
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cell Dev. Biol. , 06 January 2023
Sec. Stem Cell Research
Volume 10 - 2022 | https://doi.org/10.3389/fcell.2022.1107881
This article is part of the Research Topic Reviews in: Induced Pluripotent Stem Cells View all 7 articles
A correction has been applied to this article in:
Corrigendum: Deepening the understanding of CNVs on chromosome 15q11–13 by using hiPSCs: An overview
 Angela Maria Giada Giovenale1,2†
Angela Maria Giada Giovenale1,2† Giorgia Ruotolo1,2†
Giorgia Ruotolo1,2† Amata Amy Soriano1
Amata Amy Soriano1 Elisa Maria Turco1
Elisa Maria Turco1 Giovannina Rotundo1
Giovannina Rotundo1 Alessia Casamassa1
Alessia Casamassa1 Angela D’Anzi1
Angela D’Anzi1 Angelo Luigi Vescovi1,2*
Angelo Luigi Vescovi1,2* Jessica Rosati1*
Jessica Rosati1*The human α7 neuronal nicotinic acetylcholine receptor gene (CHRNA7) is widely expressed in the central and peripheral nervous systems. This receptor is implicated in both brain development and adult neurogenesis thanks to its ability to mediate acetylcholine stimulus (Ach). Copy number variations (CNVs) of CHRNA7 gene have been identified in humans and are genetically linked to cognitive impairments associated with multiple disorders, including schizophrenia, bipolar disorder, epilepsy, Alzheimer’s disease, and others. Currently, α7 receptor analysis has been commonly performed in animal models due to the impossibility of direct investigation of the living human brain. But the use of model systems has shown that there are very large differences between humans and mice when researchers must study the CNVs and, in particular, the CNV of chromosome 15q13.3 where the CHRNA7 gene is present. In fact, human beings present genomic alterations as well as the presence of genes of recent origin that are not present in other model systems as well as they show a very heterogeneous symptomatology that is associated with both their genetic background and the environment where they live. To date, the induced pluripotent stem cells, obtained from patients carrying CNV in CHRNA7 gene, are a good in vitro model for studying the association of the α7 receptor to human diseases. In this review, we will outline the current state of hiPSCs technology applications in neurological diseases caused by CNVs in CHRNA7 gene. Furthermore, we will discuss some weaknesses that emerge from the overall analysis of the published articles.
The major contribution to the variability of the human genome comes from DNA deletions and duplications larger than 1 Kb of one or both alleles, defined as copy number variations (CNVs) (Zarrei et al., 2015). These quantitative structural variants can be located in gene-rich or poor chromosome regions (Redon et al., 2006). CNVs can be grouped into three main categories of clinical relevance (Nowakowska, 2017): 1) Benign variants that do not have a negative effect on human phenotype (Tuzun et al., 2005), (Sharp et al., 2005) and constitute an important source for human phenotypic variability (Pinto et al., 2007). 2) Pathogenic variants that are associated with a wide spectrum of diseases. CNVs can change human phenotypes by acting through various rearrangements of genome structure that include: duplications or deletions of dosage-sensitive genes; rearranged breakpoints that interrupt and inactivate genes; generation of fusion genes at the breakpoints with a gain-of-function; unmasking of recessive mutations or of transvections, when CNV, occurring in one chromosome, affects an allele on the other chromosome at the same locus. CNVs are pathological even when they occur in non-coding regions (“position effect” variation) involved in gene expression regulation (Lupski and Stankiewicz, 2005). 3) Variants of uncertain significance (VOUS), with an ambiguous clinical relevance that does not permit classification as pathogenic or benign (Vermeesch et al., 2012).
Many of these quantitative structural variants happen in correspondence with chromosome 15q11–13, which shows a high frequency of genomic rearrangements due to its intrinsic genomic instability (Makoff and Flomen, 2007; Rosenfeld et al., 2011). The proximal region of the long arm of chromosome 15 is characterized by three segmental duplications that are subject to Non-Allelic Homologous Recombination (NAHR), due to their high homology. These chromosome regions, for their characteristics, are known as BreakPoint regions (BP3, BP4, BP5) since, after NAHR, they determine chromosomal rearrangements, such as microdeletions and microduplications. Rearrangements of chromosome 15q11-13 are associated with Prader-Willi and Angelman syndromes and neurological disorders (Makoff and Flomen, 2007; Sharp et al., 2008; Miller et al., 2009; Rosenfeld et al., 2011). Generally, CNVs affecting 15q13.3 have been reported correlated to a highly variable phenotype and different neurological manifestations (Budisteanu et al., 2021). Among these, both chromosome 15q13.3 deletions and duplications are associated with autism spectrum disorder (ASD), intellectual disability (ID)/developmental delay (DD), mood disorders, speech problems, and schizophrenia (SCZ) (Sharp et al., 2008; Miller et al., 2009; van Bon et al., 2009; Szafranski et al., 2010; Beal, 2014; Szatkiewicz et al., 2014; Bacchelli et al., 2015; Lowther et al., 2015; Pettigrew et al., 2015; Zhou et al., 2016). On the one hand, CNVs involving deletion generally exhibit severe, highly penetrant patient phenotypes; for instance, 15q13.3 microdeletions are usually associated with cognitive deficits, behavioral abnormalities, and ASD (Ben-Shachar et al., 2009; Miller et al., 2009; Pagnamenta et al., 2009; van Bon et al., 2009; Masurel-Paulet et al., 2010). On the other hand, CNVs involving duplication often cause widely variable and less penetrant phenotypic expressivity among affected subjects (Mefford et al., 2012; Newman et al., 2015). 15q13.3 microduplications are indeed correlated to milder clinical phenotypes, including borderline ID, ASD and attention deficit hyperactivity disorder (ADHD) (Williams et al., 2012; Gillentine and Schaaf, 2015; Meganathan et al., 2021). However, 15q13.3 microduplications are frequent both in clinical and non-clinical cases (symptomatic and non-symptomatic), making their contribution to pathogenicity hard to estimate (Wiśniowiecka-Kowalnik et al., 2013; Coe et al., 2014; Rehm et al., 2015; Gillentine et al., 2017). This region is important because it is here that deletions or duplications affect the human α7 nicotinic cholinergic receptor (CHRNA7) gene. The situation is further complicated by the presence of the chimeric fusion gene CHRFAM7A, specific only to human beings, responsible for variations in nAChR structure that impacts patient phenotypes in a variable way (Araud et al., 2011). Consistent with new evidence suggesting that the expression of CHRFAM7A fusion gene dominant-negatively inhibits the channel functions of α7-nAChR, accumulating data have shown an association between CHRFAM7A dosage and Alzheimer’s disease, nicotine dependence, schizophrenia, bipolar disorder, and other neuropsychiatric disorders (Ihnatovych et al., 2019; Peng et al., 2022).
Multiple animal model systems have been used to study the impact of the 15q13.3 CNVs on the human brain, but they have failed to fully recapitulate human phenotypes, perhaps due to the different genes present in this chromosome region. Thus, induced pluripotent stem cells (iPSCs) provide a useful human model to understand the pathological mechanisms involving these CNVs (Gillentine, 2022). This review will be focused both on the use of hiPSCs technology for the comprehension of 15q13.3 CNVs-dependent neurological diseases and on future applications of iPSCs in drug development for the abovementioned disorders and diseases.
The human (CHRNA7) gene includes ten exons and encodes for the α7 subunit of neuronal nicotinic Acetylcholine Receptor (α7nAChR), widely expressed both in central and peripheral nervous systems. On the cell surface, five α7nAChR monomers associate to form an oligomeric ligand-gated ion channel, belonging to the Nicotinic Acetylcholine Receptor (nAChRs) family. nAChRs are selective transmembrane ion channels for Na+, K+, and Ca2+, which bind neurotransmitters and regulate excitatory and inhibitory signaling. They can be composed of α or β subunits, creating both homo- and hetero-pentameric channels. In particular, α7nAChR is characterized by a low affinity for nicotine, a high affinity for α-bungarotoxin (α -Bgtx), high permeability to calcium (Marks and Collins, 1982), and fast desensitization (Zhang et al., 2011). The α7 receptor is expressed in presynaptic neurons and astrocytes (Vijayaraghavan et al., 1992; Patel et al., 2017) where it modulates neurotransmitter release, as well as in post-synaptic regions, influencing gene expression. Based on these functions, α7nAChR is implicated in cognitive processes and synaptic plasticity (Shen and Yakel, 2009), neurotransmitter release and immune responsiveness with consequent implications for Alzheimer’s disease, Parkinson’s disease and so on (Schaaf, 2014; Quik et al., 2015; Liu et al., 2017). Moreover, its expression in extra-neuronal tissues indicates an additional role in modulating several calcium-activated signal pathways influencing proliferation, differentiation, migration, and inflammatory response (Wang et al., 2003; Campbell et al., 2010; Mucchietto et al., 2018).
A second distinct α7nAChR gene was discovered in the human genome in 1998 (Gault et al., 1998). Close to CHRNA7, there are four repeated sequences, called FAM7A, made up of exons A, B, C, and E, copies of two exons of the Unc-51 Like Kinase 4 gene (ULK4) (Lang et al., 2014), and exons D and F, homologous to the GOLGA8B gene (Stephens et al., 2012). The duplication of exons 5–10 of CHRNA7 together with a FAM7A sequence, and the subsequent insertion of this cassette at 1.6 Mb upstream of CHRNA7, provoke the formation of the chimeric gene CHRFAM7A (Figure 1). The duplicated cassette has an inverted orientation concerning the CHRNA7 gene; furthermore, a 2bp deletion polymorphism is present, in which the CHRFAM7AΔ2bp insertion also results in an inverted orientation (Flomen et al., 2008). This new chimeric gene is largely expressed in both CNS and peripheral tissues and is present in about 90%–95% of individuals, 30% of whom have a single copy instead of two alleles (Riley et al., 2002; Sinkus et al., 2009). Homologous CHRNA7 has not been found in primates and rodents (Locke et al., 2003), indicating recent origins. CHRFAM7A presents an Open Reading Frame (ORF) in correspondence with the exon B, and the translation product results in a smaller protein than the original CHRNA7 (46 kDa instead of 57 kDa), lacking part of the N-terminal domain that contains the signal peptide and part of the agonist-binding site. A second isoform originates from the ORF in exon 6 and produces a shorter peptide (38 kDa), lacking the ligand peptide and the entire binding site. The transcription of CHRFAM7A does not affect the expression of CHRNA7, and even though CHRFAM7A has a lower transcription rate in the brain, the encoded protein (dupα7) has a dominant negative effect on the α7 subunits (de Lucas-Cerrillo et al., 2011; Chan et al., 2019). There are two possible explanations for this phenotype. The first is that the dupα7 protein preferentially localizes at the endoplasmic reticulum, where it might sequester the α7 subunits and reduce the α7nAChR activity (Araud et al., 2011). The second mechanism might involve the assembly of dupα7 with other α7 subunits in the cellular membrane. When homomeric, Dupα7 receptors are not functional, as ligands cannot bind to them, but in vitro experiments have shown the formation of heteromeric ion channels (Wang et al., 2014). The efficiency of receptors is guaranteed as long as there are three dupα7 adjacent to α7 subunits, which allow the formation of dupα7/α7 interfaces, containing the agonist binding site. The presence of CHRFAM7A in double copy, or the presence of the 2bp depleted polymorphism could influence the overall function of α7 receptors.
