- 1Department of Pharmacy, The Second Xiangya Hospital, Central South University, Changsha, China
- 2Institute of Clinical Pharmacy, Central South University, Changsha, China
- 3College of Traditional Chinese Medicine, Guangzhou University of Chinese Medicine, Guangzhou, China
- 4Department of Medical Genetics, The Second Xiangya Hospital, Central South University, Changsha, China
- 5Department of Vascular Surgery, The Second Xiangya Hospital of Central South University, Changsha, China
Bone-related diseases seriously affect the lives of patients and carry a heavy economic burden on society. Treatment methods cannot meet the diverse clinical needs of affected patients. Exosomes participate in the occurrence and development of many diseases through intercellular communication, including bone-related diseases. Studies have shown that exosomes can take-up and “package” non-coding RNAs and “deliver” them to recipient cells, thereby regulating the function of recipient cells. The exosomal non-coding RNAs secreted by osteoblasts, osteoclasts, chondrocytes, and other cells are involved in the regulation of bone-related diseases by inhibiting osteoclasts, enhancing chondrocyte activity and promoting angiogenesis. Here, we summarize the role and therapeutic potential of exosomal non-coding RNAs in the bone-related diseases osteoporosis, osteoarthritis, and bone-fracture healing, and discuss the clinical application of exosomes in patients with bone-related diseases.
Introduction
Bone is one of the most complex tissues in mammals. It undergoes continuous shaping, remodeling and repair throughout its life cycle to protect vital organs and provide rigid support for the entire body (Long and Ornitz, 2013; Riddle and Clemens, 2017). Bone-related diseases are global health problems that seriously affect the quality of life of patients, and include osteoporosis (OP), osteoarthritis (OA), and fracture, etc. (Manolagas, 2013; Liu et al., 2015b). With the aging of the population becoming serious, it is estimated that the global OP and OA patients have reached 200 million and 250 million, respectively (Vos et al., 2012; Weaver et al., 2016).
Bone homeostasis is maintained in two main parts: osteoblast-mediated bone formation and osteoclast-mediated bone resorption. In addition, bone-marrow adipocytes, stromal cells, and osteocytes in the bone microenvironment contribute to the maintenance of bone homeostasis (Lems and Raterman, 2017; Chawalitpong et al., 2018; Zhao et al., 2018). Bone homeostasis also gradually declines with aging during bone remodeling. Enhanced bone resorption or weakened bone formation can lead to disruption of bone homeostasis, which can result in severe bone loss and osteoporotic fractures in older and postmenopausal women (Matsuo and Irie, 2008; Cao, 2011). The moieties involved in osteoblast-mediated bone formation and osteoclast-mediated bone resorption are signaling proteins, and they maintain bone homeostasis. To ensure the integrity and versatility of bones, bone remodeling occurs throughout life (Zhao et al., 2006; Suchacki et al., 2017). It has also been proposed that osteocytes have essential roles in bone remodeling because they affect the activities of osteoblasts and osteoclasts (Tatsumi et al., 2007).
Exosomes are membrane-bound extracellular vesicles produced in the endosomal compartment of most eukaryotic cells. Exosomes have been shown to be important vehicles for communication between bone cells that maintain bone homeostasis (Kalluri, 2016). Recent studies have demonstrated that exosomes secreted by bone marrow mesenchymal stem cells (BMSCs), osteoclasts, and osteoblasts are involved in the regulation of bone metabolism. Osteoblast activity is inhibited by exosomes secreted by osteoclasts, thereby inhibiting the bone formation-activity of osteoblasts (Li et al., 2016; Sun et al., 2016). However, the promotion of osteoblast differentiation is regulated through exosomes secreted by osteoblasts and BMSCs (Cui et al., 2016; Xu and Wang, 2017).
Exosomes contain various biologically active molecules, which can be delivered to target cells through ligand–receptor interactions, endocytosis, direct membrane fusion, or conduction of signaling pathways (Kalamvoki et al., 2014). In particular, non-coding RNAs (ncRNAs) are present in exosomes. Research of exosomal ncRNAs in breast cancer (Naseri et al., 2018), neurologic diseases (Xin et al., 2012), nephropathy (Wang et al., 2019), and other diseases has attracted considerable attention. Different exosomal ncRNAs have different mechanisms in bone-related diseases (Figure 1). They play a vital part in bone remodeling and have an indispensable role in bone-related diseases.
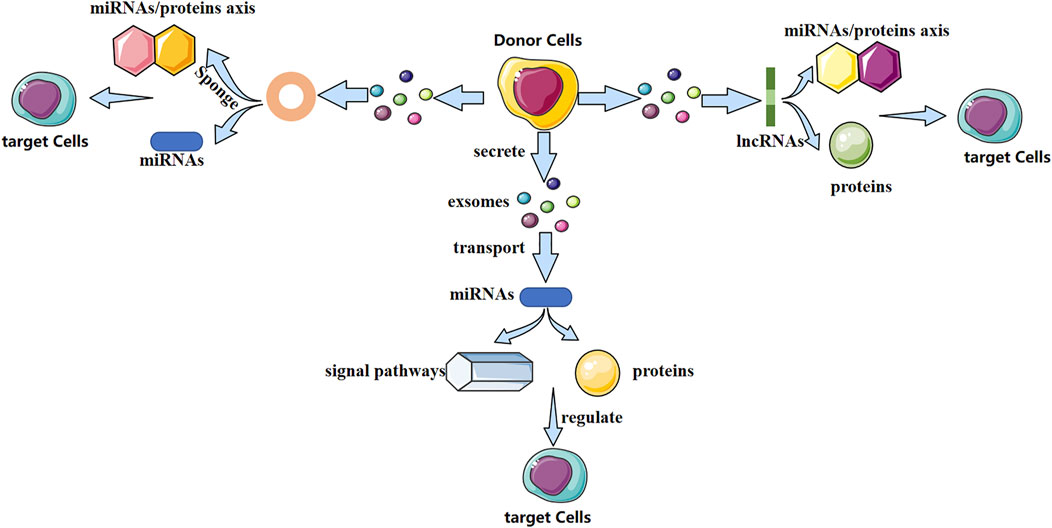
FIGURE 1. Possible mechanism of action of three exosomal non-coding RNAs (schematic). Different exosomal non-coding RNAs (miRNA, lncRNA, and circRNA) regulate the fate and differentiation of target cells through different mechanisms, thereby affecting disease progression.
The purpose of this review is to demonstrate the role of exosomal non-coding RNAs in bone-related diseases and discuss its potential clinical applications. With a view to providing new research directions and ideas for bone-related diseases in the aging process.
Functions and Contents of Exosomes
Exosomes were discovered ∼40 years ago. Their diameter is, in general, between 40 and 150 nm (Pan and Johnstone, 1983; Zhou et al., 2017). Formation of early endosomes is by invagination. Then, under the regulation of endosomal transport complexes and some related proteins, these early endosomes “sprout” and form multivesicular bodies. The latter can release vesicles after fusion with the plasma membrane, and exosomes are formed (Simons and Raposo, 2009; Soekmadji et al., 2013).
An increasing number of cells have been shown to secrete exosomes: MSCs, lymphocytes and macrophages (van Niel et al., 2001; Bourdonnay et al., 2015; Guay et al., 2019). Exosomes mediate intracellular communication through biologically active molecules such as, proteins, long non-coding (lnc)RNAs and microRNAs (miRNAs) that they contain (Thery et al., 2002; Kowal et al., 2014). Exosomes have been studied extensively in the last decade due to their multiple functions in various physiological processes and diseases. Exosomes can participate directly in signal transmission between cells, which is one of the most important factors in paracrine regulation (Dai et al., 2019).
After donor cells secrete exosomes into the extracellular matrix (ECM), exosomes will be recognized and internalized by recipient cells; then exosomes release their contents into recipient cells to regulate cell function (Kalluri and LeBleu, 2020) (Figure 2). Due to their small size, stable structure, low toxicity, and other characteristics, exosomes are being employed increasingly as “nano-medicine carriers” for tissue regeneration and disease treatment (El-Andaloussi et al., 2012; Batrakova and Kim, 2015; Yang et al., 2019a). In general, the separation method is selected based on the experimental principle and source of exosomes. Ultracentrifugation is used as the “gold standard” for separation and purification of exosomes, among which density gradient ultracentrifugation is the best method. The advantages are simple pretreatment, non-requirement of specialist knowledge, and economic affordability (Li et al., 2017). The exosomes prepared by size-exclusion chromatography have high purity, which can preserve the integrity of vesicles and prevent exosome aggregation (Patel et al., 2019). Immunoaffinity isolation, field-flow fractionation, and precipitation are also used in the isolation and extraction of exosomes (Kang et al., 2017; Gurunathan et al., 2019; Zhang and Lyden, 2019).
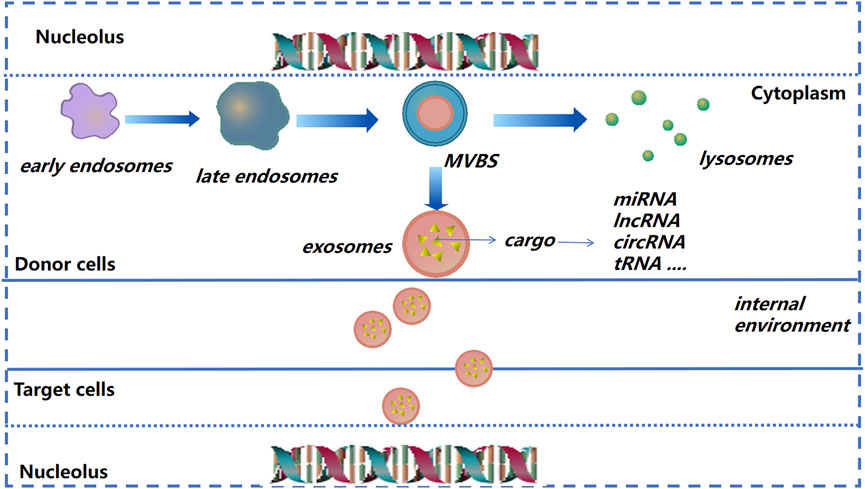
FIGURE 2. Exosome formation and cargo release (schematic). As the endocardium of the early endosome forms a vesicle into the inner bud, an exosome begins to form and then transform into multivesicular bodies. The latter can release vesicles after fusion with the plasma membrane, and are then known as “exosomes.”
Classification and Function of ncRNAs
Mammalian cellular RNAs have been studied for decades (Morris and Mattick, 2014). Advances in sequencing technology have led to the discovery of many ncRNAs. The latter include circular RNAs (circRNAs), miRNAs, lncRNAs, and transfer RNAs (tRNAs) (Sharp, 2009; Ling et al., 2013; St Laurent et al., 2015; Ebbesen et al., 2016). According to the difference in length, ncRNAs can be divided into small ncRNAs and lncRNAs (Knowling and Morris, 2011). Only ∼2% of the human genome is transcribed into proteins, most of the remainder are transcribed into ncRNAs of different sizes and functions, so a large class of RNAs does not code for proteins (Iyer et al., 2015; Gupta and Thum, 2016). ncRNAs participate in several biological processes, including regulating gene expression at the transcriptional level and directing genome rearrangement or DNA synthesis (Cech and Steitz, 2014). The lncRNAs associated with chromatin modification have been described recently. Small RNAs transcribed from promoter regions and bidirectional promoters produce ncRNAs of different sizes (Khalil et al., 2009; Neil et al., 2009; Taft et al., 2009). Under certain conditions, miRNAs expression can be used to accurately identify the origin of poorly differentiated tumors compared with that using protein-encoded messenger (m)RNAs, so ncRNAs could be “ideal” diagnostic markers (Rosenfeld et al., 2008). Besides, studies have shown that ∼200 miRNAs have been characterized sufficiently to enable classification of cancer types. Also, spectral analyses of miRNAs seem to overcome some of the difficulties of early detection associated with colon cancer and other occult cancers (Lu et al., 2005; Aslam et al., 2009; Cortez and Calin, 2009). Such large numbers of ncRNAs with diverse mechanisms constitute a huge and efficient gene-regulatory network, which is involved in many physiological and pathological processes (Ulitsky and Bartel, 2013; Zhang et al., 2019b; Wu et al., 2020).
Roles of ncRNAs in Bone-Related Diseases
Roles of Exosomal miRNAs in Bone-Related Diseases
miRNAs are small coding RNAs derived from a hairpin or double-stranded RNA precursor of length ∼22 nucleotides. miRNAs are generated from introns and exons of protein-encoded and non-coding transcripts by RNA polymerase II (Lee et al., 2003; Landthaler et al., 2004; Mattick and Makunin, 2005). miRNAs are involved in regulating the proliferation, differentiation, and apoptosis of cells, as well as embryonic development (Ambros, 2003; Giraldez et al., 2005; Hatfield et al., 2005; Naguibneva et al., 2006; Plasterk, 2006; Lee et al., 2015). Several studies have demonstrated miRNAs to be involved in the regulation of osteoclasts and osteoblasts. For example, inhibiting miR-31 expression blunts osteoclast formation and bone resorption (Mizoguchi et al., 2013). Besides, miR-140-3p can regulate osteoblast differentiation by targeting the transforming growth factor (TGF)β3 signaling pathway (Fushimi et al., 2018). Exosomal miRNAs from different cells have also been studied extensively in bone-related diseases (Figure 3).
