-independent (Hua et al., 2018). The m6A-independent mechanism by which METTL3 regulates AXL translation remains unclear (Hua et al., 2018). In contrast, in colorectal cancer cells, Mettl3 over-expression resulted in decreased migration, while decreased Mettl3 activated the p38/ERK pathways, resulting in increased migration (Deng et al., 2019). Whether regulation of p38/ERK by METTL3 is m6A-dependent was not explored in this study (Deng et al., 2019).
Another m6A writer-associated protein, WTAP, also induced migration and metastasis by increasing the mRNA expression of migration-promoting genes, Mmp7, Mmp28, Cathepsin H, and Muc1 in cholangiocarcinoma cells (Jo et al., 2013). However, this study did not investigate whether WTAP-mediated increases in Mmp7, Mmp28, Cathepsin H, and Muc1 are m6A-dependent (Jo et al., 2013).
Erasers
The role of the m6A eraser FTO in migration is not well-established and requires further study. A study in cervical cancer suggested that FTO regulates migration by promoting the protein translation of E2F1 and MYC, two regulators of cell cycle and migration, in an m6A-dependent manner (Zou et al., 2019). Furthermore, in melanoma cells, overexpression of FTO promoted migration and overall tumorigenicity in an m6A-dependent manner, while knockdown of FTO inhibited migration (Yang S. et al., 2019).
Readers
The m6A reader YTHDF2 may have inhibitory effects on migration. m6A deposition on lncRNA THOR contributes to increased migration across many different cancer cell types (Liu H. et al., 2020). Interestingly, m6A on THOR is read by YTHDF1 and YTHDF2, which can mediate the transcription or decay of THOR, respectively, and therefore influence migration through their respective effects on THOR RNA metabolism (Liu H. et al., 2020). In pancreatic cancer, YTHDF2 was involved in a “migration-proliferation dichotomy” wherein YTHDF2 promoted proliferation but inhibited migration by suppressing YAP signaling, an EMT-promoting signaling pathway (Chen et al., 2017). While YAP contains two m6A sites, it is unclear whether YTHDF2 regulates YAP expression by directly regulating mRNA stability, or whether YTHDF2 regulates upstream regulators of YAP (Chen et al., 2017).
While many studies have reported the effects of RNA modification by writers, erasers and readers on regulating migration, the unique biophysical mechanisms that underlie these transitions are not well-elucidated and remain an active area of research. For example, future studies are needed to explore the potential role of RNA modifications in regulating cytoskeletal proteins.
Metabolism
m6A mediates cellular metabolism in a cell-type dependent manner. The intersection of epitranscriptomics and metabolism remains an understudied area of research. The role of m6A in mediating cancer metabolism is further reviewed elsewhere (Han et al., 2020).
Writers
The m6A writer METTL3 promotes lipogenesis and adipogenesis across several different contexts. In HCC cell lines, METTL3-mediated m6A promoted lncRNA LINC00958 RNA stability in an m6A-dependent manner (Zuo et al., 2020). With increased RNA stability, LINC00958 promoted lipogenesis by regulating the miR-3619-5p/HDGF pathway, which, in turns, regulates lipogenesis enzymes such as SREB1, FASN, SCD1, and ACC1 (Zuo et al., 2020). As a result, increased LINC0095 RNA stability resulted in increased cholesterol and triglyceride levels and lipid droplet formation (Zuo et al., 2020). Interestingly, FTO and METTL3 may communicate to coordinate adipogenesis and fat absorption, as visceral fat taken from offspring of high-fat diet-fed mice mothers exhibited decreased FTO expression, and increased METTL3 expression, at 3 weeks of age (Li et al., 2016). Increased m6A levels were also noted at 3 weeks of age in the visceral fat of these offspring (Li et al., 2016). However, at 8 weeks of age, both FTO and METTL3 were increased in the visceral fat, despite their contradictory functions, with no changes in m6A levels noted (Li et al., 2016). These results suggest a unique coordination between FTO and METTL3 in response to a high-fat diet and within development, but the mechanism remains unclear (Li et al., 2016). In addition to lipogenesis and adipogenesis, METTL3 was found to regulate glucose metabolism in colorectal cancer, as METTL3-mediated m6A promoted the mRNA stability of Hk2 and Glut1 in an m6A and IGF2BP2/3-dependent manner (Shen et al., 2020). Increased Hk2 and Glut1 mRNA stability subsequently led to the activation of glycolysis and promoted colorectal cancer cell progression (Shen et al., 2020).
Erasers
FTO serves a pivotal role in multiple metabolic processes, including fat metabolism, gluconeogenesis, metabolic stress, and lactate production. Several seminal studies were intrinsic to establishing the role of FTO in fat metabolism. However, because FTO was only recently established as an m6A eraser, the contexts in which FTO promotes obesity in an m6A-independent or dependent manner is not entirely clear. In humans, the FTO SNP rs9939609 was found to be linked to body mass index and is believed to be one of the strongest genetic determinants of obesity propensity (Frayling et al., 2007). Additionally, the FTO SNP rs8050136 in humans decreases the binding affinity of the CUX1 isoform P110, resulting in decreased expression of FTO and leptin signaling, preventing satiety, and promoting obesity (Stratigopoulos et al., 2011).
More recent studies have suggested that the demethylase activity of FTO is indispensable for its role in mediating fat metabolism, including lipogenesis and adipogenesis. Decreased m6A levels, mediated by the demethylase activity of FTO, were found to increase triglyceride deposition in HepG2 hepatocyte cells (Kang et al., 2018). Additional studies in HepG2 cells show that FTO mediated an increase in the expression of SREBP1c and CIDEC, two transcriptional regulators of lipogenesis, by increasing their nuclear localization, thereby promoting lipogenesis (Wang Y. et al., 2015; Chen A. et al., 2018). Increases in SREBP1c protein expression were found to be m6A-dependent, as mutant FTO was did not mediate changes in SREBP1c processing or protein expression (Chen A. et al., 2018). The mechanism by which FTO-mediated m6A demethylation mediates CIDEC expression remains unclear (Chen A. et al., 2018). In 3T3-L1 preadipocytes, FTO promoted adipogenesis through regulation of cell cycle progression (Wu et al., 2018). Mitotic clonal expansion (MCE) is a pivotal prerequisite process required for adipocyte differentiation and adipogenesis (Tang et al., 2003). Within this process, differentiating adipocytes are required to enter the cell cycle and proliferate (Tang et al., 2003). In 3T3-L1 cells with Fto knockdown, increased m6A levels resulted in decreased mRNA expression of cell-cycle control genes Ccna2 and Cdk2, which regulate the S to G2 transition (Wu et al., 2018). Subsequently, increased m6A levels on Ccna2 and Cdk2 mRNA transcripts resulted in m6A-dependent/YTHDF2-mediated decay of Ccna2 and Cdk2 mRNA and decreased CCNA2 and CDK2 protein expression (Wu et al., 2018). Decreased expression of CCNA2 and CDK2 resulted in the impairment of MCE and preadipocytes were unable to progress to the G2 phase, halting preadipocyte development (Wu et al., 2018). FTO has also been found to regulate RUNX1T1, an adipocyte transcription factor (Zhao et al., 2014). Within the Runx1t1 mRNA transcript, m6A was enriched at exonic regions near 5′ and 3′ splice sites; accordingly, increased m6A levels, mediated through Fto knockdown, led to increased binding by the splicing protein SRSF2, which resulted in changes in exon splicing and inclusion in the Runx1t1 transcript (Zhao et al., 2014; Zhang B. et al., 2020). The m6A-dependent roles of FTO in lipid and adipocyte metabolism are highlighted in Figure 3.
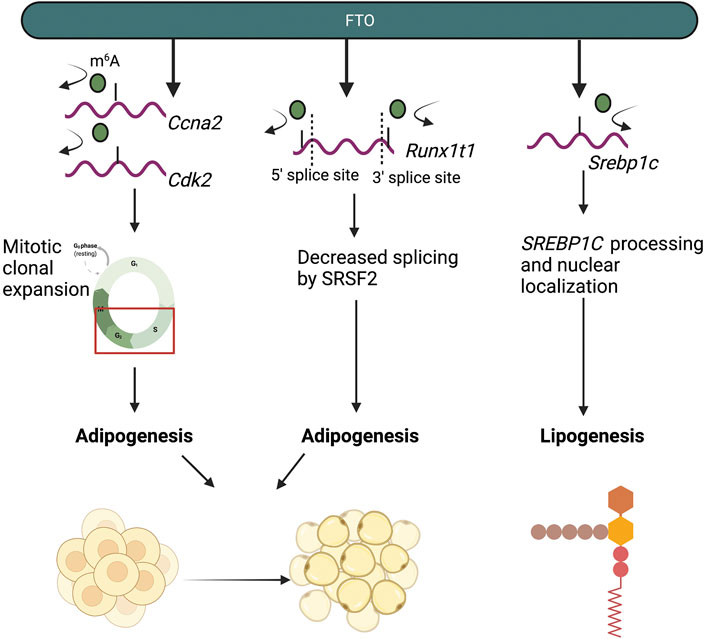
FIGURE 3. The m6A-dependent roles of FTO in adipocyte and lipid metabolism. FTO regulates and promotes adipogenesis and lipogenesis by demethylating mRNA transcripts of genes involved in adipogenesis (Ccna2, Cdk2, Runx1t1) and lipogenesis (Srebp1c).
FTO may also serve a role in regulating gluconeogenesis. Increased glucose uptake induced the expression of Fto, resulting in overall decreases in m6A (Yang Y. et al., 2019). High FTO expression was also correlated with increased mRNA expression of genes involved in glucose and lipid metabolism, including Foxo1, G6pc, Dgat2, and Fasn, upon glucose stimulation (Jensen-Urstad and Semenkovich, 2012; Kousteni, 2012; Yang Y. et al., 2019). However, the mechanism by which FTO regulates the expression of Foxo1, G6pc, Dgat2, and Fasn was not explored in this study (Yang Y. et al., 2019). In another study, FTO was found to demethylate m6A sites on Foxo1, resulting in increased FOXO1 expression, and increased gluconeogenesis (Peng et al., 2019). Interestingly, Foxo1 mRNA expression was not changed by changes in FTO expression or activity (Peng et al., 2019). Rather, mutating an internal m6A site on the Foxo1 mRNA transcript prevented FTO-mediated increases in FOXO1, establishing that the internal m6A site on the Foxo1 mRNA transcript is required for the FTO-FOXO1 axis (Peng et al., 2019).
Furthermore, FTO and ALKBH5 may regulate metabolism in response to cellular stress through interactions with ATF4, a stress-response gene and major regulator of cellular metabolism. Under stress conditions, ATF4 expression increases (Zhou et al., 2018). However, upon Fto or Alkbh5 knockdown in MEF and 293T cells, ATF4 expression failed to increase upon amino acid starvation (Zhou et al., 2018). Mechanistically, m6A methylation on the 5′-UTR of Atf4 is dynamically changed in response to stress; increased m6A methylation on the Atf4 mRNA transcript due to Fto or Alkbh5 knockdown results in decreased ATF4 translation in response to amino acid starvation (Zhou et al., 2018). Conversely, Mettl3 or Mettl14 knockdown in this context resulted in increased ATF4 translation upon amino acid starvation, suggesting that the ATF4 regulation in response to starvation is m6A-dependent (Zhou et al., 2018).
Additionally, ALKBH5 was found to regulate lactate production in melanoma and colon cancer (Li et al., 2020). Knockdown of Alkbh5 in melanoma and colon cancer cells resulted in m6A-dependent decreases in the stability of Mct4/Slc16a3 mRNA, a regulator of lactate secretion (Li et al., 2020). Accordingly, Alkbh5 knockdown resulted in decreased lactate production in the tumor interstitial fluid of the tumor microenvironment in both melanoma and colon cancer (Li et al., 2020).
Readers
The role of readers in mediating metabolism requires future study. However, one study showed that m6A on mitochondrial carrier homology 2 (Mtch2) mRNA resulted in increased MTCH2 protein expression, resulting in increased adipogenesis in longissimus dorsi muscle cells taken from both lean Landrace-breed pigs and obese Jinhua-breed pigs (Jiang et al., 2019).
The role of m6A in metabolism is an exciting new field of interest. Few studies have addressed the role of other MTC proteins, such as WTAP and VIRMA, in regulating metabolism. Other potential directions the field could address include how the cellular microenvironment influences cell-intrinsic changes in m6A and how changes in m6A can contribute to changes in cancer cell metabolism.
Autophagy
Autophagy is a conserved cellular process that is mobilized during the stress response and within normal housekeeping functions. The autophagic process involves the removal and degradation of excessive or damaged organelles or proteins, as well as other biological molecules, into membrane-bound autophagosomes (Klionsky, 2007; Mizushima et al., 2008). m6A has been found to regulate autophagy; however, the discrete mechanisms and cellular contexts with which m6A influence autophagy remain unexplored (Frankel et al., 2017; Abildgaard et al., 2020).
Writers
The m6A writer METTL3 has been found to negatively regulate autophagy across several contexts. In HCC, METTL3 inhibited autophagy by depositing m6A at the 3′-UTR of Foxo3a, a negative regulator of autophagy (Lin et al., 2020). m6A on the 3′-UTR of Foxo3a resulted in downstream YTHDF1-mediated Foxo3a mRNA stabilization and subsequently inhibited autophagy (Lin et al., 2020). Furthermore, in an ischemic heart model, METTL3-mediated m6A on the 3′-UTR of Tfeb, which promotes autophagy and lysosome biogenesis, resulted in HNRNPD-mediated decreases in Tfeb mRNA expression and decreased autophagy (Song et al., 2019).
Autophagy can also promote therapeutic resistance and cell survival. Accordingly, METTL3 promoted resistance to gefitinib in NSCLC through regulation of two core autophagy genes, Atg5 and Atg7 (Glick et al., 2010; Liu S. et al., 2020). Accordingly, Mettl3 knockdown in NSCLC cells resulted in decreased Atg5 and Atg7 mRNA expression (Liu S. et al., 2020). However, whether this regulation was m6A-dependent is unclear (Liu S. et al., 2020).
Additionally, in human keratinocytes, METTL14 abundance was found to be down-regulated by UVB exposure through NBR1-mediated selective autophagy (Yang Z. et al., 2021). Furthermore, mTORC1, a negative regulator of autophagy, promoted the stabilization of the MTC consisting of METTL3, METTL14, WTAP, and RMB15/RBM15B (Tang et al., 2021). Mechanistically, mTORC1 promoted the stabilization of the MTC by regulating the chaperonin CCT, which facilitates protein folding and stabilization of the MTC in Drosophila S2R+ and human HEK293T cells (Tang et al., 2021). The mechanism by which mTORC1 regulates CCT is detailed further elsewhere (Tang et al., 2021). Increased stabilization of the MTC led to increased m6A deposition and mRNA degradation of two downstream targets, autophagy genes Atg1 and Atg8a, resulting in the suppression of autophagy (Tang et al., 2021).
Furthermore, one study found that WTAP could regulate autophagy by mediating the phosphorylation of the positive autophagy regulator, AMPK, in HCC cells (Li G. et al., 2021). Mechanistically, WTAP decreased the mRNA stability of Lkb1, the kinase upstream of AMPK which regulates AMPK phosphorylation, in an m6A-dependent manner (Li G. et al., 2021). Subsequently, knockdown of Wtap resulted in increased autophagy (Li G. et al., 2021).
