
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Cell Dev. Biol. , 10 January 2022
Sec. Molecular and Cellular Reproduction
Volume 9 - 2021 | https://doi.org/10.3389/fcell.2021.824024
This article is part of the Research Topic Fertilization in the Spotlight: Dynamics and Mechanisms of Sperm-Egg Interaction View all 16 articles
Gamete fusion is the climax of fertilization in all sexually reproductive organisms, from unicellular fungi to humans. Similarly to other cell-cell fusion events, gamete fusion is mediated by specialized proteins, named fusogens, that overcome the energetic barriers during this process. In recent years, HAPLESS 2/GENERATIVE CELL-SPECIFIC 1 (HAP2/GCS1) was identified as the fusogen mediating sperm-egg fusion in flowering plants and protists, being both essential and sufficient for the membrane merger in some species. The identification of HAP2/GCS1 in invertebrates, opens the possibility that a similar fusogen may be used in vertebrate fertilization. HAP2/GCS1 proteins share a similar structure with two distinct families of exoplasmic fusogens: the somatic Fusion Family (FF) proteins discovered in nematodes, and class II viral glycoproteins (e.g., rubella and dengue viruses). Altogether, these fusogens form the Fusexin superfamily. While some attributes are shared among fusexins, for example the overall structure and the possibility of assembly into trimers, some other characteristics seem to be specific, such as the presence or not of hydrophobic loops or helices at the distal tip of the protein. Intriguingly, HAP2/GCS1 or other fusexins have neither been identified in vertebrates nor in fungi, raising the question of whether these genes were lost during evolution and were replaced by other fusion machinery or a significant divergence makes their identification difficult. Here, we discuss the biology of HAP2/GCS1, its involvement in gamete fusion and the structural, mechanistic and evolutionary relationships with other fusexins.
The merging of the plasma membranes of two independent cells with the subsequent formation of an individual cell containing both cytoplasmic contents mixed is known as cell-cell fusion. This biological process is mediated and finely controlled by fusion proteins, termed fusogens, which are specialized proteins capable of overcoming the energetic barriers required for the fusion to occur (Chernomordik and Kozlov, 2003) and are both necessary and sufficient to mediate membrane merging (Brukman et al., 2019). In particular, the fusion between two gametes or “gamete fusion” is one of the hallmarks of meiotic sex and its ubiquitous distribution among eukaryotes suggests an ancestral origin (Ramesh et al., 2005; Speijer et al., 2015; Radzvilavicius, 2016). The first gamete fusogen identified was HAPLESS 2/GENERATIVE CELL-SPECIFIC 1 (HAP2/GCS1) that catalyzes fertilization in flowering plants, protists and probably in some invertebrates. This protein was originally found as an essential sperm factor required for male fertility in flowering plants (Johnson et al., 2004; Mori et al., 2006; von Besser et al., 2006) being later shown that it is necessary for mating in Chlamydomonas and Plasmodium (Hirai et al., 2008; Liu et al., 2008). More recently, the Arabidopsis HAP2/GCS1 (AtHAP2/GCS1) was shown to be sufficient to induce fusion of mammalian cells in culture and the infection of enveloped virus to cells (Valansi et al., 2017). Strikingly, HAP2/GCS1 shares an overall three-dimensional structure with the somatic Fusion Family (FF) proteins discovered in nematodes (Mohler et al., 2002; Sapir et al., 2007; Pérez-Vargas et al., 2014) and with class II viral glycoproteins (e.g., dengue, rubella and zika viruses) (Fédry et al., 2017; Pinello et al., 2017; Valansi et al., 2017). This superfamily of fusion proteins essential for sexual reproduction and exoplasmic merger of plasma membranes was named Fusexin. Even though the mechanisms of cell-cell and virus-cell fusion are diverse and may be mediated by different families of fusion proteins (Segev et al., 2018; Vance and Lee, 2020), the fusexins represent a remarkable case of ancestral fusogens present across the tree of life. Here we review the recent research on HAP2/GCS1-mediated gamete fusion and its functional and structural relationships with other fusexins. Furthermore, we compare the regulation of fusion processes driven by fusexins to the fertilization process in mammals, where the presence of distant members of the Fusexin superfamily is uncertain.
Previous studies have shown FF proteins, like C. elegans’ EFF-1 (CeEFF-1), and HAP2/GCS1 are structurally homologous to viral class II fusion proteins and display a trimeric, postfusion hairpin conformation consisting of three β-sheet-rich domains (DI, DII, DIII) (Figure 1) (Pérez-Vargas et al., 2014; Fédry et al., 2017; Pinello et al., 2017; Valansi et al., 2017). The first of the three domains consist of a β-barrel, followed by a mostly β-stranded elongated domain, and an Immunoglobulin (Ig)-like domain. They are anchored to the cell surface by transmembrane domains at the C terminus. Even though these proteins share very low sequence similarities, the surprising conservation in their architecture suggests that they share a common ancestor forming the Fusexin superfamily (Figure 1). A particular region on one end of the ectodomain has been extensively studied due to functional implications (Figure 1). In viral class II fusion proteins, fusion loops or α-helices located at the tip of domain II containing hydrophobic residues have been proposed to insert into the target membrane during conformational changes, like tick-borne encephalitis virus protein E (Bressanelli et al., 2004). In CeEFF-1, the cd loop at the membrane-proximal side may play a similar structural role, however it is mainly composed of acidic residues forming an electronegative surface which is unlikely to interact with lipidic membranes (Pérez-Vargas et al., 2014). The fusion loops regions in HAP2/GCS1 orthologs are highly variable, with a single helix in AtHAP2/GCS1, three short loops in Trypanosoma cruzi (TcHAP2/GCS1) and three amphipathic helices in Chlamydomonas reinhardtii (CrHAP2/GCS1) (Baquero et al., 2019). The functional relevance of the different configuration of this region is discussed in the next section.
