- 1Gene Expression and Signaling, Institute of Cell Biochemistry, Hannover Medical School, Hannover, Germany
- 2Mount Desert Island Biological Laboratory, Bar Harbor, ME, United States
Adult stem cells depend on their niches for regulatory signaling that controls their maintenance, division, and their progeny differentiation. While communication between various types of stem cells and their niches is becoming clearer, the process of stem cell niche establishment is still not very well understood. Model genetic organisms provide simplified systems to address various complex questions, for example, how is a stem cell niche formed? What signaling cascades induce the stem cell niche formation? Are the mechanisms of stem cell niche formation conserved? Notch signaling is an evolutionarily conserved pathway first identified in fruit flies, crucial in fate acquisition and spatiotemporal patterning. While the core logic behind its activity is fairly simple and requires direct cell–cell interaction, it reaches an astonishing complexity and versatility by combining its different modes of action. Subtleties such as equivalency between communicating cells, their physical distance, receptor and ligand processing, and endocytosis can have an effect on the way the events unfold, and this review explores some important general mechanisms of action, later on focusing on its involvement in stem cell niche formation. First, looking at invertebrates, we will examine how Notch signaling induces the formation of germline stem cell niche in male and female Drosophila. In the developing testis, a group of somatic gonadal precursor cells receive Delta signals from the gut, activating Notch signaling and sealing their fate as niche cells even before larval hatching. Meanwhile, the ovarian germline stem cell niche is built later during late larval stages and requires a two-step process that involves terminal filament formation and cap cell specification. Intriguingly, double security mechanisms of Notch signaling activation coordinated by the soma or the germline control both steps to ensure the robustness of niche assembly. Second, in the vast domains of mammalian cellular signaling, there is an emerging picture of Notch being an active player in a variety of tissues in health and disease. Notch involvement has been shown in stem cell niche establishment in multiple organs, including the brain, muscle, and intestine, where the stem cell niches are essential for the maintenance of adult stem cells. But adult stem cells are not the only cells looking for a home. Cancer stem cells use Notch signaling at specific stages to gain an advantage over endogenous tissue and overpower it, at the same time acquiring migratory and invasive abilities to claim new tissues (e.g., bone) as their territory. Moreover, in vitro models such as organoids reveal similar Notch employment when it comes to the developing stem cell niches. Therefore, a better understanding of the processes regulating stem cell niche assembly is key for the fields of stem cell biology and regenerative medicines.
Dialects of Notch signaling
The Notch signaling pathway was discovered over a century ago and received its name after the notched and serrated wing phenotype displayed in Drosophila melanogaster mutants (Metz and Bridges, 1917). Since then, extensive research revealed a slew of complexity and versatility surrounding this highly conserved pathway. It was found to be involved in a wide range of behaviors and developmental key points. Canonical Notch signaling is activated by interactions between the Notch receptor and Delta- or Serrate-like ligands that are expressed by adjacent cells. Upon binding, the Notch is cleaved, which leads to the release of the Notch intracellular domain (NICD). Once freed, the NICD enters the nucleus where it forms transcriptional complexes with other important participants (e.g., nuclear effector Mastermind) to activate target genes. The targets encode basic helix–loop–helix proteins that function as nuclear effectors of Notch signaling to regulate the transcriptional activity of multiple genes. As in many other pathways, the Notch pathway has more paralogs for each of the key proteins in mammals, compared to invertebrates such as Drosophila, which has only one Notch receptor and two ligands, Delta and Serrate (Maine et al., 1995; Lissemore and Starmer, 1999). Importantly, the core logic of the pathway is preserved and operates under the same principle throughout the animal kingdom. The direct signal transducing mechanism from the membrane to the nucleus without second-messenger amplification and regulation enables Notch to function as a juxtacrine signaling pathway to effectively regulate cell fate specification depending on the inputs from the neighboring cells (Yamamoto et al., 2014; Kovall et al., 2017). Various intra- and extracellular signals and mechanistic cues modify the strength of Notch pathway activity, which dynamically guides cells along opposing developmental fates in a tissue- and time-dependent manner (Bray, 2006).
Before cells acquire a certain fate, they could concomitantly express Delta or Notch and be indecisive with regard to Notch signaling. Remarkably, in a culture of equivalent cells which do not receive any outside triggers, fate determination can happen stochastically when eventually one cell would express more of either Notch or Delta due to transcriptional noise. Its neighbor would follow through by acquiring the opposite fate, and the salt and pepper pattern of Notch signaling activation would be generated at the plane level (Sprinzak et al., 2011; Shaya et al., 2017; Nandagopal et al., 2018). Notch and Delta co-expression is also the basis of a self-inhibitory mechanism called cis-inhibition, where Notch and Delta expressed by the same cell bind each other forming an inhibitory complex (Figure 1). This intrinsically prevents Notch signaling activation in certain cells, which opens the possibility of tissue patterning by delimitating the signal’s reach (de Celis and Bray, 1997; Sprinzak et al., 2010; del Alamo et al., 2011; Palmer et al., 2014). Inhibition can also be initiated from the surrounding cells and is coined as “trans-inhibition” which occurs due to the formation of trans-inhibitory Notch–Delta complexes at the membranes of adjacent cells. Insufficient mechanical force between the membranes or defective Delta endocytosis is the culprit for lack of Notch cleavage, which prevents Notch signaling activation (Kooh et al., 1993; Seugnet et al., 1997).
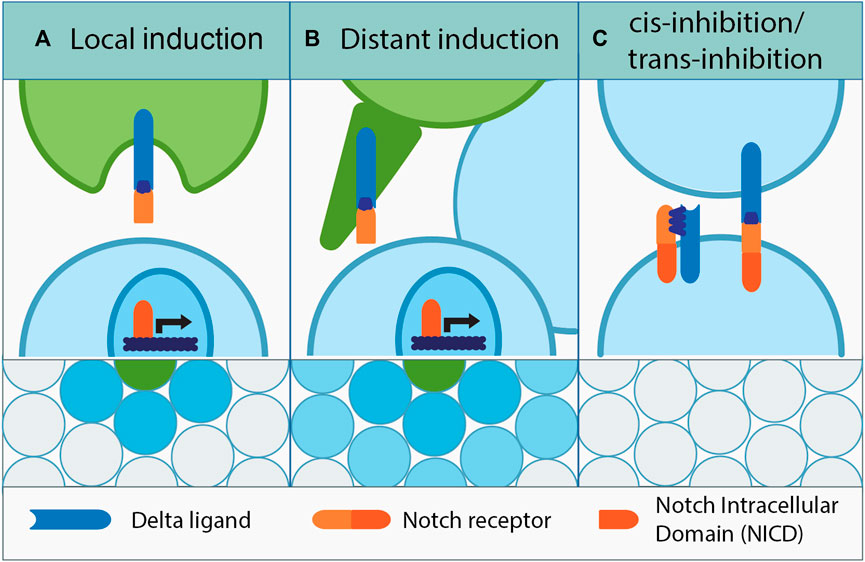
FIGURE 1. Dialects of Notch. (A) Among equipotent adjacent cells, a signal-sending cell induces Notch signaling in its neighbors through “lateral inhibition.” The same principle among non-equipotent cells is called “local induction.” Upon binding to the Delta ligand, the cleaved NICD domain of the Notch receptor travels to the nucleus and activates Notch-dependent gene expression. (B) Delta signal can also be sent further away by using cell projections, which establish direct contact with the target signal-receiving cell. This mechanism called “distant induction,” and it facilitates cell communication and Notch-dependent cell fate induction across several cell diameters. (C) Notch signaling inhibition can be intrinsic or extrinsic; cis-inhibition occurs between the Delta ligand and Notch receptor present on the same cell, while trans-inhibition occurs between adjacent cells that form the Delta–Notch complex, which fail to trigger Notch cleavage due to improper Delta processing or lack of sufficient membrane driven force.
There are several modes of Notch signaling activation. In this case, the cells are equipotent and adjacent to each other, and they can engage in “lateral inhibition” (Figure 1) (Lai, 2004; Le Borgne, 2006; Fiuza and Arias, 2007). This simply means that in the Notch-activated cell, the ligand expression or activation is suppressed, consolidating the signal sending/receiver status of the communicating cells (Irvine and Wieschaus, 1994; Cohen et al., 1997; Rodriguez-Esteban et al., 1997; Zhang and Gridley, 1998). Lateral inhibition is meant to enforce Notch signaling boundaries and has been observed in a multitude of tissues originating from evolutionarily different species (Wilkinson et al., 1994; Kushwah et al., 2014; Petrovic et al., 2014; Vanorny and Mayo, 2017). In some cases, lateral inhibition can be also used in a long range, for example, in the epithelial cells within imaginal discs of the Drosophila larva. The activity of Notch signaling is mediated by Delta-promoted planar filopodia; these dynamic structures are able to intermittently transmit Delta–Notch signals for the proper spatial organization of mechanosensory bristles (Hunter et al., 2019). This mechanism not only shapes the developmental outcome, but it also has been proposed to promote tumorigenesis driven by mammalian mesenchymal cells that were prevented from differentiating through long-range lateral inhibition (De Joussineau et al., 2003; Cohen et al., 2010; Hadjivasiliou et al., 2016).
When cells are non-equipotent, Notch signaling activation is generally regarded as “peripheral induction,” which is divided once again into short and long ranges (local and distant). “Local induction” takes place when non-equipotent cells are adjacent, and the Notch signal is activated in the neighbor of a Delta-transmitting cell (Yatsenko and Shcherbata, 2018). However, when the signal needs to reach farther away, more than one cell layer apart, this is referred to as “distant induction,” accomplished through cellular projections (Figure 1), e.g., between the germline stem cells and their somatic niches (Yatsenko and Shcherbata, 2021) or tumor and mesenchymal cells in the epithelial compartment of the tumorous wing discs in Drosophila (Boukhatmi et al., 2020). Notably, local induction is direct, whereas the distant one can lead to gradual propagation of the signal.
Importantly, what all of the mechanisms described previously have in common is the fact that the signal activation is triggered by direct membrane contacts or through cellular projections when it is between distant cells. The intensity of the trans-interactional communication is dependent on the amount of Notch and Delta presented at the membrane level, and the activation efficiency which heavily leans on the cell–cell contact geometry (Khait et al., 2016). The reasoning behind it is that with fewer contact points there will be a lesser chance of Delta–Notch interaction, resulting in a weaker Notch activity and vice versa.
As important as the strength of the Notch signaling per se is, the early events prior to its activation also need to be carried out in a precise manner for both the receptor and ligand, as their expression, processing, and endocytosis must be spatiotemporally regulated (Le Borgne et al., 2005). For the processing of Delta, the ubiquitin E3 ligases, Neuralized (in Drosophila and mammals) and Mind bomb (only mammals), are needed in the signal-sending cells to promote Delta endocytosis. This step is required for Notch signaling activation because it presents the processed ligand to the surface of the signal-sending cell. Although the critical steps in Delta processing are known, it remains unclear how they could potentially be affecting the signaling strength (Perez-Mockus and Schweisguth, 2017). The signal-receiving cell has to express the Notch receptor which can bind to the ligand, forming a complex. Upon Delta–Notch ligand–receptor interaction, Notch is cleaved twice, first by a metalloprotease (Kuzbanian in Drosophila and ADAM10/TACE in mammals) and second by the γ-secretase complex, leading to the release of the Notch intracellular domain, which translocates to the nucleus, activating Notch-dependent gene expression (Pan and Rubin, 1997; Wen et al., 1997; Qi et al., 1999; Hu et al., 2002; Lopez-Schier and St Johnston, 2002).
