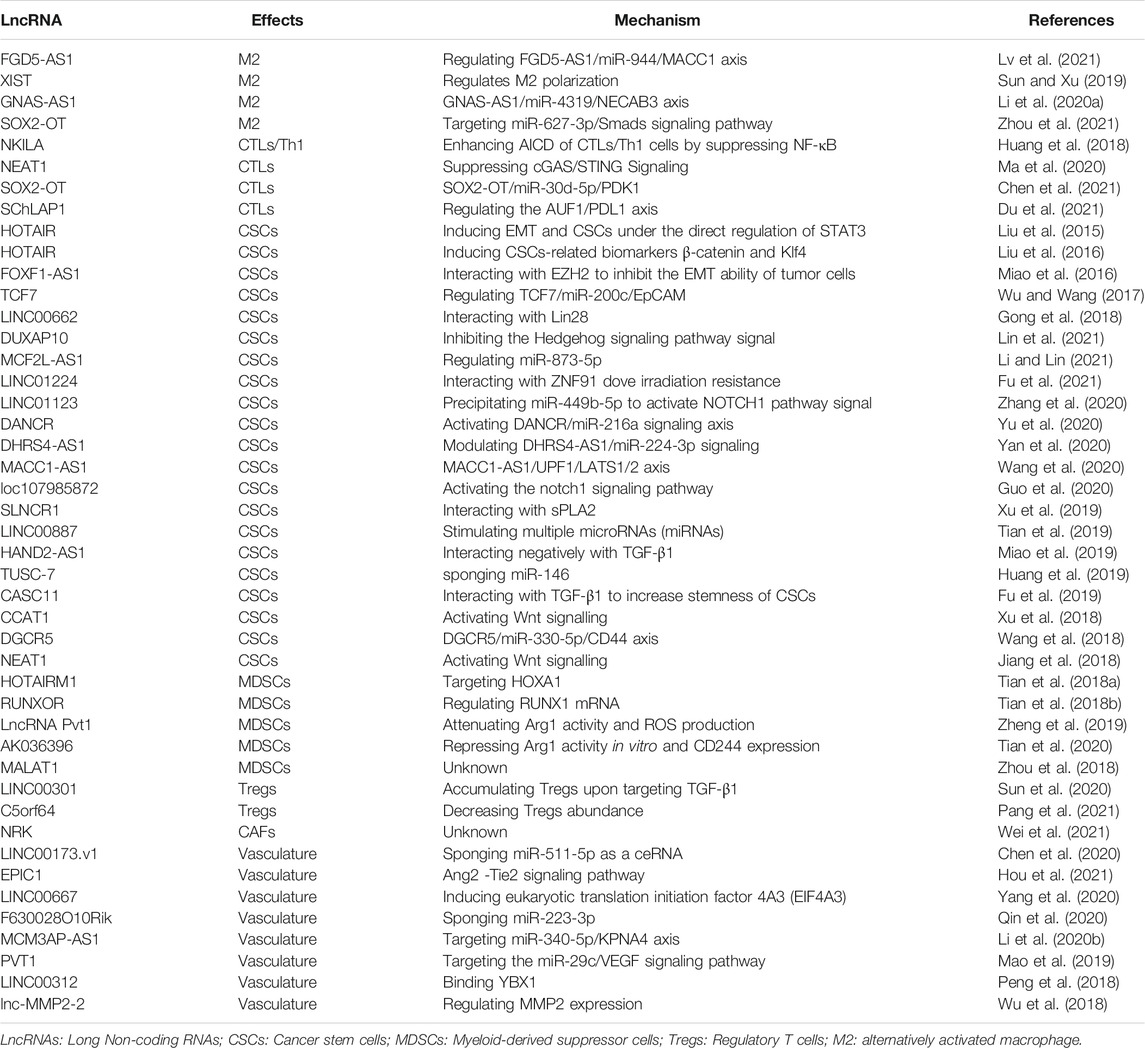
- 1Department of Medical Oncology, Lung Cancer Center, West China Hospital, Sichuan University, Chengdu, China
- 2Department of Biotherapy, Cancer Center, West China Hospital, Sichuan University, Chengdu, China
- 3Oncology Department, People’s Hospital of Deyang City, Deyang, China
- 4Department of Oncology, The First Affiliated Hospital of Chengdu Medical College, Chengdu Medical College, Chengdu, China
- 5Department of Chemoradiotherapy, Tangshan People’s Hospital, Tangshan, China
The development of various therapeutic interventions, particularly immune checkpoint inhibitor therapy, have effectively induced tumor remission for patients with advanced lung cancer. However, few cancer patients can obtain significant and long-lasting therapeutic effects for the limitation of immunological nonresponse and resistance. For this case, it’s urgent to identify new biomarkers and develop therapeutic targets for future immunotherapy. Over the past decades, tumor microenvironment (TME)-related long non-coding RNAs (lncRNAs) have gradually become well known to us. A large number of existing studies have indicated that TME-related lncRNAs are one of the major factors to realize precise diagnosis and treatment of lung cancer. Herein, this paper discusses the roles of lncRNAs in TME, and the potential application of lncRNAs as biomarkers or therapeutic targets for immunotherapy in lung cancer.
Introduction
Lung cancer ranks the most important leading cause of cancer-related deaths globally (Bray et al., 2018; Nasim et al., 2019). Non-small cell lung cancer (NSCLC) accounts for more than 80% of all lung cancers (Bender, 2014). Despite the improvements of NSCLC treatment in traditional therapies, the overall cure and survival rates for NSCLC remain low, particularly in metastatic diseases (Siegel et al., 2020). Existing research suggests that combination treatment options (using immunotherapies or targeted therapies) may be the ultimate curative option. Hence, it is quite essential to investigate the precise molecular mechanism and biomarkers to promote the effectiveness of treatment, especially immune checkpoint inhibitors (ICIs).
ICIs reactivate dysfunctional and/or exhausted T cells by targeting immune checkpoints including cytotoxic T lymphocyte related protein 4 (CLTA-4), programmed cell death protein 1 (PD-1), or its ligand, PD-L1. To date, ICIs have altered treatment paradigm in multiple indications such as melanoma, NSCLC, renal cell carcinoma (RCC) and so on (Pardoll, 2012; Hoos, 2016; Papaioannou et al., 2016; Munn and Jain, 2019; Xin Yu et al., 2019). However, the treatment response of ICIs remains unsatisfactory, and the objective response rate (ORR) for ICIs alone is only about 15–25%, and even lower in pancreatic carcinoma, triple negative breast cancer, and colorectal cancer with microsatellite stability (MSS) (Gao et al., 2019). Most patients still face the dilemmas of primary/acquired resistance of ICIs. A great number of studies have investigated the resistance mechanisms that limit the efficacy of ICIs such as disability of neoantigen presentation, activation of T cell, the impaired formation of T cell memory and the dysregulation of tumor microenvironment (TME) (Gajewski et al., 2013; Hegde et al., 2016; Jenkins et al., 2018; Yi et al., 2019; Schoenfeld and Hellmann, 2020). TME, comprised of the interaction between tumor cells, tumor-associated stromal cells as well as extracellular matrix, has been considered to be of great significance in activating the effect of ICIs. Notably, identifying abnormal TME and treatment-related biomarkers are not only the important means of antitumor therapy, but also of immune efficiency improvement (Gao et al., 2019).
LncRNAs, the most frequently expressed nonprotein-coding RNAs, have at least 200 nucleotides and are usually located in the cell nucleus, cytoplasm and exosomes where they interact with various molecules like DNA, RNA, proteins and so forth (Atianand and Fitzgerald, 2014). Multiple pathophysiological processes through the epigenetic, transcriptional and post-transcriptional regulation of gene are regulated by lncRNAs, of which lncRNAs include at least five categories including intergenic lncRNAs, intronic lncRNAs, antisense lncRNAs, sense lncRNAs and bidirectional lncRNAs (pseudogenes and retrotransposons) (Rinn and Chang, 2012). In TME, lncRNAs can directly or indirectly affect the growth of tumor cells, and play a nonnegligible role in the regulatory recircuit of the immune cells, promoting recruitment of immunosuppressive cells such as Tregs, M2-type macrophages and myeloid-derived suppressor cells (MDSCs), down-regulating the expression of adhesion molecules on endothelial cells, as well as up-regulating of immune checkpoints (PD-1/PD-L1 and CTLA4), which could contribute to tumor development and resistance to drugs or radiotherapy (Fu et al., 2021; Taheri et al., 2021). Specifically, lncRNAs are new emerging therapeutic targets and important prognostic biomarkers in multiple cancers including lung cancer (Tokgun et al., 2020). Accumulating studies have identified that lncRNAs are essential mediators of intercellular communication between tumor and stromal cells in local and distant microenvironment of lung cancer.
