
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Cell Dev. Biol., 16 January 2023
Sec. Signaling
Volume 10 - 2022 | https://doi.org/10.3389/fcell.2022.1096899
This article is part of the Research TopicHomeostatic Regulation of Protein Synthesis, Folding and Secretion by Stress Response Pathways in EukaryotesView all 5 articles
The secretory pathway is an intracellular highway for the vesicular transport of newly synthesized proteins that spans the endoplasmic reticulum (ER), Golgi, lysosomes and the cell surface. A variety of cargo receptors, chaperones, and quality control proteins maintain the smooth flow of cargo along this route. Among these is vesicular transport protein TMED9, which belongs to the p24/transmembrane emp24 domain (TMED) family of proteins, and is expressed across vertebrate species. The TMED family is comprised of structurally-related type I transmembrane proteins with a luminal N-terminal Golgi-dynamics domain, a luminal coiled-coil domain, a transmembrane domain and a short cytosolic C-terminal tail that binds COPI and COPII coat proteins. TMED9, like other members of the TMED family, was first identified as an abundant constituent of the COPI and COPII coated vesicles that mediate traffic between the ER and the Golgi. TMED9 is typically purified in hetero-oligomers together with TMED family members, suggesting that it may function as part of a complex. Recently, TMED family members have been discovered to play various roles in secretory pathway homeostasis including secreted protein processing, quality control and degradation of misfolded proteins, and post-Golgi trafficking. In particular, TMED9 has been implicated in autophagy, lysosomal sorting, viral replication and cancer, which we will discuss in this Mini-Review.
The first member of the transmembrane emp24 domain (TMED) family proteins, TMED11, was discovered in rough microsomes derived from canine endoplasmic reticulum (ER) in 1991 (Wada et al., 1991). Within a few years, TMED9 and other TMED family proteins were found to be type I transmembrane COPI and COPII coatomer binding proteins localized to the secretory pathway and conserved across mammals, yeast, and plants (Schimmoller et al., 1995; Stamnes et al., 1995; Belden and Barlowe, 1996; Elrod-Erickson and Kaiser, 1996; Sohn et al., 1996; Dominguez et al., 1998; Contreras et al., 2004a; Contreras et al., 2004b). The TMED family was originally referred to as the p24 family after their size (∼24 kDa), subfamily (ɑ, β, δ, or γ), and the order in which they were identified (1–5) (Strating et al., 2009). Each TMED protein has several aliases. Strating et al. (2009) organized the names in a useful reference table.
The secretory pathway is the major biosynthetic hub for the production, secretion, and turnover of soluble secretory and transmembrane proteins in eukaryotic cells. Traffic through the secretory pathway begins at the ER, where proteins are synthesized, folded, and processed prior to export to the Golgi for subsequent transport to the cell surface or lysosomes. Within the early secretory pathway, which is comprised of the ER, ER-Golgi intermediate compartment (ERGIC) and Golgi, protein folding is aided and monitored by chaperones and protein quality control (PQC) machinery (Anelli and Sitia, 2008; Adams et al., 2019; Sun and Brodsky, 2019). While TMED9 and TMED family members are well-characterized as regulators of homeostasis and vesicular transport within the early secretory pathway (Strating and Martens, 2009; Pastor-Cantizano et al., 2016; D'Arcangelo et al., 2015; Belden and Barlowe, 2001; Elrod-Erickson and Kaiser, 1996), their precise functions within this area are yet to be determined.
TMED family proteins promote efficient and selective secretion of diverse classes of proteins. TMED family members, including TMED10 and TMED2, facilitate ER-export of glycosylphosphatidylinositol-anchored proteins (GPI-APs) in yeast and cultured mammalian cells (Muniz et al., 2000; Marzioch et al., 1999; D'Arcangelo et al., 2015; Schimmoller et al., 1995; Belden and Barlowe, 2001; Fujita et al., 2011), and are required for ER-export of misfolded GPI-APs destined for lysosomal degradation (Satpute-Krishnan et al., 2014; Sikorska et al., 2016; Zavodszky and Hegde, 2019). TMED10 was recently shown to promote the unconventional protein secretion (UPS) of leaderless cargo including mature IL-1β (Zhang M. et al., 2020). TMED9 in particular has emerged as a major regulator of secretory pathway protein homeostasis through its involvement in protein trafficking and degradation. TMED9 has a propensity to form and function as a hetero-oligomer with other TMED family members (Belden and Barlowe, 1996; Fullekrug et al., 1999; Muniz et al., 2000; Fujita et al., 2011). Therefore, in this review we will describe TMED9 in the context of the larger TMED family of proteins.
In humans there are 11 genes annotated as TMED1-11. TMED family members are expressed throughout the body, as demonstrated in mice (Strating et al., 2009), and are highly expressed in secretory cell types (Zhang and Volchuk, 2010). Given their ubiquity, it is thus unsurprising that some TMED proteins are developmentally essential and knockout of either TMED2 or TMED10 is embryonic lethal in mice (Denzel et al., 2000; Jerome-Majewska et al., 2010) and reduces viability in cultured cells (Blomen et al., 2015).
Various studies have shown that the TMED proteins form oligomers of varying stoichiometry (Fullekrug et al., 1999; Emery et al., 2000; Jenne et al., 2002). A series of siRNA knockdown experiments revealed that knockdown of TMEDs 2, 4, 5, 9, or 10 destabilized other TMED family members while TMED7 knockdown primarily affected TMED5. Loss of TMEDs nine or 10 inhibited GPI-AP trafficking, whereas WNT trafficking was inhibited in cells lacking either TMEDs 2, 4, 9, or 10 (Tashima et al., 2022). Because of the interdependency between TMED family member expression and function, it is technically challenging to discriminate between the functions of individual TMED proteins or their oligomeric complexes.
Mammalian TMED9 and its yeast homolog, Erv25p, were first discovered as secretory pathway proteins (Dominguez et al., 1998; Marzioch et al., 1999). TMED9 localizes primarily to the ER and ERGIC, but is found in post-Golgi secretory vesicles along with other TMED family proteins (Shevchenko et al., 1997; Dominguez et al., 1998; Marzioch et al., 1999; Breuza et al., 2004). Later TMED9 was discovered to be critical for the generation of ER exit sites (ERES) in a cell-free microsome budding assay (Lavoie et al., 1999). Further emphasizing its role(s) in the secretory pathway, depletion of TMED9 leads to the fragmentation of Golgi structures and the partial dissociation of COPI from the Golgi (Mitrovic et al., 2008). The yeast homolog of TMED9, Erv25p, has been shown to be play a role in efficient ER-to-Golgi transport of the yeast GPI-AP, Gas1 (Belden and Barlowe, 1996). However teasing apart TMED9’s individual role from other TMED-family members, including TMEDs 2 and 10, is difficult because knockdown of each impacts the expression of the others (Fujita et al., 2011). Taken together, TMED9 along with its family appears to regulate multiple critical trafficking steps in the secretory pathway. Excellent reviews have been written to discuss the role of the TMED proteins in the early secretory pathway (Pastor-Cantizano et al., 2016; Aber et al., 2019).
