
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
PERSPECTIVE article
Front. Cell Dev. Biol. , 04 March 2022
Sec. Molecular and Cellular Reproduction
Volume 10 - 2022 | https://doi.org/10.3389/fcell.2022.781953
This article is part of the Research Topic The Role of Calcium Signaling in Gametogenesis and Early Embryogenesis View all 8 articles
Oocyte activation deficiency (OAD) is the basis of Total Fertilisation Failure (TFF) and is attributed to mutations in the PLCζ gene—termed male factor infertility. This derives abnormal Ca2+ oscillations and could be the main cause of primary disruptions in the gene expression of Ca2+-related proteins. Epigenetic mechanisms are universally accepted as key regulators of gene expression. However, epigenetic dysregulations have not been considered as potential mechanisms of oocyte-borne OAD. Herein, we discuss changes in the DNA methylome during oogenesis and embryogenesis. We further highlight key pathways comprising the oocyte Ca2+ toolkit, which could be targets of epigenetic alterations, especially aberrations in DNA methylation. Considering that the vast majority of epigenetic modifications examined during fertilization revolve around alterations in DNA methylation, we aim in this article to associate Ca2+-specific mechanisms with these alterations. To strengthen this perspective, we bring evidence from cancer research on the intricate link between DNA methylation and Ca2+ signaling as cancer research has examined such questions in a lot more detail. From a therapeutic standpoint, if our hypothesis is proven to be correct, this will explain the cause of TFF in idiopathic cases and will open doors for novel therapeutic targets.
Fertilization encompasses the fusion of sperm and oocyte membranes, pro-nuclei formation, and the initiation of early embryogenesis. Following membrane fusion, the crucial trigger mediating downstream events is oocyte activation, defined by cortical granule exocytosis, resumption of meiosis II, and the induction of early embryogenesis (Kashir et al., 2010). Calcium (Ca2+) is the principal mediator of mammalian oocyte activation, displaying long-lasting oscillations from membrane fusion to pronuclei formation to kickstart signaling pathways culminating in oocyte activation (Kashir et al., 2010; Saleh et al., 2020). The trigger of Ca2+ oscillations is phospholipase ζ (PLCζ)—a sperm-specific PLC isoform—which produces inositol-1,4,5-triphosphate (IP3) via hydrolysis of phosphatidylinositol-4,5-biphosphate (PIP2) (Saunders et al., 2002). IP3 binds to IP3Rs on the endoplasmic reticulum (ER) to elicit Ca2+ release from internal stores.
The discovery of PLCζ and Ca2+ oscillations have revolutionized the world of assisted reproductive technology (ART) in treating infertility, a misfortune affecting ∼15% of couples worldwide. Firstly, intracytoplasmic sperm injection (ICSI) has proven instrumental as an alternative to conventional in vitro fertilization (IVF). ICSI circumvents the numerous in vivo barriers to fertilization by introducing sperm, and thereby PLCζ, directly into the ooplasm. However, partial or total fertilization failure (TFF) affects ∼1–5% of ICSI procedures where seemingly normal sperm and eggs exhibit failed fertilization (Yeste et al., 2016). Oocyte activation deficiency (OAD) is considered the main cause of TFF and is attributed to mutations in the PLCζ gene—termed male factor infertility [readers are referred to (Kashir, 2020; Zafar et al., 2021) for more detailed reviews]. To counter this, artificial oocyte activators (OAA), such as calcium ionophores (ionomycin and calcimycin), have been produced to induce Ca2+ transients in oocytes. But these molecules elicit a solitary [Ca2+] rise rather than the repetitive oscillations seen in vivo, which negatively impacts embryogenesis (Yeste et al., 2017). Consequently, developing exogenous PLCζ to trigger oscillations better approximating the in vivo fertilization is a focus of ongoing research (Kashir et al., 2010; Kashir et al., 2011).
However, although a large proportion of TFF cases are attributed to male factor infertility, approximately 26% of TFF cases are considered idiopathic (Yeste et al., 2016). This has significantly hindered the efficacy of ART. Indeed, ∼1—5% of ICSI procedures fail, success rates per cycle of ICSI remain a meager ∼27%, and global pregnancy rates and live births following ART is ∼40% (Kashir, 2020). Mechanistic studies behind these idiopathic cases of TFF are crucial in developing novel diagnostic and therapeutic procedures to improve ART efficacy. To this end, other potential SOAFs, such as PAWP, have been put forward as alternatives to PLCζ with mutations in them manifesting as TFF. However, this was proven unlikely, as knockout (KO) mice for the gene encoding PAWP (gene name Wbp2nl) showed no demonstrable defect in oocyte-activating ability (Satouh, 2015).
Even though male factor infertility accounts for a substantial percentage of TFF cases, the role of potential oocyte factors should not be dismissed; discussion on this topic, however, is limited. It would be prudent now to appreciate the dependence of PLCζ on various ooplasmic proteins. Mutations affecting these proteins could cause an oocyte-borne OAD. Even if oocyte activation is achieved, the atypical Ca2+ profile could have deleterious effects on embryonic development. Indeed, too few oscillations compromise the embryo’s ability to implant, whereas an amplified Ca2+ response impairs post-implantation development (Ozil et al., 2006). This relates to Ca2+ oscillations affecting gene expression (Ozil et al., 2006). Consistently, analysis of TFF metaphase II (MII) oocytes relative to controls revealed substantial differences in gene expression profiles of TFF oocytes, with genes involved in meiosis, cell growth, and apoptosis being disproportionately affected (Gasca et al., 2008).
Abnormal Ca2+ oscillations alternatively could derive from primary disruptions in the gene expression of Ca2+-related proteins. Epigenetic mechanisms are universally accepted as key regulators of gene expression and are known to regulate Ca2+ signaling genes. Yet, epigenetic dysregulations have not been considered as potential mechanisms of oocyte-borne OAD. In this manuscript, we discuss epigenetic programs during oogenesis and embryogenesis, emphasizing DNA methylation; since most of the epigenetic programming events studied during fertilization typically revolve around changes in DNA methylation profiles, we feel it appropriate to focus on this particular epigenetic process and uncover the Ca2+-specific mechanisms underlying DNA methylation alterations. Lastly, we present evidence from cancer research as a model which has investigated these questions in great detail and confirmed the intricate link between DNA methylation and Ca2+ signaling in somatic cells, further warranting an investigation into this relationship in oocytes. If this hypothesis proves favorable, new mechanisms to explain supposedly idiopathic cases of TFF could emerge alongside novel therapeutic targets.
In short, epigenetic modifications regulate gene expression without altering nucleotide sequence. Epigenetic modulations include DNA methylation, histone tail post-translational modifications (PTMs), and non-coding RNAs (ncRNAs) (Le Blévec et al., 2020; Zafar et al., 2021). DNA methylation and histone PTMs regulate chromatin accessibility and packaging to control the binding of transcription factors to either facilitate or repress gene expression. On the other hand, ncRNAs are strong post-transcriptional regulators, targeting the 3′ untranslated regions (3′-UTRs) of specified mRNA molecules to induce their degradation (Le Blévec et al., 2020; Zafar et al., 2021).
While multiple epigenetic mechanisms are involved in determining the fate of gene expression profiles, the predominant player almost exclusively studied in the context of calcium release at fertilization has been methylation/demethylation of maternal and paternal DNA. This is also perhaps due to the consideration that in most mammals calcium oscillations persist for about 2–4 h post-gamete fusion, after which they cease around pronuclear formation. Additionally, while there are of course certain processes that could be influenced by the specific patterns of calcium release at fertilization, such processes are almost unique to fertilization and as such cannot be investigated in model systems such as cancer. One such example is sperm head decondensation postfertilization, which involves a protamine-to-histone change in DNA composition. During spermatogenesis, sperm nuclei are remodeled to incorporate protamine, which are small arginine-rich nuclear proteins that allow strong DNA binding to form highly stable and compacted DNA (Sassone-Corsi, 2002; Ward, 2010; Ribas-Maynou et al., 2021). However, 10–15% of the human sperm genome (∼1% in mice) remains histone-bound, suggesting that histones facilitate post-translational modifications that are transmitted to the early zygote and persist during pre-implantation embryogenesis (Hales et al., 2011). Indeed, histones play a key role in protamine transition and chromatin reorganization during spermatogenesis. It is thought that profiles of calcium release exert a significant role in the rate and quantity of protamine-to-histone transition (McLay et al., 2002), but this still requires further investigation.
