- Department of Biological Sciences, Murray State University, Murray, KY, United States
Membraneless granules assemble in different cell types and cellular loci and are the focus of intense research due to their fundamental importance for cellular organization. These dynamic organelles are commonly assembled from RNA and protein components and exhibit soft matter characteristics of molecular condensates currently characterized with biophysical approaches and super-resolution microscopy imaging. In addition, research on the molecular mechanisms of the RNA–protein granules assembly provided insights into the formation of abnormal granules and molecular aggregates, which takes place during many neurodegenerative disorders including Parkinson’s diseases (PD), Alzheimer’s disease (AD), amyotrophic lateral sclerosis (ALS), and frontotemporal dementia (FTD). While these disorders are associated with formation of abnormal granules, membraneless organelles are normally assembled in neurons and contribute to translational control and affect stability of neuronal RNAs. More recently, a new subtype of membraneless granules was identified in Drosophila glia (glial granules). Interestingly, glial granules were found to contain proteins which are the principal components of the membraneless granules in germ cells (germ granules), indicating some similarity in the functional assembly of these structures in glia and germline. This mini review highlights recent research on glial granules in the context of other membraneless organelles, including their assembly mechanisms and potential functions in the nervous system.
Introduction
Since description of characteristic membraneless granules in germ cells of over 80 animal species in eight phyla documented several decades ago (Eddy, 1975), it is now recognized that the assembly of large intracellular organelles, which lack the membrane, is one of the fundamental landmarks of cellular organization. In addition to formation of cell type-specific granules (such as germ granules in germ cells or neuronal granules in neurons), the membraneless structures are commonly assembled in nuclei and cytoplasm of most cells and include nucleoli, Cajal bodies, processing (P) bodies, and stress granules (Kiebler and Bassell, 2006; Anderson and Kedersha, 2008; Arkov and Ramos, 2010; Voronina et al., 2011; Gao and Arkov, 2013; Feric et al., 2016; Protter and Parker, 2016; Luo et al., 2018; Formicola et al., 2019; Marnik and Updike, 2019; Strom and Brangwynne, 2019; Trcek and Lehmann, 2019; Campos-Melo et al., 2021; Courchaine et al., 2021). In addition, multiple subcellular loci with crucial general functions such as silent chromatin (heterochromatin) and the RNA polymerase II-bound regions of transcriptionally active genes show properties of membraneless granules (Larson et al., 2017; Strom et al., 2017; Boehning et al., 2018; Lu et al., 2018). These soft matter properties are based on the granules’ assembly mechanisms which often can be described as protein/nucleic acid demixing (condensation) or phase separation from the rest of the cytosol (or nucleoplasm).
A recent study pointed to a number of common principles of the assembly of several membraneless ribonucleoprotein (RNP) granules. First, it was found that RNA components of the germ granules assemble in the large granule as homotypic clusters—large aggregates of the same RNA species (Little et al., 2015; Trcek et al., 2015; Trcek et al., 2020). In addition, some protein components of germ granules assemble into the granules as distinct clusters as well; however, the proteins show some overlap in the granules. Interestingly, the extent of the overlap for the same proteins and the distribution of the proteins within the granules can drastically change during development (Vo et al., 2019). Similar “docking” of two protein clusters, namely, coilin and survival of motor neuron (SMN) clusters, was recently described during the formation of Cajal body/gems granule in the nucleus (Courchaine et al., 2021). Also, a recent study provided a mechanistic view for the role of MEG-3 protein clusters, assembled at the interfaces of germ granules in Caenorhabditis elegans (P granules), in the regulation of granule condensates’ dynamics and stability (Folkmann et al., 2021). Interestingly, the function of these protein clusters is similar to the role of commonly used emulsion stabilizers (Pickering agents) adsorbed to interfaces of emulsion droplets.
Overall, for different membraneless granules, including germ granules, nucleoli, and Cajal bodies/gems, common structural features have emerged that show non-homogenous distribution of granule components within the granules, which can change during development and can be regulated by specific post-translational modifications of protein components. The functional significance and molecular mechanisms driving this non-random distribution of granule components is the subject of current research.
Many membraneless structures share landmark molecular characteristics, such as the presence of intrinsically disordered proteins (IDPs) or protein regions (IDPRs), which lack a defined folded structure, and low-complexity regions (LCRs), which may or may not have a distinct structure but show low diversity of their amino acids (Martin and Mittag, 2018). These protein regions are important components that can drive the assembly (condensation) of membraneless granules using their multivalent low-affinity associations with their interacting partners. Given the high occurrence of IDPs and IDPRs (about 51% of human proteins are either IDPs or contain IDPRs (Deiana et al., 2019)), it is surprising that different cell types use the same proteins to assemble membraneless granules. In particular, previous research works identified proteins that are found in both neuronal and germ granules, supporting the notion that neurons and germ cells utilize the same components to assemble the granules for specific similar functions in these cells (Kulkarni et al., 2020), and indicating that in addition to generally ubiquitous IDPRs, specifically structured protein domains may be selected to build the granules in different cells. Consistent with this, it was recently found that a structured Tudor (Tud) domain of SMN protein, and not its two IDPRs, is sufficient for formation of membraneless condensates (Courchaine et al., 2021).
In this mini review, the author highlights novel membraneless granules identified in glia of the adult Drosophila brain (referred to as “glial granules”) (Tindell et al., 2020), which were found to contain protein components of germ granules and discuss the potential significance of glial granules for brain functions in the context of other membraneless organelles.
Glia and Glial Granules
Glial Cells in Drosophila
Since one of the principal components of the Drosophila germ granules, Tud scaffold protein (Arkov et al., 2006; Gao et al., 2015; Vo et al., 2019), was found to be expressed not only in germline but also in the Drosophila head, distribution of Tud in the adult brain was recently examined in more detail (Tindell et al., 2020). These experiments led to the identification of Tud-containing large granules of about 0.5–2 μm in size in the cytoplasm of specific types of glial cells (but not in the neurons of the adult brain) (Tindell et al., 2020), Figure 1).