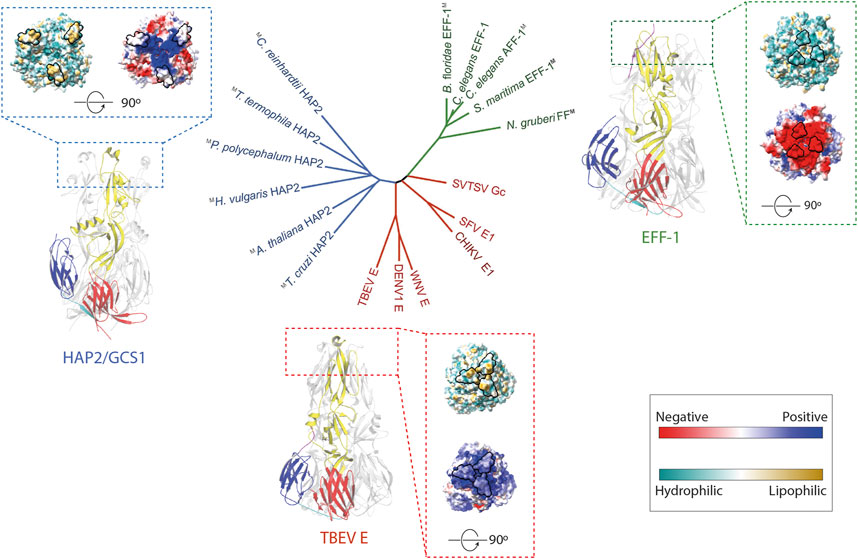
FIGURE 1. The Fusexin superfamily. Ribbon representation of the fusion protein trimers C. elegans EFF-1 (PDB 4OJC; Pérez-Vargas et al., 2014), TBEV E (PDB 1URZ; Bressanelli et al., 2004), and A. thaliana HAP2 (PDB 5OW3; Fedry et al., 2018) with domains I, II, III colored by red, yellow, blue, respectively. Surface representation of trimers viewed from the membrane was displayed. The surface is colored according to the electrostatic potential on a scale from -5 to 5 kT/e (calculated with APBS, Jurrus et al., 2018) from red (negative) to blue (positive). Solvent-excluded surfaces of trimers are colored based on molecular lipophilicity potential maps, ranging from dark cyan (hydrophilic) to dark gold (lipophilic). Proposed fusion loops at the tip of domain II are contoured in black. In the center, an unrooted tree inferred using a distance matrix extracted from (Valansi et al., 2017). Colors are HAP2/GCS1, blue; FF proteins, green; class II viral fusogens, red. M superscript represents models.
How do fusexins mediate membrane fusion? First of all, some fusogens may be required only in one of the opposing membranes, as in the case of class II viral glycoproteins (Podbilewicz, 2014), while others must be present on both sides, like the EFF-1 and AFF-1 somatic fusogens (Podbilewicz et al., 2006; Avinoam et al., 2011). These two possible mechanisms are termed unilateral and bilateral, respectively (Figure 2). However, in the case of HAP2/GCS1 the evidence is controversial. On one hand, the deletion of the Athap/gcs1 gene induces male-specific sterility (Johnson et al., 2004; von Besser et al., 2006), even though some levels of expression were detected in ovules (Borges et al., 2008), suggesting a unilateral mechanism of action. In contrast, when studied in heterologous systems, AtHAP2/GCS1 could mediate exoplasmic fusion only bilaterally (Valansi et al., 2017). Similarly to flowering plants, genetic studies in Plasmodium and Chlamyodomonas, showed that HAP2/GSC1 is absolutely required in only one of the fusing gametes (Hirai et al., 2008; Liu et al., 2008). By contrast, in the ciliated protozoan Tetrahymena thermophila, all seven mating types express HAP2/GCS1 and its absence in only 1 cell of the mating pair disrupts fusion (Cole et al., 2014; Pinello et al., 2017). The slime mold Dictyostelium represents a particular case where two homologs of HAP2/GCS1 were identified: HgrB, which is expressed in the three sex types, and HgrA, expressed only in types I and II (Okamoto et al., 2016). Genetic studies suggest that both HgrA and HgrB are essential in both membranes in crosses between types I and II, but only in one membrane when crossed with the third gamete type (Okamoto et al., 2016; Bloomfield, 2019).
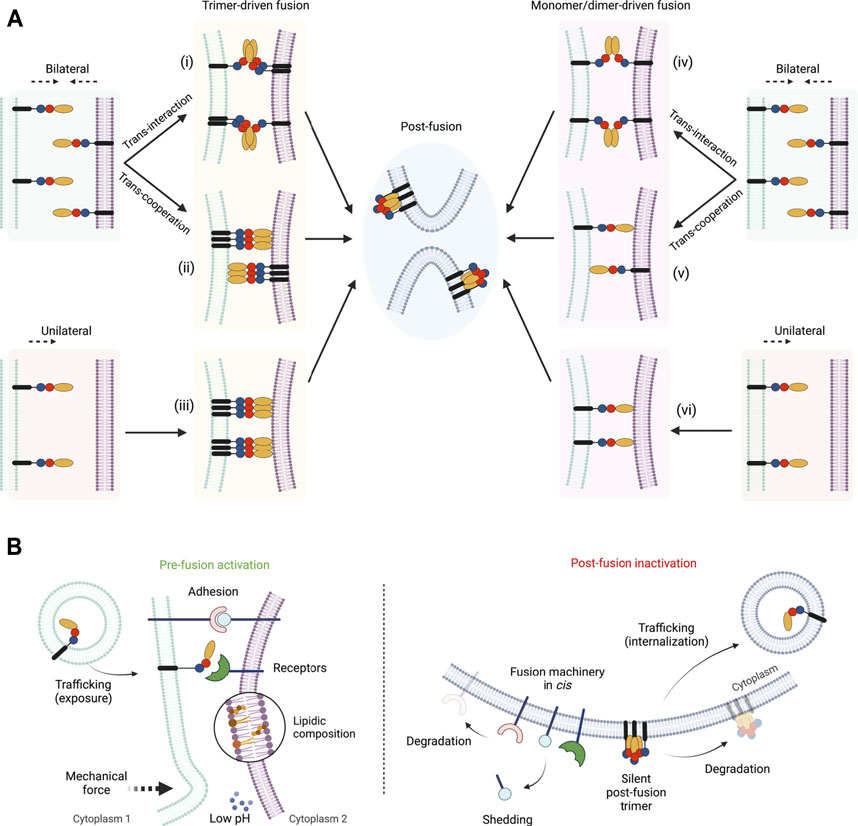
FIGURE 2. Pathways and regulations of fusexin-mediated fusion. (A) Different models for the mechanism of action of fusexins. Fusion can be induced from both merging membranes (“bilateral,” i, ii, iv and v) or from one of them (“unilateral,” iii and vi). Within the bilateral mechanism, fusexins may interact forming dimers or trimers leading to fusion (“trans-interaction,” reminiscent of trans-SNAREs, i and iv) or cooperate by binding to the opposed membrane (“trans-cooperation,” ii and v). Depending on the intermediate states that mediate membrane merging the mechanisms can be divided into trimer-driven fusion (i, ii and iii) or monomer/dimer-driven fusion (iv, v and vi). Independently of the mechanism, the post-fusion conformation is hairpin-shaped trimers. (B) Known mechanisms of pre-fusion activation (left panel) or post-fusion inactivation (right panel) of fusexins described in this review. Created with BioRender.com.
