- 1Key Laboratory of Medical Electrophysiology, Ministry of Education and Medical Electrophysiological Key Laboratory of Sichuan Province, Collaborative Innovation Center for Prevention of Cardiovascular Diseases, Institute of Cardiovascular Research, Southwest Medical University, Luzhou, China
- 2Department of Obstetrics, Sichuan Clinical Research Center for Birth Defects, The Affiliated Hospital of Southwest Medical University, Luzhou, China
- 3National Heart Research Institute of Singapore, Singapore, Singapore
In recent years, cardiovascular disease (CVD) continues to be the leading cause of global disease burden. Extensive efforts have been made across basic, translational, and clinical research domains to curb the CVD epidemic and improve the health of the population. The successful completion of the Human Genome Project catapulted sequencing technology into the mainstream and aroused the interests of clinicians and scientific researchers alike. Advances in single-cell RNA sequencing (scRNA-seq), which is based on the transcriptional phenotypes of individual cells, have enabled the investigation of cellular fate, heterogeneity, and cell–cell interactions, as well as cell lineage determination, at a single-cell resolution. In this review, we summarize recent findings on the embryological development of the cardiovascular system and the pathogenesis and treatment of cardiovascular disease, as revealed by scRNA-seq technology. In particular, we discuss how scRNA-seq can help identify potential targets for the treatment of cardiovascular diseases and conclude with future perspectives for scRNA-seq.
Introduction
Cardiovascular disease (CVD), primarily ischemic heart disease and stroke, remains the leading cause of mortality and morbidity across the world (GBD 2019 Diseases and Injuries Collaborators, 2020). Recent evidence indicates that the prevalence of total CVD has continued to increase, nearly doubling from 271 million in 1990 to 523 million in 2019. Similarly, the number of CVD-related deaths has steadily increased from 12.1 million in 1990 reaching 18.6 million in 2019. The burden of CVD continues its decade-long rise in almost all countries except high-income countries. In most low- and middle-income countries, the overall burden of CVD, in terms of morbidity and mortality, is higher than that in high-income countries (Roth et al., 2020). Therefore, the promotion of cardiovascular health remains an important task for clinicians and scientific researchers.
With the completion of the Human Genome Project and the rapid development of sequencing technologies, there has been a growing interest in single-cell genomic and transcriptomic studies. Single-cell sequencing investigates biodiversity and heterogeneity at a single-cell resolution (Grün and van Oudenaarden, 2015). Since it was rated as “The Method of the Year” by Nature Methods in 2013, multiple single-cell sequencing methods and platforms have been developed for various applications. In general, single-cell genome sequencing is used to detect copy number variations and single-nucleotide variations within the genome of individual cells (Adey, 2021). Similarly, single-cell epigenome sequencing is used to detect modifications, such as methylation of DNA and histones, in the epigenome of individual cells (Prompsy et al., 2020).
In contrast, single-cell RNA sequencing (scRNA-seq) is used to detect the mRNA level of individual cells (Paik et al., 2020). In the field of modern cardiovascular research, it is a powerful tool for elucidating cellular heterogeneity, cell types, intercellular crosstalk, and trajectory analysis of cellular dynamics (Litviňuková et al., 2020; Cheng et al., 2021; Grancharova et al., 2021; Kan et al., 2021). In this review, we summarize published research using scRNA-seq on the embryological development of the cardiovascular system and pathogenesis of CVD. This is followed by a discussion on how progress in scRNA-seq technology can enable the identification of new targets for cell-based therapy. In addition, we compare the differences and similarities between scRNA-seq and bulk RNA sequencing and discuss the potential applications of scRNA-seq in cardiovascular research.
