
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cell Dev. Biol., 06 January 2023
Sec. Nuclear Organization and Dynamics
Volume 10 - 2022 | https://doi.org/10.3389/fcell.2022.1097446
This article is part of the Research TopicMolecular Architecture and Dynamics of Meiotic ChromosomesView all 16 articles
Meiocytes organize higher-order chromosome structures comprising arrays of chromatin loops organized at their bases by linear axes. As meiotic prophase progresses, the axes of homologous chromosomes align and synapse along their lengths to form ladder-like structures called synaptonemal complexes (SCs). The entire process of meiotic recombination, from initiation via programmed DNA double-strand breaks (DSBs) to completion of DSB repair with crossover or non-crossover outcomes, occurs in the context of chromosome axes and SCs. These meiosis-specific chromosome structures provide specialized environments for the regulation of DSB formation and crossing over. In this review, we summarize insights into the importance of chromosome architecture in the regulation of meiotic recombination, focusing on cohesin-mediated axis formation, DSB regulation via tethered loop-axis complexes, inter-homolog template bias facilitated by axial proteins, and crossover regulation in the context of the SCs. We also discuss emerging evidence that the SUMO and the ubiquitin-proteasome system function in the organization of chromosome structure and regulation of meiotic recombination.
Homologous recombination during meiosis underlies biological diversity and ensures proper chromosome segregation during the first division to create haploid gametes. During meiotic prophase-I, chromosomes develop highly organized three-dimensional structures where loops of chromatin emanate from structural axes that also interconnect sister chromatids. Programmed DNA double-strand breaks (DSBs) at recombination hotspots, which initiate meiotic recombination, are localized to DNA sequences found in chromatin loops while many factors responsible for DSB formation reside on the axes, indicating that tethering of DSB sites in loops to their corresponding chromosome axes–loop-axis tethering–is a crucial step in the initiation of meiotic recombination (Blat et al., 2002; Panizza et al., 2011). Following DSB formation, homolog search of the DSB ends for homologous chromosomes leads to pairing of the structural axes of two homologous chromosomes and synapsis along their lengths to form the synaptonemal complexes (SCs). The SCs are zipper-like structures where the lateral/axial elements localized to each homolog sandwich the central region composed of transverse filaments and a central element (Figure 1). Later steps of recombination such as the formation of double-Holliday junctions and their resolution into crossover products, occur within the context of the SCs. In this review, we present key findings about the regulation of meiotic recombination in relation to chromosome architecture.
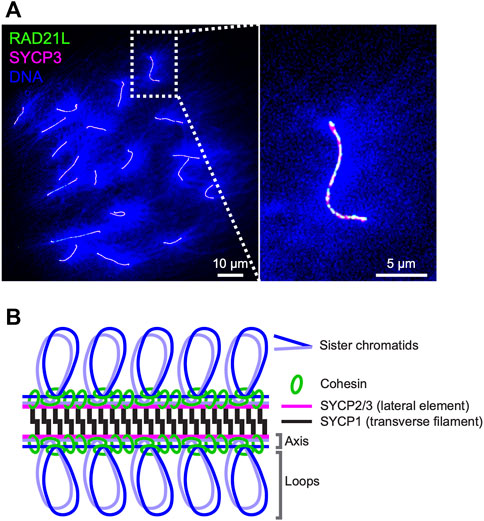
FIGURE 1. Axis-loop chromosome structure and the synaptonemal complex in mice (A) Surface spreads of mouse oocyte pachytene chromosomes immunostained for RAD21L (green), SYCP3 (magenta), and DNA (DAPI; blue). DNA is condensed on chromosome axes where cohesin complexes and axis core proteins localize and spread as loops from axes. RAD21L and SYCP3 are shown as a representative of meiotic cohesin and axis core protein, respectively (B) Schematic representation of the mouse synaptonemal complex. Cohesin complexes interconnect axes of sister chromatids and lateral elements SYCP2 and SYCP3 and a transverse filament protein SYCP1 form a ladder/zipper-like structure.
During the meiotic S-phase, cohesin complexes interconnect sister chromatids and are assumed to establish the core unit of chromosome axis via loop extrusion, likely with help of evolutionarily related axis core proteins (budding yeast Red1, mammalian SYCP2/SYCP3, and plant ASY3/ASY4; West et al., 2019; Figures 1A,B). The cohesin complexes consist of two SMCs (structure maintenance of chromosome), SMC1 and SMC3; and two non-SMC kleisin subunits, SCC3/STAG and the α-kleisin RAD21/SCC1 (Nasmyth and Haering, 2005). REC8 is a meiosis-specific α-kleisin subunit that is well-conserved from yeast to mammals and is required for the formation of chromosome axes and the SCs in budding yeast, C. elegans, and mice (Klein et al., 1999; Pasierbek et al., 2001; Xu et al., 2005). Recent Hi-C analysis of yeast meiosis revealed Rec8-dependent intra-chromosome interactions between distal chromosomal loci and high-frequency contacts between Rec8 binding sites (Muller et al., 2018; Schalbetter et al., 2019), supporting a model in which interactions between adjacent cohesin-binding sites assemble structural axes. Deletion of the budding yeast REC8 gene causes various defects in meiotic recombination; the redistribution and reduction of DSBs, impaired choice of recombination template, and persistence of joint molecule DNA intermediates (Kugou et al., 2009; Kim et al., 2010), indicating important roles of cohesin-mediated chromosome structures and/or the cohesin complexes themselves in the regulation of recombination. Recent work in fission yeast identified a rec8 separation-of-function mutant, rec8-F204S, that is proficient for sister chromatid cohesion (SCC) but deficient for axis-loop structure (Sakuno et al., 2022). This rec8 mutant was defective in meiotic recombination, revealing an essential role for Rec8-cohesin-mediated axis-loop chromosome structure and not cohesion per se in meiotic recombination.
In mice, the topologically associating domains (TADs, comprising ∼1 Mbp-intra-chromosomal interactions), characteristic of interphase chromosomes, are diminished and intra-chromosomal interactions around 2.5–4.5 Mbp became more evident during meiotic prophase-I, consistent with the formation of axis-loop structures (Alavattam et al., 2019; Wang Y. et al., 2019; Patel et al., 2019; Vara et al., 2019; Luo et al., 2020; Zuo et al., 2021). REC8, SMC1β, STAG3, and RAD21L (a second meiosis-specific α-kleisin; Figure 1A) are known meiosis-specific cohesin subunits that localize to chromosome axes in mice as six distinct complexes; three SMC1β-cohesin complexes (RAD21-SMC1β-SMC3-STAG3, RAD21L-SMC1β-SMC3-STAG3, and REC8-SMC1β-SMC3-STAG3) and three SMC1α-cohesin complexes (RAD21-SMC1α-SMC3-STAG1/2, RAD21-SMC1α-SMC3-STAG3, and RAD21L-SMC1α-SMC3-STAG3) (Revenkova et al., 2004; Ishiguro et al., 2011; Lee and Hirano, 2011; Fukuda et al., 2014). With the exception of Rad21L−/− females, all mice that are knockout mutants for the meiosis-specific cohesin components are sterile, and show defects in synapsis and compromised meiotic recombination (Revenkova et al., 2004; Herran et al., 2011; Llano et al., 2012; Fukuda et al., 2014). Axis lengths in meiocytes are shorter in all mutants, and double mutant mice such as Smc1β−/− Rec8−/− show much shorter axis lengths than the corresponding single mutants (Biswas et al., 2016; Ward et al., 2016). These observations highlight the importance of the multiple cohesin complexes in the organization of meiotic chromosome axis structure in mice.
During meiosis, cohesin plays a dual role in sister chromatid cohesion (SCC) and the formation of axis-loop structure. A recent series of studies established the loop extrusion activity of SMC complexes including the mitotic SCC1/RAD21-based cohesin (RAD21-SMC1A-SMC3-STAG1), which requires the cohesin loader complex SCC2/NIPBL-SCC4/MAU2 (Davidson et al., 2019; Kim et al., 2019; Kaur et al., 2022). This loop extrusion seems to be distinct from cohesin’s SCC activity (Davidson and Peters, 2021). It follows that meiotic chromosome structure and cohesion may be mediated by two independent ensembles of cohesin complexes. Importantly, several organisms including vertebrates and nematodes contain two distinct meiotic cohesins (Severson et al., 2009; Ishiguro et al., 2011; Lee and Hirano, 2011; Severson and Meyer, 2014). In mice, the cohesins with REC8 and RAD21L localize to non-overlapping sites along chromosome axes (Ishiguro et al., 2011). Moreover, REC8, and thus REC8-based cohesin, localizes to the chromosomes as early as meiotic S-phase and persists until metaphase-II; whereas RAD21L-cohesin appears on the chromosome later, in leptonema and disappears earlier in late prophase-I (Herran et al., 2011; Lee and Hirano, 2011; Ishiguro et al., 2014; Biswas et al., 2016). One simple idea is that REC8-cohesin functions for SCC and RAD21L-cohesin functions for loop extrusion and thus axis-loop formation. Future studies are essential to evaluate the hypothesis.
WAPL and PDS5 are highly conserved cohesin regulators that contribute to the association and dissociation of cohesin complexes from chromosomes, and thereby modulate chromosome architecture in somatic cells (Kueng et al., 2006; Tedeschi et al., 2013; Haarhuis et al., 2017; Wutz et al., 2017). In C. elegans, cytological analysis of wapl-1 null mutants indicated minor defects in the repair of meiotic DSBs (Crawley et al., 2016). Physical analysis of meiotic recombination at a well-characterized DSB hotspot in budding yeast revealed a subtle reduction in the levels of meiotic DSBs and the homolog bias of DSB repair in rad61/wpl1 deletion mutants (Challa et al., 2016; Hong et al., 2019). More severe defects were seen in pds5 meiotic null mutants with an interhomolog bias defect similar to that of a rec8 deletion mutant (Hong et al., 2019). Both rad61/wpl1 and pds5 mutants showed shortened chromosome axes in budding yeast (Challa et al., 2016; Yang et al., 2022) and in fission yeast (Ding et al., 2006; Sakuno et al., 2022). Importantly, the budding yeast pds5 mutant forms SCs between sister chromatids instead of homologs (Jin et al., 2009), which is reminiscent of the phenotypes seen in mouse Rec8, Rad21L, Stag3, and Smc1β knockout mutant spermatocytes (Xu et al., 2005; Ishiguro et al., 2011; Llano et al., 2012; Agostinho et al., 2016). Recent studies also revealed that depletion of PDS5 (both PDS5A and PDS5B) in mice leads to shortened chromosome axes, which form normal SCs between homologs, but are compromised for meiotic recombination (Viera et al., 2020). The prophase-I phenotypes of Wapl mutant mice have not been reported yet.
