
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell Dev. Biol., 21 January 2022
Sec. Cellular Biochemistry
Volume 9 - 2021 | https://doi.org/10.3389/fcell.2021.807345
This article is part of the Research TopicWaken the Silent Majority: Principles and Pathogenic Significance of Non-Acetyl Acylation and other Understudied Post-Translational ModificationsView all 11 articles
Arginylation is a post-translational modification mediated by the arginyltransferase (Ate1). We recently showed that conditional deletion of Ate1 in the nervous system leads to increased light-evoked response sensitivities of ON-bipolar cells in the retina, indicating that arginylation regulates the G-protein signaling complexes of those neurons and/or photoreceptors. However, none of the key players in the signaling pathway were previously shown to be arginylated. Here we show that Gαt1, Gβ1, RGS6, and RGS7 are arginylated in the retina and RGS6 and RGS7 protein levels are elevated in Ate1 knockout, suggesting that arginylation plays a direct role in regulating their protein level and the G-protein-mediated responses in the retina.
Arginylation is a post-translational modification that has been implicated in a large number of key physiological processes (see, e.g., Zanakis et al., 1984; Bongiovanni et al., 1999; Kwon et al., 2002; Hu et al., 2005; Karakozova et al., 2006; Decca et al., 2007; Rai et al., 2008; Saha and Kashina, 2011; Lee et al., 2012; Jiang et al., 2016; Wang et al., 2017a; Wang et al., 2017b). It was initially characterized as a post-translational modification of the protein N-termini for ubiquitin-proteasomal degradation (Ciechanover et al., 1988; Balzi et al., 1990). N-terminal arginylation can target proteins for degradation both in vitro (Davydov and Varshavsky, 2000) and in vivo (Lee et al., 2005) via ubiquitin-proteasome dependent N-end rule pathway (Varshavsky, 2011). While initially arginylation was believed to be exclusively targeting the N-terminal Asp, Glu, or Cys, as well as Asn and Gln upon deamidation, it has been later shown that arginylation can also target internal Asp and Glu residues by conjugating Arg to their acidic side chains (Wang et al., 2014). This discovery expands the scope of potentially arginylated proteins.
G-protein signaling is involved in virtually every known physiological process and plays a major role in neural signal transduction (Stewart and Fisher, 2015; Squires et al., 2018). It involves three crucial components: G-protein coupled receptors (GPCRs), G-protein heterotrimers, and Regulator of G-protein signaling (RGS) proteins. Upon stimulation, GPCRs facilitate the formation of Gα-GTP and promote the dissociation of Gα and Gβγ from each other and from GPCRs. Opposite to GPCRs, RGS proteins accelerate the termination of G-protein signaling by facilitating hydrolysis of the GTP bound to the Gα subunit of the G-protein, and thus promoting its re-association with Gβγ subunits and GPCRs (De Vries et al., 1995; Dohlman et al., 1995; Druey et al., 1996; Hunt et al., 1996; Koelle and Horvitz, 1996; Watson et al., 1996).
We recently discovered that conditional deletion of arginyltransferase (Ate1) in the nervous system affects G-protein signaling in the retina. In particular, lack of Ate1 leads to increased light-evoked response sensitivities of ON-bipolar cells, as well as their downstream neurons (Fina et al., 2021), indicating that arginylation might be involved in the three G-protein signaling complexes of the first two stages of visual signal transduction. Among the GPCRs, G-proteins, and RGSs in the mouse retina (see Table 1), RHO, Gβ5L, Gγ13, and RGS11 have the N-end rule residues, while others have the potential to be arginylated on the side chains. However, none of them are known to be arginylated.
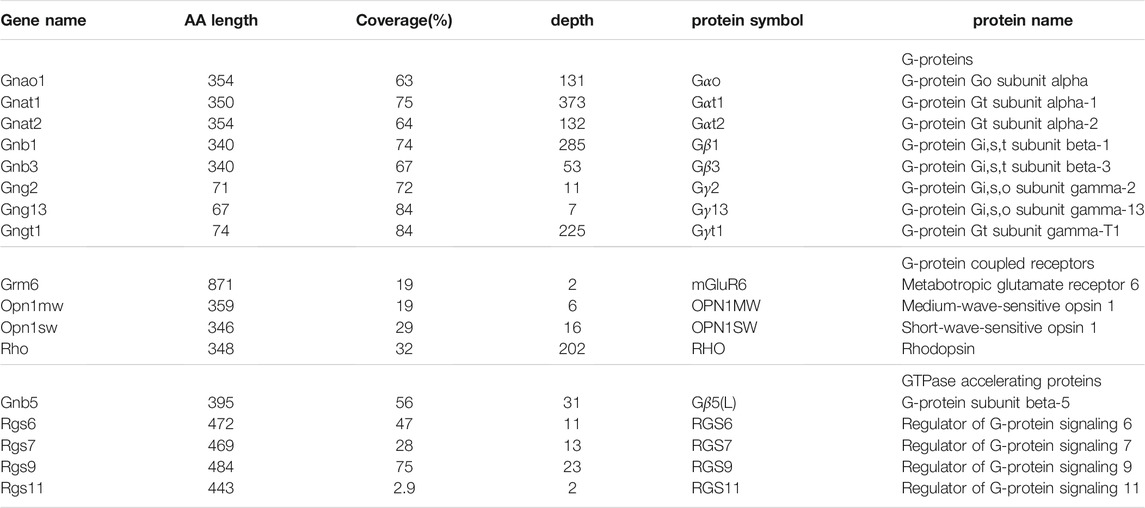
TABLE 1. Components of the G-protein signaling complexes in the retina screened for arginylation. The coverage is calculated with the MS/MS peptides identified during tryptic/P search against the whole mouse proteome without including arginylation into the search. The depth is the average number of MS/MS scans of the identified peptides over each covered amino acid (AA) base. Rgs9 (short) isoform and Gnb5 (long) isoform are listed. The latter encodes two protein isoforms: Gβ5 and Gβ5L. The three well-known G-protein signaling complexes in the retina are: RHO-Gαt1-Gβ1-Gγt1-RGS9-Gβ5L, OPN1MW/OPN1SW-Gαt2-Gβ3-Gγ2-RGS9-Gβ5L, mGluR6-Gαo-Gβ3-Gγ13-RGS7/RGS11-Gβ5.
Here we performed mass spectrometry analysis of mouse retina, using both our own data and data from the public domain, to investigate if any of the proteins involved in G-protein signaling in the retina are arginylated. We found that Gαt1, Gβ1, RGS6 and RGS7 were arginylated. Immunostaining revealed a prominent increase in the levels of RGS6 and its obligatory binding partner Gβ5 in synaptic processes of starburst amacrine cells of Ate1 knockout retina, corroborating our previous finding that RGS7 and Gβ5 increase in synaptic processes of ON-bipolar cells. We propose that arginylation regulates RGS6 and RGS7 levels and thus regulates G-protein signaling in the retina.
To obtain brain-specific Ate1 knockout mice, Ate1-floxed mice (Leu et al., 2009; Kurosaka et al., 2010) were crossed with the Jackson Laboratory strain B6.Cg-Tg(Nes-cre)1Kln/J, expressing Cre recombinase under Nesting promoter. Mice were maintained in C57BL6/129SVJ background in accordance with the University of Pennsylvania IACUC.
Retina collection and immunostaining was performed as previously described (Fina et al., 2021). Briefly, mice were dark adapted overnight and euthanized with a mixture containing ketamine/xylazine (300 μg each per g body-weight). Immediately after the euthanasia, eyes were enucleated and the cornea removed under dim red-light. Eyeballs were fixed in the dark in 4% paraformaldehyde for 60 min, rinsed in phosphate buffer (PB) 3 times, cryoprotected overnight at 4°C in 0.1M PB containing 30% sucrose and then, with the lens removed, embedded in a mixture of two parts 20% sucrose in PB and one part optimal cutting temperature compound (Tissue Tek, Electron Microscopy Sciences, Hateld, PA, United States). Radial sections were stained with antibodies and imaged using a confocal laser scanning microscope (Olympus Fluoview 1,000, Center Valley, PA, United States) under an oil-immersion objective (Tummala et al., 2016). Littermate retinas from WT and KO mice were immunostained and imaged in parallel under the same conditions and settings. To compensate the exposure difference between imaging of retinal slides, each set of images from the same retinal slide was normalized by the non-specific signal in the outer nuclear layer (ONL).
For image quantification, a region of interest (ROI, of the same 6 × 6 μm area size for WT and KO mice) was positioned on the lines drawn manually along the bands of stained starburst amacrine cell (SAC) processes in the inner plexiform layer (IPL), and the intensity measurement for each pixel was taken from the average z-stacks (of the same 0.3μm/slice thickness for WT and KO mice) using MATLAB software (MathWorks, Natick, MA, United States). The pixel intensity was thresholded by the background—the non-specific signal in ONL. The average intensity of the pixels in the ROI was calculated for each section to represent the immunostaining level in IPL, averaged along the SAC bands for each section, and then averaged over 1 to 4 sections per retina.
