
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cell Dev. Biol., 25 February 2022
Sec. Molecular and Cellular Oncology
Volume 10 - 2022 | https://doi.org/10.3389/fcell.2022.746443
This article is part of the Research TopicThe Role of ncRNAs (non-coding RNAs) in Regulating Tumor Immune MicroenvironmentView all 32 articles
 Soudeh Ghafouri-Fard1
Soudeh Ghafouri-Fard1 Sajad Najafi2
Sajad Najafi2 Bashdar Mahmud Hussen3,4
Bashdar Mahmud Hussen3,4 Aryan R. Ganjo4
Aryan R. Ganjo4 Mohammad Taheri5,6*
Mohammad Taheri5,6* Mohammad Samadian7*
Mohammad Samadian7*Long non-coding RNAs (lncRNAs) are a heterogeneous group of ncRNAs with characteristic size of more than 200 nucleotides. An increasing number of lncRNAs have been found to be dysregulated in many human diseases particularly cancer. However, their role in carcinogenesis is not precisely understood. DLX6-AS1 is an lncRNAs which has been unveiled to be up-regulated in various number of cancers. In different cell studies, DLX6-AS1 has shown oncogenic role via promoting oncogenic phenotype of cancer cell lines. Increase in tumor cell proliferation, migration, invasion, and EMT while suppressing apoptosis in cancer cells are the effects of DLX6-AS1 in development and progression of cancer. In the majority of cell experiment, mediator miRNAs have been identified which are sponged and negatively regulated by DLX6-AS1, and they in turn regulate expression of a number of transcription factors, eventually affecting signaling pathways involved in carcinogenesis. These pathways form axes through which DLX6-AS1 promotes carcinogenicity of cancer cells. Xenograft animal studies, also have confirmed enhancing effect of DLX6-AS1 on tumor growth and metastasis. Clinical evaluations in cancerous patients have also shown increased expression of DLX6-AS1 in tumor tissues compared to healthy tissues. High DLX6-AS1 expression has shown positive association with advanced clinicopathological features in cancerous patients. Survival analyses have demonstrated correlation between high DLX6-AS1 expression and shorter survival. In cox regression analysis, DLX6-AS1 has been found as an independent prognostic factor for patients with various types of cancer.
In complex organisms, genome sequencing analyses have unveiled that just a small fraction of genome (e.g., 1–2% for mammals) encodes for protein via coding RNAs or messenger RNAs (mRNAs) that are located in the middle of central dogma making connection between DNA and corresponding protein. These protein-coding regions are those which have been described as genes for more than half a century in biology literature. However, the majority of large genomes i.e., more than 80% is transcribed to non-coding RNAs (ncRNAs) for which no corresponding protein have been found, but a huge number of regulatory functions are recognized. Unlike the primary expectations which termed ncRNAs as “junk” DNA without biological importance, today it is clarified that they are involved in gene regulation at transcriptional and post-transcriptional levels, and through which they play critical roles in a vast number of biological processes such as imprinting, methylation, and silencing via several interactions with DNA, RNA, and proteins (Mattick, 2001). Based on size and function of transcripts, ncRNAs are categorized in several classes including microRNAs (miRNAs), small interfering RNAs (siRNAs), PIWI-interacting RNAs and long ncRNAs (lncRNAs). Transcripts of more than 200 nucleotide length are classified as lncRNAs which were primarily reported by Okazaki et al. in an analysis of mouse transcriptome in 2002 (Okazaki et al., 2002). RNA polymerase II is predominantly responsible for transcription of lncRNAs. They mainly endure capping, polyadenylation, splicing after transcription, and also trimethylation on histone 3 corresponding to lysine 4 (H3K4me3) (Losko et al., 2016; Bertone et al., 2004; Guttman et al., 2009). Thousands of heterogenous lncRNAs have been identified in multicellular organisms [60,000 encoding loci in human genome (Iyer et al., 2015)] showing tissues specificity which is also conserved during evolution (Necsulea et al., 2014) and acting as regulators of gene expression both in nucleus or cytoplasm (Fatica and Bozzoni, 2014) suggesting their involvement in specific biologic processes. Several databases have been created to store and provide access to an increasing number of lncRNAs. Examples of these databases are TRlnc for regulatory lncRNAs in humans (Li et al., 2020a), lncRNASNP1 and 2 for single nucleotide polymorphisms (SNPs) of human and mouse lncRNAs (Gong et al., 2014; Miao et al., 2017), LncRNA2Target v2.0 for target genes of lncRNAs (Cheng et al., 2018), CRISPRlnc for validated single guide RNAs (sgRNAs) used in clustered regularly interspaced short palindromic repeats (CRISPR)-associated protein number 9 (Cas9) gene editing technology for lncRNAs (Chen et al., 2018) and clusLnc2Cancer for effective lncRNAs in human cancers (Ning et al., 2015). They act in cis and trans modes by gathering and localizing transcription factors to a locus. Gene expression regulation at several levels including transcription, translation and splicing, epigenetic regulation in X-chromosome inactivation or dosage compensation, genomic imprinting, involvement in developmental and differential processes, neurogenesis, regulation of cell cycle, and cell transportation are among the fundamental roles which have been recognized for lncRNAs (Mattick, 2009; Wilusz et al., 2009; Wu et al., 2013; Dey et al., 2014; Fatica and Bozzoni, 2014). Accordingly, an increasing number of lncRNAs have been associated with various types of human diseases. Dysregulation in expression levels or mutation of lncRNAs are found to play role in the pathogenesis of diseases like age-related diseases, cardiovascular diseases (Uchida and Dimmeler, 2015), kidney and liver