- Department of Cell and Developmental Biology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, United States
The septin family of proteins can assemble into filaments that further organize into different higher order structures to perform a variety of different functions in different cell types and organisms. In the budding yeast Saccharomyces cerevisiae, the septins localize to the presumptive bud site as a cortical ring prior to bud emergence, expand into an hourglass at the bud neck (cell division site) during bud growth, and finally “split” into a double ring sandwiching the cell division machinery during cytokinesis. While much work has been done to understand the functions and molecular makeups of these structures, the mechanisms underlying the transitions from one structure to another have largely remained elusive. Recent studies involving advanced imaging and in vitro reconstitution have begun to reveal the vast complexity involved in the regulation of these structural transitions, which defines the focus of discussion in this mini-review.
Introduction
Septins are GTP-binding proteins that assemble into heteropolymers that can organize into various filament-containing structures such as rings, hourglasses, and gauzes in different cell types (Byers and Goetsch, 1976a; Kim et al., 1991; Frazier et al., 1998; Lippincott et al., 2001; Ihara et al., 2005; Kissel et al., 2005; Rodal et al., 2005; John et al., 2007; Sirajuddin et al., 2007; Tada et al., 2007; Xie et al., 2007; Bertin et al., 2008; Ong et al., 2014; Renshaw et al., 2014; Karasmanis et al., 2019; Wang et al., 2019). As such, they are considered the fourth cytoskeletal component along with microfilaments, intermediate filaments, and microtubules (Mostowy and Cossart, 2012). Septins play critical roles in cytokinesis, exocytosis, mitosis, ciliogenesis, and cell morphogenesis by acting as a cellular scaffold and/or diffusion barrier (Longtine et al., 1996; Gladfelter et al., 2001; Weirich et al., 2008; McMurray and Thorner, 2009; Oh and Bi, 2011; Mostowy and Cossart, 2012; Dolat et al., 2014). Not surprisingly, mutations in human septin genes have been linked to several diseases including male infertility, cancer, and neurodegenerative diseases (Hall and Russell, 2004; Roeseler et al., 2009; Lin et al., 2011; Dolat et al., 2014; Ageta-Ishihara and Kinoshita, 2021).
Septins are conserved in eukaryotes except higher land plants (Pan et al., 2007). In humans, there are 13 septin genes whose products can form several different combinations of heterooligomers (usually octamers) depending on the tissue type in which they are expressed (Kinoshita, 2003; Pan et al., 2007; Estey et al., 2010; Sellin et al., 2011; Mendonca et al., 2019; Soroor et al., 2021). Additionally, many septin genes, especially SEPT9, code for multiple isoform variants (Robertson et al., 2004; Hall et al., 2005; Estey et al., 2010; Connolly et al., 2011a; Connolly et al., 2011b; Sellin et al., 2012). Such a complexity in septin expression and assembly has hampered the rapid progress in the analysis of human septins. Model organisms have had a major impact on our understanding of septin biology with much of the emphasis placed on the budding yeast Saccharomyces cerevisiae. There are a total of seven septin genes in S. cerevisiae, five of which (CDC3, CDC10, CDC11, CDC12, and SHS1) are expressed in mitotically active cells (Hartwell, 1971; Byers and Goetsch, 1976b; Carroll et al., 1998; Mino et al., 1998) and the other two (SPR3 and SPR28) are expressed during meiosis (Ozsarac et al., 1995; De Virgilio et al., 1996; Fares et al., 1996). The limited number of septin genes, coupled with the ease of genetic manipulation and easily scored phenotypes associated with septin malfunction, make budding yeast an excellent model organism for studying the regulation of septin organization.
To understand how septin high order structures are regulated, the precise organization of their building blocks must be known. In S. cerevisiae, the mitotic septins oligomerize into hetero-octamers comprised of a core Cdc12-Cdc3-Cdc10-Cdc10-Cdc3-Cdc12 hexamer with either Cdc11 or Shs1 at the terminal ends (Bertin et al., 2008; Garcia et al., 2011; Khan et al., 2018). The presence of Cdc11 or Shs1 can influence what higher order structures form in vitro, with Cdc11-capped octamers more likely to form paired filaments from end-on-end Cdc11 interactions (Frazier et al., 1998; Bertin et al., 2008) and Shs1-capped octamers to laterally associate into curved bundles and rings (Garcia et al., 2011). With Cdc11 and Shs1 both expressed in mitotic cells, it is possible that the regulated combination of differentially capped octamers could produce the distinct structures in vivo, from the nascent ring at the presumptive bud site, to the hourglass at the bud neck, and finally the double ring surrounding the cytokinesis machinery during cell division (Bertin et al., 2012; Ong et al., 2014; Mcquilken et al., 2017; Weems and McMurray, 2017; Marquardt et al., 2019).
The septin architecture is dynamically remodeled at the division site during the cell cycle, and this involves the regulation by post-translational modifications (PTMs) and septin-associated proteins (SAPs) (Gladfelter et al., 2001; McMurray and Thorner, 2009; Hernández-Rodríguez and Momany, 2012; Alonso et al., 2015; Perez et al., 2016). Many yeast septins have PTMs such as phosphorylation, acetylation, and sumoylation added or removed at precise times, which may change their architectural organization [for full reviews on these modifications see (Hernández-Rodríguez and Momany, 2012; Perez et al., 2016; Marquardt et al., 2019)]. In fact, several protein kinases such as the LKB1-like Elm1 and the MARK/PAR1-related Gin4 not only depend on septins for their localization to the bud neck, but also influence the stability or functionality of the septin structures present there (Longtine et al., 1998; Bouquin et al., 2000; Mortensen et al., 2002; Asano et al., 2006). Besides the PTMs, different SAPs are also involved in the regulation of septin organization during the cell cycle. For example, Bni5 is associated with the septin hourglass before cytokinesis (Lee et al., 2002; Fang et al., 2010) and can bundle septin filaments in vitro (Patasi et al., 2015; Booth et al., 2016). The Rho guanine nucleotide-exchange factor (RhoGEF) Bud3 and the anillin-like protein Bud4 can stabilize the double ring structure during cytokinesis (Wloka et al., 2011; Eluere et al., 2012; Mcquilken et al., 2017).
In this review, we will summarize and draw conclusions from recent work that has begun to illustrate the regulation that occurs at the transition times between both the nascent ring to hourglass (Marquardt et al., 2020) and the hourglass to double ring structures (Chen et al., 2020). These different structures each have specific functions at their respective stages during the cell cycle and the precise transformations over a relatively short time ensure that these functions are ordered appropriately. While we are beginning to elucidate the pathways involved in the regulation of these structural transitions, much work remains to fully comprehend the mechanisms of septin assembly and remodeling in yeast and beyond.
Transforming the Nascent Septin Ring Into a Septin Hourglass
Upon starting a new cell cycle, haploid cells develop a new bud site axial to the previous division site (Chant and Pringle, 1995; Pringle et al., 1995). Through a series of feedback loops involving the Rho-like small GTPase Cdc42, its GEFs, and GTPase-activating proteins, septin recruitment, and targeted exocytosis, a nascent ring is formed at the new budding site, and the growth of the bud begins (Gladfelter et al., 2002; Caviston et al., 2003; Iwase et al., 2006; Okada et al., 2013). The septin ring then expands into an hourglass spanning the bud-neck region. This septin hourglass serves as both a scaffold for the assembly of cytokinesis machinery such as the actomyosin ring (AMR) (Bi et al., 1998; Lippincott and Li, 1998; Fang et al., 2010; Schneider et al., 2013; Finnigan et al., 2015) and numerous other protein complexes such as the morphogenesis checkpoint cascade (Barral et al., 1999; Shulewitz et al., 1999; Longtine et al., 2000; Cid et al., 2001b) and as a membrane diffusion barrier to compartmentalize the mother and bud plasma membranes (Barral et al., 2000; Luedeke et al., 2005; Shcheprova et al., 2008). As a cellular scaffold, it is not surprising that the septin hourglass is highly stable when analyzed by fluorescence recovery after photo bleaching (FRAP), which is in stark contrast to the relatively mobile nature of the nascent ring during its initial assembly process (Caviston et al., 2003; Dobbelaere et al., 2003). It remains unclear whether an altered self-assembly state of septins and/or the addition of new SAPs during hourglass expansion accounts for this increased stability.
