
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell Dev. Biol. , 29 November 2021
Sec. Epigenomics and Epigenetics
Volume 9 - 2021 | https://doi.org/10.3389/fcell.2021.782268
 Weihua Tian1
Weihua Tian1 Zhang Wang1
Zhang Wang1 Dandan Wang1
Dandan Wang1 Yihao Zhi1
Yihao Zhi1 Jiajia Dong1
Jiajia Dong1 Ruirui Jiang1,2,3
Ruirui Jiang1,2,3 Ruili Han1,2,3
Ruili Han1,2,3 Zhuanjian Li1,2,3
Zhuanjian Li1,2,3 Xiangtao Kang1,2,3
Xiangtao Kang1,2,3 Hong Li1,2,3*
Hong Li1,2,3* Xiaojun Liu1,2,3*
Xiaojun Liu1,2,3*Skeletal muscle development and intramuscular fat (IMF) content, which positively contribute to meat production and quality, are regulated by precisely orchestrated processes. However, changes in three-dimensional chromatin structure and interaction, a newly emerged mediator of gene expression, during the skeletal muscle development and IMF deposition have remained unclear. In the present study, we analyzed the differences in muscle development and IMF content between one-day-old commercial Arbor Acres broiler (AA) and Chinese indigenous Lushi blue-shelled-egg chicken (LS) and performed Hi-C analysis on their breast muscles. Our results indicated that significantly higher IMF content, however remarkably lower muscle fiber diameter was detected in breast muscle of LS chicken compared to that of AA broiler. The chromatin intra-interaction was prior to inter-interaction in both AA and LS chicken, and chromatin inter-interaction was heavily focused on the small and gene-rich chromosomes. For genomic compartmentalization, no significant difference in the number of B type compartments was found, but AA had more A type compartments versus LS. The A/B compartment switching of AA versus LS showed more A to B switching than B to A switching. There were no significant differences in the average sizes and distributions of topologically associating domains (TAD). Additionally, approximately 50% of TAD boundaries were overlapping. The reforming and disappearing events of TAD boundaries were identified between AA and LS chicken breast muscles. Among these, the HMGCR gene was located in the TAD-boundary regions in AA broilers, but in TAD-interior regions in LS chickens, and the IGF2BP3 gene was located in the AA-unique TAD boundaries. Both HMGCR and IGF2BP3 genes exhibited increased mRNA expression in one-day-old AA broiler breast muscles. It was demonstrated that the IGF2BP3 and HMGCR genes regulated by TAD boundary sliding were potential biomarkers for chicken breast muscle development and IMF deposition. Our data not only provide a valuable understanding of higher-order chromatin dynamics during muscle development and lipid accumulation but also reveal new insights into the regulatory mechanisms of muscle development and IMF deposition in chicken.
Chicken is often regarded as one of the most desirable meats worldwide due to its proteins, polyunsaturated fatty acids (PUFA), calcium, phosphorus, and iron content, as well as its potential medicinal function and low cost (Soriano-Santos, 2010). In recent decades, the genetic selection for body weight, growth rate, and feed conversion rate of broiler chickens has contributed to a prominent increase in production efficiency, such as meat yield; however, there has also been a parallel decrease in meat quality. The synchronized improvement of both meat yield and quality has become a hot topic in molecular breeding in chicken. Skeletal muscle constitutes the largest proportion and most valuable component of meat yield, and its development and composition are of extreme importance in meat production efficiency and quality, particularly breast muscle in poultry.
Skeletal muscle is mainly composed of muscle fibers derived from the first formation of primary muscle fibers at 3.5 embryos old and the subsequent formation of secondary muscle fibers at 15 embryos old until hatching during the embryonic period in chicken (Bandman and Rosser, 2000; Picard et al., 2002). Myoblasts undergo first proliferation, followed by the fusion to form myotubes, then final differentiation into muscle fibers. Muscle growth and development go through two stages, an increase in the number of proliferative myofibers (termed hyperplasia) from the embryonic stage to early growth stage, and an increase in the size of myofibers (termed hypertrophy) during late growth stages. It has been demonstrated that the total number and morphological structure of myofibers are fixed before hatching or birth in poultry (Smith, 1963). Muscle development after birth mainly profits from growing muscle fiber diameter and length, which involves complex regulatory networks and signaling pathways. Compared to slow-growing native chicken, fast-growing commercial broilers showed a significant increase in diameter, cross-sectional area, and density of breast muscle fibers at hatching. It has been proposed that heavier chicks could have enhanced satellite cell proliferation and earlier differentiation. The increased activity of muscle satellite cells in the late stage of the embryo could make breast muscle development faster (Velleman et al., 2000; Sklan et al., 2003).
It was reported that myocytes and adipocytes could affect each other during development, and meat-producing animals that have increasing muscularity displayed a reduced intramuscular fat (IMF) development (Hocquette et al., 2010). IMF refers to the lipid between muscle fibers, mainly distributed in epimysium, perimysium, and endomysium. IMF content serves as a deeply predominant factor positively underlying meat quality, including flavor, tenderness, and juiciness, since IMF is particularly rich in phospholipids, an important precursor substance carrying unsaturated fatty acid. IMF content depends upon the hyperplasia (number) and hypertrophy (size) of adipocytes in muscle. Generally, the number of adipocytes, which are mainly determined by genetic factors, is stationary during embryonic and early development stages. The size of adipocyte refers to the ability of lipid droplet accumulation, which is determined by lipid hydrolysis and traffic of both endogenous exogenous triglyceride in the blood, uptake across cytomembrane, and metabolism including fatty acids esterification, oxidation, and lipolysis in the cytoplasm (Qiu et al., 2017). IMF deposition is mainly regulated by genetic and nutritional factors with medium to high heritability. Increasing research on the difference in meat quality among chicken breeds has emphasized the comparison of fast-growing commercial broilers and slow-growing native chicken breeds, indicating that the latter has superiority and priority in meat quality and palatability (Leenstra, 1986; Chen et al., 2005; Li et al., 2006; Hocquette et al., 2010; Chen et al., 2016). It has been established that the IMF level at the beginning of the growth period was a likely key determinant of the final IMF level in cattle, indicating such significance of IMF accumulation in the early period. The chicken IMF content showed a significant increase from day 56 (fast-growth age), day 98 (marketing age), and day 140 (first egg age), but reached a peak at hatching in which the protein expression profile of breast muscles was especially varied from the other ages (Liu et al., 2016). It was similar to duck, which manifests a maximum lipid level and the relative area occupied by adipocytes on cross-sections of breast muscle (Chartrin et al., 2007). Another case in point also showed that the major period of IMF deposition in chicken was from embryo day 17 to day 1 (Liu R. et al., 2017). This evidence supports the idea that more attention should be paid to IMF deposition at the early period, especially the hatching.
It has been confirmed that no genetic conflict occurs between meat quality and meat production, implying that selection is valuable to synchronously improve them in broilers (Le Bihan-Duval et al., 2008). Both skeletal muscle development and IMF deposition are sophisticated and precisely orchestrated processes mediated by a network of regulatory factors. Although several studies have highlighted the roles of coding and noncoding genes in skeletal muscle development and IMF deposition, their precise molecular mechanism has largely remained elusive, especially three-dimensional chromatin structure and interaction, which have been gradually characterized as critical new regulators and have been recognized as an emerging mediator for triggering gene expression (de Laat and Duboule, 2013; Dekker and Mirny, 2016).
High-through chromosome conformation capture (Hi-C) technique has recently been developed to study the genome-wide spatial chromatin organization and obtain genome-wide and high-resolution chromatin intra- (cis) and inter- (trans) interaction to elucidate gene regulation mechanisms, consequently phenotype formation (Belton et al., 2012; Stadhouders et al., 2012; Liu et al., 2019). Chromatin is characterized with high conformation by hierarchical structural units organized into following chromosome territory (CT), compartments including transcriptionally active gene-rich compartment A (open euchromatin) and transcriptionally inactive gene-desert compartment B (closed heterochromatin), regions of more localized interaction known as topologically associating domain (TAD) as well as chromatin loop in the nucleus (Gibcus and Dekker, 2013). It has been largely acknowledged that compartment B has a higher interaction intensity than compartment A, and A/B compartment switch (A to B and B to A) directly aids in the activation or inhibition of gene expression. TADs represent local contact-enriched areas within the compartment, which correspond to relatively isolated packing units of gene expression regulation with the spatial proximity of genes and their regulatory elements located in a long linear region (Dixon et al., 2012). Between TADs, there are boundaries, called TAD boundaries. TAD boundaries maintain TAD independence to characterize by self-association and insulation, which respectively promotes the gene promoter interaction with regulatory elements within TAD or avoids that across TAD borders, and play a crucial role in stabilizing genome structure and regulating gene expression (de Laat and Duboule, 2013; Ibrahim and Mundlos, 2020). However, the disappearing and remodeling of TAD boundaries may lead to abnormal intergenic interactions and altered gene expression levels (Dixon et al., 2015). Chromatin loops facilitate long-range interactions of the promoter with enhancer and silencer. They can affect gene regulation to a certain extent while dynamic formation and the disappearance of the loop structure occur (Stadhouders et al., 2012).
Lushi blue-shelled-egg chicken (LS) is a Chinese indigenous breed with the strengths of crude feed tolerance, strong adaptability, high disease resistance, as well as favorable meat quality. However, this breed has a low growth rate, feed conversion ratio, and meat production. Arbor Acres broiler (AA) is a specialized fast-growing commercial broiler, characterized as having fast weight gain, a high feed conversion ratio, and massive meat production, especially breast muscle.
To determine the regulation mechanisms underlying genome organization and genome-wide chromatin interactions during skeletal muscle development and IMF deposition, we comprehensively determined the chromatin interaction, A/B compartment switch, TAD distribution, and potential genes and signaling pathways, and unveiled gene regulation mediated by chromatin interaction at the chromosome scale using Hi-C in the breast muscles from commercial broiler AA and Chinese native Lushi chicken. Our data not only reveal a connection between three-dimensional chromatin interaction with muscle development and IMF deposition but also provide a valuable genomic resource for new insights into understanding the molecular mechanisms of skeletal muscle development and IMF deposition in chicken.
All birds used in this study were obtained from the Animal Center of Henan Agricultural University. The procedures of all animal experiments were approved by the Institutional Animal Care and Use Committee of Henan Agricultural University (Permit Number: 11-0085). All efforts were made to humanely slaughter the birds and consider animal welfare.
All the experimental chickens were raised in the same feeding environment with free access to water and feed. Breast muscles tissues were collected from 10-embryonic-old (E10), 14-embryonic-old (E14), 18-embryonic-old (E18), 1-day-old (1D), 3-week-old (3W), and 5-week-old (5W) AA and LS chicken (n = 6), respectively. Subsequently, they were immediately frozen in liquid nitrogen and stored at −80°C. A total of 30 5-week-old male Lushi and AA chickens (15 individuals for each breed) were selected to measure body weight, shank length, shank circumference, sternum length, body oblique length, as well as depth, width, and angle of breast muscle. Then, blood was collected from the wing vein into a 5 ml coagulation tube, underwent an incubation for 2 h, and centrifuged at 3,000 r/min for 10 min at room temperature for serum collection. According to the manufacturer’s specifications (Huili, Changchun, China), the serum biochemical indexes including alanine transaminase (ALT), aspartate transaminase (AST), glucose (GLU), lactate dehydrogenase (LDH), triglyceride (TG), total cholesterol (T-CHO), high density lipoprotein (HDL-c), low density lipoprotein (LDL-c) and very low-density lipoprotein (VLDL-c) were tested. After being humanely slaughtered, the breast muscle was harvested and infiltrated in 4% paraformaldehyde for measuring muscle fiber diameter by hematoxylin-eosin (HE) staining, intramuscular fat content by oil red O staining, and muscle TG, T-CHO content by Tissue TG and T-CHO ELISA kits (Sinobestbio, Shanghai, China) following manufacturer’s introductions. A total of 40 one-day-old male Lushi and AA chickens (20 individuals for each breed) were used to measure the birth weight. Then, based on the high uniformity of birth weight, three one-day-old chickens each breed were selected to collect breast muscle samples and equivalently mixed into one composite sample, respectively, for Hi-C analysis.
The breast muscle fiber diameter was measured by hematoxylin-eosin staining as described by Carvalho (de Carvalho and Taboga, 1996). The paraffin sections of breast muscle were immobilized with 10% paraformaldehyde (Servicebio, Wuhan, China) for 5 min, and were washed with distilled water, then stained with hematoxylin for 15 min. Following 1% hydrochloric acid-ethanol differentiation for several seconds, the paraffin sections were subsequently stained with 0.5% eosin for 3 min and then went through gradient alcohol dehydration for 2 min and vitrification by dimethylbenzene for 2 min. The cell nucleus was stained blue, and the cytoplasm was stained pink. All reactions were performed in triplicate.