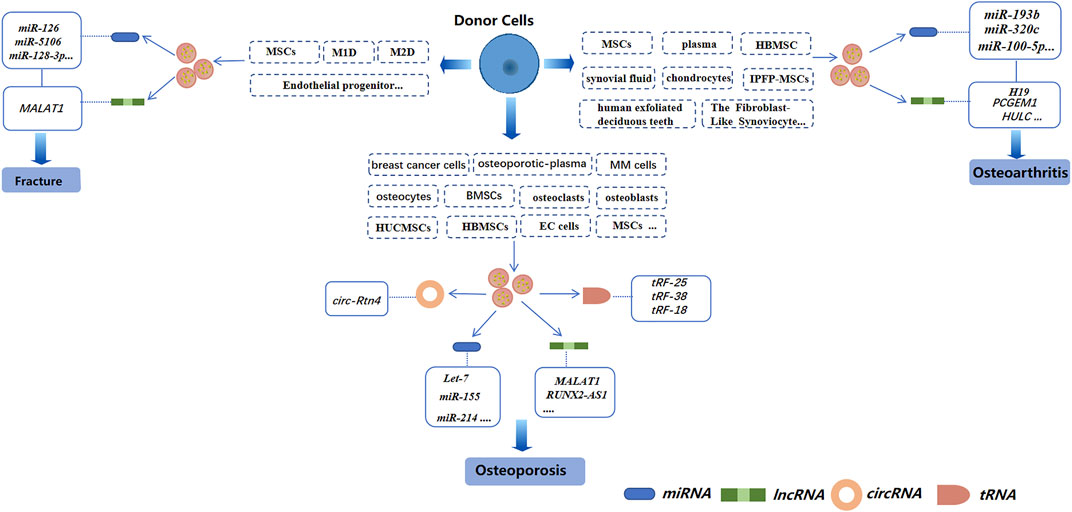
FIGURE 3. Exosomal non-coding RNAs derived from different cells in three bone-related diseases (schematic). Different sources of exosomes, such as BMSCs, serum, and cancer-cell sources, have different effects on osteoporosis, osteoarthritis, and bone fractures by regulating different exosomal non-coding RNAs.
Roles of Exosomal miRNAs in OP
OP is characterized by fragile bones and an increased risk of fracture. OP may be a systemic deterioration of bone mass and bone microstructure due to bone metabolic imbalance (Rachner et al., 2011). OP can lead to a decline in the quality of life of patients, but also bring about a huge economic burden to society (Curtis et al., 2016). miRNAs have been shown to play an important part in OP (Tang et al., 2017) (Table.1).
The pathological process of OP is caused mainly by an imbalance between osteogenic differentiation and osteoclast differentiation. Recent studies have demonstrated that such imbalance can be regulated by exosomal ncRNAs. For instance, Sun and co-workers showed that the exosomes secreted by osteoclasts contain miR-214, which was transferred to osteoblasts through Ephrin-A2/Eph-A2 recognition and inhibited osteoblast function (Sun et al., 2016). Li et al. (2016) showed the function of exosomal miR-214-3p in the communication between osteoclasts and osteoblasts. miR-214-3p can transfer from osteoclasts to osteoblasts and inhibit osteoblastic bone formation. Importantly, targeted inhibition of exosomal miR-214-3p in osteoclasts can reverse the inhibition of osteoblast activity and promote bone formation, which may be a potential treatment for bone loss. Studies have indicated that miR-30d-5p and miR-133b-3p can inhibit osteoblast differentiation by targeting the Runt-related transcription factor 2 (RUNX2) gene, and miR-30d-5p and miR-133b-3p show high expression in osteoblast-derived exosomes (Li et al., 2008; Zhang et al., 2011). Hwang et al. (2014) revealed that miR-140-3p had high expression in osteoblast exosomes. miR-140-3p inhibited the formation of osteoblasts by blunting expression of the bone morphogenetic protein 2 (BMP2) gene. Let-7 was identified in the osteoblast precursors and exosomes of mineralized osteoblasts, which promoted osteogenesis by regulating the high mobility AT-hook 2 gene and axis-like protein (AXIN)2 gene (Egea et al., 2012; Wei et al., 2014). Chen et al. (Chen et al., 2014) identified miR-503-3p in osteoblast-derived exosomes. They demonstrated that miR-503-3p could prevent osteoclast differentiation by inhibiting expression of the receptor activator of nuclear factor-kappa B (RANK) gene. miR-667-3p, miR-6769b-5p, miR-7044-5p, miR-7668-3p, and miR-874-3p which show high expression in the exosomes secreted by mineralized osteoblasts, can promote the osteogenic differentiation of osteoblast precursors by inhibiting AXIN1 expression and enhancing β-catenin expression (Cui et al., 2016). These observations indicate that exosomal miRNAs from osteocytic cells can change the differentiation trend of osteoblasts and clasts, thereby inhibiting or promoting bone formation.
Xu et al. (2014) showed that expression of miR-199b, miR-218, miR-148a and miR-135b in exosomes increased significantly and miR-221 expression was decreased during the early stage of osteogenic differentiation of human bone marrow mesenchymal stem cells (hBMSCs). Besides, miR-199b, miR-218, miR-135b, and miR-148a have been suggested to be regulators in osteoblast differentiation in HBMSCs (Schaap-Oziemlak et al., 2010; Hassan et al., 2012; Cheng et al., 2013; Lauvrak et al., 2013; Xu et al., 2013). Qin et al. (2017) established that, after myostatin treatment, miR-218 expression in osteocyte-derived exosomes was downregulated, which could integrate into osteoblastic cells and inhibit osteoblast differentiation by downregulating the wingless type (Wnt) signaling pathway. Several key miRNAs related to osteogenesis (miR-34a, miR-27a, and miR-22) and adipogenesis (miR-143 and miR-375) have been detected in osteoblasts and adipocyte-derived exosomes, respectively (Narayanan et al., 2018). To study how exosomes promote osteogenic differentiation, Qin et al. (2016) detected miRNAs in exosomes, and found miR-27a, miR-206a, and miR-196a to have high expression. Furthermore, miR-196a had the greatest functional potential. Thus, targeting such exosomal miRNAs may be an ideal treatment strategy for patients with OP.
Rong et al. (Xu et al., 2018) demonstrated that miR-31a-5p could promote osteoclastogenesis and bone resorption, and that its expression in the exosomes of BMSCs of old rats was significantly higher than that in young rats. In addition, the differentiation and function of osteoclasts could be inhibited by blunting expression of miR-31a-5p in exosomes. miR-151-5p from the exosomes of exogenous BMSCs could be spliced into endogenous BMSCs to promote osteogenic differentiation. Bone reduction can be saved by injection of exosomal miR-151-5p in vivo (Chen et al., 2017). Jiang et al. (2018) showed that miR-21 expression in MSC-derived exosomes extracted from healthy adults was significantly lower than that of miR-21 in MSC-derived exosomes extracted from patients with OP.
Yang et al. (2020a) revealed that exosomal miR-1263 derived from human umbilical cord mesenchymal stem cells (hUCMSCs) could inhibit BMSC apoptosis and prevent disuse OP in rats. Guo et al. (2019) showed that miR-20a-5p transported from breast cancer cell-derived exosomes promoted the proliferation and differentiation of osteoclasts by targeting the SRC kinase signaling inhibitor 1 gene. Song et al. (2019) confirmed that blocking the level of exosomal miR-155 secreted by vascular endothelial cells (EC) can reverse the inhibition of osteoclast differentiation of BMMS and further prevent bone resorption, indicating that exosomes miR-155 may have potential to treat osteoporosis. That was the first time that vascular endothelial cells had been found to treat OP. The low yield of exosomes hinders their clinical popularization. Therefore, Fan et al. (2020) suggested “exosome mimetics” as an alternative strategy to generate exosome-associated vesicles with high yields and enhanced regeneration capacity. Recently, Li et al. (2021) showed that that exosomal miR-186 isolated from BMSCs could promote the osteogenesis observed in postmenopausal OP through the hippo-signaling pathway. Hence, the therapeutic effect of exosomal miRNAs in OP has great potential (Figure 4).
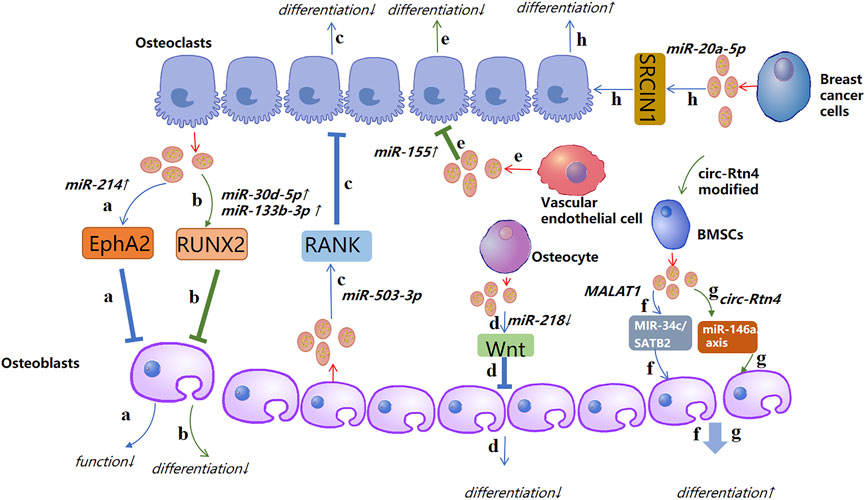
FIGURE 4. A possible mechanism of action of exosomal non-coding RNAs in osteoporosis (schematic). Different sources of exosomal non-coding RNAs, such as miRNA, lncRNA, circRNA, and tRNA, promote or inhibit the activity of osteoblasts and osteoclasts by regulating different targets, thereby affecting the course of osteoporosis. ↑: promotion; ↓: inhibition.
The literature suggests that many miRNAs have high expression in exosomes, and that exosomal miRNAs can promote or inhibit osteogenesis by regulating cellular differentiation through signaling pathways or related proteins, which has huge implications for the clinical treatment of OP. In conclusion, exosomal miRNAs from different sources have important roles in the pathological process and treatment of OP. In addition, exosomal miRNAs may be clinical markers for OP diagnosis.
Roles of Exosomal miRNAs in OA
OA is a degenerative disease of the joints that causes chronic pain, cartilage degeneration, synovitis, disability, and carries an economic burden (Zarb and Carlsson, 1999; Loeser et al., 2012; Berenbaum, 2013; Glyn-Jones et al., 2015). ECM loss and cartilage destruction are the main features of OA (Kobayashi et al., 2005). As a weightbearing joint, the knee joint is a common site for OA. Because of the poor blood supply of cartilage and the weak potential of chondrocyte proliferation/migration, the regeneration ability of articular cartilage is very limited (Yuan et al., 2017). Clinical treatment of OA is aimed at alleviating pain symptoms. If OP progresses, it can be treated only with joint-replacement surgery, but complete repair and regeneration of damaged articular cartilage is not possible (Toh et al., 2017).
In recent years, the role of exosomal miRNAs in OA has been studied (Figure 5A). For example, exosomes derived from hBMSCs overexpressing miR-26a-5p have been shown to delay damage to synovial fibroblasts in vitro and reduce OA damage in vivo. hBMSC-derived exosomes with high expression of miR-26a-5p in OA have been shown to inhibit expression of an inhibitor of synovial-fibroblast damage, prostaglandin-endoperoxide synthase 2, which is very important for OA treatment (Jin et al., 2020b). Sun et al. (2019) suggested that exosomes extracted from hBMSCs with high expression of miR-320c were better than exosomes extracted from control hBMSCs in terms of promoting the proliferation of hBMSC chondrocytes and downregulating matrix metallopeptidase (MMP)13 expression. Studies have shown that TGF-β1 regulates Sp1through MSC-exosomal miR-135b to promote chondrocyte proliferation, thereby promoting cartilage repair (Wang et al., 2018). Mao et al. (2018b) established that miR-92a-3p expression was increased in the exosomes of MSC chondrocytes. Importantly, therapy using exosomal miR-92a-3p from MSCs can promote cartilage proliferation and expression of ECM genes in MSCs. Conversely, by enhancing expression of the Wnt family member 5a gene, therapy using exosomal miR-92a-3p from MSCs inhibits cartilage differentiation and reduces ECM synthesis in cartilage. Those results suggest that exosomal miRNAs derived from MSCs and hBMSCs are involved in regulation of the pathological process of OA.
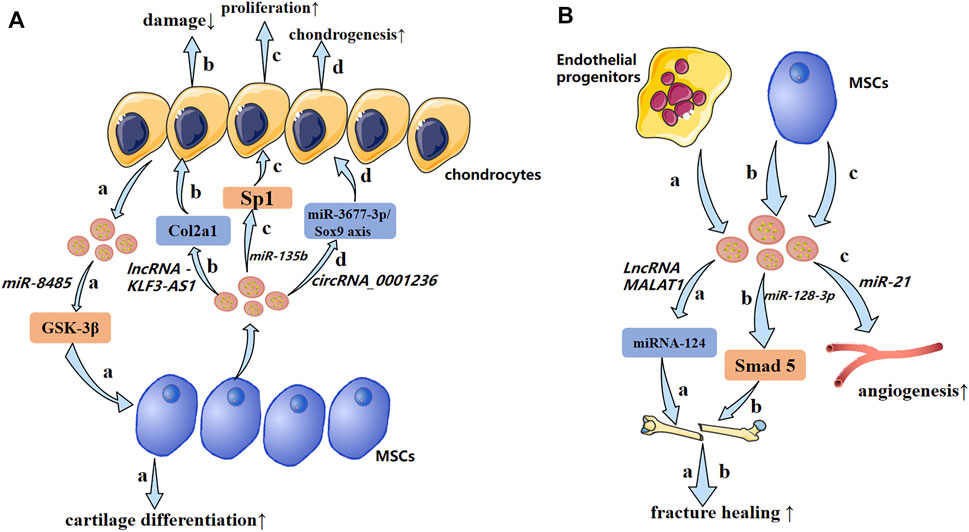
FIGURE 5. A possible mechanism of exosomal non-coding RNAs in osteoarthritis and impaired fracture healing (schematic). (A) Exosomal non-coding RNAs secreted by chondrocytes and MSCs produce pathological effects on each other through different mechanisms. (B) Endothelial progenitors and MSCs promote fracture healing and angiogenesis by transporting lncRNAs and miRNAs, respectively. ↑: promotion; ↓: inhibition.
Levels of exosomal miR-193b in the plasma of OA patients are lower than those in healthy people. miR-193b can target the histone deacetylase (HDAC)3 gene, thereby promoting acetylation of histone H3 and regulating the metabolism of primary human chondrocytes (Lin et al., 2014). Tao et al. (2017) revealed that exosomes derived from human synovial MSCs overexpressing miR-140-5p could promote cartilage regeneration and delay the progression of knee OA. miR-200C appears to have an important role in reducing interleukin (IL)-6-mediated inflammation. In addition, Withrow et al. used polymerase chain reactions (PCRs) to identify “exosomal cargoes.” Examination of exosomes in the synovial fluid of OA patients and non-OA patients showed that miR-200C expression was increased by 2.5-times in the exosomes of OA cases (Rokavec et al., 2012). Scholars have tested if miRNAs have expression differences in the exosomes of IL-1β-stimulated synovial fibroblasts compared with those of synovial fibroblasts of a control group. Fifty miRNAs had differential expression in the exosomes of synovial fibroblasts stimulated by IL-1β. Among them, miR-4454 was related to inflammatory stimulation, and miR-199b was related to cartilage formation (Zhang et al., 2012b; Zhou et al., 2014). Inflammatory factors may be links in stimulating exosomal ncRNAs to regulate OA. HDAC2/8 has been shown to inhibit cartilage development by inhibiting expression of cartilage-specific genes. Exosomes derived from miR-95-5p-overexpressing primary chondrocytes regulate cartilage development by targeting HDAC2/8 directly (Mao et al., 2017; Mao et al., 2018a). Therefore, exosomal miRNAs may be a new direction for targeted therapy of OA.