Recently, TMED9 was shown to participate in unconventional protein secretion (UPS) from the ER to the plasma membrane during ER stress in cells expressing the dominant-inhibitory form of ADP-ribosylation factor 1 (ARF1-Q71L), which blocks ER-to-Golgi transport (Park et al., 2022). TMED9 was found to participate in the assembly of a heterooligomeric trafficking complex governing SARS-Cov2 spike protein and cystic fibrosis transmembrane conductance regulator (CFTR) secretion (Park et al., 2022). Although Park et al. (2022) found that TMED9 did not bind to CFTR or Spike proteins, silencing TMED9 reduced the cell surface trafficking of these UPS cargo. These findings suggest that TMED9 may participate in a variety of yet undiscovered trafficking pathways.
The TMED proteins are structurally conserved among eukaryotes despite significant variations in sequence identity (Strating et al., 2009) (Figure 1A). Each family member contains four major regions: the GOLD domain, coiled-coil domain, transmembrane domain, and a cytoplasmic COP-binding region (Figures 1B, C). Whether these conserved domains allow the TMED proteins to act interchangeably in certain processes is unknown.
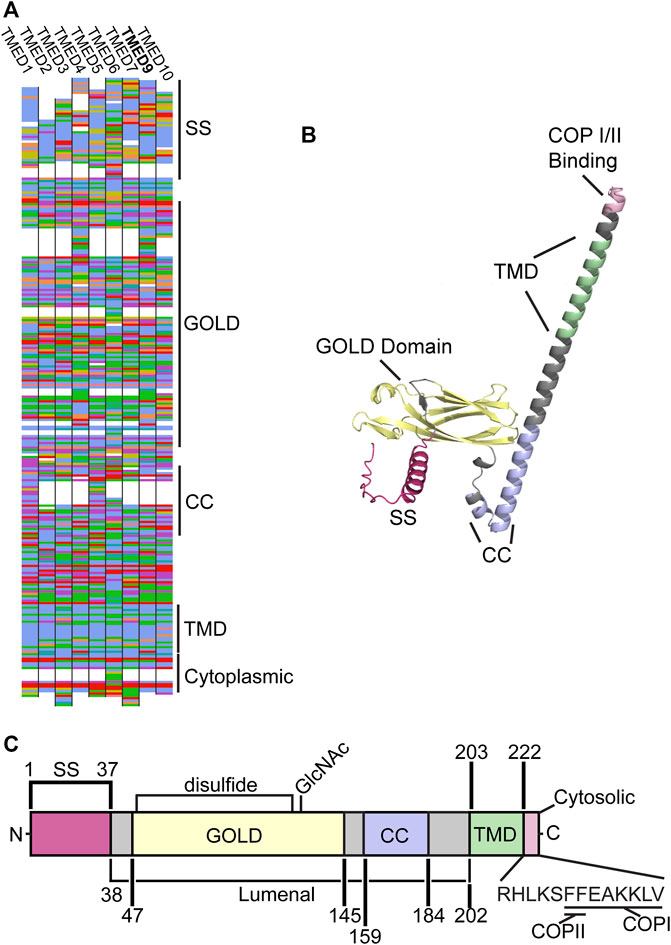
FIGURE 1. (A) The protein sequence for human TMEDs 1, 2, 3, 4, 5, 6, 7, 9, and 10 are shown. Sequences were aligned with Muscle and drawn with AlignmentViewer (alignmentviewer.org). Aligned amino acids are colored in the Clustal2 color code. Structural motifs for TMED9 are indicated (SS: signal sequence, GOLD, CC: coiled-coil, TMD: transmembrane domain, COP: COPI/II). (B) The Alphafold (Jumper et al., 2021; Varadi et al., 2022) structure for the human TMED9 protein (AF-Q9BVK6-F1). The signal sequence (SS, red), GOLD domain (yellow), coiled-coil (CC, lilac), transmembrane domain (TMD, green), and COP I/II (pink) binding sites are indicated (C) A predicted domain map of the human TMED9 protein (Q9BVK6) as compiled and annotated by Uniprot (UniProt, 2021). The structural domains from the N-terminus “N” to the C-terminus “C” in (B) are indicated, as well as a conserved disulfide bond, N-linked glycan (GlcNAc), and the COPI and COPII binding sites. Amino acid positions are given and domains are depicted to scale.
The Golgi dynamics (GOLD) domain consists of eight β-strands and one disulfide bond (Nagae et al., 2016). Despite low sequence homology, the GOLD domains found in TMEDs 1, 2, 5, and 10 are structurally similar (Nagae et al., 2016; Nagae et al., 2017; Mota et al., 2022). The GOLD domain is chiefly involved in hetero and homo-oligomerization (Nagae et al., 2016; Zhang M. et al., 2020; Mota et al., 2022). Heterodimerization occurs across a range of sites on each GOLD domain, depending on the TMED proteins involved (Nagae et al., 2016). Dimerization is dependent on solution ionic strength (Mota et al., 2022) and pH (Nagae et al., 2016) in vitro, suggesting that intracellular localization may influence dimerization. These findings are largely sourced from studies involving purified GOLD domains rather than intact TMED proteins. Beyond its role in oligomerization, the GOLD domain has also been shown to participate in substrate recognition (Park et al., 2022) and the interaction between TMEDs 9 and 10 with syntaxin 17 (Muppirala et al., 2011).
The coiled-coil (CC) domain mediates TMED oligomerization and substrate recognition. Early observations showed that CC deletion abolished post-ER transport of hetero-oligomeric complexes (Ciufo and Boyd, 2000; Emery et al., 2000) and recently the TMED7 CC was shown to participate in TMED7 homooligomerization (Liaunardy-Jopeace et al., 2014). Recent studies have also shown that the CC domain appears to mediate substrate recognition in the case of GPI-anchored proteins (Theiler et al., 2014) and TLR4 complex binding (Liaunardy-Jopeace et al., 2014).
