- 1Department of Neurology, Nara Medical University, Kashihara, Japan
- 2Department of Future Basic Medicine, Nara Medical University, Kashihara, Japan
- 3Micro-Nano Biomechanics Laboratory, Department of Mechanical Systems Engineering, Ibaraki University, Hitachi, Japan
- 4Life Science Center for Survival Dynamics, Tsukuba Advanced Research Alliance, Tsukuba, Japan
- 5V-iCliniX Laboratory, Nara Medical University, Kashihara, Japan
Proline:arginine (PR) poly-dipeptides from the GGGGCC repeat expansion in C9orf72 have cytotoxicity and bind intermediate filaments (IFs). However, it remains unknown how PR poly-dipeptides affect cytoskeletal organization and focal adhesion (FA) formation. Here, we show that changes to the cytoskeleton and FA by PR poly-dipeptides result in the alteration of cell stiffness and mechanical stress response. PR poly-dipeptides increased the junctions and branches of the IF network and increased cell stiffness. They also changed the distribution of actin filaments and increased the size of FA and intracellular calcium concentration. PR poly-dipeptides or an inhibitor of IF organization prevented cell detachment. Furthermore, PR poly-dipeptides induced upregulation of mechanical stress response factors and led to a maladaptive response to cyclic stretch. These results suggest that the effects of PR poly-dipeptides on mechanical properties and mechanical stress response may serve as a pathogenesis of C9orf72-related neurodegeneration.
Introduction
A hexanucleotide repeat expansion in C9orf72 is common in familial amyotrophic lateral sclerosis (ALS) with frontotemporal dementia (FTD) (C9-ALS/FTD) (DeJesus-Hernandez et al., 2011), which leads to the production of proline:arginine (PR), glycine:arginine (GR), glycine:alanine (GA), proline:alanine (PA), and glycine:proline (GP) poly-dipeptides (Ash et al., 2013; Kwon et al., 2014). Arginine-rich poly-dipeptides show cytotoxicity (Kwon et al., 2014; Jovičić et al., 2015; Hao et al., 2019) and cause cell deformation and degeneration (Kwon et al., 2014; Mizielinska et al., 2014), and intermediate filaments (IFs) were found to be the binding targets of arginine-rich poly-dipeptides (Lee et al., 2016; Lin et al., 2016).
IFs regulate the focal adhesion (FA) formation and cellular mechanical properties (Kim et al., 2016; Hu et al., 2019; Laly et al., 2021). FA is responsible for adhesion to the extracellular matrix, which is formed by protein complexes such as integrin, vinculin, paxillin, focal adhesion kinase (FAK), talin, zyxin, α-Actinin, vasodilator-stimulated phosphoprotein (VASP) (Abercrombie and Dunn, 1975). Cytoskeleton and FA convert intracellular and extracellular mechanical forces into intracellular signals that induce mechanical stress responses (Hurtley, 1998; Fletcher and Mullins, 2010; López-Colomé et al., 2017). The cytoskeletal dysfunction was found as a feature of neurodegenerative diseases, for example Parkinson’s disease (McLean et al., 2000; Sweers et al., 2011; Häbig et al., 2013; Lu et al., 2017), Alzheimer’s disease (Bamburg and Bloom, 2009; Levy Nogueira et al., 2016), and ALS (Wu et al., 2012; Smith et al., 2014; Nicolas et al., 2018; Zhang et al., 2022). However, it is unclear whether the mechanical stress response is associated with the pathogenesis of neurodegeneration. Therefore, it is crucial to evaluate the cytoskeletal organization, FA formation and the mechanical stress response induced by arginine-rich poly-dipeptides.
In this study, we evaluated the effects of PR poly-dipeptides, the most toxic among five different poly-dipeptides (Kwon et al., 2014), on the cytoskeletal organization, cell stiffness, and FA formation. PR poly-dipeptides induced a high-density network of IFs and increased cell stiffness in conjunction with the abnormal remodeling of actin filaments and FA. PR poly-dipeptides prevented cell detachment. Furthermore, they induced the expression of mechanical stress response factors, leading to a maladaptive mechanical stress response. These results suggest that PR poly-dipeptides alter the stability of IFs, resulting in changes to mechanical stress responses.
Materials and Methods
Peptide Synthesis
A synthetic peptide consisting of twenty repeats of the PR poly-dipeptide (PR20) with an HA tag at the carboxyl terminus was synthesized (SCRUM Inc., Tokyo, JAPAN).
Cell Culture
Human osteosarcoma cells (U2OS) and fibroblast (BJ-hTERT) cells were cultured in Dulbecco’s modified Eagle medium (DMEM) high glucose with 10% fetal bovine serum (FBS, Hyclone) and 1% penicillin-streptomycin at 37°C in 5% CO2. The cells were used for experiments after a one-hour treatment at 37°C with a synthetic peptide consisting of PR20 (final concentration 10 µM). Rat vascular smooth muscle cells (SMCs; Lonza, R-ASM-580) were grown in DMEM with 20% FBS and 1x Antibiotic-Antimyotic (Thermo Fisher Scientific).
Immunofluorescence
The U2OS cells, fibroblast BJ-hTERT cells or rat vascular SMCs were fixed in 4% paraformaldehyde in phosphate-buffered saline (PBS) at room temperature for 15 min and permeabilized with 0.1% Triton X-100 in PBS for 10 min. The fixed cells were incubated with a blocking solution (5% bovine serum albumin in PBS with 0.1% Tween20) at room temperature for 1 h. The cells were incubated with primary antibodies, vimentin (Santa Cruz Biotechnology, sc6260, 1:100), cytokeratin 14 (Abcam, ab7800, 1:250), vinculin (Abcam, ab129002, 1:500), phospho-Paxillin (Cell Signaling, 2541, 1:50), phospho-ERM (Cell Signaling, 3726, 1:200), or phospho-Cofilin (Cell Signaling, 3313, 1:100) in the blocking solution at 4°C overnight. Secondary antibodies (Thermo Fisher Scientific, A-21422, A-21429, 1:2000) were incubated at room temperature for 1 h in the blocking solution. Alexa Fluor488-phalloidin (Thermo Fisher Scientific, A12379, 1:1000) and Alexa Fluor555-β-tubulin (Abcam, ab206627, 1:1000) were also incubated at room temperature for an hour in the blocking solution. Images were captured using the confocal microscope FV3000 (Olympus, Tokyo) or LSM 710 (ZEISS). Captured images were analyzed by ImageJ (version 1.53d, National Institutes of Health (NIH), United States).
Imaging Analysis
Analysis of IF organization was performed by quantifying the junctions, branches, and alignments. The images were converted to 8-bit and binarized using fixed contrast and brightness thresholds. For junction and branch measurements, 100 μm2 regions of interest (ROI) in the cytoplasm, excluding the nucleus, were selected. At least one ROI was selected per cell. The number of junctions and branches was measured in the skeletonized images using the skeleton plugin (https://imagej.net/AnalyzeSkeleton, last accessed 1 March 2022). The bundle alignments of IFs were measured by the dispersion of the IF angles in whole cells (Laly et al., 2021), using the OrientationJ plugin (http://bigwww.epfl.ch/demo/orientation/, last accessed 1 March 2022). Fluorescence intensity of IFs in perinuclear regions was measured within 1 μm from the nucleus. Fluorescence intensity of actin filaments was evaluated by the Quimp plugin, which outlines a cell using their original algorithm (Baniukiewicz et al., 2018). Based on the outline, the plugin measures the fluorescence intensity of the whole cell and around the cell cortex. As the fluorescence intensity around the cell cortex is affected by the size of the cell, the ratio of fluorescence intensity around the cell cortex to the fluorescence intensity of the whole cell was calculated. To measure the fluorescence intensity of pERM, a region of cells was defined in the merged image. The average fluorescence intensity of this region was measured on the pERM image. The average fluorescence intensity was divided by the number of cells in the ROI to obtain the fluorescence intensity of pERM per cell.
Analysis of FA was performed by quantifying the size and the number of FA. The images were converted to 8-bit and binarized using fixed contrast and brightness thresholds. The average size and the number of FAs per cell were quantified from binary images using the “Analyze Particle” function in ImageJ (https://imagej.net/imaging/particle-analysis, last accessed 1 March 2022) with the following settings: size (pixel^2), 0.5-infinity; circularity, 0.00–1.00. The polymerized β-tubulin was measured using the Tubeness plugin (https://imagej.net/plugins/tubeness, last accessed 1 March 2022), which identified tube-like structures. The ratio of the polymerized β-tubulin to cell area was calculated.