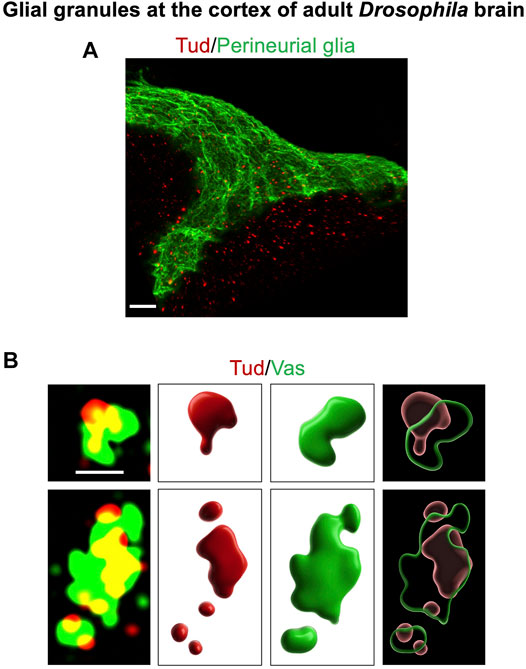
FIGURE 1. Glial granules in the Drosophila adult brain contain germline proteins, and they assemble from distinct protein clusters. This figure gives examples of glial granules recently published in Tindell et al. (2020). (A) A super-resolution microscopy image of a cortex region of the adult Drosophila brain shows multiple glial granules containing Tud protein (red channel) in surface glia (the outermost layer of surface glia, perineurial glia, is labeled with the membrane marker GFP-mCD8, green channel) and in the neighboring cortex glia. (B) Overlays of super-resolution optical sections of individual glial granules, which show Tud (red) and Vas (green) proteins (left panels), and corresponding 3D reconstructions of the individual protein clusters (middle panels) and composite granules (right panels) are shown. Scale bar in (A) is 7 μm. In (B), scale bar, shown in the first optical section (top left), is the same for the other optical section and is 1 μm.
In the adult Drosophila brain, there are several types of glial cells, including surface glia, composed of perineurial and subperineurial glia, cortex glia, astrocyte-like glia, and ensheathing glia (Freeman, 2015; Kremer et al., 2017; Bittern et al., 2021). Interestingly, Tud-containing glial granules were observed in perineurial, subperineurial, and cortex glia, which are located at the surface and cortex of the brain. Perineurial and subperineurial glia form a barrier at the brain surface, and while the perineurial glia layer is porous and permeable, subperineurial glial cells, located below perineurial glia, are tightly connected with septate junctions, which prevent paracellular diffusion of the molecules, and thereby, surface glia is analogous to the blood–brain barrier in the mammalian brain. While both perineurial and subperineurial glia together form a continuous layer covering the entire central and peripheral nervous systems (Kremer et al., 2017), these glial cells employ different developmental strategies to form the layer. In particular, during development and expansion of the nervous system, perineurial glial cells constantly divide to produce a large number of relatively small narrow and elongated cells (Awasaki et al., 2008; Avet-Rochex et al., 2012). In contrast, subperineurial glial cells keep their small numbers during development but greatly enlarge their size by endoreplication (Unhavaithaya and Orr-Weaver, 2012).
As directly connected with hemolymph, perineurial glia is involved in transportation of nutrients from the hemolymph into the nervous system through its membrane transporters. In addition, perineureal glia plays an important role in ethanol tolerance using its A-kinase anchoring protein (Akap200)-dependent structural remodeling of actin cytoskeleton (Parkhurst et al., 2018). Interestingly, perineurial glia have circadian molecular clock (Zhang et al., 2018), and it is involved in the regulation of circadian locomotion behavior (Kozlov et al., 2020).
Subperineurial glia has been implicated in male courtship (Hoxha et al., 2013) and sleep behavior (Artiushin et al., 2018). In addition, subperineurial glia’s efflux transporters are regulated by the circadian clock in perineurial glia, and they are responsible for pumping xenobiotics out into hemolymph most actively during daytime (Zhang et al., 2018).
Cortex glial cells encapsulate virtually all neuronal cell bodies in the fly brain (Figure 2) and are functionally equivalent to mammalian protoplasmic astrocytes (Pereanu et al., 2007; Awasaki et al., 2008; Freeman, 2015). Cortex glia are involved in several essential physiological and behavioral aspects such as metabolic neuronal support, control of neuronal excitability, sleep, locomotion, ethanol response, and removal of dead neurons using phagocytosis (Coutinho-Budd et al., 2017; Farca Luna et al., 2017; Delgado et al., 2018; Lee et al., 2019; Mclaughlin et al., 2019; Nakano et al., 2019; Weiss et al., 2019). In addition, defects in the membrane of the cortex glia cause light-induced seizures (photosensitive epilepsy) (Kunduri et al., 2018). Interestingly, a recent study demonstrated that fragile X mental retardation protein (FMRP), which is one of the crucial components of neuronal granules, has an additional role in glia-mediated phagocytosis that is responsible for neuronal clearance after injury and during neurodevelopment (O’Connor et al., 2017). Importantly, during development, neuronal FMRP signals to cortex and ensheathing glia to activate glial phagocytosis to carry out removal of transient neurons (Vita et al., 2021).
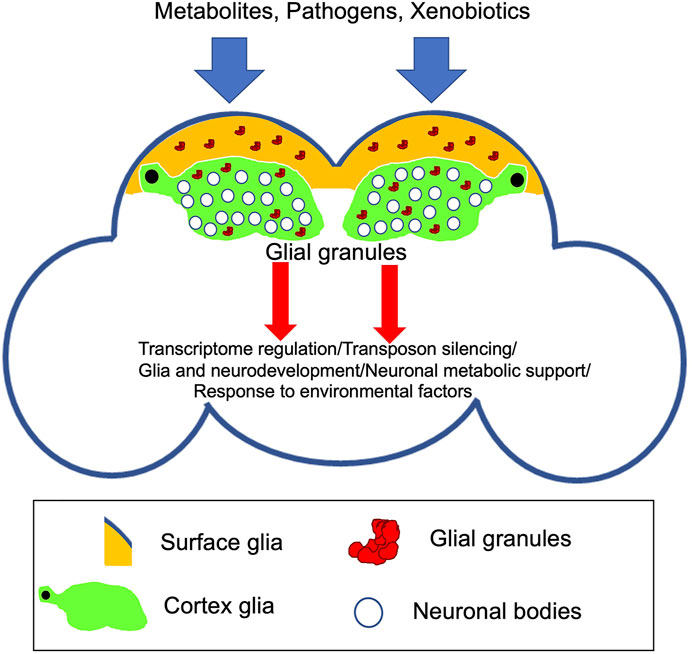
FIGURE 2. Schematic of the adult Drosophila brain indicating novel glial granules assembled in surface and cortex glia. Recent evidence points to the function of these granules in transcriptome regulation and silencing of transposable elements in the Drosophila brain (Tindell et al., 2020). Future research will test whether glial granules are involved in specific aspects of brain development and function including neuronal survival and metabolic support, behavior, and response to external/environmental factors.
