- 1Department of Basic Medical Sciences, College of Medicine, QU Health, Qatar University, Doha, Qatar
- 2Biomedical and Pharmaceutical Research Unit, QU Health, Qatar University, Doha, Qatar
- 3Institute of Cell and Neurobiology, Charité—Universitätsmedizin Berlin, Berlin, Germany
- 4Center for Chronically Sick Children (Sozialpädiatrisches Zentrum, SPZ), Charité—Universitätsmedizin Berlin, Berlin, Germany
- 5Department of Pediatric Neurology, Charité—Universitätsmedizin Berlin, Berlin, Germany
Microcephaly or reduced head circumference results from a multitude of abnormal developmental processes affecting brain growth and/or leading to brain atrophy. Autosomal recessive primary microcephaly (MCPH) is the prototype of isolated primary (congenital) microcephaly, affecting predominantly the cerebral cortex. For MCPH, an accelerating number of mutated genes emerge annually, and they are involved in crucial steps of neurogenesis. In this review article, we provide a deeper look into the microcephalic MCPH brain. We explore cytoarchitecture focusing on the cerebral cortex and discuss diverse processes occurring at the level of neural progenitors, early generated and mature neurons, and glial cells. We aim to thereby give an overview of current knowledge in MCPH phenotype and normal brain growth.
Introduction
Microcephaly is clinically defined by a significant reduction of the occipito-frontal head circumference (OFC) of more than two (microcephaly) or three (severe microcephaly) SDs below the mean for a given sex, age, and ethnicity (von der Hagen et al., 2014). The prevalence of microcephaly ranges between 1.5 and 8.7 per 10,000 births in Europe and the United States, respectively (Cragan et al., 2016; Morris et al., 2016). However, 15%–20% of children with developmental delay have microcephaly (Sassaman and Zartler, 1982; Watemberg et al., 2002; Aggarwal et al., 2013). Depending on the time of appearance, microcephaly can be classified as primary/congenital or secondary/postnatal (Passemard et al., 2013; Woods and Parker, 2013; Zaqout et al., 2017). It has been suggested that the primary causes of microcephaly lead to a reduction in the number of generated neurons, while the secondary causes mainly affect the dendritic complexity and synaptic formations (Woods, 2004). Primary microcephaly is by definition present at birth, and it can be caused by environmental and/or genetic factors (Zaqout et al., 2017; Alcantara and O'Driscoll, 2014; Kaindl et al., 2010). Various environmental factors such as infections, toxins, radiation, or alcohol result in primary microcephaly. The recent identification of epidemic infections with the Zika virus as a cause for primary microcephaly has highlighted this rare condition as a key topic in neuroscience to understand normal brain development (Kleber de Oliveira et al., 2016; Subramanian et al., 2019). This condition is an addition to the genetic prototype of isolated primary microcephaly, autosomal recessive primary microcephaly (microcephaly primary hereditary (MCPH)).
MCPH is a group of rare heterogeneous neurodevelopmental disorders characterized by intellectual disability and a significant reduction in the brain volume reflected by a reduction in the head circumference already at birth (Kaindl et al., 2010; Zaqout et al., 2017; Slezak et al., 2021). The reduction in brain volume in MCPH cases affects disproportionately the neocortex, though without obvious changes in the cortical organization (Kaindl et al., 2010; Kraemer et al., 2011; Jayaraman et al., 2018). The increasing use of whole-exome sequencing (WES) has uncovered a growing number of novel and disease-causing MCPH variants (Boycott et al., 2013). Simultaneously, further radiological and postmortem studies expand the spectrum of brain malformations reported in individuals with MCPH. The prevalence of MCPH differs from 1:10,000 in populations with a high rate of consanguineous marriage to 1:250,000 in the general population (Van Den Bosch, 1959; Cox et al., 2006). In consanguineous families, most MCPH diagnosed cases reveal homozygous variants in the disease-causing gene. However, compound heterozygous variants are increasingly discovered in MCPH patients, raising the importance of using advanced and accurate diagnosis methods for such cases (Jean et al., 2020).
Currently (December 2021), twenty-eight MCPH-related genes have been identified and tagged sequentially as MCPH1–MCPH28 (MCPH; OMIM phenotypic series: PS251200; Siskos et al., 2021) (Table 1). Still, more genetic loci are expected to exist given the fact that approximately 62% of western Europeans/North Americans and 25% of Indians/Pakistani families diagnosed with MCPH fail to show linkage to any of the MCPH loci (Verloes et al., 1993; Kaindl et al., 2010; Sajid Hussain et al., 2013). Most MCPH gene variants are nonsense, frameshift, or splice site-affecting variants leading to a production of non-functional, truncated proteins (Kaindl et al., 2010; Barbelanne and Tsang, 2014; Jean et al., 2020). Most of the MCPH genes encode centrosomal and/or pericentriolar matrix (PCM) proteins that are, in turn, ubiquitously expressed (Kaindl et al., 2010; Hussain et al., 2013; Barbelanne and Tsang, 2014). It is therefore not surprising to find that many MCPH proteins are involved in centriole biogenesis including organization, maturation, and distribution (Subramanian et al., 2019; Jean et al., 2020). Furthermore, MCPH proteins play crucial roles in microtubule dynamics, mitotic spindle formation, DNA damage responses, Wnt signaling, transcriptional regulation, and cell cycle checkpoint control (Kraemer et al., 2011; Mahmood et al., 2011; Jayaraman et al., 2018; Jean et al., 2020). Disruption of one or more of these functions during cortical neurogenesis adversely affects neuronal progenitor proliferation, differentiation, and survival leading to a severe reduction in the total number of generated neurons reflected by the microcephaly phenotype. Being highly conserved among species, ongoing research on MCPH animal models deems to be an important key for understanding the pathomechanisms behind microcephaly as well as the role of MCPH proteins during normal brain development (Gilbert et al., 2005; Woods et al., 2005; Zaqout et al., 2017). Although microcephaly found in MCPH patients simulates an evolutionary retrogression of the brain size (McHenry, 1994), human brain evolution cannot be attributed solely to the protein-coding sequences of MCPH genes (Pervaiz et al., 2021). Therefore, it has been hypothesized that complex conditional effects of human-specific coding and non-coding regulatory changes in MCPH only assist this evolution process (Pervaiz et al., 2021).
Classically, radiological investigations of patients with MCPH fail to show severe brain malformation except for simplified neocortical gyration. However, the increasing number of reported MCPH-linked mutations reveals that further deformities in brain architecture might occur (Table 2). The overall aim of this review is to explore the various effects of MCPH disease-causing genes on the cytoarchitecture of the cerebral cortex.
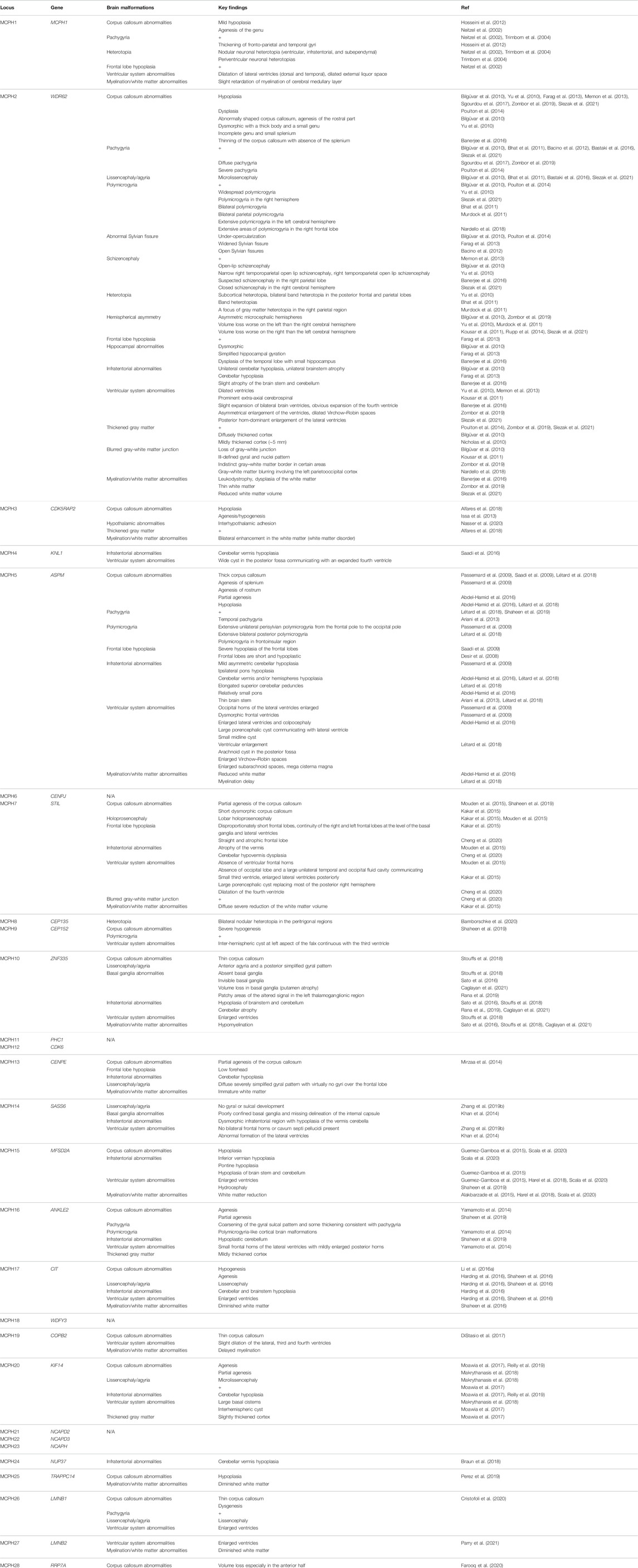
TABLE 2. Brain malformations associated with microcephaly primary hereditary (MCPH) additional to microcephaly.
Normal Corticogenesis
MCPH arises principally from a decreased production of neurons due to defects in progenitor proliferation, differentiation, and/or apoptosis during critical stages of brain development. Hence, it is important to briefly review the normal process of cortical neurogenesis before discussing the multiple facets of MCPH protein functions in maintaining a smooth running of this process.
Before the neurogenesis journey begins, the neural stem cells represented by neuroepithelial progenitors (NE) at the ventricular zone (VZ) undergo initial expansion in number through symmetrical cell divisions (Homem et al., 2015). Once the antiproliferative gene Tis21 starts to be expressed, NE cells begin to switch from proliferative division to neuronic division (Götz and Huttner, 2005). Simultaneously, NE cells transform gradually into more fate-restricted progenitors known as radial glial cells (RGCs) as an indication for their glial gene expressions (Götz and Huttner, 2005; Mori et al., 2005; Subramanian et al., 2019). RGCs possess apical processes attaching to the ventricular surface and basal processes reaching the basement membrane (future pial surface) (Homem et al., 2015). RGCs expand their number and exhibit a much higher number of asymmetrical cell divisions as compared with NE cells (Subramanian et al., 2019). During cell expansion, RGC nuclei show a characteristic interkinetic nuclear migration (INM) synchronized with the cell cycle phases during proliferation (Kosodo et al., 2011). The RGC nuclei migrate toward the basal side of the developing cortex during G1 phase and remain there during S phase before they migrate apically during G2 phase and proceed with M-phase once they reach the ventricular surface (Kosodo et al., 2011; Miyata et al., 2014). This pattern of migration during early neurogenesis requires functional microtubules and actin filaments (Götz and Huttner, 2005). It has been proposed that INM allows RGC rapid proliferation while maintaining their dense packing and determines cell fate through signaling gradients along their migration pathway (Götz and Huttner, 2005; Baye and Link, 2007; Del Bene et al., 2008). It is therefore very likely to find defects in neurogenesis involving RGC expansion and neuronal cell fate decisions when INM is disrupted (Latasa et al., 2009). Intriguingly, INM shows differences between species and might affect the total number of the generated neurons and thence the brain size (Okamoto et al., 2014).
Asymmetrical division of an RGC at the ventricular surface generates a self-renewing RG daughter cell and either a postmitotic neuron or a basal progenitor (intermediate progenitor cell (IPC)) (Homem et al., 2015; Subramanian et al., 2019). It has been earlier believed that the asymmetrical division of RGCs is principally driven by a change in mitotic spindle positions leading to a shift of the cleavage plane orientation from perpendicular (vertical) to parallel (horizontal) relative to the ventricular surface (Chenn and McConnell, 1995; Zhong et al., 1996; Kosodo et al., 2004; Fietz and Huttner, 2011; Homem et al., 2015). However, further investigations revealed that the rate of asymmetrical divisions in RGCs is not necessarily altered by the orientation of the cleavage plane (Morin et al., 2007; Konno et al., 2008; Postiglione et al., 2011). The asymmetrical RGC fate might be affected by inheriting centrioles with different maturity and primary cilium (Wang et al., 2009; Goetz and Anderson, 2010; Paridaen et al., 2013). It has been also shown that alterations in RGC cycle length control the shift from self-renewing divisions to neurogenic divisions (Calegari and Huttner, 2003; Calegari et al., 2005; Pilaz et al., 2009; Arai et al., 2011). Furthermore, it has been proposed that Notch signaling triggers neurogenic cell fate either by its distinct apicobasal gradient during INM or through asymmetric inheritance of endosomes positive for Sara (Smad anchor for receptor activation) (Del Bene et al., 2008; Nerli et al., 2020). This latter is achieved by targeting signaling endosomes to the central spindle by the action of plus-end kinesin motor (Klp98A) (Derivery et al., 2015).
Unlike RGCs, IPCs lack the connection with the ventricular surface and settle mainly in the subventricular zone (SVZ) basal to the VZ (Rakic, 2009; Homem et al., 2015; Subramanian et al., 2019; Heide and Huttner, 2021). IPCs undergo some proliferative divisions and terminate by generating two cortical neurons (Noctor et al., 2004; Pontious et al., 2008; Rakic, 2009). Asymmetrical divisions of RGCs in many mammals, especially in primates, yield an additional generation of a special type of basal progenitors known as basal RGCs (outer RGCs (oRGCs)) (Homem et al., 2015). Compared with IPCs, the oRGCs show a much higher proliferative capacity, which amplifies the total number of generated neurons and contribute to the characteristic folded cerebral cortex observed in primates, especially in humans (Reillo et al., 2011; Fietz et al., 2010; Hansen et al., 2010; Betizeau et al., 2013; Heide and Huttner, 2021). The newly established 3D in vitro human brain organoid model exhibits a considerable number of oRGCs (Lancaster et al., 2013; Zhang et al., 2019a).
Postmitotic cortical neurons are generated from both VZ and SVZ neural progenitors in an inside-out manner by which later-born neurons (superficial layers IV–II) bypass earlier-born neurons (deep layers VI–V) (Molyneaux et al., 2007; Leone et al., 2008). This pattern of neural generation is under spatiotemporal, cell cycle, and competency precise controls of neural progenitor fates (Kohwi and Doe, 2013). Eventually, six neural layers are created in the developing cerebral cortex, and the neurons start the formation of their distinctive dendrites, axons, and functional synapses. The fully developed cortical network contains 80% glutamatergic excitatory neurons produced by VZ and SVZ neural progenitors located in the dorsal telencephalon and 20% GABAergic inhibitory neurons that originated from the medial and caudal ganglionic eminence (Marín, 2013; Costa and Müller, 2014). The terminal number of generated cortical neurons is affected not only by the original number of neural progenitors but also by their starting and ending proliferation points and their cell lineage (Homem et al., 2015).
At the final stage of neurogenesis, RGCs lose their neuronal lineage and the connection with the apical surface switching to glial cell generators (Malatesta et al., 2000; Qian et al., 2000). Cortical astrocytes are firstly detected followed by oligodendrocytes, and the number of both glial cells is hugely expanded postnatally (Qian et al., 2000). Glial cells induce the development of white matter and axonal outgrowth by producing myelin and forming astrocytic branches (Subramanian et al., 2019). Taken together, forming a normal cerebral cortex requires highly organized spatiotemporal control for the neural progenitor populations to generate different neuronal and glial subtypes. Any defect during this process can lead to a major impact on brain development.
