- Department of Biological Chemistry and Pharmacology, College of Medicine, The Ohio State University, Columbus, OH, United States
In the brain, primary cilia are found on most, if not all, central neurons. The importance of neuronal cilia is underscored by the fact that human diseases caused by primary cilia dysfunction, which are known as ciliopathies, are associated with neuropathologies, including neuropsychiatric disorders and learning and memory deficits. Neuronal cilia are enriched for certain G protein-coupled receptors and their downstream effectors, suggesting they sense and respond to neuromodulators in the extracellular milieu. GPCR ciliary localization is disrupted in neurons from mouse models of the ciliopathy Bardet-Biedl syndrome, with GPCRs failing to localize to cilia, indicating the Bardet-Biedl syndrome proteins are required for trafficking of G protein-coupled receptors into neuronal cilia. Yet, dopamine receptor 1 accumulates in cilia in the absence of Bardet-Biedl syndrome proteins, suggesting Bardet-Biedl syndrome proteins are required for normal ciliary import and export. To further explore the roles of the Bardet-Biedl syndrome proteins in neuronal cilia, we examined localization of ciliary signaling proteins in a new constitutive Bbs1 knockout mouse model. Interestingly, we find that two additional ciliary G protein-coupled receptors (Gpr161 and Gpr19) abnormally accumulate in cilia on Bardet-Biedl syndrome neurons. In addition, we find that the GPCR signaling protein β-arrestin accumulates in a subset of cilia in the brain, suggesting the presence of additional unidentified ciliary G protein-coupled receptors. These results confirm the importance of the Bardet-Biedl syndrome proteins in establishing ciliary GPCR pathways and indicate that loss of Bbs1 leads to complex changes in the localization of signaling proteins in the brain.
Introduction
Primary cilia are signaling organelles that mediate specialized signal transduction pathways critical for normal development and cellular homeostasis (Anvarian et al., 2019). These pathways are established and maintained by coordinating the trafficking of proteins into and out of cilia (Nachury and Mick, 2019). The Sonic hedgehog (Shh) pathway is a well characterized example of the functional importance of dynamic localization of ciliary signaling components. In the absence of Shh, the Shh receptor Patched (Ptch) and orphan GPCR Gpr161 are enriched in cilia and maintain repression of the pathway (Rohatgi et al., 2007; Mukhopadhyay et al., 2013). Shh binding results in Ptch and Gpr161 removal from cilia and concomitant ciliary accumulation of the GPCR-like Smoothened (Smo), which is the major transducer of Shh signaling, leading to pathway activation (Corbit et al., 2005; Rohatgi et al., 2007). β-arrestins, scaffolding proteins that regulate GPCR signaling and internalization, are recruited to activated Gpr161 and mediate its export from cilia (Pal et al., 2016). Overall, the dynamic localization of Shh signaling components to cilia is critical for proper Shh signaling and defects in Shh signaling are associated with ciliopathies.
Ciliopathies are associated with neuropathologies, such as hyperphagia-induced obesity, neuropsychiatric disorders, and learning and memory deficits (Suciu and Caspary, 2021). Certain GPCRs are enriched in neuronal cilia, including somatostatin receptor 3 (Sstr3) (Handel et al., 1999), serotonin receptor 6 (Htr6) (Brailov et al., 2000), melanin-concentrating hormone receptor 1 (Mchr1) (Berbari et al., 2008a), dopamine receptor 1 (D1) (Domire et al., 2011), Neuropeptide Y receptor 2 (NPY2) (Loktev and Jackson, 2013), kisspeptin receptor 1 (Kiss1r) (Koemeter-Cox et al., 2014), and melanocortin four receptor (MC4R) (Siljee et al., 2018). Neuronal cilia also contain the GPCR effectors type 3 adenylyl cyclase (AC3) (Bishop et al., 2007) and β-arrestin (Green et al., 2016), suggesting that GPCRs signal on the ciliary membrane. Indeed, numerous studies have now shown that GPCR ciliary localization is critical for normal signaling. Ablation of cilia on Kiss1r-positive neurons reduces agonist-mediated increases in firing rate in the mouse hypothalamus (Koemeter-Cox et al., 2014). Disruption of Sstr3 ciliary signaling results in increased neuronal excitability in rat neocortical neurons (Tereshko et al., 2021). Disruption of ciliary Mchr1 leads to decreased agonist-mediated cAMP and extracellular signal-regulated kinase signaling in mouse hypothalamic neurons (Hsiao et al., 2021). Inhibition of MC4R ciliary signaling in the mouse hypothalamus results in hyperphagia-induced obesity (Wang et al., 2021). Mice with disrupted D1 ciliary localization have a reduction in D1-mediated signaling in the striatum that is associated with a decrease in locomotor activity and obesity (Stubbs et al., 2022). Recently, synapses between serotonergic axons and primary cilia of hippocampal CA1 neurons have been described in the mouse brain (Sheu et al., 2022). Importantly, serotonin release stimulates ciliary Htr6 and activates a non-canonical signaling pathway to increase chromatin accessibility (Sheu et al., 2022). Overall, ciliary GPCR signaling appears to be important for normal neuronal function.
GPCR localization to neuronal cilia is dynamic and regulated by environmental cues. For example, D1 ciliary localization is increased in response to an increase in cyclic AMP levels and decreased in response to agonist treatment (Domire et al., 2011). NPY2 and Sstr3 are also exported from neuronal cilia in response to agonist (Loktev and Jackson, 2013; Green et al., 2016). The proteins disrupted in the ciliopathy Bardet-Biedl syndrome (BBS) are important regulators of ciliary localization of GPCRs in the brain. BBS is a heterogeneous autosomal recessive disorder characterized by a wide range of phenotypes, including obesity, behavioral disturbances, and cognitive deficits (Forsythe and Beales, 2013). A group of BBS proteins (BBS1, 2, 4, 5, 7, 8, 9, and 18) form a complex called the BBSome (Nachury et al., 2007; Loktev et al., 2008). The BBSome is required for ciliary localization of Sstr3, Mchr1, and NPY2 (Berbari et al., 2008b; Loktev and Jackson, 2013). Yet, D1 accumulates in neuronal cilia in BBSome mutant mice (Domire et al., 2011; Zhang et al., 2013) and does not undergo agonist-mediated ciliary export on BBSome mutant neurons (Domire et al., 2011). Taken together, these results suggest that the BBSome is required for trafficking of some GPCRs into cilia and others out of cilia in the brain.
Previous studies characterizing the impact of Bbs1 loss in mice utilized a model where the most common human BBS mutation (M390R) was knocked into the mouse gene (Davis et al., 2007). Although M390R knock-in mice exhibit phenotypes consistent with mice lacking a BBSome subunit, we decided to generate a constitutive Bbs1 knockout mouse model, as loss of Bbs1 protein is known to disrupt BBSome formation and function (Guo et al., 2016). We then performed an analysis of the consequences of Bbs1 loss on the ciliary localization of specific signaling proteins in the brain.
