
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cell Dev. Biol., 08 November 2022
Sec. Epigenomics and Epigenetics
Volume 10 - 2022 | https://doi.org/10.3389/fcell.2022.1012179
This article is part of the Research TopicProtein Modifications in Epigenetic Dysfunctional Diseases: Mechanisms and Potential Therapeutic StrategiesView all 15 articles
As a Class II tumor suppressor, ING5 contains nuclear localization signal, plant homeodomain, novel conserved region, and leucine zipper-like domains. ING5 proteins form homodimer into a coil-coil structure, and heterodimers with ING4, histone H3K4me3, histone acetyltransferase (HAT) complex, Tip60, Cyclin A1/CDK2, INCA1 and EBNA3C for the transcription of target genes. The acetylated proteins up-regulated by ING5 are preferentially located in nucleus and act as transcription cofactors, chromatin and DNA binding functions, while those down-regulated by ING5 mostly in cytoplasm and contribute to metabolism. ING5 promotes the autoacetylation of HAT p300, p53, histone H3 and H4 for the transcription of downstream genes (Bax, GADD45, p21, p27 and so forth). Transcriptionally, YY1 and SRF up-regulate ING5 mRNA expression by the interaction of YY1-SRF-p53-ING5 complex with ING5 promoter. Translationally, ING5 is targeted by miR-196, miR-196a, miR-196b-5p, miR-193a-3p, miR-27-3p, miR-200b/200a/429, miR-1307, miR-193, miR-222, miR-331-3p, miR-181b, miR-543 and miR-196-b. ING5 suppresses proliferation, migration, invasion and tumor growth of various cancer cells via the suppression of EGFR/PI3K/Akt, IL-6/STAT3, Akt/NF-κB/NF-κB/MMP-9 or IL-6/CXCL12 pathway. ING5-mediated chemoresistance is closely linked to anti-apoptosis, overexpression of chemoresistant genes, the activation of PI3K/Akt/NF-κB and Wnt/β-catenin signal pathways. Histologically, ING5 abrogation in gastric stem-like and pdx1-positive cells causes gastric dysplasia and cancer, and conditional ING5 knockout in pdx1-positive and gastric chief cells increases MNU-induced gastric carcinogenesis. Intestinal ING5 deletion increases AOM/DSS- induced colorectal carcinogenesis and decreases high-fat-diet weight. The overexpression and nucleocytoplasmic translocation of ING5 are seen during carcinogenesis, and ING5 expression was inversely associated with aggressive behaviors and poor prognosis in a variety of cancers. These findings indicated that ING5 might be used for a molecular marker for carcinogenesis and following progression, and as a target for gene therapy if its chemoresistant function might be ameliorated.
Carcinogenesis is a complex biological process, and characterized by both frequent genetic and epigenetic changes, including the inactivation of tumor suppressor genes (TSG) and the activation of oncogenes. The chromosomal deletion, mutation and hypermethylation of TSGs result in their loss (Class I) and inactivation (Class II), finally to malignantly transform the normal cells. Inhibitor of growth (ING) family members belong to Class II TSG and play a critical role in initiation, promotion and development of cancers (Zheng et al., 2011; Dantas et al., 2019). In the review, we demonstrate that ING5 interacts with ING4, histone H3K4me3, HAT (histone acetyltransferase) complex, Tip60, Cyclin A1/CDK2, INCA1 and EBNA3C, and promotes the autoacetylation of HAT p300, p53, histone H3 and H4 for the transcription of downstream genes. Transcriptionally, YY1 and SRF up-regulate ING5 mRNA expression by the interaction of YY1-SRF-p53-ING5 complex with ING5 promoter. Translationally, ING5 is targeted by miR-196, miR-193a-3p, miR-27-3p and so forth. ING5 suppresses aggressive phenotypes of various cancer cells via EGFR/PI3K/Akt, IL-6/STAT3, Akt/NF-κB/NF-κB/MMP-9 or IL-6/CXCL12 pathway. Histologically, conditional ING5 abrogation causes gastric dysplasia and cancer, and increases chemically-induced gastrointestinal carcinogenesis. In human cancer samples, the up-regulated expression of ING5 was in gastric, breast, and colorectal cancers, but down-regulated in head and neck squamous cell carcinoma (HNSCC), lung cancer, osteosarcoma, prostate cancer, ovarian cancer, hepatocellular carcinoma (HCC), esophageal cancer, and thyroid cancer. These results suggest that ING5 might be used for a molecular marker for carcinogenesis and following progression, and as a target for gene therapy.
The ING (inhibitor of growth) family is composed of ING1–5, and involved in apoptosis, senescence and DNA damage. They function as epigenetic readers of H3K4Me3 histone, and contribute to HAT or deacetylase complex formation. Their gatekeeper functions are evidenced by their arrest of the cell cycle, stabilization of p53, or maintenance of HAT activity. In contrast, their caretaker functions are evidenced by the nucleotide excision repair (NER) and double-strand break (DSB) repair of ING1-, and ING2- and ING3-mediated accumulation of ATM, BRCA1, and 53BP1 (Archambeau et al., 2019; Dantas et al., 2019). Among them, ING5 maps to human chromosome 2q37.3 (Figure 1A), contains 11 exons, theoretically spliced into 5 forms and encodes 5 proteins (Figure 1B). The 3 isoforms of ING5 are produced according to Genbank (Figure 1C). ING5 protein contains plant homeodomain (PHD), nuclear localization signal (NLS), novel conserved region (NCR), and leucine zipper-like (LZL) domains. Their NLS favors nucleic translocation, LZL mediates interaction with leucine zipper proteins, PHD interacts with such chromatin-interacting proteins as methylated histone H3, and NCR binds to HAT complex to remodel chromatin and modulate transcription (Zhang et al., 2017a; Archambeau et al., 2019; Dantas et al., 2019).