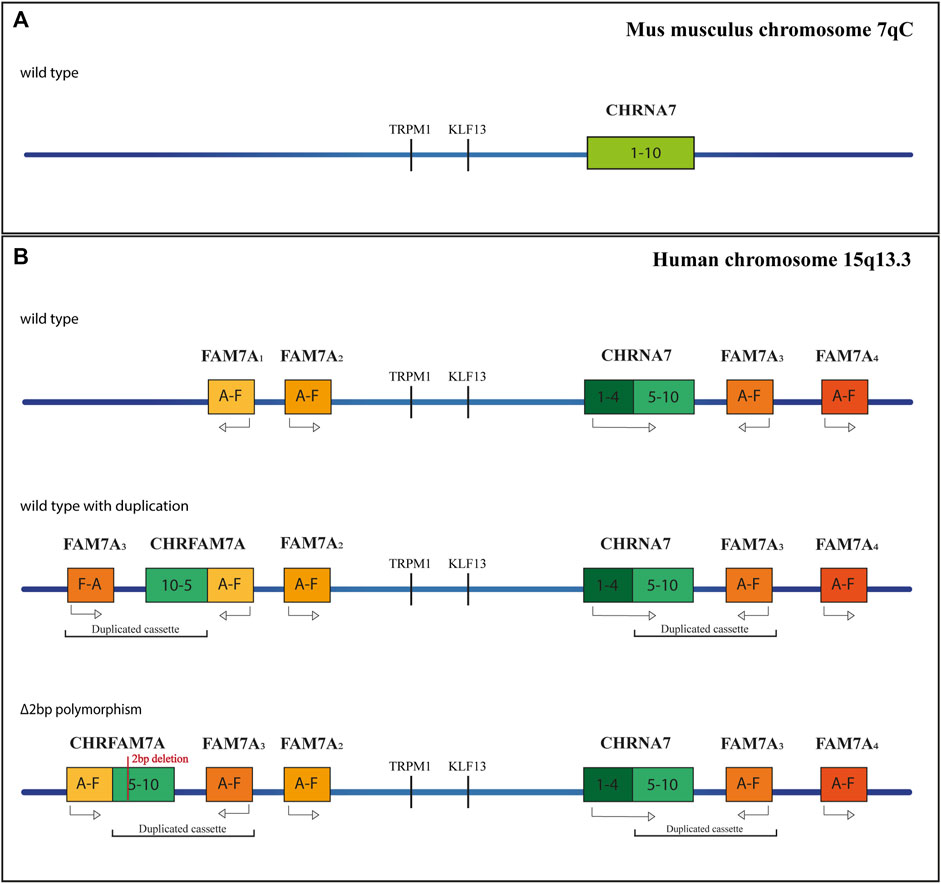
FIGURE 1. Schematic representation of the CHRNA7 locus. (A) Orthologus locus in mus musculus, at the 7qC chromosome. (B) Different genetic profiles of the human Chromosome 15q13.3, without duplication, with the chimeric gene and with the 2bp mutation.
Multiple animal model systems have been used to determine the role and function of CHRNA7 in human phenotypes. The first studies to explore putative regulatory mechanisms of α7nAChR (de Lucas-Cerrillo et al., 2011) were carried out on Xenopus laevis oocytes and rat cell lines previously transfected with two constructs carrying CHRFAM7A and CHRFAM7AΔ2bp (de Lucas-Cerrillo et al., 2011). These experiments highlighted that the chimeric gene CHRFAM7A negatively regulates α7nAChR functions, especially in the presence of partial duplication with the 2-bp deletion, CHRFAM7AΔ2bp (Araud et al., 2011). Currently, murine models are mainly used for the 15q13.3 CNV study (Fejgin et al., 2014; Lewis et al., 2018; Felix et al., 2019). The first Chrna7-deficient mice showed attention deficits, anxiety, and anhedonia (Zhang et al., 2016) but this model was found to be unsuitable for recapitulating human pathological phenotypes associated with learning, memory, and sensorimotor gating (Yin et al., 2017). Further studies on Chrna7 knockout mice highlighted altered temporal processing of the auditory brainstem response (ABR) signal that may contribute to degraded spike timing in the midbrain (Felix et al., 2019). However, experiments on Chrna7 knockout mice did not provide significant information owing to the several essential differences between humans and murine species: first, behavioral and functional defects were observed in homozygous knockout mice, while in humans, the 15q13.3 CNV are generally heterozygous; second, the Endoplasmic Reticulum (ER) chaperone RIC3 involved in nAChRs assembly in human beings is expressed differently than in mice (Halevi et al., 2003), suggesting a probable functional difference in α7nAChR assembly; third, the negative modulator CHRFAM7A is only expressed in humans, indicating that different regulatory mechanisms are probably involved in mice (Yin et al., 2017). Preliminary studies on CHRFAM7A transgenic mice have highlighted a different expression of proteins involved in the signaling pathways at the basis of PD, AD, Huntington’s disease (HD), and alcoholism such as calcium signaling, oxidative phosphorylation and others. It suggests that the CHRFAM7A gene contributes to the pathological process likely through precise control of α7-nAChR functions in the brain (Yin et al., 2017; Jiang et al., 2019). However, the role of CHRFAM7A in 15q.13.3 CNV needs to be explored more in depth, using a human cellular model such as iPSCs.
The advent of cellular reprogramming technology has proven that induced Pluripotent Stem Cells (iPSCs) can represent an extremely useful resource for studying neurodevelopmental/neuropsychiatric diseases. Human-induced Pluripotent Stem Cells (hiPSCs) are generated by reprogramming somatic cells into pluripotent stem cells, which can be further differentiated into a large variety of cell types, such as neurons, astrocytes, and oligodendrocytes (Dimos et al., 2008; Chambers et al., 2009) (Figure 2). To date, it is possible to reprogram a large number of cell types, such as fibroblasts (skin biopsies), keratinocytes (hair roots) (Piao et al., 2014; Soliman et al., 2017), cord blood endothelial cells (Haase et al., 2009), blood cells (Ye et al., 2009), T lymphocytes (Brown et al., 2010; Seki et al., 2011), melanocytes (Utikal et al., 2009), hepatocytes (Liu et al., 2010), mesangial cells (Song et al., 2011), exfoliated renal epithelial cells from urine samples (Ruiz et al., 2010; Zhou et al., 2012). A growing number of studies have demonstrated that iPSCs are an excellent cellular model for studying both syndromic and idiopathic forms of neurodevelopmental/neuropsychiatric disorders, which have their origin in the prenatal period during cell differentiation (Soliman et al., 2017). Furthermore, iPSC-derived neurons can be generated from patients carrying a specific genetic background, corresponding to a particular neuropsychiatric disease, where rare but penetrant genetic abnormalities are likely to play a role (Park et al., 2008; Marchetto et al., 2010; Dolmetsch and Geschwind, 2011; Grskovic et al., 2011; Paşca et al., 2011). Recently, the production of iPSC-derived 3D organoids has allowed researchers to investigate the interaction of multiple cell types in a more brain-like microenvironment. This model takes advantage of the capability of hiPSCs to self-organize into embryoid bodies and subsequently differentiate into organoids in response to environmental cues mimicking in vivo conditions. The use of human neural organoids allows researchers to study, in vitro, both the three-dimensional (3D) cytoarchitecture of the human brain and its development, overcoming the limitations associated with the use of animal models (Guy et al., 2021). To date, many neurodevelopmental and neurodegenerative disorders such as primary microcephaly (Lancaster et al., 2013; Li et al., 2017a; Zhang et al., 2019), macrocephaly (Li et al., 2017b), autism spectrum disorder (Mariani et al., 2015; Schafer et al., 2019), Parkinson’s disease (PD) (Kim et al., 2019; Smits et al., 2019), and Alzheimer’s disease (Raja et al., 2016; Gonzalez et al., 2018; Lin et al., 2018; Ghatak et al., 2019) were studied using this technology.
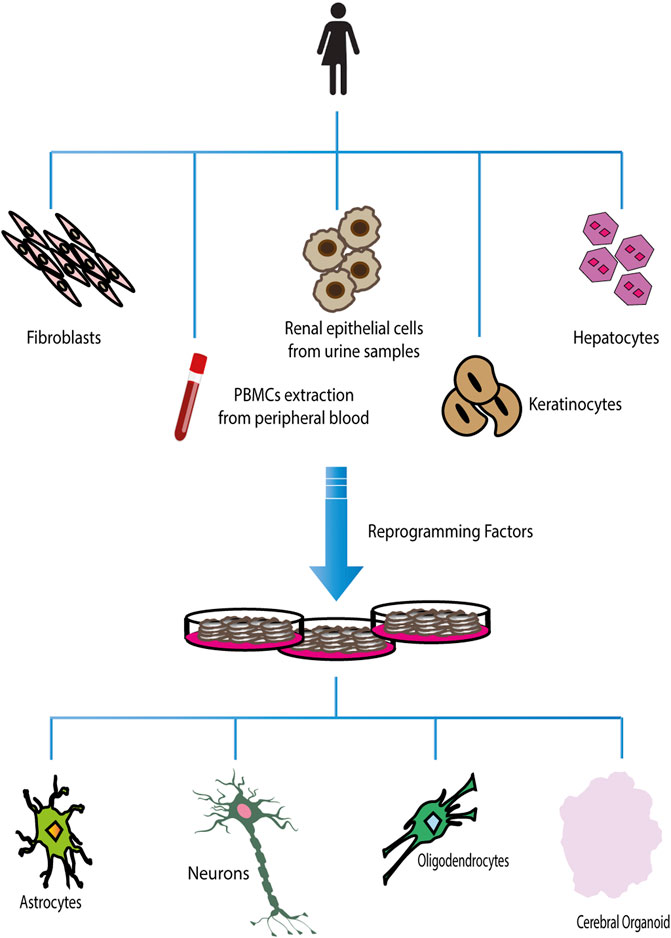
FIGURE 2. Reprogramming and differentiation of hiPSCs. Different adult somatic cells can be reprogrammed in induced Pluripotent Stem Cells. Once hiPSCs have been obtained, they can be used to generate all types of terminally differentiated cells.
