- 1Sichuan Provincial Key Laboratory for Human Disease Gene Study, Department of Laboratory Medicine, Center for Medical Genetics, Sichuan Provincial People’s Hospital, School of Medicine, University of Electronic Science and Technology of China, Chengdu, China
- 2Research Unit for Blindness Prevention of the Chinese Academy of Medical Sciences (2019RU026), Sichuan Academy of Medical Sciences and Sichuan Provincial People’s Hospital, Chengdu, China
- 3Department of Ophthalmology, First People’s Hospital of Shangqiu, Shangqiu, China
The asparaginase and isoaspartyl peptidase 1 (ASRGL1) is an L-asparaginase and beta-aspartyl peptidase enzyme that may be involved in the formation of L-aspartate, a neurotransmitter that can operate as an excitatory neurotransmitter in some brain regions. Although variants in ASRGL1 have been reported in retinitis pigmentosa (RP) patients, the in vivo functions and mechanisms of ASRGL in RP remains unknown due to the lack of suitable disease models. To explore the role of ASRGL in RP, we generated an Asrgl1 knockout mouse model (Asrgl1 KO) using the CRISPR/Cas9 technique. Asrgl1 ablation in mice led to an attenuated electroretinogram (ERG) response around 8 months. The thickness of the outer nuclei layer (ONL) started to decrease around 9 months in Asrgl1 KO mice and gradually intensified at 12 and 15 months. Immunostaining revealed thinner inner segment (IS) and thinner outer segment (OS) as well as the progressive degeneration of rod and cone cells in Asrgl1 KO mice. One hundred forty-nine transcriptional differentially expressed genes (DEGs) were found by RNA-seq in Asrgl1 KO retina. These DEGs were linked to a number of biological processes that were considerably enriched, including gastrointestinal disease and organismal injury and abnormalities. By analysis of canonical pathways, glucocorticoid receptor signaling was the most significant canonical pathway altered in Asrgl1 KO retina. Several molecules, including NFE2L2, IL-4, Foxp3, and Fos, were in the central nodes of the interaction network in Asrgl1 KO retina. In summary, our study provided a knockout mouse model for a better understanding of the molecular mechanism for ASRGL1-related RP.
Introduction
Retinitis pigmentosa is one of the most common causes of visual impairment around the world, including symptoms such as night blindness and progressive loss of peripheral vision in the early stage, and no complete yet effective treatment exists up to date (Ferrari et al., 2011). The prevalence of this disorder is approximately 1:4,000 (Hartong et al., 2006). As an inherited retinal degeneration disease, it can be inherited in three ways: autosomal recessive (50%–60%), autosomal dominant (30%–40%), or X-linked (5%–15%). The phenotype of non-syndromic retinitis pigmentosa (RP) in RP patients might be limited to the eye or it can be part of a syndrome that includes extraocular illnesses such hearing loss, obesity, and neurologic diseases (Bhatti, 2006). Over 90 genes have been associated with non-syndromic RP according to the RetNet database (https://sph.uth.edu/retnet/; date last accessed Jul 30, 2021). However, only about 50%–60% of RP cases can be explained by these mutations, and the pathological and molecular mechanisms of some of the genes remain unknown (Daiger et al., 2013; Huang et al., 2015).
Asparaginase and isoaspartyl peptidase 1 (ASRGL1) has been reported as a RP disease gene in 2016 (Biswas et al., 2016). The p.G178R mutation, which causes photoreceptor degeneration and progressive vision loss, was discovered in a large five-generation pedigree with early-onset recessive retinal degeneration utilizing linkage analysis and homozygosity mapping combined with exome sequencing. ASRGL1 encodes an enzyme L-asparaginase, has both L-asparaginase and beta-aspartyl peptidase activity, and may be involved in the production of L-aspartate (Cantor et al., 2009). However, the pathological and molecular mechanisms of ASRGL1 in causing of RP remains unknown.
To investigate the function of Asrgl1 in the mammalian retina and explore the molecular mechanism how Asrgl1 affect photoreceptors or RPE cells, we developed a novel mouse model of RP with the Asrgl1 gene knockout on a C57BL/6J genetic background by CRISPR/CAS9 technology. Asrgl1 ablation in mice resulted in a typical retinal degeneration phenotype with decreased electroretinogram (ERG) response; thinner outer nuclei layer (ONL), inner segment (IS), and outer segment (OS); and decreased rod and cone proteins. A high-throughput transcriptional sequencing analysis found 149 transcriptional differentially expressed genes (DEGs) between Asrgl1 knockout (KO) mouse retina and the normal controls. Glucocorticoid receptor signaling was the most changed pathway and might play key roles in the pathological process of ASRGL1-related RP. Our studies provide an Asrgl1 KO mouse model for improving our understanding of RP disease mechanisms.
Materials and Methods
Generation of Asrgl1 Knockout Mice
The Animal Care and Use Committee of the Sichuan Provincial People’s Hospital authorized all animal experiments, which followed the ARVO guidelines for the use of animals in ophthalmology and vision research.
Asrgl1 KO mice were generated by using the CRISPR/Cas9 system on C57BL/6J genetic background. The Cas9 mRNA and two single guide RNAs targeting a region upstream of exon 3 and downstream of exon 4 of Asrgl1 exon 3 were microinjected into mouse zygotes (Figure 1). gRNA1 sequence (matching reverse strand of gene) was as follows: TCTGTCGCTAGCAACCAACAAGG. gRNA2 sequence (matching forward strand of gene) was as follows: ATATTGCTAGACAGGCTTGGAGG. As confirmed by Sanger sequencing and PCR genotyping analysis, the conventional translation start site was successfully eliminated.
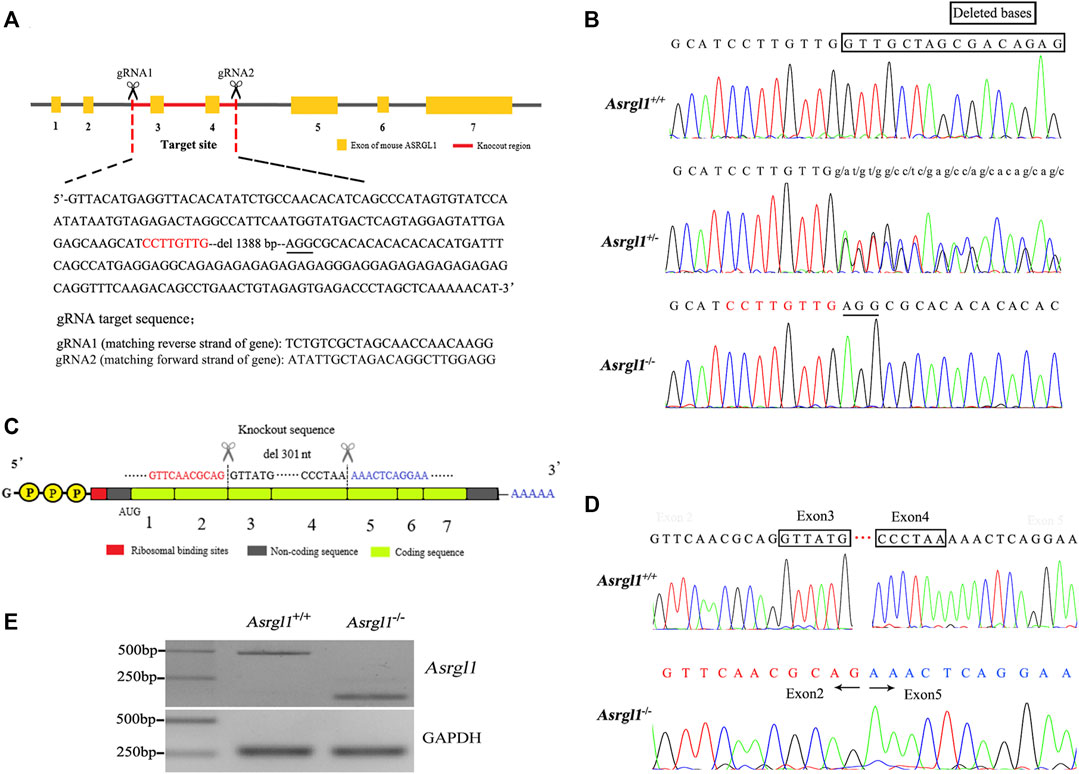
FIGURE 1. Generation of Asrgl1 knockout (KO) mice. (A) The partial genomic sequence of Asrgl1 exons was shown. The 1,388-bp genomic deletion was indicated in red. (B) Sanger sequencing of Asrgl1 genome in the wild-type (WT), Asrgl1+/− and Asrgl1−/− mice. (C) Schematic illustration of cDNA deletion in Asrgl1 KO mice. (D) Sanger sequencing of Asrgl1 cDNA in WT and Asrgl1−/− mice. A 301-bp fragment consisting of exon 3 and 4 is deleted in Asrgl1 KO allele. (E) Reverse transcription PCR analysis of Asrgl1 expression in WT and Asrgl1−/− mouse retinas at 3 months of age. Gapdh was used as a loading control.
