
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cell Dev. Biol. , 13 May 2022
Sec. Signaling
Volume 10 - 2022 | https://doi.org/10.3389/fcell.2022.803947
The tumor microenvironment encompasses various innate immune cells which regulate tumor progression. Exploiting innate immune cells is a new frontier of cancer immunotherapy. However, the classical surface markers for cell-type classification cannot always well-conclude the phenotype, which will further hinge our understanding. The innate immune cells include dendritic cells, monocytes/macrophages, natural killer cells, and innate lymphoid cells. They play important roles in tumor growth and survival, in some cases promoting cancer, in other cases negating cancer. The precise characterization of innate immune cells at the single-cell level will boost the potential of cancer immunotherapy. With the development of single-cell RNA sequencing technology, the transcriptome of each cell in the tumor microenvironment can be dissected at a single-cell level, which paves a way for a better understanding of the cell type and its functions. Here, we summarize the subtypes and functions of innate immune cells in the tumor microenvironment based on recent literature on single-cell technology. We provide updates on recent achievements and prospects for how to exploit novel functions of tumor-associated innate immune cells and target them for cancer immunotherapy.
The tumor microenvironment (TME) is a complicated structure containing diverse immune cells, stromal cells (including fibroblasts, vascular networks, mesenchymal stromal cells, pericytes, and adipocytes), extracellular matrix, and multiple signaling molecules. The immune cells consist of innate immune cells including some myeloid suppressive cells and innate lymphoid cells, and adaptive immune cells like T and B lymphocytes. The stromal cells, including CAFs (cancer-associated fibroblasts) and adipocytes, also play as immunomodulators in TME (Gunaydin, 2021; Wu et al., 2021). Other non-cellular components including the growth factors, cytokines, small regulatory RNAs, and metabolites released from the tumor or immune cells could further remodel the TME (Patel et al., 2018) (Figure 1).
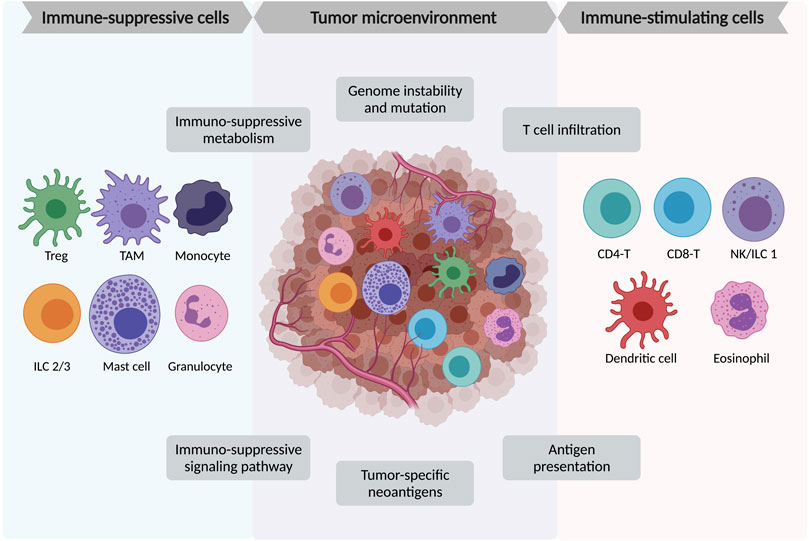
FIGURE 1. Major immune cell components in tumor micro-environment (TME). TME is a complex structure containing heterogeneous tumor cells, blood vessels, various immune cells, stromal cells, and the extracellular matrix. Among the immune cells, the immune-suppressive cells include FoxP3+ regulatory T cells (Treg), tumor-associated macrophages (TAMs), monocytes, innate lymphoid cells type 2 and 3 (ILC2/3), mast cells, and granulocytes. The immune-stimulating cells include tumor-infiltrating CD4+ and CD8+ lymphocytes, natural killer (NK) cells/innate lymphoid cells type 1 (ILC1), dendritic cells, and eosinophils.
Accumulating evidence has revealed that the relevant cellular and non-cellular components within TME can regulate the tumor initiation, growth, metastasis, and immune response to cancer therapies (Jin and Jin, 2020). Tumors are initiated due to complex biological factors, leading to hyperplasia beyond control. Within the tumor progression or invasion, TME gradually becomes hypoxic, and the acidosis structure is based on tumor cell metabolism. At the same time, various immune cells also play different roles in immune response, recognition, cytotoxicity, and interaction with each other. Understanding the role of each player in TME, especially at a single-cell level, can help us better establish the knowledge network and design effective therapeutic strategies. In this review, we will focus on the role of innate immune cells in TME. With the recent public available single-cell datasets, we will discuss their novel subtypes, the involved immune response or pathways in the tumor, and potential predictive values for therapeutic targets.
The innate immune system is the first barrel of our immunology compared with the adaptive immune system. The major functions include immune cell recruitment by cytokines, complement cascade activation, external substances identification and removal, adaptive immune system initiation, and an overall physical and chemical barrier of our body. Among these, the innate immune cells consist of myeloid cells, like dendritic cells (DCs), monocytes, macrophages, neutrophils, as well as the lymphoid cells, such as natural killer cells (NKs) and innate lymphoid cells (ILCs).
DCs are one of the most strengthened antigen-presenting cells in the mammalian immune system, bridging the activation of innate and adaptive immunology. DCs are often located in the tissues directly facing the external environment, like the skin, nose, lung, digestive system, and bloodstream. Upon the antigen-activated, they migrate to lymph nodes and launch and regulate the adaptive immune system through interaction with T and B lymphocytes. In TME, DCs infiltrate tumors and present the naïve T cells with processed tumor-derived antigens for T cell functioning. The infiltration of DCs can be a predictive marker for survival evaluation (Melaiu et al., 2020).
Monocytes are a large proportion of leukocytes and can differentiate into macrophages upon stimulation. They take part in the innate immune system as well as regulate the adaptive immune system. In cancers, monocytes play roles in both anti- and pro-tumor immunity, through cytokine secretion, phagocytosis, angiogenesis promotion. Phenotypical changes of monocytes in peripheral blood can be used for diagnosis, prediction, and prognosis (Hamm et al., 2016; Kiss et al., 2020).
Macrophages are another type of leukocyte capable of phagocytosis, antigen presentation, and anti-inflammatory function. The tissue-resident macrophages exist in nearly all tissue in our body and exhibit tissue-specific functions (Davies et al., 2013). Along with monocytes, they are composite for the mononuclear phagocyte system. In TME, macrophages are a double-edged sword, as they can directly phagocytose the tumor and remodel TME, but at the same time, they can also promote angiogenesis for tumor progression. Based on these properties, macrophages have opened a new avenue for cancer immunotherapy (Duan and Luo, 2021).
Neutrophils are the most abundant granulocytes accounting for 50–70% of the human bloodstream possessing high cell heterogeneity in homeostasis and infection (Xie et al., 2020). They are the marker of acute inflammation and are recruited to the injury sites for phagocytosis. This process is called chemotaxis, leading to the neutrophils’ migration toward sites of infection or inflammation, directed by cytokines and the complement system. In the tumor, neutrophils can be polarized into anti-tumor (N1) or pro-tumor (N2) phenotypes (Shaul and Fridlender, 2019).
NK cells are cytotoxic lymphocytes belonging to the innate immune system. It is increasingly accepted as a type of innate lymphoid cell as they develop from the common lymphoid progenitor. Upon pathogen infection or tumor formation, NK cells can rapidly respond, recognize, and attack the stressed cells without antibody or MHC recognition, enabling a fast immune reaction. In cancer, NK cells also exhibit potential for immunotherapy and prognosis (Melaiu et al., 2020; Liu et al., 2021).
ILCs are another heterogeneous lymphoid population in the innate immune system, which are mainly tissue-resident cells and rarely show up in blood circulation. They mainly secrete cytokines to regulate innate and adaptive immunity. Many of ILCs function similarly to T cells, which are likely to be the innate counterparts of T cells (Panda and Colonna, 2019). In cancer, ILCs exhibit diverse functions, with various clinical predictive values listed in this review (Bruchard and Ghiringhelli, 2019).
In cancer studies, multiple sample types have been sequenced for different purposes. For example, tumor cell lines, tumor spheroids or organoids, patient tumor resections, patient-derived xenografts, and circulating tumor cells (CTCs) from liquid biopsies are common samples for cancer research. Normally, for single-cell RNA-seq, fresh samples are preferred and followed by standard sample preparation protocol, including tissue dissociation, filtration, and RBC lysis. Dead cells and doublets removal are usually required and can be achieved by flow cytometry with optional specific cell sorting like immune cell enrichment and filtration. After this, the single-cell suspension will be sent for cell encapsulation, library construction, and in-depth sequencing. For processing frozen or hard-to-dissociate tissues, like bone, adipose, and liver, single-nucleus RNA-seq (snRNA-seq) are required for better preservation of the samples and capture of the nuclear transcripts (Slyper et al., 2020). Notably, the evaluation of CTCs at a single-cell level is usually difficult due to cell rarity and frailty but can be improved by microfluidics or immunoaffinity enrichment methods (Lim et al., 2019). It has also been suggested that the combination of tumor tissue and CTCs samples can better reflect the tumor heterogeneity and offer more clinical information (Figure 2).
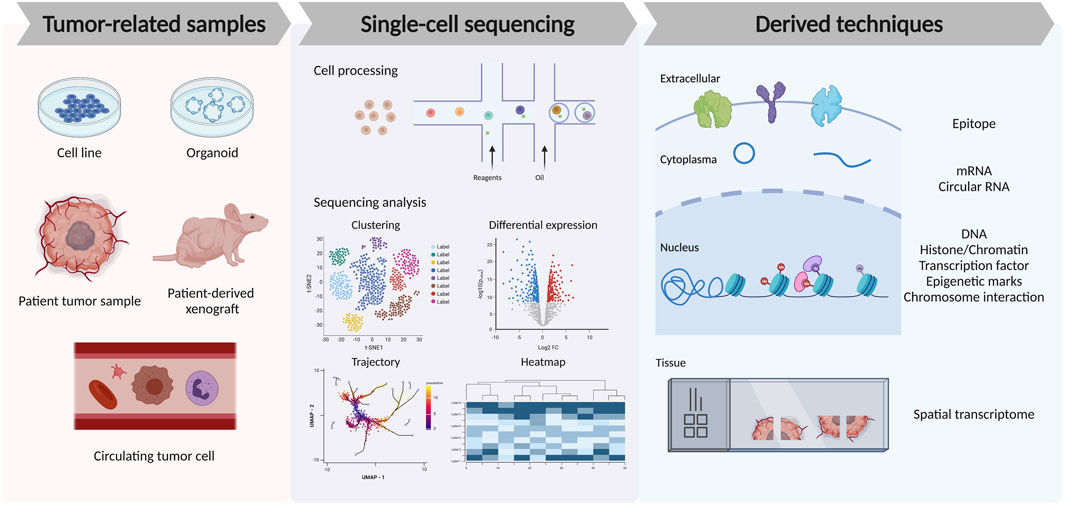
FIGURE 2. Recent advances in single-cell technologies to analyze tumor microenvironment. The left panel shows different tumor-related sample types used for single-cell TME studies. The middle panel shows the cell process of droplet-based single-cell sequencing technology and the downstream analysis from sequencing data, including the clustering and cell-type annotation, differential gene expression among samples or cell types, trajectory analysis of the cell lineage, and heatmap for gene dynamics. The right panel shows the recent derived techniques from single-cell sequencing, which allows the detection of the single-cell epitope, mRNA, circular RNA, DNA, epigenetic modification, chromosome interaction, and spatial information among the tissue.
