
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell Dev. Biol. , 12 January 2022
Sec. Molecular and Cellular Reproduction
Volume 9 - 2021 | https://doi.org/10.3389/fcell.2021.810118
This article is part of the Research Topic Fertilization in the Spotlight: Dynamics and Mechanisms of Sperm-Egg Interaction View all 16 articles
 Takafumi Matsumura1,2†
Takafumi Matsumura1,2† Taichi Noda1,3,4
Taichi Noda1,3,4 Yuhkoh Satouh1†
Yuhkoh Satouh1† Akane Morohoshi1,5
Akane Morohoshi1,5 Shunsuke Yuri6
Shunsuke Yuri6 Masaki Ogawa1,2
Masaki Ogawa1,2 Yonggang Lu1,7
Yonggang Lu1,7 Ayako Isotani1,6,7
Ayako Isotani1,6,7 Masahito Ikawa1,2,5,7,8*
Masahito Ikawa1,2,5,7,8*Fertilization occurs as the culmination of multi-step complex processes. First, mammalian spermatozoa undergo the acrosome reaction to become fusion-competent. Then, the acrosome-reacted spermatozoa penetrate the zona pellucida and adhere to and finally fuse with the egg plasma membrane. IZUMO1 is the first sperm protein proven to be essential for sperm-egg fusion in mammals, as Izumo1 knockout mouse spermatozoa adhere to but fail to fuse with the oolemma. However, the IZUMO1 function in other species remains largely unknown. Here, we generated Izumo1 knockout rats by CRISPR/Cas9 and found the male rats were infertile. Unlike in mice, Izumo1 knockout rat spermatozoa failed to bind to the oolemma. Further investigation revealed that the acrosome-intact sperm binding conceals a decreased number of the acrosome-reacted sperm bound to the oolemma in Izumo1 knockout mice. Of note, we could not see any apparent defects in the binding of the acrosome-reacted sperm to the oolemma in the mice lacking recently found fusion-indispensable genes, Fimp, Sof1, Spaca6, or Tmem95. Collectively, our data suggest that IZUMO1 is required for the sperm-oolemma binding prior to fusion at least in rat.
In sexual reproduction, the binding and fusion between the plasma membranes of spermatozoa and eggs are fundamental prerequisites for successful fertilization. In mammals, the spermatozoa ejaculated into the female reproductive tract undergo physiological, biochemical, and morphological changes (i.e., capacitation and acrosome reaction) to gain fertilization competency. The capacitated and acrosome-reacted sperm pass through the oocyte cumulus cell layer and the zona pellucida (ZP) and eventually bind to and fuse with the egg plasma membrane (Okabe, 2013). In the early days, several candidate sperm proteins involved in sperm-egg fusion have been identified using enzyme inhibitors and antibodies that block the in vitro fertilization (IVF) (Okabe, 2018). However, when their functions were challenged using the gene knockout (KO) mice, only IZUMO1 was found indispensable for the sperm-egg fusion (Okabe, 2018; Fujihara et al., 2020; Lamas-Toranzo et al., 2020; Noda et al., 2020).
IZUMO1 is an immunoglobulin (Ig)-like type I membrane protein that localizes to the acrosomal membrane and translocates to the sperm plasma membrane during the acrosome reaction (Satouh et al., 2012; Isotani et al., 2017). Izumo1 KO spermatozoa can penetrate the ZP but cannot fuse with the egg plasma membrane, resulting in male sterility (Inoue et al., 2005). Through an Avidity-based Extracellular Interaction Screen (AVEXIS) assay, IZUMO1 was found to interact with a glycosylphosphatidylinositol (GPI)-anchored protein, JUNO (also called FOLR4 or IZUMO1R), expressed on the oolemma (Bushell et al., 2008; Bianchi et al., 2014). The tertiary structures of IZUMO1, JUNO, and their complex have been intensively studied by X-ray crystallography (Aydin et al., 2016; Han et al., 2016; Kato et al., 2016; Nishimura et al., 2016; Ohto et al., 2016). The IZUMO1-JUNO, ligand-receptor interaction exists in mice and humans in a species-specific manner, implying that the two molecules might govern the gamete incompatibility among species (Bianchi and Wright, 2015).
Nevertheless, unlike other eukaryotic fusion proteins (e.g., HAP2/GCS1), neither of the two molecules possesses a fusogenic peptide or enables the cell-cell fusion in vitro by transient expression (Bianchi et al., 2014; Aydin et al., 2016). In addition, the cultured cells, such as HEK293T and COS-7 cells, transiently expressing mouse IZUMO1 can tightly bind to but not fuse with the ZP-free mouse eggs, suggesting the IZUMO1-JUNO interaction alone is insufficient for membrane fusion (Inoue et al., 2013; Ohto et al., 2016). Given these findings, researchers speculate that IZUMO1 and JUNO are implicated in sperm-egg binding rather than fusion. However, the number of Izumo1 KO mouse spermatozoa bound to the oolemma of the ZP-free eggs is comparable to the wild-type group (Inoue et al., 2005), rendering the detailed role of IZUMO1 during the sperm-egg fusion unclear.
In this study, we deleted Izumo1 in rats using the CRISPR/Cas9 system and analyzed the IZUMO1 function during rat fertilization. Combined with a reanalysis of the Izumo1 KO mice, our data indicate that IZUMO1 is required for binding the acrosome-reacted spermatozoa to oolemma prior to fusion.
All mice and rats were maintained under specific-pathogen-free conditions with ad libitum feeding. B6D2F1 mice were purchased from Japan SLC, Inc. B6D2-Tg(CAG/Su9-DsRed2,Acr3-EGFP)RBGS002Osb mice and STOCK Izumo1<tm1Osb> (Izumo1 KO or Izumo−/−) mice that are generated and maintained in our laboratory are used for generating Izumo1−/−; RBGS mice. B6D2-4930451I11Rik<em1Osb> (Fimp KO) mice, STOCK Llcfc1<em2Osb> (Sof1 KO) mice, B6D2-Spaca6<em2Osb> (Spaca6 KO) mice, and B6D2-Tmem95<em1Osb> (Tmem95 KO) mice are generated and maintained in our laboratory. WIAR rats were purchased from Institute for Animal Reproduction. Wistar rats were purchased from Charles River Laboratories Japan, Inc.
IZUMO1 sequences used for alignment are retrieved from the National Center for Biotechnology Information (NCBI) database: Homo sapiens (Human), NP_872381.2; Mus musculus (Mouse), NP_001018013.1; Rattus norvegicus (Rat), NP_001017514.1; Bos taurus (Cattle), XP_024835010.1; Strigops habroptila (Kakapo), XP_030366609.1; Terrapene carolina triunguis (Turtle), XP_026513927.1; and Danio rerio (Zebrafish), NP_001314727.1. The domain structure of IZUMO1 was derived from the NCBI database or was predicted by the Simple Modular Architecture Research Tool (SMART). Sequence alignment of IZUMO1 homologs was conducted and visualized using the Clustal Omega program (https://www.ebi.ac.uk/Tools/msa/clustalo/).