To add to this complex tango of Delta–Notch signaling determined by the differential expression of the Notch receptor and their ligands, the cell can express factors required for Delta processing in a polarized manner. In the Drosophila sensory organ lineages, in one daughter cell, Notch signaling is on, while in the other one, it is off. This is a result of the polarization of the stem cell, which ensures that one daughter cell inherits the ubiquitin E3 ligase, Neuralized, and the other the Notch inhibitor, Numb. Neuralized promotes the endocytosis of Delta, thus activating Notch expression in the first daughter cell, while Numb inhibits Notch and turns up Delta expression in the second daughter cell. In this manner, cell fate is ensured and regulated through each asymmetric division (Le Borgne and Schweisguth, 2003; Fiuza and Arias, 2007). These data show that the very early fate determination of the stem cell daughters is decided through this signaling pathway by asymmetric localization of various factors and polarized endosome dynamics (Strutt et al., 2002; Langevin et al., 2005; Benhra et al., 2010; Zhao et al., 2021).
After exploring some of the main ways in which Notch signaling can orchestrate cell fate and tissue patterning, one thing is clear: the acquisition of a particular state seals the fate of subsequent neighboring cells through mutually exclusive signaling states. This translates to an elegant and malleable subsequent activation of gene-encoding factors essential in the establishment of cell identities and by extrapolation, ensuring the proper formation of tissues and organs (Lai, 2004; Fiuza and Arias, 2007; Barad et al., 2010; Hunter et al., 2016). In contrast to the emerging picture of how Notch signaling shapes cell behavior in development, there is one scenario that received little attention, that is, stem cell niche maintenance and even less, its formation. So far, it has been shown that maintenance of multiple types of adult stem cells in many organisms depends on Notch signaling (e.g., human adult muscle, brain, mammary, and hematopoietic stem cells) (Carlson et al., 2009; Ables et al., 2011; Maillard, 2014; Chakrabarti et al., 2018), suggesting that it has a conserved role in stem cell niches. More interestingly for this review, however, is that Notch signaling has been demonstrated to play an important role in formation of all well-characterized stem cell niches in Drosophila, such as ovarian, testicular, and intestinal niches (Ward et al., 2006; Song et al., 2007; Mathur et al., 2010; Okegbe and DiNardo, 2011). Similarly, in mammals, Notch signaling is active in the formation of various niches, e.g., the neural ventricular zone (VZ) (Byun et al., 2019). Since Notch signaling in the context of stem cell niche formation appears to have great potential but is insufficiently researched, we will focus and draw the attention to this area of interest by introducing a few fascinating examples across the animal kingdom.
Notch and the stem cell niche formation
Around half a century ago, Schofield proposed the concept of the stem cell niche to describe the microenvironment capable of supporting stem cells (Schofield, 1978). Initially, it was used to describe independent anatomical sites observed to regulate the hematopoietic stem cell population, but later on, with the advancement of knowledge, it has been extended to multiple other tissues (Fuller and Spradling, 2007; Spradling et al., 2008). The first empirical evidence for the ability of the niche to support stemness came from one of the most studied animal models, Drosophila, in particular from studies of ovarian and testicular germline stem cell niches (Xie and Spradling, 2000; Tulina and Matunis, 2001). In the same vein, we will begin our exploration of Notch signaling contribution to stem cell niche establishment in the Drosophila ovary.
Stem cell niches for the Drosophila germline stem cells
Ovary
Notch signaling is one of the main pathways that control the formation of the ovarian germline stem cell (GSC) niche in Drosophila. The formation and size of the ovarian GSC niche depend on the strength of Notch signaling since its absence leads to a reduced niche size, while Notch signaling upregulation leads to the opposite (Ward et al., 2006; Song et al., 2007; Yatsenko and Shcherbata, 2018; Yatsenko and Shcherbata, 2021). In the developing Drosophila ovary, there are three main elements that need to be put together in the following order to create the ovarian germline stem cell niche unit: terminal filaments (TFs), cap cells (CpCs), and GSCs (Figure 2). Notch signaling uses several ways of coordinating the process of niche assembly.
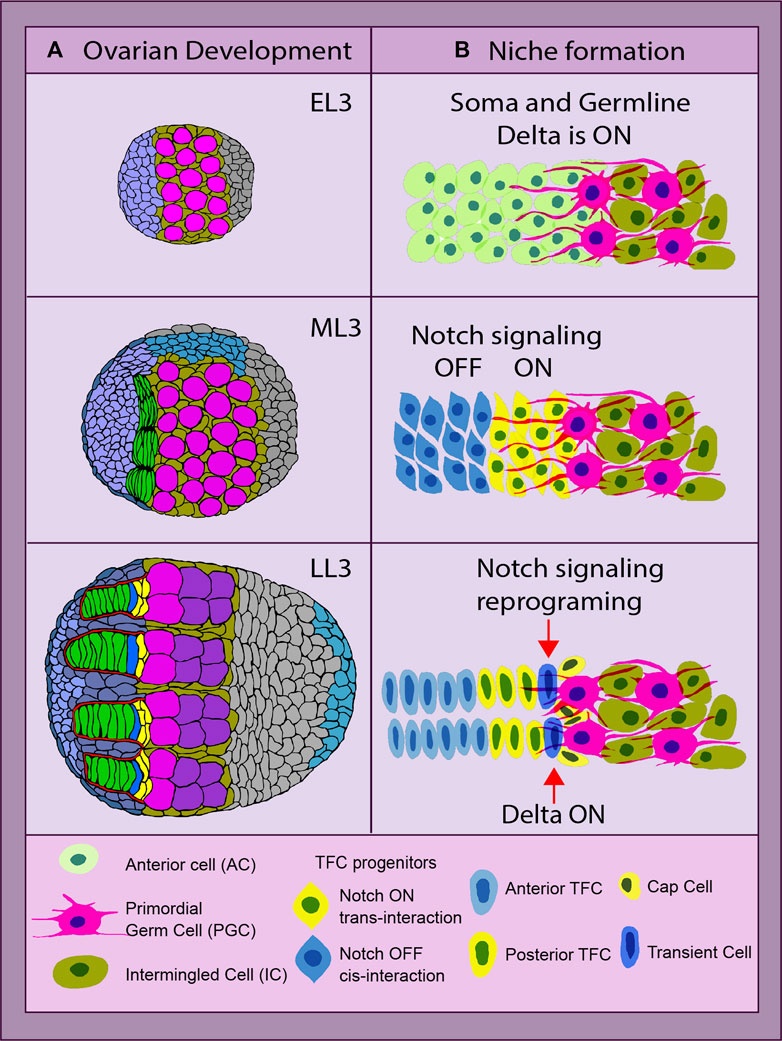
FIGURE 2. Drosophila ovarian development and niche formation. (A) depicts an overview of ovarian organogenesis throughout the early L3 (EL3) stage to late L3 (LL3), which is the last one before metamorphosis ensues. The transition at the organ level from a ball of largely undifferentiated cells to a highly complex and organized structure, with the stem cell niche consisting of the terminal filaments (TFs) (green) parallel to each other and adjacent cap cells (CpCs) (yellow). The primordial germ cells (PGCs) and germline stem cells (GSCs) are depicted in pink (Yatsenko and Shcherbata, 2018). (B) shows the key steps of the stem cell niche establishment. The anterior somatic cells (ACs, light green) are TF cell precursors. The anterior ACs have Notch signaling inhibited via cis- or trans-inhibition (blue), while the posterior ACs receive a Delta signal via the germline protrusions and become Notch activated (yellow). These two parallel mechanisms of Notch signaling status acquisition secure the first step of the stem cell niche assembly—TF formation. Later on, the most posterior of the TFC in the stalk (TC, dark blue) experiences a reprogramming of its Notch signal-receiving status, becoming a Delta-sending cell instead. With its new ability of sending the signal, it recruits the second element of the niche, the CpCs (yellow, dark nucleus), making it ready to host the stem cells. All yellow cells have Notch signaling ON, and blue cells have Notch signaling OFF (Yatsenko and Shcherbata, 2021).
There are two major cell types that form the ovarian GSC niche per se: TFCs and CpCs. Despite originating from different somatic cell precursors and forming different developmental time points, both rely heavily on proper Notch signaling. However, the acquisition of a certain Notch signaling status in both cell types varies significantly and depends on the cell type, position, and the source of the Delta ligand. The formation of the GSC niche is a sequential process, where TFCs are formed first, followed by CpC specification. While both elements are part of the niche, the CpCs are the ones in direct contact with the GSCs, acting as the signaling source for their specification and maintenance (Figure 2). First, during development, the newly specified GSCs are recruited only from primordial germ cells (PGCs) located in CpC proximity. Second, only the GSCs that are physically attached to the niche are maintained in the adult germarium (Figure 2).
TFC specification begins at the third instar larva stage in the ovary that consists of anterior cells (ACs) and intermingled cells (ICs) that are intermingled with PGCs. In order to differentiate into TFCs, a certain status of Notch signaling (ON or OFF) has to be acquired by their anterior somatic cell precursors. Notch ON or OFF status in newly formed TFCs depends on their position in regard to PGCs. The cells closer to the germline are Notch active, while the cells more distant from the germline are not (Figure 2). The activation of Notch signaling in the posterior TFCs is achieved via PGC-generated protrusions that provide the source of the Delta ligand, resulting in the long-range Notch activation in posterior TFCs (Yatsenko and Shcherbata, 2021). In contrast to the posterior, the anterior TFCs express high levels of membrane-bound Delta, which prohibits Notch signaling activation via the cis- or trans-inhibition mode (Yatsenko and Shcherbata, 2021). Therefore, the commitment to TF cell fate is achieved by both Notch signaling activation and suppression. Such a dual mechanism of achieving a certain Notch status in TFC precursors provides a double security mechanism, which reinforces the first step of the stem cell niche formation, the TF assembly (Figure 2). The presence of this double mechanism is supported by the data showing that even in ovaries without PGCs acting as a Delta ligand source, the TFs and the niche, despite not being fully normal, are still being formed (Yatsenko and Shcherbata, 2021). This indicates that Delta from PGCs only plays an instructive role in this process, and the Notch signaling cis- or trans-inhibition in Notch and Delta co-expressing TF precursors is used to reassure TF formation.
The next step of ovarian GSC niche formation is CpC differentiation. CpCs originate from another somatic cell type called intermingled somatic cells (ICs), which are mixed in between PGCs. In order for CpCs to differentiate from ICs, they need to activate Notch signaling. This is achieved by Delta ligand expression by the most posterior TFC called the transient cell (TC) (Figure 2). Since the TC already has Notch signaling activated, in order to become a Delta signal-sending cell, it needs to be reprogrammed via steroid signaling. Steroids activate miRNA-125 that targets Tom, a Neuralized inhibitor (Yatsenko and Shcherbata, 2018). As a result, the presence of the Neuralized allows Dl to be processed by endocytosis.
Processed Delta from the most posterior TFC binds to the Notch receptor present at the membranes of the adjacent somatic CpC precursors (ICs), leading to Notch signaling activation via local induction. ICs are bivalent, since they express both Notch and Delta. Notch activation via local induction performed by the TC resolves this bivalency, committing them to CpC fate for life (Yatsenko and Shcherbata, 2018). This concludes the process of the ovarian stem cell niche establishment.
If the expected reprogramming does not occur, the natural fluctuation of Delta will stochastically permit a cell to own the signal-sending status. Since CpC precursors co-express Delta and Notch, slightly higher levels of Delta are sufficient to induce Notch signaling in the neighboring equipotent cells via lateral inhibition (Yatsenko and Shcherbata, 2018). This leads to the appearance of ectopic but functional niches, underlining nature’s imperative to maintain fertility despite defects or errors.
Newly formed CpCs signal to proximal PGCs, transforming them into GSCs, which finalizes the assembly of the adult ovarian GSC niche unit. Interestingly, with various ways to activate Notch signaling in the developing ovarian GSC niche, the intensity of Notch activation is different among the cells that compose it. For example, posterior TFCs and CpCs are in close proximity but the levels of Notch activation in these cells differ, where TFCs have high Notch activity while CpCs have much lower Notch activity (Yatsenko and Shcherbata, 2021). In this case, the kinetics of Notch signaling strongly depend on the geometry of the cell contacts, the expression levels of Notch and Delta, and the range at which the Delta ligand is sent. Such various levels of Notch activity are important for the correct cell fate determination since the environment in which the niche is specified contains many bivalent cells (both ACs and ICs). Due to stochastic fluctuations, they could randomly choose Delta signal-sending status, resulting in the appearance of unnecessary ectopic niches.