In recent years, single-cell sequencing technology has developed rapidly and derived various applications. For example, the Cellular Indexing of Transcriptomes and Epitopes by Sequencing (CITE-seq) enables cell surface epitope and transcriptome measurement at the same time (Stoeckius et al., 2017). Other techniques targeting methylation, histone modification, chromatin accessibility, or the TCR repertoire at a single-cell level provide more precise genomic information for cancer studies. As just mentioned, scRNA-seq and snRNA-seq capture different cellular compositions. The choice between these two technologies depends on sample availability, storage, and biological questions (Slyper et al., 2020). Both technologies allow for sample multiplexing, which largely helps with experiment design and reducing batch effects. More recently, the emerging spatial transcriptomics allows the acquirement of cell transcriptome and cellular position information at the same time, providing informative data for TME studies, like insights into the communication between tumor and immune cells through ligand-receptor interactions at a spatial level.
In Table 1, we list publicly available datasets of TME using different sequencing techniques. Samples can be mouse tumor models or human patient samples. Some studies applied multi-omics techniques for sequencing, including normal single-cell transcriptome, CITE-seq, target V (D)J-seq, and spatial transcriptome analysis. These data are precious resources for us to better understand the underlying immune response and cellular interactions within TME. Although the existence or absence of immune cells can be analyzed through conventional FACS using specific markers, the advantage of single-cell analysis is to perform in-depth transcriptional profiling of immune cells in the tumor samples. With scRNA-seq, individual immune cells could be distinguished and further assist discovering novel subtypes. This helps facilitate our understanding of complicated TME and raise new targets for treatment. Furthermore, the exploitation on multiple scRNA-seq datasets could help define specific gene signatures or specific innate immune cell subsets that respond or resist to cancer immunotherapy, or have predictive or prognostic value in patients undergoing immunotherapy.
High-throughput sequencing techniques offer insights into the intra- and inter-tumoral heterogeneity, tumor origin and evolution, mechanisms of tumor invasion and metastasis, characterization of TME, drug resistance, and future therapeutic design (Li et al., 2021a). The most commonly used computational tools for cancer studies include (Gunaydin, 2021) Seurat and Scanpy, the most commonly used single-cell downstream analysis workflow in R and python platforms (Wolf et al., 2018; Hao et al., 2021); (Wu et al., 2021) Monocle, depicting a “pseudotime” for lineage trajectory prediction (Cao et al., 2019); (Patel et al., 2018) VeloAE, studying cellular transition under different dimension of RNA velocity (Qiao and Huang, 2021); (Jin and Jin, 2020) Cellsnp-lite, genotyping tumor cells at a single-cell level (Huang and Huang, 2021); (Melaiu et al., 2020) MQuad, identifying tumor cell populations with mitochondrial mutations (Kwok et al., 2022); (Kiss et al., 2020) CellChat, inferring cell-cell communication networks and signaling pathways (Jin et al., 2021); (Hamm et al., 2016) FlowGrid, allowing for fast clustering of very large single-cell datasets (Fang and Ho, 2021). Among these, Monocle, VeloAE, and other trajectory analysis tools can help distinguish normal and abnormal differentiation of immune cells, especially for those activated- or pathological-immune cells within tumor sites. They can also help infer the origins of newly discovered immune cell subtypes. On the other hand, Cellsnp-lite and MQuad detect the somatic mutations for tumor clonality identification, separating different tumor clones and the specific immune contexture. CellChat exploits the ligand-receptor expression to depict the crosstalk between innate immune cells and specific tumor clones. For massive tumor datasets analysis, like the pan-cancer analysis, FlowGrid can assist the large datasets combination and clustering within a relatively short time. These robust tools can assist us to analyze immune cell-cancer data from genetic and cellular aspects, providing overall insights into cancer immunology research.
Within the TME, DCs function by antigen processing and presentation to T cell surfaces to activate the cytotoxic T lymphocytes (CTLs). Conventionally, DCs can be divided into several subtypes based on their immune phenotype and functions, including CD123+ plasmacytoid DC (pDC), CLEC9A+ XCR1+ CADM1+ type1 conventional DC (cDC1), CDA1+ CD172A+ type2 conventional DC (cDC2), LAMP3+ DC, and CD14+ monocyte-derived DC (moDC) (Wylie et al., 2019). The single-cell transcriptome profiling on human advanced osteosarcoma and lung cancers in humans and mice provides similar division on DCs subtypes and finds conserved results across two species (Zhou et al., 2020; Zilionis et al., 2019). However, some of the markers that help to differentiate the DC subsets are not transcriptionally enriched but have a good protein expression level. In this scenario, the more recently developed CITE-seq method can better capture the cell surface proteome for a better phenotypical understanding of innate immune cells. In Table 2, we have listed the DCs as well as other innate immune cell transcriptome and cell surface markers to better distinguish relative sub-groups. For example, in one NSCLC (non-small cell lung cancer) scRNA-seq and CITE-seq combined dataset, the authors have also identified these five subtypes of DCs. The antibody-oligo conjugates from CITE-seq provide additional information for cell surface proteome, which is more solid than sole transcriptomic markers (Leader et al., 2021).
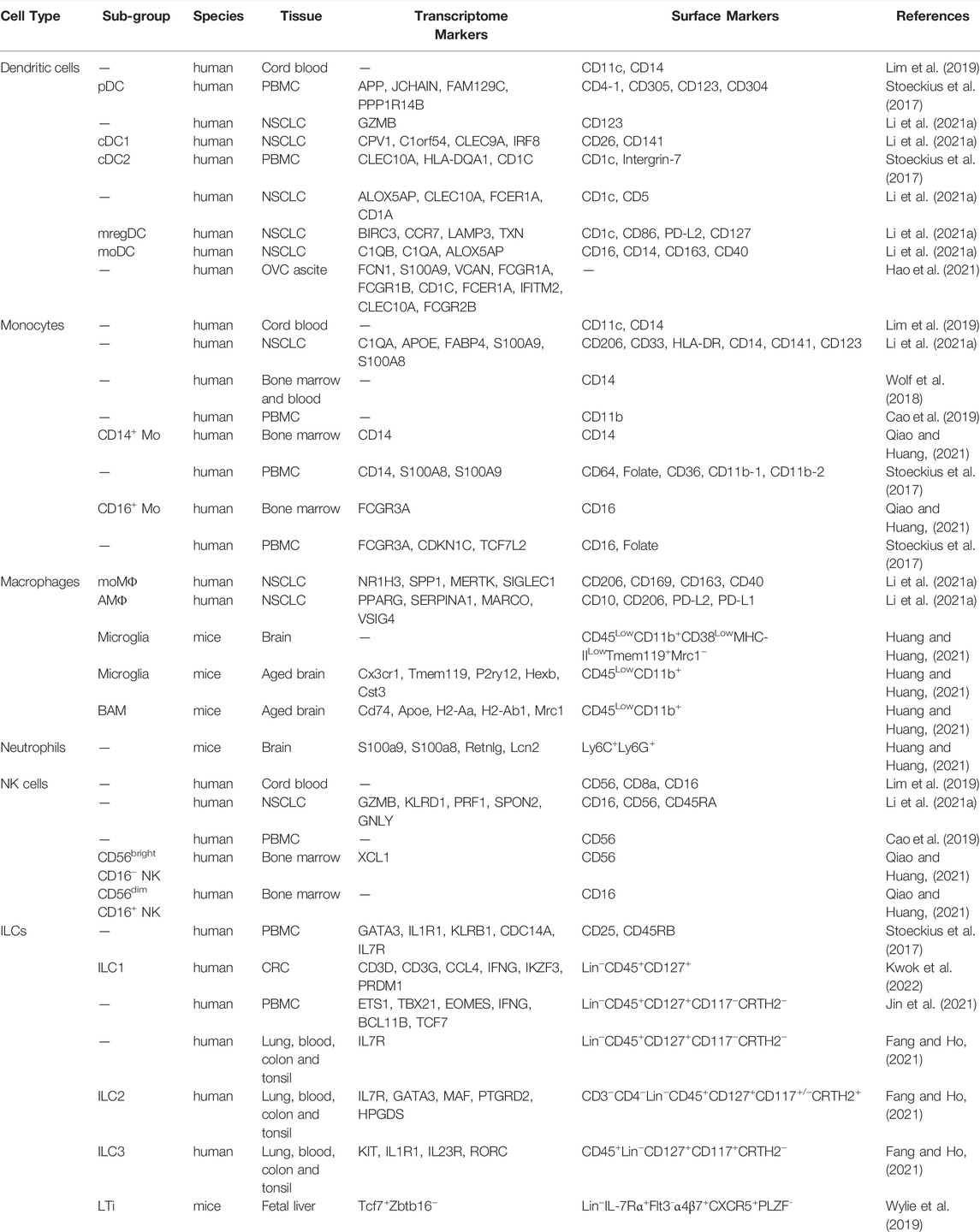
TABLE 2. Phenotypic markers of innate immune cells using CITE-seq or scRNA-seq. pDC, plasmacytoid DC. cDC1, type1 conventional DC. cDC2, type2 conventional DC. mregDC, mature DC expressing regulatory molecules. moMΦ, monocyte-derived macrophage. AMΦ, alveolar macrophage (tissue-resident macrophage). NSCLC, non-small cell lung cancer. PBMC, peripheral blood mononuclear cells. OVC, ovarian cancer. BAM, border-associated macrophages. CRC, colorectal cancer. Transcriptome markers are mainly derived from clustering results, while surface markers are collected based on antibody-derived tags (ADT) by CITE-seq or FACS antibodies.
In TME, pDCs mainly secrete type I interferons (IFN-I), which can improve anti-tumor immunity through an interaction with tumor and immune cells. However, their antigen presentation ability is much lower than cDCs and is thought to initiate immune tolerance through the secretion of tolerogenic factors including interleukin 10 (IL-10), tumor growth factor (TGF-β) (Villadangos and Young, 2008). Besides, pDCs express some T cell ligands such as programmed cell death ligand 1 (PD-L1) to bind to inhibitory T cell receptors (Matta et al., 2010). However, whether pDCs correlate with better or poor prognosis in tumors is still controversial. Various studies have pointed out the infiltrating pDCs in tumors with poor prognosis in ovarian cancer and melanoma (Labidi-Galy et al., 2012; Aspord et al., 2013). However, more recent studies have shown that higher densities of tumor-infiltrating pDCs correspond to better survival in triple-negative breast cancer and colon cancer (Oshi et al., 2020; Kießler et al., 2021). A large-sample cohort investigation on breast cancer multi-omics datasets also supported this conclusion (Tian et al., 2021). In nasopharyngeal carcinoma (NPC), the favorable prognostic value of pDCs suggests their anti-tumor effects within TME (Chen et al., 2020a). These conflicting conclusions might be due to different tumor types and can be further confirmed by performing pan-cancer analysis based on abundant publicly available datasets.
cDCs can be divided into two groups, CLEC9A+ XCR1+ cDC1 requiring BATF3 and IRF8 for maturation, and CD1A+ cDC2 dependent on IRF4 (Murphy et al., 2016). cDCs are the most powerful antigen-presenting cells to induce T cell-mediated immune responses through MHC class I cross-presentation and MHC class II presentation in humans. cDCs also infiltrate the tumors but only account for a small portion of immune cells in TME (Böttcher et al., 2018; Diao et al., 2018). In TME, cDC1 plays a key role in launching anti-tumor immunity of CTLs (Roberts et al., 2016). Tumor-associated antigens from tumor sites are transferred to tumor-draining lymph nodes by these cDC1s and followed by CD8+ T cells activation (Salmon et al., 2016). cDC1 also regulates the local immune response through chemokines XCL1 and CCL5 produced by NK cells (Böttcher et al., 2018). In single-cell pan-cancer analysis, cDC2 shows variation among different cancer types with diverse functions like development and maintenance of Langerhans cells and promoting T cell differentiation (Cheng et al., 2021). The same study has also conveyed that the CXCL9+ cDC2s subtype is more likely to transit into LAMP3+ cDC and acquired an enhanced immune-suppressive phenotype.