We generated a pX330 (#42230, Addgene, Cambridge, MA, USA) plasmid encoding a single-guide RNA (sgRNA) and the Cas9 protein to target the second exon of Izumo1 (target sequence: 5′-GGTGGCTGCAATAAAGACTT-3′). The DNA cleavage activity of this plasmid was validated as described previously (Mashiko et al., 2013). Superovulated WIAR (3–4 weeks old) females were mated with the mature males, and the two-pronuclear zygotes were collected from the oviduct. The circular pX330 plasmid was injected into one pronucleus of each zygote at concentrations of 5 ng/μl, 10 ng/μl, or 20 ng/μl (Mashiko et al., 2013). The following primer set was used for genotyping PCR of rat Izumo1 (Forward: 5′-TGTTTAGCACCCTCCTCCCTGC-3′, Reverse: 5′-AGAGAGTATGTATCTCTGCCTGTCCTGG-3′).
The total RNA was collected from the adult rat testes, purified using NucleoSpin RNA Plus (Takara, Kyoto, Japan), and reverse-transcribed into cDNA using SuperScript IV VILO Master Mix (Thermo Fisher Scientific, Waltham, MA, USA). PCR was performed using KOD DNA polymerase (Toyobo, Osaka, Japan); the PCR conditions for detecting rat Izumo1 were as follows: initial denaturing at 98°C for 30 s, followed by denaturing at 98°C for 15 s, annealing at 55°C for 5 s, and elongation 68°C for 5 s for 33 cycles. The PCR conditions for detecting rat Actb were as follows: initial denaturing at 98°C for 30 s, followed by denaturing at 98°C for 15 s, annealing at 58°C for 5 s, and elongation at 68°C for 5 s for 30 cycles. The PCR primers for rat Izumo1 were: Forward; 5′-ATGGGGCTACATTTTACACTC-3′ and Reverse; 5′-TTAGTTATCTGCTTCCTCAACC-3’. The PCR primers for rat Actb were: Forward; 5′-GTCCACCCGCGAGTACAAC-3′ and Reverse; 5′-GGATGCCTCTCTTGCTCTGG-3′.
The mouse monoclonal antibody against rat CD46 [MM1] (ab180652) was purchased from Abcam (Cambridge, UK). The rat monoclonal antibody against IZUMO1 (KS-64 125) (Ikawa et al., 2011) and rabbit polyclonal antibody against human IZUMO1 (Inoue et al., 2005) are generated in our laboratory as previously described. The goat anti-rat IgG Alexa Fluor 488-conjugated antibody (A11006), goat anti-mouse IgG antibody labeled with Alexa Fluor 568 (A11004), and goat anti-rabbit IgG Alexa Fluor Plus 488-conjugated antibody (A11034) were purchased from Thermo Fisher Scientific.
Testes were fixed in 4% paraformaldehyde in PBS and were processed for plastic embedding using Technovit® 8100 (Mitsui Chemicals, Tokyo, Japan) as described previously (Matsumura et al., 2019). Sections were made at a thickness of 5 µm and treated with 1% periodic acid (Wako, Osaka, Japan) for 10 min, followed by treatment with the Schiff’s reagent (Muto Pure Chemicals Co., Ltd., Tokyo, Japan) for 15 min. The sections were further stained with Mayer’s hematoxylin solution (Wako) and observed under a light microscope.
For detecting IZUMO1 in sperm cells, the frozen rat testes were disaggregated in PBS using BioMasher II (Nippi, Tokyo, Japan), and the germ cells were smeared on glass slides. The cells were air-dried on microscope slides, washed with PBS, and permeabilized with methanol. After treatment with image-iT FX Signal enhancer (Thermo Fisher Scientific) for 30 min at room temperature, the cells were incubated with the rabbit anti-human IZUMO1 polyclonal antibody (1:100) diluted by Can Get Signal A (Toyobo) at 4°C for overnight. After incubation with goat anti-rabbit IgG Alexa Fluor Plus 488 (1:200) and Lectin PNA Alexa Fluor 568 conjugate (Thermo Fisher Scientific) (1:200) for 1 h at room temperature, the spermatozoa were stained with Hoechst 33342 to visualize the nuclei (Dojindo, Kumamoto, Japan) and were mounted in Prolong Diamond (Thermo Fisher Scientific). For sperm morphological analysis, the cauda epididymal spermatozoa of male rats were incubated in the Human Tubal Fluid (HTF) medium (Aoto et al., 2011) for 10 min and observed under a BX50F phase-contrast microscope (Olympus, Tokyo, Japan).
Each male rat was caged with two Wistar females for 2 months. The number of delivered pups was counted. Copulation was confirmed by the formation of vaginal plugs or the presence of spermatozoa in the vaginal smears. Alternatively, superovulated Wistar females (3–4 weeks old) were mated with the male rats (3–6 months old). The eggs were collected from the oviducts of the superovulated females about 22 h after the hCG injection and mating. The eggs were observed under an inverted microscope (Olympus IX71), and the fertilization rate was recorded.
The HTF medium was prepared as described previously (Aoto et al., 2011). The cauda epididymal spermatozoa from male rats (3–6 months old) were preincubated for an hour in the HTF medium. The final sperm concentration was adjusted to 1 × 105 spermatozoa/mL in 100 µl HTF medium drops for insemination. The cumulus-oocyte complexes were collected from the oviductal ampulla of the superovulated females (3–4 weeks old), and the cumulus cells were removed by 0.03% hyaluronidase. The cumulus-free eggs were subsequently treated with 1 mg/ml collagenase to remove the ZP. To assess the sperm-egg fusion, the ZP-free eggs were fixed with 2% paraformaldehyde and stained by Hoechst 33342 after insemination for 5 h. Swollen sperm nuclei could be observed in the ooplasm when fusion occurred. The sperm-oolemma adhesion was assessed under a microscope at 0, 0.5, 2, and 5 h after insemination. To examine the acrosome status, spermatozoa were probed with anti-CD46 antibody (MM1) (1:200) and goat anti-mouse IgG Alexa Fluor 568-conjugated secondary antibody (1:200) in the HTF medium. An hour after insemination, the eggs were stained by Hoechst 33342, and the Z-stack images were captured using a BZ-X710 microscope (Keyence, Osaka, Japan). The total number of bound spermatozoa was counted. The spermatozoa in the HTF medium were smeared on microscope slides and observed under a fluorescence microscope.