Erasers
The role of the m6A eraser FTO as a regulator of autophagy has been studied in a variety of different contexts. In HeLa cells, knockdown of Fto decreased autophagic flux (Jin et al., 2018). Interestingly, only the catalytically active form of FTO was able to increase autophagic flux, evidenced by increased LC3B puncta in cells expressing wild-type, but not catalytically inactive mutant, FTO, which suggests that FTO regulates autophagy in an m6A-dependent manner (Jin et al., 2018). RNA immunoprecipitation (RIP)-qPCR revealed that FTO binds to Ulk1 mRNA, a gene involved in the initial stages of autophagy and is an important recruitment factor in autophagosome formation (Zachari and Ganley, 2017; Jin et al., 2018). The interaction between FTO and Ulk1 was further elucidated as three m6A sites were found in the 3′-UTR of the Ulk1 transcript, which were subsequently targeted for degradation by YTHDF2 (Jin et al., 2018). FTO-mediated demethylation of Ulk1 may therefore preserve Ulk1 from YTHDF2-mediated degradation (Jin et al., 2018). In addition to ULK1, FTO may also preserve core autophagy genes, Atg5 and Atg7, from YTHDF2-mediated degradation in adipocytes (Glick et al., 2010; Wang et al., 2020). Knockdown of Fto in 3T3-L1 cells increased m6A levels across the Atg5 and Atg7 mRNA transcripts, resulting in YTHDF2-mediated degradation and inhibition of autophagy (Wang et al., 2020). Interestingly, knockdown of Fto in 3T3-L1 cells did not change m6A levels on ULK1, emphasizing the cell-type dependent regulation of autophagy by FTO (Wang et al., 2020).
FTO has also been shown to be a target for p62-dependent selective autophagy. In human keratinocytes, FTO protein expression was stabilized and up-regulated by arsenic exposure through inhibition of p62-mediated autophagy (Cui et al., 2021).
Furthermore, the m6A eraser ALKBH5 was found to promote autophagy in Leydig cells (Chen Y. et al., 2020). Mechanistically, m6A promoted the translation of PPM1A, a negative AMPK regulator, in a YTHDF1-dependent manner (Chen Y. et al., 2020). Furthermore, m6A also reduced the mRNA stability of Camkk2, a positive AMPK regulator, in a YTHDF2-dependent manner, resulting in autophagy inhibition (Chen Y. et al., 2020). However, decreased m6A methylation, mediated by ALKBH5, resulted in autophagy induction by preventing Camkk2 mRNA decay (Chen Y. et al., 2020).
Readers
Few studies have examined the role of only m6A readers in autophagy. YTHDC1 was found to regulate autophagy in human keratinocytes treated with high glucose, as knockdown of Ythdc1 resulted in decreased autophagic flux (Liang et al., 2021). Mechanistically, YTHDC1 promoted mRNA stability of the autophagy receptor Sqstm1 in an m6A-dependent manner (Liang et al., 2021). Accordingly, knockdown of Ythdc1 resulted in Sqstm1 mRNA degradation, leading to decreased autophagic flux (Liang et al., 2021). Furthermore, in HCC, YTHDF1 was identified to positively regulate autophagy by promoting the translation of core autophagy proteins ATG2A and ATG14 under hypoxic conditions in an m6A-dependent manner (Li Q. et al., 2021).
As this is a relatively new field of research, future studies are needed to identify the context-dependent role of m6A at different stages in the autophagic process, from the formation of phagophore, autophagosome, and autolysosome, to cargo degradation in the lysosomes, and identify the different cellular stressors and stimuli that mediate dynamic m6A changes within this process (Glick et al., 2010).
DNA Damage Response
Elucidating the communication between m6A and DNA damage response (DDR) is an active area of study. The role of m6A in modulating these pathways will add new insights into the DDR machinery.
Writers
In response to UVC or UVA radiation, m6A and DNA Pol κ were rapidly recruited to sites of DNA damage (Xiang et al., 2017). While the detailed mechanism behind the role of DNA Pol κ in DDR is not fully understood, the catalytic activity of METTL3 was found to be required for DNA Pol κ recruitment to DNA damage sites (Xiang et al., 2017). Knockout of Mettl3 in HeLa and U2OS cells exposed to UV radiation resulted in decreased cyclobutene pyrimidine dimer (CPD) removal, a major UV damage product, (Xiang et al., 2017). Knockout of Mettl14 in human keratinocyte cell lines HaCaT and normal human epidermal keratinocytes (NHEK) cells also resulted in decreased CPD removal upon UVB irradiation (Yang Z. et al., 2021). Interestingly, another study found that m6A was recruited to DNA damage lesions only in the presence of CPDs in response to UVA or UVB radiation, suggesting that m6A and m6A-associated enzymes may specifically recognize CPDs (Svobodová Kovaříková et al., 2020).
Furthermore, in response to UVA exposure, m6A RNA modifications may serve in the nucleotide excision repair pathway, but not the non-homologous end-joining (NHEJ) pathway, as treatment with an inhibitor of SUV40H1/H2, which are NHEJ-specific enzymes, had no effect on m6A recruitment (Svobodová Kovaříková et al., 2020). Interestingly, however, knockout of other NHEJ enzymes SUV39H1/H2, did decrease the recruitment of m6A in response to UVA exposure, suggesting that the role of m6A in NHEJ may be complex (Svobodová Kovaříková et al., 2020). In response to UVB exposure, METTL14 inhibited UVB-induced skin tumorigenesis by regulating global genome repair (GGR) in human keratinocyte cell lines (Yang Z. et al., 2021). Mechanistically, METTL14 regulated the m6A deposition onto the DNA damage repair gene Ddb2 and promoted YTHDF1-mediated DDB2 translation, and subsequent knockdown of Mettl14 resulted in decreased DDB2 abundance in an m6A-dependent manner (Yang Z. et al., 2021). The discrete epitranscriptomic mechanisms that underlie the DDR in response to UVA, UVB, and UVC exposure remains unclear.
Interestingly, in response to double-stranded breaks (DSBs), which were induced by X-ray radiation or Zeocin treatment, a chemical that induces DSBs, METTL3 was activated and phosphorylated at S43 by the key DDR kinase ATM, which then localized to DNA damage sites (Zhang C. et al., 2020). At these DNA-damage sites, METTL3 deposited m6A onto DNA damage-associated RNA, forming a DNA-RNA hybrid (Zhang C. et al., 2020). Accordingly, knockdown of Mettl3 resulted in decreased homologous recombination, a key process in the double-stranded break repair process (Zhang C. et al., 2020). YTHDC1 was also recruited to the DNA-RNA hybrid sites, potentially serving to preserve m6A deposition on these RNA hybrids (Zhang C. et al., 2020). The DNA-RNA hybrids then recruited the DNA damage-associated proteins RAD51 and BRCA in order to initiate HR (Zhang C. et al., 2020).
Furthermore, in the NCCIT stem cell line, VIRMA was found to promote resistance to cisplatin through regulation of the DNA damage response (Miranda-Gonçalves et al., 2021). Accordingly, knockdown of VIRMA resulted in increased expression of DNA repair proteins, including
In addition to m6A writers, METTL16, a methyltransferase that targets non-coding RNAs, including U6 small nuclear RNA, was also recruited to sites of DNA damage at a later time point (20–30 mins post UVA/UVC micro-irradiation) (Svobodová Kovaříková et al., 2020). However, the substrates of METTL16 in response to UV radiation were not explored in this study (Svobodová Kovaříková et al., 2020).
Erasers
In response to metabolic stress, UVC, and H2O2 treatment, the m6A eraser FTO increased the mRNA stability of DNA repair pathway genes, including Hspa1a (Hsp70), in osteoblasts (Zhang Q. et al., 2019). Increased mRNA stability and expression of Hspa1a served to protect osteoblasts from NF-κ β-mediated apoptosis (Zhang Q. et al., 2019). While the Hspa1a mRNA transcript does contain m6A sites, this study did not address whether FTO promotes Hspa1a mRNA stability in an m6A-dependent manner (Zhang Q. et al., 2019).
The role of m6A writers and erasers in the DNA damage response is highlighted in Figure 4.
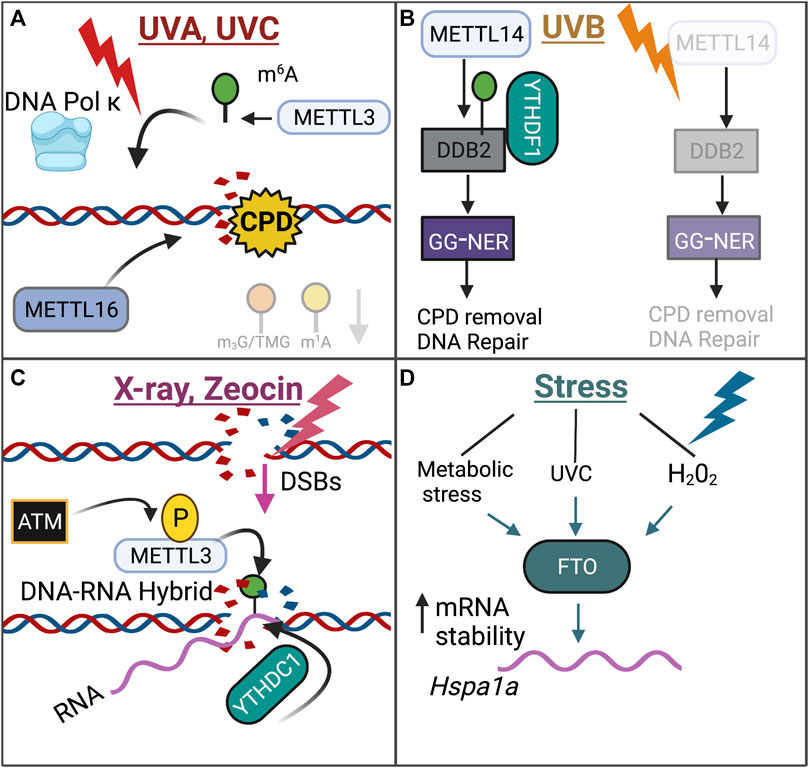
FIGURE 4. Review of the role of m6A within the DNA Damage Response. The role of m6A with the DNA Damage Response is dependent on the DNA damaging agent, highlighted through the distinct mechanisms that are employed upon exposure to UVA/UVC (A), UVB (B), X-ray/Zeocin (C), or Stress (D).
The role of m6A in mediating DNA damage response is an emerging field of interest. There are several gaps in this field that should be addressed accordingly. One major area of interest is elucidating the specific roles of m6A readers in the DNA damage response. There is limited research on this topic. While m6A readers have been implicated in DDR, it has only been in the context of their communication with the m6A writers and erasers. Furthermore, another area of interest is to further elucidate the function of the m6A machinery in response to chronic vs. acute DNA damage in response to genotoxic agents such as UV radiation, arsenic, chemotherapy, and ionizing radiation.
Liquid-Liquid Phase Separation
Liquid-liquid phase separation (LLPS) involves the formation of biological condensates containing aggregates of proteins or nucleic acids within the cell (Lyon et al., 2021). Biophysical mechanisms of liquid-liquid phase separation formation are discussed elsewhere (Lyon et al., 2021). Recent work has uncovered fascinating roles for these condensates in cellular functions, including involvement in stress responses, diseases, and aging (Alberti and Hyman, 2021; Lyon et al., 2021).
Readers
YTHDF readers YTHDF1 and YTHDF3 are believed to be critical in mediating LLPS as depletion of Ythdf1 or Ythdf3 prevented stress granule (SG) formation (Ries et al., 2019; Fu and Zhuang, 2020). There are contrasting reports on the role of YTHDF2 in LLPS and SG formation, as sodium-arsenite-induced SGs required YTHDF2, but not oxidative-stress-induced SG (Ries et al., 2019; Fu and Zhuang, 2020). These contrasting reports highlight that the role of YTHDF2 in LLPS and SG formation may be context-dependent (Ries et al., 2019; Fu and Zhuang, 2020). Biophysically, YTHDF1/3 are hypothesized to facilitate LLPS by lowering the activation energy input needed for phase separation (Fu and Zhuang, 2020). Alternatively, another hypothesis states YTHDF1/3 may serve as shell proteins that promote SG formation (Fu and Zhuang, 2020). However, the mechanism by which YTHDF1 and YTHDF3 mediate LLPs and SG formation is unclear (Fu and Zhuang, 2020). Furthermore, YTHDF3 has also been found to promote triaging of mRNAs into SGs in response to oxidative stress (Anders et al., 2018). Under these conditions, mRNA transcripts are dynamically patterned with m6A at the 5′-UTR and 5′-CDS regions and are partitioned into stress granules by YTHDF3, and are prevented from undergoing translation (Anders et al., 2018).
In addition to YTHDF1 and YTHDF3, two independent studies have established a role for YTHDC1 in LLPS. YTHDC1 is structurally made up of N or C-terminal internally-disordered regions (IDRs), which are believed to be necessary for YTHDC1’s role in LLPS (Lee J.-H. et al., 2021; Cheng et al., 2021). In MEF, 293T, and HeLa cells, m6A-eRNAs, which localize to active enhancer regions, recruited YTHDC1 to form YTHDC1-BRD4 condensates (Lee J.-H. et al., 2021). Additionally, YTHDC1 also formed m6A-YTHDC1 condensates, termed nYACs, in AML cells (Cheng et al., 2021). In this context, the number of nYACs increased in AML cells, as compared to normal hematopoietic cells, and also promoted tumorigenesis by promoting an undifferentiated state and cell survival (Cheng et al., 2021). Furthermore, nYACs can influence mRNA metabolism by preventing m6A-decorated mRNAs from being degraded by the PAXT-exosome complex (Cheng et al., 2021). The role of LLPS in tumorigenesis remains an emerging area of interest and is detailed further (Jiang et al., 2020).
It is important to note that whether m6A is critical for LLPS and stress granule formation remains controversial. One study found that m6A on mRNAs promoted YTHDF1-3 partitioning into phase-separated structures (Ries et al., 2019). However, a more recent study demonstrated that mRNAs, with or without m6A modifications, show minor differences in partitioning to stress granules and therefore argue that m6A may only play a minor role in stress granule partitioning (Khong et al., 2021). The authors of this study hypothesize that it is not only m6A-YTHDF interactions that promote stress granule partitioning, but rather, there may be several other RNA-protein interactions outside of m6A-YTHDF that promote stress granule portioning (Khong et al., 2021). The identity and nature of these RNA-protein interactions remain unclear.
The field of LLPS is rapidly expanding and research into this topic breaches disciplines in biophysics, biochemistry, disease biology, as well as epitranscriptomics. Of the many gaps of knowledge within this field, expanding the role of m6A machinery in LLPS, namely specific m6A writers and erasers, remains paramount.
A summary of the role of m6A modification in cellular responses covered in this review can be found in Figure 5.