diseases (Takahashi et al., 2014; Ignarski et al., 2019), ophthalmologic diseases (Wawrzyniak et al., 2018), neurodegenerative and other diseases affecting central nervous system (CNS) (Pastori and Wahlestedt, 2012; Wan et al., 2017), and particularly various types of cancer. Mediation of a number of cancer-associated processes like cell cycle regulation, epigenetic regulation, and involvement in signaling pathways and hormone-related pathways indicate potential roles of lncRNAs act as contributors in the development and progression of cancer (Sahu et al., 2015). MALAT1, HOTAIR, H19, HOTTIP, ANRIL, and NEAT1 are among the most famous lncRNAs which have been mostly studied in many types of cancer exhibiting dysregulation in cancer cells, tissues and body fluids of affected patients. In this review, we aim to have an overview of studies which have assessed tumorigenic effects of the lncRNA distal-less homeobox 6 antisense RNA 1 (DLX6-AS1) in three levels of cell, animal, and human studies. In humans, DLX6-AS1 gene is located on chromosome 7q21.3, primarily identified by Feng et al. (2006) to promote DLX5/6 function in trans mode. This lncRNA has been found to be up-regulated in a growing number of different types of cancerous tissues compared to normal tissues. Promoting carcinogenesis via increasing tumor cell proliferation, migration, and invasion through enhancing Epithelial–Mesenchymal Transition (EMT) along with suppression of apoptosis and chemosensitivity have been shown in cell studies of DLX6-AS1 overexpression. Enhanced tumor growth and metastasis has confirmed tumorigenic potentials of DLX6-AS1 in animal studies. Correlation between high DLX6-AS1 expression and advanced clinicopathological features and also poor prognosis and survival in cancerous patients has suggested DLX6-AS1 not only as a diagnostic and prognostic biomarker but also as a therapeutical target.
Cancer cell lines have been used to evaluate function of DLX6-AS1 in cell cycle progression, cell proliferation and apoptosis. Moreover, high throughput RNA sequencing and also confirmation via quantitative real-time polymerase chain reaction (qRT-PCR) analyses have facilitated identification of differentially expressed lncRNAs in cancer cell lines compared to controls. In vitro experiments have shown significant increase in expression levels of DLX6-AS1 in cancer cell lines. In different cell experiment, it has been demonstrated that DLX6-AS1 overexpression promotes tumor cell proliferation, migration, and invasion, while suppressing apoptosis. In cell counting, colony formation, and 5-Bromo-2-deoxyUridine (BrdU) assays, decreased proliferation of cancer cells is reported for DLX6-AS1 knockdown. Wound healing, Matrigel and Transwell assays for assessment tumor cell migration and invasion show suppressed metastatic capability of cancerous cells under DLX6-AS1 silencing. Flowcytometry also demonstrated cell cycle arrest in treated cancer cells. Furthermore, decreased cell viability and elevated apoptosis in 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-2H-tetrazolium bromide (MTT), flowcytometry, and apoptotic marker assays have unveiled increased apoptosis in DLX6-AS1-silenced cancer cells. In hepatocellular carcinoma (HCC), DLX6-AS1 has been shown to be highly expressed in human HCC cell lines versus normal liver cells, while miR-513c as its downstream microRNA exhibited down-regulation indicating DLX6-AS1 acts as sponge for this miRNA (Liu et al., 2020a). Cullin4A (Cul4A) was also known as target gene of miR-513c which showed increase in expression level following DLX6-AS1 up-regulation. In other words, DLX6-AS1 elevated Cul4A expression by binding to and sponging miR-513c. Cul4A, itself positively regulated activity of annexin A10 (ANXA10). DLX6-AS1 silencing using specific short hairpin RNA (shRNA) repressed cell viability, invasion, and migration of HCC cells. Also, Cul4A knockdown was shown to inhibit tumorigenic effects of HCC cells via inhibition of ANXA10 degradation through ubiquitin-associated pathway. The results showed that DLX6-AS1 exerts its tumorigenic role via miR-513c/Cul4A/ANXA10 axis. In a distinct study (Zhang et al., 2017), DLX6-AS1 was shown to exert same tumorigenic roles in HCC cells via miR-203a/MMP-2 axis.
In other experiments, DLX6-AS1 has been shown to sponge many other miRNAs and affect transcription factors, genes or signaling pathways which eventually promotes malignant phenotypes. miRNAs which are mainly negatively regulated by up-stream DLX6-AS1 exhibit down-regulation in cancer tissues and cells, and their overexpression reverse the malignant phenotypes of DLX6-AS1 in cancer cell lines. Downstream factors demonstrate expression changes consistent with DLX6-AS1. Overexpression of these factors drives same influences with DLX6-AS1 overexpression. In a study in ovarian cancer (Kong and Zhang, 2020), miR-195-5p was shown to be down-regulated in cancer tissues and was identified as target of up-regulated DLX6-AS1. While DLX6-AS1 promoted cell proliferation, migration, and invasion in tumor cell lines, miR-195-5p overexpression reversed malignant phenotypes. Four and a half LIM domains protein 2 (FHL2) which is known to play role in development and progression of different types of cancer via activation of androgen receptor (AR or NR3C4), Wnt/β-catenin pathway or several genes was demonstrated as target of miR-195-5p. FHL2 overexpression exhibited same results on malignant phenotypes of cancer cells. In other words, DLX6-AS1 exerted its tumorigenic effects in ovarian cancer cells via miR-195-5p/FHL2 signaling axis. In bladder cancer, miR-195-5p as target of DLX6-AS1 was shown to down-regulate the vascular endothelial growth factor A (VEGFA) and consequently inhibit malignancy phenotype in cancer cells, while miR-195-5p inhibition returned the DLX6-AS1 tumorigenic effects (Zhao et al., 2020a).
