- 1Department of Cardiology, Sun Yat-sen Memorial Hospital, Sun Yat-sen University, Guangzhou, China
- 2Laboratory of Cardiac Electrophysiology and Arrhythmia in Guangdong Province, Guangzhou, China
- 3Division of Vascular Surgery, The First Affiliated Hospital, Sun Yat-sen University, Guangzhou, China
- 4National-Guangdong Joint Engineering Laboratory for Diagnosis and Treatment of Vascular Diseases, First Affiliated Hospital, Sun Yat-sen University, Guangzhou, China
- 5Department of Respiratory Medicine, Sun Yat-Sen Memorial Hospital, Sun Yat-sen University, Guangzhou, China
Atheroclerosis refers to a chronic inflammatory disease featured by the accumulation of fibrofatty lesions in the intima of arteries. Cardiovasular events associated with atherosclerosis remain the major causes of mortality worldwide. Recent studies have indicated that ferroptosis, a novel programmed cell death, might participate in the process of atherosclerosis. However, the ferroptosis landscape is still not clear. In this study, 59 genes associated with ferroptosis were ultimately identified in atherosclerosis in the intima. Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses were performed for functional annotation. Through the construction of protein–protein interaction (PPI) network, five hub genes (TP53, MAPK1, STAT3, HMOX1, and PTGS2) were then validated histologically. The competing endogenous RNA (ceRNA) network of hub genes was ultimately constructed to explore the regulatory mechanism between lncRNAs, miRNAs, and hub genes. The findings provide more insights into the ferroptosis landscape and, potentially, the therapeutic targets of atherosclerosis.
Introduction
Coronary artery disease (CAD) and stroke, caused by atherosclerosis (AS), are considered as types of chronic inflammatory arterial diseases characterized by the accumulation of lipids and the formation of atherosclerotic plaques (Benjamin et al., 2017; Libby et al., 2019). Endothelial dysfunction and low-density lipoprotein cholesterol (LDL) infiltration into the subendothelial layer of arteries initiated atherogenesis (Peluso et al., 2012; Förstermann et al., 2017; Kattoor et al., 2017). Lipid peroxidation played a significant part in the pathogenesis of AS (Gisterå and Hansson, 2017).
Ferroptosis is a form of programmed cell death definitely modulated by iron-dependent lethal lipid peroxidation. Morphologically, ferroptosis is characterized by vanished mitochondria cristae and condensed and ruptured mitochondrial membranes (Mou et al., 2019). Iron overload or the inactivation of glutathione peroxidase 4 (GPX4) promoted reactive oxygen species (ROS), which hastened lipid peroxidation, eventually leading to ferroptosis (Tang et al., 2021). ROS was implicated in endothelial dysfunction, which was the initiating link of AS (Maiocchi et al., 2021). Recent studies have elucidated the status of ferroptosis in AS (Bai et al., 2020; Stockwell et al., 2020; Zhou et al., 2021). Ferrostatin-1 (Fer-1) alleviated the atherosclerotic lesion in high-fat diet (HFD)-fed ApoE−/− mice, probably via attenuating the endothelial dysfunction induced by oxidation-modified LDL (ox-LDL) (Bai et al., 2020). Prostaglandin-endoperoxide synthase 2 (PTGS2), a ferroptosis-related protein, positively correlated with the severity of AS (Zhou et al., 2021). However, how ferroptosis regulates the progress of AS still requires further investigation.
In the present study, datasets were downloaded from the Gene Expression Omnibus (GEO). Afterward, bioinformatics analyses were performed to select differentially expressed genes (DEGs). Combined with ferroptosis-related genes (FRGs), five hub genes—TP53, MAPK1, STAT3, HMOX1, and PTGS2—were ultimately screened and validated histologically.
Materials and Methods
Data Collection and Acquisition of Ferroptosis-Related Gene
The RNA expression data were collected from the GEO (http://www.ncbi.nlm.nih.gov/geo/) database with series numbers GSE97210, GSE125771, GSE41571, and GSE28829. FRGs that drive, suppress, or mark ferroptosis were retrieved from the public FerrDb database (http://www.zhounan.org/ferrdb). After removing repetitive genes, 149 FRGs that were validated by experiments were eventually obtained for subsequent analyses.
Identification of Differentially Expressed Ferroptosis-Related Genes
The “AnnoProbe” package was employed in the re-annotation of the series matrix. The “limma” package in R software was utilized to calibrate the microarray data and identify the DEGs between the atherosclerotic plaques and normal arterial intimae. The messenger RNAs (mRNAs) and long non-coding RNAs (lncRNAs) that meet the defined criteria, |log2FC| ≥ 1 and adjusted p < 0.05, were considered as DEGs and differentially expressed lncRNAs (DElncRNAs), respectively. Thereafter, the intersecting genes between DEGs and FRGs were defined as the differentially expressed ferroptosis-related genes (DE-FRGs). The DEGs and DElncRNAs were displayed in volcano plots based on the “ggplot2” package. The number of DE-FRGs was shown in a Venn diagram using the “Venndiagram” package. The expressions of DE-FRGs were visualized in a heatmap with the “ggplot2” package.
Gene Set Enrichment Analysis in Atherosclerotic Plaque
Gene set enrichment analysis (GSEA) was employed to detect the related signaling pathways in atherosclerotic plaque progression, which was performed using the OmicStudio online tool (http://www.omicstudio.cn/tool). The significant gene sets that conform to the nominal (NOM) p-value <0.05 and false discovery rate (FDR) <25% were shown.
Functional Annotation and Pathway Enrichment of DEGs and DE-FGRs
Gene Ontology (GO) biological process and Kyoto Encyclopedia of Genes and Genomes (KEGG) annotation were performed using the Metascape website (http://metascape.org). Biological process and KEGG pathway enrichment analyses using the “ClusterProfiler” package were performed to obtain insights into the potential functions of the DEGs and DE-FRGs. The top 20 results were shown in the enrichment scatter plots.
Construction of Protein–Protein Interaction Network of DE-FRGs
The STRING database (http://string-db.org/) was employed to analyze the interactions of the distinct DE-FRGs. Cytoscape software 3.8.1 (http://cytoscape.org/) was then utilized to construct and visualize the protein–protein interaction (PPI) network. The molecular complexes were examined using the MCODE algorithm. The top 5 genes of the PPI network were defined as the hub genes, which were calculated based on the maximum neighborhood component (MNC), degree, and edge percolated component (EPC) algorithms by utilizing the cytoHubba plug-in.