Regardless of the species, structural and biochemical studies show that the hydrophobic tip of domain II of HAP2/GCS1 (Figure 1) is critical for interactions with liposomes (Pinello et al., 2017; Fédry et al., 2018; Feng et al., 2018; Baquero et al., 2019). This HAP2/GCS1-membrane interaction is consistent with a unilateral pathway (Figures 2Aiii,vi) but also with a bilateral mechanism with trans-cooperation (Figures 2Aii,v), or even a hybrid system. More recently, this hydrophobic region was shown to be relevant for oligomerization in Chlamydomonas (Zhang et al., 2021). It was shown that the formation of a small population of SDS-resistant trimers in this species was observed even in the absence of fusion, however, the temporal and spatial resolution of these experiments is not sufficient to confirm that these trimers are responsible for membrane merging.
As mentioned before, all fusexins studied thus far can form trimers. The most studied group within the Fusexin superfamily are the viral class II fusion proteins. In mature virions of alphaviruses and flaviviruses, these proteins exist as heterodimers/trimers or homodimers, respectively, in some cases lying parallel to the membrane and forming an icosahedral lattice on its surface (Zhang et al., 2003; Voss et al., 2010). Upon internalization into the target cells, the low pH within endosomal compartments triggers the activity of class II fusogens leading to infection (White et al., 2008). It is commonly accepted that class II viral fusogens, after a transient monomeric state, trimerize before fusion and that the zippering of these trimers is the driving force for membrane merger (Baquero et al., 2013). In the post-fusion state, these proteins can be found in the already previously mentioned folded trimers of hairpins, however, the existence of the hypothetical extended trimer has not been experimentally confirmed. Therefore, the possibility that monomers rearrange into hairpins to induce fusion prior to their trimerization cannot be excluded (Figures 2Aiv,v,vi).
Even though CeEFF-1 is suggested to be monomeric on cell-derived membrane vesicles (Zeev-Ben-Mordehai et al., 2014), its ectodomain purified and crystallized as a post-fusion trimer (Pérez-Vargas et al., 2014). The monomeric (but not trimeric) soluble protein was able to inhibit cell-cell fusion in vitro (Pérez-Vargas et al., 2014) and soluble EFF-1 trimers stimulate the fusion of insect cells ectopically expressing EFF-1 (Podbilewicz et al., 2006). Similarly to viral fusexins, EFF-1-mediated fusion can be blocked by adding soluble domain III (Pérez-Vargas et al., 2014). Based on the presence of an electronegative instead of a hydrophobic patch on the tip of the domain II and the bilateral requirement of EFF-1, it was proposed that trimers/dimers are formed in trans and that a bidirectional zippering occurs from both fusing membranes (Podbilewicz, 2014, Figure 2Ai,iv). However, a trans-cooperation mechanism where cis-trimers are required from both sides is still a possible model (Figure 2Aii). In the latter scenario, the binding to the opposite membrane might be indirect, distinctly to Class II viral fusexins that involve direct hydrophobic interactions with the membranes.
Whether fusion is mediated by monomers, dimers, trimers or even higher oligomeric states is not completely elucidated and different fusexins may utilize different strategies that should not be excluded (Figure 2A).
Fusion must be a tightly-regulated process. Excessive gamete fusion can lead to non-specific fertilization or to polygamy that may produce unviable polyploid zygotes. Also, the fusion of gametes to their same type or to somatic cells must be avoided. In this sense, even after cell fate determination and gamete encounter, many regulatory mechanisms are established to regulate the activity of gamete fusogens.
Specific adhesion of the gametes seems to be a robust way of regulating fusion as fusogens require the membranes to be in close proximity (Hernández and Podbilewicz, 2017). The concept of “fertilization synapse” was introduced to describe the complex arrangement of the membranes to enable fusion (Krauchunas et al., 2016). In flowering plants, the sperm-specific protein GEX2 (Gamete EXpressed 2) contains Ig-like domains and it was shown to be essential for sperm attachment and the subsequent fertilization (Mori et al., 2014). The known Chlamydomonas factor for gamete adhesion is FUS1 (Ferris et al., 1996; Misamore et al., 2003), which also contains extracellular Ig-like domains that form a structure similar to the one of GEX2 (Pinello et al., 2021). In contrast to the plant counterpart, FUS1 is expressed in the plus mating type, the opposite membrane to HAP2/GCS1. However, no direct interaction between them was reported. Recently, MAR1 (Minus Adhesion Receptor 1) has been suggested to be the molecular partner of FUS1 in the minus gamete and a key regulator of HAP2/GCS1 localization to the fusion site (Pinello et al., 2021, Figure 2B). The only attachment couple known in mammals is the sperm-borne Izumo1 and the oocyte-specific Juno (Izumo Receptor). While the former is a type I transmembrane Ig-like protein, the latter is bound to the egg plasma membrane by a GPI anchor. The interaction between these two proteins leads to tight gamete binding that is species specific and is essential for fusion to occur (Inoue et al., 2005; Bianchi et al., 2014; Bianchi and Wright, 2015). This molecular interaction appears to be an evolutionary novelty as Juno is present only in mammals, however, Izumo1 has a broader distribution involving many lineages of vertebrates (Grayson, 2015). Interestingly, an Izumo-like gene involved in late stages of fertilization was described also in C. elegans (Nishimura et al., 2015; Takayama et al., 2021).