Notably, one single-cell RNA-seq study on human NSCLC samples has revealed a new type of mature DC expressing regulatory molecules (mregDCs) (Maier et al., 2020). The follow-up studies from the same group further characterize this new subtype with LAMP3 and PD-L1 expression (Leader et al., 2021). In another single-cell transcriptome dataset from human hepatocellular carcinoma, researchers found another small proportion of cDC cells specifically expressing maturation marker LAMP3, migration marker CCR7, lymphocyte recirculation chemokines CCL19 and CCL21 (Zhang et al., 2019). In the single-cell bladder urothelial carcinoma dataset, LAMP3+ DCs also exhibit T cell recruitment function, suggesting an immunosuppressive TME (Chen et al., 2020b). In a pan-cancer analysis, researchers further found that cDC1-derived LAMP3+ DCs and cDC2-derived LAMP3+ DCs are under different ligand-receptor control, suggesting potentially diverse functions (Cheng et al., 2021). This evidence concludes the migrating function of cDCs from tumor to lymph node and the communication with lymphoid cells, suggesting this DC subtype as a potential target for immunotherapy.
Another type of DCs is the moDC, also called inflammatory DC. They only show up in response to inflammation, infection, and cancer (Laoui et al., 2016). A single-cell data on peritoneal ascites from ovarian cancer patients have characterized moDCs and end-stage moDCs. These moDCs use the vacuolar pathway and can efficiently induce cytotoxic CD8+ T cells (Tang-Huau et al., 2018). However, in TME, the role of moDCs is still elusive. Some study has shown that they are essential for CD8+ T cells and anti-tumor response (Kuhn et al., 2015). In contrast, other studies provided some tumorigenic evidence of moDCs, like inhibitory immune response through nitric oxide release and reactive radical (Laoui et al., 2016).
In humans, monocytes can be divided into three main subtypes, CD14++ CD16− classical monocytes, CD14+ CD16++ non-classical monocytes, and CD14++ CD16+ intermediate monocytes, which have also been validated by several single-cell transcriptome datasets (Dutertre et al., 2019; Zhang et al., 2020). According to tumor patients’ clinical data, alterations of monocytes in peripheral blood are informative for diagnosis and prognosis (Kiss et al., 2020). For example, a higher number of CD14+ HLA-DRlow monocytes is linked to later cancer stages and poor survival (Okla et al., 2018). More interestingly, the number of CD14+ HLA-DRlow monocytes progressively drop in patients who received and responded to anti-CTLA4 immune checkpoint blockade, suggesting their prediction value (Gebhardt et al., 2015).
During tumorigenesis, monocytes show various functions at different stages. The classical monocytes can differentiate into pro-tumor TAMs in many tumor types. In a single-cell RNA-seq dataset on human bladder urothelial carcinoma, monocyte-TAMs polarization is observed along with the recruitment of monocytes from normal mucosal tissues (Chen et al., 2020b). In a single-cell pan-cancer analysis, tumor and adjacent normal tissue-derived monocytes express more HLA genes and macrophage-related genes, suggesting a migration of monocytes and maturation into TAMs (Cheng et al., 2021). In human colon cancer, FCN1+ monocytes enriched in tumors show a similar pattern to blood CD14+ monocytes, probably migrating to tumors and obtaining some tumor-specific transcriptional profiles (Zhang et al., 2020).
In a pan-cancer analysis, tumor-infiltrated monocytes can also promote tumor growth by suppressing T cell functions through some inflammatory cytokines and chemokines (Cheng et al., 2021). According to clinical data, intra-tumoral chemokine CCL2 produced by monocytes are negatively correlated to CD8+ T cell numbers within hepatocellular carcinoma patients (Li et al., 2017). In comparison, patients suffering from pancreatic cancer with low CCL2 and high CD8 show a better prognosis (Sanford et al., 2013). Within the TME, they also secrete chemokine CCL5 that enroll immunosuppressive regulatory T cells (Tregs) (Plitas et al., 2016). In the meantime, secretions including IL-4, IL-10, and IL-13 by Tregs are likely to further promote the differentiation of classical monocytes into activated TAMs, progressing the immunosuppression (Pommier et al., 2013). In addition to T cells interactions, monocytes also secrete chemokines CCL3, CCL4, CCL5, and IL-15, which are essential for NK cells recruitment in metastatic tumors sites (Hanna et al., 2015; Kubo et al., 2017).
Typically, macrophages can be classified into proinflammatory M1 macrophages and anti-inflammatory M2 macrophages. M1 macrophages can be induced by IFN-γ and lipopolysaccharide (LPS) and can directly attack tumor cells or pathogens and promote inflammatory effects by releasing cytokines like TNF-α, IL-6, IL-12, IL-18 (Mills et al., 2016). They can also present the antigens with MHC class II, activating the adaptive immune responses. In comparison, the M2 are induced by IL-4 or IL-13, and usually have lower MHC class II expression (Lawrence and Natoli, 2011). In TME, the tumor-associated macrophages (TAMs) are more M2-like based on their transcriptome profiling (Qian and Pollard, 2010). However, more and more evidence has shown that the simple division of M1/M2 macrophage in TME is not inclusive, as they found that TAMs share both M1 and M2 patterns (Biswas et al., 2006; Chávez-Galán et al., 2015). Macrophages are of high plasticity and can be affected by the local microenvironment. Many single-cell transcriptome datasets have also pinpointed the inaccuracy of M1 vs. M2 classification (Sathe et al., 2020; Zhang et al., 2020; Zhou et al., 2020; He et al., 2021). In some cancer like pancreatic ductal adenocarcinoma and nasopharyngeal carcinoma, an M1-M2 coupled pattern rather than separate states has been observed in single-cell data, which calls for more precise definitions of TAMs (Peng et al., 2019; Jin et al., 2020). According to single-cell pan-cancer analysis, TAMs subtypes have shown distinct transcriptomic profiles among different cancer types (Cheng et al., 2021). The SPP1+ TAMs and C1QC1+ TAMs are more like M2 phenotype, while ISG15+ TAMs express more M1-related genes. This again emphasizes the limitation of the simple division of M1/M2.
According to a human single-cell colon cancer dataset, C1QC+ and SPP1+ TAMs are likely to differentiate from FCN1+ monocytes in tumors (Zhang et al., 2020). The gene expression profile of C1QC+ TAMs is more related to phagocytosis and antigen presentation, while SPP1+ TAMs show a preferential expression pattern in angiogenesis regulation, suggesting the potential invasive function of this subtype. In the same study, researchers found that expression of C1QC and high expression of SPP1 correlate to poor prognosis of patients, implying that SPP1 depletion on TAMs might result in a better outcome. In hepatocellular carcinoma, there is another interesting subtype called tissue-resident FOLR2+ TAMs, showing similar patterns to fetal liver macrophages and support the oncofetal reprogramming concept in TME (Sharma et al., 2020). These TAMs have more interaction with other immune cells through some immune checkpoints, like CD28−CD86, SIRPA-CD47, and CD86-CTLA4. Compared with SPP1+ TAMs, FOLR2+ TAMs also exhibit more immune-suppressive interactions with Tregs.
In TME, TAMs can promote tumor progression in many aspects. They secrete the chemokines or cytokines like IL-6, IL-8, and IL-10 (Xu et al., 2014; Wang et al., 2021). IL-6 is mainly responsible for cell cycle regulation, tumor angiogenesis, and local inflammation improvement. It can also promote cancer stem cell self-renewal through STAT3 phosphorylation (Wan et al., 2014). In the meantime, IL-8 can activate a wide range of pathways like JAK2/STAT3/Snail pathways, leading to monocytes recruitment and further macrophage polarization (Fu et al., 2015). Notably, IL-8 is highly expressed by TAMs and the serum IL-8 can be used as a predictive biomarker for immune checkpoint blockade efficacy (Schalper et al., 2020). TAMs induce tumorigenesis through several immunosuppressive checkpoints. A famous ligand-receptor pair is programmed cell death protein (PD-1) and programmed cell death-ligand 1 (PD-L1). As reported in a large NSCLC cohort analysis, high expression of PD-L1 in TAMs is correlated to better overall survival (Liu et al., 2020). Other checkpoints include two “don’t eat me signaling”, CD47-SIRPα signaling and CD24-SIGLEC10 signaling, especially expressed on macrophages and tumors (Yu et al., 2016; Barkal et al., 2019). TAMs can also promote cancer metastasis by delivering small cell vesicles called exosomes, packaging the molecules like proteins and nucleic acids. Mass spectrometric analysis has shown the existence of apolipoprotein E (ApoE) within the exosomes, activating the PI3K-AKT pathway and inducing epithelial-mesenchymal transition (EMT), increasing tumor metastasis potential (Zheng et al., 2018) (Figure 3).
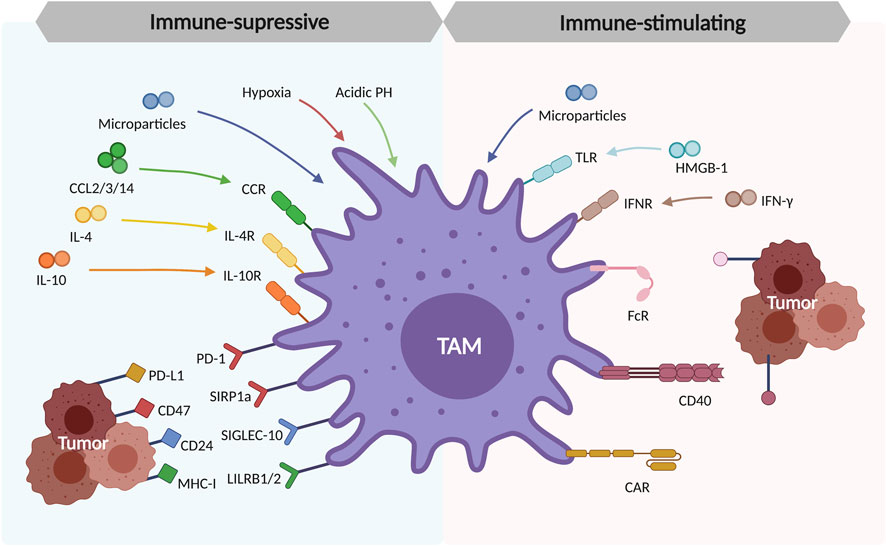
FIGURE 3. Polarization and formation of tumor-associated macrophages (TAMs) in TME. The tissue-resident macrophages and recruited macrophages from monocytes are the origins of TAMs. These TAMs are polarized in response to the cytokines produced by tumor cells in TME (CCL2/3/14, IL-4, and IL-10), the environmental factors like hypoxia and acidic PH in TME, the immunosuppressive checkpoints (PD-1/PD-L1, CD47/SIRP1a, CD24/SIGLEC-10, MHC-I/LILRB1/2).
On the other hand, TAMs enhance anti-tumor ability through the blockade of some signaling molecules on the tumor cells. Some tumor cells express MHC class I component β2-microglobulin signaling ligand. The knockout of its receptor LILRB1 on macrophages inhibited tumor growth (Barkal et al., 2018). LILRB1 inhibition combined with anti-CD47 monoclonal antibody treatment improved the macrophage phagocytosis and tumor-killing effects, without hurting normal cells (Barkal et al., 2018). Besides, TAMs can function against tumors through macrophage-mediated programmed cell removal. The toll-like receptor (TLR) pathway activated by IL-10 will further induce the BTK signaling pathway and release the “eat me” signaling, which can be a promising immunotherapeutic target (Byrne et al., 2013).
In cancer, the role of the neutrophils is still controversial. These cells are involved in many cancer types, located inside the tumor or surrounding tissues, or in the peripheral blood. In the tumor sites, neutrophils can be stimulated by cytokines and polarized into tumor-associated neutrophils (TAN), with either anti-tumor activity (N1) or a pro-tumor activity (N2), like the nomenclature of M1/M2 macrophages (Shaul and Fridlender, 2019; Veglia et al., 2021). Decades before, scientists have relied on meta-analysis to evaluate the role of neutrophils in cancer, revealing that TANs resided in tumor tissues correlated to unfavorable recurrence-free and poor overall survival (Shen et al., 2014). However, the exact anti-tumor or pro-tumor function of neutrophils in TME is hard to evaluate as there are no accurate markers for distinguishing the populations, which calls for further studies using scRNA-seq and CITE-seq to better characterize them. In one single-cell dataset on human colon tumor samples, researchers have tried Smart-seq2 and 10 × scRNA-seq platform for neutrophils enrichment (Zhang et al., 2020). However, either technique sufficiently captures this cell type, probably due to the difficulty of cell purification procedure and RNA capturing from cells.