Results
Loss of Bbs1 in the mouse brain impacts GPCR ciliary localization
The conditional Bbs1 knockout allele has loxP sites flanking exon three and Bbs1 mRNA is lost following recombination (Carter et al., 2012). We crossed conditional Bbs1 knockout mice with transgenic mice expressing Cre recombinase under control of the CMV promoter, which is expressed in all tissues, in order to generate mice carrying a germline deletion of the Bbs1 allele. We then intercrossed mice heterozygous for a wildtype and null allele (Bbs1+/−) to generate wildtype (Bbs1+/+) and constitutive knockout (Bbs1−/−) animals. To test whether loss of Bbs1 impacts ciliary GPCR localization in the brain, we colabeled brain sections from adult Bbs1+/+ and Bbs1−/− mice with antibodies against Sstr3, Mchr1, or D1 and an antibody against AC3, which serves as a marker for neuronal cilia (Bishop et al., 2007). In Bbs1+/+ sections we detected ciliary localization of both Sstr3 and Mchr1 in numerous brain regions, with especially abundant labeling in the CA3 region of the hippocampus and nucleus accumbens, respectively (Figures 1A, B). Similar to other BBSome mutant mice (Berbari et al., 2008b), we failed to detect ciliary localization of either Sstr3 or Mchr1in brain sections from Bbs1−/− mice (Figures 1A, B). The lengths of AC3-positive cilia were similar between both genotypes, indicating that loss of ciliary localization is not due to overt ciliary defects in Bbs1−/− mice (Table 1).
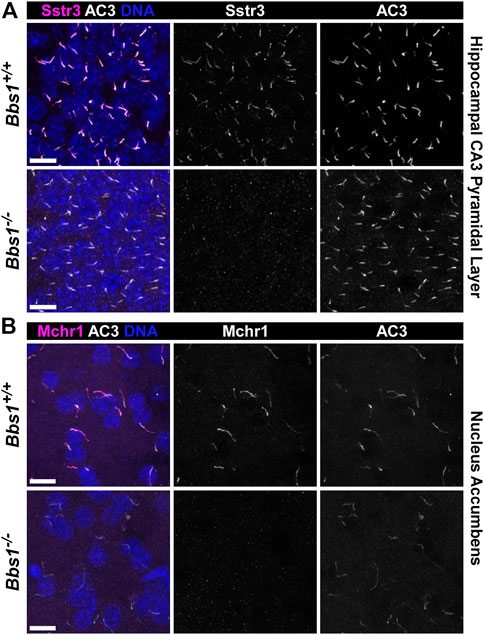
FIGURE 1. Bbs1−/− mice lack Sstr3 and Mchr1 ciliary localization in the brain (A) Representative images of the pyramidal layer of the hippocampal CA3 region from adult Bbs1+/+ (upper row) and Bbs1−/− (lower row) mice colabeled for Sstr3 (magenta) and AC3 (white). Sstr3 localized to 74% (623/846; n = 3 animals) of AC3-positive cilia in Bbs1+/+ sections and 0% (0/417; n = 3 animals) of AC3-positive cilia in Bbs1−/− sections (B) Representative images of the nucleus accumbens from adult Bbs1+/+ (upper row) and Bbs1−/− (lower row) mice colabeled for Mchr1 (magenta) and AC3 (white). Mchr1 localized to 72% (675/938; n = 3 animals) of AC3-positive cilia in Bbs1+/+ sections and 0% (0/471; n = 3 animals) of AC3-positive cilia in Bbs1−/− sections. Nuclei are stained with DRAQ5 (blue). Scale bars represent 10 µm.
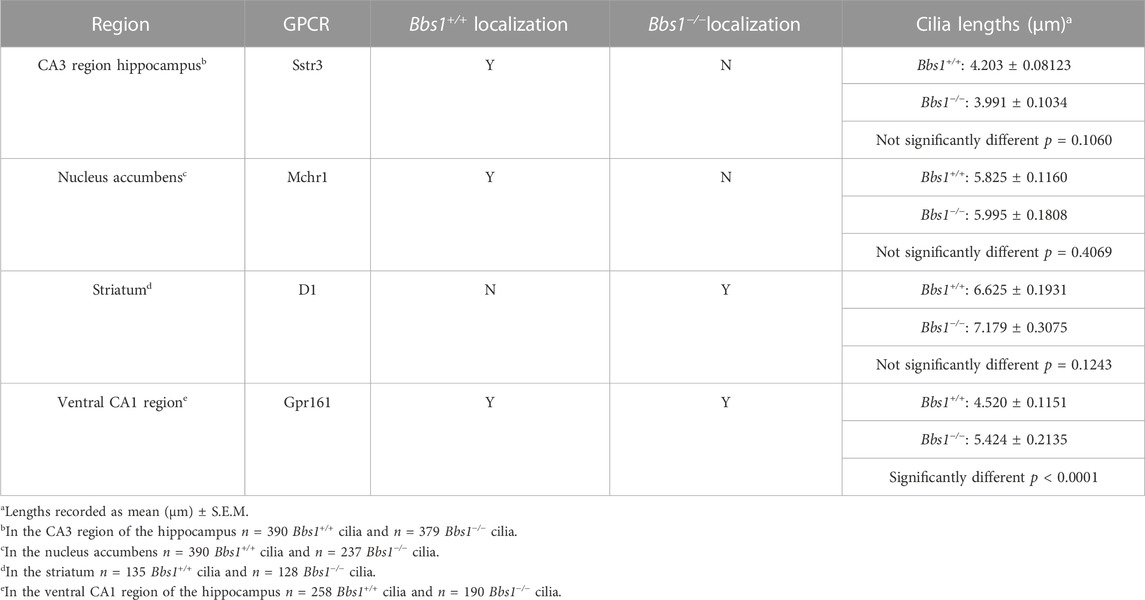
TABLE 1. Summary of GPCR ciliary localization and ciliary length in Bbs1+/+ and Bbs1−/− brain regions.
D1 ciliary localization was not detected in Bbs1+/+ sections. However, in Bbs1−/− sections we observed abundant D1 ciliary localization in several brain regions, including the striatum, amygdala, and olfactory tubercle (Figure 2A), similar to other BBSome mutant mice. Interestingly, we consistently observed a reduction in AC3 labeling in cilia in the striatum of Bbs1−/− mice compared to Bbs1+/+ mice. Quantification of mean ciliary AC3 fluorescence in D1-negative and D1-positive cilia in the striatum of Bbs1−/− sections confirmed that AC3 signal is reduced in cilia that are enriched for D1 (t = 2.271, df = 286, p < 0.05) (Figure 2B). The lengths of AC3-positive cilia were similar between both genotypes (Table 1), indicating that the reduction in AC3 fluorescence is not due to differences in cilia length. We previously reported the same observation after conditionally disrupting Bbs1 in D1-expressing neurons and proposed that the ciliary accumulation of D1 may be preventing AC3 ciliary localization or interfering with binding of the AC3 antibody (Stubbs et al., 2022). Nevertheless, together these results are consistent with findings in other BBSome mutant mice and suggest that the BBSome is required for ciliary entry or retention of Sstr3 and Mchr1 but is required for ciliary export of D1.