Germline Components of Glial Granules
Tudor and Tudor Domains
Tud protein is one of the principal components of germ granules and plays a role of molecular scaffold due to its 11 protein–protein interaction modules (Tud domains) (Arkov et al., 2006; Liu et al., 2010; Gao et al., 2015; Zheng et al., 2016). The Tud domain is a 50–55 amino acid β-barrel structure, which forms an interaction cavity referred to as “aromatic cage” since it is lined with aromatic amino acids. Aromatic cage of the Tud domain interacts with methylated amino acids of Tud partner proteins such as methylated arginines and lysines. In germ cells, Tud is required for the assembly of germ granules at the posterior pole of the egg (polar granules), and it is essential for formation of primordial germ cells during early embryogenesis (Boswell and Mahowald, 1985; Thomson and Lasko, 2004; Arkov et al., 2006). Tud domain-containing proteins are molecular landmarks of germline in many animals and important contributors to the mechanisms responsible for silencing of transposable elements (Arkov and Ramos, 2010). Similarly, glial granule Tud protein is required for silencing of transposable elements in the Drosophila adult brain (Tindell et al., 2020).
Interestingly, neuronal granule FMRP contains two tandem domains of Tud family (Ramos et al., 2006; Myrick et al., 2015). In Drosophila germ cells, FMRP homolog was also found to associate with Piwi family proteins, Piwi and Aubergine (Aub), and contribute to silencing of transposable elements (Megosh et al., 2006; Bozzetti et al., 2015).
Piwi Family Proteins
Drosophila genome encodes three Piwi family proteins (Piwi, Ago3, and Aub), which belong to a distinct clade of a larger group of argonaute (Ago) proteins (Ku and Lin, 2014; Iwasaki et al., 2015; Arkov, 2018). Similar to other Argonautes, which associate with small guide RNAs, Piwi proteins associate with 24–31 nucleotides long Piwi-interacting RNAs (piRNAs). In germline, piRNAs guide Piwi proteins to the transposon RNA targets, and thereby Piwi proteins silence these targets by either direct endonucleolytic cleavage (Nishida et al., 2015) or by repressing transcription of transposon genes by the assembly of heterochromatin at transposon genomic loci (Le Thomas et al., 2013).
In different animals, Tud domain-containing proteins interact with symmetrically dimethylated arginines (sDMAs) of Piwi proteins in germ granules (Kirino et al., 2009; Vagin et al., 2009). In particular, in Drosophila germline, methylated Aub associates with Tud domain proteins Tud and Krimper (Nishida et al., 2009; Kirino et al., 2010; Huang et al., 2021; Vrettos et al., 2021). In glial granules, both Piwi and Ago3 were identified; however, Aub protein was not detected in these granules. In addition to Ago3 assembly in glial granules, Ago3 was also expressed in a distinct population of neurons (Tindell et al., 2020). Consistent with these data, previous study showed expression of Aub and Ago3 in neurons of mushroom body, which is crucial for olfactory memory, and provided genetic evidence that, similar to the role of Aub and Ago3 in germ cells, these proteins repress transposable elements in the adult brain (Perrat et al., 2013). Interestingly, Aub principal interacting partner in germ granules, Tud protein, is not detected in neurons of the adult brain, and whether Aub is assembled in neuronal granules in mushroom body and associates with other Tud domain-containing proteins awaits further investigation.
ATP-Dependent RNA Helicase Vasa
Vasa (Vas) is a DEAD-box RNA helicase, which is required for development and specification of germline in different organisms (Raz, 2000) (see Figure 1B, which shows Vas and Tud in glial granules (Tindell et al., 2020)). Similar to Tud domain and Piwi proteins, Vas is essential for silencing of transposable elements in germline and is one of the principal components of germ granules (Dehghani and Lasko, 2017). Generally, in the granules, Vas may use its RNA helicase activity to remodel RNA in ribonucleoprotein complexes and to function as an RNA chaperone and regulator of RNA stability and translation (Sengoku et al., 2006; Arkov and Ramos, 2010). In particular, it was shown that Vas helps to dissociate target RNA fragments from Aub homolog from Bombyx mori (silkworm) (Siwi) after Siwi’s cleavage of its target RNA, thereby increasing the efficiency of transposon silencing mechanisms (Nishida et al., 2015; Arkov, 2018).
Polar Granule Component
The polar granule component (pgc) encodes a 71 amino acid protein, and in germ cells it localizes to the nuclei where it represses elongation of transcription by preventing the action of positive transcription elongation factor (P-TEFb). P-TEFb is composed of cyclin-dependent kinase 9 (Cdk9) and cyclin T (CycT), and Pgc protein binds to Cdk9 subunit of the P-TEFb complex, which interferes with association of P-TEFb with transcriptionally active regions (Hanyu-Nakamura et al., 2008). Recently, Pgc was shown to play a role in repressing production of miRNA precursors in germ cells, thereby enabling expression of germline mRNAs, which are targeted by miRNAs, thus preventing germ cell death during embryogenesis (Hanyu-Nakamura et al., 2019). While all the reported functions of Pgc in germ cells are associated with its role in transcription, identification of Pgc protein in cytoplasmic glial granules may point to a novel role of this protein in glia. Alternatively, Pgc protein may be stored in glial granules and, upon a signaling event in glia, Pgc could be released from the granules and translocated to the nucleus for transcriptional regulation.
Functional Aspects of the Membraneless Organelles
Different membraneless granules have been implicated in a variety of functions, including regulation of translation and RNA storage/localization (P-bodies, stress granules, germ granules, and neuronal granules) (Arkov and Ramos, 2010; Voronina et al., 2011; Gao and Arkov, 2013; Luo et al., 2018; Formicola et al., 2019; Marnik and Updike, 2019; Trcek and Lehmann, 2019; Campos-Melo et al., 2021), compartmentalization of distinct steps of ribosome assembly in nucleoli (Feric et al., 2016), and bringing together different piRNA pathway components to enable efficient silencing of transposable elements in germ granules (Arkov, 2018).
In particular, in the nervous system, neuronal or transport membraneless granules are assembled to control translation of neuronal mRNAs. One of the principal components of neuronal granules, FMRP, is a well characterized RNA-binding protein and translational repressor (Tsang et al., 2019; Lai et al., 2020). Upon the assembly of the granules, translation of multiple neuronal mRNAs is repressed during transport of these mRNAs from neuronal cell body to the synaptic regions. When granules arrive in the synaptic region, they release or unmask these mRNAs for local translation. Interestingly, there is evidence that this mRNA unmasking and translational activation during neuronal stimulation is promoted by disassembly of the neuronal granules which is induced by dephosphorylation and methylation of FMRP, decreasing its property to form RNA–protein condensates by phase separation (Tsang et al., 2019).