The transmembrane domain (TMD) is essential in TMED protein sorting. The TMD of TMED2 but not TMED10 binds to sphingomyelin (SM) C18, promoting TMED2 dimerization and regulating cargo transport (Brugger et al., 2000; Contreras et al., 2012; Aisenbrey et al., 2019; Pannwitt et al., 2019). It is unclear if other TMED family members interact with SM in this way. Because membrane lipid content can affect membrane thickness, TMD length and lipid binding may increase the affinity of TMED proteins for membrane microdomains that are enriched with SM. Whereas no defined sorting motif has been identified within TMED protein TMDs, lengthening the TMED10 TMD impacts that protein’s sorting (Blum and Lepier, 2008). Intriguingly, it has been reported that membrane SM content affects the formation of coatomer protein (COP)-marked vesicles (Brugger et al., 2000). It is possible this effect is mediated by TMED proteins, since they interact with membrane lipids and COP (Pannwitt et al., 2019).
The cytoplasmic tail of the TMED proteins contains a region required for COP I/II binding, which we will discuss below. In addition, the cytoplasmic domain has been shown to bind to mature IL1β for TMED10 (Zhang M. et al., 2020), and syntaxin 17 and TC48 for TMEDs 9 and 10 (Muppirala et al., 2011; Muppirala et al., 2013). Intriguingly, the TMED9 cytoplasmic domain was also recently implicated in the formation of autophagic vesicles through its interaction with Sec12, the guanine-nucleotide exchange factor for Sar1 that functions upstream of COPII coat assembly (Weissman et al., 2001; Li et al., 2022).
COPI proteins bind to dilysine (KKXX) motifs and structurally related sites in cargo proteins (Ma and Goldberg, 2013). Of the TMEDs, only TMEDs 4, 9, 10, and 11 include a canonical KKXX motif, however KXK motifs in some TMED orthologs also enable COPI binding (Teasdale and Jackson, 1996; Dominguez et al., 1998; Belden and Barlowe, 2001; Pastor-Cantizano et al., 2016; UniProt, 2021). Consequently, TMEDs 9 and 10 have been shown to bind COPI components more strongly than TMEDs 2, 3, or 7. COPI binding is important for TMED retrieval from the Golgi back to the ER (Bremser et al., 1999) and mutations of this motif in TMEDs 2, 9, and 10 alters their ER-Golgi cycling kinetics (Dominguez et al., 1998; Blum and Lepier, 2008). Interestingly, COPI components recognize TMED oligomers rather than TMED monomers (Bethune et al., 2006).
COPII binding to the TMED proteins is mediated through aromatic residues in the cytoplasmic domain which fit into a binding pocket in the SEC24 COPII coat proteins (Ma et al., 2017). While all TMED family proteins display cytoplasmic aromatic residues, variations in the polypeptide sequence enable different TMED proteins to associate with different SEC24 isoforms (Wendeler et al., 2007).
Only a handful of diseases have been directly linked to TMED9. However, TMED family proteins have been tied to a variety of human diseases. Because the TMED proteins function as heteromeric complexes, TMED9 is likely to participate in some of the diseases associated with other TMED family members. Thus, we have listed diseases associated with each of the TMED proteins in Table 1.
Elevated TMED9 expression has been observed in multiple cancer types (Ju et al., 2021). In breast cancer, elevated TMED9 levels are associated with poor prognoses (Ju et al., 2021). In head and neck squamous cell carcinoma, expression of each of the TMED proteins is elevated. In particular, high expression of TMEDs 2, 9, and 10 was found to be associated with poor prognoses, whereas high expression of TMEDs 1, 3, 4, 5, 6, and 7 was not (Gao et al., 2022). Similarly, elevated TMED9 expression is associated with reduced survival time in individuals with epithelial ovarian cancer (EOC) in vivo. TMED9 knockdown reduces EOC cell proliferation in vitro (Han et al., 2022).
TMED9 expression may regulate cancer cell proliferation through its effect on growth factor signaling (Buechling et al., 2011; Nakano et al., 2017; Zhang X. et al., 2020; Di Minin et al., 2022; Tashima et al., 2022). For example, biochemical and microscopy approaches revealed that TMED9 loss was associated with dysregulation of TGFɑ trafficking and secretion in colon cancer and hepatocellular carcinoma cells (Mishra et al., 2019; Yang et al., 2021). Furthermore, loss of TMED9 led to impaired WNT trafficking (Tashima et al., 2022) and significant changes in the expression of genes regulated by WNT signaling (Yang et al., 2021). This TMED9-WNT signaling axis has been implicated in Paneth cell function in the intestines (Goga et al., 2021).
The TMED proteins have been implicated in various neurodegenerative diseases. TMED10 associates with and is required for the clearance of artificial and prion-disease associated mutants of prion protein (PrP) (Satpute-Krishnan et al., 2014). TMED2 was found to co-immunoprecipitate with atlastin whose misfolding leads to hereditary spastic paraplegia (Namekawa et al., 2007). Although TMED9 has not been thoroughly studied in the context of neurological disease, TMED9 has been shown to interact with wild type TDP-43, whose aggregation has been associated with the development of amyotrophic lateral sclerosis (ALS) (Redler and Dokholyan, 2012; Feneberg et al., 2020), and to associate with spastin whose mutations lead to hereditary spastic paraplegia (Reid et al., 2005). The precise motifs or domains of TMED9 involved in binding to TDP-43 or spastin remain unknown.
Various studies have demonstrated that alterations in TMED9 and TMED10 (commonly referred to as TMP21 in the Alzheimer’s field) expression promote the processing of amyloid precursor protein (APP) to amyloid beta (Aβ) by γ-secretase. Alzheimer’s disease has been associated with mutations in the genes encoding subunits of γ-secretase (Zhang et al., 2011). A single nucleotide polymorphism in TMED10 that resulted in heightened TMP21 expression was found to be genetically associated with Alzheimer’s disease in patients (Zhang et al., 2018). Additionally, alterations in TMED10 expression were found to impact pathological APP processing in cell culture models (Zhang et al., 2018). TMED10/TMP21 has been shown to co-immunoprecipitate with and regulate the activity of the γ-secretase complex. Intriguingly, depletion of TMED10/TMP21, results in increased generation of Aβ (Chen et al., 2006; Dolcini et al., 2008). Similarly, TMED9 co-immunoprecipitates with the core γ-secretase components and knockdown of TMED9 mRNA induces an increase in Aβ generation (Hasegawa et al., 2010; Bai et al., 2015). Because of TMED9’s tendency to heterooligomerize with TMED10/TMP21 (Dominguez et al., 1998; Fullekrug et al., 1999), TMED9 may function in a complex with TMED10/TMP21 to regulate γ-secretase processing of APP.