The breast muscle samples (n = 3 each group) were sectioned with a freezing microtome. The frozen sections of breast muscle were immobilized with 10% paraformaldehyde (Servicebio, Wuhan, China) for 30 min, washed three times with 1× phosphate buffer saline (PBS) (Gibco, Gaithersburg, MD, United States), and then incubated with oil red O solution (Servicebio, Wuhan, China) for 15 min. Finally, the cell nuclei of breast muscle tissue were re-stained with hematoxylin (Servicebio, Wuhan, China) for 5 min. The lipid droplet was stained red, and the cell nucleus was stained blue.
The collected fresh muscle samples were cut into pieces with 2–3 mm diameter on the ice and washed in PBS (Gibco, United States). The samples were centrifugated at 4°C, 2,000 × g for 10 min and were resuspended with serum-free Dulbecco’s Modified Eagle’s medium (DMEM) (Gibco, Gaithersburg, MD, United States). The resuspended cells were DNA-protein cross-linked by a final 2% concentration of formaldehyde, incubated at room temperature for 10 min with soft shock at an interval of 2 min. The cross-linked samples were then supplemented with a final concertation of 0.2 M glycine and incubated at room temperature for 5 min, followed by 15 min on the ice to completely terminate cross-linking, and collected by centrifugation at 4°C, 2,000 × g for 10 min. Total DNA, gelatinin protein residues, and chromatin integrity were used to evaluate the crosslinking effect using the proteinase K DNA extraction method, in concert with agarose gel electrophoresis. The qualified samples were stored at −80°C for further use.
Hi-C experiment was performed as previously reported with minor modifications (Lieberman-Aiden et al., 2009). Briefly, chromatin digestion and isolation were performed by restriction enzyme MboI at 37°C, and the resulting sticky ends were filled in with nucleotides and introduced biotinylated nucleotides at ligation junctions. After that DNA fragment ends were ligated with T4 DNA ligase, purified with a ranging size from 300 to 500 bp, and sheared. Biotinylated junctions were isolated with streptavidin beads, then sequencing connectors were added to form joint products to construct the Hi-C library with an Illumina TruSeq DNA Sample Prep Kit according to the manufacturer’s instructions. Hi-C libraries were sequenced on an Illumina Novaseq 6000 platform configured for 150 bp paired-end reads. The Hi-C experiment was performed by Annoroad Gene Technology Co., Ltd. (Beijing, China).
Clean Hi-C data were generated by filtering out low-quality reads (>15% of bases with Q scores ≤ 19%), reads containing over 5% poly-N, and adapters from Hi-C raw data using Trimmomatic (Bolger et al., 2014). Clean reads were mapped to Gallus gallus 5.0 reference genome with a two-step approach embedded in HiC-Pro V2.7.8 software. The unique mapped paired-end reads were generated by filtering out the unmapped paired-end reads, paired-end reads with singleton and multi mapped paired-end reads from clean reads. Then the reads with single side mapping, self-circle ligation, dangling ends, dumped pairs and duplicated valid pairs from PCR artifacts were discarded. Only valid read pairs with unique mapped paired-ends involving two kinds of restriction fragments were used to build the contact maps. The valid paired-end reads were divided into intrachromosomal interactions (cis) reads and inter-chromosomal interactions (trans) reads, where cis rate and trans rate respectively referred to the proportion of cis and trans interaction paired-end reads in the valid paired-end reads. The raw contact maps were normalized with a sparse-based implementation of the iterative correction method by HiC-Pro and visualized by HiTC (Servant et al., 2012). Normalized interchromosomal interaction matrix was obtained from observed/expected number of contacts between all pairs of whole chromosomes to evaluate the spatial proximity of chromosomes. We performed an overall Hi-C subtraction of the Z score matrices (AA minus LS) with genomic distance using a modified LOWESS method, by which the weighted-average and weighted-standard deviation for every genomic distance were calculated to consequently normalize for genomic distance signal bias (Sanyal et al., 2012).
The A/B compartments were estimated by eigenvector analysis of the genome contact matrix after normalization using the observed/expected method (Lieberman-Aiden et al., 2009). Pearson correlation matrix responding to the correlation between each of two loci on the chromosome and the whole genome interaction were employed to dramatically sharpen the plaid pattern. In the normalized interaction matrix heat map, blue indicates a lower observed interaction frequency than expected interaction frequency, on the contrary to red. The correlation coefficient ranged from −1 to 1, and the color transited from blue to red. According to principal component analysis (PCA) analysis with dimensionality reduction of the genome contact correlation coefficient matrix, the chromatin region could be distinctly divided into two parts according to the first eigenvector, which was called the A/B compartment. The positive values and negative values in the first eigenvector respectively, determined the A and B compartments.
To determine TADs and their boundaries, we proceeded with the previously reported insulation score method (Crane et al., 2015). Briefly, an “insulation score” that reflects the aggregate of interactions occurring across each interval was assigned to genomic intervals along the chromosome to quantify TADs. Of which, the sum of interaction strength of the local chromosome regions was calculated, after smooth transformation, these valleys/minima were interpreted as TAD boundaries or areas of high local insulation, wherein that the minima of insulation profile in total local interaction located at the TAD boundary. It involves two steps, first, the interaction strength value of each bin was calculated by summing its interaction strength in the 10 kb binned Hi-C data (the number of unique mapped read pairs to these regions), subsequently, the interaction strength of each bin served as a value to draw its fluctuation curve on the chromosome to define the boundary.
The miRanda (http://www.microrna.org/microrna/home.do) and TargetScan (http://www.targetscan.org/vert_72/) were employed to predict the potential target genes of miRNA. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis of genes was conducted using DAVID (http://david.abcc.ncifcrf.gov/) with a cut-off criterion of p value ≤ 0.05.
Total RNA was extracted from the breast muscle tissues of E10, E14, E18, 1D, 3W, and 5W AA and LS chickens (n = 6 each group) using TRIzol® reagents (Invitrogen, Carlsbad, CA) according to the manufacturer’s protocol. The RNA integrity was detected using 1% TAE agarose gel electrophoresis and Agilent Bioanalyzer 2100 system (Agilent Technologies, CA, United States). The concentration, as well as purity assessed by OD260nm/280 nm and OD260nm/230nm, of RNA samples, were measured using NanoPhotometer® spectrophotometer (IMPLEN, CA, United States). The RNA samples with the ratios of 28S/18S band brightness more than 1.5, as well as 1.8–2.0 of OD260 nm/280 nm and 2.0–2.3 of OD260 nm/230 nm were reversely transcribed into cDNA using PrimerScriptTM RT reagent kit (Takara, Kyoto, Japan) following the manufacturer’s instructions.
To detect mRNA expression profiles, SYBR green-based qRT-PCRs were performed in triplicate on a LightCycler®96 Real-Time PCR system (Roche Applied Science, United States) in a 10 μl reaction volume containing 5 μl 2 × SYBR® Premix Ex Taq™ II (Takara, Kyoto, Japan), 3 μl RNase-free water, 0.5 μl each of forward and reverse primers (10 μM), and 1 μl cDNA (about 300 ng). The qRT-PCR amplification procedure comprised an initial denaturation at 95°C for 5 min; 40 cycles of denaturation at 95°C for 30 s, annealing at optimum temperature for 30 s and extension at 72°C for 30 s; followed by a final extension at 72°C for 10 min. The housekeeping gene GAPDH was served as an internal control to normalize the relative gene expression. The mRNA relative expression was calculated using the 2−∆∆Ct method. The primers were designed via NCBI Primer-BLAST (https://www.ncbi.nlm.nih.gov/tools/primer-blast/) and synthesized by Sangon Biotech (Shanghai, China). The qRT-PCR primer sequences are represented as follows: IGF2BP3-F: GCCTTGGCAGTTGGAGCTAT, IGF2BP3-R: AGCTTGGCATCTGGTCCTTC; HMGCR-F: GGTGTCCTTGTCCGGGGAG, HMGCR-R: GGCCATGCATTCGAAAAAGC; GAPDH-F: AGAACATCATCCCAGCGT, GAPDH-R: AGCCTTCACTACCCTCTTG.
All data were presented as mean ± standard deviation (SD), and the statistical significance between the two experimental groups was evaluated by t-test for comparisons using SPSS 23.0 (IBM, Chicago, IL, United States). The *p value < 0.05 was considered statistically significant, and **p value < 0.01 was considered extremely significant. Graphics were produced via GraphPad Prism 8 (GraphPad Software, San Diego, CA, United States).
Compared with Lushi chicken, AA broiler is characterized by extremely significant higher live weight (p = 3.35E-22) and body size indexes at 5 weeks old including shank length (p = 3.68E-19), shank circumference (p = 1.47E-14), sternal length (p = 2.02E-22), body length (p = 8.87E-27), chest depth (p = 3.23E-23), chest width (p = 9.19E-23) and chest angle (p = 3.31E-20) (Table 1). There was no significant difference in the serum levels of ALT (p = 0.581), AST (p = 0.367), GLU (p = 0.083), LDH (p = 0.823), and TG (p = 0.112) between 5-week-old AA broiler and Lushi chicken, although serum TG exhibited an increased trend in LS chicken. Of note, significantly higher serum T-CHO (p = 0.005), HDL-c (p = 0.005), LDL-c (p = 0.001), and VLDL-c (p = 0.047) were found in LS chicken versus AA broiler (Table 1). The remarkably increased IMF content of pectorals (Figure 1A), but remarkably decreased breast muscle fiber diameter (Figure 1B) were observed in Lushi chicken in comparison with AA broiler at 5 weeks old. Simultaneously, the conspicuously superior IMF content was uncovered in one-day-old LS chicken compared to the AA broiler (Figure 1C), but the latter exhibited a prominent breast muscle fiber diameter in pectorals (Figure 1D). Moreover, significantly increased TG level was observed in the muscle of one-day-old LS chicken compared to AA (p = 0.0010) (Figure 1E), and there was no significant difference of muscle T-CHO between them (p = 0.1048) (Figure 1F).
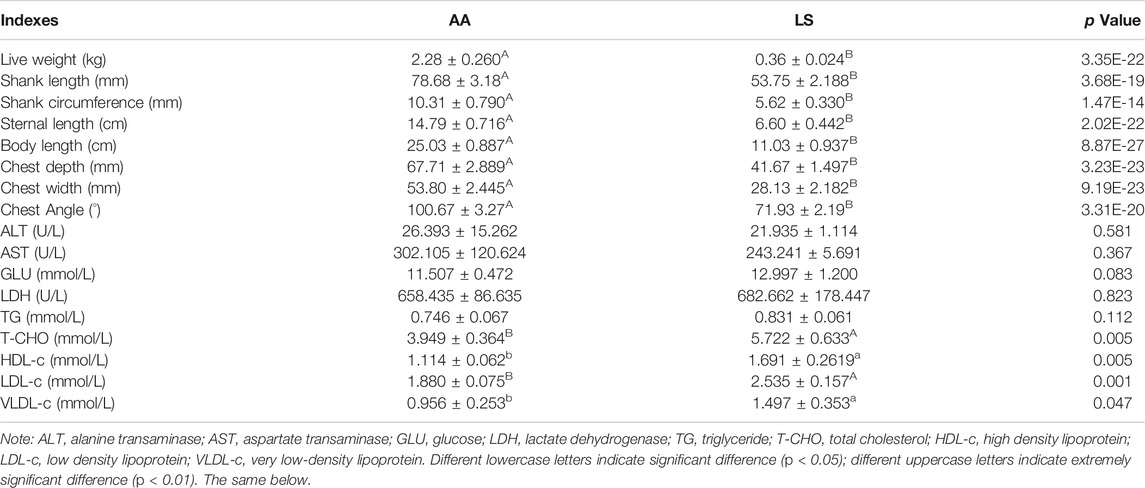
TABLE 1. The difference of body sizes and serum biochemical indexes between 5-week-old AA broiler and Lushi chicken.