The localization of the fusogens plays a crucial role to regulate fusion. For example, in C. elegans the dynamic internalization of EFF-1 into intracellular early endosomes regulates its function (Smurova and Podbilewicz, 2017). Similarly, AtHAP2/GCS1 localization to the plasma membrane increases upon interaction with EC1 (Egg Cell 1), a small cysteine-rich protein secreted by the female gamete, which is necessary for sperm cell plasma membrane to gain fusion competence (Sprunck et al., 2012). In mammals, the egg tetraspanin CD9 is proposed to be responsible for membrane organization and correct localization of adhesion- and fusion-related proteins, such as Juno (Inoue et al., 2020; Umeda et al., 2020). In the mammalian sperm, Izumo1 localizes into the acrosome, giant intracellular vesicle in the head. Only after undergoing capacitation, a maturation process in the female tract, the acrosomal content is released and Izumo1 relocalizes to the equatorial region of the plasma membrane where tight binding to the oocyte plasma membrane occurs (Satouh et al., 2012). The fusogens involved in mammalian gamete fusion may follow similar localization behavior to Juno and/or Izumo1.
Another factor affecting the activity of fusion is the lipidic composition of both membranes. For example, cholesterol in the target membrane promotes the fusion mediated by some viral class II fusexins (Umashankar et al., 2008; Osuna-Ramos et al., 2018; Pattnaik and Chakraborty, 2021). On the other hand, AtHAP2/GCS1 was reported to bind better to liposomes containing a lipid that mimics the phospholipid phosphatidylserine (Fedry et al., 2018). Phosphatidylserine is a known mediator of many fusion events (Whitlock and Chernomordik, 2021), including EFF-1-mediated neuronal repair of axons in C. elegans (Neumann et al., 2015) and mammalian fertilization (Rival et al., 2019). Albeit lipids may affect the activity of fusion proteins, the composition of the membrane by itself is known to determine the curvature of the bilayer and, therefore, influence directly the progression of hemifusion and pore opening (reviewed in Chernomordik and Kozlov, 2008).
An additional element that could contribute to the cell-cell fusion process is the membrane tension by mechanical forces. For instance, it is proposed that during muscle formation in Drosophila melanogaster and in Schizosaccharomyces pombe mating, actin-rich protrusions from 1 cell are resisted by actomyosin contractions in the other, producing membrane stress that leads to fusion (Kim and Chen, 2019; Muriel et al., 2021). In this sense, actin polymerization was shown to improve EFF-1-driven fusion of insect cells in culture (Shilagardi et al., 2013). Furthermore, EFF-1 and F-actin colocalize during seam cell daughter-hyp7 cell fusion in the C. elegans larvae, however, this interaction seems to be important for the correct localization of the fusogen rather than a mechanical induction of fusion (Yang et al., 2017). The surface of mammalian oocytes is structurally complex and its cortex is enriched with F-actin (Longo, 1985, 1987). Nonetheless, the relevance of this actin network specifically for mammalian gamete fusion is unclear since contradicting results were reported [reviewed in (Sun and Schatten, 2006)]. It is possible that it is involved in membrane merging or in the later incorporation of the sperm. In addition, the actin cytoskeleton might control the position of the sperm during fusion, distancing it from the maternal chromosomes avoiding the elimination of paternal chromosomes during the formation of the second polar body (Mori et al., 2021). Cortical actin dynamics are also required for the correct trafficking of the cortical granules (Connors et al., 1998; Vogt et al., 2019), specialized vesicles of the oocyte that contribute to the block of polyspermy after fertilization.
Mechanical stress on the membranes can also be generated by the pushing and pulling movements of the sperm after attachment. Even though it was previously thought that motility was dispensable for mammalian gamete fusion (Yanagimachi, 1988), recent studies have suggested that a specific beating mode of the flagellum is required for this process (Ravaux et al., 2016) supporting older studies using human gametes (Wolf et al., 1995). It is also possible that these kinetic perturbations are only required for the accumulation of CD9 to the fusion site (Chalbi et al., 2014).
After fusion occurs, a rapid functional silencing of the fusogens is often required. First of all, if the fusion proteins require trans-interactions to be activated (with receptors or to other fusogens), the post-fusion configuration by itself serves as a mechanism of preventing them since all the proteins end up in the same membrane. Alternatively, the fusogens might be removed from the membrane by internalization, as is reported for CeEFF-1 (Smurova and Podbilewicz, 2016), or simply by degradation, such as CrHAP2/GCS1 (Liu et al., 2010). Certainly, the adhesion molecules can suffer these changes turning the membrane into fusion incompetent. For example, Chlamydomonas FUS1 is also degraded after fertilization (Liu et al., 2010) and the mammalian Juno is shed in extracellular vesicles from the surface of the oocyte after fusion (Bianchi et al., 2014). Other biochemical changes in the gametes triggered by fertilization may also affect the activity of the fusogens and block further fusion events. Fast changes in electric potential occur in the plasma membrane of fertilized eggs of some organisms with external fertilization, like sea urchins and frogs, which prevents polyspermy (Jaffe, 1976; Cross and Elinson, 1980). More recently, the release of inorganic zinc from mammalian fertilized oocytes was described as “zinc sparks” which can induce changes in the sperm or in the membrane of the oocyte itself (Kim et al., 2011; Duncan et al., 2016). These changes may induce conformational changes in the fusogens, in the same way that low pH induces structural rearrangements in the viral fusexins, but into an inactivating form. Finally, there is biochemical evidence for EFF-1 and viral Class II proteins that trimerization is irreversible (Bressanelli et al., 2004; Modis et al., 2004; Liao and Kielian, 2005; Nayak et al., 2009; Pérez-Vargas et al., 2014), which would mean that whenever the post-fusion conformation is reached the protein remains locked. This represents a regulatory mechanism that prevents additional fusion events from occurring. Likely this irreversibility of the trimerization is also true for HAP2/GCS1, however, these studies are still missing.
Even though the overall fusexin architecture is strikingly conserved, their distribution among extremely distant organisms possibly has arisen into a diversity of mechanisms of action that might be similar or not (Figure 2). The differences in sequence, local structural features, molecular environments and interactions highlight the necessity of studying the individual biology of fusexins while avoiding overgeneralization.
On the other hand, mammals and other HAP2/GCS1-lacking organisms present an ongoing mystery. While several genes were found to be required for gamete fusion to occur (Brukman et al., 2019; Bianchi and Wright, 2020), the identity of the proteins that are both essential and sufficient for membrane merging is still unknown in fungi and vertebrates. New artificial intelligence-based tools that predict the folding of proteins (Jumper et al., 2021) may be the key to find other families of fusogens that may have replaced HAP2/GCS1, or else uncover those proteins that diverged significantly during evolution but kept the essential structure of fusexins or other families of fusogens that may have replaced HAP2/GCS1.
