- 1State Key Laboratory of Veterinary Etiological Biology, Key Laboratory of Veterinary Parasitology of Gansu Province, Lanzhou Veterinary Research Institute, CAAS, Lanzhou, China
- 2Department of Clinical Oncology, Queen Elizabeth Hospital, Hong Kong SAR, China
- 3Research Center for Hydatid Disease in Iran, Kerman University of Medical Sciences, Kerman, Iran
- 4Key Laboratory of Applied Technology on Green-Eco-Healthy Animal Husbandry of Zhejiang Province, Zhejiang International Science and Technology Cooperation Base for Veterinary Medicine and Health Management, Zhejiang Provincial Engineering Laboratory for Animal Health Inspection and Internet Technology, Zhejiang International Science and Technology Cooperation Base for Veterinary Medicine and Health Management, China-Australia Joint Laboratory for Animal Health Big Data Analytics, College of Animal Science and Technology and College of Veterinary Medicine of Zhejiang A&F University, Hangzhou, China
The larvae of Echinococcus multilocularis causes alveolar echinococcosis, which poses a great threat to the public health. However, the molecular mechanisms underlying the host and parasite interactions are still unclear. Exploring the transcriptomic maps of mRNA, miRNA and lncRNA expressed in the liver in response to E. multilocularis infection will help us to understand its pathogenesis. Using liver perfusion, different cell populations including the hepatic cells, hepatic stellate cells and Kupffer cells were isolated from mice interperitoneally inoculated with protoscoleces. Their transcriptional profiles including lncRNAs, miRNAs and mRNAs were done by RNA-seq. Among these cell populations, the most differentially-expressed (DE) mRNA, lncRNAs and miRNAs were annotated and may involve in the pathological processes, mainly including metabolic disorders, immune responses and liver fibrosis. Following the integrative analysis of 38 differentially-expressed DEmiRNAs and 8 DElncRNAs, the lncRNA-mRNA-miRNA networks were constructed, including F63-miR-223-3p-Fbxw7/ZFP36/map1b, F63-miR-27-5p-Tdrd6/Dip2c/Wdfy4 and IFNgAS1-IFN-γ. These results unveil the presence of several potential lncRNA-mRNA-miRNA axes during E. multilocularis infection, and further exploring of these axes may contribute to better understanding of the pathogenic mechanisms.
Introduction
Alveolar echinococcosis (AE), a zoonosis with an increasing concern, is caused by a canine parasite Echinococcus multilocularis (Eckert and Deplazes, 2004). At which human beings are infected by intake of food or water contaminated with the eggs shed by definitive hosts, such as dogs and foxes (Conraths et al., 2017; Liu et al., 2018; Wang et al., 2018; Kotwa et al., 2019). Once infected, the eggs penetrate the small intestine wall, finally reach the liver and lung and then grow in an infiltrative pattern (Czermak et al., 2008; Mueller et al., 2020). The clinical manifestations of human AE are diverse, including jaundice, weight loss, fever, anaemia, and abdominal pain (Kolářová et al., 2015; Massolo et al., 2019).
The main treatment strategies for AE are surgery and anti-echinococcal drugs, such as albendazole (ABZ) and mebendazole (MBZ) (Craig et al., 2007; Gottstein et al., 2015; Wang S. et al., 2021; Huang et al., 2021). However, both strategies have limitations. For surgical treatment, it is prone to recurrent due to diffuse and undetected parasite infiltration into host tissues (Huang et al., 2021). In terms of drug treatment, the poor intestinal absorption and some side effects are concerned (Gottstein et al., 2015; Craig et al., 2017). Some other treatment strategies such as traditional Chinese medicines and new formulation drug have limited treatment efficacy (Maggiore et al., 2015; Almalki et al., 2017; Pensel et al., 2017; Torabi et al., 2018). Therefore, it is urgent to study the genome-scale transcriptional background of the liver in response to the infection for discovering the potential molecular target for the effective therapeutics.
lncRNAs, with the length of more than 200 bp, have been confirmed as a momentous regulatory factor in many cellular processes, such as genomic imprinting, post transcriptional regulation, and cell differentiation (Kallen et al., 2013; Myant et al., 2013; Yang Q. et al., 2018). lncRNAs show a competing endogenous effect by acting as miRNA sponges, which is helpful to decipher the miRNA-lncRNA regulatory networks (Li et al., 2020; Zheng et al., 2020). Another non-coding RNAs, miRNAs, are members of endogenous small RNA with the length of about 18–24 nucleotides, involved in the regulation of mRNA expression. By binding to the 3′-untranslated region, miRNAs block the translation or induce the degradation of the target mRNA, thus modulating cell differentiation, growth, proliferation and apoptosis (Li et al., 2021; Mohammadi et al., 2021; Xiang et al., 2021). miRNAs have been reported as diagnostic markers and therapeutic targets for the control of diseases (Broermann et al., 2020; Tombolan et al., 2020). For instance, miR-155 was exploited as the potential target for treatment of the disease by Toxoplasma gondii (Xu et al., 2021). Another study found that the novel-miR-1 derived from Cysticercus pisiformis was released into host serum, which could be exploited for diagnosis of C. pisiformis infection (Chen et al., 2021).
Clinically, the intrahepatic lesions are the most common clinical trait of AE (Woolsey and Miller, 2021). The liver is comprised of a number of specialized cells, e,g, hepatocytes (HCs) as the hepatic parenchyma are the most abundant cell population, performing the fundamental functions in endocrine homeostasis and metabolism (Wang et al., 2015). Besides, there is a small proportion of nonparenchymal cells (NPCs), including hepatic stellate cells (HSCs), Kupffer cells (KCs), and hepatic sinusoidal endothelial cells (LSECs). Among these cells, HSCs reside in the perisinusoidal space (space of Disse) filled with thin permeable connective tissues, which have been recognized as the major source of type I and III collagens and fibronectin. Under normal condition, quiescent HSCs (qHSCs) are characterized by enrichment of vitamin A in cytoplasm, responsible for tissue homeostasis by involving in proliferation and differentiation signaling (Balmer and Blomhoff. 2002). Upon stimuli, they transfer into activated HSCs (aHSCs) that secret a-SMA and extracellular matrix (ECM) components (type I and III collagens and fibronectin). As a result, the persisting increase of ECM components will trigger liver fibrosis, possibly leading to liver cancer. However, once the stimuli disappear, the aHSCs gradually return back to the quiescent ones and the degree of fibrosis decreases (Kisseleva et al., 2012; Khomich et al., 2019). Another important component of NPCs, KCs, is the major immune cells that reside in the liver, responsible for clearing foreign materials under the normal conditions. Moreover, KCs protect liver from injury by releasing cytokines, reactive oxygen and others, playing a key role in the acute and chronic responses in the liver injury (Roberts et al., 2007).
Various liver cell populations perform distinct biological functions, and thus it is important to elucidate their role during E. multilocularis infection. In this study, we defined the transcriptomes of HCs, HSCs and KCs at two time points (2 and 3 months) post infection (p.i.) of E. multilocularis. Besides, the potential interaction networks among differentially expressed mRNAs, miRNAs and lncRNAs were identified. The current results provide the transcriptional expression of three liver cell populations in response to E. multilocularis infection and a clue for further investigation of a role of the networks in the pathogenesis.
Materials and Methods
Parasites
The protoscolices used in this study were obtained from Mongolian gerbil infected with E. multilocularis as previously described (Spiliotis and Brehm, 2009). In brief, cysts were dissected from infected Mongolian gerbil under sterile conditions. After cutting the cysts into pieces, the protoscolices were collected by gravity and washed several times in cold PBS. The purity and activity of protoscolices were checked using optical microscopy and trypan blue exclusion, respectively.
Animal Infection
100 six-week old BALB/c mice were purchased from Laboratory Animal Center of Lanzhou Veterinary Research Institute and were randomly divided two groups: experimental (E) group (60 mice) and control (C) group (40 mice). In E group, 600 protoscolices were injected into the abdominal cavity. In C group, the same volume of phosphate buffer saline (PBS) was injected. In E group, whose cysts in the liver were considered as infected, otherwise not. In order to obtain enough cells at two timepoints 2 m p.i. and 3 m p.i., by the cause of a few HSCs and KCs in the liver, we mixed cells from six mice for RNA-seq and qRT-PCR, and three batches of samples were concluded in two groups. All mice were reared under standard feeding conditions, free access to food and water.
Isolation of HCs, HSCs and KCs
We collected the HCs, HSCs and KCs samples from mice 2 m p.i. and 3 m p.i. in E and C groups, accordingly named as HC-2M-E, HC-2M-C, HC-3M-E, HC-3M-C, HSC-2M-E, HSC-2M-C, HSC-3M-E, HSC-3M-C, KC-2M-E, KC-2M-C, KC-3M-E and KC-3M-C. In each group, the cells of six mice were mixed together to obtain enough HSCs and KCs. The perfusion procedure for mouse liver was strictly followed as previously reported (Mederacke et al., 2015). In detail, 10 ml of 0.019% EGTA and 30 ml of 0.04% collagenase Ⅳ solution were sequentially perfused into every liver, followed by further liver digestion using 80 ml of 0.08% collagenase Ⅳ solution with 1% DNase. The digested liver tissues were filtered into a 50 ml tube through a 70 μm cell strainer. The mixture was centrifuged at 50 ×g for 4 min at 4°C, and the cell pellet was washed three times in the DMEM with 5% FBS to obtain HCs. The supernatant was sequentially centrifuged at 600 ×g and 500 ×g for 10 min at 4°C to obtain NPCs. The NPC-containing solution was mixed with 5 ml Gey’s Balanced Salt Solution (GBSS), then the mixture was gently overlaid onto the Optiprep solutions at different concentrations, which contained 8 ml of 11.5% Optiprep in the upper layer and 4 ml of 20% Optiprep in the bottom, followed by centrifugation at 1,400 ×g for 17 min at 4°C without break. The cells in the upper layer (HSCs) and the lower layer (KCs and ECs) were transferred into two centrifuge tubes, respectively, and washed three times in GBSS. Then, KCs and ECs were cultured in RPMI 1640 medium with 10% fetal bovine serum (FBS). After 4 h, KCs were adhered completely and collected after 0.25% trypsin digestion, while ECs were not adhered and abandoned after PBS washed (Zheng et al., 2008). The HCs, HSCs and KCs were immediately stored at −80°C for later use.