Furthermore, DLX6-AS1 has been shown to up-regulated DLK1, a regulator of cell differentiation and prognostic factor for several cancers, through sponging miR-129-5p which in turn triggers Wnt signaling, and eventually promotes stemness in osteosarcoma cell lines (Zhang et al., 2018). PI3K/AKT/mTOR signaling pathway is another critical tumorigenic pathway which is known to be activated by DLX6-AS1, promoting malignant phenotype of colorectal cancer cells (Zhang et al., 2019a).
Overall, it is demonstrated that DLX6-AS1 acts as an oncogenic lncRNA enhancing malignant phenotype of several cancer cells (Figure 1).

FIGURE 1. Oncogenic role of DLX6-AS1 in different cancer types is exerted through various mechanisms, particularly sponging miRNAs.
DLX6-AS1 is oncogenic lncRNA has been found to be up-regulated in a growing number of different types of cancerous tissues compared to normal tissues. Promoting carcinogenesis via increasing tumor cell proliferation, migration, and invasion through enhancing Epithelial–Mesenchymal Transition (EMT). miRNAs have been identified which are negatively regulated by DLX6-AS1, and they regulate expression of a number of transcription factors, eventually affecting signaling pathways involved in carcinogenesis.
Table 1 shows the findings of the studies conducted on DLX6-AS1 oncogenic role in various cancer cell lines.
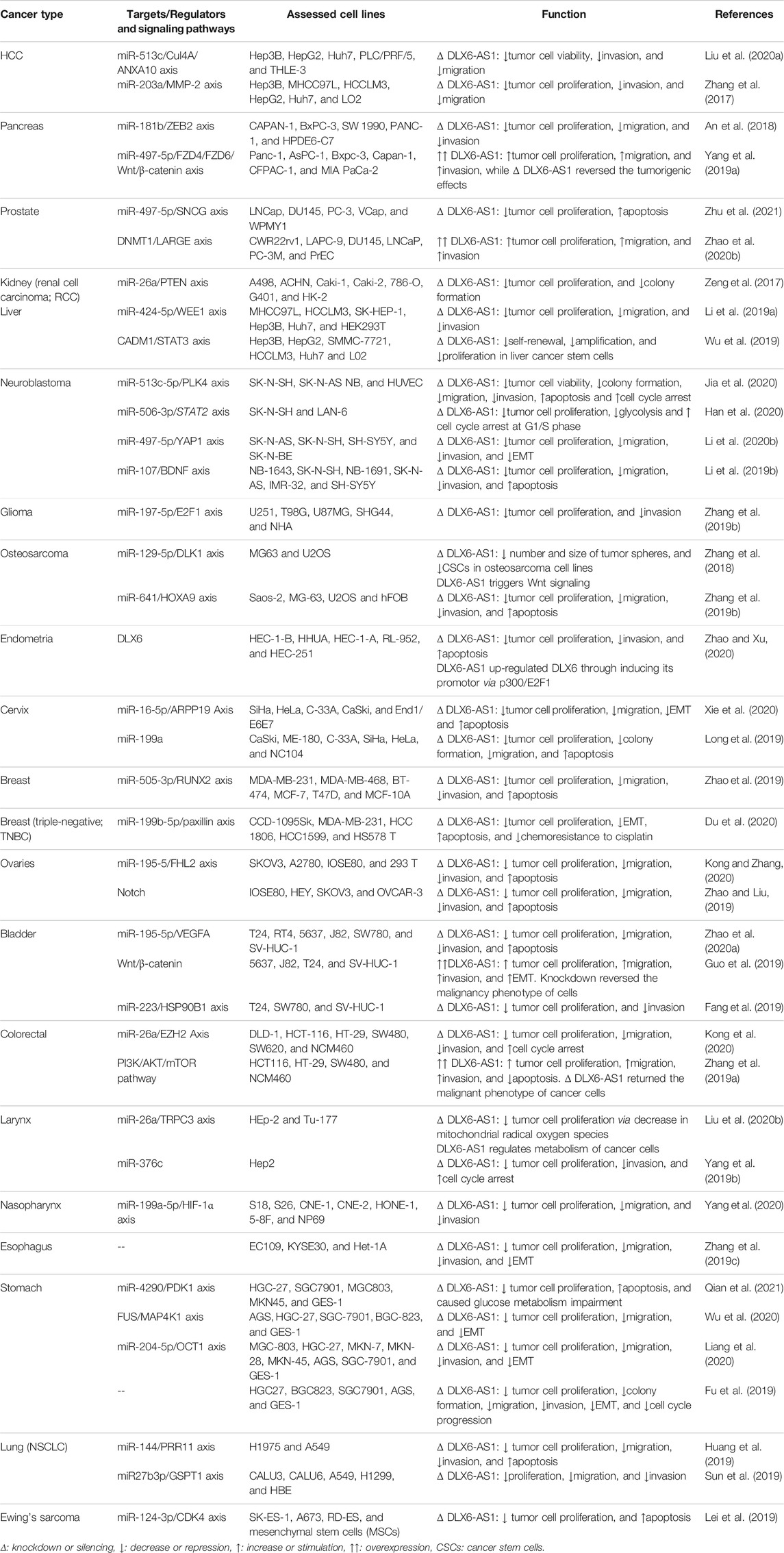
TABLE 1. an overview to the oncogenic influences of DLX6-AS1 in cell studies of different types of cancer.