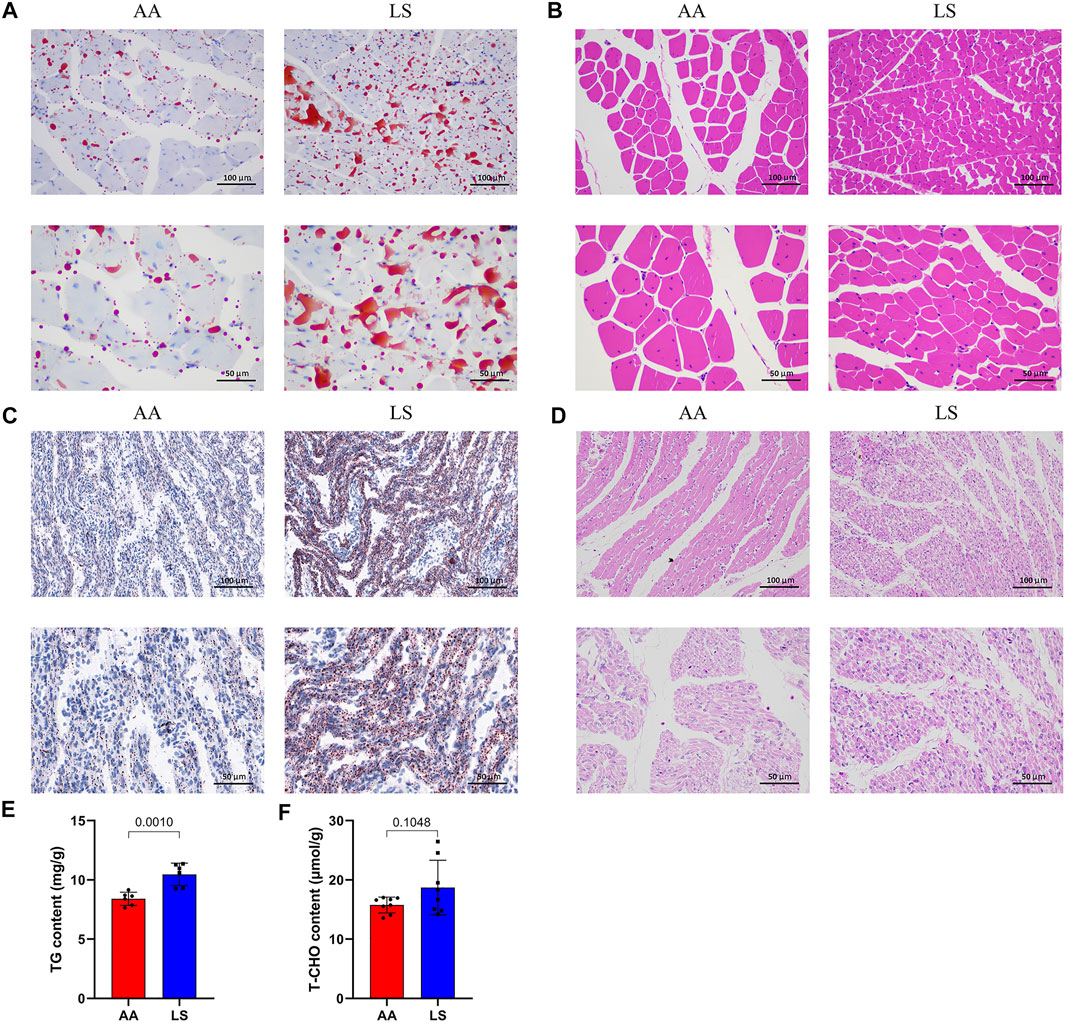
FIGURE 1. IMF deposition and fiber diameter in LS chicken breast muscle compared with AA broiler. (A) The difference in lipid droplet accumulation in the breast muscles of 5-week-old AA broiler and LS chicken as determined by oil red O staining. (B) The difference in breast muscle fiber diameter in the breast muscles of 5-week-old AA broiler and LS chicken determined by HE staining. (C) The difference in lipid droplet accumulation in the breast muscles of one-day-old AA broiler and LS chicken determined by oil red O staining. (D) The difference in breast muscle fiber diameter in the breast muscles of one-day-old AA broiler and LS chicken determined by HE staining. Scale bar: 100 μm (20X), 50 μm (40X). (E,F) The concentrations of TG and T-CHO, respectively, in the breast muscle of AA broiler and LS chicken (n = 8 for each group).
As shown in Table 2, 648,484,686 and 635,659,872 raw reads were produced from AA and LS, respectively. After filtration, clean reads totaling AA 622,163,611 (92.31%) and LS 609,409,215 (92.55%) were respectively, obtained with a Phred quality value >30 occupying 95.86 and 96.25%. Subsequently, AA 416,641,076 (66.97%) and LS 413,930,207 (67.92%) unique mapped paired-end reads from clean reads were mapped to chicken genome. After removal of dangling end paired-end reads, self-circle paired-end reads, and dumped paired-end reads, 377,935,675 (90.71%) and 314,219,395 (75.91%) unique mapped paired-end reads involving two kinds of restriction fragments, termed interaction paired-end reads, were respectively, obtained in AA and LS HiC libraries. After a final removal of reads with duplicate sequences from PCR amplification artifacts, a total of 173,384,380 (41.61% in AA unique mapped paired-end reads) and 170,953,363 (41.3% in LS unique mapped paired-end reads) valid paired-end reads were respectively, recognized for further analysis.
Whole genome-wide intra- (cis) and inter- (trans) interaction data of AA and LS chicken breast muscles were visualized as chromosome versus chromosome heat maps, where darker colors represent more frequent interaction events (Figures 2A,B). Of which, 89.98% of AA interaction data and 83.4% of LS interaction data were found as cis interaction, 10.02 and 16.6% as trans interaction (Figure 2C). Then, to evaluate whether the chromosomes clustering was altered between AA and LS chicken breast muscles, we compared the chr1-20 and chrZ genome-wide interaction differences and draw LS minus AA genome-wide heatmap of significant differential interactions. It was demonstrated that compared with AA, LS was equipped with significant increases in genome-wide interaction (Figure 2D). In order to intuitively show the trans-interaction, 1,000 bin pairs with the strongest interaction among chromosomes were used to draw circle plots. The circle plots showed that there was frequent interaction between the small, gene-rich, and high gene density chromosomes in AA and LS chicken breast muscles. Compared to LS, AA was equipped with the relatively repressed interaction frequency between large chromosomes and small chromosomes, in parallel with more frequent interaction between the large chromosomes (Figures 2E,F). Moreover, we standardized the interaction between each chromosome using the observed/expected number of contacts between all pairs of whole chromosomes to get a standardized chromosome interaction matrix in AA and LS chicken breast muscle, where the higher interaction between two chromosomes represents closer physical proximity (Figures 2G,H). As such, AA broiler breast muscle was characterized by more frequent interaction between small, gene-rich chromosomes, especially Chr25-33, in parallel with LS showing a similar trend except for the weaker interaction between Chr16 and Chr17-33. Considering that chromosome size made a difference in chromosomal interaction, we sorted all chromosomes subjected to gene density and conducted hierarchical clustering to exhibit the correlation more intuitively between chromosome size and the intensity of chromosomal interaction according to chromosomal interaction values. It was revealed that Chr30 with a size of 0.025 Mb possessed the highest gene density, following Chr32, Chr31, Chr16, Chr25, Chr33, Chr28, Chr27, Chr26, Chr23, Chr21 raging in size from 0.04 to 7 Mb (Figure 2I). Interestingly, hierarchical clustering analysis of whole chromosome positioning indicated that the chromosome trans-interaction was heavily concentrated on the small and gene-rich chromosomes in AA and LS chicken breast muscle (Figures 2J,K).
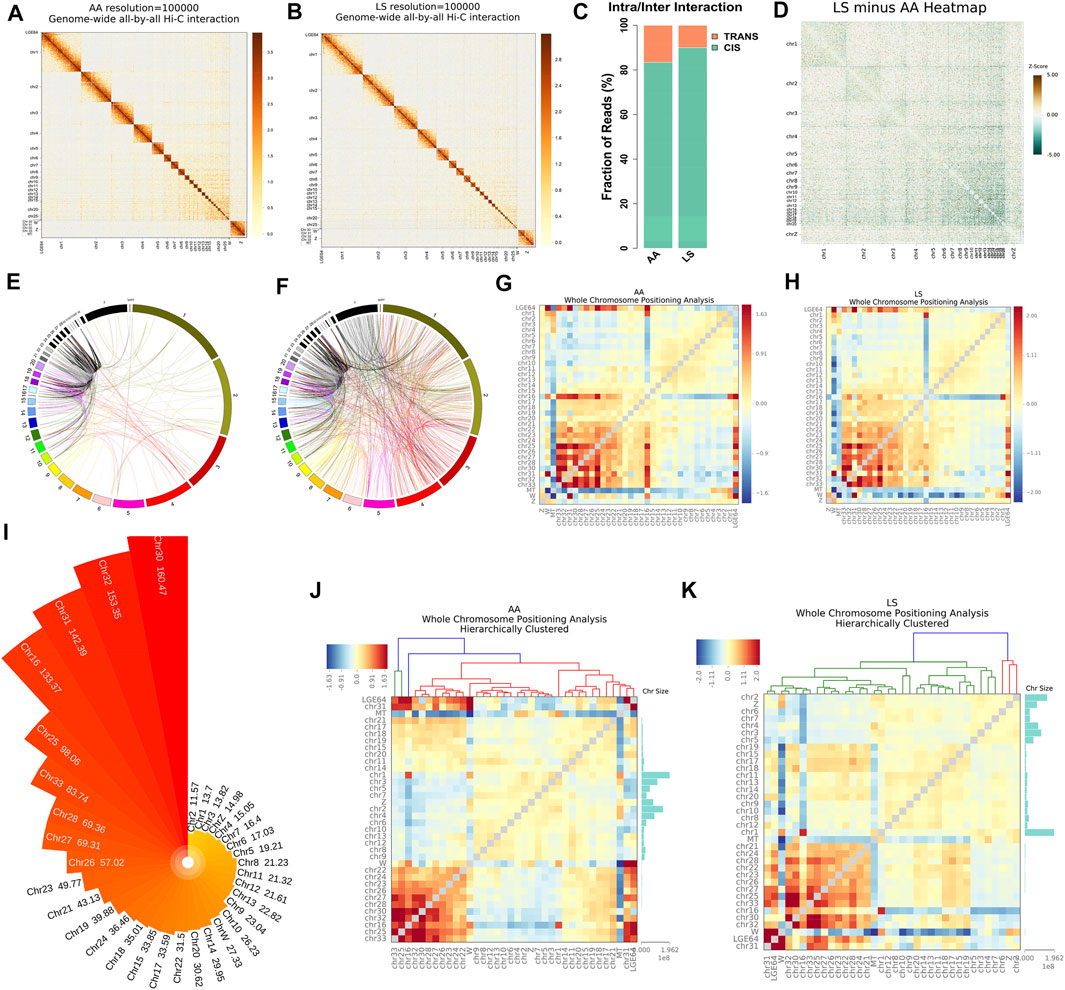
FIGURE 2. Chromatin cis/trans interaction analysis of AA and LS chicken breast muscle using Hi-C. (A,B) Genome-wide all-by-all Hi-C interaction heatmap of breast muscles in AA broiler and LS chicken, respectively. (C) Genome-wide cis/trans interaction percentage of AA and LS chicken breast muscles. (D) Genome-wide heatmap of significant differential interactions between AA and LS chicken breast muscles. Chromosomes are stacked from top left to bottom right from chr1 through chr20 and chrZ. The red color represents LS-enriched interactions and the green color represents AA-enriched interactions, the white regions are the insignificantly changed interacting regions between the two breeds. (E,F) Circle plot from 1,000 bin pairs with the strongest trans-interaction in AA and LS chicken breast muscles, respectively. (G,H) Standardized trans-interaction using observed/expected number of contacts between all pairs of whole chromosomes in AA and LS chicken breast muscles, respectively. (I) Gene density of each chromosome in chicken. (J,K) Hierarchical clustering analysis of whole chromosome positioning between chromosomes of AA and LS chicken breast muscles, respectively.
To verify whether there are any differences in the compartmentalization between the AA and LS genomes from breast muscles, we compared the A/B compartments throughout the whole genome at 100 kb resolution. There was no significant difference in the number of B type compartments between the two breeds, except more A type compartments in AA versus LS (Table 3). AA and LS chicken breast muscles shared 47.69% stable A to A compartments and 48.73% stable B to B compartments, where 2.02% of all compartments constituted a switching in genomic compartmentalization from A-type in AA to B-type in LS, referring to A to B switching, and 1.56% vice versa (Figure 3A; Supplementary Table S1). To further explore whether small and gene-rich chromosomes dominatingly contributed to the A/B compartment switching, the compartment distribution of Chr16-33 and the rest of the genomes were analyzed. A distinct increase of stable A compartments and compartments switching from A-type in AA to B-type in LS on these chromosomes were found (Figure 3B). The small, gene-rich chromosomes possessed relatively abundant A compartment and poor B compartment in proportion versus large, gene-poor chromosomes, in either LS or AA chicken genomes from breast muscle (Supplementary Figure S1). We emphasized the A/B compartment switching on Chr21-33 and observed that there was no compartment switching on Chr21, on which both AA and LS merely constituted 1.45% B to A compartment switching, together with 52.17% stable A compartments and 46.38% stable B compartments (Figure 3C; Supplementary Figure S2A). As for Chr23 and Chr26, there were powerful A-type compartments with a significant increase of compartment switching from B-type in LS to A-type in AA, in which B to A compartment switching occupied 6.90%, A to B occupied 1.72% on Chr23, as well as respectively, 7.27 and 1.82% on Chr26 (Figures 3D,E; Supplementary Figures S2B,C). On Chr22, AA and LS genomes displayed a similar distribution for the A and B compartments, which had approximately equivalent percentages, and had the same proportion of A/B compartments switching (1.89% for each of A to B and B to A switching) (Figure 3F; Supplementary Figure S2D). In terms of Chr28, both AA and LS merely constituted stable A compartments and 44% stable B compartments (Figure 3G; Supplementary Figure S2E). Only A to B compartment switching was respectively, detected on Chr24 (3.17%), Chr25 (2.86%), Chr27 (1.75%), and Chr33 (11.76%) in the breast muscles of the two kinds of chickens (Figures 3H–K; Supplementary Figures S2F,G–I).