RNA-Seq
Following the manufacturer’s instructions, total RNA was extracted using TRIzol reagent. After evaluation of RNA concentration and integrity, these samples were used to construct the libraries for sequencing (BGI, Wuhan, China). For construction of the lncRNA + mRNA library, rRNA-depleted RNA was fragmented by adding first strand master mix (Invitrogen, United States), and the first-strand and second-strand cDNA was generated separately by using random primers reverse transcription. Then, the cDNA was subjected to end-repair and was 3′ adenylated, the adapters were ligated to the end of 3′ adenylated cDNA. After PCR amplification, the cDNA fragments (lncRNA + mRNA library) were enriched and purified with Ampure XP beads.
For small RNA library, the 18–30 nt bands were excised and recovered from total RNA by using 15% urea denaturing polyacrylamide gel electrophoresis (PAGE) gel. Subsequently, the 18–30 nt small RNAs were ligated to adenylated 3′ and 5′ adapters separately and transcripted into cDNA by SuperScript Reverse Transcriptase (Invitrogen, United States). After PCR amplification, the cDNA fragments was enriched and 110–130 nt fragments (small RNA library) were selected by agarose gel electrophoresis. Last, the BGISEQ-500 platform (BGI, Wuhan, China) was used to sequence. In order to get clean reads, the reads with low quality and adaptor contaminants were removed from the raw data using SOAPnuke software (v1.5.2; -l 15 -q 0.2 -n 0.05) and the Q20, Q30 and GC content were calculated to assess the quality of the clean reads.
Data Analysis
Using HISAT2 software (v2.0.4; http://www.ccb,jhu.edu/software/hisat/index.shtml), the clean reads were aligned against the Mus musculus genome (GCF_000001635.26_GRCm38.p6). Bowtie2 (v2.2.5; http://bowtiebio.sourceforge.net/%20Bowtie2%20/index.shtml) was used to align the clean reads to known and novel, coding and noncoding transcripts. Subsequently, the expression levels of mRNA and lncRNAs were calculated by using RESM (v1.2.12; http://github.com/deweylab/RSEM) with the FPKM standardized method. After analyzing the results of the relative expression of genes in this study, we found the genes was mainly enriched near 2 in fold change. Besides, we used multiple hypothesis test correction for the p-value of the difference test, and False Discover Rate (FDR) ≤ 0.001 was considered as statistically different. Based on these, the differentially expressed lncRNAs (DElncRNAs) and differentially expressed mRNA (DEmRNAs) with a fold change ≥2 and FDR ≤0.001 were screened out.
Functional Enrichment Analysis
We predicted the potential targets of DElncRNAs by analyzing the position between genes in the genome as previously reported (Ren et al., 2018). If the relative position between genes was less than 10 kb, we defined it as cis-regulation. Otherwise, we defined it as trans-regulation. Besides, for constructing the lncRNA-mRNA-miRNA networks, target prediction of miRNAs to the DEmRNAs and DElncRNAs was performed using RNAhybrid, miRanda and TargetScan databases.
Gene Ontology (GO) was used to describe the genetic attributes with terms under the biological process, cellular component, and molecular function categories. By comparing the DElncRNAs, DEmRNAs and the targets of DEmiRNAs with the genes of mouse, the significantly enriched GO terms were determined. Similarly, the significantly enriched pathway terms were obtained using the KEGG pathway database. The GO and pathway terms with Q value ≤0.05 was defined as a term which was significantly enriched.
qRT-PCR Assay
For evaluation of the purity of liver cell populations and the expression of DEmRNAs, DElncRNAs and DEmiRNAs, qRT-PCR was performed. The genes for the cell markers (Alb, F4/80, α-SMA, Col1α1, Col3α1 and GFAP), DElncRNAs and their nearby genes located within the 10 kb upstream and downstream in the genome, and the metabolism- and inflammation-related DEmRNAs were selected for validation. 1 μg of total RNA was reversely transcribed to cDNA using HiScript Ⅲ First-strand cDNA Synthesis Kit (Vazyme, Munich, Germany) as recommended by the manufacturer. qRT-PCR was performed using All-in-OneTM qPCR Mix (GeneCopoeia) with an ABI 7500 Thermal Cycler (ThermoFisher Scientific, United States) according to the standard method. Specific primers for the selected DElncRNAs and DEmRNAs were obtained from TSINGKE (Xi’an, China) (Supplementary Table S1), and specific primers for miRNAs were purchased from GeneCopoeia (United States). The relative expression level was normalized to GAPDH for DEmRNAs and DElncRNAs or to U6 for DEmiRNAs. The 2−ΔΔCt algorithm was used to calculate the relative expression levels represented as relative fold-change (FC). All experiments were performed in triplicate.
Statistical Analysis
The statistical analysis was conducted using Prism 6 (GraphPad) software. The comparison between E and C groups was analyzed using Student’s t-test. The results were presented as mean ± standard deviation (SD). Significant differences were indicated as *p < 0.05, **p < 0.01 and ***p < 0.001.
Results
Primary HC, HSC and KC Isolation and Characterization
The primary HCs, HSCs and KCs were isolated from liver of anesthetized mice by using 0.04% collagenase Ⅳ solution, with the largest number of HCs, followed by KCs and then HSCs. By comparing the relative expression levels of cell markers (HCs: Alb, HSCs: α-SMA, GFAP, Col1α1 and Col1α3; KCs: F4/80), the results showed that Alb was predominantly expressed in HCs, with the percentage of 69.44, F4/80 predominantly expressed in KC with 73.19, and α-SMA, GFAP, Col1α1 and Col1α3 predominantly expressed in HSC (Supplementary Figure S1) with 83.09, 87.70, 93.36 and 89.55, respectively, suggesting each type of cell was enrichment.
Sequencing Data
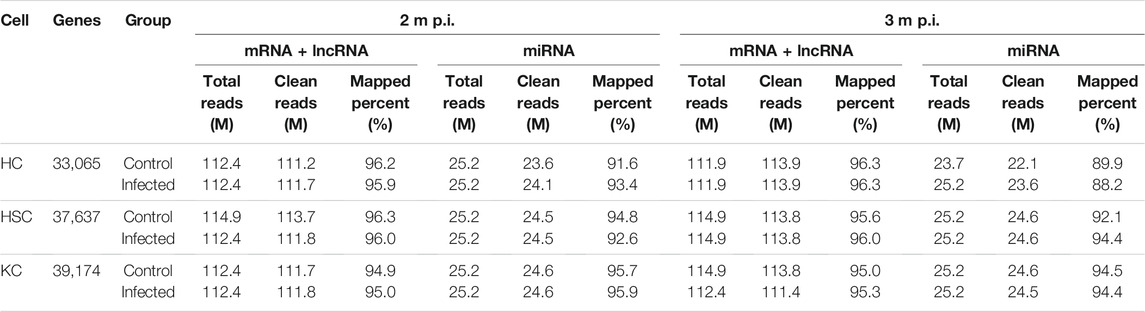
As shown in Table 1, after removal of the low quality reads, 112M–115M total reads were produced in mRNA + lncRNA libraries and 24–25M in miRNA libraries. In mRNA + lncRNA libraries, the mapped percent was ranged from 95 to 96 with the average mapped rate of 96% and a total of 43,183 genes were identified. In miRNA libraries, 88–96% clean reads were mapped with the average mapped rate of 93% and a total of 1,201 small RNAs were predicted. Additionally, a total of 33,065 genes were detected in HCs, 37,637 in HSCs and 39,174 in KCs.
LncRNA, mRNA and miRNA Profiles in HCs, HSCs and KCs
As shown in Figure 1, in 2 m p.i. HC samples, 274 upregulated and 219 downregulated mRNA were identified. Simultaneously, 1,111 upregulated and 119 downregulated mRNA were identified in 3 m p.i. samples. Additionally, 65 upregulated and 97 downregulated lncRNAs, and 12 upregulated and 23 downregulated miRNAs were found in 2 m p.i. samples, while 136 upregulated and 82 downregulated lncRNAs, and 93 upregulated and 6 downregulated miRNAs in 3 m p.i. samples (Figure 1A, Supplementary Table S2). Furthermore, DEmRNAs, DElncRNAs and DEmiRNAs of two time point were identified, with the commonly shared 151 DEmRNAs (9.6%), 28 DElncRNAs (8%) and 14 DEmiRNAs (11.7%) (Figure 1B).