High expression of miR-100-5p has been detected in stem cells from human exfoliated deciduous teeth-exosomes (SHED-Exos). miR-100-5p targets the 3′ untranslated region of mammalian target of rapamycin (mTOR) directly, and SHED-Exos miR-100-5p inhibits temporomandibular joint (TMJ) inflammation (Luo et al., 2019). miR-100-5p could be a new drug for treatment of TMJ inflammation. Liu et al. (2018a) suggested that expression of miRNAs in the exosomes of subchondral osteoblasts in patients with OA was significantly different compared with that of healthy people. Upregulated miR expression may be involved in the occurrence and progression of OA. Besides, it was worth noting that hsa-miR-4717-5p was the differentially expressed gene with the largest folding changes, which target gene was RGS2. Simultaneously, reverse transcription-quantitative polymerase chain reaction (RT-qPCR) showed that hsa-miR-4717-5p expression in the OA group was higher than that in the control group. Injection of BMSC-derived exosomal miR-9-5p in a mouse model of OA could reduce inflammation and OA-like injury. The main manifestations were downregulation of proinflammatory factors and a reduction in oxidative stress damage. Syndecan 1 was the target gene of miR-9-5p, and its upregulation led to exacerbation of inflammation and OA-like damage, contrary to that observed with exosomal miR-9-5p (Jin et al., 2020a). Exosome-like vesicles from chondrocytes of OA patients were shown to stimulate inflammasome activation and increase production of mature IL-1β by macrophages through the miR-449a-5p/ATG4B/autophagy pathway, thereby aggravating synovitis and accelerating OA progression (Ni et al., 2019). miR-8485 from the exosomes of chondrocytes could stimulate the cartilage differentiation of BMSCs by activating the Wnt/β-catenin pathway. This mechanism was related to inhibition of expression of the glycogen synthase kinase-3β gene by exosomes (Li et al., 2020). Wu et al. (2019) revealed that exosomes from infrapatellar fat-pad MSCs could promote the abnormal gait of OA mice and reduce the pathological changes of articular cartilage in vivo. RNA-sequencing of exosomes demonstrated that miR-100-5p had high expression in exosomes from infrapatellar fat-pad MSCs, and might regulate the biological behavior of chondrocytes by inhibiting the mTOR signaling pathway.
In summary, exosomal miRNAs from different sources are being discovered gradually, and may become a therapeutic method in OA (Table 2). Use of exosomal miRNAs as a means of treatment and diagnosis of OA may be an emerging direction for future clinical research.
Roles of Exosomal miRNAs in Impaired Fracture Healing
A fracture can be complete or partial fracture of bone structure caused by an external force or accumulated strain (Claes et al., 2012; Einhorn and Gerstenfeld, 2015; Zhang et al., 2018a). About 10% of patients with a fracture have delayed healing. Long-term treatment can bring physical and psychological discomfort to patients, unnecessary financial burdens to patients and their families as well as poor quality of life (Komatsu and Warden, 2010; Gomez-Barrena et al., 2015). Fracture healing is a complex process. The activation, proliferation, and differentiation of local MSCs or progenitor cells are affected by specific growth factors and cytokine cascades (Murata et al., 2014).
Researchers have focused gradually on the effect of exosomal ncRNAs on fracture healing (Figure 5B). Liu et al. (2020) demonstrated that Hypo-Exos (exosomes derived from MSCs under hypoxia) promotes angiogenesis, proliferation, and migration by transporting exosomal miR-126, thereby accelerating fracture healing. Meanwhile knockout of hypoxia-inducible factor (HIF)-1α expression resulted in a significant reduction of MSC-derived exosomal miR-126, thereby eliminating the influence of exosomes derived from MSCs under hypoxia, and suggesting that hypoxic pretreatment mediated the production of exosomal miR-126 by activating HIF-1α. Exosomal miR-128-3p of MSCs in old rats can regulate bone formation and fracture healing by targeting Smad 5. For older people, a small synthetic RNA (exosomal miR-128-3P) may be a promising strategy for fracture healing (Xu et al., 2020b). Xiong et al. (2020) revealed that miR-5106 was significantly overexpressed in M2 macrophage-derived exosomes (M2D-Exos), whereas expression was decreased in M1 macrophage-derived exosomes (M1D-Exos). They suggested that exosomal miR-5106 could target salt-induced kinase (SIK2) and SIK3 genes directly to induce osteogenic differentiation of BMSCs. More importantly, local injections of miR-5106 agonists or M2D-Exos at fracture sites could accelerate healing in vivo. Studies have shown that MSC-derived promotion of fracture healing is related to exosomal miRNAs. Differentially expressed miRNAs such as miR-21, miR-4532, miR-125b-5p, and miR-338-3p may help to enhance bone formation and angiogenesis (Furuta et al., 2016) (Table 3).
Compared with the study of exosomal miRNAs in OP and OA, the study of exosomal miRNAs in fracture healing is less deep. Recent research on exosomal miRNAs in fractures has focused mainly on bone formation and angiogenesis.
Roles of Exosomal lncRNAs in Bone-Related Diseases
lncRNAs are a family of transcripts containing >200 nucleotides that do not encode proteins (Costa, 2010; Moran et al., 2012; Geisler and Coller, 2013; Ulitsky and Bartel, 2013). Due to their different positions relative to protein-coding genes, they can be divided into five categories: antisense; long intergenic non-coding RNAs (lincRNAs); sense-overlapping; sense intronic; processed transcript (Quinn and Chang, 2016; Huynh et al., 2017). lncRNAs have been shown to be involved in nuclear structure and gene expression as regulatory factors during development. They are involved in regulation of the cell cycle, differentiation, transcription, and translation (Hu et al., 2012; Rinn and Chang, 2012; Joh et al., 2014; Quinodoz and Guttman, 2014; Vance and Ponting, 2014). lncRNAs are essential for bone formation. For example, targeted destruction of an lncRNA called HOX antisense intergenic RNA led to metacarpal deformities and allogeneic transformation of the spine. Furthermore, lncRNAs are regulators in the osteogenesis process of MSCs (Li et al., 2013b; Gu et al., 2017; Tye et al., 2018). It has been reported that lncRNAs are related to the progression of some diseases, and are being studied as new therapeutic targets (Gutschner and Diederichs, 2012; Li et al., 2013a; Wahlestedt, 2013; Hrdlickova et al., 2014).
Roles of Exosomal lncRNAs in OP
Compared with the research of exosomal miRNAs in OP, the research of exosomal lncRNAs in OP has not made much progress. Yang et al. (2019) demonstrated that a BMSC-derived gene in exosomes, metastasis-associated lung adenocarcinoma transcript (MALAT)1, promoted osteoblast activity. In addition, the in vivo experimental results of an ovariectomized mouse model showed that miR-34c reversed the effect of MALAT1, and that special AT-rich sequence-binding protein 2 reversed the effect of miR-34c in ovariectomized mice. Multiple myeloma is characterized by the reduced osteogenic potential of MSCs. Li et al. (2018) found that exosomal lncRNA RUNX2-AS1 from myeloma cells could be delivered to MSCs, thereby inhibiting the osteogenicity of MSCs. Their results suggested that osteogenic differentiation from multiple myeloma cells to MSCs was through a unique exosomal lncRNA RUNX2-AS1/RUNX2 pathway.
RUNX2-AS1 in exosomal lncRNAs may be a potential therapeutic target for the bone injury caused by multiple myeloma. Xu et al. (2020a) showed that the lncRNAs of exosomes secreted by osteoclasts affect osteogenesis during granule-induced osteolysis. They demonstrated that the exosomes of RAW264.7 cells induced by titanium particles repressed the osteogenic differentiation of MC3T3-E1 cells. According to analyses from the Gene Ontology database, Kyoto Encyclopedia of Genes and Genomes database and verification by RT-qPCR, they identified two candidate lncRNAs: NONMMUT000375.2 and NONMMUT071578. These two lncRNAs regulated expression of four important genes related to osteoclast differentiation: B-cell lymphoma 2, Wnt11, TGFβ, and 3-phosphoinositide-dependent protein kinase-1. Exosomal lncRNA H19 isolated from BMSCs has been shown to promote osteogenesis through the Angpt1/Tie2-NO signaling pathway in CBS heterozygous mice, thereby alleviating OP (Behera et al., 2021). Whether exosomal lncRNA H19 can promote osteogenesis in normal mice may become a new research direction.
Recent research has shown that exosomal lncRNAs also regulate the differentiation direction of cells through related proteins and signaling pathways, thereby affecting osteogenesis. Study of exosomal lncRNAs in bone-related diseases is an emerging research direction, but faces practical problems, such as the screening and isolation of exosomal lncRNAs.
Roles of Exosomal lncRNAs in OA
Scholars have hypothesized that selective packaging of ncRNA into exosomes could reflect the cellular response to cartilage-cell death during OA pathogenesis. Song et al. (2017) showed that expression of exosomal hepatocellular carcinoma upregulated long non-coding RNA (HULC) in OA patients was downregulated significantly, whereas exosomal miR-372-3p expression in OA patients was upregulated significantly. Besides, HULC overexpression in normal chondrocytes significantly promoted the apoptosis and inhibited the proliferation of cells. Zhao and Xu (2018) suggested that in synovial-fluid samples, exosome expression in the control group was significantly lower than that in patients with early OA or late OA. Furthermore, expression of the exosomal lncRNA prostate cancer gene expression marker (PCGEM)1 gene in late OA was significantly higher than that in early OA. And the early expression of OA was significantly higher than that of the control group. Therefore, they concluded that exosomal lncRNA PCGEM1 may be a novel indicator to distinguish early OA from late OA.
Exosomes derived from KLF3-AS1-overexpressing-MSCs (MSCKLF3-AS1-Exos) have been shown to participate in MSC-Exos-mediated induction of chondrocyte proliferation via the miR-206/G-protein-coupled receptor kinase interacting protein (GIT)1 axis. KLF3-AS1 promoted GIT1 expression by “sponging” miR-206 as a competitive endogenous RNA. In addition, MSCKLF3-AS1-Exos alleviated the chondrocyte damage induced by IL-1β (Zhang et al., 2015; Liu et al., 2018b). Liu et al. (2018c) demonstrated that lncRNA KLF3-AS1 expression was upregulated significantly in MSC-Exos, and that exosomal KLF3-AS1 inhibited IL-1β-induced chondrocyte apoptosis by upregulating expression of Col2a1 and aggrecan, and downregulating expression of MMP13 and Runx2. Exosomal KLF3-AS1 could also promote cartilage repair in a rat model of OA. Tan et al. (2020) reported that exosomal lncRNA H19 may be a therapeutic target for OA because it promotes the proliferation and migration of chondrocytes by targeting the miR-106b-5p/tissue inhibitor of metalloproteinases 2 axis and inhibits ECM degradation in OA.
Roles of Exosomal lncRNAs in Impaired Fracture Healing
Only one study has been done on the role of exosomal lncRNAs in impaired fracture healing. The reason why few studies on exosomal lncRNAs in bone-related diseases have been done may be that fewer lncRNAs are carried in exosomes, or that lncRNAs have only slight effects on bone-related diseases.
Bone repair involves two main features: 1) bone resorption caused by osteoclastogenesis; 2) endothelial progenitor cells stimulate neovascularization and osteogenic differentiation. Cui et al. (2019) showed that endothelial progenitor cell-derived exosomal MALAT1 was bound directly to miR-124. This action reduced inhibition of integrin subunit β1 and promoted the migration and osteoclast differentiation of bone marrow-derived macrophages. This phenomenon stimulated the recruitment of osteoclast precursor cells and differentiation, which led to bone repair.
Roles of Other Exosomal lncRNAs in Bone-Related Diseases
Roles of Exosomal circRNAs in OP
circRNAs are endogenous RNAs with covalent closed-loop structures. Neither 5–3 ends nor poly (A) tails are present mainly in cytoplasm. They are produced during RNA splicing and are generated by exons, introns or a combination of both (Owens et al., 2012; Li et al., 2015; Lasda and Parker, 2016). circRNAs are regarded as significant regulators of cell function, rather than a non-functional byproduct of abnormal RNA splicing (Salzman, 2016). Yang et al. (2020b) reviewed the development of circRNAs in OP. They showed that circRNAs act by targeting the major genes and signaling pathways related to osteoblast differentiation and osteoclast differentiation. For example, circRNA_0016624 has been reported to activate miR-98 and enhance expression of BMP2 (which plays an important part in induction of osteogenic differentiation), so circRNA_0016624 promoted osteoblast differentiation. However, the role of exosomal circRNAs in OP has not been explored thoroughly.
Cao et al. (2020) demonstrated that co-cultivation with exosomes derived from circ-Rtn4 modified BMSCs (Rtn4-Exos) could reduce the cytotoxicity and apoptosis of mouse cells (MC3T3-E1) induced by the tumor necrosis factor (TNF)-α gene, which was manifested by expression of caspase-3, caspase-3, and Bax protein as well as a reduction in caspase-3 activity. In particular, Rtn4-Exos exhibited its function in TNF-α-treated MC3T3-E1 cells by sponging miR-146a, which is the target of circ-RTN4. Thus, their findings indicated that Rtn4-Exos suppressed the cytotoxicity and apoptosis of MC3T3-E1 cells induced by TNF-α through sponging of miR-146a. Hence, Rtn4-Exos could be considered as a new drug candidate for OP treatment. Exosomal hsa_circ_0006859 isolated from the serum of postmenopausal women has been revealed to be a potential biomarker of postmenopausal OP. It enhances the adipogenic and osteogenic differentiation of hBMSCs by sponging miR-431-5p (Zhi et al., 2021).