To date, very few studies have been published regarding the expression and functionality of nAChRs in human iPSC-derived neurons under physiological conditions. Based on a previous study (Gill et al., 2013), in 2015 researchers profiled the electrophysiological properties of GABA neurons (catalog number R1013, Cellular Dynamics International) (Chatzidaki et al., 2015). In these iPSC-derived cells, they confirmed the expression of both CHRNA7 and CHRFAM7A genes, demonstrating that hiPSC-derived neurons recapitulated the main features of the α7nAChR: low open probability (the tendency for fewer receptors to open) and fast desensitization, characteristics that could be inverted with the addition of positive allosteric modulators (PAM type II) together with selective agonists, as observed in HEK 293 cells (DaCosta et al., 2011), Xenopus laevis Oocytes (Pałczyńska et al., 2012) and BOSC 23 Cells (Williams et al., 2011). In the second study (Larsen et al., 2019), three different clones belonging to the same reprogramming event (BIONi010-A, BIONi010-B, and BIONi010-C, European Bank of induced Pluripotent Stem Cells) were obtained from fibroblasts of a healthy subject. They observed an increase in the expression of CHRNA7 and CHRFAM7A during neuronal maturation, highlighting the player role of these genes during neural differentiation. The interesting outcome of this work was the formation of a neuronal network model that offered the possibility of evaluating α7 receptor involvement during synaptic transmission. These papers demonstrated that hiPSC-derived neurons not only highlight features of α7nAChR commonly observed in other models but represent a more specific representation of human conditions that could evidence and clarify new mechanisms not already well characterized. Nevertheless, the important outcome of these two works is that even using different reprogramming and differentiation protocols, both models recapitulate the same receptor features, confirming the reproducibility of this model.
Since the regulation of α7nAChR is complicated by the presence of the CHRFAM7A fusion gene in humans, understanding its influence on α7nAChR function is useful for preclinical studies. This motivation prompted Ihnatovych and collaborators in 2019 to undertake an interesting study in which they evaluated how the amount of CHRFAM7A influences the receptor functions (Ihnatovych et al., 2019). Fibroblasts were reprogrammed from two subjects affected by Alzheimer’s Disease (AD), carrying respectively 0 (ancestral haplotype) and 1 copy of the CHRFAM7A gene (UB068-0 copy, UB052-1 copy). These hiPSCs were differentiated into medial ganglionic eminence (MGE) progenitors and neurons. They found a stable CHRNA7 expression during differentiation over 40 days in the 0-copy line and overexpression of both CHRNA7 and CHRFAM7A mRNAs in the 1-copy cell line, perhaps as possible compensation for the negative modulation of the chimeric gene. Moreover, the presence of CHRFAM7A modified the kinetics of channel opening in pharmacological assays with PNU 120596, showing faster desensitization of the channel in 1-copy cells than in 0-copy cells. As Alzheimer’s patients accumulate amyloid beta (Aβ), which CHRNA7 binds to, and the researchers observed a decrease in the uptake of the Aβ1-42 peptide in the 1-copy line in a dosage-dependent manner, they concluded this could be a possible protective effect under physiological conditions.
The group subsequently published two other papers (Ihnatovych et al., 2020; Szigeti et al., 2020), exploring electrophysiology and Aβ uptake in the first study and the anti-inflammatory effects of microglia (MGL) in the second, in the presence and absence of the CHRFAM7A allele and presence and absence of CHRFAM7Abp. The Aβ uptake was quantified by transfecting MGE progenitors with an empty vector (EV), CHRFAM7A, and CHRFAM7Abp cDNAs. CHRFAM7A transfected cells showed a mitigated Aβ1-42 uptake compared to EV and CHRFAM7Abp transfected cells, suggesting that while CHRFAM7A is a dominant negative modulator, the inverted CHRFAM7Abp has the same functions of a null allele for the two AD relevant phenotypes analyzed. UB068 (CHRFAM7A null) hiPSCs were then genome-edited and differentiated into MGE progenitors. The comparison between UB068 (CHRFAM7A null) and the two CHRFAM7A carrier lines, UB052 and UB068_CHRFAM7A, demonstrated different responses to Acetylcholine Esterase Inhibitors (AChEIs) and encenicline exposure. In particular, the Aβ1-42 uptake was linear with a dose of AChEIs and decreased in presence of encenicline only in UB068. These data were very interesting because they highlighted how only carriers of ancestral haplotype could benefit from α7nAChR targeting therapies. The information obtained from patients’ iPSC-derived cells is highly transferable to humans: in fact, starting from a genotyping of 1,174 subjects that split the population 1:3 for non-carriers to carriers of the direct CHRFAM7A functional allele, Ihnatovych and his group studied the response to initiation of AChEI therapy and delayed disease modifying treatment (DMT) effect over a 6-year observation period. The analysis confirmed preliminary pharmacological data on cells, showing that carriers with 0 copies of the CHRFAM7A had both a superior response to first exposure and a greater DMT effect from AChEI. The complex study conducted by the research group over the years has bridged the gap in cholinergic strategies in AD, showing that not considering CHRFAM7A could lead to the rejection of drug candidates that could be beneficial for 25% of AD patients (Szigeti et al., 2020).
In the second paper, the authors analyzed the internalization of fluorescent-Aβ in all microglial cell lines (MGL) finding that cells with CHRFAM7A presented a reduction of the dose-response curve of Aβ internalization. They also treated MGL with mecamylamine (MLA, a nicotinic acetylcholine receptor antagonist) and evaluated their internalization activity of the Aβ peptide: despite an increase of internalization of Aβ in the 0-copy line, no differences were detected for lines carrying CHRFAM7A. This suggested that MLA had a pro-inflammatory effect through the activation of Microglia cells. The activation of MGL cells leads to the translocation of p65 (representing the NF-kB activation pathway) in nuclei and consequently the expression of pro-inflammatory genes. Cells carrying CHRFAM7A showed an increase of p65 with respect to the line without fusion gene; however, the treatment with MLA caused an increase of p65 only in the 0-copy line concerning basal condition. This suggested that CHRNA7 had an anti-inflammatory action while CHRFAM7A possessed a pro-inflammatory activity, as a consequence of its negative effect on CHRNA7 (Ihnatovych et al., 2020). To completely clarify the molecular mechanism of the effects of CHRFAM7A it would have been appropriate to include a control line carrying a double copy of the fusion gene, which represents the major polymorphism in the population.
Patient-derived induced Pluripotent Stem Cells (iPSCs) technology can be a potentially limitless useful source of human cells for investigating molecular mechanisms underlying neurological diseases. To better evaluate the potential implication of CHRNA7 and CHRFAM7A dosages in such processes, here we summarized a state of the art of human iPSCs and iPSC-derived models from subjects carrying CNVs at 15q13.3 (Figure 3).
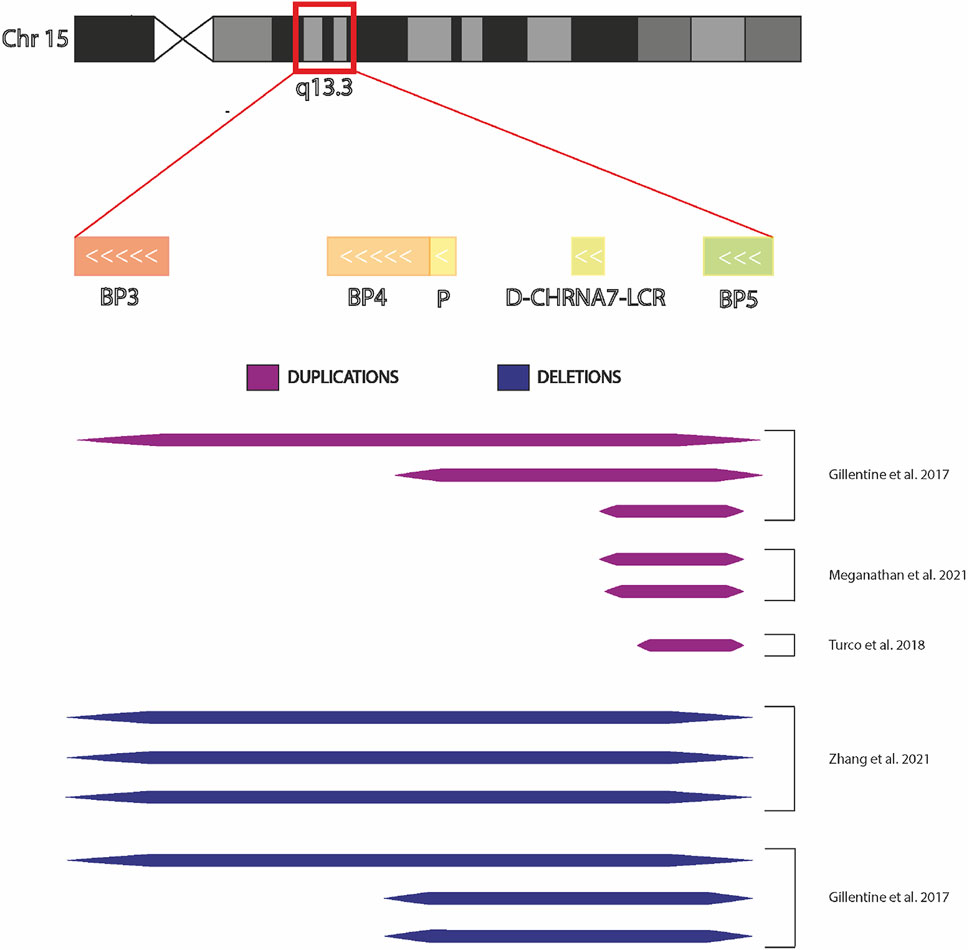
FIGURE 3. Chromosome 15q13.3 and hiPSCs derived from individuals with CNV. Graphic representation of chromosomal region 15q13.3 showing BreakPoint regions BP3, BP4, and BP5 and the extensions the microdeletions and microduplications present in the hiPSCs in the published studies.