The proteinopathy mucin-1 kidney disease (MKD) results from a frameshift mutation in the MUC1 gene. (Dvela-Levitt et al. (2019) recently demonstrated that TMED9 binds to and mediates the post-ER trafficking of MUC1 aggregates. Under steady-state conditions, this TMED9-MUC1 complex drives the accumulation of toxic MUC1 in the ERGIC. Fortuitously, the authors found that the small molecule BRD4780 was able to reduce MUC1 aggregate levels both in vivo in mice and in vitro in cell culture and human kidney organoid models by reducing TMED9 stability and accelerating clearance of TMED9-MUC1 complexes from the ER and ERGIC to lysosomes (Dvela-Levitt et al., 2019).
Over the last decade, TMED9 has emerged as an important regulator of cellular proteostasis. It has recently been shown that TMED9 contributes to autophagy and autophagosome biogenesis. TMED9 was first identified in intracellular vesicles enriched with ATG9 and Rab1 thought to participate in autophagosome assembly (Kakuta et al., 2017). A role for TMED9 in autophagosome maturation was later demonstrated by Evans et al. (2021). The authors found that TMED9 knockdown attenuated autophagic activity and reduced viral production, potentially by decreasing COPII-dependent viral transport (Delorme-Axford et al., 2014; Evans et al., 2021).
In line with these findings, TMED9 was recently shown to directly participate in autophagosome biogenesis. It has long been known that TMED9 participates in ER exit site (ERES) formation for cargo transport to the Golgi (Lavoie et al., 1999; Fujita et al., 2011). However, Li et al. (2022) found that ERES-localized Sec12 and ERGIC-localized TMED9 interact directly in trans through their cytoplasmic domains, bringing ERES into close proximity with the ERGIC. ERES-ERGIC association is important for the generation of starvation-induced autophagosomes (Li et al., 2022). These findings suggest that TMED9 may directly influence the recruitment of COPII machinery at the ERGIC to contribute membranes for autophagosome formation.
TMED9 has been found in every organelle along the secretory pathway from the ER and Golgi, to the plasma membrane, to lysosomes and autophagosomes (Hasegawa et al., 2010; Li et al., 2022). While precise mechanistic functions of TMED9 remain elusive at each point, it is clear that TMED9 wears many hats in secretory pathway homeostasis. Building upon early findings that TMED9 binds to COPI and COPII coat proteins (Lavoie et al., 1999), recent studies indicate that TMED9 regulates the initial recruitment of COP machinery to the ERGIC membrane to promote the formation of autophagic membranes in coordination with COPII machinery (Kakuta et al., 2017; Evans et al., 2021; Li et al., 2022). Furthermore, TMED9 expression correlates with the development of multiple cancer types. Roles for TMED9 in the regulation of cancer cell growth (Mishra et al., 2019; Ju et al., 2021; Yang et al., 2021), APP processing (Hasegawa et al., 2010; Bai et al., 2015), and protein degradation (Dvela-Levitt et al., 2019) underscore the importance of this cargo receptor in health and disease.
The TMED protein family field is a rapidly evolving area of research. The exciting discovery that BRD4780 targets pathological TMED9-MUC1 aggregates to lysosomes demonstrates the potential to pharmacologically target TMED proteins for the resolution of proteinopathies (Dvela-Levitt et al., 2019). It is as yet unclear how BRD4780 induces lysosomal degradation of TMED9-MUC1, but possible mechanisms may involve directly altering TMED9’s structure or by preventing its oligomerization with other TMED family members. Preventing hetero-oligomerization of TMED9 has been shown to reduce its stability and the stability of other TMED proteins (Tashima et al., 2022). Because of this interdependence, BRD4780 may potentially be exploited to modulate various PQC pathways involving TMED heterooligomers. These findings encourage future research into the role of TMED9 and the other TMED proteins in clinical proteinopathies.
Beyond their clinical implications, the TMED proteins have now been shown to participate in a variety of essential cellular processes. New empirical tools such as cryoelectron microscopy and AI based modeling tools like Alphafold2 enable future structural studies to better characterize the interaction of the TMED proteins with one another and their cargo. Furthermore, structure-driven mutagenesis strategies may be used to disrupt oligomer formation in order to reveal the independent functions of the TMED proteins in the secretory pathway. Compelling questions include: Does environmental pH affect the function and binding of TMED proteins? Does N-linked glycosylation influence TMED cargo recognition? What is the precise role of TMED9 in autophagosome biogenesis, and how is this balanced with its role in secretion? What are the specific roles of each TMED family member and their various oligomeric states? As we gain answers, we may soon understand the functions and clinical relevance of TMED9 and its stubbornly mysterious family of proteins.
BR and PS-K have substantially and directly contributed to the conception, writing and design of this work. BR prepared the figure and table with input from PS-K.
