
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell Dev. Biol. , 24 March 2022
Sec. Molecular and Cellular Pathology
Volume 10 - 2022 | https://doi.org/10.3389/fcell.2022.808212
This article is part of the Research Topic Immune Modulation in Tumor Microenvironment: New Perspectives for Cancer Immunotherapy View all 19 articles
 Junchang Zhang1,2†
Junchang Zhang1,2† Han Wang1,2†
Han Wang1,2† Cheng Yuan1
Cheng Yuan1 Jing Wu1
Jing Wu1 Jiannan Xu1,2
Jiannan Xu1,2 Songyao Chen1,2
Songyao Chen1,2 Changhua Zhang1*
Changhua Zhang1* Yulong He1,2*
Yulong He1,2*Integrin alpha L (ITGAL) is a member of the integrin family in which the abnormal expression is linked with carcinogenesis and immune regulation. However, the relation between ITGAL and the prognosis of gastric cancer (GC) and tumor-infiltrating lymphocytes (TILs) are not well understood. The differential expressions of ITGAL in human tumors and the clinical prognosis in GC were systematically analyzed via multiple databases including Gene Expression Profiling Interaction Analysis (GEPIA), UALCAN, Tumor Immune Estimation Resource (TIMER), and Kaplan–Meier (KM) plotter. TIMER, GEPIA, and TISIDB databases were used to comprehensively investigate the correlation between ITGAL and tumor infiltration immune cells. Also, further results were investigated by immunohistochemistry, qRT-PCR, and Western blot. We found that ITGAL expression in GC samples was considerably increased than in peritumor samples. Sample type, subgroup, cancer stage, lymphatic node stage, and worse survival were strongly related to high ITGAL expression. Moreover, upregulated ITGAL expression was strongly connected with immunomodulators, chemokines, and infiltrating levels of CD8+, CD4+ T cell, B cell, monocyte, neutrophil, macrophage, T-cell regulatory, NK cell, and myeloid dendritic cell in stomach adenocarcinoma (STAD). Specifically, immunohistochemistry and bioinformatic analysis showed that ITGAL expression was shown to have strong relationships with various immunological marker sets including PD1 (T-cell exhaustion marker). In conclusion, ITGAL is a prognostic biomarker for GC patients. It might regulate tumor immune microenvironment leading to poor prognosis. Furthermore, studies are essential to explore therapeutic targeting ITGAL.
Gastric cancer (GC) is the world’s third leading cause of cancer-related mortality among common fatal tumors (Bray et al., 2018). Despite considerable improvements in diagnosis and therapy about GC, the prognosis still remains challenging (Luebeck et al., 2013). At present, immunotherapy is a prominent subject in the area of cancer treatment and has been an effective treatment in various types of cancer (Larkin et al., 2015; Motzer et al., 2015; Reck et al., 2016), including gastric cancer (Lote et al., 2015; Procaccio et al., 2017). Nevertheless, not all GC patients benefit from immunotherapy, which may be related to the immune microenvironment of tumors (Davidson et al., 2015). Therefore, it is an emergent issue to look for specific immune-related biomarkers with GC and discover new immunotherapy targets.
ITGAL, also known as CD11a, encodes an integrin component of LFA-1, which expressed in immune cells (Silva et al., 2008; Kuwano et al., 2010; Sumagin et al., 2010) and regulated intercellular adhesion and the costimulation signaling of lymphocytes (Ley et al., 2007; Samatov et al., 2013). ITGAL, as a member of the integrin family, which play important roles during angiogenesis and cancer development (Silva et al., 2008; Winograd-Katz et al., 2014; Seguin et al., 2015; Xie et al., 2021), also participates in immune reactions, inflammatory processes, and construction of the tumor microenvironment, thus, contributing to the pathogenesis of diverse tumors, such as renal cancer, colorectal cancer, ovarian cancer, melanoma, prostate adenocarcinoma, and head and neck squamous cell carcinoma (Vendrell et al., 2007; Boguslawska et al., 2016; Song et al., 2019; Zhao et al., 2019; Ji et al., 2020). These researchers suggested that ITGAL might have a significant influence on cancer growth and transformation, and may be a novel target in treating a variety of malignancies. However, the possible mechanisms of ITGAL about tumor development and immune engagement with GC are still unknown.
In this present study, the ITGAL expression and its connection to GC patient prognosis were investigated utilizing diverse databases including the Gene Expression Profiling Interaction Analysis (GEPIA), Oncomine, Kaplan–Meier (KM) plotter, and UALCAN datasets. Furthermore, the Tumor Immune Estimation Resource (TIMER) and immunohistochemistry were performed to investigate the relationship of ITGAL with immune-related cells in the distinct tumor microenvironments. This study uncovered the critical involvement of ITGAL in GC and the possible connection and mechanism by which ITGAL may regulate tumor-infiltrating immune cells.
The Tumor Immune Estimation Resource (TIMER2.0) is a web-based interactive platform to analyze immune infiltration systematically in various malignancies (https://timer.cistrome.org/) (Li et al., 2020). The TIMER2.0 database applies six advanced algorithms to provide a more rigorous evaluation of tumor-infiltrating lymphocyte (TIL) levels for The Cancer Genome Atlas (TCGA) or tumor-related data. Additionally, the database can also precisely estimate tumor purity. We investigated ITGAL expression in various malignancies and the relationship between the expression of ITGAL and TILs via gene modules. Furthermore, the relationship between ITGAL expression with gene markers of TILs, including markers of CD8+/CD4+ T cells, B cells, monocytes, natural killer (NK) cells, dendritic cells (DCs), TAMs, M1macrophages, M2 macrophages, neutrophils, T cells, and related subtypes, has been analyzed via correlation modules. Expression dispersion maps were created between a pair of custom genes for GC and the statistical significance of the correlation and estimation of Spearman, by correlation module. The level of gene expression was shown as log2 RSEM.
The Gene Expression Profiling Interactive Analysis (GEPIA) (Tang et al., 2017) online database (http://gepia.cancer-pku.cn/index.html) is a comprehensive platform that obtained the analysis data from TCGA and The Genotype–Tissue Expression (GTEx) databases. In the present research, we used the GEPIA data source to assess the ITGAL levels in GC samples and healthy samples via the “DIY Expression” page.
The UALCAN website provides an extensive and interactive study of bioinformatics applying RNA-seq and clinical data of 31 malignancies from TCGA (Chandrashekar et al., 2017) (http://ualcan.path.uab.edu/). The database can compare gene expression in tumors to healthy samples, and in different tumor stages or subtypes, as well as other clinicopathological features. This research will, respectively, examine the ITGAL expression lever from major clinical features such as tissue type (healthy/tumor), GC stage (stages 1, 2, 3, and 4), lymph node stage (N0 1, 2, and 3), and cancer subgroup.
The KM plotter (http://kmplot.com/analysis/) could evaluate the survival prognosis of related genes via mapping the survival curve using 1,065 GC samples with an average follow-up of 33 months (Lánczky et al., 2016). The prognostic significance of ITGAL in GC, including overall survival (OS), first progression (FP), and post-progression survival (PPS), was investigated using this database. The hazard ratio (HR) with 95% confidence intervals was also estimated, as well as the log-rank p-value. Statistical significance was defined as p < 0.05.
TISIDB (http://cis.hku.hk/TISIDB/index.php) is an online platform that combines various heterogeneous data sources to study tumor and immune system interactions. This database may help researchers understand how tumors and immune cells interact, as well as forecast immunotherapy responses and identify new immunotherapy targets. It would become a valuable resource for cancer immunology research and therapy. In this research, TISIDB was utilized to investigate the association of ITGAL with 28 TILs, 45 immunostimulators, 24 immunoinhibitors, 41 chemokines, and 18 receptors in GC (Ru et al., 2019).
According to the instructions, the total RNA of tissue samples was extracted by using MolPure® Cell/Tissue Total RNA Kit (YEASEN CAT#19221ES50). The cDNA of samples was synthesized from 2 µg RNA via Evo M‐MLV reverse transcription master mix (Accurate Biology, CAT#AG11706). The qRT-PCR was performed using a SYBR Green Pro Taq HS premixed qPCR kit (Accurate Biology, CAT# AG11701). The ITGAL primer sequence is as follows: forward 5′-CTGCTTTGCCAGCCTCTCTGT-3′ and reverse 5′-GCTCACAGGTATCTGGCTATGG-3′. GAPDH: forward 5′-CGGAGTCAACGGATTTGGTCGT-3′ and reverse 5′-TCTCAGCCTTGACGGTGCCA-3′. The 2−ΔΔCT calculation method was used to determine the relative target gene level.
Protease inhibitors are used to lyse tissue samples in a radioimmunoprecipitation analysis (RIPA) solution. The BCA protein assay kit (KeyGEN BioTECH, cat#KGP903) was used to valuate the protein concentration. Western blot was performed as previously described (Wang et al., 2021). The following antibodies were used: anti-ITGAL (Abclonal, cat#A1644) and anti‐GAPDH (Proteintech, cat#60004‐1‐Ig).
This research was conducted on 10 paraffin-embedded GC specimens from the FAHSYSU Department of Pathology. IHC was performed to investigate the expression of ITGAL and PD1, thus, to identify the connection between ITGAL expression and TILs. IHC staining of these specimens were conducted as previously described (Yang et al., 2019). Anti-ITGAL (Abclonal, cat#A1644) and anti-PD1 (Abclonal, cat#A11973) were used for IHC staining. The staining intensity was classified as follows: negative (−), weak (+), moderate (++), and strong (+++).
The KM plots were performed to construct survival curves. For KM plots, GEPIA, and TISIDB, HR and p-values were described using log-rank test. Spearman’s correlation coefficient was calculated to analyze the connection of ITGAL expression with immune infiltration levels, immunomodulators, and chemokines. The strength of correlation was judged to be a very weak correlation if <0.2, weak if <0.4, moderate if <0.6, strong if <0.8, and very strong if <1.0. Statistical significance was defined as p < 0.05.