Few studies have been carried out on exosomal circRNAs. However, exosomal circRNAs seem to influence the fate and differentiation of target cells through sponging miRNAs.
Roles of Exosomal circRNAs in OA
Zhu et al. (2021) reported that circ_0001846 has high expression in the exosomes of an IL-1β-treated human chondrocyte cell line (CHON-001). They showed that transfer of exosomal circ_0001846 regulates IL-1β -induced chondrocyte damage through the miR-149-5p/Wnt5B axis. Guo et al. (2021) demonstrated that exosomal circ-BRWD1 isolated from CHON-001 cells can affect the progression of CHON-001 cells induced by IL-1β by regulating the miR-1277/TRAF6 axis, thereby promoting OA development. Mao et al. (2021) showed that exosomal circRNA_0001236 transported by MSCs promotes expression of cartilage-specific genes and proteins via the miR-3677-3p/SRY-box transcription factor (SOX) 9 axis. Thus, exosomes overexpressing circRNA_0001236 mitigate cartilage degradation, inhibit osteoarthritis progression, and enhance cartilage repair.
Those results suggest the mechanism of action of exosomal circRNAs in OA. They provide a new direction for the study of exosomal circRNAs in OA and other bone-related diseases.
Roles of Exosomal tRNAs in OP
The nucleotide information on the mRNA is associated with the amino acid sequence tRNA by decoding the nucleotide triad to help the ribosome synthesize proteins. Besides this routine function, tRNAs are involved in the signal transduction, survival, and apoptosis of cells, metabolism of amino acids and porphyrins and stress responses (Giege, 2008; Phizicky and Hopper, 2010; Raina and Ibba, 2014). Recent studies have shown that tRNA-derived fragments (tRFs) are involved in post-transcriptional regulation and could be considered as therapeutic targets for certain diseases (Kumar et al., 2016).
The tRFs derived from tRNA or pre-tRNA are lncRNAs (Zhang et al., 2019a). Zhang et al. (2018b) found that, compared with those in healthy controls, there were 11 upregulated tRFs and 18 downregulated tRFs in exosomes in the plasma of OP patients. Among them, expression of exosomal tRF-25-R9ODMJ6B26 (tRF-25), tRF-38-QB1MK8YUBS68BFD2 (tRF-38) and tRF-18-BS68BFD2 (tRF-18) was increased significantly. In addition, expression of tRF-25, tRF-38, and tRF-18 in the plasma of OP patients had good accuracy for OP diagnosis, and could be used as diagnostic biomarkers for OP.
In addition to the exosomal ncRNAs reviewed above, exosomal mRNAs have a role in OP. Seven genes associated with mRNAs have been detected in the exosomes of differentiated hBMSCs: ACIN1, DDX6, DGKA, DKK2, Lsm2, RPS2, and Xsox17. They showed obvious differential expression and could induce differentiation into mineralized bone cells (Xu et al., 2014).
Clinical Potential of Exosomal ncRNAs in Bone-Related Diseases
Sun et al. (2016) incubated exosomes isolated from OP patients and from the serum of healthy controls with osteoblasts. They showed that miR-214 expression was much higher upon incubation with exosomes from hFOB1.19 cells than that in non-OP individuals. Simultaneously, mRNA expression also decreased in osteogenesis-related genes. Those results suggest the clinical importance of increased miR-214 expression in the serum of OP patients, and suggest its potential use as a marker for the diagnosis and therapy of OP. Gao et al. (2019) reported that exosomal miR-320 isolated from leukemia cells could be taken up by BMSCs and bind to heterogeneous nuclear ribonucleoprotein A1 to inhibit osteogenic differentiation. Hence, heterogeneous nuclear ribonucleoprotein A1-mediated transfer of exosomal miR-320 from leukemia cells to BMSCs could be an important mediator of leukemia progression and a potential therapeutic target for chronic myelogenous leukemia.
Expression of exosomal lncRNA plasmacytoma variant translocation-1 in the serum of OA patients has been shown by Meng et al. (2020) to reduce progression of lipopolysaccharide-induced OA by mediating the high-mobility group protein 1/toll-like receptor 4/nuclear factor-kappa B pathway through miR-935p. Liang et al. (2020) reported that chondrocyte-affinity peptides in exosomes reduced OA progression in a rat model by delivering miR-140 to the deep cartilage area and inhibiting cartilage degradation, which points to organelle-based acellular OA treatment.
Apart from OP, OA, and bone fractures, we also found meaningful clinical studies of exosomal ncRNAs in other bone-related diseases. For example, Fang et al. (2020) demonstrated that, compared with healthy individuals, expression of tRNA-derived small RNA (tsRNA)-10277 in the exosomes of plasma of patients with steroid-induced osteonecrosis of the femoral head (SONFH) was downregulated significantly. Also, exosomes in BMSCs loaded with tsRNA-10277 enhanced the osteogenic differentiation of dexamethasone-induced BMSCs. That study provides new ideas for the osteogenic effects of exosomes in BMSCs carrying specific tsRNAs on SONFH. Besides, there is evidence that exosomal lncRNA nuclear enriched abundant transcript (NEAT)1 isolated from the serum of patients with rheumatoid arthritis can promote the development of rheumatoid arthritis through the miR-144-3p/ROCK2 axis (Liu et al., 2021). This NEAT1/miR-144–3p/ROCK2 regulatory pathway may become a new treatment target for rheumatoid arthritis.
Future Perspectives
Bone-related diseases not only affect quality of life, but also threaten life in severe cases. Accumulating evidence suggests that ncRNAs with regulatory effects are essential for the pathogenesis of bone-related diseases.
From the research of exosomes and ncRNAs, some common points can be found in the research of bone-related diseases. For example, exosomal miR-155 can inhibit osteoclast activity (Song et al., 2019) and miR-155 expression is upregulated significantly in osteoclasts (Zhang et al., 2012a; Zhao et al., 2017). In addition, reduced expression of miR-155 can target the gene for the leptin receptor and increase its expression through the 5′ adenosine monophosphate-activated protein kinase signaling pathway which, ultimately, represses osteoclast activation and bone resorption of osteoclasts in alendronate-treated osteoporotic mice (Mao et al., 2019). This commonality is not accidental, and merits in-depth exploration. Liu et al. (2015a) demonstrated that BMSC transplantation can rescue bone loss in Fas-deficient MRL/LPR mice by secreting exosomes. This is achieved by reducing miR-29b expression, thereby enhancing osteogenic differentiation in vitro and promoting bone formation in vivo. Petho et al. (2018) reported that miR-677-3p, miR-680, miR-3084-3p and miR-5000 had high expression in the exosomes of mineralized osteoblasts. Furthermore, Kumar et al. (2017) reviewed application of miRNAs as peripheral biomarkers in aging and age-related diseases, including OP.
Research on exosomal ncRNAs and OA has also shown a certain connection. The chondrocytes were treated with channel protein connexin 43 (Cx43) exosomes released by OA chondrocytes, osteocytes and synovial cells eventually increased cell senescence levels and senescence-associated secretory phenotypes through p53/p16 and NF-kß (Varela-Eirín et al., 2019). In general, lncRNA PCGEM1 in the synoviocytes of OA patients targets miR-770 to stimulate synoviocyte proliferation by acting as a spongy lncRNA (Kang et al., 2016). M2 macrophages polarized by lncRNA MM2P significantly strengthen chondrocyte function and promote the delivery of M2-derived exosomal SOX9 to chondrocytes (Bai et al., 2020). Future studies can explore if these lncRNAs are carried in exosomes and are involved in OA regulation. Zavatti et al. (2020) revealed the efficacy of exosomes in human amniotic-fluid SCs against cartilage damage, indicating a positive correlation with their TGF-β content. circRNAs are relatively uncharted territory in OA. Zhou et al. (2019b) established that, through sponging miR-127-5p, circRNA.33186 encouraged OA pathogenesis. MSC- and BMSC-derived exosomes make a great contribution to fracture healing, but the mechanism of action needs further exploration (Furuta et al., 2016; Hao et al., 2017). Besides, hUCMSC-derived exosomes can repair bone fractures in rats mainly through the Wnt signaling pathway (Zhou et al., 2019a). Most of the exosomes that can be used to treat bone-related diseases come from SCs. Hao et al. (2017) reviewed a promising strategy for SC-derived exosomes to heal fractures. However, Borel et al. (2020) found that prostate cancer-derived exosomes could promote the differentiation and activity of osteoblasts through the phospholipase-D2 pathway. That discovery broadens investigation of the mechanism of action of exosomes in bone-related diseases. Also, treatment of rheumatoid arthritis with exosomal ncRNAs shows great prospects for development (Ding et al., 2020; Meng and Qiu, 2020).
Although studies have shown that exosomal ncRNAs are emerging diagnostic markers and disease targets, the transformation from basic science to clinical application will be challenging. Looking for commonality between bone-related diseases may aid their treatment using exosomes. Certainly, investigation of the connection points between miR-155 in exosomes and osteoclasts could yield interesting results.
Conclusion
Study of exosomal ncRNAs in OP is more in-depth than that in OA and impaired fracture healing. As important cell communicators, exosomes (and their contents) have indispensable roles in the occurrence, development, and treatment of OP, OA, and fracture. Exploring the detailed mechanism of action of exosomal ncRNAs in bone-related diseases will help transformation from basic research to their clinical application in bone-related diseases.
Author Contributions
QL and HHe conceived this review. HL drafted the manuscript. QZ and XX designed the figures. JW and HZ designed the tables. HHu revised the manuscript. All authors were involved in critical revision of the manuscript and approved the final version for submission.
Funding
This work was supported by the Fundamental Research Funds for the Central Universities of Central South University (No. 2021zzts1074), China National Key R&D Program (Nos 2020YFC2009000 and 2020YFC2009001), Natural Science Foundation of Changsha (KQ2007051), grants awarded to QL by the Hunan Provincial Department of Finance Grant (2020-83, 2019-93 and 2018-92), Hunan Provincial Development and Reform Commission of Innovastive Research Program (2021-212-23), Hunan Innovative Province Construction Special Project (2021ZK4025), National Science and Technology Major Project (2020ZX09201-28), and Natural Science Foundation of Hunan Province (2020JJ4799).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The handling editor declared a shared affiliation with the authors.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
OP, osteoporosis; OA, osteoarthritis; BMSCs, bone marrow mesenchymal stem cells; lncRNA, long non-codingRNA; miRNA, microRNA; tRNAs, transfer RNA; circRNAs, circular RNA; hBMSCs, human bone marrow mesenchymal stem cells; hUCMSCs, human umbilical cord mesenchymal stem cells; ECM, extracellular Matrix; SHED, human exfoliated deciduous teeth; TMJ, temporomandibular joint; SIK2, salt-induced kinase 2 and 3; GIT1, G-protein-coupled receptor kinase interacting protein 1.