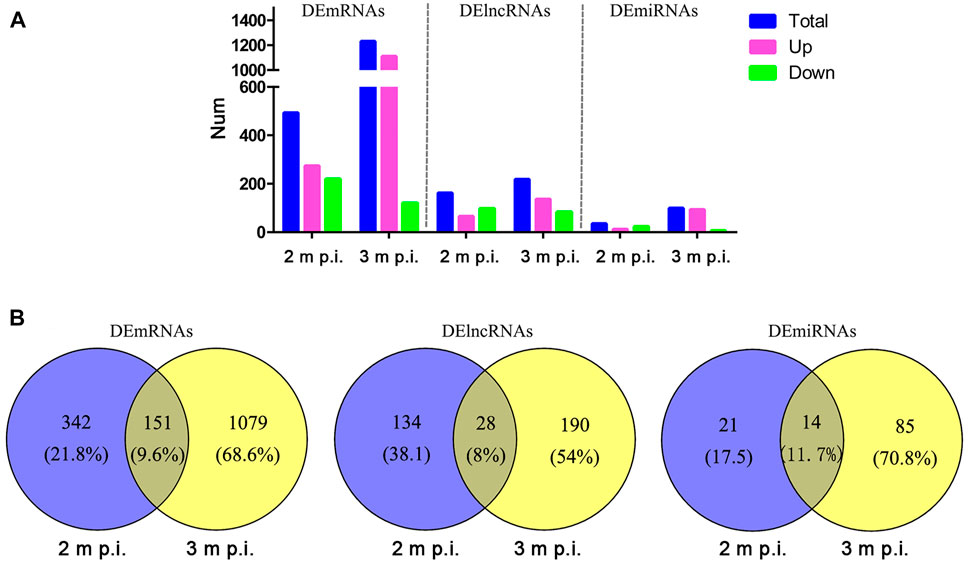
FIGURE 1. Comparisons of DElncRNAs, DEmRNAs and DEmiRNAs differentially expressed in HCs at 2 m p.i. and 3 m p.i (A) The number of DElncRNAs, DEmRNAs and DEmiRNAs at two infection stages. Sky blue represents the total number of DEgemes, red and violet represents upregulated and downregulated genes, respectively. (B) Veen diagrams showing the common and unique DElncRNAs, DEmRNAs and DEmiRNAs at two infection stages.
In 2 m p.i. HSC samples, total 293 upregulated and 335 downregulated mRNAs, 98 upregulated and 127 downregulated lncRNAs, and 11 upregulated and 20 downregulated miRNAs were found, while total 241 upregulated and 142 downregulated mRNAs, 66 upregulated and 63 downregulated lncRNAs, and 56 upregulated and 21 downregulated miRNAs were found in 3 m p.i. HSC samples (Supplementary Figure S2A, Supplementary Table S2). The 80 (8.6%), 22 (6.6%) and 18 (20%) common differentially-expressed genes (DEGs) were identified as DEmRNAs, DElncRNAs and DEmiRNAs, respectively (Supplementary Figure S2B).
In 2 m p.i. KC samples, total 102 upregulated and 1,291 downregulated mRNAs, 85 upregulated and 177 downregulated lncRNAs, and 5 upregulated and 38 downregulated miRNAs were found, while total 79 upregulated and 205 downregulated mRNAs, 69 upregulated and 32 downregulated lncRNAs, and 9 upregulated and 12 downregulated miRNAs were found in 3 m p.i. KC samples (Supplementary Figure S3A, Supplementary Table S2). The 111 (7.1%), 18 (5.2%) and 3 (4.9%) common DEGs were identified as DEmRNAs, DElncRNAs and DEmiRNAs, respectively (Supplementary Figure S3B).
GO and KEGG Analyses of DElncRNAs and DEmiRNAs
Of the 195 DEmiRNAs identified, the most was functionally annotated, such as miR-7a-5p, miR-223-3p, miR-22-3p, miR-146a-5p, miR-378a-3p, miR-467a-5p, miR-532-5p, miR-652-3p, miR-871-3p and miR-96-5p (Coffey et al., 2019; Ota et al., 2019; Chu et al., 2020; Du et al., 2020; Liang et al., 2020; Yan et al., 2020; Wang X. et al., 2021; Gajeton et al., 2021; Han et al., 2021; Kinoshita et al., 2021). Unlike the DEmiRNAs, most of the DElncRNAs were not annotated with unknown functions. Based on this, this study focused on the biological function of DElncRNAs, with the most enriched biological processes of DEmRNAs and DElncRNAs in HCs were immune system process, innate immune response, inflammatory response and cell adhesion (Figures 2A,B, Supplementary Table S4), and the enriched pathways included pancreatic secretion, protein digestion and absorption, fatty acid biosynthesis, ECM-receptor interaction and PI3K-Akt signaling pathway (Figures 2C, D, Supplementary Table S3).
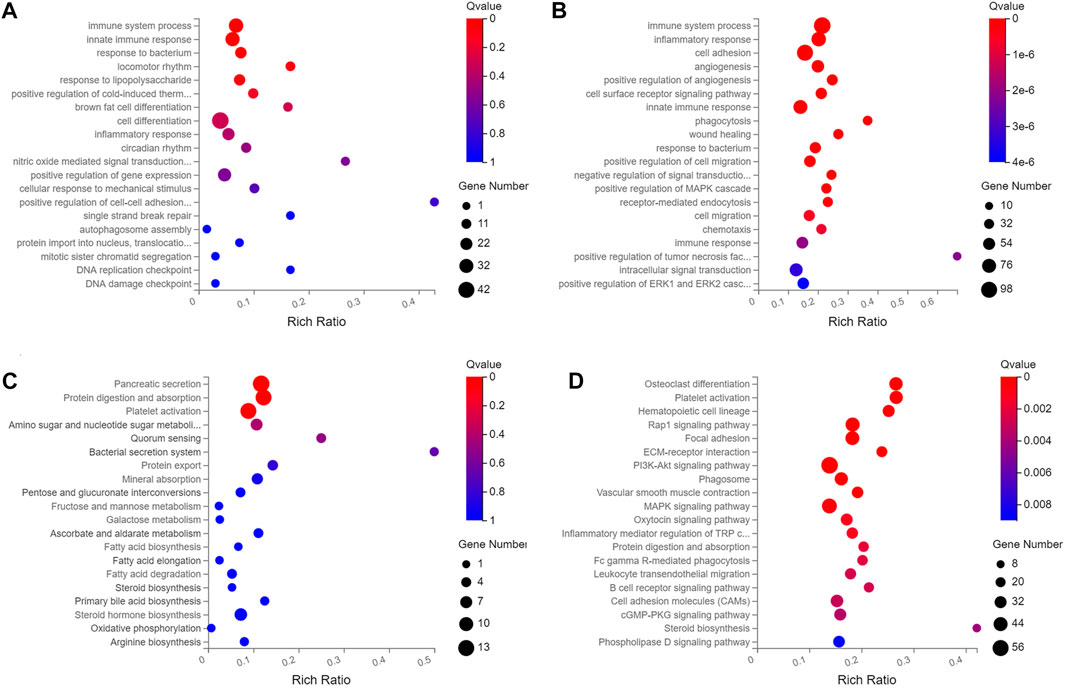
FIGURE 2. The top twenty enriched biological process and pathways of DElncRNAs and DEmRNAs in HCs at 2 m p.i. and 3 m p.i.. (A) Predicted biological process terms of DElncRNAs and DEmRNAs in HCs at 2 m p.i.. (B) Predicted biological process terms of DElncRNAs and DEmRNAs in HCs at 3 m p.i.. (C) Predicted pathways of DElncRNAs and DEmRNAs in HCs at 2 m p.i.. (D) Predicted pathways of DElncRNAs and DEmRNAs in HCs at 3 m p.i..
In HSCs, the enriched biological processes included immune system process, adaptive immune response, negative regulation of lipid biosynthetic and receptor-mediated endocytosis (Supplementary Figures S4A, B, Supplementary Table S5), and the enriched pathways included T cell receptor signaling pathway, Th1 and Th2 cell differentiation, IL-17 signaling pathway, Toll-like receptor signaling pathway and NF-κB signaling pathway (Supplementary Figures S4C, D, Supplementary Table S4), indicating that HSCs may play an immune regulatory role after E. multilocularis infection.
While in KCs, the enriched biological processes included immune system process, acute-phase response, cellular response to IFN-β and exogenous drug catabolic process (Supplementary Figures S5 A, B, Supplementary Table S6), and the enriched pathways included complement and coagulation cascades, TNF signaling pathway, cytokine-cytokine receptor interaction, IL-17 signaling pathway and PPAR signaling pathway (Supplementary Figures S5C,D, Supplementary Table S5).
LncRNA-mRNA-miRNA Networks
It has been demonstrated that lncRNAs function as miRNA “sponges”, which competitively suppress the activity of miRNAs (Alkan and Akgül, 2022). Since lncRNAs interact with miRNAs through miRNA Response Elements (MREs), combined using the free energy and score of RNAhybrid, miRanda and TargetScan databases, the potential MREs were predicted and 38 DEmiRNAs that putatively targeted 8 DElncRNAs were then identified (Supplementary Table S6). Based on these, partial lncRNA-mRNA-miRNA networks were obtained (Figure 3). In this network, some DElncRNAs were predicted to bind multiple DEmiRNAs, and DEmiRNAs were predicted to bind multiple DEmRNAs, such as F630028O10Rik (abbreviated as F63)-miR-223-3p-Fbxw7/ZFP36/map1b and F63-miR-27-5p-Tdrd6/Dip2c/Wdfy4. Considering these RNAs are differentially expressed in various cells, some important roles may play by these lncRNAs and miRNAs through these regulatory pathways, which provide some clues for further studies of DElncRNAs and DEmiRNAs.