Atomic Force Microscopy
Atomic force microscopy (AFM) measurements were performed using a NanoWizard IV AFM (JPK Instruments-AG, Germany) mounted on top of an inverted optical microscope (IX73, Olympus, Japan) equipped with a digital CMOS camera (Zyla, Andor) as described in a previous study (Nagayama et al., 2019). Prior to AFM imaging of the surface topography and mechanical properties of U2OS cells in PR20-treated cells, the cells were adapted to a CO2-independent medium (Invitrogen) for 30 min at room temperature (25 °C). AFM quantitative imaging (QI) mode was used to obtain a force–displacement curve at a resolution of 128 × 128 pixels (100 µm × 100 µm of measured area) by a precisely controlled high-speed indentation test using rectangular-shaped silicon nitride cantilevers with a cone probe (BioLever-mini, BL-AC40TS-C2, Olympus, Japan). The test was performed at a spring constant of 0.08–0.10 N/m and a nominal tip radius of 10 nm. The QI mode measurements were performed within an hour after the transfer of the specimen to the AFM. These high-speed indentations were performed until a preset force of 1 nN was reached. This typically corresponded to cell indentation depths of 300–400 nm. Cell elasticity was calculated from the obtained force–displacement curves by applying the Hertzian model (Hertz, 1881), which approximates the sample to be isotropic and linearly elastic. Young’s (elastic) modulus is extracted by fitting all force–displacement curves with the following Hertzian model approximation:
where F is the applied force, E is the elastic modulus, ν is the Poisson’s ratio (0.5 for a non-compressible biological sample), α is the opening angle of the cone of the cantilever tip, and δ is the indentation depth of the sample recorded in the force–displacement curves. Using the results of the Hertzian model approximation, we identified the Z contact points (specimen surface) and the elastic modulus of the specimens at each pixel and produced a surface topography map and elastic modulus map of the specimens.
Intracellular Calcium Imaging
The intracellular calcium concentration was analyzed using the Calbryte590™ assay kit (AAT Bioquest, United States). The U2OS cells were seeded in 96-well glass bottom microplates and cultured for 24 h. The cells were incubated for an hour at 37°C with Calbryte590™. After removal of the Calbryte590™ solution, the cells were administrated to 10 µM of PR20 for an hour and analyzed by a confocal microscopy and SpectraMax microplate reader (Molecular Devices, United States).
Cell Detachment Experiment
The U2OS cells were transfected with LifeAct-GFP plasmids (60101, ibidi GmbH) using FuGENE-HD transfection reagent (E2311, Promega). For the cell detachment experiment, transfected cells were seeded in glass bottom plates and cultured for 24 h. The cells were administered to 10 µM of PR20 for an hour, 50 μM of (-)-Blebbistatin (B0560-1MG, Sigma-Aldrich) for an hour (Korobova et al., 2014), 10 µM of Nocodazole (M1404-10MG, Sigma-Aldrich) for 30 min (Baum et al., 2014), 2 µM of Withaferin-A (ab120644, Abcam) for 3 hours (Grin et al., 2012), and observed after 30 min incubation with 5 µM of EDTA. EDTA reduces integrin-mediated adhesion, which is a Ca2+-dependent FA molecule by chelating Ca2+ and Mg2+, causing cell detachment (Mould et al., 1995). Equivalent amounts of DMSO (D8418, Sigma–Aldrich) were added as vehicle control, as Blebbistatin, Nocodazole and Withaferin-A were diluted with DMSO. Blebbistatin, Nocodazole and Withaferin-A are used as cytoskeleton inhibitors. Blebbistatin is an inhibitor of myosinⅡ and inhibits the formation of actomyosin and suppresses stress fiber formation (Kovács et al., 2004). Nocodazole binds to the growing end of β-tubulin and prevents microtubule polymerization (Chang et al., 2008). Withaferin-A binds to the vimentin rod domain and disrupts both Vim-IF and Keratin-IF network (Bargagna-Mohan et al., 2007; Grin et al., 2012). Images were captured using a confocal microscope FV3000 (Olympus, Tokyo). Captured images were analyzed by ImageJ (version 1.53d, NIH, United States). As detached cells with a round shape showed high circularity (Fuhrmann et al., 2014), degrees of cell detachment were analyzed by measuring the circularity of cells.
Cyclic Stretch Experiment
Cyclic stretch was performed using a uniaxial cell stretch system (Central Workshop Tsukuba University) as described in a previous study (Yamashiro et al., 2020). We employed rat vascular smooth muscle cells (SMCs) or U2OS cells. The cells were plated on silicon elastomer bottomed culture plates (SC4Ha, Menicon Life Science) coated with a cell attachment factor containing gelatin (Thermo Fisher Scientific, S006100) and subjected to cyclic stretch with a frequency of 1.0 Hz (60 cycles/min) and 20% strain for 6 h.
Western Blotting
Total proteins are isolated from rat vascular SMCs treated with 10 µM of PR20 using RIPA lysis buffer. After sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS-PAGE), proteins were transferred to polyvinylidene difluoride membranes (PVDF). Membranes were blocked with and incubated overnight at 4°C with the following primary antibodies: Thbs1 (NeoMarkers, MS-421), Egr1 (Cell Signaling, 4154), phospho-FAK(Y397) (Cell Signaling, 3283), FAK (Cell Signaling, 3285), phospho-ERK (Cell Signaling, 4376), ERK (Cell Signaling, 9102), phospho-ERM (Cell Signaling, 3141), ERM (Cell Signaling, 3142), GAPDH (Cell Signaling, 2118). Then, membranes were incubated with second antibodies anti-mouse or anti-rabbit HRP-conjugated antibody (1:1000, Bio-Rad). Blots were visualized using a chemiluminescence kit (Santa Cruz Biotechnology) or SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Fisher Scientific).
Statistical Analysis
All experiments are presented as means ± SD (except for Figure 6B). Statistical analysis was performed using Prism 8 (GraphPad Software, California, United States). A Shapiro-Wilk test was used to test for normality. The Mann–Whitney U test, a nonparametric test, or unpaired t test was conducted. p < 0.05 denotes statistical significance.
Results
PR Poly-Dipeptides Increase the Junctions and Branches of the Intermediate Filament Network
To investigate the effect of PR poly-dipeptides on cytoskeleton, we first examined the changes of Vimentin (Vim) and Keratin intermediate filament organization in U2OS cells. Vim-IFs were predominantly localized to the perinuclear region in the control cells (CTRL; vehicle control) (Figure 1A). After 1 h of exposure to 10 µM of PR20, the fluorescence intensity of Vim-IFs in the perinuclear region decreased (p = 0.028; 19.8 ± 2.8 a.u. in CTRL, 12.6 ± 1.5 a.u. in PR20-treated; Supplementary Figure S1A) and Vim-IFs formed a mesh network in the cytoplasm (Figure 1A). The IF organization was analyzed by the alignments and the number of branches and junctions in skeletonized images (Supplementary Figure S1B). Vim-IF alignments were comparable between CTRL and PR20-treated cells (p = 0.678; 21.0 ± 2.2 in CTRL, 21.9 ± 2.5 in PR20-treated; Supplementary Figure S1C). There were more junctions of Vim-IFs in PR20-treated cells than in CTRL (p = 0.026; 97.3 ± 18.1 in CTRL, 154.5 ± 5.9 in PR20-treated; Figure 1B). There were also more branches of Vim-IFs in PR20-treated cells than in CTRL (p = 0.016; 232.9 ± 39.5 in CTRL, 358.2 ± 21.0 in PR20-treated; Figure 1C).
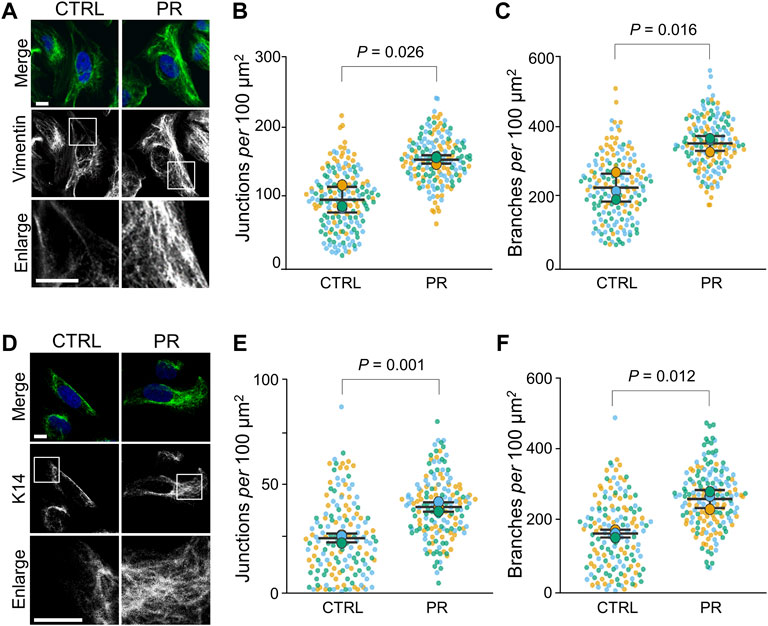
FIGURE 1. PR poly-dipeptides (PR20) change the organization of intermediate filaments (IF) of U2OS cells. (A) Fluorescent images of Vim-IFs in untreated control (CTRL) and PR20-treated (PR) cells. Vim-IFs (green) and DAPI (blue) are shown. Scale bars are 10 µm. (B,C) Quantification of Vim-IF junctions (in B) and branches (in C) per 100 μm2 in the cytoplasm in CTRL and PR cells. Bars are means ± SD. In each experiment, 50 to 60 cells were evaluated (n = 3). P value shows Welch’s t-test. (D) Fluorescent images of cytokeratin14 (K14) in CTRL and PR cells. K14 (green) and DAPI (blue) are shown. Scale bars are 10 µm. (E,F) Quantification of Keratin-IF junctions (in E) and branches (in F) per 100 μm2 in the cytoplasm in CTRL and PR cells. Bars are means ± SD. In each experiment, 50 to 58 cells were evaluated (n = 3). p value shows Welch’s t-test. In quantification graphs, small dots are individual datapoints, and large dots indicate the averages of each experiment in orange (first; n = 1), blue (second; n = 2) and green (third; n = 3), respectively.