Electroretinograms in Mice
Mouse ERG detection was performed on an Espion Visual Electrophysiology System (Diagnosys, Lowell, MA, United States). Briefly, female Asrgl1 KO mice and female controls were dark-adapted overnight 1 day before detection. In the next morning, before ERG, animals were anesthetized and the eyes were dilated with a drop of tropicamide and phenylephrine, as well as tetracaine (0.5%). Throughout the experiment, a heating platform was used to keep the body temperature at 37°C. Gold wire loops were used to record dark-adapted ERGs in response to flashes with intensities ranging from 0.003 to 20 cds/m2. After 20 min of complete light adaptation, cone-mediated ERGs were recorded with white flashes.
Retinal Hematoxylin and Eosin Staining
The eyes with the whole retina and optic nerve were chosen for imaging by hematoxylin and eosin (H&E) staining. Female wild-type (WT) and female KO mice’s eyes were enucleated, marked for orientation on the nasal side, fixed overnight in 1.22% glutaraldehyde and 0.8% paraformaldehyde in 0.08 M phosphate buffer, embedded in paraffin, and cut into 5-μm slices. H&E staining was used on sections taken from five places around the optic nerve. The rows of photoreceptors in the outer nuclear layer were counted using H&E-stained slices. Every 200 μm from the optic nerve, three measurements of the outer nuclear layer were recorded and averaged. The optic nerve was given the number 0 mm.
Immunohistochemistry Analysis
Enucleated eyeballs were removed from female WT and female Asrgl1 KO mice for immunohistochemistry, marked on the nasal side for orientation, fixed for 1 h in 4% paraformaldehyde in 100 mM phosphate buffer (pH 7.4), and cryoprotected in 30% sucrose. For sectioning, tissues were immersed in an optimal cutting temperature (OCT) solution and frozen on dry ice. For 30 min, sections were blocked and permeabilized in phosphate buffer with 10% normal goat serum and 0.2% Triton X-100, then labeled with different primary antibodies overnight at 4°C. Then, the slides were washed and incubated with secondary antibodies for 2 h. Finally, the slides were washed and covered with coverslips. The primary antibodies used in our study were as follows: rhodopsin (1D4, cat. #MA1-722, Thermo Fisher, Waltham, MA, United States), Na-K pump (cat. #MA3928, Thermo Fisher, Waltham, MA, United States), opsin (red/green, cat. #AB5405, Millipore, Burlington, MA, United States), and 594-conjugated peanut agglutinin (PNA) (cat. #RL1072, Vector laboratories, Burlingame, CA, United States). Secondary antibodies (Alexa Fluor 488 and Alexa Fluor 594) and DAPI (cat. #D8417, Sigma, St Louis, MO, United States, 1:2,000 dilution) were also used in the experiments. Confocal microscopy was used to image eye sections (LSM800, Carl Zeiss, Jena, Germany).
RNA Isolation and RT-PCR
TRIzol (Invitrogen, Austin, TX, United States) was used to extract total RNA from the retinas of female Asrgl1 KO mice and female normal controls, according to the manufacturer’s protocol. cDNA was made as previously described. An Eppendorf Mastercycler personal PCR equipment (Eppendorf, Germany) was used to amplify Asrgl1 genotyping products, which were then analyzed on a 1.5% agarose gel electrophoresis. Using the SYBR PCR Master Mix kit (Applied Biosystems, Foster City, CA) and the 7500 Fast Real-Time PCR detection machine, equal amounts of cDNA from the retinas of Asrgl1 KO mice and normal controls were submitted to PCR. Specific primers used in our study were listed in Supplementary Data S1.
RNA-Seq and Differential Expression Analysis
RNA was isolated from the retinas of female Asrgl1 KO mice and female normal controls, and the integrity of the RNA was determined using the Bioanalyzer 2100 system’s RNA Nano 6000 Assay Kit (Agilent Technologies, CA, United States). The library was then produced and tested for transcriptome sequencing using the Agilent Bioanalyzer 2100 instrument. The index-coded samples were clustered using the TruSeq PE Cluster Kit v3-cBot-HS (Illumina) on a cBot Cluster Generation System according to the manufacturer’s instructions. The library preparations were sequenced on an Illumina Novaseq device after cluster creation, yielding 150-bp paired-end reads. Raw data (raw readings) in fastq format were processed using in-house perl scripts, resulting in clean, high-quality data. The level of gene expression was measured after reads were mapped to the reference genome. All of the downstream analyses were based on clean, high-quality data. Prior to differential gene expression analysis, the read counts for each sequenced library were modified by one scaling normalization factor using the edgeR tool package. The edgeR R package was used to perform differential expression analysis of two situations. The P values were adjusted using the Benjamini and Hochberg method. The criterion for significant differential expression was set at 0.05 corrected P-value and log2 fold change. The raw data have been uploaded to the Genome Sequence Archive (https://ngdc.cncb.ac.cn/gsa/browse/CRA005193), and the assigned accession number was CRA005193.
Functional Analysis, Canonical Pathway Analysis, and Generation of Networks
Top biological functions as well as canonical pathways associated with DEGs generated from the RNA-seq between the Asrgl1 KO retina and normal controls were identified by Ingenuity pathway analysis (IPA) (Qiagen). Fisher’s exact test was used to determine the likelihood of any biological function or canonical pathway being explained only by chance. Algorithmically, molecular interaction networks are created based on the connectivity of the molecules. Fisher’s exact test was used to determine network scores, which corresponded to −log 10 (p value).
Statistical Analysis
Statistical significance was established using Prism 7.0 software and the Student’s t-test or one-way ANOVA (GraphPad Software, La Jolla, CA, United States). The data are presented as the mean SEM unless otherwise noted in the text and figure legends. The ERG datasets in Figure 2 and the ONL thickness measurement dataset in Figure 3 were subjected to ANOVA tests. The number of animals used in each experiment is indicated by n. In the figure legends, the total number of animals used in each experiment is also listed. The following p values are indicated by asterisks in the figures: *≤0.05, **≤0.01, and ***≤0.001.