We thank Karen Williams for her invaluable support.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Aber, R., Chan, W., Mugisha, S., and Jerome-Majewska, L. A. (2019). Transmembrane emp24 domain proteins in development and disease. Genet. Res. (Camb) 101, e14. doi:10.1017/S0016672319000090
Adams, B. M., Oster, M. E., and Hebert, D. N. (2019). Protein quality control in the endoplasmic reticulum. Protein J. 38, 317–329. doi:10.1007/s10930-019-09831-w
Aisenbrey, C., Kemayo-Koumkoua, P., Salnikov, E. S., Glattard, E., and Bechinger, B. (2019). Investigations of the structure, topology, and interactions of the transmembrane domain of the lipid-sorting protein p24 being highly selective for sphingomyelin-C18. Biochemistry 58, 2782–2795. doi:10.1021/acs.biochem.9b00375
Anelli, T., and Sitia, R. (2008). Protein quality control in the early secretory pathway. EMBO J. 27, 315–327. doi:10.1038/sj.emboj.7601974
Bai, X. C., Rajendra, E., Yang, G., Shi, Y., and Scheres, S. H. (2015). Sampling the conformational space of the catalytic subunit of human gamma-secretase. Elife 4, e11182. doi:10.7554/eLife.11182
Belden, W. J., and Barlowe, C. (2001). Distinct roles for the cytoplasmic tail sequences of Emp24p and Erv25p in transport between the endoplasmic reticulum and Golgi complex. J. Biol. Chem. 276, 43040–43048. doi:10.1074/jbc.M108113200
Belden, W. J., and Barlowe, C. (1996). Erv25p, a component of COPII-coated vesicles, forms a complex with Emp24p that is required for efficient endoplasmic reticulum to Golgi transport. J. Biol. Chem. 271, 26939–26946. doi:10.1074/jbc.271.43.26939
Bethune, J., Kol, M., Hoffmann, J., Reckmann, I., Brugger, B., and Wieland, F. (2006). Coatomer, the coat protein of COPI transport vesicles, discriminates endoplasmic reticulum residents from p24 proteins. Mol. Cell Biol. 26, 8011–8021. doi:10.1128/MCB.01055-06
Blomen, V. A., Majek, P., Jae, L. T., Bigenzahn, J. W., Nieuwenhuis, J., Staring, J., et al. (2015). Gene essentiality and synthetic lethality in haploid human cells. Science 350, 1092–1096. doi:10.1126/science.aac7557
Blum, R., and Lepier, A. (2008). The luminal domain of p23 (Tmp21) plays a critical role in p23 cell surface trafficking. Traffic 9, 1530–1550. doi:10.1111/j.1600-0854.2008.00784.x
Bremser, M., Nickel, W., Schweikert, M., Ravazzola, M., Amherdt, M., Hughes, C. A., et al. (1999). Coupling of coat assembly and vesicle budding to packaging of putative cargo receptors. Cell 96, 495–506. doi:10.1016/s0092-8674(00)80654-6
Breuza, L., Halbeisen, R., Jeno, P., Otte, S., Barlowe, C., Hong, W., et al. (2004). Proteomics of endoplasmic reticulum-Golgi intermediate compartment (ERGIC) membranes from brefeldin A-treated HepG2 cells identifies ERGIC-32, a new cycling protein that interacts with human Erv46. J. Biol. Chem. 279, 47242–47253. doi:10.1074/jbc.M406644200
Brugger, B., Sandhoff, R., Wegehingel, S., Gorgas, K., Malsam, J., Helms, J. B., et al. (2000). Evidence for segregation of sphingomyelin and cholesterol during formation of COPI-coated vesicles. J. Cell Biol. 151, 507–518. doi:10.1083/jcb.151.3.507
Buechling, T., Chaudhary, V., Spirohn, K., Weiss, M., and Boutros, M. (2011). p24 proteins are required for secretion of Wnt ligands. EMBO Rep. 12, 1265–1272. doi:10.1038/embor.2011.212
Chen, F., Hasegawa, H., Schmitt-Ulms, G., Kawarai, T., Bohm, C., Katayama, T., et al. (2006). TMP21 is a presenilin complex component that modulates gamma-secretase but not epsilon-secretase activity. Nature 440, 1208–1212. doi:10.1038/nature04667
Ciufo, L. F., and Boyd, A. (2000). Identification of a lumenal sequence specifying the assembly of Emp24p into p24 complexes in the yeast secretory pathway. J. Biol. Chem. 275, 8382–8388. doi:10.1074/jbc.275.12.8382
Connolly, D. J., O'neill, L. A., and Mcgettrick, A. F. (2013). The GOLD domain-containing protein TMED1 is involved in interleukin-33 signaling. J. Biol. Chem. 288, 5616–5623. doi:10.1074/jbc.M112.403899
Contreras, F. X., Ernst, A. M., Haberkant, P., Bjorkholm, P., Lindahl, E., Gonen, B., et al. (2012). Molecular recognition of a single sphingolipid species by a protein's transmembrane domain. Nature 481, 525–529. doi:10.1038/nature10742
Contreras, I., Ortiz-Zapater, E., and Aniento, F. (2004a). Sorting signals in the cytosolic tail of membrane proteins involved in the interaction with plant ARF1 and coatomer. Plant J. 38, 685–698. doi:10.1111/j.1365-313X.2004.02075.x
Contreras, I., Yang, Y., Robinson, D. G., and Aniento, F. (2004b). Sorting signals in the cytosolic tail of plant p24 proteins involved in the interaction with the COPII coat. Plant Cell Physiol. 45, 1779–1786. doi:10.1093/pcp/pch200
D'arcangelo, J. G., Crissman, J., Pagant, S., Copic, A., Latham, C. F., Snapp, E. L., et al. (2015). Traffic of p24 proteins and COPII coat composition mutually influence membrane scaffolding. Curr. Biol. 25, 1296–1305. doi:10.1016/j.cub.2015.03.029
Delorme-Axford, E., Morosky, S., Bomberger, J., Stolz, D. B., Jackson, W. T., and Coyne, C. B. (2014). BPIFB3 regulates autophagy and coxsackievirus B replication through a noncanonical pathway independent of the core initiation machinery. mBio 5, e02147. doi:10.1128/mBio.02147-14
Denzel, A., Otto, F., Girod, A., Pepperkok, R., Watson, R., Rosewell, I., et al. (2000). The p24 family member p23 is required for early embryonic development. Curr. Biol. 10, 55–58. doi:10.1016/s0960-9822(99)00266-3
Di Minin, G., Holzner, M., Grison, A., Dumeau, C. E., Chan, W., Monfort, A., et al. (2022). TMED2 binding restricts SMO to the ER and Golgi compartments. PLoS Biol. 20, e3001596. doi:10.1371/journal.pbio.3001596