DNA methylation involves covalent modification of DNA, in which a methyl group from S-adenosylmethionine (SAM) is donated to the carbon-5 of cytosine, forming 5-methylcytosine (5-mC) in a Cytosine-Guanine (CpG) dinucleotide context. The enzyme catalyzing DNA methylation are DNA methyltransferases (DMNTs), whereas methylation is erased by ten-eleven translocases (TETs) (Ito et al., 2011). Although CpG dinucleotides across the genome are methylated, CpG clusters termed CG islands—(CGIs) associated with gene promoters and enhancers (i.e., regulatory regions)—are constitutively unmethylated (Deaton and Bird, 2011). Overall, DNA methylation constitutes a repressive epigenetic mark.
In sperm, DNA methylation is initiated prenatally and completed before puberty. In contrast, oocytes before puberty are practically unmethylated. Methylation in oocytes is initiated in the cohort of oocytes recruited at the beginning of each ovarian cycle to undergo ovulation. Methylation proceeds with follicular growth from the primary to the preantral and antral stages when methylation is completed (Sendžikaitė and Kelsey, 2019; Barberet et al., 2020). The mammalian oocyte methylome is established in a transcription-dependent manner, leaving non-transcribing genes and intergenic areas hypomethylated, resulting in a bimodal and clustered methylome consisting of hyper- and hypomethylated domains (Sendžikaitė and Kelsey, 2019).
Postfertilization, both paternal and maternal DNA is demethylated via TETs. Paternal DNA is demethylated actively and rapidly, unlike the passive demethylation characteristic of maternal DNA. This event is essential to abolish gamete identity and confer cells of the growing embryo with pluripotent potential (Cantone and Fisher, 2013; Sendžikaitė and Kelsey, 2019). Following implantation, DNA methylation is regained and proceeds in a lineage-specific pattern. Primordial germ cells (PGCs) subsequently experience a second wave of DNA demethylation to completely abolish parental epigenomes, which eventually are regained in a gender-dependent manner (Sendžikaitė and Kelsey, 2019).
Some regions of parental DNA evade demethylation during embryogenesis and are termed differentially methylated regions (DMRs). Most DMRs comprise imprinted loci which feature monoallelic expression depending on the parental origin of the allele (Tucci et al., 2019). Imprinted loci contain a cluster of genes whose expression depends on the DNA methylation status of an imprinting control region (ICR). ICRs of imprinted loci contain a CG-rich sequence recognized by the KRAB zinc-finger protein 57, which subsequently recruits a set of regulatory proteins to protect these sites (Tucci et al., 2019). Therefore, epigenetic marks on imprinted loci are inherited and maintained throughout life, only being erased in PGCs.
Other epigenetic modifications such as histone-PTMs also exhibit reprogramming during early embryogenesis. Histones are closely associated with DNA to form nucleosomes that contain two each of histones H2A, H2B, H3, and H4. Each histone has N-terminal tails with amino acid residues that serve as potential sites of covalent modifications such as acetylation, methylation, or phosphorylation. Histone PTMs are found at distinct loci within the genome and serve to alter the spatial arrangement of nucleosomes and modulate chromatin structure to control binding of transcription factors, thereby regulating gene expression (Kouzarides, 2007). More layers of complexity are added by the same covalent modifications at distinct loci having varying effects; for example, tri-methylation at Lysine 9 of H3 (H3K9me3) silences gene expression, whereas methylation of lysine 4 of H3 (H3K4me) is a marker of active transcription. During early embryogenesis, a major resetting of histone PTMs occurs with clear asymmetry between paternal and maternal epigenomes. For instance, in mice, mature oocyte non-canonical H3K4me3 marks are distributed broadly in both promoters and distal regions and are inherited by the embryo and persist until zygotic genome activation (ZGA), when they are removed at the late two-cell stage. In contrast, sperm H3k3me3 and H3k27me3 are quickly removed after fertilization (Zhang et al., 2016). However, recent advances in the field have allowed the detection of histone PTM reprogramming in human pre-implantation embryogenesis (Skene and Henikoff, 2017), revealing important differences between mice and humans in this regard (Xia et al., 2019). Indeed, in human oocytes, H3K4me3 features sharp peaks at promoters, and weaker distal H3K4me3 marks are seen in pre-ZGA embryos (Xia et al., 2019). However, the function of these histone marks in humans remains to be identified.
A non-coding RNA (ncRNA) is not translated into a protein. 90% of the eukaryotic genome is transcribed, but mRNA accounts for only 1–2% of total RNA (Ponting and Grant Belgard, 2010). The remaining ∼98% are ncRNAs, which are further divided into “housekeeping” RNAs (including transfer RNA and ribosomal RNA required for translation) and “regulatory RNAs”, which are divided according to size into small ncRNAs (sncRNAs), which include PIWI-interacting RNAs (piRNAs), small interfering RNAs (siRNAs), and micro-RNAs (miRNAs), and long ncRNAs (lncRNAs) (Deniz and Erman, 2016). Recently, knowledge on the role of ncRNAs in regulating different aspects of biology led to the discovery of their role in gametogenesis and embryogenesis, which is comprehensively reviewed here (Pauli et al., 2011). piRNAs are essential for spermatogenesis and fertility in mice, presumably by preventing the accumulation of transposons (Fu and Wang, 2014). Furthermore, miRNAs and siRNAs are involved in heterochromatin formation, transcriptional silencing, and DNA repair during spermatogenesis (Hilz et al., 2016). In contrast, sncRNAs are non-essential for oogenesis and early embryogenesis pre-ZGA. However, ncRNAs are essential in embryogenesis post-ZGA, as the zygotic genome expresses miRNAs to degrade maternal mRNAs transcribed during oogenesis. Additionally, regulatory ncRNAs are responsible for maintaining pluripotency of embryonic stem cells and their differentiation, germ layer specific, cell fate specification, and morphogenesis, as is comprehensively reviewed here (Pauli et al., 2011).
However, it remains that in a developmental context, DNA methylation reprogramming is best characterized (Zhu et al., 2021). Furthermore, demethylation processes ensue immediately following fertilization; the sperm genome is fully demethylated before the first cell division, whereas the passive demethylation characterizing the maternal epigenome begins and continues over the subsequent cell divisions (Seisenberger et al., 2013). Consistent with this temporal association between Ca2+ oscillations and DNA demethylation, altered DNA methylation patterns are associated with different oocyte activation protocols utilized in ART. Indeed, the long-term epigenetic alterations observed in ART involve environmental disturbances experienced by in vitro embryos, and DNA methylation changes are largely implicated in this setting (Zhu et al., 2021). In contrast, histone PTM reprogramming begins later in preimplantation embryogenesis, decreasing the likelihood of a potential interplay with fertilization-associated Ca2+ dynamics; the same applies to ncRNAs. Therefore, this perspective highlights the potential interplay between gene expression and Ca2+ dynamics at fertilization through Ca2+-induced changes in DNA methylation patterns.
Abnormal Ca2+ oscillations postfertilization are confirmed to yield suboptimal developmental outcomes. The mechanism behind this, however, remains to be elucidated. What is known is that abnormal postfertilization Ca2+ profiles disrupt gene expression, as evidenced by experimental manipulation of Ca2+ oscillations modifying the blastocyst transcriptome (Stein et al., 2020). Calcium signaling parameters, particularly amplitude, are important determinants of postfertilization development (Ozil and Huneau, 2001). Dampened oscillations jeopardize preimplantation development, with genes of RNA processing and polymerase-II transcription particularly affected (Ozil et al., 2006). In contrast, hyperstimulation of Ca2+ oscillations compromises postimplantation development by lowering blastocyst developmental competence. In this context, genes involved in metabolism were preferentially involved (Ozil et al., 2006). A study assessing strontium-induced parthenogenetic oocyte activation—characterized by repetitive Ca2+ oscillations—exhibited higher expression of genes encoding proteins involved in cell proliferation, cell adhesion, and ion transport. In contrast, microarray analysis of gene expression profiles of embryos retrieved from oocytes activated in a Ca2+-devoid manner exhibited higher expression of genes of the cell cycle, apoptosis, and cell differentiation (Rogers et al., 2006). In addition, these embryos showed decreased developmental rates to the blastocyst stage and a reduced inner cell mass, highlighting the importance of Ca2+ in proper cell division (Rogers et al., 2006).
In short, postfertilization Ca2+ profiles are confirmed to influence the nature of subsequent cell divisions and embryonic development, with this effect possibly being mediated through alterations in embryo gene expression. Simultaneously, major epigenetic reprogramming characterized by parental DNA demethylation is being established; such programs ensue immediately following Ca2+ oscillations. Therefore, could abnormal Ca2+ oscillations disrupt these programs? Future work should address this question by evaluating the epigenetic profiles of affected genes. If such mechanisms are at play, and since epigenetic modifications are reversible, efforts to correct dysregulation of the oocyte DNA methylome could prove fruitful.
