- 1Students’ Scientific Research Center, Tehran University of Medical Sciences, Tehran, Iran
- 2Department of Pathology, Isfahan University of Medical Sciences, Isfahan, Iran
- 3Student Research Committee, Kashan University of Medical Sciences, Kashan, Iran
- 4Student Research Committee, School of Medicine, Shahid Beheshti University of Medical Sciences, Tehran, Iran
- 5Student Research Committee, School of Medicine, Mazandaran University of Medical Sciences, Sari, Iran
- 6Advanced Therapy Medicinal Product (ATMP) Department, Breast Cancer Research Center, Motamed Cancer Institute, ACECR, Tehran, Iran
- 7Proteomics Research Center, Shahid Beheshti University of Medical Sciences, Tehran, Iran
- 8School of Medicine, Kashan University of Medical Sciences, Kashan, Iran
- 9Laser Research Centre, Faculty of Health Science, University of Johannesburg, Johannesburg, South Africa
- 10Anatomical Sciences Research Center, Institute for Basic Sciences, Kashan University of Medical Sciences, Kashan, Iran
- 11Department of Pathology, School of Medicine, Center for Women’s Health Research Zahra, Tabriz University of Medical Sciences, Tabriz, Iran
- 12Research Center for Biochemistry and Nutrition in Metabolic Diseases, Institute for Basic Sciences, Kashan University of Medical Sciences, Kashan, Iran
A major terrifying ailment afflicting the humans throughout the world is brain tumor, which causes a lot of mortality among pediatric and adult solid tumors. Several major barriers to the treatment and diagnosis of the brain tumors are the specific micro-environmental and cell-intrinsic features of neural tissues. Absence of the nutrients and hypoxia trigger the cells’ mortality in the core of the tumors of humans’ brains: however, type of the cells’ mortality, including apoptosis or necrosis, has been not found obviously. Current studies have emphasized the non-coding RNAs (ncRNAs) since their crucial impacts on carcinogenesis have been discovered. Several investigations suggest the essential contribution of such molecules in the development of brain tumors and the respective roles in apoptosis. Herein, we summarize the apoptosis-related non-coding RNAs in brain tumors.
Introduction
Cancer is specified as uncontrolled cell growth and proliferation resulting in an imbalance between division and death of the cells as a result of many different factors including physicochemical or biological agents (Evan and Vousden, 2001; Jafari et al., 2021a). Today in the world, cancer is still one of the paramount health issues (Nikolaou et al., 2018; Jafari et al., 2020). Brain tumors are almost specified with great morbidity and mortality rate (Weller et al., 2015). Glioma is the most common and aggressive diagnosed tumor of the central nervous system (CNS) with low survival and high recurrence rate (Khani et al., 2019; Xu et al., 2020). Among all tumors, glioma represents just 1–2%, whereas approximately 30% of all preliminary and 80% of every malignant tumor of the brain are affiliated with glioma, which responsible for many brain disease-related deaths. It is thought neurological stem and/or progenitor cells are responsible for brain tumors. Based on morphological resemblance to the normal neuroglial cells, these heterogeneous tumors histologically are classified into oligodendrogliomas, astrocytomas, and ependymomas (Weller et al., 2015). Based on classification by WHO, there are two classes of glioma with four distinct grades including low- (I and II) and high- (III and IV) grades. Classification of glioma based upon molecular characteristics including genetics, gene expression, DNA methylation, and so on plays a pivotal role in diagnosis, prognosis, and treatment (Li et al., 2019a). Despite much research and clinical investigation in recent years, the prognosis and treatment of glioma are not so desirable (Xu et al., 2020). Great proliferation rate with high resistance to therapies, sensitivity of the CNS and low ability to repair itself, and inability of most drugs to pass the blood brain barrier (BBB) prevent impressive treatment of glioma (So et al., 2021). One of the malignant glioma’s hallmarks is its ability to infiltrate the brain, leads to avoidable recurrence post local therapy (So et al., 2021). Nowadays, existing therapeutic approaches include resection surgery, chemotherapy, radiotherapy, and combination therapies. However, contrary to considerable progress in treatment options, after initial resection, frequently quick progression and a high rate of recurrence are shown in patients with glioma (Aldape et al., 2019). Discovering new, innovative, and effective therapies for brain tumors depends on the improved perception of the molecular aspects of this disorder (Aldape et al., 2019).
Several studies have indicated the contribution of apoptosis in the pathophysiology of brain tumors (Mellai and Schiffer, 2007; Fernald and Kurokawa, 2013; Mohamed Yusoff, 2015). Apoptosis is a physiological process during aging and development which led to programmed cell death (Majtnerová and Roušar, 2018; Wang, 2020). The term of apoptosis was first used in a paper published in 1972 by Kerr, Wyllie and Currie to describe a hemostatic process which maintains cell populations by removing damaged and unnecessary cells (Majtnerová and Roušar, 2018). Apoptosis is controlled genetically and is an eventually conserved process among multicellular organisms (D’Arcy, 2019; Jafari et al., 2021b). The mediators of apoptosis are enzymes named caspases. Caspases are cytosolic proteases with the proteolytic activity and they are expressed as pro-enzymes in most cells. When a caspase is activated, it can also activate other procaspases and can induce a cascade of activated caspases which can finally led to cellular death (D’Arcy, 2019; Elmore, 2007). A wide range of conditions and stimuli can initiate apoptosis (Elmore, 2007; Majtnerová and Roušar, 2018). Today three main pathways are described in apoptosis including extrinsic, intrinsic and perforin/granzyme-mediated pathways (Majtnerová and Roušar, 2018). The intrinsic pathway is mitochondrial and initiate with death signals like DNA damage and withdrawal of trophic factors. The response to death signals is disruption of the mitochondrial membrane permeability which led to activation of a series of caspases (Wang, 2020). The extrinsic pathway is induced via death receptors placed on plasma membrane such as TNF and Fas receptors. This pathway also eventually leads to the activation of caspases (Wang, 2020). In perforin/granzyme pathway, apoptosis can be caused by each of the granzyme A or granzyme B. Both extrinsic and intrinsic pathway in addition to granzyme B pathway led to apoptosis via cleavage of caspase-3. But granzyme A pathway is initiated via single stranded DNA damage and induce apoptosis via a caspase independent pathway (Elmore, 2007). A variety of morphological and biochemical changes occurs in cells during apoptosis (Majtnerová and Roušar, 2018). Morphological changes contain cellular shrinkage, nuclear fragmentation and chromatin condensation. The cytoplasmic membrane undergoes changes including budding, bleeding, loss of integrity and transfer of phosphatidylserine (PS) to the extracellular part of membrane. The exposure of PS in outer membrane helps recognition and swallowing apoptotic cells by macrophages. These changes can be observed by light and electron microscopes but electron microscopy give more valuable information and show us more morphological changes (Elmore, 2007; Pistritto et al., 2016; Xu et al., 2019a). Some of the biochemical features which are observed in apoptotic cells contain protein cross-linking, DNA segmentation, and nuclear and cytoskeletal proteins cleavage. Apoptotic cells also form apoptotic bodies and express the ligands necessary for recognition by phagocytic cells and finally, they are identified and removed by phagocytic cells (Elmore, 2007).
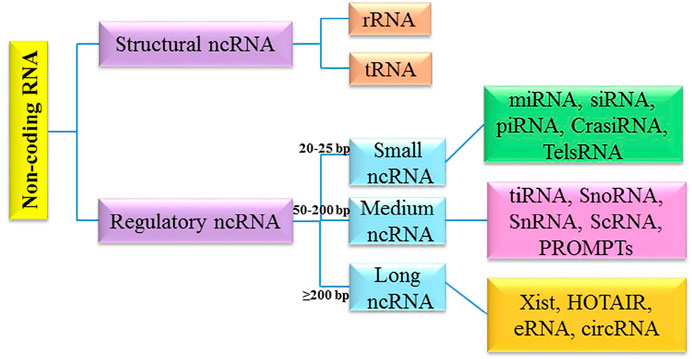
It has been shown that different cellular/molecular mechanisms are associated with apoptosis-related processes in brain tumors. Along with genetic mechanisms, epigenetic mechanisms (e.g., non-coding RNAs networks) greatly affect the regulation of apoptosis-related processes. The non-coding RNAs (ncRNAs) belong to a class of RNAs, which function at the RNA level (Nicoloso and Calin, 2008; Pop et al., 2018). NcRNAs are highly heterogeneous in length, structure and cellular function (Hashemian et al., 2020; Mirzaei and Hamblin, 2020; Balandeh et al., 2021; Dashti et al., 2021; Mahjoubin-Tehran et al., 2021). They can be classified into two main classes: structural non-coding RNA and regulatory non-coding RNA. Structural ncRNA include ribosomal RNA (rRNA) and transfer RNA (tRNA). Regulatory ncRNAs are divided into three subclasses based on the length, and include different types from small nucleolar/nuclear RNA (snoRNA/snRNA), small interfering RNAs (siRNA), microRNA (miRNA), and piwiRNA (PiRNA) to Xist and circRNA (Zhang et al., 2019a) (Figure 1).
In this study, we describe the structure and biogenesis of miRNA, lncRNA, and circRNA, then summarize the apoptosis-related non-coding RNAs in brain tumors.
Non-Coding RNAs and Brain Tumors
MicroRNAs Biogenesis
MiRNAs are small single stranded non-coding RNAs, with the mean length of nearly 22 nucleotides, which play important roles in regulating gene expression at the post-transcriptional level and RNA silencing (Shabaninejad et al., 2019). In 1993, lin-4, the discovery of the first miRNA, lin-4, by the Ambros and Ruvkun team led to a revolution in molecular biology (Lee et al., 1993; Achkar et al., 2016; Gebert and MacRae, 2019; O'Brien et al., 2018).
Generally, miRNA is critical for normal animal development and is capable of regulating a variety of biological processes and signaling pathways like metabolism and differentiation as well as rapid growth or proliferation. Increasing evidence have shown the correlation between miRNA deregulation and numerous kinds of human diseases, including heart disease, metabolic disorder, and cancer. In addition, miRNAs contribute importantly to invasion, metastasis and the tumor angiogenesis and are capable of exiting from the cells through vesicles and entering the extra-cellular fluids (O'Brien et al., 2018; Lin and Gregory, 2015).
RNA polymerase II (Pol. II) enzyme begins miRNAs biogenesis in the nucleus and approximately half of the miRNAs have been identified to be intragenic, largely derived from the introns, whereas the others to be intergenic with their specific promoters (Gebert and MacRae, 2019).
Therefore, a long transcript referred to as the primary microRNA (pri-miRNA) is generated after the transcription by Pol. II and hence Pri-miRNA may create a cluster of two or more miRNAs or a miRNA. Furthermore, it has a local stem loop structure, undergoing several steps of maturation like splicing, polyadenylation, as well as capping. Scission of Pri-miRNA to pre-miRNA is conducted by a micro-processor complex which has two parts: the double-strand RNase DROSHA and its crucial cofactor; that is, DiGeorge syndrome critical region 8 (DGCR8) (Lee et al., 1993; Ha and Kim, 2014; Lin and Gregory, 2015; Gebert and MacRae, 2019).
Research has shown a length of nearly 65 nucleotides of pre-miRNAs with a hairpin-like structure that is the same as the pri-miRNA. Then, Exportin 5 translocates pre-miRNA into cytoplasm, wherein its maturity ends. Pre-miRNA is converted to a mature ∼22 nucleotide miRNA duplex by RNase III DICER1 enzyme and its binding protein, and transactivation response element RNA-binding protein (TRBP) in the cytoplasm. A strand works as one of the guide miRNAs and the other is loaded to Argonaute (AGO) for making a complex: RNA-induced silencing complex (RISC). The complex is assembled by two steps of loading and unwinding of RNA duplex. RISC is capable of binding to the 3′-UTR of the target mRNAs from 5′-UTR of miRNA that may repress translation or degrade mRNA (Ha and Kim, 2014; Lin and Gregory, 2015). The schematic steps of biogenesis of miRNA is illustrated in Figure 2.
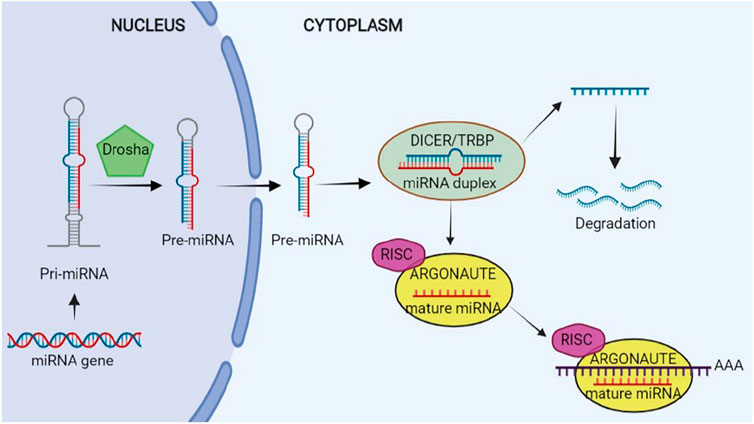
FIGURE 2. Biogenesis of microRNA (miRNA). This figure is created by www.BioRender.com.
MiRNAs may target and regulate a majority of the protein-coding genes due to the existence of one conserved binding site for miRNAs in the above genes. Additionally, numerous mRNAs may be targeted by a miRNA (Ha and Kim, 2014).
