
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
BRIEF RESEARCH REPORT article
Front. Cell Dev. Biol. , 21 September 2022
Sec. Morphogenesis and Patterning
Volume 10 - 2022 | https://doi.org/10.3389/fcell.2022.1005776
This article is part of the Research Topic Creating Asymmetry and Keeping Symmetry: the Underlying Genetic and Signaling Mechanisms View all 12 articles
 Carolina Agostini1,2†
Carolina Agostini1,2† Anja Bühler1†
Anja Bühler1† Alessandra Antico Calderone1
Alessandra Antico Calderone1 Narendar Aadepu2,3
Narendar Aadepu2,3 Cathrin Herder2
Cathrin Herder2 Felix Loosli2*‡
Felix Loosli2*‡ Matthias Carl1*‡
Matthias Carl1*‡Morphological left-right brain asymmetries are universal phenomena in animals. These features have been studied for decades, but the functional relevance is often unclear. Studies from the zebrafish dorsal diencephalon on the genetics underlying the establishment and function of brain asymmetries have uncovered genes associated with the development of functional brain asymmetries. To gain further insights, comparative studies help to investigate the emergence of asymmetries and underlying genetics in connection to functional adaptation. Evolutionarily distant isogenic medaka inbred lines, that show divergence of complex traits such as morphology, physiology and behavior, are a valuable resource to investigate intra-species variations in a given trait of interest. For a detailed study of asymmetry in the medaka diencephalon we generated molecular probes of ten medaka genes that are expressed asymmetrically in the zebrafish habenulae and pineal complex. We find expression of eight genes in the corresponding brain areas of medaka with differences in the extent of left-right asymmetry compared to zebrafish. Our marker gene analysis of the diverged medaka inbred strains revealed marked inter-strain size differences of the respective expression domains in the parapineal and the habenulae, which we hypothesize may result from strain-specific gene loss. Thus, our analysis reveals both inter-species differences but also intra-species plasticity of gene expression in the teleost dorsal diencephalon. These findings are a starting point showing the potential to identify the genetics underlying the emergence and modulations of asymmetries. They are also the prerequisite to examine whether variance in habenular gene expression may cause variation of behavioral traits.
Brain asymmetries are present throughout bilateria. From insects to humans the morphologies of multiple parts of the central nervous system differ between the two sides of the body (Frasnelli et al., 2012; Rogers, 2014; Duboc et al., 2015; Alqadah et al., 2018; Kong et al., 2018). Given the widespread existence of asymmetries in the animal kingdom, it is likely that morphological and functional asymmetries are often associated and are advantageous for neuronal computation and the animals’ performance, well-being and behavior (Vallortigara and Rogers, 2005; Rogers, 2014; Güntürkün and Ocklenburg, 2017; Miletto Petrazzini et al., 2020). However, for the majority of brain asymmetries a functional relevance remains to be identified. For instance, numerous reports find links of asymmetry alterations to mental disorders, while others do not support this idea (Mundorf et al., 2021). Putative causative genetic mechanisms underlying such differences in left-right brain asymmetry have remained largely elusive. One approach to assess this enigma is to investigate the emergence of morphological brain asymmetries in connection to functional adaptation and the underlying genetics using comparative approaches. Teleosts show the highest radiation amongst vertebrates with diverged species that adapted to habitats with widely different conditions (Wittbrodt, Meyer and Schartl, 1998; Volff, 2005; Braasch et al., 2014). Importantly, a plethora of molecular genetic tools and resources are available to study phenotypic traits and molecular mechanisms in many teleost species.
Unlike numerous rather subtle left-right brain asymmetries, the dorsal diencephalon of teleosts is a remarkable exception as evidenced in numerous studies using the zebrafish (Danio rerio). The pineal complex consists of a centrally positioned pineal organ and a group of left-sided parapineal cells (Concha and Wilson, 2001; Beretta et al., 2012). In addition, also the adjacent bilaterally formed habenulae exhibit pronounced asymmetries. The neuronal subpopulations of the dorsal habenulae (dHb), which correspond to the mammalian medial habenulae (Amo et al., 2010), differ substantially between hemispheres with respect to size, gene expression, connectivity and function (Gamse et al., 2002; Concha et al., 2003; Gamse et al., 2003; Aizawa et al., 2005; Bianco et al., 2008; Miyasaka et al., 2009; Agetsuma et al., 2010; Dreosti et al., 2014; Krishnan et al., 2014; Facchin et al., 2015; Zhang et al., 2017). How these asymmetries have emerged and facilitate functional adaptation is largely unknown.
In view of the high divergence within the teleost clade a comparative study of diverged species will provide insights into the evolution of organs and underlying genetic mechanisms (Furutani-Seiki and Wittbrodt, 2004). The Japanese Killifish medaka (Oryzias latipes) is ideally suited for such studies due to its divergence from zebrafish of more than 100 Mya of independent evolution. Furthermore, medaka is a fully established laboratory teleost model with a long history of genetic studies (Takeda and Shimada, 2010). For example, several highly inbred isogenic strains, derived from different medaka wild populations, are available. A molecular genetic toolbox has been established that allows genetic studies, detailed analysis of gene function and genome editing (Kirchmaier et al., 2015). Most of the protocols established for zebrafish can be used for medaka with minor modifications. Thus, direct comparisons of reporter gene constructs are possible between these two teleost species.
Medaka also permits studies of intra-species variation, which is the basis of population genetic studies. The inbred strains from diverged medaka populations allow to study the plasticity of quantitative traits within this species. For example, using such inbred medaka strains, a genetic basis of strain-specific craniofacial variance has been shown (Kimura et al., 2007). Furthermore, a genetic contribution to the variation of brain morphology was reported in medaka (Ishikawa at al., 1999). Also, behavioral traits have been analysed by comparing isogenic inbred strains, suggesting that startle behavior in medaka is defined by intraspecific genetic polymorphisms (Tsuboko et al., 2014). Recently, a population genetic resource has been established, consisting of a panel of 80 inbred strains derived from the same wild population (Fitzgerald et al., 2022). Thus, medaka offers unique genetic resources to further our knowledge of intra-species variation in complex traits.
In this study, we used medaka as a diverged teleost for a comparative analysis of brain asymmetry. Using whole mount in situ hybridization (WISH) we examined the expression of a select number of marker genes that have been shown to be asymmetrically expressed in the developing diencephalon of zebrafish. We isolated and analysed 10 genes, 8 of which we find expressed in the medaka dorsal diencephalon. All of the commonly used markers for the pineal/parapineal complex in zebrafish, otx5l, sox1a and gfi1ab, are similarly expressed in medaka. Regarding the habenular specific genes, we observed largely conserved expression of the habenular precursor cell marker cxcr4, while the markers for habenular neuron subpopulations, prox1a, cntn2, nrp1 and kctd12.1, differed in their extent of left-right asymmetry. We additionally revealed pronounced intraspecies variation of asymmetric gene expression by comparing two isogenic medaka strains from southern and northern populations, respectively, iCab and Kaga (Spivakov et al., 2014). In Kaga embryos, otx5l and sox1a expression in the parapineal are enlarged, while kctd12.1 is much more prominently expressed, particularly in right dorsal habenular neurons compared to iCab embryos. These findings hint at intraspecific plasticity of left-right asymmetry.