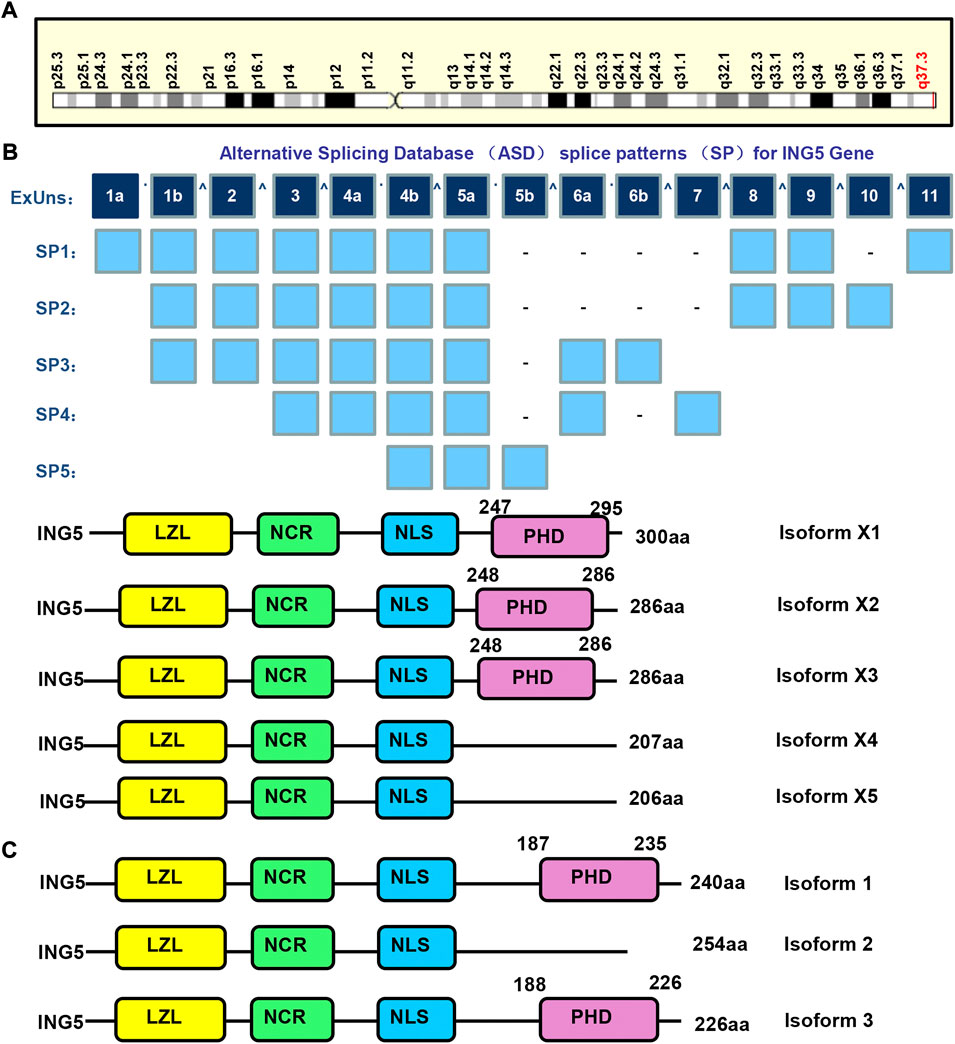
FIGURE 1. The structures of ING5 gene and protein. ING5 is localized in human chromosome 2q37.3 (A), and theoretically spliced into 5 forms and encodes 5 proteins (B). The 3 isoforms of ING5 are produced according to Genbank (C). LZL, leucine zipper-Like motif; NCR: novel conserved region; NLS, nuclear localization signal; PHD, plant homeodomain.
ING5 proteins form homodimer via their amino-terminal domain, and fold independently into a coil-coil structure. The PHD domains of the homodimer are chemically equivalent and independent of the rest of ING5. ING5 protein forms heterodimer with ING4 protein as well (Ormaza et al., 2019). ING5 binds to histone H3K4me3 and forms 2 HAT complexes, H3-MOZ-BRPF-MORF-ING5 and H4-JADE-HBO1-ING5. Both MCM and HAT complexes are involved in DNA replication because ING5 silencing abolishes DNA synthesis, and HBO1 silencing promotes the progression of S phase (Zhang et al., 2017a; Wu et al., 2018; Archambeau et al., 2019; Dantas et al., 2019; Ormaza et al., 2019). Zhang et al. (2011) identified ING5 as a partner of inhibitor of Cyclin A1 (INCA) that interacted with Cyclin A1/CDK2. Linzen et al. (2015) demonstrated that ING5 was phosphorylated at T152 by Cyclin E/CDK2 and Cyclin A/CDK2, which was repressed by CDK2 inhibitor p27KIP1. Wang et al. (2012) demonstrated ING5 suppressed the proliferation of mesenchymal stem cells by down-regulating CDK2 expression. ING5 and p53 bound to amino-terminal residues 129 to 200 of Epstein-Barr virus (EBV) EBNA3C. The conserved domain of ING5 competitively interacted with both p53 and EBNA3C. EBNA3C significantly inhibited ING5-mediated regulation of p53 transcription (Saha et al., 2011) (Figure 2).