MicroRNAs and Apoptosis in Brain Tumors
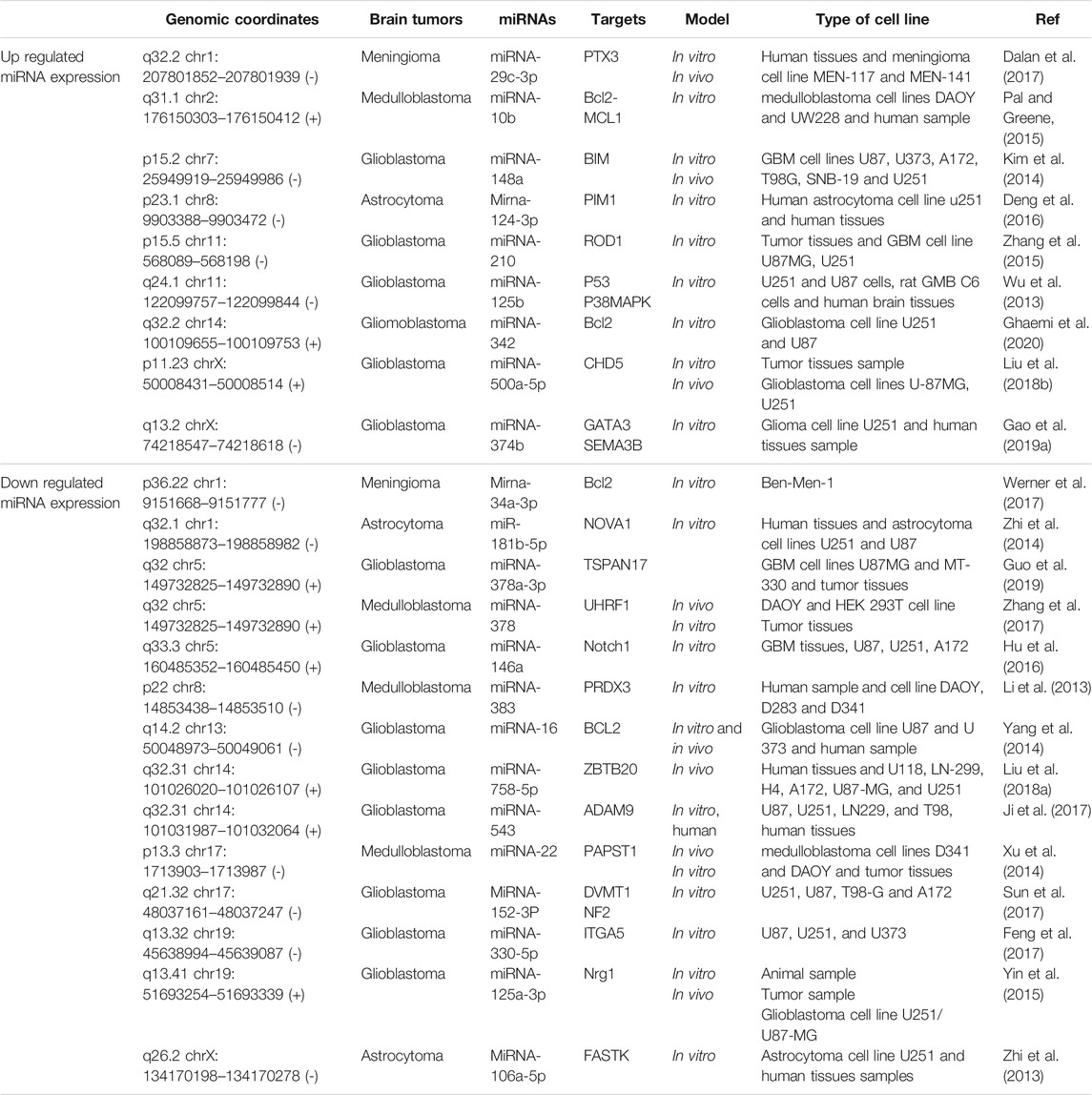
The miRNA-342 is elevated in glioblastoma cell, and could directly modulate BCL2 expression, suggesting a possible role in apoptosis induction (Ghaemi et al., 2020). The miRNA-16 can promote apoptosis by targeting BCL2 in glioblastoma cell (Yang et al., 2014). The miR-148a is elevated in glioblastoma cell. miR-148a inhibition could induce apoptosis via pro-apoptotic molecule BIM in glioblastoma (Kim et al., 2014). The elevated level of miR-330-5p in glioblastoma could induce cell apoptosis via targeting ITGA5 expression in glioblastoma cells (Feng et al., 2017). The miR-758-5p was up-regulated in glioblastoma. It can promote apoptosis by decreasing ZBTB20 in glioblastoma cell (Liu et al., 2018a). There is a downregulation of miR-543 in glioblastoma and its upregulation can induce apoptosis by targeting ADAM9 (Ji et al., 2017). MiR-152-3p affected the apoptosis of glioblastoma cells via NF2 overexpression (Sun et al., 2017). The inhibition of increased miR-210 in glioblastoma can increase ROD1 expression and apoptosis (Zhang et al., 2015). The overexpression of miR-125a-3p leads to decreased Nrg1 and increased apoptosis in glioblastoma cell (Yin et al., 2015). In another study, miR-500a-5p inhibited apoptosis in glioblastoma through targeting CHD5 mRNA 3′-UTR (Liu et al., 2018b). MiR-146a was down regulated in glioblastoma. In addition, upregulation of miR-146a suppresses Notch1 and stimulate in glioblastoma (Hu et al., 2016) MiR-125b was markedly up-regulated in glioma. MiR-125b regulates cell apoptosis by different signaling pathways (Wu et al., 2013). Elevated levels of miR-374b are reported in glioma. MiR-374b is implicated in the glioma progression by regulating apoptosis via targeting GATA3 and SEMA3B (Gao et al., 2019a). Upregulation of decreased miR-378a-3p in glioblastoma decreased the expression of TSPAN17 and increased apoptosis (Guo et al., 2019). MiR-152-3p induced glioma cell apoptosis via decreasing DNMT1 (Sun et al., 2017). In astrocytoma tumor cells, miRNA-124-3p was up-regulated and its over-expression leads to increasing apoptosis via targeting 3′-UTR of PIM1 (Deng et al., 2016). Furthermore, the over-expression of miRNA-160a-5p is able to enhance apoptosis in astrocytoma cells via decreasing Fas-activated serine/threonine kinase (FASTK), an anti-apoptotic agent (Zhi et al., 2013). The induction of an increase in MiR-181b-5p level in astrocytoma showed an increase in apoptosis through targeting NOVA1 in astrocytoma (Zhi et al., 2014). A study revealed a significant down-regulation of miR-106a-5p in astrocytoma. The Over-expression of miR-106a-5p leads to decrease in FASTK expression and increase in apoptosis (Zhi et al., 2013). Besides, miRNA-10b was up-regulated in medulloblastoma cell. miRNA-10b inhibition could also decrease the expression of BCL2 and MCL-1 protein expression in medulloblastoma cell lines (Pal and Greene, 2015). Xu et al. (2014) found that over-expression of miR-22 stimulated apoptosis via inhibiting PAPST1 in medulloblastoma cell. MiR-378 was down-regulated in medulloblastoma. The over-expression of miR-378 showed promoting apoptosis through targeting UHRF1 in medulloblastoma (Zhang et al., 2017). MiR-383 negatively regulated PRDX3. The over expression of miR-383 and inhibiting PRDX3 could leads to stimulating apoptosis in medulloblastoma (Li et al., 2013).
Meningioma cell lines show a low level miRNA-34a-3p and miRNA-34a-3p upregulation can stimulate cancer cell apoptosis via decreasing BCL2 protein (Werner et al., 2017). MiR-29c-3p was up-regulated in meningioma cell. MiR-29c-3p induces apoptosis via targeting PTX3 (Dalan et al., 2017). Apoptosis-related miRNAs in brain tumors and their expression were summarized in Table 1.
Biogenesis of lncRNAs
NcRNAs ability to regulate gene expression at transcriptional and post-transcriptional levels explains their house-keeping functions in numerous biological processes (Statello et al., 2021). Overall, researchers have described lncRNAs as long RNA transcripts of ≥200 nucleotides, which do not result in protein production (Nam et al., 2016). Moreover, they contribute importantly to the adjustment of the translation machineries and in its modulation via regulating the essential performance of other ncRNAs like small nucleolar RNA (snoRNA), miRNAs, and so forth. Several scholars presented many regulatory patterns that are under the control of numerous lncRNAs influencing different cellular activities that are correlated to the normal development and pathophysiology of some diseases like different kinds of cancers, neurological and cardio-vascular conditions, as well as metabolic and immunological dysfunctions (Maass et al., 2014). Therefore, it is important to know lncRNAs biogenesis for its differentiation from other kinds of RNA and deciphering its applicable prominence. Studies have also demonstrated transcription of numerous groups of lncRNAs from numerous DNA elements like enhancers, intergenic regions, and promoters in the eukaryotic genomes (Maass et al., 2014; Fang and Fullwood, 2016). In addition, they revealed the contribution of several mechanisms to the biogenesis of lncRNA like cleavage through ribonuclease P (RNaseP) for generating mature ends and forming protein (snoRNP) complex caps at their ends, snoRNA, and circular structures. However, there are not enough information of the action of synthesis and modulation of various lncRNAs (Dahariya et al., 2019).
Besides the dimension of the other classes of ncRNAs (e.g., siRNAs, miRNAs, and small sno/sn RNAs), lncRNAs enjoy the secondary and 3D structures that empower them for having protein-like and RNA functions (Cabili et al., 2015). Several studies determined the location of lncRNAs in the nucleus (Derrien et al., 2012), with the contribution of many lncRNAs to the cytoplasm (Cabili et al., 2015). In addition, it is possible for many lncRNAs to be transmitted to the near cells or serum via exosome trafficking. Earlier researchers have regarded lncRNAs as the by-products of transcription process. Nonetheless, lncRNAs contribute to the cell differentiation process, growth as well as pathogenesis of numerous illnesses like cancers (Maass et al., 2014). LncRNAs can modulate gene expression at the time of transcription, post-transcription, and even epigenetically (Cabili et al., 2015). Actually, lncRNAs greatly affect gene expression by modifying chromatin and remodeling, modifying histone, as well as changing the nucleosome localization. In comparison to the mRNAs, numerous lncRNAs are situated at the nucleus and researchers have shown the existence of fewer exons in the lncRNA genes in comparison to the mRNAs. Results have demonstrated the less effective splicing of lncRNAs than the mRNAs (Guo et al., 2020; Statello et al., 2021). In fact, they exhibited longer distances between and the branch point and 3′ splice site weaker internal splicing signals. Besides processing and transcription, lncRNAs frequently consist of the embedded sequence motifs with the ability of recruiting specific nuclear factors that enhance the lncRNA nuclear localization and function. Furthermore, researchers observed export of numerous lncRNAs to the cytosol with the similar export and processing pathways to the mRNAs. When the lncRNAs reached the cytoplasm, they experience particular arrangement processes for assigning various lncRNAs to the certain organelles or distribution in the cytoplasm for associating with several RNA-binding proteins (RBPs) (Statello et al., 2021). According to estimations, 50% of the pools of 70% of cytoplasmic lncRNAs occur in polysome fraction and lncRNAs at multiple level regulate the genes’ expression. (Carlevaro-Fita et al., 2016). In fact, through interactions with RNA, proteins, as well as DNA, lncRNAs may modulate the chromatin structure and functions and transcription of the distant and near genes, and influence the RNA splicing, translation, and stability. Research has showed the contribution of lncRNAs to the creation and modulation of nuclear condensates and organelles. They are capable of suppressing the genes’ expression via interferences in the transcription machinery and alter recruitment of the transcription factors or Pol II at the suppressed promoter (Pal and Greene, 2015) and histone modification (Pal and Greene, 2015; Lin et al., 2020) and declines the accessibility of chromatin (Statello et al., 2021).
One of the other kinds of lncRNAs classification has been done according to their genomic profile or correlation with the protein coding genes: 1) one or more exons of a coding gene are overlapped by sense lncRNAs, 2) full or partial complementarity to transcripts on the opposite strand exhibits anti-sense transcripts, 3) an intron of a gene produces the intronic lncRNAs, 4) there is a similar promoter in both protein-coding genes and bi-directional transcripts, though their transcription is formed in the opposite direction, 5) inter-genic lncRNAs (lincRNAs) are situated between the protein-coding genes and their transcription occurs individually, 6) enhancer RNAs (eRNAs) have been found to be created from the enhancer regions of the protein-coding genes, and finally 7) the origin of circRNAs is the splicing processes of the protein coding genes that create the covalently-closed loops (Beermann et al., 2016). lncRNA biogenesis occurs in the nucleus resembling synthesizing the protein-coding transcripts. Moreover, histone modifications occur frequently for epigenetic marking of lncRNA promoters that are modulated by transcription factors, which advocate or hinder the gene expression (Guttman and Rinn, 2012). Like mRNAs, Pol II transcribes numerous lncRNAs whereas other lncRNA promoters protect the structures transcribed by Pol III. Therefore, there is a precise spatial or temporal regulation of lncRNA expression. Generally, there are fewer lncRNAs than mRNAs, though their expression is largely limited to the certain kinds of cells (Cabili et al., 2011; Derrien et al., 2012). Based on the transcriptome-wide investigations, lncRNAs exhibited more certain expression profiles than the mRNAs (Elmore, 2007; Nikolaou et al., 2018). In other words, their expression is done in a cell tissue-, type-, developmental stage or disease state-specific way. For reaching the mature forms, the nascent RNA transcripts experience several processing steps during and after transcription, like splicing, 5ʹ-capping, chemical base modification, as well as poly-adenylation. The schematic steps of biogenesis of lncRNA is shown in Figure 3.