scRNA-Seq and Cardiovascular Development
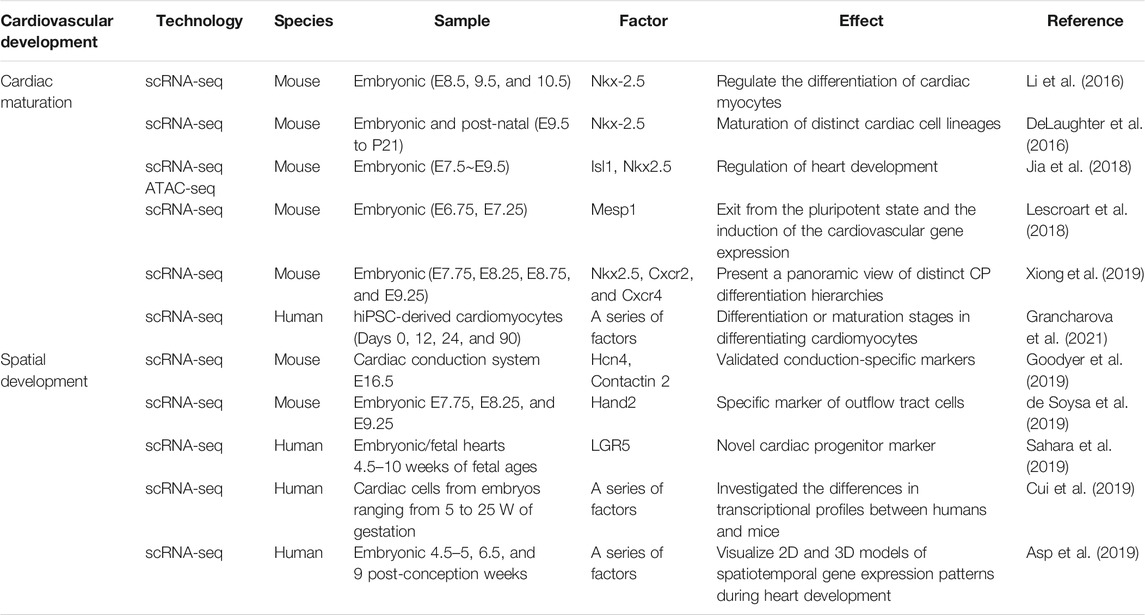
Compared to adult hearts, embryonic and neonatal cardiac tissues are easier to digest and isolate single cardiomyocytes with higher viability. The lineage and heterogeneity of various cells in the process of cardiac development can be comprehensively analyzed by scRNA-seq (Table 1). In 2016, a single-cell atlas of mouse embryonic and postnatal hearts was generated by two independent groups (DeLaughter et al., 2016; Li et al., 2016). They found that the homeobox gene Nkx-2.5 plays an important role in cardiac maturation. By a combination of scRNA-seq and Assay for Transposase-Accessible Chromatin using sequencing (ATAC-seq), Jia et al. (2018) studied mouse cardiac progenitor cells (CPCs) from E7.5 to E9.5 and found that the fate transition of cardiac progenitor cells is closely related to the chromatin state. This correlation depends on the changes of Isl1 and Nkx-2.5. Later on, Mesp1 was identified as another important factor for promoting transition from cardiac progenitors to cardiomyocytes (Lescroart et al., 2018). By single-cell transcriptomic analysis of Nkx-2.5 and Isl1 lineages, Xiong et al. (2019) presented a panoramic view of distinct CP differentiation hierarchies, supported the fact that the first and second heart fields (FHF and SHF, respectively) take different differentiation paths, and revealed the importance of Nkx-2.5 in Cxcr2-and Cxcr4-activated transcription in SHF.
In addition, scRNA-seq contributes to the identification of factors responsible for the spatial development of the heart. Recently, uniquely expressed genes responsible for the development of the cardiac conduction system were identified in an embryonic mouse (Goodyer et al., 2019). Similarly, Hand2 was found to serve as a marker for outflow tract cells but not right ventricular cells, as proposed previously (de Soysa et al., 2019). On the contrary, the LGR5+ cardiac progenitor cells were found to be located in the outflow tract region of the human embryonic heart (Cui et al., 2019; Sahara et al., 2019). By a combination of spatial transcriptomics, Asp et al. (2019) generated a 3D single-cell transcriptional atlas of the embryonic human heart.
As for human vascular cells, Su et al. (2018) used scRNA-seq to study the dynamic lineage of coronary arteries and found that COUP-TF2 blocked arterial formation in the pre-arterial stage. Grancharova et al. (2021) used scRNA-seq to profile gene expression during the differentiation of human-induced pluripotent stem cells (hiPSCs) to cardiomyocytes, revealing genetic features which could identify stages of differentiation or maturation in differentiating cardiomyocytes. Currently, scRNA-seq has become an important tool for studying embryological development of the cardiovascular system. The applications of scRNA-seq will help dissect the mechanisms underlying common CVDs.