Brain Phenotype in Individuals With Microcephaly Primary Hereditary
The morphological changes in the brain structure of MCPH individuals have been mainly identified by radiological studies. Most MCPH cases show a reduction in brain volume associated with a simplified neocortical gyration pattern. However, the increased number of reported mutations and the ongoing neuroimaging of MCPH individuals reveal further brain malformations (Table 2). Some of these structural changes point toward the causative MCPH gene (e.g., the association between malformations of basal ganglia and mutations in gene encoding zinc finger-335 protein (ZNF335; MCPH10) (Sato et al., 2016; Stouffs et al., 2018; Rana et al., 2019; Caglayan et al., 2021)). The increasing number of reported brain malformations in MCPH individuals widens its pathogenesis spectrum. This indicates that the disruption of MCPH proteins not only is affecting the generation of neurons but could additionally affect neuronal differentiation, migration, dendritic and axonal outgrowth, and synaptogenesis. This is understandable given the fact that MCPH proteins are highly expressed in various neuroprogenitor organelles, especially the centrosome. In the following sections, we will discuss the consequences of MCPH mutations on brain development.
Accidents During the Brain Development Journey in Microcephaly Primary Hereditary
Studying the molecular mechanisms behind the pathogenesis of MCPH is very limited in humans. In fact, MCPH genes are highly conserved among different species (Woods et al., 2005; Gilbert et al., 2005), and this led to the discovery of several MCPH animal models mimicking the human phenotype (Table 1). Therefore, most of our current knowledge on the role of MCPH proteins in brain development is enriched through extensive studies on MCPH animal models. However, the pronounced difference of the human brain compared with most of the studied MCPH animal models establishes a new research direction toward 3D in vitro human brain organoid systems in studying the pathogenesis of microcephaly (Muzio and Consalez, 2013; Gabriel et al., 2020). The remarkable presence of oRGCs in this model opens the door for deeper insights into their role during the course of this disease in humans (Lancaster et al., 2013; Zhang et al., 2019a). As many MCPH proteins share overlapped functions, we saw to categorize them according to their major role(s) rather than discussing each one individually (Figure 1).
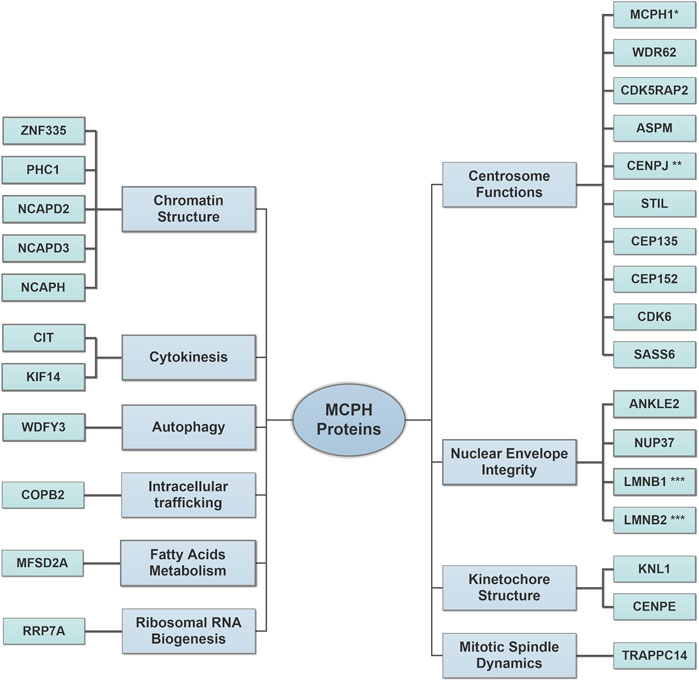
FIGURE 1. Major roles of microcephaly primary hereditary (MCPH) proteins in brain development. The increased number of discovered MCPH proteins expands the pathomechanism spectrum to include several cellular components. Centrosome Functions: the proteins of this group regulate proper centrosomal functions to balance the transition between neural progenitor cell (NPC) proliferation and differentiation by controlling cell cycle progression and cell cycle exit fraction. Nuclear Envelope Integrity: the proteins of this group affect the proper spindle alignment and cell fate determinants during NPC proliferation and protect radial glial cell (RGC) nuclei from mechanical stress injury during INM. Kinetochore Structure: the proteins of this group assure the correct alignment of chromosomes during mitosis. Mitotic Spindle Dynamics: the proteins of this group regulate the spindle dynamics and cell division. Chromatin Structure: the proteins of this group regulate gene expression during neurogenesis and assure proper DNA damage repair. Cytokinesis: the proteins of this group regulate the terminal step in the cell cycle, which leads to a physical separation between the daughter cells. Autophagy: the proteins of this group facilitate the removal of cytosolic protein aggregates and maintain mitochondrial homeostasis. Intracellular trafficking: the proteins of this group control the cellular retrograde trafficking from the Golgi to the endoplasmic reticulum. Fatty Acid Metabolisms: the proteins of this group affect the postnatal neuronal morphogenesis, which requires a normal lipogenesis process. Ribosomal RNA Biogenesis: the proteins of this group regulate ribosomal RNA processing and affect primary cilia resorption. Please refer to (Table 1) for full protein names. *MCPH1 is also involved in chromatin structure. **CENPJ is also involved in kinetochore structure. ***LMNB1 and LMNB2 are also involved in mitotic spindle dynamics.
Dysfunctional Centrosome
Almost one-third of MCPH mutations occur in centrosomal or mitotic spindle proteins. Defective centrosomes can affect cell cycle progression and cell division, leading to abnormal chromosomal numbers, cell cycle arrest, and apoptosis (Barbelanne and Tsang, 2014). It has been proposed that alterations of the cleavage plane orientation during NE proliferation increase asymmetric cell divisions (Buchman and Tsai, 2007; Knoblich, 2008; Zhong and Chia, 2008). This, in turn, leads to an early consumption of progenitor cells at the expense of a premature generation of neurons with ultimately reduced number, thence a smaller brain (Fish et al., 2006; Buchman et al., 2010; Kaindl et al., 2010; Lizarraga et al., 2010). In this notion, several MCPH mouse models show a shift in the cleavage plan orientation of NE cells favoring neurogenic cell fate (Fish et al., 2006; Lizarraga et al., 2010; Pulvers et al., 2010; Gruber et al., 2011; Xu et al., 2014). This evidence has been also supported by human brain organoid models (Lancaster et al., 2013; Zhang et al., 2019a). Intriguingly, most of the MCPH fly models with defected centrosomal proteins exhibit normal brain size (Basto et al., 2006; Lucas and Raff, 2007; Roque et al., 2012), indicating that changes in the cleavage plane orientation might only have a minor impact on brain growth. Alternatively, flies could have compensatory mechanisms bypassing the effect of the misoriented cleavage plane (Nigg and Raff, 2009). On the other hand, further studies discovered MCPH mouse models with the microcephaly phenotype, though unaffected cleavage plane (Pulvers et al., 2010; Marjanović et al., 2015). Furthermore, depletion of some other MCPH centrosomal proteins in mice does not affect brain growth at all (Malumbres et al., 2004).
The neural progenitor cell (NPC) symmetrical proliferation speed is frequently reduced in mutated MCPH genes, which encode centrosomal proteins (Lizarraga et al., 2010; Gruber et al., 2011; Sgourdou et al., 2017; Ding et al., 2019). This is much obvious toward the end of neurogenesis (Lizarraga et al., 2010; Gruber et al., 2011; Sgourdou et al., 2017) when the later-born neurons (superficial layers II–IV) start to be generated. Together with the premature generation of neurons, this explains why superficial cortical neurons are the most affected in most MCPH models (Lizarraga et al., 2010; Chen et al., 2014). This is in line with postmortem histological analysis described in a case of WD-repeat-containing protein 62 gene (WDR62; MCPH2) mutation (Yu et al., 2010). In addition, MCPH proteins are important for the normal distribution of cells between cortical zones. Knockout of abnormal spindle-like, microcephaly-associated gene (Aspm) in ferret increases the number of generated oRGCs, affecting the RGC overall proliferative capacity (Johnson et al., 2018). Likewise, knockdown of the cyclin-dependent kinase five regulatory subunit-associated protein two gene Cdk5rap2 in a mouse model alters the distribution of progenitor pool leading to more generation of basal progenitors (Buchman et al., 2010). By contrast, somatosensory cortical layer VI has been reported to be thinner in an Aspm knockout mouse model (Fujimori et al., 2014). Indeed, several in vivo and in vitro studies including human brain organoid models revealed that MCPH centrosomal genes balance the transition between NPC proliferation and differentiation by controlling cell cycle progression and cell cycle exit fraction (Buchman et al., 2010; Lizarraga et al., 2010; Bogoyevitch et al., 2012; Lancaster et al., 2013; Hainline et al., 2014; Zhang et al., 2019a). This explains, respectively, the reduced proliferation and premature neuronal differentiation detected in the respective MCPH animal models. Furthermore, using the conditional knockout mouse model, it has been shown that centromeric protein J (Cenpj) regulates NPC cell cycle progression by regulating cilium disassembly during neurogenesis (Ding et al., 2019). Similarly, depletion of WDR62 and centrosomal-P4.1-associated protein (CPAP) in human cerebral organoids impairs the cilium disassembly and cell cycle progression (Gabriel et al., 2016; Zhang et al., 2019a).
It has been reported that mutations in genes encoding MCPH centrosomal proteins alter the maturation and cellular number of centrosomes (Pfaff et al., 2007; Rodrigues-Martins et al., 2007; Yabe et al., 2007; Megraw et al., 2011; Hussain et al., 2012; McIntyre et al., 2012; Hussain et al., 2013; Arquint and Nigg, 2014). This, in turn, might affect the proper distribution of chromosomes to daughter cells leading to spindle instability and mitotic delay or arrest at metaphase checkpoint (Ripoll et al., 1985; Gonzalez et al., 1990; Megraw et al., 1999; Riparbelli et al., 2002; Matsuura et al., 2004; Pfaff et al., 2007; Lizarraga et al., 2010; Kim et al., 2011a; Vitale et al., 2011; Novorol et al., 2013; Chen et al., 2014). In most of these cases, such defect triggers the apoptotic cascade leading to cellular loss (Pfaff et al., 2007; Lizarraga et al., 2010; Vitale et al., 2011; McIntyre et al., 2012; Novorol et al., 2013; Chen et al., 2014). Remarkably, increased apoptosis of NPCs—associated with or without proliferation/differentiation defects—contributes to the microcephaly phenotype by depleting the neural stem cell pool (Izraeli et al., 1999; Pfaff et al., 2007; Lizarraga et al., 2010; Gruber et al., 2011; McIntyre et al., 2012; Sgourdou et al., 2017; Zhang et al., 2019a; Ding et al., 2019). Intriguingly, neuronal populations also seem to be vulnerable to apoptosis during later stages of development (Lizarraga et al., 2010).
Apparently, the impact of mutated MCPH centrosomal genes in brain development not only is restricted to NPC proliferation and differentiation phase but also exceeds it to affect neuronal migration, dendritic and axonal outgrowth, and synaptogenesis. The presence of gray matter heterotopia, polymicrogyria, lissencephaly, and pachygyria in several MCPH conditions points toward impaired neuronal migration (Table 2) (Leventer et al., 2010; Yu et al., 2010). The signs of this impairment have been reported in postmortem histopathological MCPH samples and various MCPH animal models (Yu et al., 2010; Buchman et al., 2011; Xu et al., 2014). Besides that, an interesting role of Cdk5rap2 in regulating dendritic development and synaptogenesis in superficial cortical layer II/III has been reported (Zaqout et al., 2019). The contribution of dendritic complexity deficits in the microcephaly phenotype points toward a progressive nature during the MCPH course (van Dyck and Morrow, 2017).
Defective Chromatin Structure
Chromatin structure is a golden stone for gene expression regulation during neurogenesis. Mutations in genes encoding chromatin-linked proteins expand the pathomechanism spectrum of the MCPH. Microcephalin (BRCT/BRIT1) mutated cells taken from MCPH1 individuals and mouse models displayed premature chromosome condensation (PCC) associated with a high frequency of prophase-like cells and defective DNA damage repair (Jackson et al., 2002; Neitzel et al., 2002; Liang et al., 2010; Trimborn et al., 2010). This feature has been considered as a diagnostic marker for individuals with MCPH1 gene mutations (Jackson et al., 2002). In flies, mcph1 mutants display embryonic lethality due to mitotic arrest and uncoordinated centrosome/nuclear cycles in early syncytial cell cycles (Brunk et al., 2007). Likewise, mutations in gene encoding polyhomeotic-like one protein (PHC1; MCPH11) are associated with aberrant DNA damage repair (Awad et al., 2013). In addition, PHC1 mutations disturb the expression of Nanog and the ubiquitination of histone H2A, in which the former maintains pluripotency and the latter affects the cell cycle progression by the accumulation of Geminin (Awad et al., 2013; Chen et al., 2021).
Complete ablation of Znf335 gene in mice is embryonically lethal, but conditional knockout leads to a reduction in the cortical size affecting the forebrain much severely (Yang et al., 2012). This has been attributed to disruptions in NPC proliferation, cell fate, and neuronal differentiation (Yang et al., 2012). Consistently, postmortem histopathological studies on brain samples taken from ZNF335 patients reveal a severe reduction in the neuronal number associated with abnormalities in neuronal morphogenesis, migration, and polarity (Yang et al., 2012).
Mutations in genes encoding condensin complex proteins NCAPD2, NCAPD3, and NCAPH have been linked to MCPH21, 22, and 23, respectively (Martin et al., 2016). One of the hallmarks of hypomorphic Ncaph2 mice is the formation of a chromatin bridge in apical NPCs (Martin et al., 2016). These bridges result from failed sister chromatid disentanglement leading to chromosome segregation errors and aneuploidy (Martin et al., 2016). Subsequently, NPCs undergo a reduced cell proliferation and an increased apoptosis without obvious alterations in spindle orientations or cell fat (Martin et al., 2016).
Deformed Kinetochore Proteins
Mutations in genes encoding kinetochore scaffold one protein (KNL1, previously known as CASC5) and centromere-associated protein E (CENPE) have been linked to MCPH4 (Jamieson et al., 1999; Genin et al., 2012; Mirzaa et al., 2014; Saadi et al., 2016; Szczepanski et al., 2016; Zarate et al., 2016). Proper function of proteins associated with centromeric kinetochore assures the correct alignment of chromosomes during mitosis; else, spindle assembly checkpoint (SAC) is activated and suspends the mitotic progression (Cleveland et al., 2003; Musacchio and Salmon, 2007; Santaguida and Musacchio, 2009; Hori and Fukagawa, 2012). Conditional knockout of Knl1 in mouse cortical NPCs results in DNA damage due to chromosomal segregation errors (Shi et al., 2019). This triggers a p53-dependent apoptotic cascade and leads to massive loss of NPCs and microcephaly (Shi et al., 2019). Similar to MCPH centrosomal proteins, the progressive loss of NPCs in Knl1 conditional knockout mice affects mainly the later-born neurons (superficial layers II–IV) (Shi et al., 2019). CENPE facilitates the transition from metaphase to anaphase during the cell cycle (Yen et al., 1991). Disruption of Cenpe function in mice and Drosophila leads to early embryonic lethality due to chromosomal instability (Yucel et al., 2000; Putkey et al., 2002).