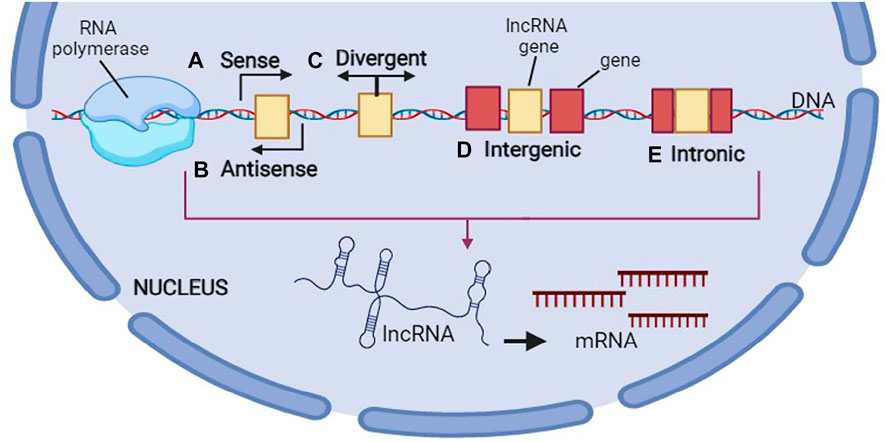
FIGURE 3. Biogenesis of long non-coding RNA (lncRNA). This figure is created by www.BioRender.com.
Researchers have found the biological contributions of lncRNAs as 1) the transcription regulators in cis or trans, 2) regulator of mRNA processing, protein activity, and posttranscriptional control, and 3) structuring the nuclear domains (Quinn and Chang, 2016). Overall, RNA polymerase II transcribe the lncRNAs, which exhibit hallmarks of the protein-coding genes, including conservation and chromatin structure of promoters, modulation of expression via morphogens and transcription factors, ranges of half-life tissue-specific expression, splicing, as well as other splice variants. Several lncRNAs enjoy a 3-helical structure at their 3′ end, created by cleavage via RnaseP, which helps protecting them against degradation (Yang et al., 2016).
Long Non-coding RNAs and Apoptosis in Brain Tumors
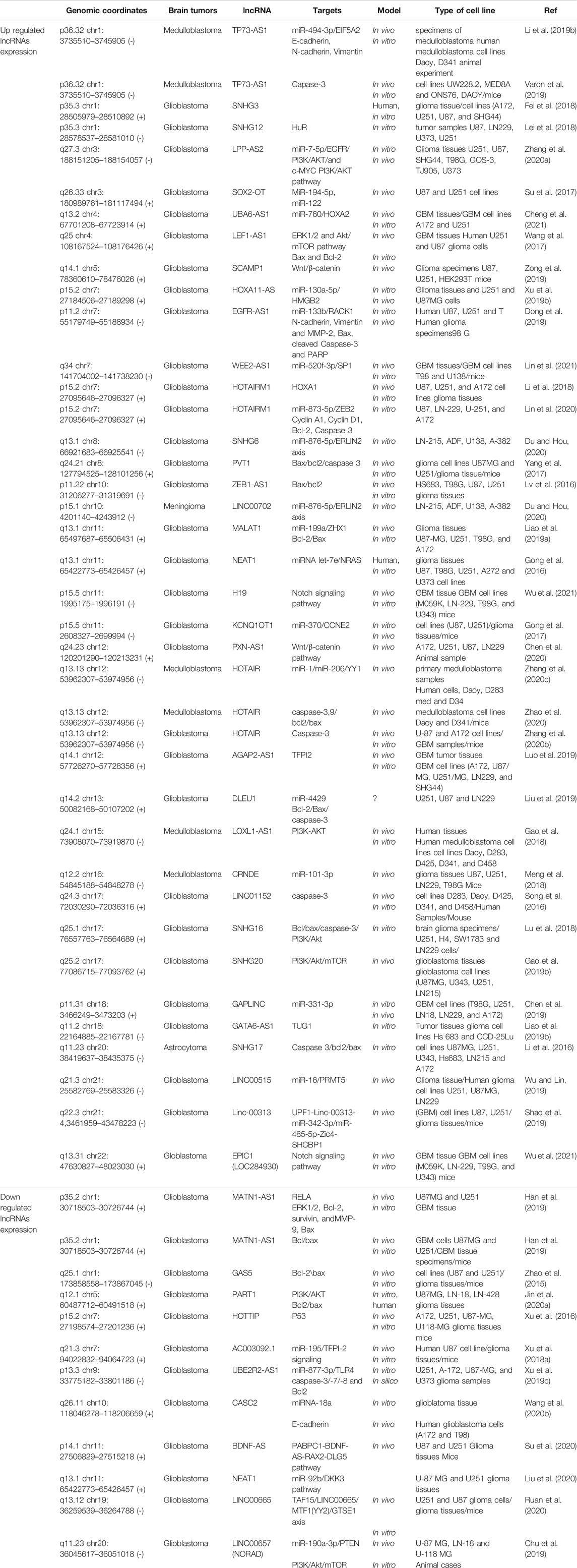
LncRNA EGFR-AS1 expression was notably up-regulated in glioma. EGFR-AS1 inhibition induced apoptosis. MiR-133b is a target of EGFR-AS1. Lnc-EGFR-AS1 knockdown increased miR-133b expression. MiR-133b was able to decrease RACK1 expression. The over-expression of miR-133b causes glioma cells to increase their apoptosis by reducing N-cadherin, Vimentin, MMP-2 and Bcl-2 and elevating Bax, cleaved Caspase-3 and PARP expression (Dong et al., 2019). There is a remarkable increase in LncRNA HOXA11-AS in glioma. The direct target of LncRNA HOXA11-AS is miRNA-130-5p, i.e., it downregulates miRNA-130-5p. Apoptosis was greatly increased upon miR-130-5p transfection. The oncogenic function of LncRNA HOXA11-AS is partly related to miR-130a-5p-HMGB2 axis. The down-regulation of LncRNA HOXA11-AS leads to increasing apoptosis in glioma (Xu et al., 2019b). Zhang et al. (2020a) showed that glioma cells apoptosis was increased by LPP-AS2 inhibition. LPP-AS2 knockdown leads to decreased EGFR the expression. LPP-AS2 acts as a molecular sponge for miR-7-5p and inhibiting miR-7-5p reverses the cell apoptosis stimulated by LPP-AS2 knockdown. LPPAS2 sponging of miR-7-5p up-regulates EGFR to activate the PI3K/AKT/c-MYC pathway (Zhang et al., 2020a). Human glioma cells have low amounts of lncRNA UBE2R2-AS1. Glioma cells growth is in part regulated by the UBE2R2-AS1/miR-877-3p/TLR4 pathway. Following upregulation of UBE2R2-AS1 in glioma cells, there was a remarkable elevation of caspase-3, caspase-7, and caspase-8, while reduction of Bcl2 (Xu et al., 2019c). LncRNA Gas5 is down-regulated in glioma. The Overexpression of Gas5 promotes the apoptosis. Gas5 and miR- 222 expressions are inversely associated. The overexpression of Gas5 upregulates Plexin C1, bmf by lowering the level of miR-222. The down-regulation of miR-222 increases the level of bmf (a pro-apoptotic agent), which increase Bax expression and decrease Bcl-2 expression in glioma (Zhao et al., 2015). lncRNA ZEB1-AS1 is elevated in glioma. Knockdown of ZEB1-AS1 induces apoptosis by increasing Bax and lowering Bcl-2 (Lv et al., 2016). LncRNA H19 expression was up-regulated in glioblastoma cells. H19 knockdown leads to increasing apoptosis in glioblastoma cells under TMZ treatment. H19 knockdown reduces pro-caspase 3 and increases cleaved caspase 3 and Bax, and decreases Bcl-2 in glioblastoma cells under TMZ treatment (Li et al., 2016). High levels of lncRNA SNHG6 are present in glioma cells. The knockdown of SNHG6 increases apoptosis. By regulating miR-101-3p expression, SNHG6 exerts its effect in glioma tumorigenesis (Meng et al., 2018). NEAT1 is downregulated in glioma. Elevating the level of NEAT1 in glioma cells can enhance apoptosis. NEAT1 exerts its tumor suppressor effects by down-regulating miR-92b and up-regulating DKK3 (Liu et al., 2020). LncRNA SNHG16 is elevated in glioma. SNHG16 inhibition leads to apoptosis via upregulating caspase-3 and Bax and, down-regulating Bcl-2, Bcl-xl and Mcl-1. In addition, SNHG16 may affect apoptosis via regulation of PI3K/Akt pathway. By binding to MiR-4518 and its inhibition, SNHG16 can regulate PRMT5 expression. SNHG16 could exerts oncogenic function through the SNHG16-miR-4518 axis in glioma cells (Lu et al., 2018). Apoptosis of glioma cells could be enhanced by inhibiting lncRNA EPIC1. The down regulation of EPIC1 reduces the Cdc20 expression which functions as an anti-apoptotic agent in glioma cells (Wang et al., 2020a). LncRNA SOX2OT is present in high levels in glioma. SOX2OT inhibition upregulates miR-194-5p and miR-122 which in turn enhance the apoptosis of glioblastoma stem cells (Su et al., 2017). Glioblastoma cells have low levels of LncRNA CASC2. LncRNA CASC2 acts as a tumor suppressor by stimulating apoptosis through CASC2-miR-18a axis in glioblastoma (Wang et al., 2020b). LncRNA GAPLINC was significantly up-regulated in glioblastoma tissues. GAPLINC inhibition promoted apoptosis. GAPLINC can target miR-331-3p. GAPLINC could be an oncogenic lncRNA via negative modulation of miR-331-3p in glioblastoma (Chen et al., 2019). The overexpression of LncRNA LINC00657 leads to miR-190a-3p inhibition; Consequently, PTEN expression is stimulated regulating caspase3 through PI3K/Akt/mTOR pathway. Thus, the overexpression of LncRNA LINC00657 induces apoptosis in glioblastoma (Chu et al., 2019). LncRNA SNHG20 was highly expressed in glioblastoma. The knockdown of SNHG20 significantly promotes cell apoptosis. PI3K/Akt/mTOR signaling pathway were inhibited by SNHG20 knockdown, while were stimulated by SNHG20 overexpression. Apoptosis is increased with IGF-1, a PI3K signaling activator, whereas apoptosis is decreased with GDC-094, a PI3K signaling inhibitor. Apoptosis induced by SNHG20 knockdown is efficiently rescued by IGF-1 (Gao et al., 2019b). Glioblastoma stem cells have high amounts of LncRNA NEAT1. LncRNA NEAT1 inhibition promotes glioblastoma stem cells apoptosis. There is a binding region between LncRNA NEAT1 and microRNA let-7e. NEAT1 inhibition increases the level of let-7e which can downregulate NRAS (a pro-oncogenic factor) as its target. In addition, the overexpression of NRAS significantly inhibit apoptosis in glioblastoma stem cells (Gong et al., 2016). LncRNA AGAP2-AS1 is elevated in glioblastoma and its inhibition leads to TFPI2 overexpression and enhanced apoptosis (Luo et al., 2019). LncRNA MANTN1-AS1 was down-regulated in glioblastoma. LncRNA MANTN1-AS1 induce glioblastoma cell apoptosis via regulation of different proteins RELA, ERK1/2, survivin, MMP-9, Bcl-2 and Bax (Han et al., 2019). LncRNA DLEU1 was significantly up-regulated in glioblastoma tissues. DLEU1 knockdown increase Bax and decrease Bcl-2. SP1 is a target of miR-4429 and DLEU1 sponged miR-4429 to induce SP1 expression. Therefore, SP1–DLEU1–miR-4429 pathway could regulate apoptosis in glioblastoma (Liu et al., 2019). LncRNA HOTAIRM1 inhibits apoptosis in glioblastoma through regulation of miR-873-5p/ZEB2 axis. By inactivating miR-873-5p, HOTAIRM1 upregulates ZEB2 (Lin et al., 2020). LncRNA MALAT1 has pivotal role in ZHX1 expression. MALAT1 induce ZHX1 expression through miR-199a in glioblastoma cells. The overexpression of ZHX1 is related to decreased Bax and increased Bcl-2 in glioblastoma (Liao et al., 2019a). In another study showed a significant up-regulation of LncRNA LEF1-AS1 expression in glioblastoma. The Knockdown of LEF1-AS1 significantly promoted cell apoptosis through regulating p27, Blc-2, and Bax expression (Wang et al., 2017). LINC01152 was up-regulated in glioblastoma. The silenced LINC01152 decreases the levels of MAML2. The apoptosis of glioblastoma cells is stimulated by MAML2 depletion. LINC01152 and MAML2 3′UTR have binding sites for miR-466. miR-466 inhibition could in part reverse the enhanced apoptosis observed after LINC01152 silencing (Wu et al., 2021). High levels of LncRNA SNHG3 are observed in glioma. SNHG3 inhibition induces cell apoptosis (Fei et al., 2018). In glioma cells, the silencing of lncRNA PVT1 inhibits EZH2 which in turn increases caspase 3 and Bax and reduces Bcl-2 and as a result, apoptosis is promoted (Yang et al., 2017). LncRNA GATA6-AS was up-regulated in glioma. GATA6-AS overexpression results