- 1Laboratory of Drosophila Neural Development, Centre for DNA Fingerprinting and Diagnostics (CDFD), Hyderabad, India
- 2Graduate Studies, Manipal Academy of Higher Education, Manipal, India
Hox genes have been known for specifying the anterior-posterior axis (AP) in bilaterian body plans. Studies in vertebrates have shown their importance in developing region-specific neural circuitry and diversifying motor neuron pools. In Drosophila, they are instrumental for segment-specific neurogenesis and myogenesis early in development. Their robust expression in differentiated neurons implied their role in assembling region-specific neuromuscular networks. In the last decade, studies in Drosophila have unequivocally established that Hox genes go beyond their conventional functions of generating cellular diversity along the AP axis of the developing central nervous system. These roles range from establishing and maintaining the neuromuscular networks to controlling their function by regulating the motor neuron morphology and neurophysiology, thereby directly impacting the behavior. Here we summarize the limited knowledge on the role of Drosophila Hox genes in the assembly of region-specific neuromuscular networks and their effect on associated behavior.
Introduction
Feeding, locomotion, and reproduction are some of the most fundamental behaviors exhibited by bilaterians. Regional specialization of the muscles, as well as the central nervous system (CNS) along the anterior-posterior (AP) axis, is a prerequisite for the successful and reproducible execution of these behaviors (Philippidou and Dasen, 2013). Role of Hox genes in assembling region-specific neural circuitry and MN diversification has been examined in vertebrates (Dasen et al., 2005; Dasen et al., 2008; di Sanguinetto et al., 2008; Philippidou and Dasen, 2013) and to some extent in insects (Dixit et al., 2008; Dutta et al., 2010; Baek et al., 2013; Garaulet et al., 2014; Enriquez et al., 2015; Picao-Osorio et al., 2015; Friedrich et al., 2016; Issa et al., 2019; Garaulet et al., 2020). Hox genes are also involved in the specification, survival, and functioning of other neuronal cell types (van den Akker et al., 1999; Pattyn et al., 2003; Gaufo et al., 2004; Holstege et al., 2008; Miguez et al., 2012; Huber et al., 2012; Bussell et al., 2014; Baek et al., 2019); however, such reports (for specific cell types) are limited. The MNs are central players in the functioning of neuromuscular networks. Their role in fine tuning muscle control is important for behavioral execution. The loss of MNs or perturbation of their function owing to a disease affects the behavior and leads to progressive muscle wasting. Therefore, studying their specification and functioning will give insights into the molecular basis of complex behaviors and disease.
In Drosophila, Hox genes are known to establish segment-specific patterns of myogenesis and neurogenesis (Michelson, 1994; Technau et al., 2014). However, the molecular basis of how Hox genes play a role in the specification and regional adjustment of the motor neuron (MN) networks is just beginning to be understood. Therefore much remains to be learned about their role in the assembly, maintenance, and functioning of segment-specific neuromuscular networks. In this regard, Drosophila as a model organism offers many unique advantages over other models (Hales et al., 2015; Schlegel et al., 2017; Yamaguchi and Yoshida, 2018). These advantages are a short life cycle, a fully sequenced genome, less redundancy than vertebrates, and a wide array of molecular genetic tools. In its short life cycle, Drosophila undergoes remarkable morphological and behavioral changes with different modes of feeding and locomotion for different stages. In just 10 days, it progresses from a static, non-feeding embryonic stage to a crawling and feeding larva, followed by an immobile and non-feeding pupal stage, eventually eclosing as a sexually active adult with an entirely different mode of navigation, locomotion, and foraging. It has a wide repertoire of simple, well-established behaviors (Nichols et al., 2012; Neckameyer and Bhatt, 2016), and many of the neuromuscular modules executing these behaviors are simple and well investigated. Compared to the vertebrates, Drosophila has a relatively less complex nervous system and musculature, and a fantastic array of molecular tools for reproducibly making subtle genetic manipulations in a cell-specific manner. The effect of these manipulations can be assayed in live and behaving animals (Korona et al., 2017; Martín and Alcorta, 2017), which is a tremendous advantage in correlating a gene to behavior.
In this review, we summarize existing Drosophila literature elucidating the role of Hox genes in the assembly and functioning of region-specific muscle-MN connections and their contribution in executing associated behaviors.
Role of Hox Genes in the Specification of Anterior-Posterior Axis in Drosophila Central Nervous System
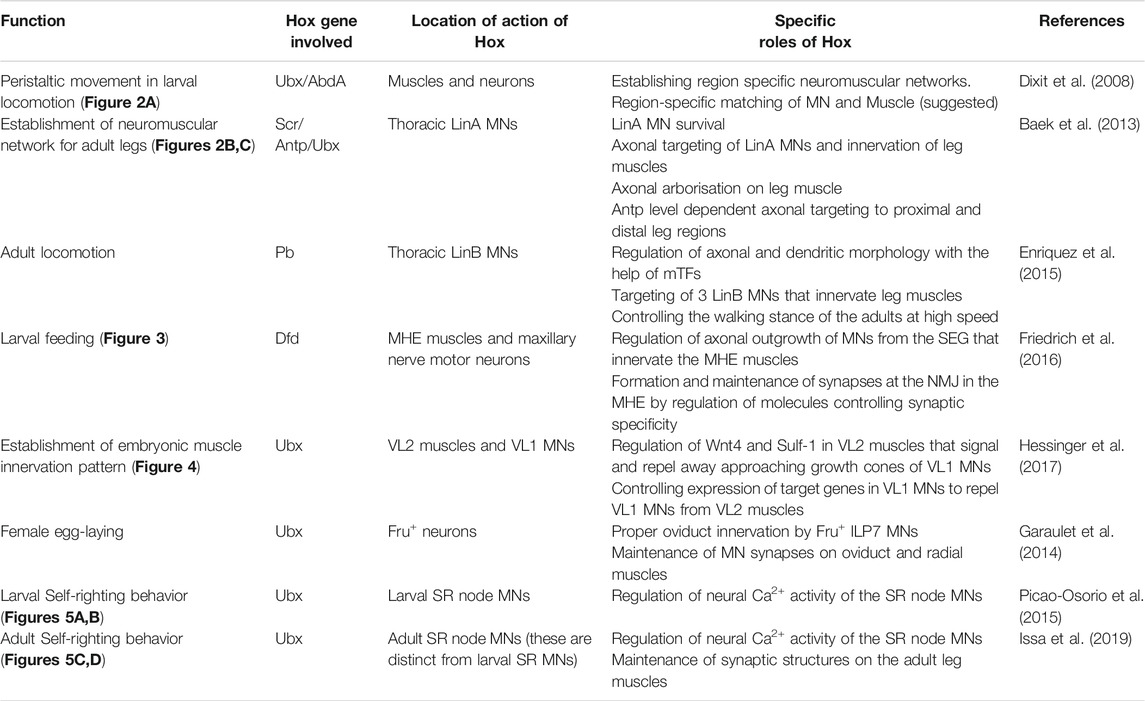
Hox genes are a family of homeodomain (HD) containing transcription factors (TFs), which play an important role in determining the anterior-posterior (AP) axis of bilaterian organisms (Hart et al., 1985; Regulski et al., 1985; Akam, 1989; Carroll, 1995). They are known to specify the AP axis by differentially regulating their downstream target genes with the help of TALE-HD containing cofactors Pbx/Exdradenticle (Exd) and Meis/Homothorax (Hth) (Mann and Chan, 1996; Mann and Affolter, 1998; Moens and Selleri, 2006; Merabet et al., 2007; Lelli et al., 2011; Saadaoui et al., 2011; Hudry et al., 2012). Hox genes execute these functions by giving the segments where they are expressed a very distinct identity, translating into divergent morphologies/properties along the AP axis of the body (including epidermal structure, CNS, and musculature). In Drosophila, there are eight Hox genes (compared to 39 Hox genes in vertebrates) which are organized into two complexes-the Antennapedia Complex (Antp-C) (Kaufman et al., 1990) [comprising the genes labial (lab), proboscipedia (pb), Deformed (Dfd), Sex combs reduced (Scr), Antennapedia (Antp)], and the Bithorax Complex (BX-C) (Sánchez-Herrero et al., 1985; Tiong et al., 1985; Maeda and Karch, 2006) [consisting of the genes Ultrabithorax (Ubx), abdominal-A (abd-A) and Abdominal-B (Abd-B)]. Drosophila CNS consists of the brain and segmented ventral nerve cord (VNC). Hox genes pattern the VNC, which is the Drosophila equivalent of the vertebrate hindbrain and spinal cord. The embryonic VNC specified by Hox factors can be broadly divided into five regions, namely: supra-esophageal ganglia (SPG) expressing pb and labial; sub-esophageal ganglia (SEG) [composed of maxillary (Mx), mandibular (Mn), and labial segments (Lb)] expressing Hox genes Dfd, Scr and Antp; thoracic ganglia (T1-T3 segments) expressing Antp and Ubx; abdominal ganglia (A1-A7 segments) expressing Ubx, abd-A, and Abd-B; and the terminal region (A8-A10 segments) expressing Abd-B (Hirth et al., 1998; Kuert et al., 2014) (Figure 1). The expression of pb has also been reported in other segments (SEG to A9) of VNC (Baek et al., 2013; Enriquez et al., 2015; Hirth et al., 1998).
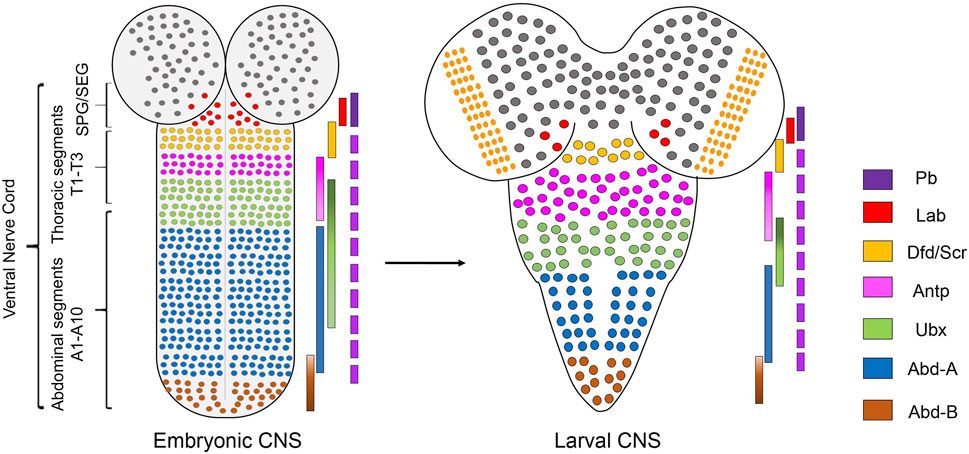
FIGURE 1. Expression of Hox genes in Drosophila CNS. Schematics of embryonic (stage 14) and second instar larval CNS show Hox genes’ expression pattern in different regions along the AP-axis. Drosophila CNS has a brain and ventral nerve cord (VNC). VNC is divided into SPG/SEG region, thoracic segments (T1-T3), abdominal (A1-A7) segments, and terminal (A8-A10) segments. The precise extent of Hox gene expression in these regions is shown by overlapping color-coded bars. Pb is expressed in all the segments from SEG to A9.
The neurogenesis in Drosophila happens in two phases, embryonic and larval, separated by a period of mitotic quiescence for the neural stem cells (called neuroblast-NB), which are the progenitors and generate all the neurons and glial cells of the CNS (Homem and Knoblich, 2012). In embryonic stages (stages 9–11), NBs delaminate from the neuroectoderm in each segment (Hartenstein and Wodarz, 2013). Five such successive waves of delamination generate 30 NBs per hemisegment of the embryo (Truman and Bate, 1988; Doe, 1992; Hartenstein and Wodarz, 2013). This blueprint of the CNS, when superimposed with the spatial genes [responsible for determining the AP and DV (Dorso-Ventral) (Skeath, 1999) axis and segment polarity genes (Bhat, 1999)], gives the NBs their specific positional identity (Schmid et al., 1999; Truman et al., 2004). This spatial identity of the NBs, in collaboration with the sequentially expressing temporal series TFs (Doe, 2017) expressed during embryogenesis, results in the generation of a segment-specific variety of cell types and cell numbers in the embryo. The transition of these temporal series TFs (Hunchback>Kruppel>Pdm>Castor >Grh) is intricately coupled to the NB cell cycle, which precisely times the expression of these factors and further contributes to specific cell type generation (Isshiki et al., 2001). The embryonic phase generates neurons required for larval CNS and eventually contributes to 10% of the adult neurons, while postembryonic neurogenesis contributes to the remaining 90% of the adult neurons (Homem and Knoblich, 2012). Hox genes contribute to the generation of the cellular variety along the AP axis in both embryonic and post-embryonic phase of neurogenesis by regulating fate specification, quiescence, proliferation, differentiation and apoptosis of NBs and their progeny (Prokop and Technau, 1994; Prokop et al., 1998; Bello et al., 2003; Miguel-Aliaga and Thor, 2004; Berger et al., 2005a; Berger et al., 2005b; Rogulja-Ortmann et al., 2008; Tsuji et al., 2008; Kannan et al., 2010; Karlsson et al., 2010; Suska et al., 2011; Kuert et al., 2012; Baek et al., 2013; Birkholz et al., 2013a; Birkholz et al., 2013b; Estacio-Gómez et al., 2013; Baumgardt et al., 2014; Kuert et al., 2014; Arya et al., 2015; Becker et al., 2016; Khandelwal et al., 2017; Monedero Cobeta et al., 2017; Yaghmaeian Salmani et al., 2018; Bahrampour et al., 2019; Ghosh et al., 2019; Bakshi et al., 2020).