By applying RNA-seq data from various cancer types in the TCGA, we investigated the differential expression of ITGAL between tumor and surrounding healthy tissues. Figure 1A shows the findings. According to the TIMER database, we discovered that ITGAL expression levels are increased in breast invasive carcinoma, kidney renal clear cell carcinoma, kidney renal papillary cell carcinoma, and stomach adenocarcinoma (STAD), but are decreased in colon adenocarcinoma and lung adenocarcinoma compared with peritumor tissues.
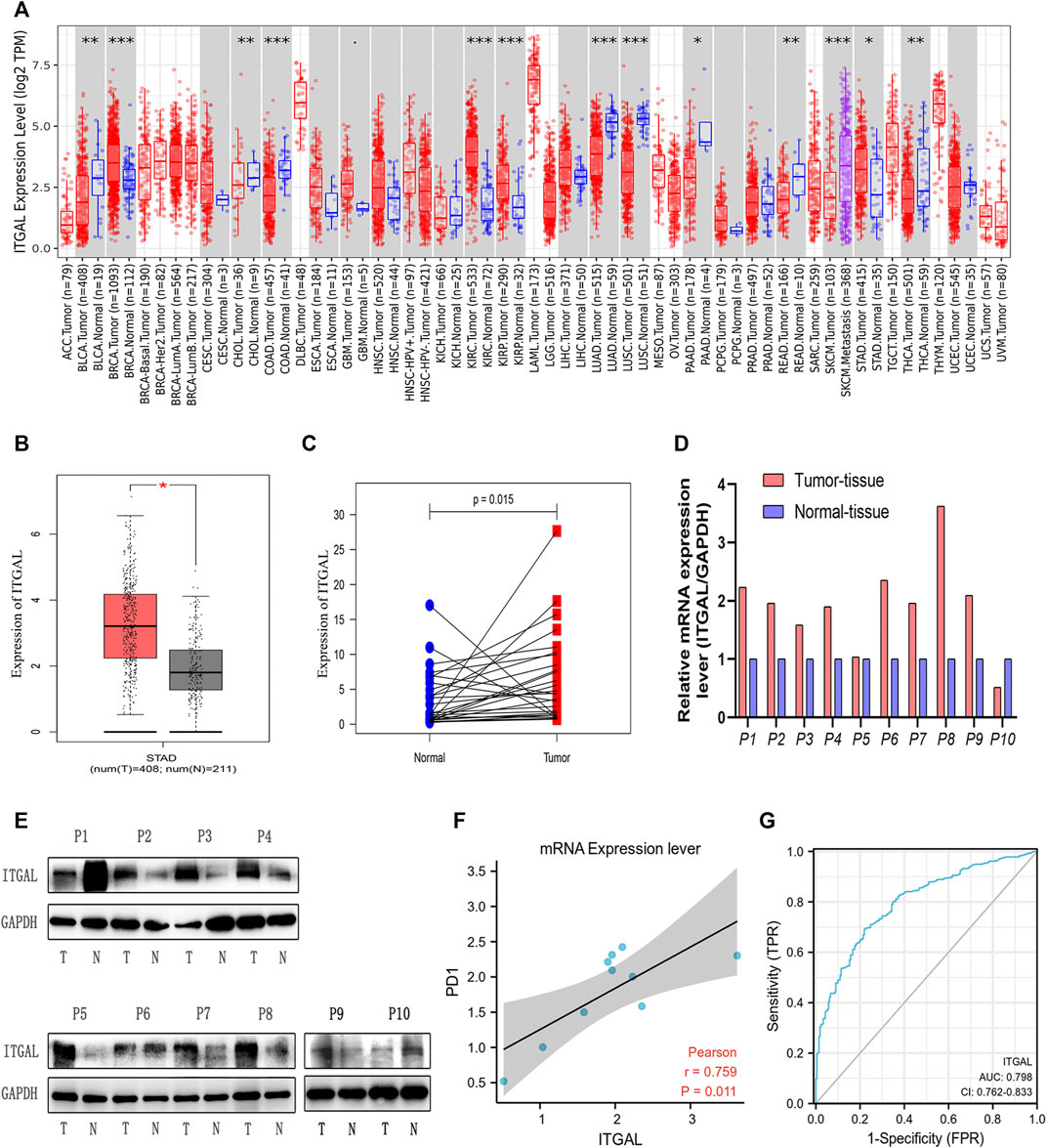
FIGURE 1. Integrin alpha L (ITGAL) expression levels in different types of human cancers. (A) Increased or decreased ITGAL in different tumor types from The Cancer Genome Atlas (TCGA) database were determined by Tumor Immune Estimation Resource (TIMER) (*p < 0.05, **p < 0.01, ***p < 0.001). (B) Increased ITGAL in gastric cancer tissues compared with normal tissues in Gene Expression Profiling Interaction Analysis (GEPIA). (C) Increased ITGAL expression in gastric cancer compared with the matching normal tissue from TCGA database (n = 27). (D) The mRNA level of ITGAL in 10 pairs of GC tissues and their paired normal adjacent tissues. (E) Western blotting was used to detect the protein level of ITGAL in 10 pairs of GC tissues and their paired adjacent normal tissues. (F) The correlation analysis between ITGAL and PD1 mRNA level. (G) The receiver-operating characteristic (ROC) curve analysis of ITGAL in gastric cancer (GC) patients.
To verify these findings in GC, 619 samples from the TCGA database were examined by GEPIA. As Figure 1B shows, we discovered that ITGAL mRNA was substantially greater in GC samples (408 cases) than that in healthy samples (211cases) (p < 0.05) from TCGA, which was similar with the TIMER database. As we can see from Figure 1C, TCGA database showed that ITGAL expression was higher than the matching normal tissue (n = 27). Meanwhile, Western blotting and qRT-PCR were performed to examine the ITGAL expression in 10 pairs of GC cases and adjacent healthy tissues. We observed that in most GC tissues, ITGAL mRNA (Figure 1D) and protein levels (Figure 1E) were higher than the paired adjacent healthy tissues. Interestingly, increased ITGAL mRNA expression was consistent with PD1 (Figure 1F), and the AUC was 0.798 (95% CI 0.762–0.833) for ITGAL in GC (Figure 1G). The above data indicated that ITGAL was strongly increased in GC tissues and may be potential diagnostic biomarker for GC.
We examined ITGAL expression in relation to several clinical–pathological parameters in patients, including sample type (healthy/primary tumor), tumor stage (stage 1, 2, 3, and 4), lymph node stage (N0 1, 2, and 3), and gastric cancer subgroup by applying UALCAN. As shown in Figure 2A, GC samples had substantially higher ITGAL expression than that in healthy samples (p = 0.0015). A study of cancer stages revealed that ITGAL in the middle and late-stage cancers was significantly higher expressed than in the early stages, suggesting a potential function for ITGAL in tumor development and migration (Figure 2B). ITGAL expression in lymph node stage samples was markedly higher in lymph nodes at all stages of cancer development than normal, indicating that ITGAL is present in malignancy (Figure 2C). Furthermore, as demonstrated in Figure 2D, ITGAL expression was significantly elevated in diffuse adenocarcinoma compared with normal tissue.
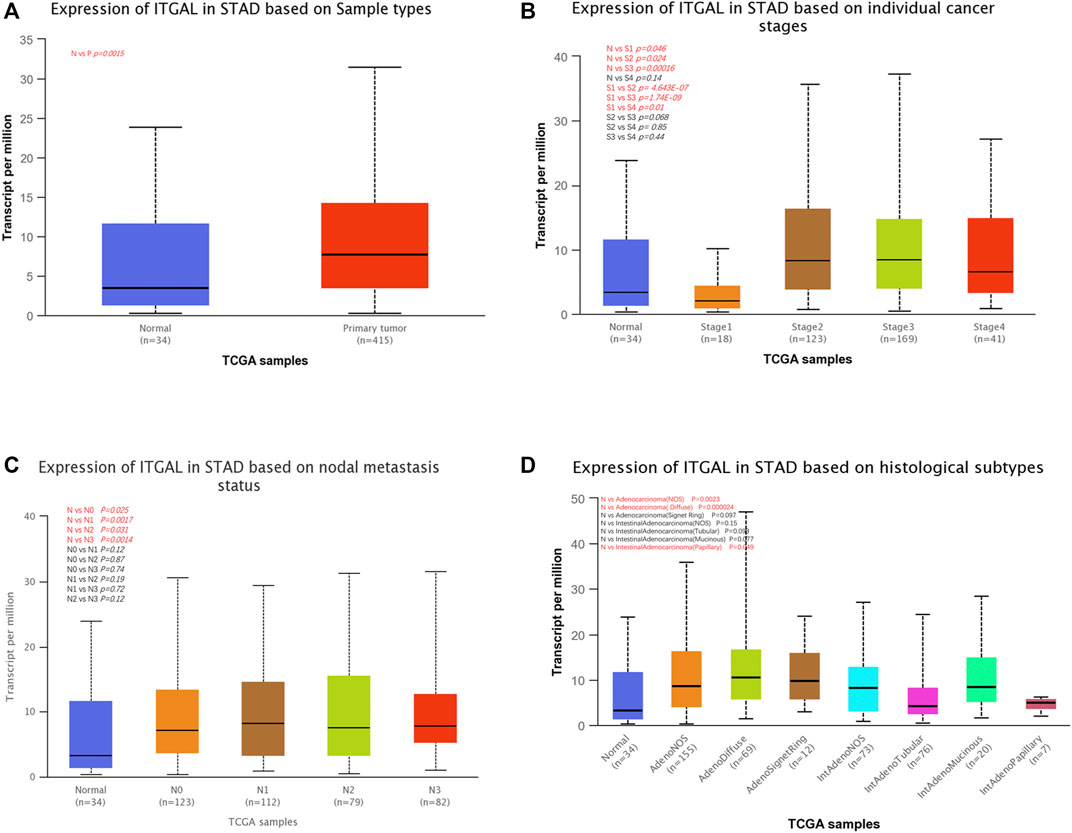
FIGURE 2. Correlation between ITGAL mRNA expression level and clinicopathological parameters of gastric cancer through the UALCAN database. (A) Sample type (normal/primary tumor). (B) Cancer stage (stage 1, 2, 3, and 4). (C) Lymph node stage (N0 1, 2, and 3). (D) Gastric cancer subgroup. N, normal; P, primary tumor; S1, stage 1; S2, stage 2; S3, stage 3; S4, stage 4; STAD, stomach adenocarcinoma.