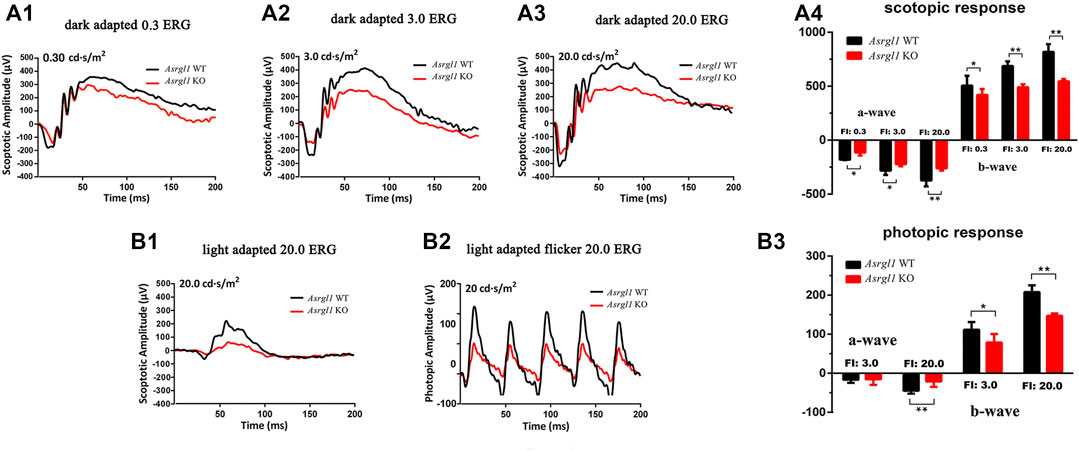
FIGURE 2. ElectroretinogramERG) examinations of Asrgl1 KO mice. (A1–A4) Asrgl1 KO mice at 8 months of age exhibited a reduced ERG scotopic response. Reduced amplitude peaks were observed for both the a-wave and the b-wave at light intensities of 0.3, 3.0, and 20.0 cds/m2 in Asrgl1 KO mice; two-tailed Student’s t-test was used. ∗∗p < 0.01; ∗∗∗p < 0.001. (B1–B3) Compared with WT mice, 8-month-old KO mice exhibited a reduced photopic response at 20.0 cds/m2; sample size n = 4. A two-tailed Student’s t-test was used. ∗∗p < 0.01; ∗∗∗p < 0.001.
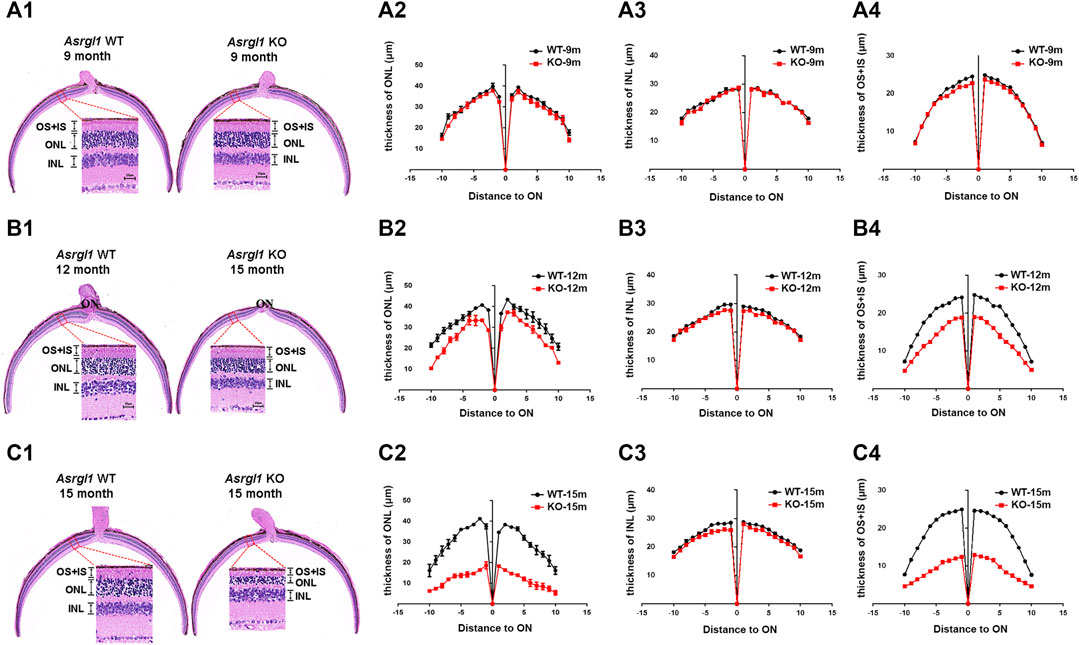
FIGURE 3. Degeneration of the outer retina in Asrgl1 KO mice. (A1,B1,C1) Hematoxylin and eosin (H&E) staining of paraffin sections of the Asrgl1 KO and corresponding control retinas at the ages of 9, 12, and 15 months. Scale bar: 25 μm. (A2,B2,C2) Quantification analysis of the outer nuclei layer (ONL) thickness of the KO (n = 6) and WT (n = 6) retinas from mice at 9, 12, and 15 months old. (A3,B3,C3) Quantification analysis of the inner nuclei layer (INL) thickness of the KO (n = 6) and WT (n = 6) retinas from mice at 9, 12, and 15 months old. (A4,B4,C4) Quantification analysis of the outer segment (OS) plus inner segment (IS) thickness of the KO (n = 6) and WT (n = 6) retinas from mice at 9, 12, and 15 months old. The Asrgl1 KO mouse retinas showed a drastically gradual degeneration of the photoreceptor cells at 15 months old.
Results
Generation of Asrgl1 Knockout Mice
ASRGL1 is widely expressed in all human tissues studied, with the highest levels of expression in the brain, female tissues including the uterine cervix and fallopian tube, and male tissues like the testis (Edqvist et al., 2015). To investigate the functions of AsrgL1 in vivo, Asrgl1 KO were generated using CRISPR/Cas9 technology. The target sites were selected as exon 3 and exon 4 (Figure 1A). Genotyping analysis using PCR primers flanking the deletion site revealed the successful generation of Asrgl1 knockout mice (Supplementary Figure S1). To validate the genomic Asrgl1 ablation, mouse DNA samples were subjected to Sanger sequencing analysis, and results showed genomic deletion of 1,388 bp in Asrgl1 KO mice (Figure 1B). Both Sanger sequencing and electrophoresis analysis of cDNA product from Asrgl1 KO retinas revealed a 301-bp deletion of exon 3 and exon 4 in Asrgl1 KO (Figure 1C–E). All the above data showed the successful generation of Asrgl1 KO by CRISPR/Cas9 technology.