Notably, budding yeast Pds5 interacts with the proteasome and the shortened chromosome axis length of pds5 mutants is rescued by reducing levels of ubiquitin, suggesting that Pds5 regulates axis length via the ubiquitin-proteasome system (Yang et al., 2022). Consistently, the proteasome is indeed localized on chromosome axes in budding yeast, C. elegans, and mice (Ahuja et al., 2017; Rao et al., 2017). Although changes in chromosome structures resulting from mutation of PDS5 might indirectly affect meiotic recombination in mice, physical interaction between PDS5 and two RAD51 mediators, BRCA2 and the SWS1-SWSAP1, has been reported. Moreover, DSB repair is defective in PDS5 mutant somatic cells from fly and human (Brough et al., 2012; Kusch, 2015; Couturier et al., 2016; Martino et al., 2019). These data support more direct roles for PDS5 in meiotic recombination, either as a component of cohesin or as an independent complex.
Meiotic recombination is initiated by programmed DSBs formed via an evolutionarily conserved topoisomerase VI-like protein, Spo11, and its partners (Bergerat et al., 1997; Keeney et al., 1997; de Massy, 2013; Robert et al., 2016). DSB sites are located in chromatin loops while Spo11 partners such as Rec114-Mer2-Mei4 in budding yeast, Rec7-Rec15-Rec24 in fission yeast, and REC114-IHO1-MEI4 in mice localize to chromosome axes where cohesin also localizes, suggesting that tethered loop-axis complexes (TLACs) form during the initiation of meiotic recombination to regulate both DSB formation and the ensuing steps of meiotic recombination (Blat et al., 2002; Kumar et al., 2010; Panizza et al., 2011; Miyoshi et al., 2012; Fowler et al., 2013; Ito et al., 2014; Kumar et al., 2015; Stanzione et al., 2016); (Figure 2).
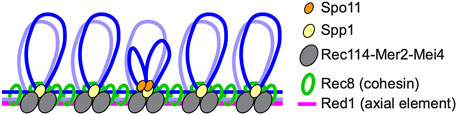
FIGURE 2. Tethered loop-axis complex (TLAC) formation to regulate DSB formation in budding yeast. Schematic representation of the budding yeast TLAC. Spo11 partner Rec114-Mer2-Mei4 complex localizes to chromosome axes where Rec8 cohesin and an axial element Red1 reside, and Spp1, a component of the Set1/COMPASS complex, tethers Spo11-bound DSB hotspots within loops to chromosome axes via the interaction with Mer2.
Molecular mechanisms of TLAC formation have been studied in yeasts. Spp1 in budding yeast and Mde2 in fission yeast are identified as proteins important for the formation of TLACs (Miyoshi et al., 2012; Acquaviva et al., 2013; Sommermeyer et al., 2013; Adam et al., 2018). In budding yeast, DSB hotspots are preferentially located in promoter regions within chromatin loops (Pan et al., 2011; Ito et al., 2014). Spp1, a component of the COMPASS/Set1 complex that catalyzes histone H3K4 trimethylation, is thought to recognize H3K4 trimethylation marks around DSB hotspots via its PHD domain, and connect these sites to chromosome axes by interacting with axis-associated Mer2. Spp1 is likely to mediate TLAC formation independently from the role in the COMPASS/Set1 complex (Karanyi et al., 2018). Although Spp1-mediated TLACs contribute to DSB formation, meiotic cells are equipped with another layer of regulation for meiotic DSB formation, since spp1 mutants still form relatively high levels of DSBs (Acquaviva et al., 2013; Sommermeyer et al., 2013; Zhang et al., 2020). Given that Mer2 itself has an ability to directly bind to nucleosomes and the association of Mer2 to chromosome axes is regulated by its interacting axis-associated protein Hop1, the Hop1-Mer2 may contribute to TLAC formation both via and independently of Spp1 (Panizza et al., 2011; Rousova et al., 2021). In fission yeast, where most DSB hotspots are in long intergenic regions (Fowler et al., 2014), DSB hotspots are marked by another epigenetic mark, H3K9 acetylation, and the H3K4 trimethylation mark is dispensable for meiotic DSB formation (Yamada et al., 2013). Mde2 expresses only after the meiotic S-phase and is thought to bridge Rec12Spo11-containing subcomplex at DSB hotspots and an axis-located subcomplex containing Rec15Mer2 (budding yeast homologs in superscript) (Miyoshi et al., 2012). Importantly, fission yeast Hop1 also physically interacts with Rec15Mer2 and promotes chromosomal localization of Rec15Mer2, suggesting significant contribution of Hop1 to TLAC formation in both yeasts (Kariyazono et al., 2019).
Whether or not the mechanism of TLAC formation is conserved remains unclear. In mice, PRDM9, a germ cell-specific H3K4 trimethylation transferase with a zinc-finger array domain, recognizes specific DNA sequences, deposits H3K4me3 and H3K36me3 marks, and directs DSB formation at its binding sites (Baudat et al., 2010; Diagouraga et al., 2018). Recent ChIP-seq analysis for meiotic cohesin components REC8 and RAD21L revealed their localization to promoter regions (Vara et al., 2019) and no overlap of meiotic cohesin binding sites with DMC1 (the meiosis-specific RAD51 homolog) and PRDM9 binding sites (Jin et al., 2021). CXXC1 is an ortholog of budding yeast Spp1, and the physical interaction of CXXC1 with PRDM9 and IHO1, an axis-associated protein considered to be the ortholog of budding yeast Mer2, suggested a similar mechanism of TLAC formation between budding yeast and mouse (Imai et al., 2017; Parvanov et al., 2017). However, depletion of CXXC1 in mouse germ cells caused no or small defects in DSB formation and the early steps of DSB repair (Tian et al., 2018; Jiang et al., 2020), suggesting that factor(s) other than CXXC1 plays a critical role in TLAC formation and meiotic DSB formation in mice. A mammalian ortholog of fission yeast Mde2 has not been identified yet.
DSB formation is followed by nuclease-mediated 5′-strand resection to form long single-stranded tails. Invasion of the resected DSB end into a template homologous duplex DNA forms a nascent D (displacement)-loop structure. At this stage, D-loop intermediates are thought to differentiate into crossover and non-crossover pathways (Hunter, 2015). The majority are matured as non-crossovers via DNA synthesis to extend the invading end, dissociation of the D-loop, and annealing of the displaced strand to seal the DSB (synthesis-dependent strand annealing) (Allers and Lichten, 2001; Hunter and Kleckner, 2001). Along the crossover pathways, D-loops differentiate into metastable D-loops called Single-End Invasions (SEIs) which then form double-Holliday junctions (dHJs) via DNA synthesis and capture of the second DSB end. dHJs are specifically resolved into crossover products. These events also occur in the context of meiotic chromosome axes and SCs. A prominent feature of meiotic recombination is that homology search and strand exchange are biased to occur between homologous chromosomes (inter-homolog) rather than between sister chromatids (inter-sister). This biased template choice is regulated by components of the axial/lateral elements of the SC and axis-associated proteins.
In budding yeast deletion mutants of axis-associated proteins Red1, Hop1, and the associated recombination-checkpoint kinase Mek1, DSBs are repaired primarily via inter-sister recombination (Kim et al., 2010; Lao and Hunter, 2010). The Hop1-Red1-Mek1 pathway, along with other factors that promote inter-homolog recombination (Zierhut et al., 2004), may mediate inter-homolog bias by inhibiting inter-sister recombination, promoting inter-homolog recombination, and/or by impeding the progression of recombination until homologs have been engaged (Lao and Hunter, 2010). Further mutant analysis suggested that meiotic cohesin Rec8 promotes inter-sister bias, which is counteracted by Red1 and Mek1/Mre4 (Kim et al., 2010). Mek1 is a meiosis-specific, axis-associated kinase that phosphorylates various targets including Rad54 and Hed1. The phosphorylation of both Rad54 and Hed1 suppresses Rad51-mediated inter-sister recombination, which partly explains the involvement of Mek1 in the suppression of inter-sister recombination (Niu et al., 2007; Niu et al., 2009; Callender et al., 2016; Kniewel et al., 2017). Importantly, the meiotic Rad51 homolog, Dmc1, bears an ability to promote inter-homolog bias (Brown and Bishop, 2014). However, the exact mechanism of inter-homolog bias and the relationship between Mek1-mediated phosphorylation and Rec8-cohesin remain to be resolved.
Hop1 is a conserved HORMA domain-containing protein that specifically localizes to unsynapsed axes and is locally depleted from sites of synapsis (Smith and Roeder, 1997), distinct from its binding partner Red1 and the cohesin complexes that appear to be constitutive components of chromosome axes before and after SC formation. Removal of Hop1 from synapsed axes is mediated by an evolutionarily conserved AAA+ ATPase Pch2, and yeast pch2Δ mutants show increased inter-sister recombination, suggesting that Pch2 also contributes to inter-homolog bias via the Hop1-Red1-Mek1 axis (Borner et al., 2008; Zanders et al., 2011). In mice, the two HORMA domain-containing proteins HORMAD1 and HORMAD2 also preferentially localize to unsynapsed axes (Wojtasz et al., 2009). In the absence of the HORMADs, the repair of radiation-induced exogenous DSBs was accelerated in Spo11-and Dmc1-deficient meiocytes in which inter-sister recombination is preferred, suggesting that, like budding yeast Hop1, mouse HORMADs may impede inter-sister recombination (Shin et al., 2013; Rinaldi et al., 2017; Carofiglio et al., 2018). The removal of HORMADs from synapsed axes is mediated by the Pch2 homolog TRIP13 (Wojtasz et al., 2009; Roig et al., 2010; Ye et al., 2017). In Trip13 mutant meiocytes, unrepaired DSBs persist (Li and Schimenti, 2007; Roig et al., 2010; Rinaldi et al., 2017), supporting the idea that HORMADs suppress inter-sister DSB repair.
Synaptonemal Complexes (SCs) are tripartite protein structures where the two lateral/axial elements of homologous chromosomes are connected along their lengths by a central region comprising tightly-packed transverse filaments and a central element. The dependency of SC formation on DSBs and recombination differs among species, with recombination-dependent synapsis in most analyzed fungi, plants, and mammals where SC formation tends to initiate at sites of recombination (SC also initiates at centromeres in budding yeast). By contrast, DSBs are dispensable for the SC formation in Drosophila and C. elegans in which synapsis initiates at centromeres and terminal pairing centers, respectively (MacQueen et al., 2005; Takeo et al., 2011). Despite these differences, SCs have a common function in the formation and/or regulation of crossing over in all organisms (with known exceptions being Schizosaccharomyces pombe and Aspergillus nidulans that have no typical SC structure).