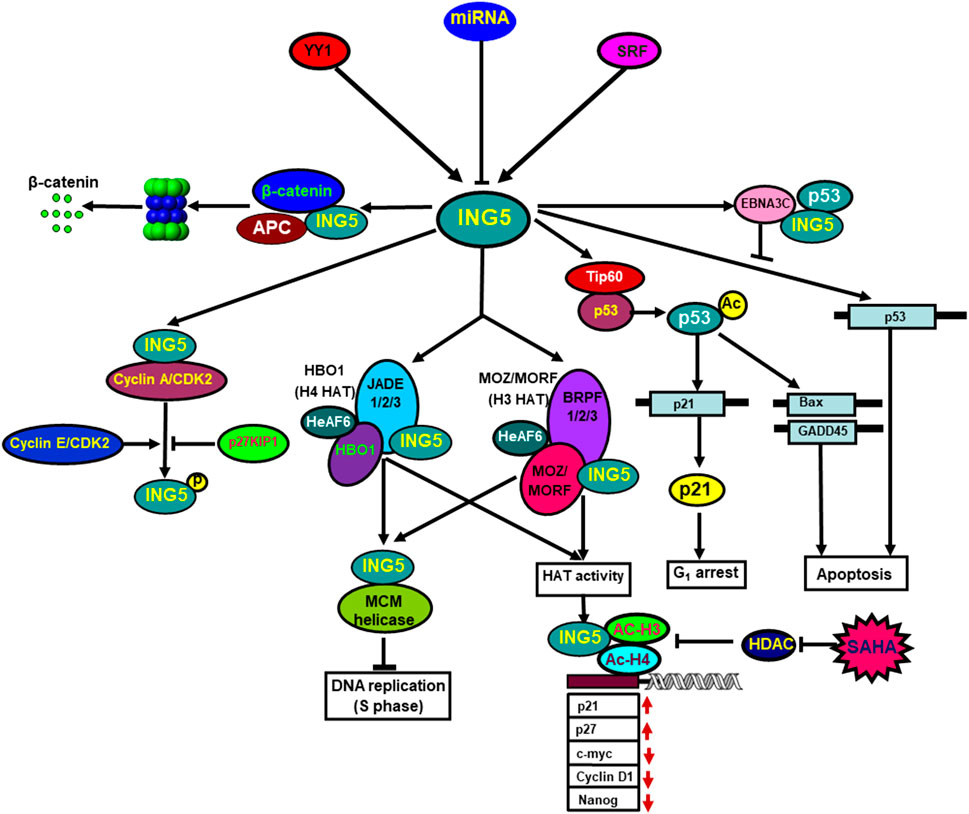
FIGURE 2. The biological functions of ING5. Transcriptionally, YY1 and SRF can up-regulate ING5 mRNA expression by the interaction of YY1-SRF-p53-ING5 complex with ING5 promoter. Translationally, ING5 expression is suppressed by as such miRNA targets as miR-196a, miR-193a-3p, miR-27-3p, miR-1307, miR-193, miR-222, miR-331-3p, miR-543 and so forth. ING5 interacts with histone H3K4me3 and is involved in the formation of two different HAT (histone acetyltransferase) complexes, (H4-HBO1-JADE-ING5 and H3-MOZ-MORF-BRPF-ING5), which contribute to DNA replication via ING5 and MCM complexes. ING5 binds to Cyclin A1/CDK2, and is phosphorylated by Cyclin E/CDK2 and Cyclin A/CDK2, which is repressed by CDK2 inhibitor p27KIP1. ING5 interacts with Epstain-Barr virus EBNA3C, which suppresses ING5-mediated transcription of p53. Additionally, ING5 assisted Tip60 to acetylate p53, and acetylated p53 at K120 binds to and activates the promoters of Bax and GADD45. ING5 also activates the p21 promoter to induce p21 expression via p53 acetylation at Lys-382. HAT and SAHA exposure recruited ING5 and acetylated histones H3 and H4 to the promoters of c-myc, nanog, Cyclin D1, p21, and p27, thereby regulating their transcription.
ING5 profoundly influences protein lysine acetylation with 122 of 163 acetylation peptides significantly up-regulated, and 72 of 100 acetylation peptides down-regulated by ING5. The acetylated proteins up-regulated by ING5 are preferentially located in nucleus, and act as transcription cofactors, chromatin and DNA binding factors, while those down-regulated by ING5 are principally in cytosol, and involved in cellular metabolism. ING5 promotes HAT p300 autoacetylation at K1555, K1558, K1560, K1647 and K1794, resulting in the activation of HAT and subsequent acetylation of p53 at K382 and histone H3 at K18. HAT inhibitor impairs ING5-mediated acetylation of H3K18 and p53K382, and subsequently down-regulates both p21 and Bax expression (Zhang et al., 2017b). Ormaza et al. (2019) found that ING5 acetylated histone H3 by HBO1 complex, and H4 by MOZ/MORF complex. Liu et al. (2013) found that ING5 facilitated Tip60 to acetylate p53 at K120 in response to DNA damage by the formation of p53-Tip60 complex. Acetylated p53 at K120 interacted with and activated the promoters of the apoptotic genes, Bax and GADD45. ING5 was reported to activate the cyclin-dependent kinase inhibitor p21 promoter and up-regulate p21 expression, and acetylate p53 protein at K382 (Shiseki et al., 2003) (Figure 2). Wang et al. (2018) found that ING5 enhanced the expression of stem cell markers (Oct4, Olig2 and Nestin) to promote self-renewal, prevent lineage differentiation and increase stem-like population of brain tumor initiating cells.
ING5 inhibits proliferation, glucose catabolism, anti-apoptosis, migration, invasion, epithelial- mesenchymal transition (EMT) or lung metastasis, and induces cell arrest, senescence, autophagy, fat accumulation, and chemoresistance in gastric, colorectal, breast, lung, ovarian cancer cells, neuroblastoma and glioma cells (Gou et al., 2015; Zhao et al., 2016; Ding et al., 2017; Zhao et al., 2017; Zheng et al., 2017; Wu et al., 2018; Yu et al., 2022; Zheng et al., 2022). ING5 inhibited tumor growth, blood supply or lung metastasis of gastric, colorectal, ovarian, breast or lung cancer cells by suppressing proliferation, and inducing autophagy and apoptosis in nude xenograft models. ING5-mediated chemoresistance was closely linked to anti-apoptosis, overexpression of chemoresistant genes, the activation of PI3K/Akt/NF-κB and Wnt/β-catenin signal pathways (Gou et al., 2015; Zhao et al., 2016; Ding et al., 2017; Zhao et al., 2017; Zheng et al., 2017; Wu et al., 2018; Yu et al., 2022). Qi and Zhang (2014) found that intact ING5 inhibited the proliferation and anti-apoptosis in tongue squamous cell carcinoma cells. In addition, 2 mutants of ING5 (aa 107-226 and 1-184) can facilitate cellular senescence with Cyclin E and CDK2 hypoexpression. In our study, we found that all wild-type and mutant ING5 had either the LZL or PHD domain, and weakened proliferation, migration and invasion of gastric cancer cells, which was closely linked to cdc-2, VEGF, and MMP-9 hypoexpression (Zheng et al., 2022).