Similar to neuronal granules, stress granules repress translation of their RNA components upon various stress conditions and can transfer the RNAs to P-bodies for degradation of these RNAs (Anderson and Kedersha, 2008). However, recently it was also shown that some RNAs in stress granules are translationally active (Mateju et al., 2020). Importantly, dysregulation of RNA–protein granules, in particular, stress granules, has been strongly implicated in development of several neurodegenerative diseases, such as Alzheimer’s, Huntington’s, ALS, and FTD, for which there is currently no cure (Ash et al., 2014; Mcaleese et al., 2017; St-Amour et al., 2018; Coudert et al., 2019; Ryan and Fawzi, 2019; Wolozin and Ivanov, 2019; Dudman and Qi, 2020). Furthermore, mutations in genes encoding stress granule RNA-binding proteins including TAR-DNA binding protein (TDP-43), fused in sarcoma/translated in liposarcoma (FUS/TLS), heterogeneous nuclear ribonucleoprotein A1 (hnRNPA1), and T cell-restricted intracellular antigen-1 (TIA-1) have been found in ALS/FTD patients pointing to direct link between abnormal regulation of stress granule formation in these neurodegenerative disorders (Benajiba et al., 2009; Kwiatkowski et al., 2009; Pesiridis et al., 2009; Gendron et al., 2013; Kim et al., 2013; Mackenzie et al., 2017). Importantly, many of these mutations in stress granule proteins are found in their LCRs, which result in the granule’s decreased dynamics and increased liquid–liquid phase separation, and ultimately, it leads to irreversibly insoluble pathological granules (Baradaran-Heravi et al., 2020).
While glial granules have been described very recently, it is possible that they are involved in post-transcriptional gene regulation in glia, including translational repression, RNA degradation, and recruitment of other RNAs for storage. It is intriguing that Tud-containing glial granules have been detected only in surface and cortex glia, which may indicate that the granules have roles important for the specific functions of these types of glia located at or close to the surface of the brain. For example, these functions may be related to the brain response to or protection from the environmental factors or pathogens present in hemolymph and metabolic support of neurons (Figure 2). In addition, to prevent a pathological cell transformation, co-expression of germline genes in somatic cells may need to be restricted to and tightly controlled in a limited group of cells when germ-like granules are needed for function since ectopic expression of several germline components of glial granules including Piwi and Vas has been observed in Drosophila larval brain tumors in lethal (3) malignant brain tumor (l(3)mbt) mutants (Janic et al., 2010). Importantly, ectopic expression of these genes contributes to tumor development in the l(3)mbt mutants indicating that the germline genes may act to drive tumorigenesis in somatic cells.
Transcriptomic data showed that glial granule component Tud is required for silencing of transposable elements in the brain (Tindell et al., 2020). Consistent with these data, Piwi proteins, Piwi, and Ago3, which are the principal factors of the transposon silencing mechanisms in the germ granules, are bona fide components of glial granules. In addition, non-transposon RNAs are likely to be regulated in glial granules also, which is consistent with the finding that tud mutant showed significant misregulation of multiple non-transposon RNAs in the Drosophila brain (Figure 2).
Future Perspectives
In conclusion, while it is intriguing that several proteins are shared by glial and germ granules, these granules are likely to include specific components needed for their functions in glia and germline, respectively. Therefore, future research will comprehensively analyze the glial granule composition and test functional roles of the granule components in the brain. In particular, future studies on glial granules should determine whether specific neuron/glia or glia/hemolymph signaling affects the assembly and potential roles of these novel granules in post-transcriptional gene regulation, response to environmental factors, and behavior (Figure 2). Furthermore, potential functional links between glial granules and other membraneless organelles, such as stress granules, should be explored. In addition, given the importance of glia in virtually every aspect of neuronal activity, it will be important to determine whether glial granules show age-related changes in dynamics and composition, which may be linked to neurodegeneration. Overall, future research works on glial granules may provide important insights into biogenesis and formation of pathological granules in the nervous system and suggest ways to reverse their assembly.
Author Contributions
AA conceived the idea and wrote the manuscript.
Funding
Work in the Arkov laboratory was supported by grants from the National Science Foundation (MCB-2130162) and NIH National Institute of General Medical Sciences (R01GM129153).
Conflict of Interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The author would like to thank Samuel J. Tindell for his help with the figures for the study.