In this review, we focus on describing how lncRNAs derived from tumor cells, immune cells or exosomes regulate the TME in lung cancer to promote tumor progression, emphasizing the role of these lncRNAs in tumor cells, lymphoid immune cells, macrophages, cancer-related fibroblasts, tumor vasculature and other components of TME (Figure 1).
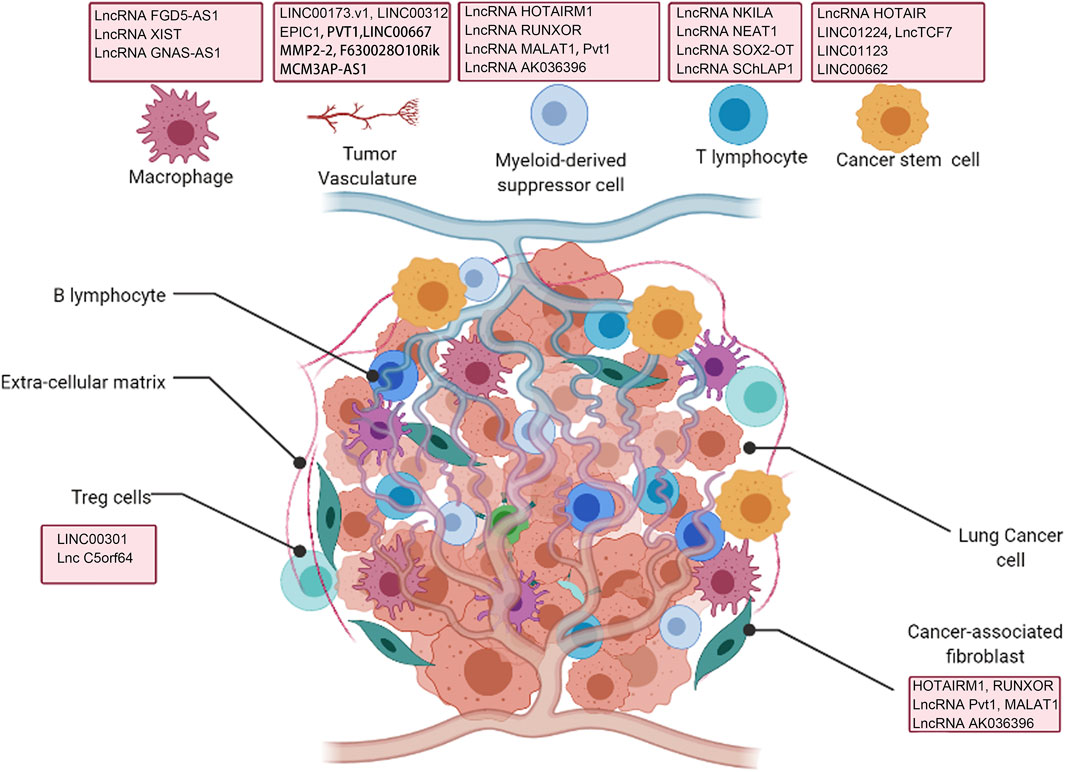
FIGURE 1. Interaction of lncRNAs with tumor microenvironment (TME) regulating lung cancer development.
LncRNAs Affect Lymphoid Immune Cells
Existing research has reported that lymphoid immune cells within tumors have two-sided (positive and negative) effects on tumorigenesis and tumor progression (Joyce and Pollard, 2009). LncRNAs as important regulator molecules influence the activity and sensitivity of tumor-infiltrating T cells. Duan et al. conducted lncRNA profiling to screen for differentially expressed lncRNAs related to CD8+ T cells and activated memory CD4+ T cells in NSCLC (Duan et al., 2020). A total of 90 DElncRNAs (differential expression lncRNA) showed different expression patterns in CD8+ T immune cells, and 48 DElncRNAs were associated with activated memory CD4+ T cells. The enrichment pathway analyses revealed that differentially expressed lncRNAs were mainly involved in cytokine–cytokine receptor interaction as well as viral protein interaction with cytokine–cytokine receptor, suggesting that T cell-specific lncRNAs mediated an immune response during NSCLC progression. This study provided a cursory but comprehensive indication of the vital role of lncRNAs in influencing the biological function of T lymphoid cells in lung cancer. Specifically, a study reported that a large proportion of CTLs/Th1 cells underwent apoptosis in NSCLC, a process that could be further facilitated by anti-CD3, while the proportion of Th2 cells/Treg cells were relatively low in TME, suggesting CTLs/Th1 cells were more sensitive to activation-induced cell death (AICD) than Tregs/Th2 cells. Further analysis indicted that STAT1-mediated transcription of NKILA caused AICD of CTLs/Th1 cells by suppressing NF-κB which was upregulated in Tregs/Th2 cells compared with CTLs/Th1 cells (Huang et al., 2018). Tang et al. also observed that the propotion of Th2 was high in peripheral blood of NSCLC, and the proportion of CTLs/Th1 cell was lower than that in normal control. LncRNA NEAT1 was also reported to be upregulated in lung cancer tissues with high TILs. NEAT1 negatively regulated CD8+ T cells in lung cancer cells via increasing CXCL10, CCL5, and IFN-β expression, which directly induced cGAS/STING Signaling, to suppress immune response (Ma et al., 2020). In addition, signaling from SOX2-OT/miR-30d-5p/PDK1 contributed to apoptosis of CD8+ T cells, which in turn promoted immune escape of NSCLC (Chen et al., 2021). The direct interaction between SChLAP1 and AUF1 antagonized the binding between AUF1 and PD-L1 mRNA 3′-UTR, resulting in improving PD-L1 mRNA stability and expression, thereby attenuating CTLs function (Du et al., 2021). Collectively, lncRNAs control lung cancer development and metastasis by affecting T cell function in TME, and targeting lncRNA may alter the activity of CTLs, thereby enhancing the effectiveness of immunotherapy.
LncRNAs in Cancer Stem Cells
Cancer stem cells (CSCs) are regarded as a population of tumor cells characterized by abilities to self-renew or to differentiate into cancer non-stem progenies, and therefore often involved in the resistance to cancer therapies, tumorigenesis, epithelial-to-mesenchymal transition (EMT) and tumor metastases (Clarke and Fuller, 2006; Zhao, 2016). CD44, CD133, OCT-4, Bmi-1, ALDH1, ABCG2 and KLF4 are common CSC biomarkers that are specifically and highly expressed on the cell surface. For example, Prior researches elucidated the determinants about the resistance to cancer treatments including growth features associated with slow division and quiescence (Vidal et al., 2014; Ajani et al., 2015), ATP-binding cassette (ABC) transporters expression levels involved in elimination of drugs (Abdullah and Chow, 2013), and the presence of increased detoxification of endogenous and exogenous aldehyde substrates via the aid of aldehyde dehydrogenases (ALDHs) (Sládek, 2003; Ahmed Laskar and Younus, 2019). As a pivotal component of the TME, the maintenance of CSCs as well as the growth and progression of tumors are inseparable from the cancer microenvironment, together with various regulatory factors. To date, lncRNAs involved in both CSCs biological functions and cancer development have received considerable research attention (Schwerdtfeger et al., 2021). Here we have reviewed relevant literatures to summarize the functions.
In regulating tumorigenesis, lncRNA HOTAIR was reported to exert pro-cancer effects by inducing CSCs and EMT formation under the direct regulation of STAT3 under cigarette smoke exposure (Liu et al., 2015). The relationship between stemness and EMT programs has been reviewed in previous literature (Wilson et al., 2020). DUXAP10 was obviously upregulated in Cd-induced lung cancer cells. DUXAP10 knockdown induced the stemness markers including KLF4, KLF5 and Nanog downregulation, weakened the capacity of spheres formation and reduced the number of stem cells marked by CD133 via inhibiting the Hedgehog signaling pathway signal, which was involved in Cd carcinogenesis in lung cells (Lin et al., 2021). As for resistance to cancer therapies, Liu et al. revealed that lncRNA HOTAIR could cause cisplatin resistance via inducing stem cell-related biomarkers β-catenin and KLF4, especially directly regulating KLF4, to promote stemness (Liu et al., 2016). Similarly, LINC01224 is obviously upregulated in NSCLC cells and associated with NSCLC radioresistance. LINC01224 knock-down dramatically promotes the abilities of self-renew by regulating the expression of ZNF91 and therefore suppresses the irradiation sensitivity of NSCLC (Fu et al., 2021). In terms of EMT and metastasis, FOXF1-AS1 acted as a protective factor and interacted with PRC2 components EZH2 to hinder self-renewal of NSCLC CSCs and reduce the number of stem-like cells, thus leading to impaired EMT capacity of tumor cells (Miao et al., 2016). LncTCF7 overexpression increased NSCLC sphere formation and expression of specific markers (EpCAM, Sox2, Oct4 and Nanog). Further study of the molecular mechanism of TCF7 regulation of CSC revealed that TCF7 may exhibit oncogenic activity by regulating EpCAM via competitively binding miR-200c, which stimulated invasive activity and enhanced the self-renewal capacity of CSCs (Wu and Wang, 2017). The interaction with LINC00662 and its RNA binding protein Lin28 can elevate CSCs stemness and invasion ability of tumor cells, contributing to the poor prognosis of NSCLC (Gong et al., 2018). The expression of LINC01123 is up-regulated in LUAD. Mechanistically, LINC01123 could precipitate miR-449b-5p to release NOTCH1, thereby promoting downstream NOTCH1 signaling and resulting in accelerating LUAD cell stemness and EMT (Zhang et al., 2020). More LncRNAs affecting CSC properties are summarized in Table 1. Taken together, all these data elucidate that dysregulation of lncRNAs affects CSCs traits and tumor invasion, and could be potential targets of tumor immunotherapy.