ART protocols are used globally to treat infertility. Current data regarding the frequency of congenital defects in children conceived through ART look reassuring. However, a small body of evidence associates certain procedures, including ICSI, with an increased risk of imprinting disorders, including Prader-Willi and Angelman syndrome (Palermo et al., 2009). This stems from studies revealing DNA methylation of certain genes and histones being impaired after ART [for more detail, review (Ciapa and Arnoult, 2011)]. The mechanism behind such disruptions, however, is currently unknown. Intriguingly, ICSI elicits Ca2+ oscillations deviating from the physiologic transients seen in vivo (Tesarik and Sousa, 1994; Kurokawa and Fissore, 2003). A study comparing the Ca2+ oscillatory pattern elicited by ICSI to normal IVF reported a longer duration of oscillations in the IVF group, with consequent reductions in cell numbers in the ICSI group and a concurrent slower hatching rate of ICSI-generated zygotes compared to IVF (Kurokawa and Fissore, 2003). Such oscillations, as highlighted above, affect gene expression profiles during embryonic development. Might the abnormal Ca2+ profile caused by ICSI be responsible for these epigenetic defects? Ca2+ oscillations regulating embryo gene expression and ICSI-induced atypical oscillations manifesting as epigenetic defects indicate a possible causal relationship between the two processes, meriting an in-depth investigation.
Calcium also leads to the production of reactive oxygen species (ROS). ROS generation plays a role in cell cycle resumption during both oocyte maturation and activation. During oocyte activation, physiologic Ca2+ transients elicit ROS levels sufficient to cause cell cycle resumption (Tiwari et al., 2017). However, in cases of abnormal Ca2+ profiles, overproduction of ROS exerts oxidative stress (OS) on oocytes, promoting apoptosis through intrinsic mitochondrial and extrinsic FAS receptor-ligand pathways (Tiwari et al., 2017). Furthermore, oxidative stress (OS) is a well-established modifier of the oocyte DNA methylome: guanine is most susceptible to OS, whereas oxidation of cytosine produces 5-hydroxymethylcytosine (5-hmC), a physiological prerequisite for DNA demethylation. In addition to initiating aberrant DNA demethylation, ROS can reduce the affinity of DMNTs for DNA [for more detailed descriptions refer to (Menezo et al., 2016)]. Hence, perhaps Ca2+ -dependent OS disrupts the oocyte DNA methylome, resulting in TFF (Figure 1). This would make sense of the aforementioned observation where TFF MII oocytes display altered expression profiles disproportionately involving genes encoding apoptosis-mediating proteins (Gasca et al., 2008).
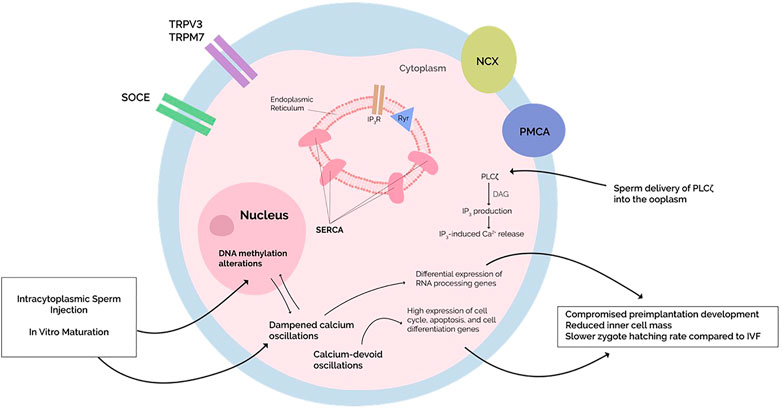
FIGURE 1. Dampening or absence of calcium oscillations is seen in intracytoplasmic sperm injection (ICSI) and cycloheximide-induced oocyte activation, respectively. This alters embryonic development by altering gene expression. The molecular mechanisms behind such changes, however, remain unknown. Since an increased risk of epigenetic defects is perceived in ISCI with simultaneous changes in DNA methylation patterns, perhaps the DNA methylation alterations and differential gene expression are due to the dampened oscillations since major epigenetic processes occur immediately after the cessation of calcium oscillations. Alternatively, primary dysregulations in the oocyte DNA methylome—as induced by ICSI or in vitro maturation—may affect calcium homeostasis, although further studies need to be conducted.
PLCζ-induced Ca2+ oscillations activate numerous pathways concluding in cortical granule exocytosis, meiosis II resumption, and pronuclear formation. The initial event of oocyte activation is cortical granule exocytosis, characterized by the exocytosis of enzymes that modify the zona pellucida and prevent further sperm entry—blocking polyspermy. Substantial evidence currently exists implicating protein kinase C as the principal mediator of cortical granule exocytosis. Calcium transients activate PKCs which translocate to the oocyte cortex, phosphorylating myristoylated alanine-C-rich proteins (MARCKS). This disrupts the actin cytoskeleton, allowing for exocytosis of cortical granules to prevent polyspermy (Ducibella and Matson, 2007; Tsaadon, 2008).
Meiosis resumption and pronuclei formation follow cortical granule exocytosis. Metaphase II (MII) arrest is maintained by cytostatic factor (CSF), an inhibitor of anaphase-promoting complex (APC). Specifically, endogenous meiosis inhibitor (Emi2)—a subunit of CSF—inhibits APC. APC degrades cyclin B1 (CCNB1) of the CCNB1/cyclin-dependent kinase 1 (CDK1) complex, collectively termed maturation promoting factor (MPF). Therefore, by inhibiting APC, CSF maintains high levels of MPF to sustain MII arrest. Emi2 is degraded by polo-like kinase (Plk1), which itself requires a preceding phosphorylation step. Plk1 is phosphorylated—and activated—by calcium/calmodulin kinase II (CAMKII), an intracellular kinase activated by Ca2+ oscillations. Therefore, acute rises in intracellular Ca2+ activate CAMKII, leading to phosphorylation of Plk1 which degrades Emi2 subunit of CSF, thereby relieving APC from inhibition which then degrades tCCNB1 of the MPF complex. A second mechanism by which CAMKII promotes meiotic resumption is by phosphorylating and activating WEE1B. WEE1B, in turn, phosphorylates CDK1, leading to inhibition of MPF and meiosis resumption. Finally, the degradation of MPF triggers a decline in the activity of mitogen-activated protein kinase (MAPK), allowing pronuclei formation.
Therefore, CAMKII is the principal transducer of Ca2+ oscillations, which was confirmed by injection of CAMKII complementary RNA (cRNA) resulting in meiosis II resumption and pronuclear formation (Knott et al., 2006). CAMKII cRNA injection into eggs also yields developmental success rates similar to physiologic activation (Knott et al., 2006). More detailed descriptions of the molecular events of egg activation are found here (Wakai et al., 2011; Sanders and Swann, 2016).
Just as the importance of Ca2+ oscillations has been emphasized, equally important are the transport proteins allowing these oscillations to occur. To support calcium signaling, somatic cells and oocytes possess a unique calcium toolkit that functions to maintain low cytosolic [calcium] ([Ca2+]I)—approximately 100 nM (Clapham, 2007). This calcium signaling toolkit comprises various ion channels, atpase pumps, and transporters on organelle membranes and the plasma membrane (PM). A complex interplay exists between these various components to generate cell- and stimulus-specific Ca2+ responses. A detailed review of the Ca2+ toolkit is found here (Carvacho et al., 2018).
Increases in [Ca2+]I primarily result from Ca2+ release from internal stores through the activation of the inositol-1,4,5-triphosphate receptor (IP3R) and ryanodine receptor (RyR) on the endoplasmic reticulum membrane (ER). The IP3R, predominantly the IP3R1 isoform in oocytes, is essential to rises in [Ca2+]I: IP3R1 activation requires both Ca2+ and IP3, resulting in Ca2+ influx at low [Ca2+]I and closure via negative feedback when [Ca2+]I is elevated (Tosti, 2006; Kashir et al., 2014; Machaty et al., 2017). The RyR shares this pattern of activity.