References
Anderson, P., and Kedersha, N. (2008). Stress Granules: The Tao of RNA Triage. Trends Biochem. Sci. 33, 141–150. doi:10.1016/j.tibs.2007.12.003
Arkov, A. L., and Ramos, A. (2010). Building RNA-Protein Granules: Insight from the Germline. Trends Cell Biol. 20, 482–490. doi:10.1016/j.tcb.2010.05.004
Arkov, A. L. (2018). RNA Selection by PIWI Proteins. Trends Biochem. Sci. 43, 153–156. doi:10.1016/j.tibs.2017.12.007
Arkov, A. L., Wang, J.-Y. S., Ramos, A., and Lehmann, R. (2006). The Role of Tudor Domains in Germline Development and Polar Granule Architecture. Development 133, 4053–4062. doi:10.1242/dev.02572
Artiushin, G., Zhang, S. L., Tricoire, H., and Sehgal, A. (2018). Endocytosis at the Drosophila Blood-Brain Barrier as a Function for Sleep. Elife 7. doi:10.7554/eLife.43326
Ash, P. E. A., Vanderweyde, T. E., Youmans, K. L., Apicco, D. J., and Wolozin, B. (2014). Pathological Stress Granules in Alzheimer’s Disease. Brain Research 1584, 52–58. doi:10.1016/j.brainres.2014.05.052
Avet-Rochex, A., Kaul, A. K., Gatt, A. P., Mcneill, H., and Bateman, J. M. (2012). Concerted Control of Gliogenesis by InR/TOR and FGF Signalling in the Drosophila Post-Embryonic Brain. Development 139, 2763–2772. doi:10.1242/dev.074179
Awasaki, T., Lai, S.-L., Ito, K., and Lee, T. (2008). Organization and Postembryonic Development of Glial Cells in the Adult Central Brain of Drosophila. J. Neurosci. 28, 13742–13753. doi:10.1523/jneurosci.4844-08.2008
Baradaran-Heravi, Y., Van Broeckhoven, C., and Van Der Zee, J. (2020). Stress Granule Mediated Protein Aggregation and Underlying Gene Defects in the FTD-ALS Spectrum. Neurobiol. Dis. 134, 104639. doi:10.1016/j.nbd.2019.104639
Benajiba, L., Le Ber, I., Camuzat, A., Lacoste, M., Thomas-Anterion, C., Couratier, P., Legallic, S., Salachas, F., Hannequin, D., Decousus, M., Lacomblez, L., Guedj, E., Golfier, V., Camu, W., Dubois, B., Campion, D., Meininger, V., Brice, A., and French, C. (2009). TARDBP Mutations in Motoneuron Disease With Frontotemporal Lobar Degeneration TARDBP Mutations in Motoneuron Disease With Frontotemporal Lobar Degeneration. Ann Neurol. 65, 470–473. doi:10.1002/ana.21612
Bittern, J., Pogodalla, N., Ohm, H., Brüser, L., Kottmeier, R., Schirmeier, S., and Klämbt, C. (2021). Neuron-Glia Interaction in the Drosophila Nervous System. Develop. Neurobiol. 81, 438–452. doi:10.1002/dneu.22737
Boehning, M., Dugast-Darzacq, C., Rankovic, M., Hansen, A. S., Yu, T., Marie-Nelly, H., Mcswiggen, D. T., Kokic, G., Dailey, G. M., Cramer, P., Darzacq, X., and Zweckstetter, M. (2018). RNA Polymerase II Clustering Through Carboxy-Terminal Domain Phase Separation. Nat Struct Mol Biol 25, 833–840. doi:10.1038/s41594-018-0112-y
Boswell, R., and Mahowald, A. P. (1985). Tudor, a Gene Required for Assembly of the Germ Plasm in Drosophila melanogaster. Cell 43, 97–104. doi:10.1016/0092-8674(85)90015-7
Bozzetti, M. P., Specchia, V., Cattenoz, P. B., Laneve, P., Geusa, A., Sahin, H. B., Di Tommaso, S., Friscini, A., Massari, S., Diebold, C., and Giangrande, A. (2015). The Drosophila Fragile X Mental Retardation Protein Participates in the piRNA Pathway. J Cell Sci 128, 2070–2084. doi:10.1242/jcs.161810
Campos-Melo, D., Hawley, Z. C. E., Droppelmann, C. A., and Strong, M. J. (2021). The Integral Role of RNA in Stress Granule Formation and Function. Front. Cell Dev. Biol. 9, 621779. doi:10.3389/fcell.2021.621779
Coudert, L., Nonaka, T., Bernard, E., Hasegawa, M., Schaeffer, L., and Leblanc, P. (2019). Phosphorylated and Aggregated TDP-43 with Seeding Properties are Induced Upon Mutant Huntingtin (mHtt) Polyglutamine Expression in Human Cellular Models. Cell. Mol. Life Sci. 76, 2615–2632. doi:10.1007/s00018-019-03059-8
Courchaine, E. M., Barentine, A. E. S., Straube, K., Lee, D.-R., Bewersdorf, J., and Neugebauer, K. M. (2021). DMA-Tudor Interaction Modules Control the Specificity of In Vivo Condensates. Cell 184, 3612–3625. doi:10.1016/j.cell.2021.05.008
Coutinho-Budd, J. C., Sheehan, A. E., and Freeman, M. R. (2017). The Secreted Neurotrophin Spätzle 3 Promotes Glial Morphogenesis and Supports Neuronal Survival and Function. Genes Dev. 31, 2023–2038. doi:10.1101/gad.305888.117
Dehghani, M., and Lasko, P. (2017). Multiple Functions of the DEAD-Box Helicase Vasa in Drosophila Oogenesis. Results Probl. Cell Differ. 63, 127–147. doi:10.1007/978-3-319-60855-6_6
Deiana, A., Forcelloni, S., Porrello, A., and Giansanti, A. (2019). Intrinsically Disordered Proteins and Structured Proteins with Intrinsically Disordered Regions Have Different Functional Roles in the Cell. PLoS One 14, e0217889. doi:10.1371/journal.pone.0217889
Delgado, M. G., Oliva, C., López, E., Ibacache, A., Galaz, A., Delgado, R., et al. (2018). Chaski, a Novel Drosophila Lactate/Pyruvate Transporter Required in Glia Cells for Survival under Nutritional Stress. Sci. Rep. 8, 1186. doi:10.1038/s41598-018-19595-5
Dudman, J., and Qi, X. (2020). Stress Granule Dysregulation in Amyotrophic Lateral Sclerosis. Front. Cel. Neurosci. 14, 598517. doi:10.3389/fncel.2020.598517
Eddy, E. M. (1975). Germ Plasm and the Differentiation of the Germ Cell Line. Int. Rev. Cytol. 43, 229–280. doi:10.1016/s0074-7696(08)60070-4
Farca Luna, A. J., Perier, M., and Seugnet, L. (2017). Amyloid Precursor Protein in Drosophila Glia Regulates Sleep and Genes Involved in Glutamate Recycling. J. Neurosci. 37, 4289–4300. doi:10.1523/jneurosci.2826-16.2017