LncRNAs Contribute to Macrophage Plasticity
Macrophages, most of which derived from blood monocytes, are involved in EMT and are present in almost all tissues such as hepatic Kupffer cells (KCs) or brain microglia (Varol et al., 2015). According to the different mechanisms of action, macrophages are roughly divided into classically activated macrophages known as “killer” macrophages (M1) activated by IFN-γ, TNF-α as well as lipopolysaccharide (LPS) and alternatively activated macrophages known as “repair” macrophage (M2) activated by IL-4, IL-10 or IL-13 (Li et al., 2019). Basic studies have indicated that in mouse models, high levels of MHC II molecules are expressed in tumor-associated macrophages (TAMs) with the hallmark of M1 during the early stages of tumor development. However, the advanced stage of tumor is mainly characterized by low-level MHC II molecules of M2, which indicates that an M1-to-M2 transformation is present in tumor progress (Mantovani et al., 2002; Wang et al., 2011). Obviously, the accumulation of macrophages is related to tumor angiogenesis, tumor invasion and immunosuppression (Condeelis and Pollard, 2006). LncRNAs expression has been implicated in many cellular and developmental processes like cell proliferation and apoptosis (Batista and Chang, 2013; Heward and Lindsay, 2014). Prior reports have suggested that lncRNAs are also involved in regulating macrophage polarization (Huang et al., 2016).
In recent years, it has been shown that lncRNAs originating from tumor cells are involved in the polarization of TAMs and result in tumor progression. Tumor cell-derived FGD5-AS1 via exosomes transportation promotes upregulation of M2 polarization markers (CD163, CD206, ARG1) and downregulation of M1 macrophage markers iNOS and IL-2 in NSCLC (Lv et al., 2021). Sun et al. summarized that the upregulated lncRNA XIST promoted macrophage conversion to M2 characterized by the deletion of specific makers like IL-10 and CD163, to affect tumor invasion and migration of lung cancer (Sun and Xu, 2019). GNAS-AS1 inhibits miR-4319 expression, and consequently activates N-terminal EF-hand calcium binding protein 3 (NECAB3) in THP-1-differentiated macrophages. Therefore, GNAS-AS1 increases the number of M2 macrophage and consequently promoting NSCLC cell growth and metastasis (Li Z. et al., 2020). Moreover, tumor-derived exosomal SOX2 overlapping transcript also play an important role in modulating the polarization of TAMs in NSCLC though regulating SOX2/miR-627-3p/Smads axis (Zhou et al., 2021). Taken together, lncRNAs directly or indirectly regulate the polarization of TAMs to affect lung cancer progression and metastasis, but the specific regulatory mechanism controlling the macrophages M2 polarization in lung cancer needs to be further studied.
LncRNAs Promote by Regulating Immune Suppressive Cells
Studies have shown that the interactions between non-tumor cells exposed to the tumor microenvironment and tumor cells contribute to tumor progression and metastasis. In addition, T cells in tumors are often dysregulated and unable to generate specific responses to tumor cells in a timely manner. Immunosuppressive cells, including regulatory T cells (Tregs) usually expressing CD4, CD25 and FOXP3 markers (T cells that are not immunosuppressive) and MDSC, can contribute directly or indirectly to immunosuppression, which are reviewed in detail below.
Myeloid-Derived Suppressor Cells
Myeloid-derived suppressor cells (MDSCs) express two specifical markers, CD11b and Gr1, and represent a heterogeneous population of myeloid origin that are activated and proliferated by growth factors and cytokines released by tumor cells. Once MDSCs are activated, they accumulate in lymphoid organs and tumors and exert immunosuppression on T cells. Currently, the cellular mechanisms by which MDSCs have been shown to be involved in immunosuppressive activity are: 1) inhibition of CTL cells activation and proliferation in an MHC-restricted or unrestricted and antigen-specific manner (Nagaraj et al., 2007; Zou et al., 2021); 2) indirectly affects T cell activation via the induction of Treg proliferation and benefiting from TGF, IL-10 production (Serafini et al., 2008); 3) Stimulation of macrophage conversion to M2 by secreting IL-10 and down-regulation of IL-12 that promotes M1 generation (Sinha et al., 2007); 4) interaction with type II iNKT, which promotes tumor progression through IL-13 production, thereby inducing aggregation of MDSCs (Zou et al., 2021). Of note, MDSCs have immunosuppressive activity only when activated. The molecules that can activate MDSCs include two main categories: 1) tumor-derived soluble factor (TDSF), which inducing the proliferation of MDSCs though activating STAT3 to stimulate the proliferation of myeloid cells and inhibit the differentiation of mature myeloid cells, including VEGF, SCF, GM-CSF, G-CSF, IL-6, IL-10, IL-12, MMP9 and CCL2 (Talmadge, 2007; Marigo et al., 2008); 2) soluble factors released by activated T cells and tumor-derived stromal cells, such as IFN-γ, TLRs ligands, IL-4, IL-13 as well as TGF-β, are responsible for the activation of different transcription factors such as STAT6, STAT1 and NFγB (Gabrilovich and Nagaraj, 2009). Recently, it has been shown that lncRNAs affect lung cancer progression by regulating the immunosuppressive function of MDSCs in TME. Tian et al. discovered that HOTAIRM1 is obviously downregulated in MDSCs and its expression reduces in peripheral blood of lung cancer. Overexpression of HOTAIRM1 can positively target HOXA1 and induce subsequent reduction of the immunosuppression function of MDSCs, as well as increase the number of Th1/CD8+ cytotoxic T lymphocyte cells (CTLs), thereby sustaining improving the antitumor immune response (Tian et al., 2018a). Moreover, they also observed that the expression of lncRNA RUNXOR is higher in the blood of lung cancer patients than the levels in healthy samples, while decreases after surgery. Further detection suggested that RUNXOR can promote the activation of MDSCs and decrease the proportion of Th1/CTL cells by regulating RUNX1 mRNA expression (Tian et al., 2018b). A study on lncRNA MALAT1 indicated that MALAT1 directly affected MDSCs differentiation in lung cancer (Zhou et al., 2018). Some studies are more detailed. Zheng et al. identified that the expression level of lncRNA Pvt1 was upregulated in G-MDSCs following induction of IL-6 and GM-CSF. In contrast, when Pvt1 was knocked down, Arg1 activity and ROS production were significantly reduced and the ability of G-MDSCs to suppress T cells turned weak. In mice injected with Lewis lung carcinoma cells, they found that the number of CTLs/Th1 cells increased compared with the normal treatment group. The function of G-MDSCs was obviously upregulated under hypoxic conditions though targeting HIF-1α, and inhibition of HIF-1α by YC-1 apparently reduced Pvt1 expression in G-MDSCs (Zheng et al., 2019). Owing to the diverse phenotype of MDSCs, MDSCs tend to be classified into CD11b+Ly6G+Ly6Clow polymorphonuclear MDSCs (PMN-MDSCs) and CD11b+Ly6G+Ly6Chi monocytic MDSCs (M-MDSCs) in mice (Youn et al., 2008). A latest study revealed that LncRNA AK036396 had a fairly high level of expression in PMN-MDSCs, whereas knockdown of AK036396 repressed Arg1 activity in vitro and CD244 expression leading to reduction of immunosuppressive effects of PMN-MDSCs (Sagiv et al., 2015). At the mechanistic level, the researchers found that lncRNA AK036396 could interact with Fcnb through abrogating its ubiquitination to enhance its stability in the cytoplasm of myeloid cells, to enhance the immunosuppression of PMN-MDSCs and attenuate Th1/CTL cells responses, and ultimately accelerating tumor progression (Tian et al., 2020). These studies have demonstrated that lncRNAs play a significant role in the aggregation and activation of MDSCs. However, the role and regulatory mechanism during tumor progression of these lncRNAs within MDSCs in lung cancer awaits further exploration.
