- 1Centro de Biología Molecular Severo Ochoa, Consejo Superior de Investigaciones Científicas, Universidad Autónoma de Madrid, Madrid, Spain
- 2Centro de Investigación Biomédica en Red de Enfermedades Raras, Instituto de Salud Carlos III, Madrid, Spain
Hedgehog (Hh) signaling is a highly regulated molecular pathway implicated in many developmental and homeostatic events. Mutations in genes encoding primary components or regulators of the pathway cause an array of congenital malformations or postnatal pathologies, the extent of which is not yet fully defined. Mosmo (Modulator of Smoothened) is a modulator of the Hh pathway, which encodes a membrane tetraspan protein. Studies in cell lines have shown that Mosmo promotes the internalization and degradation of the Hh signaling transducer Smoothened (Smo), thereby down-modulating pathway activation. Whether this modulation is essential for vertebrate embryonic development remains poorly explored. Here, we have addressed this question and show that in zebrafish embryos, the two mosmo paralogs, mosmoa and mosmob, are expressed in the head mesenchyme and along the entire ventral neural tube. At the cellular level, Mosmoa localizes at the plasma membrane, cytoplasmic vesicles and primary cilium in both zebrafish and chick embryos. CRISPR/Cas9 mediated inactivation of both mosmoa and mosmob in zebrafish causes frontonasal hypoplasia and craniofacial skeleton defects, which become evident in the adult fish. We thus suggest that MOSMO is a candidate to explain uncharacterized forms of human congenital craniofacial malformations, such as those present in the 16p12.1 chromosomal deletion syndrome encompassing the MOSMO locus.
Introduction
Communication among cells is a fundamental mechanism for the development of multicellular organisms. This communication is mostly mediated by elaborated signaling mechanisms, among which the Hedgehog (Hh) pathway represents a prototypical example. This pathway is evolutionary conserved and pleiotropically used among species (Ingham et al., 2011). Indeed, its function has been involved in a wide variety of developmental events including cell specification, proliferation, differentiation, migration, and axon guidance as well as in adult tissues’ homeostasis and regeneration (Sánchez-Camacho and Bovolenta, 2009; Briscoe and Thérond, 2013; Petrova and Joyner, 2014). These functions are exerted in different tissues and organs: among others, the central nervous system (CNS), the limbs, the vascular system, and the craniofacial structures (Abramyan, 2019; Sasai et al., 2019).
Hedgehog signaling relies on the widespread participation of core components of the pathway such as the transmembrane proteins Patched (Ptc) and Smoothened (Smo). By default, Ptc blocks the function of Smo that remains localized in endosomes. Upon Hh ligand binding, Ptc releases Smo inhibition, enabling Smo localization at the primary cilium of the targeted cell, thereby initiating the activation of specific intracellular cascades (Murone et al., 1999). The diversification and specificity of the signaling outputs is instead fostered by the participation of other components that have more restricted spatio-temporal distributions and/or can modify intracellular signaling in a context dependent manner. These include, for example, the ligands themselves [i.e., Sonic (Shh), Indian (Ihh), and Desert (Dhh) hedgehog], a number of Hh binding proteins such as Boc, Cdon, and Gas1 that can act both as positive (Cole and Krauss, 2003; Allen et al., 2007, 2011) or negative signaling regulators (Bergeron et al., 2011; Cardozo et al., 2014; Echevarría-Andino and Allen, 2020) and transcriptional or non-transcriptional effectors of the pathway (e.g., Gli1, Gli2, Gli3, PKA, and Src) (Jia et al., 2004; Sánchez-Camacho and Bovolenta, 2009; Yam et al., 2009; Hui and Angers, 2011). This diversity also explains the broad spectrum of congenital malformations (e.g., holoprosencephaly, ciliopathies, skeletal, and craniofacial defects) associated with mutations in gene encoding components of the Hh pathway (Sasai et al., 2019) or its defective postnatal function, which has been associated with a large number of cancer types (Jeng et al., 2020).
Whether we have unveiled the full extent of the Hh pathway complexity and of the pathologies associated to its dysfunction is still undetermined. Indeed, a recent genome-wide screen aimed at identifying novel modulators of Hh signaling using CRISPR/Cas9 technology in the NIH-3T3 mouse cell line, uncovered the existence of new pathway regulators, including an unannotated gene, now known as MOSMO (MOdulator of SMOothened) (Pusapati et al., 2018). In the same study, Mosmo was demonstrated to encode a membrane tetraspan protein, which promoted the endocytosis of the Hh transducer Smo, thereby lowering its levels at the cell plasma membrane (Pusapati et al., 2018). To what extent Mosmo participates in Hh signaling regulation in vivo, however, it is just beginning to be elucidated (Lasser et al., 2020; Kong et al., 2021; Pizzo et al., 2021).
Here, we have addressed this question and report that in zebrafish the two mosmo paralogs (mosmoa and mosmob) have an overlapping distribution in embryonic ventral neural tube and then in the larva head mesenchyme. Consistent with the latter distribution genetic inactivation of both paralogs causes frontonasal hypoplasia and craniofacial skeleton defects, suggesting that MOSMO is a candidate to explain uncharacterized forms of these type of human congenital malformations.
Methods
Fish Lines and Husbandry
AB/Tübingen (AB/Tue) zebrafish were maintained at 28°C on 14/10 h light/dark cycle. Embryos were raised at 28°C, collected and maintained in E3 medium (5 mM NaCl, 0.17 mM KCl, 0.33 mM CaCl, 0.33 mM MgSO4, 10–5% Methylene Blue). All used procedures were approved by the ethical committees for animal experimentation of the Consejo Superior de Investigaciones Científicas (CSIC) and Comunidad Autónoma de Madrid.
Chick Embryos Maintenance
Fertilized chick embryos (Santa Isabel Farm, Cordoba, Spain) were incubated at 38°C in a humidified incubator until the desired stage, determined according to Hamburger and Hamilton (1992).
Whole Mount in situ Hybridization
Total mRNA from AB/Tue zebrafish embryos was extracted using RNeasy Mini kit (Qiagen) according to manufacturer instructions. cDNA was synthesized using Super Script kit (Roche) following manufacturer instructions. PCR products, obtained from cDNA amplification using specific primers (Supplementary Table 1), were cloned in PCSA plasmid (Agilent Technologies), as described by the manufacturer. Plasmid DNA preparations were obtained using Genopure Plasmid Midi kit (Roche) following kit instructions. Digoxigenin-UTP-labeled antisense probes for in situ hybridization (ISH) were synthesized and purified using Super Script kit (Roche) following the manufacturer instructions. ISH was performed by standard procedures and visualized with NBT/BCIP (dark blue).
Cloning Procedures
The PCSA-mosmoa_p1 plasmid was used as a template to amplify by PCR mosmoa and further add an hemagglutinin tag (HA) and restriction sites with the following primers: Fw 5′-aatCTCGAGCCTGAGATGGATAAACTC-3′. Rv 5′-ttaGAA TTCTCAAGCGTAATCTGGAACATCGTATGGGTAGCCAGG AAGACACACTTC-3′. The PCR product was cloned in PCSA plasmid (Agilent Technologies) as described by the manufacturer. The mosmoa-HA fragment was then excised with restriction enzymes and cloned in the pCIG vector (Megason and McMahon, 2002) for chick embryo electroporation and in pCS2 for cell transfection and synthesis of mRNA to be injected in zebrafish embryos.