ING5 was also demonstrated to inhibit proliferation, glucose catabolism, migration, invasion, EMT, and tumor growth, and promote apoptosis, cell cycle arrest, senescence, or autophagy in prostate cancer cells by activating p53 and inactivating Akt (Barlak et al., 2020), in lung cancer cells by phosphorylating β-catenin at S33/37, and suppressing IL-6/STAT3 and EGFR/PI3K/Akt pathways (Zhang et al., 2015; Liu et al., 2017; Liu et al., 2019), in neuroblastoma cells by acetylating histones (Wu et al., 2018), in colorectal cancer cells through PI3K/Akt pathway (Xin et al., 2020), in osteosarcoma cells via Smad pathway (Zhang et al., 2018a), in esophageal squamous carcinoma cells through IL-6/CXCL12 (Wang et al., 2021) and Akt/NF-κB/MMP-9 (Zhang et al., 2018b) pathways, and in breast cancer cells via PI3K/Akt/NF-κB pathway (Zhao et al., 2015; Cui et al., 2017), in glioma cells (Zhao et al., 2017) and gastric cancer cells (Gou et al., 2015) via either β-catenin/TCF-4 or PI3K/Akt pathway, and thyroid cancer via c-Met/PI3K/Akt signaling pathway (Gao and Han, 2018). These findings suggest that ING5 reverses the aggressiveness mostly through PI3K/Akt signal pathway.
Wang et al. (2018) reported that ING5 promoted stemness and self-renewal, and prevented lineage differentiation in glioblastoma cells via Ca2+ and follicle-stimulating hormone. Chen et al. (2017) demonstrated that ING5 inhibited cell proliferation and chemoresistance, and promoted cell apoptosis in ovarian cancer cells. Li et al. (2015) found that ING5 silencing increased the chemoresistance and inhibited the DNA damage response pathway in bladder cancer, while it was the converse for ING5 expression in chemoresistant bladder cancer cells. Our previous work showed that ING5 overexpression caused chemoresistance and lipogenesis of cancer cells, and chemoresistant cells to 5-FU, cisplatin and sorafenib showed ING5 overexpression and lipogenesis, suggesting that ING5-mediated lipogenesis results in drug resistance (Xuan et al., 2022).
ING5 knockdown elevated cell viability and impaired cell cycle G0/G1 phase arrest and apoptosis in PC-12 and SH-SY5Y cells after oxygen-glucose deprivation (Zhang et al., 2019). Wu et al. (2018) demonstrated that HAT recruited ING5 and acetylated histones H3 and H4 to the promoters of c-myc, nanog, Cyclin D1, p21, and p27, thereby up-regulating p21 and p27 expression, and down-regulating c-myc, Cyclin D1 and nanog expression. SAHA treatment attenuated the histone deacetylase inhibitor and finally strengthened the formation of ING5-acetyl- H3-acetyl-H4 (Figure 2).
At the transcriptional level, our group found out a suppressor between −2000 and −1000 bp, and an enhancer between −800 and −100 bp although two promoter sequences were predicted from -1995 to −1690 bp and from -1973 to −1400 bp. The trans-acting factors of ING5 promoters are predicted to be SRF (−717 to −678 bp), CTCF (−48 to 0 bp), YY1 (−48 to −25 bp), Sp1 (−44 to −30 bp; 32 to 20), PPAR-γ1 (−24 to −25 bp), WT1 (−10 to 1 bp), and Pax-5 (−1 to 25 bp). EMSA and ChiP indicated that only SRF and YY1 could interact with the ING5 promoter (−50 to 0 bp). In gastric cancer cells, either SRF or YY1 silencing decreased ING5 mRNA and protein expression. SRF was found to interact with YY1, p53 and ING5, and either SFR or YY1 was co-localized with p53 or ING5 in the nuclei of cancer cells (Zheng et al., 2022). These findings indicate that SRF, YY1, ING5 and p53 form a complex to interact with the ING5 promoter and up-regulate its expression in gastric cancer cells.
At the translational level, both miR-196 and miR-196b-5p target ING5 in colorectal cancer cells (Pourdavoud et al., 2020; Xin et al., 2020). By targeting ING5, miR-27-3p promotes the G1-S phase transition in osteosarcoma cells (Ye et al., 2018) and miR-196b-5p inhibits the apoptosis of pancreatic cancer cells (Ma et al., 2021). In ovarian cancer cells, miR-200b/200a/429 targets ING5 which blocks miR-200b/200a/429-induced malignant transformation (Guan et al., 2020), and miR-1307 targets and down-regulated ING5 expression (Chen et al., 2017). miR-1307- mediated apoptosis and chemosensitivity were in vivo and vitro reversed by ING5 knockdown. CAF (cancer-associated fibroblast)-derived exosomal miR-196a confers cisplatin resistance in HNSCC cells by targeting ING5 (Qin et al., 2019), and miR-193a-3p induces the chemoresistance of breast cancer cells by targeting ING5, in line with the effects of ING5 silencing (Li et al., 2015). In triple-negative breast cancer cells, miR-193 bound to the 3′-UTR of ING5 mRNA to inhibit its expression and miR-193 inhibitor suppressed cell proliferation through ING5/PI3K/Akt pathway (Xu et al., 2020). CASC2 acted as a competitor of miR-222 to up-regulate ING5 expression in pulmonary arterial smooth muscle cells (PASMCs). CASC2- mediated inhibitory effects of hypoxia on the migration and proliferation of PASMCs could be weakened by either miR-222 inhibitor or ING5 overexpression (Han et al., 2020). miR-331-3p could promote proliferation of HCC cells by targeting ING5, which was strengthened by hepatitis B virus (HBV) (Cao et al., 2015). HBx protein of HBV accelerated the proliferation of HCC cells by up-regulating miR-181b to target ING5 (Xie et al., 2018). In neuroblastoma cells, ING5 is a negatively- regulated target gene of miR-376c-3p (Zhang et al., 2019), and SAHA down-regulates miR-196-b and miR-543 expression to facilitate the translation of ING5 protein (Wu et al., 2018). miR-193 induces proliferation and CDK2 activity in bone mesenchymal stem cells by targeting ING5 (Wang et al., 2012).
ING5 was conditionally deleted in gastric pit cells using Capn8-cre mice, in gastric parietal cells using Atp4b-cre mice, in gastric pdx-1-positive cells using pdx1-cre mice, in gastric stem-like cells using K19-cre mice, in gastric chief cells using PGC-cre mice, and in intestinal epithelial cells using villin-cre mice. The normal gastric epithelium was observed in Capn8-cre; ING5f/f mice and regenerative dysplasia was detected in PGC-cre; ING5f/f mice. However, well-, moderately-, or poorly-differentiated adenocarcinoma was observed in Atp4b-cre; ING5f/f ,pdx1-cre; ING5f/f and K19-cre; ING5f/f mice, even in pdx1-cre; ING5f/f and K19-cre; ING5f/f and wild-type (WT) mice exposed to MNU. After the treatment with MNU, chemically-induced gastric cancer and dysplasia were more common in K19-cre; ING5f/f and Pdx1-cre; ING5f/f mice than those in WT mice. These data demonstrated that ING5 knockout (KO) might cause spontaneous and chemically-induced gastric carcinogenesis (Zheng et al., 2022). After exposure to AOM/DSS, the cancer formation rate of intestinal KO mice was high in comparison to that of WT mice (Yu et al., 2022).