The Toyoda-Yokoyama-Hosi (TYH) medium was prepared as described previously (Toyoda et al., 1971) and used for sperm preincubation, egg preparation, and insemination. The cauda epididymal spermatozoa from adult male mice (3–6 months) were preincubated for 2 h. The final concentration of spermatozoa was adjusted to 1 × 105 spermatozoa/mL in 100 µl TYH medium drops for insemination. Twenty minutes after insemination, the ZP-free eggs were moved to fresh medium drops, fixed with 0.8% paraformaldehyde, and stained by Hoechst 33342. The eggs were transferred to glass-bottomed chambers to observe Acr-EGFP fluorescence using a spinning-disk confocal microscope (Olympus). The cauda epididymal spermatozoa obtained from Fimp, Sof1, Tmem95, and Spaca6 KO male mice were preincubated at a concentration of 1–2 × 105 spermatozoa/mL for 2.5 h followed by probing with the monoclonal antibody against IZUMO1 (KS-64 125) (1:100) and goat anti-rat IgG Alexa Fluor 488-conjugated antibody (1:200) for another 30 min. The sperm concentration was adjusted to 1 × 105 spermatozoa/mL in 100 µl fresh medium drops and incubated with the ZP-free eggs for 30 min. The eggs were gently washed in fresh medium drops three times and fixed with 0.2% paraformaldehyde. The spermatozoa from B6D2F1 or Sof1+/−(Sof1 Het) male mice were used as control. The eggs were observed using a BZ-X710 microscope (Keyence). For calculating the total number of spermatozoa bound to the oolemma, the sperm tails were counted using a bright field microscope. The spermatozoa with and without green fluorescence in the acrosome were recognized as acrosome-intact and acrosome-reacted, respectively.
Statistical analyses were performed using a two-tailed student’s t-test (n ≧ 3). Differences were considered significant at p < 0.05. Data represent the means ± standard deviation (SD), and error bars indicate SD.
The Izumo1 KO rat strain was deposited under the name W; WIAR-Izumo1<em1Osb>, which will be available through the National BioResource Project for the Rat in Japan (NBRP; Kyoto, Japan). The B6D2-Tg(CAG/Su9-DsRed2, Acr3-EGFP)RBGS002Osb mice, STOCK Izumo1<tm1Osb> (Izumo1 KO or Izumo1−/−) mice, B6D2-4930451I11Rik<em1Osb> (Fimp KO), STOCK Llcfc1<em2Osb> (Sof1 KO), B6D2-Spaca6<em2Osb> (Spaca6 KO), and B6D2-Tmem95<em1Osb> (Tmem95 KO) mouse lines used in this study are available through either the Riken BioResource Center (Riken BRC; Tsukuba, Japan) or the Center for Animal Resources and Development, Kumamoto University (CARD; Kumamoto, Japan).
The amino acid sequences of IZUMO1 are conserved in mammals, birds, reptiles, and fish (Supplementary Figure S1). To knockout rat Izumo1 with the CRISPR/Cas9-mediated gene disruption, we designed a single-guide RNA (sgRNA) targeting the second exon of Izumo1 (Figure 1A). By injecting the sgRNA/Cas9 expressing plasmid (pX330) into the pronuclei of 215 zygotes, we obtained a male and a female mutant rat (2/13 pups, 15.3%) (Supplementary Figures S2A,B). As the mutant male rat (Izumo1−8/−29) showed sterility (Supplementary Figure 2C), we established a mutant line harboring 7-bp deletion by pairing the mutant female with a wild-type (WT) male (Figure 1B). We confirmed that Izumo1−7/−7 (KO) rats that Izumo1 mRNA derived from the mutant allele were transcribed in the testis (Figure 1C). But the translation of IZUMO1 is expected to be interrupted by a premature termination codon introduced by the frameshift (Supplementary Figure S2D). Indeed, the loss of IZUMO1 protein in the sperm acrosome was confirmed by immunostaining (Figure 1D). When Izumo1wt/−7 (Het) or Izumo1 KO male rats were individually caged with WT females for 2 months, all Izumo1 Het males sired pups (pups/plug: 6.79 ± 1.45), whereas Izumo1 KO males did not produce any pups despite the normal mating behavior indicated by formation of the copulation plugs (Figure 1E).
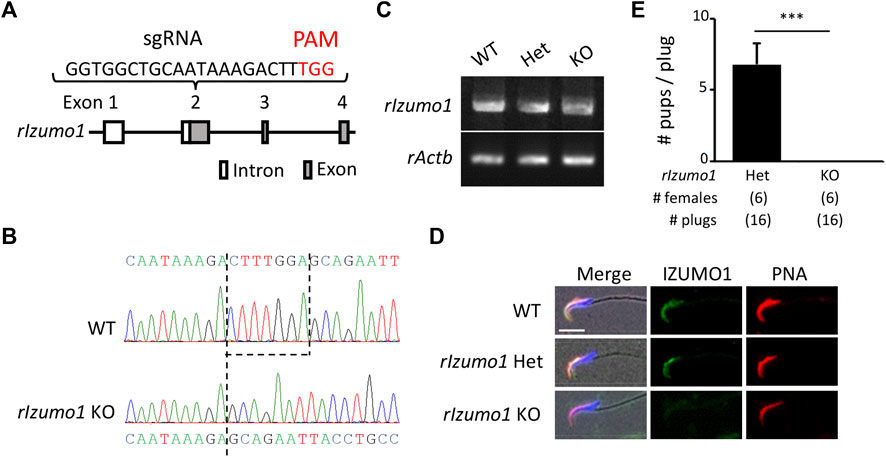
FIGURE 1. Generation of Izumo1-deficient rats using CRISPR/Cas9. (A) Design of sgRNA for disrupting Izumo1 in rats. White and gray boxes represent the untranslated and coding regions, respectively. (B) Genomic Izumo1 sequences of WT and mutant (Izumo1−7/−7) alleles. (C) Detection of rat Izumo1 mRNA in testis by RT-PCR. (D) Immunostaining of IZUMO1 in the testicular sperm from WT or mutant rats. The acrosomal caps and the nuclei were stained by the fluorophore-conjugated Lectin PNA and Hoechst 33342, respectively. Scale bar = 10 µm. (E) The average number of pups per copulatory plug. Three males each were tested for the Izumo1 Het and Izumo1 KO groups. ***p < 0.001.