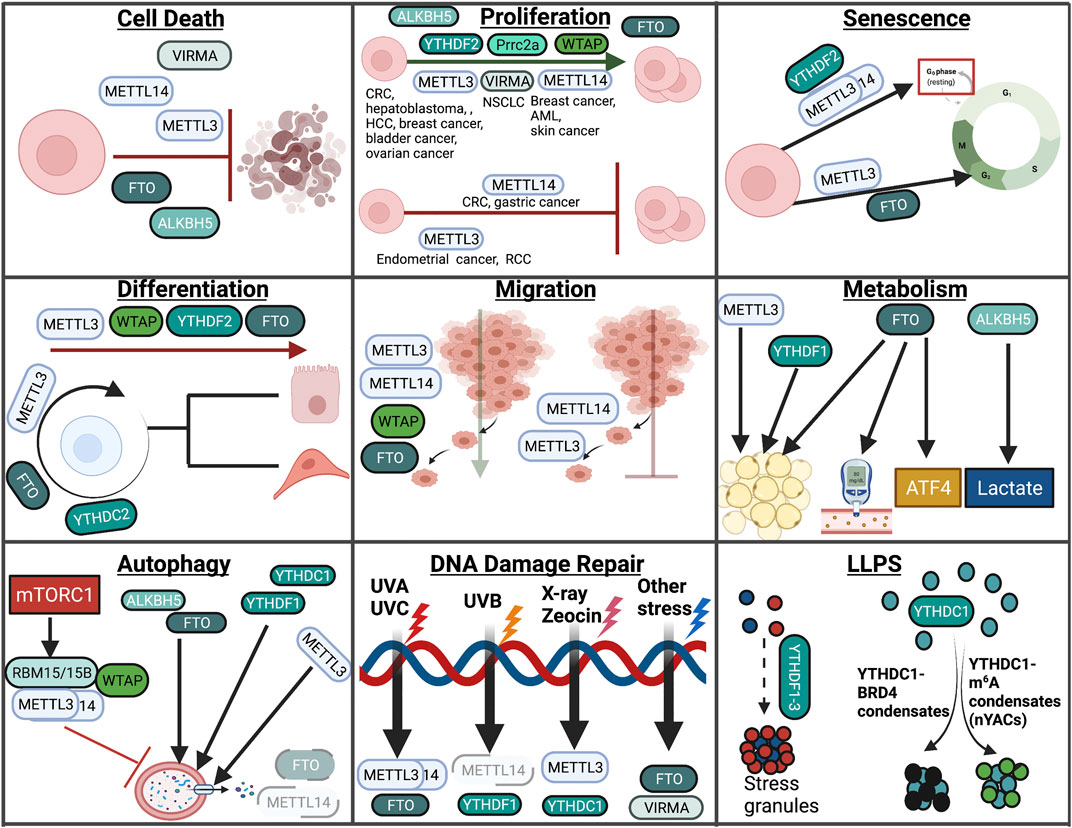
FIGURE 5. Overview of m6A in cellular processes. In this review, we highlight the role of RNA modifications in cellular processes such as cell death, proliferation, senescence, differentiation, migration, metabolism, autophagy, the DNA damage response, and LLPS. Within these processes, RNA modifications assume unique and context-dependent functions.
The Role of m5C in Diverse Cellular Functions
m5C has been implicated in several cellular contexts, including cell death, proliferation, senescence, differentiation, migration, metabolism, and DDR (Figure 6). The role of m5C in autophagy and LLPS has not been studied extensively and will therefore not be covered in this section. The role of m5C in autophagy and LLPS represents a gap of knowledge within this growing field and therefore requires further studies.
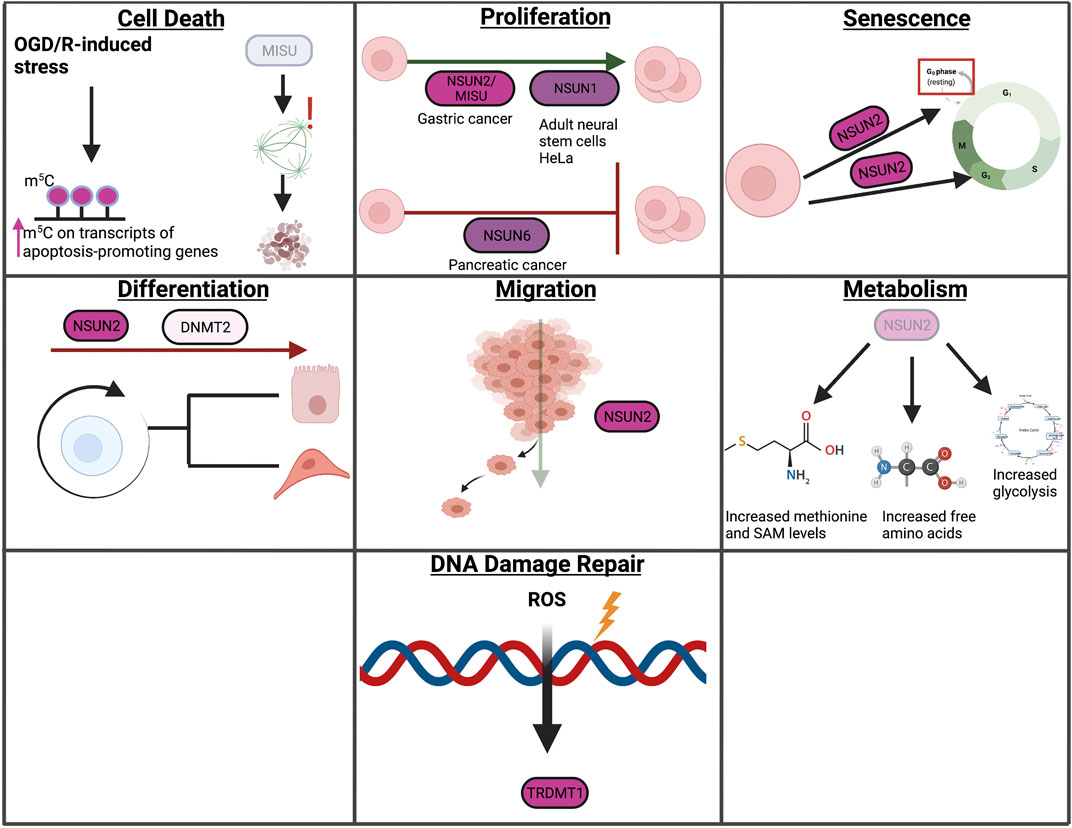
FIGURE 6. Overview of m5C in cellular processes. In this review, we highlight the role of RNA modifications in cellular processes such as cell death, proliferation, senescence, differentiation, migration, metabolism, and the DNA damage response. Within these processes, RNA modifications assume unique and context-dependent functions.
Cell Death
Writers
The role of m5C in cell death is not widely explored. Accordingly, one study employing an oxygen-glucose deprivation/reoxygenation (OGD/R) model in neurons found that m5C-methylated sites were increased upon OGD/R (Jian et al., 2021). Furthermore, RNA bisulfite sequencing revealed that m5C hypermethylated transcripts after OGD/R were functionally involved in apoptosis (Jian et al., 2021). While the functional significance of these findings remains to be explored, these results suggest that m5C methylation may be important in mediating apoptosis in response to cellular stress mediated by OGD/R (Jian et al., 2021). Furthermore, MISU, an NSUN2 homolog, was identified as a critical regulator of mitotic integrity; accordingly, MISU depletion resulted in apoptosis, potentially through decreased spindle integrity and induction of aneuploidy (Hussain et al., 2009).
Proliferation
Writers
m5C RNA methylation may also have important roles in cell proliferation. Similar to trends seen with m6A, the role of m5C RNA modifications in cell proliferation have been studied primarily in the context of cancer.
Low expression of NSUN6, an m5C methyltransferase, was found in pancreatic cancer patients, and may contribute to pancreatic cancer cell proliferation through regulation of CDK10 (Yang R. et al., 2021). While decreased NSUN6 expression was correlated with decreased CDK10 expression, resulting in increased cell proliferation, this study did not establish whether NSUN6 regulates CDK10 expression in an m5C-dependent manner (Yang R. et al., 2021).
High levels of NSUN2, another m5C methyltransferase, have been found in several different cancer types, but the functional role of NSUN2 in regulating proliferation was not clear (Okamoto et al., 2012). A recent study found in gastric cancer found that high NSUN2 levels were associated with worse overall survival, and that knockdown of Nsun2 resulted in decreased proliferation in gastric cancer cells (Hu et al., 2021). NSUN2 protein stability was found to be regulated by SUMOylation, a post-translational modification (Hu et al., 2021). Interestingly, increased proliferation was also noted upon Nsun2 overexpression in these cells, using both wild-type and catalytically inactive mutant NSUN2, suggesting that NSUN2 may have both m5C-dependent and m5C-independent functions in promoting proliferation (Hu et al., 2021). RNA bisulfite sequencing revealed that NSUN2-dependent m5C methylated transcripts were involved in oncogenic pathways, including the RAP1 pathway, as well as pathways involved in drug resistance and cell cycle (Hu et al., 2021). Furthermore, NSUN2 promoted proliferation in U2OS cells by depositing m5C onto Cdk1 transcripts, resulting in increased CDK1 translation (Xing et al., 2015). The role of NSUN2 in promoting proliferation is further detailed elsewhere (Wang, 2016).
Interestingly, MISU, a NSUN2 homolog, was identified as a MYC target and mediated MYC-induced cell growth in human epidermis cells (Frye and Watt, 2006). However, the mechanism by which MISU regulates MYC-induced proliferation is unclear (Frye and Watt, 2006).
Furthermore, the expression of NSUN1, alternatively known as NOP2, was increased in adult neural stem cells after stroke and was positively correlated with adult neural stem cell proliferation, suggesting a potential role for NSUN1 in promoting recovery after stroke (Kosi et al., 2015). Additionally, NSUN1, also known as NOL1, promoted proliferation by binding to the Ccnd1 promoter and promoting Ccnd1 transcription in HeLa cells (Hong et al., 2016). Whether NSUN1 promoted the proliferation in an m5C-dependent manner in these studies was not established.
Senescence
Writers
The m5C writer NSUN2 has been found to promote senescence in a variety of contexts. In HeLa cells, METTL3/14 and NSUN2 cooperated to increase p21 translation in response to oxidative stress, ultimately leading to the induction of cellular senescence (Li Q. et al., 2017). NSUN2 also promoted oxidative-stress-induced cellular senescence in human umbilical vein endothelial cells through m5C methylation of Shc mRNA, which led to increased SHC protein expression, activation of p38/MAPK, and increased ROS levels, thereby establishing a positive feedback loop (Cai et al., 2016). However, the role of NSUN2 in regulating senescence may be context and stimuli-dependent. In human diploid fibroblasts, NSUN2 negatively regulated senescence by methylating p27KIP1, a CDK inhibitor, at the 5′-UTR, resulting in decreased p27 translation and increased CDK1 translation (Tang et al., 2015). By indirectly promoting CDK1 translation, NSUN2 served to promote cellular proliferation and inhibit cellular senescence (Tang et al., 2015).
Differentiation
Writers
NSUN2 levels are highly expressed in undifferentiated epidermal progenitor cells (Sajini et al., 2019). Vault tRNAs (vtRNAs) are RNA POLIII-derived transcripts that make up vault ribonucleoproteins, and can be processed into smaller regulatory RNAs (svRNAs) (Stadler et al., 2009; Sajini et al., 2019). Accordingly, Sajini et al. found that processing of vtRNA VTRNA1.1 is dependent on NSUN2-dependent m5C methylation and is critical for proper epidermal cell development (Sajini et al., 2019). NSUN2-mediated m5C on tRNAs is also believed to be required for epidermal stem cell, testis, and neural stem cell differentiation (Blanco et al., 2011; Hussain et al., 2013; Flores et al., 2017). Due to the important role of NSUN2 in promoting neural stem cell differentiation, loss of Nsun2 is linked to several developmental disorders (Flores et al., 2017). DNMT2-mediated m5C on tRNA is believed to be required for hematopoiesis, as Dnmt2-deficient mice showed decreased stem and progenitor cell populations (Tuorto et al., 2015). The role of m5C in differentiation and development is summarized elsewhere (Song et al., 2021).
Migration
Writers
In addition to m6A writers, the m5C writer NSUN2 promoted the mRNA translation of autotaxin (Atx) in U87 glioma cells in an m5C-dependent manner (Xu et al., 2020). NSUN2 deposited m5C at the 3′-UTR of Atx, enhancing Atx translation and promoting the export of Atx from the nucleus to the cytoplasm through coordination with m5C reader ALYREF (Xu et al., 2020). Downstream, ATX then converts lysophosphatidylcholine to lysophosphatidic acid, a lipid that has been to promote migration and overall tumorigenicity (Valdés-Rives and González-Arenas, 2017; Xu et al., 2020). Furthermore, NSUN2 was also found to promote migration in gastric cancer cells (Hu et al., 2021).
Metabolism
Writers
Nsun2−/− mice resulted in changes in the metabolism of methionine and amino acids, and the TCA cycle (Gkatza et al., 2019). Nsun2 deletion resulted in increased methionine and S-adenosyl-methionine (SAM) levels (Gkatza et al., 2019). Furthermore, free amino acid levels were increased upon loss of Nsun2, which was hypothesized to indicate overall decreases in translation (Gkatza et al., 2019). Additionally, Nsun2 loss resulted in a metabolic shift towards glycolysis (Gkatza et al., 2019). Taken together, the authors hypothesized that Nsun2 loss results in the induction of a catabolic state, and that NSUN2 functions to promote an anabolic fate (Gkatza et al., 2019).
DNA Damage Response
Writers
Interestingly, Chen et al. found that m5C was localized to DSBs upon ROS-induced DNA damage, and is present at DNA-damage-induced DNA-RNA hybrids (Chen H. et al., 2020). Interestingly, tRNA methyltransferase TRDMT1 was also found to localize to DNA-damage-induced DNA-RNA hybrids and was hypothesized to serve as a damage-induced m5C methyltransferase (Chen H. et al., 2020). Together, TRDMT1 and m5C are believed to be necessary to mediate homologous recombination in response to DNA damage (Chen H. et al., 2020; Zhu X. et al., 2021).
The Role of m1A in Diverse Cellular Functions
m1A has been studied in several cellular contexts, including, cell death, proliferation, senescence, migration, metabolism, DDR, and LLPS (Figure 7). The role of m1A in differentiation in autophagy has not been studied extensively and will therefore not be covered in this section. The role of m1A in differentiation and autophagy requires further study.
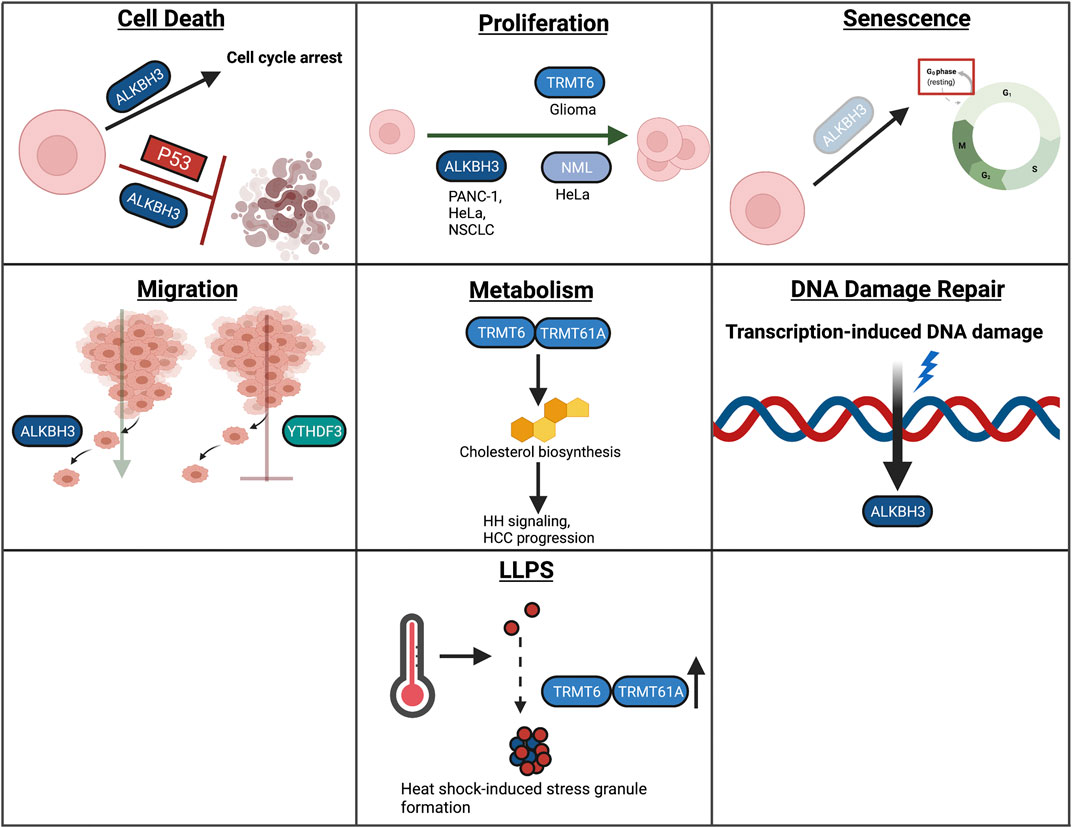
FIGURE 7. Overview of m1A in cellular processes. In this review, we highlight the role of RNA modifications in cellular processes such as cell death, proliferation, senescence, migration, metabolism, the DNA damage response, and LLPS. Within these processes, RNA modifications assume unique and context-dependent functions.