Asrgl1 Knockout Mice Exhibited Impaired Visual Function
ERG examination was first performed to test the in vivo function of Asrgl1 in the retina. In three 6-month-old Asrgl1 KO mice and littermate controls, no abnormal ERG response was observed (data not shown). However, all Asrgl1 KO mice exhibited reduced scotopic and photopic responses at 8 months of age. The mean a- and b-wave amplitudes of Asrgl1 KO were reduced by approximately 15% and 30%, respectively, in scotopic response tests (Figure 2A1–A4), which indicated altered rod cell function. When compared to WT mice, Asrgl1 KO had a somewhat flat wave under light-adapted conditions, showing that cone cell function was also impaired in Asrgl1 KO (Figure 2B1–B3). Although, by H&E staining, the retinas from Asrgl1 KO mice revealed no difference in the retinal ONL thickness at 6 months of age (Supplementary Figure S2), the thickness of the ONL started to decrease around 9 months of age (Figure 3A1, A2) and gradually intensified at 12 months (Figure 3B1, B2) and 15 months of age (Figure 3C1, C2). The photoreceptor layer including the inner segment and outer segment decreased around 9 months (Figure 3A1, A4) and intensified at 12 months (Figure 3B1, B4) and 15 months of age (Figure 3C1, C4). There was no obvious change in the inner nuclei layer (INL) of Asrgl1 KO at 9 months of age (Figure 3A1, A3), and the thickness of the INL in Asrgl1 KO was slightly decreased at 15 months of age (Figure 3C1, C3).
Loss of Asrgl1 Leads to Degeneration and Apoptosis of Photoreceptor Cells
To investigate the pathological changes underlying the Asrgl1 knockout mice’s retinal degeneration phenotype, immunostaining was performed with retinal cryosections in Asrgl1 KO and WT mice. There were no changes in rhodopsin and M-opsin expression in Asrgl1 KO at 8 months of age (Supplementary Figure S3A, B). However, decreased rhodopsin protein was observed in the photoreceptor of 12- and 15-month-old Asrgl1 KO mice, respectively (Figure 4A, C), compared to WT mice, which indicated a shortened outer segment of 12- and 15-month-old Asrgl1 KO mice. Rhodopsin is a key outer segment protein that is required for adequate vision, and its manufacture and transit are tightly regulated (Palczewski, 2006). Decreased rhodopsin is cytotoxic and leads to rod death. Decreased Na-K ATP staining in 15-month-old Asrgl1 KO mice indicated shortened inner segment. M-opsin, a cone cell marker, and PNA (which binds to the sheaths of the cone matrix in all three types of cones) were also used to stain retinal sections. The number of cone cells was reduced in 12-month-old Asrgl1 KO mice and drastically decreased in 15-month-old Asrgl1 KO mice (Figure 4B,D). A TUNEL assay was utilized to determine whether the retinal degeneration was caused by apoptosis. There were no changes in TUNEL-positive cells in 8-month-old Asrgl1 KO mice (Supplementary Figure S3C). TUNEL-positive cells were substantially more abundant at 15 months of age in Asrgl1 KO retinas than in WT retinas (Figure 4E,F), and TUNEL-labeled nuclei were mostly found in the ONL, indicating continuous photoreceptor cell death in Asrgl1 KO retinas.
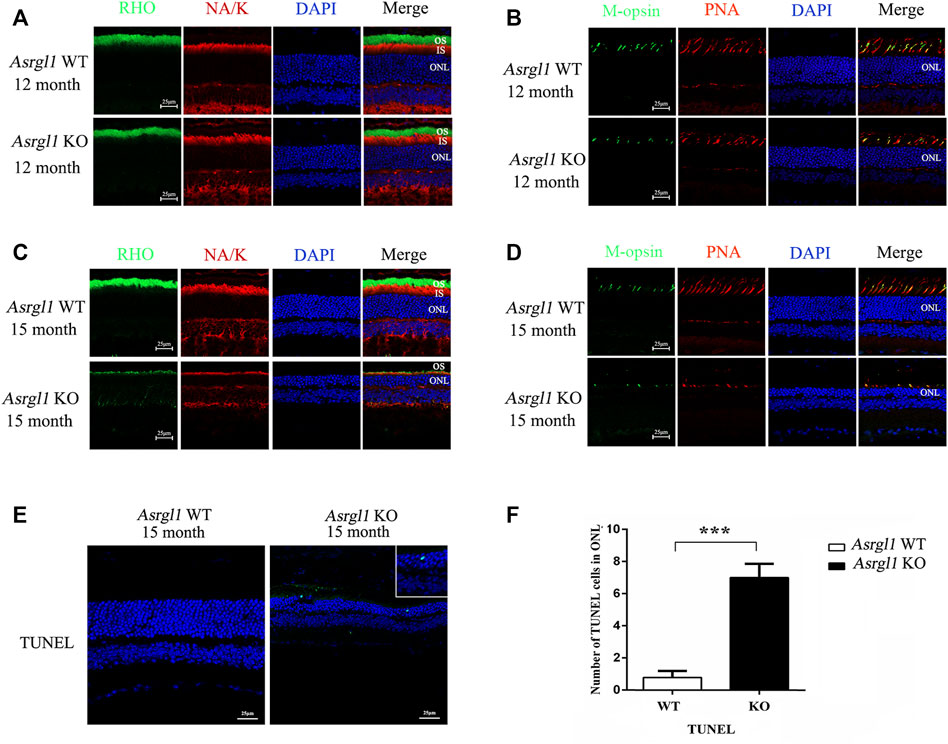
FIGURE 4. Immunofluorescence staining analysis of the Asrgl1 KO retinas. (A,C) Retinal cryosections from 12- and 15-month-old mice (n = 6) were labelled with rod-specific markers rhodopsin (outer segment) and Na-K ATPase (inner segment). (B,D) M-opsin and peanut agglutinin (PNA) was used as the cone markers in 12- and 15-month-old mice (n = 6). DAPI was used to counterstain the nuclei; scale bars: 25 μm. The data shown are representative of three independent tests. At least three slides of each retina were stained. (E) TUNEL assays in 15-month-old WT and KO mice revealed increased apoptosis. The magnified square indicated TUNEL-positive cells in the retinas. This experiment was performed three times. Sample size n = 6. (F) Quantification of TUNEL-positive cells in WT and KO retinas; sample size n = 6; ∗∗∗p < 0.001. Statistical significance was determined by a two-tailed unpaired t-test.
Differentially Expressed Genes in the Retinas Between Asrgl1 KO and Normal Controls
To investigate the transcriptional effect of Asrgl1 ablation, mouse retinal tissues from 8-month-old Asrgl1 KO (n = 3) and WT mice (n = 4) were subjected to RNA-seq analysis. There were 149 DEGs identified in Asrgl1 mice (Supplementary Table S2), among which 89 were upregulated (p ≤ 0.01 and log2 ratio ≥1) and 60 (p ≤ 0.01 and log2 ratio ≤−1) were downregulated. Volcano plots of differentially expressed genes are shown in Figure 5A. IPA was used to identify numerous highly enriched biological processes related with DEGs in order to investigate the biological functions enriched in DEGs (Table 1). Gastrointestinal disease (p value: 4.79E−02 to 2.61E−05, 13 genes assigned), organismal injury and abnormalities (p value: 4.94E−02 to 2.61E−05, 31 genes assigned), and cancer (p value: 4.79E−02 to 3.23E−04, 18 genes assigned) were discovered as the top three highly enriched terms in the perspective of diseases and disorders. Cellular development (p value: 4.16E−02 to 3.14E−05, seven genes assigned), cell morphology (p value: 4.79E−02 to 1.07E−03, six genes assigned), and cell death and survival (p value: 4.94E−02 to 1.22E−03, seven genes assigned) were found as the top three highly enriched terms in the perspective of molecular and cellular functions. Digestive system development and function (p value: 3.38E−02 to 2.61E−05, seven genes assigned), organ morphology (p value: 4.48E−02 to 2.61E−05, 13 genes assigned), and hematological system development and function (p value: 4.60E−02 to 3.14E−05, seven genes assigned) were shown as the top three enriched terms in the perspective of physiological system development and functions.