The medaka inbred strains iCab and Kaga were kept in closed stocks at the Institute of Biological and Chemical Systems, Biological Information Processing (IBCS-BIP) at the Karlsruhe Institute of Technology (KIT), as previously described (Loosli et al., 2000). Medaka fish strains were maintained and bred according to standard procedures (Kinoshita et al., 2009). Medaka embryos were raised in a Petri dish containing ERM medium for 4 days at 28°C with a 14 h light/10 h dark cycle. Developmental stages were determined according to Iwamatsu, 2004.
cDNA fragments of gfi1ab (Gene ID: 110015008), lov/kctd12.1 (Gene ID: 101161445), otx5l (Gene ID: 101164666), cntn2 (Gene ID: 101161878), prox1b (Gene ID: 100302040), cxcr4 (Gene ID: 100049444), nrp1 (Gene ID: 101161516), sox1a (Gene ID: 100304447), slc17a6b/vglut2 (Gene ID: 101175075), slc18a3b/vachtb (Gene ID: 101163117) were generated from cDNA libraries extracted from one to six dpf iCab embryos. Because zebrafish PROX1a and medaka PROX1b share the highest sequence homology with 71.80% and developmental gene expression, we renamed medaka prox1b to prox1a. The following primers were used: gfi1ab (Fw: 5′-GAGTCGGTGGATAGGTACGG; Rw: 3′-AGAAGAATCAGGCTGCCAGT; 1185 bp cDNA), lov/kctd12.1 (Fw: 5′- CCGCGTTTCCATCGAATCAT; Rv: 3′-GCTCATCTCAGTTCACAGCA; 1155 bp cDNA), otx5l (Fw: 5′-GCACTCCGAGCAGTTTGAAA; Rv: 3′-GAGAGCTGTGAATGCATGGG; 813 bp cDNA), cntn2 (Fw: 5′-GTCACGGCCATGATAACCAC; Rv: 3′-ACCTGGATCTCAAAAGCGGA; 575 bp cDNA), prox1b (new prox1a) (Fw: 5′-AACCAGCATGGGTCAGAGAG; Rv: 3′-TTTGAAAGCTGCACTGATGG; 784 bp cDNA), cxcr4 (Fw: 5′-TTCGACAACAGCTCTGAAGG-3′; Rw: 3′-GGCGTAGAGGATGGGATTCA; 951 bp cDNA), nrp1 (Fw: 5′-TCATCTCCTACACGGGCATC; Rv: 3′-TCCAACACCATCCTCCAGTC; 886 bp cDNA), sox1a (Fw: 5′-GATGATGGAAACGGACCTCC; Rv: 3′-AAATGTGCGTTAGAGGGACC; 956 bp cDNA), slc17a6b/vglut2 (Fw: 5′-TGTGGGGATGATTCATGGCT; Rv: 3′-GATAGTCCGCTAACTGGCCT; 749 bp cDNA), slc18a3b/vachtb (Fw: 5′-TTGTTTGCGTTGCTCTCCTC; Rv: 3′-ACCCAAGGCTCCATAGAACC; 896 bp cDNA). For gfi, lov/kctd12.1, otx5l, cntn2 and prox1a cDNAs the following PCR program was used: 95°C for 5 min, 35 cycles of 95°C for 45 s, 58°C for 45 s, 72°C for 45 s and a final step at 72°C for 5 min. Cloning was performed with pCR™ 2.1-TOPO TA dual promoter cloning Kit (Invitrogen), following the manufacturer’s instructions. For cxcr4, nrp1, sox1a, slc17a6b/vglut2, slc18a3b/vachtb cDNAs the PCR program was the following: 98°C for 30 s, 35 cycles of 98°C for 30 s, 56°C for 30 s, 72°C for 30 s and the final step at 72°C for 30 s. Cloning was performed with Zero Blunt™ TOPO™ PCR cloning Kit (ThermoFisher), following the manufacturer’s instructions.
Whole-mount in situ hybridization has been performed using digoxigenin-labeled antisense RNA probes following standard procedures as described in Loosli et al. (1998) with the following modification: the temperature of hybridization and washing steps was 65°C.
For anti-acetylated tubulin immunostaining, four dpf embryos were fixed overnight at room temperature in 4% PFA/2x PBST and stored at −20°C in methanol. Samples were digested with a 10 μg/ml Proteinase K (Sigma-Aldrich) treatment for 45 min, refixed in 4% PFA/2x PBST, and blocked in PBS with 0.1% Tween20 and 5% normal goat serum (NGS). For antibody labeling, primary mouse anti-acetylated tubulin (1:1000, Sigma, #T6793) and secondary goat anti-mouse Alexa Fluor 488-conjugated (1:250, Invitrogen,# A11001) antibodies were used. For nuclear staining, embryos were incubated in PBS, 0.1% Tween20 containing DAPI (1:5000, ThermoFisher Scientific).
Pigmentation was prevented by treating growing embryos with 0.003% PTU/ddH2O (500 μl PTU/ddH2O solution in about 10 ml medium), added at 24 hpf in a Petri dish with 20–30 eggs per dish. The medium and the PTU were changed every day. Before immunolabeling, pigments were removed by incubating fixed dechorionated Kaga embryos at four dpf in freshly prepared 30% H2O2 and 1% KOH solution for up to several hours at RT.
All in situ hybridization images were acquired by light microscopy using Zeiss Axio Imager M2 microscope and Keyence BZ-X810 microscope with a ×20 objective lens. Prior to acquisition, stained embryos were mounted in glycerol (Sigma-Aldrich) on microscope glass slides (Thermo Scientific).
For fluorescence confocal microscopy, embryos were mounted in 1% low-melt agarose in glass-bottom dishes (LabTek). Embryos were imaged using a TCS SP8 (Leica) inverse laser scanning microscope. Images were acquired with a ×20 air objective. Stack analysis, maximum intensity projections (MIPs), cropping and 2D-views were performed using Fiji und CS5 photoshop (Adobe) software.
Pineal complex, pineal and parapineal area measurements were performed with ImageJ-Fiji software using the Analyze Particles command. Habenula size was measured with ImageJ-Fiji by selecting and measuring the area of the habenula on confocal maximum projections stained with DAPI. The mean and standard error of the mean (SEM) were computed and plotted in a bar graph. The statistical analysis of the measured data was performed with the GraphPad Prism nine software, by applying the Mann–Whitney U test. Statistical values of p-value < 0.05 were considered significant.