Understanding Paired and Unpaired Septin Filament Assembly and Organization in vitro
To understand how a stable hourglass can form from a relatively dynamic septin ring (Figure 1A), one must first understand how septins are assembled into filaments, which are further organized into higher-order structures such as rings and hourglasses. Yeast septins were first seen at the bud neck as a membrane-associated structure made from filaments with a 10 nm width (Byers and Goetsch, 1976a). However, when expressed and purified in vitro from E. coli or budding yeast in the presence of high salt (>300 mM KCl or NaCl) only septin rods of the approximate length of an octamer were formed (Figure 1B, left) (Frazier et al., 1998; Bertin et al., 2008). When the ionic strength was lowered (50–75 mM salt) and with Cdc11 serving as the terminal septin subunit were filaments visualized (Frazier et al., 1998; Bertin et al., 2008), with the C-terminal extensions (CTEs) of Cdc3 and Cdc12 interacting on neighboring filaments to form the paired filaments (Figure 1B, right) (Bertin et al., 2008; Bertin et al., 2010). In contrast, the Shs1-capped septin rods associate laterally into curved bundles and rings in vitro under low salt condition (Garcia et al., 2011). While data from this study concerning the ability for Shs1-capped septin rods to assemble end-on-end into filaments is not as clear as that of Cdc11-capped septin rods, additional analysis by Förster Resonance Energy Transfer has more conclusively negated the possibility that two Shs1-capped rods can polymerize end-on-end (Booth et al., 2015). Given that the budding yeast cytoplasm has potassium and sodium concentrations of 200–300 and 20 mM, respectively (van Eunen et al., 2010; Cyert and Philpott, 2013), the ability of yeast septins to spontaneously generate filaments in the cytoplasm should be quite low.
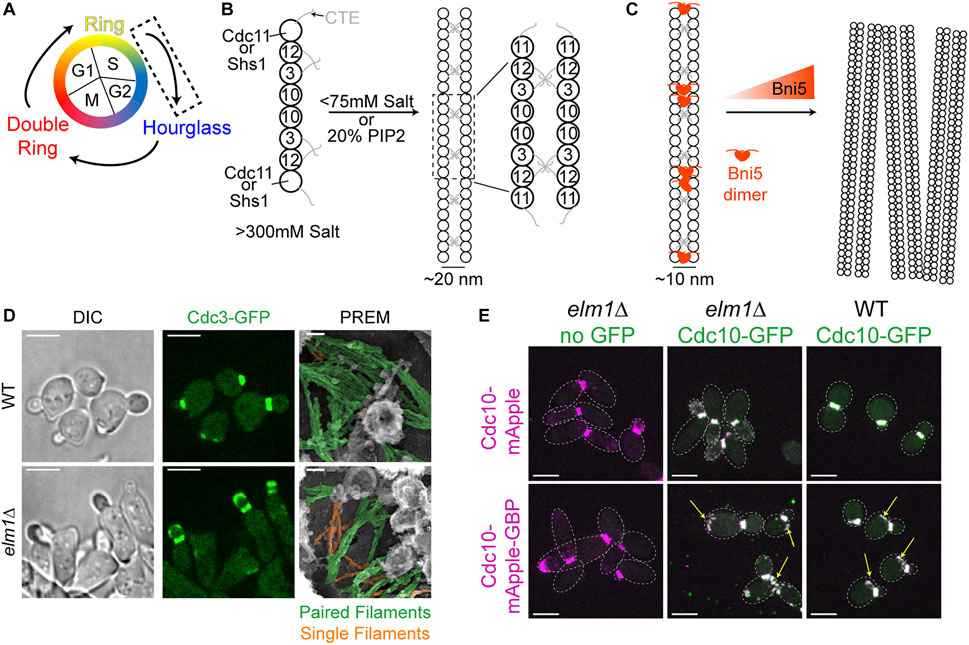
FIGURE 1. Transitioning from a dynamic ring to a stable hourglass. (A) Depiction of different septin structures visualized during the yeast cell cycle. Arrows indicate transitions occurring between two structures. Dashed box indicates that the focus of this figure is on the nascent ring-to-hourglass transition that occurs during bud emergence and expansion. (B) Left, depiction of septin rod assembly in high salt (>300 mM KCl or NaCl) in vitro (Frazier et al., 1998; Bertin et al., 2008). Right, depiction of Cdc11-capped rods forming paired filaments in either low salt (<75 mM) (Bertin et al., 2008) or on the surface of PIP2-containing lipid monolayers (Bertin et al., 2010; Bridges et al., 2014); dashed box indicates zoomed in inset shown to the right. (C) Left, depiction of two Bni5 proteins (red molecules) interacting with Cdc11-CTEs found in paired septin filaments to lower the average interfilament distance to 10 nm (Finnigan et al., 2015; Booth et al., 2016). Right, depiction of Bni5 bundling septin filaments into large clusters in vitro in a dose-dependent manner (Patasi et al., 2015). (D) Representative images of WT (top) or elm1Δ (bottom) cells synchronized in G1 by α-factor arrest and released for 1.5 h to obtain small-to medium-sized buds. Left and middle panels show DIC and fluorescence imaging of Cdc3 (green), respectively; scale bars = 5 µm. Right panels show PREM images of samples with paired filaments pseudo-colored green and single filaments pseudo-colored orange; scale bars = 50 nm. Images taken from (Marquardt et al., 2020). (E) Representative images of indicated strains with Cdc10-mApple (top row) or Cdc10-mApple-GBP (bottom row) in magenta and Cdc10-GFP, which is expressed exogenously from a CEN plasmid, in green. Images are maximum projections. Yellow arrow indicates septin ring from previous cell cycle. Dotted line is cell periphery. Scale bars = 5 µm. Images taken from (Marquardt et al., 2020).
Strikingly, in the presence of lipid monolayers containing 20% phosphatidylinositol-4,5-bisphosphate (PIP2), purified Cdc11-capped septin rods could form long paired septin filaments even at higher salt concentrations (Figure 1B, right) (Bertin et al., 2010; Bridges et al., 2014). This implies that septin octamers probably exist in the cytoplasm and are assembled into paired filaments once bound to the plasma membrane at the presumptive bud site or the bud neck. This indeed appears to be the case, as it was found by fluorescence correlation spectroscopy that septins in the cytoplasm exist as complexes and not as monomers or filaments (Bridges et al., 2014). The septin complexes appear to “sense” and favor the membrane curvature found at the bud neck, as the purified septin complexes preferentially engaged with lipid-coated beads of diameters producing a similar positive curvature as that found at the bud neck (Bridges et al., 2016; Cannon et al., 2019). The organization of the septin filaments into higher-order structures may also be governed by the changes in membrane curvature during bud growth and cytokinesis, as suggested by a recent study that examined and modeled the impact of varying degrees of positive and negative curvature on septin filament assembly and organization on lipid-covered surfaces in vitro (Beber et al., 2019).
Higher-order assemblies of septins are likely regulated by PTMs as well as SAPs (Gladfelter et al., 2001; Mcmurray and Thorner, 2009; Hernández-Rodríguez and Momany, 2012; Alonso et al., 2015; Perez et al., 2016). One such SAP, Bni5, localizes to the bud neck in a septin-dependent manner (Lee et al., 2002). Bni5 appears to interact with the CTEs of the terminal subunits Cdc11 and Shs1 in vitro and in vivo (Figure 1C) (Finnigan et al., 2015; Booth et al., 2016). Recombinant Bni5 can reduce the distance between individual filaments of a paired filament and can also bundle septin filaments, thereby contributing to a higher-order organization (Figures 1B,C) (Patasi et al., 2015; Booth et al., 2016). It is possible that a combination of phospholipid composition, PTMs, and SAPs is able to bypass the need for positive curvature during the nascent septin ring formation, as there is no discernable bud neck at this point and the membrane topology at the presumptive bud site does not match the curvature preference of the septins. Taken together, the in vitro analysis of the ability of septins to interact with membranes, recognize membrane curvature, and recruit SAPs for higher-order organization will undoubtedly help understand the mechanisms for the assembly and regulation of the plethora of septin structures in vivo.
A LKB1-Like Kinase Acts as a Regulator of Septin Filament Pairing to Control Hourglass Assembly and Stabilization
Upon bud emergence or shortly after, the septins transition from a dynamic nascent ring at the presumptive bud site to a stable septin hourglass at the bud neck (Caviston et al., 2003; Dobbelaere et al., 2003). This stabilization could be caused by filament pairing as observed by thin-section EM and EM tomography (Byers and Goetsch, 1976a; Bertin et al., 2012; Bertin and Nogales, 2012) and by polarized fluorescence microscopy (Vrabioiu and Mitchison, 2006; Demay et al., 2011). These studies are not in agreement as to the orientation of the paired filaments in relation to the mother-bud axis, but are all in agreement that the hourglass seems to be made up of mainly paired filaments. This was further confirmed by our group in cell cortices visualized by platinum-replica electron microscopy (PREM) (Ong et al., 2014). The PREM data favors the orientation of radial paired filaments (about 300–400 nm in length) in an “early hourglass” being parallel to the mother-bud axis as postulated by the groups using polarized fluorescence microscopy (Vrabioiu and Mitchison, 2006; Demay et al., 2011), because the presumed same paired filaments in a “late or transitional hourglass” lie perpendicular to the visualized AMR that runs circumferentially at the bud neck (Ong et al., 2014). Despite the consensus on the paired filaments making up the septin hourglass, whether and how filament pairing is regulated in vivo has remained elusive.