Next, in order to get a well grasp of the significance and possible molecular mechanism of ITGAL expression in tumor development, we observed the correlation between the ITGAL expression and clinical–pathological features of GC in the KM plotter. Upregulated ITGAL expression was linked with a poorer OS and PPS in male and female patients. Specifically, increased ITGAL mRNA expression was associated with poorer OS and PPS in stage 1 (OS HR = 0.22, p = 0.0062) and stage 2 (OS HR = 2.18, p = 0.022; PPS HR = 3.58, p = 0.00016) of GC patients (Table 1). Furthermore, we discovered that OS and PPS at stage N1 (OS HR = 2.2, p = 0.00082; PPS HR = 3.55, p = 2.1e−08) and N1 + 2 + 3 (OS HR = 1.55, p = 0.001; PPS HR = 1.94, p = 3.9e−06) were related to ITGAL expression at the same time. These findings suggested that the prognostic significance of ITGAL in GC patients was determined by their clinical features, particularly in early-stage and LN metastases of GC.
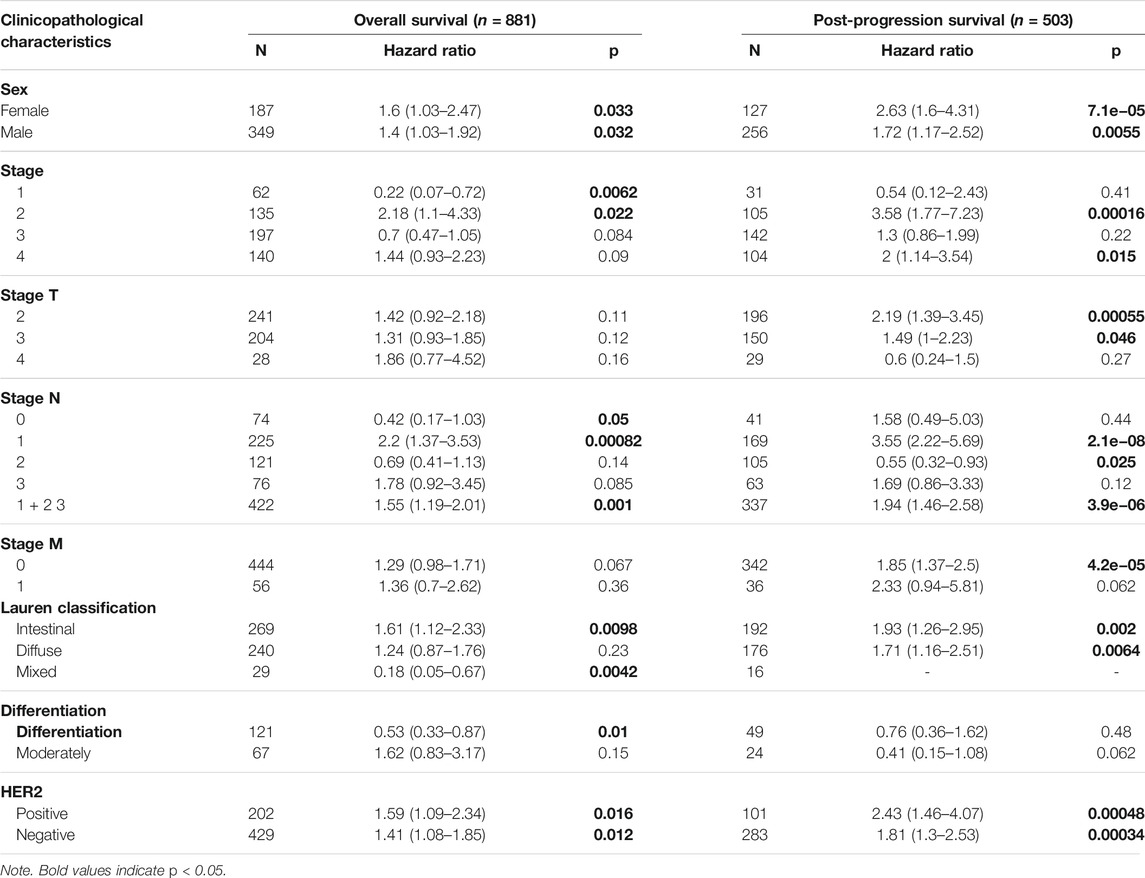
TABLE 1. Correlation of integrin alpha L (ITGAL) mRNA expression and clinical prognosis in gastric cancer with different clinicopathological factors by Kaplan–Meier plotter.
The findings indicated that GC patients had a higher degree of ITGAL mRNA expression compared with normal controls. Therefore, more investigation was necessary to determine whether ITGAL expression is associated with tumor outcome. In the present research, we examined the ITGAL expression and their relationship with prognosis via Kaplan–Meier survival curves in order to establish whether ITGAL can be used as a prognostic biomarker in GC. Notably, ITGAL expression was associated with a favorable outcome for GC, and this study showed that increased ITGAL expression was linked with a worse prognosis in the GC cohort 213475-s-at (OS: HR = 1.25, p = 0.0091) and 1554240-a-at (OS: HR = 1.47, p = 0.00057) (Figure 3).
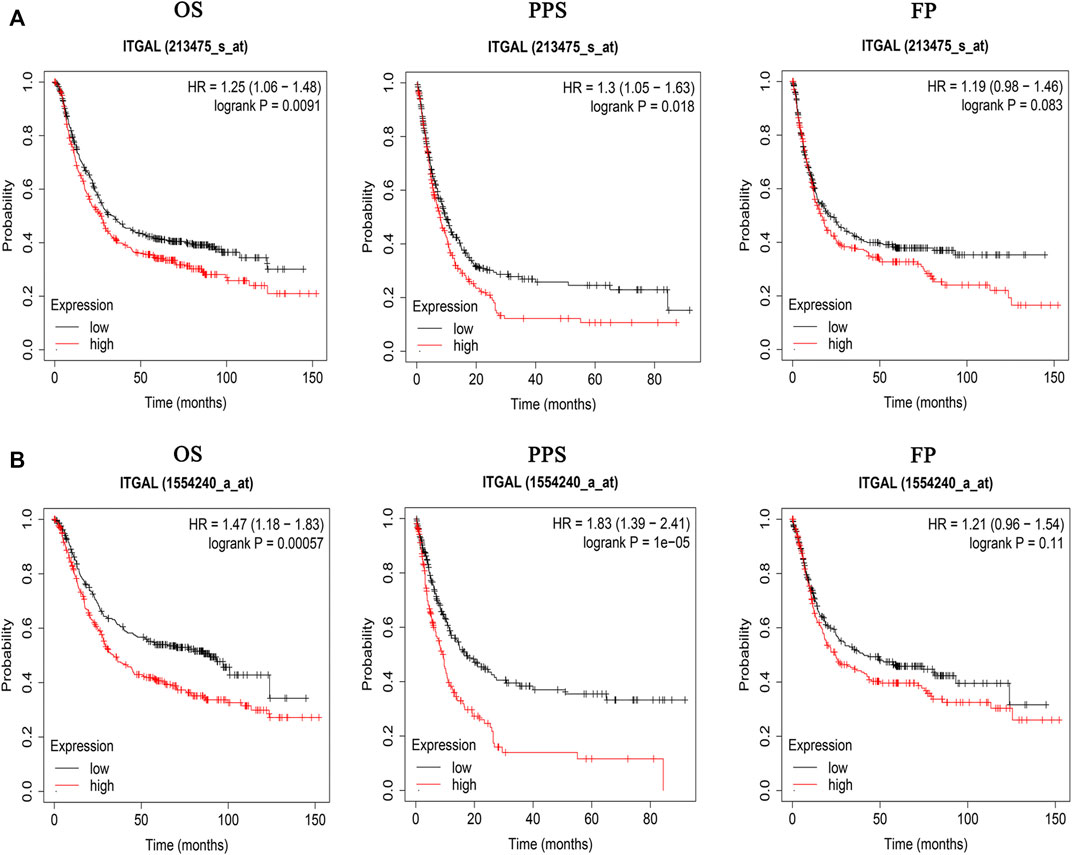
FIGURE 3. Kaplan–Meier survival curves comparing the high and low expression of ITGAL in gastric cancer in Kaplan–Meier plotter databases. (A) Survival curves of OS, PPS, and FP in the gastric cancer cohort (213475-s-at). (B) OS, PPS, and FP survival curves of gastric cancer (1554240-a-at). OS, overall survival; PPS, post-progression survival; FP, first progression.
Immune infiltration is a key factor associated with tumor progression. Therefore, TISIDB and TIMER platforms were performed to assess ITGAL expression connection to immune cell infiltration levels in STAD. The purity of tumors in clinical cancer samples has a significant impact on the study of immune infiltration using genetic techniques (Yoshihara et al., 2013). Therefore, in this study, ITGAL expression is adversely correlated with the purity of STAD (rho = -0.266, p <1.38e−7). Our results also discovered that ITGAL had a strong correlation with the abundance of TILs (Figure 4A). For instance, high-expression level of ITGAL was positively correlated with infiltrating degree of CD8+ T cell (rho = 0.732), CD4+ T cell (rho = 0.466), B cell (rho = 0.719), monocyte (rho = 0.728), neutrophil (rho = 0.574), and macrophage (rho = 0.675), T-cell regulatory (rho = 0.67), NK cell (rho = 0.618), and myeloid dendritic cell (rho = 0.8) (Figure 4B). All the p-values were far less than 0.001. These results indicate that ITGAL plays a key function in immune infiltration of GC.