Identification of the Correlation of Hub FRGs and Mitochondrial Function-Related Genes
A total of 1,262 genes related to mitochondrial function were obtained from the online database Integrated Mitochondrial Protein Index (IMPI; https://mitominer.mrc-mbu.cam.ac.uk/release-4.0/impi.do). The intersecting genes of the DEGs and mitochondrial function-related genes (MFRGs) were defined as the differentially expressed mitochondrial function-related genes (DE-MFRGs). Pearson’s correlation analysis between the hub genes and DE-MFRGs was performed utilizing the “stats” package. All results were displayed in a heatmap.
Validation of Hub Gene Expression of Atherosclerotic Plaque Datasets
The three microarray datasets of atherosclerotic plaques (GSE28829: n = 29, advanced vs. early plaques; GSE125771: n = 16, high vs. low calcified plaques; and GSE41571: n = 11, ruptured vs. stable plaques) that were retrieved from the GEO database were used to verify the expressions of the hub genes. The “limma” package was also applied to identify the DEGs with thresholds of |log2FC| ≥ 1 and adjusted p < 0.05. The results were visualized in volcano plots and the hub genes were marked.
Atherosclerosis Animal Model Procedure
The animal experiment was reviewed and approved by the Animal Ethics Committee, which was subordinate to the First Affiliated Hospital of Sun Yat-Sen University (permit no. 2021593). Five-week-old male Apoe−/− mice (purchased from GemPharmatech Co., Ltd., Nanjing, China) were subsequently randomly divided into two groups. The mice of the control group were fed with normal chow diet for 12 weeks; the mice of the high-fat group were fed a high-fat diet for 12 weeks. All the mice were sacrificed after feeding for 12 weeks, and the heart tissues, including aortic roots, were collected.
Hematoxylin and Eosin, Oil Red O, and Masson Staining
Hematoxylin and eosin (HE) and Oil Red O staining were conducted in accordance with the methods mentioned in a previous study (Jiang et al., 2020). The atherosclerotic lesions in the aortic valve of mice were observed with a microscope (CKX53; Olympus Optical Co., Ltd., Tokyo, Japan).
Immunohistochemistry Staining of Hub Gene Validation
For immunohistochemistry (IHC), the heart tissues were harvested and fixed for 24 h with 10% neutral buffered formalin before the optimal cutting temperature (OCT) embedding procedure. Then, the embedded blocks were processed as 5-μm cryosections that were fixed in 4% paraformaldehyde (PFA) for another 10 min. After washing with phosphate-buffered saline (PBS) three times, antigen retrieval was followed by the heat method for 3 min. The slides were washed in PBS for another three times, blocked in 5% bovine serum albumin (BSA) for 1 h at 37°C, and then incubated with HOMX1 antibody (GB11549; Servicebio, Wuhan, China) at 1:600 dilution, MAPK1 antibody (GB11370-1; Servicebio) at 1:200 dilution, PTGS2 antibody (GB11077-1; Servicebio) at 1:500 dilution, TP53 antibody (10442-1-AP; Proteintech, Wuhan, China) at 1:200 dilution and STAT3 antibody (ET1607-38; Huabio) at 1:100 dilution at room temperature for 60 min. Staining followed the IHC kit protocol of Servicebio, and the nuclei were visualized by hematoxylin staining and examined using the CKX53 microscope (Olympus Optical Co., Ltd.).
Construction of the lncRNA–miRNA–mRNA ceRNA Network
The co-expressions of DElncRNAs and hub genes were analyzed with Pearson’s correlation. Only DElncRAN–hub gene pairs with a correlation coefficient >0.5 and p < 0.05 were selected. Subsequently, the ENCORI dataset (starbase.sysu.edu.cn) was employed to determine potentially interacting lncRNA–microRNA (miRNA) pairs. The miRanda, TargetSacan, and ENCORI databases were chosen to identify miRNA–mRNA pairs. Finally, lncRNA–miRNA–mRNA networks, which comprised five hub FRGs, were ultimately constructed.
Results
Identification of DE-FRGs and DElncRNAs in Atherosclerotic Plaques
To investigate FRGs differentially expressed in atherosclerotic plaques, 149 FRGs were extracted from FerrDb, a database for regulators, markers, and diseases associated with ferroptosis. Through differential expression analysis of GSE97210, 4,017 lncRNAs and 6,754 genes were significantly differentially expressed in atherosclerotic plaques compared with normal intimae, with thresholds of |log2FC| ≥ 1 and adjusted p < 0.05 (Figures 1A, B). After taking the intersection of DEGs and FRGs, a total of 59 FRGs expressed differentially were defined as DE-FRGs (Figure 1C). The expressions of the 59 DE-FRGs were visualized in a heatmap (Figure 1D).
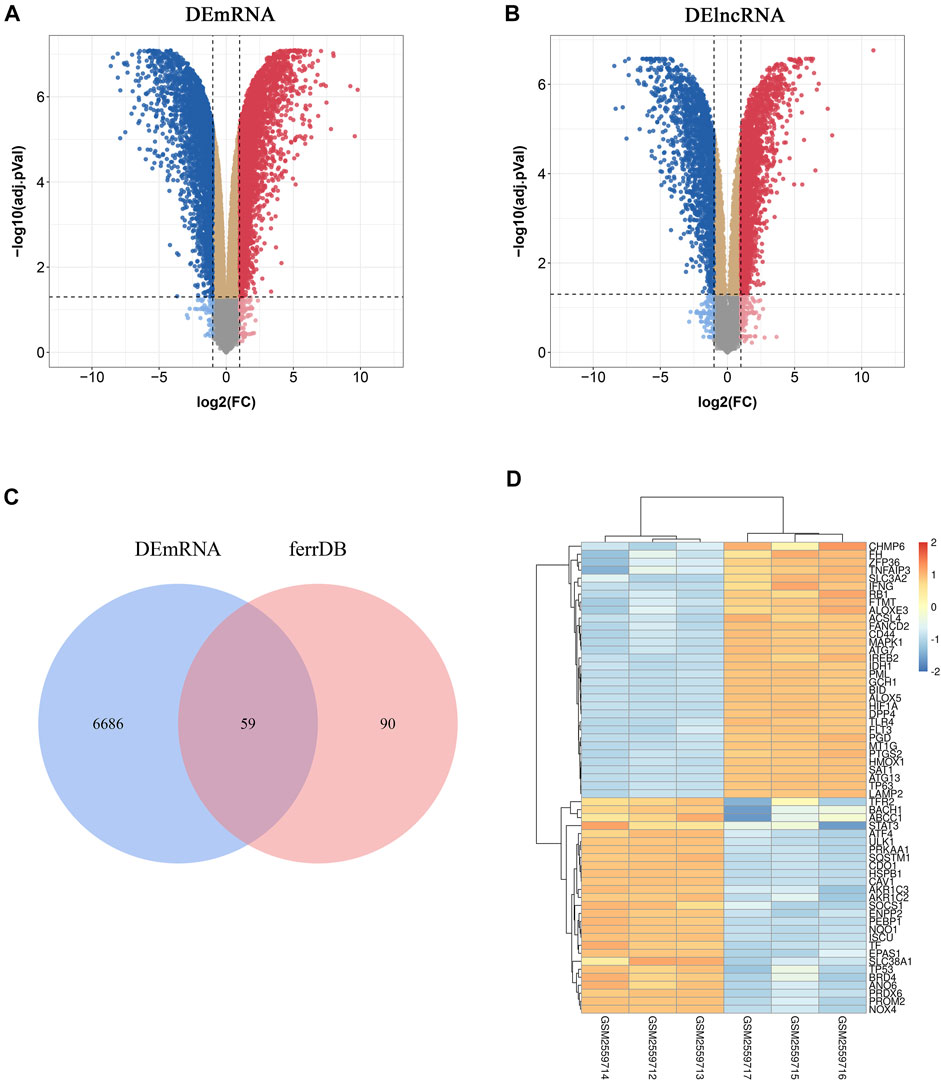
FIGURE 1. Identification of differentially expressed ferroptosis-related genes (DE-FRGs) and differentially expressed long non-coding RNAs (DElncRNAs). (A,B) Volcano plots displaying significantly differentially expressed genes (A) and lncRNAs (B) in atherosclerotic plaques. Red dots represent the upregulated genes and blue dots denote the downregulated genes, with thresholds of |log2FC| ≥ 1 and adjusted p < 0.05. (C) Venn diagram displaying the DE-FRGs. (D) Heatmap displaying the expressions of the 59 DE-FRGs in atherosclerotic plaques. Red bricks indicate the more highly expressed FRGS and blue bricks indicate lower expression.