Next, Keratin-IFs were analyzed using cells stained with anti-cytokeratin14 (K14) antibody. Keratin-IFs formed a mesh network in the cytoplasm of PR20-treated cells (Figure 1D). The fluorescence intensity of Keratin-IFs in the perinuclear region was comparable between CTRL and PR20-treated cells (p = 0.428; 12.7 ± 1.3 in CTRL, 13.9 ± 2.2 in PR20-treated; Supplementary Figure S1D). Keratin-IF alignments were also comparable between CTRL and PR20-treated cells (p = 0.919; 44.7 ± 3.6 in CTRL, 45.1 ± 5.2 in PR20-treated; Supplementary Figure S1E). There were more junctions (p = 0.001; 25.5 ± 2.1 in CTRL, 40.3 ± 2.3 in PR20-treated) and branches (p = 0.012; 165.9 ± 11.0 in CTRL, 264.3 ± 26.0 in PR20-treated) of Keratin-IFs in PR20-treated cells than in CTRL (Figures 1E,F). Together, these results indicate that PR poly-dipeptides induce a high-density network of IFs in the cytoplasm.
PR Poly-Dipeptides Increase Cell Stiffness
To evaluate the effects of PR poly-dipeptides on cellular mechanics, we analyzed the cell stiffness and cell height by atomic force microscopy (AFM) in U2OS cells (Figure 2A). The number of filamentous actin (F-actin) stress fibers across the whole cell was reduced, and F-actin stress fibers around the nucleus were thick and short in PR20-treated cells (Figure 2B, middle, arrows). We measured the average elastic modulus at the cell central region. Cell elasticity at the cell central region increased in PR20-treated cells than in CTRL (p = 0.032; 8.6 ± 5.1 kPa in CTRL, 12.9 ± 8.2 kPa in PR20-treated; Figure 2C). There were no significant differences in cell height between CTRL and PR20-treated cells (p = 0.583; 3.9 ± 1.0 µm in CTRL, 4.0 ± 0.9 µm in PR20-treated; Figure 2D). These results suggest that PR poly-dipeptides increase cell stiffness by thick and short F-actin stress fibers.
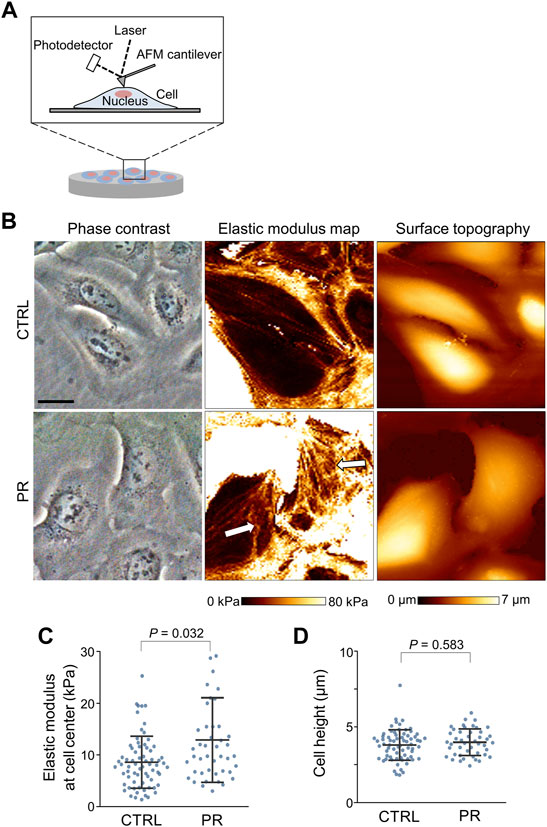
FIGURE 2. Atomic force microscopy (AFM) images of the surface of U2OS cells. (A) Schematic view of AFM measurement of cell surface, including cell stiffness and cell height. (B) Representative images of phase contrast images (left), elastic modulus maps (middle), and surface topographic images (right) of U2OS cells measured by AFM. Arrows in the middle image show thick- and short-stress fibers in PR20-treated (PR) cells. Scale bars are 20 µm. (C,D) Quantification graph shows elastic modulus at cell central region (in C) and cell height (in D) in CTRL and PR cells. Bars are means ± SD. 45 to 69. Cells were evaluated per condition. P value shows Mann–Whitney U test.
PR Poly-Dipeptides Change the Distribution of F-Actin
To investigate the effects of PR poly-dipeptides on other cytoskeletal elements, we analyzed the distribution of F-actin by fluorescence images (Supplementary Figure S2A). In CTRL, F-actin extended across the whole cell and equally distributed in the cytoplasm, whereas after PR20-treatment, F-actin was not distributed equally in the cytoplasm but mainly localized around the cell cortex in U2OS cells and BJhTERT cells (Figure 3A). PR20-treated cells showed a higher fluorescence intensity of F-actin around the cell cortex than CTRL in both cell lines (U2OS; p = 0.007; 8.4 ± 0.6%, in CTRL, 11.0 ± 0.7% in PR20-treated cells. BJhTERT; p = 0.032; 9.1 ± 0.2%, in CTRL, 10.8 ± 0.6% in PR20-treated cells; Figures 3B,C). Similar to U2OS cells and BJhTERT cells (Figure 3A), PR20 changed F-actin organization in rat vascular smooth muscle cells (SMCs) (Supplementary Figure S2B). PR20 also caused nuclear deformation and the unclear boundary between the nucleus and the endoplasmic reticulum (ER) in SMCs (Supplementary Figure S2C). These results suggest that PR poly-dipeptides change the distribution of F-actin.
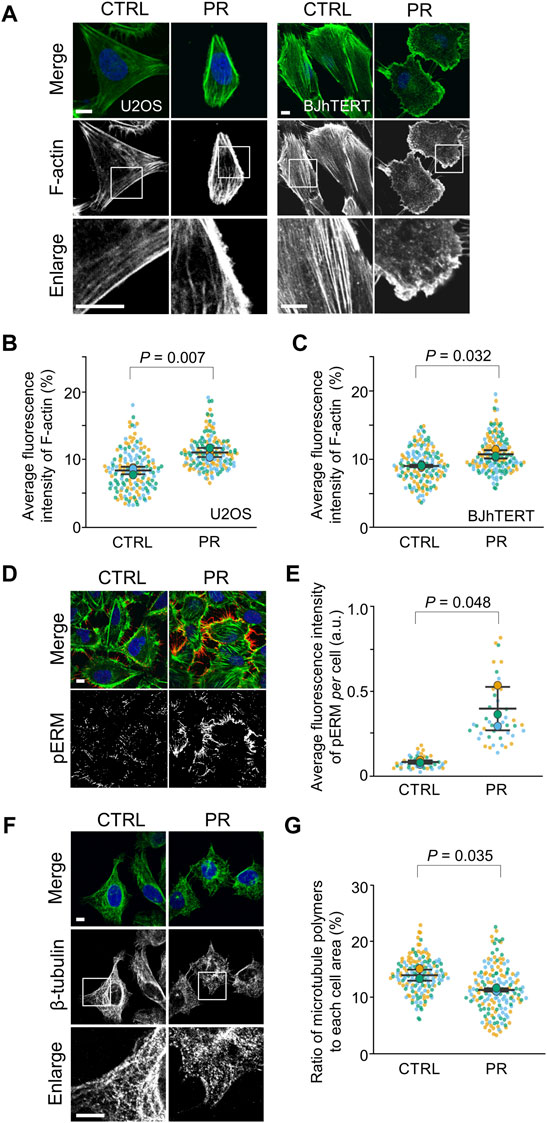
FIGURE 3. PR20 changes the organization of F-actin and β-tubulin and promotes the phosphorylation of ERM (pERM). (A) Fluorescent images of phalloidin in U2OS and BJhTERT cells after treatment with 10 µM of PR20 or CTRL. phalloidin (green) and DAPI (blue) are shown. Scale bars are 10 µm. (B,C) Quantification of average fluorescence intensity of F-actin in U2OS cells (in B) and BJhTERT cells (in C). Bars are means ± SD. In each experiment, 50 to 55 cells were evaluated (n = 3). p value shows Welch’s t-test. (D) Fluorescent images of pERM (red), phalloidin (green) and DAPI (blue) after treatment with or without 10 µM of PR20 in U2OS cells. A bar is 10 µm. (E) Average fluorescence intensity of pERM per cell after treatment with 10 µM of PR20 or CTRL. Bars are means ± SD. 15 regions were evaluated (n = 3). p value shows Welch’s t-test. (F) Fluorescent images of β-tubulin in U2OS cells after treatment with 10 µM of PR20 or CTRL. Bars are 10 µm. (G) The ratio of microtubule polymers in CTRL and PR20-treated (PR) cells. Bars are means ± SD. In each experiment, 50 to 57 cells were evaluated (n = 3). p value shows Welch’s t-test. In quantification graphs, small dots are individual datapoints, and large dots indicate the averages of each experiment in orange (first; n = 1), blue (second; n = 2) and green (third; n = 3), respectively.