We still have only witnessed the tip of the iceberg, as we know very little about the dynamics of the Notch signaling in the living cell, including aspects that are less explored, such as the role of cell mechanics in this process. Therefore, observing how Notch signaling occurs in the living organism could provide us with many new answers that will help us to better understand how the ovarian niche is formed.
On one level, distant Notch signaling governs the induction in the somatic precursors of terminal filament cell (TFC) fate by the germline-produced Delta ligand. Notch activation ensures TFC cell fate acquisition, which eventually leads to the TFC cell shape change, intercalation, and stalk formation (Figure 2). Primordial germ cells (PGCs) are able to send the Delta signal several cell diameters to the adjacent anterior somatic cells by using cellular projections (Yatsenko and Shcherbata, 2021). This makes sense, as the niche has to form in the proximity of its stem cells. What enforces the importance of the germline projections is that TFC precursors lack the ubiquitin ligase Neuralized, meaning that there will be no Delta endocytosis to apply the necessary mechanical strain to trigger the signal (Yatsenko and Shcherbata, 2018). Not only that, but also these projections could be dynamic, pulling-force generators, causing through their retraction the needed mechanical cue (Hadjivasiliou et al., 2016). This explains why Notch signaling is “ON” in the posterior TFCs touched by Delta-bearing projections, while Notch is “OFF” in the anterior TFCs that have high Delta but no Neuralized. This mechanism is particularly relevant because it has been observed in various processes, from morphogenesis to tumorigenesis (Boukhatmi et al., 2020).
Taken together, these data perfectly describe the multiple Notch signaling modes, such as spatial cellular Delta–Notch trans- and cis-interactions. In time, TFCs and CpCs eventually acquire a certain Notch signaling status because of the differential Notch signaling activation. Once the ball is rolling, higher spatial organization and fate commitment are gradually gained, until the stem cell niches are formed. In the mature ovarian niche, Notch ON/OFF signaling patterns are once again needed in the life-long maintenance of the GSCs and stem cell self-renewal which are a requirement for optimal fertility. In developing and mature tissues, Notch is indispensable, a fact that will become even clearer later in this review. It makes for a very promising target for regenerative medicine, cell therapies, and cancer treatments, and there are more excellent examples to learn from.
Testis
The ovarian stem cell niche is not the only model that can give us a glimpse into Notch involvement in niche assembly. Research in the Drosophila testicular niche has unveiled even earlier Notch signaling implication in the gonadal niche development. This is because the somatic gonadal precursors (SGPs) terminally differentiate at the late embryo stage, long before larval transition (Wawersik et al., 2005; Casper and Van Doren, 2006). They originate from the mesoderm, while the PGCs develop at the posterior pole of the embryo. Being initially on the outside of the embryo, the PGCs migrate through the endoderm to reach the mesodermal part called the posterior midgut (PM). While they are traveling, the SGPs are specified from the lateral mesoderm and meet with the PGCs at stage 11 at the PM (Sonnenblick, 1941; Brookman et al., 1992; Boyle and DiNardo, 1995; Boyle et al., 1997). They finally coalesce at stage 14, and at the end of the last embryonic stage, they are organized in two round gonads with an already specified niche, called the “hub” (Figure 3) (Boyle and DiNardo, 1995; Boyle et al., 1997; Jenkins et al., 2003; Clark et al., 2007). The hub consists of cells derived from a subgroup of the apical SGPs. Notch signaling in posterior SGPs is antagonized by the PGC-induced EGFR (epidermal growth factor receptor), a mechanism which restricts and ensures that only anterior Notch-positive SGPs become hub cells (Kitadate and Kobayashi, 2010).
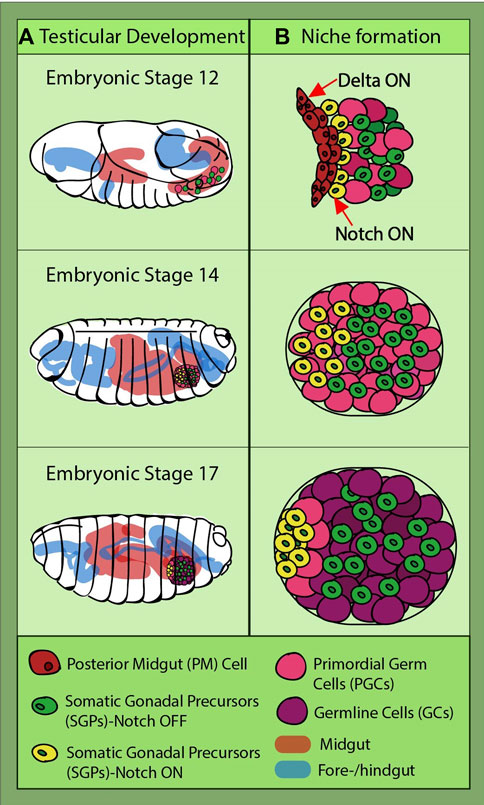
FIGURE 3. Drosophila testicular development and niche formation. (A) presents an overview of the journey of the primordial germ cells (PGCs) (pink) and somatic gonadal precursors (SGPs) (green and yellow) which takes place during embryonic stages 12–17, the last one before hatching into a larva. They are strung along together with the posterior midgut (PM); at stage 14, they leave the gut, coalesce, and go on to organize themselves into two round gonads. (B) depicts the direct interaction between the PM cells and some of the SGPs (yellow), this being the moment when their fate is sealed through Notch induction caused by the Delta-sending gut cells. Notch-active SGPs will go on to form the germline niche and recruit GSCs (light pink).
Further studies confirmed Notch activation in a subgroup of SGPs (Okegbe and DiNardo, 2011). During their embryogenic journey from the endoderm to mesoderm, SGPs receive a Notch-inducing signal from the Delta-expressing posterior gut cells (Figure 3). A drastic reduction in hub cell numbers was observed in the fog mutant embryos, in which the posterior midgut is not internalized, preventing SGP communication with the Delta-expressing gut cells (Okegbe and DiNardo, 2011). Later on, the same group identified a large Maf transcription factor, Traffic jam (Tj), as the downstream target of Notch induction in hub precursors (Wingert and DiNardo, 2015). After SGPs and PGCs coalesce, in the Notch-activated SGPs, Tj is downregulated. The relatively long time (∼6 h) between Notch activation and reduction in detectable Tj levels suggests intermediary steps in this repression cascade. Reduced Tj levels lead to a relief in unpaired and fasciclin III inhibition, allowing the cells to acquire the hub cell fate. Conversely, tj mutants yielded ectopic niches containing cells, of which some were not completely converted to hub cells. This fits with other findings which revealed that a gene called midline is needed for Tj accumulation in early-stage SGPs (Tripathy et al., 2014). Interestingly, midline has been found in other tissues to antagonize Notch signaling and is being inhibited in Notch-positive cells (Das et al., 2013). This would be an interesting avenue for further exploration to determine whether midline is regulated by Notch in SGPs.
Wingert and DiNardo (2015) concluded in their experiments that Notch activates another transcription factor, Bowl, along with Tj inhibition. Bowl activation was enough to rescue the ectopic niches in tj mutants in terms of cell morphology, number, aggregation, and hub localization. The exact relationship between Notch and Bowl has not yet been elucidated in male gonadal development, and speculating is difficult given that their interaction is context dependent (de Celis and Bray, 2003; Hao et al., 2003; Benitez et al., 2009; Greenberg and Hatini, 2009). Nevertheless, similar to the Notch–Tj relationship, understanding Notch–Bowl interaction in greater detail would be instrumental in figuring out the subtle control mechanisms behind gonadal niche formation.
A valuable lesson that can be taken from both male and female Drosophila gonadal niche formation is the important role of Tj in coordinating the events, especially considering its emerging connection with Notch. This has a great translational potential, as both mammalian Tj orthologues, c-Maf and MafB, are expressed in the somatic cells dispersed between the germline in the developing mammalian gonad (DeFalco et al., 2011), while at the same time, Notch prevents differentiation of the somatic cell progenitors (Tang et al., 2008). Overall, despite our current rudimentary understanding, data highlight Notch as one of the main coordinators of gonadal stem cell niche establishment.
To enforce that idea, a recent study identified multiple signaling pathways that are active in the developing human gonad, among which was also Notch. Although the exact make-up of mammalian spermatogonial niches has not been elucidated, their cells follow a similar trajectory during development and have contact with other germ layers (Zamboni and Upadhyay, 1982; McLaren, 1991; Satoh, 1991). Single-cell RNA-seq conducted by Li et al. (2017) provided a framework to understand Notch signaling-mediated communication between human gonadal somatic cells and their stem cells, the fetal germ cells (FGCs) (Li et al., 2017). They found that in the fetal ovary and testis, two Notch ligands are highly expressed: Delta-like ligand 3 (Dll3; in FGC in all their phases of development) and Jagged1 (specifically in oogenesis). Notch 2 receptor and its target gene, HES1, were expressed in nearly all somatic cells, making their potential interaction with FGCs extremely likely. One subgroup of the somatic cells also expresses Dll3, opening the possibility of somatic inter-communication. Concomitantly, the BMP signaling pathway seems to run in parallel with Notch (Li et al., 2017). Gradually, there is a pattern emerging, especially when compared to the aforementioned gonadal stem cell niches. Although at this point, it is only speculative, it might not be a stretch of the imagination to identify similar mechanisms in the human gonadal niche in the future.
Mammalian stem cell niches and Notch
The role of Notch signaling in the adult stem cell niches has been demonstrated in multiple systems, including mammalian models. In comparison to the straightforward Delta–Notch pathway organization in flies and worms, mammals have five Notch ligands (Delta-like 1, 3, and 4; and Jagged 1 and 2) and four Notch receptors (Bray, 2006).
For example, it has been proposed that quiescent neural stem cells (NSCs) produce their own niche cells using the Notch ligand Delta-like ligand 1 (Dll1) (Kawaguchi et al., 2013). The Dll1 protein is induced in activated NSCs and segregates to one daughter cell during mitosis. Dll1-expressing cells reside in close proximity to quiescent NSCs, which allows a feedback signal to maintain quiescent neural stem cells in the adult mouse subventricular zone, while keeping a balance between NSCs and their progeny. There are data suggesting an additional feedback mechanism via EGFR signaling in the progeny cells that cause cell non-autonomous Notch signaling reduction in NSCs and induction of neurogenesis in the progeny (Aguirre et al., 2010). As a result, the number of NSCs decreases, resulting in shrinkage of the stem cell pool having homeostatic and compositional altering effects on the overall tissue.
Another great example is the adult muscle satellite stem cells in which cell-autonomous Notch activity is able to induce the production of ECM collagens. These collagens are essential components of the muscle stem cell niche and are essential for the quiescence maintenance of the satellite cells. When activated, NCID targets its effector, recombining binding protein suppressor of hairless (RBP-J), and consequently upregulates enhancers close to important collagen genes (Col5a1, Col5a3, Col6a1, and Col6a2) (Baghdadi et al., 2018a). Activation of Notch signaling in adult muscle satellite cells is required for production of the ECM collagen V (COLV), a critical component of the quiescent stem cell niche (Baghdadi et al., 2018a). Specifically, deletion of Col5a1 leads to abnormal cell cycle entry and gradual decline in stem cells. The model proposed here has Notch as sensor of homeostatic changes and physical damage of the niche, by experiencing a sharp downregulation and pushing the stem cells out of their quiescence (Mourikis et al., 2012; Mourikis and Tajbakhsh, 2014). OFF Notch signal means less collagen V and more activated satellite cells are ready to regenerate the tissue. Interestingly, the myogenic cells only interacted with COLV when it was added to the cell medium, but not when it was a part of the coating substrate, suggesting that it acts as a signaling molecule. Searching for collagen receptors, the only cells that showed slowed down proliferation in the presence of COLV were the calcitonin receptor (CALCR)-positive cells. Subsequent experiments put CALCR downstream of the Notch-induced quiescence axis (Baghdadi et al., 2018a). When the CALCR ligand, elcatonin, was administered in Col5a1-null mice stem cells, they displayed a higher stem cell marker expression and experienced a prolongation of the G0-to-S transition (Baghdadi et al., 2018a). This means that not only satellite muscle cell state can be directly influenced, but also that CALCR adds a deeper layer of quantitative and qualitative quiescence control. Recently, it has been found that Notch is even more versatile in its activity because it ensures that the satellite cells maintain their physical position by driving the expression of miR-708, which targets the Tns3 transcripts. They code for the focal adhesion component tensin-3, and its downregulation directly affects the migratory mechanisms of the satellite cells, keeping them in place (Baghdadi et al., 2018b). Of course, there are more levels of complexity regarding Delta–Notch signaling within the mammalian stem cell niches than already presented, especially when it comes to the balance modulation between stem cell self-renewal and regeneration upon injury, for example, extensively reviewed in Kann et al. (2021). However, in the scope of the present review, we focused on the role of Notch signaling in the process of stem cell niche establishment.