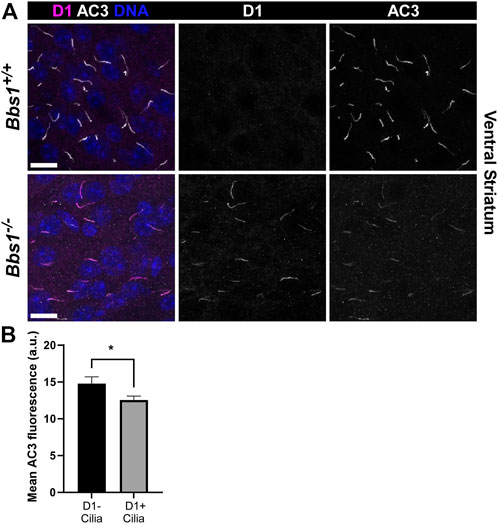
FIGURE 2. D1 accumulates in cilia in the brains of Bbs1−/− mice and is associated with a reduction in AC3 ciliary signal (A) Representative images of the ventral striatum from adult Bbs1+/+ (upper row) and Bbs1−/− (lower row) mice colabeled for D1 (magenta) and AC3 (white). D1 localized to 0% (0/739; n = 3 animals) of AC3-positive cilia in Bbs1+/+ sections and 64% (279/434; n = 3 animals) of AC3-positive cilia in Bbs1−/− sections. Nuclei are stained with DRAQ5 (blue). Scale bars represent 10 µm (B) Quantification of mean ciliary AC3 fluorescence in D1-negative (n = 100) and D1-positive (n = 194) cilia in Bbs1−/− sections. Note that the AC3 signal is significantly lower in D1-positive cilia. Values are expressed as mean ± SEM. *p < 0.05.
Gpr161 and Gpr19 accumulate in cilia on Bbs1 knockout neurons
Gpr161 ciliary localization is restricted to pyramidal neurons in the CA1/CA2 regions of the adult mouse hippocampus (Mukhopadhyay et al., 2013). To test whether loss of Bbs1 impacts Gpr161 ciliary localization in the brain, we colabeled brain sections from adult Bbs1+/+ and Bbs1−/− mice with antibodies against Gpr161 and AC3. In Bbs1+/+ sections Gpr161-positive cilia were barely detectable (Figure 3A). However, in Bbs1−/− sections Gpr161-positive cilia were much more pronounced (Figure 3A), suggesting that the BBSome is required for Gpr161 ciliary export. Quantification of Gpr161-positive cilia in Bbs1+/+ and Bbs1−/− sections confirmed that there are significantly more Gpr161-positive cilia in Bbs1−/− sections (t = 9.981, df = 27, p < 0.0001) (Figure 3B). Similar to D1 ciliary accumulation, we consistently observed a reduction in AC3 signal in Gpr161-positive cilia in Bbs1−/− sections. Quantification of mean ciliary AC3 fluorescence in Gpr161-negative and Gpr161-positive cilia in the hippocampal CA1 region of Bbs1−/− sections confirmed that AC3 signal is reduced in cilia that are enriched for Gpr161 (t = 2.779, df = 242, p < 0.01) (Figure 3C). Interestingly, quantification of the lengths of AC3-positive cilia in Bbs1+/+ and Bbs1−/− sections revealed that cilia are significantly longer in Bbs1−/− sections (Table 1), which may contribute to the reduction in AC3 fluorescence.
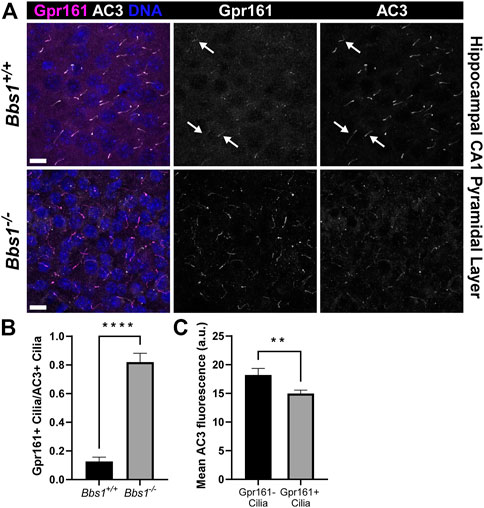
FIGURE 3. Gpr161 accumulates in cilia in the brains of Bbs1−/− mice and is associated with a reduction in AC3 ciliary signal (A) Representative images of the ventral CA1 region of the hippocampus from adult Bbs1+/+ (upper row) and Bbs1−/− (lower row) mice colabeled for Gpr161 (magenta) and AC3 (white). Note that Gpr161-positive cilia are barely detectable in the Bbs1+/+ section (indicated by arrows) but prominent in the Bbs1−/− section (middle panels). Nuclei are stained with DRAQ5 (blue). Scale bars represent 10 µm (B) Quantification of Gpr161-positive cilia relative to AC3-positive cilia reveals a significant increase in Gpr161-positive cilia in Bbs1−/− sections (889/1,093; n = 3 animals), compared to Bbs1+/+ sections (91/959; n = 3 animals) (C) Quantification of mean ciliary AC3 fluorescence in Gpr161-negative (n = 74) and Gpr161-positive (n = 170) cilia in Bbs1−/− sections. Note that the AC3 signal is significantly lower in Gpr161-positive cilia. Values are expressed as mean ± SEM. **p < 0.01 ****p < 0.0001.
To confirm our in vivo findings, we colabeled cultured Bbs1+/+ and Bbs1−/− hippocampal neurons with antibodies against Gpr161 and AC3. In Bbs1+/+ cultures, we rarely observed Gpr161-positive cilia. The proportion of Gpr161-positive cilia we observed was significantly lower than what has been previously published (Badgandi et al., 2017) and may reflect differences in culturing and/or labeling procedures. Nevertheless, in Bbs1−/− cultures Gpr161-positive cilia were noticeably more abundant (Figure 4A). Quantification of AC3-positive cilia relative to nuclei in Bbs1+/+ and Bbs1−/− cultures indicated that the proportion of cilia did not vary by genotype (t = 0.3262, df = 57, p = 0.74) (Figure 4B). However, quantification of Gpr161-positive cilia relative to AC3-positive cilia showed a significant increase in the proportion of Gpr161-positive cilia in Bbs1−/− cultures (t = 7.88, df = 57, p < 0.0001) (Figure 4C), providing further evidence that the BBSome is required for Gpr161 ciliary export. Our results are consistent with a previous study showing that Gpr161 accumulates in cilia on fibroblasts from mice homozygous for the M390R mutant Bbs1 allele (Yee et al., 2015). Gpr161 also accumulates in cilia on BBSome mutant cell lines (Goetz et al., 2017; Nozaki et al., 2018).
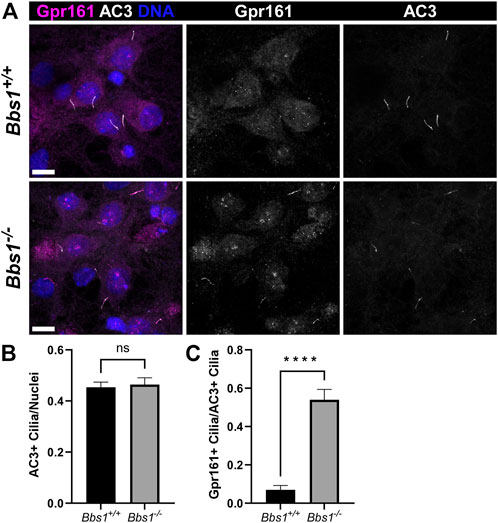
FIGURE 4. Gpr161 accumulates in cilia on Bbs1−/− hippocampal neurons (A) Representative images of cultured hippocampal neurons from Bbs1+/+ (upper row) and Bbs1−/− (lower row) mice colabeled for Gpr161 (magenta) and AC3 (white). Note that Gpr161 ciliary localization is more pronounced in the Bbs1−/− culture (middle panels). Nuclei are stained with DRAQ5 (blue). Scale bars represent 10 µm (B) Quantification of AC3-positive cilia relative to nuclei reveals a similar fraction between Bbs1+/+ (353/748; n = 3 animals) and Bbs1−/− (286/627 nuclei; n = 3 animals) cultures (C) Quantification of Gpr161-positive cilia relative to AC3-positive cilia reveals a significant increase in Gpr161-positive cilia in Bbs1−/− cultures (145/286; n = 3 animals), compared to Bbs1+/+ cultures (18/353; n = 3 animals). Values are expressed as mean ± SEM. ns = not significantly different, ****p < 0.0001.