In 2017, Gillentine and collaborators used somatic cells belonging to probands carrying 15q13.3 CNV for the first time to produce induced pluripotent stem cells to differentiate into Neural Progenitors Cells (NPCs) (Gillentine et al., 2017). These cortical-like NPCs were utilized to investigate the molecular consequences of CHRNA7 copy-number variation, overcoming the limitations of animal models. The subjects who participated in the study were three individuals with 15q13.3 deletions (two of which spanned BP4/BP5 and one spanned BP3/BP5), three individuals with 15q13.3 duplications (the first spanned BP4/BP5, the second spanned BP3/BP5 and the last duplication spanned +9 D-CHRNA7-LCR/BP5) and three copy neutral control lines. All selected patients had similar ages and phenotypic variability ranging from ID/DD, ASD to ADHD; only the last duplication (the shorter one) resulted asymptomatic.
The authors explored gene expression of CHRNA7, α7 nAChR-dependent calcium flux, and the consequences of altered calcium signaling. As expected, the qPCR analysis showed a significant decrease of about 50% in CHRNA7 expression compared to controls both in hiPSCs and NPCs carrying the deletions. To characterize the duplications, they used primers specific for the duplicated portion of CHRNA7 in the asymptomatic proband and identified an increased expression of CHRNA7 in all three lines compared to the controls.
The great potentiality of the use of human iPSCs was demonstrated through the FLIPR Tetra High-Throughput Cellular Screening System (Molecular Devices) with which the authors co-treated the NPCs with two drugs, 1 μM epibatidine and 3 μM PNU-120596 (a positive allosteric modulator), to accurately detect α7 nAChR-specific calcium flux. The analysis showed a decreased calcium flux in both groups of NPCs; a reduction was also confirmed from the downregulation of downstream effectors, such as the JAK2-PI3K pathway known for its role in modulating neuronal excitability and neurotransmitter release. Although this result was expected in the probands carrying the deletion, it was not expected in the lines with duplication. From the observation of general deregulation of all other nAChRs in these cells, the authors decided to verify two possible hypotheses. Having observed that calcium flux did not vary among CNV and controls, despite general deregulation of all other nAChRs in these cells, the authors hypothesized an altered function of the chaperons RIC3 and NACHO necessary to the nAChR receptors assembly, trafficking, and cell surface expression. Gene expression analysis demonstrated an upregulation of both mRNAs in duplicated NPCs samples while there was a downregulation in deleted NPCs samples. Since the altered expression of these two nAChRs-specific chaperons is related to the stress of Endoplasmic Reticulum (ER) (Severance and Yolken, 2007; Srinivasan et al., 2012; Lewis and Picciotto, 2013), the authors explored two pathways, PERK activation and IREα splicing of XBP1, associated with ER stress. In particular, the levels of the spliced form of XBP1 resulted increased in NPCs with duplications, but similar to controls in NPCs with deletions. The reduced expression of CHRNA7 on the cell surface and the consequent calcium flux alteration found in NPC with duplication was therefore due to the accumulation of proteins in the endoplasmic reticulum. The authors had unexpectedly found two different mechanisms with the same result, in cells carrying deletion and duplication of the 15q13.3 region.
In 2018 another study was published by Turco et al. on an iPSC line carrying a 15q13.3 duplication. Genotype analysis showed that the current duplication involved only the CHRNA7 gene. This evidence, in addition to the healthy phenotype of the subject, suggests that duplications of the single gene are not sufficient for the onset of the phenotype, by what was already observed in the asymptomatic proband (Gillentine et al., 2017). Further studies carried out on this apparently healthy duplication will be useful to clarify the mechanisms at the base of pathologic phenotypes.
Variable penetrance could explain why individuals carrying the same duplication from a partially shared genetic background present different phenotypes. In order to understand this phenomenon, Meganathan and collaborators selected a family of four individuals, including three subjects with the same 15q13.3 duplication: the mother, who had no clinical diagnosis (UM), her older son, who exhibited distinct features of autism and emotional dysregulation (the affected proband, AP), and her younger affected son, who exhibited mild ASD, ADHD, and mood disorder traits, while the father did not carry the duplication. The CNV in these three subjects was a duplication of −400 kilobase at chromosome 15, band q13.3; the only gene located in the duplicated region was CHRNA7. Three clonal hiPSC lines per subject (from UM and AP) were produced, while single clonal hiPSC lines derived from unrelated, unaffected male and female donors (UC-M and UC-F) were used as control subjects. hiPSCs were differentiated into cortical Excitatory Neurons (cExN), cortical Inhibitory interNeurons (cIN), and neural progenitor cells (cExNPCs and cINPCs), then combined at a 1:1 ratio to generate cortical neural organoids.
The authors observed disruptions of the correct cellular physiology and reduced neurodevelopment in the AP-derived model, while it was not observed in either the UM or unrelated healthy controls. Neuronal gene expression was dysregulated in the AP, including reduced expression of genes related to behavior, psychological disorders, neuritogenesis, neuronal migration, WNT pathway, axonal guidance, and GABA receptor signaling. This dysregulation influenced cellular functioning, resulting in increased neural progenitor proliferation, impaired neuronal differentiation, maturation, and migration, and increased endoplasmic reticulum (ER) stress. Both the neuronal migration deficit and elevated ER stress were selectively rescued by different pharmacologic agents. The UM model instead exhibited upregulated expression of genes in many of the same pathways, suggesting that molecular compensation could have contributed to the lack of neurodevelopmental phenotypes in this model. However, both AP- and UM-derived neurons exhibited shared alterations of neuronal function, including increased action-potential firing and elevated cholinergic activity, consistent with increased homomeric CHRNA7 channel activity (Meganathan et al., 2021).
This research presents certain limitations: in particular, despite the genotyping analysis, the expression of the CHRNA7 gene, and also of the chimeric gene, was not observed. Moreover, the electrophysiological analysis only evaluates an alteration in the flux activity when induced by Ach and choline, without using different allosteric modulators, which are essential for confirming that calcium influx depends on the α7 Receptor, excluding the participation of other nAChR members. The authors did not exclude two possible hypotheses: whether the observed alterations were directly associated with alterations of CHRNA7 activity or due to indirect mechanisms that might have affected the functionality of different nACh Receptors. Further studies will be necessary to clarify the role of CHRNA7 in these pathological phenotypes.
In 2021, Zhang and colleagues reprogrammed six fibroblast lines into hiPSC (three of the lines belonged to individuals with 15q13.3 microdeletions and the others belonged to healthy individuals), to study the effects of the 15q13.3 microdeletion on genome-wide gene expression, DNA methylation, chromatin accessibility, and sensitivity to cisplatin-induced DNA damage. In all three patients, the heterozygous 15q13.3 microdeletions were detected between breakpoints 3 and 4 (BP3/BP4). The iPSCs were differentiated into induced Neurons (iNs) using the neurogenin-2 induction method. To determine the effects of 15q13.3 microdeletions on gene expression of Chr15, they performed RNA-seq on iPSCs and iNs of 6 cell lines. Using Genome-wide analyses, they found 178 Differentially Expressed Genes (DEGs) in hiPSCs and 369 in iNs with 15q13.3 microdeletions. Some of the DEGs found in iNs are known to be involved in neuropsychiatric disorders, such as: CACNG3, SCN8A, SPATA5, and for intellectual disability. No enriched Gene Ontology (GO) terms were found in hiPSCs, while in iNs 46 significant Biological Process (GOBP) and 7 Molecular Function (GOMF) terms were identified. These enriched GO terms belong to different processes related to a neurodevelopmental disease such as WNT binding, ribosome biogenesis, and DNA repair and DNA binding. The researchers focused their attention on the DNA repair pathway, evaluating the cell survival of iNs in response to DNA damage induced by cisplatin. Cells carrying the 15q13.3 microdeletion showed a reduced survival rate compared with control iNs, demonstrating a high susceptibility to DNA damage, as previously observed in cortex derived from the murine model (Gordon et al., 2021). In addition, the authors identified: 1) A differential methylation state of protocadherins, which, modulating the WNT pathway, are implicated in various neurodevelopmental processes, including synapse assembly, neuronal differentiation, and neurotransmission. 2) reduced accessibility in the regulatory binding sites of the genes implicated in various neuropsychiatric disorders. 3) The consequences of the 15q13.3 microdeletion were not associated with the haploinsufficiency of a single gene but with the combination of various genes located in the CNV; this observation was possible using CRISPR/cas9 technique to generate cell lines carrying a homologous deletion of every single gene presents in the CNV. This could explain the variability of pathological phenotypes observed in individuals with different 15q13.3 CNVs (Zhang et al., 2021). Nevertheless, in order to correlate the pathological phenotype with the CNV and not with the patient’s background, Zhang and colleagues should have created the specific microdeletions by the CRISP/cas9 technique in iPSCs derived from healthy donors (Tai et al., 2016).
Thanks to these previous studies it has been possible to highlight many of the molecular mechanisms associated with 15q13.3 CNV: in particular, both in the presence of duplications or deletions, the cellular phenotype is comparable, characterized by a reduction of the calcium flux and its downstream effectors (Figure 4).