We recently provided the first evidence of such regulation using genetic perturbations and PREM analyses of the septin hourglass in vivo (Marquardt et al., 2020). The protein kinase Elm1 localizes to the bud neck from bud emergence to the onset of cytokinesis in a septin dependent manner (Bouquin et al., 2000; Kang et al., 2016; Marquardt et al., 2020). This localization pattern matches perfectly with the timing of septin hourglass assembly and maintenance at the bud neck. Elm1 had already been implicated in maintaining the stability of septin structures at the bud neck (Bouquin et al., 2000; Asano et al., 2006), but the underlying mechanism had remained unknown at the time. We discovered that the septin hourglass was selectively destabilized with mostly the daughter half of the hourglass mislocalized to the growing bud cortex in elm1Δ mutants in both a kinase-dependent and -independent manner (Figure 1D, left and center panels) and that these phenotypes are at least partially mediated through Bni5 (Marquardt et al., 2020). This destabilization appears to be largely due to an increase in single septin filaments comprising the septin hourglass-like structure when visualized by PREM (Figure 1D, right panels) (Marquardt et al., 2020). This was confirmed by artificially pairing septin filaments using the GFP-nanobody/binding protein (GBP) system (Rothbauer et al., 2006; Kubala et al., 2010) and witnessing a significant rescue of the septin phenotype at the hourglass stage in elm1Δ cells (Figure 1E) (Marquardt et al., 2020). The same experiment also provided evidence that septin filament pairing and unpairing are likely regulated throughout the cell cycle, because aberrant septin structures at the old division site remained even in otherwise wild-type (WT) cells when the septins were forced to stay paired (Figure 1E, arrows) (Marquardt et al., 2020). Thus, to understand the structural transitions or architectural remodeling, one must understand the mechanisms of septin filament pairing and unpairing during the cell cycle.
Transforming the Septin Hourglass Into a Double Ring
At the onset of cytokinesis in S. cerevisiae, the septin hourglass undergoes a dramatic architectural remodeling by “splitting” into two separate rings (Figure 2A), which allows the AMR to access the plasma membrane and initiate its constriction (Kim et al., 1991; Cid et al., 2001a; Lippincott et al., 2001; Tamborrini et al., 2018). A similar transition occurs in the fission yeast Schizosaccharomyces pombe (Berlin et al., 2003; Tasto et al., 2003) as well as the filamentous fungus Aspergillus nidulans (Westfall and Momany, 2002). In mammalian cells (Renshaw et al., 2014; Karasmanis et al., 2019; Wang et al., 2019), septins also undergo reorganization from an hourglass-like structure during cleavage furrow ingression to a double ring during abscission. Despite the striking similarity of septin architectural remodeling at the division site across species, the molecular mechanisms underlying this process remain largely unknown in any system.
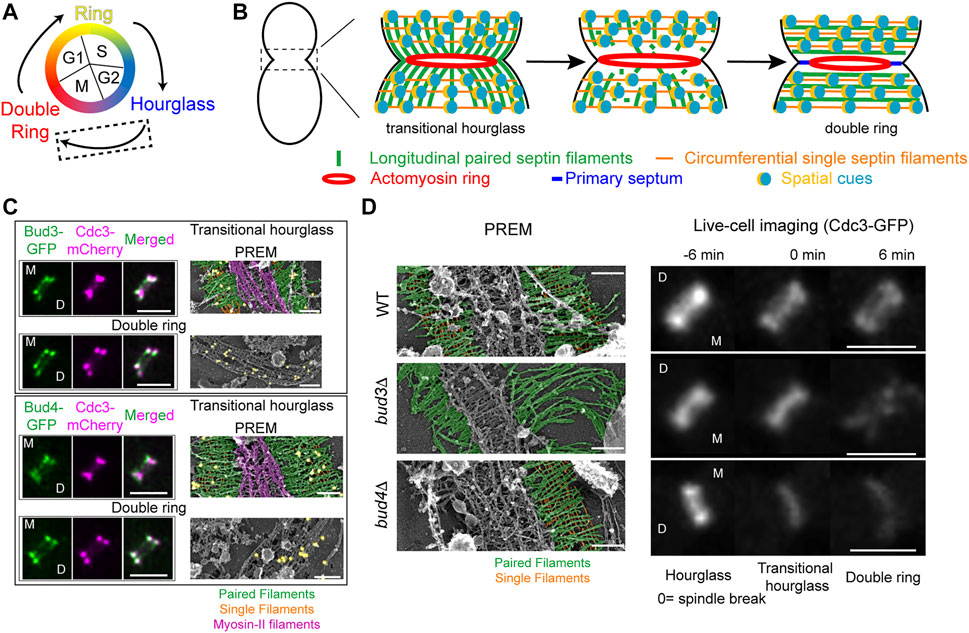
FIGURE 2. Transitioning from an hourglass to a double ring. (A) Depiction of different septin structures visualized during the yeast cell cycle. Arrows indicate transitions occurring between two structures. Dashed box indicates that the focus of this figure is on the hourglass-to-double ring transition that occurs at the onset of cytokinesis. (B) A model of septin architectural remodeling at the division site. (C) Bud3 (green, top) and Bud4 (green, bottom) localization in relation to the septin hourglass and double ring (magenta) at the bud neck by iSIM. Scale bars = 2 µm. Immunogold labeling PREM analysis of Bud3 (yellow, top) and Bud4 (yellow, bottom) localization in relation to the septin transitional hourglass and double ring (right panels). Scale bar = 200 nm. Paired filaments (green), single filaments (orange), and myosin-II filaments (purple). Images taken from (Chen et al., 2020). (D) PREM analysis of the transitional hourglass (left panels) with paired filaments pseudo-colored green and single filaments pseudo-colored orange; scale bars = 200 nm. Images taken from (Chen et al., 2020). Representative images of septin organization (Cdc3-GFP) at the bud neck during the HDR transition in WT (top), bud3Δ (middle), and bud4Δ (bottom) cells (right panels). Scale bars = 2 µm. Images taken from (Chen et al., 2020).
Spatially Controlled Septin Filament Disassembly and Reorganization During the Hourglass-to-Double Ring Transition
Two major events are associated with the septin hourglass-to-double ring (HDR) transition: septin filament disassembly and reorganization. FRAP analysis indicates that septins become more dynamic during the HDR transition (Caviston et al., 2003; Dobbelaere et al., 2003). This dynamic change is accompanied by the net loss of septins at the division site by 20–30%, implicating septin filament disassembly in the process (Caviston et al., 2003; Dobbelaere et al., 2003; Demay et al., 2011; Wloka et al., 2011). Analysis by polarized fluorescence microscopy suggests that paired septin filaments reorganize from a radial arrangement in the hourglass to a circumferential arrangement in the double ring, a 90° change in filament orientation (Vrabioiu and Mitchison, 2006; Demay et al., 2011). Despite these insights, the detailed architectures of the septin hourglass and double ring at the filament level and the molecular mechanisms controlling the HDR transition remained unknown.
By combining cell synchronization with PREM, we have determined the architecture of different septin assemblies at the division site during the cell cycle (Ong et al., 2014; Chen et al., 2020; Marquardt et al., 2020). The “early hourglass” from pre-anaphase cells contains exclusively paired filaments arranged radially and parallel to the mother-bud axis (Ong et al., 2014). This structure is converted into a “transitional hourglass” before cytokinesis, in which the paired filaments are connected perpendicularly by periodic circumferential single septin filaments on the membrane-proximal side (Ong et al., 2014). Strikingly, these intersecting septin filaments, which define a septin gauze, are located exclusively at the outer zones of the transitional hourglass whereas myosin-II filaments occupy the middle zone but exclusively on the cytoplasmic side of the hourglass (Figure 2B, left) (Ong et al., 2014; Chen et al., 2020). At the onset of cytokinesis and under the control of the mitotic exit network (Cid et al., 2001a; Lippincott et al., 2001; Ong et al., 2014; Tamborrini et al., 2018; Chen et al., 2020), this “zonal” transitional hourglass is remodeled into a double ring that consists of exclusively circumferential paired and single septin filaments (Figure 2B, right) (Ong et al., 2014; Chen et al., 2020). These distinct architectures revealed by PREM analysis provide a foundational blueprint for mechanistic analysis of septin high-order assembly and remodeling.
In addition to septin filament disassembly (Figure 2B, center) (Caviston et al., 2003; Dobbelaere et al., 2003; Demay et al., 2011; Wloka et al., 2011), careful analysis of septin behavior during the HDR transition by FRAP, photo-activation, photoconversion, and super-resolution microscopy suggests that there must be “spatial cues” present at the ends of the septin hourglass that controls septin filament reassembly and reorganization into a double ring (Figure 2B) (Ong et al., 2014). What are the spatial cues? How do they control the HDR transition? These questions remained unanswered.