Internal Ca2+ release is supported by extracellular influx. Store-operated calcium entry (SOCE), elicited by the depletion of [Ca2+]ER, occurs through an ER Ca2+ sensor stromal interaction molecule (STIM1) that recruits ORAI1 at PM-ER junctions to allow Ca2+ entry (Lewis, 1999; Carvacho et al., 2018). The voltage-gated T-type Ca2+ channel (Cav3.1–3.3) and members of the transient receptor potential (TRP) family, specifically TRPV3 and TRPM7, also contribute to extracellular Ca2+ influx (Liu et al., 2011; Carvacho et al., 2013; Bernhardt et al., 2015).
To sustain oscillations and prevent cellular toxicity [Ca2+]I must normalize and ER Ca2+ stores ([Ca2+]ER) replenished. To achieve this, sarcoplasmic reticulum Ca2+ ATPases (SERCA) and plasma membrane Ca2+ ATPases (PMCAs) pump calcium into the ER and extracellular space, respectively (Berridge et al., 2003). SERCA, specifically SERCA2b in oocytes, allows [Ca2+]I and [Ca2+]ER to display parallel but opposite oscillatory responses, where a rise in [Ca2+]i is accompanied by a simultaneous decline in [Ca2+]ER. Sodium-calcium exchangers (NCX) also participate in the extrusion of Ca2+ into the extracellular milieu in rodent oocytes (Carroll, 2000).
The mitochondria aid SERCA and PMCA in buffering [Ca2+]I by taking up Ca2+ (Rizzuto et al., 2000). Importantly, the rise in mitochondrial Ca2+ content sustains ATP levels in oocytes and eggs. Furthermore, inhibiting mitochondrial ATP production by oligomycin impairs refilling of [Ca2+]ER, indicating that Ca2+-induced ATP production during transients sustains SERCA activity (Wakai et al., 2013).
Intriguingly, these Ca2+ channels are not expressed uniformly throughout oocyte development. Rather, each channel exhibits differential expression. SOCE- and Cav3.1–3.3—mediated currents are maximal in GV oocytes and progressively diminish over the subsequent stages of oocyte maturation (Cheon et al., 2013; Bernhardt et al., 2015; Carvacho et al., 2018). In contrast, TRPV3 expression is negligible at maturation and progressively increases to maximal levels in MII oocytes (Carvacho et al., 2018). The mechanisms underlying this differential expression are unknown. However, since this process coincides with the insidious establishment of the oocyte DNA methylome, investigations into a potential interplay are warranted.
In short, various proteins maintain intracellular Ca2+ homeostasis, allowing postfertilization Ca2+ oscillations to occur. Abnormalities in such mechanisms negatively impact Ca2+ transients (Yeste et al., 2016). Research or clinical consideration of these scenarios, however, is rare. Accordingly, we propose that genes encoding Ca2+ transporters could be epigenetically silenced through DNA methylation, manifesting as abnormal Ca2+ profiles (Figure 2). Similar postulations have been made by other studies (Ciapa and Arnoult, 2011; Stein et al., 2020), although the connection between DNA methylation and Ca2+ oscillations has not been stated.
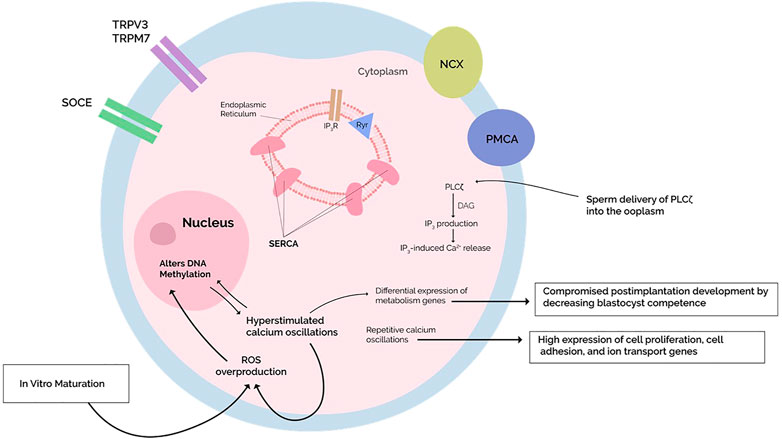
FIGURE 2. Repetitive calcium oscillations approximating in vivo fertilization feature a differential gene expression with high expression of cell cycle, cell adhesion, and ion transport genes. On the other hand, hyperstimulation of calcium oscillations compromises postimplantation development by lowering blastocyst competence with high expression of metabolism-related genes. Simultaneously, the Hyperstimulated calcium transients increase the production of reactive oxygen species (ROS), which are known to induce changes in DNA methylation through oxidative stress. The mechanisms underlying the observed differential expression are unknown, but when coupled with mounting evidence of calcium-dependent ROS generation altering the oocyte DNA methylome, perhaps these epigenetic alterations account for the differential expression. Studies investigating a potential causal relationship are warranted. In vitro maturation (IVM) protocols generate increased oxidative stress on the oocyte, altering DNA methylation programs during oocyte maturation—differential expression is indeed reported after IVM, but whether it also disrupts calcium homeostasis and what effects this has on postfertilization calcium oscillations remains investigational.
During in vitro maturation (IVM), oxidative phosphorylation in mitochondria may be disturbed along with the processing of genes pertaining to epigenetics and the cell cycle (Virant-Klun et al., 2018) [discussed comprehensively here (Zhang et al., 2021)]. A study compared the abundance of mRNA transcripts within IVM and in vivo matured oocytes, revealing substantial differences in polyadenylated mRNA transcript abundance of oocyte genes Mater and Zar1—both play important roles in postfertilization development—and gdf9, which is involved in conferring postfertilization developmental competence (Camargo et al., 2019). The changes in gene expression of these proteins may underpin the reduced developmental competence of IVM oocytes. Furthermore, the mRNA transcript abundance of heat shock proteins and peroxiredoxin is also reduced (Camargo et al., 2019). These proteins have antioxidant functions, protecting the oocyte from OS. Therefore, the reduced abundance of these genes may be associated with an increased OS on the oocyte. As discussed above, OS disrupts the oocyte DNA methylome (Camargo et al., 2019). This is one illustration of how the oocyte DNA methylome may get disrupted. But whether such disruptions involve Ca2+−homeostasis genes during oocyte development remains investigational. Although, the studies mentioned prior found no changes in the polyadenylated mRNA transcript abundance of such genes.
However, the in vivo matured oocytes were also stimulated by exogenous hormones FSH and LH in these experiments; these oocytes were not matured in natural conditions. The differences reported in this experiment might therefore not reflect the true extent of gene expression differences between in vivo and in vitro matured oocytes. Additionally, evidence from cancer research describes DNA methylation and Ca2+ homeostasis as being intricately linked in several somatic cells: studies have substantiated Ca2+ homeostasis disruptions as being predicated on alterations in the promoter methylation status of genes encoding vital Ca2+ proteins, thus leading to dysregulations in proper progression through the cell cycle (Izquierdo-Torres et al., 2020).
Fundamentally, carcinogenesis is characterized by dysregulation of the cell cycle, which is dependent on Ca2+ (Kim et al., 2020). Altered Ca2+ homeostasis underlies the acquisition of several hallmarks of cancer, including sustained proliferative signaling, replicative immortality, cell death resistance, angiogenesis induction, invasion and metastasis, and altered metabolism (Hanahan and Weinberg, 2011; Berridge, 2012; Izquierdo-Torres et al., 2020). Similarly, in the setting of fertilization, the egg is released from metaphase II arrest, which is triggered by Ca2+ by largely the same proteins. Therefore, the fertilization Ca2+ signal also controls cell cycle progression (Whitaker, 2008). Embryos derived from Ca2+-devoid fertilization consistently display a reduction in inner cell mass at the blastocyst stage (Rogers et al., 2006). CAMKII is the key protein ensuring proper cell cycle progression in both scenarios. The emergence of CAMKII in controlling cancer cell proliferation is confirmed by experiments pharmacologically inhibiting CAMKII to reduce tumor mass and proliferation (Wang et al., 2015).