Experiments in animal models have confirmed oncogenic role of DLX6-AS1. It is expected that DLX6-AS1 overexpression or silencing increases or suppresses malignant features of cancer cells in xenograft models, respectively. To examine this claim, treated cells; either overexpressing or with silenced for DLX6-AS1; have been injected to the animals; mainly BALB/c nude mice, and then tumor size or volume, and metastasis in expected organ have been checked at certain intervals. Changes in chemosensitivity have also been assessed occasionally. Decreased tumor growth and metastasis, and also chemoresistance have been reported under DLX6-AS1 knockdown conditions in animal studies. Opposite findings have been reported when DLX6-AS1 was overexpressed in injected cancer cells to the nude mice. Taken together, these findings demonstrate oncogenic role of DLX6-AS1 in tumor progression and metastasis in animal studies are consistent with the results of cell studies (Table 2).
Cancerous tissues resected from patients have shown significantly increased expression of DLX6-AS1 compared to matched normal adjacent tissues (NATs) and healthy people in microarray analysis and qRT-PCR. In non-small cell lung cancer (NSCLC), DLX6-AS1 high expression levels were found to be positively associated with advanced clinicopathological features including higher disease stage, tumor metastasis to lymph nodes and also weak differentiation of cancer cells in patients (Zhang et al., 2019c). Also, Guo et al. (2019) demonstrated high DLX6-AS1 expression in bladder cancer patients with advanced TNM stage, positive lymph node and distant metastases. Survival analysis via Kaplan-Meier curve has shown association between high DLX6-AS1 expression and shorter overall survival (OS), and/or disease-free survival (DFS) in several types of cancer like HCC (Liu et al., 2020a; Zhang et al., 2017), gastric cancer (Qian et al., 2021; Fu et al., 2019), glioma (Zhang et al., 2019b), breast cancer (Zhao et al., 2019), and several others (Table 3). Competitive endogenous RNA (ceRNA) network analysis has demonstrated reliability of DLX6-AS1 along with three other lncRNAs and two more miRNAs in a signature as prognostic biomarkers in HCC patients (Long et al., 2019). Ding et al. (2021) showed serum exosomal levels of DLX6-AS1 can act as a prognostic biomarker in cervical cancer patients. Also, multivariate cox regression has shown that DLX6-AS1 is an independent prognostic factor for survival in a number of cancers such as gastric cancer (Qian et al., 2021), osteosarcoma (Zhang et al., 2018), and ovarian cancer (Zhao and Liu, 2019). Furthermore, a value of 0.795 for area under curve (AUC) in receiver operating characteristic (ROC) curve has shown acceptable efficiency of DLX6-AS1 in diagnosis of glioma (Zhang et al., 2019b). Taken together, according to the clinical data, DLX6-AS1 is suggested as a potential prognostic biomarker for different types of human cancer and a putative factor to manage cancerous patients.
LncRNAs are a heterogeneous group of ncRNAs with characteristic size of more than 200 nucleotides. An increasing number of lncRNAs have been found to be dysregulated in many human diseases particularly cancer. However, their role in carcinogenesis is not precisely understood. DLX6-AS1 is an lncRNAs which has been unveiled to be up-regulated in a various number of cancers. In different cell studies, DLX6-AS1 has shown oncogenic role via promoting oncogenic phenotype of cancer cell lines. Increase in tumor cell proliferation, migration, invasion, and EMT while suppressing apoptosis in cancer cells are the effects of DLX6-AS1 in the development and progression of cancer. Silencing experiments using specific shRNA against DLX6-AS1 have shown suppression of tumorigenic potential. Similar pattern of expression in different types of cancer originated from various tissues not only reveals its universal function in the tumorigenesis, but also emphasizes the suitability of therapeutic modalities against this lncRNA for a wide range of human malignancies.
In the majority of cell experiments, mediator miRNAs have been identified which are negatively regulated by DLX6-AS1, and they regulate expression of a number of transcription factors, eventually affecting signaling pathways involved in carcinogenesis. These pathways form axes through which DLX6-AS1 regulates transcription factors, and/or signaling pathways eventually promotes carcinogenicity of cancer cells. Identification of functional routes of DLX6-AS1 effects in the carcinogenesis is an important step toward design of targeted therapies in cancer. It is also important to mention that these therapies should not affect pathways with crucial roles in the physiological features of normal cells.
Xenograft animal studies also have confirmed enhancing effect of DLX6-AS1 on tumor growth and metastasis. Clinical evaluations in cancerous patients have shown increased expression of DLX6-AS1 in tumor tissues compared to healthy tissues. High DLX6-AS1 expression has shown positive association with advanced clinicopathological features in cancerous patients. Survival analyses have demonstrated correlation between high DLX6-AS1 expression and shorter survival. In cox regression analysis, DLX6-AS1 has been suggested as an independent prognostic factor for patients with various types of cancer.
Animal and cell line studies have confirmed that therapeutic modalities targeting DLX6-AS1 can effectively reduce tumorigenic potential of malignant cells, induce their apoptosis and diminish tumor size and burden. However, the efficacy and safety of these methods have not been evaluated in the clinical settings.