Gene Set Enrichment Analysis
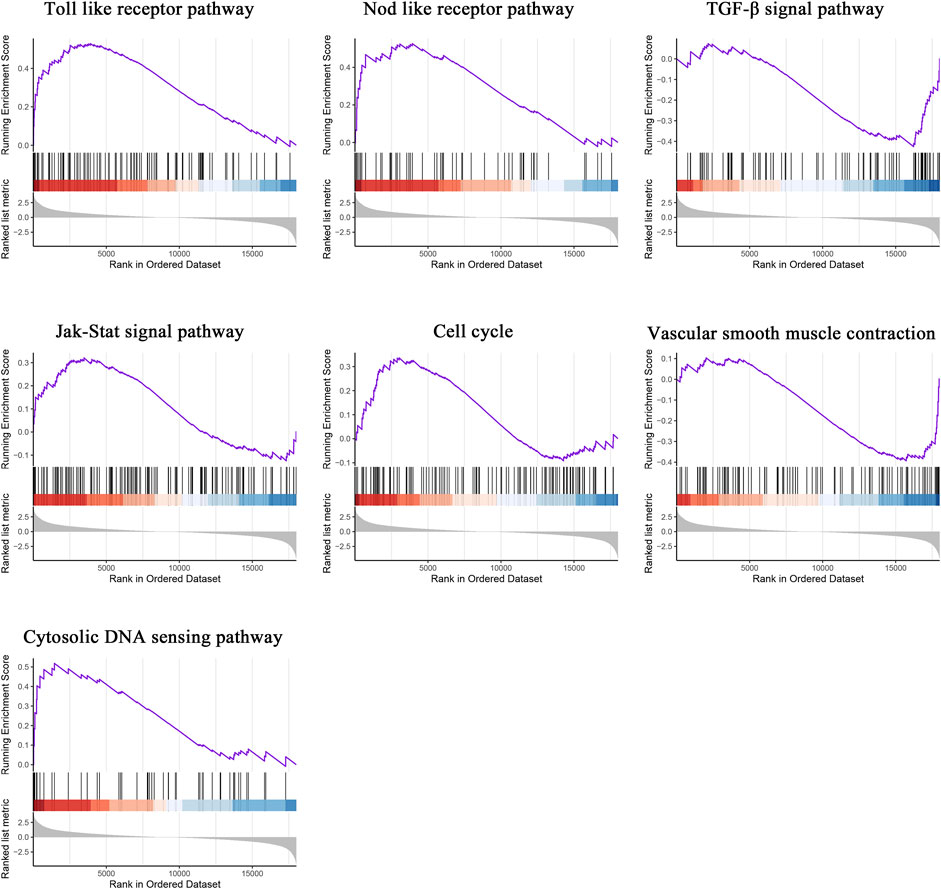
To compare the distinct pathways between the two groups, GSEA was subsequently conducted. The Toll-like receptor pathway, NOD-like receptor pathway, transforming growth factor beta (TGF-β) signal pathway, Janus kinase (JAK)/signal transducer and activator of transcription (STAT) signal pathway, cell cycle, vascular smooth muscle contraction, and cytosolic DNA sensing pathway were significantly enriched in atherosclerotic plaques (Figure 2).
Functional Enrichment Analysis of DEGs and DE-FRGs
Based on GO and KEGG pathway analyses, exploration of the potential biological functions and pathways of DEGs was conducted for the two groups. The top 20 results were shown in the enrichment scatter plots. Based on GO analysis, the DEGs were significantly enriched in related processes such as positive regulation of IκB kinase/NF-κB signaling and regulation of autophagy (Figure 3A). Investigation of the KEGG pathway analysis primarily suggested that these DEGs were involved in TGF-β signaling pathway, chemokine signaling pathway, and vascular smooth muscle contraction (Figure 3B). In addition, GO analysis was performed on DE-FRGs, for which the results indicated that the DE-FRGs were involved in iron transport, autophagy, and negative regulation of the Toll-like pathway (Figure 3C). As expected, these DE-FRGs were significantly enriched in ferroptosis, mitophagy, and autophagy, as indicated by the KEGG pathway analysis (Figure 3D).