in TUG1 downregulation and apoptosis inhibition (Liao et al., 2019b). LncRNA UBA6-AS1 expression was enhanced in glioblastoma. Through miR-760/HOXA2 regulation, UBA6-AS1 inhibition can improve apoptosis (Cheng et al., 2021). LncRNA KCNQ1OT1 significantly was Up-Regulated in glioma. LncRNA KCNQ1OT1 down-regulation induces glioma cells apoptosis. KCNQ1OT1 expression inversely correlates with miR-370 expression in glioma cells. Apoptosis is increased by miR-370 upregulation. MiR-370 exerts its anti-oncogenic effects through CCNE2 downregulation (Gong et al., 2017). LncRNA SCAMP1 is observed in high levels in glioma cells. The inhibition of SCAMP1 promotes apoptosis via molecular sponging of miR-499a-5p (Zong et al., 2019). LncRNA HOTAIR expression was up-regulated in glioblastoma. There is positively correlation between HOTAIR with the HK2 expression. HK2 depletion sensitizes the glioblastoma cells to TMZ-induced apoptosis. HK2 absence in glioblastoma cells also enhances the cleavage of caspase-3 following TMZ treatment and induces higher apoptosis Depletion of HOTAIR also suppresses the expression of HK2 increases the TMZ-induced apoptosis and cleavage caspase-3 in glioblastoma cells. The expression of miR-125 up-regulates by HOTAIR knockdown and miR-125 could be the downstream of HOTAIR for HK2 regulation (Zhang et al., 2020b). Low levels of LncRNA HOTTIP is observed in glioma. HOTTIP upregulation increases cell apoptosis. HOTTIP directly binds to BREgene and down-regulate BRE expression. Through BRE downregulation which results in cyclin A and CDK2 suppression and P53 elevation, HOTTIP reduces glioma cell growth (Xu et al., 2016). LncRNA LINC00515 was over-expressed in glioma tissues. LINC00515 deficiency increases apoptosis in glioma cells. LINC00515 regulates PRMT5 expression via sponging miR-16 (Wu and Lin, 2019). A remarkably low level of LncRNA PART1 is observed in glioma. PART1 inactivates miR-190a-3p which suppresses PTEN/AKT axis and as a result apoptosis in enhanced in glioma cells (Jin et al., 2020a). LncRNA WEE2-AS1 is elevated in glioblastoma. Through miR-520f-3p/SP1 axis, WEE2-AS1 inhibition improves apoptosis in glioblastoma (Lin et al., 2021). LncRNA Linc-00313 is elevated in glioma. Linc-00313 suppression significantly promotes apoptosis in glioma; this effect is due to miR-342-3p and miR-485- 5p upregulation (Shao et al., 2019). LncRNA AC003092.1 is lowered in glioblastoma. LncRNA AC003092.1 exerts its effects in TMZ chemosensitivity by regulating miR-195/TFPI-2 pathway (Xu et al., 2018a). Glioma cells have elevated levels of lncRNA SNHG12. The overexpression of lncRNA SNHG12 significantly inhibit the apoptosis in glioma cell via targeting Hu antigen R (HuR) (Lei et al., 2018). LncRNA HOTAIRM1 is upregulated in glioma and glioblastoma. HOTAIRM1 inhibited cell apoptosis via regulation of HOXA1 gene (Li et al., 2018). The overexpression of lncRNA PABPC1 increased apoptosis in glioblastoma cells via BDNF-AS-RAX2-DLG5 axis (Su et al., 2020). Glioblastoma cells show elevated levels of LncRNA PXN-AS1. Apoptosis of glioblastoma cells was promoted by PXN-AS1 inhibition through lowering Bcl-2 and elevating Bax (Chen et al., 2020). There are low levels of TAF15 and LncRNA LINC00665 in glioma, on the contrary, high levels of MTF1, YY2, and GTSE1 are observed. TAF15 upregulation could promotes apoptosis via targeting MTF1,YY2, and GTSE1 (Ruan et al., 2020). LncRNA MATN1-AS1 is lowered in glioblastoma. MATN1-AS1 upregulation is able to promote apoptosis through RELA, survivin, ERK1/2, MMP-9, and Bcl-2 inhibition, and Bax elevation (Han et al., 2019).
Medulloblastoma tissues are observed to have elevated levels of LncRNA HOTAIR which binds to miR-1 and miR-206 and inhibits them to upregulate YY1. Through miR-1/miR-206-YY1 pathway, HOTAIR suppression leads to increased apoptosis in medulloblastoma (Zhang et al., 2020c). Medulloblastoma tissues demonstrate high levels of LncRNA LOXL1-AS1. The proliferation and metastasis of medulloblastoma cells are improved by LOXL1-AS1 via stimulating PI3K-AKT pathway and therefore, its suppression results in increased apoptosis (Gao et al., 2018). There is an elevated level of LncRNA TP73-AS1 in medulloblastoma cells which binds to miR-494-3p to inhibit it and in turn activate EIF5A2, the pathway that explains promoted apoptosis following TP73-AS1 suppression (Li et al., 2019b). LncRNA CRNDE was up-regulated in medulloblastoma. The knockdown of lncRNA CRNDE induces apoptosis via promoting the activity of caspase-3 in medulloblastoma cells (Song et al., 2016) Cleaved-caspase-3, caspase-9, and Bax elevation, and bcl-2 reduction are the results of HOTAIR downregulation in medulloblastoma cells (Zhao et al., 2020). Varon et al. (2019) reported a significant up-regulation of lncRNA TP73-AS1 in medulloblastoma cells. Silencing TP73-AS1 stimulates apoptosis via targeting caspase 3 in medulloblastoma cells.
Malignant meningioma shows LncRNA LINC00702 to be upregulated. LINC00702 stimulates tumorigenesis by inactivating miR-4652-3p which consequently activates ZEB1. Therefore, LINC00702 downregulation induces apoptosis (Li et al., 2019c). Astrocytoma cells are shown to have high amounts of LncRNA SNHG17. SNHG17 can bind to miR-876-5p to keep it from sponging ERLIN2. This pathway justifies the enhanced apoptosis observed after SNHG17 inhibition (Du and Hou, 2020). Table 2 lists apoptosis-related lncRNAs in brain tumors and their expression.
Biogenesis of the Circular RNAs
Circular RNAs (circRNAs) characterize a type of endogenous ncRNA generated by back splicing events, with the ubiquitous presence in numerous species. They have one or more exons with the major location in the cytoplasm so that a few of the circRNAs consisting of intron originate in the nucleus (Guo et al., 2014). In spite of the linear RNAs, circRNAs make a continuous loop formation using covalent bonds and therefore do not have a 5–3′ direction or a polyadenylated tail; these features result in their higher stability in the plasma and tissues (Suzuki and Tsukahara, 2014).
Hence, the mentioned specific structural features imply the fact that they are pathologically and physiologically major transcripts. A lot of investigations have confirmed that the abnormal expression of circRNAs is one of the major regulatory elements in expansion and carcinogenesis of some cancers such as hematological malignancies, lung cancer, and liver cancer, revealing the probable targets to diagnosis and prognosis of these cancers by circRNAs. Some recent investigations performed about features and biogenesis of the circRNAs, summarized their functions and mechanisms of action in cancers, and discussed their capacity to diagnose and treat the diseases of interest (Bach et al., 2019; Zhang et al., 2020d; Lei et al., 2020; Shen et al., 2021).
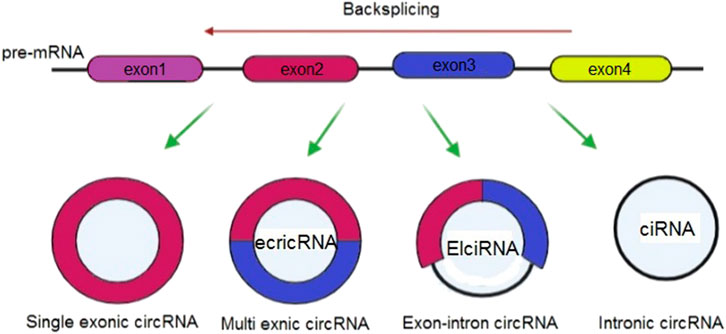
Considering their origins, it is possible to classify circRNAs into 3 groups of circular intron RNAs (ciRNAs), exonic circRNAs (ecircRNAs), and exon–intron circRNAs (EIciRNAs) (Meng et al., 2017) (Figure 4). Various circularization mechanisms regulate circRNA biogenesis, out of which ecircRNA is abundantly found so that it includes most of specified circRNAs (Bolha et al., 2017). In fact, gene transcription is regulated by cirRNAs. Overall, Pol II transcribes a pre-mRNA which consists of exons and introns as well as a seven- methylguanosine cap and poly-adenosine tail, which is added to its 5′- and 3′-ends. After that, pre-mRNA is spliced at the canonical splice sites (5′-GU & 3′-AG at introns) using spliceosomes for maturity and translatability. In addition, one of the specific splicing manners known as back-splicing generates CircRNA, where the 3′-end of an exon binds to the 5′-end of its own or an upstream exon via a 3′, 5′- phosphodiester bond, and establishes a closed structure with a back-splicing junction. Researchers viewed circRNAs as the splicing errors consisting of scrambled exons, and presented and verified two biogenesis models based on the splicing events sequence and various intermediates, including direct backsplicing model and lariat model (Zhang et al., 2020d; Zhou et al., 2020).