In the CNS, Hox genes are expressed in NBs in early embryonic stages, but their expression from the NBs is largely excluded thereafter. However, Hox genes continue to express in the neurons as they differentiate, project axons/dendrites, and form synaptic connections (Hirth et al., 1998) in embryonic and postembryonic stages. This led to the suggestion that Hox genes may have a role in the assembly and functioning of neuromuscular networks (Hirth et al., 1998; Dixit et al., 2008). More so, since the regionally distinct muscle patterns are known to be established by Hox genes early in development (Michelson, 1994). However, barring their role in regulating neuronal differentiation and apoptosis (Hirth et al., 1998; Miguel-Aliaga and Thor, 2004; Rogulja-Ortmann et al., 2008; Karlsson et al., 2010; Suska et al., 2011; Baek et al., 2013; Estacio-Gómez et al., 2013), the utility of the sustained expression of Hox genes in neurons had not been entirely clear. Therefore, the role of Hox genes in the regional specialization of the MNs, and their contribution to the assembly of functional neuromuscular networks (along the AP axis) remained unaddressed for a long time. Here, we focus on the reported roles of Drosophila Hox genes in the establishment of functional neuromuscular networks and behavior (summarized in Table 1). We also attempt to identify some common themes in the context of neuromuscular network assembly and functioning, which are independent of the conventional role of Hox genes in AP axis determination.
Role of Hox Genes in Locomotion
One of the first reports implicating the role of Hox genes in assembling the segment-specific neuromuscular networks in Drosophila was by Dixit et al. (2008) from Bate and Vijay Raghavan groups in Cambridge and Bangalore. This report established the role of Hox genes in regulating segmental peristaltic movements in larval locomotion (Dixit et al., 2008). This work, in many ways, laid the foundation for exploring the molecular basis of the genetic control that results in equivalent cells of CNS (along the AP axis) to form regionally specialized neuromuscular networks.
This study showed that the thoracic and abdominal segments of the larval body have distinct movement patterns during larval peristalsis (Figure 2A). The experiments suggested that the abdominal peristaltic movements critically relied on BX-C, specifically Ubx and abd-A (Figure 2B). The mutants for Ubx or abd-A were used to show that either of these genes was necessary for developing the neuromuscular networks coordinating these movements. Interestingly, the ubiquitous expression of either of the two genes was also sufficient for transforming the anterior segments to posterior identity (Figure 2D). This resulted in morphological transformation of the epidermal denticle belts (which had been known from earlier studies) and the anterior segments gaining the peristaltic activity like abdominal ones. However, this did not happen in the case of Antp overexpression (Figure 2C). In the absence of anatomical data, the study speculated that these movements rely on region-specific muscle architecture and their precise innervation by cognate MNs. In agreement with this, the expression of Ubx or AbdA in muscles was not sufficient for anterior segments to show a peristaltic pattern similar to posterior segments. This observation supported the idea that in the case of Hox-dependent segmental transformation, the MNs and the entire neural circuitry are reorganized to match the transformed pattern of muscle. This study also suggested that a one-to-one match in the identity of the muscle with that of the underlying neural circuitry is required for the proper execution of abdominal peristaltic movements.
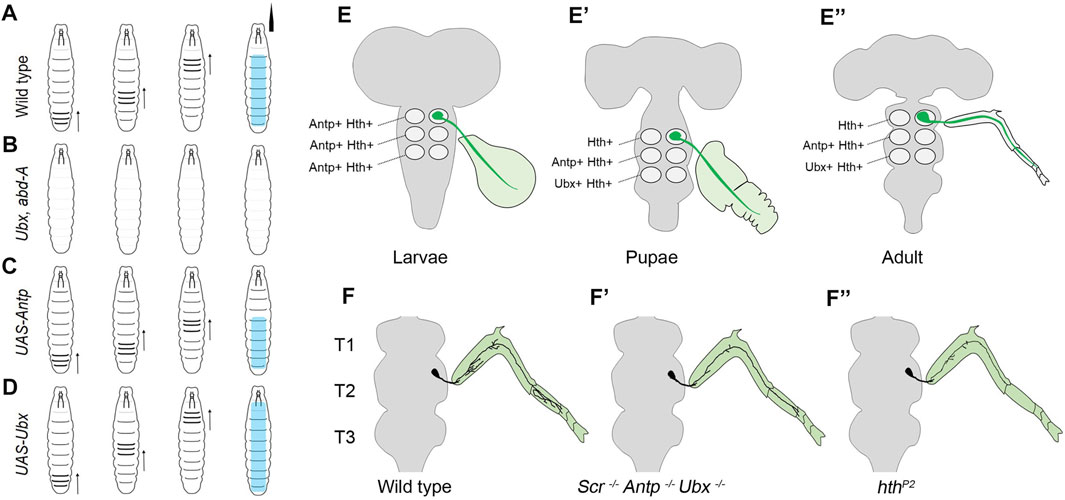
FIGURE 2. Summary of the role of Hox genes in larval peristalsis and leg innervating MNs. (A) Schematic showing abdominal peristaltic movement in wild-type larva. (B) Shows that abdominal peristaltic movements are lost in Ubx, abd-A double mutant. (C) Shows that Antp overexpression transforms anterior segments but transformed segments do not show peristaltic movements. The abdominal peristaltic movements are unaffected. (D) Shows that in the case of Ubx overexpression, thoracic and more anterior segments get transformed and gain abdominal peristaltic movements. The direction of peristalsis is shown with an arrow, and its extent is shown in cyan color. (E–E”) Shows that thoracic MNs (in green) innervate primordial leg tissue in larval and pupal stages and adult leg muscles. Also shown is the change in the expression code of Antp, Ubx, and Hth in LinA MNs of different thoracic segments (T1-T3) across different developmental stages. (F–F”) Shows the wild-type arborization pattern of thoracic MNs innervating to the adult leg muscles. This axonal arborization is affected in triple Hox triple mutants (Scr−, Antp−, Ubx−) and hthP2 mutants. This suggests that Hox/Hth is required for the survival, targeting and morphology of MNs innervating to the adult leg.
Subsequently, two studies comprehensively addressed the developmental role of Hox genes in survival, targeting, and morphology of thoracic MNs, which innervate the leg muscles responsible for adult locomotion (Baek et al., 2013; Enriquez et al., 2015). These studies were built upon previous work which had shown that 50 MNs innervating the Drosophila adult leg muscles are generated by 11 NBs located in each thoracic hemisegment (Baek and Mann, 2009; Brierley et al., 2012). Two-third of these 50 MNs are generated by two NB lineages, LinA (or Lin 15), which generate 28 MNs, and LinB (or Lin 24), which generate seven MNs (Truman et al., 2004; Baek and Mann, 2009). These studies had characterized stereotypic axonal and dendritic morphologies of all the 50 MNs (generated by LinA and LinB) at the single-cell level, down to their synaptic innervations of the 14 leg muscles on the four segments of the adult leg (Baek and Mann, 2009; Brierley et al., 2012).
The first study by Baek et al. (2013), from Mann’s group in New York focussed on LinA MNs and showed that Hox genes (Scr, Antp, and Ubx) and their TALE-HD containing cofactor Hth are required for the survival of the MNs in all the three thoracic segments. They found that all newly born thoracic MNs express Antp and Hth in all the three thoracic segments in larval stages (Figures 2E–E'). As the development progressed, this expression code transformed from being Antp+/Hth+ in all the segments to Hth+ in T1, Antp+/Hth+ in T2, and Ubx+/Hth+ in the T3 segment in the late pupal stage and adults (Figures 2E–E'). This change in TF code is suggested to specialize the MNs innervating the adult legs to execute their segment-specific functions (Szebenyi, 1969; Kaplan and Trout, 1974; Dawkins and Dawkins, 1976; Trimarchi and Schneiderman, 1993; Dickinson et al., 2000). Exd was found to express in all the segments in all the stages. Subsequent clonal analysis (Wu and Luo, 2006) done with Hox triple (Scr−, Antp−, Ubx−) and hth mutants indicated that expression of both Hox and hth genes is required autonomously within the thoracic lineages for survival, targeting, and morphology of the adult MNs (Figures 2F–F') (Baek et al., 2013). Since Antp was the only Hox factor to be expressed initially in NBs, Antp single mutant was tested. Curiously, this mutant recapitulated most of the phenotype exhibited by Hox triple mutant, except the death of MNs in the T3 segment, which was attributed to Ubx and Antp redundancy in this segment. Subsequently, the death of the MNs was blocked by the expression of p35, a viral protein commonly used to block apoptosis (LaCount et al., 2000). In this case, the surviving MNs in Hox triple (Scr−, Antp−, Ubx−) or hth mutant backgrounds targeted roughly to the same region along the proximal-distal (PD) axis of the adult leg segments, with terminal arborization defects. This suggested that Hox (and hth) genes are not needed by LinA progeny to assume the thorax-specific lineage identity or the MN fate. However, they are required for the appropriate specification of the finer morphological features of these MNs necessary for the functional muscle innervation (Figures 2F–F'). This was similar to what is known for the role of Hox genes in vertebrates MN specification, wherein MN identity is established independent of Hox genes (Jessell, 2000; Dasen et al., 2005).
The study also provided a novel alternative mechanism to diversify cell fate within a given lineage by modulating the expression level of Hox factor Antp. Usually, NB progeny rely on temporal series TFs for fate diversification (Doe, 2017). Baek et al. (2013) observed that within the same lineage, Antp is expressed at high levels in late-born MNs and low levels in early-born MNs. This variation in the Antp gene expression levels in MNs was found to have an instructional role in their axonal targeting. It was observed that high Antp expressing late-born MNs targeted the distal region, and low Antp expressing early-born MNs targeted the proximal regions of the adult leg. Expectedly, this pattern could be reversed by overexpression or the knockdown of Antp. Though, in the null allele of Antp, both distal and proximal targeting of MN was affected with no specific bias, indicating that low level gave a distinct phenotype from the absence of Antp. This variation in the expression levels of Antp had cell-autonomous consequences in MN innervation and did not show any defect in the leg muscles of the adult fly. This was in contrast to an earlier work by Dutta et al. from VijayRaghavan and Rodriguez groups at Bangalore, where Hox dysregulation in MNs resulted in muscle development defects (Dutta et al., 2010). The experiment in this study shows that knockdown of Ubx in adult MNs resulted in modest reduction and developmental deformity in adult leg muscles (Dutta et al., 2010). On the other hand, Ubx overexpression in the MNs innervating thoracic dorsolateral muscle (DLM) of adults caused a dramatic reduction in the number of DLM fibers (Dutta et al., 2010). These observations implied an active communication between the adult thoracic MNs and their muscle targets. Dutta et al. (2010) also suggested that Hox expression needs to be tightly controlled within a narrow range for the assembly of functional neuromuscular network. It is to be noted that this study relied on a chronic knockdown/overexpression of Ubx in the MNs, while Baek et al. (2013) used temporally controlled overexpression or knockdown.
The requirement of Hox genes in determining the morphology of thoracic MNs was followed up by Enriquez et al. from Mann’s group in New York (Enriquez et al., 2015). This work identified the role of Hox gene proboscipedia (pb) in determining the morphological characteristics of three thoracic MNs (Enriquez et al., 2015). The proboscipedia (pb) expresses from the supraesophageal region to the A9 segment in embryonic and larval CNS (Hirth et al., 1998; Baek et al., 2013), but its role in neurogenesis had not been tested. Previous studies had suggested that the morphology of MNs arising out of thoracic LinA and LinB lineages were under precise genetic control, which had a bearing on their function (Baek and Mann, 2009; Brierley et al., 2012). However, the genetic determinants regulating the individual neuronal morphology for these (or any neuron) were not known. LinB (or Lin 24), which produces only seven MNs in each hemisegment (of 3 thoracic segments) were an attractive system to address this problem owing to few but well-characterized MNs in this lineage (Baek and Mann, 2009; Brierley et al., 2012). Enriquez et al. screened 230 antibodies against different TFs for their expression in larval LinB lineage [marked by GFP using MARCM (Wu and Luo, 2006)]. They identified six TFs whose combinatorial expression was sufficient to uniquely classify the seven MNs of LinB lineage. These factors were Empty spiracle (Ems), the Zinc finger homeodomain factors 1 and 2 (Zfh1 and Zfh2), the Hox TF Proboscipedia (Pb), the Pax6 ortholog Twin of Eyeless (Toy), and Prospero (Pros). Further, they observed that the TFs combinations observed in each of the seven MNs were not observed in any other neuron of the CNS. After that, they used lineage tracing experiments to correlate larval LinB MNs (with unique TF code) to their adult counterparts, each of which corresponds to distinct morphology and muscle innervation. This supported the idea that the characteristic expression of these six TFs probably results in distinct axonal and dendritic morphologies of these MNs. Consequently, these factors were called morphology TFs (mTFs).