References
Ambros, V. (2003). MicroRNA Pathways in Flies and Worms. Cell 113 (6), 673–676. doi:10.1016/s0092-8674(03)00428-8
Aslam, M. I., Taylor, K., Pringle, J. H., and Jameson, J. S. (2009). MicroRNAs Are Novel Biomarkers of Colorectal Cancer. Br. J. Surg. 96 (7), 702–710. doi:10.1002/bjs.6628
Bai, J., Zhang, Y., Zheng, X., Huang, M., Cheng, W., Shan, H., et al. (2020). LncRNA MM2P-Induced, Exosome-Mediated Transfer of Sox9 from Monocyte-Derived Cells Modulates Primary Chondrocytes. Cell Death Dis 11 (9), 763. doi:10.1038/s41419-020-02945-5
Batrakova, E. V., and Kim, M. S. (2015). Using Exosomes, Naturally-Equipped Nanocarriers, for Drug Delivery. J. Controlled Release 219, 396–405. doi:10.1016/j.jconrel.2015.07.030
Behera, J., Kumar, A., Voor, M. J., and Tyagi, N. (2021). Exosomal lncRNA-H19 Promotes Osteogenesis and Angiogenesis through Mediating Angpt1/Tie2-NO Signaling in CBS-Heterozygous Mice. Theranostics 11 (16), 7715–7734. doi:10.7150/thno.58410
Berenbaum, F. (2013). Osteoarthritis as an Inflammatory Disease (Osteoarthritis Is Not Osteoarthrosis!). Osteoarthritis and Cartilage 21 (1), 16–21. doi:10.1016/j.joca.2012.11.012
Borel, M., Lollo, G., Magne, D., Buchet, R., Brizuela, L., and Mebarek, S. (2020). Prostate Cancer-Derived Exosomes Promote Osteoblast Differentiation and Activity through Phospholipase D2. Biochim. Biophys. Acta (Bba) - Mol. Basis Dis. 1866 (12), 165919. doi:10.1016/j.bbadis.2020.165919
Bourdonnay, E., Zasłona, Z., Penke, L. R. K., Speth, J. M., Schneider, D. J., Przybranowski, S., et al. (2015). Transcellular Delivery of Vesicular SOCS Proteins from Macrophages to Epithelial Cells Blunts Inflammatory Signaling. J. Exp. Med. 212 (5), 729–742. doi:10.1084/jem.20141675
Cao, G., Meng, X., Han, X., and Li, J. (2020). Exosomes Derived from circRNA Rtn4-Modified BMSCs Attenuate TNF-α-Induced Cytotoxicity and Apoptosis in Murine MC3T3-E1 Cells by Sponging miR-146a. Biosci. Rep. 40 (5). doi:10.1042/BSR20193436
Cao, X. (2011). Targeting Osteoclast-Osteoblast Communication. Nat. Med. 17 (11), 1344–1346. doi:10.1038/nm.2499
Cech, T. R., and Steitz, J. A. (2014). The Noncoding RNA Revolution-Trashing Old Rules to Forge New Ones. Cell 157 (1), 77–94. doi:10.1016/j.cell.2014.03.008
Chawalitpong, S., Chokchaisiri, R., Suksamrarn, A., Katayama, S., Mitani, T., Nakamura, S., et al. (2018). Cyperenoic Acid Suppresses Osteoclast Differentiation and Delays Bone Loss in a Senile Osteoporosis Mouse Model by Inhibiting Non-canonical NF-Κb Pathway. Sci. Rep. 8 (1), 5625. doi:10.1038/s41598-018-23912-3
Chen, C., Cheng, P., Xie, H., Zhou, H.-D., Wu, X.-P., Liao, E.-Y., et al. (2014). MiR-503 Regulates Osteoclastogenesis via Targeting RANK. J. Bone Miner Res. 29 (2), 338–347. doi:10.1002/jbmr.2032
Chen, C., Wang, D., Moshaverinia, A., Liu, D., Kou, X., Yu, W., et al. (2017). Mesenchymal Stem Cell Transplantation in Tight-Skin Mice Identifies miR-151-5p as a Therapeutic Target for Systemic Sclerosis. Cell Res 27 (4), 559–577. doi:10.1038/cr.2017.11
Cheng, P., Chen, C., He, H.-B., Hu, R., Zhou, H.-D., Xie, H., et al. (2013). miR-148a Regulates Osteoclastogenesis by Targeting V-Maf Musculoaponeurotic Fibrosarcoma Oncogene Homolog B. J. Bone Miner Res. 28 (5), 1180–1190. doi:10.1002/jbmr.1845
Claes, L., Recknagel, S., and Ignatius, A. (2012). Fracture Healing under Healthy and Inflammatory Conditions. Nat. Rev. Rheumatol. 8 (3), 133–143. doi:10.1038/nrrheum.2012.1
Cortez, M. A., and Calin, G. A. (2009). MicroRNA Identification in Plasma and Serum: a New Tool to Diagnose and Monitor Diseases. Expert Opin. Biol. Ther. 9 (6), 703–711. doi:10.1517/14712590902932889
Costa, F. F. (2010). Non-coding RNAs: Meet Thy Masters. Bioessays 32 (7), 599–608. doi:10.1002/bies.200900112
Cui, Y., Fu, S., Sun, D., Xing, J., Hou, T., and Wu, X. (2019). EPC‐derived Exosomes Promote Osteoclastogenesis through LncRNA‐MALAT1. J. Cel Mol Med 23 (6), 3843–3854. doi:10.1111/jcmm.14228
Cui, Y., Luan, J., Li, H., Zhou, X., and Han, J. (2016). Exosomes Derived from Mineralizing Osteoblasts Promote ST2 Cell Osteogenic Differentiation by Alteration of microRNA Expression. FEBS Lett. 590 (1), 185–192. doi:10.1002/1873-3468.12024
Curtis, E. M., van der Velde, R., Moon, R. J., van den Bergh, J. P. W., Geusens, P., de Vries, F., et al. (2016). Epidemiology of Fractures in the United Kingdom 1988-2012: Variation with Age, Sex, Geography, Ethnicity and Socioeconomic Status. Bone 87, 19–26. doi:10.1016/j.bone.2016.03.006
Dai, J., Escara-Wilke, J., Keller, J. M., Jung, Y., Taichman, R. S., Pienta, K. J., et al. (2019). Primary Prostate Cancer Educates Bone Stroma through Exosomal Pyruvate Kinase M2 to Promote Bone Metastasis. J. Exp. Med. 216 (12), 2883–2899. doi:10.1084/jem.20190158
Ding, Y., Wang, L., Wu, H., Zhao, Q., and Wu, S. (2020). Exosomes Derived from Synovial Fibroblasts under Hypoxia Aggravate Rheumatoid Arthritis by Regulating Treg/Th17 Balance. Exp. Biol. Med. (Maywood) 245 (14), 1177–1186. doi:10.1177/1535370220934736
Ebbesen, K. K., Kjems, J., and Hansen, T. B. (2016). Circular RNAs: Identification, Biogenesis and Function. Biochim. Biophys. Acta (Bba) - Gene Regul. Mech. 1859 (1), 163–168. doi:10.1016/j.bbagrm.2015.07.007
Egea, V., Zahler, S., Rieth, N., Neth, P., Popp, T., Kehe, K., et al. (2012). Tissue Inhibitor of Metalloproteinase-1 (TIMP-1) Regulates Mesenchymal Stem Cells through Let-7f microRNA and Wnt/-catenin Signaling. Proc. Natl. Acad. Sci. 109 (6), E309–E316. doi:10.1073/pnas.1115083109
Einhorn, T. A., and Gerstenfeld, L. C. (2015). Fracture Healing: Mechanisms and Interventions. Nat. Rev. Rheumatol. 11 (1), 45–54. doi:10.1038/nrrheum.2014.164
El-Andaloussi, S., Lee, Y., Lakhal-Littleton, S., Li, J., Seow, Y., Gardiner, C., et al. (2012). Exosome-mediated Delivery of siRNA In Vitro and In Vivo. Nat. Protoc. 7 (12), 2112–2126. doi:10.1038/nprot.2012.131
Fan, J., Lee, C.-S., Kim, S., Chen, C., Aghaloo, T., and Lee, M. (2020). Generation of Small RNA-Modulated Exosome Mimetics for Bone Regeneration. ACS Nano 14 (9), 11973–11984. doi:10.1021/acsnano.0c05122
Fang, S., He, T., Jiang, J., Li, Y., and Chen, P. (2020). Osteogenic Effect of tsRNA-10277-Loaded Exosome Derived from Bone Mesenchymal Stem Cells on Steroid-Induced Osteonecrosis of the Femoral Head. Dddt Vol. 14, 4579–4591. doi:10.2147/DDDT.S258024
Furuta, T., Miyaki, S., Ishitobi, H., Ogura, T., Kato, Y., Kamei, N., et al. (2016). Mesenchymal Stem Cell-Derived Exosomes Promote Fracture Healing in a Mouse Model. Stem Cell Transl Med 5 (12), 1620–1630. doi:10.5966/sctm.2015-0285
Fushimi, S., Nohno, T., Nagatsuka, H., and Katsuyama, H. (2018). Involvement of miR-140-3p in Wnt3a and TGFβ3 Signaling Pathways during Osteoblast Differentiation in MC3T3-E1 Cells. Genes Cells 23 (7), 517–527. doi:10.1111/gtc.12591
Gao, X., Wan, Z., Wei, M., Dong, Y., Zhao, Y., Chen, X., et al. (2019). Chronic Myelogenous Leukemia Cells Remodel the Bone Marrow Niche via Exosome-Mediated Transfer of miR-320. Theranostics 9 (19), 5642–5656. doi:10.7150/thno.34813
Geisler, S., and Coller, J. (2013). RNA in Unexpected Places: Long Non-coding RNA Functions in Diverse Cellular Contexts. Nat. Rev. Mol. Cel Biol 14 (11), 699–712. doi:10.1038/nrm3679
Giegé, R. (2008). Toward a More Complete View of tRNA Biology. Nat. Struct. Mol. Biol. 15 (10), 1007–1014. doi:10.1038/nsmb.1498
Giraldez, A. J., Cinalli, R. M., Glasner, M. E., Enright, A. J., Thomson, J. M., Baskerville, S., et al. (2005). MicroRNAs Regulate Brain Morphogenesis in Zebrafish. Science 308 (5723), 833–838. doi:10.1126/science.1109020
Glyn-Jones, S., Palmer, A. J. R., Agricola, R., Price, A. J., Vincent, T. L., Weinans, H., et al. (2015). Osteoarthritis. The Lancet 386 (9991), 376–387. doi:10.1016/S0140-6736(14)60802-3
Gómez-Barrena, E., Rosset, P., Lozano, D., Stanovici, J., Ermthaller, C., and Gerbhard, F. (2015). Bone Fracture Healing: Cell Therapy in Delayed Unions and Nonunions. Bone 70, 93–101. doi:10.1016/j.bone.2014.07.033
Gu, X., Li, M., Jin, Y., Liu, D., and Wei, F. (2017). Identification and Integrated Analysis of Differentially Expressed lncRNAs and circRNAs Reveal the Potential ceRNA Networks during PDLSC Osteogenic Differentiation. BMC Genet. 18 (1), 100. doi:10.1186/s12863-017-0569-4
Guay, C., Kruit, J. K., Rome, S., Menoud, V., Mulder, N. L., Jurdzinski, A., et al. (2019). Lymphocyte-Derived Exosomal MicroRNAs Promote Pancreatic β Cell Death and May Contribute to Type 1 Diabetes Development. Cel Metab. 29 (2), 348–361. doi:10.1016/j.cmet.2018.09.011
Guo, L., Zhu, Y., Li, L., Zhou, S., Yin, G., Yu, G., et al. (2019). Breast Cancer Cell‐derived Exosomal miR‐20a‐5p Promotes the Proliferation and Differentiation of Osteoclasts by Targeting SRCIN1. Cancer Med. 8 (12), 5687–5701. doi:10.1002/cam4.2454
Guo, Z., Wang, H., Zhao, F., Liu, M., Wang, F., Kang, M., et al. (2021). Exosomal Circ-BRWD1 Contributes to Osteoarthritis Development through the Modulation of miR-1277/TRAF6 axis. Arthritis Res. Ther. 23 (1), 159. doi:10.1186/s13075-021-02541-8
Gupta, S. K., and Thum, T. (2016). Non-coding RNAs as Orchestrators of Autophagic Processes. J. Mol. Cell Cardiol. 95, 26–30. doi:10.1016/j.yjmcc.2015.11.012
Gurunathan, S., Kang, M.-H., Jeyaraj, M., Qasim, M., and Kim, J.-H. (2019). Review of the Isolation, Characterization, Biological Function, and Multifarious Therapeutic Approaches of Exosomes. Cells 8 (4), 307. doi:10.3390/cells8040307
Gutschner, T., and Diederichs, S. (2012). The Hallmarks of Cancer. RNA Biol. 9 (6), 703–719. doi:10.4161/rna.20481
Hao, Z.-C., Lu, J., Wang, S.-Z., Wu, H., Zhang, Y.-T., and Xu, S.-G. (2017). Stem Cell-Derived Exosomes: A Promising Strategy for Fracture Healing. Cell Prolif 50 (5), e12359. doi:10.1111/cpr.12359
Hassan, M. Q., Maeda, Y., Taipaleenmaki, H., Zhang, W., Jafferji, M., Gordon, J. A. R., et al. (2012). miR-218 Directs a Wnt Signaling Circuit to Promote Differentiation of Osteoblasts and Osteomimicry of Metastatic Cancer Cells. J. Biol. Chem. 287 (50), 42084–42092. doi:10.1074/jbc.M112.377515
Hatfield, S. D., Shcherbata, H. R., Fischer, K. A., Nakahara, K., Carthew, R. W., and Ruohola-Baker, H. (2005). Stem Cell Division Is Regulated by the microRNA Pathway. Nature 435 (7044), 974–978. doi:10.1038/nature03816
Hrdlickova, B., de Almeida, R. C., Borek, Z., and Withoff, S. (2014). Genetic Variation in the Non-coding Genome: Involvement of Micro-RNAs and Long Non-coding RNAs in Disease. Biochim. Biophys. Acta (Bba) - Mol. Basis Dis. 1842 (10), 1910–1922. doi:10.1016/j.bbadis.2014.03.011
Hu, W., Alvarez‐Dominguez, J. R., and Lodish, H. F. (2012). Regulation of Mammalian Cell Differentiation by Long Non‐coding RNAs. EMBO Rep. 13 (11), 971–983. doi:10.1038/embor.2012.145
Huynh, N. P. T., Anderson, B. A., Guilak, F., and McAlinden, A. (2017). Emerging Roles for Long Noncoding RNAs in Skeletal Biology and Disease. Connect. Tissue Res. 58 (1), 116–141. doi:10.1080/03008207.2016.1194406
Hwang, S., Park, S.-K., Lee, H. Y., Kim, S. W., Lee, J. S., Choi, E. K., et al. (2014). miR-140-5p Suppresses BMP2-Mediated Osteogenesis in Undifferentiated Human Mesenchymal Stem Cells. FEBS Lett. 588 (17), 2957–2963. doi:10.1016/j.febslet.2014.05.048