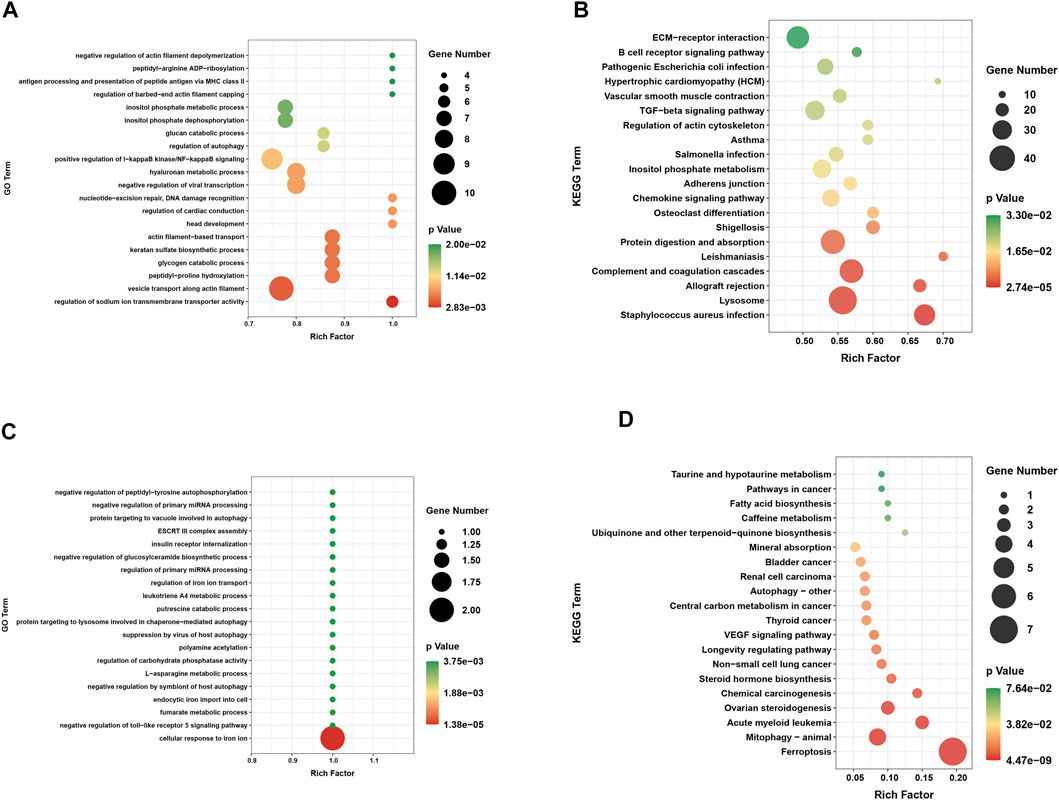
FIGURE 3. Functional annotation of DEGs and DE-FRGs. (A) GO enrichment analysis of biological processes enriched by DEGs. (B) Annotation of pathway enriched by DEGs. (C) GO enrichment analysis of biological processes enriched by DE-FRGs. (D) KEGG pathway enrichment analysis enriched by DE-FRGs.
Protein–Protein Interaction Network Construction and Visualization
For the purpose of exploring the interactions between each DE-FRG, all DE-FRGs were submitted to the STING database, which is well known for PPI. The PPI network was established and visualized using Cytoscape 3.8.1. After removing the isolated DE-FRGs, the PPI networks of DE-FRGs were displayed including 54 nodes and 190 edges (Figure 4A). According to the scores calculated using the MCODE algorithm, the PPI networks were divided into two clusters (Figure 4B). The first cluster was composed of nine genes (TP53, CD44, STAT3, CAV1, TLR4, IFNG, MAPK1, and PTGS2), whereas the second cluster consisted of six genes (LAMP2, NQO1, SQSTM1, ATG13, HMOX1, and ATF4). Subsequently, the top 5 intersecting genes analyzed based on the MNC, degree, and EPC algorithms were selected as the hub genes, which included TP53, MAPK1, STAT3, HMOX1, and PTGS2 (Figure 4C).
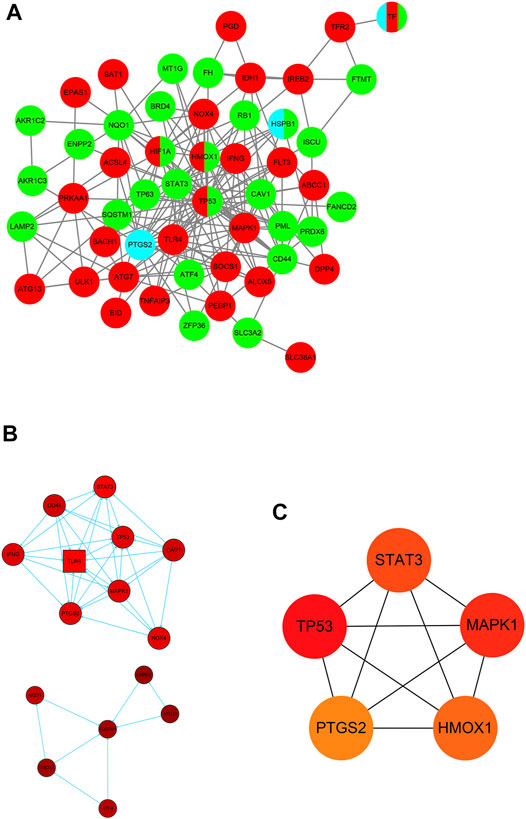
FIGURE 4. Identification of five hub ferroptosis-related genes (FRGs). (A) Protein–protein interaction (PPI) network of 59 differentially expressed FRGs (DE-FRGs). Red indicates the driver of ferroptosis, light blue indicates the marker of ferroptosis, and green indicates the suppressor of ferroptosis. (B) Two clusters of the PPI network. (C) Hub genes calculated using the maximum neighborhood component (MNC), degree, and edge percolated component (EPC) algorithms.
Identification of the Relationship Between Hub Genes and Mitochondrial Function
Since ferroptosis is always accompanied by mitochondrial dysfunction (Battaglia et al., 2020; Otasevic et al., 2021), the relationship between hub genes and MFRGs was analyzed. Among the 1,262 MFRGs, 505 genes were eventually classified as DE-MFRGs (Supplementary Figure S1A). A total of 210 DE-MFRGs were upregulated and 295 were downregulated. Pearson’s correlation analysis showed that most DE-MFRGs were highly correlated with the hub FRGs, whether positive or negative (|r| ≥ 0.5, p < 0.05) (Supplementary Figure S1B).
Validation of Hub Gene Expression in Atherosclerotic Plaque
To find out whether the hub genes were differentially expressed in other types of atherosclerotic plaques, another three microarray datasets (GSE28829, GSE125771, and GSE41571) were chosen for analysis. A total of 320 DEGs were identified between early and advanced plaques, 34 between low and highly calcified plaques, and 1,431 between ruptured and stable plaques (Figures 5A–C). Only HMOX1 among the 5 hub genes was consistently upregulated in advanced and ruptured atherosclerotic plaques, which suggested that a higher level of HMOX1 expression could predict more serious kinds of atherosclerotic plaques. HMOX1 was also highly expressed in highly calcified atherosclerotic plaques, although there was no statistical significance. To determine the expression of the five top-ranking hub genes in atherosclerotic plaques, a preclinical model of AS was generated. Compared with the control, more obvious atherosclerotic plaques and lipid accumulation were observed in mice fed the Western diet. Likewise, atherosclerotic plaques exhibited apparent fibrous caps, as revealed by Masson’s stain (Supplementary Figure S2). As expected, HMOX1 and PTGS2 were highly expressed in atherosclerotic plaques induced by the Western diet (Figure 6). However, no differences in MAPK1, TP53 and STAT3 were seen between the two groups.