Next, a clever combination of MARCM (Wu and Luo, 2006) with the Flybow technique (Hadjieconomou et al., 2011) was used to mark adult MNs where they removed or overexpressed pb to analyze its effect in MNs of LinB. The Hox gene pb was shown to be essential for the morphological identity of 3 out 7 MNs of LinB. Interestingly, loss or overexpression of pb did not affect the expression of the other five mTFs, which was in contrast to what is known for the temporal series TFs (Doe, 2017), which play a crucial role in generating neuronal diversity. When pb mutant LinB NB was analyzed, it was observed that the number of MNs generated in the lineage was unaffected. These MNs did not lose or change their identity; they remained glutaminergic, and their axons targeted the leg muscles. However, there was a reduction in the area covered by dendrites of MNs, and specific axon targeting defects were observed on adult leg muscles. Conversely, misexpression of pb in LinA MNs resulted in the relocalization of their dendrites to an area on the neuropil, where typically LinB dendrites were located. However, LinA retained many of its features and did not gain all the characteristics of LinB. Since pb mutant MNs show defective leg muscle innervation, the adults with pb mutant MNs were tested for walking behavior. Most walking parameters were normal, except that at high speed, the flies with pb mutant LinB MNs showed more wobble in walking than the control adults. This indicated a role of Pb expressing LinB MNs in stable walking at high speed. In order to establish that mTF code (of Pb with other factors) was instructive for the MN morphology, the TF code of specific LinB MNs was changed to other MNs in the LinB lineage by simultaneous knockdown and overexpression of the mTFs. It was observed that altering the mTF code resulted in the predictable transformation of the morphology, which supported the idea that different combinations of mTFs determined the MN’s morphological identity and led to the suggestion that role of pb in morphology had a critical bearing on the fly walking behavior. These results also established a genetic basis for the morphology of the MNs. They also suggested that MNs rely on a unique combination of different mTFs, which collectively give them their distinct signature morphologies. An idea proposed in the study is that temporal TFs most likely direct a stepwise change in the mTFs code for successive MNs (generated in LinB) and thus progressively change their morphology. To test this idea, it will be an important (though tedious) task to delineate the role of individual mTFs in determining the final MN morphology in the context of LinB. The results also raise the question of whether Pb plays a similar role in determining the morphology of other thoracic MNs (working with a different set of mTFs). Alternatively, considering Pb expression in other segments; it is a possibility that Pb may contribute to determining the morphologies of MNs found in other segments of VNC as well.
Moreover, since different levels of Antp have already been shown to play a role in regulating the morphology of MNs (Baek et al., 2013), one wonders if there is any interaction between pb (or other mTFs) and resident Hox gene in determining the final MN morphology. It is to be noted here that Antp expressing LinA MNs did not express Pb (Baek et al., 2013). Experimental testing of this idea will also determine whether the identity of the NB has any consequence on the choice of the mTFs employed. However, the existence of a lineage-specific combination of the mTFs has already been ruled out by Enriquez et al. (2015).
Hox Code for Neuromuscular Assembly in Embryogenesis and Larval Feeding Circuit
Feeding is a fundamental behavior necessary for the survival of an animal. In Drosophila, the feeding behavior has been investigated in larval and adult stages (Pool and Scott, 2014; Miroschnikow et al., 2020). The neurons responsible for feeding behavior and taste perception reside in the maxillary (Mx) and mandibular (Mn) neuromeres of larval SEG, which express Hox gene Dfd (McGinnis and Krumlauf, 1992; Hirth et al., 1998; Kuert et al., 2014). The Dfd loss of function mutants die during embryogenesis due to their inability to hatch. The hypomorphic alleles that survive until adulthood starve to death, owing to their inability to extend proboscis and ingest food (Merrill et al., 1987; Regulski et al., 1987; Restifo and Merrill, 1994).
Building on these observations, a study by Friedrich et al. from Lohmann’s group at Heidelberg investigated the role of Dfd in larval feeding behavior (Friedrich et al., 2016) and exploited the fact that both hatching and feeding rely on the same motor circuit responsible for the up and down movement of the larval mouth hooks (Pereanu et al., 2007; Schoofs et al., 2010; Hückesfeld et al., 2015) (Figure 3). These movements are executed by mouth hook elevator (MHE) and depressor (MHD) muscles in the larval head, which receive synaptic input from neurons in Mx, Mn, and Lb neuromeres and contribute to the larval feeding circuit (Pereanu et al., 2007; Schoofs et al., 2010; Hückesfeld et al., 2015). The authors show that the MNs from SEG expressed Dfd and innervate the elevator but not the depressor muscles (Figure 3A). Congruent to these observations, embryos mutant for Dfd were found to exhibit axonogenesis defects (Figure 3B) and consequent failure to hatch into larvae.
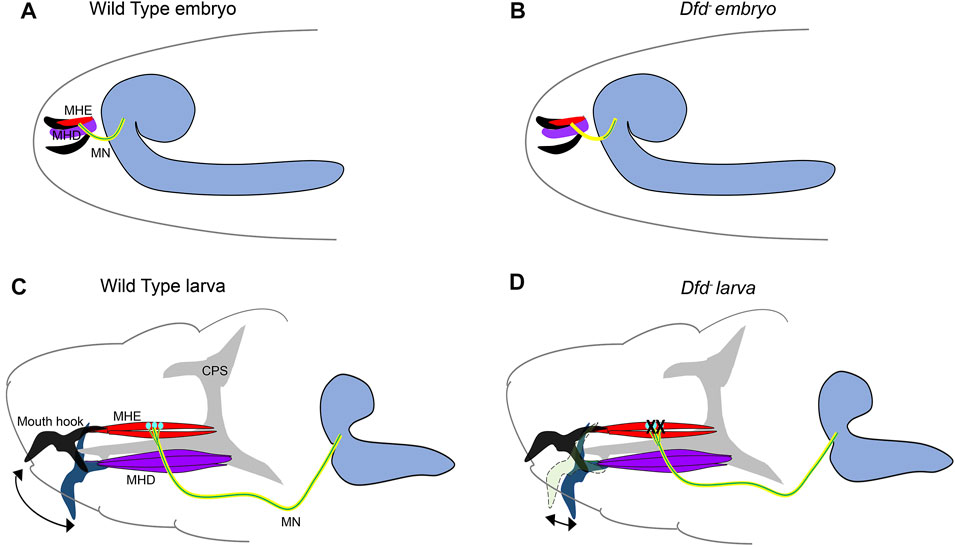
FIGURE 3. Role of Dfd in embryonic axonogenesis and larval feeding. Schematics show the major components responsible for embryonic mouth hook movement and larval feeding circuitry. The maxillary nerve (shown in yellow, originates from the SEG region of embryonic/larval CNS and innervates both MHD (shown in purple) and MHE (shown in red) muscles. The Dfd positive motorneurons (shown in green) from SEG synapse with the MHE muscles only and are crucial for the mouth hook elevation during embryonic hatching and larval feeding. The grey shaded region represents the Cephalopharyngeal Skeleton (CPS), extension and retraction of which is integral to the feeding process. The CNS is shown in blue on the right. The mouth hooks are shown in black in the embryos. In larvae, black mouth hook shows the extent of its elevation in wild-type, blue mouth hook shows the extent of its depression in wild type, and light green mouth hooks show the extent of its elevation in a Dfd3 mutant. (A,B) Shows the schematic of wild type and Dfd mutant embryos, latter show a severe restriction in axon outgrowth for the Dfd positive motor neurons resulting in failure of these neurons to project to the MHE. These mutants show hatching defects. (C,D) Shows the schematic of the wild type and Dfd mutant larvae. In the absence of Dfd, the mouth hook elevation is drastically reduced (double-sided arrow indicates the extent of mouth hook elevation and depression in wild type and Dfd mutant larvae). Dfd is required in both MHE and MNs to regulate assembly and maintenance of the feeding motor unit to execute optimum mouth hook movement.
Blocking the synaptic transmission in Dfd expressing MNs using tetanus toxin also compromised embryonic hatching. Next, using a temperature-sensitive allele (Dfd3) it was shown that Dfd is chronically required in the assembly, maintenance, and functionality of the feeding circuit. It was observed that Dfd3 embryos exhibited mouth hook movement and hatching defects when raised to non-permissive temperature in late embryogenesis (which is much after the formation of synapses). In corroboration to this, Dfd3 larvae, when shifted to non-permissive temperature as late as in the third instar stage, showed head-mouth hook movement defects, further establishing the chronic requirement for the gene (Figure 3D). Interestingly in both these cases, the innervation of the elevator muscle was found to be normal. Similar knockdown (KD) of Dfd in neurons by RNA interference or Dfd3 allele exhibited a significant change in synaptic morphology coupled with the reduction in the expression of a synaptic gene, Ankyrin-2 extra-large (Ank2-XL). However, unlike in the case of Ubx KD in MNs (Dutta et al., 2010) reported earlier by Dutta et al., muscles in the larval feeding circuit were normal in the case of Dfd KD in MNs. Since Dfd was found to be expressed both in the elevator muscles and the MNs from SEG, it was proposed that Hox expression in both these cells types provides them with a molecular code to identify each other during synaptic assembly. In agreement with this idea, a synaptic target recognition molecule “Connectin” (Con) (Nose et al., 1994) was found to be amongst the direct transcriptional targets of Dfd in CNS. Interestingly, this homophilic cell adhesion molecule Con was expressed in MNs and muscle devoid of Dfd protein, and its expression was regained in the MNs mutant for Dfd. This suggested that Dfd functioned to bring together MNs and cognate muscles by actively repressing Con in the cells of the feeding circuit. However, the identity of cell adhesion molecule(s) positively required by these cells for assembly of the neuromuscular feeding unit is yet to be determined.
The work established the role of Dfd as a critical coordinator for the formation, maintenance, and functioning of the neuromuscular network in the larval feeding circuit. The results also showed that synaptic stability and plasticity are determined by the half-life of synaptic proteins as well as the transcriptional program, which sustains the supply of synaptic components that maintains the neuromuscular junction. Lastly, it was proposed that Hox genes provide the molecular code for matching the MNs and muscles during developmental synaptic assembly through their transcriptional targets. However, even though Dfd was shown to have a role in the functioning of the feeding circuit, it remains to be investigated whether Dfd played a role in regulating the neural activity of the motor neurons to regulate the feeding behavior.
Continuing on the theme of Hox gene providing a molecular basis for matching the MNs and muscles, a subsequent study by Hessinger et al. (2017) from Rogulja-Ortmann and Technau groups at Mainz established a similar role for Hox gene Ubx in the assembly of the embryonic neuromuscular junction. This study unraveled the mechanism of how Ubx plays a role in determining the target specificity of the MN and its cognate muscle during embryogenesis. In the abdominal segments (A2-A7 segments) of embryonic CNS, ventrally projecting RP MNs innervate ventrolateral (VL) muscles on the embryonic body wall. The RP MNs 1, 3, 4, and 5 are some of the MNs known to innervate four VL muscles (VL1-4) in the abdominal segments (Bossing et al., 1996; Landgraf et al., 1997). Hessinger et al. focussed on the innervation of RP5 and V MNs (referred to as VL1-MNs) onto the VL1 muscles of abdominal segments (Figure 4). Through meticulous genetics, the study established that precise innervation of VL1 muscle by its cognate MNs (VL1-MNs) relies on Ubx mediated activation of Wnt4 signaling in VL2 muscle (Figure 4A). The authors found that AbdA had no role in this innervation, which entirely relied on Ubx dependent expression of the Wnt4 and sulfatase 1 gene (sulf1-known to be necessary for axon guidance) in the VL2 muscle. Wnt4 and Sulf1 expression in VL2 muscle played an instrumental role in repelling the axons of the MNs facilitating them to innervate their correct target, which was the VL1 muscle (Figure 4B). Wnt4 is a member of the Wnt family of signaling molecules while Sulf1 is a sulfatase implicated in regulating Wnt and BMP gradient in neuromuscular junction (Nose et al., 1994; Inaki et al., 2007). The secretion of Wnt4 and Sulf1 by VL2 was paralleled with the activation of canonical Wnt4 signaling in VL1-MNs. This facilitated the repulsion of MNs away from VL2 muscles, thereby establishing a precise neuromuscular connection (between VL1 muscle and VL1-MNs). Congruent to this, the knockdown of the canonical Wnt4 signaling pathway in the VL1-MNs resulted in their targeting defects. On the expected lines in Ubx mutants, Wnt4 and sufl1 genes were downregulated in VL2 muscles. Consequently, VL1-MNs could not go past VL2 muscles, and the innervation of VL1 muscles by these MNs was lost. Finally, as was observed in the case of larval peristalsis (Dixit et al., 2008) and feeding circuitry (Friedrich et al., 2016), it was the simultaneous expression of Ubx in both MNs and the muscles which rescued the Ubx mutant phenotype. Collectively, these studies highlight the importance of Hox genes in establishing a complementary molecular code between MN and muscles for the functional assembly of the neuromuscular networks.