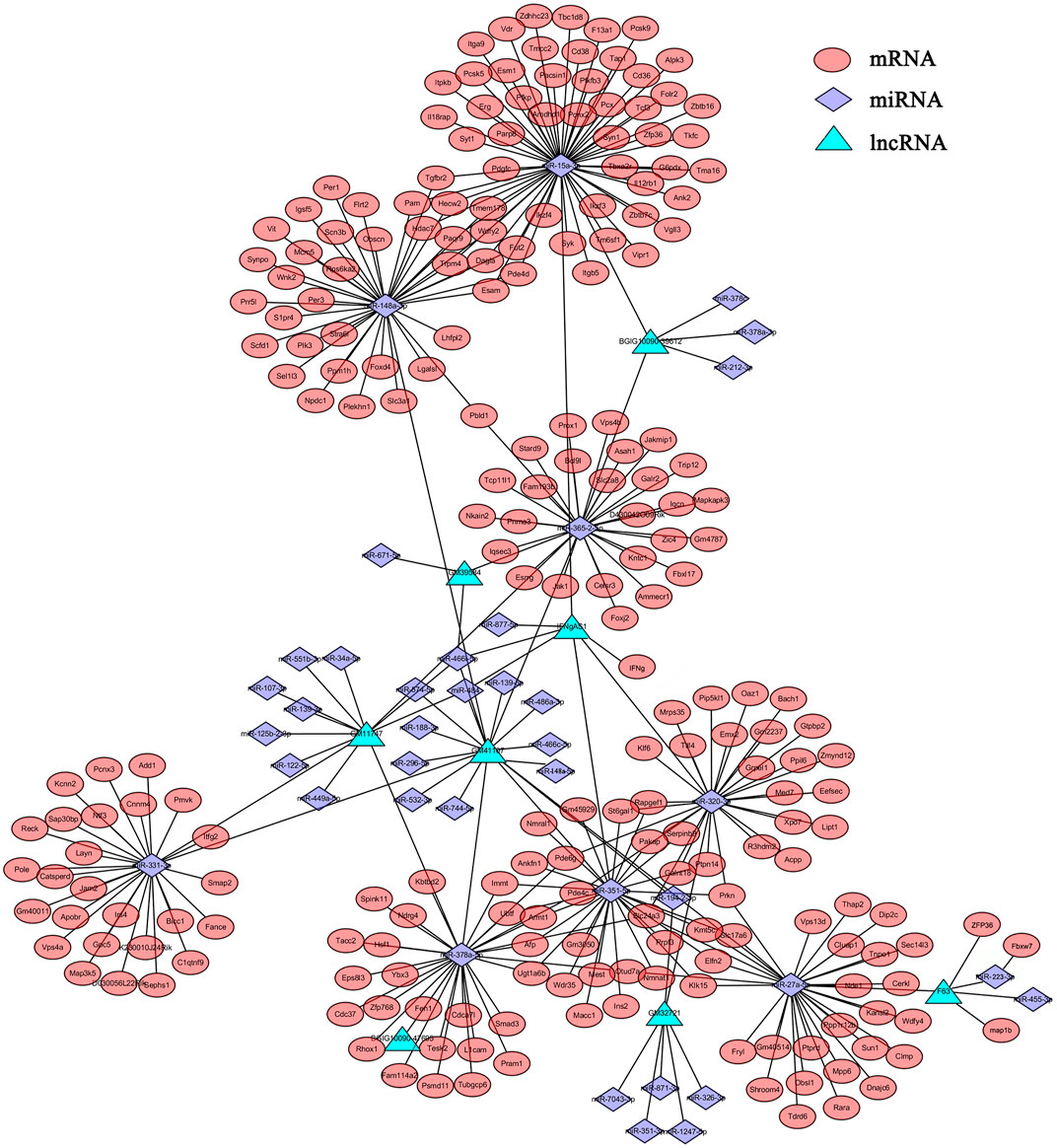
FIGURE 3. The lncRNA-mRNA-miRNA network. The relationships of lncRNAs-miRNAs-mRNAs were constructed based the results of predicted targets using Cytoscape software (v3.6.1). Different colors were used to show different genes, with green for DElncRNAs, purple for DEmiRNAs and red for DEmRNAs.
qRT-PCR Validation
To verify the lncRNA-mRNA-miRNA networks and the data of RNA-seq, qRT-PCR was used to examine the expression of 9 DElncRNAs (IFNgAS1, GM39584, F63, GM32721, GM11747, GM41107, BGIG10090-47695, BGIG10090-39612 and BGIG10090-34058) (Figure 4), 12 DEmRNAs (IFN-γ, IL-4, IL-12, IL-10, a-SMA, COL1a1, TGF-β1, ZFP36, VDR, EGFR, Fbxw7 and map1b) (Figure 5) and 5 DEmiRNAs (miR-143-3p, miR-451a, miR-146b-5p, miR-222-3p and miR-342-3p) (Figure 6). The overall expression patterns of these DElncRNAs, DEmRNAs and DEmiRNAs between qRT-PCR and RNA-seq was consistent, suggesting the reliability of RNA-seq. It was worth mentioning that the expression of inflammation related genes (IFN-γ and IL-4) were remarkably upregulated in HSCs at 2 m p.i. and 3 m p.i. However, in KCs, the IFN-γ was remarkably downregulated and IL-4 still upregulated at 2 m p.i. and 3 m p.i. Additionally, the fibrosis related factors including α-SMA and Col1α1 showed a sharp increase in 3 m p.i., suggesting the persistence of inflammatory responses in the infection and the occurrence of fibrosis. Additionally, some lncRNA-mRNA pairs in the network were identified with a similar expression trend, such as GM39584-Egfr, F63-Fbxw7/map1b and IFNgAS1-IFN-γ, suggesting the potential regulatory role of these DElncRNAs.
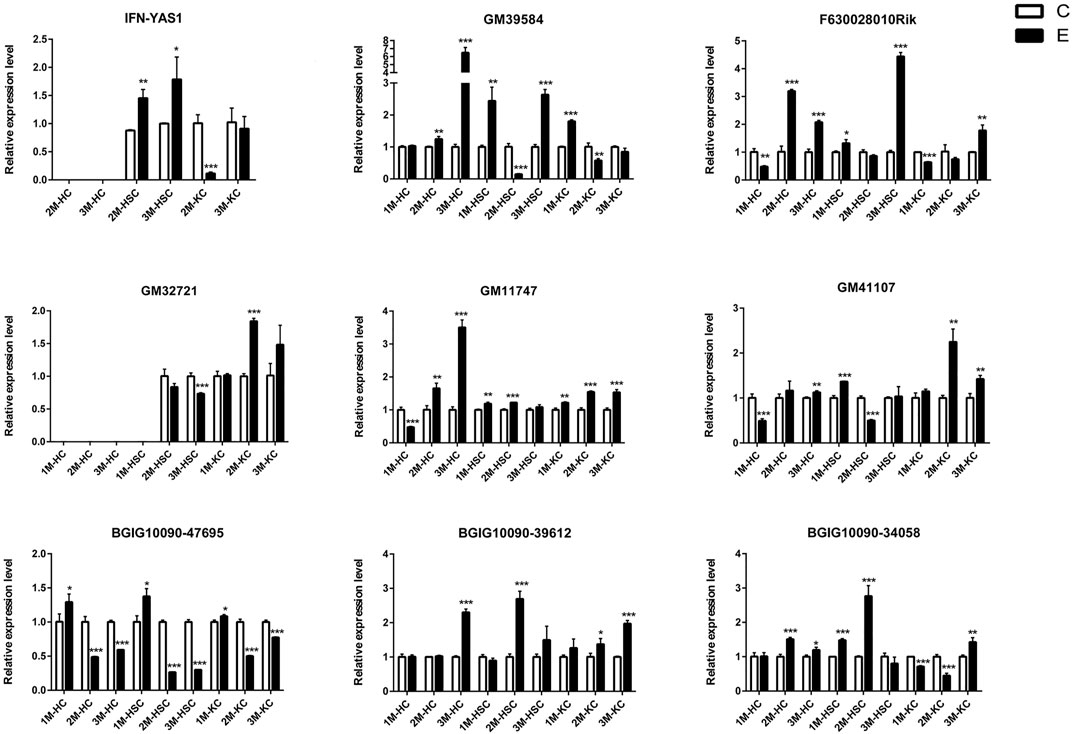
FIGURE 4. The relative expression levels of 9 lncRNAs in 2M-HC, 3M-HC, 2M-HSC, 3M-HSC, 2M-KC and 3M-KC were determined by qRT-PCR. The number of biological replicates for each experiment was 3 and the relative expression levels were normalized to the expression levels of GAPDH. Data are presented as means with SD. p-values were analyzed by Student’s t-test. ***p < 0.001, **p < 0.01 and *p < 0.05.