Interruption of Fatty Acids Uptake Into the Brain
Proper brain development and function require essential omega-3 fatty acids, which need to be obtained from the circulation via specific transporters (Alakbarzade et al., 2015). Sodium-dependent lysophosphatidylcholine (LPC) transporter (MFSD2A) is exclusively expressed in the endothelium of the blood–brain barrier (BBB) and a major transport facilitator for docosahexaenoic acid (DHA) (Ben-Zvi et al., 2014; Nguyen et al., 2014; Alakbarzade et al., 2015; Guemez-Gamboa et al., 2015). Depending on the transporter residual activity, mutations in MFSD2A (MCPH15) gene is associated with either a progressive microcephaly syndrome or a much lethal phenotype (Berger et al., 2012; Alakbarzade et al., 2015; Guemez-Gamboa et al., 2015; Harel et al., 2018; Scala et al., 2020). The progressive feature associated with the milder form of this disease raises the possibility that LPC transportation is continuously required for membrane biogenesis in the brain (Alakbarzade et al., 2015; Scala et al., 2020). Endothelial-specific deletion of Mfsd2a in mice leads to a microcephaly phenotype accompanied by a reduction in neuronal arborization and dendritic length (Chan et al., 2018). Interestingly, neuronal loss detected in Mfsd2a knockout mice was restricted to cerebellar Purkinje cells and hippocampal CA1 and CA3 regions (Nguyen et al., 2014). Taken together, these data demonstrate that, unlike other MCPHs, Mfsd2a deficiency affects the postnatal neuronal morphogenesis, which requires a normal lipogenesis process (van Deijk et al., 2017; Ziegler et al., 2017; Chan et al., 2018). Notably, variable degrees of white matter reduction have been also reported in MCPH15 individuals (Alakbarzade et al., 2015; Harel et al., 2018; Scala et al., 2020). Further studies are required to assess to which extent do white matter deficits contribute to the microcephaly phenotype (Huang and Li, 2021).
Altered Nuclear Envelope
Mutations in several genes encoding nuclear envelop components have been recently linked to MCPH conditions. Ankyrin repeat- and lem domain-containing protein 2 (Ankle2) is localized to the endoplasmic reticulum and nuclear envelope (Link et al., 2019). Drosophila dAnkle2 mutant larvae show a reduction in the brain size due to impaired nuclear envelope integrity, which eventually affects proper spindle alignment and cell fate determinants during NPC proliferation (Link et al., 2019). Another study, however, suggests that the reduction in Drosophila dAnkle2 mutant NPCs cells is due to defects in proliferation and massive apoptosis rather than an alteration in asymmetrical cell division (Yamamoto et al., 2014). In this line, Caenorhabditis elegans ANKLE2 ortholog protein (LEM-4L) plays a critical role in mitosis by facilitating nuclear envelope reassembly during mitotic exit (Asencio et al., 2012).
Individuals with mutations in genes encoding various nuclear pore complex proteins (NPC) are diagnosed with severe forms of nephrotic syndrome (Braun et al., 2018). However, mutations in NPC subunit component nucleoporin 37 (NUP37) also exhibit intellectual disability and MCPH (Braun et al., 2018). More recently and yet to be linked to a specific OMIM MCPH number, mutations in NUP85 subunit are associated with a reduction in brain volume, delayed myelination, agenesis of the corpus callosum, gray matter heterotopia, and frontal lobe cortical malformation (Ravindran et al., 2021). Fibroblasts derived from NUP85 individuals are characterized by reduced cell viability, proliferation rate, abnormal mitotic spindle apparatus, and altered cytoskeletal protein expressions (Ravindran et al., 2021). As most of the studies performed in viable animal models with NPC defects focused on the nephrotic phenotype, further investigations to understand their effects on brain growth are still warranted.
B-type lamins 1 and 2 (LMNB1/2) are intermediate filament proteins involved in nuclear envelope reassembly, in which the deficiency leads to fragile nuclei more susceptible to nuclear membrane (NM) rupture (Coffinier et al., 2011; Chen et al., 2019). In humans, mutations in LMNB1 and LMNB2 have been linked to MCPH26 and MCPH27, respectively (Cristofoli et al., 2020; Parry et al., 2021). During early neurogenesis, RGC nuclei undergo INM, which represents mechanical stress that threatens RGCs with weakened nuclear lamina (Chen et al., 2019). Therefore, lack of murine Lmnb1/2 during this critical step triggers NPC apoptosis and leads to abnormal neuronal migration reflected by disorganized cortical layering (Kim et al., 2011b; Coffinier et al., 2011; Young et al., 2012; Chen et al., 2019). This migration defect not only is confined to the cerebral cortex but also affects the hippocampal and cerebellar layering (Coffinier et al., 2010; Coffinier et al., 2011). In addition, it has been proposed that Lmnb1/2 is localized at the mitotic spindle and plays a role in INM and neuronal migration via interaction with dynein in organizing NPC spindle orientation (Tsai et al., 2006; Kim et al., 2011b). However, no abnormal metaphase spindle formation has been noticed in lymphoblastoid cells (LCLs) derived from LMNB1 individuals (Cristofoli et al., 2020).
Defective Cytokinesis
Cytokinesis is the terminal step in the cell cycle, which leads to a physical separation between the daughter cells. Defects in this process frequently result in the formation of binucleated cells, aneuploidy, chromosomal instability, cell cycle arrest, and apoptosis (Li et al., 2016a). Notably, the elevated number of binucleated cells—including pyramidal and Purkinje cells—is considered as a key feature for cytokinesis failures (Di Cunto et al., 2000; Roberts et al., 2000; Reilly et al., 2019). Citron rho-interacting kinase (CIT) midbody protein has important roles in cytokinesis, and its defect leads to MCPH17 in humans (Li et al., 2016a; Basit et al., 2016; Harding et al., 2016; Shaheen et al., 2016). Postmortem histopathological analysis of brain samples taken from MCPH17 individuals reveals a thickened neocortex with disorganized layers and unmyelinated white matter with scattered neurons (Harding et al., 2016). In addition, the cerebellar cortex and hippocampus show dysplastic and hypoplastic features, and Purkinje cells exhibit a simplified dendritic tree where many of them are multinucleated (Harding et al., 2016). Studies conducted in Cit knockout rodent models reveal that NPCs undergo massive apoptosis due to interrupted cytokinesis (Di Cunto et al., 2000; Roberts et al., 2000). In these models, binucleated neurons have been detected in several brain and spinal cord regions; however, apoptosis seems to be more pronounced in the cerebral cortex, granular layers of cerebellum, hippocampus, and olfactory bulb (Di Cunto et al., 2000; Sarkisian et al., 2002). In addition, the high rate of cell death reported at the ganglionic eminence reduces the total number of generated interneurons (Sarkisian et al., 2001; Di Cunto et al., 2000). Consistent with human brain findings, cerebellar Purkinje cells are disorganized and show underdeveloped dendritic complexity (Di Cunto et al., 2000). The presence of disorganized cortical layering and scattered neurons in the white matter should raise the possibility of an abnormal migration process even though it is yet to be confirmed by further studies.
The role of CIT in cytokinesis requires a proper function of kinesin family 14 (KIF14) microtubule motor protein (Makrythanasis et al., 2018). It is then unsurprising to realize that mutations in KIF14 also lead to MCPH by a common mechanism as CIT mutations (Moawia et al., 2017; Makrythanasis et al., 2018; Reilly et al., 2019). This is supported by several studies conducted in various animal models with depleted KIF14 homologs (Ohkura et al., 1997; Fujikura et al., 2013; Reilly et al., 2019). Mutations of Drosophila KIF14 homolog, also known as kinesin-like protein at 38B (KLP38B), affect the cell cycle progression due to cytokinesis failure (Ohkura et al., 1997). In the same notion, Laggard mice (lag), an animal model for KIF14, are characterized by microcephaly, cortical dysgenesis, and severe hypomyelination as a consequence of massive apoptosis during late neurogenesis (Fujikura et al., 2013). Consequently, Cux1-positive upper cortical neurons are much reduced in number, and some of them are displaced (Fujikura et al., 2013). Similar to Cit knockout models, lag mice show scattered cerebellar Purkinje cells with simplified dendritic trees pointing toward abnormalities in neuronal migration and neurite formation (Fujikura et al., 2013).
Disturbed Autophagy and Mitochondrial Dynamics
MCPH18 is caused by mutations in gene encoding WD repeat and FYVE domain-containing 3 (WDFY3), also known as Autophagy-Linked FYVE (ALFY) (Kadir et al., 2016). Normally, this scaffolding protein facilitates the removal of cytosolic protein aggregates, which, in turn, maintains mitochondrial homeostasis (Kadir et al., 2016; Napoli et al., 2018; Napoli et al., 2021). Wdfy3 is highly expressed in RGCs, and its loss of function prevents the transition from symmetrical proliferative divisions to asymmetrical differentiative divisions by altering the Wnt signaling cascade (Tacchelly-Benites et al., 2013; Orosco et al., 2014; Kadir et al., 2016). The imbalance in NPCs mode of cell division leads to regional differences in neocortical thickness and opposing phenotypes of micro- and macrocephaly (Orosco et al., 2014; Le Duc et al., 2019). On the other hand, the disruption of mitochondrial dynamics in Wdfy3 mutant mice decreases the synaptic density, alters the synaptic plasticity, and probably affects dendritic development (Napoli et al., 2018; Napoli et al., 2021). Remarkably, proteins involved in GABAergic neurotransmission are downregulated in Wdfy3 mutant mice (Napoli et al., 2021). Furthermore, Wdfy3 plays a role in neural migration during early neurogenesis (Orosco et al., 2014).
Interrupted Intracellular Trafficking
Coatomer Protein Complex Subunit Beta 2 (COPB2) controls the cellular retrograde trafficking from the Golgi to the endoplasmic reticulum (Orci et al., 1993; Letourneur et al., 1994). Interestingly, mutations in COPB2 interrupt brain growth and lead to MCPH19 (DiStasio et al., 2017). While the complete loss of Copb2 is incompatible with life, partial loss of Copb2 in mice interferes with the growth of the brain (DiStasio et al., 2017). This has been associated with increased cell death and a high number of proliferative cells positive for phosphorylated histone H3 (pH3) (DiStasio et al., 2017). Still, further studies are necessitated to dissect the exact role of Copb2 in controlling brain size.
Disturbed Mitotic Spindle Dynamics
Trafficking protein particle complex subunit 14 (TRAPPC14)—also known as microtubule-associated protein 11 (MAP11)—is localized to mitotic spindles and interacts with α-tubulin regulates the spindle dynamics and cell division (Perez et al., 2019). Recently, TRAPPC14 mutations have been linked to MCPH25 in human and microcephaly phenotypes in the zebrafish model (Perez et al., 2019). This has been mainly attributed to a decreased brain cell proliferation due to altered spindle dynamics affecting the mitotic progression and probably the cytokinesis, however, without increased apoptosis (Perez et al., 2019). In addition, TRAPPC14 has been implicated in ciliogenesis and cilia stability, which, in turn, could affect brain growth (Cuenca et al., 2019).
Defective Ribosome Biogenesis
The most recently diagnosed MCPH28 cases have been linked to a mutation in Ribosomal RNA Processing seven Homolog A (RRP7A) (Farooq et al., 2020). It is known that mutations in genes involved in ribosome biogenesis are associated with neurodevelopmental defects together with other abnormalities (Hetman and Slomnicki, 2019). The encoded RRP7A protein shows high expression in RGCs and cellular localizations at the centrosome, primary cilium, and nucleolus (Farooq et al., 2020). Depletion of RRP7A alters ribosomal RNA processing and affects primary cilia resorption, causing a delay in S-phase entry and progression (Farooq et al., 2020). Mutated rrp7a zebrafish embryos display a reduction in the expression pattern of some proliferation and neural differentiation markers, while TUNEL assay analysis indicates increased apoptosis (Farooq et al., 2020). These findings might result from defective rrp7a functions at the level of centrosome and/or primary cilia. MicroRNA processing has been identified as a contributing factor in temporal fate specification (Kohwi and Doe, 2013). Thus, dysregulated ribosomal RNA processing with subsequent nucleolar stress establishes a new insight into MCPH pathomechanisms.
Microcephaly Primary Hereditary Versus Infection-Induced Microcephaly
After describing the genetic component of microcephaly, we here shed some light on some infectious agents associated with microcephaly. Particularly Toxoplasma gondii, cytomegalovirus, rubella virus, and syphilis, but also the herpes simplex virus, HIV, and Zika virus, have been reported in children born with microcephaly. The severity of microcephaly depends not only on the type of the infectious agent but much importantly on the gestational age when an infection occurs (Devakumar et al., 2018). It has been shown that neural progenitors are targeted by these pathogens; however, the mechanisms by which most of these infections lead to microcephaly are not fully understood (Devakumar et al., 2018). Nevertheless, the epidemic infections with the Zika virus and its association with congenital microcephaly triggered extensive research in this field. Several studies based on various in vitro and in vivo models point toward NPC cell cycle arrest or an increase in cell death upon the infection with the Zika virus (Adams Waldorf et al., 2016; Li et al., 2016b; Cugola et al., 2016; Garcez et al., 2016; Tang et al., 2016; Wu et al., 2016). Intriguingly, several MCPH genes including Mcph1, Aspm, Cdk5rap2, Stil, and Cep135 are downregulated in brain tissues extracted from Zika-infected mice (Li et al., 2016b; Wu et al., 2016). This raises the possibility that infection-induced microcephaly might alter brain growth via altering the expression of various MCPH genes. However, the direct impact of infectious agents on the pathogenesis of microcephaly cannot be ruled out.
Conclusion
The journey during brain growth and development is impeded at specific points with crucial steps (Figure 2). Minor defects at any of these developmental points result in various neurodevelopmental disorders including MCPH. The earlier the insult during the journey, the greater the impact on brain growth. The major obstacle is faced during the rapid NPC proliferation just before the commencing generation of neurons. Either decreased proliferation or increased NPCs apoptosis depletes the neuronal stem cell pool and ultimately leads to a smaller number of generated neurons. Obviously, most MCPHs are caused by mutations in centrosomal proteins. Hence, dysfunctional centrosome alters NPC proliferation, cell cycle progression, and cell fate determination. In addition, the resulting aneuploidy and increased DNA damage response associated with some mutated MCPH genes trigger apoptosis. The accelerated number of discovered MCPH genes expands the pathomechanism spectrum of this disease beyond the centrosomal component. Similarly, the simultaneous generation of animal models mimicking the human MCPH phenotype provides a strong platform for future studies to dissect further molecular mechanisms behind the microcephaly phenotype. This will also expand our knowledge of normal brain growth and evolution.
