- 1Department of General Surgery, The First Affiliated Hospital of Nanchang University, Nanchang University, Nanchang, China
- 2Imaging Department, The First Affiliated Hospital of Nanchang University, Nanchang University, Nanchang, China
Circular RNAs (circRNAs) are non-coding RNAs (ncRNAs) without 5′ caps and 3′ tails, which are formed from precursor mRNAs (pre-mRNAs) that are inversely back-spliced by exons. CircRNAs are characterized by a covalently closed circular structure and are abundantly expressed in eukaryotic cells. With the development of RNA-sequencing, it was discovered that circRNAs play important roles in the regulation of numerous human genes and are related to the occurrence, development, and prognosis of diseases. Studies in various cancers have revealed that circRNAs have both positive and negative effects on the occurrence and development of tumors. Circ-ABCB10, a circular RNA originating from exons of ABCB10 located on chromosome 1q42, has been proven to play an important role in different types of cancers. Here, we report the primary findings of recent research studies by many contributors about the roles of circ-ABCB10 in cancer and clearly formulate its influence and functions in different aspects of cancer biology, which gives us a broad picture of circ-ABCB10. Thus, this study aimed to generalize the roles of circ-ABCB10 in the diagnosis and treatment of different types of tumors and its related miRNA genes. In this way, we wish to provide a sufficient understanding and assess the future development direction of the research on circ-ABCB10.
Introduction
Circular RNAs (circRNAs) are a class of endogenous single-stranded closed circular RNAs formed by reverse splicing and covalent binding without a 5′ cap and 3′ poly (a) tail (Sanger et al., 1976; Chen, 2016; Pamudurti et al., 2017; Kristensen et al., 2018; Qu et al., 2018). CircRNAs were first discovered in RNA viruses in 1976 (Sanger et al., 1976) and were initially considered “splicing noise” in organisms but have become a research hotspot in the meantime (Kristensen et al., 2018). With the rapid development of RNA-sequencing technology, many circRNAs have been discovered. To date, more than 30,000 cyclic RNAs with unique structures have been identified and have attracted increasing attention. Most circRNAs are evolutionarily conserved across species (Pamudurti et al., 2017). CircRNAs can originate from introns, exons, or from both introns and exons (Su et al., 2019; Yu and Kuo, 2019). Because circRNAs have a specific closed-ring structure, they are more resistant to exonucleases than linear RNAs (Salzman et al., 2013; Shang et al., 2019). In addition, most circRNAs are usually found in the cytoplasm and are derived from protein-coding genes. These genes contain one or more exons toward the 5′ cap of the gene, with long introns on both sides (Dong et al., 2017a; Nicolet et al., 2018). The long introns containing the wing region will become circRNAs, which usually contain specific sequences (Qu et al., 2015). They can induce the formation of circRNAs by mutually complementing with circRNA promoters and are usually expressed in a cell type– or tissue-specific manner (Patop and Kadener, 2018; Shi et al., 2020). Moreover, it was found that circRNAs are abnormally expressed in colorectal cancer (CRC) and osteoarthritis (Bachmayr-Heyda et al., 2015; Yang et al., 2020a). Thus, circRNAs are widely expressed in various human cell types and will perhaps become a new direction in research on disease biomarkers for aging (Bahn et al., 2015) and therapy (Hou and Zhang, 2017; Tang et al., 2017). According to gene structures and their specific molecular cycling mechanisms, circRNAs are divided into four types: exonic circRNAs (ecRNAs) (Salzman et al., 2012), circular intronic RNAs (ciRNAs) (Zhang et al., 2013), exon–intron circRNAs (eIciRNAs) (Li et al., 2015), and intergenic circRNAs (Tang and Hann, 2020). Generally speaking, ecRNAs are mainly found in the cytoplasm (Memczak et al., 2013) and regulate the expression of genes. CiRNAs and eIciRNAs tend to be localized in the nucleus and play a significant role in regulating parental genes.
Studies have shown that circRNAs produced via back-splicing feature different biogenesis than typical splicing of linear RNA. First, acting as miRNA sponges, circRNAs are more likely to bind to other miRNAs and are known as “super sponges” (Hansen et al., 2013). According to previous reports, circRNAs can inhibit miRNAs from binding to their target genes (Thomas and Sætrom, 2014). Second, by binding to proteins and RNA-binding proteins (RBPs), circRNAs can affect their function and interaction with other proteins (Zang et al., 2020). Studies demonstrate that RBPs can also regulate the formation of circRNAs by forming RNA–protein complexes (RPCs) (Conn et al., 2015). Third, circRNAs retained in the nucleus can regulate alternative splicing, transcription, or translation (Li et al., 2018a; Guarnerio et al., 2019). For example, Circ-ubr5 might undergo a specific RNA–RNA interaction by binding to the splicing regulator QKI(Qin et al., 2018). CircITGA7 could increase the transcriptional expression of integrin alpha 7 (ITGA7) by inhibiting RAS-responsive element-binding protein 1 (RREB1) (Li et al., 2018a).
Moreover, circRNAs can function as autophagy regulators, affecting tumorigenesis. For example, Circ_104075 was found to act as an autophagy regulator in glioma cells (Chi et al., 2019). In general, the abnormal expression of circRNAs is associated with the occurrence and progression of human cancer by affecting the growth, migration, invasion, proliferation, and other pathological processes of cells (Conn et al., 2015; Khan et al., 2016; Dong et al., 2019). In addition, circRNAs are also associated with clinicopathological characteristics, such as lymph node metastasis, differentiation, or distant metastasis. All of these findings provide the basis for potential biomarkers and therapeutic targets in the diagnosis and treatment of human cancers. In addition, circRNAs can act as a protein sponge to adsorb one or more proteins through binding sites, thus acting as a protein scaffold to directly mediate protein–protein interactions and regulate gene expression. Recent studies have found that the abnormal expression of Circ-ABCB10, also known as hsa_circ_000871, may be involved in the occurrence and development of many different tumors, such as esophageal squamous carcinoma cells, glioma, non–small cell lung cancer, oral squamous cell carcinoma, lung cancer, epithelial ovarian cancer, breast cancer, thyroid cancer, and hepatocellular carcinoma (Cortés-López and Miura, 2016; Wang et al., 2017). Thus, it is significant to summarize the function of Circ-ABCB10 in the occurrence and progression of human cancers.