Treg Cells
Regulatory T cells (Tregs), as a major subset of infiltrating CD4+ T cells in TME, specifically express the master transcription factor FOXP3 (Sakaguchi et al., 2020), and have been found to suppress anti-tumor immune responses in diverse ways, including: 1) targeting TGF-β, thus inhibits the anti-tumor effects promoted by CD4+ cells, CD8+ cells, and NK cells (Konkel and Chen, 2011; Sun et al., 2020); 2) disrupting metabolism by scavenging cytokines such as IL-2, or producing immunosuppressive adenshakes by extracellular enzymes CD39 and CD73 (Spolski et al., 2018); 3) inhibiting the maturation and function of DC; 4) dissolved by granulase A or B and perforation induced CD8+ lymphocytes. Several studies involving humans and mice have shown that extrinsic tissue and tumors in different tissues contain the largest Tregs, and that the absence of Treg cells can significantly improve anti-tumor immunity. A few studies reported that lncRNA affects tumor progression by regulating the biological behaviors and function of Tregs. Sun et al. identified a novel lncRNA, LINC00301, that can promote the accumulation of Tregs and decrease CD8+ T cell in NSCLC upon targeting TGF-β1 (Sun et al., 2020). Moreover, lncRNA C5orf64 expression is positively correlated with NSCLC survival, but negatively associated with Tregs levels (Pang et al., 2021).
The Role of LncRNA in Cancer-Associated Fibroblasts
Cancer-associated fibroblasts (CAFs), as one of major components in TME that derive from the differentiation of quiescent fibroblasts by activation of various external factors like cytokines/chemokines, growth factors, hypoxia factors, lncRNAs and so on, participate in the entire cancer developmental process, from tumor initiation to progression, including carcinogenesis, proliferation, migration, EMT, drug resistance, metabolic reprogramming, angiogenesis and immunosuppression (Wu et al., 2017; Shoucair et al., 2020; Yang et al., 2021). Growing studies also reveal that lncRNAs also play a nonnegligible role in cancer cells and CAFs by shuttling via exosomes and directly within CAFs or cancer cells. But, the roles of lncRNAs in CAFs during lung cancer progression indeed remain unclear and are poorly studied. Teng et al. had attempted to identify differentially expressed lncRNAs between CAFs and normal fibroblasts in NSCLC using lncRNA profiling analysis with the intention of selecting important biomarkers working in TME. They found that upregulated lncRNAs were involved in important cancer-related regulatory pathways such as NOD-like receptor signaling (Teng et al., 2019). The roles and mechanisms of action of individual lncRNAs in CAFs of lung cancer are starting to be realized.
The Tumor Vasculature is Supported by LncRNAs
In physiological environment, angiogenesis maintains a relatively dynamic homeostasis and is strictly controlled by pro-angiogenesis and anti-angiogenesis regulators (Ribatti et al., 2007). Hypoxia and acidosis in tumor bed are often attributed to a large consumption of oxygen and nutrients and an active metabolism under a disproportionate blood supply (Kerbel, 2008; Sun, 2012). In this case, it continues to induce the production of large amounts of pro-angiogenic factors in TME (Ronca et al., 2017). Meanwhile, various angiogenic factors such as Vascular endothelial growth factor (VEGF) (Ferrara et al., 2003), angiopoietin (ANGPT) (Fagiani and Christofori, 2013) and basic fibroblast growth factor (bFGF) (Zheng et al., 2018) also increase and have immunosuppressive functions. As a result, the balance of pro- and anti-angiogenesis is disturbed in cancer, leading to a shift to angiogenesis and immunosuppressive microenvironment (Ribatti et al., 2007). Some evidence has shown that lncRNAs act on tumor progression by regulation of VEGF in lung cancer. A report (Chen et al., 2020) showed that LINC00173.v1 is upregulated in lung squamous cell carcinoma (SQC) tissues and is negatively associated with SQC prognosis. Knockdown of LINC00173.v1 suppresses VEGFA expression, thus attenuating vascular endothelial cell proliferation and migration, as well as tumorigenesis of SQC cells. Mechanistic evidence has exhibited that LINC00173. v1 exerts these functions by sponging miR-511-5p as a ceRNA. Hou et al. (2021) reported that lncRNA EPIC1 is significantly upregulated in NSCLC tissues and cells. EPIC1 silence represses Ang2 -Tie2 signaling pathway-related proteins, thereby inhibiting HUVECs (human umbilical vein endothelial cell) proliferation and channel forming abilities. lncRNA LINC00667 induces eukaryotic translation initiation factor 4A3 (EIF4A3) expression and secretion, and consequently activates the mRNA and protein levels of VEGFA. Therefore, LINC00667 promotes angiogenesis of NSCLC cells and consequently results in NSCLC tumor growth and metastasis (Yang et al., 2020). Knockdown of lncRNA F630028O10Rik in lung cancer increases VEGFA and VEGFR2 expression by sponging miR-223-3p, which means that F630028O10Rik could inhibit tube formation in vascular endothelial cells, thus further influencing angiogenesis in lung cancer (Qin et al., 2020). Besides, lncRNAs, MCM3AP-AS1 (Li X. et al., 2020) and PVT1 (Mao et al., 2019) also play a key role in accelerating angiogenesis in lung cancer, respectively. These researches provide a rationale for using the anti-angiogenic effects of lncRNAs as a therapeutic option for lung cancer. Yet, the effect on tumor angiogenesis for lncRNAs still awaits further investigation.
LncRNAs as Prognostic Biomarkers
Diagnosis and therapies using lncRNAs are being developed. For example, the investigators have conducted clinical settings to identify lncRNA biomarkers from the plasma to facilitate detection of early lung cancer (NCT03830619). Studies have suggested that lncRNAs not only interact with the TME, but closely correlate with cancer prognosis. We extracted a number of gene expression profiles of lncRNAs in Lung adenocarcinoma from the TCGA database (N = 468). Based on the best cutoff value of gene expressions, we performed Kaplan-Meier survival analysis to validate the association between part of lncRNAs mentioned above and prognosis of lung cancer using the R package “Survminer”. As illustrated in Figure 2, the key lncRNAs exhibit good performance in prognostic prediction of lung cancer (p < 0.05). For instance, FDG5-AS1, HOTAIR, NKILA and LINC00662 are regarded as risk factors for survival of lung cancer (Liu et al., 2015; Gong et al., 2018; Huang et al., 2018; Lv et al., 2021), while C5orf64, DHRS4-AS1 and FOXF1-AS1 are protective factors. These genes have important effects on lung cancer (Miao et al., 2016; Yan et al., 2020; Pang et al., 2021). As far as these lncRNAs are concerned, although there is no specific clinical application in lung cancer to date, they are still potential for the comprehensive treatment and diagnosis of lung cancer.
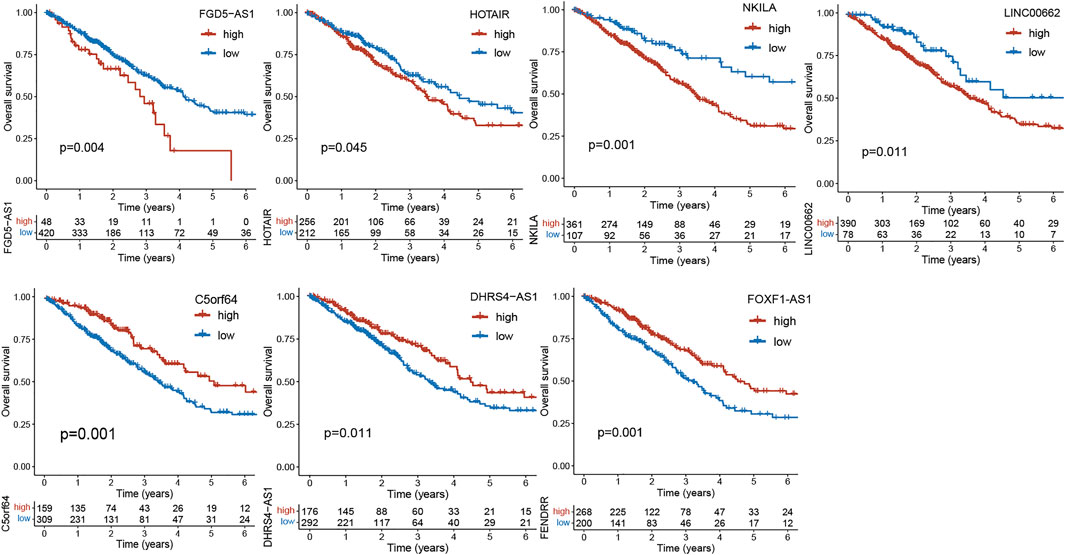
FIGURE 2. Kaplan-Meier curves of overall survival (OS) between low- and high-expression groups of lncRNAs in the TCGA cohort.