Gpr19 is an orphan GPCR that also localizes to cilia on hippocampal neurons (Badgandi et al., 2017). As we were unable to detect Gpr19 in brain sections from adult Bbs1+/+ and Bbs1−/− mice using our labeling procedures, we tested the impact of Bbs1 loss on Gpr19 ciliary localization by colabeling cultured Bbs1+/+ and Bbs1−/− hippocampal neurons with antibodies against Gpr19 and AC3. Similar to Gpr161, we rarely observed Gpr19-positive cilia in Bbs1+/+ cultures but observed abundant Gpr19-positive cilia in Bbs1−/− cultures (Figure 5A). Quantification of AC3-positive cilia, nuclei, and Gpr19-positive cilia revealed similar proportions of cilia for each genotype (t = 1.645, df = 58, p = 0.1) (Figure 5B) but a significant increase in the proportion of Gpr19-positive cilia in Bbs1−/− cultures (t = 18.68, df = 58, p < 0.0001) (Figure 5C). Thus, these results suggest the BBSome is also required for Gpr19 ciliary export.
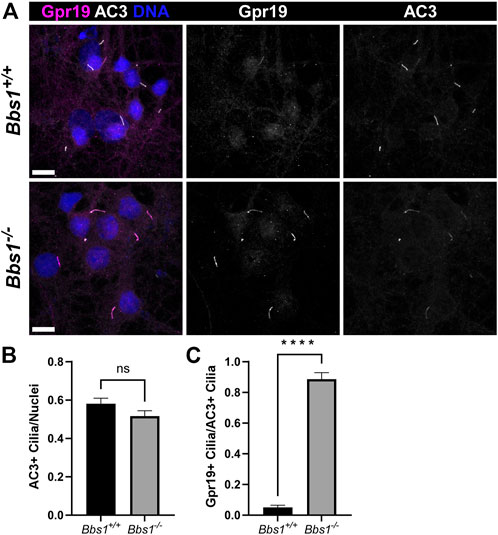
FIGURE 5. Gpr19 accumulates in cilia on Bbs1−/− hippocampal neurons (A) Representative images of cultured hippocampal neurons from Bbs1+/+ (upper row) and Bbs1−/− (lower row) mice colabeled for Gpr19 (magenta) and AC3 (white). Note that Gpr19 ciliary localization is more pronounced in the Bbs1−/− culture (middle panels). Nuclei are stained with DRAQ5 (blue). Scale bars represent 10 µm (B) Quantification of AC3-positive cilia relative to nuclei reveals a similar fraction between Bbs1+/+ (470/836; n = 3 animals) and Bbs1−/− (317/647; n = 3 animals) cultures (C) Quantification of Gpr19-positive cilia relative to AC3-positive cilia reveals a significant increase in Gpr19-positive cilia in Bbs1−/− cultures (270/317; n = 3 animals), compared to Bbs1+/+ cultures (20/470; n = 3 animals). Values are expressed as mean ± SEM. ns = not significantly different, ****p < 0.0001.
β-arrestin accumulates in subsets of cilia in the brains of Bbs1−/− mice
We previously showed that the GPCR signaling protein β-arrestin is recruited into neuronal cilia, ostensibly in response to ciliary receptor activation (Green et al., 2016). Further, β-arrestin 2 accumulates in cilia on a cell line lacking Bbs19 (Mick et al., 2015), suggesting that β-arrestin 2 requires BBS proteins for ciliary exit. To test whether loss of Bbs1 impacts β-arrestin localization to neuronal cilia, we colabeled brain sections from adult Bbs1+/+ and Bbs1−/− mice with antibodies against β-arrestin and AC3. We did not observe β-arrestin-positive cilia in Bbs1+/+ sections but observed abundant β-arrestin-positive cilia in Bbs1−/− sections (Figures 6A,B). β-arrestin-positive cilia were most abundant in the amygdala (Figure 6A) and on pyramidal neurons in the CA3 region of the hippocampus (Figure 6B) but were also found in numerous other brain regions, including the hypothalamus, thalamus, and cortex. β-arrestin did not accumulate in all cilia throughout the brain, suggesting that β-arrestin does not require the BBSome for ciliary exit or is only recruited into certain cilia.
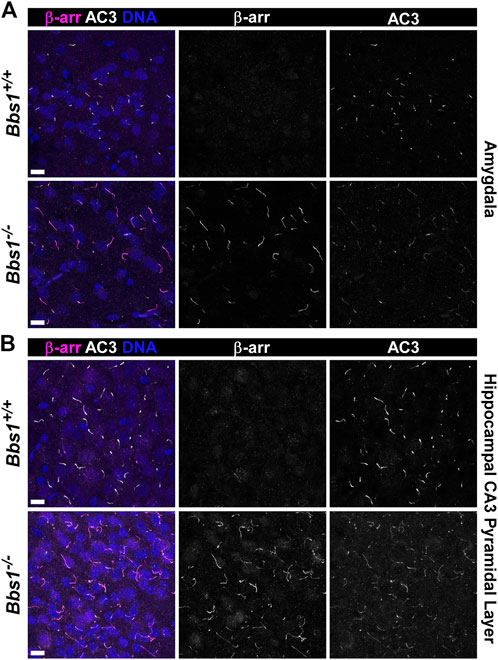
FIGURE 6. β-arrestin accumulates in cilia in the brains of Bbs1−/− mice (A) Representative images of the amygdala from adult Bbs1+/+ (upper row) and Bbs1−/− (lower row) mice colabeled for β-arrestin (β-arr; magenta) and AC3 (white). β-arrestin localized to 0% of AC3-positive cilia in Bbs1+/+ sections (0/1,540; n = 3 animals) and 90% of AC3-positive cilia in Bbs1−/− sections (712/790; n = 3 animals) (B) Representative images of the CA3 region of the hippocampus from adult Bbs1+/+ (upper row) and Bbs1−/− (lower row) mice colabeled for β-arrestin (magenta) and AC3 (white). β-arrestin localized to 0% of AC3-positive cilia in Bbs1+/+ sections (0/871; n = 3 animals) and 84% of AC3-positive cilia in Bbs1−/− sections (604/722; n = 3 animals). Nuclei are stained with DRAQ5 (blue). Scale bars represent 10 µm.
To test whether D1 ciliary accumulation leads to β-arrestin ciliary accumulation, we colabeled brain sections from adult Bbs1+/+ and Bbs1−/− mice with antibodies against D1 and β-arrestin. As expected, we did not observe D1-or β-arrestin-positive cilia in Bbs1+/+ sections (Figure 7). In Bbs1−/− sections we observed abundant cilia that were positive for D1 or β-arrestin but D1 and β-arrestin colocalized in only 0.9% (2/214; n = 3 animals) of cilia (Figure 7). This result suggests that D1 ciliary accumulation is rarely associated with β-arrestin ciliary recruitment and accumulation.