Periodic acid-Schiff (PAS) staining of testis sections showed no sign of defects in the spermatogenesis of Izumo1 KO rats (Supplementary Figure S3A). In addition, there were no apparent abnormalities in the morphology of Izumo1 KO spermatozoa collected from the cauda epididymis (Supplementary Figure S3B). To determine the fertilizing ability of Izumo1 KO spermatozoa in vivo, we collected eggs from the oviduct of wild-type females 22 h after pairing with the KO males. Though Izumo1 KO spermatozoa penetrated the ZP at a comparable rate as the Het spermatozoa, no fertilized eggs were observed (Figures 2A,B).
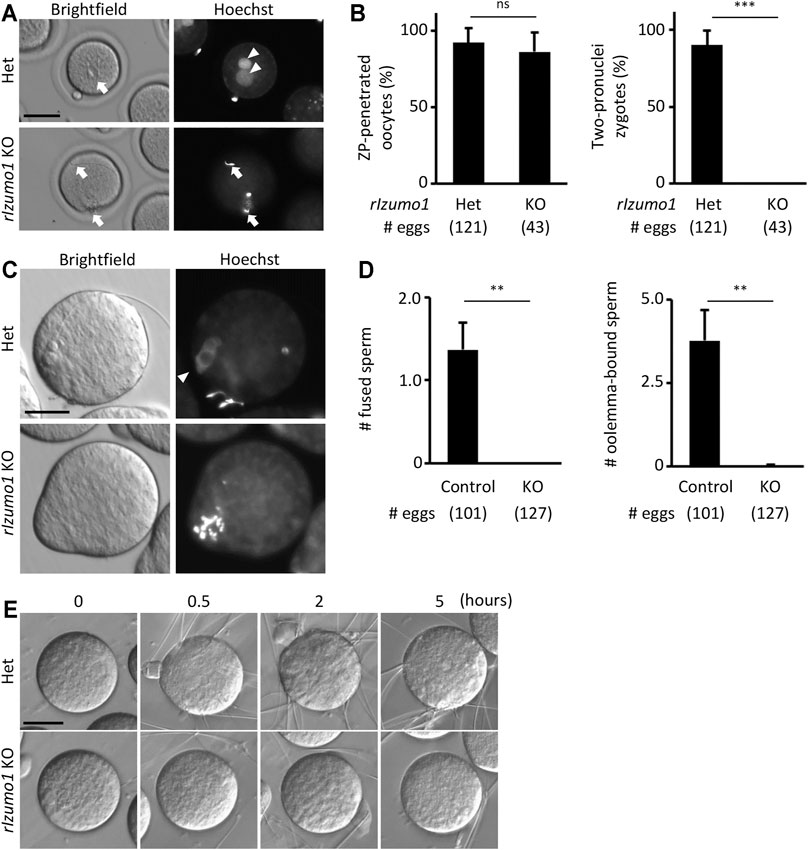
FIGURE 2. Impaired ability to bind to the oolemma in Izumo1 KO rat sperm. (A) The eggs recovered from the ampulla of the superovulated females mated with Izumo1 Het or Izumo1 KO male. The nuclei were stained by Hoechst 33342. Scale bar = 100 µm. (B) The average ratio of the ZP-penetrated eggs (left) or two-pronuclear zygotes (right) to the total recovered eggs. ns represents no statistical difference. ***p < 0.001. (C) The sperm-egg fusion was assessed by staining the nuclei with Hoechst 33342. Fused sperm are indicated by the fluorescence signals of the swollen sperm nuclei. Scale bar = 50 µm. (D) The average numbers of the fused spermatozoa (left) and oolemma-bound spermatozoa (right). Izumo1 WT and Het rat sperm were used as control. **p < 0.01 (E) The sperm-egg adhesion was captured at 0, 0.5, 2, and 5 h after insemination. Scale bar = 50 µm.
We next performed in vitro fertilization using the enzymatically ZP-removed eggs. While the WT and Het spermatozoa successfully adhered to and fused with the eggs after an hour of insemination (oolemma-bound and fused spermatozoa: 3.77 ± 0.94 and 1.36 ± 0.33 spermatozoa/egg, respectively), none of the Izumo1 KO spermatozoa adhered to or fused with the eggs (oolemma-bound and fused spermatozoa: 0.02 ± 0.03 and 0 ± 0 spermatozoa/egg, respectively) (Figures 2C,D). Even when we extended the insemination period up to 5 h, adhesion of the Izumo1 KO spermatozoa to the oolemma was not observed (Figure 2E, Supplementary Movies S1, S2). These findings suggest the sterility of Izumo1 KO rats is attributed to the inability of KO spermatozoa to bind to the oolemma.
In mice and hamsters, it is known that both the acrosome-intact and reacted spermatozoa can stick to the surface of the ZP-free eggs (Phillips and Yanagimachi, 1982; Calarco, 1991; Yanagimachi, 1994; Satouh et al., 2012). Since IZUMO1 is exposed to the sperm surface during the acrosome reaction (Satouh et al., 2012; Isotani et al., 2017), the attachment of the acrosome-intact spermatozoa to the oolemma does not involve the IZUMO1-JUNO interaction and does not lead to subsequent fusion between the two plasma membranes. Thus, in the following text, the acrosome-intact or reacted spermatozoa attachment to the oolemma is referred to as the fusion-incompetent or fusion-competent sperm adhesion, respectively. To rule out the fusion-incompetent sperm adhesion, we assessed the acrosome status of the live spermatozoa by immunostaining of CD46 (Supplementary Figure S4A). Like IZUMO1, CD46 is localized to the inner acrosomal membrane and is translocated to the sperm plasma membrane during the acrosome reaction (Mizuno et al., 2004). Since CD46 is inaccessible to the antibody before being exposed to the sperm surface, no fluorescence signals should be seen in the live acrosome-intact spermatozoa. As a result, 82.6 ± 12.4% of the spermatozoa adhered to the oolemma were acrosome-reacted, whereas 24.5 ± 11.2% of the spermatozoa in the medium were acrosome-reacted (Supplementary Figures S4B,C), indicating that the acrosome-intact spermatozoa are less prone to adhering to the oolemma compared with the acrosome-reacted spermatozoa in rats.
The lack of adhesion ability of the Izumo1 KO rat spermatozoa to the oolemma is distinct from our previous study, in which we could not see apparent defects in the adhesion of Izumo1 KO mouse spermatozoa to the oolemma (Inoue et al., 2005). Of note, it was reported that the acrosome-reacted spermatozoa hardly bind to the JUNO KO eggs in mice (Bianchi et al., 2014). Thus, we speculated that an excessive number of the acrosome-intact spermatozoa adhered to the oolemma might conceal a lack of fusion-competent adhesion by the acrosome-reacted spermatozoa. To address this concern, we generated Izumo1 KO mice carrying the RBGS (Red Body Green Spermatozoa) transgene that encodes EGFP in the acrosome and DsRed2 in the mitochondria to re-evaluate the sperm acrosome status during fertilization. The mouse spermatozoa were preincubated for 2 h and incubated with the ZP-free eggs for 20 min in the TYH medium. We did not see any differences in the average rates of the acrosome reaction in WT and Izumo1 KO spermatozoa (WT: 20.8 ± 15.8%; KO: 21.7 ± 11.0%). While the total numbers of the spermatozoa bound to the oolemma were comparable between the WT and KO groups (Figures 3A,B), the percentage of the oolemma-bound acrosome-reacted spermatozoa was drastically decreased in the Izumo1 KO mice (WT: 25.3 ± 11.3%; KO: 3.28 ± 3.78%, Figure 3C).