Taken together, these findings demonstrate carcinogenic role of DLX6-AS1 in the development and progression of different human cancers suggesting diagnostic and prognostic potentials of DLX6-AS1 in human cancers. Known role of up-regulated DLX6-AS1 in cancer tissues and clinical samples also suggest therapeutic potentials in finding treatments for different types of cancer via targeting DLX6-AS1. Further studies are required to utilize diagnostic, prognostic, and therapeutic potentials of DLX6-AS1 in clinical settings. Moreover, measurement of DLX6-AS1 levels in biofluids is an important step towards identification of non-invasive routes for diagnostic purposes.
MT and SGF wrote the draft and revised it. SN and MS collected the data and designed the figures and tables. All the authors read and approved the submitted version.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
An, Y., Chen, X.-m., Yang, Y., Mo, F., Jiang, Y., Sun, D.-l., et al. (2018). LncRNA DLX6-AS1 Promoted Cancer Cell Proliferation and Invasion by Attenuating the Endogenous Function of miR-181b in Pancreatic Cancer. Cancer Cel Int 18 (1), 143. doi:10.1186/s12935-018-0643-7
Bertone, P., Stolc, V., Royce, T. E., Rozowsky, J. S., Urban, A. E., Zhu, X., et al. (2004). Global Identification of Human Transcribed Sequences with Genome Tiling Arrays. Science 306 (5705), 2242–2246. doi:10.1126/science.1103388
Chen, W., Zhang, G., Li, J., Zhang, X., Huang, S., Xiang, S., et al. (2018). CRISPRlnc: a Manually Curated Database of Validated sgRNAs for lncRNAs. Nucleic Acids Res. 47 (D1), D63–D68. doi:10.1093/nar/gky904
Cheng, L., Wang, P., Tian, R., Wang, S., Guo, Q., Luo, M., et al. (2018). LncRNA2Target v2.0: a Comprehensive Database for Target Genes of lncRNAs in Human and Mouse. Nucleic Acids Res. 47 (D1), D140–D144. doi:10.1093/nar/gky1051
Dey, B. K., Mueller, A. C., and Dutta, A. (2014). Long Non-coding RNAs as Emerging Regulators of Differentiation, Development, and Disease. Transcription 5 (4), e944014. doi:10.4161/21541272.2014.944014
Ding, X. Z., Zhang, S. Q., Deng, X. L., and Qiang, J. H. (2021). Serum Exosomal lncRNA DLX6-AS1 Is a Promising Biomarker for Prognosis Prediction of Cervical Cancer. Technol. Cancer Res. Treat. 20, 1533033821990060. doi:10.1177/1533033821990060
Du, C., Wang, Y., Zhang, Y., Zhang, J., Zhang, L., and Li, J. (2020). LncRNA DLX6-AS1 Contributes to Epithelial-Mesenchymal Transition and Cisplatin Resistance in Triple-Negative Breast Cancer via Modulating Mir-199b-5p/Paxillin Axis. Cel Transpl. 29, 963689720929983. doi:10.1177/0963689720929983
Fang, C., Xu, L., He, W., Dai, J., and Sun, F. (2019). Long Noncoding RNA DLX6-AS1 Promotes Cell Growth and Invasiveness in Bladder Cancer via Modulating the miR-223-Hsp90b1 axis. Cell Cycle 18 (23), 3288–3299. doi:10.1080/15384101.2019.1673633
Fatica, A., and Bozzoni, I. (2014). Long Non-coding RNAs: New Players in Cell Differentiation and Development. Nat. Rev. Genet. 15 (1), 7–21. doi:10.1038/nrg3606
Feng, J., Bi, C., Clark, B. S., Mady, R., Shah, P., and Kohtz, J. D. (2006). The Evf-2 Noncoding RNA Is Transcribed from the Dlx-5/6 Ultraconserved Region and Functions as a Dlx-2 Transcriptional Coactivator. Genes Dev. 20 (11), 1470–1484. doi:10.1101/gad.1416106
Fu, X., Tian, Y., Kuang, W., Wen, S., and Guo, W. (2019). Long Non-coding RNA DLX6-AS1 Silencing Inhibits Malignant Phenotypes of Gastric Cancer Cells. Exp. Ther. Med. 17 (6), 4715–4722. doi:10.3892/etm.2019.7521
Gong, J., Liu, W., Zhang, J., Miao, X., and Guo, A.-Y. (2014). lncRNASNP: a Database of SNPs in lncRNAs and Their Potential Functions in Human and Mouse. Nucleic Acids Res. 43 (D1), D181–D186. doi:10.1093/nar/gku1000
Guo, J., Chen, Z., Jiang, H., Yu, Z., Peng, J., Xie, J., et al. (2019). The lncRNA DLX6-AS1 Promoted Cell Proliferation, Invasion, Migration and Epithelial-To-Mesenchymal Transition in Bladder Cancer via Modulating Wnt/β-Catenin Signaling Pathway. Cancer Cel Int 19, 312. doi:10.1186/s12935-019-1010-z
Guttman, M., Amit, I., Garber, M., French, C., Lin, M. F., Feldser, D., et al. (2009). Chromatin Signature Reveals over a Thousand Highly Conserved Large Non-coding RNAs in Mammals. Nature 458 (7235), 223–227. doi:10.1038/nature07672