scRNA-Seq and Cardiovascular Disease
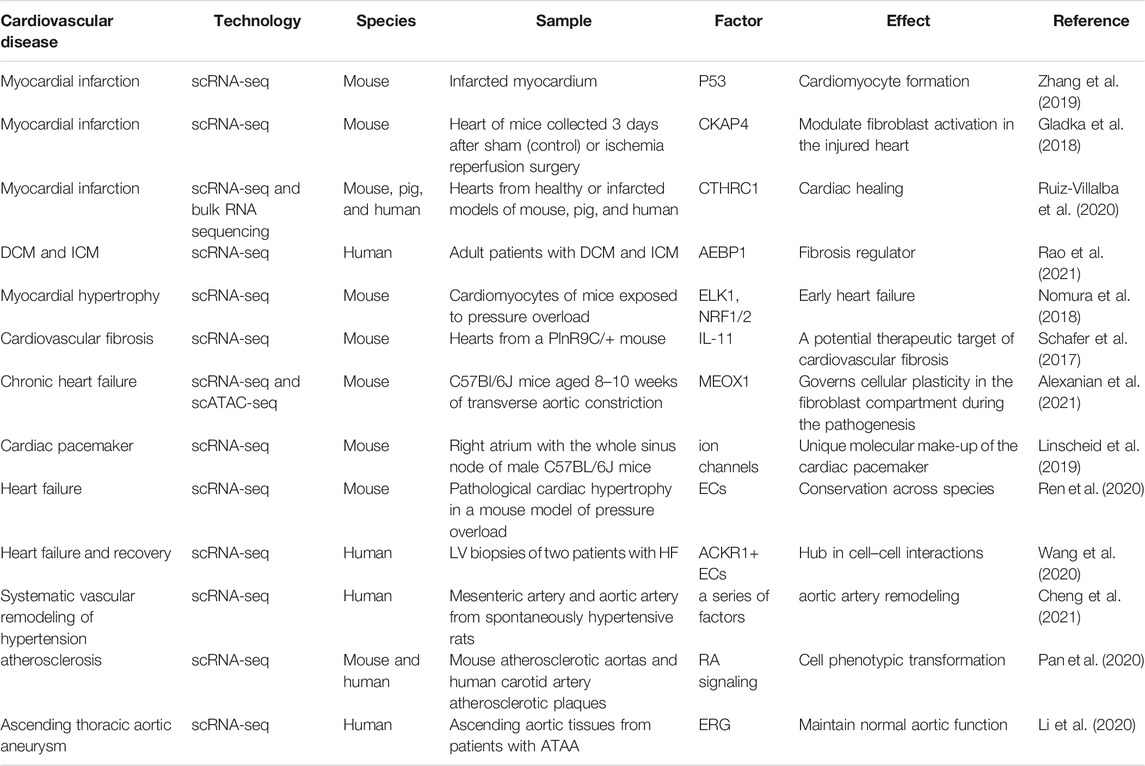
scRNA-seq can comprehensively provide mRNA information of each cell in the heart and blood vessel in disease states, which can enable the discovery of new therapeutic targets for CVDs (Table 2). In a mouse model of ischemic heart disease, including myocardial infarction (MI), Zhang et al. (2019) observed limited but measurable myogenesis. However, Gladka et al. (2018) did not find any evidence of significant cardiac proliferation but observed excessive fibrosis after MI instead. They found that CKAP4 regulates fibroblast activation in damaged hearts and can be used as a marker for activated cardiac myofibroblasts. The heterogeneity of cardiac fibroblasts was further identified in a mouse MI heart in one subpopulation, Cthrc1 was shown to be involved in the scar healing process, and can, therefore, serve as a potential therapeutic target (Ruiz-Villalba et al., 2020). By scRNA-seq of infarcted and non-infarcted regions dissected from human heart samples, one novel transcription factor, AEBP1, was identified to regulate cardiac fibrosis during MI (Rao et al., 2021).
Nomura et al. (2018) found that in pressure overload–induced cardiac hypertrophy and heart failure mouse models, ERK1/2 and NRF1/2 are involved in the regulation of early cardiac hypertrophy, while p53 is mainly involved in cardiac remodeling in the decompensated phase. Interestingly, Schafer et al. (2017) performed genome sequencing on the hearts of multiple fibrosis mice models and found that IL-11 is specifically expressed in activated cardiac fibroblasts and is a new specific marker for cardiac fibrosis. As a result, interfering with IL-11 gene expression or blocking the binding of IL-11 to receptors can effectively prevent and treat cardiac fibrosis, suggesting that IL-11 is an important therapeutic target. By combining scRNA-seq and scATAC-seq, Alexanian et al. (2021) recently identified that bromodomain and extraterminal (BET)–dependent regulation of MEOX1, a fibroblast-specific enhancer, controls fibroblast activation during chronic heart failure. In light of this, they suggested new therapeutic approaches targeting MEOX1 expression be developed instead of systemic BET inhibitors that have broad effects to alleviate the pathogenesis of fibrotic diseases.