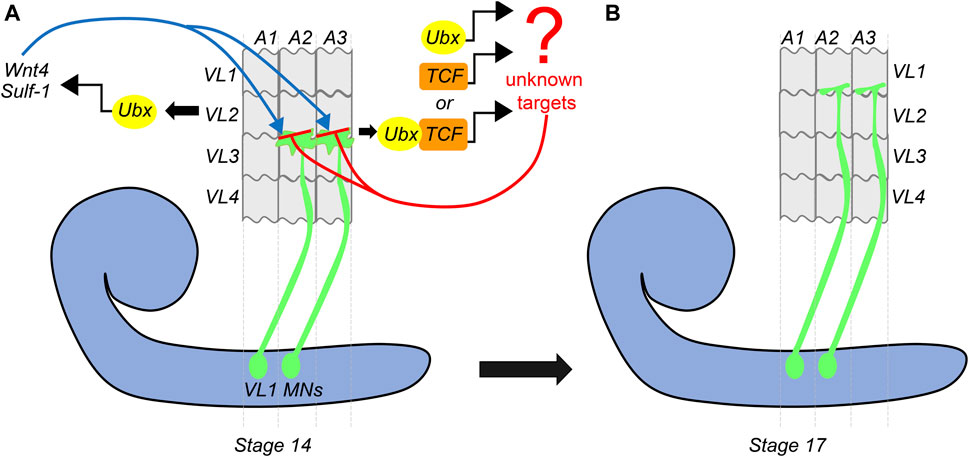
FIGURE 4. Dual role of Ubx in embryonic muscle innervation. Schematic of stage-14 and 17 embryos show the mechanism of innervation of ventrolateral muscles (VL1-4) of the embryonic body wall (shown in grey) by VL1-MNs (shown in green) coming from the embryonic CNS (shown in blue). (A) At stage-14, the approaching growth cone of the VL1 MNs is repelled by Ubx expressing VL2 muscles. Ubx mediates activation of Wnt4 and Sulf-1 in the VL2 muscles, which then interact with the Wnt receptors on the growth cone of the MNs. This leads to the activation of Wnt signaling (armadillo/TCF) in the VL1 MNs. Ubx and TCF in these MNs act together or in parallel to regulate the expression of unknown target genes, resulting in the repulsion of the VL1-MNs by VL2 muscles, thereby pushing them to their final target (VL1 muscles) by stage 17. (B) This suggests that Ubx expression in both VL2 muscles and VL1-MNs is required for establishing precise neuromuscular connections in the embryo.
Role of BX-C miRNA Mediated Hox Regulation in Behavior
BX-C has a bidirectionally transcribed microRNA (miRNA) locus with two overlapping miRNA’s on the opposite strand (iab4/8). This miRNA locus lies between abd-A and Abd-B and has been shown to target neighboring homeotic genes and results in homeotic transformation on overexpression (Ronshaugen et al., 2005; Bender, 2008; Stark et al., 2008; Tyler et al., 2008). Phenotypically, the homozygous deletion for the miRNA shows sterility and no other significant phenotype (Bender, 2008; Lemons et al., 2012). Recent studies have focussed on the role of this locus in CNS development, sterility, and adult behavior.
Role of Ubx in Egg-Laying Behavior
One of these studies by Garaulet et al. (2014) from Lai’s group in New York investigated the role of BX-C miRNA in CNS patterning and female sterility. Garaulet et al. demonstrated that in contrast to the embryonic epidermis where AbdA and AbdB repress anterior Hox gene Ubx, in larval CNS, it is the BX-C miRNA, that represses the BX-C genes outside their normal domain of expression. The deletion of this locus results in deregulation of Hox genes Ubx and abd-A and their cofactor exd and hth in posterior VNC of larval CNS. This was in agreement with what had been reported for this miRNA previously in the embryonic CNS as well (Bender, 2008; Thomsen et al., 2010; Gummalla et al., 2012). The subsequent genetic analysis shows that the sterility phenotype reported in miRNA-deleted females was substantially rescued by heterozygosity for BX-C genes (Ubx, abdA) and their cofactor hth. This effect was recapitulated by the targeted knockdown of Ubx in neurons, thereby establishing that deregulation of Hox and hth genes in neurons is critical for the sterility phenotype. Phenotypically, the ovary morphology in the mutant female flies was normal, and flies were capable of mating. Since the defect seemed to be in the egg-laying, therefore the focus shifted to the oviduct. The oviduct has two kinds of inputs, Insulin-like peptide 7 (ILP7) expressing excitatory glutaminergic MNs and inhibitory octopaminergic neurons terminating on radial muscles and epithelial linings (Rodríguez-Valentín et al., 2006; Yang et al., 2008; Castellanos et al., 2013). Significantly, the BX-C miRNA deletion did not alter the number or the transmitter identity for the ILP7+ MNs or the octopaminergic neurons. However, there was a reduction in the innervation of ILP7+ MNs on the oviduct and synaptic bouton count of the MNs on the radial muscles. These defects were substantially rescued by heterozygosity of Ubx and abd-A, but not by hth. However, the overexpression of Ubx or hth specifically in ILP7+ MNs did not recapitulate the sterility. This suggested that the broad de-repression of these genes in CNS was the cause of adult sterility. A search for functional neuronal domain responsible for the sterility was narrowed down to the Fruitless (Fru) expressing neurons (Stockinger et al., 2005) [which include ILP7+ MNs of oviduct as well (Castellanos et al., 2013)]. The Ubx and Hth overexpression in Fru+ neurons resulted in significant female sterility (90% in Ubx and 22% in Hth), suggesting that these neurons contribute to the female egg-laying program. However, other neuronal lineages from Fru expressing domain relevant for fertility and egg-laying behavior were not identified. Quite surprisingly, a subsequent study by the same group with a new deletion allele for BX-C miRNA showed that female flies were normal in their egg-laying behavior and had a functional neuromuscular control at the genital tract (Garaulet et al., 2020). Instead, this study suggested that the miRNA-deleted female had a behavioral shift from a virgin state to a post-mated state. This shift was attributed to the misregulation of hth in CNS. However, whether the misregulation for Hox genes (Ubx and abd-A) play a role in the behavioral shift was not reported. Also, this study did not investigate the innervation of ILP7+ MNs in females homozygous for new miRNA deletion. This suggests that either Hox genes of BX-C have no role in this behavioral shift for the female flies or the same is yet to be investigated.
Role of Ubx in Self Righting Behavior
Continuing on the theme of BX-C miRNA mediated repression of homeotic genes, two elegant studies from the Alonso Lab at Sussex in the UK have uncovered a role of the homeotic gene Ubx and the BX-C miRNA in self-righting (SR) motor behavior in Drosophila larvae and adults (Picao-Osorio et al., 2015; Issa et al., 2019). These studies show for the first time a post-developmental role of Hox gene and the importance of maintaining a very fine control over Hox expression in CNS to regulate neural physiology and behavior. SR is an innate reflex that corrects the body orientation when it is out of its normal upright position (Figures 5A,C). This response is evolutionarily conserved amongst all the bilaterians (Penn and Jane Brockmann, 1995; Faisal and Matheson, 2001; Jusufi et al., 2011).
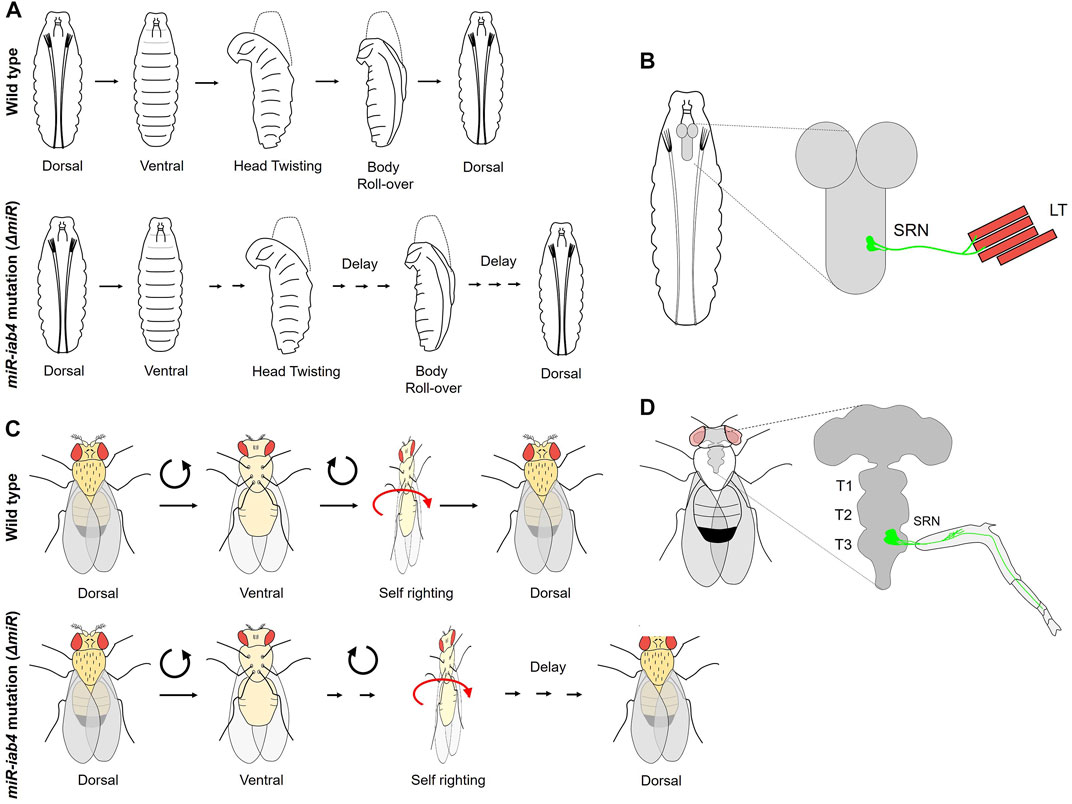
FIGURE 5. Role of BX-C miRNA in self-righting behavior. (A) The larval SR behavioral response in wild-type and miR-iab4 mutants is shown. Head twisting and body roll-over get significantly delayed in miR-iab4 mutants compared to wild-type larvae. (B) Shows that SR node (SRN) neurons originating from abdominal segments innervate to the lateral transverse (LT1/2) muscles of the larval body wall. (C) Shows adult self-righting behavioral response in wild type and miR-iab4 mutants, latter shows delayed SR behavior in adults as well. (D) Shows the innervation of the adult leg muscles by SRN neurons originating from T3 segments.
The first study by Picao-Osorio et al. (2015) established a role of BX-C miRNA iab4 in the regulation of Ubx in a defined group of MNs required to execute the SR behavior in larvae. Initially, the larvae for the deletion of BX-C miRNA were tested for different behavior assays of abdominal peristaltic waves, turning, and SR. All the behaviors were normal except for the SR behavior where miRNA-deleted larvae took a long time to turn themselves over after being put on their dorsal side (Figure 5A). Since Ubx was a known target of BX-C miRNA in VNC (Bender, 2008; Tyler et al., 2008; Thomsen et al., 2010; Garaulet et al., 2014), it was tested by targeted overexpression in its native transcriptional domain, and its role was confirmed in SR defects. Next, the cellular basis of aberrant SR behavior was narrowed down to Ubx regulation by iab4 to two metameric MNs in larval VNC (SR node neurons or SRN). The SRN innervated the lateral transverse (LT) muscles of the larval body wall, the LT1/2 (Figure 5B) (Picao-Osorio et al., 2015). Interestingly, the authors did not find any developmental consequence of Ubx dysregulation in larval CNS, and conditional expression of Ubx in the SRN in larval stages could recapitulate the SR behavior defects. This suggested that tampering with the levels of Ubx in these neurons specifically affected physiology and behavior. Similarly, a specific MN GAL4 line, which innervated LT muscles, was used for misexpressing Ubx and was shown to delay the SR in larvae. These observations were further corroborated by the differences in calcium activity traces of the SR MNs [measured using in vivo calcium sensor GCaMP (Chen et al., 2013)] in miRNA-deleted larvae compared to the wild-type controls. Artificial thermogenic activation (Hamada et al., 2008) or silencing (Kitamoto, 2001) of SR MNs also resulted in SR behavior defects, which was also reflected by the difference in calcium activity traces (Chen et al., 2013) in the test and the wild-type controls. However, it was not clear from this study whether similar SR movements in morphologically distinct organisms like larvae or adults relied on common or different genetic modules. To address this, the same group investigated and found a role of miRNA-mediated Ubx regulation in adult SR behavior (Figure 5C) (Issa et al., 2019). In this case, as well, overexpressing Ubx in its native domain could recapitulate these defects. Subsequently, Ubx was upregulated in two different subsets of adult leg MNs. However, the SR defect was reported in only in one case, further restricting the MNs responsible for SR defects in adults. These MNs were different from those required for executing SR behavior in larvae (Picao-Osorio et al., 2015). The downregulation of Ubx in adult-specific SR MNs was sufficient to rescue the behavioral defects reported in miRNA deletion. This knockdown also increased the number of synaptic varicosities on the femur muscles of the adult leg and rescued the neural activity in MNs back to the wild-type levels. These results supported a previously suggested idea that Hox genes have a role in assembling and maintaining the synaptic structures (Friedrich et al., 2016). The Drosophila larva and adult are divergent in lifestyle, behavioral properties, muscle structure, and neural constitution. Therefore, this study suggests that similar movements performed by organisms with distinct biomechanical, morphological, and neural structures could rely on the same miRNA/Hox genetic module, which can be redeployed in different developmental stages for equivalent behavior.