The more secluded intestinal crypt is a stem cell niche that, in time, attracted considerable attention, especially due to the arduous efforts of recreating it in vitro for different purposes. The very dynamic single-layered intestinal epithelium has a unique wave-like architecture, with invaginations called crypts and protrusions named villi. In the crypt resides a stem cell population made of highly proliferative Lgr5+ intestinal stem cells, active crypt base columnar (CBC) stem cells, and facultative stem cells. The stem cells ensure the replenishment of epithelial cells going through a rapid turnover of only a few days (Barker et al., 2007). Similar to other stem cells, they self-renew while generating transit amplifying cells that later on migrate up the villus and terminally differentiate into absorptive enterocytes, mucus-secreting goblet cells, and hormone-secreting enteroendocrine cells. The only stationary cells remaining next to the stem cells are the Paneth cells with a half-life of several weeks. In case of stem cell loss, the facultative quiescent stem cell population kickstarts their cell cycle, occupies the empty niche, and generates progeny (Bankaitis et al., 2018). Wnt/R-spondin and Notch signaling represent the primary pathways involved in intestinal cell renewal. Similar to the satellite muscle niche, Notch inhibition results in stem cell loss, niche collapse, and an amplification in the secretory cell type number (van Es et al., 2005; Riccio et al., 2008; Pellegrinet et al., 2011; VanDussen et al., 2012). However, when it comes to the crypt formation in vivo, Notch involvement is presently unclear. Luckily, in vitro models such as organoids have recently shed light on the process of intestinal niche establishment and uncovered a central role of Notch.
Organoids
The constantly popular intestinal organoid has been a valuable tool to understand the initial events of gut development and 3D self-organization. They are created by seeding a single Lgr5+ stem cell in Matrigel which is capable of giving rise to all required cell types (Sato et al., 2009; Spence et al., 2011). However, the principles behind this self-organization are not completely understood. At first, the stem cells organize in a sphere-shaped conformation until the first so-called symmetry breaking event occurs when the Paneth cells emerge and start secreting the Wnt3a ligand (Sato et al., 2011). As the name suggests, this event is meant to polarize the structure and trigger the niche establishment.
When it comes to the intestinal crypt and Notch, in vitro studies have taught us important lessons. Serra et al. (2019) used single-cell genomics and imaging to determine the mechanisms of crypt formation. Notch activity in the organoid has been found to have two sides: symmetry breakage and cell fate maintenance (Serra et al., 2019). The latter has been previously discussed, while the former needs further attention. It seems that until the 4-cell stage, the stem cells equally present the Hippo signaling transcription factor and mechanosensor Yap1 in the nucleus (Figure 4). During the transition to the 8-cell stage, presumably due to the increased crowding of the cells, a subset of cells translocates Yap1 to their cytosol where it is inactivated (Serra et al., 2019). It was found that this Yap1 activation pattern is the one responsible for triggering symmetry breaking. This means that the cells with remaining high levels of nuclear Yap1 start to also express Dll1, which is consistent with Dll1 being a Yap1 target in other tissues (Gregorieff et al., 2015; Totaro et al., 2017). As a result, neighboring cells start expressing the Notch target, Hes1 (Serra et al., 2019). Dll1-positive cells gradually lose the nuclear Yap1 between the 16-cell and the 32-cell stages and commit to the Paneth cell fate (Figure 4). This is backed by the consequent loss of Yap1 target gene expression. To confirm these findings, γ-secretase inhibitors were applied, which resulted in the reduced symmetry breaking, Paneth cell differentiation, and the increase in the fraction of enterocytes (Serra et al., 2019). Overall, Notch/Dll1 activation represents the necessary nudge for the Paneth cell fate acquisition for symmetry breaking and later on, for the coordination of distinct cell fate specifications. Additionally, recent findings show the importance of the Yap ON/OFF and Notch ON/OFF circuitry in the spatial organization of the organoid (Gjorevski et al., 2022). Apparently, Paneth cells exclusively differentiate, and niches are initialized at the meeting point between a Yap1 ON cell and its Yap1 OFF neighbor. Yap1 activity was also strongly and directly correlated with the physical spreading of the cultured cells (Gjorevski et al., 2022). This links the environmental mechanosensing ability of Hippo signaling with its alternate activity patterning and in the end, with Notch–Delta lateral inhibition, selective Paneth cell differentiation, and crypt formation.
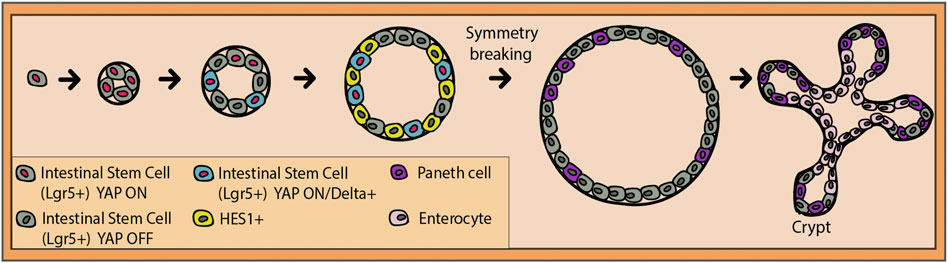
FIGURE 4. Transition from a single intestinal stem cell (gray) to an intestinal organoid. Transcription factor YAP is active in all stem cells until the four-cell stage (red nucleus). Presumably, due to physically increasing crowding of the stem cells, some of them will export YAP into the cytoplasm where it gets deactivated (dark gray nucleus). The ones that still maintain YAP ON will gradually start to express Delta (blue cells with red nucleus), and their neighbors will express a Notch target, HES1 (yellow cells with dark gray nucleus). Entering the 16–32-cell stage, the Delta-positive cells lose their nuclear YAP and are ready to commit to the Paneth cell fate. This event breaks the symmetry of the organoid and pushes the formation of the intestinal crypt (niche), where stem cells and Paneth cells reside.
Although less studied, the organoid model of cervical cancer also showed sensitivity for fluctuating Notch levels. It is mimicking the endo-ectocervical transition zones where up to 90% of cervical cancer originates (Burghardt and Ostor, 1983; Deng et al., 2018). Therefore, this model organoid was created to enable detailed studies between inflammation induction (e.g., HPV infection), the appearance of abnormal changes in the niche and cancer occurrence. In that transition region resides a population of ectocervical stem cells which maintain complex interactions with their adjacent stromal niche cell subpopulations (Lohmussaar et al., 2021). A gene expression screen revealed high expression of the Notch ligands Dll3 and manic fringe (MFNG), while differentiated cells presented high expression levels of Notch 2 and 3 receptors, and their targets, HES1 and presenilin 1 (PSEN1) (Chumduri et al., 2021). The trans-activating interaction encourages differentiation and spatial complexity in the organoid, a fact sustained by the negative effect that γ-secretase inhibition has on organoid architecture (Chumduri et al., 2021). Considering all this, it is only valid to wonder if these observations also apply to cancer stem cells and their niches.
Cancer
The concept of cancer stem cells (CSCs) has been laid out around four decades ago and describes how resident adult stem cells are pushed by various factors to assume a malignant identity and create cancerous tumors. They achieve that by hijacking the normal abilities of regeneration sustained by healthy stem cells and have been identified in many cancers (Lapidot et al., 1994; Uckun et al., 1995; Bonnet and Dick, 1997; Al-Hajj et al., 2003; Singh et al., 2004; Dalerba et al., 2007; O'Brien et al., 2007; Ricci-Vitiani et al., 2007). Despite the fact that many of these CSCs are tied to a niche, the simple act of eradicating their support as a therapeutic strategy has not yielded the expected success. Fairly recently, due to an updated and more realistic view on CSCs and their microenvironment, effective therapies have started to develop (Sun et al., 2019). Subsequent to the CSC identification, lineage tracing revealed a functional niche for some of these cells and later studies pinpointed Notch signaling as one of the active partners in the CSC dynamics across the board (Hu et al., 2012; Takebe et al., 2015).
Looking at the broader picture, early dysregulation of Notch signaling comes up as a hallmark of CSC appearance and establishment, becoming a more attractive target for all types of cancers (Farnie and Clarke, 2007; Saunders et al., 2015). Aberrant Notch induction in CSCs promotes many pro-proliferative downstream targets and helps the tumor to create a supportive environment (Androutsellis-Theotokis et al., 2006; Schreck et al., 2010; Steg et al., 2014). At times, these malignant stem cells overtake and invade the “home” of their healthy neighbors (Hu et al., 2012). We are exploring both scenarios, as Notch is a key factor that is being manipulated in cancer niche initiation and expansion.
Supported in the hypoxic environment in which tumors like to sprout, the CSCs secrete the vascular endothelial growth factor (VEGF) inducing angiogenesis (Lau et al., 2017) (Figure 5A). The vascular endothelial naturally responds to this signal, and nearby blood vessels start to bud new branches and consequently create a niche to support the tumor and its CSCs. The VEGF-mediated upregulation of Dll4 or JAG1 expression in the epithelial tip cells of the budding vessel induces Notch signal trans-activation in the adjacent epithelial cell (Mailhos et al., 2001; Patel et al., 2005; Patel et al., 2006; Cao et al., 2014). This lateral inhibition represses the tip cell fate in the neighboring cell by inhibiting Dll4 and VEGF2 expressions and allowing it to gain the alternative fate, that of the stalk cell. Both types are required for proper vessel formation, as Notch inactivation promotes tip cell overproliferation, spatial disorganization, and instability of the newly formed vasculature (Mack and Iruela-Arispe, 2018). Increasing Dll4 and Notch activity leads to a sparse vasculature network, while turning it off via blockers had the opposite effect with the added paradoxical outcome of partially non-functional blood vessels (Noguera-Troise et al., 2006). Interestingly, it has been shown that endothelial cells directly promote the stem cell features in cancer cells through Notch signaling (Zhu et al., 2011; Lu et al., 2013; Cao et al., 2014; Ghiabi et al., 2014). Under normal circumstances, the crosstalk between VEGF and Notch is meant to coordinate and ensure the proper tube morphology and appropriate organization of the developing vasculature (Blanco and Gerhardt, 2013; Mack and Iruela-Arispe, 2018). The tumor hijacks this process for its own benefit, creating a niche capable of concomitantly sustaining cancer stem cell pool and the continuous tumor growth. This phenomenon vividly illustrates one of the complex ways in which CSCs manipulate their microenvironment through Notch.