Conclusion and Future Perspectives
Lung cancer is a highly malignant tumor that poses a serious threat to human health and life. There is a lack of effective means to identify early-stage lung cancer, leading to high mortality and failure of comprehensive interventions. Meanwhile, with advances in immunotherapy, the role of TME in the lung cancer diagnosis and prognostic is becoming increasingly critical. Components of TME interacting with lncRNA have been shown to contribute to immunomodulation and cancer progress. Hence, we summarized recent advancement involving lncRNAs and their roles in the crosstalk between components of TME including infiltrated immune cells, CSCs, immune suppressive cells, macrophage, CAFs and part of the underlying molecular mechanisms. So far, only a small number of lncRNAs have been well elucidated in tumor-mesenchymal crosstalk, and in-depth studies are worthwhile to identify more lncRNAs and their specific biological functions and mechanisms of involvement. A deeper understanding of the role played by lncRNAs in the tumor microenvironment may greatly facilitate further discovery of potential biomarkers and the development of novel targeted therapies for the treatment of lung cancer. To date, the pressing issue has been the in-depth and systematic elucidation of the regulatory determinant mechanism of lncRNAs.
In conclusion, the important regulatory roles of lncRNAs in the TME have been gradually described; however, the clinical applications of lncRNAs still need to be further explored. Along with further research, tumor-associated lncRNAs crosstalk will open a new era of anti-tumor therapy.
Author Contributions
SD, TL and Y-Y L collated the data and wrote the manuscript. YH, TL, and ZX provided helpful discussion. Z-ZW and FL reviewed the manuscript. All authors reviewed the manuscript.
Funding
This study was supported by the Science and Technology Department of Sichuan Province (2018JY0389, 2019YFS0443).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
LncRNAs, Long Non-coding RNAs; LUAD, Lung adenocarcinoma; TME, Tumor microenvironment; ICIs, Immune checkpoint inhibitors; NSCLC, Non-small cell lung cancer; SQC, Lung squamous cell carcinoma; CLTA-4, cytotoxic T lymphocyte related protein 4; ORR, objective response rate; M1, Classically activated macrophages; M2, alternatively activated macrophage; MPS, monocyte macrophage system; EMT, Epithelial-to-mesenchymal transition; KCs, Kupffer cells; CTLs, Cytotoxic T lymphocyte cells; VEGF, Vascular endothelial growth factor; CSCs, Cancer stem cells; MDSCs, Myeloid-derived suppressor cells; Tregs, Regulatory T cells; VEGFA: vascular endothelial growth factor A.
References
Abdullah, L. N., and Chow, E. K. H. (2013). Mechanisms of Chemoresistance in Cancer Stem Cells. Clin. translational Med. 2, 3. doi:10.1186/2001-1326-2-3
Ahmed Laskar, A., and Younus, H. (2019). Aldehyde Toxicity and Metabolism: The Role of Aldehyde Dehydrogenases in Detoxification, Drug Resistance and Carcinogenesis. Drug Metab. Rev. 51, 42–64. doi:10.1080/03602532.2018.1555587
Ajani, J. A., Song, S., Hochster, H. S., and Steinberg, I. B. (2015). Cancer Stem Cells: The Promise and the Potential. Semin. Oncol. 42 (1), S3–S17. doi:10.1053/j.seminoncol.2015.01.001
Atianand, M. K., and Fitzgerald, K. A. (2014). Long Non-Coding RNAs and Control of Gene Expression in the Immune System. Trends Molecular Medicine 20, 623–631. doi:10.1016/j.molmed.2014.09.002
Batista, P. J., and Chang, H. Y. (2013). Long Noncoding RNAs: Cellular Address Codes in Development and Disease. Cell 152, 1298–1307. doi:10.1016/j.cell.2013.02.012
Bray, F., Ferlay, J., Soerjomataram, I., Siegel, R. L., Torre, L. A., and Jemal, A. (2018). Global Cancer Statistics 2018: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA: A Cancer J. Clinicians 68, 394–424. doi:10.3322/caac.21492
Chen, J., Liu, A., Wang, Z., Wang, B., Chai, X., Lu, W., et al. (2020). LINC00173.v1 Promotes Angiogenesis and Progression of Lung Squamous Cell Carcinoma by Sponging miR-511-5p to Regulate VEGFA Expression. Mol. Cancer 19, 98. doi:10.1186/s12943-020-01217-2
Chen, Z., Chen, Z., Xu, S., and Zhang, Q. (2021). LncRNA SOX2-OT/miR-30d-5p/PDK1 Regulates PD-L1 Checkpoint through the mTOR Signaling Pathway to Promote Non-Small Cell Lung Cancer Progression and Immune Escape. Front. Genet. 12, 674856. doi:10.3389/fgene.2021.674856
Clarke, M. F., and Fuller, M. (2006). Stem Cells and Cancer: Two Faces of Eve. Cell 124, 1111–1115. doi:10.1016/j.cell.2006.03.011
Condeelis, J., and Pollard, J. W. (2006). Macrophages: Obligate Partners for Tumor Cell Migration, Invasion, and Metastasis. Cell 124, 263–266. doi:10.1016/j.cell.2006.01.007
Du, Z., Niu, S., Wang, J., Wu, J., Li, S., and Yi, X. (2021). SChLAP1 Contributes to Non-Small Cell Lung Cancer Cell Progression and Immune Evasion through Regulating the AUF1/PD-L1 Axis. Autoimmunity 54, 1–9. doi:10.1080/08916934.2021.1913582
Duan, J., Pan, Y., Yang, X., Zhong, L., Jin, Y., Xu, J., et al. (2020). Screening of T Cell-Related Long Noncoding RNA-MicroRNA-mRNA Regulatory Networks in Non-Small-Cell Lung Cancer. Biomed. Research International 2020, 1–13. doi:10.1155/2020/5816763
Fagiani, E., and Christofori, G. (2013). Angiopoietins in Angiogenesis. Cancer Lett. 328, 18–26. doi:10.1016/j.canlet.2012.08.018
Ferrara, N., Gerber, H.-P., and LeCouter, J. (2003). The Biology of VEGF and its Receptors. Nat. Med. 9, 669–676. doi:10.1038/nm0603-669
Fu, W., Zhao, J., Hu, W., Dai, L., Jiang, Z., Zhong, S., et al. (2021). LINC01224/ZNF91 Promote Stem Cell-Like Properties and Drive Radioresistance in Non-Small Cell Lung Cancer. Cancer Manag. Res. 13, 5671–5681. doi:10.2147/cmar.s313744
Fu, Y., Zhang, P., Nan, H., Lu, Y., Zhao, J., Yang, M., et al. (2019). LncRNA CASC11 Promotes TGF-β1, Increases Cancer Cell Stemness and Predicts Postoperative Survival in Small Cell Lung Cancer. Gene 704, 91–96. doi:10.1016/j.gene.2019.04.019
Gabrilovich, D. I., and Nagaraj, S. (2009). Myeloid-derived Suppressor Cells as Regulators of the Immune System. Nat. Rev. Immunol. 9, 162–174. doi:10.1038/nri2506
Gajewski, T. F., Woo, S.-R., Zha, Y., Spaapen, R., Zheng, Y., Corrales, L., et al. (2013). Cancer Immunotherapy Strategies Based on Overcoming Barriers within the Tumor Microenvironment. Curr. Opin. Immunol. 25, 268–276. doi:10.1016/j.coi.2013.02.009
Gao, L., Yang, X., Yi, C., and Zhu, H. (2019). Adverse Events of Concurrent Immune Checkpoint Inhibitors and Antiangiogenic Agents: A Systematic Review. Front. Pharmacol. 10, 1173. doi:10.3389/fphar.2019.01173