Because of the large size of intact adult cardiomyocytes, researchers have used nuclei extracted from myocytes for scRNA-seq, so-called snRNA-seq. Linscheid et al. (2019) performed snRNA-seq of the sinoatrial node from the adult mouse heart and found that membrane clock–related channel proteins have a higher expression in the cardiomyocytes of the sinoatrial node than that of the atrium. More recently, Litviňuková et al. (2020) combined scRNA-seq and snRNA-seq methods to generate an atlas of adult human hearts. Another approach to carry out scRNA-seq of the adult human heart is to use FACS or a platform with a large-bore nozzle to pick a single myocyte. Using this technique, Ren et al. (2020) analyzed the spatiotemporal interplay among adult cardiomyocytes and non-cardiomyocytes in a pressure overload mouse model. They revealed that targeting macrophages by dapagliflozin, as well as TD139 and arglabin, could prevent cardiac hypertrophy from progressing to heart failure. For human heart failure, Wang et al. (2020) proposed that changes in heart function are most correlated with cardiac contractility and metabolism. They found that injection of ACKR1+-endothelial cells preserved cardiac function after injury.
Coming to blood vessels, Cheng et al. (2021) described the first cell atlas of resistant and conductive arteries in hypertensive rats. The dysregulated gene profile of individual cells during hypertensive vascular remodeling was characterized in artery type–specific and cell type–specific manners. For atherosclerosis, Gu et al. (2019) sequenced normal and apoE−/− mouse single cells and mapped the aortic adventitia, including endothelial cells, immune cells, adventitia mesenchymal cells, and other cell subgroups. It was revealed that the activation of macrophages triggers an inflammatory response in the blood vessels of apoE−/− mice, suggesting that the interaction between adventitial cells and immune cells is crucial in the early stages of atherosclerosis.
In addition, smooth muscle cells were reported to transition to macrophage- and fibrochondrocyte-like cells during atherosclerosis, a phenomenon regulated by the retinoic acid pathway (Pan et al., 2020). Recently, scRNA-seq was performed on patients with ascending thoracic aortic aneurysm (ATAA) and ascending aortic tissue of controls (Li et al., 2020). The authors not only described cellular composition and molecular alteration of the ascending aortic wall during the ATAA but also indicated the critical role of the ERG gene in the function of the aortic wall.
scRNA-Seq and Cell-Based Therapy
In recent years, cardiovascular cells differentiated from human embryonic stem cells or hiPSCs have provided a new source of cells for repairing or regenerating the injured heart. Transplantation of cardiovascular cells was found to improve wall stress, cardiac metabolism, and contractile performance in a porcine heart model of MI (Xiong et al., 2013; Ye et al., 2014). Genetic modification of hiPSC-CMs to overexpress angiopoietin-1 or co-administration of hiPSC-CMs with thymosin β4 (Tb4) not only improved neovascularization of the infarcted myocardium but also improved hiPSC-CM viability, proliferation, and engraftment (Tan et al., 2021; Tao et al., 2021). Similarly, hiPSC-CMs overexpressing cyclin D2 promoted myocyte proliferation in both donor cells and recipient porcine hearts (Zhao et al., 2021).
However, the insufficient maturity of cardiomyocyte differentiation in vitro and the complexity of differentiated cardiomyocyte subsets are still unsolved questions (Lundy et al., 2013; Tan and Ye, 2018). By scRNA-seq of the mouse heart at different time points during differentiation and characterization of 1) CMs derived from stem cells and 2) CMs and ECs derived from a congenital heart disease mouse model, DeLaughter et al. (2016) distinguished and contrasted the developmental lineages of the cells of the left atria, primordial ventricle, and subsequently left and right ventricles at time points spanning embryonic to post-natal cardiac development and verified that the maturity of pluripotent stem cell–derived cardiomyocytes is positively correlated with the culture duration.
Ni et al. (2021) suggested the application of glucose starvation for stem cell differentiation to obtain homologous and mature cardiomyocytes. Meanwhile, Cho et al. (2017) investigated the difference between differentiated cardiomyocytes (either simply cultured or implanted into the heart of newborn rats) and adult cardiomyocytes by scRNA-seq and found that implantation into the heart of newborn rats could promote the maturation of stem cell–derived cardiomyocytes. This experiment not only established a stem cell–based adult arrhythmic right ventricular cardiomyopathy model but also indicated the reliability of cell-based therapy.
Furthermore, Monsanto et al. (2020) found scaffold-free three-dimensional (3D) microenvironments spontaneously formed by mesenchymal stem cells, endothelial progenitor cells, and c-Kit + cardiac interstitial cells cultured together, which was termed “CardioCluster,” in which the expression of stem cell–relevant factors, adhesion/extracellular matrix molecules, and cytokines was detected by scRNA-seq profiling. Afterward, Monsanto et al. injected the CardioCluster into murine MI models that were followed for 20 weeks and found that it improved cell retention and capillary density with preservation of cardiomyocyte size and long-term cardiac function. Despite cellular therapy for cardiac diseases being the focus of intensive research efforts, progress toward cardiac structural and functional recovery remains limited. Future studies should focus on developing different strategies to boost the potency of stem cell repair.