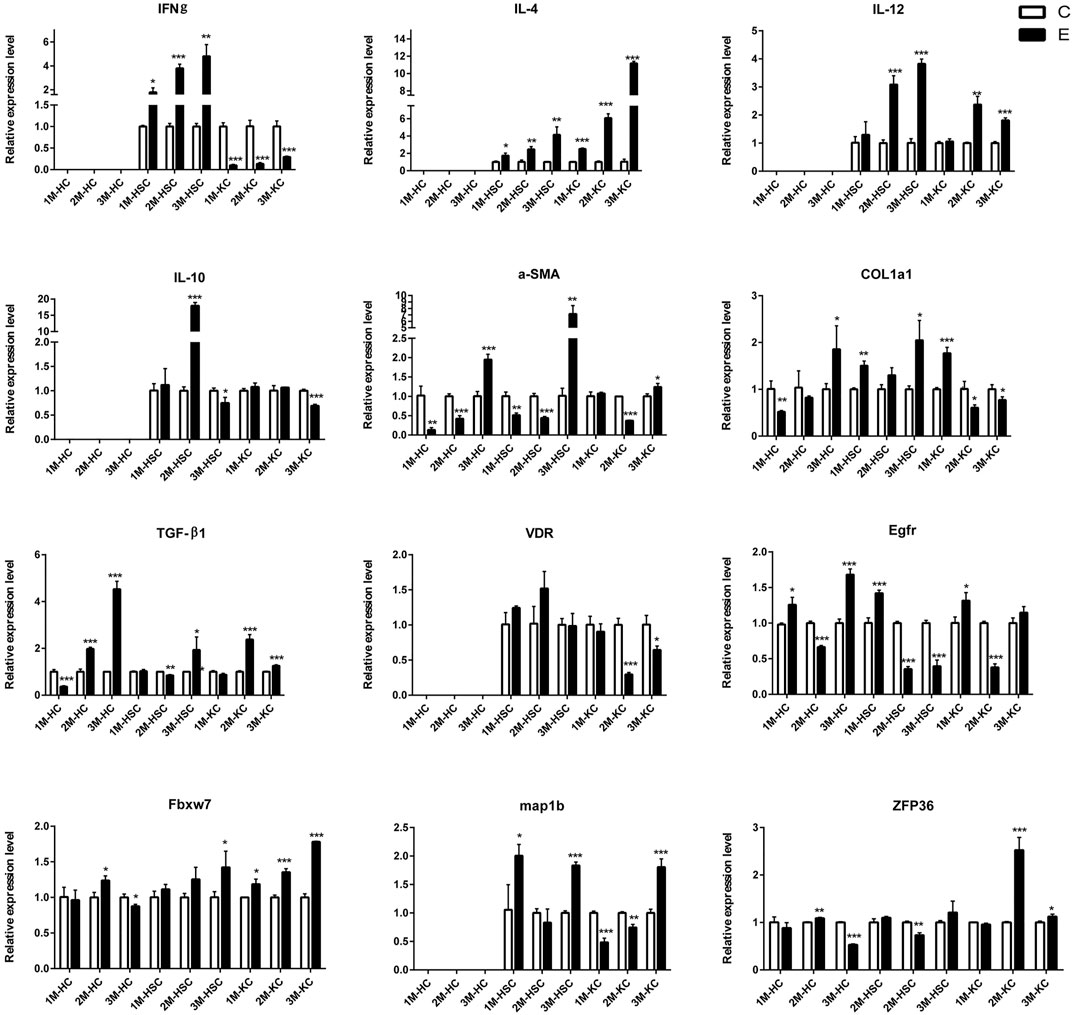
FIGURE 5. The relative expression levels of 12 mRNAs in 2M-HC, 3M-HC, 2M-HSC, 3M-HSC, 2M-KC and 3M-KC were determined by qRT-PCR. The number of biological replicates for each experiment was 3 and the relative expression levels were normalized to the expression levels of GAPDH. Data are presented as means with SD. p-values were analyzed by Student’s t-test. ***p < 0.001, **p < 0.01 and *p < 0.05.
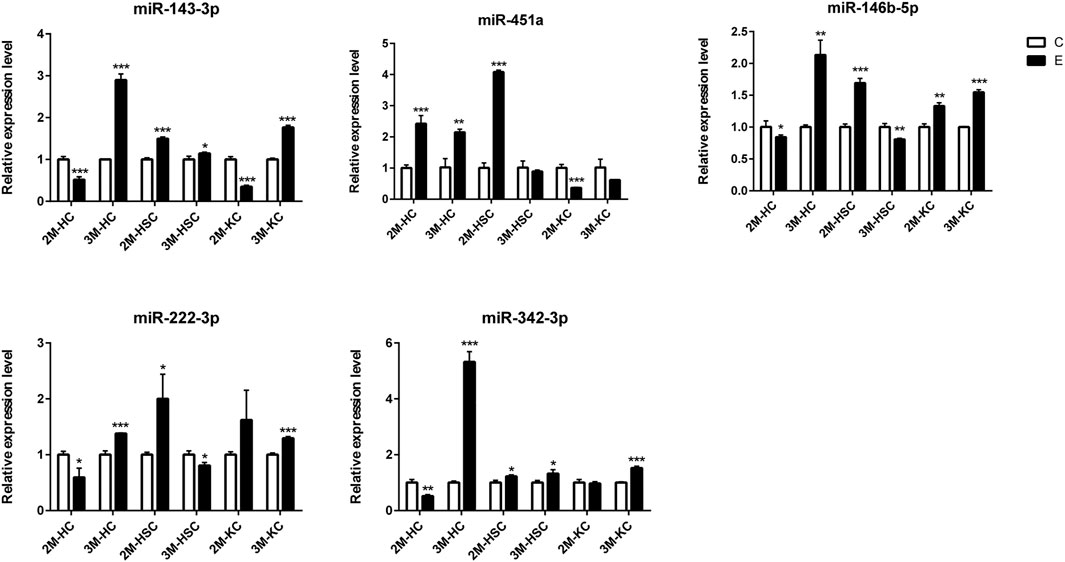
FIGURE 6. The relative expression levels of 5 miRNAs in 2M-HC, 3M-HC, 2M-HSC, 3M-HSC, 2M-KC and 3M-KC were determined by qRT-PCR. The number of biological replicates for each experiment was 3 and the relative expression levels were normalized to the expression levels of U6. Data are presented as means with SD. p-values were analyzed by Student’s t-test. ***p < 0.001, **p < 0.01 and *p < 0.05.
In addition, we identified some potential markers for HCs, HSCs and KCs (Figure 7). For instance, the expression of BGIG10090-47695, miR-146b-5p, miR-222-3p and EGFR was remarkably abundant in HCs comparing HSCs and KCs, while F63, GM41107 and GM32721 were remarkably abundant in HSCs, and BGIG10090-39612, map1b and VDR in KCs.
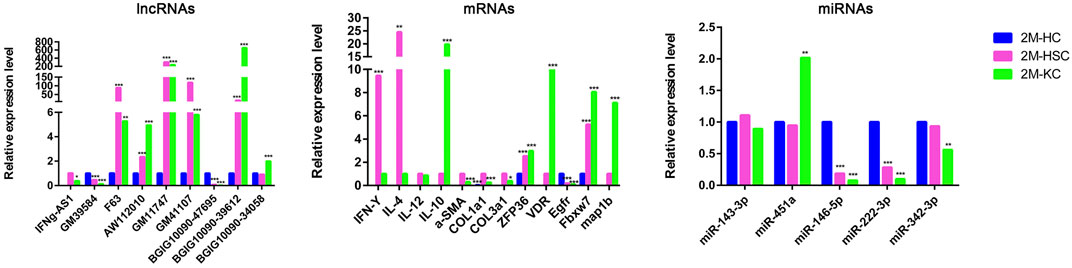
FIGURE 7. The relative expression level of DElncRNAs, DEmRNAs, and DEmiRNAs in different cells at 2 m p.i.. Sky blue represents the expression of genes in HCs, red and purple represents HSCs and KCs, respectively.
Discussion
E. multilocularis is one of zoonotic tapeworms with public health concern. Elucidating the pathogenic mechanisms of the parasite in the liver will help us to take better prevention and treatment measures. It is known that HCs, HSCs, KCs together with others constitute the liver and play divergent roles. Exploring the specific biological processes of these cells after E. multilocularis infection will systematically clarify the pathogenic mechanisms. Therefore, the current study focuses on the transcriptomic maps of mRNA, miRNAs and lncRNAs expressed in HCs, HSCs and KCs in the liver after infection, which aims to provide clues for further investigation of the pathogenic mechanisms to better control AE.
The current results showed that, after the infection, the expression of a number of mRNAs, lncRNAs and miRNAs in HCs, HSCs and KCs was changed. One of our interests is the genes related to metabolism in HCs. For example, the F-box family member F-box- and WD repeat domain-containing 7 (Fbxw7) was up- and downregulated in 2 m p.i. and 3 m p.i., respectively. A previous study showed massive lipid deposition and increasing proliferation of hepatocytes in Fbxw7-deficient mice (Onoyama et al., 2011). Therefore, the changes in the expression of Fbxw7 may reflect the host’s response to eliminate the parasite in 2 m p.i. and the parasite’s strategy for persistent infection in 3 m p.i. Consistent with the lncRNA-mRNA-miRNA networks constructed, the qRT-PCR results suggested that Fbxw7 might be the potential target gene of miR-223-3p, which was regulated by F63 (lncRNA). It was reported that in Cryptosporidium parvum infection, cirs-7 was upregulated and promoted C. parvum propagation by regulating the miR-1270-RelA axis, which provided a control strategy against C. parvum infection (Yin et al., 2021). Therefore, elucidating the regulatory mechanism of F63-miR-223-3p-Fbxw7 may provide a potential target for the development of anti-E. multilocularis drugs. Another gene enriched in HCs, epidermal growth factor receptor (EGFR), was found to be highly expressed and was predicted to participate in the PI3K/AKT signaling pathway in 3 m p.i., suggesting an essential role in host defense against the infection. It is worth mentioning that EGFR was located upstream within 10 kb of GM39584 (lncRNA). Interestingly, their expression trend was consistent, suggesting that GM39584 play a cis-regulation role. It is worth exploring whether and how the EGFR-GM39584 axis plays a role during E. multilocularis infection.