Gong, W., Su, Y., Liu, Y., Sun, P., and Wang, X. (2018). Long Non-Coding RNA Linc00662 Promotes Cell Invasion and Contributes to Cancer Stem Cell-Like Phenotypes in Lung Cancer Cells. J. Biochem. 164, 461–469. doi:10.1093/jb/mvy078
Guo, H., Feng, Y., Yu, H., Xie, Y., Luo, F., and Wang, Y. (2020). A Novel lncRNA, Loc107985872, Promotes Lung Adenocarcinoma Progression via the Notch1 Signaling Pathway with Exposure to Traffic-Originated PM2.5 Organic Extract. Environ. Pollut. 266, 115307. doi:10.1016/j.envpol.2020.115307
Hegde, P. S., Karanikas, V., and Evers, S. (2016). The Where, the When, and the How of Immune Monitoring for Cancer Immunotherapies in the Era of Checkpoint Inhibition. Clin. Cancer Res. 22, 1865–1874. doi:10.1158/1078-0432.ccr-15-1507
Heward, J. A., and Lindsay, M. A. (2014). Long Non-Coding RNAs in the Regulation of the Immune Response. Trends Immunology 35, 408–419. doi:10.1016/j.it.2014.07.005
Hoos, A. (2016). Development of Immuno-Oncology Drugs - from CTLA4 to PD1 to the Next Generations. Nat. Rev. Drug Discov. 15, 235–247. doi:10.1038/nrd.2015.35
Hou, Y., Jia, H., Cao, Y., Zhang, S., Zhang, X., Wei, P., et al. (2021). LncRNA EPIC1 Promotes Tumor Angiogenesis via Activating the Ang2/Tie2 axis in Non-Small Cell Lung Cancer. Life Sci. 267, 118933. doi:10.1016/j.lfs.2020.118933
Huang, D., Chen, J., Yang, L., Ouyang, Q., Li, J., Lao, L., et al. (2018). NKILA lncRNA Promotes Tumor Immune Evasion by Sensitizing T Cells to Activation-Induced Cell Death. Nat. Immunol. 19, 1112–1125. doi:10.1038/s41590-018-0207-y
Huang, G., Wang, M., Li, X., Wu, J., Chen, S., Du, N., et al. (2019). TUSC7 Suppression of Notch Activation through Sponging MiR-146 Recapitulated the Asymmetric Cell Division in Lung Adenocarcinoma Stem Cells. Life Sci. 232, 116630. doi:10.1016/j.lfs.2019.116630
Huang, Z., Luo, Q., Yao, F., Qing, C., Ye, J., Deng, Y., et al. (2016). Identification of Differentially Expressed Long Non-Coding RNAs in Polarized Macrophages. Sci. Rep. 6, 19705. doi:10.1038/srep19705
Jenkins, R. W., Barbie, D. A., and Flaherty, K. T. (2018). Mechanisms of Resistance to Immune Checkpoint Inhibitors. Br. J. Cancer 118 (1), 9–16. doi:10.1038/bjc.2017.434
Jiang, P., Xu, H., Xu, C., Chen, A., Chen, L., Zhou, M., et al. (2018). NEAT1 Contributes to the CSC-Like Traits of A549/CDDP Cells via Activating Wnt Signaling Pathway. Chem. Biol. Interact. 296, 154–161. doi:10.1016/j.cbi.2018.10.001
Joyce, J. A., and Pollard, J. W. (2009). Microenvironmental Regulation of Metastasis. Nat. Rev. Cancer 9, 239–252. doi:10.1038/nrc2618
Kerbel, R. S. (2008). Tumor Angiogenesis. N. Engl. J. Med. 358, 2039–2049. doi:10.1056/NEJMra0706596
Konkel, J. E., and Chen, W. (2011). Balancing Acts: The Role of TGF-β in the Mucosal Immune System. Trends Mol. Med. 17, 668–676. doi:10.1016/j.molmed.2011.07.002
Li, S., and Lin, L. (2021). Long Noncoding RNA MCF2L‐AS1 Promotes the Cancer Stem Cell‐Like Traits in Non‐Small Cell Lung Cancer Cells through Regulating miR ‐873‐5p Level. Environ. Toxicol. 36, 1457–1465. doi:10.1002/tox.23142
Li, X., Liu, R., Su, X., Pan, Y., Han, X., Shao, C., et al. (2019). Harnessing Tumor-Associated Macrophages as Aids for Cancer Immunotherapy. Mol. Cancer 18, 177. doi:10.1186/s12943-019-1102-3
Li, X., Yu, M., and Yang, C. (2020b). YY1‐Mediated Overexpression of Long Noncoding RNA MCM3AP‐AS1 Accelerates Angiogenesis and Progression in Lung Cancer by Targeting miR‐340‐5p/KPNA4 Axis. J. Cel Biochem 121, 2258–2267. doi:10.1002/jcb.29448
Li, Z., Feng, C., Guo, J., Hu, X., and Xie, D. (2020a). GNAS-AS1/miR-4319/NECAB3 Axis Promotes Migration and Invasion of Non-Small Cell Lung Cancer Cells by Altering Macrophage Polarization. Funct. Integr. Genomics 20, 17–28. doi:10.1007/s10142-019-00696-x
Lin, H.-P., Wang, Z., and Yang, C. (2021). LncRNA DUXAP10 Upregulation and the Hedgehog Pathway Activation Are Critically Involved in Chronic Cadmium Exposure-Induced Cancer Stem Cell-Like Property. Toxicol. Sci. : official J. Soc. Toxicol. 184, 33–45. doi:10.1093/toxsci/kfab099
Liu, M.-Y., Li, X.-Q., Gao, T.-H., Cui, Y., Ma, N., Zhou, Y., et al. (2016). Elevated HOTAIR Expression Associated with Cisplatin Resistance in Non-Small Cell Lung Cancer Patients. J. Thorac. Dis. 8, 3314–3322. doi:10.21037/jtd.2016.11.75
Liu, Y., Luo, F., Xu, Y., Wang, B., Zhao, Y., Xu, W., et al. (2015). Epithelial-Mesenchymal Transition and Cancer Stem Cells, Mediated by a Long Non-coding RNA, HOTAIR, Are Involved in Cell Malignant Transformation Induced by Cigarette Smoke Extract. Toxicol. Appl. Pharmacol. 282, 9–19. doi:10.1016/j.taap.2014.10.022
Lv, J., Li, Q., Ma, R., Wang, Z., Yu, Y., Liu, H., et al. (2021). Long Noncoding RNA FGD5-AS1 Knockdown Decrease Viability, Migration, and Invasion of Non-Small Cell Lung Cancer (NSCLC) Cells by Regulating the MicroRNA-944/MACC1 Axis. Technol. Cancer Res. Treat. 20, 153303382199009. doi:10.1177/1533033821990090
Ma, F., Lei, Y.-Y., Ding, M.-G., Luo, L.-H., Xie, Y.-C., and Liu, X.-L. (2020). LncRNA NEAT1 Interacted With DNMT1 to Regulate Malignant Phenotype of Cancer Cell and Cytotoxic T Cell Infiltration via Epigenetic Inhibition of P53, cGAS, and STING in Lung Cancer. Front. Genet. 11, 250. doi:10.3389/fgene.2020.00250
Mantovani, A., Sozzani, S., Locati, M., Allavena, P., and Sica, A. (2002). Macrophage Polarization: Tumor-Associated Macrophages as a Paradigm for Polarized M2 Mononuclear Phagocytes. Trends Immunology 23, 549–555. doi:10.1016/s1471-4906(02)02302-5
Mao, Z., Xu, B., He, L., and Zhang, G. (2019). PVT1 Promotes Angiogenesis by Regulating miR-29c/Vascular Endothelial Growth Factor (VEGF) Signaling Pathway in Non-Small-Cell Lung Cancer (NSCLC). Med. Sci. Monit. 25, 5418–5425. doi:10.12659/msm.917601
Marigo, I., Dolcetti, L., Serafini, P., Zanovello, P., and Bronte, V. (2008). Tumor-induced Tolerance and Immune Suppression by Myeloid Derived Suppressor Cells. Immunol. Rev. 222, 162–179. doi:10.1111/j.1600-065X.2008.00602.x
Miao, F., Chen, J., Shi, M., Song, Y., Chen, Z., and Pang, L. (2019). LncRNA HAND2-AS1 Inhibits Non-Small Cell Lung Cancer Migration, Invasion and Maintains Cell Stemness through the Interactions with TGF-β1. Biosci. Rep. 39 (1), BSR20181525. doi:10.1042/bsr20181525
Miao, L., Huang, Z., Zengli, Z., Li, H., Chen, Q., Yao, C., et al. (2016). Loss of Long Noncoding RNA FOXF1-AS1 Regulates Epithelial-Mesenchymal Transition, Stemness and Metastasis of Non-Small Cell Lung Cancer Cells. Oncotarget 7, 68339–68349. doi:10.18632/oncotarget.11630
Munn, L. L., and Jain, R. K. (2019). Vascular Regulation of Antitumor Immunity. Science 365, 544–545. doi:10.1126/science.aaw7875