Circular RNAs and Apoptosis in Brain Tumors
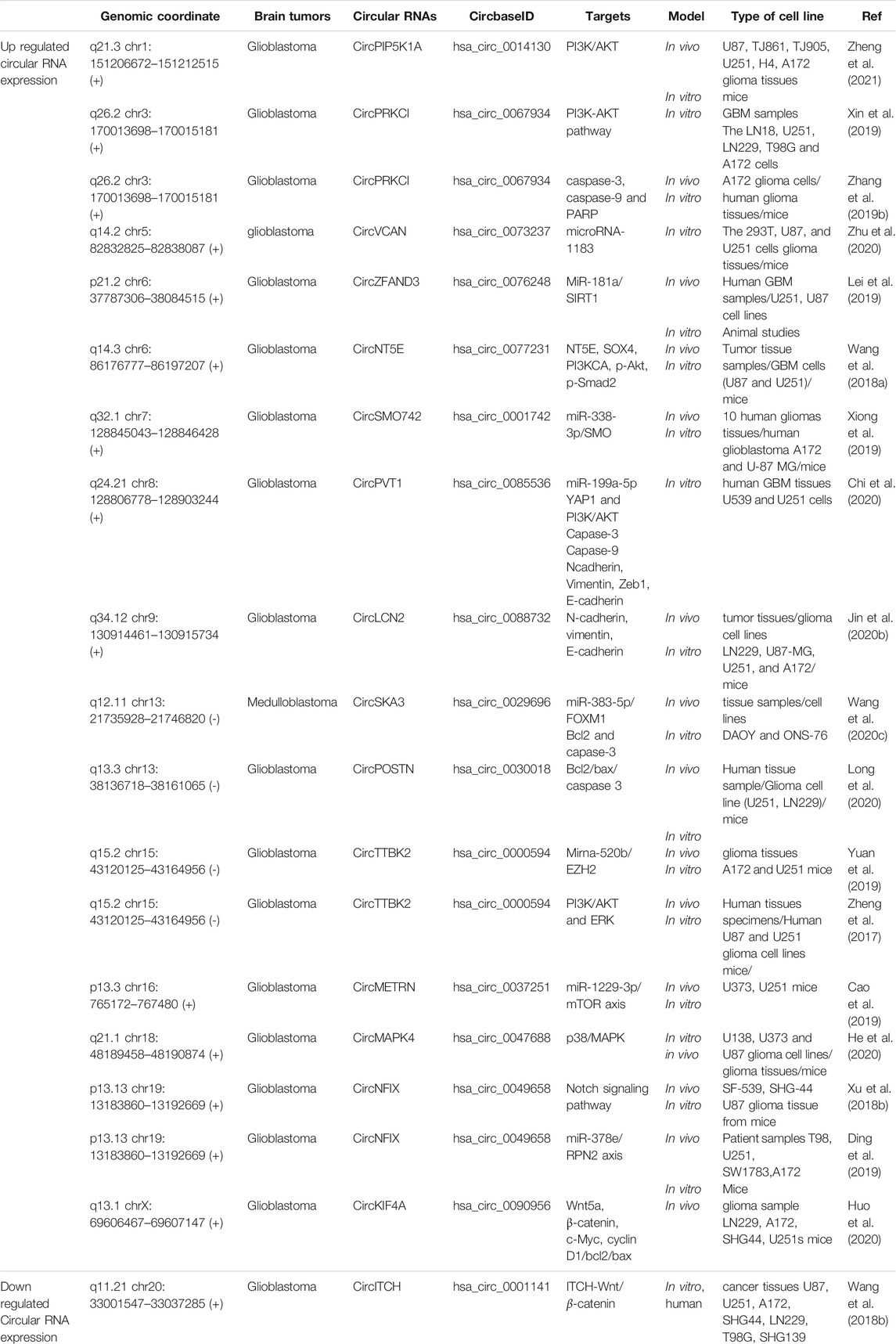
Glioblastoma cells show a remarkably high level of Circular RNA hsa_circ_0067934. The hsa_circ_0067934 silencing promotes apoptosis in glioblastoma cells via PI3K-AKT pathway (Xin et al., 2019). MiR‐181a was down-regulated in glioma, while the circ_0076248 and SIRT1 were up-regulated. The down-regulation of hsa_circ_0076248 or miR-181a overexpression upregulate p53 and SIRT1. Down regulation of hsa_circ_0076248 leads to inhibiting xenograft tumors’ growth and increasing apoptosis (Lei et al., 2019). CircPVT1 was over-expressed in glioblastoma tissues. Silencing circPVT1 increasese the number of apoptotic cells by increased cleaved-Caspase-3 and cleavedCaspase-9 expression via targeting miR-199a-5p (Chi et al., 2020). Glioblastoma cells were shown to have a greatly elevated level of circNT5E. circNT5E up-regulation inhibits apoptosis via miR-422a (Wang et al., 2018a). Glioma cells have a high level of Circ-TTBK2 and EZH2. Through miR-520b/EZH2 pathway, circ-TTBK2 suppression is able to improve apoptosis (Yuan et al., 2019). The high level of circSMO742 and SMO can inhibit miR-338-3p which results in increased proliferation, migration and invasion and decreased apoptosis of glioma cells (Xiong et al., 2019). Cir-ITCH is observed in low amounts in glioma. Through miR-214 inactivation and modulation of ITCH-Wnt/β-catenin axis in glioma cells, cir-ITCH upregulation can improve apoptosis (Wang et al., 2018b). Glioma cells have an elevated level of circPOSTN. Via targeting miR-361-5p/TPX2, circPOSTN suppression can lower Bcl-2 and elevate Bax cand caspase-3 which result in apoptosis (Long et al., 2020). CircPIP5K1A was up-regulated in glioma cells. The apoptosis increases after the knockdown of circPIP5K1A through TCF12/PI3K/AKT axis regulation by miR-515-5p inactivation (Zheng et al., 2021). Circ-MAPK4 was over-expressed in glioma tissues. Circ-MAPK4 silencing increases apoptosis by elevating caspase-3, caspase-7, caspase-9, and PARP1 via targeting miR-125a-3p and regulating p38/MAPK (Peng et al., 2020). Glioma cells show a greatly high level of circ-TTBK2. circ-TTBK2 regulates miR-217/HNF1β/Derlin-1 pathway which reduces apoptosis (Zheng et al., 2017). Glioma cells show hsa_circ_0088732 to be upregulated. hsa_circ_0088732 inhibition stimulates apoptosis through the miR-661/RAB3D pathway (Jin et al., 2020b). Glioma cells show circNFIX to be overexpressed. circNFIX impedes miR-34a-5p which inhibits apoptosis and therefore, circNFIX knockout can show the opposite effects (Xu et al., 2018b). CircKIF4A expression was increased in glioma. By targeting miR-139-3p which downregulates cyclin D1, c-Myc and Bcl2, and upregulates Bax, CircKIF4A suppression can improve apoptosis in glioma cells (Huo et al., 2020). Glioma cells show circPRKCI to be overexpressed. CircPRKCI shRNA could induce apoptosis by increasing Annexin V, and cleaving caspase-3, caspase-9 and PARP via targeting miR-545 (Zhang et al., 2019b). circNFIX is present in high amounts in glioma tissues. circNFIX suppression targets miR-378e/RPN2 pathway to induce apoptosis (Ding et al., 2019). hsa_circ_00037251 inhibition promotes apoptosis in glioma via miR-1229-3p/mTOR pathway (Cao et al., 2019). Radioresistant glioma was shown to have notably elevated levels of circ_VCAN. It exerts a carcinogenic role in glioma via regulating microRNA-1183. The knockdown of circ_VCAN stimulated apoptosis in glioma cells (Zhu et al., 2020). CircSKA3 was up-regulated in medulloblastoma tissues. sicircSKA3 reduces Bcl-2, while elevates caspase-3 in medulloblastoma cells by targeting miR-383-5p (Wang et al., 2020c). Table 3 illustrates apoptosis-related circular RNAs in brain tumors and their expression.
Conclusion
Various cellular functions reveal the contribution of ncRNAs to the cancer development at the transcriptional, translational, as well as epigenetic, level. Hence, ncRNAs can be a hopeful path of molecular medicine, though, patterns of RNAs expression in the brain tumors are not constant and stable, and thus researchers must provide proper alternatives to be utilized as the diagnostic or treatment instruments. With their stable structure, various ncRNAs have shown their capacity to function as regulatory agents. Studies illustrated a few circRNAs, lncRNAs even snoRNAs as the potent treatment, diagnostic, prognostic targets in the tumors of brain. Therefore, there are limited investigations revealing the strict correlation of RNAs with prognosis of the cases with gliomas, WHO grade, as well as their real potential diagnostic value. Nonetheless, multiple investigations largely emphasized the pathological- clinical specimens. A major difficulty is that we must be wait for those research that specify circRNAs, snoRNAs, miRNA, and lncRNAs from the bodily fluids, in particular, CSF and blood of the cases with brain tumors. Hence, it is important to focus on the particular methods about stability, abundance, and longer half-life for identifying ncRNAs molecules associated with the noninvasive diagnosis and classification of the sub-types of gliomas. Hence, such attempts may simplify the evaluation of the initial treatment with higher sensitivity, magnetic resonance imaging (MRI), and or other conventional bio-markers. Thus, expression profile of the regulator RNAs that is identified by analyzing circRNAs, snoRNAs, miRNA, and lncRNA along with studying the respective effects would be of high importance for exploring the molecular pathology of the tumors in the brain. Finally, more research must be performed for developing one of the new RNA-based approaches for treating any malignant tumor and using their diagnostic and prognostic potency, which may result in promising results for cases with the brain tumors.
Author Contributions
OT, FN, JR, RH, HJ, AR, MH, and HM contributed in data collection and manuscript drafting. All authors approved the final version for submission. MD, AJ, MM, and MT, critically revised the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Achkar, N. P., Cambiagno, D. A., and Manavella, P. A. (2016). miRNA Biogenesis: a Dynamic Pathway. Trends Plant Sci. 21 (12), 1034–1044. doi:10.1016/j.tplants.2016.09.003
Aldape, K., Brindle, K. M., Chesler, L., Chopra, R., Gajjar, A., Gilbert, M. R., et al. (2019). Challenges to Curing Primary Brain Tumours. Nat. Rev. Clin. Oncol. 16 (8), 509–520. doi:10.1038/s41571-019-0177-5
Bach, D.-H., Lee, S. K., and Sood, A. K. (2019). Circular RNAs in Cancer. Mol. Ther. - Nucleic Acids 16, 118–129. doi:10.1016/j.omtn.2019.02.005
Balandeh, E., Mohammadshafie, K., Mahmoudi, Y., Hossein Pourhanifeh, M., Rajabi, A., Bahabadi, Z. R., et al. (2021). Roles of Non-coding RNAs and Angiogenesis in Glioblastoma. Front. Cel Dev. Biol. 9, 716462. doi:10.3389/fcell.2021.716462
Beermann, J., Piccoli, M.-T., Viereck, J., and Thum, T. (2016). Non-coding RNAs in Development and Disease: Background, Mechanisms, and Therapeutic Approaches. Physiol. Rev. 96 (4), 1297–1325. doi:10.1152/physrev.00041.2015
Bolha, L., Ravnik-Glavač, M., and Glavač, D. (2017). Circular RNAs: Biogenesis, Function, and a Role as Possible Cancer Biomarkers. Int. J. Genomics 2017, 6218353. doi:10.1155/2017/6218353
Cabili, M. N., Dunagin, M. C., McClanahan, P. D., Biaesch, A., Padovan-Merhar, O., Regev, A., et al. (2015). Localization and Abundance Analysis of Human lncRNAs at Single-Cell and Single-Molecule Resolution. Genome Biol. 16 (1), 20–16. doi:10.1186/s13059-015-0586-4
Cabili, M. N., Trapnell, C., Goff, L., Koziol, M., Tazon-Vega, B., Regev, A., et al. (2011). Integrative Annotation of Human Large Intergenic Noncoding RNAs Reveals Global Properties and Specific Subclasses. Genes Dev. 25 (18), 1915–1927. doi:10.1101/gad.17446611
Cao, Q., Shi, Y., Wang, X., Yang, J., Mi, Y., Zhai, G., et al. (2019). Circular METRN RNA Hsa_circ_0037251 Promotes Glioma Progression by Sponging miR-1229-3p and Regulating mTOR Expression. Sci. Rep. 9 (1), 19791. doi:10.1038/s41598-019-56417-8
Carlevaro-Fita, J., Rahim, A., Guigó, R., Vardy, L. A., and Johnson, R. (2016). Cytoplasmic Long Noncoding RNAs Are Frequently Bound to and Degraded at Ribosomes in Human Cells. RNA 22 (6), 867–882. doi:10.1261/rna.053561.115
Chen, H. H., Zong, J., and Wang, S. J. (2019). LncRNA GAPLINC Promotes the Growth and Metastasis of Glioblastoma by Sponging miR-331-3p. Eur. Rev. Med. Pharmacol. Sci. 23 (1), 262–270. doi:10.26355/eurrev_201901_16772
Chen, H., Hou, G., Yang, J., Chen, W., Guo, L., Mao, Q., et al. (2020). SOX9‐activated PXN‐AS1 Promotes the Tumorigenesis of Glioblastoma by EZH2‐mediated Methylation of DKK1. J. Cel Mol Med 24 (11), 6070–6082. doi:10.1111/jcmm.15189
Cheng, F., Liu, J., Zhang, Y., You, Q., Chen, B., Cheng, J., et al. (2021). Long Non-coding RNA UBA6-AS1 Promotes the Malignant Properties of Glioblastoma by Competitively Binding to microRNA-760 and Enhancing Homeobox A2 Expression. Cancer Manag. Res. 13, 379–392. doi:10.2147/cmar.s287676
Chi, G., Yang, F., Xu, D., and Liu, W. (2020). Silencing hsa_circ_PVT1 (circPVT1) Suppresses the Growth and Metastasis of Glioblastoma Multiforme Cells by Up-Regulation of miR-199a-5p. Artif. Cell nanomedicine, Biotechnol. 48 (1), 188–196. doi:10.1080/21691401.2019.1699825
Chu, L., Yu, L., Liu, J., Song, S., Yang, H., Han, F., et al. (2019). Long Intergenic Non-coding LINC00657 Regulates Tumorigenesis of Glioblastoma by Acting as a Molecular Sponge of miR-190a-3p. Aging 11 (5), 1456–1470. doi:10.18632/aging.101845
Dahariya, S., Paddibhatla, I., Kumar, S., Raghuwanshi, S., Pallepati, A., and Gutti, R. K. (2019). Long Non-coding RNA: Classification, Biogenesis and Functions in Blood Cells. Mol. Immunol. 112, 82–92. doi:10.1016/j.molimm.2019.04.011
Dalan, A. B., Gulluoglu, S., Tuysuz, E. C., Kuskucu, A., Yaltirik, C. K., Ozturk, O., et al. (2017). Simultaneous Analysis of miRNA-mRNA in Human Meningiomas by Integrating Transcriptome: A Relationship between PTX3 and miR-29c. BMC cancer 17 (1), 207–209. doi:10.1186/s12885-017-3198-4
D’Arcy, M. S. (2019). Cell Death: a Review of the Major Forms of Apoptosis, Necrosis and Autophagy. Cel Biol. Int. 43 (6), 582–592.