Numerous studies have pointed out similarities between tumor and developmental biology. Preimplantation embryo cells undergo genome-wide reprogramming in the form of DNA demethylation, which confers pluripotent potential. This continues in primordial germ cells, which become undifferentiated and also display migratory capabilities (Fujimoto, 1977). When stringent vivo regulations are removed, such cells can divide indefinitely, i.e., display immortality. Furthermore, the embryo trophoblast exhibits invasive properties during implantation, characterized by invasion of the maternal uterine wall (Monk and Holding, 2001). Cancer cells are also undifferentiated, immortal, and invasive. In theory, these hallmarks of cancer may be the consequence of the re-expression of such developmental genes in an entirely inappropriate context (Manzo, 2019). Indeed, when transplanted into adult mice, embryonic cells give rise to tumors (Hogan, 1981). Several tumors accordingly feature a gene expression signature approximating a developing embryo, which is correlated with more aggressive phenotypes. A study evaluating the gene expression signature of 293 lung tumors revealed a reversion to a germ cell expression pattern in the most aggressive tumors (Rousseaux et al., 2013). Similarly, a report analyzing colorectal cancer genomic data implicated promoters of developmental genes as major targets of deregulations in their methylation status (An et al., 2015). Embryogenesis involves the precise regulation of several organized signaling pathways, which are then silenced in adult somatic cells. Examples of key developmental signaling pathways reactivated in cancer include the Wnt, Hedgehog, and Notch pathways (Aiello and Stanger, 2016). The reactivation of such pathways—by mutations or epigenetic alterations—is a hallmark feature of cancer, exemplifying the commonalities between cancer and developmental biology.
Epigenetic disruptions, including DNA methylation modifications, contribute to the oncogenic process, leading to aberrant gene activity. Both DNA hypermethylation and hypomethylation are detected in cancer cells: hypermethylation occurs on promoter regions of tumor suppressor genes (TSG), thus leading to gene silencing and tumor progression (Ehrlich, 2002; Locke et al., 2019). In contrast, hypomethylation occurs on DNA repeat sequences, promoting genome instability or oncogene activation.
The genes encoding the Ca2+ signaling toolkit have emerged as targets of aberrant DNA methylation, thus altering Ca2+ homeostasis to perpetuate the carcinogenic process. By rewiring Ca2+ signatures, cancer cells acquire a proliferative advantage compared to typical cells, allowing them to proliferate indefinitely amongst other cancer hallmarks. A study analyzing DNA methylation patterns in 12 cancer cell types showed Ca2+- toolkit genes—including NCX, CAMK, PMCA, PKC, IP3R—as being major targets of hypermethylation and downregulation (Wang et al., 2017). Likewise, alterations in expression and activity levels of SERCA, PMCA, TRPM7, ORAI/STIM1-mediated SOCE, and IP3R isoforms have been demonstrated in several cancer cell types [detailed reviews can be found here (Marchi and Pinton, 2016; Raynal et al., 2016; Izquierdo-Torres et al., 2020)]. Studies in pancreatic cancer have revealed significant rewiring of cancer cell Ca2+ signatures, with subsequent analysis on gene expression profiles implicating aberrant epigenetic programs as underlying mechanisms (Gregório et al., 2020; Kutschat et al., 2021). Similar findings are seen in hepatic cancer stem cells (CSCs), which display altered Ca2+ dynamics due to upregulation of the IP3R2 gene expression (Sun et al., 2019); whether or not these are due to epigenetic modulations are unknown. This study further implicated the altered Ca2+ profiles as underlying the self-renewal of CSCs, with IP3R2 knockdown consistently suppressing tumor formation (Sun et al., 2019). Because Ca2+ functions as a key second messenger, cancer cells—through a rewiring in their Ca2+ signaling machinery—withstand various adverse stimuli and exhibit unique cellular transcriptomes, leading to increased cell proliferation, invasion, and resistance to apoptosis. This rewiring takes place at the genetic level, with aberrant DNA methylations crucial to tumor instigation (Parkash and Asotra, 2010; Gregório et al., 2020).
Epigenetic changes are reversible: therapeutic correction of methylation status is correlated with a significantly more favorable prognosis (Kim et al., 2019; Meneses-Morales et al., 2019; Patergnani et al., 2020). For example, the SERCA3 gene (ATP2A3) exhibits hypermethylation and decreased expression in colon cancer: patients with low expression levels of SERCA3 displayed an average survival of 16.6 months, whereas patients with high expression levels survived 26.7 months (Meneses-Morales et al., 2019). This was consistent with pharmacologic administration of DMNT inhibitors (DMNTi) to induce SERCA3 expression, which decreased cell viability (Meneses-Morales et al., 2019). Collectively, pharmacologically targeting epigenetic-induced disturbances in Ca2+ signaling is currently seen as a potentially feasible and efficacious pharmacologic strategy to combat cancers. Nevertheless, in keeping with the present discussion, identifying Ca2+ channels, transporters, or signaling molecules in oocytes showing OAD and elucidating whether epigenetic dysregulations these findings would not only further our understanding of OAD and TFF but also could inform the identification of novel druggable targets to correct such ailments.
Ca2+ signaling in itself also modulates gene expression through various signal transduction pathways—the mechanism through which Ca2+ profiles regulate cell cycle progression [readers are referred to (Parkash and Asotra, 2010; Marchi and Pinton, 2016) for more detailed descriptions]. Briefly, several early cell-cycle mediating genes in G1 and phosphorylation status of retinoblastoma protein (Rb) in the G1/S transition phase are regulated by Ca2+. In addition, CaMK mediates physiologic cell cycle progression, as evidenced by inhibition of CaMK causing cell cycle arrest. Another Ca2+-dependent protein, calcineurin, mediates progression through the G1 and S phases through activating transcription factors CREB1 and NFAT, which upregulate gene expression of target genes inducing cell cycle progression (Parkash and Asotra, 2010; Cui et al., 2017). In turn, the activation of CaMK and calcineurin requires Ca2+ oscillations generated through the aforementioned Ca2+ toolkit. Therefore, perturbations in the Ca2+ signaling toolkit indirectly influence gene expression (Izquierdo-Torres et al., 2020). However, studies on Ca2+ profiles influencing epigenetic programs are limited.
To this end, a study described 11 pharmacologic agents, including cardiac glycosides, as triggering TSG reactivation—originally epigenetically silenced—in colon cancer (Cui et al., 2017). However, rather than directly modifying the epigenome, such drugs trigger CAMKII activity. Activation of CAMKII causes the release of methyl CpG binding protein from methylated promoters, inducing gene reactivation. Consistently, abolishing CAMKII activity blocked TSG reactivation (Cui et al., 2017). In short, drugs altering CAMKII activity indirectly reactivate epigenetically silenced TSGs via demethylation. Since CAMKII activity relies on Ca2+ profiles, perhaps targeting Ca2+ oscillation-generating channels and transporters represents a therapeutic pathway to reconstitute normal epigenetic patterns in cancer cells, albeit further research is required to address these issues.
In the context of oocyte activation deficiency, could aberrant Ca2+ responses—through CAMKII—lead to epigenetic modifications in genes involved in subsequent development, thus accounting for the reduced developmental success rates seen in oocytes displaying abnormal Ca2+ profiles? CAMKII mediates meiotic resumption during oocyte activation and affects preimplantation embryogenesis. Abnormal postfertilization Ca2+ oscillations-induced changes in CAMKII activity could therefore alter gene expression via epigenetic dysregulation, accounting for the impaired developmental competence of these eggs. At the same time, it is important to consider that epigenetic changes also target Ca2+ homeostasis. In this context, the antihypertensive drug hydralazine induces DNA demethylation of the SERCA2a gene, activating gene expression and thus leading to improved cardiac performance (Kao et al., 2011). Likewise, methamphetamine induces demethylation of the Cav1.2 (L-type Ca2+ channel) gene in mice cardiomyocytes, leading to overexpression and a consequent hypercontractile state as evidenced by histopathologic examination showing contraction band necrosis (Koczor et al., 2015). Such discoveries should prompt investigations into possible pharmacologic interventions aimed at epigenome correction in cancer cells and oocytes.
We investigated the potential link between Ca2+ signaling and homeostasis and DNA methylation alterations in oocytes, intending to elucidate a novel mechanism of OAD; one which involves Ca2+ homeostasis alterations secondarily disrupting the oocyte DNA methylome or vice versa. To strengthen our hypothesis, we discussed the well-established link between Ca2+ profiles and epigenome regulation in cancer cells, which given the similarities in cancer and developmental biology, constitutes a suitable model to study this process.