For HSCs, we focused on the expression patterns of fibrosis-related genes, such as α-SMA, Col1α1 and Col1α3. As expected, E. multilocularis infection induced these genes to be upregulated, suggesting the tendency of liver fibrosis. Besides, Col1α1, one of the target genes of F63, was most enriched in HSCs. F63 acts as a competing endogenous RNA (ceRNA) for the miR-1231-5p/Col1α1 axis, involved in regulating post-spinal cord injury pyroptosis by activating the PI3K/AKT pathway (Xu et al., 2020). It is therefore hypothesized that F63 regulate the expression of Col1α1 indirectly and be involved in the apoptosis of hepatocytes. By analyzing the DEgenes of HSCs at 2 m p.i. and 3 m p.i., we found the inflammation-related pathways were enriched significantly, including cytokine-cytokine receptor interaction, chemokine signaling pathway, IL-17 signaling pathway, NF-kappa B signaling pathway, T cell receptor signaling pathway and Th1, Th2 and Th17 cell differentiation. Consistently, IFN-γ, IL-4 and IL-12 were remarkable upregulated in HSCs at 1 m p.i., 2 m p.i. and 3m p.i, suggesting that HSCs may be involved in immune regulation together with KCs. However, the level of IFN-γ in HSCs was higher than that of KCs and its expression trend in both cells was just opposite during the course of infection. Previous studies identified IFNgAS1 involved in IFN-γ-mediated host defense as an important regulator of IFN-γ expression and AW112010 promoted pro-inflammatory pathways by suppressing IL10 expression, which reduced the number of IFN-γ expression after knocked the expression of AW112010 (Petermann et al., 2019; Peng et al., 2020; Yang et al., 2020). In this work, the expression of IFN-γ in HSCs and KCs was positively correlated with AW112010 and IFNgAS1. Considering the similar expression pattern between IFNgAS1, AW112010 and IFN-γ, we guess that the similar role be played in HSCs after E. multilocularis infection. Therefore, whether the expression of IFN-γ is regulated by both AW112010 and IFNgAS1 is worthy of further exploration. Besides, it was reported that miR-155 positively regulates IFN-γ expression via the Tim-3 pathway in NK cells, and miR-29b/142-5p also induces IFN-γ upregulation by targeting DNMTs (Cheng et al., 2015; Yang Y. et al., 2018). In our work, we found that both miR-155 and miR-29b were downregulated expressed in HSCs at 3 m p.i., which provides a clue for investigation of the mechanism of IFN-γ regulation.
It is known that the liver resident macrophages, KCs, are the first line of defense against inflammation/infections in the liver. It has been shown that the activation of vitamin D receptor (VDR), expressed on KCs, decreases hepatic inflammation in diet-induced model of NASH (Dong et al., 2020). In this study, the expression of VDR was downregulated continuously, suggesting that KCs may be active against E. multilocularis infection. Previous studies identified that miR-125b and miR-351-5p involved in the pathological process by targeting VDR (Mohri et al., 2009; He et al., 2018). It is possible that miR-125b and miR-351-5p may play a similar regulatory role in KCs. Moreover, the expression of IFN-γ was decreased, while IL-4 and IL-10 were increased in KCs, implying that E. multilocularis infection cause a Th2 immune response rather than Th1 immune response. Additionally, the KEGG pathway analysis revealed that the DEgenes were significantly enriched in the IL-17 signaling pathway, TNF signaling pathway and cytokine-cytokine receptor interaction. We found some metabolism-related pathways were also enriched, such as ascorbate and aldarate metabolism, steroid hormone biosynthesis, glycine, serine and threonine metabolism, tryptophan metabolism, biosynthesis of secondary metabolites, arginine biosynthesis and fatty acid biosynthesis, indicating that KCs also play a role in metabolism regulation during E. multilocularis infection.
In summary, the present study revealed the transcriptomic maps of mRNA, miRNAs and lncRNAs expressed in HCs, HSCs and KCs during E. multilocularis infection. Additionally, by integrate analyzing the RNA-seq data, we found some potential regulatory axis, such as F63-miR-223-3p-Fbxw7, GM39584-EGFR, F63-miR-1231-5p-Col1α1, IFNgAS1/AW112010-IFN-γ, and miR-125b/miR-351-5p-VDR. The future studies need to insight into clarify these regulatory axis and provide potential treatment targets for AEs.
Data Availability Statement
The original contributions presented in the study are publicly available. This data can be found here: PRJNA732233 and PRJNA770143.
Ethics Statement
The animal study was reviewed and approved by Animal Ethics Committee of Lanzhou Veterinary Research Institute, Chinese Academy of Agricultural Sciences.
Author Contributions
TL performed the animal experiments and analyzed the data. TL and YZ performed RNA-seq analysis and analyzed the data. TL wrote portions of the manuscript draft. HL, YL, LW, GC, GP, and XG participated in some experiments and data analysis. TL, YZ, and XL designed the experiments, analyzed the data, and revised the manuscript. WC, MH, YZ, and XL oversaw the overall execution of the project and gave final approval of the version. All authors contributed to the article and approved the submitted version.
Funding
This research was funded by the National Natural Science Foundation of China (No. 32072889).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2022.798551/full#supplementary-material
Supplementary Figure 1 | The expression of cell markers in HCs, HSCs and KCs.
Supplementary Figure 2 | Comparisons of DElncRNAs, DEmRNAs and DEmiRNAs differentially expressed in HSCs at 2 m p.i. and 3 m p.i.. (A) The number of DElncRNAs, DEmRNAs and DEmiRNAs at two infection stages. Sky blue represents the total number of DEgemes, red and violet represents upregulated and downregulated genes, respectively. (B) Veen diagrams showing the common and unique DElncRNAs, DEmRNAs and DEmiRNAs at two infection stages.
Supplementary Figure 3 | Comparisons of DElncRNAs, DEmRNAs and DEmiRNAs differentially expressed in KCs at 2 m p.i. and 3 m p.i.. (A) The number of DElncRNAs, DEmRNAs and DEmiRNAs at two infection stages. Sky blue represents the total number of DEgemes, red and purple represents upregulated and downregulated genes, respectively. (B) Veen diagrams showing the common and unique DElncRNAs, DEmRNAs and DEmiRNAs at two infection stages.
Supplementary Figure 4 | The top twenty enriched biological process and pathways of DElncRNAs and DEmRNAs in HSCs at 2 m p.i. and 3 m p.i.. (A) Predicted biological process terms of DElncRNAs and DEmRNAs in HSCs at 2 m p.i.. (B) Predicted biological process terms of DElncRNAs and DEmRNAs in HSCs at 3 m p.i.. (C) Predicted pathways of DElncRNAs and DEmRNAs in HSCs at 2 m p.i.. (D) Predicted pathways of DElncRNAs and DEmRNAs in HSCs at 3 m p.i..
Supplementary Figure 5 | The top twenty enriched biological process and pathways of DElncRNAs and DEmRNAs in KCs at 2 m p.i. and 3 m p.i.. (A) Predicted biological process terms of DElncRNAs and DEmRNAs in KCs at 2 m p.i.. (B) Predicted biological process terms of DElncRNAs and DEmRNAs in KCs at 3 m p.i.. (C) Predicted pathways of DElncRNAs and DEmRNAs in KCs at 2 m p.i.. (D) Predicted pathways of DElncRNAs and DEmRNAs in KCs at 3 m p.i..