The ZMM proteins are a group of meiosis-specific proteins that facilitate crossing over by promoting/stabilizing the crossover-pathway joint-molecule intermediates, SEIs and dHJs, and promoting the crossover-specific resolution of dHJs via MutLγ. Initially identified in budding yeast, the ZMMs comprise eight members that define five structures or activities: Zip1SYCP1 is the transverse filament components of SCs but also functions locally at recombination sites; Zip2SHOC1-Spo16 is related to XPF-ERCC1 and thought to bind and stabilize recombination intermediates; Zip4TEX11 is a long TPR-repeat protein that appears to bridge chromosome axes and recombination complexes by forming the ZZS complex with Zip2SHOC1-Spo16; Zip3RNF212 is inferred to be an E3-ligase for SUMO modification that promotes the localization of other ZMMs to recombination sites; Msh4-Msh5 (MutSγ), homologous to DNA mismatch-repair factor MutS, binds and stabilizes joint molecules; and Mer3HFM1 is a DNA helicase that stabilizes joint molecules and regulates the length of recombination-associated DNA synthesis (mammalian homologs in superscript) (Lynn et al., 2007; De Muyt et al., 2018; Arora and Corbett, 2019). In budding yeast, all ZMM proteins are also required for SC formation, with Zip2-Spo16-Zip4 and Zip3 being defined as synapsis initiation complexes (SICs) that assemble at synapsis initiation sites, which mature into crossover sites, indicating a close link between SC initiation and crossing over at least in budding yeast and similarly in Sordaria macrospora (Chua and Roeder, 1998; Agarwal and Roeder, 2000; Borner et al., 2004; Fung et al., 2004; Tsubouchi et al., 2006; Shinohara et al., 2008; Zhang et al., 2014a). In mice, the number of ZMM-associated recombination sites, detected as cytological foci, is in large excess relative to SC-initiation sites and crossovers. Meiocytes from mouse zmm knockouts for Hfm1, Msh4, Msh5, Shoc1, Spo16, and Tex11 show defects in synapsis and crossover formation, as seen in budding yeast. The exception is mouse knockout mutant for the ZIP3 homolog Rnf212, in which synapsis occurs efficiently but crossing over fails (de Vries et al., 1999; Kneitz et al., 2000; Yang et al., 2008; Guiraldelli et al., 2013; Reynolds et al., 2013; Zhang Q. et al., 2018; Guiraldelli et al., 2018; Zhang et al., 2019).
Like the budding yeast zip1Δ mutant, knockout mutation of components of the SC central region, SYCP1, SYCE1, SYCE2, SYCE3, TEX12, and SIX6OS1, in mice abolishes both synapsis and crossing over (de Vries et al., 2005; Bolcun-Filas et al., 2007; Hamer et al., 2008; Bolcun-Filas et al., 2009; Schramm et al., 2011; Gomez et al., 2016). In C. elegans, mutation of components of the SC central region (SYP-1, SYP-2, SYP-3, and SYP-4) also causes a severe reduction or loss of crossovers (MacQueen et al., 2002; Colaiacovo et al., 2003; Smolikov et al., 2007a; Smolikov et al., 2007b; Smolikov et al., 2009), indicating a coupling between SC formation and crossing over in most organisms. A notable exception is Arabidopsis thaliana, in which meiocytes lacking the SC central element ZYP1 are defective for synapsis but form elevated numbers of crossovers (Capilla-Perez et al., 2021). Similarly, the absence of the central element proteins Ecm11 and Gmc2 in budding yeast causes defective SC formation but increased crossing over (Voelkel-Meiman et al., 2016; Lee et al., 2021). These observations suggest that full synapsis and the SC central region are not essential for crossing over per se, but may function to control a proper number of crossovers.
Despite the close link between SC formation and crossing over in most species, uncoupling of the two events is implicated in a meiosis-specific depletion mutant of a component of SCF (Skp1-Cullin-F box) E3 ubiquitin ligase, Cdc53. The budding yeast cdc53 mutant is largely proficient in crossover formation, but is severely defective for the elongation of SCs and shows the abnormal accumulation of ZMM proteins (Zhu et al., 2021). Moreover, when Cdc53 depletion is combined with the pch2 deletion mutation, lacking the AAA+ ATPase that removes Hop1HORMAD1 from synapsed axes, the formation of full-length SCs is restored, but now DSB repair and crossing over are stalled. This uncoupling is unexpected since most yeast mutants defective for meiotic DSB repair also impair SC elongation. A possible explanation is that SCF is part of a regulatory surveillance mechanism that couples SC elongation and DSB repair in meiotic cells.
Crossovers, in concert with cohesion between sister chromatids, create connections between homologs called chiasmata that enable stable bipolar orientation of homologs on the meiosis-I spindle and consequently accurate disjunction at the first meiotic division. The number and position of crossovers, and thus chiasmata, are strictly controlled: each pair of homologous chromosomes (a bivalent) obtains at least one crossover (the obligate crossover or crossover assurance) and when multiple crossovers form between a bivalent they are evenly spaced (crossover interference). Crossover homeostasis can maintain crossover numbers at the expense of non-crossovers to buffer against variation in DSB numbers and inter-homolog bias (Martini et al., 2006; Cole et al., 2012; Lao et al., 2013). In addition, the phenomenon of crossover covariation describes the observation that within individual nuclei, crossover frequencies covary across different chromosomes, which may have adaptative advantages by balancing the cost-benefit ratio of crossing over (Wang S. et al., 2019). The precise mechanisms of these crossover control processes remain unresolved.
In budding yeast, crossover interference has been analyzed genetically by analyzing the segregation patterns of linked gene alleles and spore autonomous fluorescent makers in tetrads (Cao et al., 1990; Sym and Roeder, 1994; Shinohara et al., 2003; Thacker et al., 2011; Lao et al., 2013); and in prophase-I nuclei by analyzing the distribution of crossover-specific Zip2 and Zip3 immunostaining foci along SCs (Fung et al., 2004; Zhang et al., 2014b). Zip3 foci are evenly spaced, implying the establishment of interference patterning at or before the time of Zip3 loading, which depends on DSB formation (Zhang et al., 2014b). Mutant analysis revealed that the SUMO-targeted ubiquitin ligase (STUbL), Slx5/8 and SUMOylation of Top2 and axis protein Red1 are required for crossover interference (Zhang et al., 2014c). These and other observations support the proposal of Kleckner and colleagues that crossover interference is mediated by the imposition and relief of mechanical stress along meiotic chromosome axes (the beam-film model; Kleckner et al., 2004; Zhang et al., 2014b).
ZHP-3 is a C. elegans RING-domain protein related to Zip3 and is essential for crossover formation (Jantsch et al., 2004). ZHP-3 functions with three paralogs (ZHP-1,2,4) inferred to act as two heterodimeric complexes ZHP-1/2 and ZHP-3/4 (Zhang L. et al., 2018). ZHP-3 localizes along SCs in two phases; first as multiple foci along each SC before becoming restricted to a single crossover-specific focus in late pachynema (Bhalla et al., 2008). In C. elegans, robust crossover assurance and absolute interference ensures that each pair of homologous chromosomes obtains exactly one crossover. In vivo imaging using Fluorescence Recovery After Photobleaching (FRAP) technology revealed the dynamic properties of the SC central region and a switch from a dynamic to a stable state as pachytene progresses, the timing of which coincides with crossover designation (Pattabiraman et al., 2017). Other in vivo imaging studies support the idea that the SC has liquid crystalline properties, suggesting that the diffusion of the ZHP complexes within the SC might govern crossover patterning via a diffusion-mediated or coarsening or condensation process (Rog et al., 2017; Stauffer et al., 2019; Zhang et al., 2021).
Diffusion-mediated coarsening as a mechanism for crossover patterning is also suggested from analysis in Arabidopsis. Both plants and Sordaria encode a sole RING-domain crossover factor called HEI10 (without Zip3RNF212 orthologs). The localization pattern of HEI10 is also dynamic: forming multiple discrete foci along the SCs in early pachynema, which then reduce in number until most foci have disappeared while a few sites accumulate HEI10 and mature into crossover sites marked by MutLγ (Chelysheva et al., 2012; Wang et al., 2012; De Muyt et al., 2014). Analysis of HEI10-focus patterning in several different contexts via super-resolution structure-illumination microscopy (SIM) imaging of fixed cells combined with modeling by computational simulation is compatible with diffusion-mediated coarsening of HEI10 foci as a mechanism for crossover patterning (Morgan et al., 2021).
Mammals encode both Zip3 homolog RNF212 and HEI10, both of which are essential for crossover regulation in mice (Ward et al., 2007; Strong and Schimenti, 2010; Reynolds et al., 2013; Qiao et al., 2014). RNF212 shows dynamic localization along SCs similar to that of HEI10 in Arabidopsis and Sordaria, forming numerous discrete foci during early pachynema, which become restricted to crossover sites as pachytene progresses. By contrast, mouse HEI10 localizes only to crossover sites during mid-late pachynema and is not detected along SCs at earlier stages (Figure 3). It is suggested that RNF212-dependent SUMOylation stabilizes ZMM factors to confer crossover-competency to recombination sites, and HEI10-dependent ubiquitination subsequently licenses crossover/non-crossover differentiation by recruiting proteasomes to SCs to degrade as yet unknown factors (Rao et al., 2017). Importantly, the dosage of Rnf212 and Hei10 affects crossover rate in humans and mice, as seen for Arabidopsis Hei10 (Kong et al., 2008; Chowdhury et al., 2009; Fledel-Alon et al., 2011; Kong et al., 2014; Ziolkowski et al., 2017). This similarity in the dosage effect on crossover numbers is consistent with the possibility that crossover patterning in mammals may also involve the diffusion-mediated accumulation of RNF212 and HEI10 at crossover sites.
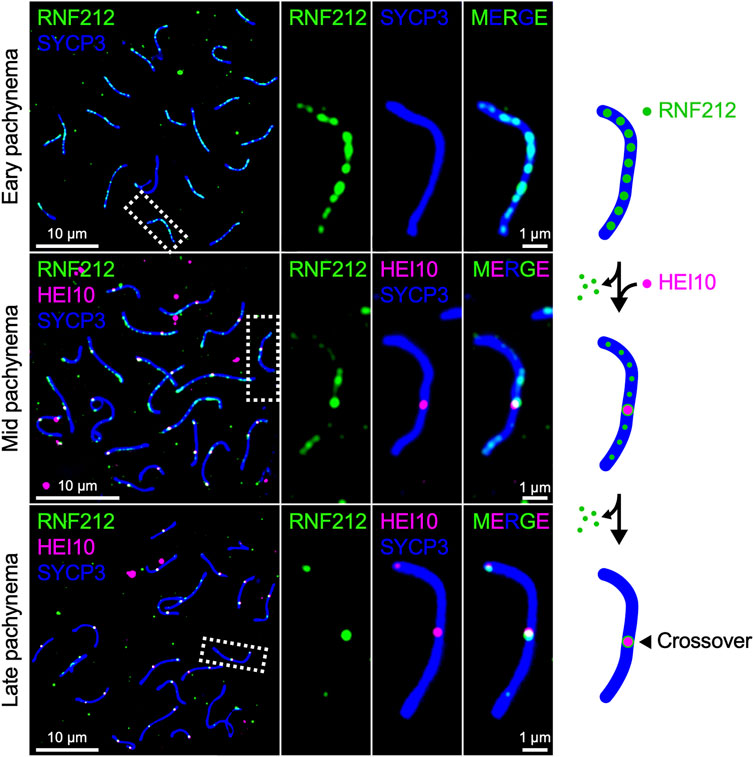
FIGURE 3. Chromosomal localization of RNF212 and HEI10 in mice. Successive stages of mouse pachytene spermatocytes immunostained for RNF212 (green), HEI10 (magenta), and SYCP3 (blue), HEI10 and SYCP3. RNF212 forms numerous discrete foci along the entire SCs (marked by SYCP3) in early pachynema before HEI10 foci emerge (top), loses most of foci but accumulates at HEI10-bound crossover sites in mid pachynema (middle), and eventually is restricted to crossover sites in late pachynema (bottom).