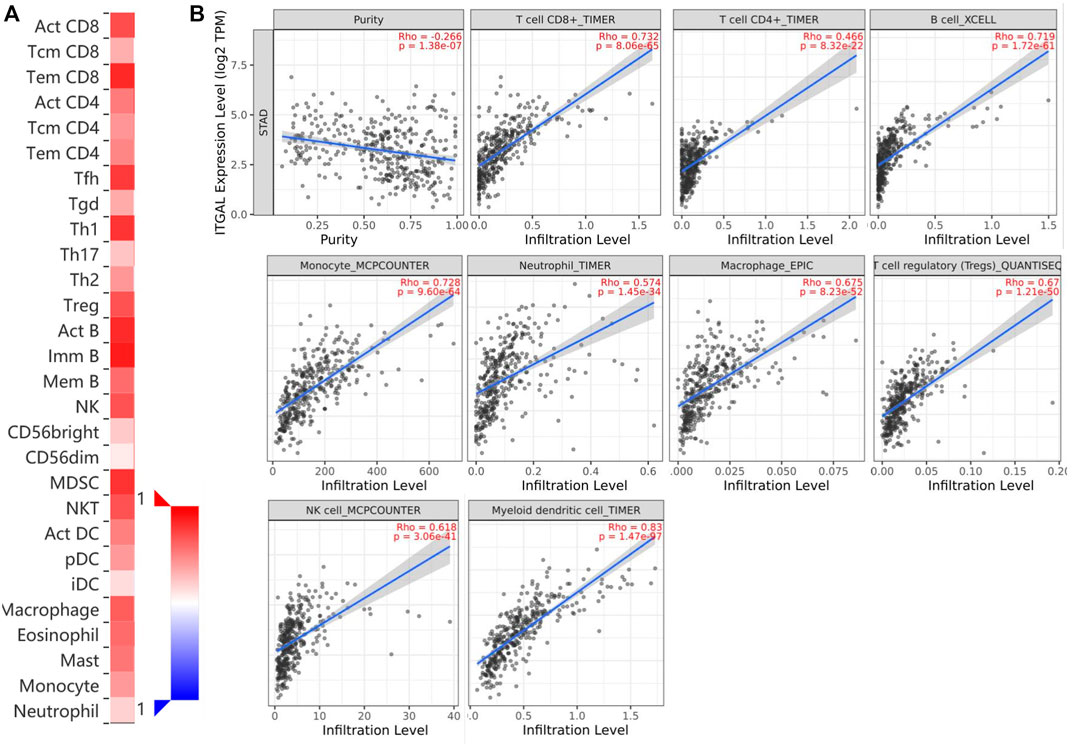
FIGURE 4. Correlation of ITGAL expression with immune infiltration in gastric cancer. (A) Correlation between the expression of ITGAL and the abundance of TILs in gastric cancer available at TISIDB database. (B) Correlation of ITGAL expression with infiltration levels of CD8 + T cell, CD4 + T cell, Treg cell, B cell, neutrophil, macrophage, myeloid dendritic cell, natural killer cell, and monocyte in gastric cancer available at TIMER2.0 database. TILs, tumor-infiltrating lymphocytes; TIMER2.0, Tumor Immune Estimation Resource. Color images are available online.
TIMER and GEPIA databases were applied to study the association among ITGAL and different biomarkers of TILs (CD8+/CD4+ T cells, NK cells, B cells, monocytes, DCs, TAMs, M1macrophages, M2 macrophages, neutrophils, T cells, and related subtypes) in STAD. We found that ITGAL was associated with the majority of TILs markers in STAD. The several functional T cells, including Th1/Th2/Th17/Tfh cells, Tregs, and exhausted T cells were also analyzed. Particularly, ITGAL was strongly linked with the majority of immune marker sets of TILs in STAD (Table 2).
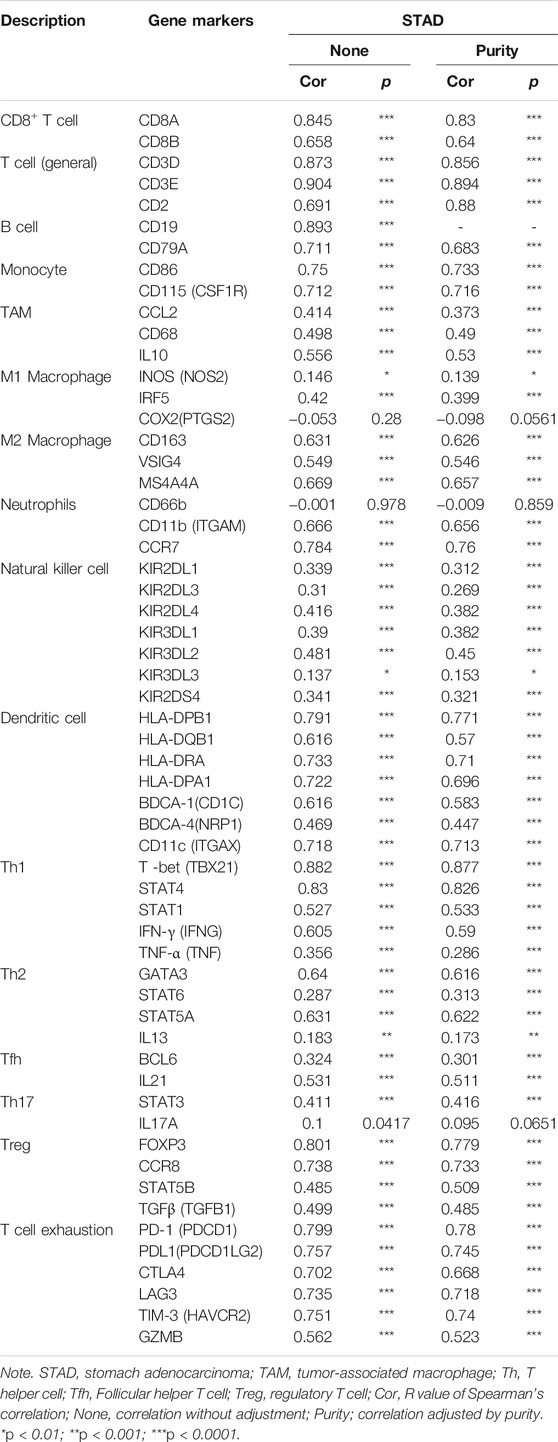
TABLE 2. Correlation analysis between ITGAL and related genes and markers of immune cells in Tumor Immune Estimation Resource (TIMER2.0).
Obviously, ITGAL had a significant association with the majority of marker sets of monocytes, TAMs, M2 macrophages, and T cell exhaustion in STAD (Table 2). Specifically, this study implicated that PDCD1, PDCD1G2, CTLA4, LAG3, HAVCR2, GZMB of T-cell exhaustion, and chemokine ligand (CCL)-2, CD68, and IL10 of TAMs are all strongly correlated with ITGAL in STAD, as well as IRF5 of M1 phenotype and CD163, VSIG4, MS4A4A of M2 phenotype (p < 0.0001; Figures 5A–E). Furthermore, according to the GEPIA database, we further assessed the connection of ITGAL expression with the aforementioned markers of TAMs, M2 macrophages, monocytes, and T-cell exhaustion, and the correlations are comparable with that in TIMER (Table 3). Therefore, ITGAL may regulate T-cell exhaustion, and macrophage polarization in STAD.
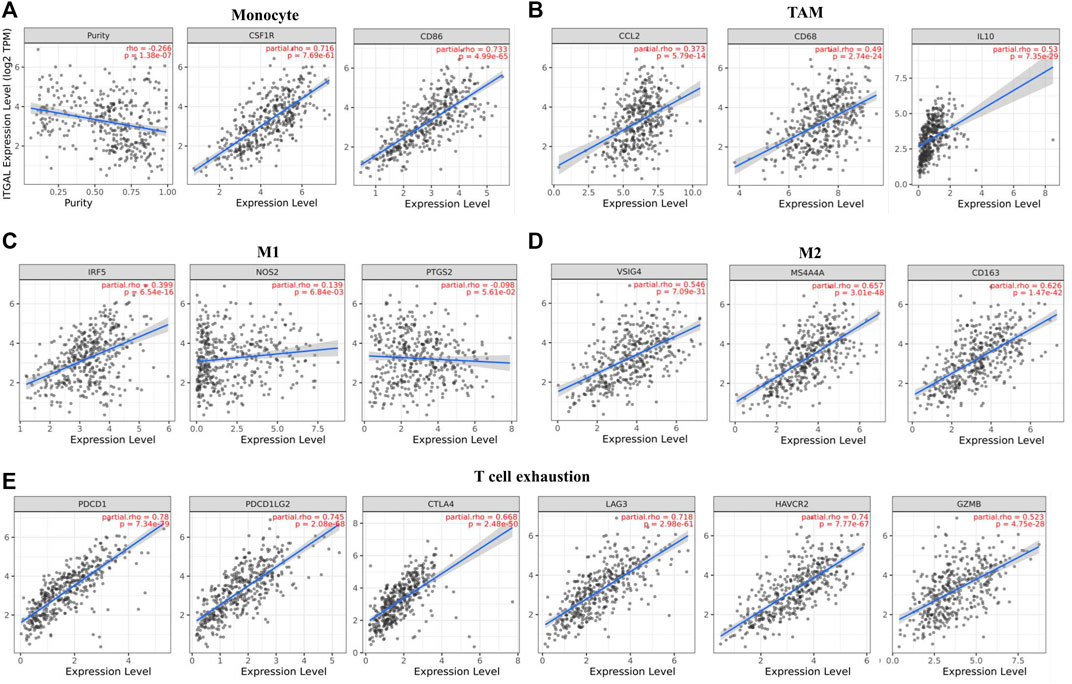
FIGURE 5. ITGAL expression correlated with monocyte, macrophages, and T cell exhaustion in stomach adenocarcinoma (STAD). Markers include CD86 and CSF1R of monocytes; CCL2, CD68, and IL10 of TAMs; NOS2, IRF5, and PTGS2 of M1 macrophages; and CD163, VSIG4, and MS4A4A of M2 macrophages. Scatterplots of correlations between ITGAL expression and gene markers of monocytes (A), TAMs (B), and M1 (C), M2 macrophages (D) and T-cell exhaustion (E) in STAD (n = 415).