Tissue collection and processing was performed as previously described (Fina et al., 2021). Briefly, freshly excised mouse brains were flash-frozen in liquid nitrogen and ground in liquid nitrogen using mortar and pestle to obtain whole brain lysates. Total RNA was extracted using Trizol, and RNA sequencing (stranded TruSeq with Ribo-Zero, Illumina, San Diego, CA, United States) was used to quantify mRNA levels in the whole brain lysates of 3 littermate pairs of 3 month old mice and analyzed using STAR software package (Dobin et al., 2013).
Freshly excised mouse retinas, with the cornea and the lens removed, were flash-frozen in liquid nitrogen and ground up in liquid nitrogen to obtain retinal lysates. The retinal proteins were pulled down with phaloidin (manuscript in preparation).
For mass spectrometry, the resulting protein samples of 4 retinas of 6 month old mice were reduced with tris(2-carboxyethyl)phosphine (TCEP), alkylated with iodoacetamide, and digested with trypsin. Tryptic digests were analyzed using a 1.5 h LC gradient on the Thermo Q Exactive Plus mass spectrometer.
Data analysis was performed on the samples described above (processed as .raw files), as well as the original raw data files from public data sets (ProteomeXchange Consortium accession number PXD003441 (Zhao et al., 2016), PXD009909 (Harman et al., 2018), PXD014459 (Sze et al., 2021), and PXD023439 (Chen et al., 2021)), all of which are wild-type mice of C57BL6 backgrounds. The raw data files were processed using MaxQuant software (Version 1.6.7.0 for PXD014459 and 1.6.17.0 for the rest) from Max-Planck Institute for Biochemistry, Martinsried, Germany (Tyanova et al., 2016). To avoid mis-assignment of arginylation to a peptide, for each raw data file, we did two complete search runs, one without arginylation and one with arginylation modifications.
In the first run (without arginylation modifications), all searches were against the mouse protein reference UP000000589 and a locally compiled list of contaminants. In the second run, the “MS/MS first search” (used for mass recalibration in MaxQuant) was the same as the first run but the “MS/MS main search” and the “Second peptide search” were against the proteins of interest (Table 1). In addition, “Match between runs” and “Dependent peptides” were enabled in the second run. Consensus identification lists were generated with false discovery rates of 1% at protein, peptide and site levels. The two parameter .xml files and the two locally compiled. fasta files are included in the Supplementary Material. We excluded the “arginylated” peptides identified in the second run if they were from the scans which have at least one peptide identified in the first run. In addition, we also inspected MS/MS spectra to exclude “arginylated” peptides which could be mistaken for N-terminal or C-terminal arginine due to missed cleavage.
Rabbit anti-Gβ5 (dilution 1:500) was a gift from Dr. C.K. Chen, Baylor College of Medicine, Houston, TX, United States; rabbit anti-RGS6/RGS7 (dilution 1:100) was a gift from Dr. T.G. Wensel, Baylor College of Medicine, Houston, TX, United States.
In the RNA sequencing and the immunostaining experiments, KO and WT littermate pairs were compared pairwise, and the p-values for these comparisons were calculated using paired Student’s t-test and paired Wilcoxon t-test (signed-rank test for median) on the 3 and 7 KO/WT pairs, respectively. For the immunostaining experiments, the average staining intensity of all samples on each slide was normalized to 1 and then the average staining intensity of all samples of each WT and KO pair was normalized to 1 in order to reduce the potential experimental variation.
We previously reported that the ON-bipolar responses and RGS7 protein levels are regulated by Ate1, however arginylation of RGS7 or in the components of the G-protein signaling complexes which give rise to ON-bipolar responses has never been directly demonstrated. To test whether any of these proteins are arginylated, we analyzed retina samples by mass spectrometry, and searched the results from LC-MS/MS runs for potential arginylation against a limited database including G-proteins, GPCRs, and RGS proteins (Table 1). These candidate proteins were chosen based on the existence of three well characterized G-protein signaling complexes in the retina. In rod and cone photoreceptors, rhodopsins and opsins are the GPCRs responding to photon stimulation in dim and bright light, respectively, and they activate G-proteins Gαt1-Gβ1-Gγt1 and Gαt2-Gβ3-Gγ2 which are deactivated by RGS9 in the obligatory heterodimer with Gβ5L (Cowan et al., 1998; He et al., 1998). In ON-bipolar cells, glutamate released by photoreceptors activates the ON-bipolar cells’ GPCR, the metabotropic glutamate receptor 6 (mGluR6) receptor, which in turn activates the Gαo-Gβ3-Gγ13 proteins that can be deactivated by RGS7-Gβ5 and RGS11-Gβ5 (Mojumder et al., 2009; Chen et al., 2010; Zhang et al., 2010; Cao et al., 2012). We also included RGS6 in this screen, since it is the closest family member to RGS7 and is widely distributed in the retina, including prominent representation in the dendrites of cholinergic starburst amacrine cells (Voigt, 1986; Haverkamp and Wässle, 2000; Song et al., 2007).
To search for arginylation of these proteins, we used MaxQuant (Tyanova et al., 2016). To avoid mis-assignment, we first searched all MS/MS scans without arginylation modifications and then searched the unidentified scans with arginylation modifications, including potential addition of unmodified as well as mono- and di-methylated Arg. The search covered the G-proteins (Table 1, top) really well with the coverage ranging between 63 and 84 percent. However, the coverage for GPCRs was low, between 19 and 32 percent (Table 1, middle). This low coverage is likely related to the fact that GPCRs are tightly membrane-bound, and thus a majority of these proteins would be excluded from the soluble retina homogenate. The coverage of RGS and Gβ5, the GTPase accelerating proteins in the retina, varied a lot (Table 1, bottom). Most surprisingly, we found only one peptide for RGS11 in two MS/MS scans, despite the fact that RGS11 has similar molar amounts to RGS7 in the retina (Sarria et al., 2015).
This search revealed several putative arginylated sites, on Gαt1, Gβ1, RGS6, and RGS7. These were the only G-protein and RGS family members identified. We found no instance of arginylation of the GPCRs. In the next sections, we detail the evidence for these arginylation sites.
In outer segments of rod photoreceptors, the transducin Gαt1 binds with Gβ1Gγt1 heterodimer in the dark. Light stimulation activates rhodopsin, which facilitates the formation of Gαt1-GTP and hence promotes the dissociation of Gαt1 from Gβ1γt1 and rhodopsin. This activated Gαt1 then interacts with the downstream phosphodiesterase (PDE6) and starts the phototransduction cascade.
We found that Gαt1 was arginylated at the site of E167 or D169 (Figure 1). This site is located in the middle of the protein and on the opposite side of the interface with PDE6 (Figure 2A). It is within the α-helical domain, near a linker with the β-sheets of the GTPase domain (Figure 2B). This site is also on the solvent-exposed surface of Gαt1Gβ1Gγt1 heterotrimer in the GDP-bound state (Lambright et al., 1996), in a region of Gαt1 α-helical domain, which interacts with Gβ1 when Gαt1 is activated by rhodopsin, thus helping to provide an open route between the α-helical and GTPase domains and facilitating GDP and GTP exchange (Gao et al., 2019). This site is shared between Gαt1, Gαt2, and Gαo (Figure 2B). Arginylation of this site can affect the rate of GDP-GTP exchange of these Gα proteins and the G-protein signaling in rods, cones, and ON-bipolar cells.
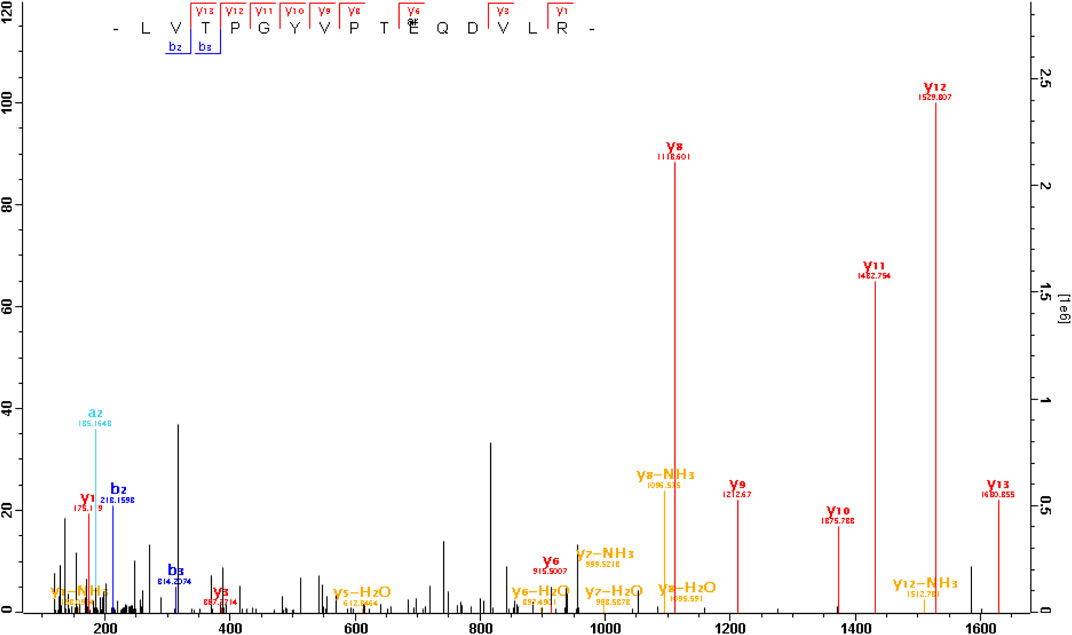
FIGURE 1. The MS/MS spectrum of an arginylated peptide of Gαt1. The peptide LVTPGYVPT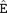 QDVLR or LVTPGYVPTEQD
QDVLR or LVTPGYVPTEQD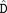 VLR (m/z = 615.007 3, mass error = − 0.3ppm, charge = 3) was arginylated at E167 or D169 of Gαt1 (capped).