scRNA-Seq and Bulk and Spatial RNA Sequencing
Although bulk RNA sequencing (bulk RNA-seq) is the basis for the development of scRNA-seq and spatial RNA sequencing (spRNA-seq), the latter two can offer transcriptional information at a single-cell level or near single-cell level (Yifan et al., 2020). In addition, data from scRNA-seq and spRNA-seq can be corroborated to reveal the spatial information of individual cells (Stark et al., 2019). For example, scRNA-seq can confirm and enrich the regional gene expression data obtained by spRNA-seq to generate a 3D atlas of the human embryonic heart (Asp et al., 2019). Similarly, by combining scRNA-seq and spRNA-seq, one subpopulation of cardiomyocytes overexpressing Nrf1 has been identified near the infarcted region of the heart (Cui et al., 2021). Thus, the activation of the Nrf1 pathway represents a new therapeutic approach for cardiac injury.
Future Perspectives
scRNA-seq provides a powerful tool for studying cell heterogeneity, tracking cell dynamics, and subsequently finding new therapeutic strategies for CVDs. It has, thus, opened a fresh chapter in the field of modern cardiovascular research. With constant improvement in scRNA-seq technology and the combination of multiomics and spatiotemporal analysis, scRNA-seq will play a greater role in the field of cardiovascular research. In particular, the comprehensive study and analysis of genomics, epigenomics, and transcriptomics at the single-cell level holds promise for the elucidation of gene regulatory mechanisms at a single-cell resolution, contributes to individualized therapy of CVD in clinical practice, improves therapeutic efficacy and clinical outcomes, and facilitates the practice of precision medicine.
Author Contributions
YF and HZ conducted the literature review and wrote the first draft of the manuscript. XL assisted in reviewing literature. JL, KX, and XF participated in the procedures. LY and GL edited and revised the manuscript. All authors have substantially contributed to the article and approved the submitted version.
Funding
This research was supported by NSFC 81870261, Sichuan Youth Science and Technology Innovation Research Team 2020JDTD0024, and Collaborative Innovation Center for the Prevention and Treatment of Cardiovascular Diseases in Sichuan Province xtcx 2019-6 and xtcx 2019-07.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Adey, A. C. (2021). Tagmentation-based Single-Cell Genomics. Genome Res. 31 (10), 1693–1705. doi:10.1101/gr.275223.121
Alexanian, M., Przytycki, P. F., Micheletti, R., Padmanabhan, A., Ye, L., Travers, J. G., et al. (2021). A Transcriptional Switch Governs Fibroblast Activation in Heart Disease. Nature 595 (7867), 438–443. doi:10.1038/s41586-021-03674-1
Asp, M., Giacomello, S., Larsson, L., Wu, C., Fürth, D., Qian, X., et al. (2019). A Spatiotemporal Organ-wide Gene Expression and Cell Atlas of the Developing Human Heart. Cell 179 (7), 1647–1660. doi:10.1016/j.cell.2019.11.025
Cheng, J., Gu, W., Lan, T., Deng, J., Ni, Z., Zhang, Z., et al. (2021). Single-cell RNA Sequencing Reveals Cell Type- and Artery Type-specific Vascular Remodelling in Male Spontaneously Hypertensive Rats. Cardiovasc. Res. 117 (4), 1202–1216. doi:10.1093/cvr/cvaa164
Cho, G.-S., Lee, D. I., Tampakakis, E., Murphy, S., Andersen, P., Uosaki, H., et al. (2017). Neonatal Transplantation Confers Maturation of PSC-Derived Cardiomyocytes Conducive to Modeling Cardiomyopathy. Cel Rep. 18 (2), 571–582. doi:10.1016/j.celrep.2016.12.040
Cui, M., Atmanli, A., Morales, M. G., Tan, W., Chen, K., Xiao, X., et al. (2021). Nrf1 Promotes Heart Regeneration and Repair by Regulating Proteostasis and Redox Balance. Nat. Commun. 12 (1), 5270. doi:10.1038/s41467-021-25653-w
