- Department of Genome Sciences, University of Washington, Seattle, WA, United States
The process of gene duplication followed by gene loss or evolution of new functions has been studied extensively, yet the role gene duplication plays in the function and evolution of fertilization proteins is underappreciated. Gene duplication is observed in many fertilization protein families including Izumo, DCST, ZP, and the TFP superfamily. Molecules mediating fertilization are part of larger gene families expressed in a variety of tissues, but gene duplication followed by structural modifications has often facilitated their cooption into a fertilization function. Repeat expansions of functional domains within a gene also provide opportunities for the evolution of novel fertilization protein. ZP proteins with domain repeat expansions are linked to species-specificity in fertilization and TFP proteins that experienced domain duplications were coopted into a novel sperm function. This review outlines the importance of gene duplications and repeat domain expansions in the evolution of fertilization proteins.
Introduction
The fertilization of oocytes by sperm is an essential function in sexual reproduction, and multiple stages of the fertilization cascade have been described (Vacquier, 1998). First the sperm is drawn to the egg through chemotaxis (Ramírez-Gómez et al., 2019), and it then binds to the egg and releases proteins stored in the acrosome. The sperm then passes through the glycoproteinaceous egg coat (Monne et al., 2008; Wilburn and Swanson, 2016) (named Zona Pellucida in mammals), and proceeds to the oocyte cell membrane to initiate fusion (Siu et al., 2021). Understanding fertilization requires knowledge of both these broad steps of the fertilization cascade and the molecular mechanism underlying them. Research into the evolution and function of gametic proteins has implications for the development of novel contraception or treatments for unexplained human infertility (Gelbaya et al., 2014).
Many fertilization proteins are members of gene families that result from whole gene duplication events, which is a common mechanism for gene birth (Hughes, 1994). There has been extensive research into the relationship between gene duplication and other aspects of reproductive biology, including the neuroendocrine control of reproduction (Dufour et al., 2020), protease activity in the female reproductive tract (Kelleher et al., 2007; Kelleher and Markow, 2009), the resolution of sexual conflict (Gallach et al., 2010, 2011; Connallon and Clark, 2011; Gallach and Betrán, 2011), and hybridization barriers (Ting et al., 2004). This review specifically focuses on our growing knowledge of duplicated protein families implicated in fertilization. These proteins include the Izumo1 and Juno pair of interacting proteins, which each arose from independent gene duplication events and are essential to gamete membrane fusion function in mammals (Bianchi et al., 2014). DCST1 and DCST2 are paralogous proteins expressed in the sperm membrane of some bilateral animals, that are essential for fertilization (Inoue et al., 2021a, 1). Other duplicated proteins that act in fertilization include ADAMs (Primakoff and Myles, 2000; Civetta, 2003; Finn and Civetta, 2010), CRISPs (Busso et al., 2007; Da Ros et al., 2008; Gibbs et al., 2011; Maldera et al., 2014), Catspers (Clapham and Garbers, 2005; Navarro et al., 2008; Speer et al., 2021), and PKDREJ on the male side (Sutton et al., 2008), and tetraspanins (CD9,CD81) (Le Naour et al., 2000, 9; Miyado et al., 2000; Frolikova et al., 2018) and EBR1 on the female side (Kamei and Glabe, 2003; Hart, 2013). Genomic resources suggests that most of these families (ADAMs, tetraspanins, EBR, PKRDEJ, Catsper) have orthologs in other bilateral animals, while CRISP has orthologs in animals and in yeast (Howe et al., 2021).
Duplicated genes can experience further structural diversification, such as the duplication of individual functional protein domains. Proteins containing tandemly duplicated domains constitute a small, but significant portion of the genome (Han et al., 2007; Nacher et al., 2010). Independent tandem duplications of individual functional domains is also a recurrent trend in some protein families (TFP,ZP) (Galindo et al., 2002; Aagaard et al., 2010; Doty et al., 2016). There are several families of reproductive proteins on both the sperm and egg that show a history of being coopted from non-reproductive functions (Figure 1). Three finger proteins (TFPs) have been frequently coopted for fertilization including SPACA4 in tetrapods, Bouncer in fish, and multiple classes of sperm proteins in plethodontid salamanders (PMF, SPFs) (Doty et al., 2016; Fujihara et al., 2021). Salamander SPFs have a duplicated three finger protein domain, and have evolved structural modifications to those domains (Doty et al., 2016). Similarly, the family of ZP proteins (named after the Zona Pellucida), essential components of egg coats across vertebrates and invertebrates (Wilburn and Swanson, 2016), show evidence of independent expansions of ZP-N domains in different lineages (Liang and Dean, 1993; Galindo et al., 2002). These highlight the role of gene duplication and repeat domain expansions in fertilization. An observed trend is rapid sequence evolution in reproductive proteins (Swanson and Vacquier, 2002), and newly duplicated domains can provide novel substrates for evolving new functions at multiple stages of the fertilization cascade.
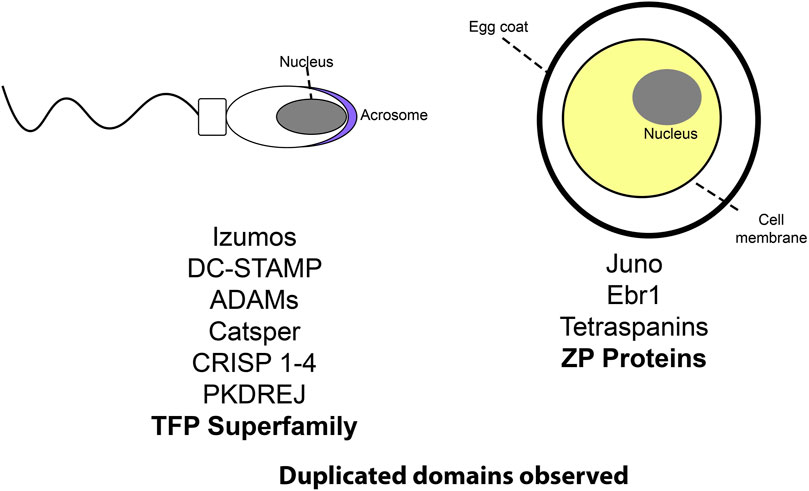
FIGURE 1. A cartoon schematic listing several protein families involved in reproduction. Those with notable repeat expansions are bolded.
The role of duplications in genome evolution is well documented across the tree of life. (Kondrashov et al., 2002; Conant and Wolfe, 2008). Gene duplication (Ponting, 2008) is an important source for new genetic material that facilitates biological innovation. The duplication and differentiation of genomic regions has been linked to the evolution of modularity in organisms (Wagner et al., 2007). Modularity is an abstract concept in which part of an organism (such as a network of protein interactions) functions largely autonomously relative to other aspects of the organisms’ biology (Wagner and Altenberg, 1996; West-Eberhard, 2005). Duplicated genes can participate in existing modular protein interaction networks, which facilitates increasing biological complexity of these networks (Wagner et al., 2007). Such increases in modular network complexity through gene duplication has been linked to adaptations in humans (Perry et al., 2007). Duplicated functional domains can similarly contribute to the evolution of biological complexity. This review will discuss both whole gene duplications and within gene domain duplications, and their role in the evolution of reproductive functions.
When genes duplicate they experience one of three possible fates: pseudogenization, subfunctionalization, and neofunctionalization (Walsh, 2003; Innan, 2009). Due to redundancies in function, the duplicated gene may no longer experience conservation and accumulate silencing mutations, resulting in a non-coding “pseudogene” (Figure 2). New mutations are frequently deleterious, so pseudogenization is hypothesized to be the most common fate of duplicated genes (Lynch and Conery, 2000). However, the other two fates of duplicated genes (subfunctionalization and neofunctionalization) are common mechanisms for biological innovation. Under neofunctionalization, one gene copy maintains its original function while the other experiences positive selection and evolves a novel function. While under subfunctionalization, both copies parse the original function, and neither gene is sufficient (Walsh, 2003; Innan, 2009).
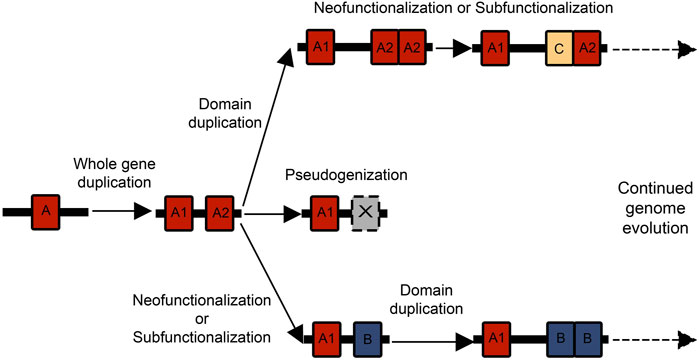
FIGURE 2. There are multiple possible combinations of whole gene and domain duplications that can birth new genes and functional domains. Often a whole gene duplication begins the process, and then one of the gene duplicates experiences a domain expansion. These genes can then act as substrates for further duplication and neofunctionalization or subfunctionalization events.