Chick Embryo Electroporation
The pCIG Mosmoa-HA plasmid (1 μg/μl) was co-injected with a pCAG-2A-Arl13b-tRFP [Arl13b-tRFP construct generated by Schmitz et al. (2017)] (1 μg/μl) into the neural tube ventricle of HH10 chick embryos followed by in ovo electroporation as previously described (Cardozo et al., 2014).
Cell Transfection, Tissue Processing, and Immunochemistry
Human embryonic kidney (HEK) cells were cultured on glass coverslips in DMEM supplemented with 10% fetal calf serum and glutamine (2 mM). The pCS2-mosmoa-HA construct was transfected using lipotransfectin (Solmeglas) following the manufacture instructions. Cells were fixed with 4% paraformaldehyde in 0.1 M phosphate buffer pH 7.2 (wt/vol) at 37°C and then washed in PBS containing 0.5% Triton-X-100. Chick embryos were fixed by immersion in 4% cold PFA overnight at 4°C, washed, incubated in a 15% sucrose-PBS solution (wt/vol), embedded and frozen in a 7.5% gelatine in 15% sucrose solution (wt/vol). Cryostat sections, whole embryos or cells samples were stained with Hoechst and α-HA antibody produced in rabbit (1:250. Sigma, H-6908) and Donkey anti-Rabbit Alexa Fluor 488 secondary antibody (Invitrogen, A-21206), following standard procedures.
Zebrafish Mutant Generation
Single guide RNAs (sgRNAs) targeting coding regions of mosmoa and mosmob for CRISPR/Cas9 deletion were designed using the tools provided by CHOPCHOP online service1 searching for potential disruption of restriction enzyme sites (Labun et al., 2019). Oligos were designed as described in Varshney et al. (2016) and their sequence is reported in Supplementary Table 1. sgRNAs were transcribed and purified using Maxi Script T7 (NEB) following the manufacturer’s instructions. sgRNAs were microinjected together with Cas9 protein (300 ng/μL; EnGen® Spy Cas9 NLS, New England Biolabs) in 1 to 2 cell stage AB/Tue using a Narishige microinjector. F0 embryos were let grown and outcrossed with wt AB/Tue fish. Genomic DNA from tail clips of F1 zebrafish embryos was amplified by PCR and digested to identify disruption of selected restriction sites. DNA from potential mutants were sequenced, those with a disrupted and truncated reading frame were selected to generate the fish lines.
Genotyping
DNA from embryos or adult fish was amplified by PCR using the primers listed in Supplementary Table 1. PCR products were digested with selected enzymes at 37°C for 2 h to distinguish among wt, heterozygous, and mutant mosmoa and/or mosmob fish.
Bone and Cartilage Staining
Cartilage staining of zebrafish larvae and adult fish bones was performed with Alcian Blue and Alizarin Red, as, respectively described in Schilling et al. (1996), Sakata-Haga et al. (2018).
Imaging and Data Processing
Embryos were immersed in 75% glycerol and whole-body images were obtained using a Leica CTR5000 stereomicroscope connected to a Leica DFC500 digital camera operated by Leica software. The adult fish stained with the Alizarin Red chromogen, which also emits red fluorescence, were photographed under fluorescent light stimulation using a Leica CTR5000 stereomicroscope connected to a Leica DFC350 FX digital camera operated by Leica software. The drawings in Figure 4G were traced in Adobe Illustrator using representative photographs of wt and mosmoa–/–;mosmob–/– adult mutants. LSM710 confocal laser scanning coupled to an AxioObserver inverted microscope (Zeiss) was used to obtain digital images of cryostat sections or cells samples. ImageJ (Fiji) software was used to process and analyze images.
Statistical Analyses
The ImageJ (Fiji) software was employed to obtain quantifications reported in Figure 3. Adult fish were anesthetized with tricaine and photographed in lateral views and the distance from the eye to the tip of the preorbital region was measured and normalized to the eye size in each one of the analyzed genotypes. Data were analyzed using GraphPad Prism 7 statistic software. One-way ANOVA test was used owing to the parametric distribution of the data, followed by Tukey’s multiple comparisons test to determine differences among groups.
Results
Mosmo Paralogs Show a Largely Overlapping Distribution in the Developing Zebrafish
The zebrafish genome carries two different paralogs of the mosmo gene: mosmoa and mosmob (ZFIN:ZDB-GENE-101203-6; ZFIN:ZDB-GENE-060929-1030). To determine their expression pattern during embryonic and larval development, we generated two different specific ISH probes for each one of the two paralogs (Figure 1A). Both mosmoa and mosmob were detected at gastrulation and bud stages as well as during somitogenesis (Figures 1H,I,O,P). At this stage shha, one of the ligands of the pathway, is expressed along the midline of the entire ventral neural tube (Figures 1B,C), from which it diffuses to pattern the adjacent cells with a mechanism highly conserved across vertebrates (Martí et al., 1995; Roelink et al., 1995). During somitogenesis, mosmoa and mosmob were also found localized along the length of the ventral neural tube (Figures 1J,Q) with an overlapping distribution that, however, was more dorsally extended than that of shha (Figures 1C–G). Specific expression was also observed in the mesenchyme surrounding the neural tube (Figures 1D–G) and in the optic vesicles (Figures 1K,R). At 2 dpf and larval stages, mosmoa and mosmob were no longer detected in the neural tube but strongly localized in the head mesenchyme (Figures 1L,M,S,T), surrounding, among others, the ethmoid plate (Figures 1N,N′,U,U′).