NB and BP conceived and drafted the manuscript. NB and XL prepared the figures. NB, XL, and BP wrote the manuscript, discussed its content, and revised the work. All authors approved the final submitted version.
Work in our lab is funded by the Israel Science Foundation (ISF grants 257/17, 2462/18, 2327/19 and 178/20). This project has received funding from the European Union’s Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie grant agreement No 844807.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The authors would like to thank Debora J. Cohen, Natalia Pascuali, Yael Iosilevskii, Clari Valansi and the members of Podbilewicz lab for the insightful discussions and comments on this review.
Avinoam, O., Fridman, K., Valansi, C., Abutbul, I., Zeev-Ben-Mordehai, T., Maurer, U. E., et al. (2011). Conserved Eukaryotic Fusogens Can Fuse Viral Envelopes to Cells. Science 332, 589–592. doi:10.1126/science.1202333
Baquero, E., Albertini, A. A., Vachette, P., Lepault, J., Bressanelli, S., and Gaudin, Y. (2013). Intermediate Conformations during Viral Fusion Glycoprotein Structural Transition. Curr. Opin. Virol. 3, 143–150. doi:10.1016/j.coviro.2013.03.006
Baquero, E., Fédry, J., Legrand, P., Krey, T., and Rey, F. A. (2019). Species-Specific Functional Regions of the Green Alga Gamete Fusion Protein HAP2 Revealed by Structural Studies. Structure 27, 113–124. doi:10.1016/j.str.2018.09.014
Bianchi, E., Doe, B., Goulding, D., and Wright, G. J. (2014). Juno Is the Egg Izumo Receptor and Is Essential for Mammalian Fertilization. Nature 508, 483–487. doi:10.1038/nature13203
Bianchi, E., and Wright, G. J. (2015). Cross-species Fertilization: The Hamster Egg Receptor, Juno, Binds the Human Sperm Ligand, Izumo1. Phil. Trans. R. Soc. B 370 (1661), 20140101. doi:10.1098/rstb.2014.0101
Bianchi, E., and Wright, G. J. (2020). Find and Fuse: Unsolved Mysteries in Sperm-Egg Recognition. Plos Biol. 18, e3000953. doi:10.1371/journal.pbio.3000953
Bloomfield, G. (2019). Sex and Macrocyst Formation in Dictyostelium. Int. J. Dev. Biol. 63, 439–446. doi:10.1387/ijdb.190183gb
Borges, F., Gomes, G., Gardner, R., Moreno, N., McCormick, S., Feijó, J. A., et al. (2008). Comparative Transcriptomics of Arabidopsis Sperm Cells. Plant Physiol. 148, 1168–1181. doi:10.1104/pp.108.125229
Bressanelli, S., Stiasny, K., Allison, S. L., Stura, E. A., Duquerroy, S., Lescar, J., et al. (2004). Structure of a Flavivirus Envelope Glycoprotein in its Low-pH-Induced Membrane Fusion Conformation. EMBO J. 23, 728–738. doi:10.1038/sj.emboj.7600064
Brukman, N. G., Uygur, B., Podbilewicz, B., and Chernomordik, L. V. (2019). How Cells Fuse. J. Cel Biol. 218, 1436–1451. doi:10.1083/jcb.201901017
Chalbi, M., Barraud-Lange, V., Ravaux, B., Howan, K., Rodriguez, N., Soule, P., et al. (2014). Binding of Sperm Protein Izumo1 and its Egg Receptor Juno Drives Cd9 Accumulation in the Intercellular Contact Area Prior to Fusion during Mammalian Fertilization. Development 141, 3732–3739. doi:10.1242/dev.111534
Chernomordik, L. V., and Kozlov, M. M. (2003). Protein-Lipid Interplay in Fusion and Fission of Biological Membranes, 175–207. doi:10.1146/annurev.biochem.72.121801.16150472
Cole, E. S., Cassidy-Hanley, D., Fricke Pinello, J., Zeng, H., Hsueh, M., Kolbin, D., et al. (2014). Function of the Male-gamete-specific Fusion Protein HAP2 in a Seven-Sexed Ciliate. Curr. Biol. 24, 2168–2173. doi:10.1016/j.cub.2014.07.064
Connors, S. A., Kanatsu-Shinohara, M., Schultz, R. M., and Kopf, G. S. (1998). Involvement of the Cytoskeleton in the Movement of Cortical Granules during Oocyte Maturation, and Cortical Granule Anchoring in Mouse Eggs. Develop. Biol. 200, 103–115. doi:10.1006/dbio.1998.8945
Cross, N. L., and Elinson, R. P. (1980). A Fast Block to Polyspermy in Frogs Mediated by Changes in the Membrane Potential. Develop. Biol. 75, 187–198. doi:10.1016/0012-1606(80)90154-2
Duncan, F. E., Que, E. L., Zhang, N., Feinberg, E. C., O'Halloran, T. V., and Woodruff, T. K. (2016). The Zinc Spark Is an Inorganic Signature of Human Egg Activation. Sci. Rep. 6 (16), 24737–24738. doi:10.1038/srep24737
Fédry, J., Forcina, J., Legrand, P., Péhau-Arnaudet, G., Haouz, A., Johnson, M., et al. (2018). Evolutionary Diversification of the HAP2 Membrane Insertion Motifs to Drive Gamete Fusion across Eukaryotes. Plos Biol. 16, e2006357. doi:10.1371/JOURNAL.PBIO.2006357
Fédry, J., Liu, Y., Péhau-Arnaudet, G., Pei, J., Li, W., Tortorici, M. A., et al. (2017). The Ancient Gamete Fusogen HAP2 Is a Eukaryotic Class II Fusion Protein. Cell 168, 904–915. doi:10.1016/j.cell.2017.01.024
Feng, J., Dong, X., Pinello, J., Zhang, J., Lu, C., Iacob, R. E., et al. (2018). Fusion Surface Structure, Function, and Dynamics of Gamete Fusogen HAP2. Elife 7, e39772. doi:10.7554/eLife.39772