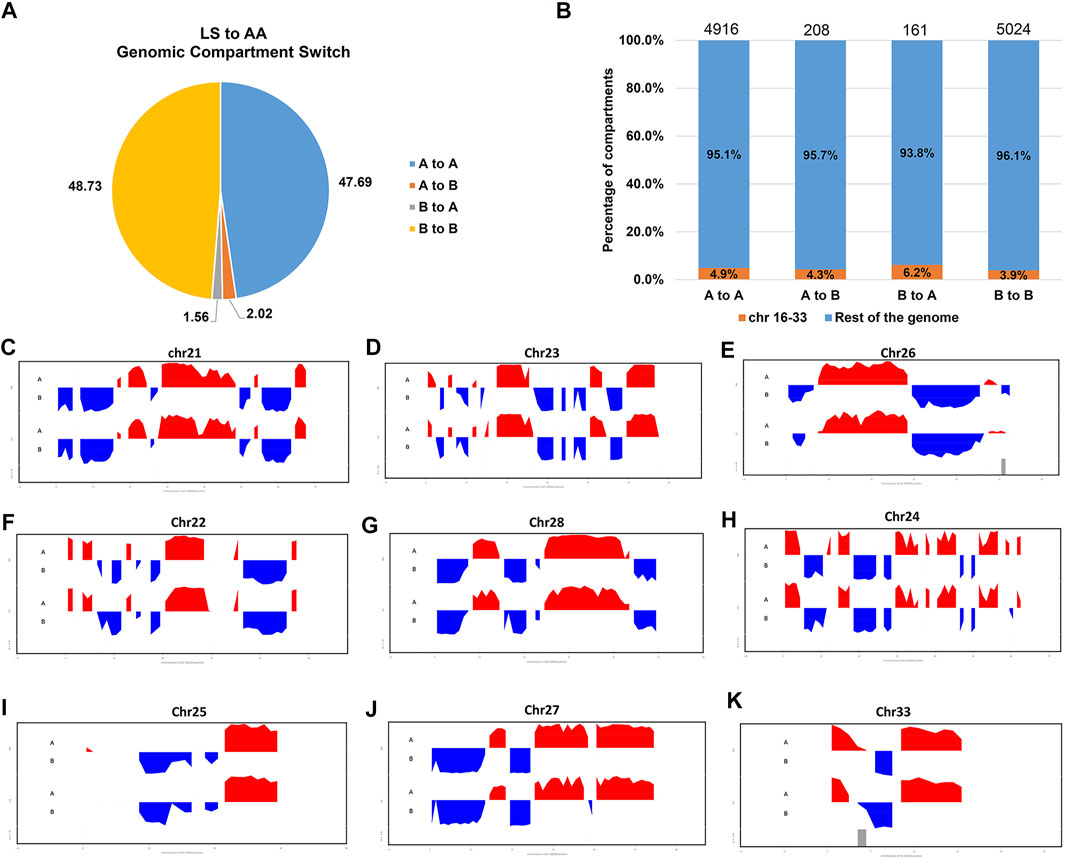
FIGURE 3. Genomic compartmentalization of AA and LS chicken breast muscles. (A) The genomic compartment changes between AA and LS chicken genomes from breast muscles. “A” and “B” mean the open and closed compartments, respectively. “A to A” means open compartments in both chickens, “B to B” represents closed compartments in both chickens, “A to B” means the compartments that are open in AA but closed in LS, and “B to A” means the compartments that are closed in AA and open in LS. (B) The percentage of compartments that have switched (A to B or B to A) or been stable (A to A or B to B) between AA and LS chicken breast muscle genomes for chr16 through chr33 (orange) and the rest of the genome (blue). (C–K) First principal component of chr21, chr23, chr26, chr22, chr28, chr24, chr25, chr27 and chr33, respectively. The upward red irregular rectangles represent open A-type compartmentalization and the downward blue irregular rectangles represent closed B-type compartmentalization.
A total of 1,226 long chain non-coding RNAs (lncRNAs), 592 microRNAs (miRNAs), and 11,980 mRNAs, were respectively, authenticated in the stable A compartments at the whole genome scale. Among those, 34 lncRNAs, 13 miRNAs, and 394 mRNAs in A to B compartment switching region, 42 lncRNAs, 6 miRNAs, and 272 mRNAs located in B to A compartment switching region, as well as 879 lncRNAs, 299 miRNAs, and 3,740 mRNAs in stable B compartments (Figure 4A; Supplementary Table S2).
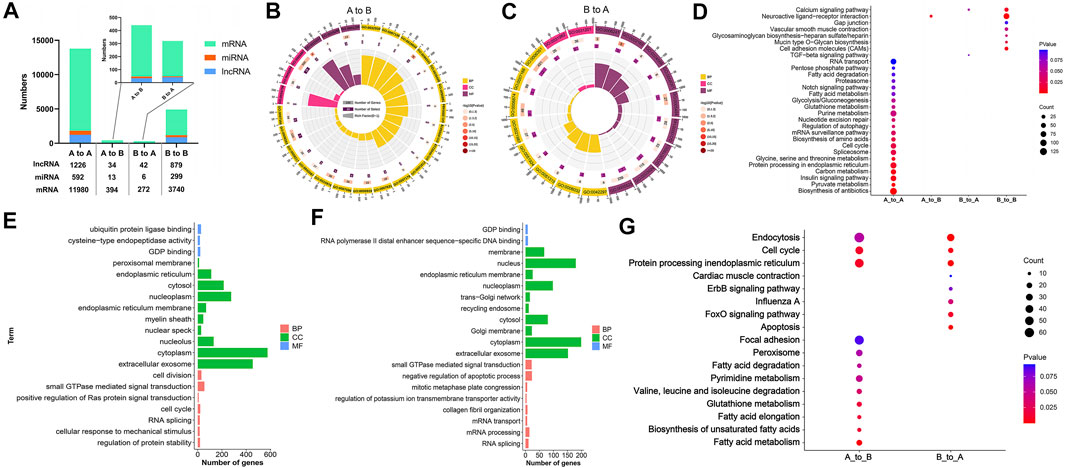
FIGURE 4. Functional annotation and signaling pathway enrichment of genes located on the A/B compartment switch region between AA and LS chicken breast muscles. (A) Distribution of lncRNA, miRNA, and mRNA in A to A, A to B, B to A, B to B compartments regions between AA and LS chicken genomes from breast muscles. (B) GO enrichment analysis of mRNAs within A to B compartment switching regions. (C) GO enrichment analysis of mRNAs within B to A compartment switching regions. (D) KEGG pathways analysis of mRNAs in A to A, A to B, B to A, B to B compartments regions. (E) GO enrichment analysis of potential targeting mRNAs of miRNAs in A to B compartment regions. (F) GO enrichment analysis of potential targeting mRNAs of miRNAs in B to A compartment regions. (G) KEGG pathways analysis of potential targeting mRNAs of miRNAs in A to B and B to A compartments regions.
To better understand the correlation of genomic compartmentalization with regulating muscle development and IMF deposition between AA and LS, we performed the functional annotation and signaling pathway enrichment analysis of genes located in the A/B switching region in their breast muscles. A total of 25 clusters based on the GO functional annotation of mRNAs in the A to B compartment switching region were validated. There were nine significant terms including negative regulation of platelet aggregation (p = 0.0030), cell proliferation (p = 0.0132), blood coagulation (p = 0.0253), positive regulation of neuron projection development (p = 0.0253), feeding behavior (p = 0.0395), negative chemotaxis (p = 0.0436), an integral component of the plasma membrane (p = 0.0123), thrombin receptor activity (p = 0.0019), and transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding (p = 0.0224) (Figure 4B; Supplementary Table S3). Moreover, 19 clusters based on the GO functional annotation of mRNAs in B to A compartment switching region were validated, including five significant terms: cellular calcium ion homeostasis (p = 0.0334), dystroglycan binding (p = 0.0016), peptidyl-dipeptidase activity (p = 0.0382), SNAP receptor activity (p = 0.0469) as well as transcription, DNA-templated (p = 0.0351) (Figure 4C; Supplementary Table S3). Moreover, the top 20 GO terms from mRNAs in stable A and B compartments according to p value were respectively, selected and showed that the majority of terms focused on cellular components, following molecular function and biological process least in both stable A and B compartments (Supplementary Figures S3A,B and Supplementary Table S3).
To identify the crucial signaling pathways that participated in muscle development and IMF deposition between AA and LS, we mapped these genes respectively, from stable and switched compartments to KEGG orthologs. The genes on A to B compartment switching region were significantly enriched in the neuroactive ligand-receptor interaction pathway (p = 0.0129). Genes in the B to A compartment switching region were enriched in the calcium signaling pathway (p = 0.0632) and TGF-β signaling pathway (p = 0.0761). Concerning mRNAs on stable A compartment, they were significantly enriched in the biosynthesis of antibiotics (p = 2.15E-05), pyruvate metabolism (p = 0.0015), insulin signaling pathway (p = 0.0052), carbon metabolism (p = 0.0055), protein processing in the endoplasmic reticulum (p = 0.0095), spliceosome (p = 0.0228), cell cycle (p = 0.0236), mRNA surveillance pathway (p = 0.0344), regulation of autophagy (p = 0.0384), nucleotide excision repair (p = 0.0410), purine metabolism (p = 0.0459) and two amino acid-related metabolism pathways including glycine, serine and threonine metabolism (p = 0.0097), and biosynthesis of amino acids (p = 0.0254). The mRNAs on the stable B compartment were significantly associated with neuroactive ligand-receptor interaction (p = 3.96E-15), cell adhesion molecules (CAMs) (p = 0.0052), calcium signaling pathway (p = 0.0116), mucin type O-Glycan biosynthesis (p = 0.0252), glycosaminoglycan biosynthesis-heparan sulfate/heparin (p = 0.0467) and vascular smooth muscle contraction (p = 0.0482) (Figure 4D; Supplementary Table S4).
Given that miRNA can act as a post-transcriptional regulator to participate in various physiological processes via complementarily binding to the 3′-untranslated region (3′UTR) of its target, to comprehensively investigate the potential effects of A/B compartment switching on muscle development and IMF deposition, the targets of miRNA from A to B compartment switching region and B to A compartment switching region were analyzed, respectively. A total of 4,722 genes were obtained from two intersecting software programs, revealing that miRanda and TargetScan were potential targets of miRNAs located on the A to B compartment switching region. They were mainly clustered in regulatory and metabolic processes, otherwise directly related terms to lipid metabolisms such as acyl-CoA dehydrogenase activity, coenzyme A biosynthetic process lipid homeostasis, and fatty-acyl-CoA binding (Figure 4E; Supplementary Table S5). Moreover, 1,479 potential targets of miRNAs located on the A to B compartment switching region were predicted and clustered in cellular activity, mRNA processing, and organelle-related terms (Figure 4F; Supplementary Table S5). The signaling pathway enrichment analysis indicated that the potential targets of miRNAs were significantly associated with fatty acid metabolism (p = 0.0029), protein processing in the endoplasmic reticulum (p = 0.0092), cell cycle (p = 0.0109), biosynthesis of unsaturated fatty acids (p = 0.0162), fatty acid elongation (p = 0.0162), glutathione metabolism (p = 0.0173), valine, leucine, and isoleucine degradation (p = 0.0352), pyrimidine metabolism (p = 0.0497) on the A to B compartment switching region, as well as endocytosis (p = 2.74E-05), apoptosis (p = 3.63E-05), protein processing in the endoplasmic reticulum (p = 0.0008), cell cycle (p = 0.0048), FoxO signaling pathway (p = 0.0115), influenza A (p = 0.0406) on the B to A compartment switching region (Figure 4G; Supplementary Table S5).