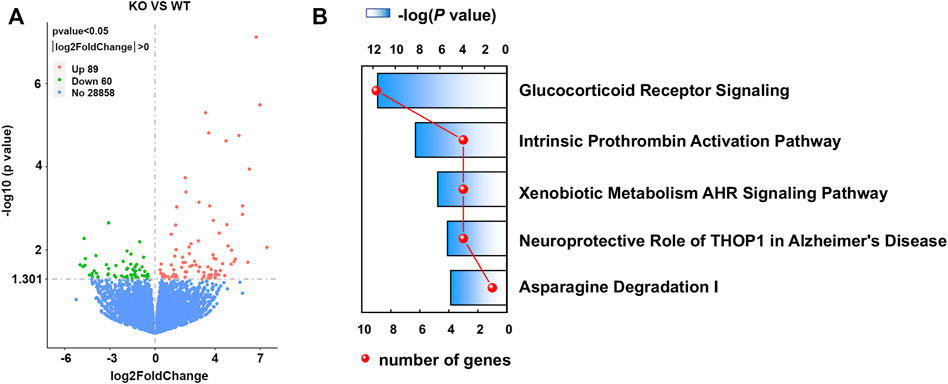
FIGURE 5. Volcano plots of differentially expressed genes (DEGs) and top canonical pathways involved in Asrgl1 KO mice. (A) Volcano plots of DEG profile. The vertical lines correspond to 2.0 FC up and down, and the horizontal line represents −log 10 (P-value). (B) Ingenuity pathway was used to analyze the total significantly differentially expressed genes and identify the canonical pathways involved in Asrgl1 KO mice. The top five significant pathways identified were displayed (blue bar). The red curve shows the total number of genes in each of these pathways.
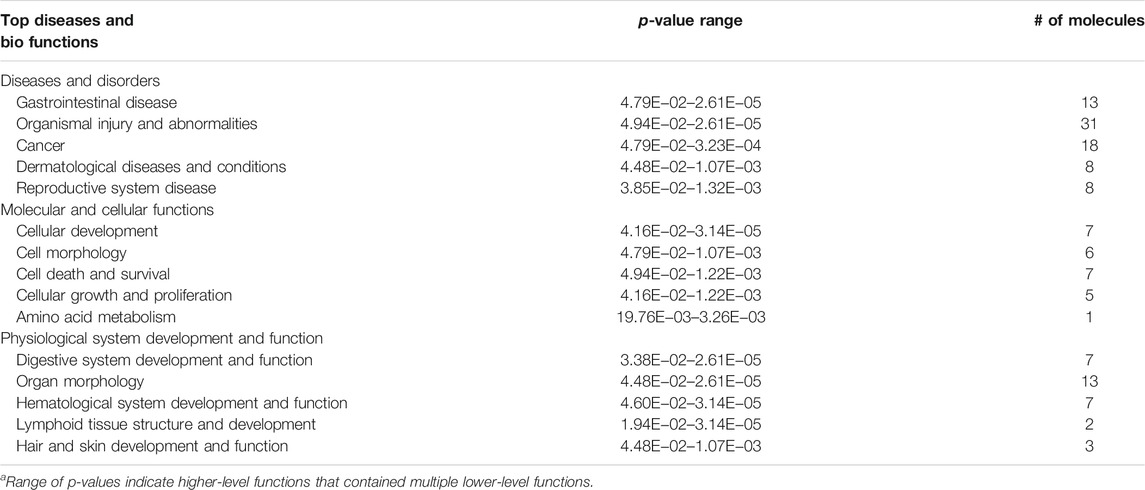
TABLE 1. Functional analysis for the DEGs in the retina between Asrgl1 knockout mice and normal controls.
Glucocorticoid Receptor Signaling Pathway Is Involved in Asrgl1 Knockout Mice
Canonical pathway analysis could inform the key metabolism and signaling pathways in which the DEGs may be involved. The main metabolism and signaling pathways in which DEGs may be engaged could be informed via canonical pathway analysis. In this study, canonical molecular pathways enriched by DEGs were explored by bioinformation analysis of IPA software. Figure 5B shows the top five canonical pathways in the retina of Asrgl1 KO mice that are significantly associated with DEGs. The glucocorticoid receptor signaling (p value = 9.36E−06), intrinsic prothrombin activation pathway (p value = 2.78E−04), and xenobiotic metabolism AHR signaling pathway (p value = 2.04E−03) were the top three major pathways altered in retinal tissues in Asrgl1 KO mouse. The molecules that were changed in the glucocorticoid receptor signaling were IL1β, KRT5, KRT7, KRT13, KRT14, KRT15, KRT19, KRT80, and KRT6B. Our results firstly suggested an association of Asrgl1 with GR signaling in the pathological process of retinitis pigmentosa.
Influenced Interaction Network by Ablation of Asrgl1 in Mouse Retina
The molecular interaction networks were then created and ranked based on the connectivity of the detected DEGs. The network of “Cell Morphology, Embryonic Development” (score = 49) (Figure 6) was the most enriched network alerted in retinal tissues of Asrgl1 KO mice. Notably, several genes including NFE2L2, IL-4, Foxp3, and Fos were the core “nodes” with the most connections in the network. To validate the expression levels of the main altered genes in glucocorticoid receptor signaling pathway and network of “Cell Morphology, Embryonic Development,” real-time PCR was performed. As Figure 7 shows, the expression of KRT5, KRT7, KRT13, KRT14, KRT15, KRT19, and KRT80 in glucocorticoid receptor signaling pathway all increased in Asrgl1 knockout mice compared to the normal controls. Interestingly, only the expression of Foxp3 was decreased among the selected “central node” genes, namely, NFE2L2, IL-4, Foxp3, and Fos. As Foxp3 is a transcriptional regulator that is required for regulatory T-cell development and inhibitory activity (Treg) (Li et al., 2015), its function in Arsgl1-related RP needs to be explored further in follow-up studies.