There are a group of cells called myeloid-derived suppressor cells (MDSCs), including CD11b+Gr1high polymorphonuclear MDSCs (PMN-MDSCs) and CD11b+Gr1low (M-MDSCs) in mice. While in human, PMN-MDSCs are often referred to as CD14−CD15+CD66b+CD16+CD11b+CD33+HLA-DR− (Shaul and Fridlender, 2019). These PMN-MDSCs are usually considered pathologic neutrophils due to shared markers with normal neutrophils (Long et al., 2021). However, another review has pointed out that PMN-MDSCs can only stand for a sub-group of neutrophils with divergence at the cell activation level, but not a whole new cell type (Shaul and Fridlender, 2019). With this debate, a recently published single-cell RNA-seq dataset on mice breast tumor models has provided an overview of neutrophils and PMN-MDSCs at the transcriptome level (Alshetaiwi et al., 2020). They have separated the neutrophils from the spleen in homeostasis and MDSCs derived from the MMTV-PyMT mouse breast cancer model and checked how PMN-MDSCs emerge as a distinct cell cluster from neutrophils. Their analysis pointed out that in TME, PMN-MDSCs diverge from neutrophil progenitors and follow an alternative maturation process.
Taken together, although the single-cell transcriptome profiling in mice can provide a better understanding of neutrophil cell fate decisions and developmental trajectory, a protein expression level measurement like CITE-seq could be the best way forward. Another tricky thing is that most of the phenotype characterization and functional tests are conducted in mice models owing to the technical difficulties to enrich neutrophils in human tumor samples. However, neutrophils in these two species are largely different in biology and functions, which calls for the need for more human studies (Masucci et al., 2019). Besides, many experimental studies focus on human circulating neutrophils for functional assessment, which cannot fully mimic the intra-tumoral ones.
NK cells can be identified with unique CD56+ CD3− markers. Conventionally, NK cells can be divided into two subtypes, CD56dim CD16+ NK cells and CD56bright CD16− NK cells. Differences between these two subtypes are that CD56dim CD16+ NK cells are the major cluster of NK cells with higher production of cytokines and robust cytotoxic activity, while the latter one is more similar to T helper cells and with less cytotoxic activity (Cooper et al., 2001). Recently, single-cell transcriptome and trajectory analysis revealed that there might be an additional NK cell type residing in bone marrow, NK0 cells, that can differentiate into CD56dim CD16+ NK cells and CD56bright CD16− NK cells (Crinier et al., 2021).
In TME, NK cells can directly attack cells with the help of cytokines like IL-12, IL-15, and IL-18. The MHC class I balances activation and inhibition states (Huntington et al., 2007). In addition, they can assist other immune cells like DCs and T cells to launch an anti-tumor response. Studies based on the mice in vivo test and single-cell transcriptome analysis on human nasopharyngeal carcinoma have revealed that NK cells recruit cDC1s by secreting chemokines CCL5, XCL1, and XCL2, which also link to the patients’ better overall survival (Böttcher et al., 2018; Gong et al., 2021). In the lung adenocarcinoma murine model, activated NK cells by tumor ligands facilitate the T cell response and lower the tumor progression (Schmidt et al., 2019). However, TME can restrain NK cell functions. For example, the TME-recruited TAMs and Tregs inhibit CD8+ T cells and NK cells through cytokines TGF-β, IL-10 and lead to higher expression of inhibitory checkpoints (Ben-Shmuel et al., 2020). As reported by a single-cell study on human melanoma samples, NK cell populations are significantly lower across the patients (de Andrade et al., 2019). In a mass cytometry by time-of-flight (CyTOF) combined with single-cell transcriptomics on the human lung adenocarcinoma dataset, NK cells are the least abundant cells compared with other immune cells (Lavin et al., 2017). These NK cells at tumor sites lowly express CD57 and IFN-γ, while tumor-infiltrated NK cells exhibit higher expression of CXCR3. Another single-cell omics study on nasopharyngeal carcinoma has revealed that they didn’t observe the exhaustion markers like KIR, NKG2A, LAG3, or HAVCR2, and pointed out the NK cells as promising TME-based targets for cancer therapy, as they keep activated and can be a positive regulator in immune response (Gong et al., 2021). Collectively, these findings have suggested that various cancer types might lead to the difference in the frequency of NK cells, their activation status, and the expression of inhibitory markers. In this case, a more thorough analysis of publicly available single-cell datasets can provide more insights into the correlation between the tumor intrinsic factors and innate immune cell phenotypes.
Innate lymphoid cells are another type of highly heterogeneous lymphocyte. They lack the antigen receptors but express diverse activating and inhibiting receptors, responsible for different functions (Artis and Spits, 2015). Based on their dependent transcription factors and cytokines for development, ILCs can be divided into three groups and five cell types, group one containing NK cells and ILC1, group two within ILC2, group three containing ILC3, and lymphoid tissue inducer (LTi) cells. However, it is not that easy to markedly separate these sub-groups because of similar transcriptome profiles and cell state plasticity. Many studies have performed scRNA-seq analysis on mouse or human tissues and obtained some conclusive markers (Ghaedi et al., 2020; Qi et al., 2021). For example, high expression of SLAMF1 in the colon and rectal tumors correlated to much higher survival in patients (Qi et al., 2021). Other phenotypic markers of ILC classification have been well-summarized in another review paper (Roma et al., 2021). The CITE-seq method also showed that these ILCs possess high plasticity in mouse brain and intestinal tissue (Golomb et al., 2020). These cells function differently within TME in promoting and suppressing tumor growth, with a correlation to poor or better clinical outcomes (Yuan et al., 2021).
In group one ILCs, NK cells, and ILC1 lineage diverge due to the required transcription factor T-bet, as ILC1 completely relies on T-bet for its development (Vivier et al., 2018). These different developmental requirements lead to the lower cytotoxic ability of ILC1 and different marker gene expression Tbet+ Eomes− (Simonetta et al., 2016). In TME, ILC1, which is similar to T helper cells, can also respond to IL-12 and IL-15 cytokine-enriched environments and produce IFN-γ. However, it is still ambiguous whether ILC1 possesses anti-tumor or pro-tumor effects. The secreted IFN-γ by ILC1 can recruit and activate monocytes and macrophages for phagocytosis function (Panda and Colonna, 2019). On the other hand, ILC1 has shown pro-tumor function and favors tumorigenesis in response to TGF-β (Gao et al., 2017). A recent single-cell transcriptome study on human colorectal cancer has confirmed its transcriptional similarity to T helper cells (Qi et al., 2021). They also detected another cluster of helper-like ILC1 subset with additional expression of TIGIT, which is described as intermediate ILC1s in methylcholanthrene-induced mouse tumor models (Gao et al., 2017). Interestingly, they also observed an ILC1 to ILC3 transition in the blood of cancer patients, but not in normal groups (Qi et al., 2021). However, the underlying mechanism is still unknown and requires further studies.
In cancer, ILC2 has anti-tumor and pro-tumor effects, depending on its cytokine secretion and communication with other immune cells. Clinical data has shown the increase of ILC2 in peripheral blood and tumor tissue in many cancer types, like breast and gastric, but none in the healthy gut samples (Bie et al., 2014; Salimi et al., 2018; Qi et al., 2021). ILC2 produces IL-4 and IL-13, which polarize the macrophages into M2 and participate in pro-tumor activities. Amphiregulin secreted by ILC2 controls the local inflammation through Tregs but also induces immune suppression and promotes the EGFR + tumor growth (Zaiss et al., 2015). However, there are also anti-tumor effects reported. IL-33 activates ILC2 and secrete CXCL1 and CXCL2, as well as the CXCR2 on the tumor surface. This engagement limits tumor growth (Kim et al., 2016).
Group three ILCs contain ILC3 and LTi cells. Single-cell analysis reveals the developmental divergency of ILC3 and LTi cells (Ishizuka et al., 2016). ILC3 is the innate counterpart of Th17, representing both anti- and pro-tumor effects. It has been reported in a human single-cell colorectal cancer dataset that this cancer type won’t affect the heterogeneity of ILC3 subtypes (Qi et al., 2021). According to the diffusion map analysis on single-cell transcriptome data, ILC3 can transdifferentiate into ILCreg under TGF-β signaling stimulation at the late stage of colorectal cancer, which promotes tumor growth (Wang et al., 2020). The secretion of IL-22 by ILC3 also promotes tumorigenesis (Ghaedi and Ohashi, 2020). In contrast, ILC3 restricts the tumor-associated T cell functions with NKp46 expression (Crome et al., 2017). In a mice melanoma model, ILC3 expressing NKp46 prevents tumor growth by helping immune cells infiltrate into tumor sites (Eisenring et al., 2010). For LTi cells, their recruitment to the tumor site is related to CCL21 expression by tumor cells, indicating immune tolerance and invasion (Shields et al., 2010).
As mentioned in the previous section, the emergence of single-cell technologies allows precise definitions of innate immune cells during the development and within TME at an overall transcriptome level rather than a single molecule. Furthermore, CITE-seq technology provides a simultaneous evaluation of the transcriptome and the surface proteome of single cells, acting as a high-throughput FACS analysis combined with general single-cell RNA-sequencing. In Table 2, the common innate immune cell transcriptome and cell surface markers allow a better understanding of relative sub-groups and their functions in TME. For example, one scRNA-seq and CITE-seq dataset containing paired tumor and normal lung tissues from 35 NSCLC patients has depicted the immune response of early-stage lung cancer (Leader et al., 2021). Notably, CITE-seq data with Antibody-Derived Tags (ADTs) can offer supplementary information for cellular classification as high-dimensional models. These integrated immune phenotypes defined by scRNA-seq and CITE-seq further determine an immune activation signature that is related to immune checkpoint blockade rather than overall immune content alterations.
The advance of single-cell technology came to the stage where we could predict the prognosis of cancer patients through the precise definition of innate immune cells in the tumor microenvironment. The conventional definition of innate immune cells depends on a limited number of markers including surface proteins and cytokines, mainly with flow cytometry. With the increase of subtypes of innate immune cells, increasing overlap of markers between different cell populations makes definition challenging. For instance, IL-17 production is one of the key features of ILC3. However, IL-17 is expressed in both ILC3 and LTi cells, requires additional markers to distinguish them (Borger et al., 2019). Heterogeneity adds up more complexity. A subtype of ILC3 labeled with NCR produces IFN-γ rather than IL-17 (Pantazi and Powell, 2019). Compelling evidence demonstrates that the use of scRNA-seq broadens available markers to define innate immune cells more precisely than flow cytometry.
The use of scRNA-seq discovered novel functions of innate immune cells. For instance, Trem2 + macrophages associate with adipocytes and regulate lipid metabolism, indicating the role of macrophages as metabolic sensors as well as potential therapeutic targets in metabolic diseases (Jaitin et al., 2019). Single-cell technology can also facilitate our understanding of the intracellular gene network and crosstalk among various cellular components in the complicated tumor immune landscape. As mentioned above, CCL5 secreted by NK cells recruits cDC1 and assists the anti-tumor function. Expression of CCL5 in NK cells also correlates with better overall survival in melanoma patients (Mgrditchian et al., 2017). However, CCL5 produced by monocytes recruit immunosuppressive Tregs, which can induce themselves into activated TAMs and progress the immunosuppression. These complicated molecular and cellular networks are hard to evaluate by conventional technologies but can be depicted by higher resolution sequencing methods like scRNA-seq and scATAC-seq.