Cell Death
Erasers
In NSCLC, knockdown of the m1A eraser Alkbh3 promoted cell cycle arrest (Kogaki et al., 2017). Interestingly, knockdown of both Alkbh3 and Tp53 resulted in the induction of apoptosis, suggesting that TP53 may be critical for shifting cell fate from cell cycle arrest to undergoing apoptosis (Kogaki et al., 2017). However, whether ALKBH3 mediates this function as an RNA or DNA methyltransferase was not elucidated (Kogaki et al., 2017).
Proliferation
Writers
Many studies have found positive associations with m1A regulators, such as the m1A methyltransferase TRMT6, and cancer (Wang et al., 2019; Shi et al., 2020; Wang B. et al., 2021). However, few studies have established the biological mechanisms by which m1A regulators assert their oncogenic function. Inhibition of Trmt6 resulted in decreased proliferation in glioma cell lines, establishing a potential oncogenic role for TRMT6 in regulating proliferation (Macari et al., 2016; Wang B. et al., 2021). Furthermore, inhibition of Alkbh3, an m1A demethylase, resulted in decreased proliferation in HeLa, PANC-1, and NSCLC cancer cells, suggesting a potential role for this demethylase in proliferation (Tasaki et al., 2011; Ueda et al., 2017; Chen et al., 2019). Furthermore, Waku et al. established that nucleomethylin (NML) can function as an m1A 28S rRNA methyltransferase, and that inhibition of NML results in decreased proliferation (Waku et al., 2016).
Senescence
Erasers
Few studies have explored the role of m1A in cellular senescence. One study, however, found that knockdown of Alkbh3 in NSCLC cells resulted in senescence induction and cell cycle arrest, followed by increased expression of cell cycle arrest proteins, p27 and p21 (Tasaki et al., 2011).
Migration
Erasers
Few studies have investigated the role of m1A and m1A regulators in migration. One study found that knockdown of Alkbh3 in HeLa cells resulted in decreased invasion (Chen et al., 2019). Furthermore, YTHDF3, serving as an m1A reader, was found to inhibit invasion in HTR8/SVneo cells by promoting the mRNA decay of Igf1r (Zheng et al., 2020). IGF1R is upstream of the pro-migratory MMP9 signaling pathway, and subsequent knockdown of Ythdf3 resulted in increased IGF1R and MMP9 expression, resulting in increased invasion and migration in these cells (Zheng et al., 2020).
Metabolism
Writers
As previously mentioned, TRMT6 has been found to be associated with oncogenesis in a variety of different cancers. In HCC, the TRMT6/TRMT61A m1A methyltransferase complex was identified to mediate m1A tRNA methylation, which resulted in increased PPAR
DNA Damage Response
Erasers
Knockdown of Alkbh3 in NSCLC resulted in increased phosphorylation of critical DDR factors ATM, ATR, and H2AX, suggesting that decreased ALKBH3 may promote single or double-stranded breaks (Kogaki et al., 2017). These DDR factors, as well as DNA-PKcs, were further upregulated in p53/Alkbh3 dual-knockout cells, establishing that p53 may be a critical regulator of ALKBH3 in mediating DDR (Kogaki et al., 2017). Whether the role of ALKBH3 in DDR is mediated through its demethylase function was not explored in this study (Kogaki et al., 2017). ALKBH3 has also been suggested to function as a DNA repair protein in response to transcription-induced DNA damage (Liefke et al., 2015). Furthermore, levels of m1A, found on small RNAs, were also noted to be decreased in UV-irradiated cells (Svobodová Kovaříková et al., 2020). However, the functional role of m1A on small RNAs in response to UV exposure was not detailed in this study (Svobodová Kovaříková et al., 2020).
LLPS
Writers
m1A methyltransferase TRMT61/61A and m1A were increased in heat-shock-induced, stress-granule-sequestered mRNAs (Alriquet et al., 2020). In response to stress, mRNAs can form irreversible protein aggregates (Alriquet et al., 2020). Conversely, m1A-patterned mRNAs were identified to be sequestered into reversible mRNA-protein aggregates, which can then undergo translation (Alriquet et al., 2020). Therefore, the authors hypothesize that m1A serves a protective role on mRNAs in response to stress (Alriquet et al., 2020).
Therapeutics Targeting RNA Modifications
Due to the pervasiveness of RNA modifications in disease, the development of targeted therapeutics remains critical and is an active area of research. Here, we will briefly summarize advances in the development of therapeutics targeting RNA modifications.
m6A-Targeted Therapeutics
Yankova et al. recently identified a small molecule inhibitor (STM2457) for METTL3 using a high throughput drug screen (Yankova et al., 2021). STM2547 was identified to be specific to METTL3 and did not disrupt the METTL3-METTL14 complex (Yankova et al., 2021). As METTL3 has been found to serve an oncogenic function in leukemia, the in vitro and in vivo efficacy of STM457 was explored as a therapeutic for AML (Vu et al., 2017). STM2457-treatment in AML cell lines resulted in decreased proliferation in a dose-dependent manner and decreased the colony forming capability of mouse AML cells (Yankova et al., 2021). Interestingly, STM2457 showed selectivity for AML cells, but did not affect CD34+ cells, hematopoietic stem and progenitor cells, or non-leukemogenic cell lines (Yankova et al., 2021). STM2457 also decreased the protein expression of oncogenic METTL3 targets, SP1 and BRD4 (Yankova et al., 2021).
Many small molecule inhibitors for FTO have been discovered, including rhein, NCDPCB, meclofenamic acid, MO-I-500, and fluorescein derivatives, among others (Chen et al., 2012; Wang T. et al., 2015; He et al., 2015; Huang et al., 2015; Singh et al., 2016). While these inhibitors inhibit FTO, clinical efficacy of FTO inhibitors has remained unclear. FTO has been found to serve as an oncogene in AML (Li Z. et al., 2017). Accordingly, two studies have developed FTO inhibitors targeting AML (Huang et al., 2019; Su et al., 2020). Huang et al. identified FB23-2 as a potential inhibitor for FTO (Huang et al., 2019). Treatment of AML cell lines for FB23-2 slightly decreased AML proliferation and promoted apoptosis, as well as promoted myeloid differentiation (Huang et al., 2019). FB23-2 treatment also resulted in minimal changes in proliferation in bone marrow cells derived from a healthy donor (Huang et al., 2019). Furthermore, FB23-2 showed promising therapeutic efficacy in mice, targeting both AML and leukemia stem cell populations (Huang et al., 2019). Furthermore, using a high throughput screen, Su et al. reported the discovery of two small molecule inhibitors targeting FTO, CS1 and CS2, with efficacy in targeting AML (Su et al., 2020). Treatment of AML cell lines with CS1 and CS2 resulted in decreased proliferation, increased apoptosis, and prevented the self-renewal capabilities of leukemia stem cells and leukemia initiating cells (Su et al., 2020). Treatment of healthy control cells showed no change (Su et al., 2020). To date, neither FB23-2 or CS1/CS2 have been employed in clinical trials. While Selberg et al. have described the development of a potential ALKBH5 inhibitor, further studies are needed to reconcile the cell-type specific effect of ALKBH5 inhibition (Selberg et al., 2021).
m5C-Targeted Therapeutics
m5C-directed therapeutics have also been explored. Few studies have explored the therapeutic potential of targeting m5C reader YBX1, but have only identified non-specific compounds that effectively target YBX1 (Shibata et al., 2020). Shibata et al. identified compounds, TAS0612 and everolimus, as potential compounds that target YBX1 phosphorylation (pYBX1) (Shibata et al., 2020). Increased YBX1 phosphorylation was found to be associated with resistance to fulvestrant, an antiestrogen commonly used to treat ER-positive breast cancer (Shibata et al., 2020). TAS0612 is a multi-kinase inhibitor that targets both the AKT/mTOR/p70S6K pathway, and pYBX1 was identified to be a downstream target of these pathways (Shibata et al., 2020). Everolimus is an mTORC1 inhibitor (Shibata et al., 2020). Accordingly, TAS0612 and everolimus treatment resulted in increased sensitivity to fulvestrant (Shibata et al., 2020). However, this study did not address whether changes in YBX1 phosphorylation changes m5C regulation (Shibata et al., 2020). Azacytidine is a well-established drug targeting DNMT2 DNA methylation (Stresemann and Lyko, 2008). However, one study, using bisulfite sequencing, argued that azacytidine may also target DNMT2-mediated tRNA methylation (Schaefer et al., 2009). However, follow-up studies are needed to identify whether azacytidine-mediated changes in tRNA methylation are due to m5C or other mechanisms (Schaefer et al., 2009).
m1A-Targeted Therapeutics
Wang et al. recently identified thiram as a potential candidate compound that selectively inhibits m1A writer complex TRMT6/TRMT61A (Wang Y. et al., 2021). Thiram treatment resulted in decreased oncosphere formation in HCC cell lines in vitro, and decreased tumor growth in vivo (Wang Y. et al., 2021). However, further pre-clinical studies are necessary to determine the safety of thiram treatment in patients, due to reported toxicities (Maita et al., 1991; Wang Y. et al., 2021).
Compound HUHS015 has been identified as an ALKBH3 inhibitor (Nakao et al., 2014). As previously mentioned, ALKBH3 has been found to serve an oncogenic role in many cancers, including prostate cancer (Liefke et al., 2015). HUHS015 has been found to decrease the growth of prostate cancer cell line DU145 and decreased tumor burden in xenograft models (Nakao et al., 2014; Mabuchi et al., 2015). To date, no clinical trial using HUHS015 has been employed.
Perspectives
While the roles of RNA modifications have been extensively studied for several cellular functions, there remain several areas of interest that are not well-established and require further examination.
Two areas of interest that remain open areas of research include evaluating the roles of RNA modifications in mediating specialized forms of cell death and within LLPS. In the area of cellular death, the role of RNA modifications has been well-studied in terms of apoptosis. However, emerging evidence suggests that RNA modifications may be important in mediating specialized forms of cell death including ferroptosis, pyroptosis, or other mechanisms of specialized cell death (Guo et al., 2020; Shen et al., 2021). Understanding the roles of RNA modifications in these specialized forms of cell death may lead to increased knowledge surrounding the cellular decisions that mediate these forms of cell death.
Furthermore, as previously mentioned, LLPS remains an emerging field of research. The field of LLPS encompasses the intersection of cell biology and biophysics; not only are the biophysical mechanisms by which these condensates form an active area of interest, but more recently, increased attention has been placed on detailing the role these condensates play within cellular processes. Increasing our understanding of RNA modifications in this process will aid in understanding the function and necessity of LLPS in mediating cellular functions.
The roles of other RNA modifications, other than m6A, within mammalian cellular processes is another gap of knowledge within the field of epitranscriptomics that remains critical to address. For example, while there are several studies that have identified cellular functions for pseudouridine in Drosophila, few studies have been done to explore the role of pseuoduridine in mammalian cellular functions (Vicidomini et al., 2015; Song et al., 2020).
Another area of interest that requires further study is understanding the cell-type specific function of the role of RNA modification in cellular functions. As demonstrated, not only do RNA modifications differ across cell types, but they can also differ across contexts, including across differentiation states and in response to stress. Understanding the relevance and pervasiveness of RNA modifications in these processes, and how different cell types adopt distinct mechanisms for RNA modifications across these functions, remains an important area of research.
In addition, due to the prevalence of RNA modifications in diverse cellular functions, the dysregulation of RNA modifications contributes to the etiology of several diseases. RNA modifications have been found to contribute to the pathologies of several diseases including cancer, diabetes, cardiovascular diseases, and developmental and neurological diseases. Increasing our understanding of the distinct roles that RNA modifications play in these cellular processes will allow for an increased understanding of disease etiology. While there are no therapeutics currently in clinical use that target RNA modifications, an increased understanding of their roles in disease etiology may contribute to the development of therapeutics that aim to selectively target this epitranscriptomic re-wiring.
Author Contributions
EW, Y-HC, and Y-YH wrote the manuscript.
Funding
Work in the authors’ laboratory was supported in part by NIH grants 5T32CA009594-33 (EW), ES031534 (Y-YH), ES024373 (Y-YH), ES030576 (Y-YH), the CACHET (NIH ES027792), the University of Chicago Comprehensive Cancer Center (NIH CA014599), the CTSA (NIH UL1 TR000430), and the University of Chicago Friends of Dermatology Endowment Fund.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We apologize to those investigators whose work could not be directly referenced owing to space limitations. We thank Ann Motten for her critical reading of the manuscript. All figures were created with Biorender.com.