To evaluate whether the redistribution of F-actin by PR poly-dipeptides result from the F-actin reorganization, we analyzed Ezrin/Radixin/Moesin (ERM) proteins and Cofilin. As cortical F-actin is formed by ERM proteins (Bretscher et al., 2002), we analyzed cortical F-actin by ERM phosphorylation in U2OS cells (Supplementary Figure S2D). Phospho-ERM (pERM) localized to protrusive structures, such as filopodia, both in CTRL and PR20-treated cells (Figure 3D). The fluorescence intensity of pERM per cell was increased in PR20-treated cells (p = 0.048; 0.1 ± 0.0 a.u. in CTRL, 0.4 ± 0.1 a.u. in PR20-treated; Figure 3E). As F-actin is depolymerized by phosphorylated Cofilin (pCofilin) (Sumi et al., 1999), we evaluated pCofilin. The level of pCofilin was comparable between CTRL and PR20-treated cells in U2OS cells and SMCs (Supplementary Figures S2E,F). These results suggest that PR poly-dipeptides enhance the reorganization of cortical F-actin through pERM.
To evaluate the effects of PR poly-dipeptides on microtubules, we analyzed the ratio of polymerized β-tubulin. In CTRL, β-tubulin showed a filamentous network in the cytoplasm, but it appeared as dots with a diffused network in PR20-treated cells (Figure 3F). Compared to CTRL, PR20-treatment reduced the ratio of polymerized β-tubulin (p = 0.035; 13.9 ± 1.0% in CTRL, 11.3 ± 0.3% in PR20-treated cells; Figure 3G). These results suggest that PR poly-dipeptides disrupt the stability of the microtubule network.
PR Poly-Dipeptides Increase FA Size and Intracellular Calcium Concentration
To investigate the effects of PR poly-dipeptides on FA formation, we analyzed the localization, the number and size of Vinculin and pPaxillin in U2OS cells (Supplementary Figure S3A). In CTRL, we found Vinculin and pPaxillin to be equally distributed in the cytoplasm (Supplementary Figure S3B; Figure 4A). On the other hand, in PR20-treated cells, Vinculin and pPaxillin were reduced in the cell central region and mainly observed in the cell periphery (Supplementary Figure S3B; Figure 4A). Although the number of Vinculin decreased in PR20-treated cells (p = 0.003; 68.9 ± 3.6 in CTRL, 36.6 ± 6.2 in PR20-treated; Supplementary Figure S3C), the size of Vinculin was comparable between CTRL and PR20-treated cells (p = 0.506; 1.6 ± 0.1 µm2 in CTRL, 1.7 ± 0.3 µm2 in PR20-treated; Supplementary Figure S3D). There was no significant difference in the fluorescence intensity of pPaxillin between CTRL and PR20-treated cells (p = 0.131; 3.3 ± 1.0 a.u. in CTRL, 6.2 ± 2.2 a.u. in PR20-treated; Figure 4B). However, the size of pPaxillin was larger in PR20-treated cells than in CTRL (p = 0.034; 1.4 ± 0.0 µm2 in CTRL, 1.6 ± 0.1 µm2 in PR20-treated; Figure 4C). These results suggest that PR poly-dipeptides induce the maturation of FA.
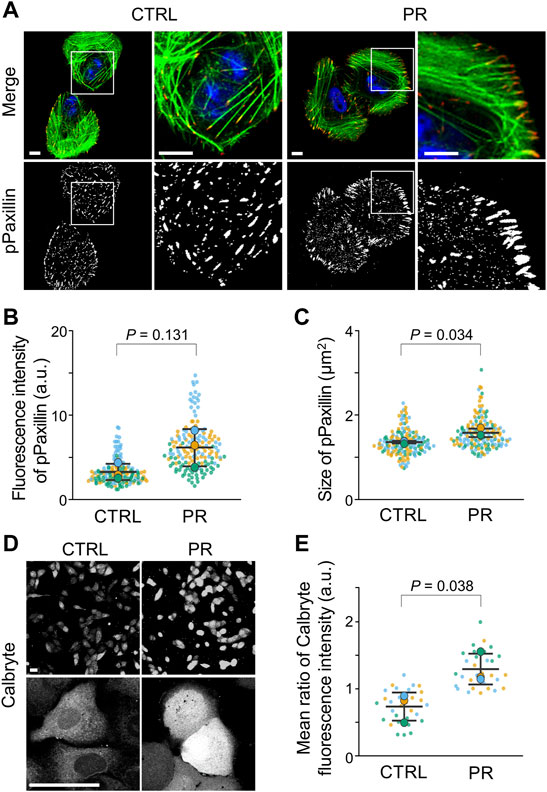
FIGURE 4. PR20 increases the size of FA and intracellular calcium concentration. (A) Fluorescent images of pPaxillin (red), phalloidin (green) and DAPI (blue) are shown. Scale bars are 10 µm. (B,C) Average fluorescence intensity of pPaxillin (in B) and size of pPaxillin (in C). Bars are means ± SD. In each experiment, 41 to 57 cells were evaluated (n = 3). p value shows Welch’s t-test. (D,E) Fluorescence images of Calbryte590™ in CTRL and PR20-treated (PR) cells. Scale bars are 50 µm. Quantification shows in E. Bars are means ± SD. In each experiment, 10 cells were evaluated (n = 3). p value shows Welch’s t-test. In quantification graphs, small dots are individual datapoints, and large dots indicate the averages of each experiment by orange (first; n = 1), blue (second; n = 2), and green (third; n = 3), respectively.
Since FA size is regulated by intracellular calcium (Huang et al., 2018), we measured the intracellular calcium concentration using calcium indicator calbryte590™ (Supplementary Figure S3E). The fluorescent images of PR20-treated cells showed a stronger fluorescence signal compared to CTRL cells (Figure 4D). The calbryte590™ fluorescence intensity using a microplate reader was higher in PR20-treated cells than in CTRL (p = 0.038; 0.7 ± 0.2 in CTRL, 1.3 ± 0.2 a.u. in PR20-treated; Figure 4E). These results suggest that PR poly-dipeptides increase intracellular calcium concentration.
PR Poly-Dipeptides Prevent Cell Detachment
To investigate the impact of cytoskeletal organization on adhesive strength, U2OS cells were transfected with Lifeact-GFP, which enables live imaging of F-actin, and challenged by cell detachment using EDTA after PR poly-dipeptides and cytoskeleton inhibitors, Withaferin-A, Blebbistatin and Nocodazole. Before EDTA treatment, F-actin stress fibers extended across the whole cell in CTRL. PR20-treated cells enhanced cortical F-actin compared to CTRL (Figure 5A), while Withaferin-A-treated cells were comparable to CTRL (Figure 5A). Blebbistatin-treated cells showed fewer F-actin stress fibers than CTRL (Figure 5A). Nocodazole-treated cells showed more F-actin stress fibers than CTRL (Figure 5A). After EDTA-treatment, CTRL and Nocodazole-treated cells showed a rounded shape and F-actin was localized in the cell cortex, whereas PR20, Withaferin-A and Blebbistatin-treated cells maintained a spread shape and formed protrusions (Figure 5A).