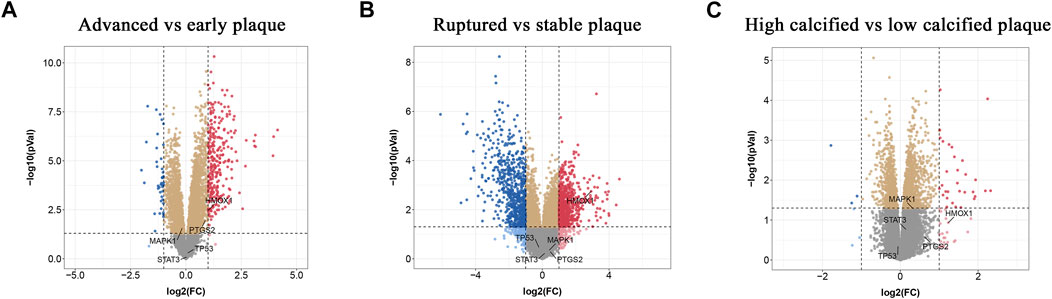
FIGURE 5. Validation of the expressions of hub ferroptosis-related genes (FRGs) in advanced, ruptured, and calcified atherosclerotic plaques. (A–C) Volcano plots displaying the expressions of hub genes in advanced, ruptured, and highly calcified plaques. Red dots indicate high relative expression and blue dots denote low relative expression.
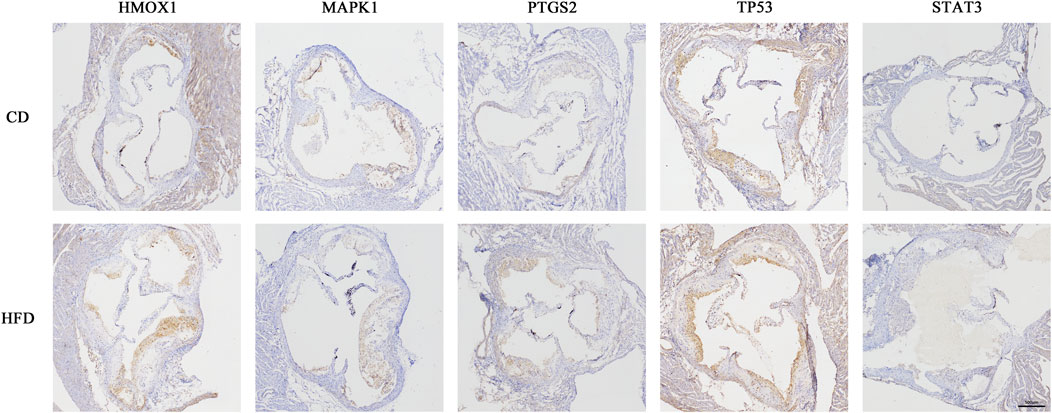
FIGURE 6. Five hub genes expressed in atherosclerotic plaques of Apoe−/− mice. Immunohistochemical staining of the aortic valve from Apoe−/− mice using HMOX1, MAPK1, PTGS2, TP53, and STAT3 antibodies.
Construction of the lncRNA–miRNA–mRNA ceRNA Network
Based on the competitive endogenous RNA hypothesis, lncRNA–miRNA–mRNA competing endogenous RNA (ceRNA) networks were constructed to explore the functions of lncRNAs acting as miRNA sponges in atherosclerotic plaques (Figure 7). The co-expressed upregulated lncRNA and hub FRG pairs were integrated into the upregulated ceRNA network with the predicted miRNAs. This ceRNA network contained 1,439 lncRNA nodes, 184 miRNA nodes, 5 hub gene nodes, and 20,292 edges.
FIGURE 7. Construction of the ceRNA Network. Red nodes indicated the predicted lncRNAs. Blue nodes indicated the predicted miRNAs. Green nodes indicated the hub genes.
Discussion
In this study, we mainly concentrated on the role of ferroptosis in the development of AS. A chronic and progressive disease of the arteries, AS is characterized by the accumulation of lipid and/or fibrous composition in the intima of arteries. Once the lipid-rich plaques rupture, a stroke or heart attack might occur, which are still the major causes of mortality worldwide (Benjamin et al., 2017). In the initial stage of AS, LDL deposition into the intima was modified by the oxidation (ox-LDL), which exhibited immunogenic and pro-inflammatory properties (Tabas et al., 2007; Tabas and Bornfeldt, 2016; Bai et al., 2020; Stockwell et al., 2020; Zhou et al., 2021). Ferroptosis was induced by excessive iron and lethal lipid peroxidation (Bai et al., 2020; Stockwell et al., 2020; Zhou et al., 2021). Excessive iron accelerated the production of ROS, which led to lipid peroxidation via Fenton reaction, which is necessary for ferroptosis (Pratt et al., 2011; Dev and Babitt, 2017; Stockwell et al., 2017). Several studies have demonstrated that iron overload bolstered the development of AS, and a low-iron diet or the administration of iron chelators reduced the severity of AS in preclinical models (Zhang et al., 2010; Hu et al., 2019; Cai et al., 2020; Vinchi et al., 2020). Consistently, recent studies have highlighted the importance of ferroptosis in AS. In vitro, ox-LDL triggered the ferroptosis of human umbilical vein endothelial cells (HUVECs) with elevated ROS generation and impaired viability, which was rescued by the activation of PDSS2/Nrf2 signaling (Yang et al., 2021). In addition, in a mouse model of AS, suppression of ferroptosis by Fer-1 improved the ROS-stimulated lipid peroxidation and endothelial dysfunction, thus attenuating the atherosclerotic lesion (Bai et al., 2020).