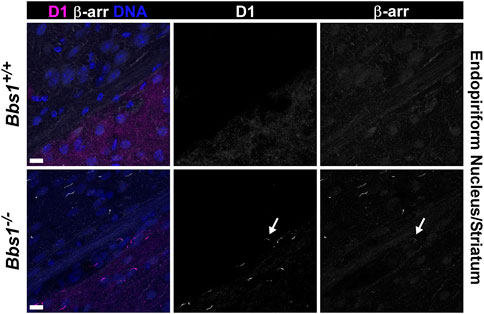
FIGURE 7. β-arrestin rarely accumulates in D1-positive cilia in the brains of Bbs1−/− mice. Representative images of the junction of the striatum (lower right corner) and the endopiriform nucleus (upper left corner) in adult Bbs1+/+ (upper row) and Bbs1−/− (lower row) mice colabeled for D1 (magenta) and β-arrestin (β-arr; white). The striatum is distinguished by the presence of D1 labeling and the endopiriform nucleus is distinguished by a lack of D1 labeling. Note the lack of D1-or β-arrestin-positive cilia in the Bbs1+/+ section but the presence of D1-positive cilia in the striatum and β-arrestin-positive cilia in the endopiriform nucleus of the Bbs1−/− section (left panels). A single cilium (indicated by an arrow) is positive for both D1 and β-arrestin (middle and right panel). β-arrestin colocalized to 0.9% (2/214; n = 3 animals) of D1-positive cilia in Bbs1−/− sections. Nuclei are stained with DRAQ5 (blue). Scale bars represent 10 µm.
Discussion
Our results showing that Sstr3 and Mchr1 fail to localize to neuronal cilia in Bbs1−/− mice are consistent with a role for the BBSome in trafficking of some GPCRs into cilia. Other mechanisms have been implicated in the trafficking of GPCRs into cilia. The ubiquitously expressed tubby family protein Tulp3 directs ciliary localization of heterologous Sstr3, Mchr1, D1, Gpr161, and Gpr19 in cultured cell lines (Mukhopadhyay et al., 2010; Mukhopadhyay et al., 2013; Badgandi et al., 2017). Disruption of Tulp3 activity in cultured hippocampal neurons reduces Gpr161 and Gpr19 ciliary localization (Badgandi et al., 2017). Yet, neurons also express the founding tubby family member Tub. Interestingly, Tub is required for localization of Sstr3 and Mchr1 to neuronal cilia (Sun et al., 2012) but not Gpr161 and Gpr19 (Badgandi et al., 2017). Thus, it appears that Sstr3 and Mchr1 require Tub for ciliary localization whereas Gpr161 and Gpr19 can be trafficked to cilia by Tulp3. The basis for this differential requirement for GPCR ciliary localization in hippocampal neurons is unclear. It is interesting that GPCRs that are Tub-dependent fail to localize to cilia on BBSome mutant neurons and GPCRs that localize independently of Tub accumulate in cilia on BBSome mutant neurons. Perhaps, the BBSome and Tub act cooperatively to traffic a subset of GPCRs to cilia on neurons.
An alternative explanation for our results is that BBSome function is confined to ciliary GPCR export and Sstr3 and Mchr1 are constitutively ectocytosed from cilia on BBSome mutant neurons. Nager et al. (2017) investigated agonist-mediated ciliary export of heterologously-expressed Sstr3 in a ciliated renal epithelial cell line lacking BBSome or β-arrestin 2 function and found that activated Sstr3 is recruited to the ciliary tip and released in extracellular vesicles named ectosomes. A similar observation was made with NPY2 (Nager et al., 2017). Based on these findings it was hypothesized that the BBSome and β-arrestin 2 cooperatively export activated GPCRs from cilia and the loss of GPCR localization on neuronal cilia in BBSome mutant mice may be due to constitutive ectocytosis of the receptors rather than a defect in trafficking into cilia (Nager et al., 2017). Yet, we found that Gpr161 and Gpr19 accumulate in cilia on BBSome mutant hippocampal neurons, indicating that receptors can accumulate when ciliary export is blocked. Why would some ciliary receptors be constitutively ectocytosed while other receptors accumulate in cilia on the same population of neurons? One possibility is that Sstr3 and Mchr1 are activated on the ciliary membrane (a requirement for ectocytosis) but Gpr161 and Gpr19 are not. However, D1 ciliary localization decreases after agonist treatment in wildtype neurons, suggesting that D1 is activated on the ciliary membrane (Domire et al., 2011). Yet, in the absence of the BBSome D1 accumulates in neuronal cilia. An alternative possibility is that the BBSome traffics additional proteins into cilia that are required for activation of certain GPCRs. Nevertheless, our results show that the regulation of GPCR ciliary localization in neurons is complex and significant gaps remain in our understanding of GPCR trafficking into and out of cilia.
It is interesting to contemplate the consequences of Gpr161 ciliary accumulation in the brains of adult BBSome mutant mice. Shh signaling components, including Ptch and Smo, are expressed in rat adult hippocampal neurons (Petralia et al., 2011a; Petralia et al., 2011b). Shh treatment of hippocampal neurons regulates presynaptic terminal structure and function (Mitchell et al., 2012) and axon elongation (Yao et al., 2015), suggesting a role in synapse formation. Shh treatment also increases the density of dendritic spines on mature cultured mouse hippocampal neurons (Sasaki et al., 2010). As cilia are required for normal dendrite outgrowth of mouse cortical neurons (Guadiana et al., 2013), it is conceivable that cilia mediate Shh signaling on adult neurons. Interestingly, Bbs4 knockout mice have a significant reduction in dendritic spines on hippocampal neurons (Haq et al., 2019). It is possible that ciliary accumulation of Gpr161 on BBSome mutant hippocampal neurons disrupts Shh signaling and results in a reduction of dendritic spines. Smo accumulates in cilia of unstimulated BBSome mutant mouse embryonic fibroblasts (Zhang et al., 2012; Goetz et al., 2017), suggesting a role for the BBSome in the export of Smo from cilia. Thus, it is feasible that Smo and Gpr161 accumulate together in cilia on BBSome mutant hippocampal neurons, which could contribute to the increase in ciliary length. Further studies are necessary to test this possibility and determine the consequences for Shh pathway activity.
One model for GPCR ciliary export posits that upon receptor activation, β-arrestin is recruited into cilia where it facilitates receptor ubiquitination to mark its removal by the BBSome (Shinde et al., 2020). We detected β-arrestin accumulating in a subset of cilia throughout the brain of Bbs1−/− mice, suggesting these cilia contain an activated GPCR that requires the BBSome for ciliary export and upon recruitment β-arrestin binds to the activated receptor and becomes trapped in the cilium. Interestingly, β-arrestin was rarely observed in D1-enriched cilia. Possible explanations for this finding are that D1 is not activated on the ciliary membrane, β-arrestin is not recruited to activated D1, or the overaccumulation of D1 in the cilium prevents β-arrestin recruitment. β-arrestin is also recruited into cilia in response to Gpr161 activation (Pal et al., 2016; Shinde et al., 2020). Unfortunately, we were unable to colabel for β-arrestin and Gpr161 to test whether they colocalize within the same cilia since the antibodies are the same species. However, we observed Gpr161 ciliary accumulation in some brain regions where there was not β-arrestin ciliary accumulation. Thus, similar to D1, Gpr161 ciliary accumulation is not always associated with β-arrestin ciliary accumulation. It is likely that β-arrestin ciliary localization indicates the presence of additional known and unknown activated GPCRs on the ciliary membrane. An important area of future study will be identifying potential novel ciliary GPCRs and determining the causes of β-arrestin ciliary recruitment.