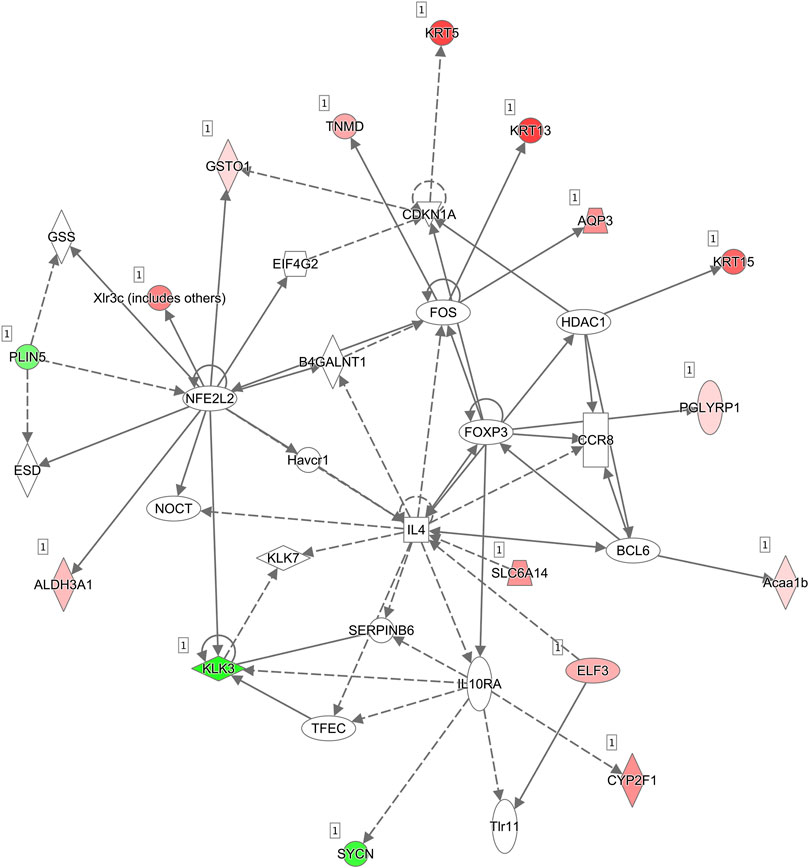
FIGURE 6. Molecular interaction networks. The most significant biological network of “Cell Morphology, Embryonic Development” was generated. Upregulated mRNAs are indicated in red, while downregulated mRNAs are in green. Solid lines represent the direct function, while the dotted lines represent the indirect function.
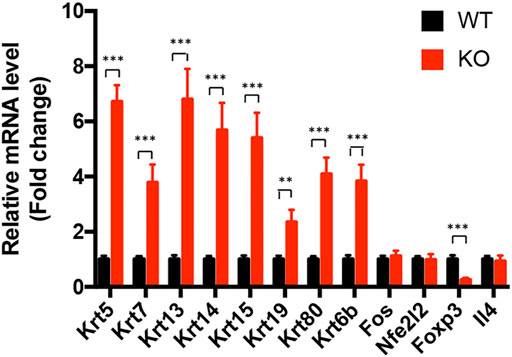
FIGURE 7. Transcriptional validation of the differentially expressed genes in Asrgl1 KO mice. The expression of Krt5, Krt7, Krt13, Krt14, Krt15, Krt19, and Krt80 in glucocorticoid receptor signaling pathway and the “central node” genes, NFE2L2, IL-4, Foxp3, and Fos, in the interaction network between WT and Asrgl1 KO mice (n = 6) was validated by real-time PCR. A two-tailed Student’s t-test was used; ∗∗p < 0.01; ∗∗∗p < 0.001. Data were from an average of four samples per group. Each test was repeated at least three times.
Discussion
RP is the leading cause of inherited blindness in the developed world. Although more than 90 genes have been found associated to RP, some of the gene functions remain unknown (Dias et al., 2018). There are numerous RP animal models available, which have contributed to a better understanding of the disease’s pathogenesis and the development of therapeutic strategies targeted at curing or delaying the hereditary illness (Petersen-Jones, 1998). In this study, we generated a novel Asrgl1 knockout mouse model to investigate the in vivo roles of ASRGL1, a new RP gene reported in 2016.
We found Asrgl1 ablation led to an attenuated ERG response around 8 months. The thickness of the ONL started to decrease at around 9 months in Asrgl1 KO mice and gradually intensified at 12 and 15 months. Thinner IS and OS as well as the diminished expression of functional markers in the rod and cone were revealed by immunostaining in Asrgl1 KO mice. Although the patients exhibited an early-onset retinal dystrophy, late-onset retinal dystrophy was observed in Asrgl1 KO mice. It is not uncommon for the phenotypes of people with Asrgl1 mutations and those of knockout mice to differ. One example is the difference between patients with CRB1 mutations and retinal degeneration-8 (rd8) mice phenotypes. CRB1 mutations led to retinitis pigmentosa 12, LCA8, or childhood- and juvenile-onset cone–rod dystrophy (den Hollander et al., 2004; Bujakowska et al., 2012). The majority of the patients exhibited severe vision loss. However, Mehalow et al. (2003) described a new mouse model, rd8, with a single-base loss in the Crb1 gene in mice. rd8 mutation resulted in a frameshift and premature stop codon. The truncated protein lost the transmembrane and cytoplasmic domains of CRB1. However, rd8 homozygous mutant mice only exhibited mild retinal degeneration. Photoreceptor degeneration was observed only within spotted regions of the retina. Furthermore, the severity of retinal pathology is influenced by genetic backgrounds (Aleman et al., 2011; Luhmann et al., 2015). Thus, our Asrgl1 knockout mouse model showed a late-onset photoreceptor degeneration phenotype, which is valuable to investigate the pathogenesis in ASRGL1-related RP and further assess the in vivo function of Asrgl1 in the retina.
The 308-amino-acid ASRGL1 protein is activated by autocleavage at amino acid 168, forming an alpha- and beta-chain that can dimerize into a heterodimer (Cantor et al., 2009; Li et al., 2012). In some brain regions, the ASRGL1 enzyme is involved in the synthesis of L-aspartate, which can operate as an excitatory neurotransmitter. In endometrioid endometrial carcinoma, loss of ASRGL1 expression was an independent biomarker for disease-specific survival, and combined ASRGL1 and p53 could be used as an independent predictor of survival (Edqvist et al., 2015; Huvila et al., 2018). Asrgl1 was reported to be trafficked by neural stem/progenitor cell (NSC)-derived extracellular vesicles (EVs) and thus enables NSC EVs as independent metabolic units with asparaginase activity (Iraci et al., 2017). Biswas et al. (2016) showed that the p.G178R mutation in ASRGL1 impaired the autocatalytic processing of ASRGL1 and resulted in the function loss of ASRGL1, which caused early-onset recessive retinal degeneration in a five-generation pedigree. Although the authors observed retinal abnormalities and loss of cone photoreceptors in a zebrafish model, the in vivo function of ASRGL1 in mammalian animals remains unknown. In our study, we firstly observed the photoreceptor degeneration in Asrgl1 knockout mouse models as well as the transcriptional molecular changes in the Asrgl1 knockout mouse retina.
Deletion of genomic fragments in Asrgl1 knockout mice was performed by CRISPR/Cas9 technology. Several available antibodies against ASRGL1 were subjected to immunoblotting and immunostaining analysis. Unfortunately, none of them worked well. Since Asrgl1 is consist of seven exons and encodes a 326-amino-acid protein in Mus musculus, the deletion of a 301-bp fragment spanning exon 3 and exon 4 in Asrgl1 knockout mice most likely led to generation of shortened proteins lacking majority of the downstream domains after exon 2 and resulted in impaired formation of heterodimer comprising an alpha- and beta-chain by autocleavage.