A RhoGEF and an Anillin Act Together as the Spatial Cues to Drive the Septin Hourglass-to-Double Ring Transition
Our recent work suggests that the RhoGEF Bud3 and the anillin-like protein Bud4 function as the spatial cues to drive the HDR transition (Chen et al., 2020). Bud3 and Bud4 were initially identified as factors essential for axial budding in S. cerevisiae (Chant et al., 1995; Sanders and Herskowitz, 1996). Bud3 also acts as a GEF for Cdc42 in early G1 phase to spatially link successive polarization or budding events in haploid cells (Kang et al., 2014). Bud3 and Bud4 interact with each other (Kang et al., 2012; Kang et al., 2013; Wu et al., 2015) and associate with the septin hourglass after S/G2 and with the septin double ring after the HDR transition in a nearly identical pattern (Figure 2C, left) (Chant et al., 1995; Sanders and Herskowitz, 1996; Wloka et al., 2011; Kang et al., 2013; Chen et al., 2020). PREM coupled with immunogold labeling indicates that Bud3 and Bud4 specifically associate with the septin gauze at the outer zones of the transitional hourglass (Figure 2C, right) (Chen et al., 2020). Both proteins have been implicated in septin organization, likely at the stage of double ring formation (Lord et al., 2000; Gladfelter et al., 2005; Guo et al., 2011; Wloka et al., 2011; Eluere et al., 2012; Kang et al., 2013; Wu et al., 2015; Mcquilken et al., 2017). Taken together, these observations suggest that Bud3 and Bud4 likely define the spatial cues that act in the right place at the right time to drive the HDR transition, but how?
Live-cell imaging and PREM analyses demonstrate that Bud3 and Bud4 play distinct and essential roles in controlling the septin HDR transition (Chen et al., 2020). As indicated above, the transitional hourglass in WT cells possesses a zonal architecture where the myosin-II filaments are sandwiched by Bud3 and Bud4-decorated septin gauze (Figure 2C; Figure 2D, top row, left). During the HDR transition, the septins in the middle of the hourglass are preferentially lost whereas those at the ends of the hourglass are preferentially stabilized (Figure 2D, top row, right), leading to the double ring formation. In the absence of Bud3, however, the circumferential single filaments at the ends of the transitional hourglass are completely lost, with many of the structures displaying elongated paired filaments at one side of the hourglass (Figure 2D, middle row, left) (Chen et al., 2020). This filament-level observation may explain the phenotype of bud3Δ cells detected by live-cell imaging that, during the HDR transition, the septin hourglass was first thinned at its edges, followed by the disintegration or fragmentation of the remaining structure in the middle (Figure 2D, middle row, right) (Chen et al., 2020). These observations suggest that Bud3 is specifically required for the stabilization of the transitional hourglass at its edges, and, furthermore, this selective stabilization mechanism must act before the cell cycle-triggered disassembly of the septin filaments in the middle of the hourglass (located between the PM and the myosin-II filaments). In the absence of Bud4, the entire transitional hourglass became destabilized, often in an asymmetric manner where one side of the structure was affected more than the other (Figure 2D, bottom row, left) (Chen et al., 2020). This is consistent with live-cell imaging analysis showing that the mother side of the septin double ring is preferentially lost during the HDR transition (Wloka et al., 2011; Chen et al., 2020). In the absence of both Bud3 and Bud4, the transitional hourglass was severely compromised and, consequently, was hardly detectable by PREM and the double ring was essentially abolished (Chen et al., 2020). Taken together, these data suggest that Bud3 and Bud4 function as the spatial cues by exclusively localizing to the outer zones of the transitional hourglass and instructing the double ring formation at the ends of the hourglass. Bud3 is essential for the circumferential single filament assembly whereas Bud4 is likely required for the stability of both paired and single filaments in the transitional hourglass, especially at the mother side of the bud neck.
Discussion
How a cell assembles a septin structure at a discrete membrane site and how the structure is remodeled in situ into a distinct architecture to perform specific functions are central questions in the septin field that remain largely unanswered. Based on the collective data presented above, it is safe to say that the transitions from the nascent ring to the hourglass and the hourglass to double ring require precise regulation by specific SAPs. Recent works have placed Elm1 as a regulator of septin filament pairing to stabilize the early septin hourglass at the onset of bud formation (Marquardt et al., 2020) and Bud3 and Bud4 as the spatial cues at the ends of a late hourglass to reorganize and stabilize both single and paired filaments during the HDR transition (Chen et al., 2020). By utilizing genetic perturbations and following septin kinetics via confocal microscopy and septin filament architecture via PREM, these studies have begun to elucidate the mechanisms of septin organization in vivo at an unprecedented resolution.
While Elm1 is known to regulate septin filament pairing during hourglass formation (Marquardt et al., 2020), the underlying mechanism remains unknown. Could Elm1 directly phosphorylate the septins or another SAP to induce the change from single to paired filaments, or does Elm1 oppose an unknown molecule that destabilizes septin filament pairing? As septins tend to form paired filaments in vitro either in solution under low-salt condition or on lipid monolayer even under high-salt condition (Frazier et al., 1998; Bertin et al., 2008; Bertin et al., 2010; Bridges et al., 2014), we favor the latter possibility of an indirect regulation by antagonizing a destabilization factor. However, other studies have shown that Elm1 can phosphorylate SAPs with known roles in septin stability including Bni5 (Patasi et al., 2015), Gin4 (Asano et al., 2006), and the morphogenetic checkpoint kinase Hsl1 (Szkotnicki et al., 2008), leaving the possibility open for non-mutually exclusive pathways of action.
Protein kinases have been found to regulate septin structural stability in other systems including at the annulus of spermatozoa and at the base of dendritic spines (Brand et al., 2012; Shen et al., 2017; Yadav et al., 2017; Lin et al., 2019). Similar to the LKB1-like kinase Elm1, LKB1 regulates glucose metabolism by phosphorylating AMP-activated protein kinase (AMPK) (Hawley et al., 2003; Hong et al., 2003; Woods et al., 2003; Shaw et al., 2004; Rubenstein et al., 2006; Mccartney et al., 2016), and has a conserved function in polarity (Watts et al., 2000; Nakano and Takashima, 2012). However, there is no evidence thus far that LKB1 or other Elm1 “homologues” regulate septin architecture in other systems.
Bud3 and Bud4 are known to localize exclusively to the gauze-like structure at the outer zones of the transitional hourglass (Chen et al., 2020). However, it remains a mystery how this unique localization is achieved and how it facilitates the HDR transition. Does the Bud3 and Bud4 complex function as the membrane anchor to stabilize the single filaments at the ends of the transitional hourglass during the paired filament disassembly, and, at the same time, also function as the spatial landmark to recruit septin complexes and/or short filaments from the disassembled paired filaments and reassemble and reorganize them into a double ring? How Bud3 and Bud4 interact with each other and with the septins also remains unknown.
The HDR transition is triggered by the activation of the mitotic exit network (Cid et al., 2001a; Lippincott et al., 2001; Tamborrini et al., 2018) and is accompanied by 90° change in the orientation of the paired filaments (Vrabioiu and Mitchison, 2006; Demay et al., 2011; Ong et al., 2014). Whether and how Bud3 and Bud4 are involved in this process remains unknown. All these questions must be addressed in order to achieve a mechanistic understanding of the HDR transition.
Like the Bud3-Bud4 module in budding yeast, a RhoGEF-anillin module appears to be involved in the coordination of septin remodeling and cytokinesis in many organisms including fission yeast (Berlin et al., 2003; Tasto et al., 2003; Munoz et al., 2014; Wang et al., 2015), Drosophila (Hickson and O’Farrell, 2008), and mammalian cells (Frenette et al., 2012; Renshaw et al., 2014; Karasmanis et al., 2019; Wang et al., 2019). Whether this module represents a conserved core mechanism for septin remodeling during cytokinesis across model systems requires further investigation.
Author Contributions
JM and XC wrote the initial draft. EB revised the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the NIH grant GM116876.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank the members of the Bi laboratory, especially Hiroki Okada, for stimulating discussions and critically reading the manuscript.