Tandem duplications of individual protein domains within a gene can add greater complexity to the duplication process. Paralogous genes experiencing relaxed selection can have greater freedom for tandem domain duplications. There is strong research interest in the mechanisms underlying domain repeat expansions and how they affect the evolution of protein families (Björklund et al., 2005, 2006; Vogel et al., 2005; Weiner et al., 2006; Moore et al., 2008; Buljan and Bateman, 2009). Repeats can experience concerted evolution where they maintain a high degree of sequence identity (Elder and Turner, 1995; Liao, 1999), through unequal recombination and gene conversion (Schimenti, 1999). Under this scenario, the repeat expansion of highly identical domains is itself an innovation that could allow proteins to evolve novel functions. A repeat domain expansion could also affect dosage or protein interaction networks. Repeated domains could similarly differentiate in amino acid sequence, leading to neofunctionalization or subfunctionalization with the original domain. There are many possible orders and combinations of whole gene duplications and domain duplications that can contribute to the expansion of gene families (Figure 2). The process by which duplicate genes are maintained and experience subfunctionalization or neofunctionalization has been characterized under the duplication-degeneration-complementation model (DDC) (Force et al., 1999). While most classical population genetics models (Walsh, 2003; Innan, 2009) primarily discuss the effect of silencing or beneficial mutations on coding regions, the DDC model focuses on the effect of mutations on regulatory regions and subfunctionalization. Essentially, mutations that can silence certain regulatory regions in a duplicate gene can lead to the two genes partitioning expression and eventually function (Force et al., 1999). Other models have suggested subfunctionalization is primarily important as a transition phase to neofunctionalization (Rastogi and Liberles, 2005). The mechanisms of subfunctionalization and neofunctionalization remain a subject of rich debate, and concepts like the DDC model could have ramifications for protein evolution.
Subfunctionalization and neofunctionalization are foundational to the evolution of increased complexity in genomes and protein networks, and it is worth examining their particular importance in fertilization. Fertilization proteins are some of the most rapidly evolving proteins in genomes, as evidenced by high amino divergence (Swanson and Vacquier, 2002). Their rapid evolution is likely driven by factors such as sexual conflict and molecular arms race dynamics between gametes, which can also contribute to the maintenance of fertilization barriers between species (Gavrilets and Waxman, 2002; Gavrilets, 2014). The general trend of rapid evolution in reproductive proteins could facilitate the subfunctionalization or neofunctionalization of domains.
Izumo/Juno
The fusion of sperm and egg is necessary for fertilization, but there are only a few known pairs of interacting gametic proteins identified at this stage (Wilburn and Swanson, 2016). After years of research the interacting pair Izumo1 and Juno were identified in mammals (Bianchi et al., 2014). Izumo1 is the sperm expressed protein that mediates fusion (Inoue et al., 2005), and it interacts with the egg surface bound folate receptor 4 (known as Juno) (Bianchi and Wright, 2014). Izumo1 and Juno are each part of protein families with multiple paralogues, but only the Izumo1/Juno pair is capable of interacting (Bianchi et al., 2014). There are four members of both the Izumo (Ellerman et al., 2009) and folate receptor families (FOLR) in mammals (Elwood, 1989; Shen et al., 1994; Spiegelstein et al., 2000; Petronella and Drouin, 2014). Despite being part of the folate receptor family, Juno does not actually bind folate, exemplifying how a single member of this gene family has been coopted for a novel reproductive function (Bianchi et al., 2014).
While Juno represents a clear cooption into fertilization, the evolution of the Izumo gene family could also present an interesting example of neofunctionalization. Izumo1-4 all have a highly structurally conserved Izumo domain, but Izumo1 and Izumo4 have a shared pair of β-strands extending from this domain. Izumo1 experienced further structural modifications, as its β-strand extensions act as a hinge between the Izumo domain and a coopted immunoglobulin-like domain (Aydin et al., 2016; Ohto et al., 2016). Such substantial structural changes could be important for the protein’s ability to bind Juno. Research into other Izumo proteins suggests their involvement in fertilization. Izumo1-3 are transmembrane testis expressed proteins (Ellerman et al., 2009), while Izumo4 lacks a transmembrane domain and is expressed in the acrosome (Guasti et al., 2020). Izumo3 shows evidence of positive selection (Grayson and Civetta, 2012), and is necessary for sperm acrosome formation (Inoue et al., 2021b). The parallel histories of structural modifications in Izumo1 and Juno allowed for this essential interaction to evolve.
The relationship between Izumo1, Juno and their paralogs is highlighted by our phylogeny (Figure 3), which contains a long branch leading to Juno (FOLR4). This could reflect the rapid accumulation of mutations in the Juno branch as it was coopted to bind Izumo1 during gametic membrane fusion. Crystal structures confirm that 1:1 binding complexes form between Izumo1 and Juno (Aydin et al., 2016; Ohto et al., 2016). The adhesion of Izumo1 and Juno is conserved in mammals, and after the adhesion event Juno is released from the egg’s surface in vesicles and may act to bind and neutralize acrosome reacted sperm (Bianchi et al., 2014). In mammals, this interaction functions as a block against polyspermy (Bianchi and Wright, 2014). Blocks to polyspermy are essential, because eggs that fuse with multiple sperm are not viable and mammalian blocks to polyspermy exist at both the cell membrane (Evans, 2020) and egg coat (Fahrenkamp et al., 2020).
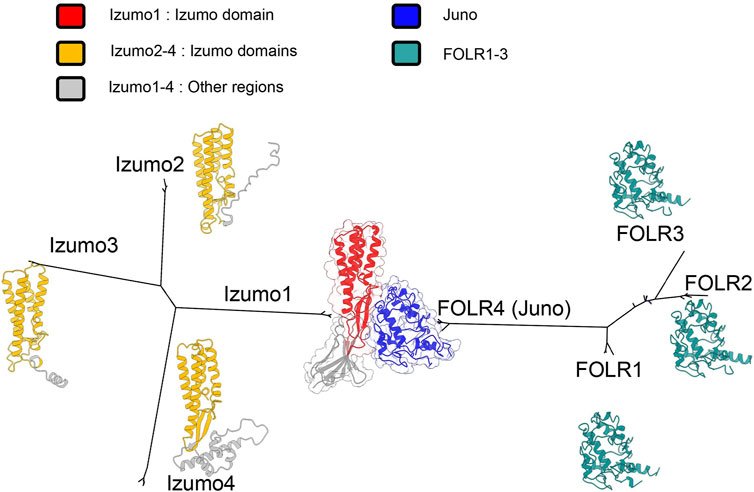
FIGURE 3. Unrooted maximum likelihood phylogenies for Izumo and FOLR gene families in a subset of primates, based on multiple sequence alignments (Katoh and Standley, 2013; Kozlov et al., 2019). Both gene families independently duplicated, but FOLR4 was coopted to bind Izumo1. Crystal structures have been obtained for the Izumo1-Juno complex (Aydin et al., 2016). For other proteins, alphafold predicted structures were used (Jumper et al., 2021). Using predictions of signal peptides and transmembrane domains, and secondary structural alignments, we identified shared izumo domains (Sonnhammer et al., 1998; Krogh et al., 2001; Almagro Armenteros et al., 2019).
Mutations to residues conserved in mammals greatly reduce binding, highlighting that particular changes to amino acid sequence and protein structure facilitated the neofunctionalization of Juno (Aydin et al., 2016). The more variable structural features (Ohto et al., 2016) in Juno may be important for the species-specificity of its binding to Izumo1 (Bianchi et al., 2014; Bianchi and Wright, 2015; Han et al., 2016). Comparative genetic analyses identify positive selection in a subset of mammals (Laurasiatheria) (Grayson and Civetta, 2012), and that Juno is likely rapidly coevolving with Izumo1, which contributes to the specificity of their interactions (Grayson, 2015). This specific binding is essential to both Juno’s function in initiating membrane fusion, and the post-fusion neutralization of acrosome-reacted sperm (Wright and Bianchi, 2016).
DCST
While Izumo1 and Juno are thought to initiate the complex molecular process of gametic membrane fusion in mammals, recent transgenic experiments and complementation studies have demonstrated that DCST1 and DCST2 are also essential (Inoue et al., 2021a). The DCST1/2 proteins are expressed on the sperm surface, and contain variable (4–6) transmembrane helical domains (DC-STAMP) (Inoue et al., 2021a, 1). DC-STAMP (dendritic cell specific transmembrane protein) refers to both the name of the domain and one of the proteins that contains this domain (Hartgers et al., 2000). The originally identified DC-STAMP protein has four transmembrane domains (Hartgers et al., 2001), and it is highly expressed in myeloid dendrocytes (Hartgers et al., 2000, 2001; Eleveld-Trancikova et al., 2005, 2008). The expression of DC-STAMP has been induced in macrophages (Staege et al., 2001) and osteoclasts (Nomiyama et al., 2005). This broad array of functions has motivated much research into the molecular mechanisms of DC-STAMP interactions, which has supported a role in osteoclast fusion (Kukita et al., 2004; Yagi et al., 2005; Jansen et al., 2009). There is also evidence of DC-STAMP related signaling in immune response (Nair et al., 2016). Along with these other diverse functions, it seem that DC-STAMP domains have been coopted into an essential role in sperm-egg membrane fusion.
DCST1/2 are the first known essential fertilization factors that are conserved in both vertebrates and invertebrates (Inoue et al., 2021a). DCST1/2 orthologues have been identified in both Caenorhabditis and Drosophila (Kroft et al., 2005; Wilson et al., 2006, 2018), which is the first known example sperm related factors being conserved this broadly across vertebrates and invertebrates (Inoue et al., 2021a, 1). However, there has been extensive structural diversification of these DCST1/2 across animals (Figure 4), especially between invertebrates and vertebrates. The low sequence identity of DCST1/2 proteins across animals, makes the conservation of reproductive function all the more remarkable. The ubiquitin ligase activity of DCST1 (Nair et al., 2016) raises questions about the function of DCST1/2 in sperm. There is intense research interest into the signal activity of long non-coding RNA produced by DCST1 and its effect on cancer cell progression (Hu et al., 2020; Ai et al., 2021, 1; Wang et al., 2021). More investigation is necessary to understand the function of DC-STAMP domains in a broad range of signaling networks, and how they were neofunctionalized in sperm DCST1/2.