We screened five ferroptosis-related genes—TP53, MAPK1, STAT3, HMOX1, and PTGS2—that are probably implicated in AS via bioinformatics analysis. The tumor suppressor p53 (TP53) is considered as a classical tumor suppressor. In response to cellular stresses such as DNA damage, hypoxia, oncogene activation, and ribosomal stress, activated TP53 could boost cell cycle arrest, DNA damage repair, various pathways of cell death, and metabolic changes (Hafner et al., 2019). It was reported that a TP53 mutation was involved in oncogenesis (Levine, 2020). TP53 has been demonstrated to promote cancer ferroptosis predominantly via regulating SLC7A11 expression and cystine uptake (Jiang et al., 2015). Importantly, enhancing TP53 activity protected against AS development in HFD-fed ApoE−/− mice via regulating smooth muscle cell proliferation and apoptosis (Mercer et al., 2005; Wu et al., 2014). TP53 is a well-known gene for the tumor suppressor protein p53 that participates in AS (Guevara et al., 1999; Merched et al., 2003), but is also involved in lipid metabolism (Goldstein et al., 2012). Surprisingly, bioinformatics analysis indicated the relatively low expression of TP53 in atherosclerotic plaques. IHC staining of TP53 showed no differences between mouse atherosclerotic plaques. It seems that the expression of TP53 could not distinguish the deteriorated plaques. This result might be due to the diversity of plaque lesions, for which further experimental evidence is needed. Heme oxygenase (HMOX1), a rate-limiting enzyme of heme degradation process, controls the generation of biliverdin, iron, and carbon monoxide (Stocker and Perrella, 2006). HMOX1, as the downstream of Nrf2, is involved in the maintenance of cellular homeostasis, but its role in ferroptosis is controversial in cancer cells and renal proximal tubule cells (Wang et al., 2021). The different effects may be related to different cells. In vascular cells, the role of HMOX1 is protective in endothelial dysfunction (Nitti et al., 2020). Our study suggested that a higher level of HMOX1 expression potentially predicts the much more serious types of atherosclerotic plaques, but independently associated with calcification. Based on this, the effect of HMOX1 involved in ferroptosis and AS in vitro was investigated to further explore the mechanism responsible. MAPK1 encoded mitogen-associated protein kinase 1 (MAPK1), a component of the mitogen-activated protein kinase (MAPK)/extracellular signal-regulated kinase (ERK) signaling pathway, which stimulated ferroptosis via increasing ROS production (Liu et al., 2021; Su et al., 2019). The downregulation of the lncRNA MALAT1 could significantly improve the cardiac function in acute myocardial infarction and hypoxia by inhibiting the ERK/MAPK pathway (Fan et al., 2019). It is known that the JAK2/STAT3 pathway is involved in AS (Wang et al., 2018) and also associated with ferroptosis (Yang et al., 2020). In Jak2 mice, hematopoietic Jak2VF expression contributed to early lesion formation and increased complexity in advanced AS, which promoted the accumulation of iron in plaques and increased necrotic core formation (Wang et al., 2018). Furthermore, Yang et al. found that Auranofin, an anti-inflammatory drug used to treat rheumatoid arthritis, mitigates systemic iron overload and induces ferroptosis (Yang et al., 2020). PTGS2 upregulation was suggested to be a downstream marker of ferroptosis, and it was confirmed that PTGS2 is a hub gene of ferroptosis in human coronary artery AS (Zhou et al., 2021). A correlation was shown between the expressions of PTGS2 and ACSL4, and caspase-1 and NLRP3(12). Although functional roles of FRGs were implied in our study, the involved signaling pathways and interaction networks within lncRNAs should be clarified and validated in further research.
Iron bioresorbable coronary scaffold (IBS) system was enrolled in a randomized phase III clinical trial on stent implantation in CAD patients. The preclinical study showed a relatively higher fibrin score in the IBS group, which means a higher risk of thrombogenicity than permanent cobalt–chromium alloy and durable polymer (Zheng et al., 2019). The high fibrin score and corrosion of stents might be related to ferroptosis and iron overload-regulated cell death. There was no evidence of iron bioresorbable stents promoting AS, but the stent was implanted in porcine non-atherosclerotic coronary arteries. This needs to be confirmed by further research.
In summary, we identified five ferroptosis-related genes—TP53, MAPK1, STAT3, HMOX1, and PTGS2—in the development of AS. However, the studies on ferroptosis and AS are still at an infant stage. More studies will reveal the limited molecular mechanism of ferroptosis, thus providing more evidence for ferroptosis in the prevention and treatment of AS. This study provides new insights into ferroptosis and AS.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Ethics Statement
The animal study was reviewed and approved by the Animal Ethics Committee of the First Affiliated Hospital of Sun Yat-Sen University.
Author Contributions
CY, JW, and YC were responsible for the concept and design of this study. KW and YY contributed to the bioinformatics analysis. KW and YL performed the animal experiments. TH, KW, and YL made the major effort of manuscript preparation. All authors read and approved the final manuscript.
Funding
This study was supported by the National Natural Science Foundation of China (grant numbers 82070495 and 82070237).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Dr. Jianming Zeng (University of Macau) and all the members of his bioinformatics team, biotrainee, for generously sharing their experience and codes.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2021.800833/full#supplementary-material
References