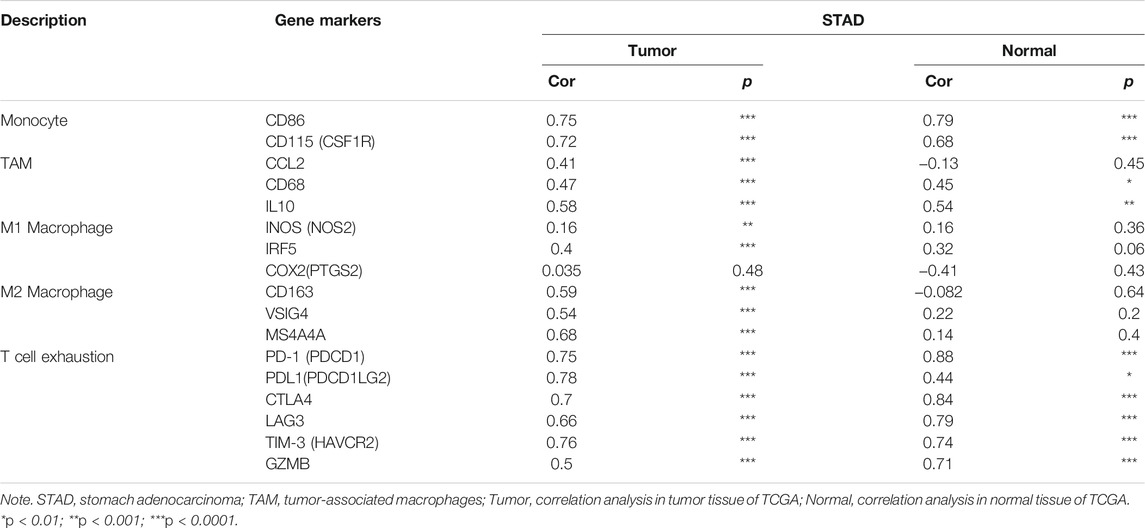
TABLE 3. Correlation analysis between ITGAL and related genes and markers of monocyte, macrophages, and T-cell exhaustion in Gene Expression Profiling Interaction Analysis (GEPIA).
Immunomodulators are substances that affect the function of the immune system. This research indicated that ITGAL was significantly connected with immunoinhibitors (p < 2.2e−16), such as BTLA (rho = 0.843), CD96 (rho = 0.89), CD274 (rho = 0.526), CSF1R (rho = 0.69), HAVCR2 (rho = 0.716), PDCD1 (rho = 0.77), and TIGIT (rho = 0.88) (Figure 6A). The expression of ITGAL was also closely associated with immunostimulators (p < 2.2e−16), including CD27 (rho = 0.849), CD28 (rho = 0.841), CD40 (rho = 0.559), CD48 (rho = 0.88), CD80 (rho = 0.665), CD86 (rho = 0.723), CXCR4 (rho = 0.638), ICOS (rho = 0.793), IL2RA (rho = 0.702), KLRK1 (rho = 0.833), and LTA (rho = 0.819) (Figure 6B). These results suggested that ITGAL is intimately engaged in the regulation of the immune interaction and may modulate tumor immune escape.
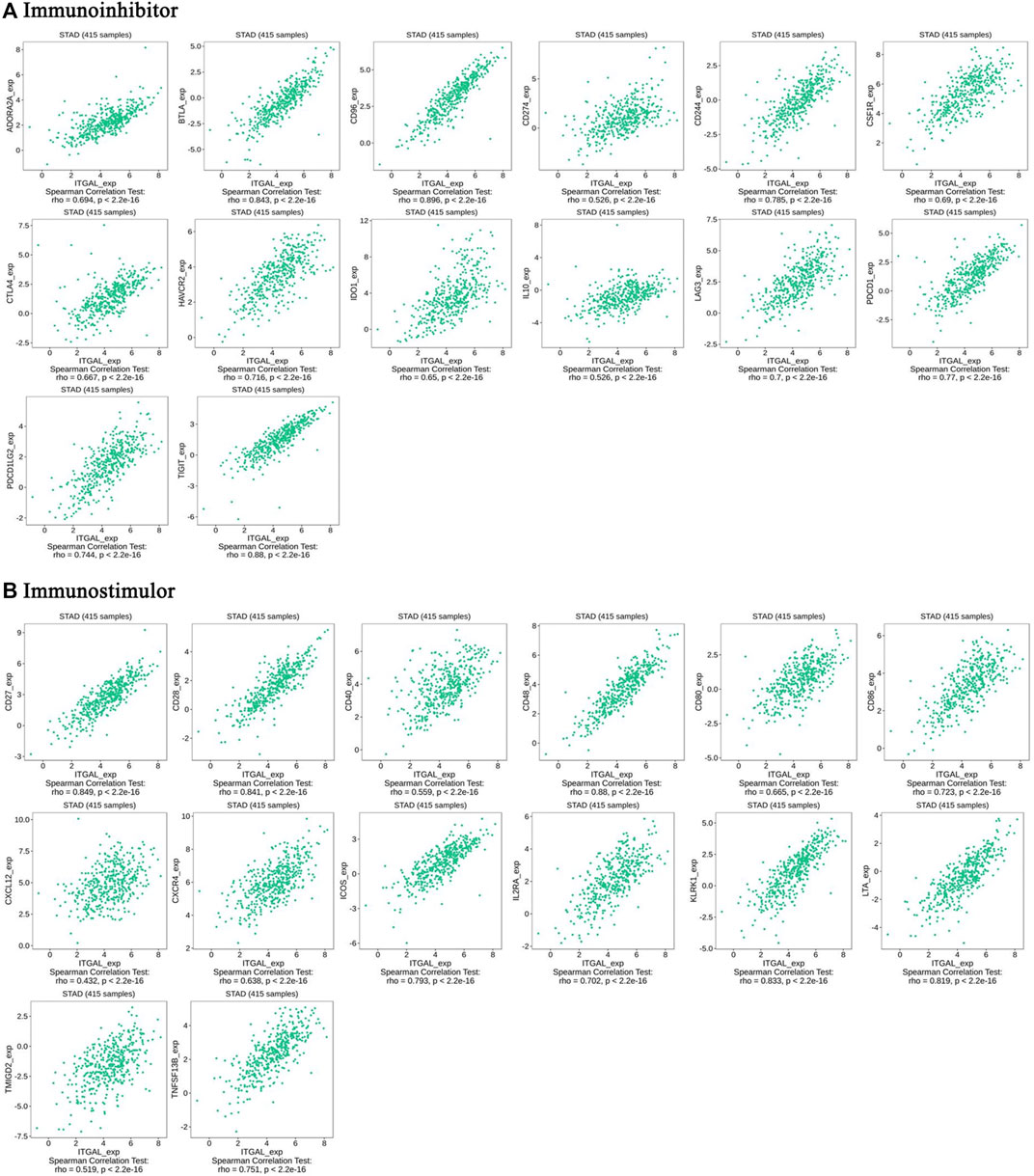
FIGURE 6. The expression of ITGAL is associated with immunomodulators in gastric cancer. (A) Correlation between ITGAL expression and immunoinhibitors in gastric cancer available at TISIDB database. (B) Correlation between ITGAL expression and immunostimulators in gastric cancer available at TISIDB database. Color images are available online.
Chemokines play a great function in controlling infiltration degree of immune cell. This research implicated the association between ITGAL expression with chemokines. For example, ITGAL expression was significantly linked with CCL2 (rho = 0.353), CCL3 (rho = 0.353), CCL4 (rho = 0.584), CCL5 (rho = 0.788), CCL8 (rho = 0.347), CCL11 (rho = 0.396), CCL13 (rho = 0.408), CCL17 (rho = 0.54), CCL18 (rho = 0.383), CCL19 (rho = 0.663), CCL21 (rho = 0.471), CCL22 (rho = 0.677), CCL23 (rho = 0.458), CX3CL1 (rho = 0.421), CXCL9 (rho = 0.673), CXCL10 (rho = 0.526), CXCL13 (rho = 0.73), and XCL2 (rho = 0.668) (Figure 7A). All the values of p were far less than 0.001. Meanwhile, we demonstrated that ITGAL expression was also significantly correlated with chemokine receptors (p < 0.001), including CCR1 (rho = 0.662), CCR2 (rho = 0.797), CCR4 (rho = 0.782), CCR5 (rho = 0.896), CCR6 (rho = 0.379), CCR7 (rho = 0.772), CCR8 (rho = 0.711), CCR9 (rho = 0.344), CCR10 (rho = 0.336), CXCR3 (rho = 0.755), CXCR4 (rho = 0.638), CXCR5 (rho = 0.707), CXCR6 (rho = 0.821), and XCR1 (rho = 0.679) (Figure 7B). These results further demonstrated the findings that ITGAL may function as an immunoregulatory factor in GC.