To this end, comparative studies evaluating the differential expression between normal oocytes and those showing OAD—and those showing TFF after ICSI or IVM—by employing microarrays would constitute a start. Using tiled DNA microarrays, potential causal DNA methylation changes can be identified. Subsequently, pathway enrichment analysis can identify families of genes targeted by such a process. If the Ca2+ signaling toolkit is involved, as is suggested by cancer research and drugs affecting the myocardium, then novel research directions and potential therapeutic and prophylactic strategies might, theoretically, result.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
AS authored the manuscript, AY conceived the idea, authored and critically reviewed the manuscript, JK conceived the idea, and authored critically reviewed the manuscript and KAK critically reviewed and authored the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Aiello, N. M., and Stanger, B. Z. (2016). Echoes of the Embryo: Using the Developmental Biology Toolkit to Study Cancer. Dis. Model. Mech. 9, 105–114. doi:10.1242/DMM.023184
An, N., Yang, X., Cheng, S., Wang, G., and Zhang, K. (2015). Developmental Genes Significantly Afflicted by Aberrant Promoter Methylation and Somatic Mutation Predict Overall Survival of Late-Stage Colorectal Cancer. Sci. Rep. 5 (5), 1–13. doi:10.1038/srep18616
Barberet, J., Barry, F., Choux, C., Guilleman, M., Karoui, S., Simonot, R., et al. (2020). What Impact Does Oocyte Vitrification Have on Epigenetics and Gene Expression? Clin. Epigenet. 12, 121. doi:10.1186/s13148-020-00911-8
Bernhardt, M. L., Zhang, Y., Erxleben, C. F., Padilla-Banks, E., McDonough, C. E., Miao, Y.-L., et al. (2015). CaV3.2 T-type Channels Mediate Ca2+ Entry during Oocyte Maturation and Following Fertilization. J. Cell Sci. 128, 4442–4452. doi:10.1242/jcs.180026
Berridge, M. J., Bootman, M. D., and Roderick, H. L. (2003). Calcium Signalling: Dynamics, Homeostasis and Remodelling. Nat. Rev. Mol. Cell Biol. 4, 517–529. doi:10.1038/nrm1155
Berridge, M. J. (2012). Calcium Signalling Remodelling and Disease. Biochem. Soc. Trans. 40, 297–309. doi:10.1042/BST20110766
Camargo, L. S. A., Munk, M., Sales, J. N., Wohlres-Viana, S., Quintão, C. C. R., and Viana, J. H. M. (2019). Differential Gene Expression between In Vivo and In Vitro Maturation: a Comparative Study with Bovine Oocytes Derived from the Same Donor Pool. JBRA Assist. Reprod. 23, 7–14. doi:10.5935/1518-0557.20180084
Cantone, I., and Fisher, A. G. (2013). Epigenetic Programming and Reprogramming during Development. Nat. Struct. Mol. Biol. 20, 282–289. doi:10.1038/nsmb.2489
Carroll, J. (2000). Na+-Ca2+ Exchange in Mouse Oocytes: Modifications in the Regulation of Intracellular Free Ca2+ during Oocyte Maturation. Reproduction. 118, 337–342. doi:10.1530/jrf.0.1180337
Carvacho, I., Lee, H. C., Fissore, R. A., and Clapham, D. E. (2013). TRPV3 Channels Mediate Strontium-Induced Mouse-Egg Activation. Cell Rep. 5, 1375–1386. doi:10.1016/j.celrep.2013.11.007
Carvacho, I., Piesche, M., Maier, T. J., and Machaca, K. (2018). Ion Channel Function during Oocyte Maturation and Fertilization. Front. Cell Dev. Biol. 6, 63. doi:10.3389/fcell.2018.00063
Cheon, B., Lee, H.-C., Wakai, T., and Fissore, R. A. (2013). Ca2+influx and the Store-Operated Ca2+entry Pathway Undergo Regulation during Mouse Oocyte Maturation. MBoC 24, 1396–1410. doi:10.1091/mbc.E13-01-0065
Ciapa, B., and Arnoult, C. (2011). Could Modifications of Signalling Pathways Activated after ICSI Induce a Potential Risk of Epigenetic Defects? Int. J. Dev. Biol. 55, 143–152. doi:10.1387/ijdb.103122bc
Cui, C., Merritt, R., Fu, L., and Pan, Z. (2017). Targeting Calcium Signaling in Cancer Therapy. Acta Pharmaceutica Sinica B 7, 3–17. doi:10.1016/J.APSB.2016.11.001
Deaton, A. M., and Bird, A. (2011). CpG Islands and the Regulation of Transcription. Genes Dev. 25, 1010–1022. doi:10.1101/gad.2037511
Deniz, E., and Erman, B. (2016). Long Noncoding RNA (lincRNA), a New Paradigm in Gene Expression Control. Funct. Integr. Genomics 17, 135–143. doi:10.1007/S10142-016-0524-X
Ducibella, T., and Matson, S. (2007). Secretory Mechanisms and Ca2+ Signaling in Gametes: Similarities to Regulated Neuroendocrine Secretion in Somatic Cells and Involvement in Emerging Pathologies. Endocr. Pathol. 18, 191–203. doi:10.1007/S12022-007-0015-7
Ehrlich, M. (2002). DNA Methylation in Cancer: Too Much, but Also Too Little. Oncogene 21, 5400–5413. doi:10.1038/sj.onc.1205651
Fujimoto, T., Miyayama, Y., and Fuyuta, M. (1977). The Origin, Migration and fine Morphology of Human Primordial Germ Cells. Anat. Rec. 188, 315–329. doi:10.1002/AR.1091880305
Gasca, S., Reyftmann, L., Pellestor, F., Rème, T., Assou, S., Anahory, T., et al. (2008). Total Fertilization Failure and Molecular Abnormalities in Metaphase II Oocytes. Reprod. Biomed. Online 17, 772–781. doi:10.1016/S1472-6483(10)60404-X
Gregório, C., Soares-Lima, S. C., Alemar, B., Recamonde-Mendoza, M., Camuzi, D., de Souza-Santos, P. T., et al. (2020). Calcium Signaling Alterations Caused by Epigenetic Mechanisms in Pancreatic Cancer: From Early Markers to Prognostic Impact. Cancers 202012, 1735. Page 1735 12. doi:10.3390/CANCERS12071735
Hales, B. F., Grenier, L., Lalancette, C., and Robaire, B. (2011). Epigenetic Programming: from Gametes to Blastocyst. Birth Defects Res. A: Clin. Mol. Teratology 91, 652–665. doi:10.1002/BDRA.20781
Hanahan, D., and Weinberg, R. A. (2011). Hallmarks of Cancer: The Next Generation. Cell 144, 646–674. doi:10.1016/j.cell.2011.02.013
Hilz, S., Modzelewski, A. J., Cohen, P. E., and Grimson, A. (2016). The Roles of microRNAs and siRNAs in Mammalian Spermatogenesis. Development 143, 3061–3073. doi:10.1242/DEV.136721
Hogan, B. (1981). From Embryo to Teratocarcinoma in Tissue Culture. Nature 292, 111–112. doi:10.1038/292111B0
Ito, S., Shen, L., Dai, Q., Wu, S. C., Collins, L. B., Swenberg, J. A., et al. (2011). Tet Proteins Can Convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science 333, 1300–1303. doi:10.1126/science.1210597
Izquierdo-Torres, E., Hernández-Oliveras, A., Fuentes-García, G., and Zarain-Herzberg, Á. (2020). Calcium Signaling and Epigenetics: A Key point to Understand Carcinogenesis. Cell Calcium 91, 102285. doi:10.1016/j.ceca.2020.102285
Kao, Y.-H., Cheng, C.-C., Chen, Y.-C., Chung, C.-C., Lee, T.-I., Chen, S.-A., et al. (2011). Hydralazine-induced Promoter Demethylation Enhances Sarcoplasmic Reticulum Ca2+-ATPase and Calcium Homeostasis in Cardiac Myocytes. Lab. Invest. 91, 1291–1297. doi:10.1038/labinvest.2011.92
Kashir, J., Heindryckx, B., Jones, C., de Sutter, P., Parrington, J., and Coward, K. (2010). Oocyte Activation, Phospholipase C Zeta and Human Infertility. Hum. Reprod. Update 16, 690–703. doi:10.1093/humupd/dmq018