VLR (m/z = 615.007 3, mass error = − 0.3ppm, charge = 3) was arginylated at E167 or D169 of Gαt1 (capped).
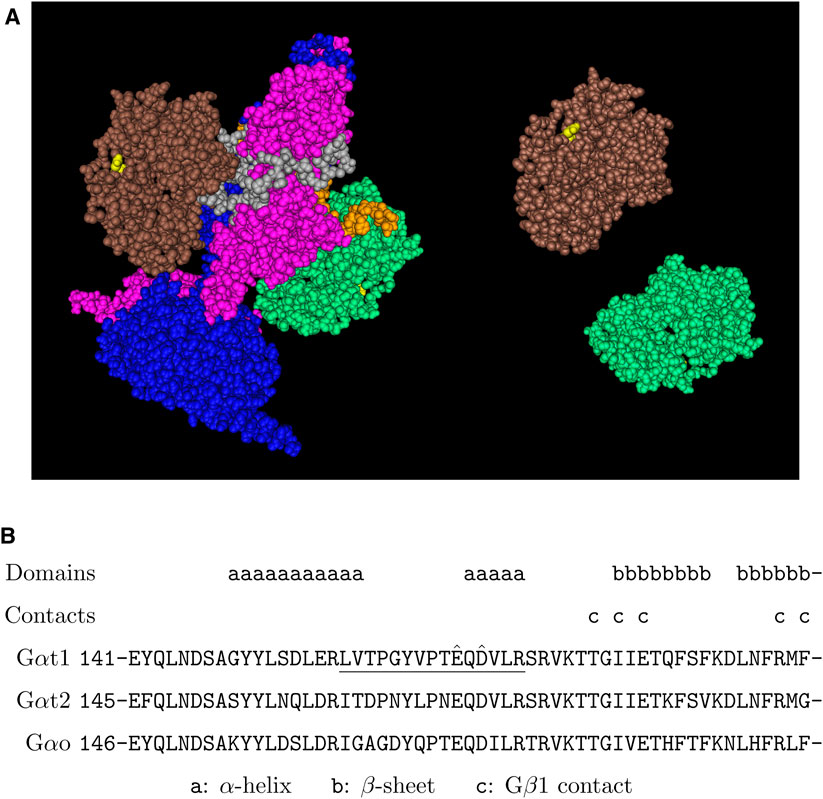
FIGURE 2. Putative arginylation sites of Gαt1. (A) The 3D structure of 2 Gαt1 in the complex with PDE6 based on the structure data 7JSN (Gao et al., 2020). Brown/green: Gαt1, blue: PDEα, pink: PDEβ, gray/orange: PDEγ, yellow: Gαt1 E167. (B) The putative arginylation sites (capped), the peptide (underlined), and the surrounding regions are shown. The putative arginylation sites E167 and D169 are within the α-helical domain of six helices, near the beginning of the 6th α-helix which is connected by a linker region to the 2nd of the six-stranded β-sheets of the Ras-like GTPase domain (Lambright et al., 1996). These sites are shared between Gαt1, Gαt2, and Gαo.
In the outer segments of the rods, Gβ1 is the important link between Gαt1 and Gγt1. Each of Gαt1 and Gγt1 has a membrane domain, which is not strong enough to anchor Gαt1 or Gβ1Gγt1 separately to the disk membrane, but both of them together anchor the heterotrimer in the GDP-bound inactive state. Recently, it has been proposed to play an important role in facilitating GDP-GTP exchange during Gαt1 activation (Gao et al., 2019). Gβ1 is also found in the synaptic terminals of the rods (Peng et al., 1992). Interestingly, Gβ1, when translocated to the synaptic terminals of the rods during light adaptation (Whelan and McGinnis, 1988; Sokolov et al., 2002) facilitates the synaptic transmission to ON-bipolar cells (Majumder et al., 2013).
We found that Gβ1 was arginylated at two sites: E12 and D27 (Figure 3). These sites are located near the N-terminus of the protein before the seven-bladed β-propellers (WD40 domains) and in particular D27 is at the junction of the Gβ1 N-terminus, seven-blades, and Gγ (Figure 4A). These sites are located within a region of high homology and is conserved between Gβ1, Gβ3, and Gβ5 (Figure 4B). Based on this homology, we propose that these sites can also be arginylated in Gβ3 and possibly in Gβ5. Gβ3 bound with Gγ2 plays a similar role in the cones as Gβ1 in the rods. Gβ3 is also found in the synaptic terminals of the cones (Peng et al., 1992). Arginylation of Gβ1 and Gβ3 can potentially affect the phototransduction in the outer segments and the signal transmission to ON-bipolar cells (OBCs) at the synaptic terminals of rods and cones, respectively. In addition, Gβ3 in complex with Gγ13 in OBCs facilitates the post-synaptic light-on responses of both rod-OBCs and cone-OBCs (Dhingra et al., 2012; Ramakrishnan et al., 2015) and thus arginylation of Gβ3, if it happens, can potentially affect post-synaptic responses to both rod and cone signals.
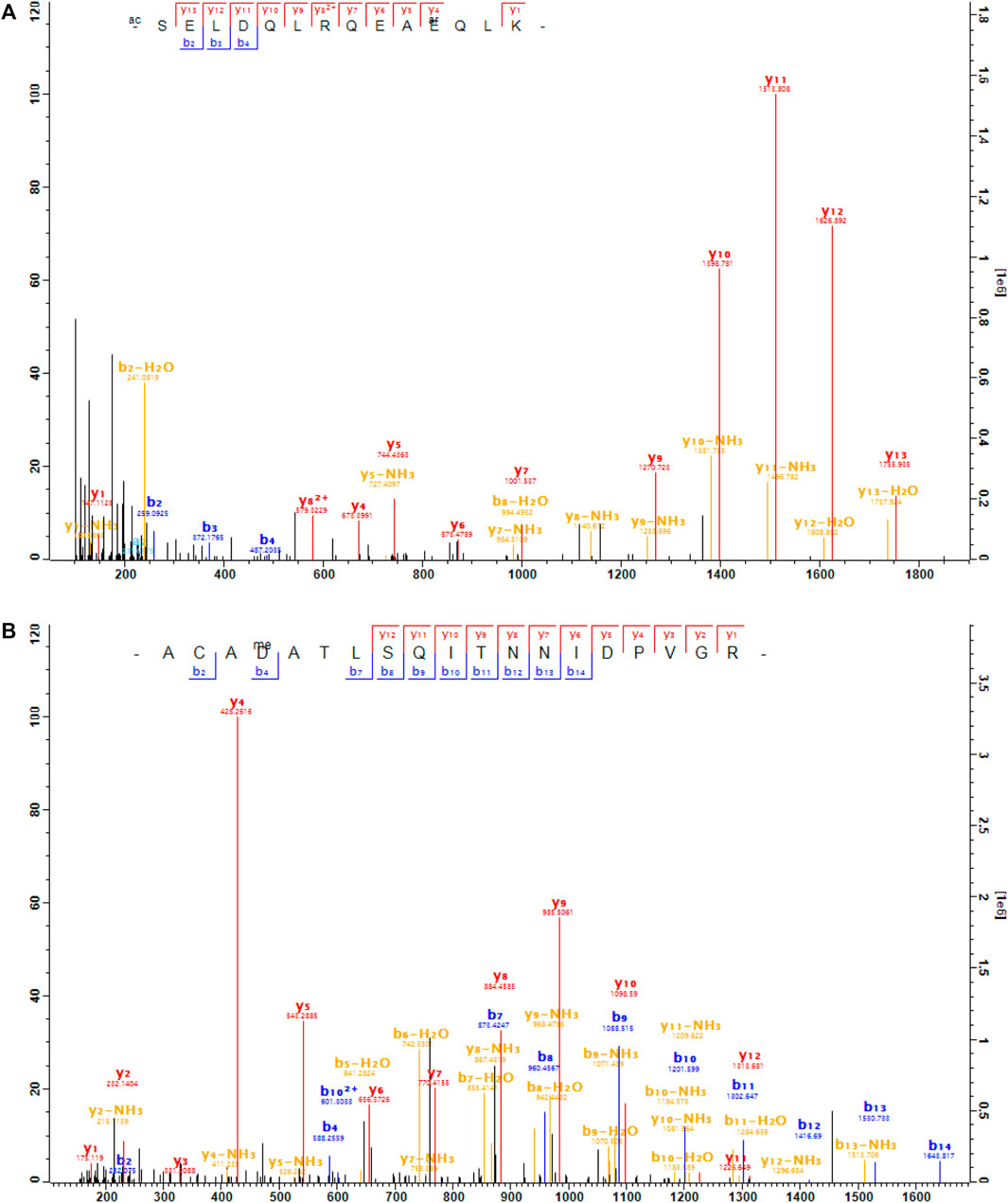
FIGURE 3. The MS/MS spectra of the arginylated peptides of Gβ1. (A), the peptide SELDQLRQEA
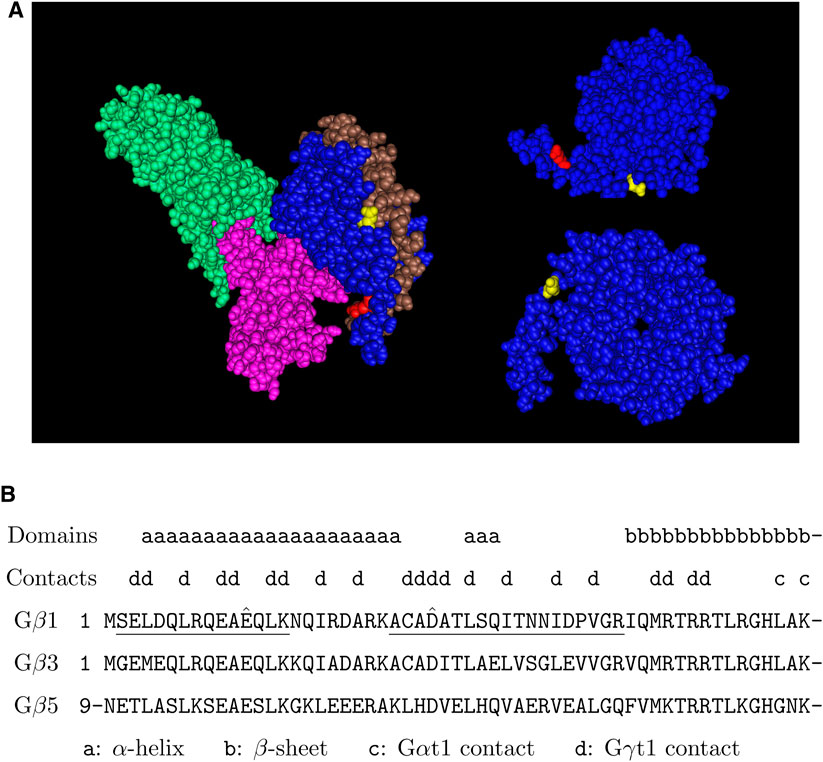