Feric, M., Vaidya, N., Harmon, T. S., Mitrea, D. M., Zhu, L., Richardson, T. M., et al. (2016). Coexisting Liquid Phases Underlie Nucleolar Subcompartments. Cell 165, 1686–1697. doi:10.1016/j.cell.2016.04.047
Folkmann, A. W., Putnam, A., Lee, C. F., and Seydoux, G. (2021). Regulation of Biomolecular Condensates by Interfacial Protein Clusters. Science 373, 1218–1224. doi:10.1126/science.abg7071
Formicola, N., Vijayakumar, J., and Besse, F. (2019). Neuronal Ribonucleoprotein Granules: Dynamic Sensors of Localized Signals. Traffic 20, 639–649. doi:10.1111/tra.12672
Freeman, M. R. (2015). Drosophila Central Nervous System Glia. Cold Spring Harb Perspect. Biol. 7. doi:10.1101/cshperspect.a020552
Gao, M., and Arkov, A. L. (2013). Next Generation Organelles: Structure and Role of Germ Granules in the Germline. Mol. Reprod. Dev. 80, 610–623. doi:10.1002/mrd.22115
Gao, M., Thomson, T. C., Creed, T. M., Tu, S., Loganathan, S. N., Jackson, C. A., et al. (2015). Glycolytic Enzymes Localize to Ribonucleoprotein Granules in Drosophila Germ Cells, Bind Tudor and Protect from Transposable Elements. EMBO Rep. 16, 379–386. doi:10.15252/embr.201439694
Gendron, T. F., Rademakers, R., and Petrucelli, L. (2013). TARDBP Mutation Analysis in TDP-43 Proteinopathies and Deciphering the Toxicity of Mutant TDP-43. J. Alzheimers Dis. 33 (Suppl. 1), S35–S45. doi:10.3233/JAD-2012-129036
Hanyu-Nakamura, K., Matsuda, K., Cohen, S. M., and Nakamura, A. (2019). Pgc Suppresses the Zygotically Acting RNA Decay Pathway to Protect Germ Plasm RNAs in the Drosophila Embryo. Development 146, 1–11. doi:10.3410/f.735366787.793558578
Hanyu-Nakamura, K., Sonobe-Nojima, H., Tanigawa, A., Lasko, P., and Nakamura, A. (2008). Drosophila Pgc Protein Inhibits P-TEFb Recruitment to Chromatin in Primordial Germ Cells. Nature 451, 730–733. doi:10.1038/nature06498
Hoxha, V., Lama, C., Chang, P. L., Saurabh, S., Patel, N., Olate, N., et al. (2013). Sex-specific Signaling in the Blood-Brain Barrier Is Required for Male Courtship in Drosophila. PLoS Genet. 9, e1003217. doi:10.1371/journal.pgen.1003217
Huang, X., Hu, H., Webster, A., Zou, F., Du, J., Patel, D. J., et al. (2021). Binding of Guide piRNA Triggers Methylation of the Unstructured N-Terminal Region of Aub Leading to Assembly of the piRNA Amplification Complex. Nat. Commun. 12, 4061. doi:10.1038/s41467-021-24351-x
Iwasaki, Y. W., Siomi, M. C., and Siomi, H. (2015). PIWI-interacting RNA: Its Biogenesis and Functions. Annu. Rev. Biochem. 84, 405–433. doi:10.1146/annurev-biochem-060614-034258
Janic, A., Mendizabal, L., Llamazares, S., Rossell, D., and Gonzalez, C. (2010). Ectopic Expression of Germline Genes Drives Malignant Brain Tumor Growth in Drosophila. Science 330, 1824–1827. doi:10.1126/science.1195481
Kiebler, M. A., and Bassell, G. J. (2006). Neuronal RNA Granules: Movers and Makers. Neuron 51, 685–690. doi:10.1016/j.neuron.2006.08.021
Kim, H. J., Kim, N. C., Wang, Y.-D., Scarborough, E. A., Moore, J., Diaz, Z., et al. (2013). Mutations in Prion-like Domains in hnRNPA2B1 and hnRNPA1 Cause Multisystem Proteinopathy and ALS. Nature 495, 467–473. doi:10.1038/nature11922
Kirino, Y., Kim, N., De Planell-Saguer, M., Khandros, E., Chiorean, S., Klein, P. S., et al. (2009). Arginine Methylation of Piwi Proteins Catalysed by dPRMT5 Is Required for Ago3 and Aub Stability. Nat. Cell Biol. 11, 652–658. doi:10.1038/ncb1872
Kirino, Y., Vourekas, A., Sayed, N., De Lima Alves, F., Thomson, T., Lasko, P., et al. (2010). Arginine Methylation of Aubergine Mediates Tudor Binding and Germ Plasm Localization. RNA 16, 70–78. doi:10.1261/rna.1869710
Kozlov, A., Koch, R., and Nagoshi, E. (2020). Nitric Oxide Mediates Neuro-Glial Interaction that Shapes Drosophila Circadian Behavior. PLoS Genet. 16, e1008312. doi:10.1371/journal.pgen.1008312
Kremer, M. C., Jung, C., Batelli, S., Rubin, G. M., and Gaul, U. (2017). The Glia of the Adult Drosophila Nervous System. Glia 65, 606–638. doi:10.1002/glia.23115
Ku, H.-Y., and Lin, H. (2014). PIWI Proteins and Their Interactors in piRNA Biogenesis, Germline Development and Gene Expression. Natl. Sci. Rev. 1, 205–218. doi:10.1093/nsr/nwu014
Kulkarni, A., Lopez, D. H., and Extavour, C. G. (2020). Shared Cell Biological Functions May Underlie Pleiotropy of Molecular Interactions in the Germ Lines and Nervous Systems of Animals. Front. Ecol. Evol. 81–22. doi:10.3389/fevo.2020.00215
Kunduri, G., Turner-Evans, D., Konya, Y., Izumi, Y., Nagashima, K., Lockett, S., et al. (2018). Defective Cortex Glia Plasma Membrane Structure Underlies Light-Induced Epilepsy in Cpes Mutants. Proc. Natl. Acad. Sci. USA 115, E8919–E8928. doi:10.1073/pnas.1808463115
Kwiatkowski, T. J., Bosco, D. A., Leclerc, A. L., Tamrazian, E., Vanderburg, C. R., Russ, C., et al. (2009). Mutations in the FUS/TLS Gene on Chromosome 16 Cause Familial Amyotrophic Lateral Sclerosis. Science 323, 1205–1208. doi:10.1126/science.1166066
Lai, A., Valdez‐Sinon, A. N., and Bassell, G. J. (2020). Regulation of RNA Granules by FMRP and Implications for Neurological Diseases. Traffic 21, 454–462. doi:10.1111/tra.12733