Two main classes of morphological asymmetries can be distinguished (Concha et al., 2012): in class I, asymmetric structures differ in size between the two sides, while class II asymmetries comprise structures present only on one side of the body. The habenulae and the pineal complex exhibit class I and class II asymmetries respectively, the former of which can readily be visualized by immunolabeling of dendritic arborisation using anti-Acetylated Tubulin (Signore et al., 2009). We corroborated this finding by showing subtle, but reproducible, left-right differences in the extent of Acetylated Tubulin expressing habenular structures (Figure 1A). In zebrafish, numerous genes have been described that are either expressed specifically or largely specifically in subnuclei of the dorsal habenulae (dHb), in all dHb neurons or in dHb neuronal precursor cells. Of these, we isolated the medaka orthologues for kctd12.1 (Gamse et al., 2003; Sato et al., 2021) and nrp1 expressed predominantly in lateral neurons of the dorsal habenulae (dHbl) (Kuan et al., 2007), cntn2 (Carl et al., 2007) and slc18a3b/vachtb as markers for medial neurons of the dorsal habenulae (dHbm) (Hong et al., 2013), slc17a6b/vglut2 expressed in all habenular neurons (Thisse et al., 2005) and cxcr4b in dorsal habenular precursors (Roussigné et al., 2009). In addition, we re-assessed the expression of medaka prox1a (formerly named prox1b), which was reported to be right-dominantly asymmetric in the habenulae (Deguchi et al., 2009).
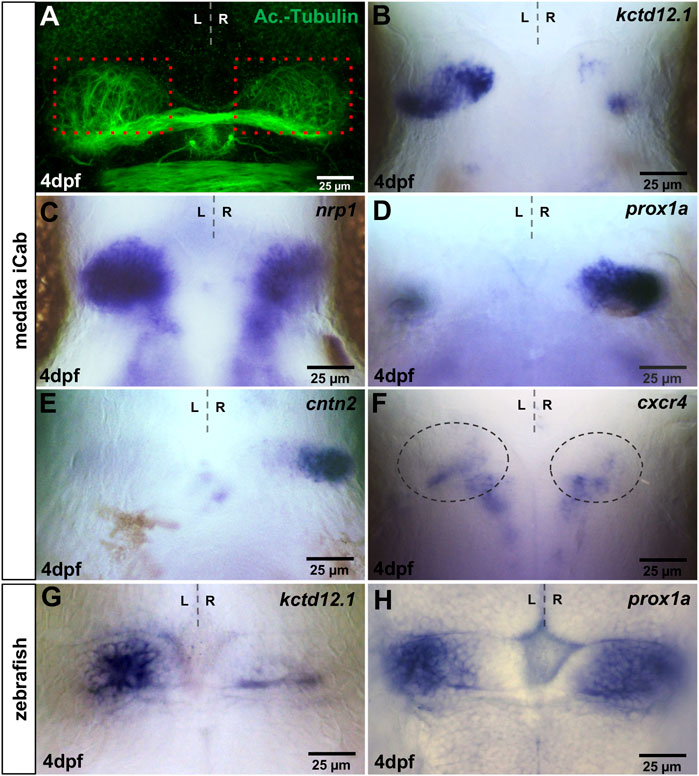
FIGURE 1. Visualisation of asymmetries in the medaka habenulae. iCab embryos at four dpf focused on the dorsal diencephalon; anterior to the top. (A) Immunostaining for anti-acetylated tubulin shows subtle left-right asymmetric dendritic arborisation in the habenula. Please compare the green signal within the same-sized red boxes. (B–H) Expression of habenula and pineal complex marker genes. (B,C) kctd12.1 and nrp1 are stronger expressed in left-sided habenular neurons. (D,E) prox1a and cntn2 are mainly expressed in right-sided habenular neurons, while (F) cxcr4b is expressed in habenular progenitors, the area of which is encircled. (G) In the zebrafish, kctd12.1 is expressed in lateral neurons of the dorsal habenulae, while (H) prox1a is expressed in medial dorsal habenular neurons.
In four dpf old medaka iCab embryos, kctd12.1 is expressed predominantly in left-sided habenular neurons, which likely correspond to the dHbl subnucleus (Figure 1B). The expression is enriched in a proximal and a distal cluster connected by cells with lower expression. On the right side, kctd12.1 is weakly expressed in two small domains in a proximo-anterior and a distal-posterior cluster of cells differently from zebrafish, in which the gene exhibits a rather weak but continuous expression (Figure 1G) (Gamse et al., 2003). Also, medaka nrp1 exhibits strong asymmetric left-sided expression in the dHb (Figure 1C). However, numerous dHb neurons express nrp1 also on the right side, unlike its zebrafish orthologue (Kuan et al., 2007). In contrast to the left-sided habenular marker genes, we find strong prox1a expression in right habenular neurons as described (Figure 1D) (Deguchi et al., 2009), which likely represent dHbm neurons, whereas only a few cells on the left express prox1a. We re-assessed zebrafish prox1a expression (Thisse et al., 2005) and observed not only right-dominant expression of this gene’s mRNA in the habenula but also a prominent expression domain of prox1a on the left side (Figure 1H). Also, cntn2 is expressed almost exclusively on the right side in four dpf iCab (Figure 1E), while in the zebrafish habenulae it is also expressed, albeit less prominently, on the left side (Carl et al., 2007). Therefore, our two chosen dHbm marker genes are more restricted to right-sided neurons in medaka compared to zebrafish. cxcr4b expression in habenular precursors in zebrafish (Roussigné et al., 2009) is downregulated during habenular maturation. Similarly, we find only a few cells expressing this gene in the iCab habenula at four dpf (Figure 1F). Our slc18a3b/vachtb probe did not detect any expression, while we found slc17a6b/vglut2 expression in various brain regions at four dpf, in particular in telencephalic structures, but not in the habenulae (Supplementary Figure S1).
Zebrafish otx5 is expressed in both pineal and parapineal cells (Figure 2A) (Gamse et al., 2002), while sox1a (Clanton et al., 2013) and gfi1 (gfi1ab) (Dufourcq et al., 2004) are expressed exclusively in zebrafish parapineal cells. We isolated the respective medaka orthologs and found conserved otx5l transcripts in both pineal and left-sided parapineal cells of iCab embryos at four dpf (Figure 2B). As described previously, analyzing the expression of a Rx2:GFP transgene (Signore et al., 2009), the medaka parapineal volume judged by otx5l labeling appears to be slightly larger than the zebrafish parapineal. In analogy to zebrafish, sox1a and gfi1ab transcripts were detected specifically in medaka parapineal cells including a few cells seemingly connecting the parapineal with the pineal (Figures 2C,D). Therefore, the specificity of pineal complex genes is evolutionarily conserved between zebrafish and medaka.