The Features and Biological Functions of Circ-ABCB10
Circ-ABCB10 originates from exons 2 and 3 of the ABCB10 gene, located on chromosome 1 (Duan et al., 2020). The antisense strand of Circ-ABCB10 undergoes back-splicing of the 5′ and 3′ ends to form circular RNA (Figure 1) With specific expression in different developmental stages and tissues, Circ-ABCB10 was first reported to promote breast cancer proliferation and migration by sponging miR-1271 (Chen et al., 2019). It was also found that Circ-ABCB10 is highly expressed in human brain regions such as the forebrain, cerebellum, occipital lobe, frontal cortex, and parietal lobe (Memczak et al., 2013). Different from linear RNAs, circRNAs are single-chain circular RNAs without 5ʹ to 3ʹ polarity or a polyadenylated tail. This blocked structure makes circ-ABCB10 more resistant to RNA degradation. Due to these unique characteristics, circ-ABCB10 is related to several characteristics of cancers (Jayson et al., 2014). Some studies have demonstrated that circ-ABCB10 could stimulate tumor growth (Liang et al., 2017; Zhao et al., 2021) and insulin resistance (IR) (Lux and Bullinger, 2018; Ouyang et al., 2018). Furthermore, the high expression of circ-ABCB10 was closely related to the pathological grade and tumor lymph node metastasis stage (Luo et al., 2018). Thus, circ-ABCB10 may be a promising diagnostic and prognostic biomarker and a target for novel treatment strategies in the future.
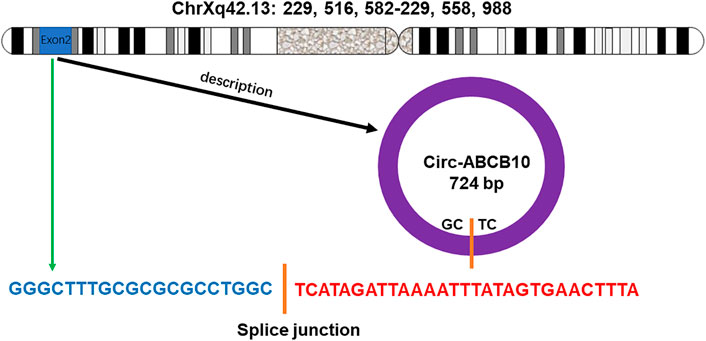
FIGURE 1. Gene encoding circ-ABCB10 is located on the chromosome Xq42.13. The antisense strand of circ-ABCB10 undergoes back-splicing of the 5′ and 3′ ends to form circular RNA. The green arrow indicates the “head-to-tail” splicing site of circ-ABCB10.
Circ-ABCB10 Functions as a miRNA Sponge
Like lncRNAs, the main role of circRNAs in molecular regulation is to act as a “sponge” to absorb functional miRNA to decrease their abundance in the cytoplasm and thereby regulate gene expression (Dong et al., 2017b; Li et al., 2017). Among the target miRNAs that can be regulated by circRNAs, it was demonstrated that miR-1252 can provide potential biomarkers and therapeutic targets in non–small cell lung cancer (Tian et al., 2019) and epithelial ovarian cancer (Chen et al., 2019). Similarly, it was also found that circ-ABCB10 can sponge miR-1271, which may have a complementary sequence, possibly regulating cell proliferation and migration of breast cancer and epithelial ovarian cancer cells (Liang et al., 2017; Lin et al., 2021; Zhao et al., 2021). Furthermore, since conserved miR-1271 target sites on circ-ABCB10 are complementary to miR-1271, these sites could be a lodging site for transport (Lin et al., 2021). In addition, circ-ABCB10 was reported to sponge miR-145-5p, affecting the miR-620/FABP5 axis, miR-1252 FOXR2, and miR-670-3p in oral squamous cell carcinoma, glioma, non–small cell lung cancer, and esophageal squamous cell carcinoma, respectively (Tian et al., 2019; Chen et al., 2020; Sun et al., 2020; Zhang et al., 2020). Furthermore, circ-ABCB10 was also found to promote glycolysis and colony formation by sponging miR-229-3p (Zhao et al., 2020). In addition, Circ-ABCB10 can promote angiogenesis via the microRNA-29b-3p/vascular endothelial growth factor A-axis (Tang et al., 2020) (Figure 2).