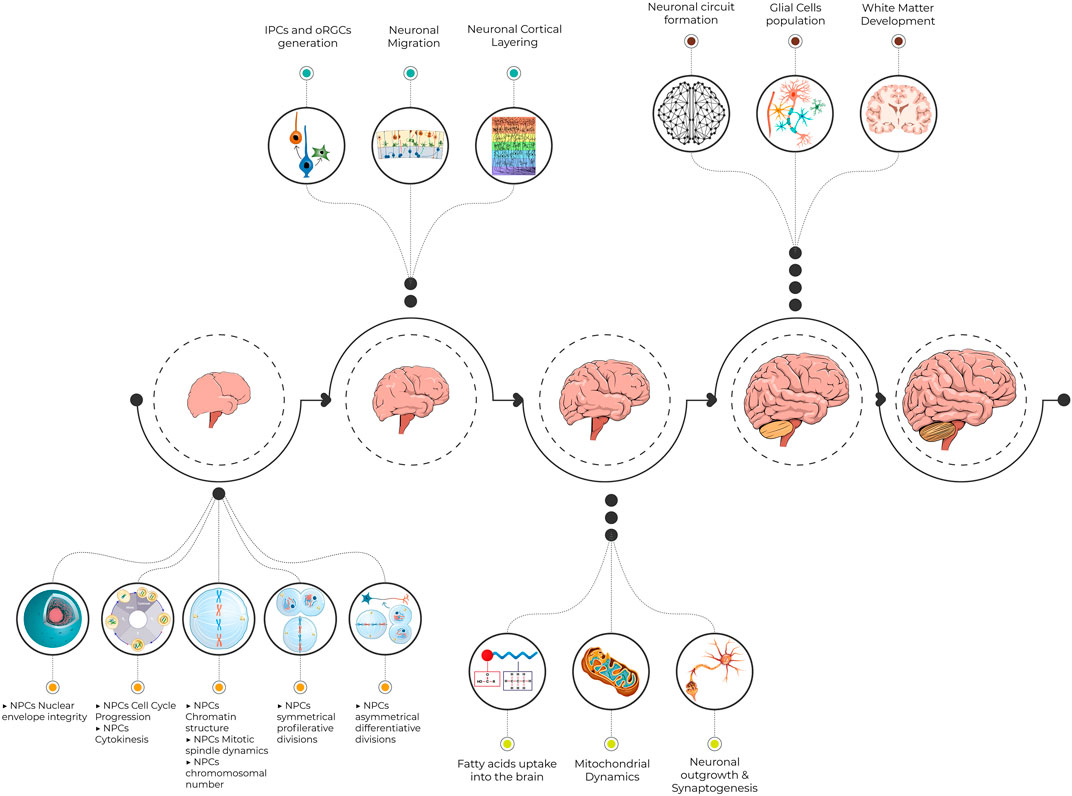
FIGURE 2. An illustrative figure demonstrating the pathway toward normal brain development. Minor defects at crucial steps in neurogenesis result in various neurodevelopmental disorders including MCPH. NPCs, neural progenitor cells; IPCs, intermediate progenitor cells; oRGCs, outer radial glial cells; MCPH, microcephaly primary hereditary.
Author Contributions
SZ wrote the initial manuscript draft and generated the figures and tables. AK revised the manuscript. Both authors approved the final article.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We acknowledge support from the Open Access Publication Fund of Charité—Universitätsmedizin Berlin. We also thank Mr. Ahmed Hany Amarah for his help in designing Figure 2.
References
Abdel-Hamid, M. S., Ismail, M. F., Darwish, H. A., Effat, L. K., Zaki, M. S., and Abdel-Salam, G. M. H. (2016). Molecular and Phenotypic Spectrum ofASPM-Related Primary Microcephaly: Identification of Eight Novel Mutations. Am. J. Med. Genet. 170, 2133–2140. doi:10.1002/ajmg.a.37724
Adams Waldorf, K. M., Stencel-Baerenwald, J. E., Kapur, R. P., Studholme, C., Boldenow, E., Vornhagen, J., et al. (2016). Fetal Brain Lesions after Subcutaneous Inoculation of Zika Virus in a Pregnant Nonhuman Primate. Nat. Med. 22, 1256–1259. doi:10.1038/nm.4193
Aggarwal, A., Mittal, H., Patil, R., Debnath, S., and Rai, A. (2013). Clinical Profile of Children with Developmental Delay and Microcephaly. J. neurosciences Rural Pract. 04, 288–291. doi:10.4103/0976-3147.118781
Alakbarzade, V., Hameed, A., Quek, D. Q. Y., Chioza, B. A., Baple, E. L., Cazenave-Gassiot, A., et al. (2015). A Partially Inactivating Mutation in the Sodium-dependent Lysophosphatidylcholine Transporter MFSD2A Causes a Non-lethal Microcephaly Syndrome. Nat. Genet. 47, 814–817. doi:10.1038/ng.3313
Alcantara, D., and O'Driscoll, M. (2014). Congenital Microcephaly. Am. J. Med. Genet. 166, 124–139. doi:10.1002/ajmg.c.31397
Alfares, A., Alhufayti, I., Alsubaie, L., Alowain, M., Almass, R., Alfadhel, M., et al. (2018). A New Association between CDK5RAP2 Microcephaly and Congenital Cataracts. Ann. Hum. Genet. 82, 165–170. doi:10.1111/ahg.12232
Arai, Y., Pulvers, J. N., Haffner, C., Schilling, B., Nüsslein, I., Calegari, F., et al. (2011). Neural Stem and Progenitor Cells Shorten S-phase on Commitment to Neuron Production. Nat. Commun. 2, 154. doi:10.1038/ncomms1155
Ariani, F., Mari, F., Amitrano, S., Di Marco, C., Artuso, R., Scala, E., et al. (2013). Exome Sequencing Overrides Formal Genetics: ASPM Mutations in a Case Study of Apparent X‐linked Microcephalic Intellectual Deficit. Clin. Genet. 83, 288–290. doi:10.1111/j.1399-0004.2012.01901.x
Arquint, C., and Nigg, E. A. (2014). STIL Microcephaly Mutations Interfere with APC/C-mediated Degradation and Cause Centriole Amplification. Curr. Biol. 24, 351–360. doi:10.1016/j.cub.2013.12.016
Asencio, C., Davidson, I. F., Santarella-Mellwig, R., Ly-Hartig, T. B. N., Mall, M., Wallenfang, M. R., et al. (2012). Coordination of Kinase and Phosphatase Activities by Lem4 Enables Nuclear Envelope Reassembly during Mitosis. Cell 150, 122–135. doi:10.1016/j.cell.2012.04.043
Awad, S., Al-Dosari, M. S., Al-Yacoub, N., Colak, D., Salih, M. A., Alkuraya, F. S., et al. (2013). Mutation in PHC1 Implicates Chromatin Remodeling in Primary Microcephaly Pathogenesis. Hum. Mol. Genet. 22, 2200–2213. doi:10.1093/hmg/ddt072
Bacino, C. A., Arriola, L. A., Wiszniewska, J., and Bonnen, P. E. (2012). WDR62 Missense Mutation in a Consanguineous Family with Primary Microcephaly. Am. J. Med. Genet. 158a, 622–625. doi:10.1002/ajmg.a.34417
Bamborschke, D., Daimagüler, H. S., Hahn, A., Hussain, M. S., Nürnberg, P., and Cirak, S. (2020). Mutation in CEP135 Causing Primary Microcephaly and Subcortical Heterotopia. Am. J. Med. Genet. 182, 2450–2453. doi:10.1002/ajmg.a.61762
Banerjee, S., Chen, H., Huang, H., Wu, J., Yang, Z., Deng, W., et al. (2016). Novel Mutations c.28G>T (p.Ala10Ser) and c.189G>T (p.Glu63Asp) in WDR62 Associated with Early Onset Acanthosis and Hyperkeratosis in a Patient with Autosomal Recessive Microcephaly Type 2. Oncotarget 7, 78363–78371.
Barbelanne, M., and Tsang, W. Y. (2014). Molecular and Cellular Basis of Autosomal Recessive Primary Microcephaly. Biomed. Res. Int. 2014, 547986. doi:10.1155/2014/547986
Basit, S., Al-Harbi, K. M., Alhijji, S. A. M., Albalawi, A. M., Alharby, E., Eldardear, A., et al. (2016). CIT, a Gene Involved in Neurogenic Cytokinesis, Is Mutated in Human Primary Microcephaly. Hum. Genet. 135, 1199–1207. doi:10.1007/s00439-016-1724-0
Bastaki, F., Mohamed, M., Nair, P., Saif, F., Tawfiq, N., Aithala, G., et al. (2016). Novel Splice-Site Mutation inWDR62revealed by Whole-Exome Sequencing in a Sudanese Family with Primary Microcephaly. Congenit. Anom. 56, 135–137. doi:10.1111/cga.12144
Basto, R., Lau, J., Vinogradova, T., Gardiol, A., Woods, C. G., Khodjakov, A., et al. (2006). Flies without Centrioles. Cell 125, 1375–1386. doi:10.1016/j.cell.2006.05.025
Baye, L. M., and Link, B. A. (2007). Interkinetic Nuclear Migration and the Selection of Neurogenic Cell Divisions during Vertebrate Retinogenesis. J. Neurosci. 27, 10143–10152. doi:10.1523/jneurosci.2754-07.2007
Ben-Zvi, A., Lacoste, B., Kur, E., Andreone, B. J., Mayshar, Y., Yan, H., et al. (2014). Mfsd2a Is Critical for the Formation and Function of the Blood-Brain Barrier. Nature 509, 507–511. doi:10.1038/nature13324
Berger, J. H., Charron, M. J., and Silver, D. L. (2012). Major Facilitator Superfamily Domain-Containing Protein 2a (MFSD2A) Has Roles in Body Growth, Motor Function, and Lipid Metabolism. PLoS One 7, e50629. doi:10.1371/journal.pone.0050629
Betizeau, M., Cortay, V., Patti, D., Pfister, S., Gautier, E., Bellemin-Ménard, A., et al. (2013). Precursor Diversity and Complexity of Lineage Relationships in the Outer Subventricular Zone of the Primate. Neuron 80, 442–457. doi:10.1016/j.neuron.2013.09.032
Bhat, V., Girimaji, S., Mohan, G., Arvinda, H., Singhmar, P., Duvvari, M., et al. (2011). Mutations in WDR62, Encoding a Centrosomal and Nuclear Protein, in Indian Primary Microcephaly Families with Cortical Malformations. Clin. Genet. 80, 532–540. doi:10.1111/j.1399-0004.2011.01686.x
Bilgüvar, K., Öztürk, A. K., Louvi, A., Kwan, K. Y., Choi, M., Tatlı, B., et al. (2010). Whole-exome Sequencing Identifies Recessive WDR62 Mutations in Severe Brain Malformations. Nature 467, 207–210. doi:10.1038/nature09327
Blachon, S., Gopalakrishnan, J., Omori, Y., Polyanovsky, A., Church, A., Nicastro, D., et al. (2008). Drosophila Asterless and Vertebrate Cep152 Are Orthologs Essential for Centriole Duplication. Genetics 180, 2081–2094. doi:10.1534/genetics.108.095141
Bogoyevitch, M. A., Yeap, Y. Y., Qu, Z., Ngoei, K. R., Yip, Y. Y., Zhao, T. T., et al. (2012). WD40-repeat Protein 62 Is a JNK-Phosphorylated Spindle Pole Protein Required for Spindle Maintenance and Timely Mitotic Progression. J. Cel Sci 125, 5096–5109. doi:10.1242/jcs.107326
Boycott, K. M., Vanstone, M. R., Bulman, D. E., and MacKenzie, A. E. (2013). Rare-disease Genetics in the Era of Next-Generation Sequencing: Discovery to Translation. Nat. Rev. Genet. 14, 681–691. doi:10.1038/nrg3555
Braun, D. A., Lovric, S., Schapiro, D., Schneider, R., Marquez, J., Asif, M., et al. (2018). Mutations in Multiple Components of the Nuclear Pore Complex Cause Nephrotic Syndrome. J. Clin. Invest. 128, 4313–4328. doi:10.1172/jci98688
Brunk, K., Vernay, B., Griffith, E., Reynolds, N. L., Strutt, D., Ingham, P. W., et al. (2007). Microcephalin Coordinates Mitosis in the syncytialDrosophilaembryo. J. Cel. Sci. 120, 3578–3588. doi:10.1242/jcs.014290
Buchman, J. J., Durak, O., and Tsai, L.-H. (2011). ASPM Regulates Wnt Signaling Pathway Activity in the Developing Brain. Genes Dev. 25, 1909–1914. doi:10.1101/gad.16830211
Buchman, J. J., and Tsai, L.-H. (2007). Spindle Regulation in Neural Precursors of Flies and Mammals. Nat. Rev. Neurosci. 8, 89–100. doi:10.1038/nrn2058
Buchman, J. J., Tseng, H.-C., Zhou, Y., Frank, C. L., Xie, Z., and Tsai, L.-H. (2010). Cdk5rap2 Interacts with Pericentrin to Maintain the Neural Progenitor Pool in the Developing Neocortex. Neuron 66, 386–402. doi:10.1016/j.neuron.2010.03.036
Caglayan, A. O., Yaghouti, K., Kockaya, T., Kemer, D., Cankaya, T., Ameziane, N., et al. (2021). Biallelic ZNF335 Mutations Cause Basal Ganglia Abnormality with Progressive Cerebral/cerebellar Atrophy. J. Neurogenet. 35, 23–28. doi:10.1080/01677063.2020.1833006
Calegari, F., Haubensak, W., Haffner, C., and Huttner, W. B. (2005). Selective Lengthening of the Cell Cycle in the Neurogenic Subpopulation of Neural Progenitor Cells during Mouse Brain Development. J. Neurosci. 25, 6533–6538. doi:10.1523/jneurosci.0778-05.2005
Calegari, F., and Huttner, W. B. (2003). An Inhibition of Cyclin-dependent Kinases that Lengthens, but Does Not Arrest, Neuroepithelial Cell Cycle Induces Premature Neurogenesis. J. Cel. Sci. 116, 4947–4955. doi:10.1242/jcs.00825
Carvalho-Santos, Z., Machado, P., Alvarez-Martins, I., Gouveia, S. M., Jana, S. C., Duarte, P., et al. (2012). BLD10/CEP135 Is a Microtubule-Associated Protein that Controls the Formation of the Flagellum central Microtubule Pair. Developmental Cel 23, 412–424. doi:10.1016/j.devcel.2012.06.001
Chan, J. P., Wong, B. H., Chin, C. F., Galam, D. L. A., Foo, J. C., Wong, L. C., et al. (2018). The Lysolipid Transporter Mfsd2a Regulates Lipogenesis in the Developing Brain. Plos Biol. 16, e2006443. doi:10.1371/journal.pbio.2006443
Chen, J.-F., Zhang, Y., Wilde, J., Hansen, K. C., Lai, F., and Niswander, L. (2014). Microcephaly Disease Gene Wdr62 Regulates Mitotic Progression of Embryonic Neural Stem Cells and Brain Size. Nat. Commun. 5, 3885. doi:10.1038/ncomms4885
Chen, L., Tong, Q., Chen, X., Jiang, P., Yu, H., Zhao, Q., et al. (2021). PHC1 Maintains Pluripotency by Organizing Genome-wide Chromatin Interactions of the Nanog Locus. Nat. Commun. 12, 2829. doi:10.1038/s41467-021-22871-0
Chen, N. Y., Yang, Y., Weston, T. A., Belling, J. N., Heizer, P., Tu, Y., et al. (2019). An Absence of Lamin B1 in Migrating Neurons Causes Nuclear Membrane Ruptures and Cell Death. Proc. Natl. Acad. Sci. USA 116, 25870–25879. doi:10.1073/pnas.1917225116
Cheng, C., Yang, Y., Zhu, X., Yu, X., Zhang, T., Yang, F., et al. (2020). Novel Compound Heterozygous Variants in the STIL Gene Identified in a Chinese Family with Presentation of Foetal Microcephaly. Eur. J. Med. Genet. 63, 104091. doi:10.1016/j.ejmg.2020.104091
Chenn, A., and McConnell, S. K. (1995). Cleavage Orientation and the Asymmetric Inheritance of Notchl Immunoreactivity in Mammalian Neurogenesis. Cell 82, 631–641. doi:10.1016/0092-8674(95)90035-7