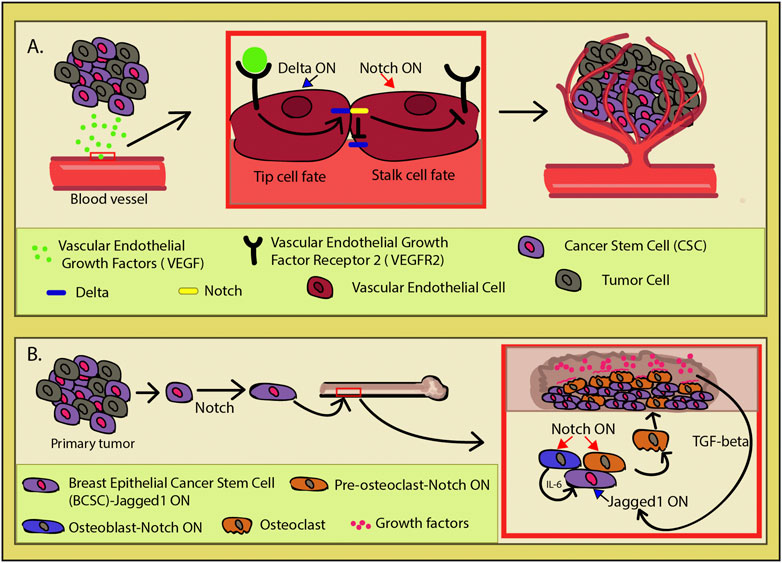
FIGURE 5. Cancer-induced angiogenesis. (A) Cancer stem cells (CSCs) secrete vascular endothelial growth factors (VEGFs) which induces in the nearby endothelium the process of vascular budding. VEGF activation upregulates Delta-like ligand 4 (Dll4) (blue) expression. In order to achieve functional vasculature during angiogenesis, endothelial cells need to acquire one of two cell fates (tip or stalk). These two phenotypes are necessary for a functional vasculature and exhibit different behaviors. The tip cell has migratory qualities and leads the nascent vasculature toward the VEGF source, while the stalk cells have a supportive role in the angiogenesis. Dll4 induction in an endothelial cell induces lateral inhibition in its neighbor through Notch (yellow) activation which in turn inhibits Dll4 and vascular endothelial growth factor receptor 2 (VEGFR2) expression (red square). This whole process is a two-way street, as the CSCs directly induce angiogenesis, while the newly formed vasculature network actively promotes the stem cell features of the CSCs, making it the perfect cancer niche. (B) In breast cancer, after the primary tumor is established, some breast epithelial cancer stem cells (BCSCs) (purple) go through biochemical changes and undergo the Notch-mediated loss of some epithelial characteristics. This transformation confers cell capacity to migrate and invade new tissues. In particular, cells have an affinity toward bone as they initiate a self-sustained program of invasion. By expressing Jagged1 (JAG1), they are able to induce Notch in the osteoblasts (blue) and osteoclasts (orange). Osteoblasts will then secrete IL-6, promoting tumor growth, while osteoclasts go into osteoclastogenesis and erode the bone, making space for the expanding mass. Bone destruction releases growth factors (pink), initiating the TGF-β pathway which upregulates JAG1 in the invading cancers cells and reinforcing the vicious cycle.
It gets even more intriguing when Notch operates as a compass for metastatic cells to find a new microenvironment to colonize. In the breast epithelium primary tumor, hypoxia induces the loss of some epithelial characteristics via Jagged1(JAG1)/Notch activation in the breast cancer stem cells (BCSCs) (Al-Hajj and Clarke, 2004; Martin et al., 2005; Leong et al., 2007; Sahlgren et al., 2008; Gangopadhyay et al., 2013; Shao et al., 2015) (Figure 5B). BCSCs undergo biochemical changes and lose their epithelial cell status, exhibiting migratory capacity, invasiveness, resistance to apoptosis, and greatly elevated production of ECM components (Al-Hajj et al., 2003; Kalluri and Neilson, 2003; Scioli et al., 2019). Multiple studies demonstrate that upon Notch activation in non-invasive breast cancer cells, they gain the invasive and migratory qualities in vivo which is correlated with metastasis and poor prognosis (Bolos et al., 2013; Kontomanolis et al., 2014; Castro et al., 2015; Lai et al., 2018; Leontovich et al., 2018). Among multiple dysregulated factors, Notch signaling activation also causes elevation of the urokinase-type plasminogen activator (uPA), β1-integrin, β-catenin, matrix metalloproteinase (MMP)-2, and MMP-9 levels, conferring the cancer cells with even greater ability to invade new tissues (Shimizu et al., 2011; Lai et al., 2018; Wagley et al., 2020).
In metastatic breast cancer cells, NOTCH4, NOTCH3, and JAG1 were found to be upregulated compared to other cancer cell types (Lawson et al., 2015). Particularly, JAG1 expression was tied with bone-tropic metastatic breast cancer. SMAD-dependent TGF-β signaling induces JAG1 upregulation in the cancer cells and activates Notch 1 in osteoblasts within the niche which then secretes IL-6, stimulating the tumor growth and affecting osteoclast differentiation (Figure 5B) (Sethi et al., 2011; Wagley et al., 2020). Osteoclastogenesis is upregulated by Notch, leading to bone erosion and supporting cancer invasion through TGF-β activation. That is due to the bone being a reservoir of growth factors that are released upon osteolysis. The newly released growth factors go on to close the vicious feedback loop between the cancer cells and their newly created niche, as one of the TGF-β targets is JAG1 (Figure 5). The cycle is reinforced by JAG1 continuous activation, which in turn amplifies its effect in the tumors’ advantage, acting as an important mediator between the bone microenvironment and freshly seeded metastatic cells (Sethi et al., 2011).
In short, the TGFβ–Notch axis enables the invasive cancer cells to discover compatible niches, and once settled in, it allows the new tumor to enhance the space it takes through perturbation and destruction of the endogenous tissue. This final point only builds on the still incomplete but fascinating ways in which this Notch signaling pathway operates in health, disease, and development.
Final thoughts
Based on the examples illustrated in this review, Notch signaling has distinguished itself as a widespread stem cell niche sculptor. It remains to be seen if Notch signaling activation is a universal requisite for stem cell niche formation, which seems to be the tendency to which the discoveries so far point. Similarly, the concept of stem cell niches and stem cells is ever evolving, and recent studies suggest a unifying neural stemness as the ground state of tumorigenicity (Cao, 2022; Zhang et al., 2022). It draws significant parallels between tumorigenicity and pluripotent differentiation potential, both of which provide evidence for common cellular and biochemical processes (e.g., cell cycle, gene expression, and metabolism), which paints a picture of fundamental and universal mechanisms that govern the development of organisms. Notch signaling fits perfectly in this universal language as illustrated in the establishment of the stem cell niches across biological systems. From niches responsible for the maintenance of species reproduction to tissue morphogenesis and tumorigenesis, it serves as a simple but efficient and versatile communication bridge between pluripotent cells and their niches. Along with other cellular signaling pathways, the Notch pathway shapes the outcome of multicellular interactions in many different scenarios, and perhaps it is time to take a more comprehensive view of disease development and onset by learning and understanding the fundamental patterns that make up the world around us.
Author contributions
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
Acknowledgments
The authors apologize to colleagues whose work they were unable to include. The authors would like to thank all members of the Shcherbata laboratory and the reviewers for helpful suggestions and critical reading of the manuscript. The authors would like to thank VolkswagenStiftung and DAAD for funding.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Ables, J. L., Breunig, J. J., Eisch, A. J., and Rakic, P. (2011). Not(ch) just development: Notch signalling in the adult brain. Nat. Rev. Neurosci. 12, 269–283. doi:10.1038/nrn3024
Aguirre, A., Rubio, M. E., and Gallo, V. (2010). Notch and EGFR pathway interaction regulates neural stem cell number and self-renewal. Nature 467, 323–327. doi:10.1038/nature09347
Al-Hajj, M., and Clarke, M. F. (2004). Self-renewal and solid tumor stem cells. Oncogene 23, 7274–7282. doi:10.1038/sj.onc.1207947
Al-Hajj, M., Wicha, M. S., Benito-Hernandez, A., Morrison, S. J., and Clarke, M. F. (2003). Prospective identification of tumorigenic breast cancer cells. Proc. Natl. Acad. Sci. U. S. A. 100, 3983–3988. doi:10.1073/pnas.0530291100
Androutsellis-Theotokis, A., Leker, R. R., Soldner, F., Hoeppner, D. J., Ravin, R., Poser, S. W., et al. (2006). Notch signalling regulates stem cell numbers in vitro and in vivo. Nature 442, 823–826. doi:10.1038/nature04940
Baghdadi, M. B., Castel, D., Machado, L., Fukada, S. I., Birk, D. E., Relaix, F., et al. (2018a). Reciprocal signalling by Notch-Collagen V-CALCR retains muscle stem cells in their niche. Nature 557, 714–718. doi:10.1038/s41586-018-0144-9
Baghdadi, M. B., Firmino, J., Soni, K., Evano, B., Di Girolamo, D., Mourikis, P., et al. (2018b). Notch-induced miR-708 antagonizes satellite cell migration and maintains quiescence. Cell Stem Cell 23, 859–868. doi:10.1016/j.stem.2018.09.017
Bankaitis, E. D., Ha, A., Kuo, C. J., and Magness, S. T. (2018). Reserve stem cells in intestinal homeostasis and injury. Gastroenterology 155, 1348–1361. doi:10.1053/j.gastro.2018.08.016
Barad, O., Rosin, D., Hornstein, E., and Barkai, N. (2010). Error minimization in lateral inhibition circuits. Sci. Signal. 3, ra51. doi:10.1126/scisignal.2000857
Barker, N., Van Es, J. H., Kuipers, J., Kujala, P., Van Den Born, M., Cozijnsen, M., et al. (2007). Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449, 1003–1007. doi:10.1038/nature06196
Benhra, N., Vignaux, F., Dussert, A., Schweisguth, F., and Le Borgne, R. (2010). Neuralized promotes basal to apical transcytosis of delta in epithelial cells. Mol. Biol. Cell 21, 2078–2086. doi:10.1091/mbc.e09-11-0926
Benitez, E., Bray, S. J., Rodriguez, I., and Guerrero, I. (2009). Lines is required for normal operation of Wingless, Hedgehog and Notch pathways during wing development. Development 136, 1211–1221. doi:10.1242/dev.021428
Blanco, R., and Gerhardt, H. (2013). VEGF and Notch in tip and stalk cell selection. Cold Spring Harb. Perspect. Med. 3, a006569. doi:10.1101/cshperspect.a006569
Bolos, V., Mira, E., Martinez-Poveda, B., Luxan, G., Canamero, M., Martinez, A. C., et al. (2013). Notch activation stimulates migration of breast cancer cells and promotes tumor growth. Breast Cancer Res. 15, R54. doi:10.1186/bcr3447
Bonnet, D., and Dick, J. E. (1997). Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat. Med. 3, 730–737. doi:10.1038/nm0797-730
Boukhatmi, H., Martins, T., Pillidge, Z., Kamenova, T., and Bray, S. (2020). Notch mediates inter-tissue communication to promote tumorigenesis. Curr. Biol. 30, 1809–1820. doi:10.1016/j.cub.2020.02.088
Boyle, M., Bonini, N., and Dinardo, S. (1997). Expression and function of clift in the development of somatic gonadal precursors within the Drosophila mesoderm. Development 124, 971–982. doi:10.1242/dev.124.5.971
Boyle, M., and Dinardo, S. (1995). Specification, migration and assembly of the somatic cells of the Drosophila gonad. Development 121, 1815–1825. doi:10.1242/dev.121.6.1815
Bray, S. J. (2006). Notch signalling: A simple pathway becomes complex. Nat. Rev. Mol. Cell Biol. 7, 678–689. doi:10.1038/nrm2009
Brookman, J. J., Toosy, A. T., Shashidhara, L. S., and White, R. A. (1992). The 412 retrotransposon and the development of gonadal mesoderm in Drosophila. Development 116, 1185–1192. doi:10.1242/dev.116.4.1185
Burghardt, E., and Ostor, A. G. (1983). Site and origin of squamous cervical cancer: A histomorphologic study. Obstet. Gynecol. 62, 117–127.