References
Abildgaard, M. H., Brynjólfsdóttir, S. H., and Frankel, L. B. (2020). The Autophagy-RNA Interplay: Degradation and beyond. Trends Biochem. Sci. 45 (10), 845–857. doi:10.1016/j.tibs.2020.07.007
Alberti, S., and Hyman, A. A. (2021). Biomolecular Condensates at the Nexus of Cellular Stress, Protein Aggregation Disease and Ageing. Nat. Rev. Mol. Cel Biol 22 (3), 196–213. doi:10.1038/s41580-020-00326-6
Alriquet, M., Calloni, G., Martínez-Limón, A., Delli Ponti, R., Hanspach, G., Hengesbach, M., et al. (2020). The Protective Role of m1A during Stress-Induced Granulation. J. Mol. Cel Biol. 12 (11), 870–880. doi:10.1093/jmcb/mjaa023
Anders, M., Chelysheva, I., Goebel, I., Trenkner, T., Zhou, J., Mao, Y., et al. (2018). Dynamic m6A Methylation Facilitates mRNA Triaging to Stress Granules. Life Sci. Alliance 1 (4), e201800113. doi:10.26508/lsa.201800113
Bansal, H., Yihua, Q., Iyer, S. P., Ganapathy, S., Proia, D., Penalva, L. O., et al. (2014). WTAP Is a Novel Oncogenic Protein in Acute Myeloid Leukemia. Leukemia 28 (5), 1171–1174. doi:10.1038/leu.2014.16
Batista, P. J., Molinie, B., Wang, J., Qu, K., Zhang, J., Li, L., et al. (2014). m6A RNA Modification Controls Cell Fate Transition in Mammalian Embryonic Stem Cells. Cell Stem Cell 15 (6), 707–719. doi:10.1016/j.stem.2014.09.019
Bergsbaken, T., Fink, S. L., and Cookson, B. T. (2009). Pyroptosis: Host Cell Death and Inflammation. Nat. Rev. Microbiol. 7 (2), 99–109. doi:10.1038/nrmicro2070
Blanco, S., Kurowski, A., Nichols, J., Watt, F. M., Benitah, S. A., and Frye, M. (2011). The RNA-Methyltransferase Misu (NSun2) Poises Epidermal Stem Cells to Differentiate. Plos Genet. 7 (12), e1002403. doi:10.1371/journal.pgen.1002403
Bohnsack, K., Höbartner, C., and Bohnsack, M. (2019). Eukaryotic 5-methylcytosine (m5C) RNA Methyltransferases: Mechanisms, Cellular Functions, and Links to Disease. Genes 10 (2), 102. doi:10.3390/genes10020102
Boissel, S., Reish, O., Proulx, K., Kawagoe-Takaki, H., Sedgwick, B., Yeo, G. S. H., et al. (2009). Loss-of-function Mutation in the Dioxygenase-Encoding FTO Gene Causes Severe Growth Retardation and Multiple Malformations. Am. J. Hum. Genet. 85 (1), 106–111. doi:10.1016/j.ajhg.2009.06.002
Cai, J., Yang, F., Zhan, H., Situ, J., Li, W., Mao, Y., et al. (2019). RNA m6A Methyltransferase METTL3 Promotes the Growth of Prostate Cancer by Regulating Hedgehog Pathway. Ott 12, 9143–9152. doi:10.2147/OTT.S226796
Cai, X., Hu, Y., Tang, H., Hu, H., Pang, L., Xing, J., et al. (2016). RNA Methyltransferase NSUN2 Promotes Stress-Induced HUVEC Senescence. Oncotarget 7 (15), 19099–19110. doi:10.18632/oncotarget.8087
Cai, X., Wang, X., Cao, C., Gao, Y., Zhang, S., Yang, Z., et al. (2018). HBXIP-elevated Methyltransferase METTL3 Promotes the Progression of Breast Cancer via Inhibiting Tumor Suppressor Let-7g. Cancer Lett. 415, 11–19. doi:10.1016/j.canlet.2017.11.018
Cao, Y., Zhuang, Y., Chen, J., Xu, W., Shou, Y., Huang, X., et al. (2019). Dynamic Effects of Fto in Regulating the Proliferation and Differentiation of Adult Neural Stem Cells of Mice. Hum. Mol. Genet. 29 (5), 727–735. doi:10.1093/hmg/ddz274
Casella, G., Tsitsipatis, D., Abdelmohsen, K., and Gorospe, M. (2019). mRNA Methylation in Cell Senescence. Wiley Interdiscip. Rev. RNA 10 (6), e1547. doi:10.1002/wrna.1547
Chen, A., Chen, X., Cheng, S., Shu, L., Yan, M., Yao, L., et al. (2018a). FTO Promotes SREBP1c Maturation and Enhances CIDEC Transcription during Lipid Accumulation in HepG2 Cells. Biochim. Biophys. Acta (Bba) - Mol. Cel Biol. Lipids 1863 (5), 538–548. doi:10.1016/j.bbalip.2018.02.003
Chen, B., Ye, F., Yu, L., Jia, G., Huang, X., Zhang, X., et al. (2012). Development of Cell-Active N6-Methyladenosine RNA Demethylase FTO Inhibitor. J. Am. Chem. Soc. 134 (43), 17963–17971. doi:10.1021/ja3064149
Chen, G., Goto, Y., Sakamoto, R., Tanaka, K., Matsubara, E., Nakamura, M., et al. (2011). GLI1, a Crucial Mediator of Sonic Hedgehog Signaling in Prostate Cancer, Functions as a Negative Modulator for Androgen Receptor. Biochem. Biophysical Res. Commun. 404 (3), 809–815. doi:10.1016/j.bbrc.2010.12.065
Chen, H., Gao, S., Liu, W., Wong, C.-C., Wu, J., Wu, J., et al. (2021). RNA N6-Methyladenosine Methyltransferase METTL3 Facilitates Colorectal Cancer by Activating the m6A-GLUT1-mTORC1 Axis and Is a Therapeutic Target. Gastroenterology 160 (4), 1284–1300. doi:10.1053/j.gastro.2020.11.013
Chen, H., Yang, H., Zhu, X., Yadav, T., Ouyang, J., Truesdell, S. S., et al. (2020a). m5C Modification of mRNA Serves a DNA Damage Code to Promote Homologous Recombination. Nat. Commun. 11 (1), 2834. doi:10.1038/s41467-020-16722-7
Chen, J., Sun, Y., Xu, X., Wang, D., He, J., Zhou, H., et al. (2017). YTH Domain Family 2 Orchestrates Epithelial-Mesenchymal Transition/proliferation Dichotomy in Pancreatic Cancer Cells. Cell Cycle 16 (23), 2259–2271. doi:10.1080/15384101.2017.1380125
Chen, M., Wei, L., Law, C.-T., Tsang, F. H.-C., Shen, J., Cheng, C. L.-H., et al. (2018b). RNA N6-Methyladenosine Methyltransferase-like 3 Promotes Liver Cancer Progression through YTHDF2-dependent Posttranscriptional Silencing of SOCS2. Hepatology 67 (6), 2254–2270. doi:10.1002/hep.29683
Chen, X., Xu, M., Xu, X., Zeng, K., Liu, X., Pan, B., et al. (2020b). METTL14-mediated N6-Methyladenosine Modification of SOX4 mRNA Inhibits Tumor Metastasis in Colorectal Cancer. Mol. Cancer 19 (1), 106. doi:10.1186/s12943-020-01220-7
Chen, Y., Wang, J., Xu, D., Xiang, Z., Ding, J., Yang, X., et al. (2020c). m6A mRNA Methylation Regulates Testosterone Synthesis through Modulating Autophagy in Leydig Cells. Autophagy, 1–19. doi:10.1080/15548627.2020.1720431
Chen, Z., Qi, M., Shen, B., Luo, G., Wu, Y., Li, J., et al. (2019). Transfer RNA Demethylase ALKBH3 Promotes Cancer Progression via Induction of tRNA-Derived Small RNAs. Nucleic Acids Res. 47 (5), 2533–2545. doi:10.1093/nar/gky1250
Cheng, Y., Luo, H., Izzo, F., Pickering, B. F., Nguyen, D., Myers, R., et al. (2019). m6A RNA Methylation Maintains Hematopoietic Stem Cell Identity and Symmetric Commitment. Cel Rep. 28 (7), 1703–1716. doi:10.1016/j.celrep.2019.07.032
Cheng, Y., Xie, W., Pickering, B. F., Chu, K. L., Savino, A. M., Yang, X., et al. (2021). N6-Methyladenosine on mRNA Facilitates a Phase-Separated Nuclear Body that Suppresses Myeloid Leukemic Differentiation. Cancer Cell 39 (7), 958–972. doi:10.1016/j.ccell.2021.04.017
Choe, J., Lin, S., Zhang, W., Liu, Q., Wang, L., Ramirez-Moya, J., et al. (2018). mRNA Circularization by METTL3-eIF3h Enhances Translation and Promotes Oncogenesis. Nature 561 (7724), 556–560. doi:10.1038/s41586-018-0538-8
Cui, Y.-H., Yang, S., Wei, J., Shea, C. R., Zhong, W., Wang, F., et al. (2021). Autophagy of the m6A mRNA Demethylase FTO Is Impaired by Low-Level Arsenic Exposure to Promote Tumorigenesis. Nat. Commun. 12 (1), 2183. doi:10.1038/s41467-021-22469-6
Dahal, U., Le, K., and Gupta, M. (2019). RNA m6A Methyltransferase METTL3 Regulates Invasiveness of Melanoma Cells by Matrix Metallopeptidase 2. Melanoma Res. 29 (4), 382–389. doi:10.1097/cmr.0000000000000580
Deng, R., Cheng, Y., Ye, S., Zhang, J., Huang, R., Li, P., et al. (2019). m6A Methyltransferase METTL3 Suppresses Colorectal Cancer Proliferation and Migration through P38/ERK Pathways. Ott 12, 4391–4402. doi:10.2147/OTT.S201052
Ding, C., Zou, Q., Ding, J., Ling, M., Wang, W., Li, H., et al. (2018). Increased N6‐methyladenosine Causes Infertility Is Associated with FTO Expression. J. Cel Physiol 233 (9), 7055–7066. doi:10.1002/jcp.26507
Einstein, J. M., Perelis, M., Chaim, I. A., Meena, J. K., Nussbacher, J. K., Tankka, A. T., et al. (2021). Inhibition of YTHDF2 Triggers Proteotoxic Cell Death in MYC-Driven Breast Cancer. Mol. Cel 81 (15), 3048–3064. doi:10.1016/j.molcel.2021.06.014
Elmore, S. (2007). Apoptosis: A Review of Programmed Cell Death. Toxicol. Pathol. 35 (4), 495–516. doi:10.1080/01926230701320337
Esteve-Puig, R., Bueno-Costa, A., and Esteller, M. (2020). Writers, Readers and Erasers of RNA Modifications in Cancer. Cancer Lett. 474, 127–137. doi:10.1016/j.canlet.2020.01.021
Fei, Q., Zou, Z., Roundtree, I. A., Sun, H.-L., and He, C. (2020). YTHDF2 Promotes Mitotic Entry and Is Regulated by Cell Cycle Mediators. Plos Biol. 18 (4), e3000664. doi:10.1371/journal.pbio.3000664
Flores, J. V., Cordero-Espinoza, L., Oeztuerk-Winder, F., Andersson-Rolf, A., Selmi, T., Blanco, S., et al. (2017). Cytosine-5 RNA Methylation Regulates Neural Stem Cell Differentiation and Motility. Stem Cel. Rep. 8 (1), 112–124. doi:10.1016/j.stemcr.2016.11.014
Frankel, L. B., Lubas, M., and Lund, A. H. (2017). Emerging Connections between RNA and Autophagy. Autophagy 13 (1), 3–23. doi:10.1080/15548627.2016.1222992
Frayling, T. M., Timpson, N. J., Weedon, M. N., Zeggini, E., Freathy, R. M., Lindgren, C. M., et al. (2007). A Common Variant in the FTO Gene Is Associated with Body Mass index and Predisposes to Childhood and Adult Obesity. Science 316 (5826), 889–894. doi:10.1126/science.1141634
Frye, M., Harada, B. T., Behm, M., and He, C. (2018). RNA Modifications Modulate Gene Expression during Development. Science 361 (6409), 1346–1349. doi:10.1126/science.aau1646
Frye, M., and Watt, F. M. (2006). The RNA Methyltransferase Misu (NSun2) Mediates Myc-Induced Proliferation and Is Upregulated in Tumors. Curr. Biol. 16 (10), 971–981. doi:10.1016/j.cub.2006.04.027
Fu, Y., and Zhuang, X. (2020). m6A-binding YTHDF Proteins Promote Stress Granule Formation. Nat. Chem. Biol. 16 (9), 955–963. doi:10.1038/s41589-020-0524-y
Fukusumi, Y., Naruse, C., and Asano, M. (2008). Wtap Is Required for Differentiation of Endoderm and Mesoderm in the Mouse Embryo. Dev. Dyn. 237 (3), 618–629. doi:10.1002/dvdy.21444
Garcia-Saez, I., Lacroix, F. B., Blot, D., Gabel, F., and Skoufias, D. A. (2011). Structural Characterization of HBXIP: the Protein that Interacts with the Anti-apoptotic Protein Survivin and the Oncogenic Viral Protein HBx. J. Mol. Biol. 405 (2), 331–340. doi:10.1016/j.jmb.2010.10.046
Gkatza, N. A., Castro, C., Harvey, R. F., Heiß, M., Popis, M. C., Blanco, S., et al. (2019). Cytosine-5 RNA Methylation Links Protein Synthesis to Cell Metabolism. Plos Biol. 17 (6), e3000297. doi:10.1371/journal.pbio.3000297
Glick, D., Barth, S., and Macleod, K. F. (2010). Autophagy: Cellular and Molecular Mechanisms. J. Pathol. 221 (1), 3–12. doi:10.1002/path.2697
Graille, M. (2022). Division of Labor in Epitranscriptomics: What Have We Learnt from the Structures of Eukaryotic and Viral Multimeric RNA Methyltransferases? Wiley Interdiscip. Rev. RNA 13 (1), e1673. doi:10.1002/wrna.1673
Guo, M., Yan, R., Ji, Q., Yao, H., Sun, M., Duan, L., et al. (2020). IFN Regulatory Factor-1 Induced Macrophage Pyroptosis by Modulating m6A Modification of Circ_0029589 in Patients with Acute Coronary Syndrome. Int. Immunopharmacology 86, 106800. doi:10.1016/j.intimp.2020.106800
Han, J., Wang, J.-z., Yang, X., Yu, H., Zhou, R., Lu, H.-C., et al. (2019). METTL3 Promote Tumor Proliferation of Bladder Cancer by Accelerating Pri-miR221/222 Maturation in m6A-dependent Manner. Mol. Cancer 18 (1), 110. doi:10.1186/s12943-019-1036-9
Han, X., Wang, L., and Han, Q. (2020). Advances in the Role of m6A RNA Modification in Cancer Metabolic Reprogramming. Cell Biosci 10 (1), 117. doi:10.1186/s13578-020-00479-z
He, H., Wu, W., Sun, Z., and Chai, L. (2019). MiR-4429 Prevented Gastric Cancer Progression through Targeting METTL3 to Inhibit m6A-Caused Stabilization of SEC62. Biochem. Biophysical Res. Commun. 517 (4), 581–587. doi:10.1016/j.bbrc.2019.07.058
He, W., Zhou, B., Liu, W., Zhang, M., Shen, Z., Han, Z., et al. (2015). Identification of A Novel Small-Molecule Binding Site of the Fat Mass and Obesity Associated Protein (FTO). J. Med. Chem. 58 (18), 7341–7348. doi:10.1021/acs.jmedchem.5b00702
He, Y., Wang, W., Xu, X., Yang, B., Yu, X., Wu, Y., et al. (2022). Mettl3 Inhibits the Apoptosis and Autophagy of Chondrocytes in Inflammation through Mediating Bcl2 Stability via Ythdf1-Mediated m6A Modification. Bone 154, 116182. doi:10.1016/j.bone.2021.116182