The TAD boundaries were responsible for maintaining TAD independence and structural integrity, which are necessary for gene regulation. To investigate whether TAD boundaries were altered by large-scale chromosomal interactions and genomic compartment switching between AA and LS chicken breast muscle genomes, the TAD boundaries were identified by calculating the insulation plot of genome-wide interaction maps at 40 kb resolution and found that 357 and 356 TAD boundaries in AA and LS genomes, respectively. Among those, there were 178 overlapping TAD boundaries in both AA and LS, 179 AA-specific TAD boundaries, and 178 LS-specific TAD boundaries (Figure 5A). As indicated in Figure 5B, AA and LS did not show a significant change in TAD score distribution on whole chromosomes. In terms of the average size of TADs, there was no significant change between AA and LS (Figure 5C), wherein TADs on Chr10 were charactered by the largest average size in length in both AA and LS genomes (Figures 5D,E). The TAD distributions on all chromosomes of AA and LS were shown in Figures 5F,G, respectively, indicating Chr1 possessed a maximum number of TAD and Chr17 possessed a minimum in these chromosomes with TADs.
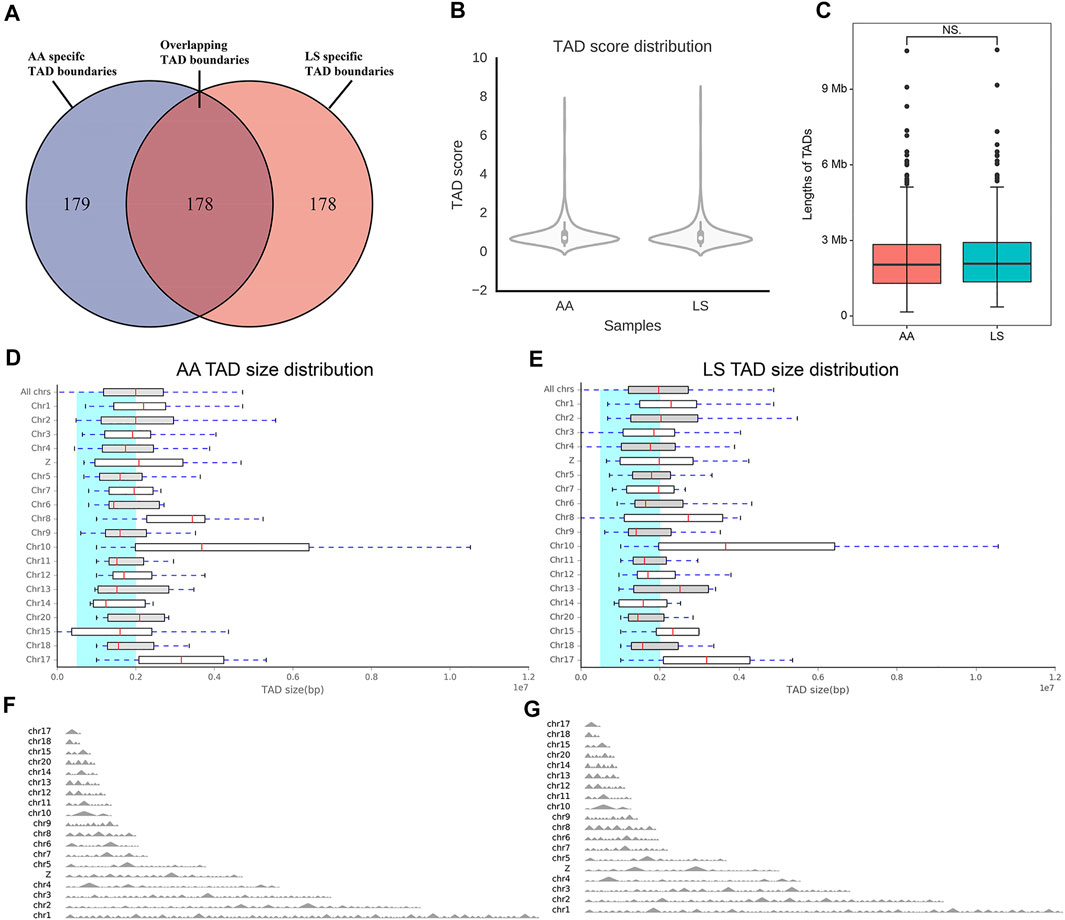
FIGURE 5. Comparative analysis of TAD between AA and LS chicken breast muscles. (A) Venn diagram showing that ∼ 50% of all the TAD boundaries between AA and LS chicken breast muscles are conserved. (B) TAD score distribution of AA and LS chicken breast muscles. (C) Differences in the lengths of TAD between AA and LS chicken breast muscles. (D,E) TAD size distribution of AA and LS chicken breast muscles, respectively. (F,G) TAD distributions on all chromosomes of AA and LS chicken breast muscles, respectively.
It has been well acknowledged that the dynamic changes of TAD boundaries give rise to the reforming and/or disappearing of TAD, resulting in the alternation of regulatory activity and following abnormal gene expression and subsequent disease phenotypes. There were 29 reforming events (Chr1, 2, 3, 4, 6, 7, 8, 14, 20, Z) and 32 disappearing events (Chr1, 2, 3, 4, 5, 6, 13, 15, Z) of TAD boundaries existing in AA versus LS chicken breast muscle. A total of 50 and 67 genes, were located on AA and LS TAD boundaries, respectively (Supplementary Table S6). And the 50 genes were clustered into two GO terms including defense response to the bacterium (p = 0.00402) and extracellular region (p = 0.00509). The 67 genes were clustered into 11 terms, including defense response to Gram-negative bacterium (p = 1.2575E-05), defense response to Gram-positive bacterium (p = 1.41986E-05), innate immune response (p = 0.00086), membrane disruption in other organisms (p = 0.00378), negative regulation of gene expression (p = 0.00672), mitochondrion (p = 0.00891), cytolysis (p = 0.02061), positive regulation of gene expression (p = 0.02377), lipopolysaccharide binding (p = 0.02779), developmental growth (p = 0.03351) as well as cell death (p = 0.03717) (Table 4). Of which, insulin like growth factor 2 mRNA binding protein 3 (IGF2BP3), a histone modification factor as m6A readers involving m6A RNA methylation were found in AA- specific TAD boundary, implying that there was probably a connection with the formation of TAD (Figure 6A). Another lipid-related gene, 3-hydroxy-3-methylglutaryl-CoA reductase (HMGCR) was located in the AA-TAD boundary, whereas in LS-TAD interior regions (Figure 6B).
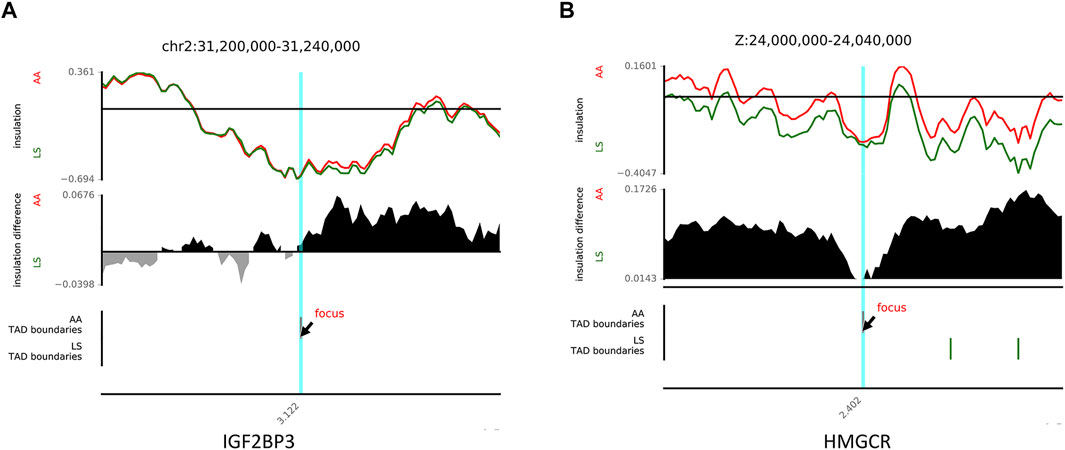
FIGURE 6. The examples of disappearing and reforming of TAD. (A) TAD disappearing showing an example of TAD boundary between AA (red) and LS (green) on chr2 (chr2:31,200,000-31,240,000). (B) TAD reforming showing an example of TAD boundary between AA (red) and LS (green) on chrZ (chrZ:24,000,000-24,040,000).
To determine the difference in the mRNA abundance of the candidates, the relative expression levels of IGF2BP3 and HMGCR genes in each developmental stage of chicken breast muscle were quantified by qRT-PCR. The expression of the IGF2BP3 gene was gradually decreased as breast muscle development was processed in both AA and LS chickens. However, a significantly increased IGF2BP3 mRNA expression was observed in the breast muscle of the AA broiler compared to LS chicken during all breast muscle development stages (p < 0.05) (Figure 7A). Similarly, the mRNA abundance of the HMGCR gene exhibited a gradually decreased trend during breast muscle development stages in LS chicken and AA broiler, while HMGCR mRNA expression was overall up-regulated in breast muscles of AA broiler than LS chicken, and was especially significantly higher at E10 (p = 0.049), E14 (p = 0.014), E18 (p = 0.020), 1D (p = 0.020), and 5W (p = 0.001) (Figure 7B).
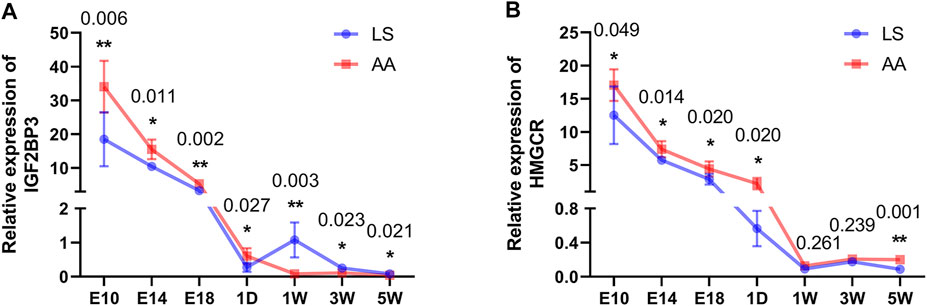
FIGURE 7. Validation of mRNA expression of IGF2BP3 and HMGCR candidates in breast muscles from AA and LS chicken by qRT-PCR. (A) IGF2BP3 mRNA expression in breast muscles from AA and LS chicken by qRT-PCR. (B) HMGCR mRNA expression in breast muscles from AA and LS chicken by qRT-PCR. The numbers over each comparison indicate p value.
Chicken has been widely accepted as a crucial research model organism for implications on phylogenetics and embryology, along with agriculture and biology, such as adipogenesis, development, immune, medicine, and cancer, owing to its convenience for operation and the extensive conservation of synteny between chicken and mammals (Burt, 2007; Beacon and Davie, 2021). Both muscle development and IMF deposition are complex and precisely orchestrated processes mediated by a network of regulatory factors directly and indirectly related to growth development and lipid metabolism. Increasing investigations are committed to elucidating these regulatory mechanisms at genomic, transcriptomic, and proteomic scales, where the genome has been deemed a linear molecular model. However, this is insufficient when it comes to revealing the connection between discrete regulatory elements, structural variations, and gene function, and consequently, it is imperative to apply three-dimensional genomics based on the spatial conformation of chromatin to explain the regulatory mechanism of gene expression. Therefore, in this study, we first identified a comparative analysis of genome-wide interactions between AA and LS chicken breast muscles upon hatching, when IMF has been proved to accumulate earliest, using Hi-C to illuminate the regulatory mechanism underlying chicken muscle development and IMF deposition at the three-dimensional chromosome scale.
Different from mammals whose lipid metabolism occurs in adipose tissue, it is the liver that principally governs lipid metabolism in chicken, where over 90% de novo fatty acids are synthesized. Endogenous lipids synthesized by the liver and exogenous sources absorbed from the diet undergo catalytic reaction into acyl-CoA and subsequently metabolization to form TG, phospholipids, and total cholesterol, which are incorporated into very low-density lipoprotein (VLDL) by apolipoprotein and then transported in the blood circulation (Leveille et al., 1968; O'Hea and Leveille, 1969). After that, VLDL is hydrolyzed into fatty acids by lipoprotein lipase (LPL) and then uptaken by fat cells in muscles, and reesterified to form TG, which is stored to enhance the IMF deposition (Qiu et al., 2017). Therefore, serum TG, T-CHO, and VLDL content mostly reflect lipid metabolism activity in chicken, and serum VLDL has been widely regarded to have a strong correlation with fat deposition (Griffin and Whitehead, 1982; Hermier, 1997). Our results showed the increased serum TG, T-CHO, and VLDL levels, together with enhanced accumulation of lipid droplets in breast muscle of LS chicken compared with that in breast muscle of AA broiler. It implied that more IMF deposition in LS chicken breast muscle might benefit from dramatically increased lipid metabolism in LS compared with AA, consistent with those signaling pathways related to lipid metabolism could contribute to the high capacity of IMF accumulation (Cui et al., 2012; Liu R. et al., 2017). Interestingly, LS chicken has markedly superior IMF content to AA broilers, while there was no significant difference of muscle T-CHO between them, suggesting a lower intramuscular cholesterol proportion of fat in breast muscle of LS chicken than that of AA broiler, indicating that the meat has lower cholesterol. Moreover, the accretion rate of IMF depends upon the muscle growth rate, that is, increasing muscularity could dilute the final fat content of muscle (Hocquette et al., 2010). Here, our study showed a larger breast muscle fiber diameter in AA than that in LS, to some extent, those might also explain why more IMF deposition existed in LS chicken breast muscle.