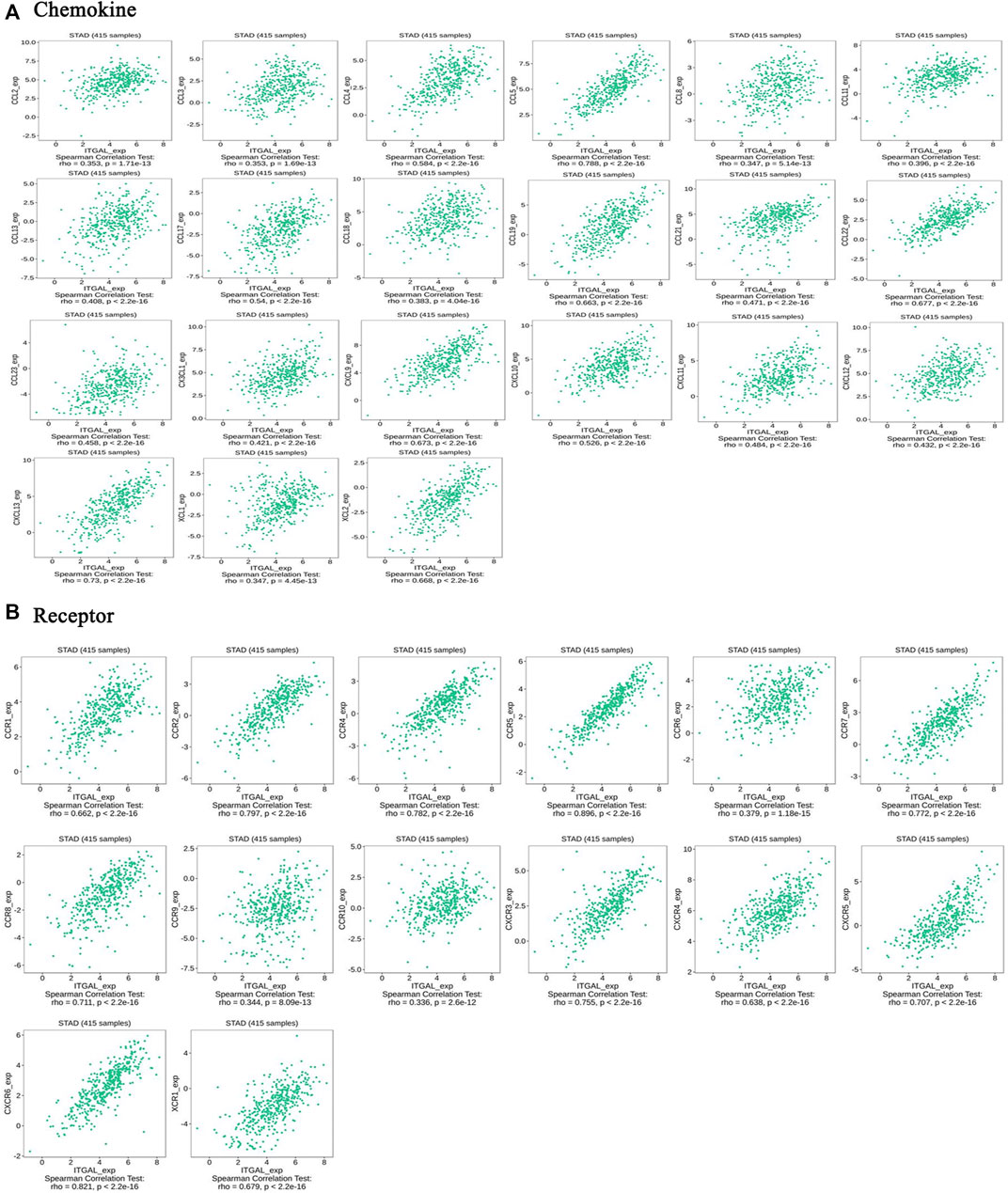
FIGURE 7. Correlation between the expression of ITGAL and chemokines in gastric cancer. (A) Correlation between ITGAL expression and chemokines in gastric cancer available at TISIDB database. (B) Correlation between ITGAL expression and chemokine receptors in gastric cancer available at TISIDB database. Color images are available online.
In this research, TIMER and GEPIA databases were performed to investigate whether ITGAL expression had a correlation with T-cell exhaustion infiltration in GC (Figure 5 and Table 3), and ITGAL expression was positively correlated with PD1 (PDCD1) in GC. Therefore, we conducted an immunohistochemical method to determine the association between ITGAL and PD1 expression, and the results are presented in Figure 8. By scoring staining intensity, we classified the levels of the expression into four groups: negative (−), weak (+), moderate (++), and strong (+ + +) staining. ITGAL showed strong expression in most GC specimens, and a few specimens showed weak staining. Obviously, PD1 was consistent with ITGAL staining intensity (Supplementary Figure S1). The expression level of PD1 was correspondingly high and low in ITGAL strong and weak expression samples. These findings further demonstrated that the upregulated ITGAL corresponds to a higher T-cell exhaustion infiltration degree.
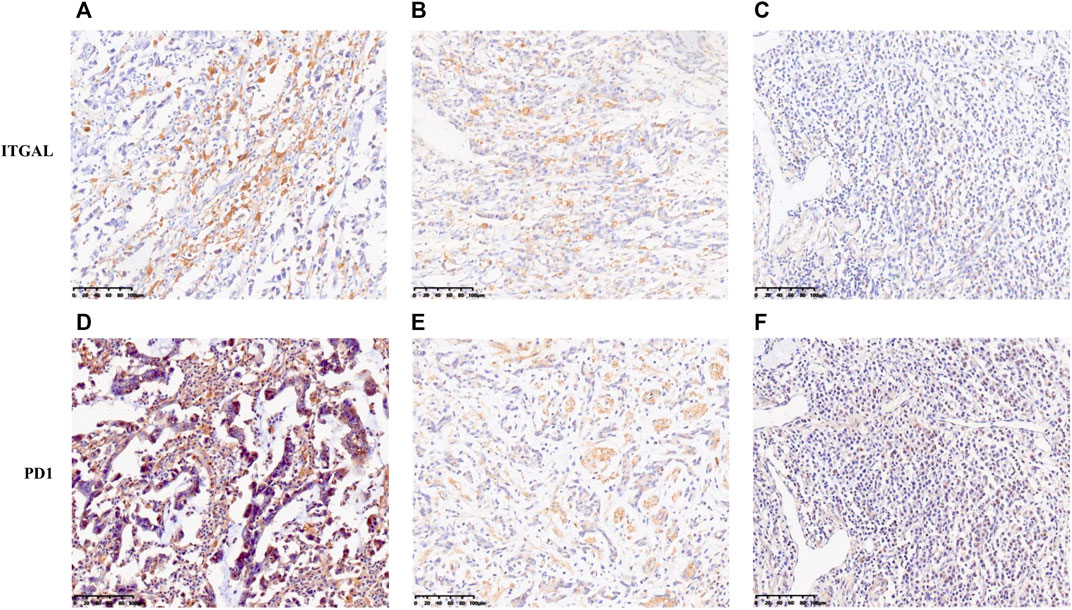
FIGURE 8. Correlation between ITGAL expression and immune infiltration levers of T-cell exhaustion in gastric cancer. The expression of ITGAL [(A) +++, (B) ++, (C) +]. The expression of PD1 [(D) +++, (E) ++, (F) +]. The expression density of ITGAL and PD1 in gastric cancer tissue was quantitated by scoring staining intensity, including negative (−) and weak (+) staining, moderate (++) and strong (+ + +) staining, respectively.
In the present research, a comprehensive bioinformatics investigation was performed to systematically analyze the clinical significance and expression lever of ITGAL in GC. Our analyses revealed poor prognosis was consistent with high expression of ITGAL in GC. Furthermore, our data also indicated that ITGAL expression had a close association with infiltration degrees of different immune cells, immunostimulators, immunoinhibitors, chemokines, and receptors in GC. Therefore, our research revealed new insights in understanding the critical function of ITGAL, and it may be a prognostic biomarker linked with immune infiltration of GC.
Integrins, the heterodimers produced by α and β subunit noncovalent binding, not only can regulate intercellular function including cellular adhesion, cell–matrix adhesion, and tumor microenvironment but also control cell proliferation and migration by recognizing specific extracellular ligands (Ruoslahti and Pierschbacher, 1987; Anikeeva et al., 2005; Schwartz et al., 2018). ITGAL, as a member of the integrin family, plays a key function in a variety of immunological processes, including leukocyte–endothelial cell interaction and cytotoxic T-cell-mediated killing (Barber et al., 2004; Giblin and Lemieux, 2006). Although ITGAL has not been thoroughly investigated, it is known that the carcinogenic potential of ITGAL, whose expression levels of ITGAL plays a crucial role in carcinogenic potential, was correlated with renal cancer, ovarian cancer, colorectal cancer, and head and neck squamous cell carcinoma (Vendrell et al., 2007; Boguslawska et al., 2016; Song et al., 2019; Zhao et al., 2019; Ji et al., 2020). However, the possible function of ITGAL in regulating tumor immunity and its clinical significance in GC are still unknown.
Thus, we evaluated ITGAL expression of GC by dependent databases including GEPIA, TIMER, TCGA, and UALCAN. We discovered that ITGAL was an aberrant expression between cancer and paracancerous tissues in various malignancies. Moreover, ITGAL was obviously increased in GC compared with paracancerous samples. These results were consistent with those from TCGA database. We also found that the ITGAL mRNA and protein levels were increased in most GC samples compared with the paired paracancerous samples. These results show that the level of ITGAL expression may serve as a potential diagnostic indicator in GC. Furthermore, to confirm whether ITGAL can be used as a prognostic biomarker, we used the KM plotter database to analyze the correlation between the ITGAL expression and OS, PPS, and FP in GC cohorts (213475-s-at and 1554240-a-at). Notably, analysis of this database indicated that the higher ITGAL expression correlated with HR for worse OS and poor PPS of GC. In addition, upregulated ITGAL expression had a significant correlation with a worse prognosis of GC in stages 1 and 2, N1 and N1 + 2 + 3 with the highest HR for worse OS and in stages 2 and 4, T2 to T3, N1, N2, and N1 + 2 + 3 for worse PPS. Together, these observations strongly support our hypothesis that ITGAL is a prognostic biomarker in GC.
Additionally, this study discovered that ITGAL is strongly related to the degree of immune infiltration in GC. In the cancer microenvironment, it has been demonstrated that immune cell infiltration plays critical roles in the development and progression of cancers (Curigliano, 2018; Klauschen et al., 2018). ITGAL is a tissue-specific integrin that plays a role in inflammatory and immune responses (Lu et al., 2002). Recent studies have revealed that ITGAL could promote T-cell migration via reaction to Rho GTPase signaling suppression caused direct CLL cell contact (Ramsay et al., 2013; Haspels et al., 2018). However, whether ITGAL expression is linked with immune infiltration in GC remains unknown. Therefore, we systematically examined the association between ITGAL expression and the degree of immune infiltration in GC. In our experience, this is the first time that ITGAL regulating immune infiltration with GC is identified. Our study showed that ITGAL expression had a strong correlation with TILs including CD8+T cell, CD4+T cell, Treg cell, B cell, neutrophil, TAM, DCs, NK cell, and monocyte. At the same time, increased ITGAL expression was associated with immunostimulators, immunoinhibitors, chemokines, and receptors. In addition, this study also demonstrated the association between ITGAL expression and the TIL marker genes of GC. Obviously, ITGAL expression had an association with M2 macrophage markers, including CD163, VSIG4, and MS4A4A, whereas M1 macrophage markers, such as NOS2 and IRF5, correlated with ITGAL expression in very weak and moderate ways correspondingly. Macrophages, a kind of specialized immune cells, are divided into M1 and M2 macrophages (Jayasingam et al., 2019), and play a crucial function in proliferation (Su et al., 2017), angiogenesis (Sammarco et al., 2018), invasion (Zhang et al., 2018), and metastasis (Song et al., 2017), and immunity of tumor (Wynn et al., 2013). These findings indicate that ITGAL has a potential function to regulate the polarization of TAMs.