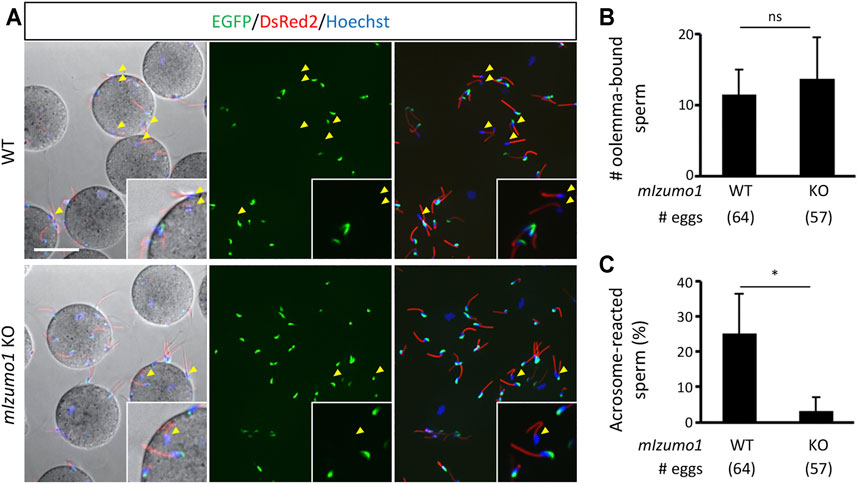
FIGURE 3. Reduced binding of the acrosome-reacted Izumo1 KO mouse sperm to the oolemma. (A) A representative image of mouse sperm adhered to egg membrane. Spermatozoa with the EGFP signals are acrosome-intact, whereas the ones without EGFP are acrosome-reacted (yellow arrowhead). DsRed2 is expressed in the sperm midpiece. The sperm nuclei were stained by Hoechst 33342. Scale bar = 50 µm (B) The total numbers of oolemma-bound spermatozoa. ns represents no statistical difference. (C) The ratios of the acrosome-reacted sperm to the total number of oolemma-bound sperm. *p < 0.05.
Recent studies uncovered novel sperm proteins necessary for sperm-egg fusion in addition to IZUMO1 (Lorenzetti et al., 2014; Fujihara et al., 2020; Lamas-Toranzo et al., 2020; Noda et al., 2020; Inoue et al., 2021; Noda et al., 2021), yet whether these molecules are involved in the gamete adhesion or fusion remains unclear. Hence, we performed the sperm-egg fusion assays using the Fimp, Sof1, Spaca6, and Tmem95 mutant mouse spermatozoa with the sperm acrosome status being unraveled using an anti-IZUMO1 antibody. As a result, we could not see apparent differences in both the total numbers of bound spermatozoa and the ratios of the acrosome-reacted spermatozoa to the total bound spermatozoa between the WT and KO groups (Figures 4A–C).
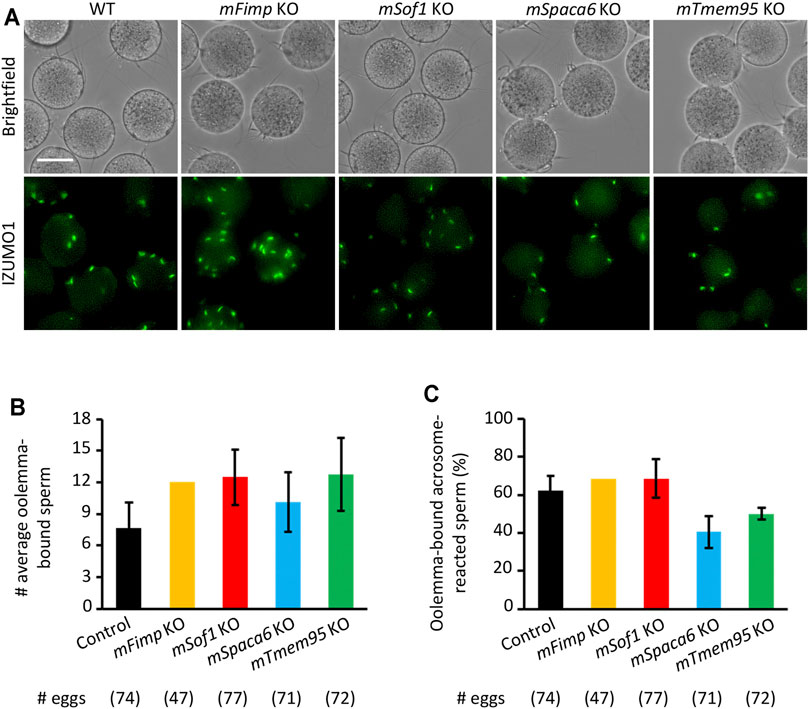
FIGURE 4. Mutant sperm lacking the novel fusion-related genes bind to the oolemma after the acrosome reaction. (A) Observation of the binding between WT eggs and WT or Fimp, Sof1, Tmem95, and Spaca6 KO spermatozoa. The acrosome-reacted sperm on the oolemma are indicated by the fluorescence signals of IZUMO1. Scale bar = 50 µm. (B) The numbers of total oolemma-bound spermatozoa. WT and Sof1 Het mouse sperm were used as control. (C) The ratio of the acrosome-reacted sperm to the total oolemma-bound sperm. WT and Sof1 Het mouse sperm were used as control.