FIGURE 4. Putative arginylation sites of Gβ1. (A) The 3D structure of Gβ1 in the Gαt1-Gβ1-Gγt1 heterotrimer in complex with rhodopsin based on the structure data 6OYE (Gao et al., 2019). Blue: Gβ1, brown: Gγt1, pink: Gαt1, green: rhodopsin, red: Gβ1 E12, yellow: Gβ1 D27. (B) The putative arginylation sites (capped), the peptides (underlined), and the surrounding regions are shown. The putative arginylation sites are on the N-terminal directly proceeding the first β-sheet of the seven WD40 domains (Sondek et al., 1996). These sites are shared between Gβ1, Gβ3, and Gβ5.
Gβ5 is a special Gβ existing as an obligatory heterodimer with all R7 family (RGS6, RGS7, RGS9, RGS11) proteins (Cabrera et al., 1998; Liang et al., 2000; Witherow et al., 2000; Zhang and Simonds, 2000). It has 2 splice variants, Gβ5 (short) and Gβ5L, which includes additional 42 N-terminal amino acid residues added to the Gβ5 (short) sequence. In the retina, the Gβ5L isoform forms an obligatory heterodimer with RGS9 (short), and the Gβ5 isoform forms obligatory heterodimers with RGS6, RGS7, or RGS11. Arginylation of Gβ5 can affect all the G-protein signaling in the retina, predominately regulated by R7 family proteins.
RGS6 is the closest family member to RGS7 and presumably shares many structural and functional properties with RGS7. We have previously found that the deletion of Arginyltransferase 1 (Ate1) in the retina leads to a prominent increase in the levels of RGS7 and its obligatory binding partner Gβ5 in the dendritic processes of ON-bipolar cells (Fina et al., 2021), but in that work we found no direct evidence of arginylation.
Here, we found that RGS6 was arginylated at the site of D15 and RGS7 was arginylated at three sites: E73, D74 and E77 (Figure 5). These sites are located in the N-terminal region which has the membrane targeting DEP domain. These sites are conserved between RGS6 and RGS7, and the surrounding regions are highly homologous between RGS6 and RGS7 (Figure 6). Thus, it is likely that these sites are arginylated in both RGS6 and RGS7, and that this arginylation can directly contribute to an increase of their levels in synaptic processes of starburst amacrine cells and ON-bipolar cells, respectively.
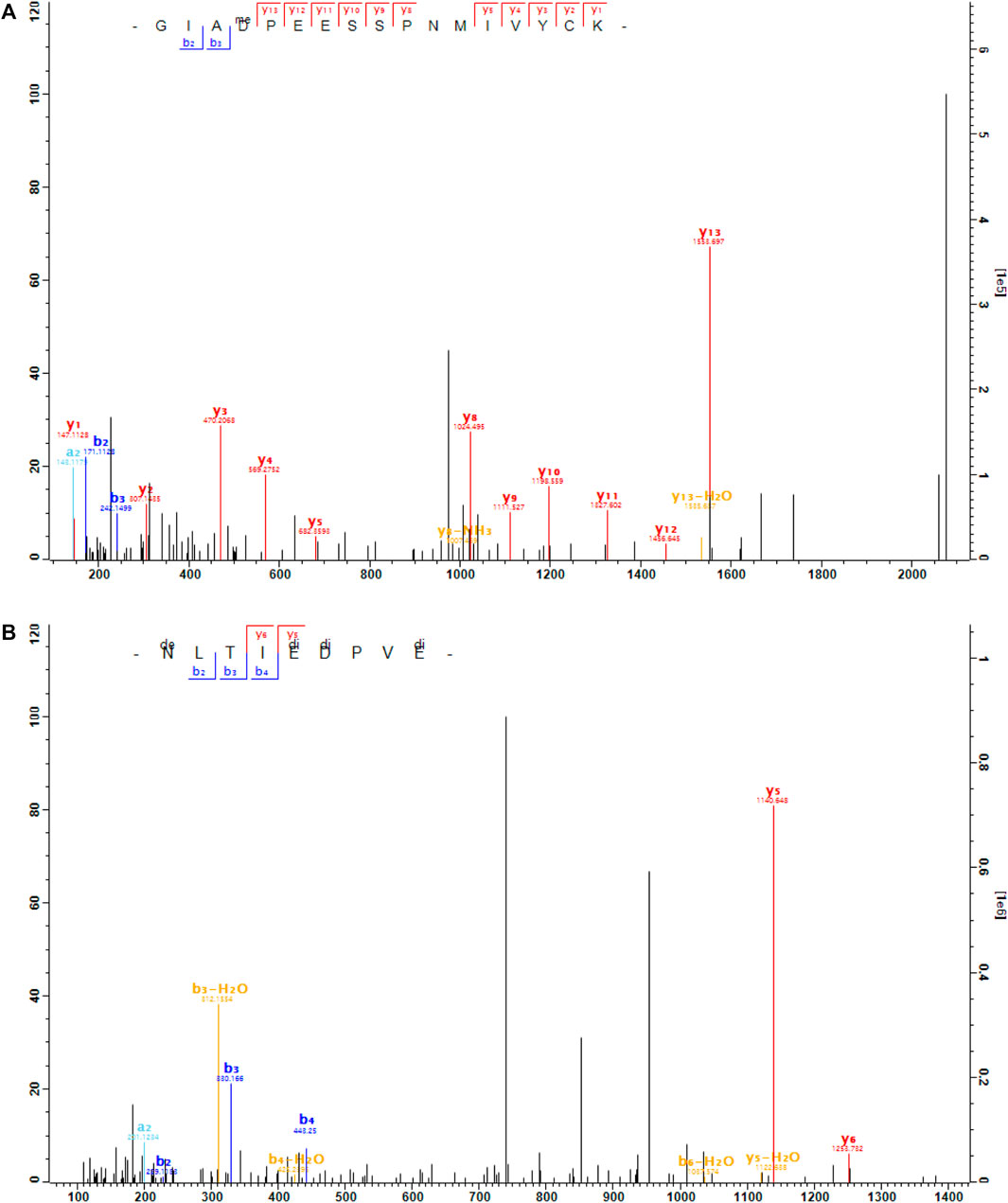
FIGURE 5. The MS/MS spectra of the arginylated peptides of RGS6 and RGS7. (A), the peptide GIAD
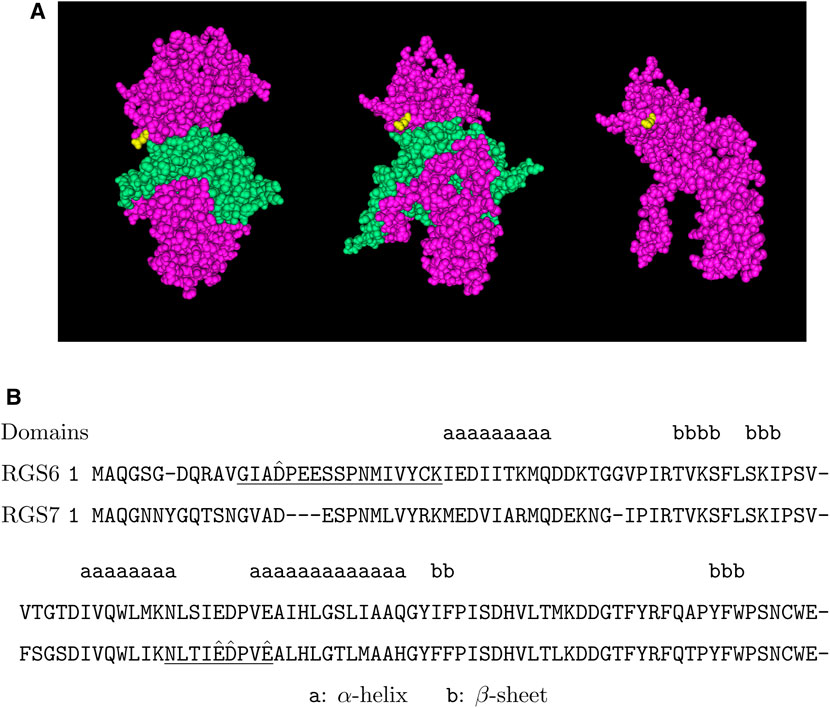
FIGURE 6. Putative arginylation sites of RGS6 and RGS7. (A) The 3D structure of RGS7 in the RGS7-Gβ5 heterodimer based on the structure data 6N9G (Patil et al., 2018). Pink: RGS7, green: Gβ5, yellow: RGS7 E73. (B) The putative arginylation sites (capped), the peptides (underlined), and the surrounding regions are shown with RGS6 and RGS7 alignment (Altschul et al., 1997). The putative arginylation sites are near/on the DEP domain, one in front of the 1st α-helix and three between the 2nd and 3rd α-helices in the secondary structures (Wong et al., 2000). These sites are shared between RGS6 and RGS7.