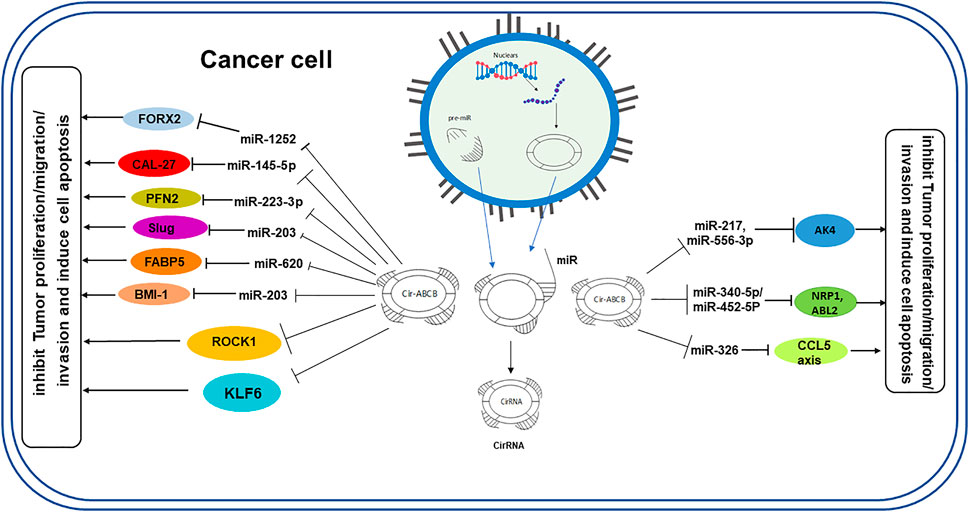
FIGURE 2. Overview of circ-ABCB10 function as a miRNA sponge in the proliferation and metastasis of various cancer cell types. Circ-ABCB10 is released from the nucleus, acting as a sponge of miRNAs that regulate target genes to promote or inhibit tumor growth.
The Roles of Circ-ABCB10 in Cancer Progression
With the progress of circRNA research, relevant studies demonstrated that Circ-ABCB10 is abnormally expressed in various cancers, such as esophageal squamous cell carcinoma (Zhang et al., 2020) and esophageal cancer (Wang et al., 2020). The upregulation of circ-ABCB10 was found to promote cell proliferation in various tumors, including esophageal squamous cell carcinoma (Zhang et al., 2020), esophageal cancer (Wang et al., 2020), glioma (Sun et al., 2020), non–small cell lung cancer (Tian et al., 2019), and oral squamous cell carcinoma (Chen et al., 2020). However, the overexpression of circ-ABCB10 was also found to inhibit the migration and invasion of lung cancer cells (Hu et al., 2020; Wu et al., 2020), hepatocellular carcinoma cells (Yang et al., 2020b), and rectal cancer cells (Xian et al., 2020). Generally speaking, these studies indicate that the expression of Circ-ABCB10 is dynamically regulated in tumor progression. The details of its diverse roles are summarized in Table 1.
1) In Proliferation and Invasion
It has been reported that circ-ABCB10 participates in tumor proliferation and migration by sponging several miRNAs via various transmitting pathways. It has been found that circ-ABCB10 can sponge miR-1252 and miR-584-5p in non–small cell lung cancer cells and accordingly stimulate the expression of the downstream targeted genes FOXR2 and E2F5 (He et al., 2018). It is worth saying that in the previous study, miR-584-5p exerts a promoting effect on gastric cancer. Thus, we can speculate that circ-ABCB10 may sponge different miRNAs to regulate the same tumor growth. At the same time, circ-ABCB10 can sponge miR-620 and upregulate the expression of the FABP5 axis to promote tumor growth and proliferation in glioma (Sun et al., 2020). In addition, circ-ABCB10 can sponge miR-203 and promote the proliferation of EC cells (Wang et al., 2020). MiR-203 can suppress slug/E-cadherin signals to inhibit cell invasion (Gao et al., 2017a). In addition, circ-ABCB10 can also promote the function of Bmi-1 by sponging miR-203 and enhance the tumor growth in osteosarcoma (Zhou et al., 2018). It indicates that there may be a correlation between circ-ABCB10 and miR-203 in the proliferation of tumor. Furthermore, the overexpression of circ-ABCB10 can stimulate tumor proliferation and invasion by sponging miR-1271, which is also a pivotal miRNA in most regulation processes of circRNAs in breast cancer (Wang et al., 2018) and promote the proliferation and progression of clear cell renal cell carcinoma by activating the target gene—miR-331-3p/miR-1228-5p (Huang et al., 2019). The interesting thing is that in rectal cancer, the number of circ-ABCB10 is decreasing but has an effect of inhibiting ferroptosis and apoptosis by regulating miR-326/CCL5 (Xian et al., 2020). This illustrates that the level of circ-ABCB10 can also have an influence on the proliferation and invasion of tumor. However, in hepatocellular carcinoma, in vivo experiments indicate that circ-ABCB10 may be an anti-oncogenic factor (Gao et al., 2017a). With the upregulation of circ-ABCB10, the downstream targeted genes of the signaling axis, namely, NRP1 and ABL2 are upregulated. In this case, it can easily be supposed that circ-ABCB10 can regulate the expression of multiple downstream genes, which has a synergistic effect on tumor growth. Coincidentally, circ-ABCB10 can also regulate the proliferation and EMT of cervical cancer by inhibiting the miR-128-3p/ZEB1 axis (Feng et al., 2021). In general, circ-ABCB10 can act as an oncogene by promoting tumor proliferation and migration in glioma, non–small cell lung cancer, esophageal cancer, breast cancer, and renal cancer (Tang et al., 2017; Deng et al., 2018; Wang et al., 2018; Wang et al., 2020; Zhao et al., 2020) and can also be an anti-oncogene by inhibiting the growth in hepatocellular carcinoma and cervical cancer (Gao et al., 2017a; Feng et al., 2021). It was also shown that circ-ABCB10 plays an obvious role in the proliferation and migration of tumors, with different effects on different cancers.