Ferris, P. J., Woessner, J. P., and Goodenough, U. W. (1996). A Sex Recognition Glycoprotein Is Encoded by the Plus Mating-type Gene Fus1 of Chlamydomonas Reinhardtii. MBoC 7, 1235–1248. doi:10.1091/mbc.7.8.1235
Grayson, P. (2015). Izumo1 and Juno: the Evolutionary Origins and Coevolution of Essential Sperm-Egg Binding Partners. R. Soc. Open Sci. 2, 150296. doi:10.1098/RSOS.150296
Hernández, J. M., and Podbilewicz, B. (2017). The Hallmarks of Cell-Cell Fusion. Development 144, 4481–4495. doi:10.1242/dev.155523
Hirai, M., Arai, M., Mori, T., Miyagishima, S.-y., Kawai, S., Kita, K., et al. (2008). Male Fertility of Malaria Parasites Is Determined by GCS1, a Plant-type Reproduction Factor. Curr. Biol. 18, 607–613. doi:10.1016/j.cub.2008.03.045
Inoue, N., Ikawa, M., Isotani, A., and Okabe, M. (2005). The Immunoglobulin Superfamily Protein Izumo Is Required for Sperm to Fuse with Eggs. Nature 434, 234–238. doi:10.1038/nature03362
Inoue, N., Saito, T., and Wada, I. (2020). Unveiling a Novel Function of CD9 in Surface Compartmentalization of Oocytes. Development 147, dev189985. doi:10.1242/DEV.189985
Jaffe, L. A. (1976). Fast Block to Polyspermy in Sea Urchin Eggs Is Electrically Mediated. Nature 261, 68–71. doi:10.1038/261068a0
Johnson, M. A., von Besser, K., Zhou, Q., Smith, E., Aux, G., Patton, D., et al. (2004). Arabidopsis Hapless Mutations Define Essential Gametophytic Functions. Genetics 168, 971–982. doi:10.1534/genetics.104.029447
Jumper, J., Evans, R., Pritzel, A., Green, T., Figurnov, M., Ronneberger, O., et al. (2021). Highly Accurate Protein Structure Prediction with AlphaFold. Nature 596, 583–589. doi:10.1038/s41586-021-03819-2
Jurrus, E., Engel, D., Star, K., Monson, K., Brandi, J., Felberg, L. E., et al. (2018). Improvements to the APBS Biomolecular Solvation Software Suite. Protein Sci. 27, 112–128. doi:10.1002/pro.3280
Kim, A. M., Bernhardt, M. L., Kong, B. Y., Ahn, R. W., Vogt, S., Woodruff, T. K., et al. (2011). Zinc Sparks Are Triggered by Fertilization and Facilitate Cell Cycle Resumption in Mammalian Eggs. ACS Chem. Biol. 6, 716–723. doi:10.1021/cb200084y
Kim, J. H., and Chen, E. H. (2019). The Fusogenic Synapse at a Glance. J. Cel Sci. 132, jcs213124. doi:10.1242/JCS.213124
Krauchunas, A. R., Marcello, M. R., and Singson, A. (2016). The Molecular Complexity of Fertilization: Introducing the Concept of a Fertilization Synapse. Mol. Reprod. Dev. 83, 376–386. doi:10.1002/mrd.22634
Liao, M., and Kielian, M. (2005). Domain III from Class II Fusion Proteins Functions as a Dominant-Negative Inhibitor of Virus Membrane Fusion. J. Cel Biol. 171, 111–120. doi:10.1083/jcb.200507075
Liu, Y., Misamore, M. J., and Snell, W. J. (2010). Membrane Fusion Triggers Rapid Degradation of Two Gamete-specific, Fusion-Essential Proteins in a Membrane Block to Polygamy inChlamydomonas. Development 137, 1473–1481. doi:10.1242/dev.044743
Liu, Y., Tewari, R., Ning, J., Blagborough, A. M., Garbom, S., Pei, J., et al. (2008). The Conserved Plant Sterility Gene HAP2 Functions after Attachment of Fusogenic Membranes in Chlamydomonas and Plasmodium Gametes. Genes Dev. 22, 1051–1068. doi:10.1101/gad.1656508
Longo, F. J. (1987). Actin-plasma Membrane Associations in Mouse Eggs and Oocytes. J. Exp. Zool. 243, 299–309. doi:10.1002/jez.1402430215
Longo, F. J. (1985). Fine Structure of the Mammalian Egg Cortex. Am. J. Anat. 174, 303–315. doi:10.1002/aja.1001740310
Misamore, M. J., Gupta, S., and Snell, W. J. (2003). TheChlamydomonasFus1 Protein Is Present on the Mating TypeplusFusion Organelle and Required for a Critical Membrane Adhesion Event during Fusion withminusGametes. MBoC 14, 2530–2542. doi:10.1091/mbc.e02-12-0790
Modis, Y., Ogata, S., Clements, D., and Harrison, S. C. (2004). Structure of the Dengue Virus Envelope Protein after Membrane Fusion. Nature 427, 313–319. doi:10.1038/nature02165
Mohler, W. A., Shemer, G., del Campo, J. J., Valansi, C., Opoku-Serebuoh, E., Scranton, V., et al. (2002). The Type I Membrane Protein EFF-1 Is Essential for Developmental Cell Fusion. Develop. Cel. 2, 355–362. doi:10.1016/s1534-5807(02)00129-6
Mori, M., Yao, T., Mishina, T., Endoh, H., Tanaka, M., Yonezawa, N., et al. (2021). RanGTP and the Actin Cytoskeleton Keep Paternal and Maternal Chromosomes Apart during Fertilization. J. Cel Biol. 220, e202012001. doi:10.1083/jcb.202012001
Mori, T., Igawa, T., Tamiya, G., Miyagishima, S.-y., and Berger, F. (2014). Gamete Attachment Requires GEX2 for Successful Fertilization in Arabidopsis. Curr. Biol. 24, 170–175. doi:10.1016/j.cub.2013.11.030
Mori, T., Kuroiwa, H., Higashiyama, T., and Kuroiwa, T. (2006). Generative Cell Specific 1 Is Essential for Angiosperm Fertilization. Nat. Cel Biol. 8, 64–71. doi:10.1038/ncb1345