To further reveal the molecular mechanism of ASRGL1 in mouse retinas, we explored differentially expressed genes, pathways, and interaction network in Asrgl1 knockout mouse retinas by transcriptional analysis. Our results suggested an important role of glucocorticoid receptor signaling in the physiology of Asrgl1-related retinal degeneration. Although glucocorticoid receptor (GR) has been found in various cell types of the eye, including the retina, little is known yet about the mechanism of glucocorticoid signaling in distinct layers of the eye (Kadmiel and Cidlowski, 2013; Gallina et al., 2014; Sulaiman et al., 2018). As GR signaling is commonly associated with anti-inflammatory responses, GR agonists are widely used to treat inflammatory diseases of the eye. Further investigation is warranted to explore how ASRG1L influence the glucocorticoid receptor signaling pathway and its role in the pathological process of RP. Interestingly, glucocorticoids have been reported to suppress the expression of a subset of the keratin family genes K5-K14, K6-K16, and K17 (Radoja et al., 2000). However, in the present study, several genes in the keratin gene family, such as Krt5, Krt7, Krt13, Krt14, Krt15, Krt19, and Krt80, were found to have increased transcriptional expression in the Asrgl1 knockout mice. The keratin gene family consists the highest number of members in humans with 54 distinct functional genes (Moll et al., 2008). The functions of several keratins in the eye have been reported. Krt8/keratin 8 was shown to protect against degeneration of retinal pigment epithelium under oxidative stress (Baek et al., 2017), and KRT3 and KRT12 gene mutations associated with Meesmann corneal dystrophy (Chen et al., 2015). However, the function of keratin family genes in the retinas still needs to be further investigated. Several molecular interaction networks were generated using the identified DEGs. In the most enriched network, “Cell Morphology, Embryonic Development,” NFE2L2, IL-4, Foxp3, and Fos were the network’s central “nodes” with the greatest number of connections, which indicated the important roles of these molecules in the Asrgl1 knockout retina. NFE2L2 (NRF2) is a transcription factor involved in the oxidative stress response, which has been linked to several ocular diseases (Ildefonso et al., 2016; Naguib et al., 2021). An anti-inflammatory cytokine, IL-4, has been reported to inhibit the proliferation of retinal cells (Silva et al., 2008). Foxp3 is a transcriptional regulator that is required for regulatory T-cell development and inhibitory activity (Treg). Foxp3+ Tregs can be recruited to the retina to repair abnormal angiogenesis (Deliyanti et al., 2017). C-fos has been linked to cell death and regeneration in retinal ganglion cells (Oshitari et al., 2002). Through c-Fos, inflammatory signals from photoreceptors influenced pathological retinal angiogenesis (Sun et al., 2017). Except the most enriched network “Cell Morphology, Embryonic Development,” several other interaction networks with lower scores were identified (not shown). Although, we revealed the transcriptional regulation of glucocorticoid receptor signaling pathway and several molecules in Asrgl1 KO mice, the intrinsic mechanism will be further explored in our future work.
In conclusion, our study firstly explored the function of Asrgl1 using a novel knockout mouse model and confirm its role in the pathological process of RP. Studies utilizing mouse models have been proven important not only in recapitulation of the disease phenotype in humans but also in improving our understanding of disease mechanisms, and this knockout model is valuable for further development of translational therapeutic approaches.
Data Availability Statement
The data presented in the study are deposited in the Genome Sequence Archive (https://ngdc.cncb.ac.cn/gsa/browse/CRA005193), accession number was CRA005193.
Ethics Statement
The animal study was reviewed and approved by the Animal Care and Use Committee of the Sichuan Provincial People’s Hospital.
Author Contributions
XZ and YZ designed the study. WT, WL, XJ, HY, ZJ, XL, DJ, KS, and YY performed the experiments and plotted the data. YZ and WT validated the data and drafted the manuscript. YZ and XZ reviewed and edited the manuscript. XZ funded the experiments for the study. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (81970825, 81970841, and 81221003), the Department of Science and Technology of Sichuan Province (22JCQN0028 and 2021YFS0386), the CAMS Innovation Fund for Medical Sciences (2019-12M-5-032), the Department of Chengdu Science and Technology (nos. 2018-YF05-01080-SN and 2021-YF05-01316-SN), and the National University Basic funding (ZYGX2021J026).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors would like to thank Chengdu LiLai Biotechnology Co., Ltd for providing technical assistance with histology analysis.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2021.783547/full#supplementary-material
References
Aleman, T. S., Cideciyan, A. V., Aguirre, G. K., Huang, W. C., Mullins, C. L., Roman, A. J., et al. (2011). HumanCRB1-Associated Retinal Degeneration: Comparison with Therd8 Crb1-Mutant Mouse Model. Invest. Ophthalmol. Vis. Sci. 52, 6898–6910. doi:10.1167/iovs.11-7701
Baek, A., Yoon, S., Kim, J., Baek, Y. M., Park, H., Lim, D., et al. (2017). Autophagy and KRT8/keratin 8 Protect Degeneration of Retinal Pigment Epithelium under Oxidative Stress. Autophagy 13, 248–263. doi:10.1080/15548627.2016.1256932
Bhatti, M. T. (2006). Retinitis Pigmentosa, Pigmentary Retinopathies, and Neurologic Diseases. Curr. Neurol. Neurosci. Rep. 6, 403–413. doi:10.1007/s11910-996-0021-z
Biswas, P., Chavali, V. R., Agnello, G., Stone, E., Chakarova, C., Duncan, J. L., et al. (2016). A Missense Mutation in ASRGL1 Is Involved in Causing Autosomal Recessive Retinal Degeneration. Hum. Mol. Genet. 25, 2483–2497. doi:10.1093/hmg/ddw113
Bujakowska, K., Audo, I., Mohand-Saïd, S., Lancelot, M.-E., Antonio, A., Germain, A., et al. (2012). CRB1 Mutations in Inherited Retinal Dystrophies. Hum. Mutat. 33, 306–315. doi:10.1002/humu.21653
Cantor, J. R., Stone, E. M., Chantranupong, L., and Georgiou, G. (2009). The Human Asparaginase-like Protein 1 hASRGL1 Is an Ntn Hydrolase with β-Aspartyl Peptidase Activity. Biochemistry 48, 11026–11031. doi:10.1021/bi901397h
Chen, J. L., Lin, B. R., Gee, K. M., Gee, J. A., Chung, D. W., Frausto, R. F., et al. (2015). Identification of Presumed Pathogenic KRT3 and KRT12 Gene Mutations Associated with Meesmann Corneal Dystrophy. Mol. Vis. 21, 1378–1386.