Larson, A. G., Elnatan, D., Keenen, M. M., Trnka, M. J., Johnston, J. B., Burlingame, A. L., et al. (2017). Liquid Droplet Formation by HP1α Suggests a Role for Phase Separation in Heterochromatin. Nature 547, 236–240. doi:10.1038/nature22822
Le Thomas, A., Rogers, A. K., Webster, A., Marinov, G. K., Liao, S. E., Perkins, E. M., et al. (2013). Piwi Induces piRNA-Guided Transcriptional Silencing and Establishment of a Repressive Chromatin State. Genes Dev. 27, 390–399. doi:10.1101/gad.209841.112
Lee, K. M., Mathies, L. D., and Grotewiel, M. (2019). Alcohol Sedation in Adult Drosophila is Regulated by Cysteine Proteinase-1 in Cortex Glia. Commun. Biol. 2, 252. doi:10.1038/s42003-019-0492-5
Little, S. C., Sinsimer, K. S., Lee, J. J., Wieschaus, E. F., and Gavis, E. R. (2015). Independent and Coordinate Trafficking of Single Drosophila Germ Plasm mRNAs. Nat. Cell Biol. 17, 558–568. doi:10.1038/ncb3143
Liu, H., Wang, J.-Y. S., Huang, Y., Li, Z., Gong, W., Lehmann, R., et al. (2010). Structural Basis for Methylarginine-dependent Recognition of Aubergine by Tudor. Genes Dev. 24, 1876–1881. doi:10.1101/gad.1956010
Lu, H., Yu, D., Hansen, A. S., Ganguly, S., Liu, R., Heckert, A., et al. (2018). Phase-separation Mechanism for C-Terminal Hyperphosphorylation of RNA Polymerase II. Nature 558, 318–323. doi:10.1038/s41586-018-0174-3
Luo, Y., Na, Z., and Slavoff, S. A. (2018). P-Bodies: Composition, Properties, and Functions. Biochemistry 57, 2424–2431. doi:10.1021/acs.biochem.7b01162
Mackenzie, I. R., Nicholson, A. M., Sarkar, M., Messing, J., Purice, M. D., Pottier, C., et al. (2017). TIA1 Mutations in Amyotrophic Lateral Sclerosis and Frontotemporal Dementia Promote Phase Separation and Alter Stress Granule Dynamics. Neuron 95, 808–816.e9. doi:10.1016/j.neuron.2017.07.025
Marnik, E. A., and Updike, D. L. (2019). Membraneless Organelles: P Granules in Caenorhabditis elegans. Traffic 20, 373–379. doi:10.1111/tra.12644
Martin, E. W., and Mittag, T. (2018). Relationship of Sequence and Phase Separation in Protein Low-Complexity Regions. Biochemistry 57, 2478–2487. doi:10.1021/acs.biochem.8b00008
Mateju, D., Eichenberger, B., Voigt, F., Eglinger, J., Roth, G., and Chao, J. A. (2020). Single-Molecule Imaging Reveals Translation of mRNAs Localized to Stress Granules. Cell 183, 1801–1812. doi:10.1016/j.cell.2020.11.010
Mcaleese, K. E., Walker, L., Erskine, D., Thomas, A. J., Mckeith, I. G., and Attems, J. (2017). TDP-43 Pathology in Alzheimer's Disease, Dementia with Lewy Bodies and Ageing. Brain Pathol. 27, 472–479. doi:10.1111/bpa.12424
Mclaughlin, C. N., Perry-Richardson, J. J., Coutinho-Budd, J. C., and Broihier, H. T. (2019). Dying Neurons Utilize Innate Immune Signaling to Prime Glia for Phagocytosis during Development. Dev. Cell 48, 506–522. doi:10.1016/j.devcel.2018.12.019
Megosh, H. B., Cox, D. N., Campbell, C., and Lin, H. (2006). The Role of PIWI and the miRNA Machinery in Drosophila Germline Determination. Curr. Biol. 16, 1884–1894. doi:10.1016/j.cub.2006.08.051
Myrick, L. K., Hashimoto, H., Cheng, X., and Warren, S. T. (2015). Human FMRP Contains an Integral Tandem Agenet (Tudor) and KH Motif in the Amino Terminal Domain. Hum. Mol. Genet. 24, 1733–1740. doi:10.1093/hmg/ddu586
Nakano, R., Iwamura, M., Obikawa, A., Togane, Y., Hara, Y., Fukuhara, T., et al. (2019). Cortex Glia clear Dead Young Neurons via Drpr/dCed-6/Shark and Crk/Mbc/dCed-12 Signaling Pathways in the Developing Drosophila Optic Lobe. Dev. Biol. 453, 68–85. doi:10.1016/j.ydbio.2019.05.003
Nishida, K. M., Iwasaki, Y. W., Murota, Y., Nagao, A., Mannen, T., Kato, Y., et al. (2015). Respective Functions of Two Distinct Siwi Complexes Assembled During PIWI-Interacting RNA Biogenesis in Bombyx Germ Cells. Cell Rep. 10, 193–203. doi:10.1016/j.celrep.2014.12.013
Nishida, K. M., Okada, T. N., Kawamura, T., Mituyama, T., Kawamura, Y., Inagaki, S., et al. (2009). Functional Involvement of Tudor and dPRMT5 in the piRNA Processing Pathway in Drosophila Germlines. EMBO J. 28, 3820–3831. doi:10.1038/emboj.2009.365
O’connor, R. M., Stone, E. F., Wayne, C. R., Marcinkevicius, E. V., Ulgherait, M., Delventhal, R., et al. (2017). A Drosophila Model of Fragile X Syndrome Exhibits Defects in Phagocytosis by Innate Immune Cells. J. Cell Biol. 216, 595–605. doi:10.1083/jcb.201607093
Parkhurst, S. J., Adhikari, P., Navarrete, J. S., Legendre, A., Manansala, M., and Wolf, F. W. (2018). Perineurial Barrier Glia Physically Respond to Alcohol in an Akap200-dependent Manner to Promote Tolerance. Cell Rep. 22, 1647–1656. doi:10.1016/j.celrep.2018.01.049
Pereanu, W., Spindler, S., Cruz, L., and Hartenstein, V. (2007). Tracheal Development in the Drosophila Brain is Constrained by Glial Cells. Dev. Biol. 302, 169–180. doi:10.1016/j.ydbio.2006.09.022
Perrat, P. N., Dasgupta, S., Wang, J., Theurkauf, W., Weng, Z., Rosbash, M., et al. (2013). Transposition-driven Genomic Heterogeneity in the Drosophila Brain. Science 340, 91–95. doi:10.1126/science.1231965
Pesiridis, G. S., Lee, V. M.-Y., and Trojanowski, J. Q. (2009). Mutations in TDP-43 Link Glycine-Rich Domain Functions to Amyotrophic Lateral Sclerosis. Hum. Mol. Genet. 18, R156–R162. doi:10.1093/hmg/ddp303
Protter, D. S. W., and Parker, R. (2016). Principles and Properties of Stress Granules. Trends Cel Biol. 26, 668–679. doi:10.1016/j.tcb.2016.05.004