Dolcini, V., Dunys, J., Sevalle, J., Chen, F., Guillot-Sestier, M. V., St George-Hyslop, P., et al. (2008). TMP21 regulates Abeta production but does not affect caspase-3, p53, and neprilysin. Biochem. Biophys. Res. Commun. 371, 69–74. doi:10.1016/j.bbrc.2008.03.151
Dominguez, M., Dejgaard, K., Fullekrug, J., Dahan, S., Fazel, A., Paccaud, J. P., et al. (1998). gp25L/emp24/p24 protein family members of the cis-Golgi network bind both COP I and II coatomer. J. Cell Biol. 140, 751–765. doi:10.1083/jcb.140.4.751
Duquet, A., Melotti, A., Mishra, S., Malerba, M., Seth, C., Conod, A., et al. (2014). A novel genome-wide in vivo screen for metastatic suppressors in human colon cancer identifies the positive WNT-TCF pathway modulators TMED3 and SOX12. EMBO Mol. Med. 6, 882–901. doi:10.15252/emmm.201303799
Dvela-Levitt, M., Kost-Alimova, M., Emani, M., Kohnert, E., Thompson, R., Sidhom, E. H., et al. (2019). Small molecule targets TMED9 and promotes lysosomal degradation to reverse proteinopathy. Cell 178, 521–535. doi:10.1016/j.cell.2019.07.002
Elrod-Erickson, M. J., and Kaiser, C. A. (1996). Genes that control the fidelity of endoplasmic reticulum to Golgi transport identified as suppressors of vesicle budding mutations. Mol. Biol. Cell 7, 1043–1058. doi:10.1091/mbc.7.7.1043
Emery, G., Rojo, M., and Gruenberg, J. (2000). Coupled transport of p24 family members. J. Cell Sci. 113 (13), 2507–2516. doi:10.1242/jcs.113.13.2507
Evans, A. S., Lennemann, N. J., and Coyne, C. B. (2021). BPIFB3 interacts with ARFGAP1 and TMED9 to regulate non-canonical autophagy and RNA virus infection. J. Cell Sci. 134, jcs251835. doi:10.1242/jcs.251835
Feneberg, E., Gordon, D., Thompson, A. G., Finelli, M. J., Dafinca, R., Candalija, A., et al. (2020). An ALS-linked mutation in TDP-43 disrupts normal protein interactions in the motor neuron response to oxidative stress. Neurobiol. Dis. 144, 105050. doi:10.1016/j.nbd.2020.105050
Fujita, M., Watanabe, R., Jaensch, N., Romanova-Michaelides, M., Satoh, T., Kato, M., et al. (2011). Sorting of GPI-anchored proteins into ER exit sites by p24 proteins is dependent on remodeled GPI. J. Cell Biol. 194, 61–75. doi:10.1083/jcb.201012074
Fullekrug, J., Suganuma, T., Tang, B. L., Hong, W., Storrie, B., and Nilsson, T. (1999). Localization and recycling of gp27 (hp24gamma3): Complex formation with other p24 family members. Mol. Biol. Cell 10, 1939–1955. doi:10.1091/mbc.10.6.1939
Gao, W., Zhang, Z. W., Wang, H. Y., Li, X. D., Peng, W. T., Guan, H. Y., et al. (2022). TMED2/9/10 serve as biomarkers for poor prognosis in head and neck squamous carcinoma. Front. Genet. 13, 895281. doi:10.3389/fgene.2022.895281
Goga, A., Yagabasan, B., Herrmanns, K., Godbersen, S., Silva, P. N., Denzler, R., et al. (2021). miR-802 regulates Paneth cell function and enterocyte differentiation in the mouse small intestine. Nat. Commun. 12, 3339. doi:10.1038/s41467-021-23298-3
Han, G. H., Yun, H., Chung, J. Y., Kim, J. H., and Cho, H. (2022). TMED9 expression level as a biomarker of epithelial ovarian cancer progression and prognosis. Cancer Genomics Proteomics 19, 692–702. doi:10.21873/cgp.20352
Hasegawa, H., Liu, L., and Nishimura, M. (2010). Dilysine retrieval signal-containing p24 proteins collaborate in inhibiting gamma-cleavage of amyloid precursor protein. J. Neurochem. 115, 771–781. doi:10.1111/j.1471-4159.2010.06977.x
Hou, W., Gupta, S., Beauchamp, M. C., Yuan, L., and Jerome-Majewska, L. A. (2017). Non-alcoholic fatty liver disease in mice with heterozygous mutation in TMED2. PLoS One 12, e0182995. doi:10.1371/journal.pone.0182995
Jenne, N., Frey, K., Brugger, B., and Wieland, F. T. (2002). Oligomeric state and stoichiometry of p24 proteins in the early secretory pathway. J. Biol. Chem. 277, 46504–46511. doi:10.1074/jbc.M206989200
Jerome-Majewska, L. A., Achkar, T., Luo, L., Lupu, F., and Lacy, E. (2010). The trafficking protein Tmed2/p24beta(1) is required for morphogenesis of the mouse embryo and placenta. Dev. Biol. 341, 154–166. doi:10.1016/j.ydbio.2010.02.019
Ju, G., Xu, C., Zeng, K., Zhou, T., and Zang, L. (2021). High expression of transmembrane P24 trafficking protein 9 predicts poor prognosis in breast carcinoma. Bioengineered 12, 8965–8979. doi:10.1080/21655979.2021.1990673
Jumper, J., Evans, R., Pritzel, A., Green, T., Figurnov, M., Ronneberger, O., et al. (2021). Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589. doi:10.1038/s41586-021-03819-2
Kakuta, S., Yamaguchi, J., Suzuki, C., Sasaki, M., Kazuno, S., and Uchiyama, Y. (2017). Small GTPase Rab1B is associated with ATG9A vesicles and regulates autophagosome formation. FASEB J. 31, 3757–3773. doi:10.1096/fj.201601052R
Lavoie, C., Paiement, J., Dominguez, M., Roy, L., Dahan, S., Gushue, J. N., et al. (1999). Roles for alpha(2)p24 and COPI in endoplasmic reticulum cargo exit site formation. J. Cell Biol. 146, 285–299. doi:10.1083/jcb.146.2.285
Li, S., Yan, R., Xu, J., Zhao, S., Ma, X., Sun, Q., et al. (2022). A new type of ERGIC-ERES membrane contact mediated by TMED9 and SEC12 is required for autophagosome biogenesis. Cell Res. 32, 119–138. doi:10.1038/s41422-021-00563-0
Liaunardy-Jopeace, A., Bryant, C. E., and Gay, N. J. (2014). The COP II adaptor protein TMED7 is required to initiate and mediate the delivery of TLR4 to the plasma membrane. Sci. Signal 7, ra70. doi:10.1126/scisignal.2005275
Liew, F. Y., Pitman, N. I., and Mcinnes, I. B. (2010). Disease-associated functions of IL-33: The new kid in the IL-1 family. Nat. Rev. Immunol. 10, 103–110. doi:10.1038/nri2692
Ma, W., Goldberg, E., and Goldberg, J. (2017). ER retention is imposed by COPII protein sorting and attenuated by 4-phenylbutyrate. Elife 6, e26624. doi:10.7554/eLife.26624