Importantly, these studies show that the miRNA-dependent post-transcriptional regulation of Hox gene Ubx can control the neural activity of MN to regulate the behavior of an animal. This function of Hox genes in neural physiology is independent of their role in development. The authors also suggest that other behavioral modules (like postural adjustment and locomotion) could also be controlled by miRNA. Furthermore, it is also possible that other adult movement-associated behaviors (like flight, walking, and jumping) (Szebenyi, 1969; Kaplan and Trout, 1974; Dawkins and Dawkins, 1976; Trimarchi and Schneiderman, 1993; Dickinson et al., 2000) may also be regulated by miRNA-mediated regulation of Ubx or Antp. For instance, Baek et al. show that Antp and Hth are the primary factors expressed in all thoracic leg MNs in larval stages. However, in the late pupal and adult stage, the T1 MNs express Hth, T2 express Antp and Hth, and T3 express Ubx and Hth (Baek et al., 2013). Therefore, one possibility worth considering is whether BX-C miRNA-mediated regulation of Antp and Hth also contributes to adult SR or other movement-associated behaviors. This is plausible considering that Hth has already been shown to be a target of BX-C miRNA in CNS (Bender, 2008; Tyler et al., 2008; Thomsen et al., 2010; Garaulet et al., 2014; Garaulet et al., 2020). Since 40% of the miRNA in the Drosophila genome were shown to affect the larval SR behavior (Picao-Osorio et al., 2017), therefore it may also be worthwhile to check whether any of these miRNA’s contribute to the regulation of SR or other movement associated behaviors through the regulation of Hox (Ubx or Antp) or Hth.
Lastly, a tempting question is whether the miRNA/Hox genetic module could also function in MNs of other behavioral circuits like feeding, mating, courtship, grooming, and virgin/mated behavioral shift. Moreover, if such control exists, it needs to be investigated whether it is executed primarily through Hox genes or other miRNA targets.
Conclusions
The survival of an organism depends on its ability to successfully and reproducibly execute a multitude of essential behaviors. This critically relies on Hox-dependent region-specific neuromuscular networks established along the AP axis of the body. Hox genes have been extensively investigated for their role in MN specification and motor circuit assembly in the hindbrain and the spinal cord of vertebrate CNS (Jessell, 2000; Philippidou and Dasen, 2013). The MNs in the hindbrain have a clustered organization, while in the spinal cord MNs are organized into longitudinal columns. At lower cervical (brachial) and lumbar levels of the spinal cord, MNs of the lateral motor column (LMC) project axons toward the forelimbs and hindlimbs (Landmesser, 2001). These columnar identities are regulated by the action of one or multiple Hox genes. Hox genes also diversify the MNs within LMC to generate approximately 50 MN pools targeting different limb muscles (Dasen et al., 2005). The cross-repressive interactions between different Hox genes set up a distinct transcriptional profile for each pool, which contributes to their clustering and peripheral muscle innervation (Dasen et al., 2008). Expectedly, individual Hox mutants in vertebrates affect the pool sizes, their position, and MN arborization. For example, in the case of HoxC6 mutants, brachial LMC size is reduced (Tiret et al., 1998; Vermot et al., 2005; Lacombe et al., 2013). Similarly, in the lumbar region where Hox10 is a major determinant of LMC identity, different mutant combinations for Hox10 result in defects in hindlimb innervation and compromise MN survival (Wahba et al., 2001; Lin and Carpenter, 2003; Shah et al., 2004; Wu et al., 2008). It has also been shown in the spinal cord MNs that acquisition of their basic MN identity and features is Hox independent (Jessell, 2000). These observations are reminiscent of the thoracic LinA/LinB lineage in Drosophila, which generate MNs innervating the adult leg muscles. In the case of LinB lineage, the Hox gene Pb and other mTFs play an instructional role in giving unique axonal and dendritic arborization to three MNs of the lineage, thereby regulating the morphological diversity of the MNs (Enriquez et al., 2015). Interestingly, Pb was not required for the survival of these MNs. This underlines the importance of Hox in determining the uniqueness of neuronal morphology. This genetic control of the morphological diversification of MNs was also shown to be critical in their functional capability for flawless walking at high speed (Baek and Mann, 2009; Baek et al., 2013; Enriquez et al., 2015). The role of Hox genes in determining the morphology of the vertebrate MNs has been reported. However, in our limited knowledge, no similar functional correlation between MN morphology and behavior has been established so far in the vertebrates. The observations made in thoracic LinA MNs are closer to what is reported in the vertebrates. In the case of Antp or the Hox triple (Scr−, Antp−, Ubx−) mutants LinA NBs, MNs were reported to undergo apoptosis. When the cell death was blocked, the surviving neurons took their fate as thoracic MNs. These MNs innervate the right target muscles and exhibit subtle arborization defects (Baek et al., 2013). This was similar to what was reported in the case of vertebrate. However, unlike vertebrates, the majority of the Drosophila LinA MNs do not show expression of more than one Hox factor, or Hox gene cross-regulation playing a central role in determining MN identity (Baek et al., 2013). Only in the case of the T3 segment, LinA MNs express Antp in larval stages and Ubx in pupal and adult stages (Figures 2E–E') (Baek et al., 2013). The mutant analysis for these MNs suggested that Ubx expression represses Antp, and these two genes function redundantly in these cells of the T3 segment (Baek et al., 2013). None of the thoracic LinA MNs expressed Hox gene Pb (Baek et al., 2013). The apparent differences in the role of Hox genes in Drosophila compared to their elaborate role in specifying MN pool identity might be due to the complex limb musculature found in the vertebrates, which need a very refined control from MNs. It is reported that 11 Hox genes are required to diversify the MN pools, which innervate the muscles of anterior limbs alone (Dasen et al., 2005). On the other hand, Drosophila leg musculature is not as complex and therefore may not require such complex transcriptional code to generate a large diversity of MNs. However, all these conclusions in Drosophila and their comparisons with vertebrates are based on studies done in LinA and LinB lineages, which constitutes only two-third of the leg MNs. It is possible that detailed analysis of other leg innervating MNs in Drosophila may give some additional insights (Truman et al., 2004; Baek and Mann, 2009).
The other Drosophila studies discussed here (summarized in Table 1) highlight the importance of Hox genes in setting up a molecular code for the functional assembly of neuromuscular networks (Friedrich et al., 2016; Hessinger et al., 2017). These studies also established that the requirement of Hox genes in the cells is not transient and restricted to the formation of the networks, but is chronic and is required for the maintenance and functioning of the networks much after they are established (Friedrich et al., 2016). At the cellular level, Hox genes have been shown to play a role in the survival of the MNs (Baek et al., 2013), their muscle innervation, and in determining their axonal and dendritic morphology (discussed above) (Baek et al., 2013; Enriquez et al., 2015). Notably, studies (with Antp) also established that the level of Hox genes in the adult MNs could regulate their axonal targeting and innervation onto the muscles, with low expressing MNs targeting proximal leg muscles and vice versa (Baek et al., 2013). The studies with BX-C miRNA emphasized the importance of maintaining a fine control over Hox expression in the MNs to establish a functional neuromuscular network and its role in executing the behavior (Garaulet et al., 2014; Picao-Osorio et al., 2015; Issa et al., 2019). More specifically, the miRNA-mediated control of Ubx expression on the neural activity of the SR MNs was the first instance where fine control over the levels of Hox gene has been shown to impact both neurophysiology and behavior. How exactly is this effect executed in MNs, and whether the miRNA-mediated regulation of Hox levels impacts other adult behaviors remains to be addressed.
Many roles discussed here go beyond the conventional developmental roles played by Hox genes in AP axis determination. These studies establish that in addition to giving the neurons their positional identity and the capacity to form the region-specific neural circuitry, Hox genes have a functional requirement in adult stages in regulating, at the very least, the morphology and neural activity of the MNs and their functions. Therefore these functions, to some extent, explain the sustained and robust neuronal expression of these genes post differentiation (Hirth et al., 1998; Technau et al., 2006). In order to further understand the role of Hox genes in the assembly of neuromuscular networks along the AP axis as well as their function beyond, there is a need to identify their targets in MNs. For instance, both Hox and Hth were similarly required for thoracic MNs to survive (Baek et al., 2013), but phenotypes like axonal and dendritic morphology differed when either Hox or Hth were individually removed (Baek et al., 2013; Enriquez et al., 2015). This supported the idea that distinct genetic programs downstream of Hox and Hth control axonal and dendritic morphology independent of each other. Therefore, identifying Hox and Exd/Hth targets specifically in MNs will be useful to understand their role in neuromuscular circuit assembly and their morphological diversification. Hox target genes have been identified in past using various approaches (Leemans et al., 2001; Barmina et al., 2005; Mohit et al., 2006; Hersh et al., 2007; Hueber et al., 2007; Agrawal et al., 2011; Choo et al., 2011; Pavlopoulos and Akam, 2011; Slattery et al., 2011; Sorge et al., 2012; Beh et al., 2016; Prasad et al., 2016; Shlyueva et al., 2016). However, none of these approaches were geared towards identifying the targets within CNS or its specific cell types. Identifying Hox targets in MNs may have been technically difficult so far, but a finer refinement of targeted DamID (TaDa) (Southall et al., 2013) to an elegant nano DAM technique (https://www.biorxiv.org/content/10.1101/2021.06.07.447332v2) may provide a useful mean for identifying MN specific targets genes downstream to these factors. Lastly, the distinct morphological phenotypes observed in MNs in Antp and hth mutants also suggest that Antp may be using cofactors other than canonical Hox cofactors (like Exd and Hth) (Baek et al., 2013). This is not unusual as Hox genes have been shown to use cooperative co-factors other than Exd/Hth (Gebelein et al., 2004; Ghosh et al., 2019), as well as novel collaborative co-factors in both neural and non-neural cell types (Gebelein et al., 2004; Mann et al., 2009; Baëza et al., 2015; Khandelwal et al., 2017; Bischof et al., 2018; Ghosh et al., 2019; Bakshi et al., 2020; Carnesecchi et al., 2020). However, the question remains whether any of these non-canonical cofactors facilitate Hox genes to carry out their conventional and newly discovered roles in MNs.
Author Contributions
RJ conceptualized, researched and wrote the manuscript. RS researched and helped in manuscript writing and made the figures. AB researched and helped in manuscript writing and made the figures.