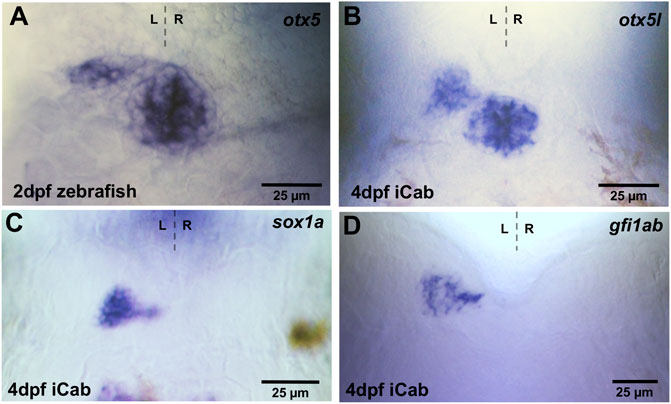
FIGURE 2. Expression of pineal and parapineal marker genes in the pineal complex. Zebrafish and iCab embryos at stages indicated focused on the dorsal diencephalon; anterior to the top. (A,B) otx5/otx5l is expressed in the pineal and parapineal in zebrafish and medaka, while (C,D) sox1a and gfi1ab are specifically expressed in the parapineal.
Northern and southern medaka populations are diverged by at least 4 million years and show differences in various traits such as morphology and behavior (Takeda and Shimada, 2010; Tsuboko et al., 2014). In particular, the latter observation suggests that neural circuits are differently wired in these populations and cause such differences. Therefore, we assessed whether variations in gene expression are detectable in the dorsal diencephalic asymmetric neural circuit. Like observed in iCab embryos, we did not detect specific slc18a3b expression in Kaga embryos, while Please change into slc17a6b/vglut2 is predominantly expressed in the telencephalon and not in the habenulae (data not shown). We find that prox1a expression in dHbm neurons is largely conserved between the southern iCab strain and the northern Kaga strain (Figures 3A,B). In contrast, kctd12.1 is expressed in a larger domain on the left in Kaga embryos when compared to iCab (Figures 3C,D). In addition, the anterior-proximal expression domain on the right hemisphere is expanded in Kaga embryos. We verified that the differences in gene expression are not due to differences in habenular sizes between the two strains by measuring the habenular size by nuclear staining in embryos at four dpf (Figures 3E–G). We did not find any significant differences in the left, right or total habenular volume between iCab and Kaga. However, we noticed that in both strains the left habenula is about 20% larger than the right. This corresponds to previous estimates from marker gene expression and neuropil labelings in zebrafish (Concha et al., 2000; Gamse et al., 2003). Our habenular size measurement shows that the observed differences of kctd12.1 expression between iCab and Kaga are not due to differences in habenular size.
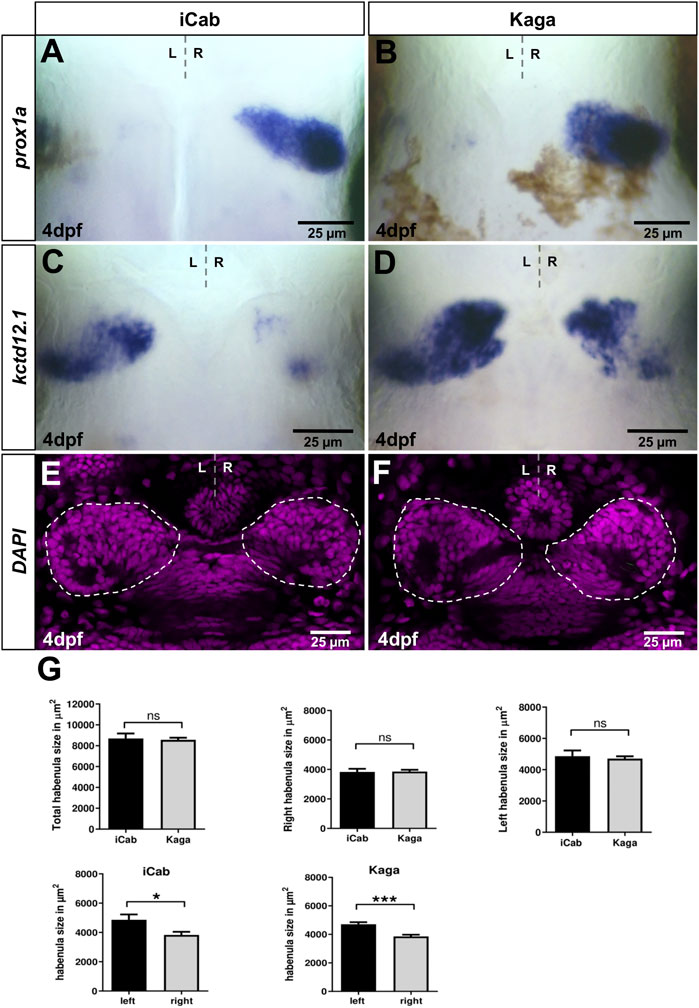
FIGURE 3. Habenular marker genes have conserved and diverged expression patterns in distant medaka strains. Dorsal views focused on the diencephalon at four dpf, anterior to the top. (A,B) prox1a expression in right-sided habenular neurons is comparable in iCab and Kaga, whereas (C,D) kctd12.1 expression is particularly enlarged in right-sided habenular neurons in Kaga. (E,F) Single plane confocal images of the diencephalon of DAPI labeled embryos (niCab = 6; nKaga = 9). (G) No difference in habenular size between iCab and Kaga embryos, but a significant difference between the left and right habenulae in both populations was found. Dashed lines indicate the habenulae in (E,F). Bar graphs show the means of the habenular areas. Error bars represent SEM (standard error of the mean).
Next, we compared gene expression in the pineal complex between the two medaka strains (Figures 4A,B). While the otx5l expression domain in the pineal alone or in the pineal/parapineal complex together does not significantly differ between iCab and Kaga, the parapineal expression domain is significantly larger in Kaga embryos (Figure 4E). Similarly, also the parapineal specific expression of sox1a is larger in Kaga embryos (Figures 4C,D). Taken together, our intra-species analysis of asymmetric diencephalic neuronal structures and associated gene expression reveals differences in the parapineal expression domains of otx5l and sox1a as well as of the dHbl marker kctd12.1, while dHbm marker gene expression appears conserved in embryos of northern and southern medaka populations.