References
Ageta-Ishihara, N., and Kinoshita, M. (2021). Developmental and Postdevelopmental Roles of Septins in the Brain. Neurosci. Res. 170, 6–12. doi:10.1016/j.neures.2020.08.006
Alonso, A., Greenlee, M., Matts, J., Kline, J., Davis, K. J., and Miller, R. K. (2015). Emerging Roles of Sumoylation in the Regulation of Actin, Microtubules, Intermediate Filaments, and Septins. Cytoskeleton. 72, 305–339. doi:10.1002/cm.21226
Asano, S., Park, J.-E., Yu, L.-R., Zhou, M., Sakchaisri, K., Park, C. J., et al. (2006). Direct Phosphorylation and Activation of a Nim1-Related Kinase Gin4 by Elm1 in Budding Yeast. J. Biol. Chem. 281, 27090–27098. doi:10.1074/jbc.m601483200
Barral, Y., Mermall, V., Mooseker, M. S., and Snyder, M. (2000). Compartmentalization of the Cell Cortex by Septins Is Required for Maintenance of Cell Polarity in Yeast. Mol. Cell. 5, 841–851. doi:10.1016/s1097-2765(00)80324-x
Barral, Y., Parra, M., Bidlingmaier, S., and Snyder, M. (1999). Nim1-Related Kinases Coordinate Cell Cycle Progression With the Organization of the Peripheral Cytoskeleton in Yeast. Genes Development. 13, 176–187. doi:10.1101/gad.13.2.176
Beber, A., Taveneau, C., Nania, M., Tsai, F.-C., Di Cicco, A., Bassereau, P., et al. (2019). Membrane Reshaping by Micrometric Curvature Sensitive Septin Filaments. Nat. Commun. 10, 420. doi:10.1038/s41467-019-08344-5
Berlin, A., Paoletti, A., and Chang, F. (2003). Mid2p Stabilizes Septin Rings During Cytokinesis in Fission Yeast. J. Cell Biol. 160, 1083–1092. doi:10.1083/jcb.200212016
Bertin, A., Mcmurray, M. A., Grob, P., Park, S.-S., Garcia, G., Patanwala, I., et al. (2008). Saccharomyces cerevisiae Septins: Supramolecular Organization of Heterooligomers and the Mechanism of Filament Assembly. Proc. Natl. Acad. Sci. 105, 8274–8279. doi:10.1073/pnas.0803330105
Bertin, A., Mcmurray, M. A., Pierson, J., Thai, L., Mcdonald, K. L., Zehr, E. A., et al. (2012). Three-Dimensional Ultrastructure of the Septin Filament Network in Saccharomyces Cerevisiae. MBoC. 23, 423–432. doi:10.1091/mbc.e11-10-0850
Bertin, A., Mcmurray, M. A., Thai, L., Garcia, G., Votin, V., Grob, P., et al. (2010). Phosphatidylinositol-4,5-Bisphosphate Promotes Budding Yeast Septin Filament Assembly and Organization. J. Mol. Biol. 404, 711–731. doi:10.1016/j.jmb.2010.10.002
Bertin, A., and Nogales, E. (2012). Septin Filament Organization in Saccharomyces cerevisiae. Communicative Integr. Biol. 5, 503–505. doi:10.4161/cib.21125
Bi, E., Maddox, P., Lew, D. J., Salmon, E. D., Mcmillan, J. N., Yeh, E., et al. (1998). Involvement of an Actomyosin Contractile Ring in Saccharomyces cerevisiae Cytokinesis. J. Cell Biol. 142, 1301–1312. doi:10.1083/jcb.142.5.1301
Booth, E. A., Sterling, S. M., Dovala, D., Nogales, E., and Thorner, J. (2016). Effects of Bni5 Binding on Septin Filament Organization. J. Mol. Biol. 428, 4962–4980. doi:10.1016/j.jmb.2016.10.024
Booth, E. A., Vane, E. W., Dovala, D., and Thorner, J. (2015). A Förster Resonance Energy Transfer (FRET)-Based System Provides Insight Into the Ordered Assembly of Yeast Septin Hetero-Octamers. J. Biol. Chem. 290, 28388–28401. doi:10.1074/jbc.m115.683128
Bouquin, N., Barral, Y., Courbeyrette, R., Blondel, M., Snyder, M., and Mann, C. (2000). Regulation of Cytokinesis by the Elm1 Protein Kinase in Saccharomyces cerevisiae. J. Cell Sci. 113, 1435–1445. doi:10.1242/jcs.113.8.1435
Brand, F., Schumacher, S., Kant, S., Menon, M. B., Simon, R., Turgeon, B., et al. (2012). The Extracellular Signal-Regulated Kinase 3 (Mitogen-Activated Protein Kinase 6 [MAPK6])-MAPK-Activated Protein Kinase 5 Signaling Complex Regulates Septin Function and Dendrite Morphology. Mol. Cell. Biol. 32, 2467–2478. doi:10.1128/mcb.06633-11
Bridges, A. A., Jentzsch, M. S., Oakes, P. W., Occhipinti, P., and Gladfelter, A. S. (2016). Micron-Scale Plasma Membrane Curvature Is Recognized by the Septin Cytoskeleton. J. Cell Biol. 213, 23–32. doi:10.1083/jcb.201512029
Bridges, A. A., Zhang, H., Mehta, S. B., Occhipinti, P., Tani, T., and Gladfelter, A. S. (2014). Septin Assemblies Form by Diffusion-Driven Annealing on Membranes. Proc. Natl. Acad. Sci. USA. 111, 2146–2151. doi:10.1073/pnas.1314138111
Byers, B., and Goetsch, L. (1976a). A Highly Ordered Ring of Membrane-Associated Filaments in Budding Yeast. J. Cell Biol. 69, 717–721. doi:10.1083/jcb.69.3.717
Byers, B., and Goetsch, L. (1976b). Loss of the Filamentous Ring in Cytokinesis-Defective Mutants of Budding Yeast. Abstr. First Internat. Congr. Cell Biol. 70, 35a.
Cannon, K. S., Woods, B. L., Crutchley, J. M., and Gladfelter, A. S. (2019). An Amphipathic Helix Enables Septins to Sense Micrometer-Scale Membrane Curvature. J. Cell Biol. 218, 1128–1137. doi:10.1083/jcb.201807211
Carroll, C. W., Altman, R., Schieltz, D., Yates, J. R., and Kellogg, D. (1998). The Septins Are Required for the Mitosis-Specific Activation of the Gin4 Kinase. J. Cell Biol. 143, 709–717. doi:10.1083/jcb.143.3.709
Caviston, J. P., Longtine, M., Pringle, J. R., and Bi, E. (2003). The Role of Cdc42p GTPase-Activating Proteins in Assembly of the Septin Ring in Yeast. MBoC. 14, 4051–4066. doi:10.1091/mbc.e03-04-0247
Chant, J., Mischke, M., Mitchell, E., Herskowitz, I., and Pringle, J. R. (1995). Role of Bud3p in Producing the Axial Budding Pattern of Yeast. J. Cell Biol. 129, 767–778. doi:10.1083/jcb.129.3.767
Chant, J., and Pringle, J. R. (1995). Patterns of Bud-Site Selection in the Yeast Saccharomyces cerevisiae. J. Cell Biol. 129, 751–765. doi:10.1083/jcb.129.3.751
Chen, X., Wang, K., Svitkina, T., and Bi, E. (2020). Critical Roles of a RhoGEF-Anillin Module in Septin Architectural Remodeling During Cytokinesis. Curr. Biol. 30, 1477–1490. e1473. doi:10.1016/j.cub.2020.02.023
Cid, V. J., Adamiková, L., Sánchez, M., Molina, M., and Nombela, C. (2001a). Cell Cycle Control of Septin Ring Dynamics in the Budding Yeast. Microbiology. 147, 1437–1450. doi:10.1099/00221287-147-6-1437
Cid, V. J., Shulewitz, M. J., Mcdonald, K. L., and Thorner, J. (2001b). Dynamic Localization of the Swe1 Regulator Hsl7 During the Saccharomyces Cerevisiae Cell Cycle. MBoC. 12, 1645–1669. doi:10.1091/mbc.12.6.1645
Connolly, D., Abdesselam, I., Verdier-Pinard, P., and Montagna, C. (2011a). Septin Roles in Tumorigenesis. Biol. Chem. 392, 725–738. doi:10.1515/bc.2011.073
Connolly, D., Yang, Z., Castaldi, M., Simmons, N., Oktay, M. H., Coniglio, S., et al. (2011b). Septin 9 Isoform Expression, Localization and Epigenetic Changes during Human and Mouse Breast Cancer Progression. Breast Cancer Res. 13, R76. doi:10.1186/bcr2924
Cyert, M. S., and Philpott, C. C. (2013). Regulation of Cation Balance in Saccharomyces cerevisiae. Genetics. 193, 677–713. doi:10.1534/genetics.112.147207
De Virgilio, C., Demarini, D. J., and Pringle, J. R. (1996). SPR28, a Sixth Member of the Septin Gene Family in Saccharomyces cerevisiae that Is Expressed Specifically in Sporulating Cells. Microbiology. 142, 2897–2905. doi:10.1099/13500872-142-10-2897
Demay, B. S., Bai, X., Howard, L., Occhipinti, P., Meseroll, R. A., Spiliotis, E. T., et al. (2011). Septin Filaments Exhibit a Dynamic, Paired Organization That Is Conserved From Yeast to Mammals. J. Cell Biol. 193, 1065–1081. doi:10.1083/jcb.201012143