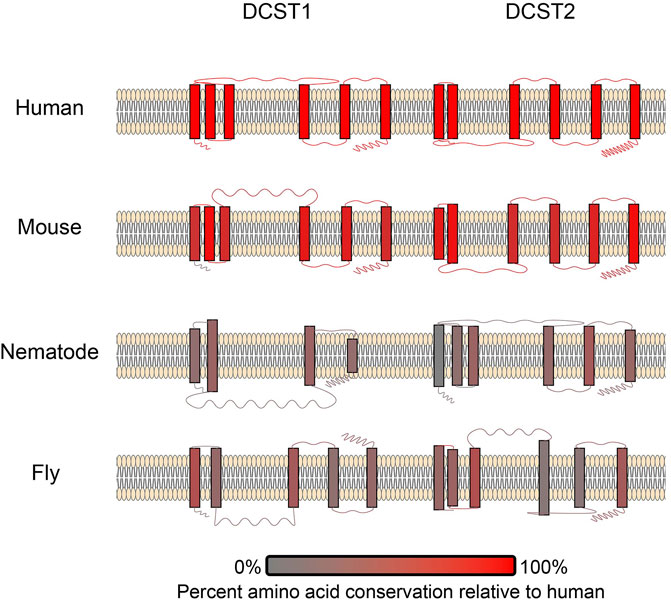
FIGURE 4. A schematic of DCST1/2 proteins in multiple species. The number of transmembrane domains and loop lengths differ across species. Transmembrane domains and loops are colored based on conservation (Pei et al., 2008), where red coloration signifies amino acid conservation relative to humans. Therefore, the human examples are all red.
ZP Domains
ZP proteins are an essential class of egg coat proteins. An important feature of ZP proteins is the ZP module that consists of two domains, ZP-N and ZP-C, named after their relative N-terminal and C-terminal positioning. ZP-N and ZP-C domain are immunoglobular domains with characteristic patterns of disulfide bonding and β-sheets (Bokhove and Jovine, 2018), and likely resulted from an ancestral domain duplication. The variability in amino acid sequence, disulfide placement, and loop structures between ZP-N and ZP-C (Lin et al., 2011) suggests differences in their biological function and evolutionary history.
ZP-N domains are of particular interest, because they form asymmetric dimers with their β-sandwich edges which are believed to promote polymerization between ZP modules (Jovine et al., 2002; Wilburn and Swanson, 2017; Bokhove and Jovine, 2018). There are several ZP proteins identified in vertebrates (ZP1-4, ZPAX and ZPD), and there appears to be a history of lineage specific gain and loss of ZP proteins among vertebrates (Galindo et al., 2002; Conner et al., 2005; Goudet et al., 2008; Claw and Swanson, 2012; Meslin et al., 2012; Shu et al., 2015; Killingbeck and Swanson, 2018). Like other families discussed in this review, there also multiple ZP proteins with non-reproductive functions (e.g., uromodulin and tectorin-alpha) (Legan et al., 1997; Brunati et al., 2015; Bokhove et al., 2016). This may be another example of domains being coopted into a reproductive function, and ZP-N polymerization domains may be important for egg coat assembly and structure.
Not only has gene duplication produced an assortment of ZP proteins, there are also examples of independent repeat expansions of ZP-N in both vertebrates and invertebrate egg coat proteins (Figure 5). Some have only one additional ZP-N domain, but there are more dramatic repeat expansion like mammalian ZP2 (4 ZP-Ns) and abalone VERL (23 ZP-Ns) (Galindo et al., 2002). This process of domain duplications helped contribute to the diversity of ZP proteins. Given the ability of ZP-N domains to dimerize (Jovine et al., 2002; Bokhove and Jovine, 2018; Litscher and Wassarman, 2020), their duplications could create opportunities to evolve novel binding functions. Proteins with duplicated ZP-N domains, such as mammalian ZP2 and abalone VERL, are thought to be essential for species-specific in fertilization (Avella et al., 2013, 2014; Raj et al., 2017). Species-specificity in abalone is associated with the coevolution between VERL and the sperm protein lysin (Galindo et al., 2003; Clark et al., 2009), suggesting a cooption of ZP-Ns in sperm-egg interactions during egg coat dissolution.
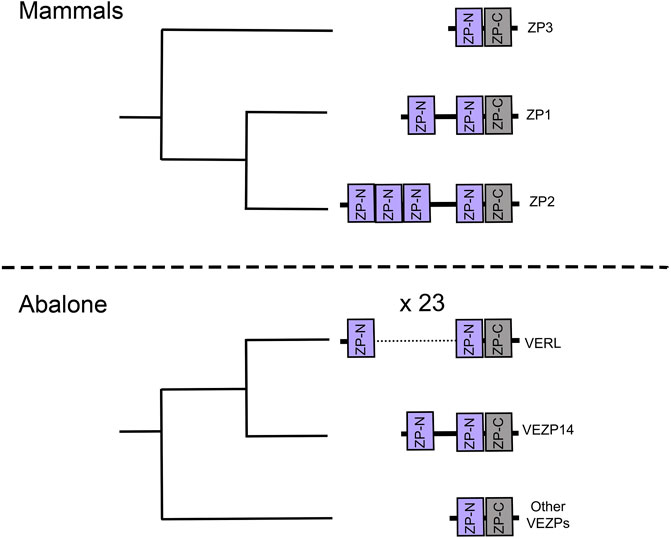
FIGURE 5. Cladograms of ZP-N proteins are based on phylogenies from the literature (Aagaard et al., 2010; Claw and Swanson, 2012). These suggest independent repeat expansion of the ZP-N domain in both abalone and human egg coat genes.
Neofunctionalization of ZP-N domains can also drive new interactions between ZP proteins, such as the evolution of essential intermolecular crosslinks (Nishimura et al., 2019), which affect the physical assemblage of proteins in the supramolecular structure of the egg coat. Indeed, mouse research has suggested the importance of egg coat supramolecular structure in fertilization (Rankin et al., 2003; Avella et al., 2013). The structure of the egg coat is also important for the oocyte’s ability to block polyspermy. Protein cleavage of ZP2 is thought to initiate other egg coat structural modifications, which “harden” the egg coat and prevent sperm binding (Bleil et al., 1981; Gahlay et al., 2010; Fahrenkamp et al., 2020). Gene and domain duplications has produced a family of ZP proteins that contribute to the egg coat supramolecular structure, and are involved in both sperm recognition and polyspermy avoidance.
TFP Superfamily
Three finger proteins are defined by their TFP domains, which have a characteristic disulfide bonding pattern and fold (Galat, 2008; Galat et al., 2008). The broader TFP protein superfamily also includes proteins with structurally modified TFP-like domains (Galat, 2015). While TFPs were originally identified in snake toxins (Low et al., 1976; Tsernoglou and Petsko, 1977), members of the TFP superfamily have been to coopted for reproductive functions into sperm (SPACA4, PMFs, and SPFs), egg (Bouncer), and pheromones (PMFs, and SPFs) (Doty et al., 2016; Fujihara et al., 2021; Wilburn et al., 2022) (Figure 6). Bouncer plays a role in species-specific sperm-egg fusion in teleost fish (Herberg et al., 2018), which raises questions about how other TFPs may function in fertilization. The TFP superfamily includes both soluble and membrane bound proteins, and has great functional diversity across many tissues and taxa (Alape-Girón et al., 1999; Tsetlin, 1999; Kini, 2002; Nirthanan et al., 2003; Kessler et al., 2017). Similar to ZP proteins, we observe a history of gene duplication, repeat expansion of domains, and functional diversification of TFP containing proteins.
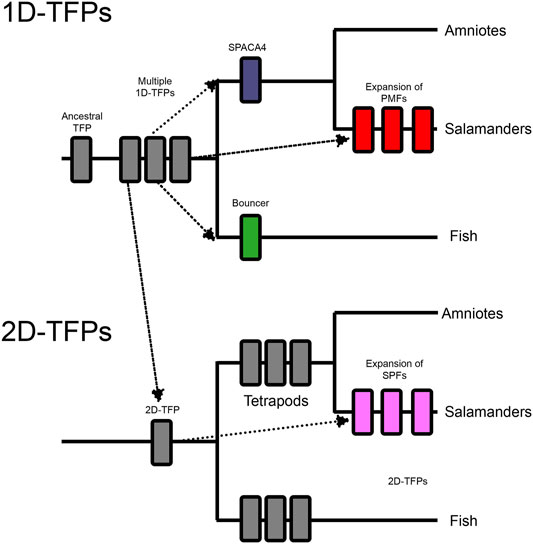
FIGURE 6. These two cladograms outline the whole gene and domain duplications within the three finger protein superfamilty (TFPs) and their expansions into reproductive systems. An ancestral single domain TFP (1D-TFP), duplicated into multiple vertebrate 1D-TFPs, and also had a domain level duplication which created a lineage of two TFP domain proteins (2D-TFPs). The 1D-TFPs produced tetrapod SPACA4, fish Bouncer, and multiple salamander PMFs. The 2D-TFPs also duplicated throughout vertebrates including salamander SPFs. Both salamander PMF and SPF protein families include both sperm and pheromone expressed members (Wilburn et al., 2022).