2) In Metastasis
Metastasis is an important step in cancer progression, and circ-ABCB10 can influence cancer metastasis in different ways (Gao et al., 2017b; Liang et al., 2017; Tang et al., 2017; Zhao et al., 2021). In oral squamous cell carcinoma, circ-ABCB10 was found to be upregulated and related to metastasis and tumor clinical staging of oral squamous cell carcinoma (OSCC) patients, aggravating the progression of OSCC by sponging miR-145-5p (Chen et al., 2020). In breast cancer, circ-ABCB10 acts as a miR-223-3p sponge and regulates the expression of the miR-223-3p targeted gene PFN2 to induce cell migration (Zhao et al., 2021). In addition, it can also promote invasion by sponging miR-1271 and reducing IR sensitivity (Tang et al., 2017). Similarly, the expression of ROCK1 is upregulated and is correlated with circ-ABCB10 expression in NPC cells (Liang et al., 2017). ROCK1 was upregulated following the overexpression of circ-ABCB10 in NPC (Liang et al., 2017). It is demonstrated that circ-ABCB10 has a positive role in cancer metastasis. Similar to proliferation and invasion, circ-ABCB10 also has an adverse effect on tumor metastasis. It is also seen in hepatocellular carcinoma that circ-ABCB10 can suppress the migration and metastasis by inhibiting the miR-340-5p/miR-452-5P—NRP1/ABL2 axis (Yang et al., 2020b). Thus, circ-ABCB10 plays a key role in tumor metastasis in different cancers.
3) In Angiogenesis
Angiogenesis, the creation of new blood vessels, plays a significant role in bone regeneration and osteoblast differentiation and provides essential nutrients and oxygen during bone formation (Hu et al., 2013; Kusumbe et al., 2014). Recently, it was found that circRNAs may be involved in the progression of angiogenesis during the occurrence and development of disease (Zheng et al., 2016; Zhong et al., 2016; Gao et al., 2017b; Deng et al., 2018; He et al., 2018). For example, there is evidence that circ-IARS in pancreatic cancer cells can influence human umbilical vein endothelial cell (HUVEC) monolayers to promote tumor metastasis (Li et al., 2018b). This fact indicates that circ-ABCB10 may also influence the angiogenesis of HUVECs. Vascular endothelial growth factor (VEGF) is another significant cytokine that promotes angiogenesis (Siveen et al., 2017). Kim et al. found that human amnion–derived mesenchymal stem cells (hAMSCs) can promote angiogenesis (Kim et al., 2012), and further research confirmed that hAMSCs can significantly increase the expression of VEGF and circ-ABCB10 (Tang et al., 2020). This implies that circ-ABCB10 may promote cell growth through related pathways. A series of experiments demonstrated that circ-ABCB10 can enhance the angiogenesis of HUVECs by sponging miR-29b-3p and VEGFA (Tang et al., 2020). It was also demonstrated that conditioned medium from hAMSCs (hAMSC-CM) can indirectly promote circ-ABCB10 transcription but not the release of hAMSC-CM by exosomes (Tang et al., 2020). However, there are still numerous questions related to the mechanisms through which circ-ABCB10 may promote angiogenesis.
Conclusion
In recent years, it has been found that circRNAs originate from the cyclization of pre-mRNAs and have many unique features not found in other RNAs. Because of their unique stable structure and tissue-specific expression, circRNAs have potential applications as novel biomarkers for assessing tumor progression (Jin et al., 2016; Zhang et al., 2017; Jakobi and Dieterich, 2019) and for the diagnosis and treatment of tumors (Lux and Bullinger, 2018; Ouyang et al., 2018). Increasing numbers of studies have found that circRNAs are abnormally expressed during tumorigenesis. In this article, we discussed and reviewed the role of circ-ABCB10 in different cancers and comprehensively summarized a list of related signaling pathways in various tumors. Circ-ABCB10 was found to promote the proliferation, migration, and metastasis of diverse cancers, such as esophageal squamous cell carcinoma, lung cancer, epithelial ovarian cancer, breast cancer, and hepatocellular carcinoma. Moreover, some studies demonstrated that circ-ABCB10 may also contribute to angiogenesis (Tang et al., 2020). From this perspective, circ-ABCB10 might be a potential biomarker for the diagnosis and prognosis of human cancers. However, the knowledge on the roles of circ-ABCB10 in cancer is still limited, and there are still many unresolved problems. For example, there is a lack of knowledge related to the relevant set of miRNA genes for each cancer. In addition, it is also necessary to identify the main sponging targets of circ-ABCB10. Understanding these processes may lead to completely new tumor therapies.
Author Contributions
RS and WW collected the related manuscript. ZH drafted and revised the manuscript. XZ designed the review. JL participated in the design of the review and helped draft and revise the manuscript. All authors read and approved the final manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Bachmayr-Heyda, A., Reiner, A. T., Auer, K., Sukhbaatar, N., Aust, S., Bachleitner-Hofmann, T., et al. (2015). Correlation of Circular RNA Abundance with Proliferation - Exemplified with Colorectal and Ovarian Cancer, Idiopathic Lung Fibrosis and normal Human Tissues. Sci. Rep. 5, 8057. doi:10.1038/srep08057
Bahn, J. H., Zhang, Q., Li, F., Chan, T.-M., Lin, X., Kim, Y., et al. (2015). The Landscape of microRNA, Piwi-Interacting RNA, and Circular RNA in Human Saliva. Clin. Chem. 61 (1), 221–230. doi:10.1373/clinchem.2014.230433
Chen, F., Li, X. H., Liu, C., Zhang, Y., and Wang, R. J. (2020). Circ-ABCB10 Accelerates the Malignant Progression of Oral Squamous Cell Carcinoma by Absorbing miRNA-145-5p. Eur. Rev. Med. Pharmacol. Sci. 24 (2), 681–690. doi:10.26355/eurrev_202001_20045
Chen, L.-L. (2016). The Biogenesis and Emerging Roles of Circular RNAs. Nat. Rev. Mol. Cel Biol 17 (4), 205–211. doi:10.1038/nrm.2015.32
Chen, Y., Ye, X., Xia, X., and Lin, X. (2019). Circular RNA ABCB10 Correlates with Advanced Clinicopathological Features and Unfavorable Survival, and Promotes Cell Proliferation while Reduces Cell Apoptosis in Epithelial Ovarian Cancer. Cbm 26 (2), 151–161. doi:10.3233/cbm-190064
Chi, G., Xu, D., Zhang, B., and Yang, F. (2019). Matrine Induces Apoptosis and Autophagy of Glioma Cell Line U251 by Regulation of circRNA-104075/BCL-9. Chemico-Biological Interactions 308, 198–205. doi:10.1016/j.cbi.2019.05.030
Conn, S. J., Pillman, K. A., Toubia, J., Conn, V. M., Salmanidis, M., Phillips, C. A., et al. (2015). The RNA Binding Protein Quaking Regulates Formation of circRNAs. Cell 160 (6), 1125–1134. doi:10.1016/j.cell.2015.02.014
Cortés-López, M., and Miura, P. (2016). Emerging Functions of Circular RNAs. Yale J. Biol. Med. 89 (4), 527–537.