Dobbelaere, J., Gentry, M. S., Hallberg, R. L., and Barral, Y. (2003). Phosphorylation-Dependent Regulation of Septin Dynamics During the Cell Cycle. Developmental Cell. 4, 345–357. doi:10.1016/s1534-5807(03)00061-3
Dolat, L., Hu, Q., and Spiliotis, E. T. (2014). Septin Functions in Organ System Physiology and Pathology. Biol. Chem. 395, 123–141. doi:10.1515/hsz-2013-0233
Eluère, R., Varlet, I., Bernadac, A., and Simon, M.-N. (2012). Cdk and the Anillin Homolog Bud4 Define a New Pathway Regulating Septin Organization in Yeast. Cell Cycle. 11, 151–158. doi:10.4161/cc.11.1.18542
Estey, M. P., Di Ciano-Oliveira, C., Froese, C. D., Bejide, M. T., and Trimble, W. S. (2010). Distinct Roles of Septins in Cytokinesis: SEPT9 Mediates Midbody Abscission. J. Cell Biol. 191, 741–749. doi:10.1083/jcb.201006031
Fang, X., Luo, J., Nishihama, R., Wloka, C., Dravis, C., Travaglia, M., et al. (2010). Biphasic Targeting and Cleavage Furrow Ingression Directed by the Tail of a Myosin II. J. Cell Biol. 191, 1333–1350. doi:10.1083/jcb.201005134
Fares, H., Goetsch, L., and Pringle, J. R. (1996). Identification of a Developmentally Regulated Septin and Involvement of the Septins in Spore Formation in Saccharomyces cerevisiae. J. Cell Biol. 132, 399–411. doi:10.1083/jcb.132.3.399
Finnigan, G. C., Booth, E. A., Duvalyan, A., Liao, E. N., and Thorner, J. (2015). The Carboxy-Terminal Tails of Septins Cdc11 and Shs1 Recruit Myosin-II Binding Factor Bni5 to the Bud Neck in Saccharomyces cerevisiae. Genetics. 200, 843–862. doi:10.1534/genetics.115.176503
Frazier, J. A., Wong, M. L., Longtine, M. S., Pringle, J. R., Mann, M., Mitchison, T. J., et al. (1998). Polymerization of Purified Yeast Septins: Evidence That Organized Filament Arrays May Not Be Required for Septin Function. J. Cell Biol. 143, 737–749. doi:10.1083/jcb.143.3.737
Frenette, P., Haines, E., Loloyan, M., Kinal, M., Pakarian, P., and Piekny, A. (2012). An Anillin-Ect2 Complex Stabilizes central Spindle Microtubules at the Cortex During Cytokinesis. PLoS One. 7, e34888. doi:10.1371/journal.pone.0034888
Garcia, G., Bertin, A., Li, Z., Song, Y., Mcmurray, M. A., Thorner, J., et al. (2011). Subunit-Dependent Modulation of Septin Assembly: Budding Yeast Septin Shs1 Promotes Ring and Gauze Formation. J. Cell Biol. 195, 993–1004. doi:10.1083/jcb.201107123
Gladfelter, A., Pringle, J. R., and Lew, D. J. (2001). The Septin Cortex at the Yeast Motherâ€"bud Neck. Curr. Opin. Microbiol. 4, 681–689. doi:10.1016/s1369-5274(01)00269-7
Gladfelter, A. S., Bose, I., Zyla, T. R., Bardes, E. S. G., and Lew, D. J. (2002). Septin Ring Assembly Involves Cycles of GTP Loading and Hydrolysis by Cdc42p. J. Cell Biol. 156, 315–326. doi:10.1083/jcb.200109062
Gladfelter, A. S., Kozubowski, L., Zyla, T. R., and Lew, D. J. (2005). Interplay Between Septin Organization, Cell Cycle and Cell Shape in Yeast. J. Cell Sci. 118, 1617–1628. doi:10.1242/jcs.02286
Guo, J., Gong, T., and Gao, X.-D. (2011). Identification of an Amphipathic Helix Important for the Formation of Ectopic Septin Spirals and Axial Budding in Yeast Axial Landmark Protein Bud3p. PLoS One. 6, e16744. doi:10.1371/journal.pone.0016744
Hall, P. A., Jung, K., Hillan, K. J., and Russell, S. H. (2005). Expression Profiling the Human Septin Gene Family. J. Pathol. 206, 269–278. doi:10.1002/path.1789
Hall, P. A., and Russell, S. H. (2004). The Pathobiology of the Septin Gene Family. J. Pathol. 204, 489–505. doi:10.1002/path.1654
Hartwell, L. (1971). Genetic Control of the Cell Division Cycle in Yeast *1IV. Genes Controlling Bud Emergence and Cytokinesis. Exp. Cell Res. 69, 265–276. doi:10.1016/0014-4827(71)90223-0
Hawley, S. A., Boudeau, J., Reid, J. L., Mustard, K. J., Udd, L., Mäkelä, T. P., et al. (2003). Complexes Between the LKB1 Tumor Suppressor, STRAD Alpha/Beta and MO25 Alpha/Beta Are Upstream Kinases in the AMP-Activated Protein Kinase Cascade. J. Biol. 2, 28. doi:10.1186/1475-4924-2-28
Hernández-Rodríguez, Y., and Momany, M. (2012). Posttranslational Modifications and Assembly of Septin Heteropolymers and Higher-Order Structures. Curr. Opin. Microbiol. 15, 660–668. doi:10.1016/j.mib.2012.09.007
Hickson, G. R., and O'farrell, P. H. (2008). Rho-Dependent Control of Anillin Behavior During Cytokinesis. J. Cell Biol. 180, 285–294.
Hong, S.-P., Leiper, F. C., Woods, A., Carling, D., and Carlson, M. (2003). Activation of Yeast Snf1 and Mammalian AMP-Activated Protein Kinase by Upstream Kinases. Proc. Natl. Acad. Sci. 100, 8839–8843. doi:10.1073/pnas.1533136100
Ihara, M., Kinoshita, A., Yamada, S., Tanaka, H., Tanigaki, A., Kitano, A., et al. (2005). Cortical Organization by the Septin Cytoskeleton Is Essential for Structural and Mechanical Integrity of Mammalian Spermatozoa. Developmental Cell. 8, 343–352. doi:10.1016/j.devcel.2004.12.005
Iwase, M., Luo, J., Nagaraj, S., Longtine, M., Kim, H. B., Haarer, B. K., et al. (2006). Role of a Cdc42p Effector Pathway in Recruitment of the Yeast Septins to the Presumptive Bud Site. MBoC. 17, 1110–1125. doi:10.1091/mbc.e05-08-0793
John, C. M., Hite, R. K., Weirich, C. S., Fitzgerald, D. J., Jawhari, H., Faty, M., et al. (2007). The Caenorhabditis elegans Septin Complex Is Nonpolar. EMBO J. 26, 3296–3307. doi:10.1038/sj.emboj.7601775
Kang, H., Tsygankov, D., and Lew, D. J. (2016). Sensing a Bud in the Yeast Morphogenesis Checkpoint: a Role for Elm1. MBoC. 27, 1764–1775. doi:10.1091/mbc.e16-01-0014
Kang, P. J., Angerman, E., Jung, C. H., and Park, H. O. (2012). Bud4 Mediates the Cell-Type-Specific Assembly of the Axial Landmark in Budding Yeast. J. Cell Sci. 125, 3840–3849. doi:10.1242/jcs.103697
Kang, P. J., Hood-Degrenier, J. K., and Park, H. O. (2013). Coupling of Septins to the Axial Landmark by Bud4 in Budding Yeast. J. Cell Sci. 126, 1218–1226. doi:10.1242/jcs.118521
Kang, P. J., Lee, M. E., and Park, H.-O. (2014). Bud3 Activates Cdc42 to Establish a Proper Growth Site in Budding Yeast. J. Cell Biol. 206, 19–28. doi:10.1083/jcb.201402040
Karasmanis, E. P., Hwang, D., Nakos, K., Bowen, J. R., Angelis, D., and Spiliotis, E. T. (2019). A Septin Double Ring Controls the Spatiotemporal Organization of the ESCRT Machinery in Cytokinetic Abscission. Curr. Biol. 29, 2174–2182. doi:10.1016/j.cub.2019.05.050
Khan, A., Newby, J., and Gladfelter, A. S. (2018). Control of Septin Filament Flexibility and Bundling by Subunit Composition and Nucleotide Interactions. MBoC. 29, 702–712. doi:10.1091/mbc.e17-10-0608
Kim, H. B., Haarer, B. K., and Pringle, J. R. (1991). Cellular Morphogenesis in the Saccharomyces cerevisiae Cell Cycle: Localization of the CDC3 Gene Product and the Timing of Events at the Budding Site. J. Cell Biol. 112, 535–544. doi:10.1083/jcb.112.4.535
Kinoshita, M. (2003). Assembly of Mammalian Septins. J. Biochem. 134, 491–496. doi:10.1093/jb/mvg182
Kissel, H., Georgescu, M.-M., Larisch, S., Manova, K., Hunnicutt, G. R., and Steller, H. (2005). The Sept4 Septin Locus Is Required for Sperm Terminal Differentiation in Mice. Developmental Cell. 8, 353–364. doi:10.1016/j.devcel.2005.01.021
Kubala, M. H., Kovtun, O., Alexandrov, K., and Collins, B. M. (2010). Structural and Thermodynamic Analysis of the GFP:GFP-nanobody Complex. Protein Sci. 19, 2389–2401. doi:10.1002/pro.519