The cell-cell membrane adhesion and fusion processes are considered independent biological events, each of which requires its suites of molecules. For instance, the gamete adhesion is potentiated by FUS1, whereas HAP2 regulates the gamete fusion in Chlamydomonas reinhardtii (Misamore et al., 2003; Liu et al., 2008; Aguilar et al., 2013; Hernandez and Podbilewicz, 2017). In mammal syngamy, studies using the cultured cell lines strongly suggest that IZUMO1 and JUNO are responsible for the sperm-egg adhesion instead of fusion per se – the HEK293T, COS-7, and K562 cells expressing IZUMO1 (Inoue et al., 2013; Chalbi et al., 2014; Ohto et al., 2016) or JUNO (Bianchi et al., 2014; Chalbi et al., 2014; Kato et al., 2016) attach to the ZP-free eggs or sperm, respectively, but never fused with them. To better understand the functions of IZUMO1 and JUNO, the plasma membrane fusion needs to be directly investigated using sperm and eggs. However, the ZP-intact eggs are not ideal for studying gamete fusion in vitro. With the presence of the ZP, sperm penetration and fusion timing is not controllable. In addition, the cortical reaction is triggered after incorporating the initial spermatozoon with the oolemma, thereby preventing additional spermatozoa from penetrating the ZP and approaching the oolemma. Therefore, the ZP should be removed mechanically by a piezo-driven micromanipulator or chemically by collagenase or acidic Tyrode before the fusion analysis (Yamagata et al., 2002; Inoue et al., 2005; Yamatoya et al., 2011; Nozawa et al., 2018). Nevertheless, the acrosomal-intact sperm, which cannot pass through the ZP or fuse with the oolemma, non-specifically bind to the egg membrane in a fusion-incompetent way in mice and hamsters (Phillips and Yanagimachi, 1982; Calarco, 1991; Yanagimachi, 1994).
Like mice and hamsters, rats have been used as a popular rodent model for spermatogenesis and reproductive toxicology (Russell et al., 1993; OECD, 2016). However, they have not been used intensively in the fertilization field due to a delay in developing a highly efficient and convenient in vitro fertilization system (Honda et al., 2019). Distinct to the previous findings using mice or hamsters, we discovered that the acrosome-intact sperm in rats are less likely to adhere to the oolemma than the acrosome-reacted sperm. Such uniqueness in the rats enables us to explicitly quantify the fusion-competent binding between the acrosome-reacted sperm and oolemma. In a previous study, the acrosome-intact human sperm adhered to the mouse egg plasma membrane but not to the human egg surface (Liu and Baker, 1994). Thus, it is tempting to speculate that the fusion-incompetent sperm adhesion in mice and hamsters might be attributed to the molecular architecture of the egg plasma membrane.
In combination with a transgene encoding Acrosin-GFP, the sperm acrosome status can be readily examined by the presence or absence of the green fluorescence signals (Nakanishi et al., 1999; Hasuwa et al., 2010). In 2014, Bianchi et al. reported the adhesion rate of the acrosome-reacted Acr-GFP spermatozoa to the JUNO-deficient egg membrane is significantly reduced compared to that of the wild-type group. Here, using a similar approach, we discovered that the IZUMO1-deficient mouse spermatozoa hardly adhere to the egg membrane after the acrosome reaction. However, a small population of the acrosome-reacted Izumo1 KO mouse spermatozoa can still adhere to the oolemma, which might be attributed to the existence of other sperm-oolemma adhesion machinery on top of the IZUMO1-JUNO interaction in mice.
Interestingly, a more recent study indicates that SPACA6 is absent in the Izumo1 KO mouse spermatozoa (Inoue et al., 2021). Therefore, it remains possible that the complete loss of the oolemma binding ability in the Izumo1 KO rat spermatozoa might be attributed to not only the loss of the IZUMO1-JUNO interaction but also the collaterally disrupted interactions between SPACA6 or other sperm surface proteins with their receptors on the egg surface. Therefore, to further investigate the complex functions of those newly identified fusion-related molecules, the fusion-competent adhesion between the Juno KO eggs and the Fimp, Sof1, Spaca6, or Tmem95 KO spermatozoa that retain IZUMO1 should also be analyzed. It should be noted that, while we are preparing this manuscript, Dcst1/2 KO spermatozoa were also added to the list (Inoue et al., 2021; Noda et al., 2021).
In conclusion, our data suggest that IZUMO1 is critical for the sperm-egg adhesion preceding the plasma membrane fusion in mice and rats. Rats have been demonstrated as a suitable model for the study of mammalian fertilization because the acrosome-intact spermatozoa are less likely to adhere to the ZP-free egg surface non-specifically. Furthermore, the CRISPR/Cas9 system simplifies genome editing in rats, thereby making the genetically modified rats more accessible for researchers. Collectively, this study highlights that the rat model could be intensively employed for efficient screening of molecules involved in the sperm-egg adhesion and fusion by either genetic approaches or inhibition analyses using recombinant proteins or antibodies.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
The animal study was reviewed and approved by the Animal Care and Use Committee of the Research Institute for Microbial Diseases, Osaka University, Japan (#Biken-AP-R03-01-0).
TM, AI, and MI designed the research. TM, TN, YS, AM, MO, and AI performed research. TN, AI, MO, and SY produced the mutant strains. TM, TN, YS, AI, and MI analyzed data. TM, YL, and MI wrote the paper. All authors reviewed and commented on the manuscript.