Funding
RJ acknowledges funding support from the Department of Science and Technology, India (CRG/2021/ 003275); Department of Biotechnology, India (BT/PR26385/MED/122/110/2017, BT/PR27455/BRB/10/1647/2018 and BT/PR41306/MED/122/259/2020); CDFD core funds and UGC, India (Fellowship award to AB) (UGC Ref No. 22/06/2014(i)EU-V, 2061430472), and ICMR, India (Fellowship award to RS) (ICMR Ref. No. 3/1/3/JRF-2012/HRD-63 (40260)).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Agrawal, P., Habib, F., Yelagandula, R., and Shashidhara, L. S. (2011). Genome-level Identification of Targets of Hox Protein Ultrabithorax in Drosophila: Novel Mechanisms for Target Selection. Sci. Rep. 1, 205. doi:10.1038/srep00205
Akam, M. (1989). Hox and HOM: Homologous Gene Clusters in Insects and Vertebrates. Cell 57, 347–349. doi:10.1016/0092-8674(89)90909-4
Arya, R., Sarkissian, T., Tan, Y., and White, K. (2015). Neural Stem Cell Progeny Regulate Stem Cell Death in a Notch and Hox Dependent Manner. Cell Death Differ 22, 1378–1387. doi:10.1038/cdd.2014.235
Baek, M., Enriquez, J., and Mann, R. S. (2013). Dual Role for Hox Genes and Hox Co-factors in Conferring Leg Motoneuron Survival and Identity in Drosophila. Development 140, 2027–2038. doi:10.1242/dev.090902
Baek, M., and Mann, R. S. (2009). Lineage and Birth Date Specify Motor Neuron Targeting and Dendritic Architecture in Adult Drosophila. J. Neurosci. 29, 6904–6916. doi:10.1523/jneurosci.1585-09.2009
Baek, M., Menon, V., Jessell, T. M., Hantman, A. W., and Dasen, J. S. (2019). Molecular Logic of Spinocerebellar Tract Neuron Diversity and Connectivity. Cel Rep. 27, 2620–2635. doi:10.1016/j.celrep.2019.04.113
Baëza, M., Viala, S., Heim, M., Dard, A., Hudry, B., Duffraisse, M., et al. (2015). Inhibitory Activities of Short Linear Motifs Underlie Hox Interactome Specificity In Vivo. eLife 4, e06034. doi:10.7554/eLife.06034
Bahrampour, S., Jonsson, C., and Thor, S. (2019). Brain Expansion Promoted by Polycomb-Mediated Anterior Enhancement of a Neural Stem Cell Proliferation Program. Plos Biol. 17, e3000163. doi:10.1371/journal.pbio.3000163
Bakshi, A., Sipani, R., Ghosh, N., and Joshi, R. (2020). Sequential Activation of Notch and Grainyhead Gives Apoptotic Competence to Abdominal-B Expressing Larval Neuroblasts in Drosophila Central Nervous System. Plos Genet. 16, e1008976. doi:10.1371/journal.pgen.1008976
Barmina, O., Gonzalo, M., McIntyre, L. M., and Kopp, A. (2005). Sex- and Segment-specific Modulation of Gene Expression Profiles in Drosophila. Develop. Biol. 288, 528–544. doi:10.1016/j.ydbio.2005.09.052
Baumgardt, M., Karlsson, D., Salmani, B. Y., Bivik, C., MacDonald, R. B., Gunnar, E., et al. (2014). Global Programmed Switch in Neural Daughter Cell Proliferation Mode Triggered by a Temporal Gene cascade. Develop. Cel. 30, 192–208. doi:10.1016/j.devcel.2014.06.021
Becker, H., Renner, S., Technau, G. M., and Berger, C. (2016). Cell-Autonomous and Non-cell-autonomous Function of Hox Genes Specify Segmental Neuroblast Identity in the Gnathal Region of the Embryonic CNS in Drosophila. Plos Genet. 12, e1005961. doi:10.1371/journal.pgen.1005961
Beh, C. Y., El-Sharnouby, S., Chatzipli, A., Russell, S., Choo, S. W., and White, R. (2016). Roles of Cofactors and Chromatin Accessibility in Hox Protein Target Specificity. Epigenetics & Chromatin 9, 1. doi:10.1186/s13072-015-0049-x
Bello, B. C., Hirth, F., and Gould, A. P. (2003). A Pulse of the Drosophila Hox Protein Abdominal-A Schedules the End of Neural Proliferation via Neuroblast Apoptosis. Neuron 37, 209–219. doi:10.1016/s0896-6273(02)01181-9
Bender, W. (2008). MicroRNAs in the Drosophila Bithorax Complex. Genes Dev. 22, 14–19. doi:10.1101/gad.1614208
Berger, C., Pallavi, S. K., Prasad, M., Shashidhara, L. S., and Technau, G. M. (2005a). A Critical Role for Cyclin E in Cell Fate Determination in the central Nervous System of Drosophila melanogaster. Nat. Cel Biol 7, 56–62. doi:10.1038/ncb1203
Berger, C., Pallavi, S. K., Prasad, M., Shashidhara, L. S., and Technau, G. M. (2005b). Cyclin E Acts under the Control of Hox-Genes as a Cell Fate Determinant in the Developing central Nervous System. Cell Cycle 4, 422–425. doi:10.4161/cc.4.3.1524
Bhat, K. M. (1999). Segment Polarity Genes in Neuroblast Formation and Identity Specification during Drosophila Neurogenesis. Bioessays 21, 472–485. doi:10.1002/(sici)1521-1878(199906)21:6<472::aid-bies4>3.0.co;2-w
Birkholz, O., Rickert, C., Berger, C., Urbach, R., and Technau, G. M. (2013). Neuroblast Pattern and Identity in the Drosophila Tail Region and Role of Doublesex in the Survival of Sex-specific Precursors. Development 140, 1830–1842. doi:10.1242/dev.090043
Birkholz, O., Vef, O., Rogulja-Ortmann, A., Berger, C., and Technau, G. M. (2013). Abdominal-B and Caudal Inhibit the Formation of Specific Neuroblasts in the Drosophila Tail Region. Development 140, 3552–3564. doi:10.1242/dev.096099
Bischof, J., Duffraisse, M., Furger, E., Ajuria, L., Giraud, G., Vanderperre, S., et al. (2018). Generation of a Versatile BiFC ORFeome Library for Analyzing Protein-Protein Interactions in Live Drosophila. eLife 7, e38853. doi:10.7554/eLife.38853
Bossing, T., Udolph, G., Doe, C. Q., and Technau, G. M. (1996). The Embryonic Central Nervous System Lineages of Drosophila Melanogaster. Develop. Biol. 179, 41–64. doi:10.1006/dbio.1996.0240
Brierley, D. J., Rathore, K., VijayRaghavan, K., and Williams, D. W. (2012). Developmental Origins and Architecture of Drosophila Leg Motoneurons. J. Comp. Neurol. 520, 1629–1649. doi:10.1002/cne.23003
Bussell, J. J., Yapici, N., Zhang, S. X., Dickson, B. J., and Vosshall, L. B. (2014). Abdominal-B Neurons Control Drosophila virgin Female Receptivity. Curr. Biol. 24, 1584–1595. doi:10.1016/j.cub.2014.06.011
Carnesecchi, J., Sigismondo, G., Domsch, K., Baader, C. E. P., Rafiee, M.-R., Krijgsveld, J., et al. (2020). Multi-level and Lineage-specific Interactomes of the Hox Transcription Factor Ubx Contribute to its Functional Specificity. Nat. Commun. 11, 1388. doi:10.1038/s41467-020-15223-x
Carroll, S. B. (1995). Homeotic Genes and the Evolution of Arthropods and Chordates. Nature 376, 479–485. doi:10.1038/376479a0
Castellanos, M. C., Tang, J. C. Y., and Allan, D. W. (2013). Female-biased Dimorphism Underlies a Female-specific Role for post-embryonic Ilp7 Neurons in Drosophila Fertility. Development 140, 3915–3926. doi:10.1242/dev.094714
Chen, T.-W., Wardill, T. J., Sun, Y., Pulver, S. R., Renninger, S. L., Baohan, A., et al. (2013). Ultrasensitive Fluorescent Proteins for Imaging Neuronal Activity. Nature 499, 295–300. doi:10.1038/nature12354
Choo, S. W., White, R., and Russell, S. (2011). Genome-wide Analysis of the Binding of the Hox Protein Ultrabithorax and the Hox Cofactor Homothorax in Drosophila. PloS one 6, e14778. doi:10.1371/journal.pone.0014778
Dasen, J. S., De Camilli, A., Wang, B., Tucker, P. W., and Jessell, T. M. (2008). Hox Repertoires for Motor Neuron Diversity and Connectivity Gated by a Single Accessory Factor, FoxP1. Cell 134, 304–316. doi:10.1016/j.cell.2008.06.019
Dasen, J. S., Tice, B. C., Brenner-Morton, S., and Jessell, T. M. (2005). A Hox Regulatory Network Establishes Motor Neuron Pool Identity and Target-Muscle Connectivity. Cell 123, 477–491. doi:10.1016/j.cell.2005.09.009
Dawkins, R., and Dawkins, M. (1976). Hierachical Organization and Postural Facilitation: Rules for Grooming in Flies. Anim. Behav. 24, 739–755.
di Sanguinetto, S. A. D. T., Dasen, J. S., and Arber, S. (2008). Transcriptional Mechanisms Controlling Motor Neuron Diversity and Connectivity. Curr. Opin. Neurobiol. 18, 36–43. doi:10.1016/j.conb.2008.04.002
Dickinson, M. H., Farley, C. T., Full, R. J., Koehl, M. A. R., Kram, R., and Lehman, S. (2000). How Animals Move: an Integrative View. Science 288, 100–106. doi:10.1126/science.288.5463.100
Dixit, R., Vijayraghavan, K., and Bate, M. (2008). Hox Genes and the Regulation of Movement inDrosophila. Devel Neurobio 68, 309–316. doi:10.1002/dneu.20589
Doe, C. Q. (1992). Molecular Markers for Identified Neuroblasts and Ganglion Mother Cells in the Drosophila central Nervous System. Development 116, 855–863. doi:10.1242/dev.116.4.855
Doe, C. Q. (2017). Temporal Patterning in the Drosophila CNS. Annu. Rev. Cel Dev. Biol. 33, 219–240. doi:10.1146/annurev-cellbio-111315-125210
Dutta, D., Umashankar, M., Lewis, E. B., Rodrigues, V., and Vijayraghavan, K. (2010). Hox Genes Regulate Muscle Founder Cell Pattern Autonomously and Regulate Morphogenesis through Motor Neurons. J. Neurogenet. 24, 95–108. doi:10.3109/01677063.2010.494317
Enriquez, J., Venkatasubramanian, L., Baek, M., Peterson, M., Aghayeva, U., and Mann, R. S. (2015). Specification of Individual Adult Motor Neuron Morphologies by Combinatorial Transcription Factor Codes. Neuron 86, 955–970. doi:10.1016/j.neuron.2015.04.011
Estacio-Gómez, A., Moris-Sanz, M., Schäfer, A.-K., Perea, D., Herrero, P., and Díaz-Benjumea, F. J. (2013). Bithorax-complex Genes Sculpt the Pattern of Leucokinergic Neurons in the Drosophila central Nervous System. Development 140, 2139–2148. doi:10.1242/dev.090423
Faisal, A. A., and Matheson, T. (2001). Coordinated Righting Behaviour in Locusts. J. Exp. Biol. 204, 637–648. doi:10.1242/jeb.204.4.637
Friedrich, J., Sorge, S., Bujupi, F., Eichenlaub, M. P., Schulz, N. G., Wittbrodt, J., et al. (2016). Hox Function Is Required for the Development and Maintenance of the Drosophila Feeding Motor Unit. Cel Rep. 14, 850–860. doi:10.1016/j.celrep.2015.12.077
Garaulet, D. L., Castellanos, M. C., Bejarano, F., Sanfilippo, P., Tyler, D. M., Allan, D. W., et al. (2014). Homeotic Function of Drosophila Bithorax-Complex miRNAs Mediates Fertility by Restricting Multiple Hox Genes and TALE Cofactors in the CNS. Develop. Cel. 29, 635–648. doi:10.1016/j.devcel.2014.04.023
Garaulet, D. L., Zhang, B., Wei, L., Li, E., and Lai, E. C. (2020). miRNAs and Neural Alternative Polyadenylation Specify the Virgin Behavioral State. Develop. Cel. 54, 410–423. doi:10.1016/j.devcel.2020.06.004
Gaufo, G. O., Wu, S., and Capecchi, M. R. (2004). Contribution of Hox Genes to the Diversity of the Hindbrain Sensory System. Development 131, 1259–1266. doi:10.1242/dev.01029
Gebelein, B., McKay, D. J., and Mann, R. S. (2004). Direct Integration of Hox and Segmentation Gene Inputs during Drosophila Development. Nature 431, 653–659. doi:10.1038/nature02946
Ghosh, N., Bakshi, A., Khandelwal, R., Rajan, S. G., and Joshi, R. (2019). The Hox Gene Abdominal-B Uses DoublesexF as a Cofactor to Promote Neuroblast Apoptosis in the Drosophila central Nervous System. Development 146, dev175158. doi:10.1242/dev.175158
Gummalla, M., Maeda, R. K., Castro Alvarez, J. J., Gyurkovics, H., Singari, S., Edwards, K. A., et al. (2012). Abd-A Regulation by the Iab-8 Noncoding RNA. Plos Genet. 8, e1002720. doi:10.1371/journal.pgen.1002720
Hadjieconomou, D., Rotkopf, S., Alexandre, C., Bell, D. M., Dickson, B. J., and Salecker, I. (2011). Flybow: Genetic Multicolor Cell Labeling for Neural Circuit Analysis in Drosophila melanogaster. Nat. Methods 8, 260–266. doi:10.1038/nmeth.1567
Hales, K. G., Korey, C. A., Larracuente, A. M., and Roberts, D. M. (2015). Genetics on the Fly: A Primer on the Drosophila Model System. Genetics 201, 815–842. doi:10.1534/genetics.115.183392
Hamada, F. N., Rosenzweig, M., Kang, K., Pulver, S. R., Ghezzi, A., Jegla, T. J., et al. (2008). An Internal thermal Sensor Controlling Temperature Preference in Drosophila. Nature 454, 217–220. doi:10.1038/nature07001
Hart, C., Awgulewitsch, A., Fainsod, A., McGinnis, W., and Ruddle, F. H. (1985). Homeo Box Gene Complex on Mouse Chromosome 11: Molecular Cloning, Expression in Embryogenesis, and Homology to a Human Homeo Box Locus. Cell 43, 9–18. doi:10.1016/0092-8674(85)90007-8
Hartenstein, V., and Wodarz, A. (2013). Initial Neurogenesis in Drosophila. Wiley Interdiscip. Rev. Dev. Biol. 2, 701–721. doi:10.1002/wdev.111
Hersh, B. M., Nelson, C. E., Stoll, S. J., Norton, J. E., Albert, T. J., and Carroll, S. B. (2007). The UBX-Regulated Network in the Haltere Imaginal Disc of D. melanogaster. Develop. Biol. 302, 717–727. doi:10.1016/j.ydbio.2006.11.011
Hessinger, C., Technau, G. M., and Rogulja-Ortmann, A. (2017). The Drosophila Hox Gene Ultrabithorax Acts in Both Muscles and Motoneurons to Orchestrate Formation of Specific Neuromuscular Connections. Development 144, 139–150. doi:10.1242/dev.143875
Hirth, F., Hartmann, B., and Reichert, H. (1998). Homeotic Gene Action in Embryonic Brain Development of Drosophila. Development 125, 1579–1589. doi:10.1242/dev.125.9.1579
Holstege, J. C., de Graaff, W., Hossaini, M., Cano, S. C., Jaarsma, D., van den Akker, E., et al. (2008). Loss of Hoxb8 Alters Spinal Dorsal Laminae and Sensory Responses in Mice. Proc. Natl. Acad. Sci. 105, 6338–6343. doi:10.1073/pnas.0802176105
Homem, C. C. F., and Knoblich, J. A. (2012). Drosophila Neuroblasts: a Model for Stem Cell Biology. Development 139, 4297–4310. doi:10.1242/dev.080515
Huber, L., Ferdin, M., Holzmann, J., Stubbusch, J., and Rohrer, H. (2012). HoxB8 in Noradrenergic Specification and Differentiation of the Autonomic Nervous System. Develop. Biol. 363, 219–233. doi:10.1016/j.ydbio.2011.12.026