References
Alkan, A. H., and Akgül, B. (2022). Endogenous miRNA Sponges. Methods Mol. Biol. 2257, 91–104. doi:10.1007/978-1-0716-1170-8_5
Almalki, E., Al-Shaebi, E. M., Al-Quarishy, S., El-Matbouli, M., and Abdel-Baki, A.-A. S. (2017). In Vitro effectiveness of Curcuma Longa and Zingiber Officinale Extracts on Echinococcus Protoscoleces. Saudi J. Biol. Sci. 24, 90–94. doi:10.1016/j.sjbs.2016.05.007
Balmer, J. E., and Blomhoff, R. (2002). Gene Expression Regulation by Retinoic Acid. J. Lipid Res. 43, 1773–1808. doi:10.1194/jlr.r100015-jlr200
Broermann, A., Schmid, R., Gabrielyan, O., Sakowski, M., Eisele, C., Keller, S., et al. (2020). Exosomal miRNAs as Potential Biomarkers to Monitor Phosphodiesterase 5 Inhibitor Induced Anti-fibrotic Effects on CCl4 Treated Rats. Ijms 22, 382. doi:10.3390/ijms22010382
Chen, G., Wang, L., Liu, T., Li, Y., Zhang, S., Li, H., et al. (2021). Identification and Expression Profiling of Circulating MicroRNAs in Serum of Cysticercus Pisiformis-Infected Rabbits. Genes 12, 1591. doi:10.3390/genes12101591
Cheng, Y. Q., Ren, J. P., Zhao, J., Wang, J. M., Zhou, Y., Li, G. Y., et al. (2015). MicroRNA-155 Regulates Interferon-Γproduction in Natural Killer Cells via Tim-3 Signalling in Chronic Hepatitis C Virus Infection. Immunology 145, 485–497. doi:10.1111/imm.12463
Chu, X., Zheng, W., Wang, J., Zhang, J., Pan, Y., and Shao, C. (2020). CDK6 Inhibition Targeted by miR-378a-3p Protects against Intestinal Injury Induced by Ionizing Radiation. Biochem. Biophysical Res. Commun. 531, 328–334. doi:10.1016/j.bbrc.2020.07.093
Coffey, A. R., Kanke, M., Smallwood, T. L., Albright, J., Pitman, W., Gharaibeh, R. Z., et al. (2019). microRNA-146a-5p Association with the Cardiometabolic Disease Risk Factor TMAO. Physiol. Genomics 51, 59–71. doi:10.1152/physiolgenomics.00079.2018
Conraths, F. J., Probst, C., Possenti, A., Boufana, B., Saulle, R., LaTorre, G., et al. (2017). Potential Risk Factors Associated with Human Alveolar Echinococcosis: Systematic Review and Meta-Analysis. Plos Negl. Trop. Dis. 11, e0005801. doi:10.1371/journal.pntd.0005801
Craig, P. S., Hegglin, D., Lightowlers, M. W., Torgerson, P. R., and Wang, Q. (2017). Echinococcosis. Adv. Parasitol. 96, 55–158. doi:10.1016/bs.apar.2016.09.002
Craig, P. S., McManus, D. P., Lightowlers, M. W., Chabalgoity, J. A., Garcia, H. H., Gavidia, C. M., et al. (2007). Prevention and Control of Cystic Echinococcosis. Lancet Infect. Dis. 7, 385–394. doi:10.1016/S1473-3099(07)70134-2
Czermak, B. V., Akhan, O., Hiemetzberger, R., Zelger, B., Vogel, W., Jaschke, W., et al. (2008). Echinococcosis of the Liver. Abdom. Imaging 33, 133–143. doi:10.1007/s00261-007-9331-0
Dong, B., Zhou, Y., Wang, W., Scott, J., Kim, K., Sun, Z., et al. (2020). Vitamin D Receptor Activation in Liver Macrophages Ameliorates Hepatic Inflammation, Steatosis, and Insulin Resistance in Mice. Hepatology 71, 1559–1574. doi:10.1002/hep.30937
Du, Y., Yang, Y. T., Tang, G., Jia, J. S., Zhu, N., and Yuan, W. J. (2020). Butyrate Alleviates Diabetic Kidney Disease by Mediating the miR‐7a‐5p/P311/TGF‐β1 Pathway. FASEB j. 34, 10462–10475. doi:10.1096/fj.202000431R
Eckert, J., and Deplazes, P. (2004). Biological, Epidemiological, and Clinical Aspects of Echinococcosis, a Zoonosis of Increasing Concern. Clin. Microbiol. Rev. 17, 107–135. doi:10.1128/CMR.17.1.107-135.2004
Gajeton, J., Krukovets, I., Yendamuri, R., Verbovetskiy, D., Vasanj,i, A., Sul, L., et al. (2021). miR‐467 Regulates Inflammation and Blood Insulin and Glucose. J. Cell. Mol. Medi 25, 2549–2562. doi:10.1111/jcmm.16224
Gottstein, B., Stojkovic, M., Vuitton, D. A., Millon, L., Marcinkute, A., and Deplazes, P. (2015). Threat of Alveolar Echinococcosis to Public Health - a challenge for Europe. Trends Parasitol. 31, 407–412. doi:10.1016/j.pt.2015.06.001
Han, Y., Zhang, J., Huang, S., Cheng, N., Zhang, C., Li, Y., et al. (2021). MicroRNA-223-3p Inhibits Vascular Calcification and the Osteogenic Switch of Vascular Smooth Muscle Cells. J. Biol. Chem. 296, 100483. doi:10.1016/j.jbc.2021.100483
He, X., Sun, Y., Lei, N., Fan, X., Zhang, C., Wang, Y., et al. (2018). MicroRNA-351 Promotes Schistosomiasis-Induced Hepatic Fibrosis by Targeting the Vitamin D Receptor. Proc. Natl. Acad. Sci. USA 115, 180–185. doi:10.1073/pnas.1715965115
Huang, G. Q., Yexie, Z. H., Guo, Y. M., and Zhao, S. Y. (2021). Treatment of End-Stage Hepatic Alveolar Echinococcosis in Qinghai: Current Status and Future Perspectives. Chin. J. Clinicians (Electronic Edition) 15, 209–212.
Kallen, A. N., Zhou, X.-B., Xu, J., Qiao, C., Ma, J., Yan, L., et al. (2013). The Imprinted H19 lncRNA Antagonizes Let-7 microRNAs. Mol. Cell 52, 101–112. doi:10.1016/j.molcel.2013.08.027
Khomich, O., Ivanov, A. V., and Bartosch, B. (2019). Metabolic Hallmarks of Hepatic Stellate Cells in Liver Fibrosis. Cells 9, 24. doi:10.3390/cells9010024
Kinoshita, C., Kikuchi-Utsumi, K., Aoyama, K., Suzuki, R., Okamoto, Y., Matsumura, N., et al. (2021). Inhibition of miR-96-5p in the Mouse Brain Increases Glutathione Levels by Altering NOVA1 Expression. Commun. Biol. 4, 182. doi:10.1038/s42003-021-01706-0
Kisseleva, T., Cong, M., Paik, Y., Scholten, D., Jiang, C., Benner, C., et al. (2012). Myofibroblasts Revert to an Inactive Phenotype during Regression of Liver Fibrosis. Proc. Natl. Acad. Sci. 109, 9448–9453. doi:10.1073/pnas.1201840109
Kolářová, L., Matějů, J., Hrdý, J., Kolářová, H., Hozáková, L., Žampachová, V., et al. (2015). Human Alveolar Echinococcosis, Czech Republic, 2007-2014. Emerg. Infect. Dis. 21, 2263–2265. doi:10.3201/eid2112.150743
Kotwa, J. D., Isaksson, M., Jardine, C. M., Campbell, G. D., Berke, O., Pearl, D. L., et al. (2019). Echinococcus Multilocularis Infection, Southern Ontario, Canada. Emerg. Infect. Dis. 25, 265–272. doi:10.3201/eid2502.180299
Li, Q., Li, H., Liang, J., Mei, J., Cao, Z., Zhang, L., et al. (2021). Sertoli Cell‐derived Exosomal MicroRNA‐486‐5p Regulates Differentiation of Spermatogonial Stem Cell through PTEN in Mice. J. Cell Mol Med 25, 3950–3962. doi:10.1111/jcmm.16347
Li, X., Rui, B., Cao, Y., Gong, X., and Li, H. (2020). Long Non-coding RNA LINC00152 Acts as a Sponge of miRNA-193b-3p to Promote Tongue Squamous Cell Carcinoma Progression. Oncol. Lett. 19, 2035–2042. doi:10.3892/ol.2020.11293
Liang, L., Su, W., Zhou, L., Cao, Y., Zhou, X., Liu, S., et al. (2020). Statin Downregulation of miR-652-3p Protects Endothelium from Dyslipidemia by Promoting ISL1 Expression. Metabolism 107, 154226. doi:10.1016/j.metabol.2020.154226
Liu, C.-N., Xu, Y.-Y., Cadavid-Restrepo, A. M., Lou, Z.-Z., Yan, H.-B., Li, L., et al. (2018). Estimating the Prevalence of Echinococcus in Domestic Dogs in Highly Endemic for Echinococcosis. Infect. Dis. Poverty 7, 77. doi:10.1186/s40249-018-0458-8
Maggiore, M., Pensel, P. E., Denegri, G., and Elissondo, M. C. (2015). Chemoprophylactic and Therapeutic Efficacy of Thymol in Murine Cystic Echinococcosis. Parasitol. Int. 64, 435–440. doi:10.1016/j.parint.2015.06.005
Massolo, A., Klein, C., Kowalewska-Grochowska, K., Belga, S., MacDonald, C., Vaughan, S., et al. (2019). European Echinococcus Multilocularis Identified in Patients in Canada. N. Engl. J. Med. 381, 384–385. doi:10.1056/NEJMc1814975
Mederacke, I., Dapito, D. H., Affò, S., Uchinami, H., and Schwabe, R. F. (2015). High-yield and High-Purity Isolation of Hepatic Stellate Cells from normal and Fibrotic Mouse Livers. Nat. Protoc. 10, 305–315. doi:10.1038/nprot.2015.017
Mohammadi, M., Spotin, A., Mahami-Oskouei, M., Shanehbandi, D., Ahmadpour, E., Casulli, A., et al. (2021). MicroRNA-365 Promotes Apoptosis in Human Melanoma Cell A375 Treated with Hydatid Cyst Fluid of Echinococcus Granulosus Sensu Stricto. Microb. Pathogenesis 153, 104804. doi:10.1016/j.micpath.2021.104804