Meiotic chromosomes organize into specialized structures that help strictly regulate the number and position of meiotic DSBs, the choice of recombination template, and the differentiation of crossovers and non-crossovers to ultimately ensure the completion of DSB repair and accurate chromosome segregation. A diversity of approaches and model species are providing major insights into this molecular basis of the chromosome structure-recombination interface. However, major questions still remain to be addressed, including: Do cohesins and associated factors have direct functions in the regulation of meiotic recombination? Which factor(s) are responsible for TLAC formation in other organisms than yeasts, and how is TLAC formation coupled to DSB formation? How is inter-homolog bias established? What mechanisms underlie crossover patterning in mammals in which both Zip3/RNF212-family and HEI10-family RING-domain proteins are present? Recently, structural analysis of axis core proteins, Hop1/HORMADs, DSB proteins and associated proteins, and SC components is providing mechanistic insights into their functions (West et al., 2018; Boekhout et al., 2019; West et al., 2019; Sanchez-Saez et al., 2020; Claeys Bouuaert et al., 2021; Dunce et al., 2021; Rousova et al., 2021; Nore et al., 2022). Further mutant analysis based on protein structure will be a key to answer these unaddressed questions.
As presented above, SUMO, ubiquitin, and proteasome are involved in the regulation of chromosome axis length and crossover interference in budding yeast, and presumptive SUMO and ubiquitin ligases, RNF212 and HEI10, are essential for crossover regulation in mice, highlighting central roles for the SUMO and ubiquitin-proteasome systems in meiotic chromosome organization and the regulation of meiotic recombination. Indeed, SUMO is enriched on chromosome axes and SCs in budding yeast, Sordaria, mice, and humans, and ubiquitin and proteasome have been localized to chromosome axes in budding yeast, C. elegans, and mice (Voelkel-Meiman et al., 2013; Klug et al., 2013; Brown et al., 2008; De Muyt et al., 2014; Ahuja et al., 2017; Rao et al., 2017; Figure 4). Numerous meiotic factors, including cohesin and recombination proteins, undergo SUMOylation in budding yeast (Bhagwat et al., 2021), and the SCF ubiquitin ligase, which regulates SC elongation in conjunction to Pch2TRIP13 in budding yeast (Zhu et al., 2021), localizes to synapsed chromosome axes and targets HORMAD1 in mouse (Guan et al., 2020; Guan et al., 2022). Future analysis will further elucidate molecular roles of SUMO and the ubiquitin-proteasome system in the regulation of meiotic recombination in conjunction with chromosome architecture.
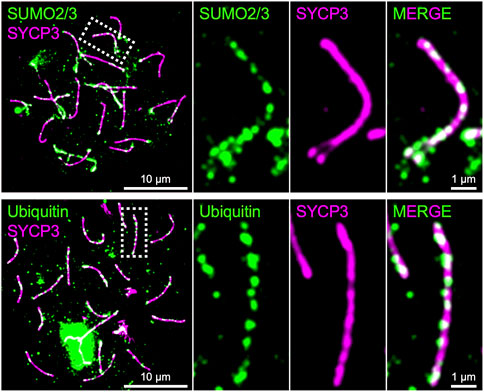
FIGURE 4. Chromosome axes decorated by SUMO and Ubiquitin in mice. Surface spreads of mouse spermatocyte chromosomes immunostained for SYCP3 (green) and SUMO2/3 (green; top) and Ubiquitin (green; bottom), respectively. Both SUMO and ubiquitin are enriched on chromosome axes where SYCP3 localizes.
Both authors wrote the manuscript and approved it for publication.
This study was supported by JSPS KAKENHI Grant Number 22125001 and 22125002 to AS and 20K15716 to MI.
We thank Neil Hunter for his insightful comments on the manuscript. We apologize for those whose work could not be cited due to the focused aspect of this study.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acquaviva, L., Szekvolgyi, L., Dichtl, B., Dichtl, B. S., de La Roche Saint Andre, C., Nicolas, A., et al. (2013). The COMPASS subunit Spp1 links histone methylation to initiation of meiotic recombination. Science 339, 215–218. doi:10.1126/science.1225739
Adam, C., Guerois, R., Citarella, A., Verardi, L., Adolphe, F., Beneut, C., et al. (2018). The PHD finger protein Spp1 has distinct functions in the Set1 and the meiotic DSB formation complexes. PLoS Genet. 14, e1007223. doi:10.1371/journal.pgen.1007223
Agarwal, S., and Roeder, G. S. (2000). Zip3 provides a link between recombination enzymes and synaptonemal complex proteins. Cell 102, 245–255. doi:10.1016/s0092-8674(00)00029-5
Agostinho, A., Manneberg, O., van Schendel, R., Hernandez-Hernandez, A., Kouznetsova, A., Blom, H., et al. (2016). High density of REC8 constrains sister chromatid axes and prevents illegitimate synaptonemal complex formation. EMBO Rep. 17, 901–913. doi:10.15252/embr.201642030
Ahuja, J. S., Sandhu, R., Mainpal, R., Lawson, C., Henley, H., Hunt, P. A., et al. (2017). Control of meiotic pairing and recombination by chromosomally tethered 26S proteasome. Science 355, 408–411. doi:10.1126/science.aaf4778
Alavattam, K. G., Maezawa, S., Sakashita, A., Khoury, H., Barski, A., Kaplan, N., et al. (2019). Attenuated chromatin compartmentalization in meiosis and its maturation in sperm development. Nat. Struct. Mol. Biol. 26, 175–184. doi:10.1038/s41594-019-0189-y
Allers, T., and Lichten, M. (2001). Differential timing and control of noncrossover and crossover recombination during meiosis. Cell 106, 47–57. doi:10.1016/s0092-8674(01)00416-0
Arora, K., and Corbett, K. D. (2019). The conserved XPF:ERCC1-like Zip2:Spo16 complex controls meiotic crossover formation through structure-specific DNA binding. Nucleic Acids Res. 47, 2365–2376. doi:10.1093/nar/gky1273
Baudat, F., Buard, J., Grey, C., Fledel-Alon, A., Ober, C., Przeworski, M., et al. (2010). PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science 327, 836–840. doi:10.1126/science.1183439
Bergerat, A., de Massy, B., Gadelle, D., Varoutas, P. C., Nicolas, A., Forterre, P., et al. (1997). An atypical topoisomerase II from Archaea with implications for meiotic recombination. Nature 386, 414–417. doi:10.1038/386414a0
Bhagwat, N. R., Owens, S. N., Ito, M., Boinapalli, J. V., Poa, P., Ditzel, A., et al. (2021). SUMO is a pervasive regulator of meiosis. Elife 10, e57720. doi:10.7554/eLife.57720
Bhalla, N., Wynne, D. J., Jantsch, V., and Dernburg, A. F. (2008). ZHP-3 acts at crossovers to couple meiotic recombination with synaptonemal complex disassembly and bivalent formation in C. elegans. PLoS Genet. 4, e1000235. doi:10.1371/journal.pgen.1000235
Biswas, U., Hempel, K., Llano, E., Pendas, A., and Jessberger, R. (2016). Distinct roles of meiosis-specific cohesin complexes in mammalian spermatogenesis. PLoS Genet. 12, e1006389. doi:10.1371/journal.pgen.1006389
Blat, Y., Protacio, R. U., Hunter, N., and Kleckner, N. (2002). Physical and functional interactions among basic chromosome organizational features govern early steps of meiotic chiasma formation. Cell 111, 791–802. doi:10.1016/s0092-8674(02)01167-4
Boekhout, M., Karasu, M. E., Wang, J., Acquaviva, L., Pratto, F., Brick, K., et al. (2019). REC114 partner ANKRD31 controls number, timing, and location of meiotic DNA breaks. Mol. Cell 74, 1053–1068. doi:10.1016/j.molcel.2019.03.023
Bolcun-Filas, E., Costa, Y., Speed, R., Taggart, M., Benavente, R., De Rooij, D. G., et al. (2007). SYCE2 is required for synaptonemal complex assembly, double strand break repair, and homologous recombination. J. Cell Biol. 176, 741–747. doi:10.1083/jcb.200610027
Bolcun-Filas, E., Hall, E., Speed, R., Taggart, M., Grey, C., de Massy, B., et al. (2009). Mutation of the mouse Syce1 gene disrupts synapsis and suggests a link between synaptonemal complex structural components and DNA repair. PLoS Genet. 5, e1000393. doi:10.1371/journal.pgen.1000393
Borner, G. V., Barot, A., and Kleckner, N. (2008). Yeast Pch2 promotes domainal axis organization, timely recombination progression, and arrest of defective recombinosomes during meiosis. Proc. Natl. Acad. Sci. U. S. A. 105, 3327–3332. doi:10.1073/pnas.0711864105
Borner, G. V., Kleckner, N., and Hunter, N. (2004). Crossover/noncrossover differentiation, synaptonemal complex formation, and regulatory surveillance at the leptotene/zygotene transition of meiosis. Cell 117, 29–45. doi:10.1016/s0092-8674(04)00292-2
Brough, R., Bajrami, I., Vatcheva, R., Natrajan, R., Reis-Filho, J. S., Lord, C. J., et al. (2012). APRIN is a cell cycle specific BRCA2-interacting protein required for genome integrity and a predictor of outcome after chemotherapy in breast cancer. EMBO J. 31, 1160–1176. doi:10.1038/emboj.2011.490
Brown, M. S., and Bishop, D. K. (2014). DNA strand exchange and RecA homologs in meiosis. Cold Spring Harb. Perspect. Biol. 7, a016659. doi:10.1101/cshperspect.a016659
Brown, P. W., Hwang, K., Schlegel, P. N., and Morris, P. L. (2008). Small ubiquitin-related modifier (SUMO)-1, SUMO-2/3 and SUMOylation are involved with centromeric heterochromatin of chromosomes 9 and 1 and proteins of the synaptonemal complex during meiosis in men. Hum. Reprod. 23, 2850–2857. doi:10.1093/humrep/den300
Callender, T. L., Laureau, R., Wan, L., Chen, X., Sandhu, R., Laljee, S., et al. (2016). Mek1 down regulates Rad51 activity during yeast meiosis by phosphorylation of Hed1. PLoS Genet. 12, e1006226. doi:10.1371/journal.pgen.1006226