After the oral administration of high-fat diet, villin-cre; ING5f/f showed a lower weight, a slighter fatty liver and a lighter fat weight than WT mice (Yu et al., 2022). He et al. (2021) found that liver-specific reconstitution of JFK expression resulted in hepatic lipid accumulation in JFK KO mice by destabilizing ING5 and inhibiting fatty acid β-oxidation. Therefore, we hypothesized that ING5 might play an important role in fat absorption and fatty acid β-oxidation.
Cengiz et al. (2007) found that 3 frequently-deleted regions at 2q37.3, 2q33-35 and 2q21-24 harboring ING5 were detected in oral cancers, and loss of heterozygosity (LOH) of chromosome 2q in 85% (33/39) of oral squamous carcinomas, indicating an important role of ING5 deletion in oral carcinogenesis (Cengiz et al., 2010). There were 5 different alternative splicing variants of ING5, which resulted in ING5 mRNA hypoexpression in 61% of the oral tumors as compared to the matched normal samples (Figure 3). These missense mutations of ING5 gene were located to NCR and LZL domains of ING5 protein (Cengiz et al., 2007; Cengiz et al., 2010).
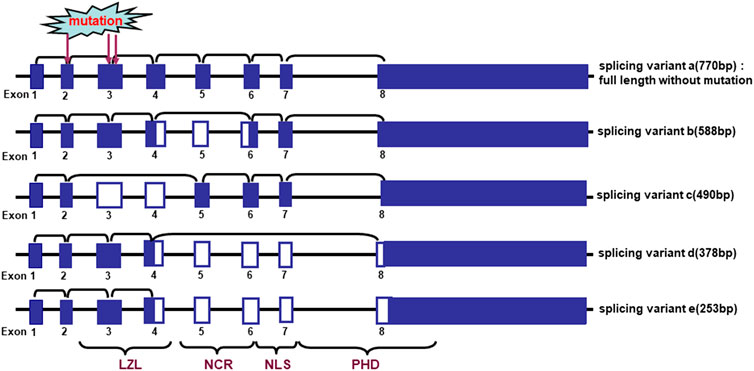
FIGURE 3. Alternative splicing of ING5 mRNA in oral squamous cell carcinoma. The schematic representation indicates the genomic structure and alternatively splicing variants of ING5. LZL, leucine zipper-Like motif; NCR: novel conserved region; NLS, nuclear localization signal; PHD, plant homeodomain.
In our previous study, ING5 protein is primarily observed the cytoplasm and nucleus in the breast, intestine, stomach, and tongue, lung. Totally, ING5 expression was detected in 40.6% (400/986) of cancer tissues, among which ING5 was more frequently expressed in colorectal (56.3%), breast (79.9%) and endometrial (50.0%) cancers, and less in pancreatic cancer (22.6%) and HCC (14.5%) (Yang et al., 2019). Although a high ING5 expression was observed in gastric, breast, and colorectal cancers (Xing et al., 2011; Zhao et al., 2015; Wang et al., 2021; Zheng et al., 2022), ING5 was found to be expressed in osteosarcoma, prostate cancer, ovarian cancer, HCC, lung cancer, esophageal cancer, and thyroid cancer at a low level (Qi and Zhang, 2014; Cao et al., 2015; Zhang et al., 2015; Cui et al., 2017; Zheng et al., 2017; Zhang et al., 2018a). The low nuclear expression of ING5 and its nucleocytoplasmic translocation were involved in the tumorigenesis of gastric cancer, breast cancer, HNSCC, and colorectal cancer (Li et al., 2010; Xing et al., 2011; Zheng et al., 2011; Ding et al., 2017). Bilgiç et al. (2018) found that ING5 was under-expressed in the atrophic gastritis and gastric cancer compared with the normal mucosa.
As for the clinicopathological and prognostic significances, ING5 mRNA expression was negatively correlated with vascular and lymphatic invasion, and clinicopathological staging of ovarian cancers. There was an adverse relationship between ING5 mRNA expression and the overall or progression-free survival of the ovarian cancer patients with stage I, Grade 3, and Grade 2, indicating the prognostic significance of ING5 depends on the cancer type, subtyping and molecular level (Zheng et al., 2017). In gastric, colorectal and breast cancers, nuclear ING5 expression was negatively associated with depth of invasion, tumor size, dedifferentiation, and TNM staging, opposite to the finding of cytoplasmic ING5 expression (Xing et al., 2011; Zheng et al., 2011; Ding et al., 2017). ING5 immunoreactivity was less expressed in ovarian mucinous and serous adenocarcinoma than miscellaneous subtypes, and negatively correlated with differentiation and low ki-67 expression of ovarian cancer (Zhang et al., 2018b). In lung cancer, ING5 protein was distinctly expressed in small cell carcinoma < large cell carcinoma < adenocarcinoma < squamous cell carcinoma (Zhao et al., 2016). The nuclear ING5 expression was positively linked to a better prognosis of gastric cancer patients, albeit not independent (Xing et al., 2011), in agreement with the report of Zhang et al. about lung cancer (Zhang et al., 2015).
In summary, ING5 protein can acetylate HAT p300, p53, histone H3 and H4 for the transcription of downstream genes via the formation of homodimer or heterodimers with ING4, histone H3K4me3, HAT complex, Tip60, Cyclin A1/CDK2, INCA1 and EBNA3C. Transcriptionally, YY1 and SRF can up-regulate ING5 mRNA expression by the interaction of YY1-SRF-p53-ING5 complex with ING5 promoter. ING5 inhibits proliferation, migration, invasion and tumor growth of cancer cells via IL-6/STAT3, EGFR/Akt/NF-κB/MMP-9 or IL-6/CXCL12 pathways. ING5-mediated lipogenesis might lead to chemoresistance. Target ING5 KO increased the sensitivity of chemically-induced gastric and colorectal carcinogenesis, and suppressed lipogenesis and fat absorption in intestine. The overexpression and nucleocytoplasmic translocation were seen and ING5 expression is inversely associated with aggressive behaviors and poor prognosis of cancers.