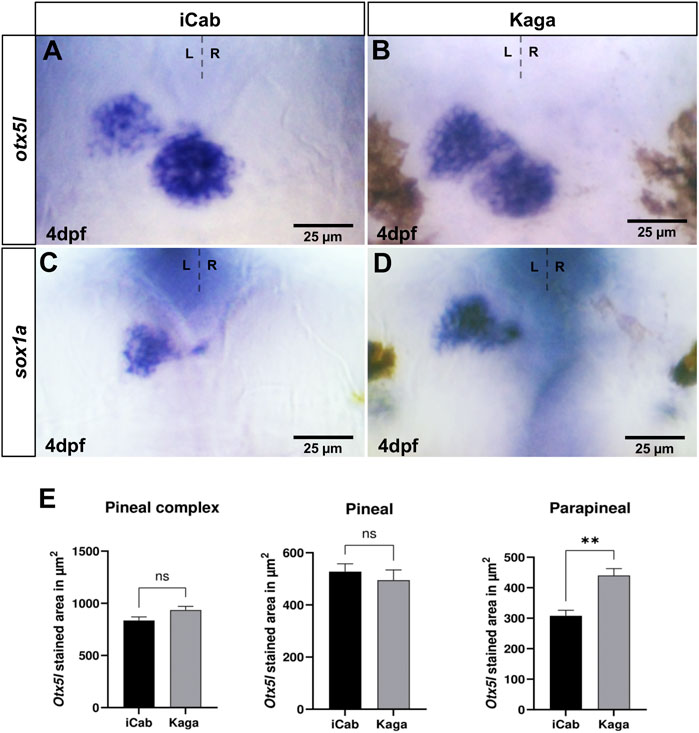
FIGURE 4. Parapineal size differs significantly between Kaga and iCab embryos. Dorsal views focused on the diencephalon of four dpf embryos with anterior to the top. (A–D) otx5l and sox1a are expressed in pineal/parapineal of both iCab and Kaga embryos. (A,B,E) The parapineal expression domain of otx5l is significantly larger in Kaga embryos compared to iCab embryos, whereas no size difference of the otx5l expression domain in the pineal alone or in the pineal/parapineal complex together was detected (niCab = 5; nKaga = 5). Bar graphs show the means of the pineal complex, pineal and parapineal areas. Error bars represent SEM (standard error of the mean).
Our study on marker gene expression in left-right asymmetric neuronal structures in the medaka diencephalon reveals conserved and species-specific differences in the teleost lineage. We find that five out of the seven genes, selected due to their specific expression in the zebrafish habenulae, are also expressed in the medaka habenulae. Notably, right dominant markers for dHbm neurons such as prox1a and cntn2 are more restricted to this side in medaka iCab embryos than in zebrafish, while left-sided nrp1 expression in dHbl neurons is more left-specific in zebrafish (Thisse et al., 2005; Carl et al., 2007; Kuan et al., 2007). Previous studies have identified roles for cntn2 and nrp1 in neuronal migration and axon fasciculation as well as axon guidance during zebrafish development, respectively (De Bruin et al., 2016; Gurung et al., 2018). Therefore, the findings of our comparative study could contribute to explain the rather subtle differences in the targeting of habenular efferent axons in the interpeduncular nucleus between medaka and zebrafish (Signore et al., 2009). Furthermore, all three pineal complex genes, otx5l, sox1a and gfi1ab show a largely conserved expression between medaka and zebrafish (Gamse et al., 2002; Dufourcq et al., 2004; Clanton et al., 2013).
Our comparison between diverged medaka inbred strains revealed different variations of gene expression in the asymmetric diencephalic structures investigated. Firstly, the parapineal, as defined by otx5l and sox1a expression, is bigger in the Kaga strain compared to iCab embryos. This is paralleled by a stronger expression of kctd12.1 in left dHbl neurons and remarkably also on the right hemisphere of Kaga embryos. In contrast, prox1a expression in dHbm neurons exhibits the same pattern in Kaga and iCab habenulae suggesting that some right habenular neurons may express both genes in Kaga. This, in turn, might implicate different fates of habenular neurons on the right side between Kaga and iCab. Future connectome and functional analyses will be needed to test this hypothesis. Another question arising is whether the differences detected in parapineal size and dHbl marker gene expression between iCab and Kaga are separated evolutionary events or whether they are linked to one another and caused by a, perhaps more likely, single event. Many years of research in zebrafish have shown that dHbl neurogenesis and the parapineal are connected, although the precise mechanism is still not fully resolved, dHb neurons develop from multipotent pools of habenular precursor cells adjacent to the pineal complex. Influenced by temporally tightly regulated Wnt/beta-catenin signaling alongside Notch signaling, they develop into dHbm neurons (Aizawa et al., 2007; Carl et al., 2007; Beretta et al., 2013; Hüsken et al., 2014; Guglielmi et al., 2020). This predominantly happens in the right hemisphere. On the left half of the brain, parapineal cells migrate away from the pineal influenced by Nodal and Fgf signaling shortly before habenular precursors become post-mitotic and differentiate (Concha et al., 2003; Regan et al., 2009; Roussigné et al., 2018). It is thought that yet unknown signals released from these parapineal cells that depend on sox1a function (Lekk et al., 2019) influence habenular neuron precursors on the left side to adopt dHbl character (Concha et al., 2003; Hüsken et al., 2014). In this process, the size of the parapineal not only appears to influence left dHb precursors but, in certain circumstances, also the right habenula. Pitx2 genes are members of the bicoid-class of paired homeodomain transcription factors and targets of Nodal signaling. In zebrafish, pitx2c is expressed in the left dorsal diencephalon and influences parapineal development (Essner et al., 2000; Garric et al., 2014). In pitx2c hypomorphic embryos, the parapineal cell number is increased. These embryos exhibit an enlarged domain of right-sided dHbl typical kctd12.1 expression, while dHbm markers remain unaffected. This phenotype is remarkably similar to our observations in Kaga embryos and raises the hypothesis that evolutionary events may have caused modulations around the genomic interval comprising the Kaga pitx2 gene. In medaka iCab embryos belonging to the southern population, the pitx2 gene has been identified and its asymmetric expression in the brain described (Jaszczyszyn et al., 2007; Soukup et al., 2018). Regarding northern populations, only the medaka HNI genome sequence is currently available. Although validating our hypothesis certainly warrants the analysis of the Kaga genome, we performed a preliminary HNI genome analysis and could only detect high sequence homologies for pitx1 and pitx3 in the genome of zebrafish and medaka HNI. We did not detect any sequence homologies representing pitx2 in the HNI genome (data not shown). Future functional experiments and the availability of a Kaga genome sequence will allow investigating whether the differences detected in parapineal size and kctd12.1 gene expression in Kaga are linked and caused by a single event during evolution, which is the loss of pitx2 in northern populations.
Our work has substantiated that medaka is particularly suitable for elucidating intra-species variations for population genetic studies. Here we used established inbred strains from diverged medaka populations to search for variance of the emergence of brain asymmetries. Based on our findings, we suggest that such intra-species comparisons can serve as a starting point to link brain asymmetries to adaptive behaviors in future studies. Behavioral tests in medaka to investigate anxiety-like behaviors and sociability have recently been reported (Lucon-Xiccato et al., 2022). Both of them are strongly influenced by habenular function and could therefore serve as a readout for habenular function (Bühler and Carl, 2021). In view of the recently established panel of inbred strains derived from a single wild population, medaka offers unique genetic resources to tackle this long-standing question in neuroscience (Fitzgerald et al., 2022).