This work was supported by the Ministry of Education, Culture, Sports, Science, and Technology (MEXT)/Japan Society for the Promotion of Science (JSPS) KAKENHI grants (JP19J01276 to TM, JP18K14612 and JP20H03172 to TN, JP19K06686 to YS, JP19J12450 to AM, JP18K16735 to YL, JP21K19267 to AI, JP19H05750, JP21H05033, JPJSBP120208403 to MI); JST, PRESTO Grant JPMJPR2148 to TN; and Takeda Science Foundation grants to TN and MI; The Nakajima Foundation to TN; and the Eunice Kennedy Shriver National Institute of Child Health and Human Development (P01HD087157 and R01HD088412 to MI); and the Bill and Melinda Gates Foundation (INV-001902 to MI). The funders had no role in study design, data collection, analysis, decision to publish, or manuscript preparation. Drs. TM and AM were JSPS research fellows. Open access charges were funded by the Bill and Melinda Gates Foundation (INV-001902 to MI).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We would like to thank the members of the Department of Experimental Genome Research, and NPO for Biotechnology Research and Development for experimental assistance and discussion.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2021.810118/full#supplementary-material
Aguilar, P. S., Baylies, M. K., Fleissner, A., Helming, L., Inoue, N., Podbilewicz, B., et al. (2013). Genetic Basis of Cell-Cell Fusion Mechanisms. Trends Genet. 29 (7), 427–437. doi:10.1016/j.tig.2013.01.011
Aoto, T., Takahashi, R.-I., and Ueda, M. (2011). A Protocol for Rat In Vitro Fertilization during Conventional Laboratory Working Hours. Transgenic Res. 20 (6), 1245–1252. doi:10.1007/s11248-011-9492-z
Aydin, H., Sultana, A., Li, S., Thavalingam, A., and Lee, J. E. (2016). Molecular Architecture of the Human Sperm IZUMO1 and Egg JUNO Fertilization Complex. Nature 534 (7608), 562–565. doi:10.1038/nature18595
Bianchi, E., Doe, B., Goulding, D., and Wright, G. J. (2014). Juno Is the Egg Izumo Receptor and Is Essential for Mammalian Fertilization. Nature 508 (7497), 483–487. doi:10.1038/nature13203
Bianchi, E., and Wright, G. J. (2015). Cross-species Fertilization: the Hamster Egg Receptor, Juno, Binds the Human Sperm Ligand, Izumo1. Phil. Trans. R. Soc. B 370 (1661), 20140101. doi:10.1098/rstb.2014.0101
Bushell, K. M., Söllner, C., Schuster-Boeckler, B., Bateman, A., and Wright, G. J. (2008). Large-scale Screening for Novel Low-Affinity Extracellular Protein Interactions. Genome Res. 18 (4), 622–630. doi:10.1101/gr.7187808
Calarco, P. G. (1991). Fertilization of the Mouse Oocyte. J. Elec. Microsc. Tech. 17 (4), 401–411. doi:10.1002/jemt.1060170404
Chalbi, M., Barraud-Lange, V., Ravaux, B., Howan, K., Rodriguez, N., Soule, P., et al. (2014). Binding of Sperm Protein Izumo1 and its Egg Receptor Juno Drives Cd9 Accumulation in the Intercellular Contact Area Prior to Fusion during Mammalian Fertilization. Development 141 (19), 3732–3739. doi:10.1242/dev.111534
Fujihara, Y., Lu, Y., Noda, T., Oji, A., Larasati, T., Kojima-Kita, K., et al. (2020). Spermatozoa Lacking Fertilization Influencing Membrane Protein (FIMP) Fail to Fuse with Oocytes in Mice. Proc. Natl. Acad. Sci. USA 117 (17), 9393–9400. doi:10.1073/pnas.1917060117
Han, L., Nishimura, K., Sadat Al Hosseini, H., Bianchi, E., Wright, G. J., and Jovine, L. (2016). Divergent Evolution of Vitamin B9 Binding Underlies Juno-Mediated Adhesion of Mammalian Gametes. Curr. Biol. 26 (3), R100–R101. doi:10.1016/j.cub.2015.12.034
Hasuwa, H., Muro, Y., Ikawa, M., Kato, N., Tsujimoto, Y., and Okabe, M. (2010). Transgenic Mouse Sperm that Have green Acrosome and Red Mitochondria Allow Visualization of Sperm and Their Acrosome Reaction In Vivo. Exp. Anim. 59 (1), 105–107. doi:10.1538/expanim.59.105
Hernández, J. M., and Podbilewicz, B. (2017). The Hallmarks of Cell-Cell Fusion. Development 144 (24), 4481–4495. doi:10.1242/dev.155523
Honda, A., Tachibana, R., Hamada, K., Morita, K., Mizuno, N., Morita, K., et al. (2019). Efficient Derivation of Knock-Out and Knock-In Rats Using Embryos Obtained by In Vitro Fertilization. Sci. Rep. 9 (1), 11571. doi:10.1038/s41598-019-47964-1
Ikawa, M., Tokuhiro, K., Yamaguchi, R., Benham, A. M., Tamura, T., Wada, I., et al. (2011). Calsperin Is a Testis-specific Chaperone Required for Sperm Fertility. J. Biol. Chem. 286 (7), 5639–5646. doi:10.1074/jbc.M110.140152
Inoue, N., Hagihara, Y., and Wada, I. (2021). Evolutionarily Conserved Sperm Factors, DCST1 and DCST2, Are Required for Gamete Fusion. eLife 10, e66313. doi:10.7554/eLife.66313
Inoue, N., Hamada, D., Kamikubo, H., Hirata, K., Kataoka, M., Yamamoto, M., et al. (2013). Molecular Dissection of IZUMO1, a Sperm Protein Essential for Sperm-Egg Fusion. Development 140 (15), 3221–3229. doi:10.1242/dev.094854
Inoue, N., Ikawa, M., Isotani, A., and Okabe, M. (2005). The Immunoglobulin Superfamily Protein Izumo Is Required for Sperm to Fuse with Eggs. Nature 434 (7030), 234–238. doi:10.1038/nature03362
Isotani, A., Matsumura, T., Ogawa, M., Tanaka, T., Yamagata, K., Ikawa, M., et al. (2017). A Delayed Sperm Penetration of Cumulus Layers by Disruption of Acrosin Gene in Rats†. Biol. Reprod. 97 (1), 61–68. doi:10.1093/biolre/iox066
Kato, K., Satouh, Y., Nishimasu, H., Kurabayashi, A., Morita, J., Fujihara, Y., et al. (2016). Structural and Functional Insights into IZUMO1 Recognition by JUNO in Mammalian Fertilization. Nat. Commun. 7, 12198. doi:10.1038/ncomms12198
Lamas-Toranzo, I., Hamze, J. G., Bianchi, E., Fernández-Fuertes, B., Pérez-Cerezales, S., Laguna-Barraza, R., et al. (2020). TMEM95 Is a Sperm Membrane Protein Essential for Mammalian Fertilization. Elife 9, e53913. doi:10.7554/eLife.53913
Liu, D. Y., and Baker, H. W. G. (1994). Acrosome Status and Morphology of Human Spermatozoa Bound to the Zona Pellucida and Oolemma Determined Using Oocytes that Failed to Fertilize In Vitro. Hum. Reprod. 9 (4), 673–679. doi:10.1093/oxfordjournals.humrep.a138570