Mohri, T., Nakajima, M., Takagi, S., Komagata, S., and Yokoi, T. (2009). MicroRNA Regulates Human Vitamin D Receptor. Int. J. Cancer 125, 1328–1333. doi:10.1002/ijc.24459
Mueller, M. C., Marx, M., Peyerl-Hoffmann, G., and Kern, W. V. (2020). Spatial Distribution and Incidence Trend of Human Alveolar Echinococcosis in Southwest Germany: Increased Incidence and Urbanization of the Disease? Infection 48, 923–927. doi:10.1007/s15010-020-01479-4
Myant, K. B., Cammareri, P., McGhee, E. J., Ridgway, R. A., Huels, D. J., Cordero, J. B., et al. (2013). ROS Production and NF-Κb Activation Triggered by RAC1 Facilitate WNT-Driven Intestinal Stem Cell Proliferation and Colorectal Cancer Initiation. Cell Stem Cell. 12, 761–773. doi:10.1016/j.stem.2013.04.006
Onoyama, I., Suzuki, A., Matsumoto, A., Tomita, K., Katagiri, H., Oike, Y., et al. (2011). Fbxw7 Regulates Lipid Metabolism and Cell Fate Decisions in the Mouse Liver. J. Clin. Invest. 121, 342–354. doi:10.1172/JCI40725
Ota, H., Ito-Matsuoka, Y., and Matsui, Y. (2019). Identification of the X-Linked Germ Cell Specific miRNAs (XmiRs) and Their Functions. PLoS One 14, e0211739. doi:10.1371/journal.pone.0211739
Peng, H., Ren, S., Liu, Y., Zhou, H., Tang, X., Yang, J., et al. (2020). Elevated Expression of the Long Noncoding RNA IFNG-AS1 in the Peripheral Blood from Patients with Rheumatoid Arthritis. J. Immunol. Res. 2020, 6401978. doi:10.1155/2020/6401978
Pensel, P. E., Elissondo, N., Gambino, G., Gamboa, G. U., Benoit, J. P., and Elissondo, M. C. (2017). Experimental Cystic Echinococcosis Therapy: In Vitro and In Vivo Combined 5-fluorouracil/albendazole Treatment. Vet. Parasitol. 245, 62–70. doi:10.1016/j.vetpar.2017.08.011
Petermann, F., Pękowska, A., Johnson, C. A., Jankovic, D., Shih, H.-Y., Jiang, K., et al. (2019). The Magnitude of IFN-γ Responses Is Fine-Tuned by DNA Architecture and the Non-coding Transcript of Ifng-As1. Mol. Cell 75, 1229–1242. doi:10.1016/j.molcel.2019.06.025
Peters, L., Burkert, S., and Grüner, B. (2021). Parasites of the Liver - Epidemiology, Diagnosis and Clinical Management in the European Context. J. Hepatol. 75, 202–218. doi:10.1016/j.jhep.2021.02.015
Ren, G.-J., Fan, X.-C., Liu, T.-L., Wang, S.-S., and Zhao, G.-H. (2018). Genome-wide Analysis of Differentially Expressed Profiles of mRNAs, lncRNAs and circRNAs during Cryptosporidium Baileyi Infection. BMC Genomics 19, 356. doi:10.1186/s12864-018-4754-2
Roberts, R. A., Ganey, P. E., Ju, C., Kamendulis, L. M., Rusyn, I., and Klaunig, J. E. (2007). Role of the Kupffer Cell in Mediating Hepatic Toxicity and Carcinogenesis. Toxicol. Sci. 96, 2–15. doi:10.1093/toxsci/kfl173
Spiliotis, M., and Brehm, K. (2009). Axenic In Vitro Cultivation of Echinococcus Multilocularis Metacestode Vesicles and the Generationof Primary Cell Cultures. Methods Mol. Biol. 470, 245–262. doi:10.1007/978-1-59745-204-5_17
Tombolan, L., Millino, C., Pacchioni, B., Cattelan, M., Zin, A., Bonvini, P., et al. (2020). Circulating miR-26a as Potential Prognostic Biomarkers in Pediatric Rhabdomyosarcoma. Front. Genet. 11, 606274. doi:10.3389/fgene.2020.606274
Torabi, N., Dobakhti, F., Faghihzadeh, S., and Haniloo, A. (2018). In Vitro and In Vivo Effects of Chitosan-Praziquantel and Chitosan-Albendazole Nanoparticles on Echinococcus Granulosus Metacestodes. Parasitol. Res. 117, 2015–2023. doi:10.1007/s00436-018-5849-z
Wang, H., Thorling, C. A., Liang, X., Bridle, K. R., Grice, J. E., Zhu, Y., et al. (2015). Diagnostic Imaging and Therapeutic Application of Nanoparticles Targeting the Liver. J. Mater. Chem. B 3, 939–958. doi:10.1039/c4tb01611d
Wang, S., Ma, Y., Wang, W., Dai, Y., Sun, H., Li, J., et al. (2021a). Status and prospect of Novel Treatment Options toward Alveolar and Cystic Echinococcosis. Acta Trop. 226, 106252. doi:10.1016/j.actatropica.2021.106252
Wang, X., Chi, J., Dong, B., Xu, L., Zhou, Y., Huang, Y., et al. (2021b). MiR‐223‐3p and miR‐22‐3p Inhibit Monosodium Urate‐induced Gouty Inflammation by Targeting NLRP3. Int. J. Rheum. Dis. 24, 599–607. doi:10.1111/1756-185X.14089
Wang, X., Liu, J., Zuo, Q., Mu, Z., Weng, X., Sun, X., et al. (2018). Echinococcus Multilocularis and Echinococcus Shiquicus in a Small Mammal Community on the Eastern Tibetan Plateau: Host Species Composition, Molecular Prevalence, and Epidemiological Implications. Parasites Vectors 11, 302. doi:10.1186/s13071-018-2873-x
Woolsey, I. D., and Miller, A. L. (2021). Echinococcus Granulosus Sensu Lato and Echinococcus Multilocularis: A Review. Res. Vet. Sci. 135, 517–522. doi:10.1016/j.rvsc.2020.11.010
Xiang, P., Yeung, Y. T., Wang, J., Wu, Q., Du, R., Huang, C., et al. (2021). miR ‐17‐3p Promotes the Proliferation of Multiple Myeloma Cells by Downregulating P21 Expression through LMLN Inhibition. Int. J. Cancer 148, 3071–3085. doi:10.1002/ijc.33528
Xu, S., Wang, J., Jiang, J., Song, J., Zhu, W., Zhang, F., et al. (2020). TLR4 Promotes Microglial Pyroptosis via lncRNA-F630028O10Rik by Activating PI3K/AKT Pathway after Spinal Cord Injury. Cell Death Dis 11, 693. doi:10.1038/s41419-020-02824-z
Xu, Y., Wu, J., Yuan, X., Liu, W., Pan, J., and Xu, B. (2021). MicroRNA-155 Contributes to Host Immunity against Toxoplasma Gondii. Parasite 28, 83. doi:10.1051/parasite/2021082
Yan, X., Zeng, D., Zhu, H., Zhang, Y., Shi, Y., Wu, Y., et al. (2020). MiRNA-532-5p Regulates CUMS-Induced Depression-like Behaviors and Modulates LPS-Induced Proinflammatory Cytokine Signaling by Targeting STAT3. Ndt Vol. 16, 2753–2764. doi:10.2147/NDT.S251152
Yang, Q., Wan, Q., Zhang, L., Li, Y., Zhang, P., Li, D., et al. (2018a). Analysis of LncRNA Expression in Cell Differentiation. RNA Biol. 15, 413–422. doi:10.1080/15476286.2018.1441665
Yang, X., Bam, M., Becker, W., Nagarkatti, P. S., and Nagarkatti, M. (2020). Long Noncoding RNA AW112010 Promotes the Differentiation of Inflammatory T Cells by Suppressing IL-10 Expression through Histone Demethylation. J.I. 205, 987–993. doi:10.4049/jimmunol.2000330
Yang, Y., Jin, Z., Dong, R., Zheng, C., Huang, Y., Zheng, Y., et al. (2018b). MicroRNA-29b/142-5p Contribute to the Pathogenesis of Biliary Atresia by Regulating the IFN-γ Gene. Cell Death Dis 9, 545. doi:10.1038/s41419-018-0605-y
Yin, Y.-L., Liu, T.-L., Yao, Q., Wang, Y.-X., Wu, X.-M., Wang, X.-T., et al. (2021). Circular RNA ciRS-7 Affects the Propagation of Cryptosporidium Parvum in HCT-8 Cells by Sponging miR-1270 to Activate the NF-Κb Signaling Pathway. Parasites Vectors 14, 238. doi:10.1186/s13071-021-04739-w
Zheng, B., Wang, H., Cui, G., Guo, Q., Si, L., Yan, H., et al. (2020). ERG-associated lncRNA (ERGAL) Promotes the Stability and Integrity of Vascular Endothelial Barrier during Dengue Viral Infection via Interaction with miR-183-5p. Front. Cel. Infect. Microbiol. 10, 477. doi:10.3389/fcimb.2020.00477
Keywords: Echinococcus multilocularis, hepatic cells, hepatic stellate cells, kupffer cells, regulatory network
Citation: Liu T, Li H, Li Y, Wang L, Chen G, Pu G, Guo X, Cho WC, Fasihi Harandi M, Zheng Y and Luo X (2022) Integrative Analysis of RNA Expression and Regulatory Networks in Mice Liver Infected by Echinococcus multilocularis. Front. Cell Dev. Biol. 10:798551. doi: 10.3389/fcell.2022.798551
Received: 20 October 2021; Accepted: 17 February 2022;
Published: 24 March 2022.
Edited by:
Natalia Martins Feitosa, Federal University of Rio de Janeiro, BrazilReviewed by:
Deborah Stroka, University of Bern, SwitzerlandMaude Giroud, Ludwig Maximilian University of Munich, Germany
Copyright © 2022 Liu, Li, Li, Wang, Chen, Pu, Guo, Cho, Fasihi Harandi, Zheng and Luo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xuenong Luo, bHVveHVlbm9uZ0BjYWFzLmNu; Yadong Zheng, emhlbmd5YWRvbmdAemFmdS5lZHUuY24=
 Tingli Liu
Tingli Liu Hong Li1
Hong Li1 Guoliang Chen
Guoliang Chen Xiaola Guo
Xiaola Guo William C. Cho
William C. Cho Majid Fasihi Harandi
Majid Fasihi Harandi Yadong Zheng
Yadong Zheng Xuenong Luo
Xuenong Luo