As expected, the normalized chicken Hi-C maps of AA and LS chicken breast muscle displayed a typical plaid-pattern and strong signals along diagonals, rooted in striking nonrandom patterns in its genomic sequence composition (Bickmore and van Steensel, 2013). The interactions occurred most frequently in cis and decay with genomic distance, in agreement with a previous report revealing that the intensity of chromatin interaction decreased as the linear distance of the genome increased, and is higher within the same chromosome than between different chromosomes (Lieberman-Aiden et al., 2009). In mammalian cell nuclei, the distribution of chromosomes is not random but closely concerned with gene density, chromosome size, GC content, and transcription activity, etc, wherein the gene-rich chromosomes concentrate at the center of the nucleus and the gene-poor chromosomes are located towards the nuclear periphery (Sun et al., 2000; Boyle et al., 2001; Parada et al., 2003; Misteli, 2007; Lieberman-Aiden et al., 2009). Our studies have revealed that, in both LS and AA breast muscle, the small gene-rich chromosomes displayed increased inter-chromosomal interaction frequency between pairs of chromosomes compared with large gene-poor chromosomes, corresponding to these chromosomes being equipped with a widely acknowledged strong physical proximity and frequent colocalization in the center of the nucleus. The formation and disappearance of chromatin interactions have been widely acknowledged to play a critical role in cell fate determination and differentiation, cooperating with gene-specific expression regulation (Bartkuhn and Renkawitz, 2008). Of note, inter-chromosomal interaction frequency showed that the large, gene-poor chromosomes in LS chicken breast muscle preferentially interacted with each other compared to AA broiler, speculating that it might contribute to explaining the difference of muscle development and IMF deposition, which needs to be further studied.
It has been acknowledged that compartment A has a more open spatial structure, higher gene density, stronger gene expression activity, and higher GC base content; on the contrary, chromatin compartment B contains centromeres and has a more closed spatial structure with lower internal gene density and lower transcriptional activity. Within the same type of compartment, chromatin interaction frequency is higher, and at the same linear distance, the interaction frequency between B type compartments is superior to A type compartments. In our study, the B compartment of either LS or AA chicken breast muscle covered approximately 60% of the chicken genome, in accordance with the point in rice (Liu C. et al., 2017). There was no significant difference in the number of B type compartments between the two breeds, more A type compartments in AA being likely to cause the dynamic regulation of gene expression related to muscle growth and development and/or lipid metabolism, resulting in a difference in muscle development and IMF deposition between AA and LS chickens. During muscle development, the programmed, controllable and orderly expression of a large number of genes leads to the specific expression of muscle cell genes, forming a complex regulatory network and signaling pathways. Simultaneously, IMF deposition is also mediated by multiple pathways and influenced by a number of genes, especially in chickens, because, unlike mammals, the liver is principally responsible for lipid metabolism in avian. According to our gene annotation analysis of mRNAs located in stable A and B compartment regions, these genes were involved in the development, enzymatic activity, transport, immune response, and other processes. The chromatin organization and stability are considered to be regulated by post-translationally histone modifications, such as methylation, acetylation, phosphorylation, and ubiquitination (Elgin and Workman, 2002; Shah, 2015). Moreover, there is a mutual dependency between chromatin organization and patterns of epigenetic marks. For example, DNA methylation is confined to the A compartment in cardiac myocytes (Nothjunge et al., 2017), unmethylated CpGs are enriched in the A compartment, and methylation levels are decreased to a greater extent in the A compartment than in the B compartment in mouse embryos (Ke et al., 2017). Moreover, both global demethylation and remethylation correlate with chromatin compartments in early embryonic development in mammals (Zhang et al., 2018), etc. Interestingly, within A compartments, there are some processes involving histone modifications, for instance, histone acetyltransferase activity, ubiquitin protein ligase activity, ubiquitin protein ligase binding, ubiquitin conjugating enzyme activity, protein dephosphorylation, ubiquitin-dependent protein catabolic process, protein autoubiquitination, ubiquinone biosynthetic process, histone H4 acetylation, suggesting that more histone modifications take place in the A compartment in chicken breast muscle. Moreover, the genes located within A compartments participate in various pathways related to energy metabolism, autophagy, fatty acid metabolism, insulin signaling pathway, and Notch signaling pathway, which have been demonstrated to be involved in muscle development and/or lipid accumulation. For example, Lactate Dehydrogenase A (LDHA), acyl-CoA synthetases (ACSs) gene family, long chain fatty acid elongases (ELOVLs) gene family, stearoyl-CoA desaturase (SCD) and histone deacetylases (HDACs) gene family, hydroxyacyl-CoA dehydrogenase (trifunctional protein), alpha subunit (HADHA), HMGCR, and mitogen-activated protein kinases (MAPKs) gene family are universal genes responsible for the development of skeletal muscle and lipid metabolism (Mashek, 2013; Li et al., 2016; Kim et al., 2017). From the late embryonic stage to hatching, a great number of genes also undergoe transcription and translation to form proteins to meet the following demands of energy at hatching. This could be the reason that the genes concerned with energy metabolism are located in the A compartment in the chicken breast muscle at hatching. The alterations of compartment types (from A to B, or from B to A) mean that changes of chromatin state (from an open active state to a closed inactive state, vice versa) directly affect the inhibition or activation of gene expression. Here, the KEGG pathways showed that the genes within A to B compartment switching regions were clustered to neuroactive ligand-receptor interaction, which has been proven to exert a role in adipocyte differentiation in human adipose-derived stem cells (Guo and Cao, 2019) and bovine muscle development (Cassar-Malek et al., 2007). Furthermore, the genes within the B to A compartment switching regions were significantly involved in the calcium signaling pathway, a highly versatile intracellular signal being capable of regulating many different processes (Berridge, 2012), as well as the TGF-β signaling pathway, which could multifunctionally regulate many cellular processes including cell growth, cell differentiation, apoptosis, morphogenesis, immune regulation, wound healing, inflammation, and cancer in both adult organisms and developing embryos (Morikawa et al., 2016). These results indicate that A/B compartment switching might yield the dynamic regulation of biological processes, leading to the difference of muscle development and IMF deposition between LS and AA chicken breast muscle. It is of note that the representative genes related to lipid metabolism, such as monoacylglycerol O-acyltransferase 1 (MOGAT1), acyl-CoA synthetase long chain family member 3 (ACSL3), lipin 1 (LPIN1), CCAAT/enhancer binding protein gamma (CEBPG), together with the representative genes related to muscle growth and development, such as insulin like growth factor 2 mRNA binding protein 1 (IGF2BP1), insulin like growth factor 1 (IGF1), myosin, and heavy chain (MYH) gene family were found within the A to B compartment regions. Regarding B to A compartment switching regions, representative genes including Wnt family member 16 (WNT16), SET and MYND domain containing 2 (SMYD2), BTG anti-proliferation factor 2 (BTG2), and PR/SET domain 6 (PRDM6) have been confirmed to exert vital functions in both muscle development and lipid metabolism in mammals. The A/B compartment switching might generate the changes of expression levels of these genes, thereby forming the distinct ability of muscle development and IMF deposition between LS and AA chickens. However, further research is needed to identify which are the dominant genes.
An increasing number of investigations have indicated that TAD is not only a structural unit of chromatin but also a functional unit involved in many biological processes, such as genome evolution, DNA replication, and gene regulation (Galupa and Heard, 2017; Szabo et al., 2019). The stability of the TAD structure plays a crucial role in maintaining the systematical and sequential physiological activities of the organism. The reconstruction and/or disappearance of TAD boundaries would directly lead to abnormal gene expression, consequently causing diseases such as human finger deformities (Lupiáñez et al., 2015) and fetal severe limb anomalies (Trimouille et al., 2019). We found no significant difference in TAD distribution and size occurred in LS and AA chicken breast muscles. Almost 50% of all the TAD boundaries between LS and AA chicken breast muscles were overlapped, indicating that the TAD positioning in breast muscles between the two breeds was moderately conserved, and that, their transcriptional expression regulation varied. This might partly be limited by sequencing technology and the quality of the chicken reference genome. Similar to a previous report, it is hard to identify TADs within several of the smallest chicken chromosomes, which are poorly represented in the current assembly of the chicken genome (Schmid et al., 2015). While further improvement of the chicken genome assembly emerges, the TAD analysis of these tiny chromosomes in the interphase nucleus could be supplemented. It has been verified that the TAD boundary was enriched with histone modification, transcriptional start sites, RNA polymerase II, and transcriptional factors, and could be associated with the different gene distribution and expression pattern from TAD-interior regions (Lieberman-Aiden et al., 2009; Mourad and Cuvier, 2015; Hong and Kim, 2017). Consistent with this opinion, our study also showed that TAD boundaries comprised histone modification factors and transcriptional factors, indicating a certain connection between transcription and local chromatin topology in AA and LS chicken breast muscles.
It is noteworthy that the IGF2BP3 gene, a family of RNA-binding proteins that also acts as an RNA N6-methyladenosine reader, is located on the AA unique TAD boundaries, which were related to mRNA stability and translation. It could regulate the production, localization, and expression of downstream genes, especially IGF2, acting as a master switch governing the initiation of skeletal muscle development (Nielsen et al., 1999; Chao and D'Amore, 2008; Huang et al., 2018). A recent study demonstrated that knockdown of the IGF2BP3 gene significantly delayed differentiation and induced proliferation of C2C12 myoblasts, and revealed that IGF2BP3 might be considered as a candidate gene in pig breeding for meat production traits (Yang et al., 2021). Additionally, IGF2BP3 has been identified as a potential candidate gene related to IMF deposition in chicken, and the mRNA expression of IGF2BP3 gene in the insulin pathway was decreased in skeletal muscle of dwarf chicken, whose IMF content was significantly increased, compared with normal chicken (Lin et al., 2012; Ye et al., 2014), which is consistent with our present study that there is lower IGF2BP3 expression in LS chicken than fast-growing AA broiler. This evidence further implies that IGF2BP3 might play indispensable roles in breast muscle development and intramuscular fat deposition in chicken. HMGCR could drive the catalyzed conversion of HMG-CoA into mevalonic acid, a rate-limiting step in cholesterol synthesis, thus playing a critical role in cholesterol homeostasis (Sharpe and Brown, 2013). It has been reported that the polymorphisms of the HMGCR gene were associated with several lipid deposition- and cholesterol-related traits, and also with IMF content of gluteus medius muscle in pigs (Canovas et al., 2010; Chalupová et al., 2012). In bovine intramuscular adipocytes, the alteration of the HMGCR gene could affect the expression abundance of AMP-activated protein kinase (AMPK) and sirtuin type 1 (SIRT1), which exert important roles in the regulation of energy metabolism (Lin et al., 2020). The polymorphisms in the 3′UTR of the HMGCR gene exhibited significant association with the total cholesterol content in chicken muscle. This evidence shows a strong correlation between HMGCR gene expression with IMF deposition in livestock and poultry (Cui et al., 2010). In our present study, the decreased mRNA expression of the HMGCR gene was detected in the breast muscle of LS chicken compared to the AA broiler, in accordance with the lower T-CHO proportion of LS chicken, leading to the speculation that LS could be managed by a slower rate of intramuscular cholesterol synthesis. It was also evidenced that anti-HMGCR could inhibit muscle cell fusion and trigger atrophy on fully differentiated myotubes in vitro primary human myoblasts/myotubes isolated from human muscle biopsies of nonmyopathic patients, suggesting that HMGCR could facilitate muscle development in humans (Arouche-Delaperche et al., 2015). Additionally, RNAi-HMGCR specifically in the corpus allatum yields dwarf flies, suggesting HMGCR could control development in Drosophila (Belgacem and Martin, 2007). In chicken, polymorphism of the HMGCR gene was reported to correlate with leg muscle weight and leg muscle fiber diameter, revealing that the HMGCR gene might also exert a crucial role in muscle development (Wei et al., 2012). The lower levels of IGF2BP3 and HMGCR mRNA expression during breast muscle development in LS chicken might contribute to the suppressive effect on breast muscle development and the stimulative effect on IMF deposition. Given that classical TAD-boundary regions were linked to the transcriptional control of mammalian genomes (Dixon et al., 2012), it has been proven that, compared to TAD-interior regions, the genes located in TAD-boundary regions were prone to being highly expressed (Dong et al., 2018). This might explain the higher expression profiles of HMGCR genes in one-day-old AA chicken breast muscles, which are located in the AA TAD-boundary regions, but in LS TAD-interior regions, as well as higher expression profiles of IGF2BP3 gene located on the AA unique TAD boundaries. Collectively, this implies that the TAD boundary disappearing or sliding in LS chicken verse AA chicken could contribute to lower transcriptional expression of IGF2BP3 and HMGCR genes in LS chicken breast muscles, and was thus responsible for slower muscle development as well as higher IMF content and lower intramuscular cholesterol proportion in LS chicken, thereby suppressing the breast muscle development but yielding excellent meat quality in LS chicken. However, the specific biological functions and regulatory mechanism at chromatin level underlying chicken breast muscle development and IMF deposition require further research.