Cao, L., Alani, E., and Kleckner, N. (1990). A pathway for generation and processing of double-strand breaks during meiotic recombination in S. cerevisiae. Cell 61, 1089–1101. doi:10.1016/0092-8674(90)90072-m
Capilla-Perez, L., Durand, S., Hurel, A., Lian, Q., Chambon, A., Taochy, C., et al. (2021). The synaptonemal complex imposes crossover interference and heterochiasmy in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 118, e2023613118. doi:10.1073/pnas.2023613118
Carofiglio, F., Sleddens-Linkels, E., Wassenaar, E., Inagaki, A., van Cappellen, W. A., Grootegoed, J. A., et al. (2018). Repair of exogenous DNA double-strand breaks promotes chromosome synapsis in SPO11-mutant mouse meiocytes, and is altered in the absence of HORMAD1. DNA Repair (Amst) 63, 25–38. doi:10.1016/j.dnarep.2018.01.007
Challa, K., Lee, M. S., Shinohara, M., Kim, K. P., and Shinohara, A. (2016). Rad61/Wpl1 (Wapl), a cohesin regulator, controls chromosome compaction during meiosis. Nucleic Acids Res. 44, 3190–3203. doi:10.1093/nar/gkw034
Chelysheva, L., Vezon, D., Chambon, A., Gendrot, G., Pereira, L., Lemhemdi, A., et al. (2012). The Arabidopsis HEI10 is a new ZMM protein related to Zip3. PLoS Genet. 8, e1002799. doi:10.1371/journal.pgen.1002799
Chowdhury, R., Bois, P. R., Feingold, E., Sherman, S. L., and Cheung, V. G. (2009). Genetic analysis of variation in human meiotic recombination. PLoS Genet. 5, e1000648. doi:10.1371/journal.pgen.1000648
Chua, P. R., and Roeder, G. S. (1998). Zip2, a meiosis-specific protein required for the initiation of chromosome synapsis. Cell 93, 349–359. doi:10.1016/s0092-8674(00)81164-2
Claeys Bouuaert, C., Pu, S., Wang, J., Oger, C., Daccache, D., Xie, W., et al. (2021). DNA-driven condensation assembles the meiotic DNA break machinery. Nature 592, 144–149. doi:10.1038/s41586-021-03374-w
Colaiacovo, M. P., MacQueen, A. J., Martinez-Perez, E., McDonald, K., Adamo, A., La Volpe, A., et al. (2003). Synaptonemal complex assembly in C. elegans is dispensable for loading strand-exchange proteins but critical for proper completion of recombination. Dev. Cell 5, 463–474. doi:10.1016/s1534-5807(03)00232-6
Cole, F., Kauppi, L., Lange, J., Roig, I., Wang, R., Keeney, S., et al. (2012). Homeostatic control of recombination is implemented progressively in mouse meiosis. Nat. Cell Biol. 14, 424–430. doi:10.1038/ncb2451
Couturier, A. M., Fleury, H., Patenaude, A. M., Bentley, V. L., Rodrigue, A., Coulombe, Y., et al. (2016). Roles for APRIN (PDS5B) in homologous recombination and in ovarian cancer prediction. Nucleic Acids Res. 44, 10879–10897. doi:10.1093/nar/gkw921
Crawley, O., Barroso, C., Testori, S., Ferrandiz, N., Silva, N., Castellano-Pozo, M., et al. (2016). Cohesin-interacting protein WAPL-1 regulates meiotic chromosome structure and cohesion by antagonizing specific cohesin complexes. Elife 5, e10851. doi:10.7554/eLife.10851
Davidson, I. F., Bauer, B., Goetz, D., Tang, W., Wutz, G., and Peters, J. M. (2019). DNA loop extrusion by human cohesin. Science 366, 1338–1345. doi:10.1126/science.aaz3418
Davidson, I. F., and Peters, J. M. (2021). Genome folding through loop extrusion by SMC complexes. Nat. Rev. Mol. Cell Biol. 22, 445–464. doi:10.1038/s41580-021-00349-7
de Massy, B. (2013). Initiation of meiotic recombination: How and where? Conservation and specificities among eukaryotes. Annu. Rev. Genet. 47, 563–599. doi:10.1146/annurev-genet-110711-155423
De Muyt, A., Pyatnitskaya, A., Andreani, J., Ranjha, L., Ramus, C., Laureau, R., et al. (2018). A meiotic XPF-ERCC1-like complex recognizes joint molecule recombination intermediates to promote crossover formation. Genes Dev. 32, 283–296. doi:10.1101/gad.308510.117
De Muyt, A., Zhang, L., Piolot, T., Kleckner, N., Espagne, E., and Zickler, D. (2014). E3 ligase Hei10: A multifaceted structure-based signaling molecule with roles within and beyond meiosis. Genes Dev. 28, 1111–1123. doi:10.1101/gad.240408.114
de Vries, F. A., de Boer, E., van den Bosch, M., Baarends, W. M., Ooms, M., Yuan, L., et al. (2005). Mouse Sycp1 functions in synaptonemal complex assembly, meiotic recombination, and XY body formation. Genes Dev. 19, 1376–1389. doi:10.1101/gad.329705
de Vries, S. S., Baart, E. B., Dekker, M., Siezen, A., de Rooij, D. G., de Boer, P., et al. (1999). Mouse MutS-like protein Msh5 is required for proper chromosome synapsis in male and female meiosis. Genes Dev. 13, 523–531. doi:10.1101/gad.13.5.523
Diagouraga, B., Clement, J. A. J., Duret, L., Kadlec, J., de Massy, B., and Baudat, F. (2018). PRDM9 methyltransferase activity is essential for meiotic DNA double-strand break formation at its binding sites. Mol. Cell 69, 853–865. doi:10.1016/j.molcel.2018.01.033
Ding, D. Q., Sakurai, N., Katou, Y., Itoh, T., Shirahige, K., Haraguchi, T., et al. (2006). Meiotic cohesins modulate chromosome compaction during meiotic prophase in fission yeast. J. Cell Biol. 174, 499–508. doi:10.1083/jcb.200605074
Dunce, J. M., Salmon, L. J., and Davies, O. R. (2021). Structural basis of meiotic chromosome synaptic elongation through hierarchical fibrous assembly of SYCE2-TEX12. Nat. Struct. Mol. Biol. 28, 681–693. doi:10.1038/s41594-021-00636-z
Fledel-Alon, A., Leffler, E. M., Guan, Y., Stephens, M., Coop, G., and Przeworski, M. (2011). Variation in human recombination rates and its genetic determinants. PLoS One 6, e20321. doi:10.1371/journal.pone.0020321
Fowler, K. R., Gutierrez-Velasco, S., Martin-Castellanos, C., and Smith, G. R. (2013). Protein determinants of meiotic DNA break hot spots. Mol. Cell 49, 983–996. doi:10.1016/j.molcel.2013.01.008
Fowler, K. R., Sasaki, M., Milman, N., Keeney, S., and Smith, G. R. (2014). Evolutionarily diverse determinants of meiotic DNA break and recombination landscapes across the genome. Genome Res. 24, 1650–1664. doi:10.1101/gr.172122.114
Fukuda, T., Fukuda, N., Agostinho, A., Hernandez-Hernandez, A., Kouznetsova, A., and Hoog, C. (2014). STAG3-mediated stabilization of REC8 cohesin complexes promotes chromosome synapsis during meiosis. EMBO J. 33, 1243–1255. doi:10.1002/embj.201387329
Fung, J. C., Rockmill, B., Odell, M., and Roeder, G. S. (2004). Imposition of crossover interference through the nonrandom distribution of synapsis initiation complexes. Cell 116, 795–802. doi:10.1016/s0092-8674(04)00249-1
Gomez, H. L., Felipe-Medina, N., Sanchez-Martin, M., Davies, O. R., Ramos, I., Garcia-Tunon, I., et al. (2016). C14ORF39/SIX6OS1 is a constituent of the synaptonemal complex and is essential for mouse fertility. Nat. Commun. 7, 13298. doi:10.1038/ncomms13298
Guan, Y., Leu, N. A., Ma, J., Chmatal, L., Ruthel, G., Bloom, J. C., et al. (2020). SKP1 drives the prophase I to metaphase I transition during male meiosis. Sci. Adv. 6, eaaz2129. doi:10.1126/sciadv.aaz2129
Guan, Y., Lin, H., Leu, N. A., Ruthel, G., Fuchs, S. Y., Busino, L., et al. (2022). SCF ubiquitin E3 ligase regulates DNA double-strand breaks in early meiotic recombination. Nucleic Acids Res. 50, 5129–5144. doi:10.1093/nar/gkac304
Guiraldelli, M. F., Eyster, C., Wilkerson, J. L., Dresser, M. E., and Pezza, R. J. (2013). Mouse HFM1/Mer3 is required for crossover formation and complete synapsis of homologous chromosomes during meiosis. PLoS Genet. 9, e1003383. doi:10.1371/journal.pgen.1003383
Guiraldelli, M. F., Felberg, A., Almeida, L. P., Parikh, A., de Castro, R. O., and Pezza, R. J. (2018). SHOC1 is a ERCC4-(HhH)2-like protein, integral to the formation of crossover recombination intermediates during mammalian meiosis. PLoS Genet. 14, e1007381. doi:10.1371/journal.pgen.1007381
Haarhuis, J. H. I., van der Weide, R. H., Blomen, V. A., Yanez-Cuna, J. O., Amendola, M., van Ruiten, M. S., et al. (2017). The cohesin release factor WAPL restricts chromatin loop extension. Cell 169, 693–707. doi:10.1016/j.cell.2017.04.013
Hamer, G., Wang, H., Bolcun-Filas, E., Cooke, H. J., Benavente, R., and Hoog, C. (2008). Progression of meiotic recombination requires structural maturation of the central element of the synaptonemal complex. J. Cell Sci. 121, 2445–2451. doi:10.1242/jcs.033233
Herran, Y., Gutierrez-Caballero, C., Sanchez-Martin, M., Hernandez, T., Viera, A., Barbero, J. L., et al. (2011). The cohesin subunit RAD21L functions in meiotic synapsis and exhibits sexual dimorphism in fertility. EMBO J. 30, 3091–3105. doi:10.1038/emboj.2011.222
Hong, S., Joo, J. H., Yun, H., Kleckner, N., and Kim, K. P. (2019). Recruitment of Rec8, Pds5 and Rad61/Wapl to meiotic homolog pairing, recombination, axis formation and S-phase. Nucleic Acids Res. 47, 11691–11708. doi:10.1093/nar/gkz903
Hunter, N., and Kleckner, N. (2001). The single-end invasion: An asymmetric intermediate at the double-strand break to double-holliday junction transition of meiotic recombination. Cell 106, 59–70. doi:10.1016/s0092-8674(01)00430-5
Hunter, N. (2015). Meiotic recombination: The essence of heredity. Cold Spring Harb. Perspect. Biol. 7, a016618. doi:10.1101/cshperspect.a016618