Ma, W., and Goldberg, J. (2013). Rules for the recognition of dilysine retrieval motifs by coatomer. EMBO J. 32, 926–937. doi:10.1038/emboj.2013.41
Marzioch, M., Henthorn, D. C., Herrmann, J. M., Wilson, R., Thomas, D. Y., Bergeron, J. J., et al. (1999). Erp1p and Erp2p, partners for Emp24p and Erv25p in a yeast p24 complex. Mol. Biol. Cell 10, 1923–1938. doi:10.1091/mbc.10.6.1923
Mishra, S., Bernal, C., Silvano, M., Anand, S., and Ruiz, I. a. A. (2019). The protein secretion modulator TMED9 drives CNIH4/TGFα/GLI signaling opposing TMED3-WNT-TCF to promote colon cancer metastases. Oncogene 38, 5817–5837. doi:10.1038/s41388-019-0845-z
Mitrovic, S., Ben-Tekaya, H., Koegler, E., Gruenberg, J., and Hauri, H. P. (2008). The cargo receptors Surf4, endoplasmic reticulum-Golgi intermediate compartment (ERGIC)-53, and p25 are required to maintain the architecture of ERGIC and Golgi. Mol. Biol. Cell 19, 1976–1990. doi:10.1091/mbc.E07-10-0989
Mota, D., Cardoso, I. A., Mori, R. M., Batista, M. R. B., Basso, L. G. M., Nonato, M. C., et al. (2022). Structural and thermodynamic analyses of human TMED1 (p24γ1) Golgi dynamics. Biochimie 192, 72–82. doi:10.1016/j.biochi.2021.10.002
Muniz, M., Nuoffer, C., Hauri, H. P., and Riezman, H. (2000). The Emp24 complex recruits a specific cargo molecule into endoplasmic reticulum-derived vesicles. J. Cell Biol. 148, 925–930. doi:10.1083/jcb.148.5.925
Muppirala, M., Gupta, V., and Swarup, G. (2013). Emerging role of tyrosine phosphatase, TCPTP, in the organelles of the early secretory pathway. Biochim. Biophys. Acta 1833, 1125–1132. doi:10.1016/j.bbamcr.2013.01.004
Muppirala, M., Gupta, V., and Swarup, G. (2011). Syntaxin 17 cycles between the ER and ERGIC and is required to maintain the architecture of ERGIC and Golgi. Biol. Cell 103, 333–350. doi:10.1042/BC20110006
Nagae, M., Hirata, T., Morita-Matsumoto, K., Theiler, R., Fujita, M., Kinoshita, T., et al. (2016). 3D structure and interaction of p24β and p24δ golgi dynamics domains: Implication for p24 complex formation and cargo transport. J. Mol. Biol. 428, 4087–4099. doi:10.1016/j.jmb.2016.08.023
Nagae, M., Liebschner, D., Yamada, Y., Morita-Matsumoto, K., Matsugaki, N., Senda, T., et al. (2017). Crystallographic analysis of murine p24γ2 Golgi dynamics domain. Proteins 85, 764–770. doi:10.1002/prot.25242
Nakano, N., Tsuchiya, Y., Kako, K., Umezaki, K., Sano, K., Ikeno, S., et al. (2017). TMED10 protein interferes with transforming growth factor (TGF)-beta signaling by disrupting TGF-beta receptor complex formation. J. Biol. Chem. 292, 4099–4112. doi:10.1074/jbc.M116.769109
Namekawa, M., Muriel, M. P., Janer, A., Latouche, M., Dauphin, A., Debeir, T., et al. (2007). Mutations in the SPG3A gene encoding the GTPase atlastin interfere with vesicle trafficking in the ER/Golgi interface and Golgi morphogenesis. Mol. Cell Neurosci. 35, 1–13. doi:10.1016/j.mcn.2007.01.012
Pannwitt, S., Stangl, M., and Schneider, D. (2019). Lipid binding controls dimerization of the coat protein p24 transmembrane helix. Biophys. J. 117, 1554–1562. doi:10.1016/j.bpj.2019.09.021
Park, H., Seo, S. K., Sim, J. R., Hwang, S. J., Kim, Y. J., Shin, D. H., et al. (2022). TMED3 complex mediates ER stress-associated secretion of CFTR, pendrin, and SARS-CoV-2 spike. Adv. Sci. (Weinh) 9, e2105320. doi:10.1002/advs.202105320
Pastor-Cantizano, N., Montesinos, J. C., Bernat-Silvestre, C., Marcote, M. J., and Aniento, F. (2016). p24 family proteins: key players in the regulation of trafficking along the secretory pathway. Protoplasma 253, 967–985. doi:10.1007/s00709-015-0858-6
Pradat, P. F., Dubourg, O., De Tapia, M., Di Scala, F., Di Scala, L., Lenglet, T., et al. (2012). Muscle gene expression is a marker of amyotrophic lateral sclerosis severity. Neurodegener. Dis. 9, 38–52. doi:10.1159/000329723
Redler, R. L., and Dokholyan, N. V. (2012). The complex molecular biology of amyotrophic lateral sclerosis (ALS). Prog. Mol. Biol. Transl. Sci. 107, 215–262. doi:10.1016/B978-0-12-385883-2.00002-3
Reid, E., Connell, J., Edwards, T. L., Duley, S., Brown, S. E., and Sanderson, C. M. (2005). The hereditary spastic paraplegia protein spastin interacts with the ESCRT-III complex-associated endosomal protein CHMP1B. Hum. Mol. Genet. 14, 19–38. doi:10.1093/hmg/ddi003
Satpute-Krishnan, P., Ajinkya, M., Bhat, S., Itakura, E., Hegde, R. S., and Lippincott-Schwartz, J. (2014). ER stress-induced clearance of misfolded GPI-anchored proteins via the secretory pathway. Cell 158, 522–533. doi:10.1016/j.cell.2014.06.026
Schimmoller, F., Singer-Kruger, B., Schroder, S., Kruger, U., Barlowe, C., and Riezman, H. (1995). The absence of Emp24p, a component of ER-derived COPII-coated vesicles, causes a defect in transport of selected proteins to the Golgi. EMBO J. 14, 1329–1339. doi:10.1002/j.1460-2075.1995.tb07119.x
Shevchenko, A., Keller, P., Scheiffele, P., Mann, M., and Simons, K. (1997). Identification of components of trans-Golgi network-derived transport vesicles and detergent-insoluble complexes by nanoelectrospray tandem mass spectrometry. Electrophoresis 18, 2591–2600. doi:10.1002/elps.1150181415
Shin, J. H., Park, S. J., Jo, D. S., Park, N. Y., Kim, J. B., Bae, J. E., et al. (2019). Down-regulated TMED10 in Alzheimer disease induces autophagy via ATG4B activation. Autophagy 15, 1495–1505. doi:10.1080/15548627.2019.1586249
Sikorska, N., Lemus, L., Aguilera-Romero, A., Manzano-Lopez, J., Riezman, H., Muniz, M., et al. (2016). Limited ER quality control for GPI-anchored proteins. J. Cell Biol. 213, 693–704. doi:10.1083/jcb.201602010