This is the first evidence that R7 family RGS proteins are arginylated on side chains, in contrast to the previous finding that R4 family RGS proteins are arginylated on N-termini (Lee et al., 2005).
To test whether abolishment of arginylation affects the level of RGS6 and RGS7 proteins in the G-protein signaling complexes, we compared the RGS6 and RGS7 protein levels in retinal synaptic processes of the wild-type (WT) and the conditional Ate1 knockout (KO) mice. In the KO mice, Ate1 deletion is driven by neuron-specific Nestin promoter, occurring in the entire nervous system during embryogenesis (Wang et al., 2017a; Wang et al., 2017b). ATE1 protein level in these mice is greatly reduced in most retinal neurons (Fina et al., 2021).
RGS6 and RGS7 in the retina are prominently expressed in the synaptic processes of starburst amacrine cells in the inner plexiform layer (IPL) and ON-bipolar cells in the outer plexiform layer (OPL), separately (Song et al., 2007). In agreement with previous studies, our immunohistochemistry staining showed that RGS6 was prominently enriched in the starburst amacrine cell (SAC) processes, which are localized within two distinct bands in the IPL (Figure 7A). We used the total sum within the 1.1μm × 1.1 μm window, centered and averaged along the SAC bands, to quantify the RGS6 protein level in the synaptic processes. The RGS6 protein level was significantly higher in KO than in WT (Figure 7B). Consistent with that, RGS6’s obligatory binding partner Gβ5 was also enriched in the two distinct bands in the IPL (Figure 7C) and its level was significantly higher in KO than in WT (Figure 7D). In addition, in a separate study, we showed that RGS7 and Gβ5 proteins levels were significantly higher in KO than in WT dendritic processes of both rod- and cone-OBCs in the OPL (Fina et al., 2021). These results are summarized in Figures 8A,B.
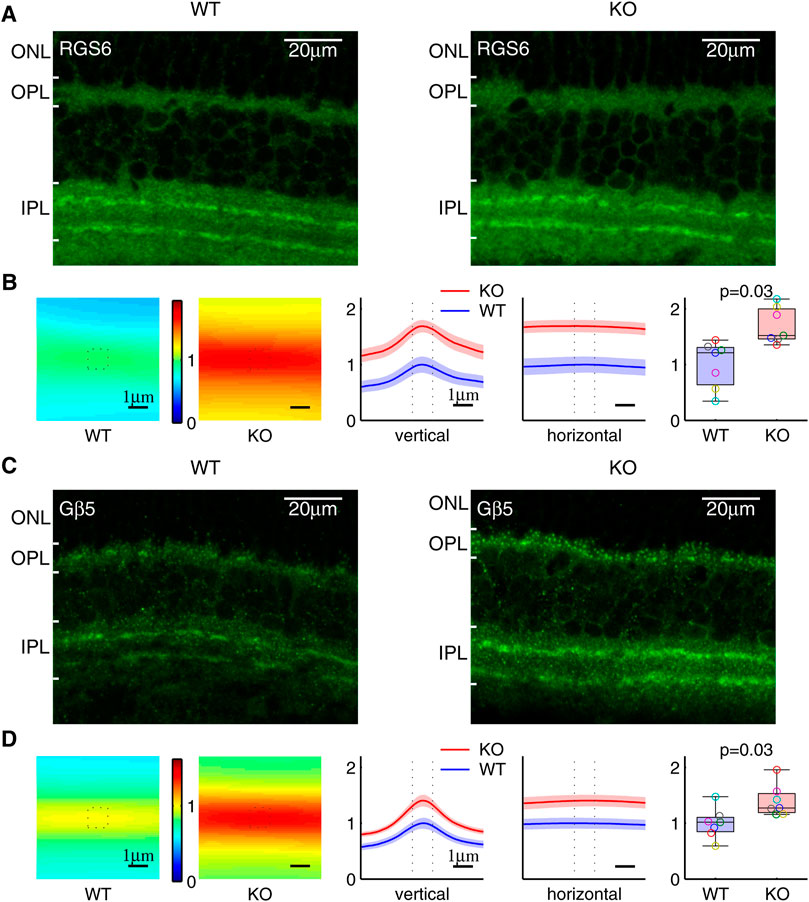
FIGURE 7. Increased RGS6 and Gβ5 protein level in the synaptic processes of Ate1 knockout (KO) mouse. (A) Immunohistochemistry of retinas stained with anti-RGS6. Both the wild-type (WT) and KO retinas have clear staining of two distinct bands of synaptic processes in the inner plexiform layer (IPL). (B) The RGS6 staining intensities averaged along the bands are shown on the left. The values along the vertical and horizontal middle lines are plotted in the middle with the SEM in shaded colors. The dotted lines mark the central 1.1 μm region surrounding the maxima. In the box plot on the right, the total sum at the central region shows a significant increase of RGS6 proteins in the synaptic processes of KO mice. (C) and (D) Immunohistochemistry of retina stained with anti-Gβ5 show the same pattern and the results as (A) and (B) for RGS6. Each pair of data points of the same color represents a WT/KO littermate pair (n = 7). The p-values are from paired Wilcoxon t-test (signed-rank test for median). The maximum length of the whiskers are 1.5 times of the inter quartile range. Note: the anti-Gβ5 and anti-RGS6 also stained puncta in ON-bipolar dendrites in the outer plexiform layer (OPL)—the latter due to a cross interaction with RGS7; however, its staining in IPL represents RGS6 only since RGS7 is not present in IPL (Song et al., 2007). ONL: the outer nuclear layer.