Ramos, A., Hollingworth, D., Adinolfi, S., Castets, M., Kelly, G., Frenkiel, T. A., et al. (2006). The Structure of the N-Terminal Domain of the Fragile X Mental Retardation Protein: a Platform for Protein-Protein Interaction. Structure 14, 21–31. doi:10.1016/j.str.2005.09.018
Raz, E. (2000). The Function and Regulation of Vasa-like Genes in Germ-Cell Development. Genome Biol. 1, REVIEWS1017. doi:10.1186/gb-2000-1-3-reviews1017
Ryan, V. H., and Fawzi, N. L. (2019). Physiological, Pathological, and Targetable Membraneless Organelles in Neurons. Trends Neurosciences 42, 693–708. doi:10.1016/j.tins.2019.08.005
Sengoku, T., Nureki, O., Nakamura, A., Kobayashi, S., and Yokoyama, S. (2006). Structural Basis for RNA Unwinding by the DEAD-Box Protein Drosophila Vasa. Cell 125, 287–300. doi:10.1016/j.cell.2006.01.054
St-Amour, I., Turgeon, A., Goupil, C., Planel, E., and Hébert, S. S. (2018). Co-occurrence of Mixed Proteinopathies in Late-Stage Huntington's Disease. Acta Neuropathol. 135, 249–265. doi:10.1007/s00401-017-1786-7
Strom, A. R., and Brangwynne, C. P. (2019). The Liquid Nucleome - Phase Transitions in the Nucleus at a Glance. J. Cell Sci. 132. doi:10.1242/jcs.235093
Strom, A. R., Emelyanov, A. V., Mir, M., Fyodorov, D. V., Darzacq, X., and Karpen, G. H. (2017). Phase Separation Drives Heterochromatin Domain Formation. Nature 547, 241–245. doi:10.1038/nature22989
Thomson, T., and Lasko, P. (2004). Drosophila Tudor is Essential for Polar Granule Assembly and Pole Cell Specification, but Not for Posterior Patterning. Genesis 40, 164–170. doi:10.1002/gene.20079
Tindell, S. J., Rouchka, E. C., and Arkov, A. L. (2020). Glial Granules Contain Germline Proteins in the Drosophila Brain, Which Regulate Brain Transcriptome. Commun. Biol. 3, 699. doi:10.1038/s42003-020-01432-z
Trcek, T., Douglas, T. E., Grosch, M., Yin, Y., Eagle, W. V. I., Gavis, E. R., et al. (2020). Sequence-Independent Self-Assembly of Germ Granule mRNAs into Homotypic Clusters. Mol. Cell 78, 941–950. doi:10.1016/j.molcel.2020.05.008
Trcek, T., Grosch, M., York, A., Shroff, H., Lionnet, T., and Lehmann, R. (2015). Drosophila Germ Granules Are Structured and Contain Homotypic mRNA Clusters. Nat. Commun. 6, 7962. doi:10.1038/ncomms8962
Trcek, T., and Lehmann, R. (2019). Germ Granules in Drosophila. Traffic 20, 650–660. doi:10.1111/tra.12674
Tsang, B., Arsenault, J., Vernon, R. M., Lin, H., Sonenberg, N., Wang, L.-Y., et al. (2019). Phosphoregulated FMRP Phase Separation Models Activity-dependent Translation Through Bidirectional Control of mRNA Granule Formation. Proc. Natl. Acad. Sci. USA 116, 4218–4227. doi:10.1073/pnas.1814385116
Unhavaithaya, Y., and Orr-Weaver, T. L. (2012). Polyploidization of Glia in Neural Development Links Tissue Growth to Blood-Brain Barrier Integrity. Genes Dev. 26, 31–36. doi:10.1101/gad.177436.111
Vagin, V. V., Wohlschlegel, J., Qu, J., Jonsson, Z., Huang, X., Chuma, S., et al. (2009). Proteomic Analysis of Murine Piwi Proteins Reveals a Role for Arginine Methylation in Specifying Interaction with Tudor Family Members. Genes Dev. 23, 1749–1762. doi:10.1101/gad.1814809
Vita, D. J., Meier, C. J., and Broadie, K. (2021). Neuronal Fragile X Mental Retardation Protein Activates Glial Insulin Receptor Mediated PDF-Tri Neuron Developmental Clearance. Nat. Commun. 12, 1160. doi:10.1038/s41467-021-21429-4
Vo, H. D. L., WahiduzzamanTindell, S. J., Tindell, S. J., Zheng, J., Gao, M., and Arkov, A. L. (2019). Protein Components of Ribonucleoprotein Granules from Drosophila Germ Cells Oligomerize and Show Distinct Spatial Organization during Germline Development. Sci. Rep. 9, 19190. doi:10.1038/s41598-019-55747-x
Voronina, E., Seydoux, G., Sassone-Corsi, P., and Nagamori, I. (2011). RNA Granules in Germ Cells. Cold Spring Harbor Perspect. Biol. 3, a002774. doi:10.1101/cshperspect.a002774
Vrettos, N, Maragkakis, M, Alexiou, P, Sgourdou, P, Ibrahim, F, Palmieri, D, Kirino, Y, and Mourelatos, Z (2021). Modulation of Aub-TDRD Interactions Elucidates piRNA Amplification and Germplasm Formation. Life Sci Alliance 4. doi:10.26508/lsa.202000912
Weiss, S, Melom, JE, Ormerod, KG, Zhang, YV, and Littleton, JT (2019). Glial Ca2+ Signaling Links Endocytosis to K+ Buffering Around Neuronal Somas to Regulate Excitability. Elife 8. doi:10.7554/eLife.44186
Wolozin, B., and Ivanov, P. (2019). Stress Granules and Neurodegeneration. Nat. Rev. Neurosci. 20, 649–666. doi:10.1038/s41583-019-0222-5
Zhang, S. L., Yue, Z., Arnold, D. M., Artiushin, G., and Sehgal, A. (2018). A Circadian Clock in the Blood-Brain Barrier Regulates Xenobiotic Efflux. Cell 173, 130–139. doi:10.1016/j.cell.2018.02.017
Keywords: membraneless organelles, glia, germ granules, stress granules, Tudor domain, PIWI, neurodegenerative disease
Citation: Arkov AL (2022) Looking at the Pretty “Phase” of Membraneless Organelles: A View From Drosophila Glia. Front. Cell Dev. Biol. 10:801953. doi: 10.3389/fcell.2022.801953
Received: 26 October 2021; Accepted: 19 January 2022;
Published: 07 February 2022.
Edited by:
Nathan Anthony Smith, Children’s National Hospital, United StatesReviewed by:
Jaeda Coutinho-Budd, University of Vermont, United StatesEkaterina Voronina, University of Montana, United States
Copyright © 2022 Arkov. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Alexey L. Arkov, YWFya292QG11cnJheXN0YXRlLmVkdQ==
 Alexey L. Arkov
Alexey L. Arkov