The precise definition of innate immune cells matters in the understanding of the tumor microenvironment. TAMs have been classically defined into M1 (pro-inflammatory) and M2 (anti-inflammatory) based on the expression of CD80, CD206. However, recent data from scRNA-seq of renal cell carcinoma demonstrated TAMs could be separated into more than 5 distinct populations based on the expression of immune checkpoint genes. TAMS of patients who responded to immune checkpoint blockades exhibited more pro-inflammatory phenotypes as well as an increase of co-inhibitory genes such as VSIR, VSIG4, PD-L2, and SIGLEC10 compared with non-responders (Bi et al., 2021). The observation indicates the potential prognostic value of scRNA-seq data in cancer patients. ILCs have been classified based on flow cytometry and the expression of key transcription factors (Spits et al., 2013). Although the definition has been standard in the field of immunology, increasing evidence suggests that ILCs could adapt their fate based on the tumor microenvironment. In the lung cancer enriched with IL-23, ILC1s were converted to ILC3s. The shift from tumor-killing ILC1s to tumor-promoting ILC3s was reflected in the poor prognosis of the patients’ cohort with the high level of IL-23 expression and the increase of tumor-promoting ILC3s in the tumor (Koh et al., 2019). A recent study with scRNA-seq of metastatic pancreatic cancer cells in the liver revealed that pancreatic cancer cells adopt the intermediate transcriptional state between the pancreas and liver. Tumor microenvironment signals such as TGF-β drove the transcriptional heterogeneity and drug response. In vitro organoid models that lack immune cells could not recapitulate the transcriptional heterogeneity observed in vivo (Raghavan et al., 2021). Not only the prognosis prediction value but also this study indicates the significance of immune cells in the tumor microenvironment to shape the phenotype of cancer cells. Indeed, TGF-β, causative to the transcriptional heterogeneity in this study, is one of the major immune-suppressive cytokines that potentially explains the organoid models lacking immune cells did not recapitulate the change (Pickup et al., 2013). With these observations, we anticipate that single-cell technology will enable the precision medicine of cancer patients with the precise definition of innate immune cells in the tumor microenvironment.
At the same time, an adequate single-cell experimental design is required to obtain in-depth immune characterization and exclude confounding factors from phenotypic correlations. The choice of technology should be based on the biomedical hypothesis. For example, the combination of single-cell transcriptomics and proteomics provides a better profile of the molecular targets and cell types. The single-cell and spatial transcriptomics aim to find novel genome-wide discoveries at a spatial level. In this way, the pathologists can play a critical role in region selection to cover higher tumor heterogeneity. Large cohort single-cell datasets can be useful for pan-cancer integrative analysis and biomarker discovery through various machine learning models to predict the response to immunotherapy and survival rate. In all, full efforts from experimental, statistical, and computational scientists are required for better study design and data mining, thus realizing the ultimate goal of translational medicine for cancer therapy.
Single-cell RNA transcriptome technology has depicted the precise gene expression landscape of TME. Various computational tools such as ligand-receptor mapping and cytokine signaling activity analysis have contributed to the understanding of innate immune cells and their functions in TME (Browaeys et al., 2020; Jiang et al., 2021). Based on these findings, we propose two main directions for immunotherapy strategies for innate immune cells.
The first is the blockade of immune checkpoints on immune cells or tumor cells. Immune checkpoints are initially for self-tolerance to avoid self-attack. However, this has been exploited by some tumor cells to evade immune response (Pardoll, 2012). As innate immune cells exhibit a quicker response to tumors than the adaptive immune cells, they are thought to have a better response to immunotherapy. At the same time, immune cells of myeloid lineages are more infiltrated into the tumors than lymphoid cells. Some pre-clinical and clinical studies have employed this checkpoint blockade concept on myeloid cells for cancer immunotherapy, including the CD47-SIRPα signaling axis, one of the hottest myeloid checkpoints in the past decade. Studies have pointed out the broad expression of CD47 on many solid tumor cell surfaces, including glioblastoma, breast, liver, colon, bladder, ovarian, and prostate cancers (Willingham et al., 2012). Anti-CD47 antibody blockade exhibits inhibitory effects on tumor growth and tumor metastasis in murine ovarian cancer model (Willingham et al., 2012). In phase I and II clinical trials, anti-CD47 antibody or SIRPαFc treatment has shown safety, tolerability, and efficacy in advanced colorectal cancer, non-Hodgkin’s lymphoma for cancer immunotherapy (Feng et al., 2019). In recent years, more and more myeloid immune checkpoints have been discovered, including CD24-SIGLEC10, MHC class I-LILRB1, and CD73−CD39 (Barkal et al., 2018; Barkal et al., 2019; Goswami et al., 2020). These promising checkpoints have paved a bright way for cancer immunotherapy.
Other attempts can be the agonists of innate immune responses. Our immune system has many common pathways in response to infection and tumors, including TLR, RIG-I-like receptors (RLRs), and cGAS-STING (Li et al., 2021b). Synthetic molecules designed to mimic these pathways provoke innate immune reactions at the tumor sites independent from tumor antigens. For example, TLR agonists can initiate both innate and adaptive immune systems and exhibit anti-tumor effects in vivo mouse models (Davis et al., 2011). Another reported small-molecule agonist targeting TLR-2 can suppress leukemia progression through cytotoxic T cell activation (Cen et al., 2019). Similarly, STING agonists have also been shown to have anti-tumor effects in the mouse melanoma model (Demaria et al., 2015). This anti-tumor immunity was further elevated through PD1 and CTLA4 blockade. Many clinical trials for agonist monotherapy and combined treatment with immune checkpoint inhibitors have been conducted in solid tumors and lymphoma (Jiang et al., 2020). However, the more in-depth research is required as the innate immune system can be a double-edged sword to the downstream initiation of adaptive immunity. For example, STING signaling can activate the innate myeloid cells, but can also cause T cell stress and Treg infiltration through indoleamine 2,3-dioxygenase (Liang et al., 2015). Thus, further studies are required for the investigation of the whole cGAS-STING cascade for future safer immunotherapy.
Innate immune cells are the first barrel of our immunology. The innate immune cells directly attack the tumor cells, while acting as a messenger to bridge the innate immune and adaptive immune system together for anti-or pro-tumor effects. In this review, we summarize key cell types in the innate immune systems and their functions in TME. We referred to the recently published single-cell omics dataset to present a precise functional conclusion at a single-cell level. We provide current and prospective biomarkers for predicting patient survival or treatment effect on immunotherapy. We also conclude the promising immunotherapy targets for further studies. As TME is a complicated structure comprising various environmental properties and cellular communication, an overall understanding of TME offers us insights into new immunotherapy investigation.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article in Table 1.
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Alshetaiwi, H., Pervolarakis, N., McIntyre, L. L., Ma, D., Nguyen, Q., Rath, J. A., et al. (2020). Defining the Emergence of Myeloid-Derived Suppressor Cells in Breast Cancer Using Single-Cell Transcriptomics. Sci. Immunol. 5 (44), eaay6017. doi:10.1126/sciimmunol.aay6017
Artis, D., and Spits, H. (2015). The Biology of Innate Lymphoid Cells. Nature 517 (7534), 293–301. doi:10.1038/nature14189
Aspord, C., Leccia, M.-T., Charles, J., and Plumas, J. (2013). Plasmacytoid Dendritic Cells Support Melanoma Progression by Promoting Th2 and Regulatory Immunity through OX40L and ICOSL. Cancer Immunol. Res. 1 (6), 402–415. doi:10.1158/2326-6066.cir-13-0114-t
Barkal, A. A., Brewer, R. E., Markovic, M., Kowarsky, M., Barkal, S. A., Zaro, B. W., et al. (2019). CD24 Signalling through Macrophage Siglec-10 Is a Target for Cancer Immunotherapy. Nature 572 (7769), 392–396. doi:10.1038/s41586-019-1456-0
Barkal, A. A., Weiskopf, K., Kao, K. S., Gordon, S. R., Rosental, B., Yiu, Y. Y., et al. (2018). Engagement of MHC Class I by the Inhibitory Receptor LILRB1 Suppresses Macrophages and Is a Target of Cancer Immunotherapy. Nat. Immunol. 19 (1), 76–84. doi:10.1038/s41590-017-0004-z
Ben-Shmuel, A., Biber, G., and Barda-Saad, M. (2020). Unleashing Natural Killer Cells in the Tumor Microenvironment-The Next Generation of Immunotherapy? Front. Immunol. 11 (275), 275. doi:10.3389/fimmu.2020.00275
Bi, K., He, M. X., Bakouny, Z., Kanodia, A., Napolitano, S., Wu, J., et al. (2021). Tumor and Immune Reprogramming during Immunotherapy in Advanced Renal Cell Carcinoma. Cancer Cell 39 (5), 649–661. doi:10.1016/j.ccell.2021.02.015
Bie, Q., Zhang, P., Su, Z., Zheng, D., Ying, X., Wu, Y., et al. (2014). Polarization of ILC2s in Peripheral Blood Might Contribute to Immunosuppressive Microenvironment in Patients with Gastric Cancer. J. Immunol. Res. 2014, 923135. doi:10.1155/2014/923135