Deng, N., Li, L., Gao, J., Zhou, J., Wang, Y., Wang, C., et al. (2018). Hsa_circ_0009910 Promotes Carcinogenesis by Promoting the Expression of miR-449a Target IL6R in Osteosarcoma. Biochem. Biophysical Res. Commun. 495 (1), 189–196. doi:10.1016/j.bbrc.2017.11.028
Dong, R., Ma, X.-K., Chen, L.-L., and Yang, L. (2017). Increased Complexity of circRNA Expression during Species Evolution. RNA Biol. 14 (8), 1064–1074. doi:10.1080/15476286.2016.1269999
Dong, W., Dai, Z.-h., Liu, F.-c., Guo, X.-g., Ge, C.-m., Ding, J., et al. (2019). The RNA-Binding Protein RBM3 Promotes Cell Proliferation in Hepatocellular Carcinoma by Regulating Circular RNA SCD-circRNA 2 Production. EBioMedicine 45, 155–167. doi:10.1016/j.ebiom.2019.06.030
Dong, Y., He, D., Peng, Z., Peng, W., Shi, W., Wang, J., et al. (2017). Circular RNAs in Cancer: an Emerging Key Player. J. Hematol. Oncol. 10 (1), 2. doi:10.1186/s13045-016-0370-2
Duan, Z. N., Dong, C. G., and Liu, J. H. (2020). Circ-ABCB10 Promotes Growth and Metastasis of Nasopharyngeal Carcinoma by Upregulating ROCK1. Eur. Rev. Med. Pharmacol. Sci. 24 (23), 12208–12215. doi:10.26355/eurrev_202012_24011
Feng, W., Guo, R., Zhang, D., and Zhang, R. (2021). Circ-ABCB10 Knockdown Inhibits the Malignant Progression of Cervical Cancer through microRNA-128-3p/ZEB1 axis. Biol. Proced. Online 23 (1), 17. doi:10.1186/s12575-021-00154-8
Gao, P., Wang, S., Jing, F., Zhan, J., and Wang, Y. (2017). microRNA-203 Suppresses Invasion of Gastric Cancer Cells by Targeting ERK1/2/Slug/E-Cadherin Signaling. Cbm 19 (1), 11–20. doi:10.3233/cbm-160167
Gao, Y.-L., Zhang, M.-Y., Xu, B., Han, L.-J., Lan, S.-F., Chen, J., et al. (2017). Circular RNA Expression Profiles Reveal that Hsa_circ_0018289 Is Up-Regulated in Cervical Cancer and Promotes the Tumorigenesis. Oncotarget 8 (49), 86625–86633. doi:10.18632/oncotarget.21257
Guarnerio, J., Zhang, Y., Cheloni, G., Panella, R., Mae Katon, J., Simpson, M., et al. (2019). Intragenic Antagonistic Roles of Protein and circRNA in Tumorigenesis. Cell Res 29 (8), 628–640. doi:10.1038/s41422-019-0192-1
Hansen, T. B., Jensen, T. I., Clausen, B. H., Bramsen, J. B., Finsen, B., Damgaard, C. K., et al. (2013). Natural RNA Circles Function as Efficient microRNA Sponges. Nature 495 (7441), 384–388. doi:10.1038/nature11993
He, Q., Zhao, L., Liu, Y., Liu, X., Zheng, J., Yu, H., et al. (2018). circ-SHKBP1 Regulates the Angiogenesis of U87 Glioma-Exposed Endothelial Cells through miR-544a/FOXP1 and miR-379/FOXP2 Pathways. Mol. Ther. - Nucleic Acids 10, 331–348. doi:10.1016/j.omtn.2017.12.014
Hou, L.-D., and Zhang, J. (2017). Circular RNAs: An Emerging Type of RNA in Cancer. Int. J. Immunopathol Pharmacol. 30 (1), 1–6. doi:10.1177/0394632016686985
Hu, N., Jiang, D., Huang, E., Liu, X., Li, R., Liang, X., et al. (2013). BMP9-regulated Angiogenic Signaling Plays an Important Role in the Osteogenic Differentiation of Mesenchymal Progenitor Cells. J. Cel Sci 126 (Pt 2), 532–541. doi:10.1242/jcs.114231
Hu, T. Y., Zhu, Q. X., Duan, Q. Y., Jin, X. Y., and Wu, R. (2020). CircABCB10 Promotes the Proliferation and Migration of Lung Cancer Cells through Down-Regulating microRNA-217 Expression. Eur. Rev. Med. Pharmacol. Sci. 24 (11), 6157–6165. doi:10.26355/eurrev_202006_21511
Huang, Y., Zhang, Y., Jia, L., Liu, C., and Xu, F. (2019). Circular RNA ABCB10 Promotes Tumor Progression and Correlates with Pejorative Prognosis in clear Cell Renal Cell Carcinoma. Int. J. Biol. Markers 34 (2), 176–183. doi:10.1177/1724600819842279
Jakobi, T., and Dieterich, C. (2019). Computational Approaches for Circular RNA Analysis. Wiley Interdiscip. Rev. RNA 10 (3), e1528. doi:10.1002/wrna.1528
Jayson, G. C., Kohn, E. C., Kitchener, H. C., and Ledermann, J. A. (2014). Ovarian Cancer. The Lancet 384 (9951), 1376–1388. doi:10.1016/s0140-6736(13)62146-7
Jin, X., Feng, C.-y., Xiang, Z., Chen, Y.-p., and Li, Y.-m. (2016). CircRNA Expression Pattern and circRNA-miRNA-mRNA Network in the Pathogenesis of Nonalcoholic Steatohepatitis. Oncotarget 7 (41), 66455–66467. doi:10.18632/oncotarget.12186