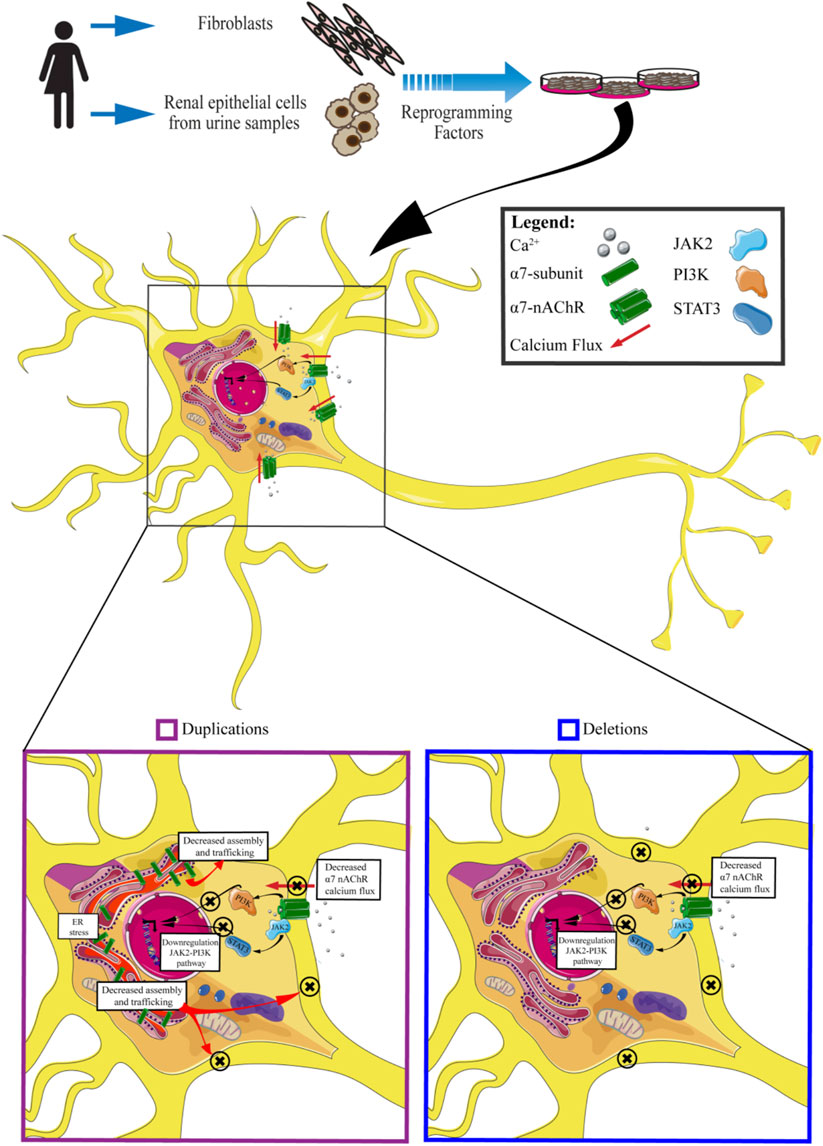
FIGURE 4. An insight into molecular effects of CNV 15q13.3. Cells carrying CNV duplications show decreased calcium flux associated with the α7 receptor, downregulation of JAK2-PI3K pathway, decreased assembly and trafficking of nAchRs, and ER stress. Cells carrying CNV deletions exhibit decreased α7nAchRs calcium flux and downregulation of JAK2-PI3K pathway.
Neuronal nAChR subtypes, such as a7, have been identified as promising targets for drug development in a variety of neurological and psychiatric disorders (Gill et al., 2013). Therefore, there has been and is a great interest from both academic laboratories and pharmaceutical companies to develop novel subtype-selective nAChR ligands (Hurst et al., 2013). For this reason, the identification of innovative cellular assays, those providing access to native human nAChRs, is an important discovery goal. The research carried out up to now has concentrated mainly on the utilization of receptor agonists and antagonists to explore the physiological behavior of α7 nAChR receptors in the presence and absence of the CNVs, using calcium flux as a readout of receptor functionality. As shown in the preceding paragraphs, the data are very interesting, but an accurate review of all of these published articles points to evidence that the perspective on developing drugs for treating these types of CNV needs to be revised in order to be able to address the mechanisms that influence α7 nAChR both upstream and downstream. Intervening upstream might mean modifying the transport of the receptor towards the membrane, reducing reticular stress, or also reducing the formation of heterodimers between CHRNA7 e CHRFAM7A subunits, while intervening downstream could involve modifying gene expression or epigenetic regulation that has been influenced by dysregulated calcium flux.
Thanks to the use of hiPSCs it will be possible to study and clarify the role of CHRNA7 and the contribution of the human fusion gene (CHRFAM7A) in neurological-neuropsychiatric diseases. Without the advent of iPSCs technology, these types of studies would have been unthinkable, due to the impossibility of collecting neural tissue from living patients. In this specific context, hiPSC have been an enormous benefit, not only for their capacity of differentiation or the maintenance of the patients’ genetic background but, above all, for the possibility of using pharmacological drugs, to understand both the functionality of the receptors and their response to drugs in physiological conditions, and in the presence of CNVs, to improve pathological functions (Table 1).
However, the phenotypic complexity and the variable penetrance associated with CNVs requires the definition of common guidelines to be followed by all researchers working with patients’ iPSC-derived cells so that the results, obtained in different laboratories, can be compared and above all, the information obtained from different experiments can be put together and analyzed globally. The findings so far published are extremely interesting, having highlighted the complexity of receptor functioning, but as can be seen in Figure 3, comparing the CNVs studied in the different articles, it is evident that it is impossible to have a complete and unified picture of the findings. When studies are carried out on deleted CNV, it is important to take in account if the truncation includes also other genes, because either the deregulation of their expression or function could influence the pathological phenotype, as some published articles have highlighted. Likewise, in the presence of duplication, it is necessary to verify the localization of it (where it starts and stops), whether it completely duplicates the gene or whether it is positioned casually inside the gene, provoking a fused protein that may be deregulated in the functionality. Putting all this information together would help researchers unify the data. Moreover, it is essential to consider the specific genetic background of the affected subjects, as well as the specific profile of healthy controls, evaluating the absence of any kind of alteration and defining the homo/heterozygosity of the CHRFAM7A locus and the presence or absence of CHRFAM7Abp. This is fundamental because studies carried out on the CHRNA7 receptor without considering the presence of CHRFAM7A would be incomplete, and potentially lead to erroneous conclusions. Without this information, subsequent evaluations of drug effects could encounter important criticalities because the results are influenced by the genetic background of the patient. Thus, using models with a specific genetic background is important not only for the molecular and functional study of the receptor but also for the transferability of the results into clinical practice, developing drugs that can act in accordance with the specific patients’ genetic profiles. For this purpose, genome editing will accelerate the understanding of the causal relationship between CNV and disease phenotype, as creating the same CNV in different genetic backgrounds, obviously characterized by the presence/absence of the main modifiers of the nicotinic receptor function, the contribution of additional risk alleles to a cell phenotype will be evident.
AMGG and GR supervised the writing of the paper, AS, EMT, GR, AC, and AD wrote sections of the paper, ALV and JR revised, read, and approved the submitted version.
This work was supported by the Italian Ministry of Health, Ricerca Corrente Program 2022-2024 and 5 × 1000 funds (to JR).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Araud, T., Graw, S., Berger, R., Lee, M., Neveu, E., Bertrand, D., et al. (2011). The chimeric gene CHRFAM7A, a partial duplication of the CHRNA7 gene, is a dominant negative regulator of α7*nAChR function Biochem. Pharmacol. 82 (8), 904–914. doi:10.1016/j.bcp.2011.06.018
Bacchelli, E., Battaglia, A., Cameli, C., Lomartire, S., Tancredi, R., Thomson, S., et al. (2015). Analysis of CHRNA7 rare variants in autism spectrum disorder susceptibility. Am. J. Med. Genet. A 167A (4), 715–723. doi:10.1002/ajmg.a.36847
Beal, J. C. (2014). Case report: Neuronal migration disorder associated with chromosome 15q13.3 duplication in a boy with autism and seizures. J. Child. Neurol29(12) 29, NP186–8. doi:10.1177/0883073813510356
Ben-Shachar, S., Lanpher, B., German, J. R., Qasaymeh, M., Potocki, L., Nagamani, S. C. S., et al. (2009). Microdeletion 15q13.3: A locus with incomplete penetrance for autism, mental retardation, and psychiatric disorders. J. Med. Genet. 46 (6), 382–388. doi:10.1136/jmg.2008.064378
Brown, M. E., Rondon, E., Rajesh, D., Mack, A., Lewis, R., Feng, X., et al. (2010). Derivation of induced pluripotent stem cells from human peripheral blood T lymphocytes. PLoS One 5 (6), e11373. doi:10.1371/journal.pone.0011373
Budisteanu, M., Papuc, S. M., Streata, I., Cucu, M., Pirvu, A., Serban-Sosoi, S., et al. (2021). The phenotypic spectrum of 15q13.3 region duplications: Report of 5 patients. Genes 1, 1025. doi:10.3390/genes12071025
Campbell, N. R., Fernandes, C. C., Halff, A. W., and Berg, D. K. (2010). Endogenous signaling through alpha7-containing nicotinic receptors promotes maturation and integration of adult-born neurons in the hippocampus. J. Neurosci. 30 (26), 8734–8744. doi:10.1523/JNEUROSCI.0931-10.2010
Chambers, S. M., Fasano, C. A., Papapetrou, E. P., Tomishima, M., Sadelain, M., and Studer, L. (2009). Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat. Biotechnol. 27 (3), 275–280. doi:10.1038/nbt.1529
Chan, T., Williams, E., Cohen, O., Eliceiri, B. P., Baird, A., and Costantini, T. W. (2019). CHRFAM7A alters binding to the neuronal alpha-7 nicotinic acetylcholine receptor Neurosci. Lett. 690, 126–131. doi:10.1016/j.neulet.2018.10.010
Chatzidaki, A., Fouillet, A., Li, J., Dage, J., Millar, N. S., Sher, E., et al. (2015). Pharmacological characterisation of nicotinic acetylcholine receptors expressed in human iPSC-derived neurons PLoS One 23, e0125116. doi:10.1371/journal.pone.0125116
Coe, B. P., Witherspoon, K., Rosenfeld, J. A., van Bon, B. W. M., Vulto-van Silfhout, A. T., Bosco, P., et al. (2014). Refining analyses of copy number variation identifies specific genes associated with developmental delay Nat. Genet. 46 (10), 1063–1071. doi:10.1038/ng.3092
DaCosta, C. J. B., Free, C. R., Corradi, J., Bouzat, C., and Sine, S. M. (2011). Single-channel and structural foundations of neuronal α7 acetylcholine receptor potentiation J. Neurosci. 31, 13870–13879. doi:10.1523/jneurosci.2652-11.2011
de Lucas-Cerrillo, A. M., Maldifassi, M. C., Arnalich, F., Renart, J., Atienza, G., Serantes, R., et al. (2011). Function of partially duplicated human α77 nicotinic receptor subunit CHRFAM7A gene: Potential implications for the cholinergic anti-inflammatory response. J. Biol. Chem. 286 (1), 594–606. doi:10.1074/jbc.M110.180067
Dimos, J. T., Rodolfa, K. T., Niakan, K. K., Weisenthal, L. M., Mitsumoto, H., Chung, W., et al. (2008). Induced pluripotent stem cells generated from patients with ALS can be differentiated into motor neurons. Science 321 (5893), 1218–1221. doi:10.1126/science.1158799