In the future, aberrant ING5 expression might be used as a molecular marker for carcinogenesis, progression and prognosis of malignancies. Additionally, ING5 functions as a tumor suppressor, but facilitates lipogenesis and subsequent chemoresistance. If ING5 will be used as a gene therapy target of cancers, the chemoresistant function should be ameliorated. Therefore, ING5 knockout or the suppression of ING5-related signal pathway would also be helpful to the control of body weight and the prevention of obesity-related diseases. If we can develop the anti-ING5 reagents, these drugs would be used to treat cancer, reverse chemoresistance and control the obesity.
Writing -original draft, H-cZ and HX; writing -review and editing, H-mJ and H-cZ; funding acquisition, H-cZ.
The present study was supported by Award for Liaoning Distinguished Professor, Natural Science Foundation of Hebei Province (21377772D), Emphasis Project of Education Department of Hebei Province (ZD2022096) and National Natural Scientific Foundation of China (81672700).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
CAF, cancer-associated fibroblast; DSB, double-strand break; EBV, Epstain-Barr virus; EMT, epithelial- mesenchymal transition; HAT, histone acetyltransferase; HBV, hepatitis B virus; HCC, hepatocellular carcinoma; HNSCC, head and neck squamous cell carcinoma; ING, inhibitor of growth; KO, knockout; LOH, loss of heterozygosity; LZL, leucine zipper-like; NER, nucleotide excision repair; NCR, novel conserved region; NLS, nuclear localization signal; PASMC, pulmonary arterial smooth muscle cell; PHD, plant homeodomain; WT, wild-type.
Archambeau, J., Blondel, A., and Pedeux, R. (2019). Focus-ING on DNA Integrity: Implication of ING proteins in cell cycle regulation and DNA repair modulation. Cancers 12 (1), 58. doi:10.3390/cancers12010058
Barlak, N., Capik, O., Sanli, F., Kilic, A., Aytatli, A., Yazici, A., et al. (2020). ING5 inhibits cancer aggressiveness by inhibiting Akt and activating p53 in prostate cancer. Cell Biol. Int. 44 (1), 242–252. doi:10.1002/cbin.11227
Bilgiç, F., Gerçeker, E., Boyacıoğlu, S. Ö., Kasap, E., Demirci, U., Yıldırım, H., et al. (2018). Potential role of chromatin remodeling factor genes in atrophic gastritis/gastric cancer risk. Turk. J. Gastroenterol. 29 (4), 427–435. doi:10.5152/tjg.2018.17350
Cao, Y., Chen, J., Wang, D., Peng, H., Tan, X., Xiong, D., et al. (2015). Upregulated in Hepatitis B virus-associated hepatocellular carcinoma cells, miR-331-3p promotes proliferation of hepatocellular carcinoma cells by targeting ING5. Oncotarget 6 (35), 38093–38106. doi:10.18632/oncotarget.5642
Cengiz, B., Gunduz, M., Nagatsuka, H., Beder, L., Gunduz, E., Tamamura, R., et al. (2007). Fine deletion mapping of chromosome 2q21-37 shows three preferentially deleted regions in oral cancer. Oral Oncol. 43 (3), 241–247. doi:10.1016/j.oraloncology.2006.03.004
Cengiz, B., Gunduz, E., Gunduz, M., Beder, L. B., Tamamura, R., Bagci, C., et al. (2010). Tumor-specific mutation and downregulation of ING5 detected in oral squamous cell carcinoma. Int. J. Cancer 127 (9), 2088–2094. doi:10.1002/ijc.25224
Chen, W. T., Yang, Y. J., Zhang, Z. D., An, Q., Li, N., Liu, W., et al. (2017). MiR-1307 promotes ovarian cancer cell chemoresistance by targeting the ING5 expression. J. Ovarian Res. 10 (1), 1. doi:10.1186/s13048-016-0301-4
Cui, S., Liao, X., Ye, C., Yin, X., Liu, M., Hong, Y., et al. (2017). ING5 suppresses breast cancer progression and is regulated by miR-24. Mol. Cancer 16 (1), 89. doi:10.1186/s12943-017-0658-z
Dantas, A., Al Shueili, B., Yang, Y., Nabbi, A., Fink, D., and Riabowol, K. (2019). Biological functions of the ING proteins. Cancers (Basel) 11 (11), 1817. doi:10.3390/cancers11111817
Ding, X. Q., Zhao, S., Yang, L., Zhao, X., Zhao, G. F., Zhao, S. P., et al. (2017). The nucleocytoplasmic translocation and up-regulation of ING5 protein in breast cancer: a potential target for gene therapy. Oncotarget 8 (47), 81953–81966. doi:10.18632/oncotarget.17918
Gao, W., and Han, J. (2018). Overexpression of ING5 inhibits HGF-induced proliferation, invasion and EMT in thyroid cancer cells via regulation of the c-Met/PI3K/Akt signaling pathway. Biomed. Pharmacother. 98 (33), 265–270. doi:10.1016/j.biopha.2017.12.045