Iyer, M. K., Niknafs, Y. S., Malik, R., Singhal, U., Sahu, A., Hosono, Y., et al. (2015). The Landscape of Long Noncoding RNAs in the Human Transcriptome. Nat. Genet. 47 (3), 199–208. doi:10.1038/ng.3192
Jiang, L. B., Tian, L., and Zhang, C. G. (2018). Bone Marrow Stem Cells-Derived Exosomes Extracted from Osteoporosis Patients Inhibit Osteogenesis via microRNA-21/SMAD7. Eur. Rev. Med. Pharmacol. Sci. 22 (19), 6221–6229. doi:10.26355/eurrev_201810_16028
Jin, Z., Ren, J., and Qi, S. (2020a). Exosomal miR-9-5p Secreted by Bone Marrow-Derived Mesenchymal Stem Cells Alleviates Osteoarthritis by Inhibiting Syndecan-1. Cell Tissue Res 381 (1), 99–114. doi:10.1007/s00441-020-03193-x
Jin, Z., Ren, J., and Qi, S. (2020b). Human Bone Mesenchymal Stem Cells-Derived Exosomes Overexpressing microRNA-26a-5p Alleviate Osteoarthritis via Down-Regulation of PTGS2. Int. Immunopharmacology 78, 105946. doi:10.1016/j.intimp.2019.105946
Joh, R. I., Palmieri, C. M., Hill, I. T., and Motamedi, M. (2014). Regulation of Histone Methylation by Noncoding RNAs. Biochim. Biophys. Acta (Bba) - Gene Regul. Mech. 1839 (12), 1385–1394. doi:10.1016/j.bbagrm.2014.06.006
Kalamvoki, M., Du, T., and Roizman, B. (2014). Cells Infected with Herpes Simplex Virus 1 export to Uninfected Cells Exosomes Containing STING, Viral mRNAs, and microRNAs. Proc. Natl. Acad. Sci. USA 111 (46), E4991–E4996. doi:10.1073/pnas.1419338111
Kalluri, R., and LeBleu, V. S. (2020). The Biology , Function , and Biomedical Applications of Exosomes. Science 367 (6478). doi:10.1126/science.aau6977
Kalluri, R. (2016). The Biology and Function of Exosomes in Cancer. J. Clin. Invest. 126 (4), 1208–1215. doi:10.1172/JCI81135
Kang, Y.-T., Kim, Y. J., Bu, J., Cho, Y.-H., Han, S.-W., and Moon, B.-I. (2017). High-purity Capture and Release of Circulating Exosomes Using an Exosome-specific Dual-Patterned Immunofiltration (ExoDIF) Device. Nanoscale 9 (36), 13495–13505. doi:10.1039/c7nr04557c
Kang, Y., Song, J., Kim, D., Ahn, C., Park, S., Chun, C.-H., et al. (2016). PCGEM1 Stimulates Proliferation of Osteoarthritic Synoviocytes by Acting as a Sponge for miR-770. J. Orthop. Res. 34 (3), 412–418. doi:10.1002/jor.23046
Khalil, A. M., Guttman, M., Huarte, M., Garber, M., Raj, A., Rivea Morales, D., et al. (2009). Many Human Large Intergenic Noncoding RNAs Associate with Chromatin-Modifying Complexes and Affect Gene Expression. Pnas 106 (28), 11667–11672. doi:10.1073/pnas.0904715106
Knowling, S., and Morris, K. V. (2011). Non-coding RNA and Antisense RNA. Nature's Trash or Treasure? Biochimie 93 (11), 1922–1927. doi:10.1016/j.biochi.2011.07.031
Kobayashi, M., Squires, G. R., Mousa, A., Tanzer, M., Zukor, D. J., Antoniou, J., et al. (2005). Role of Interleukin-1 and Tumor Necrosis Factor ? in Matrix Degradation of Human Osteoarthritic Cartilage. Arthritis Rheum. 52 (1), 128–135. doi:10.1002/art.20776
Komatsu, D. E., and Warden, S. J. (2009). The Control of Fracture Healing and its Therapeutic Targeting: Improving upon Nature. J. Cel. Biochem. 109 (2), a–n. doi:10.1002/jcb.22418
Kowal, J., Tkach, M., and Théry, C. (2014). Biogenesis and Secretion of Exosomes. Curr. Opin. Cel Biol. 29, 116–125. doi:10.1016/j.ceb.2014.05.004
Kumar, P., Kuscu, C., and Dutta, A. (2016). Biogenesis and Function of Transfer RNA-Related Fragments (tRFs). Trends Biochem. Sci. 41 (8), 679–689. doi:10.1016/j.tibs.2016.05.004
Kumar, S., Vijayan, M., Bhatti, J. S., and Reddy, P. H. (2017). MicroRNAs as Peripheral Biomarkers in Aging and Age-Related Diseases. Prog. Mol. Biol. Transl Sci. 146, 47–94. doi:10.1016/bs.pmbts.2016.12.013
Landthaler, M., Yalcin, A., and Tuschl, T. (2004). The Human DiGeorge Syndrome Critical Region Gene 8 and its D. melanogaster Homolog Are Required for miRNA Biogenesis. Curr. Biol. 14 (23), 2162–2167. doi:10.1016/j.cub.2004.11.001
Lasda, E., and Parker, R. (2016). Circular RNAs Co-precipitate with Extracellular Vesicles: A Possible Mechanism for circRNA Clearance. PLoS One 11 (2), e0148407. doi:10.1371/journal.pone.0148407
Lauvrak, S. U., Munthe, E., Kresse, S. H., Stratford, E. W., Namløs, H. M., Meza-Zepeda, L. A., et al. (2013). Functional Characterisation of Osteosarcoma Cell Lines and Identification of mRNAs and miRNAs Associated with Aggressive Cancer Phenotypes. Br. J. Cancer 109 (8), 2228–2236. doi:10.1038/bjc.2013.549
Lee, K.-P., Shin, Y. J., Panda, A. C., Abdelmohsen, K., Kim, J. Y., Lee, S.-M., et al. (2015). miR-431 Promotes Differentiation and Regeneration of Old Skeletal Muscle by Targeting Smad4. Genes Dev. 29 (15), 1605–1617. doi:10.1101/gad.263574.115
Lee, Y., Ahn, C., Han, J., Choi, H., Kim, J., Yim, J., et al. (2003). The Nuclear RNase III Drosha Initiates microRNA Processing. Nature 425 (6956), 415–419. doi:10.1038/nature01957
Lems, W. F., and Raterman, H. G. (2017). Critical Issues and Current Challenges in Osteoporosis and Fracture Prevention. An Overview of Unmet Needs. Ther. Adv. Musculoskelet. 9 (12), 299–316. doi:10.1177/1759720X17732562
Li, B., Xu, H., Han, H., Song, S., Zhang, X., Ouyang, L., et al. (2018). Exosome-mediated Transfer of lncRUNX2-AS1 from Multiple Myeloma Cells to MSCs Contributes to Osteogenesis. Oncogene 37 (41), 5508–5519. doi:10.1038/s41388-018-0359-0
Li, D., Liu, J., Guo, B., Liang, C., Dang, L., Lu, C., et al. (2016). Osteoclast-derived Exosomal miR-214-3p Inhibits Osteoblastic Bone Formation. Nat. Commun. 7, 10872. doi:10.1038/ncomms10872
Li, J.-p., Liu, L.-h., Li, J., Chen, Y., Jiang, X.-w., Ouyang, Y.-r., et al. (2013a). Microarray Expression Profile of Long Noncoding RNAs in Human Osteosarcoma. Biochem. Biophysical Res. Commun. 433 (2), 200–206. doi:10.1016/j.bbrc.2013.02.083
Li, L., Liu, B., Wapinski, O. L., Tsai, M.-C., Qu, K., Zhang, J., et al. (2013b). Targeted Disruption of Hotair Leads to Homeotic Transformation and Gene Derepression. Cel Rep. 5 (1), 3–12. doi:10.1016/j.celrep.2013.09.003
Li, L., Zhou, X., Zhang, J.-t., Liu, A.-f., Zhang, C., Han, J.-c., et al. (2021). Exosomal miR-186 Derived from BMSCs Promote Osteogenesis through Hippo Signaling Pathway in Postmenopausal Osteoporosis. J. Orthop. Surg. Res. 16 (1), 23. doi:10.1186/s13018-020-02160-0
Li, P., Kaslan, M., Lee, S. H., Yao, J., and Gao, Z. (2017). Progress in Exosome Isolation Techniques. Theranostics 7 (3), 789–804. doi:10.7150/thno.18133
Li, Z., Hassan, M. Q., Volinia, S., van Wijnen, A. J., Stein, J. L., Croce, C. M., et al. (2008). A microRNA Signature for a BMP2-Induced Osteoblast Lineage Commitment Program. Proc. Natl. Acad. Sci. 105 (37), 13906–13911. doi:10.1073/pnas.0804438105
Li, Z., Huang, C., Bao, C., Chen, L., Lin, M., Wang, X., et al. (2015). Exon-intron Circular RNAs Regulate Transcription in the Nucleus. Nat. Struct. Mol. Biol. 22 (3), 256–264. doi:10.1038/nsmb.2959
Li, Z., Wang, Y., Xiang, S., Zheng, Z., Bian, Y., Feng, B., et al. (2020). Chondrocytes-derived Exosomal miR-8485 Regulated the Wnt/β-Catenin Pathways to Promote Chondrogenic Differentiation of BMSCs. Biochem. Biophysical Res. Commun. 523 (2), 506–513. doi:10.1016/j.bbrc.2019.12.065
Liang, Y., Xu, X., Li, X., Xiong, J., Li, B., Duan, L., et al. (2020). Chondrocyte-Targeted MicroRNA Delivery by Engineered Exosomes toward a Cell-free Osteoarthritis Therapy. ACS Appl. Mater. Inter. 12 (33), 36938–36947. doi:10.1021/acsami.0c10458
Lin, X., Wu, L., Zhang, Z., Yang, R., Guan, Q., Hou, X., et al. (2014). MiR-335-5p Promotes Chondrogenesis in Mouse Mesenchymal Stem Cells and Is Regulated through Two Positive Feedback Loops. J. Bone Miner Res. 29 (7), 1575–1585. doi:10.1002/jbmr.2163
Ling, H., Fabbri, M., and Calin, G. A. (2013). MicroRNAs and Other Non-coding RNAs as Targets for Anticancer Drug Development. Nat. Rev. Drug Discov. 12 (11), 847–865. doi:10.1038/nrd4140
Liu, B., Wu, P., Mei, L., Luo, Y., Li, H., and Mao, X. (2018a). Differential Expression of Exosomal miRNAs in Osteoblasts in Osteoarthritis. Zhong Nan Da Xue Xue Bao Yi Xue Ban 43 (12), 1294–1300. doi:10.11817/j.issn.1672-7347.2018.12.003
Liu, R., Jiang, C., Li, J., Li, X., Zhao, L., Yun, H., et al. (2021). Serum-derived Exosomes Containing NEAT1 Promote the Occurrence of Rheumatoid Arthritis through Regulation of miR-144-3p/ROCK2 axis. Ther. Adv. Chronic Dis. 12, 204062232199170. doi:10.1177/2040622321991705
Liu, S., Liu, D., Chen, C., Hamamura, K., Moshaverinia, A., Yang, R., et al. (2015a). MSC Transplantation Improves Osteopenia via Epigenetic Regulation of Notch Signaling in Lupus. Cel Metab. 22 (4), 606–618. doi:10.1016/j.cmet.2015.08.018
Liu, W., Yang, L. H., Kong, X. C., An, L. K., and Wang, R. (2015b). Meta-analysis of Osteoporosis: Fracture Risks, Medication and Treatment. Minerva Med. 106 (4), 203–214.
Liu, W., Li, L., Rong, Y., Qian, D., Chen, J., Zhou, Z., et al. (2020). Hypoxic Mesenchymal Stem Cell-Derived Exosomes Promote Bone Fracture Healing by the Transfer of miR-126. Acta Biomater. 103, 196–212. doi:10.1016/j.actbio.2019.12.020
Liu, Y., Lin, L., Zou, R., Wen, C., Wang, Z., and Lin, F. (2018b). MSC-derived Exosomes Promote Proliferation and Inhibit Apoptosis of Chondrocytes via lncRNA-KLF3-AS1/miR-206/GIT1 axis in Osteoarthritis. Cell Cycle 17 (21-22), 2411–2422. doi:10.1080/15384101.2018.1526603
Liu, Y., Zou, R., Wang, Z., Wen, C., Zhang, F., and Lin, F. (2018c). Exosomal KLF3-AS1 from hMSCs Promoted Cartilage Repair and Chondrocyte Proliferation in Osteoarthritis. Biochem. J. 475 (22), 3629–3638. doi:10.1042/BCJ20180675
Loeser, R. F., Goldring, S. R., Scanzello, C. R., and Goldring, M. B. (2012). Osteoarthritis: a Disease of the Joint as an Organ. Arthritis Rheum. 64 (6), 1697–1707. doi:10.1002/art.34453
Long, F., and Ornitz, D. M. (2013). Development of the Endochondral Skeleton. Cold Spring Harbor Perspect. Biol. 5 (1), a008334. doi:10.1101/cshperspect.a008334
Lu, J., Getz, G., Miska, E. A., Alvarez-Saavedra, E., Lamb, J., Peck, D., et al. (2005). MicroRNA Expression Profiles Classify Human Cancers. Nature 435 (7043), 834–838. doi:10.1038/nature03702
Luo, P., Jiang, C., Ji, P., Wang, M., and Xu, J. (2019). Exosomes of Stem Cells from Human Exfoliated Deciduous Teeth as an Anti-inflammatory Agent in Temporomandibular Joint Chondrocytes via miR-100-5p/mTOR. Stem Cel Res Ther 10 (1), 216. doi:10.1186/s13287-019-1341-7
Manolagas, S. C. (2013). Steroids and Osteoporosis: the Quest for Mechanisms. J. Clin. Invest. 123 (5), 1919–1921. doi:10.1172/JCI68062
Mao, G., Hu, S., Zhang, Z., Wu, P., Zhao, X., Lin, R., et al. (2018a). Exosomal miR-95-5p Regulates Chondrogenesis and Cartilage Degradation via Histone Deacetylase 2/8. J. Cel Mol Med 22 (11), 5354–5366. doi:10.1111/jcmm.13808
Mao, G., Xu, Y., Long, D., Sun, H., Li, H., Xin, R., et al. (2021). Exosome-transported circRNA_0001236 Enhances Chondrogenesis and Suppress Cartilage Degradation via the miR-3677-3p/Sox9 axis. Stem Cel Res Ther 12 (1), 389. doi:10.1186/s13287-021-02431-5
Mao, G., Zhang, Z., Hu, S., Zhang, Z., Chang, Z., Huang, Z., et al. (2018b). Exosomes Derived from miR-92a-3p-Overexpressing Human Mesenchymal Stem Cells Enhance Chondrogenesis and Suppress Cartilage Degradation via Targeting WNT5A. Stem Cel Res Ther 9 (1), 247. doi:10.1186/s13287-018-1004-0
Mao, G., Zhang, Z., Huang, Z., Chen, W., Huang, G., Meng, F., et al. (2017). MicroRNA-92a-3p Regulates the Expression of Cartilage-specific Genes by Directly Targeting Histone Deacetylase 2 in Chondrogenesis and Degradation. Osteoarthritis and Cartilage 25 (4), 521–532. doi:10.1016/j.joca.2016.11.006