Khan, M. A. F., Reckman, Y. J., Aufiero, S., van den Hoogenhof, M. M. G., van der Made, I., Beqqali, A., et al. (2016). RBM20 Regulates Circular RNA Production from the Titin Gene. Circ. Res. 119 (9), 996–1003. doi:10.1161/circresaha.116.309568
Kim, S.-W., Zhang, H.-Z., Kim, C. E., An, H. S., Kim, J.-M., and Kim, M. H. (2012). Amniotic Mesenchymal Stem Cells Have Robust Angiogenic Properties and Are Effective in Treating Hindlimb Ischaemia. Cardiovasc. Res. 93 (3), 525–534. doi:10.1093/cvr/cvr328
Kristensen, L. S., Hansen, T. B., Venø, M. T., and Kjems, J. (2018). Circular RNAs in Cancer: Opportunities and Challenges in the Field. Oncogene 37 (5), 555–565. doi:10.1038/onc.2017.361
Kusumbe, A. P., Ramasamy, S. K., and Adams, R. H. (2014). Coupling of Angiogenesis and Osteogenesis by a Specific Vessel Subtype in Bone. Nature 507 (7492), 323–328. doi:10.1038/nature13145
Li, J., Li, Z., Jiang, P., Peng, M., Zhang, X., Chen, K., et al. (2018). Circular RNA IARS (Circ-IARS) Secreted by Pancreatic Cancer Cells and Located within Exosomes Regulates Endothelial Monolayer Permeability to Promote Tumor Metastasis. J. Exp. Clin. Cancer Res. 37 (1), 177. doi:10.1186/s13046-018-0822-3
Li, X., Wang, J., Zhang, C., Lin, C., Zhang, J., Zhang, W., et al. (2018). Circular RNA circITGA7 Inhibits Colorectal Cancer Growth and Metastasis by Modulating the Ras Pathway and Upregulating Transcription of its Host geneITGA7. J. Pathol. 246 (2), 166–179. doi:10.1002/path.5125
Li, Y., Dong, Y., Huang, Z., Kuang, Q., Wu, Y., Li, Y., et al. (2017). Computational Identifying and Characterizing Circular RNAs and Their Associated Genes in Hepatocellular Carcinoma. PLoS One 12 (3), e0174436. doi:10.1371/journal.pone.0174436
Li, Z., Huang, C., Bao, C., Chen, L., Lin, M., Wang, X., et al. (2015). Exon-intron Circular RNAs Regulate Transcription in the Nucleus. Nat. Struct. Mol. Biol. 22 (3), 256–264. doi:10.1038/nsmb.2959
Liang, H. F., Zhang, X. Z., Liu, B. G., Jia, G. T., and Li, W. L. (2017). Circular RNA Circ-ABCB10 Promotes Breast Cancer Proliferation and Progression through Sponging miR-1271. Am. J. Cancer Res. 7 (7), 1566–1576.
Lin, X., Chen, Y., Ye, X., and Xia, X. (2021). Circular RNA ABCB10 Promotes Cell Proliferation and Invasion, but Inhibits Apoptosis via Regulating the microRNA-1271-mediated Capn4/Wnt/β-catenin S-ignaling P-athway in E-pithelial O-varian C-ancer. Mol. Med. Rep. 23 (5), 26. doi:10.3892/mmr.2021.12026
Luo, L., Gao, Y., and Sun, X. (2018). Circ-ITCH Correlates with Small Tumor Size, Decreased FIGO Stage and Prolonged Overall Survival, and it Inhibits Cells Proliferation while Promotes Cells Apoptosis in Epithelial Ovarian Cancer. Cbm 23 (4), 505–513. doi:10.3233/cbm-181609
Lux, S., and Bullinger, L. (2018). Circular RNAs in Cancer. Adv. Exp. Med. Biol. 1087, 215–230. doi:10.1007/978-981-13-1426-1_17
Memczak, S., Jens, M., Elefsinioti, A., Torti, F., Krueger, J., Rybak, A., et al. (2013). Circular RNAs Are a Large Class of Animal RNAs with Regulatory Potency. Nature 495 (7441), 333–338. doi:10.1038/nature11928
Nicolet, B. P., Engels, S., Aglialoro, F., van den Akker, E., von Lindern, M., and Wolkers, M. C. (2018). Circular RNA Expression in Human Hematopoietic Cells Is Widespread and Cell-type Specific. Nucleic Acids Res. 46 (16), 8168–8180. doi:10.1093/nar/gky721
Ouyang, S. B., Wang, J., Zhao, S. Y., Zhang, X. H., and Liao, L. (2018). CircRNA_0109291 Regulates Cell Growth and Migration in Oral Squamous Cell Carcinoma and its Clinical Significance. Iran J. Basic Med. Sci. 21 (11), 1186–1191. doi:10.22038/IJBMS.2018.30347.7313
Pamudurti, N. R., Bartok, O., Jens, M., Ashwal-Fluss, R., Stottmeister, C., Ruhe, L., et al. (2017). Translation of CircRNAs. Mol. Cel 66 (1), 9–21. doi:10.1016/j.molcel.2017.02.021
Patop, I. L., and Kadener, S. (2018). circRNAs in Cancer. Curr. Opin. Genet. Develop. 48, 121–127. doi:10.1016/j.gde.2017.11.007