Cleveland, D. W., Mao, Y., and Sullivan, K. F. (2003). Centromeres and Kinetochores. Cell 112, 407–421. doi:10.1016/s0092-8674(03)00115-6
Coffinier, C., Chang, S. Y., Nobumori, C., Tu, Y., Farber, E. A., Toth, J. I., et al. (2010). Abnormal Development of the Cerebral Cortex and Cerebellum in the Setting of Lamin B2 Deficiency. Proc. Natl. Acad. Sci. 107, 5076–5081. doi:10.1073/pnas.0908790107
Coffinier, C., Jung, H.-J., Nobumori, C., Chang, S., Tu, Y., Barnes, R. H., et al. (2011). Deficiencies in Lamin B1 and Lamin B2 Cause Neurodevelopmental Defects and Distinct Nuclear Shape Abnormalities in Neurons. MBoC 22, 4683–4693. doi:10.1091/mbc.e11-06-0504
Costa, M. R., and Müller, U. (2014). Specification of Excitatory Neurons in the Developing Cerebral Cortex: Progenitor Diversity and Environmental Influences. Front Cel Neurosci 8, 449. doi:10.3389/fncel.2014.00449
Cottee, M. A., Muschalik, N., Wong, Y. L., Johnson, C. M., Johnson, S., Andreeva, A., et al. (2013). Crystal Structures of the CPAP/STIL Complex Reveal its Role in Centriole Assembly and Human Microcephaly. eLife 2, e01071. doi:10.7554/eLife.01071
Cox, J., Jackson, A. P., Bond, J., and Woods, C. G. (2006). What Primary Microcephaly Can Tell Us about Brain Growth. Trends Mol. Med. 12, 358–366. doi:10.1016/j.molmed.2006.06.006
Cragan, J. D., Isenburg, J. L., Parker, S. E., Alverson, C. J., Meyer, R. E., Stallings, E. B., et al. (2016). Population-based Microcephaly Surveillance in the United States, 2009 to 2013: An Analysis of Potential Sources of Variation. Birth Defects Res. A: Clin. Mol. Teratology 106, 972–982. doi:10.1002/bdra.23587
Cristofoli, F., Moss, T., Moore, H. W., Devriendt, K., Flanagan-Steet, H., May, M., et al. (2020). De Novo Variants in LMNB1 Cause Pronounced Syndromic Microcephaly and Disruption of Nuclear Envelope Integrity. Am. J. Hum. Genet. 107, 753–762. doi:10.1016/j.ajhg.2020.08.015
Cuenca, A., Insinna, C., Zhao, H., John, P., Weiss, M. A., Lu, Q., et al. (2019). The C7orf43/TRAPPC14 Component Links the TRAPPII Complex to Rabin8 for Preciliary Vesicle Tethering at the Mother Centriole during Ciliogenesis. J. Biol. Chem. 294, 15418–15434. doi:10.1074/jbc.ra119.008615
Cugola, F. R., Fernandes, I. R., Russo, F. B., Freitas, B. C., Dias, J. L. M., Guimarães, K. P., et al. (2016). The Brazilian Zika Virus Strain Causes Birth Defects in Experimental Models. Nature 534, 267–271. doi:10.1038/nature18296
David, A., Liu, F., Tibelius, A., Vulprecht, J., Wald, D., Rothermel, U., et al. (2014). Lack of Centrioles and Primary Cilia inSTIL−/−mouse Embryos. Cell Cycle 13, 2859–2868. doi:10.4161/15384101.2014.946830
Del Bene, F., Wehman, A. M., Link, B. A., and Baier, H. (2008). Regulation of Neurogenesis by Interkinetic Nuclear Migration through an Apical-Basal Notch Gradient. Cell 134, 1055–1065. doi:10.1016/j.cell.2008.07.017
Derivery, E., Seum, C., Daeden, A., Loubéry, S., Holtzer, L., Jülicher, F., et al. (2015). Polarized Endosome Dynamics by Spindle Asymmetry during Asymmetric Cell Division. Nature 528, 280–285. doi:10.1038/nature16443
Desir, J., Cassart, M., David, P., Van Bogaert, P., and Abramowicz, M. (2008). Primary Microcephaly with ASPM Mutation Shows Simplified Cortical Gyration with Antero-Posterior Gradient Pre- and post-natally. Am. J. Med. Genet. 146A, 1439–1443. doi:10.1002/ajmg.a.32312
Devakumar, D., Bamford, A., Ferreira, M. U., Broad, J., Rosch, R. E., Groce, N., et al. (2018). Infectious Causes of Microcephaly: Epidemiology, Pathogenesis, Diagnosis, and Management. Lancet Infect. Dis. 18, e1–e13. doi:10.1016/s1473-3099(17)30398-5
Di Cunto, F., Imarisio, S., Hirsch, E., Broccoli, V., Bulfone, A., Migheli, A., et al. (2000). Defective Neurogenesis in Citron Kinase Knockout Mice by Altered Cytokinesis and Massive Apoptosis. Neuron 28, 115–127. doi:10.1016/s0896-6273(00)00090-8
Ding, W., Wu, Q., Sun, L., Pan, N. C., and Wang, X. (2019). Cenpj Regulates Cilia Disassembly and Neurogenesis in the Developing Mouse Cortex. J. Neurosci. 39, 1994–2010. doi:10.1523/jneurosci.1849-18.2018
DiStasio, A., Driver, A., Sund, K., Donlin, M., Muraleedharan, R. M., Pooya, S., et al. (2017). Copb2 Is Essential for Embryogenesis and Hypomorphic Mutations Cause Human Microcephaly. Hum. Mol. Genet. 26, 4836–4848. doi:10.1093/hmg/ddx362
Farag, H. G., Froehler, S., Oexle, K., Ravindran, E., Schindler, D., Staab, T., et al. (2013). Abnormal Centrosome and Spindle Morphology in a Patient with Autosomal Recessive Primary Microcephaly Type 2 Due to Compound Heterozygous WDR62 Gene Mutation. Orphanet J. Rare Dis. 8, 178. doi:10.1186/1750-1172-8-178
Farooq, M., Lindbæk, L., Krogh, N., Doganli, C., Keller, C., Mönnich, M., et al. (2020). RRP7A Links Primary Microcephaly to Dysfunction of Ribosome Biogenesis, Resorption of Primary Cilia, and Neurogenesis. Nat. Commun. 11, 5816. doi:10.1038/s41467-020-19658-0
Fietz, S. A., and Huttner, W. B. (2011). Cortical Progenitor Expansion, Self-Renewal and Neurogenesis-A Polarized Perspective. Curr. Opin. Neurobiol. 21, 23–35. doi:10.1016/j.conb.2010.10.002
Fietz, S. A., Kelava, I., Vogt, J., Wilsch-Bräuninger, M., Stenzel, D., Fish, J. L., et al. (2010). OSVZ Progenitors of Human and Ferret Neocortex Are Epithelial-like and Expand by Integrin Signaling. Nat. Neurosci. 13, 690–699. doi:10.1038/nn.2553
Finley, K. D., Edeen, P. T., Cumming, R. C., Mardahl-Dumesnil, M. D., Taylor, B. J., Rodriguez, M. H., et al. (2003). Blue cheeseMutations Define a Novel, Conserved Gene Involved in Progressive Neural Degeneration. J. Neurosci. 23, 1254–1264. doi:10.1523/jneurosci.23-04-01254.2003
Fish, J. L., Kosodo, Y., Enard, W., Paabo, S., and Huttner, W. B. (2006). Aspm Specifically Maintains Symmetric Proliferative Divisions of Neuroepithelial Cells. Proc. Natl. Acad. Sci. 103, 10438–10443. doi:10.1073/pnas.0604066103
Fujikura, K., Setsu, T., Tanigaki, K., Abe, T., Kiyonari, H., Terashima, T., et al. (2013). Kif14 Mutation Causes Severe Brain Malformation and Hypomyelination. PLoS One 8, e53490. doi:10.1371/journal.pone.0053490
Fujimori, A., Itoh, K., Goto, S., Hirakawa, H., Wang, B., Kokubo, T., et al. (2014). Disruption of Aspm Causes Microcephaly with Abnormal Neuronal Differentiation. Brain Development 36, 661–669. doi:10.1016/j.braindev.2013.10.006
Gabriel, E., Ramani, A., Altinisik, N., and Gopalakrishnan, J. (2020). Human Brain Organoids to Decode Mechanisms of Microcephaly. Front. Cell. Neurosci. 14, 115. doi:10.3389/fncel.2020.00115
Gabriel, E., Wason, A., Ramani, A., Gooi, L. M., Keller, P., Pozniakovsky, A., et al. (2016). CPAP Promotes Timely Cilium Disassembly to Maintain Neural Progenitor Pool. Embo J. 35, 803–819. doi:10.15252/embj.201593679
Garcez, P. P., Loiola, E. C., Madeiro da Costa, R., Higa, L. M., Trindade, P., Delvecchio, R., et al. (2016). Zika Virus Impairs Growth in Human Neurospheres and Brain Organoids. Science 352, 816–818. doi:10.1126/science.aaf6116
Genin, A., Desir, J., Lambert, N., Biervliet, M., Van Der Aa, N., Pierquin, G., et al. (2012). Kinetochore KMN Network Gene CASC5 Mutated in Primary Microcephaly. Hum. Mol. Genet. 21, 5306–5317. doi:10.1093/hmg/dds386
Gilbert, S. L., Dobyns, W. B., and Lahn, B. T. (2005). Genetic Links between Brain Development and Brain Evolution. Nat. Rev. Genet. 6, 581–590. doi:10.1038/nrg1634
Goetz, S. C., and Anderson, K. V. (2010). The Primary Cilium: a Signalling centre during Vertebrate Development. Nat. Rev. Genet. 11, 331–344. doi:10.1038/nrg2774
Gonzalez, C., Saunders, R. D., Casal, J., Molina, I., Carmena, M., Ripoll, P., et al. (1990). Mutations at the Asp Locus of Drosophila lead to Multiple Free Centrosomes in Syncytial Embryos, but Restrict Centrosome Duplication in Larval Neuroblasts. J. Cel Sci 96 (Pt 4), 605–616. doi:10.1242/jcs.96.4.605
Gosling, K. M., Makaroff, L. E., Theodoratos, A., Kim, Y.-H., Whittle, B., Rui, L., et al. (2007). A Mutation in a Chromosome Condensin II Subunit, Kleisin Beta, Specifically Disrupts T Cell Development. Proc. Natl. Acad. Sci. 104, 12445–12450. doi:10.1073/pnas.0704870104
Götz, M., and Huttner, W. B. (2005). The Cell Biology of Neurogenesis. Nat. Rev. Mol. Cel. Biol. 6, 777–788.
Gruber, R., Zhou, Z., Sukchev, M., Joerss, T., Frappart, P.-O., and Wang, Z.-Q. (2011). MCPH1 Regulates the Neuroprogenitor Division Mode by Coupling the Centrosomal Cycle with Mitotic Entry through the Chk1-Cdc25 Pathway. Nat. Cel Biol 13, 1325–1334. doi:10.1038/ncb2342
Guemez-Gamboa, A., Nguyen, L. N., Yang, H., Zaki, M. S., Kara, M., Ben-Omran, T., et al. (2015). Inactivating Mutations in MFSD2A, Required for omega-3 Fatty Acid Transport in Brain, Cause a Lethal Microcephaly Syndrome. Nat. Genet. 47, 809–813. doi:10.1038/ng.3311
Hainline, S. G., Rickmyre, J. L., Neitzel, L. R., Lee, L. A., and Lee, E. (2014). The Drosophila MCPH1-B Isoform Is a Substrate of the APCCdh1 E3 Ubiquitin Ligase Complex. Biol. open 3, 669–676. doi:10.1242/bio.20148318
Hansen, D. V., Lui, J. H., Parker, P. R. L., and Kriegstein, A. R. (2010). Neurogenic Radial Glia in the Outer Subventricular Zone of Human Neocortex. Nature 464, 554–561. doi:10.1038/nature08845
Harding, B. N., Moccia, A., Drunat, S., Soukarieh, O., Tubeuf, H., Chitty, L. S., et al. (2016). Mutations in Citron Kinase Cause Recessive Microlissencephaly with Multinucleated Neurons. Am. J. Hum. Genet. 99, 511–520. doi:10.1016/j.ajhg.2016.07.003
Harel, T., Quek, D. Q. Y., Wong, B. H., Cazenave-Gassiot, A., Wenk, M. R., Fan, H., et al. (2018). Homozygous Mutation in MFSD2A, Encoding a Lysolipid Transporter for Docosahexanoic Acid, Is Associated with Microcephaly and Hypomyelination. Neurogenetics 19, 227–235. doi:10.1007/s10048-018-0556-6
Heide, M., and Huttner, W. B. (2021). Human-Specific Genes, Cortical Progenitor Cells, and Microcephaly. Cells 10, 1209. doi:10.3390/cells10051209
Hetman, M., and Slomnicki, L. P. (2019). Ribosomal Biogenesis as an Emerging Target of Neurodevelopmental Pathologies. J. Neurochem. 148, 325–347. doi:10.1111/jnc.14576
Hiraki, M., Nakazawa, Y., Kamiya, R., and Hirono, M. (2007). Bld10p Constitutes the Cartwheel-Spoke Tip and Stabilizes the 9-fold Symmetry of the Centriole. Curr. Biol. 17, 1778–1783. doi:10.1016/j.cub.2007.09.021
Homem, C. C. F., Repic, M., and Knoblich, J. A. (2015). Proliferation Control in Neural Stem and Progenitor Cells. Nat. Rev. Neurosci. 16, 647–659. doi:10.1038/nrn4021
Hori, T., and Fukagawa, T. (2012). Establishment of the Vertebrate Kinetochores. Chromosome Res. 20, 547–561. doi:10.1007/s10577-012-9289-9
Hosseini, M. M., Tonekaboni, S. H., Papari, E., Bahman, I., Behjati, F., Kahrizi, K., et al. (2012). A Novel Mutation in MCPH1 Gene in an Iranian Family with Primary Microcephaly. J. Pak Med. Assoc. 62, 1244–1247.