Hückesfeld, S., Schoofs, A., Schlegel, P., Miroschnikow, A., and Pankratz, M. J. (2015). Localization of Motor Neurons and Central Pattern Generators for Motor Patterns Underlying Feeding Behavior in Drosophila Larvae. PloS one 10, e0135011. doi:10.1371/journal.pone.0135011
Hudry, B., Remacle, S., Delfini, M.-C., Rezsohazy, R., Graba, Y., and Merabet, S. (2012). Hox Proteins Display a Common and Ancestral Ability to Diversify Their Interaction Mode with the PBC Class Cofactors. Plos Biol. 10, e1001351. doi:10.1371/journal.pbio.1001351
Hueber, S. D., Bezdan, D., Henz, S. R., Blank, M., Wu, H., and Lohmann, I. (2007). Comparative Analysis of Hox Downstream Genes inDrosophila. Development 134, 381–392. doi:10.1242/dev.02746
Inaki, M., Yoshikawa, S., Thomas, J. B., Aburatani, H., and Nose, A. (2007). Wnt4 Is a Local Repulsive Cue that Determines Synaptic Target Specificity. Curr. Biol. 17, 1574–1579. doi:10.1016/j.cub.2007.08.013
Issa, A. R., Picao-Osorio, J., Rito, N., Chiappe, M. E., and Alonso, C. R. (2019). A Single MicroRNA-Hox Gene Module Controls Equivalent Movements in Biomechanically Distinct Forms of Drosophila. Curr. Biol. 29, 2665–2675. doi:10.1016/j.cub.2019.06.082
Isshiki, T., Pearson, B., Holbrook, S., and Doe, C. Q. (2001). Drosophila Neuroblasts Sequentially Express Transcription Factors Which Specify the Temporal Identity of Their Neuronal Progeny. Cell 106, 511–521. doi:10.1016/s0092-8674(01)00465-2
Jessell, T. M. (2000). Neuronal Specification in the Spinal Cord: Inductive Signals and Transcriptional Codes. Nat. Rev. Genet. 1, 20–29. doi:10.1038/35049541
Jusufi, A., Zeng, Y., Full, R. J., and Dudley, R. (2011). Aerial Righting Reflexes in Flightless Animals. Integr. Comp. Biol. 51, 937–943. doi:10.1093/icb/icr114
Kannan, R., Berger, C., Myneni, S., Technau, G. M., and Shashidhara, L. S. (2010). Abdominal-A Mediated Repression of Cyclin E Expression during Cell-Fate Specification in the Drosophila central Nervous System. Mech. Dev. 127, 137–145. doi:10.1016/j.mod.2009.09.008
Kaplan, W. D., and Trout, W. E. (1974). Genetic Manipulation of an Abnormal Jump Response in Drosophila. Genetics 77, 721–739. doi:10.1093/genetics/77.4.721
Karlsson, D., Baumgardt, M., and Thor, S. (2010). Segment-specific Neuronal Subtype Specification by the Integration of Anteroposterior and Temporal Cues. Plos Biol. 8, e1000368. doi:10.1371/journal.pbio.1000368
Kaufman, T. C., Seeger, M. A., and Olsen, G. (1990). Molecular and Genetic Organization of the Antennapedia Gene Complex of Drosophila melanogaster. Adv. Genet. 27, 309–362. doi:10.1016/s0065-2660(08)60029-2
Khandelwal, R., Sipani, R., Govinda Rajan, S., Kumar, R., and Joshi, R. (2017). Combinatorial Action of Grainyhead, Extradenticle and Notch in Regulating Hox Mediated Apoptosis in Drosophila Larval CNS. Plos Genet. 13, e1007043. doi:10.1371/journal.pgen.1007043
Kitamoto, T. (2001). Conditional Modification of Behavior inDrosophila by Targeted Expression of a Temperature-Sensitiveshibire Allele in Defined Neurons. J. Neurobiol. 47, 81–92. doi:10.1002/neu.1018
Korona, D., Koestler, S. A., and Russell, S. (2017). Engineering the Drosophila Genome for Developmental Biology. J. Dev. Biol. 5, 16. doi:10.3390/jdb5040016
Kuert, P. A., Bello, B. C., and Reichert, H. (2012). The Labial Gene Is Required to Terminate Proliferation of Identified Neuroblasts in Postembryonic Development of the Drosophila Brain. Biol. Open 1, 1006–1015. doi:10.1242/bio.20121966
Kuert, P. A., Hartenstein, V., Bello, B. C., Lovick, J. K., and Reichert, H. (2014). Neuroblast Lineage Identification and Lineage-specific Hox Gene Action during Postembryonic Development of the Subesophageal Ganglion in the Drosophila central Brain. Develop. Biol. 390, 102–115. doi:10.1016/j.ydbio.2014.03.021
Lacombe, J., Hanley, O., Jung, H., Philippidou, P., Surmeli, G., Grinstein, J., et al. (2013). Genetic and Functional Modularity of Hox Activities in the Specification of Limb-Innervating Motor Neurons. Plos Genet. 9, e1003184. doi:10.1371/journal.pgen.1003184
LaCount, D. J., Hanson, S. F., Schneider, C. L., and Friesen, P. D. (2000). Caspase Inhibitor P35 and Inhibitor of Apoptosis Op-IAP Block In Vivo Proteolytic Activation of an Effector Caspase at Different Steps. J. Biol. Chem. 275, 15657–15664. doi:10.1074/jbc.m000791200
Landgraf, M., Bossing, T., Technau, G. M., and Bate, M. (1997). The Origin, Location, and Projections of the Embryonic Abdominal Motorneurons ofDrosophila. J. Neurosci. 17, 9642–9655. doi:10.1523/jneurosci.17-24-09642.1997
Landmesser, L. T. (2001). The Acquisition of Motoneuron Subtype Identity and Motor Circuit Formation. Int. J. Dev. Neurosci. 19, 175–182. doi:10.1016/s0736-5748(00)00090-3
Leemans, R., Loop, T., Egger, B., He, H., Kammermeier, L., Hartmann, B., et al. (2001). Identification of Candidate Downstream Genes for the Homeodomain Transcription Factor Labial in Drosophila through Oligonucleotide-Array Transcript Imaging. Genome Biol. 2, RESEARCH0015. doi:10.1186/gb-2001-2-5-research0015
Lelli, K. M., Noro, B., and Mann, R. S. (2011). Variable Motif Utilization in Homeotic Selector (Hox)-Cofactor Complex Formation Controls Specificity. Proc. Natl. Acad. Sci. 108, 21122–21127. doi:10.1073/pnas.1114118109
Lemons, D., Paré, A., and McGinnis, W. (2012). Three Drosophila Hox Complex microRNAs Do Not Have Major Effects on Expression of Evolutionarily Conserved Hox Gene Targets during Embryogenesis. PloS one 7, e31365. doi:10.1371/journal.pone.0031365
Lin, A. W., and Carpenter, E. M. (2003). Hoxa10 andHoxd10 Coordinately Regulate Lumbar Motor Neuron Patterning. J. Neurobiol. 56, 328–337. doi:10.1002/neu.10239
Maeda, R. K., and Karch, F. (2006). The ABC of the BX-C: the Bithorax Complex Explained. Development 133, 1413–1422. doi:10.1242/dev.02323
Mann, R. S., and Affolter, M. (1998). Hox Proteins Meet More Partners. Curr. Opin. Genet. Develop. 8, 423–429. doi:10.1016/s0959-437x(98)80113-5
Mann, R. S., and Chan, S.-K. (1996). Extra Specificity from Extradenticle: the Partnership between HOX and PBX/EXD Homeodomain Proteins. Trends Genet. 12, 258–262. doi:10.1016/0168-9525(96)10026-3
Mann, R. S., Lelli, K. M., and Joshi, R. (2009). Hox Specificity Unique Roles for Cofactors and Collaborators. Curr. Top. Dev. Biol. 88, 63–101. doi:10.1016/s0070-2153(09)88003-4
Martín, F., and Alcorta, E. (2017). Novel Genetic Approaches to Behavior in Drosophila. J. Neurogenet. 31, 288–299. doi:10.1080/01677063.2017.1395875
McGinnis, W., and Krumlauf, R. (1992). Homeobox Genes and Axial Patterning. Cell 68, 283–302. doi:10.1016/0092-8674(92)90471-n
Merabet, S., Saadaoui, M., Sambrani, N., Hudry, B., Pradel, J., Affolter, M., et al. (2007). A Unique Extradenticle Recruitment Mode in the Drosophila Hox Protein Ultrabithorax. Proc. Natl. Acad. Sci. 104, 16946–16951. doi:10.1073/pnas.0705832104
Merrill, V. K. L., Turner, F. R., and Kaufman, T. C. (1987). A Genetic and Developmental Analysis of Mutations in the Deformed Locus in Drosophila melanogaster. Develop. Biol. 122, 379–395. doi:10.1016/0012-1606(87)90303-4
Michelson, A. M. (1994). Muscle Pattern Diversification in Drosophila Is Determined by the Autonomous Function of Homeotic Genes in the Embryonic Mesoderm. Development 120, 755–768. doi:10.1242/dev.120.4.755
Miguel-Aliaga, I., and Thor, S. (2004). Segment-specific Prevention of pioneer Neuron Apoptosis by Cell-Autonomous, Postmitotic Hox Gene Activity. Development 131, 6093–6105. doi:10.1242/dev.01521
Miguez, A., Ducret, S., Di Meglio, T., Parras, C., Hmidan, H., Haton, C., et al. (2012). Opposing Roles for Hoxa2 and Hoxb2 in Hindbrain Oligodendrocyte Patterning. J. Neurosci. 32, 17172–17185. doi:10.1523/JNEUROSCI.0885-12.2012
Miroschnikow, A., Schlegel, P., and Pankratz, M. J. (2020). Making Feeding Decisions in the Drosophila Nervous System. Curr. Biol. 30, R831–R840. doi:10.1016/j.cub.2020.06.036
Moens, C. B., and Selleri, L. (2006). Hox Cofactors in Vertebrate Development. Develop. Biol. 291, 193–206. doi:10.1016/j.ydbio.2005.10.032
Mohit, P., Makhijani, K., Madhavi, M. B., Bharathi, V., Lal, A., Sirdesai, G., et al. (2006). Modulation of AP and DV Signaling Pathways by the Homeotic Gene Ultrabithorax during Haltere Development in Drosophila. Develop. Biol. 291, 356–367. doi:10.1016/j.ydbio.2005.12.022
Monedero Cobeta, I., Salmani, B. Y., and Thor, S. (2017). Anterior-Posterior Gradient in Neural Stem and Daughter Cell Proliferation Governed by Spatial and Temporal Hox Control. Curr. Biol. 27, 1161–1172. doi:10.1016/j.cub.2017.03.023
Neckameyer, W. S., and Bhatt, P. (2016). Protocols to Study Behavior in Drosophila. Methods Mol. Biol. 1478, 303–320. doi:10.1007/978-1-4939-6371-3_19
Nichols, C. D., Becnel, J., and Pandey, U. B. (2012). Methods to Assay Drosophila Behavior. J. visualized experiments : JoVE 61, 3795. doi:10.3791/3795
Nose, A., Takeichi, M., and Goodman, C. S. (1994). Ectopic Expression of Connectin Reveals a Repulsive Function during Growth Cone Guidance and Synapse Formation. Neuron 13, 525–539. doi:10.1016/0896-6273(94)90023-x
Pattyn, A., Vallstedt, A., Dias, J. M., Samad, O. A., Krumlauf, R., Rijli, F. M., et al. (2003). Coordinated Temporal and Spatial Control of Motor Neuron and Serotonergic Neuron Generation from a Common Pool of CNS Progenitors. Genes Dev. 17, 729–737. doi:10.1101/gad.255803
Pavlopoulos, A., and Akam, M. (2011). Hox Gene Ultrabithorax Regulates Distinct Sets of Target Genes at Successive Stages of Drosophila Haltere Morphogenesis. Proc. Natl. Acad. Sci. 108, 2855–2860. doi:10.1073/pnas.1015077108
Penn, D., and Jane Brockmann, H. (1995). Age-biased Stranding and Righting in Male Horseshoe Crabs, Limulus polyphemus. Anim. Behav. 49, 1531–1539. doi:10.1016/0003-3472(95)90074-8
Pereanu, W., Spindler, S., Im, E., Buu, N., and Hartenstein, V. (2007). The Emergence of Patterned Movement during Late Embryogenesis ofDrosophila. Devel Neurobio. 67, 1669–1685. doi:10.1002/dneu.20538
Philippidou, P., and Dasen, J. S. (2013). Hox Genes: Choreographers in Neural Development, Architects of Circuit Organization. Neuron 80, 12–34. doi:10.1016/j.neuron.2013.09.020
Picao-Osorio, J., Johnston, J., Landgraf, M., Berni, J., and Alonso, C. R. (2015). MicroRNA-encoded Behavior in Drosophila. Science 350, 815–820. doi:10.1126/science.aad0217
Picao-Osorio, J., Lago-Baldaia, I., Patraquim, P., and Alonso, C. R. (2017). Pervasive Behavioral Effects of MicroRNA Regulation in Drosophila. Genetics 206, 1535–1548. doi:10.1534/genetics.116.195776
Pool, A.-H., and Scott, K. (2014). Feeding Regulation in Drosophila. Curr. Opin. Neurobiol. 29, 57–63. doi:10.1016/j.conb.2014.05.008
Prasad, N., Tarikere, S., Khanale, D., Habib, F., and Shashidhara, L. S. (2016). A Comparative Genomic Analysis of Targets of Hox Protein Ultrabithorax Amongst Distant Insect Species. Sci. Rep. 6, 27885. doi:10.1038/srep27885
Prokop, A., Bray, S., Harrison, E., and Technau, G. M. (1998). Homeotic Regulation of Segment-specific Differences in Neuroblast Numbers and Proliferation in the Drosophila central Nervous System. Mech. Dev. 74, 99–110. doi:10.1016/s0925-4773(98)00068-9
Prokop, A., and Technau, G. M. (1994). Early Tagma-specific Commitment of Drosophila CNS Progenitor NB1-1. Development 120, 2567–2578. doi:10.1242/dev.120.9.2567
Regulski, M., Harding, K., Kostriken, R., Karch, F., Levine, M., and McGinnis, W. (1985). Homeo Box Genes of the Antennapedia and Bithorax Complexes of Drosophila. Cell 43, 71–80. doi:10.1016/0092-8674(85)90013-3
Regulski, M., McGinnis, N., Chadwick, R., and McGinnis, W. (1987). Developmental and Molecular Analysis of Deformed ; a Homeotic Gene Controlling Drosophila Head Development. EMBO J. 6, 767–777. doi:10.1002/j.1460-2075.1987.tb04819.x
Restifo, L. L., and Merrill, V. K. L. (1994). Two Drosophila Regulatory Genes, Deformed and the Broad-Complex, Share Common Functions in Development of Adult CNS, Head, and Salivary Glands. Develop. Biol. 162, 465–485. doi:10.1006/dbio.1994.1102
Rodríguez-Valentín, R., López-González, I., Jorquera, R., Labarca, P., Zurita, M., and Reynaud, E. (2006). Oviduct Contraction inDrosophilais Modulated by a Neural Network that Is Both, Octopaminergic and Glutamatergic. J. Cel. Physiol. 209, 183–198. doi:10.1002/jcp.20722
Rogulja-Ortmann, A., Renner, S., and Technau, G. M. (2008). Antagonistic Roles for Ultrabithorax and Antennapediain Regulating Segment-specific Apoptosis of Differentiated Motoneurons in the Drosophila Embryonic central Nervous System. Development 135, 3435–3445. doi:10.1242/dev.023986
Ronshaugen, M., Biemar, F., Piel, J., Levine, M., and Lai, E. C. (2005). The Drosophila microRNA Iab-4 Causes a Dominant Homeotic Transformation of Halteres to Wings. Genes Dev. 19, 2947–2952. doi:10.1101/gad.1372505
Saadaoui, M., Merabet, S., Litim-Mecheri, I., Arbeille, E., Sambrani, N., Damen, W., et al. (2011). Selection of Distinct Hox-Extradenticle Interaction Modes fine-tunes Hox Protein Activity. Proc. Natl. Acad. Sci. 108, 2276–2281. doi:10.1073/pnas.1006964108
Sánchez-Herrero, E., Vernós, I., Marco, R., and Morata, G. (1985). Genetic Organization of Drosophila Bithorax Complex. Nature 313, 108–113.