Mao, Z., Zhu, Y., Hao, W., Chu, C., and Su, H. (2019). MicroRNA‐155 Inhibition Up‐regulates LEPR to Inhibit Osteoclast Activation and Bone Resorption via Activation of AMPK in Alendronate‐treated Osteoporotic Mice. IUBMB Life 71 (12), 1916–1928. doi:10.1002/iub.2131
Matsuo, K., and Irie, N. (2008). Osteoclast-osteoblast Communication. Arch. Biochem. Biophys. 473 (2), 201–209. doi:10.1016/j.abb.2008.03.027
Mattick, J. S., and Makunin, I. V. (2005). Small Regulatory RNAs in Mammals. Hum. Mol. Genet. 141, R121–R132. doi:10.1093/hmg/ddi101
Meng, Q., and Qiu, B. (2020). Exosomal MicroRNA-320a Derived from Mesenchymal Stem Cells Regulates Rheumatoid Arthritis Fibroblast-like Synoviocyte Activation by Suppressing CXCL9 Expression. Front. Physiol. 11, 441. doi:10.3389/fphys.2020.00441
Meng, Y., Qiu, S., Sun, L., and Zuo, J. (2020). Knockdown of Exosome-mediated lnc-PVT1 A-lleviates L-ipopolysaccharide-induced O-steoarthritis P-rogression by M-ediating the HMGB1/TLR4/NF-κB P-athway via miR-93-5p. Mol. Med. Rep. 22 (6), 5313–5325. doi:10.3892/mmr.2020.11594
Mizoguchi, F., Murakami, Y., Saito, T., Miyasaka, N., and Kohsaka, H. (2013). miR-31 Controls Osteoclast Formation and Bone Resorption by Targeting RhoA. Arthritis Res. Ther. 15 (5), R102. doi:10.1186/ar4282
Moran, V. A., Perera, R. J., and Khalil, A. M. (2012). Emerging Functional and Mechanistic Paradigms of Mammalian Long Non-coding RNAs. Nucleic Acids Res. 40 (14), 6391–6400. doi:10.1093/nar/gks296
Morris, K. V., and Mattick, J. S. (2014). The Rise of Regulatory RNA. Nat. Rev. Genet. 15 (6), 423–437. doi:10.1038/nrg3722
Murata, K., Ito, H., Yoshitomi, H., Yamamoto, K., Fukuda, A., Yoshikawa, J., et al. (2014). Inhibition of miR-92a Enhances Fracture Healing via Promoting Angiogenesis in a Model of Stabilized Fracture in Young Mice. J. Bone Miner Res. 29 (2), 316–326. doi:10.1002/jbmr.2040
Naguibneva, I., Ameyar-Zazoua, M., Polesskaya, A., Ait-Si-Ali, S., Groisman, R., Souidi, M., et al. (2006). The microRNA miR-181 Targets the Homeobox Protein Hox-A11 during Mammalian Myoblast Differentiation. Nat. Cel Biol 8 (3), 278–284. doi:10.1038/ncb1373
Narayanan, K., Kumar, S., Padmanabhan, P., Gulyas, B., Wan, A. C. A., and Rajendran, V. M. (2018). Lineage-specific Exosomes Could Override Extracellular Matrix Mediated Human Mesenchymal Stem Cell Differentiation. Biomaterials 182, 312–322. doi:10.1016/j.biomaterials.2018.08.027
Naseri, Z., Kazemi Oskuee, R., Jaafari, M. R., and Forouzandeh, M. (2018). Exosome-mediated Delivery of Functionally Active miRNA-142-3p Inhibitor Reduces Tumorigenicity of Breast Cancer In Vitro and In Vivo. Ijn Vol. 13, 7727–7747. doi:10.2147/IJN.S182384
Neil, H., Malabat, C., d’Aubenton-Carafa, Y., Xu, Z., Steinmetz, L. M., and Jacquier, A. (2009). Widespread Bidirectional Promoters Are the Major Source of Cryptic Transcripts in Yeast. Nature 457 (7232), 1038–1042. doi:10.1038/nature07747
Ni, Z., Kuang, L., Chen, H., Xie, Y., Zhang, B., Ouyang, J., et al. (2019). The Exosome-like Vesicles from Osteoarthritic Chondrocyte Enhanced Mature IL-1β Production of Macrophages and Aggravated Synovitis in Osteoarthritis. Cel Death Dis 10 (7), 522. doi:10.1038/s41419-019-1739-2
Owens, R. A., Sano, T., and Duran-Vila, N. (2012). Plant Viroids: Isolation, Characterization/detection, and Analysis. Methods Mol. Biol. 894, 253–271. doi:10.1007/978-1-61779-882-5_17
Pan, B.-T., and Johnstone, R. M. (1983). Fate of the Transferrin Receptor during Maturation of Sheep Reticulocytes In Vitro: Selective Externalization of the Receptor. Cell 33 (3), 967–978. doi:10.1016/0092-8674(83)90040-5
Patel, G. K., Khan, M. A., Zubair, H., Srivastava, S. K., Khushman, M. d., Singh, S., et al. (2019). Comparative Analysis of Exosome Isolation Methods Using Culture Supernatant for Optimum Yield, Purity and Downstream Applications. Sci. Rep. 9 (1), 5335. doi:10.1038/s41598-019-41800-2
Pethő, A., Chen, Y., and George, A. (2018). Exosomes in Extracellular Matrix Bone Biology. Curr. Osteoporos. Rep. 16 (1), 58–64. doi:10.1007/s11914-018-0419-y
Phizicky, E. M., and Hopper, A. K. (2010). tRNA Biology Charges to the Front. Genes Dev. 24 (17), 1832–1860. doi:10.1101/gad.1956510
Plasterk, R. H. A. (2006). Micro RNAs in Animal Development. Cell 124 (5), 877–881. doi:10.1016/j.cell.2006.02.030
Qin, Y., Peng, Y., Zhao, W., Pan, J., Ksiezak-Reding, H., Cardozo, C., et al. (2017). Myostatin Inhibits Osteoblastic Differentiation by Suppressing Osteocyte-Derived Exosomal microRNA-218: A Novel Mechanism in Muscle-Bone Communication. J. Biol. Chem. 292 (26), 11021–11033. doi:10.1074/jbc.M116.770941
Qin, Y., Wang, L., Gao, Z., Chen, G., and Zhang, C. (2016). Bone Marrow Stromal/stem Cell-Derived Extracellular Vesicles Regulate Osteoblast Activity and Differentiation In Vitro and Promote Bone Regeneration In Vivo. Sci. Rep. 6, 21961. doi:10.1038/srep21961
Quinn, J. J., and Chang, H. Y. (2016). Unique Features of Long Non-coding RNA Biogenesis and Function. Nat. Rev. Genet. 17 (1), 47–62. doi:10.1038/nrg.2015.10
Quinodoz, S., and Guttman, M. (2014). Long Noncoding RNAs: an Emerging Link between Gene Regulation and Nuclear Organization. Trends Cel Biol. 24 (11), 651–663. doi:10.1016/j.tcb.2014.08.009
Rachner, T. D., Khosla, S., and Hofbauer, L. C. (2011). Osteoporosis: Now and the Future. The Lancet 377 (9773), 1276–1287. doi:10.1016/S0140-6736(10)62349-5
Raina, M., and Ibba, M. (2014). tRNAs as Regulators of Biological Processes. Front. Genet. 5, 171. doi:10.3389/fgene.2014.00171
Riddle, R. C., and Clemens, T. L. (2017). Bone Cell Bioenergetics and Skeletal Energy Homeostasis. Physiol. Rev. 97 (2), 667–698. doi:10.1152/physrev.00022.2016
Rinn, J. L., and Chang, H. Y. (2012). Genome Regulation by Long Noncoding RNAs. Annu. Rev. Biochem. 81, 145–166. doi:10.1146/annurev-biochem-051410-092902
Rokavec, M., Wu, W., and Luo, J.-L. (2012). IL6-mediated Suppression of miR-200c Directs Constitutive Activation of Inflammatory Signaling Circuit Driving Transformation and Tumorigenesis. Mol. Cel 45 (6), 777–789. doi:10.1016/j.molcel.2012.01.015
Rosenfeld, N., Aharonov, R., Meiri, E., Rosenwald, S., Spector, Y., Zepeniuk, M., et al. (2008). MicroRNAs Accurately Identify Cancer Tissue Origin. Nat. Biotechnol. 26 (4), 462–469. doi:10.1038/nbt1392
Salzman, J. (2016). Circular RNA Expression: Its Potential Regulation and Function. Trends Genet. 32 (5), 309–316. doi:10.1016/j.tig.2016.03.002
Schaap-Oziemlak, A. M., Raymakers, R. A., Bergevoet, S. M., Gilissen, C., Jansen, B. J. H., Adema, G. J., et al. (2010). MicroRNA Hsa-miR-135b Regulates Mineralization in Osteogenic Differentiation of Human Unrestricted Somatic Stem Cells. Stem Cell Dev. 19 (6), 877–885. doi:10.1089/scd.2009.0112
Simons, M., and Raposo, G. (2009). Exosomes - Vesicular Carriers for Intercellular Communication. Curr. Opin. Cel Biol. 21 (4), 575–581. doi:10.1016/j.ceb.2009.03.007
Soekmadji, C., Russell, P., and Nelson, C. (2013). Exosomes in Prostate Cancer: Putting Together the Pieces of a Puzzle. Cancers 5 (4), 1522–1544. doi:10.3390/cancers5041522
Song, H., Li, X., Zhao, Z., Qian, J., Wang, Y., Cui, J., et al. (2019). Reversal of Osteoporotic Activity by Endothelial Cell-Secreted Bone Targeting and Biocompatible Exosomes. Nano Lett. 19 (5), 3040–3048. doi:10.1021/acs.nanolett.9b00287
Song, J., Kang, Y., Chun, C. H., and Jin, E. J. (2017). Selective Loading of Exosomal HULC and miR-372 Is Responsible for Chondrocyte Death during OA Pathogenesis. Animal Cells & Syst. 21 (6), 397–403. doi:10.1080/19768354.2017.1406871
St. Laurent, G., Wahlestedt, C., and Kapranov, P. (2015). The Landscape of Long Noncoding RNA Classification. Trends Genet. 31 (5), 239–251. doi:10.1016/j.tig.2015.03.007
Suchacki, K. J., Roberts, F., Lovdel, A., Farquharson, C., Morton, N. M., MacRae, V. E., et al. (2017). Skeletal Energy Homeostasis: a Paradigm of Endocrine Discovery. J. Endocrinol. 234 (1), R67–R79. doi:10.1530/JOE-17-0147
Sun, H., Hu, S., Zhang, Z., Lun, J., Liao, W., and Zhang, Z. (2019). Expression of Exosomal microRNAs during Chondrogenic Differentiation of Human Bone Mesenchymal Stem Cells. J. Cel Biochem 120 (1), 171–181. doi:10.1002/jcb.27289
Sun, W., Zhao, C., Li, Y., Wang, L., Nie, G., Peng, J., et al. (2016). Osteoclast-derived microRNA-Containing Exosomes Selectively Inhibit Osteoblast Activity. Cell Discov 2, 16015. doi:10.1038/celldisc.2016.15
Taft, R. J., Glazov, E. A., Cloonan, N., Simons, C., Stephen, S., Faulkner, G. J., et al. (2009). Tiny RNAs Associated with Transcription Start Sites in Animals. Nat. Genet. 41 (5), 572–578. doi:10.1038/ng.312
Tan, F., Wang, D., and Yuan, Z. (2020). The Fibroblast-like Synoviocyte Derived Exosomal Long Non-coding RNA H19 Alleviates Osteoarthritis Progression through the miR-106b-5p/TIMP2 Axis. Inflammation 43 (4), 1498–1509. doi:10.1007/s10753-020-01227-8
Tang, L., Yin, Y., Liu, J., Li, Z., and Lu, X. (2017). MiR-124 Attenuates Osteoclastogenic Differentiation of Bone Marrow Monocytes via Targeting Rab27a. Cell Physiol Biochem 43 (4), 1663–1672. doi:10.1159/000484027
Tao, S.-C., Yuan, T., Zhang, Y.-L., Yin, W.-J., Guo, S.-C., and Zhang, C.-Q. (2017). Exosomes Derived from miR-140-5p-Overexpressing Human Synovial Mesenchymal Stem Cells Enhance Cartilage Tissue Regeneration and Prevent Osteoarthritis of the Knee in a Rat Model. Theranostics 7 (1), 180–195. doi:10.7150/thno.17133
Tatsumi, S., Ishii, K., Amizuka, N., Li, M., Kobayashi, T., Kohno, K., et al. (2007). Targeted Ablation of Osteocytes Induces Osteoporosis with Defective Mechanotransduction. Cel Metab. 5 (6), 464–475. doi:10.1016/j.cmet.2007.05.001
Théry, C., Zitvogel, L., and Amigorena, S. (2002). Exosomes: Composition, Biogenesis and Function. Nat. Rev. Immunol. 2 (8), 569–579. doi:10.1038/nri855
Toh, W. S., Lai, R. C., Hui, J. H. P., and Lim, S. K. (2017). MSC Exosome as a Cell-free MSC Therapy for Cartilage Regeneration: Implications for Osteoarthritis Treatment. Semin. Cel Dev. Biol. 67, 56–64. doi:10.1016/j.semcdb.2016.11.008
Tye, C. E., Boyd, J. R., Page, N. A., Falcone, M. M., Stein, J. L., Stein, G. S., et al. (2018). Regulation of Osteogenesis by Long Noncoding RNAs: An Epigenetic Mechanism Contributing to Bone Formation. Connect. Tissue Res. 59 (Suppl. 1), 35–41. doi:10.1080/03008207.2017.1412432
Ulitsky, I., and Bartel, D. P. (2013). lincRNAs: Genomics, Evolution, and Mechanisms. Cell 154 (1), 26–46. doi:10.1016/j.cell.2013.06.020
van Niel, G., Raposo, G., Candalh, C., Boussac, M., Hershberg, R., Cerf–Bensussan, N., et al. (2001). Intestinal Epithelial Cells Secrete Exosome-like Vesicles. Gastroenterology 121 (2), 337–349. doi:10.1053/gast.2001.26263
Vance, K. W., and Ponting, C. P. (2014). Transcriptional Regulatory Functions of Nuclear Long Noncoding RNAs. Trends Genet. 30 (8), 348–355. doi:10.1016/j.tig.2014.06.001
Varela-Eirín, M., Varela-Vázquez, A., Guitián-Caamao, A., Bravo-López, S. B., and Mayán, M. D. (2019). “Fri0529 Spread of Senescence and Joint Inflammation via Connexin43-Positive Exosomes Released by Osteoarthritic Chondrocytes,” in Annual European Congress of Rheumatology (Madrid: EULAR), 12–15.