We previously reported a reduction in AC3 signal in D1-enriched cilia (Stubbs et al., 2022). In brain sections from Bbs1−/− mice we saw a significant decrease in AC3 signal in D1-and Gpr161-enriched cilia. Although we cannot rule out the possibility that D1 and Gpr161 ciliary accumulation interferes with binding of the AC3 antibody, it seems more likely that D1 or Gp161 accumulation prevents AC3 ciliary localization. It is tempting to speculate that GPCR overaccumulation in neuronal cilia crowds out other proteins, limits the availability of ciliary signaling partners, and disrupts GPCR ciliary signaling. Functional studies utilizing live-cell imaging and genetically-encoded signaling reporters should reveal whether AC3-mediated cAMP signaling is impacted by GPCR ciliary accumulation.
In summary, we have shown that Sstr3 and Mchr1 fail to localize to cilia, while D1, Gpr161, and Gpr19 accumulate in cilia on neurons from Bbs1−/− mice. We further show that β-arrestin accumulates in a subset of cilia throughout the brain of Bbs1−/− mice but is rarely found in D1-enriched cilia. Our findings provide additional support for a role for BBS proteins in the export of ciliary GPCRs in the brain but raise questions about why some GPCRs are absent from cilia and others accumulate in cilia.
Materials and methods
Mice and tissue preparation
All mouse studies were conducted in accordance with institutional guidelines based on National Institutes of Health standards and were performed with Institutional Animal Care and Use Committee approval at the Ohio State University. All animals were maintained in a temperature and humidity-controlled vivarium with 12-h light/dark cycle at a maximum of five per cage and given access to normal chow and water ad libitum. Generation of the germline Bbs1 deleted (null) allele has been previously described (Stubbs et al., 2022). To increase the viability of Bbs1-null animals, Bbs1wt/null mice on a congenic background (C57BL/6J) were crossed with FVB/NJ mice from the Jackson Laboratory (#001800). Constitutive Bbs1null/null mice were generated by intercrossing Bbs1wt/null male and female mice on a mixed background. Fixed mouse brains were generated and processed as previously described (Berbari et al., 2008b), and sliced at a thickness of 20 µm. Neurons were cultured as previously described (Green et al., 2016), with the exception that neurons were cultured in Neurobasal plus medium containing B-27 plus supplement (Thermo-Fisher).
Immunofluorescence
Seven to 10 days after plating, neuronal cultures were fixed and processed for immunofluorescence as previously described (Green et al., 2016). Primary antibodies included anti-AC3 (mouse, EnCor MCA-1A12 1:1,000; rabbit, EnCor RPCA-ACIII 1:1,000, chicken, EnCor CPCA-ACIII 1:1,000) (Wong et al., 2000), anti-Sstr3 (goat, Santa Cruz M-18 1:250; rabbit, Gramsch ss-830 1:100) (Handel et al., 1999), anti-Mchr1 (rabbit, Invitrogen 711,649 1:250) (Jasso et al., 2021), anti-D1 (mouse, Santa Cruz sc-33660 1:500) (Domire et al., 2011), anti-pan-arrestin (rabbit, Abcam Ab2914 1:500) (Jones and Hinkle, 2008). Anti-Gpr161 (rabbit, 1:250) (Mukhopadhyay et al., 2013) and anti-Gpr19 (rabbit, 1:250) (Badgandi et al., 2017) were a kind gift from Saikat Mukhopadhyay (UT Southwestern). Secondary antibodies included Alexa fluor 488- and 546-conjugated donkey anti-mouse, Alexa fluor 488- and 546-conjugated donkey anti-rabbit, Alexa fluor 488-conjugated donkey anti-goat, and Alexa fluor 546-conjugated goat anti-chicken (Invitrogen 1:1,000). Nucleic acids were stained with DRAQ5 (Thermo-Fisher Scientific, 1:5,000). Samples were imaged on a Leica TCS SP8 laser scanning confocal microscope at the Hunt-Curtis Imaging Facility in the Department of Neuroscience at OSU. Multiple consecutive focal planes (Z-stack), spaced at 0.3–0.5 μm intervals, were captured.
Quantification and statistical analysis of neuronal cilia
Quantification of cilia in brain slices was performed using coronal sections from 5–7-week-old Bbs1+/+ and Bbs1−/− littermates (2 male and one female of each genotype). For each section, six to nine fields were imaged and the number of Sstr3-, Mchr1-, D1-, Gpr161-, arrestin-, and AC3-positive cilia was counted by an individual blind to the genotype. The results were expressed as the proportion of AC3-positive cilia that were positive for each GPCR or arrestin. To quantify relative ciliary AC3 levels, fluorescence intensity of ciliary AC3 in D1-positive and -negative cilia and Gpr161-positive and -negative cilia were measured in ImageJ. ROIs were drawn around individual cilia and the mean fluorescence intensity of the cilium compared to background was calculated. The fluorescence intensity of AC3 in D1-and Gpr161-positive cilia were averaged and compared to the average fluorescence intensity of AC3 in D1-and Gpr161-negative cilia, respectively. Quantification of ciliary lengths was measured in ImageJ by drawing lines calibrated to the scale bar along the length of AC3-positive cilia. Quantification of Gpr161-and Gpr19-positive cilia in cultured neurons was performed on three coverslips from three animals for each condition and genotype. For each coverslip at least six fields were imaged. The number of nuclei, Gpr161-or Gpr19-positive cilia, and AC3-positive cilia was counted by an individual blind to the genotype. The results were expressed as the proportion of AC3-positive cilia to nuclei and the proportion of Gpr161-or Gpr19-positive cilia to AC3-positive cilia. Data were analyzed using Prism nine software (GraphPad Software) and results were expressed as mean ± SEM. Statistical significance was tested using Student’s t-test.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics statement
The animal study was reviewed and approved by Institutional Animal Care and Use Committee approval at the Ohio State University.
Author contributions
TS, JIB, JB, and KM performed experiments. TS, JIB, JB, and KM contributed to the manuscript. TS, JIB, and KM conceptualized the project. All authors contributed, read and approved the submitted manuscript.
Funding
This work was supported by research project grants R21 MH107021 and MH121744 from the NIH/NIMH to KM.
Acknowledgments
We are grateful to Saikat Mukhopadhyay (UT Southwestern) for providing the Gpr161 and Gpr19 antibodies.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2022.1092161/full#supplementary-material.