Imai, Y., Baudat, F., Taillepierre, M., Stanzione, M., Toth, A., and de Massy, B. (2017). The PRDM9 KRAB domain is required for meiosis and involved in protein interactions. Chromosoma 126, 681–695. doi:10.1007/s00412-017-0631-z
Ishiguro, K., Kim, J., Fujiyama-Nakamura, S., Kato, S., and Watanabe, Y. (2011). A new meiosis-specific cohesin complex implicated in the cohesin code for homologous pairing. EMBO Rep. 12, 267–275. doi:10.1038/embor.2011.2
Ishiguro, K., Kim, J., Shibuya, H., Hernandez-Hernandez, A., Suzuki, A., Fukagawa, T., et al. (2014). Meiosis-specific cohesin mediates homolog recognition in mouse spermatocytes. Genes Dev. 28, 594–607. doi:10.1101/gad.237313.113
Ito, M., Kugou, K., Fawcett, J. A., Mura, S., Ikeda, S., Innan, H., et al. (2014). Meiotic recombination cold spots in chromosomal cohesion sites. Genes cells. 19, 359–373. doi:10.1111/gtc.12138
Jantsch, V., Pasierbek, P., Mueller, M. M., Schweizer, D., Jantsch, M., and Loidl, J. (2004). Targeted gene knockout reveals a role in meiotic recombination for ZHP-3, a Zip3-related protein in Caenorhabditis elegans. Mol. Cell Biol. 24, 7998–8006. doi:10.1128/MCB.24.18.7998-8006.2004
Jiang, Y., Zhang, H. Y., Lin, Z., Zhu, Y. Z., Yu, C., Sha, Q. Q., et al. (2020). CXXC finger protein 1-mediated histone H3 lysine-4 trimethylation is essential for proper meiotic crossover formation in mice. Development 147, dev183764. doi:10.1242/dev.183764
Jin, H., Guacci, V., and Yu, H. G. (2009). Pds5 is required for homologue pairing and inhibits synapsis of sister chromatids during yeast meiosis. J. Cell Biol. 186, 713–725. doi:10.1083/jcb.200810107
Jin, X., Fudenberg, G., and Pollard, K. S. (2021). Genome-wide variability in recombination activity is associated with meiotic chromatin organization. Genome Res. 31, 1561–1572. doi:10.1101/gr.275358.121
Karanyi, Z., Halasz, L., Acquaviva, L., Jonas, D., Hetey, S., Boros-Olah, B., et al. (2018). Nuclear dynamics of the Set1C subunit Spp1 prepares meiotic recombination sites for break formation. J. Cell Biol. 217, 3398–3415. doi:10.1083/jcb.201712122
Kariyazono, R., Oda, A., Yamada, T., and Ohta, K. (2019). Conserved HORMA domain-containing protein Hop1 stabilizes interaction between proteins of meiotic DNA break hotspots and chromosome axis. Nucleic Acids Res. 47, 10166–10180. doi:10.1093/nar/gkz754
Kaur, P., Shi, Z., Lu, X., Zhang, H., Finkelstein, I. J., Tao, Y. J., et al. 2022. DNA capture and loop extrusion dynamics by cohesin-NIPBL. bioRxiv[preprint]. doi:10.1101/2022.08.18.504320
Keeney, S., Giroux, C. N., and Kleckner, N. (1997). Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88, 375–384. doi:10.1016/s0092-8674(00)81876-0
Kim, K. P., Weiner, B. M., Zhang, L., Jordan, A., Dekker, J., and Kleckner, N. (2010). Sister cohesion and structural axis components mediate homolog bias of meiotic recombination. Cell 143, 924–937. doi:10.1016/j.cell.2010.11.015
Kim, Y., Shi, Z., Zhang, H., Finkelstein, I. J., and Yu, H. (2019). Human cohesin compacts DNA by loop extrusion. Science 366, 1345–1349. doi:10.1126/science.aaz4475
Kleckner, N., Zickler, D., Jones, G. H., Dekker, J., Padmore, R., Henle, J., et al. (2004). A mechanical basis for chromosome function. Proc. Natl. Acad. Sci. U. S. A. 101, 12592–12597. doi:10.1073/pnas.0402724101
Klein, F., Mahr, P., Galova, M., Buonomo, S. B., Michaelis, C., Nairz, K., et al. (1999). A central role for cohesins in sister chromatid cohesion, formation of axial elements, and recombination during yeast meiosis. Cell 98, 91–103. doi:10.1016/S0092-8674(00)80609-1
Klug, H., Xaver, M., Chaugule, V. K., Koidl, S., Mittler, G., Klein, F., et al. (2013). Ubc9 sumoylation controls SUMO chain formation and meiotic synapsis in Saccharomyces cerevisiae. Mol. Cell 50, 625–636. doi:10.1016/j.molcel.2013.03.027
Kneitz, B., Cohen, P. E., Avdievich, E., Zhu, L., Kane, M. F., Hou, H., et al. (2000). MutS homolog 4 localization to meiotic chromosomes is required for chromosome pairing during meiosis in male and female mice. Genes Dev. 14, 1085–1097. doi:10.1101/gad.14.9.1085
Kniewel, R., Murakami, H., Liu, Y., Ito, M., Ohta, K., Hollingsworth, N. M., et al. (2017). Histone H3 threonine 11 phosphorylation is catalyzed directly by the meiosis-specific kinase Mek1 and provides a molecular readout of Mek1 activity in vivo. Genetics 207, 1313–1333. doi:10.1534/genetics.117.300359
Kong, A., Thorleifsson, G., Frigge, M. L., Masson, G., Gudbjartsson, D. F., Villemoes, R., et al. (2014). Common and low-frequency variants associated with genome-wide recombination rate. Nat. Genet. 46, 11–16. doi:10.1038/ng.2833
Kong, A., Thorleifsson, G., Stefansson, H., Masson, G., Helgason, A., Gudbjartsson, D. F., et al. (2008). Sequence variants in the RNF212 gene associate with genome-wide recombination rate. Science 319, 1398–1401. doi:10.1126/science.1152422
Kueng, S., Hegemann, B., Peters, B. H., Lipp, J. J., Schleiffer, A., Mechtler, K., et al. (2006). Wapl controls the dynamic association of cohesin with chromatin. Cell 127, 955–967. doi:10.1016/j.cell.2006.09.040
Kugou, K., Fukuda, T., Yamada, S., Ito, M., Sasanuma, H., Mori, S., et al. (2009). Rec8 guides canonical Spo11 distribution along yeast meiotic chromosomes. Mol. Biol. Cell 20, 3064–3076. doi:10.1091/mbc.E08-12-1223
Kumar, R., Bourbon, H. M., and de Massy, B. (2010). Functional conservation of Mei4 for meiotic DNA double-strand break formation from yeasts to mice. Genes Dev. 24, 1266–1280. doi:10.1101/gad.571710
Kumar, R., Ghyselinck, N., Ishiguro, K., Watanabe, Y., Kouznetsova, A., Hoog, C., et al. (2015). MEI4 - a central player in the regulation of meiotic DNA double-strand break formation in the mouse. J. Cell Sci. 128, 1800–1811. doi:10.1242/jcs.165464
Kusch, T. (2015). Brca2-Pds5 complexes mobilize persistent meiotic recombination sites to the nuclear envelope. J. Cell Sci. 128, 717–727. doi:10.1242/jcs.159988
Lao, J. P., Cloud, V., Huang, C. C., Grubb, J., Thacker, D., Lee, C. Y., et al. (2013). Meiotic crossover control by concerted action of Rad51-Dmc1 in homolog template bias and robust homeostatic regulation. PLoS Genet. 9, e1003978. doi:10.1371/journal.pgen.1003978
Lao, J. P., and Hunter, N. (2010). Trying to avoid your sister. PLoS Biol. 8, e1000519. doi:10.1371/journal.pbio.1000519
Lee, J., and Hirano, T. (2011). RAD21L, a novel cohesin subunit implicated in linking homologous chromosomes in mammalian meiosis. J. Cell Biol. 192, 263–276. doi:10.1083/jcb.201008005
Lee, M. S., Higashide, M. T., Choi, H., Li, K., Hong, S., Lee, K., et al. (2021). The synaptonemal complex central region modulates crossover pathways and feedback control of meiotic double-strand break formation. Nucleic Acids Res. 49, 7537–7553. doi:10.1093/nar/gkab566
Li, X. C., and Schimenti, J. C. (2007). Mouse pachytene checkpoint 2 (trip13) is required for completing meiotic recombination but not synapsis. PLoS Genet. 3, e130. doi:10.1371/journal.pgen.0030130
Llano, E., Herran, Y., Garcia-Tunon, I., Gutierrez-Caballero, C., de Alava, E., Barbero, J. L., et al. (2012). Meiotic cohesin complexes are essential for the formation of the axial element in mice. J. Cell Biol. 197, 877–885. doi:10.1083/jcb.201201100
Luo, Z., Wang, X., Jiang, H., Wang, R., Chen, J., Chen, Y., et al. (2020). Reorganized 3D genome structures support transcriptional regulation in mouse spermatogenesis. iScience 23, 101034. doi:10.1016/j.isci.2020.101034
Lynn, A., Soucek, R., and Borner, G. V. (2007). ZMM proteins during meiosis: Crossover artists at work. Chromosome Res. 15, 591–605. doi:10.1007/s10577-007-1150-1
MacQueen, A. J., Colaiacovo, M. P., McDonald, K., and Villeneuve, A. M. (2002). Synapsis-dependent and -independent mechanisms stabilize homolog pairing during meiotic prophase in C. elegans. Genes Dev. 16, 2428–2442. doi:10.1101/gad.1011602
MacQueen, A. J., Phillips, C. M., Bhalla, N., Weiser, P., Villeneuve, A. M., and Dernburg, A. F. (2005). Chromosome sites play dual roles to establish homologous synapsis during meiosis in C. elegans. Cell 123, 1037–1050. doi:10.1016/j.cell.2005.09.034
Martini, E., Diaz, R. L., Hunter, N., and Keeney, S. (2006). Crossover homeostasis in yeast meiosis. Cell 126, 285–295. doi:10.1016/j.cell.2006.05.044
Martino, J., Brunette, G. J., Barroso-Gonzalez, J., Moiseeva, T. N., Smith, C. M., Bakkenist, C. J., et al. (2019). The human Shu complex functions with PDS5B and SPIDR to promote homologous recombination. Nucleic Acids Res. 47, 10151–10165. doi:10.1093/nar/gkz738
Miyoshi, T., Ito, M., Kugou, K., Yamada, S., Furuichi, M., Oda, A., et al. (2012). A central coupler for recombination initiation linking chromosome architecture to S phase checkpoint. Mol. Cell 47, 722–733. doi:10.1016/j.molcel.2012.06.023
Morgan, C., Fozard, J. A., Hartley, M., Henderson, I. R., Bomblies, K., and Howard, M. (2021). Diffusion-mediated HEI10 coarsening can explain meiotic crossover positioning in Arabidopsis. Nat. Commun. 12, 4674. doi:10.1038/s41467-021-24827-w