Liu, Y., Tewari, R., Ning, J., Blagborough, A. M., Garbom, S., Pei, J., et al. (2008). The Conserved Plant Sterility Gene HAP2 Functions after Attachment of Fusogenic Membranes in Chlamydomonas and Plasmodium Gametes. Genes Dev. 22 (8), 1051–1068. doi:10.1101/gad.1656508
Lorenzetti, D., Poirier, C., Zhao, M., Overbeek, P. A., Harrison, W., and Bishop, C. E. (2014). A Transgenic Insertion on Mouse Chromosome 17 Inactivates a Novel Immunoglobulin Superfamily Gene Potentially Involved in Sperm-Egg Fusion. Mamm. Genome 25 (3-4), 141–148. doi:10.1007/s00335-013-9491-x
Mashiko, D., Fujihara, Y., Satouh, Y., Miyata, H., Isotani, A., and Ikawa, M. (2013). Generation of Mutant Mice by Pronuclear Injection of Circular Plasmid Expressing Cas9 and Single Guided RNA. Sci. Rep. 3, 3355. doi:10.1038/srep03355
Matsumura, T., Endo, T., Isotani, A., Ogawa, M., and Ikawa, M. (2019). An Azoospermic Factor Gene, Ddx3y and its Paralog, Ddx3x Are Dispensable in Germ Cells for Male Fertility. J. Reprod. Develop. 65 (2), 121–128. doi:10.1262/jrd.2018-145
Misamore, M. J., Gupta, S., and Snell, W. J. (2003). TheChlamydomonasFus1 Protein Is Present on the Mating TypeplusFusion Organelle and Required for a Critical Membrane Adhesion Event during Fusion withminusGametes. MBoC 14 (6), 2530–2542. doi:10.1091/mbc.e02-12-0790
Mizuno, M., Harris, C. L., Johnson, P. M., and Morgan, B. P. (2004). Rat Membrane Cofactor Protein (MCP; CD46) Is Expressed Only in the Acrosome of Developing and Mature Spermatozoa and Mediates Binding to Immobilized Activated C31. Biol. Reprod. 71 (4), 1374–1383. doi:10.1095/biolreprod.104.030114
Nakanishi, T., Ikawa, M., Yamada, S., Parvinen, M., Baba, T., Nishimune, Y., et al. (1999). Real-time Observation of Acrosomal Dispersal from Mouse Sperm Using GFP as a Marker Protein. FEBS Lett. 449 (2-3), 277–283. doi:10.1016/s0014-5793(99)00433-0
Nishimura, K., Han, L., Bianchi, E., Wright, G. J., de Sanctis, D., and Jovine, L. (2016). The Structure of Sperm Izumo1 Reveals Unexpected Similarities with Plasmodium Invasion Proteins. Curr. Biol. 26 (14), R661–R662. doi:10.1016/j.cub.2016.06.028
Noda, T., Blaha, A., Fujihara, Y., Gert, K. R., Emori, C., Deneke, V. E., et al. (2021). Sperm Membrane Proteins DCST1 and DCST2 Are Required for the Sperm-Egg Fusion Process in Mice and Fish. bioRxiv 2004, 440256. doi:10.1101/2021.04.18.440256
Noda, T., Lu, Y., Fujihara, Y., Oura, S., Koyano, T., Kobayashi, S., et al. (2020). Sperm Proteins SOF1, TMEM95, and SPACA6 Are Required for Sperm−oocyte Fusion in Mice. Proc. Natl. Acad. Sci. USA 117 (21), 11493–11502. doi:10.1073/pnas.1922650117
Nozawa, K., Satouh, Y., Fujimoto, T., Oji, A., and Ikawa, M. (2018). Sperm-borne Phospholipase C Zeta-1 Ensures Monospermic Fertilization in Mice. Sci. Rep. 8 (1), 1315. doi:10.1038/s41598-018-19497-6
OECD (2016). Test No. 421: Reproduction/Developmental Toxicity Screening Test. Paris: OECD Publishing.
Ohto, U., Ishida, H., Krayukhina, E., Uchiyama, S., Inoue, N., and Shimizu, T. (2016). Structure of IZUMO1-JUNO Reveals Sperm-Oocyte Recognition during Mammalian Fertilization. Nature 534 (7608), 566–569. doi:10.1038/nature18596
Okabe, M. (2018). Sperm-egg Interaction and Fertilization: Past, Present, and Future. Biol. Reprod. 99 (1), 134–146. doi:10.1093/biolre/ioy028
Okabe, M. (2013). The Cell Biology of Mammalian Fertilization. Development 140 (22), 4471–4479. doi:10.1242/dev.090613
Phillips, D. M., and Yanagimachi, R. (1982). Difference in the Manner of Association of Acrosome-Intact and Acrosome-Reacted Hamster Spermatozoa with Egg Microvilli as Revealed by Scanning Electron Microscopy. (Hamster/sperm-egg Association/scanning Electron Microscopy). Dev. Growth Differ. 24 (6), 543–551. doi:10.1111/j.1440-169x.1982.00543.x
Russell, L. D., Ettlin, R. A., Hikim, A. P. S., and Clegg, E. D. (1993). Histological and Histopathological Evaluation of the Testis. Int. J. Androl. 16 (1), 83. doi:10.1111/j.1365-2605.1993.tb01156.x
Satouh, Y., Inoue, N., Ikawa, M., and Okabe, M. (2012). Visualization of the Moment of Mouse Sperm-Egg Fusion and Dynamic Localization of IZUMO1. J. Cel Sci 125 (Pt 21), 4985–4990. doi:10.1242/jcs.100867
Toyoda, Y., Yokoyama, M., and Hosi, T. (1971). Studies on the Fertilization of Mouse Eggs In Vitro. Jpn. J. Anim. Reprod. 16 (4), 152–157. doi:10.1262/jrd1955.16.152
Yamagata, K., Nakanishi, T., Ikawa, M., Yamaguchi, R., Moss, S. B., and Okabe, M. (2002). Sperm from the Calmegin-Deficient Mouse Have normal Abilities for Binding and Fusion to the Egg Plasma Membrane. Develop. Biol. 250 (2), 348–357. doi:10.1006/dbio.2002.0803
Yamatoya, K., Ito, C., Araki, M., Furuse, R., and Toshimori, K. (2011). One-step Collagenase Method for Zona Pellucida Removal in Unfertilized Eggs: Easy and Gentle Method for Large-Scale Preparation. Reprod. Med. Biol. 10 (2), 97–103. doi:10.1007/s12522-011-0075-8
Keywords: fertilization, sperm-oolemma adhesion, gamete fusion, FOLR4, knockout, Juno
Citation: Matsumura T, Noda T, Satouh Y, Morohoshi A, Yuri S, Ogawa M, Lu Y, Isotani A and Ikawa M (2022) Sperm IZUMO1 Is Required for Binding Preceding Fusion With Oolemma in Mice and Rats. Front. Cell Dev. Biol. 9:810118. doi: 10.3389/fcell.2021.810118
Received: 06 November 2021; Accepted: 21 December 2021;
Published: 12 January 2022.
Edited by:
Amber R. Krauchunas, University of Delaware, United StatesReviewed by:
Bart Gadella, Utrecht University, Netherlands Pablo Bermejo-Álvarez, Instituto Nacional de Investigación y Tecnología Agraria y Alimentaria (INIA), SpainCopyright © 2022 Matsumura, Noda, Satouh, Morohoshi, Yuri, Ogawa, Lu, Isotani and Ikawa. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Masahito Ikawa, aWthd2FAYmlrZW4ub3Nha2EtdS5hYy5qcA==
†Present address:Takafumi Matsumura, Department of Regenerative Medicine, Yokohama City University Graduate School of Medicine, Yokohama, JapanYuhkoh Satouh, Laboratory of Molecular Traffic, Institute for Molecular and Cellular Regulation, Gunma University, Maebashi, Japan
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.