Huang, B., and Li, X. (2021). The Role of Mfsd2a in Nervous System Diseases. Front. Neurosci. 15, 730534. doi:10.3389/fnins.2021.730534
Hussain, M. S., Baig, S. M., Neumann, S., Nürnberg, G., Farooq, M., Ahmad, I., et al. (2012). A Truncating Mutation of CEP135 Causes Primary Microcephaly and Disturbed Centrosomal Function. Am. J. Hum. Genet. 90, 871–878. doi:10.1016/j.ajhg.2012.03.016
Hussain, M. S., Baig, S. M., Neumann, S., Peche, V. S., Szczepanski, S., Nürnberg, G., et al. (2013). CDK6 Associates with the Centrosome during Mitosis and Is Mutated in a Large Pakistani Family with Primary Microcephaly. Hum. Mol. Genet. 22, 5199–5214. doi:10.1093/hmg/ddt374
Issa, L., Mueller, K., Seufert, K., Kraemer, N., Rosenkotter, H., Ninnemann, O., et al. (2013). Clinical and Cellular Features in Patients with Primary Autosomal Recessive Microcephaly and a Novel CDK5RAP2 Mutation. Orphanet J. Rare Dis. 8, 59–73. doi:10.1186/1750-1172-8-59
Izraeli, S., Lowe, L. A., Bertness, V. L., Good, D. J., Dorward, D. W., Kirsch, I. R., et al. (1999). The SIL Gene Is Required for Mouse Embryonic Axial Development and Left-Right Specification. Nature 399, 691–694. doi:10.1038/21429
Jackson, A. P., Eastwood, H., Bell, S. M., Adu, J., Toomes, C., Carr, I. M., et al. (2002). Identification of Microcephalin, a Protein Implicated in Determining the Size of the Human Brain. Am. J. Hum. Genet. 71, 136–142. doi:10.1086/341283
Jamieson, C. R., Govaerts, C., and Abramowicz, M. J. (1999). Primary Autosomal Recessive Microcephaly: Homozygosity Mapping of MCPH4 to Chromosome 15. Am. J. Hum. Genet. 65, 1465–1469. doi:10.1086/302640
Jayaraman, D., Bae, B.-I., and Walsh, C. A. (2018). The Genetics of Primary Microcephaly. Annu. Rev. Genom. Hum. Genet. 19, 177–200. doi:10.1146/annurev-genom-083117-021441
Jean, F., Stuart, A., and Tarailo-Graovac, M. (2020). Dissecting the Genetic and Etiological Causes of Primary Microcephaly. Front. Neurol. 11, 570830. doi:10.3389/fneur.2020.570830
Jerka-Dziadosz, M., Gogendeau, D., Klotz, C., Cohen, J., Beisson, J., and Koll, F. (2010). Basal Body Duplication in Paramecium: the Key Role of Bld10 in Assembly and Stability of the Cartwheel. Cytoskeleton (Hoboken) 67, 161–171. doi:10.1002/cm.20433
Johnson, M. B., Sun, X., Kodani, A., Borges-Monroy, R., Girskis, K. M., Ryu, S. C., et al. (2018). Aspm Knockout Ferret Reveals an Evolutionary Mechanism Governing Cerebral Cortical Size. Nature 556, 370–375. doi:10.1038/s41586-018-0035-0
Kadir, R., Harel, T., Markus, B., Perez, Y., Bakhrat, A., Cohen, I., et al. (2016). ALFY-controlled DVL3 Autophagy Regulates Wnt Signaling, Determining Human Brain Size. Plos Genet. 12, e1005919. doi:10.1371/journal.pgen.1005919
Kaindl, A. M., Passemard, S., Kumar, P., Kraemer, N., Issa, L., Zwirner, A., et al. (2010). Many Roads lead to Primary Autosomal Recessive Microcephaly. Prog. Neurobiol. 90, 363–383. doi:10.1016/j.pneurobio.2009.11.002
Kakar, N., Ahmad, J., Morris-Rosendahl, D. J., Altmüller, J., Friedrich, K., Barbi, G., et al. (2015). STIL Mutation Causes Autosomal Recessive Microcephalic Lobar Holoprosencephaly. Hum. Genet. 134, 45–51. doi:10.1007/s00439-014-1487-4
Kettleborough, R. N. W., Busch-Nentwich, E. M., Harvey, S. A., Dooley, C. M., de Bruijn, E., van Eeden, F., et al. (2013). A Systematic Genome-wide Analysis of Zebrafish Protein-Coding Gene Function. Nature 496, 494–497. doi:10.1038/nature11992
Khan, M. A., Rupp, V. M., Orpinell, M., Hussain, M. S., Altmüller, J., Steinmetz, M. O., et al. (2014). A Missense Mutation in the PISA Domain of HsSAS-6 Causes Autosomal Recessive Primary Microcephaly in a Large Consanguineous Pakistani Family. Hum. Mol. Genet. 23, 5940–5949. doi:10.1093/hmg/ddu318
Kim, H.-T., Lee, M.-S., Choi, J.-H., Jung, J.-Y., Ahn, D.-G., Yeo, S.-Y., et al. (2011). The Microcephaly Gene Aspm Is Involved in Brain Development in Zebrafish. Biochem. Biophysical Res. Commun. 409, 640–644. doi:10.1016/j.bbrc.2011.05.056
Kim, Y., Sharov, A. A., McDole, K., Cheng, M., Hao, H., Fan, C.-M., et al. (2011). Mouse B-type Lamins Are Required for Proper Organogenesis but Not by Embryonic Stem Cells. Science 334, 1706–1710. doi:10.1126/science.1211222
Kleber de Oliveira, W., Cortez-Escalante, J., De Oliveira, W. T. G. H., do Carmo, G. M. I., Henriques, C. M. P., Coelho, G. E., et al. (2016). Increase in Reported Prevalence of Microcephaly in Infants Born to Women Living in Areas with Confirmed Zika Virus Transmission during the First Trimester of Pregnancy - Brazil, 2015. MMWR Morb. Mortal. Wkly. Rep. 65, 242–247. doi:10.15585/mmwr.mm6509e2
Knoblich, J. A. (2008). Mechanisms of Asymmetric Stem Cell Division. Cell 132, 583–597. doi:10.1016/j.cell.2008.02.007
Kohwi, M., and Doe, C. Q. (2013). Temporal Fate Specification and Neural Progenitor Competence during Development. Nat. Rev. Neurosci. 14, 823–838. doi:10.1038/nrn3618
Konno, D., Shioi, G., Shitamukai, A., Mori, A., Kiyonari, H., Miyata, T., et al. (2008). Neuroepithelial Progenitors Undergo LGN-dependent Planar Divisions to Maintain Self-Renewability during Mammalian Neurogenesis. Nat. Cel Biol 10, 93–101. doi:10.1038/ncb1673
Kosodo, Y., Röper, K., Haubensak, W., Marzesco, A.-M., Corbeil, D., and Huttner, W. B. (2004). Asymmetric Distribution of the Apical Plasma Membrane during Neurogenic Divisions of Mammalian Neuroepithelial Cells. Embo J. 23, 2314–2324. doi:10.1038/sj.emboj.7600223
Kosodo, Y., Suetsugu, T., Suda, M., Mimori-Kiyosue, Y., Toida, K., Baba, S. A., et al. (2011). Regulation of Interkinetic Nuclear Migration by Cell Cycle-Coupled Active and Passive Mechanisms in the Developing Brain. EMBO J. 30, 1690–1704. doi:10.1038/emboj.2011.81
Kousar, R., Hassan, M. J., Khan, B., Basit, S., Mahmood, S., Mir, A., et al. (2011). Mutations in WDR62 Gene in Pakistani Families with Autosomal Recessive Primary Microcephaly. BMC Neurol. 11, 119. doi:10.1186/1471-2377-11-119
Kraemer, N., Issa, L., Hauck, S. C. R., Mani, S., Ninnemann, O., and Kaindl, A. M. (2011). What's the Hype about CDK5RAP2? Cell. Mol. Life Sci. 68, 1719–1736. doi:10.1007/s00018-011-0635-4
Lancaster, M. A., Renner, M., Martin, C.-A., Wenzel, D., Bicknell, L. S., Hurles, M. E., et al. (2013). Cerebral Organoids Model Human Brain Development and Microcephaly. Nature 501, 373–379. doi:10.1038/nature12517
Latasa, M. J., Cisneros, E., and Frade, J. M. (2009). Cell Cycle Control of Notch Signaling and the Functional Regionalization of the Neuroepithelium during Vertebrate Neurogenesis. Int. J. Dev. Biol. 53, 895–908. doi:10.1387/ijdb.082721ml
Le Duc, D., Giulivi, C., Hiatt, S. M., Napoli, E., Panoutsopoulos, A., Harlan De Crescenzo, A., et al. (2019). Pathogenic WDFY3 Variants Cause Neurodevelopmental Disorders and Opposing Effects on Brain Size. Brain. 142, 2617–2630. doi:10.1093/brain/awz198
Leidel, S., Delattre, M., Cerutti, L., Baumer, K., and Gönczy, P. (2005). SAS-6 Defines a Protein Family Required for Centrosome Duplication in C. elegans and in Human Cells. Nat. Cel Biol 7, 115–125. doi:10.1038/ncb1220
Leone, D. P., Srinivasan, K., Chen, B., Alcamo, E., and McConnell, S. K. (2008). The Determination of Projection Neuron Identity in the Developing Cerebral Cortex. Curr. Opin. Neurobiol. 18, 28–35. doi:10.1016/j.conb.2008.05.006
Létard, P., Drunat, S., Vial, Y., Duerinckx, S., Ernault, A., Amram, D., et al. (2018). Autosomal Recessive Primary Microcephaly Due to ASPM Mutations: An Update. Hum. Mutat. 39, 319–332.
Letourneur, F., Gaynor, E. C., Hennecke, S., Démollière, C., Duden, R., Emr, S. D., et al. (1994). Coatomer Is Essential for Retrieval of Dilysine-Tagged Proteins to the Endoplasmic Reticulum. Cell 79, 1199–1207. doi:10.1016/0092-8674(94)90011-6
Leventer, R. J., Jansen, A., Pilz, D. T., Stoodley, N., Marini, C., Dubeau, F., et al. (2010). Clinical and Imaging Heterogeneity of Polymicrogyria: a Study of 328 Patients. Brain. 133, 1415–1427. doi:10.1093/brain/awq078
Li, C., Xu, D., Ye, Q., Hong, S., Jiang, Y., Liu, X., et al. (2016). Zika Virus Disrupts Neural Progenitor Development and Leads to Microcephaly in Mice. Cell Stem Cell 19, 120–126. doi:10.1016/j.stem.2016.04.017
Li, H., Bielas, S. L., Zaki, M. S., Ismail, S., Farfara, D., Um, K., et al. (2016). Biallelic Mutations in Citron Kinase Link Mitotic Cytokinesis to Human Primary Microcephaly. Am. J. Hum. Genet. 99, 501–510. doi:10.1016/j.ajhg.2016.07.004
Li, R., Sun, L., Fang, A., Li, P., Wu, Q., and Wang, X. (2017). Recapitulating Cortical Development with Organoid Culture In Vitro and Modeling Abnormal Spindle-like (ASPM Related Primary) Microcephaly Disease. Protein Cell 8, 823–833. doi:10.1007/s13238-017-0479-2
Liang, Y., Gao, H., Lin, S.-Y., Peng, G., Huang, X., Zhang, P., et al. (2010). BRIT1/MCPH1 Is Essential for Mitotic and Meiotic Recombination DNA Repair and Maintaining Genomic Stability in Mice. Plos Genet. 6, e1000826. doi:10.1371/journal.pgen.1000826
Lim, A., and Kraut, R. (2009). The Drosophila BEACH Family Protein, Blue Cheese, Links Lysosomal Axon Transport with Motor Neuron Degeneration. J. Neurosci. 29, 951–963. doi:10.1523/jneurosci.2582-08.2009
Link, N., Chung, H., Jolly, A., Withers, M., Tepe, B., Arenkiel, B. R., et al. (2019). Mutations in ANKLE2, a ZIKA Virus Target, Disrupt an Asymmetric Cell Division Pathway in Drosophila Neuroblasts to Cause Microcephaly. Developmental Cel 51, 713–729. e716. doi:10.1016/j.devcel.2019.10.009
Lizarraga, S. B., Margossian, S. P., Harris, M. H., Campagna, D. R., Han, A.-P., Blevins, S., et al. (2010). Cdk5rap2 Regulates Centrosome Function and Chromosome Segregation in Neuronal Progenitors. Development 137, 1907–1917. doi:10.1242/dev.040410
Lucas, E. P., and Raff, J. W. (2007). Maintaining the Proper Connection between the Centrioles and the Pericentriolar Matrix Requires Drosophila Centrosomin. J. Cel Biol 178, 725–732. doi:10.1083/jcb.200704081
Mahmood, S., Ahmad, W., and Hassan, M. J. (2011). Autosomal Recessive Primary Microcephaly (MCPH): Clinical Manifestations, Genetic Heterogeneity and Mutation Continuum. Orphanet J. Rare Dis. 6, 39–54. doi:10.1186/1750-1172-6-39
Makrythanasis, P., Maroofian, R., Stray-Pedersen, A., Musaev, D., Zaki, M. S., Mahmoud, I. G., et al. (2018). Biallelic Variants in KIF14 Cause Intellectual Disability with Microcephaly. Eur. J. Hum. Genet. 26, 330–339. doi:10.1038/s41431-017-0088-9
Malatesta, P., Hartfuss, E., and Gotz, M. (2000). Isolation of Radial Glial Cells by Fluorescent-Activated Cell Sorting Reveals a Neuronal Lineage. Development 127, 5253–5263. doi:10.1242/dev.127.24.5253
Malumbres, M., Sotillo, R., Santamaría, D., Galán, J., Cerezo, A., Ortega, S., et al. (2004). Mammalian Cells Cycle without the D-type Cyclin-dependent Kinases Cdk4 and Cdk6. Cell 118, 493–504. doi:10.1016/j.cell.2004.08.002
Marín, O. (2013). Cellular and Molecular Mechanisms Controlling the Migration of Neocortical Interneurons. Eur. J. Neurosci. 38, 2019–2029.