Muller, H., Scolari, V. F., Agier, N., Piazza, A., Thierry, A., Mercy, G., et al. (2018). Characterizing meiotic chromosomes' structure and pairing using a designer sequence optimized for Hi-C. Mol. Syst. Biol. 14, e8293. doi:10.15252/msb.20188293
Nasmyth, K., and Haering, C. H. (2005). The structure and function of SMC and kleisin complexes. Annu. Rev. Biochem. 74, 595–648. doi:10.1146/annurev.biochem.74.082803.133219
Niu, H., Li, X., Job, E., Park, C., Moazed, D., Gygi, S. P., et al. (2007). Mek1 kinase is regulated to suppress double-strand break repair between sister chromatids during budding yeast meiosis. Mol. Cell Biol. 27, 5456–5467. doi:10.1128/MCB.00416-07
Niu, H., Wan, L., Busygina, V., Kwon, Y., Allen, J. A., Li, X., et al. (2009). Regulation of meiotic recombination via Mek1-mediated Rad54 phosphorylation. Mol. Cell 36, 393–404. doi:10.1016/j.molcel.2009.09.029
Nore, A., Juarez-Martinez, A. B., Clement, J., Brun, C., Diagouraga, B., Laroussi, H., et al. (2022). TOPOVIBL-REC114 interaction regulates meiotic DNA double-strand breaks. Nat. Commun. 13, 7048. doi:10.1038/s41467-022-34799-0
Pan, J., Sasaki, M., Kniewel, R., Murakami, H., Blitzblau, H. G., Tischfield, S. E., et al. (2011). A hierarchical combination of factors shapes the genome-wide topography of yeast meiotic recombination initiation. Cell 144, 719–731. doi:10.1016/j.cell.2011.02.009
Panizza, S., Mendoza, M. A., Berlinger, M., Huang, L., Nicolas, A., Shirahige, K., et al. (2011). Spo11-accessory proteins link double-strand break sites to the chromosome axis in early meiotic recombination. Cell 146, 372–383. doi:10.1016/j.cell.2011.07.003
Parvanov, E. D., Tian, H., Billings, T., Saxl, R. L., Spruce, C., Aithal, R., et al. (2017). PRDM9 interactions with other proteins provide a link between recombination hotspots and the chromosomal axis in meiosis. Mol. Biol. Cell 28, 488–499. doi:10.1091/mbc.E16-09-0686
Pasierbek, P., Jantsch, M., Melcher, M., Schleiffer, A., Schweizer, D., and Loidl, J. (2001). A Caenorhabditis elegans cohesion protein with functions in meiotic chromosome pairing and disjunction. Genes Dev. 15, 1349–1360. doi:10.1101/gad.192701
Patel, L., Kang, R., Rosenberg, S. C., Qiu, Y., Raviram, R., Chee, S., et al. (2019). Dynamic reorganization of the genome shapes the recombination landscape in meiotic prophase. Nat. Struct. Mol. Biol. 26, 164–174. doi:10.1038/s41594-019-0187-0
Pattabiraman, D., Roelens, B., Woglar, A., and Villeneuve, A. M. (2017). Meiotic recombination modulates the structure and dynamics of the synaptonemal complex during C. elegans meiosis. PLoS Genet. 13, e1006670. doi:10.1371/journal.pgen.1006670
Qiao, H., Prasada Rao, H. B., Yang, Y., Fong, J. H., Cloutier, J. M., Deacon, D. C., et al. (2014). Antagonistic roles of ubiquitin ligase HEI10 and SUMO ligase RNF212 regulate meiotic recombination. Nat. Genet. 46, 194–199. doi:10.1038/ng.2858
Rao, H. B., Qiao, H., Bhatt, S. K., Bailey, L. R., Tran, H. D., Bourne, S. L., et al. (2017). A SUMO-ubiquitin relay recruits proteasomes to chromosome axes to regulate meiotic recombination. Science 355, 403–407. doi:10.1126/science.aaf6407
Revenkova, E., Eijpe, M., Heyting, C., Hodges, C. A., Hunt, P. A., Liebe, B., et al. (2004). Cohesin SMC1 beta is required for meiotic chromosome dynamics, sister chromatid cohesion and DNA recombination. Nat. Cell Biol. 6, 555–562. doi:10.1038/ncb1135
Reynolds, A., Qiao, H., Yang, Y., Chen, J. K., Jackson, N., Biswas, K., et al. (2013). RNF212 is a dosage-sensitive regulator of crossing-over during mammalian meiosis. Nat. Genet. 45, 269–278. doi:10.1038/ng.2541
Rinaldi, V. D., Bolcun-Filas, E., Kogo, H., Kurahashi, H., and Schimenti, J. C. (2017). The DNA damage checkpoint eliminates mouse oocytes with chromosome synapsis failure. Mol. Cell 67, 1026–1036. doi:10.1016/j.molcel.2017.07.027
Robert, T., Vrielynck, N., Mezard, C., de Massy, B., and Grelon, M. (2016). A new light on the meiotic DSB catalytic complex. Semin. Cell Dev. Biol. 54, 165–176. doi:10.1016/j.semcdb.2016.02.025
Rog, O., Kohler, S., and Dernburg, A. F. (2017). The synaptonemal complex has liquid crystalline properties and spatially regulates meiotic recombination factors. Elife 6, e21455. doi:10.7554/eLife.21455
Roig, I., Dowdle, J. A., Toth, A., de Rooij, D. G., Jasin, M., and Keeney, S. (2010). Mouse TRIP13/PCH2 is required for recombination and normal higher-order chromosome structure during meiosis. PLoS Genet. 6, e1001062. doi:10.1371/journal.pgen.1001062
Rousova, D., Nivsarkar, V., Altmannova, V., Raina, V. B., Funk, S. K., Liedtke, D., et al. (2021). Novel mechanistic insights into the role of Mer2 as the keystone of meiotic DNA break formation. Elife 10, e72330. doi:10.7554/eLife.72330
Sakuno, T., Tashiro, S., Tanizawa, H., Iwasaki, O., Ding, D. Q., Haraguchi, T., et al. (2022). Rec8 Cohesin-mediated Axis-loop chromatin architecture is required for meiotic recombination. Nucleic Acids Res. 50, 3799–3816. doi:10.1093/nar/gkac183
Sanchez-Saez, F., Gomez, H. L., Dunne, O. M., Gallego-Paramo, C., Felipe-Medina, N., Sanchez-Martin, M., et al. (2020). Meiotic chromosome synapsis depends on multivalent SYCE1-SIX6OS1 interactions that are disrupted in cases of human infertility. Sci. Adv. 6, eabb1660. doi:10.1126/sciadv.abb1660
Schalbetter, S. A., Fudenberg, G., Baxter, J., Pollard, K. S., and Neale, M. J. (2019). Principles of meiotic chromosome assembly revealed in S. cerevisiae. Nat. Commun. 10, 4795. doi:10.1038/s41467-019-12629-0
Schramm, S., Fraune, J., Naumann, R., Hernandez-Hernandez, A., Hoog, C., Cooke, H. J., et al. (2011). A novel mouse synaptonemal complex protein is essential for loading of central element proteins, recombination, and fertility. PLoS Genet. 7, e1002088. doi:10.1371/journal.pgen.1002088
Severson, A. F., Ling, L., van Zuylen, V., and Meyer, B. J. (2009). The axial element protein HTP-3 promotes cohesin loading and meiotic axis assembly in C. elegans to implement the meiotic program of chromosome segregation. Genes Dev. 23, 1763–1778. doi:10.1101/gad.1808809
Severson, A. F., and Meyer, B. J. (2014). Divergent kleisin subunits of cohesin specify mechanisms to tether and release meiotic chromosomes. Elife 3, e03467. doi:10.7554/eLife.03467
Shin, Y. H., McGuire, M. M., and Rajkovic, A. (2013). Mouse HORMAD1 is a meiosis i checkpoint protein that modulates DNA double-strand break repair during female meiosis. Biol. Reprod. 89, 29. doi:10.1095/biolreprod.112.106773
Shinohara, M., Oh, S. D., Hunter, N., and Shinohara, A. (2008). Crossover assurance and crossover interference are distinctly regulated by the ZMM proteins during yeast meiosis. Nat. Genet. 40, 299–309. doi:10.1038/ng.83
Shinohara, M., Sakai, K., Shinohara, A., and Bishop, D. K. (2003). Crossover interference in Saccharomyces cerevisiae requires a TID1/RDH54- and DMC1-dependent pathway. Genetics 163, 1273–1286. doi:10.1093/genetics/163.4.1273
Smith, A. V., and Roeder, G. S. (1997). The yeast Red1 protein localizes to the cores of meiotic chromosomes. J. Cell Biol. 136, 957–967. doi:10.1083/jcb.136.5.957
Smolikov, S., Eizinger, A., Hurlburt, A., Rogers, E., Villeneuve, A. M., and Colaiacovo, M. P. (2007a). Synapsis-defective mutants reveal a correlation between chromosome conformation and the mode of double-strand break repair during Caenorhabditis elegans meiosis. Genetics 176, 2027–2033. doi:10.1534/genetics.107.076968
Smolikov, S., Eizinger, A., Schild-Prufert, K., Hurlburt, A., McDonald, K., Engebrecht, J., et al. (2007b). SYP-3 restricts synaptonemal complex assembly to bridge paired chromosome axes during meiosis in Caenorhabditis elegans. Genetics 176, 2015–2025. doi:10.1534/genetics.107.072413
Smolikov, S., Schild-Prufert, K., and Colaiacovo, M. P. (2009). A yeast two-hybrid screen for SYP-3 interactors identifies SYP-4, a component required for synaptonemal complex assembly and chiasma formation in Caenorhabditis elegans meiosis. PLoS Genet. 5, e1000669. doi:10.1371/journal.pgen.1000669
Sommermeyer, V., Beneut, C., Chaplais, E., Serrentino, M. E., and Borde, V. (2013). Spp1, a member of the Set1 Complex, promotes meiotic DSB formation in promoters by tethering histone H3K4 methylation sites to chromosome axes. Mol. Cell 49, 43–54. doi:10.1016/j.molcel.2012.11.008
Stanzione, M., Baumann, M., Papanikos, F., Dereli, I., Lange, J., Ramlal, A., et al. (2016). Meiotic DNA break formation requires the unsynapsed chromosome axis-binding protein IHO1 (CCDC36) in mice. Nat. Cell Biol. 18, 1208–1220. doi:10.1038/ncb3417
Stauffer, W. T., Zhang, L., and Dernburg, A. (2019). Diffusion through a liquid crystalline compartment regulates meiotic recombination. SPIE Proc. 2019. doi:10.1117/12.2513378
Strong, E. R., and Schimenti, J. C. (2010). Evidence implicating CCNB1IP1, a RING domain-containing protein required for meiotic crossing over in mice, as an E3 SUMO ligase. Genes (Basel) 1, 440–451. doi:10.3390/genes1030440
Sym, M., and Roeder, G. S. (1994). Crossover interference is abolished in the absence of a synaptonemal complex protein. Cell 79, 283–292. doi:10.1016/0092-8674(94)90197-x