Sohn, K., Orci, L., Ravazzola, M., Amherdt, M., Bremser, M., Lottspeich, F., et al. (1996). A major transmembrane protein of Golgi-derived COPI-coated vesicles involved in coatomer binding. J. Cell Biol. 135, 1239–1248. doi:10.1083/jcb.135.5.1239
Stamnes, M. A., Craighead, M. W., Hoe, M. H., Lampen, N., Geromanos, S., Tempst, P., et al. (1995). An integral membrane component of coatomer-coated transport vesicles defines a family of proteins involved in budding. Proc. Natl. Acad. Sci. U. S. A. 92, 8011–8015. doi:10.1073/pnas.92.17.8011
Strating, J. R., and Martens, G. J. (2009). The p24 family and selective transport processes at the ER-Golgi interface. Biol. Cell 101, 495–509. doi:10.1042/BC20080233
Strating, J. R., Van Bakel, N. H., Leunissen, J. A., and Martens, G. J. (2009). A comprehensive overview of the vertebrate p24 family: Identification of a novel tissue-specifically expressed member. Mol. Biol. Evol. 26, 1707–1714. doi:10.1093/molbev/msp099
Sun, Z., and Brodsky, J. L. (2019). Protein quality control in the secretory pathway. J. Cell Biol. 218, 3171–3187. doi:10.1083/jcb.201906047
Tashima, Y., Hirata, T., Maeda, Y., Murakami, Y., and Kinoshita, T. (2022). Differential use of p24 family members as cargo receptors for the transport of glycosylphosphatidylinositol-anchored proteins and Wnt1. J. Biochem. 171, 75–83. doi:10.1093/jb/mvab108
Teasdale, R. D., and Jackson, M. R. (1996). Signal-mediated sorting of membrane proteins between the endoplasmic reticulum and the golgi apparatus. Annu. Rev. Cell Dev. Biol. 12, 27–54. doi:10.1146/annurev.cellbio.12.1.27
Theiler, R., Fujita, M., Nagae, M., Yamaguchi, Y., Maeda, Y., and Kinoshita, T. (2014). The α-helical region in p24γ2 subunit of p24 protein cargo receptor is pivotal for the recognition and transport of glycosylphosphatidylinositol-anchored proteins. J. Biol. Chem. 289, 16835–16843. doi:10.1074/jbc.M114.568311
Uniprot, C. (2021). UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 49, D480–D489. doi:10.1093/nar/gkaa1100
Varadi, M., Anyango, S., Deshpande, M., Nair, S., Natassia, C., Yordanova, G., et al. (2022). AlphaFold protein structure database: Massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 50, D439–D444. doi:10.1093/nar/gkab1061
Wada, I., Rindress, D., Cameron, P. H., Ou, W. J., Doherty, J. J., Louvard, D., et al. (1991). SSR alpha and associated calnexin are major calcium binding proteins of the endoplasmic reticulum membrane. J. Biol. Chem. 266, 19599–19610. doi:10.1016/s0021-9258(18)55036-5
Wang, X., Yang, R., Jadhao, S. B., Yu, D., Hu, H., Glynn-Cunningham, N., et al. (2012). Transmembrane emp24 protein transport domain 6 is selectively expressed in pancreatic islets and implicated in insulin secretion and diabetes. Pancreas 41, 10–14. doi:10.1097/MPA.0b013e318223c7e4
Weissman, J. T., Plutner, H., and Balch, W. E. (2001). The mammalian guanine nucleotide exchange factor mSec12 is essential for activation of the Sar1 GTPase directing endoplasmic reticulum export. Traffic 2, 465–475. doi:10.1034/j.1600-0854.2001.20704.x
Wendeler, M. W., Paccaud, J. P., and Hauri, H. P. (2007). Role of Sec24 isoforms in selective export of membrane proteins from the endoplasmic reticulum. EMBO Rep. 8, 258–264. doi:10.1038/sj.embor.7400893
Yang, Y. C., Chien, M. H., Lai, T. C., Tung, M. C., Jan, Y. H., Chang, W. M., et al. (2021). Proteomics-based identification of TMED9 is linked to vascular invasion and poor prognoses in patients with hepatocellular carcinoma. J. Biomed. Sci. 28, 29. doi:10.1186/s12929-021-00727-5
Yang, Z., Sun, Q., Guo, J., Wang, S., Song, G., Liu, W., et al. (2019). GRSF1-mediated MIR-G-1 promotes malignant behavior and nuclear autophagy by directly upregulating TMED5 and LMNB1 in cervical cancer cells. Autophagy 15, 668–685. doi:10.1080/15548627.2018.1539590
Zavodszky, E., and Hegde, R. S. (2019). Misfolded GPI-anchored proteins are escorted through the secretory pathway by ER-derived factors. Elife 8, e46740. doi:10.7554/eLife.46740
Zhang, L., and Volchuk, A. (2010). p24 family type 1 transmembrane proteins are required for insulin biosynthesis and secretion in pancreatic beta-cells. FEBS Lett. 584, 2298–2304. doi:10.1016/j.febslet.2010.03.041
Zhang, M., Liu, L., Lin, X., Wang, Y., Li, Y., Guo, Q., et al. (2020a). A translocation pathway for vesicle-mediated unconventional protein secretion. Cell 181, 637–652. doi:10.1016/j.cell.2020.03.031
Zhang, X., Luo, Y., and Li, Q. (2020b). TMED3 promotes proliferation and migration in breast cancer cells by activating wnt/β-catenin signaling. Onco Targets Ther. 13, 5819–5830. doi:10.2147/OTT.S250766
Zhang, X., Wu, Y., Cai, F., Liu, S., Bromley-Brits, K., Xia, K., et al. (2018). A novel alzheimer-associated SNP in Tmp21 increases amyloidogenesis. Mol. Neurobiol. 55, 1862–1870. doi:10.1007/s12035-017-0459-9
Keywords: transmembrane emp24 domain, p24 family, cargo receptor, autophagy, secretory pathway homeostasis, COP Coatomer, endoplasmic reticulum, Golgi
Citation: Roberts BS and Satpute-Krishnan P (2023) The many hats of transmembrane emp24 domain protein TMED9 in secretory pathway homeostasis. Front. Cell Dev. Biol. 10:1096899. doi: 10.3389/fcell.2022.1096899
Received: 12 November 2022; Accepted: 29 December 2022;
Published: 16 January 2023.
Edited by:
Francesca Zappa, Altos labs, United StatesReviewed by:
Tiziana Anelli, San Raffaele Scientific Institute (IRCCS), ItalyCopyright © 2023 Roberts and Satpute-Krishnan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Prasanna Satpute-Krishnan, cHJhc2FubmEua3Jpc2huYW5AdXN1aHMuZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.