Byun, S. H., Kwon, M., Lee, S. M., Noh, H., and Yoon, K. (2019). PACT increases mammalian embryonic neural stem cell properties by facilitating activation of the notch signaling pathway. Biochem. Biophys. Res. Commun. 513, 392–397. doi:10.1016/j.bbrc.2019.04.010
Cao, Y. (2022). Neural is fundamental: Neural stemness as the ground state of cell tumorigenicity and differentiation potential. Stem Cell Rev. Rep. 18, 37–55. doi:10.1007/s12015-021-10275-y
Cao, Z., Ding, B. S., Guo, P., Lee, S. B., Butler, J. M., Casey, S. C., et al. (2014). Angiocrine factors deployed by tumor vascular niche induce B cell lymphoma invasiveness and chemoresistance. Cancer Cell 25, 350–365. doi:10.1016/j.ccr.2014.02.005
Carlson, M. E., Suetta, C., Conboy, M. J., Aagaard, P., Mackey, A., Kjaer, M., et al. (2009). Molecular aging and rejuvenation of human muscle stem cells. EMBO Mol. Med. 1, 381–391. doi:10.1002/emmm.200900045
Casper, A., and Van Doren, M. (2006). The control of sexual identity in the Drosophila germline. Development 133, 2783–2791. doi:10.1242/dev.02415
Castro, N. P., Fedorova-Abrams, N. D., Merchant, A. S., Rangel, M. C., Nagaoka, T., Karasawa, H., et al. (2015). Cripto-1 as a novel therapeutic target for triple negative breast cancer. Oncotarget 6, 11910–11929. doi:10.18632/oncotarget.4182
Chakrabarti, R., Celia-Terrassa, T., Kumar, S., Hang, X., Wei, Y., Choudhury, A., et al. (2018). Notch ligand Dll1 mediates cross-talk between mammary stem cells and the macrophageal niche. Science 360, eaan4153. doi:10.1126/science.aan4153
Chumduri, C., Gurumurthy, R. K., Berger, H., Dietrich, O., Kumar, N., Koster, S., et al. (2021). Opposing Wnt signals regulate cervical squamocolumnar homeostasis and emergence of metaplasia. Nat. Cell Biol. 23, 184–197. doi:10.1038/s41556-020-00619-0
Clark, I. B., Jarman, A. P., and Finnegan, D. J. (2007). Live imaging of Drosophila gonad formation reveals roles for Six4 in regulating germline and somatic cell migration. BMC Dev. Biol. 7, 52. doi:10.1186/1471-213X-7-52
Cohen, B., Bashirullah, A., Dagnino, L., Campbell, C., Fisher, W. W., Leow, C. C., et al. (1997). Fringe boundaries coincide with Notch-dependent patterning centres in mammals and alter Notch-dependent development in Drosophila. Nat. Genet. 16, 283–288. doi:10.1038/ng0797-283
Cohen, M., Georgiou, M., Stevenson, N. L., Miodownik, M., and Baum, B. (2010). Dynamic filopodia transmit intermittent Delta-Notch signaling to drive pattern refinement during lateral inhibition. Dev. Cell 19, 78–89. doi:10.1016/j.devcel.2010.06.006
Dalerba, P., Dylla, S. J., Park, I. K., Liu, R., Wang, X., Cho, R. W., et al. (2007). Phenotypic characterization of human colorectal cancer stem cells. Proc. Natl. Acad. Sci. U. S. A. 104, 10158–10163. doi:10.1073/pnas.0703478104
Das, S., Chen, Q. B., Saucier, J. D., Drescher, B., Zong, Y., Morgan, S., et al. (2013). The Drosophila T-box transcription factor Midline functions within the Notch-Delta signaling pathway to specify sensory organ precursor cell fates and regulates cell survival within the eye imaginal disc. Mech. Dev. 130, 577–601. doi:10.1016/j.mod.2013.08.001
De, C., and Bray, S. (1997). Feed-back mechanisms affecting Notch activation at the dorsoventral boundary in the Drosophila wing. Development 124, 3241–3251. doi:10.1242/dev.124.17.3241
De Celis, J. M., and Bray, S. J. (2003). Bowl is required downstream of Notch for elaboration of distal limb patterning. Development 130, 5943–5952. doi:10.1242/dev.00833
De Joussineau, C., Soule, J., Martin, M., Anguille, C., Montcourrier, P., and Alexandre, D. (2003). Delta-promoted filopodia mediate long-range lateral inhibition in Drosophila. Nature 426, 555–559. doi:10.1038/nature02157
Defalco, T., Takahashi, S., and Capel, B. (2011). Two distinct origins for Leydig cell progenitors in the fetal testis. Dev. Biol. 352, 14–26. doi:10.1016/j.ydbio.2011.01.011
Del Alamo, D., Rouault, H., and Schweisguth, F. (2011). Mechanism and significance of cis-inhibition in Notch signalling. Curr. Biol. 21, R40–R47. doi:10.1016/j.cub.2010.10.034
Deng, H., Hillpot, E., Mondal, S., Khurana, K. K., and Woodworth, C. D. (2018). HPV16-Immortalized cells from human transformation zone and endocervix are more dysplastic than ectocervical cells in organotypic culture. Sci. Rep. 8, 15402. doi:10.1038/s41598-018-33865-2
Farnie, G., and Clarke, R. B. (2007). Mammary stem cells and breast cancer--role of Notch signalling. Stem Cell Rev. 3, 169–175. doi:10.1007/s12015-007-0023-5
Fiuza, U. M., and Arias, A. M. (2007). Cell and molecular biology of Notch. J. Endocrinol. 194, 459–474. doi:10.1677/JOE-07-0242
Fuller, M. T., and Spradling, A. C. (2007). Male and female Drosophila germline stem cells: Two versions of immortality. Science 316, 402–404. doi:10.1126/science.1140861
Gangopadhyay, S., Nandy, A., Hor, P., and Mukhopadhyay, A. (2013). Breast cancer stem cells: A novel therapeutic target. Clin. Breast Cancer 13, 7–15. doi:10.1016/j.clbc.2012.09.017
Ghiabi, P., Jiang, J., Pasquier, J., Maleki, M., Abu-Kaoud, N., Rafii, S., et al. (2014). Endothelial cells provide a notch-dependent pro-tumoral niche for enhancing breast cancer survival, stemness and pro-metastatic properties. PLoS One 9, e112424. doi:10.1371/journal.pone.0112424
Gjorevski, N., Nikolaev, M., Brown, T. E., Mitrofanova, O., Brandenberg, N., Delrio, F. W., et al. (2022). Tissue geometry drives deterministic organoid patterning. Science 375, eaaw9021. doi:10.1126/science.aaw9021
Greenberg, L., and Hatini, V. (2009). Essential roles for lines in mediating leg and antennal proximodistal patterning and generating a stable Notch signaling interface at segment borders. Dev. Biol. 330, 93–104. doi:10.1016/j.ydbio.2009.03.014
Gregorieff, A., Liu, Y., Inanlou, M. R., Khomchuk, Y., and Wrana, J. L. (2015). Yap-dependent reprogramming of Lgr5(+) stem cells drives intestinal regeneration and cancer. Nature 526, 715–718. doi:10.1038/nature15382
Hadjivasiliou, Z., Hunter, G. L., and Baum, B. (2016). A new mechanism for spatial pattern formation via lateral and protrusion-mediated lateral signalling. J. R. Soc. Interface 13, 20160484. doi:10.1098/rsif.2016.0484
Hao, I., Green, R. B., Dunaevsky, O., Lengyel, J. A., and Rauskolb, C. (2003). The odd-skipped family of zinc finger genes promotes Drosophila leg segmentation. Dev. Biol. 263, 282–295. doi:10.1016/j.ydbio.2003.07.011
Hu, Y., Ye, Y., and Fortini, M. E. (2002). Nicastrin is required for gamma-secretase cleavage of the Drosophila Notch receptor. Dev. Cell 2, 69–78. doi:10.1016/s1534-5807(01)00105-8
Hu, Y. Y., Zheng, M. H., Zhang, R., Liang, Y. M., and Han, H. (2012). Notch signaling pathway and cancer metastasis. Adv. Exp. Med. Biol. 727, 186–198. doi:10.1007/978-1-4614-0899-4_14
Hunter, G. L., Hadjivasiliou, Z., Bonin, H., He, L., Perrimon, N., Charras, G., et al. (2016). Coordinated control of Notch/Delta signalling and cell cycle progression drives lateral inhibition-mediated tissue patterning. Development 143, 2305–2310. doi:10.1242/dev.134213
Hunter, G. L., He, L., Perrimon, N., Charras, G., Giniger, E., and Baum, B. (2019). A role for actomyosin contractility in Notch signaling. BMC Biol. 17, 12. doi:10.1186/s12915-019-0625-9
Irvine, K. D., and Wieschaus, E. (1994). fringe, a Boundary-specific signaling molecule, mediates interactions between dorsal and ventral cells during Drosophila wing development. Cell 79, 595–606. doi:10.1016/0092-8674(94)90545-2
Jenkins, A. B., Mccaffery, J. M., and Van Doren, M. (2003). Drosophila E-cadherin is essential for proper germ cell-soma interaction during gonad morphogenesis. Development 130, 4417–4426. doi:10.1242/dev.00639
Kalluri, R., and Neilson, E. G. (2003). Epithelial-mesenchymal transition and its implications for fibrosis. J. Clin. Invest. 112, 1776–1784. doi:10.1172/JCI20530
Kann, A. P., Hung, M., and Krauss, R. S. (2021). Cell-cell contact and signaling in the muscle stem cell niche. Curr. Opin. Cell Biol. 73, 78–83. doi:10.1016/j.ceb.2021.06.003
Kawaguchi, D., Furutachi, S., Kawai, H., Hozumi, K., and Gotoh, Y. (2013). Dll1 maintains quiescence of adult neural stem cells and segregates asymmetrically during mitosis. Nat. Commun. 4, 1880. doi:10.1038/ncomms2895
Khait, I., Orsher, Y., Golan, O., Binshtok, U., Gordon-Bar, N., Amir-Zilberstein, L., et al. (2016). Quantitative analysis of delta-like 1 membrane dynamics elucidates the role of contact geometry on notch signaling. Cell Rep. 14, 225–233. doi:10.1016/j.celrep.2015.12.040
Kitadate, Y., and Kobayashi, S. (2010). Notch and Egfr signaling act antagonistically to regulate germ-line stem cell niche formation in Drosophila male embryonic gonads. Proc. Natl. Acad. Sci. U. S. A. 107, 14241–14246. doi:10.1073/pnas.1003462107
Kontomanolis, E., Panteliadou, M., Giatromanolaki, A., Pouliliou, S., Efremidou, E., Limberis, V., et al. (2014). Delta-like ligand 4 (DLL4) in the plasma and neoplastic tissues from breast cancer patients: Correlation with metastasis. Med. Oncol. 31, 945. doi:10.1007/s12032-014-0945-0
Kooh, P. J., Fehon, R. G., and Muskavitch, M. A. (1993). Implications of dynamic patterns of Delta and Notch expression for cellular interactions during Drosophila development. Development 117, 493–507. doi:10.1242/dev.117.2.493
Kovall, R. A., Gebelein, B., Sprinzak, D., and Kopan, R. (2017). The canonical notch signaling pathway: Structural and biochemical insights into shape, sugar, and force. Dev. Cell 41, 228–241. doi:10.1016/j.devcel.2017.04.001
Kushwah, R., Guezguez, B., Lee, J. B., Hopkins, C. I., and Bhatia, M. (2014). Pleiotropic roles of Notch signaling in normal, malignant, and developmental hematopoiesis in the human. EMBO Rep. 15, 1128–1138. doi:10.15252/embr.201438842
Lai, E. C. (2004). Notch signaling: Control of cell communication and cell fate. Development 131, 965–973. doi:10.1242/dev.01074