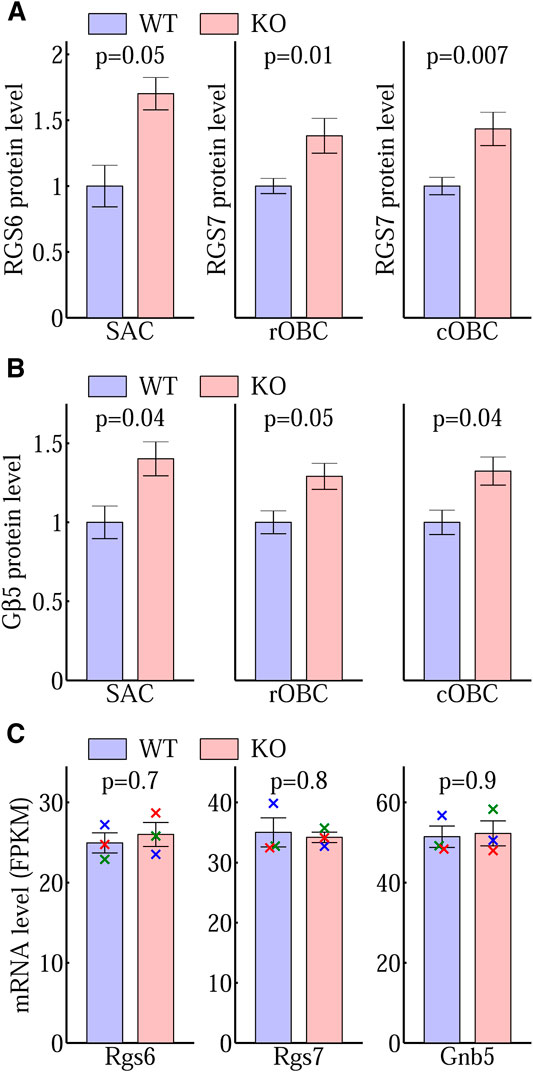
FIGURE 8. Post-translational changes of RGS6, RGS7, and Gβ5. Immunostaining quantification of (A) the protein levels of RGS6 in SAC and RGS7 in rOBC and cOBC synaptic processes and (B) the corresponding Gβ5 levels demonstrate a significantly higher level of these proteins in the Ate1 knockout (KO) than the wild-type (WT) mice. SAC: starburst amacrine cell. rOBC: rod ON-bipolar cell. cOBC: cone ON-bipolar cell. In (A) and (B), the bar plots for SAC are based on the same data of the box plots of Figure 7 and the bar plots for rOBC and cOBC are based on the data of Fina et al. (2021). The p-values are from paired Student’s t-test and the error bars represent SEM. The differences between KO and WT were also evaluated with a non-parametric statistical hypothesis test (Figure 7 for RGS6 and Gβ5 in SAC; Fina et al. (2021) for RGS7 and Gβ5 in rOBC and cOBC), which confirmed the conclusions in (A) and (B). (C) The Rgs6, Rgs7, and Gnb5 (short) mRNA encoding RGS6, RGS7, and Gβ5 proteins in the mouse brains from WT and KO littermates, expressed in Fragments Per Kilobase of transcript per Million mapped reads (FPKM) from RNA sequencing. There is no significant difference between WT and KO. Each pair of data points of the same color represents a WT/KO littermate pair (n = 3). The p-values are from paired Student’s t-test and the error bars represent SEM. The p-values from non-parametric statistical hypothesis test (paired Wilcoxon t-test) (0.8, 1, and 1, respectively) also shows no significant difference in (C).
It is interesting to know if the observed increase of RGS6 and RGS7 proteins at the synaptic processes are associated with an overall increase of their levels in the retina. In our previous study, we used Western blots to show that in total retina extracts RGS7 levels increased in Ate1 knockout, but Gβ5 did not (Fina et al., 2021). Thus, since RGS6 is the dominant binding partner with Gβ5 in the retina (Shim et al., 2012) and they form an obligatory complex in vivo, it would be highly unlikely for the total RGS6 in the retina to increase significantly in the absence of arginylation without simultaneously causing Gβ5 to do the same.
We compared the mRNA levels of Rgs6, Rgs7, and Gnb5 of Ate1 KO and WT mice. There was no significant difference for any of them (Figure 8C). This indicates that the changes of the corresponding protein levels and/or distributions are most likely post-translational.
It is conceivable that the overall increase of RGS7 protein level causes its increase at ON-bipolar dendritic processes. However, it cannot be the case for RGS6. Its redistribution to SAC synaptic processes upon arginylation could be due, e.g., to the fact that arginylation of RGS6 and/or other proteins in SAC could contribute to its targeting to the membrane and the synapses. It is known that light exposure leads to redistribution of G-proteins in the rods from the outer segments to the inner segments and the rod terminals (Whelan and McGinnis, 1988; Sokolov et al., 2002). Our ongoing study also shows that RGS7 levels at the synapses change significantly depending on the synaptic activity. Since Ate1 knockout increases the activities of ON-bipolar cells and down-stream neurons (Fina et al., 2021), RGS6 can undergo activity-dependent redistribution. Similar mechanisms could also be at work with RGS7. One possibility is that Ate1 knockout changes ON-bipolar activities pre-synaptically through Gβ1.
Gβ1 plays an important role in the rods. In addition to the rising phase of light responses in the outer segments, it also facilitates the synaptic transmission to OBCs (Majumder et al., 2013). Here, we presented evidence that Gβ1 is arginylated. However, we did not observe any significant change of the rising phase of the light responses of the rods after deletion of Ate1 (Fina et al., 2021). In our current ongoing project, we also did not see any obvious change in Gβ1 in the outer segments after Ate1 deletion (manuscript in preparation). It should be noted that lack of changes we have seen so far in Gβ1 does not exclude a possible effect of Gβ1 at the pre-synaptic terminals of photoreceptors, in contact with On-bipolar cell dendritic processes. It would be very interesting to address this possibility in a follow up study.
According to prior studies, RGS6 is not implicated in G-protein signaling in photoreceptors or ON-bipolar cells, where the major players are RGS9 (in the photoreceptors) and RGS7/RGS11 (in the ON-bipolar cells). In our experiments, the strongest RGS6 staining and the significant change due to Ate1 deletion, by far, was observed in the synaptic processes of starburst amacrine cells. This suggests that RGS6 likely plays an arginylation-dependent role in G-protein signaling in these cells. Elucidating this role is a very interesting future direction, and we hope our present findings will help future research to reveal the functional role of RGS6 in the retina.
This work represents the first comprehensive analysis of arginylation of the components of G-protein signaling in the retina, including GPCR, G-proteins, and RGS proteins. We find putative arginylation sites on the aspartate and glutamate side chains of Gαt1, Gβ1, RGS6, and RGS7, and no N-terminal arginylation among the proteins involved in the G-protein signaling in the retina. While, due to probabilistic nature of mass spectrometry analysis of post-translational modifications, as well as potential alteration of protein stability or intracellular localization of proteins after arginylation, it is still possible that other proteins in the retina are also arginylated, the targets identified here are likely more prominent and abundant.
Our results provide concrete evidence that arginylation targets proteins involved in G-protein signaling in the retina, and extend a mechanistic foundation to our previous findings that arginylation regulates retina responses in vision. We show here that abolishment of arginylation leads to an increase in RGS6 and RGS7 protein levels, suggesting that this mechanism can directly regulate the events downstream of RGS. Taken together, our findings demonstrate that arginylation regulates G-protein responses in the retina by directly targeting several key players in this signaling cascade.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
The animal study was reviewed and approved by University of Pennsylvania IACUC.
AK provided the brain-specific Ate1 knockout mice; DD and MF designed and performed the immunohistochemistry experiments; DD, AK, JW, and PV prepared samples for RNA sequencing and mass spectrometry; H-YT performed mass spectrometry; DD conceptualized the study, analyzed the data, and wrote the first draft of the manuscript; DD and AK prepared the figures and wrote the manuscript. All authors reviewed the manuscript.