Vos, T., Flaxman, A. D., Naghavi, M., Lozano, R., Michaud, C., Ezzati, M., et al. (2012). Years Lived with Disability (YLDs) for 1160 Sequelae of 289 Diseases and Injuries 1990-2010: a Systematic Analysis for the Global Burden of Disease Study 2010. Lancet 380 (9859), 2163–2196. doi:10.1016/S0140-6736(12)61729-2
Wahlestedt, C. (2013). Targeting Long Non-coding RNA to Therapeutically Upregulate Gene Expression. Nat. Rev. Drug Discov. 12 (6), 433–446. doi:10.1038/nrd4018
Wang, C., Zhu, G., He, W., Yin, H., Lin, F., Gou, X., et al. (2019). BMSCs Protect against Renal Ischemia‐reperfusion Injury by Secreting Exosomes Loaded with miR‐199a‐5p that Target BIP to Inhibit Endoplasmic Reticulum Stress at the Very Early Reperfusion Stages. FASEB j. 33 (4), 5440–5456. doi:10.1096/fj.201801821R
Wang, R., Xu, B., and Xu, H. (2018). TGF-β1 Promoted Chondrocyte Proliferation by Regulating Sp1 through MSC-Exosomes Derived miR-135b. Cell Cycle 17 (24), 2756–2765. doi:10.1080/15384101.2018.1556063
Weaver, C. M., Alexander, D. D., Boushey, C. J., Dawson-Hughes, B., Lappe, J. M., LeBoff, M. S., et al. (2016). Calcium Plus Vitamin D Supplementation and Risk of Fractures: an Updated Meta-Analysis from the National Osteoporosis Foundation. Osteoporos. Int. 27 (1), 367–376. doi:10.1007/s00198-015-3386-5
Wei, J., Li, H., Wang, S., Li, T., Fan, J., Liang, X., et al. (2014). let-7 Enhances Osteogenesis and Bone Formation while Repressing Adipogenesis of Human Stromal/mesenchymal Stem Cells by Regulating HMGA2. Stem Cell Dev. 23 (13), 1452–1463. doi:10.1089/scd.2013.0600
Wu, J., Kuang, L., Chen, C., Yang, J., Zeng, W.-N., Li, T., et al. (2019). miR-100-5p-abundant Exosomes Derived from Infrapatellar Fat Pad MSCs Protect Articular Cartilage and Ameliorate Gait Abnormalities via Inhibition of mTOR in Osteoarthritis. Biomaterials 206, 87–100. doi:10.1016/j.biomaterials.2019.03.022
Wu, Q., Liu, W., Wang, J., Zhu, L., Wang, Z., and Peng, Y. (2020). Exosomal Noncoding RNAs in Colorectal Cancer. Cancer Lett. 493, 228–235. doi:10.1016/j.canlet.2020.08.037
Xin, H., Li, Y., Buller, B., Katakowski, M., Zhang, Y., Wang, X., et al. (2012). Exosome‐Mediated Transfer of miR‐133b from Multipotent Mesenchymal Stromal Cells to Neural Cells Contributes to Neurite Outgrowth. Stem Cells 30 (7), 1556–1564. doi:10.1002/stem.1129
Xiong, Y., Chen, L., Yan, C., Zhou, W., Yu, T., Sun, Y., et al. (2020). M2 Macrophagy-Derived Exosomal miRNA-5106 Induces Bone Mesenchymal Stem Cells towards Osteoblastic Fate by Targeting Salt-Inducible Kinase 2 and 3. J. Nanobiotechnol 18 (1), 66. doi:10.1186/s12951-020-00622-5
Xu, J.-F., Yang, G.-h., Pan, X.-H., Zhang, S.-J., Zhao, C., Qiu, B.-S., et al. (2014). Altered microRNA Expression Profile in Exosomes during Osteogenic Differentiation of Human Bone Marrow-Derived Mesenchymal Stem Cells. PLoS One 9 (12), e114627. doi:10.1371/journal.pone.0114627
Xu, J., Li, D., Cai, Z., Sun, H., Su, B., Qiu, M., et al. (2020a). Exosomal lncRNAs NONMMUT000375 .2 and NONMMUT071578 .2 Derived from Titanium Particle Treated RAW264 .7 Cells Regulate Osteogenic Differentiation of MC3T3‐E1 Cells. J. Biomed. Mater. Res. 108 (11), 2251–2262. doi:10.1002/jbm.a.36983
Xu, R., Shen, X., Si, Y., Fu, Y., Zhu, W., Xiao, T., et al. (2018). MicroRNA-31a-5p from Aging BMSCs Links Bone Formation and Resorption in the Aged Bone Marrow Microenvironment. Aging Cell 17 (4), e12794. doi:10.1111/acel.12794
Xu, S., Cecilia Santini, G., De Veirman, K., Vande Broek, I., Leleu, X., De Becker, A., et al. (2013). Upregulation of miR-135b Is Involved in the Impaired Osteogenic Differentiation of Mesenchymal Stem Cells Derived from Multiple Myeloma Patients. PLoS One 8 (11), e79752. doi:10.1371/journal.pone.0079752
Xu, S., and Wang, Z. (2017). Bone Marrow Mesenchymal Stem Cell-Derived Exosomes Enhance Osteoclastogenesis during Alveolar Bone Deterioration in Rats. RSC Adv. 7 (34), 21153–21163. doi:10.1039/c6ra27931g
Xu, T., Luo, Y., Wang, J., Zhang, N., Gu, C., Li, L., et al. (2020b). Exosomal miRNA-128-3p from Mesenchymal Stem Cells of Aged Rats Regulates Osteogenesis and Bone Fracture Healing by Targeting Smad5. J. Nanobiotechnol 18 (1), 47. doi:10.1186/s12951-020-00601-w
Yang, B.-c., Kuang, M.-j., Kang, J.-y., Zhao, J., Ma, J.-x., and Ma, X.-l. (2020a). Human Umbilical Cord Mesenchymal Stem Cell-Derived Exosomes Act via the miR-1263/Mob1/Hippo Signaling Pathway to Prevent Apoptosis in Disuse Osteoporosis. Biochem. Biophysical Res. Commun. 524 (4), 883–889. doi:10.1016/j.bbrc.2020.02.001
Yang, B., Chen, Y., and Shi, J. (2019a). Exosome Biochemistry and Advanced Nanotechnology for Next-Generation Theranostic Platforms. Adv. Mater. 31 (2), 1802896. doi:10.1002/adma.201802896
Yang, X., Yang, J., Lei, P., and Wen, T. (2019b). LncRNA MALAT1 Shuttled by Bone Marrow-Derived Mesenchymal Stem Cells-Secreted Exosomes Alleviates Osteoporosis through Mediating microRNA-34c/SATB2 axis. Aging 11 (20), 8777–8791. doi:10.18632/aging.102264
Yang, Y., Yujiao, W., Fang, W., Linhui, Y., Ziqi, G., Zhichen, W., et al. (2020b). The Roles of miRNA, lncRNA and circRNA in the Development of Osteoporosis. Biol. Res. 53 (1), 40. doi:10.1186/s40659-020-00309-z
Yuan, Y., Yan, G., Gong, R., Zhang, L., Liu, T., Feng, C., et al. (2017). Effects of Blue Light Emitting Diode Irradiation on the Proliferation, Apoptosis and Differentiation of Bone Marrow-Derived Mesenchymal Stem Cells. Cel Physiol Biochem 43 (1), 237–246. doi:10.1159/000480344
Zarb, G. A., and Carlsson, G. E. (1999). Temporomandibular Disorders: Osteoarthritis. J. Orofac Pain 13 (4), 295–306.
Zavatti, M., Beretti, F., Casciaro, F., Bertucci, E., and Maraldi, T. (2020). Comparison of the Therapeutic Effect of Amniotic Fluid Stem Cells and Their Exosomes on Monoiodoacetate‐induced Animal Model of Osteoarthritis. Biofactors 46 (1), 106–117. doi:10.1002/biof.1576
Zhang, H., and Lyden, D. (2019). Asymmetric-flow Field-Flow Fractionation Technology for Exomere and Small Extracellular Vesicle Separation and Characterization. Nat. Protoc. 14 (4), 1027–1053. doi:10.1038/s41596-019-0126-x
Zhang, J., Zhao, H., Chen, J., Xia, B., Jin, Y., Wei, W., et al. (2012a). Interferon-β-induced miR-155 Inhibits Osteoclast Differentiation by Targeting SOCS1 and MITF. FEBS Lett. 586 (19), 3255–3262. doi:10.1016/j.febslet.2012.06.047
Zhang, L.-Q., Zhao, G.-Z., Xu, X.-Y., Fang, J., Chen, J.-M., Li, J.-W., et al. (2015). Integrin-β1 Regulates Chondrocyte Proliferation and Apoptosis through the Upregulation of GIT1 Expression. Int. J. Mol. Med. 35 (4), 1074–1080. doi:10.3892/ijmm.2015.2114
Zhang, L., Liu, S., Wang, J.-H., Zou, J., Zeng, H., Zhao, H., et al. (2019a). Differential Expressions of microRNAs and Transfer RNA-Derived Small RNAs: Potential Targets of Choroidal Neovascularization. Curr. Eye Res. 44 (11), 1226–1235. doi:10.1080/02713683.2019.1625407
Zhang, L., Meng, X., Zhu, X.-w., Yang, D.-c., Chen, R., Jiang, Y., et al. (2019b). Long Non-coding RNAs in Oral Squamous Cell Carcinoma: Biologic Function, Mechanisms and Clinical Implications. Mol. Cancer 18 (1), 102. doi:10.1186/s12943-019-1021-3
Zhang, N., Zhang, R. F., Zhang, A. N., Dong, G. X., Suo, N., Wu, Z. P., et al. (2018a). MiR-204 Promotes Fracture Healing via Enhancing Cell Viability of Osteoblasts. Eur. Rev. Med. Pharmacol. Sci. 22 (1 Suppl. l), 29–35. doi:10.26355/eurrev_201807_15356
Zhang, Y., Cai, F., Liu, J., Chang, H., Liu, L., Yang, A., et al. (2018b). Transfer RNA-Derived Fragments as Potential Exosome tRNA-Derived Fragment Biomarkers for Osteoporosis. Int. J. Rheum. Dis. 21 (9), 1659–1669. doi:10.1111/1756-185X.13346
Zhang, Y., Xie, R.-l., Croce, C. M., Stein, J. L., Lian, J. B., van Wijnen, A. J., et al. (2011). A Program of microRNAs Controls Osteogenic Lineage Progression by Targeting Transcription Factor Runx2. Proc. Natl. Acad. Sci. 108 (24), 9863–9868. doi:10.1073/pnas.1018493108
Zhang, Z., Kang, Y., Zhang, Z., Zhang, H., Duan, X., Liu, J., et al. (2012b). Expression of microRNAs during Chondrogenesis of Human Adipose-Derived Stem Cells. Osteoarthritis and Cartilage 20 (12), 1638–1646. doi:10.1016/j.joca.2012.08.024
Zhao, C., Irie, N., Takada, Y., Shimoda, K., Miyamoto, T., Nishiwaki, T., et al. (2006). Bidirectional ephrinB2-EphB4 Signaling Controls Bone Homeostasis. Cel Metab. 4 (2), 111–121. doi:10.1016/j.cmet.2006.05.012
Zhao, H., Zhang, J., Shao, H., Liu, J., Jin, M., Chen, J., et al. (2017). Transforming Growth Factor beta1/Smad4 Signaling Affects Osteoclast Differentiation via Regulation of miR-155 Expression. Mol. Cell 40 (3), 211–221. doi:10.14348/molcells.2017.2303
Zhao, W., Shen, G., Ren, H., LiangYu, D., Yu, X., Zhang, Z., et al. (2018). Therapeutic Potential of microRNAs in Osteoporosis Function by Regulating the Biology of Cells Related to Bone Homeostasis. J. Cel Physiol 233 (12), 9191–9208. doi:10.1002/jcp.26939
Zhao, Y., and Xu, J. (2018). Synovial Fluid-Derived Exosomal lncRNA PCGEM1 as Biomarker for the Different Stages of Osteoarthritis. Int. Orthopaedics (Sicot) 42 (12), 2865–2872. doi:10.1007/s00264-018-4093-6
Zhi, F., Ding, Y., Wang, R., Yang, Y., Luo, K., and Hua, F. (2021). Exosomal Hsa_circ_0006859 Is a Potential Biomarker for Postmenopausal Osteoporosis and Enhances Adipogenic versus Osteogenic Differentiation in Human Bone Marrow Mesenchymal Stem Cells by Sponging miR-431-5p. Stem Cel Res Ther 12 (1), 157. doi:10.1186/s13287-021-02214-y
Zhou, F., Wang, W., Xing, Y., Wang, T., Xu, X., and Wang, J. (2014). NF-κB Target microRNAs and Their Target Genes in TNFα-Stimulated HeLa Cells. Biochim. Biophys. Acta (Bba) - Gene Regul. Mech. 1839 (4), 344–354. doi:10.1016/j.bbagrm.2014.01.006
Zhou, J., Liu, H. X., Li, S. H., Gong, Y. S., Zhou, M. W., Zhang, J. H., et al. (2019a). Effects of Human Umbilical Cord Mesenchymal Stem Cells-Derived Exosomes on Fracture Healing in Rats through the Wnt Signaling Pathway. Eur. Rev. Med. Pharmacol. Sci. 23 (11), 4954–4960. doi:10.26355/eurrev_201906_18086
Zhou, L., Lv, T., Zhang, Q., Zhu, Q., Zhan, P., Zhu, S., et al. (2017). The Biology, Function and Clinical Implications of Exosomes in Lung Cancer. Cancer Lett. 407, 84–92. doi:10.1016/j.canlet.2017.08.003
Zhou, Z.-b., Huang, G.-x., Fu, Q., Han, B., Lu, J.-j., Chen, A.-m., et al. (2019b). circRNA.33186 Contributes to the Pathogenesis of Osteoarthritis by Sponging miR-127-5p. Mol. Ther. 27 (3), 531–541. doi:10.1016/j.ymthe.2019.01.006
Keywords: bone-related diseases, exosomes, non-coding RNAs, osteoporosis, osteoarthritis
Citation: Li H, Zheng Q, Xie X, Wang J, Zhu H, Hu H, He H and Lu Q (2021) Role of Exosomal Non-Coding RNAs in Bone-Related Diseases. Front. Cell Dev. Biol. 9:811666. doi: 10.3389/fcell.2021.811666
Received: 09 November 2021; Accepted: 09 December 2021;
Published: 23 December 2021.
Edited by:
Changjun Li, Central South University, ChinaCopyright © 2021 Li, Zheng, Xie, Wang, Zhu, Hu, He and Lu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hao He, aGVoYW82ODY4QGNzdS5lZHUuY24=; Qiong Lu, Y2hyaXN0eV9sdXFAY3N1LmVkdS5jbg==
 Hang Li
Hang Li Qiyue Zheng
Qiyue Zheng Xinyan Xie
Xinyan Xie Jiaojiao Wang
Jiaojiao Wang Haihong Zhu
Haihong Zhu Haoye Hu4
Haoye Hu4 Qiong Lu
Qiong Lu