Cui, Y., Zheng, Y., Liu, X., Yan, L., Fan, X., Yong, J., et al. (2019). Single-Cell Transcriptome Analysis Maps the Developmental Track of the Human Heart. Cel Rep. 26 (7), 1934–1950. doi:10.1016/j.celrep.2019.01.079
de Soysa, T. Y., Ranade, S. S., Okawa, S., Ravichandran, S., Huang, Y., Salunga, H. T., et al. (2019). Single-cell Analysis of Cardiogenesis Reveals Basis for Organ-Level Developmental Defects. Nature 572 (7767), 120–124. doi:10.1038/s41586-019-1414-x
DeLaughter, D. M., Bick, A. G., Wakimoto, H., McKean, D., Gorham, J. M., Kathiriya, I. S., et al. (2016). Single-Cell Resolution of Temporal Gene Expression during Heart Development. Developmental Cel 39 (4), 480–490. doi:10.1016/j.devcel.2016.10.001
GBD 2019 Diseases and Injuries Collaborators (2020). Global Burden of 369 Diseases and Injuries in 204 Countries and Territories, 1990-2019: A Systematic Analysis for the Global Burden of Disease Study 2019. Lancet 396, 1204–1222. doi:10.1016/s0140-6736(20)30925-9
Gladka, M. M., Molenaar, B., de Ruiter, H., van der Elst, S., Tsui, H., Versteeg, D., et al. (2018). Single-Cell Sequencing of the Healthy and Diseased Heart Reveals Cytoskeleton-Associated Protein 4 as a New Modulator of Fibroblasts Activation. Circulation 138 (2), 166–180. doi:10.1161/circulationaha.117.030742
Goodyer, W. R., Beyersdorf, B. M., Paik, D. T., Tian, L., Li, G., Buikema, J. W., et al. (2019). Transcriptomic Profiling of the Developing Cardiac Conduction System at Single-Cell Resolution. Circ. Res. 125 (4), 379–397. doi:10.1161/circresaha.118.314578
Grancharova, T., Gerbin, K. A., Rosenberg, A. B., Roco, C. M., Arakaki, J. E., DeLizo, C. M., et al. (2021). A Comprehensive Analysis of Gene Expression Changes in a High Replicate and Open-Source Dataset of Differentiating hiPSC-Derived Cardiomyocytes. Sci. Rep. 11 (1), 15845. doi:10.1038/s41598-021-94732-1
Grün, D., and van Oudenaarden, A. (2015). Design and Analysis of Single-Cell Sequencing Experiments. Cell 163 (4), 799–810. doi:10.1016/j.cell.2015.10.039
Gu, W., Ni, Z., Tan, Y.-Q., Deng, J., Zhang, S.-J., Lv, Z.-C., et al. (2019). Adventitial Cell Atlas of Wt (Wild Type) and ApoE (Apolipoprotein E)-Deficient Mice Defined by Single-Cell RNA Sequencing. Atvb 39 (6), 1055–1071. doi:10.1161/atvbaha.119.312399
Jia, G., Preussner, J., Chen, X., Guenther, S., Yuan, X., Yekelchyk, M., et al. (2018). Single Cell RNA-Seq and ATAC-Seq Analysis of Cardiac Progenitor Cell Transition States and Lineage Settlement. Nat. Commun. 9 (1), 4877. doi:10.1038/s41467-018-07307-6
Kan, H., Zhang, K., Mao, A., Geng, L., Gao, M., Feng, L., et al. (2021). Single-cell Transcriptome Analysis Reveals Cellular Heterogeneity in the Ascending Aortas of normal and High-Fat Diet-Fed Mice. Exp. Mol. Med. 53 (9), 1379–1389. doi:10.1038/s12276-021-00671-2
Lescroart, F., Wang, X., Lin, X., Swedlund, B., Gargouri, S., Sànchez-Dànes, A., et al. (2018). Defining the Earliest Step of Cardiovascular Lineage Segregation by Single-Cell RNA-Seq. Science 359 (6380), 1177–1181. doi:10.1126/science.aao4174
Li, G., Xu, A., Sim, S., Priest, J. R., Tian, X., Khan, T., et al. (2016). Transcriptomic Profiling Maps Anatomically Patterned Subpopulations Among Single Embryonic Cardiac Cells. Developmental Cel 39 (4), 491–507. doi:10.1016/j.devcel.2016.10.014
Li, Y., Ren, P., Dawson, A., Vasquez, H. G., Ageedi, W., Zhang, C., et al. (2020). Single-Cell Transcriptome Analysis Reveals Dynamic Cell Populations and Differential Gene Expression Patterns in Control and Aneurysmal Human Aortic Tissue. Circulation 142 (14), 1374–1388. doi:10.1161/circulationaha.120.046528
Linscheid, N., Logantha, S. J. R. J., Poulsen, P. C., Zhang, S., Schrölkamp, M., Egerod, K. L., et al. (2019). Quantitative Proteomics and Single-Nucleus Transcriptomics of the Sinus Node Elucidates the Foundation of Cardiac Pacemaking. Nat. Commun. 10 (1), 2889. doi:10.1038/s41467-019-10709-9