Kashir, J. (2020). Increasing Associations between Defects in Phospholipase C Zeta and Conditions of Male Infertility: Not Just ICSI Failure? J. Assist. Reprod. Genet. 37, 1273–1293. doi:10.1007/s10815-020-01748-z
Kashir, J., Jones, C., Lee, H. C., Rietdorf, K., Nikiforaki, D., Durrans, C., et al. (2011). Loss of Activity Mutations in Phospholipase C Zeta (PLC ) Abolishes Calcium Oscillatory Ability of Human Recombinant Protein in Mouse Oocytes. Hum. Reprod. 26, 3372–3387. doi:10.1093/HUMREP/DER336
Kashir, J., Nomikos, M., Lai, F. A., and Swann, K. (2014). Sperm-induced Ca2+ Release during Egg Activation in Mammals. Biochem. Biophysical Res. Commun. 450, 1204–1211. doi:10.1016/j.bbrc.2014.04.078
Kim, H. S., Choi, J. H., Lee, J. Y., Kang, J., Myung, J. K., Kim, W. H., et al. (2020). Downregulation of SMOC2 Expression in Papillary Thyroid Carcinoma and its Prognostic Significance. Sci. Rep. 10. doi:10.1038/S41598-020-61828-Z
Kim, T., Han, S., Kim, J. T., Yoo, S. M., Lee, M. S., Lee, S. H., et al. (2019). Differential Expression of Tescalcin by Modification of Promoter Methylation Controls Cell Survival in Gastric Cancer Cells. Oncol. Rep. 41, 3464–3474. doi:10.3892/OR.2019.7099
Knott, J. G., Gardner, A. J., Madgwick, S., Jones, K. T., Williams, C. J., and Schultz, R. M. (2006). Calmodulin-dependent Protein Kinase II Triggers Mouse Egg Activation and Embryo Development in the Absence of Ca2+ Oscillations. Developmental Biol. 296, 388–395. doi:10.1016/J.YDBIO.2006.06.004
Koczor, C., Torres, R., Fields, E., Jedrzejczak, M., Jiao, Z., Ludaway, T., et al. (2015). Methamphetamine Alters Calcium Channel Gene DNA Methylation and Gene Expression in the Murine Heart. FASEB j. 29, 768–815. doi:10.1096/FASEBJ.29.1_SUPPLEMENT.768.15
Kouzarides, T. (2007). Chromatin Modifications and Their Function. Cell 128, 693–705. doi:10.1016/J.CELL.2007.02.005
Kurokawa, M., and Fissore, R. A. (2003). ICSI-generated Mouse Zygotes Exhibit Altered Calcium Oscillations, Inositol 1,4,5-trisphosphate Receptor-1 Down-Regulation, and Embryo Development. Mol. Hum. Reprod. 9, 523–533. doi:10.1093/MOLEHR/GAG072
Kutschat, A. P., Johnsen, S. A., and Hamdan, F. H. (2021). Store-Operated Calcium Entry: Shaping the Transcriptional and Epigenetic Landscape in Pancreatic Cancer. Cells 10, 966. Cells 10. doi:10.3390/CELLS10050966
Le Blévec, E., Muroňová, J., Ray, P. F., and Arnoult, C. (2020). Paternal Epigenetics: Mammalian Sperm Provide Much More Than DNA at Fertilization. Mol. Cell Endocrinol. 518, 110964. doi:10.1016/j.mce.2020.110964
Lewis, R. S. (1999). “12 Store-Operated Calcium Channels,” in Advances in Second Messenger and Phosphoprotein Research (Adv Second Messenger Phosphoprotein Res), 279–307. doi:10.1016/S1040-7952(99)80014-7
Liu, W., Su, L.-T., Khadka, D. K., Mezzacappa, C., Komiya, Y., Sato, A., et al. (2011). TRPM7 Regulates Gastrulation during Vertebrate Embryogenesis. Developmental Biol. 350, 348–357. doi:10.1016/j.ydbio.2010.11.034
Locke, W. J., Guanzon, D., Ma, C., Liew, Y. J., Duesing, K. R., Fung, K. Y. C., et al. (2019). DNA Methylation Cancer Biomarkers: Translation to the Clinic. Front. Genet. 10, 1150. doi:10.3389/FGENE.2019.01150
Machaty, Z., Miller, A. R., and Zhang, L. (2017). “Egg Activation at Fertilization,” in Advances in Experimental Medicine and Biology (Springer New York LLC), 1–47. doi:10.1007/978-3-319-46095-6_1
Manzo, G. (2019). Similarities Between Embryo Development and Cancer Process Suggest New Strategies for Research and Therapy of Tumors: A New Point of View. Front. Cell Dev. Biol. 7, 20. doi:10.3389/FCELL.2019.00020
Marchi, S., and Pinton, P. (2016). Alterations of Calcium Homeostasis in Cancer Cells. Curr. Opin. Pharmacol. 29, 1–6. doi:10.1016/J.COPH.2016.03.002
McLay, D. W., Carroll, J., and Clarke, H. J. (2002). The Ability to Develop an Activity that Transfers Histones onto Sperm Chromatin Is Acquired with Meiotic Competence during Oocyte Growth. Developmental Biol. 241, 195–206. doi:10.1006/DBIO.2001.0499
Meneses‐Morales, I., Izquierdo‐Torres, E., Flores‐Peredo, L., Rodríguez, G., Hernández‐Oliveras, A., and Zarain‐Herzberg, Á. (2019). Epigenetic Regulation of the Human ATP2A3 Gene Promoter in Gastric and colon Cancer Cell Lines. Mol. Carcinog. 58, 887–897. doi:10.1002/MC.22978
Menezo, Y. J. R., Silvestris, E., Dale, B., and Elder, K. (2016). Oxidative Stress and Alterations in DNA Methylation: Two Sides of the Same coin in Reproduction. Reprod. BioMedicine Online 33, 668–683. doi:10.1016/j.rbmo.2016.09.006
Monk, M., and Holding, C. (2001). Human Embryonic Genes Re-expressed in Cancer Cells. Oncogene 20, 8085–8091. doi:10.1038/sj.onc.1205088
Ozil, J.-P., Banrezes, B., Tóth, S., Pan, H., and Schultz, R. M. (2006). Ca2+ Oscillatory Pattern in Fertilized Mouse Eggs Affects Gene Expression and Development to Term. Developmental Biol. 300, 534–544. doi:10.1016/J.YDBIO.2006.08.041
Ozil, J. P., and Huneau, D. (2001). Activation of Rabbit Oocytes: the Impact of the Ca2+ Signal Regime on Development. Development 128, 917–928. doi:10.1242/DEV.128.6.917
Palermo, G., Neri, Q., Takeuchi, T., and Rosenwaks, Z. (2009). ICSI: where We Have Been and where We Are Going. Semin. Reprod. Med. 27, 191–201. doi:10.1055/S-0029-1202309
Parkash, J., and Asotra, K. (2010). Calcium Wave Signaling in Cancer Cells. Life Sci. 87, 587–595. doi:10.1016/J.LFS.2010.09.013
Patergnani, S., Danese, A., Bouhamida, E., Aguiari, G., Previati, M., Pinton, P., et al. (2020). Various Aspects of Calcium Signaling in the Regulation of Apoptosis, Autophagy, Cell Proliferation, and Cancer. Int. J. Mol. Sci 21, 8323–8327. doi:10.3390/IJMS21218323
Pauli, A., Rinn, J. L., and Schier, A. F. (2011). Non-coding RNAs as Regulators of Embryogenesis. Nat. Rev. Genet. 12, 136–149. doi:10.1038/NRG2904
Ponting, C. P., and Belgard, T. G. (2010). Transcribed Dark Matter: Meaning or Myth? Hum. Mol. Genet. 19, R162–R168. doi:10.1093/HMG/DDQ362
Raynal, N. J.-M., Lee, J. T., Wang, Y., Beaudry, A., Madireddi, P., Garriga, J., et al. (2016). Targeting Calcium Signaling Induces Epigenetic Reactivation of Tumor Suppressor Genes in Cancer. Cancer Res. 76, 1494–1505. doi:10.1158/0008-5472.CAN-14-2391
Ribas-Maynou, J., Garcia-Bonavila, E., Hidalgo, C. O., Catalán, J., Miró, J., and Yeste, M. (2021). Species-Specific Differences in Sperm Chromatin Decondensation Between Eutherian Mammals Underlie Distinct Lysis Requirements. Front. Cell Dev. Biol. 9, 1143. doi:10.3389/FCELL.2021.669182/BIBTEX
Rizzuto, R., Bernardi, P., and Pozzan, T. (2000). Mitochondria as All‐round Players of the Calcium Game. J. Physiol. 529, 37–47. doi:10.1111/j.1469-7793.2000.00037.x
Rogers, N. T., Halet, G., Piao, Y., Carroll, J., Ko, M. S. H., and Swann, K. (2006). The Absence of a Ca2+ Signal during Mouse Egg Activation Can Affect Parthenogenetic Preimplantation Development, Gene Expression Patterns, and Blastocyst Quality. Reproduction 132, 45–57. doi:10.1530/REP.1.01059