Muriel, O., Michon, L., Kukulski, W., and Martin, S. G. (2021). Ultrastructural Plasma Membrane Asymmetries in Tension and Curvature Promote Yeast Cell Fusion. J. Cel Biol. 220, e202103142. doi:10.1083/jcb.202103142
Nayak, V., Dessau, M., Kucera, K., Anthony, K., Ledizet, M., and Modis, Y. (2009). Crystal Structure of Dengue Virus Type 1 Envelope Protein in the Postfusion Conformation and its Implications for Membrane Fusion. J. Virol. 83, 4338–4344. doi:10.1128/jvi.02574-08
Neumann, B., Coakley, S., Giordano-Santini, R., Linton, C., Lee, E. S., Nakagawa, A., et al. (2015). EFF-1-mediated Regenerative Axonal Fusion Requires Components of the Apoptotic Pathway. Nature 517, 219–222. doi:10.1038/nature14102
Nishimura, H., Tajima, T., Comstra, H. S., Gleason, E. J., and L’Hernault, S. W. (2015). The Immunoglobulin-like Gene Spe-45 Acts during Fertilization in Caenorhabditis elegans like the Mouse Izumo1 Gene. Curr. Biol. 25, 3225–3231. doi:10.1016/j.cub.2015.10.056
Okamoto, M., Yamada, L., Fujisaki, Y., Bloomfield, G., Yoshida, K., Kuwayama, H., et al. (2016). Two HAP2-GCS1 Homologs Responsible for Gamete Interactions in the Cellular Slime Mold with Multiple Mating Types: Implication for Common Mechanisms of Sexual Reproduction Shared by Plants and Protozoa and for Male-Female Differentiation. Develop. Biol. 415, 6–13. doi:10.1016/j.ydbio.2016.05.018
Osuna-Ramos, J. F., Reyes-Ruiz, J. M., and del Ángel, R. M. (2018). The Role of Host Cholesterol during Flavivirus Infection. Front. Cel. Infect. Microbiol. 8, 388. doi:10.3389/fcimb.2018.00388
Pattnaik, G. P., and Chakraborty, H. (2021). Cholesterol: A Key Player in Membrane Fusion that Modulates the Efficacy of Fusion Inhibitor Peptides. Vitam. Horm. 117, 133–155. doi:10.1016/bs.vh.2021.06.003
Pérez-Vargas, J., Krey, T., Valansi, C., Avinoam, O., Haouz, A., Jamin, M., et al. (2014). Structural Basis of Eukaryotic Cell-Cell Fusion. Cell 157, 407–419. doi:10.1016/j.cell.2014.02.020
Pinello, J. F., Lai, A. L., Millet, J. K., Cassidy-Hanley, D., Freed, J. H., and Clark, T. G. (2017). Structure-function Studies Link Class II Viral Fusogens with the Ancestral Gamete Fusion Protein HAP2. Curr. Biol. 27, 651–660. doi:10.1016/j.cub.2017.01.049
Pinello, J. F., Liu, Y., and Snell, W. J. (2021). MAR1 Links Membrane Adhesion to Membrane Merger during Cell-Cell Fusion in Chlamydomonas. Develop. Cel. 56, 3380–3392. doi:10.1016/j.devcel.2021.10.023
Podbilewicz, B., Leikina, E., Sapir, A., Valansi, C., Suissa, M., Shemer, G., et al. (2006). The C. elegans Developmental Fusogen EFF-1 Mediates Homotypic Fusion in Heterologous Cells and In Vivo. Develop. Cel. 11, 471–481. doi:10.1016/j.devcel.2006.09.004
Podbilewicz, B. (2014). Virus and Cell Fusion Mechanisms. Annu. Rev. Cel Dev. Biol. 30, 111–139. doi:10.1146/annurev-cellbio-101512-122422
Radzvilavicius, A. L. (2016). Evolutionary Dynamics of Cytoplasmic Segregation and Fusion: Mitochondrial Mixing Facilitated the Evolution of Sex at the Origin of Eukaryotes. J. Theor. Biol. 404, 160–168. doi:10.1016/j.jtbi.2016.05.037
Ramesh, M., Malik, S., and Logsdon, J. (2005). A Phylogenomic Inventory of Meiotic GenesEvidence for Sex in Giardia and an Early Eukaryotic Origin of Meiosis. Curr. Biol. 15, 185–191. doi:10.1016/s0960-9822(05)00028-x
Ravaux, B., Garroum, N., Perez, E., Willaime, H., and Gourier, C. (2016). A Specific Flagellum Beating Mode for Inducing Fusion in Mammalian Fertilization and Kinetics of Sperm Internalization. Sci. Rep. 6 (1 6), 31886–31913. doi:10.1038/srep31886
Rival, C. M., Xu, W., Shankman, L. S., Morioka, S., Arandjelovic, S., Lee, C. S., et al. (2019). Phosphatidylserine on Viable Sperm and Phagocytic Machinery in Oocytes Regulate Mammalian Fertilization. Nat. Commun. 10, 4456. doi:10.1038/S41467-019-12406-Z
Sapir, A., Choi, J., Leikina, E., Avinoam, O., Valansi, C., Chernomordik, L. V., et al. (2007). AFF-1, a FOS-1-Regulated Fusogen, Mediates Fusion of the Anchor Cell in C. elegans. Develop. Cel. 12, 683–698. doi:10.1016/j.devcel.2007.03.003
Satouh, Y., Inoue, N., Ikawa, M., and Okabe, M. (2012). Visualization of the Moment of Mouse Sperm-Egg Fusion and Dynamic Localization of IZUMO1. J. Cel Sci. 125, 4985–4990. doi:10.1242/jcs.100867
Segev, N., Avinoam, O., and Podbilewicz, B. (2018). Fusogens. Curr. Biol. 28, R378–R380. doi:10.1016/j.cub.2018.01.024
Shilagardi, K., Li, S., Luo, F., Marikar, F., Duan, R., Jin, P., et al. (2013). Actin-propelled Invasive Membrane Protrusions Promote Fusogenic Protein Engagement during Cell-Cell Fusion. Science 340, 359–363. doi:10.1126/science.1234781
Smurova, K., and Podbilewicz, B. (2017). Endocytosis Regulates Membrane Localization and Function of the Fusogen EFF-1. Small GTPases 8, 177–180. doi:10.1080/21541248.2016.1211399