Dashti, F., Mirazimi, S. M. A., Rabiei, N., Fathazam, R., Rabiei, N., Piroozmand, H., et al. (2021). The Role of Non-coding RNAs in Chemotherapy for Gastrointestinal Cancers. Mol. Ther. - Nucleic Acids 26, 892–926. doi:10.1016/j.omtn.2021.10.004
Deng, D., Wang, L., Chen, Y., Li, B., Xue, L., Shao, N., et al. (2016). MicroRNA‐124‐3p Regulates Cell Proliferation, Invasion, Apoptosis, and Bioenergetics by Targeting PIM1 in Astrocytoma. Cancer Sci. 107 (7), 899–907. doi:10.1111/cas.12946
Derrien, T., Johnson, R., Bussotti, G., Tanzer, A., Djebali, S., Tilgner, H., et al. (2012). The GENCODE V7 Catalog of Human Long Noncoding RNAs: Analysis of Their Gene Structure, Evolution, and Expression. Genome Res. 22 (9), 1775–1789. doi:10.1101/gr.132159.111
Ding, C., Wu, Z., You, H., Ge, H., Zheng, S., Lin, Y., et al. (2019). CircNFIX promotes progression of glioma through regulating miR-378e/RPN2 axis. J. Exp. Clin. Cancer Res. 38 (1), 506–512. doi:10.1186/s13046-019-1483-6
Dong, Z.-Q., Guo, Z.-Y., and Xie, J. (2019). The lncRNA EGFR-AS1 Is Linked to Migration, Invasion and Apoptosis in Glioma Cells by Targeting miR-133b/RACK1. Biomed. Pharmacother. 118, 109292. doi:10.1016/j.biopha.2019.109292
Du, F., and Hou, Q. (2020). SNHG17 Drives Malignant Behaviors in Astrocytoma by Targeting miR-876-5p/ERLIN2 axis. BMC cancer 20 (1), 839. doi:10.1186/s12885-020-07280-8
Elmore, S. (2007). Apoptosis: a Review of Programmed Cell Death. Toxicol. Pathol. 35 (4), 495–516. doi:10.1080/01926230701320337
Evan, G. I., and Vousden, K. H. (2001). Proliferation, Cell Cycle and Apoptosis in Cancer. Nature 411 (6835), 342–348. doi:10.1038/35077213
Fang, Y., and Fullwood, M. J. (2016). Roles, Functions, and Mechanisms of Long Non-coding RNAs in Cancer. Genomics, Proteomics & Bioinformatics 14 (1), 42–54. doi:10.1016/j.gpb.2015.09.006
Fei, F., He, Y., He, S., He, Z., Wang, Y., Wu, G., et al. (2018). LncRNA SNHG3 Enhances the Malignant Progress of Glioma through Silencing KLF2 and P21. Biosci. Rep. 38 (5), BSR20180420. doi:10.1042/BSR20180420
Feng, L., Ma, J., Ji, H., Liu, Y., and Hu, W. (2017). miR-330-5p Suppresses Glioblastoma Cell Proliferation and Invasiveness through Targeting ITGA5. Biosci. Rep. 37 (3), BSR20170019. doi:10.1042/BSR20170019
Fernald, K., and Kurokawa, M. (2013). Evading Apoptosis in Cancer. Trends Cell Biology 23 (12), 620–633. doi:10.1016/j.tcb.2013.07.006
Gao, J., Bai, S., Wang, Y., Zhao, S., He, Z., and Wang, R. (2019). MiR-374b Targets GATA3 to Promote Progression and Development of Glioblastoma via Regulating SEMA3B. Neoplasma 66 (4), 543–554. doi:10.4149/neo_2018_180830N659
Gao, R., Zhang, R., Zhang, C., Liang, Y., and Tang, W. (2018). LncRNA LOXL1-AS1 Promotes the Proliferation and Metastasis of Medulloblastoma by Activating the PI3K/AKT Pathway. Anal. Cel Pathol (Amst) 2018, 9275685. doi:10.1155/2018/9275685
Gao, X. F., He, H. Q., Zhu, X. B., Xie, S. L., and Cao, Y. (2019). LncRNA SNHG20 Promotes Tumorigenesis and Cancer Stemness in Glioblastoma via Activating PI3K/Akt/mTOR Signaling Pathway. Neoplasma 66 (4), 532–542. doi:10.4149/neo_2018_180829N656
Gebert, L. F. R., and MacRae, I. J. (2019). Regulation of microRNA Function in Animals. Nat. Rev. Mol. Cel Biol 20 (1), 21–37. doi:10.1038/s41580-018-0045-7
Ghaemi, S., Arefian, E., Rezazadeh Valojerdi, R., Soleimani, M., Moradimotlagh, A., and Jamshidi Adegani, F. (2020). Inhibiting the Expression of Anti-apoptotic Genes BCL2L1 and MCL1, and Apoptosis Induction in Glioblastoma Cells by microRNA-342. Biomed. Pharmacother. 121, 109641. doi:10.1016/j.biopha.2019.109641
Gong, W., Zheng, J., Liu, X., Liu, Y., Guo, J., Gao, Y., et al. (2017). Knockdown of Long Non-coding RNA KCNQ1OT1 Restrained Glioma Cells' Malignancy by Activating miR-370/CCNE2 Axis. Front. Cel. Neurosci. 11, 84. doi:10.3389/fncel.2017.00084
Gong, W., Zheng, J., Liu, X., Ma, J., Liu, Y., and Xue, Y. (2016). Knockdown of NEAT1 restrained the malignant progression of glioma stem cells by activating microRNA let-7e. Oncotarget 7 (38), 62208–62223. doi:10.18632/oncotarget.11403
Guo, C.-J., Ma, X.-K., Xing, Y.-H., Zheng, C.-C., Xu, Y.-F., Shan, L., et al. (2020). Distinct Processing of lncRNAs Contributes to Non-conserved Functions in Stem Cells. Cell 181 (3), 621–636. e22. doi:10.1016/j.cell.2020.03.006
Guo, J. U., Agarwal, V., Guo, H., and Bartel, D. P. (2014). Expanded Identification and Characterization of Mammalian Circular RNAs. Genome Biol. 15 (7), 409–414. doi:10.1186/s13059-014-0409-z
Guo, X. B., Zhang, X. C., Chen, P., Ma, L. M., and Shen, Z. Q. (2019). miR-378a-3p Inhibits Cellular Proliferation and Migration in Glioblastoma Multiforme by Targeting Tetraspanin 17. Oncol. Rep. 42 (5), 1957–1971. doi:10.3892/or.2019.7283
Guttman, M., and Rinn, J. L. (2012). Modular Regulatory Principles of Large Non-coding RNAs. Nature 482 (7385), 339–346. doi:10.1038/nature10887
Ha, M., and Kim, V. N. (2014). Regulation of microRNA Biogenesis. Nat. Rev. Mol. Cel Biol 15 (8), 509–524. doi:10.1038/nrm3838
Han, N., Yang, L., Zhang, X., Zhou, Y., Chen, R., Yu, Y., et al. (2019). LncRNA MATN1-AS1 Prevents Glioblastoma Cell from Proliferation and Invasion via RELA Regulation and MAPK Signaling Pathway. Ann. Transl Med. 7 (23), 784. doi:10.21037/atm.2019.11.36
Hashemian, S. M., Pourhanifeh, M. H., Fadaei, S., Velayati, A. A., Mirzaei, H., and Hamblin, M. R. (2020). Non-coding RNAs and Exosomes: Their Role in the Pathogenesis of Sepsis. Mol. Ther. - Nucleic Acids 21, 51–74. doi:10.1016/j.omtn.2020.05.012
He, J., Huang, Z., He, M., Liao, J., Zhang, Q., Wang, S., et al. (2020). Circular RNA MAPK4 (Circ-MAPK4) Inhibits Cell Apoptosis via MAPK Signaling Pathway by Sponging miR-125a-3p in Gliomas. Mol. Cancer 19 (1), 17. doi:10.1186/s12943-019-1120-1
Hu, H.-q., Sun, L.-g., and Guo, W.-j. (2016). Decreased miRNA-146a in Glioblastoma Multiforme and Regulation of Cell Proliferation and Apoptosis by Target Notch1. Int. J. Biol. Markers 31 (3), 270–275. doi:10.5301/jbm.5000194
Huo, L. W., Wang, Y. F., Bai, X. B., Zheng, H. L., and Wang, M. D. (2020). circKIF4A Promotes Tumorogenesis of Glioma by Targeting miR-139-3p to Activate Wnt5a Signaling. Mol. Med. 26 (1), 29–15. doi:10.1186/s10020-020-00159-1
Jafari, A., Rezaei-Tavirani, M., Farhadihosseinabadi, B., Zali, H., and Niknejad, H. (2021). Human Amniotic Mesenchymal Stem Cells to Promote/suppress Cancer: Two Sides of the Same coin. Stem Cel Res. Ther. 12 (1), 1–11. doi:10.1186/s13287-021-02196-x
Jafari, A., Rezaei-Tavirani, M., Niknejad, H., and Zali, H. (2021). Tumor Targeting by Conditioned Medium Derived from Human Amniotic Membrane: New Insight in Breast Cancer Therapy. Technol. Cancer Res. Treat. 20, 15330338211036318. doi:10.1177/15330338211036318
Jafari, A., Rezaei-Tavirani, M., Salimi, M., Tavakkol, R., and Jafari, Z. (2020). Oncological Emergencies from Pathophysiology and Diagnosis to Treatment: A Narrative Review. Soc. Work Public Health 35 (8), 689–709. doi:10.1080/19371918.2020.1824844
Ji, T., Zhang, X., and Li, W. (2017). MicroRNA-543 Inhibits Proliferation, Invasion and Induces Apoptosis of Glioblastoma Cells by Directly Targeting ADAM9. Mol. Med. Rep. 16 (5), 6419–6427. doi:10.3892/mmr.2017.7332
Jin, T., Liu, M., Liu, Y., Li, Y., Xu, Z., He, H., et al. (2020). Lcn2-derived Circular RNA (Hsa_circ_0088732) Inhibits Cell Apoptosis and Promotes EMT in Glioma via the miR-661/RAB3D Axis. Front. Oncol. 10, 170. doi:10.3389/fonc.2020.00170
Jin, Z., Piao, L., Sun, G., Lv, C., Jing, Y., and Jin, R. (2020). Long Non-coding RNA PART1 Exerts Tumor Suppressive Functions in Glioma via Sponging miR-190a-3p and Inactivation of PTEN/AKT Pathway. Onco Targets Ther. 13, 1073–1086. doi:10.2147/ott.s232848
Khani, P., Nasri, F., Khani Chamani, F., Saeidi, F., Sadri Nahand, J., Tabibkhooei, A., et al. (2019). Genetic and Epigenetic Contribution to Astrocytic Gliomas Pathogenesis. J. Neurochem. 148 (2), 188–203. doi:10.1111/jnc.14616
Kim, J., Zhang, Y., Skalski, M., Hayes, J., Kefas, B., Schiff, D., et al. (2014). microRNA-148a Is a Prognostic oncomiR that Targets MIG6 and BIM to Regulate EGFR and Apoptosis in Glioblastoma. Cancer Res. 74 (5), 1541–1553. doi:10.1158/0008-5472.can-13-1449
Lee, R. C., Feinbaum, R. L., and Ambros, V. (1993). The C. elegans Heterochronic Gene Lin-4 Encodes Small RNAs with Antisense Complementarity to Lin-14. cell 75 (5), 843–854. doi:10.1016/0092-8674(93)90529-y
Lei, B., Huang, Y., Zhou, Z., Zhao, Y., Thapa, A. J., Li, W., et al. (2019). Circular RNA Hsa_circ_0076248 Promotes Oncogenesis of Glioma by Sponging miR‐181a to Modulate SIRT1 Expression. J. Cel Biochem 120 (4), 6698–6708. doi:10.1002/jcb.27966
Lei, M., Zheng, G., Ning, Q., Zheng, J., and Dong, D. (2020). Translation and Functional Roles of Circular RNAs in Human Cancer. Mol. Cancer 19 (1), 30–39. doi:10.1186/s12943-020-1135-7
Lei, W., Wang, Z. L., Feng, H. J., Lin, X. D., Li, C. Z., and Fan, D. (2018). Long Non-coding RNA SNHG12promotes the Proliferation and Migration of Glioma Cells by Binding to HuR. Int. J. Oncol. 53 (3), 1374–1384. doi:10.3892/ijo.2018.4478
Li, B., Shen, M., Yao, H., Chen, X., and Xiao, Z. (2019). Long Noncoding RNA TP73-AS1 Modulates Medulloblastoma Progression In Vitro and In Vivo by Sponging miR-494-3p and Targeting EIF5A2. Onco Targets Ther. 12, 9873–9885. doi:10.2147/ott.s228305
Li, K. K.-W., Pang, J. C.-S., Lau, K.-M., Zhou, L., Mao, Y., Wang, Y., et al. (2013). MiR-383 Is Downregulated in Medulloblastoma and Targets Peroxiredoxin 3 (PRDX3). Brain Pathol. 23 (4), 413–425. doi:10.1111/bpa.12014
Li, Q., Dong, C., Cui, J., Wang, Y., and Hong, X. (2018). Over-expressed lncRNA HOTAIRM1 Promotes Tumor Growth and Invasion through Up-Regulating HOXA1 and Sequestering G9a/EZH2/Dnmts Away from the HOXA1 Gene in Glioblastoma Multiforme. J. Exp. Clin. Cancer Res. 37 (1), 265. doi:10.1186/s13046-018-0941-x
Li, T., Ren, J., Ma, J., Wu, J., Zhang, R., Yuan, H., et al. (2019). LINC00702/miR-4652-3p/ZEB1 axis Promotes the Progression of Malignant Meningioma through Activating Wnt/β-Catenin Pathway. Biomed. Pharmacother. 113, 108718. doi:10.1016/j.biopha.2019.108718
Li, W., Jiang, P., Sun, X., Xu, S., Ma, X., and Zhan, R. (2016). Suppressing H19 Modulates Tumorigenicity and Stemness in U251 and U87MG Glioma Cells. Cell Mol Neurobiol 36 (8), 1219–1227. doi:10.1007/s10571-015-0320-5
Li, Y., Ren, Z., Peng, Y., Li, K., Wang, X., Huang, G., et al. (2019). Classification of Glioma Based on Prognostic Alternative Splicing. BMC Med. Genomics 12 (1), 165. doi:10.1186/s12920-019-0603-7
Liao, K., Lin, Y., Gao, W., Xiao, Z., Medina, R., Dmitriev, P., et al. (2019). Blocking lncRNA MALAT1/miR-199a/ZHX1 axis Inhibits Glioblastoma Proliferation and Progression. Mol. Ther. - Nucleic Acids 18, 388–399. doi:10.1016/j.omtn.2019.09.005
Liao, Y., Zhang, B., Zhang, T., Zhang, Y., and Wang, F. (2019). LncRNA GATA6-AS Promotes Cancer Cell Proliferation and Inhibits Apoptosis in Glioma by Downregulating lncRNA TUG1. Cancer Biother. Radiopharm. 34 (10), 660–665. doi:10.1089/cbr.2019.2830
Lin, H., Zuo, D., He, J., Ji, T., Wang, J., and Jiang, T. (2021). Long Noncoding RNA WEE2-AS1 Plays an Oncogenic Role in Glioblastoma by Functioning as a Molecular Sponge for microRNA-520f-3p. Oncol. Res. Featuring Preclinical Clin. Cancer Ther. 28 (6), 591–603.