Bai, T., Li, M., Liu, Y., Qiao, Z., and Wang, Z. (2020). Inhibition of Ferroptosis Alleviates Atherosclerosis through Attenuating Lipid Peroxidation and Endothelial Dysfunction in Mouse Aortic Endothelial Cell. Free Radic. Biol. Med. 160, 92–102. PMID: 32768568. doi:10.1016/j.freeradbiomed.2020.07.026
Battaglia, A. M., Chirillo, R., Aversa, I., Sacco, A., Costanzo, F., and Biamonte, F. (2020). Ferroptosis and Cancer: Mitochondria Meet the "Iron Maiden" Cell Death. Cells 9 (6). ARTN 1505. doi:10.3390/cells9061505
Benjamin, E. J., Blaha, M. J., Chiuve, S. E., Cushman, M., Das, S. R., Deo, R., et al. (2017). Heart Disease and Stroke Statistics-2017 Update: A Report from the American Heart Association. Circulation 135 (10), e146–e603. PubMed PMID: 28122885; PubMed Central PMCID: PMCPMC5408160. doi:10.1161/CIR.0000000000000485
Cai, J., Zhang, M., Liu, Y., Li, H., Shang, L., Xu, T., et al. (2020). Iron Accumulation in Macrophages Promotes the Formation of Foam Cells and Development of Atherosclerosis. Cell Biosci 10 (1), 137. PubMed PMID: 33292517; PubMed Central PMCID: PMCPMC7691057. doi:10.1186/s13578-020-00500-5
Dev, S., and Babitt, J. L. (2017). Overview of Iron Metabolism in Health and Disease. Hemodialysis Int. 21 (Suppl. 1), S6–S20. PubMed PMID: 28296010; PubMed Central PMCID: PMCPMC5977983. doi:10.1111/hdi.12542
Fan, Y. Z., Huang, H., Wang, S., Tan, G. J., and Zhang, Q. Z. (2019). Effect of lncRNA MALAT1 on Rats with Myocardial Infarction through Regulating ERK/MAPK Signaling Pathway. Eur. Rev. Med. Pharmacol. Sci. 23 (20), 9041–9049. PubMed PMID: WOS:000495355200039. doi:10.26355/eurrev_201910_19306
Förstermann, U., Xia, N., and Li, H. (2017). Roles of Vascular Oxidative Stress and Nitric Oxide in the Pathogenesis of Atherosclerosis. Circ. Res. 120 (4), 713–735. PubMed PMID: 28209797. doi:10.1161/CIRCRESAHA.116.309326
Gisterå, A., and Hansson, G. K. (2017). The Immunology of Atherosclerosis. Nat. Rev. Nephrol. 13 (6), 368–380. PMID: WOS:000401409300008. doi:10.1038/nrneph.2017.51
Goldstein, I., Ezra, O., Rivlin, N., Molchadsky, A., Madar, S., Goldfinger, N., et al. (2012). p53, a Novel Regulator of Lipid Metabolism Pathways. J. Hepatol. 56 (3), 656–662. PubMed PMID: WOS:000301221200023. doi:10.1016/j.jhep.2011.08.022
Guevara, N. V., Kim, H.-S., Antonova, E. I., and Chan, L. (1999). The Absence of P53 Accelerates Atherosclerosis by Increasing Cell Proliferation In Vivo. Nat. Med. 5(3), 335–339. doi:10.1038/6585
Hafner, A., Bulyk, M. L., Jambhekar, A., and Lahav, G. (2019). The Multiple Mechanisms that Regulate P53 Activity and Cell Fate. Nat. Rev. Mol. Cel Biol 20 (4), 199–210. PubMed PMID: 30824861. doi:10.1038/s41580-019-0110-x
Hu, X., Cai, X., Ma, R., Fu, W., Zhang, C., and Du, X. (2019). Iron‐load Exacerbates the Severity of Atherosclerosis via Inducing Inflammation and Enhancing the Glycolysis in Macrophages. J. Cel Physiol 234 (10), 18792–18800. PubMed PMID: 30927265. doi:10.1002/jcp.28518
Jiang, L., Kon, N., Li, T., Wang, S.-J., Su, T., Hibshoosh, H., et al. (2015). Ferroptosis as a P53-Mediated Activity during Tumour Suppression. Nature 520 (7545), 57–62. PubMed PMID: 25799988; PubMed Central PMCID: PMCPMC4455927. doi:10.1038/nature14344
Jiang, L., Qiao, Y., Wang, Z., Ma, X., Wang, H., and Li, J. (2020). Inhibition of microRNA‐103 Attenuates Inflammation and Endoplasmic Reticulum Stress in Atherosclerosis through Disrupting the PTEN‐mediated MAPK Signaling. J. Cel Physiol 235 (1), 380–393. PubMed PMID: WOS:000488990600031. doi:10.1002/jcp.28979
Kattoor, A. J., Pothineni, N. V. K., Palagiri, D., and Mehta, J. L. (2017). Oxidative Stress in Atherosclerosis. Curr. Atheroscler. Rep. 19 (11), 42. PubMed PMID: 28921056. doi:10.1007/s11883-017-0678-6
Levine, A. J. (2020). p53: 800 Million Years of Evolution and 40 Years of Discovery. Nat. Rev. Cancer 20 (8), 471–480. PubMed PMID: 32404993. doi:10.1038/s41568-020-0262-1
Libby, P., Buring, J. E., Badimon, L., Hansson, G. K., Deanfield, J., Bittencourt, M. S., et al. (2019). Atherosclerosis. Nat. Rev. Dis. Primers 5 (1), 56. Epub 2019/08/20PubMed PMID: 31420554. doi:10.1038/s41572-019-0106-z
Liu, T. Y., Li, X. H., Cui, Y. T., Meng, P. P., Zeng, G. H., Wang, Q., et al. (2021). Bioinformatics Analysis Identifies Potential Ferroptosis Key Genes in the Pathogenesis of Intracerebral Hemorrhage. Front. Neurosci-switz 15. ARTN 661663. doi:10.3389/fnins.2021.661663
Maiocchi, S. L., Ku, J., Thai, T., Chan, E., Rees, M. D., and Thomas, S. R. (2021). Myeloperoxidase: A Versatile Mediator of Endothelial Dysfunction and Therapeutic Target during Cardiovascular Disease. Pharmacol. Ther. 221, 107711. PubMed PMID: 33137376. doi:10.1016/j.pharmthera.2020.107711
Mercer, J., Figg, N., Stoneman, V., Braganza, D., and Bennett, M. R. (2005). Endogenous P53 Protects Vascular Smooth Muscle Cells from Apoptosis and Reduces Atherosclerosis in ApoE Knockout Mice. Circ. Res. 96 (6), 667–674. PubMed PMID: 15746445. doi:10.1161/01.RES.0000161069.15577.ca
Merched, A. J., Williams, E., and Chan, L. (2003). Macrophage-specific P53 Expression Plays a Crucial Role in Atherosclerosis Development and Plaque Remodeling. Atvb 23 (9), 1608–1614. PubMed PMID: WOS:000185318700021. doi:10.1161/01.Atv.0000084825.88022.53
Mou, Y., Wang, J., Wu, J., He, D., Zhang, C., Duan, C., et al. (2019). Ferroptosis, a New Form of Cell Death: Opportunities and Challenges in Cancer. J. Hematol. Oncol. 12 (1), 34. PubMed PMID: 30925886; PubMed Central PMCID: PMCPMC6441206. doi:10.1186/s13045-019-0720-y
Nitti, M., Furfaro, A. L., and Mann, G. E. (2020). Heme Oxygenase Dependent Bilirubin Generation in Vascular Cells: A Role in Preventing Endothelial Dysfunction in Local Tissue Microenvironment? Front. Physiol. 11, 23. ARTN 23. doi:10.3389/fphys.2020.00023