Hong, J., Lee, J. H., and Chung, I. K. (2016). Telomerase Activates Transcription of Cyclin D1 Gene through an Interaction with NOL1. J. Cel Sci 129 (8), 1566–1579. doi:10.1242/jcs.181040
Horiuchi, K., Umetani, M., Minami, T., Okayama, H., Takada, S., Yamamoto, M., et al. (2006). Wilms' Tumor 1-associating Protein Regulates G2/M Transition through Stabilization of Cyclin A2 mRNA. Proc. Natl. Acad. Sci. 103 (46), 17278–17283. doi:10.1073/pnas.0608357103
Hu, Y., Chen, C., Tong, X., Chen, S., Hu, X., Pan, B., et al. (2021). NSUN2 Modified by SUMO-2/3 Promotes Gastric Cancer Progression and Regulates mRNA m5C Methylation. Cell Death Dis 12 (9), 842. doi:10.1038/s41419-021-04127-3
Hua, W., Zhao, Y., Jin, X., Yu, D., He, J., Xie, D., et al. (2018). METTL3 Promotes Ovarian Carcinoma Growth and Invasion through the Regulation of AXL Translation and Epithelial to Mesenchymal Transition. Gynecol. Oncol. 151 (2), 356–365. doi:10.1016/j.ygyno.2018.09.015
Huang, H., Weng, H., Deng, X., and Chen, J. (2020). RNA Modifications in Cancer: Functions, Mechanisms, and Therapeutic Implications. Annu. Rev. Cancer Biol. 4 (1), 221–240. doi:10.1146/annurev-cancerbio-030419-033357
Huang, Y., Su, R., Sheng, Y., Dong, L., Dong, Z., Xu, H., et al. (2019). Small-Molecule Targeting of Oncogenic FTO Demethylase in Acute Myeloid Leukemia. Cancer cell 35 (4), 677–691. doi:10.1016/j.ccell.2019.03.006
Huang, Y., Yan, J., Li, Q., Li, J., Gong, S., Zhou, H., et al. (2015). Meclofenamic Acid Selectively Inhibits FTO Demethylation of m6A over ALKBH5. Nucleic Acids Res. 43 (1), 373–384. doi:10.1093/nar/gku1276
Hussain, S., Benavente, S. B., Nascimento, E., Dragoni, I., Kurowski, A., Gillich, A., et al. (2009). The Nucleolar RNA Methyltransferase Misu (NSun2) Is Required for Mitotic Spindle Stability. J. Cel Biol. 186 (1), 27–40. doi:10.1083/jcb.200810180
Hussain, S., Tuorto, F., Menon, S., Blanco, S., Cox, C., Flores, J. V., et al. (2013). The Mouse Cytosine-5 RNA Methyltransferase NSun2 Is a Component of the Chromatoid Body and Required for Testis Differentiation. Mol. Cel Biol 33 (8), 1561–1570. doi:10.1128/MCB.01523-12
Jensen-Urstad, A. P. L., and Semenkovich, C. F. (2012). Fatty Acid Synthase and Liver Triglyceride Metabolism: Housekeeper or Messenger? Biochim. Biophys. Acta (Bba) - Mol. Cel Biol. Lipids 1821 (5), 747–753. doi:10.1016/j.bbalip.2011.09.017
Jian, H., Zhang, C., Qi, Z., Li, X., Lou, Y., Kang, Y., et al. (2021). Alteration of mRNA 5-Methylcytosine Modification in Neurons after OGD/R and Potential Roles in Cell Stress Response and Apoptosis. Front. Genet. 12. doi:10.3389/fgene.2021.633681
Jiang, Q., Sun, B., Liu, Q., Cai, M., Wu, R., Wang, F., et al. (2019). MTCH2 Promotes Adipogenesis in Intramuscular Preadipocytes via an M 6 A‐YTHDF1‐dependent Mechanism. FASEB j. 33 (2), 2971–2981. doi:10.1096/fj.201801393RRR
Jiang, S., Fagman, J. B., Chen, C., Alberti, S., and Liu, B. (2020). Protein Phase Separation and its Role in Tumorigenesis. eLife 9, e60264. doi:10.7554/eLife.60264
Jiang, Z.-x., Wang, Y.-n., Li, Z.-y., Dai, Z.-h., He, Y., Chu, K., et al. (2021). The m6A mRNA Demethylase FTO in Granulosa Cells Retards FOS-dependent Ovarian Aging. Cel Death Dis 12 (8), 744. doi:10.1038/s41419-021-04016-9
Jin, S., Zhang, X., Miao, Y., Liang, P., Zhu, K., She, Y., et al. (2018). m6A RNA Modification Controls Autophagy through Upregulating ULK1 Protein Abundance. Cell Res 28 (9), 955–957. doi:10.1038/s41422-018-0069-8
Jo, H.-J., Shim, H.-E., Han, M.-E., Kim, H.-J., Kim, K.-S., Baek, S., et al. (2013). WTAP Regulates Migration and Invasion of Cholangiocarcinoma Cells. J. Gastroenterol. 48 (11), 1271–1282. doi:10.1007/s00535-013-0748-7
Kang, H., Zhang, Z., Yu, L., Li, Y., Liang, M., and Zhou, L. (2018). FTO Reduces Mitochondria and Promotes Hepatic Fat Accumulation through RNA Demethylation. J. Cel. Biochem. 119 (7), 5676–5685. doi:10.1002/jcb.26746
Khong, A., Matheny, T., Huynh, T. N., Babl, V., and Parker, R. (2021). Limited Effects of m6A Modification on mRNA Partitioning into Stress Granules. bioRxiv. doi:10.1101/2021.03.19.436090
Klionsky, D. J. (2007). Autophagy: from Phenomenology to Molecular Understanding in Less Than a Decade. Nat. Rev. Mol. Cel Biol 8 (11), 931–937. doi:10.1038/nrm2245
Kogaki, T., Ohshio, I., Kawaguchi, M., Kimoto, M., Kitae, K., Hase, H., et al. (2017). TP53 Gene Status Is a Critical Determinant of Phenotypes Induced by ALKBH3 Knockdown in Non-small Cell Lung Cancers. Biochem. Biophysical Res. Commun. 488 (2), 285–290. doi:10.1016/j.bbrc.2017.05.024
Kosi, N., Alić, I., Kolačević, M., Vrsaljko, N., Jovanov Milošević, N., Sobol, M., et al. (2015). Nop2 Is Expressed during Proliferation of Neural Stem Cells and in Adult Mouse and Human Brain. Brain Res. 1597, 65–76. doi:10.1016/j.brainres.2014.11.040
Kousteni, S. (2012). FoxO1, the Transcriptional Chief of Staff of Energy Metabolism. Bone 50 (2), 437–443. doi:10.1016/j.bone.2011.06.034
Kumari, R., and Jat, P. (2021). Mechanisms of Cellular Senescence: Cell Cycle Arrest and Senescence Associated Secretory Phenotype. Front. Cel Developmental Biol.9. doi:10.3389/fcell.2021.645593
Lan, T., Li, H., Zhang, D., Xu, L., Liu, H., Hao, X., et al. (2019). KIAA1429 Contributes to Liver Cancer Progression through N6-methyladenosine-dependent post-transcriptional Modification of GATA3. Mol. Cancer 18 (1), 186. doi:10.1186/s12943-019-1106-z
Lee, H., Bao, S., Qian, Y., Geula, S., Leslie, J., Zhang, C., et al. (2019). Stage-specific Requirement for Mettl3-dependent m6A mRNA Methylation during Haematopoietic Stem Cell Differentiation. Nat. Cel Biol 21 (6), 700–709. doi:10.1038/s41556-019-0318-1
Lee, J., Wu, Y., Harada, B. T., Li, Y., Zhao, J., He, C., et al. (2021a). N6 -methyladenosine Modification of lncRNA Pvt1 Governs Epidermal Stemness. Embo j 40 (8), e106276. doi:10.15252/embj.2020106276
Lee, J.-H., Wang, R., Xiong, F., Krakowiak, J., Liao, Z., Nguyen, P. T., et al. (2021b). Enhancer RNA m6A Methylation Facilitates Transcriptional Condensate Formation and Gene Activation. Mol. Cel 81 (16), 3368–3385. doi:10.1016/j.molcel.2021.07.024
Li, G., Deng, L., Huang, N., Cui, Z., Wu, Q., Ma, J., et al. (2021a). m6A mRNA Methylation Regulates LKB1 to Promote Autophagy of Hepatoblastoma Cells through Upregulated Phosphorylation of AMPK. Genes 12 (11), 1747. doi:10.3390/genes12111747
Li, J., Han, Y., Zhang, H., Qian, Z., Jia, W., Gao, Y., et al. (2019). The m6A Demethylase FTO Promotes the Growth of Lung Cancer Cells by Regulating the m6A Level of USP7 mRNA. Biochem. Biophysical Res. Commun. 512 (3), 479–485. doi:10.1016/j.bbrc.2019.03.093
Li, N., Kang, Y., Wang, L., Huff, S., Tang, R., Hui, H., et al. (2020). ALKBH5 Regulates Anti-PD-1 Therapy Response by Modulating Lactate and Suppressive Immune Cell Accumulation in Tumor Microenvironment. Proc. Natl. Acad. Sci. USA 117 (33), 20159–20170. doi:10.1073/pnas.1918986117
Li, Q., Li, X., Tang, H., Jiang, B., Dou, Y., Gorospe, M., et al. (2017a). NSUN2-Mediated m5C Methylation and METTL3/METTL14-Mediated m6A Methylation Cooperatively Enhance P21 Translation. J. Cel. Biochem. 118 (9), 2587–2598. doi:10.1002/jcb.25957
Li, Q., Ni, Y., Zhang, L., Jiang, R., Xu, J., Yang, H., et al. (2021b). HIF-1α-induced Expression of m6A Reader YTHDF1 Drives Hypoxia-Induced Autophagy and Malignancy of Hepatocellular Carcinoma by Promoting ATG2A and ATG14 Translation. Sig Transduct Target. Ther. 6 (1), 76. doi:10.1038/s41392-020-00453-8
Li, X., Tang, J., Huang, W., Wang, F., Li, P., Qin, C., et al. (2017b). The M6A Methyltransferase METTL3: Acting as a Tumor Suppressor in Renal Cell Carcinoma. Oncotarget 8 (56), 96103–96116. doi:10.18632/oncotarget.21726
Li, X., Yang, J., Zhu, Y., Liu, Y., Shi, X. e., and Yang, G. (2016). Mouse Maternal High-Fat Intake Dynamically Programmed mRNA m6A Modifications in Adipose and Skeletal Muscle Tissues in Offspring. Ijms 17 (8), 1336. doi:10.3390/ijms17081336
Li, Z., Qian, P., Shao, W., Shi, H., He, X. C., Gogol, M., et al. (2018). Suppression of m6A Reader Ythdf2 Promotes Hematopoietic Stem Cell Expansion. Cel Res 28 (9), 904–917. doi:10.1038/s41422-018-0072-0
Li, Z., Weng, H., Su, R., Weng, X., Zuo, Z., Li, C., et al. (2017c). FTO Plays an Oncogenic Role in Acute Myeloid Leukemia as a N 6 -Methyladenosine RNA Demethylase. Cancer Cell 31 (1), 127–141. doi:10.1016/j.ccell.2016.11.017
Liang, D., Lin, W.-J., Ren, M., Qiu, J., Yang, C., Wang, X., et al. (2021). m6A Reader YTHDC1 Modulates Autophagy by Targeting SQSTM1 in Diabetic Skin. Autophagy, 1–20. doi:10.1080/15548627.2021.1974175
Liefke, R., Windhof-Jaidhauser, I. M., Gaedcke, J., Salinas-Riester, G., Wu, F., Ghadimi, M., et al. (2015). The Oxidative Demethylase ALKBH3 marks Hyperactive Gene Promoters in Human Cancer Cells. Genome Med. 7 (1), 66. doi:10.1186/s13073-015-0180-0
Lin, S., Liu, J., Jiang, W., Wang, P., Sun, C., Wang, X., et al. (2019a). METTL3 Promotes the Proliferation and Mobility of Gastric Cancer Cells. Open Med. (Wars) 14, 25–31. doi:10.1515/med-2019-0005
Lin, X., Chai, G., Wu, Y., Li, J., Chen, F., Liu, J., et al. (2019b). RNA m6A Methylation Regulates the Epithelial Mesenchymal Transition of Cancer Cells and Translation of Snail. Nat. Commun. 10 (1), 2065. doi:10.1038/s41467-019-09865-9
Lin, Z., Niu, Y., Wan, A., Chen, D., Liang, H., Chen, X., et al. (2020). RNA M6 A Methylation Regulates Sorafenib Resistance in Liver Cancer through FOXO3-Mediated Autophagy. EMBO J. 39 (12), e103181. doi:10.15252/embj.2019103181
Liu, H., Xu, Y., Yao, B., Sui, T., Lai, L., and Li, Z. (2020a). A Novel N6-Methyladenosine (m6A)-dependent Fate Decision for the lncRNA THOR. Cel Death Dis 11 (8), 613. doi:10.1038/s41419-020-02833-y
Liu, J., Eckert, M. A., Harada, B. T., Liu, S.-M., Lu, Z., Yu, K., et al. (2018). m6A mRNA Methylation Regulates AKT Activity to Promote the Proliferation and Tumorigenicity of Endometrial Cancer. Nat. Cel Biol 20 (9), 1074–1083. doi:10.1038/s41556-018-0174-4
Liu, L., Wang, J., Sun, G., Wu, Q., Ma, J., Zhang, X., et al. (2019). m6A mRNA Methylation Regulates CTNNB1 to Promote the Proliferation of Hepatoblastoma. Mol. Cancer 18 (1), 188. doi:10.1186/s12943-019-1119-7
Liu, P., Li, F., Lin, J., Fukumoto, T., Nacarelli, T., Hao, X., et al. (2021a). m6A-independent Genome-wide METTL3 and METTL14 Redistribution Drives the Senescence-Associated Secretory Phenotype. Nat. Cel Biol 23 (4), 355–365. doi:10.1038/s41556-021-00656-3
Liu, S., Li, Q., Li, G., Zhang, Q., Zhuo, L., Han, X., et al. (2020b). The Mechanism of m6A Methyltransferase METTL3-Mediated Autophagy in Reversing Gefitinib Resistance in NSCLC Cells by β-elemene. Cel Death Dis 11 (11), 969. doi:10.1038/s41419-020-03148-8
Liu, X., Xiao, M., Zhang, L., Li, L., Zhu, G., Shen, E., et al. (2021b). The m6A Methyltransferase METTL14 Inhibits the Proliferation, Migration, and Invasion of Gastric Cancer by Regulating the PI3K/AKT/mTOR Signaling Pathway. J. Clin. Lab. Anal. 35 (3), e23655. doi:10.1002/jcla.23655
Lopez-Pajares, V., Yan, K., Zarnegar, B. J., Jameson, K. L., and Khavari, P. A. (2013). Genetic Pathways in Disorders of Epidermal Differentiation. Trends Genet. 29 (1), 31–40. doi:10.1016/j.tig.2012.10.005
Luo, G., Xu, W., Zhao, Y., Jin, S., Wang, S., Liu, Q., et al. (2020). RNA M 6 A Methylation Regulates Uveal Melanoma Cell Proliferation, Migration, and Invasion by Targeting c‐Met. J. Cel Physiol 235 (10), 7107–7119. doi:10.1002/jcp.29608
Lyon, A. S., Peeples, W. B., and Rosen, M. K. (2021). A Framework for Understanding the Functions of Biomolecular Condensates across Scales. Nat. Rev. Mol. Cel Biol 22 (3), 215–235. doi:10.1038/s41580-020-00303-z
Mabuchi, M., Shimizu, T., Ueda, M., Sasakawa, Y., Nakao, S., Ueda, Y., et al. (2015). Improving the Bioavailability and Anticancer Effect of the PCA-1/ALKBH3 Inhibitor HUHS015 Using Sodium Salt. In Vivo 29 (1), 39–43.