In conclusion, the chromatin structure of the chicken breast muscle from LS and AA broiler chickens was identified and compared using large-scale chromosomal cis- and trans-interactions to examine genomic compartmentalization (A/B compartments) and TAD formation. Two genes IGF2BP3 and HMGCR regulated by TAD boundary sliding were identified as potential biomarkers for chicken breast muscle development and IMF deposition. To our best knowledge, our findings are the first to illuminate the regulatory mechanism of muscle development and IMF deposition at a three-dimensional chromosomal level in chickens, providing new insight into the functional role of chromatin organization in the formation of avian economic traits.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA757147.
The animal study was reviewed and approved by the Institutional Animal Care and Use Committee of Henan Agricultural University (Permit Number: 11-0085).
WT performed the experiments, analyzed data, and prepared the manuscript. ZW and DW collected the samples and participated in immunohistochemistry study. YZ and JD performed the RNA extraction and qRT-PCR. RJ, RH and ZL participated in data analyses and provided comments on manuscript revision. XK participated in the design of the study and critical discussion on the results. HL participated in the design of the study and helped to revise the manuscript. XL conceived the study and provided overall supervision. All authors read and approved the final version of the manuscript.
This work was supported by the National Natural Science Foundation of China (No. 31902146) and the Key Scientific Research project of Higher Education of Henan Province (No. 21A230011).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2021.782268/full#supplementary-material
Arouche-Delaperche, L., Butler-Browne, G., and Benveniste, O. (2015). Effects of Auto-Antibodies Anti-signal Recognition Particle (SRP) and Anti-Hydroxymethylglutaryl-CoA Reductase (HMGCR) on Muscle Cells. Neuromuscul. Disord. 25, S250. doi:10.1016/j.nmd.2015.06.236
Bandman, E., and Rosser, B. W. C. (2000). Evolutionary Significance of Myosin Heavy Chain Heterogeneity in Birds. Microsc. Res. Tech. 50 (6), 473–491. doi:10.1002/1097-0029(20000915)50:6<473:aid-jemt5>3.0.co;2-r
Bartkuhn, M., and Renkawitz, R. (2008). Long Range Chromatin Interactions Involved in Gene Regulation. Biochim. Biophys. Acta (Bba) - Mol. Cel Res. 1783 (11), 2161–2166. doi:10.1016/j.bbamcr.2008.07.011
Beacon, T. H., and Davie, J. R. (2021). The Chicken Model Organism for Epigenomic Research. Genome 64 (4), 476–489. doi:10.1139/gen-2020-0129
Belgacem, Y. H., and Martin, J.-R. (2007). Hmgcr in the Corpus Allatum Controls Sexual Dimorphism of Locomotor Activity and Body Size via the Insulin Pathway in Drosophila. PLoS One 2 (1), e187. doi:10.1371/journal.pone.0000187
Belton, J.-M., McCord, R. P., Gibcus, J. H., Naumova, N., Zhan, Y., and Dekker, J. (2012). Hi-C: a Comprehensive Technique to Capture the Conformation of Genomes. Methods 58 (3), 268–276. doi:10.1016/j.ymeth.2012.05.001
Berridge, M. J. (2012). Calcium Signalling Remodelling and Disease. Biochem. Soc. Trans. 40 (2), 297–309. doi:10.1042/BST20110766
Bickmore, W. A., and van Steensel, B. (2013). Genome Architecture: Domain Organization of Interphase Chromosomes. Cell 152 (6), 1270–1284. doi:10.1016/j.cell.2013.02.001
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a Flexible Trimmer for Illumina Sequence Data. Bioinformatics 30 (15), 2114–2120. doi:10.1093/bioinformatics/btu170
Boyle, S., Gilchrist, S., Bridger, J. M., Mahy, N. L., Ellis, J. A., and Bickmore, W. A. (2001). The Spatial Organization of Human Chromosomes within the Nuclei of Normal and Emerin-Mutant Cells. Hum. Mol. Genet. 10 (3), 211–219. doi:10.1093/hmg/10.3.211
Burt, D. W. (2007). Emergence of the Chicken as a Model Organism: Implications for Agriculture and Biology. Poult. Sci. 86 (7), 1460–1471. doi:10.1093/ps/86.7.1460
Cánovas, A., Quintanilla, R., Gallardo, D., Díaz, I., Noguera, J. L., Ramírez, O., et al. (2010). Functional and Association Studies on the Pig HMGCR Gene, a Cholesterol-Synthesis Limiting Enzyme. Animal 4 (2), 224–233. doi:10.1017/S1751731109991145
Cassar-Malek, I., Passelaigue, F., Bernard, C., Léger, J., and Hocquette, J.-F. (2007). Target Genes of Myostatin Loss-Of-Function in Muscles of Late Bovine Fetuses. BMC Genomics 8, 63. doi:10.1186/1471-2164-8-63
Chalupová, P., Kaplanova, K., Weisz, F., Bryndova, M., Vykoukalova, Z., Jzl, M., et al. (2012). Association of 15 Candidate Genes with Meat Quality Traits in Czech Large White Pigs. Afr. J. Agric. Res. 7 (25), 3719–3728. doi:10.5897//AJAR11.2144
Chao, W., and D’Amore, P. A. (2008). IGF2: Epigenetic Regulation and Role in Development and Disease. Cytokine Growth Factor. Rev. 19 (2), 111–120. doi:10.1016/j.cytogfr.2008.01.005
Chartrin, P., Bernadet, M. D., Guy, G., Mourot, J., Hocquette, J.-F., Rideau, N., et al. (2007). Do age and Feeding Levels Have Comparable Effects on Fat Deposition in Breast Muscle of Mule Ducks? Animal 1 (1), 113–123. doi:10.1017/S1751731107658029
Chen, J., Wen, J., Wang, S., Zhao, G., Zheng, M., and Li, X. (2005). Studies on the Characteristics of Deposition of Chicken IMP and IMF. Acta Vet. Zootech. Sin. 36 (8), 843. CNKI:SUN:XMSY.0.2005-08-017.
Chen, Y., Qiao, Y., Xiao, Y., Chen, H., Zhao, L., Huang, M., et al. (2016). Differences in Physicochemical and Nutritional Properties of Breast and Thigh Meat from Crossbred Chickens, Commercial Broilers, and Spent Hens. Asian Australas. J. Anim. Sci. 29 (6), 855–864. doi:10.5713/ajas.15.0840
Crane, E., Bian, Q., McCord, R. P., Lajoie, B. R., Wheeler, B. S., Ralston, E. J., et al. (2015). Condensin-driven Remodelling of X Chromosome Topology during Dosage Compensation. Nature 523 (7559), 240–244. doi:10.1038/nature14450
Cui, H. X., Yang, S. Y., Wang, H. Y., Zhao, J. P., Jiang, R. R., Zhao, G. P., et al. (2010). The Effect of a Mutation in the 3-UTR Region of theHMGCRGene on Cholesterol in Beijing-You Chickens. Anim. Biotechnol. 21 (4), 241–251. doi:10.1080/10495398.2010.514777
Cui, H.-X., Liu, R.-R., Zhao, G.-P., Zheng, M.-Q., Chen, J.-L., and Wen, J. (2012). Identification of Differentially Expressed Genes and Pathways for Intramuscular Fat Deposition in Pectoralis Major Tissues of Fast-And Slow-Growing Chickens. BMC Genomics 13, 213. doi:10.1186/1471-2164-13-213
Carvalho, H. F., and Taboga, S. R. (1996). Fluorescence and Confocal Laser Scanning Microscopy Imaging of Elastic Fibers in Hematoxylin-Eosin Stained Sections. Histochem. Cel Biol 106 (6), 587–592. doi:10.1007/BF02473274
de Laat, W., and Duboule, D. (2013). Topology of Mammalian Developmental Enhancers and Their Regulatory Landscapes. Nature 502 (7472), 499–506. doi:10.1038/nature12753
Dekker, J., and Mirny, L. (2016). The 3D Genome as Moderator of Chromosomal Communication. Cell 164 (6), 1110–1121. doi:10.1016/j.cell.2016.02.007
Dixon, J. R., Selvaraj, S., Yue, F., Kim, A., Li, Y., Shen, Y., et al. (2012). Topological Domains in Mammalian Genomes Identified by Analysis of Chromatin Interactions. Nature 485 (7398), 376–380. doi:10.1038/nature11082
Dixon, J. R., Jung, I., Selvaraj, S., Shen, Y., Antosiewicz-Bourget, J. E., Lee, A. Y., et al. (2015). Chromatin Architecture Reorganization during Stem Cell Differentiation. Nature 518 (7539), 331–336. doi:10.1038/nature14222
Dong, Q., Li, N., Li, X., Yuan, Z., Xie, D., Wang, X., et al. (2018). Genome-wide Hi-C Analysis Reveals Extensive Hierarchical Chromatin Interactions in rice. Plant J. 94 (6), 1141–1156. doi:10.1111/tpj.13925
Elgin, S. C. R., and Workman, J. L. (2002). A Year Dominated by Histone Modifications, Transitory and Remembered. Curr. Opin. Genet. Dev. 12 (2), 127–129. doi:10.1016/s0959-437x(02)00276-9
Galupa, R., and Heard, E. (2017). Topologically Associating Domains in Chromosome Architecture and Gene Regulatory Landscapes during Development, Disease, and Evolution. Cold Spring Harb Symp. Quant Biol. 82, 267–278. doi:10.1101/sqb.2017.82.035030
Gibcus, J. H., and Dekker, J. (2013). The Hierarchy of the 3D Genome. Mol. Cel. 49 (5), 773–782. doi:10.1016/j.molcel.2013.02.011
Griffin, H. D., and Whitehead, C. C. (1982). Plasma Lipoprotein Concentration as an Indicator of Fatness in Broilers: Development and Use of a Simple Assay for Plasma Very Low Density Lipoproteins. Br. Poult. Sci. 23(4), 307–313. doi:10.1080/00071688208447962
Guo, Z., and Cao, Y. (2019). An lncRNA-miRNA-mRNA ceRNA Network for Adipocyte Differentiation from Human Adipose-Derived Stem Cells. Mol. Med. Rep. 19 (5), 4271–4287. doi:10.3892/mmr.2019.10067
Hermier, D. (1997). Lipoprotein Metabolism and Fattening in Poultry. J. Nutr. 127 (5 Suppl. l), 805S–808S. doi:10.1093/jn/127.5.805S
Hocquette, J. F., Gondret, F., Baéza, E., Médale, F., Jurie, C., and Pethick, D. W. (2010). Intramuscular Fat Content in Meat-Producing Animals: Development, Genetic and Nutritional Control, and Identification of Putative Markers. Animal 4 (2), 303–319. doi:10.1017/s1751731109991091
Hong, S., and Kim, D. (2017). Computational Characterization of Chromatin Domain Boundary-Associated Genomic Elements. Nucleic Acids Res. 45 (18), 10403–10414. doi:10.1093/nar/gkx738
Huang, H., Weng, H., Sun, W., Qin, X., Shi, H., Wu, H., et al. (2018). Recognition of RNA N6-Methyladenosine by IGF2BP Proteins Enhances mRNA Stability and Translation. Nat. Cel Biol. 20 (3), 285–295. doi:10.1038/s41556-018-0045-z
Ibrahim, D. M., and Mundlos, S. (2020). Three-dimensional Chromatin in Disease: What Holds Us Together and what Drives Us Apart? Curr. Opin. Cel Biol. 64, 1–9. doi:10.1016/j.ceb.2020.01.003
Ke, Y., Xu, Y., Chen, X., Feng, S., Liu, Z., Sun, Y., et al. (2017). 3D Chromatin Structures of Mature Gametes and Structural Reprogramming during Mammalian Embryogenesis. Cell 170 (2), 367–381.e20. doi:10.1016/j.cell.2017.06.029
Kim, J.-H., Bae, K.-H., Byun, J.-K., Lee, S., Kim, J.-G., Lee, I. K., et al. (2017). Lactate Dehydrogenase-A Is Indispensable for Vascular Smooth Muscle Cell Proliferation and Migration. Biochem. Biophys. Res. Commun. 492 (1), 41–47. doi:10.1016/j.bbrc.2017.08.041
Le Bihan-Duval, E., Debut, M., Berri, C. M., Sellier, N., Santé-Lhoutellier, V., Jégo, Y., et al. (2008). Chicken Meat Quality: Genetic Variability and Relationship with Growth and Muscle Characteristics. BMC Genet. 9, 53. doi:10.1186/1471-2156-9-53
Leenstra, F. R. (1986). Effect of Age, Sex, Genotype and Environment on Fat Deposition in Broiler Chickens-A Review. World's Poult. Sci. J. 42 (1), 12–25. doi:10.1079/WPS19860002
Leveille, G. A., O'Hea, E. K., and Chakrabarty, K. (1968). In Vivo lipogenesis in the Domestic Chicken. Exp. Biol. Med. 128 (2), 398–401. doi:10.3181/00379727-128-33022
Li, W., Li, H., Wen, J., Chen, J., Zhao, G., and Zheng, M. (2006). Association of the H-FABP and A-FABP Gene Expression with Intramuscular Fat Content in Chickens. Acta Vet. Zootech. Sin. 37 (5), 417. CNKI:SUN:XMSY.0.2006-05-000.