Otasevic, V., Vucetic, M., Grigorov, I., Martinovic, V., and Stancic, A. (2021). Ferroptosis in Different Pathological Contexts Seen through the Eyes of Mitochondria. Oxid Med. Cel Longev 2021, 5537330. Artn 5537330. doi:10.1155/2021/5537330
Peluso, I., Morabito, G., Urban, L., Ioannone, F., and Serafi, M. (2012). Oxidative Stress in Atherosclerosis Development: the central Role of LDL and Oxidative Burst. Emiddt 12 (4), 351–360. PubMed PMID: 23061409. doi:10.2174/187153012803832602
Pratt, D. A., Tallman, K. A., and Porter, N. A. (2011). Free Radical Oxidation of Polyunsaturated Lipids: New Mechanistic Insights and the Development of Peroxyl Radical Clocks. Acc. Chem. Res. 44 (6), 458–467. PubMed PMID: 21486044; PubMed Central PMCID: PMCPMC3124811. doi:10.1021/ar200024c
Stocker, R., and Perrella, M. A. (2006). Heme Oxygenase-1. Circulation 114 (20), 2178–2189. PubMed PMID: 17101869. doi:10.1161/CIRCULATIONAHA.105.598698
Stockwell, B. R., Friedmann Angeli, J. P., Bayir, H., Bush, A. I., Conrad, M., Dixon, S. J., et al. (2017). Ferroptosis: A Regulated Cell Death Nexus Linking Metabolism, Redox Biology, and Disease. Cell 171 (2), 273–285. PubMed PMID: 28985560; PubMed Central PMCID: PMCPMC5685180. doi:10.1016/j.cell.2017.09.021
Stockwell, B. R., Jiang, X., and Gu, W. (2020). Emerging Mechanisms and Disease Relevance of Ferroptosis. Trends Cel Biol. 30 (6), 478–490. PubMed PMID: WOS:000533615500006. doi:10.1016/j.tcb.2020.02.009
Su, L., Jiang, X., Yang, C., Zhang, J., Chen, B., Li, Y., et al. (2019). Pannexin 1 Mediates Ferroptosis that Contributes to Renal Ischemia/reperfusion Injury. J. Biol. Chem. 294 (50), 19395–19404. PubMed PMID: WOS:000516527800035. doi:10.1074/jbc.RA119.010949
Tabas, I., and Bornfeldt, K. E. (2016). Macrophage Phenotype and Function in Different Stages of Atherosclerosis. Circ. Res. 118 (4), 653–667. PubMed PMID: WOS:000371335100010. doi:10.1161/Circresaha.115.306256
Tabas, I., Williams, K. J., and Borén, J. (2007). Subendothelial Lipoprotein Retention as the Initiating Process in Atherosclerosis. Circulation 116 (16), 1832–1844. PubMed PMID: 17938300. doi:10.1161/CIRCULATIONAHA.106.676890
Tang, D., Chen, X., Kang, R., and Kroemer, G. (2021). Ferroptosis: Molecular Mechanisms and Health Implications. Cell Res 31 (2), 107–125. PubMed PMID: 33268902; PubMed Central PMCID: PMCPMC8026611. doi:10.1038/s41422-020-00441-1
Vinchi, F., Porto, G., Simmelbauer, A., Altamura, S., Passos, S. T., Garbowski, M., et al. (2020). Atherosclerosis Is Aggravated by Iron Overload and Ameliorated by Dietary and Pharmacological Iron Restriction. Eur. Heart J. 41 (28), 2681–2695. PubMed PMID: 30903157. doi:10.1093/eurheartj/ehz112
Wang, W., Liu, W., Fidler, T., Wang, Y., Tang, Y., Woods, B., et al. (2018). Macrophage Inflammation, Erythrophagocytosis, and Accelerated Atherosclerosis in Jak2 V617F Mice. Circ. Res. 123 (11), E35–E47. PubMed PMID: WOS:000449493300001. doi:10.1161/Circresaha.118.313283
Wang, Y., Quan, F., Cao, Q., Lin, Y., Yue, C., Bi, R., et al. (2021). Quercetin Alleviates Acute Kidney Injury by Inhibiting Ferroptosis. J. Adv. Res. 28, 231–243. PubMed PMID: WOS:000648670300003. doi:10.1016/j.jare.2020.07.007
Wu, G., Cai, J., Han, Y., Chen, J., Huang, Z.-P., Chen, C., et al. (2014). LincRNA-p21 Regulates Neointima Formation, Vascular Smooth Muscle Cell Proliferation, Apoptosis, and Atherosclerosis by Enhancing P53 Activity. Circulation 130 (17), 1452–1465. PubMed PMID: 25156994; PubMed Central PMCID: PMCPMC4244705. doi:10.1161/CIRCULATIONAHA.114.011675
Yang, K., Song, H., and Yin, D. (2021). PDSS2 Inhibits the Ferroptosis of Vascular Endothelial Cells in Atherosclerosis by Activating Nrf2. J. Cardiovasc. Pharmacol. 77 (6), 767–776. PubMed PMID: 33929387; PubMed Central PMCID: PMCPMC8274586. doi:10.1097/FJC.0000000000001030
Yang, L., Wang, H., Yang, X., Wu, Q., An, P., Jin, X., et al. (2020). Auranofin Mitigates Systemic Iron Overload and Induces Ferroptosis via Distinct Mechanisms. Signal. Transduct Target. Ther. 5 (1), 138. ARTN 138. doi:10.1038/s41392-020-00253-0
Zhang, W.-J., Wei, H., and Frei, B. (2010). The Iron Chelator, Desferrioxamine, Reduces Inflammation and Atherosclerotic Lesion Development in Experimental Mice. Exp. Biol. Med. (Maywood) 235 (5), 633–641. PubMed PMID: 20463304; PubMed Central PMCID: PMCPMC3057189. doi:10.1258/ebm.2009.009229
Zheng, J.-F., Qiu, H., Tian, Y., Hu, X.-Y., Luo, T., Wu, C., et al. (2019). Preclinical Evaluation of a Novel Sirolimus-Eluting Iron Bioresorbable Coronary Scaffold in Porcine Coronary Artery at 6 Months. JACC: Cardiovasc. Interventions 12 (3), 245–255. PubMed PMID: WOS:000457565300008. doi:10.1016/j.jcin.2018.10.020
Zhou, Y., Zhou, H., Hua, L., Hou, C., Jia, Q., Chen, J., et al. (2021). Verification of Ferroptosis and Pyroptosis and Identification of PTGS2 as the Hub Gene in Human Coronary Artery Atherosclerosis. Free Radic. Biol. Med. 171, 55–68. PubMed PMID: WOS:000663621300001. doi:10.1016/j.freeradbiomed.2021.05.009
Keywords: atherosclerosis, ferroptosis, carotid artery, ceRNA, bioinformatics
Citation: Huang T, Wang K, Li Y, Ye Y, Chen Y, Wang J and Yao C (2022) Construction of a Novel Ferroptosis-Related Gene Signature of Atherosclerosis. Front. Cell Dev. Biol. 9:800833. doi: 10.3389/fcell.2021.800833
Received: 24 October 2021; Accepted: 15 November 2021;
Published: 05 January 2022.
Edited by:
Qingchun Zeng, Southern Medical University, ChinaReviewed by:
Hao Zhou, University of Wyoming, United StatesYong Liu, Guangdong Provincial People’s Hospital, China
Copyright © 2022 Huang, Wang, Li, Ye, Chen, Wang and Yao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yangxin Chen, dGpjeXgxOTk1QDE2My5jb20=; Jingfeng Wang, ZHJ3amZfc3lzdUAxMjYuY29t; Chen Yao, eWFvY2hlbkBtYWlsLnN5c3UuZWR1LmNu
†These authors have contributed equally to this work and share first authorship
 Tucheng Huang1,2†
Tucheng Huang1,2† Kangjie Wang
Kangjie Wang Chen Yao
Chen Yao