An ancestral TFP protein experienced gene duplication to produce an assortment of single TFP-like domain proteins (1D-TFPs). One of these TFP genes experienced a tandem domain expansion to produce the ancestor of proteins with two TFP-like domains (2D-TFPs). Three independent cooption events have produced TFPs in gametes (Figure 6). A cooption of 1D-TFPs occurred in the ancestor of tetrapods and produced both Bouncer in fish, and SPACA4 in amniotes (Figure 6). Despite their protein homology, Bouncer is egg expressed while SPACA4 is sperm expressed and it is implicated in interactions between the sperm and egg coat (Fujihara et al., 2021), highlighting the functional diversification of TFPs. Another independent cooption of 1D-TFPs resulted in the sperm expressed plethodontid modulating factor (PMFs) salamanders, which extensively duplicated producing a diverse family of reproductive molecules (Wilburn et al., 2012, 2014, 2017; Doty et al., 2016). Salamander PMFs are hypervariable proteins expressed in multiple tissues, and while they are structurally similar to other TFPs, they differ in loop length and disulfide bridge patterning, and show evidence of persistent diversification and positive selection (Palmer et al., 2010; Wilburn et al., 2012, 2014).
Among 2D-TFPs there was independent cooption into the sodefrin precursor-like factors (SPFs) of salamander sperm. SPFs then experienced their own history of gene duplications and radiation (Palmer et al., 2007). Both PMFs and SPFs experienced disulfide bond reshuffling relative to the canonical 1D-TFP and 2D-TFP binding patterns, and these changes reflect the neofunctionalization of these molecules (Doty et al., 2016). These striking examples of independent gene duplications and neofunctionalization for reproductive functions raises questions as to whether there a more additional unknown cooptions of TFPs, and whether some protein domains are more susceptible to cooption in diverse biological contexts.
Both PMFs and SPFs are highly duplicated protein families, with some members being coopted into pheromone function and others for sperm expression (Doty et al., 2016; Wilburn et al., 2022). As the sperm paralogs of PMFs and SPFs have only recently been discovered, functional studies have not yet been conducted. Male salamanders produce large number of PMFS and SPFs within their mental glands which promote ritual courtship behavior in females (Doty et al., 2016). Duplications of secreted male-expressed sperm proteins could have provided an evolutionary substrate to evolve new pheromones (Wilburn et al., 2022). Structural changes in PMFs and SPFs, such as disulfide shuffling, may contribute to new functions in both sperm and pheromones. The TFP’s superfamily’s history of gene duplication, domain duplication, and neofunctionalization provides a unique model for the evolution of large gene families involved in fertilization.
Discussion and Conclusion
Within this review we discussed examples of duplicated gene families with roles in fertilization. Gene duplication and neofunctionalization is an essential process for the evolution of greater genomic and functional complexity in organisms. Duplicated paralogous genes have been coopted into both sperm (Izumo1, DCST1/2) and egg (Juno) proteins involved in gamete membrane fusion (Bianchi et al., 2014; Inoue et al., 2021a, 1). Domain duplications within paralogs is also observed in the TFP superfamily and ZPs and has allowed both groups of genes to adopt novel functions at multiple stages of fertilization. As seen with TFPs, duplication events are often followed by notable protein structural changes (Doty et al., 2016) which may be tied to their cooption for novel fertilization functions. It is intriguing to consider hypotheses that account for these patterns of gene family expansion and diversification common in reproductive molecules.
Duplication events can facilitate the rapid evolution and neofunctionalization observed in many families of fertilization proteins. This rapid evolution can also be influenced by multiple factors such as sexual conflict, polyspermy avoidance, or genetic drift (Vacquier et al., 1997). The necessity of pathogen avoidance or blocks to polyspermy can drive oocytes to evolve reduced sperm binding ability. The sperm would then coevolutionarily “chase” the egg, which can contribute to the rapid sequence evolution of gametic proteins, and to the species-specificity of these protein interactions (Gavrilets and Waxman, 2002; Gavrilets, 2014). The rapid evolution of reproductive proteins is explored in terms of amino acid mutations, but the repeat expansion of domains could also be part of this trend. Proteins with repeated domains could experience drift resulting in ever-changing molecular target, that interacting proteins must coevolutionarily chase (Vacquier et al., 1997).
Duplications of reproductive proteins can also contribute to the phenomenon of functional redundancy, in which two duplicated genes have partially overlapping functions and can compensate for each other’s loss (Kafri et al., 2009). Functional redundancy has been observed in the CRISP family of reproductive proteins (Curci et al., 2020), and this property could emerge in other large protein families. While functional redundancy seems like it would be temporary as duplicated genes subfunctionalized or neofunctionalized, it can be a surprisingly evolutionarily stable property. Functional redundancy could confer fitness advantages by maintaining the robusticity of protein interaction networks in spite of stochasticity of expression between cells (Kafri et al., 2009). The rapid evolution of other reproductive proteins in these networks could place even greater value on robustness and stability of essential functions. Robusticity in these protein networks is believed to reduce the fitness cost of new mutations, which would increase the “evolvability” of these proteins and facilitate functional innovation (Kirschner and Gerhart, 2008). The concepts of functional redundancy and robusticity of function may also apply to domain repeat expansions like the ZP-N domains of VERL. The processes of gene duplication, repeat domain expansion, structural modification, and neofunctionalization have been fundamental to the evolution of reproductive molecules across life.
Author Contributions
Both authors were involved in the conception of this review. AR principally conducted the literature review, and the writing of the manuscript. WS provided substantial literature suggestions and editorial feedback.
Funding
The lab is funded by the NIH grant HD105025 awarded to WS.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Damien B. Wilburn for sharing his code for visualizing transmembrane proteins, and fellow lab members Jolie Carlisle and Jan Aagaard for engaging in discussions.
References
Aagaard, J. E., Vacquier, V. D., MacCoss, M. J., and Swanson, W. J. (2010). ZP Domain Proteins in the Abalone Egg Coat Include a Paralog of VERL under Positive Selection that Binds Lysin and 18-kDa Sperm Proteins. Mol. Biol. Evol. 27, 193–203. doi:10.1093/molbev/msp221
Ai, Y., Liu, S., Luo, H., Wu, S., Wei, H., Tang, Z., et al. (2021). lncRNA DCST1-AS1 Facilitates Oral Squamous Cell Carcinoma by Promoting M2 Macrophage Polarization through Activating NF-κB Signaling. J. Immunol. Res. 2021, 1–9. doi:10.1155/2021/5524231
Alape-Girón, A., Persson, B., Cederlund, E., Flores-Díaz, M., Gutiérrez, J. M., Thelestam, M., et al. (1999). Elapid Venom Toxins: Multiple Recruitments of Ancient Scaffolds. Eur. J. Biochem. 259, 225–234. doi:10.1046/j.1432-1327.1999.00021.x
Almagro Armenteros, J. J., Tsirigos, K. D., Sønderby, C. K., Petersen, T. N., Winther, O., Brunak, S., et al. (2019). SignalP 5.0 Improves Signal Peptide Predictions Using Deep Neural Networks. Nat. Biotechnol. 37, 420–423. doi:10.1038/s41587-019-0036-z
Avella, M. A., Xiong, B., and Dean, J. (2013). The Molecular Basis of Gamete Recognition in Mice and Humans. Mol. Hum. Reprod 19, 279–289. doi:10.1093/molehr/gat004