Biswas, S. K., Gangi, L., Paul, S., Schioppa, T., Saccani, A., Sironi, M., et al. (2006). A Distinct and Unique Transcriptional Program Expressed by Tumor-Associated Macrophages (Defective NF- B and Enhanced IRF-3/STAT1 Activation). Blood 107 (5), 2112–2122. doi:10.1182/blood-2005-01-0428
Borger, J. G., Lau, M., and Hibbs, M. L. (2019). The Influence of Innate Lymphoid Cells and Unconventional T Cells in Chronic Inflammatory Lung Disease. Front. Immunol. 10, 1597. doi:10.3389/fimmu.2019.01597
Böttcher, J. P., Bonavita, E., Chakravarty, P., Blees, H., Cabeza-Cabrerizo, M., Sammicheli, S., et al. (2018). NK Cells Stimulate Recruitment of cDC1 into the Tumor Microenvironment Promoting Cancer Immune Control. Cell 172 (5), 1022. doi:10.1016/j.cell.2018.01.004
Browaeys, R., Saelens, W., and Saeys, Y. (2020). NicheNet: Modeling Intercellular Communication by Linking Ligands to Target Genes. Nat. Methods 17 (2), 159–162. doi:10.1038/s41592-019-0667-5
Bruchard, M., and Ghiringhelli, F. (2019). Deciphering the Roles of Innate Lymphoid Cells in Cancer. Front. Immunol. 10, 656. doi:10.3389/fimmu.2019.00656
Byrne, J. C., Ní Gabhann, J., Stacey, K. B., Coffey, B. M., McCarthy, E., Thomas, W., et al. (2013). Bruton's Tyrosine Kinase Is Required for Apoptotic Cell Uptake via Regulating the Phosphorylation and Localization of Calreticulin. J.Immunol 190 (10), 5207–5215. doi:10.4049/jimmunol.1300057
Cao, J., Spielmann, M., Qiu, X., Huang, X., Ibrahim, D. M., Hill, A. J., et al. (2019). The Single-Cell Transcriptional Landscape of Mammalian Organogenesis. Nature 566 (7745), 496–502. doi:10.1038/s41586-019-0969-x
Cen, X., Zhu, G., Yang, J., Yang, J., Guo, J., Jin, J., et al. (2019). TLR1/2 Specific Small‐Molecule Agonist Suppresses Leukemia Cancer Cell Growth by Stimulating Cytotoxic T Lymphocytes. Adv. Sci. 6 (10), 1802042. doi:10.1002/advs.201802042
Chávez-Galán, L., Olleros, M. L., Vesin, D., and Garcia, I. (2015). Much More Than M1 and M2 Macrophages, There Are Also CD169(+) and TCR(+) Macrophages. Front. Immunol. 6, 263. doi:10.3389/fimmu.2015.00263
Chen, Y.-P., Yin, J.-H., Li, W.-F., Li, H.-J., Chen, D.-P., Zhang, C.-J., et al. (2020). Single-cell Transcriptomics Reveals Regulators Underlying Immune Cell Diversity and Immune Subtypes Associated with Prognosis in Nasopharyngeal Carcinoma. Cell Res 30 (11), 1024–1042. doi:10.1038/s41422-020-0374-x
Chen, Z., Zhou, L., Liu, L., Hou, Y., Xiong, M., Yang, Y., et al. (2020). Single-cell RNA Sequencing Highlights the Role of Inflammatory Cancer-Associated Fibroblasts in Bladder Urothelial Carcinoma. Nat. Commun. 11 (1), 5077. doi:10.1038/s41467-020-18916-5
Cheng, S., Li, Z., Gao, R., Xing, B., Gao, Y., Yang, Y., et al. (2021). A Pan-Cancer Single-Cell Transcriptional Atlas of Tumor Infiltrating Myeloid Cells. Cell 184 (3), 792–809. doi:10.1016/j.cell.2021.01.010
Cooper, M. A., Fehniger, T. A., and Caligiuri, M. A. (2001). The Biology of Human Natural Killer-Cell Subsets. Trends Immunol. 22 (11), 633–640. doi:10.1016/s1471-4906(01)02060-9
Crinier, A., Dumas, P.-Y., Escalière, B., Piperoglou, C., Gil, L., Villacreces, A., et al. (2021). Single-cell Profiling Reveals the Trajectories of Natural Killer Cell Differentiation in Bone Marrow and a Stress Signature Induced by Acute Myeloid Leukemia. Cell Mol Immunol 18 (5), 1290–1304. doi:10.1038/s41423-020-00574-8
Crome, S. Q., Nguyen, L. T., Lopez-Verges, S., Yang, S. Y. C., Martin, B., Yam, J. Y., et al. (2017). A Distinct Innate Lymphoid Cell Population Regulates Tumor-Associated T Cells. Nat. Med. 23 (3), 368–375. doi:10.1038/nm.4278
Davies, L. C., Jenkins, S. J., Allen, J. E., and Taylor, P. R. (2013). Tissue-resident Macrophages. Nat. Immunol. 14 (10), 986–995. doi:10.1038/ni.2705
Davis, M. B., Vasquez-Dunddel, D., Fu, J., Albesiano, E., Pardoll, D., and Kim, Y. J. (2011). Intratumoral Administration of TLR4 Agonist Absorbed into a Cellular Vector Improves Antitumor Responses. Clin. Cancer Res. 17 (12), 3984–3992. doi:10.1158/1078-0432.ccr-10-3262
de Andrade, L. F., Lu, Y., Luoma, A., Ito, Y., Pan, D., Pyrdol, J. W., et al. (2019). Discovery of Specialized NK Cell Populations Infiltrating Human Melanoma Metastases. JCI Insight 4 (23), e133103. doi:10.1172/jci.insight.133103
Demaria, O., De Gassart, A., Coso, S., Gestermann, N., Di Domizio, J., Flatz, L., et al. (2015). STING Activation of Tumor Endothelial Cells Initiates Spontaneous and Therapeutic Antitumor Immunity. Proc. Natl. Acad. Sci. U.S.A. 112 (50), 15408–15413. doi:10.1073/pnas.1512832112
Diao, J., Gu, H., Tang, M., Zhao, J., and Cattral, M. S. (2018). Tumor Dendritic Cells (DCs) Derived from Precursors of Conventional DCs Are Dispensable for Intratumor CTL Responses. J.Immunol 201 (4), 1306–1314. doi:10.4049/jimmunol.1701514
Duan, Z., and Luo, Y. (2021). Targeting Macrophages in Cancer Immunotherapy. Sig Transduct Target. Ther. 6 (1), 127. doi:10.1038/s41392-021-00506-6
Dutertre, C.-A., Becht, E., Irac, S. E., Khalilnezhad, A., Narang, V., Khalilnezhad, S., et al. (2019). Single-Cell Analysis of Human Mononuclear Phagocytes Reveals Subset-Defining Markers and Identifies Circulating Inflammatory Dendritic Cells. Immunity 51 (3), 573–589. doi:10.1016/j.immuni.2019.08.008
Eisenring, M., vom Berg, J., Kristiansen, G., Saller, E., and Becher, B. (2010). IL-12 Initiates Tumor Rejection via Lymphoid Tissue-Inducer Cells Bearing the Natural Cytotoxicity Receptor NKp46. Nat. Immunol. 11 (11), 1030–1038. doi:10.1038/ni.1947
Fang, X., and Ho, J. W. K. (2021). FlowGrid Enables Fast Clustering of Very Large Single-Cell RNA-Seq Data. Bioinformatics [Epub ahead of print]. doi:10.1093/bioinformatics/btab521
Feng, M., Jiang, W., Kim, B. Y. S., Zhang, C. C., Fu, Y.-X., and Weissman, I. L. (2019). Phagocytosis Checkpoints as New Targets for Cancer Immunotherapy. Nat. Rev. Cancer 19 (10), 568–586. doi:10.1038/s41568-019-0183-z
Fu, X.-T., Dai, Z., Song, K., Zhang, Z.-J., Zhou, Z.-J., Zhou, S.-L., et al. (2015). Macrophage-secreted IL-8 Induces Epithelial-Mesenchymal Transition in Hepatocellular Carcinoma Cells by Activating the JAK2/STAT3/Snail Pathway. Int. J. Oncol. 46 (2), 587–596. doi:10.3892/ijo.2014.2761
Gao, Y., Souza-Fonseca-Guimaraes, F., Bald, T., Ng, S. S., Young, A., Ngiow, S. F., et al. (2017). Tumor Immunoevasion by the Conversion of Effector NK Cells into Type 1 Innate Lymphoid Cells. Nat. Immunol. 18 (9), 1004–1015. doi:10.1038/ni.3800
Gebhardt, C., Sevko, A., Jiang, H., Lichtenberger, R., Reith, M., Tarnanidis, K., et al. (2015). Myeloid Cells and Related Chronic Inflammatory Factors as Novel Predictive Markers in Melanoma Treatment with Ipilimumab. Clin. Cancer Res. 21 (24), 5453–5459. doi:10.1158/1078-0432.ccr-15-0676
Ghaedi, M., Shen, Z. Y., Orangi, M., Martinez-Gonzalez, I., Wei, L., Lu, X., et al. (2020). Single-cell Analysis of RORα Tracer Mouse Lung Reveals ILC Progenitors and Effector ILC2 Subsets. J. Exp. Med. 217 (3), jem.20182293. doi:10.1084/jem.20182293
Ghaedi, M., and Ohashi, P. S. (2020). ILC Transdifferentiation: Roles in Cancer Progression. Cel Res 30 (7), 562–563. doi:10.1038/s41422-020-0326-5
Golomb, S. M., Guldner, I. H., Zhao, A., Wang, Q., Palakurthi, B., Aleksandrovic, E. A., et al. (2020). Multi-modal Single-Cell Analysis Reveals Brain Immune Landscape Plasticity during Aging and Gut Microbiota Dysbiosis. Cel Rep. 33 (9), 108438. doi:10.1016/j.celrep.2020.108438
Gong, L., Kwong, D. L.-W., Dai, W., Wu, P., Li, S., Yan, Q., et al. (2021). Comprehensive Single-Cell Sequencing Reveals the Stromal Dynamics and Tumor-specific Characteristics in the Microenvironment of Nasopharyngeal Carcinoma. Nat. Commun. 12 (1), 1540. doi:10.1038/s41467-021-21795-z
Goswami, S., Walle, T., Cornish, A. E., Basu, S., Anandhan, S., Fernandez, I., et al. (2020). Immune Profiling of Human Tumors Identifies CD73 as a Combinatorial Target in Glioblastoma. Nat. Med. 26 (1), 39–46. doi:10.1038/s41591-019-0694-x
Gunaydin, G. (2021). CAFs Interacting with TAMs in Tumor Microenvironment to Enhance Tumorigenesis and Immune Evasion. Front. Oncol. 11, 668349. doi:10.3389/fonc.2021.668349
Hamm, A., Prenen, H., Van Delm, W., Di Matteo, M., Wenes, M., Delamarre, E., et al. (2016). Tumour-educated Circulating Monocytes Are Powerful Candidate Biomarkers for Diagnosis and Disease Follow-Up of Colorectal Cancer. Gut 65 (6), 990–1000. doi:10.1136/gutjnl-2014-308988
Hanna, R. N., Cekic, C., Sag, D., Tacke, R., Thomas, G. D., Nowyhed, H., et al. (2015). Patrolling Monocytes Control Tumor Metastasis to the Lung. Science 350 (6263), 985–990. doi:10.1126/science.aac9407
Hao, Y., Hao, S., Andersen-Nissen, E., Mauck, W. M., Zheng, S., Butler, A., et al. (2021). Integrated Analysis of Multimodal Single-Cell Data. Cell 184 (13), 3573–3587. doi:10.1016/j.cell.2021.04.048
He, D., Wang, D., Lu, P., Yang, N., Xue, Z., Zhu, X., et al. (2021). Single-cell RNA Sequencing Reveals Heterogeneous Tumor and Immune Cell Populations in Early-Stage Lung Adenocarcinomas Harboring EGFR Mutations. Oncogene 40 (2), 355–368. doi:10.1038/s41388-020-01528-0
Huang, X., and Huang, Y. (2021). Cellsnp-lite: an Efficient Tool for Genotyping Single Cells. Bioinformatics. doi:10.1093/bioinformatics/btab358
Huntington, N. D., Vosshenrich, C. A. J., and Di Santo, J. P. (2007). Developmental Pathways that Generate Natural-Killer-Cell Diversity in Mice and Humans. Nat. Rev. Immunol. 7 (9), 703–714. doi:10.1038/nri2154
Ishizuka, I. E., Chea, S., Gudjonson, H., Constantinides, M. G., Dinner, A. R., Bendelac, A., et al. (2016). Single-cell Analysis Defines the Divergence between the Innate Lymphoid Cell Lineage and Lymphoid Tissue-Inducer Cell Lineage. Nat. Immunol. 17 (3), 269–276. doi:10.1038/ni.3344
Jaitin, D. A., Adlung, L., Thaiss, C. A., Weiner, A., Li, B., Descamps, H., et al. (2019). Lipid-Associated Macrophages Control Metabolic Homeostasis in a Trem2-dependent Manner. Cell 178 (3), 686–698. doi:10.1016/j.cell.2019.05.054