Lee, P. R., Song, S., Ro, H.-S., Park, C. J., Lippincott, J., Li, R., et al. (2002). Bni5p, a Septin-Interacting Protein, Is Required for normal Septin Function and Cytokinesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 22, 6906–6920. doi:10.1128/mcb.22.19.6906-6920.2002
Lin, C. H., Shen, Y. R., Wang, H. Y., Chiang, C. W., Wang, C. Y., and Kuo, P. L. (2019). Regulation of Septin Phosphorylation: SEPT12 Phosphorylation in Sperm Septin Assembly. Cytoskeleton. 76, 137–142. doi:10.1002/cm.21491
Lin, Y.-H., Kuo, Y.-C., Chiang, H.-S., and Kuo, P.-L. (2011). The Role of the Septin Family in Spermiogenesis. Spermatogenesis. 1, 298–302. doi:10.4161/spmg.1.4.18326
Lippincott, J., and Li, R. (1998). Sequential Assembly of Myosin II, an IQGAP-Like Protein, and Filamentous Actin to a Ring Structure Involved in Budding Yeast Cytokinesis. J. Cell Biol. 140, 355–366. doi:10.1083/jcb.140.2.355
Lippincott, J., Shannon, K. B., Shou, W., Deshaies, R. J., and Li, R. (2001). The Tem1 Small GTPase Controls Actomyosin and Septin Dynamics During Cytokinesis. J. Cell Sci. 114, 1379–1386. doi:10.1242/jcs.114.7.1379
Longtine, M. S., Demarini, D. J., Valencik, M. L., Al-Awar, O. S., Fares, H., De Virgilio, C., et al. (1996). The Septins: Roles in Cytokinesis and Other Processes. Curr. Opin. Cell Biol. 8, 106–119. doi:10.1016/s0955-0674(96)80054-8
Longtine, M. S., Fares, H., and Pringle, J. R. (1998). Role of the Yeast Gin4p Protein Kinase in Septin Assembly and the Relationship Between Septin Assembly and Septin Function. J. Cell Biol. 143, 719–736. doi:10.1083/jcb.143.3.719
Longtine, M. S., Theesfeld, C. L., Mcmillan, J. N., Weaver, E., Pringle, J. R., and Lew, D. J. (2000). Septin-dependent Assembly of a Cell Cycle-Regulatory Module in Saccharomyces cerevisiae. Mol. Cell Biol. 20, 4049–4061. doi:10.1128/mcb.20.11.4049-4061.2000
Lord, M., Yang, M. C., Mischke, M., and Chant, J. (2000). Cell Cycle Programs of Gene Expression Control Morphogenetic Protein Localization. J. Cell Biol. 151, 1501–1512. doi:10.1083/jcb.151.7.1501
Luedeke, C., Frei, S. B., Sbalzarini, I., Schwarz, H., Spang, A., and Barral, Y. (2005). Septin-Dependent Compartmentalization of the Endoplasmic Reticulum During Yeast Polarized Growth. J. Cell Biol. 169, 897–908. doi:10.1083/jcb.200412143
Marquardt, J., Chen, X., and Bi, E. (2019). Architecture, Remodeling, and Functions of the Septin Cytoskeleton. Cytoskeleton. 76, 7–14. doi:10.1002/cm.21475
Marquardt, J., Yao, L.-L., Okada, H., Svitkina, T., and Bi, E. (2020). The LKB1-Like Kinase Elm1 Controls Septin Hourglass Assembly and Stability by Regulating Filament Pairing. Curr. Biol. 30, 2386–2394. doi:10.1016/j.cub.2020.04.035
Mccartney, R. R., Garnar-Wortzel, L., Chandrashekarappa, D. G., and Schmidt, M. C. (2016). Activation and Inhibition of Snf1 Kinase Activity by Phosphorylation within the Activation Loop. Biochim. Biophys. Acta (Bba) - Proteins Proteomics. 1864, 1518–1528. doi:10.1016/j.bbapap.2016.08.007
Mcmurray, M. A., and Thorner, J. (2009). Septins: Molecular Partitioning and the Generation of Cellular Asymmetry. Cell Div. 4, 18. doi:10.1186/1747-1028-4-18
Mcquilken, M., Jentzsch, M. S., Verma, A., Mehta, S. B., Oldenbourg, R., and Gladfelter, A. S. (2017). Analysis of Septin Reorganization at Cytokinesis Using Polarized Fluorescence Microscopy. Front. Cell Dev. Biol. 5, 42. doi:10.3389/fcell.2017.00042
Mendonça, D. C., Macedo, J. N., Guimarães, S. L., Barroso Da Silva, F. L., Cassago, A., Garratt, R. C., et al. (2019). A Revised Order of Subunits in Mammalian Septin Complexes. Cytoskeleton. 76, 457–466. doi:10.1002/cm.21569
Mino, A., Tanaka, K., Kamei, T., Umikawa, M., Fujiwara, T., and Takai, Y. (1998). Shs1p: A Novel Member of Septin That Interacts with Spa2p, Involved in Polarized Growth inSaccharomyces Cerevisiae. Biochem. Biophysical Res. Commun. 251, 732–736. doi:10.1006/bbrc.1998.9541
Mortensen, E. M., Mcdonald, H., Yates, J., and Kellogg, D. R. (2002). Cell Cycle-Dependent Assembly of a Gin4-Septin Complex. MBoC. 13, 2091–2105. doi:10.1091/mbc.01-10-0500
Mostowy, S., and Cossart, P. (2012). Septins: the Fourth Component of the Cytoskeleton. Nat. Rev. Mol. Cell. Biol. 13, 183–194. doi:10.1038/nrm3284
Muñoz, S., Manjón, E., and Sánchez, Y. (2014). The Putative Exchange Factor Gef3p Interacts With Rho3p GTPase and the Septin Ring During Cytokinesis in Fission Yeast. J. Biol. Chem. 289, 21995–22007. doi:10.1074/jbc.m114.548792
Nakano, A., and Takashima, S. (2012). LKB 1 and AMP ‐Activated Protein Kinase: Regulators of Cell Polarity. Genes Cells. 17, 737–747. doi:10.1111/j.1365-2443.2012.01629.x
Oh, Y., and Bi, E. (2011). Septin Structure and Function in Yeast and beyond. Trends Cell Biol. 21, 141–148. doi:10.1016/j.tcb.2010.11.006
Okada, S., Leda, M., Hanna, J., Savage, N. S., Bi, E., and Goryachev, A. B. (2013). Daughter Cell Identity Emerges From the Interplay of Cdc42, Septins, and Exocytosis. Developmental Cell. 26, 148–161. doi:10.1016/j.devcel.2013.06.015
Ong, K., Wloka, C., Okada, S., Svitkina, T., and Bi, E. (2014). Architecture and Dynamic Remodelling of the Septin Cytoskeleton During the Cell Cycle. Nat. Commun. 5, 5698. doi:10.1038/ncomms6698
Ozsarac, N., Bhattacharyya, M., Dawes, I. W., and Clancy, M. J. (1995). The SPR3 Gene Encodes a Sporulation-Specific Homologue of the Yeast CDC3/10/11/12 Family of Bud Neck Microfilaments and Is Regulated by ABFI. Gene. 164, 157–162. doi:10.1016/0378-1119(95)00438-c
Pan, F., Malmberg, R. L., and Momany, M. (2007). Analysis of Septins Across Kingdoms Reveals Orthology and New Motifs. BMC Evol. Biol. 7, 103. doi:10.1186/1471-2148-7-103
Patasi, C., Godočíková, J., Michlíková, S., Nie, Y., Káčeriková, R., Kválová, K., et al. (2015). The Role of Bni5 in the Regulation of Septin Higher-Order Structure Formation. Biol. Chem. 396, 1325–1337. doi:10.1515/hsz-2015-0165
Perez, A. M., Finnigan, G. C., Roelants, F. M., and Thorner, J. (2016). Septin-Associated Protein Kinases in the Yeast Saccharomyces cerevisiae. Front. Cell Dev. Biol. 4, 119. doi:10.3389/fcell.2016.00119
Pringle, J. R., Bi, E., Harkins, H. A., Zahner, J. E., De Virgilio, C., Chant, J., et al. (1995). Establishment of Cell Polarity in Yeast. Cold Spring Harbor Symposia Quantitative Biol. 60, 729–744. doi:10.1101/sqb.1995.060.01.079
Renshaw, M. J., Liu, J., Lavoie, B. D., and Wilde, A. (2014). Anillin-Dependent Organization of Septin Filaments Promotes Intercellular Bridge Elongation and Chmp4B Targeting to the Abscission Site. Open Biol. 4, 130190. doi:10.1098/rsob.130190
Robertson, C., Church, S. W., Nagar, H. A., Price, J., Hall, P. A., and Russell, S. H. (2004). Properties ofSEPT9 Isoforms and the Requirement for GTP Binding. J. Pathol. 203, 519–527. doi:10.1002/path.1551
Rodal, A. A., Kozubowski, L., Goode, B. L., Drubin, D. G., and Hartwig, J. H. (2005). Actin and Septin Ultrastructures at the Budding Yeast Cell Cortex. MBoC. 16, 372–384. doi:10.1091/mbc.e04-08-0734
Roeseler, S., Sandrock, K., Bartsch, I., and Zieger, B. (2009). Septins, a Novel Group of GTP-Binding Proteins - Relevance in Hemostasis, Neuropathology and Oncogenesis. Klin. Padiatr. 221, 150–155. doi:10.1055/s-0029-1220706