Lai, X. X., Li, G., Lin, B., and Yang, H. (2018). Interference of Notch 1 inhibits the proliferation and invasion of breast cancer cells: Involvement of the β‑catenin signaling pathway. Mol. Med. Rep. 17, 2472–2478. doi:10.3892/mmr.2017.8161
Langevin, J., Le Borgne, R., Rosenfeld, F., Gho, M., Schweisguth, F., and Bellaiche, Y. (2005). Lethal giant larvae controls the localization of notch-signaling regulators numb, neuralized, and Sanpodo in Drosophila sensory-organ precursor cells. Curr. Biol. 15, 955–962. doi:10.1016/j.cub.2005.04.054
Lapidot, T., Sirard, C., Vormoor, J., Murdoch, B., Hoang, T., Caceres-Cortes, J., et al. (1994). A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature 367, 645–648. doi:10.1038/367645a0
Lau, E. Y., Ho, N. P., and Lee, T. K. (2017). Cancer stem cells and their microenvironment: Biology and therapeutic implications. Stem Cells Int. 2017, 3714190. doi:10.1155/2017/3714190
Lawson, D. A., Bhakta, N. R., Kessenbrock, K., Prummel, K. D., Yu, Y., Takai, K., et al. (2015). Single-cell analysis reveals a stem-cell program in human metastatic breast cancer cells. Nature 526, 131–135. doi:10.1038/nature15260
Le Borgne, R. (2006). Regulation of Notch signalling by endocytosis and endosomal sorting. Curr. Opin. Cell Biol. 18, 213–222. doi:10.1016/j.ceb.2006.02.011
Le Borgne, R., Bardin, A., and Schweisguth, F. (2005). The roles of receptor and ligand endocytosis in regulating Notch signaling. Development 132, 1751–1762. doi:10.1242/dev.01789
Le Borgne, R., and Schweisguth, F. (2003). Unequal segregation of Neuralized biases Notch activation during asymmetric cell division. Dev. Cell 5, 139–148. doi:10.1016/s1534-5807(03)00187-4
Leong, K. G., Niessen, K., Kulic, I., Raouf, A., Eaves, C., Pollet, I., et al. (2007). Jagged1-mediated Notch activation induces epithelial-to-mesenchymal transition through Slug-induced repression of E-cadherin. J. Exp. Med. 204, 2935–2948. doi:10.1084/jem.20071082
Leontovich, A. A., Jalalirad, M., Salisbury, J. L., Mills, L., Haddox, C., Schroeder, M., et al. (2018). NOTCH3 expression is linked to breast cancer seeding and distant metastasis. Breast Cancer Res. 20, 105. doi:10.1186/s13058-018-1020-0
Li, L., Dong, J., Yan, L., Yong, J., Liu, X., Hu, Y., et al. (2017). Single-cell RNA-seq analysis maps development of human germline cells and gonadal niche interactions. Cell Stem Cell 20, 891–892. doi:10.1016/j.stem.2017.05.009
Lissemore, J. L., and Starmer, W. T. (1999). Phylogenetic analysis of vertebrate and invertebrate Delta/Serrate/LAG-2 (DSL) proteins. Mol. Phylogenet. Evol. 11, 308–319. doi:10.1006/mpev.1998.0588
Lohmussaar, K., Oka, R., Espejo Valle-Inclan, J., Smits, M. H. H., Wardak, H., Korving, J., et al. (2021). Patient-derived organoids model cervical tissue dynamics and viral oncogenesis in cervical cancer. Cell Stem Cell 28, 1380–1396 e6. doi:10.1016/j.stem.2021.03.012
Lopez-Schier, H., and St Johnston, D. (2002). Drosophila nicastrin is essential for the intramembranous cleavage of notch. Dev. Cell 2, 79–89. doi:10.1016/s1534-5807(01)00109-5
Lu, J., Ye, X., Fan, F., Xia, L., Bhattacharya, R., Bellister, S., et al. (2013). Endothelial cells promote the colorectal cancer stem cell phenotype through a soluble form of Jagged-1. Cancer Cell 23, 171–185. doi:10.1016/j.ccr.2012.12.021
Mack, J. J., and Iruela-Arispe, M. L. (2018). NOTCH regulation of the endothelial cell phenotype. Curr. Opin. Hematol. 25, 212–218. doi:10.1097/MOH.0000000000000425
Mailhos, C., Modlich, U., Lewis, J., Harris, A., Bicknell, R., Ish-Horowicz, D., et al. (2001). Delta4, an endothelial specific notch ligand expressed at sites of physiological and tumor angiogenesis. Differentiation. 69, 135–144. doi:10.1046/j.1432-0436.2001.690207.x
Maillard, I. (2014). Notch and human hematopoietic stem cells. Blood 123, 1115–1116. doi:10.1182/blood-2014-01-546754
Maine, E. M., Lissemore, J. L., and Starmer, W. T. (1995). A phylogenetic analysis of vertebrate and invertebrate Notch-related genes. Mol. Phylogenet. Evol. 4, 139–149. doi:10.1006/mpev.1995.1014
Martin, T. A., Goyal, A., Watkins, G., and Jiang, W. G. (2005). Expression of the transcription factors snail, slug, and twist and their clinical significance in human breast cancer. Ann. Surg. Oncol. 12, 488–496. doi:10.1245/ASO.2005.04.010
Mathur, D., Bost, A., Driver, I., and Ohlstein, B. (2010). A transient niche regulates the specification of Drosophila intestinal stem cells. Science 327, 210–213. doi:10.1126/science.1181958
Mclaren, A. (1991). Development of the mammalian gonad: The fate of the supporting cell lineage. Bioessays 13, 151–156. doi:10.1002/bies.950130402
Metz, C. W., and Bridges, C. B. (1917). Incompatibility of mutant races in Drosophila. Proc. Natl. Acad. Sci. U. S. A. 3, 673–678. doi:10.1073/pnas.3.12.673
Mourikis, P., Sambasivan, R., Castel, D., Rocheteau, P., Bizzarro, V., and Tajbakhsh, S. (2012). A critical requirement for notch signaling in maintenance of the quiescent skeletal muscle stem cell state. Stem Cells 30, 243–252. doi:10.1002/stem.775
Mourikis, P., and Tajbakhsh, S. (2014). Distinct contextual roles for Notch signalling in skeletal muscle stem cells. BMC Dev. Biol. 14, 2. doi:10.1186/1471-213X-14-2
Nandagopal, N., Santat, L. A., Lebon, L., Sprinzak, D., Bronner, M. E., and Elowitz, M. B. (2018). Dynamic ligand discrimination in the notch signaling pathway. Cell 172, 869–880. doi:10.1016/j.cell.2018.01.002
Noguera-Troise, I., Daly, C., Papadopoulos, N. J., Coetzee, S., Boland, P., Gale, N. W., et al. (2006). Blockade of Dll4 inhibits tumour growth by promoting non-productive angiogenesis. Nature 444, 1032–1037. doi:10.1038/nature05355
O'Brien, C. A., Pollett, A., Gallinger, S., and Dick, J. E. (2007). A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature 445, 106–110. doi:10.1038/nature05372
Okegbe, T. C., and Dinardo, S. (2011). The endoderm specifies the mesodermal niche for the germline in Drosophila via Delta-Notch signaling. Development 138, 1259–1267. doi:10.1242/dev.056994
Palmer, W. H., Jia, D., and Deng, W. M. (2014). Cis-interactions between Notch and its ligands block ligand-independent Notch activity. Elife 3, e04415. doi:10.7554/eLife.04415
Pan, D., and Rubin, G. M. (1997). Kuzbanian controls proteolytic processing of Notch and mediates lateral inhibition during Drosophila and vertebrate neurogenesis. Cell 90, 271–280. doi:10.1016/s0092-8674(00)80335-9
Patel, N. S., Dobbie, M. S., Rochester, M., Steers, G., Poulsom, R., Le Monnier, K., et al. (2006). Up-regulation of endothelial delta-like 4 expression correlates with vessel maturation in bladder cancer. Clin. Cancer Res. 12, 4836–4844. doi:10.1158/1078-0432.CCR-06-0285
Patel, N. S., Li, J. L., Generali, D., Poulsom, R., Cranston, D. W., and Harris, A. L. (2005). Up-regulation of delta-like 4 ligand in human tumor vasculature and the role of basal expression in endothelial cell function. Cancer Res. 65, 8690–8697. doi:10.1158/0008-5472.CAN-05-1208
Pellegrinet, L., Rodilla, V., Liu, Z., Chen, S., Koch, U., Espinosa, L., et al. (2011). Dll1- and dll4-mediated notch signaling are required for homeostasis of intestinal stem cells. Gastroenterology 140, 1230–1240 e1-7. doi:10.1053/j.gastro.2011.01.005
Perez-Mockus, G., and Schweisguth, F. (2017). Cell polarity and notch signaling: Linked by the E3 ubiquitin ligase neuralized? BioEssays 39, 1700128. doi:10.1002/bies.201700128
Petrovic, J., Formosa-Jordan, P., Luna-Escalante, J. C., Abello, G., Ibanes, M., Neves, J., et al. (2014). Ligand-dependent Notch signaling strength orchestrates lateral induction and lateral inhibition in the developing inner ear. Development 141, 2313–2324. doi:10.1242/dev.108100
Qi, H., Rand, M. D., Wu, X., Sestan, N., Wang, W., Rakic, P., et al. (1999). Processing of the notch ligand delta by the metalloprotease Kuzbanian. Science 283, 91–94. doi:10.1126/science.283.5398.91
Ricci-Vitiani, L., Lombardi, D. G., Pilozzi, E., Biffoni, M., Todaro, M., Peschle, C., et al. (2007). Identification and expansion of human colon-cancer-initiating cells. Nature 445, 111–115. doi:10.1038/nature05384
Riccio, O., Van Gijn, M. E., Bezdek, A. C., Pellegrinet, L., Van Es, J. H., Zimber-Strobl, U., et al. (2008). Loss of intestinal crypt progenitor cells owing to inactivation of both Notch1 and Notch2 is accompanied by derepression of CDK inhibitors p27Kip1 and p57Kip2. EMBO Rep. 9, 377–383. doi:10.1038/embor.2008.7
Rodriguez-Esteban, C., Schwabe, J. W., Pena, J. L. D., Foys, B., Eshelman, B., and Izpisua, B. (1997). Radical fringe positions the apical ectodermal ridge at the dorsoventral boundary of the vertebrate limb. Nature 386, 360–366. doi:10.1038/386360a0
Sahlgren, C., Gustafsson, M. V., Jin, S., Poellinger, L., and Lendahl, U. (2008). Notch signaling mediates hypoxia-induced tumor cell migration and invasion. Proc. Natl. Acad. Sci. U. S. A. 105, 6392–6397. doi:10.1073/pnas.0802047105
Sato, T., Van Es, J. H., Snippert, H. J., Stange, D. E., Vries, R. G., Van Den Born, M., et al. (2011). Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature 469, 415–418. doi:10.1038/nature09637
Sato, T., Vries, R. G., Snippert, H. J., Wetering, M. V. D., Barker, N., Stange, D. E., et al. (2009). Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459, 262–265. doi:10.1038/nature07935
Satoh, M. (1991). Histogenesis and organogenesis of the gonad in human embryos. J. Anat. 177, 85–107.
Saunders, L. R., Bankovich, A. J., Anderson, W. C., Aujay, M. A., Bheddah, S., Black, K., et al. (2015). A DLL3-targeted antibody-drug conjugate eradicates high-grade pulmonary neuroendocrine tumor-initiating cells in vivo. Sci. Transl. Med. 7, 302ra136. doi:10.1126/scitranslmed.aac9459
Schofield, R. (1978). The relationship between the spleen colony-forming cell and the haemopoietic stem cell. Blood Cells 4, 7–25.