Furthermore, upregulated ITGAL expression was strongly correlated with Tregs markers (FOXP3, CCR8) and T-cell exhaustion markers (PD1, CTLA4, LAG3). Immune checkpoint blockade is the main immunotherapeutic strategy. PD1/PDL1, as the important immune checkpoint component, has been verified to regulate the function of TILs. To date, PD1/PDL1 checkpoint blockade therapy is widely performed to various malignancies including GC (Kang et al., 2017; Fuchs et al., 2018; Kim et al., 2018), but some studies discovered that PD-1 has a critical function in tumor antigen tolerance, leading to poor therapeutic effect in some patients with PD1 therapy (Woo et al., 2012; Kang et al., 2017). Therefore, the most important is to improve tumor cell response to immune checkpoint inhibitors and cytokines. Our results revealed that increased ITGAL expression was not only associated with PD1 and CTLA4 but also significantly correlated with cell response to chemokines according to the TISIDB, TIMER, and GEPIA databases. These results reflected that it may be a strategy for enhancing immunotherapy effectiveness by targeting ITGAL. In conjunction with these results, ITGAL played a vital function in recruiting and modulating TILs in GC, and it is worth to continue investigating the molecular mechanism and function of ITGAL in modulating tumor microenvironment.
However, our study has some limitations. A limitation of this study is that most data are based on the online platform databases, which are updated and expanded continuously; therefore, the results of the research may be affected. Second, the information of complications and treatment option is not included in our study. Third, the in vitro and in vivo experiment is not used to validate the function of ITGAL in GC and the molecular mechanism of ITGAL in GC immunity, but in the future study, we guarantee that we will put more emphasis to the whole baseline information of patients, and experiments will be performed to further validate the projected results.
The upregulated ITGAL expression is closely correlated with poor prognosis and enhanced immune infiltration degree including CD8+ T cells, C4+ T cells, macrophages, neutrophils, and myeloid dendritic cells in GC. Moreover, the expression of ITGAL contributes to the regulation of M2 macrophages, Treg, and T-cell exhaustion. Therefore, this study suggests that ITGAL may, as a prognosis biomarker, highlight its novel potential function in the regulation of immune cell infiltration in GC patients.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
The studies involving human participants were reviewed and approved by the ethics committee of the Seventh Affiliated Hospital of Sun Yat-sen University, Shenzhen, Guangdong, China. The patients/participants provided their written informed consent to participate in this study.
YH and CZ designed and supervised the study. JZ and HW carried out the bioinformatics analysis, performed the experiments, and analyzed the data. JZ, HW, CY and JW wrote the paper. JZ, JX, SC, YH and CZ edited the manuscript and provided critical comments. All authors read and approved the final manuscript.
This study was funded by grants from the National Natural Science Foundation of China (81772579), Guangdong Province Science and Technology Plan Projects (2014A020212693), “3&3” Project of The First Affiliated Hospital of Sun Yat-Sen University (YH), and Sanming Project of Medical in Shenzhen (SZSM201911010). All these study sponsors have no roles in the study design, in the collection, analysis, and interpretation of data.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed nor endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2022.808212/full#supplementary-material
Supplementary Figure 1 | Correlation between ITGAL and PD1 expression based on immunohistochemical score.
Anikeeva, N., Somersalo, K., Sims, T. N., Thomas, V. K., Dustin, M. L., and Sykulev, Y. (2005). Distinct Role of Lymphocyte Function-Associated Antigen-1 in Mediating Effective Cytolytic Activity by Cytotoxic T Lymphocytes. Proc. Natl. Acad. Sci. 102 (18), 6437–6442. doi:10.1073/pnas.0502467102
Barber, D. F., Faure, M., and Long, E. O. (2004). LFA-1 Contributes an Early Signal for NK Cell Cytotoxicity. J. Immunol. 173 (6), 3653–3659. doi:10.4049/jimmunol.173.6.3653
Boguslawska, J., Kedzierska, H., Poplawski, P., Rybicka, B., Tanski, Z., and Piekielko-Witkowska, A. (2016). Expression of Genes Involved in Cellular Adhesion and Extracellular Matrix Remodeling Correlates with Poor Survival of Patients with Renal Cancer. J. Urol. 195 (6), 1892–1902. doi:10.1016/j.juro.2015.11.050
Bray, F., Ferlay, J., Soerjomataram, I., Siegel, R. L., Torre, L. A., and Jemal, A. (2018). Global Cancer Statistics 2018: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA: A Cancer J. Clinicians 68 (6), 394–424. doi:10.3322/caac.21492
Chandrashekar, D. S., Bashel, B., Balasubramanya, S. A. H., Creighton, C. J., Ponce-Rodriguez, I., Chakravarthi, B. V. S. K., et al. (2017). UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 19 (8), 649–658. doi:10.1016/j.neo.2017.05.002
Curigliano, G. (2018). Gyneco-oncological Genomics and Emerging Biomarkers for Cancer Treatment with Immune-Checkpoint Inhibitors. Semin. Cancer Biol. 52 (Pt 2), 253–258. doi:10.1016/j.semcancer.2018.05.004
Davidson, M., Okines, A. F. C., and Starling, N. (2015). Current and Future Therapies for Advanced Gastric Cancer. Clin. Colorectal Cancer 14 (4), 239–250. doi:10.1016/j.clcc.2015.05.013
Fuchs, C. S., Doi, T., Jang, R. W., Muro, K., Satoh, T., Machado, M., et al. (2018). Safety and Efficacy of Pembrolizumab Monotherapy in Patients with Previously Treated Advanced Gastric and Gastroesophageal Junction Cancer. JAMA Oncol. 4 (5), e180013. doi:10.1001/jamaoncol.2018.0013
Giblin, P., and Lemieux, R. (2006). LFA-1 as a Key Regulator of Immune Function: Approaches toward the Development of LFA-1-Based Therapeutics. Cpd 12 (22), 2771–2795. doi:10.2174/138161206777947731
Haspels, H. N., Rahman, M. A., Joseph, J. V., Gras Navarro, A., and Chekenya, M. (2018). Glioblastoma Stem-like Cells Are More Susceptible Than Differentiated Cells to Natural Killer Cell Lysis Mediated through Killer Immunoglobulin-like Receptors-Human Leukocyte Antigen Ligand Mismatch and Activation Receptor-Ligand Interactions. Front. Immunol. 9, 1345. doi:10.3389/fimmu.2018.01345
Jayasingam, S. D., Citartan, M., Thang, T. H., Mat Zin, A. A., Ang, K. C., and Ch'ng, E. S. (2019). Evaluating the Polarization of Tumor-Associated Macrophages into M1 and M2 Phenotypes in Human Cancer Tissue: Technicalities and Challenges in Routine Clinical Practice. Front. Oncol. 9, 1512. doi:10.3389/fonc.2019.01512
Ji, C., Li, Y., Yang, K., Gao, Y., Sha, Y., Xiao, D., et al. (2020). Identification of Four Genes Associated with Cutaneous Metastatic Melanoma. Open Med. (Wars) 15 (1), 531–539. doi:10.1515/med-2020-0190
Kang, Y.-K., Boku, N., Satoh, T., Ryu, M.-H., Chao, Y., Kato, K., et al. (2017). Nivolumab in Patients with Advanced Gastric or Gastro-Oesophageal junction Cancer Refractory to, or Intolerant of, at Least Two Previous Chemotherapy Regimens (ONO-4538-12, ATTRACTION-2): a Randomised, Double-Blind, Placebo-Controlled, Phase 3 Trial. The Lancet 390 (10111), 2461–2471. doi:10.1016/s0140-6736(17)31827-5
Kim, S. T., Cristescu, R., Bass, A. J., Kim, K.-M., Odegaard, J. I., Kim, K., et al. (2018). Comprehensive Molecular Characterization of Clinical Responses to PD-1 Inhibition in Metastatic Gastric Cancer. Nat. Med. 24 (9), 1449–1458. doi:10.1038/s41591-018-0101-z
Klauschen, F., Müller, K.-R., Binder, A., Bockmayr, M., Hägele, M., Seegerer, P., et al. (2018). Scoring of Tumor-Infiltrating Lymphocytes: From Visual Estimation to Machine Learning. Semin. Cancer Biol. 52 (Pt 2), 151–157. doi:10.1016/j.semcancer.2018.07.001
Kuwano, Y., Spelten, O., Zhang, H., Ley, K., and Zarbock, A. (2010). Rolling on E- or P-Selectin Induces the Extended but Not High-Affinity Conformation of LFA-1 in Neutrophils. Blood 116 (4), 617–624. doi:10.1182/blood-2010-01-266122