Macari, F., El-houfi, Y., Boldina, G., Xu, H., Khoury-Hanna, S., Ollier, J., et al. (2016). TRM6/61 Connects PKCα with Translational Control through tRNAiMet Stabilization: Impact on Tumorigenesis. Oncogene 35 (14), 1785–1796. doi:10.1038/onc.2015.244
Maita, K., Tsuda, S., and Shirasu, Y. (1991). Chronic Toxicity Studies with Thiram in Wistar Rats and Beagle Dogs. Toxicol. Sci. 16 (4), 667–686. doi:10.1016/0272-0590(91)90154-v10.1093/toxsci/16.4.667
Malla, S., Melguizo-Sanchis, D., and Aguilo, F. (2019). Steering Pluripotency and Differentiation with N6-Methyladenosine RNA Modification. Biochim. Biophys. Acta (Bba) - Gene Regul. Mech. 1862 (3), 394–402. doi:10.1016/j.bbagrm.2018.10.013
Merkestein, M., Laber, S., McMurray, F., Andrew, D., Sachse, G., Sanderson, J., et al. (2015). FTO Influences Adipogenesis by Regulating Mitotic Clonal Expansion. Nat. Commun. 6 (1), 6792. doi:10.1038/ncomms7792
Meyer, K. D., Patil, D. P., Zhou, J., Zinoviev, A., Skabkin, M. A., Elemento, O., et al. (2015). 5′ UTR m6A Promotes Cap-independent Translation. Cell 163 (4), 999–1010. doi:10.1016/j.cell.2015.10.012
Miranda-Gonçalves, V., Lobo, J., Guimarães-Teixeira, C., Barros-Silva, D., Guimarães, R., Cantante, M., et al. (2021). The Component of the m6A Writer Complex VIRMA Is Implicated in Aggressive Tumor Phenotype, DNA Damage Response and Cisplatin Resistance in Germ Cell Tumors. J. Exp. Clin. Cancer Res. 40 (1), 268. doi:10.1186/s13046-021-02072-9
Mizushima, N., Levine, B., Cuervo, A. M., and Klionsky, D. J. (2008). Autophagy Fights Disease through Cellular Self-Digestion. Nature 451 (7182), 1069–1075. doi:10.1038/nature06639
Nakao, S., Mabuchi, M., Shimizu, T., Itoh, Y., Takeuchi, Y., Ueda, M., et al. (2014). Design and Synthesis of Prostate Cancer Antigen-1 (PCA-1/ALKBH3) Inhibitors as Anti-prostate Cancer Drugs. Bioorg. Med. Chem. Lett. 24 (4), 1071–1074. doi:10.1016/j.bmcl.2014.01.008
Niu, Y., Lin, Z., Wan, A., Chen, H., Liang, H., Sun, L., et al. (2019). RNA N6-Methyladenosine Demethylase FTO Promotes Breast Tumor Progression through Inhibiting BNIP3. Mol. Cancer 18 (1), 46. doi:10.1186/s12943-019-1004-4
Okamoto, M., Hirata, S., Sato, S., Koga, S., Fujii, M., Qi, G., et al. (2012). Frequent Increased Gene Copy Number and High Protein Expression of tRNA (Cytosine-5-)-methyltransferase (NSUN2) in Human Cancers. DNA Cel Biol. 31 (5), 660–671. doi:10.1089/dna.2011.1446
Peng, S., Xiao, W., Ju, D., Sun, B., Hou, N., Liu, Q., et al. (2019). Identification of Entacapone as a Chemical Inhibitor of FTO Mediating Metabolic Regulation through FOXO1. Sci. Translational Med. 11, eaau7116. doi:10.1126/scitranslmed.aau7116
Qian, J.-Y., Gao, J., Sun, X., Cao, M.-D., Shi, L., Xia, T.-S., et al. (2019). KIAA1429 Acts as an Oncogenic Factor in Breast Cancer by Regulating CDK1 in an N6-methyladenosine-independent Manner. Oncogene 38 (33), 6123–6141. doi:10.1038/s41388-019-0861-z
Regl, G., Neill, G. W., Eichberger, T., Kasper, M., Ikram, M. S., Koller, J., et al. (2002). Human GLI2 and GLI1 Are Part of a Positive Feedback Mechanism in Basal Cell Carcinoma. Oncogene 21 (36), 5529–5539. doi:10.1038/sj.onc.1205748
Ries, R. J., Zaccara, S., Klein, P., Olarerin-George, A., Namkoong, S., Pickering, B. F., et al. (2019). m6A Enhances the Phase Separation Potential of mRNA. Nature 571 (7765), 424–428. doi:10.1038/s41586-019-1374-1
Rosselló-Tortella, M., Ferrer, G., and Esteller, M. (2020). Epitranscriptomics in Hematopoiesis and Hematologic Malignancies. Blood Cancer Discov. 1 (1), 26–31. doi:10.1158/2643-3249.Bcd-20-0032
Roundtree, I. A., Evans, M. E., Pan, T., and He, C. (2017). Dynamic RNA Modifications in Gene Expression Regulation. Cell 169 (7), 1187–1200. doi:10.1016/j.cell.2017.05.045
Sajini, A. A., Choudhury, N. R., Wagner, R. E., Bornelöv, S., Selmi, T., Spanos, C., et al. (2019). Loss of 5-methylcytosine Alters the Biogenesis of Vault-Derived Small RNAs to Coordinate Epidermal Differentiation. Nat. Commun. 10 (1), 2550. doi:10.1038/s41467-019-10020-7
Schaefer, M., Hagemann, S., Hanna, K., and Lyko, F. (2009). Azacytidine Inhibits RNA Methylation at DNMT2 Target Sites in Human Cancer Cell Lines. Cancer Res. 69 (20), 8127–8132. doi:10.1158/0008-5472.Can-09-0458
Schwartz, S., Agarwala, S. D., Mumbach, M. R., Jovanovic, M., Mertins, P., Shishkin, A., et al. (2013). High-Resolution Mapping Reveals a Conserved, Widespread, Dynamic mRNA Methylation Program in Yeast Meiosis. Cell 155 (6), 1409–1421. doi:10.1016/j.cell.2013.10.047
Selberg, S., Seli, N., Kankuri, E., and Karelson, M. (2021). Rational Design of Novel Anticancer Small-Molecule RNA m6A Demethylase ALKBH5 Inhibitors. ACS omega 6 (20), 13310–13320. doi:10.1021/acsomega.1c01289
Shen, C., Xuan, B., Yan, T., Ma, Y., Xu, P., Tian, X., et al. (2020). m6A-dependent Glycolysis Enhances Colorectal Cancer Progression. Mol. Cancer 19 (1), 72. doi:10.1186/s12943-020-01190-w
Shen, L., Liang, Z., Wong, C. E., and Yu, H. (2019). Messenger RNA Modifications in Plants. Trends Plant Sci. 24 (4), 328–341. doi:10.1016/j.tplants.2019.01.005
Shen, M., Li, Y., Wang, Y., Shao, J., Zhang, F., Yin, G., et al. (2021). N6-methyladenosine Modification Regulates Ferroptosis through Autophagy Signaling Pathway in Hepatic Stellate Cells. Redox Biol. 47, 102151. doi:10.1016/j.redox.2021.102151
Shi, Q., Xue, C., Yuan, X., He, Y., and Yu, Z. (2020). Gene Signatures and Prognostic Values of m1A-Related Regulatory Genes in Hepatocellular Carcinoma. Sci. Rep. 10 (1), 15083. doi:10.1038/s41598-020-72178-1
Shibata, T., Watari, K., Kawahara, A., Sudo, T., Hattori, S., Murakami, Y., et al. (2020). Targeting Phosphorylation of Y-Box-Binding Protein YBX1 by TAS0612 and Everolimus in Overcoming Antiestrogen Resistance. Mol. Cancer Ther. 19 (3), 882–894. doi:10.1158/1535-7163.Mct-19-0690
Singh, B., Kinne, H. E., Milligan, R. D., Washburn, L. J., Olsen, M., and Lucci, A. (2016). Important Role of FTO in the Survival of Rare Panresistant Triple-Negative Inflammatory Breast Cancer Cells Facing a Severe Metabolic Challenge. PloS one 11 (7), e0159072. doi:10.1371/journal.pone.0159072
Song, H., Feng, X., Zhang, H., Luo, Y., Huang, J., Lin, M., et al. (2019). METTL3 and ALKBH5 Oppositely Regulate m6A Modification of TFEB mRNA, Which Dictates the Fate of Hypoxia/reoxygenation-Treated Cardiomyocytes. Autophagy 15 (8), 1419–1437. doi:10.1080/15548627.2019.1586246
Song, P., Tayier, S., Cai, Z., and Jia, G. (2021). RNA Methylation in Mammalian Development and Cancer. Cell Biol Toxicol 37 (6), 811–831. doi:10.1007/s10565-021-09627-8
Song, W., Ressl, S., and Tracey, W. D. (2020). Loss of Pseudouridine Synthases in the RluA Family Causes Hypersensitive Nociception in Drosophila. G3 (Bethesda, Md. 10 (12), 4425–4438. doi:10.1534/g3.120.401767
Spina, A., De Pasquale, V., Cerulo, G., Cocchiaro, P., Della Morte, R., Avallone, L., et al. (2015). HGF/c-MET Axis in Tumor Microenvironment and Metastasis Formation. Biomedicines 3 (1), 71–88. doi:10.3390/biomedicines3010071
Stadler, P. F., Chen, J. J.-L., Hackermüller, J., Hoffmann, S., Horn, F., Khaitovich, P., et al. (2009). Evolution of Vault RNAs. Mol. Biol. Evol. 26 (9), 1975–1991. doi:10.1093/molbev/msp112
Stratigopoulos, G., LeDuc, C. A., Cremona, M. L., Chung, W. K., and Leibel, R. L. (2011). Cut-like Homeobox 1 (CUX1) Regulates Expression of the Fat Mass and Obesity-Associated and Retinitis Pigmentosa GTPase Regulator-Interacting Protein-1-like (RPGRIP1L) Genes and Coordinates Leptin Receptor Signaling. J. Biol. Chem. 286 (3), 2155–2170. doi:10.1074/jbc.M110.188482
Stresemann, C., and Lyko, F. (2008). Modes of Action of the DNA Methyltransferase Inhibitors Azacytidine and Decitabine. Int. J. Cancer 123 (1), 8–13. doi:10.1002/ijc.23607
Su, R., Dong, L., Li, C., Nachtergaele, S., Wunderlich, M., Qing, Y., et al. (2018). R-2HG Exhibits Anti-tumor Activity by Targeting FTO/m6A/MYC/CEBPA Signaling. Cell 172 (1-2), 90–e23. doi:10.1016/j.cell.2017.11.031
Su, R., Dong, L., Li, Y., Gao, M., Han, L., Wunderlich, M., et al. (2020). Targeting FTO Suppresses Cancer Stem Cell Maintenance and Immune Evasion. Cancer cell 38 (1), 79–96. doi:10.1016/j.ccell.2020.04.017
Sun, T., Wu, Z., Wang, X., Wang, Y., Hu, X., Qin, W., et al. (2020). LNC942 Promoting METTL14-Mediated m6A Methylation in Breast Cancer Cell Proliferation and Progression. Oncogene 39 (31), 5358–5372. doi:10.1038/s41388-020-1338-9
Svobodová Kovaříková, A., Stixová, L., Kovařík, A., Komůrková, D., Legartová, S., Fagherazzi, P., et al. (2020). N(6)-Adenosine Methylation in RNA and a Reduced m(3)G/TMG Level in Non-coding RNAs Appear at Microirradiation-Induced DNA Lesions. Cells 9 (2), 360. doi:10.3390/cells9020360
Taketo, K., Konno, M., Asai, A., Koseki, J., Toratani, M., Satoh, T., et al. (2018). The Epitranscriptome m6A Writer METTL3 Promotes Chemo- and Radioresistance in Pancreatic Cancer Cells. Int. J. Oncol. 52 (2), 621–629. doi:10.3892/ijo.2017.4219
Tang, H.-W., Weng, J.-H., Lee, W. X., Hu, Y., Gu, L., Cho, S., et al. (2021). mTORC1-chaperonin CCT Signaling Regulates m6A RNA Methylation to Suppress Autophagy. Proc. Natl. Acad. Sci. USA 118 (10), e2021945118. doi:10.1073/pnas.2021945118
Tang, H., Fan, X., Xing, J., Liu, Z., Jiang, B., Dou, Y., et al. (2015). NSun2 Delays Replicative Senescence by Repressing P27 (KIP1) Translation and Elevating CDK1 Translation. Aging 7 (12), 1143–1155. doi:10.18632/aging.100860
Tang, J., Wang, F., Cheng, G., Si, S., Sun, X., Han, J., et al. (2018). Wilms' Tumor 1-associating Protein Promotes Renal Cell Carcinoma Proliferation by Regulating CDK2 mRNA Stability. J. Exp. Clin. Cancer Res. 37 (1), 40. doi:10.1186/s13046-018-0706-6
Tang, Q.-Q., Otto, T. C., and Lane, M. D. (2003). Mitotic Clonal Expansion: a Synchronous Process Required for Adipogenesis. Proc. Natl. Acad. Sci. 100 (1), 44–49. doi:10.1073/pnas.0137044100
Tang, X., Liu, S., Chen, D., Zhao, Z., and Zhou, J. (2019). The Role of the Fat Mass and Obesity-Associated Protein in the Proliferation of Pancreatic Cancer Cells. Oncol. Lett. 17 (2), 2473–2478. doi:10.3892/ol.2018.9873
Tasaki, M., Shimada, K., Kimura, H., Tsujikawa, K., and Konishi, N. (2011). ALKBH3, a Human AlkB Homologue, Contributes to Cell Survival in Human Non-small-cell Lung Cancer. Br. J. Cancer 104 (4), 700–706. doi:10.1038/sj.bjc.6606012
Tuorto, F., Herbst, F., Alerasool, N., Bender, S., Popp, O., Federico, G., et al. (2015). The tRNA Methyltransferase Dnmt2 Is Required for Accurate Polypeptide Synthesis during Haematopoiesis. Embo J. 34 (18), 2350–2362. doi:10.15252/embj.201591382
Ueda, Y., Ooshio, I., Fusamae, Y., Kitae, K., Kawaguchi, M., Jingushi, K., et al. (2017). AlkB Homolog 3-mediated tRNA Demethylation Promotes Protein Synthesis in Cancer Cells. Sci. Rep. 7 (1), 42271. doi:10.1038/srep42271
Valdés-Rives, S. A., and González-Arenas, A. (2017). Autotaxin-Lysophosphatidic Acid: From Inflammation to Cancer Development. Mediators Inflamm. 2017, 9173090. doi:10.1155/2017/9173090
Vasic, R., Gao, Y., Liu, C., and Halene, S. (2020). The Role of RNA Epigenetic Modification in Normal and Malignant Hematopoiesis. Curr. Stem Cel Rep 6 (4), 144–155. doi:10.1007/s40778-020-00178-y
Vicidomini, R., Di Giovanni, A., Petrizzo, A., Iannucci, L. F., Benvenuto, G., Nagel, A. C., et al. (2015). Loss of Drosophila Pseudouridine Synthase Triggers Apoptosis-Induced Proliferation and Promotes Cell-Nonautonomous EMT. Cel Death Dis 6 (3), e1705. doi:10.1038/cddis.2015.68
Vu, L. P., Pickering, B. F., Cheng, Y., Zaccara, S., Nguyen, D., Minuesa, G., et al. (2017). The N6-Methyladenosine (m6A)-Forming Enzyme METTL3 Controls Myeloid Differentiation of normal Hematopoietic and Leukemia Cells. Nat. Med. 23 (11), 1369–1376. doi:10.1038/nm.4416
Waku, T., Nakajima, Y., Yokoyama, W., Nomura, N., Kako, K., Kobayashi, A., et al. (2016). NML-mediated rRNA Base Methylation Links Ribosomal Subunit Formation to Cell Proliferation in a P53-dependent Manner. J. Cel Sci 129 (12), 2382–2393. doi:10.1242/jcs.183723
Wang, B., Niu, L., Wang, Z., and Zhao, Z. (2021a). RNA m1A Methyltransferase TRMT6 Predicts Poorer Prognosis and Promotes Malignant Behavior in Glioma. Front. Mol. Biosci. 8, 692130. doi:10.3389/fmolb.2021.692130
Wang, C.-X., Cui, G.-S., Liu, X., Xu, K., Wang, M., Zhang, X.-X., et al. (2018a). METTL3-mediated m6A Modification Is Required for Cerebellar Development. Plos Biol. 16 (6), e2004880. doi:10.1371/journal.pbio.2004880
Wang, J., Zhang, J., Ma, Y., Zeng, Y., Lu, C., Yang, F., et al. (2021b). WTAP Promotes Myocardial Ischemia/reperfusion Injury by Increasing Endoplasmic Reticulum Stress via Regulating m6A Modification of ATF4 mRNA. Aging 13 (8), 11135–11149. doi:10.18632/aging.202770