Jiang, M., Chen, P., Wang, L., Li, W., Chen, B., Liu, Y., et al. (2020). cGAS-STING, an Important Pathway in Cancer Immunotherapy. J. Hematol. Oncol. 13 (1), 81. doi:10.1186/s13045-020-00916-z
Jiang, P., Zhang, Y., Ru, B., Yang, Y., Vu, T., Paul, R., et al. (2021). Systematic Investigation of Cytokine Signaling Activity at the Tissue and Single-Cell Levels. Nat. Methods 18 (10), 1181–1191. doi:10.1038/s41592-021-01274-5
Jin, M.-Z., and Jin, W.-L. (2020). The Updated Landscape of Tumor Microenvironment and Drug Repurposing. Sig Transduct Target. Ther. 5 (1), 166. doi:10.1038/s41392-020-00280-x
Jin, S., Guerrero-Juarez, C. F., Zhang, L., Chang, I., Ramos, R., Kuan, C.-H., et al. (2021). Inference and Analysis of Cell-Cell Communication Using CellChat. Nat. Commun. 12 (1), 1088. doi:10.1038/s41467-021-21246-9
Jin, S., Li, R., Chen, M.-Y., Yu, C., Tang, L.-Q., Liu, Y.-M., et al. (2020). Single-cell Transcriptomic Analysis Defines the Interplay between Tumor Cells, Viral Infection, and the Microenvironment in Nasopharyngeal Carcinoma. Cel Res 30 (11), 950–965. doi:10.1038/s41422-020-00402-8
Kießler, M., Plesca, I., Sommer, U., Wehner, R., Wilczkowski, F., Müller, L., et al. (2021). Tumor-infiltrating Plasmacytoid Dendritic Cells Are Associated with Survival in Human colon Cancer. J. Immunother. Cancer 9 (3), e001813. doi:10.1136/jitc-2020-001813
Kim, J., Kim, W., Moon, U. J., Kim, H. J., Choi, H.-J., Sin, J.-I., et al. (2016). Intratumorally Establishing Type 2 Innate Lymphoid Cells Blocks Tumor Growth. J.I. 196 (5), 2410–2423. doi:10.4049/jimmunol.1501730
Kiss, M., Caro, A. A., Raes, G., and Laoui, D. (2020). Systemic Reprogramming of Monocytes in Cancer. Front. Oncol. 10, 1399. doi:10.3389/fonc.2020.01399
Koh, J., Kim, H. Y., Lee, Y., Park, I. K., Kang, C. H., Kim, Y. T., et al. (2019). IL23-Producing Human Lung Cancer Cells Promote Tumor Growth via Conversion of Innate Lymphoid Cell 1 (ILC1) into ILC3. Clin. Cancer Res. 25 (13), 4026–4037. doi:10.1158/1078-0432.ccr-18-3458
Kubo, H., Mensurado, S., Gonçalves-Sousa, N., Serre, K., and Silva-Santos, B. (2017). Primary Tumors Limit Metastasis Formation through Induction of IL15-Mediated Cross-Talk between Patrolling Monocytes and NK Cells. Cancer Immunol. Res. 5 (9), 812–820. doi:10.1158/2326-6066.cir-17-0082
Kuhn, S., Yang, J., and Ronchese, F. (2015). Monocyte-Derived Dendritic Cells Are Essential for CD8+ T Cell Activation and Antitumor Responses after Local Immunotherapy. Front. Immunol. 6, 584. doi:10.3389/fimmu.2015.00584
Kwok, A. W. C., Qiao, C., Huang, R., Sham, M-H., Ho, J. W. K., and Huang, Y. (2022). MQuad Enables Clonal Substructure Discovery Using Single Cell Mitochondrial Variants. Nat. Commun. 13, 1205. doi:10.1038/s41467-022-28845-0
Labidi-Galy, S. I., Treilleux, I., Goddard-Leon, S., Combes, J.-D., Blay, J.-Y., Ray-Coquard, I., et al. (2012). Plasmacytoid Dendritic Cells Infiltrating Ovarian Cancer Are Associated with Poor Prognosis. Oncoimmunology 1 (3), 380–382. doi:10.4161/onci.18801
Laoui, D., Keirsse, J., Morias, Y., Van Overmeire, E., Geeraerts, X., Elkrim, Y., et al. (2016). The Tumour Microenvironment Harbours Ontogenically Distinct Dendritic Cell Populations with Opposing Effects on Tumour Immunity. Nat. Commun. 7, 13720. doi:10.1038/ncomms13720
Lavin, Y., Kobayashi, S., Leader, A., Amir, E.-a. D., Elefant, N., Bigenwald, C., et al. (2017). Innate Immune Landscape in Early Lung Adenocarcinoma by Paired Single-Cell Analyses. Cell 169 (4), 750–765. doi:10.1016/j.cell.2017.04.014
Lawrence, T., and Natoli, G. (2011). Transcriptional Regulation of Macrophage Polarization: Enabling Diversity with Identity. Nat. Rev. Immunol. 11 (11), 750–761. doi:10.1038/nri3088
Leader, A. M., Grout, J. A., Maier, B. B., Nabet, B. Y., Park, M. D., Tabachnikova, A., et al. (2021). Single-cell Analysis of Human Non-small Cell Lung Cancer Lesions Refines Tumor Classification and Patient Stratification. Cancer Cell 39, 1594. doi:10.1016/j.ccell.2021.10.009
Li, L., Xiong, F., Wang, Y., Zhang, S., Gong, Z., Li, X., et al. (2021). What Are the Applications of Single-Cell RNA Sequencing in Cancer Research: a Systematic Review. J. Exp. Clin. Cancer Res. 40 (1), 163. doi:10.1186/s13046-021-01955-1
Li, X., Dai, H., Wang, H., and Han, W. (2021). Exploring Innate Immunity in Cancer Immunotherapy: Opportunities and Challenges. Cel Mol Immunol 18 (6), 1607–1609. doi:10.1038/s41423-021-00679-8
Li, X., Yao, W., Yuan, Y., Chen, P., Li, B., Li, J., et al. (2017). Targeting of Tumour-Infiltrating Macrophages via CCL2/CCR2 Signalling as a Therapeutic Strategy against Hepatocellular Carcinoma. Gut 66 (1), 157–167. doi:10.1136/gutjnl-2015-310514
Liang, D., Xiao-Feng, H., Guan-Jun, D., Er-Ling, H., Sheng, C., Ting-Ting, W., et al. (2015). Activated STING Enhances Tregs Infiltration in the HPV-Related Carcinogenesis of Tongue Squamous Cells via the C-jun/CCL22 Signal. Biochim. Biophys. Acta (Bba) - Mol. Basis Dis. 1852 (11), 2494–2503. doi:10.1016/j.bbadis.2015.08.011
Lim, S. B., Lim, C. T., and Lim, W.-T. (2019). Single-Cell Analysis of Circulating Tumor Cells: Why Heterogeneity Matters. Cancers 11 (10), 1595. doi:10.3390/cancers11101595
Liu, S., Galat, V., Galat4, Y., Lee, Y. K. A., Wainwright, D., and Wu, J. (2021). NK Cell-Based Cancer Immunotherapy: from Basic Biology to Clinical Development. J. Hematol. Oncol. 14 (1), 7. doi:10.1186/s13045-020-01014-w
Liu, Y., Zugazagoitia, J., Ahmed, F. S., Henick, B. S., Gettinger, S. N., Herbst, R. S., et al. (2020). Immune Cell PD-L1 Colocalizes with Macrophages and Is Associated with Outcome in PD-1 Pathway Blockade Therapy. Clin. Cancer Res. 26 (4), 970–977. doi:10.1158/1078-0432.ccr-19-1040
Long, W., Chen, J., Gao, C., Lin, Z., Xie, X., and Dai, H. (2021). Brief Review on the Roles of Neutrophils in Cancer Development. J. Leukoc. Biol. 109 (2), 407–413. doi:10.1002/jlb.4mr0820-011r
Maier, B., Leader, A. M., Chen, S. T., Tung, N., Chang, C., LeBerichel, J., et al. (2020). A Conserved Dendritic-Cell Regulatory Program Limits Antitumour Immunity. Nature 580 (7802), 257–262. doi:10.1038/s41586-020-2134-y
Masucci, M. T., Minopoli, M., and Carriero, M. V. (2019). Tumor Associated Neutrophils. Their Role in Tumorigenesis, Metastasis, Prognosis and Therapy. Front. Oncol. 9, 1146. doi:10.3389/fonc.2019.01146
Matta, B. M., Castellaneta, A., and Thomson, A. W. (2010). Tolerogenic Plasmacytoid DC. Eur. J. Immunol. 40 (10), 2667–2676. doi:10.1002/eji.201040839
Melaiu, O., Chierici, M., Lucarini, V., Jurman, G., Conti, L. A., De Vito, R., et al. (2020). Cellular and Gene Signatures of Tumor-Infiltrating Dendritic Cells and Natural-Killer Cells Predict Prognosis of Neuroblastoma. Nat. Commun. 11 (1), 5992. doi:10.1038/s41467-020-19781-y
Mgrditchian, T., Arakelian, T., Paggetti, J., Noman, M. Z., Viry, E., Moussay, E., et al. (2017). Targeting Autophagy Inhibits Melanoma Growth by Enhancing NK Cells Infiltration in a CCL5-dependent Manner. Proc. Natl. Acad. Sci. U S A. 114 (44), E9271. doi:10.1073/pnas.1703921114
Mills, C. D., Lenz, L. L., and Harris, R. A. (2016). A Breakthrough: Macrophage-Directed Cancer Immunotherapy. Cancer Res. 76 (3), 513–516. doi:10.1158/0008-5472.can-15-1737
Murphy, T. L., Grajales-Reyes, G. E., Wu, X., Tussiwand, R., Briseño, C. G., Iwata, A., et al. (2016). Transcriptional Control of Dendritic Cell Development. Annu. Rev. Immunol. 34, 93–119. doi:10.1146/annurev-immunol-032713-120204
Okla, K., Wertel, I., Wawruszak, A., Bobiński, M., and Kotarski, J. (2018). Blood-based Analyses of Cancer: Circulating Myeloid-Derived Suppressor Cells - Is a new era Coming? Crit. Rev. Clin. Lab. Sci. 55 (6), 376–407. doi:10.1080/10408363.2018.1477729
Oshi, M., Newman, S., Tokumaru, Y., Yan, L., Matsuyama, R., Kalinski, P., et al. (2020). Plasmacytoid Dendritic Cell (pDC) Infiltration Correlate with Tumor Infiltrating Lymphocytes, Cancer Immunity, and Better Survival in Triple Negative Breast Cancer (TNBC) More Strongly Than Conventional Dendritic Cell (cDC). Cancers (Basel) 12 (11), 3342. doi:10.3390/cancers12113342
Panda, S. K., and Colonna, M. (2019). Innate Lymphoid Cells in Mucosal Immunity. Front. Immunol. 10, 861. doi:10.3389/fimmu.2019.00861
Pantazi, E., and Powell, N. (2019). Group 3 ILCs: Peacekeepers or Troublemakers? What's Your Gut Telling You?!. Front. Immunol. 10, 676. doi:10.3389/fimmu.2019.00676
Pardoll, D. M. (2012). The Blockade of Immune Checkpoints in Cancer Immunotherapy. Nat. Rev. Cancer 12 (4), 252–264. doi:10.1038/nrc3239
Patel, H., Nilendu, P., Jahagirdar, D., Pal, J. K., and Sharma, N. K. (2018). Modulating Secreted Components of Tumor Microenvironment: A Masterstroke in Tumor Therapeutics. Cancer Biol. Ther. 19 (1), 3–12. doi:10.1080/15384047.2017.1394538
Peng, J., Sun, B.-F., Chen, C.-Y., Zhou, J.-Y., Chen, Y.-S., Chen, H., et al. (2019). Single-cell RNA-Seq Highlights Intra-tumoral Heterogeneity and Malignant Progression in Pancreatic Ductal Adenocarcinoma. Cel Res 29 (9), 725–738. doi:10.1038/s41422-019-0195-y
Pickup, M., Novitskiy, S., and Moses, H. L. (2013). The Roles of TGFβ in the Tumour Microenvironment. Nat. Rev. Cancer 13 (11), 788–799. doi:10.1038/nrc3603
Plitas, G., Konopacki, C., Wu, K., Bos, P. D., Morrow, M., Putintseva, E. V., et al. (2016). Regulatory T Cells Exhibit Distinct Features in Human Breast Cancer. Immunity 45 (5), 1122–1134. doi:10.1016/j.immuni.2016.10.032
Pommier, A., Audemard, A., Durand, A., Lengagne, R., Delpoux, A., Martin, B., et al. (2013). Inflammatory Monocytes Are Potent Antitumor Effectors Controlled by Regulatory CD4 + T Cells. Proc. Natl. Acad. Sci. U.S.A. 110 (32), 13085–13090. doi:10.1073/pnas.1300314110
Qi, J., Crinier, A., Escalière, B., Ye, Y., Wang, Z., Zhang, T., et al. (2021). Single-cell Transcriptomic Landscape Reveals Tumor Specific Innate Lymphoid Cells Associated with Colorectal Cancer Progression. Cel Rep. Med. 2 (8), 100353. doi:10.1016/j.xcrm.2021.100353
Qian, B.-Z., and Pollard, J. W. (2010). Macrophage Diversity Enhances Tumor Progression and Metastasis. Cell 141 (1), 39–51. doi:10.1016/j.cell.2010.03.014