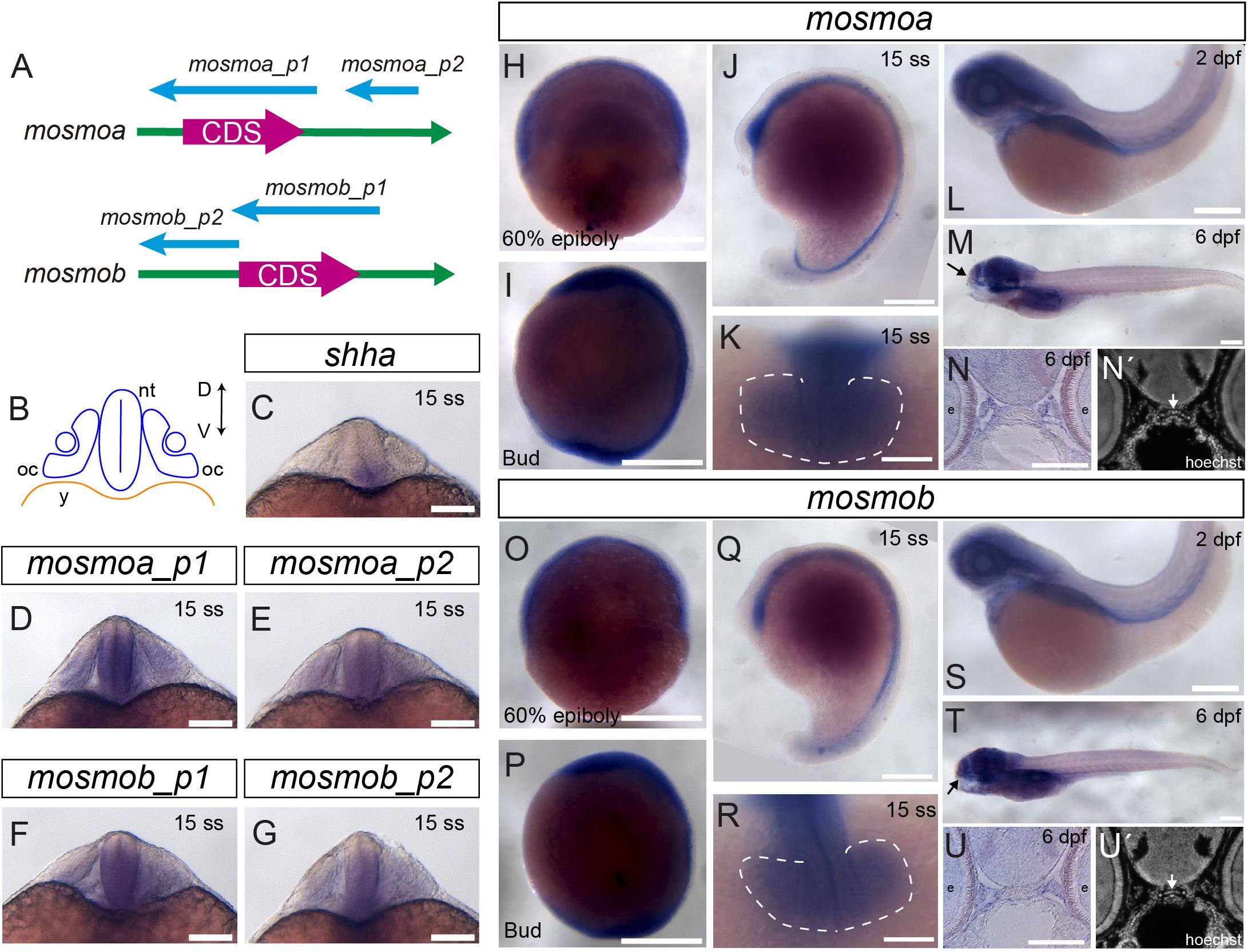
Figure 1. Mosmo paralogs show a largely overlapping distribution in zebrafish. (A) Schematic representation of mosmoa and mosmob mRNAs and probes used for in situ hybridization (ISH). p1, probe #1. p2, probe #2. (B) Schematic representation of a frontal section of a zebrafish embryo at 20 hpf at the level of the optic cup. Expression pattern of shha (C), mosmoa (D,E) and mosmob (F,G) at 20 hpf. Note that the two probes for mosmoa and mosmob show an identical distribution. Note also that both paralogs have an overlapping distribution in the ventral neural tube but more dorsally extended than that of shha. (H,U) Expression pattern of embryos hybridized with mosmoa and mosmob at 60% epiboly (H,O), bud (I,P) and 15 ss stage (J,K,Q,R), 2 dpf (L,S) as well as at 6 dpf (M,T). Note that at 60% epiboly and bud stage the expression of both genes is localized along the ventral anterior-posterior axis of the embryos. At 15 ss, a low level expression of both mosmoa and mosmob is detected in the optic vesicles (K,R, dashed line) and along the ventral neural tube from the diencephalon to the tail bud (J,Q). At 2 and 6 dpf, the expression of both genes localizes to the head region. The eyes were removed in panels (M,T). Frontal sections of 6 dpf embryos hybridized in toto for mosmoa (N) or mosmob (U) and counterstained with Hoechst (N′,U′). ISH signal for both paralogs localizes around the ethmoid cartilage (N,N′,U,U′ white arrows). D, dorsal; V, ventral; oc, optic cup; nt, neural tube; and y, yolk. Scale bars: 200 μM.
The reported patterns were consistently observed with both of the probes generated for each one of the paralogs (Figure 1), validating the reported distribution.
Mosmoa Localizes at the Plasma Membrane, Endosomes, and Primary Cilia
Attempts to determine the subcellular localization of the protein showed that, at least in NIH-3T3 cells, Mosmo localizes at the plasma membrane and endosome (Pusapati et al., 2018). To verify if this is the subcellular distribution in the developing embryo, we generated a human influenza hemagglutinin (HA) tagged version of mosmoa (mosmoa-HA). We focused on this paralog because its amino acid (aa) sequence is 100% identical to that of its human ortholog, whereas mosmob aa sequence has a lower homology (89.8% identity). We first verified the efficiency of our construct by transfecting HEK cells with the mosmoa-HA containing plasmid followed by immunostaining for HA. As reported for NIH-3T3 cells (Pusapati et al., 2018), the tagged protein was detected at the plasma membrane and endosomes (Figure 2A). When mosmoa-HA mRNA was injected in zebrafish, HA immunosignal was similarly localized at the blastomers’ plasma membrane and endosomes (Figure 2B).
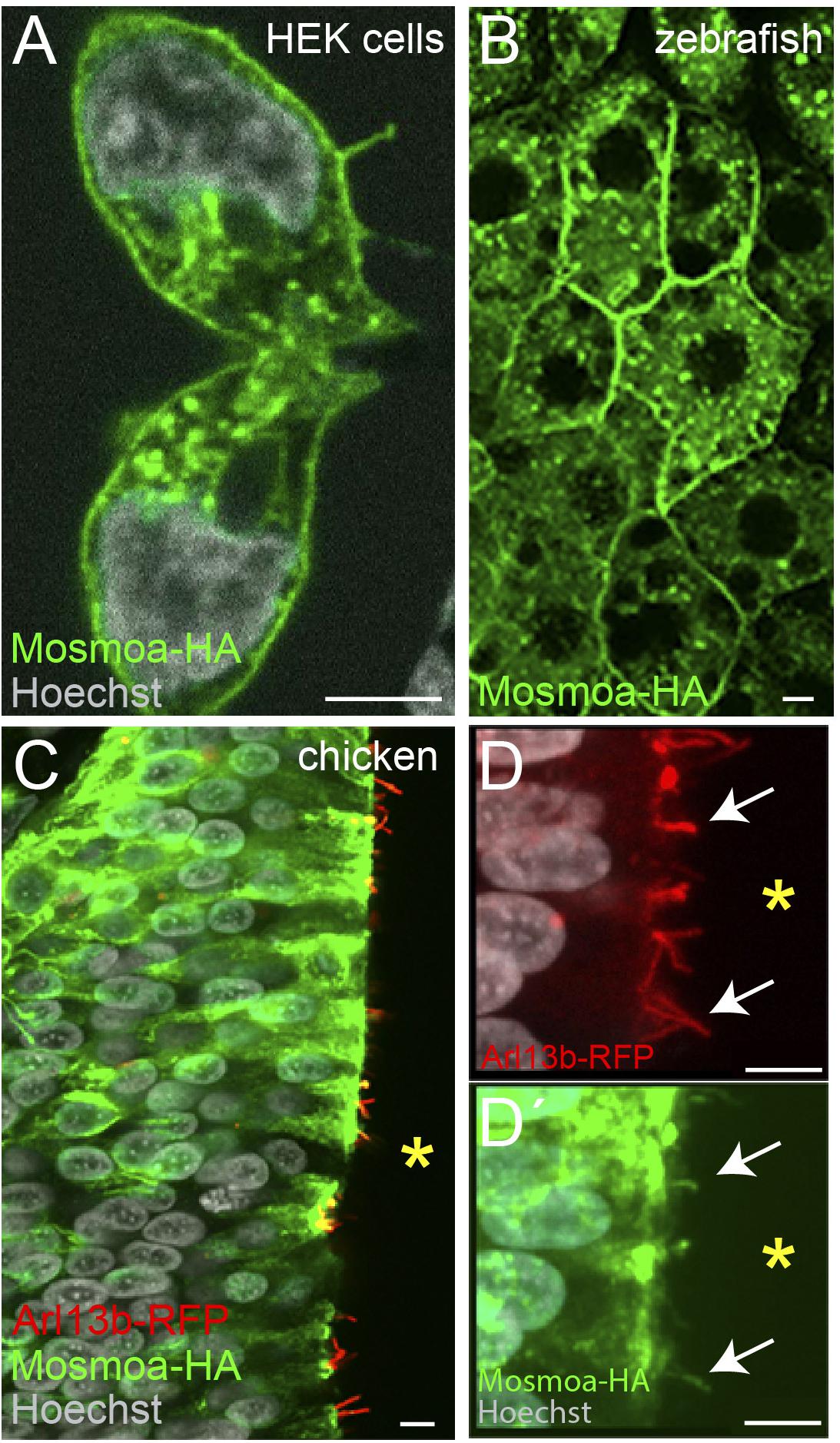
Figure 2. Mosmoa localizes at the plasma membrane, endosomes, and primary cilia. (A) Example of human embryonic kidney (HEK) cells transfected with mosmoa-HA, immunostained for α-HA (green), and counterstained with Hoechst (white). (B) Dorsal view of a zebrafish gastrula (7 hpf) injected with mosmoa-HA mRNA and immunostained for α-HA (green). (C–D′) Transversal sections of chick embryo neural tubes co-electroporated with mosmoa-HA and the cilia marker arl13b-RFP. In both HEK cells and zebrafish EVL cells Mosmoa-HA signal localizes at the plasma membrane and in endo-vesicles. In HH14 chick embryos, neural tube Mosmoa-HA is also observed in the in the Arl13b-positive cilia (white arrows) (D,D′). Yellow asterisk marks the neural tube ventricle (C–D′). Scale bars: 5 μm.