Dolmetsch, R., and Geschwind, D. H. (2011). The human brain in a dish: The promise of iPSC-derived neurons Cell 145 (6), 831–834. doi:10.1016/j.cell.2011.05.034
Fejgin, K., Nielsen, J., Birknow, M. R., Bastlund, J. F., Nielsen, V., Lauridsen, J. B., et al. (2014). A mouse model that recapitulates cardinal features of the 15q13.3 microdeletion syndrome including schizophrenia- and epilepsy-related alterations Biol. Psychiatry 76 (2), 128–137. doi:10.1016/j.biopsych.2013.08.014
Felix, R. A., Chavez, V. A., Novicio, D. M., Morley, B. J., and Portfors, C. V. (2019). Nicotinic acetylcholine receptor subunit α7-knockout mice exhibit degraded auditory temporal processing J. Neurophysiol. 122 (2), 451–465. doi:10.1152/jn.00170.2019
Flomen, R. H., Davies, A. F., Di Forti, M., La Cascia, C., Mackie-Ogilvie, C., Murray, R., et al. (2008). The copy number variant involving part of the alpha7 nicotinic receptor gene contains a polymorphic inversion Eur. J. Hum. Genet. 16 (11), 1364–1371. doi:10.1038/ejhg.2008.112
Gault, J., Robinson, M., Berger, R., Drebing, C., Logel, J., Hopkins, J., et al. (1998). Genomic organization and partial duplication of the human alpha7 neuronal nicotinic acetylcholine receptor gene (CHRNA7) Genomics 52 (2), 173–185. doi:10.1006/geno.1998.5363
Ghatak, S., Dolatabadi, N., Trudler, D., Zhang, X., Wu, Y., Mohata, M., et al. (2019). Mechanisms of hyperexcitability in Alzheimer's disease hiPSC-derived neurons and cerebral organoids vs isogenic controls Elife 29, e50333. doi:10.7554/eLife.50333
Gill, J. K., Chatzidaki, A., Ursu, D., Sher, E., and Millar, N. S. Contrasting properties of α7-selective orthosteric and allosteric agonists examined on native nicotinic acetylcholine receptors [internet]. 8, PLoS ONE. 2013, e55047. doi:10.1371/journal.pone.0055047
Gillentine, M. A. (2022). Exploring 15q13.3 copy number variants in iPSCs, Curr. Top. iPSCs Technol., 17, 333–360.
Gillentine, M. A., and Schaaf, C. P. (2015). The human clinical phenotypes of altered CHRNA7 copy number. Biochem. Pharmacol. 97 (4), 352–362. doi:10.1016/j.bcp.2015.06.012
Gillentine, M. A., Yin, J., Bajic, A., Zhang, P., Cummock, S., Kim, J. J., et al. (2017). Functional consequences of CHRNA7 copy-number alterations in induced pluripotent stem cells and neural progenitor cells Am. J. Hum. Genet. 101 (6), 874–887. doi:10.1016/j.ajhg.2017.09.024
Gonzalez, C., Armijo, E., Bravo-Alegria, J., Becerra-Calixto, A., Mays, C. E., and Soto, C. (2018). Modeling amyloid beta and tau pathology in human cerebral organoids. Mol. Psychiatry 23 (12), 2363–2374. doi:10.1038/s41380-018-0229-8
Gordon, A., Forsingdal, A., Klewe, I. V., Nielsen, J., Didriksen, M., Werge, T., et al. (2021). Transcriptomic networks implicate neuronal energetic abnormalities in three mouse models harboring autism and schizophrenia-associated mutations. Mol. Psychiatry 26 (5), 1520–1534. doi:10.1038/s41380-019-0576-0
Grskovic, M., Javaherian, A., Strulovici, B., and Daley, G. Q. (2011). Induced pluripotent stem cells–opportunities for disease modelling and drug discovery. Nat. Rev. Drug Discov. 10 (12), 915–929. doi:10.1038/nrd3577
Guy, B., Zhang, J. S., Duncan, L. H., and Johnston, R. J. (2021). Human neural organoids: Models for developmental neurobiology and disease Dev. Biol. 478, 102–121. doi:10.1016/j.ydbio.2021.06.012
Haase, A., Olmer, R., Schwanke, K., Wunderlich, S., Merkert, S., Hess, C., et al. (2009). Generation of induced pluripotent stem cells from human cord blood Cell Stem Cell 5 (4), 434–441. doi:10.1016/j.stem.2009.08.021
Halevi, S., Yassin, L., Eshel, M., Sala, F., Sala, S., Criado, M., et al. (2003). Conservation within the RIC-3 gene family. Effectors of mammalian nicotinic acetylcholine receptor expression. J. Biol. Chem. 278 (36), 34411–34417. doi:10.1074/jbc.M300170200
Hurst, R., Rollema, H., and Bertrand, D. (2013). Nicotinic acetylcholine receptors: From basic science to therapeutics. Pharmacol. Ther. 137 (1), 22–54. doi:10.1016/j.pharmthera.2012.08.012
Ihnatovych, I., Birkaya, B., Notari, E., and Szigeti, K. (2020). iPSC-derived microglia for modeling human-specific DAMP and PAMP responses in the context of Alzheimer's disease Int. J. Mol. Sci. 21, 9668. doi:10.3390/ijms21249668
Ihnatovych, I., Nayak, T. K., Ouf, A., Sule, N., Birkaya, B., Chaves, L., et al. (2019). iPSC model of CHRFAM7A effect on α7 nicotinic acetylcholine receptor function in the human context. Transl. Psychiatry 9 (1), 59. doi:10.1038/s41398-019-0375-z
Jiang, Y., Yuan, H., Huang, L., Hou, X., Zhou, R., and Dang, X. (2019). Global proteomic profiling of the uniquely human CHRFAM7A gene in transgenic mouse brain. Gene 25 (714), 143996. doi:10.1016/j.gene.2019.143996
Kim, H., Park, H. J., Choi, H., Chang, Y., Park, H., Shin, J., et al. (2019). Modeling G2019S-LRRK2 sporadic Parkinson’s disease in 3D midbrain organoids. Stem Cell Rep. 12 (3), 518–531. doi:10.1016/j.stemcr.2019.01.020
Lancaster, M. A., Renner, M., Martin, C. A., Wenzel, D., Bicknell, L. S., Hurles, M. E., et al. (2013). Cerebral organoids model human brain development and microcephaly. Nature 501 (7467), 373–379. doi:10.1038/nature12517
Lang, B., Pu, J., Hunter, I., Liu, M., Martin-Granados, C., Reilly, T. J., et al. (2014). Recurrent deletions of ULK4 in schizophrenia: A gene crucial for neuritogenesis and neuronal motility J. Cell Sci. 127, 630–640. doi:10.1242/jcs.137604
Larsen, H. M., Hansen, S. K., Mikkelsen, J. D., Hyttel, P., and Stummann, T. C. (2019). Alpha7 nicotinic acetylcholine receptors and neural network synaptic transmission in human induced pluripotent stem cell-derived neurons Stem Cell Res. 41, 101642. doi:10.1016/j.scr.2019.101642
Lewis, A. S., and Picciotto, M. R. (2013). High-affinity nicotinic acetylcholine receptor expression and trafficking abnormalities in psychiatric illness. Psychopharmacology 229 (3), 477–485. doi:10.1007/s00213-013-3126-5
Lewis, A. S., Pittenger, S. T., Mineur, Y. S., Stout, D., Smith, P. H., and Picciotto, M. R. (2018). Bidirectional regulation of aggression in mice by hippocampal alpha-7 nicotinic acetylcholine receptors Neuropsychopharmacology 43 (6), 1267–1275. doi:10.1038/npp.2017.276
Li, R., Sun, L., Fang, A., Li, P., Wu, Q., and Wang, X. (2017). Recapitulating cortical development with organoid culture in vitro and modeling abnormal spindle-like (ASPM related primary) microcephaly disease Protein Cell 8 (11), 823–833. doi:10.1007/s13238-017-0479-2
Li, Y., Muffat, J., Omer, A., Bosch, I., Lancaster, M. A., Sur, M., et al. (2017). Induction of expansion and folding in human cerebral organoids Cell Stem Cell 20 (3), 385–396. doi:10.1016/j.stem.2016.11.017
Lin, Y. T., Seo, J., Gao, F., Feldman, H. M., Wen, H. L., Penney, J., et al. (2018). APOE4 causes widespread molecular and cellular alterations associated with Alzheimer’s disease phenotypes in human iPSC-derived brain cell types. Neuron 27, 1141–1154. doi:10.1016/j.neuron.2018.05.008
Liu, H., Ye, Z., Kim, Y., Sharkis, S., and Jang, Y. Y. (2010). Generation of endoderm-derived human induced pluripotent stem cells from primary hepatocytes. Hepatology 51 (5), 1810–1819. doi:10.1002/hep.23626
Liu, Y., Hao, S., Yang, B., Fan, Y., Qin, X., Chen, Y., et al. (2017). Wnt/β-catenin signaling plays an essential role in α7 nicotinic receptor-mediated neuroprotection of dopaminergic neurons in a mouse Parkinson’s disease model. Biochem. Pharmacol. 140, 115–123. doi:10.1016/j.bcp.2017.05.017
Locke, D. P., Archidiacono, N., Misceo, D., Cardone, M. F., Deschamps, S., Roe, B., et al. (2003). Refinement of a chimpanzee pericentric inversion breakpoint to a segmental duplication cluster Genome Biol. 4 (8), R50. doi:10.1186/gb-2003-4-8-r50
Lowther, C., Costain, G., Stavropoulos, D. J., Melvin, R., Silversides, C. K., Andrade, D. M., et al. (2015). Delineating the 15q13.3 microdeletion phenotype: A case series and comprehensive review of the literature. Genet. Med. 17 (2), 149–157. doi:10.1038/gim.2014.83
Lupski, J. R., and Stankiewicz, P. (2005). Genomic disorders: Molecular mechanisms for rearrangements and conveyed phenotypes. PLoS Genet. 1 (6), e49. doi:10.1371/journal.pgen.0010049
Makoff, A. J., and Flomen, R. H. (2007). Detailed analysis of 15q11-q14 sequence corrects errors and gaps in the public access sequence to fully reveal large segmental duplications at breakpoints for Prader-Willi, Angelman, and inv dup(15) syndromes. Genome Biol. 8 (6), R114. doi:10.1186/gb-2007-8-6-r114
Marchetto, M. C. N., Carromeu, C., Acab, A., Yu, D., Yeo, G. W., Mu, Y., et al. (2010). A model for neural development and treatment of rett syndrome using human induced pluripotent stem cells. Cell. 143 (4), 527–539. doi:10.1016/j.cell.2010.10.016
Mariani, J., Coppola, G., Zhang, P., Abyzov, A., Provini, L., Tomasini, L., et al. (2015). FOXG1-Dependent dysregulation of GABA/glutamate neuron differentiation in autism spectrum disorders. Cell. 162 (2), 375–390. doi:10.1016/j.cell.2015.06.034
Marks, M. J., and Collins, A. C. (1982). Characterization of nicotine binding in mouse brain and comparison with the binding of alpha-bungarotoxin and quinuclidinyl benzilate. Mol. Pharmacol. 22 (3), 554–564.