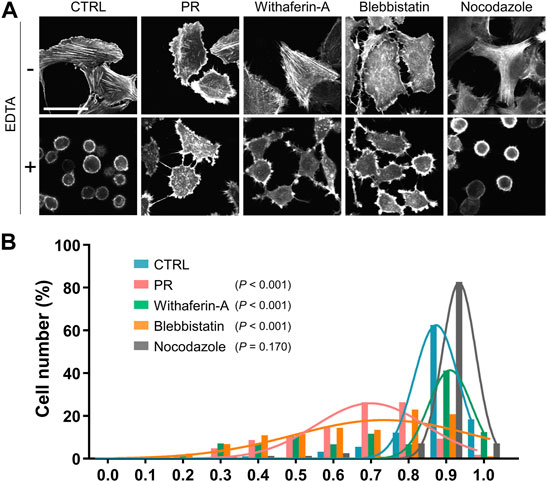
FIGURE 5. PR20 inhibits cell detachment by EDTA. (A) Representative LifeAct-GFP images of U2OS cells in CTRL and PR20-treated (PR) and the cytoskeletal inhibitors, Withaferin-A, Blebbistatin and Nocodazole treatment, following detachment by 5 µM EDTA. A bar is 50 µm. (B) A histogram of the cell circularity of U2OS cells after detachment by 5 µM EDTA in CTRL (n = 179, blue), PR20 (n = 158, red), Withaferin-A (n = 243, green), Blebbistatin (n = 316, orange), and Nocodazole (n = 242, gray). A circularity of 1.0 indicates a complete circle. As it approaches 0.0, it indicates an elongated polygon. p value shows Welch’s t-test.
As the detached cells showed a rounded shape and high circularity (Fuhrmann et al., 2014), degrees of cell detachment were analyzed by measuring the circularity of cells. The circularity of PR20-treated cells was lower than CTRL (p < 0.001; 0.9 ± 0.1 in CTRL, 0.7 ± 0.2 in PR20; Figure 5B; Supplementary Figure S4). Withaferin-A-treated cells (p < 0.001; 0.8 ± 0.0) and Blebbistatin-treated cells (p < 0.001; 0.7 ± 0.2) also decreased circularity after EDTA-treatment (Figure 5B; Supplementary Figure S4). Nocodazole-treated cells showed no significant difference in the circularity compared to CTRL after EDTA-treatment (p = 0.170; 0.9 ± 0.0 in Nocodazole; Figure 5B; Supplementary Figure S4). These results indicate that PR poly-dipeptides prevent cell detachment via the reorganization of IFs and F-actin.
PR Poly-Dipeptides Attenuate the Cyclic Stretch-Induced Reorientation of F-Actin Stress Fibers
Thus far, we have observed that PR poly-dipeptides induced changes in cytoskeletal organization and FA formation, leading to the increase in cell stiffness and the prevention of cell detachment. Therefore, we assume that PR poly-dipeptides might alter the mechanical stress response. To test this hypothesis, we used rat SMCs and U2OS cells and performed uniaxial stretch experiment. Mechanical stretch responses were evaluated by the reorientation angle and the expression of mechanical stress response factor, such as early growth response 1 (Egr1), Thbs1, phosphorylation of extracellular signal-regulated kinase (ERK) and FAK as previously reported (Yamashiro et al., 2015; Yamashiro et al., 2018; Yamashiro et al., 2020). Interestingly, in rat SMCs, PR20-treatment activated FA and upregulated the expression of mechanical stress response factors prior to stretch-stimuli (Figures 6A,B). Although CTRL cells were aligned perpendicularly to stretch direction, PR20-treated cells failed to align correctly and decreased number of cells after stretching (Figure 6C). A histogram of the percentage of the orientation angle (θ) showed that PR20 suppressed the cyclic stretch-induced reorientation of F-actin stress fibers (p < 0.001, 75.3 ± 10.5° in CTRL, 59.4 ± 23.2° in PR20; Figure 6D). U2OS cells showed similar results as rat SMCs (p < 0.001, 73.1 ± 10.7° in CTRL, 65.8 ± 8.8° in PR20; Figures 6E,F). Consequently, these results suggest that PR poly-dipeptides promote the activation of FA and induce upregulation of mechanical stress response factors, leading to sensitive and maladaptive responses to mechanical stimuli.
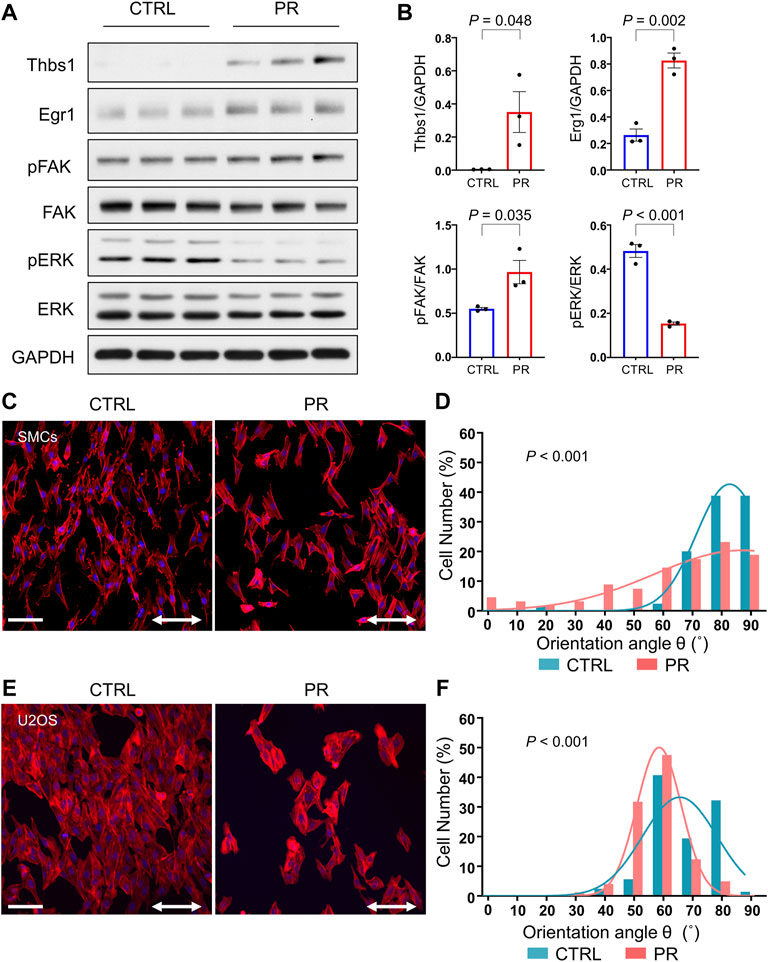
FIGURE 6. PR20 prohibits the cyclic stretch-induced reorientation of F-actin stress fibers. (A) Western blotting shows Thbs1, Egr1, pFAK, FAK, pERK and ERK levels in CTRL and PR20-treated (PR) cells (n = 3). GAPDH was used as a loading control. (B) Quantification graphs are shown. Bars are means ± SEM. p value shows unpaired t-test. (C,E) Rat vascular SMCs (in C) and U2OS cells (in E) with or without 10 µM of PR20 were subjected to cyclic stretch (20% strain, 1.0 Hz (60 cycles/min) for 6 hours. Phalloidin (red) and DAPI (blue) are shown. Scale bars are 100 µm. The two-way arrows indicate stretch direction. (D,F) Histograms of the percentage of the orientation angle (θ) for each cell in rat vascular SMCs (in D) and U2OS cells (in F). The orientation of each cell was analyzed by measuring the orientation angle (θ) of the long axis of the ellipse relative to the stretch axis in CTRL (blue) and PR cells (red). 70 to 104 cells were evaluated in each condition. p value shows Mann–Whitney U test.
Discussion
Here we show that PR poly-dipeptides increased the junctions and branches of the IF network and cell stiffness. This is in conjunction with changes to the distribution of F-actin and the enhancement of FA size. We also demonstrated that cytoskeletal reorganization induced by PR poly-dipeptides prevented cell detachment and led to maladaptive responses to cyclic stretch.
PR poly-dipeptides bind proteins with low-complexity domains and nuclear import receptors (Lee et al., 2016; Lin et al., 2016; Hayes et al., 2020; Hutten et al., 2020; Nanaura et al., 2021). Head domains of IF proteins self-associate via the formation of labile but structurally specific cross-β interactions (Zhou et al., 2021), and PR poly-dipeptides target these polymeric forms of the IF head domains (Lin et al., 2016). In this study, we show that PR poly-dipeptides increased the junctions and branches of the IF network (Figure 1). We speculate that PR poly-dipeptides stabilize these dynamic cross-β interaction of the IF head domains, and lead to the malfunction and dysregulation of IF network formation (Figure 7). However, the detailed mechanisms of how PR poly-dipeptides interfere with cytoskeleton in cells remain unknown.
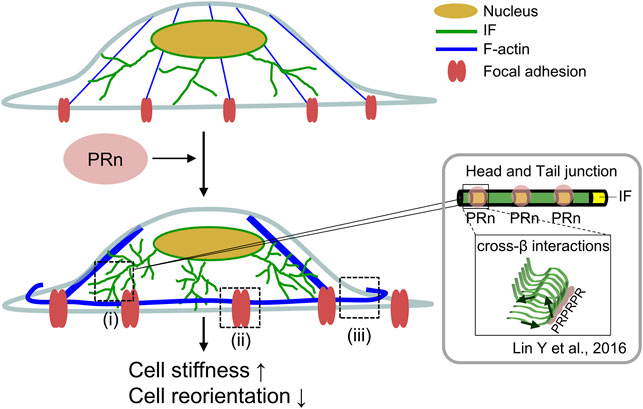
FIGURE 7. Graphical abstract. PR poly-dipeptides bind to IF head domain and induced the alterations of cellular mechanics. PR poly-dipeptides (pink) bind to the IF head domain (yellow) and dysregulate the IF dynamics. This results in an increase of IF branches and IF mesh network (green) (ⅰ). The IF network increases the size of FA (red) (ⅱ) and thickness of F-actin stress fibers (blue) (ⅲ), leading to cell stiffness. Further, the alterations of cytoskeleton and FA disrupt cell reorientation after cyclic stretch. IF: intermediate filament, PRn: PR poly-dipeptides.