Han, J. Y., Guo, S., Wei, N., Xue, R., Li, W., Dong, G., et al. (2020). ciRS-7 Promotes the Proliferation and Migration of Papillary Thyroid Cancer by Negatively Regulating the miR-7/epidermal Growth Factor Receptor axis. Biomed. Res. Int. 2020, 9875636. doi:10.1155/2020/9875636
Huang, Y., Ni, R., Wang, J., and Liu, Y. (2019). Knockdown of lncRNA DLX6-AS1 Inhibits Cell Proliferation, Migration and Invasion while Promotes Apoptosis by Downregulating PRR11 Expression and Upregulating miR-144 in Non-small Cell Lung Cancer. Biomed. Pharmacother. 109, 1851–1859. doi:10.1016/j.biopha.2018.09.151
Ignarski, M., Islam, R., and Müller, R.-U. (2019). Long Non-coding RNAs in Kidney Disease. Ijms 20 (13), 3276. doi:10.3390/ijms20133276
Iyer, M. K., Niknafs, Y. S., Malik, R., Singhal, U., Sahu, A., Hosono, Y., et al. (2015). The Landscape of Long Noncoding RNAs in the Human Transcriptome. Nat. Genet. 47 (3), 199–208. doi:10.1038/ng.3192
Jia, P., Wei, E., Liu, H., Wu, T., and Wang, H. (2020). Silencing of Long Non‐coding RNA DLX6‐AS1 Weakens Neuroblastoma Progression by the miR ‐513c‐5p/PLK4 axis. IUBMB Life 72 (12), 2627–2636. doi:10.1002/iub.2392
Kong, L., and Zhang, C. (2020). LncRNA DLX6-AS1 Aggravates the Development of Ovarian Cancer via Modulating FHL2 by Sponging miR-195-5p. Cancer Cel Int 20, 370. doi:10.1186/s12935-020-01452-z
Kong, W. Q., Liang, J. J., Du, J., Ye, Z. X., Gao, P., and Liang, Y. L. (2020). Long Noncoding RNA DLX6-AS1 Regulates the Growth and Aggressiveness of Colorectal Cancer Cells via Mediating miR-26a/EZH2 Axis. Cancer Biother. Radiopharm. 36 (9), 753–764. doi:10.1089/cbr.2020.3589
Lei, X., Yang, S., Yang, Y., Zhang, J., Wang, Y., and Cao, M. (2019). Long Noncoding RNA DLX6-AS1 Targets miR-124-3p/CDK4 to Accelerate Ewing's Sarcoma. Am. J. Transl Res. 11 (10), 6569–6576.
Li, C., Wang, S., and Yang, C. (2020). RETRACTED: Long Non-coding RNA DLX6-AS1 Regulates Neuroblastoma Progression by Targeting YAP1 via miR-497-5p. Life Sci. 252, 117657. doi:10.1016/j.lfs.2020.117657
Li, D., Tang, X., Li, M., and Zheng, Y. (2019). Long Noncoding RNA DLX6‐AS1 Promotes Liver Cancer by Increasing the Expression of WEE1 via Targeting miR‐424‐5p. J. Cel Biochem 120 (8), 12290–12299. doi:10.1002/jcb.28493
Li, X., Zhang, H., and Wu, X. (2019). Long Noncoding RNA DLX6-AS1 Accelerates the Glioma Carcinogenesis by Competing Endogenous Sponging miR-197-5p to Relieve E2F1. Gene 686, 1–7. doi:10.1016/j.gene.2018.10.065
Li, Y., Li, X., Yang, Y., Li, M., Qian, F., Tang, Z., et al. (2020). TRlnc: a Comprehensive Database for Human Transcriptional Regulatory Information of lncRNAs. Brief. Bioinformatics 22 (2), 1929–1939. doi:10.1093/bib/bbaa011
Liang, Y., Zhang, C.-D., Zhang, C., and Dai, D.-Q. (2020). DLX6-AS1/miR-204-5p/OCT1 Positive Feedback Loop Promotes Tumor Progression and Epithelial-Mesenchymal Transition in Gastric Cancer. Gastric Cancer 23 (2), 212–227. doi:10.1007/s10120-019-01002-1
Liu, X., Peng, D., Cao, Y., Zhu, Y., Yin, J., Zhang, G., et al. (2020). Upregulated lncRNA DLX6-AS1 Underpins Hepatocellular Carcinoma Progression via the miR-513c/Cul4A/ANXA10 axis. Cancer Gene Ther. 28 (5), 486–501. doi:10.1038/s41417-020-00233-0
Liu, Y., Liu, X., Zhang, X., Deng, J., Zhang, J., and Xing, H. (2020). lncRNA DLX6-AS1 Promotes Proliferation of Laryngeal Cancer Cells by Targeting the miR-26a/TRPC3 Pathway. Cmar Vol. 12, 2685–2695. doi:10.2147/cmar.s237181
Long, J., Bai, Y., Yang, X., Lin, J., Yang, X., Wang, D., et al. (2019). Construction and Comprehensive Analysis of a ceRNA Network to Reveal Potential Prognostic Biomarkers for Hepatocellular Carcinoma. Cancer Cel Int 19, 90. doi:10.1186/s12935-019-0817-y
Losko, M., Kotlinowski, J., and Jura, J. (2016). Long Noncoding RNAs in Metabolic Syndrome Related Disorders. Mediators Inflamm. 2016, 5365209. doi:10.1155/2016/5365209
Mattick, J. S. (2001). Non‐coding RNAs: the Architects of Eukaryotic Complexity. EMBO Rep. 2 (11), 986–991. doi:10.1093/embo-reports/kve230
Mattick, J. S. (2009). The Genetic Signatures of Noncoding RNAs. Plos Genet. 5 (4), e1000459. doi:10.1371/journal.pgen.1000459
Miao, Y.-R., Liu, W., Zhang, Q., and Guo, A.-Y. (2017). lncRNASNP2: an Updated Database of Functional SNPs and Mutations in Human and Mouse lncRNAs. Nucleic Acids Res. 46 (D1), D276–D280. doi:10.1093/nar/gkx1004