Lin, S., and Gregory, R. I. (2015). MicroRNA Biogenesis Pathways in Cancer. Nat. Rev. Cancer 15 (6), 321–333. doi:10.1038/nrc3932
Lin, Y.-H., Guo, L., Yan, F., Dou, Z.-Q., Yu, Q., and Chen, G. (2020). Long Non-coding RNA HOTAIRM1 Promotes Proliferation and Inhibits Apoptosis of Glioma Cells by Regulating the miR-873-5p/ZEB2 axis. Chin. Med. J. 133 (2), 174–182. doi:10.1097/cm9.0000000000000615
Liu, D., Zou, Z., Li, G., Pan, P., and Liang, G. (2020). Long Noncoding RNA NEAT1 Suppresses Proliferation and Promotes Apoptosis of Glioma Cells via Downregulating MiR-92b. Cancer Control 27 (1), 1073274819897977. doi:10.1177/1073274819897977
Liu, J., Jiang, J., Hui, X., Wang, W., Fang, D., and Ding, L. (2018). Mir-758-5p Suppresses Glioblastoma Proliferation, Migration and Invasion by Targeting ZBTB20. Cell Physiol Biochem 48 (5), 2074–2083. doi:10.1159/000492545
Liu, X., Chen, R., and Liu, L. (2019). SP1-DLEU1-miR-4429 Feedback Loop Promotes Cell Proliferative and Anti-apoptotic Abilities in Human Glioblastoma. Biosci. Rep. 39 (12), BSR20190994. doi:10.1042/BSR20190994
Liu, Z., Su, D., Qi, X., and Ma, J. (2018). MiR-500a-5p P-romotes G-lioblastoma C-ell P-roliferation, M-igration and I-nvasion by T-argeting C-hromodomain H-elicase DNA B-inding P-rotein 5. Mol. Med. Rep. 18 (3), 2689–2696. doi:10.3892/mmr.2018.9259
Long, N., Chu, L., Jia, J., Peng, S., Gao, Y., Yang, H., et al. (2020). CircPOSTN/miR-361-5p/TPX2 axis Regulates Cell Growth, Apoptosis and Aerobic Glycolysis in Glioma Cells. Cancer Cel Int 20 (1), 374. doi:10.1186/s12935-020-01454-x
Lu, Y.-F., Cai, X.-L., Li, Z.-Z., Lv, J., Xiang, Y.-a., Chen, J.-J., et al. (2018). LncRNA SNHG16 Functions as an Oncogene by Sponging MiR-4518 and Up-Regulating PRMT5 Expression in Glioma. Cel Physiol Biochem 45 (5), 1975–1985. doi:10.1159/000487974
Luo, W., Li, X., Song, Z., Zhu, X., and Zhao, S. (2019). Long Non-coding RNA AGAP2-AS1 Exerts Oncogenic Properties in Glioblastoma by Epigenetically Silencing TFPI2 through EZH2 and LSD1. Aging 11 (11), 3811–3823. doi:10.18632/aging.102018
Lv, Q.-L., Hu, L., Chen, S.-H., Sun, B., Fu, M.-L., Qin, C.-Z., et al. (2016). A Long Noncoding RNA ZEB1-AS1 Promotes Tumorigenesis and Predicts Poor Prognosis in Glioma. Int. J. Mol. Sci. 17 (9), 1431. doi:10.3390/ijms17091431
Maass, P. G., Luft, F. C., and Bähring, S. (2014). Long Non-coding RNA in Health and Disease. J. Mol. Med. 92 (4), 337–346. doi:10.1007/s00109-014-1131-8
Mahjoubin-Tehran, M., Rezaei, S., Jesmani, A., Birang, N., Morshedi, K., Khanbabaei, H., et al. (2021). New Epigenetic Players in Stroke Pathogenesis: From Non-coding RNAs to Exosomal Non-coding RNAs. Biomed. Pharmacother. 140, 111753. doi:10.1016/j.biopha.2021.111753
Majtnerová, P., and Roušar, T. (2018). An Overview of Apoptosis Assays Detecting DNA Fragmentation. Mol. Biol. Rep. 45 (5), 1469–1478.
Mellai, M., and Schiffer, D. (2007). Apoptosis in Brain Tumors: Prognostic and Therapeutic Considerations. Anticancer Res. 27 (1A), 437–448.
Meng, Q., Yang, B.-Y., Liu, B., Yang, J.-X., and Sun, Y. (2018). Long Non-coding RNA SNHG6 Promotes Glioma Tumorigenesis by Sponging miR-101-3p. Int. J. Biol. Markers 33 (2), 148–155. doi:10.1177/1724600817747524
Meng, X., Li, X., Zhang, P., Wang, J., Zhou, Y., and Chen, M. (2017). Circular RNA: an Emerging Key Player in RNA World. Brief Bioinform 18 (4), 547–557. doi:10.1093/bib/bbw045
Mirzaei, H., and Hamblin, M. R. (2020). Regulation of Glycolysis by Non-coding RNAs in Cancer: Switching on the Warburg Effect. Mol. Ther. - Oncolytics 19, 218–239. doi:10.1016/j.omto.2020.10.003
Mohamed Yusoff, A. (2015). Role of Mitochondrial DNA Mutations in Brain Tumors: A Mini-Review. J. Can. Res. Ther. 11 (3), 535. doi:10.4103/0973-1482.161925
Nam, J. W., Choi, S. W., and You, B. H. (2016). Incredible RNA: Dual Functions of Coding and Noncoding. Mol. Cell 39 (5), 367–374. doi:10.14348/molcells.2016.0039
Nicoloso, M. S., and Calin, G. A. (2008). MicroRNA Involvement in Brain Tumors: from Bench to Bedside. Brain Pathol. 18 (1), 122–129. doi:10.1111/j.1750-3639.2007.00119.x
Nikolaou, M., Pavlopoulou, A., Georgakilas, A. G., and Kyrodimos, E. (2018). The challenge of Drug Resistance in Cancer Treatment: a Current Overview. Clin. Exp. Metastasis 35 (4), 309–318. doi:10.1007/s10585-018-9903-0
O'Brien, J., Hayder, H., Zayed, Y., and Peng, C. (2018). Overview of microRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 9, 402. doi:10.3389/fendo.2018.00402
Pal, R., and Greene, S. (2015). microRNA-10b Is Overexpressed and Critical for Cell Survival and Proliferation in Medulloblastoma. Plos one 10 (9), e0137845. doi:10.1371/journal.pone.0137845
Peng, H., Tan, X., Wang, Y., Dai, L., Liang, G., Guo, J., et al. (2020). Clinical Significance of Ki67 and Circulating Tumor Cells with an Epithelial-Mesenchymal Transition Phenotype in Non-small Cell Lung Cancer. Am. J. Transl Res. 12 (6), 2916–2928.
Pistritto, G., Trisciuoglio, D., Ceci, C., Garufi, A., and D'Orazi, G. (2016). Apoptosis as Anticancer Mechanism: Function and Dysfunction of its Modulators and Targeted Therapeutic Strategies. Aging 8 (4), 603–619. doi:10.18632/aging.100934
Pop, S., Enciu, A. M., Necula, L. G., and Tanase, C. (2018). Long Non‐coding RNA S in Brain Tumours: Focus on Recent Epigenetic Findings in Glioma. J. Cel Mol Med 22 (10), 4597–4610. doi:10.1111/jcmm.13781
Quinn, J. J., and Chang, H. Y. (2016). Unique Features of Long Non-coding RNA Biogenesis and Function. Nat. Rev. Genet. 17 (1), 47–62. doi:10.1038/nrg.2015.10
Ruan, X., Zheng, J., Liu, X., Liu, Y., Liu, L., Ma, J., et al. (2020). lncRNA LINC00665 Stabilized by TAF15 Impeded the Malignant Biological Behaviors of Glioma Cells via STAU1-Mediated mRNA Degradation. Mol. Ther. - Nucleic Acids 20, 823–840. doi:10.1016/j.omtn.2020.05.003
Shabaninejad, Z., Yousefi, F., Movahedpour, A., Ghasemi, Y., Dokanehiifard, S., Rezaei, S., et al. (2019). Electrochemical-based Biosensors for microRNA Detection: Nanotechnology Comes into View. Anal. Biochem. 581, 113349. doi:10.1016/j.ab.2019.113349
Shao, L., He, Q., Liu, Y., Liu, X., Zheng, J., Ma, J., et al. (2019). UPF1 Regulates the Malignant Biological Behaviors of Glioblastoma Cells via Enhancing the Stability of Linc-00313. Cell Death Dis 10 (9), 629. doi:10.1038/s41419-019-1845-1
Shen, H., Liu, B., Xu, J., Zhang, B., Wang, Y., Shi, L., et al. (2021). Circular RNAs: Characteristics, Biogenesis, Mechanisms and Functions in Liver Cancer. J. Hematol. Oncol. 14 (1), 1–16. doi:10.1186/s13045-021-01145-8
So, J. S., Kim, H., and Han, K. S. (2021). Mechanisms of Invasion in Glioblastoma: Extracellular Matrix, Ca2+ Signaling, and Glutamate. Front Cel Neurosci 15, 663092. doi:10.3389/fncel.2021.663092
Song, H., Han, L. M., Gao, Q., and Sun, Y. (2016). Long Non-coding RNA CRNDE Promotes Tumor Growth in Medulloblastoma. Eur. Rev. Med. Pharmacol. Sci. 20 (12), 2588–2597.
Statello, L., Guo, C.-J., Chen, L.-L., and Huarte, M. (2021). Gene Regulation by Long Non-coding RNAs and its Biological Functions. Nat. Rev. Mol. Cel Biol 22 (2), 96–118. doi:10.1038/s41580-020-00315-9
Su, R., Cao, S., Ma, J., Liu, Y., Liu, X., Zheng, J., et al. (2017). Knockdown of SOX2OT Inhibits the Malignant Biological Behaviors of Glioblastoma Stem Cells via Up-Regulating the Expression of miR-194-5p and miR-122. Mol. Cancer 16 (1), 171. doi:10.1186/s12943-017-0737-1
Su, R., Ma, J., Zheng, J., Liu, X., Liu, Y., Ruan, X., et al. (2020). PABPC1-induced Stabilization of BDNF-AS Inhibits Malignant Progression of Glioblastoma Cells through STAU1-Mediated Decay. Cel Death Dis 11 (2), 81–17. doi:10.1038/s41419-020-2267-9
Sun, J., Tian, X., Zhang, J., Huang, Y., Lin, X., Chen, L., et al. (2017). Regulation of Human Glioma Cell Apoptosis and Invasion by miR-152-3p through Targeting DNMT1 and Regulating NF2 : MiR-152-3p Regulate Glioma Cell Apoptosis and Invasion. J. Exp. Clin. Cancer Res. 36 (1), 100–113. doi:10.1186/s13046-017-0567-4
Suzuki, H., and Tsukahara, T. (2014). A View of Pre-mRNA Splicing from RNase R Resistant RNAs. Int. J. Mol. Sci. 15 (6), 9331–9342. doi:10.3390/ijms15069331
Varon, M., Levy, T., Mazor, G., Ben David, H., Marciano, R., Krelin, Y., et al. (2019). The Long Noncoding RNA TP73‐AS1 Promotes Tumorigenicity of Medulloblastoma Cells. Int. J. Cancer 145 (12), 3402–3413. doi:10.1002/ijc.32400
Wang, H., Li, Y., Hou, J., Jiang, C., Li, D., Lu, J., et al. (2018). Biological Activity and Stability of New Peptide Derivative KW-WK from Bovine Lactoferricin. Shipin Kexue/Food Sci. 39 (20), 57–62.