Litviňuková, M., Talavera-López, C., Maatz, H., Reichart, D., Worth, C. L., Lindberg, E. L., et al. (2020). Cells of the Adult Human Heart. Nature 588 (7838), 466–472. doi:10.1038/s41586-020-2797-4
Lundy, S. D., Zhu, W.-Z., Regnier, M., and Laflamme, M. A. (2013). Structural and Functional Maturation of Cardiomyocytes Derived from Human Pluripotent Stem Cells. Stem Cell Development 22 (14), 1991–2002. doi:10.1089/scd.2012.0490
Monsanto, M. M., Wang, B. J., Ehrenberg, Z. R., Echeagaray, O., White, K. S., Alvarez, R., et al. (2020). Enhancing Myocardial Repair with CardioClusters. Nat. Commun. 11 (1), 3955. doi:10.1038/s41467-020-17742-z
Ni, X., Xu, K., Zhao, Y., Li, J., Wang, L., Yu, F., et al. (2021). Single-cell Analysis Reveals the Purification and Maturation Effects of Glucose Starvation in hiPSC-CMs. Biochem. Biophysical Res. Commun. 534, 367–373. doi:10.1016/j.bbrc.2020.11.076
Nomura, S., Satoh, M., Fujita, T., Higo, T., Sumida, T., Ko, T., et al. (2018). Cardiomyocyte Gene Programs Encoding Morphological and Functional Signatures in Cardiac Hypertrophy and Failure. Nat. Commun. 9 (1), 4435. doi:10.1038/s41467-018-06639-7
Paik, D. T., Cho, S., Tian, L., Chang, H. Y., and Wu, J. C. (2020). Single-cell RNA Sequencing in Cardiovascular Development, Disease and Medicine. Nat. Rev. Cardiol. 17 (8), 457–473. doi:10.1038/s41569-020-0359-y
Pan, H., Xue, C., Auerbach, B. J., Fan, J., Bashore, A. C., Cui, J., et al. (2020). Single-Cell Genomics Reveals a Novel Cell State during Smooth Muscle Cell Phenotypic Switching and Potential Therapeutic Targets for Atherosclerosis in Mouse and Human. Circulation 142 (21), 2060–2075. doi:10.1161/circulationaha.120.048378
Prompsy, P., Kirchmeier, P., Marsolier, J., Deloger, M., Servant, N., and Vallot, C. (2020). Interactive Analysis of Single-Cell Epigenomic Landscapes with ChromSCape. Nat. Commun. 11 (1), 5702. doi:10.1038/s41467-020-19542-x
Rao, M., Wang, X., Guo, G., Wang, L., Chen, S., Yin, P., et al. (2021). Resolving the Intertwining of Inflammation and Fibrosis in Human Heart Failure at Single-Cell Level. Basic Res. Cardiol. 116 (1), 55. doi:10.1007/s00395-021-00897-1
Ren, Z., Yu, P., Li, D., Li, Z., Liao, Y., Wang, Y., et al. (2020). Single-Cell Reconstruction of Progression Trajectory Reveals Intervention Principles in Pathological Cardiac Hypertrophy. Circulation 141 (21), 1704–1719. doi:10.1161/circulationaha.119.043053
Roth, G. A., Mensah, G. A., Johnson, C. O., Addolorato, G., Ammirati, E., Baddour, L. M., et al. (2020). Global Burden of Cardiovascular Diseases and Risk Factors, 1990-2019: Update from the GBD 2019 Study. J. Am. Coll. Cardiol. 76 (25), 2982–3021. doi:10.1016/j.jacc.2020.11.010
Ruiz-Villalba, A., Romero, J. P., Hernández, S. C., Vilas-Zornoza, A., Fortelny, N., Castro-Labrador, L., et al. (2020). Single-Cell RNA Sequencing Analysis Reveals a Crucial Role for CTHRC1 (Collagen Triple Helix Repeat Containing 1) Cardiac Fibroblasts after Myocardial Infarction. Circulation 142 (19), 1831–1847. doi:10.1161/circulationaha.119.044557
Sahara, M., Santoro, F., Sohlmér, J., Zhou, C., Witman, N., Leung, C. Y., et al. (2019). Population and Single-Cell Analysis of Human Cardiogenesis Reveals Unique LGR5 Ventricular Progenitors in Embryonic Outflow Tract. Developmental Cel 48 (4), 475–490. e477. doi:10.1016/j.devcel.2019.01.005
Schafer, S., Viswanathan, S., Widjaja, A. A., Lim, W.-W., Moreno-Moral, A., DeLaughter, D. M., et al. (2017). IL-11 Is a Crucial Determinant of Cardiovascular Fibrosis. Nature 552 (7683), 110–115. doi:10.1038/nature24676
Stark, R., Grzelak, M., and Hadfield, J. (2019). RNA Sequencing: the Teenage Years. Nat. Rev. Genet. 20 (11), 631–656. doi:10.1038/s41576-019-0150-2