Avella, M. A., Baibakov, B., and Dean, J. (2014). A Single Domain of the ZP2 Zona Pellucida Protein Mediates Gamete Recognition in Mice and Humans. J. Cell. Biol. 205, 801–809. doi:10.1083/jcb.201404025
Aydin, H., Sultana, A., Li, S., Thavalingam, A., and Lee, J. E. (2016). Molecular Architecture of the Human Sperm IZUMO1 and Egg JUNO Fertilization Complex. Nature 534, 562–565. doi:10.1038/nature18595
Bianchi, E., and Wright, G. J. (2014). Izumo Meets Juno. Cell Cycle 13, 2019–2020. doi:10.4161/cc.29461
Bianchi, E., and Wright, G. J. (2015). Cross-species Fertilization: the Hamster Egg Receptor, Juno, Binds the Human Sperm Ligand, Izumo1. Phil. Trans. R. Soc. B 370, 20140101. doi:10.1098/rstb.2014.0101
Bianchi, E., Doe, B., Goulding, D., and Wright, G. J. (2014). Juno Is the Egg Izumo Receptor and Is Essential for Mammalian Fertilization. Nature 508, 483–487. doi:10.1038/nature13203
Björklund, Å. K., Ekman, D., Light, S., Frey-Skött, J., and Elofsson, A. (2005). Domain Rearrangements in Protein Evolution. J. Mol. Biol. 353, 911–923. doi:10.1016/j.jmb.2005.08.067
Björklund, Å. K., Ekman, D., and Elofsson, A. (2006). Expansion of Protein Domain Repeats. Plos Comput. Biol. 2, e114. doi:10.1371/journal.pcbi.0020114
Bleil, J. D., Beall, C. F., and Wassarman, P. M. (1981). Mammalian Sperm-Egg Interaction: Fertilization of Mouse Eggs Triggers Modification of the Major Zona Pellucida Glycoprotein, ZP2. Dev. Biol. 86, 189–197. doi:10.1016/0012-1606(81)90329-8
Bokhove, M., and Jovine, L. (2018). Structure of Zona Pellucida Module Proteins. Curr. Topic Dev. Biol. 130, 413–442. doi:10.1016/bs.ctdb.2018.02.007
Bokhove, M., Nishimura, K., Brunati, M., Han, L., de Sanctis, D., Rampoldi, L., et al. (2016). A Structured Interdomain Linker Directs Self-Polymerization of Human Uromodulin. Proc. Natl. Acad. Sci. USA 113, 1552–1557. doi:10.1073/pnas.1519803113
Brunati, M., Perucca, S., Han, L., Cattaneo, A., Consolato, F., Andolfo, A., et al. (2015). The Serine Protease Hepsin Mediates Urinary Secretion and Polymerisation of Zona Pellucida Domain Protein Uromodulin. Elife 4, e08887. doi:10.7554/eLife.08887
Buljan, M., and Bateman, A. (2009). The Evolution of Protein Domain Families. Biochem. Soc. Trans. 37, 751–755. doi:10.1042/BST0370751
Busso, D., Goldweic, N. M., Hayashi, M., Kasahara, M., and Cuasnicú, P. S. (2007). Evidence for the Involvement of Testicular Protein CRISP2 in Mouse Sperm-Egg Fusion1. Biol. Reprod. 76, 701–708. doi:10.1095/biolreprod.106.056770
Civetta, A. (2003). Positive Selection within Sperm-Egg Adhesion Domains of Fertilin: An ADAM Gene with a Potential Role in Fertilization. Mol. Biol. Evol. 20, 21–29. doi:10.1093/molbev/msg002
Clapham, D. E., and Garbers, D. L. (2005). International Union of Pharmacology. L. Nomenclature and Structure-Function Relationships of CatSper and Two-Pore Channels. Pharmacol. Rev. 57, 451–454. doi:10.1124/pr.57.4.7
Clark, N. L., Gasper, J., Sekino, M., Springer, S. A., Aquadro, C. F., and Swanson, W. J. (2009). Coevolution of Interacting Fertilization Proteins. Plos Genet. 5, e1000570. doi:10.1371/journal.pgen.1000570
Claw, K. G., and Swanson, W. J. (2012). Evolution of the Egg: New Findings and Challenges. Annu. Rev. Genom. Hum. Genet. 13, 109–125. doi:10.1146/annurev-genom-090711-163745
Conant, G. C., and Wolfe, K. H. (2008). Turning a Hobby into a Job: How Duplicated Genes Find New Functions. Nat. Rev. Genet. 9, 938–950. doi:10.1038/nrg2482
Connallon, T., and Clark, A. G. (2011). The Resolution of Sexual Antagonism by Gene Duplication. Genetics 187, 919–937. doi:10.1534/genetics.110.123729
Conner, S. J., Lefièvre, L., Hughes, D. C., and Barratt, C. L. R. (2005). Cracking the Egg: Increased Complexity in the Zona Pellucida. Human Reprod. 20, 1148–1152. doi:10.1093/humrep/deh835
Curci, L., Brukman, N. G., Weigel Muñoz, M., Rojo, D., Carvajal, G., Sulzyk, V., et al. (2020). Functional Redundancy and Compensation: Deletion of Multiple Murine Crisp Genes Reveals Their Essential Role for Male Fertility. FASEB j. 34, 15718–15733. doi:10.1096/fj.202001406R
Da Ros, V. G., Maldera, J. A., Willis, W. D., Cohen, D. J., Goulding, E. H., Gelman, D. M., et al. (2008). Impaired Sperm Fertilizing Ability in Mice Lacking Cysteine-RIch Secretory Protein 1 (CRISP1). Dev. Biol. 320, 12–18. doi:10.1016/j.ydbio.2008.03.015
Doty, K. A., Wilburn, D. B., Bowen, K. E., Feldhoff, P. W., and Feldhoff, R. C. (2016). Co-option and Evolution of Non-olfactory Proteinaceous Pheromones in a Terrestrial Lungless Salamander. J. Proteomics 135, 101–111. doi:10.1016/j.jprot.2015.09.019
Dufour, S., Quérat, B., Tostivint, H., Pasqualini, C., Vaudry, H., and Rousseau, K. (2020). Origin and Evolution of the Neuroendocrine Control of Reproduction in Vertebrates, with Special Focus on Genome and Gene Duplications. Physiol. Rev. 100, 869–943. doi:10.1152/physrev.00009.2019
Elder, J. F., and Turner, B. J. (1995). Concerted Evolution of Repetitive DNA Sequences in Eukaryotes. Q. Rev. Biol. 70, 297–320. doi:10.1086/419073
Eleveld-Trancikova, D., Triantis, V., Moulin, V., Looman, M. W. G., Wijers, M., Fransen, J. A. M., et al. (2005). The Dendritic Cell-Derived Protein DC-STAMP Is Highly Conserved and Localizes to the Endoplasmic Reticulum. J. Leukoc. Biol. 77, 337–343. doi:10.1189/jlb.0804441
Eleveld-Trancikova, D., Janssen, R. A. J., Hendriks, I. A. M., Looman, M. W. G., Moulin, V., Jansen, B. J. H., et al. (2008). The DC-Derived Protein DC-STAMP Influences Differentiation of Myeloid Cells. Leukemia 22, 455–459. doi:10.1038/sj.leu.2404910
Ellerman, D. A., Pei, J., Gupta, S., Snell, W. J., Myles, D., and Primakoff, P. (2009). Izumo Is Part of a Multiprotein Family Whose Members Form Large Complexes on Mammalian Sperm. Mol. Reprod. Dev. 76, 1188–1199. doi:10.1002/mrd.21092
Elwood, P. C. (1989). Molecular Cloning and Characterization of the Human Folate-Binding Protein cDNA from Placenta and Malignant Tissue Culture (KB) Cells. J. Biol. Chem. 264, 14893–14901. doi:10.1016/S0021-9258(18)63786-X
Evans, J. P. (2020). Preventing Polyspermy in Mammalian Eggs-Contributions of the Membrane Block and Other Mechanisms. Mol. Reprod. Dev. 87, 341–349. doi:10.1002/mrd.23331
Fahrenkamp, E., Algarra, B., and Jovine, L. (2020). Mammalian Egg Coat Modifications and the Block to Polyspermy. Mol. Reprod. Dev. 87, 326–340. doi:10.1002/mrd.23320
Finn, S., and Civetta, A. (2010). Sexual Selection and the Molecular Evolution of ADAM Proteins. J. Mol. Evol. 71, 231–240. doi:10.1007/s00239-010-9382-7
Force, A., Lynch, M., Pickett, F. B., Amores, A., Yan, Y.-l., and Postlethwait, J. (1999). Preservation of Duplicate Genes by Complementary, Degenerative Mutations. Genetics 151, 1531–1545. doi:10.1093/genetics/151.4.1531
Frolikova, M., Manaskova-Postlerova, P., Cerny, J., Jankovicova, J., Simonik, O., Pohlova, A., et al. (2018). CD9 and CD81 Interactions and Their Structural Modelling in Sperm Prior to Fertilization. Ijms 19, 1236. doi:10.3390/ijms19041236
Fujihara, Y., Herberg, S., Blaha, A., Panser, K., Kobayashi, K., Larasati, T., et al. (2021). The Conserved Fertility Factor SPACA4/Bouncer Has Divergent Modes of Action in Vertebrate Fertilization. Proc. Natl. Acad. Sci. USA 118, e2108777118. doi:10.1073/pnas.2108777118
Gahlay, G., Gauthier, L., Baibakov, B., Epifano, O., and Dean, J. (2010). Gamete Recognition in Mice Depends on the Cleavage Status of an Egg's Zona Pellucida Protein. Science 329, 216–219. doi:10.1126/science.1188178
Galat, A., Gross, G., Drevet, P., Sato, A., and Ménez, A. (2008). Conserved Structural Determinants in Three-Fingered Protein Domains. FEBS J. 275, 3207–3225. doi:10.1111/j.1742-4658.2008.06473.x
Galat, A. (2008). The Three-Fingered Protein Domain of the Human Genome. Cell. Mol. Life Sci. 65, 3481–3493. doi:10.1007/s00018-008-8473-8
Galat, A. (2015). Multidimensional Drift of Sequence Attributes and Functional Profiles in the Superfamily of the Three-Finger Proteins and Their Structural Homologues. J. Chem. Inf. Model. 55, 2026–2041. doi:10.1021/acs.jcim.5b00322