Daiger, S. P., Sullivan, L. S., and Bowne, S. J. (2013). Genes and Mutations Causing Retinitis Pigmentosa. Clin. Genet. 84, 132–141. doi:10.1111/cge.12203
Deliyanti, D., Talia, D. M., Zhu, T., Maxwell, M. J., Agrotis, A., Jerome, J. R., et al. (2017). Foxp3+ Tregs Are Recruited to the Retina to Repair Pathological Angiogenesis. Nat. Commun. 8, 748. doi:10.1038/s41467-017-00751-w
den Hollander, A. I., Davis, J., van der Velde-Visser, S. D., Zonneveld, M. N., Pierrottet, C. O., Koenekoop, R. K., et al. (2004). CRB1 Mutation Spectrum in Inherited Retinal Dystrophies. Hum. Mutat. 24, 355–369. doi:10.1002/humu.20093
Dias, M. F., Joo, K., Kemp, J. A., Fialho, S. L., Cunha, A. D. S., Woo, S. J., et al. (2018). Molecular Genetics and Emerging Therapies for Retinitis Pigmentosa: Basic Research and Clinical Perspectives. Prog. Retin. Eye Res. 63, 107–131. doi:10.1016/j.preteyeres.2017.10.004
Edqvist, P.-H. D., Huvila, J., Forsström, B., Talve, L., Carpén, O., Salvesen, H. B., et al. (2015). Loss of ASRGL1 Expression Is an Independent Biomarker for Disease-specific Survival in Endometrioid Endometrial Carcinoma. Gynecol. Oncol. 137, 529–537. doi:10.1016/j.ygyno.2015.03.055
Ferrari, S., Di Iorio, E., Barbaro, V., Ponzin, D., Sorrentino, F. S., and Parmeggiani, F. (2011). Retinitis Pigmentosa: Genes and Disease Mechanisms. Curr. Genomics 12, 238–249. doi:10.2174/138920211795860107
Gallina, D., Zelinka, C., and Fischer, A. J. (2014). Glucocorticoid Receptors in the Retina, Müller Glia and the Formation of Müller Glia-Derived Progenitors. Development 141, 3340–3351. doi:10.1242/dev.109835
Hartong, D. T., Berson, E. L., and Dryja, T. P. (2006). Retinitis Pigmentosa. The Lancet 368, 1795–1809. doi:10.1016/s0140-6736(06)69740-7
Huang, X.-F., Huang, F., Wu, K.-C., Wu, J., Chen, J., Pang, C.-P., et al. (2015). Genotype-phenotype Correlation and Mutation Spectrum in a Large Cohort of Patients with Inherited Retinal Dystrophy Revealed by Next-Generation Sequencing. Genet. Med. 17, 271–278. doi:10.1038/gim.2014.138
Huvila, J., Laajala, T. D., Edqvist, P.-H., Mardinoglu, A., Talve, L., Pontén, F., et al. (2018). Combined ASRGL1 and P53 Immunohistochemistry as an Independent Predictor of Survival in Endometrioid Endometrial Carcinoma. Gynecol. Oncol. 149, 173–180. doi:10.1016/j.ygyno.2018.02.016
Ildefonso, C. J., Jaime, H., Brown, E. E., Iwata, R. L., Ahmed, C. M., Massengill, M. T., et al. (2016). Targeting the Nrf2 Signaling Pathway in the Retina with a Gene-Delivered Secretable and Cell-Penetrating Peptide. Invest. Ophthalmol. Vis. Sci. 57, 372–386. doi:10.1167/iovs.15-17703
Iraci, N., Gaude, E., Leonardi, T., Costa, A. S. H., Cossetti, C., Peruzzotti-Jametti, L., et al. (2017). Extracellular Vesicles Are Independent Metabolic Units with Asparaginase Activity. Nat. Chem. Biol. 13, 951–955. doi:10.1038/nchembio.2422
Kadmiel, M., and Cidlowski, J. A. (2013). Glucocorticoid Receptor Signaling in Health and Disease. Trends Pharmacol. Sci. 34, 518–530. doi:10.1016/j.tips.2013.07.003
Li, W., Cantor, J. R., Yogesha, S. D., Yang, S., Chantranupong, L., Liu, J. Q., et al. (2012). Uncoupling Intramolecular Processing and Substrate Hydrolysis in the N-Terminal Nucleophile Hydrolase hASRGL1 by Circular Permutation. ACS Chem. Biol. 7, 1840–1847. doi:10.1021/cb300232n
Li, Z., Li, D., Tsun, A., and Li, B. (2015). FOXP3+ Regulatory T Cells and Their Functional Regulation. Cell Mol Immunol 12, 558–565. doi:10.1038/cmi.2015.10
Luhmann, U. F. O., Carvalho, L. S., Holthaus, S.-M. k., Cowing, J. A., Greenaway, S., Chu, C. J., et al. (2015). The Severity of Retinal Pathology in Homozygous Crb1rd8/rd8 Mice Is Dependent on Additional Genetic Factors. Hum. Mol. Genet. 24, 128–141. doi:10.1093/hmg/ddu424
Mehalow, A. K., Kameya, S., Smith, R. S., Hawes, N. L., Denegre, J. M., Young, J. A., et al. (2003). CRB1 Is Essential for External Limiting Membrane Integrity and Photoreceptor Morphogenesis in the Mammalian Retina. Hum. Mol. Genet. 12, 2179–2189. doi:10.1093/hmg/ddg232
Moll, R., Divo, M., and Langbein, L. (2008). The Human Keratins: Biology and Pathology. Histochem. Cel Biol 129, 705–733. doi:10.1007/s00418-008-0435-6
Naguib, S., Backstrom, J. R., Gil, M., Calkins, D. J., and Rex, T. S. (2021). Retinal Oxidative Stress Activates the NRF2/ARE Pathway: An Early Endogenous Protective Response to Ocular Hypertension. Redox Biol. 42, 101883. doi:10.1016/j.redox.2021.101883
Oshitari, T., Dezawa, M., Okada, S., Takano, M., Negishi, H., Horie, H., et al. (2002). The Role of C-Fos in Cell Death and Regeneration of Retinal Ganglion Cells. Invest. Ophthalmol. Vis. Sci. 43, 2442–2449.
Palczewski, K. (2006). G Protein-Coupled Receptor Rhodopsin. Annu. Rev. Biochem. 75, 743–767. doi:10.1146/annurev.biochem.75.103004.142743
Petersen-Jones, S. M. (1998). Animal Models of Human Retinal Dystrophies. Eye (Lond) 12, 566–570. doi:10.1038/eye.1998.146
Radoja, N., Komine, M., Jho, S. H., Blumenberg, M., and Tomic-Canic, M. (2000). Novel Mechanism of Steroid Action in Skin through Glucocorticoid Receptor Monomers. Mol. Cel Biol 20, 4328–4339. doi:10.1128/mcb.20.12.4328-4339.2000
Silva, A., Campellocosta, P., Linden, R., and Shollfranco, A. (2008). Interleukin-4 Blocks Proliferation of Retinal Progenitor Cells and Increases Rod Photoreceptor Differentiation through Distinct Signaling Pathways. J. Neuroimmunology 196, 82–93. doi:10.1016/j.jneuroim.2008.03.003
Sulaiman, R. S., Kadmiel, M., and Cidlowski, J. A. (2018). Glucocorticoid Receptor Signaling in the Eye. Steroids 133, 60–66. doi:10.1016/j.steroids.2017.11.002
Keywords: retinitis pigmentosa, asparaginase and isoaspartyl peptidase 1, knockout mouse model, RNA-seq, photoreceptor degeneration
Citation: Zhou Y, Tian W, Jiang X, Yang H, Jiang Z, Li X, Jiang D, Sun K, Yang Y, Liu W and Zhu X (2022) Deletion of Asrgl1 Leads to Photoreceptor Degeneration in Mice. Front. Cell Dev. Biol. 9:783547. doi: 10.3389/fcell.2021.783547
Received: 26 September 2021; Accepted: 23 November 2021;
Published: 18 January 2022.
Edited by:
Qingjiong Zhang, Sun Yat-sen University, ChinaReviewed by:
Ruifang Sui, Peking Union Medical College Hospital (CAMS), ChinaMallika Valapala, Indiana University, United States
Wenmin Sun, Sun Yat-sen University, China
Copyright © 2022 Zhou, Tian, Jiang, Yang, Jiang, Li, Jiang, Sun, Yang, Liu and Zhu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yu Zhou, emhvdXl1NDIyQHlhaG9vLmNvbQ==; Wenjing Liu, bmljb2xld2psaXVAMTYzLmNvbQ==; Xianjun Zhu, eGp6aHVAdWVzdGMuZWR1LmNu
†These authors have contributed equally to this work
 Yu Zhou1,2*†
Yu Zhou1,2*† Zhilin Jiang
Zhilin Jiang Wenjing Liu
Wenjing Liu Xianjun Zhu
Xianjun Zhu