Masurel-Paulet, A., Andrieux, J., Callier, P., Cuisset, J. M., Le Caignec, C., Holder, M., et al. (2010). Delineation of 15q13.3 microdeletions Clin. Genet. 78 (2), 149–161. doi:10.1111/j.1399-0004.2010.01374.x
Mefford, H. C., Rosenfeld, J. A., Shur, N., Slavotinek, A. M., Cox, V. A., Hennekam, R. C., et al. (2012). Further clinical and molecular delineation of the 15q24 microdeletion syndrome J. Med. Genet. 49 (2), 110–118. doi:10.1136/jmedgenet-2011-100499
Meganathan, K., Prakasam, R., Baldridge, D., Gontarz, P., Zhang, B., Urano, F., et al. (2021). Altered neuronal physiology, development, and function associated with a common chromosome 15 duplication involving CHRNA7 BMC Biol. 19 (1), 147. doi:10.1186/s12915-021-01080-7
Miller, D. T., Shen, Y., Weiss, L. A., Korn, J., Anselm, I., Bridgemohan, C., et al. (2009). Microdeletion/duplication at 15q13.2q13.3 among individuals with features of autism and other neuropsychiatric disorders. J. Med. Genet. 46, 242–248. doi:10.1136/jmg.2008.059907
Mucchietto, V., Fasoli, F., Pucci, S., Moretti, M., Benfante, R., Maroli, A., et al. (2018). α9- and α7-containing receptors mediate the pro-proliferative effects of nicotine in the A549 adenocarcinoma cell line. Br. J. Pharmacol. 175 (11), 1957–1972. doi:10.1111/bph.13954
Newman, S., Hermetz, K. E., Weckselblatt, B., and Rudd, M. K. (2015). Next-generation sequencing of duplication CNVs reveals that most are tandem and some create fusion genes at breakpoints Am. J. Hum. Genet. 96, 208–220. doi:10.1016/j.ajhg.2014.12.017
Nowakowska, B. (2017). Clinical interpretation of copy number variants in the human genome. J. Appl. Genet. 58 (4), 449–457. doi:10.1007/s13353-017-0407-4
Pagnamenta, A. T., Wing, K., Akha, E. S., Knight, S. J. L., Bölte, S., Schmötzer, G., et al. (2009). A 15q13.3 microdeletion segregating with autism. Eur. J. Hum. Genet. 17, 687–692. doi:10.1038/ejhg.2008.228
Pałczyńska, M. M., Jindrichova, M., Gibb, A. J., and Millar, N. S. (2012). Activation of α7 nicotinic receptors by orthosteric and allosteric agonists: Influence on single-channel kinetics and conductance Mol. Pharmacol. 82 (5), 910–917. doi:10.1124/mol.112.080259
Park, I. H., Arora, N., Huo, H., Maherali, N., Ahfeldt, T., Shimamura, A., et al. (2008). Disease-specific induced pluripotent stem cells Cell 134 (5), 877–886. doi:10.1016/j.cell.2008.07.041
Paşca, S. P., Portmann, T., Voineagu, I., Yazawa, M., Shcheglovitov, A., Paşca, A. M., et al. (2011). Using iPSC-derived neurons to uncover cellular phenotypes associated with Timothy syndrome. Nat. Med. 17 (12), 1657–1662. doi:10.1038/nm.2576
Patel, H., McIntire, J., Ryan, S., Dunah, A., and Loring, R. (2017). Anti-inflammatory effects of astroglial α7 nicotinic acetylcholine receptors are mediated by inhibition of the NF-κB pathway and activation of the Nrf2 pathway. J. Neuroinflammation 14 (1), 192. doi:10.1186/s12974-017-0967-6
Peng, W., Mao, L., and Dang, X. (2022). The emergence of the uniquely human α7 nicotinic acetylcholine receptor gene and its roles in inflammation Gene 842, 146777. doi:10.1016/j.gene.2022.146777
Pettigrew, K. A., Reeves, E., Leavett, R., Hayiou-Thomas, M. E., Sharma, A., Simpson, N. H., et al. (2015). Copy number variation screen identifies a rare de novo deletion at chromosome 15q13.1-13.3 in a child with language impairment. PLoS One 11, 10e0134997. doi:10.1371/journal.pone.0134997
Piao, Y., Hung, S. S. C., Lim, S. Y., Wong, R. C. B., and Ko, M. S. H. (2014). Efficient generation of integration-free human induced pluripotent stem cells from keratinocytes by simple transfection of episomal vectors. Stem Cells Transl. Med. 3 (7), 787–791. doi:10.5966/sctm.2013-0036
Pinto, D., Marshall, C., Feuk, L., and Scherer, S. W. (2007). Copy-number variation in control population cohorts. Hum. Mol. Genet. 15, 16. doi:10.1093/hmg/ddm241
Quik, M., Zhang, D., McGregor, M., and Bordia, T. (2015). Alpha7 nicotinic receptors as therapeutic targets for Parkinson's disease Biochem. Pharmacol. 97 (4), 399–407. doi:10.1016/j.bcp.2015.06.014
Raja, W. K., Mungenast, A. E., Lin, Y. T., Ko, T., Abdurrob, F., Seo, J., et al. (2016). Self-organizing 3D human neural tissue derived from induced pluripotent stem cells recapitulate Alzheimer's disease phenotypes PLoS One 13, 11e0161969. doi:10.1371/journal.pone.0161969
Redon, R., Ishikawa, S., Fitch, K. R., Feuk, L., Perry, G. H., Andrews, T. D., et al. (2006). Global variation in copy number in the human genome. Nature 444 (7118), 444–454. doi:10.1038/nature05329
Rehm, H. L., Berg, J. S., Brooks, L. D., Bustamante, C. D., Evans, J. P., Landrum, M. J., et al. (2015). ClinGen–the clinical genome resource. N. Engl. J. Med. 372 (23), 2235–2242. doi:10.1056/NEJMsr1406261
Riley, B., Williamson, M., Collier, D., Wilkie, H., and Makoff, A. A. (2002). A 3-Mb map of a large Segmental duplication overlapping the alpha7-nicotinic acetylcholine receptor gene (CHRNA7) at human 15q13-q14 Genomics 79 (2), 197–209. doi:10.1006/geno.2002.6694
Rosenfeld, J. A., Stephens, L. E., Coppinger, J., Ballif, B. C., Hoo, J. J., French, B. N., et al. (2011). Deletions flanked by breakpoints 3 and 4 on 15q13 may contribute to abnormal phenotypes. Eur. J. Hum. Genet. 19 (5), 547–554. doi:10.1038/ejhg.2010.237
Ruiz, S., Brennand, K., Panopoulos, A. D., Herrerías, A., Gage, F. H., and Izpisua-Belmonte, J. C. (2010). High-efficient generation of induced pluripotent stem cells from human astrocytes PLoS One 5 (12), e15526. doi:10.1371/journal.pone.0015526
Schaaf, C. P. (2014). Nicotinic acetylcholine receptors in human genetic disease. Genet. Med. 16 (9), 649–656. doi:10.1038/gim.2014.9
Schafer, S. T., Paquola, A. C. M., Stern, S., Gosselin, D., Ku, M., Pena, M., et al. (2019). Pathological priming causes developmental gene network heterochronicity in autistic subject-derived neurons. Nat. Neurosci. 22 (2), 243–255. doi:10.1038/s41593-018-0295-x
Seki, T., Yuasa, S., and Fukuda, K. (2011). Derivation of induced pluripotent stem cells from human peripheral circulating T cells. Curr. Protoc. Stem Cell Biol. 4, Unit4A.3. Unit4A.3. doi:10.1002/9780470151808.sc04a03s18
Severance, E. G., and Yolken, R. H. (2007). Lack of RIC-3 congruence with beta2 subunit-containing nicotinic acetylcholine receptors in bipolar disorder. Neuroscience 148 (2), 454–460. doi:10.1016/j.neuroscience.2007.06.008
Sharp, A. J., Locke, D. P., McGrath, S. D., Cheng, Z., Bailey, J. A., Vallente, R. U., et al. (2005). Segmental duplications and copy-number variation in the human genome. Am. J. Hum. Genet. 77 (1), 78–88. doi:10.1086/431652
Sharp, A. J., Mefford, H. C., Li, K., Baker, C., Skinner, C., Stevenson, R. E., et al. (2008). A recurrent 15q13.3 microdeletion syndrome associated with mental retardation and seizures. Nat. Genet. 40 (3), 322–328. doi:10.1038/ng.93
Shen, J. X., and Yakel, J. L. (2009). Nicotinic acetylcholine receptor-mediated calcium signaling in the nervous system. Acta Pharmacol. Sin. 30 (6), 673–680. doi:10.1038/aps.2009.64
Sinkus, M. L., Lee, M. J., Gault, J., Logel, J., Short, M., Freedman, R., et al. (2009). A 2-base pair deletion polymorphism in the partial duplication of the alpha7 nicotinic acetylcholine gene (CHRFAM7A) on chromosome 15q14 is associated with schizophrenia. Brain Res. 29, 1–11. doi:10.1016/j.brainres.2009.07.041
Smits, L. M., Reinhardt, L., Reinhardt, P., Glatza, M., Monzel, A. S., Stanslowsky, N., et al. (2019). Modeling Parkinson's disease in midbrain-like organoids NPJ Park. Dis. 5 (5), 5. doi:10.1038/s41531-019-0078-4