This work was supported by the NIH/NIGMS R35GM122505, NIH/NINDS R01NS102435, NIH/NEI P30EY001583, the Office of Dean at School of Veterinary Medicines and the Office of the Vice Provost for Research at the University of Pennsylvania.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank Drs. Noga Vardi, Catherine CL Wong, Shuaxin Gao, and Nan Zhang for helpful discussions. We thank Stephanie Sterling and Nicolae A Leu for help with mouse colony maintenance and breeding. We thank Drs. Jason CK Chen and Theodore G Wensel for donating antibodies used in this study.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2021.807345/full#supplementary-material
Altschul, S., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Miller, W., et al. (1997). Gapped BLAST and PSI-BLAST: a New Generation of Protein Database Search Programs. Nucleic Acids Res. 25, 3389–3402. doi:10.1093/nar/25.17.3389
Balzi, E., Choder, M., Chen, W. N., Varshavsky, A., and Goffeau, A. (1990). Cloning and Functional Analysis of the Arginyl-tRNA-Protein Transferase Gene ATE1 of Saccharomyces cerevisiae. J. Biol. Chem. 265, 7464–7471. doi:10.1016/s0021-9258(19)39136-7
Bongiovanni, G., Fissolo, S., Barra, H. S., and Hallak, M. E. (1999). Posttranslational Arginylation of Soluble Rat Brain Proteins after Whole Body Hyperthermia. J. Neurosci. Res. 56, 85–92. doi:10.1002/(sici)1097-4547(19990401)56:1<85:aid-jnr11>3.0.co;2-t
Cabrera, J. L., de Freitas, F., Satpaev, D. K., and Slepak, V. Z. (1998). Identification of the Gβ5-RGS7 Protein Complex in the Retina. Biochem. Biophys. Res. Commun. 249, 898–902. doi:10.1006/bbrc.1998.9218
Cao, Y., Pahlberg, J., Sarria, I., Kamasawa, N., Sampath, A. P., and Martemyanov, K. A. (2012). Regulators of G Protein Signaling RGS7 and RGS11 Determine the Onset of the Light Response in ON Bipolar Neurons. Proc. Natl. Acad. Sci. 109, 7905–7910. doi:10.1073/pnas.1202332109
Chen, F. S., Shim, H., Morhardt, D., Dallman, R., Krahn, E., McWhinney, L., et al. (2010). Functional Redundancy of R7 RGS Proteins in ON-Bipolar Cell Dendrites. Invest. Ophthalmol. Vis. Sci. 51, 686–693. doi:10.1167/iovs.09-4084
Chen, K., Chen, C., Li, H., Yang, J., Xiang, M., Wang, H., et al. (2021). Widespread Translational Control Regulates Retinal Development in Mouse. Nucleic Acids Res. 49 (17), 9648–9664. doi:10.1093/nar/gkab749
Ciechanover, A., Ferber, S., Ganoth, D., Elias, S., Hershko, A., and Arfin, S. (1988). Purification and Characterization of Arginyl-tRNA-Protein Transferase from Rabbit Reticulocytes. Its Involvement in post-translational Modification and Degradation of Acidic NH2 Termini Substrates of the Ubiquitin Pathway. J. Biol. Chem. 263, 11155–11167. doi:10.1016/s0021-9258(18)37936-5
Cowan, C. W., Fariss, R. N., Sokal, I., Palczewski, K., and Wensel, T. G. (1998). High Expression Levels in Cones of RGS9, the Predominant GTPase Accelerating Protein of Rods. Proc. Natl. Acad. Sci. 95, 5351–5356. doi:10.1073/pnas.95.9.5351
Davydov, I. V., and Varshavsky, A. (2000). RGS4 Is Arginylated and Degraded by the N-End Rule Pathway In Vitro. J. Biol. Chem. 275 (30), 22931–22941. doi:10.1074/jbc.m001605200
De Vries, L., Mousli, M., Wurmser, A., and Farquhar, M. G. (1995). GAIP, a Protein that Specifically Interacts with the Trimeric G Protein G Alpha I3, Is a Member of a Protein Family with a Highly Conserved Core Domain. Proc. Natl. Acad. Sci. 92 (25), 11916–11920. doi:10.1073/pnas.92.25.11916
Decca, M. B., Carpio, M. A., Bosc, C., Galiano, M. R., Job, D., Andrieux, A., et al. (2007). Post-translational Arginylation of Calreticulin. J. Biol. Chem. 282 (11), 8237–8245. doi:10.1074/jbc.m608559200
Dhingra, A., Ramakrishnan, H., Neinstein, A., Fina, M. E., Xu, Y., Li, J., et al. (2012). G 3 Is Required for Normal Light ON Responses and Synaptic Maintenance. J. Neurosci. 32, 11343–11355. doi:10.1523/jneurosci.1436-12.2012
Dobin, A., Davis, C. A., Schlesinger, F., Drenkow, J., Zaleski, C., Jha, S., et al. (2013). STAR: Ultrafast Universal RNA-Seq Aligner. Bioinformatics 29 (1), 15–21. doi:10.1093/bioinformatics/bts635
Dohlman, H. G., Apaniesk, D., Chen, Y., Song, J., and Nusskern, D. (1995). Inhibition of G-Protein Signaling by Dominant Gain-Of-Function Mutations in Sst2p, a Pheromone Desensitization Factor in Saccharomyces cerevisiae. Mol. Cel Biol 15 (7), 3635–3643. doi:10.1128/mcb.15.7.3635
Druey, K. M., Blumer, K. J., Kang, V. H., and Kehrl, J. H. (1996). Inhibition of G-Protein-Mediated MAP Kinase Activation by a New Mammalian Gene Family. Nature 379 (6567), 742–746. doi:10.1038/379742a0
Fina, M. E., Wang, J., Nikonov, S. S., Sterling, S., Vardi, N., Kashina, A., et al. (2021). Arginyltransferase (Ate1) Regulates the RGS7 Protein Level and the Sensitivity of Light-Evoked ON-Bipolar Responses. Sci. Rep. 11, 9376. doi:10.1038/s41598-021-88628-3
Gao, Y., Hu, H., Ramachandran, S., Erickson, J. W., Cerione, R. A., and Skiniotis, G. (2019). Structures of the Rhodopsin-Transducin Complex: Insights into G-Protein Activation. Mol. Cel. 75 (4), 781–790. doi:10.1016/j.molcel.2019.06.007
Gao, Y., Eskici, G., Ramachandran, S., Poitevin, F., Seven, A. B., Panova, O., et al. (2020). Structure of the Visual Signaling Complex between Transducin and Phosphodiesterase 6. Mol. Cel. 80 (2), 237–245.e4. doi:10.1016/j.molcel.2020.09.013
Harman, J. C., Guidry, J. J., and Gidday, J. M. (2018). Comprehensive Characterization of the Adult ND4 Swiss Webster Mouse Retina: Using Discovery-Based Mass Spectrometry to Decipher the Total Proteome and Phosphoproteome. Mol. Vis. 24, 875–889.
Haverkamp, S., and Wässle, H. (2000). Immunocytochemical Analysis of the Mouse Retina. J. Comp. Neurol. 424, 1–23. doi:10.1002/1096-9861(20000814)424:1<1:aid-cne1>3.0.co;2-v
He, W., Cowan, C. W., and Wensel, T. G. (1998). RGS9, a GTPase Accelerator for Phototransduction. Neuron 20, 95–102. doi:10.1016/s0896-6273(00)80437-7
Hu, R.-G., Sheng, J., Qi, X., Xu, Z., Takahashi, T. T., and Varshavsky, A. (2005). The N-End Rule Pathway as a Nitric Oxide Sensor Controlling the Levels of Multiple Regulators. Nature 437 (7061), 981–986. doi:10.1038/nature04027
Hunt, T. W., Fields, T. A., Casey, P. J., and Peralta, E. G. (1996). RGS10 Is a Selective Activator of Gαi GTPase Activity. Nature 383 (6596), 175–177. doi:10.1038/383175a0
Jiang, Y., Lee, J., Lee, J. H., Lee, J. W., Kim, J. H., Choi, W. H., et al. (2016). The Arginylation branch of the N-End Rule Pathway Positively Regulates Cellular Autophagic Flux and Clearance of Proteotoxic Proteins. Autophagy 12 (11), 2197–2212. doi:10.1080/15548627.2016.1222991
Karakozova, M., Kozak, M., Wong, C. C. L., Bailey, A. O., Yates, J. R., Mogilner, A., et al. (2006). Arginylation of ß-Actin Regulates Actin Cytoskeleton and Cell Motility. Science 313 (5784), 192–196. doi:10.1126/science.1129344
Koelle, M. R., and Horvitz, H. R. (1996). EGL-10 Regulates G Protein Signaling in the C. elegans Nervous System and Shares a Conserved Domain with many Mammalian Proteins. Cell 84, 115–125. doi:10.1016/s0092-8674(00)80998-8
Kurosaka, S., Leu, N. A., Zhang, F., Bunte, R., Saha, S., Wang, J., et al. (2010). Arginylation-dependent Neural Crest Cell Migration Is Essential for Mouse Development. Plos Genet. 6 (3), e1000878. doi:10.1371/journal.pgen.1000878
Kwon, Y. T., Kashina, A. S., Davydov, I. V., Hu, R.-G., An, J. Y., Seo, J. W., et al. (2002). An Essential Role of N-Terminal Arginylation in Cardiovascular Development. Science 297 (5578), 96–99. doi:10.1126/science.1069531
Lambright, D. G., Sondek, J., Bohm, A., Skiba, N. P., Hamm, H. E., and Sigler, P. B. (1996). The 2.0 Å Crystal Structure of a Heterotrimeric G Protein. Nature 379 (6563), 311–319. doi:10.1038/379311a0
Lee, M. J., Tasaki, T., Moroi, K., An, J. Y., Kimura, S., Davydov, I. V., et al. (2005). RGS4 and RGS5 Are In Vivo Substrates of the N-End Rule Pathway. Proc. Natl. Acad. Sci. 102 (42), 15030–15035. doi:10.1073/pnas.0507533102
Lee, M. J., Kim, D. E., Zakrzewska, A., Yoo, Y. D., Kim, S.-H., Kim, S. T., et al. (2012). Characterization of Arginylation branch of N-End Rule Pathway in G-Protein-Mediated Proliferation and Signaling of Cardiomyocytes. J. Biol. Chem. 287 (28), 24043–24052. doi:10.1074/jbc.m112.364117