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Fish husbandry was performed in accordance with EU directive 2010/63/EU guidelines as well as with German animal protection regulations (Tierschutzgesetz §11, Abs. 1, no. 1; Regierungspräsidium Karlsruhe, Germany; husbandry permits AZ35-9185.64/BH KIT) and the Italian Ministry of Health - permit 151/2019-PR.
Conceptualization: MC, FL; Methodology: FL, CH, NA, MC, AB; Validation: AB, AC; Formal analysis: AC, AB; Investigation: CA, AB, AC; Resources: FL, MC; Data curation: AC, AB; Writing—original draft: MC, FL; Writing—review and editing: MC, FL, AB, CA, AC; Visualization: AB, CA, AC; Supervision: FL, MC; Project administration: MC; Funding acquisition: MC, FL, AB.
This research was funded by a Deutsche Forschungsgemeinschaft (DFG)-long term fellowship, grant number BU3753/1-1, Caritro Bando 2020 per progetti presentati da giovani ricercatori post-doc #10868 and a DMM Conference Travel Grant CTG-DMM2111555 to AB. FL, CH, and NA were supported by the Natural, Artificial and Cognitive Information Processing (NACIP) Helmholtz funding programme.
We are grateful to the Department of Cellular, Computational and Integrative Biology, CIBIO, University of Trento for financial support. The authors acknowledge Nadeshda Wolf, Natalja Kusminski and Thomas Seitz for supporting the fish husbandry, and Alicia Günthel for laboratory assistance.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2022.1005776/full#supplementary-material
Agetsuma, M., Aizawa, H., Aoki, T., Nakayama, R., Takahoko, M., Goto, M., et al. (2010). The habenula is crucial for experience-dependent modification of fear responses in zebrafish. Nat. Neurosci. 13 (11), 1354–1356. doi:10.1038/nn.2654
Aizawa, H., Bianco, I. H., Hamaoka, T., Miyashita, T., Uemura, O., Concha, M. L., et al. (2005). Laterotopic representation of left-right information onto the dorso-ventral axis of a zebrafish midbrain target nucleus. Curr. Biol. 15 (3), 238–243. doi:10.1016/j.cub.2005.01.014
Aizawa, H., Goto, M., Sato, T., and Okamoto, H. (2007). Temporally regulated asymmetric neurogenesis causes left-right difference in the zebrafish habenular structures. Dev. Cell 12 (1), 87–98. doi:10.1016/j.devcel.2006.10.004
Alqadah, A., Hsieh, Y.-W., Morrissey, Z. D., and Chuang, C.-F. (2018). Asymmetric development of the nervous system. Dev. Dyn. 247 (1), 124–137. doi:10.1002/dvdy.24595
Amo, R., Aizawa, H., Takahoko, M., Kobayashi, M., Takahashi, R., Aoki, T., et al. (2010). Identification of the zebrafish ventral habenula as a homolog of the mammalian lateral habenula. J. Neurosci. 30 (4), 1566–1574. doi:10.1523/JNEUROSCI.3690-09.2010
Beretta, C. A., Dross, N., Bankhead, P., and Carl, M. (2013). The ventral habenulae of zebrafish develop in prosomere 2 dependent on Tcf7l2 function. Neural Dev. 8, 19. doi:10.1186/1749-8104-8-19
Beretta, C. A., Dross, N., Guiterrez-Triana, J. A., Ryu, S., and Carl, M. (2012). Habenula circuit development: Past, present, and future. Front. Neurosci. 6, 51. doi:10.3389/FNINS.2012.00051
Bianco, I. H., Carl, M., Russell, C., Clarke, J. D. W., and Wilson, S. W. (2008). Brain asymmetry is encoded at the level of axon terminal morphology. Neural Dev. 3 (1), 9. doi:10.1186/1749-8104-3-9
Braasch, I., Peterson, S. M., Desvignes, T., McCluskey, B. M., Batzel, P., and Postlethwait, J. H. (2014). A new model army: Emerging fish models to study the genomics of vertebrate Evo-Devo. J. Exp. Zool. B Mol. Dev. Evol. 324 (4), 316–341. doi:10.1002/JEZ.B.22589
Bühler, A., and Carl, M. (2021). Zebrafish tools for deciphering habenular network-linked mental disorders. Biomolecules 11 (2), 324. doi:10.3390/biom11020324
Carl, M., Bianco, I. H., Bajoghli, B., Aghaallaei, N., Czerny, T., and Wilson, S. W. (2007). Wnt/Axin1/beta-catenin signaling regulates asymmetric nodal activation, elaboration, and concordance of CNS asymmetries. Neuron 55 (3), 393–405. doi:10.1016/j.neuron.2007.07.007
Clanton, J. A., Hope, K. D., and Gamse, J. T. (2013). Fgf signaling governs cell fate in the zebrafish pineal complex. Development 140 (2), 323–332. doi:10.1242/dev.083709
Concha, M. L., Burdine, R. D., Russell, C., Schier, A. F., and Wilson, S. W. (2000). A nodal signaling pathway regulates the laterality of neuroanatomical asymmetries in the zebrafish forebrain. Neuron 28 (2), 399–409. doi:10.1016/S0896-6273(00)00120-3
Concha, M. L., Russell, C., Regan, J. C., Tawk, M., Sidi, S., Gilmour, D. T., et al. (2003). Local tissue interactions across the dorsal midline of the forebrain establish CNS laterality. Neuron 39 (3), 423–438. doi:10.1016/S0896-6273(03)00437-9
Concha, M. L., Bianco, I. H., and Wilson, S. W. (2012). Encoding asymmetry within neural circuits. Nat. Rev. Neurosci. 13 (12), 832–843. doi:10.1038/nrn3371
Concha, M. L., and Wilson, S. W. (2001). Asymmetry in the epithalamus of vertebrates. J. Anat. 199 (1), 63–84. doi:10.1046/j.1469-7580.2001.19910063.x