Marjanović, M., Sánchez-Huertas, C., Terré, B., Gómez, R., Scheel, J. F., Pacheco, S., et al. (2015). CEP63 Deficiency Promotes P53-dependent Microcephaly and Reveals a Role for the Centrosome in Meiotic Recombination. Nat. Commun. 6, 7676. doi:10.1038/ncomms8676
Martin, C.-A., Murray, J. E., Carroll, P., Leitch, A., Mackenzie, K. J., Halachev, M., et al. (2016). Mutations in Genes Encoding Condensin Complex Proteins Cause Microcephaly through Decatenation Failure at Mitosis. Genes Dev. 30, 2158–2172. doi:10.1101/gad.286351.116
Matsuura, K., Lefebvre, P. A., Kamiya, R., and Hirono, M. (2004). Bld10p, a Novel Protein Essential for Basal Body Assembly in Chlamydomonas. J. Cel Biol 165, 663–671. doi:10.1083/jcb.200402022
McHenry, H. M. (1994). Tempo and Mode in Human Evolution. Proc. Natl. Acad. Sci. 91, 6780–6786. doi:10.1073/pnas.91.15.6780
McIntyre, R. E., Lakshminarasimhan Chavali, P., Ismail, O., Carragher, D. M., Sanchez-Andrade, G., Forment, J. V., et al. (2012). Disruption of Mouse Cenpj, a Regulator of Centriole Biogenesis, Phenocopies Seckel Syndrome. Plos Genet. 8, e1003022. doi:10.1371/journal.pgen.1003022
Megraw, T. L., Li, K., Kao, L. R., and Kaufman, T. C. (1999). The Centrosomin Protein Is Required for Centrosome Assembly and Function during Cleavage in Drosophila. Development 126, 2829–2839. doi:10.1242/dev.126.13.2829
Megraw, T. L., Sharkey, J. T., and Nowakowski, R. S. (2011). Cdk5rap2 Exposes the Centrosomal Root of Microcephaly Syndromes. Trends Cel Biol. 21, 470–480. doi:10.1016/j.tcb.2011.04.007
Memon, M. M., Raza, S. I., Basit, S., Kousar, R., Ahmad, W., and Ansar, M. (2013). A Novel WDR62 Mutation Causes Primary Microcephaly in a Pakistani Family. Mol. Biol. Rep. 40, 591–595. doi:10.1007/s11033-012-2097-7
Mirzaa, G. M., Vitre, B., Carpenter, G., Abramowicz, I., Gleeson, J. G., Paciorkowski, A. R., et al. (2014). Mutations in CENPE Define a Novel Kinetochore-Centromeric Mechanism for Microcephalic Primordial Dwarfism. Hum. Genet. 133, 1023–1039. doi:10.1007/s00439-014-1443-3
Miyata, T., Okamoto, M., Shinoda, T., and Kawaguchi, A. (2014). Interkinetic Nuclear Migration Generates and Opposes Ventricular-Zone Crowding: Insight into Tissue Mechanics. Front. Cel Neurosci 8, 473. doi:10.3389/fncel.2014.00473
Moawia, A., Shaheen, R., Rasool, S., Waseem, S. S., Ewida, N., Budde, B., et al. (2017). Mutations of KIF14 Cause Primary Microcephaly by Impairing Cytokinesis. Ann. Neurol. 82, 562–577. doi:10.1002/ana.25044
Molyneaux, B. J., Arlotta, P., Menezes, J. R. L., and Macklis, J. D. (2007). Neuronal Subtype Specification in the Cerebral Cortex. Nat. Rev. Neurosci. 8, 427–437. doi:10.1038/nrn2151
Mori, T., Buffo, A., and Götz, M. (2005). The Novel Roles of Glial Cells Revisited: the Contribution of Radial Glia and Astrocytes to Neurogenesis. Curr. Top. Dev. Biol. 69, 67–99. doi:10.1016/s0070-2153(05)69004-7
Morin, X., Jaouen, F., and Durbec, P. (2007). Control of Planar Divisions by the G-Protein Regulator LGN Maintains Progenitors in the Chick Neuroepithelium. Nat. Neurosci. 10, 1440–1448. doi:10.1038/nn1984
Morris, J. K., Rankin, J., Garne, E., Loane, M., Greenlees, R., Addor, M.-C., et al. (2016). Prevalence of Microcephaly in Europe: Population Based Study. Bmj 354, i4721. doi:10.1136/bmj.i4721
Mottier-Pavie, V., and Megraw, T. L. (2009). DrosophilaBld10 Is a Centriolar Protein that Regulates Centriole, Basal Body, and Motile Cilium Assembly. MBoC 20, 2605–2614. doi:10.1091/mbc.e08-11-1115
Mouden, C., de Tayrac, M., Dubourg, C., Rose, S., Carré, W., Hamdi-Rozé, H., et al. (2015). Homozygous STIL Mutation Causes Holoprosencephaly and Microcephaly in Two Siblings. PLoS One 10, e0117418. doi:10.1371/journal.pone.0117418
Murdock, D. R., Clark, G. D., Bainbridge, M. N., Newsham, I., Wu, Y.-Q., Muzny, D. M., et al. (2011). Whole-exome Sequencing Identifies Compound Heterozygous Mutations in WDR62 in Siblings with Recurrent Polymicrogyria. Am. J. Med. Genet. 155, 2071–2077. doi:10.1002/ajmg.a.34165
Musacchio, A., and Salmon, E. D. (2007). The Spindle-Assembly Checkpoint in Space and Time. Nat. Rev. Mol. Cel Biol 8, 379–393. doi:10.1038/nrm2163
Muzio, L., and Consalez, G. G. (2013). Modeling Human Brain Development with Cerebral Organoids. Stem Cel Res Ther 4, 154. doi:10.1186/scrt384
Naim, V., Imarisio, S., Di Cunto, F., Gatti, M., and Bonaccorsi, S. (2004). DrosophilaCitron Kinase Is Required for the Final Steps of Cytokinesis. MBoC 15, 5053–5063. doi:10.1091/mbc.e04-06-0536
Napoli, E., Panoutsopoulos, A. A., Kysar, P., Satriya, N., Sterling, K., Shibata, B., et al. (2021). Wdfy3 Regulates Glycophagy, Mitophagy, and Synaptic Plasticity. J. Cereb. Blood Flow Metab., 0271678X2110273. doi:10.1177/0271678x211027384
Napoli, E., Song, G., Panoutsopoulos, A., Riyadh, M. A., Kaushik, G., Halmai, J., et al. (2018). Beyond Autophagy: a Novel Role for Autism-Linked Wdfy3 in Brain Mitophagy. Sci. Rep. 8, 11348. doi:10.1038/s41598-018-29421-7
Nardello, R., Fontana, A., Antona, V., Beninati, A., Mangano, G. D., Stallone, M. C., et al. (2018). A Novel Mutation of WDR62 Gene Associated with Severe Phenotype Including Infantile Spasm, Microcephaly, and Intellectual Disability. Brain Development 40, 58–64. doi:10.1016/j.braindev.2017.07.003
Nasser, H., Vera, L., Elmaleh-Bergès, M., Steindl, K., Letard, P., Teissier, N., et al. (2020). CDK5RAP2 Primary Microcephaly Is Associated with Hypothalamic, Retinal and Cochlear Developmental Defects. J. Med. Genet. 57, 389–399. doi:10.1136/jmedgenet-2019-106474
Neitzel, H., Neumann, L. M., Schindler, D., Wirges, A., Tönnies, H., Trimborn, M., et al. (2002). Premature Chromosome Condensation in Humans Associated with Microcephaly and Mental Retardation: a Novel Autosomal Recessive Condition. Am. J. Hum. Genet. 70, 1015–1022. doi:10.1086/339518
Nerli, E., Rocha-Martins, M., and Norden, C. (2020). Asymmetric Neurogenic Commitment of Retinal Progenitors Involves Notch through the Endocytic Pathway. eLife 9. doi:10.7554/eLife.60462
Nguyen, L. N., Ma, D., Shui, G., Wong, P., Cazenave-Gassiot, A., Zhang, X., et al. (2014). Mfsd2a Is a Transporter for the Essential omega-3 Fatty Acid Docosahexaenoic Acid. Nature 509, 503–506. doi:10.1038/nature13241
Nicholas, A. K., Khurshid, M., Désir, J., Carvalho, O. P., Cox, J. J., Thornton, G., et al. (2010). WDR62 Is Associated with the Spindle Pole and Is Mutated in Human Microcephaly. Nat. Genet. 42, 1010–1014. doi:10.1038/ng.682
Nigg, E. A., and Raff, J. W. (2009). Centrioles, Centrosomes, and Cilia in Health and Disease. Cell 139, 663–678. doi:10.1016/j.cell.2009.10.036
Noctor, S. C., Martínez-Cerdeño, V., Ivic, L., and Kriegstein, A. R. (2004). Cortical Neurons Arise in Symmetric and Asymmetric Division Zones and Migrate through Specific Phases. Nat. Neurosci. 7, 136–144. doi:10.1038/nn1172
Novorol, C., Burkhardt, J., Wood, K. J., Iqbal, A., Roque, C., Coutts, N., et al. (2013). Microcephaly Models in the Developing Zebrafish Retinal Neuroepithelium point to an Underlying Defect in Metaphase Progression. Open Biol. 3, 130065. doi:10.1098/rsob.130065
Ohkura, H., Török, T., Tick, G., Hoheisel, J., Kiss, I., and Glover, D. M. (1997). Mutation of a Gene for a Drosophila Kinesin-like Protein, Klp38B, Leads to Failure of Cytokinesis. J. Cel Sci 110 ( Pt 8) (Pt 8), 945–954. doi:10.1242/jcs.110.8.945
Okamoto, M., Shinoda, T., Kawaue, T., Nagasaka, A., and Miyata, T. (2014). Ferret-mouse Differences in Interkinetic Nuclear Migration and Cellular Densification in the Neocortical Ventricular Zone. Neurosci. Res. 86, 88–95. doi:10.1016/j.neures.2014.10.006
Orci, L., Palmer, D. J., Ravazzola, M., Perrelet, A., Amherdt, M., and Rothman, J. E. (1993). Budding from Golgi Membranes Requires the Coatomer Complex of Non-clathrin Coat Proteins. Nature 362, 648–652. doi:10.1038/362648a0
Orosco, L. A., Ross, A. P., Cates, S. L., Scott, S. E., Wu, D., Sohn, J., et al. (2014). Loss of Wdfy3 in Mice Alters Cerebral Cortical Neurogenesis Reflecting Aspects of the Autism Pathology. Nat. Commun. 5, 4692. doi:10.1038/ncomms5692
Paridaen, J. T. M. L., Wilsch-Bräuninger, M., and Huttner, W. B. (2013). Asymmetric Inheritance of Centrosome-Associated Primary Cilium Membrane Directs Ciliogenesis after Cell Division. Cell 155, 333–344. doi:10.1016/j.cell.2013.08.060
Parry, D. A., Martin, C.-A., Greene, P., Marsh, J. A., Ambrose, J. C., Arumugam, P., et al. (2021). Heterozygous Lamin B1 and Lamin B2 Variants Cause Primary Microcephaly and Define a Novel Laminopathy. Genet. Med. 23, 408–414. doi:10.1038/s41436-020-00980-3
Passemard, S., Kaindl, A. M., and Verloes, A. (2013). “Chapter 13 - Microcephaly,” in Handbook of Clinical Neurology. Editors O. Dulac, M. Lassonde, and H. B. Sarnat (Elsevier), 129–141.
Passemard, S., Titomanlio, L., Elmaleh, M., Afenjar, A., Alessandri, J.-L., Andria, G., et al. (2009). Expanding the Clinical and Neuroradiologic Phenotype of Primary Microcephaly Due to ASPM Mutations. Neurology 73, 962–969. doi:10.1212/wnl.0b013e3181b8799a
Perez, Y., Bar-Yaacov, R., Kadir, R., Wormser, O., Shelef, I., Birk, O. S., et al. (2019). Mutations in the Microtubule-Associated Protein MAP11 (C7orf43) Cause Microcephaly in Humans and Zebrafish. Brain. 142, 574–585. doi:10.1093/brain/awz004
Pervaiz, N., Kang, H., Bao, Y., and Abbasi, A. A. (2021). Molecular Evolutionary Analysis of Human Primary Microcephaly Genes. BMC Ecol. Evo 21, 76. doi:10.1186/s12862-021-01801-0
Pfaff, K. L., Straub, C. T., Chiang, K., Bear, D. M., Zhou, Y., and Zon, L. I. (2007). The Zebra Fish Cassiopeia Mutant Reveals that SIL Is Required for Mitotic Spindle Organization. Mol. Cel Biol 27, 5887–5897. doi:10.1128/mcb.00175-07
Pilaz, L.-J., Patti, D., Marcy, G., Ollier, E., Pfister, S., Douglas, R. J., et al. (2009). Forced G1-phase Reduction Alters Mode of Division, Neuron Number, and Laminar Phenotype in the Cerebral Cortex. Proc. Natl. Acad. Sci. 106, 21924–21929. doi:10.1073/pnas.0909894106
Pontious, A., Kowalczyk, T., Englund, C., and Hevner, R. F. (2008). Role of Intermediate Progenitor Cells in Cerebral Cortex Development. Dev. Neurosci. 30, 24–32. doi:10.1159/000109848
Postiglione, M. P., Jüschke, C., Xie, Y., Haas, G. A., Charalambous, C., and Knoblich, J. A. (2011). Mouse Inscuteable Induces Apical-Basal Spindle Orientation to Facilitate Intermediate Progenitor Generation in the Developing Neocortex. Neuron 72, 269–284. doi:10.1016/j.neuron.2011.09.022
Poulton, C. J., Schot, R., Seufert, K., Lequin, M. H., Accogli, A., Annunzio, G. D., et al. (2014). Severe Presentation ofWDR62mutation: Is There a Role for Modifying Genetic Factors? Am. J. Med. Genet. 164, 2161–2171. doi:10.1002/ajmg.a.36611
Pulvers, J. N., Bryk, J., Fish, J. L., Wilsch-Brauninger, M., Arai, Y., Schreier, D., et al. (2010). Mutations in Mouse Aspm (Abnormal Spindle-like Microcephaly Associated) Cause Not Only Microcephaly but Also Major Defects in the Germline. Proc. Natl. Acad. Sci. 107, 16595–16600. doi:10.1073/pnas.1010494107
Putkey, F. R., Cramer, T., Morphew, M. K., Silk, A. D., Johnson, R. S., McIntosh, J. R., et al. (2002). Unstable Kinetochore-Microtubule Capture and Chromosomal Instability Following Deletion of CENP-E. Developmental Cel 3, 351–365. doi:10.1016/s1534-5807(02)00255-1
Qian, X., Shen, Q., Goderie, S. K., He, W., Capela, A., Davis, A. A., et al. (2000). Timing of CNS Cell Generation. Neuron 28, 69–80. doi:10.1016/s0896-6273(00)00086-6
Rakic, P. (2009). Evolution of the Neocortex: a Perspective from Developmental Biology. Nat. Rev. Neurosci. 10, 724–735. doi:10.1038/nrn2719
Rana, R., Gwasikoti, N., Vaswani, N. D., and Kaushik, J. S. (2019). Phenomenology of Epilepsy in a Child with a ZNF335 Encephalopathy. Indian J. Pediatr. 86, 967–968. doi:10.1007/s12098-019-02991-8
Ravindran, E., Jühlen, R., Vieira-Vieira, C. H., Ha, T., Salzberg, Y., Fichtman, B., et al. (2021). Expanding the Phenotype of NUP85 Mutations beyond Nephrotic Syndrome to Primary Autosomal Recessive Microcephaly and Seckel Syndrome Spectrum Disorders. Hum. Mol. Genet. 30, 2068–2081. doi:10.1093/hmg/ddab160
Reillo, I., de Juan Romero, C., García-Cabezas, M. Á., and Borrell, V. (2011). A Role for Intermediate Radial Glia in the Tangential Expansion of the Mammalian Cerebral Cortex. Cereb. Cortex 21, 1674–1694. doi:10.1093/cercor/bhq238
Reilly, M. L., Stokman, M. F., Magry, V., Jeanpierre, C., Alves, M., Paydar, M., et al. (2019). Loss-of-function Mutations inKIF14cause Severe Microcephaly and Kidney Development Defects in Humans and Zebrafish. Hum. Mol. Genet. 28, 778–795. doi:10.1093/hmg/ddy381
Riparbelli, M. G., Callaini, G., Glover, D. M., and Avides, M. d. C. (2002). A Requirement for the Abnormal Spindle Protein to Organise Microtubules of the central Spindle for Cytokinesis inDrosophila. J. Cel. Sci. 115, 913–922. doi:10.1242/jcs.115.5.913
Ripoll, P., Pimpinelli, S., Valdivia, M. M., and Avila, J. (1985). A Cell Division Mutant of Drosophila with a Functionally Abnormal Spindle. Cell 41, 907–912. doi:10.1016/s0092-8674(85)80071-4
Roberts, M. R., Bittman, K., Li, W.-W., French, R., Mitchell, B., LoTurco, J. J., et al. (2000). TheFlatheadMutation Causes CNS-specific Developmental Abnormalities and Apoptosis. J. Neurosci. 20, 2295–2306. doi:10.1523/jneurosci.20-06-02295.2000
Rodrigues-Martins, A., Bettencourt-Dias, M., Riparbelli, M., Ferreira, C., Ferreira, I., Callaini, G., et al. (2007). DSAS-6 Organizes a Tube-like Centriole Precursor, and its Absence Suggests Modularity in Centriole Assembly. Curr. Biol. 17, 1465–1472. doi:10.1016/j.cub.2007.07.034