Leu, N. A., Kurosaka, S., and Kashina, A. (2009). Conditional Tek Promoter-Driven Deletion of Arginyltransferase in the Germ Line Causes Defects in Gametogenesis and Early Embryonic Lethality in Mice. PLoS One 4 (11), e7734. doi:10.1371/journal.pone.0007734
Liang, J.-J., Chen, H. H. D., Jones, P. G., and Khawaja, X. Z. (2000). RGS7 Complex Formation and Colocalization with the Gbeta5 Subunit in the Adult Rat Brain and Influence on Gbeta5gamma2-mediated PLCbeta Signaling. J. Neurosci. Res. 60, 58–64. doi:10.1002/(sici)1097-4547(20000401)60:1<58:aid-jnr6>3.0.co;2-l
Majumder, A., Pahlberg, J., Boyd, K. K., Kerov, V., Kolandaivelu, S., Ramamurthy, V., et al. (2013). Transducin Translocation Contributes to Rod Survival and Enhances Synaptic Transmission from Rods to Rod Bipolar Cells. Proc. Natl. Acad. Sci. USA 110, 12468–12473. doi:10.1073/pnas.1222666110
Mojumder, D. K., Qian, Y., and Wensel, T. G. (2009). Two R7 Regulator of G-Protein Signaling Proteins Shape Retinal Bipolar Cell Signaling. J. Neurosci. 29, 7753–7765. doi:10.1523/jneurosci.1794-09.2009
Patil, D. N., Rangarajan, E. S., Novick, S. J., Pascal, B. D., Kojetin, D. J., Griffin, P. R., et al. (2018). Structural Organization of a Major Neuronal G Protein Regulator, the RGS7-Gβ5-R7BP Complex. eLife 7, e42150. doi:10.7554/eLife.42150
Peng, Y. W., Robishaw, J. D., Levine, M. A., and Yau, K. W. (1992). Retinal Rods and Cones Have Distinct G Protein Beta and Gamma Subunits. Proc. Natl. Acad. Sci. 89, 10882–10886. doi:10.1073/pnas.89.22.10882
Rai, R., Wong, C. C. L., Xu, T., Leu, N. A., Dong, D. W., Guo, C., et al. (2008). Arginyltransferase Regulates Alpha Cardiac Actin Function, Myofibril Formation and Contractility during Heart Development. Development 135 (23), 3881–3889. doi:10.1242/dev.022723
Ramakrishnan, H., Dhingra, A., Tummala, S. R., Fina, M. E., Li, J. J., Lyubarsky, A., et al. (2015). Differential Function of Gγ13 in Rod Bipolar and On Cone Bipolar Cells. J. Physiol. 593, 1531–1550. doi:10.1113/jphysiol.2014.281196
Saha, S., and Kashina, A. (2011). Posttranslational Arginylation as a Global Biological Regulator. Dev. Biol. 358 (1), 1–8. doi:10.1016/j.ydbio.2011.06.043
Sarria, I., Pahlberg, J., Cao, Y., Kolesnikov, A. V., Kefalov, V. J., Sampath, A. P., et al. (2015). Sensitivity and Kinetics of Signal Transmission at the First Visual Synapse Differentially Impact Visually-Guided Behavior. Elife 4, e06358. doi:10.7554/eLife.06358
Shim, H., Wang, C.-T., Chen, Y.-L., Chau, V. Q., Fu, K. G., Yang, J., et al. (2012). Defective Retinal Depolarizing Bipolar Cells in Regulators of G Protein Signaling (RGS) 7 and 11 Double Null Mice. J. Biol. Chem. 287, 14873–14879. doi:10.1074/jbc.m112.345751
Sokolov, M., Lyubarsky, A. L., Strissel, K. J., Savchenko, A. B., Govardovskii, V. I., Pugh, E. N., et al. (2002). Massive Light-Driven Translocation of Transducin between the Two Major Compartments of Rod Cells. Neuron 34 (1), 95–106. doi:10.1016/s0896-6273(02)00636-0
Sondek, J., Bohm, A., Lambright, D. G., Hamm, H. E., and Sigler, P. B. (1996). Crystal Structure of a GA Protein βγdimer at 2.1 Å Resolution. Nature 379 (6563), 369–374. doi:10.1038/379369a0
Song, J. H., Song, H., Wensel, T. G., Sokolov, M., and Martemyanov, K. A. (2007). Localization and Differential Interaction of R7 RGS Proteins with Their Membrane Anchors R7BP and R9AP in Neurons of Vertebrate Retina. Mol. Cel. Neurosci. 35, 311–319. doi:10.1016/j.mcn.2007.03.006
Squires, K. E., Montañez-Miranda, C., Pandya, R. R., Torres, M. P., and Hepler, J. R. (2018). Genetic Analysis of Rare Human Variants of Regulators of G Protein Signaling Proteins and Their Role in Human Physiology and Disease. Pharmacol. Rev. 70 (3), 446–474. doi:10.1124/pr.117.015354
Stewart, A., and Fisher, R. A. (2015). Introduction: G Protein-Coupled Receptors and RGS Proteins. Prog. Mol. Biol. Transl. Sci. 133, 1–11. doi:10.1016/bs.pmbts.2015.03.002
Sze, Y. H., Zhao, Q., Cheung, J. K. W., Li, K. K., Tse, D. Y. Y., To, C. H., et al. (2021). High-pH Reversed-Phase Fractionated Neural Retina Proteome of normal Growing C57BL/6 Mouse. Sci. Data 8 (1), 27. doi:10.1038/s41597-021-00813-1
Tummala, S. R., Dhingra, A., Fina, M. E., Li, J. J., Ramakrishnan, H., and Vardi, N. (2016). Lack of mGluR6-Related cascade Elements Leads to Retrograde Trans-synaptic Effects on Rod Photoreceptor Synapses via Matrix-Associated Proteins. Eur. J. Neurosci. 43 (11), 1509–1522. doi:10.1111/ejn.13243
Tyanova, S., Temu, T., and Cox, J. (2016). The MaxQuant Computational Platform for Mass Spectrometry-Based Shotgun Proteomics. Nat. Protoc. 11 (12), 2301–2319. doi:10.1038/nprot.2016.136
Varshavsky, A. (2011). The N-End Rule Pathway and Regulation by Proteolysis. Protein Sci. 20 (8), 1298–1345. doi:10.1002/pro.666
Voigt, T. (1986). Cholinergic Amacrine Cells in the Rat Retina. J. Comp. Neurol. 248, 19–35. doi:10.1002/cne.902480103
Wang, J., Han, X., Wong, C. C. L., Cheng, H., Aslanian, A., Xu, T., et al. (2014). Arginyltransferase ATE1 Catalyzes Midchain Arginylation of Proteins at Side Chain Carboxylates In Vivo. Chem. Biol. 21 (3), 331–337. doi:10.1016/j.chembiol.2013.12.017
Wang, J., Han, X., Leu, N. A., Sterling, S., Kurosaka, S., Fina, M., et al. (2017a). Protein Arginylation Targets Alpha Synuclein, Facilitates normal Brain Health, and Prevents Neurodegeneration. Sci. Rep. 7 (1), 11323. doi:10.1038/s41598-017-11713-z
Wang, J., Pavlyk, I., Vedula, P., Sterling, S., Leu, N. A., Dong, D. W., et al. (2017b). Arginyltransferase ATE1 Is Targeted to the Neuronal Growth Cones and Regulates Neurite Outgrowth during Brain Development. Dev. Biol. 430 (1), 41–51. doi:10.1016/j.ydbio.2017.08.027
Watson, N., Linder, M. E., Druey, K. M., Kehrl, J. H., and Blumer, K. J. (1996). RGS Family Members: GTPase-Activating Proteins for Heterotrimeric G-Protein α-subunits. Nature 383 (6596), 172–175. doi:10.1038/383172a0
Whelan, J. P., and McGinnis, J. F. (1988). Light-dependent Subcellular Movement of Photoreceptor Proteins. J. Neurosci. Res. 20 (2), 263–270. doi:10.1002/jnr.490200216
Witherow, D. S., Wang, Q., Levay, K., Cabrera, J. L., Chen, J., Willars, G. B., et al. (2000). Complexes of the G Protein Subunit Gβ5 with the Regulators of G Protein Signaling RGS7 and RGS9. J. Biol. Chem. 275, 24872–24880. doi:10.1074/jbc.m001535200
Wong, H. C., Mao, J., Nguyen, J. T., Srinivas, S., Zhang, W., Liu, B., et al. (2000). Structural Basis of the Recognition of the Dishevelled DEP Domain in the Wnt Signaling Pathway. Nat. Struct. Biol. 7, 1178–1184. doi:10.1038/82047
Zanakis, M. F., Chakraborty, G., Sturman, J. A., and Ingoglia, N. A. (1984). Posttranslational Protein Modification by Amino Acid Addition in Intact and Regenerating Axons of the Rat Sciatic Nerve. J. Neurochem. 43, 1286–1294. doi:10.1111/j.1471-4159.1984.tb05385.x
Zhang, J.-H., and Simonds, W. F. (2000). Copurification of Brain G-Protein β5 with RGS6 and RGS7. J. Neurosci. 20 (3), RC59. doi:10.1523/jneurosci.20-03-j0004.2000
Zhang, J., Jeffrey, B. G., Morgans, C. W., Burke, N. S., Haley, T. L., Duvoisin, R. M., et al. (2010). RGS7 and -11 Complexes Accelerate the ON-Bipolar Cell Light Response. Invest. Ophthalmol. Vis. Sci. 51, 1121–1129. doi:10.1167/iovs.09-4163
Keywords: arginylation, G-protein signaling, RGS, retina, mass spectrometry
Citation: Fina ME, Wang J, Vedula P, Tang H-Y, Kashina A and Dong DW (2022) Arginylation Regulates G-protein Signaling in the Retina. Front. Cell Dev. Biol. 9:807345. doi: 10.3389/fcell.2021.807345
Received: 02 November 2021; Accepted: 17 December 2021;
Published: 21 January 2022.
Edited by:
Fangliang Zhang, University of Miami, United StatesReviewed by:
Marta E. Hallak, National University of Cordoba, ArgentinaCopyright © 2022 Fina, Wang, Vedula, Tang, Kashina and Dong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Anna Kashina, YWthc2hpbmFAdXBlbm4uZWR1; Dawei W. Dong, ZGRvbmdAdXBlbm4uZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.