Galindo, B. E., Moy, G. W., Swanson, W. J., and Vacquier, V. D. (2002). Full-Length Sequence of VERL, the Egg Vitelline Envelope Receptor for Abalone Sperm Lysin. Gene 288, 111–117. doi:10.1016/s0378-1119(02)00459-6
Galindo, B. E., Vacquier, V. D., and Swanson, W. J. (2003). Positive Selection in the Egg Receptor for Abalone Sperm Lysin. Proc. Natl. Acad. Sci. 100, 4639–4643. doi:10.1073/pnas.0830022100
Gallach, M., and Betrán, E. (2011). Intralocus Sexual Conflict Resolved through Gene Duplication. Trends Ecol. Evol. 26, 222–228. doi:10.1016/j.tree.2011.02.004
Gallach, M., Chandrasekaran, C., and Betrán, E. (2010). Analyses of Nuclearly Encoded Mitochondrial Genes Suggest Gene Duplication as a Mechanism for Resolving Intralocus Sexually Antagonistic Conflict in Drosophila. Genome Biol. Evol. 2, 835–850. doi:10.1093/gbe/evq069
Gallach, M., Domingues, S., and Betrán, E. (2011). Gene Duplication and the Genome Distribution of Sex-Biased Genes. Int. J. Evol. Biol. 2011, 1–20. doi:10.4061/2011/989438
Gavrilets, S., and Waxman, D. (2002). Sympatric Speciation by Sexual Conflict. Proc. Natl. Acad. Sci. 99, 10533–10538. doi:10.1073/pnas.152011499
Gavrilets, S. (2014). Is Sexual Conflict an "Engine of Speciation". Cold Spring Harbor Perspect. Biol. 6, a017723. doi:10.1101/cshperspect.a017723
Gelbaya, T. A., Potdar, N., Jeve, Y. B., and Nardo, L. G. (2014). Definition and Epidemiology of Unexplained Infertility. Obstet. Gynecol. Surv. 69(2), 109–115. doi:10.1097/OGX.0000000000000043
Gibbs, G. M., Orta, G., Reddy, T., Koppers, A. J., Martinez-Lopez, P., Luis de la Vega-Beltran, J., et al. (2011). Cysteine-rich Secretory Protein 4 Is an Inhibitor of Transient Receptor Potential M8 with a Role in Establishing Sperm Function. Proc. Natl. Acad. Sci. 108, 7034–7039. doi:10.1073/pnas.1015935108
Goudet, G., Mugnier, S., Callebaut, I., and Monget, P. (2008). Phylogenetic Analysis and Identification of Pseudogenes Reveal a Progressive Loss of Zona Pellucida Genes during Evolution of Vertebrates1. Biol. Reprod. 78, 796–806. doi:10.1095/biolreprod.107.064568
Grayson, P., and Civetta, A. (2012). Positive Selection and the Evolution of Izumo Genes in Mammals. Int. J. Evol. Biol. 2012, 1–7. doi:10.1155/2012/958164
Grayson, P. (2015). Izumo1 and Juno: the Evolutionary Origins and Coevolution of Essential Sperm-Egg Binding Partners. R. Soc. Open Sci. 2, 150296. doi:10.1098/rsos.150296
Guasti, P. N., Souza, F. F., Scott, C., Papa, P. M., Camargo, L. S., Schmith, R. A., et al. (2020). Equine Seminal Plasma and Sperm Membrane: Functional Proteomic Assessment. Theriogenology 156, 70–81. doi:10.1016/j.theriogenology.2020.06.014
Han, J.-H., Batey, S., Nickson, A. A., Teichmann, S. A., and Clarke, J. (2007). The Folding and Evolution of Multidomain Proteins. Nat. Rev. Mol. Cel. Biol. 8, 319–330. doi:10.1038/nrm2144
Han, L., Nishimura, K., Sadat Al Hosseini, H., Bianchi, E., Wright, G. J., and Jovine, L. (2016). Divergent Evolution of Vitamin B9 Binding Underlies Juno-Mediated Adhesion of Mammalian Gametes. Curr. Biol. 26, R100–R101. doi:10.1016/j.cub.2015.12.034
Hart, M. W. (2013). Structure and Evolution of the Sea star Egg Receptor for Sperm Bindin. Mol. Ecol. 22, 2143–2156. doi:10.1111/mec.12251
Hartgers, F. C., Vissers, J. L. M., Looman, M. W. G., Zoelen, C. v., Huffine, C., Figdor, C. G., et al. (2000). DC-STAMP, a Novel Multimembrane-Spanning Molecule Preferentially Expressed by Dendritic Cells. Eur. J. Immunol. 30, 3585–3590. doi:10.1002/1521-4141(200012)30:12<3585:aid-immu3585>3.0.co;2-y
Hartgers, F. C., Looman, M. W. G., van der Woning, B., Merkx, G. F. M., Figdor, C. G., and Adema, G. J. (2001). Genomic Organization, Chromosomal Localization, and 5′ Upstream Region of the Human DC-STAMP Gene. Immunogenetics 53, 145–149. doi:10.1007/s002510100302
Herberg, S., Gert, K. R., Schleiffer, A., and Pauli, A. (2018). The Ly6/uPAR Protein Bouncer Is Necessary and Sufficient for Species-specific Fertilization. Science 361, 1029–1033. doi:10.1126/science.aat7113
Howe, K. L., Achuthan, P., Allen, J., Allen, J., Alvarez-Jarreta, J., Amode, M. R., et al. (2021). Ensembl 2021. Nucleic Acids Res. 49, D884–D891. doi:10.1093/nar/gkaa942
Hu, S., Yao, Y., Hu, X., and Zhu, Y. (2020). LncRNA DCST1-AS1 Downregulates miR-29b through Methylation in Glioblastoma (GBM) to Promote Cancer Cell Proliferation. Clin. Transl. Oncol. 22, 2230–2235. doi:10.1007/s12094-020-02363-1
Hughes, A. L. (1994). The Evolution of Functionally Novel Proteins after Gene Duplication. Proc. R. Soc. Lond. B 256, 119–124. doi:10.1098/rspb.1994.0058
Innan, H. (2009). Population Genetic Models of Duplicated Genes. Genetica 137, 19–37. doi:10.1007/s10709-009-9355-1
Inoue, N., Ikawa, M., Isotani, A., and Okabe, M. (2005). The Immunoglobulin Superfamily Protein Izumo Is Required for Sperm to Fuse with Eggs. Nature 434, 234–238. doi:10.1038/nature03362
Inoue, N., Hagihara, Y., and Wada, I. (2021a). Evolutionarily Conserved Sperm Factors, DCST1 and DCST2, Are Required for Gamete Fusion. Elife 10, e66313. doi:10.7554/eLife.66313
Inoue, N., Satouh, Y., and Wada, I. (2021b). IZUMO Family Member 3, IZUMO3, Is Involved in Male Fertility through the Acrosome Formation. Mol. Reprod. Dev. 88, 479–481. doi:10.1002/mrd.23520
Jansen, B. J. H., Eleveld-Trancikova, D., Sanecka, A., van Hout-Kuijer, M., Hendriks, I. A. M., Looman, M. G. W., et al. (2009). OS9 Interacts with DC-STAMP and Modulates its Intracellular Localization in Response to TLR Ligation. Mol. Immunol. 46, 505–515. doi:10.1016/j.molimm.2008.06.032
Jovine, L., Qi, H., Williams, Z., Litscher, E., and Wassarman, P. M. (2002). The ZP Domain Is a Conserved Module for Polymerization of Extracellular Proteins. Nat. Cell Biol. 4, 457–461. doi:10.1038/ncb802
Jumper, J., Evans, R., Pritzel, A., Green, T., Figurnov, M., Ronneberger, O., et al. (2021). Highly Accurate Protein Structure Prediction with AlphaFold. Nature 596, 583–589. doi:10.1038/s41586-021-03819-2
Kafri, R., Springer, M., and Pilpel, Y. (2009). Genetic Redundancy: New Tricks for Old Genes. Cell 136, 389–392. doi:10.1016/j.cell.2009.01.027
Kamei, N., and Glabe, C. G. (2003). The Species-specific Egg Receptor for Sea Urchin Sperm Adhesion Is EBR1,a Novel ADAMTS Protein. Genes Dev. 17, 2502–2507. doi:10.1101/gad.1133003
Katoh, K., and Standley, D. M. (2013). MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 30, 772–780. doi:10.1093/molbev/mst010
Kelleher, E. S., and Markow, T. A. (2009). Duplication, Selection and Gene Conversion in a Drosophila mojavensis Female Reproductive Protein Family. Genetics 181, 1451–1465. doi:10.1534/genetics.108.099044
Kelleher, E. S., Swanson, W. J., and Markow, T. A. (2007). Gene Duplication and Adaptive Evolution of Digestive Proteases in Drosophila Arizonae Female Reproductive Tracts. Plos Genet. 3, e148. doi:10.1371/journal.pgen.0030148
Kessler, P., Marchot, P., Silva, M., and Servent, D. (2017). The Three-finger Toxin Fold: a Multifunctional Structural Scaffold Able to Modulate Cholinergic Functions. J. Neurochem. 142, 7–18. doi:10.1111/jnc.13975
Killingbeck, E. E., and Swanson, W. J. (2018). Egg Coat Proteins Across Metazoan Evolution. Curr. Top. Dev. Biol. 130, 443–488. doi:10.1016/bs.ctdb.2018.03.005
Kini, R. M. (2002). Molecular Moulds with Multiple Missions: Functional Sites in Three-finger Toxins. Clin. Exp. Pharmacol. Physiol. 29, 815–822. doi:10.1046/j.1440-1681.2002.03725.x
Kirschner, M. W., and Gerhart, J. C. (2008). The Plausibility of Life: Resolving Darwin's Dilemma. New Haven: Yale University Press. doi:10.12987/9780300128673
Kondrashov, F. A., Rogozin, I. B., Wolf, Y. I., and Koonin, E. V. (2002). Selection in the Evolution of Gene Duplications. Genome Biol. 3, research0008. doi:10.1186/gb-2002-3-2-research0008
Kozlov, A. M., Darriba, D., Flouri, T., Morel, B., and Stamatakis, A. (2019). RAxML-NG: a Fast, Scalable and User-Friendly Tool for Maximum Likelihood Phylogenetic Inference. Bioinformatics 35, 4453–4455. doi:10.1093/bioinformatics/btz305
Kroft, T. L., Gleason, E. J., and L'Hernault, S. W. (2005). The Spe-42 Gene Is Required for Sperm-Egg Interactions during C. elegans Fertilization and Encodes a Sperm-specific Transmembrane Protein. Dev. Biol. 286, 169–181. doi:10.1016/j.ydbio.2005.07.020