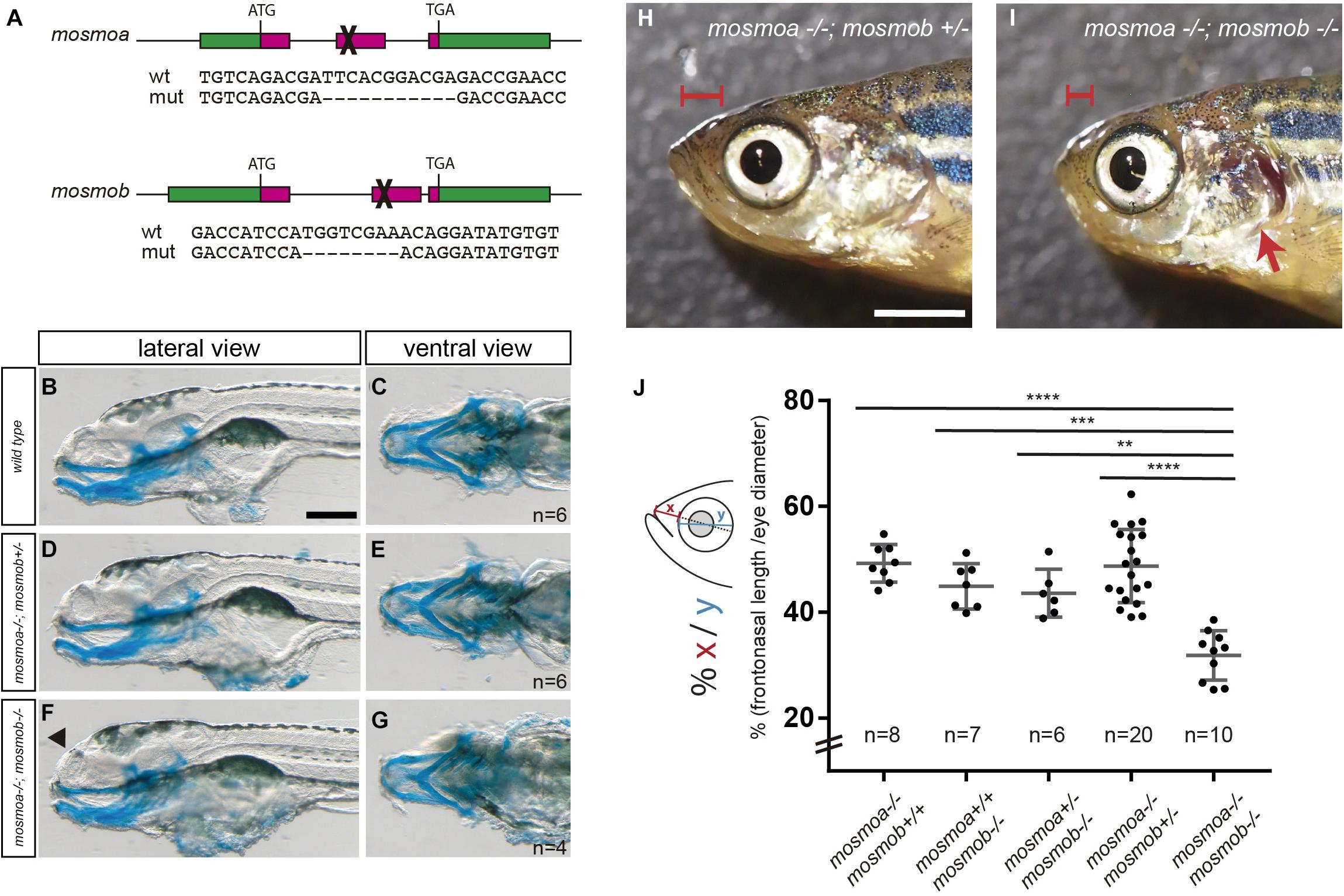
Figure 3. mosmoa–/–;mosmob–/– double mutants display facial abnormalities. (A) Schematic representation of the strategy used to inactivate mosmoa and mosmob zebrafish genes using CRISPR-Cas9 technology and sequence of the selected mutants. (B–G) Lateral and ventral views of 5 dpf wild type (B,C), mosmoa–/–;mosmob± (D,E) and mosmoa–/–;mosmob–/– (F,G) zebrafish larvae stained with Alcian Blue to detect cartilage head organization. The rostral tip of the head appeared flatter in some of the double mutants (F, arrowhead). Eyes were removed for better staining visualization. The number of animals analyzed for each genotype is indicated in the right bottom corner in panels (C,E,G). Scale bar 150 μm. (H,I) Lateral view of adult mosmoa–/–;mosmob± (H) and mosmoa–/–;mosmob–/– double mutants (I). Note that in double mutants the head is flatter and shorter (I, red brackets) than in mosmoa–/–;mosmob± fish and the operculum is abnormal exposing the gills (I, red arrow). (J) Quantification of the distance from the eye to the tip of the preorbital region (x) in relation to the eye size (y) in adult fish of different genotypes. mosmoa–/–;mosmob–/– double mutants show a shorter fronto-nasal length than their siblings. One-way ANOVA followed by Tukey’s multiple comparison tests to analyze differences among groups. **P < 0.01, ***P < 0.001, and ****P < 0.0001. Scale bar 20 mm.