Nagaraj, S., Gupta, K., Pisarev, V., Kinarsky, L., Sherman, S., Kang, L., et al. (2007). Altered Recognition of Antigen Is a Mechanism of CD8+ T Cell Tolerance in Cancer. Nat. Med. 13, 828–835. doi:10.1038/nm1609
Nasim, F., Sabath, B. F., and Eapen, G. A. (2019). Lung Cancer. Med. Clin. North America 103, 463–473. doi:10.1016/j.mcna.2018.12.006
Pang, Z., Chen, X., Wang, Y., Wang, Y., Yan, T., Wan, J., et al. (2021). Long Non-coding RNA C5orf64 Is a Potential Indicator for Tumor Microenvironment and Mutation Pattern Remodeling in Lung Adenocarcinoma. Genomics 113, 291–304. doi:10.1016/j.ygeno.2020.12.010
Papaioannou, N. E., Beniata, O. V., Vitsos, P., Tsitsilonis, O., and Samara, P. (2016). Harnessing the Immune System to Improve Cancer Therapy. Ann. Transl. Med. 4, 261. doi:10.21037/atm.2016.04.01
Pardoll, D. M. (2012). The Blockade of Immune Checkpoints in Cancer Immunotherapy. Nat. Rev. Cancer 12, 252–264. doi:10.1038/nrc3239
Peng, Z., Wang, J., Shan, B., Li, B., Peng, W., Dong, Y., et al. (2018). The Long Noncoding RNA LINC00312 Induces Lung Adenocarcinoma Migration and Vasculogenic Mimicry through Directly Binding YBX1. Mol. Cancer 17, 167. doi:10.1186/s12943-018-0920-z
Qin, L., Zhong, M., Adah, D., Qin, L., Chen, X., Ma, C., et al. (2020). A Novel Tumour Suppressor lncRNA F630028O10Rik Inhibits Lung Cancer Angiogenesis by Regulating miR‐223‐3p. J. Cel Mol Med 24, 3549–3559. doi:10.1111/jcmm.15044
Ribatti, D., Nico, B., Crivellato, E., Roccaro, A. M., and Vacca, A. (2007). The History of the Angiogenic Switch Concept. Leukemia 21, 44–52. doi:10.1038/sj.leu.2404402
Rinn, J. L., and Chang, H. Y. (2012). Genome Regulation by Long Noncoding RNAs. Annu. Rev. Biochem. 81, 145–166. doi:10.1146/annurev-biochem-051410-092902
Ronca, R., Benkheil, M., Mitola, S., Struyf, S., and Liekens, S. (2017). Tumor Angiogenesis Revisited: Regulators and Clinical Implications. Med. Res. Rev. 37, 1231–1274. doi:10.1002/med.21452
Sagiv, J. Y., Michaeli, J., Assi, S., Mishalian, I., Kisos, H., Levy, L., et al. (2015). Phenotypic Diversity and Plasticity in Circulating Neutrophil Subpopulations in Cancer. Cel Rep. 10, 562–573. doi:10.1016/j.celrep.2014.12.039
Sakaguchi, S., Mikami, N., Wing, J. B., Tanaka, A., Ichiyama, K., and Ohkura, N. (2020). Regulatory T Cells and Human Disease. Annu. Rev. Immunol. 38, 541–566. doi:10.1146/annurev-immunol-042718-041717
Schoenfeld, A. J., and Hellmann, M. D. (2020). Acquired Resistance to Immune Checkpoint Inhibitors. Cancer cell 37 (4), 443–455. doi:10.1016/j.ccell.2020.03.017
Schwerdtfeger, M., Desiderio, V., Kobold, S., Regad, T., Zappavigna, S., and Caraglia, M. (2021). Long Non-Coding RNAs in Cancer Stem Cells. Translational Oncol. 14, 101134. doi:10.1016/j.tranon.2021.101134
Serafini, P., Mgebroff, S., Noonan, K., and Borrello, I. (2008). Myeloid-Derived Suppressor Cells Promote Cross-Tolerance in B-Cell Lymphoma by Expanding Regulatory T Cells. Cancer Res. 68, 5439–5449. doi:10.1158/0008-5472.can-07-6621
Shoucair, I., Weber Mello, F., Jabalee, J., Maleki, S., and Garnis, C. (2020). The Role of Cancer-Associated Fibroblasts and Extracellular Vesicles in Tumorigenesis. Int. J. Mol. Sci. 21, 6837. doi:10.3390/ijms21186837
Siegel, R. L., Miller, K. D., and Jemal, A. (2020). Cancer Statistics, 2020. CA A. Cancer J. Clin. 70, 7–30. doi:10.3322/caac.21590
Sinha, P., Clements, V. K., Bunt, S. K., Albelda, S. M., and Ostrand-Rosenberg, S. (2007). Cross-Talk between Myeloid-Derived Suppressor Cells and Macrophages Subverts Tumor Immunity toward a Type 2 Response. J. Immunol. 179, 977–983. doi:10.4049/jimmunol.179.2.977
Sládek, N. E. (2003). Human Aldehyde Dehydrogenases: Potential Pathological, Pharmacological, and Toxicological Impact. J. Biochem. Mol. Toxicol. 17, 7–23. doi:10.1002/jbt.10057
Spolski, R., Li, P., and Leonard, W. J. (2018). Biology and Regulation of IL-2: from Molecular Mechanisms to Human Therapy. Nat. Rev. Immunol. 18, 648–659. doi:10.1038/s41577-018-0046-y
Sun, C.-C., Zhu, W., Li, S.-J., Hu, W., Zhang, J., Zhuo, Y., et al. (2020). FOXC1-Mediated LINC00301 Facilitates Tumor Progression and Triggers an Immune-Suppressing Microenvironment in Non-Small Cell Lung Cancer by Regulating the HIF1α Pathway. Genome Med. 12, 77. doi:10.1186/s13073-020-00773-y
Sun, W. (2012). Angiogenesis in Metastatic Colorectal Cancer and the Benefits of Targeted Therapy. J. Hematol. Oncol. 5, 63. doi:10.1186/1756-8722-5-63
Sun, Y., and Xu, J. (2019). TCF-4 Regulated lncRNA-XIST Promotes M2 Polarization of Macrophages and Is Associated With Lung Cancer. Onco Targets Ther. 12, 8055–8062. doi:10.2147/ott.s210952
Taheri, M., Shoorei, H., Tondro Anamag, F., Ghafouri-Fard, S., and Dinger, M. E. (2021). LncRNAs and miRNAs Participate in Determination of Sensitivity of Cancer Cells to Cisplatin. Exp. Mol. Pathol. 123, 104602. doi:10.1016/j.yexmp.2021.104602
Talmadge, J. E. (2007). Pathways Mediating the Expansion and Immunosuppressive Activity of Myeloid-Derived Suppressor Cells and Their Relevance to Cancer Therapy. Clin. Cancer Res. 13, 5243–5248. doi:10.1158/1078-0432.ccr-07-0182
Teng, C., Huang, G., Luo, Y., Pan, Y., Wang, H., Liao, X., et al. (2019). Differential Long Noncoding RNAs Expression in Cancer-Associated Fibroblasts of Non-small-cell Lung Cancer. Pharmacogenomics 20, 143–153. doi:10.2217/pgs-2018-0102
Tian, X., Ma, J., Wang, T., Tian, J., Zhang, Y., Mao, L., et al. (2018a). Long Non-Coding RNA HOXA Transcript Antisense RNA Myeloid-Specific 1-HOXA1 Axis Downregulates the Immunosuppressive Activity of Myeloid-Derived Suppressor Cells in Lung Cancer. Front. Immunol. 9, 473. doi:10.3389/fimmu.2018.00473
Tian, X., Ma, J., Wang, T., Tian, J., Zheng, Y., Peng, R., et al. (2018b). Long Non-Coding RNA RUNXOR Accelerates MDSC-Mediated Immunosuppression in Lung Cancer. BMC cancer 18, 660. doi:10.1186/s12885-018-4564-6
Tian, X., Zheng, Y., Yin, K., Ma, J., Tian, J., Zhang, Y., et al. (2020). LncRNAAK036396Inhibits Maturation and Accelerates Immunosuppression of Polymorphonuclear Myeloid-Derived Suppressor Cells by Enhancing the Stability of Ficolin B. Cancer Immunol. Res. 8, 565–577. doi:10.1158/2326-6066.cir-19-0595
Tian, Y., Yu, M., Sun, L., Liu, L., Huo, S., Shang, W., et al. (2019). Long Non-Coding RNA00887 Reduces the Invasion and Metastasis of Non-Small Cell Lung Cancer by Causing the Degradation of miRNAs. Oncol. Rep. 42, 1173–1182. doi:10.3892/or.2019.7228
Tokgun, O., Tokgun, P. E., Inci, K., and Akca, H. (2020). lncRNAs as Potential Targets in Small Cell Lung Cancer: MYC -Dependent Regulation. Anticancer Agents Med. Chem. 20, 2074–2081. doi:10.2174/1871520620666200721130700