Krogh, A., Larsson, B., von Heijne, G., and Sonnhammer, E. L. L. (2001). Predicting Transmembrane Protein Topology with a Hidden Markov Model: Application to Complete genomes11Edited by F. Cohen. J. Mol. Biol. 305, 567–580. doi:10.1006/jmbi.2000.4315
Kukita, T., Wada, N., Kukita, A., Kakimoto, T., Sandra, F., Toh, K., et al. (2004). RANKL-induced DC-STAMP Is Essential for Osteoclastogenesis. J. Exp. Med. 200, 941–946. doi:10.1084/jem.20040518
Le Naour, F., Rubinstein, E., Jasmin, C., Prenant, M., and Boucheix, C. (2000). Severely Reduced Female Fertility in CD9-Deficient Mice. Science 287, 319–321. doi:10.1126/science.287.5451.319
Legan, P. K., Rau, A., Keen, J. N., and Richardson, G. P. (1997). The Mouse Tectorins: Modular Matrix Proteins of the Inner Ear Homologous To Components of the Sperm-Egg Adhesion System. J. Biol. Chem. 272, 8791–8801. doi:10.1074/jbc.272.13.8791
Liang, L. F., and Dean, J. (1993). Conservation of Mammalian Secondary Sperm Receptor Genes Enables the Promoter of the Human Gene to Function in Mouse Oocytes. Dev. Biol. 156, 399–408. doi:10.1006/dbio.1993.1087
Liao, D. (1999). Concerted Evolution: Molecular Mechanism and Biological Implications. Am. J. Hum. Genet. 64, 24–30. doi:10.1086/302221
Lin, S. J., Hu, Y., Zhu, J., Woodruff, T. K., and Jardetzky, T. S. (2011). Structure of Betaglycan Zona Pellucida (ZP)-C Domain Provides Insights into ZP-Mediated Protein Polymerization and TGF- Binding. Proc. Natl. Acad. Sci. 108, 5232–5236. doi:10.1073/pnas.1010689108
Litscher, E. S., and Wassarman, P. M. (2020). Zona Pellucida Proteins, Fibrils, and Matrix. Annu. Rev. Biochem. 89, 695–715. doi:10.1146/annurev-biochem-011520-105310
Low, B. W., Preston, H. S., Sato, A., Rosen, L. S., Searl, J. E., Rudko, A. D., et al. (1976). Three Dimensional Structure of Erabutoxin B Neurotoxic Protein: Inhibitor of Acetylcholine Receptor. Proc. Natl. Acad. Sci. 73, 2991–2994. doi:10.1073/pnas.73.9.2991
Lynch, M., and Conery, J. S. (2000). The Evolutionary Fate and Consequences of Duplicate Genes. Science 290, 1151–1155. doi:10.1126/science.290.5494.1151
Maldera, J. A., Weigel Munoz, M., Chirinos, M., Busso, D., Ge Raffo, F., Battistone, M. A., et al. (2014). Human Fertilization: Epididymal hCRISP1 Mediates Sperm-Zona Pellucida Binding through its Interaction with ZP3. Mol. Hum. Reprod. 20, 341–349. doi:10.1093/molehr/gat092
Meslin, C., Mugnier, S., Callebaut, I., Laurin, M., Pascal, G., Poupon, A., et al. (2012). Evolution of Genes Involved in Gamete Interaction: Evidence for Positive Selection, Duplications and Losses in Vertebrates. PLOS ONE 7, e44548. doi:10.1371/journal.pone.0044548
Miyado, K., Yamada, G., Yamada, S., Hasuwa, H., Nakamura, Y., Ryu, F., et al. (2000). Requirement of CD9 on the Egg Plasma Membrane for Fertilization. Science 287, 321–324. doi:10.1126/science.287.5451.321
Monné, M., Han, L., Schwend, T., Burendahl, S., and Jovine, L. (2008). Crystal Structure of the ZP-N Domain of ZP3 Reveals the Core Fold of Animal Egg coats. Nature 456, 653–657. doi:10.1038/nature07599
Moore, A. D., Björklund, Å. K., Ekman, D., Bornberg-Bauer, E., and Elofsson, A. (2008). Arrangements in the Modular Evolution of Proteins. Trends Biochem. Sci. 33, 444–451. doi:10.1016/j.tibs.2008.05.008
Nacher, J. C., Hayashida, M., and Akutsu, T. (2010). The Role of Internal Duplication in the Evolution of Multi-Domain Proteins. Biosystems 101, 127–135. doi:10.1016/j.biosystems.2010.05.005
Nair, S., Bist, P., Dikshit, N., and Krishnan, M. N. (2016). Global Functional Profiling of Human Ubiquitome Identifies E3 Ubiquitin Ligase DCST1 as a Novel Negative Regulator of Type-I Interferon Signaling. Sci. Rep. 6, 36179. doi:10.1038/srep36179
Navarro, B., Kirichok, Y., Chung, J. J., and Clapham, D. E. (2008). Ion Channels that Control Fertility in Mammalian Spermatozoa. Int. J. Dev. Biol. 52, 607–613. doi:10.1387/ijdb.072554bn
Nirthanan, S., Gopalakrishnakone, P., Gwee, M. C. E., Khoo, H. E., and Kini, R. M. (2003). Non-conventional Toxins from Elapid Venoms. Toxicon 41, 397–407. doi:10.1016/S0041-0101(02)00388-4
Nishimura, K., Dioguardi, E., Nishio, S., Villa, A., Han, L., Matsuda, T., et al. (2019). Molecular Basis of Egg Coat Cross-Linking Sheds Light on ZP1-Associated Female Infertility. Nat. Commun. 10, 3086. doi:10.1038/s41467-019-10931-5
Nomiyama, H., Egami, K., Wada, N., Tou, K., Horiuchi, M., Matsusaki, H., et al. (2005). Short Communication: Identification of Genes Differentially Expressed in Osteoclast-like Cells. J. Interferon Cytokine Res. 25, 227–231. doi:10.1089/jir.2005.25.227
Ohto, U., Ishida, H., Krayukhina, E., Uchiyama, S., Inoue, N., and Shimizu, T. (2016). Structure of IZUMO1-JUNO Reveals Sperm-Oocyte Recognition during Mammalian Fertilization. Nature 534, 566–569. doi:10.1038/nature18596
Palmer, C. A., Watts, R. A., Houck, L. D., Picard, A. L., and Arnold, S. J. (2007). Evolutionary Replacement of Components in a Salamander Pheromone Signaling Complex: More Evidence for Phenotypic-Molecular Decoupling. Evolution 61, 202–215. doi:10.1111/j.1558-5646.2007.00017.x
Palmer, C. A., Watts, R. A., Hastings, A. P., Houck, L. D., and Arnold, S. J. (2010). Rapid Evolution of Plethodontid Modulating Factor, a Hypervariable Salamander Courtship Pheromone, Is Driven by Positive Selection. J. Mol. Evol. 70, 427–440. doi:10.1007/s00239-010-9342-2
Pei, J., Kim, B. H., and Grishin, N. V. (2008). PROMALS3D: a Tool for Multiple Protein Sequence and Structure Alignments. Nucleic Acids Res. 36, 2295–2300. doi:10.1093/nar/gkn072
Perry, G. H., Dominy, N. J., Claw, K. G., Lee, A. S., Fiegler, H., Redon, R., et al. (2007). Diet and the Evolution of Human Amylase Gene Copy Number Variation. Nat. Genet. 39, 1256–1260. doi:10.1038/ng2123
Petronella, N., and Drouin, G. (2014). Purifying Selection against Gene Conversions in the Folate Receptor Genes of Primates. Genomics 103, 40–47. doi:10.1016/j.ygeno.2013.10.004
Ponting, C. P. (2008). The Functional Repertoires of Metazoan Genomes. Nat. Rev. Genet. 9, 689–698. doi:10.1038/nrg2413
Primakoff, P., and Myles, D. G. (2000). The ADAM Gene Family: Surface Proteins with Adhesion and Protease Activity. Trends Genet. 16, 83–87. doi:10.1016/S0168-9525(99)01926-5
Raj, I., Sadat Al Hosseini, H., Dioguardi, E., Nishimura, K., Han, L., Villa, A., et al. (2017). Structural Basis of Egg Coat-Sperm Recognition at Fertilization. Cell 169, 1315–1326.e17. doi:10.1016/j.cell.2017.05.033
Ramírez-Gómez, H. V., Tuval, I., Guerrero, A., and Darszon, A. (2019). Analysis of Sperm Chemotaxis. Methods Cell Biol. 151, 473–486. doi:10.1016/bs.mcb.2018.12.002
Rankin, T. L., Coleman, J. S., Epifano, O., Hoodbhoy, T., Turner, S. G., Castle, P. E., et al. (2003). Fertility and Taxon-Specific Sperm Binding Persist after Replacement of Mouse Sperm Receptors with Human Homologs. Dev. Cel. 5, 33–43. doi:10.1016/s1534-5807(03)00195-3
Rastogi, S., and Liberles, D. A. (2005). Subfunctionalization of Duplicated Genes as a Transition State to Neofunctionalization. BMC Evol. Biol. 5, 28. doi:10.1186/1471-2148-5-28
Schimenti, J. C. (1999). Mice and the Role of Unequal Recombination in Gene-Family Evolution. Am. J. Hum. Genet. 64, 40–45. doi:10.1086/302220
Shen, F., Ross, J. F., Wang, X., and Ratnam, M. (1994). Identification of a Novel Folate Receptor, a Truncated Receptor, and Receptor Type .Beta. In Hematopoietic Cells: cDNA Cloning, Expression, Immunoreactivity, and Tissue Specificity. Biochemistry 33, 1209–1215. doi:10.1021/bi00171a021
Shu, L., Suter, M. J.-F., and Räsänen, K. (2015). Evolution of Egg coats: Linking Molecular Biology and Ecology. Mol. Ecol. 24, 4052–4073. doi:10.1111/mec.13283
Siu, K. K., Serrão, V. H. B., Ziyyat, A., and Lee, J. E. (2021). The Cell Biology of Fertilization: Gamete Attachment and Fusion. J. Cell Biol. 220, e202102146. doi:10.1083/jcb.202102146
Sonnhammer, E. L., von Heijne, G., and Krogh, A. (1998). A Hidden Markov Model for Predicting Transmembrane Helices in Protein Sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 6, 175–182.