As the IF network contributes to cell stiffness (Laly et al., 2021), the high-density network of IFs induced by PR poly-dipeptides might increase cell stiffness. PR poly-dipeptides also reduced the fluorescence intensity of Vim-IFs in the perinuclear region but not Keratin-IFs (Figures 1A,D). These data might result from a difference in IF localization—Vim-IFs diffuse in the perinuclear region and the cytoplasm, whereas Keratin-IFs mainly distribute in the cytoplasm (Grin et al., 2012; Li et al., 2019).
PR poly-dipeptides induced the formation of cortical F-actin and the maturation of FA (Figures 3, 4). The formation of cortical F-actin and the maturation of FA are regulated by IF organization (Helfand et al., 2011). The IF organization by PR poly-dipeptides might alter the formation of cortical F-actin and the maturation of FA. Cortical F-actin is formed by ERM proteins (Bretscher et al., 2002). ERM proteins also induce FAK activation via signaling pathways independent of FA (Poullet et al., 2001). PR poly-dipeptides might induce the maturation of FA via ERM phosphorylation. Nonetheless, it is still unclear how PR poly-dipeptides affect the formation of cortical F-actin and the maturation of FA.
The dysregulation of intracellular calcium is reported as a cause of cell death in C9-ALS/FTD (Li et al., 2020; Choi et al., 2019; Dafinca et al., 2016; Dafinca et al., 2020; Baughman et al., 2011). PR poly-dipeptides increased the intracellular calcium concentration (Figures 4D,E). The increase in intracellular calcium concentration promotes cytoskeletal reorganization and FA maturation (Price et al., 2003; Hartzell et al., 2016; O'Brien et al., 1997). Cytoskeletal reorganization and FA maturation by PR poly-dipeptides might be related to the increase in intracellular calcium. Further investigation is required to understand how the increase in intracellular calcium by PR poly-dipeptides is related to cytoskeletal reorganization and FA maturation.
PR poly-dipeptides, Blebbistatin, and Withaferin-A prevented cell detachment (Figure 5). Cell detachment depends on the adhesive strength and cell stiffness (Sen and Kumar, 2009; Tee et al., 2011). While Blebbistatin decreases cell stiffness by myosin II inhibition (Sen and Kumar, 2009), Withaferin-A increases cell stiffness by inhibition of IF organization (Bargagna-Mohan et al., 2007). Taken together, the alteration of IF organization by PR poly-dipeptides might prevent cell detachment through increasing cell stiffness.
Further, we found that PR poly-dipeptides induced upregulation of Thbs1, a mechanical stress response factor (Figures 6A,B). Thbs1 is induced by cyclic stretch and is related to mechanical stress response (Yamashiro et al., 2020; Laly et al., 2021), suggesting that PR poly-dipeptides induce a maladaptive mechanical stress response in a Thbs1-dependent manner. For the adaptive mechanical stress response, the cells first reduce the adhesive strength of FA, and then they reorient to a perpendicular position and reorganize F-actin stress fibers and FA (Nagayama et al., 2012; Livne et al., 2014). The maturation of FA by PR poly-dipeptides increases adhesive strength and might consequently suppress cell reorientation after cyclic stretch. The mechanical stress response regulates a wide range of cellular functions involved in cell death (King et al., 2011). Hence, further research is required to investigate the effect of maladaptive mechanical stress responses induced by PR poly-dipeptides on intracellular signaling.
PR poly-dipeptides reduced the number of cells after cyclic stretch (Figures 6C,E). PR poly-dipeptides cause cell death in a time-dependent manner (Kwon et al., 2014), resulting in the reduction of the number of cells. In addition, by FA maturation, cells treated with PR poly-dipeptides are expected to be exposed to higher continuous mechanical stress during cyclic stretch, resulting in the reduction of number of cells.
This study revealed that PR poly-dipeptides change the IF organization, leading to an alteration of the mechanical stress response. These results suggest that the pathogenesis by arginine-rich poly-dipeptides might be partially explained by the changes in cellular mechanical properties and mechanical stress responses.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author Contributions
TS, RN, YY, and EM designed the research. TS, RN, KN, and YY performed research. TS, KN, and YY analyzed the data. TS, YY, and EM wrote the paper. SKi, HN, MM, MN, SKo, NI, TK, KN, and KS helped to analyze and interpret the data, and critically revise the manuscript. YY and EM conceptualized the study, developed the study design, supervised the authors throughout the study, and provided expertise in manuscript preparation. All authors read and approved the final manuscript.
Funding
This work was supported by the grants from AMED (JP20dm0307032 and JP21wm0425004 to EM, JP21ek0210158 to YY, JP20gm0810005 to KN), JSPS KAKENHI (JP20H03199 to EM, JP21H02677 and JP21K19881 to YY, JP19K17043 to TS, JP21K15032 to SOK, JP19K21306, JP20K16583 to HN, JP19K23976 to MN, JP19K08150 to SHK, JP19K07978 to TK, JP21H03804 to KN), Uehara Memorial Foundation to EM and SOK, Naito Foundation to EM, MSD Life Science Foundation to EM and YY, Tokyo Biochemical Research Foundation to SOK and by unrestricted funds provided to EM from Dr. Taichi Noda (KTX Corp., Aichi, Japan) and Dr. Yasuhiro Horii (Koseikai, Nara, Japan).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors thank Dr. Akira Nagafuchi for his advice on experimental design and useful discussions. The authors thank Keren-Happuch E Fan Fen for her critical reading of the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2022.750829/full#supplementary-material
References
Abercrombie, M., and Dunn, G. A. (1975). Adhesions of Fibroblasts to Substratum during Contact Inhibition Observed by Interference Reflection Microscopy. Exp. Cel Res. 92 (1), 57–62. doi:10.1016/0014-4827(75)90636-9
Ash, P. E. A., Bieniek, K. F., Gendron, T. F., Caulfield, T., Lin, W.-L., Dejesus-Hernandez, M., et al. (2013). Unconventional Translation of C9ORF72 GGGGCC Expansion Generates Insoluble Polypeptides Specific to c9FTD/ALS. Neuron 77 (4), 639–646. doi:10.1016/j.neuron.2013.02.004
Bamburg, J. R., and Bloom, G. S. (2009). Cytoskeletal Pathologies of Alzheimer Disease. Cell Motil. Cytoskeleton 66 (8), 635–649. doi:10.1002/cm.20388
Baniukiewicz, P., Collier, S., and Bretschneider, T. (2018). QuimP: Analyzing Transmembrane Signalling in Highly Deformable Cells. Bioinformatics 34 (15), 2695–2697. doi:10.1093/bioinformatics/bty169
Bargagna-Mohan, P., Hamza, A., Kim, Y.-e., Khuan Ho, Y., Mor-Vaknin, N., Wendschlag, N., et al. (2007). The Tumor Inhibitor and Antiangiogenic Agent Withaferin A Targets the Intermediate Filament Protein Vimentin. Chem. Biol. 14 (6), 623–634. doi:10.1016/j.chembiol.2007.04.010