Wang, H. (2020). MicroRNAs and Apoptosis in Colorectal Cancer. Int. J. Mol. Sci. 21 (15), 5353. doi:10.3390/ijms21155353
Wang, J., Qin, C., Zhong, C., Wen, Y., Ke, S., and Liao, B. (2020). Long Non-coding RNA CASC2 Targeting miR-18a Suppresses Glioblastoma Cell Growth, Metastasis and EMT In Vitro and In Vivo. J. Biosciences 45 (1), 1–14. doi:10.1007/s12038-020-00077-8
Wang, J., Liu, X., Yan, C., Liu, J., Wang, S., Hong, Y., et al. (2017). LEF1-AS1, a Long Non-coding RNA, Promotes Malignancy in Glioblastoma. Onco Targets Ther. 10, 4251–4260. doi:10.2147/ott.s130365
Wang, J., Yang, S., Ji, Q., Li, Q., Zhou, F., Li, Y., et al. (2020). Long Non-coding RNA EPIC1 Promotes Cell Proliferation and Motility and Drug Resistance in Glioma. Mol. Ther. - Oncolytics 17, 130–137. doi:10.1016/j.omto.2020.03.011
Wang, R., Zhang, S., Chen, X., Li, N., Li, J., Jia, R., et al. (2018). CircNT5E Acts as a Sponge of miR-422a to Promote Glioblastoma Tumorigenesis. Cancer Res. 78 (17), 4812–4825. doi:10.1158/0008-5472.can-18-0532
Wang, X., Xu, D., Pei, X., Zhang, Y., Zhang, Y., Gu, Y., et al. (2020). CircSKA3 Modulates FOXM1 to Facilitate Cell Proliferation, Migration, and Invasion while Confine Apoptosis in Medulloblastoma via miR-383-5p. Cancer Manag. Res. 12, 13415–13426. doi:10.2147/cmar.s272753
Weller, M., Wick, W., Aldape, K., Brada, M., Berger, M., Pfister, S. M., et al. (2015). Glioma. Nat. Rev. Dis. Primers 1 (1), 15017–15018. doi:10.1038/nrdp.2015.17
Werner, T. V., Hart, M., Nickels, R., Kim, Y.-J., Menger, M. D., Bohle, R. M., et al. (2017). MiR-34a-3p Alters Proliferation and Apoptosis of Meningioma Cells In Vitro and Is Directly Targeting SMAD4, FRAT1 and BCL2. Aging 9 (3), 932–954. doi:10.18632/aging.101201
Wu, J., Wang, N., Yang, Y., Jiang, G., Mu, Q., Zhan, H., et al. (2021). LINC01152 Upregulates MAML2 Expression to Modulate the Progression of Glioblastoma Multiforme via Notch Signaling Pathway. Cel Death Dis. 12 (1), 1–14. doi:10.1038/s41419-021-04106-8
Wu, N., Lin, X., Zhao, X., Zheng, L., Xiao, L., Liu, J., et al. (2013). MiR-125b Acts as an Oncogene in Glioblastoma Cells and Inhibits Cell Apoptosis through P53 and p38MAPK-independent Pathways. Br. J. Cancer 109 (11), 2853–2863. doi:10.1038/bjc.2013.672
Wu, Z., and Lin, Y. (2019). Long Noncoding RNA LINC00515 Promotes Cell Proliferation and Inhibits Apoptosis by Sponging miR-16 and Activating PRMT5 Expression in Human Glioma. Onco Targets Ther. 12, 2595–2604. doi:10.2147/ott.s198087
Xin, J., Zhang, X. Y., Sun, D. K., Tian, L. Q., and Xu, P. (2019). Up-regulated Circular RNA Hsa_circ_0067934 Contributes to Glioblastoma Progression through Activating PI3K-AKT Pathway. Eur. Rev. Med. Pharmacol. Sci. 23 (8), 3447–3454. doi:10.26355/eurrev_201904_17709
Xiong, Z., Zhou, C., Wang, L., Zhu, R., Zhong, L., Wan, D., et al. (2019). Circular RNA SMO Sponges miR-338-3p to Promote the Growth of Glioma by Enhancing the Expression of SMO. Aging 11 (24), 12345–12360. doi:10.18632/aging.102576
Xu, C. H., Xiao, L. M., Liu, Y., Chen, L. K., Zheng, S. Y., Zeng, E. M., et al. (2019). The lncRNA HOXA11-AS Promotes Glioma Cell Growth and Metastasis by Targeting miR-130a-5p/HMGB2. Eur. Rev. Med. Pharmacol. Sci. 23 (1), 241–252. doi:10.26355/eurrev_201901_16770
Xu, H., Zhang, Y., Qi, L., Ding, L., Jiang, H., and Yu, H. (2018). NFIX Circular RNA Promotes Glioma Progression by Regulating miR-34a-5p via Notch Signaling Pathway. Front. Mol. Neurosci. 11, 225. doi:10.3389/fnmol.2018.00225
Xu, L. M., Chen, L., Li, F., Zhang, R., Li, Z. Y., Chen, F. F., et al. (2016). Over-expression of the Long Non-coding RNA HOTTIP Inhibits Glioma Cell Growth by BRE. J. Exp. Clin. Cancer Res. 35 (1), 162. doi:10.1186/s13046-016-0431-y
Xu, N., Liu, B., Lian, C., Doycheva, D. M., Fu, Z., Liu, Y., et al. (2018). Long Noncoding RNA AC003092.1 Promotes Temozolomide Chemosensitivity through miR-195/TFPI-2 Signaling Modulation in Glioblastoma. Cel Death Dis 9 (12), 1139. doi:10.1038/s41419-018-1183-8
Xu, Q.-F., Pan, Y.-W., Li, L.-C., Zhou, Z., Huang, Q.-L., Pang, J. C.-s., et al. (2014). MiR-22 Is Frequently Downregulated in Medulloblastomas and Inhibits Cell Proliferationviathe Novel TargetPAPST1. Brain Pathol. 24 (6), 568–583. doi:10.1111/bpa.12136
Xu, S., Tang, L., Li, X., Fan, F., and Liu, Z. (2020). Immunotherapy for Glioma: Current Management and Future Application. Cancer Lett. 476, 1–12. doi:10.1016/j.canlet.2020.02.002
Xu, W., Hu, G.-Q., Da Costa, C., Tang, J.-H., Li, Q.-R., Du, L., et al. (2019). Long Noncoding RNA UBE2R2-AS1 Promotes Glioma Cell Apoptosis via Targeting the miR-877-3p/TLR4 axis. Onco Targets Ther. 12, 3467–3480. doi:10.2147/ott.s201732
Xu, X., Lai, Y., and Hua, Z. C. (2019). Apoptosis and Apoptotic Body: Disease Message and Therapeutic Target Potentials. Biosci. Rep. 39 (1), BSR20180992. doi:10.1042/BSR20180992
Yang, A., Wang, H., and Yang, X. (2017). Long Non-coding RNA PVT1 Indicates a Poor Prognosis of Glioma and Promotes Cell Proliferation and Invasion via Target EZH2. Biosci. Rep. 37 (6), BSR20170871. doi:10.1042/BSR20170871
Yang, J. X., Rastetter, R. H., and Wilhelm, D. (2016). Non-coding RNAs: An Introduction. Adv. Exp. Med. Biol. 886, 13–32. doi:10.1007/978-94-017-7417-8_2
Yang, T. Q., Lu, X. J., Wu, T. F., Ding, D. D., Zhao, Z. H., Chen, G. L., et al. (2014). Micro RNA ‐16 Inhibits Glioma Cell Growth and Invasion through Suppression of BCL 2 and the Nuclear factor‐κB1/MMP 9 Signaling Pathway. Cancer Sci. 105 (3), 265–271. doi:10.1111/cas.12351
Yin, F., Zhang, J. N., Wang, S. W., Zhou, C. H., Zhao, M. M., Fan, W. H., et al. (2015). MiR-125a-3p Regulates Glioma Apoptosis and Invasion by Regulating Nrg1. PloS one 10 (1), e0116759. doi:10.1371/journal.pone.0116759
Yuan, D. H., Zhao, J., and Shao, G. F. (2019). Circular RNA TTBK2 Promotes the Development of Human Glioma Cells via miR-520b/EZH2 axis. Eur. Rev. Med. Pharmacol. Sci. 23 (24), 10886–10898. doi:10.26355/eurrev_201912_19792
Zhang, C., Ma, L., Niu, Y., Wang, Z., Xu, X., Li, Y., et al. (2020). Circular RNA in Lung Cancer Research: Biogenesis, Functions, and Roles. Int. J. Biol. Sci. 16 (5), 803–814. doi:10.7150/ijbs.39212
Zhang, J., Chen, G., Gao, Y., and Liang, H. (2020). HOTAIR/miR‐125 axis‐mediated Hexokinase 2 Expression Promotes Chemoresistance in Human Glioblastoma. J. Cel Mol Med 24 (10), 5707–5717. doi:10.1111/jcmm.15233
Zhang, J., Li, N., Fu, J., and Zhou, W. (2020). Long Noncoding RNA HOTAIR Promotes Medulloblastoma Growth, Migration and Invasion by Sponging miR-1/miR-206 and Targeting YY1. Biomed. Pharmacother. 124, 109887. doi:10.1016/j.biopha.2020.109887
Zhang, P., Wu, W., Chen, Q., and Chen, M. (2019). Non-coding RNAs and Their Integrated Networks. J. Integr. Bioinform 16 (3). doi:10.1515/jib-2019-0027
Zhang, S., Lai, N., Liao, K., Sun, J., and Lin, Y. (2015). MicroRNA-210 Regulates Cell Proliferation and Apoptosis by Targeting Regulator of Differentiation 1 in Glioblastoma Cells. fn 3 (3), 236–244. doi:10.5114/fn.2015.54424
Zhang, X., Niu, W., Mu, M., Hu, S., and Niu, C. (2020). Long Non-coding RNA LPP-AS2 Promotes Glioma Tumorigenesis via miR-7-5p/EGFR/PI3K/AKT/c-MYC Feedback Loop. J. Exp. Clin. Cancer Res. 39 (1), 196. doi:10.1186/s13046-020-01695-8
Zhang, X., Yang, H., Zhao, L., Li, G., and Duan, Y. (2019). Circular RNA PRKCI Promotes Glioma Cell Progression by Inhibiting microRNA-545. Cel Death Dis 10 (8), 616. doi:10.1038/s41419-019-1863-z
Zhang, Z.-Y., Zhu, B., Zhao, X.-W., Zhan, Y.-B., Bao, J.-J., Zhou, J.-Q., et al. (2017). Regulation of UHRF1 by microRNA-378 Modulates Medulloblastoma Cell Proliferation and Apoptosis. Oncol. Rep. 38 (5), 3078–3084. doi:10.3892/or.2017.5939
Zhao, L., Chen, T., Tang, X., Li, S., Liang, R., and Wang, Y. (2020). Medulloblastoma Malignant Biological Behaviors Are Associated with HOTAIR/miR-483-3p/CDK4 axis. Ann. Transl Med. 8 (14), 886. doi:10.21037/atm-20-5006
Zhao, X., Wang, P., Liu, J., Zheng, J., Liu, Y., Chen, J., et al. (2015). Gas5 Exerts Tumor-Suppressive Functions in Human Glioma Cells by Targeting miR-222. Mol. Ther. 23 (12), 1899–1911. doi:10.1038/mt.2015.170
Zheng, J., Liu, X., Xue, Y., Gong, W., Ma, J., Xi, Z., et al. (2017). TTBK2 Circular RNA Promotes Glioma Malignancy by Regulating miR-217/HNF1β/Derlin-1 Pathway. J. Hematol. Oncol. 10 (1), 52–19. doi:10.1186/s13045-017-0422-2
Zheng, K., Xie, H., Wu, W., Wen, X., Zeng, Z., and Shi, Y. (2021). CircRNA PIP5K1A Promotes the Progression of Glioma through Upregulation of the TCF12/PI3K/AKT Pathway by Sponging miR-515-5p. Cancer Cel Int 21 (1), 27–13. doi:10.1186/s12935-020-01699-6
Zhi, F., Wang, Q., Deng, D., Shao, N., Wang, R., Xue, L., et al. (2014). MiR-181b-5p Downregulates NOVA1 to Suppress Proliferation, Migration and Invasion and Promote Apoptosis in Astrocytoma. PLoS One 9 (10), e109124. doi:10.1371/journal.pone.0109124
Zhi, F., Zhou, G., Shao, N., Xia, X., Shi, Y., Wang, Q., et al. (2013). miR-106a-5p Inhibits the Proliferation and Migration of Astrocytoma Cells and Promotes Apoptosis by Targeting FASTK. PLoS One 8 (8), e72390. doi:10.1371/journal.pone.0072390
Zhou, W-Y., Cai, Z-R., Liu, J., Wang, D-S., Ju, H-Q., and Xu, R-H. (2020). Circular RNA: Metabolism, Functions and Interactions with Proteins. Mol. Cancer 19 (1), 1–19. doi:10.1186/s12943-020-01286-3
Zhu, C., Mao, X., and Zhao, H. (2020). The Circ_VCAN with Radioresistance Contributes to the Carcinogenesis of Glioma by Regulating microRNA-1183. Medicine (Baltimore) 99 (8), e19171. doi:10.1097/MD.0000000000019171
Keywords: brain tumors, non-coding RNAs, apoptosis, microRNA, long non-coding (lnc) RNA
Citation: Tamtaji OR, Derakhshan M, Rashidi Noshabad FZ, Razaviyan J, Hadavi R, Jafarpour H, Jafari A, Rajabi A, Hamblin MR, Mahabady MK, Taghizadieh M and Mirzaei H (2022) Non-Coding RNAs and Brain Tumors: Insights Into Their Roles in Apoptosis. Front. Cell Dev. Biol. 9:792185. doi: 10.3389/fcell.2021.792185
Received: 09 October 2021; Accepted: 08 December 2021;
Published: 17 January 2022.
Edited by:
Jing Zhang, Shanghai Jiao Tong University, ChinaReviewed by:
Youliang Wang, Beijing Institute of Technology, ChinaGiovanni Nigita, The Ohio State University, United States
Copyright © 2022 Tamtaji, Derakhshan, Rashidi Noshabad, Razaviyan, Hadavi, Jafarpour, Jafari, Rajabi, Hamblin, Mahabady, Taghizadieh and Mirzaei. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mahmood Khaksary Mahabady, bWtoYWtzYXJ5bWFoYWJhZHlAZ21haWwuY29t; Mohammad Taghizadieh, TW9oYW1tYWRUYWdoaXphZGllaEBnbWFpbC5jb20=; Hamed Mirzaei, aC5taXJ6YWVpMjAwMkBnbWFpbC5jb20=
 Omid Reza Tamtaji1
Omid Reza Tamtaji1 Hamed Mirzaei
Hamed Mirzaei