Gou, W. F., Shen, D. F., Yang, X. F., Zhao, S., Liu, Y. P., Sun, H. Z., et al. (2015). ING5 suppresses proliferation, apoptosis, migration and invasion, and induces autophagy and differentiation of gastric cancer cells: a good marker for carcinogenesis and subsequent progression. Oncotarget 6 (23), 19552–19579. doi:10.18632/oncotarget.3735
Guan, W., Cui, H., Huang, P., Chun, W. J., Lee, J. W., Kim, H., et al. (2020). miR-200b/200a/429 cluster stimulates ovarian cancer development by targeting ING5. J. Oncol. 2020, 3404059. doi:10.1155/2020/3404059
Han, Y., Liu, Y., Yang, C., Gao, C., Guo, X., and Cheng, J. (2020). LncRNA CASC2 inhibits hypoxia- induced pulmonary artery smooth muscle cell proliferation and migration by regulating the miR-222/ING5 axis. Cell. Mol. Biol. Lett. 25, 21. doi:10.1186/s11658-020-00215-y
He, L., Yan, R., Yang, Z., Zhang, Y., Liu, X., Yang, J., et al. (2021). SCF JFK is functionally linked to obesity and metabolic syndrome. EMBO Rep. 22 (7), e52036. doi:10.15252/embr.202052036
Li, X., Nishida, T., Noguchi, A., Zheng, Y., Takahashi, H., Yang, X., et al. (2010). Decreased nuclear expression and increased cytoplasmic expression of ING5 may be linked to tumorigenesis and progression in human head and neck squamous cell carcinoma. J. Cancer Res. Clin. Oncol. 136 (10), 1573–1583. doi:10.1007/s00432-010-0815-x
Li, Y., Deng, H., Lv, L., Zhang, C., Qian, L., Xiao, J., et al. (2015). The miR-193a-3p-regulated ING5 gene activates the DNA damage response pathway and inhibits multi-chemoresistance in bladder cancer. Oncotarget 6 (12), 10195–10206. doi:10.18632/oncotarget.3555
Linzen, U., Lilischkis, R., Pandithage, R., Schilling, B., Ullius, A., Lüscher-Firzlaff, J., et al. (2015). ING5 is phosphorylated by CDK2 and controls cell proliferation independently of p53. PLoS One 10, e0123736. doi:10.1371/journal.pone.0123736
Liu, N., Wang, J., Wang, J., Wang, R., Liu, Z., Yu, Y., et al. (2013). ING5 is a Tip60 cofactor that acetylates p53 in response to DNA damage. Cancer Res. 73 (12), 3749–3760. doi:10.1158/0008-5472.CAN-12-3684
Liu, X. L., Zhang, X. T., Meng, J., Zhang, H. F., Zhao, Y., Li, C., et al. (2017). ING5 knockdown enhances migration and invasion of lung cancer cells by inducing EMT via EGFR/PI3K/Akt and IL-6/STAT3 signaling pathways. Oncotarget 8 (33), 54265–54276. doi:10.18632/oncotarget.17346
Liu, X. L., Meng, J., Zhang, X. T., Liang, X. H., Zhang, F., Zhao, G. R., et al. (2019). ING5 inhibits lung cancer invasion and epithelial-mesenchymal transition by inhibiting the WNT/β-catenin pathway. Thorac. Cancer 10 (4), 848–855. doi:10.1111/1759-7714.13013
Ma, D., Chen, S., Wang, H., Wei, J., Wu, H., Gao, H., et al. (2021). Baicalein induces apoptosis of pancreatic cancer cells by regulating the expression of miR-139-3p and miR-196b-5p. Front. Oncol. 11, 653061. doi:10.3389/fonc.2021.653061
Ormaza, G., Rodríguez, J. A., Ibáñez de Opakua, A., Merino, N., Villate, M., Gorroño, I., et al. (2019). The tumor suppressor ING5 is a dimeric, bivalent recognition molecule of the histone H3K4me3 mark. J. Mol. Biol. 431 (12), 2298–2319. doi:10.1016/j.jmb.2019.04.018
Pourdavoud, P., Pakzad, B., Mosallaei, M., Saadatian, Z., Esmaeilzadeh, E., Alimolaie, A., et al. (2020). MiR-196: emerging of a new potential therapeutic target and biomarker in colorectal cancer. Mol. Biol. Rep. 47 (12), 9913–9920. doi:10.1007/s11033-020-05949-8
Qi, L., and Zhang, Y. (2014). Truncation of inhibitor of growth family protein 5 effectively induces senescence, but not apoptosis in human tongue squamous cell carcinoma cell line. Tumour Biol. 35 (4), 3139–3144. doi:10.1007/s13277-013-1410-y
Qin, X., Guo, H., Wang, X., Zhu, X., Yan, M., Wang, X., et al. (2019). Exosomal miR-196a derived from cancer-associated fibroblasts confers cisplatin resistance in head and neck cancer through targeting CDKN1B and ING5. Genome Biol. 20 (1), 12. doi:10.1186/s13059-018-1604-0
Saha, A., Bamidele, A., Murakami, M., and Robertson, E. S. (2011). EBNA3C attenuates the function of p53 through interaction with inhibitor of growth family proteins 4 and 5. J. Virol. 85 (5), 2079–2088. doi:10.1128/JVI.02279-10
Shiseki, M., Nagashima, M., Pedeux, R. M., Kitahama-Shiseki, M., Miura, K., Okamura, S., et al. (2003). p29ING4 and p28ING5 bind to p53 and p300, and enhance p53 activity. Cancer Res. 63 (10), 2373–2378.