Schreck, K. C., Taylor, P., Marchionni, L., Gopalakrishnan, V., Bar, E. E., Gaiano, N., et al. (2010). The notch target Hes1 directly modulates Gli1 expression and hedgehog signaling: A potential mechanism of therapeutic resistance. Clin. Cancer Res. 16, 6060–6070. doi:10.1158/1078-0432.CCR-10-1624
Scioli, M. G., Storti, G., D'Amico, F., Gentile, P., Fabbri, G., Cervelli, V., et al. (2019). The role of breast cancer stem cells as a prognostic marker and a target to improve the efficacy of breast cancer therapy. Cancers (Basel) 11, 1021. doi:10.3390/cancers11071021
Serra, D., Mayr, U., Boni, A., Lukonin, I., Rempfler, M., Challet Meylan, L., et al. (2019). Self-organization and symmetry breaking in intestinal organoid development. Nature 569, 66–72. doi:10.1038/s41586-019-1146-y
Sethi, N., Dai, X., Winter, C. G., and Kang, Y. (2011). Tumor-derived JAGGED1 promotes osteolytic bone metastasis of breast cancer by engaging notch signaling in bone cells. Cancer Cell 19, 192–205. doi:10.1016/j.ccr.2010.12.022
Seugnet, L., Simpson, P., and Haenlin, M. (1997). Requirement for dynamin during Notch signaling in Drosophila neurogenesis. Dev. Biol. 192, 585–598. doi:10.1006/dbio.1997.8723
Shao, S., Zhao, X., Zhang, X., Luo, M., Zuo, X., Huang, S., et al. (2015). Notch1 signaling regulates the epithelial-mesenchymal transition and invasion of breast cancer in a Slug-dependent manner. Mol. Cancer 14, 28. doi:10.1186/s12943-015-0295-3
Shaya, O., Binshtok, U., Hersch, M., Rivkin, D., Weinreb, S., Amir-Zilberstein, L., et al. (2017). Cell-cell contact area affects notch signaling and notch-dependent patterning. Dev. Cell 40, 505–511. doi:10.1016/j.devcel.2017.02.009
Shimizu, M., Cohen, B., Goldvasser, P., Berman, H., Virtanen, C., and Reedijk, M. (2011). Plasminogen activator uPA is a direct transcriptional target of the JAG1-Notch receptor signaling pathway in breast cancer. Cancer Res. 71, 277–286. doi:10.1158/0008-5472.CAN-10-2523
Singh, S. K., Hawkins, C., Clarke, I. D., Squire, J. A., Bayani, J., Hide, T., et al. (2004). Identification of human brain tumour initiating cells. Nature 432, 396–401. doi:10.1038/nature03128
Song, X., Call, G. B., Kirilly, D., and Xie, T. (2007). Notch signaling controls germline stem cell niche formation in the Drosophila ovary. Development 134, 1071–1080. doi:10.1242/dev.003392
Sonnenblick, B. P. (1941). Germ cell movements and sex differentiation of the gonads in the Drosophila embryo. Proc. Natl. Acad. Sci. U. S. A. 27, 484–489. doi:10.1073/pnas.27.10.484
Spence, J. R., Mayhew, C. N., Rankin, S. A., Kuhar, M. F., Vallance, J. E., Tolle, K., et al. (2011). Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature 470, 105–109. doi:10.1038/nature09691
Spradling, A. C., Nystul, T., Lighthouse, D., Morris, L., Fox, D., Cox, R., et al. (2008). Stem cells and their niches: Integrated units that maintain Drosophila tissues. Cold Spring Harb. Symp. Quant. Biol. 73, 49–57. doi:10.1101/sqb.2008.73.023
Sprinzak, D., Lakhanpal, A., Lebon, L., Garcia-Ojalvo, J., and Elowitz, M. B. (2011). Mutual inactivation of Notch receptors and ligands facilitates developmental patterning. PLoS Comput. Biol. 7, e1002069. doi:10.1371/journal.pcbi.1002069
Sprinzak, D., Lakhanpal, A., Lebon, L., Santat, L. A., Fontes, M. E., Anderson, G. A., et al. (2010). Cis-interactions between Notch and Delta generate mutually exclusive signalling states. Nature 465, 86–90. doi:10.1038/nature08959
Steg, A. D., Burke, M. R., Amm, H. M., Katre, A. A., Dobbin, Z. C., Jeong, D. H., et al. (2014). Proteasome inhibition reverses hedgehog inhibitor and taxane resistance in ovarian cancer. Oncotarget 5, 7065–7080. doi:10.18632/oncotarget.2295
Strutt, D., Johnson, R., Cooper, K., and Bray, S. (2002). Asymmetric localization of frizzled and the determination of notch-dependent cell fate in the Drosophila eye. Curr. Biol. 12, 813–824. doi:10.1016/s0960-9822(02)00841-2
Sun, H. R., Wang, S., Yan, S. C., Zhang, Y., Nelson, P. J., Jia, H. L., et al. (2019). Therapeutic strategies targeting cancer stem cells and their microenvironment. Front. Oncol. 9, 1104. doi:10.3389/fonc.2019.01104
Takebe, N., Miele, L., Harris, P. J., Jeong, W., Bando, H., Kahn, M., et al. (2015). Targeting notch, hedgehog, and Wnt pathways in cancer stem cells: Clinical update. Nat. Rev. Clin. Oncol. 12, 445–464. doi:10.1038/nrclinonc.2015.61
Tang, H., Brennan, J., Karl, J., Hamada, Y., Raetzman, L., and Capel, B. (2008). Notch signaling maintains Leydig progenitor cells in the mouse testis. Development 135, 3745–3753. doi:10.1242/dev.024786
Totaro, A., Castellan, M., Battilana, G., Zanconato, F., Azzolin, L., Giulitti, S., et al. (2017). YAP/TAZ link cell mechanics to Notch signalling to control epidermal stem cell fate. Nat. Commun. 8, 15206. doi:10.1038/ncomms15206
Tripathy, R., Kunwar, P. S., Sano, H., and Renault, A. D. (2014). Transcriptional regulation of Drosophila gonad formation. Dev. Biol. 392, 193–208. doi:10.1016/j.ydbio.2014.05.026
Tulina, N., and Matunis, E. (2001). Control of stem cell self-renewal in Drosophila spermatogenesis by JAK-STAT signaling. Science 294, 2546–2549. doi:10.1126/science.1066700
Uckun, F. M., Sather, H., Reaman, G., Shuster, J., Land, V., Trigg, M., et al. (1995). Leukemic cell growth in SCID mice as a predictor of relapse in high-risk B-lineage acute lymphoblastic leukemia. Blood 85, 873–878. doi:10.1182/blood.v85.4.873.bloodjournal854873
Van, J. H. E., Van, G. I. J. N., Riccio, O., Born, M. V. D., Vooijs, M., et al. (2005). Notch/gamma-secretase inhibition turns proliferative cells in intestinal crypts and adenomas into goblet cells. Nature 435, 959–963. doi:10.1038/nature03659
Vandussen, K. L., Carulli, A. J., Keeley, T. M., Patel, S. R., Puthoff, B. J., Magness, S. T., et al. (2012). Notch signaling modulates proliferation and differentiation of intestinal crypt base columnar stem cells. Development 139, 488–497. doi:10.1242/dev.070763
Vanorny, D. A., and Mayo, K. E. (2017). The role of Notch signaling in the mammalian ovary. Reproduction 153, R187–R204. doi:10.1530/REP-16-0689
Wagley, Y., Chesi, A., Acevedo, P. K., Lu, S., Wells, A. D., Johnson, M. E., et al. (2020). Canonical Notch signaling is required for bone morphogenetic protein-mediated human osteoblast differentiation. Stem Cells 38, 1332–1347. doi:10.1002/stem.3245
Ward, E. J., Shcherbata, H. R., Reynolds, S. H., Fischer, K. A., Hatfield, S. D., and Ruohola-Baker, H. (2006). Stem cells signal to the niche through the Notch pathway in the Drosophila ovary. Curr. Biol. 16, 2352–2358. doi:10.1016/j.cub.2006.10.022
Wawersik, M., Milutinovich, A., Casper, A. L., Matunis, E., Williams, B., and Van Doren, M. (2005). Somatic control of germline sexual development is mediated by the JAK/STAT pathway. Nature 436, 563–567. doi:10.1038/nature03849
Wen, C., Metzstein, M. M., and Greenwald, I. (1997). SUP-17, a Caenorhabditis elegans ADAM protein related to Drosophila KUZBANIAN, and its role in LIN-12/NOTCH signalling. Development 124, 4759–4767. doi:10.1242/dev.124.23.4759
Wilkinson, H. A., Fitzgerald, K., and Greenwald, I. (1994). Reciprocal changes in expression of the receptor lin-12 and its ligand lag-2 prior to commitment in a C. elegans cell fate decision. Cell 79, 1187–1198. doi:10.1016/0092-8674(94)90010-8
Wingert, L., and Dinardo, S. (2015). Traffic jam functions in a branched pathway from Notch activation to niche cell fate. Development 142, 2268–2277. doi:10.1242/dev.124230
Xie, T., and Spradling, A. C. (2000). A niche maintaining germ line stem cells in the Drosophila ovary. Science 290, 328–330. doi:10.1126/science.290.5490.328
Yamamoto, S., Schulze, K. L., and Bellen, H. J. (2014). Introduction to notch signaling. Methods Mol. Biol. 1187, 1–14. doi:10.1007/978-1-4939-1139-4_1
Yatsenko, A. S., and Shcherbata, H. R. (2021). Distant activation of Notch signaling induces stem cell niche assembly. PLoS Genet. 17, e1009489. doi:10.1371/journal.pgen.1009489
Yatsenko, A. S., and Shcherbata, H. R. (2018). Stereotypical architecture of the stem cell niche is spatiotemporally established by miR-125-dependent coordination of Notch and steroid signaling. Development 145, dev159178. doi:10.1242/dev.159178
Zamboni, L., and Upadhyay, S. (1982). The contribution of the mesonephros to the development of the sheep fetal testis. Am. J. Anat. 165, 339–356. doi:10.1002/aja.1001650309
Zhang, M., Liu, Y., Shi, L., Fang, L., Xu, L., and Cao, Y. (2022). Neural stemness unifies cell tumorigenicity and pluripotent differentiation potential. J. Biol. Chem. 298, 102106. doi:10.1016/j.jbc.2022.102106
Zhang, N., and Gridley, T. (1998). Defects in somite formation in lunatic fringe-deficient mice. Nature 394, 374–377. doi:10.1038/28625
Zhao, X., Garcia, J. Q., Tong, K., Chen, X., Yang, B., Li, Q., et al. (2021). Polarized endosome dynamics engage cytoplasmic Par-3 that recruits dynein during asymmetric cell division. Sci. Adv. 7, eabg1244. doi:10.1126/sciadv.abg1244
Zhu, T. S., Costello, M. A., Talsma, C. E., Flack, C. G., Crowley, J. G., Hamm, L. L., et al. (2011). Endothelial cells create a stem cell niche in glioblastoma by providing NOTCH ligands that nurture self-renewal of cancer stem-like cells. Cancer Res. 71, 6061–6072. doi:10.1158/0008-5472.CAN-10-4269
Keywords: stem cell niche, cell differentiation, Delta, Notch, cancer, organoids, cell reprogramming
Citation: Zamfirescu A-M, Yatsenko AS and Shcherbata HR (2022) Notch signaling sculpts the stem cell niche. Front. Cell Dev. Biol. 10:1027222. doi: 10.3389/fcell.2022.1027222
Received: 24 August 2022; Accepted: 02 December 2022;
Published: 20 December 2022.
Edited by:
Acaimo González-Reyes, Andalusian Centre for Developmental Biology (CABD) (CSIC), SpainReviewed by:
Patricia Rojas Ríos, University of Seville, SpainSalvador C. Herrera, Andalusian Center for Development Biology (CSIC), Spain
Copyright © 2022 Zamfirescu, Yatsenko and Shcherbata. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Halyna R. Shcherbata, c2hjaGVyYmF0YS5oYWx5bmFAbWgtaGFubm92ZXIuZGU=
 Ana-Maria Zamfirescu
Ana-Maria Zamfirescu Andriy S. Yatsenko
Andriy S. Yatsenko Halyna R. Shcherbata
Halyna R. Shcherbata