Soliman, M. A., Aboharb, F., Zeltner, N., and Studer, L. (2017). Pluripotent stem cells in neuropsychiatric disorders. Mol. Psychiatry 22 (9), 1241–1249. doi:10.1038/mp.2017.40
Song, B., Niclis, J. C., Alikhan, M. A., Sakkal, S., Sylvain, A., Kerr, P. G., et al. (2011). Generation of induced pluripotent stem cells from human kidney mesangial cells. J. Am. Soc. Nephrol. 22 (7), 1213–1220. doi:10.1681/ASN.2010101022
Srinivasan, R., Richards, C. I., Xiao, C., Rhee, D., Pantoja, R., Dougherty, D. A., et al. (2012). Pharmacological chaperoning of nicotinic acetylcholine receptors reduces the endoplasmic reticulum stress response. Mol. Pharmacol. 81 (6), 759–769. doi:10.1124/mol.112.077792
Stephens, S. H., Franks, A., Berger, R., Palionyte, M., Fingerlin, T. E., Wagner, B., et al. (2012). Multiple genes in the 15q13-q14 chromosomal region are associated with schizophrenia Psychiatr. Genet. 22 (1), 1–14. doi:10.1097/YPG.0b013e32834c0c33
Szafranski, P., Schaaf, C. P., Person, R. E., Gibson, I. B., Xia, Z., Mahadevan, S., et al. (2010). Structures and molecular mechanisms for common 15q13.3 microduplications involving CHRNA7: Benign or pathological? Hum mutat. Hum. Mutat. 31 (7), 840–850. doi:10.1002/humu.21284
Szatkiewicz, J. P., O’Dushlaine, C., Chen, G., Chambert, K., Moran, J. L., Neale, B. M., et al. (2014). Copy number variation in schizophrenia in Sweden. Mol. Psychiatry 19 (7), 762–773. doi:10.1038/mp.2014.40
Szigeti, K., Ihnatovych, I., Birkaya, B., Chen, Z., Ouf, A., Indurthi, D. C., et al. (2020). CHRFAM7A: A human specific fusion gene, accounts for the translational gap for cholinergic strategies in Alzheimer's disease EBioMedicine 59, 102892. doi:10.1016/j.ebiom.2020.102892
Tai, D. J. C., Ragavendran, A., Manavalan, P., Stortchevoi, A., Seabra, C. M., Erdin, S., et al. (2016). Engineering microdeletions and microduplications by targeting segmental duplications with CRISPR Nat. Neurosci. 19 (3), 517–522. doi:10.1038/nn.4235
Tuzun, E., Sharp, A. J., Bailey, J. A., Kaul, R., Morrison, V. A., Pertz, L. M., et al. (2005). Fine-scale structural variation of the human genome. Nat. Genet. 37 (7), 727–732. doi:10.1038/ng1562
Utikal, J., Maherali, N., Kulalert, W., and Hochedlinger, K. (2009). Sox2 is dispensable for the reprogramming of melanocytes and melanoma cells into induced pluripotent stem cells J. Cell Sci. 122, 3502–3510. doi:10.1242/jcs.054783
van Bon, B. W. M., Mefford, H. C., Menten, B., Koolen, D. A., Sharp, A. J., Nillesen, W. M., et al. (2009). Further delineation of the 15q13 microdeletion and duplication syndromes: A clinical spectrum varying from non-pathogenic to a severe outcome. J. Med. Genet. 46 (8), 511–523. doi:10.1136/jmg.2008.063412
Vermeesch, J. R., Brady, P. D., Sanlaville, D., Kok, K., and Hastings, R. J. (2012). Genome-wide arrays: Quality criteria and platforms to be used in routine diagnostics. Hum. Mutat. 33 (6), 906–915. doi:10.1002/humu.22076
Vijayaraghavan, S., Pugh, P. C., Zhang, Z. W., Rathouz, M. M., and Berg, D. K. (1992). Nicotinic receptors that bind alpha-bungarotoxin on neurons raise intracellular free Ca2+ Neuron 8 (2), 353–362. doi:10.1016/0896-6273(92)90301-s
Wang, H., Yu, M., Ochani, M., Amella, C. A., Tanovic, M., Susarla, S., et al. (2003). Nicotinic acetylcholine receptor alpha7 subunit is an essential regulator of inflammation. Nature 421 (6921), 384–388. doi:10.1038/nature01339
Wang, Y., Xiao, C., Indersmitten, T., Freedman, R., Leonard, S., and Lester, H. A. (2014). The duplicated α7 subunits assemble and form functional nicotinic receptors with the full-length α7 J. Biol. Chem. 289 (38), 26451–26463. doi:10.1074/jbc.M114.582858
Williams, D. K., Wang, J., and Papke, R. L. (2011). Investigation of the molecular mechanism of the α7 nicotinic acetylcholine receptor positive allosteric modulator PNU-120596 provides evidence for two distinct desensitized states. Mol. Pharmacol. 80 (6), 1013–1032. doi:10.1124/mol.111.074302
Williams, N. M., Franke, B., Mick, E., Anney, R. J. L., Freitag, C. M., Gill, M., et al. (2012). Genome-wide analysis of copy number variants in attention deficit hyperactivity disorder: The role of rare variants and duplications at 15q13.3 Am. J. Psychiatry 169, 195–204. doi:10.1176/appi.ajp.2011.11060822
Wiśniowiecka-Kowalnik, B., Kastory-Bronowska, M., Bartnik, M., Derwińska, K., Dymczak-Domini, W., Szumbarska, D., et al. (2013). Application of custom-designed oligonucleotide array CGH in 145 patients with autistic spectrum disorders. Eur. J. Hum. Genet. 21 (6), 620–625.
Ye, Z., Zhan, H., Mali, P., Dowey, S., Williams, D. M., Jang, Y. Y., et al. (2009). Human-induced pluripotent stem cells from blood cells of healthy donors and patients with acquired blood disorders. Blood 114 (27), 5473–5480. doi:10.1182/blood-2009-04-217406
Yin, J., Chen, W., Yang, H., Xue, M., and Schaaf, C. P. (2017). Chrna7 deficient mice manifest no consistent neuropsychiatric and behavioral phenotypes Sci. Rep. 3 (7), 39941. doi:10.1038/srep39941
Zarrei, M., MacDonald, J. R., Merico, D., and Scherer, S. W. (2015). A copy number variation map of the human genome. Nat. Rev. Genet. 16 (3), 172–183. doi:10.1038/nrg3871
Zhang, J., Xue, F., Whiteaker, P., Li, C., Wu, W., Shen, B., et al. (2011). Desensitization of alpha7 nicotinic receptor is governed by coupling strength relative to gate tightness J. Biol. Chem., 286, 25331–25340. doi:10.1074/jbc.m111.221754
Zhang, J. C., Yao, W., Ren, Q., Yang, C., Dong, C., Ma, M., et al. (2016). Depression-like phenotype by deletion of α7 nicotinic acetylcholine receptor: Role of BDNF-TrkB in nucleus accumbens. Sci. Rep. 8, 36705. doi:10.1038/srep36705
Zhang, S., Zhang, X., Purmann, C., Ma, S., Shrestha, A., Davis, K. N., et al. (2021). Network effects of the 15q13.3 microdeletion on the transcriptome and epigenome in human-induced neurons. Biol. Psychiatry 89 (5), 497–509. doi:10.1016/j.biopsych.2020.06.021
Zhang, W., Yang, S. L., Yang, M., Herrlinger, S., Shao, Q., Collar, J. L., et al. (2019). Modeling microcephaly with cerebral organoids reveals a WDR62-CEP170-KIF2A pathway promoting cilium disassembly in neural progenitors. Nat. Commun. 13, 2612. doi:10.1038/s41467-019-10497-2
Zhou, D., Gochman, P., Broadnax, D. D., Rapoport, J. L., and Ahn, K. (2016). 15q13.3 duplication in two patients with childhood-onset schizophrenia. Am. J. Med. Genet. B Neuropsychiatr. Genet. 171 (6), 777–783. doi:10.1002/ajmg.b.32439
Keywords: neurodevelopmental disorders, neuropsychiatric disorders, 15q11-13, CHRNA7, nicotinic acetylcholine receptor, copy number variation, CNV
Citation: Giovenale AMG, Ruotolo G, Soriano AA, Turco EM, Rotundo G, Casamassa A, D’Anzi A, Vescovi AL and Rosati J (2023) Deepening the understanding of CNVs on chromosome 15q11–13 by using hiPSCs: An overview. Front. Cell Dev. Biol. 10:1107881. doi: 10.3389/fcell.2022.1107881
Received: 25 November 2022; Accepted: 16 December 2022;
Published: 06 January 2023.
Edited by:
Gabriella Minchiotti, National Research Council (CNR), ItalyReviewed by:
Massimiliano Caiazzo, Utrecht University, NetherlandsCopyright © 2023 Giovenale, Ruotolo, Soriano, Turco, Rotundo, Casamassa, D’Anzi, Vescovi and Rosati. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jessica Rosati, ai5yb3NhdGlAY3NzLW1lbmRlbC5pdA==; Angelo Luigi Vescovi, YW5nZWxvLnZlc2NvdmlAdW5pbWliLml0
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.