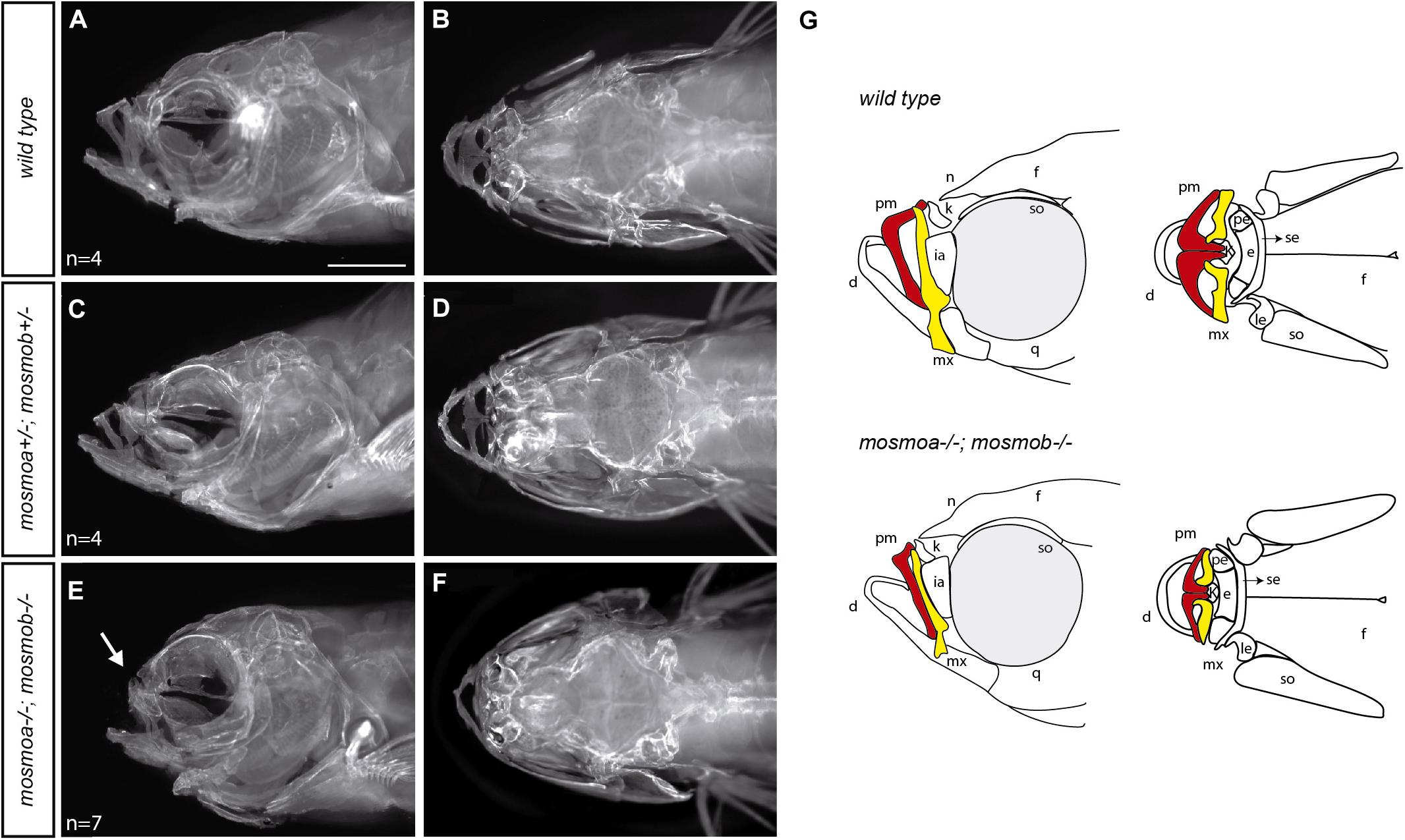
Figure 4. Mosmo paralogs are required for head bone formation in zebrafish. (A–F) Lateral (A,C,E) and dorsal (B,D,F) views of the head of wt (A,B), mosmoa±;mosmob± (C,D) and mosmoa–/–;mosmob–/– (E,F) adult zebrafish stained with alizarin red to label bone tissue. Note the bone malformation in the frontonasal region of mosmoa–/–;mosmob–/– double mutants (E, arrow) in comparison to heterozygous and wt fish (A,C). (G) Cartoons of the craniofacial appearance of wt and mosmoa–/–;mosmob–/– adult mutants, from lateral and dorsal views, highlighting the maxillary (yellow) and premaxillary (red) bones. The number of animals analyzed for each genotype is indicated in the left bottom corner in panels (A,C,E). d, dentary; e, ethmoid; f, frontal; ia, infraorbital; k, kinethmoid; le, lateral ethmoid; mx, maxillary; n, nasal; pe, pre-ethmoid; pm, premaxillary; q, quadrate; se, supraethmoid; and so, supraorbital. Scale bar, 2 mm.
When Hh signaling is active, Smo localizes at the primary cilium of the targeted cells. Notably, Mosmo was also observed in the primary cilia of NIH-3T3 cells (Pusapati et al., 2018). We thus asked if this localization could be observed also in vivo. The primary cilium can be easily detected in the chick neural tube as this organelle protrudes in the rather wide ventricle of chicken embryos. We thus co-electroporated two plasmids carrying mosmoa-HA and arl13b-RFP, respectively. The latter is a primary cilium specific protein, widely used to visualize this structure (Schmitz et al., 2017). Indeed, 24 h after electroporation, at HH14, HA, and RFP fluorescent signals co-localized in the cilium of a subset of the electroporated cells (Figures 2C–D′).
Taken together these data indicate that in vivo Mosmoa localizes at the plasma membrane, endosomes, and the primary cilia, suggesting that it may favors Smo translocation to this organelle, thus influencing signaling activation.
Mosmo Paralogs Are Required for Zebrafish Craniofacial Formation
To explore the possible roles of mosmoa and mosmob, we inactivated the two genes using CRISPR-Cas9 technology. We selected founders at the F1 generation that carried frameshift mutations in either mosmoa or mosmob gene and generated stable mosmoa–/– and mosmob–/– mutant lines (Figure 3A). Mosmoa–/– and mosmob–/– mutant embryos show no gross morphological defects and grew to adulthood without evident defects. This was perhaps not surprising given that both genes share expression pattern and their respective proteins present a high degree of sequence homology, suggesting that the two paralogs may compensate each other activity. To overcome this possible compensatory effect, we intercrossed the mosmoa–/– and mosmob–/– mutant lines obtaining a mosmoa–/–;mosmob–/– double mutant fish. At first glance, double mutant embryos showed no major gross alterations or histological defects along the neural tube and their size was similar to that of their sibling (not shown).
Other than in the neural tube, the two mosmo paralogs are expressed with a largely overlapping pattern also in different regions of the larva head. We thus used Alcian blue staining to label the cranio-facial cartilage of the larva. There were no obvious differences in the cartilaginous elements when wt, mosmoa–/–;mosmob–/+ and mosmoa–/–;mosmob–/– were compared (Figures 3B–G), although the rostral tip of the head appeared flatter at least in part of the double mutants (Figure 3F, arrowhead). To determine if this abnormality was only transient, we analyzed the morphology of the head in the adult fish. Mosmoa–/–;mosmob–/– double mutants consistently exhibited a significantly shorter frontonasal region (Figures 3H,I), which was not observed in their sibling of other genotypes (Figure 3J). Furthermore, the operculum was reduced in size, leaving the gills exposed (Figure 3I).