References
Anvarian, Z., Mykytyn, K., Mukhopadhyay, S., Pedersen, L. B., and Christensen, S. T. (2019). Cellular signalling by primary cilia in development, organ function and disease. Nat. Rev. Nephrol. 15, 199–219. doi:10.1038/s41581-019-0116-9
Badgandi, H. B., Hwang, S. H., Shimada, I. S., Loriot, E., and Mukhopadhyay, S. (2017). Tubby family proteins are adapters for ciliary trafficking of integral membrane proteins. J. Cell Biol. 216, 743–760. doi:10.1083/jcb.201607095
Berbari, N. F., Johnson, A. D., Lewis, J. S., Askwith, C. C., and Mykytyn, K. (2008a). Identification of ciliary localization sequences within the third intracellular loop of G protein-coupled receptors. Mol. Biol. Cell 19, 1540–1547. doi:10.1091/mbc.e07-09-0942
Berbari, N. F., Lewis, J. S., Bishop, G. A., Askwith, C. C., and Mykytyn, K. (2008b). Bardet-Biedl syndrome proteins are required for the localization of G protein-coupled receptors to primary cilia. Proc. Natl. Acad. Sci. U. S. A. 105, 4242–4246. doi:10.1073/pnas.0711027105
Bishop, G. A., Berbari, N. F., Lewis, J. S., and Mykytyn, K. (2007). Type III adenylyl cyclase localizes to primary cilia throughout the adult mouse brain. J. Comp. Neurol. 505, 562–571. doi:10.1002/cne.21510
Brailov, I., Bancila, M., Brisorgueil, M. J., Miquel, M. C., Hamon, M., and Verge, D. (2000). Localization of 5-HT(6) receptors at the plasma membrane of neuronal cilia in the rat brain. Brain Res. 872, 271–275. doi:10.1016/s0006-8993(00)02519-1
Carter, C. S., Vogel, T. W., Zhang, Q., Seo, S., Swiderski, R. E., Moninger, T. O., et al. (2012). Abnormal development of NG2+PDGFR-α+ neural progenitor cells leads to neonatal hydrocephalus in a ciliopathy mouse model. Nat. Med. 18, 1797–1804. doi:10.1038/nm.2996
Corbit, K. C., Aanstad, P., Singla, V., Norman, A. R., Stainier, D. Y., and Reiter, J. F. (2005). Vertebrate Smoothened functions at the primary cilium. Nature 437, 1018–1021. doi:10.1038/nature04117
Davis, R. E., Swiderski, R. E., Rahmouni, K., Nishimura, D. Y., Mullins, R. F., Agassandian, K., et al. (2007). A knockin mouse model of the Bardet-Biedl syndrome 1 M390R mutation has cilia defects, ventriculomegaly, retinopathy, and obesity. Proc. Natl. Acad. Sci. U. S. A. 104, 19422–19427. doi:10.1073/pnas.0708571104
Domire, J. S., Green, J. A., Lee, K. G., Johnson, A. D., Askwith, C. C., and Mykytyn, K. (2011). Dopamine receptor 1 localizes to neuronal cilia in a dynamic process that requires the Bardet-Biedl syndrome proteins. Cell Mol. Life Sci. 68, 2951–2960. doi:10.1007/s00018-010-0603-4
Forsythe, E., and Beales, P. L. (2013). Bardet-Biedl syndrome. Eur. J. Hum. Genet. 21, 8–13. doi:10.1038/ejhg.2012.115
Goetz, S. C., Bangs, F., Barrington, C. L., Katsanis, N., and Anderson, K. V. (2017). The Meckel syndrome-associated protein MKS1 functionally interacts with components of the BBSome and IFT complexes to mediate ciliary trafficking and hedgehog signaling. PLoS One 12, e0173399. doi:10.1371/journal.pone.0173399
Green, J. A., Schmid, C. L., Bley, E., Monsma, P. C., Brown, A., Bohn, L. M., et al. (2016). Recruitment of beta-arrestin into neuronal cilia modulates somatostatin receptor subtype 3 ciliary localization. Mol. Cell Biol. 36, 223–235. doi:10.1128/MCB.00765-15
Guadiana, S. M., Semple-Rowland, S., Daroszewski, D., Madorsky, I., Breunig, J. J., Mykytyn, K., et al. (2013). Arborization of dendrites by developing neocortical neurons is dependent on primary cilia and type 3 adenylyl cyclase. J. Neurosci. 33, 2626–2638. doi:10.1523/JNEUROSCI.2906-12.2013
Guo, D. F., Cui, H., Zhang, Q., Morgan, D. A., Thedens, D. R., Nishimura, D., et al. (2016). The BBSome controls energy homeostasis by mediating the transport of the leptin receptor to the plasma membrane. PLoS Genet. 12, e1005890. doi:10.1371/journal.pgen.1005890
Handel, M., Schulz, S., Stanarius, A., Schreff, M., Erdtmann-Vourliotis, M., Schmidt, H., et al. (1999). Selective targeting of somatostatin receptor 3 to neuronal cilia. Neuroscience 89, 909–926. doi:10.1016/s0306-4522(98)00354-6
Haq, N., Schmidt-Hieber, C., Sialana, F. J., Ciani, L., Heller, J. P., Stewart, M., et al. (2019). Loss of Bardet-Biedl syndrome proteins causes synaptic aberrations in principal neurons. PLoS Biol. 17, e3000414. doi:10.1371/journal.pbio.3000414
Hsiao, Y. C., Munoz-Estrada, J., Tuz, K., and Ferland, R. J. (2021). The transition zone protein AHI1 regulates neuronal ciliary trafficking of MCHR1 and its downstream signaling pathway. J. Neurosci. 41, 3932–3943. doi:10.1523/JNEUROSCI.2993-20.2021
Jasso, K. R., Kamba, T. K., Zimmerman, A. D., Bansal, R., Engle, S. E., Everett, T., et al. (2021). An N-terminal fusion allele to study melanin concentrating hormone receptor 1. Genesis 59, e23438. doi:10.1002/dvg.23438
Jones, B. W., and Hinkle, P. M. (2008). Arrestin binds to different phosphorylated regions of the thyrotropin-releasing hormone receptor with distinct functional consequences. Mol. Pharmacol. 74, 195–202. doi:10.1124/mol.108.045948
Koemeter-Cox, A. I., Sherwood, T. W., Green, J. A., Steiner, R. A., Berbari, N. F., Yoder, B. K., et al. (2014). Primary cilia enhance kisspeptin receptor signaling on gonadotropin-releasing hormone neurons. Proc. Natl. Acad. Sci. U. S. A. 111, 10335–10340. doi:10.1073/pnas.1403286111
Loktev, A. V., and Jackson, P. K. (2013). Neuropeptide Y family receptors traffic via the Bardet-Biedl syndrome pathway to signal in neuronal primary cilia. Cell Rep. 5, 1316–1329. doi:10.1016/j.celrep.2013.11.011
Loktev, A. V., Zhang, Q., Beck, J. S., Searby, C. C., Scheetz, T. E., Bazan, J. F., et al. (2008). A BBSome subunit links ciliogenesis, microtubule stability, and acetylation. Dev. Cell 15, 854–865. doi:10.1016/j.devcel.2008.11.001
Mick, D. U., Rodrigues, R. B., Leib, R. D., Adams, C. M., Chien, A. S., Gygi, S. P., et al. (2015). Proteomics of primary cilia by proximity labeling. Dev. Cell 35, 497–512. doi:10.1016/j.devcel.2015.10.015
Mitchell, N., Petralia, R. S., Currier, D. G., Wang, Y. X., Kim, A., Mattson, M. P., et al. (2012). Sonic hedgehog regulates presynaptic terminal size, ultrastructure and function in hippocampal neurons. J. Cell Sci. 125, 4207–4213. doi:10.1242/jcs.105080