Speer, K. F., Allen-Waller, L., Novikov, D. R., and Barott, K. L. (2021). Molecular Mechanisms of Sperm Motility Are Conserved in an Early-Branching Metazoan. Proc. Natl. Acad. Sci. USA 118, e2109993118. doi:10.1073/pnas.2109993118
Spiegelstein, O., Eudy, J. D., and Finnell, R. H. (2000). Identification of Two Putative Novel Folate Receptor Genes in Humans and Mouse. Gene 258, 117–125. doi:10.1016/S0378-1119(00)00418-2
Staege, H., Brauchlin, A., Schoedon, G., and Schaffner, A. (2001). Two Novel Genes FIND and LIND Differentially Expressed in Deactivated and Listeria -infected Human Macrophages. Immunogenetics 53, 105–113. doi:10.1007/s002510100306
Sutton, K. A., Jungnickel, M. K., and Florman, H. M. (2008). A Polycystin-1 Controls Postcopulatory Reproductive Selection in Mice. Proc. Natl. Acad. Sci. 105, 8661–8666. doi:10.1073/pnas.0800603105
Swanson, W. J., and Vacquier, V. D. (2002). The Rapid Evolution of Reproductive Proteins. Nat. Rev. Genet. 3, 137–144. doi:10.1038/nrg733
Ting, C. T., Tsaur, S.-C., Sun, S., Browne, W. E., Chen, Y.-C., Patel, N. H., et al. (2004). Gene Duplication and Speciation in Drosophila: Evidence from the Odysseus Locus. Proc. Natl. Acad. Sci. 101, 12232–12235. doi:10.1073/pnas.0401975101
Tsernoglou, D., and Petsko, G. A. (1977). Three-dimensional Structure of Neurotoxin a from Venom of the Philippines Sea Snake. Proc. Natl. Acad. Sci. 74, 971–974. doi:10.1073/pnas.74.3.971
Tsetlin, V. (1999). Snake Venom Alpha-Neurotoxins and Other 'three-finger' Proteins. Eur. J. Biochem. 264, 281–286. doi:10.1046/j.1432-1327.1999.00623.x
Vacquier, V. D., Swanson, W. J., and Lee, Y.-H. (1997). Positive Darwinian Selection on Two Homologous Fertilization Proteins: what Is the Selective Pressure Driving Their Divergence. J. Mol. Evol. 44, S15–S22. doi:10.1007/PL00000049
Vacquier, V. D. (1998). Evolution of Gamete Recognition Proteins. Science 281, 1995–1998. doi:10.1126/science.281.5385.1995
Vogel, C., Teichmann, S. A., and Pereira-Leal, J. (2005). The Relationship between Domain Duplication and Recombination. J. Mol. Biol. 346, 355–365. doi:10.1016/j.jmb.2004.11.050
Wagner, G. P., and Altenberg, L. (1996). Perspective: Complex Adaptations and the Evolution of Evolvability. Evolution 50, 967–976. doi:10.1111/j.1558-5646.1996.tb02339.x
Wagner, G. P., Pavlicev, M., and Cheverud, J. M. (2007). The Road to Modularity. Nat. Rev. Genet. 8, 921–931. doi:10.1038/nrg2267
Walsh, B. (2003). Population-Genetic Models of the Fates of Duplicate Genes. Genetica 118, 279–294. doi:10.1007/978-94-010-0229-5_16
Wang, J., Lei, C., Shi, P., Teng, H., Lu, L., Guo, H., et al. (2021). LncRNA DCST1-AS1 Promotes Endometrial Cancer Progression by Modulating the MiR-665/HOXB5 and MiR-873-5p/CADM1 Pathways. Front. Oncol. 11, 3112. doi:10.3389/fonc.2021.714652
Weiner, J., Beaussart, F., and Bornberg-Bauer, E. (2006). Domain Deletions and Substitutions in the Modular Protein Evolution. FEBS J. 273, 2037–2047. doi:10.1111/j.1742-4658.2006.05220.x
West-Eberhard, M. J. (2005). Developmental Plasticity and the Origin of Species Differences. Proc. Natl. Acad. Sci. 102, 6543–6549. doi:10.1073/pnas.0501844102
Wilburn, D. B., and Swanson, W. J. (2016). From Molecules to Mating: Rapid Evolution and Biochemical Studies of Reproductive Proteins. J. Proteomics 135, 12–25. doi:10.1016/j.jprot.2015.06.007
Wilburn, D. B., and Swanson, W. J. (2017). The “ZP Domain” Is Not One, but Likely Two Independent Domains. Mol. Reprod. Dev. 84, 284–285. doi:10.1002/mrd.22781
Wilburn, D. B., Bowen, K. E., Gregg, R. G., Cai, J., Feldhoff, P. W., Houck, L. D., et al. (2012). Proteomic and UTR Analyses of a Rapidly Evolving Hypervariable Family of Vertebrate Pheromones. Evolution 66, 2227–2239. doi:10.1111/j.1558-5646.2011.01572.x
Wilburn, D. B., Bowen, K. E., Doty, K. A., Arumugam, S., Lane, A. N., Feldhoff, P. W., et al. (2014). Structural Insights into the Evolution of a Sexy Protein: Novel Topology and Restricted Backbone Flexibility in a Hypervariable Pheromone from the Red-Legged Salamander, Plethodon shermani. PLOS ONE 9, e96975. doi:10.1371/journal.pone.0096975
Wilburn, D. B., Arnold, S. J., Houck, L. D., Feldhoff, P. W., and Feldhoff, R. C. (2017). Gene Duplication, Co-option, Structural Evolution, and Phenotypic Tango in the Courtship Pheromones of Plethodontid Salamanders. Herpetologica 73, 206–219. doi:10.1655/Herpetologica-D-16-00082.1
Wilburn, D. B., Kunkel, C. L., Feldhoff, R. C., Feldhoff, P. W., and Searle, B. C. (2022). Recurrent Co-option and Recombination of Cytokine and Three finger Proteins in Multiple Reproductive Tissues throughout Salamander Evolution. bioRxiv. doi:10.1101/2022.01.04.475003
Wilson, K. L., Fitch, K. R., Bafus, B. T., and Wakimoto, B. T. (2006). Sperm Plasma Membrane Breakdown during Drosophila Fertilization Requires Sneaky, an Acrosomal Membrane Protein. Development 133, 4871–4879. doi:10.1242/dev.02671
Wilson, L. D., Obakpolor, O. A., Jones, A. M., Richie, A. L., Mieczkowski, B. D., Fall, G. T., et al. (2018). The Caenorhabditis elegans Spe‐49 Gene Is Required for Fertilization and Encodes a Sperm‐specific Transmembrane Protein Homologous to SPE‐42. Mol. Reprod. Dev. 85, 563–578. doi:10.1002/mrd.22992
Wright, G. J., and Bianchi, E. (2016). The Challenges Involved in Elucidating the Molecular Basis of Sperm-Egg Recognition in Mammals and Approaches to Overcome Them. Cell Tissue Res. 363, 227–235. doi:10.1007/s00441-015-2243-3
Keywords: gene duplication, fertilization, subfunctionalization, neofunctionalization, sperm, egg, reproduction
Citation: Rivera AM and Swanson WJ (2022) The Importance of Gene Duplication and Domain Repeat Expansion for the Function and Evolution of Fertilization Proteins. Front. Cell Dev. Biol. 10:827454. doi: 10.3389/fcell.2022.827454
Received: 02 December 2021; Accepted: 12 January 2022;
Published: 27 January 2022.
Edited by:
Enrica Bianchi, University of York, United KingdomReviewed by:
Shunsuke Nishio, Karolinska Institutet (KI), SwedenEsther Betran, University of Texas at Arlington, United States
Copyright © 2022 Rivera and Swanson. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Alberto M. Rivera, YWxiZXJ0b21hcmNvc3JpdmVyYUBnbWFpbC5jb20=, YW1yaXZlcmFAdXcuZWR1
 Alberto M. Rivera
Alberto M. Rivera Willie J. Swanson
Willie J. Swanson