Schlegel, P., Costa, M., and Jefferis, G. S. (2017). Learning from Connectomics on the Fly. Curr. Opin. Insect Sci. 24, 96–105. doi:10.1016/j.cois.2017.09.011
Schmid, A., Chiba, A., and Doe, C. Q. (1999). Clonal Analysis of Drosophila Embryonic Neuroblasts: Neural Cell Types, Axon Projections and Muscle Targets. Development 126, 4653–4689. doi:10.1242/dev.126.21.4653
Schoofs, A., Niederegger, S., van Ooyen, A., Heinzel, H.-G., and Spiess, R. (2010). The Brain Can Eat: Establishing the Existence of a central Pattern Generator for Feeding in Third Instar Larvae of Drosophila virilis and Drosophila melanogaster. J. Insect Physiol. 56, 695–705. doi:10.1016/j.jinsphys.2009.12.008
Shah, V., Drill, E., and Lance-Jones, C. (2004). Ectopic Expression of Hoxd10 in Thoracic Spinal Segments Induces Motoneurons with a Lumbosacral Molecular Profile and Axon Projections to the Limb. Dev. Dyn. 231, 43–56. doi:10.1002/dvdy.20103
Shlyueva, D., Meireles-Filho, A. C. A., Pagani, M., and Stark, A. (2016). Genome-Wide Ultrabithorax Binding Analysis Reveals Highly Targeted Genomic Loci at Developmental Regulators and a Potential Connection to Polycomb-Mediated Regulation. PloS one 11, e0161997. doi:10.1371/journal.pone.0161997
Skeath, J. B. (1999). At the Nexus between Pattern Formation and Cell-type Specification: the Generation of Individual Neuroblast Fates in the Drosophila Embryonic central Nervous System. Bioessays 21, 922–931. doi:10.1002/(sici)1521-1878(199911)21:11<922::aid-bies4>3.0.co;2-t
Slattery, M., Ma, L., Négre, N., White, K. P., and Mann, R. S. (2011). Genome-wide Tissue-specific Occupancy of the Hox Protein Ultrabithorax and Hox Cofactor Homothorax in Drosophila. PloS one 6, e14686. doi:10.1371/journal.pone.0014686
Sorge, S., Ha, N., Polychronidou, M., Friedrich, J., Bezdan, D., Kaspar, P., et al. (2012). Thecis-regulatory Code of Hox Function inDrosophila. EMBO J. 31, 3323–3333. doi:10.1038/emboj.2012.179
Southall, T. D., Gold, K. S., Egger, B., Davidson, C. M., Caygill, E. E., Marshall, O. J., et al. (2013). Cell-type-specific Profiling of Gene Expression and Chromatin Binding without Cell Isolation: Assaying RNA Pol II Occupancy in Neural Stem Cells. Develop. Cel. 26, 101–112. doi:10.1016/j.devcel.2013.05.020
Stark, A., Bushati, N., Jan, C. H., Kheradpour, P., Hodges, E., Brennecke, J., et al. (2008). A Single Hox Locus in Drosophila Produces Functional microRNAs from Opposite DNA Strands. Genes Dev. 22, 8–13. doi:10.1101/gad.1613108
Stockinger, P., Kvitsiani, D., Rotkopf, S., Tirián, L., and Dickson, B. J. (2005). Neural Circuitry that Governs Drosophila Male Courtship Behavior. Cell 121, 795–807. doi:10.1016/j.cell.2005.04.026
Suska, A., Miguel-Aliaga, I., and Thor, S. (2011). Segment-specific Generation of Drosophila Capability Neuropeptide Neurons by Multi-Faceted Hox Cues. Develop. Biol. 353, 72–80. doi:10.1016/j.ydbio.2011.02.015
Szebenyi, A. L. (1969). Cleaning Behaviour in Drosophila melanogaster. Anim. Behav. 17, 641–651. doi:10.1016/s0003-3472(69)80006-0
Technau, G. M., Berger, C., and Urbach, R. (2006). Generation of Cell Diversity and Segmental Pattern in the Embryonic central Nervous System ofDrosophila. Dev. Dyn. 235, 861–869. doi:10.1002/dvdy.20566
Technau, G. M., Rogulja-Ortmann, A., Berger, C., Birkholz, O., and Rickert, C. (2014). Composition of a Neuromere and its Segmental Diversification under the Control ofHoxGenes in the Embryonic CNS ofDrosophila. J. Neurogenet. 28, 171–180. doi:10.3109/01677063.2013.868459
Thomsen, S., Azzam, G., Kaschula, R., Williams, L. S., and Alonso, C. R. (2010). Developmental RNA Processing of 3′UTRs in Hox mRNAs as a Context-dependent Mechanism Modulating Visibility to microRNAs. Development 137, 2951–2960. doi:10.1242/dev.047324
Tiong, S., Bone, L. M., and Whittle, J. R. S. (1985). Recessive Lethal Mutations within the Bithorax-Complex in Drosophila. Mol. Gen. Genet. 200, 335–342. doi:10.1007/bf00425445
Tiret, L., Le Mouellic, H., Maury, M., and Brulet, P. (1998). Increased Apoptosis of Motoneurons and Altered Somatotopic Maps in the Brachial Spinal Cord of Hoxc-8-Deficient Mice. Development 125, 279–291. doi:10.1242/dev.125.2.279
Trimarchi, J. R., and Schneiderman, A. M. (1993). Giant Fiber Activation of an Intrinsic Muscle in the Mesothoracic Leg of Drosophila melanogaster. J. Exp. Biol. 177, 149–167. doi:10.1242/jeb.177.1.149
Truman, J. W., and Bate, M. (1988). Spatial and Temporal Patterns of Neurogenesis in the central Nervous System of Drosophila melanogaster. Develop. Biol. 125, 145–157. doi:10.1016/0012-1606(88)90067-x
Truman, J. W., Schuppe, H., Shepherd, D., and Williams, D. W. (2004). Developmental Architecture of Adult-specific Lineages in the Ventral CNS of Drosophila. Development 131, 5167–5184. doi:10.1242/dev.01371
Tsuji, T., Hasegawa, E., and Isshiki, T. (2008). Neuroblast Entry into Quiescence Is Regulated Intrinsically by the Combined Action of Spatial Hox Proteins and Temporal Identity Factors. Development 135, 3859–3869. doi:10.1242/dev.025189
Tyler, D. M., Okamura, K., Chung, W.-J., Hagen, J. W., Berezikov, E., Hannon, G. J., et al. (2008). Functionally Distinct Regulatory RNAs Generated by Bidirectional Transcription and Processing of microRNA Loci. Genes Dev. 22, 26–36. doi:10.1101/gad.1615208
van den Akker, E., Reijnen, M., Korving, J., Brouwer, A., Meijlink, F., and Deschamps, J. (1999). Targeted Inactivation of Hoxb8 Affects Survival of a Spinal Ganglion and Causes Aberrant Limb Reflexes. Mech. Dev. 89, 103–114. doi:10.1016/s0925-4773(99)00212-9
Vermot, J., Schuhbaur, B., Mouellic, H. L., McCaffery, P., Garnier, J.-M., Hentsch, D., et al. (2005). Retinaldehyde Dehydrogenase 2 and Hoxc8 Are Required in the Murine Brachial Spinal Cord for the Specification of Lim1+ Motoneurons and the Correct Distribution of Islet1+ Motoneurons. Development 132, 1611–1621. doi:10.1242/dev.01718
Wahba, G. M., Hostikka, S. L., and Carpenter, E. M. (2001). The Paralogous Hox Genes Hoxa10 and Hoxd10 Interact to Pattern the Mouse Hindlimb Peripheral Nervous System and Skeleton. Develop. Biol. 231, 87–102. doi:10.1006/dbio.2000.0130
Wu, J. S., and Luo, L. (2006). A Protocol for Mosaic Analysis with a Repressible Cell Marker (MARCM) in Drosophila. Nat. Protoc. 1, 2583–2589. doi:10.1038/nprot.2006.320
Wu, Y., Wang, G., Scott, S. A., and Capecchi, M. R. (2008). Hoxc10 and Hoxd10 Regulate Mouse Columnar, Divisional and Motor Pool Identity of Lumbar Motoneurons. Development 135, 171–182. doi:10.1242/dev.009225
Yaghmaeian Salmani, B., Monedero Cobeta, I., Rakar, J., Bauer, S., Curt, J. R., Starkenberg, A., et al. (2018). Evolutionarily Conserved Anterior Expansion of the central Nervous System Promoted by a Common PcG-Hox Program. Development 145, dev160747. doi:10.1242/dev.160747
Yamaguchi, M., and Yoshida, H. (2018). Drosophila as a Model Organism. Adv. Exp. Med. Biol. 1076, 1–10. doi:10.1007/978-981-13-0529-0_1
Keywords: Hox, Drosophila, motor neuron (MN), behavior, feeding, locomotion, self righting behavior, neuromuscular network
Citation: Joshi R, Sipani R and Bakshi A (2022) Roles of Drosophila Hox Genes in the Assembly of Neuromuscular Networks and Behavior. Front. Cell Dev. Biol. 9:786993. doi: 10.3389/fcell.2021.786993
Received: 30 September 2021; Accepted: 14 December 2021;
Published: 07 January 2022.
Edited by:
Ernesto Sánchez-Herrero, Centre for Molecular Biology Severo Ochoa (CSIC), SpainReviewed by:
Claudio Alonso, University of Sussex, United KingdomJimena Berni, University of Sussex, United Kingdom
Copyright © 2022 Joshi, Sipani and Bakshi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rohit Joshi, cm9oaXRAY2RmZC5vcmcuaW4=, cmoyMTUyQGdtYWlsLmNvbQ==
†These authors have contributed equally to this work
 Rohit Joshi
Rohit Joshi Rashmi Sipani
Rashmi Sipani Asif Bakshi
Asif Bakshi