De Bruin, A., Cornelissen, P. W. A., Kirchmaier, B. C., Mokry, M., Iich, E., Nirmala, E., et al. (2016). Genome-wide analysis reveals NRP1 as a direct HIF1α-E2F7 target in the regulation of motorneuron guidance in vivo. Nucleic Acids Res. 44 (8), 3549–3566. doi:10.1093/NAR/GKV1471
Deguchi, T., Fujimori, Kazuhiro E., Kawasaki, T., Ohgushi, H., and Yuba, S. (2009). Molecular cloning and gene expression of the prox1a and prox1b genes in the medaka, Oryzias latipes’. Gene Expr. Patterns 9, 341–347. doi:10.1016/j.gep.2009.02.004
Dreosti, E., Vendrell Llopis, N., Carl, M., Yaksi, E., and Wilson, S. W. (2014). Left-right asymmetry is required for the habenulae to respond to both visual and olfactory stimuli. Curr. Biol. 24 (4), 440–445. doi:10.1016/j.cub.2014.01.016
Duboc, V., Dufourcq, P., Blader, P., and Roussigné, M. (2015). Asymmetry of the brain: Development and implications. Annu. Rev. Genet. 49 (1), 647–672. doi:10.1146/annurev-genet-112414-055322
Dufourcq, P., Rastegar, S., Strähle, U., and Blader, P. (2004). Parapineal specific expression of gfi1 in the zebrafish epithalamus. Gene Expr. Patterns 4 (1), 53–57. doi:10.1016/s1567-133x(03)00148-0
Essner, J. J., Branford, W. W., Zhang, J., and Yost, H. J. (2000). Mesendoderm and left-right brain, heart and gut development are differentially regulated by pitx2 isoforms. Development 127 (5), 1081–1093. doi:10.1242/DEV.127.5.1081
Facchin, L., Duboué, E. R., and Halpern, M. E. (2015). Disruption of epithalamic left–right asymmetry increases anxiety in zebrafish. J. Neurosci. 35 (48), 15847–15859. doi:10.1523/JNEUROSCI.2593-15.2015
Fitzgerald, T., Brettell, I., Leger, A., Wolf, N., Kusminski, N., Monahan, J., et al. (2022). The medaka inbred kiyosu-karlsruhe (MIKK) panel. Genome Biol. 23 (1), 59. doi:10.1186/s13059-022-02623-z
Frasnelli, E., Vallortigara, G., and Rogers, L. J. (2012). Left–right asymmetries of behaviour and nervous system in invertebrates. Neurosci. Biobehav. Rev. 36 (4), 1273–1291. doi:10.1016/j.neubiorev.2012.02.006
Furutani-Seiki, M., and Wittbrodt, J. (2004). Medaka and zebrafish, an evolutionary twin study. Mech. Dev. 121 (7), 629–637. doi:10.1016/j.mod.2004.05.010
Gamse, J. T., Shen, Y.-C., Thisse, C., Thisse, B., Raymond, P. A., Halpern, M. E., et al. (2002). Otx5 regulates genes that show circadian expression in the zebrafish pineal complex. Nat. Genet. 30, 117. doi:10.1038/ng793
Gamse, J. T., Thisse, C., Thisse, B., and Halpern, M. E. (2003). The parapineal mediates left-right asymmetry in the zebrafish diencephalon. Development 130 (6), 1059–1068. doi:10.1242/dev.00270
Garric, L., Ronsin, B., Roussigné, M., Booton, S., Gamse, J. T., Dufourcq, P., et al. (2014). Pitx2c ensures habenular asymmetry by restricting parapineal cell number. Dev. Camb. 141 (7), 1572–1579. doi:10.1242/dev.100305
Guglielmi, L., Bühler, A., Moro, E., Argenton, F., Poggi, L., and Carl, M. (2020). Temporal control of Wnt signaling is required for habenular neuron diversity and brain asymmetry. Development 147 (6), dev182865. doi:10.1242/dev.182865
Güntürkün, O., and Ocklenburg, S. (2017). Ontogenesis of lateralization. Neuron 94 (2), 249–263. doi:10.1016/j.neuron.2017.02.045
Gurung, S., Asante, E., Hummel, D., Williams, A., Feldman-Schultz, O., Halloran, M. C., et al. (2018). Distinct roles for the cell adhesion molecule Contactin2 in the development and function of neural circuits in zebrafish. Mech. Dev. 152, 1–12. doi:10.1016/J.MOD.2018.05.005
Hong, E., Santhakumar, K., Akitake, C. A., Ahn, S. J., Thisse, C., Thisse, B., et al. (2013). Cholinergic left-right asymmetry in the habenulo-interpeduncular pathway. Proc. Natl. Acad. Sci. U. S. A. 110 (52), 21171–21176. doi:10.1073/pnas.1319566110
Hüsken, U., Stickney, H. L., Gestri, G., Bianco, I. H., Faro, A., Young, R. M. M., et al. (2014). Tcf7l2 is required for left-right asymmetric differentiation of habenular neurons. Curr. Biol. 24 (19), 2217–2227. doi:10.1016/J.CUB.2014.08.006
Ishikawa, Y., Yoshimoto, M., Yamamoto, N., and Ito, H. (1999). Different brain morphologies from different genotypes in a single teleost species, the medaka (Oryzias latipes). Brain Behav. Evol. 53 (1), 2–9. doi:10.1159/000006577
Iwamatsu, T. (2004). Stages of normal development in the medaka Oryzias latipes. Mech. Dev. 121 (7), 605–618. doi:10.1016/j.mod.2004.03.012
Jaszczyszyn, Y., Haeussler, M., Heuzé, A., Debiais-Thibaud, M., Casane, D., Bourrat, F., et al. (2007). Comparison of the expression of medaka (Oryzias latipes) pitx genes with other vertebrates shows high conservation and a case of functional shuffling in the pituitary. Gene 406 (1–2), 42–50. doi:10.1016/J.GENE.2007.05.024
Kimura, T., Shimada, A., Sakai, N., Mitani, H., Naruse, K., Takeda, H., et al. (2007). Genetic analysis of craniofacial traits in the medaka. Genetics 177 (4), 2379–2388. doi:10.1534/genetics.106.068460
Kinoshita, M., Murata, K., Naruse, K., and Tanaka, M. (2009). Medaka: Biology, management, and experimental protocols. Iowa: Wiley-Blackwell.