Hedgehog signaling is essential for the development of the anterior neurocranium (Wada et al., 2005) and disruption of smo activity in zebrafish affects the craniofacial skeleton (Eberhart et al., 2006; Swartz et al., 2012). Thus, the frontonasal hypoplasia observed in the double mutants could be the consequence of alterations in the osseous components of the craniofacial skeleton. To determine this possibility, we stained the skeleton of wt, mosmoa±;mosmob± and mosmoa–/–;mosmob–/– adult fish with Alizarin red (Figures 4A–F). The bones of the frontonasal region, especially the maxillary and premaxillary bones of mosmoa–/–;mosmob–/– double mutants were altered as compared to those of wt or heterozygous fish (Figures 4A–F) as highlighted in the schematic drawings reporting the phenotypes (Figure 4G).
Taken together these data indicate that mosmoa and mosmob have an overlapping function, which is required for the acquisition of a proper craniofacial structure in zebrafish.
Discussion
Modulators of Hh signaling play crucial roles in diversifying the output of Hh signaling (Gallardo and Bovolenta, 2018). The present study reinforces this idea and shows that in zebrafish the combined activity of the two mosmo paralogs, mosmoa and mosmob, are required for the proper craniofacial formation in zebrafish.
This apparently restricted effect is somewhat surprising as both mosmoa and mosmob are expressed with an overlapping pattern not only in the craniofacial mesenchyme of the larvae but also along the ventral region of the embryonic neural tube. The latter distribution overlaps with that of a number of Hh signaling components, including the ligand shha, shhb (Ekker et al., 1995), or the receptor ptch2 (Concordet et al., 1996), and the transducer smo (Varga et al., 2001). In line with the idea that Mosmo acts on Smo promoting its endocytosis (Pusapati et al., 2018), we found Mosmoa localized in endocytic vesicles and the plasma membrane as well as the primary cilium, where Smo translocate when Hh signaling is activated. Thus and as previously proposed (Pusapati et al., 2018), the combined activity of the two mosmo paralogs could modulate Hh signaling activation in different contexts during development. However, loss of mosmo function in zebrafish seems to be mostly linked to the formation of the cranio-facial skeleton, with an evident head hypoplasia in the adult mutant fish but no other obvious defects. Indeed, Mosmo double mutants grow to adulthood and do not seem to have obvious behavioral problems, supporting a non-essential role of mosmo paralogs for zebrafish growth, survival, and reproduction. Consistent with this idea, we have not observed neural tube defects or gross abnormalities in other organs of the mutants at least upon histological analysis. Nevertheless, we cannot rule out the possibility that subtle defects may be found with a more in-depth analysis. Indeed, a recent study shows that, in mouse, Mosmo contributes to embryonic development and its loss of function causes skeletal, heart, and lung anomalies leading to embryonic lethality (Kong et al., 2021). However, coinciding with our observations, no defects in neural tube patterning were, however, found (Kong et al., 2021). Knock-down of mosmo in Xenopus instead shows a craniofacial phenotype in which both craniofacial and cartilage development appears affected, in association with alteration of neural crest cell proliferation and migration (Lasser et al., 2020). Interestingly, the coexistence of neurodevelopmental and craniofacial defects were observed in experiments performed in both Drosophila and Xenopus aimed at testing the importance of “a two-hit model” as trigger of neurodevelopmental disorders (Pizzo et al., 2021). Notably the study demonstrated a synergistic interaction between mutated mosmo and setd5 (Pizzo et al., 2021), a gene encoding a histone methyltransferase, which has been associated with intellectual disability (Grozeva et al., 2014). This functional interaction observed in both Drosophila and Xenopus seems to be present also in humans (Pizzo et al., 2021). In this respect the mosmoa–/–;mosmob–/– double mutants, could be an additional model in which to explore how setd5 and mosmo synergize causing more severe congenital malformations.
The Hh ligands Shh and Ihh are osteogenic regulators and both are expressed in craniofacial elements (Chiang et al., 1996; Pan et al., 2013). In both mouse and zebrafish, Ihh secreted by chondrocytes stimulates the ossification of the perichondrial cell layer that surrounds the developing cartilage (St-Jacques et al., 1999; Hammond and Schulte-Merker, 2009). In mice, conditional inactivation of ihh in cranial neural crest cells causes skeletal malformations, including a markedly hypoplastic nasomaxillary complex (Amano et al., 2020). Furthermore, zebrafish mutants lacking enzymes involved in proteoglycans synthesis (fam20b–/– and xylt1–/–) exhibit an accelerated ihh expression and premature bone formation, resulting in an adult fish with midface hypoplasia among other malformations (Eames et al., 2011). These features resemble those we observe in the mosmoa–/–; mosmob–/– double mutants. Thus, it is tempting to speculate that mosmo paralogs may participate in signaling response triggered by ihh during osteogenesis, perhaps with an accelerated bone formation in the absence of Mosmo activity.
Intraflagellar proteins (IFTs) in the primary cilia, such as IFT80, affect Hh signaling and are required for osteoblast differentiation (Yuan et al., 2016). The craniofacial/skeletal abnormalities linked to Mosmo function in Xenopus (Lasser et al., 2020; Pizzo et al., 2021), mouse (Kong et al., 2021), and zebrafish (this study) together with MOSMO protein subcellular localization, suggest that MOSMO homologs could be key controllers of SMO translocation to the primary cilia during osteogenesis, thereby modulating signal transduction. Although worthwhile testing, this possibility remains at the moment a speculation, given the lack appropriate genetic tools that enable following protein movements within the cilium.
Independently of the precise pathway components with which Mosmo may function, the coincidence of some phenotypic features observed upon inactivation of Mosmo in Xenopus (Lasser et al., 2020; Pizzo et al., 2021), mouse (Kong et al., 2021), and zebrafish (this study) suggests that defective function of the human MOSMO may have similar consequences. Notably, a deletion in chromosome 16, encompassing the human MOSMO among others genes, causes a rare disease known as recurrent 16p12.1 deletion syndrome. Patients present developmental delay, intellectual disability, and other anomalies which may vary from individual to individual. Among these anomalies, craniofacial and skeletal are among the most frequently found defects (Girirajan et al., 2010), including microcephaly and flat face (Ballif et al., 2007; Girirajan et al., 2010), resembling, at some point, the phenotype observed in adult mosmoa–/–;mosmob–/– mutants.
Conclusion
In conclusion, our study shows a restricted and overlapping distribution of mosmo genes in zebrafish revealing the subcellular localization of the Mosmoa protein during development in endosomes, plasma membrane, and primary cilia. More importantly, the generation of mosmoa–/–;mosmob–/– zebrafish mutants provides support for the idea that the human MOSMO might be a candidate gene underlying uncharacterized forms of rare congenital craniofacial malformations. The double mutants further provide the opportunity to dissect the contribution of MOSMO to the phenotype associated with the human 16p12.1 deletion syndrome.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Ethics Statement
The animal study was reviewed and approved by the Consejo Superior de Investigaciones Científicas (CSIC) and Comunidad Autónoma de Madrid.
Author Contributions
MC and PB conceptualized and designed the research study and wrote the manuscript. MS and CC-M performed the ISH analysis and mutant generation and characterization. NT and MC generated data reported in Figures 2, 3 and IG in Figure 4. PB obtained the financial support. All authors read and approved the manuscript.