Su, T., Stanley, G., Sinha, R., D’Amato, G., Das, S., Rhee, S., et al. (2018). Single-cell Analysis of Early Progenitor Cells that Build Coronary Arteries. Nature 559 (7714), 356–362. doi:10.1038/s41586-018-0288-7
Tan, S. H., Loo, S. J., Gao, Y., Tao, Z. H., Su, L. P., Wang, C. X., et al. (2021). Thymosin β4 Increases Cardiac Cell Proliferation, Cell Engraftment, and the Reparative Potency of Human Induced-Pluripotent Stem Cell-Derived Cardiomyocytes in a Porcine Model of Acute Myocardial Infarction. Theranostics 11 (16), 7879–7895. doi:10.7150/thno.56757
Tan, S. H., and Ye, L. (2018). Maturation of Pluripotent Stem Cell-Derived Cardiomyocytes: a Critical Step for Drug Development and Cell Therapy. J. Cardiovasc. Trans. Res. 11 (5), 375–392. doi:10.1007/s12265-018-9801-5
Tao, Z., Loo, S., Su, L., Tan, S., Tee, G., Gan, S. U., et al. (2021). Angiopoietin-1 Enhanced Myocyte Mitosis, Engraftment, and the Reparability of hiPSC-CMs for Treatment of Myocardial Infarction. Cardiovasc. Res. 117 (6), 1578–1591. doi:10.1093/cvr/cvaa215
Wang, L., Yu, P., Zhou, B., Song, J., Li, Z., Zhang, M., et al. (2020). Single-cell Reconstruction of the Adult Human Heart during Heart Failure and Recovery Reveals the Cellular Landscape Underlying Cardiac Function. Nat. Cel Biol. 22 (1), 108–119. doi:10.1038/s41556-019-0446-7
Xiong, H., Luo, Y., Yue, Y., Zhang, J., Ai, S., Li, X., et al. (2019). Single-Cell Transcriptomics Reveals Chemotaxis-Mediated Intraorgan Crosstalk during Cardiogenesis. Circ. Res. 125 (4), 398–410. doi:10.1161/circresaha.119.315243
Xiong, Q., Ye, L., Zhang, P., Lepley, M., Tian, J., Li, J., et al. (2013). Functional Consequences of Human Induced Pluripotent Stem Cell Therapy. Circulation 127 (9), 997–1008. doi:10.1161/circulationaha.112.000641
Ye, L., Chang, Y.-H., Xiong, Q., Zhang, P., Zhang, L., Somasundaram, P., et al. (2014). Cardiac Repair in a Porcine Model of Acute Myocardial Infarction with Human Induced Pluripotent Stem Cell-Derived Cardiovascular Cells. Cell Stem Cell 15 (6), 750–761. doi:10.1016/j.stem.2014.11.009
Yifan, C., Fan, Y., and Jun, P. (2020). Visualization of Cardiovascular Development, Physiology and Disease at the Single-Cell Level: Opportunities and Future Challenges. J. Mol. Cell Cardiol. 142, 80–92. doi:10.1016/j.yjmcc.2020.03.005
Zhang, Y., Gago-Lopez, N., Li, N., Zhang, Z., Alver, N., Liu, Y., et al. (2019). Single-cell Imaging and Transcriptomic Analyses of Endogenous Cardiomyocyte Dedifferentiation and Cycling. Cell Discov. 5, 30. doi:10.1038/s41421-019-0095-9
Zhao, M., Nakada, Y., Wei, Y., Bian, W., Chu, Y., Borovjagin, A. V., et al. (2021). Cyclin D2 Overexpression Enhances the Efficacy of Human Induced Pluripotent Stem Cell-Derived Cardiomyocytes for Myocardial Repair in a Swine Model of Myocardial Infarction. Circulation 144 (3), 210–228. doi:10.1161/circulationaha.120.049497
Keywords: single-cell RNA sequencing, cellular heterogeneity, cardiovascular system, therapeutic target, cell based therapy
Citation: Fan Y, Zhou H, Liu X, Li J, Xu K, Fu X, Ye L and Li G (2022) Applications of Single-Cell RNA Sequencing in Cardiovascular Research. Front. Cell Dev. Biol. 9:810232. doi: 10.3389/fcell.2021.810232
Received: 06 November 2021; Accepted: 14 December 2021;
Published: 31 January 2022.
Edited by:
Qiang Wu, Macau University of Science and Technology, Macao SAR, ChinaReviewed by:
David T. Paik, Stanford Cardiovascular Institute, United StatesCopyright © 2022 Fan, Zhou, Liu, Li, Xu, Fu, Ye and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lei Ye, ye.lei@nhcs.com.sg; Guang Li, liguang@swmu.edu.cn
†These authors have contributed equally to this work and share first authorship
 Yu Fan
Yu Fan