Rousseaux, S., Debernardi, A., Jacquiau, B., Vitte, A.-L., Vesin, A., Nagy-Mignotte, H., et al. (2013). Ectopic Activation of Germline and Placental Genes Identifies Aggressive Metastasis-Prone Lung Cancers. Sci. Transl. Med. 5. 186ra66. doi:10.1126/SCITRANSLMED.3005723
Saleh, A., Kashir, J., Thanassoulas, A., Safieh-Garabedian, B., Lai, F. A., and Nomikos, M. (2020). Essential Role of Sperm-specific PLC-Zeta in Egg Activation and Male Factor Infertility: An Update. Front. Cell Dev. Biol. 8, 28. doi:10.3389/fcell.2020.00028
Sanders, J. R., and Swann, K. (2016). Molecular Triggers of Egg Activation at Fertilization in Mammals. Reproduction 152, R41–R50. doi:10.1530/REP-16-0123
Sassone-Corsi, P. (2002). Unique Chromatin Remodeling and Transcriptional Regulation in Spermatogenesis. Science 296, 2176–2178. doi:10.1126/SCIENCE.1070963
Satouh, Y., Nozawa, K., and Ikawa, M. (2015). Sperm Postacrosomal WW Domain-Binding Protein Is Not Required for Mouse Egg Activation1. Biol. Reprod. 93, 1. doi:10.1095/BIOLREPROD.115.131441
Saunders, C. M., Larman, M. G., Parrington, J., Cox, L. J., Royse, J., Blayney, L. M., et al. (2002). PLCζ: a Sperm-specific Trigger of Ca2+ Oscillations in Eggs and Embryo Development. Development 129, 3533–3544. doi:10.1242/dev.129.15.3533
Seisenberger, S., Peat, J. R., Hore, T. A., Santos, F., Dean, W., and Reik, W. (2013). Reprogramming DNA Methylation in the Mammalian Life Cycle: Building and Breaking Epigenetic Barriers. Phil. Trans. R. Soc. B 368, 20110330. doi:10.1098/RSTB.2011.0330
Sendžikaitė, G., and Kelsey, G. (2019). The Role and Mechanisms of DNA Methylation in the Oocyte. Essays Biochem. 63, 691–705. doi:10.1042/EBC20190043
Skene, P. J., and Henikoff, S. (2017). An Efficient Targeted Nuclease Strategy for High-Resolution Mapping of DNA Binding Sites. Elife 6, e21856. doi:10.7554/ELIFE.21856
Stein, P., Savy, V., Williams, A. M., and Williams, C. J. (2020). Modulators of Calcium Signalling at Fertilization. Open Biol. 10, 200118. doi:10.1098/rsob.200118
Sun, C., Shui, B., Zhao, W., Liu, H., Li, W., Lee, J. C., et al. (2019). Central Role of IP3R2-Mediated Ca2+ Oscillation in Self-Renewal of Liver Cancer Stem Cells Elucidated by High-Signal ER Sensor. Cell Death Dis 10, 1. doi:10.1038/s41419-019-1613-2:
Tesarik, J., and Sousa, M. (1994). Comparison of Ca2+ Responses in Human Oocytes Fertilized by Subzonal Insemination and by Intracytoplasmic Sperm Injection. Fertil. Sterility 62, 1197–1204. doi:10.1016/S0015-0282(16)57185-4
Tiwari, M., Prasad, S., Shrivastav, T. G., and Chaube, S. K. (2017). Calcium Signaling During Meiotic Cell Cycle Regulation and Apoptosis in Mammalian Oocytes. Available at: https://pubmed.ncbi.nlm.nih.gov/27791263/(Accessed May 22, 2021).
Tosti, E. (2006). Calcium Ion Currents Mediating Oocyte Maturation Events. Reprod. Biol. Endocrinol. 4, 26. doi:10.1186/1477-7827-4-26
Tsaadon, L., Kaplan-Kraicer, R., and Shalgi, R. (2008). Myristoylated Alanine-Rich C Kinase Substrate, but Not Ca2+/calmodulin-dependent Protein Kinase II, Is the Mediator in Cortical Granules Exocytosis. Reproduction 135, 613–624. doi:10.1530/REP-07-0554
Tucci, V., Isles, A. R., Kelsey, G., Ferguson-Smith, A. C., Tucci, V., Bartolomei, M. S., et al. (2019). Genomic Imprinting and Physiological Processes in Mammals. Cell 176, 952–965. doi:10.1016/j.cell.2019.01.043
Virant-Klun, I., Bauer, C., Ståhlberg, A., Kubista, M., Skutella, T., I, V.-K., et al. (2018). Human Oocyte Maturation In Vitro Is Improved by Co-culture with Cumulus Cells from Mature Oocytes. Reprod. BioMedicine Online 36, 508–523. doi:10.1016/J.RBMO.2018.01.011
Wakai, T., Vanderheyden, V., and Fissore, R. A. (2011). Ca2+ Signaling During Mammalian Fertilization: Requirements, Players, and Adaptations. Cold Spring Harbor Perspect. Biol. 3, a006767. doi:10.1101/CSHPERSPECT.A006767
Wakai, T., Zhang, N., Vangheluwe, P., and Fissore, R. A. (2013). Regulation of Endoplasmic Reticulum Ca2+ Oscillations in Mammalian Eggs. J. Cell Sci. 126, 5714–5724. doi:10.1242/jcs.136549
Wang, X.-X., Xiao, F.-H., Li, Q.-G., Liu, J., He, Y.-H., and Kong, Q.-P. (2017). Large-scale DNA Methylation Expression Analysis across 12 Solid Cancers Reveals Hypermethylation in the Calcium-Signaling Pathway. Oncotarget 8, 11868–11876. doi:10.18632/ONCOTARGET.14417
Wang, Y.-Y., Zhao, R., and Zhe, H. (2015). The Emerging Role of CaMKII in Cancer. Available at: www.impactjournals.com/oncotarget/.
Ward, W. S. (2010). Function of Sperm Chromatin Structural Elements in Fertilization and Development. Mol. Hum. Reprod. 16, 30–36. doi:10.1093/MOLEHR/GAP080
Whitaker, M. (2008). Calcium Signalling in Early Embryos. Phil. Trans. R. Soc. B 363, 1401–1418. doi:10.1098/RSTB.2008.2259
Xia, W., Xu, J., Yu, G., Yao, G., Xu, K., Ma, X., et al. (2019). Resetting Histone Modifications during Human Parental-To-Zygotic Transition. Science 365, 353–360. doi:10.1126/SCIENCE.AAW5118
Yeste, M., Jones, C., Amdani, S. N., and Coward, K. (2017). Oocyte Activation and Fertilisation: Crucial Contributors from the Sperm and Oocyte. Results Probl. Cell Differ 59, 213–239. doi:10.1007/978-3-319-44820-6_8
Yeste, M., Jones, C., Amdani, S. N., Patel, S., and Coward, K. (2016). Oocyte Activation Deficiency: A Role for an Oocyte Contribution? Hum. Reprod. Update 22, 23–47. doi:10.1093/HUMUPD/DMV040
Zafar, M. I., Lu, S., and Li, H. (2021). Sperm-oocyte Interplay: an Overview of Spermatozoon's Role in Oocyte Activation and Current Perspectives in Diagnosis and Fertility Treatment. Cell Biosci 11. doi:10.1186/s13578-020-00520-1
Zhang, B., Zheng, H., Huang, B., Li, W., Xiang, Y., Peng, X., et al. (2016). Allelic Reprogramming of the Histone Modification H3K4me3 in Early Mammalian Development. Nature 537, 553–557. doi:10.1038/NATURE19361
Zhang, H.-L., Xu, Y., Ju, J.-Q., Pan, Z.-N., Liu, J.-C., and Sun, S.-C. (2021). Increased Environment-Related Metabolism and Genetic Expression in the In Vitro Matured Mouse Oocytes by Transcriptome Analysis. Front. Cell Dev. Biol. 9, 171. doi:10.3389/FCELL.2021.642010
Keywords: oocyte activation, DNA methylation, calcium, cancer, fertlization
Citation: Shafqat A, Kashir J, Alsalameh S, Alkattan K and Yaqinuddin A (2022) Fertilization, Oocyte Activation, Calcium Release and Epigenetic Remodelling: Lessons From Cancer Models. Front. Cell Dev. Biol. 10:781953. doi: 10.3389/fcell.2022.781953
Received: 23 September 2021; Accepted: 14 February 2022;
Published: 04 March 2022.
Edited by:
Rafael A. Fissore, University of Massachusetts Amherst, United StatesCopyright © 2022 Shafqat, Kashir, Alsalameh, Alkattan and Yaqinuddin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ahmed Yaqinuddin, YXlhcWludWRkaW5AYWxmYWlzYWwuZWR1
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.