Necsulea, A., Soumillon, M., Warnefors, M., Liechti, A., Daish, T., Zeller, U., et al. (2014). The Evolution of lncRNA Repertoires and Expression Patterns in Tetrapods. Nature 505 (7485), 635–640. doi:10.1038/nature12943
Ning, S., Zhang, J., Wang, P., Zhi, H., Wang, J., Liu, Y., et al. (2015). Lnc2Cancer: a Manually Curated Database of Experimentally Supported lncRNAs Associated with Various Human Cancers. Nucleic Acids Res. 44 (D1), D980–D985. doi:10.1093/nar/gkv1094
Okazaki, Y., Furuno, M., Kasukawa, T., Adachi, J., Bono, H., Kondo, S., et al. (2002). Analysis of the Mouse Transcriptome Based on Functional Annotation of 60,770 Full-Length cDNAs. Nature 420 (6915), 563–573. doi:10.1038/nature01266
Pastori, C., and Wahlestedt, C. (2012). Involvement of Long Noncoding RNAs in Diseases Affecting the central Nervous System. RNA Biol. 9 (6), 860–870. doi:10.4161/rna.20482
Qian, Y., Song, W., Wu, X., Hou, G., Wang, H., Hang, X., et al. (2021). DLX6 Antisense RNA 1 Modulates Glucose Metabolism and Cell Growth in Gastric Cancer by Targeting microRNA-4290. Dig. Dis. Sci. 66 (2), 460–473. doi:10.1007/s10620-020-06223-4
Sahu, A., Singhal, U., and Chinnaiyan, A. M. (2015). Long Noncoding RNAs in Cancer: From Function to Translation. Trends Cancer 1 (2), 93–109. doi:10.1016/j.trecan.2015.08.010
Sun, W., Zhang, L., Yan, R., Yang, Y., and Meng, X. (2019). LncRNA DLX6-AS1 Promotes the Proliferation, Invasion, and Migration of Non-small Cell Lung Cancer Cells by Targeting the miR-27b-3p/GSPT1 axis. Ott Vol. 12, 3945–3954. doi:10.2147/ott.s196865
Takahashi, K., Yan, I., Haga, H., and Patel, T. (2014). Long Noncoding RNA in Liver Diseases. Hepatology 60 (2), 744–753. doi:10.1002/hep.27043
Uchida, S., and Dimmeler, S. (2015). Long Noncoding RNAs in Cardiovascular Diseases. Circ. Res. 116 (4), 737–750. doi:10.1161/circresaha.116.302521
Wan, P., Su, W., and Zhuo, Y. (2017). The Role of Long Noncoding RNAs in Neurodegenerative Diseases. Mol. Neurobiol. 54 (3), 2012–2021. doi:10.1007/s12035-016-9793-6
Wawrzyniak, O., Zarębska, Ż., Rolle, K., and Gotz-Więckowska, A. (2018). Circular and Long Non-coding RNAs and Their Role in Ophthalmologic Diseases. Acta Biochim. Pol. 65 (4), 497–508. doi:10.18388/abp.2018_2639
Wilusz, J. E., Sunwoo, H., and Spector, D. L. (2009). Long Noncoding RNAs: Functional Surprises from the RNA World. Genes Dev. 23 (13), 1494–1504. doi:10.1101/gad.1800909
Wu, D. M., Zheng, Z. H., Zhang, Y. B., Fan, S. H., Zhang, Z. F., Wang, Y. J., et al. (2019). Down-regulated lncRNA DLX6-AS1 Inhibits Tumorigenesis through STAT3 Signaling Pathway by Suppressing CADM1 Promoter Methylation in Liver Cancer Stem Cells. J. Exp. Clin. Cancer Res. 38 (1), 237. doi:10.1186/s13046-019-1239-3
Wu, P., Zuo, X., Deng, H., Liu, X., Liu, L., and Ji, A. (2013). Roles of Long Noncoding RNAs in Brain Development, Functional Diversification and Neurodegenerative Diseases. Brain Res. Bull. 97, 69–80. doi:10.1016/j.brainresbull.2013.06.001
Wu, Q., Ma, J., Meng, W., and Hui, P. (2020). DLX6-AS1 Promotes Cell Proliferation, Migration and EMT of Gastric Cancer through FUS-Regulated MAP4K1. Cancer Biol. Ther. 21 (1), 17–25. doi:10.1080/15384047.2019.1647050
Xie, F., Xie, G., and Sun, Q. (2020). Long Noncoding RNA DLX6-AS1 Promotes the Progression in Cervical Cancer by Targeting miR-16-5p/ARPP19 Axis. Cancer Biother. Radiopharm. 35 (2), 129–136. doi:10.1089/cbr.2019.2960
Yang, B., Jia, L., Ren, H., Jin, C., Ren, Q., Zhang, H., et al. (2020). LncRNA DLX6-AS1 Increases the Expression of HIF-1α and Promotes the Malignant Phenotypes of Nasopharyngeal Carcinoma Cells via Targeting MiR-199a-5p. Mol. Genet. Genomic Med. 8 (1), e1017. doi:10.1002/mgg3.1017
Yang, J., Ye, Z., Mei, D., Gu, H., and Zhang, J. (2019). Long Noncoding RNA DLX6-AS1 Promotes Tumorigenesis by Modulating miR-497-5p/FZD4/FZD6/Wnt/β-Catenin Pathway in Pancreatic Cancer. Cmar Vol. 11, 4209–4221. doi:10.2147/cmar.s194453
Yang, Q., Sun, J., Ma, Y., Zhao, C., and Song, J. (2019). LncRNA DLX6-AS1 Promotes Laryngeal Squamous Cell Carcinoma Growth and Invasion through Regulating miR-376c. Am. J. Transl Res. 11 (11), 7009–7017.