Li, Z., Ouyang, H., Zheng, M., Cai, B., Han, P., Abdalla, B. A., et al. (2016). Integrated Analysis of Long Non-coding RNAs (LncRNAs) and mRNA Expression Profiles Reveals the Potential Role of LncRNAs in Skeletal Muscle Development of the Chicken. Front. Physiol. 7, 687. doi:10.3389/fphys.2016.00687
Lieberman-Aiden, E., van Berkum, N. L., Williams, L., Imakaev, M., Ragoczy, T., Telling, A., et al. (2009). Comprehensive Mapping of Long-Range Interactions Reveals Folding Principles of the Human Genome. Science 326 (5950), 289–293. doi:10.1126/science.1181369
Lin, S., Li, H., Mu, H., Luo, W., Li, Y., Jia, X., et al. (2012). Let-7b Regulates the Expression of the Growth Hormone Receptor Gene in Deletion-type Dwarf Chickens. BMC Genomics 13, 306. doi:10.1186/1471-2164-13-306
Lin, H., Wei, C., Zhang, X., You, W., Jin, Q., Tan, X., et al. (2020). HMGR Overexpression and Interference Affect the Expression of Steroidogenic Genes and Cholesterol Content in Bovine Intramuscular Adipocytes. Sci. Rep. 10 (1), 16606. doi:10.1038/s41598-020-73626-8
Liu, J., Fu, R., Liu, R., Zhao, G., Zheng, M., Cui, H., et al. (2016). Protein Profiles for Muscle Development and Intramuscular Fat Accumulation at Different Post-Hatching Ages in Chickens. PLoS One 11 (8), e0159722. doi:10.1371/journal.pone.0159722
Liu, C., Cheng, Y.-J., Wang, J.-W., and Weigel, D. (2017a). Prominent Topologically Associated Domains Differentiate Global Chromatin Packing in rice from Arabidopsis. Nat. Plants 3 (9), 742–748. doi:10.1038/s41477-017-0005-9
Liu, R., Wang, H., Liu, J., Wang, J., Zheng, M., Tan, X., et al. (2017b). Uncovering the Embryonic Development-Related Proteome and Metabolome Signatures in Breast Muscle and Intramuscular Fat of Fast-And Slow-Growing Chickens. BMC Genomics 18 (1), 816. doi:10.1186/s12864-017-4150-3
Liu, L., Li, Q.-Z., Jin, W., Lv, H., and Lin, H. (2019). Revealing Gene Function and Transcription Relationship by Reconstructing Gene-Level Chromatin Interaction. Comput. Struct. Biotechnol. J. 17, 195–205. doi:10.1016/j.csbj.2019.01.011
Lupiáñez, D. G., Kraft, K., Heinrich, V., Krawitz, P., Brancati, F., Klopocki, E., et al. (2015). Disruptions of Topological Chromatin Domains Cause Pathogenic Rewiring of Gene-Enhancer Interactions. Cell 161 (5), 1012–1025. doi:10.1016/j.cell.2015.04.004
Mashek, D. G. (2013). Hepatic Fatty Acid Trafficking: Multiple forks in the Road. Adv. Nutr. 4 (6), 697–710. doi:10.3945/an.113.004648
Misteli, T. (2007). Beyond the Sequence: Cellular Organization of Genome Function. Cell 128 (4), 787–800. doi:10.1016/j.cell.2007.01.028
Morikawa, M., Derynck, R., and Miyazono, K. (2016). TGF-β and the TGF-β Family: Context-Dependent Roles in Cell and Tissue Physiology. Cold Spring Harb Perspect. Biol. 8 (5), a021873. doi:10.1101/cshperspect.a021873
Mourad, R., and Cuvier, O. (2015). Predicting the Spatial Organization of Chromosomes Using Epigenetic Data. Genome Biol. 16 (1), 182. doi:10.1186/s13059-015-0752-8
Nielsen, J., Christiansen, J., Lykke-Andersen, J., Johnsen, A. H., Wewer, U. M., and Nielsen, F. C. (1999). A Family of Insulin-like Growth Factor II mRNA-Binding Proteins Represses Translation in Late Development. Mol. Cel Biol. 19 (2), 1262–1270. doi:10.1128/MCB.19.2.1262
Nothjunge, S., Nührenberg, T. G., Grüning, B. A., Doppler, S. A., Preissl, S., Schwaderer, M., et al. (2017). DNA Methylation Signatures Follow Preformed Chromatin Compartments in Cardiac Myocytes. Nat. Commun. 8 (1), 1667. doi:10.1038/s41467-017-01724-9
O'Hea, E. K., and Leveille, G. A. (1969). Lipid Biosynthesis and Transport in the Domestic Chick (Gallus domesticus). Comp. Biochem. Physiol. 30 (1), 149–159. doi:10.1016/0010-406x(69)91309-7
Parada, L. A., Roix, J. J., and Misteli, T. (2003). An Uncertainty Principle in Chromosome Positioning. Trends Cel Biol. 13 (8), 393–396. doi:10.1016/s0962-8924(03)00149-1
Picard, B., Lefaucheur, L., Berri, C. c., and Duclos, M. J. (2002). Muscle Fibre Ontogenesis in Farm Animal Species. Reprod. Nutr. Dev. 42 (5), 415–431. doi:10.1051/rnd:2002035
Qiu, F., Xie, L., Ma, J.-e., Luo, W., Zhang, L., Chao, Z., et al. (2017). Lower Expression of SLC27A1 Enhances Intramuscular Fat Deposition in Chicken via Down-Regulated Fatty Acid Oxidation Mediated by CPT1A. Front. Physiol. 8, 449. doi:10.3389/fphys.2017.00449
Sanyal, A., Lajoie, B. R., Jain, G., and Dekker, J. (2012). The Long-Range Interaction Landscape of Gene Promoters. Nature 489 (7414), 109–113. doi:10.1038/nature11279
Schmid, M., Smith, J., Burt, D. W., Aken, B. L., Antin, P. B., Archibald, A. L., et al. (2015). Third Report on Chicken Genes and Chromosomes 2015. Cytogenet. Genome Res. 145 (2), 78–179. doi:10.1159/000430927
Servant, N., Lajoie, B. R., Nora, E. P., Giorgetti, L., Chen, C.-J., Heard, E., et al. (2012). HiTC: Exploration of High-Throughput 'C' Experiments. Bioinformatics 28 (21), 2843–2844. doi:10.1093/bioinformatics/bts521
Shah, N. (2015). Regulation of Chromosome Dynamics by Histone Modifications and Histone Variants. Ann Arbor: ProQuest LLC Press.
Sharpe, L. J., and Brown, A. J. (2013). Controlling Cholesterol Synthesis beyond 3-Hydroxy-3-Methylglutaryl-CoA Reductase (HMGCR). J. Biol. Chem. 288 (26), 18707–18715. doi:10.1074/jbc.R113.479808
Sklan, D., Heifetz, S., and Halevy, O. (2003). Heavier Chicks at Hatch Improves Marketing Body Weight by Enhancing Skeletal Muscle Growth. Poult. Sci. 82 (11), 1778–1786. doi:10.1093/ps/82.11.1778
Smith, J. H. (1963). Relation of Body Size to Muscle Cell Size and Number in the Chicken ,. Poult. Sci. 42 (2), 283–290. doi:10.3382/ps.0420283
Soriano-Santos, J. (2010). “Chemical Composition and Nutritional Content of Raw Poultry Meat,” in Handbook of Poultry Science and Technology. Editor I. Guerrero-Legarreta (Hoboken: John Wiley & Sons Press), 467–489. doi:10.1002/9780470504451.ch25
Stadhouders, R., van den Heuvel, A., Kolovos, P., Jorna, R., Leslie, K., Grosveld, F., et al. (2012). Transcription Regulation by Distal Enhancers. Transcription 3 (4), 181–186. doi:10.4161/trns.20720
Sun, H. B., Shen, J., and Yokota, H. (2000). Size-dependent Positioning of Human Chromosomes in Interphase Nuclei. Biophys. J. 79 (1), 184–190. doi:10.1016/S0006-3495(00)76282-5
Szabo, Q., Bantignies, F., and Cavalli, G. (2019). Principles of Genome Folding into Topologically Associating Domains. Sci. Adv. 5 (4), eaaw1668. doi:10.1126/sciadv.aaw1668
Trimouille, A., Tingaud-Sequeira, A., Pennamen, P., André, G., Bouron, J., Boucher, C., et al. (2019). Deletion in 2q35 Excluding the IHH Gene Leads to Fetal Severe Limb Anomalies and Suggests a Disruption of Chromatin Architecture. Eur. J. Hum. Genet. 27 (3), 384–388. doi:10.1038/s41431-018-0290-4
Velleman, S. G., Liu, X., Nestor, K. E., and McFarland, D. C. (2000). Heterogeneity in Growth and Differentiation Characteristics in Male and Female Satellite Cells Isolated from turkey Lines with Different Growth Rates. Comp. Biochem. Physiol. A: Mol. Integr. Physiol. 125 (4), 503–509. doi:10.1016/s1095-6433(00)00178-1
Wei, Y., Zhu, S. K., Zhang, S., Han, R. L., Tian, Y. D., Sun, G. R., et al. (2012). Two Novel SNPs of the 3-Hydroxy-3-Methylglutaryl Coenzyme A Reductase Gene Associated with Growth and Meat Quality Traits in the Chicken. Genet. Mol. Res. 11 (4), 4765–4774. doi:10.4238/2012.November.12.10
Yang, Y., Fan, X., Yan, J., Chen, M., Zhu, M., Tang, Y., et al. (2021). A Comprehensive Epigenome Atlas Reveals DNA Methylation Regulating Skeletal Muscle Development. Nucleic Acids Res. 49 (3), 1313–1329. doi:10.1093/nar/gkaa1203
Ye, Y., Lin, S., Mu, H., Tang, X., Ou, Y., Chen, J., et al. (2014). Analysis of Differentially Expressed Genes and Signaling Pathways Related to Intramuscular Fat Deposition in Skeletal Muscle of Sex-Linked dwarf Chickens. Biomed. Res. Int. 2014, 1–7. doi:10.1155/2014/724274
Keywords: Hi-C, A/B compartment, TAD, muscle development, IMF deposition, chicken
Citation: Tian W, Wang Z, Wang D, Zhi Y, Dong J, Jiang R, Han R, Li Z, Kang X, Li H and Liu X (2021) Chromatin Interaction Responds to Breast Muscle Development and Intramuscular Fat Deposition Between Chinese Indigenous Chicken and Fast-Growing Broiler. Front. Cell Dev. Biol. 9:782268. doi: 10.3389/fcell.2021.782268
Received: 24 September 2021; Accepted: 08 November 2021;
Published: 29 November 2021.
Edited by:
Alaguraj Veluchamy, St. Jude Children’s Research Hospital, United StatesReviewed by:
Sandra G. Velleman, The Ohio State University, United StatesCopyright © 2021 Tian, Wang, Wang, Zhi, Dong, Jiang, Han, Li, Kang, Li and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaojun Liu, eGpsaXUyMDA4QGhvdG1haWwuY29t; Hong Li, bGlob25nMTk4NzEyMDJAMTYzLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.