Wang, J., Huang, W., Wu, Y., Hou, J., Nie, Y., Gu, H., et al. (2012). MicroRNA-193 pro-proliferation effects for bone mesenchymal stem cells after low-level laserirradiation treatment through inhibitor of growth family, member 5. Stem Cells Dev. 21 (13), 2508–2519. doi:10.1089/scd.2011.0695
Wang, F., Wang, A. Y., Chesnelong, C., Yang, Y., Nabbi, A., Thalappilly, S., et al. (2018). ING5 activity in self-renewal of glioblastoma stem cells via calcium and follicle stimulating hormone pathways. Oncogene 37 (3), 286–301. doi:10.1038/onc.2017.324
Wang, Y., Tan, J., Li, J., Chen, H., and Wang, W. (2021). ING5 inhibits migration and invasion of esophageal cancer cells by downregulating the IL-6/CXCL12 signaling pathway. Technol. Cancer Res. Treat. 20, 15330338211039940. doi:10.1177/15330338211039940
Wu, J. C., Jiang, H. M., Yang, X. H., and Zheng, H. C. (2018). ING5-mediated antineuroblastoma effects of suberoylanilide hydroxamic acid. Cancer Med. 7, 4554–4569. doi:10.1002/cam4.1634
Xie, X., Xu, X., Sun, C., and Yu, Z. (2018). Hepatitis B virus X protein promotes proliferation of hepatocellular carcinoma cells by upregulating miR-181b by targeting ING5. Biol. Chem. 399 (6), 611–619. doi:10.1515/hsz-2018-0178
Xin, H., Wang, C., Chi, Y., and Liu, Z. (2020). MicroRNA-196b-5p promotes malignant progression of colorectal cancer by targeting ING5. Cancer Cell Int. 20, 119. doi:10.1186/s12935-020-01200-3
Xing, Y. N., Yang, X., Xu, X. Y., Zheng, Y., Xu, H. M., Takano, Y., et al. (2011). The altered expression of ING5 protein is involved in gastric carcinogenesis and subsequent progression. Hum. Pathol. 42 (1), 25–35. doi:10.1016/j.humpath.2010.05.024
Xu, J. H., Zhao, J. X., Jiang, M. Y., Yang, L. P., Sun, M. L., and Wang, H. W. (2020). MiR-193 promotes cell proliferation and invasion by ING5/PI3K/AKT pathway of triple-negative breast cancer. Eur. Rev. Med. Pharmacol. Sci. 24 (6), 3122–3129. doi:10.26355/eurrev_202003_20679
Xuan, Y., Shi, S., Xue, H., Liu, K., Han, W. L., Li, B. C., et al. (2022). Lamp2a facilitates lipid droplet production and chemoresistance of colorectal cancer cells by maintaining ING5 protein stability. BMC Cancer. in press. doi:10.21203/rs.3.rs-1373107/v1
Yang, X. F., Shen, D. F., Zhao, S., Ren, T. R., Gao, Y., Shi, S., et al. (2019). Expression pattern and level of ING5 protein in normal and cancer tissues. Oncol. Lett. 17 (1), 63–68. doi:10.3892/ol.2018.9581
Ye, P., Ke, X., Zang, X., Sun, H., Dong, Z., Lin, J., et al. (2018). Up-regulated miR-27-3p promotes the G1-S phase transition by targeting inhibitor of growth family member 5 in osteosarcoma. Biomed. Pharmacother. 101, 219–227. doi:10.1016/j.biopha.2018.02.066
Yu, Q., Xue, H., Sun, H. Z., Ha, M. W., Yu, D. Y., Zhang, R., et al. (2022). The suppressive effects of ING5 on tumorigenesis and aggressiveness of colorectal cancer via inhibition of NF-κB and β-catenin pathways. Tumor Biol. in press.
Zhang, F., Bäumer, N., Rode, M., Ji, P., Zhang, T., Berdel, W. E., et al. (2011). The inhibitor of growth protein 5 (ING5) depends on INCA1 as a co-factor for its antiproliferative effects. PLoS One 6, e21505. doi:10.1371/journal.pone.0021505
Zhang, F., Zhang, X., Meng, J., Zhao, Y., Liu, X., Liu, Y., et al. (2015). ING5 inhibits cancer aggressiveness via preventing EMT and is a potential prognostic biomarker for lung cancer. Oncotarget 6 (18), 16239–16252. doi:10.18632/oncotarget.3842
Zhang, H., Zhou, J., Zhang, M., Yi, Y., and He, B. (2019). Upregulation of miR-376c-3p alleviates oxygen-glucose deprivation-induced cell injury by targeting ING5. Cell. Mol. Biol. Lett. 24, 67. doi:10.1186/s11658-019-0189-2
Zhang, G. J., Zhao, J., Jiang, M. L., and Zhang, L. C. (2018b). ING5 inhibits cell proliferation and invasion in esophageal squamous cell carcinoma through regulation of the Akt/NF-κB/MMP-9 signaling pathway. Biochem. Biophys. Res. Commun. 496 (2), 387–393. doi:10.1016/j.bbrc.2018.01.045
Zhang, R., Jin, J., Shi, J., and Hou, Y. (2017a). INGs are potential drug targets for cancer. J. Cancer Res. Clin. Oncol. 143 (2), 189–197. doi:10.1007/s00432-016-2219-z
Zhang, T., Meng, J., Liu, X., Zhang, X., Peng, X., Cheng, Z., et al. (2017b). ING5 differentially regulates protein lysine acetylation and promotes p300 autoacetylation. Oncotarget 9 (2), 1617–1629. doi:10.18632/oncotarget.22176
Zhang, X., Xu, Z. H., Xie, H., Sun, Y. W., Liu, J., and Zhao, Y. B. (2018a). ING5 is a potential target for osteosarcoma therapy. Technol. Cancer Res. Treat. 17, 1533033818762680. doi:10.1177/1533033818762680
Zhao, Q. Y., Ju, F., Wang, Z. H., Ma, X. Z., and Zhao, H. (2015). ING5 inhibits epithelial-mesenchymal transition in breast cancer by suppressing PI3K/Akt pathway. Int. J. Clin. Exp. Med. 8 (9), 15498–15505.
Zhao, S., Yang, X. F., Shen, D. F., Gao, Y., Shi, S., Wu, J. C., et al. (2016). The down-regulated ING5 expression in lung cancer: a potential target of gene therapy. Oncotarget 7 (34), 54596–54615. doi:10.18632/oncotarget.10519
Zhao, S., Zhao, Z. J., He, H. Y., Wu, J. C., Ding, X. Q., Yang, L., et al. (2017). The roles of ING5 in gliomas: a good marker for tumorigenesis and a potential target for gene therapy. Oncotarget 8 (34), 56558–56568. doi:10.18632/oncotarget.17802
Zheng, H. C., Xia, P., Xu, X. Y., Takahashi, H., and Takano, Y. (2011). The nuclear to cytoplasmic shift of ING5 protein during colorectal carcinogenesis with their distinct links to pathologic behaviors of carcinomas. Hum. Pathol. 42 (3), 424–433. doi:10.1016/j.humpath.2009.12.018
Zheng, H. C., Zhao, S., Song, Y., and Ding, X. Q. (2017). The roles of ING5 expression in ovarian carcinogenesis and subsequent progression: a target of gene therapy. Oncotarget 8 (61), 103449–103464. doi:10.18632/oncotarget.21968
Keywords: signal pathway, biological function, ING5, cancer, tumor suppressor
Citation: Zheng H-c, Xue H and Jiang H-m (2022) The roles of ING5 in cancer: A tumor suppressor. Front. Cell Dev. Biol. 10:1012179. doi: 10.3389/fcell.2022.1012179
Received: 05 August 2022; Accepted: 05 October 2022;
Published: 08 November 2022.
Edited by:
Wei Liu, Arizona State University, United StatesReviewed by:
Qing Feng, Nanjing Medical University, ChinaCopyright © 2022 Zheng, Xue and Jiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hua-chuan Zheng, emhlbmdfaHVhY2h1YW5AaG90bWFpbC5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.