Lánczky, A., Nagy, Á., Bottai, G., Munkácsy, G., Szabó, A., Santarpia, L., et al. (2016). miRpower: a Web-Tool to Validate Survival-Associated miRNAs Utilizing Expression Data from 2178 Breast Cancer Patients. Breast Cancer Res. Treat. 160 (3), 439–446. doi:10.1007/s10549-016-4013-7
Larkin, J., Chiarion-Sileni, V., Gonzalez, R., Grob, J. J., Cowey, C. L., Lao, C. D., et al. (2015). Combined Nivolumab and Ipilimumab or Monotherapy in Untreated Melanoma. N. Engl. J. Med. 373 (1), 23–34. doi:10.1056/NEJMoa1504030
Ley, K., Laudanna, C., Cybulsky, M. I., and Nourshargh, S. (2007). Getting to the Site of Inflammation: the Leukocyte Adhesion cascade Updated. Nat. Rev. Immunol. 7 (9), 678–689. doi:10.1038/nri2156
Li, T., Fu, J., Zeng, Z., Cohen, D., Li, J., Chen, Q., et al. (2020). TIMER2.0 for Analysis of Tumor-Infiltrating Immune Cells. Nucleic Acids Res. 48 (W1), W509–W514. doi:10.1093/nar/gkaa407
Lote, H., Cafferkey, C., and Chau, I. (2015). PD-1 and PD-L1 Blockade in Gastrointestinal Malignancies. Cancer Treat. Rev. 41 (10), 893–903. doi:10.1016/j.ctrv.2015.09.004
Lu, Q., Ray, D., Gutsch, D., and Richardson, B. (2002). Effect of DNA Methylation and Chromatin Structure onITGAL Expression. Blood 99 (12), 4503–4508. doi:10.1182/blood.v99.12.4503
Luebeck, E. G., Curtius, K., Jeon, J., and Hazelton, W. D. (2013). Impact of Tumor Progression on Cancer Incidence Curves. Cancer Res. 73 (3), 1086–1096. doi:10.1158/0008-5472.Can-12-2198
Motzer, R. J., Escudier, B., McDermott, D. F., George, S., Hammers, H. J., Srinivas, S., et al. (2015). Nivolumab versus Everolimus in Advanced Renal-Cell Carcinoma. N. Engl. J. Med. 373 (19), 1803–1813. doi:10.1056/NEJMoa1510665
Procaccio, L., Schirripa, M., Fassan, M., Vecchione, L., Bergamo, F., Prete, A. A., et al. (2017). Immunotherapy in Gastrointestinal Cancers. Biomed. Res. Int. 2017, 4346576. doi:10.1155/2017/4346576
Ramsay, A. G., Evans, R., Kiaii, S., Svensson, L., Hogg, N., and Gribben, J. G. (2013). Chronic Lymphocytic Leukemia Cells Induce Defective LFA-1-Directed T-Cell Motility by Altering Rho GTPase Signaling that Is Reversible with Lenalidomide. Blood 121 (14), 2704–2714. doi:10.1182/blood-2012-08-448332
Reck, M., Rodríguez-Abreu, D., Robinson, A. G., Hui, R., Csőszi, T., Fülöp, A., et al. (2016). Pembrolizumab versus Chemotherapy for PD-L1-Positive Non-small-cell Lung Cancer. N. Engl. J. Med. 375 (19), 1823–1833. doi:10.1056/NEJMoa1606774
Ru, B., Wong, C. N., Tong, Y., Zhong, J. Y., Zhong, S. S. W., Wu, W. C., et al. (2019). TISIDB: an Integrated Repository portal for Tumor-Immune System Interactions. Bioinformatics 35 (20), 4200–4202. doi:10.1093/bioinformatics/btz210
Ruoslahti, E., and Pierschbacher, M. D. (1987). New Perspectives in Cell Adhesion: RGD and Integrins. Science 238 (4826), 491–497. doi:10.1126/science.2821619
Samatov, T. R., Tonevitsky, A. G., and Schumacher, U. (2013). Epithelial-mesenchymal Transition: Focus on Metastatic cascade, Alternative Splicing, Non-coding RNAs and Modulating Compounds. Mol. Cancer 12 (1), 107. doi:10.1186/1476-4598-12-107
Sammarco, G., Gadaleta, C. D., Zuccalà, V., Albayrak, E., Patruno, R., Milella, P., et al. (2018). Tumor-Associated Macrophages and Mast Cells Positive to Tryptase Are Correlated with Angiogenesis in Surgically-Treated Gastric Cancer Patients. Int. J. Mol. Sci. 19, 19. doi:10.3390/ijms19041176
Schwartz, A. D., Hall, C. L., Barney, L. E., Babbitt, C. C., and Peyton, S. R. (2018). Integrin α6 and EGFR Signaling Converge at Mechanosensitive Calpain 2. Biomaterials 178, 73–82. doi:10.1016/j.biomaterials.2018.05.056
Seguin, L., Desgrosellier, J. S., Weis, S. M., and Cheresh, D. A. (2015). Integrins and Cancer: Regulators of Cancer Stemness, Metastasis, and Drug Resistance. Trends Cel Biol. 25 (4), 234–240. doi:10.1016/j.tcb.2014.12.006
Silva, R., D'Amico, G., Hodivala-Dilke, K. M., and Reynolds, L. E. (2008). Integrins. Atvb 28 (10), 1703–1713. doi:10.1161/atvbaha.108.172015
Song, W., Mazzieri, R., Yang, T., and Gobe, G. C. (2017). Translational Significance for Tumor Metastasis of Tumor-Associated Macrophages and Epithelial-Mesenchymal Transition. Front. Immunol. 8, 1106. doi:10.3389/fimmu.2017.01106
Song, Y., Pan, Y., and Liu, J. (2019). The Relevance between the Immune Response-Related Gene Module and Clinical Traits in Head and Neck Squamous Cell Carcinoma. Cmar 11, 7455–7472. doi:10.2147/cmar.S201177
Su, C., Jia, S., and Liu, H. (2017). Immunolocalization of CD163+ Tumor-Associated Macrophages and Symmetric Proliferation of Ki-67 as Biomarkers to Differentiate New Different Grades of Laryngeal Dysplasia. Am. J. Clin. Pathol. 149 (1), 8–16. doi:10.1093/ajcp/aqx107
Sumagin, R., Prizant, H., Lomakina, E., Waugh, R. E., and Sarelius, I. H. (2010). LFA-1 and Mac-1 Define Characteristically Different Intralumenal Crawling and Emigration Patterns for Monocytes and Neutrophils In Situ. J.Immunol. 185 (11), 7057–7066. doi:10.4049/jimmunol.1001638
Tang, Z., Li, C., Kang, B., Gao, G., Li, C., and Zhang, Z. (2017). GEPIA: a Web Server for Cancer and normal Gene Expression Profiling and Interactive Analyses. Nucleic Acids Res. 45 (W1), W98–w102. doi:10.1093/nar/gkx247
Vendrell, E., Ribas, M., Valls, J., Solé, X., Grau, M., Moreno, V., et al. (2007). Genomic and Transcriptomic Prognostic Factors in R0 Dukes B and C Colorectal Cancer Patients. Int. J. Oncol. 30 (5), 1099–1107. doi:10.3892/ijo.30.5.1099
Wang, X. K., Zhang, Y. W., Wang, C. M., Li, B., Zhang, T. Z., Zhou, W. J., et al. (2021). METTL16 Promotes Cell Proliferation by Up‐regulating Cyclin D1 Expression in Gastric Cancer. J. Cel Mol Med 25 (14), 6602–6617. doi:10.1111/jcmm.16664
Winograd-Katz, S. E., Fässler, R., Geiger, B., and Legate, K. R. (2014). The Integrin Adhesome: from Genes and Proteins to Human Disease. Nat. Rev. Mol. Cel Biol 15 (4), 273–288. doi:10.1038/nrm3769
Woo, S.-R., Turnis, M. E., Goldberg, M. V., Bankoti, J., Selby, M., Nirschl, C. J., et al. (2012). Immune Inhibitory Molecules LAG-3 and PD-1 Synergistically Regulate T-Cell Function to Promote Tumoral Immune Escape. Cancer Res. 72 (4), 917–927. doi:10.1158/0008-5472.Can-11-1620
Wynn, T. A., Chawla, A., and Pollard, J. W. (2013). Macrophage Biology in Development, Homeostasis and Disease. Nature 496 (7446), 445–455. doi:10.1038/nature12034
Xie, J., Guo, T., Zhong, Z., Wang, N., Liang, Y., Zeng, W., et al. (2021). ITGB1 Drives Hepatocellular Carcinoma Progression by Modulating Cell Cycle Process through PXN/YWHAZ/AKT Pathways. Front. Cel Dev. Biol. 9, 711149. doi:10.3389/fcell.2021.711149
Yang, L., Chen, Z., Xiong, W., Ren, H., Zhai, E., Xu, K., et al. (2019). High Expression of SLC17A9 Correlates with Poor Prognosis in Colorectal Cancer. Hum. Pathol. 84, 62–70. doi:10.1016/j.humpath.2018.09.002
Yoshihara, K., Shahmoradgoli, M., Martínez, E., Vegesna, R., Kim, H., Torres-Garcia, W., et al. (2013). Inferring Tumour Purity and Stromal and Immune Cell Admixture from Expression Data. Nat. Commun. 4, 2612. doi:10.1038/ncomms3612
Zhang, D., Qiu, X., Li, J., Zheng, S., Li, L., and Zhao, H. (2018). TGF-β Secreted by Tumor-Associated Macrophages Promotes Proliferation and Invasion of Colorectal Cancer via miR-34a-VEGF axis. Cell Cycle 17 (24), 2766–2778. doi:10.1080/15384101.2018.1556064
Keywords: gastric cancer, integrin alpha L (ITGAL), PD-1, tumor-associated macrophages, prognosis, tumor immune microenvironment
Citation: Zhang J, Wang H, Yuan C, Wu J, Xu J, Chen S, Zhang C and He Y (2022) ITGAL as a Prognostic Biomarker Correlated With Immune Infiltrates in Gastric Cancer. Front. Cell Dev. Biol. 10:808212. doi: 10.3389/fcell.2022.808212
Received: 03 November 2021; Accepted: 14 February 2022;
Published: 24 March 2022.
Edited by:
Zimu Deng, Shanghai Institute of Biochemistry and Cell Biology (CAS), ChinaReviewed by:
Shentong Fang, China Pharmaceutical University, ChinaCopyright © 2022 Zhang, Wang, Yuan, Wu, Xu, Chen, Zhang and He. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Changhua Zhang, emNoMjEwOEAxNjMuY29t; Yulong He, aHlsMTgyMUAxNjMuY29t
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.