Funding
This work was supported by grants from the Spanish MINECO (BFU2016-75412-R with FEDER funds); AEI (PID2019-104186RB-I00 and RED2018-102553-T) and a grant from the Fundación Ramon Areces to PB. CC-M and MC are supported by predoctoral contract from the CIBERER and a Juan de la Cierva postdoctoral contract from the AEI (IJCI-2016-27683), respectively. IG is supported by a contract (CAM20_CBM_AI_06) from the 2020 program “Garantía Juvenil” of the Comunidad Autónoma de Madrid. A CBM Institutional grant from the Fundación Ramon Areces is also acknowledged.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We acknowledged the excellent technical assistance of Alfonso Gutierrez Garcia in the fish facilities and the staff of the CBMSO Image analysis facility. We also acknowledged Elisa Martí for the pCAG-2A-Arl13b-tRFP vector.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2021.767048/full#supplementary-material
Footnotes
References
Abramyan, J. (2019). Hedgehog signaling and embryonic craniofacial disorders. J. Dev. Biol. 7:9. doi: 10.3390/jdb7020009
Allen, B. L., Song, J. Y., Izzi, L., Althaus, I. W., Kang, J.-S., Charron, F., et al. (2011). Overlapping roles and collective requirement for the coreceptors GAS1, CDO, and BOC in SHH pathway function. Dev. Cell 20, 775–787. doi: 10.1016/j.devcel.2011.04.018
Allen, B. L., Tenzen, T., and McMahon, A. P. (2007). The Hedgehog-binding proteins Gas1 and Cdo cooperate to positively regulate Shh signaling during mouse development. Genes Dev. 21, 1244–1257. doi: 10.1101/gad.1543607
Amano, K., Okuzaki, D., Aikawa, T., and Kogo, M. (2020). Indian hedgehog in craniofacial neural crest cells links to skeletal malocclusion by regulating associated cartilage formation and gene expression. FASEB J. 34, 6791–6807. doi: 10.1096/fj.201903269R
Ballif, B. C., Hornor, S. A., Jenkins, E., Madan-Khetarpal, S., Surti, U., Jackson, K. E., et al. (2007). Discovery of a previously unrecognized microdeletion syndrome of 16p11.2–p12.2. Nat. Genet. 39, 1071–1073. doi: 10.1038/ng2107
Bergeron, S. A., Tyurina, O. V., Miller, E., Bagas, A., and Karlstrom, R. O. (2011). brother of cdo (umleitung) is cell-autonomously required for Hedgehog-mediated ventral CNS patterning in the zebrafish. Development 138, 75–85. doi: 10.1242/dev.057950
Briscoe, J., and Thérond, P. P. (2013). The mechanisms of Hedgehog signalling and its roles in development and disease. Nat. Rev. Mol. Cell Biol. 14, 416–429. doi: 10.1038/nrm3598
Cardozo, M. J., Sánchez-Arrones, L., Sandonis, Á, Sánchez-Camacho, C., Gestri, G., Wilson, S. W., et al. (2014). Cdon acts as a Hedgehog decoy receptor during proximal-distal patterning of the optic vesicle. Nat. Commun. 5:4272. doi: 10.1038/ncomms5272
Chiang, C., Litingtung, Y., Lee, E., Young, K. E., Corden, J. L., Westphal, H., et al. (1996). Cyclopia and defective axial patterning in mice lacking Sonic hedgehog gene function. Nature 383, 407–413. doi: 10.1038/383407a0
Cole, F., and Krauss, R. S. (2003). Microform holoprosencephaly in mice that lack the Ig superfamily member Cdon. Curr. Biol. 13, 411–415. doi: 10.1016/s0960-9822(03)00088-5
Concordet, J. P., Lewis, K. E., Moore, J. W., Goodrich, L. V., Johnson, R. L., Scott, M. P., et al. (1996). Spatial regulation of a zebrafish patched homologue reflects the roles of sonic hedgehog and protein kinase A in neural tube and somite patterning. Development 122, 2835–2846.
Eames, B. F., Yan, Y.-L., Swartz, M. E., Levic, D. S., Knapik, E. W., Postlethwait, J. H., et al. (2011). Mutations in fam20b and xylt1 reveal that cartilage matrix controls timing of endochondral ossification by inhibiting chondrocyte maturation. PLoS Genet. 7:e1002246. doi: 10.1371/journal.pgen.1002246
Eberhart, J. K., Swartz, M. E., Crump, J. G., and Kimmel, C. B. (2006). Early hedgehog signaling from neural to oral epithelium organizes anterior craniofacial development. Development 133, 1069–1077. doi: 10.1242/dev.02281
Echevarría-Andino, M. L., and Allen, B. L. (2020). The hedgehog co-receptor BOC differentially regulates SHH signaling during craniofacial development. Development 147:dev.189076. doi: 10.1242/dev.189076
Ekker, S. C., Ungar, A. R., Greenstein, P., von Kessler, D. P., Porter, J. A., Moon, R. T., et al. (1995). Patterning activities of vertebrate hedgehog proteins in the developing eye and brain. Curr. Biol. 5, 944–955. doi: 10.1016/S0960-9822(95)00185-0
Gallardo, V., and Bovolenta, P. (2018). Positive and negative regulation of Shh signalling in vertebrate retinal development. F1000Res 7:F1000 Faculty Rev-1934.
Girirajan, S., Rosenfeld, J. A., Cooper, G. M., Antonacci, F., Siswara, P., Itsara, A., et al. (2010). A recurrent 16p12.1 microdeletion supports a two-hit model for severe developmental delay. Nat. Genet. 42, 203–209. doi: 10.1038/ng.534
Grozeva, D., Carss, K., Spasic-Boskovic, O., Parker, M. J., Archer, H., Firth, H. V., et al. (2014). De Novo loss-of-function mutations in SETD5, encoding a methyltransferase in a 3p25 microdeletion syndrome critical region, cause intellectual disability. Am. J. Hum. Genet. 94, 618–624. doi: 10.1016/j.ajhg.2014.03.006
Hamburger, V., and Hamilton, H. L. (1992). A series of normal stages in the development of the chick embryo. Dev. Dyn. 195, 231–272. doi: 10.1002/aja.1001950404
Hammond, C. L., and Schulte-Merker, S. (2009). Two populations of endochondral osteoblasts with differential sensitivity to Hedgehog signalling. Development 136, 3991–4000. doi: 10.1242/dev.042150
Hui, C.-C., and Angers, S. (2011). Gli proteins in development and disease. Annu. Rev. Cell Dev. Biol. 27, 513–537. doi: 10.1146/annurev-cellbio-092910-154048
Ingham, P. W., Nakano, Y., and Seger, C. (2011). Mechanisms and functions of Hedgehog signalling across the metazoa. Nat. Rev. Genet. 12, 393–406. doi: 10.1038/nrg2984
Jeng, K.-S., Chang, C.-F., and Lin, S.-S. (2020). Sonic hedgehog signaling in organogenesis, tumors, and tumor microenvironments. Int. J. Mol. Sci. 21:758. doi: 10.3390/ijms21030758
Jia, J., Tong, C., Wang, B., Luo, L., and Jiang, J. (2004). Hedgehog signalling activity of Smoothened requires phosphorylation by protein kinase A and casein kinase I. Nature 432, 1045–1050. doi: 10.1038/nature03179
Kong, J. H., Young, C. B., Pusapati, G. V., Espinoza, F. H., Patel, C. B., Beckert, F., et al. (2021). Gene-teratogen interactions influence the penetrance of birth defects by altering Hedgehog signaling strength. Development 148. doi: 10.1242/dev.199867
Labun, K., Montague, T. G., Krause, M., Torres Cleuren, Y. N., Tjeldnes, H., and Valen, E. (2019). CHOPCHOP v3: expanding the CRISPR web toolbox beyond genome editing. Nucleic Acids Res. 47, W171–W174. doi: 10.1093/nar/gkz365
Lasser, M., Bolduc, J., Murphy, L., O’Brien, C., Lee, S., Girirajan, S., et al. (2020). 16p12.1 deletion orthologs are expressed in motile neural crest cells and are important for regulating craniofacial development in Xenopus laevis. bioRxiv [Preprint]. doi: 10.1101/2020.12.11.421347
Martí, E., Takada, R., Bumcrot, D. A., Sasaki, H., and McMahon, A. P. (1995). Distribution of Sonic hedgehog peptides in the developing chick and mouse embryo. Development 121, 2537–2547.