Baughman, J. M., Perocchi, F., Girgis, H. S., Plovanich, M., Belcher-Timme, C. A., Sancak, Y., et al. (2011). Integrative Genomics Identifies MCU as an Essential Component of the Mitochondrial Calcium Uniporter. Nature 476 (7360), 341–345. doi:10.1038/nature10234
Baum, M., Erdel, F., Wachsmuth, M., and Rippe, K. (2014). Retrieving the Intracellular Topology from Multi-Scale Protein Mobility Mapping in Living Cells. Nat. Commun. 5, 4494. doi:10.1038/ncomms5494
Bretscher, A., Edwards, K., and Fehon, R. G. (2002). ERM Proteins and merlin: Integrators at the Cell Cortex. Nat. Rev. Mol. Cel Biol 3, 586–599. doi:10.1038/nrm882
Chang, Y.-C., Nalbant, P., Birkenfeld, J., Chang, Z.-F., and Bokoch, G. M. (2008). GEF-H1 Couples Nocodazole-Induced Microtubule Disassembly to Cell Contractility via RhoA. MBoC 19 (5), 2147–2153. doi:10.1091/mbc.e07-12-1269
Choi, S. Y., Lopez-Gonzalez, R., Krishnan, G., Phillips, H. L., Li, A. N., Seeley, W. W., et al. (2019). C9ORF72-ALS/FTD-associated Poly(GR) Binds Atp5a1 and Compromises Mitochondrial Function In Vivo. Nat. Neurosci. 22 (6), 851–862. doi:10.1038/s41593-019-0397-0
Dafinca, R., Barbagallo, P., Farrimond, L., Candalija, A., Scaber, J., Ababneh, N. a. A., et al. (2020). Impairment of Mitochondrial Calcium Buffering Links Mutations in C9ORF72 and TARDBP in iPS-Derived Motor Neurons from Patients with ALS/FTD. Stem Cel Rep. 14 (5), 892–908. doi:10.1016/j.stemcr.2020.03.023
Dafinca, R., Scaber, J., Ababneh, N. a., Lalic, T., Weir, G., Christian, H., et al. (2016). C9orf72 Hexanucleotide Expansions Are Associated with Altered Endoplasmic Reticulum Calcium Homeostasis and Stress Granule Formation in Induced Pluripotent Stem Cell-Derived Neurons from Patients with Amyotrophic Lateral Sclerosis and Frontotemporal Dementia. Stem Cells 34 (8), 2063–2078. doi:10.1002/stem.2388
DeJesus-Hernandez, M., Mackenzie, I. R., Boeve, B. F., Boxer, A. L., Baker, M., Rutherford, N. J., et al. (2011). Expanded GGGGCC Hexanucleotide Repeat in Noncoding Region of C9ORF72 Causes Chromosome 9p-Linked FTD and ALS. Neuron 72 (2), 245–256. doi:10.1016/j.neuron.2011.09.011
Fletcher, D. A., and Mullins, R. D. (2010). Cell Mechanics and the Cytoskeleton. Nature 463 (7280), 485–492. doi:10.1038/nature08908
Fuhrmann, A., Li, J., Chien, S., and Engler, A. J. (2014). Cation Type Specific Cell Remodeling Regulates Attachment Strength. PLoS One 9 (7), e102424. doi:10.1371/journal.pone.0102424
Grin, B., Mahammad, S., Wedig, T., Cleland, M. M., Tsai, L., Herrmann, H., et al. (2012). Withaferin a Alters Intermediate Filament Organization, Cell Shape and Behavior. PLoS One 7 (6), e39065. doi:10.1371/journal.pone.0039065
Häbig, K., Gellhaar, S., Heim, B., Djuric, V., Giesert, F., Wurst, W., et al. (2013). LRRK2 Guides the Actin Cytoskeleton at Growth Cones Together with ARHGEF7 and Tropomyosin 4. Biochim. Biophys. Acta 1832, 2352–2367. doi:10.1016/j.bbadis.2013.09.009
Hao, Z., Liu, L., Tao, Z., Wang, R., Ren, H., Sun, H., et al. (2019). Motor Dysfunction and Neurodegeneration in a C9orf72 Mouse Line Expressing Poly-PR. Nat. Commun. 10 (1), 2906. doi:10.1038/s41467-019-10956-w
Hartzell, C. A., Jankowska, K. I., Burkhardt, J. K., and Lewis, R. S. (2016). Calcium Influx through CRAC Channels Controls Actin Organization and Dynamics at the Immune Synapse. Elife 5. doi:10.7554/eLife.14850
Hayes, L. R., Duan, L., Bowen, K., Kalab, P., and Rothstein, J. D. (2020). C9orf72 Arginine-Rich Dipeptide Repeat Proteins Disrupt Karyopherin-Mediated Nuclear Import. Elife 9. doi:10.7554/eLife.51685
Helfand, B. T., Mendez, M. G., Murthy, S. N. P., Shumaker, D. K., Grin, B., Mahammad, S., et al. (2011). Vimentin Organization Modulates the Formation of Lamellipodia. MBoC 22 (8), 1274–1289. doi:10.1091/mbc.e10-08-0699
Hu, J., Li, Y., Hao, Y., Zheng, T., Gupta, S. K., Parada, G. A., et al. (2019). High Stretchability, Strength, and Toughness of Living Cells Enabled by Hyperelastic Vimentin Intermediate Filaments. Proc. Natl. Acad. Sci. U.S.A. 116 (35), 17175–17180. doi:10.1073/pnas.1903890116
Huang, H., Sun, Z., Hill, M. A., and Meininger, G. A. (2018). A Calcium Mediated Mechanism Coordinating Vascular Smooth Muscle Cell Adhesion during KCl Activation. Front. Physiol. 9, 1810. doi:10.3389/fphys.2018.01810
Hurtley, S. M. (1998). Cell Biology of the Cytoskeleton. Science 279 (5350), 459. doi:10.1126/science.279.5350.459a
Hutten, S., Usluer, S., Bourgeois, B., Simonetti, F., Odeh, H. M., Fare, C. M., et al. (2020). Nuclear Import Receptors Directly Bind to Arginine-Rich Dipeptide Repeat Proteins and Suppress Their Pathological Interactions. Cel Rep. 33 (12), 108538. doi:10.1016/j.celrep.2020.108538
Jovičić, A., Mertens, J., Boeynaems, S., Bogaert, E., Chai, N., Yamada, S. B., et al. (2015). Modifiers of C9orf72 Dipeptide Repeat Toxicity Connect Nucleocytoplasmic Transport Defects to FTD/ALS. Nat. Neurosci. 18 (9), 1226–1229. doi:10.1038/nn.4085
Kim, J., Jang, J., Yang, C., Kim, E. J., Jung, H., and Kim, C. (2016). Vimentin Filament Controls Integrin α5β1-mediated Cell Adhesion by Binding to Integrin through its Ser38 Residue. FEBS Lett. 590 (20), 3517–3525. doi:10.1002/1873-3468.12430
King, J. S., Veltman, D. M., and Insall, R. H. (2011). The Induction of Autophagy by Mechanical Stress. Autophagy 7 (12), 1490–1499. doi:10.4161/auto.7.12.17924
Korobova, F., Gauvin, T. J., and Higgs, H. N. (2014). A Role for Myosin II in Mammalian Mitochondrial Fission. Curr. Biol. 24 (4), 409–414. doi:10.1016/j.cub.2013.12.032
Kovács, M., Tóth, J., Hetényi, C., Málnási-Csizmadia, A., and Sellers, J. R. (2004). Mechanism of Blebbistatin Inhibition of Myosin II. J. Biol. Chem. 279, 35557–35563. doi:10.1074/jbc.M405319200
Kwon, I., Xiang, S., Kato, M., Wu, L., Theodoropoulos, P., Wang, T., et al. (2014). Poly-dipeptides Encoded by the C9orf72 Repeats Bind Nucleoli, Impede RNA Biogenesis, and Kill Cells. Science 345 (6201), 1139–1145. doi:10.1126/science.1254917
Laly, A. C., Sliogeryte, K., Pundel, O. J., Ross, R., Keeling, M. C., Avisetti, D., et al. (2021). The Keratin Network of Intermediate Filaments Regulates Keratinocyte Rigidity Sensing and Nuclear Mechanotransduction. Sci. Adv. 7 (5). doi:10.1126/sciadv.abd6187
Lee, K.-H., Zhang, P., Kim, H. J., Mitrea, D. M., Sarkar, M., Freibaum, B. D., et al. (2016). C9orf72 Dipeptide Repeats Impair the Assembly, Dynamics, and Function of Membrane-Less Organelles. Cell 167 (3), 774–788. doi:10.1016/j.cell.2016.10.002
Levy Nogueira, M., Epelbaum, S., Steyaert, J. M., Dubois, B., and Schwartz, L. (2016). Mechanical Stress Models of Alzheimer's Disease Pathology. Alzheimer's Demen. 12, 324–333. doi:10.1016/j.jalz.2015.10.005
Li, J., Gao, W., Zhang, Y., Cheng, F., Eriksson, J. E., Etienne‐Manneville, S., et al. (2019). Engagement of Vimentin Intermediate Filaments in Hypotonic Stress. J. Cel Biochem 120 (8), 13168–13176. doi:10.1002/jcb.28591
Li, S., Wu, Z., Li, Y., Tantray, I., De Stefani, D., Mattarei, A., et al. (2020). Altered MICOS Morphology and Mitochondrial Ion Homeostasis Contribute to Poly(GR) Toxicity Associated with C9-ALS/FTD. Cel Rep. 32 (5), 107989. doi:10.1016/j.celrep.2020.107989