Qin, M., Wei, G., and Sun, X. (2018). Circ-UBR5: An Exonic Circular RNA and Novel Small Nuclear RNA Involved in RNA Splicing. Biochem. Biophysical Res. Commun. 503 (2), 1027–1034. doi:10.1016/j.bbrc.2018.06.112
Qu, S., Liu, Z., Yang, X., Zhou, J., Yu, H., Zhang, R., et al. (2018). The Emerging Functions and Roles of Circular RNAs in Cancer. Cancer Lett. 414, 301–309. doi:10.1016/j.canlet.2017.11.022
Qu, S., Yang, X., Li, X., Wang, J., Gao, Y., Shang, R., et al. (2015). Circular RNA: A New star of Noncoding RNAs. Cancer Lett. 365 (2), 141–148. doi:10.1016/j.canlet.2015.06.003
Salzman, J., Chen, R. E., Olsen, M. N., Wang, P. L., and Brown, P. O. (2013). Cell-type Specific Features of Circular RNA Expression. Plos Genet. 9 (9), e1003777. doi:10.1371/journal.pgen.1003777
Salzman, J., Gawad, C., Wang, P. L., Lacayo, N., and Brown, P. O. (2012). Circular RNAs Are the Predominant Transcript Isoform from Hundreds of Human Genes in Diverse Cell Types. PLoS One 7 (2), e30733. doi:10.1371/journal.pone.0030733
Sanger, H. L., Klotz, G., Riesner, D., Gross, H. J., and Kleinschmidt, A. K. (1976). Viroids Are Single-Stranded Covalently Closed Circular RNA Molecules Existing as Highly Base-Paired Rod-like Structures. Proc. Natl. Acad. Sci. U.S.A. 73 (11), 3852–3856. doi:10.1073/pnas.73.11.3852
Shang, Q., Yang, Z., Jia, R., and Ge, S. (2019). The Novel Roles of circRNAs in Human Cancer. Mol. Cancer 18 (1), 6. doi:10.1186/s12943-018-0934-6
Shi, X., Wang, B., Feng, X., Xu, Y., Lu, K., and Sun, M. (2020). circRNAs and Exosomes: A Mysterious Frontier for Human Cancer. Mol. Ther. - Nucleic Acids 19, 384–392. doi:10.1016/j.omtn.2019.11.023
Siveen, K. S., Prabhu, K., Krishnankutty, R., Kuttikrishnan, S., Tsakou, M., Alali, F. Q., et al. (2017). Vascular Endothelial Growth Factor (VEGF) Signaling in Tumour Vascularization: Potential and Challenges. Curr. Vasc. Pharmacol. 15 (4), 339–351. doi:10.2174/1570161115666170105124038
Su, M., Xiao, Y., Ma, J., Tang, Y., Tian, B., Zhang, Y., et al. (2019). Circular RNAs in Cancer: Emerging Functions in Hallmarks, Stemness, Resistance and Roles as Potential Biomarkers. Mol. Cancer 18 (1), 90. doi:10.1186/s12943-019-1002-6
Sun, W. Y., Lu, Y. F., Cai, X. L., Li, Z. Z., Lv, J., Xiang, Y. A., et al. (2020). Circ-ABCB10 Acts as an Oncogene in Glioma Cells via Regulation of the miR-620/FABP5 axis. Eur. Rev. Med. Pharmacol. Sci. 24 (12), 6848–6857. doi:10.26355/eurrev_202006_21674
Tang, Q., and Hann, S. S. (2020). Biological Roles and Mechanisms of Circular RNA in Human Cancers. Ott Vol. 13, 2067–2092. doi:10.2147/ott.s233672
Tang, W., Ji, M., He, G., Yang, L., Niu, Z., Jian, M., et al. (2017). Silencing CDR1as Inhibits Colorectal Cancer Progression through Regulating microRNA-7. Ott Vol. 10, 2045–2056. doi:10.2147/ott.s131597
Tang, Z., Tan, J., Yuan, X., Zhou, Q., Yuan, Z., Chen, N., et al. (2020). Circular RNA-ABCB10 Promotes Angiogenesis Induced by Conditioned Medium from Human Amnion-Derived Mesenchymal Stem Cells via the microRNA-29b-3p/vascular Endothelial Growth Factor A axis. Exp. Ther. Med. 20 (3), 2021–2030. doi:10.3892/etm.2020.8939
Thomas, L. F., and Sætrom, P. (2014). Circular RNAs Are Depleted of Polymorphisms at microRNA Binding Sites. Bioinformatics 30 (16), 2243–2246. doi:10.1093/bioinformatics/btu257
Tian, X., Zhang, L., Jiao, Y., Chen, J., Shan, Y., and Yang, W. (2019). CircABCB10 Promotes Nonsmall Cell Lung Cancer Cell Proliferation and Migration by Regulating the miR‐1252/FOXR2 axis. J. Cel Biochem 120 (3), 3765–3772. doi:10.1002/jcb.27657
Wang, M., Gao, W., Lu, D., and Teng, L. (2018). MiR-1271 Inhibits Cell Growth in Prostate Cancer by Targeting ERG. Pathol. Oncol. Res. 24 (2), 385–391. doi:10.1007/s12253-017-0254-y
Wang, T., Wang, J., Ren, W., Chen, S., Cheng, Y.-F., and Zhang, X.-M. (2020). CircRNA-0008717 Promotes Cell Proliferation, Migration, and Invasion by Regulating miR-203/Slug in Esophageal Cancer Cells. Ann. Transl Med. 8 (16), 999. doi:10.21037/atm-20-5205