Varol, C., Mildner, A., and Jung, S. (2015). Macrophages: Development and Tissue Specialization. Annu. Rev. Immunol. 33, 643–675. doi:10.1146/annurev-immunol-032414-112220
Vidal, S. J., Rodriguez-Bravo, V., Galsky, M., Cordon-Cardo, C., and Domingo-Domenech, J. (2014). Targeting Cancer Stem Cells to Suppress Acquired Chemotherapy Resistance. Oncogene 33, 4451–4463. doi:10.1038/onc.2013.411
Wang, B., Li, Q., Qin, L., Zhao, S., Wang, J., and Chen, X. (2011). Transition of Tumor-Associated Macrophages from MHC Class IIhi to MHC Class IIlow Mediates Tumor Progression in Mice. BMC Immunol. 12, 43. doi:10.1186/1471-2172-12-43
Wang, R., Dong, H. X., Zeng, J., Pan, J., and Jin, X. Y. (2018). LncRNA DGCR5 Contributes to CSC‐Like Properties via Modulating miR‐330‐5p/CD44 in NSCLC. J. Cel Physiol 233, 7447–7456. doi:10.1002/jcp.26590
Wang, X., Yu, X., Wei, W., and Liu, Y. (2020). Long Noncoding RNA MACC1‐AS1 Promotes the Stemness of Nonsmall Cell Lung Cancer Cells through Promoting UPF1 ‐Mediated Destabilization of LATS1/2. Environ. Toxicol. 35, 998–1006. doi:10.1002/tox.22936
Wei, T., Song, J., Liang, K., Li, L., Mo, X., Huang, Z., et al. (2021). Identification of a Novel Therapeutic Candidate, NRK, in Primary Cancer-Associated Fibroblasts of Lung Adenocarcinoma Microenvironment. J. Cancer Res. Clin. Oncol. 147, 1049–1064. doi:10.1007/s00432-020-03489-z
Wilson, M. M., Weinberg, R. A., Lees, J. A., and Guen, V. J. (2020). Emerging Mechanisms by Which EMT Programs Control Stemness. Trends Cancer 6, 775–780. doi:10.1016/j.trecan.2020.03.011
Wu, D.-M., Deng, S.-H., Liu, T., Han, R., Zhang, T., and Xu, Y. (2018). TGF-β-Mediated Exosomal Lnc-MMP2-2 Regulates Migration and Invasion of Lung Cancer Cells to the Vasculature by Promoting MMP2 Expression. Cancer Med. 7, 5118–5129. doi:10.1002/cam4.1758
Wu, D., Zhuo, L., and Wang, X. (2017). Metabolic Reprogramming of Carcinoma-Associated Fibroblasts and its Impact on Metabolic Heterogeneity of Tumors. Semin. Cel Develop. Biol. 64, 125–131. doi:10.1016/j.semcdb.2016.11.003
Wu, J., and Wang, D. (2017). Long Noncoding RNA TCF7 Promotes Invasiveness and Self-Renewal of Human Non-Small Cell Lung Cancer Cells. Hum. Cel. 30, 23–29. doi:10.1007/s13577-016-0147-5
Xin Yu, J., Hubbard-Lucey, V. M., and Tang, J. (2019). Immuno-Oncology Drug Development Goes Global. Nat. Rev. Drug Discov. 18, 899–900. doi:10.1038/d41573-019-00167-9
Xu, C., Xiao, G., Zhang, B., Wang, M., Wang, J., Liu, D., et al. (2018). CCAT1 Stimulation of the Symmetric Division of NSCLC Stem Cells through Activation of the Wnt Signalling cascade. Gene Ther. 25, 4–12. doi:10.1038/gt.2017.98
Xu, W., Xu, Q., Kuang, D., Wang, Z., Lu, Q., Lin, Q., et al. (2019). Long Non-Coding RNA SLNCR1 Regulates Non-Small Cell Lung Cancer Migration, Invasion and Stemness through Interactions with Secretory Phospholipase A2. Mol. Med. Rep. 20, 2591–2596. doi:10.3892/mmr.2019.10518
Yan, F., Zhao, W., Xu, X., Li, C., Li, X., Liu, S., et al. (2020). LncRNA DHRS4-AS1 Inhibits the Stemness of NSCLC Cells by Sponging miR-224-3p and Upregulating TP53 and TET1. Front. Cel Dev. Biol. 8, 585251. doi:10.3389/fcell.2020.585251
Yang, H., Yang, W., Dai, W., Ma, Y., and Zhang, G. (2020). LINC00667 Promotes the Proliferation, Migration, and Pathological Angiogenesis in Non-Small Cell Lung Cancer through Stabilizing VEGFA by EIF4A3. Cell Biol Int 44, 1671–1680. doi:10.1002/cbin.11361
Yang, J., Shi, X., Yang, M., Luo, J., Gao, Q., Wang, X., et al. (2021). Glycolysis Reprogramming in Cancer-Associated Fibroblasts Promotes the Growth of Oral Cancer through the lncRNA H19/miR-675-5p/PFKFB3 Signaling Pathway. Int. J. Oral Sci. 13, 12. doi:10.1038/s41368-021-00115-7
Yi, M., Jiao, D., Qin, S., Chu, Q., Wu, K., and Li, A. (2019). Synergistic Effect of Immune Checkpoint Blockade and Anti-Angiogenesis in Cancer Treatment. Mol. Cancer 18, 60. doi:10.1186/s12943-019-0974-6
Youn, J.-I., Nagaraj, S., Collazo, M., and Gabrilovich, D. I. (2008). Subsets of Myeloid-Derived Suppressor Cells in Tumor-Bearing Mice. J. Immunol. 181, 5791–5802. doi:10.4049/jimmunol.181.8.5791
Yu, J. E., Ju, J. A., Musacchio, N., Mathias, T. J., and Vitolo, M. I. (2020). Long Noncoding RNA DANCR Activates Wnt/β-Catenin Signaling through MiR-216a Inhibition in Non-Small Cell Lung Cancer. Biomolecules 10, 1646. doi:10.3390/biom10121646
Zhang, M., Han, Y., Zheng, Y., Zhang, Y., Zhao, X., Gao, Z., et al. (2020). ZEB1-activated LINC01123 Accelerates the Malignancy in Lung Adenocarcinoma through NOTCH Signaling Pathway. Cell Death Dis 11, 981. doi:10.1038/s41419-020-03166-6
Zhao, J. (2016). Cancer Stem Cells and Chemoresistance: The Smartest Survives the Raid. Pharmacol. Ther. 160, 145–158. doi:10.1016/j.pharmthera.2016.02.008
Zheng, X., Liu, Q., Yi, M., Qin, S., and Wu, K. (2018). The Regulation of Cytokine Signaling by Retinal Determination Gene Network Pathway in Cancer. Onco Targets Ther. 11, 6479–6487. doi:10.2147/ott.s176113
Zheng, Y., Tian, X., Wang, T., Xia, X., Cao, F., Tian, J., et al. (2019). Long Noncoding RNA Pvt1 Regulates the Immunosuppression Activity of Granulocytic Myeloid-Derived Suppressor Cells in Tumor-Bearing Mice. Mol. Cancer 18, 61. doi:10.1186/s12943-019-0978-2
Zhou, D., Xia, Z., Xie, M., Gao, Y., Yu, Q., and He, B. (2021). Exosomal Long Non-Coding RNA SOX2 Overlapping Transcript Enhances the Resistance to EGFR-TKIs in Non-Small Cell Lung Cancer Cell Line H1975. Hum. Cel. 34, 1478–1489. doi:10.1007/s13577-021-00572-6
Zhou, Q., Tang, X., Tian, X., Tian, J., Zhang, Y., Ma, J., et al. (2018). LncRNA MALAT1 Negatively Regulates MDSCs in Patients with Lung Cancer. J. Cancer 9, 2436–2442. doi:10.7150/jca.24796
Keywords: lncRNA, non-small cell lung cancer, immunotherapy, tumor microenvironment, biomaker
Citation: Dai S, Liu T, Liu Y-Y, He Y, Liu T, Xu Z, Wang Z-W and Luo F (2022) Long Non-Coding RNAs in Lung Cancer: The Role in Tumor Microenvironment. Front. Cell Dev. Biol. 9:795874. doi: 10.3389/fcell.2021.795874
Received: 15 October 2021; Accepted: 08 December 2021;
Published: 03 January 2022.
Edited by:
Zong Sheng Guo, Roswell Park Comprehensive Cancer Center, United StatesReviewed by:
Mario Cioce, Campus Bio-Medico University, ItalyClaudia De Vitis, Sapienza University, Italy
Copyright © 2022 Dai, Liu, Liu, He, Liu, Xu, Wang and Luo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Feng Luo, aHhsdW9mZW5nQDE2My5jb20=; Zhi-Wu Wang, dGNtMjAwMEAxNjMuY29t
†These authors have contributed equally to this work
 Shuang Dai
Shuang Dai Ting Liu
Ting Liu Yan-Yang Liu1†
Yan-Yang Liu1† Tao Liu
Tao Liu Zhi-Wu Wang
Zhi-Wu Wang Feng Luo
Feng Luo