Roque, H., Wainman, A., Richens, J., Kozyrska, K., Franz, A., and Raff, J. W. (2012). Drosophila Cep135/Bld10 Maintains Proper Centriole Structure but Is Dispensable for Cartwheel Formation. J. Cel. Sci. 125, 5881–5886. doi:10.1242/jcs.113506
Rupp, V., Rauf, S., Naveed, I., Windpassinger, C., and Mir, A. (2014). A Novel Single Base Pair Duplication in WDR62 Causes Primary Microcephaly. BMC Med. Genet. 15, 107. doi:10.1186/s12881-014-0107-4
Saadi, A., Borck, G., Boddaert, N., Chekkour, M. C., Imessaoudene, B., Munnich, A., et al. (2009). Compound Heterozygous ASPM Mutations Associated with Microcephaly and Simplified Cortical Gyration in a Consanguineous Algerian Family. Eur. J. Med. Genet. 52, 180–184. doi:10.1016/j.ejmg.2009.03.013
Saadi, A., Verny, F., Siquier-Pernet, K., Bole-Feysot, C., Nitschke, P., Munnich, A., et al. (2016). Refining the Phenotype Associated with CASC5 Mutation. Neurogenetics 17, 71–78. doi:10.1007/s10048-015-0468-7
Sajid Hussain, M., Marriam Bakhtiar, S., Farooq, M., Anjum, I., Janzen, E., Reza Toliat, M., et al. (2013). Genetic Heterogeneity in Pakistani Microcephaly Families. Clin. Genet. 83, 446–451. doi:10.1111/j.1399-0004.2012.01932.x
Santaguida, S., and Musacchio, A. (2009). The Life and Miracles of Kinetochores. Embo J. 28, 2511–2531. doi:10.1038/emboj.2009.173
Sarkisian, M. R., Frenkel, M., Li, W., Oborski, J. A., and LoTurco, J. J. (2001). Altered Interneuron Development in the Cerebral Cortex of the Flathead Mutant. Cereb. Cortex 11, 734–743. doi:10.1093/cercor/11.8.734
Sarkisian, M. R., Li, W., Di Cunto, F., D'Mello, S. R., and LoTurco, J. J. (2002). Citron-kinase, a Protein Essential to Cytokinesis in Neuronal Progenitors, Is Deleted in the Flathead Mutant Rat. J. Neurosci. 22, Rc217. doi:10.1523/JNEUROSCI.22-08-j0001.2002
Sassaman, E. A., and Zartler, A. S. (1982). Mental Retardation and Head Growth Abnormalities. J. Pediatr. Psychol. 7, 149–156. doi:10.1093/jpepsy/7.2.149
Sato, R., Takanashi, J., Tsuyusaki, Y., Kato, M., Saitsu, H., Matsumoto, N., et al. (2016). Association between Invisible Basal Ganglia and ZNF335 Mutations: A Case Report. Pediatrics 138, 138. doi:10.1542/peds.2016-0897
Scala, M., Chua, G. L., Chin, C. F., Alsaif, H. S., Borovikov, A., Riazuddin, S., et al. (2020). Biallelic MFSD2A Variants Associated with Congenital Microcephaly, Developmental Delay, and Recognizable Neuroimaging Features. Eur. J. Hum. Genet. 28, 1509–1519. doi:10.1038/s41431-020-0669-x
Sgourdou, P., Mishra-Gorur, K., Saotome, I., Henagariu, O., Tuysuz, B., Campos, C., et al. (2017). Disruptions in Asymmetric Centrosome Inheritance and WDR62-Aurora Kinase B Interactions in Primary Microcephaly. Sci. Rep. 7, 43708. doi:10.1038/srep43708
Shaheen, R., Hashem, A., Abdel-Salam, G. M. H., Al-Fadhli, F., Ewida, N., and Alkuraya, F. S. (2016). Mutations in CIT, Encoding Citron Rho-Interacting Serine/threonine Kinase, Cause Severe Primary Microcephaly in Humans. Hum. Genet. 135, 1191–1197. doi:10.1007/s00439-016-1722-2
Shaheen, R., Maddirevula, S., Ewida, N., Alsahli, S., Abdel-Salam, G. M. H., Zaki, M. S., et al. (2019). Genomic and Phenotypic Delineation of Congenital Microcephaly. Genet. Med. 21, 545–552. doi:10.1038/s41436-018-0140-3
Shi, L., Qalieh, A., Lam, M. M., Keil, J. M., and Kwan, K. Y. (2019). Robust Elimination of Genome-Damaged Cells Safeguards against Brain Somatic Aneuploidy Following Knl1 Deletion. Nat. Commun. 10, 2588. doi:10.1038/s41467-019-10411-w
Singh, P., Ramdas Nair, A., and Cabernard, C. (2014). The Centriolar Protein Bld10/Cep135 Is Required to Establish Centrosome Asymmetry in Drosophila Neuroblasts. Curr. Biol. 24, 1548–1555. doi:10.1016/j.cub.2014.05.050
Siskos, N., Stylianopoulou, E., Skavdis, G., and Grigoriou, M. E. (2021). Molecular Genetics of Microcephaly Primary Hereditary: An Overview. Brain Sci. 11. doi:10.3390/brainsci11050581
Slezak, R., Smigiel, R., Obersztyn, E., Pollak, A., Dawidziuk, M., Wiszniewski, W., et al. (2021). Further Delineation of Phenotype and Genotype of Primary Microcephaly Syndrome with Cortical Malformations Associated with Mutations in the WDR62 Gene. Genes 12, 594. doi:10.3390/genes12040594
Stouffs, K., Stergachis, A. B., Vanderhasselt, T., Dica, A., Janssens, S., Vandervore, L., et al. (2018). Expanding the Clinical Spectrum of Biallelic ZNF335 Variants. Clin. Genet. 94, 246–251. doi:10.1111/cge.13260
Subramanian, L., Calcagnotto, M. E., and Paredes, M. F. (2019). Cortical Malformations: Lessons in Human Brain Development. Front. Cel Neurosci 13, 576. doi:10.3389/fncel.2019.00576
Szczepanski, S., Hussain, M. S., Sur, I., Altmüller, J., Thiele, H., Abdullah, U., et al. (2016). A Novel Homozygous Splicing Mutation of CASC5 Causes Primary Microcephaly in a Large Pakistani Family. Hum. Genet. 135, 157–170. doi:10.1007/s00439-015-1619-5
Tacchelly-Benites, O., Wang, Z., Yang, E., Lee, E., and Ahmed, Y. (2013). Toggling a Conformational Switch in Wnt/β-Catenin Signaling: Regulation of Axin Phosphorylation. BioEssays 35, 1063–1070. doi:10.1002/bies.201300101
Tang, H., Hammack, C., Ogden, S. C., Wen, Z., Qian, X., Li, Y., et al. (2016). Zika Virus Infects Human Cortical Neural Progenitors and Attenuates Their Growth. Cell Stem Cell 18, 587–590. doi:10.1016/j.stem.2016.02.016
Trimborn, M., Bell, S. M., Felix, C., Rashid, Y., Jafri, H., Griffiths, P. D., et al. (2004). Mutations in Microcephalin Cause Aberrant Regulation of Chromosome Condensation. Am. J. Hum. Genet. 75, 261–266. doi:10.1086/422855
Trimborn, M., Ghani, M., Walther, D. J., Dopatka, M., Dutrannoy, V., Busche, A., et al. (2010). Establishment of a Mouse Model with Misregulated Chromosome Condensation Due to Defective Mcph1 Function. PLoS One 5, e9242. doi:10.1371/journal.pone.0009242
Tsai, M.-Y., Wang, S., Heidinger, J. M., Shumaker, D. K., Adam, S. A., Goldman, R. D., et al. (2006). A Mitotic Lamin B Matrix Induced by RanGTP Required for Spindle Assembly. Science 311, 1887–1893. doi:10.1126/science.1122771
van Deijk, A.-L. F., Camargo, N., Timmerman, J., Heistek, T., Brouwers, J. F., Mogavero, F., et al. (2017). Astrocyte Lipid Metabolism Is Critical for Synapse Development and Function In Vivo. Glia 65, 670–682. doi:10.1002/glia.23120
Van Den Bosch, J. (1959). Microcephaly in the Netherlands: a Clinical and Genetical Study. Ann. Hum. Genet. 23, 91–116. doi:10.1111/j.1469-1809.1958.tb01455.x
van Dyck, L. I., and Morrow, E. M. (2017). Genetic Control of Postnatal Human Brain Growth. Curr. Opin. Neurol. 30, 114–124. doi:10.1097/wco.0000000000000405
Varmark, H., Llamazares, S., Rebollo, E., Lange, B., Reina, J., Schwarz, H., et al. (2007). Asterless Is a Centriolar Protein Required for Centrosome Function and Embryo Development in Drosophila. Curr. Biol. 17, 1735–1745. doi:10.1016/j.cub.2007.09.031
Vergnes, L., Peterfy, M., Bergo, M. O., Young, S. G., and Reue, K. (2004). Lamin B1 Is Required for Mouse Development and Nuclear Integrity. Proc. Natl. Acad. Sci. 101, 10428–10433. doi:10.1073/pnas.0401424101
Verloes, A., Drunat, S., Gressens, P., and Passemard, S. (1993). “Primary Autosomal Recessive Microcephalies and Seckel Syndrome Spectrum Disorders,” in GeneReviews(R). Editors R. A. Pagon, M. P. Adam, H. H. Ardinger, S. E. Wallace, A. Amemiya, L. J. H. Beanet al. (Seattle (WA).
Vitale, I., Galluzzi, L., Castedo, M., and Kroemer, G. (2011). Mitotic Catastrophe: a Mechanism for Avoiding Genomic Instability. Nat. Rev. Mol. Cel Biol 12, 385–392. doi:10.1038/nrm3115
von der Hagen, M., Pivarcsi, M., Liebe, J., von Bernuth, H., Didonato, N., Hennermann, J. B., et al. (2014). Diagnostic Approach to Microcephaly in Childhood: a Two-center Study and Review of the Literature. Dev. Med. Child. Neurol. 56, 732–741. doi:10.1111/dmcn.12425
Wang, X., Tsai, J.-W., Imai, J. H., Lian, W.-N., Vallee, R. B., and Shi, S.-H. (2009). Asymmetric Centrosome Inheritance Maintains Neural Progenitors in the Neocortex. Nature 461, 947–955. doi:10.1038/nature08435
Watemberg, N., Silver, S., Harel, S., and Lerman-Sagie, T. (2002). Significance of Microcephaly Among Children with Developmental Disabilities. J. Child. Neurol. 17, 117–122. doi:10.1177/088307380201700205
Wong, B. H., Chan, J. P., Cazenave-Gassiot, A., Poh, R. W., Foo, J. C., Galam, D. L. A., et al. (2016). Mfsd2a Is a Transporter for the Essential ω-3 Fatty Acid Docosahexaenoic Acid (DHA) in Eye and Is Important for Photoreceptor Cell Development. J. Biol. Chem. 291, 10501–10514. doi:10.1074/jbc.m116.721340
Woods, C. G., Bond, J., and Enard, W. (2005). Autosomal Recessive Primary Microcephaly (MCPH): A Review of Clinical, Molecular, and Evolutionary Findings. Am. J. Hum. Genet. 76, 717–728. doi:10.1086/429930
Woods, C. G. (2004). Human Microcephaly. Curr. Opin. Neurobiol. 14, 112–117. doi:10.1016/j.conb.2004.01.003
Woods, C. G., and Parker, A. (2013). Investigating Microcephaly. Arch. Dis. Child. 98, 707–713. doi:10.1136/archdischild-2012-302882
Wu, K.-Y., Zuo, G.-L., Li, X.-F., Ye, Q., Deng, Y.-Q., Huang, X.-Y., et al. (2016). Vertical Transmission of Zika Virus Targeting the Radial Glial Cells Affects Cortex Development of Offspring Mice. Cell Res 26, 645–654. doi:10.1038/cr.2016.58
Xu, D., Zhang, F., Wang, Y., Sun, Y., and Xu, Z. (2014). Microcephaly-associated Protein WDR62 Regulates Neurogenesis through JNK1 in the Developing Neocortex. Cel Rep. 6, 104–116. doi:10.1016/j.celrep.2013.12.016
Yabe, T., Ge, X., and Pelegri, F. (2007). The Zebrafish Maternal-Effect Gene Cellular Atoll Encodes the Centriolar Component Sas-6 and Defects in its Paternal Function Promote Whole Genome Duplication. Developmental Biol. 312, 44–60. doi:10.1016/j.ydbio.2007.08.054
Yamamoto, S., Jaiswal, M., Charng, W.-L., Gambin, T., Karaca, E., Mirzaa, G., et al. (2014). A drosophila Genetic Resource of Mutants to Study Mechanisms Underlying Human Genetic Diseases. Cell 159, 200–214. doi:10.1016/j.cell.2014.09.002
Yang, Y. J., Baltus, A. E., Mathew, R. S., Murphy, E. A., Evrony, G. D., Gonzalez, D. M., et al. (2012). Microcephaly Gene Links Trithorax and REST/NRSF to Control Neural Stem Cell Proliferation and Differentiation. Cell 151, 1097–1112. doi:10.1016/j.cell.2012.10.043
Yen, T. J., Compton, D. A., Wise, D., Zinkowski, R. P., Brinkley, B. R., Earnshaw, W. C., et al. (1991). CENP-E, a Novel Human Centromere-Associated Protein Required for Progression from Metaphase to Anaphase. EMBO J. 10, 1245–1254. doi:10.1002/j.1460-2075.1991.tb08066.x
Young, S. G., Jung, H.-J., Coffinier, C., and Fong, L. G. (2012). Understanding the Roles of Nuclear A- and B-type Lamins in Brain Development. J. Biol. Chem. 287, 16103–16110. doi:10.1074/jbc.r112.354407
Yu, T. W., Mochida, G. H., Tischfield, D. J., Sgaier, S. K., Flores-Sarnat, L., Sergi, C. M., et al. (2010). Mutations in WDR62, Encoding a Centrosome-Associated Protein, Cause Microcephaly with Simplified Gyri and Abnormal Cortical Architecture. Nat. Genet. 42, 1015–1020. doi:10.1038/ng.683
Yucel, J. K., Marszalek, J. D., McIntosh, J. R., Goldstein, L. S. B., Cleveland, D. W., and Philp, A. V. (2000). CENP-meta, an Essential Kinetochore Kinesin Required for the Maintenance of Metaphase Chromosome Alignment in Drosophila. J. Cel Biol 150, 1–12. doi:10.1083/jcb.150.1.1a
Zaqout, S., Morris-Rosendahl, D., and Kaindl, A. M. (2017). Autosomal Recessive Primary Microcephaly (MCPH): An Update. Neuropediatrics 48, 135–142. doi:10.1055/s-0037-1601448
Zaqout, S., Blaesius, K., Wu, Y.-J., Ott, S., Kraemer, N., Becker, L.-L., et al. (2019). Altered Inhibition and Excitation in Neocortical Circuits in Congenital Microcephaly. Neurobiol. Dis. 129, 130–143. doi:10.1016/j.nbd.2019.05.008
Zarate, Y. A., Kaylor, J. A., Bosanko, K., Lau, S., Vargas, J., and Gao, H. (2016). First Clinical Report of an Infant with Microcephaly andCASC5mutations. Am. J. Med. Genet. 170, 2215–2218. doi:10.1002/ajmg.a.37726
Zhang, W., Yang, S.-L., Yang, M., Herrlinger, S., Shao, Q., Collar, J. L., et al. (2019). Modeling Microcephaly with Cerebral Organoids Reveals a WDR62-Cep170-KIF2A Pathway Promoting Cilium Disassembly in Neural Progenitors. Nat. Commun. 10, 2612. doi:10.1038/s41467-019-10497-2
Zhang, Y., Li, H., Pang, J., Peng, Y., Shu, L., and Wang, H. (2019). Novel SASS6 Compound Heterozygous Mutations in a Chinese Family with Primary Autosomal Recessive Microcephaly. Clinica Chim. Acta 491, 15–18. doi:10.1016/j.cca.2019.01.007
Zhong, W., and Chia, W. (2008). Neurogenesis and Asymmetric Cell Division. Curr. Opin. Neurobiol. 18, 4–11. doi:10.1016/j.conb.2008.05.002
Zhong, W., Feder, J. N., Jiang, M.-M., Jan, L. Y., and Jan, Y. N. (1996). Asymmetric Localization of a Mammalian Numb Homolog during Mouse Cortical Neurogenesis. Neuron 17, 43–53. doi:10.1016/s0896-6273(00)80279-2
Ziegler, A. B., Thiele, C., Tenedini, F., Richard, M., Leyendecker, P., Hoermann, A., et al. (2017). Cell-Autonomous Control of Neuronal Dendrite Expansion via the Fatty Acid Synthesis Regulator SREBP. Cel Rep. 21, 3346–3353. doi:10.1016/j.celrep.2017.11.069
Keywords: MCPH genes, microcephaly, brain, intellectual disability, neuronal differentiation, animal models, brain malformation
Citation: Zaqout S and Kaindl AM (2022) Autosomal Recessive Primary Microcephaly: Not Just a Small Brain. Front. Cell Dev. Biol. 9:784700. doi: 10.3389/fcell.2021.784700
Received: 28 September 2021; Accepted: 01 December 2021;
Published: 17 January 2022.
Edited by:
Annette Hammes, Max Delbrück Center for Molecular Medicine (MDC), GermanyCopyright © 2022 Zaqout and Kaindl. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sami Zaqout, c2FtaS56YXFvdXRAcXUuZWR1LnFh, c2FtaS56YXFvdXRAY2hhcml0ZS5kZQ==
 Sami Zaqout
Sami Zaqout Angela M. Kaindl
Angela M. Kaindl