Kirchmaier, S., Naruse, K., Wittbrodt, J., and Loosli, F. (2015). The genomic and genetic toolbox of the teleost medaka ( Oryzias latipes ). Genetics 199 (4), 905–918. doi:10.1534/genetics.114.173849
Kong, X.-Z., Mathias, S. R., Guadalupe, T., Working Group, L., Glahn, D. C., Franke, B., et al. (2018). Mapping cortical brain asymmetry in 17,141 healthy individuals worldwide via the ENIGMA Consortium. Proc. Natl. Acad. Sci. USA. 115, E5154. doi:10.1073/pnas.1718418115
Krishnan, S., Mathuru, A. S., Kibat, C., Rahman, M., Lupton, C. E., Stewart, J., et al. (2014). The right dorsal habenula limits attraction to an odor in zebrafish. Curr. Biol. 24 (11), 1167–1175. doi:10.1016/j.cub.2014.03.073
Kuan, Y.-S., Yu, H.-H., Moens, C. B., and Halpern, M. E. (2007). Neuropilin asymmetry mediates a left-right difference in habenular connectivity. Dev. Camb. Engl. 134 (5), 857–865. doi:10.1242/dev.02791
Lekk, I., Duboc, V., Faro, A., Nicolaou, S., Blader, P., and Wilson, S. W. (2019). Sox1a mediates the ability of the parapineal to impart habenular left-right asymmetry. eLife 8, e47376. doi:10.7554/eLife.47376
Loosli, F., Köster, R. W., Carl, M., Krone, A., and Wittbrodt, J. (1998). Six3, a medaka homologue of the Drosophila homeobox gene sine oculis is expressed in the anterior embryonic shield and the developing eye. Mech. Dev. 74 (1), 159–164. doi:10.1016/S0925-4773(98)00055-0
Loosli, F., Köster, R. W., Carl, M., Kühnlein, R., Henrich, T., Mücke, M., et al. (2000). A genetic screen for mutations affecting embryonic development in medaka fish (Oryzias latipes). Mech. Dev. 97 (1), 133–139. doi:10.1016/S0925-4773(00)00406-8
Lucon-Xiccato, T., Loosli, F., Conti, F., Foulkes, N. S., and Bertolucci, C. (2022). Comparison of anxiety-like and social behaviour in medaka and zebrafish. Sci. Rep. 12, 10926. doi:10.1038/s41598-022-14978-1
Miletto Petrazzini, M. E., Sovrano, V. A., Vallortigara, G., and Messina, A. (2020). Brain and behavioral asymmetry: A lesson from fish. Front. Neuroanat. 14, 11. doi:10.3389/fnana.2020.00011
Miyasaka, N., Morimoto, K., Tsubokawa, T., Higashijima, S., Okamoto, H., and Yoshihara, Y. (2009). From the olfactory bulb to higher brain centers: Genetic visualization of secondary olfactory pathways in zebrafish. J. Neurosci. 29 (15), 4756–4767. doi:10.1523/JNEUROSCI.0118-09.2009
Mundorf, A., Peterburs, J., and Ocklenburg, S. (2021). Asymmetry in the central nervous system: A clinical neuroscience perspective. Front. Syst. Neurosci. 15, 733898. doi:10.3389/fnsys.2021.733898
Regan, J. C., Concha, M. L., Roussigne, M., Russell, C., and Wilson, S. W. (2009). An fgf8-dependent bistable cell migratory event establishes CNS asymmetry. Neuron 61 (1), 27–34. doi:10.1016/j.neuron.2008.11.030
Rogers, L. J. (2014). Asymmetry of brain and behavior in animals: Its development, function, and human relevance. genesis 52 (6), 555–571. doi:10.1002/dvg.22741
Roussigné, M., Bianco, I. H., Wilson, S. W., and Blader, P. (2009). Nodal signalling imposes left-right asymmetry upon neurogenesis in the habenular nuclei. Development 136 (9), 1549–1557. doi:10.1242/dev.034793
Roussigné, M., Wei, L., Tsingos, E., Kuchling, F., Alkobtawi, M., Tsalavouta, M., et al. (2018). Left/right asymmetric collective migration of parapineal cells is mediated by focal FGF signaling activity in leading cells. Proc. Natl. Acad. Sci. U. S. A. 115 (42), E9812–E9821. doi:10.1073/pnas.1812016115
Sato, K., Khine, |, Nwe, N., and Ohuchi, H. (2021). The opsin 3/teleost multiple tissue opsin system: mRNA localization in the retina and brain of medaka (Oryzias latipes). J. Comp. Neurol. 529, cne.25106–2516. doi:10.1002/cne.25106
Signore, I. A., Guerrero, N., Loosli, F., Colombo, A., Villalón, A., Wittbrodt, J., et al. (2009). Zebrafish and medaka: Model organisms for a comparative developmental approach of brain asymmetry. Phil. Trans. R. Soc. B 364 (1519), 991–1003. doi:10.1098/rstb.2008.0260
Soukup, V., Mrstakova, S., and Kozmik, Z. (2018). Asymmetric pitx2 expression in medaka epithalamus is regulated by nodal signaling through an intronic enhancer. Dev. Genes Evol. 228 (2), 131–139. doi:10.1007/S00427-018-0611-1
Spivakov, M., Auer, T. O., Peravali, R., Dunham, I., Dolle, D., Fujiyama, A., et al. (2014). Genomic and phenotypic characterization of a wild medaka population: Towards the establishment of an isogenic population genetic resource in fish. G3 (Bethesda, Md.) 4 (3), 433–445. doi:10.1534/g3.113.008722
Takeda, H., and Shimada, A. (2010). The art of medaka genetics and genomics: What makes them so unique? Annu. Rev. Genet. 44, 217–241. doi:10.1146/annurev-genet-051710-151001
Thisse, B., Heyer, V., Lux, A., Alunni, V., Degrave, A., Seiliez, I., et al. (2005). Spatial and temporal expression of the zebrafish genome by large-scale in situ hybridization screening. Methods Cell Biol. 77, 505–519. doi:10.1016/S0091-679X(04)77027-2
Tsuboko, S., Kimura, T., Shinya, M., Suehiro, Y., Okuyama, T., Shimada, A., et al. (2014). Genetic control of startle behavior in medaka fish. PLOS ONE 9 (11), e112527. doi:10.1371/journal.pone.0112527
Vallortigara, G., and Rogers, L. J. (2005). Survival with an asymmetrical brain: advantages and disadvantages of cerebral lateralization. Behav. Brain Sci. 28 (4), 575–589. doi:10.1017/S0140525X05000105
Volff, J.-N. (2005). Genome evolution and biodiversity in teleost fish. Heredity 94, 280–294. doi:10.1038/sj.hdy.6800635
Wittbrodt, J., Meyer, A., and Schartl, M. (1998). More genes in fish? BioEssays 20, 511. doi:10.1002/(SICI)1521-1878(199806)20:6<511::AID-BIES10>3.0.CO;2-3
Keywords: brain asymmetry, medaka, zebrafish, habenula, pineal, parapineal, population genetics, kctd12.1
Citation: Agostini C, Bühler A, Antico Calderone A, Aadepu N, Herder C, Loosli F and Carl M (2022) Conserved and diverged asymmetric gene expression in the brain of teleosts. Front. Cell Dev. Biol. 10:1005776. doi: 10.3389/fcell.2022.1005776
Received: 28 July 2022; Accepted: 29 August 2022;
Published: 21 September 2022.
Edited by:
Maria Dominguez, Spanish National Research Council (CSIC), SpainReviewed by:
Gareth Powell, University College London, United KingdomCopyright © 2022 Agostini, Bühler, Antico Calderone, Aadepu, Herder, Loosli and Carl. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Felix Loosli, ZmVsaXgubG9vc2xpQGtpdC5lZHU=; Matthias Carl, bWF0dGhpYXMuY2FybEB1bml0bi5pdA==
†These authors have contributed equally to this work and share first authorship
‡These authors have contributed equally to this work and share last authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.