Megason, S. G., and McMahon, A. P. (2002). A mitogen gradient of dorsal midline Wnts organizes growth in the CNS. Development 129, 2087–2098. doi: 10.1242/dev.129.9.2087
Murone, M., Rosenthal, A., and de Sauvage, F. J. (1999). Sonic hedgehog signaling by the Patched–Smoothened receptor complex. Curr. Biol. 9, 76–84. doi: 10.1016/S0960-9822(99)80018-9
Pan, A., Chang, L., Nguyen, A., and James, A. W. (2013). A review of hedgehog signaling in cranial bone development. Front. Physiol. 4:61. doi: 10.3389/fphys.2013.00061
Petrova, R., and Joyner, A. L. (2014). Roles for Hedgehog signaling in adult organ homeostasis and repair. Development 141, 3445–3457. doi: 10.1242/dev.083691
Pizzo, L., Lasser, M., Yusuff, T., Jensen, M., Ingraham, P., Huber, E., et al. (2021). Functional assessment of the “two-hit” model for neurodevelopmental defects in Drosophila and X. laevis. PLoS Genet. 17:e1009112. doi: 10.1371/journal.pgen.1009112
Pusapati, G. V., Kong, J. H., Patel, B. B., Krishnan, A., Sagner, A., Kinnebrew, M., et al. (2018). CRISPR screens uncover genes that regulate target cell sensitivity to the morphogen sonic hedgehog. Dev. Cell 44:271. doi: 10.1016/j.devcel.2018.01.002
Roelink, H., Porter, J. A., Chiang, C., Tanabe, Y., Chang, D. T., Beachy, P. A., et al. (1995). Floor plate and motor neuron induction by different concentrations of the amino-terminal cleavage product of sonic hedgehog autoproteolysis. Cell 81, 445–455. doi: 10.1016/0092-8674(95)90397-6
Sakata-Haga, H., Uchishiba, M., Shimada, H., Tsukada, T., Mitani, M., Arikawa, T., et al. (2018). A rapid and nondestructive protocol for whole-mount bone staining of small fish and Xenopus. Sci. Rep. 8:7453. doi: 10.1038/s41598-018-25836-4
Sánchez-Camacho, C., and Bovolenta, P. (2009). Emerging mechanisms in morphogen-mediated axon guidance. BioEssays 31, 1013–1025. doi: 10.1002/bies.200900063
Sasai, N., Toriyama, M., and Kondo, T. (2019). Hedgehog signal and genetic disorders. Front. Genet. 10:1103. doi: 10.3389/fgene.2019.01103
Schilling, T. F., Piotrowski, T., Grandel, H., Brand, M., Heisenberg, C. P., Jiang, Y. J., et al. (1996). Jaw and branchial arch mutants in zebrafish I: branchial arches. Development 123, 329–344.
Schmitz, F., Burtscher, I., Stauber, M., Gossler, A., and Lickert, H. (2017). A novel Cre-inducible knock-in ARL13B-tRFP fusion cilium reporter. Genesis 55:e23073. doi: 10.1002/dvg.23073
St-Jacques, B., Hammerschmidt, M., and McMahon, A. P. (1999). Indian hedgehog signaling regulates proliferation and differentiation of chondrocytes and is essential for bone formation. Genes Dev. 13, 2072–2086.
Swartz, M. E., Nguyen, V., McCarthy, N. Q., and Eberhart, J. K. (2012). Hh signaling regulates patterning and morphogenesis of the pharyngeal arch-derived skeleton. Dev. Biol. 369, 65–75. doi: 10.1016/j.ydbio.2012.05.032
Varga, Z. M., Amores, A., Lewis, K. E., Yan, Y.-L., Postlethwait, J. H., Eisen, J. S., et al. (2001). Zebrafish smoothened functions in ventral neural tube specification and axon tract formation. Development 128, 3497–3509. doi: 10.1242/dev.128.18.3497
Varshney, G. K., Carrington, B., Pei, W., Bishop, K., Chen, Z., Fan, C., et al. (2016). A high-throughput functional genomics workflow based on CRISPR/Cas9-mediated targeted mutagenesis in zebrafish. Nat. Protoc. 11, 2357–2375. doi: 10.1038/nprot.2016.141
Wada, N., Javidan, Y., Nelson, S., Carney, T. J., Kelsh, R. N., and Schilling, T. F. (2005). Hedgehog signaling is required for cranial neural crest morphogenesis and chondrogenesis at the midline in the zebrafish skull. Development 132, 3977–3988. doi: 10.1242/dev.01943
Yam, P. T., Langlois, S. D., Morin, S., and Charron, F. (2009). Sonic hedgehog guides axons through a noncanonical, Src-family-kinase-dependent signaling pathway. Neuron 62, 349–362. doi: 10.1016/j.neuron.2009.03.022
Keywords: hedgehog signaling (Hh), Smoothened (Smo), tetraspan transmembrane protein, craniofacial abnormalities, Mosmo
Citation: Camacho-Macorra C, Sintes M, Tabanera N, Grasa I, Bovolenta P and Cardozo MJ (2021) Mosmo Is Required for Zebrafish Craniofacial Formation. Front. Cell Dev. Biol. 9:767048. doi: 10.3389/fcell.2021.767048
Received: 30 August 2021; Accepted: 05 October 2021;
Published: 22 October 2021.
Edited by:
Rosa Barrio, CIC bioGUNE, SpainReviewed by:
Laura Anne Lowery, Boston University, United StatesSanthosh Girirajan, The Pennsylvania State University, United States
Copyright © 2021 Camacho-Macorra, Sintes, Tabanera, Grasa, Bovolenta and Cardozo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Paola Bovolenta, cGJvdm9sZW50YUBjYm0uY3NpYy5lcw==; Marcos J. Cardozo, bWNhcmRvem9AY2JtLmNzaWMuZXM=
†These authors have contributed equally to this work and share first authorship
‡These authors have contributed equally to this work and share senior authorship
 Carlos Camacho-Macorra
Carlos Camacho-Macorra Marcos Sintes
Marcos Sintes Noemí Tabanera
Noemí Tabanera Irene Grasa1
Irene Grasa1 Paola Bovolenta
Paola Bovolenta Marcos J. Cardozo
Marcos J. Cardozo