Smurova, K., and Podbilewicz, B. (2016). RAB-5- and DYNAMIN-1-Mediated Endocytosis of EFF-1 Fusogen Controls Cell-Cell Fusion. Cel Rep. 14, 1517–1527. doi:10.1016/j.celrep.2016.01.027
Speijer, D., Lukeš, J., and Eliáš, M. (2015). Sex Is a Ubiquitous, Ancient, and Inherent Attribute of Eukaryotic Life. Proc. Natl. Acad. Sci. USA 112, 8827–8834. doi:10.1073/pnas.1501725112
Sprunck, S., Rademacher, S., Vogler, F., Gheyselinck, J., Grossniklaus, U., and Dresselhaus, T. (2012). Egg Cell-Secreted EC1 Triggers Sperm Cell Activation during Double Fertilization. Science 338, 1093–1097. doi:10.1126/science.1223944
Sun, Q.-Y., and Schatten, H. (2006). Regulation of Dynamic Events by Microfilaments during Oocyte Maturation and Fertilization. Reproduction 131, 193–205. doi:10.1530/rep.1.00847
Takayama, J., Tajima, T., Onami, S., and Nishimura, H. (2021). C. elegans Spermatozoa Lacking Spe-45 Are Incapable of Fusing with the Oocyte Plasma Membrane. microPublication Biol. doi:10.17912/micropub.biology.000372
Umashankar, M., Sánchez-San Martín, C., Liao, M., Reilly, B., Guo, A., Taylor, G., et al. (2008). Differential Cholesterol Binding by Class II Fusion Proteins Determines Membrane Fusion Properties. J. Virol. 82, 9245–9253. doi:10.1128/jvi.00975-08
Umeda, R., Satouh, Y., Takemoto, M., Nakada-Nakura, Y., Liu, K., Yokoyama, T., et al. (2020). Structural Insights into Tetraspanin CD9 Function. Nat. Commun. 11 (1 11), 1606–1611. doi:10.1038/s41467-020-15459-7
Valansi, C., Moi, D., Leikina, E., Matveev, E., Graña, M., Chernomordik, L. V., et al. (2017). Arabidopsis HAP2/GCS1 Is a Gamete Fusion Protein Homologous to Somatic and Viral Fusogens. J. Cel Biol. 216, 571–581. doi:10.1083/jcb.201610093
Vance, T. D. R., and Lee, J. E. (2020). Virus and Eukaryote Fusogen Superfamilies. Curr. Biol. 30, R750–R754. doi:10.1016/j.cub.2020.05.029
Vogt, E.-J., Tokuhiro, K., Guo, M., Dale, R., Yang, G., Shin, S.-W., et al. (2019). Anchoring Cortical Granules in the Cortex Ensures Trafficking to the Plasma Membrane for post-fertilization Exocytosis. Nat. Commun. 10, 2271. doi:10.1038/s41467-019-10171-7
von Besser, K., Frank, A. C., Johnson, M. A., and Preuss, D. (2006). Arabidopsis HAP2(GCS1) Is a Sperm-specific Gene Required for Pollen Tube Guidance and Fertilization. Development 133, 4761–4769. doi:10.1242/dev.02683
Voss, J. E., Vaney, M.-C., Duquerroy, S., Vonrhein, C., Girard-Blanc, C., Crublet, E., et al. (2010). Glycoprotein Organization of Chikungunya Virus Particles Revealed by X-ray Crystallography. Nature 468, 709–712. doi:10.1038/nature09555
White, J. M., Delos, S. E., Brecher, M., and Schornberg, K. (2008). Structures and Mechanisms of Viral Membrane Fusion Proteins: Multiple Variations on a Common Theme, 189–219. doi:10.1080/1040923080205832043
Whitlock, J. M., and Chernomordik, L. V. (2021). Flagging Fusion: Phosphatidylserine Signaling in Cell-Cell Fusion. J. Biol. Chem. 296, 100411. doi:10.1016/j.jbc.2021.100411
Wolf, J. P., Feneux, D., Ducot, B., Rodrigues, D., and Jouannet, P. (1995). Influence of Sperm Movement Parameters on Human Sperm-Oolemma Fusion. Reproduction 105, 185–192. doi:10.1530/jrf.0.1050185
Yanagimachi, R. (1988). “Chapter 1 Sperm-Egg Fusion,” in Current Topics In Membranes And Transport. Editor F. Bronner (San Diego, CA: Academic Press), 3–43. doi:10.1016/s0070-2161(08)60129-x
Yang, Y., Zhang, Y., Li, W.-J., Jiang, Y., Zhu, Z., Hu, H., et al. (2017). Spectraplakin Induces Positive Feedback between Fusogens and the Actin Cytoskeleton to Promote Cell-Cell Fusion. Develop. Cel 41, 107–120. doi:10.1016/j.devcel.2017.03.006
Zeev-Ben-Mordehai, T., Vasishtan, D., Siebert, C. A., and Grünewald, K. (2014). The Full-Length Cell-Cell Fusogen EFF-1 Is Monomeric and Upright on the Membrane. Nat. Commun. 5 (1 5), 3912–3919. doi:10.1038/ncomms4912
Zhang, J., Pinello, J. F., Fernández, I., Baquero, E., Fedry, J., Rey, F. A., et al. (2021). Species-specific Gamete Recognition Initiates Fusion-Driving Trimer Formation by Conserved Fusogen HAP2. Nat. Commun. 12 (1 12), 1–12. doi:10.1038/s41467-021-24613-8
Keywords: sperm, oocyte, fusexins, gamete fusion, fertilization, HAP2/GCS1, eff-1, class II viral fusion proteins
Citation: Brukman NG, Li X and Podbilewicz B (2022) Fusexins, HAP2/GCS1 and Evolution of Gamete Fusion. Front. Cell Dev. Biol. 9:824024. doi: 10.3389/fcell.2021.824024
Received: 28 November 2021; Accepted: 21 December 2021;
Published: 10 January 2022.
Edited by:
Amber R. Krauchunas, University of Delaware, United StatesReviewed by:
Jeff Lee, University of Toronto, CanadaCopyright © 2022 Brukman, Li and Podbilewicz. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nicolas G. Brukman, bmJydWttYW5AdGVjaG5pb24uYWMuaWw=; Benjamin Podbilewicz, cG9kYmlsZXdAdGVjaG5pb24uYWMuaWw=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.