Lin, Y., Mori, E., Kato, M., Xiang, S., Wu, L., Kwon, I., et al. (2016). Toxic PR Poly-Dipeptides Encoded by the C9orf72 Repeat Expansion Target LC Domain Polymers. Cell 167 (3), 789–802. doi:10.1016/j.cell.2016.10.003
Livne, A., Bouchbinder, E., and Geiger, B. (2014). Cell Reorientation under Cyclic Stretching. Nat. Commun. 5, 3938. doi:10.1038/ncomms4938
López-Colomé, A. M., Lee-Rivera, I., Benavides-Hidalgo, R., and López, E. (2017). Paxillin: a Crossroad in Pathological Cell Migration. J. Hematol. Oncol. 10 (1), 50. doi:10.1186/s13045-017-0418-y
Lu, Z., Li, H., Hou, C., Peng, Y., Long, J., and Liu, J. (2017). Endogenously Generated Amyloid-β Increases Stiffness in Human Neuroblastoma Cells. Eur. Biophys. J. 46, 415–424. doi:10.1007/s00249-016-1185-3
McLean, P. J., Kawamata, H., Ribich, S., and Hyman, B. T. (2000). Membrane Association and Protein Conformation of Alpha-Synuclein in Intact Neurons. Effect of Parkinson's Disease-Linked Mutations. J. Biol. Chem. 275, 8812–8816. doi:10.1074/jbc.275.12.8812
Mizielinska, S., Grönke, S., Niccoli, T., Ridler, C. E., Clayton, E. L., Devoy, A., et al. (2014). C9orf72 Repeat Expansions Cause Neurodegeneration in Drosophila through Arginine-Rich Proteins. Science 345 (6201), 1192–1194. doi:10.1126/science.1256800
Mould, A. P., Akiyama, S. K., and Humphries, M. J. (1995). Regulation of Integrin α5β1-Fibronectin Interactions by Divalent Cations. J. Biol. Chem. 270 (44), 26270–26277. doi:10.1074/jbc.270.44.26270
Nagayama, K., Kimura, Y., Makino, N., and Matsumoto, T. (2012). Strain Waveform Dependence of Stress Fiber Reorientation in Cyclically Stretched Osteoblastic Cells: Effects of Viscoelastic Compression of Stress Fibers. Am. J. Physiol. Cel Physiol 302, C1469–C1478. doi:10.1152/ajpcell.00155.2011
Nagayama, K., Uchida, K., and Sato, A. (2019). A Novel Micro-grooved Collagen Substrate for Inducing Vascular Smooth Muscle Differentiation through Cell Tissue Arrangement and Nucleus Remodeling. J. Mech. Behav. Biomed. Mater. 90, 295–305. doi:10.1016/j.jmbbm.2018.10.005
Nanaura, H., Kawamukai, H., Fujiwara, A., Uehara, T., Aiba, Y., Nakanishi, M., et al. (2021). C9orf72-derived Arginine-Rich Poly-Dipeptides Impede Phase Modifiers. Nat. Commun. 12 (1), 5301. doi:10.1038/s41467-021-25560-0
Nicolas, A., Kenna, K. P., Renton, A. E., Ticozzi, N., Faghri, F., Chia, R., et al. (2018). Genome-wide Analyses Identify KIF5A as a Novel ALS Gene. Neuron 97 (6), 1268–e6. doi:10.1016/j.neuron.2018.02.027
O'Brien, E. T., Salmon, E. D., and Erickson, H. P. (1997). How Calcium Causes Microtubule Depolymerization. Cell Motil Cytoskeleton 36, 125–135. doi:10.1002/(SICI)1097-0169(1997)36:2<125::AID-CM3>3.0.CO;2-8
Poullet, P., Gautreau, A., Kadaré, G., Girault, J. A., Louvard, D., and Arpin, M. (2001). Ezrin Interacts with Focal Adhesion Kinase and Induces its Activation Independently of Cell-Matrix Adhesion. J. Biol. Chem. 276, 37686–37691. doi:10.1074/jbc.M106175200
Price, L. S., Langeslag, M., ten Klooster, J. P., Hordijk, P. L., Jalink, K., and Collard, J. G. (2003). Calcium Signaling Regulates Translocation and Activation of Rac. J. Biol. Chem. 278, 39413–39421. doi:10.1074/jbc.M302083200
Sen, S., and Kumar, S. (2009). Cell-Matrix De-adhesion Dynamics Reflect Contractile Mechanics. Cel. Mol. Bioeng. 2 (2), 218–230. doi:10.1007/s12195-009-0057-7
Smith, B. N., Ticozzi, N., Fallini, C., Gkazi, A. S., Topp, S., Kenna, K. P., et al. (2014). Exome-wide Rare Variant Analysis Identifies TUBA4A Mutations Associated with Familial ALS. Neuron 84 (2), 324–331. doi:10.1016/j.neuron.2014.09.027
Sumi, T., Matsumoto, K., Takai, Y., and Nakamura, T. (1999). Cofilin Phosphorylation and Actin Cytoskeletal Dynamics Regulated by Rho- and Cdc42-Activated LIM-Kinase 2. J. Cel Biol 147 (7), 1519–1532. doi:10.1083/jcb.147.7.1519
Sweers, K., van der Werf, K., Bennink, M., and Subramaniam, V. (2011). Nanomechanical Properties of α-synuclein Amyloid Fibrils: a Comparative Study by Nanoindentation, Harmonic Force Microscopy, and Peakforce QNM. Nanoscale Res. Lett. 6 (1), 270. doi:10.1186/1556-276x-6-270
Tee, S.-Y., Fu, J., Chen, C. S., and Janmey, P. A. (2011). Cell Shape and Substrate Rigidity Both Regulate Cell Stiffness. Biophysical J. 100 (5), L25–L27. doi:10.1016/j.bpj.2010.12.3744
Wu, C.-H., Fallini, C., Ticozzi, N., Keagle, P. J., Sapp, P. C., Piotrowska, K., et al. (2012). Mutations in the Profilin 1 Gene Cause Familial Amyotrophic Lateral Sclerosis. Nature 488 (7412), 499–503. doi:10.1038/nature11280
Yamashiro, Y., Papke, C. L., Kim, J., Ringuette, L. J., Zhang, Q. J., Liu, Z. P., et al. (2015). Abnormal Mechanosensing and Cofilin Activation Promote the Progression of Ascending Aortic Aneurysms in Mice. Sci. Signal. 8 (399), ra105. doi:10.1126/scisignal.aab3141
Yamashiro, Y., Thang, B. Q., Ramirez, K., Shin, S. J., Kohata, T., Ohata, S., et al. (2020). Matrix Mechanotransduction Mediated by Thrombospondin-1/integrin/YAP in the Vascular Remodeling. Proc. Natl. Acad. Sci. U.S.A. 117 (18), 9896–9905. doi:10.1073/pnas.1919702117
Yamashiro, Y., Thang, B. Q., Shin, S. J., Lino, C. A., Nakamura, T., Kim, J., et al. (2018). Role of Thrombospondin-1 in Mechanotransduction and Development of Thoracic Aortic Aneurysm in Mouse and Humans. Circ. Res. 123 (6), 660–672. doi:10.1161/circresaha.118.313105
Zhang, S., Cooper-Knock, J., Weimer, A. K., Shi, M., Moll, T., Marshall, J. N. G., et al. (2022). , S0896-6273, 01036–01039. doi:10.1016/j.neuron.2021.12.019Genome-wide Identification of the Genetic Basis of Amyotrophic Lateral SclerosisNeuron
Keywords: C9orf72, PR poly-dipeptides, cytoskeleton, Focal Adhesion (FA), actin
Citation: Shiota T, Nagata R, Kikuchi S, Nanaura H, Matsubayashi M, Nakanishi M, Kobashigawa S, Isozumi N, Kiriyama T, Nagayama K, Sugie K, Yamashiro Y and Mori E (2022) C9orf72-Derived Proline:Arginine Poly-Dipeptides Modulate Cytoskeleton and Mechanical Stress Response. Front. Cell Dev. Biol. 10:750829. doi: 10.3389/fcell.2022.750829
Received: 31 July 2021; Accepted: 07 March 2022;
Published: 23 March 2022.
Edited by:
Rafael Fridman, Wayne State University, United StatesReviewed by:
Fernando G. Vieira, Amyotrophic Lateral Sclerosis Therapy Development Institute, United StatesKenichi Funamoto, Tohoku University, Japan
Copyright © 2022 Shiota, Nagata, Kikuchi, Nanaura, Matsubayashi, Nakanishi, Kobashigawa, Isozumi, Kiriyama, Nagayama, Sugie, Yamashiro and Mori. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yoshito Yamashiro, eWFtYXlvc2hpdG9AdGFyYS50c3VrdWJhLmFjLmpw; Eiichiro Mori, ZW1vcmlAbmFyYW1lZC11LmFjLmpw
†These authors have contributed equally to this work
 Tomo Shiota
Tomo Shiota Riko Nagata2†
Riko Nagata2† Hitoki Nanaura
Hitoki Nanaura Masaya Matsubayashi
Masaya Matsubayashi Shinko Kobashigawa
Shinko Kobashigawa Takao Kiriyama
Takao Kiriyama Kazuaki Nagayama
Kazuaki Nagayama Kazuma Sugie
Kazuma Sugie Yoshito Yamashiro
Yoshito Yamashiro Eiichiro Mori
Eiichiro Mori