Takeo, S., Lake, C. M., Morais-de-Sa, E., Sunkel, C. E., and Hawley, R. S. (2011). Synaptonemal complex-dependent centromeric clustering and the initiation of synapsis in Drosophila oocytes. Curr. Biol. 21, 1845–1851. doi:10.1016/j.cub.2011.09.044
Tedeschi, A., Wutz, G., Huet, S., Jaritz, M., Wuensche, A., Schirghuber, E., et al. (2013). Wapl is an essential regulator of chromatin structure and chromosome segregation. Nature 501, 564–568. doi:10.1038/nature12471
Thacker, D., Lam, I., Knop, M., and Keeney, S. (2011). Exploiting spore-autonomous fluorescent protein expression to quantify meiotic chromosome behaviors in Saccharomyces cerevisiae. Genetics 189, 423–439. doi:10.1534/genetics.111.131326
Tian, H., Billings, T., and Petkov, P. M. (2018). CXXC1 is not essential for normal DNA double-strand break formation and meiotic recombination in mouse. PLoS Genet. 14, e1007657. doi:10.1371/journal.pgen.1007657
Tsubouchi, T., Zhao, H., and Roeder, G. S. (2006). The meiosis-specific zip4 protein regulates crossover distribution by promoting synaptonemal complex formation together with zip2. Dev. Cell 10, 809–819. doi:10.1016/j.devcel.2006.04.003
Vara, C., Paytuvi-Gallart, A., Cuartero, Y., Le Dily, F., Garcia, F., Salva-Castro, J., et al. (2019). Three-dimensional genomic structure and cohesin occupancy correlate with transcriptional activity during spermatogenesis. Cell Rep. 28, 352–367. doi:10.1016/j.celrep.2019.06.037
Viera, A., Berenguer, I., Ruiz-Torres, M., Gomez, R., Guajardo, A., Barbero, J. L., et al. (2020). PDS5 proteins regulate the length of axial elements and telomere integrity during male mouse meiosis. EMBO Rep. 21, e49273. doi:10.15252/embr.201949273
Voelkel-Meiman, K., Cheng, S. Y., Morehouse, S. J., and MacQueen, A. J. (2016). Synaptonemal complex proteins of budding yeast define reciprocal roles in MutSγ-mediated crossover formation. Genetics 203, 1091–1103. doi:10.1534/genetics.115.182923
Voelkel-Meiman, K., Taylor, L. F., Mukherjee, P., Humphryes, N., Tsubouchi, H., and Macqueen, A. J. (2013). SUMO localizes to the central element of synaptonemal complex and is required for the full synapsis of meiotic chromosomes in budding yeast. PLoS Genet. 9, e1003837. doi:10.1371/journal.pgen.1003837
Wang, K., Wang, M., Tang, D., Shen, Y., Miao, C., Hu, Q., et al. (2012). The role of rice HEI10 in the formation of meiotic crossovers. PLoS Genet. 8, e1002809. doi:10.1371/journal.pgen.1002809
Wang, S., Veller, C., Sun, F., Ruiz-Herrera, A., Shang, Y., Liu, H., et al. (2019a). Per-nucleus crossover covariation and implications for evolution. Cell 177, 326–338. doi:10.1016/j.cell.2019.02.021
Wang, Y., Wang, H., Zhang, Y., Du, Z., Si, W., Fan, S., et al. (2019b). Reprogramming of meiotic chromatin architecture during spermatogenesis. Mol. Cell 73, 547–561. doi:10.1016/j.molcel.2018.11.019
Ward, A., Hopkins, J., McKay, M., Murray, S., and Jordan, P. W. (2016). Genetic interactions between the meiosis-specific cohesin components, STAG3, REC8, and RAD21L. G3 (Bethesda) 6, 1713–1724. doi:10.1534/g3.116.029462
Ward, J. O., Reinholdt, L. G., Motley, W. W., Niswander, L. M., Deacon, D. C., Griffin, L. B., et al. (2007). Mutation in mouse hei10, an e3 ubiquitin ligase, disrupts meiotic crossing over. PLoS Genet. 3, e139. doi:10.1371/journal.pgen.0030139
West, A. M., Rosenberg, S. C., Ur, S. N., Lehmer, M. K., Ye, Q., Hagemann, G., et al. (2019). A conserved filamentous assembly underlies the structure of the meiotic chromosome axis. Elife 8, e40372. doi:10.7554/eLife.40372
West, A. M. V., Komives, E. A., and Corbett, K. D. (2018). Conformational dynamics of the Hop1 HORMA domain reveal a common mechanism with the spindle checkpoint protein Mad2. Nucleic Acids Res. 46, 279–292. doi:10.1093/nar/gkx1196
Wojtasz, L., Daniel, K., Roig, I., Bolcun-Filas, E., Xu, H., Boonsanay, V., et al. (2009). Mouse HORMAD1 and HORMAD2, two conserved meiotic chromosomal proteins, are depleted from synapsed chromosome axes with the help of TRIP13 AAA-ATPase. PLoS Genet. 5, e1000702. doi:10.1371/journal.pgen.1000702
Wutz, G., Varnai, C., Nagasaka, K., Cisneros, D. A., Stocsits, R. R., Tang, W., et al. (2017). Topologically associating domains and chromatin loops depend on cohesin and are regulated by CTCF, WAPL, and PDS5 proteins. EMBO J. 36, 3573–3599. doi:10.15252/embj.201798004
Xu, H., Beasley, M. D., Warren, W. D., van der Horst, G. T., and McKay, M. J. (2005). Absence of mouse REC8 cohesin promotes synapsis of sister chromatids in meiosis. Dev. Cell 8, 949–961. doi:10.1016/j.devcel.2005.03.018
Yamada, S., Ohta, K., and Yamada, T. (2013). Acetylated Histone H3K9 is associated with meiotic recombination hotspots, and plays a role in recombination redundantly with other factors including the H3K4 methylase Set1 in fission yeast. Nucleic Acids Res. 41, 3504–3517. doi:10.1093/nar/gkt049
Yang, F., Gell, K., van der Heijden, G. W., Eckardt, S., Leu, N. A., Page, D. C., et al. (2008). Meiotic failure in male mice lacking an X-linked factor. Genes Dev. 22, 682–691. doi:10.1101/gad.1613608
Yang, X., Song, M., Wang, Y., Tan, T., Tian, Z., Zhai, B., et al. (2022). The ubiquitin-proteasome system regulates meiotic chromosome organization. Proc. Natl. Acad. Sci. U. S. A. 119, e2106902119. doi:10.1073/pnas.2106902119
Ye, Q., Kim, D. H., Dereli, I., Rosenberg, S. C., Hagemann, G., Herzog, F., et al. (2017). The AAA+ ATPase TRIP13 remodels HORMA domains through N-terminal engagement and unfolding. EMBO J. 36, 2419–2434. doi:10.15252/embj.201797291
Zanders, S., Sonntag Brown, M., Chen, C., and Alani, E. (2011). Pch2 modulates chromatid partner choice during meiotic double-strand break repair in Saccharomyces cerevisiae. Genetics 188, 511–521. doi:10.1534/genetics.111.129031
Zhang, L., Espagne, E., de Muyt, A., Zickler, D., and Kleckner, N. E. (2014a). Interference-mediated synaptonemal complex formation with embedded crossover designation. Proc. Natl. Acad. Sci. U. S. A. 111, E5059–E5068. doi:10.1073/pnas.1416411111
Zhang, L., Kohler, S., Rillo-Bohn, R., and Dernburg, A. F. (2018a). A compartmentalized signaling network mediates crossover control in meiosis. Elife 7, e30789. doi:10.7554/eLife.30789
Zhang, L., Liang, Z., Hutchinson, J., and Kleckner, N. (2014b). Crossover patterning by the beam-film model: Analysis and implications. PLoS Genet. 10, e1004042. doi:10.1371/journal.pgen.1004042
Zhang, L., Stauffer, W., Zwicker, D., and Dernburg, A. F. 2021. Crossover patterning through kinase-regulated condensation and coarsening of recombination nodules. bioRxiv[preprint]. doi:10.1101/2021.08.26.457865
Zhang, L., Wang, S., Yin, S., Hong, S., Kim, K. P., and Kleckner, N. (2014c). Topoisomerase II mediates meiotic crossover interference. Nature 511, 551–556. doi:10.1038/nature13442
Zhang, Q., Ji, S. Y., Busayavalasa, K., and Yu, C. (2019). SPO16 binds SHOC1 to promote homologous recombination and crossing-over in meiotic prophase I. Sci. Adv. 5, eaau9780. doi:10.1126/sciadv.aau9780
Zhang, Q., Shao, J., Fan, H. Y., and Yu, C. (2018b). Evolutionarily-conserved MZIP2 is essential for crossover formation in mammalian meiosis. Commun. Biol. 1, 147. doi:10.1038/s42003-018-0154-z
Zhang, Y., Suzuki, T., Li, K., Gothwal, S. K., Shinohara, M., and Shinohara, A. (2020). Genetic interactions of histone modification machinery Set1 and PAF1C with the recombination complex rec114-mer2-mei4 in the formation of meiotic DNA double-strand breaks. Int. J. Mol. Sci. 21, 2679. doi:10.3390/ijms21082679
Zhu, Z., Bani Ismail, M., Shinohara, M., and Shinohara, A. (2021). SCF(Cdc4) ubiquitin ligase regulates synaptonemal complex formation during meiosis. Life Sci. Alliance 4, e202000933. doi:10.26508/lsa.202000933
Zierhut, C., Berlinger, M., Rupp, C., Shinohara, A., and Klein, F. (2004). Mnd1 is required for meiotic interhomolog repair. Curr. Biol. 14, 752–762. doi:10.1016/j.cub.2004.04.030
Ziolkowski, P. A., Underwood, C. J., Lambing, C., Martinez-Garcia, M., Lawrence, E. J., Ziolkowska, L., et al. (2017). Natural variation and dosage of the HEI10 meiotic E3 ligase control Arabidopsis crossover recombination. Genes Dev. 31, 306–317. doi:10.1101/gad.295501.116
Keywords: cohesin, axis-loop structure, synaptonemal complex, meiotic recombination, crossover
Citation: Ito M and Shinohara A (2023) Chromosome architecture and homologous recombination in meiosis. Front. Cell Dev. Biol. 10:1097446. doi: 10.3389/fcell.2022.1097446
Received: 13 November 2022; Accepted: 22 December 2022;
Published: 06 January 2023.
Edited by:
Ricardo Benavente, Julius Maximilian University of Würzburg, GermanyReviewed by:
Kevin Corbett, University of California, San Diego, United StatesCopyright © 2023 Ito and Shinohara. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Masaru Ito, bXNyaXRvMkBwcm90ZWluLm9zYWthLXUuYWMuanA=; Akira Shinohara, YXNoaW5vQHByb3RlaW4ub3Nha2EtdS5hYy5qcA==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.