Mukhopadhyay, S., Wen, X., Chih, B., Nelson, C. D., Lane, W. S., Scales, S. J., et al. (2010). TULP3 bridges the IFT-A complex and membrane phosphoinositides to promote trafficking of G protein-coupled receptors into primary cilia. Genes Dev. 24, 2180–2193. doi:10.1101/gad.1966210
Mukhopadhyay, S., Wen, X., Ratti, N., Loktev, A., Rangell, L., Scales, S. J., et al. (2013). The ciliary G-protein-coupled receptor Gpr161 negatively regulates the Sonic hedgehog pathway via cAMP signaling. Cell 152, 210–223. doi:10.1016/j.cell.2012.12.026
Nachury, M. V., Loktev, A. V., Zhang, Q., Westlake, C. J., Peranen, J., Merdes, A., et al. (2007). A core complex of BBS proteins cooperates with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell 129, 1201–1213. doi:10.1016/j.cell.2007.03.053
Nachury, M. V., and Mick, D. U. (2019). Establishing and regulating the composition of cilia for signal transduction. Nat. Rev. Mol. Cell Biol. 20, 389–405. doi:10.1038/s41580-019-0116-4
Nager, A. R., Goldstein, J. S., Herranz-Perez, V., Portran, D., Ye, F., Garcia-Verdugo, J. M., et al. (2017). An actin network dispatches ciliary GPCRs into extracellular vesicles to modulate signaling. Cell 168, 252–263. doi:10.1016/j.cell.2016.11.036
Nozaki, S., Katoh, Y., Kobayashi, T., and Nakayama, K. (2018). BBS1 is involved in retrograde trafficking of ciliary GPCRs in the context of the BBSome complex. PLoS One 13, e0195005. doi:10.1371/journal.pone.0195005
Pal, K., Hwang, S. H., Somatilaka, B., Badgandi, H., Jackson, P. K., Defea, K., et al. (2016). Smoothened determines beta-arrestin-mediated removal of the G protein-coupled receptor Gpr161 from the primary cilium. J. Cell Biol. 212, 861–875. doi:10.1083/jcb.201506132
Petralia, R. S., Schwartz, C. M., Wang, Y. X., Mattson, M. P., and Yao, P. J. (2011a). Subcellular localization of Patched and Smoothened, the receptors for Sonic hedgehog signaling, in the hippocampal neuron. J. Comp. Neurol. 519, 3684–3699. doi:10.1002/cne.22681
Petralia, R. S., Wang, Y. X., Mattson, M. P., and Yao, P. J. (2011b). Sonic hedgehog distribution within mature hippocampal neurons. Commun. Integr. Biol. 4, 775–777. doi:10.4161/cib.17832
Rohatgi, R., Milenkovic, L., and Scott, M. P. (2007). Patched1 regulates hedgehog signaling at the primary cilium. Science 317, 372–376. doi:10.1126/science.1139740
Sasaki, N., Kurisu, J., and Kengaku, M. (2010). Sonic hedgehog signaling regulates actin cytoskeleton via Tiam1-Rac1 cascade during spine formation. Mol. Cell Neurosci. 45, 335–344. doi:10.1016/j.mcn.2010.07.006
Sheu, S. H., Upadhyayula, S., Dupuy, V., Pang, S., Deng, F., Wan, J., et al. (2022). A serotonergic axon-cilium synapse drives nuclear signaling to alter chromatin accessibility. Cell 185, 3390–3407 e18. doi:10.1016/j.cell.2022.07.026
Shinde, S. R., Nager, A. R., and Nachury, M. V. (2020). Ubiquitin chains earmark GPCRs for BBSome-mediated removal from cilia. J. Cell Biol. 219, e202003020. doi:10.1083/jcb.202003020
Siljee, J. E., Wang, Y., Bernard, A. A., Ersoy, B. A., Zhang, S., Marley, A., et al. (2018). Subcellular localization of MC4R with ADCY3 at neuronal primary cilia underlies a common pathway for genetic predisposition to obesity. Nat. Genet. 50, 180–185. doi:10.1038/s41588-017-0020-9
Stubbs, T., Koemeter-Cox, A., Bingman, J. I., Zhao, F., Kalyanasundaram, A., Rowland, L. A., et al. (2022). Disruption of dopamine receptor 1 localization to primary cilia impairs signaling in striatal neurons. J. Neurosci. 42, 6692–6705. doi:10.1523/JNEUROSCI.0497-22.2022
Suciu, S. K., and Caspary, T. (2021). Cilia, neural development and disease. Semin. Cell Dev. Biol. 110, 34–42. doi:10.1016/j.semcdb.2020.07.014
Sun, X., Haley, J., Bulgakov, O. V., Cai, X., Mcginnis, J., and Li, T. (2012). Tubby is required for trafficking G protein-coupled receptors to neuronal cilia. Cilia 1, 21. doi:10.1186/2046-2530-1-21
Tereshko, L., Gao, Y., Cary, B. A., Turrigiano, G. G., and Sengupta, P. (2021). Ciliary neuropeptidergic signaling dynamically regulates excitatory synapses in postnatal neocortical pyramidal neurons. Elife 10, e65427. doi:10.7554/eLife.65427
Wang, Y., Bernard, A., Comblain, F., Yue, X., Paillart, C., Zhang, S., et al. (2021). Melanocortin 4 receptor signals at the neuronal primary cilium to control food intake and body weight. J. Clin. Invest. 131, e142064. doi:10.1172/JCI142064
Wong, S. T., Trinh, K., Hacker, B., Chan, G. C., Lowe, G., Gaggar, A., et al. (2000). Disruption of the type III adenylyl cyclase gene leads to peripheral and behavioral anosmia in transgenic mice. Neuron 27, 487–497. doi:10.1016/s0896-6273(00)00060-x
Yao, P. J., Petralia, R. S., Ott, C., Wang, Y. X., Lippincott-Schwartz, J., and Mattson, M. P. (2015). Dendrosomatic sonic hedgehog signaling in hippocampal neurons regulates axon elongation. J. Neurosci. 35, 16126–16141. doi:10.1523/JNEUROSCI.1360-15.2015
Yee, L. E., Garcia-Gonzalo, F. R., Bowie, R. V., Li, C., Kennedy, J. K., Ashrafi, K., et al. (2015). Conserved genetic interactions between ciliopathy complexes cooperatively support ciliogenesis and ciliary signaling. PLoS Genet. 11, e1005627. doi:10.1371/journal.pgen.1005627
Zhang, Q., Nishimura, D., Vogel, T., Shao, J., Swiderski, R., Yin, T., et al. (2013). BBS7 is required for BBSome formation and its absence in mice results in Bardet-Biedl syndrome phenotypes and selective abnormalities in membrane protein trafficking. J. Cell Sci. 126, 2372–2380. doi:10.1242/jcs.111740
Keywords: cilia, Bardet-Biedl syndrome, ciliopathy, Gpr161, GPR19, beta arrestin
Citation: Stubbs T, Bingman JI, Besse J and Mykytyn K (2023) Ciliary signaling proteins are mislocalized in the brains of Bardet-Biedl syndrome 1-null mice. Front. Cell Dev. Biol. 10:1092161. doi: 10.3389/fcell.2022.1092161
Received: 07 November 2022; Accepted: 22 December 2022;
Published: 09 January 2023.
Edited by:
Inna Nechipurenko, Worcester Polytechnic Institute, United StatesReviewed by:
Jeremy C. McIntyre, University of Florida, United StatesAlexandre Benmerah, INSERM U1163 Institut Imagine, France
Copyright © 2023 Stubbs, Bingman, Besse and Mykytyn. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kirk Mykytyn, TXlreXR5bi4xQG9zdS5lZHU=
 Toneisha Stubbs
Toneisha Stubbs James I. Bingman
James I. Bingman Kirk Mykytyn
Kirk Mykytyn