Wang, Y., Mo, Y., Gong, Z., Yang, X., Yang, M., Zhang, S., et al. (2017). Circular RNAs in Human Cancer. Mol. Cancer 16 (1), 25. doi:10.1186/s12943-017-0598-7
Wu, Z., Gong, Q., Yu, Y., Zhu, J., and Li, W. (2020). Knockdown of Circ-ABCB10 Promotes Sensitivity of Lung Cancer Cells to Cisplatin via miR-556-3p/AK4 axis. BMC Pulm. Med. 20 (1), 10. doi:10.1186/s12890-019-1035-z
Xian, Z. Y., Hu, B., Wang, T., Cai, J. L., Zeng, J. Y., Zou, Q., et al. (2020). CircABCB10 Silencing Inhibits the Cell Ferroptosis and Apoptosis by Regulating the miR-326/CCL5 axis in Rectal Cancer. Neoplasma 67 (5), 1063–1073. doi:10.4149/neo_2020_191024N1084
Yang, W., Ju, H. Y., and Tian, X. F. (2020). Circular RNA-ABCB10 Suppresses Hepatocellular Carcinoma Progression through Upregulating NRP1/ABL2 via Sponging miR-340-5p/miR-452-5p. Eur. Rev. Med. Pharmacol. Sci. 24 (5), 2347–2357. doi:10.26355/eurrev_202003_20501
Yang, Y., Yujiao, W., Fang, W., Linhui, Y., Ziqi, G., Zhichen, W., et al. (2020). The Roles of miRNA, lncRNA and circRNA in the Development of Osteoporosis. Biol. Res. 53 (1), 40. doi:10.1186/s40659-020-00309-z
Yu, C.-Y., and Kuo, H.-C. (2019). The Emerging Roles and Functions of Circular RNAs and Their Generation. J. Biomed. Sci. 26 (1), 29. doi:10.1186/s12929-019-0523-z
Zang, J., Lu, D., and Xu, A. (2020). The Interaction of circRNAs and RNA Binding Proteins: An Important Part of circRNA Maintenance and Function. J. Neurosci. Res. 98 (1), 87–97. doi:10.1002/jnr.24356
Zhang, S., Zhu, D., Li, H., Li, H., Feng, C., and Zhang, W. (2017). Characterization of circRNA-Associated-ceRNA Networks in a Senescence-Accelerated Mouse Prone 8 Brain. Mol. Ther. 25 (9), 2053–2061. doi:10.1016/j.ymthe.2017.06.009
Zhang, W. Q., Liu, K. Q., Pei, Y. X., Tan, J., Ma, J. B., and Zhao, J. (2020). Circ-ABCB10 Promotes Proliferation and Invasion of Esophageal Squamous Cell Carcinoma Cells by Modulating microRNA-670-3p. Eur. Rev. Med. Pharmacol. Sci. 24 (11), 6088–6096. doi:10.26355/eurrev_202006_21504
Zhang, Y., Zhang, X.-O., Chen, T., Xiang, J.-F., Yin, Q.-F., Xing, Y.-H., et al. (2013). Circular Intronic Long Noncoding RNAs. Mol. Cel 51 (6), 792–806. doi:10.1016/j.molcel.2013.08.017
Zhao, Y., Zhong, R., Deng, C., and Zhenlin, Z. (2020). Circle RNA circABCB10 Modulates PFN2 to Promote Breast Cancer Progression, as Well as Aggravate Radioresistance through Facilitating Glycolytic Metabolism via miR-223-3p. Cancer Biother. Radiopharm. 36 (6), 477–490. doi:10.1089/cbr.2019.3389
Zhao, Y., Zhong, R., Deng, C., and Zhou, Z. (2021). Circle RNA circABCB10 Modulates PFN2 to Promote Breast Cancer Progression, as Well as Aggravate Radioresistance through Facilitating Glycolytic Metabolism via miR-223-3p. Cancer Biother. Radiopharm. 36 (6), 477–490. doi:10.1089/cbr.2019.3389
Zheng, Q., Bao, C., Guo, W., Li, S., Chen, J., Chen, B., et al. (2016). Circular RNA Profiling Reveals an Abundant circHIPK3 that Regulates Cell Growth by Sponging Multiple miRNAs. Nat. Commun. 7, 11215. doi:10.1038/ncomms11215
Zhong, Z., Lv, M., and Chen, J. (2016). Screening Differential Circular RNA Expression Profiles Reveals the Regulatory Role of circTCF25-miR-103a-3p/miR-107-CDK6 Pathway in Bladder Carcinoma. Sci. Rep. 6, 30919. doi:10.1038/srep30919
Keywords: circRNA, circ-ABCB10, cancer, microRNA, tumor biology
Citation: Huang Z, Shan R, Wen W, Li J, Zeng X and Wan R (2022) The Emerging Roles of Circ-ABCB10 in Cancer. Front. Cell Dev. Biol. 10:782938. doi: 10.3389/fcell.2022.782938
Received: 25 September 2021; Accepted: 30 March 2022;
Published: 13 May 2022.
Edited by:
Zong Sheng Guo, Roswell Park Comprehensive Cancer Center, United StatesReviewed by:
Thasni Karedath, Qatar Biomedical Research Institute, QatarDemitrios Vynios, University of Patras, Greece
Carlo V. Bruschi, University of Salzburg, Austria
Copyright © 2022 Huang, Shan, Wen, Li, Zeng and Wan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaohong Zeng, WmVuZzc3NTExMjg0MEBxcS5jb20=; Renhua Wan, V2Fuend3NzI2Njk2QHNpbmEuY29t
 Zhenjun Huang1
Zhenjun Huang1 Renhua Wan
Renhua Wan