Zeng, X., Hu, Z., Ke, X., Tang, H., Wu, B., Wei, X., et al. (2017). Long Noncoding RNA DLX6-AS1 Promotes Renal Cell Carcinoma Progression via miR-26a/PTEN axis. Cell Cycle 16 (22), 2212–2219. doi:10.1080/15384101.2017.1361072
Zhang, J. J., Xu, W. R., Chen, B., Wang, Y. Y., Yang, N., Wang, L. J., et al. (2019). The Up-Regulated lncRNA DLX6-AS1 in Colorectal Cancer Promotes Cell Proliferation, Invasion and Migration via Modulating PI3K/AKT/mTOR Pathway. Eur. Rev. Med. Pharmacol. Sci. 23 (19), 8321–8331. doi:10.26355/eurrev_201910_19143
Zhang, L., He, X., Jin, T., Gang, L., and Jin, Z. (2017). Long Non-coding RNA DLX6-AS1 Aggravates Hepatocellular Carcinoma Carcinogenesis by Modulating miR-203a/MMP-2 Pathway. Biomed. Pharmacother. 96, 884–891. doi:10.1016/j.biopha.2017.10.056
Zhang, N., Meng, X., Mei, L., Zhao, C., and Chen, W. (2019). LncRNA DLX6‐AS1 Promotes Tumor Proliferation and Metastasis in Osteosarcoma through Modulating miR‐641/HOXA9 Signaling Pathway. J. Cel Biochem 120 (7), 11478–11489. doi:10.1002/jcb.28426
Zhang, R. M., Tang, T., Yu, H. M., and Yao, X. D. (2018). LncRNA DLX6-AS1/miR-129-5p/DLK1 axis Aggravates Stemness of Osteosarcoma through Wnt Signaling. Biochem. Biophys. Res. Commun. 507 (1), 260–266. doi:10.1016/j.bbrc.2018.11.019
Zhang, X., Guo, H., Bao, Y., Yu, H., Xie, D., and Wang, X. (2019). Exosomal Long Non-coding RNA DLX6-AS1 as a Potential Diagnostic Biomarker for Non-small Cell Lung Cancer. Oncol. Lett. 18 (5), 5197–5204. doi:10.3892/ol.2019.10892
Zhao, H., and Xu, Q. (2020). Long Non‐coding RNA DLX6‐AS1 Mediates Proliferation, Invasion and Apoptosis of Endometrial Cancer Cells by Recruiting p300/E2F1 in DLX6 Promoter Region. J. Cel. Mol. Med. 24 (21), 12572–12584. doi:10.1111/jcmm.15810
Zhao, J., and Liu, H. R. (2019). Down-regulation of Long Noncoding RNA DLX6-AS1 Defines Good Prognosis and Inhibits Proliferation and Metastasis in Human Epithelial Ovarian Cancer Cells via Notch Signaling Pathway. Eur. Rev. Med. Pharmacol. Sci. 23 (8), 3243–3252. doi:10.26355/eurrev_201904_17684
Zhao, P., Guan, H., Dai, Z., Ma, Y., Zhao, Y., and Liu, D. (2019). Long Noncoding RNA DLX6-AS1 Promotes Breast Cancer Progression via miR-505-3p/RUNX2 axis. Eur. J. Pharmacol. 865, 172778. doi:10.1016/j.ejphar.2019.172778
Zhao, Y.-H., Wang, Z., Zhang, N., Cui, T., and Zhang, Y.-H. (2020). Effect of ciRS-7 Expression on clear Cell Renal Cell Carcinoma Progression. Chin. Med. J. 133 (17), 2084–2089. doi:10.1097/cm9.0000000000000867
Zhao, Z., Liang, S., and Sun, F. (2020). LncRNA DLX6-AS1 Promotes Malignant Phenotype and Lymph Node Metastasis in Prostate Cancer by Inducing LARGE Methylation. Front. Oncol. 10, 1172. doi:10.3389/fonc.2020.01172
Keywords: DLX6-AS1, non-coding RNA, lncRNA, cancer, miRNA 3
Citation: Ghafouri-Fard S, Najafi S, Hussen BM, Ganjo AR, Taheri M and Samadian M (2022) DLX6-AS1: A Long Non-coding RNA With Oncogenic Features. Front. Cell Dev. Biol. 10:746443. doi: 10.3389/fcell.2022.746443
Received: 23 July 2021; Accepted: 04 February 2022;
Published: 25 February 2022.
Edited by:
Ri Cui, Wenzhou Medical University, ChinaReviewed by:
Hamed Shoorei, Birjand University of Medical Sciences, IranCopyright © 2022 Ghafouri-Fard, Najafi, Hussen, Ganjo, Taheri and Samadian. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mohammad Taheri, TW9oYW1tYWQudGFoZXJpQHVuaS1qZW5hLmRl; Mohammad Samadian, bWRzYW1hZGlhbkBnbWFpbC5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.