Rothbauer, U., Zolghadr, K., Tillib, S., Nowak, D., Schermelleh, L., Gahl, A., et al. (2006). Targeting and Tracing Antigens in Live Cells With Fluorescent Nanobodies. Nat. Methods. 3, 887–889. doi:10.1038/nmeth953
Rubenstein, E. M., Mccartney, R. R., and Schmidt, M. C. (2006). Regulatory Domains of Snf1-Activating Kinases Determine Pathway Specificity. Eukaryot. Cell. 5, 620–627. doi:10.1128/ec.5.4.620-627.2006
Sanders, S. L., and Herskowitz, I. (1996). The BUD4 Protein of Yeast, Required for Axial Budding, Is Localized to the Mother/BUD Neck in a Cell Cycle-Dependent Manner. J. Cell Biol. 134, 413–427. doi:10.1083/jcb.134.2.413
Schneider, C., Grois, J., Renz, C., Gronemeyer, T., and Johnsson, N. (2013). Septin Rings Act as a Template for Myosin Higher-Order Structures and Inhibit Redundant Polarity Establishment. J. Cell Sci. 126, 3390–3400. doi:10.1242/jcs.125302
Sellin, M. E., Sandblad, L., Stenmark, S., and Gullberg, M. (2011). Deciphering the Rules Governing Assembly Order of Mammalian Septin Complexes. MBoC. 22, 3152–3164. doi:10.1091/mbc.e11-03-0253
Sellin, M. E., Stenmark, S., and Gullberg, M. (2012). Mammalian SEPT9 Isoforms Direct Microtubule-Dependent Arrangements of Septin Core Heteromers. MBoC. 23, 4242–4255. doi:10.1091/mbc.e12-06-0486
Shaw, R. J., Kosmatka, M., Bardeesy, N., Hurley, R. L., Witters, L. A., Depinho, R. A., et al. (2004). The Tumor Suppressor LKB1 Kinase Directly Activates AMP-Activated Kinase and Regulates Apoptosis in Response to Energy Stress. Proc. Natl. Acad. Sci. 101, 3329–3335. doi:10.1073/pnas.0308061100
Shcheprova, Z., Baldi, S., Frei, S. B., Gonnet, G., and Barral, Y. (2008). A Mechanism for Asymmetric Segregation of Age During Yeast Budding. Nature. 454, 728–734. doi:10.1038/nature07212
Shen, Y.-R., Wang, H.-Y., Kuo, Y.-C., Shih, S.-C., Hsu, C.-H., Chen, Y.-R., et al. (2017). SEPT12 Phosphorylation Results in Loss of the Septin Ring/Sperm Annulus, Defective Sperm Motility and Poor Male Fertility. Plos Genet. 13, e1006631. doi:10.1371/journal.pgen.1006631
Shulewitz, M. J., Inouye, C. J., and Thorner, J. (1999). Hsl7 Localizes to a Septin Ring and Serves as an Adapter in a Regulatory Pathway that Relieves Tyrosine Phosphorylation of Cdc28 Protein Kinase in Saccharomyces cerevisiae. Mol. Cell. Biol. 19, 7123–7137. doi:10.1128/mcb.19.10.7123
Sirajuddin, M., Farkasovsky, M., Hauer, F., Kühlmann, D., Macara, I. G., Weyand, M., et al. (2007). Structural Insight Into Filament Formation by Mammalian Septins. Nature 449, 311–315. doi:10.1038/nature06052
Soroor, F., Kim, M. S., Palander, O., Balachandran, Y., Collins, R. F., Benlekbir, S., et al. (2021). Revised Subunit Order of Mammalian Septin Complexes Explains Their In Vitro Polymerization Properties. MBoC. 32, 289–300. doi:10.1091/mbc.e20-06-0398
Szkotnicki, L., Crutchley, J. M., Zyla, T. R., Bardes, E. S. G., and Lew, D. J. (2008). The Checkpoint Kinase Hsl1p Is Activated by Elm1p-Dependent Phosphorylation. MBoC. 19, 4675–4686. doi:10.1091/mbc.e08-06-0663
Tada, T., Simonetta, A., Batterton, M., Kinoshita, M., Edbauer, D., and Sheng, M. (2007). Role of Septin Cytoskeleton in Spine Morphogenesis and Dendrite Development in Neurons. Curr. Biol. 17, 1752–1758. doi:10.1016/j.cub.2007.09.039
Tamborrini, D., Juanes, M. A., Ibanes, S., Rancati, G., and Piatti, S. (2018). Recruitment of the Mitotic Exit Network to Yeast Centrosomes Couples Septin Displacement to Actomyosin Constriction. Nat. Commun. 9, 4308. doi:10.1038/s41467-018-06767-0
Tasto, J. J., Morrell, J. L., and Gould, K. L. (2003). An Anillin Homologue, Mid2p, Acts During Fission Yeast Cytokinesis to Organize the Septin Ring and Promote Cell Separation. J. Cell Biol. 160, 1093–1103. doi:10.1083/jcb.200211126
Van Eunen, K., Bouwman, J., Daran-Lapujade, P., Postmus, J., Canelas, A. B., Mensonides, F. I. C., et al. (2010). Measuring Enzyme Activities Under Standardized In Vivo-Like Conditions for Systems Biology. FEBS J. 277, 749–760. doi:10.1111/j.1742-4658.2009.07524.x
Vrabioiu, A. M., and Mitchison, T. J. (2006). Structural Insights Into Yeast Septin Organization From Polarized Fluorescence Microscopy. Nature. 443, 466–469. doi:10.1038/nature05109
Wang, K., Wloka, C., and Bi, E. (2019). Non-Muscle Myosin-II Is Required for the Generation of a Constriction Site for Subsequent Abscission. iScience. 13, 69–81. doi:10.1016/j.isci.2019.02.010
Wang, N., Wang, M., Zhu, Y.-H., Grosel, T. W., Sun, D., Kudryashov, D. S., et al. (2015). The Rho-GEF Gef3 Interacts With the Septin Complex and Activates the GTPase Rho4 During Fission Yeast Cytokinesis. MBoC. 26, 238–255. doi:10.1091/mbc.e14-07-1196
Watts, J. L., Morton, D. G., Bestman, J., and Kemphues, K. J. (2000). The C. elegans Par-4 Gene Encodes a Putative Serine-Threonine Kinase Required for Establishing Embryonic Asymmetry. Development. 127, 1467–1475. doi:10.1242/dev.127.7.1467
Weems, A., and Mcmurray, M. (2017). The Step-wise Pathway of Septin Hetero-Octamer Assembly in Budding Yeast. eLife. 6, e23689. doi:10.7554/eLife.23689
Weirich, C. S., Erzberger, J. P., and Barral, Y. (2008). The Septin Family of GTPases: Architecture and Dynamics. Nat. Rev. Mol. Cell Biol. 9, 478–489. doi:10.1038/nrm2407
Westfall, P. J., and Momany, M. (2002). Aspergillus nidulansSeptin AspB Plays Pre- and Postmitotic Roles in Septum, Branch, and Conidiophore Development. MBoC. 13, 110–118. doi:10.1091/mbc.01-06-0312
Wloka, C., Nishihama, R., Onishi, M., Oh, Y., Hanna, J., Pringle, J. R., et al. (2011). Evidence that a Septin Diffusion Barrier Is Dispensable for Cytokinesis in Budding Yeast. Biol. Chem. 392, 813–829. doi:10.1515/bc.2011.083
Woods, A., Johnstone, S. R., Dickerson, K., Leiper, F. C., Fryer, L. G. D., Neumann, D., et al. (2003). LKB1 Is the Upstream Kinase in the AMP-Activated Protein Kinase cascade. Curr. Biol. 13, 2004–2008. doi:10.1016/j.cub.2003.10.031
Wu, H., Guo, J., Zhou, Y.-T., and Gao, X.-D. (2015). The Anillin-Related Region of Bud4 Is the Major Functional Determinant for Bud4's Function in Septin Organization During Bud Growth and Axial Bud Site Selection in Budding Yeast. Eukaryot. Cell. 14, 241–251. doi:10.1128/ec.00268-14
Xie, Y., Vessey, J. P., Konecna, A., Dahm, R., Macchi, P., and Kiebler, M. A. (2007). The GTP-Binding Protein Septin 7 Is Critical for Dendrite Branching and Dendritic-Spine Morphology. Curr. Biol. 17, 1746–1751. doi:10.1016/j.cub.2007.08.042
Keywords: septins, septin-associated proteins, RhoGEF, anillin, Elm1, Bud3, Bud4
Citation: Marquardt J, Chen X and Bi E (2021) Septin Assembly and Remodeling at the Cell Division Site During the Cell Cycle. Front. Cell Dev. Biol. 9:793920. doi: 10.3389/fcell.2021.793920
Received: 12 October 2021; Accepted: 08 November 2021;
Published: 25 November 2021.
Edited by:
Matthias Gaestel, Hannover Medical School, GermanyReviewed by:
Michael McMurray, University of Colorado Denver, United StatesSimonetta Piatti, Délégation Languedoc Roussillon (CNRS), France
Copyright © 2021 Marquardt, Chen and Bi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Erfei Bi, ZWJpQHBlbm5tZWRpY2luZS51cGVubi5lZHU=
†These authors have contributed equally to this work
 Joseph Marquardt
Joseph Marquardt Xi Chen
Xi Chen Erfei Bi
Erfei Bi