Wang, T., Hong, T., Huang, Y., Su, H., Wu, F., Chen, Y., et al. (2015a). Fluorescein Derivatives as Bifunctional Molecules for the Simultaneous Inhibiting and Labeling of FTO Protein. J. Am. Chem. Soc. 137 (43), 13736–13739. doi:10.1021/jacs.5b06690
Wang, W. (2016). mRNA Methylation by NSUN2 in Cell Proliferation. WIREs RNA 7 (6), 838–842. doi:10.1002/wrna.1380
Wang, X., Wu, R., Liu, Y., Zhao, Y., Bi, Z., Yao, Y., et al. (2020). m6A mRNA Methylation Controls Autophagy and Adipogenesis by Targeting Atg5 and Atg7. Autophagy 16 (7), 1221–1235. doi:10.1080/15548627.2019.1659617
Wang, Y., Huang, Q., Deng, T., Li, B.-H., and Ren, X.-Q. (2019). Clinical Significance of TRMT6 in Hepatocellular Carcinoma: A Bioinformatics-Based Study. Med. Sci. Monit. 25, 3894–3901. doi:10.12659/msm.913556
Wang, Y., Li, Y., Yue, M., Wang, J., Kumar, S., Wechsler-Reya, R. J., et al. (2018b). N6-methyladenosine RNA Modification Regulates Embryonic Neural Stem Cell Self-Renewal through Histone Modifications. Nat. Neurosci. 21 (2), 195–206. doi:10.1038/s41593-017-0057-1
Wang, Y., Viscarra, J., Kim, S.-J., and Sul, H. S. (2015b). Transcriptional Regulation of Hepatic Lipogenesis. Nat. Rev. Mol. Cel Biol 16 (11), 678–689. doi:10.1038/nrm4074
Wang, Y., Wang, J., Li, X., Xiong, X., Wang, J., Zhou, Z., et al. (2021c). N1-methyladenosine Methylation in tRNA Drives Liver Tumourigenesis by Regulating Cholesterol Metabolism. Nat. Commun. 12 (1), 6314. doi:10.1038/s41467-021-26718-6
Wanna-Udom, S., Terashima, M., Lyu, H., Ishimura, A., Takino, T., Sakari, M., et al. (2020). The m6A Methyltransferase METTL3 Contributes to Transforming Growth Factor-Beta-Induced Epithelial-Mesenchymal Transition of Lung Cancer Cells through the Regulation of JUNB. Biochem. Biophysical Res. Commun. 524 (1), 150–155. doi:10.1016/j.bbrc.2020.01.042
Wei, W., Huo, B., and Shi, X. (2019). miR-600 Inhibits Lung Cancer via Downregulating the Expression of METTL3. Cmar 11, 1177–1187. doi:10.2147/CMAR.S181058
Weng, H., Huang, H., Wu, H., Qin, X., Zhao, B. S., Dong, L., et al. (2018). METTL14 Inhibits Hematopoietic Stem/Progenitor Differentiation and Promotes Leukemogenesis via mRNA m6A Modification. Cell Stem Cell 22 (2), 191–205. doi:10.1016/j.stem.2017.11.016
Wilkinson, E., Cui, Y. H., and He, Y. Y. (2021). Context-Dependent Roles of RNA Modifications in Stress Responses and Diseases. Int. J. Mol. Sci. 22. doi:10.3390/ijms22041949
Wu, R., Li, A., Sun, B., Sun, J.-G., Zhang, J., Zhang, T., et al. (2019). A Novel m6A Reader Prrc2a Controls Oligodendroglial Specification and Myelination. Cel Res 29 (1), 23–41. doi:10.1038/s41422-018-0113-8
Wu, R., Liu, Y., Yao, Y., Zhao, Y., Bi, Z., Jiang, Q., et al. (2018). FTO Regulates Adipogenesis by Controlling Cell Cycle Progression via m6A-YTHDF2 Dependent Mechanism. Biochim. Biophys. Acta (Bba) - Mol. Cel Biol. Lipids 1863 (10), 1323–1330. doi:10.1016/j.bbalip.2018.08.008
Wu, Z., Shi, Y., Lu, M., Song, M., Yu, Z., Wang, J., et al. (2020). METTL3 Counteracts Premature Aging via m6A-dependent Stabilization of MIS12 mRNA. Nucleic Acids Res. 48 (19), 11083–11096. doi:10.1093/nar/gkaa816
Xia, T., Wu, X., Cao, M., Zhang, P., Shi, G., Zhang, J., et al. (2019). The RNA m6A Methyltransferase METTL3 Promotes Pancreatic Cancer Cell Proliferation and Invasion. Pathol. - Res. Pract. 215 (11), 152666. doi:10.1016/j.prp.2019.152666
Xiang, Y., Laurent, B., Hsu, C.-H., Nachtergaele, S., Lu, Z., Sheng, W., et al. (2017). RNA m6A Methylation Regulates the Ultraviolet-Induced DNA Damage Response. Nature 543 (7646), 573–576. doi:10.1038/nature21671
Xing, J., Yi, J., Cai, X., Tang, H., Liu, Z., Zhang, X., et al. (2015). NSun2 Promotes Cell Growth via Elevating Cyclin-dependent Kinase 1 Translation. Mol. Cel Biol 35 (23), 4043–4052. doi:10.1128/MCB.00742-15
Xiong, X., Li, X., and Yi, C. (2018). N1-methyladenosine Methylome in Messenger RNA and Non-coding RNA. Curr. Opin. Chem. Biol. 45, 179–186. doi:10.1016/j.cbpa.2018.06.017
Xu, X., Zhang, Y., Zhang, J., and Zhang, X. (2020). NSun2 Promotes Cell Migration through Methylating Autotaxin mRNA. J. Biol. Chem. doi:10.1074/jbc.RA119.012009
Xu, Y., Chen, Y., Yao, Y., Xie, H., Lu, G., Du, C., et al. (2021). VIRMA Contributes to Non-small Cell Lung Cancer Progression via N6-methyladenosine-dependent DAPK3 post-transcriptional Modification. Cancer Lett. 522, 142–154. doi:10.1016/j.canlet.2021.08.027
Xue, C., Zhao, Y., and Li, L. (2020). Advances in RNA Cytosine-5 Methylation: Detection, Regulatory Mechanisms, Biological Functions and Links to Cancer. Biomark Res. 8 (1), 43. doi:10.1186/s40364-020-00225-0
Yan, G., Elbadawi, M., and Efferth, T. (2020). Multiple Cell Death Modalities and Their Key Features (Review). World Acad. Sci. J. 2 (2), 39–48. doi:10.3892/wasj.2020.40
Yang, F., Jin, H., Que, B., Chao, Y., Zhang, H., Ying, X., et al. (2019a). Dynamic m6A mRNA Methylation Reveals the Role of METTL3-m6A-CDCP1 Signaling axis in Chemical Carcinogenesis. Oncogene 38 (24), 4755–4772. doi:10.1038/s41388-019-0755-0
Yang, R., Liang, X., Wang, H., Guo, M., Shen, H., Shi, Y., et al. (2021a). The RNA Methyltransferase NSUN6 Suppresses Pancreatic Cancer Development by Regulating Cell Proliferation. EBioMedicine 63, 103195. doi:10.1016/j.ebiom.2020.103195
Yang, S., Wei, J., Cui, Y.-H., Park, G., Shah, P., Deng, Y., et al. (2019b). m6A mRNA Demethylase FTO Regulates Melanoma Tumorigenicity and Response to Anti-PD-1 Blockade. Nat. Commun. 10 (1), 2782. doi:10.1038/s41467-019-10669-0
Yang, X., Zhang, S., He, C., Xue, P., Zhang, L., He, Z., et al. (2020). METTL14 Suppresses Proliferation and Metastasis of Colorectal Cancer by Down-Regulating Oncogenic Long Non-coding RNA XIST. Mol. Cancer 19 (1), 46. doi:10.1186/s12943-020-1146-4
Yang, Y., Hsu, P. J., Chen, Y.-S., and Yang, Y.-G. (2018). Dynamic Transcriptomic m6A Decoration: Writers, Erasers, Readers and Functions in RNA Metabolism. Cel Res 28 (6), 616–624. doi:10.1038/s41422-018-0040-8
Yang, Y., Shen, F., Huang, W., Qin, S., Huang, J.-T., Sergi, C., et al. (2019c). Glucose Is Involved in the Dynamic Regulation of m6A in Patients with Type 2 Diabetes. J. Clin. Endocrinol. Metab. 104 (3), 665–673. doi:10.1210/jc.2018-00619
Yang, Z., Yang, S., Cui, Y. H., Wei, J., Shah, P., Park, G., et al. (2021b). METTL14 Facilitates Global Genome Repair and Suppresses Skin Tumorigenesis. Proc. Natl. Acad. Sci. U S A. 118. doi:10.1073/pnas.2025948118
Yankova, E., Blackaby, W., Albertella, M., Rak, J., De Braekeleer, E., Tsagkogeorga, G., et al. (2021). Small-molecule Inhibition of METTL3 as a Strategy against Myeloid Leukaemia. Nature 593 (7860), 597–601. doi:10.1038/s41586-021-03536-w
Ye, D., Ma, S., Xiong, Y., and Guan, K.-L. (2013). R-2-hydroxyglutarate as the Key Effector of IDH Mutations Promoting Oncogenesis. Cancer cell 23 (3), 274–276. doi:10.1016/j.ccr.2013.03.005
Yoon, K.-J., Ringeling, F. R., Vissers, C., Jacob, F., Pokrass, M., Jimenez-Cyrus, D., et al. (2017). Temporal Control of Mammalian Cortical Neurogenesis by m6A Methylation. Cell 171 (4), 877–889. doi:10.1016/j.cell.2017.09.003
Yue, H., Nie, X., Yan, Z., and Weining, S. (2019). N6‐methyladenosine Regulatory Machinery in Plants: Composition, Function and Evolution. Plant Biotechnol. J. 17 (7), 1194–1208. doi:10.1111/pbi.13149
Zachari, M., and Ganley, I. G. (2017). The Mammalian ULK1 Complex and Autophagy Initiation. Essays Biochem. 61 (6), 585–596. doi:10.1042/ebc20170021
Zhang, B., Jiang, H., Dong, Z., Sun, A., and Ge, J. (2020a). The Critical Roles of m6A Modification in Metabolic Abnormality and Cardiovascular Diseases. Genes Dis. doi:10.1016/j.gendis.2020.07.011
Zhang, C., Chen, L., Peng, D., Jiang, A., He, Y., Zeng, Y., et al. (2020b). METTL3 and N6-Methyladenosine Promote Homologous Recombination-Mediated Repair of DSBs by Modulating DNA-RNA Hybrid Accumulation. Mol. Cel 79 (3), 425–442. doi:10.1016/j.molcel.2020.06.017
Zhang, C., Chen, Y., Sun, B., Wang, L., Yang, Y., Ma, D., et al. (2017a). m6A Modulates Haematopoietic Stem and Progenitor Cell Specification. Nature 549 (7671), 273–276. doi:10.1038/nature23883
Zhang, C., Zhang, M., Ge, S., Huang, W., Lin, X., Gao, J., et al. (2019a). Reduced m6A Modification Predicts Malignant Phenotypes and Augmented Wnt/PI3K‐Akt Signaling in Gastric Cancer. Cancer Med. 8 (10), 4766–4781. doi:10.1002/cam4.2360
Zhang, M., Zhang, Y., Ma, J., Guo, F., Cao, Q., Zhang, Y., et al. (2015). The Demethylase Activity of FTO (Fat Mass and Obesity Associated Protein) Is Required for Preadipocyte Differentiation. PLOS ONE 10, e0133788. doi:10.1371/journal.pone.0133788
Zhang, Q., Riddle, R. C., Yang, Q., Rosen, C. R., Guttridge, D. C., Dirckx, N., et al. (2019b). The RNA Demethylase FTO Is Required for Maintenance of Bone Mass and Functions to Protect Osteoblasts from Genotoxic Damage. Proc. Natl. Acad. Sci. USA 116 (36), 17980–17989. doi:10.1073/pnas.1905489116
Zhang, S., Zhao, B. S., Zhou, A., Lin, K., Zheng, S., Lu, Z., et al. (2017b). m 6 A Demethylase ALKBH5 Maintains Tumorigenicity of Glioblastoma Stem-like Cells by Sustaining FOXM1 Expression and Cell Proliferation Program. Cancer Cell 31 (4), 591–606. doi:10.1016/j.ccell.2017.02.013
Zhang, X., Li, D., Jia, C., Cai, H., Lv, Z., and Wu, B. (2021). METTL14 Promotes Tumorigenesis by Regulating lncRNA OIP5-AS1/miR-98/ADAMTS8 Signaling in Papillary Thyroid Cancer. Cel Death Dis 12 (6), 617. doi:10.1038/s41419-021-03891-6
Zhao, X., Yang, Y., Sun, B.-F., Shi, Y., Yang, X., Xiao, W., et al. (2014). FTO-dependent Demethylation of N6-Methyladenosine Regulates mRNA Splicing and Is Required for Adipogenesis. Cel Res 24 (12), 1403–1419. doi:10.1038/cr.2014.151
Zheng, G., Dahl, J. A., Niu, Y., Fedorcsak, P., Huang, C.-M., Li, C. J., et al. (2013). ALKBH5 Is a Mammalian RNA Demethylase that Impacts RNA Metabolism and Mouse Fertility. Mol. Cel 49 (1), 18–29. doi:10.1016/j.molcel.2012.10.015
Zheng, Q., Gan, H., Yang, F., Yao, Y., Hao, F., Hong, L., et al. (2020). Cytoplasmic m1A Reader YTHDF3 Inhibits Trophoblast Invasion by Downregulation of m1A-Methylated IGF1R. Cell Discov 6, 12. doi:10.1038/s41421-020-0144-4
Zhou, J., Wan, J., Shu, X. E., Mao, Y., Liu, X.-M., Yuan, X., et al. (2018). N6-Methyladenosine Guides mRNA Alternative Translation during Integrated Stress Response. Mol. Cel 69 (4), 636–647. doi:10.1016/j.molcel.2018.01.019
Zhou, L., Yang, C., Zhang, N., Zhang, X., Zhao, T., and Yu, J. (2020). Silencing METTL3 Inhibits the Proliferation and Invasion of Osteosarcoma by Regulating ATAD2. Biomed. Pharmacother. 125, 109964. doi:10.1016/j.biopha.2020.109964
Zhu, H., Sun, B., Zhu, L., Zou, G., and Shen, Q. (2021a). N6-Methyladenosine Induced miR-34a-5p Promotes TNF-α-Induced Nucleus Pulposus Cell Senescence by Targeting SIRT1. Front. Cel Developmental Biol. 9. doi:10.3389/fcell.2021.642437
Zhu, R., Ji, X., Wu, X., Chen, J., Li, X., Jiang, H., et al. (2020). Melatonin Antagonizes Ovarian Aging via YTHDF2-MAPK-NF-Κb Pathway. Genes Dis. doi:10.1016/j.gendis.2020.08.005
Zhu, W., Wang, J.-Z., Wei, J.-F., and Lu, C. (2021b). Role of m6A Methyltransferase Component VIRMA in Multiple Human Cancers (Review). Cancer Cel Int 21 (1), 172. doi:10.1186/s12935-021-01868-1
Zhu, X., Wang, X., Yan, W., Yang, H., Xiang, Y., Lv, F., et al. (2021c). Ubiquitination-mediated Degradation of TRDMT1 Regulates Homologous Recombination and Therapeutic Response. NAR Cancer 3. doi:10.1093/narcan/zcab010
Zona, S., Bella, L., Burton, M. J., Nestal de Moraes, G., and Lam, E. W.-F. (2014). FOXM1: An Emerging Master Regulator of DNA Damage Response and Genotoxic Agent Resistance. Biochim. Biophys. Acta (Bba) - Gene Regul. Mech. 1839 (11), 1316–1322. doi:10.1016/j.bbagrm.2014.09.016
Zou, D., Dong, L., Li, C., Yin, Z., Rao, S., and Zhou, Q. (2019). The m6A Eraser FTO Facilitates Proliferation and Migration of Human Cervical Cancer Cells. Cancer Cel Int 19, 321. doi:10.1186/s12935-019-1045-1
Keywords: m6A, m5C, m1A, epitranscriptomics, cellular functions, therapeutics
Citation: Wilkinson E, Cui Y-H and He Y-Y (2022) Roles of RNA Modifications in Diverse Cellular Functions. Front. Cell Dev. Biol. 10:828683. doi: 10.3389/fcell.2022.828683
Received: 03 December 2021; Accepted: 14 February 2022;
Published: 08 March 2022.
Edited by:
Chengqi Yi, Peking University, ChinaReviewed by:
Zhe Liang, Chinese Academy of Agricultural Sciences, ChinaMichael James Holmes, Indiana University, United States
Adam Heck, Fred Hutchinson Cancer Research Center, United States
Copyright © 2022 Wilkinson, Cui and He. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yu-Ying He, eXloZUBtZWRpY2luZS5ic2QudWNoaWNhZ28uZWR1
 Emma Wilkinson
Emma Wilkinson Yan-Hong Cui
Yan-Hong Cui Yu-Ying He
Yu-Ying He