Qiao, C., and Huang, Y. (2021). Representation Learning of RNA Velocity Reveals Robust Cell Transitions. PNAS 118 (49), e2105859118. doi:10.1073/pnas.2105859118
Raghavan, S., Winter, P. S., Navia, A. W., Williams, H. L., DenAdel, A., Lowder, K. E., et al. (2021). Microenvironment Drives Cell State, Plasticity, and Drug Response in Pancreatic Cancer. Cell 184 (25), 6119–6137. doi:10.1016/j.cell.2021.11.017
Roberts, E. W., Broz, M. L., Binnewies, M., Headley, M. B., Nelson, A. E., Wolf, D. M., et al. (2016). Critical Role for CD103+/CD141+ Dendritic Cells Bearing CCR7 for Tumor Antigen Trafficking and Priming of T Cell Immunity in Melanoma. Cancer Cell 30 (2), 324–336. doi:10.1016/j.ccell.2016.06.003
Roma, S., Carpen, L., Raveane, A., and Bertolini, F. (2021). The Dual Role of Innate Lymphoid and Natural Killer Cells in Cancer. From Phenotype to Single-Cell Transcriptomics, Functions and Clinical Uses. Cancers (Basel) 13 (20), 5042. doi:10.3390/cancers13205042
Salimi, M., Wang, R., Yao, X., Li, X., Wang, X., Hu, Y., et al. (2018). Activated Innate Lymphoid Cell Populations Accumulate in Human Tumour Tissues. BMC Cancer 18 (1), 341. doi:10.1186/s12885-018-4262-4
Salmon, H., Idoyaga, J., Rahman, A., Leboeuf, M., Remark, R., Jordan, S., et al. (2016). Expansion and Activation of CD103 + Dendritic Cell Progenitors at the Tumor Site Enhances Tumor Responses to Therapeutic PD-L1 and BRAF Inhibition. Immunity 44 (4), 924–938. doi:10.1016/j.immuni.2016.03.012
Sanford, D. E., Belt, B. A., Panni, R. Z., Mayer, A., Deshpande, A. D., Carpenter, D., et al. (2013). Inflammatory Monocyte Mobilization Decreases Patient Survival in Pancreatic Cancer: a Role for Targeting the CCL2/CCR2 axis. Clin. Cancer Res. 19 (13), 3404–3415. doi:10.1158/1078-0432.ccr-13-0525
Sathe, A., Grimes, S. M., Lau, B. T., Chen, J., Suarez, C., Huang, R. J., et al. (2020). Single-Cell Genomic Characterization Reveals the Cellular Reprogramming of the Gastric Tumor Microenvironment. Clin. Cancer Res. 26 (11), 2640–2653. doi:10.1158/1078-0432.ccr-19-3231
Schalper, K. A., Carleton, M., Zhou, M., Chen, T., Feng, Y., Huang, S.-P., et al. (2020). Elevated Serum Interleukin-8 Is Associated with Enhanced Intratumor Neutrophils and Reduced Clinical Benefit of Immune-Checkpoint Inhibitors. Nat. Med. 26 (5), 688–692. doi:10.1038/s41591-020-0856-x
Schmidt, L., Eskiocak, B., Kohn, R., Dang, C., Joshi, N. S., DuPage, M., et al. (2019). Enhanced Adaptive Immune Responses in Lung Adenocarcinoma through Natural Killer Cell Stimulation. Proc. Natl. Acad. Sci. U.S.A. 116 (35), 17460–17469. doi:10.1073/pnas.1904253116
Sharma, A., Seow, J. J. W., Dutertre, C.-A., Pai, R., Blériot, C., Mishra, A., et al. (2020). Onco-fetal Reprogramming of Endothelial Cells Drives Immunosuppressive Macrophages in Hepatocellular Carcinoma. Cell 183 (2), 377–394. doi:10.1016/j.cell.2020.08.040
Shaul, M. E., and Fridlender, Z. G. (2019). Tumour-associated Neutrophils in Patients with Cancer. Nat. Rev. Clin. Oncol. 16 (10), 601–620. doi:10.1038/s41571-019-0222-4
Shen, M., Hu, P., Donskov, F., Wang, G., Liu, Q., and Du, J. (2014). Tumor-associated Neutrophils as a New Prognostic Factor in Cancer: a Systematic Review and Meta-Analysis. PLoS One 9 (6), e98259. doi:10.1371/journal.pone.0098259
Shields, J. D., Kourtis, I. C., Tomei, A. A., Roberts, J. M., and Swartz, M. A. (2010). Induction of Lymphoidlike Stroma and Immune Escape by Tumors that Express the Chemokine CCL21. Science 328 (5979), 749–752. doi:10.1126/science.1185837
Simonetta, F., Pradier, A., and Roosnek, E. (2016). T-bet and Eomesodermin in NK Cell Development, Maturation, and Function. Front. Immunol. 7, 241. doi:10.3389/fimmu.2016.00241
Slyper, M., Porter, C. B. M., Ashenberg, O., Waldman, J., Drokhlyansky, E., Wakiro, I., et al. (2020). A Single-Cell and Single-Nucleus RNA-Seq Toolbox for Fresh and Frozen Human Tumors. Nat. Med. 26 (5), 792–802. doi:10.1038/s41591-020-0844-1
Spits, H., Artis, D., Colonna, M., Diefenbach, A., Di Santo, J. P., Eberl, G., et al. (2013). Innate Lymphoid Cells - a Proposal for Uniform Nomenclature. Nat. Rev. Immunol. 13 (2), 145–149. doi:10.1038/nri3365
Stoeckius, M., Hafemeister, C., Stephenson, W., Houck-Loomis, B., Chattopadhyay, P. K., Swerdlow, H., et al. (2017). Simultaneous Epitope and Transcriptome Measurement in Single Cells. Nat. Methods 14 (9), 865–868. doi:10.1038/nmeth.4380
Tang-Huau, T.-L., Gueguen, P., Goudot, C., Durand, M., Bohec, M., Baulande, S., et al. (2018). Human In Vivo-generated Monocyte-Derived Dendritic Cells and Macrophages Cross-Present Antigens through a Vacuolar Pathway. Nat. Commun. 9 (1), 2570. doi:10.1038/s41467-018-04985-0
Tian, S., Yan, L., Fu, L., Zhang, Z., Zhang, J., Meng, G., et al. (2021). A Comprehensive Investigation to Reveal the Relationship between Plasmacytoid Dendritic Cells and Breast Cancer by Multiomics Data Analysis. Front. Cel Develop. Biol. 9 (634), 640476. doi:10.3389/fcell.2021.640476
Veglia, F., Sanseviero, E., and Gabrilovich, D. I. (2021). Myeloid-derived Suppressor Cells in the Era of Increasing Myeloid Cell Diversity. Nat. Rev. Immunol. 21 (8), 485–498. doi:10.1038/s41577-020-00490-y
Villadangos, J. A., and Young, L. (2008). Antigen-presentation Properties of Plasmacytoid Dendritic Cells. Immunity 29 (3), 352–361. doi:10.1016/j.immuni.2008.09.002
Vivier, E., Artis, D., Colonna, M., Diefenbach, A., Di Santo, J. P., Eberl, G., et al. (2018). Innate Lymphoid Cells: 10 Years on. Cell 174 (5), 1054–1066. doi:10.1016/j.cell.2018.07.017
Wan, S., Zhao, E., Kryczek, I., Vatan, L., Sadovskaya, A., Ludema, G., et al. (2014). Tumor-associated Macrophages Produce Interleukin 6 and Signal via STAT3 to Promote Expansion of Human Hepatocellular Carcinoma Stem Cells. Gastroenterology 147 (6), 1393–1404. doi:10.1053/j.gastro.2014.08.039
Wang, L., He, T., Liu, J., Tai, J., Wang, B., Chen, Z., et al. (2021). Pan-cancer Analysis Reveals Tumor-Associated Macrophage Communication in the Tumor Microenvironment. Exp. Hematol. Oncol. 10 (1), 31. doi:10.1186/s40164-021-00226-1
Wang, S., Qu, Y., Xia, P., Chen, Y., Zhu, X., Zhang, J., et al. (2020). Transdifferentiation of Tumor Infiltrating Innate Lymphoid Cells during Progression of Colorectal Cancer. Cel Res 30 (7), 610–622. doi:10.1038/s41422-020-0312-y
Willingham, S. B., Volkmer, J. P., Gentles, A. J., Sahoo, D., Dalerba, P., Mitra, S. S., et al. (2012). The CD47-Signal Regulatory Protein Alpha (SIRPa) Interaction Is a Therapeutic Target for Human Solid Tumors. Proc. Natl. Acad. Sci. U S A. 109 (17), 6662–6667. doi:10.1073/pnas.1121623109
Wolf, F. A., Angerer, P., and Theis, F. J. (2018). SCANPY: Large-Scale Single-Cell Gene Expression Data Analysis. Genome Biol. 19 (1), 15. doi:10.1186/s13059-017-1382-0
Wu, Q., Li, B., Li, J., Sun, S., Yuan, J., and Sun, S. (2021). Cancer-associated Adipocytes as Immunomodulators in Cancer. Biomark Res. 9 (1), 2. doi:10.1186/s40364-020-00257-6
Wylie, B., Macri, C., Mintern, J., and Waithman, J. (2019). Dendritic Cells and Cancer: From Biology to Therapeutic Intervention. Cancers 11 (4), 521. doi:10.3390/cancers11040521
Xie, X., Shi, Q., Wu, P., Zhang, X., Kambara, H., Su, J., et al. (2020). Single-cell Transcriptome Profiling Reveals Neutrophil Heterogeneity in Homeostasis and Infection. Nat. Immunol. 21 (9), 1119–1133. doi:10.1038/s41590-020-0736-z
Xu, H., Lai, W., Zhang, Y., Liu, L., Luo, X., Zeng, Y., et al. (2014). Tumor-associated Macrophage-Derived IL-6 and IL-8 Enhance Invasive Activity of LoVo Cells Induced by PRL-3 in a KCNN4 Channel-dependent Manner. BMC Cancer 14, 330. doi:10.1186/1471-2407-14-330
Yu, Y., Tsang, J. C. H., Wang, C., Clare, S., Wang, J., Chen, X., et al. (2016). Single-cell RNA-Seq Identifies a PD-1hi ILC Progenitor and Defines its Development Pathway. Nature 539 (7627), 102–106. doi:10.1038/nature20105
Yuan, X., Rasul, F., Nashan, B., and Sun, C. (2021). Innate Lymphoid Cells and Cancer: Role in Tumor Progression and Inhibition. Eur. J. Immunol. 51 (9), 2188–2205. doi:10.1002/eji.202049033
Zaiss, D. M. W., Gause, W. C., Osborne, L. C., and Artis, D. (2015). Emerging Functions of Amphiregulin in Orchestrating Immunity, Inflammation, and Tissue Repair. Immunity 42 (2), 216–226. doi:10.1016/j.immuni.2015.01.020
Zhang, L., Li, Z., Skrzypczynska, K. M., Fang, Q., Zhang, W., O’Brien, S. A., et al. (2020). Single-Cell Analyses Inform Mechanisms of Myeloid-Targeted Therapies in Colon Cancer. Cell 181 (2), 442–459. doi:10.1016/j.cell.2020.03.048
Zhang, Q., He, Y., Luo, N., Patel, S. J., Han, Y., Gao, R., et al. (2019). Landscape and Dynamics of Single Immune Cells in Hepatocellular Carcinoma. Cell 179 (4), 829–845. doi:10.1016/j.cell.2019.10.003
Zheng, P., Luo, Q., Wang, W., Li, J., Wang, T., Wang, P., et al. (2018). Tumor-associated Macrophages-Derived Exosomes Promote the Migration of Gastric Cancer Cells by Transfer of Functional Apolipoprotein E. Cell Death Dis 9 (4), 434. doi:10.1038/s41419-018-0465-5
Zhou, Y., Yang, D., Yang, Q., Lv, X., Huang, W., Zhou, Z., et al. (2020). Single-cell RNA Landscape of Intratumoral Heterogeneity and Immunosuppressive Microenvironment in Advanced Osteosarcoma. Nat. Commun. 11 (1), 6322. doi:10.1038/s41467-020-20059-6
Keywords: ScRNA-seq, tumor microenvironment, innate immune cell, data mining, coding
Citation: Sugimura R and Chao Y (2022) Deciphering Innate Immune Cell-Tumor Microenvironment Crosstalk at a Single-Cell Level. Front. Cell Dev. Biol. 10:803947. doi: 10.3389/fcell.2022.803947
Received: 28 October 2021; Accepted: 12 April 2022;
Published: 13 May 2022.
Edited by:
Andrea Graziani, University of Turin, ItalyReviewed by:
Kamalakannan Rajasekaran, Genentech, Inc., United StatesCopyright © 2022 Sugimura and Chao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ryohichi Sugimura, cmlvc0Boa3UuaGs=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.